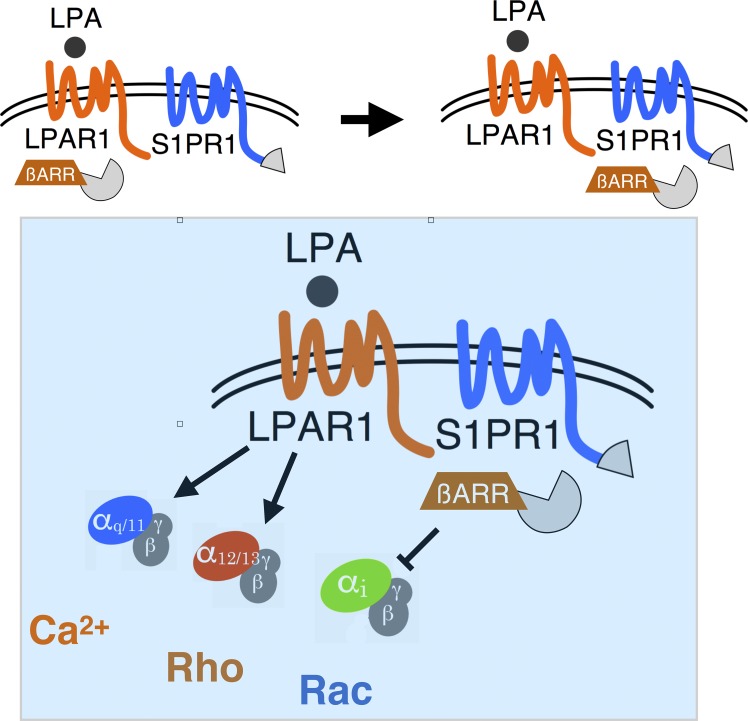
Sphingosine 1-phosphate receptor-1 signaling via the Gi pathway is suppressed by lysophosphatidic acid receptor-1, which recruits β-arrestin to the heterotypic receptor complex. This mechanism occurs at sinus lining lymphatic endothelial cells of lymph nodes and modulates complex cell–cell junction structure.
Abstract
Sphingosine 1-phosphate (S1P) and lysophosphatidic acid (LPA) activate G protein–coupled receptors (GPCRs) to regulate biological processes. Using a genome-wide CRISPR/dCas9–based GPCR signaling screen, LPAR1 was identified as an inducer of S1PR1/β-arrestin coupling while suppressing Gαi signaling. S1pr1 and Lpar1-positive lymphatic endothelial cells (LECs) of lymph nodes exhibit constitutive S1PR1/β-arrestin signaling, which was suppressed by LPAR1 antagonism. Pharmacological inhibition or genetic loss of function of Lpar1 reduced the frequency of punctate junctions at sinus-lining LECs. Ligand activation of transfected LPAR1 in endothelial cells remodeled junctions from continuous to punctate structures and increased transendothelial permeability. In addition, LPAR1 antagonism in mice increased lymph node retention of adoptively transferred lymphocytes. These data suggest that cross-talk between LPAR1 and S1PR1 promotes the porous junctional architecture of sinus-lining LECs, which enables efficient lymphocyte trafficking. Heterotypic inter-GPCR coupling may regulate complex cellular phenotypes in physiological milieu containing many GPCR ligands.
Graphical Abstract
Introduction
Membrane phospholipids are rapidly metabolized by lipases and synthases to maintain the integrity of biological membranes (Shimizu, 2009). Lysophospholipids, metabolic intermediates of membrane phospholipids, have unique geometry and biophysical properties that facilitate membrane topology, vesicle budding, and fusion (Holthuis and Menon, 2014). However, lysophospholipids evolved as extracellular lipid mediators in vertebrates (Hla, 2005). The best characterized are sphingosine 1-phosphate (S1P) and lysophosphatidic acid (LPA), structurally related lysophospholipids that were originally identified as major regulators of cellular cytoskeletal dynamics (Blaho and Hla, 2011; Moolenaar and Hla, 2012; Mutoh et al., 2012). S1P is synthesized largely in the intracellular environment and secreted via specific transporters SPNS2 and MFSD2B (Hisano et al., 2011; Proia and Hla, 2015; Vu et al., 2017; Kobayashi et al., 2018). Extracellular chaperone-bound S1P activates five G protein–coupled receptors (GPCRs) in the endothelial differentiation gene subfamily that are widely expressed (Proia and Hla, 2015). On the other hand, LPA, which is synthesized in the extracellular environment by autotaxin-mediated hydrolysis of lysophosphatidyl choline, activates six GPCRs in the endothelial differentiation gene and purinergic subfamilies (Aikawa et al., 2015).
Both S1P and LPA were originally identified as bioactive lipid mediators due to their ability to modulate cytoskeletal dynamics, neurite retraction, cell migration, cell proliferation, and intracellular ion fluxes (Moolenaar and Hla, 2012). Such activity depends on the ability of S1P and LPA to regulate Rho family GTPases (Hall, 2012). After the discovery of the GPCRs for S1P and LPA, genetic loss-of-function studies in mice have identified their essential roles in embryonic development and physiological processes of multiple organ systems (Chun et al., 2010). For example, both lysophospholipids were shown to be critical for early vascular development, since mice that lack autotaxin (Enpp2) as well as sphingosine kinases (Sphk1 and Sphk2) were embryonic lethal at early stages of gestation (Mizugishi et al., 2005; Tanaka et al., 2006; van Meeteren et al., 2006). Similarly, compound S1P and LPA receptor KOs also exhibit severe vascular development defects (Kono et al., 2004; Sumida et al., 2010). Similar studies have implicated the critical roles of S1P and LPA signaling in neuronal and immune systems (Skoura and Hla, 2009). A key question that is raised by such findings is whether S1P and LPA are redundant in their biological functions. Data available so far suggest that while some redundant functions are mediated by both lysophospholipids, some unique functions do exist. For example, naive T cell egress from secondary lymphoid organs is largely dependent on S1P signaling via lymphocyte S1PR1 (Cyster and Schwab, 2012), whereas both S1P and LPA induce fibrotic responses in the lung (Shea and Tager, 2012) and regulate cardiac development in zebrafish (Nakanaga et al., 2014). Whether S1P and LPA signaling mechanisms regulate each other (i.e., cross-talk mechanisms) is not known.
The S1PR1 receptor signals primarily via the Gi family of heterotrimeric G proteins (Lee et al., 1999). However, its activity is antagonized by β-arrestin–mediated receptor down-regulation, which involves GRK2-dependent phosphorylation, β-arrestin binding, and dynamin-regulated endocytosis (Oo et al., 2007, 2011; Arnon et al., 2011; Willinger et al., 2014). In addition, direct binding to the membrane-bound C-type lectin CD69 also induces receptor down-regulation (Shiow et al., 2006; Bankovich et al., 2010). Further, extracellular presence of ApoM-containing high density lipoprotein, which chaperones S1P, permits sustained plasma membrane signaling without inducing efficient endocytosis (Swendeman et al., 2017). We hypothesized that novel factors modulate its signaling and residency in the plasma membrane to mediate its multiple biological functions. In this report, we searched for novel regulators of S1PR1 coupling to the β-arrestin pathway. Specifically, we used the TANGO system, which uses tobacco etch virus (TEV) protease/β-arrestin fusion protein and S1PR1-TEV site tetracycline transcriptional activator as a readout (Barnea et al., 2008). Coupled with the single guide RNA (sgRNA) library-directed, CRISPR/dCas9-induced endogenous genes (Shalem et al., 2015), we screened for novel modulators of S1PR1. Surprisingly, the top hit from this unbiased, whole-genome screen was LPAR1. We validated this interaction in a luciferase complementation system that quantifies GPCR coupling to β-arrestin. Our results suggest that LPAR1’s interaction with S1PR1 attenuates S1P signaling in endothelial cells, modulates lymphatic sinus adherens junctions, and provides a permissive niche for lymphocyte trafficking.
Results
Unbiased, genome-wide search for S1PR1 modulators
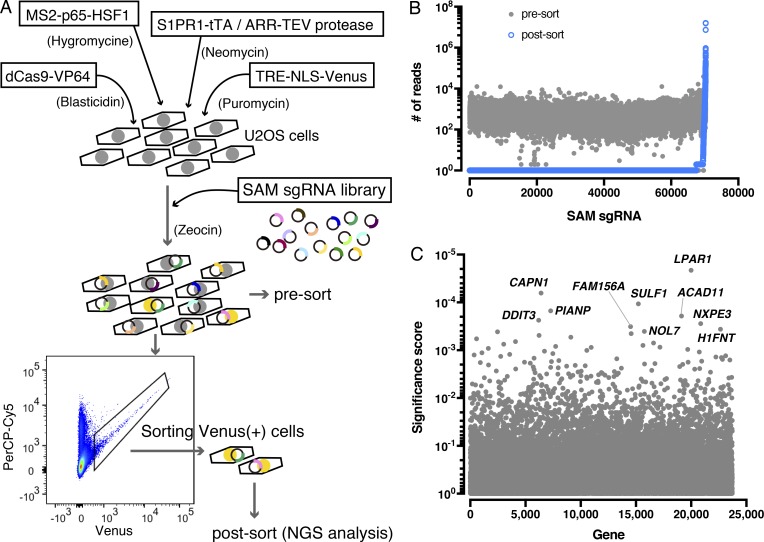
S1PR1 signaling can be readily monitored by the TANGO system, which records ligand-activated β-arrestin coupling to the GPCR, leading to nuclear fluorescent protein expression in vitro and in vivo (Kono et al., 2014). Since the receptor/β-arrestin signal is cumulative due to the stability of the nuclear fluorescent protein, we adapted this system to U2OS osteosarcoma cells that are adaptable to high-throughput screening. Previous work has shown that direct activators of S1PR1, such as CD69, regulate receptor signaling and function (Shiow et al., 2006; Bankovich et al., 2010). To search for other endogenous modulators of S1PR1 signaling, we turned to the synergistic activation mediator (SAM) system that uses CRISPR/dCas9-based, sgRNA-dependent transcriptional activation of endogenous genes (Konermann et al., 2015).
The SAM system turns on endogenous gene expression by sgRNA-dependent recruitment of multiple transcriptional activators (VP64, p65, and HSF1) upstream of transcription start sites via MS2 bacteriophage coat proteins and mutated, nuclease-deficient dCas9. This screening system was validated by the SAM sgRNA targeting SPNS2, an S1P transporter that functions upstream of S1P receptors (Kawahara et al., 2009; Hisano et al., 2012, 2013). The designed SPNS2 SAM sgRNA induced an 180-fold increase in its mRNA expression and strongly activated the S1PR1-TANGO signal (Fig. S1, A and B).
To carry out unbiased search for S1PR1-signaling modulators, the SAM sgRNA library was introduced into the S1PR1-TANGO system, in which β-arrestin2 coupling of S1PR1 can be monitored as nuclear expression of Venus fluorescent protein. Venus-positive cells (S1PR1/β-arrestin2 signaling positive) were sorted and expanded twice, and genomic DNAs were purified and sequenced by Illumina next-generation sequencing (NGS; Fig. 1 A). Bioinformatic analysis indicated that some SAM sgRNA sequences are highly enriched in the Venus-positive cells after sorting (Fig. 1 B), suggesting that target genes of these sgRNAs encode proteins that enable S1PR1 coupling to β-arrestin. The LPAR1 gene was identified as one of the top hits (Fig. 1 C). Top 10 candidates were examined individually by specific SAM sgRNAs that were enriched after sorting Venus-positive cells. The SAM sgRNA specific for LPAR1 induced its expression and turned on Venus expression, thus confirming the results from the genome-wide sgRNA screen that identified LPAR1 as a modulator of S1PR1 coupling to β-arrestin (Fig. S1, C and D).
Figure 1.
Unbiased whole genome-wide search for S1PR1 modulators. (A) Schematic of the S1PR1 modulator screening system. Four lentiviral vectors were transduced into a U2OS cell line to enable gene activation by SAM and monitor S1PR1 activation by the TANGO system. The cells introduced with a SAM sgRNA library were starved with 0.5% charcoal–treated FBS, and then the Venus-positive population was sorted, and NGS analysis was performed to identify the enriched SAM sgRNA sequences. (B) Scatterplot showing enrichment of sgRNAs after sorting. Most sgRNAs are equally distributed in the presort sample (closed gray circles), while after sorting, a small fraction of sgRNAs (2,770 out of 70,290 sgRNAs) were enriched (open blue circles). The y axis shows the number of NGS reads of sgRNAs. (C) Identification of top candidate genes using the MAGeCK method (Li et al., 2014). The names of top 10 candidate genes are indicated. TRE, tetracycline-responsive element; NLS, nuclear localization signal.
LPAR1 activation induces β-arrestin recruitment to S1PR1
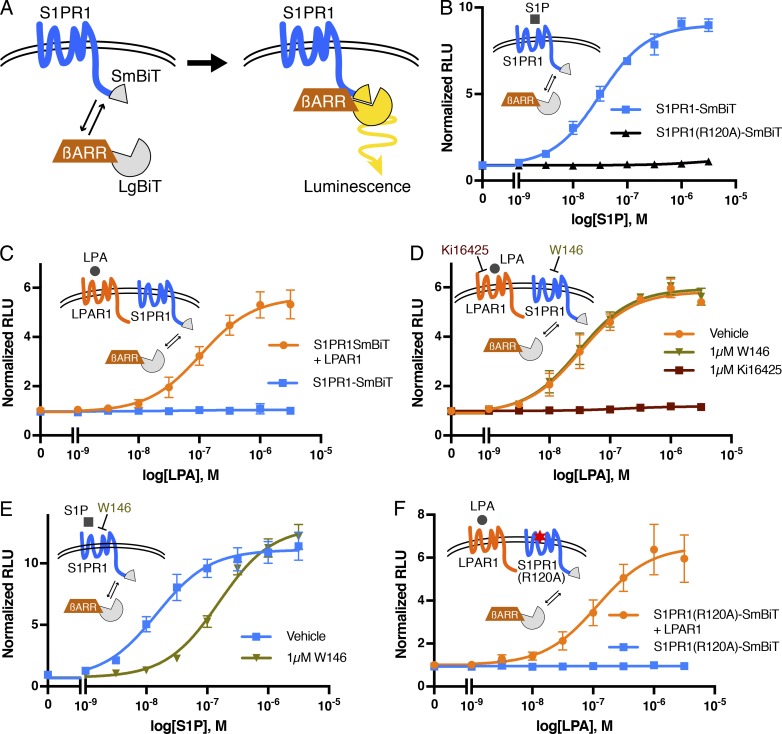
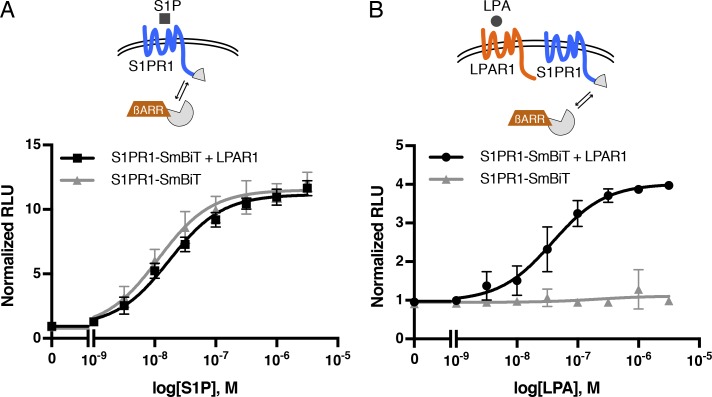
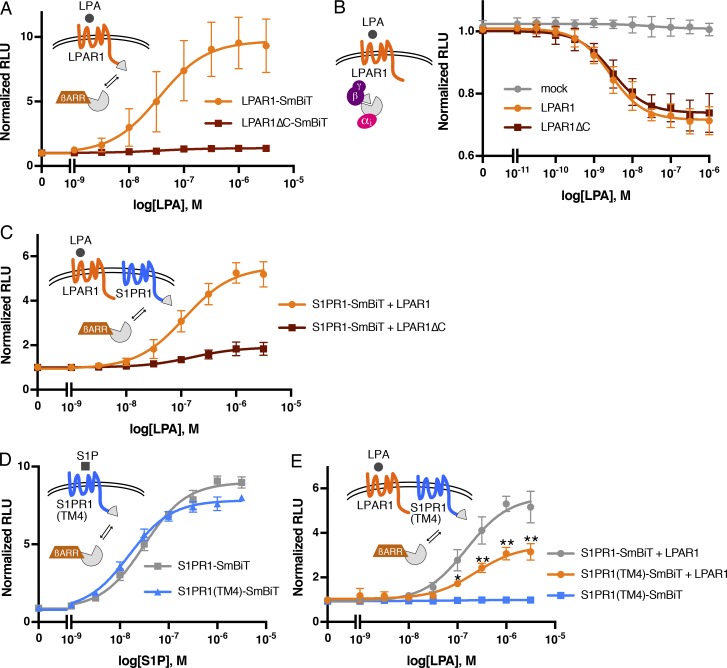
To further investigate the mechanisms involved in the regulation of S1PR1 signaling by LPAR1, we used the NanoBiT system (Dixon et al., 2016). This system is based on the structural complementation of NanoLuc luciferase and allows one to monitor the protein–protein interactions in real time. NanoLuc luciferase is split into a small subunit (SmBiT; 11 amino acids) and a large subunit (LgBiT; 18 kD) that are fused with S1PR1 and β-arrestin1 with mutations in AP-2/clathrin-binding motif (to reduce endocytosis), respectively (Fig. 2 A). S1P dose-dependently stimulated β-arrestin1 recruitment to S1PR1 in HEK293A cells transfected with S1PR1-SmBiT and LgBiT-β-arrestin1. However, the S1P ligand–binding mutant, S1PR1 (R120A), did not recruit β-arrestin upon treatment with S1P (Fig. 2 B). LPA treatment did not induce β-arrestin1 recruitment to S1PR1, consistent with the fact that LPA is not a high-affinity ligand for S1PR1 (Lee et al., 1998a; Liu et al., 1999). However, in cells coexpressing LPAR1 and S1PR1-SmBiT, LPA treatment induced β-arrestin1 recruitment to S1PR1 with a 50% effective concentration of ∼10−7 M, a physiologically relevant concentration of LPA (Fig. 2 C).
Figure 2.
Activated LPAR1 induces S1PR1/β-arrestin coupling. (A) Schematic of NanoBiT system to measure S1PR1 and the β-arrestin1 interaction. SmBiT and LgBiT were fused to the C-terminus of S1PR1 and the N-terminus of β-arrestin1, respectively. S1PR1 and β-arrestin1 coupling can be detected as luminescence signal emitted by complementation of SmBiT and LgBiT. (B) S1PR1-SmBiT or S1PR1(R120A)-SmBiT was transfected with LgBiT-β-arrestin1, and luminescence was measured 15–20 min after S1P stimulation. (C) LPAR1 or empty vector was transfected with S1PR1-SmBiT and LgBiT-β-arrestin1, and luminescence was measured 15–20 min after LPA stimulation. (D and E) Cells were incubated with 1 µM Ki16425 or W146 for 30 min before stimulation, and luminescence was measured 15–20 min after LPA (D) or S1P (E) stimulation. (F) LPAR1 or empty vector was transfected with S1PR1(R120A)-SmBiT and LgBiT-β-arrestin1, and luminescence was measured 15–20 min after LPA stimulation. Experiments were repeated (i.e., three to eight independent experiments), and results are expressed as mean ± SD. βARR, β-arrestin; RLU, relative light units.
The effect of LPA was completely blocked by Ki16425, an LPAR1 antagonist (Ohta et al., 2003), indicating that the β-arrestin1 coupling of S1PR1 is dependent on LPAR1 activation by the ligand (Fig. 2 D). W146, an S1PR1 antagonist, inhibited S1P-mediated β-arrestin1 recruitment to S1PR1 but failed to inhibit LPA/LPAR1–mediated β-arrestin1 coupling of S1PR1 (Fig. 2, D and E), suggesting that S1PR1 activation with S1P is not necessary for the LPA/LPAR1–mediated stimulation of S1PR1 coupling to β-arrestin1. Furthermore, the S1PR1 ligand–binding mutant (R120A) behaved similarly to wild-type S1PR1 by allowing LPAR1-induced β-arrestin1 coupling (Fig. 2 F). Simultaneous administration of both LPA and S1P induced an additive effect on S1PR1 coupling to β-arrestin (Fig. S2). These experiments confirm that LPAR1 activation induced inter-GPCR coupling of β-arrestin to S1PR1 independently of activated S1PR1-induced β-arrestin recruitment.
G proteins are not required for LPA/LPAR1–induced S1PR1/β-arrestin coupling
LPAR1 couples to three families of G protein α subunits (Gαi, Gα12/13, and Gαq/11), while S1PR1 is a Gαi-coupled receptor (Fukushima et al., 1998; Lee et al., 1998b; Windh et al., 1999; Ishii et al., 2000). To examine whether LPAR1-induced inter-GPCR coupling of β-arrestin1 to S1PR1 requires heterotrimeric G proteins, we used HEK293 cells lacking GNAS, GNAL, GNAQ, GNA11, GNA12, GNA13, GNAI1, GNAI2, GNAI3, GNAO1, GNAZ, GNAT1, and GNAT2 (fullΔGα) generated with CRISPR/Cas9 system (Fig. S3). Even in the HEK293 fullΔGα cells, S1P activation of S1PR1 induced β-arrestin1 coupling to a similar extent as wild-type cells, suggesting that GPCR/β-arrestin1 coupling is G-protein independent (Figs. 2 B and 3 A), a finding that was reported previously (Grundmann et al., 2018). We observed that LPA stimulation of LPAR1 induced S1PR1/β-arrestin1 coupling in the HEK293 fullΔGα cells (Fig. 3 B), indicating that heterotrimeric G protein coupling is not required for inter-GPCR β-arrestin coupling.
Figure 3.
LPAR1-mediated S1PR1/β-arrestin coupling in G protein–deficient cells. LPAR1 or empty vector was transfected with S1PR1-SmBiT and LgBiT-β-arrestin1 into HEK293 fullΔGα cells lacking all Gαs. (A and B) Luminescence was measured 15–20 min after S1P (A) or LPA (B) stimulation. Experiments were repeated (i.e., n = 3 independent experiments), and results are expressed as mean ± SD. βARR, β-arrestin; RLU, relative light units.
The LPAR1 C-terminal domain is necessary for the β-arrestin coupling of S1PR1
β-Arrestin primarily interacts with the intracellular C-terminal tail region of GPCRs, even though the third intracellular loop may also be involved (Ranjan et al., 2017). The LPAR1ΔC mutant, which lacks the C-terminal domain, did not recruit β-arrestin1 in response to LPA (Fig. 4 A). In contrast, both LPAR1 and LPAR1ΔC receptors couple to the heterotrimeric Gαi protein in an equivalent manner, which was assessed as dissociation of heteromeric G proteins using LgBiT-GNAI2/SmBiT-GNG (Fig. 4 B). However, LPAR1ΔC mutant was unable to induce β-arrestin1 recruitment to S1PR1 in response to LPA (Fig. 4 C), suggesting that initial β-arrestin1 recruitment to LPAR1 is required for the LPA-mediated inter-GPCR coupling of β-arrestin to S1PR1.
Figure 4.
The C-terminus of LPAR1 and TM4 of S1PR1 is important for LPAR1-induced inter-GPCR β-arrestin coupling. (A) LPAR1-SmBiT or LPAR1ΔC-SmBiT was transfected with LgBiT-β-arrestin1, and luminescence was measured 15–20 min after LPA stimulation. (B) A G-protein dissociation assay was performed by transfecting LgBiT-GNAI2, GNB1, and SmBiT-GNGT1 plasmids with LPAR1 or LPAR1ΔC. Luminescence was measured 6–9 min after LPA stimulation. (C) LPAR1 or LPAR1ΔC was transfected with S1PR1-SmBiT and LgBiT-β-arrestin1, and luminescence was measured 15–20 min after LPA stimulation. (D) S1PR1-SmBiT or S1PR1(TM4)-SmBiT was transfected with LgBiT-β-arrestin1, and luminescence was measured 15–20 min after S1P stimulation. (E) LPAR1 or empty vector was transfected with S1PR1-SmBiT or S1PR1(TM4)-SmBiT and LgBiT-β-arrestin1, and luminescence was measured 15–20 min after LPA stimulation. Experiments were repeated (i.e., three to five independent experiments), and the results are expressed as mean ± SD. P values were determined by two-way ANOVA followed by Sidak’s multiple comparisons test comparing “S1PR1(TM4)-SmBiT + LPAR1” to “S1PR1-SmBiT + LPAR1”; *, P = 0.0018; **, P ≤ 0.001. βARR, β-arrestin; RLU, relative light units.
Transmembrane helix 4 of S1PR1 is important for the β-arrestin coupling of S1PR1
We next examined the hypothesis that direct interactions between S1PR1 and LPAR1 are needed for inter-GPCR β-arrestin coupling. The transmembrane helix 4 of S1PR1 was reported to interact directly with CD69, a transmembrane C-type lectin (Bankovich et al., 2010). The S1PR1(TM4) mutant in which transmembrane helix 4 is replaced with that of S1PR3 decreased the association with CD69, suggesting that it is the domain involved in intermolecular association with GPCR modulators. We therefore examined the role of the transmembrane helix 4 of S1PR1 in LPAR1-mediated inter-GPCR β-arrestin coupling to S1PR1. S1PR1(TM4)-SmBiT can be expressed at same level as S1PR1-SmBiT (Fig. S4 A) and recruits β-arrestin1 after S1P stimulation (Fig. 4 D). However, the LPAR1-mediated β-arrestin1 coupling of S1PR1(TM4) was significantly attenuated (Fig. 4 E), indicating that the transmembrane helix 4 of S1PR1 is important for the LPAR1-mediated β-arrestin1 coupling of S1PR1.
LPAR1-induced inter-GPCR β-arrestin coupling attenuates S1PR1/Gi signaling
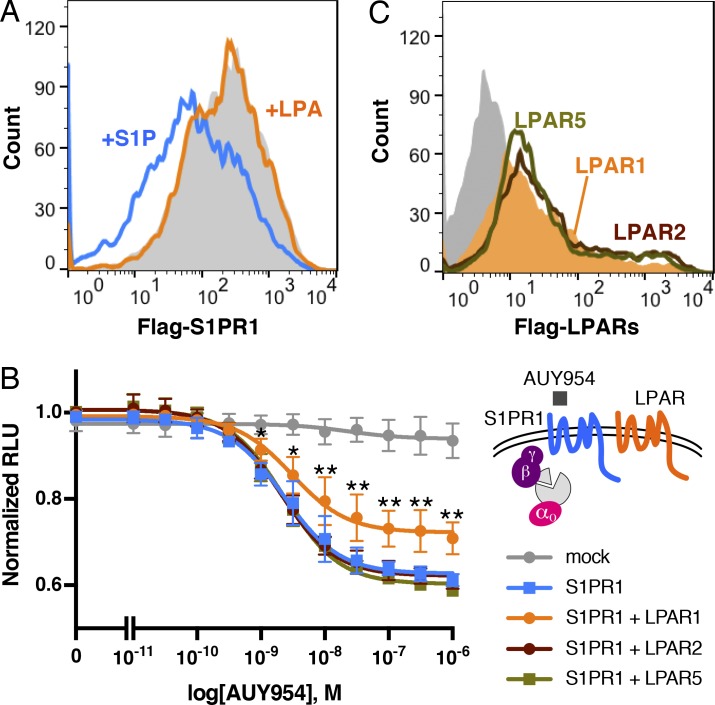
In many GPCRs, β-arrestin recruitment is an initial trigger for receptor internalization by facilitating the interaction with AP-2 and clathrin to recruit GPCRs to the endocytic machinery (Tian et al., 2014). S1PR1 with N-terminal Flag tag was expressed in HEK293A cells, and LPAR1 cell-surface receptor expression was analyzed by flow cytometry. Surprisingly, Flag-S1PR1 surface expression was not altered by LPA stimulation while S1P stimulation induced Flag-S1PR1 internalization (Fig. 5 A). Immunofluorescence analysis confirmed these conclusions (Fig. S4 B). These results suggest that while LPAR1-induced inter-GPCR β-arrestin coupling to S1PR1, this event in and of itself is not sufficient to induce S1PR1 endocytosis.
Figure 5.
LPAR1 inhibits S1PR1/G-protein signaling. (A) Flow cytometric analysis showing surface Flag-S1PR1 expression after stimulation with 1 µM S1P (blue line) or LPA (orange line) for 1 h or without simulation (gray) in HEK293A cells stably expressing Flag-S1PR1 and LPAR1. (B) S1PR1 and LPAR1, LPAR2, or LPAR5 were transfected with LgBiT-GNAO1, GNB1, and SmBiT-GNGT1 plasmids. Luminescence was measured 6–9 min after AUY954 stimulation. Data are derived from three to seven independent experiments and are expressed as mean ± SD. P values were determined by two-way ANOVA followed by Sidak’s multiple comparisons test comparing “S1PR1 + LPAR1” to S1PR1 alone; *, P ≤ 0.01; **, P ≤ 0.0001. (C) Flow cytometric analysis of HEK293A cells transfected with LPAR1 (orange), LPAR2 (brown line), or LPAR3 (dark green line) tagged with Flag at the N-terminus or empty vector (gray). RLU, relative light units.
Next, we examined whether LPAR1 activation modulates the S1PR1 signal transduction. Coupling of S1PR1 to the heterotrimeric G protein pathway was assessed using LgBiT-GNAO1/SmBiT-GNG and AUY954, an S1PR1 selective agonist (Pan et al., 2006). AUY954 induced S1PR1-mediated heteromeric G protein dissociation in a dose-dependent manner, which was significantly suppressed by coexpression with LPAR1 (Fig. 5 B). Other LPA receptors (LPAR2 and LPAR5) expressed at similar levels as LPAR1 failed to suppress S1PR1-mediated Gαi protein activation (Fig. 5, B and C). These results indicate that LPAR1 specifically induces inter-GPCR β-arrestin coupling to suppress S1PR1 heterotrimeric Gαi protein signaling output without inducing receptor endocytosis.
Endogenous LPAR1 stimulates S1PR1/β-arrestin coupling in vivo at lymphatic sinuses
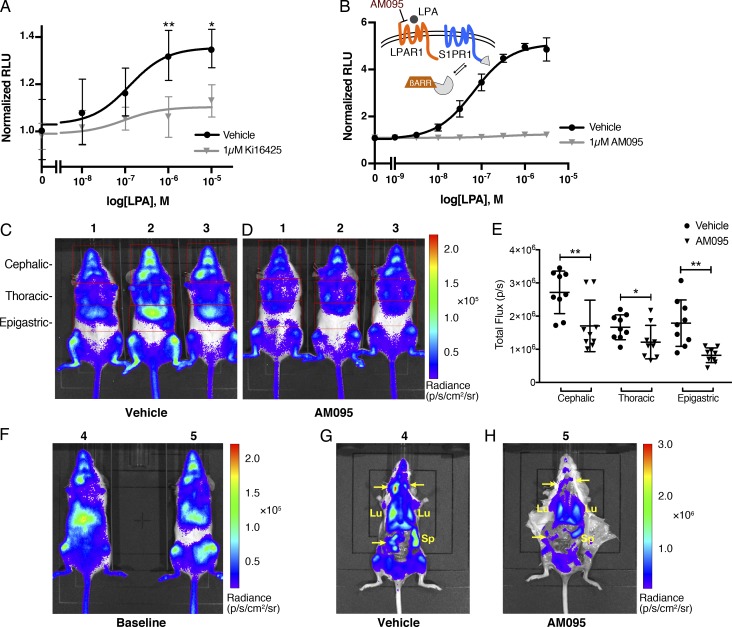
Next, to examine whether endogenously expressed LPAR1 induces inter-GPCR β-arrestin coupling to S1PR1, we isolated MEF cells from S1PR1 luciferase signaling mice, in which endogenous S1PR1/β-arrestin2 coupling can be monitored via the firefly split luciferase fragment complementation system (Kono et al., 2017). As shown in Fig. 6 A, LPA induced S1PR1/β-arrestin2 coupling in a dose-dependent manner that was blocked by Ki16425, indicating that the activation of endogenously expressed LPAR1 induces inter-GPCR β-arrestin coupling to S1PR1.
Figure 6.
Endogenous LPAR1-induced inter-GPCR β-arrestin coupling in vivo. (A) MEFs isolated from S1PR1 luciferase signaling mice were added with luciferin and then stimulated with LPA at various concentration in the presence or absence of 1 µM Ki16425. Luminescence was measured 8–12 min after LPA stimulation. Data were derived from four independent experiments and are expressed as mean ± SD. P values were determined by two-way ANOVA followed by Sidak’s multiple comparisons test comparing vehicle to Ki16425; *, P = 0.0104; **, P = 0.0021. (B) LPAR1 was transfected with S1PR1-SmBiT and LgBiT-β-arrestin1. The cells were incubated with 1 µM AM095 for 30 min before stimulation, and luminescence was measured 15–20 min after LPA stimulation. (C and D) Representative bioluminescence images of mice comparing the effects of vehicle (C) or AM095 (30 mg/kg; D) 2 h after gavage. Red open rectangles were positioned around cephalic, thoracic, and epigastric regions. (E) The bioluminescence activity was quantified by determining the total flux (photons per second [p/s]) in each region. n = 9 for each group; expressed as mean ± SD. P value was determined by paired t test. *, P ≤ 0.05; **, P ≤ 0.001. (F–H) Mice were subjected to imaging before administration (F) and then dissected in order to image internal organs after vehicle (G) or AM095 (30 mg/kg; H) administration. Arrow points to a lymph node. Lu, lung; Sp, spleen; βARR, β-arrestin; RLU, relative light units.
S1PR1 luciferase signaling mice were used to determine if LPAR1-induced inter-GPCR β-arrestin coupling to S1PR1 occurs in vivo. As previously observed, significant S1PR1 coupling to β-arrestin is seen in several organs in normal mice under homeostatic conditions (Kono et al., 2017). AM095, an orally available LPAR1 selective antagonist with desirable in vivo pharmacokinetic features (Swaney et al., 2011), completely blocked LPA/LPAR1–mediated β-arrestin1 coupling of S1PR1 in vitro (Fig. 6 B). Administration of AM095 to S1PR1 luciferase signaling mice significantly decreased bioluminescence signals (Fig. 6, C–E). Detailed imaging of dissected mice showed that S1PR1 coupling to β-arrestin in lung, spleen, and lymph nodes was significantly attenuated by AM095 treatment (Fig. 6, F–H).
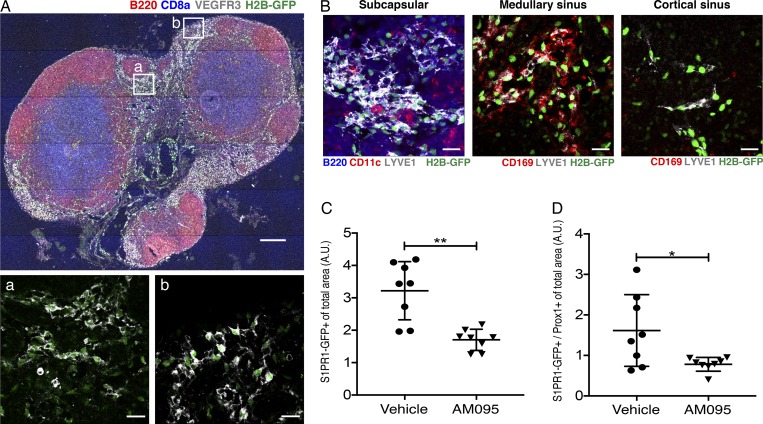
Since lymphatic endothelial cells (LECs) express both LPAR1 and S1PR1 (Heng et al., 2008), we further examined the in vivo relevance of LPAR1-induced inter-GPCR β-arrestin coupling to S1PR1 in murine lymph nodes under homeostatic conditions. For this, we used the S1PR1-GFP signaling mouse, which records cumulative S1PR1 coupling to β-arrestin while allowing high-resolution imaging studies (Kono et al., 2014). Immunofluorescence and confocal microscopy of brachial lymph node sections in adult mice showed strong S1PR1 coupling to β-arrestin in LECs that make up the cortical, medullary, and subcapsular sinuses (Fig. 7, A and B). As previously reported (Kono et al., 2014), high endothelial venules (HEVs) also exhibit S1PR1 coupling to β-arrestin (Fig. S5 A). When mice were treated with the LPAR1 inhibitor AM095 for 5 d, S1PR1 coupling to β-arrestin in sinus-lining LECs of lymph nodes was suppressed (Fig. 7, B–D). These data are consistent with quantitative imaging data using S1PR1 luciferase signaling mice shown above.
Figure 7.
S1PR1/β-arrestin coupling in LPAR1 antagonist–treated lymph nodes. (A and B) Brachial lymph node sections from S1PR1-GFP signaling mice treated with vehicle or AM095 were stained with B220 (red, B cells), CD8a (blue, T cells), and VEGFR3 (white, LECs; A); B220 (blue), CD11c (red, dendritic cells), and LYVE1 (white, LECs); or CD169 (red, macrophages) and LYVE1 (white; B). LYVE1+ lymphatics were identified as subcapsular sinuses if they were found in the subcapsular space and contained B cells and dendritic cells. Medullary sinuses contain CD169+ macrophages, and cortical sinuses are macrophage-free. (C and D) Quantification of the total (C) and Prox1–double-positive (D) GFP signal (S1PR1-β-arrestin coupling) of brachial, inguinal, and mesenteric lymph nodes of vehicle- or AM095-treated mice. Bars represent 200 µm in A and 20 µm in a, b, and B. Data were derived from eight sections for each group and are expressed as mean ± SD. P value was determined using an unpaired t test with Welch’s correction comparing vehicle to AM095; **, P ≤ 0.0021; *, P ≤ 0.0332. A.U., arbitrary unit.
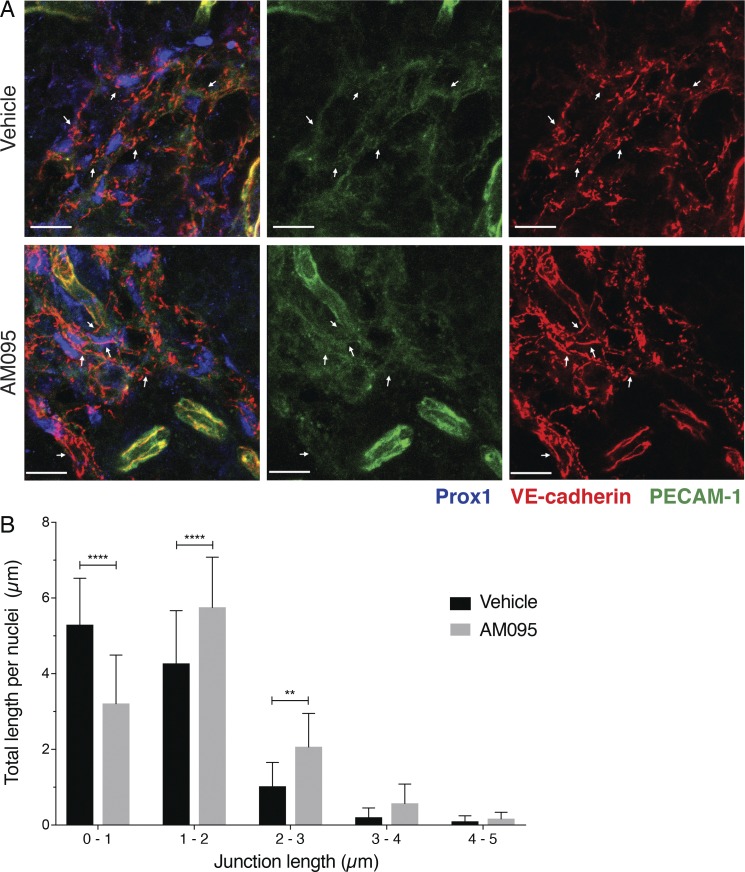
High-resolution images of cell–cell junctions in sinus-lining LECs of lymph nodes is shown in Fig. 8 A. The junctional structure is complex and contains both continuous and punctate vascular endothelial (VE)–cadherin– and PECAM-1–positive structures. We compared the junctional architecture of sinus-lining LECs from lymph nodes (lumbar, popliteal, brachial, and mesenteric) of vehicle- and AM095-treated mice. Both punctate and continuous VE-cadherin–positive LEC junctions were quantified by image analysis. As shown in Fig. 8 B, the frequency of punctate junctions was decreased and continuous junctions increased in LECs of AM095-treated mice. Similarly junctional architectural change was seen in sinus-lining LECs of brachial lymph nodes from Lpar1 KO mice (Fig. S5, B and C). Together, these data suggest that signaling of LPAR1 suppresses the formation of continuous junctions and thus enhances the sinus LEC junctional porosity.
Figure 8.
LPAR1 inhibits the formation of continuous junctions in LECs. (A) Lymph node sections (35 µm) from vehicle or 8-h AM095–treated mice were stained with Prox1 (blue), VE-cadherin (red), and PECAM-1 (green). Arrows indicate VE-cadherin–positive adherens junctions. Bars, 10 µm. (B) Quantification of junctional length in the above confocal images. Confocal images (n = 16) from three to five lymph nodes from two mice for each group were analyzed as described, and junctions of various lengths were quantified. Data are expressed as mean ± SD. P values were determined using a two-way ANOVA followed by Sidak’s multiple comparisons test comparing vehicle to AM095; **, P ≤ 0.0021; ****, P ≤ 0.0001.
LPAR1 remodels the sinus-lining LEC junctional architecture and lymphocyte retention in lymph nodes
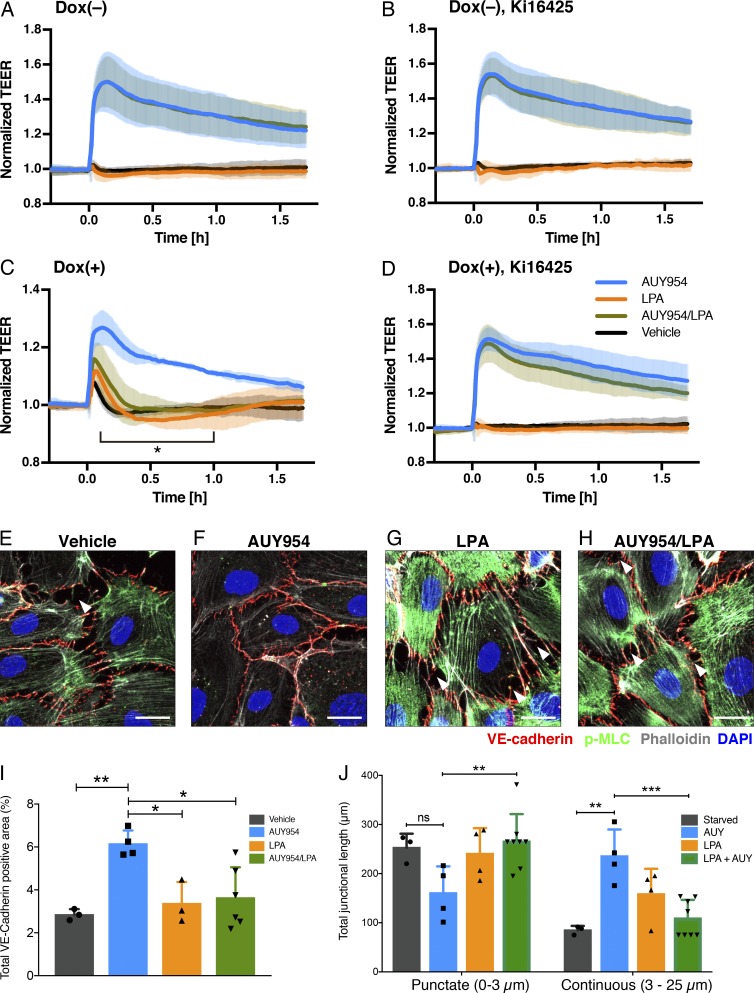
To examine whether LPAR1 modulates S1PR1-dependent barrier function in an in vitro model of endothelial cells, LPAR1 was expressed in HUVECs using an inducible lentiviral system (Fig. S5 D), and barrier function was quantified by measuring transendothelial electrical resistance (Stolwijk et al., 2015). As expected, the S1PR1 agonist AUY954 induced a sustained increase in vascular barrier function (Fig. 9 A). LPA itself did not influence barrier function in the presence or absence of AUY954 (Fig. 9 A). However, in HUVECs expressing LPAR1, LPA induced a small and transient increase in barrier function (Fig. 9 C). In sharp contrast, LPA inhibited the AUY954-induced vascular barrier increase significantly (Fig. 9 C). This was completely reversed by Ki16425, an antagonist of LPAR1 (Fig. 9, B and D), suggesting that LPAR1 induces inter-GPCR β-arrestin coupling to attenuate S1PR1-induced barrier function and thereby enhance the porosity of the endothelial monolayer.
Figure 9.
LPA/LPAR1 attenuates S1PR1-mediated barrier function. (A–D) HUVECs were analyzed for barrier function by real-time measurement of transendothelial electrical resistance (TEER) in the absence (A and B) or presence (C and D) of doxycycline (Dox), which can induce LPAR1 expression by the Tet-On system. 1 d after seeding, the cells were starved with 0.5% charcoal-treated FBS in the absence (A and C) or presence (B and D) of 1 µM Ki16425. At time 0, 100 nM AUY954 (blue), LPA (orange), AUY954 with LPA (dark green), or vehicle (black) was added. Data are from n = 3 independent experiments and expressed as mean ± SD. P values were determined by two-way ANOVA followed by Sidak’s multiple comparisons test comparing “AUY954 + LPA” to AUY954 alone; *, P ≤ 0.0001. (E–H) HUVECs expressing LPAR1 were starved with 0.5% charcoal–treated FBS for 2 h and then treated with 100 nM AUY954 and/or LPA for 30 min. Cells were fixed and stained for VE-cadherin (red) and p-MLC (green). F-actin and nuclei were stained with phalloidin (white) and DAPI (blue), respectively. Arrowheads indicate intercellular gaps. Bars, 20 µm. (I) Quantification of VE-cadherin–positive area in above confocal images expressed as mean ± SD. P value was determined by one-way ANOVA followed by Holm–Sidak’s multiple comparisons test; **, P ≤ 0.0021; *, P ≤ 0.0332. (J) Quantification of junctional length in above confocal images expressed as mean ± SD. P value was determined by two-way ANOVA followed by Tukey’s multiple comparisons test; ***, P ≤ 0.0002; **, P ≤ 0.0021; *, P < 0.0332. ns, not significant. AUY, AUY954.
To determine the cellular changes induced by LPAR1 and S1PR1 inter-GPCR β-arrestin coupling, we examined the status of VE-cadherin, a major junctional protein. F-actin and phospho-myosin light chain (p-MLC) were also examined to determine the role of Rho-coupled actin/myosin architecture, which is known to be downstream of LPAR1 (Knipe et al., 2015). As anticipated, S1PR1 activation by AUY954 strongly induced junctional VE-cadherin (Fig. 9, E and F). In S1PR1-activated HUVECs, minimal intercellular gaps were observed and VE-cadherin appeared as continuous, zipper-like structures at cell–cell borders (Fig. 9 F). Cortical F-actin was induced, and p-MLC staining was attenuated, suggesting an increase in Rac GTPase activity and a decrease in Rho GTPase activity, respectively (Fig. 9, E and F). LPA treatment strongly induced intercellular gaps that punctuate continuous VE-cadherin staining, strong F-actin staining, and stress fibers and a marked increase in p-MLC staining (Fig. 9 G). In the presence of both LPA and AUY954, junctional architecture was modulated to contain a hybrid of continuous cell–cell border staining interspersed with punctate VE-cadherin localization at the termini of actin stress fibers (Fig. 9 H). p-MLC and F-actin at stress fibers were slightly attenuated (Fig. 9, G and H). However, intercellular gaps were induced when compared with S1PR1-activated HUVECs (Fig. 9, F and H). Quantification of VE-cadherin–positive junctions is shown in Fig. 9, I and J. Total VE-cadherin staining intensity was increased by AUY954 treatment, which was blocked by LPAR1 activation. Continuous junctions (3–25 µm) were stimulated by AUY954, which activates S1PR1. In contrast, LPAR1 activation induced punctate junctions while suppressing continuous junctions. Thus, LPAR1 activation, which induces inter-GPCR β-arrestin coupling to S1PR1, helps form complex cell–cell adherens junction architecture and decreased vascular barrier function. Similar cellular mechanisms may occur in lymphatic endothelial sinuses to regulate junctional porosity.
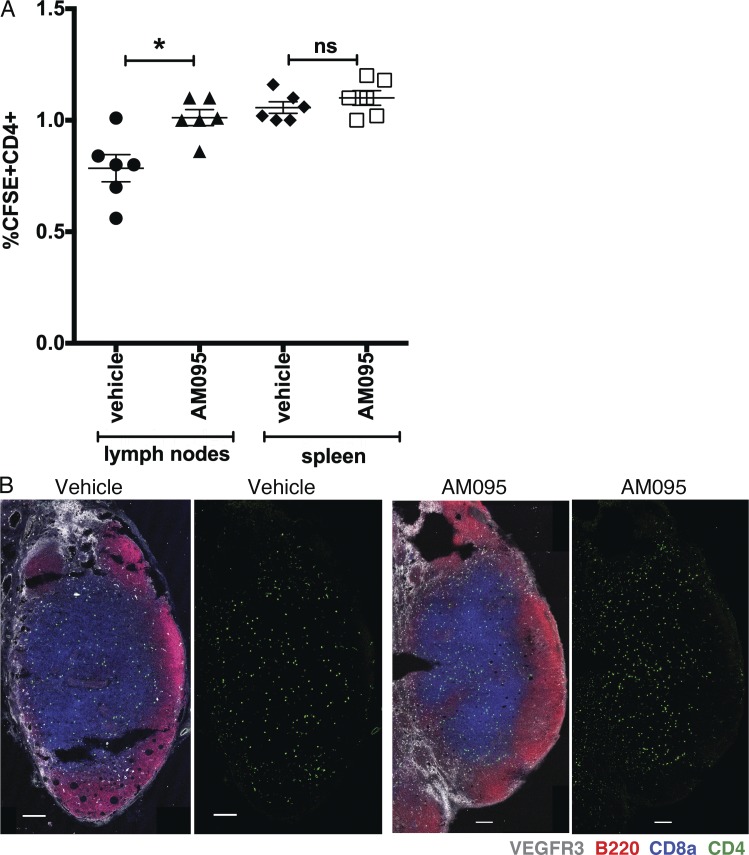
Lymphatic sinus junctional porosity may permit efficient lymphocyte egress from lymph nodes. We therefore examined the retention of adoptively transferred lymphocytes when LPAR1 is inhibited by AM095. As shown in Fig. 10 A, intravenously injected lymphocytes accumulated more in lymph nodes, but not in spleen, when LPAR1 is inhibited. Lymph node sections showed that adoptively transferred lymphocytes accumulated in T cell–rich areas of lymph nodes (Fig. 10 B), suggesting that lymph node retention is enhanced when LPAR1 is inhibited.
Figure 10.
LPAR1 regulates transferred lymphocyte trafficking at lymph nodes. (A) CFSE-labeled lymphocytes were injected into mice treated with vehicle or AM095 (30 mg/kg, gavage), and then the number of CD4- and CFSE-positive cells in lymph nodes and spleen was counted. n = 6 for each group; expressed as mean ± SD. P values were determined using an unpaired Student’s t test; *, P ≤ 0.0048. (B) 35-µm lymph node sections from vehicle- or AM095-treated mice were stained with VEGFR3 (white, LECs), B220 (red, B cells), CD8a (blue, T cells), and CFSE-CD4 (green, T cells). Bars, 100 µm. ns, not significant.
Discussion
A major finding of this study is that LPAR1 directly regulates S1PR1 function. This constitutes a heretofore undescribed cross-talk mechanism between LPA and S1P, two lysophospholipids that acquired extracellular functions as vertebrates evolved (Hla, 2005). As vertebrates acquired closed vascular systems, immune cells, which are now faced with the challenge of navigating in and out of the circulatory system, used S1P, an abundant circulatory lipid mediator with defined spatial gradients for lymphocyte trafficking (Cyster and Schwab, 2012). Our present results suggest that LPA signaling modulates S1PR1 signaling in specific contexts. The S1PR1 receptor is expressed abundantly in endothelial cells, and its cell-surface expression is controlled by multiple processes (Yanagida and Hla, 2017). For example, the lymphocyte activation–induced molecule CD69 directly interacts with S1PR1 to induce its ligand-dependent endocytosis, a process that dictates whether lymphocytes egress (Shiow et al., 2006; Bankovich et al., 2010). Indeed, tissue residency of various T cells is controlled by CD69 (Shiow et al., 2006). In endothelial cells, cell-surface signaling of S1PR1 regulates vascular barrier function (Lee et al., 1999; Oo et al., 2007). However, S1PR2, which activates the Rho GTPase, disrupts the endothelial barrier (Sanchez et al., 2007), and its function in collecting LECs is needed for lymphocyte trafficking from tissues to the lymph nodes (Xiong et al., 2019). Thus, our finding that LPAR1 modulates S1PR1 directly suggests functional cross-talk between LPA and S1P.
Our study also provides a method to discover novel regulators of GPCR signaling. By adapting a receptor reporter that induces Venus expression downstream of GPCR/β-arrestin coupling with a whole-genome–wide CRISPR/dCas9–dependent transcriptional activation system, we identified LPAR1 as a regulator of S1PR1 function. This system could be adapted to other GPCRs or signaling pathways. Given the modularity and flexibility of the CRISPR/dCas9 system, which can both activate and repress genes (Shalem et al., 2015), we suggest that many novel signaling proteins that modulate GPCRs could be identified using similar screens.
We also describe, in detail, mechanistic insight into interactions between S1PR1 and LPAR1. S1PR1 and LPAR1 interaction requires the TM4 domain of S1PR1, which was previously identified to be critical for direct interaction with CD69, an event critical for lymphocyte egress (Shiow et al., 2006). Activated LPAR1 recruits β-arrestin, which is then transferred to S1PR1, a phenomenon that we refer to as inter-GPCR β-arrestin coupling. Recent structural studies indicate that both the C-terminal tail and the third intracellular loop of GPCRs are involved in direct interaction with β-arrestin (Ranjan et al., 2017). Since the third intracellular loop of S1PR1 interacts directly with Gαi family of heterotrimeric G proteins (Lee et al., 1996), inter-GPCR β-arrestin signaling resulted in attenuation of S1PR1/Gαi signaling. However, this mechanism, in and of itself, is not sufficient to induce S1PR1 endocytosis. Thus, we suggest that LPAR1-induced inter-GPCR β-arrestin coupling results in suppression of signaling by plasma membrane–localized S1PR1. This may allow rapid reversal of S1PR1 inhibitory activity and thus acute regulation of S1PR1 GPCR.
A key issue we addressed in this study is whether this phenomenon occurs in vivo. For this, we turned to the recently developed real-time S1PR1 luciferase signaling reporter mice, which induce luciferase activity upon S1PR1/β-arrestin coupling (Kono et al., 2017). Our data show that a constitutive luciferase signal in several organs of adult S1PR1 luciferase signaling reporter mice is LPAR1 dependent. In particular, cervical and mesenteric lymph nodes showed strong luciferase activity that was suppressed by the LPAR1 antagonist AM095. High-resolution confocal microscopy studies show that sinus-lining LECs in cortical and medullary sinuses of lymph nodes are the cells in which inter-GPCR β-arrestin coupling between LPAR1 and S1PR1 occurs. Such structures are the sites at which many lymphocytes egress from the lymph node parenchyma into the lumen of the sinuses (Baluk et al., 2007; Randolph et al., 2017). In addition, lymph from afferent lymphatics that permeate through the lymph node parenchyma flow through these sinus walls to ultimately drain from the efferent lymphatic vessels. Our data suggest that inter-GPCR β-arrestin coupling between LPAR1 and S1PR1 regulates the specialized properties of lymph node sinus-lining LECs.
It is noteworthy that S1P-dependent lymphocyte egress occurs at cortical and medullary sinuses (Grigorova et al., 2009). S1P that is enriched in lymph that is secreted from LECs via SPNS2-dependent processes (Hisano et al., 2012; Mendoza et al., 2012), together with low S1P in the lymphatic parenchymal spaces, provides the spatial S1P gradient needed for efficient lymphocyte egress (Cyster and Schwab, 2012). Cell-surface S1PR1 on lymphocytes detects this gradient for a spatial cue for the egress process, which involves traverse of the lymphocyte through the sinus-lining LECs (Pham et al., 2010). Once the lymphocytes have entered the lumen of the cortical and medullary sinuses, ensuing lymph flow helps drain them into efferent lymphatic vessels (Grigorova et al., 2009), thus ensuring efficient lymphocyte trafficking. Our results suggest that LPAR1-dependent inter-GPCR β-arrestin coupling keeps LEC S1PR1 in an inactive state. It is noteworthy that LPA is generated in the lymphoid tissue parenchyma (Nakasaki et al., 2008) and regulates lymphocyte motility and traffic within the lymph node (Zhang et al., 2012; Bai et al., 2013). That LPA and S1P treatment induces additive effects on β-arrestin coupling to S1PR1 suggests that these processes are independent and provide graded responses.
We addressed the role of LPAR1-induced inter-GPCR β-arrestin coupling in endothelial cell adherens junctions and barrier function. Our results show that this mechanism alters the junctional architecture and decreases endothelial barrier function. Specifically, junctions were remodeled from continuous structures at cell–cell borders to punctate structures at the termini of actin-rich stress fibers. This results in the formation of abundant intercellular gaps, which explains the decreased vascular barrier function. Increased LPAR1-induced Rho GTPase pathways and decreased S1PR1-induced Rac GTPase pathways are likely involved, as determined by the analysis of the downstream targets p-MLC and F-actin at the cell cortex and stress fibers, respectively (Knipe et al., 2015; Burg et al., 2018). These results suggest that junctional remodeling may be a factor in the high permeability and lymphocyte egress seen in sinus-lining LECs of lymph nodes. Previous studies focused on junctions have described the presence of button-like junctions in collecting LECs of trachea lymphatics (Baluk et al., 2007), sinus-lining LECs of lymph nodes (Pham et al., 2010), and lymphatic capillaries of the small intestinal villi (Zhang et al., 2018). Such junctional specialization was hypothesized to allow lymph fluid flow. Whether such cell–cell junctions are important for efficient lymphocyte egress is not known. Our data using LPAR1 inhibitor (AM095) and the analysis of lymph nodes from Lpar1 KO mice is consistent with the notion that LPAR1/S1PR1 cross-talk is important in LEC junctional specialization and provision of a permissive environment for efficient lymphocyte egress. However, further studies using blocking antibodies to inhibit lymphocyte entry followed by detailed analysis of lymphocyte egress kinetics in situations in which LEC LPAR1 activity is modified are needed to unequivocally determine the relevance of this mechanism in lymphocyte trafficking.
In summary, we have described a mechanism by which LPAR1 suppresses cell-surface S1PR1/Gαi signaling by inter-GPCR β-arrestin coupling. This process regulates the LEC junctional architecture and barrier function at sinus-lining endothelial cells and may provide an optimal environment for efficient lymphocyte egress. Cross-talk between LPA and S1P receptors regulates complex functions of circulatory and immune systems. Pharmacologic modulation of this mechanism may be useful in lymphatic and immune disorders.
Materials and methods
Reagents
Primary antibodies used in this study include the following: PE rat monoclonal anti-Flag tag (L5), Alexa Fluor 647 mouse monoclonal anti-hemagglutinin (HA; 16B12), Alexa Fluor 647 rat monoclonal CD8a (53–6.7), Alexa Fluor 647 rat monoclonal CD169 (3D6.112), Alexa Fluor 594 rat monoclonal B220 (RA3-6B2), Alexa Fluor 647 Armenian hamster monoclonal CD11c (N418), rabbit polyclonal anti-Prox1, Brilliant violet 421 rat monoclonal CD4 (GK1.5; BioLegend); rabbit polyclonal anti–S1PR1 (H60), mouse monoclonal anti–VE-cadherin (F-8; Santa Cruz Biotechnology); rabbit polyclonal anti–p-MLC2 (Cell Signaling Technology); biotin-conjugated rat monoclonal anti-LYVE1 (ALY7), allophycocyanin rat monoclonal CD8a (53–6.7; eBioscience); goat polyclonal anti-VEGFR3, Goat polyclonal anti–VE-cadherin (R&D Systems); rat monoclonal anti-PECAM-1 (MEC13.3; BD Pharmingen); and rabbit monoclonal anti-ERG (EPR3864; Abcam). The secondary antibody used for Western blotting was HRP-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch). The secondary antibodies used for immunofluorescence were Alexa Fluor 405 donkey anti-goat IgG (Abcam), Alexa Fluor 647 donkey anti-mouse and anti-goat IgG (Invitrogen), Alexa Fluor 488 goat anti-rabbit IgG (Invitrogen), DyLight 550 donkey anti-rat IgG (Invitrogen), and DyLight 405 donkey anti-rabbit IgG (Jackson ImmunoResearch). Alexa Fluor 405 streptavidin and Alexa Fluor 546 Phalloidin were from Invitrogen. S1P and LPA were from Avanti Polar Lipids. Ki16425 and AM095 were from Sigma. W146 was from Cayman. AUY954 was from Cellagen Technology. CSFE was purchased from Molecular Probes.
Cell culture
HEK293A, HEK293T, and MEF cells were cultured in DMEM with L-glutamine, high glucose, and sodium pyruvate medium (Corning) supplemented with 10% FBS and penicillin–streptomycin (Corning) in a 37°C incubator with 5% CO2. U2OS cells were cultured in McCoy’s 5A medium (Corning) supplemented with 10% FBS and 1% penicillin–streptomycin in a 37°C incubator with 5% CO2. HUVECs were cultured in EGM-2 medium (Lonza) supplemented with 10% FBS or M199 medium (Corning) supplemented with 10% FBS, penicillin–streptomycin, endothelial cell growth factor from sheep brain extract, and 5 U/ml heparin on human fibronectin–coated dishes in a 37°C incubator with 5% CO2.
Generation of the U2OS cell line for library screening
The U2OS cells transduced with dCas9-VP64 (a gift from Feng Zhang; 61425; Addgene) and MS2-P65-HSF (a gift from Feng Zhang; 61426; Addgene; Konermann et al., 2015) were selected with 6 µg/ml Blasticidin (Gibco) and 200 µg/ml Hygromycin (Gibco), respectively. For the S1PR1-TANGO system, mouse S1pr1 linked to the tetracycline transcriptional activator via a TEV protease cleavage site and mouse β-arrestin2 linked to TEV protease were designed to be cloned in a single vector using a bicistronic internal ribosome entry site as described previously (Shalem et al., 2015), and the PCR amplicon from this vector was cloned into pCDH-CMV-MCS-EF1α-Neo lentivector (System Biosciences) with NheI and NotI digestion sites. The nuclear localization signal–Venus (a gift from Karel Svoboda; 15753; Addgene; Petreanu et al., 2007) with PEST degradation sequence at C-terminal was cloned into downstream of the tetracycline-responsive element site on pLVX-TetOn lentivector (Clontech). 600 µg/ml Geneticin (G418; Gibco) and 1 µg/ml Puromycin (Gibco) were used for selecting the cells transduced with these constructs.
To produce lentiviral particles, HEK293T cells were seeded on 10-cm dishes 1 d before transfection. On the following day, when they had reached 80–90% confluency, medium was replaced by fresh 10% FBS/DMEM medium 1 h before transfection. 20 µg lentiviral plasmid, 12.6 µg pMDL/pRRE, 9.6 µg pVSV-G, and 6 µg pRSV-REV were diluted with water and mixed with 85.25 µl of 2 M CaCl2 solution, and then 688 µl of 2 × HBS solution (274 mM NaCl, 1.5 mM Na2HPO4-7H2O, and 55 mM Hepes, pH 7.0) was slowly added into the plasmid solution while vortexing. After incubation at room temperature for 20 min, the solution mixture was added drop-wise directly to cells. Medium was replaced by 10% FBS/McCoy’s 5A medium 12–16 h after transfection. Lentiviral particle–containing supernatant was harvested 2 d after the medium change and filtered with a 0.45-µm syringe filter (Corning). PEG-it Virus Precipitation Solution (System Biosciences) was used when concentration was needed. U2OS cells were seeded 1 d before infection. On the following day, when they had reached 20–30% confluency, medium was replaced by 10% FBS/McCoy’s 5A medium containing lentiviral particles. Medium was renewed 1 d after infection, and antibiotics were added the following day. The single clones were isolated from antibiotic-resistant cells by limiting dilution and then introduced with the SAM sgRNA library (a gift from Feng Zhang; 1000000057; Addgene) at a low multiplicity of infection.
Library screening and sgRNA sequence analysis
The U2OS cells transduced with the SAM sgRNA library were cultured in 400 µg/ml Zeocin (Gibco) to select cells harboring SAM sgRNAs. Zeocin-resistant cells were allowed to grow (presort cells) or starved with 0.5% charcoal-treated FBS for 2 d. Then, starved cells were harvested, and Venus-positive cells were sorted by FACS (post-sort cells) as shown in Fig. 1 A. The sorted cells were seeded and expanded to repeat sorting. After a second expansion, genomic DNAs were harvested from 10 × 107 pre- and post-sort cells using Quick-gDNA MidiPrep (Zymo Research) according to the manufacturer’s protocol. Amplification and purification of genomic DNAs for NGS analysis was performed as described previously (Joung et al., 2017). After quality control with Agilent 2200 TapeStation, libraries were subjected to single-end sequencing on an Illumina NextSeq to generate ≥50 million reads for both pre- and post-sort cells. Reads were assigned to target genes using the previously described Python script “count_spacers.py” with default parameters (Joung et al., 2017). The resultant count table was used as input for the script “mageck” to generate significance scores for each target gene (Li et al., 2014).
RNA isolation and quantitative real-time PCR
Total RNA was isolated using TRI reagent (Zymo Research) and further purified with the Direct-zol RNA MicroPrep kit (Zymo Research), treated with DNase (30 U/µg total RNA, QIAGEN), and reverse transcribed using qScript XLT cDNA SuperMix (Quanta Bioscience). Expression of mRNA was quantitated using PerfeCTa SYBR Green FastMix Reaction Mixes (Quanta Bioscience) and the StepOnePlus Real-Time PCR System (Applied Biosystems) with cDNA equivalent to 7.5 ng total RNA.
Primers used for real-time PCR include the following (5′–3′): HPRT-Fw, TGACACTGGCAAAACAATGCA; HPRT-Rv, GGTCCTTTTCACCAGCAAGCT; SPNS2-Fw, AACGTGCTCAACTACCTGGAC; SPNS2-Rv, ATGAACACTGACTGCAGCAG; LPAR1-Fw, ACTGTGGTCATTGTGCTTGG; LPAR1-Rv, ACAGCACACGTCTAGAAGTAAC; FAM156A-Fw, TATGCTGTTGGGAGGGAAGC; and FAM156A-Rv, GCAGTATCGACATTCACATCGG.
NanoBiT assay
HEK293A cells were seeded at a density of 8 × 108 cells per 6-cm dish 1 d before transfection. The following day, expression vectors and polyethylenimine (PEI; pH 7.0; Polysciences, Inc.) were diluted in 200 µl Opti-MEM (Gibco). 300 ng LgBiT-β-arrestin1(EE) and 600 ng GPCR-SmBiT expression vectors were used for the β-arrestin recruitment assay, and 200 ng LgBiT-GNA, 1,000 ng GNB1, 1,000 ng SmBiT-GNGT1, and 400 ng GPCR expression vectors were used for the G-protein dissociation assay. 10 µl of 1 mg/ml PEI was incubated in Opti-MEM for 5 min at room temperature, and then diluted vectors and PEI were combined and mixed by vortexing and incubated for 20 min at room temperature. After incubation, the solution mixture was added drop-wise directly to cells. The following day, transfected cells were detached with 0.5 mM EDTA/PBS. After centrifugation at 190 g for 5 min, cells were suspended in 4 ml of 0.01% fatty acid–free BSA (Sigma)/HBSS (Corning) supplemented with 5 mM Hepes (Corning) and seeded on a white 96-well plate at 80 µl/well. 20 µl of 50 µM coelenterazine (Cayman) was added and incubated for 2 h at room temperature in the dark. Initial luminescence was measured as baseline using SpectraMax L (Molecular Devices), and then cells were stimulated with ligands and incubated at room temperature. Luminescence after stimulation was measured and normalized with initial reads. Development and validation of the NanoBiT G-protein dissociation assay is described elsewhere (Inoue et al., 2019).
Split firefly luciferase complementation assay in MEFs
MEFs isolated from S1PR1 luciferase signaling mice (Kono et al., 2017) were seed on a white 96-well plate. The following day, medium was replaced by 80 µl of 0.01% fatty acid–free BSA/HBSS supplemented with 5 mM Hepes and incubated for 2 h at room temperature. 20 µl of 40 mg/ml Luciferin (Perkin Elmer) was added, and initial luminescence was measured. After stimulation with LPA, luminescence was measured and normalized with initial reads. Bioluminescence in live mice and internal organs was measured as described previously (Kono et al., 2017).
Generation of Gα-depleted HEK293 cells using the CRISPR/Cas9 system
Gα-depleted HEK293 cells were generated by mutating genes encoding members of the Gαi family from previously established HEK293 cells devoid of three Gα families (Gαs, Gαq, and Gα12; Grundmann et al., 2018) using the CRISPR/Cas9 system as described previously (Ran et al., 2013; O'Hayre et al., 2017), with minor modifications. sgRNA constructs targeting the GNAI1, GNAI2, GNAI3, GNAO1, GNAT1, GNAT2, and GNAZ genes, whose mRNA was expressed in HEK293 cells (Atwood et al., 2011), were designed using a CRISPR design tool (http://crispr.mit.edu) so that a SpCas9-mediated DNA cleavage site (3 bp upstream of the protospacer adjacent motif [PAM] sequence [NGG]) encompasses a restriction enzyme–recognizing site. Designed sgRNA-targeting sequences including the SpCas9 PAM sequences were as follows: 5′-CTTTGGTGACTCAGCCCGGGCGG-3′ (GNAI1; where the restriction enzyme site [Sma I in this case] is underlined and the PAM sequence is in bold), 5′-CGTAAAGACCACGGGGATCGTGG-3′ (GNAI2; Mbo I), 5′-AGCTTGCTTCAGCAGATCCAGGG-3′ (GNAI3; Mbo I), 5′-AATCGCCTTGCTCCGCTCGAGGG-3′ (GNAO1; Xho I), 5′-TTTCAGGTGCCGGTGAGTCCGGG-3′ (GNAT1; Hinf I), 5′-AACCATGCCTCCTGAGCTCGTGG-3′ (GNAT2; Sac I), and 5′-GATGCGGGTCAGCGAGTCGATGG-3′ (GNAZ; Hinf I). The designed sgRNA-targeting sequences were inserted into the BbsI site of the pSpCas9(BB)-2A-GFP (PX458) vector (a gift from Feng Zhang; 42230; Addgene) using the following set of synthesized oligonucleotides: 5′-CACCGCTTTGGTGACTCAGCCCGGG-3′ and 5′-AAACCCCGGGCTGAGTCACCAAAGC-3′ (GNAI1; note that a guanine nucleotide [G] was introduced at the −21 position of the sgRNA (underlined), which enhances transcription of the sgRNA); 5′-CACCGCGTAAAGACCACGGGGATCG-3′ and 5′-AAACCGATCCCCGTGGTCTTTACGC-3′ (GNAI2); 5′-CACCGAGCTTGCTTCAGCAGATCCA-3′ and 5′-AAACTGGATCTGCTGAAGCAAGCTC-3′ (GNAI3); 5′-CACCGAATCGCCTTGCTCCGCTCGA-3′ and 5′-AAACTCGAGCGGAGCAAGGCGATTC-3′ (GNAO1); 5′-CACCGTTTCAGGTGCCGGTGAGTCC-3′ and 5′-AAACGGACTCACCGGCACCTGAAAC-3′ (GNAT1); 5′-CACCGAACCATGCCTCCTGAGCTCG-3′ and 5′-AAACCGAGCTCAGGAGGCATGGTTC-3′ (GNAT2); 5′-CACCGATGCGGGTCAGCGAGTCGA-3′ and 5′-AAACTCGACTCGCTGACCCGCATC-3′ (GNAZ). Correctly inserted sgRNA-encoding sequences were verified with a Sanger sequencing (Fasmac) using the primer 5′-ACTATCATATGCTTACCGTAAC-3′.
To achieve successful selection of all-allele mutant clone, we performed an iterative CRISPR/Cas9–mediated mutagenesis. Specifically, in the first round, mutations were introduced in the GNAZ gene. In the second round, the GNAI2, GNAI3, and GNAO1 genes were simultaneously mutated. In the last round, the GNAI1, GNAT1, and GNAT2 genes were targeted. Briefly, HEK293 cells devoid of three Gα families (Grundmann et al., 2018) were seeded into a 6-well culture plate and incubated for 1 d before transfection. A plasmid encoding sgRNA and SpCas9-2A-GFP was transfected into the cells using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s protocol. 3 d later, cells were harvested and processed for isolation of GFP-positive cells (∼6% of cells) using a fluorescence-activated cell sorter (SH800; Sony). After expansion of clonal cell colonies with a limiting dilution method, clones were analyzed for mutations in the targeted genes by restriction enzyme digestion as described previously (O'Hayre et al., 2017; Grundmann et al., 2018). PCR primers that amplify the sgRNA-targeting sites were as follows: 5′-AGCTGGTTATTCAGAAGAGGAGTG-3′ and 5′-TGGTCCTGATAGTTGACAAGCC-3′ (GNAI1); 5′-AAATGGCATGGGAGGGAAGG-3′ and 5′-TAAAACCTCAGTGGGGCTGG-3′ (GNAI2); 5′-AGCTGGCAGTGCTGAAGAAG-3′ and 5′-TCATACAAATGACCAAGGGCTC-3′ (GNAI3); 5′-GGTCCTTACCGAGCAGGAG-3′ and 5′-CGACATTTTTGTTTCCAGCCC-3′ (GNAO1); 5′-TAGGTGTGGCTACGGGGTC-3′ and 5′-GCACTCTTCCAGCGAGTACC-3′ (GNAT1); 5′-ACTGCTTCCATCTTAGGTCTTCG-3′ and 5′-CATCAACCCACCCTCTCACC-3′ (GNAT2); 5′-CGAAATCAAGCTGCTCCTGC-3′ and 5′-TGTCCTCCAGGTGGTACTCG-3′ (GNAZ). Candidate clones that harbored restriction enzyme–resistant PCR fragments were further assessed for their genomic DNA alterations by direct sequencing or TA cloning as described previously (O'Hayre et al., 2017; Grundmann et al., 2018).
Measurement of endothelial barrier function in vitro
Endothelial barrier function was evaluated by measuring the resistance of a cell-covered electrode using an endothelial cell impedance system Zθ device (Applied BioPhysics) in accordance with the manufacturer’s instructions. Briefly, arrays were cleaned with 10 mM L-cysteine, washed with sterile water, coated with fibronectin for 30 min at 37°C, and incubated with complete cell culture medium to run electrical stabilization. HUVECs were seeded on a 96-well electrode array (96W10idf) at a density of 2.5 × 104 cell per well in the presence or absence of 1 µg/ml doxycycline. The following day, confluent cells were starved for 2–3 h in EBM-2 (Lonza) supplemented with 0.5% charcoal–treated FBS and then stimulated with AUY954 and/or LPA. Resistance was monitored and expressed as fractional resistance, normalizing to the baseline at time 0.
Imaging studies in mice
All animal procedures were approved by the Animal Care and Use Committees of the National Institute of Diabetes and Digestive and Kidney Diseases, Boston Children’s Hospital, and University of Maryland School of Medicine and performed in accordance with National Institutes of Health guidelines. S1PR1-GFP and S1PR1-luciferase signaling mice have been previously described (Kono et al., 2014, 2017). Bioluminescence images were acquired 2 h after injection with vehicle (10 µM Na2CO3 and 20% 2-hydroxypropyl-β-cyclodextrin) by gavage. 3 h after the first imaging for vehicle, the AM095 (30 mg/kg) was administrated to the mice through gavage, and bioluminescence images were acquired 2 h later. S1PR1-GFP signaling mice were treated with vehicle or AM095 (20 mg/kg twice a day) for 5 d by gavage, and lymph nodes were collected. C57BL/6J mice were treated with AM095 (30 mg/kg twice by gavage, every 4 h), and lymph nodes were collected 8 h after the first treatment. Lymph nodes from Lpar1+/− and Lpar1−/− (Contos et al., 2000) were harvested and sectioned for confocal microscopy imaging studies.
Immunofluorescence staining
HUVECs were washed with cold PBS and fixed with 2% paraformaldehyde (PFA) for 10 min at room temperature. U2OS cell were washed with cold PBS and fixed with cold methanol for 10 min on ice. Lymph nodes were fixed in 4% PFA in PBS at 4°C, washed in PBS and embedded in optimal cutting temperature (OCT) compound (Tissue-Tek, Thermo Fisher Scientific) for frozen section. Cryosections (35 µm) were permeabilized with PBS-0.1% Triton at room temperature for 30 min and then blocked with PBS containing 75 mM NaCl, 18 mM Na3 citrate, 2% FBS, 1% BSA, and 0.05% Triton X-100. Incubation with primary antibodies was performed overnight at 4°C, followed by three washes with PBS and incubation with secondary antibodies for 3 h at room temperature. LEC images were obtained with costaining of rat anti-CD31 antibody (BD Pharmigen), goat anti-VE-cadherin, and rabbit anti-Prox1. Confocal images were taken using a Zeiss LSM 800 with Airyscan confocal microscope. The three- dimensional reconstructions of z-stack (xy projection) images are shown. Image processing and quantification was performed by using Adobe Photoshop, ImageJ, or Fiji software (National Institutes of Health). GFP, CD31, VE-cadherin, and Prox1-positive immunofluorescent signals were subjected to threshold processing, and areas occupied by their signal were quantified using Fiji software as described previously (Yanagida et al., 2017). The junction lengths were measured using Fiji software. Total lymph nodes from CFSE-CD4–injected mice were frozen in OCT, sectioned (35 µm), and stained with B220, CD8, and VEGRF3 antibodies.
Immunoblot analysis
Cells were washed with cold PBS and lysed in modified RIPA buffer (50 mM Tris, pH 7.4, 100 mM sodium chloride, 2 mM EDTA, 1% Triton X-100, 0.5% Fos-Choline, and 10 mM sodium azide) containing phosphatase inhibitors (1 mM sodium orthovanadate, 1 mM sodium fluoride, and 5 mM β-glycerophosphate) and protease inhibitor cocktail (Sigma). After incubation on ice for 30 min and a freeze/thaw cycle, protein concentrations in supernatant from centrifugation at 10,000 g (15 min at 4°C) were determined by bicinchoninic acid assay (Pierce) and denatured for 30 min at room temperature in Laemmli’s sample buffer supplemented with 10% β-mercaptoethanol. An equal amount of protein was loaded and separated on an SDS-polyacrylamide gel and transferred electrophoretically to a polyvinylidene difluoride membrane (Millipore). Transferred proteins were then probed with rabbit polyclonal anti-S1PR1 (Santa Cruz Biotechnology) and HRP-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch).
Flow cytometry analysis
U2OS cells, HEK293A cells, and HUVECs were detached with 0.05% Trypsin (Corning), 0.5 mM EDTA, and Accutase (Innovatice Cell Technologies), respectively. The harvested cells were fixed with 1% PFA for 10 min on ice and labeled with PE anti-Flag and Alexa Fluor 647 anti-HA antibodies for detecting cell–surface expression. The samples were analyzed using a BD Calibur FACS system, and FlowJo software was used for data analysis.
In vivo T cell migration assay
Mouse (C57BL/6; The Jackson Laboratory) CD4 T cells from spleen and lymph nodes were prepared (CD4 enrichment kit; STEMCELL Technologies) and CFSE labeled (Molecular Probes). Recipient mice were given vehicle or 30 mg/kg AM095 by gavage. 2 h later, 106 labeled CD4 T cells were adoptively transferred i.v. 16 h later, spleen or lymph nodes were collected and stained with anti-CD4-BV421 (clone GK1.5; BioLegend) and anti–CD8-APC (clone 53–6.7; eBioscience). The samples were analyzed with FACSAria flow cytometer (BD Biosciences). Data were analyzed with FlowJo software v 8.8.7 (Tree Star). Values are expressed as the percentage of CSFE+ CD4+ cells out of total CD4+ cells.
Spleen and lymph nodes were collected from above recipient mice and immediately submerged in OCT compound (Sakura Finetek). Tissues in OCT were quickly frozen using dry ice and then kept at −80°C for long-term storage. Sections of the lymph nodes were stained as above and imaged by confocal microscopy.
Statistical analysis
Data are expressed as means ± SD. Statistical analysis was performed as mentioned using Prism software (GraphPad). P values <0.05 were considered statistically significant.
Online supplemental material
Fig. S1 shows how SPNS2 and LPAR1 SAM sgRNAs activate target genes and Venus expression. Fig. S2 shows how LPAR1 stimulation with LPA causes an additive effect in S1P-stimulated S1PR1-SmBiT/β-arrestin coupling. Fig. S3 shows DNA sequences of the Gα genomic loci in mutant HEK293 cells. Fig. S4 shows S1PR1 protein expression and localization. Fig. S5 shows how LPAR1-mediated S1PR1/β-arrestin coupling regulates LEC junctions.
Supplementary Material
Acknowledgments
The authors thank Drs. Hiroko Kishikawa, Yuji Shinjo, and Kumiko Makide for technical assistance with FACS. The SAM sgRNA library, dCas9-VP64, MS2-P65-HSF, and pSpCas9(BB)-2A-GFP (PX458) plasmids were provided by Professor Feng Zhang (Broad Institute of Harvard and MIT, Cambridge, MA). The pCAGGS-ChR2-Venus plasmid was provided by Karel Svoboda (Howard Hughes Medical Institute, Maryland, MA, and Cold Spring Harbor Laboratory, Cold Spring Harbor, NY).
This work was supported by National Institutes of Health grants R35 HL135821 (T. Hla), RO1AI062765, and 1RO1AI14496 (J.S. Bromberg), a Fondation Leducq transatlantic network grant (SphingoNet; T. Hla and R.L. Proia), the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health intramural program (M. Kono and R.L. Proia), American Heart Association postdoctoral fellowships (A. Cartier and A. Kuo), Japan Society for the Promotion of Science KAKENHI grant 17K08264 (A. Inoue), and Japan Agency for Medical Research and Development grants PRIME JP17gm5910013 (A. Inoue) and LEAP JP17gm0010004 (A. Inoue and J. Aoki). Y. Hisano and K. Yanagida were supported in part by postdoctoral fellowships from the Japan Society for the Promotion of Science Overseas Research Fellowships. Y. Hisano was also supported by the Uehara Memorial Foundation.
T. Hla received research funding from the ONO Pharmaceutical Corporation; consulted for Steptoe and Johnson, LLP, and Bridge Medicine Inc.; and is an inventor of ApoM+HDL, S1P chaperones, and S1P receptor antagonists. The remaining authors declare no competing financial interests.
Author contributions: Y. Hisano contributed to conception of the screen, conducting the screen, validation studies in vitro, lymph node experiments, writing the manuscript, analysis of data, and figure preparations. M. Kono and R.L. Proia contributed to studies of luciferase signaling mice. A. Cartier contributed to studies of GFP signaling mice and junction analysis, quantitation in vitro and in vivo, figure preparation, and confocal microscopy. E. Engelbrecht contributed to analysis of NGS data for guide RNAs and targets. K. Kano and J. Aoki contributed to Lpar1 KO mice studies. K. Kawakami and S. Galvani contributed to NanoBiT assays. Y. Xiong, W. Piao, and J.S. Bromberg contributed to T cell adoptive transfer experiments. K. Yanagida and A. Kuo contributed to analysis of GFP signaling mice. Y. Ono, S. Ishida, and A. Inoue contributed to the analysis and setup of NanoBiT signaling assays. T. Hla contributed to conception of the screen, overall direction of the project, analysis of data, and writing/editing of the manuscript.
References
- Aikawa S., Hashimoto T., Kano K., and Aoki J.. 2015. Lysophosphatidic acid as a lipid mediator with multiple biological actions. J. Biochem. 157:81–89. 10.1093/jb/mvu077 [DOI] [PubMed] [Google Scholar]
- Arnon T.I., Xu Y., Lo C., Pham T., An J., Coughlin S., Dorn G.W., and Cyster J.G.. 2011. GRK2-dependent S1PR1 desensitization is required for lymphocytes to overcome their attraction to blood. Science. 333:1898–1903. 10.1126/science.1208248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood B.K., Lopez J., Wager-Miller J., Mackie K., and Straiker A.. 2011. Expression of G protein-coupled receptors and related proteins in HEK293, AtT20, BV2, and N18 cell lines as revealed by microarray analysis. BMC Genomics. 12:14 10.1186/1471-2164-12-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Z., Cai L., Umemoto E., Takeda A., Tohya K., Komai Y., Veeraveedu P.T., Hata E., Sugiura Y., Kubo A., et al. . 2013. Constitutive lymphocyte transmigration across the basal lamina of high endothelial venules is regulated by the autotaxin/lysophosphatidic acid axis. J. Immunol. 190:2036–2048. 10.4049/jimmunol.1202025 [DOI] [PubMed] [Google Scholar]
- Baluk P., Fuxe J., Hashizume H., Romano T., Lashnits E., Butz S., Vestweber D., Corada M., Molendini C., Dejana E., and McDonald D.M.. 2007. Functionally specialized junctions between endothelial cells of lymphatic vessels. J. Exp. Med. 204:2349–2362. 10.1084/jem.20062596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankovich A.J., Shiow L.R., and Cyster J.G.. 2010. CD69 suppresses sphingosine 1-phosophate receptor-1 (S1P1) function through interaction with membrane helix 4. J. Biol. Chem. 285:22328–22337. 10.1074/jbc.M110.123299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnea G., Strapps W., Herrada G., Berman Y., Ong J., Kloss B., Axel R., and Lee K.J.. 2008. The genetic design of signaling cascades to record receptor activation. Proc. Natl. Acad. Sci. USA. 105:64–69. 10.1073/pnas.0710487105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaho V.A., and Hla T.. 2011. Regulation of mammalian physiology, development, and disease by the sphingosine 1-phosphate and lysophosphatidic acid receptors. Chem. Rev. 111:6299–6320. 10.1021/cr200273u [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burg N., Swendeman S., Worgall S., Hla T., and Salmon J.E.. 2018. Sphingosine 1-Phosphate Receptor 1 Signaling Maintains Endothelial Cell Barrier Function and Protects Against Immune Complex-Induced Vascular Injury. Arthritis Rheumatol. 70:1879–1889. 10.1002/art.40558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun J., Hla T., Lynch K.R., Spiegel S., and Moolenaar W.H.. 2010. International Union of Basic and Clinical Pharmacology. LXXVIII. Lysophospholipid receptor nomenclature. Pharmacol. Rev. 62:579–587. 10.1124/pr.110.003111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contos J.J.A., Fukushima N., Weiner J.A., Kaushal D., and Chun J.. 2000. Requirement for the lpA1 lysophosphatidic acid receptor gene in normal suckling behavior. Proc. Natl. Acad. Sci. USA. 97:13384–13389. 10.1073/pnas.97.24.13384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyster J.G., and Schwab S.R.. 2012. Sphingosine-1-phosphate and lymphocyte egress from lymphoid organs. Annu. Rev. Immunol. 30:69–94. 10.1146/annurev-immunol-020711-075011 [DOI] [PubMed] [Google Scholar]
- Dixon A.S., Schwinn M.K., Hall M.P., Zimmerman K., Otto P., Lubben T.H., Butler B.L., Binkowski B.F., Machleidt T., Kirkland T.A., et al. . 2016. NanoLuc Complementation Reporter Optimized for Accurate Measurement of Protein Interactions in Cells. ACS Chem. Biol. 11:400–408. 10.1021/acschembio.5b00753 [DOI] [PubMed] [Google Scholar]
- Fukushima N., Kimura Y., and Chun J.. 1998. A single receptor encoded by vzg-1/lpA1/edg-2 couples to G proteins and mediates multiple cellular responses to lysophosphatidic acid. Proc. Natl. Acad. Sci. USA. 95:6151–6156. 10.1073/pnas.95.11.6151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorova I.L., Schwab S.R., Phan T.G., Pham T.H., Okada T., and Cyster J.G.. 2009. Cortical sinus probing, S1P1-dependent entry and flow-based capture of egressing T cells. Nat. Immunol. 10:58–65. 10.1038/ni.1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundmann M., Merten N., Malfacini D., Inoue A., Preis P., Simon K., Rüttiger N., Ziegler N., Benkel T., Schmitt N.K., et al. . 2018. Lack of beta-arrestin signaling in the absence of active G proteins. Nat. Commun. 9:341 10.1038/s41467-017-02661-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. 2012. Rho family GTPases. Biochem. Soc. Trans. 40:1378–1382. 10.1042/BST20120103 [DOI] [PubMed] [Google Scholar]
- Heng T.S.P., and Painter M.W.. Immunological Genome Project Consortium . 2008. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9:1091–1094. 10.1038/ni1008-1091 [DOI] [PubMed] [Google Scholar]
- Hisano Y., Kobayashi N., Kawahara A., Yamaguchi A., and Nishi T.. 2011. The sphingosine 1-phosphate transporter, SPNS2, functions as a transporter of the phosphorylated form of the immunomodulating agent FTY720. J. Biol. Chem. 286:1758–1766. 10.1074/jbc.M110.171116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisano Y., Kobayashi N., Yamaguchi A., and Nishi T.. 2012. Mouse SPNS2 functions as a sphingosine-1-phosphate transporter in vascular endothelial cells. PLoS One. 7:e38941 10.1371/journal.pone.0038941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisano Y., Ota S., Takada S., and Kawahara A.. 2013. Functional cooperation of spns2 and fibronectin in cardiac and lower jaw development. Biol. Open. 2:789–794. 10.1242/bio.20134994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hla T. 2005. Genomic insights into mediator lipidomics. Prostaglandins Other Lipid Mediat. 77:197–209. 10.1016/j.prostaglandins.2005.06.008 [DOI] [PubMed] [Google Scholar]
- Holthuis J.C., and Menon A.K.. 2014. Lipid landscapes and pipelines in membrane homeostasis. Nature. 510:48–57. 10.1038/nature13474 [DOI] [PubMed] [Google Scholar]
- Inoue A., Raimondi F., Kadji F.M.N., Singh G., Kishi T., Uwamizu A., Ono Y., Shinjo Y., Ishida S., Arang N., et al. . 2019. Cell. 177:1–15. 10.1016/j.cell.2019.04.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii I., Contos J.J.A., Fukushima N., and Chun J.. 2000. Functional comparisons of the lysophosphatidic acid receptors, LP(A1)/VZG-1/EDG-2, LP(A2)/EDG-4, and LP(A3)/EDG-7 in neuronal cell lines using a retrovirus expression system. Mol. Pharmacol. 58:895–902. 10.1124/mol.58.5.895 [DOI] [PubMed] [Google Scholar]
- Joung J., Konermann S., Gootenberg J.S., Abudayyeh O.O., Platt R.J., Brigham M.D., Sanjana N.E., and Zhang F.. 2017. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat. Protoc. 12:828–863. 10.1038/nprot.2017.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara A., Nishi T., Hisano Y., Fukui H., Yamaguchi A., and Mochizuki N.. 2009. The sphingolipid transporter spns2 functions in migration of zebrafish myocardial precursors. Science. 323:524–527. 10.1126/science.1167449 [DOI] [PubMed] [Google Scholar]
- Knipe R.S., Tager A.M., and Liao J.K.. 2015. The Rho kinases: critical mediators of multiple profibrotic processes and rational targets for new therapies for pulmonary fibrosis. Pharmacol. Rev. 67:103–117. 10.1124/pr.114.009381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi N., Kawasaki-Nishi S., Otsuka M., Hisano Y., Yamaguchi A., and Nishi T.. 2018. MFSD2B is a sphingosine 1-phosphate transporter in erythroid cells. Sci. Rep. 8:4969 10.1038/s41598-018-23300-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S., Brigham M.D., Trevino A.E., Joung J., Abudayyeh O.O., Barcena C., Hsu P.D., Habib N., Gootenberg J.S., Nishimasu H., et al. . 2015. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 517:583–588. 10.1038/nature14136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono M., Mi Y., Liu Y., Sasaki T., Allende M.L., Wu Y.P., Yamashita T., and Proia R.L.. 2004. The sphingosine-1-phosphate receptors S1P1, S1P2, and S1P3 function coordinately during embryonic angiogenesis. J. Biol. Chem. 279:29367–29373. 10.1074/jbc.M403937200 [DOI] [PubMed] [Google Scholar]
- Kono M., Tucker A.E., Tran J., Bergner J.B., Turner E.M., and Proia R.L.. 2014. Sphingosine-1-phosphate receptor 1 reporter mice reveal receptor activation sites in vivo. J. Clin. Invest. 124:2076–2086. 10.1172/JCI71194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono M., Conlon E.G., Lux S.Y., Yanagida K., Hla T., and Proia R.L.. 2017. Bioluminescence imaging of G protein-coupled receptor activation in living mice. Nat. Commun. 8:1163 10.1038/s41467-017-01340-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.J., Evans M., and Hla T.. 1996. The inducible G protein-coupled receptor edg-1 signals via the G(i)/mitogen-activated protein kinase pathway. J. Biol. Chem. 271:11272–11279. 10.1074/jbc.271.19.11272 [DOI] [PubMed] [Google Scholar]
- Lee M.-J., Thangada S., Liu C.H., Thompson B.D., and Hla T.. 1998a Lysophosphatidic acid stimulates the G-protein-coupled receptor EDG-1 as a low affinity agonist. J. Biol. Chem. 273:22105–22112. 10.1074/jbc.273.34.22105 [DOI] [PubMed] [Google Scholar]
- Lee M.-J., Van Brocklyn J.R., Thangada S., Liu C.H., Hand A.R., Menzeleev R., Spiegel S., and Hla T.. 1998b Sphingosine-1-phosphate as a ligand for the G protein-coupled receptor EDG-1. Science. 279:1552–1555. 10.1126/science.279.5356.1552 [DOI] [PubMed] [Google Scholar]
- Lee M.-J., Thangada S., Claffey K.P., Ancellin N., Liu C.H., Kluk M., Volpi M., Sha’afi R.I., and Hla T.. 1999. Vascular endothelial cell adherens junction assembly and morphogenesis induced by sphingosine-1-phosphate. Cell. 99:301–312. 10.1016/S0092-8674(00)81661-X [DOI] [PubMed] [Google Scholar]
- Li W., Xu H., Xiao T., Cong L., Love M.I., Zhang F., Irizarry R.A., Liu J.S., Brown M., and Liu X.S.. 2014. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15:554 10.1186/s13059-014-0554-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.H., Thangada S., Lee M.-J., Van Brocklyn J.R., Spiegel S., Hla T., and Guidotti G.. 1999. Ligand-induced trafficking of the sphingosine-1-phosphate receptor EDG-1. Mol. Biol. Cell. 10:1179–1190. 10.1091/mbc.10.4.1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza A., Bréart B., Ramos-Perez W.D., Pitt L.A., Gobert M., Sunkara M., Lafaille J.J., Morris A.J., and Schwab S.R.. 2012. The transporter Spns2 is required for secretion of lymph but not plasma sphingosine-1-phosphate. Cell Reports. 2:1104–1110. 10.1016/j.celrep.2012.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizugishi K., Yamashita T., Olivera A., Miller G.F., Spiegel S., and Proia R.L.. 2005. Essential role for sphingosine kinases in neural and vascular development. Mol. Cell. Biol. 25:11113–11121. 10.1128/MCB.25.24.11113-11121.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moolenaar W.H., and Hla T.. 2012. SnapShot: Bioactive lysophospholipids. Cell. 148:378–378.e2. 10.1016/j.cell.2012.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutoh T., Rivera R., and Chun J.. 2012. Insights into the pharmacological relevance of lysophospholipid receptors. Br. J. Pharmacol. 165:829–844. 10.1111/j.1476-5381.2011.01622.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanaga K., Hama K., Kano K., Sato T., Yukiura H., Inoue A., Saigusa D., Tokuyama H., Tomioka Y., Nishina H., et al. . 2014. Overexpression of autotaxin, a lysophosphatidic acid-producing enzyme, enhances cardia bifida induced by hypo-sphingosine-1-phosphate signaling in zebrafish embryo. J. Biochem. 155:235–241. 10.1093/jb/mvt114 [DOI] [PubMed] [Google Scholar]
- Nakasaki T., Tanaka T., Okudaira S., Hirosawa M., Umemoto E., Otani K., Jin S., Bai Z., Hayasaka H., Fukui Y., et al. . 2008. Involvement of the lysophosphatidic acid-generating enzyme autotaxin in lymphocyte-endothelial cell interactions. Am. J. Pathol. 173:1566–1576. 10.2353/ajpath.2008.071153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hayre M., Eichel K., Avino S., Zhao X., Steffen D.J., Feng X., Kawakami K., Aoiki J., Messer K., Sunahara R., et al. . 2017. Genetic evidence that β-arrestins are dispensable for the initiation of β2-adrenergic receptor signaling to ERK. Sci. Signal. 10:eaal3395 10.1126/scisignal.aal3395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta H., Sato K., Murata N., Damirin A., Malchinkhuu E., Kon J., Kimura T., Tobo M., Yamazaki Y., Watanabe T., et al. . 2003. Ki16425, a subtype-selective antagonist for EDG-family lysophosphatidic acid receptors. Mol. Pharmacol. 64:994–1005. 10.1124/mol.64.4.994 [DOI] [PubMed] [Google Scholar]
- Oo M.L., Thangada S., Wu M.T., Liu C.H., Macdonald T.L., Lynch K.R., Lin C.Y., and Hla T.. 2007. Immunosuppressive and anti-angiogenic sphingosine 1-phosphate receptor-1 agonists induce ubiquitinylation and proteasomal degradation of the receptor. J. Biol. Chem. 282:9082–9089. 10.1074/jbc.M610318200 [DOI] [PubMed] [Google Scholar]
- Oo M.L., Chang S.H., Thangada S., Wu M.T., Rezaul K., Blaho V., Hwang S.I., Han D.K., and Hla T.. 2011. Engagement of S1P1-degradative mechanisms leads to vascular leak in mice. J. Clin. Invest. 121:2290–2300. 10.1172/JCI45403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan S., Mi Y., Pally C., Beerli C., Chen A., Guerini D., Hinterding K., Nuesslein-Hildesheim B., Tuntland T., Lefebvre S., et al. . 2006. A monoselective sphingosine-1-phosphate receptor-1 agonist prevents allograft rejection in a stringent rat heart transplantation model. Chem. Biol. 13:1227–1234. 10.1016/j.chembiol.2006.09.017 [DOI] [PubMed] [Google Scholar]
- Petreanu L., Huber D., Sobczyk A., and Svoboda K.. 2007. Channelrhodopsin-2-assisted circuit mapping of long-range callosal projections. Nat. Neurosci. 10:663–668. 10.1038/nn1891 [DOI] [PubMed] [Google Scholar]
- Pham T.H., Baluk P., Xu Y., Grigorova I., Bankovich A.J., Pappu R., Coughlin S.R., McDonald D.M., Schwab S.R., and Cyster J.G.. 2010. Lymphatic endothelial cell sphingosine kinase activity is required for lymphocyte egress and lymphatic patterning. J. Exp. Med. 207:17–27. 10.1084/jem.20091619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proia R.L., and Hla T.. 2015. Emerging biology of sphingosine-1-phosphate: its role in pathogenesis and therapy. J. Clin. Invest. 125:1379–1387. 10.1172/JCI76369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8:2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randolph G.J., Ivanov S., Zinselmeyer B.H., and Scallan J.P.. 2017. The Lymphatic System: Integral Roles in Immunity. Annu. Rev. Immunol. 35:31–52. 10.1146/annurev-immunol-041015-055354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan R., Dwivedi H., Baidya M., Kumar M., and Shukla A.K.. 2017. Novel Structural Insights into GPCR-β-Arrestin Interaction and Signaling. Trends Cell Biol. 27:851–862. 10.1016/j.tcb.2017.05.008 [DOI] [PubMed] [Google Scholar]
- Sanchez T., Skoura A., Wu M.T., Casserly B., Harrington E.O., and Hla T.. 2007. Induction of vascular permeability by the sphingosine-1-phosphate receptor-2 (S1P2R) and its downstream effectors ROCK and PTEN. Arterioscler. Thromb. Vasc. Biol. 27:1312–1318. 10.1161/ATVBAHA.107.143735 [DOI] [PubMed] [Google Scholar]
- Shalem O., Sanjana N.E., and Zhang F.. 2015. High-throughput functional genomics using CRISPR-Cas9. Nat. Rev. Genet. 16:299–311. 10.1038/nrg3899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea B.S., and Tager A.M.. 2012. Role of the lysophospholipid mediators lysophosphatidic acid and sphingosine 1-phosphate in lung fibrosis. Proc. Am. Thorac. Soc. 9:102–110. 10.1513/pats.201201-005AW [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T. 2009. Lipid mediators in health and disease: enzymes and receptors as therapeutic targets for the regulation of immunity and inflammation. Annu. Rev. Pharmacol. Toxicol. 49:123–150. 10.1146/annurev.pharmtox.011008.145616 [DOI] [PubMed] [Google Scholar]
- Shiow L.R., Rosen D.B., Brdicková N., Xu Y., An J., Lanier L.L., Cyster J.G., and Matloubian M.. 2006. CD69 acts downstream of interferon-alpha/beta to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature. 440:540–544. 10.1038/nature04606 [DOI] [PubMed] [Google Scholar]
- Skoura A., and Hla T.. 2009. Lysophospholipid receptors in vertebrate development, physiology, and pathology. J. Lipid Res. 50(Suppl):S293–S298. 10.1194/jlr.R800047-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolwijk J.A., Matrougui K., Renken C.W., and Trebak M.. 2015. Impedance analysis of GPCR-mediated changes in endothelial barrier function: overview and fundamental considerations for stable and reproducible measurements. Pflugers Arch. 467:2193–2218. 10.1007/s00424-014-1674-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumida H., Noguchi K., Kihara Y., Abe M., Yanagida K., Hamano F., Sato S., Tamaki K., Morishita Y., Kano M.R., et al. . 2010. LPA4 regulates blood and lymphatic vessel formation during mouse embryogenesis. Blood. 116:5060–5070. 10.1182/blood-2010-03-272443 [DOI] [PubMed] [Google Scholar]
- Swaney J.S., Chapman C., Correa L.D., Stebbins K.J., Broadhead A.R., Bain G., Santini A.M., Darlington J., King C.D., Baccei C.S., et al. . 2011. Pharmacokinetic and pharmacodynamic characterization of an oral lysophosphatidic acid type 1 receptor-selective antagonist. J. Pharmacol. Exp. Ther. 336:693–700. 10.1124/jpet.110.175901 [DOI] [PubMed] [Google Scholar]
- Swendeman S.L., Xiong Y., Cantalupo A., Yuan H., Burg N., Hisano Y., Cartier A., Liu C.H., Engelbrecht E., Blaho V., et al. . 2017. An engineered S1P chaperone attenuates hypertension and ischemic injury. Sci. Signal. 10:eaal2722 10.1126/scisignal.aal2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Okudaira S., Kishi Y., Ohkawa R., Iseki S., Ota M., Noji S., Yatomi Y., Aoki J., and Arai H.. 2006. Autotaxin stabilizes blood vessels and is required for embryonic vasculature by producing lysophosphatidic acid. J. Biol. Chem. 281:25822–25830. 10.1074/jbc.M605142200 [DOI] [PubMed] [Google Scholar]
- Tian X., Kang D.S., and Benovic J.L.. 2014. β-arrestins and G protein-coupled receptor trafficking. Handb. Exp. Pharmacol. 219:173–186. 10.1007/978-3-642-41199-1_9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meeteren L.A., Ruurs P., Stortelers C., Bouwman P., van Rooijen M.A., Pradère J.P., Pettit T.R., Wakelam M.J., Saulnier-Blache J.S., Mummery C.L., et al. . 2006. Autotaxin, a secreted lysophospholipase D, is essential for blood vessel formation during development. Mol. Cell. Biol. 26:5015–5022. 10.1128/MCB.02419-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu T.M., Ishizu A.N., Foo J.C., Toh X.R., Zhang F., Whee D.M., Torta F., Cazenave-Gassiot A., Matsumura T., Kim S., et al. . 2017. Mfsd2b is essential for the sphingosine-1-phosphate export in erythrocytes and platelets. Nature. 550:524–528. 10.1038/nature24053 [DOI] [PubMed] [Google Scholar]
- Willinger T., Ferguson S.M., Pereira J.P., De Camilli P., and Flavell R.A.. 2014. Dynamin 2-dependent endocytosis is required for sustained S1PR1 signaling. J. Exp. Med. 211:685–700. 10.1084/jem.20131343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windh R.T., Lee M.J., Hla T., An S., Barr A.J., and Manning D.R.. 1999. Differential coupling of the sphingosine 1-phosphate receptors Edg-1, Edg-3, and H218/Edg-5 to the G(i), G(q), and G(12) families of heterotrimeric G proteins. J. Biol. Chem. 274:27351–27358. 10.1074/jbc.274.39.27351 [DOI] [PubMed] [Google Scholar]
- Xiong Y., Piao W., Brinkman C.C., Li L., Kulinski J.M., Olivera A., Cartier A., Hla T., Hippen K.L., Blazar B.R., et al. . 2019. CD4 T cell sphingosine 1-phosphate receptor (S1PR)1 and S1PR4 and endothelial S1PR2 regulate afferent lymphatic migration. Sci. Immunol. 4:eaav1263 10.1126/sciimmunol.aav1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagida K., and Hla T.. 2017. A dark side to omega-3 fatty acids. Nature. 552:180–181. 10.1038/d41586-017-07678-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagida K., Liu C.H., Faraco G., Galvani S., Smith H.K., Burg N., Anrather J., Sanchez T., Iadecola C., and Hla T.. 2017. Size-selective opening of the blood-brain barrier by targeting endothelial sphingosine 1-phosphate receptor 1. Proc. Natl. Acad. Sci. USA. 114:4531–4536. 10.1073/pnas.1618659114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Zarkada G., Han J., Li J., Dubrac A., Ola R., Genet G., Boyé K., Michon P., Künzel S.E., et al. . 2018. Lacteal junction zippering protects against diet-induced obesity. Science. 361:599–603. 10.1126/science.aap9331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen Y.C., Krummel M.F., and Rosen S.D.. 2012. Autotaxin through lysophosphatidic acid stimulates polarization, motility, and transendothelial migration of naive T cells. J. Immunol. 189:3914–3924. 10.4049/jimmunol.1201604 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.