Abstract
Ribbon synapses are tonically active synapses in the retina and inner ear with intense vesicle traffic. How this traffic is organized and regulated is still unknown. Synaptic ribbons, large presynaptic structures associated with numerous synaptic vesicles, appear to be essential for this process. The base of the synaptic ribbon is anchored at the active zone and is a hotspot of exocytosis. The synaptic ribbon complex is also important for vesicle replenishment. RIBEYE is a unique and major component of synaptic ribbons. It consists of a unique A-domain and an NAD(H)-binding, C-terminal B-domain. In the present study, we show that the Arf-GTPase activating protein-3 (ArfGAP3), a well characterized regulator of vesicle formation at the Golgi apparatus, is also a component of the synaptic ribbon complex in photoreceptor synapses of the mouse retina and interacts with RIBEYE as shown by multiple, independent approaches. ArfGAP3 binds to RIBEYE(B)-domain in an NAD(H)-dependent manner. The interaction is redox sensitive because NADH is more efficient than the oxidized NAD+ in promoting ArfGAP3-RIBEYE interaction. RIBEYE competes with the GTP-binding protein Arf1 for binding to ArfGAP3. Thus, binding of RIBEYE(B) to ArfGAP3 could prevent inactivation of Arf1 by ArfGAP3 and provides the synaptic ribbon with the possibility to control Arf1 function. The interaction is relevant for endocytic vesicle trafficking because overexpression of ArfGAP3 in photoreceptors strongly inhibited endocytotic uptake of FM1–43.
Keywords: ArfGAP3, endocytosis, photoreceptor synapse, ribbon synapse, RIBEYE, synaptic ribbon
Introduction
Ribbon synapses, e.g., retinal photoreceptor synapses, are continuously active synapses with a high vesicle turnover. How this vesicle traffic is organized and regulated is still largely unknown. Synaptic ribbons, large presynaptic structures associated with numerous synaptic vesicles, appear to play a central role in this process. The basal end of the synaptic ribbon is anchored at the active zone where L-type voltage-gated channels are clustered. This site is a hotspot of exocytosis (Zenisek et al., 2000; Frank et al., 2010; Chen et al., 2013). The synaptic ribbon complex is also relevant for vesicle recycling (Spassova et al., 2004; Griesinger et al., 2005; Jackman et al., 2009; Babai et al., 2010; Frank et al., 2010; Schnee et al., 2011; Snellman et al., 2011; Tian et al., 2012; Wahl et al., 2013). In photoreceptor synapses, synaptic vesicle retrieval occurs in the periactive zone, in close proximity to the synaptic ribbon (Wahl et al., 2013). RIBEYE is a major and unique component of synaptic ribbons (Schmitz et al., 2000; for review, see Schmitz, 2009). It consists of an N-terminal A-domain and a C-terminal B-domain. The B-domain is largely identical with the protein CtBP2 and binds NAD(H) (for review, see Schmitz, 2009; Schmitz et al., 2012).
Small GTP-binding proteins of the Arf family are important regulators of intracellular membrane traffic (for review, see Gillingham and Munro, 2007). Arf proteins switch between a GTP-bound, active form and a GDP-bound, inactive form. Which nucleotide is bound is regulated by the activity of two classes of proteins, the ArfGEFs (Arf GTP-exchange factors) and the ArfGAPs (Arf-GTPase-activating proteins; for review, see Inoue and Randazzo, 2007; Kahn et al., 2008; Spang et al., 2010). ArfGAPs enhance the low intrinsic GTPase activity of Arfs. A large number of ArfGAPs have been identified in higher organisms. Conserved hallmark of ArfGAPs is an ∼136 aa long, catalytically active GAP-domain (AGD) with a central Zn-finger motif that stimulates GTPase activity of the attached Arf (Cukierman et al., 1995; Goldberg, 1999). The regions outside of the AGD are divergent in different ArfGAPs. The prototypical yeast ArfGAPs gcs1p and glo3p possess mammalian orthologs in ArfGAP1 (for gcs1p) and ArfGAP2/ArfGAP3 (for glo3p). In these ArfGAPs, the conserved AGD is located at the N terminus. Both classes of ArfGAPs possess distinct motifs in their C terminus, which mediate membrane binding (Bigay et al., 2005; Kliouchnikov et al., 2009) as well as other functions (for review, see Spang et al., 2010).
In the present study, we identified ArfGAP3 as a new component of the synaptic ribbon complex in photoreceptor synapses of the mammalian retina. ArfGAP3 is a well known component of the Golgi apparatus where it regulates retrograde trafficking from the Golgi to the endoplasmic reticulum (for review, see Spang et al., 2010). We provide evidence that ArfGAP3 is important for controlling Arf1 activity at the synaptic ribbon complex and for regulating endocytic membrane traffic.
Materials and Methods
Plasmids
Bacterial expression constructs.
These include ArfGAP3(AGD)-pGEX-KG, which encodes the ArfGAP-domain (AGD) (amino acids 1–136) of bovine ArfGAP3. The insert was excised from GAP-dom(ArfGAP3)pACT2 with EcoRI and XhoI and cloned into the EcoRI/XhoI sites of pGEX-KG. ArfGAP3(AGD)-pMal-C2, encoding the ArfGAP-domain of bovine ArfGAP3. The insert was amplified from bovine ArfGAP3 cDNA (BC118087) using the following forward primer AAAAGAATTCATGGGGGACCCCAG and reverse primer AAAAGTCGACGCTATCAAGCCAGAG and cloned into the EcoRI/SalI sites of pMal-C2. Also included is ArfGAP3(AGD-extended)-pMal-C2, which encodes the extended ArfGAP-domain (AGD extended, amino acids 1–225 of bovine ArfGAP3). The insert was amplified from bovine ArfGAP3 cDNA (BC118087) using forward primer AAAAGAATTCATG GGGGACCCCAG reverse primer AAACTCGAGAAGTCCT CTTTTAGC and cloned into the EcoRI/SalI sites of pMal-C2. ArfGAP3(AGD extended)-pSNAPtagT7 encodes the extended ArfGAP-domain. The insert was amplified from bovine ArfGAP3 cDNA (BC118087) using forward primer AAAAGGATCCATGGGGGACCCCA and reverse primer AAACTCGAGAAGTCC TCTTTTAGC and cloned into the BamHI/XhoI sites of pSNAPtagT7 (NEB). ArfGAP3Cterm2-pGEX-KG encodes amino acids 226–335 of bovine ArfGAP3. The insert was amplified from bovine ArfGAP3 cDNA using forward primer AAAGAATTCGGGCCAAAAAAGGAAGT and reverse primer AAACTCGAGCGTGA TTGGTGTTTC and cloned into the EcoRI/XhoI sites of pGEX-KG. ArfGAP3Cterm3-pGEX-KG encodes amino acids 332–460 of bovine ArfGAP3. The insert was amplified from bovine ArfGAP3 cDNA using forward primer AAAGAATTCAAACACCAATCACG GCG and reverse primer AAACTCGAG AGCTGAGCTGATGGA and cloned into the EcoRI/XhoI sites of pGEX-KG. RIBEYE(B)-MBP (Magupalli et al., 2008). The plasmid pMal-C2 corresponds to the commercially available pMal-C2 vector (NEB) to which multiple STOP codons have been added in all reading frames at the end of the multiple cloning site using standard methods. RE(B)pGEX-KG (Schmitz et al., 2000). RE(B)G730ApGEX-KG (Alpadi et al., 2008; Venkatesan et al., 2010). Arf1-pGEX-KG. The insert (∼0.55 kb) was amplified from a bovine cDNA library using forward primer AAAACCATGGCGAATATCTTTGCAAAC and reverse primer AAAACTCGAGTCATTT CTGGTTC and cloned into the NcoI/XhoI sites of pGEX-KG. Plasmid constructs were verified by sequencing.
Eukaryotic expression constructs.
ArfGAP3-mCherry encodes amino acids 1–517 of bovine ArfGAP3. Full-length ArfGAP3 was amplified by PCR using forward primer AAACTCGAGGCCACCATGGGGGA CCCCAGCAAG, reverse primer AAAGAATTCCGGAACCGTAGCGATC, and ArfGAP3 cDNA as template. The ∼1.5 kb PCR product was cloned into the XhoI/EcoRI sites of pCherry-N1 (Alpadi et al., 2008). RE(B)-EGFP (Schmitz et al., 2000).
Yeast vectors
ArfGAP3-pACT2 encoding full-length bovine ArfGAP3 was obtained by yeast two-hybrid (YTH) screening with RIBEYE as bait construct. ArfGAP3cDNA-pACT2, encoding amino acids1–517 of bovine ArfGAP3, was amplified from the full-length ArfGAP3 IMAGE clone #8081904 (BC118087) using forward primer AAAGAATTCTGA TCATGGGGAC and reverse primer AAACTCGAGTTAGGAAC CGTAGCG and cloned into the EcoRI/XhoI sites of pACT2. ArfGAP3(AGD)pACT2 encodes the ArfGAP-domain of ArfGAP3. The insert was amplified by PCR using forward primer AAAGAATTCTGATCATGGGGAC, reverse primer AAACTCGAGTTAGCTATC AAGCCA, and bovine ArfGAP3 cDNA (BC118087) as a template. The PCR product was cloned into the EcoRI/XhoI sites of pACT2. CtermArfGAP3pACT2, encoding amino acids 127–517 of bovine ArfGAP3, was amplified from bovine ArfGAP3 cDNA (BC118087) using forward primer AAAGAATTCAGCACGGGCACTGAC and reverse primer AAACTCGAGTTAGGAACCGTAGCG and cloned into the EcoRI/XhoI sites of pACT2. The insert of Arf1-pGBKT7 (∼0.55 kb) was amplified from a bovine cDNA library (Alpadi et al., 2008) using forward primer AAAACCATGGCGAATATCTTTGCAAAC and reverse primer AAAACTCGAGTCATTTCTGGTTC and cloned into the NcoI/SalI sites of pGBKT7. RE(AB)pGBK-T7 (Magupalli et al., 2008). RE(B)pGBK-T7 (Magupalli et al., 2008). RE(B)NBD-pGBK-T7 (Alpadi et al., 2008). RE(B)SBD-pGBK-T7 (Alpadi et al., 2008). RE(B)G730A-pGBK-T7 (Alpadi et al., 2008). RE(A)pACT2 (Magupalli et al., 2008). pGBK-T7 (empty bait plasmid) (Tai et al., 1999; Magupalli et al., 2008). pSE1112 (control bait plasmid) (Tai et al., 1999; Magupalli et al., 2008). pACT2 (empty prey plasmid) (Magupalli et al., 2008). pSE1111 (control prey plasmid) (Tai et al., 1999; Magupalli et al., 2008). Munc119pACT2 (Alpadi et al., 2008). Munc119 is known to interact with RIBEYE(B)-domain and was used as a positive control for yeast matings. Bassoon pGAD-T7 (encoding amino acids1638–2081 of rat bassoon, NP062019.2) was cloned by reverse-transcriptase -PCR using cDNA isolated from rat R28 cells (Alpadi et al., 2008), forward primer TTTTCATATGTGCCGGATCTCCTCTGTCCCT, and reverse primer TTTTGAATTCC TGGGCCAGGCTGGCCTCCTG and cloned into the NdeI/EcoRI sites of pGADT7. Bassoon pGAD-T7 was used as a positive control mating for RIBEYE(B) (tom Dieck et al., 2005). Plasmid constructs were verified by sequencing.
Antibodies
Primary antibodies.
The following primary antibodies were used in the present study: mouse monoclonal anti-GST (Sigma; Alpadi et al., 2008) used at 1:10,000 dilution for Western blotting; mouse monoclonal anti-MBP (NEB; Alpadi et al., 2008) used at 1:10,000 dilution for Western blotting; rabbit polyclonal anti-RIBEYE(B)-domain (U2656; Schmitz et al., 2000) used at 1:10,000 for Western blotting for immunofluorescence microscopy at a 1:1000 dilution; mouse monoclonal antibodies against RIBEYE(B)-domain/CtBP2 (BD Transduction Laboratories; Alpadi et al., 2008; Schwarz et al., 2011; Wahl et al., 2013) used at a 1:1000 dilution; mouse monoclonal anti-Bassoon (Stressgen, VAM-PS003) used at a 1:100 dilution for immunofluorescence microscopy (Wahl et al., 2013); mouse monoclonal antibody against Arf1 (ARFS 1A9/5; Santa Cruz Biotechnology, sc-53168) used at a 1:500 dilution; and anti-dynamin (hudy-1; Millipore) used at a 1:50 dilution. The DyLight 650 directly labeled mouse monoclonal antibody against RIBEYE(B)/CtBP2 was diluted 1:2.
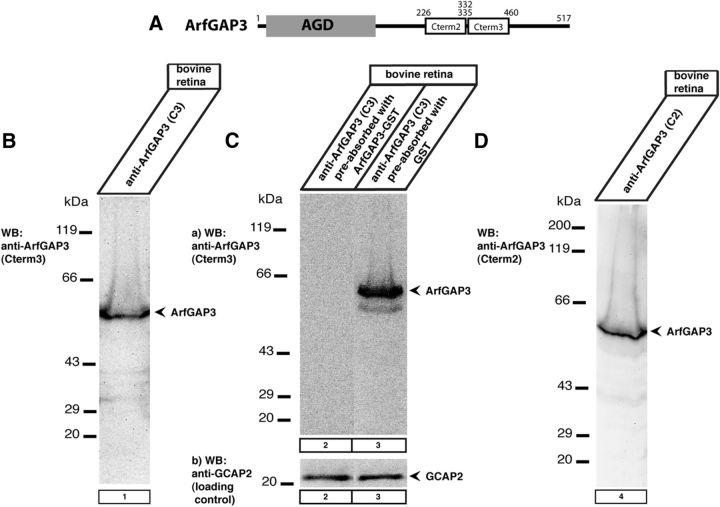
We generated two different polyclonal antisera against two different regions in the C terminus of ArfGAP3 (Cterm2 and Cterm3; see Fig. 7A). ArfGAP3Cterm2-pGEX and ArfGAP3Cterm3-pGEX were electroporated into Bl21(DE3) and fusion proteins were expressed and purified as previously described (Schmitz et al., 2000). For antibody production, the purified fusion proteins were injected into rabbits multiple times. Immune sera were screened for reactivity against the fusion protein and the endogenous protein. Pre-immune serum was used as control serum. Both ArfGAP3 antibodies (named ArfGAP3Cterm2- and ArfGAP3Cterm3-antisera) were used for Western blotting in a 1:3000 dilution for immunofluorescence microscopy in a dilution of 1:20. Affinity-purified ArfGAP3 antibodies were used at a concentration of 10 μg/ml for immunofluorescence microscopy. ArfGAP3Cterm2 and ArfGAP3Cterm3 antibodies worked well for Western blotting and immunofluorescence microscopy. Unfortunately, they did not work at the electron microscopic level, both in pre-embedding and postembedding procedures with immunogold- and immunoperoxidase-based techniques with the procedures available in our laboratory. Also the following commercially available antibodies did not work in postembedding immunogold electron microscopy: rabbit and goat polyclonal anti-ArfGAP3 antibodies (AAS68618C and ASA34060; Antibody Verify), rabbit polyclonal anti-ArfGAP3 (NBP1-18921; Novus Biologicals), rabbit polyclonal anti-ArfGAP3 (HPA000638; Sigma), and rabbit polyclonal anti-ArfGAP3 (A302-032A; Biomol).
Figure 7.
Western blot analyses of two antibodies that were generated against the C terminus of ArfGAP3. A, Schematic drawing denotes the areas against which the two polyclonal ArfGAP3 antibodies (Cterm2 and Cterm3) were generated. B–D, In Western blot analyses, both antibodies (Cterm2 and Cterm3) detected a single band at the expected running position of ArfGAP3 at ∼55 kDa. This ArfGAP3 Western blot band could be blocked by pre-absorption of the antibody with the respective ArfGAP3-GST fusion protein (lane 2; C; data not shown) but not by pre-incubation with GST alone (C, lane 3; data not shown). Cb, Loading control (immunoblotting of the same blot as shown in Ca) after stripping of the blot and reprobing with an antibody against GCAP2 (Venkatesan et al., 2010), demonstrating equal protein loading.
Secondary antibodies (for immunofluorescence labeling).
The following secondary antibodies were used: chicken anti-mouse Alexa 488, donkey anti-rabbit Alexa 568, goat anti-mouse Alexa 488. All fluorophore-conjugated secondary antibodies were purchased from Invitrogen and used at a 1:1000 dilution for 1 h at room temperature (RT) for immunolabeling experiments.
Methods
YTH analyses.
YTH assays were performed largely as previously described (Alpadi et al., 2008; Magupalli et al., 2008). The Gal4-based Matchmaker Yeast Two-Hybrid System (Clontech) was used according to manufacturer's instructions. For the YTH screening we used a bovine retinal YTH cDNA library from the retina (Tai et al., 1999; Alpadi et al., 2008; Venkatesan et al., 2010). The cDNA of the respective bait proteins were cloned in frame with the Gal4-DNA-binding domain of pGBKT7. The cDNA of the indicated prey proteins were cloned in frame with the Gal4-activation domain of pACT2 or pGADT7. The bait and prey plasmids confer tryptophan (W) and leucine (L) prototrophy to the respective auxotrophic yeast strains. Two yeast strains, Y187 and Y2HGold (Clontech), were used that contain distinct auxotrophic marker genes. (1) Y2HGold (Clontech): MATa, trp1-901, leu2-3, 112, ura3-52, his3-200, gal4, gal80, LYS2::GAL1UAS–Gal1TATA–His3,GAL2UAS–Gal2TATA–Ade2,URA3::MEL1UAS–Mel1TATA, AU R1-C, MEL1. This strain contains distinct ADE2, HIS3, MEL1, and AUR1-C reporter constructs that are only expressed in the presence of GAL4-based protein interactions (Clontech). (2) Y187: MATα, ura3-52, his3-200, ade2-101, trp1-901, leu2-3,112, gal4, met, gal80, URA3::GAL1UASGAL1TATA-lacZ (Clontech) (Harper et al., 1993). Bait plasmids were electroporated into Y2HGold yeast and prey plasmids into Y187 yeast (Clontech). Preparation of electrocompetent yeasts and electroporation of yeasts were done as described previously (Magupalli et al., 2008). For identifying transformants, yeasts were plated on the respective selective plates to identify the resulting convertents to the respective prototrophy (dropout media; Clontech/ICN). For interaction analyses, Y2HGold yeasts containing the respective bait plasmid were mated with Y187 yeasts containing the respective prey plasmid. Mating was performed for 5 h at 30°C in 1 ml of YPD medium with heavy vortexing in a thermoshaker. For assessing mating efficiency, half of the mated sample was streaked on –LW plates, the other half was plated on –ALWH selective plates with 10 mm aminotriazole (3-amino 1,2,4-triazole) and 60 ng/ml aureobasidin. Growth of mated yeasts on –LW selective medium demonstrates presence of both bait and prey plasmids; growth on –ALWH selective medium [and expression of β-galactosidase (β-gal) activity] indicates interaction of bait and prey proteins taking place. For the matings, pSE1111 and pSE1112 that encode irrelevant proteins (Magupalli et al., 2008) as well as the empty bait and prey vectors were used as negative controls (auto-activation controls). Expression of β-gal marker gene activity was qualitatively analyzed by filter assays as described previously (Magupalli et al., 2008).
Fusion protein pull-down assay.
For fusion protein pull-down experiments, either GST-tagged or maltose-binding protein (MBP)-tagged fusion proteins were used as immobilized bait proteins. If GST-tagged proteins were used as immobilized bait protein, the MBP-tagged protein was used as solubilized prey protein and vice versa. Bait and prey proteins were used in equimolar amounts [0.3 μm in incubation buffer containing 100 mm Tris, pH 8.0, 150 mm NaCl, 1 mm EDTA, 0.25% (w/v) Triton X-100 (Tx-100), and 1 mm β-mercaptoethanol (βME) if not denoted otherwise]. GST and MBP alone served as control proteins. Protein concentrations were determined using the Bradford method (Bradford, 1976). For pull-down experiments, fusion protein eluates were precleared with 10 μl of empty Sepharose beads (per 1 ml of eluate) for 1 h at 4°C. Incubations were typically done in a volume of 500 μl. After overnight incubation at 4°C, immobilized beads were allowed to settle (20 min, at 4°C). Samples were washed by repeated centrifugation of the beads (3000 rpm, 2 min, 4°C) and subsequent resuspension with binding buffer. This procedure was repeated three times. Afterward, the final pellets were boiled with SDS-sample buffer (96°C, 10 min) and subjected to SDS-PAGE and/or Western blotting.
Pre-absorption experiments.
Pre-absorption experiments were performed exactly as previously described (Wahl et al., 2013) using 50 μg of the respective GST-fusion protein. Pre-absorbed ArfGAP3Cterm2 and ArfGAP3Cterm3 immunosera were used at a 1:20 dilution for immunofluorescence microscopy and at a 1:3000 dilution for Western blotting.
Affinity purification of antibodies.
Antibodies were affinity purified by the method of Olmsted (1981). In brief, ∼50 μg of fusion protein was loaded on a 10% SDS-PAGE and transferred to nitrocellulose. The ArfGAP3Cterm-GST/ArfGAP3Cterm2 fusion protein bands at ∼35 kDa were cut out with a scalpel blade. These fusion protein-loaded nitrocellulose strips were used for affinity purification of the antisera and treated with 5% skim milk powder in PBS (for ∼30 min at RT). Next, nitrocellulose strips were incubated with the respective antisera (diluted 1:10 in 5% skim milk dissolved in PBS) and incubated overnight at 4°C. The nitrocellulose strips were washed several times with PBS. Bound antibodies were eluted from the nitrocellulose strips with a minimal volume (typically 200 μl) of 0.2 m glycine, pH 2.7, for 3–4 min (at 4°C). The antibody eluate was neutralized by the addition of 50 μl of 1 m Tris, pH 8.5. Antibody was diluted to a concentration of 0.1 mg/ml and complemented with BSA (0.1 mg/ml) for stabilization.
Direct labeling of primary antibodies (mouse anti-RIBEYE(B)-domain/CtBP2) with fluorophores (DyLight 650).
Direct labeling of mouse monoclonal antibodies against RIBEYE(B)/CtBP2 (BD) was performed exactly as previously described (Wahl et al., 2013) using the Thermo Scientific DyLight 650 Microscale Antibody Labeling Kit (Thermo Scientific #84536) according to the manufacturer's instructions.
Heterologous protein expression in eukaryotic cell lines.
For heterologous expression, COS7 cells were used that were transfected with lipofection, as previously described (Magupalli et al., 2008; Schwarz et al., 2011). Transfected cells were typically analyzed 24 h after transfection.
Coimmunoprecipitation from bovine retina.
For each immunoprecipitation, a single isolated bovine retina was incubated in 1 ml lysis buffer, containing 100 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm EDTA, and 1%Tx-100 for 45 min on vertical rotator at 4°C. The samples were mechanically cracked by forcefully ejecting the retinal lysates through a 20 gauge needle (20×). The samples were sonicated (Bandelin; Sonoplus) at 1% output for 20 half-second pulse ON/OFF cycles on ice. Afterward, the extracts were centrifuged at 13,000 rpm for 30 min at 4°C. The supernatants were transferred into new Eppendorf tubes. The centrifugation step was repeated one time to remove all cell debris. The resulting lysate was precleared by the addition of 15 μl of pre-immune serum and 20 μl of washed protein A-Sepharose beads (2 h incubation at 4°C with an overhead rotator). Next, samples were centrifuged at 13,000 rpm for 30 min at 4°C (Biofuge Fresco; Heraeus; ##3328 rotor). The supernatant was split in two equal volumes–for the control and experimental assays. For negative control immunoprecipitation, 15 μl of ArfGAP3-Cterm3 pre-immune serum was added; for the experimental immunoprecipitation, 15 μl of ArfGAP3-Cterm3 immune serum was added. Samples were incubated overnight at 4°C using an overhead rotator. Afterward, beads were allowed to settle (for 20 min, on ice). The supernatants were removed and saved; the bead pellets were resuspended in 1.0 ml of lysis buffer and washed thrice by repeated centrifugation (3000 rpm, 1 min, 4°C). The final pellet was boiled in 10 μl of SDS sample buffer, subjected to SDS-PAGE, and analyzed by Western blotting with the indicated antibodies.
Immunofluorescence microscopy of transfected COS cells.
Conventional fluorescence microscopy of transfected cells was done as previously described using a Zeiss Axiovert 200M equipped with the respective filter blocks (Schmitz et al., 2000; Magupalli et al., 2008; Wahl et al., 2013).
Immunolabeling of 0.5-μm-thick retinal resin sections.
Epon-embedded samples of mouse and bovine retinas were prepared as previously described (Wahl et al., 2013). From the tissue blocks, 0.5-μm-thick sections were cut with a Reichert ultramicrotome. Epon was removed as described previously (Wahl et al., 2013). Afterward, sections were incubated with the respective primary and secondary antibodies as described previously (Schmitz et al., 2000; Alpadi et al., 2008; Wahl et al., 2013). Immunolabeled sections were either analyzed by conventional epifluorescence microscopy or by super-resolution structured illumination microscopy (SR-SIM) as indicated. The immunofluorescence data shown in Figures 9–13 were obtained from incubations of mouse retinal sections. Qualitatively similar images were obtained from incubations of thin sections of the bovine retina (data not shown).
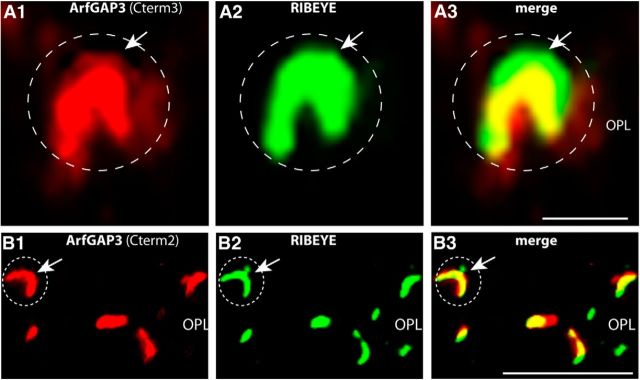
Figure 9.
ArfGAP3 is strongly enriched at synaptic ribbons of photoreceptor synapses in situ (conventional imaging). ArfGAP3 colocalizes with synaptic ribbons. The 0.5-μm-thick retinal sections are double immunolabeled with antibodies against ArfGAP3 and monoclonal antibodies against RIBEYE(B)/CtBP2 (A, B). ArfGAP3 Cterm3-antibody was used in A, and ArfGAP3 Cterm2 was used for immunolabeling of ArfGAP3 in B. Strong ArfGAP3 immunosignals were found in an identical manner with both ArfGAP3 antibodies at the RIBEYE-immunolabeled synaptic ribbons and in close vicinity to synaptic ribbons. The dashed circles denote single immunolabeled photoreceptor presynaptic terminals. Arrows in B and C point to single immunolabeled synaptic ribbons. No immunosignals were observed in the presynaptic terminals if pre-immune serum was used (data not shown). A and B were obtained by conventional imaging. OPL, Outer plexiform layer. Scale bars: A, 1 μm; B, 5 μm.
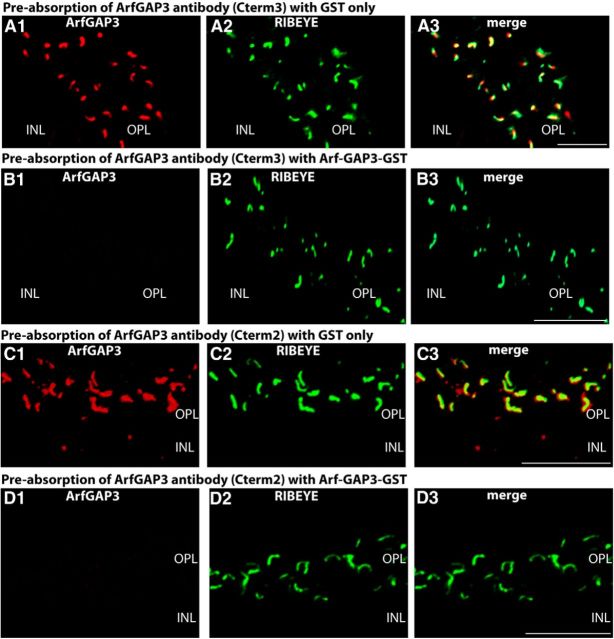
Figure 10.
A, B, Pre-absorption control experiments for the immunolabeling analyses shown in Figure 9A. Double immunolabeling of 0.5-μm-thick mouse retinal sections with ArfGAP3 (Cterm3) antibody pre-absorbed either with ArfGAP3-GST-fusion protein (B) or with GST alone (A). To visualize ribbon synapses, sections were coimmunolabeled with mouse monoclonal antibodies against RIBEYE(B)-domain/CtBP2. Pre-absorption with ArfGAP3Cterm3-GST fusion protein completely blocked the ArfGAP3 immunosignals at the synaptic ribbon (B), whereas GST alone had no influence on the ArfGAP3 immunosignals (A), demonstrating the specificity of the previous immunolabeling results. C, D, Pre-absorption control experiments for the immunolabeling analyses shown in Figure 9B. Double immunolabeling of 0.5-μm-thick mouse retinal sections with ArfGAP3 (Cterm2) antibody pre-absorbed with either ArfGAP3Cterm2-GST-fusion protein (D) or with GST alone (C). Synaptic ribbons were coimmunolabeled with mouse monoclonal antibodies against RIBEYE(B)-domain/CtBP2. Pre-absorption with the specific fusion protein completely blocked the ArfGAP3 immunosignals at the synaptic ribbon (D), whereas GST alone had no influence on the ArfGAP3 immunosignals (C), demonstrating the specificity of the previous immunolabeling results. A–D were obtained by conventional imaging. OPL, Outer plexiform layer; INL, inner nuclear layer. Scale bars: A–D, 5 μm.
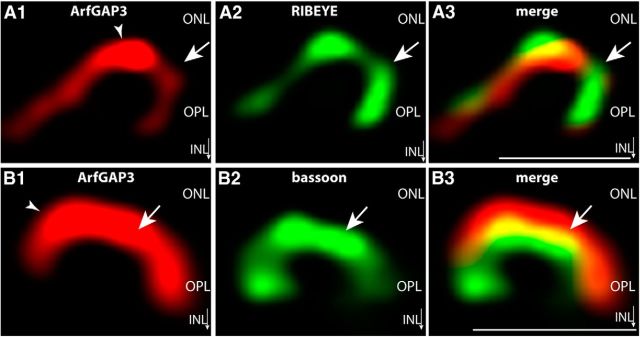
Figure 11.
ArfGAP3 is strongly enriched at the synaptic ribbon of photoreceptor synapses in situ (SR-SIM imaging with ArfGAP3 Cterm3 antibody). A, 0.5-μm-thick retinal sections double immunolabeled with affinity-purified rabbit antibodies against ArfGAP3 (ArfGAP3 Cterm3 antibody) and mouse monoclonal antibodies against RIBEYE(B)/CtBP2. B, Shows 0.5-μm-thick retinal sections double immunolabeled with affinity-purified rabbit antibodies against ArfGAP3 (Cterm3 antibody) and mouse monoclonal antibodies against the active zone protein bassoon. Arrows in A and B point to single immunolabeled synaptic ribbons. Arrowheads denote ArfGAP3 immunoreactivity at the synaptic ribbon. A and B were obtained by SR-SIM imaging. ONL, Outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer. Scale bars: A, B, 1 μm.
Figure 12.
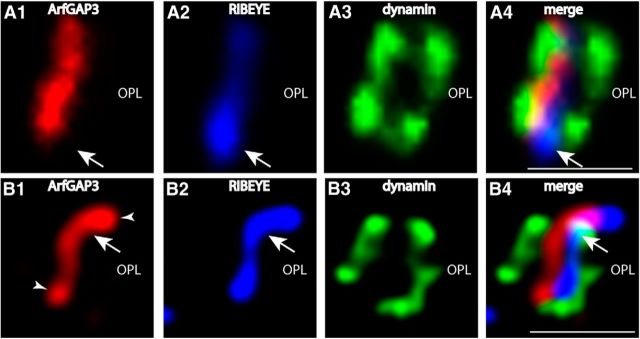
Localization of ArfGAP3 in the presynaptic photoreceptor terminal in relation to other presynaptic proteins. The 0.5-μm-thick retinal sections were triple immunolabeled with affinity-purified rabbit polyclonal antibodies against ArfGAP3 (Cterm3), mouse monoclonal antibodies against dynamin (hudy-1), and DyLight 650 directly labeled primary antibodies against RIBEYE(B)/CtBP2. ArfGAP3 and RIBEYE are located very close to each other (A, B). The ArfGAP3 immunosignal is located within the ring-like dynamin immunosignal that demarcates the presynaptic plasma membrane of the periactive zone that surrounds the synaptic ribbon (Wahl et al., 2013). A was obtained by conventional imaging; B is a micrograph obtained by SR-SIM imaging. Arrows in A and B point to immunolabeled single synaptic ribbons. Arrowheads indicate ArfGAP3 immunoreactivity at the synaptic ribbon. OPL, Outer plexiform layer. Scale bars: A, B, 1 μm.
Figure 13.
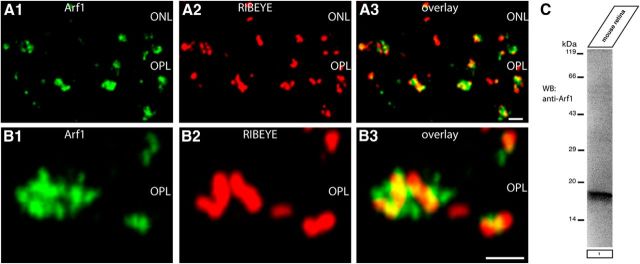
The ArfGAP3 effector Arf1 is enriched at the synaptic ribbon complex of photoreceptor synapses. A, B, Double immunolabeling experiments of 0.5-μm-thick retinal sections with mouse monoclonal antibodies against Arf1 and rabbit polyclonal antibodies against RIBEYE (U2656) demonstrated a close enrichment of Arf1 around the synaptic ribbon complex. ONL, Outer nuclear layer; OPL, outer plexiform layer. Scale bars: A, B, 1 μm. (C) Western blot analyses demonstrated that Arf1 is strongly expressed in the mouse retina.
SR-SIM.
To further improve the spatial resolution of our immunolabeling analyses, we applied multicolor SR-SIM analyses (Schermelleh et al., 2008, 2010; Wahl et al., 2013). SR-SIM was performed exactly as previously described using the Elyra PS1 setup (Wahl et al., 2013). Images were taken with a 63× Plan-Apochromat objective (NA 1.4) with excitation light wavelengths of 488, 561, and 650 nm, and then processed for SR-SIM to obtain higher resolutions (Gustafsson et al., 2008; for review, see Schermelleh et al., 2008, 2010), as previously described (Wahl et al., 2013). The Zeiss setup used for SR-SIM was checked for chromatic aberration in X-,Y-, and Z-direction using multicolor beads. For acquisition and processing, the Zen2010 software (Zeiss) was used.
Triple immunolabeling of thin retinal sections.
Triple immunolabeling experiments were performed as previously described (Wahl et al., 2013). We used a directly labeled mouse monoclonal antibody against RIBEYE(B)-domain/CtBP2 (BD) conjugated with DyLight 650, and two other primary antibodies (one from mouse, the second from rabbit [as indicated in the respective experiments]), which were not directly fluorophore labeled. First, sections were incubated with the two unlabeled primary antibodies at the same time overnight (at the dilutions given above). On the next day, sections were washed three times with PBS and afterward incubated with the respective secondary antibodies (donkey anti-rabbit Alexa 568 and chicken anti-mouse Alexa 488). After 1 h incubation, sections were washed again three times with PBS and finally incubated with the directly DyLight 650-labeled CtBP2 primary antibody (in the dilutions summarized above) overnight at 4°C. After overnight incubation, sections were washed three times with PBS and embedded with anti-fade solution containing n-propyl gallate (NPG) as previously described (Schmitz et al., 2000).
Control incubation.
Control incubations for immunolabeling experiments were done by omitting the primary antibody and only incubating with secondary antibody. No immunofluorescent signal was observed in photoreceptor synapses in these control incubations. In further control experiments, antibodies were pre-absorbed with the respective antigen as described below and processed for immunolabeling.
All experiments were done with mouse and bovine retinas of either sexes. Mice were killed in the early afternoon. Mouse eyes were collected at environmental daylight conditions (luminance of ∼2 cd/m2). Bovine eyes were obtained from a local slaughterhouse. Similar data as shown for the mouse retina were also obtained with the bovine retina (of either sexes; data not shown).
Preparation of retinas from the adult mouse for electroporation.
Retinas were isolated from adult mice within 5 min postmortem (in dim ambient light). The enucleated eyes were bisected at the equatorial plane and the posterior eye cup was transferred into ice-cold artificial CSF (ACSF) containing the following (in mm): 119 NaCl, 2.5 KCL, 2.5 CaCl2, 1.3 MgCl2, 1 NaH2PO4, 20 glucose, and 11 HEPES, pH 7.4, osmolarity ∼300 mOsm/L. ACSF was saturated with 5% CO2/95% O2 (carbogen) before use. From the posterior eyecup, the retina was gently peeled off from the pigment epithelium. Isolated retinas were transferred to black-gridded nitrocellulose filter membranes (Millipore, #HABG01300) with the ganglion cell side facing the nitrocellulose membrane. Thus, photoreceptors were facing the free surface and were in direct contact with the DNA plasmid solutions added to them in the electroporation experiments (see below). The filters with the attached retinas were transferred to sterile Petri dishes (3 cm diameter) containing ∼1.0 ml of AMES' medium, pH 7.4 (A1420, Sigma-Aldrich) pre-incubated at 37°C.
Electroporation of isolated mouse retinas and retinal explant culture.
Electroporation of adult mouse retinas was performed largely as previously described (Donovan and Dyer, 2006; Briggman and Euler, 2011; Vergara et al., 2013). Electroporation was performed with a square wave pulse electroporator (ECM 830; BTX) and a tweezertrode electroporation device (BTX; #45-0118, #45-0204). Both electrodes of tweezertrode were dipped in ACSF buffer to obtain a good electrical connection. Excess of ACSF was removed with filter paper to avoid dilution of the DNA. Before electroporation, DNA was column purified. For each electroporation, 50 μg of purified DNA was used. Immediately after isolation, the retina was placed in between the tweezertrode. The retina attached to the nitrocellulose was facing the positive electrode with the ganglion cell side while the plasmid DNA solution was added to the photoreceptor side at the negative electrode in hanging drop manner. The distance between the two electrodes was optimized to ∼3–4 mm. Electroporation was done at 20 V, 950 ms OFF, 20 ms ON (10 pulses). Electroporated retinas were transferred back to Petri dishes containing 1.0 ml of AMES' medium, pH 7.4. Retinas were transferred to open Petri dishes that were placed in an incubation chamber, as previously described (Morgan et al., 2011; Williams et al., 2013). Temperature was continuously maintained at 32°C using a feedback temperature controller (TC324B; Warner Instruments) and a transparent ITO heater attached to the incubation chamber (HI-25Dp; MicroControls). The incubation chamber was filled with distilled H2O to the lower edge of stage where the Petri dish with the retina was placed and continuously gassed with carbogen (5% CO2/95% O2). Retinas were incubated inside the light-protected incubation chamber typically for 18–24 h.
Loading of photoreceptors with FM1–43.
Isolated, electroporated retinas were incubated for 15 min in LCS containing 20 μm FM1–43 (fixable FM1–43: FM1–43FX; Invitrogen, #F35355) at 37°C in the dark, similar to the procedure described by Rea et al. (2004). We used this method, because it favors specific FM1–43 uptake in photoreceptor synaptic terminals (Rea et al., 2004). After labeling, the retinas were rinsed thrice with LCS and processed for the isolation of photoreceptor cells (see below).
Dissociation of electroporated retinas and isolation of photoreceptors.
Twenty-four hours after electroporation, photoreceptors were isolated from the retina with a papain digestion procedure, largely as previously described (Wahl et al., 2013). The papain solution containing 6 U/ml papain (Sigma, #76220-25G) in low Ca2+-containing saline solution (LCS solution; containing 132 mm NaCl, 3 mm KCl, 1 mm MgCl2 × 6H2O, 0.5 mm CaCl2, 10 mm sodium pyruvate, 10 mm glucose, and 10 mm HEPES, pH 7.4, ∼300 mOsm/L) was activated with 2.7 mm l-cysteine at 37°C for 20 min before the experiments. Isolated retinas were incubated in 1 ml of the cysteine-activated papain solution (containing 6 U papain/ml LCS) for 10 min at 25°C. LCS was saturated with 5% CO2/95% O2 before use. After removal of the papain solution, the retina was gently washed three times with 1ml of LCS solution containing 2% FCS and 0.01 mg/ml DNase (Sigma, #DN25-110MG). To dissociate photoreceptor cells, papain-treated retinas were very gently triturated (1–2 times) with a wide-bore plastic Pasteur pipette. The resulting cell suspension was plated on Concanavalin A (Sigma, #C7275-250 mg)-coated coverslips. For coating of a 25 mm round coverslip, ∼200 μl of 1 mg/ml Concanavalin A (in LCS solution) was added for 1 h at RT. Unbound Concanavalin A was removed by three washes with LCS before the addition of the dissociated cells. Cells were allowed to settle on the coverslips for 30 min at 37°C for tight attachment. Unbound retinal cells were removed by gentle washes with LCS. Photoreceptors were identified based on their typical morphology. Photoreceptors were fixed with 4% PFA in PBS for 15 min at RT. After several washes with PBS, cells were mounted with NPG antifade as previously described (Wahl et al., 2013).
Miscellaneous procedures.
SDS-PAGE and Western blotting were performed as previously described (Schmitz et al., 2000). Fusion proteins were expressed in BL21(DE3) bacteria for pGEX and pMal-C2 constructs as previously described (Schmitz et al., 2000). For expression of SNAP-tagged fusion proteins, Escherichia coli T7 Express bacteria (NEB; #C2566) were used. Expression and purification of SNAP-tagged fusion proteins were performed according to the manufacturer's instructions (NEB). SNAP-tagged fusion proteins were visualized SNAP-Vista Green (NEB; #S9147S) and covalently immobilized with SNAP-capture pull-down resin according to the manufacturer's instructions. Conventional immunofluorescence microscopy was performed as previously described (Schmitz et al., 2000) using a Zeiss Axiovert 200M equipped with the respective filter blocks.
Results
ArfGAP3 binds to RIBEYE(B)-domain in the YTH system
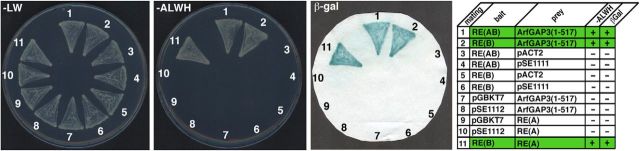
Using RIBEYE(B) as bait, we obtained full-length ArfGAP3 as a RIBEYE-interacting YTH prey clone. The original ArfGAP3 prey clone contained a deletion of 8 aa in the Zn-finger motif of the ArfGAP-domain (data not shown). To exclude an artificial interaction of these proteins in the YTH system, we recloned full-length ArfGAP3 cDNA from an IMAGE clone (IMAGE #8081904, BC118087; encoding full-length bovine ArfGAP3 with no deletion in the Zn-finger) into the pACT2 prey vector. Then, we retested whether this entirely full-length ArfGAP3 prey interacts with RIBEYE(B) in the YTH system. Similar as observed for the original ArfGAP3 prey clone, we obtained a strong interaction between RIBEYE(B) and the recloned full-length ArfGAP3, as judged by growth on –ALWH selective medium and expression of β-gal activity (Fig. 1, matings 1 and 2) indicating that the interaction between ArfGAP3 and RIBEYE(B) is real in the YTH system. Negative control matings (Fig. 1, matings 3–10) demonstrated that ArfGAP3/RIBEYE interaction is not due to auto activation.
Figure 1.
RIBEYE(B) and RIBEYE(AB) interact with ArfGAP3 in the YTH system. Summary plates of YTH analyses obtained with the indicated bait and prey plasmids. For convenience, experimental bait–prey pairs are underlayered in color (green in case of interacting bait–prey pairs; control matings are noncolored). RIBEYE(B) and also full-length RIBEYE [RIBEYE(AB)] interact with ArfGAP3 in the YTH system (matings 1 and 2). Mating 11 denotes an unrelated positive control (Magupalli et al., 2008). pSE1111 is an irrelevant prey vector and pSE1112 is an irrelevant bait vector (Tai et al., 1999; Magupalli et al., 2008). Negative control matings of the ArfGAP3 prey clone with empty bait clones (mating 7) or irrelevant bait clones (mating 8) demonstrate that the ArfGAP3 clone is not auto activating. The other matings represent negative control matings for the RIBEYE bait clones (matings 3–6) or RIBEYE prey clones (matings 9 and 10), demonstrating that these constructs are also not auto activating in the YTH system. RE(AB), full-length RIBEYE, containing both RIBEYE(A)- and RIBEYE(B)-domain; RE(A), RIBEYE(A)-domain; RE(B), RIBEYE(B)-domain; AGD, ArfGAP-domain of ArfGAP3.
Mapping of RIBEYE-ArfGAP3 interaction in the YTH system: the NAD(H)-binding subdomain RIBEYE(B) binds to the AGD-domain of ArfGAP3
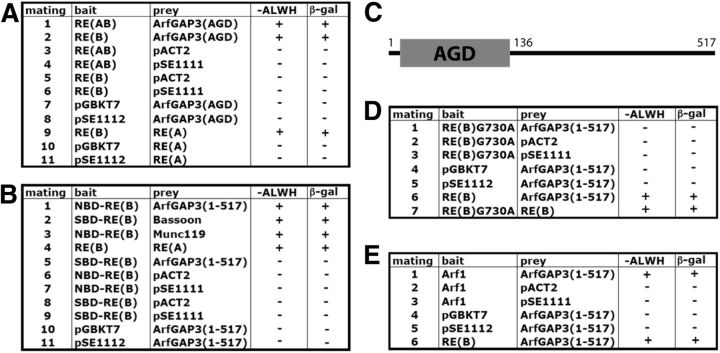
Next, we tested with the YTH system which part of ArfGAP3 mediates the interaction with RIBEYE(B). We found that the AGD encoding amino acids 1–136 of ArfGAP3 is responsible for the interaction with RIBEYE(B) (Fig. 2A, matings 1 and 2). The C terminus of ArfGAP3 did not interact with RIBEYE(B) (data not shown). RIBEYE(B) consists of an NADH-binding subdomain (NBD) and a substrate-binding subdomain (SBD; Kumar et al., 2002; Nardini et al., 2003; for review, see Schmitz, 2009; Schmitz et al., 2012). Further YTH analyses demonstrated that the NBD of RIBEYE(B) interacts with ArfGAP3 (Fig. 2B, mating 1). The SBD did not promote ArfGAP3/RIBEYE(B) interaction in the YTH system (Fig. 2B, mating 5). Negative control matings (Fig. 2A, matings 3–8, 10, and 11; B, matings 6–11) demonstrated that bait and prey clones were not auto activating.
Figure 2.
Summary tables of YTH matings. A, RIBEYE interacts with the ArfGAP-domain (AGD) of ArfGAP3. RIBEYE(B) and also full-length RIBEYE [RIBEYE(AB)] interact with the ArfGAP-domain (AGD) of ArfGAP3 in the YTH system (matings 1 and 2). Mating 9 denotes an unrelated positive control (Magupalli et al., 2008). The other matings are auto-activation controls. None of the yeast constructs is auto activating. B, The NAD(H)-binding subdomain of RIBEYE(B) interacts with ArfGAP3. The NAD(H)-binding subdomain of RIBEYE(B), the NBD, interacts with ArfGAP3 (mating 1), but not the substrate-binding subdomain of RIBEYE(B), the SBD (mating 5). Matings 2–4 are positive control matings–(Bassoon for SBD-RE(B); tom Dieck et al., 2005), Munc119 for NBD-RE(B) (Alpadi et al., 2008), RE(A) for RE(B) (Magupalli et al., 2008)–and matings 6–11 are negative controls (auto-activation controls). C, Schematic domain structures of ArfGAP3. D, An NAD(H)-binding-deficient mutant of RIBEYE(B), RE(B)G730A, does not interact with ArfGAP3 (mating 1), while wild-type RIBEYE does (mating 6). Mating 7 is a positive control mating for RE(B)G730A. E, Arf1 interacts with ArfGAP3 (mating 1). All constructs are non-auto activating as demonstrated by the negative control matings (2–5); mating 6 is a positive control mating. RE(A), RIBEYE(A)-domain; RE(B), RIBEYE(B)-domain; RE(B)G730A, RIBEYE(B)G730A; GAP-dom, GAP-domain (AGD) of ArfGAP3; NBD, NAD(H)-binding subdomain of RIBEYE(B); SBD, substrate-binding subdomain of RIBEYE(B).
RIBEYE(B) interacts with ArfGAP3 in fusion protein pull-down assays
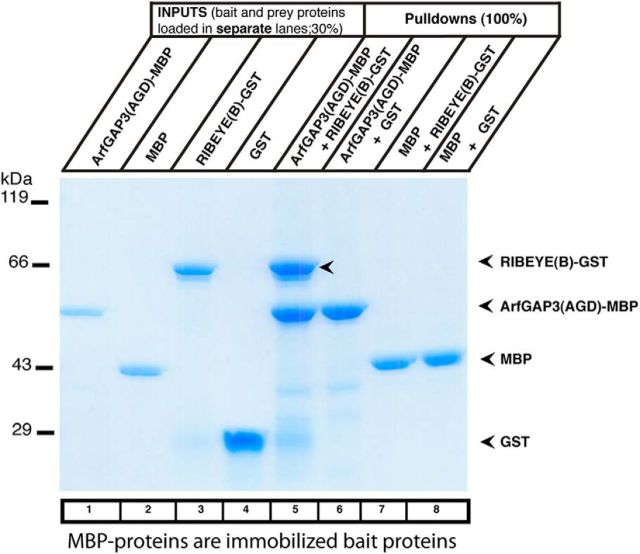
We used various independent approaches to verify the interaction between RIBEYE(B)-domain and the ArfGAP-domain (AGD) of ArfGAP3. First, we performed pull-down experiments using bacterially expressed and purified fusion protein (Fig. 3). We used MBP-tagged fusion proteins (ArfGAP3(AGD)-MBP and MBP alone) as immobilized bait proteins and GST-tagged proteins (RIBEYE(B)-GST and GST alone) as soluble prey proteins. For the pull-down experiments demonstrated in Figures 3 and 4, the buffer contained 1 mm βME (see Materials and Methods). ArfGAP3(AGD)-MBP (lane 5; but not MBP alone, lane 7) interacted with RIBEYE(B)-GST (but not with GST alone, lanes 6 and 8) as judged by protein pull-down analyses in SDS-PAGE (Fig. 3). Specificity of interaction was consistently shown both by SDS-PAGE (Fig. 3) and by Western blot analyses (Fig. 4; and data not shown). Identical results were obtained when tags were switched. In Figure 4, RIBEYE(B) was tagged with MBP and ArfGAP3(AGD) with GST. Also with these switched tags, a strong interaction was observed between RIBEYE(B) and ArfGAP3. Typically, >∼30% of the input fraction of the RIBEYE(B) prey protein was bound to the immobilized ArfGAP3 bait fusion protein.
Figure 3.
RIBEYE(B) specifically interacts with ArfGAP3 in fusion protein pull-down assays (SDS-PAGE analyses). Pull-down analyses of RIBEYE(B)/ArfGAP3 complexes were analyzed by Coomassie blue-stained polyacrylamide gel after SDS-PAGE. Lanes 1–4 show the indicated purified fusion proteins (input fractions). All input lanes represent 30% of the input fraction. Input proteins were loaded in separate lanes to demonstrate that the input fusion proteins display only a single, main protein band. In lanes 5–8, 100% of the pull-down reactions were loaded. MBP-tagged fusion proteins were used as immobilized bait proteins and GST-tagged proteins as soluble prey proteins. Only ArfGAP3-MBP pulled down RE(B)-GST (lane 5, arrowhead) but not MBP alone (lane 7). Neither MBP alone nor ArfGAP3-MBP pulled down GST alone (lanes 6 and 8). SDS-PAGE demonstrated that ArfGAP3-MBP specifically pulled down RIBEYE(B)-GST, demonstrating interaction of the two proteins in this assay system.
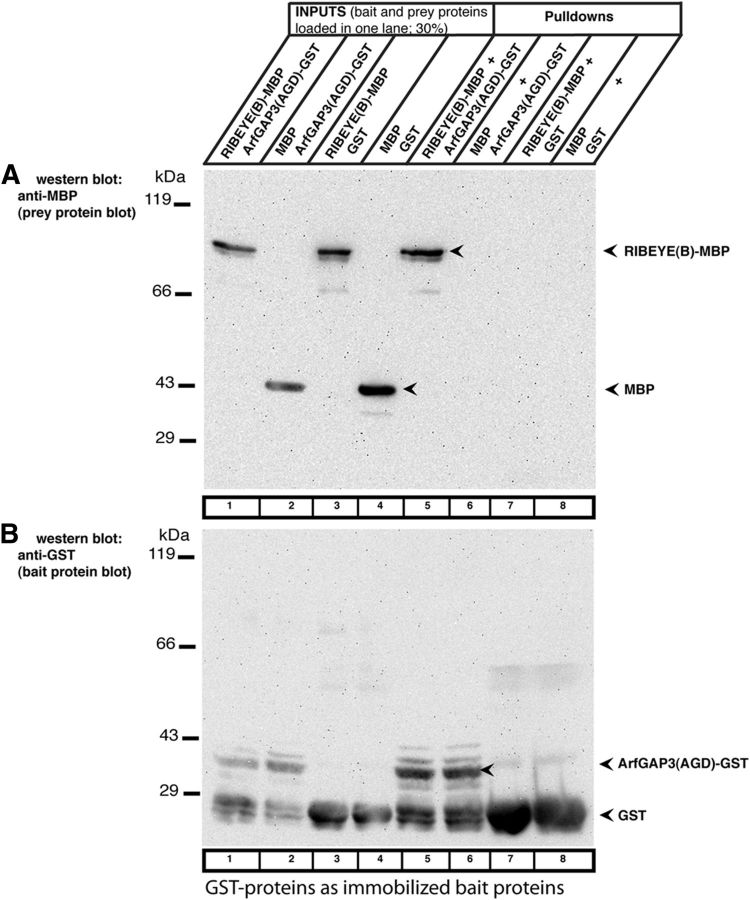
Figure 4.
RIBEYE(B) specifically interacts with ArfGAP3 in fusion protein pull-down assays (Western blot analyses). To exclude that the tag has an importance for the pull-down results and to further exclude that any prey protein is unspecifically pulled down by bait-GST, we also analyzed the results of the pull-down assays by Western blotting with anti-MBP and anti-GST antibodies. The reaction buffer used for these experiments contained 1 mm βME. GST-tagged fusion proteins were used as immobilized bait proteins and eluted MBP-tagged proteins as soluble prey proteins. Similar to the experiments described in Figure 3, only RIBEYE(B)-MBP (lane 5) and not MBP alone (lane 6) is pulled down by the ArfGAP3(AGD)-GST. GST alone (lane 7) and MBP alone (lane 8) do not pull down RIBEYE(B)-MBP, as shown by Western blotting with antibodies against MBP (Fig. 4A), demonstrating the specificity of the interaction. The Western blot data fully confirm the results shown in Figure 3 that were obtained by SDS-PAGE analyses. In Figure 4B, the same blot as analyzed in Figure 4A was reprobed (after stripping of the blot) with antibodies against GST to show equal loading of the bait proteins. RE(B)-MBP, RIBEYE(B)-MBP; ArfGAP3(AGD)-MBP, MBP-tagged ArfGAP-domain (AGD) of ArfGAP3.
ArfGAP3 interacts with RIBEYE(B) in transfected COS cells
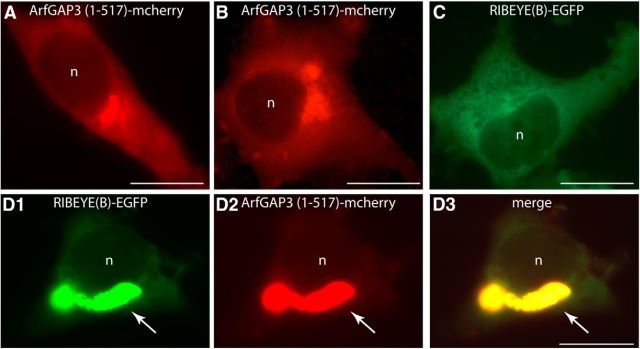
Next, we tested whether RIBEYE(B) and ArfGAP3 would interact with each other in transfected COS cells (Fig. 5). ArfGAP3 and RIBEYE(B) were tagged with different fluorescent proteins, i.e., ArfGAP3 with mCherry and RIBEYE(B) with EGFP. When ArfGAP3 was transfected alone, ArfGAP3 was enriched at a perinuclear region that represents the Golgi apparatus (Fig. 5A,B) similarly as previously described (Dogic et al., 1999; Eugster et al., 2000; Lewis et al., 2004; Watson et al., 2004; Frigerio et al., 2007; Kliouchnikov et al., 2009). When RIBEYE(B)-EGFP was transfected alone (Fig. 5C), it was largely diffusely distributed throughout the entire cell (as previously described; Schmitz et al., 2000). In contrast, when RIBEYE(B) was cotransfected with ArfGAP3, RIBEYE(B) was nearly completely recruited to the ArfGAP3-typical Golgi-like perinuclear localization (Fig. 5D) indicating interaction of RIBEYE(B) and ArfGAP3 in cotransfected COS cells.
Figure 5.
RIBEYE(B) is recruited by ArfGAP3 into a Golgi-like distribution in transfected COS cells. COS7 cells were transfected with the indicated mcherry-tagged ArfGAP3 or EGFP-tagged RIBEYE(B) constructs. Transfected cells were analyzed for the intracellular distribution of the respective proteins via direct epifluorescence microscopy. Cells transfected with ArfGAP3-mcherry alone show the typical enrichment at the Golgi apparatus in a perinuclear localization (A, B), as already previously shown (Dogic et al., 1999; Eugster et al., 2000; Lewis et al., 2004; Watson et al., 2004; Frigerio et al., 2007; Kartberg et al., 2010; Yu et al., 2012). In contrast, RIBEYE(B) is diffusely distributed in single-transfected cells (C; Schmitz et al., 2000). If RIBEYE(B)-EGFP is cotransfected with ArfGAP3-mcherry, RIBEYE(B) virtually completely redistributed from a diffuse distribution into the Golgi-like, perinuclear localization indicating interaction between RIBEYE(B) and ArfGAP3 (D). n, nucleus. The arrow in D points to the Golgi-like localization to which the RIBEYE(B)-EGFP signal is translocated in ArfGAP3-mcherry-transfected cells. Scale bars: A–D, 10 μm.
Interaction between ArfGAP3(AGD) and RIBEYE(B) is regulated by NAD(H) in a redox-sensitive manner
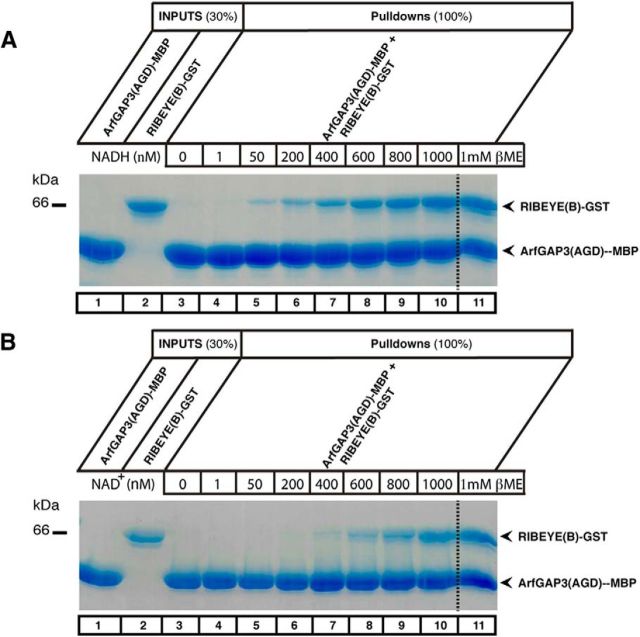
The fusion protein pull-down binding experiments shown above in Figures 3 and 4 were done with buffer that contained 1 mm βME. If βME was omitted, there was no binding between ArfGAP3 and RIBEYE(B) (Fig. 6A,B, compare lane 3 with lane 11). Previously, it has been shown that 1 mm βME promotes a conformation of RIBEYE(B) that can be also induced by the addition of NAD(H) (Venkatesan et al., 2010). Furthermore, we demonstrated that the NAD(H)-binding subdomain of RIBEYE(B), the NBD, is responsible for the interaction with ArfGAP3 (see above). Therefore, we tested whether the addition of NAD(H) could replace βME in promoting ArfGAP3/RIBEYE(B) interaction. Indeed, NAD(H) was very efficient in promoting RIBEYE(B)/ArfGAP3 interaction (Fig. 6A). The reduced form, NADH, was more efficient in promoting RIBEYE(B)/ArfGAP3 interaction in the absence of βME than its oxidized form, NAD+ (Fig. 6A,B; Kd ∼450 nm for NADH; Kd ∼700 nm for NAD+). From these experiments we concluded that NAD(H), particularly the reduced NADH, promotes RIBEYE(B)/ArfGAP3 interaction. In line with this proposal is our finding that an NAD(H)-binding-deficient RIBEYE(B) point mutant, RIBEYE(B)G730A (Alpadi et al., 2008; Schwarz et al., 2011), did not interact with ArfGAP3(AGD) in the YTH system (Fig. 2D, mating 1).
Figure 6.
The binding between RIBEYE(B)-domain and the ArfGAP-domain (AGD) of ArfGAP3 is stimulated by NAD(H) in a redox-sensitive manner. A, B, Pull-down experiments were performed as in Figure 3 both in the absence (lanes 3—10; A, B) or presence (lane 11; A, B) of 1 mm βME. RIBEYE binds to the ArfGAP-domain (AGD) of ArfGAP3 only in the presence of 1 mm βME (lane 11; A, B) but not in the absence of 1 mm βME (lane 3; A, B). Lanes 1 and 2 show the respective input fractions (30% input). We tested whether addition of NADH (A) or NAD+ (B) could substitute for the presence of 1 mm βME in promoting ArfGAP3/RIBEYE(B) interaction. As a matter of fact, increasing concentrations of both NADH (A) as well as NAD+ (B) could promote binding of RIBEYE(B)-GST to the AGD of ArfGAP3 even in the absence of 1 mm βME (lanes 4—10; A, B). The reduced form of the dinucleotide, NADH (A), was more effective than the oxidized form, NAD+ (B), to promote RIBEYE(B)/ArfGAP3 interaction. Semiquantitative evaluation of the binding experiments (n = 4) demonstrated that ∼450 nm NADH and ∼700 nm NAD+ are promoting half-maximal binding of ArfGAP3 to RIBEYE(B) in the pull-down assays.
ArfGAP3 can be coimmunoprecipitated with RIBEYE from the bovine retina
We generated two different polyclonal antibodies against ArfGAP3 to analyze the relation between RIBEYE and ArfGAP3 in situ. The two different antibodies against ArfGAP3 were directed against two different portions in the divergent C terminus of ArfGAP3 (ArfGAP Cterm2 and ArfGAP Cterm3; see Fig. 7A). Both antibodies immunodetected a typical single, major band at ∼55 kDa in crude bovine retinal homogenates in Western blot analyses (Fig. 7; data not shown) that was absent in the pre-immune serum and that could be specifically blocked by the respective ArfGAP3-GST fusion protein but not by GST alone (Fig. 7C; data not shown).
Next, we tested whether antibodies against ArfGAP3 coimmunoprecipitated endogenous retinal RIBEYE. ArfGAP3 (Cterm3) immune serum (lane 3, but not ArfGAP3 pre-immune serum, lane 2) coimmunoprecipitated RIBEYE (Fig. 8A) together with ArfGAP3 (Fig. 8B), showing interaction of these proteins also in the retina in situ (Fig. 8). Both RIBEYE and ArfGAP3 were strongly enriched in the experimental (Fig. 8, lane 3) but not in the control (Fig. 8, lane 2) immunoprecipitates. Since RIBEYE is exclusively present at synaptic ribbons in the mature retina (Schmitz et al., 2000), the coimmunoprecipitation experiments suggested that ArfGAP3 is a component of the synaptic ribbon complex in situ.
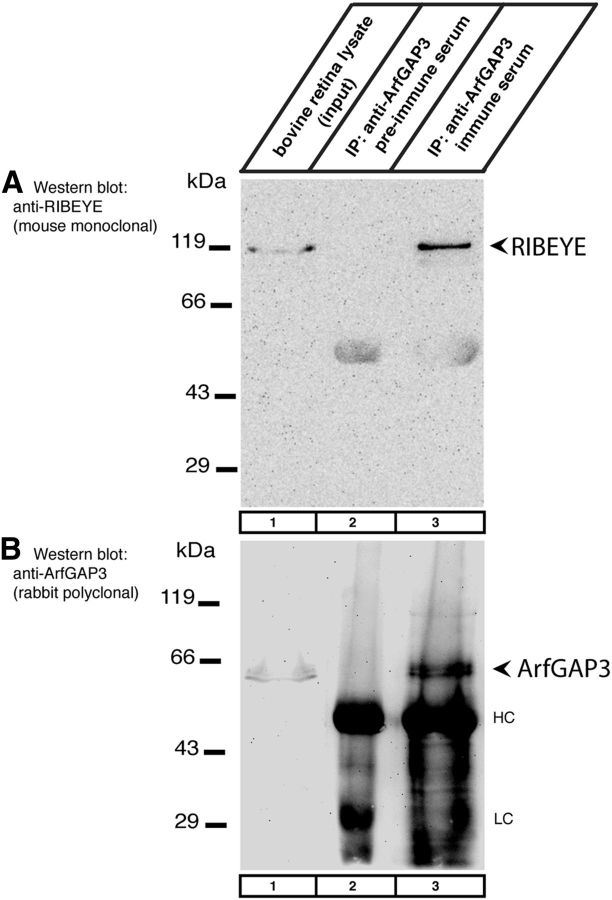
Figure 8.
Coimmunoprecipitation of RIBEYE and ArfGAP3 from the bovine retina (Western blot analyses). A, ArfGAP3 immune serum (lane 3) and ArfGAP3 pre-immune serum (lane 2) were tested for their capability to coimmunoprecipitate RIBEYE from the bovine retina. Lane 1 shows the input fraction (1% of total input). RIBEYE is coimmunoprecipitated by ArfGAP3 immune serum (Cterm3 antiserum; lane 3) but not by ArfGAP3 pre-immune serum (lane 2). B, Shows the same blot as in A but reprobed with anti-ArfGAP3 antibodies (after stripping of the blot). This blot shows that ArfGAP3 was successfully immunoprecipitated by the immune serum (lane 3) but not by the control pre-immune serum (lane 2). HC and LC indicate the Ig heavy and light chains, respectively.
ArfGAP3 is present at the photoreceptor synaptic ribbon complex
To get further insights about the in situ localization of ArfGAP3 in the retina, we performed immunolabeling analyses with both ArfGAP3 antibodies (ArfGAP3 Cterm3 and ArfGAP Cterm2; Fig. 9). Both ArfGAP3 antibodies strongly labeled the outer plexiform layer (OPL) where photoreceptor ribbon synapses are localized (Fig. 9; data not shown). Identical results were obtained independent of whether crude antisera or affinity-purified antibodies were used (compare Figs. 9, 11, 12). To define the relation between ArfGAP3 and synaptic ribbons, we performed double immunolabelings with rabbit polyclonal antibodies against ArfGAP3 and mouse monoclonal antibodies against RIBEYE(B)-domain/CtBP2 (Fig. 9). The ArfGAP3 immunosignals largely overlapped at the photoreceptor ribbon synapse with the RIBEYE immunosignals indicating that ArfGAP3 is strongly enriched at the synaptic ribbon complex (Fig. 9A,B). Identical immunolabeling results were obtained with both ArfGAP3 antibodies (Fig. 9), further demonstrating that the immunoreactivity at the RIBEYE-immunolabeled synaptic ribbon is due to ArfGAP3 and not due to an unrelated protein. The described ArfGAP3 immunosignals obtained with both ArfGAP3 antibodies could be specifically blocked by pre-absorption with the respective ArfGAP3-GST fusion protein but not by GST alone, demonstrating the specificity of the immunolabeling results (Fig. 10). To further corroborate these findings, we performed SR-SIM of thin retinal sections double immunolabeled with affinity-purified rabbit polyclonal antibodies against ArfGAP3 and mouse monoclonal antibodies against RIBEYE(B)/CtBP2 (Fig. 11). Also, SR-SIM analyses demonstrated a strong enrichment of the ArfGAP3 immunosignals at the synaptic ribbons (Fig. 11). In the SR-SIM analyses, the ArfGAP3 immunosignal was slightly shifted toward the inner nuclear layer (INL) compared with the RIBEYE immunosignal, which could indicate a localization of ArfGAP3 more toward the base of the synaptic ribbon (Fig. 11A; see also discussion). The ArfGAP3 immunosignals also showed a strong colocalization with the active zone protein bassoon (Fig. 11B). In the SR-SIM analyses, the ArfGAP3 immunosignal was slightly shifted toward the ONL compared with the bassoon immunosignal, also indicating an ArfGAP3 localization in the basal portions at the synaptic ribbon (Fig. 11B; see also discussion). Bassoon is localized to the arciform density and anchors the base of the synaptic ribbon (tom Dieck et al., 2005).
Previously, we have shown that dynamin is highly enriched at the presynaptic plasma membrane in close vicinity to the synaptic ribbon (Wahl et al., 2013). The dynamin immunosignal surrounds the synaptic ribbon in a ring-like manner in a very short distance (Wahl et al., 2013). Immunogold electron microscopy previously demonstrated that this ring of dynamin-1 immunoreactivity corresponded to the plasma membrane of the periactive zone (Wahl et al., 2013). We used dynamin as a landmark protein to further define the localization of ArfGAP3 at the synaptic ribbon complex using high-resolution fluorescence microscopy. We performed triple immunolabeling analyses with rabbit polyclonal antibodies against ArfGAP3, mouse monoclonal antibodies against dynamin (hudy-1), and Alexa 650 directly labeled primary mouse monoclonal antibodies against RIBEYE(B)-domain/CtBP2 (Fig. 12). In these triple immunolabeling analyses, the ArfGAP3 immunosignals were located very close to the RIBEYE-labeled synaptic ribbon and within the ring-like dynamin immunosignals that surrounded the synaptic ribbon (Fig. 12). Unfortunately, all the ArfGAP3 antibodies that we generated as well as various commercially available antibodies did not work at the ultrastructural level, so the ultrastructural localization remains to be elucidated by future examinations. ArfGAP3 is only weakly expressed in the inner plexiform layer (IPL) of the retina, which is probably due to the smaller size of the synaptic ribbons in bipolar cell terminals (data not shown).
RIBEYE(B) competes with Arf1 for binding to ArfGAP3
To get functional insights into the importance of the ArfGAP3/RIBEYE interaction we determined which Arf protein is interacting with ArfGAP3. In agreement with previous reports (Liu et al., 2001; Kartberg et al., 2010), we found that ArfGAP3 interacts with Arf1 (but not with Arf6; Fig. 2E, mating 1; data not shown). Western blot analyses demonstrated that Arf1 is strongly expressed in the retina and immunofluorescence microscopy documented enrichment of Arf1 in close vicinity to the synaptic ribbon complex (Fig. 13A–C).
Similar to RIBEYE(B), Arf1 also binds to the ArfGAP-domain of ArfGAP3 (data not shown). Therefore, we tested whether Arf1 and RIBEYE(B) can bind simultaneously to ArfGAP3 or whether they compete with each other in binding to ArfGAP3 (Fig. 14). To address this question, we used fusion protein pull-down experiments. We tested whether increasing concentrations of Arf1 (Fig. 14A) added to a fixed concentration of immobilized ArfGAP3 (0.15 μm) would inhibit binding of RIBEYE(B) to ArfGAP3. RIBEYE(B) was kept at a constant concentration in these experiments (0.15 μm). Similarly, we also tested whether increasing concentrations of RIBEYE(B) (Fig. 14B) added to a fixed concentration of immobilized ArfGAP3 (0.15 μm) would inhibit binding of Arf1 to ArfGAP3. Arf1 was kept at a constant concentration in these latter experiments (0.15 μm). In both sets of experiments, we observed a competitive behavior between Arf1 and RIBEYE(B) in binding to ArfGAP3: if Arf1 was increased, binding of RIBEYE(B) to ArfGAP3 was diminished and, vice versa, increasing concentrations of RIBEYE(B) inhibit binding of Arf1 to ArfGAP3. These data demonstrate that RIBEYE(B) competes with Arf1 for binding to a common binding site on ArfGAP3, indicating that binding of RIBEYE(B) and Arf1 to ArfGAP3 is mutually exclusive.
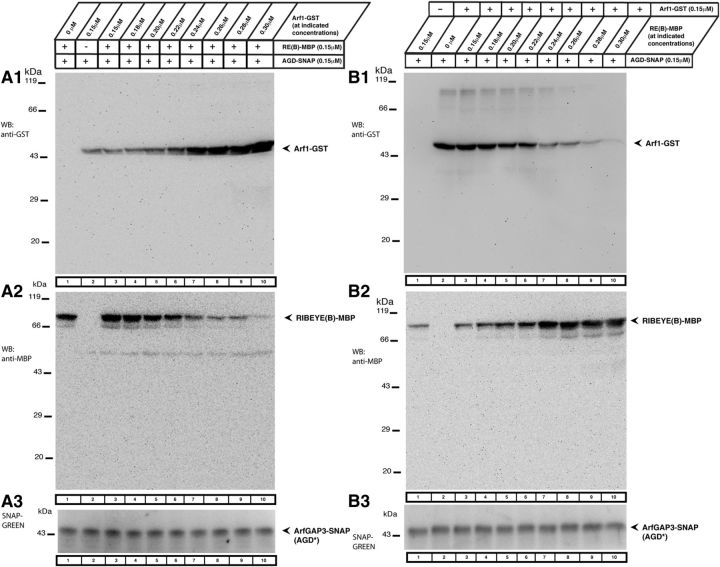
Figure 14.
RIBEYE(B) and Arf1 compete with each other for binding to ArfGAP3. We tested in fusion protein pull-down experiments whether Arf1 and RIBEYE(B) can bind simultaneously to ArfGAP3 or whether they compete with each other for ArfGAP3-binding. A, B, Show representative Westerns blots incubated with the indicated antibodies to test for the binding of the respective fusion proteins. After detection of the GST-tagged protein (A1, B1), blots were stripped and re-incubated with antibodies against MBP (A2, B2). A3, B3, SNAP-tagged immobilized ArfGAP3 bait protein was visualized with SNAP-Vista Green (NEB). A, We tested whether increasing concentrations of Arf1 added to a fixed concentration of immobilized ArfGAP3 would inhibit binding of RIBEYE(B) to ArfGAP3. RIBEYE(B) was kept at a constant concentration in these experiments. B, We tested whether increasing concentrations of RIBEYE(B) added to a fixed concentration of immobilized ArfGAP3 would inhibit binding of Arf1 to ArfGAP3. Arf1 was kept at a constant concentration in these latter experiments. In both sets of experiments, we observed a competitive behavior between Arf1 and RIBEYE(B) in binding to ArfGAP3. These data demonstrate that RIBEYE(B) competes with Arf1 for binding to ArfGAP3. indicating that binding of RIBEYE(B) and Arf1 to ArfGAP3 is mutually exclusive. Abbreviations: AGD*, extended ArfGAP-domain of ArfGAP3.
ArfGAP3 is involved in endocytic vesicle retrieval at the photoreceptor synapse
Next, we wanted to find out for which synaptic process ArfGAP3 is relevant at the synaptic ribbon. The synaptic ribbon is a site of intense membrane retrieval (Jackman et al., 2009; Snellman et al., 2011; Chen et al., 2013, Wahl et al., 2013). Therefore, we tested whether ArfGAP3 is involved in this process (Fig. 15). We added FM1–43 to the extracellular medium to compare endocytic uptake in photoreceptors that were either electroporated with ArfGAP3-mcherry or mcherry alone. In mcherry-electroporated photoreceptors, there was an intense uptake of FM1–43 (Fig. 15A,B). The uptake of FM1–43 in mcherry-transfected photoreceptors was similar to the FM1–43 uptake in nontransfected photoreceptors (data not shown). In contrast to mcherry-transfected photoreceptors, ArfGAP3-overexpressing transfected photoreceptors showed a very strong inhibition of FM1–43 uptake (Fig. 15C,D) indicating that ArfGAP3 is essentially involved in endocytosis at the photoreceptor synapse (see discussion).
Figure 15.
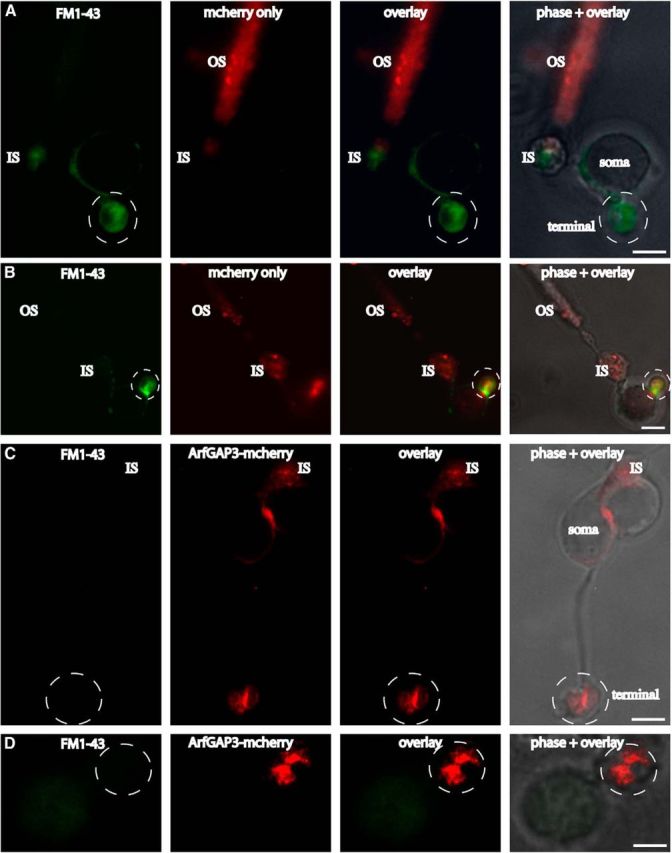
Overexpression of ArfGAP3 in mouse photoreceptors inhibits endocytic uptake of FM1–43. FM1–43 was used to compare endocytic uptake in photoreceptors that were either electroporated with mcherry alone (A, B) or ArfGAP3-mcherry (C, D). In mcherry-electroporated photoreceptors, there was an intense uptake of FM1–43 in the synaptic terminals (A, B). The uptake of FM1–43 in mcherry-transfected photoreceptors was similar to the FM1–43 uptake in nontransfected photoreceptors (data not shown). In contrast to mcherry-transfected photoreceptors, ArfGAP3-mcherry-overexpressing photoreceptors showed a strong inhibition of FM1–43 uptake in the synaptic terminals (C, D), indicating that ArfGAP3 is essential involved in endocytosis at the photoreceptor synapse. OS, Outer segment; IS, inner segment; Scale bars: A–C, 1 μm; D, 0.75 μm.
Discussion
In the present study, we demonstrated that the Arf-GTPase-activating protein-3, ArfGAP3, is a novel component of the photoreceptor synaptic ribbon complex using various independent assays. Conventional immunofluorescence microscopy as well as SR-SIM on thin retinal resin sections demonstrated that ArfGAP3 is highly enriched at the photoreceptor synaptic ribbon complex in situ. ArfGAP3 is more weakly expressed in the IPL than in photoreceptor synapses of the OPL (data not shown). This is most likely because synaptic ribbons in the IPL are smaller than in photoreceptor synapses of the OPL and because the IPL is dominated by conventional, nonribbon-containing synapses.
ArfGAP3 has been previously characterized as a component of the Golgi apparatus (for review, see Spang et al., 2010). At the Golgi apparatus, ArfGAP3 regulates vesicle trafficking in an Arf1-dependent manner (Dogic et al., 1999; Eugster et al., 2000; Lewis et al., 2004; Watson et al., 2004; Frigerio et al., 2007; Saitoh et al., 2009; Kartberg et al., 2010; Spang et al., 2010; Yu et al., 2012). The localization of ArfGAP3 at the synaptic ribbon complex in situ was confirmed by two different antibodies directed against different regions of the ArfGAP3 C terminus. Both antibodies showed the same enrichment of ArfGAP3 at the photoreceptor synaptic ribbon complex. In agreement with these morphological data, ArfGAP3 and RIBEYE were coimmunoprecipitated from the bovine retina, further demonstrating enrichment of ArfGAP3 at the photoreceptor synaptic ribbon complex.
We propose that the interaction with RIBEYE recruits ArfGAP3 to the synaptic ribbons where ArfGAP3 subsequently exerts its activity. YTH data demonstrated that the catalytically active, highly conserved ArfGAP-domain of ArfGAP3 is responsible for the binding to RIBEYE. The recruitment of ArfGAP3 to RIBEYE(B) is unlikely to require a functional catalytic activity of the AGD-domain because the initial ArfGAP3 prey clone obtained from our YTH screen that strongly interacted with RIBEYE(B) displayed a deletion in the central Zn-finger motif of the AGD-domain (Goldberg, 1999; for review, see Gillingham and Munro, 2007; Kahn et al., 2008; Donaldson and Jackson, 2011). This finding does not exclude that ArfGAP3 exerts a catalytic activity at the synaptic ribbon. More likely, the ArfGAP3-RIBEYE complex is part of a larger catalytically active multiprotein complex that functions at the synaptic ribbon. In support of this proposal, the Arf GTP/GDP exchange factor IQ-ArfGEF BRAG1, which catalyzes exchange of GDP by GTP, has been shown to be associated with synaptic ribbons (Katsumata et al., 2009).
Interestingly, the interaction between RIBEYE(B) and ArfGAP3 is stimulated by NAD(H) in a redox-sensitive manner. NADH is more efficient than the oxidized NAD+ in promoting RIBEYE(B)/ArfGAP3 interaction. Such a metabolism-dependent interaction is also known for the nuclear corepressor CtBP, which regulates transcription in a redox-sensitive manner (Zhang et al., 2002, 2006; Fjeld et al., 2003). A main function of this redox-sensitive interaction in ribbon synapses appears to be controlling Arf1 function. The small GTP-binding protein Arf1, an ArfGAP3 effector, is enriched at the synaptic ribbon. Arf1 is an important regulator of vesicle traffic at various intracellular compartments, including the Golgi apparatus and endosomal compartments (Gillingham and Munro, 2007; Kahn et al., 2008; Donaldson and Jackson, 2011) and might perform a similar role at the photoreceptor synaptic ribbon complex. We demonstrated that Arf1 and RIBEYE cannot bind at the same time. When RIBEYE is bound, e.g., at high levels of NADH, Arf1 can no longer bind to ArfGAP3. As a consequence, its GTPase activity will remain low and Arf1 will stay in its active, GTP-bound form.
Unfortunately, the antibodies against ArfGAP3 were not suitable for ultrastructural analyses. Super-resolution immunofluorescent microscopy indicated a particularly high enrichment of ArfGAP3 at the base of synaptic ribbon complex. Similarly, ArfGAP3 was also found close to bassoon, which is located at the base of the ribbon, and within the ring-like dynamin signal that surrounds the synaptic ribbon in the periactive zone. Dynamin was previously shown to be enriched at the presynaptic plasma membrane at the periactive zone (Wahl et al., 2013). Therefore, ArfGAP3 probably exerts its function at the base of the ribbon and in close proximity to the plasma membrane of the periactive zone. Clearly, future electron microscopic analyses need to demonstrate whether ArfGAP3 and Arf1 function directly at the presynaptic plasma membrane or on an endosomal compartment close to the plasma membrane.
Overexpression of ArfGAP3 in electroporated photoreceptors resulted in a strong inhibition of endocytic membrane retrieval as judged by an inhibition of uptake of FM1–43. Therefore, we propose that the RIBEYE/ArfGAP3 complex is involved in endocytic membrane retrieval at the synaptic ribbon. Recent studies demonstrated that the synaptic ribbon complex is important for endocytic membrane traffic in the tonically active ribbon synapse (Spassova et al., 2004; Griesinger et al., 2005; Khimich et al., 2005; Jackman et al., 2009; Babai et al., 2010; Frank et al., 2010; Schnee et al., 2011; Snellman et al., 2011; Tian et al., 2012; Chen et al., 2013; Wahl et al., 2013). Still, the molecular details and mechanisms used to accomplish this remain to be elucidated.
CtBP1/BARS, a close relative of RIBEYE(B)-domain/CtBP2 and component of the Golgi complex, is also localized to the synaptic ribbon complex (tom Dieck et al., 2005; for a review, see Corda et al., 2006). At the Golgi apparatus, CtBP1/BARS interacts with ArfGAP1 (Yang et al., 2005). This interaction has been proposed to be essential for vesicle formation and vesicle scission at the Golgi complex (Yang et al., 2002, 2005, 2006; Corda et al., 2006). The function of CtBP1/BARS at the synaptic ribbon is still unclear (Vaithianathan et al., 2013). There is a remarkable difference between the CtBP1/ArfGAP1 interaction compared with the RIBEYE/ArfGAP3 interaction. Interaction between CtBP1 and ArfGAP1 at the Golgi apparatus is mediated by the SBD of CtBP1, while the interaction between RIBEYE(B)domain and ArfGAP3 at the synaptic ribbons is mediated by the NBD of RIBEYE(B)-domain.
The recruitment of ArfGAP3 to the synaptic ribbon via an inducible, redox-dependent manner provides the synaptic ribbon with the possibility to regulate endocytic vesicle trafficking. The catalytically active ArfGAP-domain could control Arf1 activity in a redox-switchable manner by a competitive interaction with RIBEYE. The C terminus of ArfGAP3 could be involved in different tasks, e.g., the cargo sorting. The C terminus of ArfGAP3 has been shown to be involved in membrane-binding and cargo-binding/coat protein-binding at the Golgi apparatus (Rein et al., 2002; for review, see Nie and Randazzo, 2006; Schindler and Spang, 2007; Kliouchnikov et al., 2009; Schindler et al., 2009; Spang et al., 2010). A similar function might also apply in the ribbon synapse and would provide the RIBEYE-ArfGAP3 complex a central position in the vesicle recycling machinery.
Footnotes
This work was supported by the German research community Deutsche Forschungsgemeinschaft (SFB894 and GRK1326). We thank Dr. Ching-Hwa Sung (Weill Medical College, Cornell University, New York) for the bovine retina YTH cDNA library, Dr. Rachel Wong (University of Washington, Seattle) for the kind gift of a retina incubation chamber and for her support and advice on retinal explant cultures, Dr. Andreas Beck (Saarland University, Department of Pharmacology) for advice for quantification of binding experiments, Dr. Kannan Alpadi for cloning of the bassoon-pGAD-T7 construct, Dr. Elmar Krause (platform project P1, SFB894) for help with the SIM microscope, and Dr. Jutta Schmitz-Kraemer for critically reading this manuscript.
The authors declare no competing financial interests.
References
- Alpadi K, Magupalli VG, Käppel S, Köblitz L, Schwarz K, Seigel GM, Sung CH, Schmitz F. RIBEYE recruits a mammalian ortholog of the C. elegans protein unc119 to synaptic ribbons of photoreceptor synapses. J Biol Chem. 2008;283:26461–26467. doi: 10.1074/jbc.M801625200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babai N, Bartoletti TM, Thoreson WB. Calcium regulates vesicle replenishment at the cone ribbon synapse. J Neurosci. 2010;30:15866–15877. doi: 10.1523/JNEUROSCI.2891-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigay J, Casella JF, Drin G, Mesmin B, Antonny B. ArfGAP1 responds to membrane curvature through the folding of a lipid packing sensor motif. EMBO J. 2005;24:2244–2253. doi: 10.1038/sj.emboj.7600714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Briggman KL, Euler T. Bulk electroporation and population calcium imaging in the adult mammalian retina. J Neurophysiol. 2011;105:2601–2609. doi: 10.1152/jn.00722.2010. [DOI] [PubMed] [Google Scholar]
- Chen M, Van Hook MJ, Zenisek D, Thoreson WB. Properties of ribbon and non-ribbon release from rod photoreceptors revealed by visualizing individual synaptic vesicles. J Neurosci. 2013;33:2071–2086. doi: 10.1523/JNEUROSCI.3426-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corda D, Colanzi A, Luini A. The multiple activities of CtBP/BARS proteins: the Golgi view. Trends Cell Biol. 2006;16:167–173. doi: 10.1016/j.tcb.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Cukierman E, Huber I, Rotman M, Cassel D. The Arf1 GTPase-activating protein: zinc finger motif and Golgi complex localization. Science. 1995;270:1999–2002. doi: 10.1126/science.270.5244.1999. [DOI] [PubMed] [Google Scholar]
- Dogic D, de Chassey B, Pick E, Cassel D, Lefkir Y, Hennecke S, Cosson P, Letourneur F. The ADP-ribosylation factor GTPase-activating protein glo3p is involved in ER retrieval. Eur J Cell Biol. 1999;78:305–310. doi: 10.1016/S0171-9335(99)80064-8. [DOI] [PubMed] [Google Scholar]
- Donaldson JG, Jackson CL. Arf family G proteins and their regulators: roles in membrane transport, development and disease. Nat Rev Mol Cell Biol. 2011;12:362–375. doi: 10.1038/nrm3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan SL, Dyer MA. Preparation and square wave electroporation of retinal explant cultures. Nat Protoc. 2006;1:2710–2718. doi: 10.1038/nprot.2006.454. [DOI] [PubMed] [Google Scholar]
- Eugster A, Frigerio G, Dale M, Duden R. COP I domains required for coatomer integrity and novel interactions with Arf and ArfGAP. EMBO J. 2000;19:3905–3917. doi: 10.1093/emboj/19.15.3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjeld CC, Birdsong WT, Goodman RH. Differential binding of NAD+ and NADH allows the transcriptional corepressor C-terminal binding protein to serve as a metabolic sensor. Proc Natl Acad Sci U S A. 2003;100:9202–9207. doi: 10.1073/pnas.1633591100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank T, Rutherford MA, Strenzke N, Neef A, Pangrsic T, Khimich D, Fejtova A, Gundelfinger ED, Liberman MC, Harke B, Bryan KE, Lee A, Egner A, Riedel D, Moser T. Bassoon and the synaptic ribbon organize Ca2+-channels and vesicles to add release sites and promote refilling. Neuron. 2010;68:724–738. doi: 10.1016/j.neuron.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigerio G, Grimsey N, Dale M, Majoul I, Duden R. Two human ArfGAPs associated with COP-I-coated vesicles. Traffic. 2007;8:1644–1655. doi: 10.1111/j.1600-0854.2007.00631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham AK, Munro S. The small g proteins of the Arf family and their regulators. Annu Rev Cell Dev Biol. 2007;23:579–611. doi: 10.1146/annurev.cellbio.23.090506.123209. [DOI] [PubMed] [Google Scholar]
- Goldberg J. Structural and functional analysis of the Arf1-Arfgap complex reveals a role for coatamer in GTP hydrolysis. Cell. 1999;96:893–902. doi: 10.1016/S0092-8674(00)80598-X. [DOI] [PubMed] [Google Scholar]
- Griesinger CB, Richards CD, Ashmore JF. Fast vesicle replenishment allows indefatigable signalling at the first auditory synapse. Nature. 2005;435:212–215. doi: 10.1038/nature03567. [DOI] [PubMed] [Google Scholar]
- Gustafsson MG, Shao L, Carlton PM, Wang CJ, Golubovskaya IN, Cande WZ, Agard DA, Sedat JW. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys J. 2008;94:4957–4970. doi: 10.1529/biophysj.107.120345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-G. [DOI] [PubMed] [Google Scholar]
- Inoue H, Randazzo PA. Arf GAPs and their interacting proteins. Traffic. 2007;8:1465–1475. doi: 10.1111/j.1600-0854.2007.00624.x. [DOI] [PubMed] [Google Scholar]
- Jackman SL, Choi SY, Thoreson WB, Rabl K, Bartoletti TM, Kramer RH. Role of the synaptic ribbon in transmitting the cone light response. Nat Neurosci. 2009;12:303–310. doi: 10.1038/nn.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RA, Bruford E, Inoue H, Logsdon JM, Jr, Nie Z, Premont RT, Randazzo PA, Satake M, Theibert AB, Zapp ML, Cassel D. Consensus nomenclature of the human ArfGAP domain-containing proteins. J Cell Biol. 2008;182:1039–1044. doi: 10.1083/jcb.200806041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartberg F, Asp L, Dejgaard SY, Smedh M, Fernandez-Rodriguez J, Nilsson T, Presley JF. ArfGAP2 and ArfGAP3 are essential for COP I coat assembly on the Golgi membrane of living cells. J Biol Chem. 2010;285:36709–36720. doi: 10.1074/jbc.M110.180380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsumata O, Ohara N, Tamaki H, Niimura T, Naganuma H, Watanabe M, Sakagami H. IQ-ArfGEF/BRAG1 is associated with synaptic ribbons in the mouse retina. Eur J Neurosci. 2009;30:1509–1516. doi: 10.1111/j.1460-9568.2009.06943.x. [DOI] [PubMed] [Google Scholar]
- Khimich D, Nouvian R, Pujol R, Tom Dieck S, Egner A, Gundelfinger ED, Moser T. Hair cell synaptic ribbons are essential for synchronous auditory signalling. Nature. 2005;434:889–894. doi: 10.1038/nature03418. [DOI] [PubMed] [Google Scholar]
- Kliouchnikov L, Bigay J, Mesmin B, Parnis A, Rawet M, Goldfeder N, Antonny B, Cassel D. Discrete determinants in ArfGAP23/3 conferring Golgi localization and regulation by the COPI coat. Mol Biol Cell. 2009;20:859–869. doi: 10.1091/mbc.E08-10-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Carlson JE, Ohgi KA, Edwards TA, Rose DW, Escalante CR, Rosenfeld MG, Aggarwal AK. Transcription corepressor CtBP is an NAD(+) regulated dehydrogenase. Mol Cell. 2002;10:857–869. doi: 10.1016/S1097-2765(02)00650-0. [DOI] [PubMed] [Google Scholar]
- Lewis SM, Poon PP, Singer RA, Johnston GC, Spang A. The ArfGAP Glo3 is required for the generation of COP I vesicles. Mol Biol Cell. 2004;15:4064–4072. doi: 10.1091/mbc.E04-04-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhang C, Xing G, Chen Q, He F. Functional characterization of novel human ArfGAP3. FEBS Lett. 2001;490:79–83. doi: 10.1016/S0014-5793(01)02134-2. [DOI] [PubMed] [Google Scholar]
- Magupalli VG, Schwarz K, Alpadi K, Natarajan S, Seigel GM, Schmitz F. Multiple RIBEYE-RIBEYE interactions create a dynamic scaffold for the formation of synaptic ribbons. J Neurosci. 2008;28:7954–7967. doi: 10.1523/JNEUROSCI.1964-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JL, Soto F, Wong RO, Kerschensteiner D. Development of cell type-specific connectivity patterns of converging excitatory axons in the retina. Neuron. 2011;71:1014–1021. doi: 10.1016/j.neuron.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardini M, Spanò S, Cericola C, Pesce A, Massaro A, Millo E, Luini A, Corda D, Bolognesi M. CtBP/BARS: a dual-function protein involved in transcriptional corepression and Golgi membrane fission. EMBO J. 2003;22:3122–3130. doi: 10.1093/emboj/cdg283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z, Randazzo PA. Arf GAPs and membrane traffic. J Cell Sci. 2006;119:1203–1211. doi: 10.1242/jcs.02924. [DOI] [PubMed] [Google Scholar]
- Olmsted JB. Affinity purification of antibodies from diazotized paper blots of heterogeneous protein samples. J Biol Chem. 1981;256:11955–11957. [PubMed] [Google Scholar]
- Rea R, Li J, Dharia A, Levitan ES, Sterling P, Kramer RH. Streamlined synaptic vesicle cycle in cone photoreceptor terminals. Neuron. 2004;41:755–766. doi: 10.1016/S0896-6273(04)00088-1. [DOI] [PubMed] [Google Scholar]
- Rein U, Andag U, Duden R, Schmitt HD, Spang A. ArfGAP-mediated interaction between ER-Golgi v-SNAREs and the COP I coat. J Cell Biol. 2002;157:395–404. doi: 10.1083/jcb.200112092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh A, Shin HW, Yamada A, Waguri S, Nakayama K. Three homologous ArfGAPs participate in coat protein I-mediated transport. J Biol Chem. 2009;284:13948–13957. doi: 10.1074/jbc.M900749200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L, Carlton PM, Haase S, Shao L, Winoto L, Kner P, Burke B, Cardoso MC, Agard DA, Gustafsson MG, Leonhardt H, Sedat JW. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 2008;320:1332–1336. doi: 10.1126/science.1156947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L, Heintzmann R, Leonhardt H. A guide to superresolution fluorescence microscopy. J Cell Biol. 2010;190:165–175. doi: 10.1083/jcb.201002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler C, Spang A. Interaction of SNAREs with ArfGAPs precedes recruitment of sec18p/NSF. Mol Biol Cell. 2007;18:2852–2863. doi: 10.1091/mbc.E06-08-0756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler C, Rodriguez F, Poon PP, Singer RA, Johnston GC, Spang A. The GAP domain and the SNARE, coatomer and cargo interaction region of the ArfGAP2/3 glo3 are sufficient for glo3 function. Traffic. 2009;10:1362–1375. doi: 10.1111/j.1600-0854.2009.00952.x. [DOI] [PubMed] [Google Scholar]
- Schmitz F. The making of synaptic ribbons: how they are built and what they do. Neuroscientist. 2009;15:611–624. doi: 10.1177/1073858409340253. [DOI] [PubMed] [Google Scholar]
- Schmitz F, Königstorfer A, Südhof TC. RIBEYE, a component of ribbon synapses: a protein's journey through evolution provides insight into synaptic ribbon function. Neuron. 2000;28:857–872. doi: 10.1016/S0896-6273(00)00159-8. [DOI] [PubMed] [Google Scholar]
- Schmitz F, Natarajan S, Venkatesan JK, Wahl S, Schwarz K, Grabner CP. EF hand-mediated Ca and cGMP signaling in photoreceptor synaptic terminals. Front Mol Neurosci. 2012;5:26. doi: 10.3389/fnmol.2012.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnee ME, Santos-Sacchi J, Castellano-Muñoz M, Kong JH, Ricci AJ. Calcium-dependent synaptic vesicle trafficking underlies indefatigable release of the hair cell afferent fiber synapse. Neuron. 2011;70:326–338. doi: 10.1016/j.neuron.2011.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz K, Natarajan S, Kassas N, Vitale N, Schmitz F. The synaptic ribbon is a site of phosphatidic acid generation in ribbon synapses. J Neurosci. 2011;31:15996–16011. doi: 10.1523/JNEUROSCI.2965-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snellman J, Mehta B, Babai N, Bartoletti TM, Akmentin W, Francis A, Matthews G, Thoreson W, Zenisek D. Acute destruction of the synaptic ribbon reveals a role for the ribbon in vesicle priming. Nat Neurosci. 2011;14:1135–1141. doi: 10.1038/nn.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Shiba Y, Randazzo PA. ArfGAPs: gatekeepers of vesicle generation. FEBS Lett. 2010;584:2646–2651. doi: 10.1016/j.febslet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassova MA, Avissar M, Furman AC, Crumling MA, Saunders JC, Parsons TD. Evidence that rapid replenishment of the synaptic ribbon mediates recovery from short-term adaptation at the hair cell afferent synapse. J Assoc Res Otolaryngol. 2004;5:376–390. doi: 10.1007/s10162-004-5003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai AW, Chuang JZ, Bode C, Wolfrum U, Sung CH. Rhodopsin's carboxyterminal cytoplasmic tail acts as a membrane receptor for cytoplasmic dynein by binding to the dynein light chain Ttctex-1. Cell. 1999;97:877–887. doi: 10.1016/S0092-8674(00)80800-4. [DOI] [PubMed] [Google Scholar]
- Tian M, Xu CS, Montpetit R, Kramer RH. Rab3a mediates vesicle delivery at photoreceptor ribbon synapses. J Neurosci. 2012;32:6931–6936. doi: 10.1523/JNEUROSCI.0265-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- tom Dieck S, Altrock WD, Kessels MM, Qualmann B, Regus H, Brauner D, Fejtová A, Bracko O, Gundelfinger ED, Brandstätter JH. Molecular dissection of the photoreceptor ribbon synapse: physical interaction of Bassoon and RIBEYE is essential for the assembly of the ribbon complex. J Cell Biol. 2005;168:825–836. doi: 10.1083/jcb.200408157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaithianathan T, Akmentin W, Henry D, Matthews G. The ribbon-associated protein C-terminal-binding protein 1 is not essential for the structure and function of retinal ribbon synapses. Mol Vis. 2013;19:917–926. [PMC free article] [PubMed] [Google Scholar]
- Venkatesan JK, Natarajan S, Schwarz K, Mayer SI, Alpadi K, Magupalli VG, Sung CH, Schmitz F. Nicotinamide adenine dinucleotide-dependent binding of the neuronal Ca2+- sensor protein GCAP2 to photoreceptor synaptic ribbons. J Neurosci. 2010;30:6559–6576. doi: 10.1523/JNEUROSCI.3701-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara MN, Gutierrez C, O'Brien DR, Canto-Soler MV. Ex vivo electroporation of retinal cells: a novel, high efficiency method for functional studies in primary retinal cultures. Exp Eye Res. 2013;109:40–50. doi: 10.1016/j.exer.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl S, Katiyar R, Schmitz F. A local, periactive zone endocytic machinery at photoreceptor synapses in close vicinity to photoreceptor synaptic ribbons. J Neurosci. 2013;33:10278–10300. doi: 10.1523/JNEUROSCI.5048-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson PJ, Frigerio G, Collins BM, Duden R, Owen DJ. gamma-COP appendage domain-structure and function. Traffic. 2004;5:79–88. doi: 10.1111/j.1600-0854.2004.00158.x. [DOI] [PubMed] [Google Scholar]
- Williams PR, Morgan JL, Kerschensteiner D, Wong RO. In vitro imaging of retinal whole mounts. Cold Spring Harb Protoc. 2013;2013 doi: 10.1101/pdb.prot072645. pii:pdb.prot072645. [DOI] [PubMed] [Google Scholar]
- Yang JS, Lee SY, Gao M, Bourgoin S, Randazzo PA, Premont RT, Hsu VW. ArfGAP1 promotes the formation of COPI vesicles, suggesting function as a component of the coat. J Cell Biol. 2002;159:69–78. doi: 10.1083/jcb.200206015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Lee SY, Spanò S, Gad H, Zhang L, Nie Z, Bonazzi M, Corda D, Luini A, Hsu VW. A role for BARS at the fission step of COP I vesicle formation from the Golgi membrane. EMBO J. 2005;24:4133–4143. doi: 10.1038/sj.emboj.7600873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Zhang L, Lee SY, Gad H, Luini A, Hsu VW. Key components of the fission machinery are interchangeable. Nat Cell Biol. 2006;8:1376–1382. doi: 10.1038/ncb1503. [DOI] [PubMed] [Google Scholar]
- Yu X, Breitman M, Goldberg J. A structure-based mechanism for Arf1-dependent recruitment of coatomer to membranes. Cell. 2012;148:530–542. doi: 10.1016/j.cell.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenisek D, Steyer JA, Almers W. Transport, capture and exocytosis of single synaptic vesicles at active zones. Nature. 2000;406:849–854. doi: 10.1038/35022500. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Piston DW, Goodman RH. Regulation of corepressor function by nuclear NADH. Science. 2002;295:1895–1897. doi: 10.1126/science.1069300. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Wang SY, Nottke AC, Rocheleau JV, Piston DW, Goodman RH. Redox sensor CtBP mediates hypoxia-induced tumor cell migration. Proc Natl Acad Sci U S A. 2006;103:9029–9033. doi: 10.1073/pnas.0603269103. [DOI] [PMC free article] [PubMed] [Google Scholar]