Abstract
Except for drinking water, most beverages taste bitter or sweet. Taste perception and preferences are heritable and determinants of beverage choice and consumption. Consumption of several bitter- and sweet-tasting beverages has been implicated in development of major chronic diseases. We performed a genome-wide association study (GWAS) of self-reported bitter and sweet beverage consumption among ~370 000 participants of European ancestry, using a two-staged analysis design. Bitter beverages included coffee, tea, grapefruit juice, red wine, liquor and beer. Sweet beverages included artificially and sugar sweetened beverages (SSBs) and non-grapefruit juices. Five loci associated with total bitter beverage consumption were replicated (in/near GCKR, ABCG2, AHR, POR and CYP1A1/2). No locus was replicated for total sweet beverage consumption. Sub-phenotype analyses targeting the alcohol, caffeine and sweetener components of beverages yielded additional loci: (i) four loci for bitter alcoholic beverages (GCKR, KLB, ADH1B and AGBL2); (ii) five loci for bitter non-alcoholic beverages (ANXA9, AHR, POR, CYP1A1/2 and CSDC2); (iii) 10 loci for coffee; six novel loci (SEC16B, TMEM18, OR8U8, AKAP6, MC4R and SPECC1L-ADORA2A); (iv) FTO for SSBs. Of these 17 replicated loci, 12 have been associated with total alcohol consumption, coffee consumption, plasma caffeine metabolites or BMI in previous GWAS; none was involved in known sweet and bitter taste transduction pathways. Our study suggests that genetic variants related to alcohol consumption, coffee consumption and obesity were primary genetic determinants of bitter and sweet beverage consumption. Whether genetic variants related to taste perception are associated with beverage consumption remains to be determined.
Introduction
Beverages such as coffee, tea, soft drinks, beer and wine are widely consumed and important sources of energy, nutrients and phytochemicals (1–4). Beverage consumption has been implicated in numerous diseases. Coffee or tea consumption has been inversely associated with cardiovascular disease and mortality, type 2 diabetes, cognitive decline and specific cancers, but positively associated with adverse pregnancy outcomes (1,2,5). Alcohol consumption is related to more than 200 diseases and accounts for ~6% of death globally (3). Sugar sweetened beverages (SSBs) are a major source of added sugar and a well-known risk factor for cardiometabolic disease (6,7).
Given the widespread consumption of beverages and their substantial health implications, understanding environmental and genetic factors contributing to beverage choice and consumption level has important nutritional and broader public health implications. Knowledge of genetic determinants may inform the causal role beverages have in health and the potential population subgroups most susceptible to the health consequences of regular consumption. However, our current genetic understanding of beverage consumption is limited to coffee, alcohol and milk, for which primary genetic determinants are related to metabolism of specific constituents (8–10).
Taste perception and preferences are heritable and determinants of food and beverage choices and consumption (11,12). A recent Mendelian randomization analysis reported that bitter perception was causally associated with coffee, tea and alcohol consumption (13). Genetic variation in sweet taste receptors (e.g. TAS1R2) has been related to dietary sugar intake (14,15). Except for drinking water, most common beverages taste predominantly bitter (e.g. coffee, tea, beer, red wine, liquor, grapefruit juice) or sweet [e.g. SSBs, artificially sweetened beverages (ASBs)] (16). Targeting one inherent factor (i.e. taste) common to all beverages may minimize the impact of beverage-specific non-taste factors (e.g. social influences, fluid texture) and reveal whether genetically determined sensitivity to bitter and sweet taste is associated with beverage consumption. Accordingly, we conducted a genome-wide association study (GWAS) of consumption of bitter- and sweet-tasting beverages by leveraging genetic and phenotypic data from over 370 000 adults of European ancestry.
Results
Supplementary Material, Figure S1 summarizes the two-staged study design beginning with a discovery analysis in the UK Biobank followed by replication in three independent US cohorts. Phenotype definitions are shown in Supplementary Material, Table S1. UK Biobank participants were slightly older and presented with higher body mass index (BMI), higher energy consumption, higher total bitter beverage consumption and lower total sweet beverage consumption, compared to participants from the replication cohorts (Table 1). Stage 1 manhattan and quantile-quantile (QQ) plots are shown in Supplementary Material Figures S2–S16. Genomic inflation factors (λGC) ranged from 1.02 to 1.29 for bitter phenotypes and from 1.01 to 1.06 for sweet phenotypes. Linkage disequilibrium (LD) score regression intercepts were between 1.003 and 1.018 for all bitter and sweet beverage phenotypes. Figure 1 and Table 2 summarize all replicated single nucleotide polymorphism (SNP)-phenotype associations that are introduced in detail below. Corresponding regional plots are presented in Supplementary Material, Figures S17–S43. Detailed results of stage 1 and 2 analyses for all the independent SNPs selected for replication are presented in Supplementary Material, Table S2. Models without BMI adjustment generated similar results to those with BMI adjustment (Supplementary Material, Table S3). BMI adjusted results are presented as primary findings to reduce variance of phenotypes and to account for genetic effects mediated through body composition.
Table 1.
Demographic and dietary characteristics of the study participants
| Stage 1: Discovery | Stage 2: Replication | ||||
|---|---|---|---|---|---|
| UK Biobank 24 h recall data (n = 85 852) | UK Biobank assessment center data (n = 336 448) | Nurses’ Health Study (n = 10 675) | Health Professionals Follow-Up Study (n = 6618) | Women’s Genome Health Study (n = 22 691) | |
| Age, years | 56.5 (7.7) | 56.9 (8.0) | 52.5 (6.7) | 55.2 (8.7) | 54.7 (7.1) |
| Women | 47 786 (55.7%) | 181 271 (53.7%) | 10 675 (100%) | 0 (0%) | 22 691 (100%) |
| BMI, kg/m2 | 26.7 (4.6) | 27.4 (4.7) | 25.5 (4.9) | 25.8 (3.2) | 25.3 (6.7) |
| Total energy, kcal | 2044.0 (432.8) | .. | 1773.7 (489.3) | 1931.4 (551.5) | 1732.4 (524.9) |
| Bitter beverages, servings/day | |||||
| Total | 5.33 (4.00–6.60) | .. | 3.29 (2.11–4.71) | 3.00 (1.43–4.71) | 1.45 (1.12–1.80) |
| Bitter alcoholic beverages | 0.38 (0–1.47) | 0.43 (0–1.29) | 0.07 (0–0.39) | 0.21 (0–0.93) | 0.07 (0–0.19) |
| Bitter non-alcoholic beverages | 4.50 (3.50–5.50) | .. | 2.90 (1.86–4.29) | 2.50 (1.00–3.50) | 1.37 (1.02–1.73) |
| Coffee | 1.50 (0.50–2.67) | 1.10 (0.41–1.39) | 2.50 (1.00–3.50) | 2.00 (0.43–2.64) | 1.25 (0.69–1.50) |
| Tea | 2.67 (1.25–4.00) | 1.39 (0.69–1.79) | 0.22 (0.04–0.90) | 0.07 (0–0.43) | 0.13 (0–0.69) |
| Grapefruit juice | 0 (0–0) | .. | 0 (0–0.07) | 0 (0–0.07) | 0 (0–0.07) |
| Sweet beverages, servings/day | |||||
| Total | 0.83 (0.33–1.50) | .. | 1.22 (0.71–1.93) | 1.21 (0.64–1.93) | 0.86 (0.54–1.29) |
| SSBs | 0 (0–0.50) | .. | 0.07 (0–0.25) | 0.07 (0–0.35) | 0.07 (0–0.24) |
| ASBs | 0 (0–0) | .. | 0.21 (0–0.79) | 0.14 (0–0.71) | 0.36 (0–0.73) |
| Pure non-grapefruit juices | 0.25 (0–0.67) | .. | 0.54 (0.14–1.00) | 0.50 (0.14–1.00) | 0.36 (0.07–0.69) |
ASBs, artificially sweetened beverages; BMI, body mass index; SSBs, sugar sweetened beverages. Values are n (%) or mean (SD) or median (25th percentile to 75th percentile). One serving of a beverage can be 1 cup (e.g. coffee, tea), 1 medium glass (e.g. fruit juice, wine), 1 shot/measure (e.g. liquor/spirit) or 1 pint/bottle/can (e.g. beer, carbonated beverages).
Figure 1.
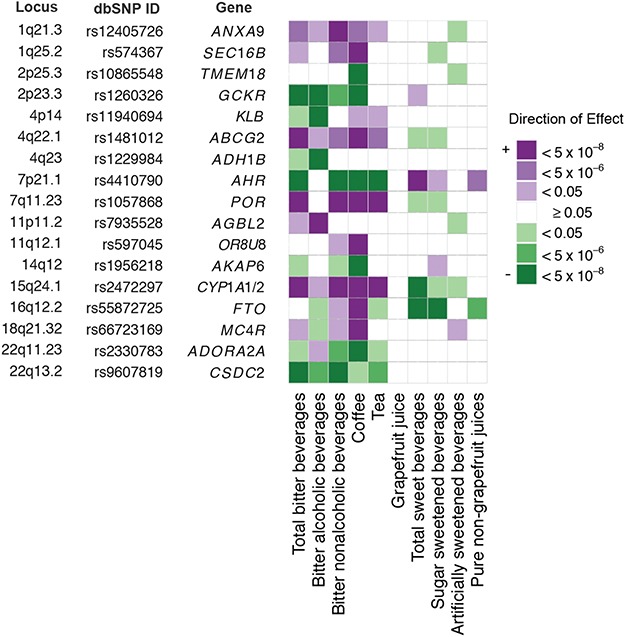
Pleiotropic associations of beverage-based loci with different total and sub-types of beverages. Shown are the P values and direction of the associations between the most significant replicated independent SNP in each locus and each of the primary phenotypes and sub-phenotypes, based on joint meta-analysis of stage 1 and stage 2 samples. These SNPs were replicated for at least one primary phenotype or sub-phenotype. We chose the minor alleles as the effect alleles. Information on locus, SNP number and closest gene is presented here and further details can be found in Table 2. The direction and strength of the associations are indicated by color. Results with P ≥ 0.05 are not colored. Of note, a genome-wide significant SNP-phenotype association may be seen due to the very small P from stage 1 analysis, although this SNP did not satisfy our replication criteria as specified in methods (e.g. CYP1A1/2 for total sweet beverages).
SNPs associated with primary bitter and sweet phenotypes
A total of 2038 SNPs were nominally (P < 5 × 10−6) associated with total bitter beverage consumption in the UK Biobank and 37 were independent and followed up for replication. Five independent SNPs mapping to five loci were replicated in/near 2p23.3 (GCKR), 4q22.1 (ABCG2), 7p21.1 (AHR), 7q11.23 (POR) and 15q24.1 (CYP1A1/2) (Table 2). SNPs in/near 16q12.2 (FTO) and AHR reached genome-wide significance for total sweet beverage consumption in the UK Biobank, but failed to replicate in stage 2 (Supplementary Material, Table S2).
SNPs associated with bitter sub-phenotypes
Seven independent SNPs mapping to five loci were replicated for bitter non-alcoholic beverage consumption. Two loci were distinct from our primary bitter phenotype, 1q21.3 (ANXA9) and 22q13.2 (CSDC2). We replicated four loci for bitter alcoholic beverage consumption including 2p23.3 (GCKR), 4p14 (KLB), 4q23 (ADH1B) and 11p11.2 (AGBL2). Fifteen independent SNPs spanning 10 loci were replicated for coffee consumption. Six loci were novel, 1q25.2 (SEC16B), 2p25.3 (TMEM18), 11q12.1 (OR8U8), 14q12 (AKAP6), 18q21.32 (MC4R) and 22q11.23 (SPECC1L-ADORA2A). To further determine whether these six novel loci can be replicated in a published GWAS of coffee consumption based on HapMap R22 (8), we re-ran the stage 1 meta-analysis in this published study after excluding NHS, HPFS and WGHS cohorts to avoid overlap with the current study (N = ~ 61 000). Only TMEM18 and MC4R reached a P < 0.01. No SNP was replicated for grapefruit juice and tea consumption.
Coffee, tea and bitter alcoholic beverage consumption were available from both assessment center visit and 24 h recalls. The direction of effect was consistent between two data sources for 334 (~98.5%) of all the 339 SNP-phenotype associations (Supplementary Material, Table S4). Post hoc analysis found that loci (e.g. GCKR, ADH1B and KLB) associated with bitter alcoholic beverage consumption were also associated with non-bitter alcoholic beverage consumption that included white wine, rose wine and fortified wine.
SNPs associated with sweet sub-phenotypes
Fifty-one SNPs mapping to 47 loci were nominally (P < 5 × 10−6) associated with one or more sweet sub-phenotypes in stage 1. Only the association between the SNP rs55872725 in FTO and SSB consumption fulfilled our replication criteria. All other 96 BMI loci had a P value of 0.002 or larger based on joint meta-analysis (Supplementary Material, Table S5).
Pleiotropic associations of beverage-based loci with different total and sub-types of beverages
A heat map was created using the most significant independent SNP from each locus that was replicated for at least one primary phenotype or sub-phenotype to compare SNP-phenotype associations across all phenotypes. Strengths and directions of SNP-phenotype associations were similar across total bitter beverages, bitter non-alcoholic beverages and coffee, based on joint meta-analysis (Fig. 1). SNPs in three loci were associated with bitter and sweet phenotypes at genome-wide significance level but in opposition directions: (i) AHR at 7p21.1 (rs4410790) and CYP1A1/2 at 15p24.1 (rs2472297) for total bitter and total sweet beverages; (ii) FTO at 16q12.2 (rs55872725) for coffee and SSBs. Post hoc analysis of UK Biobank data was conducted to assess a potential substitution effect. We observed a moderate inverse correlation between consumption of total bitter and total sweet beverages (energy-adjusted Pearson r = −0.24, P < 0.0001; Supplementary Material, Table S6). Further adjusting for total sweet beverage consumption did not materially alter the replicated associations of AHR and CYP1A1/2 with total bitter beverage consumption (Supplementary Material, Table S7). Coffee and SSB consumption were weakly correlated with an energy-adjusted Pearson correlation coefficient of −0.04 (P < 0.0001). Adjusting for coffee consumption minimally changed the replicated FTO-SSB consumption association.
Functional annotation and gene expression
Functional annotation, mapping and gene expression analyses of the full stage 1 GWAS results were conducted with FUMA (17). Among the five phenotypes with replicated loci, independent SNPs and those in LD (r2 ≥ 0.3) within 500 kb were mostly located in intergenic (36.6%, n = 1943) and intronic areas (45.1%, n = 2395). Stage 1 GWAS results for bitter alcoholic beverages were significantly enriched for gene candidates expressed in the brain and pituitary (Supplementary Material, Figures S44–S48). GWAS results for coffee were enriched for candidate genes expressed in the brain and adrenal gland. No gene expression enrichment was found for other phenotypes. Of the 32 independent SNPs or their proxies (r2 ≥ 0.8) shown in Table 2, 8 were non-synonymous (Supplementary Material, Table S8).
Candidate SNP analyses
Using the stage 1 GWAS results, we looked up SNPs mapping to 68 candidate genes based on the sweet and bitter taste transduction pathways as defined by Kegg (18). SNPs in five (TAS2R38, TAS2R31, TAS2R1, TAS2R10, TAS2R42) of the 68 candidate taste genes were at least nominally associated with consumption of bitter alcoholic beverages, coffee or tea in stage 1 (3.8 × 10−14 < P < 5 × 10−7). None of these associations were replicated in stage 2 (Supplementary Material, Table S9), although a SNP in TAS2R38 (rs713598) had a replication P value of 0.03 for tea consumption, close to the pre-specified threshold of 0.01.
An inverse association between BMI increasing T allele of FTO (rs55872725) and SSB consumption was identified even among those with BMI < 30 kg/m2 (beta = −0.014 per T allele, P < 0.0001) and BMI <25 kg/m2 (beta = −0.0121 per T allele, P = 0.002).
SNP-heritability estimates (hg2) and proportion of variance explained (r2) for bitter and sweet phenotypes
SNP-heritability (hg2) of bitter and sweet phenotypes in the UK Biobank was estimated using BOLT-REML v2.3.2 (19) and LD score regression (20). Phenotype hg2 estimates from individual-level data (BOLT, no covariates, genotyped SNPs) averaged 1.9-fold higher than corresponding hg2 estimates from summary statistics (LD score, covariate adjusted, imputed SNPs) (Supplementary Material, Table S10). BOLT hg2 for total bitter and total sweet beverages were 0.15 (0.01) and 0.07 (0.01), respectively. The highest SNP-heritability was observed for bitter non-alcoholic beverages (hg2 = 0.16 [0.01]) and the lowest was for grapefruit juice (hg2 = 0.02 [0.01]). LD score estimates were similar for summary statistics with or without BMI adjustment. Proportion of variance explained (r2) for individual lead SNPs ranged from 0.0001 (rs7935528, rs574367, rs597045, rs1956218 and rs2330783) to 0.0053 (rs2472297) (Supplementary Material, Table S11). r2 for all the lead SNPs individually added to the model for each beverage ranged from 0.0003 for SSB consumption and 0.0114 for bitter non-alcoholic beverage consumption.
Table 2.
Independent SNPs associated with beverage consumption that met pre-specified replication criteria1
| Locus | Chr:pos | dbSNP ID | Closest gene | EA/RA | EAF | Stage 1 (n = 85 852) | Stage 2 (n = 39 924) | Joint meta-analysis | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta2 | SE2 | P | Beta2 | SE2 | P | Beta2 | SE2 | P | N | ||||||
| Total bitter beverages | |||||||||||||||
| 2p23.3 | 2:27730940 | rs1260326 | GCKR | C/T | 0.61 | 1.05 | 0.18 | 2.87E-09 | 1.63 | 0.45 | 3.46E-04 | 1.13 | 0.16 | 8.33E-12 | 125 776 |
| 4q22.1 | 4:89039082 | rs1481012 | ABCG2 | A/G | 0.89 | 1.42 | 0.27 | 2.09E-07 | 2.10 | 0.71 | 3.49E-03 | 1.50 | 0.25 | 3.82E-09 | 125 776 |
| 7p21.1 | 7:17284577 | rs4410790 | AHR | C/T | 0.63 | 2.58 | 0.18 | 2.36E-46 | 3.71 | 0.46 | 3.36E-15 | 2.73 | 0.17 | 7.09E-59 | 125 776 |
| 7q11.23 | 7:75616105 | rs17685 | POR | A/G | 0.28 | 1.35 | 0.19 | 2.71E-12 | 2.50 | 0.50 | 7.64E-07 | 1.50 | 0.18 | 1.02E-16 | 125 776 |
| 15q24.1 | 15:75027880 | rs2472297 | CYP1A1/2 | T/C | 0.26 | 3.42 | 0.19 | 6.58E-67 | 3.42 | 0.54 | 4.74E-10 | 3.43 | 0.18 | 1.81E-75 | 125 776 |
| Bitter non-alcoholic beverages | |||||||||||||||
| 1q21.3 | 1:150956128 | rs12405726 | ANXA9 | A/G | 0.35 | 1.05 | 0.18 | 5.90E-09 | 1.49 | 0.45 | 1.01E-03 | 1.12 | 0.17 | 3.50E-11 | 125 776 |
| 7p21.1 | 7:17284577 | rs4410790 | AHR | C/T | 0.63 | 2.97 | 0.18 | 2.94E-61 | 3.87 | 0.44 | 1.16E-17 | 3.10 | 0.16 | 1.39E-76 | 125 776 |
| 7p21.1 | 7:17401027 | rs7791070 | KCCAT333 | T/C | 0.76 | 1.67 | 0.20 | 2.44E-16 | 2.33 | 0.52 | 8.63E-06 | 1.76 | 0.19 | 2.09E-20 | 125 776 |
| 7q11.23 | 7:73047551 | rs35855035 | MLXIPL | T/C | 0.22 | 1.05 | 0.21 | 4.71E-07 | 1.47 | 0.53 | 5.43E-03 | 1.11 | 0.19 | 1.15E-08 | 125 776 |
| 7q11.23 | 7:75615006 | rs1057868 | POR | T/C | 0.29 | 1.71 | 0.19 | 5.17E-19 | 2.41 | 0.47 | 5.50E-07 | 1.81 | 0.18 | 3.95E-24 | 125 776 |
| 15q24.1 | 15:75027880 | rs2472297 | CYP1A1/2 | T/C | 0.26 | 3.93 | 0.19 | 1.30E-87 | 3.88 | 0.52 | 3.04E-13 | 3.93 | 0.18 | 2.20E-99 | 125 776 |
| 22q13.2 | 22:41958862 | rs9607819 | CSDC2 | G/C | 0.81 | 1.29 | 0.22 | 4.56E-09 | 1.45 | 0.54 | 7.43E-03 | 1.31 | 0.20 | 1.20E-10 | 125 776 |
| Bitter alcoholic beverages 3 | |||||||||||||||
| 2p23.3 | 2:27730940 | rs1260326 | GCKR | C/T | 0.61 | 1.41 | 0.11 | 2.78E-34 | 0.87 | 0.24 | 2.87E-04 | 1.31 | 0.10 | 2.52E-36 | 376 372 |
| 4p14 | 4:39414993 | rs11940694 | KLB | G/A | 0.60 | 1.34 | 0.12 | 1.07E-30 | 0.92 | 0.24 | 1.37E-04 | 1.26 | 0.10 | 2.25E-33 | 376 372 |
| 4q23 | 4:100239319 | rs1229984 | ADH1B | C/T | 0.98 | 10.43 | 0.38 | 1.08E-151 | 3.60 | 0.64 | 2.48E-08 | 8.60 | 0.32 | 2.00E-142 | 376 372 |
| 11p11.2 | 11:47722819 | rs7935528 | AGBL2 | A/G | 0.45 | 0.75 | 0.11 | 2.83E-11 | 0.71 | 0.24 | 3.12E-03 | 0.74 | 0.10 | 3.33E-13 | 376 372 |
| Coffee 4 | |||||||||||||||
| 1q25.2 | 1:177873210 | rs574367 | SEC16B | T/G | 0.21 | 0.99 | 0.19 | 2.61E-07 | 1.62 | 0.58 | 5.19E-03 | 1.05 | 0.18 | 8.06E-09 | 375 833 |
| 2p25.3 | 2:631606 | rs10865548 | TMEM18 | G/A | 0.83 | 1.52 | 0.21 | 2.35E-13 | 1.66 | 0.59 | 5.11E-03 | 1.54 | 0.19 | 4.46E-15 | 375 833 |
| 2p23.3 | 2:27730940 | rs1260326 | GCKR | C/T | 0.61 | 1.31 | 0.16 | 2.92E-16 | 1.79 | 0.46 | 1.22E-04 | 1.36 | 0.15 | 2.62E-19 | 375 833 |
| 7p21.1 | 7:172776925 | rs117692895 | AHR | C/G | 0.01 | 4.01 | 0.71 | 2.60E-08 | 7.12 | 2.23 | 1.86E-03 | 4.29 | 0.67 | 4.13E-10 | 374 046 |
| 7p21.1 | 7:17284577 | rs4410790 | AHR | C/T | 0.63 | 3.85 | 0.16 | 4.73E-121 | 4.63 | 0.47 | 4.20E-22 | 3.94 | 0.15 | 5.59E-141 | 375 833 |
| 7p21.1 | 7:17334899 | rs4719497 | AHR | T/C | 0.87 | 1.12 | 0.23 | 1.10E-06 | 1.81 | 0.67 | 7.27E-03 | 1.20 | 0.22 | 4.23E-08 | 375 833 |
| 7p21.1 | 7:17443199 | rs12699844 | KCCAT333 | C/T | 0.47 | 1.22 | 0.16 | 1.16E-14 | 1.37 | 0.46 | 2.93E-03 | 1.24 | 0.15 | 1.35E-16 | 375 833 |
| 7p21.1 | 7:17562952 | rs73073176 | LOC101927630 | C/T | 0.87 | 2.30 | 0.23 | 1.54E-22 | 2.34 | 0.69 | 8.47E-04 | 2.31 | 0.22 | 5.56E-25 | 375 833 |
| 7q11.23 | 7:73037956 | rs34060476 | MLXIPL | G/A | 0.13 | 1.79 | 0.23 | 8.05E-15 | 2.78 | 0.68 | 5.23E-05 | 1.89 | 0.22 | 5.06E-18 | 375 833 |
| 7q11.23 | 7:75615006 | rs1057868 | POR | T/C | 0.29 | 1.88 | 0.17 | 2.44E-27 | 2.72 | 0.50 | 1.09E-07 | 1.97 | 0.16 | 5.26E-33 | 375 833 |
| 11q12.1 | 11:56272114 | rs597045 | OR8U8 | A/T | 0.69 | 1.02 | 0.17 | 2.86E-09 | 1.41 | 0.50 | 4.98E-03 | 1.07 | 0.16 | 6.62E-11 | 375 833 |
| 14q12 | 14:33075243 | rs1956218 | AKAP6 | G/A | 0.56 | 0.77 | 0.16 | 1.27E-06 | 1.30 | 0.45 | 4.52E-03 | 0.82 | 0.15 | 3.62E-08 | 375 833 |
| 15q24.1 | 15:75027880 | rs2472297 | CYP1A1/2 | T/C | 0.27 | 4.54 | 0.18 | 8.28E-141 | 4.57 | 0.56 | 8.69E-16 | 4.54 | 0.17 | 5.19E-155 | 375 833 |
| 18q21.32 | 18:57808978 | rs66723169 | MC4R | A/C | 0.23 | 1.41 | 0.19 | 3.32E-14 | 1.91 | 0.54 | 4.82E-04 | 1.47 | 0.18 | 9.88E-17 | 375 833 |
| 22q11.23 | 22:24747031 | rs2330783 | SPECC1L-ADORA2A | G/T | 0.99 | 4.44 | 0.67 | 6.20E-11 | 5.25 | 1.91 | 6.72E-03 | 4.53 | 0.63 | 1.57E-12 | 375 833 |
| SSBs | |||||||||||||||
| 16q12.2 | 16:53809123 | rs55872725 | FTO | C/T6 | 0.59 | 0.96 | 0.15 | 1.29E-10 | 1.28 | 0.21 | 6.36E-10 | 1.07 | 0.12 | 1.05E-18 | 125 776 |
Chr:pos, chromosome:position. EA, effect allele; EAF, effect allele frequency; RA, reference allele; SE, standard error.
1Independent SNPs were identified using conditional analysis. Independent SNPs with P < 5 × 10−6 in stage 1 were replicated if (i) stage 2 P < 0.01; (ii) joint stage 1 and stage 2 meta-analysis P < 5 × 10−8; and (iii) consistent direction of effect.
2Betas and SEs are interpreted as % change per allele.
3 n = 336 448 in stage 1.
4 n = 335 909 in stage 1.
5 n = 38 137 in stage 2.
6T allele is the BMI increasing allele.
Comparing to the previous GWAS literature
Of the 17 replicated loci in our study (Supplementary Material, Table S12), five have been associated with coffee consumption or plasma caffeine metabolites (GCKR, ABCG2, AHR, POR and CYP1A1/2) (8,21); three have been associated with total alcohol consumption (GCKR, KLB and ADH1B) (9,22); and five have been associated with BMI (SEC16B, TMEM18, AKAP6, FTO and MC4R) (23,24). For AKAP6, we found a different independent SNP compared to the previous GWAS literature, which may be due to the use of a recent reference panel.
Discussion
Beverages are a frequently consumed part of the human diet and their health benefits and risks have been extensively studied. Taste perception and preferences are heritable and determinants of food/beverage choice and consumption (11,12). The current study, however, suggests that genetic variants related to coffee consumption, alcohol consumption and obesity were predominant determinants of bitter and sweet beverage consumption, as defined by our groupings.
A few taste loci (e.g. TAS2R31, TAS2R38) were associated with bitter beverage consumption in the UK Biobank (P < 5 × 10−6) including bitter alcoholic beverages, coffee and tea. However, none was replicated, although TAS2R38 had a replication P value of 0.03 for tea consumption, close to the pre-specified threshold of 0.01. These findings are consistent with a recent GWAS (n = 4611), which reported that none of the 15 loci associated with 12 specific food liking (including coffee) were related to any known taste gene (25). The available genome-wide discovery efforts targeting taste perception commonly use chemical compounds that are not found in real-world beverages to assess perceived taste intensity (e.g. propylthiouracil) (26). The extent to which these findings translate to actual consumption is unclear. Leveraging data on beverage consumption, as opposed to perception or liking, has more direct public health implications. Our study is the largest and most comprehensive GWAS to identify genetic variants related to consumption of real-world bitter and sweet beverages in a general population.
When all bitter beverages were considered together, the associated loci mapped to genes implicated in caffeine pharmacology (8,21). Indeed, when our primary bitter phenotype was parsed into sub-phenotypes, some of these loci (e.g. AHR, POR, CYP1A1/A2) emerged for coffee (a major and concentrated source of caffeine), which is consistent with GWAS of coffee/caffeine-related traits (8,21). However, six novel loci for coffee consumption were additionally identified. ADORA2A and OR8U8 are biologically plausible candidates. ADORA2A encodes the adenosine 2A receptor, a key target for caffeine (27). OR8U8 encodes a member of the olfactory receptor family and thus functions in smell perception (28). Smell and taste are correlated sensory characteristics that impact food intake (29). SEC16B, TMEM18 and MC4R are confirmed BMI-loci (23). The BMI-increasing variants were associated with higher coffee consumption in our study. A recent Mendelian randomization study also reported that a BMI-increasing allele score comprising of FTO, MC4R and TMEM18 variants was associated with higher coffee consumption (30). FTO, TMEM18 and MC4R are highly expressed in hypothalamus, a brain region regulating reward and motivation aspects of eating behavior (31). AKAP6 has been associated with a number of traits such as BMI in a trans-ancestral meta-analysis and cognitive function (24,32). Our study, which focused on beverage consumption, also contributes to the growing evidence that a subset of BMI loci contribute to behavioral aspects of obesity by altering food intake and preference (33,34). Our study did not replicate previously reported coffee loci at 11p13 (BDNF), 12q24.1 (HECTD4) and 17q11.2 (EFCAB5) (8,35). This may be attributable to the study differences in coffee phenotype definition, sample characteristics and modeling approach.
Tea is generally a caffeine-containing beverage, although the caffeine content is much lower than coffee (36). SNPs near caffeine-related loci such as AHR and CYP1A1/2 were associated with tea consumption (P < 5 × 10−8) in the UK Biobank, but they were not replicated in the US cohorts. Tea is a major contributor to caffeine in UK, but not US, populations. Replication may have been impeded by the strong cultural differences for tea drinking behaviors between the two countries.
We found that variants in FTO previously linked to higher BMI were associated with lower SSB consumption regardless of BMI adjustment. This inverse association remained even among those with BMI < 25 kg/m2. FTO is the only locus that was replicated among 97 BMI loci (23). The inverse association between BMI increasing allele of FTO and SSB consumption is consistent with a previous candidate gene study (37). This ‘counterintuitive’ inverse association requires further verification in future studies. An interaction between SSB consumption and a genetic score comprising of 32 BMI-related SNPs including FTO in relation to obesity was previously identified, but the direct FTO-SSB consumption association was not explored (38). Accumulating evidence has suggested that the effect of FTO on beverage choice might depend on the form (liquid versus solid) and energy density (high-caloric versus low-caloric), independent from FTO’s effect on BMI (37,39,40). Nonetheless, our findings reiterate the complexity surrounding the FTO-BMI association.
Heritability of bitter and sweet taste perception or preferences ranges from ~0.3 to ~0.7 and varies by compound, food/beverage and study sample based on twin studies (n < 10 000) (41–43). SNP-heritability estimates presented in the current study cannot directly compare to heritability estimates reported in twin studies (44), but we can discuss relative to other traits wherein similar heritability estimates are available. SNP-heritability estimates for bitter and sweet beverage consumption were ≤0.16, lower than other common traits in UK Biobank participants such as height (0.58), BMI (0.31) and blood pressure (0.27) but similar to type 2 diabetes (0.07) and cardiovascular disease (0.16) (19). The explained phenotypic variation by replicated loci in our study was ~1% or less, which is comparable to complex behavioral traits such as smoking and alcohol-related behaviors (45,46). Our low estimates may be related to categorization of beverage taste based on self-recalled taste intensity in a different study (16), variation in taste intensity within each type of beverages, and other limitations discussed below. Also, strong environment effects (e.g. demographic, social, occupational, cultural and others) may have masked genetic effects (47).
We performed a comprehensive assessment of genetic architecture of common sweet and bitter beverage consumption. Replication of results from a UK population in a US population enhances generalizability and reduces false positive results. However, limitations in the replication sample including smaller sample size, different dietary assessment tools than those used in the UK Biobank as well as cultural differences in beverage consumption behaviors (e.g. tea) between these two countries may have reduced our ability to replicate more loci. Further, measurement error of self-reported dietary intake is inevitable, although the dietary assessment tools implemented in participating cohorts have been validated (48,49). Nonetheless, evidence has shown that beverages including coffee, tea, alcohol and carbonated beverages were among the most accurately recalled and most reproducible aspects of the diet; the corrected correlation coefficients were between 0.34–0.94 in the NHS comparing dietary records and food frequency questionnaire (FFQ) (50). Sex-specific analysis was not performed due to the smaller stage 2 sample. Lastly, one should be cautious in generalizing our results to other populations of non-European ancestry.
In conclusion, genetic variants related to coffee consumption, alcohol consumption and obesity were critical genetic determinants of bitter and sweet beverage consumption. Whether genetic variants related to taste perception are associated with beverage consumption remains to be determined.
Materials and Methods
Stage 1: discovery
Study population
The UK Biobank is a prospective cohort study that recruited over 500 000 participants aged 37–73 years from 22 centers across England, Wales and Scotland in 2006–2010. All study participants provided informed consent and the study was approved by the North West Multi-center Research Ethics Committee, the Community Health Index Advisory Group, the Patient Information Advisory Group and the National Health Service National Research Ethics Service.
Diet data collection and phenotype definitions.
Dietary intake was collected using a 24 h recall questionnaire (Oxford WebQ) in a subset of UK Biobank participants (48). We used the mean intake from participants who completed at least two dietary recalls. We defined two phenotypes for primary analysis. The first, total bitter beverages, included coffee, tea, grapefruit juice and bitter tasting alcoholic beverages (beer/cider, red wine and liquor) (Supplementary Material, Table S1). The second, total sweet beverages, included SSBs, ASBs, pure non-grapefruit juices, flavored milk and hot chocolate. The grouping rationale was based on a previous study finding that recalled taste intensity was associated with self-reported liking and habitual consumption (16). To further inform the origination and interpretation of primary results, we subsequently conducted GWAS of ‘beverage-sets’ (sub-phenotypes; Supplementary Material, Table S1). These sub-phenotypes targeted other components including caffeine, sugar, artificial sweeteners and alcohol; these are components with well-known genetic determinants or distinct tastes (8,9,47).
Self-reported consumption of alcohol, coffee and tea was also collected from all participants at baseline using a touchscreen questionnaire at each assessment center. These data were only used for sub-phenotype analysis to maximize the power for finding novel loci. Participants were asked about their alcohol drinking status (never, former, current, unknown), frequency (weekly or monthly) and types (e.g. red wine, white wine, beer/cider, spirit). Total coffee consumption was based on the question ‘How many cups of coffee do you drink each day (include decaffeinated coffee)?’ A similar question was asked about tea.
Genotyping, imputation and quality control
All participants of UK Biobank were genotyped using Affymetrix UK BiLEVE Axiom and Affymetrix UK Biobank Axiom® arrays. Quality control and imputation to the Haplotype Reference Consortium (HRC) v1.1 and UK10K reference panels were performed centrally by the Wellcome Trust Centre for Human Genetics and University of Oxford (51). Sample outliers based on heterozygosity and missingness were excluded, as were participants with sex discrepancy between self-reported and inferred sex (using X-chromosome heterozygosity). Also excluded were participants with relatedness based on kinship coefficient of >0.0442. Finally, we restricted our analysis to those who self-reported as ‘British’ and who had very similar ancestral backgrounds based on results of principal component analysis (n = 337 542). The current analysis was restricted to the autosomal SNPs from the HRC v1.1 reference panel because UK10K imputation was not valid at time of analyses. SNPs with minor allele frequency <1% or with low imputation quality score ≤0.3 were excluded.
Statistical analysis
PLINK 2.0 (52) was used to conduct genome-wide analyses of our two primary phenotypes and eight sub-phenotypes using linear regressions assuming an additive genetic model. All phenotypes were natural log-transformed after adding one to each value and further winsorized at four standard deviations. Statistical models for phenotypes based on 24 h recall data were adjusted for age, sex, BMI, total energy, proportion of 24 h recalls self-reported as capturing ‘typical intake’ and top 20 principal components to account for population sub-structure. The same adjustment was applied to the three sub-phenotypes based on the touchscreen questionnaire with the exception of total energy and ‘typical intake’. A sensitivity analysis without BMI adjustment was performed to rule out the potential collider bias (53).
Chromosome-level conditional analysis was performed to identify independent SNPs. We re-ran the aforementioned model by additionally conditioning on the most significant SNP from each chromosome that had a P < 5 × 10−6. We repeated this analysis by conditioning all previously identified SNPs from each run until no SNP had a conditioned P < 5 × 10−6. The resulting independent SNPs were followed-up in stage 2 for replication.
Stage 2: replication
Replication was carried out in three independent populations of European ancestry: Nurses’ Health Study (n = 10 675), Health Professionals Follow-up Study (n = 6618) and Women’s Genome Health Study (n = 22 691) (Supplementary File). For these three replication cohorts, dietary data were collected by the same FFQ.
A meta-analysis of these three studies was conducted with a fixed-effect model and inverse-variance weighting using METAL (54). No genomic control was applied. A joint meta-analysis of stage 1 and stage 2 studies was also conducted using METAL as described above. Genome-wide significance was defined by the following criteria: (i) stage 2 P < 0.01; (ii) joint stage 1 and stage 2 meta-analysis P < 5 × 10−8; (iii) consistent direction of effect. The effects were interpreted as percent change in consumption level per allele: ([eβ-1]*100%).
We also conducted (i) functional annotation, mapping and gene expression analyses; (ii) candidate SNP analysis; and (iii) analysis to estimate SNP-heritability and proportion of variance explained for beverage phenotypes. The methodological details are presented in the Supplementary File. Finally, we searched the NHGRI-EBI Catalog of published GWAS for all genome-wide significant associations with our replicated independent SNPs and those in high LD (r2 ≥ 0.8) (55).
Data availability
Full stage 1 GWAS results are available at https://digitalhub.northwestern.edu/users/mcc340.
Supplementary Material
Acknowledgements
This research has been conducted using the UK Biobank Resource (Application #21394). Computations in this paper were run on the Quest cluster supported in part through the computational resources and staff contributions provided for the Quest high performance computing facility at Northwestern University, which is jointly supported by the Office of the Provost, the Office for Research, and Northwestern University Information Technology.
Conflict of Interest statement. The authors declare that they have no conflicts of interest related to the study.
Funding
American Heart Association Strategically Focused Research Networks [14SFRN20480260 to V.W.Z.], National Institute on Deafness and Other Communication Disorders [R03DC01337301A1 to M.C.C.], National Institute of Health grants [P01CA87969, P01CA055075, P01DK070756, U01HG004728, UM1CA186107, UM1CA167552, R01CA49449, R01CA50385, R01HL034594, R01HL088521, R01HL35464, R01EY015473, R01EY022305, P30EY014104, R03DC013373, R03CA165131 to the Nurses’ Health Study (NHS) and Health Professionals Follow-up Study (HPFS)], the National Heart, Lung, and Blood Institute [HL04385, HL080467 to The Women’s Genome Health Study (WGHS)] and the National Cancer Institute [CA047988, UM1CA182913 to The Women’s Genome Health Study (WGHS)], with funding for genotyping in the WGHS provided by Amgen.
References
- 1. Poole R., Kennedy O.J., Roderick P., Fallowfield J.A., Hayes P.C. and Parkes J. (2017) Coffee consumption and health: umbrella review of meta-analyses of multiple health outcomes. BMJ, 359, j5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vuong Q.V. (2014) Epidemiological evidence linking tea consumption to human health: a review. Crit. Rev. Food Sci. Nutr., 54, 523–536. [DOI] [PubMed] [Google Scholar]
- 3. World Health Organization (2014) Global Status Report on Alcohol and Health 2014. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 4. Vartanian L.R., Schwartz M.B. and Brownell K.D. (2007) Effects of soft drink consumption on nutrition and health: a systematic review and meta-analysis. Am. J. Public Health, 97, 667–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Okubo H., Miyake Y., Tanaka K., Sasaki S. and Hirota Y. (2015) Maternal total caffeine intake, mainly from Japanese and Chinese tea, during pregnancy was associated with risk of preterm birth: the Osaka maternal and child health study. Nutr. Res., 35, 309–316. [DOI] [PubMed] [Google Scholar]
- 6. Imamura F., O'Connor L., Ye Z., Mursu J., Hayashino Y., Bhupathiraju S.N. and Forouhi N.G. (2015) Consumption of sugar sweetened beverages, artificially sweetened beverages, and fruit juice and incidence of type 2 diabetes: systematic review, meta-analysis, and estimation of population attributable fraction. BMJ, 351, h3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Malik V.S., Pan A., Willett W.C. and Hu F.B. (2013) Sugar-sweetened beverages and weight gain in children and adults: a systematic review and meta-analysis. Am. J. Clin. Nutr., 98, 1084–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Coffee and Caffeine Genetics Consortium, Cornelis M.C., Byrne E.M., Esko T., Nalls M.A., Ganna A., Paynter N., Monda K.L., Amin N., Fischer K. et al. (2015) Genome-wide meta-analysis identifies six novel loci associated with habitual coffee consumption. Mol. Psychiatry, 20, 647–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Clarke T.K., Adams M.J., Davies G., Howard D.M., Hall L.S., Padmanabhan S., Murray A.D., Smith B.H., Campbell A., Hayward C. et al. (2017) Genome-wide association study of alcohol consumption and genetic overlap with other health-related traits in UK biobank (N=112 117). Mol. Psychiatry, 22, 1376–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Deng Y., Misselwitz B., Dai N. and Fox M. (2015) Lactose intolerance in adults: biological mechanism and dietary management. Nutrients, 7, 8020–8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Garcia-Bailo B., Toguri C., Eny K.M. and El-Sohemy A. (2009) Genetic variation in taste and its influence on food selection. OMICS, 13, 69–80. [DOI] [PubMed] [Google Scholar]
- 12. Hayes J.E., Feeney E.L. and Allen A.L. (2013) Do polymorphisms in chemosensory genes matter for human ingestive behavior? Food. Qual. Prefer., 30, 202–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ong J.S., Hwang D.L.D., Zhong V.W., An J.Y., Gharahkhani P., Breslin P.A.S., Wright M.J., Lawlor D.A., Whitfield J., MacGregor S. et al. (2018) Understanding the role of bitter taste perception in coffee, tea and alcohol consumption through Mendelian randomization. Sci. Rep., 8, 16414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Eny K.M., Wolever T.M., Corey P.N. and El-Sohemy A. (2010) Genetic variation in TAS1R2 (Ile191Val) is associated with consumption of sugars in overweight and obese individuals in 2 distinct populations. Am. J. Clin. Nutr., 92, 1501–1510. [DOI] [PubMed] [Google Scholar]
- 15. Dias A.G., Eny K.M., Cockburn M., Chiu W., Nielsen D.E., Duizer L. and El-Sohemy A. (2015) Variation in the TAS1R2 gene, sweet taste perception and intake of sugars. J. Nutrigenet. Nutrigenomics, 8, 81–90. [DOI] [PubMed] [Google Scholar]
- 16. Cornelis M.C., Tordoff M.G., El-Sohemy A. and van Dam R.M. (2017) Recalled taste intensity, liking and habitual intake of commonly consumed foods. Appetite, 109, 182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Watanabe K., Taskesen E., van Bochoven A. and Posthuma D. (2017) Functional mapping and annotation of genetic associations with FUMA. Nat. Commun., 8, 1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kanehisa M. and Goto S. (2000) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res., 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Loh P.R., Kichaev G., Gazal S., Schoech A.P. and Price A.L. (2018) Mixed-model association for biobank-scale datasets. Nat. Genet., 50, 906–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bulik-Sullivan B.K., Loh P.R., Finucane H.K., Ripke S., Yang J., Schizophrenia Working Group of the Psychiatric Genomics Consortium, Patterson N., Daly M.J., Price A.L. and Neale B.M. (2015) LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet., 47, 291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cornelis M.C., Kacprowski T., Menni C., Gustafsson S., Pivin E., Adamski J., Artati A., Eap C.B., Ehret G., Friedrich N. et al. (2016) Genome-wide association study of caffeine metabolites provides new insights to caffeine metabolism and dietary caffeine-consumption behavior. Hum. Mol. Genet., 25, 5472–5482. [DOI] [PubMed] [Google Scholar]
- 22. Jorgenson E., Thai K.K., Hoffmann T.J., Sakoda L.C., Kvale M.N., Banda Y., Schaefer C., Risch N., Mertens J., Weisner C. et al. (2017) Genetic contributors to variation in alcohol consumption vary by race/ethnicity in a large multi-ethnic genome-wide association study. Mol. Psychiatry, 22, 1359–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Locke A.E., Kahali B., Berndt S.I., Justice A.E., Pers T.H., Day F.R., Powell C., Vedantam S., Buchkovich M.L., Yang J. et al. (2015) Genetic studies of body mass index yield new insights for obesity biology. Nature, 518, 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Akiyama M., Okada Y., Kanai M., Takahashi A., Momozawa Y., Ikeda M., Iwata N., Ikegawa S., Hirata M., Matsuda K. et al. (2017) Genome-wide association study identifies 112 new loci for body mass index in the Japanese population. Nat. Genet., 49, 1458–1467. [DOI] [PubMed] [Google Scholar]
- 25. Pirastu N., Kooyman M., Traglia M., Robino A., Willems S.M., Pistis G., Amin N., Sala C., Karssen L.C., Van Duijn C. et al. (2016) A genome-wide association study in isolated populations reveals new genes associated to common food likings. Rev. Endocr. Metab. Disord., 17, 209–219. [DOI] [PubMed] [Google Scholar]
- 26. Hwang L.D., Gharahkhani P., Breslin P.A.S., Gordon S.D., Zhu G., Martin N.G., Reed D.R. and Wright M.J. (2018) Bivariate genome-wide association analysis strengthens the role of bitter receptor clusters on chromosomes 7 and 12 in human bitter taste. BMC Genomics, 19, 678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee C.F., Lai H.L., Lee Y.C., Chien C.L. and Chern Y. (2014) The A2A adenosine receptor is a dual coding gene: a novel mechanism of gene usage and signal transduction. J. Biol. Chem., 289, 1257–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Malnic B., Godfrey P.A. and Buck L.B. (2004) The human olfactory receptor gene family. Proc. Natl. Acad. Sci. U. S. A., 101, 2584–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McCrickerd K. and Forde C.G. (2016) Sensory influences on food intake control: moving beyond palatability. Obes. Rev., 17, 18–29. [DOI] [PubMed] [Google Scholar]
- 30. Nordestgaard A.T., Thomsen M. and Nordestgaard B.G. (2015) Coffee intake and risk of obesity, metabolic syndrome and type 2 diabetes: a Mendelian randomization study. Int. J. Epidemiol., 44, 551–565. [DOI] [PubMed] [Google Scholar]
- 31. Ahima R.S. and Antwi D.A. (2008) Brain regulation of appetite and satiety. Endocrinol. Metab. Clin. North Am., 37, 811–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Andrews S.J., Das D., Anstey K.J. and Easteal S. (2017) Association of AKAP6 and MIR2113 with cognitive performance in a population-based sample of older adults. Genes Brain Behav., 16, 472–478. [DOI] [PubMed] [Google Scholar]
- 33. Willer C.J., Speliotes E.K., Loos R.J., Li S., Lindgren C.M., Heid I.M., Berndt S.I., Elliott A.L., Jackson A.U., Lamina C. et al. (2009) Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat. Genet., 41, 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Loos R.J. and Yeo G.S. (2014) The bigger picture of FTO: the first GWAS-identified obesity gene. Nat. Rev. Endocrinol., 10, 51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nakagawa-Senda H., Hachiya T., Shimizu A., Hosono S., Oze I., Watanabe M., Matsuo K., Ito H., Hara M., Nishida Y. et al. (2018) A genome-wide association study in the Japanese population identifies the 12q24 locus for habitual coffee consumption: the J-MICC study. Sci. Rep., 8, 1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mitchell D.C., Knight C.A., Hockenberry J., Teplansky R. and Hartman T.J. (2014) Beverage caffeine intakes in the U.S. Food Chem. Toxicol., 63, 136–142. [DOI] [PubMed] [Google Scholar]
- 37. Brunkwall L., Ericson U., Hellstrand S., Gullberg B., Orho-Melander M. and Sonestedt E. (2013) Genetic variation in the fat mass and obesity-associated gene (FTO) in association with food preferences in healthy adults. Food Nutr. Res., 57. doi: 10.3402/fnr.v57i0.20028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Qi Q., Chu A.Y., Kang J.H., Jensen M.K., Curhan G.C., Pasquale L.R., Ridker P.M., Hunter D.J., Willett W.C., Rimm E.B. et al. (2012) Sugar-sweetened beverages and genetic risk of obesity. N. Engl. J. Med., 367, 1387–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hasselbalch A.L., Angquist L., Christiansen L., Heitmann B.L., Kyvik K.O. and Sorensen T.I. (2010) A variant in the fat mass and obesity-associated gene (FTO) and variants near the melanocortin-4 receptor gene (MC4R) do not influence dietary intake. J. Nutr., 140, 831–834. [DOI] [PubMed] [Google Scholar]
- 40. Sobczyk-Kopciol A., Broda G., Wojnar M., Kurjata P., Jakubczyk A., Klimkiewicz A. and Ploski R. (2011) Inverse association of the obesity predisposing FTO rs9939609 genotype with alcohol consumption and risk for alcohol dependence. Addiction, 106, 739–748. [DOI] [PubMed] [Google Scholar]
- 41. Hwang L.D., Breslin P.A.S., Reed D.R., Zhu G., Martin N.G. and Wright M.J. (2016) Is the association between sweet and bitter perception due to genetic variation? Chem. Senses, 41, 737–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Treur J.L., Boomsma D.I., Ligthart L., Willemsen G. and Vink J.M. (2016) Heritability of high sugar consumption through drinks and the genetic correlation with substance use. Am. J. Clin. Nutr., 104, 1144–1150. [DOI] [PubMed] [Google Scholar]
- 43. Vink J.M., Staphorsius A.S. and Boomsma D.I. (2009) A genetic analysis of coffee consumption in a sample of Dutch twins. Twin Res. Hum. Genet., 12, 127–131. [DOI] [PubMed] [Google Scholar]
- 44. Furlotte N.A., Heckerman D. and Lippert C. (2014) Quantifying the uncertainty in heritability. J. Hum. Genet., 59, 269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ware J.J. and Munafo M.R. (2015) Genetics of smoking behaviour. Curr. Top. Behav. Neurosci., 23, 19–36. [DOI] [PubMed] [Google Scholar]
- 46. Mies G.W., Verweij K.J.H., Treur J.L., Ligthart L., Fedko I.O., Hottenga J.J., Willemsen G., Bartels M., Boomsma D.I. and Vink J.M. (2018) Polygenic risk for alcohol consumption and its association with alcohol-related phenotypes: do stress and life satisfaction moderate these relationships? Drug Alcohol Depend., 183, 7–12. [DOI] [PubMed] [Google Scholar]
- 47. Feeney E., O'Brien S., Scannell A., Markey A. and Gibney E.R. (2011) Genetic variation in taste perception: does it have a role in healthy eating? Proc. Nutr. Soc., 70, 135–143. [DOI] [PubMed] [Google Scholar]
- 48. Galante J., Adamska L., Young A., Young H., Littlejohns T.J., Gallacher J. and Allen N. (2016) The acceptability of repeat internet-based hybrid diet assessment of previous 24-h dietary intake: administration of the Oxford WebQ in UK biobank. Br. J. Nutr., 115, 681–686. [DOI] [PubMed] [Google Scholar]
- 49. Rimm E.B., Giovannucci E.L., Stampfer M.J., Colditz G.A., Litin L.B. and Willett W.C. (1992) Reproducibility and validity of an expanded self-administered Semiquantitative food frequency questionnaire among male health-professionals. Am. J. Epidemiol., 135, 1114–1126. [DOI] [PubMed] [Google Scholar]
- 50. Salvini S., Hunter D.J., Sampson L., Stampfer M.J., Colditz G.A., Rosner B. and Willett W.C. (1989) Food-based validation of a dietary questionnaire: the effects of week-to-week variation in food consumption. Int. J. Epidemiol., 18, 858–867. [DOI] [PubMed] [Google Scholar]
- 51. Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O'Connell J. et al. (2018) The UK biobank resource with deep phenotyping and genomic data. Nature, 562, 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M. and Lee J.J. (2015) Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience, 4, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Aschard H., Vilhjalmsson B.J., Joshi A.D., Price A.L. and Kraft P. (2015) Adjusting for heritable covariates can bias effect estimates in genome-wide association studies. Am. J. Hum. Genet., 96, 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Willer C.J., Li Y. and Abecasis G.R. (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics, 26, 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E., Junkins H., McMahon A., Milano A., Morales J. et al. (2017) The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res., 45, D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Full stage 1 GWAS results are available at https://digitalhub.northwestern.edu/users/mcc340.


