ABSTRACT
The Pitx2 gene encodes a homeobox transcription factor that is required for mammalian development. Disruption of PITX2 expression in humans causes congenital heart diseases and is associated with atrial fibrillation; however, the cellular and molecular processes dictated by Pitx2 during cardiac ontogeny remain unclear. To characterize the role of Pitx2 during murine heart development we sequenced over 75,000 single cardiac cell transcriptomes between two key developmental timepoints in control and Pitx2 null embryos. We found that cardiac cell composition was dramatically altered in mutants at both E10.5 and E13.5. Interestingly, the differentiation dynamics of both anterior and posterior second heart field-derived progenitor cells were disrupted in Pitx2 mutants. We also uncovered evidence for defects in left-right asymmetry within atrial cardiomyocyte populations. Furthermore, we were able to detail defects in cardiac outflow tract and valve development associated with Pitx2. Our findings offer insight into Pitx2 function and provide a compilation of gene expression signatures for further detailing the complexities of heart development that will serve as the foundation for future studies of cardiac morphogenesis, congenital heart disease and arrhythmogenesis.
KEY WORDS: Pitx2, Left-right asymmetry, Single cell RNA-seq, Cardiac development
Summary: Using single cell transcriptomics to inspect Pitx2 function in cardiac development and left-right cellular specification, this study characterizes all deviations in cell composition, cellular state and differentiation trajectories.
INTRODUCTION
Pitx2 encodes a paired related homeodomain transcription factor that is essential for both human and mouse development. Investigations aimed at dissecting the biological role of Pitx2 are important, especially given that Pitx2 has been implicated in several human diseases, including Rieger syndrome, ocular dysgenesis with glaucoma, acute appendicitis and atrial fibrillation (AF), the most common sustained human arrhythmia (Ellinor et al., 2010; Gudbjartsson et al., 2007; Lin et al., 1999; Lu et al., 1999; Semina et al., 1996; Syeda et al., 2017). In postnatal cardiomyocytes (CMs), Pitx2 regulates genes that are important for the cellular response to reactive oxygen species (ROS), and is itself a target of Nrf2 (also known as Nfe2l2), a master transcriptional regulator of the cellular antioxidant response (Tao et al., 2016). However, less is known about the cell type-specific direct targets for Pitx2 during cardiac development.
Early during embryogenesis, Pitx2 is directly regulated by the Nodal-mediated left-right asymmetry (LRA) pathway, which confers left-sided morphogenesis onto all organs in the body (Logan et al., 1998; Piedra et al., 1998; Yoshioka et al., 1998). Nodal is a Tgfβ family signaling molecule that participates in the early break in symmetry in mammalian embryos and Nodal-mediated regulation of Pitx2 takes place via an asymmetric cis-regulatory element located within the Pitx2 gene body. As a downstream effector of LRA signaling, Pitx2 plays an essential function at the late stages of LRA through mechanisms that remain poorly understood, particularly in the developing heart.
During heart development, Pitx2 has two main functions: morphogenesis of the outflow tract (OFT) and left-right specification of the atria. Pitx2 is required for complete OFT septation (Liu et al., 2001). Conditional mutagenesis revealed that Pitx2 functions in the second heart field (SHF) to regulate proliferation of OFT myocardium, and that Pitx2 was dispensable in the cardiac neural crest (Ai et al., 2006). In the left atrium, Pitx2 confers left atrial morphology (Liu et al., 2001). Pitx2 null mutant left atria have right-sided morphologic characteristics including venous valves and trabeculated myocardium (Liu et al., 2001). Moreover, Pitx2-deficient embryos also possess bilateral or ectopic sinoatrial nodes (SANs), which may explain the predisposition to AF that is observed in adult animals with decreased Pitx2 expression (Ammirabile et al., 2012; Mommersteeg et al., 2007; Wang et al., 2010). In addition to OFT morphogenesis, Pitx2 has also been implicated in atrioventricular valve development. Further, morphogenesis of both the AV cushions and the dorsal mesenchymal protrusion are defective in Pitx2 null embryos, suggesting an essential function for Pitx2 during ventricular septation.
Here, we used single cell transcriptomics to inspect Pitx2 function in cardiac development and left-right cellular specification. Deployment of a high-throughput single cell RNA-seq (scRNA-seq) platform on cardiac tissue dissected from both control and Pitx2 null embryos at embryonic day (E)10.5 and E13.5 was carried out to characterize all deviations in cell composition, cellular state and differentiation trajectories. Our data revealed that the cell fates of SHF progenitors in Pitx2-deficient hearts are severely affected at the transcriptional level. We also uncovered the transcriptional consequences associated with the loss of proper left-right specification in atrial CM populations. Finally, we detailed the transcriptional and cellular compositional shifts associated with abnormal cardiac cushion remodeling, valvulogenesis and vasculature development observed in Pitx2-deficient hearts.
RESULTS
Profiling of control and Pitx2-deficient E10.5 cardiac tissue
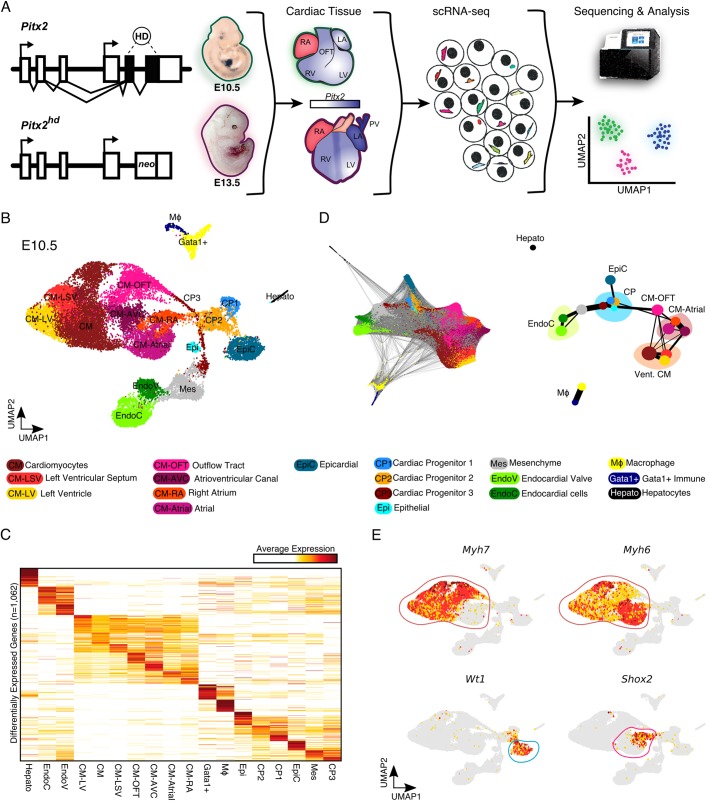
To characterize the precise cellular and molecular events dictated by Pitx2 during cardiac ontogeny, we first focused on E10.5, when Pitx2 is highly expressed and atrial septation, valvulogenesis, atrioventricular junction formation and OFT remodeling begin to occur. We performed droplet-based scRNA-seq on E10.5 murine cardiac tissue derived from control and Pitx2 null (Pitx2hd−/−) animals (Fig. 1A). After computational processing, we carried out graph-based clustering followed by dimensionality reduction using Uniform Manifold Approximation and Projection (UMAP) (McInnes and Healy, 2018). Overall, we captured 22,797 cells which belonged to 18 clusters that we assigned cell identities to based on their top differentially expressed genes (Fig. 1B,C, Fig. S1A and Table S1). Overall, we were able to identify a total of 1062 differentially expressed genes and several interesting markers for all cardiac cell types and cell states present at E10.5. We detected all major cardiac cell types, including CMs, endocardial cells (EndoCs), mesenchymal cells (Mes), epicardial cells (EpiC), macrophages (MΦ), and cardiac progenitor cells (CPs). Interestingly, the cardiac progenitors were heterogenous and were more similar to epicardial cells. Moreover, we detected several CM subtypes, which we categorized according to previously detailed markers for E10.5 CM heterogeneity (Li et al., 2016), including outflow tract CMs (CM-OFT), atrial CMs (CM-Atrial), CMs of the left ventricular septum (CM-LSV), left ventricle (CM-LV), and atrioventricular canal (CM-AVC). We also discovered a population of immune cells expressing high levels of Gata1 (Fig. S1A).
Fig. 1.
Single cell profiling of Pitx2hd−/− cardiac tissue at E10.5. (A) Schematic of the study. (B) UMAP representation of single cell transcriptomes derived from E10.5 control and Pitx2 null cardiac tissue. (C) Heatmap showing the average expression for the top differentially expressed genes between E10.5 cardiac cell clusters (n=1062). (D) Force-directed graph of PAGA-initialized embedding (left). PAGA graph showing the relationships between all the E10.5 clusters (right). (E) UMAP feature plots showing the expression of individual genes. High expression is dark red, medium expression is yellow and no expression is gray. Encircled clusters are colored according to Fig. 1B. CM-AVC, atrioventricular cardiomyocytes; CM-LSV, cardiomyocytes of left ventricular septum; CM-LV, cardiomyocytes of left ventricle; CM-OFT, cardiomyocytes of outflow tract; CM-RA, right atrial cardiomyocytes; CP, cardiac progenitors; EndoC, endocardial cells; EndoV, endocardial valve cells; Epi, epithelial cells; EpiC, epicardial cells; Gata1+, Gata1-expressing immune cells; LA, left atrium; LV, left ventricle; Mes, mesenchymal cells; MΦ, macrophage; OFT, outflow tract; PV, pulmonary vein; RA, right atrium; RV, right ventricle.
To further explore the global topology of this scRNA-seq dataset, we applied partition-based graph abstraction (PAGA), an algorithm that maps discrete connected and continuous connected cell-to-cell variation (Wolf et al., 2019). PAGA is well suited for visualizing complex developmental trajectories (Pijuan-Sala et al., 2019). The PAGA-initialized cell embeddings and resulting PAGA graphs were consistent with our UMAP results (Fig. 1D). Indeed, the Myh7-expressing ventricular CMs clearly separated from the Myh6-expressing atrial CM clusters (Fig. 1E). The similar transcriptional relationships of the CPs and Wt1-expressing EpiC cells was also evident. Moreover, the connection of the CPs with the endocardial and myocardial branches is consistent with lineage contributions of SHF progenitors (Cai et al., 2003).
We next grouped and quantified all major cardiac cell types to determine the cellular composition of the E10.5 heart (Fig. S1B). The CMs were the most prevalent cell type profiled, making up ∼67% percent of our dataset. Endothelial cells (9.8%), mesenchymal cells (9.7%), CPs (5.5%), EpiCs (4.9%) and immune cells (2.7%) comprised the remainder of the heart tissue. These results are consistent with a recent scRNA-seq study detailing a comparable stage of human ontogeny (Cui et al., 2019). However, they differ from a Fluidigm-based scRNA-seq study carried out on the E10.5 mouse heart, in which CMs were reported to comprise close to 50% of the heart (Li et al., 2016). Additional studies beyond the scope of this paper are required to comprehensively detail the composition of the developing heart and determine the cell type biases associated with various scRNA-seq platforms.
Cardiac tissue composition is altered in Pitx2 null E10.5 embryonic hearts
Next, we wanted to discern the cellular differences between control and Pitx2 null E10.5 embryonic cardiac tissue (Fig. S2A). We found that several clusters of cells displayed unequal composition between the two genotypes. To determine which clusters were statistically different we performed a chi-square-based cluster composition test on the scRNA-seq dataset (Li et al., 2018; Xiao et al., 2018). We found that EpiCs, EndoCs, CPs, MΦs and CM-LV cells were more prevalent in Pitx2-deficient animals, whereas CM-OFT, mesenchymal cells, atrial CMs and epithelial-mesenchymal transition (EMT) cells decreased in Pitx2 mutants compared with controls (Fig. S2B). Overall, we were able to characterize the putative cellular composition shifts present in Pitx2-deficient embryonic cardiac tissue.
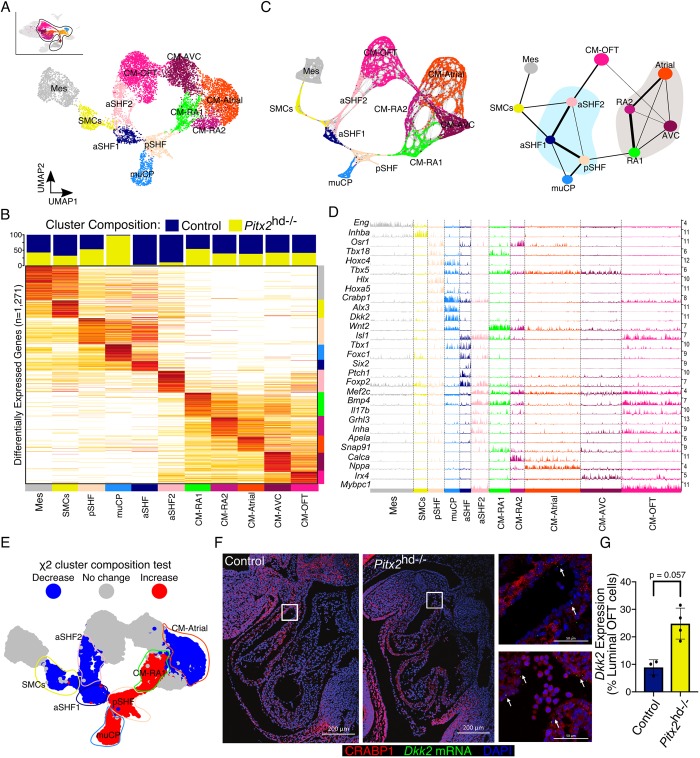
We next examined Pitx2 expression across these clusters and found that many CM and CP clusters expressed significant levels of Pitx2 (Fig. S2C). Thus, we subset these cell populations along with closely interconnected clusters and performed iterative clustering before UMAP dimensionality reduction to gain further insight into the mutant phenotype (Fig. 2A). Differential expression analysis of these clusters provided us with a gene list from which we were able to more accurately assign cell identities to CPs (Fig. 2B, and Table S2) (reviewed by Meilhac and Buckingham, 2018). And again, the PAGA-initialized cell embeddings were in close agreement with our UMAP results (Fig. 2C). Indeed, we found Tbx18-positive posterior second heart field (pSHF) progenitors, and two clusters of Isl1-expressing cells that resembled anterior second heart field (aSHF) progenitors (Fig. 2D and Fig. S2D). Strikingly, the aSHF2 population, which appeared to be actively differentiating into CM-OFT cells, expressed several unique transcripts, including the cytokine interleukin 17b (Il17b), inhibin subunit alpha (Inha), grainyhead like transcription factor 3 (Grhl3) and the peptide hormone apelin receptor early endogenous ligand (Apela). Interestingly, we identified a cluster of cells that was predominantly derived from Pitx2 mutant tissue and was transcriptionally proximal to SHF progenitors (Fig. 2B,C). These mutant CP-like cells, which we term muCPs, expressed several neural crest markers, including, Ngfr (P75), Crabp1, Gja1 (Cx43) and Twist1 (Fig. 2D and Fig. S2D). Moreover, they appeared to possess several features attributed to altered Wnt signaling, including the expression of Wnt2 (Fig. 2D). Markers for the pharyngeal mesoderm (PM), such as Lhx2 and Tcf21, were also present in the pSHF cluster (Fig. S2D). Consistent with the role of Pitx2 in the PM gene regulatory network (Harel et al., 2012), the expression of these markers was absent in the muCP population.
Fig. 2.
Pitx2-dependent cardiac cell composition at E10.5. (A) UMAP plot of subclustered E10.5 cells. (B) Cluster composition colored by genotype (top). Average gene expression heatmap showing iterative clustering results from Fig. 2A (bottom). Clusters are colored according to χ2 cluster composition analysis of E10.5 Pitx2 null cardiac tissue compared with control. (C) Force-directed graph of PAGA-initialized embedding derived from cells in Fig. 2A (left). PAGA graph showing the relationships between the subclusters (right). (D) Track plot showing gene expression (rows) for cells in highlighted clusters (columns). Track height represents gene expression. Numbers (right) indicate maximum detected expression. (E) χ2 cluster composition analysis of subclustered E10.5 Pitx2 null cardiac tissue compared with control. (F) CRABP1 IF (red) and Dkk2 FISH (green) of E10.5 embryos. Arrows indicate cells expressing Dkk2. Images on right are magnifications of the boxed areas in Control (top) and Pitx2 null (bottom) embryos. (G) Quantification of Fig. 2F. Data are mean±s.e.m. (Mann–Whitney U test). aSHF, anterior second heart field progenitors; CM-Atrial, atrial cardiomyocytes; CM-AVC, atrioventricular cardiomyocytes; CM-OFT, cardiomyocytes of outflow tract; CM-RA, right atrial cardiomyocytes; Mes, mesenchymal cells; muCP, progenitors enriched in Pitx2 null tissue; pSHF, posterior second heart field progenitors; SMCs, smooth muscle cells.
The iterative clustering of E10.5 cells also uncovered further heterogeneity in the CM populations (Fig. 2A-C). Among the two right atrial (RA) clusters identified, CM-RA1 possessed transcriptional features of the SAN, including the expression of Isl1, Tbx18, Shox2 and Hcn4 (Fig. S2D) among several additional markers (Fig. 2C). We next performed chi-square cluster composition analysis and found that muCP, pSHF and CM-RA1 clusters were more enriched in the Pitx2 mutants compared with controls (Fig. 2E). Conversely, the CM-Atrial, aSHF1, aSHF2 and smooth muscle cell (SMC) clusters possessed fewer Pitx2 mutant cells than control cells. The putative bifurcating developmental trajectory of aSHF cells to SMCs and CM-OFT also appeared to be compromised in the mutant outflow tract (Fig. 2E).
To validate the presence of the muCP cluster in the Pitx2-deficient heart, we performed immunofluorescence (IF) and fluorescence in situ hybridization (FISH) co-labeling experiments. Dkk2 (dikkopf Wnt signaling pathway inhibitor 2), which encodes for a secreted Wnt signaling modulator that has been previously shown to regulate myocardial proliferation (Phillips et al., 2011), is highly enriched in the muCPs (Fig. 2D). Crabp1 expression marked the muCP cluster, in addition to other cell populations in the OFT (Fig. S2D). In the hearts of Pitx2hd−/− animals, we observed increased expression of Dkk2 mRNA in CRABP1-positive cells localized to the endocardial portion of the OFT compared with controls (Fig. 2F,G and Fig. S2E). Furthermore, IF for ISL1, a marker for aSHF1 and aSHF2 clusters, indicated that that the number of ISL1-expressing cells in the left OFT was diminished in Pitx2 null animals (Fig. S3A). Overall, these data support our scRNA-seq findings.
Profiling of control and Pitx2-deficient E13.5 cardiac tissue
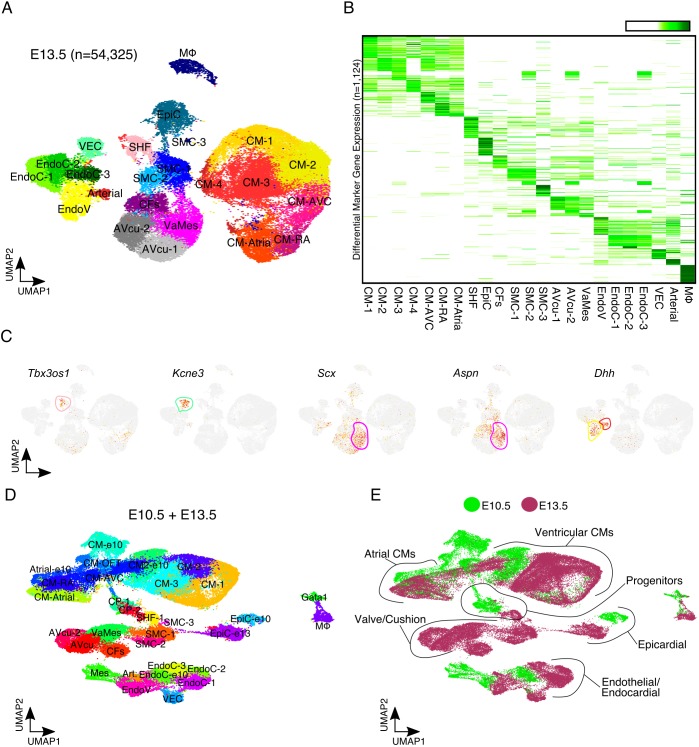
Pitx2 has previously been implicated in OFT and cardiac valve formation (Ai et al., 2006; Ma et al., 2013). To more accurately detail the role of Pitx2 in OFT development and valvulogenesis we performed scRNA-seq on control and Pitx2 null cardiac tissue at E13.5, a crucial timepoint for cardiac valve maturation and OFT septation. We captured a total of 54,325 E13.5 cells from control and Pitx2-deficient embryonic hearts (Fig. 3A). After average differential expression analysis between clusters, we identified 1124 markers (Fig. 3B and Table S3). We detected several cell types with transcriptional signatures of cardiac cushions, valves, vascular endothelial cells (VECs) and arterial cells, in addition to CMs, SMCs and EndoCs (Fig. 3A,B). As expected, E13.5 cardiac tissue composition differed greatly from E10.5 cardiac tissue (Fig. S4A). The proportion of CMs in control hearts was reduced to 44%, mesenchymal cells expanded to 21% and endothelial cells increased to 16%. These results are consistent with E13.5-E14.5 cardiac Drop-seq data we have previously reported (Xiao et al., 2018).
Fig. 3.
Single cell profiling of Pitx2hd−/− cardiac tissue at E13.5. (A) UMAP plot of 54,325 single cell transcriptomes captured from control and Pitx2 null E13.5 embryonic cardiac tissue. (B) Average expression heatmap for each E13.5 cell cluster (n=1124). High expression is indicated in green. (C) UMAP feature plots showing expression of indicated individual genes at E13.5. High expression is indicated in red, moderate expression in yellow and no expression is shown as gray. Outlines indicate clusters colored according to A. (D) UMAP plot showing individual cardiac clusters from merged E10.5 and E13.5 data. (E) UMAP plot showing the developmental timepoint identity of each individual transcriptome. Green indicates E10.5 cells, and maroon denotes cells from the E13.5 heart. Arterial, arterial endothelial cells; AVcu, atrioventricular cushion cells; CFs, cardiac fibroblasts; CM, cardiomyocyte; CM-Atrial, atrial cardiomyocytes; CM-AVC, atrioventricular cardiomyocytes; CM-RA, right atrial cardiomyocyte; EndoC, endocardial cells; EndoV, endocardial valve cells; EpiC, epicardial cell; Gata1, Gata1-expressing immune cells; Mes, mesenchymal cells; MΦ, macrophage; SHF, second heart field progenitor; SMC, smooth muscle cells; VaMes, valve mesenchyme; VEC, vascular endothelial cells.
Cells from the valve mesenchyme cluster (VaMes) expressed scleraxis (Scx), an important transcription factor required for valvulogenesis (Lincoln et al., 2006). In addition, they expressed asporin (Aspn), which encodes a cartilage extracellular protein and heart valve enriched gene (Chakraborty et al., 2008). Endocardial valve cells (EndoV) and arterial cells expressed several markers, including desert hedgehog (Dhh) (Fig. 3C). We found that VaMes, SMC, EndoV, VEC and arterial cell clusters were less numerous in Pitx2-deficient cardiac tissue (Fig. S4B). Moreover, we identified a cluster of SHF-like cells expressing high levels of the T-box 3 opposite strand 1 (Tbx3os1) antisense lncRNA, which were more numerous in control animals (Fig. 3C).
Pseudotime analysis uncovers defective SHF differentiation dynamics in Pitx2 null hearts
To further explore the phenotypic defects present in Pitx2 null embryos, we merged the E10.5 and E13.5 scRNA-seq datasets (Fig. 3D,E). The majority of cardiac cell types differed greatly across the two timepoints (Fig. 1 and Fig. 3). In order to characterize the potential defects in SHF progenitor cell differentiation we subset the merged data to include aSHF1, aSHF2, pSHF, muCP, CM-OFT, CM-Atrial and CM-RA clusters. After re-clustering and UMAP dimensionality reduction, we uncovered considerable atrial and RA CM heterogeneity (Fig. 4A). The subset of single cell transcriptomes differed distinctively by developmental timepoint (Fig. S5A). Consistent with our previous composition analysis, we found that Pitx2 null tissue displayed decreased OFT contribution and altered atrial CM composition (Fig. S5B).
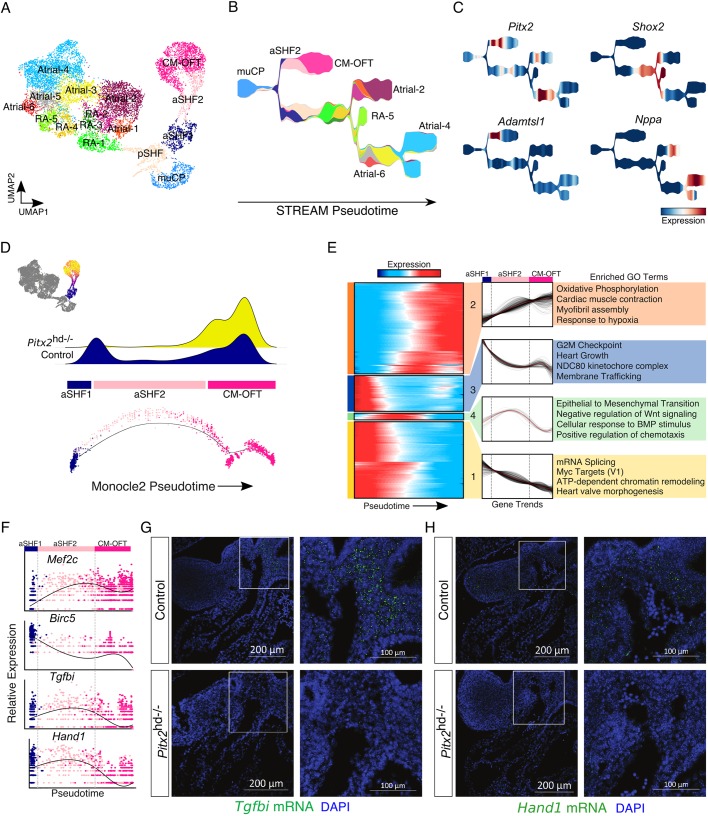
Fig. 4.
Defective cardiac outflow tract differentiation dynamics in Pitx2 null embryonic hearts. (A) UMAP plot showing subclusters derived from merged E10.5 and E13.5 embryonic datasets. (B) STREAM plot showing cluster identities. Colored according to Fig. 4A. (C) STREAM plots showing gene expression. (D) Pseudotime score for each individual outflow tract-associated transcriptome projected onto the UMAP plot (top left). Cells not included in analysis are colored dark gray. Density plot of control and Pitx2 mutant cells across the OFT ontological trajectory (top). Pseudotime minimal spanning tree (MST) plots displaying pseudotime cell identity (bottom). (E) Heatmap showing clusters of dynamic gene expression trends across pseudotime (left). Results of hierarchical clustering of gene expression trends shown in heatmap (middle). Gene ontology (GO) analysis of genes from each indicated dynamic cluster (right). (F) Individual gene expression trends plotted across pseudotime. Cells are colored according to their identity (Fig. 4A). (G) FISH for Tgfbi (green) at E10.5. Nuclei are stained blue (DAPI). (H) FISH for Hand1 (green) at E10.5. Nuclei are stained blue (DAPI). In G and H, images on right are magnifications of the boxed areas in Control (top) and Pitx2 null (bottom) embryos. aSHF, anterior second heart field progenitors; CM-OFT, cardiomyocytes of outflow tract; muCP, progenitors enriched in Pitx2 null tissue; pSHF, posterior second heart field progenitors; RA, right atrial cells.
Next, to reconstruct the cell differentiation and cell maturation trajectories of these clusters in more detail we applied the STREAM (Single cell Trajectories Reconstruction, Exploration and Mapping) analysis pipeline (Chen et al., 2019). All progenitor clusters (muCP, aSHF1, aSHF2 and pSHF) were located at the root of the branching STREAM trajectory, whereas the tips of the branches were occupied by cells from more differentiated clusters (Fig. 4B). The STREAM output was consistent with our UMAP results and suggested that Atrial-4 is the most mature atrial cluster, whereas RA-5 is the most mature RA cluster. Further, the CM-OFT branch halted approximately midway along the observed developmental axis, consistent with the loss of this cell identity by E13.5. Along the trajectory, Pitx2 expression was enriched at the aSHF2-to-CM-OFT transition as well as within a segment of the atrial CM branch (Fig. 4C).
Among the atrial clusters we annotated, Atrial-6 appeared to express the highest levels of Pitx2. In addition, Atrial-6 cells co-expressed Ramp1 and Osr1, but lacked expression of Shox2, Calca and Sema3a (Fig. S5C). Importantly, Atrial-6 cells were almost entirely absent in Pitx2-deficient hearts (Fig. S5B). Thus, Atrial-6 cells resembled left atrial CMs. Furthermore, the nearby clusters, Atrial-5 and RA-5, expressed high levels of Shox2 (Fig. 4C and Fig. S5C). The expansion of Shox2 expression in mutant atrial and RA clusters is consistent with our previous work, in which we found that Shox2 expression is repressed by PITX2 (Wang et al., 2010).
Next, to characterize the Pitx2-dependent developmental dynamics of the OFT myocardium we employed density peak clustering within the Monocle2 framework for pseudotemporal analysis (Qiu et al., 2017) on aSHF cells and the interconnected CM-OFT cluster. We found that the Monocle2 pseudotime results were consistent with our UMAP projection and STREAM analysis (Fig. 4B,D), in which cells progressed along a developmental trajectory progressing from aSHF1 cells to the intermediate aSHF2 cluster, and ultimately CM-OFT cells (Fig. 4D). Cells derived from control cardiac tissue occupied considerably more of the early and intermediate positions along the CM-OFT trajectory than did cells derived from Pitx2 null embryos (Fig. 4D and Fig. S5B). These results suggest that Pitx2 may dictate second lineage differentiation trajectories. To determine the transcriptional dynamics associated with the differentiation of SHF progenitors to CMs of the OFT myocardium, we determined all genes for which expression changes as a function of progressing pseudotime (q-value <1e-5), and then performed hierarchical clustering (nclust=4) to capture the key dynamic gene expression trends (Fig. 4E). The genes for which expression increased linearly across pseudotime, including Mef2c and Nkx2-5, belonged to gene ontology (GO) terms associated with oxidative phosphorylation, myofibril assembly and cardiac muscle contraction, (Fig. 4E,F). GO analysis also uncovered that cell cycle genes (e.g. Ccnb1), mRNA splicing genes (e.g. Srsf1), Myc target genes, chromatin remodelers and heart valve morphogenesis-associated terms decreased across pseudotime. Interestingly, the genes that were expressed at the highest level at the trajectory midpoint were those most significantly associated with epithelial-to-mesenchymal transition (EMT), including Col1a2 and chemokine C-X-C motif ligand 12 (Cxcl12) (Fig. 4E).
The intermediate cells of the CM-OFT differentiation trajectory, belonging principally to cluster aSHF2, expressed high levels of Pitx2 and were significantly depleted in mutant embryonic cardiac tissue. Importantly, we found that these cells co-expressed high levels of the matricellular protein transforming growth factor beta induced (Tgfbi), and the basic helix-loop-helix (bHLH) transcription factor Hand1 (Fig. 4F). We used FISH to assess the expression of Tgfbi and Hand1 in the E10.5 heart and found that the expression of both genes was reduced in the OFT of Pitx2-deficient animals (Fig. 4G,H, Fig. S5D,E). Overall, these data suggest that the Pitx2 gene regulatory network is required for the proper differentiation dynamics of anterior SHF progenitors.
Right atrial cardiomyocyte identity enrichment in Pitx2 null cardiac tissue
We then sought to determine the transcriptional dynamics associated with the disruption of left-right asymmetry characteristic of Pitx2 null embryos. Importantly, previous studies have detailed the ability of left pSHF cells to give rise to myocardial cells of the left atria and AVC, whereas progenitors of right pSHF contribute to the right atria (Domínguez et al., 2012). Given that RA identity was more prevalent among CMs in Pitx2 mutant embryonic cardiac tissue (Fig. S5B), we subset out RA CMs and the connected SHF progenitors (pSHF and muCP) and then performed pseudotime analysis as described above. The Monocle2 pseudotime results were consistent with our UMAP projection (Fig. 5A,B). The single cell transcriptomes analyzed proceeded along a trajectory progressing from SHF progenitors to RA CM clusters 1-5 (Fig. 5B). To better visualize the trajectory we applied STREAM (Fig. 5C). The results from the STREAM analysis were consistent with the Monocle2 output. Next, we determined all genes for which expression changes as a function of pseudotime (q-value<1e-5), and performed hierarchical clustering (nclust=4) (Fig. 5D). Genes associated with RA CM identity belonged to GO terms such as oxidative phosphorylation, cardiac conduction, SA node cell activity, mesenchymal migration, cardiac neural crest development and SHF specification (Fig. 5D,E). Conversely, genes that decreased as pseudotime progressed were associated with EMT, interleukin 12 signaling, neural crest differentiation and endocardial cushion development. In summary, these results detail the transcriptional dynamics and putative cellular compositional shifts associated with cardiac atrial asymmetry. Further work is essential to clarify these findings.
Fig. 5.
Left-right atrial cardiomyocyte identity defects in Pitx2 null cardiac tissue. (A) Pseudotime score for each individual outflow tract-associated transcriptome projected onto the UMAP plot. Cells not included in analysis are colored dark gray. (B) Pseudotime minimal spanning tree (MST) plots displaying pseudotime score (top) and cell identity (bottom). Cell clusters are colored according to Fig. 4A. (C) STREAM plot for indicated cell clusters. (D) Heatmap showing clusters of dynamic gene expression trends across pseudotime (left). Results of hierarchical clustering of gene expression trends shown in heatmap (middle). Gene ontology (GO) analysis of genes from each indicated dynamic cluster (right). (E) Individual gene expression trends plotted across pseudotime. Cells are colored according to their identity (Fig. 4A). muCP, progenitors enriched in Pitx2 null tissue; pSHF, posterior second heart field progenitors; RA, right atrial cells.
Pitx2-dependent cardiac valve development
From our E13.5 data we found that many of the cell clusters associated with valve development, SMC identity and coronary vasculature were abnormally dispersed in Pitx2 mutant cardiac tissue (Fig. S4B). To gain further insight into the cellular composition shifts and transcriptional changes dependent on Pitx2 in cardiac development, we subclustered the key endocardial and mesenchymal cell types implicated in cardiac valve development (Fig. 6A). The graph abstraction results were in agreement with our UMAP results (Fig. 6B). Endocardial cells in Pitx2-deficient embryos contributed more to the cellular composition of the heart than they did in controls (Fig. S6A). Conversely, subclusters of EndoV and VaMes cells were significantly diminished in Pitx2 null hearts. These results suggest that Pitx2 may be required for the proper differentiation or specification of endocardial cells during valve maturation.
Fig. 6.
Pitx2-dependent valvulogenesis. (A) UMAP of cardiac valve-associated cell clusters from E10.5 and E13.5. (B) Force-directed graph of PAGA-initialized embedding derived from cells in Fig. 6A (left). PAGA graph showing the relationships between the subclusters (right). (C) Individual gene expression feature plots. Outlines indicate clusters colored according to A. (D) Dotplot showing the expression of individual marker genes across the distinct cardiac valve-associated clusters. Low average expression is colored as blue, and high average expression is denoted with red. The size of the dot represent the percent of cells within that cluster expressing the indicated gene. AVcu, atrioventricular cushion cells; CFs, cardiac fibroblasts; EndoC, endocardial cells; EndoV, endocardial valve cells; Mes, mesenchymal cells; VaMes, valve mesenchyme.
We were able to identify a population of E10.5 Mes cells that clustered proximally to the endocardial cells (Fig. 2A). These Mes cells expressed low levels of endothelial markers such as VE-cadherin (Cdh5) and CD31 (Pecam1), but expressed high levels of Sox9, Snai1 and the Yap target gene Ptx3 (Fig. 6C). Notably, Sox9 is a transcription factor which is activated in endocardial cells once they undergo EMT and begin to migrate into the cardiac jelly during endocardial cushion formation (Akiyama et al., 2004), and Snai1 is a Notch-induced transcriptional repressor of VE-cadherin and a central mediator of EMT (Timmerman et al., 2004). Hence, the Mes cluster resembles a cluster of cells undergoing EMT. Moreover, these cells expressed several additional markers, including Dusp9, Klf1, Akr1b8, Tspan8 and Erbb3 (Fig. 6D). The EndoV-3 cluster, the cardiac composition of which was diminished in Pitx2 null animals, expressed markers associated with mature endocardial valve leaflets, including Wnt4 and Wnt9b (Goddard et al., 2017; Xiao et al., 2018). Moreover, we identified several additional molecular markers for cardiac valve development, including Bmp15, Il2rg, Adamts8, Ptprr, Nbl1 and Rassf5 for EndoV-3 (Fig. 6D). Additional studies beyond the scope of this paper are required to investigate the role played by Pitx2 during endocardial development.
DISCUSSION
Pitx2 expression is tightly spatiotemporally controlled during mammalian embryonic development, as it is required in several distinct cell lineages. Here, we characterized Pitx2-dependent cardiac development at E10.5 and E13.5 using single cell transcriptomics. We were able to detail cardiac cell identities and compositional shifts that resulted from a global loss of Pitx2 expression. Importantly, we found cells with cardiac progenitor-like expression profiles which displayed altered composition and transcriptional differentiation dynamics toward myocardial cell identities. Our results also further characterize the role for Pitx2 in dictating left atrial CM identity and provide expression signatures for normal and ectopic SAN-like cells. Overall, our data provide a comprehensive transcriptional profiling of Pitx2-dependent cardiac morphogenesis, as well as a resource for future investigations pertaining to the role of gene regulatory networks in organ development.
Although droplet-based scRNA-seq is a simple, high-throughput and relatively cost-effective approach, it is not without its limitations. The input derived from a single cell is small and the capture efficiency for transcripts is low, leading to dropouts and sparse data. Indeed, the technology can only resolve a fraction of a single cell's transcriptome (Hrvatin et al., 2018). Thus, we may not have detected all of the gene regulatory events that occur as a consequence of Pitx2 disruption. Improving the sensitivity of scRNA-seq platforms and applying imputation algorithms may be able to drastically improve data quality in the future (van Dijk et al., 2018). Another limitation with scRNA-seq is the loss of spatial information, which would greatly aid in cell type classification or annotation. Although we were able to easily annotate all the major cardiac cell types, we were unable to validate all the heterogeneous clusters detailed in this study. Future work characterizing these transcriptionally distinct cell types are required. And despite the large numbers of cells profiled, we may be missing rare cardiac cell types. Finally, the addition of more cells, timepoints and Pitx2-expressing tissues (e.g. craniofacial) could potentially further advance our understanding of organogenesis and LRA.
Previous work that was carried out in our lab established that Pitx2 asymmetrically patterns the SHF progenitor pool, and is required for the proper expansion and remodeling of the OFT myocardium (Ai et al., 2006). Moreover, more recent work found that Pitx2 dictates cardiac OFT development via mesoderm progenitor cells (Ma et al., 2013). Consistent with these studies, we found evidence for defective SHF-to-OFT myocardial differentiation, as well as apparent defects in the differentiation or proliferation of SMCs. Interestingly, the expression of the bHLH Hand1 was diminished in the OFT of Pitx2 null embryos. In Hand1 knockout embryonic stem cells, CM differentiation is enhanced, whereas Hand1 overexpression promotes the maintenance of proliferating precursors at the expense of CM differentiation (Risebro et al., 2006). Moreover, the conditional removal of Hand1 in the heart causes severe structural cardiac defects, including OFT defects (McFadden et al., 2005). Taken together, these studies support a model to explain the observed findings in which the lack of Hand1 expression in the Pitx2 null aSHF pool reduces the population size of this cell type while simultaneously promoting the differentiation of these cells to the default mesodermal CM cell fate. The Pitx2-expressing aSHF2 cluster also appeared to be a prominent signaling source, expressing many potent secreted molecules, including Bmp4. Consistent with the decrease of Bmp4-expressing SHF cells in Pitx2-deficient embryos, the disruption of Bmp4 expression in the SHF causes OFT defects, reduction in cardiac neural crest cell (CNCC) ingress and defective EMT (Bai et al., 2013; Ma et al., 2013). Finally, Pitx2 has previously been implicated as an upstream component of the Isl1 and Tbx1 gene regulatory networks (Harel et al., 2012; Ma et al., 2013). Although our work expands our understanding of Pitx2 function in this progenitor field, further molecular insight into the regulation of the pioneer transcription factor Isl1 (Gao et al., 2019) and other regulators of heart development should be pursued.
We found a cluster of cells (muCP) with transcriptional similarities to CNCCs present in the E10.5 Pitx2-deficient heart. Although Pitx2 is not required in the neural crest lineage for heart development, CNCC migration and localization in Pitx2-deficient animals has been shown to be disrupted (Ma et al., 2013). We believe it likely that the muCP cluster represent these undifferentiated CNCCs, however, lineage tracing and further experimental validation is necessary to definitively classify this cell type. We found that the muCP cluster expressed high levels of Wnt signaling antagonist Dkk2. Interestingly, Dkk2 expression is positively regulated by Pitx2 in the neural crest during eye development (Gage et al., 2008). It may be that Pitx2 represses the expression of Dkk2 in the CNCC, or that its expression is being dictated in a non-cell-autonomous fashion. The muCPs also expressed the PITX2 target gene and Wnt signaling ligand Wnt2 (Basu and Roy, 2013). Wnt2 has been shown to promote CM differentiation in embryonic stem cell cultures, similar to Hand1-knockouts (Onizuka et al., 2012). Thus, the loss of Pitx2 expression appears to alter the extracellular cardiac signaling apparatus, which dramatically alters the cell fate trajectories of cardiac progenitor cells.
The role of Pitx2 in the development, maturation and maintenance of atrial CMs has garnered a lot of recent attention, given that genome-wide association studies have uncovered a significant statistical association of PITX2 with AF (Ellinor et al., 2010). We found transcriptional evidence for an increase in the composition of RA CMs, as well as the expansion of cells with signatures of the SAN. Although the ectopic expansion of cells of the cardiac conduction system have been reported previously in Pitx2-deficient mouse models (Ammirabile et al., 2012; Mommersteeg et al., 2007; Wang et al., 2010), the single cell transcriptional signatures of this loss of left atrial identity and patterning with the concomitant expansion of right atrial CM identity have not yet been reported. Moreover, we found that pSHF cells were still present in Pitx2 mutant hearts and appeared to maintain their differentiation capacity to RA CMs at the transcriptional level. Thus, disruption of left-right asymmetry through the loss of Pitx2 in the pSHF progenitors does not appear to affect the size of the pSHF cell population, but likely skews their cellular identities such that without the expression of Pitx2 these cells may adopt a right atrial fate by default. In the future, it will be important to understand how defects in the proper spatiotemporal expression of Pitx2 in these cell populations contribute to the pathogenesis of AF.
MATERIALS AND METHODS
Mouse models
The Pitx2hd−/− allele has been described previously (Lu et al., 1999). All animals were maintained in a pathogen-free Baylor College of Medicine Transgenic Mouse Facility. All animal protocols and procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Baylor College of Medicine in Houston, Texas.
scRNA-seq and analysis
Homozygous Pitx2hd−/− mutant E10.5 and E13.5 embryos were visually identified and their hearts were dissected. For controls, wild-type and heterozygous embryos were used. All genotypes were confirmed using PCR for the Pitx2hd allele. After hearts were removed from mutant and control embryos, we generated single cell suspensions by treating individual hearts with 0.25% Trypsin (HyClone) for 20-30 min at 37°C with end-over-end rotation. Digestion was quenched with DMEM containing 10% fetal bovine serum and cells were pelleted. The pellets were resuspended in 1× PBS with 0.04% bovine serum albumin and counted before being loaded onto the 10x Genomics Chromium instrument. For the E10.5 timepoint, we pooled hearts of the same genotype whenever possible in order to acquire sufficient cells for loading onto the 10x instrument. For the E13.5 timepoint, we used individual hearts for each experiment. In total, we sequenced three control scRNA-seq libraries and five Pitx2hd−/− libraries for E10.5. For E13.5, we sequenced two control libraries and two Pitx2hd−/− libraries. The scRNA-seq libraries were generated using the 10x Chromium Single Cell 3′ v2 reagent kit, according to the manufacturer's instructions, and were sequenced on an Illumina Nextseq500.
The scRNA data was analyzed as previously described with minor modifications (Li et al., 2018). Briefly, raw sequencing data were handled using the 10x Genomics Cell Ranger software (www.10xgenomics.com). Fastq files were mapped to the mm10 genome, and gene counts were quantified using the Cellranger count function. For E10.5, we averaged 117,673 reads per cell, and detected an average of 4104 genes per cell across all eight experiments. For E13.5 we averaged 40,470 reads per cell, and detected 2646 genes per cell on average across four experiments. Subsequently, expression matrices from each experiment were merged and were then imported into Seurat (version 2.3.4, https://satijalab.org/seurat/) for log normalization. We corrected for batch effects by regressing out the number of molecules per cell, mitochondrial read percentage and the batch identity with the Seurat ‘RegressOut’ function. Next, we performed a principal components analysis (PCA), and significant principal components (PCs) were used as input for graph-based clustering. We used UMAP for 2-dimensional visualization of the multi-dimensional dataset (McInnes and Healy, 2018). Differential expression of the individual clusters was achieved using the Wilcoxon rank sum test (FindMarkers, default). To avoid overclustering, we merged clusters that were not transcriptionally distinct into a single cluster. Clusters composed of doublets (two different cell types within a single droplet) were removed from the dataset. And clusters that were not obviously of cardiac origin were also removed from the final analysis, including hepatocytes and lung epithelial cells. GO analysis was performed using Metascape (www.metascape.org). For pseudotemporal analysis, the normalized data from selected clusters were then passed directly into Monocle2, in which density peak clustering and downstream analysis were performed. Chi-square statistical analysis between clusters was performed and visualized as previously described (Li et al., 2018). PAGA was implemented through Scanpy (version 1.4.1) with the threshold set to 0.1 (Wolf et al., 2018). STREAM (version 0.3.8) analysis was performed through python; we used the top 15 principal components calculated from the top 2000 variably expressed genes as features.
Immunofluorescence and fluorescence in situ hybridization
Embryos were fixed in 4% paraformaldehyde overnight at 4°C and then dehydrated in an ethanol series and xylene incubation procedure before paraffin embedding and preparation of 7 μm paraffin sections. For immunofluorescent analysis, antigen presentation was performed by boiling the sections in Antigen Unmasking Solution (Vector Laboratories, H-3300) for 15 minutes. Sections were then permeabilized in PBS containing 0.5% Triton X-100 for 15 minutes at room temperature. Primary antibodies used for immunofluorescence were Isl1 (1:10, Developmental Studies Hybridoma Bank, 40.2D6) and Crabp1 (1:200, Cell Signaling Technology, D7F9T). Signal was detected using fluorophore-conjugated secondary antibodies: Donkey anti-rabbit Alexa Fluor 488 (1:200, Thermo Fisher Scientific, A-21206) and donkey anti-rabbit Alexa Fluor 647 (1:200, Thermo Fisher Scientific, A-31573). To detect mouse primary antibodies, the Mouse on Mouse (M.O.M) Detection Kit (Vector Laboratories, BMK-2202) was used followed by a Streptavidin, Alexa Fluor 647 conjugate (1:100, Thermo Fisher Scientific, S-32357) for visualization. For FISH, we used RNAscope and performed all labeling according to the manufacturer’s instructions in the RNAscope 2.5 HD Assay-RED protocol (ACD). The FISH probes used in this study were Dkk2 (ACD, 404841), Hand1 (ACD, 429651) and Tgfbi (ACD, 494551). Nuclei counterstaining was performed using DAPI (1:1000) for 10 minutes at room temperature. Images were taken using a Zeiss LSM 780 confocal microscope. Quantification of puncta was performed on Zen Blue (Zeiss) software, in which puncta were assigned to the nearest nuclei and then counting was manually performed of the area of interest.
Supplementary Material
Acknowledgements
We thank Elzbieta Klysik (Baylor College of Medicine) for performing RNA ISH. We also thank Dr Yuka Morikawa (Texas Heart Institute), Dr Ge Tao (Medical University of South Carolina), Dr Jun Wang (The University of Texas Health Science Center at Houston) and Dr Yang Xiao (University of Pennsylvania) for their technical assistance.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: M.C.H., J.F.M.; Methodology: M.C.H., L.L., T.T.T.; Validation: M.C.H., Z.A.K., L.L., T.T.T., J.D.W.; Formal analysis: M.C.H., Z.A.K., L.L.; Investigation: M.C.H., Z.A.K., T.T.T., J.D.W., J.F.M.; Resources: M.C.H., J.D.W., J.F.M.; Data curation: M.C.H.; Writing - original draft: M.C.H., J.F.M.; Writing - review & editing: M.C.H., Z.A.K., J.F.M.; Visualization: M.C.H., Z.A.K.; Supervision: J.F.M.; Project administration: J.F.M.; Funding acquisition: M.C.H., J.D.W., J.F.M.
Funding
This work was supported by grants from the National Institutes of Health (DE023177, HL127717, HL130804, HL118761 to J.F.M.; F31HL136065 to M.C.H.; F30HL14590801 to Z.A.K.), the Vivian L. Smith Foundation (to J.F.M.), a Fondation Leducq Transatlantic Networks of Excellence in Cardiovascular Research grant (14CVD01: Defining the genomic topology of atrial fibrillation, to J.F.M.), the Baylor College of Medicine Research Advocates for Student Scientists (to Z.A.K.), a Eunice Kennedy Shriver National Institute of Child Health & Human Development grant to the Intellectual and Developmental Disabilities Research Center (1U54 HD083092), and the Mouse Phenotyping Core at Baylor College of Medicine (U54 HG006348). Deposited in PMC for release after 12 months.
Data availability
The scRNA-seq from this study have been deposited in GEO under accession number GSE131181.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.180398.supplemental
References
- Ai D., Liu W., Ma L., Dong F., Lu M.-F. F., Wang D., Verzi M. P., Cai C., Gage P. J., Evans S. et al. (2006). Pitx2 regulates cardiac left-right asymmetry by patterning second cardiac lineage-derived myocardium. Dev. Biol. 296, 437-449. 10.1016/j.ydbio.2006.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama H., Chaboissier M.-C., Behringer R. R., Rowitch D. H., Schedl A., Epstein J. A. and de Crombrugghe B. (2004). Essential role of Sox9 in the pathway that controls formation of cardiac valves and septa. Proc. Natl. Acad. Sci. USA 101, 6502-6507. 10.1073/pnas.0401711101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammirabile G., Tessari A., Pignataro V., Szumska D., Sutera Sardo F., Benes J., Balistreri M., Bhattacharya S., Sedmera D. and Campione M. (2012). Pitx2 confers left morphological, molecular, and functional identity to the sinus venosus myocardium. Cardiovasc. Res. 93, 291-301. 10.1093/cvr/cvr314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Wang J., Morikawa Y., Bonilla-Claudio M., Klysik E. and Martin J. F. (2013). Bmp signaling represses Vegfa to promote outflow tract cushion development. Development 140, 3395-3402. 10.1242/dev.097360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu M. and Roy S. S. (2013). Wnt/β-catenin pathway is regulated by PITX2 homeodomain protein and thus contributes to the proliferation of human ovarian adenocarcinoma cell, SKOV-3. J. Biol. Chem. 288, 4355-4367. 10.1074/jbc.M112.409102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C.-L., Liang X., Shi Y., Chu P.-H., Pfaff S. L., Chen J. and Evans S. (2003). Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev. Cell 5, 877-889. 10.1016/S1534-5807(03)00363-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S., Cheek J., Sakthivel B., Aronow B. J. and Yutzey K. E. (2008). Shared gene expression profiles in developing heart valves and osteoblast progenitor cells. Physiol. Genomics 35, 75-85. 10.1152/physiolgenomics.90212.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Albergante L., Hsu J. Y., Lareau C. A., Lo Bosco G., Guan J., Zhou S., Gorban A. N., Bauer D. E., Aryee M. J. et al. (2019). Single-cell trajectories reconstruction, exploration and mapping of omics data with STREAM. Nat. Commun. 10, 1903 10.1038/s41467-019-09670-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Zheng Y., Liu X., Yan L., Fan X., Yong J., Hu Y., Dong J., Li Q., Wu X. et al. (2019). Single-cell transcriptome analysis maps the developmental track of the human heart. Cell Rep. 26, 1934-1950.e5. 10.1016/j.celrep.2019.01.079 [DOI] [PubMed] [Google Scholar]
- Domínguez J. N., Meilhac S. M. M., Bland Y. S., Buckingham M. E. and Brown N. A. (2012). Asymmetric fate of the posterior part of the second heart field results in unexpected left/right contributions to both poles of the heart. Circ. Res. 111, 1323-1335. 10.1161/CIRCRESAHA.112.271247 [DOI] [PubMed] [Google Scholar]
- Ellinor P. T., Lunetta K. L., Glazer N. L., Pfeufer A., Alonso A., Chung M. K., Sinner M. F., Bakker P. I. de, Mueller M. et al. (2010). Common variants in KCNN3 are associated with lone atrial fibrillation. Nat. Genet. 42, 240-244. 10.1038/ng.537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage P. J., Qian M., Wu D. and Rosenberg K. I. (2008). The canonical Wnt signaling antagonist DKK2 is an essential effector of PITX2 function during normal eye development. Dev. Biol. 317, 310-324. 10.1016/j.ydbio.2008.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R., Liang X., Cheedipudi S., Cordero J., Jiang X., Zhang Q., Caputo L., Günther S., Kuenne C., Ren Y. et al. (2019). Pioneering function of Isl1 in the epigenetic control of cardiomyocyte cell fate. Cell Res. 0, 1-16. 10.1038/s41422-019-0168-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard L. M., Duchemin A.-L. L., Ramalingan H., Wu B., Chen M., Bamezai S., Yang J., Li L., Morley M. P., Wang T. et al. (2017). Hemodynamic forces sculpt developing heart valves through a KLF2-WNT9B paracrine signaling axis. Dev. Cell 43, 274-289.e5. 10.1016/j.devcel.2017.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson D. F., Arnar D. O., Helgadottir A., Gretarsdottir S., Holm H., Sigurdsson A., Jonasdottir A., Baker A., Thorleifsson G., Kristjansson K. et al. (2007). Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature 448, 353-357. 10.1038/nature06007 [DOI] [PubMed] [Google Scholar]
- Harel I., Maezawa Y., Avraham R., Rinon A., Ma H.-Y., Cross J. W., Leviatan N., Hegesh J., Roy A., Jacob-Hirsch J. et al. (2012). Pharyngeal mesoderm regulatory network controls cardiac and head muscle morphogenesis. Proc. Natl. Acad. Sci. USA 109, 18839-18844. 10.1073/pnas.1208690109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrvatin S., Hochbaum D. R., Nagy M. A., Cicconet M., Robertson K., Cheadle L., Zilionis R., Ratner A., Borges-Monroy R., Klein A. M. et al. (2018). Single-cell analysis of experience-dependent transcriptomic states in the mouse visual cortex. Nat. Neurosci. 21, 120-129. 10.1038/s41593-017-0029-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Xu A., Sim S., Priest J. R., Tian X., Khan T., Quertermous T., Zhou B., Tsao P. S., Quake S. R. et al. (2016). Transcriptomic profiling maps anatomically patterned subpopulations among single embryonic cardiac cells. Dev. Cell 39, 491-507. 10.1016/j.devcel.2016.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Tao G., Hill M. C., Zhang M., Morikawa Y. and Martin J. F. (2018). Pitx2 maintains mitochondrial function during regeneration to prevent myocardial fat deposition. Development 145, dev168609 10.1242/dev.168609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. R., Kioussi C., O'Connell S., Briata P., Szeto D., Liu F., Izpisúa-Belmonte J. C. and Rosenfeld M. G. (1999). Pitx2 regulates lung asymmetry, cardiac positioning and pituitary and tooth morphogenesis. Nature 401, 279-282. 10.1038/45803 [DOI] [PubMed] [Google Scholar]
- Lincoln J., Alfieri C. M. and Yutzey K. E. (2006). BMP and FGF regulatory pathways control cell lineage diversification of heart valve precursor cells. Dev. Biol. 292, 292-302. 10.1016/j.ydbio.2006.03.027 [DOI] [PubMed] [Google Scholar]
- Liu C., Liu W., Lu M. F., Brown N. A. and Martin J. F. (2001). Regulation of left-right asymmetry by thresholds of Pitx2c activity. Development 128, 2039-2048. [DOI] [PubMed] [Google Scholar]
- Logan M., Pagán-Westphal S. M., Smith D. M., Paganessi L. and Tabin C. J. (1998). The transcription factor Pitx2 mediates situs-specific morphogenesis in response to left-right asymmetric signals. Cell 94, 307-317. 10.1016/S0092-8674(00)81474-9 [DOI] [PubMed] [Google Scholar]
- Lu M. F., Pressman C., Dyer R., Johnson R. L. and Martin J. F. (1999). Function of Rieger syndrome gene in left-right asymmetry and craniofacial development. Nature 401, 276-278. 10.1038/45797 [DOI] [PubMed] [Google Scholar]
- Ma H.-Y., Xu J., Eng D., Gross M. K. and Kioussi C. (2013). Pitx2-mediated cardiac outflow tract remodeling. Dev. Dyn. 242, 456-468. 10.1002/dvdy.23934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden D. G., Barbosa A. C., Richardson J. A., Schneider M. D., Srivastava D. and Olson E. N. (2005). The Hand1 and Hand2 transcription factors regulate expansion of the embryonic cardiac ventricles in a gene dosage-dependent manner. Development 132, 189-201. 10.1242/dev.01562 [DOI] [PubMed] [Google Scholar]
- McInnes L. and Healy J. (2018). UMAP: uniform manifold approximation and projection for dimension reduction. J. Open Source Softw. 3, 861 10.21105/joss.00861 [DOI] [Google Scholar]
- Meilhac S. M. M. and Buckingham M. E. (2018). The deployment of cell lineages that form the mammalian heart. Nat Rev Cardiol 15, 705-724. 10.1038/s41569-018-0086-9 [DOI] [PubMed] [Google Scholar]
- Mommersteeg M. T., Brown N. A., Prall O. W., de Gier-de Vries C., Harvey R. P., Moorman A. F. and Christoffels V. M. (2007). Pitx2c and Nkx2-5 are required for the formation and identity of the pulmonary myocardium. Circ. Res. 101, 902-909. 10.1161/CIRCRESAHA.107.161182 [DOI] [PubMed] [Google Scholar]
- Onizuka T., Yuasa S., Kusumoto D., Shimoji K., Egashira T., Ohno Y., Kageyama T., Tanaka T., Hattori F., Fujita J. et al. (2012). Wnt2 accelerates cardiac myocyte differentiation from ES-cell derived mesodermal cells via non-canonical pathway. J. Mol. Cell. Cardiol. 52, 650-659. 10.1016/j.yjmcc.2011.11.010 [DOI] [PubMed] [Google Scholar]
- Phillips M., Mukhopadhyay M., Poscablo C. and Westphal H. (2011). Dkk1 and Dkk2 regulate epicardial specification during mouse heart development. Int. J. Cardiol. 150, 186-192. 10.1016/j.ijcard.2010.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piedra M. E., Icardo J. M., Albajar M., Rodriguez-Rey J. C. and Ros M. A. (1998). Pitx2 participates in the late phase of the pathway controlling left-right asymmetry. Cell 94, 319-324. 10.1016/S0092-8674(00)81475-0 [DOI] [PubMed] [Google Scholar]
- Pijuan-Sala B., Griffiths J. A., Guibentif C., Hiscock T. W., Jawaid W., Calero-Nieto F. J., Mulas C., Ibarra-Soria X., Tyser R. C. V., Ho D. L. L. et al. (2019). A single-cell molecular map of mouse gastrulation and early organogenesis. Nature 566, 490-495. 10.1038/s41586-019-0933-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H. A. and Trapnell C. (2017). Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979-982. 10.1038/nmeth.4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risebro C. A., Smart N., Dupays L., Breckenridge R., Mohun T. J. and Riley P. R. (2006). Hand1 regulates cardiomyocyte proliferation versus differentiation in the developing heart. Development 133, 4595-4606. 10.1242/dev.02625 [DOI] [PubMed] [Google Scholar]
- Semina E. V., Reiter R., Leysens N. J., Alward W. L., Small K. W., Datson N. A., Siegel-Bartelt J., Bierke-Nelson D., Bitoun P., Zabel B. U. et al. (1996). Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nat. Genet. 14, 392-399. 10.1038/ng1296-392 [DOI] [PubMed] [Google Scholar]
- Syeda F., Kirchhof P. and Fabritz L. (2017). PITX2-dependent gene regulation in atrial fibrillation and rhythm control. J. Physiol. (Lond.) 595, 4019-4026. 10.1113/JP273123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao G., Kahr P. C., Morikawa Y., Zhang M., Rahmani M., Heallen T. R., Li L., Sun Z., Olson E. N., Amendt B. A. et al. (2016). Pitx2 promotes heart repair by activating the antioxidant response after cardiac injury. Nature 534, 119-123. 10.1038/nature17959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmerman L. A., Grego-Bessa J., Raya A., Bertrán E., Pérez-Pomares J. M. M., Díez J., Aranda S., Palomo S., McCormick F., Izpisúa-Belmonte J. C. et al. (2004). Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 18, 99-115. 10.1101/gad.276304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk D., Sharma R., Nainys J., Yim K., Kathail P., Carr A. J., Burdziak C., Moon K. R., Chaffer C. L., Pattabiraman D. et al. (2018). Recovering gene interactions from single-cell data using data diffusion. Cell 174, 716-729.e27. 10.1016/j.cell.2018.05.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Klysik E., Sood S., Johnson R. L., Wehrens X. H. T. and Martin J. F. (2010). Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proc. Natl. Acad. Sci. U.S.A. 107, 9753-9758. 10.1073/pnas.0912585107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf F. A., Angerer P. and Theis F. J. (2018). SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 10.1186/s13059-017-1382-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf F. A., Hamey F. K., Plass M., Solana J., Dahlin J. S., Göttgens B., Rajewsky N., Simon L. and Theis F. J. (2019). PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol. 20, 59 10.1186/s13059-019-1663-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Hill M. C., Zhang M., Martin T. J., Morikawa Y., Wang S., Moise A. R., Wythe J. D. and Martin J. F. (2018). Hippo signaling plays an essential role in cell state transitions during cardiac fibroblast development. Dev. Cell 45, 153-169.e6. 10.1016/j.devcel.2018.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka H., Meno C., Koshiba K., Sugihara M., Itoh H., Ishimaru Y., Inoue T., Ohuchi H., Semina E. V., Murray J. C. et al. (1998). Pitx2, a bicoid-type homeobox gene, is involved in a lefty-signaling pathway in determination of left-right asymmetry. Cell 94, 299-305. 10.1016/S0092-8674(00)81473-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.