To better understand the diversity of primary producers in the Arctic Ocean, we targeted a nitrogen cycle gene, NR, which is required for phytoplankton to assimilate nitrate into organic forms of nitrogen macromolecules. We compared this to the more detailed taxonomy from ice-influenced stations using a general taxonomic gene (18S rRNA). NR genes were ubiquitous and could be classified as belonging to diatoms, dinoflagellates, other flagellates, chlorophytes, and unknown microbial eukaryotes, suggesting novel diversity of both species and metabolism in arctic phytoplankton.
KEYWORDS: Arctic, amplicon sequencing, chlorophytes, diatoms, metagenomes, nitrate reductase
ABSTRACT
For photosynthetic microbial eukaryotes, the rate-limiting step in NO3− assimilation is its reduction to nitrite (NO2−), which is catalyzed by assimilatory nitrate reductase (NR). Oceanic productivity is primarily limited by available nitrogen and, although nitrate is the most abundant form of available nitrogen in oceanic waters, little is known about the identity of microbial eukaryotes that take up nitrate. This lack of knowledge is especially severe for ice-covered seas that are being profoundly affected by climate change. To address this, we examined the distribution and diversity of NR genes in the Arctic region by way of clone libraries and data mining of available metagenomes (total of 4.24 billion reads). We directly compared NR clone phylogenies with the V4 region of the 18S rRNA gene (DNA pool) and 18S rRNA (RNA pool) at two ice-influenced stations in the Canada Basin (Beaufort Sea). The communities from the two nucleic acid templates were similar at the level of major groups, and species identified by way of NR gene phylogeny and microscopy were a subset of the 18S results. Most NR genes from arctic clone libraries matched diatoms and chromist nanoflagellates, including novel clades, while the NR genes in arctic eukaryote metagenomes were dominated by chlorophyte NR, in keeping with the ubiquitous occurrence of Mamiellophyceae in the Arctic Ocean. Overall, these data suggest that a dynamic and mixed eukaryotic community utilizes nitrate across the Arctic region, and they show the potential utility of NR as a tool to identify ongoing changes in arctic photosynthetic communities.
IMPORTANCE To better understand the diversity of primary producers in the Arctic Ocean, we targeted a nitrogen cycle gene, NR, which is required for phytoplankton to assimilate nitrate into organic forms of nitrogen macromolecules. We compared this to the more detailed taxonomy from ice-influenced stations using a general taxonomic gene (18S rRNA). NR genes were ubiquitous and could be classified as belonging to diatoms, dinoflagellates, other flagellates, chlorophytes, and unknown microbial eukaryotes, suggesting novel diversity of both species and metabolism in arctic phytoplankton.
INTRODUCTION
Primary production over many of the world’s oceans, especially the Arctic Ocean, is limited by nitrogen (1) and light (2) availability. Light is limiting in the Arctic Ocean due to solar inclination and ice cover for much of the year. Nutrients, especially nitrogen as nitrate, are limiting in surface waters due to strong salinity stratification (3); in the Beaufort Sea and Canada Basin, nutrients are linked to relatively fresher, but nitrate-poor, Pacific Ocean-origin waters lying atop nutrient-rich Atlantic Ocean-derived deep water. This stratification, which persists even in winter, inhibits vertical mixing, making surface waters oligotrophic year-round, with little in the way of any source of nitrogen (4, 5). While nitrate and chlorophyll a (chl a) concentrations remain very low in the surface waters, a subsurface chlorophyll maximum (SCM) layer forms at the top of the halocline as soon as light becomes available when seasonal ice melts. Climate change is already having a large impact in the Arctic Ocean, compared to the global average (6, 7), and there has been a dramatic loss of multiyear sea ice over the past few decades (8), with summer ice minima being below long-term averages from September 2007 to the present (9). Multiyear ice melt has also added freshwater to the upper Arctic Ocean, increasing stratification (10, 11) and potentially further limiting the transfer of nutrients from deep waters into the euphotic zone (12). Arctic phytoplankton thus face two challenges, namely, a need to be adapted to low light levels and the ability to use nitrate as a source of nitrogen.
The majority of biologically available nitrogen in the ocean is in the form of nitrate (NO3−), and assimilation of NO3− constitutes a major pathway for the formation of organic nitrogen (13). Nitrogen and carbon cycles are tightly coupled; the assimilation of NO3− requires its reduction to NO2− by nitrate reductase (NR) and then to ammonium by NO2− reductase. Ammonium is then incorporated into organic matter in the cell via glutamine synthetase and glutamate synthase as part of the glutamine synthetase/glutamate synthase cycle, which is linked to carbon metabolism. In the surface ocean, NO3− uptake is frequently used to estimate carbon export in the world’s oceans (14) and hence the fluxes of carbon into and out of the atmosphere. Based on empirical observations in coastal systems (15), diatoms are mostly responsible for nitrate uptake. Further, since diatoms are readily lost from the upper euphotic zone directly by sedimentation or through incorporation into zooplankton fecal pellets, the presence of diatoms normally indicates high carbon export potential (13). However, other phytoplankton with lower export potential are also able to use nitrate as a nitrogen source (16, 17). Because of this, knowledge of the species that take up upper-ocean NO3− is critical for understanding global carbon budgets (18).
Assimilatory NR catalyzes the limiting step in the reduction of nitrate in the cell and consequently controls the rate of this pathway. Therefore, earlier studies focused on NR as a marker of nitrogen status in algal cultures and different ecosystems (15, 19–21). Eukaryotic assimilatory NR (EC 1.6.6.1-3) forms are homologs and separate from bacterial forms that enable nitrate respiration. The eukaryotic NR genes tend to reproduce general eukaryotic phylogeny (16, 22); for example, diatoms are monophyletic and divergent from other algae, higher plants, and fungi. Taking advantage of these characteristics, Allen et al. (22) developed pan-eukaryotic primers, which have been used to investigate gene diversity in freshwater and marine eukaryotic cultures (16, 23) and nearshore marine phytoplankton communities, with an emphasis on diatoms (22, 24). Ward (17), using a microarray populated with (among other targets) sequences generated from the aforementioned NR primers, noted large NR signals from diverse uncultivated marine phytoplankton and remarked on the paucity of protist sequences in the public databases to which to compare them. In general, little is known regarding the diversity and distribution of NR genes in offshore marine waters, and there have been no previous studies from polar waters. It is generally assumed that diatoms at the ice edge and other arctic phytoplankton with cell sizes of >20 μm are responsible for most of the NO3− uptake (25, 26). However, smaller diatoms and flagellates (5 to 20 μm) are abundant throughout the Arctic Ocean euphotic zone (27–30) and may also contribute substantially to uptake.
Many 18S rRNA gene surveys in the Arctic Ocean have mostly targeted particles of 0.2 to 3 μm or 0.8 to 10 μm collected on filters (31–33) or selected by flow cytometry (34), with few studies specifically targeting >3-μm taxa (which would be expected to include diatoms). In addition, most surveys have used DNA as a template and, since DNA can persist in the environment (35), it may not accurately reflect which taxa are actively engaged in protein synthesis at the time of sampling. Recent studies comparing 18S rRNA in environmental RNA (cDNA) and 18S rRNA genes in environmental DNA (herein referred to collectively as simply 18S) have highlighted differences between these two approaches (36–38). For arctic marine waters, Terrado et al. (32) noted that 18S clone libraries based on RNA contained more diatoms than DNA libraries during certain seasons, highlighting the utility of using both DNA and RNA as templates in gene surveys.
The main objectives of this study were to investigate the diversity and phylogeny of NR genes using cloning and Sanger sequencing of products from the aforementioned eukaryotic primers and to compare these to NR genes from non-PCR-based, publicly available, arctic metagenomes. Given that the sampling occurred in August 2007 in a region of the Arctic Ocean previously covered year-round by multiyear ice, another overarching goal was to place the NR gene distribution and taxonomic affiliations into a wider arctic context. To fill a gap in knowledge regarding the overall taxonomic composition of communities in one of the last remaining high-summer-ice regions in the Canada Basin, we targeted the V4 region of 18S from both DNA and RNA of arctic nanoplankton communities using high-throughput sequencing (HTS). Additionally, Sanger sequencing of larger 18S fragments was undertaken for comparison with the HTS survey.
RESULTS
Sampling strategy and overall station profiles.
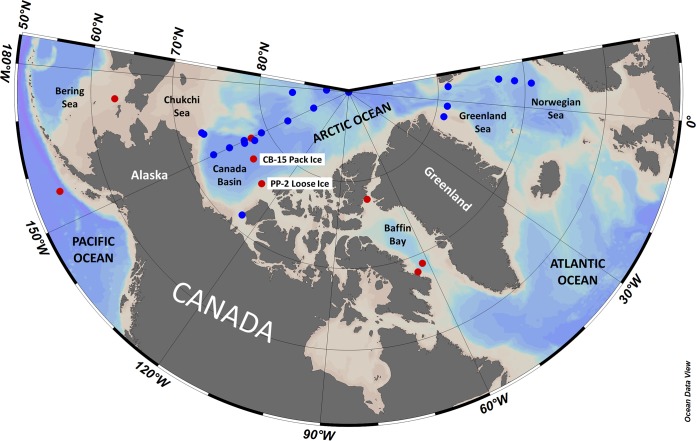
Samples were collected as part of three oceanographic expeditions on board Canadian Coast Guard icebreakers; samples from the SCM at eight stations, as well as one surface sample, were collected (Table 1 and Fig. 1). Canada Basin samples were collected in August 2007, just prior to the then-record September 2007 ice minimum in a region not previously subjected to seasonal ice melt. The surface sample, which was collected from the pack-ice-covered Canada Basin (CB-15) station, was relatively fresh (salinity of 24.11). Salinity values for the SCM samples from the North Pacific to Baffin Bay ranged from 30.13 to 32.71. Nutrient concentrations were lowest in the Canada Basin surface sample and greatest in the Bering Sea sample. The total chl a levels were 0.03 μg liter−1 in the surface sample and 0.13 to 2.25 μg liter−1 in the SCM samples, with the highest values being from Baffin Bay (BB-8-SCM) and northern Baffin Bay (L9-SCM); these two stations also had the greatest proportions of large phytoplankton (Table 1).
TABLE 1.
Sample and station characteristics of the locations physically visited in this study
| Characteristic | CB-15-surfa | CB-15-SCM | PP-2-SCM | CB-9-SCMb | L9-SCMb | BB-5-SCMb | BB-8-SCMb | NP-12-SCMb | SLIP-4-SCMb |
|---|---|---|---|---|---|---|---|---|---|
| Sampling characteristics | |||||||||
| Region | CB | CB | CB | CB | NBB | BB | BB | AGyre | BerS |
| Date of sampling | 14 August 2007 | 14 August 2007 | 20 August 2007 | 10 August 2007 | 19 August 2005 | 12 July 2007 | 13 July 2007 | 10 July 2008 | 16 July 2008 |
| Latitude (°N) | 76.969 | 76.969 | 75.845 | 77.934 | 77.833 | 68.831 | 68.091 | 53.712 | 63.003 |
| Longitude (°E) | −140.075 | −140.075 | −128.642 | −149.826 | −75.346 | −61.760 | −64.006 | −156.126 | −173.456 |
| Depth of sampling (m) | 5 | 57 | 70 | 47 | 27 | 28 | 40 | 50 | 28 |
| Physicochemical characteristics | |||||||||
| Salinity | 24.11 | 31.13 | 30.91 | 30.71 | 31.88 | 32.70 | 32.44 | 32.71 | 32.70 |
| Temperature (°C) | −0.83 | +0.27 | −1.15 | −0.41 | +1.11 | −1.56 | −1.18 | +5.14 | −1.39 |
| Nitrate level (μM) | bd | 5.6 | 4.3 | 0.7 | —c | 5.4 | 1.4 | 12.3 | 10.4 |
| Phosphate level (μM) | 0.55 | 1.27 | 1.19 | 0.63 | — | 0.97 | 0.78 | 1.31 | 1.77 |
| Silicate level (μM) | 2.4 | 12.4 | 9.9 | 5.6 | — | 9.2 | 2.9 | 24.5 | 26.7 |
| Biological characteristics | |||||||||
| Total chl a level (μg liter−1) | 0.03 | 0.32 | 0.13 | 0.29 | 1.87 | 0.68 | 2.25 | 0.23 | 0.45 |
| Chl a large fraction (>3 μm) (%) | 29 | 42 | 10 | 27 | 87 | 25 | 95 | — | 24 |
| DNA 18S pyrosequencing (no. of OTU0.98nt) | 1,063 | 1,018 | 1,173 | — | — | — | — | — | — |
| RNA 18S pyrosequencing (no. of OTU0.98nt) | 1,296 | 1,087 | — | — | — | — | — | — | — |
| DNA 18S clones (no. of OTU0.99nt) | 16 | 12 | 15 | — | — | — | — | — | — |
| RNA 18S clones (no. of OTU0.99nt) | 18 | 14 | 7 | — | — | — | — | — | — |
| DNA NR clones (no. of OTU0.94aa) | 8 | 2 | 3 | 2 | 4 | 5 | 10 | 5 | 4 |
| RNA NR clones (no. of OTU0.94aa) | 3 | 2 | 5 | 4 | — | 4 | 3 | 5 | 2 |
CB, Canada Basin; surf, surface waters; AGyre, Alaskan Gyre; BerS, Bering Sea; (N)BB, (northern) Baffin Bay; bd, below detection; NR, nitrate reductase; OTU0.98nt, 18S pyrosequencing OTUs clustered at 98% nucleotide identity; OTU0.99nt, 18S clone OTUs clustered at 99% nucleotide identity; OTU0.94aa, NR clone OTUs clustered at 94% amino acid identity; SCM, subsurface chlorophyll a maximum.
NR-only samples.
—, missing data.
FIG 1.
Map of stations that were sampled in this study for 18S and NR (red) and that were the sources of metagenomes mined from public databases (blue) across the Arctic region and the North Pacific Ocean. NR clones were sequenced from all stations in red, whereas 18S diversity surveys (clones and HTS) were conducted at the two highlighted stations (CB-15 and PP-2) in the Canada Basin (see Table 1 for details). Metagenomes from stations in blue were screened for NR genes; one station was analyzed from the Amundsen Gulf, six stations in the Greenland Sea and Norwegian Sea were part of the Joint Genome Institute (JGI) Sea of Change study, four stations in the Canada Basin were part of the JGI Western Arctic Ocean study, and nine stations across the Canada Basin and larger Arctic Ocean were part of the University of Alaska—Fairbanks GEOTRACES study (see Table S2 in the supplemental material for details). Map created with Ocean Data View (R. Schlitzer, Ocean Data View, https://odv.awi.de, 2018).
18S diversity in the Canada Basin.
The 18S diversity survey was carried out at two Canada Basin stations (specifically labeled in Fig. 1); station PP-2 was in 80% unconsolidated ice with patches of open water, whereas station CB-15 was in solid pack (multiyear) ice. HTS resulted in 1,018 to 1,296 operational taxonomic units (OTUs) (clustered at 98% similarity) per sample (Table 1), from the 6,237 equalized reads for each, with a nonredundant total of 2,923 OTUs for the entire data set. For convenience, we refer to the communities according to their origin (i.e., pack ice [CB-15] versus loose ice [PP-2]) and source template (DNA versus RNA). Overall richness was high, with individual samples having only 273 to 667 OTUs shared with any other sample (full pairwise comparisons). The Bray-Curtis community similarity tree, in which branch lengths were long and tight clusters did not form, reflected this (see Fig. S1A in the supplemental material). However, SCM samples clustered apart from the pack ice surface communities. The topologies of the trees were similar for HTS and the more limited 18S clones that were obtained. OTU and sequence counts (Fig. S1B) between the templates showed that, overall, the RNA communities were more variable (less shared between stations for RNA than for DNA). For both templates, however, the shared OTUs (24% to 57%) contained much of the sequence space (31% to 81%), and the taxonomic groups showed very good overall correlations between them at both the coarser subgroup level and the finer genus level (Fig. S2A and B).
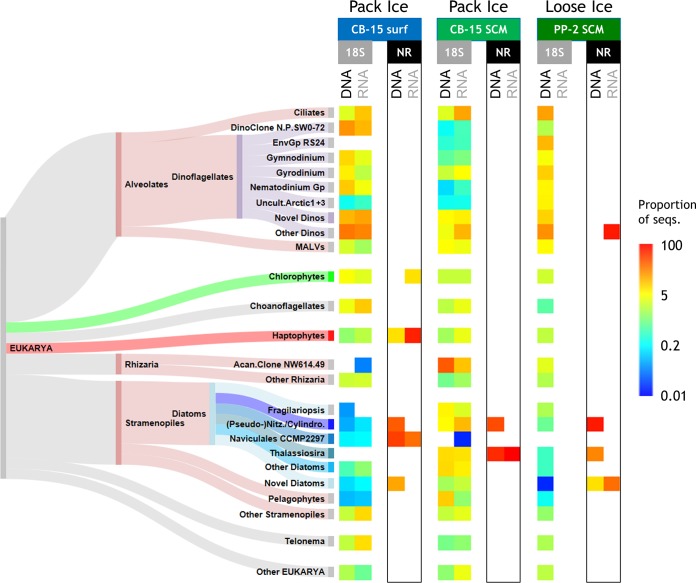
As for individual taxon differences, alveolates accounted for a large proportion of reads (42% to 81%) in the pack ice surface and loose ice SCM samples (Fig. 2). Within the alveolates, phylogenetically varied dinoflagellates were predominant; three major cultured genera (Gymnodinium, Gyrodinium [mainly Gyrodinium helveticum], and Nematodinium) and three environmental groups known by sequence only (environmental group DinoRS24, North Pole SW0-72 clade, and uncultured arctic clade 1 + 3) each comprised at least 5% of the sequences in at least one sample (see Materials and Methods for the availability of uncultured reference sequences). The putative parasites classified as dinoflagellate-related marine alveolates (MALVs) were present preferentially in DNA-sourced communities. Ciliate reads tended to outnumber dinoflagellate reads in the two RNA communities. Apart from the major taxa, there were many minor dinoflagellate contributors (35 total genus-equivalent taxa among all samples) and substantial proportions of novel or unclassified dinoflagellates (average of ∼35% of all dinoflagellates). Additionally, we found that choanoflagellate and Telonemia sequences were common in the surface water and were present in greater proportions in the RNA-sourced communities, whereas Rhizaria were more common at the SCMs and in DNA pools. For example, the pack ice SCM DNA pool had ∼34% of all reads belonging to one environmental Acantharea clone (clone NW614.49). Among stramenopiles, uncultivated marine stramenopiles (MASTs) and chrysophytes (Fig. 2, other stramenopiles) were common in the surface water RNA pool and pelagophytes were relatively abundant in the pack ice SCM DNA pool. Diatoms represented the majority of stramenopiles in the two SCM samples and a large proportion in the pack ice SCM (∼27% of all reads), with a total of 23 genera detected. The most common genera were Pseudo-nitzschia (more in RNA), Thalassiosira (similar in DNA and RNA), and Fragilariopsis (more in DNA) (Fig. 2). These three genera alone accounted for more than 66% of the diatoms in all samples, followed by nonnegligible contributions from (in descending order) Eucampia, uncultured Baltic Sea ice clone 8-90, Chaetoceros spp., and Amphora. Several other groups were present at proportions just below 5% of the total (Fig. 2), including chlorophytes (Micromonas strain CCMP2099 and Pyramimonas), haptophytes (Chrysochromulina and Phaeocystis), Picozoa, and cryptophytes. The results from microscopy of the three samples used in the 18S survey generally showed smaller proportions of dinoflagellates and larger proportions of diatoms, compared to the HTS data (Fig. 3). However, the ordering of samples from smallest to largest proportions of these two major groups was the same for the two techniques (Fig. 3).
FIG 2.
Taxonomic distributions of 18S and NR sequences. Log-scale proportions of 18S HTS and NR cloned sequences, in both DNA- and RNA-sourced communities, are represented. Other than the “novel” (unclassified) and “other” categories, genus-equivalent taxa are shown if they represented at least 5% of the 18S sequences in at least one of the samples; due to their limited number, all NR sequences are shown. Note that no RNA remained for HTS analysis of PP-2 and the Sankey diagram of the taxa is not proportional to any sequence values but simply shows the relationships of the various taxa to the eukaryotic major groups and subgroups. Color schemes for the taxa match those in Fig. 3, 5, and 6; all dinoflagellates are in mauve, diatoms in shades of blue, chlorophytes in green, haptophytes in red, and the higher-rank SAR group in rose. Acan., Acantharea; Cylindro., Cylindrotheca; Dino(s), dinoflagellate(s); EnvGp, environmental (clone) group; Gp, group; MALVs, uncultured marine alveolates; Nitz., Nitzschia; N.P., North Pole; Uncult.Arctic1 + 3, uncultured arctic clades 1 and 3.
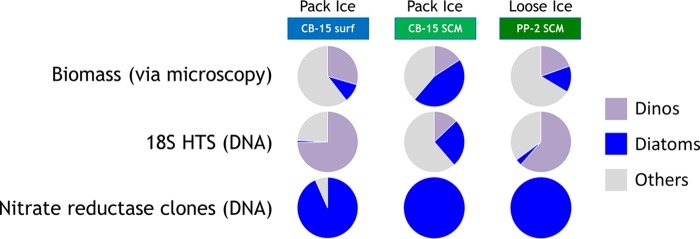
FIG 3.
Proportions of dinoflagellates (dinos), diatoms, or other eukaryotes detected via biomass analysis, 18S HTS, or NR clone sequencing. Data for the two sequencing methods are from the DNA pool since it is most comparable to microscopy (for biomass determination), as the latter also enumerates cells whether they are active/live or not.
NR clone diversity throughout the Arctic region.
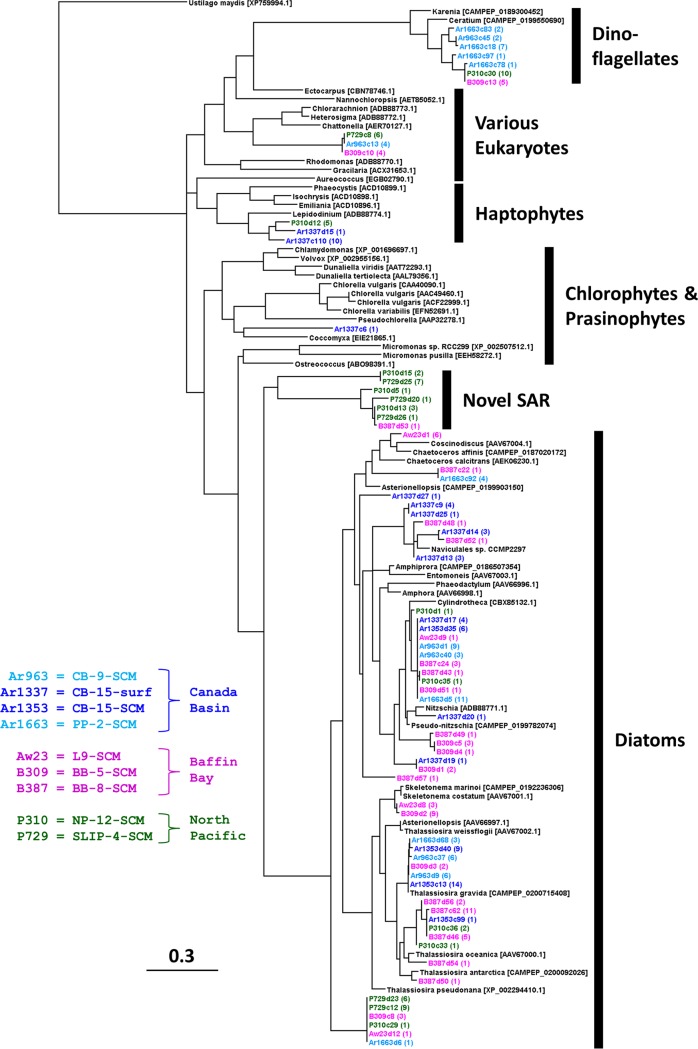
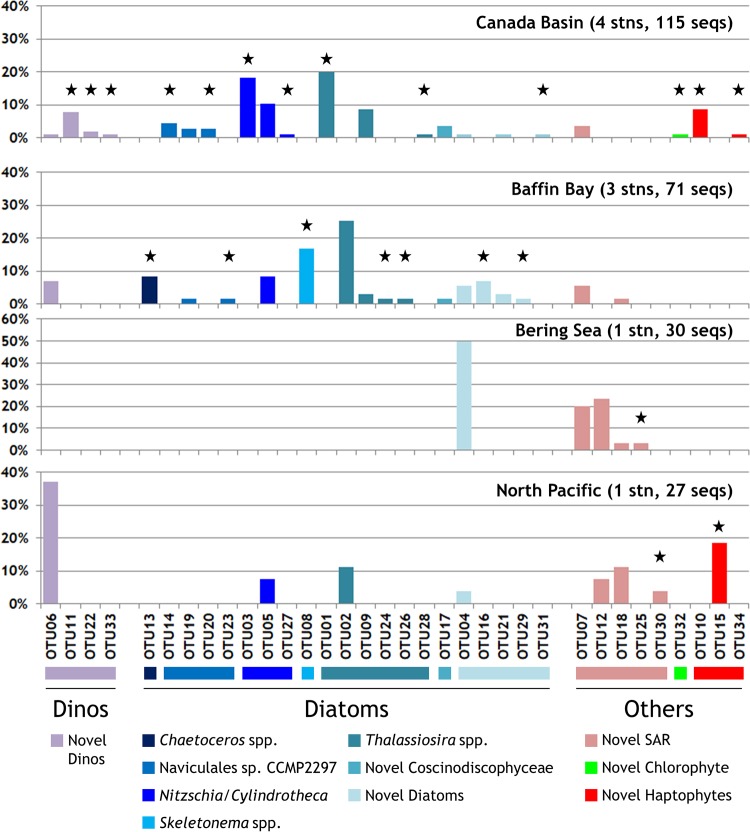
In contrast, clone taxonomy generated by PCR using the current generation of NR primers was heavily skewed toward diatoms for almost all samples. Overall, 243 individual clones were obtained from the nine DNA and eight RNA libraries. Of these, 23 different NR OTUs were retrieved from the three Canada Basin samples examined above (stations CB-15 and PP-2) and an additional 48 OTUs were identified from other clone libraries of Arctic and North Pacific samples (Table 1). The phylogenetic analysis showed that many of the NR OTUs might be Arctic Ocean-specific clades, with representatives from both the Canada Basin and Baffin Bay (Fig. 4). Other OTUs were more cosmopolitan, with representatives from the North Pacific Alaska Gyre and the Bering Sea (Fig. 5). Importantly, many of the OTUs were novel, sharing <85% protein identity with, and/or clustering away from, known taxa. For example, 1 OTU (Ar1337c6) was a distant match to an NR gene from a trebouxian chlorophyte, and another 3 divergent OTUs (Ar963c13, B309c10, and P729c8) were within a cluster made up of various raphidophytes and chlorarachniophytes. There was one large divergent cluster of 7 OTUs, primarily from the North Pacific region, which were about equally distant from various stramenopiles and alveolates (Fig. 4 and 5, novel stramenopiles-alveolates-rhizarians [SAR]). One cluster of novel haptophytes, related to Chrysochromulina sp. strain CCMP291 and Isochrysis, contained one clone (Ar1337c110) that represented 66% of all NR sequences in the pack ice surface RNA pool (Fig. 2). Another large cluster of novel OTUs were related to the newly sequenced dinoflagellates Ceratium and Karenia; 4 of the 7 OTUs in this cluster represented ∼73% of the NR sequences in the loose ice SCM RNA pool (Fig. 2).
FIG 4.
Maximum-likelihood tree of NR clones (∼100 amino acids). Sequences from the Canada Basin are in shades of blue, Baffin Bay in purple, and the North Pacific and Bering Strait in green. Reference sequences (in black) are from either the NCBI database or the MMETSP database (CAMPEP accession numbers). Clones originating from DNA are marked dX and cDNA (RNA) marked cX, where X is the clone number. The numbers in parentheses are the numbers of similar clones obtained, and the scale bar equals 0.3 amino acid substitutions per site. Note that reference sequence ADB88774.1 still retains its NCBI name of “Lepidodinium” (i.e., dinoflagellate) from the authors’ original annotation, although it is probably a novel haptophyte.
FIG 5.
Taxonomic distributions of NR clone OTUs from each region. Represented are the proportions of each of the 34 OTUs (clustered at 94% amino acid identity) in each region (DNA and RNA combined) and the taxonomic group to which each belongs (color scheme matches those in Fig. 2 and 6). OTUs were assigned a genus- or family-level label only if the amino acid identity to corresponding sequences in the NCBI database or the MMETSP database was ≥85%. Black stars indicate OTUs that were unique to the respective region. Note the larger scale for the Bering Sea panel. Dinos, dinoflagellates; SAR, stramenopiles-alveolates-rhizarians; seqs, sequences; stn(s), station(s).
As mentioned above, however, the largest contributors to NR diversity were the diatoms (50 of 71 OTUs), representing the majority of sequences in all sample DNA-sourced communities and the pack ice SCM RNA pool (Fig. 2). There was a large cluster of OTUs at the base of the well-supported diatom branch, which may represent unusual diatoms or closely related heterokonts such as the Parmales. In contrast to these (and a few more) unknowns, the main diatom clade consisted of many OTUs similar to the known genera of Pseudo-nitzschia, Nitzschia, Cylindrotheca, Naviculales sp. strain CCMP2297 (originally isolated from northern Baffin Bay), and several Thalassiosira spp. (Fig. 4 and 5). Other less-common OTUs were related to Skeletonema and Chaetoceros or uncharacterized Coscinodiscophyceae. There was also a tendency toward fewer diatom NR clones through the Bering Sea toward the North Pacific region; novel SAR, haptophytes, and dinoflagellates represented 7 of 10 OTUs in these regions (Fig. 5), although the clone libraries were small and this may be a reflection of the specific samples analyzed.
NR diversity from metagenomes throughout the Arctic region.
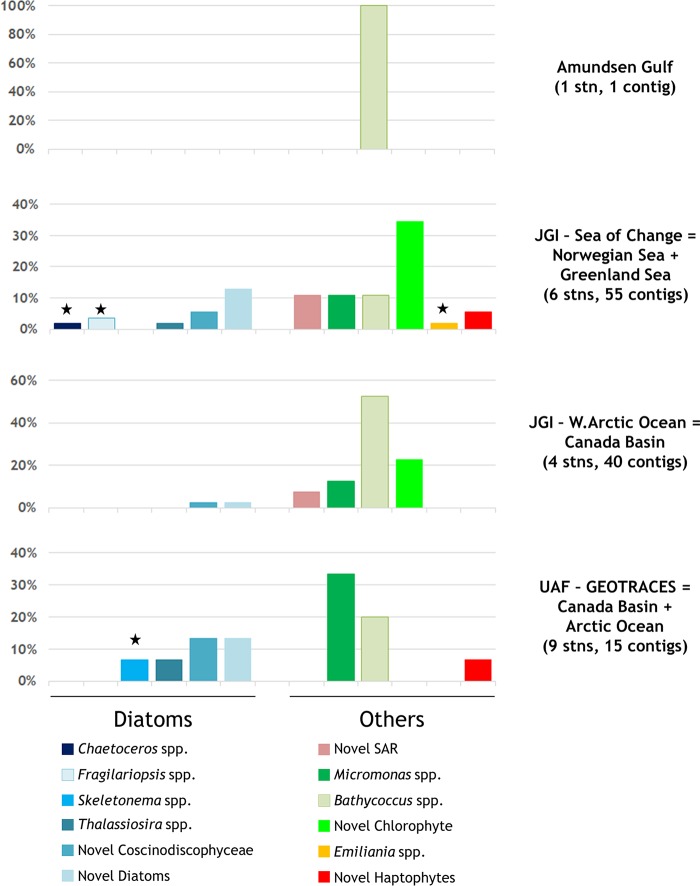
To obtain a potentially less-PCR-biased view of NR diversity and broader Arctic coverage, we screened publicly available, non-PCR-based, arctic eukaryote metagenomes for NR genes. According to our selection criteria (see Materials and Methods), this resulted in four eligible studies, most of which were not size fractionated (suggesting likely biases toward smaller cells). Overall, we searched 32 individual metagenomes (Table S1) from 20 different stations across the Arctic region (Fig. 1). These studies varied in sequencing efforts from 93 million to ∼2.2 billion reads, for a total screened data amount of 4.24 billion reads. After GenSeed-HMM processing using the known protist NR sequences in Fig. 4 as “bait” sequences, 1 to 55 contigs per study were true-positive contigs for NR genes, with up to 23 eukaryote and 53 prokaryote false-positive contigs per sample. The former false hits were nearly all sulfite oxidases, and the latter were a mixture of sulfite oxidases, sulfite dehydrogenases, and glycosyl hydrolases, all of which shared partial protein motifs with NR. A large range of NR protein identities to existing sequences was observed (45% to 100%), and many could be matched to known groups. In contrast to the more diatom-dominated PCR clone libraries in our study (targeting the large size fraction), these metagenomic contigs were dominated by chlorophytes (Fig. 6). Some of the diatom contigs did match taxa found in the clone libraries, especially in the similarly located Canada Basin samples, where they represented 40% of the contigs; however, cosmopolitan marine picoeukaryote chlorophytes totaled 53% to 100% of the contigs per sample. Contigs matching Bathycoccus and Micromonas (often the arctic strain CCMP2099, sequenced in the Marine Microbial Eukaryote Transcriptome Sequencing Project [MMETSP]) were the most abundant NR contigs with matches to known genera, followed by large numbers of chlorophyte contigs originating from unknown organisms. Many of these latter novel contigs were quite close (≥70% amino acid identity) to the identification cutoff value we employed for naming sequences, and they often had various Micromonas species/strains as their closest relatives. Full contig annotations are available in Table S2.
FIG 6.
Taxonomic distributions of NR contigs from metagenomic studies. Represented are the proportions of each of the NR-positive assembled contigs in each study and the taxonomic group to which each belongs (color scheme matches those in Fig. 2 and 5). Contigs were assigned a genus- or family-level label only if the amino acid identity to corresponding sequences in the NCBI database was ≥85%. Black stars indicate taxa that were unique to the respective study. Note the different scales in each panel. JGI, Joint Genome Institute; SAR, stramenopiles-alveolates-rhizarians; seqs, sequences; stn(s), station(s); UAF, University of Alaska—Fairbanks.
DISCUSSION
Community 18S profiles.
Samples were collected from nutrient-poor surface waters, which are typical over much of the open waters of the Beaufort Sea, and from SCMs showing typical Pacific Ocean halocline water intrusions (39), where strong salinity stratification prevents deep-water nutrients from penetrating the upper euphotic zone (3). Although the larger size fraction was targeted, we found several 18S sequences belonging to organisms of <3 μm, such as Micromonas polaris (strain CCMP2099 [40]), Picozoa (41), and some small MAST (42) clades. Small cell trapping is not unanticipated, however, because size fractionation is never perfect (i.e., pore sizes are nominal) and smaller cells may be associated with larger cells, adsorbed to particles, or retained on filters due to some pores being blocked. However, the majority of the sequences were from groups such as diatoms, dinoflagellates, and ciliates, for which microplankton- and nanoplankton-sized representatives are known and were previously reported from arctic clone libraries and HTS studies in Arctic Ocean waters (31, 43, 44). The use of RNA as the source template for 18S surveys is attractive since RNA is ephemeral and rapidly turned over in the cell, providing a means of identifying the most active members of a community. Analyzing RNA can also potentially help reduce some of the biases stemming from large-genome/high-DNA-content organisms (especially dinoflagellates [45, 46]), which can overwhelm signals from smaller organisms in DNA analyses. Although they were fairly well correlated overall (see Fig. S2 in the supplemental material), communities identified from DNA and RNA did show some differences, with certain taxonomic groups being more numerous in the RNA communities (implying greater activity) than their DNA proportions (Fig. 2). The greater proportions of ciliates, chrysophytes, haptophytes, Telonemia, and group III MALVs (as defined by Guillou et al. [47]) in RNA communities and greater proportions of rhizarians and group I MALVs in DNA samples seen here were also noted in an arctic clone library comparison of DNA versus RNA (32), following seasonal changes in the Amundsen Gulf (48), and a 24-h study in northern Baffin Bay (43). However, perhaps not surprisingly, the partitioning of the sequences (Fig. S1B) indicated that many of the differences between the DNA-sourced and RNA-sourced communities resided in the tail ends of the rank abundance curves (i.e., in the rarer OTUs).
Functional NR diversity and links to community 18S diversity.
It has been suggested that NR provides higher phylogenetic resolution than other functional genes, such as ribulose-1,5-bisphosphate carboxylase/oxygenase, potentially making NR a valuable indicator of microbial adaptation and diversity in marine environments (17, 23). However, progress toward this goal has been hampered because very few studies have targeted this gene either in natural samples (24) or in relevant culture collections (23). While there are now well over 75,000 environmental sequences for marine eukaryotic 18S (search terms: 18S rRNA or 18S rDNA, not metazoan; searched January 2019) in the NCBI database, there are very few NR sequences from marine samples available for comparison. Ward and Van Oostende (49) highlighted the magnitude of this problem when they demonstrated that the major signals captured on a phytoplankton functional gene microarray during the North Atlantic spring bloom were mostly from uncharacterized NR and nitrate transporter genes. The NR primers used in this study were originally developed by Allen et al. (22) using the very few diatom, green alga, and higher plant sequences that were available at the time. Due to the aforementioned paucity of new marine isolate NR sequences determined in the intervening decade, there has been no redesign to compensate for the assumed diatom bias of the primers. Although acknowledging this bias, some of the original authors of the primers, in their subsequent analysis of NR PCR clones from the English Channel and Monterey Bay (23), noted that a variety of other relevant phytoplankton groups are amplified by these primers, indicating their continued usefulness in NR ecological surveys. The PCR-based portion of our study most probably suffered from this same bias, as seen in the large NR clone versus microscopic diatom skew (Fig. 3), but also achieved the same success in amplifying such divergent groups as chlorophytes, haptophytes, and dinoflagellates (Fig. 5). We anticipate that the time is nearing when enough novel NR sequences will be obtained, especially via non-PCR-based metagenomic screening, that a redesign (or at least verification) of the current NR primers to yield substantial non-Metazoan coverage will be feasible.
In this NR Arctic Ocean survey, we found novel clades within known groups (Fig. 4), but many other sequences were only distantly related to known diatoms and chlorophytes or were not assignable (only 42% to 47% amino acid identity) to any current major group for which NR sequences are available. Similarly, the largest proportion of metagenomic NR sequences was from novel taxa, especially chlorophytes (Fig. 6). The dominance of the metagenomes by these latter smaller organisms is most probably a direct result of the inclusion of both size fractions in these studies, whereas picoeukaryotes would have been less common in our large-fraction clone libraries. Overall, these results indicate that there is a need for additional studies of NR in described organisms if NR is to become a useful proxy for identification of the functional distribution of marine plankton. Along those lines, it is worth noting that the MMETSP (https://www.imicrobe.us/#/projects/104) (50) allowed us to identify a previously unknown cluster of dinoflagellate OTUs, to detect arctic Micromonas sequences, and to further refine the species identities of numerous Thalassiosira OTUs. The remaining unassignable sequences may also be a consequence of the poor representation of open-ocean and/or arctic clades in culture collections.
Although the numbers of NR sequences were small, we found some differences between the DNA and RNA analyses (Fig. 2). All samples had larger proportions of diatom sequences in the DNA profiles, while the RNA profiles had similar relative abundances of diatoms (pack ice SCM), more haptophytes (pack ice surface), or more dinoflagellates (loose ice SCM). This finding suggests that the latter two can contribute more to the transcript levels than diatoms at certain times or depths, as was also noted in the nearly year-long record from the Canadian Flaw Lead Study, in which a ship sampled open leads near the Amundsen Gulf over the winter (48, 51). It is important to note, however, that this discrepancy may simply be due to different primer efficiencies, extraction differences, or cDNA synthesis biases. That being said, functional gene expression is variable and specific to different conditions, whereas ribosomal genes and other constitutive genes are required for the basic functions of the cell and are expressed when the cell is active in any capacity. NR is a highly regulated protein (52, 53) and many factors, such as NO3− concentrations, light, temperature, carbon fixation, and NH4+ concentrations, can influence its activity (16, 20, 53–56); therefore, we may be detecting true differences in transcripts versus genomic DNA.
Before our study, there have been no reports comparing 18S diversity (let alone using HTS) with the corresponding NR diversity from the same samples. Among marine phytoplankton that possess the NR gene, diatoms are considered to be among the best competitors for NO3− when concentrations are high, and they often dominate phytoplankton assemblages in upwelling areas and continental margins (22, 57). In our samples, diatoms represented 10% to 45% of the biomass (estimated by microscopy) but represented only 1% to 25% of the 18S DNA HTS proportions, even though this group dominated the NR DNA libraries (Fig. 3) due to the previously mentioned possible primer bias. On the other hand, the samples with haptophyte NR also showed, albeit only slightly, more haptophytes in 18S reads. Similarly, the loose ice RNA library concurred well with 18S taxonomy in general, showing a greater contribution by dinoflagellates (Fig. 2). Dinoflagellates are always present in polar waters and dominate 18S DNA libraries (27), probably due to large DNA content/copy number differences; therefore, they often mask the true contributions of groups such as diatoms and haptophytes, due to the compositional nature of sequencing data.
Conclusion.
Our study examining NR diversity in polar marine plankton with clone library and metagenomic screening indicated clear contributions by diatoms, dinoflagellates, and chlorophytes, with many NR sequences of uncertain taxonomic affinity. We found that SCM communities from samples from different ice covers were diverse and moderately different (∼50% shared OTUs), with diatoms being more frequent under the more densely ice-covered station. Due to the differences in success rates and biases for the various molecular techniques presented here, we suggest that more work of this kind is required to arrive at a fuller understanding of NR ecology within the larger scheme of arctic primary production based on nitrate uptake. It is clear from our results that a phylogenetically diverse array of microbial eukaryotes can use nitrate as a nitrogen source, with implications for the arctic carbon cycle.
MATERIALS AND METHODS
Sampling strategy.
Samples were collected as part of three oceanographic expeditions on board Canadian Coast Guard icebreakers (Table 1 and Fig. 1). The North Water (northern Baffin Bay) sample was collected on board the CCGS Amundsen in July 2005, in conjunction with the ArcticNet program. Samples from Baffin Bay and the Canada Basin were collected aboard the CCGS Louis S. St. Laurent in July and August 2007, as part of the International Polar Year Canada’s Three Oceans (C3O) project. North Pacific samples were collected aboard the CCGS Sir Wilfrid Laurier in July 2008, also as part of the C3O project. Seawater was obtained from the SCM at all eight stations, and an additional sample was taken at station CB-15 (Canada Basin) from surface waters (5 m). For the two Canada Basin stations where 18S surveys were also conducted (CB-15 and PP-2), the SCM layers are seasonally modified, Pacific Ocean-origin, halocline waters (all terminology was adapted from the study by McLaughlin et al. [39]). Sample CB-15-SCM was collected from the bottom of the summer mixed layer, and sample PP-2-SCM was collected from the top of the winter mixed layer. The remaining CB-15 sample (CB-15-surf) represents the seasonal, shallow, persistent surface fresh plume, where surface water is freshened by melting sea ice. Ice conditions at station CB-15 consisted of solid pack ice, whereas station PP-2 had significant ice cover (∼80%) but with patches of open water.
Sample collection and processing.
Sampling at all stations was performed using a rosette system equipped with 24 ten-liter Niskin bottles (CCGS Louis S. St. Laurent and CCGS Sir Wilfred Laurier) or 20 twelve-liter Niskin bottles (CCGS Amundsen). All rosette systems were equipped with conductivity-temperature-depth (CTD) profilers (SBE 9+; Sea-bird Electronics, Bellevue, WA) and nitrate (ISUS; Satlantic, Halifax, Canada), oxygen (SBE-43; Sea-bird Electronics), chlorophyll fluorescence (Seapoint Sensors, Exeter, NH), and photosynthetically available radiation (Li-Cor, Lincoln, NE) sensors. Seawater samples were collected for DNA and RNA analyses, microscopy, and determination of nutrient and chl a concentrations on the up-casts of the rosette. Water was collected directly from the Niskin bottles into 7-liter carboys that had been acid cleaned once, rinsed three times with Milli-Q water, and then rinsed three times with sample water before filling. Seawater for chl a analysis was collected in a separate clean 2-liter bottle that had been rinsed three times with seawater prior to filling.
For DNA analysis, approximately 6 liters of seawater was gently prefiltered through a 50-μm mesh and then filtered through a 3-μm-pore-size, 47-mm-diameter, polycarbonate filter (AMD Manufacturing, Mississauga, Canada) under positive pressure, using a peristaltic pump. The filters were then placed in 2-ml cryovials, to which 1.8 ml of lysis buffer (40 mM EDTA [pH 8.0], 50 mM Tris-HCl [pH 8.3], 0.75 M sucrose) was added. For RNA collection, samples were filtered as indicated above, except that only 4 liters of seawater was filtered for each sample and filters were covered with 600 μl of buffer RLT (Qiagen, Mississauga, Canada) with 1% β-mercaptoethanol (Sigma-Aldrich, Oakville, Canada) in cryovials. All cryovials were then immersed in liquid nitrogen for 1 h and stored at −80°C until DNA/RNA extractions were performed.
Samples for chl a determinations were filtered onto glass fiber filters (AMD), either directly (for total chl a determinations) or after a prefiltration step through a 3-μm polycarbonate filter (representing the <3-μm fraction). The >3-μm fraction concentrations were then determined by subtraction of the <3-μm fraction concentrations from the total chl a concentrations. Concentrations were determined on board following extraction with 90% acetone (EPA method 445.0; https://cfpub.epa.gov/si/si_public_record_report.cfm?Lab=NERL&dirEntryId=309417). Samples for analysis of nutrient [NO3−, PO43−, and Si(OH)4] levels were collected directly from the Niskin bottles and immediately frozen at −20°C until analysis on shore with an Astoria 2 nutrient analyzer, according to the method described by Barwell-Clarke and Whitney (58).
Microscopy counts of eukaryotic microorganisms.
Nanoplankton were enumerated using an Olympus 1X71 microscope (Olympus, Markham, Canada), at a magnification of ×1,000, under both blue and UV excitation to visualize chlorophyll fluorescence of chloroplasts and 4′,6-diamidino-2-phenylindole (DAPI)-stained nuclear DNA. Cell counts were grouped as either phototrophs (visible chloroplasts) or heterotrophs (no visible chloroplasts) and by size (1 to 3, 3 to 6, or 6 to 10 μm). Diatoms and dinoflagellates were enumerated separately. Cell sizes were estimated using a calibrated grid and an ocular micrometer. Biovolumes were also determined for each category (59). These values were then used to convert the counts (cells per milliliter) to biomass using the relationships determined by Menden-Deuer and Lessard (60), i.e., picograms of carbon per cell = 0.216 × V0.939 for protist plankton (excluding diatoms) and picograms of carbon per cell = 0.288 × V0.811 for diatoms.
Nucleic acid extraction and cDNA synthesis.
DNA was extracted from the filters as described by Harding et al. (61), using a combination of enzymatic digestion and salt-based separation. Total RNA was extracted with the RNeasy kit (Qiagen) as instructed by the manufacturer (including the optional on-column digestion with the RNase-free DNase kit [Qiagen]) after disruption of the cells using a bead beater (Disruptor Genie; Scientific Industries, Bohemia, NY), as described by Church et al. (62). The RNA extract was immediately transformed into cDNA using a high-capacity reverse transcriptase kit (Applied Biosystems, Foster City, CA), and samples were then stored at −80°C.
18S and NR gene amplification, cloning, and Sanger sequencing.
18S (stations CB-15 and PP-2 only) and NR (all stations) gene fragments were amplified using the primers listed in Table S3. Total reaction volumes (25 μl) contained 1× ThermoPol buffer (NEB, Ipswich, MA) (for 18S) or 1× Advantage 2 SA buffer (Clontech, Mountain View, CA) (for NR), 0.2 mM deoxynucleoside triphosphates (dNTPs) (Feldan Bio, Québec, Canada), 0.4 to 1.0 μM each primer, 10 μg of bovine serum albumin (BSA) (Fermentas Life Sciences, Burlington, Canada), 2 to 4 μl of template (DNA or cDNA), and 0.625 U of either Taq polymerase (NEB) or Advantage 2 polymerase (Clontech). Multiple reactions were run for each sample, using the following cycling conditions: initial denaturation at 94°C for 3 to 5 min, 30 cycles of denaturation at 94°C for 30 to 45 s, annealing at 58°C for 0.5 to 1 min, and extension at 72°C for 3 min, and a final extension at 72°C for 5 to 7 min. A second-round, nested PCR was performed for NR as described above except that annealing was performed at 55°C, extension was for only 30 s, and 2 μl of the first-round product was used as the template. For each sample, PCR products were pooled, purified using the QIAquick PCR purification kit (Qiagen), and then cloned using the Topo TA cloning kit (Invitrogen, Burlington, Canada), following the manufacturer’s instructions. Transformed clones were picked into 96-well plates and cultured overnight at 37°C in LB broth with ampicillin (Fermentas) and 7% glycerol. Positive clones were confirmed by PCR amplification as for the original reactions (NR using second-round conditions), omitting BSA and using 1.5 μl of the transformed cell culture as the template for the reaction.
For each NR clone library, 16 to 25 positive clones, as confirmed by agarose gel electrophoresis, were randomly selected to be sequenced using the vector M13 primer sites. For 18S libraries, unique clones were identified by restriction fragment length polymorphism (RFLP) screening of all clone libraries except for PP-2 cDNA, for which all clones were sequenced. Amplicons were digested with HaeIII (NEB) overnight at 37°C and visualized on a 2.5% low-melting-point agarose gel. Clones with the same RFLP patterns were considered members of the same phylotype (63). The PCR products from 1 to 3 clones from each phylotype were sequenced with the internal primer Euk528F (Table S3). All products were sequenced with an ABI 3730XL system at the Plateforme de Sequençage et Genotypage des Génomes at the Centre de Recherche du Centre Hospitalier de l’Université Laval.
18S and NR cloned gene analyses.
Sequences were manually trimmed and edited with BioEdit (www.mbio.ncsu.edu/bioedit/bioedit.html). The 18S sequences were verified against the GenBank nonredundant nucleotide database and were checked for chimeras by separate BLASTn searches of two or more sequence fragments. Poor-quality sequences, likely chimeras, and sequences with <750 bp were not used for further analysis. Translated NR sequences were compared to those in the GenBank nonredundant protein database; non-NR and spurious sequences were removed at that point, and introns found in some of the NR DNA sequences were eliminated using the criteria described by Adhitya et al. (24). The remaining quality sequences were aligned using ClustalX (www.clustal.org), and OTUs were defined using DOTUR (64). For 18S clones, nucleotide sequences of ≥99% identity were considered to represent the same OTU. For NR clones, we used the criteria given by Adhitya et al. (24), in which clones with sequence identity of ≥94% were clustered into the same OTU. One representative per clone library for each of these NR OTUs was used to construct a protein maximum-likelihood phylogenetic tree using RAxML v.8.1.1 (65). Reference sequences were selected from the report by Song and Ward (54), a BLAST search of the NCBI database, and the MMETSP database (50); the complete final sequence set in aligned format is available in File S1 in the supplemental material. Bootstrap analyses of 1,000 replications were performed to estimate the confidence limits of tree topologies.
V4 18S gene amplification and HTS using 454 pyrosequencing.
Extracted DNA and cDNA were amplified using 18S V4-region-specific 454 primers, as described by Comeau et al. (66), for stations CB-15 (surface and SCM) and PP-2 (SCM). The 5 sample-coded amplicons (3 samples × 2 nucleic acids = 6; 6 − 1 [missing cDNA for PP-2] = 5) were mixed in equal quantities, and one-eighth of the plate was sequenced on a Roche 454 GS-FLX Titanium platform at the Institut de Biologie Intégrative et des Systèmes (IBIS)/Université Laval Plateforme d’Analyses Génomiques.
HTS read preprocessing, quality control, and analysis.
Raw 454 reads were quality controlled, aligned, and clustered into OTUs at the ≥98% identity level using mothur (67, 68) (http://www.mothur.org), as described by Comeau et al. (69), changing only the minimum read size (350 bp) and adding a chimera detection step using the mothur implementation of UCHIME (70). Read equalization in this study resulted in 6,237 reads per barcode (sample) being randomly resampled for the downstream analyses. Eukarya OTUs and total sequences were taxonomically identified using a user-designed, V4-region-trimmed taxonomy outline and reference sequence database, as introduced by Comeau et al. (66, 69) and updated by Onda et al. (51). The latter study resulted in version 1.1 of the database (71), and further small changes were made to the cryptophytes in February 2018, yielding the final version of the database used in this study (available upon request). Consult the report by Comeau et al. (69) for further details and justifications regarding the expected overall error rates (<0.2%), the choice of identity level for OTUs (98% versus preferred 99%), and taxonomic identification accuracy.
Metagenome NR screening and analysis.
The NCBI SRA and EBI ENA databases were searched for any arctic (>60°N latitude) metagenomes that either targeted eukaryotes or did not specifically exclude them. Samples from these studies were retained if they fulfilled the following criteria: (i) originated from the upper 100 m of the water column (no ice or sediments); (ii) were sequenced with Illumina technology (better for subsequent assembly); and (iii) contained at least 1 million raw reads per sample. Replicate samples from the same stations and sampling times were concatenated prior to assessment of the final criterion and remained combined for subsequent assembly and analysis.
Raw reads were downloaded via the command line and screened for the presence of NR genes using GenSeed-HMM (72), which avoids a computationally exhausting complete assembly of all reads (and subsequent screening) by using hidden Markov model (HMM) profiles to assemble only contigs containing the target gene(s) of interest. The required HMM protein profile was generated with HMMER v.3.1b2 (73), using the same NR alignment file as used for tree building above (see File S1 in the supplemental material), minus the outgroup sequence, as input for hmmbuild with default options. The resulting profile was used as the seed file for GenSeed-HMM v.1.0.14, performing one run per sample set of concatenated paired-end raw metagenomic reads, with 20 as the length of extension seeds (required parameter) and 10 kb as the maximum contig length (optional parameter). Analyses were run on a local cluster with 40 cores, and the choice of a mandatory back-end assembler was ABySS v.2.0.3 (74). Assembled contigs of less than 200 bp were discarded, and the remaining contigs were annotated using manual BLASTx on the NCBI website (https://blast.ncbi.nlm.nih.gov) or locally against the MMETSP data using BioEdit. Contigs were assigned a genus- or family-level label only if the amino acid identity to corresponding sequences in GenBank was ≥85%.
Statistical and visualization techniques.
Nonparametric (nonnormal) Mann-Whitney and Spearman rank tests were used to verify differences between groups and to perform correlations, respectively. F tests and t tests (equal or unequal variance, as was the case) and linear regressions (on log-transformed data) were used with data that fulfilled the conditions of normality. All statistical analyses were carried out with Past (folk.uio.no/ohammer/past). Maps were generated with Ocean Data View (odv.awi.de), trees were beautified in Dendroscope (dendroscope.org), and the Sankey diagram was generated using SankeyMATIC (www.sankeymatic.com).
Accession number(s).
The nonredundant 18S and NR clone sequences have been deposited in GenBank under accession numbers FJ971784 to FJ971832 and FJ971900 to FJ971934 (18S) as well as GQ352559 to GQ352630 (NR). The raw pyrosequencing reads have been deposited in the NCBI Sequence Read Archive with accession number PRJNA217362.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by ongoing Natural Science and Engineering Research Council of Canada (NSERC) Discovery financing to C.L. and D.E.V. Additional funds were provided by Fonds de Recherche du Québec-Nature et Technologies to Québec-Océan, by NSERC and Department of Fisheries and Oceans (Canada) funding for the C3O project to D.E.V., and by the Faculty of Graduate Studies at the University of Victoria to M.G.L. This work is a contribution to the research programs of the C3O study, the International Polar Year (IPY-2007–2008), and ArcticNet, a Canadian Network of Centers of Excellence.
We thank our colleagues at the IBIS/Université Laval Plateforme d’Analyses Génomiques for the DNA pyrosequencing and at the IBIS Centre de Bio-informatique et de Biologie Computationnelle for bioinformatics support. We also thank Eric Collins, Kara Martin, Thomas Mock, and David Walsh for metadata associated with the Joint Genome Institute (JGI) and GEOTRACES metagenomes and Arthur Gruber and João Alves for discussions regarding GenSeed-HMM. We are indebted to the crews of the CCGS Amundsen during the ArcticNet expedition and the CCGS Louis S. St. Laurent and CCGS Sir Wilfrid Laurier during the C3O expeditions for logistics and sampling support. We specifically thank Ian Wrohan for sampling and nutrient analysis during the latter cruise.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00247-19.
REFERENCES
- 1.Tremblay J-É, Simpson K, Martin J, Miller L, Gratton Y, Barber D, Price NM. 2008. Vertical stability and the annual dynamics of nutrients and chlorophyll fluorescence in the coastal, southeast Beaufort Sea. J Geophys Res 113:C07S90. doi: 10.1029/2007JC004547. [DOI] [Google Scholar]
- 2.Monier A, Comte J, Babin M, Forest A, Matsuoka A, Lovejoy C. 2015. Oceanographic structure drives the assembly processes of microbial eukaryotic communities. ISME J 9:990–1002. doi: 10.1038/ismej.2014.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carmack EC. 2007. The alpha/beta ocean distinction: a perspective on freshwater fluxes, convection, nutrients and productivity in high-latitude seas. Deep Sea Res II Top Stud Oceanogr 54:2578–2598. doi: 10.1016/j.dsr2.2007.08.018. [DOI] [Google Scholar]
- 4.Lee SH, Joo HM, Liu Z, Chen J, He J. 2012. Phytoplankton productivity in newly opened waters of the Western Arctic Ocean. Deep Sea Res II Top Stud Oceanogr 81–84:18–27. doi: 10.1016/j.dsr2.2011.06.005. [DOI] [Google Scholar]
- 5.Varela DE, Crawford DW, Wrohan IA, Wyatt SN, Carmack EC. 2013. Pelagic primary productivity and upper ocean nutrient dynamics across Subarctic and Arctic Seas. J Geophys Res Oceans 118:7132–7152. doi: 10.1002/2013JC009211. [DOI] [Google Scholar]
- 6.Holland MM, Bitz CM. 2003. Polar amplification of climate change in coupled models. Clim Dynam 21:221–232. doi: 10.1007/s00382-003-0332-6. [DOI] [Google Scholar]
- 7.Morison J, Kwok R, Peralta-Ferriz C, Alkire M, Rigor I, Andersen R, Steele M. 2012. Changing Arctic Ocean freshwater pathways. Nature 481:66–70. doi: 10.1038/nature10705. [DOI] [PubMed] [Google Scholar]
- 8.Perovich DK, Richter-Menge JA. 2009. Loss of sea ice in the Arctic. Annu Rev Mar Sci 1:417–441. doi: 10.1146/annurev.marine.010908.163805. [DOI] [PubMed] [Google Scholar]
- 9.Zhang J, Lindsay R, Schweiger A, Steele M. 2013. The impact of an intense summer cyclone on 2012 Arctic Sea ice retreat. Geophys Res Lett 40:720–726. doi: 10.1002/grl.50190. [DOI] [Google Scholar]
- 10.Yamamoto-Kawai M, McLaughlin FA, Carmack EC, Nishino S, Shimada K, Kurita N. 2009. Surface freshening of the Canada Basin, 2003–2007: river runoff versus sea ice meltwater. J Geophys Res 114:C00A05. doi: 10.1029/2008JC005000. [DOI] [Google Scholar]
- 11.Carmack EC, Yamamoto-Kawai M, Haine TWN, Bacon S, Bluhm BA, Lique C, Melling H, Polyakov IV, Straneo F, Timmermans M-L, Williams WJ. 2016. Freshwater and its role in the Arctic marine system: sources, disposition, storage, export, and physical and biogeochemical consequences in the Arctic and global oceans. J Geophys Res Biogeosci 121:675–717. doi: 10.1002/2015JG003140. [DOI] [Google Scholar]
- 12.Hill VJ, Matrai PA, Olson E, Suttles S, Steele M, Codispoti LA, Zimmerman RC. 2013. Synthesis of integrated primary production in the Arctic Ocean. II. In situ and remotely sensed estimates. Prog Oceanogr 110:107–125. doi: 10.1016/j.pocean.2012.11.005. [DOI] [Google Scholar]
- 13.Glibert PM, Wilkerson FP, Dugdale RC, Raven JA, Dupont CL, Leavitt PR, Parker AE, Burkholder JM, Kana TM. 2016. Pluses and minuses of ammonium and nitrate uptake and assimilation by phytoplankton and implications for productivity and community composition, with emphasis on nitrogen-enriched conditions. Limnol Oceanogr 61:165–197. doi: 10.1002/lno.10203. [DOI] [Google Scholar]
- 14.Dugdale RC, Goering JJ. 1967. Uptake of new and regenerated forms of nitrogen in primary productivity. Limnol Oceanogr 12:196–206. doi: 10.4319/lo.1967.12.2.0196. [DOI] [Google Scholar]
- 15.Hutchings L, Pitcher GC, Probyn TA, Bailey GW. 1995. The chemical and biological consequences of coastal upwelling, p 65–81. In Summerhayes CP, Emeis K-C, Angel MV, Smith RI, Zeitschel B (ed), Upwelling in the oceans: modern processes and ancient records. Wiley, New York, NY. [Google Scholar]
- 16.Song B, Ward BB. 2004. Molecular characterization of the assimilatory nitrate reductase gene and its expression in the marine green alga Dunaliella tertiolecta (Chlorophyceae). J Phycol 40:721–731. doi: 10.1111/j.1529-8817.2004.03078.x. [DOI] [Google Scholar]
- 17.Ward BB. 2008. Phytoplankton community composition and gene expression of functional genes involved in carbon and nitrogen assimilation. J Phycol 44:1490–1503. doi: 10.1111/j.1529-8817.2008.00594.x. [DOI] [PubMed] [Google Scholar]
- 18.Clark DR, Rees AP, Joint I. 2008. Ammonium regeneration and nitrification rates in the oligotrophic Atlantic Ocean: implications for new production estimates. Limnol Oceanogr 53:52–62. doi: 10.4319/lo.2008.53.1.0052. [DOI] [Google Scholar]
- 19.Hong H, Li D, Lin W, Li W, Shi D. 2017. Nitrogen nutritional condition affects the response of energy metabolism in diatoms to elevated carbon dioxide. Mar Ecol Prog Ser 567:41–56. doi: 10.3354/meps12033. [DOI] [Google Scholar]
- 20.Brown KL, Twing KI, Robertson DL. 2009. Unraveling the regulation of nitrogen assimilation in the marine diatom Thalassiosira pseudonana (Bacillariophyceae): diurnal variations in transcript levels for five genes involved in nitrogen assimilation. J Phycol 45:413–426. doi: 10.1111/j.1529-8817.2009.00648.x. [DOI] [PubMed] [Google Scholar]
- 21.Berges JA, Harrison PJ. 1995. Nitrate reductase activity quantitatively predicts the rate of nitrate incorporation under steady state light limitation: a revised assay and characterization of the enzyme in three species of marine phytoplankton. Limnol Oceanogr 40:82–93. doi: 10.4319/lo.1995.40.1.0082. [DOI] [Google Scholar]
- 22.Allen AE, Ward BB, Song B. 2005. Characterization of diatom (Bacillariophyceae) nitrate reductase genes and their detection in marine phytoplankton communities. J Phycol 41:95–104. doi: 10.1111/j.1529-8817.2005.04090.x. [DOI] [Google Scholar]
- 23.Bhadury P, Ward BB. 2009. Molecular diversity of marine phytoplankton communities based on key functional genes. J Phycol 45:1335–1347. doi: 10.1111/j.1529-8817.2009.00766.x. [DOI] [PubMed] [Google Scholar]
- 24.Adhitya A, Thomas FIM, Ward BB. 2007. Diversity of assimilatory nitrate reductase genes from plankton and epiphytes associated with a seagrass bed. Microb Ecol 54:587–597. doi: 10.1007/s00248-006-9175-0. [DOI] [PubMed] [Google Scholar]
- 25.Ardyna M, Gosselin M, Michel C, Poulin M, Tremblay J. 2011. Environmental forcing of phytoplankton community structure and function in the Canadian High Arctic: contrasting oligotrophic and eutrophic regions. Mar Ecol Prog Ser 442:37–57. doi: 10.3354/meps09378. [DOI] [Google Scholar]
- 26.Simpson KG, Tremblay J-É, Gratton Y, Price NM. 2008. An annual study of inorganic and organic nitrogen and phosphorus and silicic acid in the southeastern Beaufort Sea. J Geophys Res 113:C07016. doi: 10.1029/2007JC004462. [DOI] [Google Scholar]
- 27.Lovejoy C, Carmack EC, Legendre L, Price NM. 2002. Water column interleaving: a new physical mechanism determining protist communities and bacterial states. Limnol Oceanogr 47:1819–1831. doi: 10.4319/lo.2002.47.6.1819. [DOI] [Google Scholar]
- 28.Martin J, Tremblay J, Gagnon J, Tremblay G, Lapoussière A, Jose C, Poulin M, Gosselin M, Gratton Y, Michel C. 2010. Prevalence, structure and properties of subsurface chlorophyll maxima in Canadian Arctic waters. Mar Ecol Prog Ser 412:69–84. doi: 10.3354/meps08666. [DOI] [Google Scholar]
- 29.Ardyna M, Babin M, Devred E, Forest A, Gosselin M, Raimbault P, Tremblay J-É. 2017. Shelf-basin gradients shape ecological phytoplankton niches and community composition in the coastal Arctic Ocean (Beaufort Sea): phytoplankton niches of the Beaufort Sea. Limnol Oceanogr 62:2113–2132. doi: 10.1002/lno.10554. [DOI] [Google Scholar]
- 30.Crawford DW, Cefarelli AO, Wrohan IA, Wyatt SN, Varela DE. 2018. Spatial patterns in abundance, taxonomic composition and carbon content of phytoplankton assemblages in Subarctic and Arctic Seas. Prog Oceanogr 162:132–159. doi: 10.1016/j.pocean.2018.01.006. [DOI] [Google Scholar]
- 31.Lovejoy C, Massana R, Pedrós-Alió C. 2006. Diversity and distribution of marine microbial eukaryotes in the Arctic Ocean and adjacent seas. Appl Environ Microbiol 72:3085–3095. doi: 10.1128/AEM.72.5.3085-3095.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terrado R, Medrinal E, Dasilva C, Thaler M, Vincent WF, Lovejoy C. 2011. Protist community composition during spring in an Arctic flaw lead polynya. Polar Biol 34:1901–1914. doi: 10.1007/s00300-011-1039-5. [DOI] [Google Scholar]
- 33.Marquardt M, Vader A, Stübner EI, Reigstad M, Gabrielsen TM. 2016. Strong seasonality of marine microbial eukaryotes in a high-Arctic fjord (Isfjorden, in West Spitsbergen, Norway). Appl Environ Microbiol 82:1868–1880. doi: 10.1128/AEM.03208-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balzano S, Marie D, Gourvil P, Vaulot D. 2012. Composition of the summer photosynthetic pico and nanoplankton communities in the Beaufort Sea assessed by T-RFLP and sequences of the 18S rRNA gene from flow cytometry sorted samples. ISME J 6:1480–1498. doi: 10.1038/ismej.2011.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pawlowski J, Christen R, Lecroq B, Bachar D, Shahbazkia HR, Amaral-Zettler L, Guillou L. 2011. Eukaryotic richness in the abyss: insights from pyrotag sequencing. PLoS One 6:e18169. doi: 10.1371/journal.pone.0018169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coolen MJL, Shtereva G. 2009. Vertical distribution of metabolically active eukaryotes in the water column and sediments of the Black Sea. FEMS Microbiol Ecol 70:525–539. doi: 10.1111/j.1574-6941.2009.00756.x. [DOI] [PubMed] [Google Scholar]
- 37.Koid A, Nelson WC, Mraz A, Heidelberg KB. 2012. Comparative analysis of eukaryotic marine microbial assemblages from 18S rRNA gene and gene transcript clone libraries by using different methods of extraction. Appl Environ Microbiol 78:3958–3965. doi: 10.1128/AEM.06941-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Logares R, Audic S, Santini S, Pernice MC, de Vargas C, Massana R. 2012. Diversity patterns and activity of uncultured marine heterotrophic flagellates unveiled with pyrosequencing. ISME J 6:1823–1833. doi: 10.1038/ismej.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McLaughlin F, Shimada K, Carmack E, Itoh M, Nishino S. 2005. The hydrography of the southern Canada Basin, 2002. Polar Biol 28:182–189. doi: 10.1007/s00300-004-0701-6. [DOI] [Google Scholar]
- 40.Simon N, Foulon E, Grulois D, Six C, Desdevises Y, Latimier M, Le Gall F, Tragin M, Houdan A, Derelle E, Jouenne F, Marie D, Le Panse S, Vaulot D, Marin B. 2017. Revision of the genus Micromonas Manton et Parke (Chlorophyta, Mamiellophyceae), of the type species M. pusilla (Butcher) Manton & Parke and of the species M. commoda van Baren, Bachy and Worden and description of two new species based on the genetic and phenotypic characterization of cultured isolates. Protist 168:612–635. doi: 10.1016/j.protis.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 41.Seenivasan R, Sausen N, Medlin LK, Melkonian M. 2013. Picomonas judraskeda gen. et sp. nov.: the first identified member of the Picozoa phylum nov., a widespread group of picoeukaryotes, formerly known as ‘picobiliphytes.’ PLoS One 8:e59565. doi: 10.1371/journal.pone.0059565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Massana R, Castresana J, Balagué V, Guillou L, Romari K, Groisillier A, Valentin K, Pedrós-Alió C. 2004. Phylogenetic and ecological analysis of novel marine stramenopiles. Appl Environ Microbiol 70:3528–3534. doi: 10.1128/AEM.70.6.3528-3534.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joli N, Gosselin M, Ardyna M, Babin M, Onda DF, Tremblay J-E, Lovejoy C. 2018. Need for focus on microbial species following ice melt and changing freshwater regimes in a Janus Arctic Gateway. Sci Rep 8:9405. doi: 10.1038/s41598-018-27705-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bachy C, López-García P, Vereshchaka A, Moreira D. 2011. Diversity and vertical distribution of microbial eukaryotes in the snow, sea ice and seawater near the North Pole at the end of the polar night. Front Microbiol 2:106. doi: 10.3389/fmicb.2011.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hou Y, Lin S. 2009. Distinct gene number-genome size relationships for eukaryotes and non-eukaryotes: gene content estimation for dinoflagellate genomes. PLoS One 4:e6978. doi: 10.1371/journal.pone.0006978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Galluzzi L, Bertozzini E, Penna A, Perini F, Garcés E, Magnani M. 2010. Analysis of rRNA gene content in the Mediterranean dinoflagellate Alexandrium catenella and Alexandrium taylori: implications for the quantitative real-time PCR-based monitoring methods. J Appl Phycol 22:1–9. doi: 10.1007/s10811-009-9411-3. [DOI] [Google Scholar]
- 47.Guillou L, Viprey M, Chambouvet A, Welsh RM, Kirkham AR, Massana R, Scanlan DJ, Worden AZ. 2008. Widespread occurrence and genetic diversity of marine parasitoids belonging to Syndiniales (Alveolata). Environ Microbiol 10:3349–3365. doi: 10.1111/j.1462-2920.2008.01731.x. [DOI] [PubMed] [Google Scholar]
- 48.Joli N, Monier A, Logares R, Lovejoy C. 2017. Seasonal patterns in Arctic prasinophytes and inferred ecology of Bathycoccus unveiled in an Arctic winter metagenome. ISME J 11:1372–1385. doi: 10.1038/ismej.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ward BB, Van Oostende N. 2016. Phytoplankton assemblage during the North Atlantic spring bloom assessed from functional gene analysis. J Plankton Res 38:1135–1150. doi: 10.1093/plankt/fbw043. [DOI] [Google Scholar]
- 50.Keeling PJ, Burki F, Wilcox HM, Allam B, Allen EE, Amaral-Zettler LA, Armbrust EV, Archibald JM, Bharti AK, Bell CJ, Beszteri B, Bidle KD, Cameron CT, Campbell L, Caron DA, Cattolico RA, Collier JL, Coyne K, Davy SK, Deschamps P, Dyhrman ST, Edvardsen B, Gates RD, Gobler CJ, Greenwood SJ, Guida SM, Jacobi JL, Jakobsen KS, James ER, Jenkins B, John U, Johnson MD, Juhl AR, Kamp A, Katz LA, Kiene R, Kudryavtsev A, Leander BS, Lin S, Lovejoy C, Lynn D, Marchetti A, McManus G, Nedelcu AM, Menden-Deuer S, Miceli C, Mock T, Montresor M, Moran MA, Murray S, et al. 2014. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol 12:e1001889. doi: 10.1371/journal.pbio.1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Onda DFL, Medrinal E, Comeau AM, Thaler M, Babin M, Lovejoy C. 2017. Seasonal and interannual changes in ciliate and dinoflagellate species assemblages in the Arctic Ocean (Amundsen Gulf, Beaufort Sea, Canada). Front Mar Sci 4:16. doi: 10.3389/fmars.2017.00016. [DOI] [Google Scholar]
- 52.Berges J. 1997. Miniview: algal nitrate reductases. Eur J Phycol 32:3–8. doi: 10.1017/S0967026296001084. [DOI] [Google Scholar]
- 53.Giordano M, Chen Y-B, Koblizek M, Falkowski PG. 2005. Regulation of nitrate reductase in Chlamydomonas reinhardtii by the redox state of the plastoquinone pool. Eur J Phycol 40:345–352. doi: 10.1080/09670260500334263. [DOI] [Google Scholar]
- 54.Song B, Ward BB. 2007. Molecular cloning and characterization of high-affinity nitrate transporters in marine phytoplankton. J Phycol 43:542–552. doi: 10.1111/j.1529-8817.2007.00352.x. [DOI] [Google Scholar]
- 55.Terrado R, Monier A, Edgar R, Lovejoy C. 2015. Diversity of nitrogen assimilation pathways among microbial photosynthetic eukaryotes. J Phycol 51:490–506. doi: 10.1111/jpy.12292. [DOI] [PubMed] [Google Scholar]
- 56.Gao Y, Smith GJ, Alberte RS. 2001. Temperature dependence of nitrate reductase activity in marine phytoplankton: biochemical analysis and ecological implications. J Phycol 36:304–313. doi: 10.1046/j.1529-8817.2000.99195.x. [DOI] [Google Scholar]
- 57.Takabayashi M, Wilkerson FP, Robertson D. 2005. Response of glutamine synthetase gene transcription and enzyme activity to external nitrogen sources in the diatom Skeletonema costatum (Bacillariophyceae). J Phycol 41:84–94. doi: 10.1111/j.1529-8817.2005.04115.x. [DOI] [Google Scholar]
- 58.Barwell-Clarke J, Whitney F. 1996. Institute of Ocean Sciences nutrient methods and analysis. Can Tech Rep Hydrogr Ocean Sci 182:1–50. [Google Scholar]
- 59.Hillebrand H, Dürselen C-D, Kirschtel D, Pollingher U, Zohary T. 1999. Biovolume calculation for pelagic and benthic microalgae. J Phycol 35:403–424. doi: 10.1046/j.1529-8817.1999.3520403.x. [DOI] [Google Scholar]
- 60.Menden-Deuer S, Lessard EJ. 2000. Carbon to volume relationships for dinoflagellates, diatoms, and other protist plankton. Limnol Oceanogr 45:569–579. doi: 10.4319/lo.2000.45.3.0569. [DOI] [Google Scholar]
- 61.Harding T, Jungblut AD, Lovejoy C, Vincent WF. 2011. Microbes in High Arctic snow and implications for the cold biosphere. Appl Environ Microbiol 77:3234–3243. doi: 10.1128/AEM.02611-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Church MJ, Short CM, Jenkins BD, Karl DM, Zehr JP. 2005. Temporal patterns of nitrogenase gene (nifH) expression in the oligotrophic North Pacific Ocean. Appl Environ Microbiol 71:5362–5370. doi: 10.1128/AEM.71.9.5362-5370.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Díez B, Pedrós-Alió C, Massana R. 2001. Study of genetic diversity of eukaryotic picoplankton in different oceanic regions by small-subunit rRNA gene cloning and sequencing. Appl Environ Microbiol 67:2932–2941. doi: 10.1128/AEM.67.7.2932-2941.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schloss PD, Handelsman J. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol 71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Comeau AM, Li WKW, Tremblay J-É, Carmack EC, Lovejoy C. 2011. Arctic Ocean microbial community structure before and after the 2007 record sea ice minimum. PLoS One 6:e27492. doi: 10.1371/journal.pone.0027492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schloss PD. 2009. A high-throughput DNA sequence aligner for microbial ecology studies. PLoS One 4:e8230. doi: 10.1371/journal.pone.0008230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Comeau AM, Philippe B, Thaler M, Gosselin M, Poulin M, Lovejoy C. 2013. Protists in Arctic drift and land-fast sea ice. J Phycol 49:229–240. doi: 10.1111/jpy.12026. [DOI] [PubMed] [Google Scholar]
- 70.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lovejoy C, Comeau A, Thaler M. 2016. Curated reference database of SSU rRNA for northern marine and freshwater communities of archaea, bacteria and microbial eukaryotes, v.1.1 (2002–2008). Nordicana D23. doi: 10.5885/45409XD-79A199B76BCC4110. [DOI] [Google Scholar]
- 72.Alves JMP, de Oliveira AL, Sandberg TOM, Moreno-Gallego JL, de Toledo MAF, de Moura EMM, Oliveira LS, Durham AM, Mehnert DU, Zanotto PM, Reyes A, Gruber A. 2016. GenSeed-HMM: a tool for progressive assembly using profile HMMs as seeds and its application in Alpavirinae viral discovery from metagenomic data. Front Microbiol 7:269. doi: 10.3389/fmicb.2016.00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol 7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jackman SD, Vandervalk BP, Mohamadi H, Chu J, Yeo S, Hammond SA, Jahesh G, Khan H, Coombe L, Warren RL, Birol I. 2017. ABySS 2.0: resource-efficient assembly of large genomes using a Bloom filter. Genome Res 27:768–777. doi: 10.1101/gr.214346.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.