Abstract
Background
Dysbiotic vaginal microbiota have been implicated as contributors to persistent HPV-mediated cervical carcinogenesis and genital inflammation with mechanisms unknown. Given that cancer is a metabolic disease, metabolic profiling of the cervicovaginal microenvironment has the potential to reveal the functional interplay between the host and microbes in HPV persistence and progression to cancer.
Methods
Our study design included HPV-negative/positive controls, women with low-grade and high-grade cervical dysplasia, or cervical cancer (n = 78). Metabolic fingerprints were profiled using liquid chromatography-mass spectrometry. Vaginal microbiota and genital inflammation were analysed using 16S rRNA gene sequencing and immunoassays, respectively. We used an integrative bioinformatic pipeline to reveal host and microbe contributions to the metabolome and to comprehensively assess the link between HPV, microbiota, inflammation and cervical disease.
Findings
Metabolic analysis yielded 475 metabolites with known identities. Unique metabolic fingerprints discriminated patient groups from healthy controls. Three-hydroxybutyrate, eicosenoate, and oleate/vaccenate discriminated (with excellent capacity) between cancer patients versus the healthy participants. Sphingolipids, plasmalogens, and linoleate positively correlated with genital inflammation. Non-Lactobacillus dominant communities, particularly in high-grade dysplasia, perturbed amino acid and nucleotide metabolisms. Adenosine and cytosine correlated positively with Lactobacillus abundance and negatively with genital inflammation. Glycochenodeoxycholate and carnitine metabolisms connected non-Lactobacillus dominance to genital inflammation.
Interpretation
Cervicovaginal metabolic profiles were driven by cancer followed by genital inflammation, HPV infection, and vaginal microbiota. This study provides evidence for metabolite-driven complex host-microbe interactions as hallmarks of cervical cancer with future translational potential.
Fund
Flinn Foundation (#1974), Banner Foundation Obstetrics/Gynecology, and NIH NCI (P30-CA023074).
Keywords: Lactobacillus abundance, Lipids and nucleotides, Amino acid degradation, Cervical dysplasia and cancer, Vaginal dysbiosis, Host-microbe interactions, Genital inflammation
Graphical abstract
Research in context.
Evidence before this study
Cervical cancer is a common gynaecological malignancy in women caused by persistent oncogenic HPV infection. Recent studies have demonstrated that dysbiotic microbiota is involved in HPV persistence, progression of cervical neoplasia, and genital inflammation. Metabolomic studies of the cervicovaginal microenvironment in bacterial vaginosis and pre-term birth suggest that vaginal microbiota drastically alter the metabolome. However, metabolic interactions between host and microbial communities in cervical carcinogenesis remain to be elucidated.
Added value of this study
In the present study, we identified metabolic signatures that effectively distinguish-HPV infected individuals, women with cervical dysplasia and cancer from HPV-negative women. Membrane lipids distinguished the cancer group from the HPV negative group and were highly associated with genital inflammation. In low-grade and principally high-grade dysplasia groups, we observed polymicrobial communities perturb amino acid and nucleotide metabolism in a similar manner to bacterial vaginosis. Our integrative and comprehensive bioinformatic approach pinpointed the microbial or host origin of specific amino acid metabolites and nucleotides. In addition, a novel aspect of our multi-omic analyses was to reveal the level to which specific metabolites were associated with Lactobacillus abundance as well as genital inflammation. An increase in the Lactobacillus abundance was positively associated with the levels of anti-inflammatory nucleotides. Collectively, we revealed that features of the cervicovaginal microenvironment (HPV, genital inflammation and vaginal microbiota) profoundly impact cervicovaginal metabolomes.
Implications of all the available evidence
This first report on cervicovaginal metabolomes in HPV-mediated cervical dysplasia and cancer demonstrate the predictive value of metabolic fingerprinting in abnormal cell metabolism. Observed perturbations in lipids, an emerging hallmark of cancer, link inflammation and cervical cancer. Additionally, we identified specific metabolites that were associated with Lactobacillus abundance and genital inflammation independently from the cancer. Amino acid and nucleotide metabolism connect vaginal dysbiosis to cervical dysplasia. The cervicovaginal metabolome can be a useful target for future diagnosis, prevention strategies and therapeutic intervention to positively impact women's health outcomes.
Alt-text: Unlabelled Box
1. Introduction
Persistent human papillomavirus (HPV) infection, the most common sexually transmitted disease, is the etiologic agent for the development of precancerous cervical dysplasia and cervical cancer [1,2]. Even though screening and HPV vaccination have significantly reduced the number of cases in developed countries, cervical cancer is still the fourth most commonly diagnosed cancer globally among females [3]. After HPV infection, carcinogenesis does not progress uniformly; precancerous dysplasia may regress without treatment for reasons that are uncertain [4]. Regardless, persistent HPV infection initiates a chain of reactions that suppresses the cell-mediated immune response and hinders clearance of the infection and destruction of abnormal cells [5]. Additionally, in spite of the rapidly accumulating data on host immune responses and vaginal microbiome in patients across cervical carcinogenesis [[6], [7], [8], [9], [10], [11]], our understanding of the molecular and metabolic changes in the cervicovaginal microenvironment relevant to microbiome has only recently begun to be elucidated [12,13].
The advances in the metabolomics field during the last decade have proven that cancer is a metabolic disease [14] and led to the rediscovery of the metabolome as an emerging target for cancer detection and therapies. Early studies have shown that cervical cancer patients harbour a unique collection of molecules ranging from amino acids to nucleic acids in the serum [15], tumour tissues [16], and faeces [17]. Moreover, research on cancer sniffing dogs indicated that there are cancer-specific volatile organic compounds being released in cervical cancer [18]. In comparison to serum or faeces, metabolomes of the cervicovaginal microenvironment have the added benefit of reflecting the metabolic processes occurring in the mucosal environment instead of metabolic outcomes of the entire body or other systems. However, to the best of our knowledge, metabolomes of the local microenvironment in patients with HPV, cervical dysplasia, or invasive cervical carcinoma (ICC), have not been previously reported.
Cervicovaginal metabolomes throughout cervical carcinogenesis can be influenced by several factors that are host-, microbiome-, and environment-associated. For example, HPV suppresses cytokine production [19], which alters the microbial dynamics in the cervical microenvironment. Our group and others have shown a strong association between genital immune mediators and a decrease in Lactobacillus abundance in cervical carcinogenesis [6,8]. Shifts in cervicovaginal microbial communities due to dysbiosis or environmental factors can alter the cervicovaginal metabolome. For instance, a shift from health-associated Lactobacillus-dominated microbiota towards complex anaerobic and microaerophilic communities in bacterial vaginosis (BV) is highly correlated with an increase in amino acid catabolites and polyamines [12,13]. A similar metabolic shift was also observed in smokers in comparison to non-smokers in connection to imbalances in the vaginal microbiota [20]. Additionally, alterations in cell physiology caused by HPV infection and neoplastic disease could transform the dynamics of the cervicovaginal microenvironment. For example, cancer cells with a drastically increased glucose demand ferment glucose into lactate instead of carbon dioxide, in contrast to healthy differentiated cells [21]. As such, secretions from cancer cells, as well as microbial fermentation products, could ultimately modulate the microenvironment and affect both host and microbial metabolism. In the context of HPV infection and cervical carcinogenesis, these metabolic shifts in the cervical microenvironment warrant further investigation. We hypothesized that cervicovaginal metabolic profiling would reveal HPV, inflammation, host and microbe driven metabolic signatures across cervical carcinogenesis.
Herein, we characterized differences in cervicovaginal metabolomes in HPV-negative and HPV-positive individuals, women with low-grade and high-grade cervical dysplasia, and newly diagnosed ICC using a global metabolomics approach. We report unique differences in metabolic profiles of participants with HPV infection, cervical dysplasia and ICC that discriminated these patient groups from healthy HPV-negative controls. The magnitude of the differences observed among the patient groups was on the metabolic super pathway level rather than on the individual metabolite level. However, we identified 3-hydroxybutyrate, eicosenoate, and oleate/vaccenate as key metabolites that robustly discriminated patients with ICC from healthy HPV-negative individuals, and many amino acid metabolism products such as glutamine, pyroglutamine, and N-acetyltaurine as discriminators of healthy HPV-negative individuals from participants that were HPV-positive or with cervical dysplasia.
We also interrogated the host- and microbiota-associated factors that shape the local microenvironment and found that genital inflammation and vaginal microbiota composition are strong drivers of the cervicovaginal metabolomes. Our comprehensive bioinformatic analysis of microbial contributions to the cervicovaginal metabolome pinpointed the metabolic effects of certain BV-associated microorganisms including Sneathia, Streptococcus, Prevotella, and Gardnerella on catabolism of amino acids, which differentiated high-grade cervical dysplasia from ICC. Genital inflammation, mainly in the ICC group was highly associated with various groups of lipids. Nucleotides with anti-inflammatory properties positively correlated with Lactobacillus abundance and negatively correlated with genital inflammation. Overall, our results provide metabolic insights into complex virus-host-microbiome interactions in the context of a common gynaecologic malignancy, cervical cancer that are essential for the development of future metabolome targeted diagnostics and therapies.
2. Materials and methods
2.1. Study subjects
We recruited 78 premenopausal, non-pregnant women and grouped as follows: healthy HPV-negative [Ctrl HPV (−), n = 18] and HPV-positive participants [Ctrl HPV (+), n = 11] as controls, participants with low-grade squamous intraepithelial lesions (LSIL, n = 12), high-grade squamous intraepithelial lesions (HSIL, n = 27) and newly diagnosed invasive cervical carcinoma (ICC, n = 10) at three clinical sites in Phoenix, AZ: St. Joseph's Hospital and Medical Center, University of Arizona Cancer Center and Maricopa Integrated Health System. Our study was approved by the Institutional Review Boards of each participating site and written informed consent was attained from each study participant. The inclusion criteria and participant classification for each of the groups were described previously [6]. Briefly, women that were pregnant, post-menopausal, or currently menstruating, women that were on antibiotics and antifungals within the previous three months of the study and women with the following conditions: vaginal infection, bacterial vaginosis, vulvar infection, urinary tract infection or sexually transmitted infections including chlamydia, gonorrhoea, trichomoniasis, and genital herpes, women with type I or II diabetes, or that were HIV positive, women with unusual or foul-smelling vaginal discharge, and women who use douching substances, within the previous three weeks of the study, were also excluded from the study. Sexual intercourse or vaginal lubricant use within 48 h of the sample collection also excluded women from participation. Physician's pelvic exam, medical records and surveys were used to verify exclusion criteria.
2.2. Sample collection and processing
Cervicovaginal lavages (CVL) and vaginal swabs were collected from study participants. Vaginal pH measurements, HPV genotyping, and microbiome analysis from vaginal swabs were described previously [6]. Briefly, CVLs were collected by the clinicians using 10 mL of sterile 0.9% saline solution (Teknova, Hollister, CA), immediately placed on ice and frozen at −80 °C. The CVLs were thawed on ice and clarified by centrifugation (700 ×g for 10 min at 4 °C) prior to aliquoting and aliquots were sent to Metabolon, Inc. (Durham, NC) for global metabolomics identification and quantification. Samples were randomized across the extraction and instrumentation platforms. Briefly, upon receipt, samples were inventoried, and each sample identifier was accessioned into the Metabolon LIMS system and assigned a unique identifier. Sample preparation was achieved using the MicroLab STAR® system from Hamilton company. Aliquots of 100 μL were mixed with ethanol and vigorously shaken for 2 min (Glen Mills GenoGrinder 200) to precipitate the proteins and dissociate the small molecules bound to proteins. The metabolome extract was divided into five aliquots and four of them were analysed. Samples were placed briefly on a TurboVap® (Zymark) to remove organic solvent. The extracts were stored overnight under nitrogen before preparation for analysis. The sample's extract was dried then reconstituted in solvents compatible to each of the four methods described below.
2.3. Metabolome analysis
All methods utilized a Waters ACQUITY ultra-performance liquid chromatography (UPLC) and a Thermo Scientific Q-Exactive high resolution/accurate mass spectrometer interfaced with a heated electrospray ionization (HESI-II) source and Orbitrap mass analyser operated at 35,000 mass resolution. Two of the aliquots were analysed with two separate reverse phase UPLC/MS/MS methods with positive ion mode electrospray ionization (ESI) optimized for hydrophilic compounds. In this method, the extract was gradually eluted from a C18 column (Waters UPLC BEH C18–2.1 × 100 mm, 1.7 μm) using water and methanol containing 0.05% perfluoropentanoic acid (PFPA) and 0.1% formic acid (FA). The second extracts were evaluated for more hydrophobic compounds by eluting the sample with methanol, acetonitrile, water, 0.05 PFPA, and 0.1% FA. The third aliquots were analysed using basic negative ion conditions by conditioning the sample with 6.5 mM ammonium bicarbonate at pH 8.0 and eluting with methanol and water through a dedicated separate C18 column. The last aliquots were analysed using a negative ionization following elution from HILIC column (Waters UPLC BEH Amide 2.1 × 150 mm, 1.7 μm) using a gradient consisting of water and acetonitrile with 10 mM ammonium formate, pH 10.8. Those four analytical methods were optimized according to the polarity or hydrophilicity of the compounds. The MS analysis alternated between MS and data-dependent MSn scans using dynamic exclusion. The scan range varied slightly between methods but covered 70–100 m/z.
Variability and performance of extraction and instrumentation were validated by the utilization of recovery and internal standards. A pooled composite sample consisting of a small volume of each experimental sample served as a technical replicate and extracted water samples served as process blanks. Those control samples were spaced evenly among the injections to monitor instrument stability.
2.4. Metabolome data collection
Data extraction was performed using Metabolon's Laboratory Information Management System (LIMS). Compounds were identified by comparison to library entries, which contain over 5000 commercially available purified standard or mass spectral entries of recurrent unknown entries. Mass-spectrometry based levels of metabolites were calculated based on area-under-the-curve of the peaks, that allows for quantitative analysis between the groups but not for the absolute quantification. Additionally, each biochemical's ionization coefficient depends on the environment. Hence, the threshold of detection is experiment, matrix, and compound dependent. However, average detection limits of the compounds were in the nanomolar range, and for some compounds, the detection limits were in the picomolar range. The threshold of detection is compound specific and determined by Metabolon's algorithm.
2.5. Statistical analysis
Welch's two-sided two-sample t-test was used to perform a pairwise comparison of the means of the metabolites among the groups [Ctrl HPV (−), Ctrl HPV (+), LSIL, HSIL, and ICC]. The p values below 0.05 were corrected using false discovery rate (FDR) method and q values were reported. The fold changes observed among the groups were visualized using Cytoscape network analysis [22]. A trend analysis was performed to visualize the differences observed in number of metabolites between the groups. The metabolites that distinguished the patients in the ICC group from Ctrl HPV (−) group and Ctrl HPV (+) group from LSIL and HSIL groups were identified using Receiver Operating Characteristics (ROC) analysis and the strength of the discriminators were measured with the Area Under the Curve (AUC) values. AUC values above 0.8 were considered as good and above 0.9 were considered as excellent discriminators. To measure the strength and direction of the linear associations between the metabolites and other sample data including the inflammation scores, vaginal pH, and Lactobacillus dominance, Spearman's rank correlation coefficients were calculated, unadjusted and bootstrapped p values were reported. Based on the sample size (all samples (n = 78) versus without the ICC samples (n = 68)), Spearman's rho (ρ) critical value of 0.223 and 0.239 were selected.
The dissimilarities in the metabolome dataset among the groups were analysed using principal component analysis (PCA). Multivariate analysis of variance (MANOVA) analysis was performed to determine the composite differences for the first two components among the clusters based on participant characteristics, Lactobacillus dominance, and genital inflammation. Additionally, the distances among the groups on PC1 and PC2 axes were calculated using Mann-Whitney's U test. Using R software and ggplot2 library [23], we overlaid shaded ellipses on clusters to reflect 95% confidence intervals from the centroids of the clusters. A hierarchical clustering analysis (HCA) was performed based on Euclidean distances observed among the metabolomes. The differences in the patient clusters identified by HCA were tested using the chi-square test and p values were reported. The participant demographics, features of the cervicovaginal environment, genital inflammation and pH of the patient clusters were compared using Mann-Whitney's U test and p values were reported. PCA and HCA were performed using metabolome composition of 475 metabolites that were identified by Metabolon Inc.
2.6. Bioinformatic analysis to integrate metabolomes with microbiomes
Quantitative Insights into Microbial Ecology (QIIME) v1.9 pipeline [24] was used for preparing 16S rRNA gene sequences for metagenome predictions. Operational Taxonomic Units (OTUs) with 97% sequence similarity were picked using a closed OTU picking method with Uclust [25] against the Greengenes database [26]. OTUs that were normalized based on gene copy numbers using QIIME v1.9 [24] were used for prediction of the metagenomes using the Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) [27]. Relative contribution of the human and microbial genomes to the expression of metabolites detected were calculated using Annotation of Metabolite Origins via Networks (AMON) [28]. Briefly, using the cervicovaginal metabolome datasets, host genome and microbial genomes downloaded from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (https://www.genome.jp/kegg/), and PICRUSt predicted metagenomes, we revealed microbial and host contribution to the metabolites. Homo sapiens (has) was used for the host genome and Sneathia amnii (sns), Prevotella denticola (dns), Gardnerella vaginalis ATCC 1409 (gdv), Atopobium parvulum (apv), Streptococcus sanguinis (sng), Lactobacillus crispatus (lcr) were used for the microbial genomes to represent the six most abundant bacteria observed in our dataset. Model-based Integration of Metabolite Observations and Species Abundances (MIMOSA) [29] was used to reveal the potential contribution of microbial taxa to the expression of metabolites that distinguish groups. For the MIMOSA analysis, PICRUSt-predicted metagenomes and the metabolome datasets were used. MIMOSA utilizes a predicted relative metabolic turnover (PRMT) model to estimate gene abundances from PICRUSt-predicted metagenome data to produce community-wide metabolic potential (CMP) scores for each metabolite and sample using metabolites observed, predicted metagenomes and datasets from KEGG database including genomes, reactions, and pathways. Comparing CMP scores to metabolite variations, the metabolites were classified as well predicted by certain taxa or anti-predicted. For the visualization of the dataset, only well-predicted metabolites with false discovery rate < 0.1 were presented.
2.7. Data and software availability
16S rRNA gene sequences were deposited in the National Center for Biotechnology Information (NCBI) database on BioProject accession number: PRJNA518153 and under Sequence Read Archive (SRA) accession IDs SAMN10855179 to SAMN10855255. De-identified metabolomics data is available in Supplementary files.
3. Results
3.1. Cervicovaginal metabolite diversity was reduced in Ctrl HPV (+) and cervical dysplasia groups whereas diversity was increased in cervical cancer patients compared to Ctrl HPV (−) controls
We characterized cervicovaginal metabolomes in premenopausal non-pregnant healthy HPV-negative individuals [Ctrl HPV (−)] (n = 18), healthy HPV-positive individuals [Ctrl HPV (+)] (n = 11), women with low-grade squamous intraepithelial lesions (LSIL) (n = 12), high-grade squamous intraepithelial lesions (HSIL) (n = 27), and newly diagnosed ICC (n = 10) using a global metabolomics approach. The patient characteristics and demographic information are summarized in Supplementary Table 1. Participants were assigned to the patient groups based on histology of colposcopy-directed biopsy samples that were evaluated by a pathologist. If histology was not available, cytology results were evaluated. As described in our previous publication [6], a two-tiered system of cervical intraepithelial neoplasia (CIN) was used: CIN1 and CIN2/3 were categorized as LSIL and HSIL, respectively. Our participant group consisted of 37 Hispanic and 41 non-Hispanic women and there was no significant difference in race/ethnicity among the patient groups (Fisher's exact test, p = .15). Sixty-eight percent of the participants had body mass index (BMI) >25, however BMI did not significantly differ among the groups (ANOVA, p = .47). The mean age across the groups was 38 ± 8 years and age distribution among the groups was not significantly different (ANOVA, p = .46). Vaginal pH was significantly different among the groups, pH 4.9 ± 0.6 for Ctrl HPV (−), pH 5.7 ± 1.09 for Ctrl HPV (+),pH 5.2 ± 0.6 for LSIL, pH 5.8 ± 0.9 for HSIL and pH 6.6 ± 0.9 for ICC (mean ± standard deviation, 5.6 ± 0.9) (ANOVA, p = .003). Predominant HPV genotypes in our study, determined with Linear Array HPV Genotyping Tests (Roche) on DNA extracts from vaginal swabs, were HPV 16 (n = 43), HPV 31 (n = 13), and HPV 45 (n = 13). Fifty seven out of 60 HPV (+) participants [Ctrl HPV (+), LSIL, HSIL, and ICC] were positive for multiple HPV genotypes.
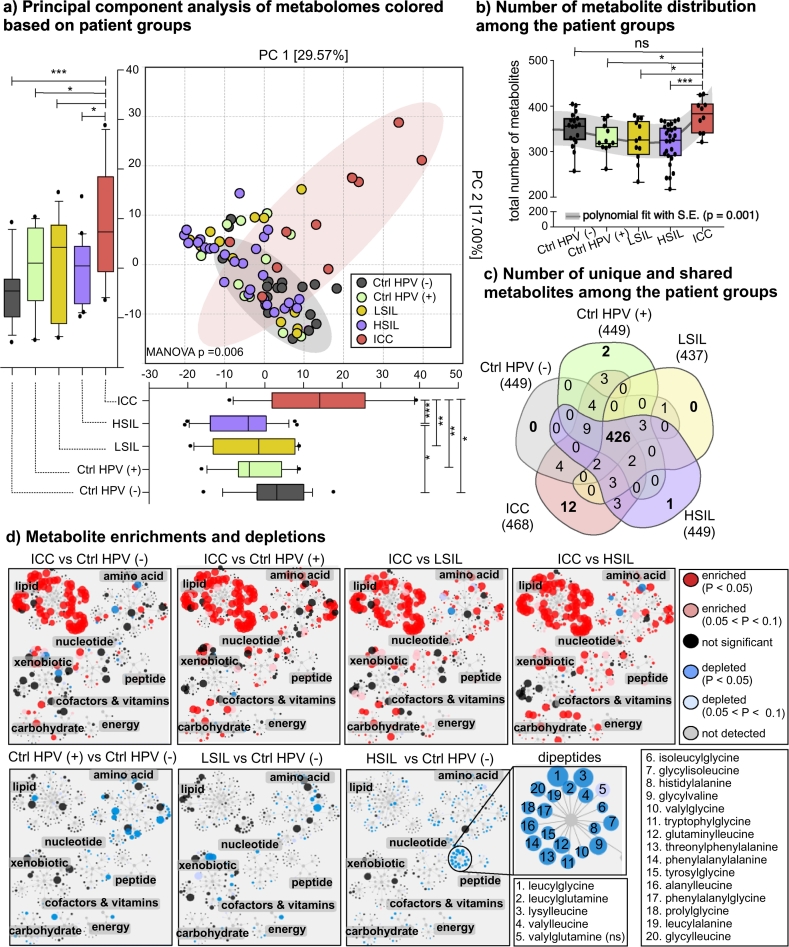
Our metabolome dataset was composed of 475 compounds of known identity from eight different metabolic pathways; 127 lipid, 147 amino acid, 34 nucleotide, 82 xenobiotic, 17 cofactor and vitamin, 10 energy, 24 carbohydrate, 32 peptide, and 2 partially-characterized molecules. Principal component analysis (PCA) of cervicovaginal lavages (CVLs) illustrated that global metabolomic profiles of ICC patients compared to other patient groups were distinct on principal component 1 (PC1) and PC2, which explained 46.57% of variation in the dataset (Fig. 1A; MANOVA, Wilk's Lambda test, p = .006; Mann-Whitney U test, p = .0009 and p = .0225, for PC1 and PC2, respectively). Due to the complex nature of the cervicovaginal lavages, the unexplained variation (53.43%) can be accounted for by the heterogeneity in participant genetic background, diet, lifestyle, etc. However, other collected participant metadata, for example, age, ethnicity, and BMI did not have a statistically significant effect on clustering (MANOVA p ≥ .05, Wilk's Lambda test, Supplementary Fig. 1). Samples from the HSIL group also formed a cluster that was significantly distinct from the Ctrl HPV (−) group samples, but only on PC1 (Mann-Whitney U test, p = .046). As shown in Fig. 1B, differences in the metabolic profiles were driven by the number of different metabolites observed in the groups (Mann-Whitney U test, p = .001). ICC patients had a significantly greater number of metabolites (378 ± 36) than the Ctrl HPV (+) (326 ± 33, p = .037), LSIL (324 ± 45, p = .022), and HSIL (316 ± 43, p = .0009) groups, but not the Ctrl HPV (−) group (348 ± 36, p = .363). Overall, Ctrl HPV (+), LSIL, and HSIL groups had a lower number of metabolites in the CVLs compared to Ctrl HPV (−) group; however, this finding did not reach significance possibly due to the limited number of samples per group. However, when we performed a trend analysis, we found that due to a reduction in the number of metabolites present in samples from HPV positive participants with and without cervical dysplasia [Ctrl HPV (+), LSIL, and HSIL], and an increase in the number of metabolites in ICC, the trend was significantly non-linear (p = .0004). Based on curve analysis, we found that the metabolite diversity trend was polynomial (p = .01).
Fig. 1.
HPV infection, dysplasia, and invasive cervical carcinoma (ICC) divergently impacted the cervicovaginal metabolic profile. A) Principal component analysis (PCA) showed a distinct cervicovaginal metabolome profile in ICC patients compared to Ctrl HPV (−), Ctrl HPV (+), LSIL, and HSIL patient groups. The difference was significant on both PC1 and PC2 axes. HSIL group was significantly different than Ctrl HPV (−) group on PC2 axis. Boxplots represent median, first and third quartile, minimum and maximum values in the dataset. Shaded ellipses represent 95% confidence intervals of the cluster centroids. Mann-Whitney U test p values represented as *p < .05, **p < .01, ***p < .001. B) Number of metabolites detected in each group. ICC patients had significantly greater number of metabolites in comparison to patients in the Ctrl HPV (+), LSIL, and HSIL groups. However, there was no significant difference between the ICC and Ctrl HPV (−) groups. ns = not significant. The trend analysis showed that the trend was polynomial (p = .01). C) Number of unique and shared metabolites among the groups visualized on a Venn diagram. Majority of the metabolites were detected in the all groups. ICC group had the greatest number of metabolites that were not detected in any of the participants in other groups. Supplementary Table 2 contains the list of metabolites that are different in each group. D) Enrichment and depletions of metabolites among patient groups visualized by Cytoscape metabolic network analysis. The node size is proportional to the magnitude of differences observed among the groups. Red and blue nodes represent enriched and depleted metabolites, respectively. In comparison to all other patient groups, ICC patients had an enrichment of metabolites that belong to lipid, amino acid, carbohydrate, and xenobiotic metabolism. Amino acids and their metabolites were depleted in Ctrl HPV (+), LSIL, and HSIL groups. Dipeptides were significantly depleted in the HSIL group compared to Ctrl HPV (−). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In order to evaluate similarities and dissimilarities in the datasets, we compared the number of shared and unique metabolites among the groups. Patient groups shared 86% of the identified metabolites (426/475), and the ICC group had the greatest number of unique metabolites (n = 12) (Fig. 1C). The unique metabolites from ICC belonged to lipid and xenobiotic super-pathways, especially from xenobiotic/drug metabolism (See Supplementary Table 2). When we visualized significantly enriched or depleted metabolites on Cytoscape networks and performed pairwise comparisons, we observed a significant enrichment of metabolites in lipid, xenobiotics, and carbohydrate super-pathways in the ICC group compared to Ctrl HPV (−), Ctrl HPV (+), LSIL, and HSIL groups. Additionally, the ICC group had an enrichment of amino acid metabolites in comparison to other groups that were HPV positive with and without cervical dysplasia [Ctrl HPV (+), LSIL, and HSIL] (Fig. 1D). Compared to the Ctrl HPV (−) group, Ctrl HPV (+), LSIL, and HSIL groups showed a depletion in metabolites mainly in the amino acid super-pathway. Interestingly, the sub-pathways under the amino acid super-pathway and the level of depletion was different for Ctrl HPV (+), LSIL, and HSIL groups when compared to the Ctrl HPV (−) group. For example, Ctrl HPV (+) participants had metabolites mainly depleted in lysine, polyamine, and phenylalanine metabolism, while LSIL participants had metabolites depleted in histidine and the urea cycle. The HSIL group had depletions in many amino acid metabolites; however, the depletions were at a lower magnitude. Additionally, all the dipeptide metabolites were depleted in the HSIL group, which might explain the differences observed on PCA (Fig. 1A). This comparative analysis revealed that HPV infection, cervical dysplasia, and ICC had a divergent impact on cervicovaginal metabolomes.
3.2. HPV infection depleted metabolites in specific amino acid and nucleotide super pathways and ICC enriched metabolites from multiple lipid pathways
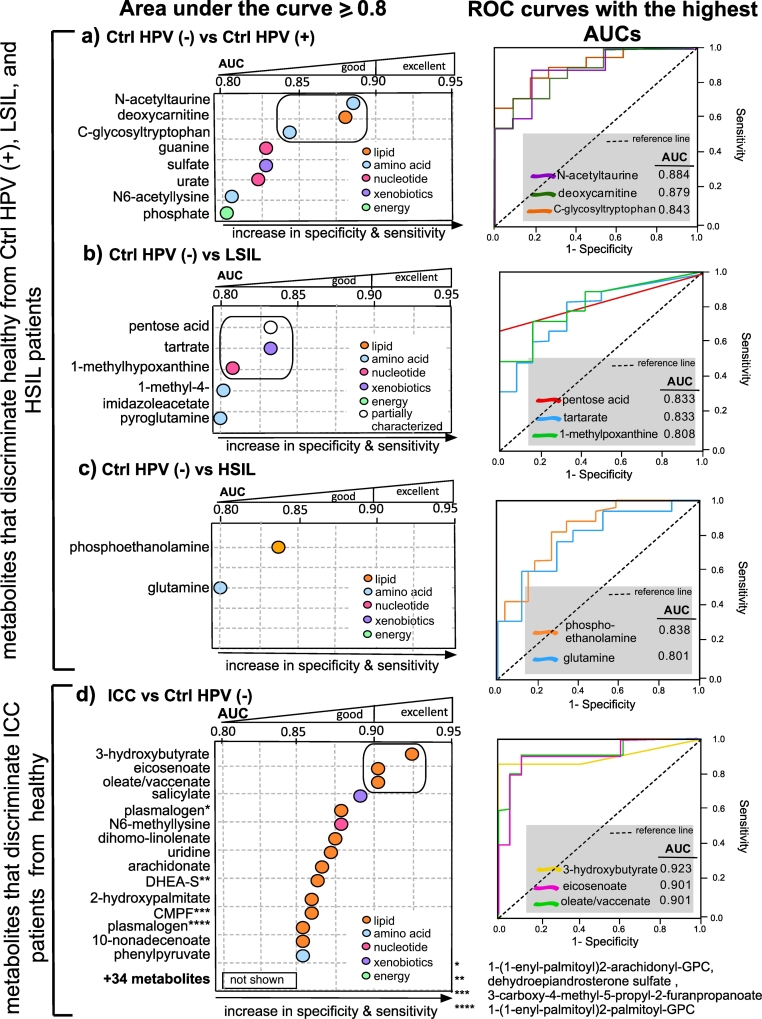
In order to explore the potential effect of HPV infection on the cervicovaginal metabolome, we performed receiver operating characteristics (ROC) analysis using samples from Ctrl HPV (−) and Ctrl HPV (+) individuals. The analysis did not reveal metabolites that discriminate Ctrl HPV (+) samples from the Ctrl HPV (−) samples (AUC < 0.8). However, when Ctrl HPV (−) samples were compared to Ctrl HPV (+) samples, we observed three amino acids (N-acetyltaurine, N6-acetyllysine, C-glycosyltryptophan), two nucleotides (guanine and urate), phosphate and deoxycarnitine had discriminatory ability with AUC values >0.8 as shown in Fig. 2A, suggesting a significant depletion of these metabolites in the Ctrl HPV (+) group. Indeed, the concentrations of these metabolites were significantly lower in the Ctrl HPV (+) group compared to Ctrl HPV (−) and ICC groups (Supplementary Table 3). When we performed ROC analysis to identify metabolites that discriminate Ctrl HPV (−) from LSIL, we found three amino acid metabolism products (pentose acid, 1-methyl-4-imidazoleacetate, pyroglutamine), one nucleotide product (1-methylhypoxanthine), and a xenobiotic (tartrate) as discriminators, as shown in Fig. 2B. Interestingly, the discriminators of Ctrl HPV (−) from HSIL were not the same metabolites as identified for LSIL. Only a phospholipid (phosphoethanolamine) and amino acid (glutamine) had >0.8 AUC (Fig. 2C). In summary, HPV infection and cervical dysplasia led to depletion of metabolites from certain amino acid and nucleotide metabolisms. These amino acid and nucleotide metabolites distinguished Ctrl HPV (−) group from the Ctrl HPV (+) and cervical dysplasia (LSIL and HSIL) groups.
Fig. 2.
Multiple metabolites from diverse pathways robustly discriminated healthy, dysplasia, and ICC patient groups. The cut-off value for the Receiver Operating Characteristics (ROC) analysis was 0.8 to include only good (0.8 ≤ AUC < 0.9) and excellent (AUC ≥ 0.9) biosignatures. A) ROC analysis comparing Ctrl HPV (−) to Ctrl HPV (+). ROC showed that amino acid, lipid, nucleotide, and xenobiotic metabolism products were indicators of absence of HPV infection. B) ROC comparing Ctrl HPV (−) to LSIL. Depletions in the amino acid metabolism products in LSIL discriminated Ctrl HPV (−) from LSIL. C) ROC comparing Ctrl HPV (−) to HSIL. Phosphoethanolamine and glutamine were discriminators of Ctrl HPV (−) from HSIL. D) ROC analysis performed between ICC and Ctrl HPV (−) groups revealed metabolites from different lipid classes with >0.9 area under the curve (AUC) values serve as strong discriminators of ICC. Those lipids were associated with cancer related inflammation. Supplementary Table 3 contains the median levels of the metabolites and Supplementary Table 4 contains the q values of Welch's t-test for group comparisons listed in this figure.
Since PCA analysis showed distinct metabolomes of ICC patients compared to the other groups, we next investigated the signatures of ICC using ROC analysis. As illustrated in Fig. 2D, 50 metabolites were either equal to or greater than AUC value of 0.80 indicating ICC significantly changes the expression of metabolomes in the cervicovaginal microenvironment and these metabolites can accurately distinguish participants with ICC from the Ctrl HPV (−). When ICC was compared to Ctrl HPV (+), most of the metabolites were similar to those that discriminated ICC from Ctrl HPV (−). Interestingly, many of the metabolites discriminated ICC group from the Ctrl HPV (−) group belonged to the lipid super-pathway (Fig. 2D). These metabolites were mainly long chain fatty acids, ketone bodies, steroids, ceramides, and plasmalogens. Among the detected molecules as discriminators of ICC, oleate/vaccinate, eicosenoate, and 3-hydroxybutyrate had AUC values >0.9, which provided more accurate discriminator and better predictive estimates than any other metabolite that was detected. As visualized in Fig. 2D, the abundance of these metabolites in the ICC group was not only significantly greater than the Ctrl HPV (−) and Ctrl HPV (+) groups, but also from the LSIL and HSIL groups (q < 0.05). When we calculated the AUC based on ICC compared to HSIL group, 71 metabolites had AUC >0.80 that distinguished ICC from HSIL. Similar to ICC and HPV comparisons, the majority of the metabolites were long chain saturated fatty acids, ketone bodies, plasmalogens, ceramides, and sphingomyelins (See Supplementary Table 3 and Supplementary Table 4). Overall, ROC analysis showed that ICC patients harbour elevated levels of lipid metabolites from different subclasses that can accurately distinguish them from all the other groups. Furthermore, those signature molecules did not show any gradation among the Ctrl HPV (+) and dysplasia groups when compared to Ctrl HPV (−) group (See Supplementary Table 3).
3.3. Microbially produced metabolites explained the dynamic shifts observed in the peptide and amino acid metabolisms
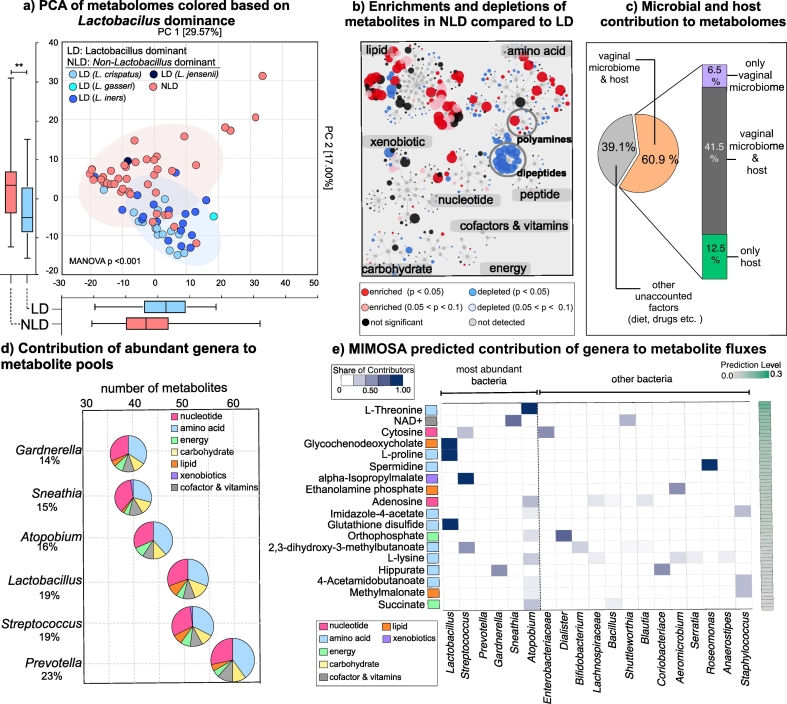
Microorganisms colonizing the cervicovaginal microenvironment produce a broad range of metabolites that are important for cervicovaginal health. Therefore, we investigated whether differences in the vaginal microbiota, in connection to the severity of cervical neoplasm, explain variations observed in the metabolome dataset. When we colour coded samples on PCA plots based on microbiota composition, metabolomes from patients with Lactobacillus-dominated (LD) vaginal microbiota formed a cluster significantly different than the metabolomes from patients with non-Lactobacillus dominance (NLD) (Fig. 3A). In accordance with the Lactobacillus dominance, vaginal pH revealed a similar clustering pattern on PCA (MANOVA, Wilk's Lambda test, p < .001, Supplementary Fig. 2). Grouping based on Lactobacillus dominance showed significance when PC1 and PC2 were analysed together (MANOVA, Wilk's Lambda test, p < .001). The difference was more apparent on PC2 (Mann-Whitney U test, p = .002), which explained 17% of the variation in the dataset. This observation provided further proof that vaginal microbiomes significantly modulate cervicovaginal metabolomes.
Fig. 3.
Vaginal microbiota composition profoundly impacted amino acid and nucleotide metabolisms. A) Principal component analysis of metabolomes visualized based on Lactobacillus dominance. Lactobacillus dominant (LD) and non-Lactobacillus dominant (NLD) metabolomes were significantly different on PC2. Boxplots represent median, first and third quartile, minimum and maximum values in the dataset. Shaded ellipses represent 95% confidence intervals of the cluster centroids. Mann-Whitney U test p values represented as *p < .05, **p < .01, ***p < .001. B) Metabolic enrichments and depletions in NLD vs LD visualized on Cytoscape networks. Metabolomes of patients that had NLD vaginal microbial communities had enrichments in lipid and amino acid metabolism. Dipeptides were depleted in patients with NLD communities. Some metabolites from polyamines were enriched whereas some were depleted in patients with NLD communities. C) Prediction of vaginal microbiota or host origins of the metabolites. PICRUSt predicted metagenomes annotated with AMON explained 60% of the detected metabolites in cervicovaginal lavages in all groups. D) The six most abundant genera within the data set were predicted to contribute 14–23% of the metabolites that were detected. The microbially produced metabolites mainly belonged to amino acid and nucleotide metabolism. E) Well-predicted (FDR < 0.1) metabolites based on MIMOSA analysis and relative contribution of microbial phylotypes to the production or depletion of metabolites. The six most dominant genera within our dataset explained the observed concentrations of many amino acids and their metabolic end-products.
Considering that metabolomes of patients with LD microbiomes diverged from metabolomes of patients with NLD microbiomes, we examined their impact on metabolic super-pathways. Fig. 3B shows metabolic networks visualizing NLD vs LD metabolomes using Cytoscape. Many metabolites from lipid, amino acid, and nucleotide pathways were significantly enriched in the NLD metabolomes compared to LD. Additionally, all metabolites from dipeptide metabolism were significantly depleted, similar to our observation in the HSIL group compared to Ctrl HPV (−) group (Fig. 1D).
We also evaluated the origins of the cervicovaginal metabolites using Annotation of Metabolite Origins via Networks (AMON) software [28]. Based on PICRUSt metagenome predictions and quantified metabolites in our dataset that have Kyoto Encyclopedia of Genes and Genomes (KEGG) identifiers, 48% of the identified metabolites were predicted to be produced by the cervicovaginal microorganisms, which is slightly less than the metabolites that can be produced by the human metabolism (54.4%) (Fig. 3C). We found that 39.1% of the metabolites could not be explained by the host or the vaginal microbiota. We did not observe any differences in number of only microbially-produced metabolites among Ctrl HPV (−), Ctrl HPV (+), LSIL, HSIL, and ICC groups, since this analysis strictly depends on presence or absence of microbial phylotypes rather than their abundances. We also investigated the individual contribution of the most abundant microbial genera detected in our dataset: Lactobacillus, Prevotella, Sneathia, Atopobium, Streptococcus, and Gardnerella [6]. Microbial genomes downloaded from KEGG showed that Prevotella was the genus capable of producing most of the metabolites in our dataset (Fig. 3D) (n = 60; 23% of metabolites detected and exist in the KEGG database) and Gardnerella was the genus capable of producing the least number of metabolites (n = 38; 14% of metabolites detected and exist in the KEGG database) compared to the other genera tested. The metabolites predicted to be produced by the most abundant bacteria were mainly amino acids, nucleic acids, or their metabolic end-products (Fig. 3D). Microbial contribution analysis did not explain the differences observed in lipid metabolism when LD and NLD groups were compared. Samples that belong to the NLD group are composed of mainly cervical dysplasia and ICC patients, hence enrichments in the lipid metabolism in NLD could be coincidental.
In order to mechanistically associate cervicovaginal metabolites in our dataset to microbiome composition, we performed Model Based Integration of Metabolite Observations and Species Abundances (MIMOSA) [29] analysis instead of correlation analysis which could lead to misinterpretation of the microbe-metabolite interactions. Correlation analysis between microbial taxa and host-associated metabolomes were found to lead to a poor explanation of the interactions and inaccurate interpretations [30]. Based on MIMOSA analyses, illustrated in Fig. 3E, of 127 metabolites that were predicted to be produced by the microbiota, 36 of them were significantly consistent with the metabolic potential (FDR < 0.1). As expected, mainly amino acids and amino acid degradation products, such as polyamines, were well-predicted to be modulated by the microorganisms. This model revealed that amino acids and their degradation products were well-predicted, and there were shifts in the products that can be produced by BV-associated microorganisms in comparison to Lactobacillus species. Lactobacillus species were the sole predictors of a secondary bile acid (glycochenodeoxycholate), amino acid (L-proline), and an amino acid product (glutathione disulphide), molecules that exert beneficial effects on the host based on metabolite fluxes. On the other hand, BV-associated bacteria explained the metabolism of specific metabolites. Atopobium contributed to the metabolism of L-threonine, adenosine, imidazole-4 acetate, glutathione disulphide, orthophosphate, l-lysine, 4-acetamidobutanoate, methylmalonate, and succinate. Gardnerella was the main contributor to the hippurate metabolism. NAD+ flux was mainly explained by the presence of Sneathia in our data set. Streptococcus contributed to 2,3-dihydroxy-3-methylbutanoate, a secondary metabolite in leucine, isoleucine, and valine metabolism, alpha-isopropylmalate, and cytosine. Interestingly, non-abundant genera including Dialister, Roseomonas, Staphylococcus, and Shuttleworthia were predicted to contribute to metabolism of a diverse set of metabolites such as orthophosphate, adenosine, l-lysine, succinate, methylmalonate, and imidazole-4 acetate (Fig. 3E). Overall, our comprehensive analysis of microbiome-metabolome relationships indicated that displacement of LD with NLD potentially induced various shifts in the amino acid metabolism that can be mostly explained by six most abundant genera detected in our dataset.
3.4. Lipid and xenobiotic metabolites were highly associated with genital inflammation
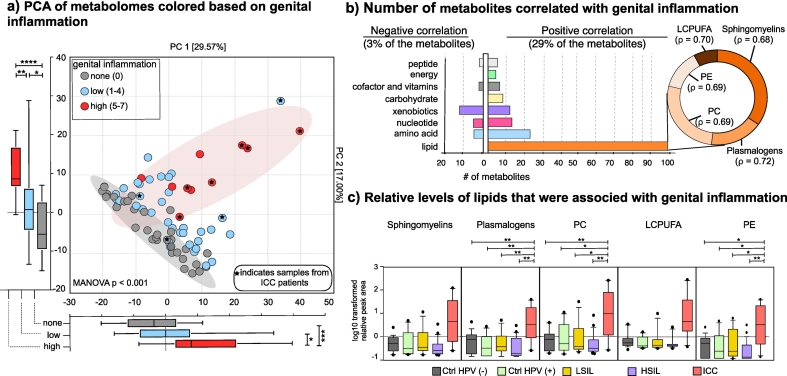
Since inflammation is a critical component of tumour progression, we investigated the link between cervicovaginal metabolomes and genital inflammation. Genital inflammation scoring strategy was previously reported [6,9,31,32]. Briefly, expression levels of seven cytokines (IL-1α, IL-1β, IL-8, MIP-1β, CCL20, RANTES, and TNFα) were evaluated in CVLs and the patients were assigned a cumulative score based on whether the level of each cytokine was in the upper quartile. Patients that scored five to seven were considered to have high genital inflammation, whereas one to five was assigned low genital inflammation and patients with a score of 0 were categorized as no inflammation. We overlaid genital inflammation scores on metabolomes and visualized on PCA as shown in Fig. 4A. Based on distances visualized on PCA, samples showed a gradation based on the genital inflammation category. Patients in the high genital inflammation category formed a cluster significantly distant from the none and low genital inflammation groups on both PC1 (Mann-Whitney U test, p values 0.0004 and 0.0143, respectively) and PC2 (p < .0001 and p = .0016, respectively). Based on MANOVA results, genital inflammation led to significant grouping on PCA (Wilk's Lambda p < .001). The difference between the low and no genital inflammation groups on PC2 was also significant (Mann-Whitney U test, p = .0228). Additionally, as indicated in Fig. 4A, the samples with high genital inflammation were mainly ICC patients (6/10). When we performed correlation analysis, 32% (187/575) of the metabolites significantly correlated with genital inflammation scores with Spearman's rho greater than the critical value (0.23 for n = 78, and bootstrapped p < .05). About 29% (168/575) and 3% (19/575) of the metabolites showed positive and negative correlation with genital inflammation, respectively (Fig. 4B). Metabolites that positively correlated with genital inflammation were mainly from lipids (n = 99) followed by amino acids (n = 23), nucleotides (n = 14) and xenobiotics (n = 11). Lipids that positively correlated (ρ > 0.68, p < .05) with genital inflammation were sphingolipids, plasmalogens, phosphatidylcholines, phospatidylethanolamines and long-chain-poly-unsaturated fatty acids (Fig. 4B, Supplementary Table 4), which are essential components of host and bacterial cellular membranes. The relative concentrations of the metabolites from these five lipid sub-families were significantly higher in the ICC group compared to other groups as shown in Fig. 4C. These data strengthen the link between genital inflammation, cancer, and lipids. The correlation between genital inflammation and lipids was stronger (ρ > 0. 74 for the first 20 highest ranked correlations) than the correlation between other metabolites and genital inflammation (Supplementary Table 4). Most metabolites that negatively correlated with genital inflammation were xenobiotics (n = 11), amino acids (n = 4), and nucleotides (n = 2).
Fig. 4.
Levels of genital inflammation highly correlated with metabolite profiles and patient groups. A) Principal component analysis (PCA) demonstrated a gradual separation of metabolomes based on presence or intensity of genital inflammation. Metabolomes of patient with high genital inflammation (inflammation score of 5–7) formed a separate cluster from patients without (inflammation score = 0) or low genital inflammation (inflammation score of 1–4) on PC1 and PC2. Majority of the high genital inflammation samples belonged to ICC patients. Boxplots represent median, first and third quartile, minimum and maximum values in the dataset. Shaded ellipses represent 95% confidence intervals of the cluster centroids. Mann-Whitney U test p values represented as *p < .05, **p < .01, ***p < .001. B) Number of metabolites positively and negatively correlated with genital inflammation. Spearman's rho correlation coefficient greater than the critical value (0.23 for n = 78) with a bootstrapped p < .05 were considered significant. Metabolites from lipid metabolism positively correlated with genital inflammation whereas xenobiotics negatively correlated with the genital inflammation. Lipids that correlated with genital inflammation belonged to sphingomyelins, plasmalogens, phosphatidylcholines (PC), long-chain-polyunsaturated fatty acids, and phosp hatidylethanolamines (PE). Supplementary Table 5 includes the list of Spearman's correlation coefficients reflecting the strength of correlation between genital inflammation and metabolites. C) Relative levels of plasmalogens, sphingomyelins, phosphatidylcholines, phosphatidylethanolamines, and long-chain polyunsaturated fatty acid metabolites were greater in the ICC group compared to the others. Boxplots represent median, first and third quartile, minimum and maximum values in the dataset. Welch's t-test p values represented as *p < .05, **p < .01, ***p < .001.
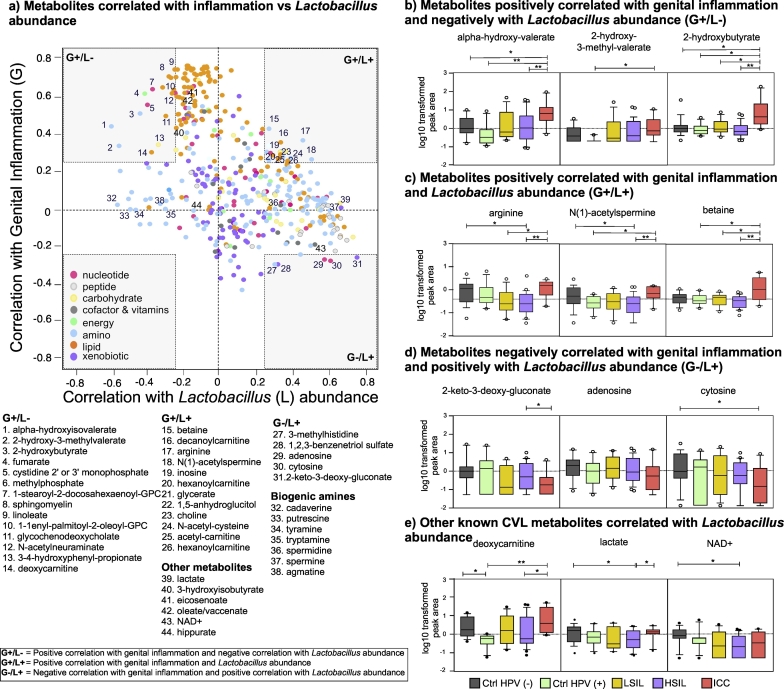
Since genital inflammation is linked to vaginal microbial community composition, we investigated the relationship between the metabolites that correlated with genital inflammation and Lactobacillus abundance. Fig. 5A illustrates Spearman's rho coefficients between the metabolites and genital inflammation versus metabolites and Lactobacillus abundance. Many of the lipids that highly correlated with genital inflammation did not correlate with Lactobacillus abundance. However, one of the sphingomyelins, linoleate, glycochenodeoxycholate and deoxycarnitine were negatively correlated with Lactobacillus abundance. Glutathione synthesis intermediate, 2-hydroxybutyrate, branched chain amino acid metabolism product, alpha-hydroxy-isovalerate, and L-isoleucine metabolism product, 2-hydroxy-3-methyl-valerate moderately and positively correlated with genital inflammation and negatively correlated with Lactobacillus abundance. These amino acid metabolites were significantly higher in the ICC group compared to other groups (Welch's two-sided t-test, q < 0.05, Fig. 5B). Metabolites that discriminated ICC from the Ctrl HPV (−), eicosenoate, 3-hydroxybutyrate, and oleate/vaccinate highly correlated with genital inflammation (Fig. 5A). Metabolites that positively correlated with Lactobacillus abundance and genital inflammation included amino acids/products such as arginine and betaine, a polyamine, N(1)-acetylspermine, and lipids mainly involved in carnitine metabolism such as decanoylcarnitine, hexanoylcarnitine, and acetylcarnitine. The levels of these metabolites were depleted in the Ctrl HPV (+), LSIL, and HSIL groups compared to Ctrl HPV (−) and ICC groups as shown in Fig. 5C and E. There were only a few metabolites that negatively correlated with genital inflammation but positively correlated with Lactobacillus abundance. These metabolites included nucleotides adenosine and cytosine and xenobiotics such as 2-keto-3-deoxy-gluconate and 1,2,3-benzenetriol. The relative levels of these nucleotides or xenobiotics were lower in the ICC group compared to other groups and the difference in cytosine levels between Ctrl HPV (−) and ICC group and 2-keto-3-deoxy-gluconate between HSIL and ICC were statistically significant (Welch's two-sided t-test, q < 0.05) (Fig. 5D).
Fig. 5.
A) Metabolites that connected genital inflammation to Lactobacillus abundance. Scatter plot of metabolites that correlated with genital inflammation and Lactobacillus abundance. Lipids (plasmalogens, sphingomyelin, phosphatidylcholine, and long chain polyunsaturated fatty acids (LCPUFA)) strongly and positively correlated with genital inflammation but not with Lactobacillus abundance. A few amino acids negatively correlated with Lactobacillus abundance positively correlated with genital inflammation. Carnitine metabolites positively correlated with both Lactobacillus abundance and genital inflammation. Biogenic amines negatively correlated with Lactobacillus abundance but did not correlate with genital inflammation. B) Relative levels of metabolites that positively correlated with genital inflammation and negatively correlated with Lactobacillus abundance among the patient groups. The concentrations were relatively higher in the ICC group. C) Relative levels of metabolites that positively correlated with genital inflammation and Lactobacillus abundance among the patient groups. The concentrations were relatively higher in the ICC group. D) Relative levels of metabolites that negatively correlated with genital inflammation and positively with Lactobacillus abundance among the patient groups. The concentrations were relatively lower in the ICC group. Metabolites with the three highest ρ values were visualized on graphs B, C, and D. E) Relative levels of metabolites that had significant associations with Lactobacillus-dominance or dysbiotic microbiota. Welch's t-test p values represented as *p < .05, **p < .01, ***p < .001.
We also observed many biogenic amines (cadaverine, putrescine, tyramine, tryptamine, and agmatine) negatively correlated with Lactobacillus abundance as predicted but did not correlate with genital inflammation. In general, levels of the biogenic amines did not significantly differ among the groups (See Supplementary Fig. 3). Additionally, lactate levels strongly and positively correlated with Lactobacillus abundance. Interestingly, non-Lactobacillus dominated HSIL group had significantly lower levels of lactate compared to Ctrl HPV (−) and ICC groups. A major Gardnerella associated metabolite, hippurate, did not correlate with either genital inflammation or Lactobacillus abundance. A metabolite that was mainly predicted to be modulated by Sneathia was NAD+ based on MIMOSA analysis. Additionally, NAD+ levels positively correlated with Lactobacillus abundance. Interestingly, NAD+ levels were significantly lower in HSIL compared to Ctrl HPV (−). To better understand the associations independent from cancer, we performed the correlation analysis by excluding all ICC samples. As shown in Supplementary Fig. 4, we observed the same trend between metabolites that were associated with inflammation and Lactobacillus when the ICC samples were excluded. However, we observed minor differences in correlation coefficients of key metabolites, for example, ρ for lactate and Lactobacillus abundance was greater (ρ = 0.65 versus ρ = 0.69) whereas ρ for alpha-hydroxy-isovalerate and Lactobacillus abundance was slightly smaller (ρ = −0.59 versus ρ = −0.58) when the ICC samples were excluded.
3.5. Metabolomes formed three distinct clusters mainly driven by cervical cancer, vaginal pH and microbial communities
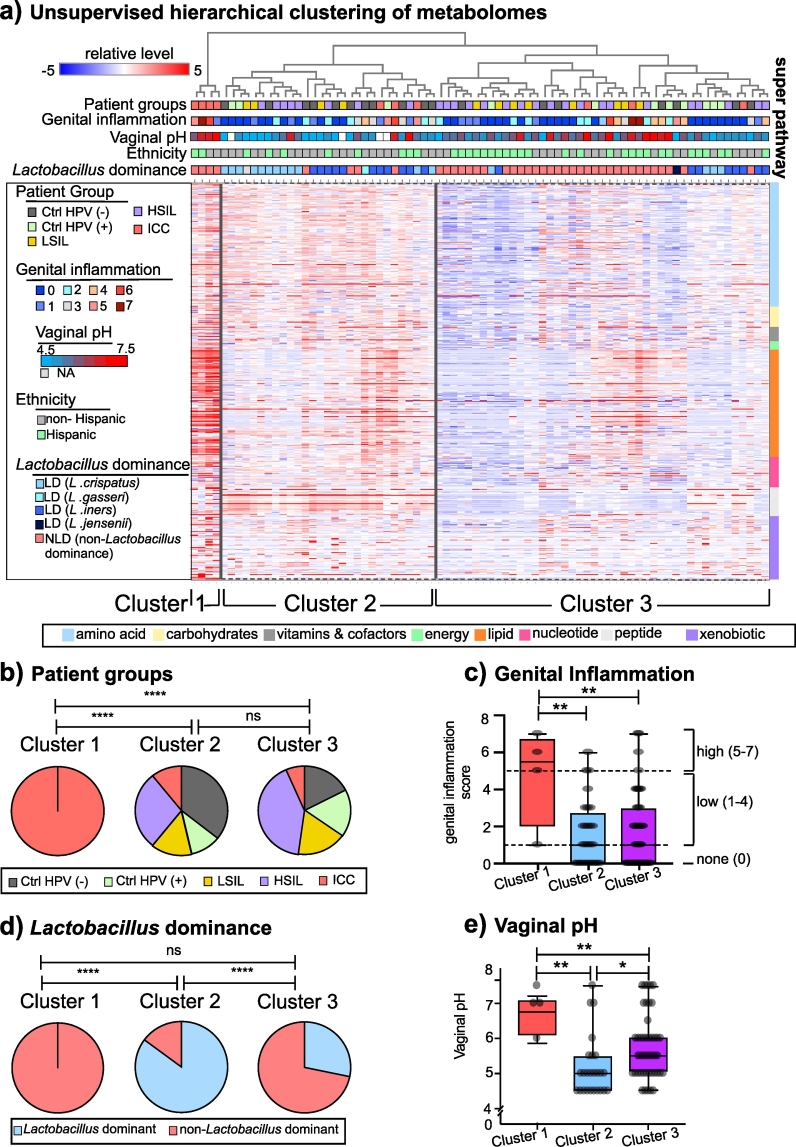
Metabolites detected in the CVLs can be impacted by many biological variables including viruses, microbial communities, host-derived factors, and outcomes of host-microbe interactions including genital inflammation and vaginal pH. We performed unsupervised hierarchical clustering using all identified metabolites to reveal whether any of these covariates drive the similarities between the metabolomes. Based on Euclidean distances among the metabolomes, samples were assembled into three clusters as shown in Fig. 6A. Cluster 1 (n = 4) composed of only ICC patients, whereas Clusters 2 (n = 29) and Cluster 3 (n = 45) were not dominated by any particular participant group. The participant group composition of Cluster 1 was significantly different than Cluster 2 (Chi-Square test, p = .002) or Cluster 3 (chi-square test, p < .0001) (Fig. 6B). As ICC explained only Cluster 1, we evaluated other factors that might be defining Cluster 2 and 3. The genital inflammation score was greater in Cluster 1 compared to Clusters 2 and 3 as shown in Fig. 6C, which did not distinguish Cluster 2 from Cluster 3. Next, we evaluated Lactobacillus dominance. As in depicted in Fig. 6D, clusters had significantly different vaginal microbiota composition. Cluster 1 was solely dominated by NLD microbiota, whereas Cluster 2 was mainly consisted of participants with LD microbiota. Similar to Cluster 1, Cluster 3 mainly consisted of patients with NLD microbiota as well. Cluster 2 was dominated by Lactobacillus crispatus and Lactobacillus iners. On the other hand, Cluster 3 was dominated by polymicrobial communities followed by L. iners. Since vaginal pH and microbial composition are linked [6], as expected, vaginal pH of the patients in each cluster differed significantly as well. Individuals in Cluster 1, that was dominated by multiple non-Lactobacillus genera, had significantly higher vaginal pH compared to the individuals in Clusters 2 and 3. Cluster 2 was composed of participants with significantly lower vaginal pH (Fig. 6E) in accordance with Lactobacillus dominance, when compared to Cluster 1 (Mann-Whitney U test, p = .002) or Cluster 3 (Mann-Whitney U test, p = .046). Consistent with our previous observations, Lactobacillus dominance and vaginal pH better explained the clustering than the other variables. In summary, these results indicate that cancer, vaginal microbiota, and genital inflammation are robust drivers of cervicovaginal metabolomes.
Fig. 6.
Metabolome profiles were driven by the features of the cervicovaginal microenvironment – HPV infection, cancer, non-Lactobacillus dominated microbiota, and genital inflammation. A) Hierarchical clustering (HCA) dendrogram constructed using top-down approach presented with patient characteristics including HPV status, genital inflammation score, vaginal pH, and microbial community based on Lactobacillus abundance. HCA analysis revealed three main clusters. B) Distribution of patient groups in the clusters. Cluster 1 only contained ICC patients whereas Cluster 2 and 3 contained patients from all the groups. The patient group composition of Cluster 1 was significantly different than Cluster 2 based on chi-square test. C) Genital inflammation scores among the clusters. Genital inflammation score of Cluster 1 was significantly higher than the scores of Clusters 2 and 3. Boxplot plots demonstrate median, 25th and 75th quartiles. D) Percentage of samples belonging to patients with Lactobacillus dominant and non-Lactobacillus dominant microbiota. Cluster 1 and 3 were dominated by non-Lactobacillus dominated communities whereas Cluster 2 had greater number of samples that were Lactobacillus dominated. Based on chi-square test, Cluster 2 was significantly different than Cluster 1 and Cluster 3. E) Vaginal pH measurements among the clusters. Cluster 1 and 3 had samples from patients with higher vaginal pH compared to Cluster 2 patients. * p < .5, **p < .01, ***p < .001, ****p < .0001.
4. Discussion
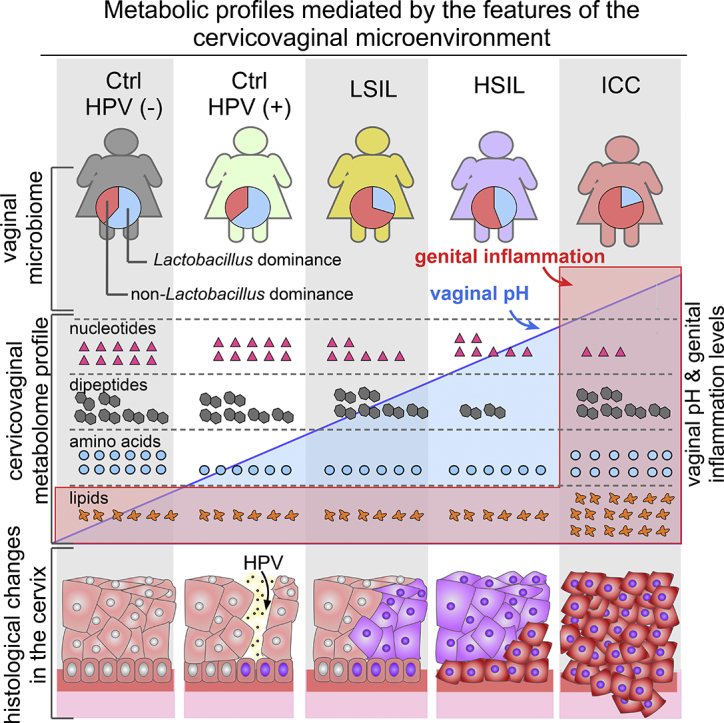
The metabolomic profiling offers a unique understanding of the complex interactions between the host and microbes in carcinogenesis. In addition, dysregulation of metabolism is an emerging hallmark of cancer [33]. A number of studies conducted on humans have shown a distinct metabolic fingerprint of cervical cancer in plasma [15], tumour tissues [16], faeces [17], and urine [34]. However, the impacts of HPV infection, cervical dysplasia and cancer on the metabolic profiles of the cervicovaginal microenvironment, a unique ecosystem where host epithelial and immune cells directly interact with viruses, bacteria and other environmental factors, have not been previously addressed. Unquestionably, metabolic fingerprinting of cervicovaginal lavages more accurately reflects the interactions between host and the microorganisms within the local microenvironment. Additionally, metabolic fingerprinting can provide insights into the interplay between persistent HPV infection, cervical neoplasm, and other features of the cervicovaginal microenvironment (i.e., genital inflammation, vaginal microbiome and vaginal pH) in the development of cervical dysplasia and progression to ICC. Here, we evaluated these metabolic interactions and characteristics of the cervicovaginal microenvironment (vaginal pH, vaginal microbiome, genital inflammation) in Ctrl HPV (−), Ctrl HPV (+), LSIL, HSIL, and newly diagnosed ICC patient groups. We show that unique metabolic profiles, impacted by genital inflammation, vaginal pH, and the vaginal microbiome, predict patient groups and reflect profound differences in the host and microbe co-metabolism during HPV infection, cervical dysplasia, and ICC.
Cervical cancer, dysplasia, and HPV infection exhibited divergent effects on the cervicovaginal metabolomes. A depletion of amino acid metabolites in HPV-positive and cervical dysplasia groups (LSIL and HSIL), and depletion of dipeptides in the HSIL group was replaced in ICC by enrichment of xenobiotics and lipids from plasmalogens, sphingomyelins, phosphatidylcholines, and long chain polyunsaturated fatty acids. Observation of xenobiotics, mainly anti-inflammatory and analgesic drugs such as ibuprofen, acetaminophen, nafloxin, and their metabolites in the CVLs of ICC patients indicated a possible translocation of these drugs from blood to the cervix and vagina. A higher abundance of lipid metabolites in ICC patients compared to healthy individuals can be explained by an increase in cell proliferation and cell membrane synthesis through activation of oncogenic pathways in the tumour microenvironment [35]. Hence, enrichment of lipids in the cervicovaginal microenvironment relevant to nutrient acquisition validated one of the emerging hallmarks of cancer metabolism [33].
Using a well-characterized and robust biomarker discovery analysis (ROC) [36], we identified three lipid molecules, 3-hydroxybutyrate, eicosenoate, and oleate/vaccenate with excellent discrimination capacity (AUC > 0.9) of ICC patients from healthy participants. A ketone body, 3-hydroxybutyrate, was elevated in ovarian cancer and was associated with invasion and migration of cancer cells [37,38]. Additionally, 3-hydroxybutyrate augments tumour growth in-vitro epithelial cell models [39]. A systematic review of 23 metabolome studies in ovarian cancer highlighted abnormally high levels of lipids including, oleate/vaccenate and eicosenoate in the serum [40], an observation consistent with the metabolic profiles we observed in the CVLs of ICC patients. In addition, elevated levels of oleic acid have been previously observed in cervical cancer biopsies [41] and oleic acid has been shown to promote cervical cancer cell growth in-vitro [42], thus validating our findings. Interestingly, these markers are not only specific to cancer, but also linked to inflammation and obesity, two comorbidities related to cancer [43]. Genital inflammation and high BMI are also characteristics of our patients although BMI did not significantly differ among the patient groups.
ROC analysis also revealed that metabolites from multiple metabolic pathways were significantly depleted in HPV positive and cervical dysplasia groups. Interestingly, we observed unique discriminators that distinguished the Ctrl HPV (−) group from the Ctrl HPV (+), LSIL or HSIL groups. Depletion of metabolites from taurine, glutamine and lysine metabolism, key amino acids that are involved in cellular growth [44], in HPV-positive women and cervical dysplasia patients indicates disruption of healthy cell metabolism as a functional consequence of potential HPV-mediated dysbiosis. Glutamine, a critical amino acid for tumour cell growth [33], and phosphoethanolamine, a well-established anti-tumour agent [45], were found to be strong discriminators of healthy patients vs. the HSIL patients. The impact of HPV on metabolite depletions serves as an example of changes in cervicovaginal microenvironment due to virus and host interactions.
Vaginal dysbiosis has emerged as a key risk factor in reproductive health [46], inflammation [47] and HPV acquisition, persistence, and cervical carcinogenesis [48]. However, the biological mechanisms and host-microbe interactions that drive persistence and carcinogenesis have not been elucidated. Thus, we integrated microbiome and metabolome data sets using the state-of-the-art bioinformatic tools (MIMOSA and AMON) to reveal a functional impact of microbiomes on metabolic fingerprints. Using a predicted relative metabolic turnover model on our measured community metabolome, we provide unique insights into the microbial contribution to observed metabolomes beyond correlations between microbial genera and metabolites. Our analysis based on microbial genomes indicated that differences in non-Lactobacillus dominated (NLD) communities compared to Lactobacillus-dominated (LD) communities specifically affect the flux of dipeptides, amino acid and nucleotide metabolites, which was consistent with a previous report that employed the same modelling technique on metabolomes in bacterial vaginosis (BV) [29]. Our NLD communities were enriched in multiple microbial genera including Gardnerella, Prevotella, Streptococcus and Atopobium, that were typically found in BV patients [13,[49], [50], [51]]. A decrease in the abundance of vaginal Lactobacillus species in BV patients was previously associated with an increase in the biogenic amine levels, amino acid degradation products and depletion of dipeptides [12,13,51,52]. Hence our findings agree with the observation that microbial diversity in the vaginal milieu alters amino acid metabolism.
Our study design excluded symptomatic BV patients, however we observed several metabolic signatures of BV in our dataset. For example, deoxycarnitine [12] and alpha-hydroxyisovalerate [52], previously identified as signatures of BV, were negatively correlated with Lactobacillus abundance. Interestingly, other acylcarnitines that are known pro-inflammatory cytokine inducers [53] were significantly associated with genital inflammation and Lactobacillus abundance. We also observed BV-associated Gardnerella was the main contributor to hippurate metabolism. Hippurate degradation is not only used as a test for Gardnerella identification [54], but also used in pathogen detection. Another important metabolic signature of BV is presence of biogenic amines. Three out of ten polyamines were significantly enriched in the NLD metabolomes and five BV-associated biogenic amines (cadaverine, putrescine, tyramine, tryptamine, and agmatine) [13,55] negatively correlated with Lactobacillus abundance, however their levels were not significantly different between LD and NLD communities. Notably, none of these biogenic amines were positively or negatively associated with genital inflammation. Arginine and its polyamine product N(1)-acetylspermine are indicative of rapidly proliferating cells [56] and positively correlated with Lactobacillus abundance and genital inflammation. Previously, arginine metabolism was previously highlighted as an emerging hallmark of cancer [33]. Interestingly, N(1)-acetylspermine levels are increased in severely inflamed sites in colorectal cancer patients [57], indicating a potential role of this polyamine in severe inflammation in accordance with our genital inflammation findings. However, other BV-associated biogenic amines did not correlate with genital inflammation. Metabolites that are precursors of biogenic amines including choline and betaine have anti-inflammatory properties [58,59]. Betaine enhances survival and provides protection to Lactobacillus species facing osmotic stress [60]. Future comparative studies that incorporate microbiomes to metabolomes in symptomatic and asymptomatic BV patients will further enhance the understanding of the role of microbes and microbe-host interactions in the context of vaginal dysbiosis and BV.
Our analysis using a well-characterized prediction model (MIMOSA) also revealed that Lactobacillus species are the key contributors to the metabolism of glycochenodeoxycholate (GCDC) in our dataset. GCDC, a key metabolic product of host-microbe co-metabolism [61], can inhibit several pathogens that are also observed in vaginal dysbiosis and BV [62]. Similar to other bile acids, GCDC induces inflammatory and toxic responses and exhibits carcinogenic effects on host epithelial cells [63]. Our observation of negative correlation with Lactobacillus abundance and positive correlation with genital inflammation further strengthened the link between GCDC, inflammation, and Lactobacillus abundance. As such, future studies are needed to investigate the role of GCDC in maintaining Lactobacillus-mediated homeostasis, as well as pathogen exclusion in the lower reproductive tract.
It is important to note that the microbiome-based data integration was limited only to the metabolites and genomes available in the KEGG database. Overall, differences in the vaginal microbial communities in cervical dysplasia and ICC were reflected upon metabolomes. Microbe-microbe and host-microbe interactions through metabolic networks can shape the local tumour environment [64], hence dysbiotic vaginal microbiome-mediated metabolome has the potential to become a hallmark of cervical cancer. Additionally, the metabolites produced by the cancer microenvironment itself, including lipids, can enhance the crosstalk between microbiota and the host, and therefore the pro-carcinogenic activities of the microbiota.
Genital inflammation, another key feature of the cervicovaginal microenvironment, co-shaped the metabolic profiles along with the microbial composition in our dataset. Notably, the lipids that discriminated the ICC group from the healthy group showed a strong positive correlation to genital inflammation. However, our additional analysis (Supplementary Fig. 4) excluding ICC patients showed that lipids remained associated with genital inflammation independent from cancer. An increase in lipid metabolites from plasmalogens and long chain polyunsaturated fatty acids in ICC, not only indicate abnormality in cellular metabolism, but also highlight the significance of lipids in cancer development, due to their roles as precursors of pro-inflammatory cytokines, inducers of abnormal gene expression and ability to dysregulate cytokine production [65]. Interestingly, phosphatidylcholines, another group of lipids that positively correlated with genital inflammation, are typically known for their anti-inflammatory properties [66], but also contribute to cellular proliferation and programmed cell death [67].
Adenosine and cytosine, nucleotides, that were well-predicted with MIMOSA analysis, negatively correlated with genital inflammation and positively correlated with Lactobacillus abundance. These well-recognized anti-inflammatory molecules [68] were also well-predicted to be modulated by the vaginal microbiome in our dataset. For instance, adenosine, an anti-inflammatory agent, has therapeutic potential because it inhibits phagocytosis, adhesion, and production of reactive oxygen species [69]. Additionally, animal and in vitro cell models demonstrated anti-inflammatory effects of adenosine through its impact on macrophages [70] and mast cells [71]. Taken together these studies indicate that the potent anti-inflammatory function of microbially modulated adenosine could be a molecule associated with genital tract health.
A key metabolite that strongly correlated with Lactobacillus abundance was lactate. When the correlation analysis was performed without the ICC patients included, the correlation between lactate and Lactobacillus was stronger and there was a trend towards negative correlation with genital inflammation (Supplementary Fig. 4). Lactic acid is the main end-product of Lactobacillus-mediated fermentation [72], and is a critical metabolite to maintain low vaginal pH (<4.5) and homeostasis in the vaginal microenvironment [73]. Higher levels of lactate and lower vaginal pH observed in Lactobacillus dominated communities compared to non-Lactobacillus-dominated communities are in agreement with other studies relating this molecule to vaginal health and homeostasis [12,73,74]. In a murine model, lactate was shown to inhibit the pro-inflammatory response of macrophages [75], hence our data highlight a potential anti-inflammatory role of lactate in the lower reproductive tract. Additionally, lactic acid can induce secretion of anti-inflammatory IL-10, reduce the production of proinflammatory IL-12 in dendritic cells, and diminish the cytotoxicity of natural killer cells [76]. On the other hand, lactate is also one of the main cellular respiration products of cancer cells, previously established as a biomarker of cervical cancer tumours [77] and is associated with angiogenesis and immunotolerance [76]. Therefore, the dual role of lactate in health and disease, its origin, and relationship to genital inflammation in the context of gynaecologic cancer warrants further investigation.
Metabolic profiling of the cervicovaginal microenvironment offers three important benefits to cervical cancer research. First, it provides a snapshot of metabolic communications between the host, microbiome, and HPV. Correspondingly, metabolic profiling is the first step to understand host-microbiota interplay in modulating the hallmarks of cancer in the context of HPV. Second, CVL collection is non-invasive method that may provide more biological insights into the mechanisms of viral persistence and disease progression. Finally, the limitations and challenges in murine and non-human primate models are avoided since these models do not faithfully recapitulate the composition of vaginal microbiota in humans [78]. A limitation of our study is the relatively small sample size and future studies with larger sample sizes are needed to validate and extend our findings. However, the results presented herein generate additional questions on virus-host epithelia and immune system interactions that can be addressed by metabolic profiling in future clinical studies. In addition, studies employing physiologically relevant 3-dimensional human FRT models [[78], [79], [80], [81]] could reveal biological mechanisms into how these microbes are dysregulating host metabolism and contributing to the hallmarks of cancer.
Multiple features of the cervicovaginal microenvironment, genital inflammation, vaginal pH, and vaginal microbiota composition explained differences in the metabolic profiles of the participant groups across cervical carcinogenesis. Our observations on genital inflammation- and microbiota-associated metabolic signatures connect the microbiome to inflammation, dysplasia, and cancer. Collectively, our analysis revealed that cancer had the greatest impact on metabolic profiles and genital inflammation, and vaginal microbiota had a secondary impact that explained metabolic profiles of healthy HPV-negative, HPV-positive individuals, and cervical dysplasia patients. Our analysis clearly demonstrated that metabolic fingerprinting of the cervicovaginal microenvironment is an excellent approach to discriminate between individuals with HPV infection, cervical dysplasia and ICC, which may facilitate future discoveries of new therapeutic and prevention avenues for patients. This integrative analysis of the cervicovaginal metabolome, microbiome and immunoproteome revealed multiple emerging hallmarks of cancer in cervical carcinogenesis, including abnormalities in nitrogen and energy metabolism, increased cellular proliferation due to changes in amino acid and carbohydrate fluxes, and dysbiotic microbiota and host-microbe co-metabolism [33,82]. In conclusion, the complex virus-host-microbe interplay within the cervicovaginal microenvironment lead to unique metabolic fingerprints that may be exploited for future development of diagnostics, preventatives or therapeutics to positively impact women's health outcomes.
The following are the supplementary data related to this article.
Principal component analysis (PCA colour-coded based on participant demographics. PCA did not demonstrate significant separation of metabolomes based on age, BMI and ethnicity. Mann-Whitney U test P values represented as *p < .05, **p < .01, ***p < .001.
Principal component analysis (PCA) colour-coded based on vaginal pH. PCA demonstrated a gradual separation of patients based on their vaginal pH measurements. Mann-Whitney U test P values represented as *p < .05, **p < .01, ***p < .001.
Relative levels of biogenic amines in the participant groups. Overall biogenic amine levels were similar among the participant groups. HSIL group has significantly lower levels of spermine in comparison to Ctrl HPV (−) and higher levels of tyramine compared to ICC. Tryptamine levels were significantly lower in the ICC group compared to Ctrl HPV (−). The Welch's t-test p values represented as *p < .05, **p < .01, ***p < .001.
Metabolites that connected genital inflammation to Lactobacillus abundance when ICC samples were excluded. Scatter plot of metabolites that correlated with genital inflammation and Lactobacillus abundance independent from cervical cancer.
Patient demographics and characteristics.
Supplementary Table 2 Metabolites that are unique and shared among groups that were listed in Fig. 1C.
Supplementary Table 3 Median levels of metabolites that discriminated from Ctrl HPV (+), LSIL, and HSIL groups and metabolites that discriminated ICC from Ctrl HPV (−) group.
Supplementary Table 4 The q values calculated based on Welch's two-sided test for metabolites that discriminated Ctrl HPV (−) from Ctrl HPV (+), LSIL, and HSIL groups and metabolites that discriminated ICC from Ctrl HPV (−) group.
Supplementary Table 5 Metabolites that positively and negatively correlated with genital inflammation with Spearman's rho critical value of >0.23.
Funding sources
This work was supported by the Flinn Foundation (#1974) to D.M.C. and M.M.H·K, Banner Foundation in Obstetrics and Gynecology Research to M.M.H.K., and National Institutes of Health NCI Grant P30 CA023074. The funds were used for patient recruitment, data collection, and analysis.
Declaration of interests
Drs. Ilhan, Laniewski, Herbst-Kralovetz and Ms. Thomas report grants from Flinn Foundation, grants from National Institute of Health, grants from Banner Foundation in Obstetrics and Gynecology Research, during the conduct of the study; Dr. Chase reports grants from Flinn Foundation, during the conduct of the study; Dr. Roe reports grants from Flinn Foundation, grants from National Institute of Health, during the conduct of the study. The authors declare no other competing interests.
Author contributions
Conceptualization, M.M.H.K. and D.M.C.; Methodology, Z.E.I.; Formal Analysis, P.L., Z.E.I., N.T., Investigation, P.L., Z.E.I., Resources, D.M.C.; Data curation, Z.E.I., P.L., N.T.; Writing Original Draft, Z.E.I., P.L., and M.M.H.K.; Writing – Review & Editing, P.L., M.M.H.K., D.M.C., D.R., N.T.; Visualization, Z.E.I., P.L., M.M.H.K.; Supervision, M.M.H.K.; Project Administration, M.M.H.K.; Funding acquisition, D.M.C. and M.M.H.K.
Acknowledgements
The authors would like to thank Kelli Williamson, Ann De Jong, Eileen Molzen, Liane Fales, and Maureen Sutton for the kind assistance in patient recruitment and sample collection, the patients that enrolled in this study, Drs. Dominique Barnes, Alison Goulder, and David Greenspan for their assistance with clinical specimen and data collection, Carol Haussler for her critical review, and Cecilia Noecker for her assistance with MIMOSA analysis.
References
- 1.Schiffman M., Doorbar J., Wentzensen N., De Sanjosé S., Fakhry C., Monk B.J. Carcinogenic human papillomavirus infection. Nat Rev Dis Prim. 2016;2 doi: 10.1038/nrdp.2016.86. [DOI] [PubMed] [Google Scholar]
- 2.Koshiol J., Lindsay L., Pimenta J.M., Poole C., Jenkins D., Smith J.S. Persistent human papillomavirus infection and cervical neoplasia: a systematic review and meta-analysis. Am J Epidemiol. 2008;168(2):123–137. doi: 10.1093/aje/kwn036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bray F., Jacques F., Soerjomataram I., Siegel R., Torre L., Jemal A. Global Cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 4.Carter J.R., Ding Z., Rose B.R. HPV infection and cervical disease: a review. Aust New Zeal J Obstet Gynaecol. 2011;51(2):103–108. doi: 10.1111/j.1479-828X.2010.01269.x. [DOI] [PubMed] [Google Scholar]
- 5.Moerman-Herzog A., Nakagawa M. Early defensive mechanisms against human papillomavirus infection. Clin Vaccine Immunol. 2015;22(8):850–857. doi: 10.1128/CVI.00223-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Łaniewski P., Barnes D., Goulder A., Cui H., Roe D.J. Linking cervicovaginal immune signatures, HPV and microbiota composition in cervical carcinogenesis in non-Hispanic and Hispanic women. Sci Rep. 2018;8(1):7593. doi: 10.1038/s41598-018-25879-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brotman R.M., Shardell M.D., Gajer P., Tracy J.K., Zenilman J.M., Ravel J. Interplay between the temporal dynamics of the vaginal microbiota and human papillomavirus detection. J Infect Dis. 2014;210(11):1723–1733. doi: 10.1093/infdis/jiu330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Audirac-Chalifour A., Torres-Poveda K., Bahena-Román M., Téllez-Sosa J., Martínez-Barnetche J., Cortina-Ceballos B. Cervical microbiome and cytokine profile at various stages of cervical cancer: a pilot study. PLoS One. 2016;11(4) doi: 10.1371/journal.pone.0153274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shannon B., Gajer P., Yi T.J., Ma B., Humphrys M.S., Thomas-Pavanel J. Distinct effects of the cervicovaginal microbiota and herpes simplex type 2 infection on female genital tract immunology. J Infect Dis. 2017;215(9):1366–1375. doi: 10.1093/infdis/jix088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitra A., MacIntyre D.A., Lee Y.S., Smith A., Marchesi J.R., Lehne B. Cervical intraepithelial neoplasia disease progression is associated with increased vaginal microbiome diversity. Sci Rep. 2015;5:1–11. doi: 10.1038/srep16865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Passmore J.-A.S., Williamson A.-L. Host immune responses associated with clearance or persistence of human papillomavirus infections. Curr Obstet Gynecol Rep. 2016;5(3):177–188. http://link.springer.com/10.1007/s13669-016-0163-1 Current Obstetrics and Gynecology Reports. Available from: [Google Scholar]
- 12.Srinivasan S., Morgan M.T., Fiedler T.L., Djukovic D., Hoffman N.G., Raftery D. Metabolic signatures of bacterial vaginosis. MBio. 2015;6(2):1–16. doi: 10.1128/mBio.00204-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson T.M., Borgogna J.L.C., Brotman R.M., Ravel J., Walk S.T., Yeoman C.J. Vaginal biogenic amines: biomarkers of bacterial vaginosis or precursors to vaginal dysbiosis? Front Physiol. 2015;6:1–15. doi: 10.3389/fphys.2015.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wishart D.S., Mandal R., Stanislaus A., Ramirez-Gaona M. Cancer metabolomics and the human metabolome database. Metabolites. 2016;6(1) doi: 10.3390/metabo6010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang K., Xia B., Wang W., Cheng J., Yin M., Xie H. A comprehensive analysis of metabolomics and transcriptomics in cervical cancer. Sci Rep. 2017;7:1–11. doi: 10.1038/srep43353. Nature Publishing Group. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sitter B., Bathen T., Hagen B., Arentz C., Skjeldestad F.E., Gribbestad I.S. Cervical cancer tissue characterized by high-resolution magic angle spinning MR spectroscopy. Magn Reson Mater Physics, Biol Med. 2004;16(4):174–181. doi: 10.1007/s10334-003-0025-5. [DOI] [PubMed] [Google Scholar]
- 17.Chai Y., Wang J., Wang T., Yang Y., Su J., Shi F. Application of1H NMR spectroscopy-based metabonomics to feces of cervical cancer patients with radiation-induced acute intestinal symptoms. Radiother Oncol. 2015;117(2):294–301. doi: 10.1016/j.radonc.2015.07.037. Elsevier Ireland Ltd. Available from: [DOI] [PubMed] [Google Scholar]
- 18.Guerrero-Flores H., Apresa-García T., Garay-Villar Ó., Sánchez-Pérez A., Flores-Villegas D., Bandera-Calderón A. A non-invasive tool for detecting cervical cancer odor by trained scent dogs. BMC Cancer. 2017;17(1):1–8. doi: 10.1186/s12885-016-2996-4. BMC Cancer. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karim R., Tummers B., Meyers C., Biryukov J.L., Alam S., Backendorf C. Human papillomavirus (HPV) Upregulates the cellular Deubiquitinase UCHL1 to suppress the Keratinocyte's innate immune response. PLoS Pathog. 2013;9(5) doi: 10.1371/journal.ppat.1003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson T.M., Borgogna J.C., Michalek R.D., Roberts D.W., Rath J.M., Glover E.D. Cigarette smoking is associated with an altered vaginal tract metabolomic profile. Sci Rep. 2018;8(1):1–13. doi: 10.1038/s41598-017-14943-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vender Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science (80- ) 2009;324(May):1029–1033. doi: 10.1126/science.1160809. Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11) doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wickman H. ggplot2: Elegant graphics for data analysis. Springer; 2016. [Google Scholar]
- 24.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 26.DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langille M.G.I., Zaneveld J., Caporaso J.G., McDonald D., Knights D., Reyes J.A. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31(9):814. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaffer M., Kevin Q., Doenges K., Zhang N., Bokatzian S., Reisdorph N. AMON: Annotation of metabolite origins via networks. bioRvix. 2018 doi: 10.1186/s12859-019-3176-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noecker C., Eng A., Srinivasan S., Theriot C.M., Young V.B., Jansson J.K. Metabolic model-based integration of microbiome taxonomic and metabolomic profiles elucidates mechanistic links between ecological and metabolic variation. mSystems. 2016;1(1):e00013–e00015. doi: 10.1128/mSystems.00013-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noecker C., Chiu H.-C., McNally C.P., Borenstein E. 2018. Defining and evaluating microbial contributions to metabolite variation in microbiome-metabolome association studies; p. 402040.https://www.biorxiv.org/content/early/2018/08/28/402040 bioRxiv. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arnold K.B., Burgener A., Birse K., Romas L., Dunphy L.J., Shahabi K. Increased levels of inflammatory cytokines in the female reproductive tract are associated with altered expression of proteases, mucosal barrier proteins, and an influx of HIV-susceptible target cells. Mucosal Immunol. 2016;9(1):194–205. doi: 10.1038/mi.2015.51. Nature Publishing Group. Available from: [DOI] [PubMed] [Google Scholar]
- 32.Shannon B., Yi T.J., Perusini S., Gajer P., Ma B., Humphrys M.S. Association of HPV infection and clearance with cervicovaginal immunology and the vaginal microbiota. Mucosal Immunol. 2017;10(5):1310–1319. doi: 10.1038/mi.2016.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pavlova N.N., Thompson C.B. The emerging hallmarks of Cancer metabolism. Cell Metab. 2016;23(1):27–47. doi: 10.1016/j.cmet.2015.12.006. Elsevier Inc. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye N., Liu C., Shi P. Metabolomics analysis of cervical cancer, cervical intraepithelial neoplasia and chronic cervicitis by 1H NMR spectroscopy. Eur J Gynaecol Oncol. 2015;36(2):174–180. [PubMed] [Google Scholar]
- 35.Santos C.R., Schulze A. Lipid metabolism in cancer. FEBS J. 2012;279(15):2610–2623. doi: 10.1111/j.1742-4658.2012.08644.x. [DOI] [PubMed] [Google Scholar]
- 36.Daskivich T., Luu M., Noah B., Fuller G., Anger J., Spiegel B. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 37.Vettukattil R., Hetland T.E., Flørenes V.A., Kærn J., Davidson B., Bathen T.F. Proton magnetic resonance metabolomic characterization of ovarian serous carcinoma effusions: chemotherapy-related effects and comparison with malignant mesothelioma and breast carcinoma. Hum Pathol. 2013;44(9):1859–1866. doi: 10.1016/j.humpath.2013.02.009. Elsevier Inc. Available from: [DOI] [PubMed] [Google Scholar]
- 38.Schmitt W.D., Hilvo M., Gopalacharyulu P., Budczies J., Denkert C., Darb-Esfahani S. Accumulated metabolites of hydroxybutyric acid serve as diagnostic and prognostic biomarkers of ovarian high-grade serous carcinomas. Cancer Res. 2016;76(4):796–804. doi: 10.1158/0008-5472.CAN-15-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonuccelli G., Tsirigos A., Whitaker-Menezes D., Pavlides S., Pestell R.G., Chiavarina B. Ketones and lactate “fuel” tumor growth and metastasis: evidence that epithelial cancer cells use oxidative mitochondrial metabolism. Cell Cycle. 2010;9(17):3506–3514. doi: 10.4161/cc.9.17.12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turkoglu O., Zeb A., Graham S., Szyperski T., Szender J.B., Odunsi K. Metabolomics of biomarker discovery in ovarian cancer: a systematic review of the current literature. Metabolomics. 2016;12(4):1–16. doi: 10.1007/s11306-016-0990-0. Springer US. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zietkowski D., Davidson R.L., Eykyn T.R., De Silva S.S., DeSouza N.M., Payne G.S. Detection of cancer in cervical tissue biopsies using mobile lipid resonances measured with diffusion-weighted1H magnetic resonance spectroscopy. NMR Biomed. 2010;23(4):382–390. doi: 10.1002/nbm.1472. [DOI] [PubMed] [Google Scholar]
- 42.Zhao L., Ruan X.Z., Zeng H., Wei L., Varghese Z., Chen Y. Dietary oleic acid-induced CD36 promotes cervical cancer cell growth and metastasis via up-regulation Src/ERK pathway. Cancer Lett. 2018;438:76–85. doi: 10.1016/j.canlet.2018.09.006. Elsevier. Available from: [DOI] [PubMed] [Google Scholar]
- 43.Meikle P.J., Summers S.A. Sphingolipids and phospholipids in insulin resistance and related metabolic disorders. Nat Rev Endocrinol. 2017;13(2):79–91. doi: 10.1038/nrendo.2016.169. Nature Publishing Group. Available from: [DOI] [PubMed] [Google Scholar]
- 44.Wang W.W., Qiao S.Y., Li D.F. Amino acids and gut function. Amino Acids. 2009;37(1):105–110. doi: 10.1007/s00726-008-0152-4. [DOI] [PubMed] [Google Scholar]
- 45.Ferreira A.K., Meneguelo R., Pereira A., Filho O.M.R., Chierice G.O., Maria D.A. Anticancer effects of synthetic phosphoethanolamine on Ehrlich ascites tumor: an experimental study. Anticancer Res. 2012;32(1):95–104. [PubMed] [Google Scholar]
- 46.Anahtar M.N., Gootenberg D.B., Mitchell C.M., Kwon D.S. Cervicovaginal microbiota and reproductive health: the virtue of simplicity. Cell Host Microbe. 2018;23(2):159–168. doi: 10.1016/j.chom.2018.01.013. Elsevier Inc. Available from. [DOI] [PubMed] [Google Scholar]
- 47.Ness R.B., Kip K.E., Hillier S.L., Soper D.E., Stamm C.A., Sweet R.L. A cluster analysis of bacterial vaginosis-associated microflora and pelvic inflammatory disease. Am J Epidemiol. 2005;162(6):585–590. doi: 10.1093/aje/kwi243. [DOI] [PubMed] [Google Scholar]
- 48.Brusselaers N., Shrestha S., Van De Wijgert J., Verstraelen H. Vaginal dysbiosis, and the risk of human papillomavirus and cervical cancer: systematic review and meta-analysis. Am J Obstet Gynecol. Dec 12 2018 doi: 10.1016/j.ajog.2018.12.011. https://linkinghub.elsevier.com/retrieve/pii/S000293781832221X Elsevier Inc. Available from. (pii: S0002-9378(18)32221-X, [Epub ahead of print] Review. PMID: 30550767) [DOI] [PubMed] [Google Scholar]
- 49.Onderdonk A.B., Delaney M.L., Fichorova N. The human microbiome during bacterial vaginosis. Clin Microbiol Rev. 2016;29(2):223–238. doi: 10.1128/CMR.00075-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fredricks D.N., Fiedler T.L., Thomas K.K., Oakley B.B., Marrazzo J.M. Targeted PCR for detection of vaginal bacteria associated with bacterial vaginosis. J Clin Microbiol. 2007;45(10):3270–3276. doi: 10.1128/JCM.01272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vitali B., Cruciani F., Picone G., Parolin C., Donders G., Laghi L. Vaginal microbiome and metabolome highlight specific signatures of bacterial vaginosis. Eur J Clin Microbiol Infect Dis. 2015;34(12):2367–2376. doi: 10.1007/s10096-015-2490-y. [DOI] [PubMed] [Google Scholar]
- 52.McMillan A., Rulisa S., Sumarah M., Macklaim J.M., Renaud J., Bisanz J.E. A multi-platform metabolomics approach identifies highly specific biomarkers of bacterial diversity in the vagina of pregnant and non-pregnant women. Sci Rep. 2015;5:1–14. doi: 10.1038/srep14174. Nature Publishing Group. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rutkowsky J.M., Schneider D., McCoin C.S., Huang S., Adams S.H., Ono-Moore K.D. Acylcarnitines activate proinflammatory signaling pathways. Am J Physiol Metab. 2014;306(12):E1378–E1387. doi: 10.1152/ajpendo.00656.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Briselden A.M., Hillier S.L. Longitudinal study of the biotypes of Gardnerella vaginalis. J Clin Microbiol. 1990;28(12):2761–2764. doi: 10.1128/jcm.28.12.2761-2764.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sujatha K., Mahalakshmi A., Solaiman D.K.Y., Shenbagarathai R. Sequence analysis, structure prediction, and functional validation of phaC1/phaC2 genes of Pseudomonas sp LDC-25 and its importance in Polyhydroxyalkanoate accumulation. J Biomol Struct Dyn. 2009;26(6):771–779. doi: 10.1080/07391102.2009.10507289. [DOI] [PubMed] [Google Scholar]
- 56.Nakajima T., Katsumata K., Kuwabara H., Soya R., Enomoto M., Ishizaki T. Urinary polyamine biomarker panels with machine-learning differentiated colorectal cancers, benign disease, and healthy controls. Int J Mol Sci. 2018;19(3):1–14. doi: 10.3390/ijms19030756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weiss T.S., Herfarth H., Obermeier F., Ouart J., Vogl D., Schölmerich J. Intracellular polyamine levels of intestinal epithelial cells in inflammatory bowel disease. Inflamm Bowel Dis. 2004;10(5):529–535. doi: 10.1097/00054725-200409000-00006. [DOI] [PubMed] [Google Scholar]
- 58.Zhao G., He F., Wu C., Li P., Li N., Deng J. Betaine in inflammation: mechanistic aspects and applications. Front Immunol. 2018;9:1–13. doi: 10.3389/fimmu.2018.01070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rowley T.J., McKinstry A., Greenidge E., Smith W., Flood P. Antinociceptive and anti-inflammatory effects of choline in a mouse model of postoperative pain. Br J Anaesth. 2010;105(2):201–207. doi: 10.1093/bja/aeq113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kets E., Bont J. Protective effect of betaine on survival of Lactobacillus plantarum subjected to drying. FEMS Microbiol Lett. 1994;116(3):251–256. [Google Scholar]
- 61.Ridlon J.M., Harris S.C., Bhowmik S., Kang D.J., Hylemon P.B. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes. 2016;7(1):22–39. doi: 10.1080/19490976.2015.1127483. Taylor & Francis. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fiorucci S., Distrutti E. Bile acid-activated receptors, intestinal microbiota, and the treatment of metabolic disorders. Trends Mol Med. 2015;21(11):702–714. doi: 10.1016/j.molmed.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 63.Tatsugami M., Ito M., Tanaka S., Yoshihara M., Matsui H., Haruma K. Bile acid promotes intestinal metaplasia and gastric carcinogenesis. Cancer Epidemiol Biomarkers Prev. 2012;21(11):2101–2107. doi: 10.1158/1055-9965.EPI-12-0730. [DOI] [PubMed] [Google Scholar]
- 64.Fulbright L.E., Ellermann M., Arthur J.C. The microbiome and the hallmarks of cancer. PLoS Pathog. 2017;13(9):e1006480. doi: 10.1371/journal.ppat.1006480. Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wallner S., Orso E., Grandl M., Konovalova T., Liebisch G., Schmitz G. Phosphatidylcholine and phosphatidylethanolamine plasmalogens in lipid loaded human macrophages. PLoS One. 2018;13(10):1–21. doi: 10.1371/journal.pone.0205706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Treede I., Braun A., Sparla R., Kühnel M., Giese T., Jerrold R. Anti-inflammatory effects of phosphatidylcholine. J. Biol. Chem. 2007;282(37):27155–27164. doi: 10.1074/jbc.M704408200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ridgway N.D. The role of phosphatidylcholine and choline metabolites to cell proliferation and survival. Crit Rev Biochem Mol Biol. 2013;48(1):20–38. doi: 10.3109/10409238.2012.735643. [DOI] [PubMed] [Google Scholar]
- 68.Hall I.H., Burnham B.S., Elkins A., Sood A., Powell W., Tomasz J. Boronated Pyrimidines and purines as cytotoxic, Hypolipidemic and anti-inflammatory agents. Met Based Drugs. 1996;3(3):155–160. doi: 10.1155/MBD.1996.155. http://www.hindawi.com/archive/1996/694271/abs/ Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cronstein BN. Adenosine, an endogenous anti-inflammatory agentJ Appl Physiol. 1994;76(1):5–13. doi: 10.1152/jappl.1994.76.1.5. http://www.ncbi.nlm.nih.gov/pubmed/8175547 Available from. [DOI] [PubMed] [Google Scholar]
- 70.Fukuda T., Majumder K., Zhang H., Matsui T., Mine Y. Adenine has an anti-inflammatory effect through the activation of adenine receptor signaling in mouse macrophage. J Funct Foods. 2017;28:235–239. doi: 10.1016/j.jff.2016.11.013. Elsevier Ltd. Available from. [DOI] [Google Scholar]
- 71.Feng C., Mery a.G., Beller E.M., Favot C., J a Boyce. Adenine nucleotides inhibit cytokine generation by human mast cells through a Gs-coupled receptor. J Immunol. 2004;173(12):7539–7547. doi: 10.4049/jimmunol.173.12.7539. http://www.jimmunol.org/cgi/doi/10.4049/jimmunol.173.12.7539 Available from. [DOI] [PubMed] [Google Scholar]
- 72.France M.T., Mendes-soares H., Forney L.J. Genomic comparisons of LActobacillus crispatus and Lactobacillus iners reveal potential ecological drivers of community composition in the vagina. 2016;82(24):7063–7073. doi: 10.4049/jimmunol.173.12.7539. Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.O'Hanlon D.E., Moench T.R., Cone R.A. Vaginal pH and Microbicidal lactic acid when lactobacilli dominate the microbiota. PLoS One. 2013;8(11) doi: 10.1371/journal.pone.0080074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Boskey E.R., Cone R.A., Whaley K.J., Moench T.R. Origins of vaginal acidity: high D/L lctate ratio is consistent with bacteria being the primary source. Hum Reprod. 2001;16(9):1809–1813. doi: 10.1093/humrep/16.9.1809. http://www.crime-expertise.org/17eme-congres-danthropologie-medico-legale-2017/ Available from. [DOI] [PubMed] [Google Scholar]
- 75.Errea A., Cayet D., Marchetti P., Tang C., Kluza J., Offermanns S. Lactate inhibits the pro-inflammatory response and metabolic reprogramming in Murine macrophages in a GPR81-independent manner. PLoS One. 2016;11(11):1–11. doi: 10.1371/journal.pone.0163694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sun S., Li H., Chen J., Qian Q. Lactic acid: no longer an inert and end-product of glycolysis. Physiology. 2017;32(6):453–463. doi: 10.1152/physiol.00016.2017. http://physiologyonline.physiology.org/lookup/doi/10.1152/physiol.00016.2017 Available from. [DOI] [PubMed] [Google Scholar]
- 77.Walenta S., Wetterling M., Lehrke M., Schwickert G., Sundfør K., Rofstad E.K. High lactate levels predict likelihood of metastases, tumor recurrence, and restricted patient survival in human cervical cancers. Cancer Res. 2000;60(4):916–921. [PubMed] [Google Scholar]
- 78.Herbst-Kralovetz M.M., Pyles R.B., Ratner A.J., Sycuro L.K., Mitchell C. New systems for studying intercellular interactions in bacterial vaginosis. J Infect Dis. 2016;214:S6–13. doi: 10.1093/infdis/jiw130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Radtke A.L., Herbst-Kralovetz M.M. Culturing and applications of rotating wall vessel bioreactor derived 3D epithelial cell models. Jove-J Vis Exp. 2012;(62) doi: 10.3791/3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Doerflinger S.Y., Throop A.L., Herbst-Kralovetz M.M. Bacteria in the vaginal microbiome alter the innate immune response and barrier properties of the human vaginal epithelia in a species-specific manner. J Infect Dis. 2014;209(12):1989–1999. doi: 10.1093/infdis/jiu004. [DOI] [PubMed] [Google Scholar]
- 81.Radtke A.L., Quayle A.J., Herbst-Kralovetz M.M. Microbial products alter the expression of membrane-associated mucin and antimicrobial peptides in a three-dimensional human endocervical epithelial cell model1. Biol Reprod. 2012;87(6):1–10. doi: 10.1095/biolreprod.112.103366. https://academic.oup.com/biolreprod/article-lookup/doi/10.1095/biolreprod.112.103366 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Goodman B., Gardner H. The microbiome and cancer. J Pathol. Nature Publishing Group. 2018;244(5):667–676. doi: 10.1038/nrc3610. Available from: [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Principal component analysis (PCA colour-coded based on participant demographics. PCA did not demonstrate significant separation of metabolomes based on age, BMI and ethnicity. Mann-Whitney U test P values represented as *p < .05, **p < .01, ***p < .001.
Principal component analysis (PCA) colour-coded based on vaginal pH. PCA demonstrated a gradual separation of patients based on their vaginal pH measurements. Mann-Whitney U test P values represented as *p < .05, **p < .01, ***p < .001.
Relative levels of biogenic amines in the participant groups. Overall biogenic amine levels were similar among the participant groups. HSIL group has significantly lower levels of spermine in comparison to Ctrl HPV (−) and higher levels of tyramine compared to ICC. Tryptamine levels were significantly lower in the ICC group compared to Ctrl HPV (−). The Welch's t-test p values represented as *p < .05, **p < .01, ***p < .001.
Metabolites that connected genital inflammation to Lactobacillus abundance when ICC samples were excluded. Scatter plot of metabolites that correlated with genital inflammation and Lactobacillus abundance independent from cervical cancer.
Patient demographics and characteristics.
Supplementary Table 2 Metabolites that are unique and shared among groups that were listed in Fig. 1C.
Supplementary Table 3 Median levels of metabolites that discriminated from Ctrl HPV (+), LSIL, and HSIL groups and metabolites that discriminated ICC from Ctrl HPV (−) group.
Supplementary Table 4 The q values calculated based on Welch's two-sided test for metabolites that discriminated Ctrl HPV (−) from Ctrl HPV (+), LSIL, and HSIL groups and metabolites that discriminated ICC from Ctrl HPV (−) group.
Supplementary Table 5 Metabolites that positively and negatively correlated with genital inflammation with Spearman's rho critical value of >0.23.
Data Availability Statement
16S rRNA gene sequences were deposited in the National Center for Biotechnology Information (NCBI) database on BioProject accession number: PRJNA518153 and under Sequence Read Archive (SRA) accession IDs SAMN10855179 to SAMN10855255. De-identified metabolomics data is available in Supplementary files.