Abstract
The availability of whole-genome sequences and associated multi-omics data sets, combined with advances in gene knockout and knockdown methods, has enabled large-scale annotation and exploration of gene and protein functions in eukaryotes. Knowing which genes are essential for the survival of eukaryotic organisms is paramount for an understanding of the basic mechanisms of life, and could assist in identifying intervention targets in eukaryotic pathogens and cancer. Here, we studied essential gene orthologs among selected species of eukaryotes, and then employed a systematic machine-learning approach, using protein sequence-derived features and selection procedures, to investigate essential gene predictions within and among species. We showed that the numbers of essential gene orthologs comprise small fractions when compared with the total number of orthologs among the eukaryotic species studied. In addition, we demonstrated that machine-learning models trained with subsets of essentiality-related data performed better than random guessing of gene essentiality for a particular species. Consistent with our gene ortholog analysis, the predictions of essential genes among multiple (including distantly-related) species is possible, yet challenging, suggesting that most essential genes are unique to a species. The present work provides a foundation for the expansion of genome-wide essentiality investigations in eukaryotes using machine learning approaches.
Keywords: Machine-learning, Essential genes, Essentiality prediction, Eukaryotes
Abbreviations: ML, Machine-learning; RNAi, RNA interference; CRISPR, Clustered regularly interspaced short palindromic repeats; PPI, Protein-protein interaction; GI, Genetic interaction; SPLS, Sparse partial least squares; OGEE, Online GEne essentiality database; GO, Gene ontology; GLM, Generalised linear model; NN, Artificial neural network; GBM, Gradient boosting method; SVM, Support-Vector machine; RF, Random Forest; ROC-AUC, Area under the receiver operating characteristic curve; PR-AUC, Area under the precision-recall curve
Graphical Abstract
1. Introduction
The sequencing and annotation of whole-genomes of eukaryotic ‘model organisms’, including the budding and fission yeasts (Saccharomyces cerevisiae and Schizosaccharomyces pombe), the elegant worm (Caenorhabditis elegans), the vinegar fly (Drosophila melanogaster), the house mouse (Mus musculus) and human (Homo sapiens) between 1995 and 2002 [[1], [2], [3], [4], [5], [6]] provided a solid foundation for structural and functional genomics explorations of these organisms. The integration of genomic and associated functional data sets as well as transcriptomic and proteomic information into specialised databases, including the Saccharomyces Genome Database [7], PomBase [8], FlyBase [9], WormBase [10], Mouse Genome Database [11] and Ensembl [12], has paved the way for large-scale comparative genomic and multi-omics investigations of these organisms. Combined with the development of gene knockdown methods, such as double-stranded RNA interference (RNAi) as well as gene-editing and -disruption technologies, including chemical and transposon mutagenesis, homologous recombination and CRISPR/Cas9, these advances have enabled genome-wide evidence-based gene annotation and the identification of genes that are crucial (i.e. essential) for life [13]. The curation of functional genomics data for essential genes, made available through specialised gene essentiality databases, has facilitated the prediction of essential homologs in both prokaryotes and eukaryotes by comparative genomics [[14], [15], [16], [17]]. Moreover, characteristics extrinsic to a gene sequence, such as gene transcription, protein function, subcellular localisation, phyletic retention and gene copy number variation, have been considered as predictors of essentiality [18,19].
The recent popularisation and expansion of high-throughput sequencing and bioinformatics tools have facilitated large-scale genomic-phenomic investigations and comparisons between or among species [20,21]. In particular, machine-learning (ML) algorithms are enhancing essentiality predictions and comparative analyses by exploring features that differentiate essential from non-essential genes [22]. For example, based on the assumption that essential genes are likely to have more molecular interaction partners [23], studies have used protein-protein interaction (PPI) network centrality measures as features for genome-scale essentiality classification using ML algorithms. However, the validity of the relationship between centrality and essentiality in PPI networks has been questioned due to a possible experimental bias [24,25], although a recent study [26] has indicated or suggested that this relationship is valid based on results from genetic interaction (GI) network analyses. Until recently, most studies of eukaryotes have applied ML methods, trained with centrality measures derived from interaction networks, focussing primarily on yeast (reviewed by [22]). However, experimental interaction studies are laborious, costly and challenging, particularly in non-model eukaryotic organisms that cannot be produced in sufficient quantities in vitro. In this context, alternative, informatic methods for essential gene prediction using features derived directly from sequence data would be advantageous, given the increasing availability of genomes and predicted proteomes. Therefore, showing that it is possible to predict essential genes within and among model species using ML algorithms, trained with features extracted directly from protein sequences (intrinsic), would significantly accelerate gene essentiality predictions in non-model species. The bioinformatic prediction of essential genes using ML models trained with features derived from gene/protein sequences has been employed and assessed in S. pombe, M. musculus, H. sapiens and Arabidopsis thaliana [[27], [28], [29], [30]]. Although some amino acid sequence composition features appear to be suitable predictors of gene essentiality within a species [17], systematic predictions and evaluations among species are lacking. While most genome-wide studies of essential genes have usually focused on single species of model eukaryotes, ML algorithms have the potential to be employed for predicting essential genes between or among species. However, no published study has yet systematically assessed or compared the performance of multiple ML algorithms for the prediction of essential genes employing protein-sequence derived features using publicly available functional genomics data, curated for essentiality. Here, we trained and evaluated the prediction performance of five classical ML models, with a focus on essentiality classification within and among eukaryotic species using intrinsic protein sequence features.
2. Materials and Methods
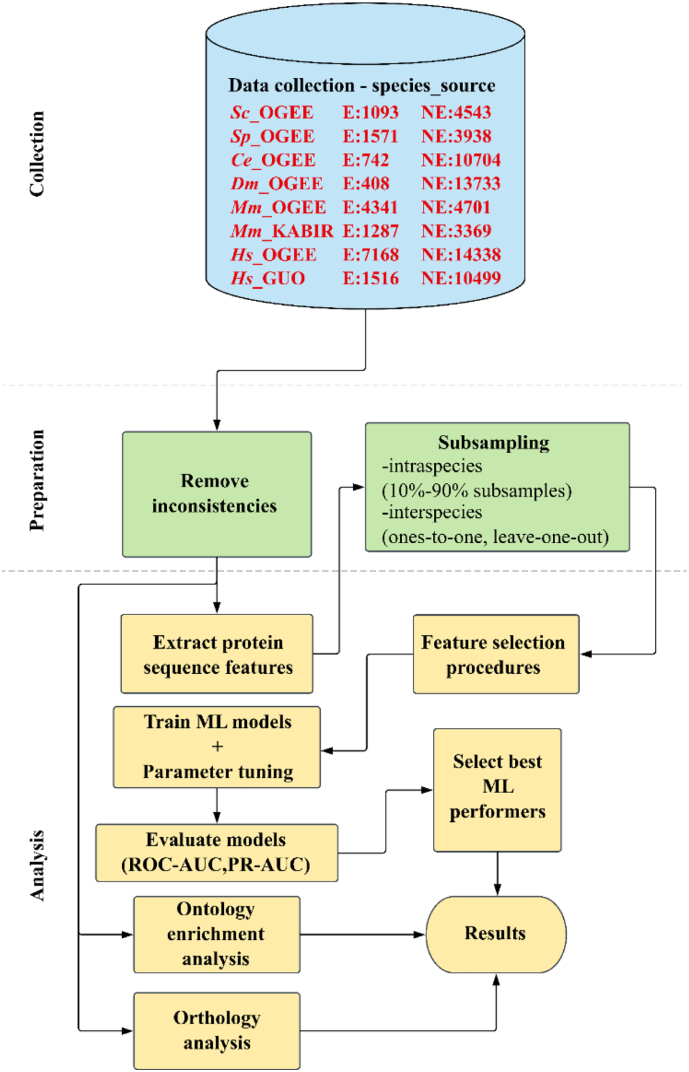
The workflow for data collection, preparation steps and analysis are depicted in Fig. 1. The data analysis was conducted in R (https://www.r-project.org), and the session information (containing software packages and versions) used here are included in the “Sessioninfo” file available in the Supplementary material.
Fig. 1.
Bioinformatic workflow for essential gene classification and evaluation using protein sequence-derived features and machine-learning methods.
2.1. Collection and Filtering of Data
In the present study, we used eukaryotic essential and non-essential genes obtained from a reference gene-essentiality database and two independent curations. Initially, protein sequences (FASTA) representing essential and non-essential genes derived from large-scale functional genomics experiments six model eukaryotic species were obtained from the Online GEne Essentiality (OGEE) database [16,31]. Species for which >80% of genes in their genome had been tested for essentiality and curated by OGEE were included, namely: S. cerevisiae (Sc_OGEE), S. pombe (Sp_OGEE), C. elegans (Ce_OGEE), D. melanogaster (Dm_OGEE), M. musculus (Mm_OGEE) and a data set representing H. sapiens cancer cell lines (Hs_OGEE). Additionally, an independent curation of the data for the same human cancer cell lines (Hs_GUO) [29] and another of essential genes from mouse (Mm_KABIR) [32] were included to investigate the effect of different curation strategies within a species on downstream analysis. Protein sequences with ambiguous entries regarding gene essentiality in OGEE (designated “inconsistent”) or containing <50 amino acids, stop or ambiguous amino acid characters were removed from the sequences.
2.2. Assigning and Comparing Essential Gene Orthologs
We assigned ortholog groups based on The Orthologous Matrix (OMA) database [33]. Briefly, we used the “oma-groups” and “oma-ensembl” files to map the Ensembl identifiers of essential genes included to their corresponding ortholog groups in the OMA database. Using the lists of ortholog groups identified in each data set, a diagram of common ortholog identifiers was generated using the “UpSetR” library for R. Additionally, a bar plot, showing pairwise essential gene orthology, was produced using the overLapper function from the “systemPipeR” package in R. We also conducted this orthology analysis using the complete gene sets for selected species for comparisons.
2.3. Gene Ontology Enrichment of Molecular Function of Essential Genes
We calculated the ‘molecular function’ enrichment of essential genes using the “clusterProfiler” [34] and “AnnotationHub” packages for R. Briefly, essential gene identifiers were first mapped to Gene Ontology (GO) identifiers using “AnnotationHub”. Then, enrichment analysis of molecular function was performed for each data set using “clusterProfiler::enrichGO”, and plots containing the top-five most enriched molecular functions were generated using “clusterProfiler::dotplot”.
2.4. Feature Extraction and Selection Procedures
We extracted 9920 intrinsic features from all individual protein sequences using the “protr” package for R [35] (see Table 1 for the full set of features). This package implements several protein feature extraction methodologies that have been used widely in ML approaches (see [35]). These features are based on amino acid composition, autocorrelation and chemical properties of individual protein sequences. For each data set, a design matrix, containing the multiple features extracted from individual protein sequences, was created with labels assigned to differentiate essential from non-essential genes. Next, we performed a standardised feature-selection approach prior to ML training. Firstly, we performed ElasticNet (alpha parameter = 0.5) feature selection using the “glmnet” package for R with cross-validation (cv.glmnet) [36], aiming to maximise the area under the receiver operating characteristic curve (ROC-AUC). Secondly, the “cv.enspls” method from the “enpls” package was used to perform Ensemble Sparse Partial Least Squares (SPLS) feature selection with cross-validation [37]. Finally, relevant features, identified by an intersection of the ElasticNet and the Ensemble SPLS methods, were selected for ML training.
Table 1.
Protein sequence-derived features utilised in the present study.
| Description | Number of features |
|---|---|
| Amino acid composition | 20 |
| Dipeptide composition | 400 |
| Tripeptide composition | 8000 |
| Protein autocorrelation features | 720 |
| Conjoint triad | 343 |
| Composition/Transition/Distribution | 147 |
| Quasi-Sequence-Order | 160 |
| Pseudo amino acid composition | 130 |
| Total | 9920 |
2.5. Subsampling, ML Training and Performance Evaluation within a Species
To estimate the prediction performance fluctuation using different data set sizes, we generated random subsamples (bootstraps; [38]) containing 10% to 90% (stepwise 10% increments) of the sequences of essential and non-essential genes in each data set for training, using the remaining data for testing (test sets). Then, we trained the following (classical) ML algorithms: Generalised Linear Model (GLM), Artificial Neural Network (NN), Gradient Boosting Method (GBM), Support-Vector Machine (SVM) and Random Forest (RF) using the “caret” package for R, performing hyperparameter tuning (for a list of parameters tested, see code provided at https://bitbucket.org/tuliocampos/essential or the static version referring to this publication at https://doi.org/10.6084/m9.figshare.8063069. For comparison, we also created a default classifier (DF), which randomly classified the test sets using the probability of essentiality calculated from the training sets defined as the ratio between the number of essential genes and the total number of reported genes for each data set. At the end of each incremental training iteration, performance evaluation metrics, including ROC-AUC and area under the precision-recall curve (PR-AUC), were obtained using the “PRROC” package in R.
2.6. ML Training and Performance Evaluation among Species
We also trained data sets containing all available data for each individual species and data set, to then perform and evaluate pairwise predictions among data sets (one-to-one), and to rank the feature importance (varImp function from “caret”) of each ML method trained with each data set. For the leave-one-out (species) approach, we used Sc_OGEE, Sp_OGEE, Ce_OGEE, Dm_OGEE, Mm_KABIR, and Hs_GUO to prepare the data sets. Six new data sets of protein sequences representing essential and non-essential genes were created, each leaving out one of the species for testing. Finally, we carried out feature selection using the ElasticNet and Ensemble SPLS consensus, followed by ML training. The performance of prediction was evaluated in the left-out species using ROC-AUC and PR-AUC metrics, as described in Subsection 2.5.
3. Results
3.1. Comparing Proportions and Ratios of Essential Genes
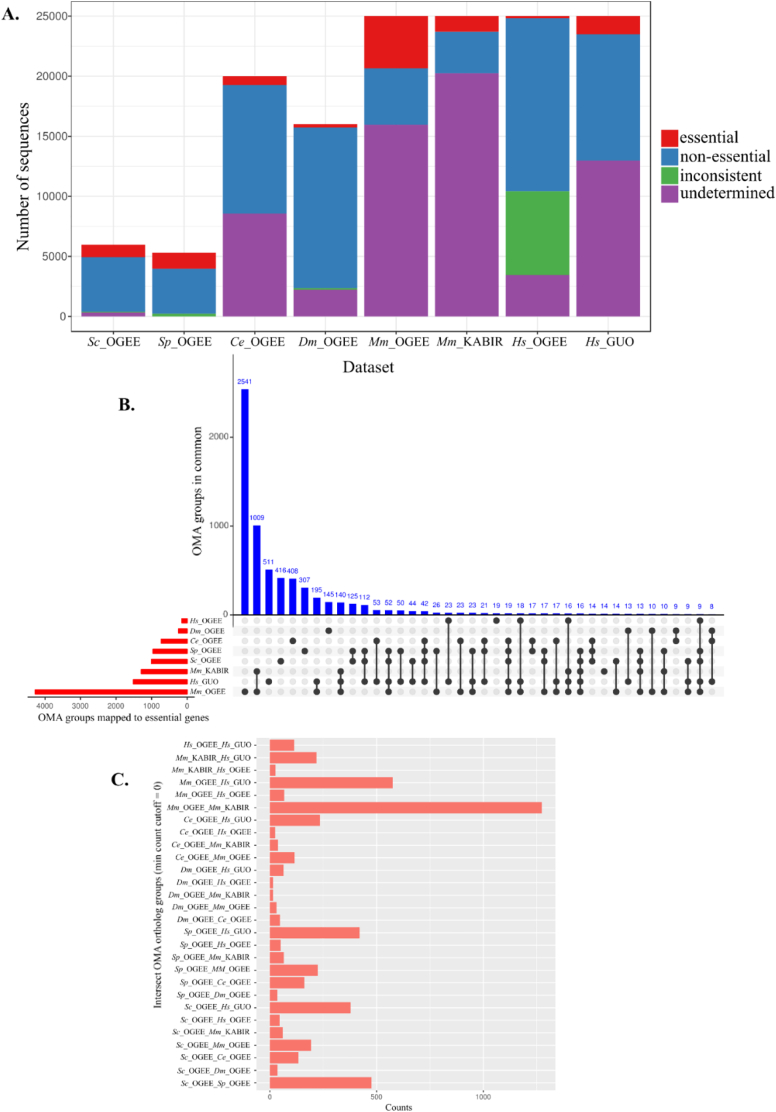
For each annotated data set obtained and used here, we summarised and compared the numbers of essential, non-essential, inconsistent, and undetermined (i.e. essentiality not reported) genes as a proportion of the available gene complement for individual species (Fig. 2A). We observed that the proportions of essential genes were considerably smaller (< 20%) than those of non-essential genes (> 80%) in most data sets, except in Mm_OGEE (~50%). In addition, the proportions of genes with inconsistent phenotypes in OGEE were low for all data sets (< 5%), except for Hs_OGEE (~28%). Almost all reference genes of S. cerevisiae and S. pombe genomes were present in the Sc_OGEE and Sp_OGEE data sets. For C. elegans, ~60% of the reference genes were present in Ce_OGEE. Hs_OGEE and Dm_OGEE contained the smallest total gene count (〈300), and the ratios of essential to non-essential genes were small (<1%). The Mm_OGEE data set contained approximately three times more essential genes than did the Mm_KABIR data set, and both had high proportions of undetermined genes (~65% and ~82%, respectively) (Fig. 2A). After filtering, the number of essential genes was considerably lower in Hs_OGEE (n = 182) than in Hs_GUO (n = 1516), whereas the number of non-essential genes was higher (n = 14362) and (n = 10499), respectively.
Fig. 2.
A. Summary of gene essentiality data obtained from different sources and used in the present study. Included are the number of genes found with multiple conflicting entries (inconsistent) as well as genes not reported as either essential or non-essential, complementing the predicted proteomes. B. Diagram exhibiting the total (red) and shared (blue) ortholog identifiers of essential genes from the OrthoOMA database used in the present study (selected species and data sets). C. Pairwise essential gene orthologs identified using the OrthoOMA ortholog groups (format: species1_source1_species2_source2). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Analyses for Orthologous Genes and Functional Enrichment for Essential Genes
An analysis among data sets revealed that approximately half of the orthologs of essential genes were exclusive to individual data sets (Fig. 2B), except between Mm_OGEE and Mm_KABIR, for which the number of orthologs (n = 1274) was almost the same as the total number of essential genes in Mm_KABIR (n = 1287), of which most (n = 1009 ortholog identifiers) were shared between these two data sets representing mouse. Hs_GUO had ~ 500 orthologs with Mm_OGEE, Sp_OGEE or Sc_OGEE. A similar number of orthologs was shared between Sp_OGEE and Sc_OGEE (Fig. 2C). Although many pairwise orthologs were identified between or among species/data sets, no essential genes were shared among all data sets used here. In three occurrences, with 19, 16 and 9 genes, orthologs were shared among five data sets, respectively, and other genes were shared among ≤4 species (Fig. 2C). When performing a similar analysis of orthologs using complete gene sets of the species studied here, we observed that most genes were exclusive to individual species and that 536 were shared among all six species (see Fig. S1).
Overall, the five most enriched functions in each species related to DNA/RNA binding and processing (Fig. S2). In total, enriched functions represented >50% of the essential genes in the Ce_OGEE, Dm_OGEE and Mm_KABIR data sets. The same functions were enriched when Mm_OGEE and Mm_KABIR were compared. By contrast, enriched molecular functions of essential genes for Sc_OGEE and Sp_OGEE accounted for <30% of the respective essential genes, and these data sets shared enrichments for “catalytic activity on RNA” and “snoRNA binding”. The top 5 enriched functions for Hs_OGEE and Hs_GUO included <30% of their respective essential genes, but these data sets did not share the same enriched functions.
3.3. Performances of Essentiality Classification Inferred by ML Models within a Species
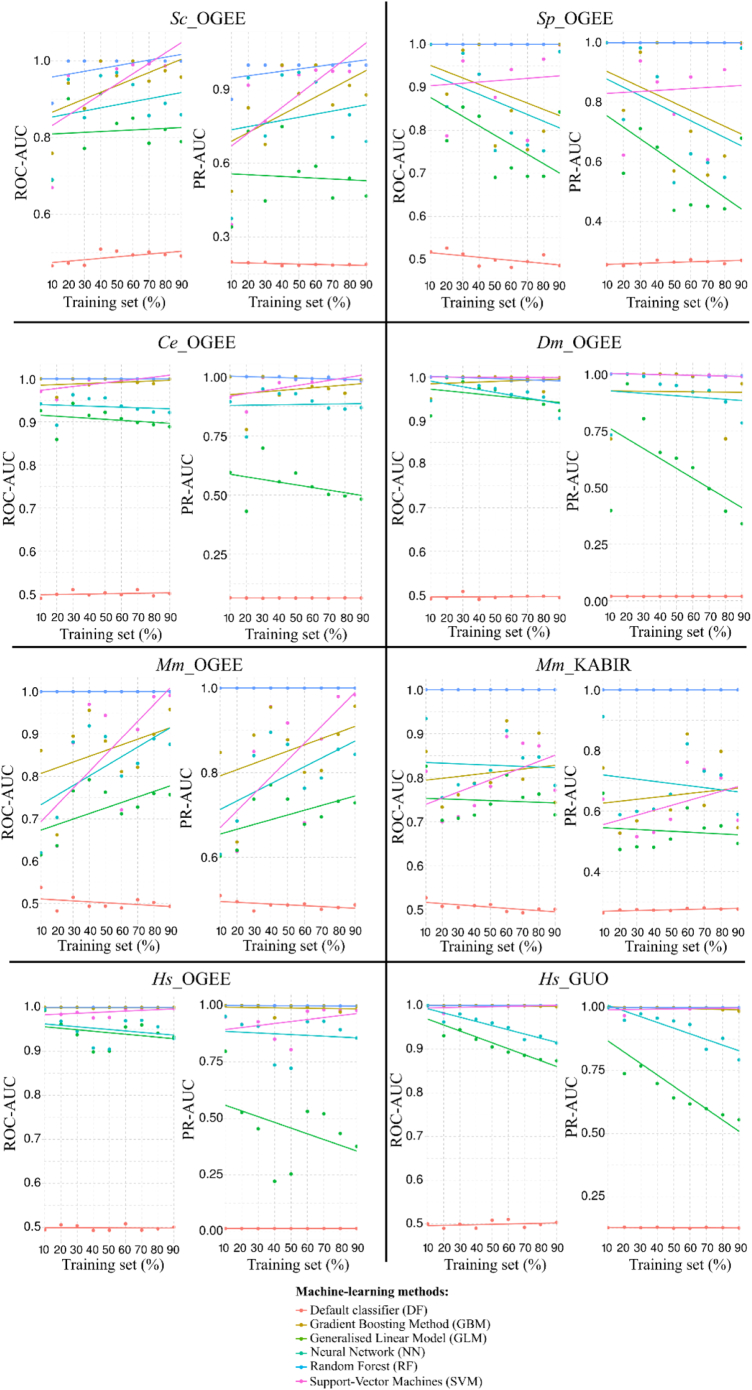
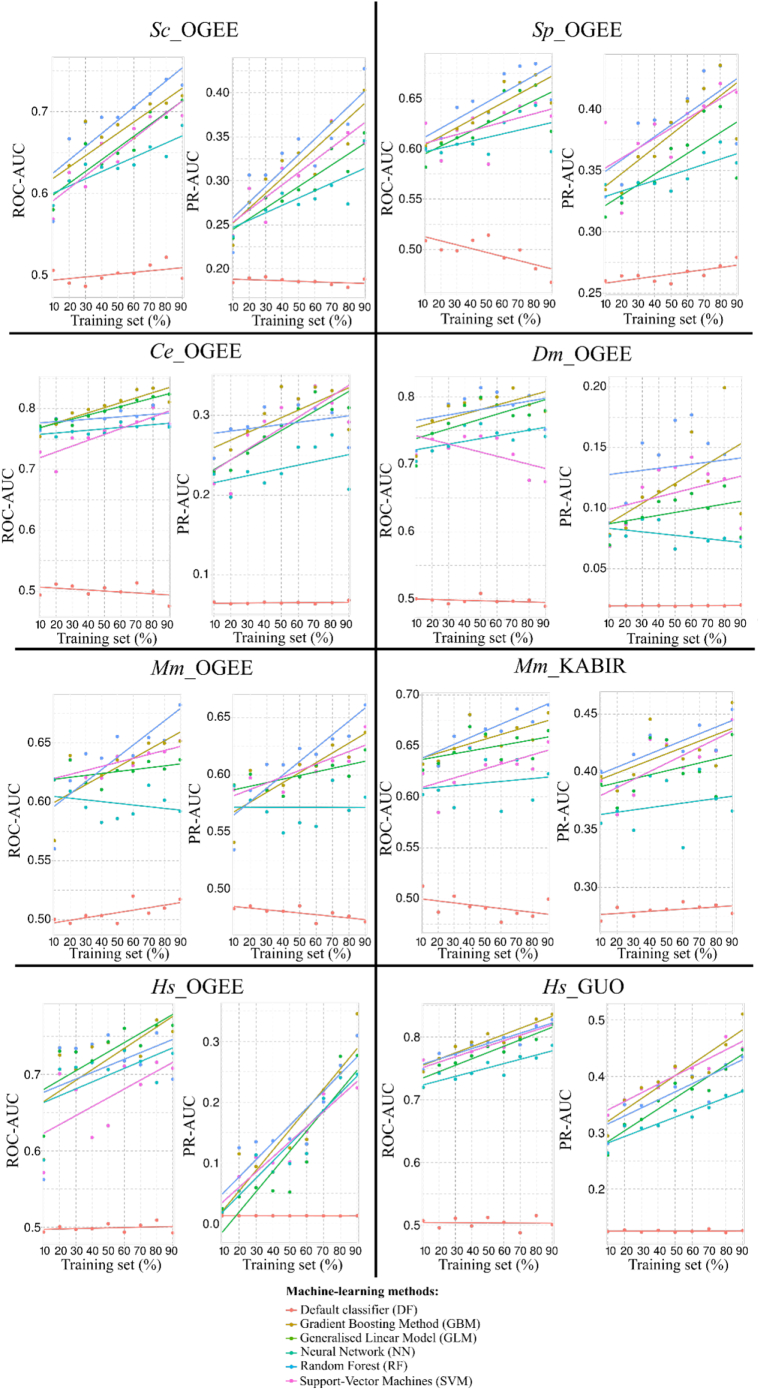
The performances of the ML models for essential gene predictions on training sets are shown in Fig. 3. Feature selection procedures were employed at each training/test step (10% to 90%), but only the final set of selected features when using 100% of each data set are reported here (see Subsection 3.4 and Fig. S4). Overall, all ML methods out-performed the default classifier (DF), in terms of both ROC-AUC and PR-AUC metrics, showing that they performed better than random classification based on known probability of essentiality. RF achieved ROC-AUC of ~ 1, and PR-AUC of ~ 1 for all data sets tested. SVM and GBM exhibited similar performances to RF using the Ce_OGEE, Dm_OGEE and human data sets. As more data were included in training sets, SVM and GBM models rapidly achieved ROC-AUC of >0.9 and PR-AUC of >0.8 for predicting Sc_OGEE, Sp_OGEE, and Mm_OGEE. The PR-AUC calculated for Mm_KABIR improved slowly when the amount of training data increased, and ranged from ~0.55 to 0.7, with ROC-AUC values of ~0.75 to 0.85. PR-AUC of GLM decreased as more data were added to the training sets, while NN performance decreased for most data sets, but increased for Sc_OGEE, Ce_OGEE and Mm_OGEE. GLM and NN, however, achieved ROC-AUC of >0.8 using small training sets (10%).
Fig. 3.
Performance evaluation of essential gene classification of training sets (self-predictions) within selected eukaryotic species using Area Under Receiver Operating Characteristic and Precision-Recall Curves (ROC-AUC and PR-AUC; training set sizes between 10 and 90%, using 10% increments). The dots represent the calculated ROC-AUC/PR-AUC values, and linear models fit dots representing the performances of each machine-learning algorithm. Feature selection procedures were performed for each subsample.
Subsequently, we evaluated the performance of the ML models for essentiality predictions on test sets within a species (Fig. 4). Again, the trained ML methods outperformed random classification (DF), and both ROC-AUC and PR-AUC of all ML models increased as more data were added to the training sets. In most cases, the performance of NN models improved slower compared with other ML models. In the fungal species, ROC-AUC using RF and GBM increased from ~0.6 to 0.75 (Sc_OGEE) and to ~0.67 (Sp_OGEE). PR-AUC increased from ~0.25 to 0.40 (Sc_OGEE) and from ~0.33 to 0.42 (Sp_OGEE). Applying GBM to Ce_OGEE and Dm_OGEE, ROC-AUC values ranged from ~0.75 to >0.80, while PR-AUC improved from ~0.25 to 0.32 (Ce_OGEE) and from ~0.1 to 0.15 (Dm_OGEE). Using each of the data sets for mouse, RF and GBM achieved ROC-AUC values ranging from ~0.6 to 0.70, and the highest PR-AUC was achieved using RF (~0.65 for Mm_OGEE, and ~0.45 for Mm_KABIR). For the human data sets, GBM achieved ROC-AUC values ranging from ~0.67 to 0.75 (Hs_OGEE) and from ~0.75 to 0.82 (Hs_GUO), while PR-AUC values ranged from ~0 to 0.26 (Hs_OGEE) and ~0.32 to 0.45 (Hs_GUO).
Fig. 4.
Performance evaluation of essential gene classification of test sets within selected eukaryotic species using Area Under Receiver Operating Characteristic and Precision-Recall Curves (ROC-AUC and PR-AUC; training set sizes between 10 and 90%, using 10% increments). The dots represent the calculated ROC-AUC/PR-AUC values, and linear models fit dots representing the performances of each machine-learning algorithm performances. Feature selection procedures were performed for each subsample.
3.4. Selected Features and Prediction Performance of ML Models Using One-to-One and Leave-One-Species-out Approaches
Using each complete essentiality data set, the number of features selected by both ElasticNet and Ensemble SPLS methods ranged from 44 for Mm_KABIR to 251 for Hs_GUO (Fig. S3). By comparing the features selected among data sets, no feature was common among all. Only one feature (CTriad_VS666, a feature related to the composition of negatively-charged amino acid triplets in a protein sequence – see “protr” for R documentation) was shared among most data sets, except for Ce_OGEE and Dm_OGEE, and 12 distinct features were shared among 4 or 5 data sets (Fig. S3). The importance of the selected features of each data set on gene essentiality prediction varied, depending on the data set and the ML method used (see Table S1).
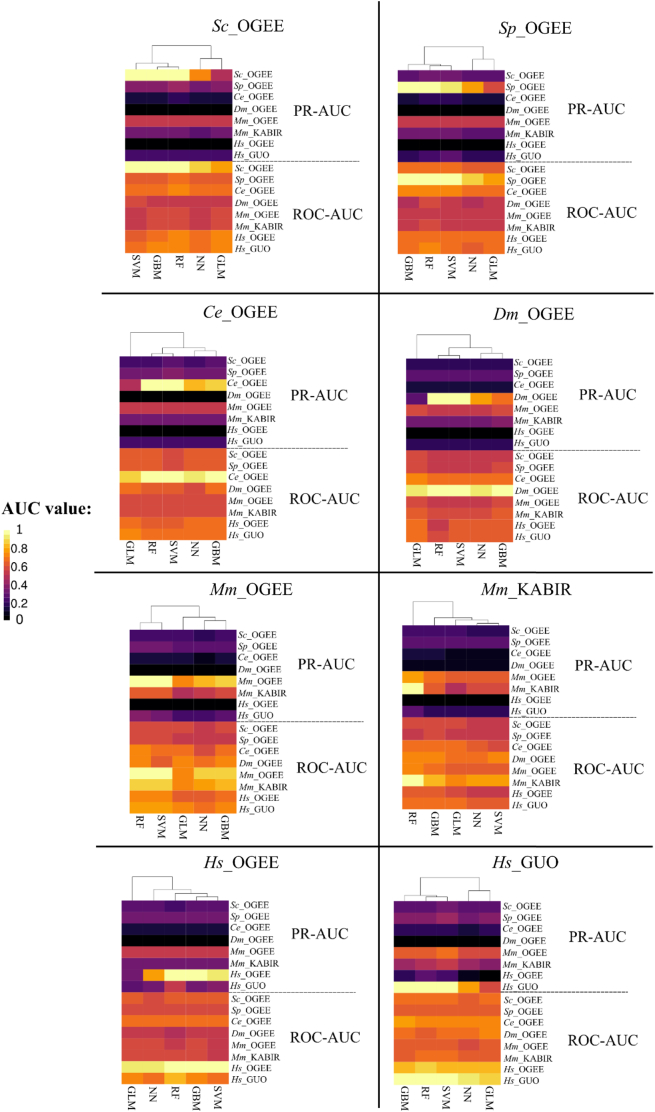
Regarding model performance in our pairwise training/prediction approach (Fig. 5), ROC-AUC of ~ 1, and PR-AUC of ~ 1 were consistently obtained with RF when predictions were performed and evaluated on training sets. SVM also achieved similar performances, except for Mm_KABIR (ROC-AUC of <0.8 and PR-AUC of <0.6). GBM achieved ROC-AUC values of ~1 for most data sets, except for Mm_OGEE (~0.85 to 0.9) and Mm_KABIR (~0.75 to 0.8). Finally, GLM and NN achieved similar and more variable ROC-AUC values (~0.65 to 0.9) for predictions from training sets, while PR-AUC varied from ~0.6 to 0.7 for NN and from ~0.35 to 0.65 for GLM.
Fig. 5.
Heatmaps depicting the prediction performances (y-axis: ROC-AUC and PR-AUC for each test set) of five machine-learning models (x-axis) trained using multiple essentiality data sets (labels on top of the heatmaps represent each of the training sets).
When models were trained with a data set to predict independent data (e.g., training with Ce_OGEE and predicting for Dm_OGEE; Fig. 5), the ROC-AUC values varied from ~0.6 to 0.75, whereas PR-AUC ranged from ~0.1 to 0.65. In addition, ML models trained with Hs_GUO and Mm_OGEE data sets achieved overall ROC-AUC values of >0.7. PR-AUC values of >0.5 were achieved for Mm_OGEE predictions, regardless of the training set used. Regarding ROC-AUC, gene essentiality in Ce_OGEE seems to be partially and consistently predicted by any other data set (~0.70 to 0.80). Interestingly, Sc_OGEE and Sp_OGEE are reasonable predictors of gene essentiality for the two human data sets (~0.65 to 0.8), considering the ROC-AUC metric, but not for the mouse and Dm_OGEE data sets (<0.65).
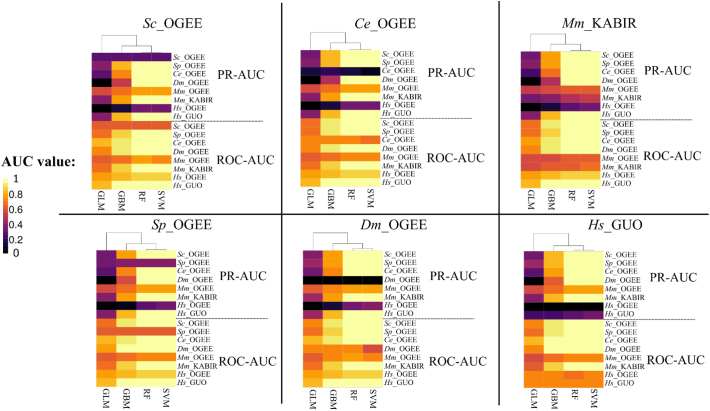
Finally, we evaluated the performance metrics using the leave-one-species-out approach (Fig. 6). The performance of the essentiality predictions on the training sets achieved ROC-AUC and PR-AUC values of >0.9. Overall, predictions for the left-out species achieved ROC-AUC values of >0.7, and PR-AUC values were variable (~0.1 to 0.6). We observed that the PR-AUC metric was penalised more when the external target data set was highly imbalanced (i.e. the number of non-essential was markedly greater than that of essential genes), as observed for Ce_OGEE and Dm_OGEE (Fig. 1). The numbers of selected features common to leave-one-out-data sets were as follows: 190 shared by six data sets, 184 by five and 126 by four (Table S1).
Fig. 6.
Heatmaps depicting the prediction performances (y-axis: ROC-AUC and PR-AUC) of four machine-learning models (x-axis) using a leave-one-species-out approach. Labels on top of each heatmap represent the species that was excluded from the training set. The Mm_OGEE and Hs_OGEE data sets were not included in any of the training sets.
4. Discussion
This study showed that, using selected features from protein sequences linked to functional genomics data sets, ML methods can predict essential genes in eukaryotes. ML-based predictions within a species were reliable, and those between or among species were better than random guessing by a default classifier. Integral to prediction performance were: (i) the nature, extent and curation of data sets, (ii) the selection of features and/or (iii) the algorithm/approach used.
ML prediction performance, measured by ROC-AUC and PR-AUC, and the selected best predictive features varied, depending on algorithm used and species studied, but RFs out-performed other methods in most scenarios. The ML methods used here consistently outperformed random guessing based on true probabilities, showing that they can successfully learn and enhance the classification of essential genes. Random Forests are known to be robust, even when features exhibit non-linear relationships with the response variable, in the presence of correlated features and/or with high-dimensional data [39]. The systematic ML approach using data subsets of variable sizes (10% to 90%) within a species revealed that, in most cases, the prediction performance increased as more data were added to the training set(s). However, the rate of improvement was variable among ML models and data sets. For C. elegans and D. melanogaster data sets, essentiality predictions employing ML methods trained with protein sequence features achieved high ROC-AUC (> 0.80), with PR-AUC values between >0.30 and >0.10, respectively. For Hs_GUO, ML performance (ROC-AUC > 0.80, PR-AUC > 0.45) was comparable with that of a published study using an SVM model trained with nucleotide composition features (ROC-AUC = 0.88 [29];. Compared with our study, [30] improved ML performance for S. pombe (ROC-AUC = 0.84) using nucleotide sequence features to train an RF model, although their study used equal numbers of essential and non-essential genes for training and performance assessments, and thus under-sampled non-essential-genes. By contrast, [27] collected sequence features from curated data from mice, performed feature selection and trained an RF method for essentiality predictions, achieving a ROC-AUC value of 0.73, which is comparable with the results obtained here (ROC-AUC of ~ 0.68, PR-AUC of >0.4) for both Mm_KABIR and Mm_OGEE using the same algorithm. In the same study [27], complemented sequence features PPI and transcription data, and the ROC-AUC value increased to 0.78, showing the complementation of intrinsic and extrinsic features can achieve improved results. In the present study, predictions between or among species, using either one-to-one or leave-one-out approaches, also performed considerably better than random guessing, but the ML methods and features used here were insufficient to achieve results of high confidence (i.e. ROC-AUC of ~ 1 and PR-AUC of ~ 1). However, this outcome might reflect a large evolutionary distance between some of the taxa studied here. Indeed, predictions among closely related species deserve detailed and critical evaluation in the future.
Here, we elected to include features that can be extracted directly from the protein sequences without performing sequence similarity comparisons or using any extrinsic data. Feature selection procedures identified the best predictors for individual data sets, markedly reducing ML model complexity, while maximising prediction performance. In agreement with previous studies, we showed that ElasticNet and Ensemble SPLS methods were highly effective at selecting the best predictive features [37]. An alternative feature selection and reduction method commonly used in ML-based essentiality studies is LASSO [27,29,40]; this method was not used herein, as it tends to discard variables unnecessarily [41]. A comparison of the many, alternative methods and approaches for feature selection [42] could be evaluated in future systematic studies - this was not within the scope of the present study. Results from the comparison of selected protein features for individual data sets revealed that gene essentiality appears to be partly species-specific, because no feature was shared among all data sets. Conversely, by comparing the features among the leave-one-out data sets, more features were selected from individual data sets, and many of them were shared among data sets, suggesting that, although the ML model complexity increased, there are protein features that might be generic predictors of essential genes in eukaryotes. In this study, we selected features using intrinsic protein sequence characteristics, but in the future, nucleotide sequence features and extrinsic features, such as expression levels, gene ontology and network centrality measures [22,43,44], might be included to improve the performance of our models. From a biological perspective, it is challenging to infer the reasons why certain sequence features are predictors of essentiality, although a previous investigation [45] has shown the relationship between codon or amino acid usage and increased gene expression as well as translation efficiency. The present study presents the most predictive protein features for each species and data set. Understanding why these features are associated with gene essentiality remains unclear, and warrants further investigation.
Previous large-scale orthology analyses have shown that only a small number of genes is conserved across the Tree of Life, meaning that many essential genes can be specific to each species [46]. Here, we showed that most essential genes were inferred to be species-specific or were shared only by closely-related species, indicating challenges associated with homology-based comparisons. This information suggests that the sets of essential genes of distantly related species appear to be markedly different, and that essential orthologs comprise a small fraction of all orthologous genes. Although this finding contradicts previous assumptions [47], it should be considered that the methods used by the OMA Orthology database to define orthologs appear to be highly stringent, which may inhibit the detection of evolutionarily distant orthologs [48]. However, the present results indicate the potential limitations of sequence alignment approaches to define orthologs between or among distantly related species, with implications for gene essentiality studies. In a previous investigation of essential genes, it had been observed that orthologs of genes linked to lethality in at least one model species were more likely to be essential in another [49]. However, essentiality predictions based solely on orthology can impose challenges on the identification of non-conserved essential genes. Moreover, the assumption that orthologs have the same function may not always be true [50]. These inferences may also have implications for studies that use orthology data to identify features for ML-training and -predictions. By evaluating functional enrichment of essential genes in each data set, we established that molecular functions usually related to conserved cellular functions such as DNA and RNA processing, but the top-five enriched functions did not include most essential genes of a respective data set. This information shows that other unknown functions might be enriched, or that there is a weak relationship between essential genes and functional enrichment. However, the variable results found among species may be, to some extent, a consequence of incomplete or inconsistent essentiality data curation.
The nature and extent of curation of functional genomics data and criteria used to predict essential genes can affect both ML- and orthology-based approaches. In this study, many genes of a species were excluded from analysis, either because functional genomics data were lacking and/or because there were multiple conflicting entries in OGEE. This aspect affected ML performance and evaluation as well as the essential gene orthology analysis. Data sets Sc_OGEE and Sp_OGEE contained most genes of their respective species and were the most complete data sets, whereas the other data sets contained many genes that remain to be validated functionally and/or curated regarding essentiality before being integrated into OGEE.
When defining essential genes from phenotypic data in the curation process, it is important to make decisions about genes that exhibit variable essentiality, which can impact subsequent analysis using ML approaches. To highlight the implications of incomplete and inconsistent gene essentiality curations, we elected to include two additional curated data sets [29,32] external to OGEE and showed that subsequent analyses were affected. For instance, Mm_KABIR is based on the analysis of multiple functional genomics studies available in the best-curated mouse database (MGD) [32], which contrasts with Mm_OGEE - a data set derived from a single large-scale study. We showed that essential genes in Mm_OGEE are almost entirely within Mm_KABIR, which means that there is consistency between these data sets. Conversely, the number of essential genes in Hs_OGEE was markedly lower, because it excluded “inconsistent” data from the OGEE database, thus sharing only a small number of orthologs with Hs_GUO, which had undergone a more thorough curation of data derived from functional genomics in cancer cell lines. The similarities and differences observed among curations for the same species are also reflected in ML performances using these data sets. Moreover, Ce_OGEE data is derived from a single, large-scale study, although many genes have been tested by multiple studies and have been available in WormBase [10]. Data sets Dm_OGEE and Sc_OGEE were each derived from two studies, Sp_OGEE from seven, and the human data sets from 18 studies [16,31].
Currently, the same eukaryotic essentiality-related data are present in both OGEE [16] and DEG [14]. Clearly, a wealth of gene essentiality information derived from multiple functional genomics investigations is accessible from species-specific databases [[9], [10], [11]] and remains to be integrated into available essentiality databases. However, given the challenges associated with inconsistent data by multiple experiments and the lack of standardised essentiality annotations among these databases, the present work did not involve data curation. When curating functional genomics data for gene essentiality [51], there are multiple aspects that need to be considered. For instance, in unicellular organisms, the essentiality of a gene is defined by its influence on organismal growth. In multicellular organisms, genes can be essential/non-essential for embryonic development, for other developmental stages or for reproduction. Essentiality in cell culture (in vitro) or in specific tissues may not translate into the lethality of a whole organism (in vivo), and different functional genomics methods might identify distinct sets of essential genes. Some genes are essential or non-essential in or to an organism, depending on certain genetic and environmental backgrounds or conditions. This context needs careful consideration. Moreover, some functional genomics methods can more effectively block the activity of genes than others [52]. Indeed, functional genomics studies using multiple methods should be undertaken to verify the specificity of gene essentiality and exclude off-target effects and technical biases [53]. In addition, some organisms are more amenable to functional genomic experimentation than others [54,55]. For instance, it has been shown that the characterisation of essential genes by RNAi and CRISPR may not always concur, but a combination of results from multiple methods can improve performance [49]. A recent study compared functional genomics data for human cell lines using mouse knockout genes, highlighting that different biological systems and experimental methods may lead to discrepant inferences or conclusions, and should be compared with caution [56]. Therefore, gene essentiality investigations by multiple studies and functional genomics platforms, followed by careful curation for essentiality are central to identifying essential and non-essential genes, in addition to genes that are essential under specific experimental/developmental/environmental conditions (i.e. “conditionally essential” genes). It should also be considered that essentiality might be a quantitative trait rather than a simplistic essential/non-essential classification, which would require standard methods for quantification [57]. Considering all of these aspects, criteria for the inclusion/exclusion of genes to train ML models for essentiality predictions should be defined with caution, depending on the purpose of a study.
Clearly, much remains to be discovered regarding the characteristics that underpin gene essentiality in eukaryotic organisms, and to what extent these characteristics can be explored to predict essential genes within and among species. The current and future availability of genomic data and functional genomics platforms for non-model organisms should allow the discovery of common and specific essential genes, ultimately contributing to our understanding of eukaryotic cells and organisms. Whether there is a minimum set of genes that is essential for the survival of a cell is one of the most fundamental and unresolved questions in biology [[58], [59], [60]]. If there is a minimum set, it would be present and essential to all or most cells and organisms. Although comparative analyses of homologs/orthologs are often used to predict conserved essential genes, which, in most cases, share similar functions in different species [48], computational methods using ML approaches and feature selection technologies should now facilitate explorations of large data sets, enabling the prioritisation of essential gene candidates for functional genomic verification. Rigorous and consistent curation of essentiality information from functional genomics data are needed for both orthology- and ML-based approaches, and adequate consideration needs to be given to the essential roles of genes in different cell types, tissues, developmental stages and environments, and their characterisation in different experimental platforms, both in vitro and in vivo.
5. Conclusion
We believe that the present study provides a basis for essential gene predictions using ML approaches, which can be extended to include other intrinsic or extrinsic features, and for evaluating other ML methods such as deep-learning [61]. We share the source code for the systematic analysis used in our study with the scientific community and suggest that future work should focus on identifying novel features and improving ML approaches to enhance the prediction of essentiality. We are confident that predictions, experimental validation and comparative analysis of essential genes will contribute to understanding the biology and evolution of eukaryotes.
Declarations of Interests
The authors declare no competing interests.
Authors' Contributions
Conceived and designed the study: TLC, NDY and PKK. Undertook the study and data analysis: TLC. Wrote the paper: TLC, NDY and RBG. Contributed to interpretation of findings and supervised the project: NDY, RBG and PKK. All authors read and approved the final version of the manuscript.
Acknowledgements
This research was funded by grants from the National Health and Medical Research Council (NHMRC) and the Australian Research Council (ARC) to RBG and NDY. Other support from the Yourgene Bioscience and Melbourne Water Corporation is gratefully acknowledged (RBG). NDY is supported by a Career Development Fellowship, and PKK by an Early Career Research Fellowship from NHMRC. TLC is a recipient of a Research Training Program Scholarship from the Australian Government and is also supported by the Oswaldo Cruz Foundation (Fiocruz/Brazil).
Footnotes
A manuscript submitted for publication in the Special Issue entitled “Machine Learning and Pattern Recognition Techniques in Molecular Function and Structure Analysis” - Guest Editors: Professors Qin Ma and Leyi Wei.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2019.05.008.
Contributor Information
Robin B. Gasser, Email: robinbg@unimelb.edu.au.
Neil D. Young, Email: nyoung@unimelb.edu.au.
Appendix A. Supplementary data
Supplementary Figures and additional information
Supplementary Table S1
Session info
References
- 1.Waterston R., Silston J. The genome of Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1995;92(24):10836–10840. doi: 10.1073/pnas.92.24.10836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goffeau A., Barrell B.G., Bussey H., Davis R.W., Dujon B., Feldmann H. Life with 6000 genes. Sci. 1996;274(5287) doi: 10.1126/science.274.5287.546. [546, 563–7] [DOI] [PubMed] [Google Scholar]
- 3.Adams M.D., Celniker S.E., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G. The genome sequence of Drosophila melanogaster. Sci. 2000;287(5461):2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 4.Wood V., Gwilliam R., Rajandream M.A., Lyne M., Lyne R., Stewart A. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415(6874):871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- 5.International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 6.Mouse Genome Sequencing Consortium Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420(6915):520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 7.Cherry J.M., Adler C., Ball C., Chervitz S.A., Dwight S.S., Hester E.T. SGD: Saccharomyces genome database. Nucleic Acids Res. 1998;26(1):73–79. doi: 10.1093/nar/26.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wood V., Harris M.A., McDowall M.D., Rutherford K., Vaughan B.W., Staines D.M. PomBase: a comprehensive online resource for fission yeast. Nucleic Acids Res. 2012;40(Database issue):D695–D699. doi: 10.1093/nar/gkr853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drysdale R., FlyBase Consortium FlyBase: a database for the Drosophila research community. Methods Mol Biol. 2008;420:45–59. doi: 10.1007/978-1-59745-583-1_3. [DOI] [PubMed] [Google Scholar]
- 10.Stein L., Sternberg P., Durbin R., Thierry-Mieg J., Spieth J. WormBase: network access to the genome and biology of Caenorhabditis elegans. Nucleic Acids Res. 2001;29(1):82–86. doi: 10.1093/nar/29.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blake J.A., Richardson J.E., Bult C.J., Kadin J.A., Eppig J.T. Mouse genome database group MGD: the mouse genome database. Nucleic Acids Res. 2003;31(1):193–195. doi: 10.1093/nar/gkg047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Birney E., Andrews T.D., Bevan P., Caccamo M., Chen Y., Clarke L. An overview of Ensembl. Genome Res. 2004;14(5):925–928. doi: 10.1101/gr.1860604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhan T., Boutros M. Towards a compendium of essential genes–from model organisms to synthetic lethality in cancer cells. Crit Rev Biochem Mol Biol. 2016;51(2):74–85. doi: 10.3109/10409238.2015.1117053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang R., Ou H.Y., Zhang C.T. DEG: a database of essential genes. Nucleic Acids Res. 2004;32:D271–D272. doi: 10.1093/nar/gkh024. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang C.T., Zhang R. Gene essentiality analysis based on DEG, a database of essential genes. Methods Mol Biol. 2007;416:391–400. doi: 10.1007/978-1-59745-321-9_27. [DOI] [PubMed] [Google Scholar]
- 16.Chen W.H., Minguez P., Lercher M.J., Bork P. OGEE: an online gene essentiality database. Nucleic Acids Res. 2012;40(Database issue):D901–D906. doi: 10.1093/nar/gkr986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seringhaus M., Paccanaro A., Borneman A., Snyder M., Gerstein M. Predicting essential genes in fungal genomes. Genome Res. 2006;16(9):1126–1135. doi: 10.1101/gr.5144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saha S., Heber S. In silico prediction of yeast deletion phenotypes. Genet Mol Res. 2006;5(1):224–232. [PubMed] [Google Scholar]
- 19.Gustafson A.M., Snitkin E.S., Parker S.C., DeLisi C., Kasif S. Towards the identification of essential genes using targeted genome sequencing and comparative analysis. BMC Genomics. 2006;7:265. doi: 10.1186/1471-2164-7-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang G., Bhardwaj N., Robilotto R., Gerstein M.B. Getting started in gene orthology and functional analysis. PLoS Comput Biol. 2010;6(3) doi: 10.1371/journal.pcbi.1000703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bodenreider O., Burgun A. A framework for comparing phenotype annotations of orthologous genes. Stud Health Technol Inform. 2010;160:1309–1313. [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X., Acencio M.L., Lemke N. Predicting essential genes and proteins based on machine learning and network topological features: a comprehensive review. Front Physiol. 2016;7:75. doi: 10.3389/fphys.2016.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeong H., Mason S.P., Barabási A.L., Oltvai Z.N. Lethality and centrality in protein networks. Nature. 2000;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 24.Coulomb S., Bauer M., Bernard D., Marsolier-Kergoat M.C. Gene essentiality and the topology of protein interaction networks. Proc Biol Sci. 2005;272(1573):1721–1725. doi: 10.1098/rspb.2005.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zotenko E., Mestre J., O'Leary D.P., Przytycka T.M. Why do hubs in the yeast protein interaction network tend to be essential: reexamining the connection between the network topology and essentiality. PLoS Comput Biol. 2008;4(8) doi: 10.1371/journal.pcbi.1000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Costanzo M., VanderSluis B., Koch E.N., Baryshnikova A., Pons C., Tan G. A global genetic interaction network maps a wiring diagram of cellular function. Sci. 2016;353(6306) doi: 10.1126/science.aaf1420. [pii: aaf1420] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan Y., Xu Y., Xu J., Ball R.L., Liang H. Predicting the lethal phenotype of the knockout mouse by integrating comprehensive genomic data. Bioinformatics. 2012;28(9):1246–1252. doi: 10.1093/bioinformatics/bts120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lloyd J.P., Seddon A.E., Moghe G.D., Simenc M.C., Shiu S.H. Characteristics of plant essential genes allow for within- and between-species prediction of lethal mutant phenotypes. Plant Cell. 2015;27(8):2133–2147. doi: 10.1105/tpc.15.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo F.B., Dong C., Hua H.L., Liu S., Luo H., Zhang H.W. Accurate prediction of human essential genes using only nucleotide composition and association information. Bioinformatics. 2017;33(12):1758–1764. doi: 10.1093/bioinformatics/btx055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nigatu D., Sobetzko P., Yousef M., Henkel W. Sequence-based information-theoretic features for gene essentiality prediction. BMC Bioinformatics. 2017;18(1):473. doi: 10.1186/s12859-017-1884-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen W.H., Lu G., Chen X., Zhao X.M., Bork P. OGEE v2: an update of the online gene essentiality database with special focus on differentially essential genes in human cancer cell lines. Nucleic Acids Res. 2017;45(D1):D940–D944. doi: 10.1093/nar/gkw1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kabir M., Barradas A., Tzotzos G.T., Hentges K.E., Doig A.J. Properties of genes essential for mouse development. PLoS One. 2017;12(5) doi: 10.1371/journal.pone.0178273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altenhoff A.M., Glover N.M., Train C.M., Kaleb K., Warwick Vesztrocy A., Dylus D. The OMA orthology database in 2018: retrieving evolutionary relationships among all domains of life through richer web and programmatic interfaces. Nucleic Acids Res. 2018;46(D1):D477–D485. doi: 10.1093/nar/gkx1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiao N., Cao D.S., Zhu M.F., Xu Q.S. Protr/ProtrWeb: R package and web server for generating various numerical representation schemes of protein sequences. Bioinformatics. 2015;31(11):1857–1859. doi: 10.1093/bioinformatics/btv042. [DOI] [PubMed] [Google Scholar]
- 36.Ma S., Huang J. Penalized feature selection and classification in bioinformatics. Brief Bioinform. 2008;9(5):392–403. doi: 10.1093/bib/bbn027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chun H., Keleş S. Sparse partial least squares regression for simultaneous dimension reduction and variable selection. J R Stat Soc Series B Stat Methodology. 2010;72(1):3–25. doi: 10.1111/j.1467-9868.2009.00723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kulesa A., Krzywinski M., Altman N. Sampling distributions and the bootstrap. Nat Methods. 2015;12(6):477–478. doi: 10.1038/nmeth.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen X., Ishwaran H. Random forests for genomic data analysis. Genomics. 2012;99(6):323–329. doi: 10.1016/j.ygeno.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu X., Wang B.J., Xu L., Tang H.L., Xu G.Q. Selection of key sequence-based features for prediction of essential genes in 31 diverse bacterial species. PLoS One. 2017;12(3) doi: 10.1371/journal.pone.0174638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kirpich A., Ainsworth E.A., Wedow J.M., Newman J.R.B., Michailidis G., McIntyre L.M. Variable selection in omics data: a practical evaluation of small sample sizes. PLoS One. 2018;13(6) doi: 10.1371/journal.pone.0197910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saeys Y., Inza I., Larrañaga P. A review of feature selection techniques in bioinformatics. Bioinformatics. 2007;23(19):2507–2517. doi: 10.1093/bioinformatics/btm344. [DOI] [PubMed] [Google Scholar]
- 43.Lei X., Zhao J., Fujita H., Zhang A. Predicting essential proteins based on RNA-Seq, subcellular localization and GO annotation datasets. Knowl-Based Syst. 2018;151:136–148. [Google Scholar]
- 44.Lee I., Lehner B., Crombie C., Wong W., Fraser A.G., Marcotte E.M. A single gene network accurately predicts phenotypic effects of gene perturbation in Caenorhabditis elegans. Nat Genet. 2008;40(2):181–188. doi: 10.1038/ng.2007.70. [DOI] [PubMed] [Google Scholar]
- 45.Qian W., Yang J.R., Pearson N.M., Maclean C., Zhang J. Balanced codon usage optimizes eukaryotic translational efficiency. PLoS Genet. 2012;8(3) doi: 10.1371/journal.pgen.1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koonin E.V. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat Rev Microbiol. 2003;1(2):127–136. doi: 10.1038/nrmicro751. [DOI] [PubMed] [Google Scholar]
- 47.Koonin E.V., Fedorova N.D., Jackson J.D., Jacobs A.R., Krylov D.M., Makarova K.S. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004;5(2):R7. doi: 10.1186/gb-2004-5-2-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trachana K., Larsson T.A., Powell S., Chen W.H., Doerks T., Muller J. Orthology prediction methods: a quality assessment using curated protein families. Bioessays. 2011;33(10):769–780. doi: 10.1002/bies.201100062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doyle M.A., Gasser R.B., Woodcroft B.J., Hall R.S., Ralph S.A. Drug target prediction and prioritization: using orthology to predict essentiality in parasite genomes. BMC Genomics. 2010;11:222. doi: 10.1186/1471-2164-11-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verster A.J., Ramani A.K., McKay S.J., Fraser A.G. Comparative RNAi screens in C. elegans and C. briggsae reveal the impact of developmental system drift on gene function. PLoS Genet. 2014;10(2) doi: 10.1371/journal.pgen.1004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rancati G., Moffat J., Typas A., Pavelka N. Emerging and evolving concepts in gene essentiality. Nat Rev Genet. 2018;19(1):34–49. doi: 10.1038/nrg.2017.74. [DOI] [PubMed] [Google Scholar]
- 52.Evers B., Jastrzebski K., Heijmans J.P., Grernrum W., Beijersbergen R.L., Bernards R. CRISPR knockout screening outperforms shRNA and CRISPRi in identifying essential genes. Nat Biotechnol. 2016;34(6):631–633. doi: 10.1038/nbt.3536. [DOI] [PubMed] [Google Scholar]
- 53.Morgens D.W., Deans R.M., Li A., Bassik M.C. Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat Biotechnol. 2016;34(6):634–636. doi: 10.1038/nbt.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hagen J., Lee E.F., Fairlie W.D., Kalinna B.H. Functional genomics approaches in parasitic helminths. Parasite Immunol. 2012;34(2–3):163–182. doi: 10.1111/j.1365-3024.2011.01306.x. [DOI] [PubMed] [Google Scholar]
- 55.Rinaldi G., Morales M.E., Cancela M., Castillo E., Brindley P.J., Tort J.F. Development of functional genomic tools in trematodes: RNA interference and luciferase reporter gene activity in Fasciola hepatica. PLoS Negl Trop Dis. 2008;2(7):e260. doi: 10.1371/journal.pntd.0000260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bartha I., di Iulio J., Venter J.C., Telenti A. Human gene essentiality. Nat Rev Genet. 2018;19(1):51–62. doi: 10.1038/nrg.2017.75. [DOI] [PubMed] [Google Scholar]
- 57.Liu G., Yong M.Y., Yurieva M., Srinivasan K.G., Liu J., Lim J.S. Gene essentiality is a quantitative property linked to cellular evolvability. Cell. 2015;163(6):1388–1399. doi: 10.1016/j.cell.2015.10.069. [DOI] [PubMed] [Google Scholar]
- 58.Koonin E.V. How many genes can make a cell: the minimal-gene-set concept. Annu Rev Genomics Hum Genet. 1998;1:99–116. doi: 10.1146/annurev.genom.1.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Juhas M., Eberl L., Glass J.I. Essence of life: essential genes of minimal genomes. Trends Cell Biol. 2011;21(10):562–568. doi: 10.1016/j.tcb.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 60.Glass J.I., Merryman C., Wise K.S., Hutchison C.A., 3rd, Smith H.O. Minimal cells - real and imagined. Cold Spring Harb Perspect Biol. 2017;9(12) doi: 10.1101/cshperspect.a023861. [pii: a023861] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eraslan G., Avsec Ž., Gagneur J., Theis F.J. Deep learning: new computational modelling techniques for genomics. Nat Rev Genet. 2019;20(7):389–403. doi: 10.1038/s41576-019-0122-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures and additional information
Supplementary Table S1
Session info