Abstract
During vertebrate embryogenesis, the neuroectoderm is induced from dorsal ectoderm and then partitioned into anterior and posterior neuroectodermal domains by posteriorizing signals, such as Wnt and fibroblast growth factor. However, little is known about epigenetic regulation of posteriorizing gene expression. Here, we report a requirement of the chromatin remodeling protein Bptf for neuroectodermal posteriorization in zebrafish embryos. Knockdown of bptf leads to an expansion of the anterior neuroectoderm at the expense of the posterior ectoderm. Bptf functionally and physically interacts with p-Smad2, which is activated by non-Nodal TGF-β signaling, to promote the expression of wnt8a, a critical gene for neural posteriorization. Bptf and Smad2 directly bind to and activate the wnt8a promoter through recruiting NURF remodeling complex. When bptf function or TGF-β signal transduction is inhibited, the nucleosome density on the wnt8a promoter is increased. We propose that Bptf and TGF-β/Smad2 mediate nucleosome remodeling to regulate wnt8a expression and hence neural posteriorization.
Keywords: bptf, neural posteriorization, nucleosome remodeling, smad2, wnt8a
Introduction
In vertebrates, the early development of the CNS during gastrulation is accomplished via a two-step mechanism. As the first step, the neuroectoderm with anterior character is induced from the dorsal ectoderm by antagonists of bone morphogenetic proteins, such as Chordin, Noggin, and Follistatin, which act to prevent the ventrally derived bone morphogenetic proteins to use the default neural fate (Lamb et al., 1993; Hemmati-Brivanlou et al., 1994; Sasai et al., 1995; Hammerschmidt et al., 1996; Schulte-Merker et al., 1997; Dal-Pra et al., 2006). As the second step, graded posteriorizing signals, including Wnt, fibroblast growth factor, and retinoic acid signals, transform the newly induced neuroectoderm into the posterior neuroectoderm, which ultimately gives rise to the posterior midbrain, the hindbrain, and the spinal cord (Cox and Hemmati-Brivanlou, 1995; Sasai and De Robertis, 1997; Niehrs, 1999; Sirotkin et al., 2000; Lekven et al., 2001; Erter et al., 2001; Rentzsch et al., 2004; Maden, 2006; Stern, 2006).
TGF-β/nodal/activin-related factors have pivotal roles in mesendoderm induction and dorsal axis determination (Zhou et al., 1993; Feldman et al., 1998), but their function in neural posteriorization is not well established. Overexpression of Antivin in zebrafish embryos, the potent antagonist of Activin ligands, results in obvious loss of posterior neural fates, but this effect may be indirect because mesendoderm induction is also abolished (Thisse et al., 2000). Although Smad2/3 activity is required for anterior–posterior (AP) patterning of the neuroectoderm in zebrafish, cell tracing experiments showed that the neuroectoderm of Nodal-deficient embryos undergoes an apparent anterior-to-posterior transformation (Erter et al., 2001; Jia et al., 2009).
Bptf is the largest subunit of the nucleosome remodeling factor (NURF) complex, which was first purified from Drosophila embryo extracts and recognized to catalyze nucleosome sliding in an ATP-dependent manner to assist transcriptional activation (Tsukiyama and Wu, 1995; Mizuguchi et al., 1997). The developmental roles of Bptf in vertebrates remain largely ambiguous. The anterior visceral endoderm and the primitive streak do not form correctly in Bptf mouse mutants, suggesting a possible role of Bptf in anterior–posterior patterning (Landry et al., 2008; Goller et al., 2008).
In this study, we find that bptf is a maternally and zygotically expressed gene during early stages of zebrafish embryonic development. Loss-of-function analyses indicate that bptf and smarca1are both required for neural posteriorization. Bptf interacts with endogenous Smad2, and their cooperative action under the control of non-Nodal TGF-β signaling is essential for establishing a proper AP neural pattern. Furthermore, we identify that wnt8a is the key downstream target of Bptf and TGF-β/Smad2. Bptf and Smad2 bind to adjacent binding motifs and recruit other NURF components, such as Smarca1, to decrease nucleosome density in the wnt8a promoter. Our data indicate that Bptf- and TGF-β/Smad2-regulated nucleosome remodeling events on cis-regulatory elements of the wnt8a promoter is indispensable for zebrafish neural posteriorization.
Materials and Methods
Zebrafish strains.
Tuebingen strain of zebrafish was used to obtain wild-type embryos. Embryos were maintained in Holtfreter's solution at 28.5°C and staged by morphology as previously described (Kimmel et al., 1995). MZoep mutant embryos were generated by crossing homozygotic male and female oeptz257/tz257 adult mutants, which were rescued by injection of oep mRNA (Gritsman et al., 1999). Homozygous p53(M214K) mutant fish line (abbreviated as p53−/−) carrying a loss-of-function p53 point mutation was kindly provided by Prof. Jinrong Peng at College of Life Sciences, Zhejiang University.
Embryonic treatment.
To inhibit TGF-β signal transduction, embryos at 16 cell stage were treated with 50 μm SB431542 under dark conditions and harvested at 75% epiboly stage for Western blot or in situ hybridization.
RNA synthesis, morpholinos, and whole-mount in situ hybridization.
The mRNAs encoding wnt8a, constitutively active smad2 (casmad2), dominant-negative smad2 (dnasmad2), dominant-negative TGF-β Receptor II (ΔkTβRII), smarca1, and dominant-negative smarca1 (smarca1K174R) were synthesized in vitro using the mMessage mMachine kit (Ambion). Digoxigenin-UTP-labeled antisense RNA probes were in vitro transcribed using MEGAscript Kit (Ambion) according to the manufacturer's instructions. siRNAs targeting all the isoforms from zebrafish bptf gene (gene ID: 324479, NCBI) were designed by and purchased from GenePharma. The sequences of siRNAs were as follows: negative control, sense 5′-ACGUGACACGUUCGGAGAATT-3′, antisense 5′-UUCUUCGAACGUGUCACGUTT-3′; siRNA1, sense 5′- UUCGGUUUCAAGCUUCGGCTT-3′, antisense 5′- GCCGAAGCUUGAAACCGAATT-3′; siRNA2, sense 5′- AACACUGGAACUGAGCACCTT-3′, antisense 5′- GGUGCUCAGUUCCAGUGUUTT-3′.
Antisense morpholinos were designed by and purchased from Gene Tools and have the following sequences: bptf translation-blocking morpholino (bptf MO1, positioning around the translation start site [TSS] of bptf), 5′-GCGGCCTGCCTCGTCTCCCCCTCAT-3′; bptf splicing-blocking morpholino (bptf MO2, targeting the splicing region between exon1 and intron1), 5′-TCCGACGAAGCGTCCGTACCTGTGT-3′; reverse control morpholino of bptf MO1 (cMO), 5′-TGTGTCCATGCCTGCGAAGCAGCCT-3′. For testing the effectiveness of bptf MO1, the expression vector bptf-GFP was generated by fusing the 23 bp upstream flanking region and the first 27 bp of the bptf open reading frame into a pEGFP-N1 vector. Microinjection and whole-mount in situ hybridization was performed as previously described (Jia et al., 2008).
Western blot and coimmunoprecipitation.
For Western blot, we used affinity-purified anti-Smad2/3 (3102, Cell Signaling Technology), anti-p-Smad2 (9510S, Cell Signaling Technology), and anti-β-actin (sc-1615, Santa Cruz Biotechnology) antibodies. For coimmunoprecipitation assays to study protein-protein interaction, embryos or HEK293T cells were harvested and lysed with TNE lysis buffer (10 mm Tris-HCl, pH 7.5, 150 mm NaCl, 2 mm EDTA, and 0.5% Nonidet P-40) containing a protease inhibitor mixture. The lysates were incubated with protein A Sepharose beads and either anti-Smad2/3 or anti-Myc (562–5, MBL) antibody at 4°C for 4 h. The beads were washed four times with TNE buffer. The bound proteins were separated by SDS-PAGE and visualized by Western blot.
Antibody generation.
The rabbit polyclonal anti-Bptf antibody was generated by our laboratory. An epitope corresponding to residues RGRRGRPPKAQLVQEC of zebrafish Bptf was chosen for immunization. This polyclonal antibody was affinity purified and validated for specificity by peptide competition assays. The purified antibody (concentration, 200 μg/ml) was used at a dilution of 1:2000 for Western blot and 1:100 for protein coimmunoprecipitation and chromatin immunoprecipitation (ChIP) assays in this study.
Dual-luciferase reporter assays.
For detection and quantification of Smad2 activity in zebrafish embryos, the Smad2-specific ARE luciferase reporter construct DNA was mixed with Renilla luciferase reporter DNA in a ratio of 10:1. Wild-type and MZoep mutant embryos were injected with 100 pg of the DNA mixture at the one-cell stage. To inhibit TGF-β signal transduction, the injected MZoep mutant embryos were incubated with 50 μm SB431542 (S4317, Sigma) from 16-cell stage under dark conditions. For analysis of promoter activity, serial truncations of the wnt8a promoter reporter were generated and injected into embryos together with indicated MOs and RNAs. A group of 20 embryos for each sample were harvested at 75% epiboly stage and lysed with passive lysis buffer (Promega) for detecting luciferase activities.
For the luciferase reporter assays performed in cell culture, HEK293 cells were transfected with the indicated constructs together with Renilla luciferase reporter DNA (20 ng) as the internal control. Cells were stimulated with TGF-β1 (1 ng/ml) or Activin A (10 ng/ml) for 16 h before harvest for luciferase assay.
Each experiment was performed in triplicate, and the data represent the mean ± SD of three independent experiments after normalized to Renilla luciferase activity.
ChIP.
Embryos were injected with indicated MOs or RNAs at one-cell stage and ∼400 embryos were harvested at 75% epiboly stage for each assay. Then these embryos were crosslinked with 1.85% for 15 min at room temperature and lysed. DNA/protein complex-containing lysate was sonicated to an apparent length of 300–1000 bp and then centrifuged at 14,000 rpm for 10 min. The indicated antibody (anti-Smad2/3 or anti-BPTF) was incubated with the diluted chromatin supernatant and BSA-blocked protein A beads overnight at 4°C. The immunocomplex was washed sequentially with salt buffers and then eluted in elution buffer. The immunoprecipitated complexes and input fraction were reverse-crosslinked by overnight incubation at 65°C. DNA was extracted with phenol:chloroform and precipitated with ethanol after treatment with RNaseA and proteinase K. Input and immunoprecipitated DNA were subjected to PCR cycles with the following specific primers: wnt8a up forward primer, 5′-CTACACATTTCATACACATCGTTGA-3′ (−1072 bp to −1048 bp, upstream region of wnt8a TSS); wnt8a up reverse primer, 5′-TGGGAAAAGCTCTGGTGTGAAAC-3′ (−914 bp to −892 bp, upstream region of wnt8a TSS); wnt8a down forward primer, 5′-CAAGCACGGAAGTTGGAGATGGATA-3′ (643 bp to 667 bp, downstream region of wnt8a TSS); wnt8a down reverse primer, 5′-TTCCCGCTTTGTAGACATTCCCT-3′ (820–842 bp, downstream region of wnt8a TSS).
MNase mapping assay.
Wild-type and bptf MOs or ΔkTβRII mRNA injected embryos were harvested at 75% epiboly stage, and mononucleosomes were prepared using Nucleosome Preparation Kit (5333, TaKaRa) according to the manufacturer's instructions. Then the resulting mononucleosome-sized DNA samples were analyzed by qRT-PCR with overlapping primer pairs covering the wnt8a promoter region from −1449 to −416 upstream of wnt8a TSS. Each set of primer pairs spanned across ∼100 bp region and were located 30 ± 10 bp away from neighboring primer pairs as previously described (Li et al., 2010).
Results
bptf is ubiquitously expressed during early embryo development
We found in a previous study that the DNA-binding motif recognized by the chromatin remodeling protein Bptf coexists at a high frequency with Smad2/4-binding sites in the zebrafish genome (Liu et al., 2011). Zebrafish bptf encodes a large protein of >2800 amino acid residues, which shares a high sequence similarity to its human and mouse orthologs (56.6% and 55.8%, respectively). The syntenic analysis of zebrafish bptf showed that its neighbor genes are also closely linked in other species, indicating that it is the authentic ortholog of mammalian Bptf (data not shown). In addition, like other vertebrate orthologs, zebrafish Bptf also has several conserved domains: an N-terminal DDT domain; two plant homeodomain finger domains, one of which mediates a direct association with H3K4me3; a glutamine-rich transcriptional activation domain; and a carboxy-terminal bromodomain that acts as an acetyl-lysine binding domain and preferentially recognizes H4K16ac (Escher et al., 2000; Zeng and Zhou, 2002; Wysocka et al., 2006; Ruthenburg et al., 2011).
We next examined the spatiotemporal expression of bptf in zebrafish embryos from one-cell stage to 30 h post-fertilization (hpf) by whole-mount in situ hybridization. As shown in Figure 1A, A′, bptf mRNA was ubiquitously distributed before mid-segmentation stages and then retained at high levels only in the head region from 24 hpf onward. We also generated zebrafish Bptf antibody to detect its protein expression at different stages. Western blotting revealed the existence of Bptf protein from blastula to mid-segmentation stages (Fig. 1B). These data indicates that bptf is both maternally and zygotically expressed during early embryo development.
Figure 1.
Zebrafish bptf is expressed during early embryonic development and involved in head patterning. A, A′, Expression pattern of zebrafish bptf at indicated stages, detected by in situ hybridization with antisense (A, A′) or sense (A) probe. From one-cell stage to 32-cell stage, animal views; from sphere to bud stages, lateral views with dorsal to the right; 10-somite stage, dorsal view with anterior to the top; 24 and 30 hpf, lateral views with anterior to the left. B, Western blot analysis of endogenous Bptf protein expression during zebrafish embryo development. C, Effectiveness of bptf MO1. Embryos coinjected with 50 pg bptf-GFP plasmid DNA and 4 ng cMO or MO1 at one-cell stage. Green fluorescence was detected at 75% epiboly stage. MO1-injected embryos showed a noticeable decrease in green fluorescence compared with control embryos. D, Effectiveness of bptf MO2. The specific splicing-interfering MO (bptf MO2, targeting the splicing region between exon1 and intron1) interfered bptf mRNA product is only detected in morphants, whereas endogenous mRNA level was significantly deceased. β-actin is used as a control. E, Embryos were injected with 4 ng bptf MO1, 4 ng bptf MO2, or their mix (bptf MOs, each 4 ng) and harvested for Western blotting at 75% epiboly stage. UIC, Uninjected control. Band densities were analyzed using Quantity One software. The numbers on top of each lane indicate the relative band densities and the mean ± SD of Bptf after normalization to β-actin from 4 independent experiments. F, Morphological effects of bptf MO-injected wild-type or p53 mutant embryos at 24 hpf. Note the enlarged forebrain in bptf morphants (arrowhead). The ratio of embryos with the representative phenotypes was indicated. Scale bar, 200 μm.
Then we adopted knockdown approach using two antisense morpholinos: the translation blocker bptf-MO1 and the splicing blocker bptf-MO2. We found that injection of 4 ng MO1 efficiently blocked the production of the Bptf-GFP fusion protein in embryos, whereas the reverse control MO (cMO) had no effect (Fig. 1C). Furthermore, the expression of endogenous bptf mRNA was eliminated by injection of 4 ng MO2, and the interfered mRNA product was only detected in morphants (Fig. 1D). We also examined whether the expression of endogenous Bptf protein was inhibited in morphants. Based on sequence information, the predicted molecular mass of zebrafish Bptf is ∼320 kDa. The Bptf antibody detected a dominant band slightly lower than expected size (∼250 kDa) in wild-type zebrafish embryo lysate. Various other bands were also detected from 80 to 130 kDa (Fig. 1E). Interestingly, all of the immunoactive bands were clearly weakened in embryo lysates from bptf morphants, suggesting the high specificity of this antibody (Fig. 1E). These faint secondary bands may represent Bptf degradation products as Bptf protein is sensitive to proteolysis (Xiao et al., 2001; Wysocka et al., 2006). More importantly, coinjection of MO1 (4 ng) and MO2 (4 ng) reduced Bptf production more efficiently (Fig. 1E). Then, coinjection of both MOs was used to knock down bptf in the subsequent experiments.
To determine the phenotypic consequences of loss of bptf function, we first examined the morphology of bptf morphants at 24 hpf. Injection bptf MOs to wild-type embryos resulted in obvious cell death in the head, a reduction in the size of the trunk and an enlargement of the forebrain (Fig. 1F). p53 mutant embryos injected with bptf MOs showed a more enlarged forebrain due to decreased cell apoptosis, excluding the possibility that the observed defects were caused by nonspecific p53 activation by morpholinos (Fig. 1F). These results indicate that bptf is involved in head patterning.
bptf is required for zebrafish neural posteriorization
To characterize the function of bptf during neural patterning along the AP axis, we examined the anterior neural markers otx2 and sox2, and the posterior neural marker hoxb1b respectively, at mid-gastrulation stages. As expected, bptf morphants showed a broader anterior neuroectoderm (Fig. 2A,B) and a much narrower and smaller posterior neuroectoderm (Fig. 2C). The increased expression of otx2 and sox2 in bptf morphants was further confirmed by RT-PCR (Fig. 2D). The expression pattern of sox3, which is expressed in both anterior and posterior neuroectoderm, was also examined in embryos at mid-gastrulation stages. The expression of sox3 in the anterior neuroectoderm was obviously expanded posteriorly, but its expression in the posterior neuroectoderm was shrunk in bptf morphants compared with control embryos (Fig. 2E). siRNA-based gene silencing has been proved to be an alternative technique for knockdown of specific gene expression in zebrafish (Dodd et al., 2004; Liu et al., 2005). To exclude the off-target effects of MO injection, we designed and injected two different siRNAs (siRNA1 and siRNA2) for targeting zebrafish bptf. As shown in Figure 2F, injection of siRNA2, although not siRNA1, dramatically decreased endogenous bptf expression. Embryos injected with siRNA2 exhibited obvious neural patterning defects similar to bptf morphants (Fig. 2G,H). In addition, injection bptf MOs into p53 mutant embryos also led to profound reduction of posterior neuroectoderm (Fig. 2I). Together, these results conclusively indicate that the neural patterning defects in bptf morphants are specific effects of bptf knockdown.
Figure 2.
bptf has crucial functions in neural posteriorization. A–E, Knockdown of bptf by morpholino injection blocks neural posteriorization. In situ hybridization of the anterior neural markers otx2 and sox2 (A, B, dorsal view with anterior at the top), the posterior neural marker hoxb1b, and the pan-neural marker sox3 (C, E, lateral views with dorsal to the right) at 75% epiboly stage. The expression of otx2 and sox2 in embryos injected with control MO or bptf MOs was analyzed at 75% epiboly stage by real-time PCR. *p < 0.05 (Student's t test). ***p < 0.001 (Student's t test). D, F–H, Knockdown of bptf by siRNA injection results in severe neural patterning defects. Embryos injected with 1.2 ng of indicated siRNAs at one-cell stage and harvested at 75% epiboly stage for real-time PCR (F) and in situ hybridization (G, H) analysis. NS, Nonsignificant. *p < 0.05 (Student's t test). F, I, The expression pattern of hoxb1b was analyzed by in situ hybridization in control MO and bptf MO-injected p53 mutant embryos at 75% epiboly stage. J, K, Abrogation of bptf function results in enlarged forebrain and loss of spinal cord. The expression pattern of six3 and shh was analyzed by in situ hybridization in control MO and bptf MO-injected embryos at the bud stage. J, Anterior view with dorsal at the top. K, Dorsal view, anterior is upward. L, M, Rescue of bptf morphants by hBptf. The expression of sox2 (L) and hoxb1b (M) at 75% epiboly stage in wild-type embryos injected with bptf MOs alone or together with 50 pg plasmid construct expressing hBptf. Black arrowheads indicate the representative spotted expression of hoxb1b.
Analysis of more differentiated anterior and posterior neural tissues with probes for six3 (forebrain marker) and shha (spinal cord marker) transcripts revealed the enlarged forebrain and severe loss of spinal cord in bptf morphants (Fig. 2J,K). As we were unable to synthesize bptf mRNA in vitro due to its large size (nearly 9000 nt), 50 pg plasmid construct expressing human Bptf (hBptf) in which the MO target sequences do not exist was coinjected with MOs into embryos at one-cell stage in rescue experiments (Loosli et al., 2003). As shown in Figure 2L, M, the expansion of anterior neuroectoderm was repressed and the decrease of posterior neuroectoderm was obviously restored by overexpression hBptf in bptf morphants. Interestingly, because of the inhomogeneous distribution of injected plasmid DNA, spotted expression of hoxb1 was observed in hBptf injected embryos (Fig. 2M). These results suggest that bptf is required for anterior-to-posterior transformation of the nascent neuroectoderm.
Bptf and Smad2 coregulate neural patterning in a Nodal-independent manner
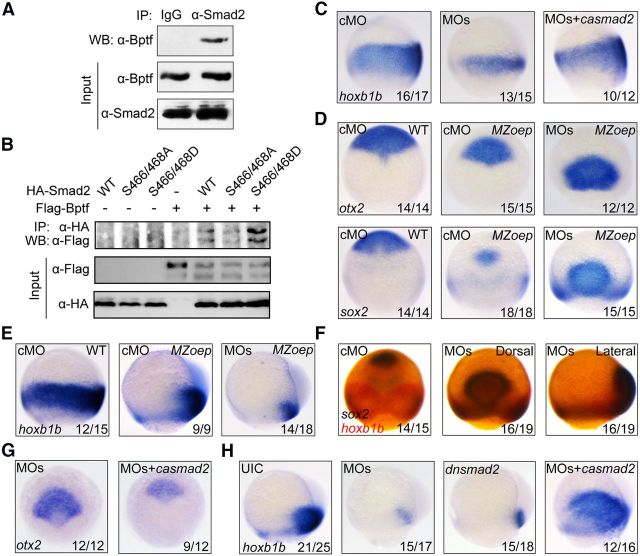
In the zebrafish embryo, Smad2/3 activities are demonstrated to posteriorize the neuroectoderm (Jia et al., 2009). We therefore asked whether Bptf physically and functionally interacts with Smad2 during the zebrafish neural patterning. The coimmunoprecipitation assays confirmed an association of endogenous Bptf and Smad2 in zebrafish embryos at mid-gastrulation stages (Fig. 3A). Importantly, coexpressed Bptf has much higher affinity for the phospho-mimetic Smad2(S466/468D) mutant compared with the wild-type Smad2 and phospho-resistant Smad2(S466/468A) mutant (Fig. 3B). Furthermore, bptf MOs-induced reduction of the hoxb1b domain could be rescued by coinjection of constitutively active smad2 (casmad2) mRNA (Fig. 3C). These results support an idea that Bptf cooperates with Smad2 to regulate neural patterning.
Figure 3.
Bptf associates with Smad2 to posteriorize the neuroectoderm independent of Nodal signaling. A, Zebrafish Bptf interacts with Smad2. Embryos were harvested and lysed at the 75% epiboly stage for coimmunoprecipitation using indicated antibodies. B, Bptf shows a much stronger binding affinity for activated Smad2. HEK293T cells were transfected with indicated expression plasmids encoding Flag-tagged Bptf, HA-tagged wild-type (WT) Smad2, phospho-resistant Smad2(S466/468A) mutant, and phospho-mimetic Smad2(S466/468D) mutant. Cells were harvested 48 h after transfection for immunoprecipitation with anti-HA antibody. C–E, Expression of neuroectodermal markers at 75% epiboly stage. Wild-type or MZoep embryos were injected with cMO or bptf MOs alone or together with casmad2 mRNA at the one-cell stage and harvested later for probing with indicated probes. Embryos were shown in lateral view (C,E) or dorsal view (D) with anterior to the top. F, Double in situ hybridization detection of sox2 (dark blue) and hoxb1b (red) in MZoep mutants injected with cMO or bptf MOs. The first two embryos were shown in dorsal view with anterior to the top and the last one in lateral view with dorsal to the right. G, H, The expression pattern of otx2 (G) and hoxb1b (H) in MZoep embryos. Injection doses: cMO, 8 ng; bptf MOs, 8 ng; casmad2 mRNA, 50 pg; dnsmad2 mRNA, 600 pg. Dorsal (G) or lateral views (H) are shown.
Smad2 is the intracellular mediator of TGF-β and Nodal signaling (Massagué et al., 2005; Wu and Hill, 2009) and is activated by Nodal signaling in early vertebrate embryos to induce mesendoderm and to promote body axis patterning (Nomura and Li, 1998; Tian and Meng, 2006; Jia et al., 2008). To determine whether Bptf and Smad2 coregulate neural patterning under the control of Nodal signaling, we first analyzed the formation of the anterior and the posterior neuroectoderm in Nodal-deficient MZoep mutants. We observed that the expression domains of both anterior (otx2 and sox2) and posterior (hoxb1b) neuroectoderm markers were decreased, but relatively well patterned alone the AP axis in mutant embryos compared with the wild-type embryos (Fig. 3D,E). We next addressed the functions of Bptf in MZoep embryos. When injected with bptf MOs, MZoep mutant embryos exhibited a marked expansion and a caudal shift of otx2 and sox2 expression in the anterior neuroectoderm with a much smaller domain of hoxb1b in the posterior neuroectoderm (Fig. 3D,E). Interestingly, a persistent expression of sox2 was detected in the ventrolateral hoxb1b-positive cells in bptf MO-injected MZoep mutants (Fig. 3D,F). It is possible that the residual posterior neuroectodermal cells may not have been fully transformed from the anterior neural fate or these cells may be in a transitional state with bipotency for differentiation at the examined stage.
Next, we wanted to determine whether Smad2 still coregulates neural patterning with Bptf in MZoep mutants. The posterior neuroectoderm was obviously reduced by overexpression of dominant-negative smad2 (dnsmad2) in MZoep mutants (Fig. 3H), suggesting that Smad2 still plays a role in neuroectoderm posteriorization. Coinjection of casmad2 mRNA repressed the expansion of the anterior neuroectoderm (Fig. 3G) and restored the posterior neuroectoderm (Fig. 3H) in bptf MO-injected MZoep mutants. These results imply that Bptf associates with Smad2 to promote posteriorization of the neuroectoderm in a Nodal-independent manner.
Non-Nodal TGF-β signaling plays a critical role in neural posteriorization
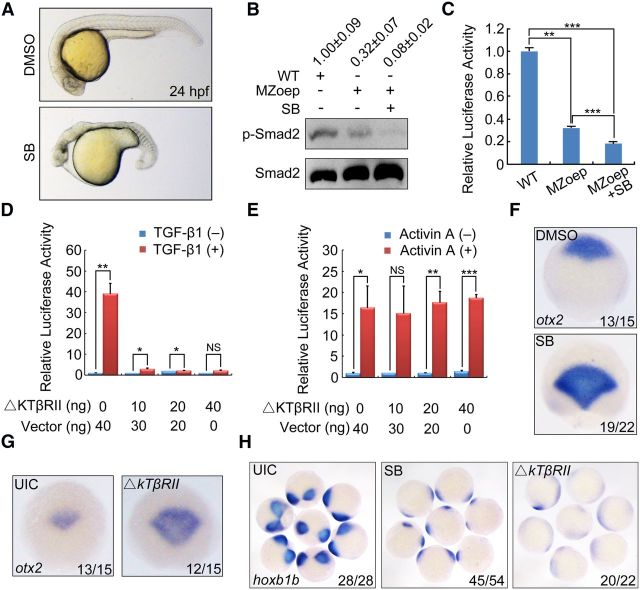
SB431542 is a specific inhibitor of TGF-β superfamily Type I activin receptor-like kinase (ALK) receptors, including ALK4, ALK5, and ALK7 (Inman et al., 2002). Embryos treated with 50 μm SB431542 from 16-cell stage to 24 hpf were missing most of mesendodermal tissues and had fused eyes, a phenotype resembling MZoep mutants (Fig. 4A) (Gritsman et al., 1999). We then examined phosphorylated-Smad2 (p-Smad2) by Western blotting to determine whether Smad2 is activated in MZoep mutants. Result indicated that p-Smad2 was indeed detected in MZoep mutants though much lower than in wild-type control (Fig. 4B). Importantly, the residual p-Smad2 in MZoep mutants could be largely reduced by treatment with SB431542 (Fig. 4B). The activation of Smad2 in MZoep embryos was also confirmed by Smad2-specific ARE luciferase reporter assays (Fig. 4C). These results suggest that Smad2 is activated by non-Nodal TGF-β signaling, which is consistent with the previous findings (Sun et al., 2006; Hagos et al., 2007).
Figure 4.
Inactivation of TGF-β signaling in MZoep mutant embryos leads to severe defects in neural AP patterning. A, Live embryos at 24 hpf. Embryos were treated with DMSO or 50 μm SB431542 (SB) from 16-cell stage to 24 hpf. The anterior is to the left. B, Detection of p-Smad2 by Western blotting in wild-type and MZoep mutant embryos. Embryos (WT and MZoep) were treated with DMSO or 50 μm SB431542 at 16-cell stage and harvested at 75% epiboly stage for Western blotting. Band densities were analyzed using Quantity One software. The numbers on top of each lane indicate the relative band densities and the mean ± SD of p-Smad2 after normalization to β-actin from 3 independent experiments. C, Reduction of ARE-luciferase reporter expression in MZoep mutant embryos treated with SB431542. WT or MZoep embryos were injected with the reporter plasmids at one-cell stage, treated with DMSO or SB431542 at 16-cell stage, and harvested at 75% epiboly stage for luciferase activity analysis. **p < 0.01 (Student's t test). ***p < 0.001 (Student's t test). D, E, ΔkTβRII overexpression specifically attenuates TGF-β-induced expression of ARE-luciferase. HEK293 cells were cotransfected with ARE-luciferase and FoxH1 constructs along with increasing amounts of ΔkTβRII construct, and treated with TGF-β1 (1 ng/ml) (D) or Activin A (10 ng/ml) (E) for 16 h before harvest for luciferase assay. *p < 0.05 (Student's t test), **p < 0.01 (Student's t test), ***p < 0.001 (Student's t test); NS, nonsignificant. F–H, Change of expression pattern of otx2 and hoxb1b at the 75% epiboly stage in MZoep embryos treated with 50 μm SB431542 or injected with 400 pg ΔkTβRII mNRA. Single embryos were shown in dorsal view for otx2 expression (F, G) and a group of embryos were shown in dorsal view mostly with posterior to the middle for hoxb1b expression (H).
Zebrafish tgfβ1a, tgfβ1b, tgfβ2, and tgfβ3 have been characterized (Kohli et al., 2003; Cheah et al., 2005; Cannon et al., 2013). tgfβ2 and tgfβ3, but not tgfβ1, have been previously shown to be expressed during gastrulation (Cheah et al., 2005; Meng A, Tsinghua University, unpublished data), which raises the possibility that TGF-β signal may have potent physiological roles in early embryonic development. Because a kinase domain truncated TGF-β Type II receptor (ΔkTβRII) selectively blocks TGF-β signaling as a dominant-negative mutant (Herskowitz, 1987; Chen et al., 1993; Brand et al., 1993), we constructed zebrafish ΔkTβRII mutant plasmid by deleting the cytoplasmic serine/threonine kinase domain. Luciferase reporter assay in HEK293 cells revealed that overexpression of ΔkTβRII significantly attenuated the TGF-β-induced expression of the ARE luciferase reporter (Fig. 4D), but had no obvious effects on Activin-induced luciferase activities (Fig. 4E), showing its functional specificity for inhibiting TGF-β signaling. SB431542-treated or ΔkTβRII mRNA-injected MZoep mutants showed greatly enlarged anterior neuroectoderm and abrogated posterior neuroectoderm (Fig. 4F–H). Consistently, similar results were observed in wild-type embryos injected with ΔkTβRII mRNA (data not shown). Therefore, we propose that it may be canonical TGF-β/Smad signaling that has an important function in transforming anterior neuroectoderm into posterior neuroectoderm.
The lateral margin mesodermal expression of wnt8a is regulated by bptf and TGF-β/Smad2
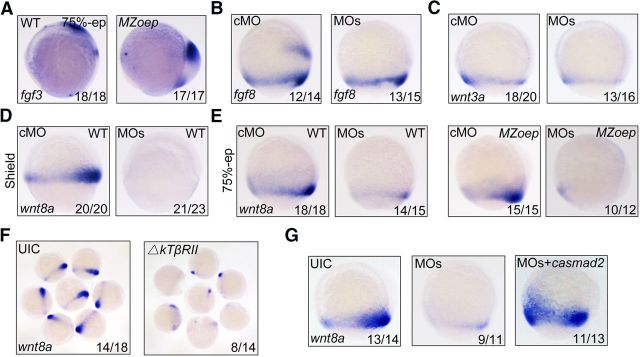
In zebrafish, the lateral/paraxial mesodermal precursors provide crucial neural transformers, such as Wnt and fibroblast growth factor ligands, to posteriorize the nascent anterior neuroectoderm during gastrulation (Woo and Fraser, 1997; Erter et al., 2001; Lekven et al., 2001; Koshida et al., 2002). To test whether the expression of these neural transformers is regulated by bptf and TGF-β/Smad2, we analyzed the expression patterns of fgf3, fgf8a, wnt3a, and wnt8a, which are prominently expressed during gastrulation (Buckles et al., 2004; Shimizu et al., 2005), in wild-type, MZoep mutant and bptf MO-injected embryos. The potential downstream target genes of bptf and TGF-β/Smad2 during neural patterning process should meet the following conditions: expressed in lateral margin mesoderm at mid-gastrulation stages and coregulated by bptf and TGF-β/Smad2 signaling, although not by Nodal signaling. We observed that fgf3 transcript was not expressed in the blastoderm margin at 75% epiboly stage in wild-type embryos and MZoep mutants (Fig. 5A). The expression of fgf8a and wnt3a was observed in the blastoderm margin at mid-gastrula stage, but their expression levels were not obviously downregulated in bptf morphants (Fig. 5B,C). In wild-type and MZoep mutant embryos, wnt8a showed a similar expression pattern in ventrolateral mesodermal precursors during gastrulation (Fig. 5E). Unlike fgf8a and wnt3a, however, wnt8a expression was abolished in bptf MO-injected wild-type and MZoep embryos (Fig. 5D,E), suggesting that its expression is regulated by bptf but not Nodal signaling. Ectopic expression of ΔkTβRII effectively inhibited wnt8a expression in MZoep mutants (Fig. 5F), and overexpression of casmad2 well restored wnt8a expression in bptf MO-injected MZoep mutants (Fig. 5G), which further supported an important role of TGF-β/Smad2 signaling for wnt8a expression. Hence, wnt8a is a target gene of bptf and TGF-β/Smad2.
Figure 5.
Identification of Bptf and TGF-β/Smad2 coregulated genes during neural posteriorization. A–C, The expression pattern of fgf3, fgf8a, and wnt3a at 75% epiboly stages in wild-type, MZoep mutant, and bptf MOs injected embryos. All embryos were shown in lateral view with dorsal to the right. D, E, The expression of wnt8a is regulated by bptf, but not Nodal, signaling. wnt8a expression was assessed at shield (D) and 75% epiboly stages (E) by in situ hybridization in wild-type (D, E) and MZoep (E) embryos injected with cMO or bptf MOs. F, Reduction of wnt8a expression in ΔkTβRII mRNA-injected MZoep mutant embryos. A group of embryos at 75% epiboly stage were shown. G, Ectopic expression of casmad2 restores bptf MOs-induced decrease of wnt8a expression. MZoep mutant embryos were injected with 8 ng bptf MOs alone or together with 50 pg casmad2 mRNA at the one-cell stage and harvested at 75% epiboly stage for in situ hybridization.
wnt8a acts at downstream of bptf and TGF-β/Smad2 to induce the posterior neural fates
To detect whether Wnt signaling is affected by bptf knockdown during neural posteriorization, we analyzed gfp reporter mRNA expression in TOPdGFP transgenic embryos, which mirrors endogenous Wnt/β-catenin activity (Dorsky et al., 2002). Knockdown of bptf caused a drastic decrease of dGFP expression in ventrolateral mesoderm from early to mid-gastrula stages (Fig. 6A), demonstrating a requirement of bptf for efficiently activating Wnt/β-catenin signaling. We then asked whether the loss of wnt8a expression accounted for bptf knockdown- or TGF-β signal inhibition-induced AP patterning defects of the neuroectoderm. Coinjection of wnt8a mRNA largely normalized otx2 and hoxb1b expression in bptf MO-injected wild-type embryos (Fig. 6B). Moreover, overexpression of wnt8a rescued the neural patterning defects caused by coinjection of bptf MOs and ΔkTβRII mRNA into wild-type embryos (Fig. 6C). Similar results were observed in further experiments using MZoep mutants (Fig. 6D). These data substantiate that function of bptf and TGF-β/Smad2 in posteriorizing the neuroectoderm is exerted by the downstream target wnt8a.
Figure 6.

Bptf and TGF-β/Smad2 posteriorize the neuroectoderm via wnt8a. A, Wnt/β-catenin signaling is reduced in bptf morphants. gfp expression was examined at shield and 75% epiboly stages by in situ hybridization in TOPdGFP transgenic fish embryos injected with cMO or bptf MOs. Top, Animal-pole views with dorsal to the right. Bottom, Lateral views with dorsal to the right. B, Expression of otx2 and hoxb1b at 75% epiboly stage in wild-type embryos injected with bptf MOs alone or together with 25 pg wnt8a mRNA. Injection of wnt8a mRNA could compromise the expansion of the anterior neuroectoderm and the loss of posterior neuroectoderm in bptf morphants. C, D, Neural patterning defects resulting from inactivation of both bptf and TGF-β signaling were rescued by coinjection of wnt8a mRNA. Wild-type embryos (C) or MZoep mutants (D) were injected with indicated MOs or RNAs at the one-cell stage and harvested at 75% epiboly stage for in situ hybridization using otx2 and hoxb1b probes. Injection doses were as follows: bptf MOs, 8 ng; ▵kTβRII mRNA, 400 pg; wnt8a mRNA, 25 pg.
Bptf and Smad2 bind to and activate the wnt8a promoter
Zebrafish wnt8a produces a bicistronic transcript, containing two open reading frames (wnt8a.1 and wnt8a.2) (Lekven et al., 2001; Ramel and Lekven, 2004). The expression of wnt8a transcripts in ventrolateral blastodermal margin is first detected at the 30% epiboly stage and maintained during gastrulation (Kelly et al., 1995; Ramel and Lekven, 2004). To further investigate the transcriptional regulation of wnt8a by Bptf and Smad2, the putative 1101-bp wnt8a promoter (1101-P-wnt8a), which possesses a margin-specific enhancer activity sufficient to drive wnt8a expression in ventrolateral mesoderm during neural AP patterning (Narayanan and Lekven, 2012), was cloned into pGL3-Basic luciferase reporter plasmid to make the construct 1101-P-luc (Fig. 7A). The transcriptional activity of 1101-P-luc was reduced by nearly 10-fold in bptf morphants, which suggests that the 1101 bp wnt8a promoter responds well to Bptf activity (Fig. 7B). Next, serial promoter truncations were generated and injected into embryos together with cMO or bptf MOs (Fig. 7A). Results showed that the transcriptional activity of the truncated promoters lacking the distal region of 177 bp was much lower and lost the response to bptf knockdown (Fig. 7B). As Bptf can regulate transcription through direct binding to DNA (Jordan-Sciutto et al., 1999; Jones et al., 2000), we searched consensus Bptf binding motifs (5′-NNMACAACAMN-3′) in that 177 bp region using Genomatix MatInspector (Genomatix database; http://www.genomatix.de), and found a candidate site (5′-ACAAAAACACT-3′, positioning from −961 to −951). The AACA>GGAG mutation within this putative Bptf-binding site of 1101-P-luc construct, generating the mutant construct 1101-mb-P-luc (Fig. 7A), led to a remarkable reduction of reporter expression in wild-type embryos, which was not deteriorated by coinjection of bptf MOs (Fig. 7C). These results indicate that the identified Bptf binding site is critical for wnt8a transcription.
Figure 7.
Bptf and Smad2 regulate wnt8a expression through distinct cis-elements. A, Schematic illustration of wnt8a promoter-driven luciferase reporters. B, C, The Bptf-binding element is essential for wnt8a transcription. Wild-type embryos were injected with indicated MOs and wnt8a promoter-driven reporter constructs at one-cell stage and then harvested and lysed at 75% epiboly stage for luciferase activity analysis. The −1101 to −924 promoter region is important for wnt8a transcription (B). 1101-mb-P-luc showed a much lower luciferase activity than the corresponding wild-type reporter (C). D, The −924 to −657 region of the wnt8a promoter is essential for responding to casmad2 overexpression. The indicated constructs and casmad2 mRNA were injected into embryos at one-cell stage, and the relative luciferase activity was measured at 75% epiboly stage. E, The Smad2/4-binding site is essential for responding to casmad2 overexpression. 1101-ms-P-luc or the related wild-type reporter 1101-P-luc was injected into embryos, and their responsiveness to overexpressed casmad2 was measured. Data represent the mean ± SD for triplicate experiments. *p < 0.05 (Student's t test). **p < 0.01 (Student's t test). ***p < 0.001 (Student's t test). F, Bptf and Smad2 specifically bind to the wnt8a promoter. ChIP assays were performed with control IgG and antibodies against Bptf or Smad2/3 in wild-type embryos. The immunoprecipitated DNA was amplified by semiqPCR with the primers recognizing specific regions denoted in the top. wnt8a up, a region upstream of the wnt8a TSS, which contains Bptf and Smad2/4 binding motifs; wnt8a down, a region downstream of the wnt8a TSS. G, ChIP experiments were performed in wild-type, ΔkTβRII mRNA-injected, or SB431542-treated embryos using anti-Bptf antibody. The immunoprecipitated DNA was amplified by qPCR with primers to detect the region upstream of the wnt8a TSS. NS, Nonsignificant. H, I, Wild-type embryos were injected with cMO or bptf MOs at the one-cell stage and harvested at 75% epiboly stage for ChIP experiments with anti-Bptf (H) or anti-Smad2/3 (I) antibodies. The immunoprecipitated DNA was subjected to qPCR with primers indicated in G.
Similar analysis was performed to search Smad2 binding sites in the wnt8a promoter. Luciferase reporter assays in embryos revealed that the region of −924 to −657 was required for response to stimulation by casmad2 overexpression (Fig. 7D). Subsequent sequence analysis and reporter assays identified the Smad2/4 binding site (5′-CAGAC-3′) from −921 to −917 as an essential regulatory element (Fig. 7E).
To verify the in vivo binding of Bptf and Smad2 to the wnt8a promoter, we performed ChIP assays in zebrafish embryos using Bptf and Smad2 antibodies respectively. As shown in Figure 7F, endogenous Bptf and Smad2 were bound to the −1072 to −892 region of the wnt8a promoter, but not to a control region (643 to 842), indicating that wnt8a is a direct target of Bptf and TGF-β/Smad2 in zebrafish embryos. We further tested whether activated Smad2 and Bptf were mutually required for associating with the wnt8a promoter. As shown in Figure 7G, inhibition of TGF-β signaling by SB435412 treatment or ΔkTβRII mRNA overexpression did not affect binding of Bptf to the wnt8a promoter. bptf knockdown obviously decreased the binding of Bptf to wnt8a promoter (Fig. 7H) but did not disrupt binding of Smad2 (Fig. 7I). These results suggest that Bptf and Smad2 independently bind to their own binding sites in the wnt8a promoter but can function synergistically to regulate wnt8a transcription.
Chromatin remodeling complex NURF is necessary for Bptf and Smad2 regulation of wnt8a expression and neural posteriorization
NURF is an ATP-dependent chromatin remodeling complex that contains a NURF301 homolog (BPTF in humans), an ATPase subunit of the ISWI family (Smarca1/SNF2L in mammals), and additional subunits. To understand the relationship between Bptf and NURF complex during neural posteriorization, we tried to interfere with the endogenous chromatin remodeling activity of NURF complex in zebrafish embryos by overexpressing smarca1K174R, a dominant-negative zebrafish SNF2L homolog, which contains a point mutation in the nucleotide-binding P-loop motif resulting in loss of the ATPase activity (Corona et al., 1999; Barak et al., 2003). Similar to bptf morphants, embryos injected with smarca1K174R mRNA exhibited a marked reduction of wnt8a expression (Fig. 8A) and inefficient posteriorization of the neuroectoderm (Fig. 8B–D). These observations revealed that the catalytic subunit of NURF complex is indispensable for wnt8a expression and neuroectodermal posteriorization.
Figure 8.
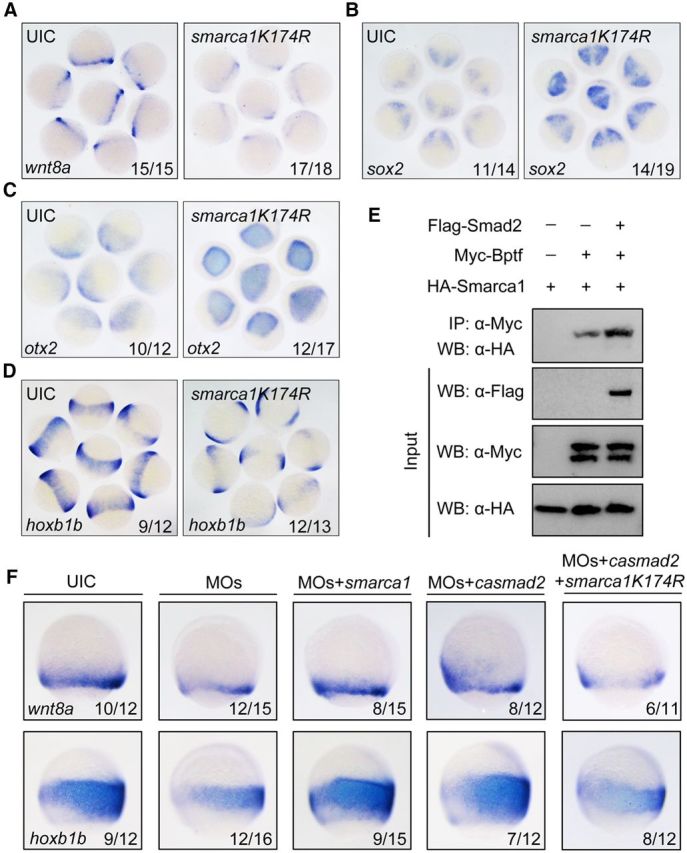
Smarca1 is required for wnt8a expression and neural posteriorization. A, Interference in smarca1 inhibits wnt8a expression. Uninjected and smarca1K174R mRNA-injected (125 pg) embryos were probed at 75% epiboly stage for wnt8a expression by in situ hybridization. B–D, smarca1K174R overexpression disrupts neuroectodermal AP patterning. One-cell stage embryos were injected with 125 pg smarca1K174R mRNA and harvested at 75% epiboly stage for in situ hybridization to detect the anterior neural markers sox2 (B) and otx2 (C) and the posterior neural marker hoxb1b (D). E, Smad2 enhances the association of Bptf and Smarca1. HEK293T cells were transfected with combinations of plasmids expressing Myc-Bptf, HA-Smarca1, and Flag-Smad2 as indicated. After 48 h, cells were lysed for immunoprecipitation with anti-Myc antibody. F, Smarca1 functions on Bptf- and Smad2-mediated wnt8a expression and neural posteriorization. Embryos were injected with indicated MOs and mRNAs at the one-cell stage and harvested at 75% epiboly stage for in situ hybridization to detect alteration of wnt8a and hoxb1b expression. Embryos were orientated laterally with dorsal to the right. Injection doses are as follows: bptf MOs, 8 ng; smarca1 mRNA, 250 pg; smarca1K174R mRNA, 125 pg; casmad2 mRNA, 50 pg.
NURF is known to be directly recruited by site-specific transcription factors (Xiao et al., 2001; Badenhorst et al., 2005). We hypothesized that the interaction of Bptf with Smad2 can enhance the recruitment of other NURF components, including Smarca1. To verify this hypothesis, coimmunoprecipitation assays were performed to assess the impact of Smad2 on the recruitment of Smarca1 to Bptf in HEK293 cells. There was a basal level interaction between Bptf and Smarca1, which was considerably enhanced in the presence of Smad2 (Fig. 8E). We then analyzed the functional importance of interactions among Bptf, Smad2, and Smarca1 in zebrafish embryos. Coinjection of wild-type smarca1 mRNA restored both wnt8a and hoxb1b expression in bptf morphants (Fig. 8F). Meanwhile, overexpression of smarca1K174R partially prevented the Smad2-mediated recovery of wnt8a and hoxb1b expression in bptf morphants (Fig. 8F). Together, these results indicate that interaction of Bptf with Smad2 facilitates the recruitment of NURF complex to regulate wnt8a expression and neural posteriorization.
Inhibited bptf expression or TGF-β signaling increases nucleosome density in the wnt8a promoter
Our data thus far demonstrated the physical association and functional link between Bptf-containing NURF complex and Smad2 on wnt8a expression during neural posteriorization. To analyze nucleosome positioning pattern in the wnt8a promoter, we performed MNase mapping assay as previously described (Li et al., 2010, 2013). In this assay, chromatin isolated from pooled embryos was digested into mononucleosomes with MNase (Fig. 9A), and resulted DNA fragments were extracted and amplified using 30 separate overlapping primer pairs, spanning ∼1000 bp DNA stretch around the identified Bptf and Smad2 binding sites in the wnt8a promoter. We observed five nucleosome binding sites, named N1, N2, N3, N4, and N5, respectively, in the examined region in wild-type embryos. Interestingly, either of the Bptf and Smad2 binding sites was not located in the nucleosome-free areas but instead fallen into the N3 site (Fig. 9B). We speculate that nucleosome packaging may be relaxed only in lateral/paraxial mesodermal precursors within which wnt8a is expressed. Indeed, a slight but steady increase in DNA amount was detected at N2 and N3 sites in bptf morphants, indicating a nucleosome repositioning event occurred in the wnt8a promoter (Fig. 9B). We further assayed histone occupancy on the wnt8a promoter through ChIP assays with antibody against histone H3 and found that the occupancy of histone H3 was enhanced in the region containing Bptf- and Smad2-binding sites in bptf morphants (Fig. 9B′). Consistent with the pivotal role of TGF-β/Smad2 in the recruitment of Bptf-containing NURF complex, interference with TGF-β signaling by overexpression of ΔkTβRII in embryos caused a solid increase of histone density at the N3 site (Fig. 9C,C′). Interestingly, increased nucleosome occupancy was also observed at the N2 site in bptf morphants but not in ΔkTβRII mRNA-injected embryos (Fig. 9B,C′), indicating a subtle difference between Bptf and TGF-β/Smad2 signaling in regulating wnt8a transcription. Together, these data suggest that both Bptf and TGF-β/Smad2 are required for nucleosome remodeling at wnt8a promoter during neural patterning.
Figure 9.
Inactivation of bptf or TGF-β signaling induces nucleosome repositioning within the wnt8a promoter. A, MNase digestion of chromatin isolated from embryos at 75% epiboly stage. Digestion with 320 units per milliliters of MNase for 30 min was appropriate to produce mononucleosome-sized DNAs. B, C, The dynamic changes of nucleosomal positions at the wnt8a promoter in bptf morphants (B) or ΔkTβRII-overexpressing embryos (C). There were five positioned nucleosomes (N1, N2, N3, N4, and N5) within the −1449 to −416 region of the wnt8a promoter in cMO-injected embryos. Bptf (green) and Smad2 (red) binding motifs were located in the DNA sequences occupied by N3. A solid increase in DNA amount was detected at N3 positioning site in bptf morphants and ΔkTβRII-overexpressing embryos. NS, Nonsignificant. **p < 0.01 (Student's t test). ***p < 0.001 (Student's t test). B′, C′, Injection of bptf MOs or ΔkTβRII mRNA results in an increase in histone density over the positioned N3 within the wnt8a promoter. Embryos were injected with 8 ng bptf MOs or 400 pg ΔkTβRII mRNA at one-cell stage and harvested at 75% epiboly stage. The resultant chromatin was extracted and subjected to ChIP assays with anti-H3 antibody followed by qPCR. NS, Nonsignificant. *p < 0.05 (Student's t test).
Discussion
The neuroectoderm patterning along the body axis is an important step of the CNS development. So far, little has been known about implications of epigenetic factors in this neural patterning process. In the present study, we find that zebrafish bptf has a crucial function in transforming the anterior neuroectoderm fate to the posterior neuroectoderm fate. The effect of bptf on posteriorization of the neuroectoderm is closely linked to the chromatin remodeling activity of NURF complex. This is supported by the finding that inhibition of endogenous chromatin remodeling activities of NURF complex by injection of dominant-negative smarca1 mRNA results in neural patterning defects similar to bptf knockdown. Consistently, coinjection of wild-type smarca1 mRNA well rescued the neural posteriorization defects caused by bptf knockdown. More importantly, loss of bptf function could enhance the nucleosome density in the wnt8a promoter and subsequently restrain transcription. Lan et al. (2007) previously reported that inactivation of the histone demethylase UTX and JMJD led to an imbalance of H3K27 methylation, mis-regulation of hox genes, and a striking posterior developmental defect, which provides an indirect evidence for involvement of H3K27 methylation in neural patterning.
Smad2 and Smad3 are intracellular effectors of TGF-β, Activin, and Nodal signals (Massagué et al., 2005; Wu and Hill, 2009). One previous report shows that Bptf, the largest subunit of NURF complex, directly interacts with and activates target gene transcription of Smad2/3 (Landry et al., 2008). Our previous work indicates that the consensus binding motif of Bptf exists most frequently in Smad2-bound promoter regions (Liu et al., 2011). In agreement with these previous findings, we reveal that zebrafish Bptf physically and functionally interacts with Smad2 during neural AP patterning. The contribution of Bptf to the Smad2-regulated neuroectodermal posteriorization is supported by the findings that the interaction of Bptf with Smad2 facilitates the recruitment of Smarca1/SNF2L, the ATPase subunit of NURF complex, and that overexpression of dominant-negative smarca1 inhibits the Smad2-mediated recovery of neural patterning defects in bptf morphants.
Zebrafish wnt8a is required during gastrulation to generate the AP pattern of the nascent neuroectoderm (Erter et al., 2001; Lekven et al., 2001). There is a margin-specific enhancer located to the −989 to −497 region upstream of the wnt8a TSS. The expression of wnt8a can be divided into two phases: Phase I, from 30% epiboly to 60% epiboly (4.7–6.5 hpf), depending on Nodal activation through the margin enhancer; and Phase II, from 70% epiboly until the end of gastrulation (7–10 hpf), requiring ntl activation of the margin enhancer independently of Nodal signaling (Narayanan and Lekven, 2012). Transplantation experiments indicate that the presumptive hindbrain precursors acquire a regional identity between shield stage and mid-gastrulation stages (6–8 hpf) (Woo and Fraser, 1998; Appel, 2000). We find that wnt8a is normally expressed in MZoep mutant embryos at 75% epiboly stage, which support the idea that the wnt8a expression during neural patterning is not regulated by Nodal signal. The inhibition of wnt8a expression by specific interference with bptf activity or TGF-β signal transduction in wild-type and MZoep mutant embryos indicates that wnt8a is a downstream target gene of bptf and TGF-β/Smad2. In contrast to wnt8a, the ventrolateral mesodermal expression of fgf8 and wnt3a remains almost unchanged in bptf morphants, suggesting the specificity of bptf in regulating wnt8a expression.
To our surprise, the expression of wnt8a is also significantly decreased in bptf morphants at shield stage. As the expression of wnt8a depending on Nodal activation at this stage (Narayanan and Lekven, 2012) and Bptf is necessary for Nodal/Smad-mediated mesendoderm differentiation of mouse embryonic stem cells (Landry et al., 2008), we speculate that the expression of wnt8a at shield stage is regulated by Bptf and Nodal/Smad but not the TGF-β/Smad pathway. The ventrolaterally expressed wnt8a functions in restricting the size of the dorsal organizer (Lekven et al., 2001; Ramel and Lekven, 2004), but the decreased expression of wnt8a in bptf morphants at early stage did not result in visible dorsoventral patterning defects. We think that loss of bptf may inhibit the activity of Nodal/Smad signaling or the expression of some unknown bptf target genes, which is essential in dorsoventral axis determination to counteract the dorsalization effects induced by the decreased expression of wnt8a.
With luciferase reporter assays and ChIP-PCR experiments, we demonstrate that Bptf and Smad2 bind to their respective consensus binding motifs in the wnt8a promoter and elevate wnt8a transcription. Interestingly, the identified Bptf and Smad2 binding motifs lie in the margin-specific enhancer, which is essential for the Phase II expression of wnt8a (Narayanan and Lekven, 2012). Unlike another bromodomain and plant homeodomain finger-containing protein TRIM33, which is essential for Smad2/3 binding to AREs in poised promoters during stem cell differentiation (Xi et al., 2011), Bptf and Smad2 independently bind to the wnt8a promoter. The adjacent location of their binding motifs may provide an appropriate space for their interaction and cooperative function. In addition to interact with Bptf, Smad2 also physically associates with Smarca1/SNF2L as previously reported (Landry et al., 2008; Xi et al., 2008). Our coimmunoprecipitation results indicate that the interaction of Bptf with Smad2 could enhance the recruitment of Smarca1/SNF2L to build up the functional NURF complex. Depletion of bptf expression or blocking TGF-β signal transduction causes an increase of the nucleosome density around Bptf and Smad2 binding motifs within the wnt8a promoter, suggesting a role in chromatin remodeling. Therefore, we propose that Bptf functions as a coactivator of Smad2 to recruit other components of chromatin remodeling complex NURF, resembling the histone acetyltransferase p300, which is required for Smad2-mediated transcription to acetylate nucleosomal histones on TGF-β responsive promoters (Ross et al., 2006).
In conclusion, our study discovers that zebrafish bptf and TGF-β/Smad2 signaling cooperatively promote neural posteriorization. TGF-β-activated Smad2 binds to wnt8a promoter and facilitates the recruitment of Bptf-containing NURF complex to drive wnt8a expression and neural patterning. The model proposed here provides a mechanism whereby bptf and TGF-β/Smad2 coregulate neural patterning in zebrafish embryos. In the future, it will be interesting to investigate whether this model applies to the neuroectoderm patterning in other animals.
Footnotes
This work was supported by National Natural Science Foundation of China Grants 31322035 and 31271532, Major Science Programs of China Grants 2011CB943904 and 2011CBA01101, and Strategic Priority Research Program of the Chinese Academy of Sciences Grant XDA01010104. We thank Professor Guohong Li (Institute of Biophysics of the Chinese Academy of Sciences) for assistance with high-resolution MNase mapping assays.
The authors declare no competing financial interests.
References
- Appel B. Zebrafish neural induction and patterning. Dev Dyn. 2000;219:155–168. doi: 10.1002/1097-0177(2000). [DOI] [PubMed] [Google Scholar]
- Badenhorst P, Xiao H, Cherbas L, Kwon SY, Voas M, Rebay I, Cherbas P, Wu C. The Drosophila nucleosome remodeling factor NURF is required for Ecdysteroid signaling and metamorphosis. Genes Dev. 2005;19:2540–2545. doi: 10.1101/gad.1342605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak O, Lazzaro MA, Lane WS, Speicher DW, Picketts DJ, Shiekhattar R. Isolation of human NURF: a regulator of Engrailed gene expression. EMBO J. 2003;22:6089–6100. doi: 10.1093/emboj/cdg582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand T, MacLellan WR, Schneider MD. A dominant-negative receptor for type beta transforming growth factors created by deletion of the kinase domain. J Biol Chem. 1993;268:11500–11503. [PubMed] [Google Scholar]
- Buckles GR, Thorpe CJ, Ramel MC, Lekven AC. Combinatorial Wnt control of zebrafish midbrain-hindbrain boundary formation. Mech Dev. 2004;121:437–447. doi: 10.1016/j.mod.2004.03.026. [DOI] [PubMed] [Google Scholar]
- Cannon JE, Place ES, Eve AM, Bradshaw CR, Sesay A, Morrell NW, Smith JC. Global analysis of the haematopoietic and endothelial transcriptome during zebrafish development. Mech Dev. 2013;130:122–131. doi: 10.1016/j.mod.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah FS, Jabs EW, Chong SS. Genomic, cDNA, and embryonic expression analysis of zebrafish transforming growth factor beta 3 (tgfbeta3) Dev Dyn. 2005;232:1021–1030. doi: 10.1002/dvdy.20282. [DOI] [PubMed] [Google Scholar]
- Chen RH, Ebner R, Derynck R. Inactivation of the type II receptor reveals two receptor pathways for the diverse TGF-beta activities. Science. 1993;260:1335–1338. doi: 10.1126/science.8388126. [DOI] [PubMed] [Google Scholar]
- Corona DF, Längst G, Clapier CR, Bonte EJ, Ferrari S, Tamkun JW, Becker PB. ISWI is an ATP-dependent nucleosome remodeling factor. Mol Cell. 1999;3:239–245. doi: 10.1016/S1097-2765(00)80314-7. [DOI] [PubMed] [Google Scholar]
- Cox WG, Hemmati-Brivanlou A. Caudalization of neural fate by tissue recombination and bFGF. Development. 1995;121:4349–4358. doi: 10.1242/dev.121.12.4349. [DOI] [PubMed] [Google Scholar]
- Dal-Pra S, Fürthauer M, Van-Celst J, Thisse B, Thisse C. Noggin1 and Follistatin-like2 function redundantly to Chordin to antagonize BMP activity. Dev Biol. 2006;298:514–526. doi: 10.1016/j.ydbio.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Dodd A, Chambers SP, Love DR. Short interfering RNA-mediated gene targeting in the zebrafish. FEBS Lett. 2004;561:89–93. doi: 10.1016/S0014-5793(04)00129-2. [DOI] [PubMed] [Google Scholar]
- Dorsky RI, Sheldahl LC, Moon RT. A transgenic Lef1/beta-catenin-dependent reporter is expressed in spatially restricted domains throughout zebrafish development. Dev Biol. 2002;241:229–237. doi: 10.1006/dbio.2001.0515. [DOI] [PubMed] [Google Scholar]
- Erter CE, Wilm TP, Basler N, Wright CV, Solnica-Krezel L. Wnt8 is required in lateral mesendodermal precursors for neural posteriorization in vivo. Development. 2001;128:3571–3583. doi: 10.1242/dev.128.18.3571. [DOI] [PubMed] [Google Scholar]
- Escher D, Bodmer-Glavas M, Barberis A, Schaffner W. Conservation of glutamine-rich transactivation function between yeast and humans. Mol Cell Biol. 2000;20:2774–2782. doi: 10.1128/MCB.20.8.2774-2782.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman B, Gates MA, Egan ES, Dougan ST, Rennebeck G, Sirotkin HI, Schier AF, Talbot WS. Zebrafish organizer development and germ-layer formation require nodal-related signals. Nature. 1998;395:181–185. doi: 10.1038/26013. [DOI] [PubMed] [Google Scholar]
- Goller T, Vauti F, Ramasamy S, Arnold HH. Transcriptional regulator BPTF/FAC1 is essential for trophoblast differentiation during early mouse development. Mol Cell Biol. 2008;28:6819–6827. doi: 10.1128/MCB.01058-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritsman K, Zhang J, Cheng S, Heckscher E, Talbot WS, Schier AF. The EGF-CFC protein one-eyed pinhead is essential for nodal signaling. Cell. 1999;97:121–132. doi: 10.1016/S0092-8674(00)80720-5. [DOI] [PubMed] [Google Scholar]
- Hagos EG, Fan X, Dougan ST. The role of maternal Activin-like signals in zebrafish embryos. Dev Biol. 2007;309:245–258. doi: 10.1016/j.ydbio.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt M, Pelegri F, Mullins MC, Kane DA, van Eeden FJ, Granato M, Brand M, Furutani-Seiki M, Haffter P, Heisenberg CP, Jiang YJ, Kelsh RN, Odenthal J, Warga RM, Nüsslein-Volhard C. dino and mercedes, two genes regulating dorsal development in the zebrafish embryo. Development. 1996;123:95–102. doi: 10.1242/dev.123.1.95. [DOI] [PubMed] [Google Scholar]
- Hemmati-Brivanlou A, Kelly OG, Melton DA. Follistatin, an antagonist of activin, is expressed in the Spemann organizer and displays direct neuralizing activity. Cell. 1994;77:283–295. doi: 10.1016/0092-8674(94)90320-4. [DOI] [PubMed] [Google Scholar]
- Herskowitz I. Functional inactivation of genes by dominant negative mutations. Nature. 1987;329:219–222. doi: 10.1038/329219a0. [DOI] [PubMed] [Google Scholar]
- Inman GJ, Nicolás FJ, Callahan JF, Harling JD, Gaster LM, Reith AD, Laping NJ, Hill CS. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol. 2002;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- Jia S, Ren Z, Li X, Zheng Y, Meng A. smad2 and smad3 are required for mesendoderm induction by transforming growth factor-beta/nodal signals in zebrafish. J Biol Chem. 2008;283:2418–2426. doi: 10.1074/jbc.M707578200. [DOI] [PubMed] [Google Scholar]
- Jia S, Wu D, Xing C, Meng A. Smad2/3 activities are required for induction and patterning of the neuroectoderm in zebrafish. Dev Biol. 2009;333:273–284. doi: 10.1016/j.ydbio.2009.06.037. [DOI] [PubMed] [Google Scholar]
- Jones MH, Hamana N, Shimane M. Identification and characterization of BPTF, a novel bromodomain transcription factor. Genomics. 2000;63:35–39. doi: 10.1006/geno.1999.6070. [DOI] [PubMed] [Google Scholar]
- Jordan-Sciutto KL, Dragich JM, Rhodes JL, Bowser R. Fetal Alz-50 clone 1, a novel zinc finger protein, binds a specific DNA sequence and acts as a transcriptional regulator. J Biol Chem. 1999;274:35262–35268. doi: 10.1074/jbc.274.49.35262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly GM, Greenstein P, Erezyilmaz DF, Moon RT. Zebrafish wnt8 and wnt8b share a common activity but are involved in distinct developmental pathways. Development. 1995;121:1787–1799. doi: 10.1242/dev.121.6.1787. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kohli G, Hu S, Clelland E, Di Muccio T, Rothenstein J, Peng C. Cloning of transforming growth factor-beta 1 (TGF-beta 1) and its type II receptor from zebrafish ovary and role of TGF-beta 1 in oocyte maturation. Endocrinology. 2003;144:1931–1941. doi: 10.1210/en.2002-0126. [DOI] [PubMed] [Google Scholar]
- Koshida S, Shinya M, Nikaido M, Ueno N, Schulte-Merker S, Kuroiwa A, Takeda H. Inhibition of BMP activity by the FGF signal promotes posterior neural development in zebrafish. Dev Biol. 2002;244:9–20. doi: 10.1006/dbio.2002.0581. [DOI] [PubMed] [Google Scholar]
- Lamb TM, Knecht AK, Smith WC, Stachel SE, Economides AN, Stahl N, Yancopolous GD, Harland RM. Neural induction by the secreted polypeptide noggin. Science. 1993;262:713–718. doi: 10.1126/science.8235591. [DOI] [PubMed] [Google Scholar]
- Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, Roberts TM, Chang HY, Shi Y. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- Landry J, Sharov AA, Piao Y, Sharova LV, Xiao H, Southon E, Matta J, Tessarollo L, Zhang YE, Ko MS, Kuehn MR, Yamaguchi TP, Wu C. Essential role of chromatin remodeling protein Bptf in early mouse embryos and embryonic stem cells. PLoS Genet. 2008;4:e1000241. doi: 10.1371/journal.pgen.1000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekven AC, Thorpe CJ, Waxman JS, Moon RT. Zebrafish wnt8 encodes two wnt8 proteins on a bicistronic transcript and is required for mesoderm and neurectoderm patterning. Dev Cell. 2001;1:103–114. doi: 10.1016/S1534-5807(01)00007-7. [DOI] [PubMed] [Google Scholar]
- Li G, Margueron R, Hu G, Stokes D, Wang YH, Reinberg D. Highly compacted chromatin formed in vitro reflects the dynamics of transcription activation in vivo. Mol Cell. 2010;38:41–53. doi: 10.1016/j.molcel.2010.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Zhang Y, Fu J, Han L, Xue L, Lv C, Wang P, Li G, Tong T. FOXA1 mediates p16(INK4a) activation during cellular senescence. EMBO J. 2013;32:858–873. doi: 10.1038/emboj.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WY, Wang Y, Sun YH, Wang Y, Wang YP, Chen SP, Zhu ZY. Efficient RNA interference in zebrafish embryos using siRNA synthesized with SP6 RNA polymerase. Dev Growth Differ. 2005;47:323–331. doi: 10.1111/j.1440-169X.2005.00807.x. [DOI] [PubMed] [Google Scholar]
- Liu Z, Lin X, Cai Z, Zhang Z, Han C, Jia S, Meng A, Wang Q. Global identification of SMAD2 target genes reveals a role for multiple co-regulatory factors in zebrafish early gastrulas. J Biol Chem. 2011;286:28520–28532. doi: 10.1074/jbc.M111.236307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loosli F, Staub W, Finger-Baier KC, Ober EA, Verkade H, Wittbrodt J, Baier H. Loss of eyes in zebrafish caused by mutation of chokh/rx3. EMBO Rep. 2003;4:894–899. doi: 10.1038/sj.embor.embor919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden M. Retinoids and spinal cord development. J Neurobiol. 2006;66:726–738. doi: 10.1002/neu.20248. [DOI] [PubMed] [Google Scholar]
- Massagué J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- Mizuguchi G, Tsukiyama T, Wisniewski J, Wu C. Role of nucleosome remodeling factor NURF in transcriptional activation of chromatin. Mol Cell. 1997;1:141–150. doi: 10.1016/S1097-2765(00)80015-5. [DOI] [PubMed] [Google Scholar]
- Narayanan A, Lekven AC. Biphasic wnt8a expression is achieved through interactions of multiple regulatory inputs. Dev Dyn. 2012;241:1062–1075. doi: 10.1002/dvdy.23787. [DOI] [PubMed] [Google Scholar]
- Niehrs C. Head in the WNT: the molecular nature of Spemann's head organizer. Trends Genet. 1999;15:314–319. doi: 10.1016/S0168-9525(99)01767-9. [DOI] [PubMed] [Google Scholar]
- Nomura M, Li E. Smad2 role in mesoderm formation, left-right patterning and craniofacial development. Nature. 1998;393:786–790. doi: 10.1038/31693. [DOI] [PubMed] [Google Scholar]
- Ramel MC, Lekven AC. Repression of the vertebrate organizer by Wnt8 is mediated by Vent and Vox. Development. 2004;131:3991–4000. doi: 10.1242/dev.01277. [DOI] [PubMed] [Google Scholar]
- Rentzsch F, Bakkers J, Kramer C, Hammerschmidt M. Fgf signaling induces posterior neuroectoderm independently of Bmp signaling inhibition. Dev Dyn. 2004;231:750–757. doi: 10.1002/dvdy.20244. [DOI] [PubMed] [Google Scholar]
- Ross S, Cheung E, Petrakis TG, Howell M, Kraus WL, Hill CS. Smads orchestrate specific histone modifications and chromatin remodeling to activate transcription. EMBO J. 2006;25:4490–4502. doi: 10.1038/sj.emboj.7601332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Milne TA, Dewell S, McGinty RK, Yuen M, Ueberheide B, Dou Y, Muir TW, Patel DJ, Allis CD. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell. 2011;145:692–706. doi: 10.1016/j.cell.2011.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai Y, De Robertis EM. Ectodermal patterning in vertebrate embryos. Dev Biol. 1997;182:5–20. doi: 10.1006/dbio.1996.8445. [DOI] [PubMed] [Google Scholar]
- Sasai Y, Lu B, Steinbeisser H, De Robertis EM. Regulation of neural induction by the Chd and Bmp-4 antagonistic patterning signals in Xenopus. Nature. 1995;376:333–336. doi: 10.1038/376333a0. [DOI] [PubMed] [Google Scholar]
- Schulte-Merker S, Lee KJ, McMahon AP, Hammerschmidt M. The zebrafish organizer requires chordino. Nature. 1997;387:862–863. doi: 10.1038/43092. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Bae YK, Muraoka O, Hibi M. Interaction of Wnt and caudal-related genes in zebrafish posterior body formation. Dev Biol. 2005;279:125–141. doi: 10.1016/j.ydbio.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Sirotkin HI, Dougan ST, Schier AF, Talbot WS. bozozok and squint act in parallel to specify dorsal mesoderm and anterior neuroectoderm in zebrafish. Development. 2000;127:2583–2592. doi: 10.1242/dev.127.12.2583. [DOI] [PubMed] [Google Scholar]
- Stern CD. Neural induction: 10 years on since the ‘default model.’. Curr Opin Cell Biol. 2006;18:692–697. doi: 10.1016/j.ceb.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Sun Z, Jin P, Tian T, Gu Y, Chen YG, Meng A. Activation and roles of ALK4/ALK7-mediated maternal TGFbeta signals in zebrafish embryo. Biochem Biophys Res Commun. 2006;345:694–703. doi: 10.1016/j.bbrc.2006.04.148. [DOI] [PubMed] [Google Scholar]
- Thisse B, Wright CV, Thisse C. Activin- and Nodal-related factors control antero-posterior patterning of the zebrafish embryo. Nature. 2000;403:425–428. doi: 10.1038/35000200. [DOI] [PubMed] [Google Scholar]
- Tian T, Meng AM. Nodal signals pattern vertebrate embryos. Cell Mol Life Sci. 2006;63:672–685. doi: 10.1007/s00018-005-5503-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- Woo K, Fraser SE. Specification of the zebrafish nervous system by nonaxial signals. Science. 1997;277:254–257. doi: 10.1126/science.277.5323.254. [DOI] [PubMed] [Google Scholar]
- Woo K, Fraser SE. Specification of the hindbrain fate in the zebrafish. Dev Biol. 1998;197:283–296. doi: 10.1006/dbio.1998.8870. [DOI] [PubMed] [Google Scholar]
- Wu MY, Hill CS. Tgf-beta superfamily signaling in embryonic development and homeostasis. Dev Cell. 2009;16:329–343. doi: 10.1016/j.devcel.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- Xiao H, Sandaltzopoulos R, Wang HM, Hamiche A, Ranallo R, Lee KM, Fu D, Wu C. Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol Cell. 2001;8:531–543. doi: 10.1016/S1097-2765(01)00345-8. [DOI] [PubMed] [Google Scholar]
- Xi Q, He W, Zhang XH, Le HV, Massagué J. Genome-wide impact of the BRG1 SWI/SNF chromatin remodeler on the transforming growth factor beta transcriptional program. J Biol Chem. 2008;283:1146–1155. doi: 10.1074/jbc.M707479200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Q, Wang Z, Zaromytidou AI, Zhang XH, Chow-Tsang LF, Liu JX, Kim H, Barlas A, Manova-Todorova K, Kaartinen V, Studer L, Mark W, Patel DJ, Massagué J. A poised chromatin platform for TGF-beta access to master regulators. Cell. 2011;147:1511–1524. doi: 10.1016/j.cell.2011.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Zhou MM. Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 2002;513:124–128. doi: 10.1016/S0014-5793(01)03309-9. [DOI] [PubMed] [Google Scholar]
- Zhou X, Sasaki H, Lowe L, Hogan BL, Kuehn MR. Nodal is a novel TGF-beta-like gene expressed in the mouse node during gastrulation. Nature. 1993;361:543–547. doi: 10.1038/361543a0. [DOI] [PubMed] [Google Scholar]