Abstract
Prostate cancer (PCa) is the second most common cause of cancer related mortality in men in the United States. As a member of the nuclear steroid receptor family, the androgen receptor (AR), which is a transcription factor with three distinct functional domains (ligand binding [LBD], DNA binding [DBD], and transactivation [TAD]) in its structure, is central to prostate cancer growth and survival. All clinically approved drugs for PCa ultimately targets the AR LBD. Clinically active drugs that target the DBD and TAD have not yet been developed due to multiple factors. Despite such limitations, the last several years have seen a rise in the discovery of molecules that could successfully target these domains. This review aims to present and comprehensively discuss such molecules that have been shown to affect AR signaling through direct or indirect interactions with the AR TAD or the DBD. The compounds discussed here include hairpin polyamides, niclosamide, marine sponge natural products (e.g. EPI compounds), mahanine, VPC compounds, JN compounds, and BET inhibitors. We highlight the significant in vitro and in vivo data found for each compound, and the apparent limitations and/or potential for further development of these agents as PCa therapies.
Keywords: Prostate cancer, CRPC, Androgen receptor, AR signaling axis, AR-TAD, AR-DBD, AR degradation
1 |. INTRODUCTION
1.1 |. Physiologic Role and Regulation of the Androgen Receptor
The androgen receptor (AR) is a ligand-activated DNA-binding transcription factor of a 110 kDa molecular weight, which facilitates the expression of androgen dependent gene products (Figure 1). As a member of the steroid and nuclear hormone receptor super family, it shares many structural and functional features with other receptors such as the glucocorticoid receptor (GR), estrogen receptor (ER), mineralocorticoid receptor, progesterone receptor (PR), and the vitamin D receptor.1,2 The nuclear steroid receptor family consists of three principal domains: 1) the carboxy-terminal ligand binding domain (LBD), 2), the central DNA binding domain (DBD), and 3) the N-terminal transactivation domain (NTD or TAD). Endogenous androgens, such as testosterone and dihydrotestosterone (DHT) bind the LBD, which results in dissociation of heat shock proteins, homodimerization of the AR in a head-to-tail fashion3, translocation to the nucleus, and recognition of and binding to palindromic cis-acting elements in target genes, which are known as androgen response elements (AREs). Transcriptional co-regulators, including both transcriptional activators and repressors, are co-recruited with the AR to ARE sites; the basal transcriptional machinery including RNA polymerase II (RNAP2) and its cofactors, also form a complex with the AR and its coregulators, the net effect of which is gene regulation.
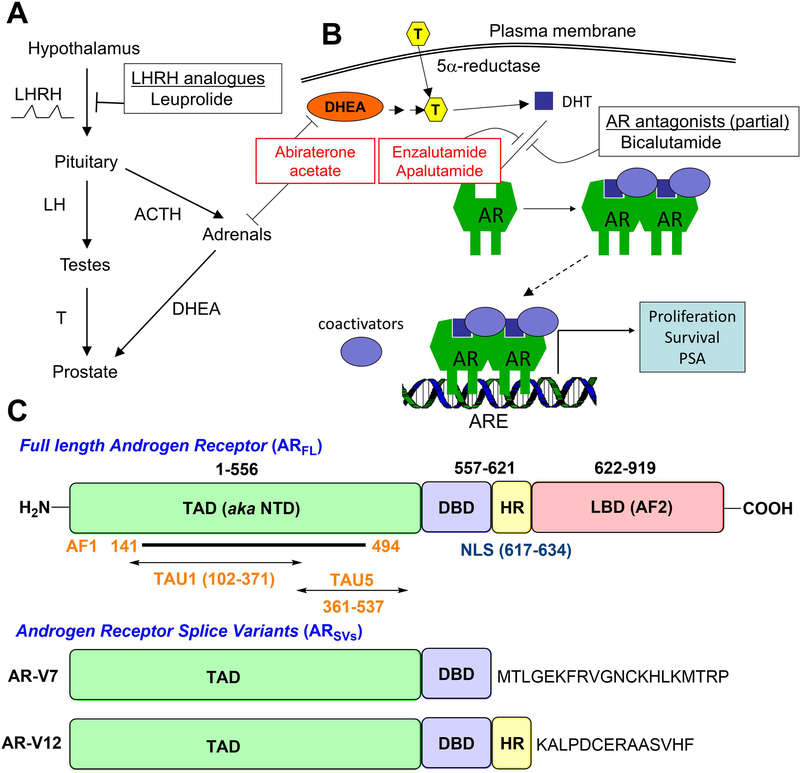
Figure 1.
A) Hormonal regulation of androgen production by the hypothalamus B) AR-dependent gene expression and effect of AR antagonists C) Full length androgen receptor (ARFL) and the clinically relevant splice variants (ARSVs) AR-V7 and AR-V12.10,11 Full length receptor has three distinct domains (C-terminal ligand binding – LBD, DNA binding – DBD, and N-terminal transactivation – TAD/NTD) while the splice variants lack a functional ligand binding domain. Most splice variants such as AR-V7 are constitutively active. (HR = hinge region, NLS = nuclear localization signal)
The principal source of androgens in an adult male is the testes, from where 90% of circulating androgens are derived. Most of the circulating androgens are represented by testosterone, which can be intracellularly converted into the more potent androgen, DHT, by 5α-reductase isoenzymes. Induction of gonadal testosterone synthesis is regulated by production of luteinizing hormone (LH) by the anterior pituitary, which in turn is stimulated by the pulsatile secretion of luteinizing hormone releasing hormone (LHRH) by the hypothalamus. Testosterone has a negative feedback effect on the anterior pituitary and hypothalamus to maintain physiologic levels of serum testosterone. Surgical or medical castration thus prevents production of the main source of androgens. However, about 10% of serum androgens are derived from the adrenal glands, which can synthesize the weak androgens, dehydroepiandrosterone and androstenedione, which in turn can be peripherally converted in target tissues, such as prostatic epithelium, to testosterone. Adrenal androgen production is under the regulation of andrenocorticotropin hormone (ACTH) by the anterior pituitary, which in turn is regulated by hypothalamic secretion of corticotropin releasing hormone (CRH). In addition to weak androgens, steroids produced by the adrenals include mineralocorticoids and glucocorticoids, the latter of which results in negative feedback to the anterior pituitary and hypothalamus to control physiologic adrenal steroid production.
1.2 |. The Role of the AR in Prostate Cancer
Prostate cancer (PCa) is the second most common cause of cancer related mortality in men in the United States. The estimated number of new US cases diagnosed will be 164,690 for the year 2018 with an estimated 29,430 deaths due to PCa.4 The androgen receptor (AR) and the physiological pathways it regulates are central to the initiation and progression of PCa.5 The binding of androgens to the AR initiates AR regulated gene expression that drives PCa growth.
Prostate cancer is most commonly clinically localized at the time of diagnosis, although about 10% of patients present with advanced, metastatic disease (Figure 2)6. Surgery and/or radiation therapy (primary local therapy [PLT]) can effectively treat clinically localized disease, although about one-third of patients relapse after PLT. Whether patients present with metastatic disease or it arises in the context of a recurrence after PLT, the mainstay of treatment for metastatic prostate cancer is endocrine therapy aimed at inhibiting the production or action of androgens that engage and activate the AR. Endocrine therapy is most commonly delivered through surgical or medical castration and is termed androgen deprivation therapy (ADT), which effectively inhibits the androgen production from the testes. The median duration of response to ADT is 18–24 months. Historically, first generation AR competitive antagonists (e.g. flutamide, bicalutamide, and nilutamide) have been combined with ADT (so called combined androgen blockade, CAB), although CAB has not yielded clinically meaningful improvements in PCa outcomes.
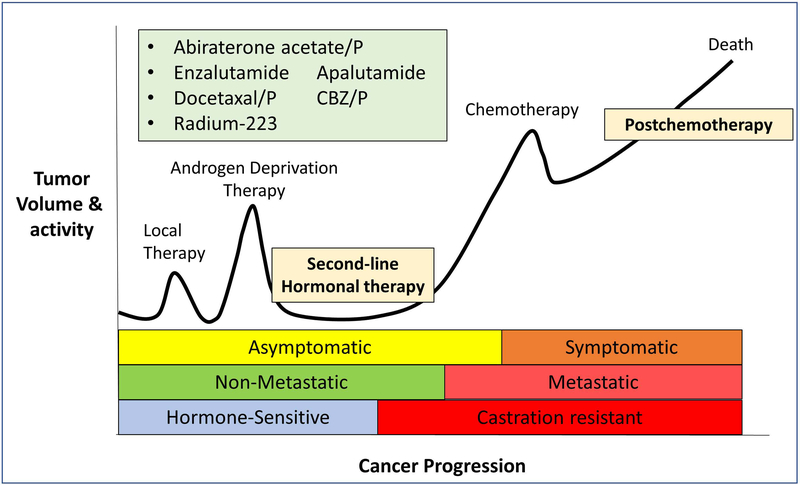
Figure 2.
Progression and the different stages of prostate cancer. While the currently available therapies (commonly used ones indicated in the figure) are quite responsive at the hormone-sensitive stages, metastatic castration-resistant disease has a poor prognosis. Figure updated and redrawn from reference.6 (P = prednisone, CBZ = cabazitaxel)
When PCa progresses despite ADT, it is termed castration resistant prostate cancer (CRPC), which is the lethal form of the disease. Interestingly, CRPC is most often still dependent upon the activation of the AR for its continued progression.7,8 Although the AR has non-genotropic effects, reactivation of AR transcriptional activity represents the principal biochemical driving force that is necessary and sufficient for castration resistance. Multiple non-mutually exclusive mechanisms account for ongoing AR transcriptional activity despite castrate levels of serum testosterone: 1) AR gene amplification, 2) AR mutations that confer agonistic activity of non-traditional ligands (e.g. progesterone, corticosteroids), 3) adrenal androgens, 4) intratumoral androgen production, 5) increased ratio of AR transcriptional activators to repressors, 6) somatic mosaicism, and 7) ligand-independent AR activation through post-translational modification of the AR (e.g. phosphorylation). Another important and more recently identified mechanism underlying castration resistance relates to the expression of constitutively active AR variants that lack a functional LBD.9–11 These AR variants arise from aberrant splicing of AR mRNA and are thus termed AR splice variants (ARSVs). Because the LBD is inhibitory (i.e. the LBD is disinhibited upon ligand binding), ARSVs that lack a functional LBD are rendered constitutively active. In addition to AR-dependent mechanisms of castration resistance, truly AR-independent pathways also exist, although treatments that target these pathways have not yet reached the clinic, and the reader is referred to reviews on this topic.12–14
1.3 |. Current Management of CRPC
Non-endocrine approaches for CRPC have been approved and include cytotoxic chemotherapy such as the taxanes docetaxel and cabazitaxel, systemic radiation in the form of radium-223 (a calcium mimetic that targets the metastases to the bone, the most common site of distant organ involvement), and a cellular vaccine known as Sipuleucel-T. While each of these treatments can improve median overall survival by approximately 2–4 months, none is curative and treatment resistance is inevitable.
Based on the pathophysiologic role of continued AR signaling in CRPC, new drugs that target the AR signaling axis have been brought to the clinic. Abiraterone acetate, an inhibitor of CYP17, an enzyme that governs androgen production, effectively inhibits androgen production from non-gonadal sources including both the adrenals and the tumor tissue itself. These non-gonadal sources of androgen can drive AR activation in mCRPC. Based on its ability to prolong progression free and overall survival, abiraterone acetate in combination with the glucocorticoid, prednisone, has received regulatory approval for metastatic CRPC (mCRPC) for patients who have undergone chemotherapy or are chemotherapy-naïve. More potent, second generation AR competitive antagonists, including enzalutamide and apalutamide, have likewise received approval for CRPC based on improvements in survival. Despite these clinical advancements for the treatment of CRPC, patients still manifest primary and secondary drug resistance to these therapies.
1.4 |. Compounds that Target the AR TAD and DBD
Since the clinical implementation of the aforementioned second-generation endocrine therapies, pre-clinical models as well as sequencing studies of cohorts of mCRPC patients have demonstrated ongoing AR expression and signaling in post-abiraterone/post-enzalutamide mCRPC.15 In fact, the AR is the most frequently mutated gene, and an AR-dependent transcriptional program is reactivated in this context.15 Thus, the AR represents a key driver of castration resistant growth in both newly developed CRPC and post-abiraterone/post-enzalutamide CRPC.
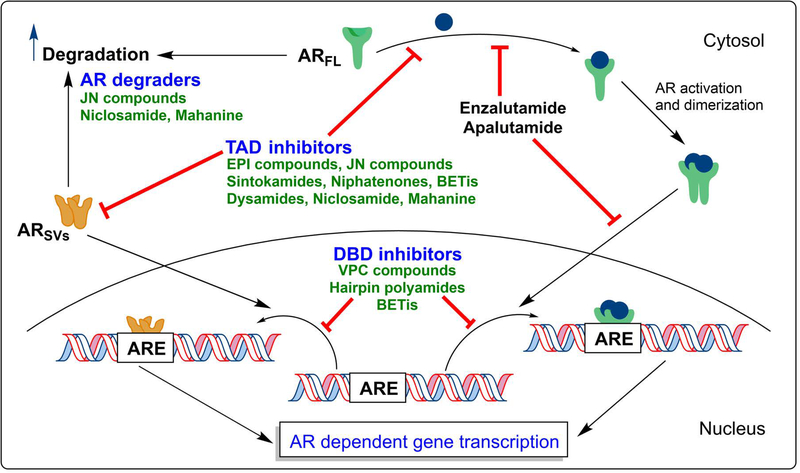
Importantly, all existing endocrine therapies approved for clinical application to PCa mechanistically function through the LBD.16,17 Specifically, these therapies either inhibit ligand production (e.g. castration or abiraterone acetate) or ligand action (e.g. AR competitive antagonists). There have also been some exciting recent developments in targeting the AR-signaling axis by the degradation18 of AR protein with ‘enzalutamide-like’ or ‘enzalutamide-like molecule conjugated’ compounds (e.g. PROTACs [proteolysis-targeting chimeras], SNIPER(AR)s [specific and nongenetic inhibitor of apoptosis protein dependent protein erasers]) in the past few years.19–25 However, therapies that target other domains of the AR, namely the TAD and DBD, have not yet been developed for clinical application nor extensively researched (compared to the targeting of the LBD). Two principal explanations account for this gap in pharmaceutical development. First, the TAD is an intrinsically disordered protein (IDP), so its crystal structure has not been resolved and therefore structure-based drug design is not currently feasible. Second, the DBD shares extreme homology to that of other nuclear steroid receptors, so specificity of drugs for the AR DBD has been considered a challenge. Nonetheless, recent drug development projects premised on either actual or in silico drug screens have resulted in potential candidate compounds that can inhibit the AR activity, through either the TAD or DBD. While there have been several reviews published on this area over the last few years, many have not gone in to comprehensive detail.26–30 In the literature review presented here, we discuss the recent developments in molecules that have shown prominent effects toward the AR-signaling axis through direct or indirect interaction with the AR-TAD or the AR-DBD. We provide an unprecedently comprehensive analysis of the subject area, detailing the associated/impacted biochemical targets/processes, the experimental tools employed to probe the targets, and present a perspective on the future of targeting the AR-signaling axis beyond the AR-LBD.
2 |. HAIRPIN POLYAMIDE ANTAGONISTS OF AR-DNA BINDING
An approach developed by Peter Dervan and co-workers over the last two decades31 for targeting CRPC is to inhibit the AR mediated transcription processes at the DNA level, with direct antagonism of AR-DNA binding using pyrrole-imidazole containing polyamides. The idea was inspired by the function of the natural product distamycin A, a polyamide DNA minor groove binder, first isolated in 1962 from cultures of Streptomyces distallicus.32 Using the concept of differentiating nucleotide base pairs through specific positions of hydrogen bond donor or acceptor sites and through complementary geometrical flexibility to associate with DNA, these hairpin polyamides have evolved as a highly efficient means of specific recognition of DNA fragments.33–35 This sequence specific association of the polyamide disrupts: 1) the association ability of transcription factors, such as the AR, to bind their respective binding site(s) at the DNA, 2) RNA polymerase II activity, and 3) replicative helicase activity.31
2.1 |. Binding Specificity
The ligand induced AR homodimers usually function in binding the AREs by identifying specific DNA half-sites (5′-AGAACA-3′) organized as inverted repeats separated by three nucleotides (IR-3 sequences).2,36 Nickols et al. showed the use of a DNA-binding polyamide PA1 (Figure 3)37 that targets AREs.36 PA1 is a N-methylimidazole (Im) and N-methylpyrrole (Py) derived polyamide to recognize and bind these base pair sequences on the ARE half-sites. The short (<6 amino acid pairs) polyamides show optimal geometrical ability to align with and bind the helical DNA strands. The multiple bifurcated hydrogen bonds between the polyamide backbone’s amide hydrogens to the purine N3 and the pyrimidine O2 provide favorable binding affinity.34,38
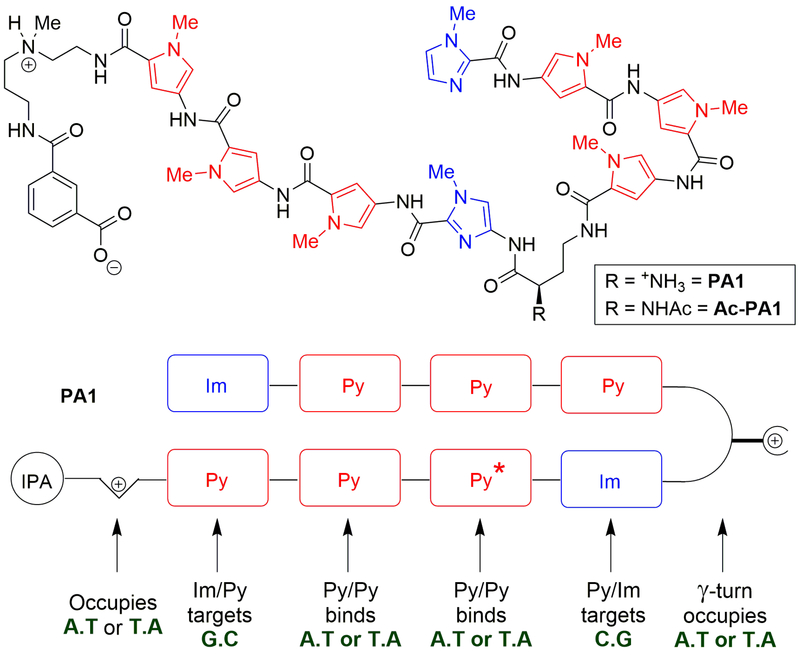
Figure 3.
Structures of hairpin polyamides PA1, PA2, and Ac-PA1. PA1 is designed to bind the ARE sequence 5′-AGAACA-3′, while PA2 has a mismatch (where Py* is substituted by an Im) that should render the binding to be weak to that sequence. Acylation of the γ-turn amino group yields an acetamide (Ac-PA1) with an improved in vivo toxicity profile.37
The specificity of binding is established through the different heterocyclic pairs in the polyamide that can recognize specific nuclear base pairs to bind in a complimentary manner (Figures 3 and 4). Py/Py pair binds both the A.T and the T.A nucleotide pairs non-selectively via hydrogen bonding interactions. In contrast the Im/Py pair provides far more specific and directional binding/recognition towards binding its target. The critical interaction that results in such specific recognition lies in the hydrogen bond formation between the lone pair on the imidazole nitrogen and the exocyclic amino group of guanines.33,38 This interaction brought about by the Im/Py pair, while recognizing the G/C base pairs over the A/T base pairs, also specifically distinguishes between G.C versus a C.G pairing. The unfavorable angle to form a thermodynamically efficient39 hydrogen bond from the cytosine side of a G.C pair makes the imidazole recognize the guanine via hydrogen bonding from the proximal side to the guanine.33 Hence the Im/Py pair has been shown to carry a 100 fold greater affinity for a G.C base pair than shown by a Py/Im pair.40
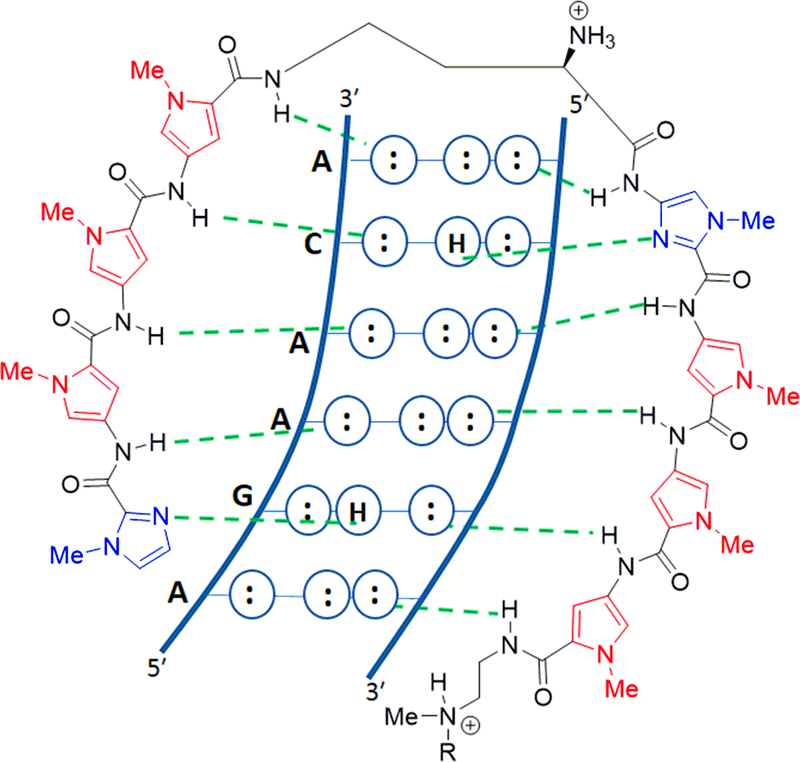
Figure 4.
Recognition of the ARE DNA half-site by PA1. Im/Py pair recognizes G.C, Py/Py pair binds A/T T/A, Py/Im pair recognizes C.G. Figure redrawn from reference.33
This binding specificity has been proven via x-ray crystallographic analysis of a polyamide of the structure ImImPyPy-β-Dp (where β = beta alanine, and Dp is dimethylaminopropylamide) bound as a dimer to its target sequence of 5′-WGGCCW-3′ (W = A or T).38 The antiparallel head-to-tail type binding of this dimer was also shown to match the adjacent DNA strands 5′ to 3′ directionality with respect to its N- to C-terminal orientation within each polyamide.38 The β-alanine end groups were accommodated in the smooth minor grooves of the A.T and the T.A base pairs flanking the GGCC recognition sequence. The Im/Im pairings are considered to be energetically unfavorable40, which prevents the slipped binding modes of the peptides from occurring.
Utilizing the knowledge about these heterocyclic pairs of Im/Py and Py/Py, PA1 was designed as a cell permeable hairpin polyamide that targets the gene sequence 5′-WGWWCW-3′ (W = A or T), which is found in the consensus ARE (Figure 4)33. The antiparallel peptide sequences were connected via a chiral γ-diaminobutyric acid hairpin turn to prevent slipped binding modes, and to give improved affinity and selectivity compared with unlinked elements.41 The γ-turns show preference to occupy A.T base pairs over G.C base pairs, owing to steric clash with the exocyclic amine of the guanine.33,41,42 The presence of the isophthalic acid (IPA) moiety in the C-terminal tail has been shown to facilitate improved nuclear translocation of such polypeptides.43,44 The aminoalkyl linker connecting the polyamide to the IPA unit also has binding preference towards A/T pairs versus G/C pairs due to steric reasons.41
2.2 |. In Vitro Effectiveness
The binding of the polyamide PA1 to the proximal PSA promotor that contains the ARE 5′-AGAACAGCAAGTGCT-3′ was evaluated via quantitative DNAse I footprint titrations using a 5′−32P-labelled PCR fragment of pAR-PSA.36 PA1 showed an association affinity, Ka, of 8.3 × 109 M−1 for the ARE consensus half-site 5′-AGAACA-3′. The other half-site sequence 5′AGTGCT-3′ was shown to bind PA1 with a much lower affinity due to the single base pair mismatch in that sequence at the 4th position. Another polyamide, PA2, which was designed as a negative control (Figure 3) to target the alternative sequence 5′-WGWCGW-3′ did not result in any measurable (Ka < 107 M−1) binding to either of the ARE half-sites under the conditions utilized. This clearly highlights the ability of these polyamides to bind specific gene sequences.
Electrophilic mobility shift assays revealed PA1 at a concentration of 10 nM inhibited the binding of factors from nuclear extracts isolated from DHT-stimulated LNCaP cells to the ARE site in the PSA promoter.36 The binding of PA1 to the prostate specific antigen (PSA) promoter ARE down-regulated the androgen induced expression of PSA. The down regulation of PSA by PA1 was comparable to the effects brought about by similar concentrations of the AR antagonist bicalutamide.36 This dose dependent down-regulation was measured to be ~70% at concentrations of 10 μM of either compound in comparison to non-treated cells or cells treated with PA2. Decreased occupancy of the AR at the PSA promotor and enhancer, as well as the intronic enhancer of the gene FKBP5 in the presence of 10 μM of PA1 was shown using ChIP (chromatin immunoprecipitation) assays. The inhibition of the AR induced gene FKBP5 by PA1 was ~60% at this concentration, as compared to ~95% by bicalutamide. Other direct AR target genes such as KLK2, KLK3, and TMPRSS2 were also demonstrated to be 2–3 fold down-regulated by PA1 as compared to the DHT-induced controls.36 Evaluation of androgen induced gene expression using DHT-stimulated LNCaP cells, revealed that PA1 at a concentration of 10 μM affected the expression of 1,053 transcripts by at least 2-fold compared to the controls.
Using LNCaP cells, Yang et al. has shown that the anti-tumor activity of PA1 can also be linked to its effects on the inhibition of RNAP2.45 This observation is consistent with previous reports that DNA binding molecules would have significant inhibitory effects on the function of RNAP2.46 The antitumor activity was blocked by the co-treatment with MG132, a proteasome inhibitor. Hence the inhibitory effect was shown to be originated in the enhanced degradation of RNAP2 large subunit RPB1, a phenomenon that triggers cellular apoptosis mechanisms.31,45 PA1 activated p53 genes although no significant cellular markers of DNA damage were observed upon extended treatment.45
2.3 |. In Vivo Effectiveness and Further Optimization
PA1 treatment of mice with LNCaP xenografts have shown its ability to act as an anti-tumor agent in vivo, resulting in up to 64% inhibition of tumor growth at a dose of 1 mg/kg.45 However, the early experiments did show some weight loss in tumor-bearing mice upon treatment with PA1.45 Complete removal of the chiral amine unit was shown to reduce in vivo toxicity, albeit at the cost of losing therapeutic effectiveness.31 Later studies showed that the acylation of the α-amino unit at the hairpin turn, yields a derivative (Ac-PA1) with prominently less in vivo toxicity, while retaining the activity profile.37 The differential toxicity could have a relation to the higher liver tissue localization of PA1 than Ac-PA1 (33% less), which was measured using radio-labelled (14C) polyamides in a LNCaP xenograft mouse model.31,47 From the same experiments, the accumulation at the tumor was found to be better with the acylated derivative. With repeated injections (three injections over 7 days) of Ac-PA1, ~2-fold accumulation was seen at both the tumor and the host-organs. The organ-accumulation is a disturbing factor given the chance for higher levels of toxicity. Interestingly in another study, LNCaP xenograft bearing mice were shown to have greater liver accumulation of Ac-PA1, than mice with A549 (lung) or U251 (brain) xenografts.48 Here, LNCaP xenografts were found to have much greater localization (up to 5×) of the polyamide than the non-PCa xenografts. The liver clearance of Ac-PA1 in the mice with LNCaP xenografts was found to be impaired.48
Given the homology between the AR and other nuclear hormone receptor DNA binding domains, a polyamide designed to target 5′-WGWWCW-3′ half-site is expected to have promiscuity in binding. Using an enzalutamide-resistant LREX′ PCa cell line, Kurmis et al. demonstrated the acetamide derivative Ac-PA1 was able to interfere with both AR- and GR- (glucocorticoid receptor) driven gene expression.49 In instances where GR up-regulation is the primary resistance mechanism to overcome AR-antagonists, this effect could be beneficial to develop an efficient dual-targeting approach. Ac-PA1 significantly reduced the growth of VCaP and LREX′ cells in vitro even upon up-regulation of AR- (by DHT) and GR- (by dexamethasone) driven transcription.49 AR- and GR-driven gene expression was also reduced upon Ac-PA1 treatment.49 Treatment of VCaP xenografts with Ac-PA1 showed a dose dependent reduction in tumor volumes up to 70% (5 mg/kg) at 6 weeks. Importantly, enzalutamide-resistant (GR-driven) LREX′ xenografts showed 80% reduction of tumor growth at the co-treatment of enzalutamide and Ac-PA1. Authors indicated a 6% weight loss in mice when treated with Ac-PA1 at 30 mg/kg, which was recovered upon treatment withdrawal (5 days).49 Arguably, some amount of toxicity would have to be expected (and perhaps accepted depending on the severity of the PCa treatment resistance) in a therapy that has the potential to hit more than one nuclear hormone receptor.
2.4 |. Outlook
The promising results detailed above, has established that hairpin polyamide compounds can be utilized successfully to target ARE (and GRE) in a broader perspective. If specific AR dependent genes were to be targeted, these polyamides would have to be programmed via changes in sequences/amino acids to bind specific ARE fragments, given the subtle degenerate nature of different AREs. Perhaps incorporating the thiamine-selective recognition element N-methyl-3-hydroxypyrrole33,35 in place of the Py unit, could provide increased binding affinities and selectivities in analogs of PA1. However, Dervan et al. have demonstrated that even though some heterocycle replacements enhance DNA binding affinity, the ability to permeate the cell or reach the nucleus is compromised by such modification.31 Padroni et al. has shown recently that thiazole derivatives could be used to substitute the imidazole units in PA1 type hairpin polyamides.50 Although the double stranded DNA binding affinity was demonstrated to be somewhat higher for the 5-alkyl thiazole containing polyamides, G-recognition selectivity was found to be diminished for the thiazole units when compared to the Im units.50
Being able to target both AR- and GR- driven transcription, Ac-PA1 derivatives may translate to particularly effective therapeutics against enzalutamide-resistant PCa’s that have GR up-regulation as the major pathway of resistance. The high molecular weight and the hydrophobic nature of the constructs have made these hairpin polyamides have poor aqueous solubility.51 That may hinder an oral drug delivery approach and negatively affect the pharmacokinetic/pharmacodynamic profile moving forward. Besides the efforts to find an optimal formulation stratergy,31 minimizing the off-target effects due to DBD homology between nuclear hormone receptors stand as the major challenge ahead for these hairpin polyamides.
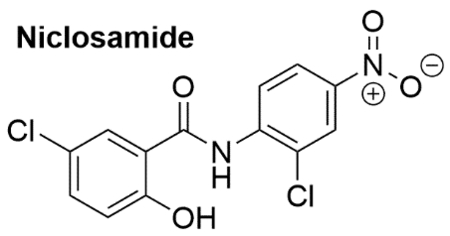
3 |. NICLOSAMIDE
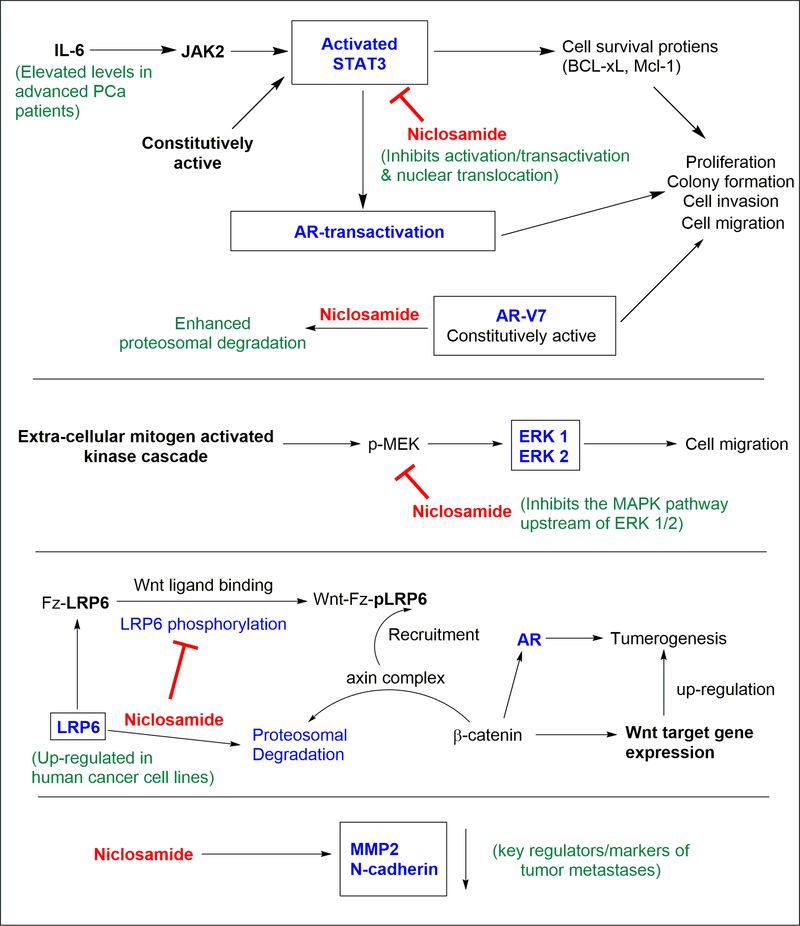
Niclosamide (Table 1) is a FDA-approved (1982) anthelminthic drug (Niclocide™) that has been used for treating tapeworm infections.52,53 Structurally it is a salicylanilide, which has two aromatic chlorine substituents and an aromatic nitro group. Niclosamide has shown to be well tolerated in humans, which presents a distinct advantage in adapting it for a novel therapeutic use. In fact, utilization of an previously approved drug like niclosamide, provides a rapid path towards clinical trials.53–55 The mechanism of action of niclosamide against tape worms involves inhibition of oxidative phosphorylation and the stimulation of ATP activity in the mitochondria.53,56 Given the recent findings that tumor related malignancies often involve deficits of oxidative phosphorylation and decreased availability of ATP in the mitochondrial activity of cancer cells57, the effect of niclosamide against such cells became an interesting topic to investigate. Niclosamide has been shown to demonstrate antineoplastic effects against many cancers including myelogenous leukemia58,59, lung cancer60,61, breast cancer62–68, colon cancer69–72, ovarian cancer73–75, prostate cancer64,76–82, adrenocortical carcinoma83, hepatocellular carcinoma84,85, multiple myeloma86, glioblastoma87, and osteosarcoma.88 In these extensive studies, niclosamide has shown remarkable ability to eradicate cancer stem cells, inhibit metastasis, and/or induce/reestablish apoptosis mechanisms. The effects of niclosamide at the cellular level involve multiple signaling pathways that are prominent in cancer progression (Figure 5). It has shown inhibitory effects towards Wnt/β-catenin, mTORC1, STAT3, NF-κB, and the Notch pathways,53,89 establishing niclosamide as a multi-pathway inhibitor of cancer progression.
Table 1.
Compounds that affect the AR signaling axis without interacting with AR-LBD.
| Compound | Mechanism(s) of action and other details |
|---|---|
| Hairpin polyamides (see figures 3 and 4 for structure) | • Antagonism of AR-DNA association by binding to DNA androgen response elements (AREs) • Enhanced degradation of RNAP2 large subunit RPB1, triggering cellular apoptosis mechanisms • Disrupts replicative helicase activity • Activates p53 genes |
 |
• FDA approved human anthelminthic • Multi pathway (e.g. IL6-JAK-STAT, MAPK, Wnt/β-catenin) inhibitor of cancer • Promotes AR-V7 degradation via a ubiquitin-proteasome pathway • Affects the IL6-STAT3 mediated AR-TAD transactivation, AR nuclear translocation, and AR-DNA binding activity • Phase I clinical trials with enzalutamide co-treatment on going (NCT02532114, NCT03123978) |
 |
• Peptidic polychlorinated marine natural products • Inhibits AR-TAD transactivation • Analogs with higher degree of chlorination shows better activity than less chlorinated analogs • Sintokamide A primarily inhibits AR-AF1 TAU-1 domain |
 | |
 |
• Marine natural products • R-Niphatenone-B shown to bind the AR-TAD AF1 region • Inhibits AR-TAD transactivation • Further development abandoned due to binding specificity issues. |
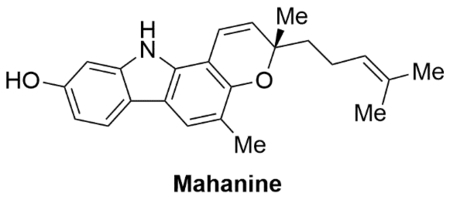 |
• Carbazole alkaloid natural product • Multi-pathway anticancer compound • Inhibits AR transactivation • Induces degradation of full length and splice variant AR via ubiquitin-proteasome pathway • Reduces AR nuclear translocation |
 |
• AR-TAD inhibitor • Isolated from a marine sponge. • Derived from bisphenol A (BPA) • Covalently binds the AR-TAD-AF1 • Primarily Inhibits AR-AF1 TAU-5 domain • PPARγ modulation effects leading to AR inhibition • Clinical trials (NCT02606123) terminated at end of Phase 1; excessive high pill burden (18 capsules/day) Anti-tumor effects at >2400 mg/kg doses • Further development abandoned |
| JN compounds(Structures not yet disclosed) | • AR-TAD inhibitors and AR degraders • 10 to 30-fold greater potency as compared to EPI-002 in cellular and functional assays. • Significant control in tumor growth in xenografts with full length and/or splice variant AR |
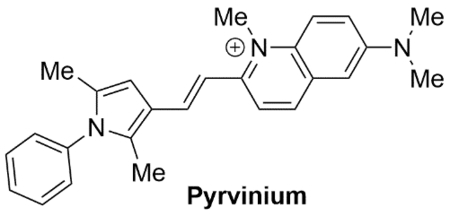 |
• Quinoline derived cyanine dye • FDA approved human anthelminthic • AR-DBD inhibitor • Cross reactivity towards other nuclear hormone receptors |
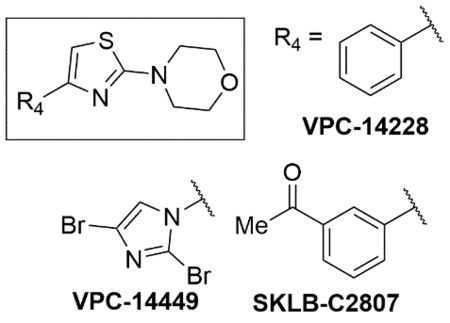 |
• Thiazolyl morpholine derivatives initially found through an in-silico drug design approach • AR-DBD inhibitor • Key H-bonding interaction of morpholine O with Tyr-594 of AR-DBD • Does not impede AR nuclear translocation |
 |
• BET inhibitors (BETi’s) • Most BETi are triazolodiazepines • AR-Chromatin binding inhibitors • Direct interaction with AR-TAD shown • In vivo and in vitro activity against PCa • Clinical Trials ongoing for multiple cancers including CRPC (NCT02711956, NCT02607228, NCT02259114) |
Figure 5.
Multi-pathway anti-cancer effects of niclosamide. Niclosamide has been shown to 1) affect the STAT, Wnt/β-catenin, and the MAPK pathways, 2) to enhance degradation of AR-V7 and LRP6, and 3) significantly lower key regulators/markers of tumor cell metastasis.
3.1 |. Niclosamide Monotherapy
3.1.1 |. Effects on the IL6/JAK2/STAT3 pathway
Of the above pathways, “JAK (Janus kinase) - STAT3 (signal transducer and activator of transcription 3)” is a major pathway through which niclosamide has shown the ability to target CRPC.80–82 Elevated levels of STAT3 has been found in prostatic carcinomas and normal tissues adjacent to the such tumors.90 Hyperactivation of the STAT pathways91,92 induces cell proliferation and prevents apoptosis mechanisms in human cancer cells through dysregulation of key proteins.80 STAT3 in particular has been shown to regulate the expression of genes that control factors central to cancer progression.93 STAT3 can be activated by growth factor receptors, non-receptor tyrosine kinases, or cytokine receptors such as the interleukin-6 (IL-6) receptor.94 Serum levels of IL-6 are often found to be elevated in advanced PCa patients.95,96 Upon ligand binding to the IL-6 receptor complex, an intracellular signaling cascade is activated where the receptor-associated JAKs phosphorylate STAT3 at Tyr-705. Dunn et al. showed that JAK2 in particular is responsible for this phosphorylation employing LNCaP cells that lack JAK1 due to epigenetic silencing.97,98 This phosphorylation activates the STAT3, which results in dimerization, nuclear translocation, and induction of specific target gene expression. JAK2 can also phosphorylate and activate STAT5 which occurs in 61% of metastatic PCa.98,99 Importantly, STAT3 signaling has been shown to interact with the AR-TAD and thereby facilitate AR transactivation.100
In view of the role of the IL6/JAK2/STAT3 pathway on induction of gene expression, growth promotion and activation of the AR, this pathway has been implicated as a major target for prostate cancer treatment.7,98,101 In an attempt to find non-peptidic small molecule inhibitors of the STAT3 signaling pathway via high-throughput screening Ren et al. found niclosamide as a hit compound.80 Treatment of DU145 PCa cells that carry constitutively active STAT3 with niclosamide resulted in the dose dependent inhibition of the STAT3 phosphorylation at Tyr-705.80 These results are in agreement with the work by Mora et al. where the inhibition of constitutively active STAT3 signaling in DU145 cells using antisense STAT3 oligonucleotides induced growth inhibition and apoptosis.102 Niclosamide was able to block the STAT3 induced luciferase activity in HeLa cells at a drug concentration of 5.0 μM.80
Niclosamide’s targeting has shown selectivity towards the STAT3 pathway, without obvious inhibitory effects against the activation of other STAT homologues, STAT1 and STAT5.80 This effect was deemed not to be exerted by the inhibition of upstream kinase activity of JAKs, since niclosamide did not affect the kinase protein levels over the course of treatment.80 Using an immunofluorescence assay, it was shown that niclosamide (1.0 μM) blocked the EGF induced nuclear translocation of STAT3 after a 2 h treatment.80 Electrophoretic mobility assays revealed that the activity of niclosamide did not result from a direct binding/interaction of niclosamide with the STAT protein’s consensus DNA elements.80 In aggregate, these results show that the activity of niclosamide is found in the inhibition of the activation/transactivation and the nuclear translocation of STAT3, although it did not directly bind to the SH2 domain of the STAT3 protein.80
Niclosamide strongly inhibited the proliferation and colony formation (IC50 = 0.7 and 0.1 μM) of DU145 PCa cells while the effect was not that pronounced in PC3 PCa cells that had a lower level of constitutively active STAT3.80 Flow cytometric analysis revealed that the compound induced G0/G1 phase arrest and the apoptosis of DU145 cells, which may have been a consequence of the down-regulation of cell survival proteins (BCL-xL, Mcl-1) and cell cycle regulators (cyclin D, c-Myc) in a dose dependent manner, as shown by a Western blot analysis.80 Similar effects of niclosamide towards the STAT3 inhibition, and the inhibition of STAT3 target genes in LNCaP, C4–2B, and DU145 cells were shown by Liu et al..78
A wound healing assay (employed to measure the migratory properties of cells) using DU145 cells showed niclosamide was able to inhibit wound healing by ~20, ~60, ~70% at drug concentrations of 0.2, 1.0, and 5.0 μM respectively.81 When the same assay was done on cells transfected with STAT3 siRNA, niclosamide showed a significantly diminished ability to inhibit wound healing, indicating the importance of the STAT3 pathway as its mechanism of action.81 Wound healing assays conducted by Liu et al. on LNCaP-s17, LNCaP-STAT3, and DU145 cells carrying constitutively active STAT3 has also showed niclosamide inhibited (80 – 90% inhibition) wound healing in a dose dependent manner.78
3.1.2 |. Effects on the MAPK pathway
Given the possibility for other pathways to be involved in the wound healing process, the MAPK (mitogen-activated protein kinase) pathway was also probed by Ren et al. via monitoring the ERK 1/2 (extracellular signal–regulated kinases) expression in DU145 cells after niclosamide treatment.81 Like STAT3, ERK 1/2 activates via initial phosphorylation (pERK) and then exerts a downstream effects to promote metastasis. At an initial 4 h treatment period, niclosamide did not show a discernable effect on the levels of pERK. However, at 24 h, pronounced inhibition of pERK was seen at concentrations of niclosamide > 1.0 μM.81 This delayed response on the ERK related pathway is in contradistinction to the effects on pSTAT3 levels, which were significantly impacted at similar doses even at the 4 h mark.81 Hence the authors postulated that niclosamide might be targeting upstream regulators of the MAPK pathway, which consequently affects the ERK 1/2 activation. To demonstrate this effect of the MAPK pathway on the cancer cell motility, the wound healing assay was conducted in the presence of PD98059 a selective MAPK inhibitor. The niclosamide (1.0 μM) induced inhibition of wound closure was significantly lowered in the cells co-treated with PD98059 (43%) compared to the control group (63%).81 This showed that the down-regulation of the MAPK pathway by niclosamide also contributes towards the motility of the DU145 PCa cell line.81 With the knowledge that MAPK pathway is involved in some AR-independent bypass pathways that drive PCa,13,103 niclosamide’s ability to affect this signaling axis adds to the impact it could have in a co-therapy with an AR-antagonist.
3.1.3 |. Effects on the Wnt/β-catenin pathway
Wnt/β-catenin pathway is another cellular pathway that has shown importance in targeting the AR-signaling axis.104,105 Wnt Signaling can interact with the AR-signaling axis and AR gene transcription processes, leading to elevated prostatic tumor growth, cell migration, and invasion properties.104,106–108 Wnt signaling promotes AR gene transcription while AR-signaling is inhibitory towards the Wnt-pathway in hormone-naïve PCa cells.104 However, the two pathways promote each other in CRPC, which leads to androgen independent PCa progression.104 In 2011, Lu et al. showed that the inhibition of low-density lipoprotein receptor related protein 6 (LRP6) mediated Wnt/β-catenin activation by niclosamide induced anticancer effects against prostate and breast cancers. Niclosamide displayed anticancer activity against DU145 and PC3 PCa cells with IC50 values less than 1 μM, and an ability to induce apoptosis at niclosamide concentrations of 1.2 μM.64 The Wnt family of glycoproteins regulates fundamental processes that direct cell proliferation, cell polarity, and cell fate determination during embryonic development and tissue homeostasis.109 A major component in Wnt signaling associated with the above functions is the transcriptional co-activator β-catenin. With abnormal upregulation, the Wnt/β-catenin pathway can lead to tumorigenesis of multiple types of cancers, including PCa. LRP6 is a co-receptor for Wnt ligands109 that is expressed and upregulated in human cancer cell lines.64 Upon Wnt ligand binding to the frizzled (Fz) receptor and its co-receptor LRP6, the LRP6 gets activated and phosphorylated (pLRP6) on the cytosolic side.109 This Wnt-Fz-pLRP6 complex recruits the axin complex from the cytosol to the receptors. The axin complex in the absence of such Wnt interference is responsible for keeping the β-catenin levels down regulated in the cells via continuous proteasomal degradation of the cytosolic β-catenin.109 The recruitment of the axin complex to the activated Wnt receptor system disrupts this regulation, and cytosolic β-catenin accumulates. β-catenin then travels to the nucleus where it functions as a coactivator of multiple transcription factors, including the TCR-LEF complex which has regulatory effects towards the AR.109,110 In addition, β-catenin has been shown to perform as a co-activator of ligand-dependent AR function in PCa cells.111,112
In experiments conducted by Lu et al., treatment of PC3 PCa cells with niclosamide concentrations > 0.3 μM induced significant reduction of free β-catenin levels as evidenced by a GST-E-cadherin binding assay.64 In PC3 cells transiently transfected with the Wnt/β-catenin signaling reporter TOPFlash luciferase, niclosamide treatment reduced luciferase activity by ~70%.64 The total cellular levels of axin2 and cyclin D1, which are transcriptional targets of the Wnt/β-catenin pathway were significantly reduced in PC3 and DU145 PCa cells by niclosamide treatment at concentrations < 1 μM.64 Even more importantly, niclosamide was able to suppress LRP6 expression and phosphorylation at concentrations of 0.3 μM.64 Treatment with niclosamide of PC3 cells pretreated with cycloheximide (protein synthesis inhibitor) revealed a half-life of 2.3 h for LRP6, as compared to the control group (without niclosamide) which showed a half-life of 6.9 h.64 However, the total LRP6 mRNA levels did not change upon niclosamide treatment as judged by RT-PCR. These results indicate that the LRP6 suppression was not at a transcriptional level, but rather mediated by enhanced LRP6 degradation.64 Niclosamide was shown not to have any effect towards the levels of cytosolic Dvl2 (Dishevelled-2), another regulator of the Wnt/β-catenin pathway.64
3.1.4 |. Effects on cellular markers of tumor metastases
A Boyden chamber assay (employed to mimic the in vivo invasion process of cancer cells) by Ren et al. using DU145 cells revealed the ability of niclosamide to inhibit the migration of cancer cells through an ECM (extra cellular matrix) membrane up to ~90% when treated for 24 hours at drug concentrations up 2.0 μM.81 Similar results were obtained by Liu et al. using LNCaP-STAT3 and DU145 cells carrying constitutively active STAT3 where the invasion was reduced by ~90% at a niclosamide concentration of 0.5 μM.78 To further explore the ability of niclosamide to inhibit tumor cell metastases, the effect of niclosamide on the levels of key proteins which are associated with tumor metastasis (MMP2, MMP9, cadherins, and catenins) were evaluated by Ren et al. in DU145 cells.81 MMP2 and MMP9 are key enzymes mediating ECM degradation that promotes metastases, while the cadherins/catenins are key factors in endothelial mesenchymal transition (EMT).81 The effect of niclosamide on MMP2 was very pronounced, resulting in almost complete eradication of MMP2 at a drug concentration of 5.0 μM after a 24 h treatment.81 While there was also an inhibitory effect towards the level of MMP9, the effect was not as pronounced as for MMP2. The effect of niclosamide on catenins was less distinct as well, a finding that suggests that showing any interference with EMT of DU145 cells is primarily via the regulation of cadherins. In corroboration of this point, the protein level of N-cadherin (a mesenchymal marker) was significantly reduced with the treatment with niclosamide.81 A repression of the levels of E-cadherin (an epithelial marker), which conventionally is seen as a sign of EMT promotion,113 was also seen. Some research suggests that the loss of E-cadherin levels alone might not be predictive of EMT.114
3.1.5 |. Effects on AR degradation
One of the most significant findings related to niclosamide effects in CRPC models is the downregulation ARSVs.79 Androgen receptor splice variant AR-V7, in particular, has been linked to CRPC and enzalutamide type AR antagonist resistance to second generation AR signaling axis inhibitors such as enzalutamide and abiraterone acetate.77–79,115 High-throughput screening of HEK293 cells stably transfected with AR-V7 with a PSA-luciferase reporter system identified niclosamide as a possible AR-V7 targeting compound.79 In LNCaP PCa cells transiently transfected with AR-V7, niclosamide was shown to inhibit the non-DHT dependent transcriptional activity (of AR-V7) while enzalutamide could not.79 DHT induced transcriptional activity in the same system however was knocked down by both niclosamide and enzalutamide. A ChIP assay showed niclosamide significantly reduced the AR-V7 recruitment to the PSA promotor in CR-2 AR-V7 cells, in which enzalutamide had no effect.79 Treatment of CWR22Rv1 cells with niclosamide inhibited the endogenous AR-V7 expression in a dose dependent manner. At lower doses (0.5 μM), the inhibition effect was significantly more prominent towards AR-V7 than the ARFL, showing a preferential inhibition.79 This effect was found to be due to more rapid AR protein degradation in the presence of niclosamide, rather than from a transcriptional level of inhibition.79 AR-V7 degradation monitored in the presence of cycloheximide (protein synthesis inhibitor) revealed niclosamide enhanced the AR protein degradation.79 MG132 (26S proteasome inhibitor) was able to reduce this AR-V7 degradation, indicting the involvement of the ubiquitin-proteasome pathway for niclosamide induced AR-V7 degradation.79 Niclosamide had minimal effects on the expression of full length AR. Niclosamide (0.5 μM) exerted significant inhibition of C4–2 neo, C4–2 AR-V7, and CWR22Rv1 PCa cell growth and induced cell apoptosis, while not effecting the growth of normal prostate epithelial cells PZ-HPV-7.79
3.2 |. Niclosamide Combination Therapies
Given the aforementioned results of niclosamide as a monotherapy, combinations of niclosamide and other compounds have been tested for PCa treatment. In human PCa tissues, AR down-regulation induced STAT3 activation has been shown to lead to the development of PCa stem-like cells.94 Such activation could result in rapid resistance to therapies like enzalutamide and result in lethal metastatic disease. As such, a potent STAT3 inhibitor like niclosamide in combination therapy with a direct or indirect AR-LBD antagonist could result in prolonged treatment effectiveness.
A study by Liu et al. corroborated the previously stated findings about the association of PCa and IL-6/JAK/STAT pathway by showing that the inhibition of constitutively active STAT3 reverses the enzalutamide resistance in LNCaP derivative PCa cells.95 It was shown that enzalutamide (20 μM) was able to exert a ~60% inhibition of the growth of LNCaP PCa cells, while the effect was modest (<20% inhibition) for LNCaP-IL6+ cells and LNCaP-s17 cells that overexpressed IL-6.95 These LNCaP-s17 cells were found to carry constitutively active STAT3, and as such had elevated STAT3 signaling resulting in elevated levels of AR, c-Myc, survivin, and Bcl-2 proteins than the control LNCaP-neo cells.78,95 The use of AG490 (a JAK2/STAT3 inhibitor) or the use of STAT3-siRNA (knocks down STAT3 expression) reversed the enzalutamide resistance in the LNCaP-s17 cells.95 ChIP assays revealed that the recruitment of AR to the proximal and the distal enhancer binding sites of the PSA promotor were significantly enhanced in the LNCaP-s17 and LNCaP-STAT3C cells carrying constitutively active STAT3 as compared to the LNCaP-neo control cells.95 These results were in agreement with previous findings that IL-6 overexpression led to enhanced AR nuclear translocation and AR-ARE DNA binding activity116, and resulted in the upregulation of intracrine androgen levels in the absence of exogenous steroid precursors.117 Enzalutamide was able to significantly inhibit the recruitment of AR to AREs in the LNCaP-neo cells, but failed to have much effect on AR recruitment to AREs in LNCaP-s17 and LNCaP-STAT3 cells with elevated STAT3 activity.95 ChIP assays conducted in a later study using the LNCaP-s17 cells showed that the knock-down of STAT3 activity using STAT3-siRNA re-established the enzalutamide sensitivity, significantly reducing the AR–ARE recruitment as well as the PSA expression.78 Such results collectively showed that the concurrent use of a STAT3 pathway inhibitor with enzalutamide (or other antiandrogens such as abiraterone) could be used for the treatment of enzalutamide resistant advanced PCa.
The colony formation activity of an enzalutamide resistant AR variant expressing C4–2B cell line was found to be dramatically inhibited by enzalutamide (20 μM) and niclosamide (0.25 μM) co-treatment.79 This effect was less pronounced using niclosamide treatment alone. The success of the combination therapy on anti-colonogenic activity was also validated in castration resistant CWR22Rv1 cells (expressing ARSVs and ARFL) as well as LNCaP-STAT3 and LNCaP-s17 cells.78,79 Again, the combination therapy was able to exert greater effect than the individual treatments in a time dependent fashion.78,79 Evaluation of the combination treatment in vivo using xenografts generated from CWR22Rv1 cells showed a significant difference in tumor weight after 3 weeks of treatment, where the combination treated tumors had a 70% less weight than the control.79 The synergistic effect was evident in considering that neither the enzalutamide treatment (non-responsive) nor the niclosamide treatment (~50% less weight compared to the control) alone were able to achieve robust inhibition.79 The effect of enzalutamide plus niclosamide on the STAT3 downstream target genes was also more pronounced than either of the individual treatments in LNCaP-s17 and LNCaP-STAT3 cells. Combination therapy was able to significantly inhibit the STAT3 phosphorylation and the expression of survivin and c-Myc compared to individual treatments.78 Evaluation of the effect on AR recruitment was done using a ChIP assay on LNCaP-s17 cells, showing the combination treatment was superior in inhibiting AR-ARE recruitment compared to enzalutamide or niclosamide alone.78 PSA secretion was downregulated (>50%) by the enzalutamide/niclosamide co-therapy than niclosamide treatment alone. Enzalutamide alone was not able to exert much inhibitory effect on the PSA levels of these cells with constitutively active STAT3. Analysis of the Ki67 (a cellular marker for cell proliferation) protein levels in CWR22Rv1 xenograft tumor samples revealed that while niclosamide moderately inhibited (~30% inhibition) the Ki67 expression, the combination treatment with enzalutamide far more prominently decreased the Ki67 levels (~80% inhibition).79
Enzalutamide treatment, while being quite efficient at early PCa treatment, has recently been shown to have pro-metastatic effects in pre-clinical models.78,118,119 The IL-6/STAT3 feed forward loop has been reported to be a major pathway through which enhanced cell motility and EMT occur in PCa metastasis.78,120,121 Evaluation of cell migration through a wound healing assay using LNCaP-s17 and LNCaP-STAT3C cells (both with constitutively active STAT3), revealed a 20 μM concentration of enzalutamide had little effect on wound healing (i.e. cell migration) inhibition, whereas total inhibition was achieved when niclosamide (0.25 μM) was used in co-treatment.78 A Boyden chamber based cell invasion assay using LNCaP-STAT3 cells showed similar enhancements in the combination treatment (20 μM enzalutamide plus 0.25 μM niclosamide, 90% reduction of invasive cells) as compared to the individual treatments of enzalutamide (no reduction) or niclosamide (50% reduction).78 These concentrations of enzalutamide are quite high and may not be achievable in vivo.
Abiraterone acetate (abiraterone) is a steroidogenesis inhibitor, that primarily blocks the CYP17A1 activity, resulting in the inhibition of androgen production from cholesterol. The presence of AR variants such as AR-V7 renders PCa cells resistant to the effect of inhibiting ligand production by abiraterone. Similar to the case with enzalutamide co-treatment, niclosamide has also shown the ability to re-sensitize abiraterone resistant PCa cells expressing AR-V7 in both in vitro and in vivo experiments.77 C4–2B AbiR cells expressing significantly high levels of AR-V7 overcame abiraterone (5 μM) resistance in the presence of si-AR-V7 or niclosamide (0.5 μM). Through oral administration, niclosamide (500 mg/kg) synergized abiraterone treatment (200 mg/kg) in a CWR22Rv1 xenograft model resulting in dramatically reduced tumor sizes in the co-treated mice.77 Similar demonstrations of co-treatment effectiveness has been done with bicalutamide, a nonsteroidal antiandrogen drug.122
3.3 |. Clinical Trials and Outlook
While niclosamide does not have an ideal pharmacokinetic profile based on the anthelminthic treatments56,89, the potency with which it inhibits the STAT3 pathway and induces apoptosis of PCa cells made it a promising drug candidate to find a viable treatment towards CRPC.80 The poor bioavailability of niclosamide, that mostly results from the sparingly soluble nature of the compound in aqueous media, could possibly be overcome by the utilization of more water-soluble analogs, preparations, or prodrugs.89,123 Given that niclosamide affects numerous signaling pathways other than of the AR and can inhibit the growth of AR null PCa cells as well as non-prostate cancers, it is not clear that niclosamide mediates its anti-PCa effects primarily through the AR. The lack of effect on full-length AR expression and its modest effects on tumor growth as a monotherapy suggest that niclosamide may not serve as a stand-alone treatment for PCa. Moreover, the applicability of niclosamide may be limited as only a minority of CRPCs express AR-V7. As such, major focus has shifted rather towards the development of co-treatments of niclosamide.
The ability of niclosamide to act as an AR splice variant inhibitor, cell invasion/migration inhibitor, and an IL-6/STAT3/AR axis inhibitor while being already an FDA approved drug, made it an attractive target to pursue as a co-drug to re-sensitize antiandrogen therapies that have succumbed to resistance mechanisms.124 Two clinical trials (Phase I, NCT02532114, NCT03123978) were initiated recently to investigate the co-treatment of AR spice variant positive mCRPC using enzalutamide and niclosamide. Furthermore, another clinical trial (Phase 1b/II, NCT02807805) to evaluate the side effects of niclosamide treatment in patients with CRPC is now in the recruitment phase for phase II. One of the aforementioned clinical trials, NCT02532114 – a dose escalating study for enzalutamide/niclosamide co-treatment, was concluded recently.125 The findings from this study appear rather unfavorable for further use of niclosamide in CRPC clinical trials. The minimum effective plasma concentrations relevant to the preclinical response data could not be achieved at the highest tolerable dose (500 mg three-times-a-day [TID]) in this study.125 While an ideal concentration level above the 82 – 330 ng/mL (0.25 – 1 μM) range was desired, only a maximum plasma concentration of 35.7 – 82 ng/mL (0.11 – 0.25 μM) was achievable at 500 mg TID dosing.125 Although previous use of the drug as an anthelminthic was deemed safe at 2 g / day as a single dose (continued for 1–7 days), here in mCRPC patients dose-limiting toxicities were found at 1000 mg TID dosing.56,125 The toxicity is likely the effect of exceeding a daily tolerable maximum (e.g. patient receives 3000 mg of the drug per day with the 1000 mg TID dosing) and/or the reduced ability of CRPC patients to tolerate the drug compared to an otherwise healthy individual with just a helminthic infection. Lack of clinical activity at tolerable doses resulted in the premature termination of this study by its data safety monitoring board.125 However, these results are in contradiction with the reported initial results from the phase 1b findings of the trial NCT02807805, where a 1600 mg TID dosing cohort of niclosamide was reported to be safely tolerated.126 This report claims only a trough serum level of 0.1 μM would be sufficient for anti-cancer activity, and two patients analyzed (as of the report date) had trough levels of 0.305 and 0.496 μM of niclosamide.126 It remains to be seen whether final results from NCT02807805 will continue to contradict the findings of the completed study NCT02532114.
While the findings from NCT02532114 are disappointing, the study does establish important groundwork for re-purposing of niclosamide as a drug for PCa, as well as other malignancies. The preclinical concerns about the specificity of the effect of niclosamide and the underlying mechanisms that overcome resistance to abiraterone and enzalutamide, seem to agree with the findings of this clinical study. Effective SAR (structure-activity relationship) optimization of niclosamide to improve it’s oral-bioavailability and increased efficacy will be essential for its further development as an anti-tumor agent.
4 |. MARINE NATURAL PRODUCTS
4.1 |. EPI Compounds
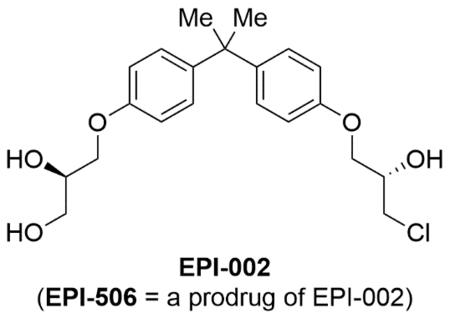
The EPI compounds developed by Marianne Sadar and co-workers are the most well-characterized publicly disclosed AR-TAD inhibitors to date. Based on a bisphenol A (BPA) derived structure, these compounds show the ability to inhibit androgen-dependent and androgen-independent AR activation.127 BPA in general is considered as an endocrine disruptor128,129 with accumulation potential in mitochondrial membranes that leads to oxidative stress induced cell death/damage.130 BPA has been shown to disrupt nuclear hormone receptor signaling, acting as an AR-antagonist (IC50 = 1–2 μM) and as an ERα agonist (IC50 = 10–100 nM).131,132 First isolated from a marine sponge (Geodia lindgreni) extract,133 EPI-001 is likely traceable to BPA derivatives used in industrial processes that were present in contaminated sea-water. On one end of the molecule, the BPA core of EPI-001 is attached to a propane-1,2-diol and on the other end to a chlorohydrin unit via ether linkages. The latter functionality provides the ability for EPI compounds to act as covalent binders. EPI-002, synthesized as a single stereoisomer (2R, 20S), has somewhat better performance characteristics in vitro and in vivo than EPI-001, which was a mixture of four diastereomers.134
4.1.1 |. Initial in vitro and in vivo efficacy
In the initial experiments, EPI-001 inhibited the ligand (R1881) induced, forskolin induced, or IL-6 induced activation of the AR to baseline levels.127 Constitutively active versions of the AR as well as DBD swapped versions were shown to be inhibited by EPI-001. These findings established that EPI-001 effects are mediated through the AR-TAD. EPI-001 blocked the androgen-regulated gene expression of some (e.g. PSA, TMPRSS2) but not all (e.g. BLVRB) genes.127 Androgen induced AR interaction at the chromatin level was reduced by EPI-001. This effect was proven not to be a result of decreased levels of AR protein, general prevention of serine phosphorylation of AR, or prevention of AR nuclear translocation.127 EPI-001 was shown not to affect GR nor PR activity at the concentrations used to inhibit AR.127 EPI-001 did not prevent AR ligand binding but inhibited the N/C interaction upon activation.127 Interaction of EPI-001 at the AR-TAD was shown to induce a conformational change as evidenced by steady state fluorescence spectra. However, no such interaction was observed at the GR-AF1.127,135 EPI-001 blocked the interaction of the transcriptional coactivator CBP (CREB binding protein) with the AR-TAD.127 A similar study with EPI-002 did not inhibit the association of p160 SRC family of coactivators with the AR, but showed consistent AR transcriptional inhibition even at elevated SRC levels.136 EPI-001 inhibited the AR-driven proliferation of LNCaP, PCa2B, and 22Rv1 cells in vitro, but did not affect the growth of RKO human colon cancer cells or MG63 osteosarcoma cells.127
Intravenous injections of EPI-001 at 50 mg/kg doses to mice demonstrated significant reduction in the weight of the prostates, LNCaP subcutaneous xenografts (start volume = 100 mm3, 14 d treatment, reduced to 73 mm3 in EPI treated, 148 mm3 in control), and serum PSA levels.127 Intratumoral injections at 20 mg/kg reduced the LNCaP xenograft tumor sizes to 35 mm3 at 25 days (start = 100 mm3, control = 436 mm3). Staining experiments on the harvested xenografts revealed reduced proliferation (Ki67 staining) and increased apoptosis (TUNEL screening).127 Similar demonstrations were made on castrated mice bearing orthotopic LNCaP xenografts. Conversely, EPI-001 did not affect the growth of AR-null PC3 xenografts. Further experiments of EPI-002 treatment on LNCaP95 derived tumors expressing AR-V7 showed growth attenuation and decreased AR-regulated gene expression.137 These in vitro and in vivo effects of EPI-001 and its specific stereoisomers against PCa cell lines bearing ARFL and ARSVs was further corroborated by Myung et al..134
An independent study by Brand et al. showed general agreement to the findings by Sadar et al. on EPI-001’s ability to affect AR activity, albeit with possible secondary effects (see section 4.1.4).138 The multi-level effects of EPI-001 led to the inhibition of TAU-1 (transcriptional activation units) and TAU-5 of the AR-TAD, reduced AR expression, and inhibition of growth of AR-positive and AR-negative PCa cell lines.138 In vitro domain swap experiments that tethered Gal4DBD to AR-TAD, TAU1, or TAU5 proteins showed the ability of EPI-001 to inhibit both the TAU domains.138 EPI-001 treatment reduced the expression of ARFL (LNCaP, VCaP, LAPC4, C4–2 cells) and ARSVs (22Rv1 cells), independent of proteasomal degradation. However, AR mRNA and protein expression of CWR-R1 cells were not affected by EPI-001 at the doses utilized.138 The rate of nascent AR mRNA synthesis in LNCaP cells was reduced by 50 μM treatment of EPI-001. The cell growth inhibition in C4–2 and 22Rv1 cells were shown to require >50 μM dose of EPI-001, while lower concentrations (>5 μM) were sufficient to achieve growth inhibition in LNCaP cells.138 However, at the higher doses (>50 μM) growth inhibitory effects were also seen in AR-negative PC3 (PCa), DU145 (PCa), and T47D (breast cancer) cell lines.138
Buchanan et al. showed that the variable length of the polyglutamine tract within the TAD is known to be inversely associated with the AR transcriptional activity.139 The inhibition of AR transcriptional activity by EPI-002 was not affected by polymorphic lengths (0, 12, 20, 40, 49) of the glutamine tract in full length human AR.136 EPI-002 was able to inhibit AR isoforms with gain-of-function mutations of the TAD (E255K, W435L) and the LBD (V715M, R761G, H874Y, T877A). AR-V7 driven expression of UBE2C, CDC20, and AKT1 were shown to be significantly reduced upon EPI-002 treatment.136
4.1.2 |. Stability and covalent binding
The chlorohydrin unit in EPI-001 can be converted to an epoxide spices at neutral (pH = 7.4) and basic (pH = 9.4) pH values. However, Brand et al. showed that had no significant detrimental effect towards the stability of EPI-001, with 91% and 87% of EPI-001 left after 12 h in pH = 7.4 and pH = 9.4 media respectively.138 Under acidic conditions EPI-001 was shown to have excellent stability, while nucleophilic additions at the chlorohydrin could happen depending on the pH of the media and the concentration(s) of the nucleophile(s). At substantially basic conditions (pH=9.4), EPI-001 formed adducts with glutathione, 2-mercaptoethanol, and cysteamine resulting in 2%, 0%, and 14% of EPI-001 remaining after 12 h of reaction time with 10 eq of nucleophiles.138 At a more physiologically relevant pH (7.4), 71% (glutathione), 88% (2-mercaptoethanol), and 100% (cysteamine) of EPI-001 was found remaining when exposed to the same nucleophilic substitution conditions.138 In a separate study by Myung et al., no discernable nucleophilic addition to EPI-001 was found when exposed to 5 eq glutathione or 3 eq of 2-mercaptoethaol at pH = 7.4 up to 7 days.134 Hence it is clear that the thiol alkylating ability of EPI compounds is dependent on the local nucleophile concentration, and that it shows good stability at acidic and neutral pH levels.
Evidence of direct and covalent binding of EPI compounds to the AR-TAD was shown by Myung et al. in 2013 through click chemistry experiments.134 Here, PCa cells were incubated with modified EPI-probes bearing an alkyne functionality. The cells were then lysed, biotin tags were attached to the alkyne functionality using click chemistry, and the subsequent mixtures analyzed by Western blots with antibodies for biotin and AR. All EPI probes bearing the chlorohydrin moiety were shown to covalently bind the AR regardless of compound chirality.134 These results showed that the chlorohydrin unit was essential for the binding at the AR, where compounds that had a hydroxy group in place of the chloro substituent did not show binding activity. Following further experimentation in cell-free conditions using purified recombinant AF-1 protein, the binding mechanism of EPI compounds at the AR-AF1 was hypothesized to be: 1) an initial reversible binding (fast) at the binding site, 2) an epoxide formation step (slow) at the chlorohydrin moiety (facilitated by active site amino acids), and 3) covalent binding (fast) to an active site nucleophile with the epoxide.134
4.1.3 |. Binding site at the AR-TAD
Given that AR-TAD is an intrinsically disordered protein domain, it is not trivial to study which sub domains or amino acid residues of it are involved in the covalent binding of EPI compounds. Using solution phase NMR studies, De Mol et al. explored this further to identify regions of AR-AF1 (AA 141–494) that undergo structural changes to facilitate selective binding of the EPI compounds.140 Two main regions of interest exist in the AF-1 that are critical for the transactivation of the AR-TAD, transcription activation units 1 and 5 (Tau-1, Tau-5). Tau-1 (AA 102–371) is important for the androgen dependent activation of AR, while Tau-5 (AA 361–537) has been associated with androgen-independent AR activation mechanisms.140 Using a predicted model for disorder propensity in the AR-TAD, a 306-residue portion of the IDP that had lower disorder (AF-1*, AA 142–448) was constructed and then studied using NMR to reveal partial folding characteristics.140 Using heteronuclear-multidimensional NMR experiments and a secondary structure prediction algorithm141, 50% helical propensity was found at the Tau-1 (185–200) and the Tau-5 (390–410) region residues.140 This secondary structure formation was independent of the inter-domain long-range interactions. Study of the transverse relaxation rates of the 15N nuclei in the AF-1* backbone further corroborated these findings.140 MS experiments revealed sufficiently slow reversible interaction of EPI-001 with AR-AF1* before undergoing covalent attachment. Studying this interaction by NMR revealed that distinct 15N chemical shift changes occurred in the Tau-5 residues 354–448 upon EPI-001 interaction. In comparison, the effect on the Tau-1 region was minimal. Using smaller polypeptides of Tau-5 interaction region (341–371, 391–414, 426–446) such interactions could not be demonstrated. Hence it is evident that the entire length of the interaction sequence is necessary, which presumably contributes to the adaptation of a partially folded structure (either naturally occurring or induced by EPI-001) that allows specific drug binding. This binding interaction was non-stereoselective with similar effects seen in the presence of all four diastereomers of EPI-001.140
4.1.4 |. PPARγ modulation
Nuclear receptor PPARγ (peroxisome-proliferator-activated receptor-gamma) has important regulatory involvement in multiple disease conditions including cancers, inflammation, and metabolic disorders.142 Most notable therapeutic use of PPARγ has been in the treatment of type-2 diabetes mellitus, via activation by thiazolidinedione (TZD) drugs. PPARγ agonists has been also generally associated with an ability to reduce tumor progression, including PCa. However, multiple mechanistic studies have found that the observed antiproliferative effects of the PPARγ agonists, occur via PPARγ-independent pathways.143,144 Perhaps the most important of these pathways in PCa is the enhancement of proteasomal degradation of Sp1, a transcriptional factor essential for the expression of multiple genes including the AR.143–146 Knockdown of Sp1 by siRNA has been shown to reduce the mRNA levels of AR, and attenuate AR-dependent gene transcription.144 Furthermore, Sp1 undergoes non-transcriptional inhibition by activated AR.146,147
Brand et al. hypothesized EPI-001 could modulate the PPARγ function, as a secondary mechanism to exert inhibitory effects on AR expression and activity in PCa.138 In agreement with their hypothesis, Brand et al. showed that EPI-001 had PPARγ modulation effects similar to PPARγ agonists such as troglitazone.138 Dose dependent induction of cyclin dependent kinase inhibitors p21 and p27 was seen upon EPI-001 treatment, in addition to the inhibitory effects on AR-protein expression.138 In comparison, troglitazone treatment was shown to inhibit the activity of AR-GAL4, Gal4-tethered AR-TAD, TAU1 and TAU5 analogous to the effects shown by EPI-001.138 Comparative, AR independent, selective PPARγ modulation activity was demonstrated with both EPI-001 and troglitazone, with both inducing mRNA expression of PPARγ target (CIDEC, TXNIP, PDK4) genes.138 siRNA mediated knock down of PPARγ activation partially rescued the AR-transcriptional inhibition by EPI-001, indicating a possible involvement of PPARγ in EPI-AR inhibition.138 This knockdown did not affect troglitazone mediated AR inhibition, which as mentioned before, occurs via PPARγ-independent pathways.138,144
Based on the traditional paradigm of PPARγ playing a tumor suppressive role, the above effects of EPI-001 appear beneficial. However, some recent findings suggest PPARγ signaling may actually be positively contributing to the development and progression of PCa.143 Increasingly enhanced expression levels of PPARγ has been shown in later stage PCa tissues in recent research143,148–150, although there is reason151 to argue it is also dependent on ethnic/hereditary backgrounds. Inhibition of PPARγ by antagonists such as GW9662 or warfarin has shown the ability to inhibit AR activity.152 Inverse regulatory effects have also been demonstrated recently by Olokpa et al., where the AR was shown to regulate the expression and the subsequent activity of PPARγ in PCa cells.153 AR activation by DHT (≥ 1 nM) was shown to reduce the levels and the activity of PPARγ in VCaP and C4–2 CRPC cell lines.153 Additionally, the use of siRNA to knock down AR protein resulted in the upregulation of PPARγ activity in CR-2 cells.153
With the above findings, it is apparent that there is some contrasting evidence about the role of PPARγ in PCa. The anti-proliferative effects of EPI-001 in the study by Brand et al. most certainly seem to indicate effects beyond simple inhibition of AR function, supported by the fact that EPI-001 treatment was shown to also inhibit the growth of AR-null cell lines.138 Interestingly, some research has shown that PPARγ activity is enhanced in AR-null (or low) cell lines.153,154 Hence with the PPARγ agonist functionality of the EPI compounds, they have the ability to exert enhanced PPARγ -dependent (and -independent) in vitro effects in such cells.
4.1.5 |. Effects on the PI3K-Akt-mTOR pathway
PI3K-Akt-mTOR (PI3K = phosphatidylinositol-3 kinase, Akt = protein kinase B [PKB], mTOR = mammalian target of rapamycin) signaling pathway has been demonstrated to have importance in PCa biology.8,155,156 Loss of proper function of the tumor suppressor gene PTEN (phosphatase and tensin homolog) is considered the major up-regulation mechanism of Akt signaling in human PCa.157 PTEN gene is reported to be altered in 40–60% of advance PCa cases.8,158 The inhibition of the PI3K pathway has shown in vitro antiproliferative effects on androgen induced growth of LNCaP cells, despite the upregulation of AR target gene expression.155 The upregulation of AR gene expression upon PI3K pathway inhibition has been linked to the relieving of feedback inhibition of HER kinases.159 Similar effects were seen in CWR22 PCa xenografts in vivo. Studies with wild type AR and mutant AR species showed, rapamycin (mTOR inhibitor) mediated upregulation of AR activity required a functional LBD.155 The co-treatment with bicalutamide and rapamycin showed synergistic, potentiated, antiproliferative effects on LNCaP cell growth.155 Marques et al. have also shown crosstalk between the PI3K pathway and the AR signaling axis, where the growth of PC346C xenografts were significantly reduced by PI3K and Akt inhibitors, despite the up-regulation of AR-target gene expression.160 This crosstalk between the pathways is reciprocal, given that AR inhibition has been shown to activate Akt signaling by reducing cellular Akt phosphatase PHLPP levels.159 Wu et al. has shown that this inter-pathway communication may be dependent on the levels of testosterone.161 Under low testosterone conditions, AR expression was up-regulated in response to sub-baseline mTOR activity, and vice versa.161 Reciprocal communication between AR and the mTOR signaling has also been shown in other cancers such as breast cancer and hepatocellular carcinoma.162,163 In hepatocellular carcinoma cells, mTOR signaling has been shown to reduce AR protein degradation and increase AR nuclear-translocation.163 Using this knowledge, dual inhibition of AR and the PI3K pathways has been validated as an efficient approach for the treatment of PCa in vitro and in vivo.159
In an attempt to explore this co-targeting approach in PCa driven by ARSVs, Kato et al. evaluated the therapeutic efficacy of a combination of EPI-002 and BEZ235 (PI3K and mTOR inhibitor) in LNCaP95 (enzalutamide resistant and PTEN-null) CRPC models.164 In the absence of androgen, BEZ235 increased the expression of ARFL and AR-V7 consistent with the reciprocal feedback mechanism. BEZ235 or everolimus (mTOR inhibitor) both reduced the expression of the AR-V7 regulated gene UBE2C. However, this unexpected effect was seen at longer (48 h) exposure to the mTOR inhibitors, and not so much at 24 h. EPI-002 was able to reduce the expression of UBE2C by AR-V7 as expected, but could not significantly decrease the expression of androgen promoted FKBP5 gene by ARFL at the concentrations (25 μM) employed.164 The latter lack of effectiveness can probably be attributed to the lower potency of the EPI compounds. BEZ235 (15 nM) and EPI-002 both inhibited the phosphorylation of pS6, a ribosomal protein regulated by mTOR signaling. This suggests some cross-reactivity of the covalent inhibitor EPI-002 towards the mTOR pathway. EPI-002 or enzalutamide co-treatment was able to diminish the BEZ235 induced increase of ARFL and AR-V7 in LNCaP cells, and the expression of AR driven genes in LNCaP95 cells.164 IL-6 or Forskolin induced AR-TAD activation was lowered by EPI-002, with no further advantage seen by EPI plus BEZ co-therapy. Co-treatment with BEZ235 (5mg/kg) and EPI-002 (100 mg/kg) showed greater reduction (over 14-days) of LNCaP95 (PTEN-null, enzalutamide resistant) xenograft volumes, than the treatment with each compound alone.164 While this is a promising in vivo result to establish mTOR and AR dual inhibition is viable in AR-V7 driven PCa,165 the large amount of EPI compound required to elicit such effect may not be easy to replicate in clinical development.
4.1.6 |. Co-treatment with Docetaxel
Microtubule targeting taxane drugs, such as docetaxel and cabazitaxel, are the most prominently used treatment at the metastatic castration-resistant stage of PCa. Microtubules play important roles within the cytoskeleton, facilitating intracellular transport functions in the interphase of the cell cycle and in playing a key role in the formation of the mitotic spindle prior to cell division.166,167 Taxanes primarily function by binding to the β-tubulin units in cellular microtubules, stabilizing the polymerized structures, which disrupts the microtubule dynamics required for proper activity.166,168–170 The inhibition of proper mitotic spindle assembly in the cells activates the “spindle assembly check point”, which would eventually lead to apoptosis through the onco-suppressive mechanism, called “mitotic catastrophe”.166,168 Apart from this non-AR involved anti-tumor mechanism of action, taxanes have few other ways it can act against PCa.171 The ligand-activated AR nuclear translocation is microtubule-network driven with the assistance of microtubule-traversing motor proteins such as dynein.166 Taxanes effects significantly hinder this process, sequestering AR to the cytoplasm, and hence reducing AR-gene transcription.166,172 Taxanes can drive FOXO1 (Forkhead box protein O1, an AR suppressive nuclear transcription factor) nuclear localization, that also results in the suppression of AR mediated transcription.166,173 Despite these multi-pathway inhibitory functions, native or acquired resistance to taxane therapy is seen in mCRPC patients. Among other methods,166 a primary avenue of resistance is believed to stem from the presence of splice variant forms of the AR.174,175 In particular, some AR-V7 driven PCa’s have shown marked resistance to docetaxel treatment, both in vitro and in vivo.174,175 AR-V7, which lacks a hinge region, was shown not to have significant association with microtubules or dynein.174,175 Although some evidence suggest that the AR-TAD was important for tubulin-AR association,176 AR-V7 nuclear translocation mechanism is likely to be independent of the microtubule network, leading to docetaxel resistance.
To evaluate the effectiveness in using a N-terminal domain inhibitor of the AR to mitigate this resistance mechanism, Martin et al. treated CRPC tumor models with both EPI (−001 or −002) and docetaxel.177 Docetaxel (1 μM) treatment resulted in significant reduction of 22Rv1 (AR-V7 driven) CRPC cell viability.177 EPI-002 mono treatment showed a much smaller effect on cell viability despite the 25 μM concentration used. The co-treatment of EPI plus docetaxel was found to have an additive effect towards reducing cell viability. In 22Rv1 xenografts, a more discernable synergism towards tumor growth suppression was seen at 11 days with EPI-docetaxel co-treatment (200 mg/kg/day EPI-001, Docetaxel 15 mg/kg/day).177 EPI-001 alone was not able to suppress the growth of 22Rv1 xenografts compared to the control group even at the relatively high dose utilized. The number of apoptotic cells was shown (TUNEL assays) to be far greater in the co-treated xenografts, than in either mono-treatment regimen. Tumor vascularity between the co-treated and the mono-treated groups was not that different.177
In agreement with previous findings,174,175 the cellular distribution of ARSVs was not significantly affected by docetaxel treatment.177 AR-driven reporter activity of probasin, PSA, and ARR3 genes were all significantly reduced by the co-treatments, though the advantage compared to single agent treatment was not universally evident.177 Expression of AR proteins (FL and SVs) was increased in 22Rv1 cells when treated with EPI compound or docetaxel alone. This effect was attenuated by the co-treatment.177
In 22Rv1 cells, the expression of N-cadherin (mesenchymal marker) was significantly up-regulated by EPI treatment indicative of EMT initiation. Docetaxel co-treatment was able to attenuate that effect.177 No significant change in the levels of cellular E-cadherin (epithelial marker) was seen with any treatment combination in vitro.177 In 22Rv1 tumor xenografts, single agent treatments increased the levels of E-cadherin and lowered the level of N-cadherin. Unfortunately, co-treatment with EPI and docetaxel reversed this effect, suggesting possible EMT initiation.177
Although the suppression of 22Rv1 xenograft growth was promising via the EPI-docetaxel co-treatment, the synergistic effectiveness of this approach is not clear. The use of EPI in this context is further hampered by the need to use a significantly high dosage (200 mg/kg/day) in xenograft studies. A more potent inhibitor of the AR-TAD might be able to exert better synergistic effects to mitigate ARSV driven taxane-resistance in CRPC.
4.1.7 |. EPI compounds as imaging agents
Based on the selective covalent binding ability of the parent compound to the AR-TAD, Sadar and co-workers developed 123I- labelled analogs of EPI-002 as potential tools for the imaging of PCa’s that express AR isoforms.178,179 Iodine substituted (at the carbon-15) EPI-002 was synthesized first as the cold version (I-EPI-002), to test the binding efficacy before moving on to incorporation of the radiolabel. The IC50 of I-EPI-002 was given as 1.17 μM for the inhibition of androgen induced transcriptional activity of endogenous AR in LNCaP cells.178,179 The authors indicated this to be as potent an effect as enzalutamide treatment and ten times as potent as EPI-002, with reference to previously reported IC50 values for those compounds. The inhibition of AR activity in reporter gene assays was achieved in similar levels (~75%) with the treatment of 25 μM of EPI-002 or 2 μM of I-EPI-002.178 Luciferase reporter assays were employed to show the iodinated version maintained specificity to the ARE without significant effects on GRE, PRE, nor ERE at the concentrations employed.178,179 I-EPI-002 was shown to inhibit the proliferation of LNCaP95 cells (AR-V7 driven) with an IC50 of 6.9 μM. Here, EPI analogs were shown to cause G0/G1 cell cycle arrest in the tumor cells.178 Binding of the radiolabeled probe (123I-EPI-002) to endogenous ARFL in LNCaP95 cells was shown by the use of western blotting and phosphorimaging. The binding of 123I-EPI-002 was found to be reduced when the cells were co-treated with EPI-002 reaffirming that they both bind the same target site(s).178
In vivo time-dependent biodistribution analysis (with the use of a gamma-spectrometer) conducted after the administration (tail-vein) of the radio probe, revealed the compound to be present in many organs. Highest levels of accumulations were observed in the intestines, gall bladder, and the liver.178 Authors indicated that this effect matches with the fact EPI compounds having lipophilic structures (123I-EPI-002 cLogP = 4.2) are expected to be eliminated by the hepatobiliary system.178 The variation of EPI compound biodistribution in tissues over time (e.g. percentage of “injected dose/gram” in the large intestine: 1.2% at 1 h – 66% at 4 h)178, may indicate that the binding of EPI compounds at the tissues is sufficiently reversible at physiological conditions. Maximum tissue concentrations (2.2% injected dose/gram) in the LNCaP95 xenograft was shown to occur at 2 h, which could be blocked up to 74% with the co-administration of cold EPI-002 (50 mg/kg).178 This blocking effect was not seen at PC3 xenografts or muscle tissue, indicating that the blocking was specific to AR containing tissue.178 Micro-SPECT/CT imaging at 2 h was able to corroborate these findings, where the radio probe was able to specifically visualize the LNCaP95 xenograft vs. the PC3 xenograft.178 While the above observations were quite promising, the stability of the radio-probe was found to be questionable due to the observation of accumulation of radioactivity in the thyroid (2.3% of injected dose/gram). If the 123I dislodging from the probe is a possibility, then further concerns do arise about the reliability of data on the long-term in vivo bio-distribution of 123I-EPI-002, as well as about the concomitant toxicological effects. Nevertheless, this study established the first proof-of-concept experiments on using an AR-TAD targeted compound to visualize AR-driven PCa tissues.
4.1.8 |. Clinical trials and outlook
First-in-human phase 1/2 clinical trials (Clinicaltrials.gov, NCT02606123) of EPI-506 (Ralaniten acetate) were initiated by ESSA Pharma in 2016–2017. EPI-506 is reported to be an acetate prodrug of EPI-002.29,180 The study was directed to evaluate the safety, pharmacokinetics, maximum tolerated dose, and anti-tumor activity of EPI-506 in men with end-stage mCRPC who have progressed after prior enzalutamide and/or abiraterone treatment and may have received one prior line of chemotherapy.181 In the phase 1 study, 28 patients were treated at EPI-506 doses ranging from 80 – 3600 mg/day doses.181 The drug was found to be generally well tolerated at doses up to 2400 mg/day. Consistent with the pre-clinical observations, somewhat higher doses (higher than 2400 mg/day) were required to see any effects on serum PSA levels. Modest lowering of PSA levels (4–29%) were seen in five men at doses ≥ 1280 mg/day. No reductions of PSA by 50% or more, a standard for evaluation of early phase clinical trial activity, was observed. In prioritizing their efforts to develop a different class of AR-TAD inhibitors with increased potency than EPI-506, ESSA Pharma Inc. announced in September 2017 that they will discontinue further development efforts towards EPI-506. This newer “Aniten program” compounds are also stated to have structural similarities to the EPI series of compounds.181
4.2 |. Polychlorinated Small Peptides: Sintokamides and Dysamides
Sintokamides are a class of natural products that were isolated from marine sponges Dysidea sp. via extraction by MeOH.182 Structurally sintokamides are polychlorinated peptides, capable of undergoing nucleophilic additions at the enone site, and nucleophilic substitution reactions at the chlorinated carbons. Sintokamide A was able to block the AR activity induced by the potent AR agonist R1881 (methyltrienolone)183 as measured by PSA-luciferase reporter assays.182,184 The effect of sintokamide was specific towards the AR, with no discernable activity against the PR or the GR mediated transcription.184 Cell viability assays conducted on LNCaP cells revealed no discernable cytotoxicity on the cells at a concentration of 10 μg/ml of Sintokamide A, showing the AR activity inhibition was not due to cytotoxicity. Similar to EPI-002 treatment, Sintokamide A was able to inhibit the proliferation of both LNCaP (ARFL driven) and LNCaP95 (AR-V7 driven) cell lines.184
Sintokamide A was found to block the androgen induced proliferation of androgen sensitive LNCaP cells at a level comparable to the AR antagonist bicalutamide.182 Similar treatments of Sintokamide A on PC3 human PCa cells lacking AR expression revealed no effects towards cell proliferation, indicating the effects of Sintokamide A was based on AR activity.182,185 Furthermore, Sintokamide A was shown not to bind the AR-LBD in exerting its activity. At concentrations of 0.5 – 50 μM it was unable to compete off Fluoromone binding to the AR-LBD.184 Sintokamide A did not affect the AR cellular distribution with or without androgen stimulation.184
Sintokamide A inhibited the transactivation of the AR-TAD stimulated by forskolin at a concentration of 5 μg/ml of Sintokamide A.182 This was demonstrated through transfecting LNCaP cells with the AR TAD-Gal4DBD chimera protein that would show its activity via a Gal4-luciferase reporter system. A sample of these cells were treated with forskolin and incubated for 24 h. Forskolin is an AR-TAD transactivation facilitator which activates the AR through a protein kinase A mediated pathway.182,186 Forskolin-induced dephosphorylation of the AR has been shown to lead to impaired ligand binding,187 which suggests the increased activity comes possibly via N-terminal transactivation. The forskolin treated cells showed significant increase in luciferase mediated luminescence indicating AR-TAD transactivation.182 In comparison, cells that were pre-treated with Sintokamide A prior to forskolin treatment, had the AR-TAD transactivation significantly diminished.182,184 However, Sintokamide A was less active than EPI-002 in asserting this effect.184 Furthermore, unlike EPI-002, Sintokamide A was not able to inhibit the IL-6 induced AR-TAD transactivation.184 Use of Sintokamide A and EPI-002 in combination was shown to have additive effects in inhibiting the AR-mediated gene expression in luciferase reporter assays.184 With these observations there is postulation that Sintokamide A and EPI-002 are likely bind two different regions in the AR-TAD-AF1.184
Biological evaluation of structurally related polychlorinated small molecule peptide analogs called dysamides has shown similar AR inhibitory activity.185 Like the sintokamides, dysamides were also isolated from marine sponges in the Dysidea sp. family.188 In a comparative activity study, sintokamides A, B, C, & E as well as dysamide A demonstrated inhibition of R1881 induced PSA-luciferase signal.185 Compared to the inhibition effected by bicalutamide, the sintokamides showed inhibition levels of ~ 60 – 100 % of the luciferase signal. Sintokamide B, which has the highest degree of chlorination in its structure, showed the greatest inhibition potency. Dysamide A was also able to exert an inhibition level of ~50%. Interestingly, the non-chlorinated analog of sintokamide was only able to show a modest percentage of inhibition (~20%) as compared to the parent compounds.185 In a separate study, isopropyl substitutions in place of chlorinated carbon groups in Sintokamide A, resulted in a compound (LYP19) with negligible activity (~40 μM in an AR reporter assay).184 Hence these compounds were postulated to also have a mechanism of action similar to EPI-002, involving a nucleophilic attack at the chlorinated carbons, at the AR-TAD binding site.
Using click chemistry experiments analogous to those done with EPI compounds,134 evidence was shown to establish covalent binding of Sintokamide A at the AR-TAD.184 LNCaP cells were incubated overnight with an analog of Sintokamide A (with an alkyne handle), the cells lysed, and then a biotin tag attached to the alkyne handles. Western blot analysis of this mixture with anti-biotin and anti-AR antibodies revealed compound bound AR.184 Streptavidin mediated AR immune precipitation confirmed this finding.184 Similar binding effects were demonstrated using purified recombinant AF-1 protein under cell free conditions.184
In a subcutaneous LNCaP xenograft model, sintokamide A treatment was shown to reduce the tumor volume with time, as compared to tumors treated with DMSO that continued to grow.184,185 However, the metabolic stability of sintokamide A was found to be poor. Following an intravenous dose of 50 mg/kg, a Cmax (8 μM) lower than the in vitro IC50 values was achieved in plasma, with a t1/2 of 1.16 h.184 Antitumoral effects of Sintokamide A (30 mg/kg/day every three days) against LNCaP95 xenografts (driven by AR-V7) were demonstrated by the ability to inhibit (~36%) tumor growth up to 15 days.184 PSA levels and the number of Ki67 (proliferation marker) positive cells in the harvested LNCaP xenografts were also found to be significantly decreased after undergoing Sintokamide A treatment.184
Unfortunately, the clinical relevance and translation ability of the in vivo data for Sintokamide A are uncertain, because the compound had to undergo intratumoral delivery due to its metabolically unstable184 nature. For any further development of sintokamides as viable therapy against PCa, SAR studies to improve the in vivo and the in vitro qualities of the compound are particularly necessary. To this end, the attempts already made to establish synthetic routes to produce Sintokamides via organic synthesis will provide a useful starting-point.185,189
4.3 |. Niphatenones
Analogous to sintokamides, niphatenones are marine natural products, that have been isolated from the marine sponge Niphates digitalis through continuous extraction of sponge samples with MeOH.190 Key structural features of niphatenones include a Michael acceptor enone moiety, and an EPI like glycerol ether moiety along with a long hydrocarbon chain. Naturally occurring niphatenones A & B both carry the stereogenic carbon in the “S” conformation.190 After initial structure elucidation, both stereoisomers of niphatenones A and B were manually synthesized on a larger scale to carry out further analysis.190 The compounds were initially evaluated on an AR mediated luciferase reporter assay on LNCaP human PCa cells. S-niphatenone-B was significantly more active in this assay than S-niphatenone-A. S-niphatenone-A was able to reduce the PSA-luciferase activity by 25 % (with the control designated 100%), while S-niphatenone-B reduced it by ~ 50%. The “R” isomers of both compounds showed somewhat better activity than the naturally occurring “S” isomers, with R-niphatenone-A showing PSA-luciferase activity of ~35%, and R-niphatenone-B showing only 25%. Hence R-niphatenone-B was the best inhibitor of the PSA-luciferase activity that was unveiled through this assay.190
Hydrogenation of the double bond of R-niphatenone-B resulted in loss of half of the activity of the original enone compound. However, this R-dihydroniphatenone-B form still functioned better than S-niphatenone-A, showing that the enone function, while important, was not an absolute necessity to show activity. Removing the glycerol moiety (akin to the EPI compounds) from the molecule via other functional group transformations also resulted in similar loss of activity. Shortening the long alkyl chain to a methyl group, resulted in complete loss of activity. Non-specific toxicity of these compounds was initially ruled out by comparing the cell proliferation of the AR dependent LNCaP cells and the AR-independent PC3 cells in the presence of the niphatenones. Niphatenone-B (S or R) did not affect the proliferation of PC3 cells that do not express the AR, while they did inhibit the AR dependent proliferation of the LNCaP cells.190 In another series of experiments involving an AR-driven PB-luciferase reporter assay, both the S and R niphatenone-B compounds were shown to have IC50 values around 6 μM towards blocking AR driven reporter responses.191
Click chemistry experiments carried out on recombinant AR-TAD AF1 proteins, showed the binding of an alkyne functionalized analog of R-niphatenone-B to the AR-TAD AF1. After allowing 50 min of interaction time with the AF1 protein, a fluorescent tag was attached using click chemistry. The material was then analyzed using SDS-PAGE and the band corresponding to AF-1 protein was found to be labelled with the fluorescent tag, validating that the R-niphatenone-B analog can covalently bind the AR-TAD-AF1 protein.190 Banuelos et al. evaluated the effect on AR-TAD transactivation of both enantiomers of niphatenone-B using LNCaP cells co-transfected with Gal4UAS-TATA-luciferase and AR-(1–558)-Gal4DBD which encodes the full AR-TAD.191 The transactivation was induced by pre-treatment with IL-6 for 24 h. Transactivation driven AR-TAD-Gal4-luciferase activity was reduced to 76% in the presence of R-niphatenone-B, while S-niphatenone-B reduced it to 50% as compared to the control (100%).191 EPI-002, which was used as a positive control, was able to reduce the activity to 40%. The activity of the constitutively active AR splice variant ARvar567, was also lowered to ~65% by both of the enantiomers of niphatenone-B.191
In assessing the cross reactivity with other nuclear hormone receptors, Banuelos et al. showed that S-niphatenone-B inhibited (lowered to 19%) the steroidal transcriptional activity of full-length AR, but not that of the PR. However, here S-niphatenone-B also lowered the transcriptional activity of GR down to 80% as measured using a GRE-luciferase reporter.191 Using fluorescence polarization assays, it was shown that S-niphatenone-B did not interfere with ligand binding to AR, PR, or GR at the concentrations less than 30 μM.191 S-niphatenone-B was able to significantly reduce (~50%) the N/C terminal interaction192–194 in the AR upon ligand binding.191 S-niphatenone-B also was able to inhibit the androgen induced expression of AR regulated genes PSA (70% inhibition) and KLK2. However, it did not affect the subcellular localization of AR nor the levels of AR protein in LNCaP cells in the absence of androgens.191 The translocation of AR in to the nucleus in the presence of androgens was not affected by S-niphatenone-B either.
Extending the work by Meimetis et al., further click chemistry experiments were done by Banuelos et al. on AR-AF1 and GR-AF1, which showed S-niphatenone-B covalently bound both these proteins. Even with the small sequence identity between the two receptor types195–197, this result raised questions about the specificity of the niphatenone binding.191 On the premise that the enone functionality might be leading to promiscuous binding, alkylation reactions of niphatenone were done using glutathione. Both the niphatenone-B enantiomers ended up being readily alkylated with glutathione.191 The negative control EPI-002, which has also been shown to be an AR-TAD-AF1 binder, did not alkylate under the same conditions. Hence the authors concluded that S-niphatenone-B was not worth pursuing further as a potential drug given its tendency to be a random alkylating agent.191 Analogs with lower reactivity that lack the enone functionality or in which it is replaced by a less reactive homologue may be worth pursuing in the future.
5 |. MAHANINE
Mahanine is a carbazole alkaloid present in the leaves and the edible parts of the Thai vegetable Micromelum minutum198,199 and the southeast-Asian curry leaf plant Murraya Koenigii.200 Mahanine has been found to exhibit antimicrobial and anti-inflammatory effects.199–201 It is a potent apoptosis-inducing agent against leukemic cells via mitochondrial pathways,198,202 and against pancreatic cancer cells via the induction of reactive oxygen species production.203 Mahanine has shown the ability to restore the activity of epigenetically silenced tumor suppressor gene RASSF1A in human PCa cells resulting in the down regulation of the key cell cycle regulator cyclin D1.204
Exploring the promising activity of mahanine against PCa cells, Amin et al. have shown that mahanine can inhibit ligand-dependent and -independent transactivation of AR, as well as initiate AR-degradation and inactivate CDK1 in PCa cells.205 DHT-induced AR-transactivation in LNCaP cells with full length AR was significantly reduced with mahanine treatment shown via luciferase reporter systems carrying either a ARR3-TK- or a PSA-promoter.205 LNCaP cells transfected with a human telomerase reverse transcriptase (hTERT)-promoter luciferase construct did not show any effect towards similar mahanine treatment, showing that mahanine affected only the AR driven promoters. Within the assay times and the mahanine doses used (up to 10 μM), the cellular AR levels did not significantly decrease. The expression of DHT-induced AR target genes GREB1, NDRG1, PSA, PMEPA1, and SGK1 were also shown to be repressed in the presence of mahanine. Using LNCaP and VCaP cells transfected with an expression vector of AR-TAD-Gal4DBD and a luciferase reporter vector, the effect of mahanine on ligand-independent AR transactivation involving AR-AF1 was measured.205 Mahanine did not affect the luciferase activity of a constitutively active VP16-Gal4DBD vector, showing the Gal4DBD was not involved in the inhibitory effect of mahanine towards the AR-TAD.205 Forskolin or IL-6 induced luciferase activity via AR-TAD transactivation was reduced by ~80% in the mahanine treated cells as compared to the control.205
At a dose of 10 μM, mahanine was shown (via western blot analysis) to decrease the levels of AR in LNCaP, VCaP, and 22Rv1 cells over three days. Mahanine induced not only the degradation of the full-length AR, but the 80-kDa splice variant AR-V7 found in the 22Rv1 cells.205 Further studies involving the pretreatment of LNCaP cells with cycloheximide to inhibit protein biosynthesis, followed by the treatment of mahanine, reaffirmed that mahanine induces AR degradation. Such degradation was significantly reduced when mahanine treatment was done in the presence of the proteasome inhibitor MG132, indicating the presence of a ubiquitin-proteasome pathway for the degradation of AR by mahanine. A two-fold increase in ubiquitinated-AR was found in LNCaP cells treated with MG132 (5 μM) and mahanine (20 μM) over 12 hours.205
The DHT induced AR nuclear translocation in LNCaP cells was greatly diminished by mahanine as shown by immunofluorescence assays. Similar experiments done on 22Rv1 cells showed that the AR-V7 splice variant was also distributed more in the cytoplasm in the presence of mahanine. Monitoring of the nuclear AR localization over 12 h showed that the AR content in the nucleus was progressively depleted as the AR migrated into the cytoplasm. By conducting the experiments in the presence of cycloheximide and MG132, the migratory effect was further confirmed.205 When the LNCaP cells were grown in CS media, mahanine was not able to prevent the DHT-induced nuclear translocation of AR. But, the translocated AR was transcriptionally inactive as judged by the PSA expression levels.205
Mahanine was able to inhibit the DHT-induced phosphorylation of AR Ser-81 which is considered an important post-translational modification206 for AR transcriptional activity.205 To evaluate a possible pathway by which this inhibition occurs, LNCaP cells were synchronized to the G2-M phase of the cell cycle by treatment with nocodazole (100 ng/mL, 24 h) where maximal CDK1 activity occurs. CDK1 is known to phosphorylate the AR at the Ser-81 site in an androgen dependent manner. While the untreated cells showed induction of AR Ser-81 phosphorylation caused by the activation of CDK1 by nocodazole, cells treated with mahanine (10 μM) showed significantly decreased phosphorylation.205 DHT-induced AR Ser-81 phosphorylation in LNCaP cells co-transfected with a constitutively active version of CDK1 (i.e. non-androgen dependent) was not significantly affected by mahanine. While these data do not indicate the inhibition of AR signaling by mahanine is exclusively dependent on a CDK1 mediated pathway, it is noteworthy since CDK1 activity is commonly elevated in CRPC. Given the ready natural availability and the possibility to devise an efficient total synthesis207, mahanine and its derivatives holds some promise to be developed further.208 Its effects on multiple signaling pathways however raise questions about specificity and underlying mechanism(s) of action.
6 |. VPC-14228, VPC-14449, SKLB-C2807, and PYRVINIUM
AR-DBD is also a less explored site as an alternative target for AR signaling axis inhibition. Binding of the activated AR (both FL and SVs) to DNA to initiate transcription is achieved through the AR-DBD. Key interactions in this association are made through the DNA recognition α-helix that consists of a P-box region209 that inserts in to the major groove of the DNA.210 Unlike the case with the TAD, crystallographic data has been obtained for the DBD (rat, PDB 1R4I) with the use of an AR-DBD dimer bound to a steroid DR3 response element.1 Based on this structural information, an in silico drug design/screening approach by Li et al. established a plausible binding site (Ser-579 to Lys-610) under the P-box region of the AR-DBD.210–214 Virtual screening of drugs at this site and hit optimization through SAR led to the discovery of thiazolyl morpholine derivatives VPC-14228 and VPC-14449. VPC-14449 showed in vitro inhibition of R1881 stimulated AR activity (IC50 = 0.12 μM) and PSA suppression (IC50 = 0.17 μM) in LNCaP eGFP cells, comparable to enzalutamide treatment under the same conditions.210 Activity inhibition, albeit with less potency, was also demonstrated later in enzalutamide-resistant 22Rv1 cells.215 There was no discernable effect seen on the cell viability of non-AR driven PC3 cell growth by VPC-14449.215 Furthermore, it showed no significant effects towards 31 genotoxin-responsive genes (e.g. CASP1, XPC, ATF3), indicating no discernable cytotoxicity.211 The structure initially reported210 for VPC-14449 in the 2014 disclosure was later found to be mis-assigned, and subsequently corrected in 2017.212
Pyrvinium, a quinoline derived cyanine dye, was tested along with the VPC compounds due to reports of its function as an AR-DBD inhibitor.216,217 While it was also able to inhibit AR transcriptional activity (IC50 = 0.194 μM), pyrvinium induced a non-PCa selective strong apoptotic response evidenced by PARP (poly (ADP-ribose) polymerase) cleaved products.211,215 Such non-selective PARP cleavage was not seen for VPC-14449.215 In a report by Lim et al., pyrvinium showed significant non-selective inhibition towards other nuclear hormone receptors as well, albeit in prostatic cells.216 This cross reactivity could be explained by modelling studies that show the interaction of pyrvinium with the AR-DBD to occur at the conserved area of Lys-610 to Pro-613.211,216 Nevertheless, there has been much interest in exploring pyrvinium pamoate as an anti-cancer therapeutic due to the fact it was once used as an anthelminthic drug with FDA approval. Given that it affects a large number of key cellular pathways and cancer types,218–220 there is a need to alter its structure/function before it can be further developed as a potential therapeutic.
Important coordinating interactions with VPC-14228 at the AR-DBD binding site were found at Ser-579, Val-582, Phe-583, Arg-586, Gln-592, Tyr-594, and Lys-610 through docking studies on a human AR-DBD homology model.210 Gln-592 (not conserved across other NR) and Tyr-594 in particular were initially proposed to be important in the selective binding to AR-DBD which has a highly conserved structure with other human nuclear hormone receptors.210,211 Site directed mutagenesis experiments (Gln592Asp, Lys593Asp, Tyr594Asp) confirmed the proposed target site, and further in vitro experiments showed VPC compounds ability to inhibit the activity of ARSVs.210,211,215 Y594D mutation (H bonding site removed) greatly reduced the activity of the VPC compounds in ARFL and ARSVs, while pyrvinium could still strongly inhibit the AR transcriptional activity.211 Using docking studies, Tyr-594 was shown to form a hydrogen bond with the VPC compound when bound to a human AR-DBD homology model.210 Later mutagenesis studies (K593D) revealed that Lys-593 was also an essential residue to facilitate VPC compound interaction at the AR-DBD.215
VPC compounds showed inhibition of ER transcriptional activity at higher doses (> 5 μM).211 But this effect was several folds lower than for AR inhibition. Similar selectivity between AR-DBD and GR-DBD was shown using a chimera construct, where the KQKYL sequence of the AR-DBD was replaced by QHNYL.215 Compared to the wild-type AR (IC50 = 0.291 μM), the chimera with GR-DBD (IC50 = 11 μM) showed resistance to VPC-14449 inhibition of R1881 induced activity.215 VPC compounds showed no significant activity in the inhibition of GR or PR activity at concentrations equal to or less than 25 μM.211 Using YFP (yellow fluorescent protein) tagged ARFL and AR-V7, Dalal et al. showed that the inhibition of the DBD by VPC compounds did not impede the nuclear translocation of the AR.211 While the chromatin binding of nuclear localized ARFL was suppressed by >1 μM concentrations, no significant inhibition was seen for AR-V7 even with the use of 50 μM concentrations of VPC-14449.215 Similar need for higher concentrations (>10 μM) of VPC-14449 to suppress chromatin binding interactions were seen for ARv567es in R1-D567 cells, and for AR in MR49F (enzalutamide resistant) and C4–2 (androgen insensitive) cell lines.215 ChIP assays showed that the association of the translocated AR with PSA and FKBP5 AREs were reduced in the presence of VPC compounds.211 VPC-14449 was able to inhibit the association of ARv567es to FASN and FKBP5 AREs at a concentration of >50 μM. But lower concentration (>10 μM) of the compound was less affective.215
Dalal et al. showed that VPC-14449 was able to inhibit clinically relevant AR-LBD mutants activity and show additive in vitro effectiveness in co-treatment with enzalutamide.215 R1881 induced AR-dependent gene expression (TMPRSS2, KLK3, FKBP5), in cell lines carrying AR with wildtype or mutant LBD, was significantly suppressed by the treatment of 5 μM VPC-14449.215 A greater concentration (50 μM) of VPC-14449 was required to induce a significant change in AR-V7 driven UBE2C gene expression.215 Expression of the GR driven gene FKBP52 was not affected under these conditions.215 Only a modest inhibitory effect on ARv567es driven expression of FASN and FKBP5 gene were seen even at 50 μM treatment with VPC-14449.215 These observations collectively suggests that these compounds do not robustly inhibit ARSV driven gene expression, contrary to the design principle of these compounds. Dalal et al. postulate this may derive from the fact the ARSVs conformational arrangement may differ significantly enough from the ARFL resulting in a different mode of association with the AREs and/or a DBD interacting compound such as VPC-14449.215 A higher dosing (intraperitoneal injection, twice daily, 4 weeks) of VPC-14449 (100 mg/kg) was shown to be as effective as enzalutamide (10 mg/kg) in blocking androgen signaling in vivo as evidenced by reductions in tumor volume and serum PSA levels in mice with LNCaP tumor xenografts.211
Further SAR analysis of VPC-14228 by Xu et al. established that the thiazole and the morpholine rings were essential to the activity of the VPC compounds.221 Introduction of an acyl group at the phenyl ring of VPC-14228 gave rise to a lead compound, SKLB-C2807, that showed antiproliferative effects (IC50 = 0.38 μM) on a LNCaP-AR PCa cell line.221 Docking studies on a human AR-DBD homology model showed SKLB-C2807 forming key H-bonding interactions (between the morpholine-O and Tyr-594, and between the carbonyloxy of the benzene ring with Arg-609) and hydrophobic interactions (benzene ring with Lys-610).221 The recent studies with SKLB-C2807 corroborated the previous findings with the VPC compounds, and showed selective in vitro activity against AR positive PCa cell lines and no impediment towards AR nuclear translocation.221
The dosing used215 to obtain significant in vivo effects seems to be quite large when considering the lower IC50 (100–200 nM) values generally reported for the VPC compounds. Metabolic stability would have to be evaluated more closely to see if there is a correlation. Furthermore, the effectiveness of these compounds against ARSV–chromatin association seems to be minimal. Redesign of these compounds to increase the in vivo efficacy and to increase the ability to affect ARSVs will be critical for the further development222 of these compounds.
7 |. JN COMPOUNDS
Our own program to develop inhibitors of the AR-TAD have resulted in a series of compounds that selectively inhibit the AR-mediated transcription in AR-positive cells in vitro and attenuate PCa xenograft growth in vivo.223,224 The initial hit compound (JN018) was identified through a high throughput screening assay in which reporter gene expression in yeast is dependent upon the constitutive transcriptional activity of an ARSV. Extensive SAR studies led to a series of compounds (termed the “JN series”) with over 150 analogs synthesized to date. The active compounds show growth inhibition of AR expressing PCa cells, but not AR null PCa and non-PCa cells. They were shown to directly bind the AR and function through inhibition of the TAD. Interestingly these compounds also show potent and selective degradation effects towards both full length and splice variant AR. The in vitro anti-tumor effects of the JN compounds are significantly more specific towards AR-positive cell lines than the in vitro effects recently shown for the AR-degradation enhancer ASC-J9®.225 Furthermore, the active JN series compounds show 10 to 30-fold higher potency in comparison to EPI-002, in cell viability and functional assays. While the structures nor the specific data of JN series compounds have been disclosed publicly yet, the lead compound along with few other analogs have moved on to in vivo testing in relevant PCa xenograft models in which oral administration resulted in significant control in tumor growth of 22Rv1 (enzalutamide-resistant) and LNCaP-AR castration resistant xenografts. Based on these promising results, further development of these compounds towards preclinical evaluation is currently underway in partnership with a prominent pharmaceutical company.
8 |. BET INHIBITORS
The Bromodomain and Extra-Terminal domain (BET) family of proteins have found profound recent interest in being a target for small molecule inhibition. These proteins are characterized by the presence of two tandem bromodomains and one extra-terminal protein domain.226 They are ~110 amino acid containing protein domains that primarily function as readers of lysine acetylation codes in histones, facilitating epigenetic regulation of gene transcription.226–228 The histone code arising from post-translational modifications translate to important information that are potentially hereditary and may result in non-DNA derived phenotypic changes.229 Enrichment of the H3K27Ac acetylation marker has been shown at the proximal sites of the AR-gene.230 Several AR co-factors (e.g. Lysine specific demethylase 1 [LSD1]) are known to control the expression of AR target genes with modifications to the histone proteins.231
The primary members of the mammalian BET protein family are BRD2, BRD3, BRD4, and BRDT. Bromodomain containing proteins are involved in the regulation of oncogenes such as Myc. Overexpression of BRD4 in particular, has been shown in multiple types of cancer.232 As such BRD4 has been a subject of extensive research as a drug discovery target in the past decade.233 BET proteins also serve as key co-regulators of other transcription factors such as the AR. Direct association of BRD4 to AR has been demonstrated.234 Reasonable understanding of the function of BET proteins led to a marked research interest resulting in many pharmaceutical companies attempting to identify BET inhibitors as anti-cancer therapies.232,235,236 Availability of crystal structures have amplified this interest due to the ability to adopt in silico design/screening approaches to accelerate hit discovery.237
8.1 |. BET inhibitor JQ1
Targeting co-activators of AR gene transcription as a method to disrupt AR gene transcription at the chromatin level is an evolving method for targeting CRPC.238 Out of the BET family of proteins, BRD4 in particular has shown the ability to interact with the AR-TAD in facilitating AR gene transcription.239 Building on the reported ability of thienodiazepines to bind bromodomains,240 JQ1, an efficient cell-permeable small molecule inhibitor of BET proteins, was found through in silico design approaches utilizing the apo crystal structure (PDB: 2OSS) of the first bromodomain of BRD4.241 It is a pan-BET inhibitor (BETi) due to the highly conserved nature of the BET acylated lysine (Kac) binding pocket. The S-(+) isomer was the active stereoisomer of the compound, with the (R)-(−) showing no binding ability.230,241 Chemically JQ1 has a thienotriazolodiazepine core structure. Hence it has structural similarity to allosteric modulators of the GABAA receptor, such as benzodiazepines (e.g. diazepam) and triazolobenzodiazepines (e.g. alprazolam). Using an ExpresSProfile assay (with 53 receptor proteins), Filippakopoulos et al. showed JQ1 (1 μM) does not affect the radioligand binding at the GABAA receptor benzodiazepine site.241 Showing further specificity in binding, JQ1 showed partial inhibition of ligand binding only in neurokinin NK2 and adenosine A3 receptors, out of 52 other receptor proteins probed.241
JQ1 inhibition of bromodomains was shown to be specific to the BET family out of all the human bromodomain containing proteins.241 This binding specificity is believed to be in part derived from the conserved gate-keeper residue Ile-146 at the bromodomain 1 of BRD proteins 1–4.242 Other bromodomain containing proteins having a bulkier gate-keeper residue (e.g. Tyr in PCAF and GCN5) does not allow for the efficient binding of JQ1 like compounds that have a pendent aryl group.242 Binding of JQ1 at the BRD3 and BRD4 bromodomains (Kd = 50, 90 nM) were about threefold better than at the BRD2 and BRDT.241 Luminescence proximity homogeneous assays have established the ability of JQ1 to inhibit the binding of acylated lysines at the BET bromodomains. JQ1 inhibited the binding of a tetraacetylated histone H4 peptide to BRD4 (33, 77 nM), but not the binding of an acetylated H3 peptide to CREBBP (CREB-binding protein).241 High resolution crystal structures and docking studies revealed a perfect fit for the geometrical shape of the (+) isomer at the bromodomain acylated lysine (Kac) binding sites. In contrast, binding of the (−) isomers to the Kac binding site in docking studies resulted in high energy distortions due to steric clashes.241 Binding of JQ1 was shown to stabilize the Kac binding site flexibility, with significant hydrophobic interactions forming between the ligand and the binding site.241 JQ1 (500 nM) was able to competitively inhibit BRD4-chromatin association, as determined by FARP (fluorescence recovery after photobleaching) experiments.241
A landmark study in 2014 by Asangani et al. established that JQ1 was able to selectively inhibit the growth and colony formation of AR-driven VCaP, LNCaP, and 22Rv1 PCa cell lines at nanomolar (50–200) IC50 values.239 Knockdown of BRD2–4 proteins by targeted siRNA resulted in similar effects on cell proliferation and invasion, phenocopying JQ1 treatment.239 JQ1 treatment of AR-positive cell lines further showed G0-G1 arrest and apoptosis, with a dose dependent increase in cleaved PARP (apoptosis marker).239 Similar to the usual BETi effects, JQ1 showed downregulation of the anti-apoptotic protein BCL-xl (B-cell lymphoma-extra large) in these cell lines in a dose dependent manner.239 AR-regulated target proteins were also seen to be down-regulated by JQ1 in the AR-positive cell lines. This down-regulation was not recovered by co-treatment with a proteasomal inhibitor (bortezomib), indicating the down-regulation to be at the transcriptional level.239
JQ1 (0.5 – 2.5 μM) showed the inhibition of MYC protein expression in AR-positive PCa cells (VCaP, LNCaP, 22Rv1), but not in the AR negative PCa cells (DU145, PC3).239 MYC is a critical proto-onco gene that acts as a regulator of cell growth and proliferation. It has been shown to be upregulated in multiple cancers, with its inactivation having been shown to result in tumor regression.243 Ligand-independent regulation of MYC by the AR has been shown in several PCa cell lines.244 In experiments by Gao et al., treatment with R1881 did not affect the expression levels of c-MYC in 22Rv1 or LNCaP PCa cells.244 Attenuation of c-MYC levels were seen when treated with JQ1, but not enzalutamide.244 BET inhibition has generally being linked to the direct knockdown of MYC gene transcription243, although contrasting evidence245 has also emerged in recent years. In support of the lack of a direct link between BET inhibition and MYC regulation, knock down of MYC by siRNA was unable to attenuate the cell invasion properties of VCaP cells, while JQ1 treatment inhibited cell invasion.239 Furthermore, exogenous expression of MYC was unable to rescue the cell growth inhibition effects of JQ1 in AR-positive cells.239
A study by Chan et al. demonstrated the ability of JQ1 to reduce the expression and the androgen mediated ARE recruitment of ARFL and ARSVs.230 Dose dependent reductions of ARFL and ARSV were demonstrated in CRPC cell lines (C4–2, 22Rv1, VCaP) upon JQ1 (0.1 to 10 μM) treatment.230 JQ1 treatment (0.5 μM) was shown to inhibit the binding of BRD2 at the proximal H3K27Ac sites at the AR gene in cells (VCaP, R1-AD1 and R1-D567).230 This effect was less prominent (in VCaP cells) towards BRD3 and BRD4 recruitment.230 Probing this effect further, Chan et al. demonstrated that the JQ1 mediated attenuation of AR-chromatin binding was not necessarily dependent on the involvement of H3K27Ac or BET family proteins.230 JQ1 treatment was able to reduce AR-chromatin binding in BRD4-involved (e.g. FASN-ARBSI) and BRD4-noninvolved (e.g. intron 5 of FKBP5) sites to a similar extent (~50%) in VCaP cells.230 These findings suggest that the anticancer effects seen in PCa via BET inhibitor treatment, are not mediated through the disruption of AR-BET protein interactions alone.
Using gel filtration chromatography experiments, Asangani et al. showed the possibility of the formation of a large functional multi-protein complex with BRD4, AR and RNAP2.239 The association ability to AR was also present in BRD proteins 2 and 3. Supporting the notion of JQ1 being a pan-BETi, ChiP-seq assays revealed a 62–86% overlap in genome wide effect on BRD2–4 towards inhibition by JQ1.239 BRD4 was able to bind the AR-TAD with a Kd of 70 nM.239 Using halo tagged AR in in vitro pull down assays, the interaction with BRD4 (primarily through bromodomain 1) with the AR-TAD was mapped out to occur at the region of amino acids 120 – 160.239
Reduction of DHT-induced AR gene expression by JQ1 was shown to be comparable to or lower than the effects seen upon enzalutamide treatment in LNCaP and VCaP cells.239 Analysis of genome wide AR-localization and recruitment at DNA via ChIP assays showed the ability of JQ1 to inhibit the DHT-induced recruitment of AR to target loci at an equivalent potency to enzalutamide.239 Recruitment of AR and BRD4 at shared loci (2031 sites identified) were differentially affected by enzalutamide and JQ1. At these loci AR-recruitment was better lowered by enzalutamide than JQ1, while JQ1 was able to completely inhibit the DHT-induced BRD4 recruitment at such sites.239
TMPRSS-ERG (Transmembrane protease serine 2 – ETS [erythroblast transformation-specific] related gene) fusion-gene is the most common (~50%) oncogenic genetic alteration found in PCa.239,246,247 It drives prostatic tumor progression with the expression of PSA as well as ERG (transcriptional regulator) protein. ERG overexpression has been implicated to be involved in developing novel super-enhancer regions, affecting the histone acetylation code, and thus driving the upregulation of specific genes that could contribute to PCa progression.248 PSA and the ERG expression in VCaP cells showed significant inhibition (at 48 h) by the treatment of JQ1 in a dose dependent manner.239 These effects were traced to de-recruitment of RNAP2 at the ERG gene, and AR/BRD4 at the TMPRSS2 promotor/enhancer regions by JQ1.239
ChIP-seq analysis revealed that DHT-treatment resulted in increased AR binding at the MYC distal enhancer, while reducing the recruitment of RNAP2 at the gene.239 Enzalutamide treatment removed this inhibitory effect towards the MYC locus. Hence, Asangani et al. postulated that the de-repression of MYC gene expression might be a mechanism in enzalutamide-resistance in CRPC.239 JQ1 treatment in contrast, had no such up-regulatory effects on c-MYC expression.239
Mice bearing VCaP xenografts showed significantly higher tumor growth inhibition by the treatment of JQ1 (50 mg/kg) than by enzalutamide (10 mg/kg).239 Similar to previous reports, the in vivo treatment by JQ1 also reduced the testis size in the treated mice.239,249 Enzalutamide treated mice were shown to have pro-metastatic (to liver and femur) effects, while JQ1 treated mice showed no discernable metastases.239 A castration-resistant VCaP xenograft model also showed ~50% inhibition in tumor volume upon JQ1 (50 mg/kg) treatment.239
I-BET762, a triazolobenzodiazepine analog of JQ1, has shown similar in vitro and in vivo BETi functions in models of PCa.250 Other recent drug development programs have uncovered non-diazepine type compounds that can still function as BETi’s. Y08060, with a reported BRD4 bromodomain-1 inhibiting IC50 of 302 nM is an example.251 This compound is a 2H-benzo[b][1,4]oxazin-3(4H)-one derivative with a pendent aryl group connected to the core via a sulfonamide linkage.251 Unfortunately, the cell growth inhibitory ability of this lead compound was still 17 to 60 fold less potent than JQ1 in parallel assessment in PCa cell lines (C4–2B, LNCaP, 22Rv1).251 Nevertheless the ability to obtain BETi’s beyond a diazepine structure may allow one to circumvent off-target effects that may occur at high-doses of JQ1 like compounds. A comprehensive review about BRD4 inhibitors with varying structures can be found elsewhere.233 One of the more interesting of these is the sulfonamide derivative PFI-1, which has been shown to occupy the Kac binding site in BRD4 and BRD2.252,253 A recent study has shown the ability of PFI-1 to inhibit the transactivation of constitutively active AR species (AR-V7 and non-sense mutant Q641X) and to attenuate the growth of AR-positive cell lines (LNCaP, 22Rv1).254 However, at the higher doses (20 μM) AR-null PC3 cells also demonstrated growth inhibition similar to LNCaP cells.254
Recent evidence have shown AR-overexpression in PCa cells can lead to increased expression of bromodomain containing proteins, including BRD4.255 Furthermore, AR-overexpression has also been linked to increased histone acetylation.256 Together these factors allow for a genome-wide increase in epigenetic DNA accessibility. Such chromatin relaxation leads to abnormally increased transcription factor binding and subsequent gene expression.255 BET inhibitor (JQ1) treatment has been shown to attenuate this chromatin opening effect, particularly in AR-overexpressing cell lines.255 Experiments by Urbanucci et al. had shown that a combination-treatment of enzalutamide and JQ1 led to apoptotic effects in the AR-overexpressing VCaP cell line, but not in LNCaP cells.255
8.2 |. Resistance to BET Inhibition
Acquired resistance to BETi’s in tumors including PCa is an emerging topic of discussion. A particular mechanism of resistance may be more important in a certain tumor type treatment than another. A report by Power et al. indicated that the attenuation of BRD4 chromatin binding can bring about reactivation of AR signaling and a silencing of DNA damage response genes DDR2.257 In BET-resistant cultured LNCaP and 22Rv1 cells, BRD4 inhibition either by the use of JQ1 or a BET-PROTAC degrader conferred no significant anti-proliferative effects. Despite the lack of downstream effects, BRD4 was present and did bind JQ1, as evidenced by cellular thermal shift assays.257 Neither JQ1 nor the PROTAC treatments impacted the MYC expression in these BETi-resistant cells.257 Probing the BETi-resistant cell lines by Gene SET Enrichment Analysis revealed a positive enrichment of AR target genes.257 Despite the overexpression of multiple canonical AR target genes, the AR transcript levels were found to not be increased in the BETi-resistant cells.257 The cause of this was traced to increased CDK9 activity and an apparent increase in AR stability.257 CDK9 is a known facilitator of AR-chromatin binding via the phosphorylation of Ser89 in the AR.206 This phosphorylation strengthens the AR-chromatin binding interaction, allowing for enhanced AR activity. Inhibition of CDK-9 activity led to a significantly larger decrease in cell viability in the BETi-resistant cells than in the sensitive ones.257 Additionally the response to enzalutamide treatment was enhanced in these cells owing to the upregulation of the AR mediated transcription.257
The DNA damage markers γH2A.X and 53BP1 have shown elevated levels in BETi-resistant cells.257 In the absence of BRD4, signaling of DNA damage is known to be enhanced.258 However, BRD4 levels are not linked to the kinetics of repair nor to the generation of DNA damage.258 COMET assays established further evidence of enhanced DNA damage in the BETi-resistant cells.257 Additionally, transcriptional silencing was observed in DDR (DNA damage repair) genes.257 BRD4 recruitment at the DDR genes was found to be reduced in the cells chronically exposed to BET inhibitors.257 Despite the transcriptional change in DDR-genes, no significant cell cycle arrest was observed in the BETi-resistant cells.257 Down-regulation of homologous recombination (HR) genes was also found in these BETi-resistant cells.257 In such HR-deficient environments, PARP mediated DNA repair becomes critical for avoiding DNA damage and cell survival.259 In response to single-stranded DNA breakage, PARP initiates corrective action through base-excision repair.259 Supportive of this notion, Pawar et al. found that the BETi-resistant cells had high PARP activity, and hence enhanced sensitivity to PARP-inhibition by Olaparib.257 Similar observations in BETi induced reduction of HR efficiency and increased sensitivity to PARP-inhibitors was recently shown in ovarian cancer by Wilson et al..260 A phase 2 clinical trial (NCT03047135) to evaluate the efficacy of Olaparib in patients with high-risk biochemically-recurrent PCa is currently underway. Preliminary efficacy for mCRPC treatment with Olaparib has been seen, with a 11/49 (ten of whom had mutations in DNA repair genes) patient PSA response. Due to their impaired DNA-repair ability the BETi resistant PCa cell lines have also shown increased sensitivity to cisplatin treatment when compared to the wildtype cell lines. The sensitivity enhancement was two-fold in LNCaP cells, while it was 20-fold in 22Rv1 cells.257
Another major mechanism of resistance to BETi demonstrated in PCa therapy arises from SPOP (speckle-type POZ protein) – mutations.234,261 This gene is reported to be frequently mutated in PCa patients with F133 being the most frequently mutated site.261 The resistance is believed to occur via increased stability of BET proteins and Akt/mTORK1 activation.261 SPOP is a cullin (CUL) based E3 ligase substrate adaptor protein, involved in the cellular protein degradation machinery.234 It was shown to promote the ubiquitination of BET proteins in a dose dependent manner.234 Gene ontology assays and coimmunoprecipitation assays have revealed BET proteins to be major binding partners of SPOP.261 Knockdown or knockout of SPOP in PCa cell lines (C4–2, 22Rv1) showed increased levels of its substrate proteins, including BET proteins and AR, without affecting BET protein mRNA expression.234 BRD4 in particular was shown to be stabilized by the lack of SPOP activity.234 Enhanced in vitro colony formation and proliferation was seen in C4–2 cells with SPOP mutations, compared to the cultures with wild-type SPOP.234,261 Knockdown of BRD4 showed significantly higher reduction in the SPOP-mutated cell proliferation.234,261 ShRNA knockdown of BRD4 resulted in reduced tumor growth in mice with 22Rv1 xenografts bearing SPOP mutations.234 Zhang et al. has reported that such enhanced stability and the resultant elevated levels of BRD4 leads to an up-regulation of cellular Akt-mTORC1 pathway.261 They also showed the upregulation of several genes in the cholesterol biosynthesis pathway (FDFT1, DHCR24, DHCR7 and MVD), and the Rho GTPase family member RAC1, in SPOP-mutated tumors.261 PTEN loss and Akt/mTORC activation is well described to be a major-occurrence in PCa cell survival.262,263 Additionally, caveolin-1 containing cholesterol-rich lipid-rafts have been associated in tumor development and metastases in PCa.264 Increased levels of RAC1 were shown to increase the phosphorylation of Akt. This phosphorylation could be reversed by the knock down of RAC1, re-sensitizing the SPOP-mutated cells to JQ1 treatment.261 Similarly, combined depletion of the cholesterol synthesis genes resulted in attenuation of the Akt/mTORC signaling and re-sensitized the C4–2 cells to JQ1 treatment.261
Non-PCa cell lines have also shown acquired resistance to BETi’s, indicating that this will be a major topic of discussion ahead in the development of BETi’s. JQ1 mediated anti-proliferative effects in hepatocellular carcinoma were overcome by the cancer cells via the upregulation of mcl-1.265 Use of a CDK inhibitor in co-treatment to reduce mcl-1 expression overcame the JQ1 resistance in HCCLM3 and BEL7402 cell lines.265
8.3 |. Clinical Trials
A BETi currently in clinical trials for CRPC, developed by Zenith Epigenetics, is ZEN-3694.266 It has concluded a dose escalation and dose confirmation phase 1 clinical trial (NCT02705469) in mCRPC patients. Based on the first clinical trial, the company announced that the drug has a good safety profile and PK properties, and they have identified a maximum tolerable dose.267 ZEN-3694 is reported to be able to bind BET proteins at a >20 fold higher potency compared to other human bromodomain containing proteins.266 Synergistic anti-proliferative effects with enzalutamide/apalutamide have been shown in VCaP cells.266 Sub-micromolar growth inhibition IC50’s have been shown against several ARFL and ARSV driven PCa cell lines (22Rv1 = 0.19 μM, VCaP = 0.9 μM, LNCaP = 0.40 μM), while not showing any discernable effect against the AR null PC3 PCa cell line.266 GR-upregulation in enzalutamide-resistant LNCaP cells was significantly inhibited by Zen-3694.266,268 In monotherapy, Zen-3694 (100 mg/kg) showed comparable in vivo activity to enzalutamide (10 mg/kg) in VCaP cells.266 At 2 h postdosing measurement, both PSA and the c-MYC expression were seen to be attenuated by ZEN-3694 treatment.266 Better in vivo tumor efficacy of Zen-3694 than enzalutamide was seen in a 22Rv1 xenograft model at the same doses.266 With these findings and the results of the first clinical trial, ZEN-3694 has moved on to another phase 1/2 clinical trial (in CRPC patients) for co-treatment with enzalutamide (NCT02711956). Other notable BET-inhibitors progressed to PCa clinical trials269 include GS-5829 (Gilead, for mCRPC, NCT02607228) and MK-8628 (Merck, for advanced solid tumors including CRPC, NCT02259114). GS-5829 is being evaluated both as a single-agent and as a co-treatment with enzalutamide. MK-8628 has completed the NCT02259114 phase 1 trial, though full information is not yet publicly available.
8.4 |. Targeting Complexity and Outlook
Being key elements to the cellular cross-talk mechanism, BET proteins interact with many cellular-signaling pathways.232,238,270 Beyond the AR-TAD, the major pathways and factors impacted include MYC, JAK/STAT pathway, PI3K/Akt pathway, p53 acetylation, and the NF-kB pathway.228,232,233,236,238,271 With the impact on several tumor-related pathways, even the BET inhibitors in clinical trials for a single ailment such as CRPC, are also being tested for treatment efficacy in other solid tumors and lymphomas. To date, most reproducible successes found with BET inhibitors lie in the treatment of hematological cancers and as a treatment towards NUT midline carcinoma.229,235,272 To increase the therapeutic impact, co-treatment methods have also been adopted or proposed to combine BET inhibitors with an existing therapeutic (e.g. enzalutamide).232,271 Additionally, PROTAC like technologies to utilize the cellular protein degradation machinery to degrade BET proteins have emerged as another potential CRPC therapeutic approach.24,273,274 ARV-771, a pan-BET degrader has shown tumor regression in 22Rv1 mice xenografts (up to 15 days) at a 30 mg/kg dosing.24
It is yet unclear why some tumor cell lines do not respond to BETi’s despite the generally accepted ability of BETi’s to affect the c-MYC expression (and other oncogenic pathways too). Even in the absence of AR function, one could assume PCa cells such as PC3 and DU145 may be affected by micromolar treatment of a BETi like JQ1. But this was not the case as demonstrated by Asangani et al.239, which has led to scrutiny about whether we know enough about the complex associations between the cancer epigenetics, MYC-regulated functions, and the overall effect of BET inhibitors.275 In regular cells, BET proteins are involved in critical processes, including cytokine gene transcription, T cell differentiation, adipogenesis, insulin production, and suppression of latent viruses.270 Andrieu et al. in a recent study276 has shown the involvement of BET proteins in the EMT process in breast cancer models. BRD3 and BRD4 were shown to have inhibitory action against this process, while BRD2 was shown to promote EMT. Additional complexity in BETi treatment is found in the fact that JQ1 has shown to confer interference to the SPOP mediated proteolytic degradation of BET proteins and increase their half-lives.261 This could mean a situation where a continued treatment with a BETi would suddenly lead to an opposite-therapeutic effect when the SPOP gene gets mutated, much like how enzalutamide becomes an agonist with AR-LBD mutations.
Being readers of the histone-acetylation code, they may have heretofore unknown effects towards translating the genetic code as well. Given the above factors, major concerns remain about the specificity and the long-term side-effects that could occur with BET inhibition, despite the promising therapeutic potential. As such, there is some belief that the undertaking of clinical trials of BET inhibitors has been premature.270
9 |. PERSPECTIVE
Over the last couple of decades, we have learned that the androgen receptor remains a critical driver of growth of castration resistant PCa. This AR dependence exists at the outset of castration resistance as well as after treatment with novel AR signaling axis inhibitors like abiraterone and enzalutamide. All of the AR targeting therapies that have received regulatory approval for clinical use directly or indirectly target the AR through its ligand binding domain. It is unlikely that additional compounds that target the LBD will produce clinically meaningful results given the frequent cross-resistance observed between currently available compounds that target the LBD. Hence the recent developments that has led to the emergence of promising compounds that can affect the AR-signaling axis through effects beyond the AR-LBD is of great interest (Figure 6). Historically, drug development efforts aimed at the N-terminus of the AR have been hampered by lack of structural knowledge of the transactivation domain and the extensive homology of the AR DNA binding domain with that of other nuclear steroid receptors. However, over the last several years, multiple groups have begun to identify compounds that target the AR TAD or DBD, although the specificity and activity of these compounds may hamper their clinical development. Nonetheless, the AR TAD and DBD remain potentially viable drug targets if some of the pitfalls of existing compounds can be overcome. Given the closer homology between the short DBD fragments between nuclear hormone receptors, the AR-TAD is perhaps the more promising of these two target domains to achieve AR selective effects.
Figure 6.
The effect on the AR-Signaling axis by recently emerged small-molecules that have direct or indirect interaction(s) with the AR N-terminal transactivation domain (TAD) or the DNA binding domain (DBD).
A large majority of the reported proof-of-principle work done to target the AR-TAD has employed compounds isolated from marine sponge extracts. In this regard, Prof. Marianne Sadar and her co-workers have established some fundamental experimental techniques in evaluating the binding of small molecule compounds at the AR-TAD. All the marine sponge derived AR-TAD inhibitors thus far reported, function as covalent drugs. Although a number of therapeutics do have covalent warheads, the reactivity of the covalent moiety is an important factor to consider in reducing the off-target effects. For example, the enone moiety in the niphatenones was too reactive to sustain it as a possible therapeutic for further development. The only compound to have progressed in to clinical trials thus far from this genre, EPI-506, met the end of further development owing to a low clinical response rate. The reported data for the bioactivity of Sintokamide A, another marine sponge isolate, suggests that it may also need concentrations similar to EPI-002 for eliciting a clinically relevant effect. Systemic bioavailability of Sintokamide A has been shown to be poor, with an elimination half-life of 1.16 h. For any further development, further SAR optimization will be necessary to improve its in vitro and in vivo characteristics.184 The poor bioavailability of sintokamides may also have a correlation to the reactive electrophilic centers in its structure.
A small molecule compound binding at the AR-TAD in particular might not have the same binding constraints that an ordered domain such as the LBD would impose. Given the IDP nature of the TAD and the fact that it is inherently designed to interact with many binding partners in the cellular environment suggests that the stereochemistry of any chiral centers in a small molecule compound binding the AR-TAD may have less functional significance, unless it imposes a large steric difference. This notion is supported by the data for EPI compounds, where all four diastereomers of EPI-001 were found to bind the AR-TAD and produce reasonably similar AR-inhibitory effects.
Niclosamide, mahanine, and BET inhibitors, while reported to have direct or indirect interactions with the AR-TAD, do not seem to exert their anti-PCa effects based on such interactions alone. Niclosamide, primarily affects the AR gene expression through the inhibition of the MAPK and the IL6/STAT3 pathways, and the enhancement of the degradation of AR-V7. The clinical trials of niclosamide with co-administration of existing AR antagonists has put greater value on this ARSV degradation effect, since substantial in vivo success in reducing ARSV expression was observed with relatively low niclosamide co-treatment doses. However, a major bottle-neck for the further development of niclosamide was found in the recently concluded dose escalation study for niclosamide (NCT02532114, in co-treatment with enzalutamide).125 The general concerns about the specificity of niclosamide’s effects due to its multi-pathway impact, and concerns about its poor oral bioavailability were found to be true in the outcome of this study. The concluded NCT02532114 study establishes the fact that it’s essential to do SAR optimization of niclosamide to improve its biological properties prior to further evaluation, and advice caution against the development of therapies that may have too many multi-pathway effects. In the event of a drug with poor bio-availability such as niclosamide, the ability for dose-escalation would be severely hindered if it has multi-pathway impact. Mahanine also has AR degradation effects. Inhibition of AR signaling by mahanine seems to be at least partly derived from its inhibition of CDK1 activity.205 AR degradation effects of mahanine appears to be slow but does affect both the ARFL and ARSVs.205 Given the involvement of BET proteins in many physiological pathways, BET inhibitors have also demonstrated multiple anti-tumor effects. BET proteins are possibly more important in the androgen dependent transactivation of the AR, and the consequent recruitment at the AREs. However, some data also show that the inhibition of AR-ARE recruitment by BET inhibitor treatment is not necessarily dependent on the BET proteins.230 Attenuation of GR upregulation in enzalutamide resistant cells by the clinical candidate Zen-3694 (a BETi) is also supportive of the notion that the anti-tumor effects by BETi’s in PCa may primarily be founded in processes independent of the AR.266,268
Interesting preferential inhibition/degradation effects of the ARFL versus ARSVs has been observed with some of these compounds. Lower doses of niclosamide were shown to enhance the degradation of AR-V7 significantly more than that of ARFL.79 In spite of its design principle, VPC-14449 was shown to have significantly less inhibition potency towards ARSV driven gene expression, than what was seen for ARFL.215 Hence the conformational arrangements of the ARFL and ARSVs may have significant differences in three dimensional shape as well as in accessibility to different sites, that would have to be taken in to account when applying in silico drug design approaches. In this context, NMR studies done on smaller fragments of the AR-TAD to find the binding locations of compounds (EPI-002, Sintokamides) may not present a conclusive determination of binding site or binding efficacy. The transient, partially folded binding conformations adopted by the AR-TAD are likely to differ between the smaller fragments (AR-AF1, TAU-1, TAU-5) of the TAD, full length AR-TAD, and ARFL. Therefore, accompanying biochemical evaluations are essential to complement any receptor-fragment based NMR finding. Following such analysis, EPI compounds are believed to bind the TAU-5 (AA 102–371) region of the AR-TAD while sintokamides bind the TAU-1 region (AA 361–537).184 TAU-5 in particular is considered necessary for androgen independent transactivation of the AR. Constitutive activity in ARSVs has been shown to have significantly low dependence on the amino acid region proximal to the N-terminus of the AR-TAD.277 This means that the binding of BRD4 at the amino acids 120–160 of the AR-TAD, as demonstrated by Asangani et al.,239 may have minimal impact towards regulatory functions of the androgen independent AR transactivation, and in PCa driven by ARSVs. BET inhibitors are therefore unlikely to have direct functional consequences towards constitutively active AR isoforms.
Hydrogen bonding is an important factor that governs the intrinsic organization and the intramolecular associations of DNA. Hence it is no surprise that the design principles of the VPC compounds as well as of the DNA binding hairpin polyamides are strongly rooted in the utilization of H-bonding to provide critical binding interactions as well as directional recognition. Homology between the DBD’s of the nuclear hormone receptors (NHRs) and between the NHRs association sites at the DNA, hamper the utilization of these two therapies to selectively target the AR or ARE’s. At the current juncture, the practical use of these compounds may be limited to advanced disease conditions where enzalutamide resistance is driven by GR overexpression. While in vitro studies have shown that VPC compounds manifest a few fold higher binding selectivity for the AR-DBD than that for other nuclear hormone receptor DBDs, in vivo work has required higher dosing that may still result in off-target effects when translated to clinical studies.
Despite the prominent extension of life expectancy granted to PCa patients by recent development of drugs like enzalutamide and apalutamide, the emergence of inevitable resistance to such therapies raise a need for a continuous search for better therapeutics. As outlined in this review, the interplay and cross-talk between multiple oncogenic pathways with the AR-signaling axis makes this an uphill task. Theories that challenge the conventional belief that AR inhibition/degradation represents the most efficacious way to target PCa have also emerged. A recent study has shown that the inhibition of AR signaling in PCa cells results in the de-suppression of a PCa promoting gene, ZBTB46, leading to EMT initiation.278 Here, Chen et al. postulate that simple targeting of AR signaling may predispose PCa to progress to a metastatic castration resistant state.278 For this reason, even the complete abolition of AR protein, as promised by the emerging PROTAC type technologies, might not be an optimal choice for long term treatment of PCa. Eradication of AR could simply be met by tumor cells in an unforeseen escalation of a reciprocal oncogenic pathway that is suppressed by AR signaling. Hence it is important to continue the study of fundamental processes in PCa to identify not only how to best target the AR signaling axis but also novel targets that may arise as a consequence of AR inhibition. In the search for such targets, Fong et al. has recently described the polycomb group protein EZH2 as a novel target in CRPC therapy, inhibition of which can attenuate AR signaling and inhibit PCa cell/xenograft growth via the matricellular protein CCN3.279 CCN3 was shown to have direct association with the AR-TAD as a part of a negative feedback loop that controls AR activity.279 Use of small molecule spliceosome inhibitors to suppress the alternative splicing derived production of AR-V7 has been also shown recently to have reasonable efficacy towards PCa growth both in vitro and in vivo.280 Finding tissue-selective AR modulators has also re-emerged as a potential approach to improve the specificity of AR directed therapies.281
The next phase of PCa drug development is likely to have a greater emphasis on accurately identifying resistance mechanisms to current and emerging monotherapies and devising co-treatment options to prolong the effectiveness of the therapeutic. Even with the concerns highlighted in the above paragraph, the AR continues to remain the best target of interest for developing PCa therapies based on the current knowledge of PCa physiology. The intrinsically disordered nature of the AR-TAD, while providing a challenge for drug discovery, can also prove to be advantageous for the chronic use of a selectively binding inhibitor. In principle, when compared to the LBD and the DBD, therapies targeted at the AR-TAD hold the greatest chance of long term success in being less susceptible to resistance mechanisms such as point mutations. Further study of this IDP domain is critical for raising the effectiveness of future targeted therapeutics of the AR-TAD. We believe that compounds with appropriate performance characteristics will make their way to clinical trials, where proof-of-principle studies will be established to show that effective targeting of the AR TAD or DBD is possible and can lead to clinically relevant improvements in the outcome of patients with mCRPC, the lethal form of PCa.
ACKNOWLEDGMENTS
This work and the development of the JN series of compounds was supported by the National Institutes of Health (CA164331 and CA12861) and the U.S. Department of Veterans Affairs. Authors thank Jiabin An for her work in the development of the JN series compounds.
ABBREVIATIONS
- ADT
androgen deprivation therapy
- Akt
protein kinase B [PKB]
- AR
androgen receptor
- ARE
androgen response element
- ARFL
full length AR
- ARSV
splice variant AR
- BET
bromodomain and Extra-Terminal domain
- BETi
bromodomain and Extra-Terminal domain protein inhibitor
- BPA
bisphenol A
- CRPC
castration resistant prostate cancer
- DBD
DNA binding domain
- DHT
dihydrotestosterone
- ECM
extra cellular matrix
- EMT
endothelial mesenchymal transition
- ER
estrogen receptor
- ERK
extracellular signal–regulated kinases
- ETS
erythroblast transformation-specific
- GR
glucocorticoid receptor
- GRE
glucocorticoid response element
- HR
homologous recombination
- IDP
intrinsically disordered protein
- IL-6
interleukin 6
- Im
N-methylimidazole
- JAK
Janus kinase
- Kac
acylated lysine
- LBD
ligand binding domain
- LRP6
low-density lipoprotein receptor related protein 6
- MAPK
mitogen-activated protein kinase
- mCRPC
metastatic castration resistant prostate cancer
- mTOR
mammalian target of rapamycin
- PARP
poly (ADP-ribose) polymerase
- PCa
prostate cancer
- PI3K
phosphatidylinositol-3 kinase
- PPARγ
peroxisome-proliferator-activated receptor-gamma
- PR
progesterone receptor
- PSA
prostate specific antigen
- PTEN
phosphatase and tensin homolog
- Py
N-methylpyrrole
- RNAP2
RNA polymerase II
- SAR
structure-activity relationship
- SPOP
speckle-type POZ protein
- STAT
signal transducer and activator of transcription
- TAD
transactivation domain
- TMPRSS
transmembrane protease serine 2
AUTHOR BIOSKETCHES
Dayan Elshan received his B.Sc. degree in Chemistry from the University of Colombo (Sri Lanka) in 2008 with first class honors. He conducted his doctoral research on the multivalent targeting of melanocortin and cholecystokinin receptors under the guidance of Prof. Eugene A. Mash at the University of Arizona and obtained his Ph.D. degree in 2014. In 2015, he joined UCLA as a postdoctoral scholar working with Prof. Michael E. Jung and Prof. Matthew B. Rettig, to develop novel prostate cancer therapies that function by targeting the androgen receptor transactivation domain (AR-TAD). His current/future research interests lie in further drug development work as a medicinal chemist, and in probing how the intrinsically disordered AR-TAD protein domain specifically interacts with its binding partners.
Matthew Rettig MD received his undergraduate degree from Wesleyan University (1986) and his medical degree from Duke University (1990). He completed internal medicine training at University of Washington (Seattle, WA, 1993) and hematology-oncology fellowship at UCLA (1996). Subsequently, he has remained as faculty at the David Geffen School of Medicine at UCLA, where he is Professor of Medicine and Urology and Medical Director of the Prostate Cancer Program. As a physician-scientist, Dr. Rettig has an active laboratory program aimed at drug development of inhibitors of growth promoting signals in prostate cancer; he also directs the clinical trials program in prostate cancer. In these capacities Dr. Rettig performs forward and reverse translational research to bridge the gap between the lab and the clinic. Dr. Rettig also serves at the Chief of Hematology-Oncology at the West Los Angeles VA, where he directs a nationwide program of biomarker driven prostate cancer clinical trials known as POPCAP (Precision Oncology Program Cancer of the Prostate).
Michael E. Jung was born in New Orleans, LA, in 1947. He received his PhD from Columbia University in 1973 working as a synthetic organic chemist with Gilbert Stork and continued his postdoctoral training as a NATO Postdoctoral Fellow in 1973–1974 at the Eidgenössische Technische Hochschule (ETH) in Zürich with Albert Eschenmoser. He joined the Department of Chemistry and Biochemistry at the University of California, Los Angeles (UCLA), in 1974 and is now a Distinguished Professor. In the past few years, the Jung group has become involved in the design of new drugs for the treatment of human diseases. Indeed, two compounds from his lab – enzalutamide (Xtandi) and apalutamide (Erleada) – have been approved for the treatment of castration-resistant prostate cancer. Professor Jung has published over 350 articles and is an inventor on over 80 patents and/or patent applications. He has given nearly 620 invited lectures and is a synthetic consultant for more than 15 industrial research sites.
Footnotes
CONFLICT OF INTEREST STATEMENT: Authors declare no conflicts interest.
REFERENCES
- 1.Shaffer PL, Jivan A, Dollins DE, Claessens F, Gewirth DT. Structural basis of androgen receptor binding to selective androgen response elements. Proc Natl Acad Sci U S A 2004;101(14):4758–4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. The nuclear receptor superfamily: The second decade. Cell 1995;83(6):835–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Royen ME, van Cappellen WA, de Vos C, Houtsmuller AB, Trapman J. Stepwise androgen receptor dimerization. J Cell Sci 2012;125(Pt 8):1970–1979. [DOI] [PubMed] [Google Scholar]
- 4.Cancer.gov. Cancer of the Prostate: SEER Stat Fact Sheets.
- 5.Tan MH, Li J, Xu HE, Melcher K, Yong EL. Androgen receptor: structure, role in prostate cancer and drug discovery. Acta Pharmacol Sin 2015;36(1):3–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heidenreich A, Pfister D, Merseburger A, Bartsch G, German Working Group on Castration-Resistant Prostate C. Castration-resistant prostate cancer: where we stand in 2013 and what urologists should know. Eur Urol 2013;64(2):260–265. [DOI] [PubMed] [Google Scholar]
- 7.Ojo D, Lin X, Wong N, Gu Y, Tang D. Prostate Cancer Stem-like Cells Contribute to the Development of Castration-Resistant Prostate Cancer. Cancers (Basel) 2015;7(4):2290–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schweizer MT, Yu EY. Persistent androgen receptor addiction in castration-resistant prostate cancer. J Hematol Oncol 2015;8:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wadosky KM, Koochekpour S. Androgen receptor splice variants and prostate cancer: From bench to bedside. Oncotarget 2017;8(11):18550–18576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao S, Zhan Y, Dong Y. Emerging data on androgen receptor splice variants in prostate cancer. Endocr Relat Cancer 2016;23(12):T199–T210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Azoitei A, Merseburger AS, Godau B, Hoda MR, Schmid E, Cronauer MV. C-terminally truncated constitutively active androgen receptor variants and their biologic and clinical significance in castration-resistant prostate cancer. J Steroid Biochem Mol Biol 2017;166:38–44. [DOI] [PubMed] [Google Scholar]
- 12.Cattrini C, Zanardi E, Vallome G, Cavo A, Cerbone L, Di Meglio A, Fabbroni C, Latocca MM, Rizzo F, Messina C, Rubagotti A, Barboro P, Boccardo F. Targeting androgen-independent pathways: new chances for patients with prostate cancer? Crit Rev Oncol Hematol 2017;118:42–53. [DOI] [PubMed] [Google Scholar]
- 13.Brennen WN, Isaacs JT. Cellular Origin of Androgen Receptor Pathway-Independent Prostate Cancer and Implications for Therapy. Cancer Cell 2017;32(4):399–401. [DOI] [PubMed] [Google Scholar]
- 14.Crona DJ, Whang YE. Androgen Receptor-Dependent and -Independent Mechanisms Involved in Prostate Cancer Therapy Resistance. Cancers (Basel) 2017;9(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, Montgomery B, Taplin ME, Pritchard CC, Attard G, Beltran H, Abida W, Bradley RK, Vinson J, Cao X, Vats P, Kunju LP, Hussain M, Feng FY, Tomlins SA, Cooney KA, Smith DC, Brennan C, Siddiqui J, Mehra R, Chen Y, Rathkopf DE, Morris MJ, Solomon SB, Durack JC, Reuter VE, Gopalan A, Gao J, Loda M, Lis RT, Bowden M, Balk SP, Gaviola G, Sougnez C, Gupta M, Yu EY, Mostaghel EA, Cheng HH, Mulcahy H, True LD, Plymate SR, Dvinge H, Ferraldeschi R, Flohr P, Miranda S, Zafeiriou Z, Tunariu N, Mateo J, Perez-Lopez R, Demichelis F, Robinson BD, Schiffman M, Nanus DM, Tagawa ST, Sigaras A, Eng KW, Elemento O, Sboner A, Heath EI, Scher HI, Pienta KJ, Kantoff P, de Bono JS, Rubin MA, Nelson PS, Garraway LA, Sawyers CL, Chinnaiyan AM. Integrative clinical genomics of advanced prostate cancer. Cell 2015;161(5):1215–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imamura Y, Sadar MD. Androgen receptor targeted therapies in castration-resistant prostate cancer: Bench to clinic. Int J Urol 2016;23(8):654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito Y, Sadar MD. Enzalutamide and blocking androgen receptor in advanced prostate cancer: lessons learnt from the history of drug development of antiandrogens. Res Rep Urol 2018;10:23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Narayanan R, Ponnusamy S, Miller DD. Destroying the androgen receptor (AR)-potential strategy to treat advanced prostate cancer. Oncoscience 2017;4(11–12):175–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ponnusamy S, Coss CC, Thiyagarajan T, Watts K, Hwang DJ, He Y, Selth LA, McEwan IJ, Duke CB, Pagadala J, Singh G, Wake RW, Ledbetter C, Tilley WD, Moldoveanu T, Dalton JT, Miller DD, Narayanan R. Novel Selective Agents for the Degradation of Androgen Receptor Variants to Treat Castration-Resistant Prostate Cancer. Cancer Res 2017;77(22):6282–6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu Z, Cai C, Gao S, Simon NI, Shen HC, Balk SP. Galeterone prevents androgen receptor binding to chromatin and enhances degradation of mutant androgen receptor. Clin Cancer Res 2014;20(15):4075–4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Narayanan RC, (TN, US: ), Miller Duane D. (Collierville, TN, US: ), Thamarai Ponnusamy (Memphis, TN, US: ), Dong-jin Hwang (Arlington, TN, US: ), Duke Charles B. (Memphis, TN, US: ), Coss Christopher C. (Upper Arlington, OH, US: ), Amanda Jones (Silver Spring, MI, US: ), Dalton James T. (Ann Arbor, MI, US: ); GTX, INC. (Memphis, TN, US: ),University of Tennessee Research Foundation; (Knoxville, TN, US: ), assignee. Selective androgen receptor degrader (SARD) ligands and methods of use thereof. United States2017. [Google Scholar]
- 22.Shibata N, Nagai K, Morita Y, Ujikawa O, Ohoka N, Hattori T, Koyama R, Sano O, Imaeda Y, Nara H, Cho N, Naito M. Development of Protein Degradation Inducers of Androgen Receptor by Conjugation of Androgen Receptor Ligands and Inhibitor of Apoptosis Protein Ligands. J Med Chem 2018;61(2):543–575. [DOI] [PubMed] [Google Scholar]
- 23.BERLIN MHR, Flemington, NJ, 08822, US: ), Kurt ZIMMERMAN (5 Skeetfield Point Road, Durham, CT, CT, US: ), Lawrence SNYDER (85 Laurel Ridge Trail, Killingworth, CT, 06419, US: ), CREW Andrew, P. (2 Winterberry Road, Guilford, CT, 06437, US: ), CREWS Craig, M. (286 Livingston Street, New Haven, CT, 06511, US: ), Xin CHEN (12 Red Fox Lane, Trumbull, CT, 06611, US: ), Hanqing DONG (248 Opening Hill Rd, Madison, CT, 06443, US: ), Caterina FERRARO (119 Towne Street, Apt. 256Stamford, CT, 06902, US: ), Meizhong JIN (6 Diane Lane, East Northport, NY, 11731, US: ), Yimin QIAN (5 Camas Court, Plainsboro, NJ, 08536, US: ), Kam SIU (42 Magnolia Road, Milford, CT, 06461, US: ), Jing WANG (14 Seaside Avenue, Milford, CT, 06460, US: ); ARVINAS, INC. (5 Science Park, New Haven, CT, 06511, US: ),YALE UNIVERSITY; (Office of the Cooperative Research, 433 Temple AvenueNew Haven, CT, 06511, US: ),Michael BERLIN (14 Hendrick Road, Flemington, NJ, 08822, US: ),Kurt ZIMMERMAN (5 Skeetfield Point Road, Durham, CT, CT, US: ),Lawrence SNYDER (85 Laurel Ridge Trail, Killingworth, CT, 06419, US: ), assignee. COMPOUNDS AND METHODS FOR THE ENHANCED DEGRADATION OF TARGETED PROTEINS2016. [Google Scholar]
- 24.Raina K, Crews CM. Targeted protein knockdown using small molecule degraders. Curr Opin Chem Biol 2017;39:46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Churcher I Protac-Induced Protein Degradation in Drug Discovery: Breaking the Rules or Just Making New Ones? J Med Chem 2018;61(2):444–452. [DOI] [PubMed] [Google Scholar]
- 26.Hupe MC, Offermann A, Perabo F, Chandhasin C, Perner S, Merseburger AS, Cronauer MV. [Inhibitors of the androgen receptor Nterminal domain : Therapies targeting the Achilles’ heel of various androgen receptor molecules in advanced prostate cancer]. Urologe A 2018;57(2):148–154. [DOI] [PubMed] [Google Scholar]
- 27.Sadar MD. Small molecule inhibitors targeting the “achilles’ heel” of androgen receptor activity. Cancer Res 2011;71(4):1208–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore TW, Mayne CG, Katzenellenbogen JA. Minireview: Not picking pockets: nuclear receptor alternate-site modulators (NRAMs). Mol Endocrinol 2010;24(4):683–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Antonarakis ES, Chandhasin C, Osbourne E, Luo J, Sadar MD, Perabo F. Targeting the N-Terminal Domain of the Androgen Receptor: A New Approach for the Treatment of Advanced Prostate Cancer. Oncologist 2016;21(12):1427–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo C, Yeh S, Niu Y, Li G, Zheng J, Li L, Chang C. Targeting androgen receptor versus targeting androgens to suppress castration resistant prostate cancer. Cancer Lett 2017;397:133–143. [DOI] [PubMed] [Google Scholar]
- 31.Dervan PB, Kurmis AA, Finn PB. Molecular Recognition of DNA by Py–Im Polyamides: From Discovery to Oncology In: Waring MJ, editor. DNA-targeting Molecules as Therapeutic Agents2018. pp 298–332. [Google Scholar]
- 32.Khalaf AI, Al-Kadhimi AAH, Ali JH. DNA Minor Groove Binders-Inspired by Nature. Acta Chimica Slovenica 2016:689–704. [DOI] [PubMed] [Google Scholar]
- 33.Dervan PB, Edelson BS. Recognition of the DNA minor groove by pyrrole-imidazole polyamides. Current Opinion in Structural Biology 2003;13(3):284–299. [DOI] [PubMed] [Google Scholar]
- 34.Wade WS, Mrksich M, Dervan PB. Design of peptides that bind in the minor groove of DNA at 5’-(A,T)G(A,T)C(A,T)-3’ sequences by a dimeric side-by-side motif. Journal of the American Chemical Society 1992;114(23):8783–8794. [Google Scholar]
- 35.White S, Szewczyk JW, Turner JM, Baird EE, Dervan PB. Recognition of the four Watson-Crick base pairs in the DNA minor groove by synthetic ligands. Nature 1998;391(6666):468–471. [DOI] [PubMed] [Google Scholar]
- 36.Nickols NG, Dervan PB. Suppression of androgen receptor-mediated gene expression by a sequence-specific DNA-binding polyamide. Proc Natl Acad Sci U S A 2007;104(25):10418–10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang F, Nickols NG, Li BC, Szablowski JO, Hamilton SR, Meier JL, Wang CM, Dervan PB. Animal toxicity of hairpin pyrrole-imidazole polyamides varies with the turn unit. J Med Chem 2013;56(18):7449–7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kielkopf CL, Baird EE, Dervan PB, Rees DC. Structural basis for G•C recognition in the DNA minor groove. Nature Structural Biology 1998;5(2):104–109. [DOI] [PubMed] [Google Scholar]
- 39.Pilch DS, Poklar N, Baird EE, Dervan PB, Breslauer KJ. The thermodynamics of polyamide-DNA recognition: hairpin polyamide binding in the minor groove of duplex DNA. Biochemistry 1999;38(7):2143–2151. [DOI] [PubMed] [Google Scholar]
- 40.White S, Baird EE, Dervan PB. On the pairing rules for recognition in the minor groove of DNA by pyrrole-imidazole polyamides. Chemistry & Biology 1997;4(8):569–578. [DOI] [PubMed] [Google Scholar]
- 41.Hsu CF, Phillips JW, Trauger JW, Farkas ME, Belitsky JM, Heckel A, Olenyuk BZ, Puckett JW, Wang CC, Dervan PB. Completion of a Programmable DNA-Binding Small Molecule Library. Tetrahedron 2007;63(27):6146–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mrksich M, Parks ME, Dervan PB. Hairpin Peptide Motif. A New Class of Oligopeptides for Sequence-Specific Recognition in the Minor Groove of Double-Helical DNA. Journal of the American Chemical Society 1994;116(18):7983–7988. [Google Scholar]
- 43.DERVAN P B (1235 St. Albans Rd, San Marino, CA, 91108, US: ), JACOBS Claire, S. (937 E. California Blvd, #3Pasedena, CA, 91106, US: ), NICKOLS Nicholas, G. (15128 Vose St, Van Nuys, CA, 91405, US: ), Daniel HARKI (297 South Madison Av, #3Pasedena, CA, 91101, US: ); CALIFORNIA INSTITUTE OF TECHNOLOGY; (1200 East California Boulevard, MC 201–85Pasedena, CA, 91125, US: ),DERVAN Peter, B. (1235 St. Albans Rd, San Marino, CA, 91108, US: ),JACOBS Claire, S. (937 E. California Blvd, #3Pasedena, CA, 91106, US: ),NICKOLS Nicholas, G. (15128 Vose St, Van Nuys, CA, 91405, US: ),Daniel HARKI (297 South Madison Av, #3Pasedena, CA, 91101, US: ), assignee. POLYAMIDES WITH TAIL STRUCTURES2008. [Google Scholar]
- 44.Nickols NG, Jacobs CS, Farkas ME, Dervan PB. Improved nuclear localization of DNA-binding polyamides. Nucleic Acids Res 2007;35(2):363–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang F, Nickols NG, Li BC, Marinov GK, Said JW, Dervan PB. Antitumor activity of a pyrrole-imidazole polyamide. Proc Natl Acad Sci U S A 2013;110(5):1863–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dickinson LA, Gulizia RJ, Trauger JW, Baird EE, Mosier DE, Gottesfeld JM, Dervan PB. Inhibition of RNA polymerase II transcription in human cells by synthetic DNA-binding ligands. Proceedings of the National Academy of Sciences 1998;95(22):12890–12895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raskatov JA, Puckett JW, Dervan PB. A C-14 labeled Py–Im polyamide localizes to a subcutaneous prostate cancer tumor. Bioorganic & Medicinal Chemistry 2014;22(16):4371–4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raskatov JA, Szablowski JO, Dervan PB. Tumor xenograft uptake of a pyrrole-imidazole (Py-Im) polyamide varies as a function of cell line grafted. J Med Chem 2014;57(20):8471–8476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kurmis AA, Yang F, Welch TR, Nickols NG, Dervan PB. A Pyrrole-Imidazole Polyamide Is Active against Enzalutamide-Resistant Prostate Cancer. Cancer Res 2017;77(9):2207–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Padroni G, Parkinson JA, Fox KR, Burley GA. Structural basis of DNA duplex distortion induced by thiazole-containing hairpin polyamides. Nucleic Acids Res 2018;46(1):42–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hargrove AE, Raskatov JA, Meier JL, Montgomery DC, Dervan PB. Characterization and solubilization of pyrrole-imidazole polyamide aggregates. J Med Chem 2012;55(11):5425–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Andrews P, Thyssen J, Lorke D. The biology and toxicology of molluscicides, bayluscide. Pharmacology & Therapeutics 1982;19(2):245–295. [DOI] [PubMed] [Google Scholar]
- 53.Li Y, Li PK, Roberts MJ, Arend RC, Samant RS, Buchsbaum DJ. Multi-targeted therapy of cancer by niclosamide: A new application for an old drug. Cancer Lett 2014;349(1):8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kraljevic S, Stambrook PJ, Pavelic K. Accelerating drug discovery. EMBO Rep 2004;5(9):837–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chong CR, Sullivan DJ Jr. New uses for old drugs. Nature 2007;448(7154):645–646. [DOI] [PubMed] [Google Scholar]
- 56.Al‐Hadiya BMH. Niclosamide: Comprehensive Profile. 2005;32:67–96. [DOI] [PubMed] [Google Scholar]
- 57.Solaini G, Sgarbi G, Baracca A. Oxidative phosphorylation in cancer cells. Biochim Biophys Acta 2011;1807(6):534–542. [DOI] [PubMed] [Google Scholar]
- 58.Jin Y, Lu Z, Ding K, Li J, Du X, Chen C, Sun X, Wu Y, Zhou J, Pan J. Antineoplastic mechanisms of niclosamide in acute myelogenous leukemia stem cells: inactivation of the NF-kappaB pathway and generation of reactive oxygen species. Cancer Res 2010;70(6):2516–2527. [DOI] [PubMed] [Google Scholar]
- 59.Jin B, Wang C, Li J, Du X, Ding K, Pan J. Anthelmintic niclosamide disrupts the interplay of p65 and FOXM1/beta-catenin and eradicates leukemia stem cells in chronic myelogenous leukemia. Clin Cancer Res 2016. [DOI] [PubMed] [Google Scholar]
- 60.Li R, Hu Z, Sun SY, Chen ZG, Owonikoko TK, Sica GL, Ramalingam SS, Curran WJ, Khuri FR, Deng X. Niclosamide overcomes acquired resistance to erlotinib through suppression of STAT3 in non-small cell lung cancer. Mol Cancer Ther 2013;12(10):2200–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu M, Ye W, Li J, Zhou P, Chu Z, Huang W. The salicylanilide derivatives inhibit signal transducer and activator of transcription 3 pathways in A549 lung cancer cells. Anti-Cancer Drugs 2016;27(1):41–47. [DOI] [PubMed] [Google Scholar]
- 62.Liu J, Chen X, Ward T, Mao Y, Bockhorn J, Liu X, Wang G, Pegram M, Shen K. Niclosamide inhibits epithelial-mesenchymal transition and tumor growth in lapatinib-resistant human epidermal growth factor receptor 2-positive breast cancer. Int J Biochem Cell Biol 2016;71:12–23. [DOI] [PubMed] [Google Scholar]
- 63.Liu J, Chen X, Ward T, Pegram M, Shen K. Combined niclosamide with cisplatin inhibits epithelial-mesenchymal transition and tumor growth in cisplatin-resistant triple-negative breast cancer. Tumour Biol 2016. [DOI] [PubMed] [Google Scholar]
- 64.Lu W, Lin C, Roberts MJ, Waud WR, Piazza GA, Li Y. Niclosamide suppresses cancer cell growth by inducing Wnt co-receptor LRP6 degradation and inhibiting the Wnt/beta-catenin pathway. PLoS One 2011;6(12):e29290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Londono-Joshi AI, Arend RC, Aristizabal L, Lu W, Samant RS, Metge BJ, Hidalgo B, Grizzle WE, Conner M, Forero-Torres A, Lobuglio AF, Li Y, Buchsbaum DJ. Effect of niclosamide on basal-like breast cancers. Mol Cancer Ther 2014;13(4):800–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Balgi AD, Fonseca BD, Donohue E, Tsang TC, Lajoie P, Proud CG, Nabi IR, Roberge M. Screen for chemical modulators of autophagy reveals novel therapeutic inhibitors of mTORC1 signaling. PLoS One 2009;4(9):e7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim SY, Kang JW, Song X, Kim BK, Yoo YD, Kwon YT, Lee YJ. Role of the IL-6-JAK1-STAT3-Oct-4 pathway in the conversion of non-stem cancer cells into cancer stem-like cells. Cell Signal 2013;25(4):961–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang YC, Chao TK, Chang CC, Yo YT, Yu MH, Lai HC. Drug screening identifies niclosamide as an inhibitor of breast cancer stem-like cells. PLoS One 2013;8(9):e74538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Monin MB, Krause P, Stelling R, Bocuk D, Niebert S, Klemm F, Pukrop T, Koenig S. The anthelmintic niclosamide inhibits colorectal cancer cell lines via modulation of the canonical and noncanonical Wnt signaling pathway. J Surg Res 2016;203(1):193–205. [DOI] [PubMed] [Google Scholar]
- 70.Suliman MA, Zhang Z, Na H, Ribeiro AL, Zhang Y, Niang B, Hamid AS, Zhang H, Xu L, Zuo Y. Niclosamide inhibits colon cancer progression through downregulation of the Notch pathway and upregulation of the tumor suppressor miR-200 family. Int J Mol Med 2016;38(3):776–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sack U, Walther W, Scudiero D, Selby M, Kobelt D, Lemm M, Fichtner I, Schlag PM, Shoemaker RH, Stein U. Novel effect of antihelminthic Niclosamide on S100A4-mediated metastatic progression in colon cancer. J Natl Cancer Inst 2011;103(13):1018–1036. [DOI] [PubMed] [Google Scholar]
- 72.Osada T, Chen M, Yang XY, Spasojevic I, Vandeusen JB, Hsu D, Clary BM, Clay TM, Chen W, Morse MA, Lyerly HK. Antihelminth compound niclosamide downregulates Wnt signaling and elicits antitumor responses in tumors with activating APC mutations. Cancer Res 2011;71(12):4172–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.King ML, Lindberg ME, Stodden GR, Okuda H, Ebers SD, Johnson A, Montag A, Lengyel E, MacLean Ii JA, Hayashi K. WNT7A/beta-catenin signaling induces FGF1 and influences sensitivity to niclosamide in ovarian cancer. Oncogene 2015;34(26):3452–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yo YT, Lin YW, Wang YC, Balch C, Huang RL, Chan MW, Sytwu HK, Chen CK, Chang CC, Nephew KP, Huang T, Yu MH, Lai HC. Growth inhibition of ovarian tumor-initiating cells by niclosamide. Mol Cancer Ther 2012;11(8):1703–1712. [DOI] [PubMed] [Google Scholar]
- 75.Lin CK, Bai MY, Hu TM, Wang YC, Chao TK, Weng SJ, Huang RL, Su PH, Lai HC. Preclinical evaluation of a nanoformulated antihelminthic, niclosamide, in ovarian cancer. Oncotarget 2016;7(8):8993–9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Circu ML, Dykes SS, Carroll J, Kelly K, Galiano F, Greer A, Cardelli J, El-Osta H. A Novel High Content Imaging-Based Screen Identifies the Anti-Helminthic Niclosamide as an Inhibitor of Lysosome Anterograde Trafficking and Prostate Cancer Cell Invasion. PLoS One 2016;11(1):e0146931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu C, Armstrong C, Zhu Y, Lou W, Gao AC. Niclosamide enhances abiraterone treatment via inhibition of androgen receptor variants in castration resistant prostate cancer. Oncotarget 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu C, Lou W, Armstrong C, Zhu Y, Evans CP, Gao AC. Niclosamide suppresses cell migration and invasion in enzalutamide resistant prostate cancer cells via Stat3-AR axis inhibition. Prostate 2015;75(13):1341–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu C, Lou W, Zhu Y, Nadiminty N, Schwartz CT, Evans CP, Gao AC. Niclosamide inhibits androgen receptor variants expression and overcomes enzalutamide resistance in castration-resistant prostate cancer. Clin Cancer Res 2014;20(12):3198–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ren X, Duan L, He Q, Zhang Z, Zhou Y, Wu D, Pan J, Pei D, Ding K. Identification of Niclosamide as a New Small-Molecule Inhibitor of the STAT3 Signaling Pathway. ACS Med Chem Lett 2010;1(9):454–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ren X, Zhang Z, Duan L, Ding K. Identification of Niclosamide as a New Lead Compound to Suppress the Metastasis of Prostate Cancer Cells. Medicinal Chemistry 2014;4(7). [Google Scholar]
- 82.NCIthesaurus. Niclosamide (Code C66240).
- 83.Satoh K, Zhang L, Zhang Y, Chelluri R, Boufraqech M, Nilubol N, Patel D, Shen M, Kebebew E. Identification of Niclosamide as a Novel Anticancer Agent for Adrenocortical Carcinoma. Clin Cancer Res 2016;22(14):3458–3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Weng S, Zhou L, Deng Q, Wang J, Yu Y, Zhu J, Yuan Y. Niclosamide induced cell apoptosis via upregulation of ATF3 and activation of PERK in Hepatocellular carcinoma cells. BMC Gastroenterol 2016;16:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tomizawa M, Shinozaki F, Motoyoshi Y, Sugiyama T, Yamamoto S, Ishige N. Niclosamide suppresses migration of hepatocellular carcinoma cells and downregulates matrix metalloproteinase-9 expression. Oncol Lett 2015;10(6):3515–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Khanim FL, Merrick BA, Giles HV, Jankute M, Jackson JB, Giles LJ, Birtwistle J, Bunce CM, Drayson MT. Redeployment-based drug screening identifies the anti-helminthic niclosamide as anti-myeloma therapy that also reduces free light chain production. Blood Cancer J 2011;1(10):e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wieland A, Trageser D, Gogolok S, Reinartz R, Hofer H, Keller M, Leinhaas A, Schelle R, Normann S, Klaas L, Waha A, Koch P, Fimmers R, Pietsch T, Yachnis AT, Pincus DW, Steindler DA, Brustle O, Simon M, Glas M, Scheffler B. Anticancer effects of niclosamide in human glioblastoma. Clin Cancer Res 2013;19(15):4124–4136. [DOI] [PubMed] [Google Scholar]
- 88.Li Z, Yu Y, Sun S, Qi B, Wang W, Yu A. Niclosamide inhibits the proliferation of human osteosarcoma cell lines by inducing apoptosis and cell cycle arrest. Oncol Rep 2015;33(4):1763–1768. [DOI] [PubMed] [Google Scholar]
- 89.Pan JX, Ding K, Wang CY. Niclosamide, an old antihelminthic agent, demonstrates antitumor activity by blocking multiple signaling pathways of cancer stem cells. Chin J Cancer 2012;31(4):178–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dhir R, Ni Z, Lou W, DeMiguel F, Grandis JR, Gao AC. Stat3 activation in prostatic carcinomas. Prostate 2002;51(4):241–246. [DOI] [PubMed] [Google Scholar]
- 91.Darnell JE Jr. STATs and Gene Regulation. Science 1997;277(5332):1630–1635. [DOI] [PubMed] [Google Scholar]
- 92.Bromberg J, Darnell JE Jr. The role of STATs in transcriptional control and their impact on cellular function. Oncogene 2000;19(21):2468–2473. [DOI] [PubMed] [Google Scholar]
- 93.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer 2009;9(11):798–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schroeder A, Herrmann A, Cherryholmes G, Kowolik C, Buettner R, Pal S, Yu H, Muller-Newen G, Jove R. Loss of androgen receptor expression promotes a stem-like cell phenotype in prostate cancer through STAT3 signaling. Cancer Res 2014;74(4):1227–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu C, Zhu Y, Lou W, Cui Y, Evans CP, Gao AC. Inhibition of constitutively active Stat3 reverses enzalutamide resistance in LNCaP derivative prostate cancer cells. Prostate 2014;74(2):201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shariat SF, Andrews B, Kattan MW, Kim J, Wheeler TM, Slawin KM. Plasma levels of interleukin-6 and its soluble receptor are associated with prostate cancer progression and metastasis. Urology 2001;58(6):1008–1015. [DOI] [PubMed] [Google Scholar]
- 97.Dunn GP, Sheehan KC, Old LJ, Schreiber RD. IFN unresponsiveness in LNCaP cells due to the lack of JAK1 gene expression. Cancer Res 2005;65(8):3447–3453. [DOI] [PubMed] [Google Scholar]
- 98.Pencik J, Pham HT, Schmoellerl J, Javaheri T, Schlederer M, Culig Z, Merkel O, Moriggl R, Grebien F, Kenner L. JAK-STAT signaling in cancer: From cytokines to non-coding genome. Cytokine 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gu L, Vogiatzi P, Puhr M, Dagvadorj A, Lutz J, Ryder A, Addya S, Fortina P, Cooper C, Leiby B, Dasgupta A, Hyslop T, Bubendorf L, Alanen K, Mirtti T, Nevalainen MT. Stat5 promotes metastatic behavior of human prostate cancer cells in vitro and in vivo. Endocr Relat Cancer 2010;17(2):481–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ueda T, Mawji NR, Bruchovsky N, Sadar MD. Ligand-independent activation of the androgen receptor by interleukin-6 and the role of steroid receptor coactivator-1 in prostate cancer cells. J Biol Chem 2002;277(41):38087–38094. [DOI] [PubMed] [Google Scholar]
- 101.Culig Z, Puhr M. Interleukin-6: a multifunctional targetable cytokine in human prostate cancer. Mol Cell Endocrinol 2012;360(1–2):52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mora LB, Buettner R, Seigne J, Diaz J, Ahmad N, Garcia R, Bowman T, Falcone R, Fairclough R, Cantor A, Muro-Cacho C, Livingston S, Karras J, Pow-Sang J, Jove R. Constitutive activation of Stat3 in human prostate tumors and cell lines: direct inhibition of Stat3 signaling induces apoptosis of prostate cancer cells. Cancer Res 2002;62(22):6659–6666. [PubMed] [Google Scholar]
- 103.Bluemn EG, Coleman IM, Lucas JM, Coleman RT, Hernandez-Lopez S, Tharakan R, Bianchi-Frias D, Dumpit RF, Kaipainen A, Corella AN, Yang YC, Nyquist MD, Mostaghel E, Hsieh AC, Zhang X, Corey E, Brown LG, Nguyen HM, Pienta K, Ittmann M, Schweizer M, True LD, Wise D, Rennie PS, Vessella RL, Morrissey C, Nelson PS. Androgen Receptor Pathway-Independent Prostate Cancer Is Sustained through FGF Signaling. Cancer Cell 2017;32(4):474–489 e476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pakula H, Xiang D, Li Z. A Tale of Two Signals: AR and WNT in Development and Tumorigenesis of Prostate and Mammary Gland. Cancers (Basel) 2017;9(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chesire D Beta-catenin signaling in prostate cancer: an early perspective. Endocrine Related Cancer 2003;10(4):537–560. [DOI] [PubMed] [Google Scholar]
- 106.Uysal-Onganer P, Kawano Y, Caro M, Walker MM, Diez S, Darrington RS, Waxman J, Kypta RM. Wnt-11 promotes neuroendocrine-like differentiation, survival and migration of prostate cancer cells. Mol Cancer 2010;9:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhu H, Mazor M, Kawano Y, Walker MM, Leung HY, Armstrong K, Waxman J, Kypta RM. Analysis of Wnt gene expression in prostate cancer: mutual inhibition by WNT11 and the androgen receptor. Cancer Res 2004;64(21):7918–7926. [DOI] [PubMed] [Google Scholar]
- 108.Verras M, Brown J, Li X, Nusse R, Sun Z. Wnt3a growth factor induces androgen receptor-mediated transcription and enhances cell growth in human prostate cancer cells. Cancer Res 2004;64(24):8860–8866. [DOI] [PubMed] [Google Scholar]
- 109.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell 2009;17(1):9–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yang X, Chen MW, Terry S, Vacherot F, Bemis DL, Capodice J, Kitajewski J, de la Taille A, Benson MC, Guo Y, Buttyan R. Complex regulation of human androgen receptor expression by Wnt signaling in prostate cancer cells. Oncogene 2006;25(24):3436–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yang F, Li X, Sharma M, Sasaki CY, Longo DL, Lim B, Sun Z. Linking beta-catenin to androgen-signaling pathway. J Biol Chem 2002;277(13):11336–11344. [DOI] [PubMed] [Google Scholar]
- 112.Truica CI, Byers S, Gelmann EP. β-Catenin Affects Androgen Receptor Transcriptional Activity and Ligand Specificity. Cancer Research 2000;60(17):4709–4713. [PubMed] [Google Scholar]
- 113.Conacci-Sorrell M, Zhurinsky J, Ben-Ze’ev A. The cadherin-catenin adhesion system in signaling and cancer. Journal of Clinical Investigation 2002;109(8):987–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hollestelle A, Peeters JK, Smid M, Timmermans M, Verhoog LC, Westenend PJ, Heine AA, Chan A, Sieuwerts AM, Wiemer EA, Klijn JG, van der Spek PJ, Foekens JA, Schutte M, den Bakker MA, Martens JW. Loss of E-cadherin is not a necessity for epithelial to mesenchymal transition in human breast cancer. Breast Cancer Res Treat 2013;138(1):47–57. [DOI] [PubMed] [Google Scholar]
- 115.Watson PA, Arora VK, Sawyers CL. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer 2015;15(12):701–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lee SO, Lou W, Hou M, de Miguel F, Gerber L, Gao AC. Interleukin-6 promotes androgen-independent growth in LNCaP human prostate cancer cells. Clin Cancer Res 2003;9(1):370–376. [PubMed] [Google Scholar]
- 117.Chun JY, Nadiminty N, Dutt S, Lou W, Yang JC, Kung HJ, Evans CP, Gao AC. Interleukin-6 regulates androgen synthesis in prostate cancer cells. Clin Cancer Res 2009;15(15):4815–4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lin TH, Lee SO, Niu Y, Xu D, Liang L, Li L, Yeh SD, Fujimoto N, Yeh S, Chang C. Differential androgen deprivation therapies with anti-androgens casodex/bicalutamide or MDV3100/Enzalutamide versus anti-androgen receptor ASC-J9(R) Lead to promotion versus suppression of prostate cancer metastasis. J Biol Chem 2013;288(27):19359–19369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lin TH, Izumi K, Lee SO, Lin WJ, Yeh S, Chang C. Anti-androgen receptor ASC-J9 versus anti-androgens MDV3100 (Enzalutamide) or Casodex (Bicalutamide) leads to opposite effects on prostate cancer metastasis via differential modulation of macrophage infiltration and STAT3-CCL2 signaling. Cell Death Dis 2013;4:e764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Abdulghani J, Gu L, Dagvadorj A, Lutz J, Leiby B, Bonuccelli G, Lisanti MP, Zellweger T, Alanen K, Mirtti T, Visakorpi T, Bubendorf L, Nevalainen MT. Stat3 promotes metastatic progression of prostate cancer. Am J Pathol 2008;172(6):1717–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gu L, Dagvadorj A, Lutz J, Leiby B, Bonuccelli G, Lisanti MP, Addya S, Fortina P, Dasgupta A, Hyslop T, Bubendorf L, Nevalainen MT. Transcription factor Stat3 stimulates metastatic behavior of human prostate cancer cells in vivo, whereas Stat5b has a preferential role in the promotion of prostate cancer cell viability and tumor growth. Am J Pathol 2010;176(4):1959–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liu C, Armstrong CM, Lou W, Lombard AP, Cucchiara V, Gu X, Yang JC, Nadiminty N, Pan CX, Evans CP, Gao AC. Niclosamide and Bicalutamide Combination Treatment Overcomes Enzalutamide- and Bicalutamide-Resistant Prostate Cancer. Mol Cancer Ther 2017;16(8):1521–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Devarakonda B, Hill RA, Liebenberg W, Brits M, de Villiers MM. Comparison of the aqueous solubilization of practically insoluble niclosamide by polyamidoamine (PAMAM) dendrimers and cyclodextrins. Int J Pharm 2005;304(1–2):193–209. [DOI] [PubMed] [Google Scholar]
- 124.GAO ACRP Davis, California, 95616, US: ), Chengfei LIU (3157 Occidental Drive, Apt. 1Sacramento, California, 95826, US: ), Wei LOU (611 Robin Place, Davis, California, 95616, US: ); THE REGENTS OF THE UNIVERSITY OF CALIFORNIA; (Office of Technology Transfer, 1111 Franklin Street 12th Floo, Oakland California, 94607–5200, US: ), assignee. TREATMENT OF METASTATIC PROSTATE CANCER2015. [Google Scholar]
- 125.Schweizer MT, Haugk K, McKiernan JS, Gulati R, Cheng HH, Maes JL, Dumpit RF, Nelson PS, Montgomery B, McCune JS, Plymate SR, Yu EY. A phase I study of niclosamide in combination with enzalutamide in men with castration-resistant prostate cancer. PLoS One 2018;13(6):e0198389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pan C-x, Lara P, Evans CP, Parikh M, Dall’era M, Liu C, Robles D, Gao A. Niclosamide in combination with abiraterone and prednisone in men with castration-resistant prostate cancer (CRPC): initial results from a phase Ib/II trial. Journal of Clinical Oncology 2018;36(6_suppl):192–192. [Google Scholar]
- 127.Andersen RJ, Mawji NR, Wang J, Wang G, Haile S, Myung JK, Watt K, Tam T, Yang YC, Banuelos CA, Williams DE, McEwan IJ, Wang Y, Sadar MD. Regression of castrate-recurrent prostate cancer by a small-molecule inhibitor of the amino-terminus domain of the androgen receptor. Cancer Cell 2010;17(6):535–546. [DOI] [PubMed] [Google Scholar]
- 128.Tri TM, Anh DH, Hoai PM, Minh NH, Nam VD, Viet PH, Minh TB. Emerging Endocrine Disrupting Chemicals and Pharmaceuticals in Vietnam: A Review of Environmental Occurrence and Fate in Aquatic and Indoor Environments. 2016;1244:223–253. [Google Scholar]
- 129.Szczepanska N, Kudlak B, Namiesnik J. Assessing ecotoxicity and the endocrine potential of selected phthalates, BADGE and BFDGE derivatives in relation to environmentally detectable levels. Sci Total Environ 2018;610–611:854–866. [DOI] [PubMed] [Google Scholar]
- 130.Ooe H, Taira T, Iguchi-Ariga SM, Ariga H. Induction of reactive oxygen species by bisphenol A and abrogation of bisphenol A-induced cell injury by DJ-1. Toxicol Sci 2005;88(1):114–126. [DOI] [PubMed] [Google Scholar]
- 131.Teng C, Goodwin B, Shockley K, Xia M, Huang R, Norris J, Merrick BA, Jetten AM, Austin CP, Tice RR. Bisphenol A affects androgen receptor function via multiple mechanisms. Chem Biol Interact 2013;203(3):556–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Luccio-Camelo DC, Prins GS. Disruption of androgen receptor signaling in males by environmental chemicals. J Steroid Biochem Mol Biol 2011;127(1–2):74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Andersen RJ. Sponging off nature for new drug leads. Biochem Pharmacol 2017;139:3–14. [DOI] [PubMed] [Google Scholar]
- 134.Myung JK, Banuelos CA, Fernandez JG, Mawji NR, Wang J, Tien AH, Yang YC, Tavakoli I, Haile S, Watt K, McEwan IJ, Plymate S, Andersen RJ, Sadar MD. An androgen receptor N-terminal domain antagonist for treating prostate cancer. J Clin Invest 2013;123(7):2948–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Reid J, Murray I, Watt K, Betney R, McEwan IJ. The androgen receptor interacts with multiple regions of the large subunit of general transcription factor TFIIF. J Biol Chem 2002;277(43):41247–41253. [DOI] [PubMed] [Google Scholar]
- 136.Yang YC, Banuelos CA, Mawji NR, Wang J, Kato M, Haile S, McEwan IJ, Plymate S, Sadar MD. Targeting androgen receptor activation function-1 with EPI to overcome resistance mechanisms in castration-resistant prostate cancer. Clin Cancer Res 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Antonarakis ES, Armstrong AJ, Dehm SM, Luo J. Androgen receptor variant-driven prostate cancer: clinical implications and therapeutic targeting. Prostate Cancer Prostatic Dis 2016:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Brand LJ, Olson ME, Ravindranathan P, Guo H, Kempema AM, Andrews TE, Chen X, Raj GV, Harki DA, Dehm SM. EPI-001 is a selective peroxisome proliferator-activated receptor-gamma modulator with inhibitory effects on androgen receptor expression and activity in prostate cancer. Oncotarget 2015;6(6):3811–3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Buchanan G, Yang M, Cheong A, Harris JM, Irvine RA, Lambert PF, Moore NL, Raynor M, Neufing PJ, Coetzee GA, Tilley WD. Structural and functional consequences of glutamine tract variation in the androgen receptor. Hum Mol Genet 2004;13(16):1677–1692. [DOI] [PubMed] [Google Scholar]
- 140.De Mol E, Fenwick RB, Phang CT, Buzon V, Szulc E, de la Fuente A, Escobedo A, Garcia J, Bertoncini CW, Estebanez-Perpina E, McEwan IJ, Riera A, Salvatella X. EPI-001, A Compound Active against Castration-Resistant Prostate Cancer, Targets Transactivation Unit 5 of the Androgen Receptor. ACS Chem Biol 2016;11(9):2499–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Marsh JA, Singh VK, Jia Z, Forman-Kay JD. Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: implications for fibrillation. Protein Sci 2006;15(12):2795–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yousefnia S, Momenzadeh S, Seyed Forootan F, Ghaedi K, Nasr Esfahani MH. The influence of peroxisome proliferator-activated receptor gamma (PPARgamma) ligands on cancer cell tumorigenicity. Gene 2018;649:14–22. [DOI] [PubMed] [Google Scholar]
- 143.Elix C, Pal SK, Jones JO. The role of peroxisome proliferator-activated receptor gamma in prostate cancer. Asian J Androl 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yang CC, Wang YC, Wei S, Lin LF, Chen CS, Lee CC, Lin CC, Chen CS. Peroxisome proliferator-activated receptor gamma-independent suppression of androgen receptor expression by troglitazone mechanism and pharmacologic exploitation. Cancer Res 2007;67(7):3229–3238. [DOI] [PubMed] [Google Scholar]
- 145.Yuan H, Young CY, Tian Y, Liu Z, Zhang M, Lou H. Suppression of the androgen receptor function by quercetin through protein-protein interactions of Sp1, c-Jun, and the androgen receptor in human prostate cancer cells. Mol Cell Biochem 2010;339(1–2):253–262. [DOI] [PubMed] [Google Scholar]
- 146.Hunter I, Hay CW, Esswein B, Watt K, McEwan IJ. Tissue control of androgen action: The ups and downs of androgen receptor expression. Mol Cell Endocrinol 2018;465:27–35. [DOI] [PubMed] [Google Scholar]
- 147.Verras M, Lee J, Xue H, Li TH, Wang Y, Sun Z. The androgen receptor negatively regulates the expression of c-Met: implications for a novel mechanism of prostate cancer progression. Cancer Res 2007;67(3):967–975. [DOI] [PubMed] [Google Scholar]
- 148.Segawa Y, Yoshimura R, Hase T, Nakatani T, Wada S, Kawahito Y, Kishimoto T, Sano H. Expression of peroxisome proliferator-activated receptor (PPAR) in human prostate cancer. Prostate 2002;51(2):108–116. [DOI] [PubMed] [Google Scholar]
- 149.ROGENHOFER S, ELLINGER J, KAHL P, STOEHR C, HARTMANN A, ENGEHAUSEN D, WIELAND W-F, MÜLLER SC, HOFSTÄDTER F, WALTER B. Enhanced Expression of Peroxisome Proliferate-activated Receptor Gamma (PPAR-γ) in Advanced Prostate Cancer. Anticancer Research 2012;32(8):3479–3483. [PubMed] [Google Scholar]
- 150.Ahmad I, Mui E, Galbraith L, Patel R, Tan EH, Salji M, Rust AG, Repiscak P, Hedley A, Markert E, Loveridge C, van der Weyden L, Edwards J, Sansom OJ, Adams DJ, Leung HY. Sleeping Beauty screen reveals Pparg activation in metastatic prostate cancer. Proc Natl Acad Sci U S A 2016;113(29):8290–8295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Park HK, Kim H, Kim HG, Cho YM, Jung WY, Han HS, Hwang TS, Kwon GY, Lim SD. Expression of Peroxisome Proliferator Activated Receptor gamma in Prostatic Adenocarcinoma. J Korean Med Sci 2015;30(5):533–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tew BY, Hong TB, Otto-Duessel M, Elix C, Castro E, He M, Wu X, Pal SK, Kalkum M, Jones JO. Vitamin K epoxide reductase regulation of androgen receptor activity. Oncotarget 2017;8(8):13818–13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Olokpa E, Bolden A, Stewart LV. The Androgen Receptor Regulates PPARgamma Expression and Activity in Human Prostate Cancer Cells. J Cell Physiol 2016;231(12):2664–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Moss PE, Lyles BE, Stewart LV. The PPARgamma ligand ciglitazone regulates androgen receptor activation differently in androgen-dependent versus androgen-independent human prostate cancer cells. Exp Cell Res 2010;316(20):3478–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kaarbo M, Mikkelsen OL, Malerod L, Qu S, Lobert VH, Akgul G, Halvorsen T, Maelandsmo GM, Saatcioglu F. PI3K-AKT-mTOR pathway is dominant over androgen receptor signaling in prostate cancer cells. Cell Oncol 2010;32(1–2):11–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Audet-Walsh E, Dufour CR, Yee T, Zouanat FZ, Yan M, Kalloghlian G, Vernier M, Caron M, Bourque G, Scarlata E, Hamel L, Brimo F, Aprikian AG, Lapointe J, Chevalier S, Giguere V. Nuclear mTOR acts as a transcriptional integrator of the androgen signaling pathway in prostate cancer. Genes Dev 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ciccarese C, Massari F, Iacovelli R, Fiorentino M, Montironi R, Di Nunno V, Giunchi F, Brunelli M, Tortora G. Prostate cancer heterogeneity: Discovering novel molecular targets for therapy. Cancer Treat Rev 2017;54:68–73. [DOI] [PubMed] [Google Scholar]
- 158.Ferraldeschi R, Welti J, Luo J, Attard G, de Bono JS. Targeting the androgen receptor pathway in castration-resistant prostate cancer: progresses and prospects. Oncogene 2015;34(14):1745–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, Arora VK, Le C, Koutcher J, Scher H, Scardino PT, Rosen N, Sawyers CL. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell 2011;19(5):575–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Marques RB, Aghai A, de Ridder CMA, Stuurman D, Hoeben S, Boer A, Ellston RP, Barry ST, Davies BR, Trapman J, van Weerden WM. High Efficacy of Combination Therapy Using PI3K/AKT Inhibitors with Androgen Deprivation in Prostate Cancer Preclinical Models. Eur Urol 2015;67(6):1177–1185. [DOI] [PubMed] [Google Scholar]
- 161.WU Y, CHHIPA RR, CHENG J, ZHANG H, MOHLER JL, IP C. Androgen Receptor-mTOR Crosstalk is Regulated by Testosterone Availability: Implication for Prostate Cancer Cell Survival. Anticancer Research 2010;30(10):3895–3901. [PMC free article] [PubMed] [Google Scholar]
- 162.Gordon MA, D’Amato NC, Gu H, Babbs B, Wulfkuhle J, Petricoin EF, Gallagher I, Dong T, Torkko K, Liu B, Elias A, Richer JK. Synergy between Androgen Receptor Antagonism and Inhibition of mTOR and HER2 in Breast Cancer. Mol Cancer Ther 2017;16(7):1389–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zhang H, Li X, Yang Y, Zhang Y, Wang HY, Zheng XFS. Significance and Mechanism of Androgen Receptor (AR) Overexpression and AR-mTOR Crosstalk in Hepatocellular Carcinoma. Hepatology 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kato M, Banuelos CA, Imamura Y, Leung JK, Caley DP, Wang J, Mawji NR, Sadar MD. Cotargeting Androgen Receptor Splice Variants and mTOR Signaling Pathway for the Treatment of Castration-Resistant Prostate Cancer. Clin Cancer Res 2016;22(11):2744–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sadar MDWV, CA: ), Jun Wang (Surrey, CA: ), Mawji Nasrin R. (Burnaby, CA: ), Minoru Kato (Ikoma, JP: ); British Columbia Cancer Agency Branch; (Vancouver, CA: ),British Columbia Cancer Agency Branch; (Vancouver, CA: ), assignee. CO-TARGETING ANDROGEN RECEPTOR SPLICE VARIANTS AND MTOR SIGNALING PATHWAY FOR THE TREATMENT OF CASTRATION-RESISTANT PROSTATE CANCER. United States: 2017. [Google Scholar]
- 166.Visconti R, Grieco D. Fighting tubulin-targeting anticancer drug toxicity and resistance. Endocr Relat Cancer 2017;24(9):T107–T117. [DOI] [PubMed] [Google Scholar]
- 167.Forth S, Kapoor TM. The mechanics of microtubule networks in cell division. J Cell Biol 2017;216(6):1525–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.van Vuuren RJ, Visagie MH, Theron AE, Joubert AM. Antimitotic drugs in the treatment of cancer. Cancer Chemother Pharmacol 2015;76(6):1101–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hornick JE, Bader JR, Tribble EK, Trimble K, Breunig JS, Halpin ES, Vaughan KT, Hinchcliffe EH. Live-cell analysis of mitotic spindle formation in taxol-treated cells. Cell Motil Cytoskeleton 2008;65(8):595–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Cassinello J, Carballido Rodriguez J, Anton Aparicio L. Role of taxanes in advanced prostate cancer. Clin Transl Oncol 2016;18(10):972–980. [DOI] [PubMed] [Google Scholar]
- 171.Fitzpatrick JM, de Wit R. Taxane mechanisms of action: potential implications for treatment sequencing in metastatic castration-resistant prostate cancer. Eur Urol 2014;65(6):1198–1204. [DOI] [PubMed] [Google Scholar]
- 172.Darshan MS, Loftus MS, Thadani-Mulero M, Levy BP, Escuin D, Zhou XK, Gjyrezi A, Chanel-Vos C, Shen R, Tagawa ST, Bander NH, Nanus DM, Giannakakou P. Taxane-induced blockade to nuclear accumulation of the androgen receptor predicts clinical responses in metastatic prostate cancer. Cancer Res 2011;71(18):6019–6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Gan L, Chen S, Wang Y, Watahiki A, Bohrer L, Sun Z, Wang Y, Huang H. Inhibition of the androgen receptor as a novel mechanism of taxol chemotherapy in prostate cancer. Cancer Res 2009;69(21):8386–8394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zhang G, Liu X, Li J, Ledet E, Alvarez X, Qi Y, Fu X, Sartor O, Dong Y, Zhang H. Androgen receptor splice variants circumvent AR blockade by microtubule-targeting agents. Oncotarget 2015;6(27):23358–23371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Thadani-Mulero M, Portella L, Sun S, Sung M, Matov A, Vessella RL, Corey E, Nanus DM, Plymate SR, Giannakakou P. Androgen receptor splice variants determine taxane sensitivity in prostate cancer. Cancer Res 2014;74(8):2270–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zhu ML, Horbinski CM, Garzotto M, Qian DZ, Beer TM, Kyprianou N. Tubulin-targeting chemotherapy impairs androgen receptor activity in prostate cancer. Cancer Res 2010;70(20):7992–8002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Martin SK, Banuelos CA, Sadar MD, Kyprianou N. N-terminal targeting of androgen receptor variant enhances response of castration resistant prostate cancer to taxane chemotherapy. Mol Oncol 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Imamura Y, Tien AH, Pan J, Leung JK, Banuelos CA, Jian K, Wang J, Mawji NR, Fernandez JG, Lin KS, Andersen RJ, Sadar MD. An imaging agent to detect androgen receptor and its active splice variants in prostate cancer. JCI Insight 2016;1(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.RJWnA ANDERSEN, Vancouver, BC V6S 1Z6, CA: ), Javier GARCIAFERNANDEZ (Las Palmeras 2, 1B Colunga, Asturias, E-33320, ES: ), Kunzhong JIAN (2510 Melfa Lane, Vancouver, BC V6T 2C6, CA: ), Dorothy SADAR, Marianne (4091 Bayridge Avenue, West Vancouver, BC V7V 3J9, CA: ), MAWJI Nasrin R. (2 Curie Avenue, Burnaby, BC V5G 4P4, 03–3421, CA: ), Adriana BANUELOS, Carmen (6 Moffatt Road, Richmond, BC V6Y 3G9, 9–7151, CA: ), Jun WANG (3542 Rosemary Heights Crescent, Surrey, BC V3S 0P2, CA: ), Yusuke IMAMURA (140–1 Chibadera-cho, Chu-o-kuChiba City, Chiba, JP: ); BRITISH COLUMBIA CANCER AGENCY BRANCH; (600 West 10th Avenue, Vancouver, British Columbia V5Z 4E6, CA: ),THE UNIVERSITY OF BRITISH COLUMBIA; (#103 – 6190 Agronomy Road, Vancouver, British Columbia V6T 1Z3, CA: ), assignee. HALOGENATED COMPOUNDS FOR CANCER IMAGING AND TREATMENT AND METHODS FOR THEIR USE2015. [Google Scholar]
- 180.NRWtA MAWJI, British Columbia V5Z 4E6, CA: ), Jun WANG (600 West 10th AvenueVancouver, British Columbia V5Z 4E6, CA: ), Adriana BANUELOS, Carmen (600 West 10th AvenueVancouver, British Columbia V5Z 4E6, CA: ), John ANDERSEN, Raymond (# Agronomy RoadVancouver, British Columbia V6T 1Z3, 103–6190, CA: ), Garcia FERNANDEZ, Javier (# Agronomy RoadVancouver, British Columbia V6T 1Z3, 103–6190, CA: ), Dorothy SADAR, Marianne (600 West 10th AvenueVancouver, British Columbia V5Z 4E6, CA: ); BRITISH COLUMBIA CANCER AGENCY BRANCH; (600 West 10th Avenue, Vancouver, British Columbia V5Z 4E6, CA: ),THE UNIVERSITY OF BRITISH COLUMBIA UNIVERSITY-INDUSTRY LIAISON OFFICE; (# Agronomy Road, Vancouver, British Columbia V6T 1Z3, 103–6190, CA: ), assignee. ESTER DERIVATIVES OF ANDROGEN RECEPTOR MODULATORS AND METHODS FOR THEIR USE2014. [Google Scholar]
- 181.ESSA_Pharma_Inc; ESSA pharma announces results from the phase 1 clinical trial of EPI-506 for treatment of mCRPC and updates clinical and stratergic plans. 2017. [Google Scholar]
- 182.Sadar MD, Williams DE, Mawji NR, Patrick BO, Wikanta T, Chasanah E, Irianto HE, Soest RV, Andersen RJ. Sintokamides A to E, chlorinated peptides from the sponge Dysidea sp. that inhibit transactivation of the N-terminus of the androgen receptor in prostate cancer cells. Org Lett 2008;10(21):4947–4950. [DOI] [PubMed] [Google Scholar]
- 183.Takeda AN, Pinon GM, Bens M, Fagart J, Rafestin-Oblin ME, Vandewalle A. The synthetic androgen methyltrienolone (r1881) acts as a potent antagonist of the mineralocorticoid receptor. Mol Pharmacol 2007;71(2):473–482. [DOI] [PubMed] [Google Scholar]
- 184.Banuelos CA, Tavakoli I, Tien AH, Caley DP, Mawji NR, Li Z, Wang J, Yang YC, Imamura Y, Yan L, Wen JG, Andersen RJ, Sadar MD. Sintokamide A Is a Novel Antagonist of Androgen Receptor That Uniquely Binds Activation Function-1 in Its Amino-terminal Domain. J Biol Chem 2016;291(42):22231–22243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.MDBA SADAR, West Vancouver, British Columbia V7V 3J9, CA: ), MAWJI Nasrin R. (2 Curle Avenue, Burnaby, British Columbia V5G 4P4, 03–3421, CA: ), Jun WANG (#25 - th Street, New Westminster, British Columbia V3M 3X9, 230 10, CA: ), ANDERSEN Raymond J. (4048 West 32nd Avenue, Vancouver, British Columbia V6S 1Z6, CA: ), WILLIAMS David E. (367 East 16th Avenue, Vancouver, British Columbia V5T 2T7, CA: ), Mike LEBLANC (#102 – 1065 East 8th Avenue, Vancouver, British Columbia V5T 1V1, CA: ), Lu-Ping YAN (Box 1540, 2205 Lower MallVancouver, British Columbia V6T 1Z4, CA: ); BRITISH COLUMBIA CANCER AGENCY BRANCH; (675 West 10th Avenue, Vancouver, British Columbia V5Z 1L3, CA: ),THE UNIVERSITY OF BRITISH COLUMBIA; (University - Industry Liaison Office, #103 – 6190 Agronomy RoadVancouver, British Columbia V6T 1Z3, CA: ),SADAR Marianne D. (4091 Bayridge Avenue, West Vancouver, British Columbia V7V 3J9, CA: ),MAWJI Nasrin R. (2 Curle Avenue, Burnaby, British Columbia V5G 4P4, 03–3421, CA: ),Jun WANG (#25 - th Street, New Westminster, British Columbia V3M 3X9, 230 10, CA: ),ANDERSEN Raymond J. (4048 West 32nd Avenue, Vancouver, British Columbia V6S 1Z6, CA: ),WILLIAMS David E. (367 East 16th Avenue, Vancouver, British Columbia V5T 2T7, CA: ),Mike LEBLANC (#102 – 1065 East 8th Avenue, Vancouver, British Columbia V5T 1V1, CA: ),Lu-Ping YAN (Box 1540, 2205 Lower MallVancouver, British Columbia V6T 1Z4, CA: ), assignee. SMALL MOLECULE INHIBITORS OF N-TERMINUS ACTIVATION OF THE ANDROGEN RECEPTOR2010. [Google Scholar]
- 186.Nazareth LV, Weigel NL. Activation of the Human Androgen Receptor through a Protein Kinase A Signaling Pathway. Journal of Biological Chemistry 1996;271(33):19900–19907. [DOI] [PubMed] [Google Scholar]
- 187.Blok LJ, de Ruiter PE, Brinkmann AO. Forskolin-induced dephosphorylation of the androgen receptor impairs ligand binding. Biochemistry 1998;37(11):3850–3857. [DOI] [PubMed] [Google Scholar]
- 188.Fu X, Ferreira ML, Schmitz FJ, Kelly-Borges M. New diketopiperazines from the sponge Dysidea chlorea. J Nat Prod 1998;61(10):1226–1231. [DOI] [PubMed] [Google Scholar]
- 189.Gu Z, Zakarian A. Concise total synthesis of sintokamides A, B, and E by a unified, protecting-group-free strategy. Angew Chem Int Ed Engl 2010;49(50):9702–9705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Meimetis LG, Williams DE, Mawji NR, Banuelos CA, Lal AA, Park JJ, Tien AH, Fernandez JG, de Voogd NJ, Sadar MD, Andersen RJ. Niphatenones, glycerol ethers from the sponge Niphates digitalis block androgen receptor transcriptional activity in prostate cancer cells: structure elucidation, synthesis, and biological activity. J Med Chem 2012;55(1):503–514. [DOI] [PubMed] [Google Scholar]
- 191.Banuelos CA, Lal A, Tien AH, Shah N, Yang YC, Mawji NR, Meimetis LG, Park J, Kunzhong J, Andersen RJ, Sadar MD. Characterization of niphatenones that inhibit androgen receptor N-terminal domain. PLoS One 2014;9(9):e107991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Langley E, Kemppainen JA, Wilson EM. Intermolecular NH2-/Carboxyl-terminal Interactions in Androgen Receptor Dimerization Revealed by Mutations That Cause Androgen Insensitivity. Journal of Biological Chemistry 1998;273(1):92–101. [DOI] [PubMed] [Google Scholar]
- 193.He B, Kemppainen JA, Voegel JJ, Gronemeyer H, Wilson EM. Activation Function 2 in the Human Androgen Receptor Ligand Binding Domain Mediates Interdomain Communication with the NH2-terminal Domain. Journal of Biological Chemistry 1999;274(52):37219–37225. [DOI] [PubMed] [Google Scholar]
- 194.He B, Kemppainen JA, Wilson EM. FXXLF and WXXLF sequences mediate the NH2-terminal interaction with the ligand binding domain of the androgen receptor. J Biol Chem 2000;275(30):22986–22994. [DOI] [PubMed] [Google Scholar]
- 195.Lavery DN, McEwan IJ. Structure and function of steroid receptor AF1 transactivation domains: induction of active conformations. Biochem J 2005;391(Pt 3):449–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.McEwan IJ, Lavery D, Fischer K, Watt K. Natural disordered sequences in the amino terminal domain of nuclear receptors: lessons from the androgen and glucocorticoid receptors. Nucl Recept Signal 2007;5:e001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.He B, Bai S, Hnat AT, Kalman RI, Minges JT, Patterson C, Wilson EM. An androgen receptor NH2-terminal conserved motif interacts with the COOH terminus of the Hsp70-interacting protein (CHIP). J Biol Chem 2004;279(29):30643–30653. [DOI] [PubMed] [Google Scholar]
- 198.Roy MK, Thalang VN, Trakoontivakorn G, Nakahara K. Mahanine, a carbazole alkaloid from Micromelum minutum, inhibits cell growth and induces apoptosis in U937 cells through a mitochondrial dependent pathway. Br J Pharmacol 2005;145(2):145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Nakahara K, Trakoontivakorn G, Alzoreky NS, Ono H, Onishi-Kameyama M, Yoshida M. Antimutagenicity of Some Edible Thai Plants, and a Bioactive Carbazole Alkaloid, Mahanine, Isolated fromMicromelum minutum. Journal of Agricultural and Food Chemistry 2002;50(17):4796–4802. [DOI] [PubMed] [Google Scholar]
- 200.Ramsewak RS, Nair MG, Strasburg GM, DeWitt DL, Nitiss JL. Biologically Active Carbazole Alkaloids fromMurrayakoenigii. Journal of Agricultural and Food Chemistry 1999;47(2):444–447. [DOI] [PubMed] [Google Scholar]
- 201.Tachibana Y, Kikuzaki H, Lajis NH, Nakatani N. Antioxidative Activity of Carbazoles fromMurraya koenigiiLeaves. Journal of Agricultural and Food Chemistry 2001;49(11):5589–5594. [DOI] [PubMed] [Google Scholar]
- 202.Roy MK, Thalang VN, Trakoontivakorn G, Nakahara K. Mechanism of mahanine-induced apoptosis in human leukemia cells (HL-60). Biochemical Pharmacology 2004;67(1):41–51. [DOI] [PubMed] [Google Scholar]
- 203.Sarkar S, Dutta D, Samanta SK, Bhattacharya K, Pal BC, Li J, Datta K, Mandal C, Mandal C. Oxidative inhibition of Hsp90 disrupts the super-chaperone complex and attenuates pancreatic adenocarcinoma in vitro and in vivo. Int J Cancer 2013;132(3):695–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Jagadeesh S, Sinha S, Pal BC, Bhattacharya S, Banerjee PP. Mahanine reverses an epigenetically silenced tumor suppressor gene RASSF1A in human prostate cancer cells. Biochem Biophys Res Commun 2007;362(1):212–217. [DOI] [PubMed] [Google Scholar]
- 205.Amin KS, Jagadeesh S, Baishya G, Rao PG, Barua NC, Bhattacharya S, Banerjee PP. A naturally derived small molecule disrupts ligand-dependent and ligand-independent androgen receptor signaling in human prostate cancer cells. Mol Cancer Ther 2014;13(2):341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Chen S, Gulla S, Cai C, Balk SP. Androgen receptor serine 81 phosphorylation mediates chromatin binding and transcriptional activation. J Biol Chem 2012;287(11):8571–8583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Hou S, Liu Y, Kong Y, Brown ML. Total Synthesis of 7-Hydroxymurrayazolinine, Murrayamine D, and Mahanine via m-Nitro Group Activated Pyran Annulation. Org Lett 2015;17(10):2298–2301. [DOI] [PubMed] [Google Scholar]
- 208.SIIoCB SINHA, 4 Raja S.c. Mallick Road Jadavpur, Kolkata 2, 700 03, IN: ), Chandra PAL, Bikas (Indian Institute of Chemical Biology, 4 Raja S.c. Mallick Road Jadavpur, Kolkata 2, 700 03, IN: ), Samir BHATTACHARYA (School of Life Sciences, Visva-Bharati Santineketan, West Bengal 5, 731 23, IN: ); COUNCIL OF SCIENTIFIC AND INDUSTRIAL RESEARCH; (Rafi Marg, New Delhi 1, 110 01, IN: ),Swati SINHA (Indian Institute of Chemical Biology, 4 Raja S.c. Mallick Road Jadavpur, Kolkata 2, 700 03, IN: ),Chandra PAL, Bikas (Indian Institute of Chemical Biology, 4 Raja S.c. Mallick Road Jadavpur, Kolkata 2, 700 03, IN: ),Samir BHATTACHARYA (School of Life Sciences, Visva-Bharati Santineketan, West Bengal 5, 731 23, IN: ), assignee. A PHARMACEUTICAL COMPOSITION COMPRISING MAHANINE USEFUL FOR THE TREATMENT OF PROSTATE CANCER2007. [Google Scholar]
- 209.Nelson CC, Hendy SC, Romaniuk PJ. Relationship between P-box Amino Acid Sequence and DNA Binding Specificity of the Thyroid Hormone Receptor. Journal of Biological Chemistry 1995;270(28):16981–16987. [DOI] [PubMed] [Google Scholar]
- 210.Li H, Ban F, Dalal K, Leblanc E, Frewin K, Ma D, Adomat H, Rennie PS, Cherkasov A. Discovery of small-molecule inhibitors selectively targeting the DNA-binding domain of the human androgen receptor. J Med Chem 2014;57(15):6458–6467. [DOI] [PubMed] [Google Scholar]
- 211.Dalal K, Roshan-Moniri M, Sharma A, Li H, Ban F, Hassona MD, Hsing M, Singh K, LeBlanc E, Dehm S, Tomlinson Guns ES, Cherkasov A, Rennie PS. Selectively targeting the DNA-binding domain of the androgen receptor as a prospective therapy for prostate cancer. J Biol Chem 2014;289(38):26417–26429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Li H, Ban F, Dalal K, Leblanc E, Frewin K, Ma D, Adomat H, Rennie PS, Cherkasov A. Correction to Discovery of Small-Molecule Inhibitors Selectively Targeting the DNA-Binding Domain of the Human Androgen Receptor. J Med Chem 2017;60(3):1225. [DOI] [PubMed] [Google Scholar]
- 213.Dalal K, Roshan-Moniri M, Sharma A, Li H, Ban F, Hassona MD, Hsing M, Singh K, LeBlanc E, Dehm S, Tomlinson Guns ES, Cherkasov A, Rennie PS. Selectively targeting the DNA-binding domain of the androgen receptor as a prospective therapy for prostate cancer. J Biol Chem 2017;292(10):4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Ban F, Dalal K, LeBlanc E, Morin H, Rennie PS, Cherkasov A. Cheminformatics Driven Development of Novel Therapies for Drug Resistant Prostate Cancer. Mol Inform 2018. [DOI] [PubMed] [Google Scholar]
- 215.Dalal K, Che M, Que NS, Sharma A, Yang R, Lallous N, Borgmann H, Ozistanbullu D, Tse R, Ban F, Li H, Tam KJ, Roshan-Moniri M, LeBlanc E, Gleave ME, Gewirth DT, Dehm SM, Cherkasov A, Rennie PS. Bypassing Drug Resistance Mechanisms of Prostate Cancer with Small Molecules that Target Androgen Receptor-Chromatin Interactions. Mol Cancer Ther 2017;16(10):2281–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Lim M, Otto-Duessel M, He M, Su L, Nguyen D, Chin E, Alliston T, Jones JO. Ligand-independent and tissue-selective androgen receptor inhibition by pyrvinium. ACS Chem Biol 2014;9(3):692–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Jones JO, Bolton EC, Huang Y, Feau C, Guy RK, Yamamoto KR, Hann B, Diamond MI. Non-competitive androgen receptor inhibition in vitro and in vivo. Proc Natl Acad Sci U S A 2009;106(17):7233–7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Momtazi-Borojeni AA, Abdollahi E, Ghasemi F, Caraglia M, Sahebkar A. The novel role of pyrvinium in cancer therapy. J Cell Physiol 2017. [DOI] [PubMed] [Google Scholar]
- 219.MtA DIAMOND, San Francisco, CA, 94122, US: ), Jeremy JONES (1327 Church St, San Francisco, CA, 94114, US: ), RENSLO Adam (5359 Shafter Ave, Oakland, CA, 94618, US: ); THE REGENTS OF THE UNIVERSITY OF CALIFORNIA; (1111 Franklin Street, 12th FloorOakland, CA, 94607, US: ),Marc DIAMOND (1426 12th Ave, San Francisco, CA, 94122, US: ),Jeremy JONES (1327 Church St, San Francisco, CA, 94114, US: ),Adam RENSLO (5359 Shafter Ave, Oakland, CA, 94618, US: ), assignee. SMALL-MOLECULE INHIBITORS OF THE ANDROGEN RECEPTOR2009. [Google Scholar]
- 220.Esumi H, Lu J, Kurashima Y, Hanaoka T. Antitumor activity of pyrvinium pamoate, 6‐(dimethylamino)‐2‐[2‐(2,5‐dimethyl‐1‐phenyl‐1H‐pyrrol‐3‐yl)ethenyl]‐1‐methyl‐quinolinium pamoate salt, showing preferential cytotoxicity during glucose starvation. Cancer Science 2004;95(8):685–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Xu R, Tian Y, Huang S, Yu J, Deng Y, Zhan M, Zhang T, Wang F, Zhao L, Chen Y. Synthesis and evaluation of novel thiazole-based derivatives as selective inhibitors of DNA-binding domain of the androgen receptor. Chem Biol Drug Des 2017. [DOI] [PubMed] [Google Scholar]
- 222.AB-WtA TCHERKASSOV, Vancouver, British Columbia V6H 1B3, CA: ), RENNIE Paul, S. (3731 Ullsmore Avenue, Richmond, British Columbia V7C 1S3, CA: ), Fuqiang BAN (17 Timbermill Crescent, Markham, Ontario L3P 6X1, CA: ), Huifang LI (2730 Acadia Road, Vancouver BC, V6T 1R9, CA: ), Jean LEBLANC, Eric Joseph (408–569 West 16th Avenue, Vancouver, British Columbia V5Z 4N7, CA: ); THE UNIVERSITY OF BRITISH COLUMBIA; (#103 – 6190 Agronomy Road, Vancouver, British Columbia V6T 1Z3, CA: ), assignee. HUMAN ANDROGEN RECEPTOR DNA-BINDING DOMAIN (DBD) COMPOUNDS AS THERAPEUTICS AND METHODS FOR THEIR USE2015. [Google Scholar]
- 223.Rettig MB, Jung ME, Ralalage NGDE, An J; The Regents of the University of California, assignee. Inhibitors of the N-terminal Domain of the Androgen Receptor. United States patent PCT/US2018/014516 2018 01/19/2018.
- 224.Elshan NGRD, An J, Wang T, Kobayashi N, Kono E, Reiter RE, Jung ME, Rettig MB. A Novel Class of Androgen Receptor Inhibitors for Treatment of Castration-Resistant Prostate Cancer. Science (in preparation) 2018. [Google Scholar]
- 225.Cheng MA, Chou FJ, Wang K, Yang R, Ding J, Zhang Q, Li G, Yeh S, Chang C, Xu D. Androgen receptor (AR) degradation enhancer ASC-J9((R)) in an FDA-approved formulated solution suppresses castration resistant prostate cancer cell growth. Cancer Lett 2018;417:182–191. [DOI] [PubMed] [Google Scholar]
- 226.Taniguchi Y The Bromodomain and Extra-Terminal Domain (BET) Family: Functional Anatomy of BET Paralogous Proteins. Int J Mol Sci 2016;17(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Ntranos A, Casaccia P. Bromodomains: Translating the words of lysine acetylation into myelin injury and repair. Neurosci Lett 2016;625:4–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Shi J, Vakoc CR. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol Cell 2014;54(5):728–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Jain AK, Barton MC. Bromodomain Histone Readers and Cancer. J Mol Biol 2017;429(13):2003–2010. [DOI] [PubMed] [Google Scholar]
- 230.Chan SC, Selth LA, Li Y, Nyquist MD, Miao L, Bradner JE, Raj GV, Tilley WD, Dehm SM. Targeting chromatin binding regulation of constitutively active AR variants to overcome prostate cancer resistance to endocrine-based therapies. Nucleic Acids Res 2015;43(12):5880–5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Eisermann K, Fraizer G. The Androgen Receptor and VEGF: Mechanisms of Androgen-Regulated Angiogenesis in Prostate Cancer. Cancers (Basel) 2017;9(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Ramadoss M, Mahadevan V. Targeting the cancer epigenome: synergistic therapy with bromodomain inhibitors. Drug Discov Today 2018;23(1):76–89. [DOI] [PubMed] [Google Scholar]
- 233.Liu Z, Wang P, Chen H, Wold EA, Tian B, Brasier AR, Zhou J. Drug Discovery Targeting Bromodomain-Containing Protein 4. J Med Chem 2017;60(11):4533–4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Dai X, Gan W, Li X, Wang S, Zhang W, Huang L, Liu S, Zhong Q, Guo J, Zhang J, Chen T, Shimizu K, Beca F, Blattner M, Vasudevan D, Buckley DL, Qi J, Buser L, Liu P, Inuzuka H, Beck AH, Wang L, Wild PJ, Garraway LA, Rubin MA, Barbieri CE, Wong KK, Muthuswamy SK, Huang J, Chen Y, Bradner JE, Wei W. Prostate cancer-associated SPOP mutations confer resistance to BET inhibitors through stabilization of BRD4. Nat Med 2017;23(9):1063–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Theodoulou NH, Tomkinson NC, Prinjha RK, Humphreys PG. Clinical progress and pharmacology of small molecule bromodomain inhibitors. Curr Opin Chem Biol 2016;33:58–66. [DOI] [PubMed] [Google Scholar]
- 236.Urbanucci A, Mills IG. Bromodomain-containing proteins in prostate cancer. Mol Cell Endocrinol 2018;462(Pt A):31–40. [DOI] [PubMed] [Google Scholar]
- 237.Spiliotopoulos D, Caflisch A. Fragment-based in silico screening of bromodomain ligands. Drug Discov Today Technol 2016;19:81–90. [DOI] [PubMed] [Google Scholar]
- 238.Fernandez-Salas E, Wang S, Chinnaiyan AM. Role of BET proteins in castration-resistant prostate cancer. Drug Discov Today Technol 2016;19:29–38. [DOI] [PubMed] [Google Scholar]
- 239.Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, Escara-Wilke J, Wilder-Romans K, Dhanireddy S, Engelke C, Iyer MK, Jing X, Wu YM, Cao X, Qin ZS, Wang S, Feng FY, Chinnaiyan AM. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature 2014;510(7504):278–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Umehara T, Akiko Tanaka, Kazuhito Sato, Shigeyuki Yokoyama; INSTITUTE OF PHYSICAL CHEMICAL RESEARCH, assignee. BRD2 BROMODOMAIN BINDER2008.
- 241.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N, French CA, Wiest O, Kung AL, Knapp S, Bradner JE. Selective inhibition of BET bromodomains. Nature 2010;468(7327):1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Smith SG, Sanchez R, Zhou MM. Privileged diazepine compounds and their emergence as bromodomain inhibitors. Chem Biol 2014;21(5):573–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, Chesi M, Schinzel AC, McKeown MR, Heffernan TP, Vakoc CR, Bergsagel PL, Ghobrial IM, Richardson PG, Young RA, Hahn WC, Anderson KC, Kung AL, Bradner JE, Mitsiades CS. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011;146(6):904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Gao L, Schwartzman J, Gibbs A, Lisac R, Kleinschmidt R, Wilmot B, Bottomly D, Coleman I, Nelson P, McWeeney S, Alumkal J. Androgen receptor promotes ligand-independent prostate cancer progression through c-Myc upregulation. PLoS One 2013;8(5):e63563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Baker EK, Taylor S, Gupte A, Sharp PP, Walia M, Walsh NC, Zannettino AC, Chalk AM, Burns CJ, Walkley CR. BET inhibitors induce apoptosis through a MYC independent mechanism and synergise with CDK inhibitors to kill osteosarcoma cells. Sci Rep 2015;5:10120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Tanase CP, Codrici E, Popescu ID, Mihai S, Enciu AM, Necula LG, Preda A, Ismail G, Albulescu R. Prostate cancer proteomics: Current trends and future perspectives for biomarker discovery. Oncotarget 2017;8(11):18497–18512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Zoni E, Karkampouna S, Thalmann GN, Kruithof-de Julio M, Spahn M. Emerging aspects of microRNA interaction with TMPRSS2-ERG and endocrine therapy. Mol Cell Endocrinol 2018;462(Pt A):9–16. [DOI] [PubMed] [Google Scholar]
- 248.Babu D, Fullwood MJ. Expanding the effects of ERG on chromatin landscapes and dysregulated transcription in prostate cancer. Nat Genet 2017;49(9):1294–1295. [DOI] [PubMed] [Google Scholar]
- 249.Matzuk MM, McKeown MR, Filippakopoulos P, Li Q, Ma L, Agno JE, Lemieux ME, Picaud S, Yu RN, Qi J, Knapp S, Bradner JE. Small-molecule inhibition of BRDT for male contraception. Cell 2012;150(4):673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Wyce A, Degenhardt Y, Bai Y, Le B, Korenchuk S, Crouthame MC, McHugh CF, Vessella R, Creasy CL, Tummino PJ, Barbash O. Inhibition of BET bromodomain proteins as a therapeutic approach in prostate cancer. Oncotarget 2013;4(12):2419–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Xiang Q, Zhang Y, Li J, Xue X, Wang C, Song M, Zhang C, Wang R, Li C, Wu C, Zhou Y, Yang X, Li G, Ding K, Xu Y. Y08060: A Selective BET Inhibitor for Treatment of Prostate Cancer. ACS Med Chem Lett 2018;9(3):262–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Picaud S, Da Costa D, Thanasopoulou A, Filippakopoulos P, Fish PV, Philpott M, Fedorov O, Brennan P, Bunnage ME, Owen DR, Bradner JE, Taniere P, O’Sullivan B, Muller S, Schwaller J, Stankovic T, Knapp S. PFI-1, a highly selective protein interaction inhibitor, targeting BET Bromodomains. Cancer Res 2013;73(11):3336–3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Fish PV, Filippakopoulos P, Bish G, Brennan PE, Bunnage ME, Cook AS, Federov O, Gerstenberger BS, Jones H, Knapp S, Marsden B, Nocka K, Owen DR, Philpott M, Picaud S, Primiano MJ, Ralph MJ, Sciammetta N, Trzupek JD. Identification of a chemical probe for bromo and extra C-terminal bromodomain inhibition through optimization of a fragment-derived hit. J Med Chem 2012;55(22):9831–9837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Hupe MC, Hoda MR, Zengerling F, Perner S, Merseburger AS, Cronauer MV. The BET-inhibitor PFI-1 diminishes AR/AR-V7 signaling in prostate cancer cells. World J Urol 2018. [DOI] [PubMed] [Google Scholar]
- 255.Urbanucci A, Barfeld SJ, Kytola V, Itkonen HM, Coleman IM, Vodak D, Sjoblom L, Sheng X, Tolonen T, Minner S, Burdelski C, Kivinummi KK, Kohvakka A, Kregel S, Takhar M, Alshalalfa M, Davicioni E, Erho N, Lloyd P, Karnes RJ, Ross AE, Schaeffer EM, Vander Griend DJ, Knapp S, Corey E, Feng FY, Nelson PS, Saatcioglu F, Knudsen KE, Tammela TLJ, Sauter G, Schlomm T, Nykter M, Visakorpi T, Mills IG. Androgen Receptor Deregulation Drives Bromodomain-Mediated Chromatin Alterations in Prostate Cancer. Cell Rep 2017;19(10):2045–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Urbanucci A, Marttila S, Janne OA, Visakorpi T. Androgen receptor overexpression alters binding dynamics of the receptor to chromatin and chromatin structure. Prostate 2012;72(11):1223–1232. [DOI] [PubMed] [Google Scholar]
- 257.Pawar A, Gollavilli PN, Wang S, Asangani IA. Resistance to BET Inhibitor Leads to Alternative Therapeutic Vulnerabilities in Castration-Resistant Prostate Cancer. Cell Rep 2018;22(9):2236–2245. [DOI] [PubMed] [Google Scholar]
- 258.Floyd SR, Pacold ME, Huang Q, Clarke SM, Lam FC, Cannell IG, Bryson BD, Rameseder J, Lee MJ, Blake EJ, Fydrych A, Ho R, Greenberger BA, Chen GC, Maffa A, Del Rosario AM, Root DE, Carpenter AE, Hahn WC, Sabatini DM, Chen CC, White FM, Bradner JE, Yaffe MB. The bromodomain protein Brd4 insulates chromatin from DNA damage signalling. Nature 2013;498(7453):246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Dulaney C, Marcrom S, Stanley J, Yang ES. Poly(ADP-ribose) polymerase activity and inhibition in cancer. Semin Cell Dev Biol 2017;63:144–153. [DOI] [PubMed] [Google Scholar]
- 260.Wilson AJ, Stubbs M, Liu P, Ruggeri B, Khabele D. The BET inhibitor INCB054329 reduces homologous recombination efficiency and augments PARP inhibitor activity in ovarian cancer. Gynecol Oncol 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Zhang P, Wang D, Zhao Y, Ren S, Gao K, Ye Z, Wang S, Pan CW, Zhu Y, Yan Y, Yang Y, Wu D, He Y, Zhang J, Lu D, Liu X, Yu L, Zhao S, Li Y, Lin D, Wang Y, Wang L, Chen Y, Sun Y, Wang C, Huang H. Intrinsic BET inhibitor resistance in SPOP-mutated prostate cancer is mediated by BET protein stabilization and AKT-mTORC1 activation. Nat Med 2017;23(9):1055–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Jamaspishvili T, Berman DM, Ross AE, Scher HI, De Marzo AM, Squire JA, Lotan TL. Clinical implications of PTEN loss in prostate cancer. Nat Rev Urol 2018;15(4):222–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Wise HM, Hermida MA, Leslie NR. Prostate cancer, PI3K, PTEN and prognosis. Clin Sci (Lond) 2017;131(3):197–210. [DOI] [PubMed] [Google Scholar]
- 264.Zhuang L, Lin J, Lu ML, Solomon KR, Freeman MR. Cholesterol-rich Lipid Rafts Mediate Akt-regulated Survival in Prostate Cancer Cells. Cancer Research 2002;62(8):2227–2231. [PubMed] [Google Scholar]
- 265.Zhang HP, Li GQ, Zhang Y, Guo WZ, Zhang JK, Li J, Lv JF, Zhang SJ. Upregulation of Mcl-1 inhibits JQ1-triggered anticancer activity in hepatocellular carcinoma cells. Biochem Biophys Res Commun 2018;495(4):2456–2461. [DOI] [PubMed] [Google Scholar]
- 266.Zenith_Epigenetics. The clinical candidate ZEN-3694, a BET bromodomain inhibitor, is efficacious in the treatment of a variety of solid tumor and hematological malignancies, alone or in combination with several standard of care therapies. (04/11/2018).
- 267.Zenith_Epigenetics. Zenith Annual Meeting and Clinical Advancements Update – December 12, 2017. (04/12/2018).
- 268.Attwell S, Campeau E, Jahagirdar R, Kharenko O, Norek K, Tsujikawa L, Calosing C, Patel R, Gesner E, Lakhotia S, Hansen H. Abstract C86: The clinical candidate ZEN-3694, a novel BET bromodomain inhibitor, is efficacious in the treatment of a variety of solid tumor and hematological malignancies, alone or in combination with several standard of care and targeted therapies. Molecular Cancer Therapeutics 2016;14(12 Supplement 2):C86–C86. [Google Scholar]
- 269.Perez-Salvia M, Esteller M. Bromodomain inhibitors and cancer therapy: From structures to applications. Epigenetics 2017;12(5):323–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Andrieu G, Belkina AC, Denis GV. Clinical trials for BET inhibitors run ahead of the science. Drug Discov Today Technol 2016;19:45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Markowski MC, De Marzo AM, Antonarakis ES. BET inhibitors in metastatic prostate cancer: therapeutic implications and rational drug combinations. Expert Opin Investig Drugs 2017;26(12):1391–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Esteve-Arenys A, Valero JG, Chamorro-Jorganes A, Gonzalez D, Rodriguez V, Dlouhy I, Salaverria I, Campo E, Colomer D, Martinez A, Rymkiewicz G, Perez-Galan P, Lopez-Guillermo A, Roue G. The BET bromodomain inhibitor CPI203 overcomes resistance to ABT-199 (venetoclax) by downregulation of BFL-1/A1 in in vitro and in vivo models of MYC+/BCL2+ double hit lymphoma. Oncogene 2018;37(14):1830–1844. [DOI] [PubMed] [Google Scholar]
- 273.Chan KH, Zengerle M, Testa A, Ciulli A. Impact of Target Warhead and Linkage Vector on Inducing Protein Degradation: Comparison of Bromodomain and Extra-Terminal (BET) Degraders Derived from Triazolodiazepine (JQ1) and Tetrahydroquinoline (I-BET726) BET Inhibitor Scaffolds. J Med Chem 2018;61(2):504–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.QIAN YCC, Plainsboro, NJ, 08536, US: ), Hanqing DONG (248 Opening Hill Rd, Madison, CT, 06443, US: ), Jing WANG (25 Elm Street, Milford, CT, 06460, US: ), Michael BERLIN (14 Hendirck Road, Flemington, NJ, 08822, US: ), Kam SIU (42 Magnolia Road, Milford, CT, 06461, US: ), CREW Andrew, P. (2 Winterberry Road, Guilford, CT, 06437, US: ), CREWS Craig, M. (286 Livingston Street, New Haven, CT, 06511, US: ); ARVINAS, INC. (5 Science Park, New Haven, CT, 06511, US: ),YALE UNIVERSITY; (Office of Cooperative Research, 433 Temple StreetNew Haven, CT, 06511, US: ), assignee. COMPOUNDS AND METHODS FOR THE TARGETED DEGRADATION OF BROMODOMAIN-CONTAINING PROTEINS2017. [Google Scholar]
- 275.Sun L, Gao P. Small molecules remain on target for c-Myc. Elife 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Andrieu GP, Denis GV. BET Proteins Exhibit Transcriptional and Functional Opposition in the Epithelial-to-Mesenchymal Transition. Mol Cancer Res 2018;16(4):580–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Jenster G, van der Korput HAGM, Trapman J, Brinkmann AO. Identification of Two Transcription Activation Units in the N-terminal Domain of the Human Androgen Receptor. Journal of Biological Chemistry 1995;270(13):7341–7346. [DOI] [PubMed] [Google Scholar]
- 278.Chen WY, Tsai YC, Siu MK, Yeh HL, Chen CL, Yin JJ, Huang J, Liu YN. Inhibition of the androgen receptor induces a novel tumor promoter, ZBTB46, for prostate cancer metastasis. Oncogene 2017;36(45):6213–6224. [DOI] [PubMed] [Google Scholar]
- 279.Fong KW, Zhao JC, Kim J, Li S, Yang YA, Song B, Rittie L, Hu M, Yang X, Perbal B, Yu J. Polycomb-Mediated Disruption of an Androgen Receptor Feedback Loop Drives Castration-Resistant Prostate Cancer. Cancer Res 2017;77(2):412–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Wang B, Lo UG, Wu K, Kapur P, Liu X, Huang J, Chen W, Hernandez E, Santoyo J, Ma SH, Pong RC, He D, Cheng YQ, Hsieh JT. Developing new targeting strategy for androgen receptor variants in castration resistant prostate cancer. Int J Cancer 2017;141(10):2121–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Narayanan R, Coss CC, Dalton JT. Development of selective androgen receptor modulators (SARMs). Mol Cell Endocrinol 2018;465:134–142. [DOI] [PMC free article] [PubMed] [Google Scholar]