Abstract
Phytohormones regulate a large variety of physiological processes in plants. In addition, salicylic acid (SA), jasmonic acid (JA), and ethylene (ET) are responsible for primary defense responses against abiotic and biotic stresses, while plant growth regulators, such as auxins, brassinosteroids (BRs), cytokinins (CKs), abscisic acid (ABA), and gibberellins (GAs), also contribute to plant immunity. To successfully colonize plants, filamentous pathogens like fungi and oomycetes have evolved diverse strategies to interfere with phytohormone pathways with the help of secreted effectors. These include proteins, toxins, polysaccharides as well as phytohormones or phytohormone mimics. Such pathogen effectors manipulate phytohormone pathways by directly altering hormone levels, by interfering with phytohormone biosynthesis, or by altering or blocking important components of phytohormone signaling pathways. In this review, we outline the various strategies used by filamentous phytopathogens to manipulate phytohormone pathways to cause disease.
Keywords: filamentous plant pathogen, effector, virulence, phytohormone, plant defense signaling
Introduction
Filamentous plant pathogens, like fungi and oomycetes, cause severe crop yield losses annually (Fisher et al., 2012). To protect themselves against pathogens, plants have evolved a multilayered defense network (Jones and Dangl, 2006). This immune system is activated when membrane localized pattern recognition receptors (PRRs) recognize microbe-associated molecular patterns (MAMPs) or host-derived damage-associated molecular patterns (DAMPs), leading to pattern triggered immunity (PTI) (Couto and Zipfel, 2016). The activation of PTI results in an array of cellular responses, including the generation of extracellular reactive oxygen species (ROS), cytosolic ion-flux changes, calcium-dependent or mitogen-activated protein kinase cascade activation, reinforcement of physical barriers, and the production of numerous defense-related molecules (Macho and Zipfel, 2014; Couto and Zipfel, 2016). In these complex immune responses, phytohormones play pivotal regulatory roles. Classical defense phytohormones are salicylic acid (SA), jasmonic acid (JA), and ethylene (ET). More recently, growth-related phytohormones, such as auxins, cytokinins (CKs), brassinosteroids (BRs), abscisic acid (ABA), and gibberellins (GAs) are also shown to modulate plant immune defenses (Pieterse et al., 2009, 2012; Tsuda and Katagiri, 2010; Berens et al., 2017).
Filamentous plant pathogens which can successfully colonize plants secrete an arsenal of effector proteins to interfere with plant defenses and facilitate pathogen colonization (Jones and Dangl, 2006; Dangl et al., 2013; Lo Presti et al., 2015). These effectors are categorized into two groups: apoplastic effectors that reside and function in the apoplast and cytoplasmic effectors that are taken up by plant cells to target various intracellular processes (Kamoun, 2006). In oomycetes, many of the cytoplasmic effectors are so called RxLR and Crinkler (CRN) effectors possessing an N-terminal RxLR motif or LxLFLAK motif, respectively, which are implicated in effector uptake (Jiang et al., 2008; Schornack et al., 2010; McGowan and Fitzpatrick, 2017). Fungal effectors which are taken up by host cells lack such a consensus motif. Given the importance of phytohormone pathways in plant immunity, it is no surprise that filamentous plant pathogens have evolved protein or toxin effectors targeting hormonal pathways. In addition, filamentous plant pathogens can also produce phytohormones and derivatives as host mimicry to manipulate or hijack host hormone homeostasis (Chanclud and Morel, 2016). In this communication, we review recent findings illustrating how this is achieved and discuss how such molecules enhance parasite fitness.
Effectors Targeting the Salicylic Acid Pathway
The phytohormone SA is a phenolic compound involved in various plant processes including growth, flowering, thermogenesis, senescence, and responses against abiotic and biotic stress (Raskin, 1992; Vlot et al., 2009; Dempsey et al., 2011). SA has been extensively studied for its role in local and systemic acquired resistance (LAR and SAR) against biotrophic and hemibiotrophic pathogens (Malamy et al., 1990; Metraux et al., 1990; Raskin, 1992; Klessig and Malamy, 1994; Glazebrook, 2005; Vlot et al., 2009; Dempsey et al., 2011).
SA is synthesized from chorismate, the end product of the shikimate pathway, via two distinct biosynthetic pathways. The phenylalanine ammonia lyase (PAL) pathway starts with the Claisen rearrangement of chorismate to prephenate catalyzed by chorismate mutase, followed by the formation of phenylalanine. Subsequently, PAL catalyzes the conversion of phenylalanine to cinnamate, which can be converted to SA in a series of enzymatic steps (Klessig and Malamy, 1994; Metraux, 2002; Dempsey et al., 2011). In the isochorismate (IC) pathway, chorismate is converted to SA in the chloroplast via two reactions catalyzed by isochorismate synthase (ICS) and isochorismate pyruvate lyase (IPL), respectively (Figure 1A; Wildermuth et al., 2001; Strawn et al., 2007; Garcion et al., 2008; Dempsey et al., 2011).
Figure 1.
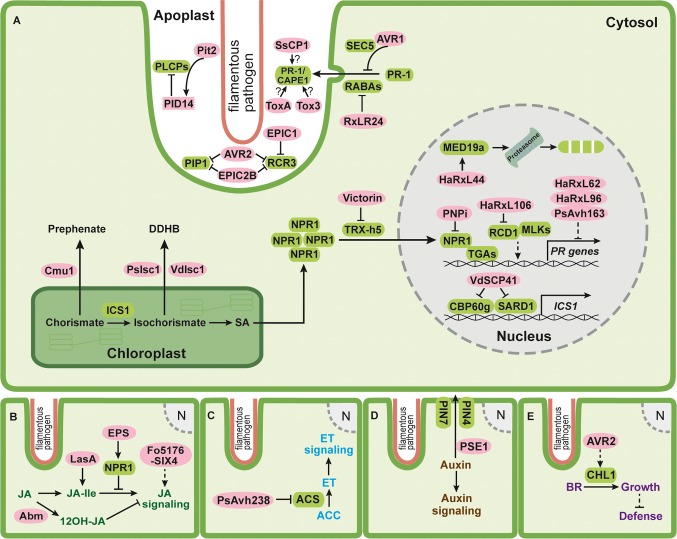
Schematic overview of effectors of filamentous phytopathogens targeting phytohormone pathways. (A) SA (salicylic acid) pathway; (B) JA (jasmonic acid) pathway; (C) ET (ethylene) pathway; (D) auxin pathway; and (E) BR (brassinosteroid) pathway. Infection structures of filamentous pathogens penetrating a plant cell are lined with salmon color. This structure or specialized feeding structures (not indicated) are the sites for secretion of pathogen effectors. The plant plasma membrane is shown in green, the plant cytosol is shown in light green, the chloroplast is outlined with dark green, and the plant nucleus is shown in gray. In (A), the apoplastic space between pathogen and plant plasma membrane is enlarged. Pathogen effectors residing either in the apoplast or in the plant cytosol are indicated by pink ovals. Plant components targeted by effectors are depicted as rounded green rectangles. Solid lines represent characterized reactions or direct interactions and dashed lines represent indirect interactions. Arrows indicate activation and bar headed lines indicate inhibition. Question marks indicate that the underlying mechanism is not yet clear.
To interfere with SA-mediated defenses, a direct and efficient way is to prevent the formation of SA. This strategy has been exploited by several filamentous plant pathogens. The biotrophic fungus Ustilago maydis, which is the causative agent of corn smut disease, secretes an active chorismate mutase Cmu1 converting chorismate to prephenate (Figure 1A; Djamei et al., 2011). Plants infected with cmu1 deletion mutants displayed significantly higher SA levels, and such mutants were less virulent than wild type strains (Djamei et al., 2011). Cmu1 is a cytoplasmic effector and it is postulated that translocated Cmu1 increases cytosolic chorismate mutase activity and diverts the flow of chorismate into the phenylpropanoid pathway, thus hindering SA biosynthesis and immunity against this biotrophic pathogen (Figure 1A; Djamei et al., 2011). Secreted chorismate mutases are not only found in several other smut fungi but also in the necrotrophic fungus Sclerotinia sclerotiorum. This could suggest that lowering SA levels might be a common strategy to suppress host defenses (Djamei et al., 2011; Kabbage et al., 2013; Derbyshire et al., 2017). However, Cmu1 was recently also found to interact with the maize kiwellin ZmKWL1 in the apoplast (Han et al., 2019). ZmKWL1 was shown to be a defense-related protein, which significantly inhibited the chorismate mutase activity of Cmu1 (Han et al., 2019). The interaction of Cmu1 with ZmKWL1 suggests that Cmu1 has an additional role in the apoplast besides its metabolic reprogramming activity. For the secreted chorismate mutase of S. sclerotiorum, it still needs to be investigated whether this protein affects SA metabolism or inhibits apoplastic defense responses. An alternative way to lower SA levels is employed by the oomycete pathogen Phytophthora sojae and the fungus Verticillium dahliae, which secrete isochorismatases PsIsc1 and VdIsc1, respectively, via an unconventional route (Liu et al., 2014). PsIsc1 was shown to function inside plant cells (Figure 1A; Liu et al., 2014). Isochorismatases convert isochorismate to 2,3-dihydro-2,3-dihydroxybenzoate (DDHB) and pyruvate, making isochorismate unavailable for SA biosynthesis. Silencing of PsIsc1 in P. sojae or inactivation of VdIsc1 in V. dahliae increased SA levels in infected plant tissue and led to the induction of the SA marker gene pathogenesis-related protein 1 (PR-1) (Liu et al., 2014). Interestingly, some fungi also produce salicylate hydroxylases that degrade SA to catechol in the fungal cytosol, which could potentially contribute to lowering SA levels in infected tissue. However, so far, salicylate hydroxylases are not yet implicated in virulence.
SA biosynthesis is tightly regulated by a complex transcriptional network (Vlot et al., 2009; Dempsey et al., 2011). Two closely related transcription factors calmodulin-binding protein 60 g (CBP60g) and SAR deficient1 (SARD1) positively regulate the SA-induced defense response through binding to promoter region of the SA biosynthetic gene ICS1 (Figure 1A; Wang et al., 2009, 2011a; Sun et al., 2015, 2018). cbp60g/sard1 double mutants of Arabidopsis thaliana were more susceptible to V. dahliae infection, illustrating that CBP60g and SARD1 promote immunity against V. dahliae (Qin et al., 2018). The nuclear effector VdSCP41 of V. dahliae was recently shown to target A. thaliana CBP60g and SARD1 (Figure 1A; Qin et al., 2018). Biochemical assays revealed that VdSCP41 bound to the transcription activation domain in the C-terminus of CBP60g, compromising its transcription activity required for the induction of ICS1 (Figure 1A; Qin et al., 2018). The deletion of VdSCP41 in V. dahliae reduced virulence, whereas A. thaliana plants expressing VdSCP41 exhibited compromised PTI-triggered expression of ICS1 and showed increased disease symptoms after infection with V. dahliae (Qin et al., 2018).
Nonexpressor of PR genes1 (NPR1) is the master regulator of SA-mediated plant immune responses (Cao et al., 1994, 1997; Shah et al., 1997; Dong, 2004; Wang et al., 2006). In uninfected plants, NPR1 oligomers reside in the cytosol in an inactive state. SA production in response to pathogen attack leads to NPR1 phosphorylation and subsequent monomerization, allowing its translocation into the nucleus to activate PR gene expression (Figure 1A; Kinkema et al., 2000; Mou et al., 2003; Lee et al., 2015). NPR1 regulates the expression of PR genes through interaction with several TGA transcription factors (Figure 1A; Despres et al., 2000; Dong, 2004; Kesarwani et al., 2007; Fu and Dong, 2013). Due to its essential role in plant immunity, NPR1 presents an interesting effector target to subvert SA-mediated defenses (Lorang et al., 2012; Kazan and Lyons, 2014).
A yeast two-hybrid screen with a library from wheat infected by the yellow stripe rust Puccinia striiformis f. sp. tritici identified a conserved rust protein PNPi (for Puccinia NPR1 interactor) as an NPR1 interaction partner (Figure 1A; Wang et al., 2016). PNPi compromised the interaction between NPR1 and TGA2.2 in a yeast three-hybrid assay (Figure 1A; Wang et al., 2016). Overexpression of PNPi in barley reduced the expression of several PR genes normally induced upon biotic stress, suggesting that PNPi might reduce PR gene expression via blocking the binding of NPR1 to TGA transcription factors also in vivo (Wang et al., 2016). The necrotrophic fungus Cochliobolus victoriae causes Victoria blight on oat and A. thaliana by secreting the cyclic peptide toxin effector victorin (Lorang et al., 2004, 2007). Victorin sensitivity in A. thaliana requires both the locus orchestrating victorin effects1 (LOV1) belonging to the nucleotide-binding leucine rich repeat (NLR) protein family and the defense-associated thioredoxin TRX-h5 (Lorang et al., 2007; Sweat and Wolpert, 2007). TRX-h5 is upregulated in response to pathogen challenge and catalyzes the critical conformational change of NPR1 from the oligomeric to monomeric state (Figure 1A; Laloi et al., 2004; Tada et al., 2008). In A. thaliana plants lacking LOV1, victorin binds to the active site of TRX-h5 and inhibits its activity, thus blocking monomerization of NPR1 and subsequent SA-mediated defense responses (Figure 1A; Lorang et al., 2012). When LOV1 is present, it acts as the guard of TRX-h5. The binding of victorin to TRX-h5 activates LOV1 and leads to hypersensitive response (HR)-like cell death, which favors the necrotrophic lifestyle of C. victoriae (Lorang et al., 2012). This illustrates that C. victoriae hijacks the guard function of LOV1 to evoke cell death and facilitate necrotrophic development.
PR-1 gene expression has been extensively employed as an SA marker due to its strong induction during SA-mediated plant immune responses (Lotan et al., 1989; Van Loon and Van Strien, 1999; Dong, 2004). PR-1 proteins are delivered to the apoplastic space and their successful secretion is prerequisite for their functions (Carr et al., 1987; Wang et al., 2005). PR-1 overexpression as well as in vitro studies using spore germination or infection structure differentiation as readouts indicated that PR-1 proteins might show antimicrobial activity (Alexander et al., 1993; Niderman et al., 1995; Rauscher et al., 1999; Sarowar et al., 2005; Kiba et al., 2007). However, very high concentrations were needed to observe such an activity, putting into question the biological relevance of such observations (Breen et al., 2017). Recently, PR-1 was shown to bind sterols and inhibit the growth of Phytophthora brassicae, whereas no effect was seen on growth of the fungal species Aspergillus niger and Botrytis cinerea with the same treatment (Gamir et al., 2017). The inhibition of P. brassicae by purified PR-1 protein P14c from tomato could be alleviated when cholesterol was added, suggesting a link between sterol-binding activity and growth inhibition (Gamir et al., 2017). These authors speculate that the selective growth inhibition of the oomycete by P14c may result from the sterol auxotrophy of P. brassicae, i.e. P. brassicae relies on environmental sterols, and P14c may deplete this supply by binding sterols. The RxLR effector RxLR24 from P. brassicae binds several A. thaliana RABA GTPases that are required for vesicle-mediated secretion of proteins (Figure 1A; Tomczynska et al., 2018). This inhibited the secretion of PR-1 proteins and presumably other defense proteins, in line with the need to reduce PR-1 levels in the apoplast for disease development of this hemibiotrophic oomycete. A similar situation exists in the potato blight oomycete Phytophthora infestans. This hemibiotroph secretes the RxLR effector AVR1 which interacts with and stabilizes Sec5, a subunit of exocyst complex (Figure 1A; Du et al., 2015). Since the secretion of PR-1 requires Sec5, the authors speculate that a stabilized exocyst by AVR1 may block focal secretion of PR-1 and other defense compounds (Figure 1A; Du et al., 2015, 2018). While the suppression of PR-1 levels by oomycete pathogen effectors is in line with their sterol auxotrophy and the sterol-binding activity of PR-1, it is presently not evident why the necrotrophic fungus S. sclerotiorum, a sterol prototroph, should secrete the cerato-platanin-like SsCP1 effector that directly interacts with A. thaliana PR-1 in the apoplast (Figure 1A; Yang et al., 2018b). ScCP1 also promotes virulence, induces necrosis-like cell death at high concentrations, and activates the SA pathway (Yang et al., 2018b). In addition, the necrotrophic wheat pathogen Parastagonospora nodorum secretes two effectors ToxA and Tox3 which target certain PR-1 isoforms including PR-1-5 (Figure 1A; Lu et al., 2014; Breen et al., 2016). PR-1-5 enhanced the necrosis inducing ability of purified ToxA on wheat leaves harboring the toxin sensitivity gene Tsn1 (Lu et al., 2014). In later studies, CAP-derived peptides (CAPE) were detected in some PR-1 proteins (Chen et al., 2014; Breen et al., 2017). CAPE1 peptides can be proteolytically liberated from PR-1b after wounding, classifying them as DAMPs (Chen et al., 2014). Infiltration with CAPE1 peptides enhanced wheat cell death caused by Tox3 in a wheat line carrying Snn3 (Breen et al., 2016). It is therefore likely that the positive biological function of PR-1 proteins toward filamentous pathogens might be connected to the liberation of defense signaling CAPE peptides.
During SA-triggered defense responses papain-like cysteine proteases (PLCPs) are secreted to the apoplastic space and play prominent roles in plant immunity against biotrophic and hemibiotrophic filamentous pathogens (Shabab et al., 2008; van der Linde et al., 2012; Misas-Villamil et al., 2016). It was shown that treatment of tomato with the SA analog benzothiadiazole (BTH) specifically induced the transcription of two PLCPs, PIP1, and RCR3 (Shabab et al., 2008). PIP1 and RCR3 are both inhibited by apoplastic effectors from evolutionarily unrelated pathogens, namely AVR2 from the tomato leaf mold fungus Cladosporium fulvum and extracellular cystatin-like protease inhibitor 2b (EPIC2B) from the oomycete P. infestans (Figure 1A; Tian et al., 2007; Shabab et al., 2008; Song et al., 2009). In addition, RCR3 is also inhibited by EPIC1 from P. infestans (Figure 1A; Song et al., 2009). While cystatin-like protease inhibitors like EPIC2B inhibit PLCPs via a conserved QxVxG motif, a distinct inhibition mechanism is speculated for AVR2 lacking an QxVxG motif (Shabab et al., 2008; Kaschani et al., 2010; Kaschani and Van der Hoorn, 2011).
Maize PLCPs, such as CP1, CP2, and XCP2, are also activated upon SA treatment as part of the SA-mediated defense response (van der Linde et al., 2012). Recently, an immune signaling peptide, Zea mays immune signaling peptide 1 (Zip1), was shown to be released from its propeptide precursor by SA-induced PLCPs and demonstrated to activate downstream SA defense signaling (Ziemann et al., 2018). The maize cystatin CC9 strongly induced upon infection by U. maydis was shown to inhibit apoplastic PLCP activity and this promoted U. maydis colonization (van der Linde et al., 2012). The virulence-promoting apoplastic effector Pit2 of U. maydis inhibits maize PLCPs via a novel 14 amino acid long motif (PID14) initially defined by mutational analyses and synthetic peptides (Figure 1A; Doehlemann et al., 2011; Mueller et al., 2013). Recent analyses demonstrated that Pit2 is processed by maize PLCPs, and as a result, a released inhibitory portion inside the PID14 domain remains bound to the PLCP and blocks its activity (Misas Villamil et al., 2019). The U. maydis Pit2 effector thus functions as a substrate mimicking molecule. The PID14 core motif is present in proteins of several plant associated fungi and bacteria, indicating the existence of a conserved microbial inhibitor motif of proteases (Misas Villamil et al., 2019).
In systems not allowing reverse genetics, effectors are often expressed constitutively in plants and effects on SA signaling are then inferred by treating such plants with SA and observing changes in PR-1 expression. With such an approach, the effector HaRxL44 from the oomycete Hyaloperonospora arabidopsidis (Hpa) was shown to suppress PR-1 expression after SA treatment (Caillaud et al., 2013). HaRxL44 is a nuclear effector interacting with and promoting the proteasomal degradation of the Mediator subunit MED19a likely by acting as an adaptor protein for E3 ligases (Figure 1A; Caillaud et al., 2013). After SA treatment, PR-1 expression was elevated in Arabidopsis plants overexpressing MED19a and med19a mutants showed reduced PR-1 transcript levels. In transgenic Arabidopsis plants overexpressing HaRxL44, PR-1 expression was strongly reduced (Caillaud et al., 2013). Suppression of PR-1 expression by Hpa occurred specifically in cells containing haustoria, sites of delivery of RxLR effectors (Caillaud et al., 2013; Whisson et al., 2016). This indicates that Hpa colonization requires HaRxL44-induced destabilization of MED19a to decrease SA-mediated defense responses. Another Hpa effector, HaRxL106, was also recently shown to dampen SA-mediated defenses. HaRxL106-expressing lines displayed significantly reduced PR-1 expression compared to wild type plants after SA treatment (Wirthmueller et al., 2018). PR-1 expression in Arabidopsis plants overexpressing NPR1 was suppressed by HaRxL106, even though neither protein levels nor subcellular localization of NPR1 were affected. Intriguingly, HaRxL106 interacted with radical-induced cell death 1 (RCD1), a nuclear protein shown to activate SA-mediated PR-1 gene expression (Figure 1A; Wirthmueller et al., 2018). In addition, RCD1 interacted with MUT9-like kinases (MLKs), which phosphorylate photoreceptor cryptochrome 2 (CRY2), phytochrome interacting factor 3 (PIF3) and histone H3 Thr3 (H3T3ph) (Wang et al., 2015; Liu et al., 2017; Ni et al., 2017). The authors speculate that HaRxL106 might act downstream of NPR1 and influence the transcriptional activity of the RCD1/MLK complex to prevent the activation of SA signaling (Wirthmueller et al., 2018). Three additional RxLR effector proteins HaRxL62, HaRxL96 and PsAvh163 from Hpa and P. sojae, respectively, were also able to suppress SA mediate defenses (Figure 1A; Anderson et al., 2012; Asai et al., 2014). Transgenic Arabidopsis plants expressing these effectors individually showed elevated susceptibility to Hpa compared to wild type plants. In addition, when challenged with SA, the transgenic plants exhibited reduced PR-1 expression, suggesting that these effectors compromise SA-triggered immunity (Anderson et al., 2012; Asai et al., 2014). In most of these cases, it remains open at which level PR-1 gene expression is affected, and further investigations are needed to elucidate the molecular targets of these effectors.
Collectively, these studies demonstrate that filamentous pathogens produce a cocktail of effectors not only to directly disrupt SA homeostasis but also to target more selectively diverse components like NPR1, PLCPs, and PR-1 in the SA signaling pathway. The deployment of effectors for SA pathway interference in fungi and oomycetes indicates convergent evolution to target this important hormone pathway.
Effectors Targeting the Jasmonic Acid Pathway
The JA pathway has long been thought to allow plants to cope with various environmental stresses including attack by necrotrophic pathogens and herbivores (Thomma et al., 1998, 1999; Glazebrook, 2005). More recently, it has also been shown that JA-mediated defenses contribute to resistance against some biotrophic or hemibiotrophic pathogens (Thaler et al., 2004; Riemann et al., 2013; Lemarie et al., 2015). In rice, JA-mediated defenses conferred immunity against the hemibiotrophic rice blast fungus Magnaporthe oryzae (Riemann et al., 2013). To overcome JA-mediated defenses, M. oryzae secretes the hydroxylated JA molecule 12OH-JA during the initial biotrophic stage (Patkar et al., 2015). The conversion of JA to 12OH-JA is catalyzed by a secreted fungal monooxygenase Abm (Figure 1B; Miersch et al., 2008; Patkar et al., 2015). It is likely that M. oryzae employs Abm to convert both fungal and host-derived JA to 12OH-JA to avoid triggering host JA-mediated immunity (Patkar et al., 2015). Indeed, an abm mutant of M. oryzae failed to produce blast symptoms on rice and accumulated large amounts of methyl JA (MeJA) in infected tissue, which provoked strong host defense responses (Patkar et al., 2015). This shows that 12OH-JA acts as a metabolite effector blocking JA-triggered defense responses.
By contrast, the hemibiotrophic ascomycete fungus Fusarium oxysporum causing root wilt is reported to produce JAs (Cole et al., 2014). JA is precursor of JA-isoleucine (JA-Ile), the ligand of the F-box protein coronatine insensitive 1-jasmonate ZIM-domain (COI1-JAZ) co-receptor complex (Yan et al., 2009). A. thaliana coi1 plants that are defective in JA signaling exhibited higher resistance against F. oxysporum than wild-type plants, indicating that F. oxysporum requires COI1-mediated JA signaling to promote virulence (Thatcher et al., 2009). Surprisingly, plant endogenous JA biosynthesis appeared dispensable for COI1-mediated JA signaling during F. oxysporum colonization (Thatcher et al., 2009; Cole et al., 2014), making it likely that JA molecules produced by F. oxysporum are used in place of plant JA to activate JA signaling (Cole et al., 2014). In addition, it has also been demonstrated that the virulence-promoting secreted in xylem (SIX) effector Fo5176-SIX4 activates JA signaling (Figure 1B; Thatcher et al., 2012). Whether this is direct and at which stage this occurs remain to be determined. The necrotrophic grapevine pathogen Lasiodiplodia mediterranea also activates JA signaling by producing the JA ester lasiojasmonate A (LasA) (Figure 1B; Chini et al., 2018). LasA can be converted to JA-Ile, a strong activator of JA signaling and inducer of cell death. LasA is thus proposed to act as a metabolite effector in late stages of infection to activate JA-mediated cell death and facilitate necrotrophy (Chini et al., 2018).
Effectors Targeting the Ethylene Pathway
The gaseous phytohormone ET is well known for its role in fruit ripening and plant senescence (Burg and Burg, 1965; Grbić and Bleecker, 1995; Bleecker and Kende, 2000). ET-insensitive A. thaliana and soybean plants are more susceptible to some pathogens and activation of ET signaling confers plant resistance upon pathogen attack, suggesting that ET signaling also plays a role in plant defense (Hoffman et al., 1999; Thomma et al., 1999; Berrocal-Lobo et al., 2002; Yang et al., 2017b).
The precursor for ET biosynthesis is 1-aminocyclopropane-1-carboxylic acid (ACC), which is derived from S-adenosylmethionine (S-AdoMet) in a reaction catalyzed by ACC synthase (ACS) (Figure 1C; Wang et al., 2002). ET production is directly correlated with ACS activity (Christians et al., 2009; Skottke et al., 2011; Li et al., 2012; Helliwell et al., 2016). The polymorphic RxLR effector PsAvh238 of P. sojae is strongly upregulated during early infection and essential for virulence (Figure 1C; Wang et al., 2011b; Yang et al., 2017a). A recent study uncovered that PsAvh238 interacts with soybean Type2 ACSs (GmACSs). By destabilizing GmACSs, PsAvh238 suppresses ET biosynthesis and this facilitates P. sojae infection (Figure 1C; Yang et al., 2018a). Silencing of GmACSs as well as inhibition of ET signaling or synthesis with chemical antagonists increased virulence of P. sojae, whereas overexpression of GmACSs in Nicotiana benthamiana leaves enhanced resistance (Yang et al., 2018a). A PsAvh238 mutant was unable to inhibit ET signaling and showed reduced virulence (Yang et al., 2018a), consistent with the notion that ET-mediated defenses have to be downregulated in this hemibiotrophic pathosystem.
Conversely, the necrotrophic fungal pathogen Cochliobolus miyabeanus causing brown spot of rice requires ET signaling for pathogenesis (Van Bockhaven et al., 2015). While exogenous application of ethephon, which is quickly converted to ET in planta, promoted disease development, ET-insensitive rice plants were more resistant to C. miyabeanus (De Vleesschauwer et al., 2010). Furthermore, C. miyabeanus was shown to produce ET, and the fungus-derived ET constituted the prevalent source of ET in infected tissues (Van Bockhaven et al., 2015). Blocking fungal ET synthesis with specific chemical inhibitors significantly compromised C. miyabeanus colonization of rice leaves (Van Bockhaven et al., 2015). This makes it likely that C. miyabeanus uses ET as a metabolite effector to promote virulence (Van Bockhaven et al., 2015).
Effectors Targeting the Auxin Pathway
Auxins constitute a group of indolic molecules that have long been recognized for their multiple roles in plant growth, development, and pathogen-host interactions (Teale et al., 2006; Barbier et al., 2017). A. thaliana mutants defective in auxin signaling were more susceptible to the necrotrophic fungi Plectosphaerella cucumerina and B. cinerea, and the application of the auxin transport inhibitor 2,3,5-triiodobenzoic acid (TIBA) rendered Arabidopsis more susceptible to P. cucumerina (Llorente et al., 2008).
Several microorganisms induce plant galls/tumors with high auxin levels. These structures likely provide the environment for pathogen differentiation and/or pathogen dissemination (Yamada, 1993; Kazan and Manners, 2009). One such example where tumors were shown to contain high IAA (indole-3-acetic acid) auxin levels are those induced by U. maydis (Turian and Hamilton, 1960). U. maydis is able to produce IAA from tryptophan (Reineke et al., 2008). Deleting two IAA dehydrogenases and a transaminase genes resulted in significantly reduced IAA production of U. maydis in axenic culture, but tumor formation was unaltered and IAA levels in in tumors were indistinguishable from those in wild type infections (Reineke et al., 2008). This suggested that fungal IAA does neither act as effector for tumor induction nor for elevating IAA levels in tumor tissue (Reineke et al., 2008).
The stem rust fungus Puccinia graminis f. sp. tritici (Pgt) produces a putative tryptophan 2-monooxygenase (Pgt-IaaM) that generates indole-3-acetamide (IAM), the precursor for IAA biosynthesis (Yin et al., 2014). In Pgt-infected wheat leaves, the expression of Pgt-IaaM was strongly induced in haustorial cells, and higher IAA levels were observed. Silencing of Pgt-IaaM during Pgt infection on wheat via host-induced gene silencing (HIGS) compromised the virulence of Pgt, whereas transgenic Arabidopsis plants constitutively expressing Pgt-IaaM displayed increased accumulation of IAA and susceptibility to biotic stress. This suggests that Pgt produced IAA acts as a virulence promoting effector in this pathosystem (Yin et al., 2014).
Directional auxin transport is controlled by an efflux carrier complex containing the PIN-formed (PIN) family proteins (Robert and Friml, 2009). The RxLR effector penetration-specific effector 1 (PSE1) of the oomycete Phytophthora parasitica infecting Arabidopsis is transiently upregulated during penetration of host roots (Evangelisti et al., 2013). The overexpression of PSE1 in Arabidopsis reduced auxin accumulation and increased susceptibility to P. parasitica. In addition, PSE1 expressing Arabidopsis plants displayed significantly enhanced accumulation of auxin exporters PIN4 and PIN7 at the root apex (Figure 1D; Evangelisti et al., 2013). It is therefore proposed that PSE1 promotes infection by altering auxin physiology (Evangelisti et al., 2013). How PSE1 achieves this mechanistically remains to be determined.
Effectors Targeting the Brassinosteroid Pathway
Brassinosteroids (BRs) are a class of polyhydroxylated steroidal phytohormones that are implicated in a wide range of plant physiological and developmental processes as well as plant defense responses (Clouse, 2011; Wang et al., 2012). It was shown that activation of BR signaling increases the susceptibility of A. thaliana and rice to Hpa and Pythium graminicola, respectively (Belkhadir et al., 2012; De Vleesschauwer et al., 2012). BR insensitive 1 (BRI1) is a leucine-rich repeat-receptor-like kinase (LRR-RLK), which perceives and transduces BR signals (Wang et al., 2001). BR signaling also involves the Kelch-repeat containing protein phosphatase BRI1 suppressor 1 (BSU1) (Mora-García et al., 2004). In A. thaliana, BR signaling regulates the tradeoff between plant growth and defense via modulating the transcription factors brassinazole-resistant 1 (BZR1) and homolog of brassinosteroid enhanced expression2 interacting with ibh1 (HBI1), two negative regulators of plant defense responses (Lozano-Durán and Zipfel, 2015). The oomycete P. infestans secretes the RxLR effector AVR2 during its biotrophic stage of potato colonization (Figure 1E; Gilroy et al., 2011). AVR2 interacts with the phosphatase BSU-like protein1 (BSL1), which is homologous to A. thaliana BSU1, suggesting a possible link between AVR2 and BR signaling (Saunders et al., 2012). Indeed, overexpression of AVR2 in potato constitutively activated the expression of several BR responsive genes including StCHL1, a basic helix-loop-helix (bHLH) transcription factor homologous to HBI1 in A. thaliana (Figure 1E; Turnbull et al., 2017). Transient overexpression of either AVR2 or StCHL1 facilitated disease development of P. infestans on N. benthamiana, whereas silencing of the CHL1 ortholog of N. benthamiana compromised susceptibility to P. infestans (Turnbull et al., 2017). It is hypothesized that AVR2 hijacks StCHL1 to activate BR signaling and suppress plant immunity (Turnbull et al., 2017). This highlights a new strategy to suppress plant immunity via exploiting the tradeoff between plant growth and immunity.
Effectors Targeting the Cytokinin Pathway
Cytokinins (CKs) are N6-substituted adenine derivatives playing important roles in plant development and stress responses (Kieber and Schaller, 2018). The CK biosynthetic pathway involves adenylate isopentenyltransferase (IPT), cytochrome P450 monooxygenase and LONELY GUY (LOG) enzymes (Takei et al., 2001, 2004; Sakakibara, 2006; Kurakawa et al., 2007). CKs can also originate from degradation of modified tRNAs in a reaction catalyzed by tRNA-IPT (Miyawaki et al., 2006). CK biosynthetic genes are detected in many plant pathogens, suggesting that CKs might be produced by pathogens to promote disease (Chanclud and Morel, 2016; Sorensen et al., 2018).
The ergot fungus Claviceps purpurea produces large amount of CKs in axenic culture and during early infection. The deletion of CptRNA-IPT abolished the production of cis-zeatin (cZ) and reduced virulence while mutants lacking either a bifunctional CpIPT-LOG or CpP450 were unaffected in virulence (Hinsch et al., 2015, 2016). However, double mutants lacking CpIPT-LOG and CptRNA-IPT were severely attenuated in virulence, illustrating that both fungal CK production pathways contribute to virulence in this pathosystem (Hinsch et al., 2016). U. maydis also produces cZ-type CKs in axenic culture and in infected plant tissues (Bruce et al., 2011; Morrison et al., 2015). U. maydis mutants lacking tRNA-IPT were deficient in cZ synthesis and showed reduced virulence in seedling infection in comparison to wild type strains (Morrison et al., 2017). Similarly, the deletion of CKS1 encoding tRNA-IPT in M. oryzae also led to a significant reduction of rice blast symptoms and increased plant defense responses (Chanclud et al., 2016). The virulence defect of the CKS1 mutant could be restored when CK was exogenously applied, reinforcing the link between CK production and pathogenicity (Chanclud et al., 2016). Recently, a new class of cytokinins called Fusarium cytokinins was shown to be produced by the cereal pathogen Fusarium pseudograminearum (Sorensen et al., 2018). Fusarium cytokinins acts as cytokinin agonists and could activate a histidine kinase 3 cytokinin receptor in a heterologous bacterial system. The expression of the biosynthetic gene cluster for Fusarium cytokinins was induced during F. pseudograminearum infection of barley, but a role in virulence has not yet been demonstrated (Sorensen et al., 2018).
It is thus likely that in these fungal pathosystems CKs function as effectors to suppress host immune responses during colonization.
Effectors Targeting the Gibberellin Pathway
Gibberellins (GAs) were initially identified from Fusarium fujikuroi, which is causative for “bakanae” disease of rice seedlings (Wulff et al., 2010). Later, GAs were also found to be produced by plants and shown to play vital roles in regulating plant growth and development (Yamaguchi, 2008). GA biosynthetic genes of F. fujikuroi are strongly induced in rice roots colonized by F. fujikuroi. In comparison to the GA-producing wild type strain, GA-nonproducing mutants lacking the entire GA biosynthetic gene cluster showed comparable root penetration and apoplastic growth but were compromised in further invasion of rice tissue, suggesting that secreted GA is used as effector for “bakanae” disease development (Wiemann et al., 2013). However, the GA biosynthetic gene cluster is restricted to Fusarium species but GA production is only found in F. fujikuroi (Wiemann et al., 2013). It remains unclear how GA contributes mechanistically to the virulence of F. fujikuroi. In the M. oryzae/rice pathosystem, it has been demonstrated that mutants lacking GA inactivating enzyme elongated uppermost internode (EUI) were more susceptible to infection by M. oryzae, whereas mutants in gibberellin 20-oxidase (GA20OX3) involved in GA synthesis showed increased rice blast resistance. This indicates that the GA pathway also plays a positive role in this pathosystem (Yang et al., 2008; Qin et al., 2013).
Effectors Targeting the Abscisic Acid Pathway
Abscisic acid (ABA) is a sesquiterpenoid synthesized via two distinct pathways involving the proteins ABA1, ABA4, NCED, ABA2, and ABA3 (Endo et al., 2014). It is an essential hormone regulating plant developmental processes and adaptive responses to diverse abiotic and biotic stresses (Cutler et al., 2010). ABA has a negative role on plant resistance against some biotrophic filamentous pathogens, such as Hpa, Fusarium graminearum, M. oryzae, and Golovinomyces cichoracearum (Fan et al., 2009; Jiang et al., 2010; Buhrow et al., 2016; Xiao et al., 2017). On the other hand, plants require the ABA pathway for resistance against several necrotrophic pathogens, including Pythium irregulare, P. cucumerina, C. miyabeanus, and Alternaria brassicicola (Ton and Mauch-Mani, 2004; Adie et al., 2007; Fan et al., 2009; De Vleesschauwer et al., 2010). Fungi like M. oryzae, U. maydis, and B. cinerea are able to synthesize ABA (Siewers et al., 2006; Bruce et al., 2011; Spence et al., 2015). An aba4 mutant of M. oryzae lacking one of the biosynthetic genes for ABA biosynthesis and producing reduced ABA levels was severely affected in appressoria formation on a hydrophobic surface in comparison to the wild type strain. Exogenous ABA application largely restored this defect (Spence et al., 2015). Furthermore, the aba4 mutant lost the ability to infect rice. However, the aba4 mutant also displayed a strong growth defect and morphological abnormalities which could not be complemented by external ABA (Spence et al., 2015). So far, it has not been possible to separate the endogenous function from a function of ABA as virulence-promoting effector in this system.
Effectors Targeting Phytohormone Crosstalk
Hormonal signaling pathways are often interconnected, and this can lead to synergistic or antagonistic functions (Weiss and Ori, 2007; Choi et al., 2010; Jiang et al., 2010; Argueso et al., 2012; Pieterse et al., 2012; Naseem et al., 2014, 2015; Berens et al., 2017). Examples are the antagonism between SA and JA pathway (Kunkel and Brooks, 2002; Takahashi et al., 2004; Spoel and Dong, 2008), and the synergism between JA and ET pathways (Xu et al., 1994; Lorenzo et al., 2003; Zhu et al., 2011) in defense signaling. Furthermore, growth-promoting hormones rely on crosstalk with defense-related hormones to balance growth-defense tradeoffs (Pieterse et al., 2009; Huot et al., 2014; Berens et al., 2017). In this context, fungal-derived phytohormones can be effectors directly influencing the crosstalk with other hormones. Interference with phytohormone crosstalk can also involve polysaccharide: the necrotrophic fungus B. cinerea secretes the exopolysaccharide (EPS) β-(1,3)(1,6)-D-glucan to promote infection of tomato (Figure 1B; Stahmann et al., 1995; El Oirdi et al., 2011). Tomato leaves pretreated with EPS were more susceptible to B. cinerea and showed enhanced SA accumulation but decreased expression of JA-marker genes PI I and PI II (El Oirdi et al., 2011). The expression of NPR1 was also induced after B. cinerea infection and knockdown of NPR1 led to significantly increased expression of these two JA-marker genes. This suggests that EPS functions as an effector which activates SA signaling and inhibit the JA signaling pathway via NPR1 (Figure 1B; El Oirdi et al., 2011).
Conclusions and Outlook
Studies in recent years have uncovered that most filamentous plant pathogens use interference with hormonal pathways as an effective strategy to promote colonization. Nearly, all phytohormone pathways can be targeted with beneficial consequences for the pathogens. The mechanisms how filamentous plant pathogens induce changes in phytohormone levels and/or signaling have become more complex by realizing that many filamentous phytopathogens can also produce phytohormones or derivatives for host phytohormone mimicry. The regulation of hormone production and deployment by fungal plant pathogens is only beginning to surface and indicates that phytohormones can be used as effectors during plant colonization. It is currently unknown why phytohormone effectors have so far not been detected in oomycete pathogens, which are phylogenetically related to brown algae but distinct from fungal lineages.
Currently, we do not understand why some pathogens use protein effectors to target hormone biosynthesis or signaling, while others shift hormonal balances by producing hormones or hormone mimics. It is at least conceivable that the use of microbial hormones as virulence factors may make it difficult for the plant to mount defense responses as these would also target the respective endogenous plant pathways. This could suggest that phytohormone effectors might be rather new additions to the battle between phytopathogens and their hosts. A comprehensive study on the evolution of such microbial traits will be needed to settle this point in future.
Given the large effector repertoire of filamentous pathogens, it is only a small number of effectors that target phytohormone pathways. Currently, it appears that the SA pathway is most extensively exploited by filamentous plant pathogens. We consider that this might be due to the fact that simple and sensitive readouts such as cell death or PR-1 expression have been developed that provide an easy way to visualize even subtle effects. Presently, only a small number of effectors have been shown to target other phytohormone pathways and for strigolactone signaling so far no modulating pathogen effectors have been detected. To uncover pathogen effectors which affect these pathways, it might be helpful to develop biosensors using promoter-fluorescent protein fusions for the activity of these pathways and employ such biosensors in high-throughput effectoromic studies to uncover effectors up or downregulating these reporter genes. This would also allow to uncover effectors with redundant functions in phytohormone signaling. With the advent of CRISPR-Cas9 technologies in filamentous pathogens, it should then become feasible to relate such redundant effectors with virulence. The fast-growing progress in multi-omics of filamentous plant pathogens and techniques for identifying protein interactions will further accelerate the discovery of filamentous pathogen effector targets and the underlying mechanism for manipulating phytohormone pathways.
Given the prominent involvement of phytohormones in defense, it is not surprising that engineering phytohormone pathways has potential for field applications. However, the opposing effects of phytohormones on disease caused by filamentous pathogens with different life styles do not allow to follow a straightforward strategy. Therefore, rather than simply increasing or decreasing hormone levels, it may be safer to modify effector targets by plant genome editing approaches like TILLING or CRISPR-Cas9 so that they can evade effector interference. This might also allow to get around the problem that higher levels of certain growth-related hormones will kick-off a trade-off between growth and defense. In addition, for real field situations, it needs to be kept in mind that plants are associated with a complex microbiome. So far, we largely lack studies that address the importance of phytohormone signaling pathways with respect to defense when plants growing in the field receive a cocktail of different signals. Evidence is accumulating that the innate immune system of plants as a whole serves both pathogen elimination and controlled accommodation of beneficial microbes (Hacquard et al., 2017). This will make it necessary to explore the role of phytohormones in plant-microbe interactions in the context of filamentous plant pathogens living in more complex natural environments.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. Our work was supported by generous funds from the Max Planck Society. XH was recipient of a scholarship from the China Scholarship Council (CSC).
References
- Adie B. A., Perez-Perez J., Perez-Perez M. M., Godoy M., Sanchez-Serrano J. J., Schmelz E. A., et al. (2007). ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19, 1665–1681. 10.1105/tpc.106.048041, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander D., Goodman R. M., Gut-Rella M., Glascock C., Weymann K., Friedrich L., et al. (1993). Increased tolerance to two oomycete pathogens in transgenic tobacco expressing pathogenesis-related protein 1a. Proc. Natl. Acad. Sci. USA 90, 7327–7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R. G., Casady M. S., Fee R. A., Vaughan M. M., Deb D., Fedkenheuer K., et al. (2012). Homologous RXLR effectors from Hyaloperonospora arabidopsidis and Phytophthora sojae suppress immunity in distantly related plants. Plant J. 72, 882–893. 10.1111/j.1365-313X.2012.05079.x, PMID: [DOI] [PubMed] [Google Scholar]
- Argueso C. T., Ferreira F. J., Epple P., To J. P., Hutchison C. E., Schaller G. E., et al. (2012). Two-component elements mediate interactions between cytokinin and salicylic acid in plant immunity. PLoS Genet. 8:e1002448. 10.1371/journal.pgen.1002448, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai S., Rallapalli G., Piquerez S. J., Caillaud M. C., Furzer O. J., Ishaque N., et al. (2014). Expression profiling during arabidopsis/downy mildew interaction reveals a highly-expressed effector that attenuates responses to salicylic acid. PLoS Pathog. 10:e1004443. 10.1371/journal.ppat.1004443, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier F. F., Dun E. A., Beveridge C. A. (2017). Apical dominance. Curr. Biol. 27, R864–R865. 10.1016/j.cub.2017.05.024, PMID: [DOI] [PubMed] [Google Scholar]
- Belkhadir Y., Jaillais Y., Epple P., Balsemao-Pires E., Dangl J. L., Chory J. (2012). Brassinosteroids modulate the efficiency of plant immune responses to microbe-associated molecular patterns. Proc. Natl. Acad. Sci. USA 109, 297–302. 10.1073/pnas.1112840108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berens M. L., Berry H. M., Mine A., Argueso C. T., Tsuda K. (2017). Evolution of hormone signaling networks in plant defense. Annu. Rev. Phytopathol. 55, 401–425. 10.1146/annurev-phyto-080516-035544, PMID: [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo M., Molina A., Solano R. (2002). Constitutive expression of ethylene-response-factor1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 29, 23–32. 10.1046/j.1365-313x.2002.01191.x, PMID: [DOI] [PubMed] [Google Scholar]
- Bleecker A. B., Kende H. (2000). Ethylene: a gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 16, 1–18. 10.1146/annurev.cellbio.16.1.1, PMID: [DOI] [PubMed] [Google Scholar]
- Breen S., Williams S. J., Outram M., Kobe B., Solomon P. S. (2017). Emerging insights into the functions of pathogenesis-related protein 1. Trends Plant Sci. 22, 871–879. 10.1016/j.tplants.2017.06.013, PMID: [DOI] [PubMed] [Google Scholar]
- Breen S., Williams S. J., Winterberg B., Kobe B., Solomon P. S. (2016). Wheat PR-1 proteins are targeted by necrotrophic pathogen effector proteins. Plant J. 88, 13–25. 10.1111/tpj.13228, PMID: [DOI] [PubMed] [Google Scholar]
- Bruce S. A., Saville B. J., Emery R. N. (2011). Ustilago maydis produces cytokinins and abscisic acid for potential regulation of tumor formation in maize. J. Plant Growth Regul. 30, 51–63. 10.1007/s00344-010-9166-8 [DOI] [Google Scholar]
- Buhrow L. M., Cram D., Tulpan D., Foroud N. A., Loewen M. C. (2016). Exogenous abscisic acid and gibberellic acid elicit opposing effects on Fusarium graminearum infection in wheat. Phytopathology 106, 986–996. 10.1094/PHYTO-01-16-0033-R, PMID: [DOI] [PubMed] [Google Scholar]
- Burg S. P., Burg E. A. (1965). Ethylene action and the ripening of fruits. Science 148, 1190–1196. 10.1126/science.148.3674.1190, PMID: [DOI] [PubMed] [Google Scholar]
- Caillaud M. C., Asai S., Rallapalli G., Piquerez S., Fabro G., Jones J. D. (2013). A downy mildew effector attenuates salicylic acid-triggered immunity in Arabidopsis by interacting with the host mediator complex. PLoS Biol. 11:e1001732. 10.1371/journal.pbio.1001732, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Bowling S. A., Gordon A. S., Dong X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6, 1583–1592. 10.1105/tpc.6.11.1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Glazebrook J., Clarke J. D., Volko S., Dong X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–63. 10.1016/S0092-8674(00)81858-9, PMID: [DOI] [PubMed] [Google Scholar]
- Carr J. P., Dixon D. C., Nikolau B. J., Voelkerding K. V., Klessig D. F. (1987). Synthesis and localization of pathogenesis-related proteins in tobacco. Mol. Cell. Biol. 7, 1580–1583. 10.1128/MCB.7.4.1580, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanclud E., Kisiala A., Emery N. R., Chalvon V., Ducasse A., Romiti-Michel C., et al. (2016). Cytokinin production by the rice blast fungus is a pivotal requirement for full virulence. PLoS Pathog. 12:e1005457. 10.1371/journal.ppat.1005457, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanclud E., Morel J. B. (2016). Plant hormones: a fungal point of view. Mol. Plant Pathol. 17, 1289–1297. 10.1111/mpp.12393, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. L., Lee C. Y., Cheng K. T., Chang W. H., Huang R. N., Nam H. G., et al. (2014). Quantitative peptidomics study reveals that a wound-induced peptide from PR-1 regulates immune signaling in tomato. Plant Cell 26, 4135–4148. 10.1105/tpc.114.131185, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A., Cimmino A., Masi M., Reveglia P., Nocera P., Solano R., et al. (2018). The fungal phytotoxin lasiojasmonate A activates the plant jasmonic acid pathway. J. Exp. Bot. 69, 3095–3102. 10.1093/jxb/ery114, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J., Huh S. U., Kojima M., Sakakibara H., Paek K. H., Hwang I. (2010). The cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in Arabidopsis. Dev. Cell 19, 284–295. 10.1016/j.devcel.2010.07.011, PMID: [DOI] [PubMed] [Google Scholar]
- Christians M. J., Gingerich D. J., Hansen M., Binder B. M., Kieber J. J., Vierstra R. D. (2009). The BTB ubiquitin ligases ETO1, EOL1 and EOL2 act collectively to regulate ethylene biosynthesis in Arabidopsis by controlling type-2 ACC synthase levels. Plant J. 57, 332–345. 10.1111/j.1365-313X.2008.03693.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse S. D. (2011). Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development. Plant Cell 23, 1219–1230. 10.1105/tpc.111.084475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S. J., Yoon A. J., Faull K. F., Diener A. C. (2014). Host perception of jasmonates promotes infection by Fusarium oxysporum formae speciales that produce isoleucine- and leucine-conjugated jasmonates. Mol. Plant Pathol. 15, 589–600. 10.1111/mpp.12117, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couto D., Zipfel C. (2016). Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 16, 537–552. 10.1038/nri.2016.77 [DOI] [PubMed] [Google Scholar]
- Cutler S. R., Rodriguez P. L., Finkelstein R. R., Abrams S. R. (2010). Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679. 10.1146/annurev-arplant-042809-112122, PMID: [DOI] [PubMed] [Google Scholar]
- Dangl J. L., Horvath D. M., Staskawicz B. J. (2013). Pivoting the plant immune system from dissection to deployment. Science 341, 746–751. 10.1126/science.1236011, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vleesschauwer D., Van Buyten E., Satoh K., Balidion J., Mauleon R., Choi I.-R., et al. (2012). Brassinosteroids antagonize gibberellin- and salicylate-mediated root immunity in rice. Plant Physiol. 158, 1833–1846. 10.1104/pp.112.193672, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vleesschauwer D., Yang Y., Cruz C. V., Hofte M. (2010). Abscisic acid-induced resistance against the brown spot pathogen Cochliobolus miyabeanus in rice involves MAP kinase-mediated repression of ethylene signaling. Plant Physiol. 152, 2036–2052. 10.1104/pp.109.152702, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey D. A., Vlot A. C., Wildermuth M. C., Klessig D. F. (2011). Salicylic acid biosynthesis and metabolism. Arabidopsis Book 9:e0156. 10.1199/tab.0156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire M., Denton-Giles M., Hegedus D., Seifbarghy S., Rollins J., van Kan J., et al. (2017). The complete genome sequence of the phytopathogenic fungus Sclerotinia sclerotiorum reveals insights into the genome architecture of broad host range pathogens. Genome Biol. Evol. 9, 593–618. 10.1093/gbe/evx030, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despres C., DeLong C., Glaze S., Liu E., Fobert P. R. (2000). The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12, 279–290. 10.1105/tpc.12.2.279, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djamei A., Schipper K., Rabe F., Ghosh A., Vincon V., Kahnt J., et al. (2011). Metabolic priming by a secreted fungal effector. Nature 478, 395–398. 10.1038/nature10454, PMID: [DOI] [PubMed] [Google Scholar]
- Doehlemann G., Reissmann S., Assmann D., Fleckenstein M., Kahmann R. (2011). Two linked genes encoding a secreted effector and a membrane protein are essential for Ustilago maydis-induced tumour formation. Mol. Microbiol. 81, 751–766. 10.1111/j.1365-2958.2011.07728.x, PMID: [DOI] [PubMed] [Google Scholar]
- Dong X. (2004). NPR1, all things considered. Curr. Opin. Plant Biol. 7, 547–552. 10.1016/j.pbi.2004.07.005, PMID: [DOI] [PubMed] [Google Scholar]
- Du Y., Mpina M. H., Birch P. R., Bouwmeester K., Govers F. (2015). Phytophthora infestans RXLR effector AVR1 interacts with exocyst component Sec5 to manipulate plant immunity. Plant Physiol. 169, 1975–1990. 10.1104/pp.15.01169, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Overdijk E. J. R., Berg J. A., Govers F., Bouwmeester K. (2018). Solanaceous exocyst subunits are involved in immunity to diverse plant pathogens. J. Exp. Bot. 69, 655–666. 10.1093/jxb/erx442, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Oirdi M., El Rahman T. A., Rigano L., El Hadrami A., Rodriguez M. C., Daayf F., et al. (2011). Botrytis cinerea manipulates the antagonistic effects between immune pathways to promote disease development in tomato. Plant Cell 23, 2405–2421. 10.1105/tpc.111.083394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo A., Okamoto M., Koshiba T. (2014). “ABA biosynthetic and catabolic pathways” in Abscisic acid: Metabolism, transport and signaling. ed. Zhang D.-P. (Dordrecht, Netherlands: Springer; ), 21–45. [Google Scholar]
- Evangelisti E., Govetto B., Minet-Kebdani N., Kuhn M. L., Attard A., Ponchet M., et al. (2013). The Phytophthora parasitica RXLR effector penetration-specific effector 1 favours Arabidopsis thaliana infection by interfering with auxin physiology. New Phytol. 199, 476–489. 10.1111/nph.12270, PMID: [DOI] [PubMed] [Google Scholar]
- Fan J., Hill L., Crooks C., Doerner P., Lamb C. (2009). Abscisic acid has a key role in modulating diverse plant-pathogen interactions. Plant Physiol. 150, 1750–1761. 10.1104/pp.109.137943, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher M. C., Henk D. A., Briggs C. J., Brownstein J. S., Madoff L. C., McCraw S. L., et al. (2012). Emerging fungal threats to animal, plant and ecosystem health. Nature 484, 186–194. 10.1038/nature10947, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z. Q., Dong X. (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. 10.1146/annurev-arplant-042811-105606, PMID: [DOI] [PubMed] [Google Scholar]
- Gamir J., Darwiche R., Van’t Hof P., Choudhary V., Stumpe M., Schneiter R., et al. (2017). The sterol-binding activity of pathogenesis-related protein 1 reveals the mode of action of an antimicrobial protein. Plant J. 89, 502–509. 10.1111/tpj.13398, PMID: [DOI] [PubMed] [Google Scholar]
- Garcion C., Lohmann A., Lamodiere E., Catinot J., Buchala A., Doermann P., et al. (2008). Characterization and biological function of the isochorismate synthase2 gene of Arabidopsis. Plant Physiol. 147, 1279–1287. 10.1104/pp.108.119420, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy E. M., Breen S., Whisson S. C., Squires J., Hein I., Kaczmarek M., et al. (2011). Presence/absence, differential expression and sequence polymorphisms between PiAVR2 and PiAVR2-like in Phytophthora infestans determine virulence on R2 plants. New Phytol. 191, 763–776. 10.1111/j.1469-8137.2011.03736.x, PMID: [DOI] [PubMed] [Google Scholar]
- Glazebrook J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. 10.1146/annurev.phyto.43.040204.135923, PMID: [DOI] [PubMed] [Google Scholar]
- Grbić V., Bleecker A. B. (1995). Ethylene regulates the timing of leaf senescence in Arabidopsis. Plant J. 8, 595–602. 10.1046/j.1365-313X.1995.8040595.x [DOI] [Google Scholar]
- Hacquard S., Spaepen S., Garrido-Oter R., Schulze-Lefert P. (2017). Interplay between innate immunity and the plant microbiota. Annu. Rev. Phytopathol. 55, 565–589. 10.1146/annurev-phyto-080516-035623, PMID: [DOI] [PubMed] [Google Scholar]
- Han X., Altegoer F., Steinchen W., Binnebesel L., Schuhmacher J., Glatter T., et al. (2019). A kiwellin disarms the metabolic activity of a secreted fungal virulence factor. Nature. 565, 650–653. 10.1038/s41586-018-0857-9, PMID: [DOI] [PubMed] [Google Scholar]
- Helliwell E. E., Wang Q., Yang Y. (2016). Ethylene biosynthesis and signaling is required for rice immune response and basal resistance against Magnaporthe oryzae infection. Mol. Plant-Microbe Interact. 29, 831–843. 10.1094/MPMI-06-16-0121-R, PMID: [DOI] [PubMed] [Google Scholar]
- Hinsch J., Galuszka P., Tudzynski P. (2016). Functional characterization of the first filamentous fungal tRNA-isopentenyltransferase and its role in the virulence of Claviceps purpurea. New Phytol. 211, 980–992. 10.1111/nph.13960, PMID: [DOI] [PubMed] [Google Scholar]
- Hinsch J., Vrabka J., Oeser B., Novak O., Galuszka P., Tudzynski P. (2015). De novo biosynthesis of cytokinins in the biotrophic fungus Claviceps purpurea. Environ. Microbiol. 17, 2935–2951. 10.1111/1462-2920.12838, PMID: [DOI] [PubMed] [Google Scholar]
- Hoffman T., Schmidt J. S., Zheng X., Bent A. F. (1999). Isolation of ethylene-insensitive soybean mutants that are altered in pathogen susceptibility and gene-for-gene disease resistance. Plant Physiol. 119, 935–950. 10.1104/pp.119.3.935, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huot B., Yao J., Montgomery B. L., He S. Y. (2014). Growth-defense tradeoffs in plants: a balancing act to optimize fitness. Mol. Plant 7, 1267–1287. 10.1093/mp/ssu049, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C. J., Shimono M., Sugano S., Kojima M., Yazawa K., Yoshida R., et al. (2010). Abscisic acid interacts antagonistically with salicylic acid signaling pathway in rice-Magnaporthe grisea interaction. Mol. Plant-Microbe Interact. 23, 791–798. 10.1094/MPMI-23-6-0791, PMID: [DOI] [PubMed] [Google Scholar]
- Jiang R. H., Tripathy S., Govers F., Tyler B. M. (2008). RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc. Natl. Acad. Sci. USA 105, 4874–4879. 10.1073/pnas.0709303105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. D., Dangl J. L. (2006). The plant immune system. Nature 444, 323–329. 10.1038/nature05286, PMID: [DOI] [PubMed] [Google Scholar]
- Kabbage M., Williams B., Dickman M. B. (2013). Cell death control: the interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum. PLoS Pathog. 9:e1003287. 10.1371/journal.ppat.1003287, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun S. (2006). A catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Phytopathol. 44, 41–60. 10.1146/annurev.phyto.44.070505.143436, PMID: [DOI] [PubMed] [Google Scholar]
- Kaschani F., Shabab M., Bozkurt T., Shindo T., Schornack S., Gu C., et al. (2010). An effector-targeted protease contributes to defense against Phytophthora infestans and is under diversifying selection in natural hosts. Plant Physiol. 154, 1794–1804. 10.1104/pp.110.158030, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaschani F., Van der Hoorn R. A. (2011). A model of the C14-EPIC complex indicates hotspots for a protease-inhibitor arms race in the oomycete-potato interaction. Plant Signal. Behav. 6, 109–112. 10.4161/psb.6.1.14190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K., Lyons R. (2014). Intervention of phytohormone pathways by pathogen effectors. Plant Cell 26, 2285–2309. 10.1105/tpc.114.125419, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K., Manners J. M. (2009). Linking development to defense: auxin in plant-pathogen interactions. Trends Plant Sci. 14, 373–382. 10.1016/j.tplants.2009.04.005, PMID: [DOI] [PubMed] [Google Scholar]
- Kesarwani M., Yoo J., Dong X. (2007). Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis. Plant Physiol. 144, 336–346. 10.1104/pp.106.095299, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba A., Nishihara M., Nakatsuka T., Yamamura S. (2007). Pathogenesis-related protein 1 homologue is an antifungal protein in Wasabia japonica leaves and confers resistance to Botrytis cinerea in transgenic tobacco. Plant Biotech. 24, 247–253. 10.5511/plantbiotechnology.24.247 [DOI] [Google Scholar]
- Kieber J. J., Schaller G. E. (2018). Cytokinin signaling in plant development. Development 145:dev149344. 10.1242/dev.149344, PMID: [DOI] [PubMed] [Google Scholar]
- Kinkema M., Fan W., Dong X. (2000). Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12, 2339–2350. 10.1105/tpc.12.12.2339, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klessig D. F., Malamy J. (1994). The salicylic acid signal in plants. Plant Mol. Biol. 26, 1439–1458. 10.1007/BF00016484, PMID: [DOI] [PubMed] [Google Scholar]
- Kunkel B. N., Brooks D. M. (2002). Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5, 325–331. 10.1016/S1369-5266(02)00275-3, PMID: [DOI] [PubMed] [Google Scholar]
- Kurakawa T., Ueda N., Maekawa M., Kobayashi K., Kojima M., Nagato Y., et al. (2007). Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445, 652–655. 10.1038/nature05504, PMID: [DOI] [PubMed] [Google Scholar]
- Laloi C., Mestres-Ortega D., Marco Y., Meyer Y., Reichheld J. P. (2004). The Arabidopsis cytosolic thioredoxin h5 gene induction by oxidative stress and its W-box-mediated response to pathogen elicitor. Plant Physiol. 134, 1006–1016. 10.1104/pp.103.035782, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. J., Park Y. J., Seo P. J., Kim J. H., Sim H. J., Kim S. G., et al. (2015). Systemic immunity requires SnRK2.8-mediated nuclear import of NPR1 in Arabidopsis. Plant Cell 27, 3425–3438. 10.1105/tpc.15.00371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemarie S., Robert-Seilaniantz A., Lariagon C., Lemoine J., Marnet N., Jubault M., et al. (2015). Both the jasmonic acid and the salicylic acid pathways contribute to resistance to the biotrophic clubroot agent Plasmodiophora brassicae in Arabidopsis. Plant Cell Physiol. 56, 2158–2168. 10.1093/pcp/pcv127, PMID: [DOI] [PubMed] [Google Scholar]
- Li G., Meng X., Wang R., Mao G., Han L., Liu Y., et al. (2012). Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. PLoS Genet. 8:e1002767. 10.1371/journal.pgen.1002767, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Song T., Zhang X., Yuan H., Su L., Li W., et al. (2014). Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nat. Commun. 5:4686. 10.1038/ncomms5686, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Wang Q., Deng W., Wang X., Piao M., Cai D., et al. (2017). Molecular basis for blue light-dependent phosphorylation of Arabidopsis cryptochrome 2. Nat. Commun. 8:15234. 10.1038/ncomms15234, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente F., Muskett P., Sanchez-Vallet A., Lopez G., Ramos B., Sanchez-Rodriguez C., et al. (2008). Repression of the auxin response pathway increases Arabidopsis susceptibility to necrotrophic fungi. Mol. Plant 1, 496–509. 10.1093/mp/ssn025, PMID: [DOI] [PubMed] [Google Scholar]
- Lo Presti L., Lanver D., Schweizer G., Tanaka S., Liang L., Tollot M., et al. (2015). Fungal effectors and plant susceptibility. Annu. Rev. Plant Biol. 66, 513–545. 10.1146/annurev-arplant-043014-114623, PMID: [DOI] [PubMed] [Google Scholar]
- Lorang J. M., Carkaci-Salli N., Wolpert T. J. (2004). Identification and characterization of victorin sensitivity in Arabidopsis thaliana. Mol. Plant-Microbe Interact. 17, 577–582. 10.1094/MPMI.2004.17.6.577, PMID: [DOI] [PubMed] [Google Scholar]
- Lorang J., Kidarsa T., Bradford C. S., Gilbert B., Curtis M., Tzeng S. C., et al. (2012). Tricking the guard: exploiting plant defense for disease susceptibility. Science 338, 659–662. 10.1126/science.1226743, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorang J. M., Sweat T. A., Wolpert T. J. (2007). Plant disease susceptibility conferred by a “resistance” gene. Proc. Natl. Acad. Sci. USA 104, 14861–14866. 10.1073/pnas.0702572104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Piqueras R., Sanchez-Serrano J. J., Solano R. (2003). Ethylene response factor1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15, 165–178. 10.1105/tpc.007468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan T., Ori N., Fluhr R. (1989). Pathogenesis-related proteins are developmentally regulated in tobacco flowers. Plant Cell 1, 881–887. 10.1105/tpc.1.9.881, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Durán R., Zipfel C. (2015). Trade-off between growth and immunity: role of brassinosteroids. Trends Plant Sci. 20, 12–19. 10.1016/j.tplants.2014.09.003, PMID: [DOI] [PubMed] [Google Scholar]
- Lu S., Faris J. D., Sherwood R., Friesen T. L., Edwards M. C. (2014). A dimeric PR-1-type pathogenesis-related protein interacts with ToxA and potentially mediates ToxA-induced necrosis in sensitive wheat. Mol. Plant Pathol. 15, 650–663. 10.1111/mpp.12122, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho A. P., Zipfel C. (2014). Plant PRRs and the activation of innate immune signaling. Mol. Cell 54, 263–272. 10.1016/j.molcel.2014.03.028 [DOI] [PubMed] [Google Scholar]
- Malamy J., Carr J. P., Klessig D. F., Raskin I. (1990). Salicylic acid: a likely endogenous signal in the resistance response of tobacco to viral infection. Science 250, 1002–1004. 10.1126/science.250.4983.1002, PMID: [DOI] [PubMed] [Google Scholar]
- McGowan J., Fitzpatrick D. A. (2017). Genomic, network, and phylogenetic analysis of the oomycete effector arsenal. mSphere 2, e00408–e00417. 10.1128/mSphere.00408-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metraux J. P. (2002). Recent breakthroughs in the study of salicylic acid biosynthesis. Trends Plant Sci. 7, 332–334. 10.1016/S1360-1385(02)02313-0, PMID: [DOI] [PubMed] [Google Scholar]
- Metraux J. P., Signer H., Ryals J., Ward E., Wyss-Benz M., Gaudin J., et al. (1990). Increase in salicylic acid at the onset of systemic acquired resistance in cucumber. Science 250, 1004–1006. 10.1126/science.250.4983.1004, PMID: [DOI] [PubMed] [Google Scholar]
- Miersch O., Neumerkel J., Dippe M., Stenzel I., Wasternack C. (2008). Hydroxylated jasmonates are commonly occurring metabolites of jasmonic acid and contribute to a partial switch-off in jasmonate signaling. New Phytol. 177, 114–127. 10.1111/j.1469-8137.2007.02252.x, PMID: [DOI] [PubMed] [Google Scholar]
- Misas Villamil J. C., Mueller A. N., Demir F., Meyer U., Okmen B., Schulze Huynck J., et al. (2019). A fungal substrate mimicking molecule suppresses plant immunity via an inter-kingdom conserved motif. Nat. Commun. 10:1576. 10.1038/s41467-019-09472-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misas-Villamil J. C., van der Hoorn R. A., Doehlemann G. (2016). Papain-like cysteine proteases as hubs in plant immunity. New Phytol. 212, 902–907. 10.1111/nph.14117, PMID: [DOI] [PubMed] [Google Scholar]
- Miyawaki K., Tarkowski P., Matsumoto-Kitano M., Kato T., Sato S., Tarkowska D., et al. (2006). Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc. Natl. Acad. Sci. USA 103, 16598–16603. 10.1073/pnas.0603522103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-García S., Vert G., Yin Y., Caño-Delgado A., Cheong H., Chory J. (2004). Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 18, 448–460. 10.1101/gad.1174204, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison E. N., Emery R. J., Saville B. J. (2015). Phytohormone involvement in the Ustilago maydis-Zea mays pathosystem: relationships between abscisic acid and cytokinin levels and strain virulence in infected cob tissue. PLoS One 10:e0130945. 10.1371/journal.pone.0130945, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison E. N., Emery R. J. N., Saville B. J. (2017). Fungal derived cytokinins are necessary for normal Ustilago maydis infection of maize. Plant Pathol. 66, 726–742. 10.1111/ppa.12629 [DOI] [Google Scholar]
- Mou Z., Fan W., Dong X. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113, 935–944. 10.1016/S0092-8674(03)00429-X, PMID: [DOI] [PubMed] [Google Scholar]
- Mueller A. N., Ziemann S., Treitschke S., Assmann D., Doehlemann G. (2013). Compatibility in the Ustilago maydis-maize interaction requires inhibition of host cysteine proteases by the fungal effector Pit2. PLoS Pathog. 9:e1003177. 10.1371/journal.ppat.1003177, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseem M., Kaltdorf M., Dandekar T. (2015). The nexus between growth and defence signalling: auxin and cytokinin modulate plant immune response pathways. J. Exp. Bot. 66, 4885–4896. 10.1093/jxb/erv297, PMID: [DOI] [PubMed] [Google Scholar]
- Naseem M., Wolfling M., Dandekar T. (2014). Cytokinins for immunity beyond growth, galls and green islands. Trends Plant Sci. 19, 481–484. 10.1016/j.tplants.2014.04.001, PMID: [DOI] [PubMed] [Google Scholar]
- Ni W., Xu S.-L., González-Grandío E., Chalkley R. J., Huhmer A. F. R., Burlingame A. L., et al. (2017). PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3. Nat. Commun. 8:15236. 10.1038/ncomms15236, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niderman T., Genetet I., Bruyere T., Gees R., Stintzi A., Legrand M., et al. (1995). Pathogenesis-related PR-1 proteins are antifungal. Isolation and characterization of three 14-kilodalton proteins of tomato and of a basic PR-1 of tobacco with inhibitory activity against Phytophthora infestans. Plant Physiol. 108, 17–27. 10.1104/pp.108.1.17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patkar R. N., Benke P. I., Qu Z., Chen Y. Y., Yang F., Swarup S., et al. (2015). A fungal monooxygenase-derived jasmonate attenuates host innate immunity. Nat. Chem. Biol. 11, 733–740. 10.1038/nchembio.1885, PMID: [DOI] [PubMed] [Google Scholar]
- Pieterse C. M., Leon-Reyes A., Van der Ent S., Van Wees S. C. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316. 10.1038/nchembio.164, PMID: [DOI] [PubMed] [Google Scholar]
- Pieterse C. M., Van der Does D., Zamioudis C., Leon-Reyes A., Van Wees S. C. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521. 10.1146/annurev-cellbio-092910-154055, PMID: [DOI] [PubMed] [Google Scholar]
- Qin X., Liu J. H., Zhao W. S., Chen X. J., Guo Z. J., Peng Y. L. (2013). Gibberellin 20-oxidase gene OsGA20ox3 regulates plant stature and disease development in rice. Mol. Plant-Microbe Interact. 26, 227–239. 10.1094/MPMI-05-12-0138-R, PMID: [DOI] [PubMed] [Google Scholar]
- Qin J., Wang K., Sun L., Xing H., Wang S., Li L., et al. (2018). The plant-specific transcription factors CBP60g and SARD1 are targeted by a Verticillium secretory protein VdSCP41 to modulate immunity. elife 7:e34902. 10.7554/eLife.34902, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin I. (1992). Role of salicylic acid in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43, 439–463. 10.1146/annurev.pp.43.060192.002255 [DOI] [Google Scholar]
- Rauscher M., Adam A. L., Wirtz S., Guggenheim R., Mendgen K., Deising H. B. (1999). PR-1 protein inhibits the differentiation of rust infection hyphae in leaves of acquired resistant broad bean. Plant J. 19, 625–633. 10.1046/j.1365-313x.1999.00545.x, PMID: [DOI] [PubMed] [Google Scholar]
- Reineke G., Heinze B., Schirawski J., Buettner H., Kahmann R., Basse C. W. (2008). Indole-3-acetic acid (IAA) biosynthesis in the smut fungus Ustilago maydis and its relevance for increased IAA levels in infected tissue and host tumour formation. Mol. Plant Pathol. 9, 339–355. 10.1111/j.1364-3703.2008.00470.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemann M., Haga K., Shimizu T., Okada K., Ando S., Mochizuki S., et al. (2013). Identification of rice allene oxide cyclase mutants and the function of jasmonate for defence against Magnaporthe oryzae. Plant J. 74, 226–238. 10.1111/tpj.12115, PMID: [DOI] [PubMed] [Google Scholar]
- Robert H. S., Friml J. (2009). Auxin and other signals on the move in plants. Nat. Chem. Biol. 5, 325–332. 10.1038/nchembio.170, PMID: [DOI] [PubMed] [Google Scholar]
- Sakakibara H. (2006). Cytokinins: activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 57, 431–449. 10.1146/annurev.arplant.57.032905.105231, PMID: [DOI] [PubMed] [Google Scholar]
- Sarowar S., Kim Y. J., Kim E. N., Kim K. D., Hwang B. K., Islam R., et al. (2005). Overexpression of a pepper basic pathogenesis-related protein 1 gene in tobacco plants enhances resistance to heavy metal and pathogen stresses. Plant Cell Rep. 24, 216–224. 10.1007/s00299-005-0928-x, PMID: [DOI] [PubMed] [Google Scholar]
- Saunders D. G., Breen S., Win J., Schornack S., Hein I., Bozkurt T. O., et al. (2012). Host protein BSL1 associates with Phytophthora infestans RXLR effector AVR2 and the Solanum demissum immune receptor R2 to mediate disease resistance. Plant Cell 24, 3420–3434. 10.1105/tpc.112.099861, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schornack S., van Damme M., Bozkurt T. O., Cano L. M., Smoker M., Thines M., et al. (2010). Ancient class of translocated oomycete effectors targets the host nucleus. Proc. Natl. Acad. Sci. USA 107, 17421–17426. 10.1073/pnas.1008491107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabab M., Shindo T., Gu C., Kaschani F., Pansuriya T., Chintha R., et al. (2008). Fungal effector protein AVR2 targets diversifying defense-related cys proteases of tomato. Plant Cell 20, 1169–1183. 10.1105/tpc.107.056325, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J., Tsui F., Klessig D. F. (1997). Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol. Plant-Microbe Interact. 10, 69–78. 10.1094/MPMI.1997.10.1.69, PMID: [DOI] [PubMed] [Google Scholar]
- Siewers V., Kokkelink L., Smedsgaard J., Tudzynski P. (2006). Identification of an abscisic acid gene cluster in the grey mold Botrytis cinerea. Appl. Environ. Microbiol. 72, 4619–4626. 10.1128/AEM.02919-05, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skottke K. R., Yoon G. M., Kieber J. J., DeLong A. (2011). Protein phosphatase 2A controls ethylene biosynthesis by differentially regulating the turnover of ACC synthase isoforms. PLoS Genet. 7:e1001370. 10.1371/journal.pgen.1001370, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Win J., Tian M., Schornack S., Kaschani F., Ilyas M., et al. (2009). Apoplastic effectors secreted by two unrelated eukaryotic plant pathogens target the tomato defense protease Rcr3. Proc. Natl. Acad. Sci. USA 106, 1654–1659. 10.1073/pnas.0809201106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen J. L., Benfield A. H., Wollenberg R. D., Westphal K., Wimmer R., Nielsen M. R., et al. (2018). The cereal pathogen Fusarium pseudograminearum produces a new class of active cytokinins during infection. Mol. Plant Pathol. 19, 1140–1154. 10.1111/mpp.12593, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence C. A., Lakshmanan V., Donofrio N., Bais H. P. (2015). Crucial roles of abscisic acid biogenesis in virulence of rice blast fungus Magnaporthe oryzae. Front. Plant Sci. 6:1082. 10.3389/fpls.2015.01082, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel S. H., Dong X. (2008). Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe 3, 348–351. 10.1016/j.chom.2008.05.009, PMID: [DOI] [PubMed] [Google Scholar]
- Stahmann K. P., Monschau N., Sahm H., Koschel A., Gawronski M., Conrad H., et al. (1995). Structural properties of native and sonicated cinerean, a beta- (1–>3) (1–>6)-D-glucan produced by Botrytis cinerea. Carbohydr. Res. 266, 115–128. 10.1016/0008-6215(94)00245-B, PMID: [DOI] [PubMed] [Google Scholar]
- Strawn M. A., Marr S. K., Inoue K., Inada N., Zubieta C., Wildermuth M. C. (2007). Arabidopsis isochorismate synthase functional in pathogen-induced salicylate biosynthesis exhibits properties consistent with a role in diverse stress responses. J. Biol. Chem. 282, 5919–5933. 10.1074/jbc.M605193200 [DOI] [PubMed] [Google Scholar]
- Sun T., Busta L., Zhang Q., Ding P., Jetter R., Zhang Y. (2018). TGACG-binding factor 1 (TGA1) and TGA4 regulate salicylic acid and pipecolic acid biosynthesis by modulating the expression of systemic acquired resistance deficient 1 (SARD1) and calmodulin-binding protein 60g (CBP60g). New Phytol. 217, 344–354. 10.1111/nph.14780, PMID: [DOI] [PubMed] [Google Scholar]
- Sun T., Zhang Y., Li Y., Zhang Q., Ding Y., Zhang Y. (2015). ChIP-seq reveals broad roles of SARD1 and CBP60g in regulating plant immunity. Nat. Commun. 6:10159. 10.1038/ncomms10159, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweat T. A., Wolpert T. J. (2007). Thioredoxin h5 is required for victorin sensitivity mediated by a CC-NBS-LRR gene in Arabidopsis. Plant Cell 19, 673–687. 10.1105/tpc.106.047563, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada Y., Spoel S. H., Pajerowska-Mukhtar K., Mou Z., Song J., Wang C., et al. (2008). Plant immunity requires conformational changes [corrected] of NPR1 via S-nitrosylation and thioredoxins. Science 321, 952–956. 10.1126/science.1156970, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H., Kanayama Y., Zheng M. S., Kusano T., Hase S., Ikegami M., et al. (2004). Antagonistic interactions between the SA and JA signaling pathways in Arabidopsis modulate expression of defense genes and gene-for-gene resistance to cucumber mosaic virus. Plant Cell Physiol. 45, 803–809. 10.1093/pcp/pch085, PMID: [DOI] [PubMed] [Google Scholar]
- Takei K., Sakakibara H., Sugiyama T. (2001). Identification of genes encoding adenylate isopentenyltransferase, a cytokinin biosynthesis enzyme, in Arabidopsis thaliana. J. Biol. Chem. 276, 26405–26410. 10.1074/jbc.M102130200, PMID: [DOI] [PubMed] [Google Scholar]
- Takei K., Yamaya T., Sakakibara H. (2004). Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-Zeatin. J. Biol. Chem. 279, 41866–41872. 10.1074/jbc.M406337200, PMID: [DOI] [PubMed] [Google Scholar]
- Teale W. D., Paponov I. A., Palme K. (2006). Auxin in action: signalling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell Biol. 7, 847–859. 10.1038/nrm2020, PMID: [DOI] [PubMed] [Google Scholar]
- Thaler J. S., Owen B., Higgins V. J. (2004). The role of the jasmonate response in plant susceptibility to diverse pathogens with a range of lifestyles. Plant Physiol. 135, 530–538. 10.1104/pp.104.041566, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher L. F., Gardiner D. M., Kazan K., Manners J. M. (2012). A highly conserved effector in Fusarium oxysporum is required for full virulence on Arabidopsis. Mol. Plant-Microbe Interact. 25, 180–190. 10.1094/MPMI-08-11-0212, PMID: [DOI] [PubMed] [Google Scholar]
- Thatcher L. F., Manners J. M., Kazan K. (2009). Fusarium oxysporum hijacks COI1-mediated jasmonate signaling to promote disease development in Arabidopsis. Plant J. 58, 927–939. 10.1111/j.1365-313X.2009.03831.x, PMID: [DOI] [PubMed] [Google Scholar]
- Thomma B. P., Eggermont K., Penninckx I. A., Mauch-Mani B., Vogelsang R., Cammue B. P., et al. (1998). Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA 95, 15107–15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma B. P., Eggermont K., Tierens K. F.-J., Broekaert W. F. (1999). Requirement of functional ethylene-insensitive 2gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant Physiol. 121, 1093–1101. 10.1104/pp.121.4.1093, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M., Win J., Song J., van der Hoorn R., van der Knaap E., Kamoun S. (2007). A Phytophthora infestans cystatin-like protein targets a novel tomato papain-like apoplastic protease. Plant Physiol. 143, 364–377. 10.1104/pp.106.090050, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomczynska I., Stumpe M., Mauch F. (2018). A conserved RxLR effector interacts with host RABA-type GTPases to inhibit vesicle-mediated secretion of antimicrobial proteins. Plant J. 95, 187–203. 10.1111/tpj.13928, PMID: [DOI] [PubMed] [Google Scholar]
- Ton J., Mauch-Mani B. (2004). Beta-amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABA-dependent priming for callose. Plant J. 38, 119–130. 10.1111/j.1365-313X.2004.02028.x, PMID: [DOI] [PubMed] [Google Scholar]
- Tsuda K., Katagiri F. (2010). Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 13, 459–465. 10.1016/j.pbi.2010.04.006, PMID: [DOI] [PubMed] [Google Scholar]
- Turian G., Hamilton R. H. (1960). Chemical detection of 3-indolylacetic acid in Ustilago zeae tumors. Biochim. Biophys. Acta 41, 148–150. 10.1016/0006-3002(60)90381-4, PMID: [DOI] [PubMed] [Google Scholar]
- Turnbull D., Yang L., Naqvi S., Breen S., Welsh L., Stephens J., et al. (2017). RXLR effector AVR2 Up-regulates a brassinosteroid-responsive bHLH transcription factor to suppress immunity. Plant Physiol. 174, 356–369. 10.1104/pp.16.01804, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bockhaven J., Spichal L., Novak O., Strnad M., Asano T., Kikuchi S., et al. (2015). Silicon induces resistance to the brown spot fungus Cochliobolus miyabeanus by preventing the pathogen from hijacking the rice ethylene pathway. New Phytol. 206, 761–773. 10.1111/nph.13270, PMID: [DOI] [PubMed] [Google Scholar]
- van der Linde K., Hemetsberger C., Kastner C., Kaschani F., van der Hoorn R. A., Kumlehn J., et al. (2012). A maize cystatin suppresses host immunity by inhibiting apoplastic cysteine proteases. Plant Cell 24, 1285–1300. 10.1105/tpc.111.093732, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Loon L., Van Strien E. (1999). The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 55, 85–97. 10.1006/pmpp.1999.0213 [DOI] [Google Scholar]
- Vlot A. C., Dempsey D. A., Klessig D. F. (2009). Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. 10.1146/annurev.phyto.050908.135202, PMID: [DOI] [PubMed] [Google Scholar]
- Wang D., Amornsiripanitch N., Dong X. (2006). A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2:e123. 10.1371/journal.ppat.0020123, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. Y., Bai M. Y., Oh E., Zhu J. Y. (2012). Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 46, 701–724. 10.1146/annurev-genet-102209-163450, PMID: [DOI] [PubMed] [Google Scholar]
- Wang Z., Casas-Mollano J. A., Xu J., Riethoven J. J., Zhang C., Cerutti H. (2015). Osmotic stress induces phosphorylation of histone H3 at threonine 3 in pericentromeric regions of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 112, 8487–8492. 10.1073/pnas.1423325112, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Han C., Ferreira A. O., Yu X., Ye W., Tripathy S., et al. (2011b). Transcriptional programming and functional interactions within the Phytophthora sojae RXLR effector repertoire. Plant Cell 23, 2064–2086. 10.1105/tpc.111.086082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. L., Li H., Ecker J. R. (2002). Ethylene biosynthesis and signaling networks. Plant Cell 14(Suppl.), S131–S151. 10.1105/tpc.001768, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. Y., Seto H., Fujioka S., Yoshida S., Chory J. (2001). BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410, 380–383. 10.1038/35066597, PMID: [DOI] [PubMed] [Google Scholar]
- Wang L., Tsuda K., Sato M., Cohen J. D., Katagiri F., Glazebrook J. (2009). Arabidopsis CaM binding protein CBP60g contributes to MAMP-induced SA accumulation and is involved in disease resistance against Pseudomonas syringae. PLoS Pathog. 5:e1000301. 10.1371/journal.ppat.1000301, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Tsuda K., Truman W., Sato M., Nguyen le V., Katagiri F., et al. (2011a). CBP60g and SARD1 play partially redundant critical roles in salicylic acid signaling. Plant J. 67, 1029–1041. 10.1111/j.1365-313X.2011.04655.x [DOI] [PubMed] [Google Scholar]
- Wang D., Weaver N. D., Kesarwani M., Dong X. (2005). Induction of protein secretory pathway is required for systemic acquired resistance. Science 308, 1036–1040. 10.1126/science.1108791, PMID: [DOI] [PubMed] [Google Scholar]
- Wang X., Yang B., Li K., Kang Z., Cantu D., Dubcovsky J. (2016). A conserved Puccinia striiformis protein interacts with wheat NPR1 and reduces induction of pathogenesis-related genes in response to pathogens. Mol. Plant-Microbe Interact. 29, 977–989. 10.1094/MPMI-10-16-0207-R, PMID: [DOI] [PubMed] [Google Scholar]
- Weiss D., Ori N. (2007). Mechanisms of cross talk between gibberellin and other hormones. Plant Physiol. 144, 1240–1246. 10.1104/pp.107.100370, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisson S. C., Boevink P. C., Wang S., Birch P. R. (2016). The cell biology of late blight disease. Curr. Opin. Microbiol. 34, 127–135. 10.1016/j.mib.2016.09.002, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemann P., Sieber C. M., von Bargen K. W., Studt L., Niehaus E. M., Espino J. J., et al. (2013). Deciphering the cryptic genome: genome-wide analyses of the rice pathogen Fusarium fujikuroi reveal complex regulation of secondary metabolism and novel metabolites. PLoS Pathog. 9:e1003475. 10.1371/journal.ppat.1003475, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth M. C., Dewdney J., Wu G., Ausubel F. M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414, 562–565. 10.1038/35107108, PMID: [DOI] [PubMed] [Google Scholar]
- Wirthmueller L., Asai S., Rallapalli G., Sklenar J., Fabro G., Kim D. S., et al. (2018). Arabidopsis downy mildew effector HaRxL106 suppresses plant immunity by binding to radical-induced cell death1. New Phytol. 220, 232–248. 10.1111/nph.15277, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff E. G., Sorensen J. L., Lubeck M., Nielsen K. F., Thrane U., Torp J. (2010). Fusarium spp. associated with rice Bakanae: ecology, genetic diversity, pathogenicity and toxigenicity. Environ. Microbiol. 12, 649–657. 10.1111/j.1462-2920.2009.02105.x, PMID: [DOI] [PubMed] [Google Scholar]
- Xiao X., Cheng X., Yin K., Li H., Qiu J. L. (2017). Abscisic acid negatively regulates post-penetration resistance of Arabidopsis to the biotrophic powdery mildew fungus. Sci. China Life Sci. 60, 891–901. 10.1007/s11427-017-9036-2, PMID: [DOI] [PubMed] [Google Scholar]
- Xu Y., Chang P.-F. L., Liu D., Narasimhan M. L., Raghothama K. G., Hasegawa P. M., et al. (1994). Plant defense genes are synergistically induced by ethylene and methyl jasmonate. Plant Cell 6, 1077–1085. 10.1105/tpc.6.8.1077, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T. (1993). The role of auxin in plant-disease development. Annu. Rev. Phytopathol. 31, 253–273. 10.1146/annurev.py.31.090193.001345 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59, 225–251. 10.1146/annurev.arplant.59.032607.092804, PMID: [DOI] [PubMed] [Google Scholar]
- Yan J., Zhang C., Gu M., Bai Z., Zhang W., Qi T., et al. (2009). The Arabidopsis coronatine insensitive1 protein is a jasmonate receptor. Plant Cell 21, 2220–2236. 10.1105/tpc.109.065730, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Li W., Cao J., Meng F., Yu Y., Huang J., et al. (2017b). Activation of ethylene signaling pathways enhances disease resistance by regulating ROS and phytoalexin production in rice. Plant J. 89, 338–353. 10.1111/tpj.13388 [DOI] [PubMed] [Google Scholar]
- Yang D. L., Li Q., Deng Y. W., Lou Y. G., Wang M. Y., Zhou G. X., et al. (2008). Altered disease development in the eui mutants and Eui overexpressors indicates that gibberellins negatively regulate rice basal disease resistance. Mol. Plant 1, 528–537. 10.1093/mp/ssn021, PMID: [DOI] [PubMed] [Google Scholar]
- Yang G., Tang L., Gong Y., Xie J., Fu Y., Jiang D., et al. (2018b). A cerato-platanin protein SsCP1 targets plant PR1 and contributes to virulence of Sclerotinia sclerotiorum. New Phytol. 217, 739–755. 10.1111/nph.14842 [DOI] [PubMed] [Google Scholar]
- Yang B., Wang Y., Guo B., Jing M., Zhou H., Li Y., et al. (2018a). The Phytophthora sojae RXLR effector Avh238 destabilizes soybean type2 GmACSs to suppress ethylene biosynthesis and promote infection. New Phytol. 222, 425–437. 10.1111/nph.15581 [DOI] [PubMed] [Google Scholar]
- Yang B., Wang Q., Jing M., Guo B., Wu J., Wang H., et al. (2017a). Distinct regions of the Phytophthora essential effector Avh238 determine its function in cell death activation and plant immunity suppression. New Phytol. 214, 361–375. 10.1111/nph.14430 [DOI] [PubMed] [Google Scholar]
- Yin C., Park J. J., Gang D. R., Hulbert S. H. (2014). Characterization of a tryptophan 2-monooxygenase gene from Puccinia graminis f. sp. tritici involved in auxin biosynthesis and rust pathogenicity. Mol. Plant-Microbe Interact. 27, 227–235. 10.1094/MPMI-09-13-0289-FI, PMID: [DOI] [PubMed] [Google Scholar]
- Zhu Z., An F., Feng Y., Li P., Xue L., Mu A., et al. (2011). Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. USA 108, 12539–12544. 10.1073/pnas.1103959108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemann S., van der Linde K., Lahrmann U., Acar B., Kaschani F., Colby T., et al. (2018). An apoplastic peptide activates salicylic acid signalling in maize. Nat. Plants 4, 172–180. 10.1038/s41477-018-0116-y, PMID: [DOI] [PubMed] [Google Scholar]