Abstract
Background
Germline DNA copy number variation (CNV) is a ubiquitous source of genetic variation and remains largely unexplored in association with epithelial ovarian cancer (EOC) risk.
Methods
CNV was quantified in the DNA of approximately 3500 cases and controls genotyped with the Illumina 610k and HumanOmni2.5M arrays. We performed a genome-wide association study of common (>1%) CNV regions (CNVRs) with EOC and high-grade serous (HGSOC) risk and, using The Cancer Genome Atlas (TCGA), performed in silico analyses of tumor-gene expression.
Results
Three CNVRs were associated (P<0.01) with EOC risk: two large (~100kb) regions within the 610k set and one small (<5kb) region with the higher resolution 2.5M data. Large CNVRs included a duplication at LILRA6 (OR=2.57, P=0.001) and a deletion at CYP2A7 (OR=1.90, P=0.007) that was strongly associated with HGSOC risk (OR=3.02, P=8.98×10−5). Somatic CYP2A7 alterations correlated with EGLN2 expression in tumors (P=2.94×10−47). An intronic ERBB4/HER4 deletion was associated with reduced EOC risk (OR=0.33, P=9.5×10−2) and somatic deletions correlated with ERBB4 downregulation (P=7.05×10−5). Five CNVRs were associated with HGSOC including two reduced-risk deletions: one at 1p36.33 (OR=0.28, P=0.001) that correlated with lower CDKIIA expression in TCGA tumors (P=2.7×10−7), and another at 8p21.2 (OR=0.52, P=0.002) that was present somatically where it correlated with lower GNRH1 expression (P=5.9×10−5).
Conclusions
Though CNV appears to not contribute largely to EOC susceptibility, a number of low-to-common frequency variants may influence the risk of EOC and tumor gene expression.
Impact
Further research on CNV and EOC susceptibility is warranted, particularly with CNVs estimated from high-density arrays.
Keywords: Ovarian Cancer, Genetic Susceptibility, Copy Number Variations, Copy Number Alterations, Single Nucleotide Polymorphisms
Introduction
Epithelial ovarian cancer (EOC) is the fifth most common cause of cancer death among women in North America (1). Since most women are diagnosed at advanced stages, better early detection and intervention is needed (1). Genome-wide association studies (GWAS) have identified thirty-nine common allelic variations associated with EOC susceptibility (2), but these variants explain only a modest fraction of heritability (3), thus more such loci likely exist. Exploration of other sources of DNA variation is warranted.
High-throughput genome technologies have revealed that the human genome contains substantial structural variation. An estimated 10-13% of DNA content can be spanned by copy number variation (CNV), segments of DNA one kilobase or larger in length, that differ from a reference genome (4, 5). Germline CNV can be inherited or occur de novo (6) and predispose to an array of complex diseases including familial and sporadic types of cancer (7), The contribution of CNV to EOC risk remains largely unexplored.
Our group has previously evaluated whether inherited CNVs were associated with overall survival among 1,056 women with EOC; no associations achieved statistical significance after adjustment for multiple comparisons (8). Almost all studies evaluating CNV and EOC risk have been conducted among BRCA1 carriers or women with hereditary breast-ovarian cancer syndrome (9–11). The largest study to-date included 357 EOC cases and 1962 non-ovarian cancer affected BRCA1 carriers from The Consortium of Investigators of Modifiers of BRCA1/BRCA2 (CIMBA), where a validated deletion in CYP2A7 was associated with decreased EOC risk (10). An analysis of The Cancer Genome Atlas (TCGA) compared the germline-somatic landscape in exomes of 429 high grade serous (HGSOC) cases to 557 controls (12). However, copy number analysis was limited to BRCA1, BRCA2, and TP53 (12). The current study represents the first large-scale genome-wide analysis of germline CNV evaluating associations with EOC risk among unselected women from the general population.
Methods
Study Population
Our GWAS of EOC utilized two genotyping platforms. Four case-control studies from Mayo Clinic, Duke University, University of Toronto and Moffitt Cancer Center used the Illumina 610-quad Beadchip Array© (“610k”). An independent sample of cases and controls from Mayo Clinic was genotyped on the Illumina HumanOmni2.5M-8 Beadchip© (“2.5M”). Both GWAS sets included patients with incident, pathologically confirmed primary EOC, either borderline or invasive, aged 20 or above. DNA samples from women having less than 80% European ancestry were excluded (13). Full study details have been previously published (14, 15).
CNV Calling and Quality Control (QC)
CNV segmentation was performed with PennCNV software (16) on probe-level B allele frequency (BAF) and log2 R ratio (LRR) for autosomal SNPs mapped to GRCh37 (hg19), with adjustment for local GC content responsible for signal fluctuations (17). Segments spanning at least 3 probes and confidence scores >10 were retained. To reduce possible batch effects or poor quality intensity data, we excluded samples with outliers (>median+1.5 interquartile range [IQR]) for LRR standard deviation, BAF drift, GC wave factor, and number of CNV calls. In total, 856 (23%) of the 610k array samples and 219 (22%) of the 2.5M array samples were excluded. The dataset used for this analysis will be made available through dbGAP under study accession phs001133.v1.p1.
Common CNV Regions and Association Testing
CNV regions (CNVRs) were defined using the CNVruler tool (18) that constructs CNVRs by merging CNV segments that overlap by at least 1 bp and trims any rare, long CNV. Logistic regression was used to compare CNVR status (deletion/no deletion; duplication/no duplication) between cases and controls that occurred with >1% frequency among all samples in a set. To adjust for population stratification, eigenvectors were calculated from a matrix of CNVR status and the first principal component was included as a covariate of regression (Supplemental Figure 1). Site, age, and experimental batch are known sources of bias (19) but none affected CNVR estimates and these were not retained in the risk model. As a sensitivity analysis for the CNVR merge method, we also employed ParseCNV (20) which performs SNP-level association testing and merges significant SNPs into risk-associated CNVRs. CNV mapping studies suggest that deletions are poorly tolerated and under negative selection whereas duplications are less likely to be pathogenic and are often under positive selection, which drives evolution of many gene families (21). Thus, for both analytic approaches, deletions and duplications were analyzed separately. Risk associations are reported for CNVRs that reached P<0.01 significance threshold. We excluded T-cell receptor (TCR) and immunoglobulin heavy (IGH) chain genomic regions from analyses as these undergo V-(D)-J recombination in lymphocytes and can result in detection of somatic copy number alterations rather than inherited, germline CNV which is the focus of this study (22, 23). These regions included TCR alpha and delta of chromosome 14 (chr14:22090057-23021075 and chr14:22891537-22935569, respectively), beta and gamma on chromosome 7 (chr7:141998851-142510972 and chr7:38279625-38407656, respectively), and IGH regions on chromosomes 14 and 16 (chr14:106032614-107288051 and chr16: 33740716-33741266).
Integration of CNV and Tumor Transcriptome using TCGA
To explore the correlation of copy number and gene expression, we obtained copy number segments, gene-level Fragments Per Kilobase of transcript per Million mapped reads (FPKM) values from RNA sequencing, and CpG island methylation data for 571 HGSOC cases from TCGA (24) that had germline CNV quantified from either blood or normal tissue samples. CNV segments were estimated from SNP 6.0 array using circular binary segmentation and were limited to those with a minimum of three probes and <10 MB in size. Samples with a high number of calls (>median+1.5 IQRs) were excluded. We defined deletions as segments with mean copy number <-0.3 and amplifications as those >0.3. We employed multivariate linear regression to model both CNV and somatic copy number alterations (SCNA) in the cancer genome (diploid, deletion, duplication) on mRNA expression level (log2 transformed) in tumor tissue. P-values were calculated with the likelihood ratio statistic. For each gene, the effect of CpG methylation was regressed out, as previously described (25). Statistical analyses were performed in R (www.r-project.org).
Results
Table 1 summarizes the clinical characteristics and copy number distribution of 2,818 subjects (1,368 cases) genotyped with the 610k array and 792 subjects (449 cases) genotyped with the 2.5M array after applying QC exclusions. Cases were slightly older than controls on average and the majority had serous tumors. Most (75%) of the CNV calls in the 610k array data were deletions while the majority (59%) in the 2.5M set were duplications. The average number and length of deletions and duplications were similar between cases and controls (Table 1). The distribution of CNV in the 610k set was largely similar across the five main histotypes of EOC and by stage at diagnosis although advanced stage endometrioid cases (n=50) averaged a significantly higher number of deletions (P=0.0003; Supplemental Table 1). Though 2.5M sample sizes were too small to investigate stage of disease, low-grade serous cases (n=43) did average a significantly lower number of duplications (P=0.002; Supplemental Table 2).
Table 1.
Study participant characteristics and Copy Number Variant (CNV) distributions
| 610k Array N=2818 |
2.5M Array N=792 |
|||||
|---|---|---|---|---|---|---|
| Case | Control | Pvaluea | Case | Control | Pvaluea | |
| N | 1368 | 1450 | 449 | 343 | ||
| Age | ||||||
| Mean (range) | 60.1 (26-91) | 57.7 (20-90) | <0.001 | 62.2 (20-88) | 58.2 (22-93) | <0.001 |
| Study Site | ||||||
| Minnesota region (Mayo) | 325 | 467 | 449 | 343 | ||
| North Carolina (Duke) | 406 | 553 | 0 | 0 | ||
| Tampa Bay (Moffitt) | 152 | 139 | 0 | 0 | ||
| Toronto (U of Toronto) | 485 | 291 | 0 | 0 | ||
| Histotype (%) | ||||||
| Serous | 825 (60%) | 346 (77%) | ||||
| High grade | 410 (30%) | 303 (67%) | ||||
| Low grade/LMP | 121 (9%) | 43 (10%) | ||||
| Unknown | 294 (21%) | 0 | ||||
| Endometrioid | 241 (18%) | 30 (7%) | ||||
| Mucinous | 63 (5%) | 28 (6%) | ||||
| Clear cell | 112 (8%) | 15 (3%) | ||||
| Mixed cell | 38 (3%) | 25 (6%) | ||||
| Other | 89 (7%) | 5 (1%) | ||||
| CNV segments (No.) | ||||||
| All | 30877 | 33626 | 28578 | 21929 | ||
| Deletions | 23532 (76%) | 25028 (74%) | 11947 (42%) | 8821 (40%) | ||
| Duplications | 7345 (24%) | 8598 (26%) | 16631 (58%) | 13108 (60%) | ||
| CNV segments (Mean) | ||||||
| All | 22.6 | 23.2 | 0.93 | 63.7 | 63.9 | 0.63 |
| Deletions | 17.2 | 17.3 | 0.56 | 26.6 | 25.7 | 0.06 |
| Duplications | 5.4 | 5.9 | 1.00 | 37.0 | 38.2 | 0.92 |
| Average CNV length (Kb) | ||||||
| All | 77 | 80 | 0.99 | 48 | 49 | 0.93 |
| Deletions | 55 | 54 | 0.34 | 25 | 26 | 0.73 |
| Duplications | 144 | 146 | 0.64 | 65 | 66 | 0.77 |
Mean age was compared between cases and controls with the student t-test. Empirical p-values are reported for difference in CNV average number of segments and difference in average CNV length based on 10000 permutations.
CNVRs were constructed by merging overlapping CNV calls across individuals in a study set, separately for deletions and duplications. In the 610k set, there were 7,384 CNVRs that included 348 regions occurring in ≥ 1% of subjects (denoted as common CNVRs henceforth), 3,105 rare regions (<1%), and 3,931 regions detected in a single individual (singletons). In the 2.5M set, CNV calls merged into 3,732 CNVRs: 624 common regions, 972 rare regions, and 2,136 singletons. Notably, the majority (>80%) of CNV was rare (<1%) or singletons. Rare CNV calls trended higher for cases than controls in both array sets but the results were not statistically significant (Supplemental Table 3).
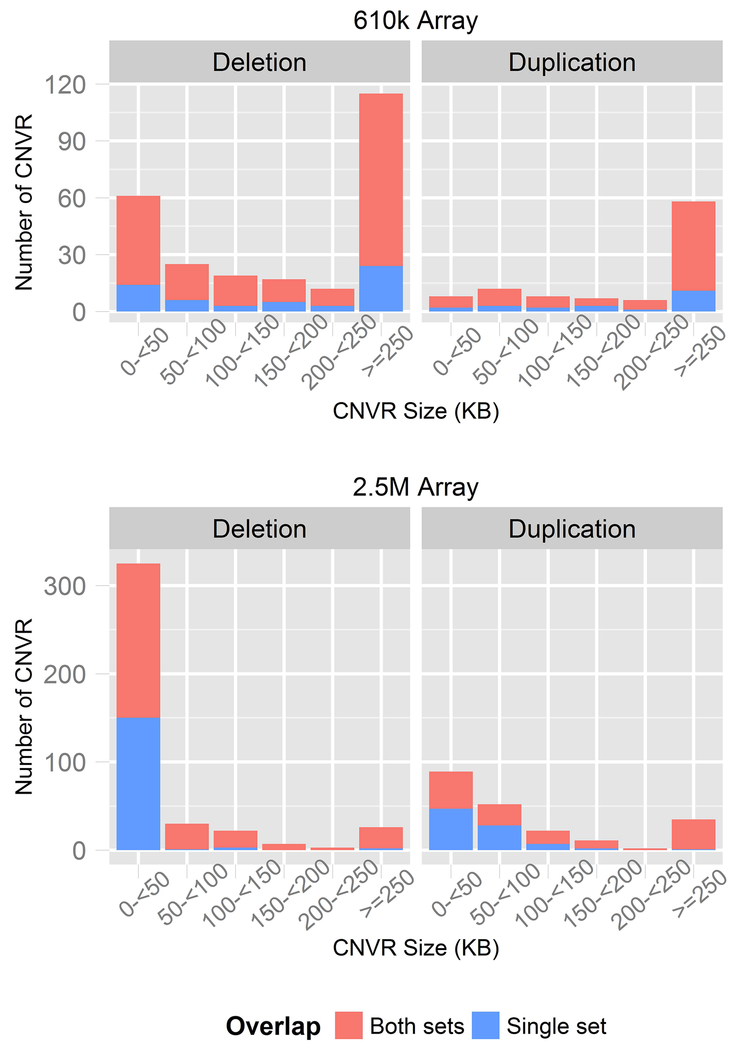
CNVR size distributions differed between array sets, likely reflecting differences in probe density (Figure 1). CNVRs in the 2.5M set spanned shorter genomic regions (median=23kb) compared to the 610k set (median=246kb) where regions >200kb in size comprised the majority (70%) of CNVRs. Most of the common CNVRs in the 610k set (N=271, 78%) were detected at least once within the higher resolution 2.5M array set and, conversely, 383 (61%) in the 2.5M set were detected at least once in the 610k set. We limited CNVRs to those detected within both array sets and excluded genomic regions that are somatically deleted in lymphocytes and are not likely to be inherited, germline CNV (see Methods). In total, 189 deletion and 74 duplication CNVRs in the 610k set and 252 deletion and 125 duplication CNVRs in the 2.5M set were analyzed for association with EOC risk.
Figure 1. Common CNV regions generated from two SNP-array data sets.
Copy number variation regions (CNVR) were compared between 2.5M and 610k array sets. Bar charts depict the number of deletion and duplication CNVRs that were common in each set. The overlap between sets is denoted by stacked bars.
Common CNV regions and EOC risk
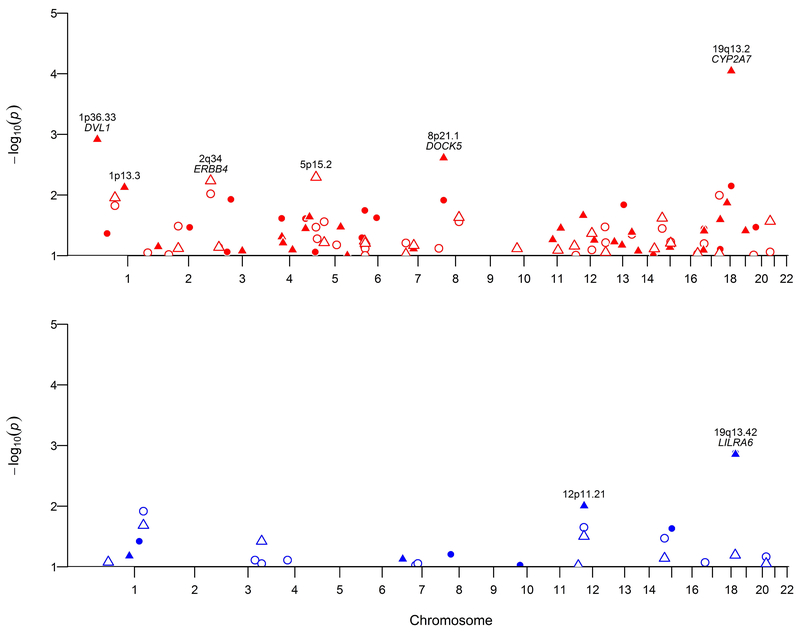
CNVR associations with EOC risk overall and with HGSOC are shown in Figure 2. Differences in copy number between all EOC cases and controls were detected (P<0.01) at three common CNVRs (Table 2). Two CNVRs spanning approximately 100kb each were associated with EOC risk within the 610k population and a third, substantially smaller CNVR <5kb in length was associated with risk in the 2.5M analysis. HGSOC-specific analysis showed that all three EOC risk-associated CNVRs were also associated with HGSOC (Table 3). An additional four CNVR in the 610k analysis and one CNVR in the 2.5M analysis were associated with HGSOC risk (P<0.01) that were not identified in the overall EOC analysis. All CNVRs were detected in both array sets albeit frequencies varied between sets and comparable regions were substantially smaller in the 2.5M set (Supplemental Table 4). Additionally, risk-associated CNVRs were compared with the Database of Genomic Variants and shown to overlap gold standard copy number variants (Supplemental Table 5).
Figure 2. Manhattan plots for association analyses between common CNVR and EOC risk.
Logistic regression was conducted for common CNVRs that were detected by both arrays. Separate analyses for deletions (top figure; red) and duplications (bottom figure; blue) in both the 610k (filled points) and 2.5M (outline points) array sets were conducted for all epithelial risk (circle) and high-grade serous specific risk (triangle).
Table 2.
Association of common germline Copy Number Variation Regions (CNVRs) with EOC susceptibility
| Merged CNVR |
SNP-level CNVR |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Array (N cases) | Locus | CNV Type | Gene | CNVR (KB)a | N (%) Case/Control | P-value | OR (95% CI) | Tag SNP | Total SNPs | P-value b | OR (95% CI) |
| 610k (n=1368) | 19q13.2 | Del | CYP2A7 | chr19:41341589-41433931 (92) | 49 (4)/29 (2) | 0.007 | 1.90 (1.19-3.03) | rs2545754 | 9 | 0.006 | 2.03 (1.22-3.36) |
| 19q13.42 | Dup | LILRA6 | chr19:54731679-54845802 (114) | 39 (3)/17 (1) | 0.001 | 2.57 (1.44-4.57) | rs11672654 | 11 | 0.01 | 5.87 (1.53-22.57) | |
| 2.5M (n=449) | 2q34 | Del | ERBB4 | chr2:213187034-213191389 (4) | 8 (2)/18 (5) | 0.0095 | 0.33 (0.14-0.76) | kgp5655115 | 12 | 0.008 | 0.33 (0.15-0.75) |
CNVR was defined by overlapping CNV segments across subjects. Coordinates are mapped to human genome build 37 (hg19). P-values are from logistic regression adjusted for the first principal component.
CNVR was defined as region with significant SNP-level statistics and p-value reported is from fisher exact test comparing deletion/no deletion or duplication/no duplication. All epithelial OC vs. controls reported.
Table 3.
Association of common germline Copy Number Variation Regions (CNVRs) with HGSOC susceptibility
| Merged CNVR |
SNP-level CNVR |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Array (N cases) | Locus | CNV Type | Gene(s) | CNVR (KB)a | N (%) Case/Control | P-value | OR (95% CI) | Tag SNP | Total SNPs | P-value | OR (95% CI) |
| 610k (n=410) | 1p36.33 | Del | ACAP3, CPSF3L, DVL1, GLTPD1, MXRA8, PUSL1, TAS1R3 b | Chr1:943468-1706160 (763) | 7 (2) 84 (6) |
0.001 | 0.28 (0.13-0.60) | rs3737717 | 13 | 0.001 | 0.31 (0.15-0.62) |
| 1p13.3 | Del | Intergenic | Chr1:111370372-111391381 (21) | 23 (6) 40 (3) |
0.007 | 2.07 (1.21-3.51) | rs6677356 | 6 | 0.008 | 2.09 (1.21-3.60) | |
| 8p21.2 | Del | DOCK5 | Chr8:24931313-25101936 (171) | 27 (7) 168 (12) |
0.002 | 0.52 (0.34-0.79) | cnvi0001694 | 7 | 0.003 | 0.54 (0.36-0.81) | |
| 12p11.21 | Dup | RP11-428G5.5 | Chr12:31975730-32068877 (93) | 19 (5) 32 (2) |
0.0098 | 2.14 (1.20-3.82) | rs1259725 | 27 | 0.015 | 2.15 (1.16-3.98) | |
| 19q13.2 | Del | CYP2A7 | chr19:41341589-41433931 (92) | 24 (6) 29 (2) |
8.98E-05 | 3.02 (1.74-5.25) | rs2545754 | 9 | 2.79E-05 | 3.54 (1.96-6.39) | |
| 19q13.42 | Dup | LILRA6 | chr19:54731679-54845802 (114) | 15 (4) 17 (1) |
0.001 | 3.16 (1.56-6.39) | rs11672654 | 8 | 0.004 | 2.98 (1.42-6.27) | |
| 2.5M (n=303) | 2q34 | Del | ERBB4 | chr2:213187034-213191389 (4) | 3 (1) 18 (5) |
0.005 | 0.18 (0.05-0.60) | kgp5655115 | 12 | 0.003 | 0.18 (0.06-0.56) |
| 5p15.2 | Del | Intergenic | Chr5:12812336-12888815 (76) | 55 (18) 36 (10) |
0.005 | 1.91 (1.22-3.01) | kgp22267001 | 3 | 0.009 | 1.85 (1.17-2.94) | |
CNVR was defined by overlapping CNV segments across subjects. P-values are from logistic regression adjusted for the first principal component.
CNV segments centered on the reported genes which fell within the SNP-level significance region.
Within the analysis of all invasive EOC, a duplication region at 19q13.42 within the human leukocyte receptor cluster (LRC) was associated with increased risk of EOC in the 610k set (P = 0.001, OR = 2.57). Duplications within this CNVR spanned leukocyte immunoglobulin like receptor A6 (LILRA6) and occurred with low-frequency (2%). The CNVR was more common in 2.5M subjects (28%) but frequency did not differ by disease status (P = 0.89, OR = 0.98). A second CNVR identified in the 610k set was a deletion at 19q13.2 spanning cytochrome P450 family 2 subfamily A member 7 (CYP2A7) and the downstream, intergenic region (P = 0.007, OR = 1.90). The deletion region was similarly common in the 2.5M population (6%) but not associated with EOC risk (P = 0.27, OR = 0.71). Finally, a small, 4kb deletion CNVR within intron 1 of erb-b2 receptor tyrosine kinase 4 (ERBB4/HER4) at 2q34 was associated with reduced EOC risk (P = 0.0095, OR = 0.33) in the 2.5M data. CNV within the boundaries of this region were rare (<1%) in the 610k analysis and not associated with risk (P = 0.47, OR = 1.60).
The HGSOC risk analysis was limited to a subset of 410 cases in the 610k array set and 303 cases in the 2.5M array set. The increase in EOC risk for CYP2A7 deletion carriers in the 610k set was stronger for HGSOC-specific risk (P = 8.98×10−5, OR = 3.02) though it was not associated with HGSOC risk in the 2.5M analysis (P = 0.18). The CNVRs at LILRA6 and ERBB4 showed slightly stronger risk associations with HGSOC (OR = 3.16 and 0.18, respectively).
Two relatively large deletion regions were associated with lower risk of HGSOC: one spanning much of 1p36.33 (P = 0.001, OR = 0.28) and the other a 171kb deletion at 8p21.2 (P= 0.002, OR = 0.52). The CNVR at 1p36.33 spanned a ~750kb region though most deletions were located within a smaller ~60kb region where multiple genes reside including disheveled segment polarity protein 1 (DVL1). While large, the 8p21.2 deletion region solely contained the transcription start site (TSS) and sequence for dedicator of cytokinesis 5 (DOCK5) and no other coding features. Both deletions were also detected with the 2.5M array set with the 1p36.33 CNVR smaller and centered on ATPase family, AAA domain containing 3B (ATAD3B) rather than DVL1 and the 8p21.2 CNVR concurring with the TSS region of DOCK5. Both CNVR associations were not statistically significant (1p36.33: P = 0.71, OR = 0.80; 8p21.2: P = 0.61, OR = 1.13).
Three other CNVRs were associated with increased risk of HGSOC. First, duplications of a 100kb region at 12p11.21 that contained lincRNA RP11-428G5.5 occurred in twice as many HGSOC cases as controls (P = 9.8×10−3, OR = 2.14). This CNVR was also detected in 3% of 2.5M subjects but was not associated with risk (P = 0.36, OR = 0.62). A smaller 21kb intergenic region at 1p13.3 revealed deletions among 6% of HGSOC cases and 3% of controls (P = 0.007, OR = 2.07). These deletions were 22kb upstream to ubiquitously expressed cell-surface protein CD53 molecule. The same deletion region was detected with the 2.5M array but only present in one control and no HGSOC cases. Finally, a more common intergenic deletion CNVR at 5p15.2 was associated with increased HGSOC risk in the 2.5M set (18% in cases, 10% in controls, P = 0.005, OR = 1.91). Deletions within this region were detected in ten HGSOC cases and five controls in the 610k set (P = 0.30, OR = 1.76).
Our analysis of CNVRs was based on the assignment of a consensus boundary defined by merging individual segments. To determine whether this affected downstream association analyses, we conducted a sensitivity analysis that identified regions where SNP-level copy number was significantly associated with EOC risk rather than solely testing in pre-defined regions. All CNVRs associated with EOC risk in our primary analysis were also detected using the SNP-based CNVR approach, with notably higher risk estimates at the CNVR containing LILRA6 (Table 2), and more similar estimates for HGSOC-specific risk associations (Table 3). Additionally, CNVR boundaries in our primary analysis were established by merging deletions and duplications separately. As an alternative approach, we merged segments irrespective of CNV type which defined gain only, loss only, and mixed type regions. Consequently, CNVR boundaries were altered from our primary analysis; however, risk associations remained significant for all regions and was strengthened for the 8p21.2 (DOCK5) association with EOC risk (P = 0.004, OR = 0.69) (Supplemental Table 6 and Supplemental Table 7). Analysis of mixed type CNV (deletion or duplication vs. diploid) did not identify any additional common CNVR associated with EOC risk.
Association of risk CNVRs with transcription levels in tumor tissue
With the multi-level data available from TCGA, we sought to determine whether germline CNV within the risk-associated CNVRs correlated with primary tumor mRNA expression levels. This required careful consideration of SCNA as they are the most prevalent alteration in the cancer genome and are known to influence oncogene activation and tumor suppressor gene inactivation in tumor tissues (26). Thus, within seven CNVRs, we quantified (i.e., deletion/diploid/duplication) CNV of the germline DNA and focal SCNA and estimated their independent effects on cis-mRNA gene expression in 382 HGSOC cases from TCGA having both CNV and RNA-sequencing data. We excluded 5p15 as no mRNA sequences were within 500kb of the CNVR.
CNVs were detected with common frequencies (1-41%) within the risk-associated regions for the TCGA set of HGSOC cases which was derived from a separate platform and segmentation algorithm, increasing our confidence in their validity (Figure 3A). These regions also contained a high frequency of somatic alterations in the tumor genome (16-26%), excluding the 1p36 region that was diploid in all HGSOC tumors. CNV at 1p36 was the only region significantly associated with expression of mRNA after adjustment for SCNA (Figure 3B). Eleven percent (N=42) of TCGA cases had germline deletions at the 1p36 CNVR and tumor expression in these subjects was significantly downregulated for cyclin dependent kinase 11A (CDK11A) compared to non-carriers (fold-change [FC] = −1.8, P = 2.68 × 10−7).
Figure 3. Correlation of risk-associated CNVR with mRNA expression in 382 primary ovarian tumors.
Linear regression was used to estimate associations of both germline copy number variation (CNV) and somatic copy number alterations (SCNA) in the tumor genome with gene expression in tumor tissue. Figures show (A) barplots for frequency of CNV and SCNA by type of alteration (deletion vs. duplication) and boxplots for expression of genes that were (B) associated with CNV at 1p36 and C) most correlated with SCNA at six risk-associated CNVR.
SCNA at the risk CNVRs generally spanned large segments of chromosomal bands but only a subset (44 out of 110) of amplified/deleted genes correlated (P<0.05 and FC>1.5) with altered gene expression (Supplemental Table 8 and Figure 3C). Four regions (1p13, 2q34, 19q13.2, and 19q13.42) exhibited both deletions and duplications in cancer genomes while 8p21.2 had only deletions and 12p11.21 had only duplications. Across all characterized genes, the most statistically significant association was between tumor copy number at 19q13.2 and egl-9 family hypoxia inducible factor 2 (EGLN2) expression (FCDel = 1.7, P = 2.94×10−47); twenty other genes were also correlated with copy number at this region. Notably, deletion of CYP2A7, the location of the risk-associated CNVR, was not associated with CYP2A7 or CYP2A6 expression (P=0.09 and 0.08, respectively). Based on a public catalogue of enhancers, the CYP2A7 CNVR overlaps an enhancer region in normal ovarian tissue predicted to affect the expression of EGLN2 (27). Somatic deletion of the enhancer region co-occurred with deletion of EGLN2 in all SCNA carriers except one. SCNA at the 19q13.42 region was correlated with 12 genes and most significantly with pre-mRNA processing factor 31 (PRPF31), a component of spliceosome complex, and TCF3 fusion partner (TFPT) which were significantly overexpressed when duplicated (FCDup = 2.0, P = 1.21×10−30 and FCDup = 2.5, P = 7.1×10−27, respectively). Expression of the immunoglobulin superfamily of genes clustered at this region that include leukocyte immunoglobulin-like receptors (LIR) and killer cell inhibitory receptors (KIR) were not associated with copy number (P>0.05). The intergenic 21kb CNVR at 1p13.3 was somatically altered in 16% of tumors and associated with expression of four genes. Tumors with somatic deletions averaged approximately 2-fold lower expression for choline/ethanolamine phosphotransferase 1 (CEPT1; FCDel = −2.02, P = 9.58×10−22), DNA damage regulated autophagy modulator 2 (DRAM2; FCDel = 1.9, P = 1.18×10−13), and DENN domain containing 2D (DENN2D; FCDel = 2.1, P = 2.75×10−11). Risk-associated deletions at 2q34 were located within the first intron of ERBB4 whose entire sequence spans >1MB in length. No other mRNAs are located within 500kb of the CNVR. ERBB4 somatic deletions were associated with a four-fold decrease in ERBB4 expression (FCDel = 4.2, P = 4.67×10−5).
SCNA at 8p21.2 and 12p11.21 were also common but showed specificity for one type of alteration. Amplifications at the 12p11 CNVR occurred in 17% of tumors and were associated with higher expression levels for four genes including two guanine exchange factors (GEFs), FYVE, RhoGEF and PH domain containing 4 (FGD4; FCDup = 1.5, P = 3.0×10−8) and DENN domain containing 5B (DENND5; FCDup = 1.7, P = 1.0×10−3), that display highest expression in ovarian tissue (28). Only two tumors contained deletions within the 12p11.21 CNVR. The 8p21.2 region was deleted more frequently (23%) than all other risk regions but duplications rarely occurred (N=4). Deletions at 8p21.2 were associated with downregulation of gonadotropin releasing hormone (GNRH1; FCDel = −1.7, P = 5.86×10−5) which is located 175kb from the germline CNVR. The 8p21.2 germline deletion region spans both the TSS of DOCK5 and upstream histone modifications consistent with an enhancer element in ovarian tissue (27).
Discussion
Copy number variation is a major source of human genetic variation that contributes as much to inter-individual differences as the more frequently studied SNP (4). Here, we describe a large genome-wide association study of CNV with EOC risk that used a comprehensive dual array design and supplemented with in silico functional follow-up. Two SNP array datasets provided complementary strengths; the 610k array set contributed discovery power with its large sample size while the 2.5M set provided considerably higher resolution. Accordingly, we identified six relatively large CNV regions associated with EOC or HGSOC risk (P<0.01) within the 610k array set and two smaller regions within the 2.5M set. In addition to limited power, the fewer detected differences and lack of replication with the 2.5M set may be due to the low frequency of variants, chance and sampling variation in the populations, and differential platform/probe CNV calling performance; it is probably a combination of these factors. By requiring CNVRs to be called by both platforms, our findings more likely reflect true variation rather than technical artifact though type I error remains possible. Thus, we further detected and functionally characterized risk-associated CNVRs through analysis of TCGA data. The integration of both germline and somatic copy number with tumor transcription revealed associations that provided insight into the potential biological consequence of genomic copy number.
A large deletion at 1p36.33 was the only CNV independently associated with tumor transcription. Carriers were estimated to have an approximately 70% lower risk of EOC (P = 0.001) and corresponding analysis of tumor tissue showed lower expression of the cyclin-dependent kinase (CDK) CDK11A in carriers. CDK11 has three isoforms involved in cell cycle control (p58), transcriptional regulation (p110), and apoptotic signaling (p46) (29). CDK11-p58 is a centrosome-associated kinase expressed during the G2 to M transition and inhibition induces cell cycle arrest and apoptosis (30) while CDK11-p110 positively regulates hedgehog signaling and the Wnt/β-catenin signaling cascade (29). Accordingly, CDK overexpression is a common feature of many cancer types and in vitro and in vivo CDK11A/B knockdown induces apoptosis in EOC cells (31). It is therefore plausible that the reduced risk of EOC observed for 1p36.33 deletion carriers is conferred through reduced CDK11-associated oncogenic signaling. Potentially complicating this theory, this CNVR was notably the only region that remained diploid in all HGSOC tumors. CDK11-p58 promotes degradation of several steroid receptors such as androgen (32), vitamin D (33), and estrogen receptors (34) which inhibits migration and invasion of ERα-positive breast cancer cells (35). A similar suppressive role in progression of EOC may explain the lack of somatic amplification at 1p36.33.
The increased risk associated with a deletion at 19q13.2 containing CYP2A7 (OR = 1.90) was the only finding that remained significant after adjustment for multiple hypothesis testing (Bonferroni corrected P = 0.02, false discovery rate=0.02). This same deletion region was recently identified in association with lower ovarian cancer risk among 2,500 BRCA1 mutation carriers (CIMBA RR = 0.50, P = 0.007) (10). CYP2A7 is a pseudogene largely expressed in the liver where it promotes expression of CYP2A6 (36) involved in the metabolism of nicotine and the tobacco-related procarcinogen nitrosamine (37). Genetic variation of CYP2A6, including a deletion, has been linked to a poor metabolizer phenotype and reduced risk of lung cancer in smokers (38). While altered enzymatic activity may similarly explain the reduced risk observed in BRCA1 carriers, the current study suggests a more complex relationship. Our in silico analyses identified EGLN2, an enzyme (aka PHD1) involved in cellular response to hypoxia (39), as the mRNA most significantly associated with CYP2A7 SCNA (P = 1.17×10−49) but CYP2A7 expression was not (P = 0.09). These data support regulation of EGLN2 by an enhancer element at CYP2A7 (27). Numerous other genes were also associated with SCNA, such as melanoma inhibitory activity (MIA), which was upregulated in polyps of a germline CYP2A7 deletion carrier in a recent study of familial adenomatous polyposis (40). MIA is a novel class of secreted proteins that interact with the extracellular matrix to promote the development, invasiveness, and metastases in melanoma as well as in pancreatic and gastric carcinomas (41, 42). Thus, germline CYP2A7 deletions could have a role in promoting tumorigenesis and progression through epigenetic regulation of cis-genes such as EGLN2 and MIA and this role may act secondarily to a separate, distinct role in metabolism that may be beneficial for BRCA1 carriers. Altogether, the consistent detection of a CYP2A7 locus deletion and its association with EOC risk warrants further investigation.
We identified five other CNVRs at nominal statistical significance but functional characterization of SCNA identified biological pathways pertinent to EOC risk. Of particular interest was the reduced EOC risk (OR = 0.52) associated with deletions at 8p21.2 where somatic deletions corresponded with lower expression of gonadotropin releasing hormone (GnRH). GnRH induces pituitary synthesis and secretion of follicle stimulating hormone (FSH) and luteinizing hormone (LH), both of which are hypothesized to have an etiologic role in EOC (43). Though we did not observe an effect of germline CNV on tumor expression of GnRH, it is tempting to hypothesize that deletions in this region may reduce systemic GnRH and thus mediate EOC risk associated with FSH/LH ‘excessive stimulation’. We also observed frequent (23%) deletion of 8p21.2 in TCGA tumors yet rare occurrence of amplifications (<1%) which is consistent with previous studies reporting common 8p21.2-p21.3 deletion and loss of heterozygosity in ovarian tumors, particularly for serous histology and high-grade and chemoresistant disease (44–47). Published analyses of TCGA data identified 8p21.2 as one of the 40 most common focal deletions in ovarian cancer genomes and the deletions correlated with GnRH expression (48). This indication of 8p21 as a tumor suppressor gene locus coincides with strong evidence that the extrapituitary, autocrine function of GnRH – involved in follicular development in the ovary (49) - counteracts growth factor receptor signaling and exerts antiproliferative and antimotility effects in ovarian and other tumors (50). Our group previously observed SNPs within GnRH that exhibited gene-level associations with increased HGSOC risk (51); we now report the first indication of an association between HGSOC risk and germline CNV at this region.
Other CNV included deletion of ERBB4 (HER4), a receptor tyrosine kinase in the epidermal growth factor receptor family (e.g. EGFR, HER2) that is commonly mutated and highly expressed in many solid tumors including ovarian (52–54) where it portends chemotherapy resistance and poor survival (55–57). Consistent with these findings, ERBB4 deletions associated with decreased EOC risk (OR=0.33). Although germline deletions were intronic and their consequence on gene function is unknown, intronic SNPs in ERBB4 affect its expression (58) and intronic CNV may also demonstrate this capability. SCNA at several risk CNVRs (1p13.3, 12p11.21, and 19q13.42) had multiple genic associations in pathways relevant to ovarian carcinogenesis including choline metabolism (CEPT1-1p13) (59), regulation of autophagy and apoptosis (DRAM2-1p13, TFPT-19q13.2) (60–62), and activation of cellular motility/migration in tumorigenesis (FGD4-12p11.21) (63). Interestingly, somatic duplications at 19q13.42 were associated with expression of PRPF3 which has been previously associated with early HGSOC relapse (64). While these transcriptome correlations are suggestive of tumor progression mechanisms, their implication in EOC risk is uncertain.
Though our study has by far the largest sample size to explore disease-associated CNV (7), analyses of GWAS for CNV suffer from reduced statistical power due to the rarity of CNV compared to SNPs and the statistically challenging detection of CNV from SNP arrays (65). Low frequency CNVRs (<5%) represented the majority (~82%) of variation identified in this study and this distribution has also been observed in a study of over 190,000 European adults where 92.4% of the CNVs were present in <1 in 1000 samples and 99.4% of them occurred with <1% frequency (65). While a meta-analysis of the two array sets may improve statistical power, we opted for a stratified analysis with comparative evaluations given the large discrepancy in probe coverage and CNV detection between arrays. High false negative and false positive CNV calls can also limit statistical power. Though multiple detection algorithms are often used to increase sensitivity, we opted to use PennCNV alone which called ~90% of all variants detected using four algorithms in a previous study (10). We controlled for false positives at multiple stages in our analytic pipeline, including stringent QC control of logRatio, BAF, and sample outliers which excluded approximately 25% of samples. Future studies should include technical validation of CNV such as quantitative PCR and this may also allow more permissive QC criterion to be used to increase sample size. Considering these limitations, we reported EOC risk associations that reached a P<0.01 threshold and did not adjust for multiple testing. As a discovery study, it was preferential to reduce type II error – and avoid missing possibly important findings – at the expense of increased type I error.
In summary, this large genome-wide study identified common CNV events in genomic regions that frequently undergo somatic alterations in ovarian tumors to promote progression. The risk associations together with in silico functional analyses highlight several novel genomic regions with biologically plausible mechanisms for EOC predisposition and pathogenesis. Replication of the findings in a larger study population profiled on the same platform is warranted. Since the initiation of this study, SNP array data from the Oncoarray (3) have become available and presents opportunity for future CNV studies.
Supplementary Material
Acknowledgements
We thank all of the women who participated along with all of the researchers, clinicians, and staff who have contributed to the participating studies.
Funding:
This work was supported by National Institutes of Health R01 CA114343 and U19 CA148112 to T. A. Sellers, R01 CA122443 to E. L. Goode, R01 CA76016 to J. M. Schildkraut, R01 CA106414 to R. Sutphen, P30 CA15083 for the Mayo Clinic Genotyping Shared Resource, and Mayo Clinic Ovarian Cancer SPORE grant P50 CA136393.
Footnotes
Conflict of Interest: The authors declare no potential conflicts of interest.
References
- 1.American Cancer Society. Cancer facts & figures 2018. Atlanta: American Cancer Society; 2018. [Google Scholar]
- 2.Jones MR, Kamara D, Karlan BY, Pharoah PDP, Gayther SA. Genetic epidemiology of ovarian cancer and prospects for polygenic risk prediction. Gynecologic oncology. 2017;147:705–13. [DOI] [PubMed] [Google Scholar]
- 3.Phelan CM, Kuchenbaecker KB, Tyrer JP, Kar SP, Lawrenson K, Winham SJ, et al. Identification of 12 new susceptibility loci for different histotypes of epithelial ovarian cancer. Nature genetics. 2017;49:680–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacDonald JR, Ziman R, Yuen RKC, Feuk L, Scherer SW. The database of genomic variants: A curated collection of structural variation in the human genome. Nucleic acids research. 2014;42:D986–D92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nature reviews Genetics. 2009;10:551–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krepischi AC, Pearson PL, Rosenberg C. Germline copy number variations and cancer predisposition. Future oncology. 2012;8:441–50. [DOI] [PubMed] [Google Scholar]
- 8.Fridley BL, Chalise P, Tsai YY, Sun Z, Vierkant RA, Larson MC, et al. Germline copy number variation and ovarian cancer survival. Front Genet 2012;3:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuusisto KM, Akinrinade O, Vihinen M, Kankuri-Tammilehto M, Laasanen SL, Schleutker J. Copy number variation analysis in familial brca1/2-negative finnish breast and ovarian cancer. PloS one. 2013;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walker LC, Marquart L, Pearson JF, Wiggins GAR, O’Mara TA, Parsons MT, et al. Evaluation of copy-number variants as modifiers of breast and ovarian cancer risk for brca1 pathogenic variant carriers. European Journal of Human Genetics. 2017;25:432–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshihara K, Tajima A, Adachi S, Quan J, Sekine M, Kase H, et al. Germline copy number variations in brca1-associated ovarian cancer patients. Genes, chromosomes & cancer. 2011;50:167–77. [DOI] [PubMed] [Google Scholar]
- 12.Kanchi KL, Johnson KJ, Lu C, McLellan MD, Leiserson MDM, Wendl MC, et al. Integrated analysis of germline and somatic variants in ovarian cancer. Nature communications. 2014;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paschou P, Drineas P, Lewis J, Nievergelt CM, Nickerson DA, Smith JD, et al. Tracing sub-structure in the european american population with pca-informative markers. PLoS genetics. 2008;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Permuth-Wey J, Chen YA, Tsai YY, Chen ZH, Qu XT, Lancaster JM, et al. Inherited variants in mitochondrial biogenesis genes may influence epithelial ovarian cancer risk. Cancer Epidem Biomar. 2011;20:1131–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pharoah PD, Tsai YY, Ramus SJ, Phelan CM, Goode EL, Lawrenson K, et al. Gwas meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nature genetics. 2013;45:362–70, 70e1–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang K, Li MY, Hadley D, Liu R, Glessner J, Grant SFA, et al. Penncnv: An integrated hidden markov model designed for high-resolution copy number variation detection in whole-genome snp genotyping data. Genome research. 2007;17:1665–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diskin SJ, Li MY, Hou CP, Yang SZ, Glessner J, Hakonarson H, et al. Adjustment of genomic waves in signal intensities from whole-genome snp genotyping platforms. Nucleic acids research. 2008;36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim JH, Hu HJ, Yim SH, Bae JS, Kim SY, Chung YJ. Cnvruler: A copy number variation-based case-control association analysis tool. Bioinformatics. 2012;28:1790–2. [DOI] [PubMed] [Google Scholar]
- 19.Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu FL, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glessner JT, Li J, Hakonarson H. Parsecnv integrative copy number variation association software with quality tracking. Nucleic acids research. 2013;41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zarrei M, MacDonald JR, Merico D, Scherer SW. A copy number variation map of the human genome. Nature reviews Genetics. 2015;16:172–83. [DOI] [PubMed] [Google Scholar]
- 22.Schwienbacher C, De Grandi A, Fuchsberger C, Facheris MF, Svaldi M, Wjst M, et al. Copy number variation and association over t-cell receptor genes--influence of DNA source. Immunogenetics. 2010;62:561–7. [DOI] [PubMed] [Google Scholar]
- 23.Tomlinson IM, Cook GP, Carter NP, Elaswarapu R, Smith S, Walter G, et al. Human immunoglobulin vh and d segments on chromosomes 15q11.2 and 16p11.2. Human molecular genetics. 1994;3:853–60. [DOI] [PubMed] [Google Scholar]
- 24.Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li QY, Seo JH, Stranger B, McKenna A, Pe’er I, LaFramboise T, et al. Integrative eqtl-based analyses reveal the biology of breast cancer risk loci. Cell. 2013;152:633–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, et al. Pan-cancer patterns of somatic copy number alteration. Nature genetics. 2013;45:1134–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. [DOI] [PubMed] [Google Scholar]
- 29.Zhou Y, Shen JK, Hornicek FJ, Kan Q, Duan Z. The emerging roles and therapeutic potential of cyclin-dependent kinase 11 (cdk11) in human cancer. Oncotarget. 2016;7:40846–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petretti C, Savoian M, Montembault E, Glover DM, Prigent C, Giet R. The pitslre/cdk11(p58) protein kinase promotes centrosome maturation and bipolar spindle formation. Embo Rep 2006;7:418–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu XZ, Gao Y, Shen JS, Yang W, Choy E, Mankin H, et al. Cyclin-dependent kinase 11 (cdk11) is required for ovarian cancer cell growth in vitro and in vivo, and its inhibition causes apoptosis and sensitizes cells to paclitaxel. Mol Cancer Ther 2016;15:1691–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zong HL, Chi YY, Wang YL, Yang YZ, Zhang L, Jiang JH, et al. Cyclin d3/cdk11(p58) complex is involved in the repression of androgen receptor. Mol Cell Biol 2007;27:7125–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chi YY, Hong Y, Zong HL, Wang YL, Zou WY, Yang JW, et al. Cdk11(p58) represses vitamin d receptor-mediated transcriptional activation through promoting its ubiquitin-proteasome degradation. Biochem Bioph Res Co 2009;386:493–8. [DOI] [PubMed] [Google Scholar]
- 34.Wang YL, Zong HL, Chi YY, Hong Y, Yang YZ, Zou WY, et al. Repression of estrogen receptor alpha by cdk11(p58) through promoting its ubiquitinproteasome degradation. J Biochem 2009;145:331–43. [DOI] [PubMed] [Google Scholar]
- 35.Chi Y, Huang S, Wang L, Zhou R, Wang L, Xiao X, et al. Cdk11p58 inhibits eralpha-positive breast cancer invasion by targeting integrin beta3 via the repression of eralpha signaling. Bmc Cancer. 2014;14:577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakano M, Fukushima Y, Yokota S, Fukami T, Takamiya M, Aoki Y, et al. Cyp2a7 pseudogene transcript affects cyp2a6 expression in human liver by acting as a decoy for mir-126(star). Drug Metab Dispos. 2015;43:703–12. [DOI] [PubMed] [Google Scholar]
- 37.Rendic S. Summary of information on human cyp enzymes: Human p450 metabolism data. Drug Metab Rev 2002;34:83–448. [DOI] [PubMed] [Google Scholar]
- 38.Liu YL, Xu Y, Li F, Chen H, Guo SL. Cyp2a6 deletion polymorphism is associated with decreased susceptibility of lung cancer in asian smokers: A meta-analysis. Tumor Biol 2013;34:2651–7. [DOI] [PubMed] [Google Scholar]
- 39.Ortmann B, Bensaddek D, Carvalhal S, Moser SC, Mudie S, Griffis ER, et al. Cdk-dependent phosphorylation of phd1 on serine 130 alters its substrate preference in cells. J Cell Sci 2016;129:191–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thean LF, Wong YH, Lo M, Loi C, Chew MH, Tang CL, et al. Chromosome 19q13 disruption alters expressions of cyp2a7, mia and mia-rab4b lncrna and contributes to fap-like phenotype in apc mutation-negative familial colorectal cancer patients. PloS one. 2017;12:e0173772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riechers A, Bosserhoff AK. Melanoma inhibitory activity in melanoma diagnostics and therapy - a small protein is looming large. Experimental dermatology. 2014;23:12–4. [DOI] [PubMed] [Google Scholar]
- 42.El Fitori J, Kleeff J, Giese NA, Guweidhi A, Bosserhoff AK, Buchler MW, et al. Melanoma inhibitory activity (mia) increases the invasiveness of pancreatic cancer cells. Cancer cell international. 2005;5:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi JH, Wong AS, Huang HF, Leung PC. Gonadotropins and ovarian cancer. Endocrine reviews. 2007;28:440–61. [DOI] [PubMed] [Google Scholar]
- 44.Engler DA, Gupta S, Growdon WB, Drapkin RI, Nitta M, Sergent PA, et al. Genome wide DNA copy number analysis of serous type ovarian carcinomas identifies genetic markers predictive of clinical outcome. PloS one. 2012;7:e30996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim SW, Kim JW, Kim YT, Kim JH, Kim S, Yoon BS, et al. Analysis of chromosomal changes in serous ovarian carcinoma using high-resolution array comparative genomic hybridization: Potential predictive markers of chemoresistant disease. Gene Chromosome Canc 2007;46:1–9. [DOI] [PubMed] [Google Scholar]
- 46.Dimova I, Orsetti B, Negre V, Rouge C, Ursule L, Lasorsa L, et al. Genomic markers for ovarian cancer at chromosomes 1, 8 and 17 revealed by array cgh analysis. Tumori. 2009;95:357–66. [DOI] [PubMed] [Google Scholar]
- 47.Brown MR, Chuaqui R, Vocke CD, Berchuck A, Middleton LP, Emmert-Buck MR, et al. Allelic loss on chromosome arm 8p: Analysis of sporadic epithelial ovarian tumors. Gynecologic oncology. 1999;74:98–102. [DOI] [PubMed] [Google Scholar]
- 48.Broad Institute TCGA Genome Data Analysis Center. Snp6 copy number analysis (gistic2). Broad Institute of MIT and Harvard; 2016. [Google Scholar]
- 49.Maggi R, Cariboni AM, Marelli MM, Moretti RM, Andre V, Marzagalli M, et al. Gnrh and gnrh receptors in the pathophysiology of the human female reproductive system. Human reproduction update. 2016;22. [DOI] [PubMed] [Google Scholar]
- 50.Grundker C, Emons G. The role of gonadotropin-releasing hormone in cancer cell proliferation and metastasis. Frontiers in endocrinology. 2017;8:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee AW, Tyrer JP, Doherty JA, Stram DA, Kupryjanczyk J, Dansonka-Mieszkowska A, et al. Evaluating the ovarian cancer gonadotropin hypothesis: A candidate gene study. Gynecologic oncology. 2015;136:542–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prickett TD, Agrawal NS, Wei X, Yates KE, Lin JC, Wunderlich JR, et al. Analysis of the tyrosine kinome in melanoma reveals recurrent mutations in erbb4. Nature genetics. 2009;41:1127–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Soung YH, Lee JW, Kim SY, Wang YP, Jo KH, Moon SW, et al. Somatic mutations of the erbb4 kinase domain in human cancers. Int J Cancer. 2006;118:1426–9. [DOI] [PubMed] [Google Scholar]
- 54.Davies S, Holmes A, Lomo L, Steinkamp MP, Kang H, Muller CY, et al. High incidence of erbb3, erbb4, and met expression in ovarian cancer. International journal of gynecological pathology : official journal of the International Society of Gynecological Pathologists. 2014;33:402–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim JY, Jung HH, Do IG, Bae S, Lee SK, Kim SW, et al. Prognostic value of erbb4 expression in patients with triple negative breast cancer. Bmc Cancer. 2016;16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saglam O, Xiong Y, Marchion DC, Strosberg C, Wenham RM, Johnson JJ, et al. Erbb4 expression in ovarian serous carcinoma resistant to platinum-based therapy. Cancer control : journal of the Moffitt Cancer Center. 2017;24:89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paatero I, Lassus H, Junttila TT, Kaskinen M, Butzow R, Elenius K. Cyt-1 isoform of erbb4 is an independent prognostic factor in serous ovarian cancer and selectively promotes ovarian cancer cell growth in vitro. Gynecologic oncology. 2013;129:179–87. [DOI] [PubMed] [Google Scholar]
- 58.Law AJ, Kleinman JE, Weinberger DR, Weickert CS. Disease-associated intronic variants in the erbb4 gene are related to altered erbb4 splice-variant expression in the brain in schizophrenia. Human molecular genetics. 2007;16:129–41. [DOI] [PubMed] [Google Scholar]
- 59.Glunde K, Bhujwalla ZM, Ronen SM. Choline metabolism in malignant transformation. Nature reviews Cancer. 2011;11:835–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoon JH, Her S, Kim M, Jang IS, Park J. The expression of damage-regulated autophagy modulator 2 (dram2) contributes to autophagy induction. Molecular biology reports. 2012;39:1087–93. [DOI] [PubMed] [Google Scholar]
- 61.Park SM, Kim K, Lee EJ, Kim BK, Lee TJ, Seo T, et al. Reduced expression of dram2/tmem77 in tumor cells interferes with cell death. Biochem Biophys Res Commun 2009;390:1340–4. [DOI] [PubMed] [Google Scholar]
- 62.Franchini C, Fontana F, Minuzzo M, Babbio F, Privitera E. Apoptosis promoted by up-regulation of tfpt (tcf3 fusion partner) appears p53 independent, cell type restricted and cell density influenced. Apoptosis : an international journal on programmed cell death. 2006;11:2217–24. [DOI] [PubMed] [Google Scholar]
- 63.Liu HP, Chen CC, Wu CC, Huang YC, Liu SC, Liang Y, et al. Epstein-barr virus-encoded lmp1 interacts with fgd4 to activate cdc42 and thereby promote migration of nasopharyngeal carcinoma cells. PLoS pathogens. 2012;8:e1002690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hartmann LC, Lu KH, Linette GP, Cliby WA, Kalli KR, Gershenson D, et al. Gene expression profiles predict early relapse in ovarian cancer after platinum-paclitaxel chemotherapy. Clin Cancer Res 2005;11:2149–55. [DOI] [PubMed] [Google Scholar]
- 65.Mace A, Tuke MA, Deelen P, Kristiansson K, Mattsson H, Noukas M, et al. Cnv-association meta-analysis in 191,161 european adults reveals new loci associated with anthropometric traits. Nature communications. 2017;8:744. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.