Abstract
The establishment of cell-type-specific dendritic arbors is fundamental for proper neural circuit formation. Here, using temporal- and cell-specific knock-down, knock-out, and overexpression approaches, we show that multiple aspects of the dendritic organization of cerebellar Purkinje cells (PCs) are controlled by a single transcriptional factor, retinoic acid-related orphan receptor-alpha (RORα), a gene defective in staggerer mutant mice. As reported earlier, RORα was required for regression of primitive dendrites before postnatal day 4 (P4). RORα was also necessary for PCs to form a single Purkinje layer from P0 to P4. The knock-down of RORα from P4 impaired the elimination of perisomatic dendrites and maturation of single stem dendrites in PCs at P8. Filopodia and spines were also absent in these PCs. The knock-down of RORα from P8 impaired the formation and maintenance of terminal dendritic branches of PCs at P14. Finally, even after dendrite formation was completed at P21, RORα was required for PCs to maintain dendritic complexity and functional synapses, but their mature innervation pattern by single climbing fibers was unaffected. Interestingly, overexpression of RORα in PCs at various developmental stages did not facilitate dendrite development, but had specific detrimental effects on PCs. Because RORα deficiency during development is closely related to the severity of spinocerebellar ataxia type 1, delineating the specific roles of RORα in PCs in vivo at different time windows during development and throughout adulthood would facilitate our understanding of the pathogenesis of cerebellar disorders.
SIGNIFICANCE STATEMENT The genetic programs by which each neuron subtype develops and maintains dendritic arbors have remained largely unclear. This is partly because dendrite development is modulated dynamically by neuronal activities and interactions with local environmental cues in vivo. In addition, dendrites are formed and maintained by the balance between their growth and regression; the effects caused by the disruption of transcription factors during the early developmental stages could be masked by dendritic growth or regression in the later stages. Here, using temporal- and cell-specific knock-down, knock-out, and overexpression approaches in vivo, we show that multiple aspects of the dendritic organization of cerebellar Purkinje cells are controlled by a single transcriptional factor, retinoic acid-related orphan receptor alpha.
Keywords: dendrite, migration, Purkinje cell, retinoic acid-related orphan receptor a, staggerer
Introduction
Dendritic arbors of every neuron grow in a specific branching pattern to properly integrate synaptic inputs in neuronal circuits. Dendrite malformation is reportedly associated with several neuropsychiatric and neurodevelopmental disorders, such as Down, Rett, and Fragile X syndromes (Kaufmann and Moser, 2000; Kulkarni and Firestein, 2012). However, the genetic programs by which each neuron subtype develops and maintains dendritic arbors have remained largely unclear. This is partly because dendrite development is not only encoded by intrinsic programs, but is also modulated dynamically by neuronal activities and interactions with local environmental cues (Sotelo and Dusart, 2009; Tavosanis, 2012). In addition, dendrites are formed and maintained by the balance between their growth and regression (Emoto, 2012). Therefore, the effects caused by the disruption of transcription factors during the early developmental stages could be masked by dendritic growth or regression in the later stages.
Cerebellar Purkinje cells (PCs) have served as a useful model with which to study dendrite development because these cells develop one of the most elaborate dendritic trees in the mammalian CNS. In mice, PCs arise from the ventricular zone during embryonic days 10–13 (E10–E13) and migrate to the cerebellar cortex by E16–E17. Postmigratory PCs exhibit fusiform shapes with a few primitive apical dendrites. These dendrites rapidly retract and the PCs enter the “stellate cell” stage by postnatal day 4 (P4) (see Fig. 1A). By ∼P8, PCs enter the “young PC” stage by losing their perisomatic dendrites and forming single-stem dendrites, which give rise to branches and branchlets with numerous spines. Retinoic acid-related orphan receptor alpha (RORα), which belongs to a superfamily of nuclear receptors, was originally identified as the gene responsible for the ataxic mouse staggerer (Sidman et al., 1962; Gold et al., 2007). Staggerer PCs exhibit atrophic, fusiform-like dendrites lacking spiny branchlets (Landis and Sidman, 1978; Soha and Herrup, 1995). Interestingly, overexpression of RORα in wild-type PCs at P0 accelerated dendrite regression in organotypic cultures, whereas dendritic growth was not affected when RORα was overexpressed after P7 (Boukhtouche et al., 2006). Although these findings indicate that RORα specifically mediates the regression of dendrites in the early phase of development, its effect on dendrite maturation at later stages is unclear in organotypic cultures in vitro. Indeed, RORα expression is not only limited to early development, but is also maintained throughout adult life. The deletion of RORα in PCs at P10–P21 has also been reported to cause dendrite atrophy at P28 in vivo (Chen et al., 2013). However, because RORα was knocked out during the late developmental stage, it is not completely clear whether RORα regulates dendrite growth or maintenance in the adult. Therefore, the role of endogenous RORα at specific time windows during development and after maturation of the PCs remains unclear. Here, using tamoxifen-induced gene knock-down, knock-out, and the overexpression method in vivo, we show that RORα plays multiple roles in PC dendritogenesis, including alignment of somata, dendrite regression, stem dendrite formation, spine formation, and maintenance of mature dendrites, at specific time windows during development and throughout adulthood in vivo.
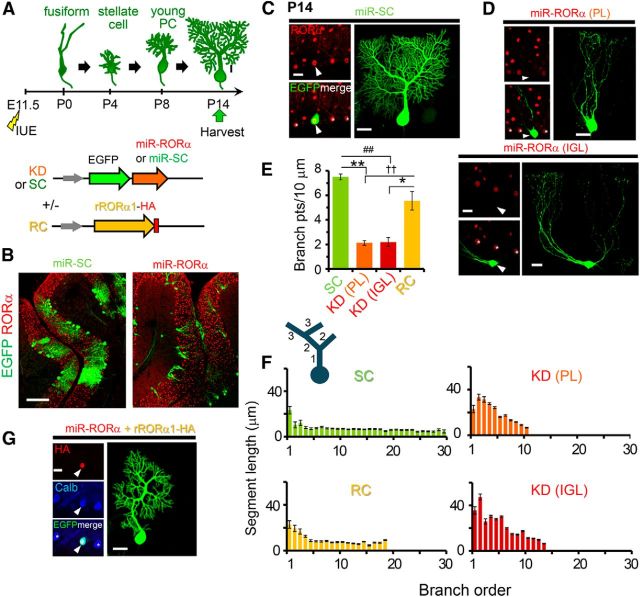
Figure 1.
The knock-down of RORα in PCs recapitulates staggerer-like dendritic morphology in vivo. A, Schematic diagram of the developmental stages of PCs in vivo and the experimental procedure. IUE was performed at E11.5 and the cerebella were fixed at P14. The plasmid vectors are described in the bottom panel. B–D, Confocal images of sagittal slices from electroporated cerebella (B) and single PCs expressing control (C, miR-SC) and knock-down (D, miR-RORα) miR at P14. Endogenous RORα immunoreactivities are shown in red together with the EGFP signals. Arrowheads in C and D indicate the transfected PC somata. The somata of 55.6% of the PCs expressing miR-RORα were located in the PL, but the others were mislocalized to the IGL, as shown in B and the upper and bottom of D. E, Number of dendritic branch points of single PCs per 10 μm of the total dendritic length. The bars represent the mean ± SEM n = 4 (SC), 5 (PL: KD), 4 (IGL:KD), and 10 (RC) cells from 2 or 3 mice, *p = 0.0144, **p = 0.000681, ##p = 0.00132, and ††p = 0.00681, respectively (ANOVA followed by Tukey's test). F, Length of the dendritic segments at each branch order. The dendritic segments are numbered in a centrifugal order as shown in the diagram on the upper left. The bars represent the mean ± SEM segment lengths of 4–10 cells from 2 or 3 mice. G, Confocal images of PCs expressing RC constructs at P14. Exogenous RORα1 immunoreactivities are shown in red (HA) and calbindin immunoreactivity is shown in blue together with the EGFP signals. Arrowheads and asterisks in C, D, and G indicate the transfected PC somata and nontransfected neighboring PCs, respectively. Scale bars: B, 100 μm; C, D, G, 20 μm.
Materials and Methods
Procedures.
All procedures related to animal care and treatment were performed in accordance with the guidelines set by the Animal Resource Committee of Keio University. The animals were killed by decapitation after anesthetization with pentobarbital (Somnopentyl; Kyoritsu Seiyaku).
cDNA constructs.
miR knock-down vectors were generated using BLOCK-iT Pol II miR RNAi Expression Vector Kits (Invitrogen). The fragments were subcloned into a pCAGGS-enhanced green fluorescent (EGFP) vector. The 21 bp target sequence of RORα is as follows: AGC TGT GTG CCA TCA AGA TTA. For the control experiments, we used two different scrambled sequences: GGC TTC AAC TGA CGT GTA TAA and GTT AAT TGA GAC GTC CAT ACG. The pCL20-L7-ERT2CreERT2 vector was described previously (Nishiyama et al., 2012). The cDNA fragment encoding full-length murine RORα1 (Nishiyama et al., 2012) was subcloned into the pCAGGS vector. cDNA encoding a hemagglutinin (HA) tag was added to the 3′ end of RORα1. To obtain the plasmid vector encoding the miR-resistant form of RORα1 (rRORα1), silent mutations were introduced to the miR target sequence by PCR. The mutated nucleotide sequence of the knock-down target is as follows: AAC TTT GCG CAA TTA AAA TCA.
For the inducible expression of miR-RORα, control miR (miR-SC), or rRORα1-HA, the fragments were subcloned into a “flex” vector (CAG-flex) which was produced by inserting two mutant loxP sites (2272loxP and 5171loxP) (Lee and Saito, 1998) into the multicloning sites of the pCAGGS vector. For the inducible CRISPR/Cas9 system, the SpCas9 cDNA was amplified by PCR from pX330-U6-Chimeric_BB-CBh-hSpCas9 plasmid (Addgene #42230; Cong et al., 2013) and subcloned into the CAG-flex vector. For construction of gRNA-expressing vector, SpCas9 fragment was removed from the pX330 plasmid and then the oligonucleotide encoding gR-LacZ or gR-RORα was inserted into BbsI sites. We used one of two different gRNA targeting sequences for RORα. The following oligonucleotide sets were annealed and inserted into the pX330 plasmid: 5′-CAC CGC ATT GAT GTC CAA TCC CGA C-3′ and 5′-AAA CGT CGG GAT TGG ACA TCA ATG C-3′ or 5′-CAC CGA GTC GGC CTT GCT GCC CTC-3′ and 5′-AAA CGA GGG CAG CAA GGC CGA CTC-3′ for RORα; 5′-CAC CTG CGA ATA CGC CCA CGC GAT-3′ and 5′-AAA CAT CGC GTG GGC GTA TTC GCA-3′ for LacZ.
In utero electroporation.
In utero electroporation (IUE) was performed on E11.5 in the cerebella of ICR mice as described previously (Nishiyama et al., 2012). Briefly, plasmid DNA was injected into the fourth ventricle by a glass pipette and a set of electrical pulses was applied two to four times. Positive electrodes were placed onto the rostral edge of the cerebellar ventricular zone and electroporated mice of either sex were analyzed. Plasmid DNA was resolved in HEPES-buffered saline (21 mm HEPES, 137 mm NaCl, 5 mm KCl, and 0.7 mm Na2HPO4). To inject a single vector, the plasmids were dissolved at a concentration of 2 mg/ml. For the double transfection of EGFP-miR and rRORα1-HA, the two plasmids were mixed in a 1:1 ratio in a final concentration of 4 mg/ml. For the overexpression of rRORα1-HA, the CAG-EGFP and CAG-rRORα1-HA vectors were mixed in a 1:2 ratio at a final concentration of 3 mg/ml. For the inducible knock-down, CAG-DsRed2, L7-ERT2CreERT2, and CAG-flex-EGFP-miR vectors were mixed in a 1:2:3 ratio for a total concentration of 5 mg/ml. Using this protocol, 4-hydroxytamoxifen (4-OHT) injection induced EGFP expression in 49 ± 7% of DsRed2-positive PCs (n = 9 experiments). The rescue experiment was performed by using CAG-flex-rRORα1-HA in place of CAG-DsRed2. For the inducible overexpression, CAG-EGFP, L7-ERT2CreERT2, and CAG-flex-rRORα1-HA were mixed in a 1:2:3 ratio for a total concentration of 5 mg/ml. For inducible CRISPR/Cas9-based gene editing, CAG-DsRed2, CAG-flex-EGFP, L7-ERT2CreERT2, U6-gRNA, and CAG-flex-Cas9 vectors were mixed in a 0.5:0.5:2:3:3 ratio or CAG-flex-EGFP, L7-ERT2CreERT2, U6-gRNA, and CAG-flex-Cas9 vectors were mixed in a 1:2:3:3 ratio for a total concentration of 5 mg/ml.
4-OHT administration.
4-OHT (Sigma-Aldrich) was dissolved in ethanol at a concentration of 20 mg/ml and diluted with 9 volumes of corn oil (Sigma-Aldrich). The diluted 4-OHT was injected intraperitoneally with 26 gauge needles into P4, P8, and P21 mice. The dosage was 200 μl at P4, 200–300 μl at P8, and 500 μl at P21.
Immunohistochemistry and fluorescence image acquisition.
For immunohistochemical analyses of the PCs at P4 or later, mice were fixed by cardiac perfusion under sodium pentobarbital anesthesia with 0.1 m sodium phosphate buffer (PB), pH 7.4, containing 4% paraformaldehyde (4% PFA/PB). The brains were then removed and soaked in 4% PFA/PB overnight. After rinsing the specimens with PBS, the cerebellum was isolated and parasagittal slices (100 μm thick) were prepared using a microslicer (DTK-2000; Dosaka). For immunohistochemical analyses of the pups at P0, the mice were decapitated and the heads were soaked in 4% PFA/PB overnight and then in 30% sucrose/PB until the heads sank. Samples were embedded in Tissue-Tek OCT compound (Sakura) and frozen. Parasagittal slices (30 μm thick) were prepared using a cryostat (Leica).
Sections were permeabilized with 0.1% Triton X-100 in PBS, blocked with 10% donkey serum in PBS, and incubated for 2–4 h or overnight with the following primary antibodies: guinea pig anti-calbindin (1 mg/ml; Frontier Institute), rabbit anti-calbindin (1:500; Millipore), mouse anti-HA (1:500; Covance), goat anti-RORα (1:500; Santa Cruz Biotechnology), and guinea pig anti-vGluT2 (1 mg/ml; Frontier Institute). After staining with the primary antibodies, the sections were washed and incubated with Alexa Fluor-546, Alexa Fluor-555, or Alexa Fluor-647 (Invitrogen), Cy3, or DyLight-649 (Jackson ImmunoResearch Laboratories)-conjugated secondary antibodies for 1–2 h. Sections were then washed and mounted onto glass slides and the fluorescent signals of the EGFP or secondary antibodies were observed using confocal microscopes, as described below.
Image analysis.
We analyzed EGFP-positive cells located in cerebellar lobules I- IV/V of the vermis. Many PC markers, such as calbidin, are reported as candidate genes with transcription that is regulated by RORα (Gold et al., 2003). Therefore, PCs were identified by EGFP signals because ∼96% of transfected cells were PCs in our IUE method (Nishiyama et al., 2012). For accurate PC identification, cells in which EGFP levels were >5 times greater than the background were included and cells having cytosolic EGFP signals brighter than their nuclear signals were excluded from the analyses. To analyze the dendritic length using IMARIS software (Zeiss), z-stack images of the EGFP-labeled single PCs were obtained at an optimal z-interval with an FV-1000 using a 40× objective [numerical aperture (NA) 1.0; Olympus], except for Figure 4, in which an LSM710 with a 63× objective (NA 1.4; Zeiss) was used. The gain was adjusted to allow signal saturation in the soma and primary dendrite only. The images were reconstructed using IMARIS and the dendrites were traced manually with the IMARIS FilamentTracer software using AutoDepth and Manual mode. The dendritic starting points were determined manually at the base of each primary dendrite on the surface of the soma. When multiple primary dendrites were observed, every primary dendrite was traced and the total dendritic length or the number of branch points was summed. For analyses of dendrites of PCs expressing miR-RORα in Figure 4, perisomatic dendrites shorter than 20 μm in length were excluded from analyses.
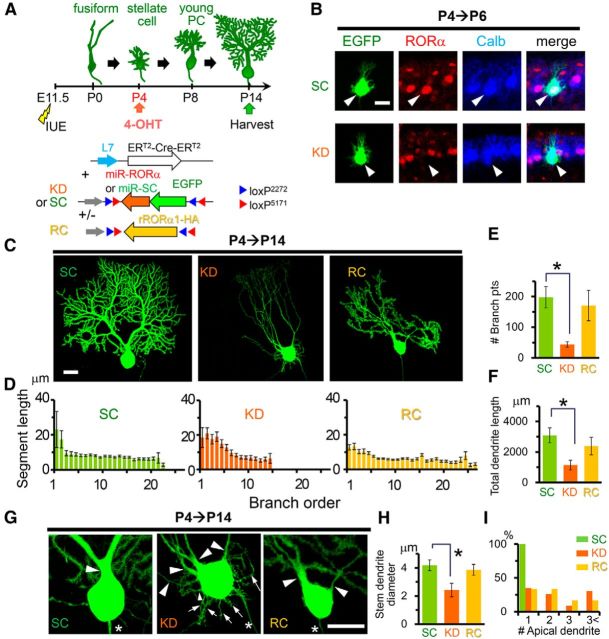
Figure 4.
Knock-down of RORα from the stellate cell stage impairs dendritic pruning and branching. A, Schematic diagram of the experimental procedure. The mice were electroporated at E11.5 and the expression of EGFP-miR-RORα (KD) or scrambled miR (SC) was induced by 4-OHT injection at P4. In the rescue (RC) experiments, the expression of rRORα1-HA was also induced. B, Application of 4-OHT at P4 induces a reduction in endogenous RORα in PCs expressing miR-RORα at P6. Immunofluorescent signals for endogenous RORα (red) and a PC marker calbindin (Calb, blue) are shown. The arrowheads indicate transfected PCs (EGFP, green). Scale bar, 20 μm. C, Dendritic branching of PCs expressing miR-SC (SC), miR-RORα (KD), or miR-RORα plus RORα1-HA (RC) at P14. Representative stacked images of EGFP fluorescence are shown. Scale bar, 20 μm. D, Length of the dendritic segment at each branch order. The bars represent the mean ± SEM segment lengths of n = 4 cells from 2 mice (SC), n = 6 cells from 3 mice (KD), and n = 6 cells from 2 mice (RC). E, Number of dendritic branch points. The bars represent the mean ± SEM branch points. n = 4 cells from 2 mice (SC), n = 6 cells from 3 mice (KD), and n = 6 cells from 2 mice (RC), *p = 0.0344 (ANOVA followed by Tukey's test). F, Total dendritic length per single PCs. The bars represent the mean ± SEM total dendritic length. n = 4 cells from 2 mice (SC), n = 6 cells from 3 mice (KD), and n = 6 cells from 2 mice (RC), *p = 0.0404 (ANOVA followed by Tukey's test). G, Enlarged images of the somata shown in C. Arrowheads and arrows indicate apical and perisomatic dendrites, respectively. Asterisks indicate axons. Scale bar, 20 μm. H, Diameters of the thickest primary dendrites. Bars represent the mean ± SEM diameters of the independent experiments. n = 6 cells from 2 mice (SC), n = 8 cells from 3 mice (KD), and n = 9 cells from 3 mice (RC), *p = 0.0328 (ANOVA followed by Tukey's test). I, Number of apical primary dendrites per single PCs. The percentages of PCs showing 1, 2, 3, and more apical primary dendrites are shown. n = 16 cells from 3 mice (SC), n = 23 cells from 7 mice (KD), n = 22 cells from 4 mice (RC), ***p = 1.14 × 10−4 (SC vs KD) and 1.50 × 10−5 (SC vs Rescue) (χ2 test followed by Bonferroni's correction).
The quantification of RORα levels was performed using ImageJ software version 1.49. A circular selection of the same size was made on the brightest region in the nucleus of an EGFP-positive cell and the mean gray value of the selection was obtained. For each EGFP-positive cell, control gray values were obtained from multiple untransfected cells with RORα signals that were brightest in the same z-plane. The obtained values were subtracted by the background value. The percentage obtained by comparing the values from the EGFP-positive cells to the values from the untransfected cells represented the relative RORα level. The background value of interest z-plane was determined by the mean gray value in an equivalent circular selection made on a dark area near the EGFP-positive cells where no soma exists. The distance between the edge of the external granular layer (EGL) and PC soma and the longest neurite length was measured using ImageJ. The diameter of the stem dendrite was determined from 2D images using the line tool in ImageJ. The width of the primary dendrite at its middle portion was measured. If there were multiple primary dendrites, then the thickest primary dendrite was analyzed.
For the spine number analyses, z-stack images of the single distal dendritic segments were obtained using the LSM710 with a 63× objective (NA 1.4; Zeiss) or the FV1000 with a 100× objective (NA 1.4; Olympus). The dendrite was reconstructed in the 3D surpass view in IMARIS. The heads of dendritic protrusions oriented in both the x–y plane and the z-direction were counted. The number of protrusions was divided by the length of the analyzed dendritic segment, which was measured with IMARIS FilamentTracer software.
The head width of dendritic spines was measured using the line tool in ImageJ. Only the protrusions oriented in the x–y direction were analyzed. The longest width within the distal half-region of each protrusion was measured.
For quantification of number of vGluT2 puncta on the soma, z-stack images of whole cell bodies of EGFP-positive PCs were obtained using the FV1000 with a 60× objective (NA 1.40; Olympus). The number of vGluT2-immuopositive puncta on the EGFP-positive soma was counted from each z-plane and combined.
Electrophysiology.
Parasagittal cerebellar slices (200 μm thick) were prepared from ICR mice in which miR or CRISPR/Cas9 constructs were transfected by IUE, as described previously (Kakegawa et al., 2008). Whole-cell patch-clamp recordings were made from EGFP-positive PCs using a 60× water-immersion objective attached to an upright microscope (BX51WI; Olympus) at room temperature. The resistance of the patch pipettes was 1.5–3 MΩ when filled with an intracellular solution composed of (in mm): 150 Cs-gluconate, 10 HEPES, 4 MgCl2, 4 Na2ATP, 1 Na2GTP, 0.4 EGTA, and 5 lidocaine N-ethyl bromide (QX-314) (pH 7.25, 295 mOsm/kg). The solution used for the slice storage and recording consisted of the following (in mm): 125 NaCl, 2.5 KCl, 2 CaCl2, 1 MgCl2, 1.25 NaH2PO4, 26 NaHCO3 and 10 d-glucose, which bubbled continuously with a mixture of 95% O2 and 5% CO2. Picrotoxin (100 μm; Sigma) was always present in the saline to block the inhibitory inputs.
To evoke the EPSCs derived from the climbing fiber (CF-EPSCs) and parallel fiber (PF-EPSCs) inputs onto the PCs, square pulses were applied through a stimulating electrode placed on the granular layer (10 μs, 0–200 μA) and the molecular layer (∼50 μm away from the pial surface; 10 μs, 0–200 μA), respectively. Selective stimulation of the CFs and PFs was confirmed by paired-pulse depression and paired-pulse facilitation, respectively, of EPSC amplitudes at a 50 ms interstimulus interval. Current responses were recorded with an Axopatch 200B amplifier (Molecular Devices) and the pCLAMP system (version 9.2; Molecular Devices) was used for data acquisition and analysis. The signals were filtered at 1 kHz and digitized at 4 kHz.
Results
Embryonic RORα knock-down recapitulates staggerer phenotypes
To characterize the roles of RORα at different developmental time points, we introduced a plasmid vector encoding miR against RORα (miR-RORα) together with EGFP into the PCs at E11.5 by IUE (Fig. 1A,B). Using this protocol, we previously showed selective transfection of the PCs in vivo (∼96% of the transfected cells in the cerebellar cortex; Nishiyama et al., 2012). In the present study, immunostaining of electroporated cerebella at P14 revealed that the protein levels of RORα in EGFP-expressing cells were significantly reduced [knock-down (KD): 9.3 ± 3% of untransfected control, n = 10 cells from 2 mice] compared with PCs transfected with a control vector encoding miR targeting a scrambled sequence (miR-SC: 106 ± 6% of untransfected control, n = 10 cells from 3 mice; KD vs SC p = 3.84 × 10−9 Student's t test). PCs expressing miR-SC exhibited the typical morphology of young PCs, with one thick stem dendrite that gave rise to many branches studded with spines in the sagittal plane (Fig. 1C). In contrast, PCs expressing miR-RORα exhibited thin, curvy dendrites with a few branches that lacked dendritic spines (Fig. 1D, top). The somata of 44.4% of the PCs expressing miR-RORα (n = 27 cells from 4 mice) were mislocalized in the internal granular layer (IGL) (Fig. 1B,D, bottom). The number of branch points per 10 μm of dendrites was significantly reduced in the PCs expressing miR-RORα, regardless of their soma position in the IGL or the Purkinje layer (PL) compared with PCs expressing miR-SC (Fig. 1E). As reported in wild-type PCs (Berry and Bradley, 1976; Bradley and Berry, 1978; Fujishima et al., 2012), control PCs expressing miR-SC exhibited longer proximal (first and second order) than distal (≥ third order) dendrites, which remained relatively constant at higher orders (Fig. 1F). In contrast, PCs expressing miR-RORα lacked highly branched (>15th branch order) dendrites (Fig. 1F). Dendrite segments at branch orders 1–6 were significantly longer in PCs expressing miR-RORα than control PCs expressing miR-SC regardless of their soma position (SC: 8.1 ± 0.5 μm, n = 180 from 3 mice vs knock-down PL: 25 ± 2 μm, n = 293 from 2 mice, p = 1.30 × 10−6 and vs knock-down IGL: 29 ± 2 μm, n = 206 from 2 mice, p = 1.30 × 10−6, ANOVA followed by Tukey's test; Fig. 1F). To rule out the off-target effect of the miR, we cotransfected PCs with miR-RORα and a cDNA encoding HA-tagged RORα1, which was resistant to the miR (rRORα1-HA; Fig. 1G). The number of branch points (Fig. 1E) and length of each dendritic segment (Fig. 1F) were restored by the coexpression of rRORα1-HA. These results indicate that the miR-based knock-down of RORα at E11.5 recapitulates staggerer phenotypes, including the mislocalization of PC somata and their primitive dendrites.
Role for RORα in postnatal cellular migration and primitive dendritic regression
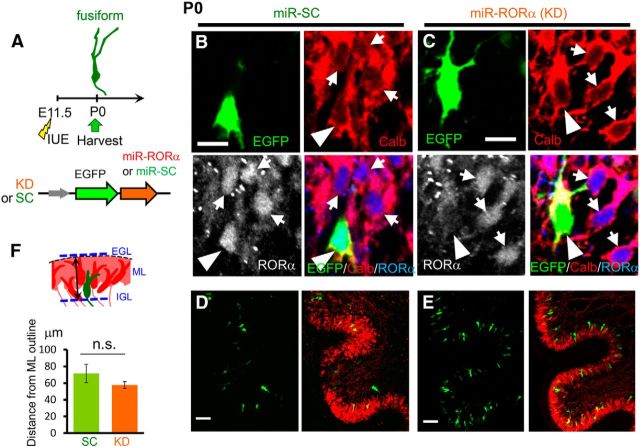
Postmitotic PCs migrate to the cerebellar cortex by E16 in a manner dependent on Reelin signaling to form a few-cell layer called the “Purkinje plate” before forming a single-cell PL (Sotelo and Dusart, 2009; Miyata et al., 2010). To clarify at which stage RORα regulates the positioning of PCs, we analyzed the effects of RORα knock-down at P0 after introducing miR-RORα at E11.5 (Fig. 2A). Immunohistochemical analyses of endogenous RORα revealed that miR-RORα effectively reduced RORα expression compared with miR-SC (KD: 24 ± 1% of untransfected control, n = 10 cells from 2 mice; SC: 75 ± 11% of untransfected control, n = 12 cells from 2 mice; KD vs SC p = 0.00103 Student's t test; Fig. 2B,C) at P0. Nevertheless, the location of PC somata expressing miR-RORα and miR-SC was indistinguishable (Fig. 2D,E) and the distance between the PC somata and the upper outline of the calbindin-immunopositive molecular layer was similar between the PCs expressing miR-SC and miR-RORα (Fig. 2F). These results indicate that RORα is unlikely to be involved in the regulation of initial PC migration and the formation of Purkinje plates, which was completed by P0.
Figure 2.
Normal migration of RORα-deficient PCs at P0. A, Schematic diagram of the experimental procedure. The mice were electroporated with plasmid vectors encoding KD or SC miR at E11.5 and fixed at P0. B–E, Immunohistochemical analyses of the cerebellar slices at P0. The cerebellar slices electroporated with miR-SC (B, D) or miR-RORα (C, E) were immunostained with anti-calbindin antibody (Calb, red) and anti-RORα antibody (grayscale or blue). EGFP fluorescence is shown in green. Low-magnification images are shown in D and E. Arrowheads and arrows indicate transfected and nontransfected PCs, respectively. F, Position of transfected PC somata. The schematic diagram indicates how the position of the PC somata from the upper outline of the ML was measured. The bars in the lower graph represent the mean ± SEM distances. n = 13 cells from 2 mice (SC) and 22 cells from 2 mice (KD); n.s., not significant (p = 0.252 by Student's t test). Scale bars: B, C, 10 μm; D, E, 50 μm.
Next, we analyzed the effects of RORα knock-down at P4 after introducing miR-RORα at E11.5 (Fig. 3A). Control PCs expressing miR-SC were aligned in a monolayer, whereas most PCs expressing miR-RORα were located in the IGL (Fig. 3B,C). In addition, the neurites of PCs expressing miR-RORα were significantly longer than those expressing miR-SC (Fig. 3D–F). PCs that expressed miR-RORα but reached the PL tended to have shorter neurites than PCs mislocalized in the IGL; the expression level of endogenous RORα in PCs transfected with miR-RORα was inversely correlated with neurite length (Fig. 3G). In addition, PCs expressing low levels of RORα were located mostly in the IGL (Fig. 3G). These results indicate that cell-intrinsic RORα signaling is necessary for PCs to form a single layer in the PL after the Purkinje plate is formed. Because mislocalized PCs tended to have longer neurites, the RORα-dependent regression of dendrites may be necessary for PCs to align in a straight line. Alternatively, the reduction of neurites length may not occur in the mislocalized environment.
Figure 3.
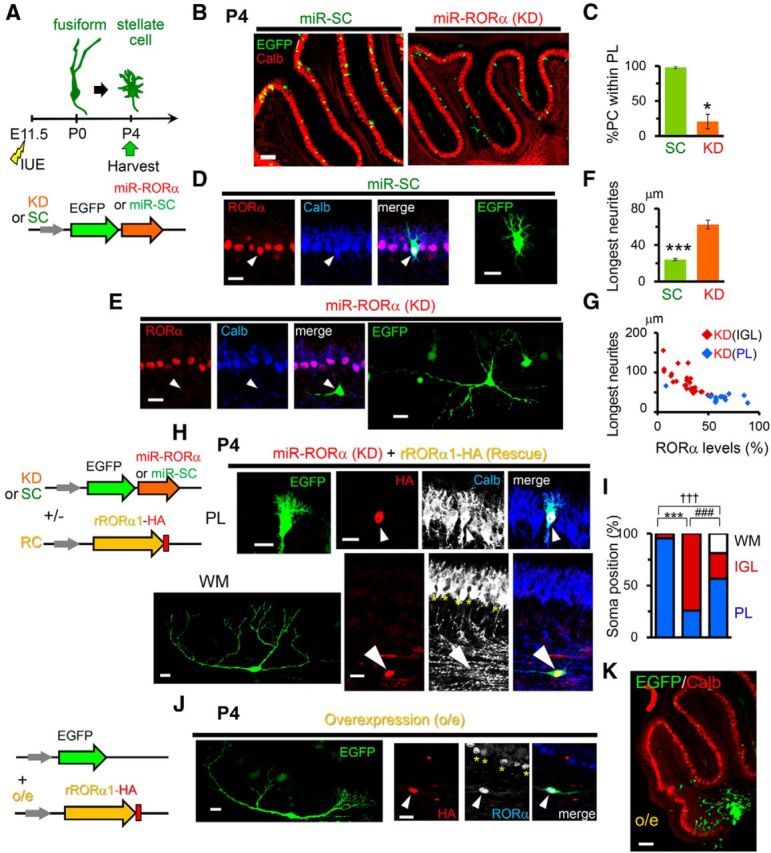
RORα regulates PC migration and positioning by P4. A, Schematic diagram of the experimental procedure. The mice were electroporated at E11.5 and fixed at P4. B, Location of PCs expressing miR-SC or miR-RORα at P4. The positions of transfected PCs (EGFP, green) were compared with other PCs immunostained with anti-calbindin (Calb, red) antibodies. Scale bar, 100 μm. C, Percentage of transfected PCs located in the PL. Bars represent the mean ± SEM of three independent experiments (*p = 0.0181, Student's t test). D, E, Confocal images of the PCs expressing miR-SC (D) and miR-RORα (E). Immunofluorescent signals of RORα (red) and calbindin (Calb, blue) together with stacked EGFP images are shown. The arrowheads indicate transfected PCs. Scale bars, 20 μm. F, Length of the longest neurites of the PCs expressing miR-SC (SC) and miR-RORα (KD). The bars represent the mean ± SEM lengths. n = 27 cells from 3 mice (SC) and n = 41 cells from 3 mice (KD), ***p = 4.52 × 10−10 (Student's t test). G, Relationship between RORα expression levels and the longest neurite lengths of PCs expressing miR-RORα (KD). Red and blue dots indicate PCs located within the IGL and PL, respectively. H, Partial rescue of RORα knock-down by the coexpression of miR-resistant rRORα1-HA in PCs. Exogenous RORα1 (HA, red) and a PC marker calbindin (Calb, blue or grayscale) are immunostained at P4. The upper and bottom indicate EGFP-positive PCs (green) located in the PL and white matter (WM), respectively. The arrowheads indicate transfected PCs. The asterisks in the bottom indicate nontransfected PCs. Scale bars, 20 μm. The schematic diagram on the left shows the plasmids used. I, Location of the PCs expressing miR-SC (SC, left bar), miR-RORα (KD, middle bar), and miR-RORα plus rRORα1-HA (RC, right bar). The percentages of PCs located in the WM, IGL, and PL are shown. n = 70 cells from 3 mice (SC), n = 58 cells from 3 mice (KD), and n = 85 cells from 3 mice (RC), ***p = 7.27 × 10−16, †††p = 8.25 × 10−8, and ###p = 8.83 × 10−4, respectively (χ2 test followed by Bonferroni's correction). J, K, Overexpression of RORα from E11.5 impaired PC migration from the WM. Immunofluorescent signals of exogenous (HA, red) and both exogenous and endogenous RORα (blue or grayscale) are shown together with a transfection marker EGFP (green). The arrowheads and asterisks indicate transfected and nontransfected PCs, respectively. The low-magnification view is shown in K. The schematic diagram on the left shows the plasmids used. Scale bars: J, 20 μm; K, 100 μm.
To rule out the off-target effect of miR, we cotransfected PCs with miR-RORα and rRORα1-HA cDNA (Fig. 3H). Most PCs coexpressing miR-RORα and rRORα1-HA were localized to the PL (Fig. 3H, top). Unexpectedly, some PCs cotransfected with miR-RORα and rRORα1-HA were mislocalized to the white matter and displayed very long neurites (Fig. 3H, bottom, I). When RORα1-HA was overexpressed without miR-RORα, most PCs exhibited long neurites (Fig. 3J) located in the white matter (Fig. 3K). These results indicate that the incomplete rescue by rRORα1 (Fig. 3I) was caused by the overexpression of rRORα1. In addition, these results indicate that low expression levels of endogenous RORα during the early developmental stages is necessary for the initial migration of PCs to reach the cerebellar cortex; after this initial migration, RORα is likely required for terminal translocation of PCs to align in the PL and pruning of neurites by P4.
RORα regulates reorganization of dendrites after the stellate cell stage
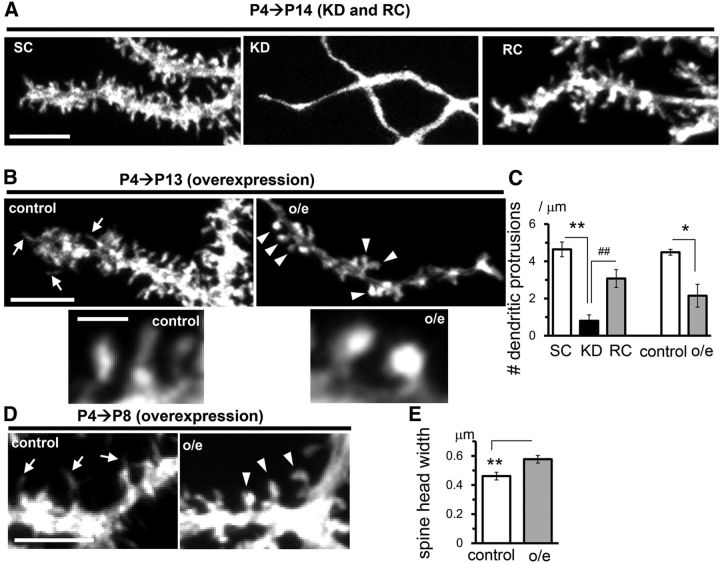
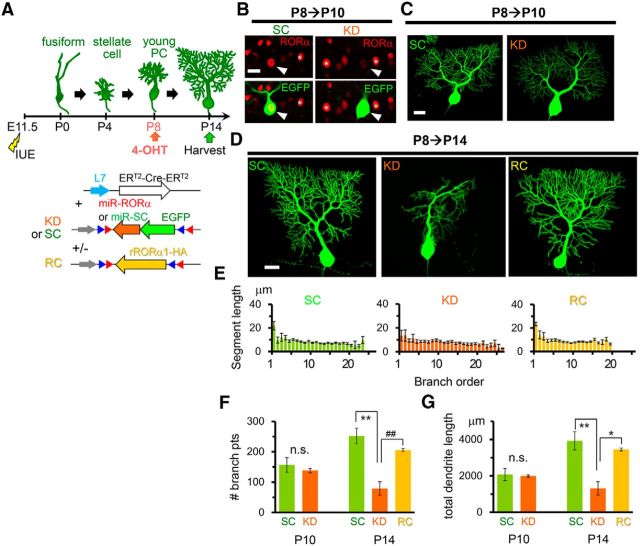
Although RORα is expressed in the PCs during development and throughout adulthood, the role of RORα beyond the fusiform stage is unclear. To address this, we used an inducible Cre-based gene knock-down system. The PC-specific L7 promoter (Oberdick et al., 1990) was used to express the conditionally active form of Cre recombinase ERT2-Cre-ERT2 (Matsuda and Cepko, 2007). By coexpressing a plasmid encoding EGFP-tagged miR flanked by two incompatible sets of flox sequences (Stern et al., 2008), miR was designed to be expressed in a 4-OHT- and L7-dependent manner (CAG-flex-miR constructs; Fig. 4A). When 4-OHT was administered from P4, endogenous RORα expression was already reduced in EGFP-positive PCs at P6 (Fig. 4B). At this stellate cell stage, no differences in dendrite morphology between PCs expressing miR-SC and miR-RORα were observed (Fig. 4B). However, at P14, PCs expressing miR-RORα from P4 displayed thin, curvy, branchless dendrites (Fig. 4C) that resembled staggerer PCs and PCs in which miR-RORα was introduced at E11.5 (Fig. 1C). Indeed, PCs expressing miR-RORα had significantly longer dendrite segments at branch orders 1–6 than PCs expressing miR-SC (KD: 17 ± 1 μm, n = 281 from 3 mice vs SC: 9.3 ± 0.6 μm, n = 195 from 2 mice, p = 1.29 × 10−6 and vs rescue: 9.5 ± 0.4 μm, n = 411 from 2 mice, p = 1.29 × 10−6, ANOVA followed by Tukey's test), but lacked highly branched (>15th branch order) dendrites (Fig. 4D). As a result, the number of dendritic branch points (Fig. 4E) and the total dendritic length were dramatically reduced in PCs expressing miR-RORα (Fig. 4F). Furthermore, although control PCs treated with miR-SC typically had thick, single dendrites (Fig. 4G), PCs expressing miR-RORα often had multiple thin, apical dendrites (Fig. 4H,I) in addition to numerous short, perisomatic dendrites (SC: 0 ± 0, n = 4 cells from 2 mice vs KD: 11 ± 1.1, n = 7 cells from 3 mice, p = 0.0169, Kruskal–Wallis test followed by Steel–Dwass test; Fig. 4G). These results indicate that RORα is required, not only for dendrite pruning at the fusiform stage (Boukhtouche et al., 2006), but also for the remodeling of dendrites during the transition from the stellate cell stage to the young PC stage.
To rule out the off-target effect, we coexpressed miR-RORα and rRORα1-HA cDNA in a 4-OHT- and L7-dependent manner (Fig. 4A) from P4. At P14, the branching pattern (Fig. 4D), number of branches (Fig. 4E), total dendrite length (Fig. 4F), diameter of the thickest stem dendrites (Fig. 4G,H), and number of perisomatic dendrites [rescue (RC):0.4 ± 0.3, n = 7 cells from 2 mice; RC vs SC p = 0.497; RC vs KD p = 0.00365, Kruskal–Wallis test followed by Steel–Dwass test; Fig. 4G] were restored by the coexpression of rRORα1-HA (Fig. 4C), confirming the specific effect of miR-RORα.
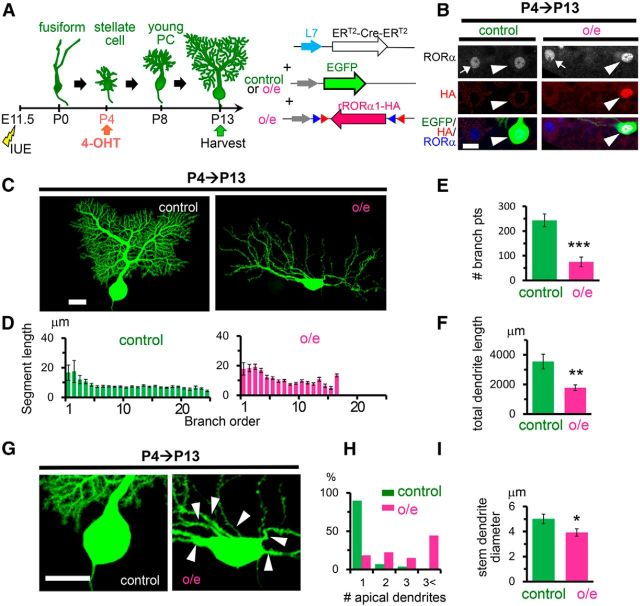
In contrast, the number of apical dendrites (Fig. 4G,I) was not restored by rRORα1-HA. Therefore, to examine the effect of the overexpression of RORα, we expressed rRORα1-HA alone in a 4-OHT- and L7-dependent manner from P4 (Fig. 5A). Immunohistochemical analyses revealed that RORα is overexpressed in the nucleus of PCs transfected with rRORα1-HA (Fig. 5B). Interestingly, PCs overexpressing rRORα1-HA also resembled staggerer PCs at P13 (Fig. 5C); these PCs had significantly longer dendrite segments at branch orders 1–6 than PCs expressing EGFP only (control: 9.3 ± 0.7 μm, n = 162 vs overexpression: 15.2 ± 0.7 μm, n = 467, 3 mice each, p = 3.93 × 10−10, Student's t test), but lacked highly branched (>15th branch order) dendrites (Fig. 5D). The number of dendritic branch points (Fig. 5E) and total dendritic length were dramatically reduced in PCs overexpressing rRORα1-HA (Fig. 5F). PCs overexpressing rRORα1-HA also had multiple stem dendrites (Fig. 5G,H), which were thinner than control PCs (Fig. 5I). In contrast, the overexpression of RORα1 reportedly accelerated the regression of primitive dendrites at the fusiform stage in cultured cerebellar slices in vitro (Boukhtouche et al., 2006). These results indicate that appropriate levels of RORα are necessary for the formation of stem dendrite and dendritic branches during the transition from the stellate cell stage to the young PC stage in vivo.
Figure 5.
Overexpression of RORα from the stellate cell stage impairs dendritic pruning and branching. A, Schematic diagram of the experimental procedure. The mice were electroporated at E11.5 and the expression of rRORα1-HA (overexpression) was induced by an injection 4-OHT at P4. B, Application of 4-OHT at P4 induced the overexpression of exogenous RORα. Immunofluorescent signals for exogenous RORα (HA, red) and both exogenous and endogenous RORα (blue or grayscale) at P13 are shown. The arrows and arrowheads indicate untransfected and transfected PCs (EGFP, green), respectively. Scale bar, 20 μm. C, Dendritic branching of PCs expressing EGFP (control) with or without RORα1-HA (overexpression) at P13. Representative stacked images of EGFP fluorescence are shown. Scale bar, 20 μm. D, Length of dendritic segment at each branching order. Bars represent the mean ± SEM segment lengths of n = 4 (control) and n = 7 (overexpression) cells from 3 mice. E, Number of dendritic branch points. Bars represent the mean ± SEM branch points. n = 4 (control) and n = 7 (overexpression) cells from 3 mice, ***p = 0.000629 (Student's t test). F, Total dendritic length per single PCs. Bars represent the mean ± SEM total dendritic length. n = 4 (control) and n = 7 (o/e) cells from 3 mice, **p = 0.00347 (Student's t test). G, Enlarged images of the somata shown in C, Arrowheads indicate the apical dendrites. Scale bar, 20 μm. H, Number of apical primary dendrites per single PC. The percentages of PCs showing 1, 2, 3, and more apical primary dendrites are shown. n = 29 (control) and n = 27 (overexpression) cells from 3 mice, ***p = 8.76 × 10−8 (χ2 test). I, Diameters of the thickest primary dendrites. Bars represent the mean ± SEM diameters of nine PCs, *p = 0.0375 (Student's t test).
RORα regulates dendritic spine formation after the stellate cell stage
Unlike in other cell types, dendritic spines are cell autonomously formed in PCs in the absence of PF inputs (Yuste and Bonhoeffer, 2004; Sotelo and Dusart, 2009). To investigate the role of RORα in spinogenesis, we examined the dendritic protrusions in PCs in which RORα was knocked down or overexpressed from P4. Although PCs expressing miR-SC were studded with spines and filopodia at P14, PCs expressing miR-RORα completely lacked protrusions on their dendrites (Fig. 6A), indicating that RORα is indispensable for spine formation after the stellate cell stage. Interestingly, PCs overexpressing rRORα1 from P4 had stubby or mushroom-like shapes (Fig. 6B, arrowheads, E) at P13, in contrast to the thin or filopodia-like spines observed in control PCs expressing only EGFP (Fig. 6B, arrows). However, the number of spines was reduced in PCs overexpressing rRORα1 (Fig. 6B,C). As a result, the coexpression of rRORα1 did not completely restore the reduced number of dendritic protrusions induced by miR-RORα (Fig. 6A,C). The overexpression of rRORα1 from P4 increased the occurrence of stubby or mushroom-shaped spines even at P8 (Fig. 6D, arrowheads), when almost all dendritic protrusions were filopodia or thin spines in the control PCs (Fig. 6D, arrows). Dendritic filopodia in PCs are not only thought to serve as the precursors to spines, but also to be involved in dendritic growth (Berry and Bradley, 1976; Bradley and Berry, 1978). Therefore, RORα overexpression in PCs during the stellate cell stage may have restricted dendrite formation by promoting the maturation of filopodia. Together, these results indicate that the appropriate expression levels of RORα are important for the proper development of dendrites and spines at the stellate cell stage.
Figure 6.
Knock-down and overexpression of RORα at the stellate cell stage cause distinct effects on the dendritic spines. The expression of miR-SC (SC), miR-RORα (KD), miR-RORα plus rRORα1-HA (RC), or rRORα1-HA (overexpression) was induced by 4-OHT at P4. A, A deficiency in RORα causes the loss of spines at the distal dendrites of PCs at P14. Representative confocal images of EGFP signals are shown. Scale bar, 5 μm. B, Excess RORα causes the loss of spines but facilitates the maturation of spines in PCs at P13. Representative confocal images of EGFP signals in the distal dendrites (top) and their enlarged views (bottom) are shown. Scale bars, 5 (top) and 1 μm (bottom). C, Densities of the dendritic protrusions per length. Bars represent the mean ± SEM number of protrusions (per micrometer) of the independent experiments, n = 4 cells from 2 mice (SC), n = 6 cells from 3 mice (KD), and n = 5 cells from 2 mice (RC), **p = 0.0000615 and ##p = 0.00309 (ANOVA followed by Tukey's test); n = 3 cells from 2 mice (control), n = 5 cells from 2 mice (overexpression), *p = 0.0144 by Student's t test. D, Overexpression of RORα from P4 facilitates the maturation of dendritic spines at P8. Representative confocal images of EGFP signals in the distal dendrites are shown. Scale bar, 5 μm. Arrows indicate dendritic protrusions. E, Width of the distal dendritic spines at P13. Bars represent the mean ± SEM spine head widths, n = 47 spines from 2 mice (control) and n = 101 spines from 2 mice (overexpression), **p = 0.0385 by Student's t test.
RORα regulates dendritic growth and maintenance after the young PC stage
To investigate the role of RORα after the young PC stage, when apical dendrites form further branches after the perisomatic dendrites are removed, RORα was knocked down from P8 (Fig. 7A). At P10, endogenous RORα expression was already reduced in PCs expressing miR-RORα compared with PCs expressing miR-SC (KD: 30 ± 4% of untransfected control, n = 6 PCs; SC: 96 ± 12% of untransfected control, n = 6 PCs; 3 mice each; KD vs SC, p = 0.00229 Student's t test; Fig. 7B), whereas no differences in dendrite morphology between PCs expressing miR-SC and miR-RORα were observed (Fig. 7C). However, at P14, PCs expressing miR-RORα displayed atrophic dendrites (Fig. 7D). Although the length of the first-order branch tended to be reduced in PCs expressing miR-RORα, the length of each second- and higher-order dendritic branch was similar between PCs expressing miR-SC and miR-RORα (Fig. 7E). Nevertheless, PCs expressing miR-RORα had significantly fewer dendritic branch points (Fig. 7F) and shorter dendrites (Fig. 7G) than PCs expressing miR-SC, indicating that the number of terminal dendritic segments was selectively reduced in PCs expressing miR-RORα. The coexpression of rRORα1-HA successfully rescued the abnormal dendritic morphology caused by miR-RORα (Fig. 7D–G), ruling out the off-target effect of miR-RORα. The dendrites of PCs expressing miR-SC gained ∼800 μm in length and 100 branch points between P10 and P14, whereas the dendrites of PCs expressing miR-RORα lost ∼700 μm in length and 60 branch points (Fig. 7F,G). Therefore, the loss of RORα not only interfered with normal dendritic growth, but also caused a reduction in the existing terminal dendrites. These results indicate that RORα is required for the formation and maintenance of growing mature dendrites during the young PC stage.
Figure 7.
Knock-down of RORα after the young PC stage impairs the formation and maintenance of dendrites. A, Schematic diagram of the experimental procedure. The expression of miR-SC (SC), miR-RORα (KD), and miR-RORα plus rRORα1-HA (rescue: RC) was induced by 4-OHT at P8. B, Application of 4-OHT at P8 induces the reduction of endogenous RORα in PCs expressing miR-RORα at P10. Immunofluorescent signals for endogenous RORα (red) were examined in EGFP-positive PCs (green, arrowheads). The asterisks indicate neighboring untransfected PCs. Scale bar, 20 μm. C, Gross dendritic arborization of PCs expressing miR-SC and miR-RORα is indistinguishable at P10. Representative stacked EGFP images of PCs are shown. Scale bar, 20 μm. D, Knock-down of RORα from P8 induces atrophic PC dendrites at P14. Representative stacked EGFP images of PCs expressing miR-SC (SC), miR-RORα (KD), and miR-RORα plus rRORα1-HA (RC) are shown. Scale bar, 20 μm. E, Length of the dendritic segment of PCs at each branch order at P14. Bars represent the mean ± SEM segment lengths of n = 3 cells from 2 mice (SC and RC) or n = 5 cells from 2 mice (KD). F, Total number of dendritic branch points in PCs at P10 and P14. Bars represent the mean ± SEM branch points. P10, n = 3 cells from 3 mice (SC and KD), n.s., not significant (p = 0.575 by Student's t test); P14, n = 3 cells from 2 mice (SC and RC) and n = 5 cells from 2 mice (KD), **p = 0.00123 and ##p = 0.00800, respectively (ANOVA followed by Tukey's test). G, Total dendritic length of single PCs at P10 and P14. Bars represent the mean ± SEM total dendritic lengths. P10, n = 3 cells from 3 mice (SC and KD); n.s., not significant (p = 0.818 by Student's t test); P14, n = 3 cells from 2 mice(SC and RC) and n = 5 cells from 2 mice (KD), *p = 0.0105 and **p = 0.00332, respectively (ANOVA followed by Tukey's test).
RORα is crucial for the maintenance of dendrites and spines in adult PCs
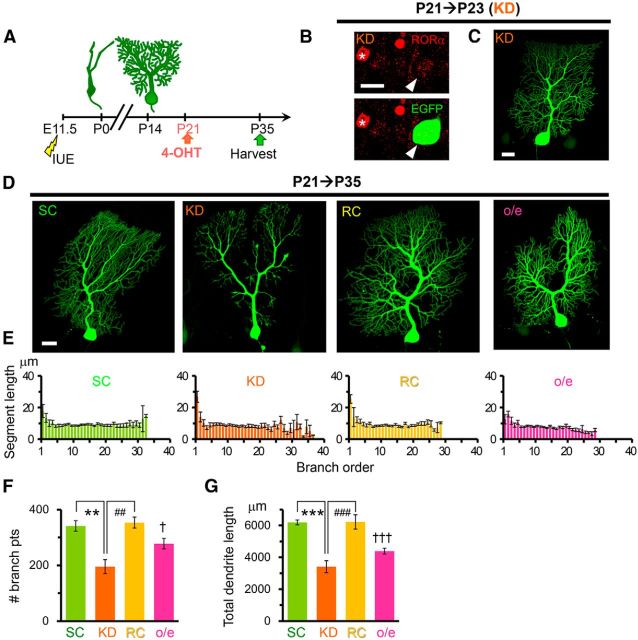
Finally, to further clarify the role of RORα in PCs after the completion of dendritic development, we knocked down RORα by injecting 4-OHT at P21 (Fig. 8A). At P23, endogenous RORα expression was reduced in PCs expressing miR-RORα compared with untransfected PCs (22 ± 3% of untransfected control, n = 8 cells from 2 mice; Fig. 8B), whereas no abnormalities in the dendrite morphology were observed (Fig. 8C). At P35, PCs expressing miR-RORα exhibited atrophic dendrites (Fig. 8D). Although the length of each dendritic branch was comparable between PCs expressing miR-SC and miR-RORα (Fig. 8E), the number of dendritic branch points (Fig. 8F) and total dendritic length (Fig. 8G) were significantly decreased in PCs expressing miR-RORα. The coexpression of rRORα1-HA successfully rescued the abnormal dendritic morphology caused by miR-RORα (Fig. 8D–G), ruling out the off-target effect of miR-RORα. These results further indicate that RORα is required for maintenance of the terminal dendrites of PCs, not only at the young PC stage (P8; Fig. 7), but also after PCs enter the mature stage.
Figure 8.
Knock-down and overexpression of RORα in mature PCs disturb the dendritic morphology. A, Schematic diagram of the experimental procedure. The expression of miR-SC (SC), miR-RORα (KD), miR-RORα plus rRORα1-HA (RC), or rRORα1-HA (overexpression) was induced by 4-OHT at P21. B, Application of 4-OHT at P21 reduced the expression of endogenous RORα in PCs expressing miR-RORα at P23. Immunofluorescent signals for endogenous RORα (red) were examined in EGFP-positive PCs (green, arrowheads). The asterisk indicates a neighboring untransfected PC. Scale bar, 20 μm. C, Gross dendritic arborization of EGFP-positive PCs expressing miR-RORα at P23. Scale bar, 20 μm. D, Knock-down of RORα from P21 induces atrophic PC dendrites at P35. Representative stacked EGFP images of PCs expressing miR-SC (SC), miR-RORα (KD), miR-RORα plus rRORα1-HA (RC), and rRORα1-HA (overexpression) are shown. Scale bar, 20 μm. E, Length of the dendritic segment at each branch order. Bars represent the mean ± SEM segment lengths of n = 5 cells from 3 mice (SC), n = 4 cells from 2 mice (KD), n = 3 cells from 3 mice (RC), and n = 4 cells from 2 mouse (overexpression). F, Total number of dendritic branch points in PCs at P35. Bars represent the mean ± SEM branch points. n = 5 cells from 3 mice (SC), n = 4 cells from 2 mice (KD), n = 3 cells from 3 mice (RC), **p = 0.00129 and ##p = 0.00184, respectively (ANOVA followed by Tukey's test); n = 4 cells from 2 mice (overexpression), †p = 0.0397 (Student's t test vs SC). G, Total dendritic length of single PCs at P35. Bars represent the mean ± SEM total dendritic lengths of the independent experiments. n = 5 cells from 3 mice (SC), n = 4 cells from 2 mice (KD), n = 3 cells from 3 mice (RC), ***p = 0.000227 and ###p = 0.000550, respectively (ANOVA followed by Tukey's test); n = 4 cells from 2 mice (overexpression), †††p = 0.000136 (Student's t test vs SC).
The number of dendritic spines on the remaining dendrites was also significantly reduced in PCs at P35 when RORα was knocked down from P21 (Fig. 9A,B). We also observed extraordinarily thin distal dendrites that completely lacked spiny protrusions (13 ± 1 spineless dendrites per PC, n = 4 PCs from 2 mice; Fig. 9A); such spineless dendrites were never observed in control PCs expressing miR-SC. These phenotypes in RORα-deficient PCs were completely rescued by the coexpression of RORα1-HA (Fig. 9A,B), indicating that the maintenance of spines also requires RORα in mature PCs.
Figure 9.
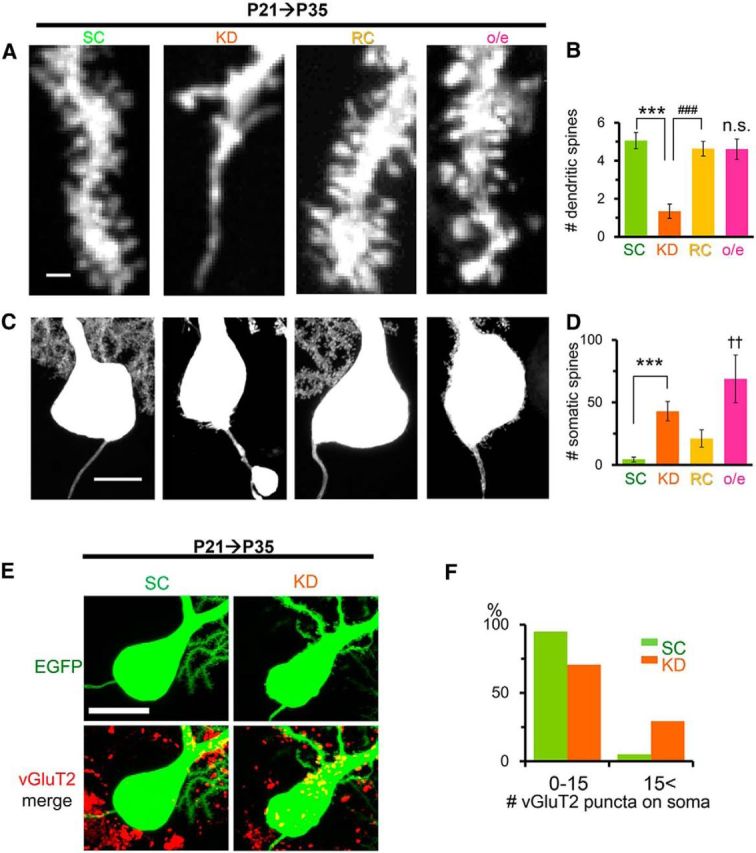
The distinct effects of knock-down and overexpression of RORα on dendritic and somatic spines of mature PCs. The expression of miR-SC (SC), miR-RORα (KD), miR-RORα plus rRORα1-HA (RC), or rRORα1-HA (overexpression) was induced by 4-OHT at P21. A, Representative confocal images of the distal dendrites of PCs at P35. Scale bar, 1 μm. B, Densities of dendritic spines per length. Bars represent the mean ± SEM number of protrusions (per micrometer). n = 6 cells from 3 mice (SC), n = 9 cells from 3 mice (KD), n = 5 cells from 3 mice (RC), ***p = 0.0000133 and ###p = 0.000100, respectively (ANOVA followed by Tukey's test); n = 12 PCs from 2 mice (overexpression), n.s., not significant vs SC (p = 0.566, Student's t test). C, Knock-down and overexpression of RORα from P21 induces somatic spiny protrusions in PCs at P35. Representative confocal images of EGFP signals at the somata are shown. Scale bar, 10 μm. D, Number of somatic spiny protrusions. Bars represent the mean ± SEM number of protrusions per PCs of the independent experiments. n = 8 (SC and KD) and n = 9 (RC) cells from 3 mice each; ***p = 0.000827 (ANOVA followed by Tukey's test), and n = 16 cells from 2 mice (overexpression); ††p = 0.00476 (Mann-Whitney U test vs SC). E, Representative confocal z-stacked images of the vGluT2 immunoreactivity (red) on EGFP-positive PC somata (green) at P35. Scale bar, 20 μm. F, Histograms showing the percentage of PCs contacted by >15 vGluT2-immunopositive puncta. The number of vGluT2 puncta on the somata of PCs expressing miR-SC (SC) or miR-RORα (KD) was counted at P35. n = 39 cells from 3 mice (SC) and n = 41 cells from 4 mice (KD), **p = 0.00451 (χ2 test).
In contrast to spineless dendrites, the soma of RORα-deficient PCs exhibited spine-like protrusions (Fig. 9C,D). Immunohistochemical analyses revealed that more vGluT2-immunopositive puncta were located on the somata of these RORα-deficient PCs (Fig. 9E,F), suggesting that these spines were at least partly innervated by CFs. The abnormal spines on the soma were not rescued completely by the coexpression of RORα1-HA (Fig. 9C,D). Therefore, we examined the effect of the overexpression of RORα1-HA from P21 in the absence of miR-RORα. Although the spines in PCs overexpressing RORα were significantly wider than that in control PCs (overexpression: 0.57 ± 0.02 μm, n = 119 spines from 2 mice vs SC: 0.49 ± 0.01 μm, n = 58 spines from 3 mice, p = 0.000152, Student's t test), the number of spines on the dendrites was comparable (Fig. 9A,B). In contrast, PCs overexpressing RORα exhibited many spine-like somatic protrusions (Fig. 9C,D), indicating that appropriate levels of RORα are necessary to prevent emergence of somatic spines in mature PCs. Some axons of the RORα-deficient PCs exhibited abnormal torpedo-like swelling (Fig. 9C), which is often observed in degenerating neurons (Coleman, 2005). Such axon swellings were never observed in PCs overexpressing RORα with or without miR-RORα. Together, these results indicate that RORα is crucial for the maintenance of mature cellular morphology, including the maintenance of dendritic spines, the prevention of somatic protrusions, and axon swellings.
RORα is required for the maintenance of synaptic functions, but not innervation patterns in mature PCs
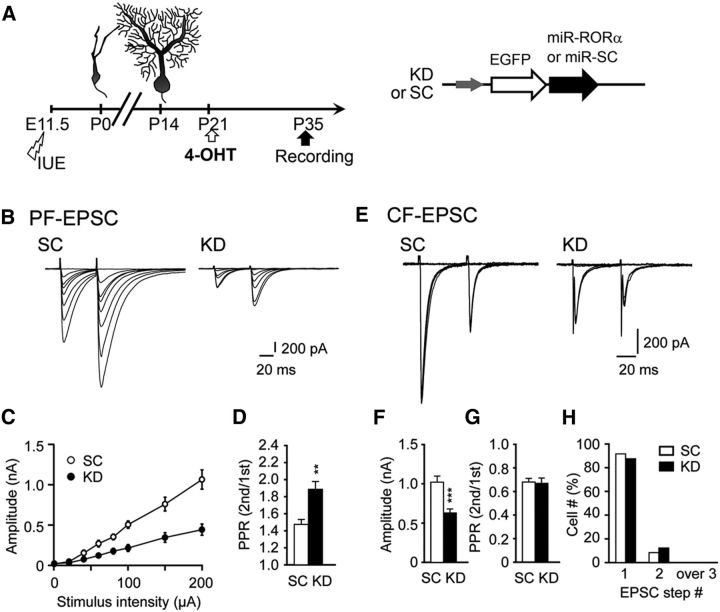
Functional abnormalities at the PF–PC synapses have been reported in staggerer PCs (Mitsumura et al., 2011). To examine the roles of RORα in the maintenance of synaptic functions in mature PCs, we performed whole-cell patch-clamp recordings in P35 slices from PCs in which RORα was knocked down from P21 (Fig. 10A). PF-evoked EPSCs were markedly decreased in PCs expressing miR-RORα (Fig. 10B,C). In addition, the paired-pulse ratio of PF-EPSCs was significantly increased in PCs expressing miR-RORα, indicating that the release probability was decreased in PFs (Fig. 10D). These results are similar to those observed in staggerer PCs (Mitsumura et al., 2011), indicating that RORα is required for the maintenance of normal PF synaptic functions. Similarly, the CF-evoked EPSCs were significantly decreased in PCs expressing miR-RORα (Fig. 10E,F). In contrast to PF-EPSCs, the paired-pulse ratio of CF-EPSCs was unaffected (Fig. 10G), indicating that the loss of RORα in PCs differentially affected the presynaptic release probability of PFs and CFs.
Figure 10.
Knock-down of RORα impairs synaptic functions, but not innervation patterns, in mature PCs. A, Schematic diagram of the experimental procedure. The expression of miR-SC (SC) or miR-RORα (KD) was induced by 4-OHT at P21 and whole-cell patch-clamp recordings were performed from PCs in cerebellar slices at P35. B–D. PF-EPSCs recorded from PCs clamped at −80 mV. Representative PF-EPSCs in PCs expressing miR-SC and miR-RORα at various stimulus intensities (B). Shown is the input–output relationship of PF-EPSCs (C, n = 9 cells for SC and n = 12 cells for KD, 3 mice each). D, Paired-pulse ratio (PPR) of PF-EPSCs at an interstimulus interval (ISI) of 50 ms (n = 22 cells for SC and n = 20 cells for KD, 3 mice each; **p = 0.003 by the Mann–Whitney U test). The PPR is defined as the amplitude of the second EPSC divided by that of the first EPSC. Error bars indicate SEM. E–H, CF-EPSCs recorded from PCs clamped at −10 mV. E, Representative CF-EPSCs in PCs expressing miR-SC and miR-RORα at various stimulus intensities. F, G, Histograms showing the mean amplitude (F, ***p = 0.0001 by the Mann–Whitney U test) and the PPR of CF-EPSCs (G; at ISI = 50 ms, p = 0.90 by the Mann–Whitney U test). Error bars indicate SEM. H, Percentage of PCs innervated by single and multiple CFs. The number of CF-EPSCs induced by different stimuli thresholds (0–200 μA) was counted. n = 17 cells from 3 mice for SC and n = 35 cells from 4 mice for KD (E–H).
Immature PCs are innervated by several CFs originating from the inferior olivary nucleus, but most wild-type PCs become innervated by single CFs by the end of the third postnatal week. In contrast, staggerer PCs remain innervated by multiple CFs even in adulthood (Crepel et al., 1980; Mariani and Changeux, 1980). To investigate the role of RORα in maintaining a single CF innervation onto mature PCs, we examined the stimulus thresholds required to evoke CF-EPSCs in PCs expressing miR-RORα from P21. At P35, we found that CF-EPSCs were evoked in an all-or-none manner in most PCs expressing miR-RORα, as well as in control PCs expressing miR-SC (Fig. 10H). Therefore, like wild-type PCs, PCs remain innervated by single CFs when RORα is knocked down from P21.
Gene deletion in mature PCs using CRISPR/Cas9 genome editing system replicated the effects of miR-based knock-down of RORα
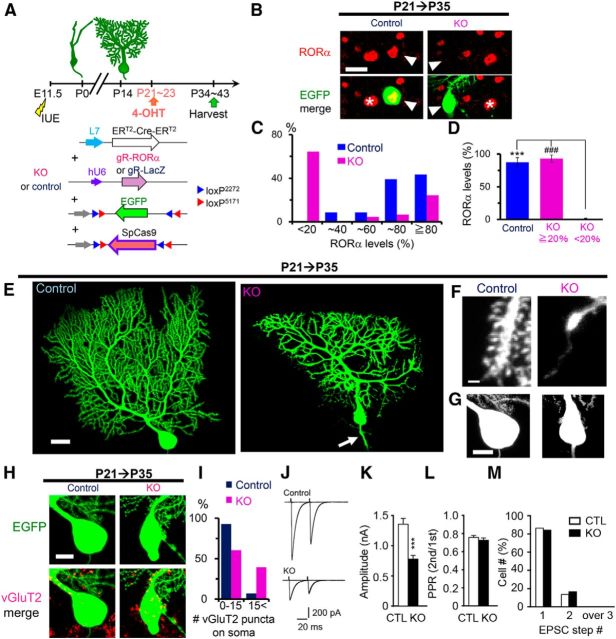
In contrast to our study, multiple functional CF innervation was reported to be reestablished on mature PCs in Cre-mediated conditional RORα knock-out mice (Chen et al., 2013). To investigate the possibility that the discrepant result was caused by remaining levels of RORα in PCs expressing miR-RORα, we knocked out RORα in mature PCs using an IUE-based tamoxifen-inducible palindromic repeats (CRISPR)/CRISPR-associated (Cas) 9 (CRISPR/Cas9) genome editing technique. We introduced Cre-dependent CAG-flex-SpCas9 and CAG-flex-EGFP vectors, together with an L7-ERT2-Cre-ERT2 plasmid and a plasmid vector encoding a guide RNA (gRNA) against RORα, into E11.5 cerebella by IUE (Fig. 11A). Single 4-OHT administration at P21 induced almost complete loss of RORα immunoreactivity in 64% of EGFP-positive PCs at P35 (Fig. 11B,C). Mean RORα levels of these PCs were significantly lower than those in the rest of PCs or PCs expressing a control gRNA against the LacZ gene (Fig. 11D). These results indicate that Cas9-gRNA exerted frame-shift null mutations in RORα in vivo.
Figure 11.
Tamoxifen-induced RORα knock-out in mature PCs by CRISPR/Cas9. A, Schematic diagram of the experimental procedure. L7-ERT2-Cre-ERT2 and chimeric guide RNA against LacZ (gR-LacZ) or RORα (gR-RORα) are cotransfected with CAG-flex-EGFP and CAG-flex-SpCas9. Expression of SpCas9 was induced by 4-OHT at P21–P23 and mice were killed for analyses at P34–P43. B, Representative confocal images of immunofluorescent signals for endogenous RORα (red) of the EGFP-positive PCs (green, arrowheads) cotransfected with gR-LacZ (control) or gR-RORα (KO) at P35. 4-OHT was administered at P21. The asterisk indicates a neighboring untransfected PCs. Scale bar, 20 μm. C, Distribution of relative RORα levels in the PCs cotransfected with control gR-LacZ (blue, control) or gR-RORα (magenta, KO). n = 23 cells from 2 mice (control) and n = 45 cells from 4 mice (KO). D, Averaged RORα levels in control PCs and PCs expressing gR-RORα. According to the RORα fluorescent intensity, PCs were classified into low (when the RORα intensity was < 20% of the nearby EGFP-negative PCs) and high (≥ 20%) groups. The RORα fluorescent intensity was averaged in each group. All control PCs fall into the high group, but KO PCs were divided into low and high groups. Bars represent the mean ± SEM. ***p = 1.27 × 10−6 and ###p = 1.27 × 10−6 (ANOVA followed by Tukey's test). E–H, Representative confocal z-stacked EGFP images of control and RORα-knock-out (KO) PCs at P35. Cas9-dependent RORα deletion in mature PCs caused dendritic atrophy and abnormal spine formation. E, Dendritic morphology. The arrow indicates axon swelling. Scale bar, 20 μm. F, Spines on distal dendrites. Knock-out PCs showed extraordinarily thin spineless dendrites. Scale bar, 1 μm. G, Abnormal spines on knock-out PC soma. Scale bar, 10 μm. H, Confocal z-stacked images of immunofluorescent signals for vGluT2 (red) on the EGFP-positive control or knock-out PC soma (green) at P35. Note that somal and axonal swelling is observed in the knock-out PC. Scale bar, 10 μm. I, Histograms showing the percentage of PCs contacted by >15 vGluT2-immunopositive puncta. n = 28 cells from 2 mice (control) and n = 33 cells from 3 mice (KO), **p = 0.00509 (χ2 test). J–M, CF-EPSCs recorded from PCs clamped at −10 mV. J, Representative CF-EPSCs in control PCs and knock-out PCs at various stimulus intensities (J). L, Histograms showing the mean amplitude (***p = 0.000249 by the Mann–Whitney U test) and the PPR (at ISI = 50 ms, p = 0.541 by the Mann–Whitney U test) of CF-EPSCs. Error bars indicate SEM. M, Percentage of PCs innervated by single and multiple CFs. The number of CF-EPSCs induced by different stimuli thresholds (0–200 μA) was counted. n = 22 cells from 2 mice for control and n = 30 cells from 3 mice for KO (J–M).
Cas9-induced knock-out of RORα induced morphological phenotypes similar to those observed in miR-induced RORα knock-down: dendritic branches (Fig. 11E) and spines (Fig. 11F) were degenerated, abnormal spines appeared on soma (Fig. 11G), and axonal were swollen (Fig. 11E, arrow). We also observed an increased number of vGluT2-immunopositive puncta (Fig. 11H) on the soma of Cas9-based RORα knock-out PCs (Fig. 11I). Furthermore, electrophysiological analyses revealed that CF-EPSCs were significantly decreased (Fig. 11J,K) without changes in the paired-pulse ratio (Fig. 11J,L). Importantly, RORα knock-out PCs, as well as PCs expressing control gRNA, were mostly innervated by single functional CFs at P34–P43 (Fig. 11J,M). These results indicate that miR-induced RORα knock-down largely reflected null phenotypes and that the maintenance of a mature single CF innervation pattern does not require RORα.
Discussion
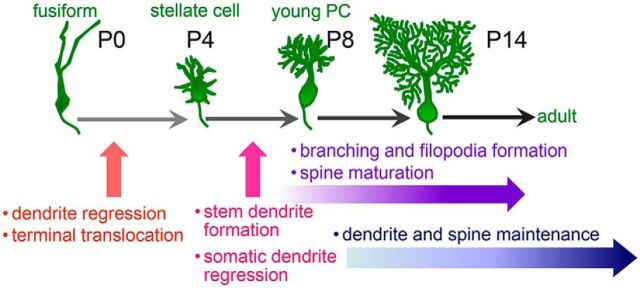
Using cell-specific and temporally controlled gene knock-down, knock-out, and induction methods, we investigated the roles of RORα at each developmental stage of PCs in vivo. We revealed that RORα is required, not only for the regression of primitive dendrites, but also for the alignment of the soma in the PL during the transition from the fusiform to the stellate cell stage (P0–P4; Fig. 12). During the transition from the stellate cell to the young PC stage (P4–P8), RORα is required for the elimination of somatic dendrites, leaving single apical stem dendrites; RORα is also necessary for dendritic branching, filopodia formation, and spine maturation (Fig. 12). Furthermore, after PCs reach a mature morphology, RORα is required for dendrite and spine maintenance. Therefore, endogenous RORα cell-autonomously plays multiple crucial roles in the morphogenesis of PCs during development and throughout adulthood.
Figure 12.
Roles for RORα at different developmental stages of PCs. Based on temporally controlled knock-down experiments, the major roles of RORα at each PC developmental stage (Sotelo and Dusart, 2009) are shown. During the transition from the fusiform to stellate cell stages (P0–P4), RORα is required for the regression of primitive dendrites and terminal translocation of PCs to align in a single PL. During the transition from the stellate cell to young PC stage (P4–P8), RORα regulates the elimination of somatic dendrites and the maturation of single-stem dendrites. RORα is crucial for the dendritic branching, filopodia formation, and spine maturation that occur from these stages. After the PCs reach mature morphology, RORα is still required for the maintenance of dendrites and spines.
Multiple roles for RORα in the early postnatal development of PC dendrites
Previous findings from organotypic cultures in vitro suggested that the main role of RORα was to regulate dendrite regression to enter the stellate cell stage (Boukhtouche et al., 2006). Our in vivo analyses confirmed that RORα mediates the elimination of primitive dendrites before P4. However, our analyses also revealed that RORα is involved in the alignment of the soma in the PL between P0 and P4. Although ectopic PC positioning is known to be caused by a loss of RORα in staggerer PCs (Gold et al., 2007), the stage at which RORα regulates PC positioning had been unknown. Furthermore, although endogenous RORα is highly expressed in PCs as early as E12.5 (Gold et al., 2007), the overexpression of RORα from E11.5 severely inhibited the migration of PCs from the white matter (Fig. 3K). These results indicate that PL formation consists of at least two different steps: migration to the cerebellar cortex and terminal translocation. Too high RORα expression is detrimental to the former step, whereas too low expression inhibits the latter step.
Mature PCs are characterized by a single stem dendrite. However, the mechanism by which a single stem dendrite is formed and selectively strengthened whereas other dendrites are eliminated is unknown. Our RORα knock-down experiments from P4 revealed that RORα plays crucial roles in such dendrite remodeling from the stellate cell to the young PC stages (Fig. 12). Interestingly, the overexpression of RORα from P4 also inhibited the pruning of surplus dendrites and the strengthening of stem dendrites. In contrast, overexpression of RORα did not cause such detrimental effects on the pruning process in organotypic cultures (Boukhtouche et al., 2006). Although several morphological features in PC dendrites are conserved in cultured PCs, there are a number of significant differences, such as the small arboreal size and delayed maturation of dendrites, probably due to reduced electrical activities and trophic support from neighboring cells and synaptic input from inferior olivary neurons (Baptista et al., 1994; Kawaguchi et al., 2010). Indeed, wild-type PCs retain three or four stem dendrites even at fully developed stages in a dissociated culture (Fujishima et al., 2012). These results indicate that the function of RORα in eliminating somatic dendrites is likely under the effect of extrinsic factors in vivo.
Analyses of Cre-mediated conditional RORα knock-out mice have previously indicated that multiple functional CF innervation was reestablished on mature PCs, together with the relocation of CF terminals to the PC soma (Chen et al., 2013). We confirmed that RORα knock-down (Fig. 9) and knock-out (Fig. 11) after P21 induced relocation of CF terminals on PC somata, indicating that RORα is required to maintain CF synapses on PC dendrites. In contrast, our electrophysiological analyses revealed that PCs remained innervated by single CFs when RORα was knocked down (Fig. 10) or knocked out (Fig. 11) after P21. Because RORα was deleted from P10 to P21 in the previous study (Chen et al., 2013), the mature CF single innervation pattern may not have been established properly during development. Our observations indicate that RORα is essential for the maintenance of dendritic complexity and functional PF and CF synapses, but not CF single innervation patterns, in mature PCs.
RORα is likely involved in the survival of PCs directly or indirectly. Indeed, RORα knock-down (Fig. 9C) and knock-out (Fig. 11E) PCs exhibited axonal swelling, which is often observed in degenerating neurons (Coleman, 2005). However, it is very difficult to analyze the survival phenotype in our experimental conditions because the number of transfected PCs and the efficiency of RORα knock-down were not always constant among experiments. Further studies using genetically modified mice and/or in vivo imaging are warranted to address this important question.
Insights into RORα-mediated PC morphological changes
How does RORα regulate multiple aspects of dendrite formation at different times during development and throughout adulthood? Since the discovery that staggerer is caused by a defective transcriptional factor RORα, many genes that target the control of RORα have been identified. Such genes are classified into two categories: genes that stimulate the proliferation of granule cells and genes involved in synaptic transmission between PCs and granule cells (Gold et al., 2003; Serra et al., 2006). Therefore, RORα may coordinate reciprocal signaling at PF–PC synapses in the developing cerebellum (Gold et al., 2007). Indeed, granule cell loss is caused by reduced PF–PC synapses (Selimi et al., 2000; Hirai et al., 2005). In addition, proper CF pruning depends on functional PF–PC synapses (Watanabe and Kano, 2011). Furthermore, terminal filopodia on PC dendrites may determine the final dendrite morphology by forming synaptic contacts with PFs during development (Berry and Bradley, 1976). Finally, PCs in weaver mice, in which most granule cells die in the EGL during the first 2 postnatal weeks, exhibit a reduced number of dendritic branches oriented randomly (Rakic and Sidman, 1973). Therefore, impaired PF–PC synaptic signaling is likely to be responsible, at least in part, for these staggerer phenotypes.
In contrast, Cbln1-null PCs develop almost normal dendritic arborization and spines despite the fact that PF–PC synapse formation is reduced by ∼80% (Hirai et al., 2005). Furthermore, PCs lacking RORα target genes, such as metabotropic glutamate receptor 1 (Aiba et al., 1994), inositol trisphosphate receptor (Matsumoto et al., 1996), and excitatory amino acid transporter 4 (Huang et al., 2004), are aligned in the PL and exhibit normal dendritic trees with many spines. These findings indicate that, although contacts between PF and PCs affect dendrite organization, previously indicated genes targeted by RORα cannot fully account for major staggerer phenotypes, such as multiple stem dendrites and severely reduced dendritic branches without any spines.
RORα regulated the elimination of primitive dendrites by P4 before PF–PC synapses were formed (Boukhtouche et al., 2006; Fig. 3), indicating a role of RORα independent from PF–PC synapses. RORα knock-down experiments at P4 further revealed that perisomatic dendrite elimination was achieved by RORα during the stellate cell to the young PC stages. Although the mechanisms underlying axon repulsion and retraction have been studied extensively, little is known about dendrite elimination. Recently, a putative transcription factor BTB/POZ domain-containing 3 (BTBD3) was reported to control transcriptional programs that trim excess dendrites in the developing mouse somatosensory cortex (Matsui et al., 2013). Therefore, RORα may regulate yet to be identified transcriptional programs leading to the pruning of PC dendrites at multiple developmental stages.
After the normal formation of PF–PC synapses, knock-down of RORα at P21 reduced the number of dendritic branches and total dendritic length without affecting the average segment length of each branch (Fig. 8). These results indicate that terminal dendrites were selectively lost when RORα was deficient in mature PCs. Furthermore, the loss of RORα eliminated essentially all spines, whereas the gain of RORα enhanced the maturation of spines at P21 (Fig. 9). In contrast, although the loss of Cbln1 in the adult cerebellum severely reduces PF–PC synapses, the dendritic arborization and spines are unaffected (Hirai et al., 2005; Uemura et al., 2010). Terminal dendrites and dendritic spines are dynamic structures largely dependent on the actin cytoskeleton. Indeed, small GTPase M-Ras was reported to regulate dendrite pruning in developing cortical neurons by affecting actin cytoskeleton dynamics (Tasaka et al., 2012). In Drosophila sensory neurons, raw specifically regulates terminal dendrite growth by modulating the actin cytoskeleton (Lee et al., 2015). Therefore, we propose that a major role of RORα in mature PCs is to stabilize spines and terminal dendrites by modulating actin cytoskeleton dynamics.
The knock-down of RORα at P4 completely prevented the formation of filopodia, whereas its overexpression enhanced the maturation of dendritic spines at P13 (Fig. 6). Furthermore, the knock-down of RORα at P8 also reduced the number of dendritic branches and total dendritic length at P10 without affecting the average segment length of each branch (Fig. 7), indicating that the formation of terminal dendritic branches also requires RORα at the young PC stage. Terminal filopodia, which are often indistinguishable from terminal dendritic branches of immature PCs (Berry and Bradley, 1976), are proposed to mediate dendritic growth and branching. A recent mathematical modeling study based on live imaging of dendrite outgrowth in cultured PCs also indicated that the dendrite pattern was formed by the stochastic addition of branches to the terminal dendrites (Fujishima et al., 2012). Therefore, we propose that a major role of RORα during development after the stellate cell and young PC stages is to form terminal filopodia that develop into branches in terminal dendrites.
A decrease of RORα during development is reported to mediate the severity of hereditary spinocerebellar ataxia type 1 (Serra et al., 2006). However, our observations indicate that RORα overexpression is detrimental to normal PC development. Therefore, future studies are warranted to clarify the precise molecular mechanism by which RORα regulates different aspects of dendrite development at each stage of PC development.
Footnotes
This work was supported by CREST, Japan Science and Technology Corporation (M.Y.), the Ministry of Education, Culture, Sports, Science and Technology of Japan (Grant-in-Aid to W.K. and M.Y.), the Japan Society for the Promotion of Science (Y.-H.T., W.K., M.Y.), the Kanae Foundation (W.K.), the Nakajima Foundation (W.K.), the Keio Gijuku Fukuzawa Memorial Fund for the Advancement of Education and Research (W.K.), and the Takeda Science Foundation (W.K. and M.Y.). We thank S. Narumi and the Collaborative Research Resources at Keio University School of Medicine for technical assistance.
The authors declare no competing financial interests.
References
- Aiba A, Kano M, Chen C, Stanton ME, Fox GD, Herrup K, Zwingman TA, Tonegawa S. Deficient cerebellar long-term depression and impaired motor learning in mGluR1 mutant mice. Cell. 1994;79:377–388. doi: 10.1016/0092-8674(94)90205-4. [DOI] [PubMed] [Google Scholar]
- Baptista CA, Hatten ME, Blazeski R, Mason CA. Cell-cell interactions influence survival and differentiation of purified Purkinje cells in vitro. Neuron. 1994;12:243–260. doi: 10.1016/0896-6273(94)90268-2. [DOI] [PubMed] [Google Scholar]
- Berry M, Bradley P. The growth of the dendritic trees of Purkinje cells in the cerebellum of the rat. Brain Res. 1976;112:1–35. doi: 10.1016/0006-8993(76)90331-0. [DOI] [PubMed] [Google Scholar]
- Boukhtouche F, Janmaat S, Vodjdani G, Gautheron V, Mallet J, Dusart I, Mariani J. Retinoid-related orphan receptor alpha controls the early steps of Purkinje cell dendritic differentiation. J Neurosci. 2006;26:1531–1538. doi: 10.1523/JNEUROSCI.4636-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley P, Berry M. The Purkinje cell dendritic tree in mutant mouse cerebellum: a quantitative Golgi study of Weaver and Staggerer mice. Brain Res. 1978;142:135–141. doi: 10.1016/0006-8993(78)90182-8. [DOI] [PubMed] [Google Scholar]
- Chen XR, Heck N, Lohof AM, Rochefort C, Morel MP, Wehrlé R, Doulazmi M, Marty S, Cannaya V, Avci HX, Mariani J, Rondi-Reig L, Vodjdani G, Sherrard RM, Sotelo C, Dusart I. Mature Purkinje cells require the retinoic acid-related orphan receptor-alpha (RORalpha) to maintain climbing fiber mono-innervation and other adult characteristics. J Neurosci. 2013;33:9546–9562. doi: 10.1523/JNEUROSCI.2977-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman M. Axon degeneration mechanisms: commonality amid diversity. Nat Rev Neurosci. 2005;6:889–898. doi: 10.1038/nrn1788. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crepel F, Delhaye-Bouchaud N, Guastavino JM, Sampaio I. Multiple innervation of cerebellar Purkinje cells by climbing fibres in staggerer mutant mouse. Nature. 1980;283:483–484. doi: 10.1038/283483a0. [DOI] [PubMed] [Google Scholar]
- Emoto K. Signaling mechanisms that coordinate the development and maintenance of dendritic fields. Curr Opin Neurobiol. 2012;22:805–811. doi: 10.1016/j.conb.2012.04.005. [DOI] [PubMed] [Google Scholar]
- Fujishima K, Horie R, Mochizuki A, Kengaku M. Principles of branch dynamics governing shape characteristics of cerebellar Purkinje cell dendrites. Development. 2012;139:3442–3455. doi: 10.1242/dev.081315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold DA, Baek SH, Schork NJ, Rose DW, Larsen DD, Sachs BD, Rosenfeld MG, Hamilton BA. RORalpha coordinates reciprocal signaling in cerebellar development through sonic hedgehog and calcium-dependent pathways. Neuron. 2003;40:1119–1131. doi: 10.1016/S0896-6273(03)00769-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold DA, Gent PM, Hamilton BA. ROR alpha in genetic control of cerebellum development: 50 staggering years. Brain Res. 2007;1140:19–25. doi: 10.1016/j.brainres.2005.11.080. [DOI] [PubMed] [Google Scholar]
- Hirai H, Pang Z, Bao D, Miyazaki T, Li L, Miura E, Parris J, Rong Y, Watanabe M, Yuzaki M, Morgan JI. Cbln1 is essential for synaptic integrity and plasticity in the cerebellum. Nat Neurosci. 2005;8:1534–1541. doi: 10.1038/nn1576. [DOI] [PubMed] [Google Scholar]
- Huang YH, Dykes-Hoberg M, Tanaka K, Rothstein JD, Bergles DE. Climbing fiber activation of EAAT4 transporters and kainate receptors in cerebellar Purkinje cells. J Neurosci. 2004;24:103–111. doi: 10.1523/JNEUROSCI.4473-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakegawa W, Miyazaki T, Emi K, Matsuda K, Kohda K, Motohashi J, Mishina M, Kawahara S, Watanabe M, Yuzaki M. Differential regulation of synaptic plasticity and cerebellar motor learning by the C-terminal PDZ-binding motif of GluRdelta2. J Neurosci. 2008;28:1460–1468. doi: 10.1523/JNEUROSCI.2553-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann WE, Moser HW. Dendritic anomalies in disorders associated with mental retardation. Cereb Cortex. 2000;10:981–991. doi: 10.1093/cercor/10.10.981. [DOI] [PubMed] [Google Scholar]
- Kawaguchi K, Habara T, Terashima T, Kikkawa S. GABA modulates development of cerebellar Purkinje cell dendrites under control of endocannabinoid signaling. J Neurochem. 2010;114:627–638. doi: 10.1111/j.1471-4159.2010.06793.x. [DOI] [PubMed] [Google Scholar]
- Kulkarni VA, Firestein BL. The dendritic tree and brain disorders. Mol Cell Neurosci. 2012;50:10–20. doi: 10.1016/j.mcn.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Landis DM, Sidman RL. Electron microscopic analysis of postnatal histogenesis in the cerebellar cortex of staggerer mutant mice. J Comp Neurol. 1978;179:831–863. doi: 10.1002/cne.901790408. [DOI] [PubMed] [Google Scholar]
- Lee G, Saito I. Role of nucleotide sequences of loxP spacer region in Cre-mediated recombination. Gene. 1998;216:55–65. doi: 10.1016/S0378-1119(98)00325-4. [DOI] [PubMed] [Google Scholar]
- Lee J, Peng Y, Lin WY, Parrish JZ. Coordinate control of terminal dendrite patterning and dynamics by the membrane protein Raw. Development. 2015;142:162–173. doi: 10.1242/dev.113423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani J, Changeux JP. Multiple innervation of Purkinje cells by climbing fibers in the cerebellum of the adult staggerer mutant mouse. J Neurobiol. 1980;11:41–50. doi: 10.1002/neu.480110106. [DOI] [PubMed] [Google Scholar]
- Matsuda T, Cepko CL. Controlled expression of transgenes introduced by in vivo electroporation. Proc Natl Acad Sci U S A. 2007;104:1027–1032. doi: 10.1073/pnas.0610155104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui A, Tran M, Yoshida AC, Kikuchi SS, U M, Ogawa M, Shimogori T. BTBD3 controls dendrite orientation toward active axons in mammalian neocortex. Science. 2013;342:1114–1118. doi: 10.1126/science.1244505. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Nakagawa T, Inoue T, Nagata E, Tanaka K, Takano H, Minowa O, Kuno J, Sakakibara S, Yamada M, Yoneshima H, Miyawaki A, Fukuuchi Y, Furuichi T, Okano H, Mikoshiba K, Noda T. Ataxia and epileptic seizures in mice lacking type 1 inositol 1,4,5-trisphosphate receptor. Nature. 1996;379:168–171. doi: 10.1038/379168a0. [DOI] [PubMed] [Google Scholar]
- Mitsumura K, Hosoi N, Furuya N, Hirai H. Disruption of metabotropic glutamate receptor signalling is a major defect at cerebellar parallel fibre-Purkinje cell synapses in staggerer mutant mice. J Physiol. 2011;589:3191–3209. doi: 10.1113/jphysiol.2011.207563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Ono Y, Okamoto M, Masaoka M, Sakakibara A, Kawaguchi A, Hashimoto M, Ogawa M. Migration, early axonogenesis, and Reelin-dependent layer-forming behavior of early/posterior-born Purkinje cells in the developing mouse lateral cerebellum. Neural Dev. 2010;5:23. doi: 10.1186/1749-8104-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama J, Hayashi Y, Nomura T, Miura E, Kakegawa W, Yuzaki M. Selective and regulated gene expression in murine Purkinje cells by in utero electroporation. Eur J Neurosci. 2012;36:2867–2876. doi: 10.1111/j.1460-9568.2012.08203.x. [DOI] [PubMed] [Google Scholar]
- Oberdick J, Smeyne RJ, Mann JR, Zackson S, Morgan JI. A promoter that drives transgene expression in cerebellar Purkinje and retinal bipolar neurons. Science. 1990;248:223–226. doi: 10.1126/science.2109351. [DOI] [PubMed] [Google Scholar]
- Rakic P, Sidman RL. Sequence of developmental abnormalities leading to granule cell deficit in cerebellar cortex of weaver mutant mice. J Comp Neurol. 1973;152:103–132. doi: 10.1002/cne.901520202. [DOI] [PubMed] [Google Scholar]
- Selimi F, Vogel MW, Mariani J. Bax inactivation in lurcher mutants rescues cerebellar granule cells but not purkinje cells or inferior olivary neurons. J Neurosci. 2000;20:5339–5345. doi: 10.1523/JNEUROSCI.20-14-05339.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra HG, Duvick L, Zu T, Carlson K, Stevens S, Jorgensen N, Lysholm A, Burright E, Zoghbi HY, Clark HB, Andresen JM, Orr HT. RORalpha-mediated Purkinje cell development determines disease severity in adult SCA1 mice. Cell. 2006;127:697–708. doi: 10.1016/j.cell.2006.09.036. [DOI] [PubMed] [Google Scholar]
- Sidman RL, Lane PW, Dickie MM. Staggerer, a new mutation in the mouse affecting the cerebellum. Science. 1962;137:610–612. doi: 10.1126/science.137.3530.610. [DOI] [PubMed] [Google Scholar]
- Soha JM, Herrup K. Stunted morphologies of cerebellar Purkinje cells in lurcher and staggerer mice are cell-intrinsic effects of the mutant genes. J Comp Neurol. 1995;357:65–75. doi: 10.1002/cne.903570107. [DOI] [PubMed] [Google Scholar]
- Sotelo C, Dusart I. Intrinsic versus extrinsic determinants during the development of Purkinje cell dendrites. Neuroscience. 2009;162:589–600. doi: 10.1016/j.neuroscience.2008.12.035. [DOI] [PubMed] [Google Scholar]
- Stern P, Astrof S, Erkeland SJ, Schustak J, Sharp PA, Hynes RO. A system for Cre-regulated RNA interference in vivo. Proc Natl Acad Sci U S A. 2008;105:13895–13900. doi: 10.1073/pnas.0806907105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasaka G, Negishi M, Oinuma I. Semaphorin 4D/Plexin-B1-mediated M-Ras GAP activity regulates actin-based dendrite remodeling through Lamellipodin. J Neurosci. 2012;32:8293–8305. doi: 10.1523/JNEUROSCI.0799-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavosanis G. Dendritic structural plasticity. Dev Neurobiol. 2012;72:73–86. doi: 10.1002/dneu.20951. [DOI] [PubMed] [Google Scholar]
- Uemura T, Lee SJ, Yasumura M, Takeuchi T, Yoshida T, Ra M, Taguchi R, Sakimura K, Mishina M. Trans-synaptic interaction of GluRdelta2 and Neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell. 2010;141:1068–1079. doi: 10.1016/j.cell.2010.04.035. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Kano M. Climbing fiber synapse elimination in cerebellar Purkinje cells. Eur J Neurosci. 2011;34:1697–1710. doi: 10.1111/j.1460-9568.2011.07894.x. [DOI] [PubMed] [Google Scholar]
- Yuste R, Bonhoeffer T. Genesis of dendritic spines: insights from ultrastructural and imaging studies. Nat Rev Neurosci. 2004;5:24–34. doi: 10.1038/nrn1300. [DOI] [PubMed] [Google Scholar]