Abstract
Background
The influence of amyloid protein-binding protein 2 (APPBP2) on lung cancer is unknown.
Methods
The function and mechanisms of APPBP2 were investigated in the NSCLC cell lines A549 and H1299. The ectopic expression of APPBP2, PPM1D and SPOP in NSCLS were examined in samples collected from ten pairs of human lung adenocarcinoma cancer tissues and adjacent normal lung tissues. shRNA vector was used for APPBP2 knockdown. Quantitative PCR and western blot assays quantified the mRNA and protein level of APPBP2, PPM1D, and SPOP. Cell proliferation was measured with BrdU, MTT, colony formation assays, and xenograft tumour growth experiments. Cell migration and invasion were analysed with transwell and wound healing assays. Co-Immunoprecipitation assay detected protein–protein interactions.
Findings
APPBP2 was upregulated in NSCLC tissues. Silencing APPBP2 in A549 and H1299 cells resulted in the inhibition of cell proliferation, migration, and invasion, enhancement of apoptosis, and a significant decrease in the expression of PPM1D and SPOP. Overexpression of PPM1D and SPOP attenuated the APPBP2-knockdown inhibition of NSCLC cells. Co-IP assay showed that PPM1D interacted with APPBP2.
Interpretation
The expression level of APPBP2 positively correlates with NSCLC cell proliferation, migration, and invasiveness. APPBP2 contributes to NSCLC progression through regulating the PPM1D and SPOP signalling pathway. This novel molecular mechanism, underlying NSCLC oncogenesis, suggests APPBP2 is a potential target for diagnosis and therapeutic intervention in NSCLC.
Fund
Key Program of Natural Science Research of Higher Education of Anhui Province (No. KJ2017A241), the National Natural Science Foundation of China (No. 81772493).
Keywords: APPBP2, Lung cancer, Non-small cell lung cancer, PPM1D, SPOP
Research in context.
Evidence before this study
APPBP2 interacts with microtubules and is functionally associated with beta-amyloid precursor protein (APP) transport and/or processing. Microtubules participate in the formation of the spindle during cell division (mitosis) responsible for cell proliferation. APP is a cell surface protein with signal-transducing properties and controls cells viability, proliferation, migration, and aggressiveness in various cancers. Based on the regulation of microtubules and APP, APPBP2 is found to be involved in the oncogenesis of various types of cancers, such as breast cancer, ovarian clear cell adenocarcinomas, desmoplastic medulloblastomas and neuroblastomas. However, the effects of APPBP2 on non-small cell lung cancer (NSCLC) remains unclear.
Added value of this study
In this study, the investigators first demonstrate that APPBP2 expression is significantly enhanced in NSCLC tumours relative to tumour-adjacent normal tissues. Then the investigators provide evidence that APPBP2 controls NSCLC cell proliferation, apoptosis, migration, and invasiveness. Moreover, the investigators discovered that PPM1D and SPOP participate in the molecular mechanism underlying the roles of APPBP2 in NSCLC. Taken together, these findings suggest that APPBP2 contributes to NSCLC progression through regulating the PPM1D and SPOP signalling pathways.
Implications of all the available evidence
Targeted therapies show great promise in effectively treating lung cancer patients. Therefore, characterizing and targeting the functionally-relevant molecular aberrations in lung cancer helps to identify new approaches to manage this disease. This research suggests that APPBP2 has a close relationship with NSCLC and contributes to the initiation and progression of NSCLC through regulating the PPM1D and SPOP pathways. Although the implications of APPBP2 in other cancers has been reported, we are the first to clarify the role of APPBP2 in NSCLC and the underlying molecular mechanisms. Therefore, this study provides a novel molecular mechanism underlying the oncogenesis of NSCLC and supports APPBP2 as a potential valuable molecular target suitable for diagnosis and therapeutic intervention in NSCLC.
Alt-text: Unlabelled Box
1. Introduction
Lung cancer is the most common cause of malignant tumours worldwide [1].Of the different types of lung cancer, non-small cell lung cancer (NSCLC) accounts for over 80% of all lung cancer cases. The majority of NSCLC cases are diagnosed at later stages with local invasion or distal metastases, consequently leading to poor effectiveness of surgical or radiotherapeutic interventions [2]. Therefore, there is an urgent need for further understanding of the mechanism underlying NSCLC oncogenesis to support the development of novel therapeutic interventions.
Cancer is the uncontrolled growth of abnormal cells anywhere in the body. Proteins that regulate cell proliferation, apoptosis, and invasion are critically involved in the pathogenesis of cancers. Amyloid protein-binding protein 2 (APPBP2) interacts with microtubules and is functionally associated with beta-amyloid precursor protein transport and/or processing [3,4].Studies have demonstrated that APPBP2 plays a key role in the oncogenesis of numerous types of cancer. For instance, Hirasawa et al. demonstrated that APPBP2 is closely associated with malignant phenotypes of ovarian adenocarcinomas [5]. In breast cancer, APPBP2 expression is significantly upregulated, prompting tumour cell invasion and metastasis [6].In desmoplastic medulloblastomas and neuroblastomas, the gene of APPBP2 is amplified with links to cancer initiation and progression [7,8]. These findings suggest that APPBP2 is implicated in the occurrence and development of cancers. However, the influence of APPBP2 on lung cancer remains unclear. Elucidating the roles of APPBP2 in NSCLC cells can improve our understanding of the molecular mechanism underlying lung cancer while benefiting the development of targeted therapies to combat NSCLC.
This study confirms the elevated expression of APPBP2 in NSCLC tissue compared with normal tumour-adjacent lung tissue. On a cellular level, the investigators demonstrate that APPBP2 promotes cell proliferation, migration, and invasiveness but inhibits cell apoptosis in NSCLC. In this study APPBP2 knockdown consistently reduces the growth of NSCLC xenograft. Moreover, the investigators prove that the level of APPBP2 expression positively correlates with the level of PPM1D and SPOP expression. Also the overexpression of PPM1D as well as SPOP can significantly attenuate the APPBP2-knockdown inhibition of cell proliferation, migration, and invasiveness in NSCLC cells. The investigators provide evidence for the interaction of APPBP2 with PPM1D. In short, this study's findings indicate that APPBP2 facilitates the oncogenesis of NSCLC through modulating the PPM1D and SPOP signalling pathway.
2. Materials and methods
2.1. Cell culture and transfection
The immortalized lung adenocarcinoma cell line A549 (RRID: CVCL_0023), non-small cell lung cancer cell line H1299 (RRID: CVCL_0060), and human embryotic cell line HEK293T were purchased from the cell line bank of Bioon (Shanghai, China). Cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% foetal bovine serum (FBS, HyClone), 10 mg/ml streptomycin and 10u/ml penicillin. The cell lines were incubated at 37 °C in a 5% CO2 atmosphere.
2.2. Patients and tissue collection
Between September 2017 and October 2018, ten pairs of lung adenocarcinoma and adjacent normal lung tissues were collected at the First Affiliated Hospital of Bengbu Medical College (Anhui province, China) from patients who underwent surgical resection of lung cancer. The protocol of sample collection was approved by the Ethics Committee of the First Affiliated Hospital of Bengbu Medical College (Bengbu, China). All patients provided informed consent for use of their samples in this study.
2.3. Immunohistochemistry
After fixing with 4% formalin and embedding in optimal cutting temperature compound (OCT), tissues were cut into 10 um sections using a microtome. Subsequently, the slides were treated with 3% H2O2 to block the endogenous peroxidase activity and heated (80 °C) for 10 min in citrate buffer (10 mM; pH 6·0) for antigen retrieval. To reduce nonspecific binding, 10% BSA + 0·4% triton-x100 was applied for 1 h at room temperature (RT). Subsequently, the slides were incubated with primary antibodies (anti-APPBP2, Sigma, RRID: ab151305, 1:500; anti-PPM1D, Sigma, RRID: ab31270, 1:500; anti-SPOP, Abcam, RRID: ab168619, 1:500 at 4 °C overnight, followed by incubating with appropriate second antibodies. After that, DAPI (abcam, RRID: b104139) was applied for nucleus staining for 10 min at RT.
2.3.1. Extraction of total RNA, reverse transcription, and real-time polymerase chain reaction analysis
Total RNA was extracted from the cell lines and the paired tissue from ten patients with lung adenocarcinoma using Trizol reagent (Takara, 9108). Quality of the total RNA was determined using RNA gel electrophoresis and NanoDrop. Then, a total of 1 μg of high-quality total RNA were directly processed for synthesizing cDNA using PrimeScript™ RT reagent Kit (Takara, RR047A). The real-time polymerase chain reaction analyses were performed in a 20 μl volume system using SYBR (Takara, R820A, 10 μl) according to the manufacture's instruction as follows: Primer mix (0·5 μl, 10 μM/each), template cDNA (1 μl), and sterile water (8·5 μl). The RT-PCR primers for APPBP2, PPM1D, SPOP, and GAPDH were summarized in Supplementary table 1.
2.4. Western blot
Total protein from the human lung adenocarcinoma cancer samples and NSCLC cells were prepared with ice-cold radioimmuno-precipitation assay buffer, quantified using BCA protein assay reagent (Beyotime, P0009, China), and separated by SDS-PAGE. Following electrophoresis, proteins in the gel were transferred to polyvinylidene difluoride membranes (Merck Millipore). After blocking for 1 h with 5% non-fat milk, the membranes were incubated overnight at 4 °C with antibodies (anti-APPBP2, Sigma, RRID: ab151305, 1:2000; anti-PPM1D, Sigma, ab31270, 1:2000; anti-SPOP, Abcam, RRID: ab168619, 1:2000), antibody against GAPDH (abcam, RRID: ab181602, 1:5000), or antibody against β-actin (abcam, RRID: ab8226, 1:5000) which was set as the internal standard. This was followed by incubation with a horseradish peroxidase-labelled goat anti-rabbit IgG secondary antibody (abcam, RRID: ab205718, 1:5000) or horseradish peroxidase-labelled goat anti-mouse IgG secondary antibody (abcam, RRID: ab6728, 1:5000) at room temperature for 1 h. Signals were detected with ECL Western Blotting Detection Reagent (GE Healthcare Life Sciences) and visualized using a Chemical Gel Documentation System (Tanon).
2.5. Gene silencing and overexpression
The knockdown of human APPBP2 was performed by shRNA [9]. The investigators set sh-NC and sh-APPBP2 as the control and experimental groups, respectively. The target sequences of sh-NC and sh-APPBP2 are presented in supplementary table 1. sh-NC and sh-APPBP2 were cloned into pLKO.1 (Addgene,8453) plasmid, respectively. All the plasmids were sequenced. The investigators transfected shRNA-pLKO.1 with helper plasmids (VSVG and delta8·9) into HEK293T cells for 72 h to package lentivirus. The titer of the viruses was measured by examining the infectious ability of HEK293T cells. A549 and H1299 cells were infected with the sh-NC and sh-APPBP2 viruses. The investigators screened the stable transfected cell lines with 2 μg/ml puromycin, then RT-PCR and western blotting were applied for biological studies.
For overexpression purposes, human APPBP2, PPM1D, and SPOP CDS were cloned into pCDNA 3·1, which was delivered into the cell lines by lipo-3000 (Invitrogen, L3000008). The expression of the genes was driven under CMV promoter.
2.6. BrdU and MTT
For BrdU in A549 (or H1299): 5 × 105 sh-NC infected cells and sh-APPBP2 infected cells were cultured into 12-well plates. When the cells were attached to the wall, the investigators added BrdU to each well with a final concentration of 10 μM and cultured these for another 8 h. They then replaced the BrdU containing medium with normal medium (10% FBA in DMEM) and further cultured for 24 h. The cells were harvested and underwent immunostaining of BrdU (abcam, RRID: ab142567, 1:500) and DAPI.
For MTT in A549 (or H1299), a modification of the method described by Mossman [10] was used in our experiments: The stable cells of sh-APPBP2 and sh-NC (transfected with plasmids or not) were cultured for 48 h. The investigators then transplanted the cells into 96-well at the appropriate seeding density of 5000 cells/well. Cells were incubated with 10 μl MTT (5 mg/ml) for 3 h, then the medium was replaced with 150 μl 100% dimethylsulfoxide (DMSO) in each well. Plates were then agitated on a plate shaker for 5 min, following which, spectrophotometric absorbance at 570 nm was immediately determined using a scanner.
2.7. Colony formation assay
A549 (or H1299) cells were treated with sh-APPBP2 or sh-NC lentivirus (10 x cell number/viral titer) and cultured for 48 h. The cells were then disassociated and seeded for colony formation in 6-well plates at two hundred viable cells (the density was measured by trypan blue assay) per well. After 14–21 days (medium exchanged every three days), colonies were scored using a microscope by adding crystal violet staining at a concentration of 0·4%. Colonies were counted only if a single clone contained >100 cells. Each assay was performed in triplicate on two independent occasions. For functional rescue experiments, after cells were transfected with PPM1D or SPOP plasmids for 48 h (Fig. 5a), 500 viable cells were seeded for colony formation in 6-well plates.
Fig. 5.
PPM1D and SPOP attenuated the inhibition of cell proliferation induced by APPBP2 knockdown in NSCLC cells. (a) Ectopic expression of PPM1D or SPOP promoted colon formation on APPBP2 silenced A549 cells. Control was set as a blank group without protein overexpression. (b) The statistical results of colony formation indicate the impact of PPM1D and SPOP on NSCLC stable cells. A one-way ANOVA revealed a significant effect of group (F[4,10] = 324.5, p < 0.001 in A549 and F[4,10] = 63.45, p < 0.001 in H1299). (c) The proliferation effects of PPM1D or SPOP expression on A549 stable cells was measured by MTT assay. A one-way ANOVA revealed a significant effect of group (F[6,14] = 106.2, p < 0.001). (d) The proliferation effects of PPM1D or SPOP expression on H1299 stable cells was measured by MTT assay. A one-way ANOVA revealed a significant effect of group (F[6,14] = 393.9, p < .001). (e) PPM1D and SPOP had a significant impact on cell apoptosis of A549 stable cells by Annexin V and flow cytometric analysis. A one-way ANOVA revealed a significant effect of group (F[6,14] = 338.4, p < 0.001). (f) PPM1D and SPOP had a significant impact on cell apoptosis of H1299 stable cells by Annexin V and flow cytometric analysis. A one-way ANOVA revealed a significant effect of group (F[6,14] = 310.9, p < 0.001). The addition or non-addition of plasmid or virus is present as “+” and”-”, respectively. Data are presented as mean ± SEM. The difference between means was compared by Tukey's Multiple Comparison test. 0.01 < *p < 0.05 and ***p < 0.001 for indicated comparison. n.s. refers as not significant.
2.8. Cell sorting for apoptosis and cell cycle assay
To study the impact of APPBP2 on apoptosis, 106 stable cells of A549 (or H1299) were stained with Annexin V-APC and 7-AAD (Biolegend, 640930, USA) according to the manufacturer's instructions. The apoptotic cell proportion was assessed with flow cytometry (BD, Calibur, USA). To study the impact of APPBP2 on the cell cycle, cells (4 × 105) were fixed in 70% ethanol for 1 h at 4 °C. Then the cells were washed twice with PBS followed by adding 10 mg/ml RNase A. The investigators then added propidium iodide (BD, 550825) at a final concentration of 0·05 mg/ml. The samples were incubated at 4 °C for 30 min in a dark environment. Finally, samples were analysed by flow cytometry. All the assays were performed in triplicate. To study the rescue function of PPM1D or SPOP on apoptosis, A549 (or H1299) stable cell lines were transfected with 1 μg PPM1D or SPOP in a 6-well plate to culture for 48-h, and then underwent apoptotic assessment.
2.9. Wound healing assay
Wound healing assay was performed according to a previously described method with some modifications [11]. A549 (or H1299) stable cells were transplanted into 6-well plates at the appropriate seeding density of 1 × 106 cells/well. While confluent, the cell monolayer was mechanically scarred with a sterile 200 μl pipette tip and incubation was continued for an additional 12 h and 24 h. Images were taken at 0, 12, and 24 h after scarring.
2.10. Transwell assay
In vitro invasion capability was measured using 24-well chambers (Millipore, 8 um, USA) pre-coated with Matrigel (BD, 256234) on the polyethylene terephthalate membrane. In the upper compartment of the transwell chambers, the investigators seeded 5 × 104 stable cells suspended in 500 μl of serum-free medium and filled the lower compartment with complete medium. After incubation at 37 °C under 5% CO2 for 6 to 10 h, some of the cells had migrated through the porous membrane, whereas the non-migrating cells remained on the upper surface of the filter. The chamber was then immersed in methanol for 10 min to fix the cells, and then scrubbed gently to remove the non-metastatic cells. The migrated cells were stained with 0·5% crystal violet for 10 min and counted under a light microscope. Each experiment was independently performed three times. For Fig. 6a,b, the A549 (or H1299) stable cells were further transfected with PPM1D or SPOP plasmid by lipo-3000. The cells were cultured for 48 h and then examined.
Fig. 6.
PPM1D and SPOP attenuated the inhibition of cell migration and invasiveness induced by APPBP2 knockdown in NSCLC cells. (a) Effects of PPM1D and SPOP on cell migration and invasion of A549 stable cells by transwell, n = 3. (b) Effects of PPM1D and SPOP on cell migration and invasion of H1299 stable cells by transwell, n = 3. (c) The statistical results of PPM1D and SPOP on cell migration and invasiveness of A549 stable cells by transwell. A one-way ANOVA revealed a significant effect of group (F[6,14] = 35.7, p < 0.001). (d) The statistical results of PPM1D and SPOP on cell migration and invasiveness of H1299 stable cells by transwell. A one-way ANOVA revealed a significant effect of group (F[6,14] = 72.58, p < 0.001). The addition or non-addition of plasmid or virus is present as “+” and “-”, respectively. Data are presented as mean ± SEM. The difference between means was compared by Tukey's Multiple Comparison test. 0.001 < **p < 0.01 and ***p < 0.001 for indicated comparison. n.s. refers as not significant. Scale bars: 50 μm.
2.11. Establishment of an adenocarcinoma xenograft mouse model
All animal experiments were conducted according to the institutional ethics and safety guidelines (Institutional Animal Welfare and Ethics Committee, First Affiliated Hospital of Bengbu medical college, Anhui, China). In total, forty 8-week-old BALB/c-Foxn1null mice were used in the experiments (Model Animal Research Centre of Nanjing University, Nanjing, China). The mice were randomly divided into four groups. APPBP2 silenced and control xenograft model sets from A549 (or H1299) were established using 5 × 106 cells injected into the right flanks of the subcutaneous tissue of the mice. Tumour length, width and tumour weight were measured every three to four days until the experiment ended. The tumour volume was calculated based on the conventional formula: V = (L × W2)/2, where L and W are the biggest and smallest diameters of the tumour.
2.12. Coimmunoprecipitation (co-IP)
To conduct the experiments, four additional plasmids were constructed. Flag, Flag-APPBP2, His, and His-PPM1D were cloned into pCDNA 3·1, respectively. For APPBP2 co-IP in A549 (or H1299) cells, the cells transformed by lipo-3000, expressing Flag-APPBP2, PPM1D, and SPOP, were cultured for 72 h. Meanwhile, the transformed cells expressing Flag, APPBP2, PPM1D, and SPOP were set as the negative control group. For PPM1D co-IP in the A549 (or H1299) cells, the transformed cells expressing His-APPBP2, PPM1D, and SPOP were cultured for 72 h. The transformed cells expressing His, APPBP2, PPM1D, and SPOP were set as the control group. The cells were lysed for co-IP and were incubated with beads (pierce, 6149) which covalently pre-couple Flag antibody (abcam,ab205606) or His antibody (abcam, ab18184). Some cell lysis, which was incubated with the beads covalently pre-coupled with human IgG, was set as another negative control. The protein binding complex was isolated by centrifuging. The precipitates were diluted with SDS sample buffer, separated on a 10% SDS–PAGE gel and subjected to immunoblotting with the corresponding antibodies.
2.13. Reanalysing the TCGA database
The investigators explored the mRNA level of APPBP2 in The Cancer Genome Atlas Lung Adenocarcinoma (TCGA-LUAD) cohort. The RNA Seq V2 RSEM data of TCGA-LUAD was obtained using TCGAbiolinks (https://github.com/BioinformaticsFMRP/TCGAbiolinks, R version 3.5). The investigators identified candidate genes which may correlate with the APPBP2 mRNA level using cBioPortal tool [12,13].
2.14. Statistical analysis
Data are presented as mean ± SEM and two-tailed t-test was used to determine statistical significance difference between two groups (except Supplementary Fig. s3b, which adopted one-tailed t-test). One-way ANOVA followed by a Tukey's Multiple Comparison test was applied for the comparison among groups in the same graph. A p-value of <0.05 was considered statistically significant.
3. Results
3.1. The elevated expression of APPBP2 in human NSCLC
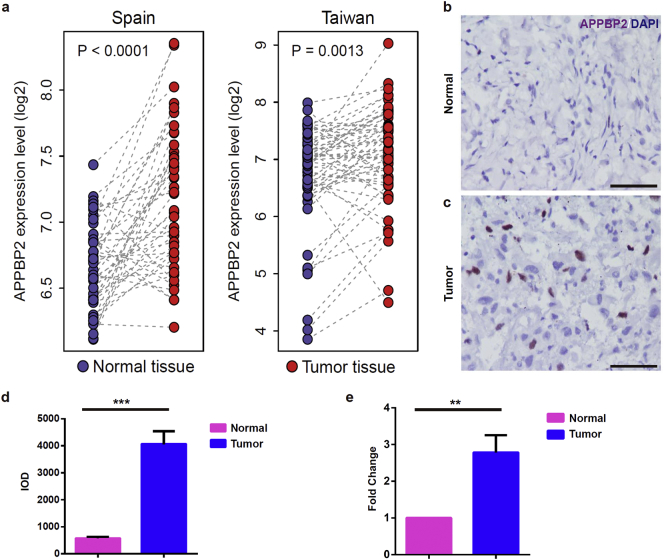
To explore the correlation of APPBP2 with NSCLC, we compared the microarray-based expression data of APPBP2 between the paired normal and tumour tissues in two NSCLC cohorts from Spain (GEO accession: GSE18842) [14] and Taiwan (GEO accession: GSE19804) [15]. In both cohorts, APPBP2 was significantly up-regulated in the tumour tissues compared with the paired normal tissues (paired t-test: p < 0.0001 for the Spain cohort and p = 0·0013 for the Taiwan cohort) (Fig. 1a). In addition, primary cancers and the paired normal tumour-adjacent lung tissues were collected from ten NSCLC patients and subsequently subjected to immunohistochemical and quantitative real-time PCR (qPCR)analysis. As expected, an elevated expression of APPBP2was observed in NSCLC tumours relative to normal lung tissues (Fig. 1b–e).
Fig. 1.
The expression of APPBP2 was elevated in human NSCLC tissues. (a) The APPBP2 expression profile of the NSCLC tissues and the adjacent normal tissues from the GEO database. The left and right panels represent samples from Spain and Taiwan, respectively. (b,c) Immunostaining of APPBP2 of the NSCLC tumour tissues (lower) and the matched normal tissues (upper) from our hospital. Blue signals represent DAPI and brown signals represent APPBP2. (d) Image grey integral optical density (IOD) from immunohistochemistry results. Pink colour and blue colour represent normal tissues and NSCLC tissues, respectively. (e) The APPBP2 mRNA expression levels of ten paired LUAD patients were measured by RT-PCR. Pink colour and blue colour represent normal tissues and LUAD tumour tissues, respectively. Data are presented as mean ± SEM. (Two-tailed t-test for group comparison) 0.001 < **p < 0.01, ***p < 0.001 for indicated comparison. Scale bar: 50 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. APPBP2-knockdown suppresses cells proliferation, but induces apoptosis in NSCLC in vitro
As unlimited growth is one of the key hallmarks of cancers [16], we attempted to assess the influence of APPBP2 on cell proliferation and apoptosis in NSCLC. First, lentivirus mediated sh-APPBP2 was applied to silence the APPBP2 expression in both A549 and H1299 cells, with a scrambled shRNA (sh-NC) as the negative control (Fig. S1a). Following the transfection of cells, qPCR and western blotting assays were performed to examine the endogenous APPBP2 expression. The results showed that the expression levels of APPBP2 dropped by about 70% (Fig. S1b-d).
Then BrdU, MTT, colony formation and flow cytometry assays were performed on the stable transfected NSCLC cell lines to detect the proliferation and/or apoptosis. BrdU assays indicated that the silencing of APPBP2 significantly suppressed the proliferation of both A549 (sh-NC: 0·65 ± 0·07, n = 3; sh-APPBP2: 0·28 ± 0·03, n = 3; Fig. 2a,b,e)and H1299 (sh-NC: 0·66 ± 0·06, n = 3; sh-APPBP2: 0·41 ± 0·03, n = 3; Fig. 2c,d,e) cells. This was consistent with the results from MTT (Fig. 2f) and colony formation assays (Fig. 2 g–i). Flow cytometry assays revealed that cell apoptosis was dramatically enhanced in two cell lines transfected with sh-APPBP2 compared with sh-NC (Fig. 2j and Fig. S2a, b), but no significant differences were observed on cell cycles (Fig. 2k and Fig. S2c, d).
Fig. 2.
APPBP2 silencing suppressed proliferation and induced apoptosis in NSCLC cells.(a-d) The immunostaining results of BrdU indicates the proliferation of A549 cells (a,b) and H1299 cells (c,d) treated with sh-NC (a,c) or sh-APPBP2 (b,d). Blue signal represents DAPI. (e) The percentages of BrdU+DAPI+cells in DAPI+ cells. 189 cells, 221 cells, 135 cells, and 250 cells were counted from left to right column, respectively. (f) Effects of APPBP2 silencing on proliferation of A549 and H1299 stable cells by MTT assay (n = 3). (g,h) Effects of APPBP2 silencing on the colony formation of H549 (g) and H1299(h) stable cells, respectively. (i) The statistical results of colony formation of NSCLC stable cells, the colon numbers are 220, 130, 296, and 136 from the left to right column, respectively. (j) Knockdown of APPBP2 increased cell apoptosis. Cell death was determined by Annexin V and flow cytometric analysis. (k) The cell cycle of A549 (or H1299) stable cells was analysed by propidium iodide staining and flow cytometry. (l–n) APPBP2 silencing inhibited tumour growth of A549 stable cells in vivo. (l) Tumour pictures from mice treated with sh-NC (upper) and sh-APPBP2 (lower). (m) Growth curve of tumour volume was measured on indicated days and (n) tumour weight at the end of experiment. (o-q) APPBP2 silencing inhibited tumour growth of H1299 stable cells in vivo. (o) Tumour pictures from mice treated with sh-NC (upper) and sh-APPBP2 (lower). (p) Growth curve of tumour volume was measured on indicated days and (q) tumour weight at the end of experiment. Data are presented as mean ± SEM. (Two-tailed t-test for group comparison) 0.01 < *p < 0.05,0.001 < **p < 0.01, ***p < 0.001 for indicated comparison. Scale bars: 25 μm.
3.3. Silencing APPBP2 reduces NSCLC growth in vivo
Xenografts are models of cancer where the tissue or cell lines are implanted into an immunodeficient or humanized mouse, which are used to create an environment that allows for the natural growth of cancer. To assess the action of APPBP2 on NSCLC growth in vivo, we constructed xenograft tumours of A549 and H1299 cells transfected with sh-APPBP2 and sh-NC respectively, followed by the investigation of tumour growth. Interestingly, we found that silencing APPBP2 reduced both the size (Fig. 2m, p) and weight (Fig. 2n, q) of xenografts.
3.4. APPBP2 silencing inhibits cell migration and invasiveness in NSCLC
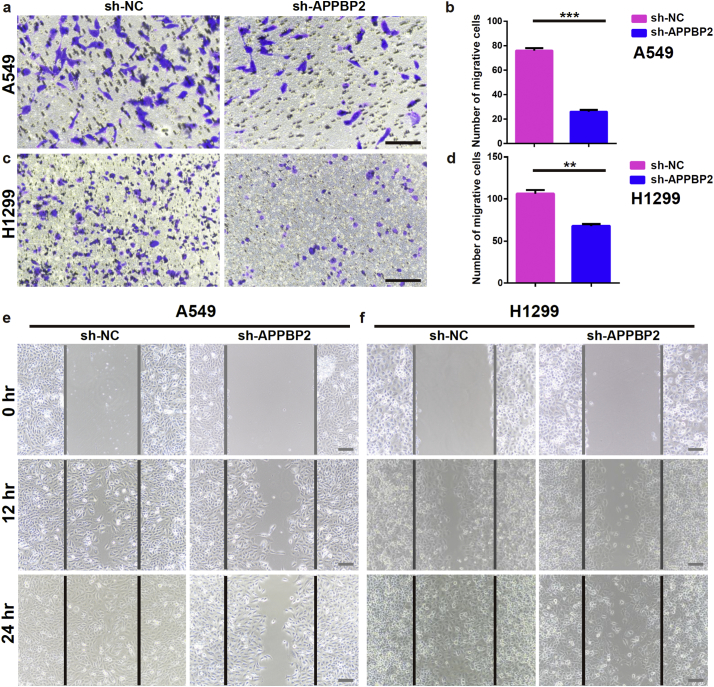
Metastasis is the leading cause of NSCLC-related mortality, with about 90% of NSCLC patients succumbing to metastasis as opposed to their primary tumours [17].To elucidate the molecular mechanism(s) underlying NSCLC metastasis, wound healing and transwell assays were performed to assess the roles of APPBP2 in the migration and invasiveness of NSCLC cells. Transwell assays showed that the invasiveness of both A549 and H1299 cells was substantially suppressed by APPBP2 silencing (Fig. 3a–d). Similarly, wound healing assays suggested that the APPBP2 knockdown obviously impaired the migratory activity in both cell lines (Fig. 3e, f).
Fig. 3.
APPBP2 silencing inhibits cells migration and invasiveness in NSCLC. (a,b) Effects of APPBP2 silencing on migration and invasion of A549 stable cells evaluated by transwell assays, n = 3. (c,d) Effects of APPBP2 silencing on the migration and invasion of H1299 cells evaluated by transwell assays, n = 3. (e) Effects of APPBP2 silencing on migration of A549 stable cells evaluated by wound healing at 0 h, 12 h, and 24 h. (f) Effects of APPBP2 silencing on migration of H1299 cells evaluated by wound healing assay at 0 h, 12 h, and 24 h. The lines define the areas migrating cells. Data are presented as mean ± SEM. (Two-tailed t-test for group comparison) 0.001 < **p < 0.01, ***p < 0.001 for indicated comparison. Scale bars: 50 μm.
3.5. APPBP2-knockdown reduced the expression of PPM1D and SPOP in NSCLC
To clarify the molecular mechanism underlying the action of APPBP2 in NSCLC, the investigators analysed a heatmap of potential correlations which synchronized well with APPBP2 expression in human NSCLC samples from the TCGA cohort (TCGA-LUAD). The heatmap revealed several cancerous genes (Fig. 4a), among which the expression of PPM1D and SPOP may be highly correlated with the APPBP2 levels in the TCGA-LUAD cohort (spearman ρ = 0·65 for PPM1D and 0·54 for SPOP; Fig. 4b,c). The investigators performed western blotting assays to explore the correlations of APPBP2 with PPM1D and SPOP in ten paired NSCLC tissues and found that three proteins of APPBP2, PPM1D, and SPOP were all enhanced in tumours relative to normal tissues (Fig. S3a,b). This was consistent with the results from the immunohistochemistry and qPCR assays (Fig. 4d–f). Moreover, the investigators found that APPBP2-knockdown significantly reduced the expression of PPM1D and SPOP in both A549 and H1299 cells, as determined by qPCR and western-blot assays (Fig. 4g, h).
Fig. 4.
APPBP2 regulated the expression of PPM1D and SPOP in NSCLC tissues and cells. (a) The heatmap of gene expression profiling organized by APPBP2 expression level by reanalysing the TCGA data. Samples were derived from human LUAD patients. Red circles indicate the genes to be studied. (b,c) PPM1D mRNA level (b) and SPOP mRNA level (c) had a positive linear relationship with the expression of APPBP2. (d) Immunostaining of PPM1D (brown colour) of human LUAD tissues and adjacent normal tissues. (e) Immunostaining of SPOP (brown colour) of human LUAD tissues and adjacent normal tissues. DAPI in (d) and (e) is indicated as blue colour. (f) PPM1D mRNA and SPOP mRNA expression level of human LUAD tissues and adjacent normal tissues were measured by RT-PCR. Yellow colour and cyan colour represent normal and tumour samples, respectively. n = 3. (g) RT-PCR results showed PPM1D mRNA and SPOP mRNA expression level in APPBP2 silenced cells of A549 or H1299 (n = 3). (h) PPM1D and SPOP were examined by western blotting on APPBP2 silenced A549 and H1299 stable cells. GAPDH acts as the internal standard. (i) co-IP between the APPBP2-flag and PPM1D or SPOP on A549 or H1299 cell contexts. (j) co-IP between PPM1D-His and APPBP2 or SPOP on A549 or H1299 cell contexts. Data are presented as mean ± SEM. (Two-tailed t-test for group comparison) 0.01 < *p < 0.05, 0.001 < **p < 0.01, ***p < 0.001 for indicated comparison. Scale bars: 50 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.6. Interaction of APPBP2 with PPM1D and SPOP in NSCLC
Co-immunoprecipitation (co-IP) is a popular technique to identify physiologically relevant protein–protein interactions by using target protein-specific antibodies to indirectly capture proteins that are bound to a specific target protein. To assess the protein interactions among APPBP2, PPM1D, and SPOP, co-IP experiments were performed on NSCLC cells using antibodies anti-Flag (fusion with APPBP2) as well as anti-His (fusion with PPM1D) and revealed that APPBP2 could bind with PPM1D (but not SPOP) in both A549 and H1299 cell contexts (Fig. 4i, j).
3.6.1. Overexpression of PPM1D and SPOP attenuate APPBP2-knockdown inhibition of cell proliferation and invasiveness in NSCLC
To evaluate the mediation of PPM1D and SPOP on the effect of APPBP2 on NSCLC, plasmids containing PPM1D and SPOP expression cassette were transfected into APPBP2-silenced A549 and H1299 cells to enhance the expression of PPM1D and SPOP. This was followed by the examination of cell proliferation, apoptosis, migration, and invasiveness using colony formation, MTT, flow cytometry and transwell assays. APPBP2-knockdown inhibition of biological behaviours in NSCLC cells was strongly attenuated by the overexpression of PPM1D and SPOP. (Fig. 5, Fig. 6 and Fig. S3c–e).
4. Disscussion
Lung cancer remains the leading cause of cancer related deaths worldwide and targeted therapies are showing great promise in effectively treating certain patients [18,19]. Therefore, new targeted molecular therapeutic strategies for lung cancer are needed to aid patients with this disease. Characterizing and targeting the functionally-relevant molecular aberrations in lung cancer helps to identify new approaches to managing this disease. The investigators report on the oncogenic role of APPBP2 in NSCLC.
APPBP2 interacts with microtubules and is functionally associated with beta-amyloid precursor protein (APP) transport and/or processing [3]. Microtubules participate in the formation of the spindle during cell division (mitosis) responsible for cell proliferation. APP is a membrane protein expressed in many tissues and has been implicated as a regulator of synapse formation [20], neural plasticity [21], and iron export [22]. Interestingly, studies have suggested that APP controls cells viability, proliferation, migration, and aggressiveness in numerous types of cancers [[23], [24], [25], [26]]. Based on these findings, we hypothesized that APPBP2 could participate in the oncogenesis of cancers through modulating microtubules and/or APP. Actually, the oncogenic roles of APPBP2 in numerous types of cancer has been reported [[5], [6], [7], [8],27]. However, the roles of APPBP2 in NSCLC are still poorly understood.
Molecular biology studies have demonstrated that cancers are a complexity of multi-stage biological process with the involvement of various factors. This includes the activation of oncogene and the inactivation of tumour suppressor gene, leading to functionally relevant molecular aberrations. In the current study, the investigators compared the expression of APPBP2 mRNA between paired tumour and the adjacent normal tissues in two NSCLC cohorts, discovering a significantly elevated expression of APPBP2 in tumours relative to normal tissues. This result was confirmed in the samples of NSCLC and paired adjacent normal tissues. Similar to our findings in NSCLC, overexpression/amplification of the APPBP2 gene has been reported in other cancers, such as breast cancer [27], ovarian clear cell adenocarcinomas [5], desmoplastic medulloblastomas [7], andneuroblastomas [8]. Taken together, these findings indicate that APPBP2 correlates with the oncogenesis of cancers, including NSCLC.
Uncontrolled proliferation and invasiveness are the key hallmarks of cancer cells and are the leading cause of NSCLC-related mortality. Therefore, the investigators undertook this study to shed light on the association of APPBP2 with the malignant phenotype of NSCLC cells. First, the investigators demonstrated that APPBP2 silencing suppressed cell proliferation and viability while inducing apoptosis in NSCLC cells. Next, the investigators confirmed that knockdown of APPBP2 inhibited the migration as well as the invasiveness of A549 and H1299 cells. Finally, the investigators provided evidence that APPBP2-knockdown reduces NSCLC xenograft growth. Taken together, the findings suggest that APPBP2 plays an oncogenic role in NSCLC.
In the investigation of the molecular mechanism underlying APPBP2 biologic effects on NSCLC, the investigators discovered that the level of APPBP2 expression positively correlated with PPM1D and SPOP expression as represented in Fig. 7. Notably, the overexpression of PPM1D and SPOP effectively attenuated APPBP2-silencing inhibition in NSCLC cell proliferation, migration, and invasiveness, indicating PPM1D and SPOP responsible for the oncogenic roles of APPBP2 in NSCLC. To explore the interaction of APPBP2 with PPM1D and SPOP, we performed co-IP assays and found that PPM1D (but not SPOP) could strongly bind to APPBP2 in both A549 and H1299 cell contexts, indicating the physiologically relevant interactions of APPBP2 with PPM1D.
Fig. 7.

Graphical signalling pathway of the effect of APPBP2 on NSCLC via PPM1D and SPOP.
The blue lines indicate the activation of the targets according to our experiments or published researches. The red lines indicate the possible inhibition of targets according to our reasonable speculation. AR: androgen receptor. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
APPBP2 contains a tetratricopeptide repeat (TPR) structural motif that frequently mediates protein-protein interactions [28,29], so the binding of APPBP2 with PPM1D is potentially mediated by the TPR structural motif. Since APPBP2 (PAT1) phosphorylates Mei2 and Ste11 in the controls of sex differentiation [30] and functions as a positive regulator of Kinesin motility, resulting in cargo transport [31], the modulation of PPM1D by APPBP2 in NSCLC is probably through phosphorylation and/or translocation.
Previous studies have proved the oncogenic roles of PPM1D in numerous types of cancers [5,6,8,32], mostly in a p38MAPK/p53/p16 pathway dependent manner [33,34]. Notably, H1299 cells have a homozygous partial deletion of the TP53 gene [35], suggesting the existence of a P53-independent manner of PPM1D affecting NSCLC cells. Consistently, a P53-independent manner of PPM1D action has been reported in clonal haematopoiesis and myelodysplastic syndrome [36,37].
Our findings suggest that SPOP is also a key molecule responsible for the effects of APPBP2 on NSCLC. Although SPOP mutations were identified in numerous types of tumours, especially in prostatic carcinoma [38], no SPOP mutations were observed in the analysis of human NSCLC samples from the TCGA cohort in the current study (Fig. 4c). Consistent with our findings, earlier study revealed that somatic mutation of SPOP is rare in lung cancer [39]. Interestingly, we found the elevated expression of SPOP in NSCLC (Fig. 4e,f and Fig. S3a,b). APPBP2 promotes a cytoplasmic retention of androgen receptor (AR) [40], leading to the dysfunction of AR upregulating the of CDK4 expression in prostatic cancer [41]. CDK4 induces the phosphorylation as well as the degradation of SPOP in multiple cancers [38,42]. In renal cell carcinoma, SPOP can be activated by HIF and SPOP has very important oncogenic role suppressing PTEN and other genes, as well as activating AKT and ERK [43].Consistently, researchers also found that SPOP, as a strong oncogenic protein, acted as a diagnostic biomarker in renal cell carcinoma [[44], [45], [46]]. Based on these reports, it is reasonable to hypothesize that APPBP2 enhances NSCLC proliferation and invasiveness partially through AR/CDK4/SPOP pathway (Fig. 7).
Collectively, APPBP2 is strongly associated with NSCLC and promotes the initiation and progression of tumours, which is, at least partly, mediated by PPM1D as well as SPOP. Our work provides a novel molecular mechanism underlying the oncogenesis of NSCLC and supports APPBP2 as a potential valuable marker suitable for diagnosis and therapeutic intervention in NSCLC.
Funding sources
This work was supported by the Key Program of Natural Science Research of Higher Education of Anhui Province (grant no. KJ2017A241), the National Natural Science Foundation of China (grant no. 81772493) and the Science and Technology Program of Anhui Province (grant nos. 2017070503B037 and YDZX20183400002554). The funders had no role in the study design, data collection, data analysis, interpretation, or writing of the paper.
Declarations of interest
All co-authors agree with the content of this manuscript and report no conflicts of interest related to this study.
Author contributions
Conceived and designed the study: X.W., X.G.L., H.G. and F.L.;Performed the experimental procedures: All authors; Analysed the data: F.L. and X.W.; Drafted the Manuscript: F.L. and H.G.; Revised the Manuscript: X. W.; Financial support: X.W.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.05.028.
Contributor Information
Xuegang Liu, Email: xgliu99@126.com.
Xiaojing Wang, Email: wangxiaojing8888@163.com.
Appendix A. Supplementary data
Supplementary material 1
Supplementary material 2
References
- 1.Torre L.A., Siegel R.L., Jemal A. Lung Cancer Statistics. Adv. Exp. Med. Biol. 2016;893:1–19. doi: 10.1007/978-3-319-24223-1_1. [DOI] [PubMed] [Google Scholar]; Torre LA, Siegel RL, Jemal A. Lung Cancer Statistics. Adv Exp Med Biol. 2016;893:1–19. [DOI] [PubMed]
- 2.Perlikos F., Harrington K.J., Syrigos K.N. Key molecular mechanisms in lung cancer invasion and metastasis: a comprehensive review. Crit Rev Oncol Hematol. 2013;87(1):1–11. doi: 10.1016/j.critrevonc.2012.12.007. [DOI] [PubMed] [Google Scholar]; Perlikos F, Harrington KJ, Syrigos KN. Key molecular mechanisms in lung cancer invasion and metastasis: a comprehensive review. Crit Rev Oncol Hematol. 2013;87(1):1–11. [DOI] [PubMed]
- 3.Zheng P., Eastman J., Vande Pol S., Pimplikar S.W. PAT1, a microtubule-interacting protein, recognizes the basolateral sorting signal of amyloid precursor protein. Proc. Natl. Acad. Sci. U. S. A. 1998;95(25):14745–14750. doi: 10.1073/pnas.95.25.14745. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zheng P, Eastman J, Vande Pol S, Pimplikar SW. PAT1, a microtubule-interacting protein, recognizes the basolateral sorting signal of amyloid precursor protein. Proc Natl Acad Sci U S A. 1998;95(25):14745–50. [DOI] [PMC free article] [PubMed]
- 4.Gao Y., Pimplikar S.W. The gamma -secretase-cleaved C-terminal fragment of amyloid precursor protein mediates signaling to the nucleus. Proc. Natl. Acad. Sci. U. S. A. 2001;98(26):14979–14984. doi: 10.1073/pnas.261463298. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gao Y, Pimplikar SW. The gamma -secretase-cleaved C-terminal fragment of amyloid precursor protein mediates signaling to the nucleus. Proc Natl Acad Sci U S A. 2001;98(26):14979–84. [DOI] [PMC free article] [PubMed]
- 5.Hirasawa A., Saito-Ohara F., Inoue J., Aoki D., Susumu N., Yokoyama T. Association of 17q21-q24 gain in ovarian clear cell adenocarcinomas with poor prognosis and identification of PPM1D and APPBP2 as likely amplification targets. Clin. Cancer Res. 2003;9(6):1995–2004. [PubMed] [Google Scholar]; Hirasawa A, Saito-Ohara F, Inoue J, Aoki D, Susumu N, Yokoyama T, et al. Association of 17q21-q24 gain in ovarian clear cell adenocarcinomas with poor prognosis and identification of PPM1D and APPBP2 as likely amplification targets. Clin Cancer Res. 2003;9(6):1995–2004. [PubMed]
- 6.Li J., Yang Y., Peng Y., Austin R.J., van Eyndhoven W.G., Nguyen K.C. Oncogenic properties of PPM1D located within a breast cancer amplification epicenter at 17q23. Nat. Genet. 2002;31(2):133–134. doi: 10.1038/ng888. [DOI] [PubMed] [Google Scholar]; Li J, Yang Y, Peng Y, Austin RJ, van Eyndhoven WG, Nguyen KC, et al. Oncogenic properties of PPM1D located within a breast cancer amplification epicenter at 17q23. Nat Genet. 2002;31(2):133–4. [DOI] [PubMed]
- 7.Ehrbrecht A., Muller U., Wolter M., Hoischen A., Koch A., Radlwimmer B. Comprehensive genomic analysis of desmoplastic medulloblastomas: identification of novel amplified genes and separate evaluation of the different histological components. J. Pathol. 2006;208(4):554–563. doi: 10.1002/path.1925. [DOI] [PubMed] [Google Scholar]; Ehrbrecht A, Muller U, Wolter M, Hoischen A, Koch A, Radlwimmer B, et al. Comprehensive genomic analysis of desmoplastic medulloblastomas: identification of novel amplified genes and separate evaluation of the different histological components. J Pathol. 2006;208(4):554-63. [DOI] [PubMed]
- 8.Saito-Ohara F., Imoto I., Inoue J., Hosoi H., Nakagawara A., Sugimoto T. PPM1D is a potential target for 17q gain in neuroblastoma. Cancer Res. 2003;63(8):1876–1883. [PubMed] [Google Scholar]; Saito-Ohara F, Imoto I, Inoue J, Hosoi H, Nakagawara A, Sugimoto T, et al. PPM1D is a potential target for 17q gain in neuroblastoma. Cancer Res. 2003;63(8):1876-83. [PubMed]
- 9.Stewart S.A., Dykxhoorn D.M., Palliser D., Mizuno H., Yu E.Y., An D.S. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9(4):493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]; Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9(4):493-501. [DOI] [PMC free article] [PubMed]
- 10.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods. 1983;65(1–2):55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]; Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65(1-2):55-63. [DOI] [PubMed]
- 11.Liang C.C., Park A.Y., Guan J.L. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2007;2(2):329–333. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]; Liang CC, Park AY, Guan JL. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc. 2007;2(2):329-33. [DOI] [PubMed]
- 12.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. [DOI] [PMC free article] [PubMed]
- 13.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401-4. [DOI] [PMC free article] [PubMed]
- 14.Sanchez-Palencia A., Gomez-Morales M., Gomez-Capilla J.A., Pedraza V., Boyero L., Rosell R. Gene expression profiling reveals novel biomarkers in nonsmall cell lung cancer. Int. J. Cancer. 2011;129(2):355–364. doi: 10.1002/ijc.25704. [DOI] [PubMed] [Google Scholar]; Sanchez-Palencia A, Gomez-Morales M, Gomez-Capilla JA, Pedraza V, Boyero L, Rosell R, et al. Gene expression profiling reveals novel biomarkers in nonsmall cell lung cancer. Int J Cancer. 2011;129(2):355-64. [DOI] [PubMed]
- 15.Lu T.P., Tsai M.H., Lee J.M., Hsu C.P., Chen P.C., Lin C.W. Identification of a novel biomarker, SEMA5A, for non-small cell lung carcinoma in nonsmoking women. Cancer Epidemiol. Biomark. Prev. 2010;19(10):2590–2597. doi: 10.1158/1055-9965.EPI-10-0332. [DOI] [PubMed] [Google Scholar]; Lu TP, Tsai MH, Lee JM, Hsu CP, Chen PC, Lin CW, et al. Identification of a novel biomarker, SEMA5A, for non-small cell lung carcinoma in nonsmoking women. Cancer Epidemiol Biomarkers Prev. 2010;19(10):2590-7. [DOI] [PubMed]
- 16.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]; Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646-74. [DOI] [PubMed]
- 17.Sharieff W., Okawara G., Tsakiridis T., Wright J. Predicting 2-year survival for radiation regimens in advanced non-small cell lung cancer. Clin Oncol (R Coll Radiol) 2013;25(12):697–705. doi: 10.1016/j.clon.2013.07.011. [DOI] [PubMed] [Google Scholar]; Sharieff W, Okawara G, Tsakiridis T, Wright J. Predicting 2-year survival for radiation regimens in advanced non-small cell lung cancer. Clin Oncol (R Coll Radiol). 2013;25(12):697-705. [DOI] [PubMed]
- 18.Malvezzi M., Bertuccio P., Rosso T., Rota M., Levi F., La Vecchia C. European cancer mortality predictions for the year 2015: does lung cancer have the highest death rate in EU women? Ann. Oncol. 2015;26(4):779–786. doi: 10.1093/annonc/mdv001. [DOI] [PubMed] [Google Scholar]; Malvezzi M, Bertuccio P, Rosso T, Rota M, Levi F, La Vecchia C, et al. European cancer mortality predictions for the year 2015: does lung cancer have the highest death rate in EU women? Ann Oncol. 2015;26(4):779-86. [DOI] [PubMed]
- 19.Printz C. Lung cancer mortality highest for black individuals in the most segregated counties. Cancer. 2013;119(11):1927. doi: 10.1002/cncr.28173. [DOI] [PubMed] [Google Scholar]; Printz C. Lung cancer mortality highest for black individuals in the most segregated counties. Cancer. 2013;119(11):1927. [DOI] [PubMed]
- 20.Priller C., Bauer T., Mitteregger G., Krebs B., Kretzschmar H.A., Herms J. Synapse formation and function is modulated by the amyloid precursor protein. J. Neurosci. 2006;26(27):7212–7221. doi: 10.1523/JNEUROSCI.1450-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]; Priller C, Bauer T, Mitteregger G, Krebs B, Kretzschmar HA, Herms J. Synapse formation and function is modulated by the amyloid precursor protein. J Neurosci. 2006;26(27):7212-21. [DOI] [PMC free article] [PubMed]
- 21.Turner P.R., O'Connor K., Tate W.P., Abraham W.C. Roles of amyloid precursor protein and its fragments in regulating neural activity, plasticity and memory. Prog. Neurobiol. 2003;70(1):1–32. doi: 10.1016/s0301-0082(03)00089-3. [DOI] [PubMed] [Google Scholar]; Turner PR, O'Connor K, Tate WP, Abraham WC. Roles of amyloid precursor protein and its fragments in regulating neural activity, plasticity and memory. Prog Neurobiol. 2003;70(1):1-32. [DOI] [PubMed]
- 22.Duce J.A., Tsatsanis A., Cater M.A., James S.A., Robb E., Wikhe K. Iron-export ferroxidase activity of beta-amyloid precursor protein is inhibited by zinc in Alzheimer's disease. Cell. 2010;142(6):857–867. doi: 10.1016/j.cell.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; Duce JA, Tsatsanis A, Cater MA, James SA, Robb E, Wikhe K, et al. Iron-export ferroxidase activity of beta-amyloid precursor protein is inhibited by zinc in Alzheimer's disease. Cell. 2010;142(6):857-67. [DOI] [PMC free article] [PubMed]
- 23.Gegelashvili G., Bock E., Schousboe A., Linnemann D. Two types of amyloid precursor protein (APP) mRNA in rat glioma cell lines: upregulation via a cyclic AMP-dependent pathway. Brain Res. Mol. Brain Res. 1996;37(1–2):151–156. doi: 10.1016/0169-328x(95)00302-9. [DOI] [PubMed] [Google Scholar]; Gegelashvili G, Bock E, Schousboe A, Linnemann D. Two types of amyloid precursor protein (APP) mRNA in rat glioma cell lines: upregulation via a cyclic AMP-dependent pathway. Brain Res Mol Brain Res. 1996;37(1-2):151-6. [DOI] [PubMed]
- 24.Hansel D.E., Rahman A., Wehner S., Herzog V., Yeo C.J., Maitra A. Increased expression and processing of the Alzheimer amyloid precursor protein in pancreatic cancer may influence cellular proliferation. Cancer Res. 2003;63(21):7032–7037. [PubMed] [Google Scholar]; Hansel DE, Rahman A, Wehner S, Herzog V, Yeo CJ, Maitra A. Increased expression and processing of the Alzheimer amyloid precursor protein in pancreatic cancer may influence cellular proliferation. Cancer Res. 2003;63(21):7032-7. [PubMed]
- 25.Ko S.Y., Chang K.W., Lin S.C., Hsu H.C., Liu T.Y. The repressive effect of green tea ingredients on amyloid precursor protein (APP) expression in oral carcinoma cells in vitro and in vivo. Cancer Lett. 2007;245(1–2):81–89. doi: 10.1016/j.canlet.2005.12.029. [DOI] [PubMed] [Google Scholar]; Ko SY, Chang KW, Lin SC, Hsu HC, Liu TY. The repressive effect of green tea ingredients on amyloid precursor protein (APP) expression in oral carcinoma cells in vitro and in vivo. Cancer Lett. 2007;245(1-2):81-9. [DOI] [PubMed]
- 26.Pandey P., Sliker B., Peters H.L., Tuli A., Herskovitz J., Smits K. Amyloid precursor protein and amyloid precursor-like protein 2 in cancer. Oncotarget. 2016;7(15):19430–19444. doi: 10.18632/oncotarget.7103. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pandey P, Sliker B, Peters HL, Tuli A, Herskovitz J, Smits K, et al. Amyloid precursor protein and amyloid precursor-like protein 2 in cancer. Oncotarget. 2016;7(15):19430-44. [DOI] [PMC free article] [PubMed]
- 27.Monni O., Barlund M., Mousses S., Kononen J., Sauter G., Heiskanen M. Comprehensive copy number and gene expression profiling of the 17q23 amplicon in human breast cancer. Proc. Natl. Acad. Sci. U. S. A. 2001;98(10):5711–5716. doi: 10.1073/pnas.091582298. [DOI] [PMC free article] [PubMed] [Google Scholar]; Monni O, Barlund M, Mousses S, Kononen J, Sauter G, Heiskanen M, et al. Comprehensive copy number and gene expression profiling of the 17q23 amplicon in human breast cancer. Proc Natl Acad Sci U S A. 2001;98(10):5711-6. [DOI] [PMC free article] [PubMed]
- 28.Lin H.C., Yeh C.W., Chen Y.F., Lee T.T., Hsieh P.Y., Rusnac D.V. C-terminal end-directed protein elimination by CRL2 ubiquitin ligases. Mol. Cell. 2018;70(4):602–613. doi: 10.1016/j.molcel.2018.04.006. [e3] [DOI] [PMC free article] [PubMed] [Google Scholar]; Lin HC, Yeh CW, Chen YF, Lee TT, Hsieh PY, Rusnac DV, et al. C-Terminal End-Directed Protein Elimination by CRL2 Ubiquitin Ligases. Mol Cell. 2018;70(4):602-13 e3. [DOI] [PMC free article] [PubMed]
- 29.Lamb J.R., Tugendreich S., Hieter P. Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem. Sci. 1995;20(7):257–259. doi: 10.1016/s0968-0004(00)89037-4. [DOI] [PubMed] [Google Scholar]; Lamb JR, Tugendreich S, Hieter P. Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem Sci. 1995;20(7):257-9. [DOI] [PubMed]
- 30.Kitamura K., Katayama S., Dhut S., Sato M., Watanabe Y., Yamamoto M. Phosphorylation of Mei2 and Ste11 by Pat1 kinase inhibits sexual differentiation via ubiquitin proteolysis and 14-3-3 protein in fission yeast. Dev. Cell. 2001;1(3):389–399. doi: 10.1016/s1534-5807(01)00037-5. [DOI] [PubMed] [Google Scholar]; Kitamura K, Katayama S, Dhut S, Sato M, Watanabe Y, Yamamoto M, et al. Phosphorylation of Mei2 and Ste11 by Pat1 kinase inhibits sexual differentiation via ubiquitin proteolysis and 14-3-3 protein in fission yeast. Dev Cell. 2001;1(3):389-99. [DOI] [PubMed]
- 31.Loiseau P., Davies T., Williams L.S., Mishima M., Palacios I.M. Drosophila PAT1 is required for Kinesin-1 to transport cargo and to maximize its motility. Development. 2010;137(16):2763–2772. doi: 10.1242/dev.048108. [DOI] [PMC free article] [PubMed] [Google Scholar]; Loiseau P, Davies T, Williams LS, Mishima M, Palacios IM. Drosophila PAT1 is required for Kinesin-1 to transport cargo and to maximize its motility. Development. 2010;137(16):2763-72. [DOI] [PMC free article] [PubMed]
- 32.Cardoso M., Paulo P., Maia S., Teixeira M.R. Truncating and missense PPM1D mutations in early-onset and/or familial/hereditary prostate cancer patients. Genes Chromosomes Cancer. 2016;55(12):954–961. doi: 10.1002/gcc.22393. [DOI] [PubMed] [Google Scholar]; Cardoso M, Paulo P, Maia S, Teixeira MR. Truncating and missense PPM1D mutations in early-onset and/or familial/hereditary prostate cancer patients. Genes Chromosomes Cancer. 2016;55(12):954-61. [DOI] [PubMed]
- 33.Yang S., Dong S., Qu X., Zhong X., Zhang Q. Clinical significance of Wip1 overexpression and its association with the p38MAPK/p53/p16 pathway in NSCLC. Mol. Med. Rep. 2017;15(2):719–723. doi: 10.3892/mmr.2016.6032. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yang S, Dong S, Qu X, Zhong X, Zhang Q. Clinical significance of Wip1 overexpression and its association with the p38MAPK/p53/p16 pathway in NSCLC. Mol Med Rep. 2017;15(2):719-23. [DOI] [PMC free article] [PubMed]
- 34.Oghabi Bakhshaiesh T., Majidzadeh A.K., Esmaeili R. Wip1: a candidate phosphatase for cancer diagnosis and treatment. DNA Repair (Amst) 2017;54:63–66. doi: 10.1016/j.dnarep.2017.03.004. [DOI] [PubMed] [Google Scholar]; Oghabi Bakhshaiesh T, Majidzadeh AK, Esmaeili R. Wip1: A candidate phosphatase for cancer diagnosis and treatment. DNA Repair (Amst). 2017;54:63-6. [DOI] [PubMed]
- 35.Lin D.L., Chang C. p53 is a mediator for radiation-repressed human TR2 orphan receptor expression in MCF-7 cells, a new pathway from tumor suppressor to member of the steroid receptor superfamily. J. Biol. Chem. 1996;271(25):14649–14652. doi: 10.1074/jbc.271.25.14649. [DOI] [PubMed] [Google Scholar]; Lin DL, Chang C. p53 is a mediator for radiation-repressed human TR2 orphan receptor expression in MCF-7 cells, a new pathway from tumor suppressor to member of the steroid receptor superfamily. J Biol Chem. 1996;271(25):14649-52. [DOI] [PubMed]
- 36.Hsu J.I., Dayaram T., Tovy A., De Braekeleer E., Jeong M., Wang F. PPM1D mutations drive clonal Hematopoiesis in response to cytotoxic chemotherapy. Cell Stem Cell. 2018;23(5):700–713. doi: 10.1016/j.stem.2018.10.004. [e6] [DOI] [PMC free article] [PubMed] [Google Scholar]; Hsu JI, Dayaram T, Tovy A, De Braekeleer E, Jeong M, Wang F, et al. PPM1D Mutations Drive Clonal Hematopoiesis in Response to Cytotoxic Chemotherapy. Cell Stem Cell. 2018;23(5):700-13 e6. [DOI] [PMC free article] [PubMed]
- 37.Lindsley R.C., Saber W., Mar B.G., Redd R., Wang T., Haagenson M.D. Prognostic mutations in Myelodysplastic syndrome after stem-cell transplantation. N. Engl. J. Med. 2017;376(6):536–547. doi: 10.1056/NEJMoa1611604. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lindsley RC, Saber W, Mar BG, Redd R, Wang T, Haagenson MD, et al. Prognostic Mutations in Myelodysplastic Syndrome after Stem-Cell Transplantation. N Engl J Med. 2017;376(6):536-47. [DOI] [PMC free article] [PubMed]
- 38.Zhang J., Bu X., Wang H., Zhu Y., Geng Y., Nihira N.T. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature. 2018;553(7686):91–95. doi: 10.1038/nature25015. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhang J, Bu X, Wang H, Zhu Y, Geng Y, Nihira NT, et al. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature. 2018;553(7686):91-5. [DOI] [PMC free article] [PubMed]
- 39.Kim M.S., Kim M.S., Yoo N.J., Lee S.H. Somatic mutation of SPOP tumor suppressor gene is rare in breast, lung, liver cancers, and acute leukemias. APMIS. 2014;122(2):164–166. doi: 10.1111/apm.12108. [DOI] [PubMed] [Google Scholar]; Kim MS, Kim MS, Yoo NJ, Lee SH. Somatic mutation of SPOP tumor suppressor gene is rare in breast, lung, liver cancers, and acute leukemias. APMIS. 2014;122(2):164-6. [DOI] [PubMed]
- 40.Devlin H.L., Mudryj M. Progression of prostate cancer: multiple pathways to androgen independence. Cancer Lett. 2009;274(2):177–186. doi: 10.1016/j.canlet.2008.06.007. [DOI] [PubMed] [Google Scholar]; Devlin HL, Mudryj M. Progression of prostate cancer: multiple pathways to androgen independence. Cancer Lett. 2009;274(2):177-86. [DOI] [PubMed]
- 41.Lu S., Tsai S.Y., Tsai M.J. Regulation of androgen-dependent prostatic cancer cell growth: androgen regulation of CDK2, CDK4, and CKI p16 genes. Cancer Res. 1997;57(20):4511–4516. [PubMed] [Google Scholar]; Lu S, Tsai SY, Tsai MJ. Regulation of androgen-dependent prostatic cancer cell growth: androgen regulation of CDK2, CDK4, and CKI p16 genes. Cancer Res. 1997;57(20):4511-6. [PubMed]
- 42.Chaikovsky A.C., Sage J. Beyond the cell cycle: enhancing the immune surveillance of Tumors via CDK4/6 inhibition. Mol. Cancer Res. MCR. 2018;16(10):1454–1457. doi: 10.1158/1541-7786.MCR-18-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chaikovsky AC, Sage J. Beyond the Cell Cycle: Enhancing the Immune Surveillance of Tumors Via CDK4/6 Inhibition. Molecular cancer research : MCR. 2018;16(10):1454-7. [DOI] [PMC free article] [PubMed]
- 43.Li G., Ci W., Karmakar S., Chen K., Dhar R., Fan Z. SPOP promotes tumorigenesis by acting as a key regulatory hub in kidney cancer. Cancer Cell. 2014;25(4):455–468. doi: 10.1016/j.ccr.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; Li G, Ci W, Karmakar S, Chen K, Dhar R, Fan Z, et al. SPOP promotes tumorigenesis by acting as a key regulatory hub in kidney cancer. Cancer Cell. 2014;25(4):455-68. [DOI] [PMC free article] [PubMed]
- 44.Chauhan A., Bhattacharyya S., Ojha R., Mandal A.K., Singh S.K. Speckle-type POZ protein as a diagnostic biomarker in renal cell carcinoma. J. Cancer Res. Ther. 2018;14(5):977–982. doi: 10.4103/jcrt.JCRT_942_15. [DOI] [PubMed] [Google Scholar]; Chauhan A, Bhattacharyya S, Ojha R, Mandal AK, Singh SK. Speckle-type POZ protein as a diagnostic biomarker in renal cell carcinoma. J Cancer Res Ther. 2018;14(5):977-82. [DOI] [PubMed]
- 45.Zhao W., Zhou J., Deng Z., Gao Y., Cheng Y. SPOP promotes tumor progression via activation of beta-catenin/TCF4 complex in clear cell renal cell carcinoma. Int. J. Oncol. 2016;49(3):1001–1008. doi: 10.3892/ijo.2016.3609. [DOI] [PubMed] [Google Scholar]; Zhao W, Zhou J, Deng Z, Gao Y, Cheng Y. SPOP promotes tumor progression via activation of beta-catenin/TCF4 complex in clear cell renal cell carcinoma. Int J Oncol. 2016;49(3):1001-8. [DOI] [PubMed]
- 46.Ding M., Lu X., Wang C., Zhao Q., Ge J., Xia Q. The E2F1-miR-520/372/373-SPOP Axis modulates progression of renal carcinoma. Cancer Res. 2018;78(24):6771–6784. doi: 10.1158/0008-5472.CAN-18-1662. [DOI] [PubMed] [Google Scholar]; Ding M, Lu X, Wang C, Zhao Q, Ge J, Xia Q, et al. The E2F1-miR-520/372/373-SPOP Axis Modulates Progression of Renal Carcinoma. Cancer Res. 2018;78(24):6771-84. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2