Abstract
Background
Cutaneous melanoma is the most aggressive form of skin cancer. It accounts for approximately 5% of all cutaneous malignancies and is currently responsible for the majority of skin cancer-related deaths. However, the exact mechanisms responsible for the occurrence of melanoma, in particular the invasive growth in normal skin or muscle tissue, remain unknown.
Materials and methods
miR-3662, a microRNA is a potential tumor suppressor targeting zinc finger E-box binding homeobox 1 (ZEB1), which functions as a key regulator of the epithelial-mesenchymal transition (EMT) process. This microRNA was identified using an online database (miRDB) and expression was confirmed by Western blot analysis. Quantitative polymerase chain reaction (qPCR) was used to examine whether miR-3662 inhibits the EMT process in the aggressive melanoma cell line, A375, through the modification of the expression of invasion-related genes in A375 cells. The effects of miR-3662 on the in vivo growth of A375 cells were examined in a nude mouse model.
Results
Using virtual screening of the miRDB database, miR-3662 was shown to target the 3ʹ untranslated region (UTR) of the ZEB1 gene. Expression of miR-3662 via a lentivirus vector significantly decreased protein levels of ZEB1 and inhibited the growth of A375 cells in vitro and in vivo. The reduction in ZEB1 expression induced by miR-3662 resulted in EMT inhibition in A375 cells and decreased the relative expression of metastasis genes.
Conclusion
Down-regulation of ZEB1’s expression via miR-3662 lentivirus vectors significantly decreased the in vitro and in vivo growth of the highly aggressive melanoma cell line A375.
Keywords: melanoma; microRNA; proliferation, invasive growth; zinc finger E-box binding homeobox 1
Introduction
Cutaneous melanoma is the most aggressive form of skin cancer, resulting in over fifty thousand deaths each year worldwide.1,2 Early stage diagnosis of cutaneous melanoma and effective primary care could significantly improve the incidence and mortality rates for this cancer.3,4 Cutaneous melanoma originates in melanocytes that colonize the skin.5,6 When exposed to risk factors, such as ultraviolet radiation (UV), mutations can accumulate within the cell genome of melanocytes, leading to their transformation to cutaneous melanoma.7,8 Researchers to date have focused largely on the alteration of tumor suppressors including PTEN, CDKN2A or TP53, or tumor promoters such as TERT, Ras, or Raf, in melanocytes or cutaneous melanoma cells.9,10 Few treatment options are available following the metastasis of cutaneous melanoma to a second site and consequently this stage of the disease then becomes life-threatening.11,12 Thus, it is important that we develop an increased understanding of the underlying mechanisms governing the invasive growth of cutaneous melanoma cells and their metastasis, to enable the development of more effective therapeutic strategies.
The human proto-oncogene zinc finger E-box binding homeobox 1 (ZEB1) can function as a transcription factor and plays additional important roles in the regulation of normal physiological functions, including cell fate decisions, conferment of cell stemness properties, and cell plasticity.13,14 ZEB1 is a key regulator of the epithelial-mesenchymal transition (EMT) process, which is responsible for the partial or complete transition of cancer cells with epithelial features to a mesenchymal state.15,16 High expression levels of ZEB1 have been identified in a variety of human cancers, such as lung cancer, hepatocellular carcinoma, and breast cancer, and research supports a role for ZEB1 in mediating the migration and invasion of cancer cells, via the induction of the EMT process.17,18 As such, ZEB1 is a promising potential therapeutic target, the inhibition of which may attenuate the proliferation or metastasis of cancer cells. Expression of ZEB1 has been identified in melanoma, though its function in the regulation of these cells remains unclear.19–21
A microRNA (miRNA) is a small noncoding RNA molecule that can mediate the silencing of targeted gene expression, by binding preferential sequences in the 3ʹ-untranslated regions (UTRs) of target mRNAs. miRNAs exhibit regulatory functions in several important cellular processes, such as differentiation and development. The identification of miRNAs targeted to human cancer-related genes and their transfection may offer a promising approach for the treatment of such tumors. This work aimed to identify a miRNA targeting ZEB1, miR-3662, and further examine the effects of this miRNA on the proliferation and invasive growth of A375, a highly aggressive melanoma cell line used to mimic the spread of cutaneous melanoma.
Materials and methods
Clinical specimens
The methods and collection of clinical specimens in this study were approved by the review board in accordance with ethical norms (Ethics Committee) at the Peking Union Medical College Hospital with written informed consent of all patients. All clinical investigations were conducted according to the principles expressed in the Declaration of Helsinki. This prospective study consisted of 80 paired tumor/non-tumor melanoma specimens between March 2014 and June 2017. Tumor pathology was confirmed and tumors staged22 as Clark I (8 cases); Clark II (12cases); Clark III (17 cases); Clark IV (19 cases); or Clark V (24 cases).
Cell line, plasmids and reagents
Expression vectors with full length ZEB1 genes or ZEB1 gene cDNA sequences with mutated miR-3662 binding sites were purchased from Vigene Corporation (Jinan, China). Lentivirus particles expressing miR-3662 were also obtained (Vigene Corporation). miR-3662 examination kits (Cat. No. 478844_mir-A25576) and inhibitors (Cat. No. 4464066-AM17000-MH19546) were purchased from Thermo Fisher (Waltham, MA, USA). Cell-based experiments and the use of cell lines were approved by the Ethics Committee of Beijing Union Hospital. Both a highly aggressive melanoma cell line, A375, and a lowly aggressive melanoma cell line, OCM-1A, were obtained from the National Infrastructure of Cell Line Resource, (Chinese Academy of Medical Sciences, Beijing, China). The usage of these cell lines was approved by the Ethics Committee of the Chinese Academy of Medical Sciences & Peking Union Medical College. The cells were maintained in Dulbecco’s modified eagle medium (DMEM; Thermo Fisher, Waltham, MA, USA) with 10% fetal bovine serum (FBS; Thermo Fisher, Waltham, MA, USA) at 37 °C with 5% CO2. Regions of the ZEB1 mRNA 3ʹUTR (1561–1740, 1681–1860 or 1861–2040) containing miR-3662 targeting sequence (TCATTTT) were cloned into pGL4.26 plasmids to form Luciferase reporters (Figure S1).
Cell proliferation and metastasis
A375 cells were initially transfected with miR-3662 or a control miRNA (expression vector with a non-targeting random sequence) and then subject to either cell proliferation or metastasis analysis. To analyze cell proliferation, the transfected cells were analyzed via 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. For the analysis of cell metastasis, the transfected cells were analyzed by transwell experiments to measure the in vitro invasion and migration of cells.23,24
Quantitative polymerase chain reaction (qPCR)
A375 cells transfected with miR-3662, miR-3662+ ZEB1Mut (an expression vector containing a full length sequence of the ZEB1 gene with mutated miR-3662 targets), miR-3662+ inhibitor (inhibitor of miR-3662), or control miRNA, were harvested 24 h after transfection and total RNA was extracted. qPCR experiments were performed using a kit following the manufacturer’s instructions and methods described in references.23,24 Primers used in qPCR experiments are listed in Table 1.
Table 1.
Premiers used in qPCR experiemnts
| Genes | Primer sequence (5ʹ-3ʹ) |
|---|---|
| E-cadherin | Forward: 5′-AAGGCACGCCTGTCGAAGCA-3′ |
| Reverse: 5′-ACGTTGTCCCGGGTGTCATCCT-3′ | |
| N-cadherin | Forward: 5′-TGCGCGTGAAGGTTTGCCAGT-3′ |
| Reverse: 5′-TGGCGTTCTTTATCCCGGCGT-3′ | |
| Vimentin | Forward: 5′-ACCGCACACAGCAAGGCGAT-3′ |
| Reverse: 5′-CGATTGAGGGCTCCTAGCGGTT-3′ | |
| TIMP-1 | Forward: 5ʹ-ACTTCCACAGGTCCCACAAC-3′ |
| Reverse: 5ʹ-CACTGTGCATTCCTCACAGC-3′ | |
| MMP3 | Forward: 5′-TGCTTTGTCCTTTGATGCTG-3′ |
| Reverse: 5′-GGAAAGAGATGGCCAAAATGA-3′ | |
| MMP9 | Forward: 5ʹ-TGGGCTACGTGACCTATGAC-3′ |
| Reverse: 5ʹ-CAAAGGTGAGAAGAGAGGGC-3′ | |
| ZEB1 | Forward: 5ʹ-GATGACCTGCCAACAGACCA-3′ |
| Reverse: 5ʹ-CCCCAGGATTTCTTGCCCTT-3′ | |
| GAPDH | Forward: 5′-CTCTCTGCTCCTCCTGTTCG-3′ |
| Reverse: 5′-ACGACCAAATCCGTTGACTC-3′ |
Luciferase
A375 cells transfected with relevant plasmids were harvested for luciferase activation using a luciferase kit (Promega, Madison, WI, USA) following the protocol provided by manufacturer (Promega, Madison, WI, USA). Transfection was performed using Lipofection2000 transfection reagent (Thermo Fisher, Waltham, MA, USA). A total of 20μg of each plasmid was transfected into each well of a 24-well plate containing cells by using the standard transfection method. Luciferase activation was calculated as fold-improvement according to (treatment group luciferase activation)/(control group luciferase activation).
Antibodies and Western blot
Antibodies to ZEB1 (Cat. No. ab181451), E-cadherin (Cat. No. ab1416), N-Cadherin (Cat. No. ab98952), Vimentin (Cat. No. ab20346), MMP3 (Cat. No. ab137659), TIMP1 (Cat. No. ab61224), MMP9 (Cat. No. ab73734) and Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Cat. No. ab8245) were obtained from Abcam (Cambridge, UK).
A375 cells transfected with miR-3662, miR-3662+ ZEB1Mut or miR-3662+ inhibitor or control miRNA were harvested 48 h following transfection and total protein samples extracted for Western blot experiments. OCM-1A cells transfected with empty vector or ZEB1 vectors were also harvested for Western blot assays. Western blots were performed following a standard Western blotting protocol. Briefly, protein samples were separated using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Following electrophoresis, proteins were transferred onto polyvinylidene fluoride (PVDF) membranes and blocked using 5% bovine serum albumin (BSA) diluted in tris-buffered saline with Tween 20 (TBS-T). Membranes were subsequently incubated with the relevant primary and secondary antibodies prior to detection by chemiluminescence.
Subcutaneous tumor model of melanoma cells
Animal experiments were approved by the Institutional Animal Care and Use Committee of the Chinese Academy of Medical Sciences & Peking Union Medical College. Animal studies were conducted in accordance with the UK Animals (Scientific Procedures) Act of 1986 and the associated guidelines. Nude mice (BALB/c nude) were purchased from Si-Bei-Fu Biotechnology (Beijing, China). For the subcutaneous tumor experiments,25,26 A375 cells that had been stably infected with miR-3662, miR-3662+ ZEB1Mut or control miRNA or OCM-1A cells stably transfected with miR-3662 or control miRNA were injected into four- to six-week-old nude mice. Each nude mouse was seeded with 1×106 cells. After six weeks of growth subcutaneous tumors were harvested and the volume and weight of the tumors were measured.
In vivo invasive growth of melanoma cells in muscle tissue
A375 cells stably infected with miR-3662, miR-3662+ ZEB1Mut, or control miRNA or OCM-1A cells stably transfected with ZEB1 vector or empty vector were mixed with biological-medical gel (Cai-Hong-Yi-Xue-She-Bei, Kunming, China) and injected into the muscle tissue in the legs of nude mice. Each mouse was seeded with approximately 5×105 cells. After four weeks’ growth, the mice were harvested and the muscle tissue containing A375 cells was taken for analysis by hematoxylin-eosin (H&E) staining.27 Images of the H&E staining were quantitatively analyzed via Image J software according to previously detailed methods.28,29 Results were shown as relative invasive growth of A375 cells: (treatment group’s total area of tumor tissues)/(control miRNA group’s total area of tumor tissues). The inhibition rate of each group was calculated as follows: [(control group’s relative invasive growth of A375 cells) - (treatment group’s relative invasive growth of A375 cells)]/(control group’s relative invasive growth of A375 cells) x 100%.
Statistical analysis
Western blot bands were analyzed using Image J software (version 1.51j8; National Institutes of Health, Bethesda, MD, USA). The relative expression of the targeted proteins was calculated as follows: (the indicated group intensity of the targeted protein bands)/(the indicated group intensity of the loading control bands). Statistical analysis was performed using the Bonferroni correction with or without two-way analysis of variance, with SPSS software (version 16.0; IBM, Armonk, New York, USA). P-values <0.05 were considered statistically significant.
Results
miR-3662 targets the 3ʹ UTR of the ZEB1 mRNA
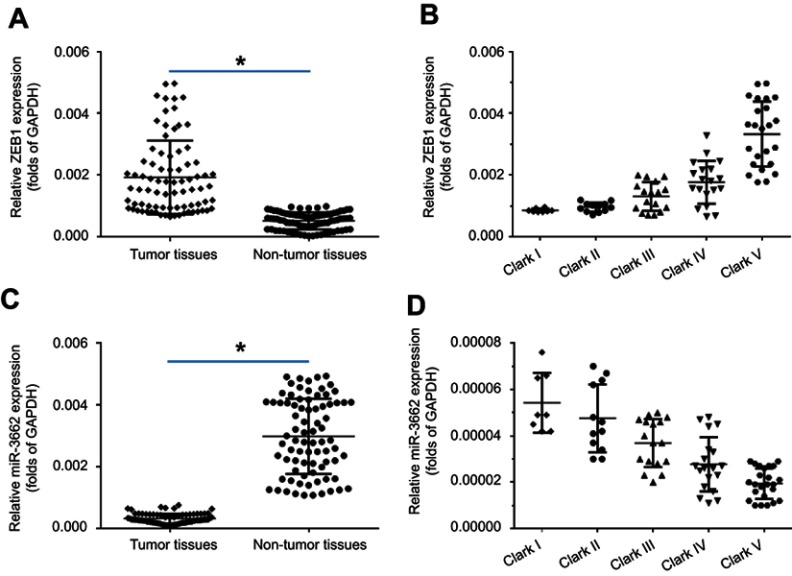
To investigate the potential role of ZEB1 in regulating melanoma, the expression of ZEB1 mRNA in clinical specimens was examined via qPCR. Results from 80 paired tumor and non-tumor specimens indicated that ZEB1 was expressed at higher levels in melanoma compared to non-tumor specimens (Figure 1A). Moreover, a higher level of ZEB1 expression was detected in the more aggressive stages of melanoma (eg Clark V or Clark IV) compared to the less aggressive stages (Clark I or Clark II) (Figure 1B).
Figure 1.
The expression of ZEB1 mRNA in melanoma specimens. A total of 80 paired melanoma and non-tumor clinical specimens were harvested for qPCR experiments. (A) ZEB1 mRNA levels in 80 paired melanoma and non-tumor clinical specimens. (B) ZEB1 mRNA levels in samples at different melanoma stages (Clark I-V). (C) miR-3662 levels in 80 paired melanoma and non-tumor clinical specimens. (D) miR-3662 levels in samples at different melanoma stages (Clark I-V). *P<0.05.
A miRNA potentially targeted to ZEB1 mRNA’s 3ʹ UTR, was identified by screening the online database, miRDB (http://www.mirdb.org/). Analysis of the expression of miR-3662 in both tumor and non-tumor tissues revealed a higher level of miR-3662 expression in non-tumor specimens than that in tumor specimens (Figure 1C). Additionally, a higher level of miR-3662 expression was detected in the less aggressive stages of melanoma (Clark I or Clark II) compared to the more aggressive stages of melanoma (such as Clark IV or Clark V) (Figure 1D).
The expression of ZEB1 and its associated miRNA indicate the potential involvement of this protein in regulating the invasive growth of melanoma tumors. To understand the role of ZEB1 in proliferation or metastasis of melanoma, MTT and transwell experiments were performed to reveal that the knockdown of ZEB1 inhibited the proliferation, in vitro invasion and migration of A375 cells (Figures S2–S4), demonstrating a role for ZEB1 in the function of melanoma cells.
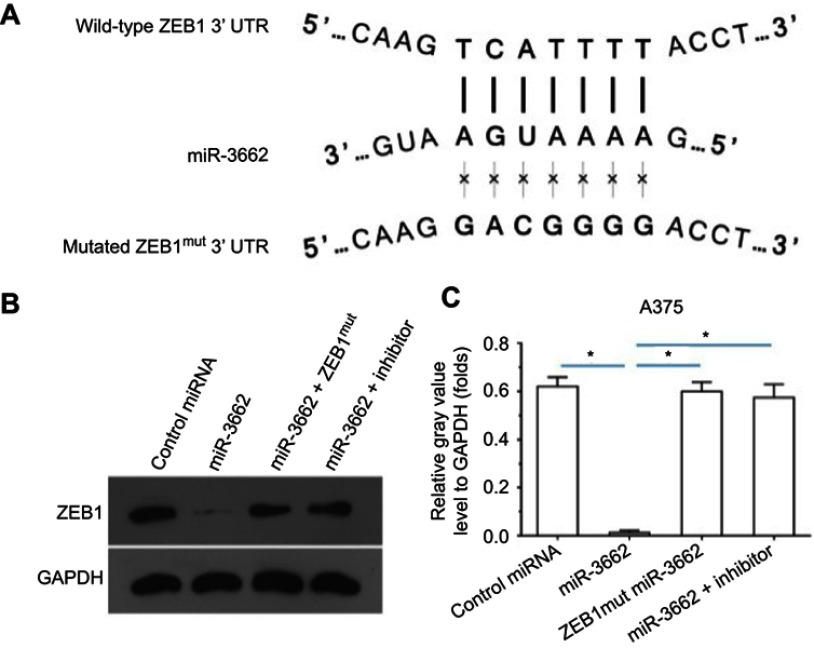
miR-3662 target sequences within the ZEB1 transcript were identified and mutated to create control miRNA, with which to transfect cells (Figures 2 and 3A). The transfection of miR-3662, but not control miRNA, resulted in a significant decrease in the protein levels of ZEB1 in A375 cells (Figure 3B and C). Moreover, transfection of miR-3662’s inhibitor or mutated ZEB1 was sufficient to restore the expression of ZEB1 within these cells. The interaction between ZEB1’s 3ʹUTR sequence and miR-3662 was further confirmed by luciferase experiments (Figure S5). Western blot analysis revealed that the decrease in ZEB1 levels resulting from miRNA-3662 expression were able to be restored with the over-expression of the mutated ZEB1 (Figure S6). Taken together, these data suggest that miR-3662 acts to repress the expression of ZEB1 protein by directly targeting the ZEB1 mRNA’s 3ʹ UTR in A375 cells.
Figure 2.
miR-3662 binding targets in ZEB1 mRNA’s 3ʹ UTR. The three binding sites of miR-3662 ZEB1 mRNA’s 3ʹ UTR.
Figure 3.
miR-3662 targets the 3ʹ UTR of ZEB1 mRNA. (A) Sequences of potential binding sites of miR-3662 (bold) and the mutated binding site on 3ʹ UTR of ZEB1 mRNA (italicized). (B) Western blot analysis of ZEB1 and GAPDH control levels in A375 cells transfected with control miRNA, miR-3662, miR-3662+ ZEB1Mut or miR-3662+ miR-miR-3662 inhibitor. (C) Quantification of ZEB1 in A375 cells transfected with control miRNA, miR-3662, miR-3662+ ZEB1Mut or miR-3662+ miR-miR-3662 inhibitor. *P<0.05.
mRNA-3662 inhibits the EMT indicators of melanoma cells and expression of invasion related factors
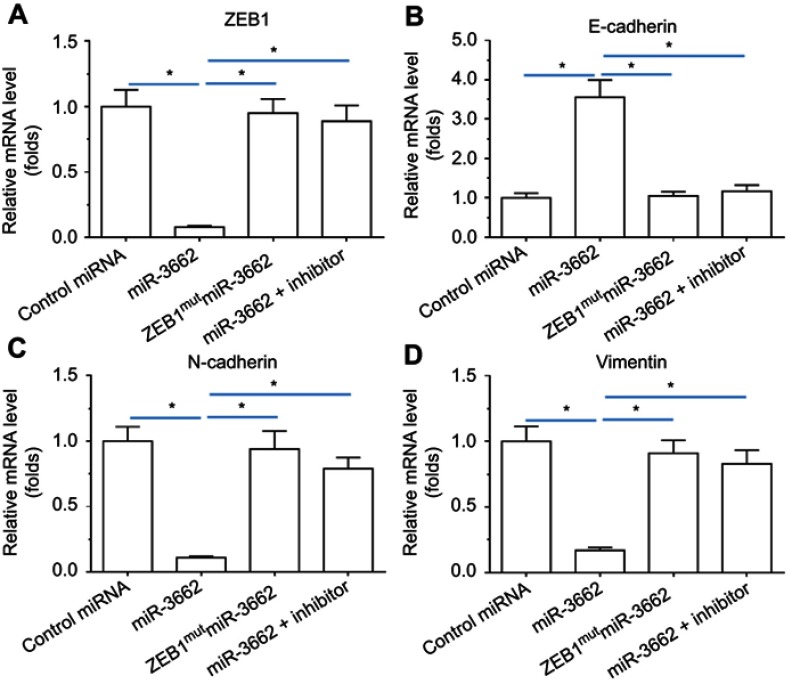
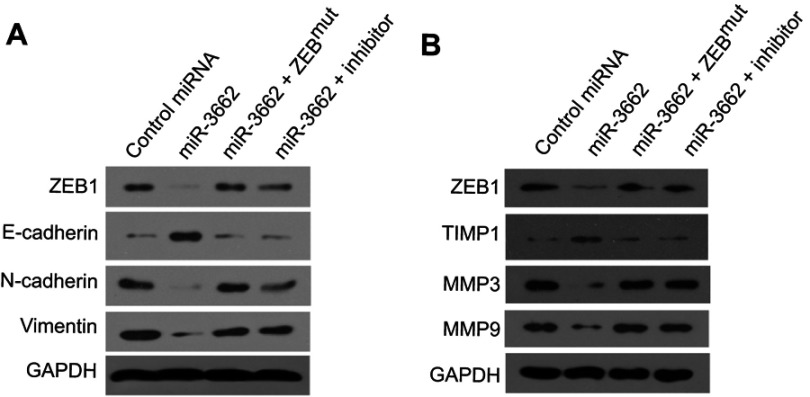
To investigate whether miR-3662 impacts on the invasive growth of A375 cells, the expression of EMT and invasion related factors was examined by qPCR and Western blot assays. Transfection of miR-3662 enhanced the expression of E-cadherin, an epithelial marker, and decreased the expression of N-cadherin and vimentin, two mesenchymal markers (Figure 4). Conversely, transfection of an inhibitor of miR-3662 or ZEB1Mut decreased the observed inhibitory effect of miR-3662 on the EMT process in A375 cells (Figure 4). These results suggest that miR-3662 acts to inhibit the EMT process of A375 cells by targeting ZEB1.
Figure 4.
miR-3662 inhibits the EMT process of A375 cells. mRNA levels of (A) ZEB1, (B) E-cadherin, (C) N-cadherin, and (D) vimentin shown as mean ± SD, in A375 cells transfected with control miRNA, miR-3662, miR-3662+ zeb1Mut, or miR-3662+ miR-miR-3662 inhibitor. *P<0.05.
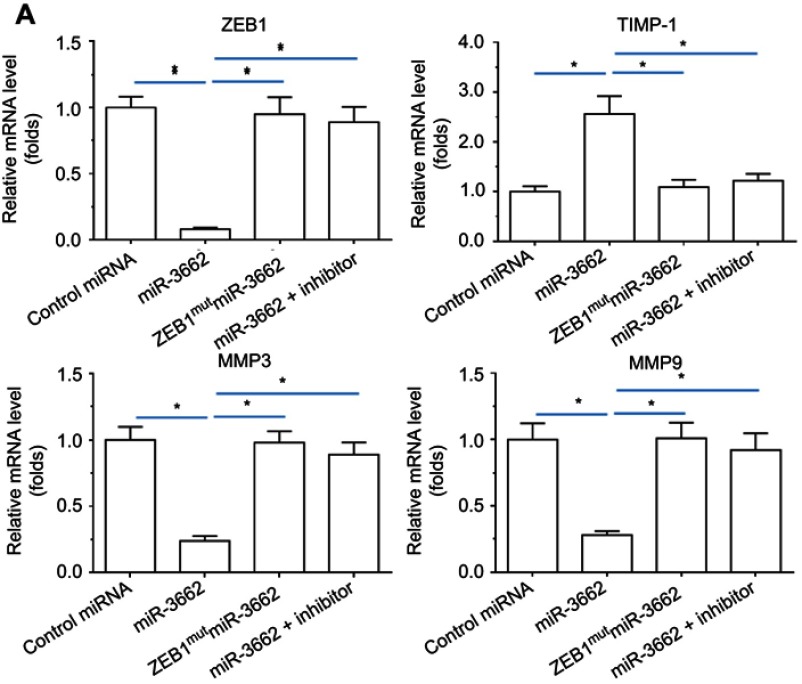
qPCR analysis of the effect of miR-3662 on the expression of invasive growth-related genes in A375 cells, demonstrated that miR-3662 expression resulted in the inhibition of mmp3 and mmp9, two matrix metalloproteinases that mediate the degradation of extracellular matrix (ECM) during the invasive growth of cancer cells (Figure 5). Furthermore, miR-3662 was also observed to increase the expression of timp1 (tissue inhibitor of metalloproteinase 1), an inhibitor of matrix metalloproteinases in tissues (Figure 5). Transfection of the miR-3662 inhibitor or ZEB1Mut inhibited the effects of miR-3662 on these invasive growth-related genes. Similar results were obtained from Western blot analysis (Figure 6), suggesting that miR-3662 functions to inhibit the expression of invasion related proteins in A375 cells by targeting ZEB1.
Figure 5.
miR-3662 inhibits invasive growth-related genes’ mRNA expression in A375 cells. mRNA levels of (A) ZEB1, (B) TIMP-1, (C) MMP3, or (D) MMP9 shown as mean ± SD, in A375 cells infected with control miRNA, miR-3662, miR-3662+ ZEB1Mut, or miR-3662+ miR-miR-3662 inhibitor. *P<0.05.
Figure 6.
miR-3662 inhibits EMT-related or invasive growth-related gene protein expression in A375 cells. (A) ZEB1, E-cadherin, N-Cadherin and Vimentin protein levels expressed in A375 cells transfected with control miRNA, miR-3662, miR-3662+ ZEB1Mut, or miR-3662+ miR-miR-3662 inhibitor determined by Western blot analysis. (B) ZEB1, TIMP-1, MMP3 and MMP9 protein levels expressed in A375 cells transfected with control miRNA, miR-3662, miR-3662+ ZEB1Mut, or miR-3662+ miR-miR-3662 inhibitor determined by Western blot analysis. *P<0.05.
To further examine the effects of ZEB1 on melanoma cells, ZEB1 was overexpressed in OCM-1A cells, a lowly aggressive melanoma cell line, and the expression of EMT and metastasis-related factors subsequently examined (Figure S7). Overexpression of ZEB1 in OCM-1A cells enhanced the EMT process in these cells and increased the expression of metastasis-related factors (Figure S7), confirming the effects of miR-3662 on ZEB1 function in melanoma cells.
miR-3662 inhibits the subcutaneous growth of melanoma cells in nude mice
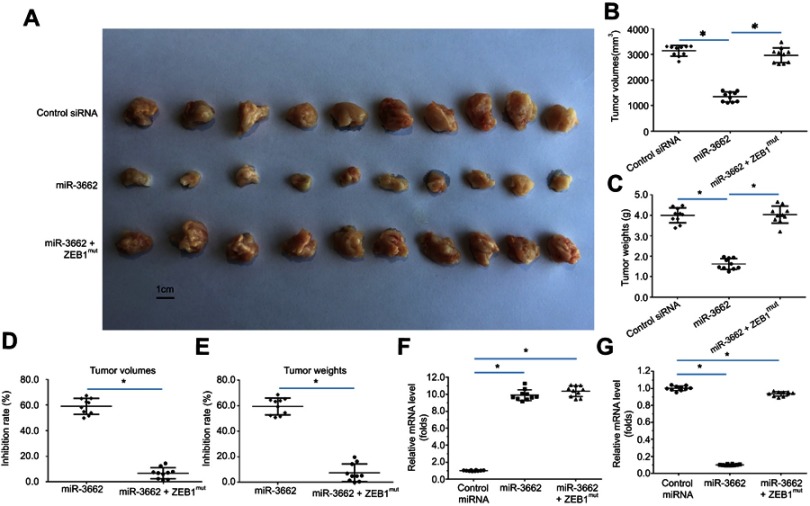
The in vivo function of miR-3662 was investigated using a nude mouse model injected subcutaneously with A375 cells. In these animals the transfection of miR-3662 gave rise to a decrease in the subcutaneous growth of A375 cells, supporting the role of miR-3662 as an inhibitor of tumor progression. Indeed, transfection of ZEB1,Mut which is unresponsive to the miRNA, almost blocked the effects of miR-3662 on A375 subcutaneous cell growth (Figure 7). Analysis of subcutaneous tumors from the murine models demonstrated reductions in tumor volumes (Figure 7B) and tumor weights (Figure 7C) with the expression of miR-3662, supporting its role in decreasing the tumorigenic potential of ZEB1. Additionally, the growth inhibition rates calculated of tumor volume (Figure 7D) or tumor weight (Figure 7E), show a reduction in growth with ZEB1 inhibition by miR-3662. Expression of miR-3662 (Figure 7F) and ZEB1 (Figure 7G) in subcutaneous tumor tissues were determined by qPCR. The injection of a lowly aggressive tumor cell line, OCM-1A into the nude mouse model, was assessed to determine if the observed results were cell line dependent. Subcutaneous tumors were observed in OCM-1A injected mice and these tumors showed increased size and weight in cells over-expressing ZEB1 (Figure S8).
Figure 7.
miR-3662 inhibits the subcutaneous growth of A375 cells. A375 cells transfected with control miRNA, miR-3662, or miR-3662+ ZEB1Mut were seeded into nude mice to form subcutaneous tumors. (A) Representative image of subcutaneous tumors, (B) volume of subcutaneous tumors and (C) weight of subcutaneous tumors. (D, E) Inhibition rates were calculated by tumor volume (D) or tumor weight (E) RNA levels of miR-3662 and ZEB1 were shown in Figure 7G. The RNA levels of miR-3662 (F) and ZEB1 (G) were shown as scatter diagrams in Figure 7F and G. *P<0.05.
These data demonstrate that miR-3662 acts to inhibit the subcutaneous growth tumor cells in vivo by targeting ZEB1.
miR-3662 inhibits in vivo invasive growth of melanoma cells in the muscle tissue of nude mice
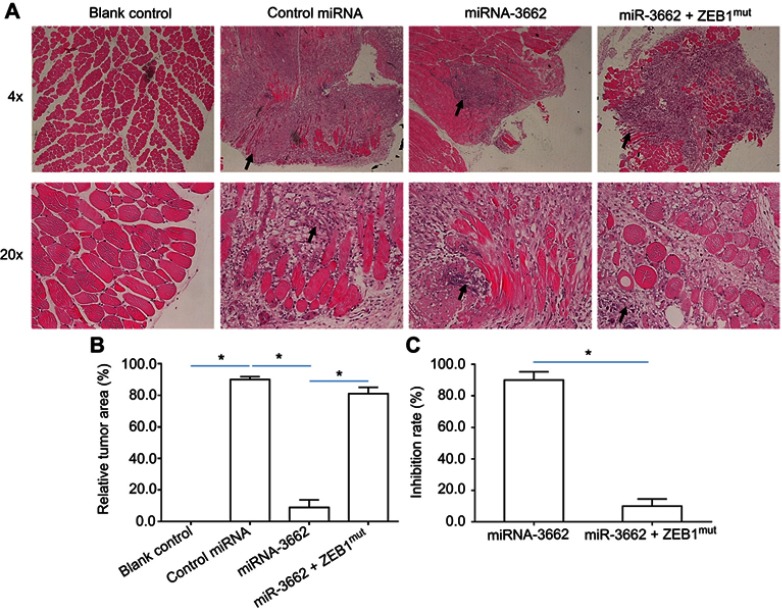
Tumorigenic cells were injected into the muscle tissue of nude mice to assess the effects of miR-3662 on the in vivo invasive growth of these cells. Injection of A375 cells into the muscle tissue resulted in the formation of nodules and muscle degradation (Figure 8). The transfection of miR-3662 in these cells, inhibited their invasive capacity in murine muscle tissue (Figure 8); whereas transfection of ZEB1Mut in these cells almost blocked the effect of miR-3662 on A375 cells’ invasive growth (Figure 8). In contrast, the injection of OCM-1A cells resulted in an expected reduction in invasive capacity in the muscle tissue of the animals. However, over-expression of ZEB1 in these cells resulted in a promotion of the growth of invasive tumors (Figure S9), confirming the role of ZEB1 in promoting tumor invasion and the inhibition of this capacity by miR-3662.
Figure 8.
miR-3662 inhibits the invasive growth of A375 in the muscle tissue of nude mice. A375 cells transfected with control miRNA, miR-3662, or miR-3662+ ZEB1Mut were seeded into the muscle tissue of nude mice. (A) Representative image of H&E stained muscle tissue with tumor-tissue formed by A375 cells, (B) relative invasive growth of A375 cells, (C) inhibition rates calculated from the relative invasive growth of A375 cells. The black arrows indicate the tumor lesions in muscle tissues formed by A375 cells. *P<0.05.
Discussion
Melanoma arises through the malignant transformation of melanocytes into metastatic melanoma, a complex process considered a neural crest neoplasia.30 Although screening for melanoma by total body skin examination could benefit people at risk for melanoma, current regular screening approaches do not effectively diagnose thin melanoma and thus promote the development of invasive melanoma, negatively impacting upon patient survival.31 The invasive growth of advanced stage melanoma (Clark III to V) or recurrence of melanoma following surgery is a threat to patients and the subsequent therapeutic options are limited.32,33 Three BRAF-MEK inhibitor combinations are presently approved for BRAFv600-mutated melanoma treatment: vemurafenib plus cobimetinib, dabrafenib plus trametinib, and encorafenib plus binimetinib.34,35 Oral administration of these small-molecule kinase inhibitors are considered first-line therapy for advanced and metastatic melanoma treatment. However, response rates of patients to small-molecule kinase inhibitors were reported to be as low as 16% of patients.1,36 Moreover, options for targeted therapies are scarce for patients suffering from BRAF wild-type melanoma.1 Therefore, novel therapeutic approaches are urgently needed for these patients.
In the present work, the expression of ZEB1, which is confirmed as a key regulator of human cancer cells’ EMT process, was found to be much higher in advanced melanoma (Clark IV or Clark V) than in the early stages (Clark I or Clark II) of melanoma. The identification of miR-3662 as potentially targeting ZEB1 through the 3ʹ-UTR of this transcript was determined via an online database (miRDB.org). miR-3662 is a novel tumor suppressor with only 5 reports. Among them, Chen et al (2018) suggested that miR-3662 suppresses hepatocellular carcinoma growth through inhibition of the HIF-1α-mediated Warburg effect.37 Therefore, targeting the ZEB1 gene by miR-3662 is a promising approach to attenuate the invasive growth of melanoma cells. Here we confirmed the expression of miR-3662 in tumorigenic cell lines by qPCR and Western blot assays, indicating a potential role for this miRNA in these cells. Three 7mer targeted sites of miR-3662 were identified in the ZEB1 mRNA 3ʹ UTR (Figure 2). Consequently, transfection of miR-3662 was assumed to be an efficient strategy to decrease ZEB1 expression. Indeed, transfection of miR-3662 inhibited the EMT process of A375 cells, decreased invasion-related gene expression, and inhibited the invasive growth of A375 cells in nude mice by decreasing the expression of ZEB1.
The BRAF mutation is widely used as an indicator or risk-related factor for melanoma’s malignant progress. The results described in this study preliminarily reveal ZEB1 may be associated with the progress or invasion of melanoma and may serve as a potential biomarker, especially for BRAF wild-type melanoma.
Invasion is an important factor that influences the prognosis of patients with malignant tumors, especially in the metastatic or invasive stage of cutaneous melanoma.38,39 The EMT process is a physical process that mediates the loss of epithelial characteristics from cancer cells, such as polarity, and the development of mesenchymal charactersitics.16,40 During the EMT process, cancer cells gain migratory properties. The ECM can function as an important barrier to confine cancer cells within their primary origin site.41,42 However, during the EMT process ECM degradation is mediated by matrix metalloproteinases (MMPs), a family of zinc-dependent enzymes that proteolytically degrade various ECM components.43,44 In the present work, transfection of miR-3662 inhibited the EMT process and the expression of matrix metalloproteinases, MMP3 or MMP9. These results reveal a potential mechanism of action for miR-3662 in inhibiting A375 cell invasion.
A375 cells are a highly aggressive melanoma cell line.45 Richard et al (2013), Pal et al (2016) and Caramel et al (2013) demonstrated that a high level of ZEB1 expression in epithelial cells is related to BRAF mutation in melanoma.46–48 Our results support this theory, indicating the invasive growth capacity of ZEB1-expressing cells in muscle tissues. The transwell invasion/migration assay is widely used to demonstrate the invasion of cancer cells by mimicking the degradation of ECM, however, it can only offer information on in vitro performance. This study, revealed both the in vitro and in vivo invasion of A375 cells by injecting cells into the muscle tissue of nude mice. Following the initial location of the A375 cells within the muscle tissue in these animals, tumor nodules were observed to form through degradation of the normal muscle tissue. The inhibitory activation of miR-3662 has been observed on A375 cells in muscle, where it is related to the decreased EMT process and reduced MMP activity. The tumor model utilized in this study mimics the invasion of melanoma in skin/muscle and highlights the potential application of miR-3662 in attenuating melanoma cells invasive growth.
In mammalian cells, miRNA is an important non-coding RNA serving as an epigenetic mechanism of gene expression silencing by post-transcriptional regulation.49–52 In this work, our results demonstrated that miR-3662 inhibited the invasion of A375 cells by targeting ZEB1, and offers a potential therapeutic target and treatment via lentiviral delivery. The expression of miRNAs in human cancer cells via the preparation of lentivirus particles is a promising new approach that may achieve significant patient benefits through antitumor activation.
Conclusion
The present work demonstrated that ZEB1 is targeted by miR-3662. The stable overexpression of miR-3662 in A375, a highly aggressive melanoma cell line, through lentiviral transfection, decreased ZEB1’s expression and inhibited the invasion of A375 cells in the muscle tissue of nude mice.
Author contributions
All authors made substantial contributions to the design and conception; acquisition, analysis or interpretation of data. Authors took part in either drafting or revising the manuscript. At the same time, authors gave final approval of the version to be published; and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Schadendorf D, van Akkooi ACJ, Berking C, et al. Melanoma. Lancet. 2018;392:971–984. doi: 10.1016/S0140-6736(18)31559-9 [DOI] [PubMed] [Google Scholar]
- 2.Carvajal RD, Piperno-Neumann S, Kapiteijn E, et al. Selumetinib in combination with dacarbazine in patients with metastatic uveal melanoma: a phase III, Multicenter, Randomized Trial (SUMIT). J Clin Oncol. 2018;36:1232–1239. doi: 10.1200/JCO.2017.74.1090 [DOI] [PubMed] [Google Scholar]
- 3.Makino E, Gutmann V, Kosnopfel C, et al. Melanoma cells resistant towards MAPK inhibitors exhibit reduced TAp73 expression mediating enhanced sensitivity to platinum-based drugs. Cell Death Dis. 2018;9:930. doi: 10.1038/s41419-018-1111-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zingg D, Debbache J, Peña-Hernández R, et al. EZH2-mediated primary cilium deconstruction drives metastatic melanoma formation. Cancer Cell. 2018;34:69–84, e14. doi: 10.1016/j.ccell.2018.06.001 [DOI] [PubMed] [Google Scholar]
- 5.Kwiatkowska-Borowczyk E, Czerwińska P, Mackiewicz J, et al. Whole cell melanoma vaccine genetically modified to stem cells like phenotype generates specific immune responses to ALDH1A1 and long-term survival in advanced melanoma patients. Oncoimmunology. 2018;7:e1509821. doi: 10.1080/2162402X.2018.1490854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cerezo M, Guemiri R, Druillennec S, et al. Translational control of tumor immune escape via the eIF4F-STAT1-PD-L1 axis in melanoma. Nat Med. 2018. [Epub ahead of print]. doi: 10.1038/s41591-018-0217-1 [DOI] [PubMed] [Google Scholar]
- 7.Marchetti P, Trinh A, Khamari R, Kluza J. Melanoma metabolism contributes to the cellular responses to MAPK/ERK pathway inhibitors. Biochim Biophys Acta Gen Subj. 2018;1862:999–1005. doi: 10.1016/j.bbagen.2018.01.018 [DOI] [PubMed] [Google Scholar]
- 8.Kuske M, Westphal D, Wehner R, et al. Immunomodulatory effects of BRAF and MEK inhibitors: implications for melanoma therapy. Pharmacol Res. 2018;136:151–159. doi: 10.1016/j.phrs.2018.08.019 [DOI] [PubMed] [Google Scholar]
- 9.Redmer T. Deciphering mechanisms of brain metastasis in melanoma - the gist of the matter. Mol Cancer. 2018;17:106. doi: 10.1186/s12943-018-0854-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alioui A, Dufour J, Leoni V, et al. Retraction Note: liver X receptors constrain tumor development and metastasis dissemination in PTEN-deficient prostate cancer. Nat Commun. 2018;9:2660. doi: 10.1038/s41467-018-04653-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ibfelt EH, Steding-Jessen M, Dalton SO, Lundstrøm SL, Osler M, Hölmich LR. Influence of socioeconomic factors and region of residence on cancer stage of malignant melanoma: a Danish nationwide population-based study. Clin Epidemiol. 2018;10:799–807. doi: 10.2147/CLEP.S160357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ross KC, Chin KF, Kim D, Marion CD, Yen TJ, Bhattacharjee V. Methotrexate sensitizes drug-resistant metastatic melanoma cells to BRAF V600E inhibitors dabrafenib and encorafenib. Oncotarget. 2018;9:13324–13336. doi: 10.18632/oncotarget.24341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, Zhang Z, Zhang Q, et al. ZEB1 confers chemotherapeutic resistance to breast cancer by activating ATM. Cell Death Dis. 2018;9:57. doi: 10.1038/s41419-018-1111-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Passacantilli I, Panzeri V, Bielli P, et al. Alternative polyadenylation of ZEB1 promotes its translation during genotoxic stress in pancreatic cancer cells. Cell Death Dis. 2017;8:e3168. doi: 10.1038/cddis.2017.518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gulei D, Mehterov N, Ling H, Stanta G, Braicu C, Berindan-Neagoe I. The “good-cop bad-cop” TGF-beta role in breast cancer modulated by non-coding RNAs. Biochim Biophys Acta Gen Subj. 2017;1861:1661–1675. doi: 10.1016/j.bbagen.2017.04.007 [DOI] [PubMed] [Google Scholar]
- 16.Chen L, Mai W, Chen M, et al. Arenobufagin inhibits prostate cancer epithelial-mesenchymal transition and metastasis by down-regulating β-catenin. Pharmacol Res. 2017;123:130–142. doi: 10.1016/j.phrs.2017.07.009 [DOI] [PubMed] [Google Scholar]
- 17.Nagaraja SS, Nagarajan D. Radiation-induced pulmonary epithelial-mesenchymal transition: a review on targeting molecular pathways and mediators. Curr Drug Targets. 2018;19:1191–1204. doi: 10.2174/1389450119666180207092234 [DOI] [PubMed] [Google Scholar]
- 18.Caramel J, Ligier M, Puisieux A. Pleiotropic roles for ZEB1 in cancer. Cancer Res. 2018;78:30–35. doi: 10.1158/0008-5472.CAN-17-2476 [DOI] [PubMed] [Google Scholar]
- 19.Wu Q, Xiang S, Ma J, et al. Long non-coding RNA CASC15 regulates gastric cancer cell proliferation, migration and epithelial mesenchymal transition by targeting CDKN1A and ZEB1. Mol Oncol. 2018;12:799–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Viswanathan VS, Ryan MJ, Dhruv HD, et al. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature. 2017;547:453–457. doi: 10.1038/nature23007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y, Lu X, Montoya-Durango DE, et al. ZEB1 regulates multiple oncogenic components involved in uveal melanoma progression. Sci Rep. 2017;7:45. doi: 10.1038/s41598-017-00079-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matheson JA, Te Marvelde L, Mailer S, et al. Prospective evaluation of prognostic indicators for early recurrence of cutaneous melanoma. Melanoma Res. 2017;27:43–49. doi: 10.1097/CMR.0000000000000302 [DOI] [PubMed] [Google Scholar]
- 23.Ji Q, Xu X, Li L, et al. miR-216a inhibits osteosarcoma cell proliferation, invasion and metastasis by targeting CDK14. Cell Death Dis. 2017;8:e3103. doi: 10.1038/cddis.2017.518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang Y, Xu X, Wang T, et al. The EGFR/miR-338-3p/EYA2 axis controls breast tumor growth and lung metastasis. Cell Death Dis. 2017;8:e2928. doi: 10.1038/cddis.2017.518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.An L, Li DD, Chu HX, et al. Terfenadine combined with epirubicin impedes the chemo-resistant human non-small cell lung cancer both in vitro and in vivo through EMT and Notch reversal. Pharmacol Res. 2017;124:105–115. doi: 10.1016/j.phrs.2017.07.021 [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Li D, Jiang Q, et al. Novel ADAM-17 inhibitor ZLDI-8 enhances the in vitro and in vivo chemotherapeutic effects of sorafenib on hepatocellular carcinoma cells. Cell Death Dis. 2018;9:743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng F, Jiang Q, Cao S, et al. Pregnane X receptor mediates sorafenib resistance in advanced hepatocellular carcinoma. Biochim Biophys Acta Gen Subj. 2018;1862:1017–1030. doi: 10.1016/j.bbagen.2018.01.011 [DOI] [PubMed] [Google Scholar]
- 28.Shao Z, Li Y, Dai W, et al. ETS-1 induces sorafenib-resistance in hepatocellular carcinoma cells via regulating transcription factor activity of PXR. Pharmacol Res. 2018;135:188–200. doi: 10.1016/j.phrs.2018.08.003 [DOI] [PubMed] [Google Scholar]
- 29.Xie H, Tian S, Yu H, et al. A new apatinib microcrystal formulation enhances the effect of radiofrequency ablation treatment on hepatocellular carcinoma. Onco Targets Ther. 2018;11:3257–3265. doi: 10.2147/OTT.S165000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tagliabue E, Gandini S, Bellocco R, et al. MC1R variants as melanoma risk factors independent of at-risk phenotypic characteristics: a pooled analysis from the M-SKIP project. Cancer Manag Res. 2018;10:1143–1154. doi: 10.2147/CMAR.S155283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mar VJ, Scolyer RA, Long GV. Computer-assisted diagnosis for skin cancer: have we been outsmarted? Lancet. 2017;389:1962–1964. doi: 10.1016/S0140-6736(17)31285-0 [DOI] [PubMed] [Google Scholar]
- 32.Hammouda MB, Riahi-Chebbi I, Souid S, et al. Macrovipecetin, a C-type lectin from macrovipera lebetina venom, inhibits proliferation migration and invasion of SK-MEL-28 human melanoma cells and enhances their sensitivity to cisplatin. Biochim Biophys Acta Gen Subj. 2018;1862:600–614. doi: 10.1016/j.bbagen.2017.11.019 [DOI] [PubMed] [Google Scholar]
- 33.Smalley KSM, Forsyth PA. The blood brain barrier and BRAF inhibitors: implications for patients with melanoma brain metastases. Pharmacol Res. 2018;135:265–267. doi: 10.1016/j.phrs.2017.11.013 [DOI] [PubMed] [Google Scholar]
- 34.Hauschild A, Larkin J, Ribas A, et al. Modeled prognostic subgroups for survival and treatment outcomes in BRAF V600-mutated metastatic melanoma: pooled analysis of 4 randomized clinical trials. JAMA Oncol. 2018;4:1382–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McQuade JL, Daniel CR, Hess KR, et al. Association of body-mass index and outcomes in patients with metastatic melanoma treated with targeted therapy, immunotherapy, or chemotherapy: a retrospective, multicohort analysis. Lancet Oncol. 2018;19:310–322. doi: 10.1016/S1470-2045(18)30078-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Hiel B, Haanen JBAG, Stokkel MPM, et al. REPOSIT study group. Vemurafenib plus cobimetinib in unresectable stage IIIc or stage IV melanoma: response monitoring and resistance prediction with positron emission tomography and tumor characteristics (REPOSIT): study protocol of a phase II, open-label, multicenter study. BMC Cancer. 2017;17:649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Z, Zuo X, Zhang Y, et al. MiR-3662 suppresses hepatocellular carcinoma growth through inhibition of HIF-1α-mediated warburg effect. Cell Death Dis. 2018;9:549. doi: 10.1038/s41419-018-1111-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma S, Deng X, Yang Y, Zhang Q, Zhou T, Liu Z. The lncRNA LINC00675 regulates cell proliferation, migration, and invasion by affecting Wnt/β-catenin signaling in cervical cancer. Biomed Pharmacother. 2018;108:1686–1693. doi: 10.1016/j.biopha.2018.10.011 [DOI] [PubMed] [Google Scholar]
- 39.Shi K, Qiu X, Zheng W, Yan D, Peng W. MiR-203 regulates keloid fibroblast proliferation, invasion, and extracellular matrix expression by targeting EGR1 and FGF2. Biomed Pharmacother. 2018;108:1282–1288. doi: 10.1016/j.biopha.2018.09.152 [DOI] [PubMed] [Google Scholar]
- 40.Lu Y, Lian S, Ye Y, et al. S-nitrosocaptopril prevents cancer metastasis in vivo by creating the hostile bloodstream microenvironment against circulating tumor cells. Pharmacol Res. 2018. pii: S1043-6618(18)30400-6 [Epub ahead of print]. doi: 10.1016/j.phrs.2018.10.020 [DOI] [PubMed] [Google Scholar]
- 41.Kurata T, Fushida S, Kinoshita J, et al. Low-dose eribulin mesylate exerts antitumor effects in gastric cancer by inhibiting fibrosis via the suppression of epithelial-mesenchymal transition and acts synergistically with 5-fluorouracil. Cancer Manag Res. 2018;10:2729–2742. doi: 10.2147/CMAR.S167846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cao G, Chen D, Liu G, Pan Y, Liu Q. CPEB4 promotes growth and metastasis of gastric cancer cells via ZEB1-mediated epithelial- mesenchymal transition. Onco Targets Ther. 2018;11:6153–6165. doi: 10.2147/OTT.S175428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kyriakopoulou K, Kefali E, Piperigkou Z, Bassiony H, Karamanos NK. Advances in targeting epidermal growth factor receptor signaling pathway in mammary cancer. Cell Signal. 2018;51:99–109. doi: 10.1016/j.cellsig.2018.07.010 [DOI] [PubMed] [Google Scholar]
- 44.Zhang C, Zhang Y, Zhu H, Hu J, Xie Z. MiR-34a/miR-93 target c-Ski to modulate the proliferaton of rat cardiac fibroblasts and extracellular matrix deposition in vivo and in vitro. Cell Signal. 2018;46:145–153. doi: 10.1016/j.cellsig.2018.03.005 [DOI] [PubMed] [Google Scholar]
- 45.Li X, Li Z, Li X, Liu B, Liu Z. Mechanisms of Tanshinone II a inhibits malignant melanoma development through blocking autophagy signal transduction in A375 cell. BMC Cancer. 2017;17(1):357. doi: 10.1186/s12885-017-3329-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richard G, Dalle S, Monet MA, et al. ZEB1-mediated melanoma cell plasticity enhances resistance to MAPK inhibitors. EMBO Mol Med. 2016;8(10):1143–1161. doi: 10.15252/emmm.201505971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pal HC, Diamond AC, Strickland LR, et al. Fisetin, a dietary flavonoid, augments the anti-invasive and anti-metastatic potential of sorafenib in melanoma. Oncotarget. 2016;7(2):1227–1241. doi: 10.18632/oncotarget.6237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caramel J, Papadogeorgakis E, Hill L, et al. A switch in the expression of embryonic EMT-inducers drives the development of malignant melanoma. Cancer Cell. 2013;24(4):466–480. doi: 10.1016/j.ccr.2013.08.018 [DOI] [PubMed] [Google Scholar]
- 49.Shahid S, Kim G, Johnson NR, et al. MicroRNAs from the parasitic plant cuscuta campestris target host messenger RNAs. Nature. 2018;553(7686):82–85. doi: 10.1038/nature25027 [DOI] [PubMed] [Google Scholar]
- 50.Li L, Kang L, Zhao W, et al. miR-30a-5p suppresses breast tumor growth and metastasis through inhibition of LDHA-mediated warburg effect. Cancer Lett. 2017;400:89–98. doi: 10.1016/j.canlet.2017.04.034 [DOI] [PubMed] [Google Scholar]
- 51.Li J, Zhao J, Wang H, et al. MicroRNA-140-3p enhances the sensitivity of hepatocellular carcinoma cells to sorafenib by targeting pregnenolone X receptor. Onco Targets Ther. 2018;11:5885–5894. doi: 10.2147/OTT.S179509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meng D, Lei M, Han Y, et al. MicroRNA −645 targets urokinase plasminogen activator and decreases the invasive growth of MDA-MB-231 triple-negative breast cancer cells. Onco Targets Ther. 2018;11:7733–7743. doi: 10.2147/OTT.S187221 [DOI] [PMC free article] [PubMed] [Google Scholar]