Abstract
Atomistic molecular dynamics simulations of membrane proteins have been shown to be extremely useful for characterizing the molecular features underlying their function, but require high computational power, limiting the understanding of complex events in membrane proteins, e.g. ion channels gating, GPCRs activation. To overcome this issue, it has been shown that coarse-grained approaches, although requiring less computational power, are still capable of correctly describing molecular events underlying big conformational changes in biological systems. Here, we present the Martini coarse-grained membrane protein dynamics (MERMAID), a publicly available web interface that allows the user to prepare and run coarse-grained molecular dynamics (CGMD) simulations and to analyse the trajectories.
INTRODUCTION
A lipid membrane isolates the cells from the external world. It has also a fundamental role in cellular communication because it is the target of all the extracellular stimuli acting on the cell. Several different families of proteins, i.e. integral membrane proteins, are specialized in detecting these signals and in internalizing this information to the cell. The original human genome sequencing project estimated 20% of the total gene count of 31 778 genes to code for membrane proteins (1). This big number shows the importance of membrane proteins for the different organisms. Unfortunately, and principally due to the difficulties at the experimental level, just a small percentage of them have been completely characterized. These difficulties can be overcome by an extensive combination of experimental and in silico studies. In this regard, atomistic molecular dynamics (MD) simulations have been extensively used to study membrane proteins at different time and space scales (2,3). Although the recent advancements in the development of force fields, algorithms and hardware, running long MD simulations to explore large conformational changes is still extremely expensive in terms of time and computational power. Indeed, exploring events that occur in the micro-milliseconds time-scale, like ion channels gating or receptor activation is prohibitive. Different techniques have been developed and implemented over the years in order to accelerate MD simulation studies (4). Among them, coarse-grained molecular dynamics (CGMD) simulations approaches have evolved very rapidly during the last few years and reached important accuracy levels (5). CGMD is computationally less expensive and can explore longer time-scales, still being able to accurately explain molecular events. Indeed, it ultimately results in reducing the degrees of freedom and increase the speed of the simulations. Thus CGMD helps to overcome the experimental limitations to study large scale biomolecular processes over longer time scales.
Due to the advancement in the simulation techniques, in recent years, there is a significant increase in the number of coarse-grained studies (6). Indeed, CG force fields like MARTINI (7), SIRAH (8), Shinoda-Devane-Klein (SDK) (9) and ELBA FFs (10), can be run routinely, within several MD simulations packages. Here, we present our Martini coarse-grained membrane proteIn dynamics, MERMAID web server, aimed at preparing and running CGMD simulations directly from a publicly available web interface (Figure 1). This interface is directed to expert and non-expert users as it allows the preparation of files and allows a heavy interaction with the users. Finally, the user can choose either to run and analyse CGMD simulations directly through our servers or to download the files to be run in local machines.
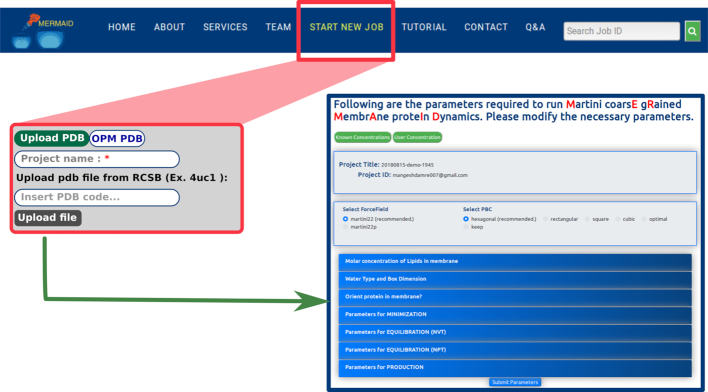
Figure 1.
MERMAID web server interface (Start page, Parameter page): MERMAID provides two ways to submit a protein structure: a) from OPM database, or b) the user can upload the protein structure from a local system. After uploading the protein structure, MERMAID will redirect the user to parameter page.
MATERIALS AND METHODS
Implementation
MERMAID workflow can be divided in two different stages that include the user interface and the backend of the server. In detail:
Web interface
The user can interact with the client web interface to submit a membrane protein structure (Figure 1). It can be either a custom PDB structure or a structure directly obtained from the Protein Data Bank (PDB) (http://www.rcsb.org/) (11). Naturally, most of the PDB structures are not aligned to the z-axis thus, in order to align these protein structures to the membrane normal axis, the user can use the PPM server (https://opm.phar.umich.edu/ppm_server). Moreover, the user is suggested to input a protein structure from the database ‘Orientations of Proteins in Membranes (OPM)’ (https://opm.phar.umich.edu/) (12). As the protein secondary structure information is needed to run MARTINI CGMD simulations, MERMAID will calculate it automatically, by using a locally installed version of the Dictionary of Protein Secondary Structure (DSSP) program (13). The user is then funneled to a series of online forms that help in the creation of the parameters files. The user can choose either to keep the default parameters unmodified or to modify them. We implemented our protocol in the GROMACS package with the use of MARTINI force fields. The processes will then run on our server for successful completion of the simulation. MARTINI force fields implemented in this protocol are freely available from http://cgmartini.nl/.
Backend of the server
All the simulations and calculations are performed in the backend of the server (Figure 2). In particular it uses: (i) a local version of Gromacs 4.6 (14) to perform simulations; (ii) a locally installed version of DSSP (13); (iii) all the programs needed for preparing the files for Martini, including martinize.py (15) and insane.py (16); (iv) GNUPLOT (http://gnuplot.info/) for data plotting and (v) NGL viewer (17) to provide an interactive molecular viewer on the web page.
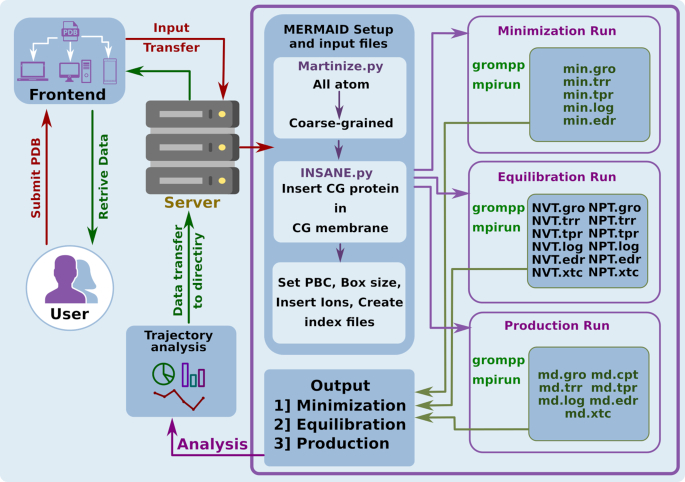
Figure 2.
Workflow of the web server. The submitted protein structure is then converted into coarse-grained structure and inserted into CG membrane to form a complex structure. The structure then undergoes minimization, equilibration and production run respectively. Every phase of the coarse-grained simulation is analysed and can be visualised. Lastly, all the results can be downloaded from our server.
Protocol
After submission of all the files and parameters, the web server creates an user directory, where all operations will be performed. During the initial setup of the system, the atomistic protein structure is converted into a coarse-grained representation with the help of the martinize.py python script (15). During this process, all the ligands, ions and atoms not belonging to the protein are removed. MERMAID performs simulations in absence of ligands due to unavailability of appropriate martini forcefield parameters. By default, disulfide bonds present in the proteins are considered during the system preparation. The CG structure is then embedded into a user-defined coarse-grained lipid membrane with the help of the python script insane.py (16). Furthermore, the MERMAID server allows the user to select different kind of lipid membranes of known composition. Some of the offered membranes include: (a) plasma membrane (Human erythrocytes) with a composition of POPC:20, POPE:18, CHOL:20, POPS:7, POPI:3, POSM:18, CDL0:0 and DPG1:3; (b) myelin membrane (Human neurons) with POPC:11, POPE:17, CHOL:28, POPS:9, POPI:1, POSM:8, CDL0:0 and DPG1:20; (c) nuclear membrane with POPC:44, POPE:17, CHOL:10, POPS:4, POPI:6, POSM:3, CDL0:1 and DPG1:1; (d) endoplasmic reticulum membrane (Rat) with POPC:48, POPE:19, CHOL:6, POPS:4, POPI:8, POSM:5, CDL0:0 and DPG1:1 and (e) Golgi membrane (rat) with POPC:25, POPE:9, CHOL:8, POPS:3, POPI:5, POSM:7, CDL0:0 and DPG1:0 (18). Moreover, the user can model a customized symmetric or asymmetric membrane varying the concentration and type of lipids. Normal or polarized water molecules are added during system preparation. Counterions can also be added at this stage of the system preparation. The user has access to all the generated input and output files for each CGMD process at any time.
As the simulations may take severral minutes to be completed, while the simulation is running, it will show a progress bar and the page will refresh continuously until the job is completed. Otherwise, it will show an error message. Thus, the user is constantly updated on the status of the simulation.
The data can be accessed either from the bookmarked web link or directly from the web server user directory by providing the required credentials in the MERMAID search bar. A typical CGMD run within our server consists of four phases:
Minimization run;
Equilibration run in two different ensembles namely constant NVT and constant NPT;
Production run continued in constant NPT ensemble;
Analysis of all the trajectories produced during simulation.
Server architecture
The MERMAID server is equipped with the web interface written in HTML 4/5 (https://www.w3.org/html/), PHP (http://www.php.net/), JavaScript (https://www.javascript.com/), Bash shell (https://www.gnu.org/software/bash/), jQuery (https://jquery.com/), and Bootstrap 4 (https://getbootstrap.com/). For the submitted jobs, the web server prepares MERMAID input files on our local cluster. At last, the results are post-processed (which includes analysis of CGMD trajectory, construction of various data plots) and displayed graphically by using Highcharts (https://www.highcharts.com/) and Plotly (https://plot.ly). The web server can be accessed from any media device and the jobs can be monitored and followed up.
RESULTS
The server has been intensively used by the members of our group and the most representative cases are available as examples to the users. The examples can be retrieved and analysed from Q&A page of MERMAID, through the ‘examples' section (http://molsim.sci.univr.it/mangesh/questions.html). They include different multimeric states of membrane proteins, i.e. (i) a monomer of the Chloride pumping rhodopsin (PDB accession code: 5B2N); (ii) a dimer of the translocator protein 18 kDa (TSPO) (PDB accession code: 4UC1); (iii) a trimer of the Escherichia coli OmpF porin (PDB accession code: 3POX) and (iv) the tetrameric conformation of the crystal structure of Aquaporin AqpM (PDB accession code: 2F2B). These simulations were generated automatically by just retrieving the structures and running the simulations with default parameters. Here, we describe more in detail case iii), the CGMD simulations of the trimeric structure of the E. coli OmpF porin. In this case, the structure was retrieved from OPM database and funneled through our pipeline (Figure 2).
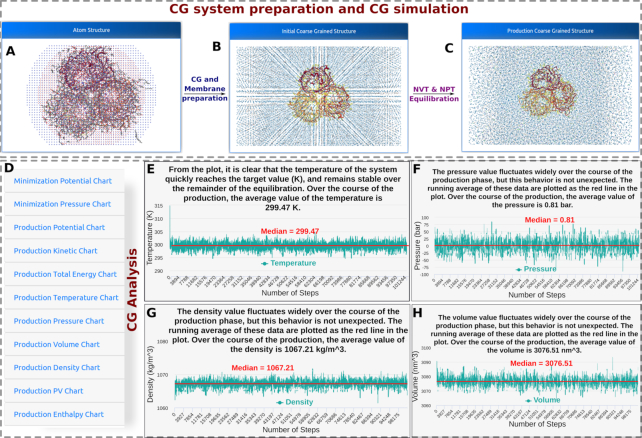
We run the protocol (see section Protocol A)_D)) of the protein embedded in a human plasma membrane with a composition of POPC:20, POPE:18, CHOL:20, POPS:7, POPI:3, POSM:18, CDL0:0 and DPG1:3 lipids respectively. The production run was of 100 ns. The system reached the temperature and pressure of equilibrium within the first 10 ns of simulation and maintained the principal structural features along the entire simulation (Figure 3E–H) (details of this simulation can be appreciated here: (http://molsim.sci.univr.it/mangesh/RESULT.php?Email=trimer&jobid=20190309-CG_User-0222).
Figure 3.
MERMAID server output. The upper panel shows the trimeric protein structure of the E. coli OmpF porin (PDB accession code: 3POX) in (A) atomistic description, (B) initial coarse-grained structure embedded in a coarse-grained membrane, (C) coarse-grained structure from the last frame of production run. A list of the analysis performed on the CG simulation (D). (E–H) shows the plots for temperature, pressure, system density, and system volume respectively.
Details of the analysis at the different stages of the simulation are enlisted in Table 1. The initially generated input data and output data can be retrieved from the server. To the best of our knowledge, it is the first server for generating input files as well as performing CGMD using Martini force fields through an online service. The MERMAID web server is freely accessible at http://molsim.sci.univr.it/mermaid/ and an extensive tutorial is available at http://molsim.sci.univr.it/mermaid/tutorial.php.
Table 1.
Analysis performed on CGMD trajectories
| Analysis | |
|---|---|
| RMSD | Kinetic energy |
| Enthalpy | Potential energy |
| Pressure | Total energy |
| PV analysis | Temperature |
| Mean square displacement | Density |
| Radial distribution function | |
LIMITATIONS
Some coarse-grained simulation jobs may fail due to missing atoms in the residues of a protein. To overcome this issue, it is recommended to the users to check the protein structures for missing atoms with the help of third-party softwares or with the use of our in-house web server (PRUDE: http://molsim.sci.univr.it/mermaid/prude.php) to add the missing atoms. After addition of missing atoms in the protein structure, the user can upload the complete protein structure to MERMAID. The other limitation resides in orienting the protein to the z-axis of the membrane. The users are strongly suggested to download the protein already aligned from the OPM server (https://opm.phar.umich.edu/) or, if not present, to align it using the PPM server (https://opm.phar.umich.edu/ppm_server). Among the results files, we offer the necessary scripts and programs to prepare the atomistic structure using backmapping resources (http://cgmartini.nl/index.php/back).
CONCLUSION
Computational techniques such as atomistic and CGMD simulations are key to gain deeper insights into the mechanisms underlying the functioning of membrane proteins. Due to the increasing interest and to the availability of more powerful algorithms and hardware, during the last years, several web servers have been developed with the scope of preparing and/or performing different resolution levels of molecular dynamics simulations (Table 2). In particular, the existing web servers (Table 2) provide a web-based graphical user interface to generate various molecular simulation systems and input files to facilitate the usage of common simulation techniques. They are widely used to prepare the input files needed to be run on local clusters, and in some of them to run short simulations. In general, they are not specialized in membrane proteins and they do not allow neither the selection of different membranes nor to run CGMD simulations. Both features, at our advice, are fundamental to get a quick initial idea on the stability of the membrane systems and to facilitate the generation of the different steps and protocols of the simulation. Our MERMAID web server implements several novel elements in the ambit of CG simulations, such as the possibility of choosing different membrane composition, but also allows the user to run the initial part of the simulation protocol. The user is free to change the parameters and to choose the different protocols for running simulations. Then the users can choose to run the simulations or to download all the input files to be run locally. If the simulations are run through our servers, a principal set of analysis are carried out in order to follow the evolution of fundamental features, such as temperature, pressure and different energy terms during the simulation (Table 1). Summarizing, the MERMAID server is a user-friendly web server, available to all the users without any login requirement. MERMAID is the only online protocol specifically designed for preparing and running CGMD simulations of membrane proteins through a web server.
Table 2.
Known web servers to perform MD simulations in all-atom (AA) form and/or in coarse-grained (CG) form in presence or absence of membrane
| Input | Membrane | Running | ||||
|---|---|---|---|---|---|---|
| Name | Preparation | Preparation | AA | CG | Level | References |
| CHARMM-GUI | ✓ | ✓ | ✗ | ✗ | MM, CG | http://www.charmm-gui.org/ (19) |
| CABS-flex | ✓ | ✗ | ✓ | ✗ | MM | http://biocomp.chem.uw.edu.pl/CABSflex/ (20) |
| locPREFMD | ✓ | ✗ | ✓ | ✗ | MM | http://feig.bch.msu.edu/locprefmd/ (21) |
| MDWeb | ✓ | ✗ | ✓ | ✗ | MM | http://mmb.irbbarcelona.org/MDWeb/ (22) |
| PREFMD | ✓ | ✗ | ✓ | ✗ | MM | http://feig.bch.msu.edu/prefmd/ (23) |
| ProBLM | ✓ | ✗ | ✗ | ✗ | MM | http://compbio.clemson.edu/sapp/problm_webserver (24) |
| SMOG | ✓ | ✗ | ✓ | ✗ | MM, C-alpha | http://smog-server.org/ (25) |
| UNRES | ✓ | ✗ | ✓ | ✗ | MM, CG | https://www.unres.pl/web_server (26) |
| Vienna-PTM | ✓ | ✗ | ✓ | ✗ | MM | http://vienna-ptm.univie.ac.at (27) |
DATA AVAILABILITY
MERMAID is freely accessible at http://molsim.sci.univr.it/mermaid/.
ACKNOWLEDGEMENTS
We deeply acknowledge the contribution of ‘Centro Piattaforme Tecnologiche dell' Università degli Studi di Verona’.
FUNDING
Department of Biotechnology, Università degli Studi di Verona (to A.G.). Funding for open access charge: Department of Biotechnology, Università degli Studi di Verona (to A.G.).
Conflict of interest statement. None declared.
REFERENCES
- 1. Lander E.S., Linton L.M., Birren B., Nusbaum C., Zod M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W. et al.. Initial sequencing and analysis of the human genome. Nature. 2001; 409:860–921. [DOI] [PubMed] [Google Scholar]
- 2. Harpole T.J., Delemotte L.. Conformational landscapes of membrane proteins delineated by enhanced sampling molecular dynamics simulations. Biochim. Biophys. Acta Biomembr. 2018; 1860:909–926. [DOI] [PubMed] [Google Scholar]
- 3. Pluhackova K., Wassenaar T.A., Bockmann R.A.. Molecular dynamics simulations of membrane proteins. Methods Mol. Biol. 2013; 1033:85–101. [DOI] [PubMed] [Google Scholar]
- 4. Marrink S.J., Corradi V., Souza P.C.T., Ingolfsson H.I., Tieleman D.P., Sansom M.S.P.. Computational modeling of realistic cell membranes. Chem. Rev. 2019; 119:6184–6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Provasi D., Boz M.B., Johnston J.M., Filizola M.. Preferred supramolecular organization and dimer interfaces of opioid receptors from simulated self-association. PLoS Comput. Biol. 2015; 11:e1004148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Takahashi K., Oda T., Naruse K.. Coarse-grained molecular dynamics simulations of biomolecules. AIMS Biophys. 2014; 1:1–15. [Google Scholar]
- 7. Marrink S.J., Risselada H.J., Yefimov S., Tieleman D.P., de Vries A.H.. The MARTINI force field: coarse grained model for biomolecular simulations. J. Phys. Chem. B. 2007; 111:7812–7824. [DOI] [PubMed] [Google Scholar]
- 8. Darre L., Machado M.R., Brandner A.F., Gonzalez H.C., Ferreira S., Pantano S.. SIRAH: a structurally unbiased coarse-grained force field for proteins with aqueous solvation and long-range electrostatics. J. Chem. Theory Comput. 2015; 11:723–739. [DOI] [PubMed] [Google Scholar]
- 9. Shinoda W., DeVane R., Klein M.L.. Zwitterionic lipid assemblies: molecular dynamics studies of monolayers, bilayers, and vesicles using a new coarse grain force field. J. Phys. Chem. B. 2010; 114:6836–6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Orsi M., Essex J.W.. The ELBA force field for coarse-grain modeling of lipid membranes. PLoS ONE. 2011; 6:e28637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E.. The Protein Data Bank. Nucleic Acids Res. 2000; 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lomize M.A., Lomize A.L., Pogozheva I.D., Mosberg H.I.. OPM: orientations of proteins in membranes database. Bioinformatics. 2006; 22:623–625. [DOI] [PubMed] [Google Scholar]
- 13. Kabsch W., Sander C.. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983; 22:2577–2637. [DOI] [PubMed] [Google Scholar]
- 14. Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A.E., Berendsen H.J.. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005; 26:1701–1718. [DOI] [PubMed] [Google Scholar]
- 15. de Jong D.H., Singh G., Bennett W.F., Arnarez C., Wassenaar T.A., Schafer L.V., Periole X., Tieleman D.P., Marrink S.J.. Improved parameters for the martini Coarse-Grained protein force field. J. Chem. Theory Comput. 2013; 9:687–697. [DOI] [PubMed] [Google Scholar]
- 16. Wassenaar T.A., Ingolfsson H.I., Bockmann R.A., Tieleman D.P., Marrink S.J.. Computational lipidomics with insane: A versatile tool for generating custom membranes for molecular simulations. J. Chem. Theory Comput. 2015; 11:2144–2155. [DOI] [PubMed] [Google Scholar]
- 17. Rose A.S., Hildebrand P.W.. NGL Viewer: a web application for molecular visualization. Nucleic Acids Res. 2015; 43:W576–W579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alberts B., Johnson A., Lewis J., Raff M., Roberts K., Walter P.. 2007; Membrane Structure, Garland Science, Taylor and Francis Group, LLC, an informa business, 270 Madison Avenue, New York NY 10016, USA, and 2 Park Square, Milton Park, Abingdon, OX14 4RN, UK.
- 19. Jo S., Kim T., Iyer V.G., Im W.. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008; 29:1859–1865. [DOI] [PubMed] [Google Scholar]
- 20. Kuriata A., Gierut A.M., Oleniecki T., Ciemny M.P., Kolinski A., Kurcinski M., Kmiecik S.. CABS-flex 2.0: a web server for fast simulations of flexibility of protein structures. Nucleic Acids Res. 2018; 46:W338–W343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Feig M. Local protein structure refinement via molecular dynamics simulations with locPREFMD. J. Chem. Inf. Model. 2016; 56:1304–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hospital A., Andrio P., Fenollosa C., Cicin-Sain D., Orozco M., Gelpi J.L.. MDWeb and MDMoby: an integrated web-based platform for molecular dynamics simulations. Bioinformatics. 2012; 28:1278–1279. [DOI] [PubMed] [Google Scholar]
- 23. Heo L., Feig M.. PREFMD: a web server for protein structure refinement via molecular dynamics simulations. Bioinformatics. 2018; 34:1063–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kimmett T., Smith N., Witham S., Petukh M., Sarkar S., Alexov E.. ProBLM web server: protein and membrane placement and orientation package. Comput. Math. Methods Med. 2014; 2014:838259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Noel J.K., Levi M., Raghunathan M., Lammert H., Hayes R.L., Onuchic J.N., Whitford P.C.. SMOG 2: a versatile software package for generating structure-based models. PLoS Comput. Biol. 2016; 12:e1004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Czaplewski C., Karczynska A., Sieradzan A.K., Liwo A.. UNRES server for physics-based coarse-grained simulations and prediction of protein structure, dynamics and thermodynamics. Nucleic Acids Res. 2018; 46:W304–W309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Margreitter C., Petrov D., Zagrovic B.. Vienna-PTM web server: a toolkit for MD simulations of protein post-translational modifications. Nucleic Acids Res. 2013; 41:W422–W426. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
MERMAID is freely accessible at http://molsim.sci.univr.it/mermaid/.