Toxoplasma gondii cysts reactivate during immune deficiency and cause fatal encephalitis. Parasite molecules that coordinate the development of acute and chronic infection are poorly characterized. Here, we show that many intravacuolar network membrane and parasitophorous vacuole membrane-associated dense granule (GRA) proteins orchestrate the development of chronic cysts in vivo. A subset of these GRA proteins also modulate acute virulence, and one protein that associates with the intravacuolar network membranes, namely GRA12, was identified as a major virulence factor required for parasite resistance to host gamma interferon (IFN-γ). Our results revealed that many parasitophorous vacuole membrane and intravacuolar network membrane-associated GRA proteins are essential for successful chronic infection.
KEYWORDS: Toxoplasma gondii, chronic infection, dense granule, innate immunity, membrane proteins, virulence factors
ABSTRACT
Toxoplasma gondii evades host immunity to establish a chronic infection. Here, we assessed the role of parasitophorous vacuole (PV) membrane (PVM)- and intravacuolar network (IVN) membrane-localized dense granule (GRA) proteins in the development of acute and chronic Toxoplasma infection. Deletion of PVM-associated GRA3, GRA7, GRA8, and GRA14 or IVN membrane-associated GRA2, GRA9, and GRA12 in the low-virulence type II Prugniaud (Pru) strain induced severe defects in the development of chronic-stage cysts in vivo without affecting the parasite growth rate or the ability to differentiate into cysts in vitro. Acute virulence of the PruΔgra2, PruΔgra3, and PruΔgra4 mutants was reduced but not abolished. In contrast, the PruΔgra12 mutant was avirulent in mice and PruΔgra12 parasites failed to establish a chronic infection. High-virulence type I strain RHΔgra12 parasites also exhibited a major defect in acute virulence. In gamma interferon (IFN-γ)-activated macrophages, type I RHΔgra12 and type II PruΔgra12 parasites resisted the coating of the PVM with host immunity-related GTPases as effectively as the parental type I RHΔku80 and type II PruΔku80 strains, respectively. Despite this resistance, Δgra12 PVs ultimately succumbed to IFN-γ-activated host cell innate immunity. Our findings uncover a key role for GRA12 in mediating resistance to host IFN-γ and reveal that many other IVN membrane-associated GRA proteins, as well as PVM-localized GRA proteins, play important roles in establishing chronic infection.
INTRODUCTION
A third of the world’s human population is currently infected by Toxoplasma gondii (1). Toxoplasma infection is acquired by incidental consumption of oocysts shed from cats in contaminated water or vegetables or by ingestion of tissue cysts present in undercooked infected meat (2). While immunocompetent hosts typically control the infection, Toxoplasma can cause severe pathology in the eye or in the developing fetus (3), particularly when the infection is triggered by a virulent strain (4). Three major strain types (type I, type II, and type III) of Toxoplasma predominate in North America and Europe and exhibit different virulence profiles in laboratory strains of mice: type I strains are highly virulent, type II strains exhibit low virulence, and type III strains are essentially avirulent (5).
Toxoplasma invades host cells and replicates within a parasitophorous vacuole (PV). Following dissemination and replication in endothelial cells at the blood brain barrier, parasites penetrate into the central nervous system (6). Primarily within neurons, tachyzoites differentiate into quiescent bradyzoites inside a modified PV termed the cyst (7, 8), which is delimited by a thick cyst wall (7). Persistence of these cysts in immune-privileged organs (9) characterizes a successful chronic infection (10, 11). If host immunity wanes, cyst reactivation causes toxoplasmic encephalitis (12, 13). The biology of cyst formation and chronic infection remains poorly understood.
Toxoplasma enters its host cells through an active motility-based invasion mechanism. Invasion and PV formation require temporally coordinated secretion of specialized parasite proteins from Apicomplexa-specific organelles, namely, the micronemes and the rhoptries (14). Soon after PV formation, the parasite secretes the contents of the dense granules into the nascent PV (15). The secreted dense granule (GRA and GRA-like) proteins then either remain soluble within the PV or traffic to the intravacuolar network (IVN) membranes, to the limiting PV membrane (PVM), to the host cell cytoplasm, or to the host cell nucleus (16).
GRA3, GRA7, GRA8, GRA14, and GRA15 are PVM associated (16). GRA3 is a type I PVM-associated transmembrane protein (17). GRA7 associates with ROP2 and ROP4 (18) and functions in concert (19, 20) with ROP18 protein complexes that resist gamma interferon (IFN-γ)-activated host immunity-related (p47) guanosine triphosphatases (IRGs) (21–33). ROP18 also resists host IFN-γ-activated guanylate binding proteins (GBPs) (34, 35). Recombinant GRA7 signals tumor necrosis factor (TNF) receptor-associated factor 6 (TRAF6) to activate mitogen-activated protein kinases in macrophages (36). GRA7 acts as a garroting protein that sequesters host endolysosomes within the PV (37). While GRA7 deforms liposomes into tubular membranes in vitro (37), Δgra7 parasites induce a hyperformation of the IVN membranes (38). The role of GRA8 at the PVM is currently unknown, but the protein was recently shown to play an independent role in organization of the subpellicular cytoskeleton and motility (39). GRA14 localizes to membranous extensions (PV extensions [PVE]) that radiate from the PVM into the host cell cytoplasm (40). GRA14 can also be exchanged between different PVs through PVE (40). GRA15 activates the host nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κβ) pathway to drive classical activation of macrophages (41–43).
The membranes of the IVN, previously dubbed the membranous nanotubular network (MNN) (16), form a highly curved tubulovesicular network of elongated nanotubules that connect the intravacuolar tachyzoites to each other and to the PVM (44, 45). F-actin filaments of 5 nm in diameter and over 100 nm in length were recently shown to form within the membranous nanotubules of 50 to 60 nm in diameter that likely constitute the IVN (46). These F-actin filaments were shown to regulate the architecture of the IVN, the formation of the residual body, parasite organization within the PV, and the movement of vesicles between parasites within the PV (46). GRA2 (47, 48), GRA4 (49), GRA6 (50), GRA9 (51), and GRA12 (52) are IVN associated. The IVN in Δgra6 parasites consists of disconnected small vesicles instead of nanotubular membranes (38, 53, 54), while the IVN in Δgra2 parasites consists of fine aggregated material (38, 53–55). GRA2 and GRA6, through lipid binding, reshape large unilamellar vesicles into nanotubular membranes in vitro (53). GRA2 and GRA6 are thus key molecules that establish the characteristic nanotubular morphology of the IVN membranes of the PV. GRA6 was also recently identified as a type I strain-dependent activator of a host transcription factor, nuclear factor of activated T cells 4 (NFAT4) (56). Type I Δgra2 parasites exhibit a slight reduction of acute virulence in mice (54, 57), altered presentation of parasite PV antigens by host major histocompatibility complex I (MHCI) (53), and decreased antitumor T cell responses in a model of Toxoplasma-induced ovarian cancer immunotherapy (58, 59).
The PV ingests host membranes to satisfy the parasites’ appetite for lipids (60). These host lipids are dynamically incorporated into the IVN membranes (61). Toxoplasma uses PV membranes to sequester and internalize host proteins, lipid droplets, membrane vesicles, and organelles through heterophagy (37, 60, 62, 63). Δgra2 PVs that lack IVN nanotubular membrane structures exhibit decreased heterophagic ingestion of host cell proteins (64), Rab-positive (Rab+) host cell vesicles (63), and Rab7-positive (Rab7+) host lipid droplets (62). Moreover, acquisition of host cell cargo by heterophagy in chronic-stage cysts is also essential for successful chronic infection (65).
We previously reported that two IVN membrane-associated GRA proteins, namely, GRA4 and GRA6, are essential for successful chronic infection (66). Several PVM and IVN-associated GRA proteins have been reported to localize to the cyst wall (7, 67, 68). Δcst2 parasites, which fail to express the cyst wall GRA protein CST2, were recently shown to have a virulence defect, and this mutant exhibited a loss of detectable cyst burdens (68). In addition, a recent gene regulatory network analysis of Toxoplasma identified a small cluster of parasite genes associated with the pathogenesis community (69). This cluster includes the GRA1, GRA2, GRA6, and GRA12 genes and is predicted to represent a master regulatory node common to all parasite life stages. While GRA2, GRA6, and GRA12 associate intimately with the IVN membranes as integral membrane proteins (16), GRA1 only associates weakly with the IVN through peripheral interactions (45).
We investigated 10 type II parasite mutants that lack expression of PVM-associated GRA proteins (GRA3, GRA7, GRA8, GRA14, and GRA15) or IVN-associated GRA proteins (GRA2, GRA4, GRA6, GRA9, and GRA12). Our results identified GRA12 as a key Toxoplasma virulence factor that resists host IFN-γ-activated innate immunity. PVM and IVN localized GRA proteins play important roles in establishing chronic infection.
RESULTS
Genetic deletion of PVM- and IVN-associated dense granule proteins.
We targeted the genetic deletion of IVN-associated GRA proteins (GRA2, GRA9, and GRA12) and several PVM-associated GRA proteins (GRA3, GRA7, GRA8, GRA10, GRA14, and GRA15) in the low-virulence type II Prugniaud (Pru)Δku80 genetic background (66). Targeted GRA gene deletion (Fig. S1A in the supplemental material) was successful at each targeted gene locus, except for GRA10 (Table S1). Absence of the targeted GRA protein was confirmed in immunofluorescence assays (IFA) (Fig. S2A).
GRA knockout and complementation strategies. (A) Knockout strategy to insert HXGPRT at deleted GRA gene loci by selection with mycophenolic acid (MPA) and xanthine (X). Double crossovers occurred at the GRA GOI locus when the PruΔku80Δhxgprt strain allowed the replacement of the targeted GRA coding sequence with that of the HXGPRT resistance marker, leading to the isolation of the PruΔku80Δgra(GOI)::HXGPRT strain. Location of the genotype validation PCRs is shown above the schematized PruΔku80Δgra(GOI)::HXGPRT genome. (B) Complementation strategy at the UPRT locus through selection with 5-fluorodeoxyuridine (FUDR). A double crossover at the UPRT locus allowed replacement of the UPRT coding sequence by the GRA12 5′ UTR followed by the GRA12 coding sequence; leading to the isolation of PruΔku80Δgra(GOI)::HXGPRT Δuprt::GRA12. Locations of the genotype validation PCRs are shown above the schematized PruΔku80Δgra(GOI)::HXGPRT genome. Download FIG S1, TIF file, 0.5 MB (493.3KB, tif) .
Copyright © 2019 Fox et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Validation of the GRA knockouts by IFA and immunoblotting. (A) Validation of GRA gene deletion in type II Δgra parasite mutant strains by IFA using specific anti-GRA antibodies. (B) Validation of GRA12 deletion in RHΔgra12 parasites by immunoblotting using rat anti-GRA12 polyclonal antibodies. (C) Validation of the GRA12 deletion in RHΔgra12 parasites by IFA using rat anti-GRA12 polyclonal antibodies. Download FIG S2, TIF file, 1.0 MB (1,022.1KB, tif) .
Copyright © 2019 Fox et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Parasite strains used or developed in this study. Download Table S1, DOC file, 0.1 MB (55KB, doc) .
Copyright © 2019 Fox et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
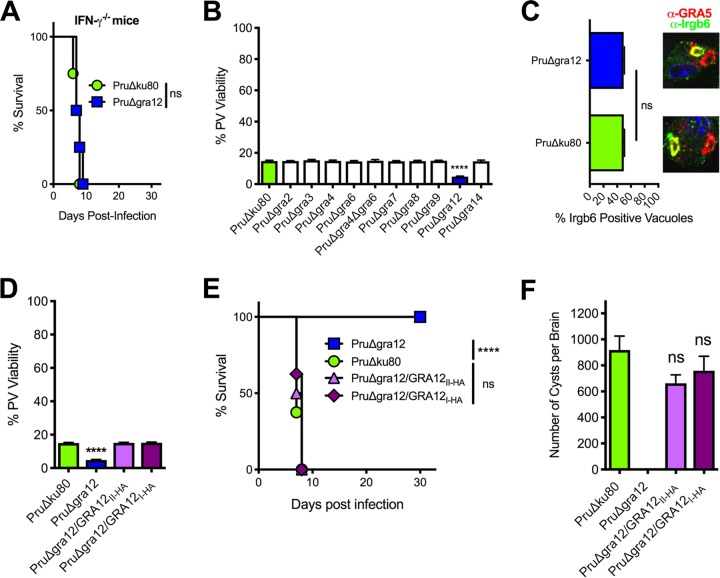
PruΔgra2, PruΔgra3, PruΔgra4, and PruΔgra12 parasites exhibit significant defects in virulence.
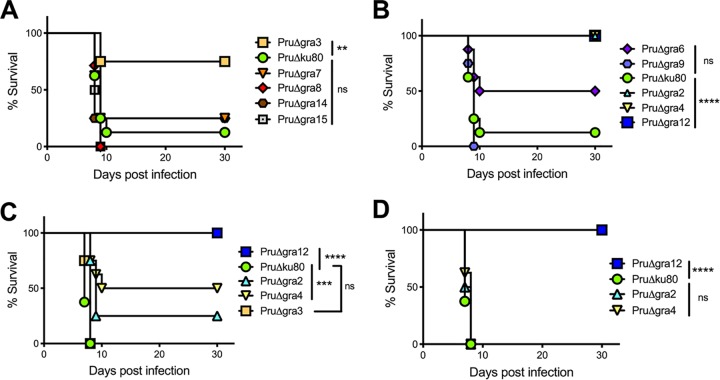
Acute-virulence phenotypes of each PruΔgra mutant were examined by challenging mice intraperitoneally (i.p.) with different parasite doses. Several parasite mutants with deletion mutations of PVM-localized (PruΔgra3) (Fig. 1A) or IVN-localized (PruΔgra2, PruΔgra4, and PruΔgra12) GRA proteins (Fig. 1B) exhibited significant virulence defects after infection with 2 × 105 tachyzoites. While PruΔgra6 parasites trended toward a reduced virulence (Fig. 1B), this difference (P = 0.09) did not achieve statistical significance. However, only PruΔgra2, PruΔgra4, and PruΔgra12 parasites exhibited significant virulence defects after infection with 2 × 106 tachyzoites, whereas PruΔgra3 parasites (Fig. 1C) and, as expected, PruΔgra6, PruΔgra7, PruΔgra8, PruΔgra9, PruΔgra14, and PruΔgra15 parasites did not exhibit any virulence defect after this challenge dose (Fig. S3B). The virulence defect of PruΔgra4 parasites was rescued by complementation with the wild-type GRA4 gene (Fig. S3C). Only PruΔgra12 parasites exhibited a significant virulence defect after infection with 2 × 107 tachyzoites (Fig. 1D). No significant defect was observed in the in vitro replication rate of any type II PruΔgra mutant (Fig. S4), which confirmed the results previously reported for both the PruΔgra4 and PruΔgra6 mutants (66). Finally, while moderate virulence defects were associated with PruΔgra2, PruΔgra3, and PruΔgra4 parasites, PruΔgra12 parasites were, surprisingly, avirulent.
FIG 1.
PruΔgra12 parasites exhibited a major defect in acute virulence. The virulence of PruΔgra parasites was evaluated in mice. All data shown are from two independent experiments with 4 mice per group. The P values were calculated by log rank Mantel-Cox test, and a P value of <0.05 was considered significant. (A) C57BL/6 mice were infected intraperitoneally (i.p.) with 2 × 105 tachyzoites of the PVM-associated GRA mutants PruΔgra3, PruΔgra7, PruΔgra8, PruΔgra14, and PruΔgra15 or the PruΔku80 parent strain. (B) C57BL/6 mice were infected i.p. with 2 × 105 tachyzoites of the IVN-associated GRA mutants PruΔgra2, PruΔgra4, PruΔgra6, PruΔgra9, and PruΔgra12 or the PruΔku80 parent strain. (C) C57BL/6 mice were infected i.p. with 2 × 106 tachyzoites of the PruΔgra2, PruΔgra3, PruΔgra4, or PruΔgra12 parasites or the PruΔku80 parental strain. (D) C57BL/6 mice were infected i.p. with 2 × 107 tachyzoites of the PruΔgra2, PruΔgra4, or PruΔgra12 parasites or the PruΔku80 parental strain. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
Virulence of Δgra parasites and rescue of Δgra4 virulence. (A) PruΔgra4 and PruΔgra6 parasites differentiate to GFP+ cysts in vitro at high pH. (B) C57BL/6 mice were infected i.p. with 2 × 106 tachyzoites of various PruΔgra parasite strains, and virulence was measured. (C) The GRA4 gene rescues PruΔgra4 virulence. ***, P < 0.005. Download FIG S3, TIF file, 0.4 MB (447.3KB, tif) .
Copyright © 2019 Fox et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Replication rates. Replication rates of PruΔgra parasites were measured in infected HFF cells in a 45-h assay (see Materials and Methods). Download FIG S4, TIF file, 0.2 MB (222.2KB, tif) .
Copyright © 2019 Fox et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
PVM- and IVN-associated dense granule proteins are essential for the development of chronic cyst burdens.
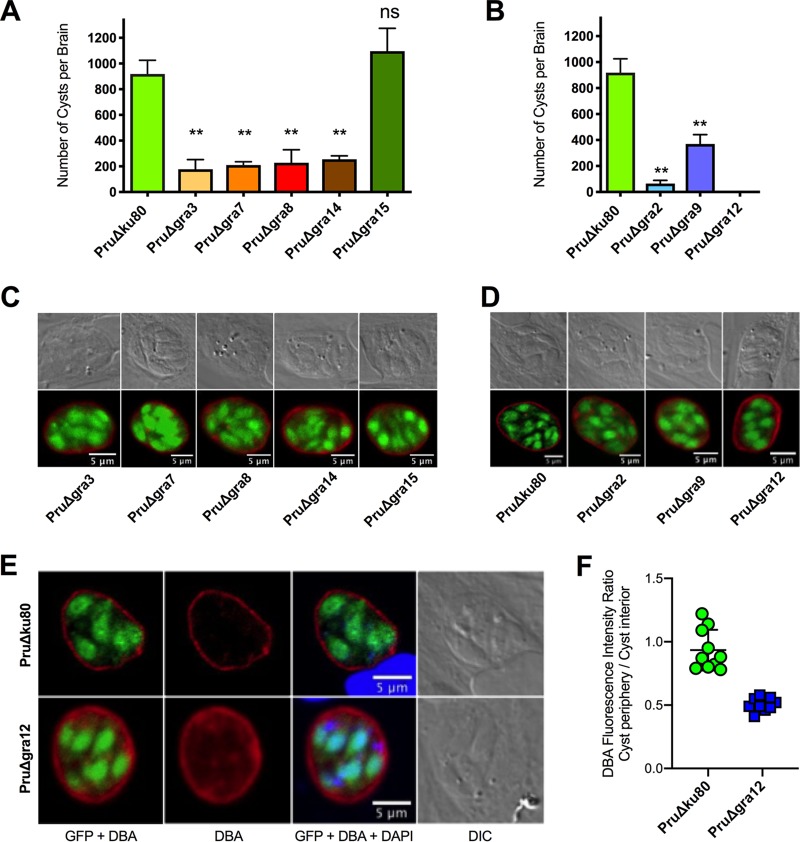
Each PruΔgra mutant was tested for its ability to establish a chronic infection in mice. Major defects in the development of brain cyst burdens were observed in the PruΔgra mutants that had abrogated expression of PVM-associated (PruΔgra3, PruΔgra7, PruΔgra8, and PruΔgra14) (Fig. 2A) or IVN-associated (PruΔgra2, PruΔgra9, and PruΔgra12) proteins (Fig. 2B). In contrast, cyst burdens were normal in PruΔgra15 parasites (Fig. 2A). Cyst burdens were reduced by more than 90% in PruΔgra2 parasite infections, similar to the >90% cyst reductions we previously reported in PruΔgra4 and PruΔgra6 parasite infections (66). In contrast, cyst burdens were reduced by ∼60% to 80% in PruΔgra3, PruΔgra7, PruΔgra8, PruΔgra9, and PruΔgra14 parasite infections. Remarkably, cysts were not detected in mice infected with PruΔgra12 parasites (Fig. 2B). Major defects in cyst burdens were associated with virulence defects in the PruΔgra2, PruΔgra3, PruΔgra4, and PruΔgra12 mutants but not in the PruΔgra6, PruΔgra7, PruΔgra8, PruΔgra9, and PruΔgra14 mutants.
FIG 2.
Cyst burdens were markedly reduced or abolished in parasites that lack expression of PVM- or IVN-associated GRA proteins. (A, B) C57BL/6 mice were infected i.p. with 200 tachyzoites of GRA knockout strains, and brain cyst burdens were counted 21 days after infection. The data presented are the cumulative results from 1 to 3 independent experiments for each strain tested and are shown as the mean values plus or minus standard errors of the means (± SEM). The P values were calculated with Student’s t test, with a P value of <0.05 considered significant. **, P < 0.001; ns, not significant. (A) PVM-associated GRA mutants PruΔgra3 (1 experiment; n = 4 mice), PruΔgra7 (1 experiment; n = 4 mice), PruΔgra8 (1 experiment; n = 4 mice), PruΔgra14 (1 experiment; n = 4 mice), and PruΔgra15 (3 experiments; n = 10 mice) and the PruΔku80 parent strain (3 experiments; n = 12 mice). (B) IVN-associated GRA mutants PruΔgra2 (1 experiment; n = 4 mice), PruΔgra9 (2 experiments; n = 5 mice), and PruΔgra12 (2 experiments; n = 8 mice) and the PruΔku80 parent strain (3 experiments; n = 12 mice). (C, D) HFF cells were infected with the designated PVM- or IVN-associated GRA knockout strain. Infected host cells were treated under bradyzoite-inducing conditions (pH 8.1 and CO2 depletion in ambient air) for 3 days. The cyst wall was stained with Dolichos biflorus agglutinin coupled to Texas red (DBA) (shown in red). Bradyzoites were visualized by their GFP expression (shown in green), which is under the control of the bradyzoite stage-specific LDH2 promoter. Samples were imaged by confocal microscopy, and PVs were located using differential interference contrast (DIC) microscopy. Representative results are shown for each strain. (E) Comparison of PruΔgra12 and PruΔku80 cysts showing images for GFP plus DBA, DBA only, GFP plus DBA plus DAPI, and DIC. Cysts shown in this panel were cysts that scored at the mean values shown in panel F. (F) DBA fluorescence intensity was measured as the ratio of DBA fluorescence intensity at the cyst periphery compared to that in the cyst interior for PruΔgra12 cysts versus parental strain PruΔku80 cysts. Each cyst measurement is shown in the graph. ****, P < 0.0001.
While reduced virulence can potentially explain reduced cyst burdens, reduced cyst burdens can also arise from a failure to build a resistant cyst wall or from a deficiency in tachyzoite-to-bradyzoite stage differentiation. Consequently, we examined the ability of the PruΔgra mutant tachyzoites to differentiate into chronic-stage bradyzoites inside a cyst wall after in vitro exposure to alkaline switch conditions. After alkaline switching, all of the PruΔgra mutants and the parental PruΔku80 strain differentiated into bradyzoites expressing green fluorescent protein (GFP) under the control of the bradyzoite-specific LDH2 promoter. These GFP-positive (GFP+) bradyzoites were visible within an apparently intact cyst wall structure revealed with fluorescently labeled Dolichos biflorus agglutinin (DBA) (Fig. 2C and D, Fig. S3A) (70).
The Golgi complex-associated aspartyl protease 5 (ASP5) is required for processing and targeting of many secreted GRA proteins. Since genetic deletion of ASP5 in type II parasites (PruΔasp5) severely impairs cyst wall development (71), we further evaluated the pattern of DBA staining in PruΔgra12 cysts. DBA almost exclusively targets and stains the major cyst wall protein CST1, which is modified by N-acetylgalactosamine residues that are specifically recognized and bound by DBA (8). Relative to the results for PruΔku80 cysts, PruΔgra12 cysts appeared to exhibit increased intensity of DBA staining inside the cyst (Fig. 2E). To quantify this observation, we randomly selected 12 PruΔgra12 and 10 PruΔku80 3-day-old in vitro cysts and measured the ratio of DBA fluorescence intensity at the cyst periphery, reflecting CST1 cargo delivered to the cyst wall, compared to the fluorescence intensity detected in the cyst interior, which highlights CST1 cargo not yet delivered to the cyst wall. In comparison to the parental PruΔku80 strain that expressed GRA12, PruΔgra12 cysts exhibited a significant decrease in the DBA fluorescence intensity ratio (cyst periphery/cyst interior) (Fig. 2F), suggesting that GRA12 could be involved in the delivery of CST1 to the cyst wall.
GRA12 localizes at the IVN.
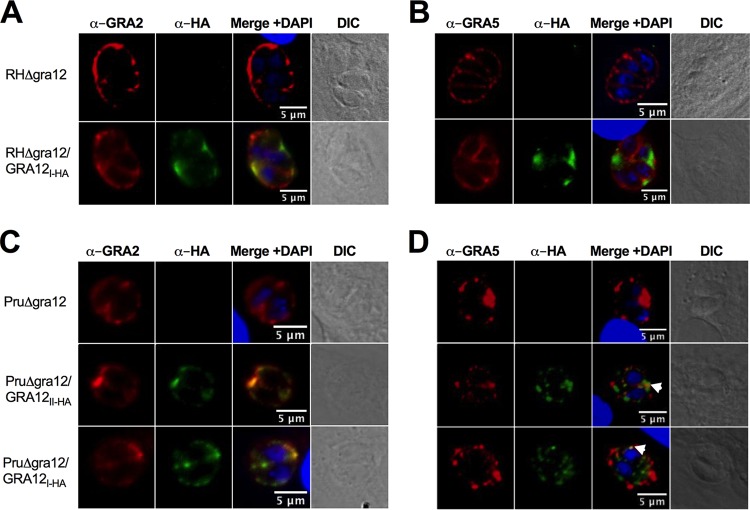
In the virulent type I RH strain, GRA12 predominantly colocalized with GRA2 at the IVN membranes (52). To confirm this GRA12 localization, we also deleted the GRA12 gene from the highly virulent type I RHΔku80 strain (Table S1, Fig. S2B and C) (72). The GRA12 protein is highly conserved between type I, type II, and type III strains (Fig. S5). The type I and type II Δgra12 mutants were complemented with the type I RH strain GRA12 gene (TGGT1_288650), thus generating the strains RHΔgra12/GRA12I-HA and PruΔgra12/GRA12I-HA, respectively, while type II PruΔgra12 parasites were complemented with the type II Pru GRA12 gene (TGME49_288650), generating the strain PruΔgra12/GRA12II-HA. All of these GRA12 complementing genes were tagged with the hemagglutinin (HA) epitope placed in-frame at the C terminus of GRA12 (Fig. S1B, Table S1) to permit the visualization of GRA12 by immunofluorescence and immunoblot experiments using antibodies specific to the HA tag. As expected, GRA12 predominantly colocalized with GRA2 and the IVN membranes in type I RH (Fig. 3A) and type II Pru strains (Fig. 3C). GRA12 also sporadically colocalized with the PVM-specific marker GRA5 (Fig. 3D; white arrowheads). Sporadic colocalization of GRA12 with the PVM-specific marker GRA3 was also previously reported (52). It is not surprising that a predominantly IVN-localized protein like GRA12 could occasionally localize with PVM-localized proteins because, within the PV, the IVN membranes have been shown to form direct connections with the PVM (45).
FIG 3.
GRA12 in the acute-stage tachyzoite PV predominantly colocalizes with IVN-associated GRA2. (A to D) HFF cells were infected for 24 h and fixed with paraformaldehyde, and the PVM was permeabilized with 0.01% saponin. Antibodies specific to the HA tag positioned at the C terminus of GRA12 were used to localize GRA12 (shown in green) in the PV relative to the IVN-associated marker GRA2 (detected by monoclonal antibodies to GRA2 and shown in red) (A, C) or the PVM-associated GRA5 (detected by monoclonal antibodies to GRA5 and shown in red) (B, D). Host cell and parasite nuclei were stained with DAPI (blue). The location of PVs in the infected host cell was confirmed by differential interference contrast (DIC) microscopy. White arrowheads indicate isolated points of colocalization of GRA12 and GRA5 near the periphery of the PV. Representative images are shown.
Amino acid alignments of GRA12 expressed by type I (GT1), type II (ME49), and type III (VEG) strains. Protein alignments and amino acid similarity and identity scores are shown for GRA12 expressed by the type I strain (TGGT_288650), type II strain ME49 (TGME49_288650), and type III strain VEG (TGVEG_288650). Download FIG S5, DOCX file, 0.1 MB (99.4KB, docx) .
Copyright © 2019 Fox et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The mature IVN forms in the absence of GRA12 expression.
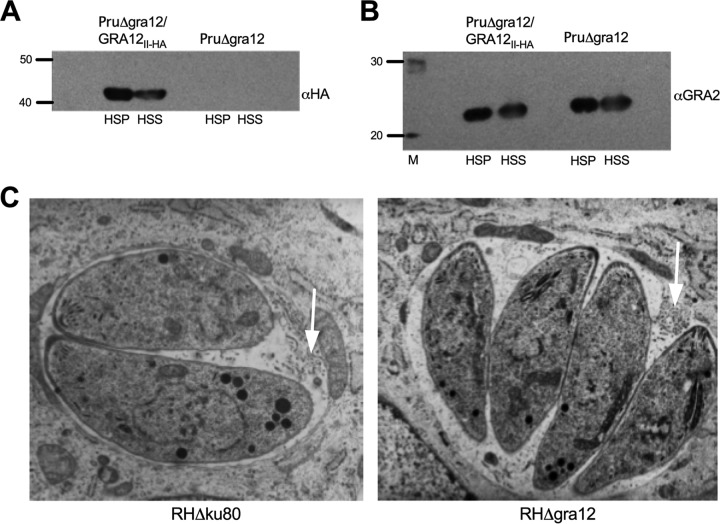
We confirmed that GRA12 is an IVN-associated protein using biochemical fractionation of human foreskin fibroblast (HFF) cells that had been infected for 24 h. Typical of IVN membrane-localized GRA proteins, GRA12 partitioned into the IVN membrane fraction (high-speed pellet [HSP]) and the vacuole soluble fraction (high-speed supernatant [HSS]) (Fig. 4A). GRA2 (38, 53–55) and GRA6 (38, 53, 54) organize the nanotubular morphology of the IVN membranes. Since GRA12 colocalizes with GRA2 and GRA6 when the IVN membranes are being organized within the PV space soon after dense granule secretion, it was previously proposed by Michelin et al. that GRA12 could also play a role in organizing the nanotubular morphology of the IVN membranes (52). To determine whether other IVN GRA proteins normally localized to the IVN in the PV in the absence of GRA12 expression, we examined the biochemical fractionation of GRA2 into the HSP (IVN membrane) and HSS (PV soluble) fractions. Fractionation of HFF cells infected by Δgra12 parasites revealed that GRA2 still partitioned normally between the HSP and HSS fractions of the PV (Fig. 4B). In addition, using transmission electron microscopy (TEM) imaging, we did not detect any significant Δgra12 tachyzoite abnormality. Furthermore, we observed typical PVs decorated with normal-looking IVN membranes in Δgra12 PVs (Fig. 4C). Collectively, these results suggested that GRA12 was not necessary for the shaping of mature IVN membranes.
FIG 4.
Intravacuolar network membranes form in the absence of GRA12 expression. (A) To confirm IVN membrane localization of GRA12, HFF cells containing mature PruΔgra12/GRA12II-HA PV or mature PruΔgra12 PV were fractionated into a high-speed soluble fraction (HSS) and a PV membrane-associated high-speed pellet (HSP) 24 h after infection. Proteins in the HSP or the HSS fractions were separated on a 10% SDS-PAGE gel under reducing conditions in parallel to a protein molecular-weight ladder (40- and 50-kDa positions are shown), transferred to nitrocellulose membranes, incubated with rabbit anti-HA tag antibodies specific to GRA12II-HA, revealed with goat anti-rabbit IgG coupled to peroxidase, and visualized by chemiluminescence. (B) To assess IVN membrane localization of GRA2 in the absence of GRA12 expression, HFF cells containing the fully developed PruΔgra12/GRA12II-HA PV or PruΔgra12 PV were fractionated into a high-speed soluble fraction (HSS) and a PV membrane-associated high-speed pellet (HSP) 24 h after infection as described in the legend to panel A. Western blots were incubated with anti-GRA2 mouse monoclonal antibodies, revealed with anti-mouse IgG coupled to peroxidase, and visualized by chemiluminescence. Twenty- and 30-kDa markers are shown. (C) Transmission electron microscopy of the type I RHΔgra12 mutant revealed the presence of a normal mature IVN (white arrows) and no tachyzoite or PV abnormalities.
GRA12 expression is essential for type II strain resistance to IFN-γ.
The cyst burden and acute-virulence phenotypes of PruΔgra12 parasites closely resembled phenotypes we previously reported for PruΔrop5 and PruΔrop18 mutants (27). We initially evaluated whether type II PruΔgra12 parasites were virulence attenuated in mice deficient in the production of IFN-γ. IFN-γ−/− knockout mice succumbed to a low infection dose of PruΔgra12 parasites, with kinetics similar to those in IFN-γ−/− mice infected by parental type II PruΔku80 parasites (Fig. 5A), suggesting that IFN-γ was required to control PruΔgra12 infection. We next evaluated the PV viability of all the type II PruΔgra mutants compared to that of the parental PruΔku80 strain in IFN-γ-activated host cells. Compared with the results for the parental PruΔku80 strain, the only PruΔgra mutant that exhibited an enhanced loss of PV viability in IFN-γ-activated mouse embryonic fibroblasts (MEFs) was PruΔgra12 (Fig. 5B).
FIG 5.
Type II PruΔgra12 parasites exhibit a loss of PV viability in IFN-γ-activated host cells. (A) Survival of C57BL/6 IFN-γ−/− knockout mice infected intraperitoneally with 2 × 102 tachyzoites of PruΔgra12 or PruΔku80. ns, not significant. (B) Mouse embryonic fibroblasts (MEFs) were stimulated with IFN-γ, and PV viability (measured as PFU) was determined in comparison to the results for nonstimulated (no IFN-γ) MEFs. The results from at least 3 independent experiments are shown as mean values plus or minus standard errors of the means (± SEM). Significant P values were calculated with Student’s t test (****, P < 0.0001). (C) Quantification of Irgb6 coating of PVs 45 min after infection of IFN-γ-stimulated bone marrow-derived macrophages (BMDM) by PruΔgra12 or PruΔku80 parasites. Representative images of PVs stained with anti-Irgb6 antibody (green) and anti-GRA5 antibody (red) are shown above the graph. At least 500 PVs were scored to determine the significance. Significant P values were calculated with Student’s t test (ns, not significant). (D) MEFs were stimulated in vitro with IFN-γ and infected with PruΔgra12 or PruΔku80 mutant parasites or complemented PruΔgra12/GRA12I-HA or PruΔgra12/GRA12II-HA parasites. PV viability (measured as PFU) was determined in comparison to the results for nonstimulated (no IFN-γ) MEFs. Results from at least 4 independent experiments are shown as mean values ± SEM. Significant P values were calculated with Student’s t test (****, P < 0.0001). (E) Survival of C57BL/6 mice infected i.p. with 2 × 106 tachyzoites of PruΔgra12, parental PruΔku80, or complemented PruΔgra12/GRA12I-HA or PruΔgra12/GRA12II-HA strains. The data presented are the combined results of 2 independent experiments, each with 4 mice per group. The P value was calculated by log rank Mantel-Cox test, and a P value of <0.05 was considered significant (****, P < 0.0001; ns, not significant). (F) CD1 mice were infected i.p. with 2 × 102 tachyzoites of each designated strain, and chronic-stage cyst burdens were counted 21 days postinfection. The data are cumulative results from 2 to 3 independent experiments for each strain tested, as follows: PruΔku80 (3 experiments; n = 12 mice), PruΔgra12 (2 experiments; n = 8 mice), PruΔgra12/GRA12I-HA (2 experiments; n = 8 mice), and PruΔgra12/GRA12II-HA (2 experiments; n = 8 mice). ns, not significant.
PruΔrop5 and PruΔrop18 PVs exhibit a loss of PV viability in IFN-γ-activated murine host cells due to a failure to resist increased coating of the PVM by IFN-γ-inducible effector immunity-related GTPase (IRG) proteins (such as Irgb6) and subsequent rupture of the PV (27). We measured PVM coating by the pioneer IRG, Irgb6 (73). Irgb6 coating of the type II strain PruΔgra12 PVM was not increased in IFN-γ-activated murine bone marrow-derived macrophages (BMDM) (Fig. 5C). In addition, type II PruΔgra12 PV viability in IFN-γ-activated host cells (Fig. 5D) and the acute-virulence phenotypes (Fig. 5E) were rescued by both the type II (strain PruΔgra12/GRA12II-HA) and the type I (strain PruΔgra12/GRA12I-HA) GRA12 genes. Correspondingly, normal levels of cyst burdens were also rescued in GRA12-complemented type II PruΔgra12 strains (Fig. 5F), suggesting that GRA12 could be a significant virulence factor that mediates resistance to host IFN-γ in both low- and high-virulence strains.
GRA12 expression is essential for type I strain resistance to host IFN-γ.
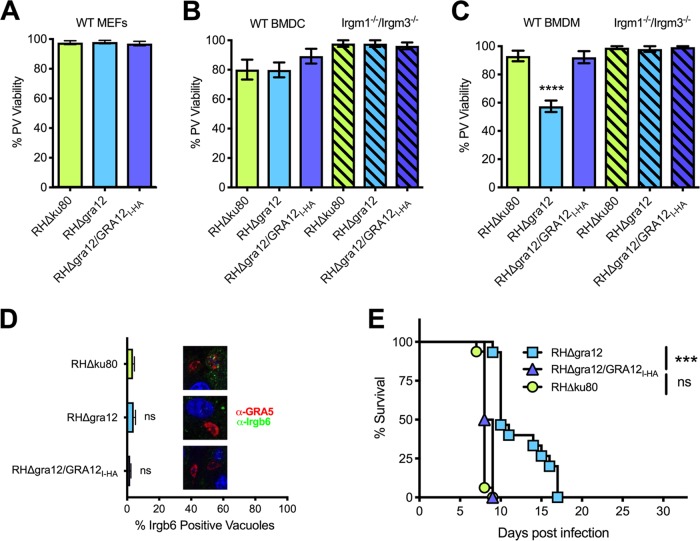
We next examined the PV viability phenotype associated with the expression of GRA12 in the virulent type I RH strain. In contrast to type II PruΔgra12 PVs that lost PV viability in MEFs (Fig. 5D), PV viability was not affected in IFN-γ-activated MEFs (Fig. 6A) or in IFN-γ-activated bone marrow-derived dendritic cells (BMDC) (Fig. 6B) infected with type I RHΔgra12 parasites. In contrast, PV viability was significantly decreased in IFN-γ-activated murine BMDM infected with RHΔgra12 parasites, and this macrophage-specific loss of PV viability was rescued by complementation with a type I RH strain GRA12 gene (strain RHΔgra12/GRA12I-HA) (Fig. 6C). The loss of PV viability of type I RHΔgra12 parasites in IFN-γ-activated BMDM was dependent on host IRGM1 and IRGM3, since a decrease in PV viability was not observed in macrophages derived from Irgm1−/−/Irgm3−/− double knockout mice (Fig. 6C) (74, 75) . In addition, PV resistance to Irgb6 PVM coating was not affected in IFN-γ-activated BMDM infected with RHΔgra12 parasites (Fig. 6D). A major virulence defect was observed after infection of CD1 mice with type I RHΔgra12 parasites, and this virulence defect was rescued by complementation with the type I GRA12 gene (Fig. 6E). Thus, in both low- and high-virulence parasite strain types, GRA12 determined the ability of the PV to resist host IFN-γ.
FIG 6.
Type I RHΔgra12 parasites exhibit a loss of PV viability in IFN-γ-activated macrophages. (A) MEFs were stimulated with IFN-γ, and PV viability (measured as PFU) was determined in comparison to the results for nonstimulated (no IFN-γ) MEFs. The results from at least 3 independent experiments are shown as mean values plus or minus standard errors of the means (± SEM). Significant P values were calculated with Student’s t test. (B) Bone marrow-derived dendritic cells (BMDC) from wild-type C57BL/6 mice or Irgm1−/−/Irgm3−/− knockout C57BL/6 mice were stimulated with IFN-γ, and PV viability (measured as PFU) was determined in comparison to the results for nonstimulated (no IFN-γ) BMDC. Results from at least 4 independent experiments are shown as mean values ± SEM. Significant P values were calculated with Student’s t test. (C) Bone marrow-derived macrophages (BMDM) from wild-type C57BL/6 mice or Irgm1−/−/Irgm3−/− knockout C57BL/6 mice were stimulated with IFN-γ and PV viability (measured as PFU) was determined in comparison to the results for nonstimulated (no IFN-γ) BMDM. Results from at least 4 independent experiments are shown as mean values ± SEM. Significant P values were calculated with Student’s t test (****, P < 0.0001). (D) Quantification of PV coating by Irgb6 45 min after infection of IFN-γ-stimulated bone marrow-derived macrophages (BMDM). At least 500 PVs were observed to determine significance. P values were calculated with Student’s t test. On the right side of the graph, representative images of PVs incubated with anti-Irgb6 (green) and anti-GRA5 (red) antibodies are shown. ns, not significant. (E) CD1 mice were infected i.p. with 1 × 102 tachyzoites of RHΔgra12, parental RHΔku80, or complemented RHΔgra12/GRA12I. The survival data shown are from 2 independent experiments, each with 4 mice per group. The P value was calculated using the log rank Mantel-Cox test, and a P value of <0.05 was considered significant (***, P < 0.001; ns, not significant).
DISCUSSION
The presence of cysts in the central nervous system characterizes the chronic stage of Toxoplasma infection (6, 10, 11). A limiting membrane termed the cyst membrane surrounds the cyst wall (7, 8), and while this cyst membrane is hypothesized to arise from the modified PVM (7, 8, 76), the fate and trafficking of PVM and IVN membrane proteins during differentiation and cyst development still remain to be investigated. Similar to the IVN membranes present in acute-stage tachyzoite PVs, mature cysts contain membrane tubules that, within an intracyst matrix, link the bradyzoites to each other and to the cyst wall and, possibly, to the cyst membrane (7). Our study showed that all the IVN-associated GRA proteins (GRA2, GRA4, GRA6, GRA9, and GRA12) and all the PVM-associated GRA proteins (GRA3, GRA7, GRA8, and GRA14) we investigated, except GRA15, were crucial for successful chronic infection.
PruΔgra parasite mutants retained a normal replication rate and differentiated to establish GFP+ bradyzoites encased by a cyst wall. In contrast to PruΔgra2, RHΔgra2, and RHΔgra6 parasites (38, 53, 54), which failed to establish typical elongated IVN nanotubular membrane structures in their PV, our data showed that RHΔgra12 parasites still established IVN nanotubular membranes within their PV and that GRA2 still partitioned normally into the IVN membrane and the PV soluble fractions. Previous results had shown that PruΔasp5 parasites, which are deficient in the Golgi complex-associated aspartyl protease 5 (ASP5) required for the processing and targeting of many secreted GRA proteins, do differentiate to establish GFP+ bradyzoites. However, their cyst wall development is severely impaired when differentiation is induced by in vitro alkaline switching (71). Our data showed that PruΔgra12 parasites exhibited a relative delay in the accumulation of the CST1 major cyst wall protein at the cyst periphery, but this defect was mild in comparison to the severe defects previously reported in PruΔasp5 cysts. Together, these findings suggested that the nanotubular IVN membrane structures present within the PV do not appear to be strictly necessary for the development of the cyst wall. These findings should be interpreted with caution, because (i) certain IVN membrane-associated GRA proteins, for example, GRA12, may participate in cyst wall development and (ii) these in vitro results may not truly reflect what happens in vivo, where the development of cysts during toxoplasmosis is triggered by host immune responses (77–79).
In mature cysts isolated from mice chronically infected with a type II strain, PVM-associated (GRA3 and GRA7) and IVN-associated (GRA2 and GRA6) GRA proteins were observed at the cyst periphery (67). Proteomic analysis of the isolated in vitro cyst wall has recently revealed that many GRA proteins localize at the cyst wall (68), including the GRA2, GRA3, GRA4, GRA5, GRA7, GRA8, GRA9, GRA12, and GRA14 proteins evaluated in this study. Our hypothesis is that IVN and PV membrane-associated GRA proteins could play a structural or organizational role during cyst development or in cyst maintenance. Deletion of the major cyst wall protein CST1 (8), as well as gene knockout of a nucleotide sugar transporter or the glycosylation pathway that glycosylates CST1 and other cyst wall proteins, increased cyst fragility and decreased cyst burdens in vivo (80, 81). We did not observe any clear increase in the frequency of broken cysts after mechanical disruption of the mouse brains infected with our PruΔgra mutants. Our finding that PruΔgra12 cysts exhibited a reduced intensity of DBA fluorescence at the cyst periphery compared to the intensity at the cyst interior suggested that GRA12 could regulate delivery or retention of the major cyst wall protein CST1 during the development of the cyst wall.
Defects in cyst burdens were not associated with a decrease in acute virulence in PruΔgra9 parasites, trended toward an association with reduced virulence in PruΔgra6 parasites, and were associated with markedly reduced virulence in PruΔgra2, PruΔgra4, and PruΔgra12 parasites. While our data identified a defect in acute virulence in PruΔgra3 parasites, we did not identify any acute virulence defect in PruΔgra7, PruΔgra8, or PruΔgra14 parasites. Our findings are consistent with previous reports that identified acute virulence defects in type I RHΔgra2 (57) and type II PruΔgra3 (82) parasites. While GRA6 and GRA7 were previously linked with virulence mechanisms in type I strains (19, 56), these virulence phenotypes were absent in type II strains. GRA6 was previously identified as a type I strain-dependent virulence factor that activates NFAT4 (56). The IRG resistance mechanisms in virulent type I strains also involved the GRA7 protein that was previously shown to prepare IRGs, through turnover, for neutralization by the PVM-localized ROP5/ROP17/ROP18 protein complexes (19, 20). Our finding that virulence is not associated with GRA7 in type II strains suggested that either GRA7 is a strain type-dependent virulence factor or the virulence phenotype, as in type I strains (19), is not visible in this type II mutant when ROP5 and ROP18 virulence functions are expressed. In addition to the defects observed in cyst wall development and in IVN nanotubular morphology, PruΔasp5 parasites also exhibit reduced virulence in mice (71, 83). Since the IVN nanotubular membrane structures are present in RHΔgra4 parasites (38) and, as we show here, in both RHΔgra12 and PruΔgra12 parasites, virulence phenotypes do not appear to be specifically correlated with the presence or absence of distinct IVN nanotubular membrane structures.
The severity of the virulence defect observed in both type I and type II Δgra12 parasites mirrored the severity of the virulence defect previously observed in high-virulence type I RHΔrop18 (25) and low-virulence type II PruΔrop18 parasites (27). Notably, the loss of PV viability of type I RHΔgra12 parasites was cell type dependent, since it was observed specifically in IFN-γ-activated macrophages but not in IFN-γ-activated dendritic cells or MEFs. These data are consistent with previous findings that showed that macrophages harbor the most effective IFN-γ-activated mechanisms that disrupt the viability of the PV (84). The loss of viability of type II PruΔgra12 PVs observed in IFN-γ-activated MEFs is most likely explained by the inherently less active ROP5 and ROP18 mechanisms that resist host IFN-γ in low-virulence type II strains (27). In such low-virulence strains, GRA12 was crucial to maintain resistance to host IFN-γ, whereas the essentiality of GRA12 in high-virulence type I strains was visible only in IFN-γ-activated macrophages that launch the most destructive attacks on the PV.
Type I RHΔgra12 and type II PruΔgra12 parasites exhibited no defect in their ability to resist increased coating of the PVM with Irgb6 in IFN-γ-activated macrophages. ROP5 and ROP18 virulence factors directly resist host effector IRGs, including the pioneer Irgb6 (73), by neutralizing them and preventing an increase in their PVM coating (23–25, 27). ROP18 also resists the recruitment of GBP1 and ubiquitin to the PV (34, 35). In addition to ROP18, other PVM-localized proteins resist the recruitment of GBPs to the PV. Type II parasites that do not express the PVM-localized rhoptry pseudokinase ROP54 failed to resist host GBP2 coating of the PVM, and PruΔrop54 parasites were attenuated in their virulence (85). The targeting of the Toxoplasma PV by IRGs and GBPs is coordinated with the functions of several host autophagy proteins and, together, they lead ultimately to the destruction of the PV (86). Our results demonstrated that the loss of Δgra12 PV viability was not observed in Irgm1−/−/Irgm3−/− macrophages that fail to express IRGM1 and IRGM3 functions. IRGM1 and IRGM3 are regulatory IRG proteins that are essential for the activation of host effector IRGs that can disrupt the PV (74, 75, 87). Regulatory IRGM proteins also regulate autophagic flux and influence the localization of GBP2 by modulating macroautophagy (88). In addition, loss of IRGM1 and IRGM3 expression induces a failure to target both IRGs and GBPs to the Toxoplasma PV (89). However, while regulatory IRGM1 and IRGM3 proteins have been shown to control the pathogen-killing mechanisms of IRG effectors (74, 75, 87), it is currently unknown whether regulatory IRGM proteins specifically regulate functions of GBP proteins that can restrict the PV. Therefore, additional studies are necessary to elucidate the specific roles of regulatory IRGM1 and IRGM3, effector IRGs, GBPs, and autophagic machinery in the restriction of PVs that lack expression of GRA12.
Interestingly, GRA12 was previously identified in proteomic analysis of ROP5 and ROP18 high-molecular-weight protein complexes but was not found associated with ROP17 protein complexes (GRA12 is shown in Table 1 of reference 24 as TGGT1_034740). In addition, BirA* proximity localization studies using a PVM-localized GRA17-BirA* fusion protein also identified GRA12 as one of the interacting partners of GRA17 (90). These proteomic studies suggested that GRA12 closely interacted with several protein complexes that localized at the PVM, which agrees with our observations that GRA12 sporadically colocalized with PVM-associated GRA5 and with previously reported data showing that GRA12 sporadically colocalized with GRA3 at the PVM (52). Taking into account that (i) GRA12 predominantly localized at the IVN rather than the PVM, (ii) GRA12 sporadically localized with PVM-localized proteins GRA3 (52) and GRA5, (iii) GRA12 was identified in PVM-localized protein complexes in association with GRA17 (90) or ROP5/ROP18 (24), (iv) within the PV, the IVN membranes form direct connections with the PVM (45), (v) Δgra12 PVs resisted early coating of the PV by host IRGs in IFN-γ-activated macrophages, (vi) Δgra12 PVs succumbed in an IRGM1/IRGM3- and IFN-γ-dependent fashion, and (vii) Δgra12 cysts accumulated less CST1 cyst wall protein at the cyst periphery relative to the amount in the cyst interior in vitro, we hypothesized that GRA12 is likely to be involved in the delivery or exchange of protein or lipid cargo between the IVN membranes and the PVM to support PVM resistance to host IFN-γ.
The PVM is a dynamic molecular interface separating the PV lumen from the host cell cytosol. The PVM is decorated with important protein complexes, such as the GRA17/GRA23 pore protein complexes (91), the ROP5/ROP18 protein complexes that allow resistance to IFN-γ-activated IRGs (25) and GBPs (34, 35), and the MYR1/MYR2/MYR3 protein complexes that translocate parasite GRA proteins across the PVM to modulate host gene transcription (92–94). GRA12 is mostly localized to the IVN and does not appear to be translocated past the PVM, suggesting it is unlikely that GRA12-mediated defense against IFN-γ is achieved through direct GRA12 protein interactions with host cell molecules. Since host cell IRGM1 and IRGM3 functions were necessary to make PVs that lack GRA12 expression nonviable, we can infer that GRA12 is required to support the functions of PVM-localized protein complexes in resisting host IFN-γ to maintain PV viability. Collectively, our results demonstrated the importance of many PV membrane GRA proteins in establishing chronic infection and revealed a virulence mechanism mediated by IVN membrane-associated GRA12 that underpins parasitophorous vacuole resistance to host IFN-γ.
MATERIALS AND METHODS
Parasite and host cell cultures.
Human foreskin fibroblasts (HFF) were amplified in Eagle’s modified essential medium (EMEM) complemented with 10% fetal bovine serum (FBS) (HyClone), 2 mM glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. Confluent HFF monolayers were infected with T. gondii strains RHΔku80 (72, 95) and PruΔku80 (66) and knockout or mutant strains derived from these parental strains in EMEM containing 1% FBS, 2 mM glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin, as previously described (96, 97). Mouse embryonic fibroblasts (MEFs) derived from C57BL/6 mice (ATCC) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 15% FBS, 2 mM glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. Bone marrow-derived dendritic cells (BMDC) and macrophages (BMDM) were isolated from the femur and tibia of C57BL/6 mice or from C57BL/6 mice that lack functional Irgm1 and Irgm3 genes (Irgm1−/−/Irgm3−/− knockout mice [74]). BMDM were differentiated in DMEM supplemented with 10% FBS, 1× minimal essential medium with nonessential amino acids, 1 mM sodium pyruvate (Life Technologies), 100 units/ml penicillin, 100 μg/ml streptomycin, and 30% L929 culture supernatant, as previously described (98). BMDC were differentiated in Roswell Park Memorial Institute (RPMI) 1640 medium (Fisher Scientific) supplemented with 10% FBS, 100 units/ml penicillin, 100 μg/ml streptomycin, 50 μM 2-mercaptoethanol (Sigma), and 20 ng/ml murine granulocyte-macrophage colony–stimulating factor (GM-CSF) (Peprotech), as previously described (99). BMDC were harvested for experiments 9 days after differentiation, while BMDM were harvested after 5 days of differentiation.
Mice.
Female 7- to 9-week-old C57BL/6 mice and IFN-γ−/− C57BL/6 mice were obtained from Jackson Laboratories (Bar Harbor, ME). Female 7- to 9-week-old CD1 mice were obtained from Charles River Laboratories (Wilmington, MA).
Acute virulence assay.
High-viability type I or type II tachyzoites were isolated from 3-day-infected HFF cultures as previously described (66, 100). Parasites were centrifuged at 900 × g for 7 min, washed, and counted in Dulbecco’s modified phosphate-buffered saline (DPBS). Parasite viability was confirmed in HFF plaque assays (101). Groups of 4 C57BL/6 mice (type II infection) were injected intraperitoneally (i.p.) with 2 × 105, 2 × 106, or 2 × 107 tachyzoites, or groups of 4 CD1 mice (type I infection) were injected intraperitoneally with 2 × 102 tachyzoites. In experiments using mice deficient in the production of IFN-γ (IFN-γ−/− mice), groups of 4 mice were injected intraperitoneally with 2 × 102 parasites. Mice were monitored for symptoms of infection, weight loss, and mortality for 30 days, and survival was evaluated using the Kaplan-Meier curve.
Cyst burden assays.
High-viability type II tachyzoites were obtained as previously described (66, 100). Tachyzoites were centrifuged at 900 × g for 7 min, washed, and counted in DPBS. Groups of 4 mice were infected by intraperitoneal (i.p.) injection with 2 × 102 parasites, and parasite viability was confirmed in a plaque assay. Mice were monitored for symptoms of infection, weight loss, and mortality for 21 days. The brains from mice infected with type II strains were harvested at 3 weeks postinfection and homogenized using a Dounce homogenizer in 2 ml of sterile DPBS. The cysts from a minimum of 10% of each brain were scored. Since Pru strain background cysts can vary in size (66, 102), they were scored with dark-field microscopy using an inverted phase-contrast fluorescence microscope (Olympus CKX41). The PruΔku80 parent strain expresses GFP under the control of the bradyzoite stage-specific LDH2 promoter (66). GFP+ cysts were scored using a total magnification power of ×150 because this magnification provided the highest sensitivity for the detection of GFP+ bradyzoites within latent cysts. GFP+ cysts were then observed in bright-field microscopy at ×300 total magnification to verify that these cysts possessed a thick translucent cyst wall that completely surrounded the GFP+ bradyzoites (66, 100).
Deletion of GRA genes.
Targeted GRA gene of interest (GOI) deletions (Table S1) were developed using the Δku80 knockout strain of the type II Prugniaud strain (PruΔku80) or the type I RH strain (RHΔku80), as previously described (see Table S1 in the supplemental material) (27, 66, 72). Briefly, GRA gene locus knockout-targeting plasmids were assembled in the yeast shuttle vectors pRS416 or pRS426 using yeast recombination cloning to fuse, in the following order, 3 distinct PCR products with 31- to 34-bp crossovers: a 5′ GRA GOI target-flanking sequence, the HXGPRT selectable marker, and a 3′ GRA GOI target-flanking sequence (Fig. S1A) (100). Knockout plasmids were engineered to delete at least 200 nucleotides of the 5′ untranslated region (UTR) and the complete coding sequence of the GRA GOI locus as defined in the ToxoDB.org database (103). All oligonucleotide primers used to construct the knockout-targeting plasmids and the ToxoDB nucleotide definitions of deleted GRA gene loci are listed in Table S2. The targeting plasmids were validated by DNA sequencing and were linearized at restriction sites inserted at the 5′ end of the 5′ target-flanking sequence (Fig. S1A). The linearized targeting plasmids were transfected by electroporation into tachyzoites of the PruΔku80 strain. GRA GOI knockouts were selected with 50 μg/ml mycophenolic acid and 50 μg/ml xanthine. Drug-selected parasite mutants were cloned by limiting dilution 30 days after transfection. GRA GOI knockouts were validated by genotype analysis using the PCR strategy shown in Fig. S1A, as follows: (i) PCR 1 verified targeted deletion of the coding region of the targeted gene (DF and DR primers), (ii) PCR 2 verified the correct targeting of the 5′-end integration (CXF and 5′DHFRCXR primers), and (iii) PCR 3 verified the correct targeting of the 3′-end integration (3′DHFRCXF and CXR primers). All the knockout validation primers are shown in Table S2.
Primers used to construct and validate GRA knockouts. Download Table S2, DOCX file, 0.04 MB (42.4KB, docx) .
Copyright © 2019 Fox et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Complementation of Δgra12 parasite strains.
Complementation plasmids were designed to complement the PruΔgra12 and RHΔgra12 strains through targeted chromosomal integration and expression of wild-type or mutant gene alleles at the uracil phosphoribosyltransferase (UPRT) chromosomal locus (TGME49_312480), as previously described (Fig. S1B) (27, 66). Complementation plasmids were developed in the pRS416 or pRS426 yeast shuttle vectors using yeast recombination to fuse, in this order, a 5′ UPRT target-flanking sequence, the complementing gene of interest with its native 5′ UTR, and the 3′ UPRT target-flanking sequence (Fig. S1B). Oligonucleotide DNA primers (Table S3) were used to generate the complementing genes, synthesized as one PCR product. Following plasmid assembly by yeast recombinational cloning, the targeting plasmids were validated by DNA sequencing. Prior to transfection, plasmids were linearized at the unique restriction site PmeI. Following transfection, parasites were cultured for 2 days in normal infection medium, and the cultures were then switched to selection medium containing 2 μM 5-fluorodeoxyuridine (FUDR). Thirty days after transfection, parasites were subcloned by limiting dilution. Accurate targeting of complementing genes into the UPRT locus was validated by genotype analysis using 4 PCRs (strategy shown in Fig. S1B), of which (i) PCR 4 verified the deletion of the UPRT coding region, (ii) PCR 5 verified correctly targeted 5′-end integration, and (iii) PCR 6 verified correctly targeted 3′-end integration of the complementing transgene at the UPRT locus. All the oligonucleotide DNA validation primers are shown in Table S3.
Primers used to construct plasmids for complementation and validation of GRA mutant strains. Download Table S3, DOCX file, 0.03 MB (30.7KB, docx) .
Copyright © 2019 Fox et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Validation of knockouts by indirect IFA and immunoblotting.
Parasites were isolated from freshly lysed cultures and suspended in Laemmli buffer. Proteins were separated by 13% SDS-PAGE (nonreduced conditions), transferred to nitrocellulose membranes, and detected using rat anti-GRA12 antibodies (1:400) (52). Proteins were detected with horseradish peroxidase (HRP)-conjugated secondary antibodies (1:20,000; Jackson Immunoresearch Laboratories), and the peroxidase activity was visualized by chemiluminescence using the SuperSignal enhanced chemiluminescence (ECL) system (Pierce Chemical). For immunofluorescence assay (IFA), confluent HFF cells were grown on glass coverslips and infected for 24 h. Infected cells were fixed in 5% paraformaldehyde for 20 min, permeabilized for 10 min in 0.002% saponin, and blocked in 5% goat serum and 5% FBS in PBS. Cells were incubated with primary antibodies for 1 h in DPBS containing 1% FBS and 0.002% saponin. The primary antibodies used in IFA validation of GRA knockouts included the following mouse monoclonal antibodies (MAbs): anti-GRA2 MAb TG17.179 (1:500) (47), anti-GRA3 MAb T6.2H11 (1:500) (104), rabbit anti-GRA4 MAb (1:500) (49), rabbit anti-GRA6 MAb (1:500) (49), anti-GRA7 MAb BATO 214 (1:500) (105), anti-GRA8 MAb 3.2 (1:500) (106), rabbit anti-GRA9 MAb (1:250) (51), rat anti-GRA12 MAb (1:400) (52), mouse polyclonal anti-GRA14 MAb (1:500) (40), rabbit antiactin MAb (1:500) (107), or anti-SAG1 MAb TG054 (1:500) (108). (Antibodies were purchased from the Biotem company, Apprieu, France, or kindly provided by L. D. Sibley, Washington University School of Medicine, Saint-Louis, MO, D. Jacobs, Innogenetics-Fujirebio Europe N.V., Ghent, Belgium, G.E. Ward, University of Vermont College of Medicine, Burlington, VT, W. Daübener, Heinrich Heine Universität, Düsseldorf, Germany, M. Lebrun, Montpellier, France, and Peter Bradley, University of California, Los Angeles, CA). Infected cells were washed and incubated for 1 h with goat anti-mouse IgG(H+L)-Alexa Fluor 488 (1:500) (Jackson), goat anti-mouse IgG(H+L)-Alexa Fluor 594 (1:500) (Jackson), goat anti-rat IgG(H+L)-Alexa Fluor 488 (1:500) (Jackson), or goat anti-rabbit IgG(H+L)-Alexa Fluor 488 (1:500) (Jackson) secondary antibody. Coverslips were then incubated in 5 μg/ml Hoechst 33342 to stain nuclei, mounted in Mowiol, and observed using an Axioplan II microscope (Zeiss) equipped with a 100× objective. Images were acquired with a black-and-white camera (Zeiss) and AxioVision version 4.7.1.
Intracellular replication rate assay.
The parasite growth rate was determined using a previously described method that scores the number of parasites per vacuole (66). Briefly, triplicate monolayers of HFF cells were infected at a multiplicity of infection (MOI) of ∼0.2 and parasites were let to invade cells for 1 h. Monolayers were then washed 3 times in PBS to remove extracellular parasites. The number of tachyzoites per vacuole was scored in at least 50 PVs randomly encountered on the coverslip at 45 h postinfection for type II parasite strains or at 30 h postinfection for type I parasite strains.
PV viability assay.
BMDC or BMDM were collected and seeded overnight into 6-well plates at a density of 5 × 106 cells per well. Triplicate sets of BMDC or BMDM cultures were incubated for 4 to 6 h in complete medium or in activation medium containing murine IFN-γ (100 U/ml; Peprotech) and TNF-α (10 U/ml; Peprotech). Cells were infected with 100 parasites per well, and cultures were left undisturbed for 6 days at 37°C. The medium was aspirated from each well, and the BMDC and BMDM monolayers were fixed in 50% methanol and 10% acetic acid and stained with 0.25% Coomassie brilliant blue to visualize plaques. The number of plaques in each well was scored. MEFs were seeded into 24-well plates at a density of 1 × 105 cells per well and reached confluence in ∼24 h. MEF control monolayers were incubated without IFN-γ or MEFs were stimulated in activation medium containing 200 U/ml IFN-γ (Peprotech) 24 h prior to parasite infection. Triplicate wells were infected with 200 or 1,000 parasites, and plaques were let to develop for 5 to 7 days. The total number of PFU per well was counted microscopically, scoring all the PFU in each well. The percentage of PV viability was calculated to represent the number of PFU in IFN-γ-activated cells divided by the number of PFU scored in control not-activated cells.
IRG coating assay.
BMDM and BMDC were harvested, seeded onto circular glass coverslips (Electron Microscopy Sciences), and incubated overnight. BMDM and BMDC were then activated with 100 U/ml IFN-γ and 10 U/ml TNF-α for 6 h and infected for 45 min with parasites at an MOI of 4. The coverslips were washed in DPBS, and cells were fixed with 4% paraformaldehyde (Electron Microscopy Sciences), permeabilized with 0.1% saponin (Sigma), and blocked in 10% FBS. For visualization, cultures were incubated with mouse anti-GRA5 antibody (Ab) (TG 17.113 at 1:2,000; Biotem) and rabbit anti-Irgb6 Ab (1:1,000) (74), washed, and incubated with anti-mouse IgG(H+L) coupled to Alexa Fluor 568 and anti-rabbit IgG(H+L) coupled to Alexa Fluor 488 as secondary antibodies (Invitrogen). Coverslips were mounted in ProLong gold with DAPI (4′,6-diamidino-2-phenylindole; Invitrogen) and imaged at ×63 magnification with a Nikon A1R SI confocal microscope (Nikon, Inc.). All images were processed using the FIJI program (109). A minimum of 500 PVs of each strain was scored for quantification of Irgb6 coating of the PVM, as previously described (27).
IFA colocalization assays.
HFF cells were cultured on circular glass coverslips (Electron Microscopy Sciences) and infected with parasites for 24 h. For visualization of dense granule proteins in the PV, cultures were fixed in 4% paraformaldehyde and permeabilized for 10 min with 0.01% saponin (Sigma). All samples were blocked for 1 h with 10% FBS and incubated for 1 h with a 1:500 dilution of primary rabbit anti-HA tag MAb (Cell Signaling) and a 1:1,000 dilution of primary mouse anti-GRA2 MAb or anti-GRA5 MAb (Biotem, France). Preparations were washed 3 times with DPBS supplemented with Ca2+ and Mg2+ and incubated for 1 h at room temperature with a 1:1,000 dilution of goat anti-rabbit and goat anti-mouse IgG secondary antibodies conjugated to Alexa Fluor 488 and Alexa Fluor 594, respectively. Samples were mounted in SlowFade gold antifade with DAPI (Life Technologies) and imaged with a Nikon A1R SI confocal microscope (Nikon, Inc.). PVs were located using differential interference contrast microscopy (DIC). Confocal images were processed using the FIJI program (109).
In vitro cyst differentiation assay.
Tachyzoite-to-bradyzoite differentiation using in vitro alkaline switch was performed as described by Knoll and colleagues (110). The differentiation medium consists of RPMI 1640 without bicarbonate and supplemented with 2.05 mM l-glutamine (HyClone), 20 mM HEPES free acid (IBI Scientific), 1% Glutamine XL (a stable form of glutamine; VWR), 1% FBS, and 1% penicillin-streptomycin. The differentiation medium pH was adjusted to 8.1 with sodium hydroxide. Monolayers of HFF cells were cultured on circular glass coverslips until confluent and infected with type II parasite strains at an MOI of ∼0.5. Infected cells were washed once 3 h later with DPBS supplemented with Ca2+ and Mg2+, and cultures were returned to high-pH differentiation medium for 3 days at 37°C in ambient air to differentiate cysts. Infected cells were fixed in 4% paraformaldehyde, and the excess of paraformaldehyde was quenched with 0.1 M glycine. Infected cells were simultaneously permeabilized and blocked for 30 min at room temperature in 3% FBS–0.2% Triton X-100 and incubated for 1 h at room temperature with a 1:250 dilution of rhodamine-labeled Dolichos biflorus agglutinin (DBA) (Vector Laboratories). Preparations were washed three times with DPBS, mounted in SlowFade gold antifade with DAPI (Life Technologies), and imaged with a Nikon A1R SI confocal microscope (Nikon, Inc.).
In vitro cyst DBA fluorescence intensity quantification assay.
Cysts (≥10) matured in vitro for 3 days were located by DIC microscopy. Images were imported into FIJI to evaluate the fluorescence intensity of rhodamine-labeled DBA in cysts. All measurements were taken at the zoom magnification of 300% to allow more accurate manual drawing separation of the cyst periphery (cyst wall) from the cyst interior (that does not include the cyst wall). First, we defined the region of interest by using the freehand selection tool to draw a circle around the entire cyst and to capture the total DBA signal. To measure fluorescence, both the DBA color channel and the DIC channel (to define the area being measured) were selected. Under the set measurements tab, the following parameters were selected: area, integrated density, and mean gray value. The region of interest was analyzed by selecting measure, and measurements for area, integrated density, and mean gray value were recorded in a table. This was repeated, independently, 3 times for each cyst. In a second step using the freehand selection tool, the region inside the cyst was defined by drawing, beneath the cyst wall, a circle that excluded the DBA fluorescence of the cyst wall. Similarly, this was repeated, independently, 3 times for each cyst. In a third step, to measure the background fluorescence, a circle was drawn close to the cyst and measured, and this was repeated 3 independent times, each time rotating the cyst aspect before each measurement. The mean fluorescence of background readings was obtained by calculating the average of the three mean gray value numbers collected. In the fourth step, the corrected total cyst fluorescence (CTCF) was calculated for each cyst region measurement by using the equation CTCF = integrated density − (area of selected cyst × mean fluorescence of background readings). To capture an estimate of the cyst wall, the CTCF of the cyst periphery region was calculated by subtracting the average CTCF of the cyst interior region from the average CTCF of the entire cyst. Finally, for each cyst, the cyst periphery CTCF was divided by the cyst interior CTCF to quantify the DBA fluorescence intensity at the cyst periphery relative to that of the cyst interior. All the ratios (10 PruΔku80 cysts and 12 PruΔgra12 cysts) were graphed using the Prism program, and significance was evaluated using the unpaired t test.
TEM.
HFF monolayers were grown to confluence on Permanox slides in DMEM supplemented with 10% FBS and 1% sodium pyruvate (D10 medium). Cells were infected for 24 h in D10 medium, rinsed with PBS, fixed for 2 h with glutaraldehyde diluted in 0.2 M NaPO4, pH 7.4, and processed for transmission electron microscopy (TEM) as previously described (54). The infected cells were flat embedded and sectioned en face to enhance the preservation of the IVN structures.
PV fractionation of infected host cells.
PV membranes were physically separated from PV soluble molecules in infected HFF cells using a previously described method (45). Briefly, HFF monolayers in 150-cm2 flasks were infected for 24 h at an MOI of 3. Infected HFFs were gently washed once with DPBS supplemented with Ca2+ and Mg2+ to remove residual cell culture medium. Infected cell monolayers were dislodged in cold DPBS supplemented with Ca2+ and Mg2+ in the presence of a cocktail of protease inhibitors (Roche). Cells were recovered by low-speed centrifugation, and infected cells were mechanically disrupted by syringing through a 27-gauge needle to break host cells and PVs and to release still-intact parasites. Parasites and large host cell debris were pelleted by low-speed centrifugation at 2,500 × g for 10 min. The supernatants containing the soluble components of the infected cells and the PV membranes were then fractionated by 1 h of ultracentrifugation at 100,000 × g into a soluble high-speed supernatant (HSS) and a high-speed pellet (HSP) containing the PV membranes. Equal fractions of the HSP and the HSS were boiled in Tris-glycine SDS sample buffer (Novex) containing 2-β-mercaptoethanol and separated on 10% Tris-glycine WedgeWell gels (Novex). Gels included lanes with a prestained protein ladder (PageRuler) and a biotinylated protein ladder (Cell Signaling) and were later visualized using antibiotin antibodies (1:1,000) conjugated to horseradish peroxidase. The proteins were transferred to nitrocellulose membranes using a semidry Trans-Blot turbo transfer system (Bio-Rad). Membranes were blocked for 1 h with 5% (wt/vol) nonfat dry milk in 1× Tris-buffered saline (10 mM Tris HCl and 150 mM NaCl at pH 7.4) containing 0.1% Tween 20 (TBS-T buffer). Membranes were incubated overnight at 4°C with primary rabbit anti-HA antibodies (1:1,000) (Cell signaling) or anti-GRA2 MAb (1:1,000) (Biotem). After 3 washes in 1× TBS-T buffer for 5 min, membranes were incubated for 1 h with secondary goat anti-rabbit IgG (1:2,000) or goat anti-mouse IgG (1:2,000) conjugated to horseradish peroxidase. After 3 washes in 1× TBS-T buffer for 5 min each, signals were detected with LumiGlo chemiluminescence reagent (Cell signaling) and exposed to X-ray film.
Statistical analyses.
Statistical analyses were performed using the GraphPad Prism 6.0 software. Statistical data for bar graphs were calculated using 2-tailed unpaired Student’s t tests, which were conducted using the assumption of equal variance. The standard errors of the means (SEM) from independent samples assayed within the represented experiments are reported. Survival experiments were analyzed using the log rank Mantel-Cox test.
Ethics statement.
All animal experiments were conducted in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals (111) and the Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC) guidelines. Animals were housed in an AAALAC-certified facility, and animal protocols were approved by the Dartmouth College Committee on the Use and Care of Animals.
ACKNOWLEDGMENTS
We are grateful to the developers of the ToxoDB.org Genome Resource. ToxoDB and EuPathDB are part of the National Institutes of Health/National Institutes of Allergy and Infectious Diseases (NIH/NIAID)-funded Bioinformatics Resource Center. This work was sponsored by the grants AI041930, AI075931, AI084570, AI104514, AI105563, AI108489, AI129869, AI131630, and AI137118 from the National Institutes of Health (NIH) to D.J.B., the grant VA I01 BX002369 from the Veterans Administration and NIH grant AI135398 to G.A.T., Labex Parafrap (ANR-11-LABX-0024) and Fondation pour la Recherche Médicale to M.F.C.D., and Cluster 10, Région Rhone-Alpes and ANR 11 EMMA 03201 to C.M. R.B.G. and L.M.R. were predoctoral trainees on NIH training grants 5T32AI007363 and 2T32AI007519. V.B. was supported by a PhD fellowship from the Parafrap Labex. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We are grateful to all the scientists and laboratories that contributed to the antibodies used in this study (L.D. Sibley, D. Jacobs, Innogenetics-Fujirebio Europe, G.E. Ward, W. Daübener, M. Lebrun, P. Bradley).
Footnotes
Citation Fox BA, Guevara RB, Rommereim LM, Falla A, Bellini V, Pètre G, Rak C, Cantillana V, Dubremetz J-F, Cesbron-Delauw M-F, Taylor GA, Mercier C, Bzik DJ. 2019. Toxoplasma gondii parasitophorous vacuole membrane-associated dense granule proteins orchestrate chronic infection and GRA12 underpins resistance to host gamma interferon. mBio 10:e00589-19. https://doi.org/10.1128/mBio.00589-19.
REFERENCES
- 1.Tenter AM, Heckeroth AR, Weiss LM. 2000. Toxoplasma gondii: from animals to humans. Int J Parasitol 30:1217–1258. doi: 10.1016/S0020-7519(00)00124-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dubey JP. 2007. The history and life cycle of Toxoplasma gondii, p 1–18. In Weiss LM, Kim K (ed), Toxoplasma gondii: the model apicomplexan parasite: perspectives and methods. Elsevier, London, United Kingdom. [Google Scholar]
- 3.Montoya JG, Remington JS. 2008. Management of Toxoplasma gondii infection during pregnancy. Clin Infect Dis 47:554–566. doi: 10.1086/590149. [DOI] [PubMed] [Google Scholar]
- 4.Behnke MS, Dubey JP, Sibley LD. 2016. Genetic mapping of pathogenesis determinants in Toxoplasma gondii. Annu Rev Microbiol 70:63–81. doi: 10.1146/annurev-micro-091014-104353. [DOI] [PubMed] [Google Scholar]
- 5.Sibley LD, Ajioka JW. 2008. Population structure of Toxoplasma gondii: clonal expansion driven by infrequent recombination and selective sweeps. Annu Rev Microbiol 62:329–351. doi: 10.1146/annurev.micro.62.081307.162925. [DOI] [PubMed] [Google Scholar]
- 6.Konradt C, Ueno N, Christian DA, Delong JH, Pritchard GH, Herz J, Bzik DJ, Koshy AA, McGavern DB, Lodoen MB, Hunter CA. 2016. Endothelial cells are a replicative niche for entry of Toxoplasma gondii to the central nervous system. Nat Microbiol 1:16001. doi: 10.1038/nmicrobiol.2016.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lemgruber L, Lupetti P, Martins-Duarte ES, De Souza W, Vommaro RC. 2011. The organization of the wall filaments and characterization of the matrix structures of Toxoplasma gondii cyst form. Cell Microbiol 13:1920–1932. doi: 10.1111/j.1462-5822.2011.01681.x. [DOI] [PubMed] [Google Scholar]
- 8.Tomita T, Bzik DJ, Ma YF, Fox BA, Markillie LM, Taylor RC, Kim K, Weiss LM. 2013. The Toxoplasma gondii cyst wall protein CST1 is critical for cyst wall integrity and promotes bradyzoite persistence. PLoS Pathog 9:e1003823. doi: 10.1371/journal.ppat.1003823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weiss LM, Kim K. 2000. The development and biology of bradyzoites of Toxoplasma gondii. Front Biosci 5:D391–D405. doi: 10.2741/A521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cabral CM, Tuladhar S, Dietrich HK, Nguyen E, MacDonald WR, Trivedi T, Devineni A, Koshy AA. 2016. Neurons are the primary target cell for the brain-tropic intracellular parasite Toxoplasma gondii. PLoS Pathog 12:e1005447. doi: 10.1371/journal.ppat.1005447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melzer TC, Cranston HJ, Weiss LM, Halonen SK. 2010. Host cell preference of Toxoplasma gondii cysts in murine brain: a confocal study. J Neuroparasitolog 1:N100505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luft BJ, Remington JS. 1992. Toxoplasmic encephalitis in AIDS. Clin Infect Dis 15:211–222. doi: 10.1093/clinids/15.2.211. [DOI] [PubMed] [Google Scholar]
- 13.Weiss LM, Dubey JP. 2009. Toxoplasmosis: a history of clinical observations. Int J Parasitol 39:895–901. doi: 10.1016/j.ijpara.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sibley LD. 2004. Intracellular parasite invasion strategies. Science 304:248–253. doi: 10.1126/science.1094717. [DOI] [PubMed] [Google Scholar]
- 15.Carruthers VB, Sibley LD. 1997. Sequential protein secretion from three distinct organelles of Toxoplasma gondii accompanies invasion of human fibroblasts. Eur J Cell Biol 73:114–123. [PubMed] [Google Scholar]
- 16.Mercier C, Cesbron-Delauw MF. 2015. Toxoplasma secretory granules: one population or more? Trends Parasitol 31:60–71. doi: 10.1016/j.pt.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 17.Henriquez FL, Nickdel MB, McLeod R, Lyons RE, Lyons K, Dubremetz JF, Grigg ME, Samuel BU, Roberts CW. 2005. Toxoplasma gondii dense granule protein 3 (GRA3) is a type I transmembrane protein that possesses a cytoplasmic dilysine (KKXX) endoplasmic reticulum (ER) retrieval motif. Parasitology 131(Pt 2):169–179. doi: 10.1017/S0031182005007559. [DOI] [PubMed] [Google Scholar]
- 18.Dunn JD, Ravindran S, Kim SK, Boothroyd JC. 2008. The Toxoplasma gondii dense granule protein GRA7 is phosphorylated upon invasion and forms an unexpected association with the rhoptry proteins ROP2 and ROP4. Infect Immun 76:5853–5861. doi: 10.1128/IAI.01667-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alaganan A, Fentress SJ, Tang K, Wang Q, Sibley LD. 2014. Toxoplasma GRA7 effector increases turnover of immunity-related GTPases and contributes to acute virulence in the mouse. Proc Natl Acad Sci U S A 111:1126–1131. doi: 10.1073/pnas.1313501111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hermanns T, Muller UB, Konen-Waisman S, Howard JC, Steinfeldt T. 2016. The Toxoplasma gondii rhoptry protein ROP18 is an Irga6-specific kinase and regulated by the dense granule protein GRA7. Cell Microbiol 18:244–259. doi: 10.1111/cmi.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Behnke MS, Fentress SJ, Mashayekhi M, Li LX, Taylor GA, Sibley LD. 2012. The polymorphic pseudokinase ROP5 controls virulence in Toxoplasma gondii by regulating the active kinase ROP18. PLoS Pathog 8:e1002992. doi: 10.1371/journal.ppat.1002992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Behnke MS, Khan A, Lauron EJ, Jimah JR, Wang Q, Tolia NH, Sibley LD. 2015. Rhoptry proteins ROP5 and ROP18 are major murine virulence factors in genetically divergent South American strains of Toxoplasma gondii. PLoS Genet 11:e1005434. doi: 10.1371/journal.pgen.1005434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Behnke MS, Khan A, Wootton JC, Dubey JP, Tang K, Sibley LD. 2011. Virulence differences in Toxoplasma mediated by amplification of a family of polymorphic pseudokinases. Proc Natl Acad Sci U S A 108:9631–9636. doi: 10.1073/pnas.1015338108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Etheridge RD, Alaganan A, Tang K, Lou HJ, Turk BE, Sibley LD. 2014. The Toxoplasma pseudokinase ROP5 forms complexes with ROP18 and ROP17 kinases that synergize to control acute virulence in mice. Cell Host Microbe 15:537–550. doi: 10.1016/j.chom.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fentress SJ, Behnke MS, Dunay IR, Mashayekhi M, Rommereim LM, Fox BA, Bzik DJ, Taylor GA, Turk BE, Lichti CF, Townsend RR, Qiu W, Hui R, Beatty WL, Sibley LD. 2010. Phosphorylation of immunity-related GTPases by a Toxoplasma gondii-secreted kinase promotes macrophage survival and virulence. Cell Host Microbe 8:484–495. doi: 10.1016/j.chom.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fleckenstein MC, Reese ML, Konen-Waisman S, Boothroyd JC, Howard JC, Steinfeldt T. 2012. A Toxoplasma gondii pseudokinase inhibits host IRG resistance proteins. PLoS Biol 10:e1001358. doi: 10.1371/journal.pbio.1001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fox BA, Rommereim LM, Guevara RB, Falla A, Hortua Triana MA, Sun Y, Bzik DJ. 2016. The Toxoplasma gondii rhoptry kinome is essential for chronic infection. mBio 7:e00193-16. doi: 10.1128/mBio.00193-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niedelman W, Gold DA, Rosowski EE, Sprokholt JK, Lim D, Farid Arenas A, Melo MB, Spooner E, Yaffe MB, Saeij JP. 2012. The rhoptry proteins ROP18 and ROP5 mediate Toxoplasma gondii evasion of the murine, but not the human, interferon-gamma response. PLoS Pathog 8:e1002784. doi: 10.1371/journal.ppat.1002784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reese ML, Shah N, Boothroyd JC. 2014. The Toxoplasma pseudokinase ROP5 is an allosteric inhibitor of the immunity-related GTPases. J Biol Chem 289:27849–27858. doi: 10.1074/jbc.M114.567057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reese ML, Zeiner GM, Saeij JP, Boothroyd JC, Boyle JP. 2011. Polymorphic family of injected pseudokinases is paramount in Toxoplasma virulence. Proc Natl Acad Sci U S A 108:9625–9630. doi: 10.1073/pnas.1015980108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saeij JP, Boyle JP, Coller S, Taylor S, Sibley LD, Brooke-Powell ET, Ajioka JW, Boothroyd JC. 2006. Polymorphic secreted kinases are key virulence factors in toxoplasmosis. Science 314:1780–1783. doi: 10.1126/science.1133690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shwab EK, Jiang T, Pena HF, Gennari SM, Dubey JP, Su C. 2016. The ROP18 and ROP5 gene allele types are highly predictive of virulence in mice across globally distributed strains of Toxoplasma gondii. Int J Parasitol 46:141–146. doi: 10.1016/j.ijpara.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 33.Taylor S, Barragan A, Su C, Fux B, Fentress SJ, Tang K, Beatty WL, Hajj HE, Jerome M, Behnke MS, White M, Wootton JC, Sibley LD. 2006. A secreted serine-threonine kinase determines virulence in the eukaryotic pathogen Toxoplasma gondii. Science 314:1776–1780. doi: 10.1126/science.1133643. [DOI] [PubMed] [Google Scholar]
- 34.Foltz C, Napolitano A, Khan R, Clough B, Hirst EM, Frickel EM. 2017. TRIM21 is critical for survival of Toxoplasma gondii infection and localises to GBP-positive parasite vacuoles. Sci Rep 7:5209. doi: 10.1038/s41598-017-05487-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haldar AK, Foltz C, Finethy R, Piro AS, Feeley EM, Pilla-Moffett DM, Komatsu M, Frickel EM, Coers J. 2015. Ubiquitin systems mark pathogen-containing vacuoles as targets for host defense by guanylate binding proteins. Proc Natl Acad Sci U S A 112:E5628–E5637. doi: 10.1073/pnas.1515966112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang CS, Yuk JM, Lee YH, Jo EK. 2016. Toxoplasma gondii GRA7-induced TRAF6 activation contributes to host protective immunity. Infect Immun 84:339–350. doi: 10.1128/IAI.00734-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coppens I, Dunn JD, Romano JD, Pypaert M, Zhang H, Boothroyd JC, Joiner KA. 2006. Toxoplasma gondii sequesters lysosomes from mammalian hosts in the vacuolar space. Cell 125:261–274. doi: 10.1016/j.cell.2006.01.056. [DOI] [PubMed] [Google Scholar]
- 38.Rommereim LM, Bellini V, Fox BA, Petre G, Rak C, Touquet B, Aldebert D, Dubremetz JF, Cesbron-Delauw MF, Mercier C, Bzik DJ. 2016. Phenotypes associated with knockouts of eight dense granule gene loci (GRA2-9) in virulent Toxoplasma gondii. PLoS One 11:e0159306. doi: 10.1371/journal.pone.0159306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Díaz-Martín RD, Mercier C, Gómez de León CT, González RM, Pozos SG, Ríos-Castro E, García RA, Fox BA, Bzik DJ, Flores RM. 2019. The dense granule protein 8 (GRA8) is a component of the sub-pellicular cytoskeleton in Toxoplasma gondii. Parasitol Res 118:1899–1918. doi: 10.1007/s00436-019-06298-7. [DOI] [PubMed] [Google Scholar]
- 40.Rome ME, Beck JR, Turetzky JM, Webster P, Bradley PJ. 2008. Intervacuolar transport and unique topology of GRA14, a novel dense granule protein in Toxoplasma gondii. Infect Immun 76:4865–4875. doi: 10.1128/IAI.00782-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jensen KD, Hu K, Whitmarsh RJ, Hassan MA, Julien L, Lu D, Chen L, Hunter CA, Saeij JP. 2013. Toxoplasma gondii rhoptry 16 kinase promotes host resistance to oral infection and intestinal inflammation only in the context of the dense granule protein GRA15. Infect Immun 81:2156–2167. doi: 10.1128/IAI.01185-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jensen KD, Wang Y, Wojno ED, Shastri AJ, Hu K, Cornel L, Boedec E, Ong YC, Chien YH, Hunter CA, Boothroyd JC, Saeij JP. 2011. Toxoplasma polymorphic effectors determine macrophage polarization and intestinal inflammation. Cell Host Microbe 9:472–483. doi: 10.1016/j.chom.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosowski EE, Lu D, Julien L, Rodda L, Gaiser RA, Jensen KD, Saeij JP. 2011. Strain-specific activation of the NF-kappaB pathway by GRA15, a novel Toxoplasma gondii dense granule protein. J Exp Med 208:195–212. doi: 10.1084/jem.20100717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Souza W, Attias M. 2015. New views of the Toxoplasma gondii parasitophorous vacuole as revealed by helium ion microscopy (HIM). J Struct Biol 191:76–85. doi: 10.1016/j.jsb.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 45.Sibley LD, Niesman IR, Parmley SF, Cesbron-Delauw MF. 1995. Regulated secretion of multi-lamellar vesicles leads to formation of a tubulo-vesicular network in host-cell vacuoles occupied by Toxoplasma gondii. J Cell Sci 108(Pt 4):1669–1677. [DOI] [PubMed] [Google Scholar]
- 46.Periz J, Whitelaw J, Harding C, Gras S, Del Rosario Minina MI, Latorre-Barragan F, Lemgruber L, Reimer MA, Insall R, Heaslip A, Meissner M. 2017. Toxoplasma gondii F-actin forms an extensive filamentous network required for material exchange and parasite maturation. Elife 6:e24119. doi: 10.7554/eLife.24119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Charif H, Darcy F, Torpier G, Cesbron-Delauw MF, Capron A. 1990. Toxoplasma gondii: characterization and localization of antigens secreted from tachyzoites. Exp Parasitol 71:114–124. doi: 10.1016/0014-4894(90)90014-4. [DOI] [PubMed] [Google Scholar]
- 48.Mercier C, Cesbron-Delauw MF, Sibley LD. 1998. The amphipathic alpha helices of the toxoplasma protein GRA2 mediate post-secretory membrane association. J Cell Sci 111(Pt 15):2171–2180. [DOI] [PubMed] [Google Scholar]
- 49.Labruyere E, Lingnau M, Mercier C, Sibley LD. 1999. Differential membrane targeting of the secretory proteins GRA4 and GRA6 within the parasitophorous vacuole formed by Toxoplasma gondii. Mol Biochem Parasitol 102:311–324. doi: 10.1016/S0166-6851(99)00092-4. [DOI] [PubMed] [Google Scholar]
- 50.Lecordier L, Moleon-Borodowsky I, Dubremetz JF, Tourvieille B, Mercier C, Deslee D, Capron A, Cesbron-Delauw MF. 1995. Characterization of a dense granule antigen of Toxoplasma gondii (GRA6) associated to the network of the parasitophorous vacuole. Mol Biochem Parasitol 70:85–94. doi: 10.1016/0166-6851(95)00010-X. [DOI] [PubMed] [Google Scholar]
- 51.Adjogble KDZ, Mercier C, Dubremetz J-F, Hucke C, Mackenzie CR, Cesbron-Delauw M-F, Däubener W. 2004. GRA9, a new Toxoplasma gondii dense granule protein associated with the intravacuolar network of tubular membranes. Int J Parasitol 34:1255–1264. doi: 10.1016/j.ijpara.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 52.Michelin A, Bittame A, Bordat Y, Travier L, Mercier C, Dubremetz JF, Lebrun M. 2009. GRA12, a Toxoplasma dense granule protein associated with the intravacuolar membranous nanotubular network. Int J Parasitol 39:299–306. doi: 10.1016/j.ijpara.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 53.Lopez J, Bittame A, Massera C, Vasseur V, Effantin G, Valat A, Buaillon C, Allart S, Fox BA, Rommereim LM, Bzik DJ, Schoehn G, Weissenhorn W, Dubremetz JF, Gagnon J, Mercier C, Cesbron-Delauw MF, Blanchard N. 2015. Intravacuolar membranes regulate CD8 T cell recognition of membrane-bound Toxoplasma gondii protective antigen. Cell Rep 13:2273–2286. doi: 10.1016/j.celrep.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 54.Mercier C, Dubremetz JF, Rauscher B, Lecordier L, Sibley LD, Cesbron-Delauw MF. 2002. Biogenesis of nanotubular network in Toxoplasma parasitophorous vacuole induced by parasite proteins. Mol Biol Cell 13:2397–2409. doi: 10.1091/mbc.e02-01-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Travier L, Mondragon R, Dubremetz JF, Musset K, Mondragon M, Gonzalez S, Cesbron-Delauw MF, Mercier C. 2008. Functional domains of the Toxoplasma GRA2 protein in the formation of the membranous nanotubular network of the parasitophorous vacuole. Int J Parasitol 38:757–773. doi: 10.1016/j.ijpara.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 56.Ma JS, Sasai M, Ohshima J, Lee Y, Bando H, Takeda K, Yamamoto M. 2014. Selective and strain-specific NFAT4 activation by the Toxoplasma gondii polymorphic dense granule protein GRA6. J Exp Med 211:2013–2032. doi: 10.1084/jem.20131272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mercier C, Howe DK, Mordue D, Lingnau M, Sibley LD. 1998. Targeted disruption of the GRA2 locus in Toxoplasma gondii decreases acute virulence in mice. Infect Immun 66:4176–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baird JR, Fox BA, Sanders KL, Lizotte PH, Cubillos-Ruiz JR, Scarlett UK, Rutkowski MR, Conejo-Garcia JR, Fiering S, Bzik DJ. 2013. Avirulent Toxoplasma gondii generates therapeutic antitumor immunity by reversing immunosuppression in the ovarian cancer microenvironment. Cancer Res 73:3842–3851. doi: 10.1158/0008-5472.CAN-12-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fox BA, Sanders KL, Rommereim LM, Guevara RB, Bzik DJ. 2016. Secretion of rhoptry and dense granule effector proteins by nonreplicating Toxoplasma gondii uracil auxotrophs controls the development of antitumor immunity. PLoS Genet 12:e1006189. doi: 10.1371/journal.pgen.1006189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Romano JD, Sonda S, Bergbower E, Smith ME, Coppens I. 2013. Toxoplasma gondii salvages sphingolipids from the host Golgi through the rerouting of selected Rab vesicles to the parasitophorous vacuole. Mol Biol Cell 24:1974–1995. doi: 10.1091/mbc.E12-11-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Caffaro CE, Boothroyd JC. 2011. Evidence for host cells as the major contributor of lipids in the intravacuolar network of Toxoplasma-infected cells. Eukaryot Cell 10:1095–1099. doi: 10.1128/EC.00002-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nolan SJ, Romano JD, Coppens I. 2017. Host lipid droplets: an important source of lipids salvaged by the intracellular parasite Toxoplasma gondii. PLoS Pathog 13:e1006362. doi: 10.1371/journal.ppat.1006362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Romano JD, Nolan SJ, Porter C, Ehrenman K, Hartman EJ, Hsia RC, Coppens I. 2017. The parasite Toxoplasma sequesters diverse Rab host vesicles within an intravacuolar network. J Cell Biol 216:4235–4254. doi: 10.1083/jcb.201701108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dou Z, McGovern OL, Di Cristina M, Carruthers VB. 2014. Toxoplasma gondii ingests and digests host cytosolic proteins. mBio 5:e01188. doi: 10.1128/mBio.01188-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Di Cristina M, Dou Z, Lunghi M, Kannan G, Huynh MH, McGovern OL, Schultz TL, Schultz AJ, Miller AJ, Hayes BM, van der Linden W, Emiliani C, Bogyo M, Besteiro S, Coppens I, Carruthers VB. 2017. Toxoplasma depends on lysosomal consumption of autophagosomes for persistent infection. Nat Microbiol 2:17096. doi: 10.1038/nmicrobiol.2017.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fox BA, Falla A, Rommereim LM, Tomita T, Gigley JP, Mercier C, Cesbron-Delauw MF, Weiss LM, Bzik DJ. 2011. Type II Toxoplasma gondii KU80 knockout strains enable functional analysis of genes required for cyst development and latent infection. Eukaryot Cell 10:1193–1206. doi: 10.1128/EC.00297-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ferguson DJ. 2004. Use of molecular and ultrastructural markers to evaluate stage conversion of Toxoplasma gondii in both the intermediate and definitive host. Int J Parasitol 34:347–360. doi: 10.1016/j.ijpara.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 68.Tu V, Mayoral J, Sugi T, Tomita T, Han B, Ma YF, Weiss LM. 2019. Enrichment and proteomic characterization of the cyst wall from in vitro Toxoplasma gondii cysts. mBio 10:e00469-19. doi: 10.1128/mBio.00469-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alonso AM, Corvi MM, Diambra L. 2019. Gene target discovery with network analysis in Toxoplasma gondii. Sci Rep 9:646. doi: 10.1038/s41598-018-36671-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Boothroyd JC, Black M, Bonnefoy S, Hehl A, Knoll LJ, Manger ID, Ortega-Barria E, Tomavo S. 1997. Genetic and biochemical analysis of development in Toxoplasma gondii. Philos Trans R Soc Lond B Biol Sci 352:1347–1354. doi: 10.1098/rstb.1997.0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hammoudi PM, Jacot D, Mueller C, Di Cristina M, Dogga SK, Marq JB, Romano J, Tosetti N, Dubrot J, Emre Y, Lunghi M, Coppens I, Yamamoto M, Sojka D, Pino P, Soldati-Favre D. 2015. Fundamental roles of the Golgi-associated Toxoplasma aspartyl protease, ASP5, at the host-parasite interface. PLoS Pathog 11:e1005211. doi: 10.1371/journal.ppat.1005211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fox BA, Ristuccia JG, Gigley JP, Bzik DJ. 2009. Efficient gene replacements in Toxoplasma gondii strains deficient for nonhomologous end joining. Eukaryot Cell 8:520–529. doi: 10.1128/EC.00357-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Khaminets A, Hunn JP, Konen-Waisman S, Zhao YO, Preukschat D, Coers J, Boyle JP, Ong YC, Boothroyd JC, Reichmann G, Howard JC. 2010. Coordinated loading of IRG resistance GTPases on to the Toxoplasma gondii parasitophorous vacuole. Cell Microbiol 12:939–961. doi: 10.1111/j.1462-5822.2010.01443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Henry SC, Daniell XG, Burroughs AR, Indaram M, Howell DN, Coers J, Starnbach MN, Hunn JP, Howard JC, Feng CG, Sher A, Taylor GA. 2009. Balance of Irgm protein activities determines IFN-gamma-induced host defense. J Leukoc Biol 85:877–885. doi: 10.1189/jlb.1008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Maric-Biresev J, Hunn JP, Krut O, Helms JB, Martens S, Howard JC. 2016. Loss of the interferon-gamma-inducible regulatory immunity-related GTPase (IRG), Irgm1, causes activation of effector IRG proteins on lysosomes, damaging lysosomal function and predicting the dramatic susceptibility of Irgm1-deficient mice to infection. BMC Biol 14:33. doi: 10.1186/s12915-016-0255-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tu V, Yakubu R, Weiss LM. 2017. Observations on bradyzoite biology. Microbes Infect 20:466–476.doi: 10.1016/j.micinf.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bohne W, Heesemann J, Gross U. 1993. Coexistence of heterogeneous populations of Toxoplasma gondii parasites within parasitophorous vacuoles of murine macrophages as revealed by a bradyzoite-specific monoclonal antibody. Parasitol Res 79:485–487. doi: 10.1007/BF00931588. [DOI] [PubMed] [Google Scholar]
- 78.Bohne W, Heesemann J, Gross U. 1994. Reduced replication of Toxoplasma gondii is necessary for induction of bradyzoite-specific antigens: a possible role for nitric oxide in triggering stage conversion. Infect Immun 62:1761–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Miller CM, Smith NC, Johnson AM. 1999. Cytokines, nitric oxide, heat shock proteins and virulence in Toxoplasma. Parasitol Today 15:418–422. doi: 10.1016/S0169-4758(99)01515-X. [DOI] [PubMed] [Google Scholar]
- 80.Caffaro CE, Koshy AA, Liu L, Zeiner GM, Hirschberg CB, Boothroyd JC. 2013. A nucleotide sugar transporter involved in glycosylation of the Toxoplasma tissue cyst wall is required for efficient persistence of bradyzoites. PLoS Pathog 9:e1003331. doi: 10.1371/journal.ppat.1003331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tomita T, Sugi T, Yakubu R, Tu V, Ma Y, Weiss LM. 2017. Making home sweet and sturdy: Toxoplasma gondii ppGalNAc-Ts glycosylate in hierarchical order and confer cyst wall rigidity. mBio 8:e02048-16. doi: 10.1128/mBio.02048-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Craver MP, Knoll LJ. 2007. Increased efficiency of homologous recombination in Toxoplasma gondii dense granule protein 3 demonstrates that GRA3 is not necessary in cell culture but does contribute to virulence. Mol Biochem Parasitol 153:149–157. doi: 10.1016/j.molbiopara.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 83.Coffey MJ, Sleebs BE, Uboldi AD, Garnham A, Franco M, Marino ND, Panas MW, Ferguson DJ, Enciso M, O'Neill MT, Lopaticki S, Stewart RJ, Dewson G, Smyth GK, Smith BJ, Masters SL, Boothroyd JC, Boddey JA, Tonkin CJ. 2015. An aspartyl protease defines a novel pathway for export of Toxoplasma proteins into the host cell. Elife 4:e10809. doi: 10.7554/eLife.10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Butcher BA, Greene RI, Henry SC, Annecharico KL, Weinberg JB, Denkers EY, Sher A, Taylor GA. 2005. p47 GTPases regulate Toxoplasma gondii survival in activated macrophages. Infect Immun 73:3278–3286. doi: 10.1128/IAI.73.6.3278-3286.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim EW, Nadipuram SM, Tetlow AL, Barshop WD, Liu PT, Wohlschlegel JA, Bradley PJ. 2016. The Rhoptry pseudokinase ROP54 modulates Toxoplasma gondii virulence and host GBP2 loading. mSphere 1:e00045-16. doi: 10.1128/mSphere.00045-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Evans RJ, Sundaramurthy V, Frickel EM. 2018. The interplay of host autophagy and eukaryotic pathogens. Front Cell Dev Biol 6:118. doi: 10.3389/fcell.2018.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Coers J, Brown HM, Hwang S, Taylor GA. 2018. Partners in anti-crime: how interferon-inducible GTPases and autophagy proteins team up in cell-intrinsic host defense. Curr Opin Immunol 54:93–101. doi: 10.1016/j.coi.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Traver MK, Henry SC, Cantillana V, Oliver T, Hunn JP, Howard JC, Beer S, Pfeffer K, Coers J, Taylor GA. 2011. Immunity-related GTPase M (IRGM) proteins influence the localization of guanylate-binding protein 2 (GBP2) by modulating macroautophagy. J Biol Chem 286:30471–30480. doi: 10.1074/jbc.M111.251967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Haldar AK, Saka HA, Piro AS, Dunn JD, Henry SC, Taylor GA, Frickel EM, Valdivia RH, Coers J. 2013. IRG and GBP host resistance factors target aberrant, “non-self” vacuoles characterized by the missing of “self” IRGM proteins. PLoS Pathog 9:e1003414. doi: 10.1371/journal.ppat.1003414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nadipuram SM, Kim EW, Vashisht AA, Lin AH, Bell HN, Coppens I, Wohlschlegel JA, Bradley PJ. 2016. In vivo biotinylation of the Toxoplasma parasitophorous vacuole reveals novel dense granule proteins important for parasite growth and pathogenesis. mBio 7:e00808-16. doi: 10.1128/mBio.00808-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gold DA, Kaplan AD, Lis A, Bett GC, Rosowski EE, Cirelli KM, Bougdour A, Sidik SM, Beck JR, Lourido S, Egea PF, Bradley PJ, Hakimi MA, Rasmusson RL, Saeij JP. 2015. The Toxoplasma dense granule proteins GRA17 and GRA23 mediate the movement of small molecules between the host and the parasitophorous vacuole. Cell Host Microbe 17:642–652. doi: 10.1016/j.chom.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Franco M, Panas MW, Marino ND, Lee MC, Buchholz KR, Kelly FD, Bednarski JJ, Sleckman BP, Pourmand N, Boothroyd JC. 2016. A novel secreted protein, MYR1, is central to Toxoplasma's manipulation of host cells. mBio 7:e02231. doi: 10.1128/mBio.02231-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Marino ND, Panas MW, Franco M, Theisen TC, Naor A, Rastogi S, Buchholz KR, Lorenzi HA, Boothroyd JC. 2018. Identification of a novel protein complex essential for effector translocation across the parasitophorous vacuole membrane of Toxoplasma gondii. PLoS Pathog 14:e1006828. doi: 10.1371/journal.ppat.1006828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Naor A, Panas MW, Marino N, Coffey MJ, Tonkin CJ, Boothroyd JC. 2018. MYR1-dependent effectors are the major drivers of a host cell’s early response to Toxoplasma, including counteracting MYR1-independent effects. mBio 9:e02401-17. doi: 10.1128/mBio.02401-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fox BA, Bzik DJ. 2010. Avirulent uracil auxotrophs based on disruption of orotidine-5′-monophosphate decarboxylase elicit protective immunity to Toxoplasma gondii. Infect Immun 78:3744–3752. doi: 10.1128/IAI.00287-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fox BA, Gigley JP, Bzik DJ. 2004. Toxoplasma gondii lacks the enzymes required for de novo arginine biosynthesis and arginine starvation triggers cyst formation. Int J Parasitol 34:323–331. doi: 10.1016/j.ijpara.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 97.Gigley JP, Fox BA, Bzik DJ. 2009. Cell-mediated immunity to Toxoplasma gondii develops primarily by local Th1 host immune responses in the absence of parasite replication. J Immunol 182:1069–1078. doi: 10.4049/jimmunol.182.2.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kim L, Butcher BA, Denkers EY. 2004. Toxoplasma gondii interferes with lipopolysaccharide-induced mitogen-activated protein kinase activation by mechanisms distinct from endotoxin tolerance. J Immunol 172:3003–3010. doi: 10.4049/jimmunol.172.5.3003. [DOI] [PubMed] [Google Scholar]
- 99.Schneider AG, Abi Abdallah DS, Butcher BA, Denkers EY. 2013. Toxoplasma gondii triggers phosphorylation and nuclear translocation of dendritic cell STAT1 while simultaneously blocking IFNgamma-induced STAT1 transcriptional activity. PLoS One 8:e60215. doi: 10.1371/journal.pone.0060215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rommereim LM, Hortua Triana MA, Falla A, Sanders KL, Guevara RB, Bzik DJ, Fox BA. 2013. Genetic manipulation in Deltaku80 strains for functional genomic analysis of Toxoplasma gondii. J Vis Exp 2013:e50598. doi: 10.3791/50598:e50598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fox BA, Bzik DJ. 2002. De novo pyrimidine biosynthesis is required for virulence of Toxoplasma gondii. Nature 415:926–929. doi: 10.1038/415926a. [DOI] [PubMed] [Google Scholar]
- 102.Aldebert D, Hypolite M, Cavailles P, Touquet B, Flori P, Loeuillet C, Cesbron-Delauw MF. 2011. Development of high-throughput methods to quantify cysts of Toxoplasma gondii. Cytometry A 79:952–958. doi: 10.1002/cyto.a.21138. [DOI] [PubMed] [Google Scholar]
- 103.Gajria B, Bahl A, Brestelli J, Dommer J, Fischer S, Gao X, Heiges M, Iodice J, Kissinger JC, Mackey AJ, Pinney DF, Roos DS, Stoeckert CJ Jr, Wang H, Brunk BP. 2007. ToxoDB: an integrated Toxoplasma gondii database resource. Nucleic Acids Res 36:D553–D556. doi: 10.1093/nar/gkm981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Achbarou A, Mercereau-Puijalon O, Sadak A, Fortier B, Leriche MA, Camus D, Dubremetz JF. 1991. Differential targeting of dense granule proteins in the parasitophorous vacuole of Toxoplasma gondii. Parasitology 103(Pt 3):321–329. doi: 10.1017/S0031182000059837. [DOI] [PubMed] [Google Scholar]
- 105.Saavedra R, De Meuter F, Hérion P. 1990. Monoclonal antibodies identify new Toxoplasma gondii soluble antigens. Hybridoma 9:453–463. doi: 10.1089/hyb.1990.9.453. [DOI] [PubMed] [Google Scholar]
- 106.Carey KL, Donahue CG, Ward GE. 2000. Identification and molecular characterization of GRA8, a novel, proline-rich, dense granule protein of Toxoplasma gondii. Mol Biochem Parasitol 105:25–37. doi: 10.1016/S0166-6851(99)00160-7. [DOI] [PubMed] [Google Scholar]
- 107.Dobrowolski JM, Sibley LD. 1996. Toxoplasma invasion of mammalian cells is powered by the actin cytoskeleton of the parasite. Cell 84:933–939. doi: 10.1016/S0092-8674(00)81071-5. [DOI] [PubMed] [Google Scholar]
- 108.Rodriguez C, Afchain D, Capron A, Dissous C, Santoro F. 1985. Major surface protein of Toxoplasma gondii (p30) contains an immunodominant region with repetitive epitopes. Eur J Immunol 15:747–749. doi: 10.1002/eji.1830150721. [DOI] [PubMed] [Google Scholar]
- 109.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tobin C, Pollard A, Knoll L. 2010. Toxoplasma gondii cyst wall formation in activated bone marrow-derived macrophages and bradyzoite conditions. J Vis Exp 2010:2091. doi: 10.3791/2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed. National Academies Press, Washington, DC. doi: 10.17226/12910. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GRA knockout and complementation strategies. (A) Knockout strategy to insert HXGPRT at deleted GRA gene loci by selection with mycophenolic acid (MPA) and xanthine (X). Double crossovers occurred at the GRA GOI locus when the PruΔku80Δhxgprt strain allowed the replacement of the targeted GRA coding sequence with that of the HXGPRT resistance marker, leading to the isolation of the PruΔku80Δgra(GOI)::HXGPRT strain. Location of the genotype validation PCRs is shown above the schematized PruΔku80Δgra(GOI)::HXGPRT genome. (B) Complementation strategy at the UPRT locus through selection with 5-fluorodeoxyuridine (FUDR). A double crossover at the UPRT locus allowed replacement of the UPRT coding sequence by the GRA12 5′ UTR followed by the GRA12 coding sequence; leading to the isolation of PruΔku80Δgra(GOI)::HXGPRT Δuprt::GRA12. Locations of the genotype validation PCRs are shown above the schematized PruΔku80Δgra(GOI)::HXGPRT genome. Download FIG S1, TIF file, 0.5 MB (493.3KB, tif) .
Copyright © 2019 Fox et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Validation of the GRA knockouts by IFA and immunoblotting. (A) Validation of GRA gene deletion in type II Δgra parasite mutant strains by IFA using specific anti-GRA antibodies. (B) Validation of GRA12 deletion in RHΔgra12 parasites by immunoblotting using rat anti-GRA12 polyclonal antibodies. (C) Validation of the GRA12 deletion in RHΔgra12 parasites by IFA using rat anti-GRA12 polyclonal antibodies. Download FIG S2, TIF file, 1.0 MB (1,022.1KB, tif) .
Copyright © 2019 Fox et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Parasite strains used or developed in this study. Download Table S1, DOC file, 0.1 MB (55KB, doc) .
Copyright © 2019 Fox et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Virulence of Δgra parasites and rescue of Δgra4 virulence. (A) PruΔgra4 and PruΔgra6 parasites differentiate to GFP+ cysts in vitro at high pH. (B) C57BL/6 mice were infected i.p. with 2 × 106 tachyzoites of various PruΔgra parasite strains, and virulence was measured. (C) The GRA4 gene rescues PruΔgra4 virulence. ***, P < 0.005. Download FIG S3, TIF file, 0.4 MB (447.3KB, tif) .
Copyright © 2019 Fox et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Replication rates. Replication rates of PruΔgra parasites were measured in infected HFF cells in a 45-h assay (see Materials and Methods). Download FIG S4, TIF file, 0.2 MB (222.2KB, tif) .
Copyright © 2019 Fox et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Amino acid alignments of GRA12 expressed by type I (GT1), type II (ME49), and type III (VEG) strains. Protein alignments and amino acid similarity and identity scores are shown for GRA12 expressed by the type I strain (TGGT_288650), type II strain ME49 (TGME49_288650), and type III strain VEG (TGVEG_288650). Download FIG S5, DOCX file, 0.1 MB (99.4KB, docx) .
Copyright © 2019 Fox et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used to construct and validate GRA knockouts. Download Table S2, DOCX file, 0.04 MB (42.4KB, docx) .
Copyright © 2019 Fox et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used to construct plasmids for complementation and validation of GRA mutant strains. Download Table S3, DOCX file, 0.03 MB (30.7KB, docx) .
Copyright © 2019 Fox et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.