Abstract
The phylogeny of Isopoda, a speciose order of crustaceans, remains unresolved, with different data sets (morphological, nuclear, mitochondrial) often producing starkly incongruent phylogenetic hypotheses. We hypothesized that extreme diversity in their life histories might be causing compositional heterogeneity/heterotachy in their mitochondrial genomes, and compromising the phylogenetic reconstruction. We tested the effects of different data sets (mitochondrial, nuclear, nucleotides, amino acids, concatenated genes, individual genes, gene orders), phylogenetic algorithms (assuming data homogeneity, heterogeneity, and heterotachy), and partitioning; and found that almost all of them produced unique topologies. As we also found that mitogenomes of Asellota and two Cymothoida families (Cymothoidae and Corallanidae) possess inversed base (GC) skew patterns in comparison to other isopods, we concluded that inverted skews cause long-branch attraction phylogenetic artifacts between these taxa. These asymmetrical skews are most likely driven by multiple independent inversions of origin of replication (i.e., nonadaptive mutational pressures). Although the PhyloBayes CAT-GTR algorithm managed to attenuate some of these artifacts (and outperform partitioning), mitochondrial data have limited applicability for reconstructing the phylogeny of Isopoda. Regardless of this, our analyses allowed us to propose solutions to some unresolved phylogenetic debates, and support Asellota are the most likely candidate for the basal isopod branch. As our findings show that architectural rearrangements might produce major compositional biases even on relatively short evolutionary timescales, the implications are that proving the suitability of data via composition skew analyses should be a prerequisite for every study that aims to use mitochondrial data for phylogenetic reconstruction, even among closely related taxa.
Keywords: base composition skew, GC skew, mitochondrial phylogenomics, Cymothoida, replication origin inversion, compositional heterogeneity
Introduction
Significant taxonomic and phylogenetic uncertainty permeates the entire order of Isopoda, a highly speciose (>10,000) order of crustaceans (class Malacostraca) that exhibit a remarkable diversity in their life histories and occupy almost all habitats on the planet Earth. The traditional morphology-based taxonomic classification and identification of isopods is further (aside from the speciosity) hampered by great intraspecific morphological variation, sexual dimorphism, sequential hermaphroditism, relatively flexible host preference, and global distribution of many species (Wetzer 2002; Wilson 2008; Lins et al. 2012, 2017; Joca et al. 2015; Shen et al. 2017; Rudy et al. 2018). However, molecular data also appear to be an unreliable tool for the task, as different data sets (mitochondrial genes, mitochondrial genomes, nuclear genes, combined mitonuclear data) often produce very different topologies (Brusca 1981; Wetzer 2002; Wilson 2009; Kilpert et al. 2012; Poore and Bruce 2012; Martin et al. 2016; Hata et al. 2017; Lins et al. 2017; Yu et al. 2018; Zou et al. 2018). As a result, even the identity of the basal isopod clade (defined as the sister-clade to all other isopod lineages; Krell and Cranston 2004) remains debated. Traditionally (morphology and single gene-based studies), Phreatoicidea was regarded as the basal clade (Wetzer 2002; Wilson 2009; Kilpert et al. 2012). However, some studies resolved Phreatoicidea+Aselotta at the base (Wilson 1999, 2009; Shen et al. 2017; Yu et al. 2018), one study found Limnoriidea (Lins et al. 2017) at the base, whereas a few studies even resolved parasitic Cymothoidae and Corallanidae (suborder Cymothoida) at the base (Wilson 2009; Lins et al. 2012, 2017; Hua et al. 2018; Zou et al. 2018). As the suborder Cymothoida was traditionally regarded as the most derived isopod clade (Brusca 1981; Wetzer 2002; Wilson 2009; Kilpert et al. 2012), these alternative hypotheses cannot be described as a minor topological instability. The monophyly of the Cymothoida is generally supported by the morphological data, but rejected by the molecular data (Brandt and Poore 2003; Wilson 2009; Kilpert et al. 2012; Lins et al. 2017; Shen et al. 2017; Hua et al. 2018; Yu et al. 2018; Zou et al. 2018). Among a number of other unresolved phylogenetic issues permeating this order are the monophyly of the suborder Oniscidea (supported by morphology, sometimes rejected by the molecular data) and the existence of several “rogue” species/taxa, such as Ligia oceanica (Ligiidae), Eurydice pulchra (Cirolanidae), and Limnoria quadripunctata (Limnoriidae), whose positions in the isopod clade often vary among studies (Schmidt 2008; Wilson 2009; Kilpert et al. 2012; Wetzer et al. 2013; Lins et al. 2017; Shen et al. 2017; Yu et al. 2018).
Historically, variation found within gene sequences was typically considered to accumulate under a neutral equilibrium model, so commonly used phylogenetic reconstruction algorithms assume homogeneity in mutational rates. As this paradigm began to change during the last few decades (Wolff et al. 2014), this was accompanied by a growing amount of evidence that compositional heterogeneity can compromise phylogenetic reconstruction in some taxa. Therefore, the evolutionary models operating under that prerequisite may not be suitable for all phylogenetic studies (Kolaczkowski and Thornton 2004; Hassanin 2006; Phillips et al. 2006; Lartillot et al. 2007; Sheffield et al. 2009; Zhong et al. 2011; Morgan et al. 2013; Cameron 2014). However, the feud about the most suitable methodological approach to account for this heterogeneity remains unresolved, with most prominent contenders currently being the CAT models (Feuda et al. 2017), and partitioning schemes, that is, different evolutionary models assigned to different character blocks, assuming homogeneity within each block (CAT models to not allow partitioned data) (Whelan and Halanych 2017). Although these two approaches account for rate heterogeneity across sites, they still assume that substitution rates for sites are constant across all included lineages. From the evolutionary perspective, this is not a likely scenario, since substitution rates are likely to be both site- and lineage-specific (Crotty et al. 2017). Indeed, heterotachy, variations in lineage-specific evolutionary rates over time (Lopez et al. 2002), is widespread in eukaryotes (Baele et al. 2006).
Mitochondrial genomes (mitogenomes) generally provide much higher phylogenetic resolution than traditionally used morphological and single gene-based molecular markers (Nie et al. 2018), so mitochondrial phylogenomics is increasingly used to tackle phylogenetic controversies (Cameron 2014; Der Sarkissian et al. 2015; Lan et al. 2017; Li et al. 2017; Bourguignon et al. 2018). Although the resolution of this approach is still limited by a very small number of available mitogenomes in isopods, overview of published studies shows that they also failed to produce results congruent with other approaches and to resolve the rogue taxa issues (Kilpert et al. 2012; Lins et al. 2017; Shen et al. 2017; Hua et al. 2018; Yu et al. 2018; Zou et al. 2018). The evolutionary history of Isopoda abounds in independent (major) life history innovations, such as free-living to parasitic lifestyle (Jones et al. 2008; Ketmaier et al. 2008; Poore and Bruce 2012; Hata et al. 2017), and radical habitat expansions (Lins et al. 2012), such as sea to freshwater, or even water to land (Wilson 2008; Broly et al. 2013; Hata et al. 2017). It has been proposed that the Cymothoida may have originated in deep seas, subsequently expanded to shallow seas, and then to brackish and freshwater (likely on several independent occasions) (Hata et al. 2017). Mitochondrial genes are central to the energy production via the oxidative phosphorylation (Gawryluk et al. 2016), and signals of adaptation to high altitude (Mishmar et al. 2003; Hassanin et al. 2009; Scott et al. 2011), deep-sea environment (Almeida et al. 2015), and shifts in physiological demands (Hassanin 2006; Botero-Castro et al. 2018), have been identified in mitogenomes of a range of animals. It is therefore highly likely that radical adaptations to life in different environments, from the anoxic environment of deep sea-inhabiting isopod species (Lins et al. 2012) to terrestrial species, would produce strikingly different evolutionary pressures on genomes of species, and result in disparate evolutionary rates of mitochondrial genes. In agreement with this hypothesis are uneven evolutionary rates (dN/dS) observed among isopod mitogenomes (Shen et al. 2017) and different mutational rates of protein-coding genes (PCGs) encoded on the majority (or plus) strand among different lineages of isopods (Lloyd et al. 2015). Conflicting phylogenetic signals among different mitochondrial regions have been reported in a number of metazoan groups, which indicates that different mitochondrial regions can accumulate substitutions in ways that are difficult to model, and thus produce biased estimates of phylogeny (Meiklejohn et al. 2014). There is evidence that this compositional heterogeneity may be comparatively highly pronounced in mitogenomes of some arthropod taxa (Hassanin 2006; Cameron 2014; Liu et al. 2017). Despite these limitations of molecular data (Ballard and Whitlock 2004; Hassanin et al. 2005; Rubinoff et al. 2005; Talavera et al. 2011; Grechko 2013; Edwards et al. 2016; Willis 2017), most previous phylogenetic studies of Isopoda ignored those limitations, or attempted to ameliorate them by using such strategies as combined data sets (mtDNA, nuclear DNA, morphology) (Wilson 2009; Lins et al. 2017), amino acid sequences (Kilpert et al. 2012; Lins et al. 2017), or applying different models to each codon position (Hata et al. 2017). However, none of those studies attempted to use algorithms designed specifically to account for compositional heterogeneity/tachy, nor studied this problem directly. We hypothesized that the extreme life history diversity of isopods might be causing pronounced compositional heterogeneity/heterotachy in their mitogenomes, and interfere with the reconstruction of their phylogeny. To test this hypothesis, we used a number of different data sets: mitochondrial DNA (single genes, genomes, nucleotides, amino acids, gene orders) and nuclear DNA (18S); and methodological approaches: data set partitioning, maximum likelihood (ML), Bayesian inference (BI), parsimony, PhyloBayes (PB) CAT-GTR model (heterogeneous), and GHOST (heterotachous).
Materials and Methods
Data Sets
As a majority of available isopod mitogenomes are incomplete, we were faced with a trade-off between the amount of data used in the analysis and the number of species used: after removing 6 (too few genes and duplicates) of the 27 available isopod mitogenomes (Oct, 2018), the data set comprised 8 complete and 13 partial sequences (table 1 and supplementary file S1, Supplementary Material online). Out of 11 valid isopod suborders, our data set covers 7. To attempt to resolve the debated issue of the basal Isopod clade with maximum resolution, we used a relatively large number of outgroups for phylogenetic analyses: a basal arthropod, Limulus polyphemus (Lavrov et al. 2000), and a number of nonisopod Malacostraca: three Decapoda, two Stomatopoda, two Amphipoda, one Mysida, and one Euphausiacea species (Kilpert and Podsiadlowski 2006; Wilson 2009). We conducted phylogenetic analyses using the following data sets: NUC—nucleotides of concatenated 13 PCGs and 2 rRNA genes (rrnL and rrnS), AAs—concatenated amino acid sequences of 13 PCGs, 15 single-gene data sets (13 PCGs + 2 rRNAs), and gene orders. We also tested the performance of data partitioning, by conducting the same analyses on both nonpartitioned and partitioned data sets.
Table 1.
Taxonomy, Length (bp), Base Composition (%), and Skews of Mitogenomes Used in the Analysis
| Species | Suborder | Family | Acc. No. | Length | A + T | A | C | AT Skew | GC Skew |
|---|---|---|---|---|---|---|---|---|---|
| Isopoda | |||||||||
| Asellus aquaticus | Asellota | Asellidae | GU130252 | 13,639 | 61.9 | 31 | 21.3 | 0.002 | −0.122 |
| Cymothoa indica | Cymothoida | Cymothoidae | MH396438 | 14,475 | 63.8 | 36 | 26.1 | 0.129 | −0.442 |
| Ichthyoxenos japonensis | Cymothoida | Cymothoidae | MF419233 | 15,440 | 72.7 | 37 | 18.7 | 0.026 | −0.375 |
| Tachaea chinensis | Cymothoida | Corallanidae | MF419232 | 14,616 | 72.8 | 38 | 18.5 | 0.055 | −0.354 |
| Eurydice pulchra | Cymothoida | Cirolanidae | GU130253 | 13,055 | 55.9 | 27 | 17.7 | −0.052 | 0.198 |
| Bathynomus sp. | Cymothoida | Cirolanidae | KU057374 | 14,965 | 58.7 | 27 | 17 | −0.093 | 0.175 |
| Gyge ovalis | Cymothoida | Bopyridae | NC_037467 | 14,268 | 59.6 | 27 | 17.8 | −0.093 | 0.118 |
| Eophreatoicus sp. | Phreatoicidea | Amphisopidae | NC_013976 | 14,994 | 69.6 | 31 | 11.4 | −0.104 | 0.25 |
| Sphaeroma serratum | Sphaeromatidea | Sphaeromatidae | GU130256 | 13,467 | 54.4 | 25 | 17.8 | −0.069 | 0.219 |
| Limnoria quadripunctata | Limnoriidea | Limnoriidae | NC_024054 | 16,515 | 66.3 | 30 | 13.6 | −0.104 | 0.188 |
| Idotea balthica | Valvifera | Idoteidae | DQ442915 | 14,247 | 61 | 28 | 16.3 | −0.076 | 0.163 |
| Glyptonotus antarcticus | Valvifera | Chaetiliidae | GU130254 | 13,809 | 65.4 | 32 | 16.6 | −0.033 | 0.038 |
| Trachelipus rathkii | Oniscidea | Trachelipodidae | MF187612 | 14,080 | 67.3 | 33 | 13.3 | −0.029 | 0.184 |
| Porcellio dilatatus | Oniscidea | Porcellionidae | KX289582 | 14,103 | 65.6 | 31 | 13.4 | −0.067 | 0.224 |
| Cylisticus convexus | Oniscidea | Cylisticidae | KR013002 | 14,154 | 67.8 | 33 | 12.9 | −0.035 | 0.194 |
| Porcellionides pruinosus | Oniscidea | Porcellionidae | KX289584 | 14,078 | 60.5 | 28 | 14.9 | −0.09 | 0.248 |
| Armadillidium album | Oniscidea | Armadillidiidae | KX289585 | 13,812 | 69.7 | 33 | 12.5 | −0.045 | 0.172 |
| Armadillidium nasatum | Oniscidea | Armadillidiidae | MF187611 | 13,943 | 68.1 | 33 | 13.4 | −0.043 | 0.16 |
| Armadillidium vulgare | Oniscidea | Armadillidiidae | MF187614 | 13,932 | 71.5 | 34 | 11.8 | −0.043 | 0.174 |
| Armadillidium vulgare | Oniscidea | Armadillidiidae | MF187613 | 13,955 | 71.3 | 34 | 11.8 | −0.039 | 0.179 |
| Ligia oceanica | Oniscidea | Ligiidae | NC_008412 | 15,289 | 60.9 | 29 | 17 | −0.041 | 0.134 |
|
| |||||||||
| Species | Order | Suborder | Acc. No. | Length | A+T | A | C | AT Skew | GC Skew |
|
| |||||||||
| Nonisopod Malacostraca and Limulus polyphemus | |||||||||
| Atergatis floridus | Decapoda | Pleocyemata | NC_037201 | 16,180 | 69.3 | 33 | 20.3 | −0.036 | −0.319 |
| Penaeus vannamei | Decapoda | Dendrobranchiata | NC_009626 | 15,990 | 67.7 | 33 | 19.2 | −0.026 | −0.192 |
| Typhlatya miravetensis | Decapoda | Pleocyemata | NC_036335 | 15,865 | 66.2 | 36 | 22.5 | 0.076 | −0.332 |
| Squilla mantis | Stomatopoda | Unipeltata | NC_006081 | 15,994 | 70.2 | 35 | 16.8 | −0.001 | −0.13 |
| Lysiosquillina maculata | Stomatopoda | Unipeltata | NC_007443 | 16,325 | 63.9 | 33 | 21.4 | 0.026 | −0.185 |
| Metacrangonyx repens | Amphipoda | Senticaudata | NC_019653 | 14,355 | 76.9 | 38 | 11.7 | −0.025 | −0.014 |
| Neomysis japonica | Mysida | NA | NC_027510 | 17,652 | 74.5 | 37 | 13.8 | −0.021 | −0.085 |
| Eulimnogammarus cyaneus | Amphipoda | Senticaudata | NC_033360 | 14,370 | 67.6 | 33 | 20.3 | −0.019 | −0.251 |
| Euphausia pacifica | Euphausiacea | NA | NC_016184 | 16,898 | 72 | 36 | 16 | 0.004 | −0.145 |
| Limulus polyphemus | Xiphosurida | NA | NC_003057 | 14,985 | 67.6 | 38 | 22.7 | 0.111 | −0.399 |
To test a signal from the nuclear data, we used partial 18S rRNA gene (1,223 bp aligned data set; supplementary file S2, Supplementary Material online). As some of the species from the mitogenomic data set were not available, we made sure to include representatives of all isopod suborders in the mitogenomic data set (55 species in total; supplementary file S1, Supplementary Material online). To obtain a more comparable data set, we sequenced the 18S gene (≈1,930 bp) of three parasitic Cymothoida species from the mtDNA data set: Cymothoa indica (Cymothoidae; GenBank accession number MK079664), Ichthyoxenos japonensis (Cymothoidae; MK542857), and Tachaea chinensis (Corallanidae; MK542858). To improve the resolution for the “problematic” Cymothoidae, we sequenced an additional species: Asotana magnifica (MK542856).
Data Manipulation and Analyses
PhyloSuite (Zhang et al. 2018) was used to batch-download all selected molecular data from the GenBank, extract genomic features, translate genes into amino acid sequences, semiautomatically reannotate ambiguously annotated tRNA genes with the help of the ARWEN (Laslett and Canbäck 2008) output, automatically replace the GenBank taxonomy with the WoRMS database taxonomy, as the latter tends to be more up to date (Costello et al. 2013), generate comparative genome statistics tables, and conduct phylogenetic analyses (Flowchart mode) using a number of incorporated plug-in programs.
We used MAFFT (Katoh and Toh 2008; Katoh and Standley 2013) to align sequences: nucleotide and amino acid sequences of PCGs were aligned in batches (using codon and normal-alignment modes, respectively) with “–auto” strategy, whereas rRNA genes (including the 18S) were aligned using Q-INS-i algorithm, which takes secondary structure information into account. Gblocks (Castresana 2000; Talavera and Castresana 2007) was used to remove ambiguously aligned regions from the concatenated alignments (default PhyloSuite parameter settings). After concatenating the alignments with PhyloSuite (all alignments in supplementary file S2, Supplementary Material online), PartitionFinder2 (Lanfear et al. 2012) was used to find the best data partitioning scheme and to select the best-fit evolutionary models for each partition (Akaike Information Criterion; supplementary file S2, Supplementary Material online), whereas ModelFinder (Kalyaanamoorthy et al. 2017) was used to select models for nonpartitioned data sets (Bayesian information criterion). χ2 test for the homogeneity of character composition of aligned sequences was performed using IQ-TREE 1.6.8 (Trifinopoulos et al. 2016).
We tested the performance of two standard homogeneous models, ML, BI, both on nonpartitioned and partitioned data, and two nonstandard heterogeneous (CAT-GTR) and heterotachous (GHOST) models (these two require the input data to be nonpartitioned). We also tested the performance of Parsimony model implemented in PAUP* 4.0 on nonpartitioned data sets (heuristic searching, TBR branch swapping, and 500 random addition sequence replicates, bootstrap: 1,000 pseudoreplicate data sets) (Swofford 2002). BI analyses were conducted using MrBayes 3.2.6 (Ronquist et al. 2012) (as PhyloSuite plug-in), with default settings until the stationarity was reached (3.8–8.3 × 106 generations depending on the data set, stationarity = average SD of split frequencies <0.01, estimated sample size >200, PSRF index ≈1). ML analyses were mostly carried out using a ML+rapid bootstrap algorithm with 1,000 replicates in RAxML (Stamatakis 2014), with the exception of ML analyses of 15 single-gene data sets and 18S, which were conducted in PhyloSuite batch mode using IQ-TREE plug-in in “TESTNEW” mode, which selects the best-fit evolution mode and conducts the phylogenetic analysis separately for each data set (Trifinopoulos et al. 2016). The CAT-GTR site mixture model implemented in PhyloBayes-MPI 1.7a (Lartillot et al. 2007) allows for site-specific rates of mutation, which is considered to be a more realistic model of amino acid evolution, especially for large multigene alignments (Maddock et al. 2016). PhyloBayes was run on the beta version of the Cipres server (https://cushion3.sdsc.edu/portal2/tools.action) (Miller et al. 2010), with default parameters (burnin = 500, invariable sites automatically removed from the alignment, two MCMC chains), and the analysis was stopped when the conditions considered to indicate a good run were reached (PhyloBayes manual: maxdiff <0.1 and minimum effective size >300). GHOST model is an edge-unlinked mixture model consisting of several site classes with separate sets of model parameters and edge lengths on the same tree topology, thus naturally accounting for heterotachous evolution (Crotty et al. 2017); analyses were run with 50,000 ultrafast bootstrap replicates (Minh et al. 2013). MLGO (Hu et al. 2014) was used to reconstruct the phylogeny using the gene-order (GO) data, with 1,000 bootstrap replicates, and an input file generated by PhyloSuite. Phylograms and gene orders were visualized in iTOL (Letunic and Bork 2007), and annotated using files generated by PhyloSuite. Skews were calculated and plotted using PhyloSuite and GraphDNA (Thomas et al. 2007).
Results
Models, Partitioning, and Compositional Heterogeneity Tests
Best-fit model for the nonpartitioned NUC data set was GTR+I+G4, mtZOA+F + I+G4 for the AAs, and SYM+R4 for the 18S. Best inferred partitioning strategy differed between NUC (12 partitions) and AAs (6 partitions) data sets (see supplementary file S1, Supplementary Material online, for details). The χ2 compositional homogeneity test denotes a sequence as “failed” if its nucleotide composition significantly deviates from the average composition of the alignment; in the nonpartitioned NUC data set only Penaeus vannamei (outgroup) passed the test (30 sequences failed); in the AAs data set 11 species failed; and in the 18S data set only Gammarus troglophilus (outgroup) failed.
NUC Data Set
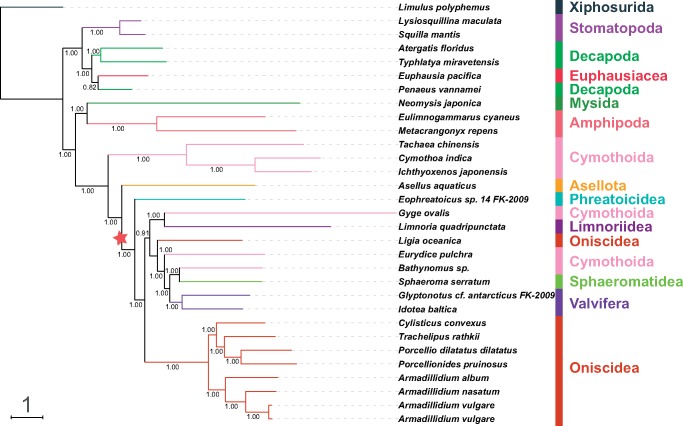
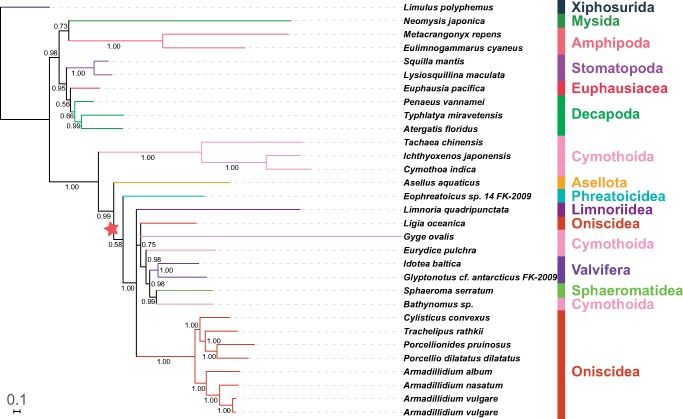
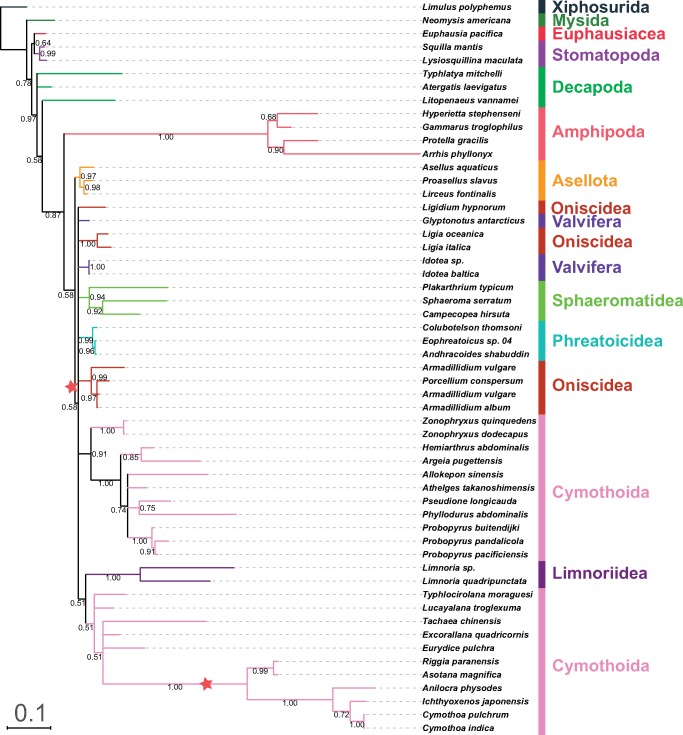
GHOST, BI, and ML analyses (both partitioned and nonpartitioned) of the NUC data set produced highly congruent topologies (referred to as the “NUC-consensus” topology henceforth). Statistical support values were very high in BI (fig. 1; all inferred topologies available in the supplementary file S3, Supplementary Material online), relatively high in GHOST, and intermediate in ML analyses (low to high). Decapoda were rendered paraphyletic by the Euphausiacea nested within the clade (not in the ML-partitioned tree), and Mysida+Amphipoda were resolved as the sister-clade to Isopoda (see table 2 for the overview of some key features). In the isopod clade, Cymothoidae+Corallanidae families (Cymothoida) formed the basal clade, followed by Asellota and Phreatoicidea branches. The remaining isopods were divided into two sister-clades: Oniscidea and a “catch-all” clade comprising Limnoriidea, Valvifera, Sphaeromatidea, L. oceanica (a “rogue” Oniscidea species), and the remaining three Cymothoida species (Gyge ovalis, Bathynomus sp., and E.pulchra), which did not cluster together (fig. 1). Parsimony analysis produced a slightly rearranged topology, but crucial features were identical (table 2). Notable differences were: Asellota forming a sister-clade with G. ovalis + Lim. quadripunctata, and Phreatoicidea at the base of the catch-all clade. The PB analysis produced a notably different overall topology, with all nonisopod lineages forming a sister-clade to the isopods (monophyletic Decapoda), but the topology of the isopod clade was relatively similar to the NUC-consensus, apart from the rogue G.ovalis (fig. 2).
Fig. 1.
—Mitochondrial phylogenomics of Isopoda (suborder information shown) reconstructed using partitioned nucleotide sequences of PCGs and rRNAs (NUC data set) and BI algorithm. A set of nine nonisopod Malacostraca species and Limulus polyphemus were used as outgroups (order information shown). The scale bar corresponds to the estimated number of substitutions per site. Bayesian posterior support values are shown next to corresponding nodes. Star sign indicates a putative origin of replication inversion scenario implied by the topology (see Discussion).
Table 2.
Summary of Some Key Features of Phylogenetic Analyses
| NUC | BI_P | BI_NP | ML_P | ML_NP | GHOST | Parsimony | PB_CAT |
|---|---|---|---|---|---|---|---|
| Basal | C+C | C+C | C+C | C+C | C+C | C+C | C+C |
| Cymothoida | Par | Par | Par | Par | Par | Par | Par |
| Sister-gr. | M+A | M+A | M+A | M+A | M+A | M+A | Malacostraca |
| Cym+Asel | + | + | + | + | + | + | + |
| Limnoriidea | G.o. | G.o. | G.o. | G.o. | G.o. | G.o. | * |
|
| |||||||
| AAs | BI_P | BI_NP | ML_P | ML_NP | GHOST | Parsimony | PB_CAT |
|
| |||||||
| Basal | Phreat. | C+C | Phreat. | Phreat. | Phreat. | C+C | Phreat. |
| Cymothoida | Par | Par | Par | Par | Par | Par | Par |
| Sister-gr. | M+A | M+A | Mysida | Mysida | Mysida | M+A | Mysida |
| Cym+Asel | +(s) | +(s) | +(s) | +(s) | +(s) | + | − |
| Limnoriidea | G.o. | G.o. | G.o. | G.o. | G.o. | G.o. | G.o.* |
|
| |||||||
| 18S | BI | ML | GHOST | Parsimony | PB_CAT | ||
|
| |||||||
| Basal | Asellota | Asellota | Asellota | Limnoriidea | Asellota | ||
| Cymothoida | Par | Par | Par | Mon | Par | ||
| Sister-gr. | Decapodaa | Decapodaa | Decapodaa | Amphipoda | Amphipoda | ||
| Cym+Asel | − | − | − | − | − | ||
| Limnoriidea | Amphipodaa | Amphipodaa | Corallanidae | Basal | Cymothoida | ||
Note.—Basal, basal isopod clade; Cymothoida, monophyletic or paraphyletic; Sister-gr., sister-group to the Isopoda; Cym+Asel, does the topology exhibit the artifact of a close relationship of Cymothoidae+Corallanidae and Asellota (isopods with congruent skews), where +(s) indicates a sister-group relationship; Limnoriidea, sister-group to this taxon, where G.o. is Gyge ovalis and * indicates polyphyly; P, partitioned; NP, nonpartitioned data set; C + C, Cymothoidae+Corallanidae; M + A, sister-group Mysida+Amphipoda; Phreat., Phreatoicidea.
Amphipoda clustered within the Isopoda clade (paraphyletic isopods).
Fig. 2.
—A phylogram reconstructed using nonpartitioned NUC data set and an algorithm designed to address compositional heterogeneity: CAT-GTR (PB). Posterior Bayesian support values are shown. See figure 1 for other details.
Single-Gene Mitochondrial Data Sets
As we hypothesized that the topological instability may be driven by conflicting signals produced by different genes (Meiklejohn et al. 2014), we conducted ML phylogenetic analyses on 15 single-gene data sets (13 PCGs + 2 rRNAs). All these produced unique topologies (supplementary file S3, Supplementary Material online). Atp6 resolved Asellota+Cymothoidae+Corallanidae as the basal isopod clade; atp8 (a very small gene) produced an almost nonsensical topology (defined as: in stark disagreement with any reasonable phylogenetic hypothesis); nad1 resolved Asellota at the base; nad2 produced a slightly rearranged NUC-consensus topology; nad3 (small gene) a nonsensical topology; nad4 produced a rearranged topology (compared with NUC-consensus); nad4L (small gene) produced a highly rearranged isopod clade; despite its large size (≈1,700 bp), nad5 produced a noncanonical isopod topology; nad6 produced a nonsensical topology (paraphyletic Isopoda, rogue G. ovalis); cox1 produced a highly rearranged topology, including both the nonisopod malacostraca and isopod clades, with Phreatoicidea+Cymothoidae+Corralanidae as the basal isopod clade, and noncanonical Valvifera position; cox2 produced a slightly rearranged NUC-consensus topology; cox3 a highly rearranged topology, with a unique Asellota+Corallanidae clade at the isopod base; cytb produced basal Phreatoicidea, and the remaining taxa divided into Oniscidea and “catch all” sister-clades, where the rogue Cymothoidae+Corallanidae (along with Asellota) were on a long branch in the derived part of the clade; rrnS (or 12S) topology was in some aspects similar to the cytb, but with Phreatoicidea+Limnoriidea as the basal isopod clade; rrnL (or 16S) produced an almost nonsensical topology, with paraphyletic Isopoda and rogue G. ovalis.
Amino Acids Data Set
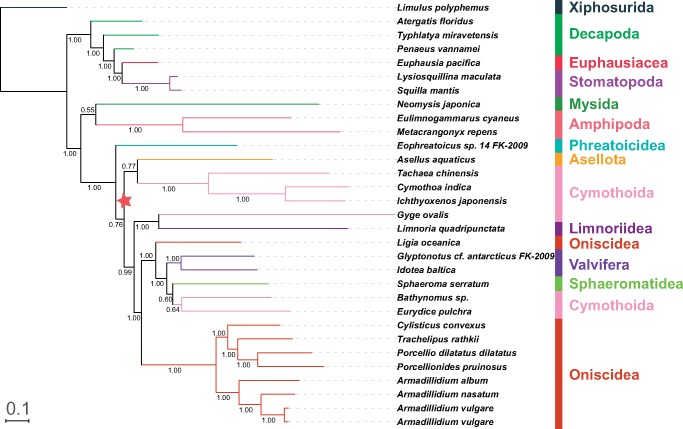
Amino acids (concatenated 13 PCGs) produced topologies that differed from the NUC-consensus one, with instable topology of the nonisopod Malacostraca, including paraphyletic Eucarida (Decapoda+Euphausiacea) in five (BI/ML, nonpartitioned/partitioned, and GHOST; fig. 3) out of six (PB; fig. 4) analyses. Sister-clade to Isopoda also varied: Mysida+Amphipoda or Mysida (table 2). In the isopod clade, partitioning had a major effect on the BI analysis: nonpartitioned data set resolved Cymothoidae+Corallanidae at the base, followed by Phreatoicidea+Asellota. Parsimony analysis produced a similar topology, but Phreatoicidea and G. ovalis + Lim. quadripunctata clades switched places. The remaining five analyses (including the partitioned BI data set) resolved Phreatoicidea as the basal clade (table 2), and the sister-group to the remaining isopods (minus Phreatoicidea) was Asellota+Cymothoidae+Corallanidae in four analyses, and Asellota in PB. Five analyses (minus PB) produced a topology of the remainder of the isopod clade that was partially congruent with the NUC-consensus topology, but instead of it being divided into Oniscidea + all other taxa, here G. ovalis+Lim. quadripunctata were at the base (except in Parsimony: Phreatoicidea). Oniscidea (rendered paraphyletic by L. oceanica) clade topology was stable in all six, but the topology of the catch-all clade was different from the NUC-consensus topology.
Fig. 3.
—A phylogram reconstructed using amino acid data set (AAs; 13 PCGs) in combination with data partitioning strategy and BI algorithm. See figure 1 for other details.
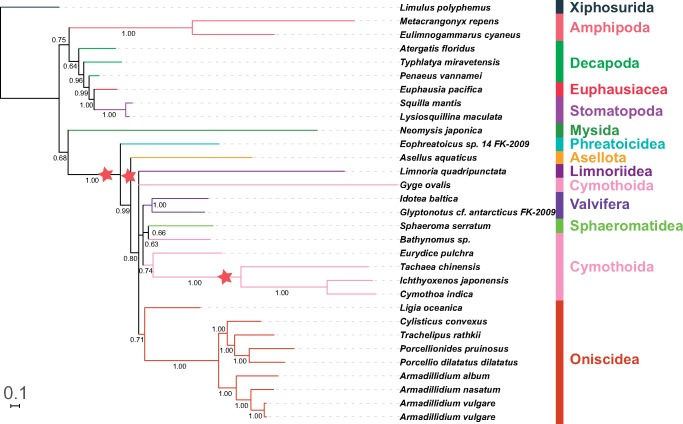
Fig. 4.
—A phylogram reconstructed using AAs data set and CAT-GTR algorithm designed for heterogeneous data sets (PB). See figure 1 for other details.
PB produced the only topology (fig. 4) with monophyletic Oniscidea, with the rogue L. oceanica at the base of the clade. Also importantly, Cymothoidae+Corallanidae clade was placed on a long branch within the catch-all clade, together with E.pulchra (all Cymothoida). The suborder was still rendered paraphyletic by the positions of Bathynomus sp. and rogue (polytomy) G. ovalis.
Gene Order-Based Phylogeny
As gene orders are unlikely to exhibit homoplasy, it has been hypothesized that they may be able to resolve difficult (deep) phylogenies in some cases (Boore 2006), so we tested this approach. As many tRNA genes were missing or we suspected that they may be misannotated, to test for the presence of false signals, we used two data sets: one excluding all tRNAs (PCGs+rRNAs) and one (PCGs+rRNAs+tRNAs) containing all (identified) tRNAs apart from the ambiguously annotated trnL genes. Both data sets produced a number of paraphyletic major clades and largely nonsensical topologies with very low bootstrap support values (supplementary file S3, Supplementary Material online: GO trees). Furthermore, the two data sets produced incongruent topologies, and different runs of the PCGs+rRNAs data set produced different topologies (supplementary file S3, Supplementary Material online).
Nuclear Marker-Based Phylogeny (18S)
To view the phylogeny of Isopoda from a nonmitochondrial perspective, we used the nuclear 18S gene. This approach can also help us test whether the underlying reason for the conflicting phylogenetic signals between different studies might be mitochondrial introgression, which, recent evidence shows, is more widespread than previously thought (Jakovlić et al. 2013; Edwards et al. 2016; Mallet et al. 2016). Results were also unstable, that is, different runs and data sets (addition and removal of taxa) would often produce different topologies, but most of them produced Asellota at the base (Limnoriidea in Parsimony, but with very low support); highly derived Cymothoida, rendered paraphyletic by the nested Limnoriidea (monophyletic in Parsimony); and Oniscidea rendered paraphyletic by the rogue Ligiidae clade (Ligia and Ligidium genera). BI, ML, and GHOST analyses produced relatively congruent topologies, with Isopoda rendered paraphyletic by the Amphipoda nested within the Cymothoida (supplementary file S3, Supplementary Material online). As PB and Parsimony analyses produced monophyletic Isopoda, with Amphipoda as the sister-clade (fig. 5), we can confidently reject this as a compositional heterogeneity artifact. Cymothoida were monophyletic only in the Parsimony analysis, and divided into two clades in all topologies: a stable monophyletic clade comprising Dajidae and Bopyridae; and a large instable (exhibiting pervasive paraphyly) clade comprising Cirolanidae, Corallanidae, and Cymothoidae families, and aforementioned “intruders” (Limnoriidea and Amphipoda). Corallanidae and Cirolanidae were mostly paraphyletic (somewhat erratic behavior of E.pulchra), whereas Cymothoidae (monophyletic) were highly derived and exhibited disproportionately long branches.
Fig. 5.
—A phylogram inferred using the nuclear 18S gene and CAT-GTR algorithm (PB). See figure 1 for other details.
Discussion
Apart from the heterotachous GHOST model, which tended to produce results identical to the common ML analysis, all other variables produced notable impacts on the topology (table 2), with unique topologies by far outnumbering identical topologies. As regards the major unresolved issue of the isopod phylogeny, the sister-clade to all other isopods (basal clade) and the monophyly of Cymothoida, mitochondrial nucleotides (NUC) consistently produced Cymothoidae and Corallanidae as the basal clade, first followed by Asellota, then by Phreatoicidea, whereas other Cymothoida (paraphyletic) tended to be scattered throughout the central “catch-all” clade. AAs, however, largely resolved Phreatoicidea as the basal isopod clade, with all remaining isopods split into two sister-clades: 1) Asellota + Cymothoidae/Corallanidae and 2) all remaining taxa. The Parsimony method produced Cymothoidae+Corallanidae at the base, followed by Asellota + (G. ovalis + Lim. quadripunctata) using both data sets. PB analysis of AAs data set produced a remarkably different topology, with Phreatoicidea at the base, followed by Asellota, but Cymothoidae+Corallanidae were relatively derived, and Cymothoida not so deeply paraphyletic. Nuclear (18S gene) topology was also strongly affected by the methodology, but mostly resolved Asellota as the basal isopod clade (Limnoriidea in Parsimony), and Cymothoida as paraphyletic, with nested Limnoriidea (not in Parsimony), but highly derived. Although it appears that we did not manage to reach a conclusion, we did manage to identify a feature of isopod mitogenomes that may explain this instability.
Base Composition Skews
Organellar genomes often exhibit a phenomenon known as strand asymmetry, or strand compositional bias, where positive AT skew values indicate more A than T on the strand, positive GC skews indicate more G than C, and vice versa (Reyes et al. 1998; Wei et al. 2010). This is believed to be caused by hydrolytic deamination of bases on the leading strand when it is single stranded, that is, during replication and/or transcription (Reyes et al. 1998; Bernt et al. 2013; Fonseca et al. 2014). Whereas other crustacean taxa usually exhibit positive overall AT skews for genes located on the plus (majority) strand and negative GC skews for genes on the minus (minority) strand (Hassanin 2006; Wei et al. 2010), isopod mitogenomes usually exhibit an inverted skew pattern (Kilpert and Podsiadlowski 2006; Kilpert et al. 2012; Yu et al. 2018). This is believed to be a consequence of an inversion of the replication origin (RO), where the changed replication order of two mitochondrial DNA strands consequently resulted in an inversed strand asymmetry (Hassanin et al. 2005; Kilpert and Podsiadlowski 2006; Wei et al. 2010; Kilpert et al. 2012; Bernt et al. 2013).
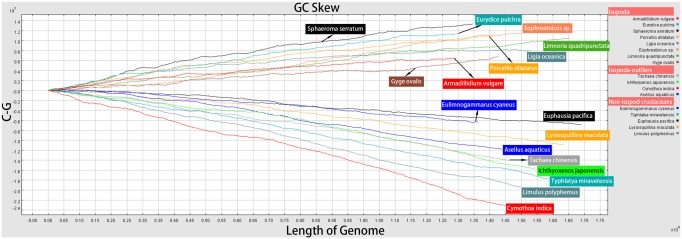
It is known that Asellus aquaticus (Asellota) possesses an inversed skew in comparison to other isopod taxa (Kilpert and Podsiadlowski 2006), but here we found that the three available Cymothoidae and Corallanidae species (C. indica, T. chinensis, and I. japonensis) also exhibit inversed skew patterns (fig. 6 and table 1). As regards nonisopod mitogenomes in the data set, they exhibit GC skews comparable to the isopod outliers, from −0.014 in Metacrangonyx repens to −0.332 in Typhlatya miravetensis, and mixed AT skews (table 1). The inversed skew in A.aquaticus led Kilpert and Podsiadlowski (2006) to speculate that this taxon branched off first in the isopod phylogeny, but a few years later Kilpert et al. (2012) noticed that the basal position of Phreatoicidea causes a conflict in explaining the inversed isopod GC skew (present in Eophreatoicus sp.; fig. 4—red stars). Our NUC data set would mostly support a modified version of the first scenario, with the RO inversion occurring in isopods after Cymothoidae+Corallanidae and Asellota branched off (figs. 1 and 2—red stars), whereas AAs would support an RO inversion in the common ancestor of Asellota+(Cymothoidae+Corallanidae) clade (fig. 3). Parsimony analyses further complicate this by Asellota forming a sister-clade with G. ovalis and Lim. quadripunctata. However, there is no support for either of these scenarios from nuclear (18S) or morphological (Wilson 2009) data, which relatively consistently indicate that Asellota (18S) or Asellota+Phreatoicidea (morphology) are the basal clade.
Fig. 6.
—Cumulative GC skews of the majority strands of a selected subset of mitogenomes used for phylogenetic analyses.
Variations in base composition can bias phylogenetic analyses (Romiguier and Roux 2017), and skew-driven LBA phylogenetic artifacts have been reported in arthropods (Hassanin et al. 2005; Hassanin 2006) and other metazoans (Sun et al. 2018). Most mitochondrial analyses produced topologies where taxa with congruent skews clustered together: outgroups+(Cymothoidae+Corallanidae)+Asellota (negative GC skews taxa), followed by the remaining Isopod taxa (positive GC skews). Therefore, we can conclude with high confidence that mitochondrial data produce artifactual clustering of branches exhibiting similar skews: the inversed skew of highly derived Cymothoidae+Corallanidae results in their clustering at the base of the isopod clade, phylogenetically close to other taxa with similar (homoplastic) skew patterns: Asellota and nonisopod Malacostraca. This explains the topological instability, incongruent phylogenetic hypotheses, and it rejects the above RO inversion scenarios. We can conclude that inversed skews of Asellota and Cymothoidae+Corallanidae are nonsynapomorphic, and that highly derived Cymothoidae+Corallanidae underwent an additional RO inversion, which eventually resulted in a double-inverted skew (homoplastic with Asellota and other Malacostraca).
In support of this, the PB (heterogeneous) analysis of the AAs data set produced an mtDNA topology that exhibited notable similarity to the 18S and morphology-based topologies, where Cymothoidae+Corallanidae clustered with E. pulchra (Cirolanidae) in the relatively derived part of the isopod clade. This indicates that a combination of the AAs data set, which is expected to be less affected by skews than nucleotides, and a heterogeneous CAT-GTR model, was the most successful in attenuating the phylogenetic artifacts caused by compositional biases. It also confutes the hypothesis that conforming skews between Asellota and Cymothoidae+Corallanidae may be a result of a mitochondrial introgression event.
Collation of Evolutionary Hypotheses
Having established the fact that taxa with conforming homoplastic skews cluster together in both AAs and NUC mitochondrial data sets (table 2), we can infer the most parsimonious hypothesis for the course of events in the evolutionary history of Isopoda. First, we can reject with confidence the basal position of Cymothoidae+Corallanidae as an artifact. This indicates that the basal isopod taxon is either Asellota (Wilson 2009), Phreatoicidea (Brusca and Wilson 1991), or Asellota+Phreatoicidea sister-clade (Wilson 1999; Dreyer and Wägele 2001; Kilpert et al. 2012). The latter two scenarios are less parsimonious, as they would require at least three independent RO inversions in the evolutionary history of Isopoda (in the ancestral isopod, in Asellota, and in Cymothoidae+Corallanidae), whereas the first scenario is more parsimonious, as it requires only two (in the ancestral isopod after the split of Asellota and in Cymothoidae+Corallanidae; fig. 5—red stars). Additionally, the 18S data set relatively consistently resolved Asellota as the basal branch (disregarding paraphyletic Isopoda and Parsimony analysis; table 2). Therefore, we can tentatively conclude that multiple evidence supports the original hypothesis of Kilpert and Podsiadlowski (2006): Asellota is the oldest isopod branch and RO inversion in isopods occurred after the Asellota branched off.
Although this resolves the issue of the deep paraphyly of Cymothoida, that is, places the rogue Cymothoidae+Corallanidae clade back within the remaining Cymothoida, 18S data still resolve the Cymothoida paraphyletic, with Limnoriidea nested between two large clades (Dajidae+Bopyridae and Cirolanidae+Corallanidae+ Cymothoidae). As Limnoriidea was resolved as the basal isopod taxon in the 18S Parsimony analysis, and Corallanidae+Cymothoidae exhibit a disproportionately long branch, we suspect that this is an LBA between two taxa exhibiting elevated evolutionary rates. The monophyly of Corallanidae is unsupported by our 18S analyses, so it will be necessary to sequence further mitogenomic (to identify skews) and nuclear data for this group of cymothoid families. This combination of skews and nuclear data would enable us to identify the exact point in the evolutionary history where the RO inversion occurred in these taxa, and infer the most parsimonious topology and/or taxonomy, that is, the one that supports a single RO inversion (or introgression event), as opposed to those that would require multiple events.
Our analyses further corroborated the existence of several rogue taxa that exhibit somewhat erratic topological behavior. The position of L.oceanica (nominally Oniscidea: Ligiidae), a recognized rogue taxon (Wilson 2009; Lins et al. 2017; Shen et al. 2017; Yu et al. 2018), was mostly resolved at the base of the “catch-all” clade in mtDNA analyses. However, in the (putatively) most reliable mitochondrial topology AAs+PB, it was resolved as the basal Oniscidea species. Although this is in perfect agreement with morphological data, which resolve Ligiidae as the most primitive Oniscidea clade (Schmidt 2008), we cannot claim that this issue is fully resolved, since the entire Ligiidae family exhibited rogue behavior in the 18S data set as well. Three Cymothoida taxa in the mtDNA data set that exhibit standard isopod skews, Bathynomus sp., E. pulchra (both Cirolanidae), and G. ovalis (Bopyridae), also exhibited rather instable topological behavior, and we did not find support for their monophyly using the mtDNA data. Although the Cirolanidae are believed to be ancient (Wetzer 2002) and highly plesiomorphic within this suborder (Brandt and Poore 2003), this is not supported by the 18S data set. As all three species exhibit highly rearranged gene orders (supplementary file S3, Supplementary Material online: Gene arrangements), and as there is evidence of a close positive correlation between the mitogenomic architectural instability and the mutation rate (Shao et al. 2003; Hassanin 2006; Xu et al. 2006), we hypothesize that frequent genome rearrangements may have resulted in an accelerated mutational rate in these species. In agreement with this hypothesis, another rogue species, Lim. quadripunctata (Limnoriidea) (Wilson 2009; Lloyd et al. 2015; Lins et al. 2017; Zou et al. 2018), also exhibits a highly rearranged gene order (supplementary file S3, Supplementary Material online: Gene arrangements), and E. pulchra and Lim. quadripunctata exhibit some of the highest evolutionary rates among the isopod (and decapod) mitogenomes (Shen et al. 2017). There are other indications that Limnoriidea is evolving under unique evolutionary pressures: Lim. quadripunctata clustered with G. ovalis in most of our mitochondrial analyses (table 2) and it was resolved as the basal isopod clade in a previous mitogenomic study (Cymothoidae+Corallanidae unavailable at the time) (Lins et al. 2017). Limnoriidea was the basal isopod clade in our 18S Parsimony analysis, and it nested within the Cymothoida in the remaining 18S analyses (sometimes together with Decapoda) (table 2). Furthermore, inverted mitogenomic skews also do not explain the disproportionately long branch of Cymothoidae in the 18S data set. This is an indication that there are other factors (aside from the RO inversions) that cause accelerated substitution rates in Cymothoidae, Corallanidae, and Limnoriidae, so we urge further studies of these taxa, as they appear to be exceptionally interesting from the molecular evolution perspective.
The topology of nonisopod Malacostraca was rather instable as well, with PB producing notably different topologies from other analyses. As nonisopod Malacostraca also exhibit notable variability in skews (table 1), we conclude that compositional heterogeneity also interfered with phylogenetic reconstruction. The relevant question for this study is that of the sister-group to Isopoda: Mysida+Amphipoda in most of our mtNUC analyses (except PB), BI+AAs, and all Parsimony topologies; Mysida in most AAs analyses (ML+GHOST+PB); and Amphipoda in the PB 18S analysis (Amphipoda were nested within the Isopoda in ML and BI 18S topologies) (table 2). As Amphipoda are considered to be the most prominent contender for this position (Wilson 2009), we can conclude that our analyses support this close relationship.
Methodological Implications
Our findings show that mitochondrial sequence data are producing artifactual LBA relationships in isopods, and thus are a poor tool for the reconstruction of their phylogeny. Intriguingly, these did not affect only the nucleotide data set, but also the amino acid data set, which should be less affected by nonadaptive compositional biases, as nonsynonymous mutations are likely to be affected by the purifying selection (as opposed to synonymous mutations). This corroborates that mitochondrial strand asymmetry (skews) can have very pronounced effects on the composition of encoded proteins (Min and Hickey 2007; Botero-Castro et al. 2018). It should be noted that a more thorough sampling of mitogenomes may have resulted in somewhat different artifacts; for example, had we included only one outgroup and one Asellota mitogenome, and a large number of Cymothoidae+Corallanidae, it is likely that we would have obtained a topology with the derived C + C, and Asellota (and possibly even the outgroup as well), clustering within the Cymothoida (inversed direction of the skew “gravity”). Regardless of the data set, it remains highly likely that attraction between the homoplastic skews of these two (distant) clades would confound the phylogenetic analysis.
All individual genes produced unique topologies, which has important implications for the interpretation of previous results inferred using single-gene data sets. This phenomenon has been observed in isopods before on a much smaller scale (Wetzer 2002), and it is in agreement with the proposed mosaic nature of (mitochondrial) genomes (Pollard et al. 2006; Degnan and Rosenberg 2009; Romiguier et al. 2013; Romiguier and Roux 2017). Intriguingly, although 12S gene produced a topology that placed Cymothoidae+Corallanidae in the derived part of the clade, Asellota clustered within the Cymothoida clade (supplementary file S3, Supplementary Material online), which supports the observation that taxa with conforming skews tend to cluster together regardless of the direction of artifactual “gravity-pull.” As gene orders produced topological instability, very low support, and almost nonsensical topologies, with highly rearranged orders at the base of the isopod clade, we hypothesize that discontinuous evolution of mitogenomic architecture evolution (Zou et al. 2017) produces phylogenetic artifacts such as LBA, and renders them useless for the task.
All of the tested standard models (BI, ML, and Parsimony) were very sensitive to compositional biases, and produced strong artifacts. This included the new heterotachous model, GHOST, which mostly produced results identical to the ML algorithm. Importantly, as regards the aforementioned feud about the most suitable methodological approach to account for compositional heterogeneity (Feuda et al. 2017; Whelan and Halanych 2017), our results indicate that (in isopods) the PB CAT-GTR model by far outperforms the partitioning (assigning different evolutionary models to different partitions). Although we discourage the use of mitochondrial data as a phylogenetic tool for the reconstruction in isopods (even in combination with nuclear and morphological data), they may still be of some use in studies focused only on the isopod suborders that exhibit congruent skews. It should be mentioned that there are other available methodological approaches designed to account for compositional heterogeneity (p4, PHASE, nhPhyML and so on) (Foster 2004; Boussau and Gouy 2006; Hassanin 2006; Gowri-Shankar and Rattray 2007; Sheffield et al. 2009; Richards et al. 2018; Yang et al. 2018), so future studies may attempt to test their performance as well.
Conclusions
With respect to our working hypothesis, we can accept the first part of it: asymmetrical mutational pressures do generate compositional heterogeneity in isopod mitogenomes and interfere with phylogenetic reconstruction. However, we were mistaken in assuming that these pressures are primarily adaptive, that is, caused by their radically diverse life histories. Instead, they appear to be driven largely by the origin of replication inversions. These appear to generate very strong compositional biases, which render the mitogenomic sequence data a very poor tool for reconstructing the evolutionary history of Isopoda. None of the tools used here managed to fully revolve these compositional biases, but PB CAT-GTR algorithm outperformed partitioning, and best results were achieved by combining it with the amino acids data set. As mtDNA data have played a major role in our understanding of the evolutionary history of life on Earth (Rubinoff et al. 2005), implications of this study are much broader than its original scope. As our findings indicate that architectural rearrangements might produce major compositional biases even on comparatively short evolutionary timescales, proving the suitability of data via GC and AT skew analyses should be a prerequisite for any study that aims to use mitochondrial data for phylogenetic reconstruction, even among the closely related taxa. These findings should not discourage scientists from sequencing further isopod mitogenomes (especially interesting are nonrepresented Asellota and Cymothoida families), as their architectural hypervariability still makes them a useful tool for unraveling the conundrums of evolution of mitochondrial architecture, and as mitochondrial skews can be used as an additional phylogenetic tool to infer the most parsimonious phylogenetic hypotheses.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
The authors would like to express their sincere appreciation to Chen Rong (Bio-Transduction Lab) for helping us to conduct some of the wet lab experiments, and the Deanship of Scientific Research at King Saud University for funding this research. They would also like to thank the anonymous reviewers for investing time and expertise into reviewing our article. This work was supported by the Earmarked Fund for China Agriculture Research System (CARS-45-15), the National Natural Science Foundation of China (31872604, 31572658), and the Deanship of Scientific Research at King Saud University (RGP 1435-012). The funders had no role in the design of the study, collection, analysis and interpretation of data, and in writing the article.
Data deposition: This project has been deposited at GenBank under the accession numbers MK079664, MK542856, MK542857 and MK542858. The remaining data are included within the article and its supplementary files.
Literature Cited
- Almeida D, Maldonado E, Vasconcelos V, Antunes A.. 2015. Adaptation of the mitochondrial genome in cephalopods: enhancing proton translocation channels and the subunit interactions. PLoS One 10:1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baele G, Raes J, Van De PY, Vansteelandt S.. 2006. An improved statistical method for detecting heterotachy in nucleotide sequences. Mol Biol Evol. 23(7):1397–1405. [DOI] [PubMed] [Google Scholar]
- Ballard JWO, Whitlock MC.. 2004. The incomplete natural history of mitochondria. Mol Ecol. 13(4):729–744. [DOI] [PubMed] [Google Scholar]
- Bernt M, Braband A, Schierwater B, Stadler PF.. 2013. Genetic aspects of mitochondrial genome evolution. Mol Phylogenet Evol. 69(2):328–338. [DOI] [PubMed] [Google Scholar]
- Boore JL. 2006. The use of genome-level characters for phylogenetic reconstruction. Trends Ecol Evol. 21(8):439–446. [DOI] [PubMed] [Google Scholar]
- Botero-Castro F, et al. 2018. In cold blood: compositional bias and positive selection drive the high evolutionary rate of vampire bats mitochondrial genomes. Genome Biol Evol. 10(9):2218–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourguignon T, et al. 2018. Transoceanic dispersal and plate tectonics shaped global cockroach distributions: evidence from mitochondrial phylogenomics. Mol Biol Evol. 35(4):970–983. [DOI] [PubMed] [Google Scholar]
- Boussau B, Gouy M.. 2006. Efficient likelihood computations with nonreversible models of evolution. Syst Biol. 55(5):756–768. [DOI] [PubMed] [Google Scholar]
- Brandt A, Poore G.. 2003. Higher classification of the flabelliferan and related Isopoda based on a reappraisal of relationships. Invert Systematics. 17(6):893–923. [Google Scholar]
- Broly P, Deville P, Maillet S.. 2013. The origin of terrestrial isopods (Crustacea: Isopoda: Oniscidea). Evol Ecol. 27(3):461–476. [Google Scholar]
- Brusca RC. 1981. A monograph on the Isopoda Cymothoidae (Crustacea) of the eastern Pacific. Zool J Linn Soc. 73(2):117–199. [Google Scholar]
- Brusca RC, Wilson GDF.. 1991. A phylogenetic analysis of the Isopoda with some classificatory recommendations. Mem Qld Mus. 31:143–204. [accessed 2019 Jan 18] Available from: http://www.vliz.be/en/imis? module=ref&refid=140503 (Accessed January 18, 2019). [Google Scholar]
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59(1):95–117. [DOI] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552. [DOI] [PubMed] [Google Scholar]
- Costello MJ, et al. 2013. Global coordination and standardisation in marine biodiversity through the World Register of Marine Species (WoRMS) and related databases. PLoS One 8(1):e51629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty SM, et al. 2017. GHOST: recovering historical signal from heterotachously-evolved sequence alignments. bioRxiv. doi: 10.1101/174789. [DOI] [PubMed] [Google Scholar]
- Degnan JH, Rosenberg NA.. 2009. Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol Evol. 24(6):332–340. [DOI] [PubMed] [Google Scholar]
- Der Sarkissian C, et al. 2015. Mitochondrial genomes reveal the extinct Hippidion as an outgroup to all living equids. Biol Lett. 11(3):20141058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyer H, Wägele J-W.. 2001. Parasites of crustaceans (Lsopoda: Bopyridae) evolved from fish parasites: molecular and morphological evidence. Zoology 103:157–178. [Google Scholar]
- Edwards SV, Potter S, Schmitt CJ, Bragg JG, Moritz C.. 2016. Reticulation, divergence, and the phylogeography–phylogenetics continuum. Proc Natl Acad Sci U S A. 113(29):8025–8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuda R, et al. 2017. Improved modeling of compositional heterogeneity supports sponges as sister to all other animals. Curr Biol. 27(24):3864–3870.e4. [DOI] [PubMed] [Google Scholar]
- Fonseca MM, Harris DJ, Posada D.. 2014. The inversion of the control region in three mitogenomes provides further evidence for an asymmetric model of vertebrate mtDNA replication. PLoS One 9(9):e106654.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PG. 2004. Modeling compositional heterogeneity. Syst Biol. 53(3):485–495. [DOI] [PubMed] [Google Scholar]
- Gawryluk RMR, et al. 2016. The earliest stages of mitochondrial adaptation to low oxygen revealed in a novel Rhizarian. Curr Biol. 26(20):2729–2738. [DOI] [PubMed] [Google Scholar]
- Gowri-Shankar V, Rattray M.. 2007. A reversible jump method for Bayesian phylogenetic inference with a nonhomogeneous substitution model. Mol Biol Evol. 24(6):1286–1299. [DOI] [PubMed] [Google Scholar]
- Grechko VV. 2013. The problems of molecular phylogenetics with the example of squamate reptiles: mitochondrial DNA markers. Mol Biol. 47(1):55–74. [DOI] [PubMed] [Google Scholar]
- Hassanin A. 2006. Phylogeny of Arthropoda inferred from mitochondrial sequences: strategies for limiting the misleading effects of multiple changes in pattern and rates of substitution. Mol Phylogenet Evol. 38(1):100–116. [DOI] [PubMed] [Google Scholar]
- Hassanin A, Léger N, Deutsch J.. 2005. Evidence for multiple reversals of asymmetric mutational constraints during the evolution of the mitochondrial genome of metazoa, and consequences for phylogenetic inferences. Syst Biol. 54(2):277–298. [DOI] [PubMed] [Google Scholar]
- Hassanin A, Ropiquet A, Couloux A, Cruaud C.. 2009. Evolution of the mitochondrial genome in mammals living at high altitude: new insights from a study of the tribe Caprini (Bovidae, Antilopinae). J Mol Evol. 68(4):293–310. [DOI] [PubMed] [Google Scholar]
- Hata H, et al. 2017. Molecular phylogeny of obligate fish parasites of the family Cymothoidae (Isopoda, Crustacea): evolution of the attachment mode to host fish and the habitat shift from saline water to freshwater. Mar Biol. 164:1–15. [Google Scholar]
- Hu F, Lin Y, Tang J.. 2014. MLGO: phylogeny reconstruction and ancestral inference from gene-order data. BMC Bioinformatics 15:354.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua CJ, et al. 2018. Basal position of two new complete mitochondrial genomes of parasitic Cymothoida (Crustacea: Isopoda) challenges the monophyly of the suborder and phylogeny of the entire order. Parasite Vectors. 11(1):628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakovlić I, Wu Q-J, Treer T, Šprem N, Gui J-F.. 2013. Introgression evidence and phylogenetic relationships among three (Para)Misgurnus species as revealed by mitochondrial and nuclear DNA markers. Arch Biol Sci. 65:1463–1467. [Google Scholar]
- Joca LK, Leray VL, Zigler KS, Brusca RC.. 2015. A new host and reproduction at a small size for the ‘snapper-choking isopod’ Cymothoa excisa (Isopoda: Cymothoidae). J Crustac Biol. 35(2):292–294. [Google Scholar]
- Jones CM, Miller TL, Grutter AS, Cribb TH.. 2008. Natatory-stage cymothoid isopods: description, molecular identification and evolution of attachment. Int J Parasitol. 38(3–4):477–491. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, Von Haeseler A, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Toh H.. 2008. Improved accuracy of multiple ncRNA alignment by incorporating structural information into a MAFFT-based framework. BMC Bioinformatics 9:212.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketmaier V, Joyce DA, Horton T, Mariani S.. 2008. A molecular phylogenetic framework for the evolution of parasitic strategies in cymothoid isopods (Crustacea). J Zool Syst Evol Res. 46:19–23. [Google Scholar]
- Kilpert F, Held C, Podsiadlowski L.. 2012. Multiple rearrangements in mitochondrial genomes of Isopoda and phylogenetic implications. Mol Phylogenet Evol. 64(1):106–117. [DOI] [PubMed] [Google Scholar]
- Kilpert F, Podsiadlowski L.. 2006. The complete mitochondrial genome of the common sea slater, Ligia oceanica (Crustacea, Isopoda) bears a novel gene order and unusual control region features. BMC Genomics 7(1):241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaczkowski B, Thornton JW.. 2004. Performance of maximum parsimony and likelihood phylogenetics when evolution is heterogeneous. Nature 431(7011):980.. [DOI] [PubMed] [Google Scholar]
- Krell F-T, Cranston PS.. 2004. Which side of the tree is more basal? Syst Entomol. 29(3):279–281. [Google Scholar]
- Lan T, et al. 2017. Evolutionary history of enigmatic bears in the Tibetan Plateau-Himalaya region and the identity of the yeti. Proc R Soc B. 284(1868):20171804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Calcott B, Ho SYW, Guindon S.. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29(6):1695–1701. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Brinkmann H, Philippe H.. 2007. Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol Biol. 7(Suppl 1):S4.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslett D, Canbäck B.. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 24(2):172–175. [DOI] [PubMed] [Google Scholar]
- Lavrov DV, Boore JL, Brown WM.. 2000. The complete mitochondrial DNA sequence of the horseshoe crab Limulus polyphemus. Mol Biol Evol. 17(5):813–824. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P.. 2007. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23(1):127–128. [DOI] [PubMed] [Google Scholar]
- Li H, et al. 2017. Mitochondrial phylogenomics of Hemiptera reveals adaptive innovations driving the diversification of true bugs. Proc R Soc B Biol Sci. 284(1862):20171223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lins LSF, Ho SYW, Lo N.. 2017. An evolutionary timescale for terrestrial isopods and a lack of molecular support for the monophyly of Oniscidea (Crustacea: isopoda). Org Divers Evol. 17(4):813–820. [Google Scholar]
- Lins LSF, Ho SYW, Wilson GDF, Lo N.. 2012. Evidence for Permo-Triassic colonization of the deep sea by isopods. Biol Lett. 8(6):979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F-F, Li Y-P, Jakovlić I, Yuan X-Q.. 2017. Tandem duplication of two tRNA genes in the mitochondrial genome of Tagiades vajuna (Lepidoptera: Hesperiidae). Eur J Entomol. 114:407–415. [Google Scholar]
- Lloyd RE, et al. 2015. The complete mitochondrial genome of Limnoria quadripunctata Holthuis (Isopoda Limnoriidae). Mitochondrial DNA 26(6):825–826. [DOI] [PubMed] [Google Scholar]
- Lopez P, Casane D, Philippe H.. 2002. Heterotachy, an important process of protein evolution. Mol Biol Evol. 19(1):1–7. [DOI] [PubMed] [Google Scholar]
- Maddock ST, et al. 2016. Next-generation mitogenomics: a comparison of approaches applied to caecilian amphibian phylogeny. PLoS One 11(6):e0156757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet J, Besansky N, Hahn MW.. 2016. How reticulated are species? BioEssays 38(2):140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MB, Bruce NL, Nowak BF.. 2016. Review of the fish-parasitic genus Cymothoa Fabricius, 1793 (Crustacea: Isopoda: Cymothoidae) from Australia. Zootaxa 4119(1):1–72. [DOI] [PubMed] [Google Scholar]
- Meiklejohn KA, et al. 2014. Incongruence among different mitochondrial regions: a case study using complete mitogenomes. Mol Phylogenet Evol. 78:314–323. [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T.. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE). New Orleans. p. 1–8.
- Min XJ, Hickey DA.. 2007. DNA asymmetric strand bias affects the amino acid composition of mitochondrial proteins. DNA Res. Int J Rapid Publ Rep Genes Genomes. 14:201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, Nguyen MAT, von Haeseler A.. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 30(5):1188–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishmar D, et al. 2003. Natural selection shaped regional mtDNA variation in humans. Proc Natl Acad Sci U S A. 100(1):171–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan CC, et al. 2013. Heterogeneous models place the root of the placental mammal phylogeny. Mol Biol Evol. 30(9):2145–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie RE, et al. 2018. The phylogeny of Galerucinae (Coleoptera: Chrysomelidae) and the performance of mitochondrial genomes in phylogenetic inference compared to nuclear rRNA genes. Cladistics 34(2):113–130. [DOI] [PubMed] [Google Scholar]
- Phillips M, McLenachan P, Down C, Gibb G, Penny D.. 2006. Combined mitochondrial and nuclear DNA sequences resolve the interrelations of the major Australasian marsupial radiations. Syst Biol. 55:122–137. [DOI] [PubMed] [Google Scholar]
- Pollard DA, Iyer VN, Moses AM, Eisen MB.. 2006. Widespread discordance of gene trees with species tree in Drosophila: evidence for incomplete lineage sorting. PLoS Genet. 2(10):e173.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poore GCB, Bruce NL.. 2012. Global diversity of marine isopods (except Asellota and Crustacean symbionts). PLoS One 7(8):e43529.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A, Gissi C, Pesole G, Saccone C.. 1998. Asymmetrical directional mutation pressure in the mitochondrial genome of mammals. Mol Biol Evol. 15(8):957–966. [DOI] [PubMed] [Google Scholar]
- Richards EJ, Brown JM, Barley AJ, Chong RA, Thomson RC.. 2018. Variation across mitochondrial gene trees provides evidence for systematic error: How much gene tree variation is biological? Syst Biol. 67(5):847–860. [DOI] [PubMed] [Google Scholar]
- Romiguier J, Ranwez V, Delsuc F, Galtier N, Douzery E.. 2013. Less is more in mammalian phylogenomics: AT-rich genes minimize tree conflicts and unravel the root of placental mammals. Mol Biol Evol. 30(9):2134–2144. [DOI] [PubMed] [Google Scholar]
- Romiguier J, Roux C.. 2017. Analytical biases associated with GC-content in molecular evolution. Front Genet. 8:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, et al. 2012. Mrbayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinoff D, Holland BS, Savolainen V.. 2005. Between two extremes: mitochondrial DNA is neither the panacea nor the nemesis of phylogenetic and taxonomic inference. Syst Biol. 54(6):952–961. [DOI] [PubMed] [Google Scholar]
- Rudy J, Rendoš M, Ľuptáčik P, Mock A.. 2018. Terrestrial isopods associated with shallow underground of forested scree slopes in the Western Carpathians (Slovakia). ZooKeys 801:323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt C. 2008. Phylogeny of the terrestrial Isopoda (Oniscidea): a review. Arthropod Syst Phylogeny. 66:191–226. [Google Scholar]
- Scott GR, et al. 2011. Molecular evolution of cytochrome c oxidase underlies high-altitude adaptation in the bar-headed goose. Mol Biol Evol. 28(1):351–363. [DOI] [PubMed] [Google Scholar]
- Shao R, Dowton M, Murrell A, Barker SC.. 2003. Rates of gene rearrangement and nucleotide substitution are correlated in the mitochondrial genomes of insects. Mol Biol Evol. 20(10):1612–1619. [DOI] [PubMed] [Google Scholar]
- Sheffield NC, Song H, Cameron SL, Whiting MF.. 2009. Nonstationary evolution and compositional heterogeneity in beetle mitochondrial phylogenomics. Syst Biol. 58(4):381–394. [DOI] [PubMed] [Google Scholar]
- Shen Y, et al. 2017. The first complete mitogenome of the South China deep-sea giant isopod Bathynomus sp. (Crustacea: Isopoda: Cirolanidae) allows insights into the early mitogenomic evolution of isopods. Ecol Evol. 7(6):1869–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Li Q, Kong L, Yu H.. 2018. Multiple reversals of strand asymmetry in molluscs mitochondrial genomes, and consequences for phylogenetic inferences. Mol Phylogenet Evol. 118:222–231. [DOI] [PubMed] [Google Scholar]
- Swofford DL. 2002. PAUP. Phylogenetic analysis using parsimony (and other methods). Sunderland (MA: ): Sinauer Associates. [Google Scholar]
- Talavera G, Castresana J.. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56(4):564–577. [DOI] [PubMed] [Google Scholar]
- Talavera G, et al. 2011. What is the phylogenetic signal limit from mitogenomes? The reconciliation between mitochondrial and nuclear data in the Insecta class phylogeny. BMC Evol Biol. 11(1):315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JM, Horspool D, Brown G, Tcherepanov V, Upton C.. 2007. GraphDNA: a Java program for graphical display of DNA composition analyses. BMC Bioinformatics 8(1):21.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ.. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–W235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei SJ, et al. 2010. New views on strand asymmetry in insect mitochondrial genomes. PLoS One 5:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzer R. 2002. Mitochondrial genes and isopod phylogeny (Peracarida: Isopoda). J Crustac Biol. 22(1):1–14. [Google Scholar]
- Wetzer R, Pérez-Losada M, Bruce N.. 2013. Phylogenetic relationships of the family Sphaeromatidae Latreille, 1825 (Crustacea: Peracarida: Isopoda) within Sphaeromatidea based on 18S-rDNA molecular data. Zootaxa 3599:161–177. [DOI] [PubMed] [Google Scholar]
- Whelan NV, Halanych KM.. 2017. Who let the CAT out of the bag? Accurately dealing with substitutional heterogeneity in phylogenomic analyses. Syst Biol. 66(2):232–255. [DOI] [PubMed] [Google Scholar]
- Willis SC. 2017. One species or four? Yes!… and, no. Or, arbitrary assignment of lineages to species obscures the diversification processes of Neotropical fishes. PLoS One 12(2):e0172349.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson G. 1999. Some of the deep-sea fauna is ancient. Crustac 72(8):1019–1030. [Google Scholar]
- Wilson G. 2008. Global diversity of Isopod crustaceans (Crustacea; Isopoda) in freshwater. Hydrobiologia 595(1):231–240. [Google Scholar]
- Wilson GDF. 2009. The phylogenetic position of the Isopoda in the Peracarida (Crustacea: Malacostraca). Arthropod Syst Phylogeny. 67:159–198. [Google Scholar]
- Wolff JN, Ladoukakis ED, Enríquez JA, Dowling DK.. 2014. Mitonuclear interactions: evolutionary consequences over multiple biological scales. Philos Trans R Soc B Biol Sci. 369(1646):20130443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Jameson D, Tang B, Higgs PG.. 2006. The relationship between the rate of molecular evolution and the rate of genome rearrangement in animal mitochondrial genomes. J Mol Evol. 63:375–392. [DOI] [PubMed] [Google Scholar]
- Yang H, Li T, Dang K, Bu W.. 2018. Compositional and mutational rate heterogeneity in mitochondrial genomes and its effect on the phylogenetic inferences of Cimicomorpha (Hemiptera: Heteroptera). BMC Genomics 19:264.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, An J, Li Y, Boyko CB.. 2018. The first complete mitochondrial genome of a parasitic isopod supports Epicaridea Latreille, 1825 as a suborder and reveals the less conservative genome of isopods. Syst Parasitol. 95(5):465–478. doi: 10.1007/s11230-018-9792-2. [DOI] [PubMed] [Google Scholar]
- Zhang D, et al. 2018. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. bioRxiv. doi: 10.1101/489088. [DOI] [PubMed] [Google Scholar]
- Zhong B, et al. 2011. Systematic error in seed plant phylogenomics. Genome Biol Evol. 3:1340–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H, et al. 2017. The complete mitochondrial genome of parasitic nematode Camallanus cotti: extreme discontinuity in the rate of mitogenomic architecture evolution within the Chromadorea class. BMC Genomics 18(1):840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H, et al. 2018. The complete mitochondrial genome of Cymothoa indica has a highly rearranged gene order and clusters at the very base of the Isopoda clade. PLoS One 13(9):e0203089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.