Abstract
Recent evidence suggests that altered expression and epigenetic modification of the glucocorticoid receptor gene (NR3C1) are related to the risk of post-traumatic stress disorder (PTSD). The underlying mechanisms, however, remain unknown. Because glucocorticoid receptor signaling is known to regulate emotional memory processes, particularly in men, epigenetic modifications of NR3C1 might affect the strength of traumatic memories. Here, we found that increased DNA methylation at the NGFI-A (nerve growth factor-induced protein A) binding site of the NR3C1 promoter was associated with less intrusive memory of the traumatic event and reduced PTSD risk in male, but not female survivors of the Rwandan genocide. NR3C1 methylation was not significantly related to hyperarousal or avoidance symptoms. We further investigated the relationship between NR3C1 methylation and memory functions in a neuroimaging study in healthy subjects. Increased NR3C1 methylation–which was associated with lower NR3C1 expression–was related to reduced picture recognition in male, but not female subjects. Furthermore, we found methylation-dependent differences in recognition memory-related brain activity in men. Together, these findings indicate that an epigenetic modification of the glucocorticoid receptor gene promoter is linked to interindividual and gender-specific differences in memory functions and PTSD risk.
Keywords: DNA methylation, GR, memory, PTSD
Introduction
Post-traumatic stress disorder (PTSD) is characterized by intrusive memories of traumatic events, avoidance, and hyperarousal symptoms [Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition (DSM-IV), American Psychiatric Association, 2000]. In contrast to a state of chronic stress, PTSD is not paralleled by increased glucocorticoid levels (Meewisse et al., 2007), but rather by enhanced hypothalamus–pituitary–adrenal (HPA) axis feedback (Yehuda, 2002; Rohleder et al., 2004; Pitman et al., 2012). Moreover, there is evidence suggesting that altered HPA axis regulation and a high number of glucocorticoid receptors (GRs) represent a pretrauma risk factor for the disorder (Yehuda, 2009; van Zuiden et al., 2011, 2013). The reason for interindividual differences in GR number and the mechanism by which GRs influence the risk for PTSD, however, remain largely unknown.
Timed activation of GRs is of crucial importance in memory consolidation of learned information, especially of emotionally arousing information (Roozendaal et al., 2006). Human studies have shown that the memory-enhancing effects of stress-induced elevations of glucocorticoids are usually more pronounced in men (Andreano and Cahill, 2006; Preuss and Wolf, 2009; Cornelisse et al., 2011). Furthermore, there is evidence that glucocorticoids impair the retrieval of memory processes (de Quervain et al., 2009). Because fear learning and memory processes play an important role in the pathogenesis of PTSD (de Quervain, 2006; Brewin, 2011; Pitman et al., 2012; Wilker et al., 2013), GRs may be implicated in PTSD risk by influencing memory processes (de Quervain et al., 2009). Accordingly, it has been shown that the BclI polymorphism of NR3C1 (nuclear receptor subfamily 3, group C, member 1), the gene encoding the GR, is associated with sensitivity to glucocorticoids (van Rossum et al., 2003), emotional memory in healthy individuals (Ackermann et al., 2013), traumatic memories and PTSD symptoms in critically ill patients (Hauer et al., 2011).
Epigenetic mechanisms may contribute to interindividual differences in GR signaling. DNA hypermethylation of the exon NR3C1-17 promoter in rodents and of its human ortholog, the exon NR3C1-1F promoter (Turner and Muller, 2005), was associated with low maternal care, maternal depression, and perinatal stress (Weaver et al., 2007; Oberlander et al., 2008). In line with the enhanced GR feedback reported in PTSD cases (Yehuda, 2009), it has been shown that increased NR3C1 expression in peripheral tissue is related to PTSD risk (van Zuiden et al., 2011). Furthermore, it appears that these changes are partially epigenetically controlled. Two recent studies suggested that DNA methylation of the GR gene promoter are inversely correlated with lifetime PTSD risk (Labonté et al., 2014; Yehuda et al., 2014). Moreover, allele-specific DNA demethylation of FKBP5, a regulator of the GR complex, has been found to mediate gene–childhood trauma interactions (Klengel et al., 2013).
Given the evidence that GR signaling influences both memory processes and PTSD risk, we investigated whether epigenetic differences in the human NR3C1 gene promoter are related to traumatic memory and the risk for PTSD in 152 survivors of the Rwandan genocide. Furthermore, we investigated the relationship of NR3C1 promoter DNA methylation with memory processes in a functional neuroimaging study in 72 healthy subjects.
Materials and Methods
Subjects: Rwanda sample.
We included 152 survivors from the 1994 Rwandan genocide (69 females, 83 males; median age, 35 years; range, 30–41 years) who lived as refugees in the Nakivale settlement in Uganda. The sample consisted of 93 subjects fulfilling the diagnostic criteria of the DSM-IV for lifetime PTSD, and 59 individuals who did not meet the DSM-IV diagnostic criteria for PTSD (61.2% with PTSD lifetime diagnosis; 31.2% subjects with current PTSD according to DSM-IV (American Psychiatric Association, 2000). Additionally, to exclude genetic relatives in the samples, only one person per household was interviewed. Candidates exhibiting current alcohol abuse and acute psychotic symptoms were excluded. All subjects had experienced highly aversive traumatic situations and were examined in 2006/2007 (de Quervain et al., 2007). The Post-Traumatic Diagnostic Scale (PDS; Foa et al., 1997) and event list (Ertl et al., 2010) were administered as a structured interview by expert psychologists from the University of Konstanz, Germany, as well as by trained local interviewers. Interviewers first went through a six-week course on principles of quantitative data collection and interviewing techniques. The translation of the instruments into Kinyarwanda was done using several steps of translations, blind back-translations, and subsequent corrections by independent groups of translators (Neuner et al., 2008). Following the translations, the psychometric properties of the translated scales were investigated in a validation study that included a retest spanning a two-week period and a cross-validation with expert rating (Neuner et al., 2008). The PDS was used to assess intrusions, avoidance, and hyperarousal. A checklist of 36 war-related and nonwar-related traumatic event types (e.g., injury by weapon, rape, accident) was used to assess traumatic events (de Quervain et al., 2007; Neuner et al., 2008). Traumatic load was estimated by assessing the number of different traumatic event types experienced or witnessed. This measure is considered more reliable than assessing the frequency of traumatic events (Neuner et al., 2008). Study procedures were approved by the ethics committees of the University of Konstanz, Germany, and the Mbarara University of Science and Technology, Mbarara, Uganda. Before the interview, all participants provided written informed consent.
Subjects were selected to have experienced ≤19 traumatic event types, to avoid known ceiling effects on PTSD risk (Kolassa et al., 2010). Saliva samples from all subjects were collected using Oragene DNA Kits (DNA Genotek).
Subjects: Swiss sample.
A total of 72 healthy young subjects (47 females, 25 males; median age, 23 years; range, 18–34 years) were included in the functional magnetic resonance imaging (fMRI) study. fMRI data from one male and two female subjects were corrupted and therefore were not included in the study. Subjects were free of any neurological or psychiatric illness, and did not take any medication at the time of the experiment (except hormonal contraceptives). The ethics committees of the Canton of Basel and Baselland approved the experiments.
Subjects were tested in the late morning and afternoon hours (mean time, 2:30 P.M.; SD ± 3 h). All participants received general information about the study and gave their written informed consent for participation. After completing the training outside of the scanner, subjects performed two different consecutive tasks in the scanner. The first two tasks, i.e., the picture-encoding task and the working-memory task (n-back), were described in detail previously (de Quervain et al., 2007). Briefly, stimuli in the picture-encoding task consisted of 72 pictures selected from the International Affective Picture System (Lang et al., 1999) as well as from in-house standardized picture sets that allowed us to equate the pictures for visual complexity and content (e.g., human presence). On the basis of normative valence scores (from 1 to 9), pictures were assigned to emotionally negative (2.3 ± 0.6), emotionally neutral (5.0 ± 0.3), and emotionally positive (7.6 ± 0.4) conditions, resulting in 24 pictures for each emotional valence. Four additional pictures showing neutral objects were used to control for primacy and recency effects in memory. Two of these pictures were presented in the beginning and two at the end of the picture task and the working memory task. In addition, 24 scrambled pictures were used. The background of the scrambled pictures contained the color information of all pictures used in the experiment (except primacy and recency pictures), overlaid with a crystal and distortion filter (Adobe Photoshop CS3). In the foreground, a mostly transparent geometrical object (rectangle or ellipse of different sizes and orientations) was shown. Participants were instructed and then trained on the picture-encoding task. After training, they were positioned in the scanner. The picture-encoding task, which included the arousal and valence ratings of the pictures, lasted for ∼20 min. Immediately afterward, subjects performed the working-memory task for 10 additional minutes. After leaving the scanner, participants gave a free recall of the pictures in a separate room (no time limit was set for this task). Forty to 50 min after the presentation of the last picture in the encoding task, participants were repositioned in the scanner and performed a recognition task for 20 min. The recognition task consisted of two sets of stimuli that were either new (i.e., not presented before) or old (i.e., presented during the picture-encoding task). Each of the two sets contained 72 pictures (24 pictures for each emotional valence).
Using an Oragene DNA and Oragene RNA Kit (DNA Genotek), saliva samples were collected from all subjects. Participants received 25 Swiss francs per hour for participation.
DNA isolation and bisulfite conversion.
Saliva DNA was initially extracted from the Oragene DNA Kit (DNA Genotek) using the precipitation protocol recommended by the producer. To obtain high-purity DNA before bisulfite conversion, samples were additionally repurified. For this purpose, 2 μg of DNA isolated via the Oragene recommended procedure, was incubated overnight at 50°C with proteinase K (lysis buffer: 30 mm Tris-Cl, 10 mm EDTA, 1% SDS, pH∼8.0, 150 ng/μl proteinase K), agitated by gentle orbital shaking. Next, the DNA was purified using a Genomic DNA Clean & Concentrator Kit (Zymo Research). The quality and concentration were assessed using gel electrophoresis and fluorometry (Qubit dsDNA BR Assay Kit, Invitrogen), respectively. Five hundred nanograms of high-purity, intact DNA was used for bisulfite conversion using an EZ DNA Methylation-Gold kit (Zymo Research) by following standard protocols. Bisulfite DNA quality and concentration was determined using an RNA Pico 6000 Kit on a Bioanalyzer 2100 instrument (Agilent Technologies) and Nanodrop 2000 (ThermoScientific). An external control sample was always converted in parallel to assess possible variations in conversion reactions. All samples were bisulfite converted on two separate occasions, under the identical conditions. Bisulfite-converted (BSC) samples were normalized to 10 ng/μl.
All samples were analyzed in quadruplicates from two independent bisulfite conversions.
Pyrosequencing analysis.
The DNA methylation status of the CpG-rich region of the human NR3C1 gene, including the exon 1F promoter (NR3C1-1F) with the NGFI-A binding site, previously reported as important for epigenetic regulation of GR gene expression (Weaver et al., 2004), was quantified by direct bisulfite pyrosequencing (Tost and Gut, 2007). Primers were designed according to recommendations of Wojdacz et al. (2009): NR3C1_Q-CpG_FW, 5′-GTTATTCGTAGGGGTATTG-3′ and NR3C1_Q-CpG_RV, 5′-biot-CAACTCCCCAAAAAAAAAAA-3′ (Microsynth). A 206 bp NR3C1 promoter fragment was amplified using an AmplyTaq Gold Kit from Applied Biosystems (Life Technologies). PCR was done in 30 μl reactions containing the following: 1× PCR buffer II, 300 μm deoxynucleotide triphosphates, final 3.5 mm MgCl2, 200 μm of each primer, 20 ng of BSC DNA. We used the following cycling conditions: 95°C, 15 min–50 × (95°C, 30 s; 55°C, 30 s; 72°C, 30 s)–72°C, 10 min. PCR products were purified and sequenced using a PyroMark ID System (Biotage) following the manufacturer's suggested protocol and two sequencing primers: NR3C1_S2, 5′-GAGTGGGTTTGGAGT-3′ and NR3C1_S3, 5′-AGAAAAGAAATTGGAGAAATT-3′ (Microsynth) were used as in Oberlander et al. (2008). Testing for PCR temperature bias and calibration was done by introducing a series of calibrator samples with known methylation levels. Briefly, we prepared unmethylated standards by using two rounds of linear whole-genome amplification with an Ovation WGA System Kit (Nugene), starting from 10 ng of DNA, as recommended by the manufacturer. Methylated standards were made using CpG methyltransferase assay with M.SssI (New England Biolabs) starting from 2 μg of purified DNA, following the standard protocol. Bisulfite conversion of standard samples was done as described above.
RNA isolation and expression analysis.
For the purpose of DNA methylation–RNA expression correlation experiments, an additional 24 (12 females, 12 males; median age, 25 years; range, 21–29 years) subjects were recruited from the Swiss sample, as described above. RNA was isolated from Oragene RNA Self-Collection Kits from saliva. Promptly after collection, samples were mixed for 10 s by vortexing, stabilized at 50°C in a water bath for 2.5 h, and stored at −80°C until further analysis. For the isolation procedure, samples were processed in 250 μl aliquots. Briefly, samples were incubated in a water bath at 90°C for 15 min, and then cooled to room temperature. Following this, 750 μl of TRIReagent LS (Ambion, Life Technologies) was added to each sample and homogenization was done for 1 min at 28 Hz using TissueLyser II (Qiagen). After homogenization, 1000 μl of absolute ethanol was added to the sample and mixed. Finally, the mixture was loaded on a Zymo-Spin IIC column and RNA was further purified using a Direct-zol kit (ZymoResearch) following the recommended procedure, including DNase I treatment. The concentration and quality of the RNA was determined using Nanodrop 2000 (ThermoScientific) and an RNA Nano 6000 Kit on a Bioanalyzer 2100 instrument (Agilent).
For reverse transcription, 350 ng of total RNA was denatured for 8 min at 70°C followed by ice incubation in the presence of 25 ng of anchored oligo(dT)20 primer (Invitrogen) and 75 ng of random decamers primers (Ambion). In the reverse transcription reaction, cDNA was generated in 25 μl reaction using a Super RT kit (HT Biotechnology). Upon completion of the reaction, the volume was adjusted to 200 μl in Lambda DNA solution (5 ng/μl final concentration; Promega). The primers were designed against splice variants that contain exon 1F of the NR3C1 gene: NR3C1_1F_FW, 5′-GTTGATATTCACTGATGGA-3′ and NR3C1_1F_RV, 5′-CTTGGAGTCTGATTGAGA-3′ (Microsynth). Additionally total expression of the GR gene was determined against exon 2: NR3C1_2_FW, 5′-CTTCAGAACAGCAACATT-3′ and NR3C1_2_RV, 5′-GACTCTCATTCGTCTCTT-3′ (Microsynth). The expression levels were normalized to the RPLPO gene (human large ribosomal protein) using the following primers: RPLP0-Ex3-4_FW, 5′-CTCTGGAGAAACTGCTGC-3′ and RPLP0-Ex3-4_RV, 5′-CTGATCTCAGTGAGGTCC-3′ (Sigma-Aldrich). qPCR was performed using the Power SYBR Green PCR Master Mix (Life Technologies) according to standard recommendations, in 12 μl final volume of reaction, using 2 μl of cDNA template, on a RotorGene 6000A instrument (Corbett Research). Cycling conditions were as follows: 95°C, 60 s–40× (95°C, 3 s; 56°C, 10 s; 72°C, 4 s). This was followed by a melting curve analysis (61–95°C, rising by 0.7°C/3 s) to attest amplification specificity. Threshold cycles (crossing point) were determined using Rotor-Gene software version 6.1 (Corbett Research). RPLPO was selected as reference gene for normalization after we tested several candidate reference genes, as had been previously described (Movassagh et al., 2010). Expression levels were normalized using a geometric mean level of expression (Vandesompele et al., 2002). Fold differences were calculated using the delta-delta-Ct method (Pfaffl, 2001) with the help of qBasePlus software (Biogazelle).
fMRI study design.
During the encoding task, the pictures were presented for 2.5 s in a quasirandomized order so that ≤4 pictures of the same category occurred consecutively. A fixation cross appeared on the screen for 500 ms before each picture presentation. Trials were separated by a variable intertrial period of 9–12 s (jitter) that was equally distributed for each stimulus category. During the intertrial period, participants subjectively rated the picture showing scenes according to valence (negative, neutral, positive) and arousal (high, medium, and low) on a three-point scale (self-assessment manikin) by pressing a button with a finger of their dominant hand. For scrambled pictures, participants rated form (vertical, symmetric, or horizontal) and size (large, medium, small) of the geometrical object in the foreground.
During the recognition task, the pictures were presented for 1 s in a quasirandomized order so that ≤4 pictures of the same category (i.e., negative new, negative old, neutral new, neutral old, positive new, positive old) occurred consecutively. A fixation cross appeared on the screen for 500 ms before each picture presentation. Trials were separated by a variable intertrial period of 6–12 s (jitter) that was equally distributed for each stimulus category. During the intertrial period, participants subjectively rated the picture as either remembered, familiar, or new on a three-point scale by pressing a button with a finger of their dominant hand. Recognition performance was assessed as a number of correctly recognized previously seen pictures, corrected for the number of false positives.
fMRI methods.
Measurements were performed on a Siemens Magnetom Verio 3 T whole-body MR unit equipped with a 12-channel head coil (Siemens Healthcare). Functional time series were acquired with a single-shot echo-planar sequence using parallel imaging [GRAPPA (generalized autocalibrating partially parallel acquisition)]. We used the following acquisition parameters: TE, 35 ms; field of view, 22 cm; acquisition matrix, 80 × 80, interpolated to 128 × 128; voxel size, 2.75 × 2.75 × 4 mm3; GRAPPA acceleration factor r, 2.0. Using a midsagittal scout image, 32 contiguous axial slices placed along the anterior–posterior commissure plane covering the entire brain with a TR = 3000 ms (α = 82°) were acquired using an ascending interleaved sequence.
Participants received earplugs and headphones to reduce scanner noise. The head of each participant was fixated in the coil using small cushions, and participants were told not to move their head. Pictures were presented in the scanner using MR-compatible liquid crystal display goggles (VisualSystem; NordicNeuroLab). Eye correction was used when necessary.
Preprocessing and data analysis was performed using SPM8 (Statistical Parametric Mapping, Wellcome Trust Centre for Neuroimaging; http://www.fil.ion.ucl.ac.uk/spm/) implemented in Matlab R2011b (Mathworks). Volumes were slice-time corrected to the first slice and realigned using the “register to mean” option. A mean image was generated from the realigned series and coregistered to the structural image. This ensured that functional and structural images were spatially aligned.
The functional images and the structural images were spatially normalized by applying DARTEL (diffeomorphic anatomical registration through exponentiated lie algebra), which leads to an improved registration between subjects. Normalization incorporated the following steps: (1) structural images of each subject were segmented using the “New Segment” procedure in SPM8; (2) the resulting gray and white matter images were used to derive a study-specific group template computed from a large population of 612 subjects, including the 69 subjects from this study where methylation data were available; (3) an affine transformation was applied to map the group template to MNI space; and (4) subject-to-template and template-to-MNI transformations were combined to map the functional images to MNI space. The functional images were smoothed with an isotropic 8 mm full width at half maximum Gaussian filter.
Intrinsic autocorrelations were accounted for by AR(1) and low-frequency drifts were removed via high-pass filter (time constant, 128 s). For each subject, evoked hemodynamic responses to event types were modeled with a delta function convolved with a canonical hemodynamic response function within the context of a general linear model. Button presses and rating scale presentation during ratings were modeled separately. In addition, six movement parameters from spatial realigning were included as regressors of no interest.
Encoding task.
The effect of picture encoding was investigated by constructing separate regressors representing the event types for positive, negative, neutral, and scrambled pictures. We contrasted brain activity during the encoding of pictures against brain activity during the presentation of scrambled pictures (positive, negative, and neutral pictures vs scrambled pictures).
Recognition task.
Eighteen different picture event types were modeled by crossing each stimulus category with the subject-specific rating of each picture while differentiating between correct and incorrect recognition ratings with respect to the new or old picture set: (negative, positive, neutral) × (remembered, familiar, new) × (correct, incorrect). We contrasted brain activity between items that were correctly recognized as seen before and new pictures (i.e., pictures seen in the encoding task and rated as remembered vs pictures not seen before and rated as new). Items rated as familiar were not included in the analysis.
The contrasts of both the encoding and the recognition task were calculated individually using a fixed-effects model (first-level analysis). The resulting contrast parameters were then used for methylation-dependent brain activity analyses in a random-effects, linear regression model where we entered the methylation level as a covariate (second-level analysis).
Construction of a population-average anatomical probabilistic atlas.
Automatic segmentation of the subjects' T1-weighted images was used to build a population-average probabilistic anatomical atlas. More precisely, each participant's T1-weighted image was first automatically segmented into cortical and subcortical structures using FreeSurfer (Fischl et al., 2002; version 4.5, http://surfer.nmr.mgh.harvard.edu/). Labeling of the cortical gyri was based on the Desikan-Killiany Atlas (Desikan et al., 2006), yielding 35 regions per hemisphere.
The segmented T1 image was then normalized to the study-specific anatomical template space using the subject's previously computed warp field. After this, the image was affine-registered to the MNI space. Nearest-neighbor interpolation was applied to preserve labeling of the different structures. The normalized segmentations were finally averaged across subjects to create a population-average probabilistic atlas. Each voxel of the template could consequently be assigned a probability of belonging to a given anatomical structure, based on the individual information from all subjects.
Statistical analyses.
Statistical analyses were done in R (version 12.15.2; R Development Core Team 2012). Due to non-normal distribution of methylation data in the Rwanda sample, we applied nonparametric tests. We assessed DNA methylation, trauma load, and age differences between two genders by Kruskal–Wallis one-way ANOVA and correlations of DNA methylation data with PTSD symptom clusters scores by Spearman's rank correlation (ρs). To account for trauma load and ceiling effects (Kolassa et al., 2010), PTSD symptom cluster scores were divided by the sum of lifetime traumatic event types (i.e., symptoms scores per traumatic event type; de Quervain et al., 2007). The relationship between DNA methylation at NR3C1 promoter and lifetime PTSD status was assessed using binary logistic regression, with NR3C1 CpG3 DNA methylation as quantitative predictor and the sum of life traumatic event types as a covariate.
Due to normal distribution of the methylation data, statistical analyses of behavioral data for the Swiss sample were performed using linear models. The relationship of NR3C1 DNA methylation to memory performance and NR3C1 expression was assessed using linear models. To account for methylation-dependent differences in arousal during recognition of previously seen pictures, both arousal and NR3C1 CpG3 DNA methylation were included in the linear model: we modeled valence-specific recognition performance as dependent on NR3C1 CpG3 DNA methylation and valence-specific arousal.
Correlation between NR3C1 CpG3 DNA methylation and NR3C1 expression was examined by linear regression model, corrected for age independently in two genders.
A comparison of methylation levels of NR3C1 CpG3 site between the Rwandan and the Swiss population was done by Kruskal–Wallis one-way ANOVA. Furthermore, we additionally assessed the equality of distributions and variability between two populations: Kolmogorov–Smirnov two-sample test and Siegel–Tukey test, respectively.
Bonferroni correction was implemented to account for multiple testing procedures. The significance threshold was set to p < 0.05.
All laboratory procedures were conducted in a blind, randomized order, including DNA and RNA isolations, bisulfite conversion, PCR, pyrosequencing, and expression analysis. Only after performing all procedures and excluding samples with low quality controls and outliers, further analysis with phenotypic data was performed.
Results
DNA methylation of the NR3C1 promoter in traumatized survivors of the Rwandan genocide
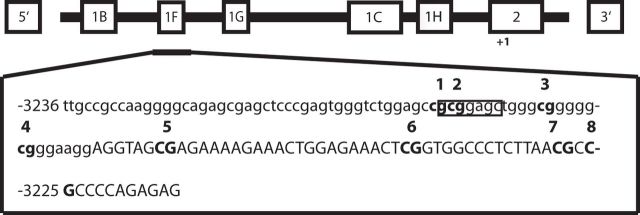
The promoter region of the NR3C1 gene studied herein is illustrated in Figure 1. It spans the exon 1F region, a human orthologue of the rat exon 1-7 (Turner and Muller, 2005), previously reported as differentially methylated (Oberlander et al., 2008; McGowan et al., 2009). The analysis covered eight CpGs, with the NGFI-A consensus binding sequence encompassing CpG positions 3 and 4. Because glucocorticoid-related changes in memory formation were repeatedly found to be more pronounced in men (Andreano and Cahill, 2006; Preuss and Wolf, 2009), we also considered possible gender differences in the present study. We first compared DNA methylation between genders and found significant differences between men and women at several CpG positions (p < 0.05, e.g., methylation levels at NR3C1_CpG3 were significantly higher in women than in men, p = 0.01). Therefore, we proceeded with gender-specific analyses. To account for trauma load, PTSD symptom cluster scores were corrected by the sum of lifetime traumatic event types (de Quervain et al., 2007). Methylation levels at the CpG3 site, which is embedded in the NGFI-A consensus binding sequence, were significantly negatively correlated with the severity of symptoms related to re-experiencing traumatic events (intrusive memories) in men (ρs = −0.355, pnominal = 0.001, pBonferroni corrected = 0.008 for eight CpG comparisons; Fig. 2, Table 1). No such association was found in women (ρs = −0.073, pnominal = 0.553; Table 2). Correlations of DNA methylation at CpG3 with the severity of symptoms related to avoidance and hyperarousal were not significant after Bonferroni correction for eight CpGs (p ≥ 0.05), indicating that the methylation changes at this site were preferentially related to the memory aspects of PTSD. None of the other examined NR3C1 promoter CpG sites were significantly associated with any of the PTSD symptom scores, after accounting for multiple testing (Tables 1, 2), indicating a pronounced role of methylation at the CpG3 site.
Figure 1.
Human NR3C1 gene and the CpG region analyzed by bisulfite pyrosequencing. The 5′ region of the NR3C1 gene contains multiple first exons, corresponding to multiple transcriptional start sites and multiple mRNA splice variants. The region analyzed by pyrosequencing is illustrated by enlarged box, below the exon 1F. Numbering is relative to the alternative transcription start site in exon 2. The CpG3 and CpG4 sites are encompassed by the potential NGFI-A binding site. Adapted from Oberlander et al. (2008).
Figure 2.
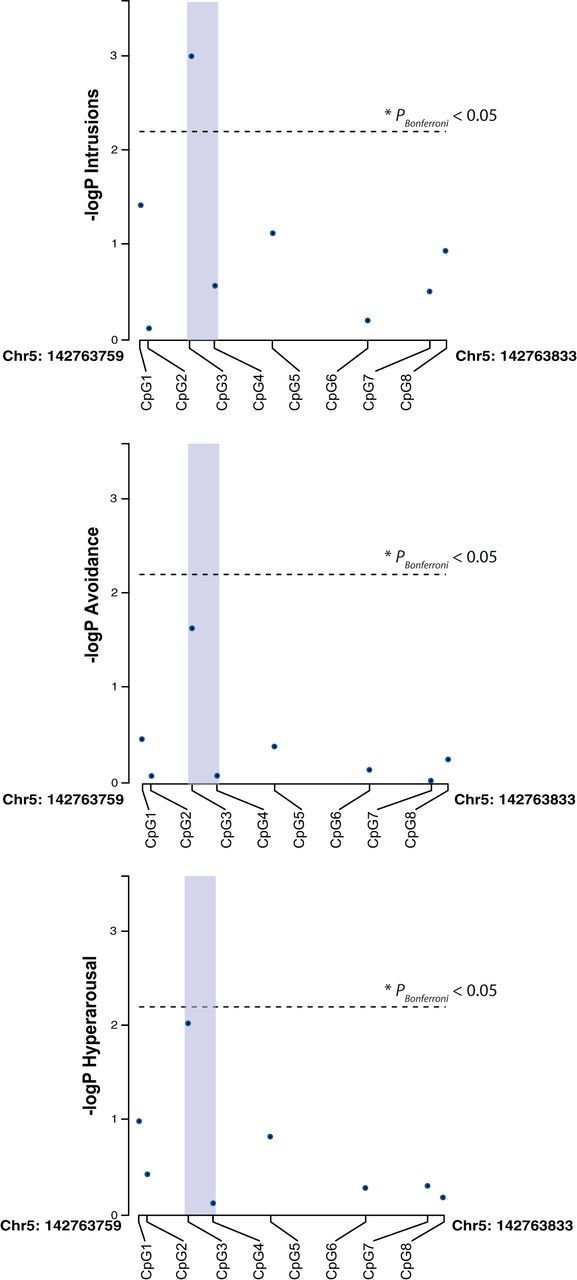
DNA methylation of NR3C1 promoter and PTSD symptoms in male genocide survivors. Association (Spearman's rank correlation, −log10p) of DNA methylation marks across eight CpG positions in the human NR3C1 gene promoter with PTSD symptoms clusters intrusions, avoidance, and hyperarousal. The association between CpG3 and intrusions reached Bonferroni-corrected statistical significance (after correcting for 8 tests; PBonferroni = 0.008). The CpGs are aligned by genomic position. The position of the potential NGFI-A binding site is shaded in purple. The horizontal dashed line indicates the p < 0.05 Bonferroni-corrected significance threshold.
Table 1.
Association of PTSD symptom clusters and DNA methylation of NR3C1 gene promoter in Rwandan mena
| PTSD symptom clusters (nmales = 83) | NR3C1_CpG1 | NR3C1_CpG2 | NR3C1_CpG3 | NR3C1_CpG4 | NR3C1_CpG5 | NR3C1_CpG6 | NR3C1_CpG7 | NR3C1 _CpG8 |
|---|---|---|---|---|---|---|---|---|
| Intrusionsb | ||||||||
| Spearman's ρ | −0.235 | −0.039 | −0.355 | −0.128 | −0.203 | −0.059 | −0.118 | −0.18 |
| p value | ||||||||
| Nominal | 0.037* | 0.733 | 0.001*** | 0.261 | 0.073 | 0.606 | 0.3 | 0.112 |
| Bonferroni correction for 8 CpGs | 0.296 | 1 | 0.008 | 1 | 0.584 | 1 | 1 | 0.896 |
| Bonferroni correction for 8 CpGs, sex, and 3 PTSD symptom clusters | 1 | 1 | 0.048 | 1 | 1 | 1 | 1 | 1 |
| Avoidanceb | ||||||||
| Spearman's ρ | −0.115 | −0.033 | −0.260 | −0.033 | −0.101 | −0.049 | −0.019 | −0.074 |
| p value | ||||||||
| Nominal | 0.315 | 0.772 | 0.021* | 0.77 | 0.376 | 0.666 | 0.87 | 0.517 |
| Bonferroni correction for 8 CpGs | 1 | 1 | 0.168 | 1 | 1 | 1 | 1 | 1 |
| Bonferroni correction for 8 CpGs, sex, and 3 PTSD symptom clusters | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Hyperarousalb | ||||||||
| Spearman's ρ | −0.187 | −0.097 | −0.346 | −0.038 | −0.165 | −0.074 | −0.08 | −0.053 |
| p value | ||||||||
| Nominal | 0.099 | 0.396 | 0.009** | 0.74 | 0.145 | 0.514 | 0.483 | 0.645 |
| Bonferroni correction for 8 CpGs | 0.792 | 1 | 0.072 | 1 | 1 | 1 | 1 | 1 |
| Bonferroni correction for 8 CpGs, sex, and 3 PTSD symptom clusters | 1 | 1 | 0.432 | 1 | 1 | 1 | 1 | 1 |
| PDS scoreb,c | ||||||||
| Spearman's ρ | −0.207 | −0.056 | −0.355 | −0.081 | −0.172 | −0.036 | −0.078 | −0.112 |
| p value | ||||||||
| Nominal | 0.067 | 0.627 | 0.001*** | 0.476 | 0.131 | 0.756 | 0.496 | 0.324 |
| Bonferroni correction for 8 CpGs | 0.536 | 1 | 0.008 | 1 | 1 | 1 | 1 | 1 |
| Bonferroni correction for 8 CpGs, sex, and 3 PTSD symptom clusters | 1 | 1 | 0.048 | 1 | 1 | 1 | 1 | 1 |
| NR3C1 DNA methylation | ||||||||
| Mean | 2.86 | 1.52 | 2.84 | 1.01 | 1.77 | 3.52 | 4.88 | 1.83 |
| SD | 1.32 | 1.55 | 1.56 | 1.45 | 1.58 | 1.81 | 1.82 | 1.56 |
aCorrelation of NR3C1 gene promoter DNA methylation with sum of PTSD-specific symptom subscores according to PDS and DSM-IV. To account for trauma load, PTSD symptom cluster scores were divided by the sum of lifetime traumatic event types. Spearman's correlation ρs coefficients and the corresponding nominal and Bonferroni-corrected p values are shown for each symptom cluster.
bSum of specific symptom clusters corrected for total number of lifetime traumatic events.
cPDS score is not an additional symptom measure, as it represents the sum of the subscores of Intrusions, Avoidance, and Hyperarousal.
***Correlation is nominally significant at the 0.001 level (2-tailed, Spearman's ρ).
**Correlation is nominally significant at the 0.01 level (2-tailed, Spearman's ρ).
*Correlation is nominally significant at the 0.05 level (2-tailed, Spearman's ρ).
Table 2.
Association of PTSD symptom clusters and DNA methylation of NR3C1 gene promoter in Rwandan womena
| PTSD symptom clusters (nfemales = 69) | NR3C1_CpG1 | NR3C1_CpG2 | NR3C1_CpG3 | NR3C1_CpG4 | NR3C1_CpG5 | NR3C1_CpG6 | NR3C1_CpG7 | NR3C1_CpG8 |
|---|---|---|---|---|---|---|---|---|
| Intrusionsb | ||||||||
| Spearman's ρ | −0.166 | 0.013 | −0.073 | 0.037 | 0.065 | 0.033 | −0.071 | 0.019 |
| p value | ||||||||
| Nominal | 0.173 | 0.916 | 0.553 | 0.763 | 0.598 | 0.785 | 0.561 | 0.876 |
| Bonferroni correction for 8 CpGs | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Bonferroni correction for 8 CpGs, sex, and 3 PTSD symptom clusters | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Avoidanceb | ||||||||
| Spearman's ρ | −0.116 | −0.118 | −0.139 | 0.006 | −0.059 | −0.058 | −0.045 | 0.092 |
| p value | ||||||||
| P-value (nominal) | 0.344 | 0.333 | 0.256 | 0.963 | 0.633 | 0.635 | 0.714 | 0.45 |
| Bonferroni correction for 8 CpGs | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Bonferroni correction for 8 CpGs, sex, and 3 PTSD symptom clusters | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Hyperarousalb | ||||||||
| Spearman's ρ | −0.003 | −0.014 | −0.052 | 0.053 | 0.002 | −0.04 | −0.057 | −0.033 |
| p value | ||||||||
| Nominal | 0.983 | 0.912 | 0.672 | 0.667 | 0.99 | 0.746 | 0.642 | 0.79 |
| Bonferroni correction for 8 CpGs | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Bonferroni correction for 8 CpGs, sex, and 3 PTSD symptom clusters | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| PDS scoreb,c | ||||||||
| Spearman's ρ | −0.114 | −0.046 | −0.089 | 0.011 | 0.003 | −0.006 | −0.063 | 0.054 |
| p value | ||||||||
| Nominal | 0.352 | 0.706 | 0.469 | 0.928 | 0.982 | 0.959 | 0.605 | 0.657 |
| Bonferroni correction for 8 CpGs | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Bonferroni correction for 8 CpGs, sex, and 3 PTSD symptom clusters | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| NR3C1 DNA methylation | ||||||||
| Mean | 3.23 | 1.90 | 3.55 | 1.10 | 2.42 | 4.16 | 5.43 | 2.29 |
| SD | 1.62 | 1.81 | 1.85 | 1.59 | 1.83 | 2.32 | 1.96 | 1.90 |
aCorrelation of NR3C1 gene promoter DNA methylation with sum of PTSD-specific symptom subscores according to PDS and DSM-IV. To account for trauma load, PTSD symptom cluster scores were divided by the sum of lifetime traumatic event types. Spearman's correlation ρ coefficients and the corresponding nominal and Bonferroni-corrected p values are shown for each symptom cluster.
bSum of specific symptom clusters corrected for total number of traumatic events in a lifetime.
cPDS score is not an additional symptom measure, as it represents the sum of the subscores of Intrusions, Avoidance and Hyperarousal.
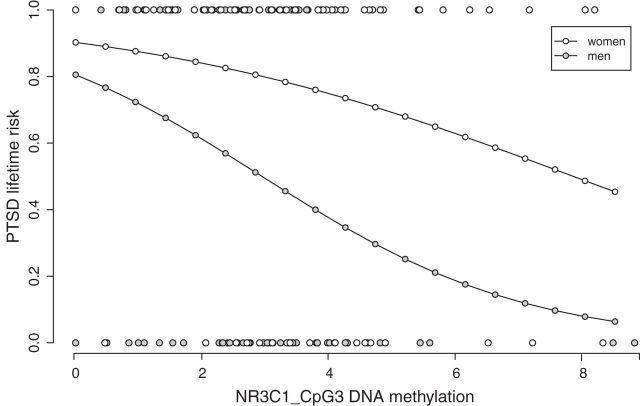
Furthermore, we investigated whether methylation levels at CpG3 were significantly associated with lifetime PTSD risk (taking into account the total number of experienced traumatic event types; see Materials and Methods). We found that higher methylation levels were significantly associated with a lower lifetime PTSD risk in men (binary logistic regression model, β = −0.519, Wald χ2 = 7.0, p = 0.008; Fig. 3), but not in women (β = −0.170, Wald χ2 = 1.4, p = 0.243; Fig. 3).
Figure 3.
Fitted values of probability for lifetime PTSD risk against DNA methylation at the NGFI-A binding site of the NR3C1 promoter for males and females. Lifetime PTSD risk was assessed against NR3C1 CpG3 DNA methylation and the number of lifetime traumatic event types via binary logistic regression (nmales = 83, pnominal_males = 0.008, βmales = −0.519; nfemales = 69, pnominal_females = 0.243, βfemales = −0.170). This graph also contains the raw CpG methylation values obtained in participants without (bottom line) and with (top line) PTSD.
Interindividual differences in methylation levels at CpG3 were not significantly associated with the number of traumatic life event types (p = 0.23 and 0.76 for men and women, respectively). Furthermore, age was not significantly associated with DNA methylation in either gender (p = 0.10 and 0.14 for men and women, respectively).
DNA methylation at the NR3C1 promoter, memory performance, and fMRI activity during picture recognition
Because in genocide survivors NR3C1 CpG3 methylation was strongly associated with the intrusive memory aspect of PTSD and given the role of glucocorticoid signaling in memory processes (de Quervain et al., 2009), we further examined the relationship of NR3C1 DNA methylation variation to memory processes in healthy humans. Specifically, we investigated whether NR3C1 CpG3 methylation levels in healthy individuals were associated with memory performance and brain activation during a picture-recognition task in the fMRI scanner.
NR3C1 CpG3 DNA methylation was negatively correlated with the correct recognition of previously seen pictures in men but not in women (men: β = −0.455, p = 0.012; women: β = −0.065, p = 0.667; Table 3). Furthermore, DNA methylation at this site in men was significantly associated with arousal during encoding of negative pictures, but not of neutral or positive pictures (Table 3). After correcting for the valence-specific arousal rating, recognition of previously seen pictures was significantly negatively correlated with NR3C1 CpG3 DNA methylation independently of the valence category in men but not in women (Table 3). Free-recall performance was not related to methylation levels at NR3C1 CpG3 (men: β = −0.291, p = 0.16; women: β = −0.048, p = 0.74).
Table 3.
NR3C1 CpG3 DNA methylation and recognition performance in the Swiss samplea
|
NR3C1 CpG3 |
||
|---|---|---|
| Women (nfemales = 47) | Men (nmales = 25) | |
| Recognition of all pictures | ||
| β | 0.065 | −0.455 |
| p value | 0.667 | 0.012* |
| Arousal for positive pictures | ||
| β | −0.148 | 0.343 |
| p value | 0.320 | 0.094 |
| Arousal for negative pictures | ||
| β | −0.009 | 0.431 |
| p value | 0.954 | 0.031* |
| Arousal for neutral pictures | ||
| β | −0.135 | 0.266 |
| p value | 0.366 | 0.199 |
| Recognition of positive pictures corrected for arousal | ||
| β | −0.036 | −0.447 |
| p value | 0.817 | 0.041* |
| Recognition of negative pictures corrected for arousal | ||
| β | 0.289 | −0.481 |
| p value | 0.050* | 0.010** |
| Recognition of neutral pictures corrected for arousal | ||
| β | −0.014 | −0.643 |
| p value | 0.927 | 0.002** |
aRecognition corrected for arousal. To account for methylation-dependent differences in arousal during recognition of previously seen pictures, both arousal and NR3C1 CpG3 DNA methylation were included in the linear model. We modeled valence-specific recognition performance as dependent on NR3C1 DNA methylation and valence-specific arousal. β Coefficients and the corresponding p values are shown for each recognition and arousal category (DNA methylation mean ± SD: μNR3C1 CpG3 Females = 3.20 ± 1.29, μNR3C1 CpG3 Males = 3.28 ± 1.21).
**Correlation is significant at the 0.01 level.
*Correlation is significant at the 0.05 level.
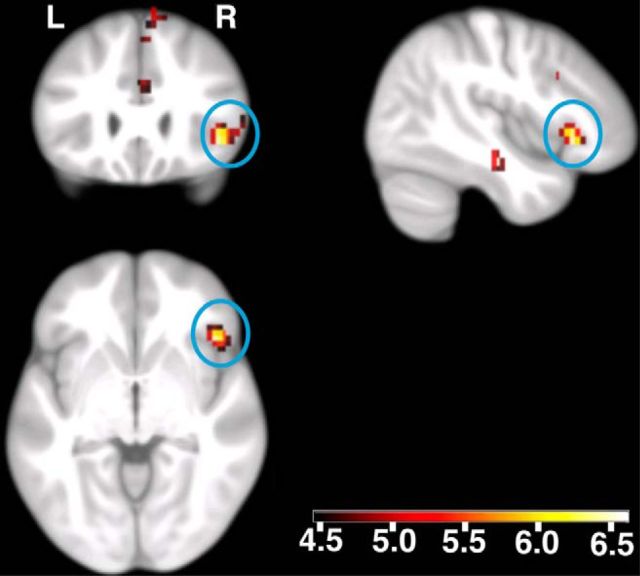
Next, we performed a methylation-dependent fMRI analysis during successful memory recognition (see Materials and Methods). In men, we found a whole-brain, family-wise error (FWE) multiple-comparison corrected (pFWE corrected < 0.05, puncorrected < 0.001) positive correlation between methylation levels at the CpG3 site of the NR3C1 promoter and brain activity related to the successful recognition of previously seen pictures in the pars triangularis and pars orbitalis of the inferior frontal gyrus, in the cuneus, and in the vicinity of the superior temporal and superior frontal cortices (Fig. 4, Table 4). No voxels correlated significantly with methylation values in the opposite direction. We did not observe significant associations between methylation levels at the CpG3 site and brain activity in females. Moreover, we did not find significant correlations of individual methylation levels with encoding-related brain activity.
Figure 4.
Methylation-dependent differences in brain activity related to successful recognition of previously seen pictures in healthy men. Displayed are voxels with a positive correlation between methylation values (at NR3C1_CpG3) and activity, using color-coded t values. The blue circles show the activation in the pars triangularis and pars orbitalis of the inferior frontal gyrus (centered at 44, 25, −4; peak pFWE corrected < 0.05; displayed at puncorrected < 0.001; a random-effects, linear-regression model). Activations are overlaid on coronal (upper left), sagittal (upper right), and axial sections of the study-specific group template (see main text). L, Left side of the brain; R, right side of the brain.
Table 4.
Methylation-dependent differences in brain activity related to the successful recognition of previously seen pictures in healthy male individualsa
| Maximum t value within cluster | Regional correspondence of the maximum | MNI coordinates at maximum |
||
|---|---|---|---|---|
| X | Y | Z | ||
| 6.4174 | Cortex-right hemisphere-pars orbitalis (36%), cortex-right hemisphere-pars triangularis (31%) of the inferior frontal gyrus | 44 | 24.75 | −4 |
| 6.5307 | Near superior temporal cortex | 44 | −16.5 | −16 |
| 6.3278 | Cortex-left hemisphere-cuneus (48%), cortex-left hemisphere-pericalcarine (10%) | 0 | −85.25 | 12 |
| 6.4978 | Near superior frontal cortex | 19.25 | 38.5 | 32 |
apFWE whole-brain corrected < 0.05. Swiss sample, nmales = 24. Regions and probabilities in accordance to the in-house atlas (see Materials and Methods).
Finally, we compared methylation levels of NR3C1 promoter CpG3 site between the Rwandan and the Swiss sample. Importantly, we did not find a significant difference in median, distribution, or variability between the samples (Kruskal–Wallis χ2 = 1.392, p = 0.24; Kolmogorov–Smirnov D = 0.150, p = 0.22; Siegel–Tukey W = 4829.5, p = 0.29). These findings suggest that methylation at this specific site is a stable trait and likely pre-existed the traumatic event in the Rwandan population.
Expression of the GR gene and DNA methylation at the NR3C1 gene promoter
Finally, we tested whether DNA methylation on the CpG3 site of the NR3C1 promoter was associated with NR3C1 gene expression in a subset of 24 healthy (12 females and 12 males) subjects. DNA methylation at the CpG3 site correlated significantly with both exon 1F and total NR3C1 expression (β = −0.812, p = 0.001 and β = −0.759, p = 0.003, respectively; in both males and females, p ≤ 0.05). Thus, DNA methylation at the NGFI-A transcriptional factor-binding site of NR3C1 correlated negatively with NR3C1 expression in saliva.
Discussion
The present study indicates that DNA methylation at the NR3C1 exon 1F promoter is linked to traumatic memories and PTSD risk in male, but not in female genocide survivors. These methylation differences are located at the NGFI-A transcriptional factor binding sequence and the methylation levels correlated negatively with the expression of NR3C1, indicating the functional relevance of the methylation status at this site.
Studies in rodents have shown that specific 5′ NR3C1-17 promoter sites are targets of DNA methylation, leading to suppressed NGFI-A binding and consequently decreased NR3C1 gene expression (Weaver et al., 2007). DNA hypermethylation at these sites has been associated with low quality of received maternal care in adult male offspring (female offspring were not examined; Weaver et al., 2004), as well as prenatal stress exposure (Mueller and Bale, 2008). Hypermethylation at the human ortholog NR3C1-1F promoter NGFI-A binding site was likewise associated to perinatal stress and maternal depression (Oberlander et al., 2008). A recent study in male combat veterans reported that DNA methylation at NR3C1-1F in peripheral blood mononuclear cells was inversely correlated with PTSD risk, NR3C1 gene expression, cortisol decline in response to dexamethasone, and clinical measures of PTSD, including poorer sleep (Yehuda et al., 2014). Another study reported that DNA methylation of NR3C1-1B and NR3C1-1C promoters in sorted T-lymphocytes is also inversely correlated with PTSD risk and morning cortisol levels in individuals with different trauma background (Labonté et al., 2014).
Epigenetic marks at the NR3C1 locus are set during a sensitive period around birth and seem to remain stable throughout life in rodents (Weaver et al., 2004; Mueller and Bale, 2008; Suderman et al., 2012) and humans (Oberlander et al., 2008; McGowan et al., 2009; Radtke et al., 2011; Suderman et al., 2012). These findings suggest that, in the present study, NR3C1 methylation differences may have existed before the traumatic events. Alternatively, trauma exposure might have triggered mechanisms that lead to altered epigenetic patterns. In the present study, interindividual differences in methylation levels at the NR3C1-1F promoter were not related to traumatic load. Likewise, Yehuda et al. also reported no significant association between trauma exposure and NR3C1-1F promoter DNA methylation (Yehuda et al., 2014). Furthermore, we did not observe significant differences in medians and interindividual variability of NR3C1-1F promoter methylation between Rwandan genocide survivors and healthy individuals not exposed to trauma. Together, these findings suggest that the differences in methylation levels pre-existed the traumatic events, supporting the idea that alterations in the HPA axis regulation represent at least partially a pretrauma risk factor for the disorder (Yehuda, 2009; de Quervain et al., 2009). Pre-existing differences in DNA methylation at NR3C1 may be a result of interplay between selected types of early adversity and specific timing of exposure. They may also reflect other types of risks associated with the HPA axis inhibition (Yehuda, 2009). Finally, one possibility is that NR3C1-1F promoter DNA methylation differences are a result of development/early life-associated stochastic epigenetic variation leading to differences in disease susceptibility (Feinberg and Irizarry, 2010). Characterization of the causal pathways involved in epigenetic modifications and the relative stability and reversibility of such changes require more research (Zhang et al., 2010; Hunter and McEwen, 2013).
Previous studies investigated epigenetic modifications associated with PTSD in peripheral blood (Chang et al., 2012; Klengel et al., 2013; Mehta et al., 2013; Labonté et al., 2014; Yehuda et al., 2014). In this study we used DNA isolated from saliva. The source of DNA in saliva is a mixture of ectodermal-origin epithelial cells and white blood cells (Zhou et al., 2011). Several studies suggest that DNA methylation signatures in genomic regions rich in cytosine-guanine dinucleotides (as it is the case with NR3C1-1F promoter) generally show stable epigenetic signatures across brain and nonbrain tissues (Ladd-Acosta et al., 2007; Mill et al., 2008; Lister et al., 2009; Dempster et al., 2011; Davies et al., 2012). Indeed, it has been shown that childhood adversity is related to NR3C1-1F promoter methylation both in peripheral tissue (Oberlander et al., 2008; Radtke et al., 2011) and in the brain (McGowan et al., 2009). Moreover, we have found that interindividual differences in methylation of the NR3C1-1F promoter in DNA isolated from saliva were associated with differences in brain activity. Together, these findings suggest that with regard to NR3C1-1F methylation, similar patterns may be present across tissues. Thus, it may be possible to a certain extent to use nonbrain tissue for the investigation of certain CNS traits, such as psychiatric disorders.
As expected, we found that increased DNA methylation at the NR3C1-1F promoter negatively correlated with expression of NR3C1. It is known that an acute and timed activation of GRs is crucially involved in memory formation (Roozendaal, 2000; Roozendaal et al., 2006). Thus, increased NR3C1 promoter methylation might be related to reduced memory formation, including traumatic memory after a traumatic event. Indeed, we found that increased NR3C1-1F CpG3 methylation was associated with less intrusive memory (genocide survivors) and less picture-recognition memory (healthy population), but only in males. It is unlikely that gender differences in methylation or expression may have accounted for the lack of relationship between methylation and memory in women, because the correlation between CpG3 methylation and NR3C1 expression was independent of gender. We hypothesize that the gender-specific findings reported herein occur post-transcriptionally, possibly reflecting gender-specific effects of glucocorticoids on memory. Indeed, the finding that NR3C1-1F methylation was related to memory only in men is in line with reports that found glucocorticoid effects on memory to be more pronounced in men than in women, or even restricted to men (Andreano and Cahill, 2006; Preuss and Wolf, 2009; Cornelisse et al., 2011). The reason for this gender-dependent difference in glucocorticoid effects is not well understood. A possible interaction between glucocorticoids and sex hormones has been proposed (Preuss and Wolf, 2009), and further efforts are necessary to address sex-related differences in glucocorticoid signaling and its molecular signatures.
The present findings point to a relationship of NR3C1-1F promoter methylation with memory processes also in healthy humans. Specifically, we found that methylation levels at the NR3C1-1F correlated negatively with recognition memory in men, but not in women. This relationship was independent of valence, which is in line with previous findings indicating that the memory-modulating effects of elevated glucocorticoid levels can also affect neutral material when the learning context is emotionally arousing (Preuss and Wolf, 2009). The neuroimaging findings indicated that male subjects with higher methylation levels had increased activation in the right ventrolateral prefrontal cortex (VLPFC) and the cuneus. Interestingly, the right VLPFC has been discussed as a brain region involved in retrieval attempt and effort rather than retrieval success (Taylor et al., 2004; Badre and Wagner, 2007), which is in line with the present finding of increased activation in this region in subjects with higher methylation levels and less successful recognition performance. Moreover, a recent meta-analysis has pointed to an involvement of the VLPFC in the neurocircuitry of PTSD (Hayes et al., 2012). Specifically, patients with PTSD show decreased activity in the inferior frontal gyrus compared with trauma-exposed controls without PTSD when reliving one's traumatic event.
The relationship between glucocorticoids, memory, and PTSD is complex and not fully understood. Whereas high numbers of GRs before trauma have been found to be a risk factor for developing PTSD (van Zuiden et al., 2011), a pharmacological elevation of glucocorticoid levels during or after a traumatic event seems to prevent and reduce PTSD symptoms (Schelling et al., 2001, 2004; Aerni et al., 2004). This may have to do with differential effects of glucocorticoids depending on timing and the memory phase affected (de Quervain et al., 2009; Joëls et al., 2011). A timed activation of GRs is important for memory consolidation and may lead to strong aversive memories in case of a traumatic event. On the other hand, elevated glucocorticoid levels have been shown to impair memory retrieval of emotionally arousing information, which may lead to reduced traumatic memories (de Quervain et al., 2009). In addition to the glucocorticoid effects on emotional memory, evidence indicates that glucocorticoids can have effects on emotional processing: a recent study suggested that the presence of elevated levels of glucocorticoids at the time of acute stress confers protection against the delayed enhancing effect of stress on amygdala synaptic connectivity and anxiety-like behavior (Rao et al., 2012). Thus, altered glucocorticoid signaling may have differential effects on the development and symptoms of PTSD, depending on the emotional and cognitive processes affected.
In conclusion, we provide evidence that DNA methylation at the NR3C1-1F promoter is preferentially related to the memory aspects of PTSD symptomatology and PTSD risk in men. Moreover, we found that the same epigenetic modification was related to memory functions in healthy subjects, suggesting that this modification possibly affects traumatic memory and PTSD via a modulation of memory processes. The present findings may add to the understanding of individual PTSD risk factors and suggest that NR3C1 DNA methylation may represent a biological marker for traumatic memories and PTSD.
Footnotes
This work was supported by the Swiss National Science Foundation (Sinergia Grants CRSIK0_122691, CRSI33_130080 to D.J.-F.D.Q. and A.P.) and by the German Research Foundation (grants to I.-T.K. and T.E.). V.V. was supported by a Werner Siemens Foundation (Zug, Switzerland) PhD grant and Opportunities for Excellence PhD Program of the Biozentrum.
The authors declare no competing financial interest.
References
- Ackermann S, Heck A, Rasch B, Papassotiropoulos A, de Quervain DJ. The BclI polymorphism of the glucocorticoid receptor gene is associated with emotional memory performance in healthy individuals. Psychoneuroendocrinology. 2013;38:1203–1207. doi: 10.1016/j.psyneuen.2012.10.009. [DOI] [PubMed] [Google Scholar]
- Aerni A, Traber R, Hock C, Roozendaal B, Schelling G, Papassotiropoulos A, Nitsch RM, Schnyder U, de Quervain DJ. Low-dose cortisol for symptoms of posttraumatic stress disorder. Am J Psychiatry. 2004;161:1488–1490. doi: 10.1176/appi.ajp.161.8.1488. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association. Diagnostical and statistical manual of mental disorders, fourth revised edition (DSM-IV-TR) Washington, DC: American Psychiatric Association; 2000. [Google Scholar]
- Andreano JM, Cahill L. Glucocorticoid release and memory consolidation in men and women. Psychol Sci. 2006;17:466–470. doi: 10.1111/j.1467-9280.2006.01729.x. [DOI] [PubMed] [Google Scholar]
- Badre D, Wagner AD. Left ventrolateral prefrontal cortex and the cognitive control of memory. Neuropsychologia. 2007;45:2883–2901. doi: 10.1016/j.neuropsychologia.2007.06.015. [DOI] [PubMed] [Google Scholar]
- Brewin CR. The nature and significance of memory disturbance in posttraumatic stress disorder. Annu Rev Clin Psychol. 2011;7:203–227. doi: 10.1146/annurev-clinpsy-032210-104544. [DOI] [PubMed] [Google Scholar]
- Chang SC, Koenen KC, Galea S, Aiello AE, Soliven R, Wildman DE, Uddin M. Molecular variation at the SLC6A3 locus predicts lifetime risk of PTSD in the Detroit Neighborhood Health Study. PLoS One. 2012;7:e39184. doi: 10.1371/journal.pone.0039184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelisse S, van Stegeren AH, Joëls M. Implications of psychosocial stress on memory formation in a typical male versus female student sample. Psychoneuroendocrinology. 2011;36:569–578. doi: 10.1016/j.psyneuen.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Davies MN, Volta M, Pidsley R, Lunnon K, Dixit A, Lovestone S, Coarfa C, Harris RA, Milosavljevic A, Troakes C, Al-Sarraj S, Dobson R, Schalkwyk LC, Mill J. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol. 2012;13:R43. doi: 10.1186/gb-2012-13-6-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempster EL, Pidsley R, Schalkwyk LC, Owens S, Georgiades A, Kane F, Kalidindi S, Picchioni M, Kravariti E, Toulopoulou T, Murray RM, Mill J. Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Hum Mol Genet. 2011;20:4786–4796. doi: 10.1093/hmg/ddr416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Quervain DJ. Glucocorticoid-induced inhibition of memory retrieval: implications for posttraumatic stress disorder. Ann NY Acad Sci. 2006;1071:216–220. doi: 10.1196/annals.1364.016. [DOI] [PubMed] [Google Scholar]
- de Quervain DJ, Kolassa IT, Ertl V, Onyut PL, Neuner F, Elbert T, Papassotiropoulos A. A deletion variant of the alpha2b-adrenoceptor is related to emotional memory in Europeans and Africans. Nat Neurosci. 2007;10:1137–1139. doi: 10.1038/nn1945. [DOI] [PubMed] [Google Scholar]
- de Quervain DJ, Aerni A, Schelling G, Roozendaal B. Glucocorticoids and the regulation of memory in health and disease. Front Neuroendocrinol. 2009;30:358–370. doi: 10.1016/j.yfrne.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, Albert MS, Killiany RJ. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Ertl V, Pfeiffer A, Saile R, Schauer E, Elbert T, Neuner F. Validation of a mental health assessment in an African conflict population. Psychol Assess. 2010;22:318–324. doi: 10.1037/a0018810. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Irizarry RA. Stochastic epigenetic variation as a driving force of development, evolutionary adaptation, and disease. Proc Natl Acad Sci U S A. 2010;107:1757–1764. doi: 10.1073/pnas.0906183107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM. Whole brain segmentation. Neuron. 2002;33:341–355. doi: 10.1016/S0896-6273(02)00569-X. [DOI] [PubMed] [Google Scholar]
- Foa EB, Cashman L, Jaycox L, Perry K. The validation of a self-report measure of posttraumatic stress disorder: The Posttraumatic Diagnostic Scale. Psychol Assess. 1997;9:445–451. doi: 10.1037/1040-3590.9.4.445. [DOI] [Google Scholar]
- Hauer D, Weis F, Papassotiropoulos A, Schmoeckel M, Beiras-Fernandez A, Lieke J, Kaufmann I, Kirchhoff F, Vogeser M, Roozendaal B, Briegel J, de Quervain D, Schelling G. Relationship of a common polymorphism of the glucocorticoid receptor gene to traumatic memories and posttraumatic stress disorder in patients after intensive care therapy. Crit Care Med. 2011;39:643–650. doi: 10.1097/CCM.0b013e318206bae6. [DOI] [PubMed] [Google Scholar]
- Hayes JP, Hayes SM, Mikedis AM. Quantitative meta-analysis of neural activity in posttraumatic stress disorder. Biol Mood Anxiety Disord. 2012;2:9. doi: 10.1186/2045-5380-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter RG, McEwen BS. Stress and anxiety across the lifespan: structural plasticity and epigenetic regulation. Epigenomics. 2013;5:177–194. doi: 10.2217/epi.13.8. [DOI] [PubMed] [Google Scholar]
- Joëls M, Fernandez G, Roozendaal B. Stress and emotional memory: a matter of timing. Trends Cogn Sci. 2011;15:280–288. doi: 10.1016/j.tics.2011.04.004. [DOI] [PubMed] [Google Scholar]
- Klengel T, Mehta D, Anacker C, Rex-Haffner M, Pruessner JC, Pariante CM, Pace TW, Mercer KB, Mayberg HS, Bradley B, Nemeroff CB, Holsboer F, Heim CM, Ressler KJ, Rein T, Binder EB. Allele-specific FKBP5 DNA demethylation mediates gene–childhood trauma interactions. Nat Neurosci. 2013;16:33–41. doi: 10.1038/nn.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolassa I-T, Ertl V, Eckart C, Kolassa S, Onyut LP, Elbert T. Spontaneous remission from PTSD depends on the number of traumatic event types experienced. Psychol Trauma. 2010;2:169–174. doi: 10.1037/a0019362. [DOI] [Google Scholar]
- Labonté B, Azoulay N, Yerko V, Turecki G, Brunet A. Epigenetic modulation of glucocorticoid receptors in posttraumatic stress disorder. Transl Psychiatry. 2014;4:e368. doi: 10.1038/tp.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd-Acosta C, Pevsner J, Sabunciyan S, Yolken RH, Webster MJ, Dinkins T, Callinan PA, Fan JB, Potash JB, Feinberg AP. DNA methylation signatures within the human brain. Am J Hum Genet. 2007;81:1304–1315. doi: 10.1086/524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang PJ, Bradley MM, Cuthbert BN. International affective picture system (IAPS): technical manual and affective ratings. In: Lang PJ, Bradley MM, Cuthbert BN, editors. Technical report A-8. Gainesville, FL: University of Florida; 1999. [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan PO, Sasaki A, D'Alessio AC, Dymov S, Labonté B, Szyf M, Turecki G, Meaney MJ. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meewisse ML, Reitsma JB, de Vries GJ, Gersons BP, Olff M. Cortisol and post-traumatic stress disorder in adults: systematic review and meta-analysis. Br J Psychiatry. 2007;191:387–392. doi: 10.1192/bjp.bp.106.024877. [DOI] [PubMed] [Google Scholar]
- Mehta D, Klengel T, Conneely KN, Smith AK, Altmann A, Pace TW, Rex-Haffner M, Loeschner A, Gonik M, Mercer KB, Bradley B, Müller-Myhsok B, Ressler KJ, Binder EB. Childhood maltreatment is associated with distinct genomic and epigenetic profiles in posttraumatic stress disorder. Proc Natl Acad Sci U S A. 2013;110:8302–8307. doi: 10.1073/pnas.1217750110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mill J, Tang T, Kaminsky Z, Khare T, Yazdanpanah S, Bouchard L, Jia P, Assadzadeh A, Flanagan J, Schumacher A, Wang SC, Petronis A. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet. 2008;82:696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Movassagh M, Choy MK, Goddard M, Bennett MR, Down TA, Foo RS. Differential DNA methylation correlates with differential expression of angiogenic factors in human heart failure. PLoS One. 2010;5:e8564. doi: 10.1371/journal.pone.0008564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller BR, Bale TL. Sex-specific programming of offspring emotionality after stress early in pregnancy. J Neurosci. 2008;28:9055–9065. doi: 10.1523/JNEUROSCI.1424-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuner F, Onyut PL, Ertl V, Odenwald M, Schauer E, Elbert T. Treatment of posttraumatic stress disorder by trained lay counselors in an African refugee settlement: a randomized controlled trial. J Consult Clin Psychol. 2008;76:686–694. doi: 10.1037/0022-006X.76.4.686. [DOI] [PubMed] [Google Scholar]
- Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S, Devlin AM. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics. 2008;3:97–106. doi: 10.4161/epi.3.2.6034. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitman RK, Rasmusson AM, Koenen KC, Shin LM, Orr SP, Gilbertson MW, Milad MR, Liberzon I. Biological studies of post-traumatic stress disorder. Nat Rev Neurosci. 2012;13:769–787. doi: 10.1038/nrn3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss D, Wolf OT. Post-learning psychosocial stress enhances consolidation of neutral stimuli. Neurobiol Learn Mem. 2009;92:318–326. doi: 10.1016/j.nlm.2009.03.009. [DOI] [PubMed] [Google Scholar]
- Radtke KM, Ruf M, Gunter HM, Dohrmann K, Schauer M, Meyer A, Elbert T. Transgenerational impact of intimate partner violence on methylation in the promoter of the glucocorticoid receptor. Transl Psychiatry. 2011;1:e21. doi: 10.1038/tp.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao RP, Anilkumar S, McEwen BS, Chattarji S. Glucocorticoids protect against the delayed behavioral and cellular effects of acute stress on the amygdala. Biol Psychiatry. 2012;72:466–475. doi: 10.1016/j.biopsych.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohleder N, Joksimovic L, Wolf JM, Kirschbaum C. Hypocortisolism and increased glucocorticoid sensitivity of pro-inflammatory cytokine production in Bosnian war refugees with posttraumatic stress disorder. Biol Psychiatry. 2004;55:745–751. doi: 10.1016/j.biopsych.2003.11.018. [DOI] [PubMed] [Google Scholar]
- Roozendaal B. 1999 Curt P. Richter award. Glucocorticoids and the regulation of memory consolidation. Psychoneuroendocrinology. 2000;25:213–238. doi: 10.1016/S0306-4530(99)00058-X. [DOI] [PubMed] [Google Scholar]
- Roozendaal B, Okuda S, de Quervain DJ, McGaugh JL. Glucocorticoids interact with emotion-induced noradrenergic activation in influencing different memory functions. Neuroscience. 2006;138:901–910. doi: 10.1016/j.neuroscience.2005.07.049. [DOI] [PubMed] [Google Scholar]
- Schelling G, Briegel J, Roozendaal B, Stoll C, Rothenhäusler HB, Kapfhammer HP. The effect of stress doses of hydrocortisone during septic shock on posttraumatic stress disorder in survivors. Biol Psychiatry. 2001;50:978–985. doi: 10.1016/S0006-3223(01)01270-7. [DOI] [PubMed] [Google Scholar]
- Schelling G, Roozendaal B, De Quervain DJ. Can posttraumatic stress disorder be prevented with glucocorticoids? Ann NY Acad Sci. 2004;1032:158–166. doi: 10.1196/annals.1314.013. [DOI] [PubMed] [Google Scholar]
- Suderman M, McGowan PO, Sasaki A, Huang TC, Hallett MT, Meaney MJ, Turecki G, Szyf M. Conserved epigenetic sensitivity to early life experience in the rat and human hippocampus. Proc Natl Acad Sci U S A. 2012;109:17266–17272. doi: 10.1073/pnas.1121260109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SF, Welsh RC, Wager TD, Phan KL, Fitzgerald KD, Gehring WJ. A functional neuroimaging study of motivation and executive function. Neuroimage. 2004;21:1045–1054. doi: 10.1016/j.neuroimage.2003.10.032. [DOI] [PubMed] [Google Scholar]
- Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat Protoc. 2007;2:2265–2275. doi: 10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]
- Turner JD, Muller CP. Structure of the glucocorticoid receptor (NR3C1) gene 5′ untranslated region: identification, and tissue distribution of multiple new human exon 1. J Mol Endocrinol. 2005;35:283–292. doi: 10.1677/jme.1.01822. [DOI] [PubMed] [Google Scholar]
- van Rossum EF, Koper JW, van den Beld AW, Uitterlinden AG, Arp P, Ester W, Janssen JA, Brinkmann AO, de Jong FH, Grobbee DE, Pols HA, Lamberts SW. Identification of the BclI polymorphism in the glucocorticoid receptor gene: association with sensitivity to glucocorticoids in vivo and body mass index. Clin Endocrinol (Oxf) 2003;59:585–592. doi: 10.1046/j.1365-2265.2003.01888.x. [DOI] [PubMed] [Google Scholar]
- van Zuiden M, Geuze E, Willemen HL, Vermetten E, Maas M, Heijnen CJ, Kavelaars A. Pre-existing high glucocorticoid receptor number predicting development of posttraumatic stress symptoms after military deployment. Am J Psychiatry. 2011;168:89–96. doi: 10.1176/appi.ajp.2010.10050706. [DOI] [PubMed] [Google Scholar]
- van Zuiden M, Kavelaars A, Geuze E, Olff M, Heijnen CJ. Predicting PTSD: pre-existing vulnerabilities in glucocorticoid-signaling and implications for preventive interventions. Brain Behav Immun. 2013;30:12–21. doi: 10.1016/j.bbi.2012.08.015. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Weaver IC, D'Alessio AC, Brown SE, Hellstrom IC, Dymov S, Sharma S, Szyf M, Meaney MJ. The transcription factor nerve growth factor-inducible protein A mediates epigenetic programming: altering epigenetic marks by immediate-early genes. J Neurosci. 2007;27:1756–1768. doi: 10.1523/JNEUROSCI.4164-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilker S, Kolassa S, Vogler C, Lingenfelder B, Elbert T, Papassotiropoulos A, de Quervain DJ, Kolassa IT. The role of memory-related gene WWC1 (KIBRA) in lifetime posttraumatic stress disorder: evidence from two independent samples from African conflict regions. Biol Psychiatry. 2013;74:664–671. doi: 10.1016/j.biopsych.2013.02.022. [DOI] [PubMed] [Google Scholar]
- Wojdacz TK, Borgbo T, Hansen LL. Primer design versus PCR bias in methylation independent PCR amplifications. Epigenetics. 2009;4:231–234. doi: 10.4161/epi.9020. [DOI] [PubMed] [Google Scholar]
- Yehuda R. Post-traumatic stress disorder. NEJM. 2002;346:108–114. doi: 10.1056/NEJMra012941. [DOI] [PubMed] [Google Scholar]
- Yehuda R. Status of glucocorticoid alterations in post-traumatic stress disorder. Ann NY Acad Sci. 2009;1179:56–69. doi: 10.1111/j.1749-6632.2009.04979.x. [DOI] [PubMed] [Google Scholar]
- Yehuda R, Flory JD, Bierer LM, Henn-Haase C, Lehrner A, Desarnaud F, Makotkine I, Daskalakis NP, Marmar CR, Meaney MJ. Lower methylation of glucocorticoid receptor gene promoter 1F in peripheral blood of veterans suffering from post-traumatic stress disorder. Biol Psychiatry. 2014 doi: 10.1016/j.biopsych.2014.02.006. pii: S0006-3223(14)00100–0. [DOI] [PubMed] [Google Scholar]
- Zhang TY, Hellstrom IC, Bagot RC, Wen X, Diorio J, Meaney MJ. Maternal care and DNA methylation of a glutamic acid decarboxylase 1 promoter in rat hippocampus. J Neurosci. 2010;30:13130–13137. doi: 10.1523/JNEUROSCI.1039-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Li S, Zhou J, Wang L, Song X, Lu X, Wang J, Ye Y, Ying B, Jia Y. DNA profiling in blood, buccal swabs and hair follicles of patients after allogeneic peripheral blood stem cells transplantation. Legal Med. 2011;13:47–51. doi: 10.1016/j.legalmed.2010.09.005. [DOI] [PubMed] [Google Scholar]