Short abstract
Review on the relationship between eosinophils, endogenous microbial flora, and probiotic bacteria, to encourage research in this field.
Keywords: Lactobacillus, Bifidobacterium, inflammation, cytokines, mucosal
Abstract
There is currently substantial interest in the therapeutic properties of probiotic microorganisms as recent research suggests that oral administration of specific bacterial strains may reduce inflammation and alter the nature of endogenous microflora in the gastrointestinal tract. Eosinophils are multifunctional tissue leukocytes, prominent among the resident cells of the gastrointestinal mucosa that promote local immunity. Recent studies with genetically altered mice indicate that eosinophils not only participate in maintaining gut homeostasis, but that the absence of eosinophils may have significant impact on the nature of the endogenous gut microflora and responses to gut pathogens, notably Clostridium difficile. Furthermore, in human subjects, there is an intriguing relationship between eosinophils, allergic inflammation, and the nature of the lung microflora, notably a distinct association between eosinophil infiltration and detection of bacteria of the phylum Actinobacteria. Among topics for future research, it will be important to determine whether homeostatic mechanisms involve direct interactions between eosinophils and bacteria or whether they involve primarily eosinophil‐mediated responses to cytokine signaling in the local microenvironment. Likewise, although is it clear that eosinophils can and do interact with bacteria in vivo, their ability to discern between pathogenic and probiotic species in various settings remains to be explored.
Abbreviations
- COPD
= chronic obstructive pulmonary disease
- DC
= dendritic cell
- EET
= eosinophil extracellular trap
- EoE
= eosinophilic esophagitis
- GRAS
= generally recognized as safe
- NOD
= nucleotide‐binding oligomerization domain
- PRR
= pattern recognition receptor
- SAMP
= senescence‐accelerated mouse prone
- WAO
= World Allergy Organization
Introduction
There is a large and growing interest in the promise of probiotic microorganisms, defined by the World Health Organization as “live micro‐organisms which, when administered in adequate amounts, confer a health benefit on the host” [1]. Probiotics, which are typically select species from the genera Lactobacillus or Bifidobacterium, currently represent a multibillion dollar commercial component of the dietary supplements industry. Nonetheless, at this time, the health benefits conferred by probiotic supplements are not completely clear, nor are they universally substantiated upon review of multiple clinical trials ([2, 3, 4, 5, 6, 7, 8, 9, 10–11]).
Probiotics are typically administered orally, and most of our current understanding and information comes from mouse model studies and clinical evaluation of their mechanisms of action at the gastrointestinal tract. Among these mechanisms, probiotic bacteria may restore balance to the endogenous gut microflora [12, 13–14], the community of microbes known as the gut microbiome. Other, related mechanisms under exploration include interactions between probiotic bacteria and the epithelial cells lining the small intestines, as well as direct and indirect interactions with macrophages, DCs, and T and B lymphocytes present in the gut mucosa (reviews in [15, 16, 17–18]).
Interestingly, and frequently overlooked, eosinophils are also among the resident cells of the gut mucosa [19, 20–21] ( Fig. 1 ). Although eosinophils circulate in small numbers in the bloodstream and can be recruited to somatic tissues, largely in response to the eosinophil chemoattractant eotaxin‐1 (CCL11), working in concert with Th2 cytokine provocation [22, 23], eosinophils are present in the gut at homeostasis [24, 25]. As such, they are poised to interact with both endogenous microflora and exogenous probiotic bacteria alone or in concert with other resident leukocytes.
Figure 1.
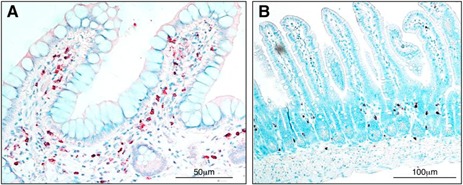
Eosinophils in the small intestines at homeostasis. Anti‐hEPX‐mAb (A)– and Anti‐mMBP‐mAb (B)–based immunohistochemistry provides evidence of tissue eosinophils in normal small‐intestinal tissue (ileum) of a human pediatric subject and a 10‐wk‐old AKR strain mice, staining red and brown, respectively. The University of Colorado School of Medicine Institutional Animal Care and Use Committee and The Institutional Review Board at the University of Colorado (COMIRB) approved these studies. Anti‐hEPX and anti‐mMBP were generous gifts from the laboratories of Dr. James J. Lee and Dr. Nancy A. Lee.
Much of the literature on probiotics and their relationship with eosinophils focuses on amelioration of allergic disease, notably allergic inflammation of the respiratory tract and skin. Among the confounding issues, there is no clear understanding of what features of probiotic organisms are crucial to elicit the desired response (e.g., bacterial species or strain that is most efficacious, use as prevention vs. therapy, duration of application, mix and composition of specific strains, length of therapy, and/or mechanism of action). Although the World Allergy Organization (WAO) has recently presented recommendations on probiotics for the prevention and therapy of eczema [26], there remains no consensus opinion on the impact of probiotics in allergic disease.
The intent of this review is to examine more closely our current understanding of the relationship between eosinophils, endogenous microbial flora, and probiotic bacteria, with the hopes of opening the doors to more interest and research in this field.
EOSINOPHILS ARE TISSUE LEUKOCYTES THAT PROMOTE LOCAL IMMUNITY
Eosinophils are granulocytic leukocytes that were first identified in 1879 by Paul Ehrlich, who remarked on their unusual staining properties, including large, refractile cytoplasmic granules that stained red with coal‐tar dyes [27]. Until recently, eosinophils were considered end‐stage, primarily cytotoxic effector cells, with limited flexibility or biosynthetic capability. However, new insights into their structure and function, notably regarding specific chemotactic and degranulation responses, together with a larger appreciation of the complex and vast array of their preformed granule cytokines has had a profound impact on this limited view. Recent studies have indicated that eosinophils are capable of coordinating local interactions with microorganisms, as well as with endothelial cells, epithelial cells, and other tissue leukocytes [22, 23, 28].
Most eosinophils are derived from a self‐replenishing source of hematopoietic precursor cells in the bone marrow and develop on a continuum through the promyelocytic stage to maturity in response to cytokines GM‐CSF (CSF‐2), IL‐3, and IL‐5 ( Fig. 2 ). Eosinophils are then released in morphologically mature form into circulation; after a brief stay (1–2 d), they are recruited to peripheral tissues. Notably, eosinophils respond to inflammatory and allergic stimulation and traffic to the lung, skin, esophagus, liver, gastrointestinal tract, and skeletal muscle.
Figure 2.
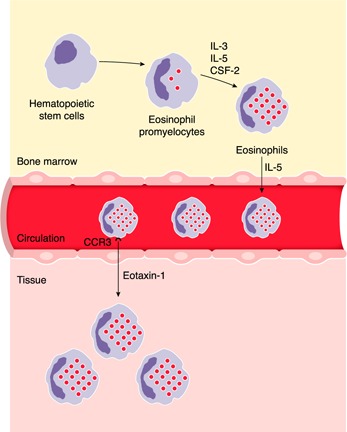
Eosinophil development and trafficking. Eosinophils develop in the bone marrow from hematopoietic stem cells in response to cytokines IL‐3, IL‐5, and CSF‐2. Morphologically mature eosinophils exit the bone marrow and circulate for a brief time until they exit into somatic tissues. Eosinophils respond to chemotactic cytokines, such as eotaxin‐1 (CCL11), via its interaction with CCR3 on the eosinophil surface, and exit circulation. Eosinophils move into the somatic tissue (such as the gastrointestinal tract) where they can reside for weeks to months. See text and references [19, 21, 25, 55].
As prominent components of the gastrointestinal mucosa at homeostasis, and in response to acute inflammatory stimuli, eosinophils are situated so as to be capable of interacting directly with bacteria and bacterial components, both those derived from commensal microflora and from administered probiotic bacteria. Eosinophils can present Ags, express PRRs, respond to pro‐ and anti‐inflammatory mediators, and release preformed granule proteins and cytokines that can modulate the local inflammatory response [28, 29, 30, 31, 32–33] ( Fig. 3 ). Selected topics on eosinophils and local immunity as they relate to interactions with commensal and pathogenic bacteria are considered in greater detail below. Numerous reviews and a comprehensive textbook featuring the basic and clinical biology of eosinophils in health and disease are available for the interested reader [22, 23, 34].
Figure 3.
Eosinophils coordinate local immunomodulatory responses. Eosinophils impart nuanced responses to their local environment and have the means to interact both directly and indirectly with probiotic bacteria and commensal microflora. See text and references [28, 29, 30, 31, 32–33].
ENDOGENOUS MICROFLORA AND PROBIOTIC MICROORGANISMS: BASIC CONCEPTS
Although the concept of helpful microorganisms probably dates to Biblical times (discussed in Soccol et al. [35]), Elie Metchnikoff [36] was among the first to publish observations on the relationship between health and the ingestion of active cultures of the “Bulgarian bacillus.” Interestingly, the text of this work, entitled The Prolongation of Life also includes a provocative discussion of the relationship between lifespan and the nature of the gut microflora in vertebrate species. More recent thoughts on endogenous flora and prolongation of life can be found in an essay by Michael Spector [37] entitled “Germs Are Us” in the “Annals of Science” series in the October 2012 issue of The New Yorker magazine.
Nearly a century after the publication of the Metchnikoff [36] treatise on the nature of mammalian gut flora, Lederberg and McCray [38] introduced the term microbiome as defining “the ecological community of commensal, symbiotic, and pathogenic microorganisms that literally share our body space.” Hooper et al. [39] were among the first to demonstrate this concept at the molecular level, characterizing the broad‐reaching impact of colonization in germ‐free mice with the commensal microorganism Bacteroides thetaiotaomicron. Because many of the organisms found among the normal microflora cannot be readily cultivated, efforts toward mass‐scale identification of normal, commensal microflora became possible only upon development of deep‐sequencing methodology, with top‐down identifications based on highly conserved sequences of 16S RNA. In 2008, the U.S. National Institutes of Health embarked on an effort to characterize the normal human microflora within multiple accessible body cavities. In 2012, the project group reported [40], among other findings, that the healthy human gastrointestinal tract included bacteria primarily of the phyla Firmicutes and Bacteroidetes, with Proteobacteria as a relatively minor component. Among the most intriguing findings of the report was the observation that there appears to be no “standard” or “defined” normal gut microbiome, even within their carefully selected sample population; composition varies significantly among individuals, although the metabolic functions supported remain constant.
Probiotic microorganisms are defined as those that confer health benefits on the recipient host [1]. This definition, on its own, is functional by nature and highly inclusive, and most recent clinical and mouse model studies feature one or more gram‐positive bacterial strains from the genera Lactobacillus or Bifidobacteria. These are not the only bacteria that are likely to be “probiotic” per se, but specific species from these genera are among those generally regarded as safe (GRAS) by the U.S. Food and Drug Administration (http://www.fda.gov). Although there is not yet a unified synthesis or hierarchy that defines probiotic action, there are several principles that are used as a basis for understanding probiotic action, including that the organism (1) may alter the balance of the gut microflora via colonization and competition for space, competition for nutrients, and secretion of favorable (or unfavorable) metabolites and secretory products; (2) may have a direct impact on intestinal epithelial cells, including inducing production of protective mucin glyco‐conjugates, limiting production of proinflammatory cytokines, stabilizing tight junctions, and preventing apoptosis; and (3) may elicit immune modulation via interactions with resident leukocytes as components of gut‐associated lymphoid tissue (GALT) in the subepithelial mucosal tissue and may interact with macrophages, DCs, and T and B lymphocytes at this site, resulting in the production of immune‐modulatory cytokines, including IL‐12, IL‐10, and TGF‐β; DCs can also sample microbial Ags directly from the intestinal lumen. These subjects are covered in depth in several recent reviews [15, 16, 17–18].
Among features common to all gram‐positive bacteria, the cell walls of both Lactobacillus and Bifidobacteria species consist of multilayered, cross‐linked peptidoglycan [41]. The cell walls and membranes of various Lactobacillus species contain polymers of d‐alanine or glycosyl‐substituted wall and membrane teichoic acids, respectively; the specific nature of the polymers vary among species and can be influenced by growth conditions [42, 43].
Among the most interesting questions currently under study include the following: What features of an individual species or of an individual strain within a bacterial species confer probiotic function? Can these features be predicted, engineered, and enhanced [44]? The teichoic acids of Lactobacillus species, which interact with the host PRR TLR‐2, have been identified as important modulators of probiotic potential. Several groups have focused on the teichoic acids and have introduced mutations into the Lactobacillus dltD gene to limit the esterification of d‐alanine to the cell‐surface teichoic acids. Interestingly, this manipulation enhances anti‐inflammatory, probiotic properties of these bacteria in mouse colitis models [45, 46] while impairing colonization [47].
PROBIOTIC BACTERIA AND THEIR IMPACT ON ALLERGIC DISEASES
As of this writing, there are scores of independent clinical studies and numerous reviews and meta‐analyses that focus on the efficacy of probiotic therapy for treatment and/or prevention of allergic disease [3, 4, 5, 6–7]. For example, following up on the 2007 Cochrane Database Systems meta‐analysis, which identified a reduction in infant eczema among those in probiotic treatment groups [3], 2 recent meta‐analyses, each considering multiple independent trials involving several thousand subjects, came to similar conclusions. Specifically, Zuccotti et al. [48] reported that infants receiving probiotic supplementation had a statistically lower risk of developing eczema than did controls. Likewise, Cuello‐Garcia et al. [4] provided a more extended analysis and reported that probiotics used by pregnant women, breast‐feeding mothers, and/or infants likewise reduced the risk of developing eczema, although they cautioned that the quality of the evidence was low. In contrast, no supporting evidence emerged when probiotic therapy was evaluated for prevention of asthma or childhood wheezing [7, 49]. In 2015, the WAO noted specifically that currently available evidence provided no strong support for probiotic supplementation as contributing toward reducing the risk of developing allergy in children overall. However, the committee highlighted the use of probiotics by infants and pregnant and breast‐feeding mothers, noting a net benefit to those infants at high risk for developing eczema, albeit via evidence rated as “very low quality” [26]. Among the confounding issues faced by these analyses, the individual clinical study designs were often complex and difficult to compare with one another; it is not clear how long probiotic administration might be necessary or which probiotic strain or strains might be most efficacious. At this point, it is not at all clear what the full impact of probiotic therapy—currently focused on oral administration of live microorganisms—might be vis à vis therapy for allergic disease; further research will certainly serve to clarify this point.
EOSINOPHILS AND MICROFLORA IN THE GASTROINTESTINAL TRACT
There is evidence from experimental mouse models indicating that eosinophils have an influence on the nature of the normal resident gastrointestinal microflora. Gastrointestinal eosinophils emerge during fetal life, before the acquisition of gut microflora [19, 20], and are maintained in the tissue by homeostatic levels of eotaxin‐1 and cytokines from type 2 innate lymphoid cells [19, 20–21, 24, 25].
Among the most interesting data in support of a role for eosinophils modulating microbial disease in the gut are recent findings from Buonomo et al. [50], who demonstrated a significant role for eosinophils in lethal infection with Clostridium difficile. Specifically, mice treated with the microbiota‐regulated cytokine IL‐25 were protected from lethal infection, an effect that was dependent on the influx of CD11b+SiglecF+ eosinophils to the gut. Eosinophil‐deficient PHIL mice [51] were not protected from C. difficile infection by IL‐25. Interestingly, mice lacking eosinophils suffered profound epithelial destruction, unrelated to levels of IL‐4, mucin, or IgA. The results suggested that eosinophils maintained epithelial barrier function via mechanisms yet to be clarified.
An intriguing counterpoint to these findings are those of DeSalvo et al. [52], who explored the interplay of IL‐33, eosinophils, and normal microflora in colitis‐prone SAMP mice. The SAMP strain featured in this study (SAMP1/YitFc [53]) carries a spontaneous mutation and has elevated levels of intestinal IL‐33. In this study, IL‐33–mediated eosinophil infiltration contributed to intestinal pathology, including profound loss of intact villous structure. Furthermore, minimal levels of IL‐33 were detected in intestinal tissue of germ‐free SAMP mice; introduction of normal microflora via fecal transplant resulted in induction of IL‐33, Th2 cytokine production, eosinophil infiltration, and inflammatory pathology.
Although there are many differences that can be accounted for between these 2 studies, among the most prominent is the fact that the responses, actions, and impact of eosinophils were clearly distinct and were influenced by the nature of the local microenvironment. One cannot see eosinophils in tissue and state a priori that the outcome will be positive or negative; eosinophil responses to their environment are clearly highly nuanced and context dependent [22, 23].
Toward this end, we are still seeking a clearer understanding of the way in which eosinophils modulate the nature of the microbiome. In their study featuring mice devoid of eosinophils (ΔdblGATA), Chu et al. [54] found that the gut microflora in these mice was highly irregular, including more bacteria per milligram of feces, along with a skewed ratio favoring gram‐negative to gram‐positive species, with more bacteria of the phylum Bacteroidetes and fewer Firmicutes than in their wild‐type BALB/c counterparts. Interestingly, Jung et al. [55] reported the reverse; in a series of experiments with the same mouse strains (BALB/c and eosinophil‐deficient ΔdblGATA) under similar conditions (cohoused for at least 3 wk), the completely opposite results were observed, including a profound decrease in the proportion of Bacteroidetes and an increase in the fraction of Firmicutes in the same strain. As we begin to understand more about factors that may have a profound impact on the results from any individual microbiome study, it would be helpful as we move forward to have specific information on the diet provided to these mice, the specific age and gender, the frequency of cage changes [56], and factors such as temperature and dark–light cycles in the vivarium [57]. Apart from the opposite results, it is not clear whether eosinophils had a direct impact (e.g., via their granule proteins and cytokines) on the nature of the microbiome or whether the differences observed were more indirect, (i.e., eosinophils modulating survival of IgA+ plasma cells, the latter having a direct impact on gut homeostasis [54] or via support of Peyer's patches and production of IL‐7 [55]). Indeed, as ΔdblGATA mice are fully devoid of eosinophils, it cannot be said for certain that the gut eosinophils per se were the only cells of this lineage contributing to this response.
Other studies suggest that the gut microbiome may have an impact on eosinophils at sites beyond the immediate gut microenvironment. Specifically, Suarez‐Zamorano et al. [58] reported that elimination of the endogenous microflora using either antibiotic treatment or generation of germ‐free mice resulted in expression of Th2 cytokines (IL‐4, IL‐5, and IL‐13) and recruitment of eosinophils and macrophages to inguinal subcutaneous (s.c.) tissue in association with the development of functional beige‐fat, a process known to be dependent on eosinophil‐mediated macrophage polarization [59, 60]. The larger mechanisms linking microflora depletion and eosinophil accumulation at this more distant site remain unclear.
Moving beyond the small intestines, Fillon et al. [61, 62–63] explored the microbiome of the esophagus in healthy individuals and in patients with EoE and gastroesophageal reflux disease, the latter 2 disorders associated with increased numbers of eosinophils in the esophagus, a tissue ordinarily devoid of these cells. Among the findings, they noted that absolute bacterial load was increased in association with both diseases, although eosinophil infiltration in terms of absolute numbers was not directly proportional to bacterial load. Bacterial phyla detected included primarily Firmicutes, Bacteroidetes, and Proteobacteria; bacteria of the genus Haemophilus were significantly increased in individuals with untreated EoE compared with healthy controls. Benitez et al. [64] also examined the esophageal flora and noted specific enrichment of Proteobacteria in patients with EoE. The role of the esophageal microflora and its potential manipulation toward amelioration of disease awaits future study.
EOSINOPHILS, PROBIOTICS, AND MICROFLORA IN THE RESPIRATORY TRACT
Lung tissue is subject to constant airflow from the exterior as well as microaspiration of bacteria present in oral secretions and, as such, is not free of microbes [65, 66]; the healthy lung microbiome includes primarily bacteria of the phyla Bacteroidetes and Firmicutes, with prominent contributions from genera Prevotella, Veillonella, and Streptococcus [65]. As in the gastrointestinal tract, respiratory diseases, including asthma, COPD, cystic fibrosis, and pneumonia, are all associated with alterations in the lung microflora [67, 68–69]. However, specifically with respect to our focus on eosinophils, a recent study by Huang et al. [70] found that bacterial numbers (total 16S RNA copy numbers) in brush extractions from patients with severe asthma were negatively correlated with total biopsy eosinophil numbers, results suggesting that higher bacterial burdens were associated with fewer numbers of eosinophils in the lung tissue. As such, most of the bacterial phyla identified (primarily Proteobacteria and Firmicutes) followed similar patterns, with the exception of 2 members of the Actinobacteria (Streptomyces and Propionicimonas), which correlated positively with eosinophil numbers in tissue. Of interest, these relationships did not extend to include correlations of bacterial numbers with eosinophils in blood or sputum.
A study by Denner et al. [71] compared lung microbiomes in asthmatics and controls and reported increased species diversity correlating directly with the percentage of eosinophils in bronchoalveolar lavage samples. In a similar study featuring COPD, Sze et al. [72] found that the size of the microbiome was reduced in association with decreasing lung surface area; as in the aforementioned study, a strong, positive correlation between the presence of Actinobacteria and eosinophil infiltration was reported. However, with a focus on the upper airways, Aurora et al. [73] characterized the microflora of the nasal cavities and found a greater bacterial burden, but no specific correlations with eosinophil numbers, in the tissue in their study of controls vs. patients with chronic rhinosinusitis. By contrast, in a study of seasonal allergic rhinitis, Choi et al. [74] reported a significant correlation between bacterial diversity in the middle meatus with nasal eosinophil count.
In all of these cases, the focus is the interaction between eosinophils, eosinophil activity, and the organisms comprising the local tissue microbiome. With this in mind, our group has shown that administration of probiotic Lactobacillus species (phylum Firmicutes) to the lower airways of mice resulted in a profound reduction in inflammation associated with acute respiratory virus infection [75, 76]; the long‐ and short‐term changes to the airway microbiome have not yet been characterized. Similar manipulations may prevent eosinophil recruitment in response to allergen provocation [77, 78].
EOSINOPHILS AND BACTERIA: DIRECT INTERACTIONS IN VIVO?
Because eosinophils are present at highest concentration in the small intestines, immediately juxtaposed to the epithelial cells lining the lumen, the potential for direct interactions among eosinophils, microflora, orally administered probiotic bacteria, and/or their components certainly exists.
Toward this end, several groups have explored the expression of PRRs on human and mouse eosinophils (reviewed in [33]). Kvarnhammar et al. [79] found that agonists of both NOD1 and NOD2, the cytosolic receptors for bacterial peptidoglycan fragments, could activate isolated human eosinophils. By contrast, it was initially unclear whether human eosinophils express TLR2 and/or under what specific conditions [33], although recently, Driss et al. [80] used TLR2‐neutralizing Abs to define the role of this PRR in eosinophil‐mediated interactions with Mycobacterium bovis, suggesting that TLR2 not only exists but has relevant function. Likewise, to the best of our knowledge, no one has determined whether eosinophils from the gastrointestinal tract express different PRRs and/or different levels of these same PRRs than those detected on eosinophils from peripheral blood and/or whether PRR expression on eosinophils in this locale changes in response to different states of inflammation or in disease.
Likewise, many groups have characterized interactions of eosinophils with bacteria in vitro, primarily those typically regarded as pathogens (e.g., Escherichia coli, Staphylococcus aureus); most of these studies predate our understanding of PRRs and probiotic bacteria. Among these findings, isolated human eosinophils are capable of direct interactions with bacteria, including phagocytosis of bacterial pathogens, release of granule proteins, and generation of reactive oxygen species [81, 82, 83–84].
However, among the more recent findings and perhaps most relevant to this topic, Yousefi et al. [85] examined the interactions between eosinophils and bacteria in the gut as well as ex vivo and found that eosinophils responded by releasing atypical “nets” known as eosinophil extracellular traps (EETs), consisting of mitochondrial DNA complexed with granule cationic proteins. Release was not accompanied by cytolysis or cell death and could be recapitulated by stimulation of cytokine‐primed, peripheral blood eosinophils with bacterial products, such as LPS, from gram‐negative pathogens. EETs have also been detected in vivo in EoE, likely in response to barrier dysfunction and perhaps related to direct exposure of eosinophils to bacteria in the esophageal lumen [86, 87].
Among the open questions: might eosinophils interact differently with bacteria that are health promoting, or probiotic? Might the presence of probiotic bacteria provide signals that modulate EET formation as well as the release of granule proteins, cytokines, and reactive oxygen species? Although we might assume that EET formation relates directly to interactions of bacteria (both probiotic and pathogenic) with eosinophil cell surface PRRs, we are not aware of any studies that have addressed this question directly.
Likewise, although there are no studies that address questions regarding interactions of eosinophils with probiotic bacteria in vivo, Hosoki et al. [88] found that isolated human eosinophils released granule stores of eosinophil‐derived neurotoxin in response to the pathogen C. difficile but not in response to the probiotic bacterium Bifidobacterium bifidum. The mechanisms underlying this distinction, as well as the way in which all eukaryotic cells distinguish between these bacterial species, remains to be elucidated. Furthermore, we have found that isolated mouse eosinophils can phagocytose the probiotic bacterial species Lactobacillus reuteri, an action that is associated with the release of anti‐inflammatory cytokine IL‐10 (Kraemer and Rosenberg, unpublished results).
CONCLUSIONS
Eosinophilic leukocytes are unique effector cells with complex roles in promoting health and homeostasis. They are among the leukocytes that are prominent in the gastrointestinal mucosa; data from mouse models suggest that they participate in maintaining the normal gut microflora and responding to gastrointestinal pathogens. As such, eosinophils may also respond to orally administered probiotic microorganisms and contribute to the resolution of gastrointestinal inflammation. Similarly, the relationship among eosinophils, lung microflora, and respiratory inflammation is open to further exploration, notably the unique association of bacteria of the phylum Actinobacteria with the degree of eosinophil infiltration in this setting. Future research will provide information on whether eosinophil‐mediated homeostatic and anti‐inflammatory mechanisms involve direct interactions with bacteria and/or responses to cytokine signaling from epithelial cells and leukocytes in the local microenvironment.
AUTHORSHIP
H.F.R. and G.T.F. wrote the manuscript. J.C.M. prepared Fig. 1 and Fig. 1 legend.
DISCLOSURES
The authors declare no conflicts of interest.
ACKNOWLEDGMENTS
Supported by U.S. National Institutes of Health (NIH) National Institute of Allergy and Infectious Diseases (NIAID) intramural funding (AI000941 to H.F.R.), by NIH (1K24DK100303 to G.T.F.), and by the Consortium for Gastrointestinal Eosinophilic Researchers (CEGIR). CEGIR (Grant U54 AI117804) is part of the Rare Diseases Clinical Research Network (RDCRN), an initiative of the Office of Rare Diseases Research (ORDR), National Center for Advancing Translational Sciences (NCATS), and is funded through collaboration between NIAID, the NIH National Institute of Diabetes and Digestive Kidney Disease, and NCATS (to G.T.F.). The authors thank Dr. James J. Lee and Dr. Nancy A. Lee (Mayo Clinic, Scottsdale, AZ, USA) for the generous gift of anti‐hEPX and anti‐mMBP used for the immunohistochemical staining images.
References
- 1. World Health Organization, Food and Agriculture Organization of the United Nations . (2001) Report of a Joint FAO/WHO expert consultation of evaluations of health and nutritional properties of probiotics in food including powder milk and live lactic acid bacteria in Cordoba, Argentina. In Probiotics in Food: Health and Nutritional Properties and Guidelines for Evaluation. WHO and FAO, Rome, Italy. FAO Food and Nutrition Paper 85.
- 2. Goldenberg, J. Z. , Ma, S. S. , Saxton, J. D. , Martzen, M. R. , Vandvik, P. O. , Thorlund, K. , Guyatt, G. H. , Johnston, B. C. (2013) Probiotics for the prevention of Clostridium difficile‐associated diarrhea in adults and children. Cochrane Database Syst. Rev. 5, CD006095. [DOI] [PubMed] [Google Scholar]
- 3. Osborn, D. A. , Sinn, J. K. H. (2007) Probiotics in infants for prevention of allergic disease and food hypersensitivity. Cochrane Database Syst. Rev. 4:CD006475. [DOI] [PubMed] [Google Scholar]
- 4. Cuello‐Garcia, C. A. , BrozDek, J. L. , Fiocchi, A. , Pawankar, R. , Yepes‐NunTez, J. J. , Terracciano, L. , Gandhi, S. , Agarwal, A. , Zhang, Y. , Schünemann, H. J. (2015) Probiotics for the prevention of allergy: A systematic review and meta‐analysis of randomized controlled trials. J. Allergy Clin. Immunol. 136, 952–961. [DOI] [PubMed] [Google Scholar]
- 5. Lee, J. , Seto, D. , Bielory, L. (2008) Meta‐analysis of clinical trials of probiotics for prevention and treatment of pediatric atopic dermatitis. J. Allergy Clin. Immunol. 121, 116–121.e11. [DOI] [PubMed] [Google Scholar]
- 6. Pelucchi, C. , Chatenoud, L. , Turati, F. , Galeone, C. , Moja, L. , Bach, J. F. , La Vecchia, C. (2012) Probiotics supplementation during pregnancy or infancy for the prevention of atopic dermatitis: a meta‐analysis. Epidemiology 23, 402–414. [DOI] [PubMed] [Google Scholar]
- 7. Azad, M. B. , Coneys, J. G. , Kozyrskyj, A. L. , Field, C. J. , Ramsey, C. D. , Becker, A. B. , Friesen, C. , Abou‐Setta, A. M. , Zarychanski, R. (2013) Probiotic supplementation during pregnancy or infancy for the prevention of asthma and wheeze: systematic review and meta‐analysis. BMJ 347, f6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mallon, P. , McKay, D. , Kirk, S. , Gardiner, K. (2007) Probiotics for induction of remission in ulcerative colitis. Cochrane Database Syst. Rev. 4, CD005573. [DOI] [PubMed] [Google Scholar]
- 9. Allen, S. J. (2015) The potential of probiotics to prevent Clostridium difficile infection. Infect. Dis. Clin. North Am. 29, 135–144. [DOI] [PubMed] [Google Scholar]
- 10. McFarland, L. V. (2015) Probiotics for the primary and secondary prevention of C. difficile infections: a meta‐analysis and systematic review. Antibiotics (Basel) 4, 160–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hell, M. , Bernhofer, C. , Stalzer, P. , Kern, J. M. , Claassen, E. (2013) Probiotics in Clostridium difficile infection: reviewing the need for a multistrain probiotic. Benef. Microbes 4, 39–51. [DOI] [PubMed] [Google Scholar]
- 12. Reid, G. , Younes, J. A. , Van der Mei, H. C. , Gloor, G. B. , Knight, R. , Busscher, H. J. (2011) Microbiota restoration: natural and supplemented recovery of human microbial communities. Nat. Rev. Microbiol. 9, 27–38. [DOI] [PubMed] [Google Scholar]
- 13. Turroni, F. , Ventura, M. , Buttó, L. F. , Duranti, S. , O'Toole, P. W. , Motherway, M. O. , van Sinderen, D. (2014) Molecular dialogue between the human gut microbiota and the host: a Lactobacillus and Bifidobacterium perspective. Cell. Mol. Life Sci. 71, 183–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gareau, M. G. , Sherman, P. M. , Walker, W. A. (2010) Probiotics and the gut microbiota in intestinal health and disease. Nat. Rev. Gastroenterol. Hepatol. 7, 503–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bron, P. A. , van Baarlen, P. , Kleerebezem, M. (2011) Emerging molecular insights into the interaction between probiotics and the host intestinal mucosa. Nat. Rev. Microbiol. 10, 66–78. [DOI] [PubMed] [Google Scholar]
- 16. Klaenhammer, T. R. , Kleerebezem, M. , Kopp, M. V. , Rescigno, M. (2012) The impact of probiotics and prebiotics on the immune system. Nat. Rev. Immunol. 12, 728–734. [DOI] [PubMed] [Google Scholar]
- 17. Lebeer, S. , Vanderleyden, J. , De Keersmaecker, S. C. (2010) Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat. Rev. Microbiol. 8, 171–184. [DOI] [PubMed] [Google Scholar]
- 18. Thaiss, C. A. , Zmora, N. , Levy, M. , Elinav, E. (2016) The microbiome and innate immunity. Nature 535, 65–74. [DOI] [PubMed] [Google Scholar]
- 19. Schroeder, S. , Masterson, J. C. , Fillon, S. , Furuta, G. T. (2013) Eosinophils as regulators of gastrointestinal physiological homeostasis, 1st ed. In Eosinophils in Health and Disease. (Lee J. J., Rosenberg H. F., eds.) Academic Press, Waltham, MA, 341–345. [Google Scholar]
- 20. Jung, Y. , Rothenberg, M. E. (2014) Roles and regulation of gastrointestinal eosinophils in immunity and disease. J. Immunol. 193, 999–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fulkerson, P. C. , Rothenberg, M. E. (2008) Origin, regulation and physiological function of intestinal oeosinophils. Best Pract. Res. Clin. Gastroenterol. 22, 411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rosenberg, H. F. , Dyer, K. D. , Foster, P. S. (2013) Eosinophils: changing perspectives in health and disease. Nat. Rev. Immunol. 13, 9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Furuta, G. T. , Atkins, F. D. , Lee, N. A. , Lee, J. J. (2014) Changing roles of eosinophils in health and disease. Ann. Allergy Asthma Immunol. 113, 3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nussbaum, J. C. , Van Dyken, S. J. , von Moltke, J. , Cheng, L. E. , Mohapatra, A. , Molofsky, A. B. , Thornton, E. E. , Krummel, M. F. , Chawla, A. , Liang, H. E. , Locksley, R. M. (2013) Type 2 innate lymphoid cells control eosinophil homeostasis. Nature 502, 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mishra, A. , Hogan, S. P. , Lee, J. J. , Foster, P. S. , Rothenberg, M. E. (1999) Fundamental signals that regulate eosinophil homing to the gastrointestinal tract. J. Clin. Invest. 103, 1719–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fiocchi, A. , Pawankar, R. , Cuello‐Garcia, C. , Ahn, K. , Al‐Hammadi, S. , Agarwal, A. , Beyer, K. , Burks, W. , Canonica, G. W. , Ebisawa, M. , Gandhi, S. , Kamenwa, R. , Lee, B. W. , Li, H. , Prescott, S. , Riva, J. J. , Rosenwasser, L. , Sampson, H. , Spigler, M. , Terracciano, L. , Vereda‐Ortiz, A. , Waserman, S. , Yepes‐NunTez, J. J. , BrozDek, J. L. , Schünemann, H. J. (2015) World Allergy Organization‐Mcmaster University guidelines for allergic disease prevention (GLAD‐P): probiotics. World Allergy Organ. J. 8, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kay, A. B. (2015) The early history of the eosinophil. Clin. Exp. Allergy 45, 575–582. [DOI] [PubMed] [Google Scholar]
- 28. Lee, J. J. , Jacobsen, E. A. , McGarry, M. P. , Schleimer, R. P. , Lee, N. A. (2010) Eosinophils in health and disease: the LIAR hypothesis. Clin. Exp. Allergy 40, 563–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Akuthota, P. , Wang, H. , Weller, P. F. (2010) Eosinophils as antigen‐presenting cells in allergic upper airway disease. Curr. Opin. Allergy Clin. Immunol. 10, 14–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kambayashi, T. , Laufer, T. M. (2014) Atypical MHC class II‐expressing antigen‐presenting cells: can anything replace a dendritic cell? Nat. Rev. Immunol. 14, 719–730. [DOI] [PubMed] [Google Scholar]
- 31. Lee, J. J. , Lee, N. A. (2005) Eosinophil degranulation: an evolutionary vestige or a universally destructive effector function? Clin. Exp. Allergy 35, 986–994. [DOI] [PubMed] [Google Scholar]
- 32. Davoine, F. , Lacy, P. (2014) Eosinophil cytokines, chemokines, and growth factors: emerging roles in immunity. Front. Immunol. 5, 570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kvarnhammar, A. M. , Cardell, L. O. (2012) Pattern‐recognition receptors in human eosinophils. Immunology 136, 11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee, J. J. , Rosenberg, H. F. (2013). Eosinophils in Health and Disease. Elsevier Press, Amsterdam. [Google Scholar]
- 35. Soccol, C. R. , Prado, M. R. M. , Garcia, L. M. B. , Rodrigues, C. , Medeiros, A. B. P. , Soccol, V. T. (2014) Current developments in probiotics. J. Microb. Biochem. Technol. 7, 11–20. [Google Scholar]
- 36. Metchnikoff, E. (1908). The Prolongation of Life: Optimistic Studies, (Mitchell P. C., ed.) G P Putnam & Sons, New York. [Google Scholar]
- 37. Specter, M. (2012) Germs are us. Bacteria make us sick. Do they also keep us alive? New Yorker 22, 32–39. [Google Scholar]
- 38. Lederberg, J. , McCray, A. T. (2001) ‘Ome Sweet ‘Omics—a genealogical treasury of words [commentary]. Scientist 15, 8. [Google Scholar]
- 39. Hooper, L. V. , Wong, M. H. , Thelin, A. , Hansson, L. , Falk, P. G. , Gordon, J. I. (2001) Molecular analysis of commensal host‐microbial relationships in the intestine. Science 291, 881–884. [DOI] [PubMed] [Google Scholar]
- 40. Human Microbiome Project Consortium . (2012) Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vollmer, W. , Blanot, D. , de Pedro, M. A. (2008) Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32, 149–167. [DOI] [PubMed] [Google Scholar]
- 42. Lebeer, S. , Vanderleyden, J. , De Keersmaecker, S. C. J. (2008) Genes and molecules of lactobacilli supporting probiotic action. Microbiol. Mol. Biol. Rev. 72, 728–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brown, S. , Santa Maria, J. P., Jr. , Walker, S. (2013) Wall teichoic acids of gram‐positive bacteria. Annu. Rev. Microbiol. 67, 313–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lee, I.‐C. , Tomita, S. , Kleerebezem, M. , Bron, P. A. (2013) The quest for probiotic effector molecules–unraveling strain specificity at the molecular level. Pharmacol. Res. 69, 61–74. [DOI] [PubMed] [Google Scholar]
- 45. Grangette, C. , Nutten, S. , Palumbo, E. , Morath, S. , Hermann, C. , Dewulf, J. , Pot, B. , Hartung, T. , Hols, P. , Mercenier, A. (2005) Enhanced antiinflammatory capacity of a Lactobacillus plantarum mutant synthesizing modified teichoic acids. Proc. Natl. Acad. Sci. USA 102, 10321–10326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Claes, I. J. , Lebeer, S. , Shen, C. , Verhoeven, T. L. , Dilissen, E. , De Hertogh, G. , Bullens, D. M. , Ceuppens, J. L. , Van Assche, G. , Vermeire, S. , Rutgeerts, P. , Vanderleyden, J. , De Keersmaecker, S. C. (2010) Impact of lipoteichoic acid modification on the performance of the probiotic Lactobacillus rhamnosus GG in experimental colitis. Clin. Exp. Immunol. 162, 306–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Walter, J. , Loach, D. M. , Alqumber, M. , Rockel, C. , Hermann, C. , Pfitzenmaier, M. , Tannock, G. W. (2007) D‐alanyl ester depletion of teichoic acids in Lactobacillus reuteri 100‐23 results in impaired colonization of the mouse gastrointestinal tract. Environ. Microbiol. 9, 1750–1760. [DOI] [PubMed] [Google Scholar]
- 48. Zuccotti, G. , Meneghin, F. , Aceti, A. , Barone, G. , Callegari, M. L. , Di Mauro, A. , Fantini, M. P. , Gori, D. , Indrio, F. , Maggio, L. , Morelli, L. , Corvaglia, L. ; Italian Society of Neonatology . (2015) Probiotics for prevention of atopic diseases in infants: systematic review and meta‐analysis. Allergy 70, 1356–1371. [DOI] [PubMed] [Google Scholar]
- 49. Ta, V. , Laubach, S. (2014) Probiotic administration in early life, atopy, and asthma: a meta‐analysis of clinical trials. Pediatrics 134 (Suppl 3), S141. [DOI] [PubMed] [Google Scholar]
- 50. Buonomo, E. L. , Cowardin, C. A. , Wilson, M. G. , Saleh, M. M. , Pramoonjago, P. , Petri, W. A., Jr. (2016) Microbiota‐regulated IL‐25 increases eosinophil number to provide protection during Clostridium difficile infection. Cell Reports 16, 432–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lee, J. J. , Dimina, D. , Macias, M. P. , Ochkur, S. I. , McGarry, M. P. , O'Neill, K. R. , Protheroe, C. , Pero, R. , Nguyen, T. , Cormier, S. A. , Lenkiewicz, E. , Colbert, D. , Rinaldi, L. , Ackerman, S. J. , Irvin, C. G. , Lee, N. A. (2004) Defining a link with asthma in mice congenitally deficient in eosinophils. Science 305, 1773–1776. [DOI] [PubMed] [Google Scholar]
- 52. De Salvo, C. , Wang, X. M. , Pastorelli, L. , Mattioli, B. , Omenetti, S. , Buela, K. A. , Chowdhry, S. , Garg, R. R. , Goodman, W. A. , Rodriguez‐Palacios, A. , Smith, D. E. , Abbott, D. W. , Cominelli, F. , Bamias, G. , Xin, W. , Lee, J. J. , Vecchi, M. , Pizarro, T. T. (2016) IL‐33 drives eosinophil infiltration and pathogenic type 2 helper T‐cell immune responses leading to chronic experimental ileitis. Am. J. Pathol. 186, 885–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pizarro, T. T. , Pastorelli, L. , Bamias, G. , Garg, R. R. , Reuter, B. K. , Mercado, J. R. , Chieppa, M. , Arseneau, K. O. , Ley, K. , Cominelli, F. (2011) SAMP1/YitFc mouse strain: a spontaneous model of Crohn's disease‐like ileitis. Inflamm. Bowel Dis. 17, 2566–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chu, V. T. , Beller, A. , Rausch, S. , Strandmark, J. , Zänker, M. , Arbach, O. , Kruglov, A. , Berek, C. (2014) Eosinophils promote generation and maintenance of immunoglobulin‐A‐expressing plasma cells and contribute to gut immune homeostasis. Immunity 40, 582–593. [DOI] [PubMed] [Google Scholar]
- 55. Jung, Y. , Wen, T. , Mingler, M. K. , Caldwell, J. M. , Wang, Y. H. , Chaplin, D. D. , Lee, E. H. , Jang, M. H. , Woo, S. Y. , Seoh, J. Y. , Miyasaka, M. , Rothenberg, M. E. (2015) IL‐1β in eosinophil‐mediated small intestinal homeostasis and IgA production. Mucosal Immunol. 8, 930–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ebino, K. Y. , Yoshinaga, K. , Saito, T. R. , Takahashi, K. W. (1988) A simple method for prevention of coprophagy in the mouse. Lab. Anim. 22, 1–4. [DOI] [PubMed] [Google Scholar]
- 57. Voigt, R. M. , Forsyth, C. B. , Green, S. J. , Mutlu, E. , Engen, P. , Vitaterna, M. H. , Turek, F. W. , Keshavarzian, A. (2014) Circadian disorganization alters intestinal microbiota. PLoS One 9, e97500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Suárez‐Zamorano, N. , Fabbiano, S. , Chevalier, C. , Stojanović, O. , Colin, D. J. , Stevanović, A. , Veyrat‐Durebex, C. , Tarallo, V. , Rigo, D. , Germain, S. , Ilievska, M. , Montet, X. , Seimbille, Y. , Hapfelmeier, S. , Trajkovski, M. (2015) Microbiota depletion promotes browning of white adipose tissue and reduces obesity. Nat. Med. 21, 1497–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Qiu, Y. , Nguyen, K. D. , Odegaard, J. I. , Cui, X. , Tian, X. , Locksley, R. M. , Palmiter, R. D. , Chawla, A. (2014) Eosinophils and type 2 cytokine signaling in macrophages orchestrate development of functional beige fat. Cell 157, 1292–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rao, R. R. , Long, J. Z. , White, J. P. , Svensson, K. J. , Lou, J. , Lokurkar, I. , Jedrychowski, M. P. , Ruas, J. L. , Wrann, C. D. , Lo, J. C. , Camera, D. M. , Lachey, J. , Gygi, S. , Seehra, J. , Hawley, J. A. , Spiegelman, B. M. (2014) Meteorin‐like is a hormone that regulates immune‐adipose interactions to increase beige fat thermogenesis. Cell 157, 1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Harris, J. K. , Fang, R. , Wagner, B. D. , Choe, H. N. , Kelly, C. J. , Schroeder, S. , Moore, W. , Stevens, M. J. , Yeckes, A. , Amsden, K. , Kagalwalla, A. F. , Zalewski, A. , Hirano, I. , Gonsalves, N. , Henry, L. N. , Masterson, J. C. , Robertson, C. E. , Leung, D. Y. , Pace, N. R. , Ackerman, S. J. , Furuta, G. T. , Fillon, S. A. (2015) Esophageal microbiome in eosinophilic esophagitis. PLoS One 10, e0128346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fillon, S. A. , Harris, J. K. , Wagner, B. D. , Kelly, C. J. , Stevens, M. J. , Moore, W. , Fang, R. , Schroeder, S. , Masterson, J. C. , Robertson, C. E. , Pace, N. R. , Ackerman, S. J. , Furuta, G. T. (2012) Novel device to sample the esophageal microbiome—the esophageal string test. PLoS One 7, e42938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fang, R. , Wagner, B. D. , Harris, J. K. , Fillon, S. A. (2016) Zero‐inflated negative binomial mixed model: an application to two microbial organisms important in oesophagitis. Epidemiol. Infect. 144, 2447–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Benitez, A. J. , Hoffmann, C. , Muir, A. B. , Dods, K. K. , Spergel, J. M. , Bushman, F. D. , Wang, M. L. (2015) Inflammation‐associated microbiota in pediatric eosinophilic esophagitis. Microbiome 3, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Dickson, R. P. , Erb‐Downward, J. R. , Martinez, F. J. , Huffnagle, G. B. (2016) The microbiome and the respiratory tract. Annu. Rev. Physiol. 78, 481–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gollwitzer, E. S. , Marsland, B. J. (2014) Microbiota abnormalities in inflammatory airway diseases ‐ Potential for therapy. Pharmacol. Ther. 141, 32–39. [DOI] [PubMed] [Google Scholar]
- 67. Hilty, M. , Burke, C. , Pedro, H. , Cardenas, P. , Bush, A. , Bossley, C. , Davies, J. , Ervine, A. , Poulter, L. , Pachter, L. , Moffatt, M. F. , Cookson, W. O. (2010) Disordered microbial communities in asthmatic airways. PLoS One 5, e8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sze, M. A. , Dimitriu, P. A. , Hayashi, S. , Elliott, W. M. , McDonough, J. E. , Gosselink, J. V. , Cooper, J. , Sin, D. D. , Mohn, W. W. , Hogg, J. C. (2012) The lung tissue microbiome in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 185, 1073–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Pragman, A. A. , Kim, H. B. , Reilly, C. S. , Wendt, C. , Isaacson, R. E. (2012) The lung microbiome in moderate and severe chronic obstructive pulmonary disease. PLoS One 7, e47305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Huang, Y. J. , Nariya, S. , Harris, J. M. , Lynch, S. V. , Choy, D. F. , Arron, J. R. , Boushey, H. (2015) The airway microbiome in patients with severe asthma: Associations with disease features and severity. J. Allergy Clin. Immunol. 136, 874–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Denner, D. R. , Sangwan, N. , Becker, J. B. , Hogarth, D. K. , Oldham, J. , Castillo, J. , Sperling, A. I. , Solway, J. , Naureckas, E. T. , Gilbert, J. A. , White, S. R. (2016) Corticosteroid therapy and airflow obstruction influence the bronchial microbiome, which is distinct from that of bronchoalveolar lavage in asthmatic airways. J. Allergy Clin. Immunol. 137, 1398–1405.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sze, M. A. , Dimitriu, P. A. , Suzuki, M. , McDonough, J. E. , Campbell, J. D. , Brothers, J. F. , Erb‐Downward, J. R. , Huffnagle, G. B. , Hayashi, S. , Elliott, W. M. , Cooper, J. , Sin, D. D. , Lenburg, M. E. , Spira, A. , Mohn, W. W. , Hogg, J. C. (2015) Host response to the lung microbiome in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 192, 438–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Aurora, R. , Chatterjee, D. , Hentzleman, J. , Prasad, G. , Sindwani, R. , Sanford, T. (2013) Contrasting the microbiomes from healthy volunteers and patients with chronic rhinosinusitis. JAMA Otolaryngol. Head Neck Surg. 139, 1328–1338. [DOI] [PubMed] [Google Scholar]
- 74. Choi, C. H. , Poroyko, V. , Watanabe, S. , Jiang, D. , Lane, J. , de Tineo, M. , Baroody, F. M. , Naclerio, R. M. , Pinto, J. M. (2014) Seasonal allergic rhinitis affects sinonasal microbiota. Am. J. Rhinol. Allergy 28, 281–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gabryszewski, S. J. , Bachar, O. , Dyer, K. D. , Percopo, C. M. , Killoran, K. E. , Domachowske, J. B. , Rosenberg, H. F. (2011) Lactobacillus‐mediated priming of the respiratory mucosa protects against lethal pneumovirus infection. J. Immunol. 186, 1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Percopo, C. M. , Dyer, K. D. , Garcia‐Crespo, K. E. , Gabryszewski, S. J. , Shaffer, A. L. , III, Domachowske, J. B. , Rosenberg, H. F. (2014) B cells are not essential for Lactobacillus‐mediated protection against lethal pneumovirus infection. J. Immunol. 192, 5265–5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Di Felice, G. , Barletta, B. , Butteroni, C. , Corinti, S. , Tinghino, R. , Colombo, P. , Boirivant, M. (2008) Use of probiotic bacteria for prevention and therapy of allergic diseases: studies in mouse model of allergic sensitization. J. Clin. Gastroenterol. 42 (Suppl 3 Pt 1), S130–S132. [DOI] [PubMed] [Google Scholar]
- 78. Nembrini, C. , Sichelstiel, A. , Kisielow, J. , Kurrer, M. , Kopf, M. , Marsland, B. J. (2011) Bacterial‐induced protection against allergic inflammation through a multicomponent immunoregulatory mechanism. Thorax 66, 755–763. [DOI] [PubMed] [Google Scholar]
- 79. Kvarnhammar, A. M. , Petterson, T. , Cardell, L. O. (2011) NOD‐like receptors and RIG‐I‐like receptors in human eosinophils: activation by NOD1 and NOD2 agonists. Immunology 134, 314–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Driss, V. , Legrand, F. , Hermann, E. , Loiseau, S. , Guerardel, Y. , Kremer, L. , Adam, E. , Woerly, G. , Dombrowicz, D. , Capron, M. (2009) TLR2‐dependent eosinophil interactions with mycobacteria: role of α‐defensins. Blood 113, 3235–3244. [DOI] [PubMed] [Google Scholar]
- 81. Cline, M. J. , Hanifin, J. , Lehrer, R. I. (1968) Phagocytosis by human eosinophils. Blood 32, 922–934. [PubMed] [Google Scholar]
- 82. Baehner, R. L. , Johnston, R. B., Jr. (1971) Metabolic and bactericidal activities of human eosinophils. Br. J. Haematol. 20, 277–285. [DOI] [PubMed] [Google Scholar]
- 83. Cohen, S. G. , Sapp, T. M. (1969) Phagocytosis of bacteria by eosinophils in infectious‐related asthma. J. Allergy 44, 113–117. [DOI] [PubMed] [Google Scholar]
- 84. Jong, E. C. , Henderson, W. R. , Klebanoff, S. J. (1980) Bactericidal activity of eosinophil peroxidase. J. Immunol. 124, 1378–1382. [PubMed] [Google Scholar]
- 85. Yousefi, S. , Gold, J. A. , Andina, N. , Lee, J. J. , Kelly, A. M. , Kozlowski, E. , Schmid, I. , Straumann, A. , Reichenbach, J. , Gleich, G. J. , Simon, H. U. (2008) Catapult‐like release of mitochondrial DNA by eosinophils contributes to antibacterial defense. Nat. Med. 14, 949–953. [DOI] [PubMed] [Google Scholar]
- 86. Simon, D. , Radonjic‐Hösli, S. , Straumann, A. , Yousefi, S. , Simon, H. U. (2015) Active eosinophilic esophagitis is characterized by epithelial barrier defects and eosinophil extracellular trap formation. Allergy 70, 443–452. [DOI] [PubMed] [Google Scholar]
- 87. Ueki, S. , Tokunaga, T. , Fujieda, S. , Honda, K. , Hirokawa, M. , Spencer, L. A. , Weller, P. F. (2016) Eosinophil ETosis and DNA traps: a new look at eosinophilic inflammation. Curr. Allergy Asthma Rep. 16, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Hosoki, K. , Nakamura, A. , Nagao, M. , Hiraguchi, Y. , Tokuda, R. , Wada, H. , Nobori, T. , Fujisawa, T. (2010) Differential activation of eosinophils by ‘probiotic’ Bifidobacterium bifidum and ‘pathogenic’ Clostridium difficile . Int. Arch. Allergy Immunol. 152 (Suppl 1), 83–89. [DOI] [PubMed] [Google Scholar]


