Summary
Invasive Salmonella infections in adults are commonly treated with fluoroquinolones, a critically important antimicrobial class. Historically, quinolone resistance was the result of chromosomal mutations, but plasmid-mediated quinolone resistance (PMQR) has emerged and is increasingly being reported in Enterobacteriaceae worldwide. PMQR may facilitate the spread of quinolone resistance, lead to higher-level quinolone resistance, and make infections harder to treat. To better understand the epidemiology of PMQR in nontyphoidal Salmonella causing human infections in the United States, we looked at trends in quinolone resistance among isolates submitted to the Centers for Disease Control and Prevention. We reviewed demographic, exposure, and outcome information for patients with isolates having a PMQR-associated phenotype during 2008–2014 and tested isolates for quinolone resistance mechanisms. We found that PMQR is emerging among nontyphoidal Salmonella causing human infections in the United States and that international travel, reptile and amphibian exposure, and food are likely sources of human infection.
Keywords: antibiotic resistance, foodborne diseases, quinolones, reptiles, Salmonella, travel
INTRODUCTION
Quinolones constitute a critically important class of antibacterial drugs (WHO, 2017). The initial member of the class, nalidixic acid, had limited clinical use. Chemical modifications, including the addition of fluorine, yielded fluoroquinolones. Fluoroquinolones (e.g., ciprofloxacin, levofloxacin) have greater potency, broader spectra of activity, improved pharmacokinetics, and a lower frequency of resistance development (Hooper & Jacoby, 2015). Fluoroquinolones have been widely used to treat various infections (Yanat et al., 2017), including invasive nontyphoidal Salmonella infections, which can be life-threatening (Crump et al., 2003). They have also been used in animals to prevent, control, and treat infections (FDA, n.d.; Yanat et al., 2017). Since quinolones were introduced into human and veterinary medicine, resistance to them has emerged and increased (Van, Nguyen, Smooker, & Coloe, 2012; Yanat et al., 2017).
Fluoroquinolones target 2 essential bacterial enzymes, DNA gyrase and topoisomerase IV. In Enterobacteriaceae including Salmonella, quinolone resistance typically develops from the accumulation of chromosomal mutations in the quinolone resistance-determining region (QRDR) of target enzyme genes, primarily gyrA and parC (Cavaco & Aarestrup, 2009; Yanat et al., 2017); one mutation generally mediates resistance to nalidixic acid and decreases susceptibility to fluoroquinolones, while additional mutations lead to full fluoroquinolone resistance (Aarestrup et al., 2003). Since the late 1990s, 3 types of plasmid-mediated quinolone resistance (PMQR) mechanisms have been identified: qnr genes, which protect target enzymes; aac(6’)-Ib-cr gene, which mediates acetylation of certain quinolones; and oqxAB and qepA genes, which produce mobile efflux pumps (Jacoby et al., 2014). A PMQR gene alone usually confers decreased susceptibility to fluoroquinolones and has less effect on nalidixic acid susceptibility (Hooper & Jacoby, 2015; Rodríguez-Martínez et al., 2016). PMQR genes are particularly frequent among Enterobacteriaceae with decreased susceptibility to fluoroquinolones combined with nalidixic acid susceptibility (Rodríguez-Martínez et al., 2016); consequently, this phenotype is a clue to the presence of PMQR (Hooper & Jacoby, 2015). During the last decade, PMQR has been increasingly reported in Enterobacteriaceae worldwide (Yanat et al., 2017).
PMQR has important implications for the treatment of infections and the dissemination of antimicrobial resistance. Plasmids harboring quinolone resistance genes can be transferred horizontally to other bacteria, thus spreading resistance. These plasmids often encode resistance to additional drug classes, especially cephalosporins (also used for treating salmonellosis). When this occurs, use of a single drug can co-select for resistance to drugs in multiple antimicrobial classes (Gay et al., 2006; Robicsek et al., 2006). While PMQR genes generally cause increases in fluoroquinolone minimum inhibitory concentrations (MICs) that are below the resistance breakpoint, they may facilitate the emergence of higher-level quinolone resistance by increasing the mutant prevention concentration (lowest concentration of drug needed to prevent the growth of quinolone-resistant mutants) and by acting additively or synergistically with other quinolone resistance mechanisms (Jacoby et al., 2014; Lin et al., 2015; Robicsek et al., 2006). The number of resistance mechanisms (QRDR mutations or PMQR genes) is correlated with ciprofloxacin MICs (Rodríguez-Martínez et al., 2016). Studies suggest that even small increases in quinolone MICs can adversely impact response to treatment (Crump et al., 2003; Humphries et al., 2012; Rodríguez-Martínez et al., 2016).
In the United States, the National Antimicrobial Resistance Monitoring System (NARMS) tracks resistance among Salmonella and other enteric bacteria from humans, retail meat, and food animals. In a series of NARMS studies, PMQR was detected in 34 nontyphoidal Salmonella strains isolated from humans during 1996–2007, and a higher proportion of isolates were found to have PMQR in 2004–2006 (Sjölund-Karlsson et al., 2009) and 2007 (Sjölund-Karlsson, 2010) than in 1996–2003 (Gay et al., 2006). In 2015, NARMS reported an increase in the proportion of ciprofloxacin-nonsusceptible isolates lacking nalidixic acid resistance (CDC, 2015), a finding that suggests that PMQR is increasing. To better understand the epidemiology of PMQR among nontyphoidal Salmonella causing human infections in the United States, we analyzed quinolone MIC data for isolates submitted to NARMS, reviewed demographic, exposure, and outcome information for patients with Salmonella isolates with a PMQR-associated phenotype, and tested a sample of isolates for quinolone-resistance mechanisms.
MATERIALS AND METHODS
Isolate Submission and Antimicrobial Susceptibility Testing
NARMS nontyphoidal Salmonella surveillance began at the Centers for Disease Control and Prevention (CDC) in 1996 with 14 sites and was nationwide by 2003. Health departments submitted every 10th (1996–2002) or 20th (2003–2014) isolate received from clinical laboratories to NARMS for surveillance purposes. This frequency-based sampling included isolates from both sporadic and outbreak cases. Health departments submitted additional isolates to NARMS through enhanced testing of outbreak isolates; data for these isolates are reported separately in the results section. Health departments serotyped isolates and performed pulsed-field gel electrophoresis (PFGE) using standardized methods (Ribot et al., 2006; Strockbine et al., 2015). MICs were determined at CDC using broth microdilution (Sensititre™, Trek Diagnostics, Oakwood Village, OH) for the quinolones ciprofloxacin and nalidixic acid in 1996–2014 and for 11 additional agents in 7 other classes in 2008–2014. Additionally, the aminoglycosides amikacin and kanamycin were tested through 2010 and 2013, respectively, and the macrolide azithromycin was tested beginning in 2011. Breakpoints are listed in the 2014 NARMS report (CDC, 2016).
Definitions
We defined the study phenotype as ciprofloxacin MIC ≥0.25 μg/mL with nalidixic acid susceptibility. We used this phenotype to identify a group of isolates likely to have PMQR (Hopkins et al., 2007; Hopkins et al., 2008; Veldman et al., 2011) so we could then genetically test those isolates and collect corresponding patient exposure and outcome data. We defined multidrug resistance (MDR) as resistance to ≥1 agent in ≥3 classes. Isolates resistant (MIC ≥1 μg/mL) or intermediate (MIC 0.12–0.5 μg/mL) to ciprofloxacin were considered ciprofloxacin nonsusceptible. An outbreak was defined as ≥2 cases of similar illness associated with a common exposure. Hereafter, the term Salmonella refers to nontyphoidal Salmonella.
Data Collection and Analysis
We analyzed historical quinolone MIC data for NARMS surveillance isolates from 1996 through 2014. For study-phenotype isolates from the period 2008–2014, we also requested patient exposure and outcome information collected by health departments; analyzed antimicrobial susceptibility patterns and patient demographic data; and reviewed summary foodborne outbreak investigation data collected by the Foodborne Disease Outbreak Surveillance System, CDC’s Outbreak Response and Prevention Branch, and health departments. We used Microsoft Access 2013 and Microsoft Excel 2013 for data analysis.
Detection of Quinolone Resistance Mechanisms
The presence of PMQR genes and QRDR mutations was determined for a subset of study-phenotype isolates from the period 2008–2014 using polymerase chain reaction (PCR) and/or whole genome sequencing (WGS). PCR primers used for aac(6’)-Ib-cr, gyrA, oqxA, oqxB, parC, qepA, qnrA, qnrB, qnrC, qnrD, and qnrS are presented in the Supplemental Information online, along with more detailed laboratory methods. Direct sequencing of PCR products confirmed the presence of aac(6’)-Ib-cr and gyrA/parC mutations (Gay et al., 2006). WGS was performed using MiSeq or HiSeq sequencers (Ilumina, San Diego, CA). Raw reads were assembled de novo using CLC Genomics Workbench 8.5 (Qiagen Inc., Germantown, MD) and assemblies were analyzed using tools developed by the Center of Genomic Epidemiology (http://cge.cbs.dtu.dk/services/). ResFinder was used with a 90% threshold for percent identity and 60% gene coverage. Sequence reads were submitted to the National Center for Biotechnology Information (NCBI) Sequence Read Archive. Isolates lacking PMQR in assemblies were confirmed to be negative for these genes using read-based approaches. Reads were examined using ARIBA v0.1 (Hunt et al., 2017) and ResFinder and by mapping reads to a recently discovered qnrE gene (Albornoz et al., 2017) using CLC Genomics Workbench.
RESULTS
Quinolone Resistance Trends and Isolate Characteristics
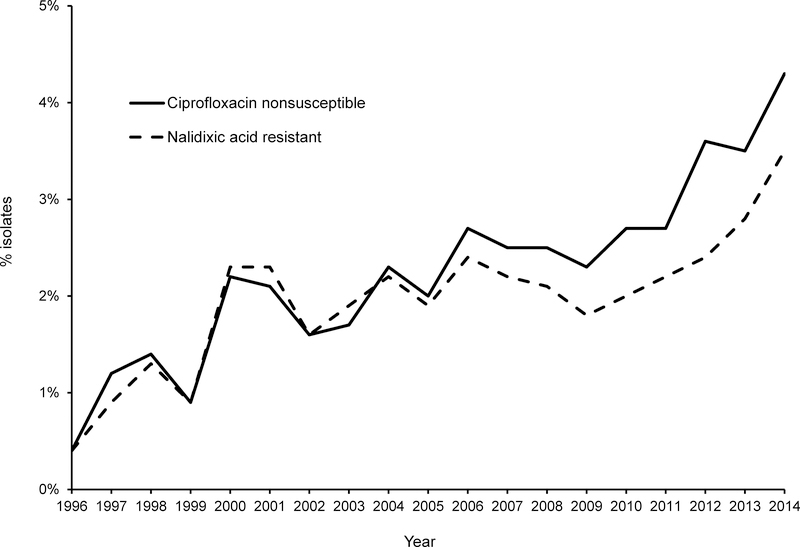
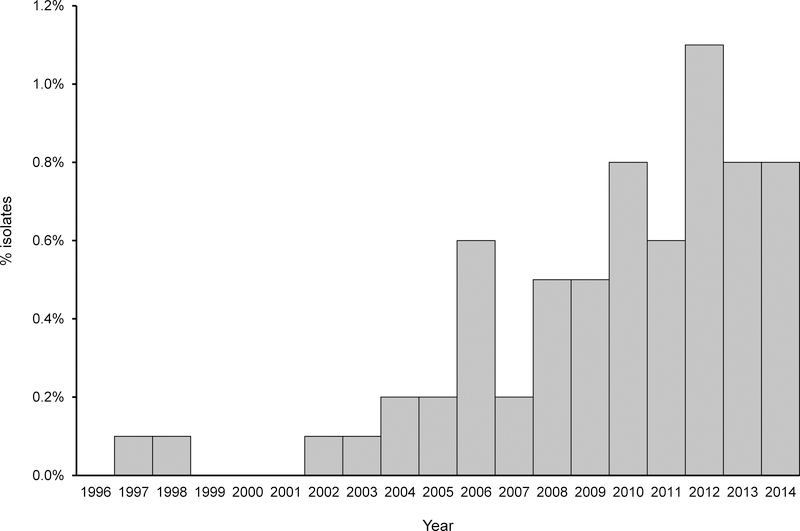
During 1996–2014, both ciprofloxacin nonsusceptibility and nalidixic acid resistance increased among Salmonella surveillance isolates; they were closely correlated until 2006, when ciprofloxacin nonsusceptibility began to exceed nalidixic acid resistance (Figure 1). Ciprofloxacin nonsusceptibility increased from 0.4% (5/1318) in 1996 to 4.3% (92/2127) in 2014. The proportion of Salmonella isolates with the study phenotype increased 18-fold from 0.05% (2/4,069) in 1996–1998 to ≈0.9% (61/6,538) in 2012–2014 (Figure 2).
FIGURE 1.
Ciprofloxacin nonsusceptibility and nalidixic acid resistance among nontyphoidal Salmonella surveillance isolates, United States, 1996–2014. Ciprofloxacin nonsusceptibility includes ciprofloxacin resistant (MIC ≥1 μg/mL) and intermediate (MIC 0.12–0.5 μg/mL).
FIGURE 2.
Percentage of nontyphoidal Salmonella surveillance isolates with the study phenotype (ciprofloxacin MIC ≥0.25 μg/mL with nalidixic acid susceptibility), United States, 1996–2014.
Eighty percent (117/147) of isolates with the study phenotype were from the period 2008–2014. During this time, 3.1% (489/15,897) of Salmonella surveillance isolates were ciprofloxacin nonsusceptible and 24% (117/489) of these isolates had the study phenotype. Enteritidis, Typhimurium, and Newport were the most common Salmonella serotypes overall and the ones with the greatest number of ciprofloxacin-nonsusceptible isolates during these years; 38% (186/489) of ciprofloxacin-nonsusceptible isolates were serotype Enteritidis (Table 1). The proportion of ciprofloxacin-nonsusceptible isolates with the study phenotype varied greatly by serotype; it was 1%, 32%, and 65% for serotypes Enteritidis, Typhimurium, and Newport, respectively. The 117 isolates with the study phenotype comprised 42 serotypes; Typhimurium (17), Newport (13), Litchfield (9), Bareilly (8), Saintpaul (6), Corvallis (5), and Stanley (5) were most common and accounted for more than half of the isolates with the study phenotype. Most isolates with the study phenotype had a ciprofloxacin MIC of 0.5 μg/mL (72) or 0.25 μg/mL (36), categorizing them as ciprofloxacin intermediate. The remaining 9 study-phenotype isolates had an MIC of 1 μg/mL, making them ciprofloxacin resistant; 7 of the 9 ciprofloxacin-resistant isolates were serotype Litchfield.
TABLE 1.
Ciprofloxacin nonsusceptibility† among nontyphoidal Salmonella surveillance isolates and plasmid-mediated quinolone resistance (PMQR) genes detected among isolates with the study phenotype‡, by serotype, United States, 2008–2014
| No. isolates |
PMQR genes detected among study
phenotype isolates tested§ |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Serotype | Total | Ciprofloxacin nonsusceptible† | Study phenotype‡ | Tested for PMQR | qnrA | qnrB | qnrS | aac(6’)-lb-cr | None |
| Typhimurium | 2,331 | 53 | 17 | 5 | 2 | 3 | |||
| Newport | 1,791 | 20 | 13 | 13 | 13 | ||||
| Litchfield | 118 | 9 | 9 | 4 | 3¶ | 1 | 3¶ | ||
| Bareilly | 145 | 8 | 8 | 2 | 2 | ||||
| Saintpaul | 419 | 14 | 6 | 4 | 4 | ||||
| Corvallis | 8 | 5 | 5 | 2 | 2 | ||||
| Stanley | 88 | 6 | 5 | 3 | 2 | 1 | |||
| I 4,[5],12:i:- | 670 | 15 | 4 | 2 | 1 | 1# | |||
| Muenchen | 355 | 4 | 4 | 2 | 2 | ||||
| Braenderup | 329 | 3 | 3 | 2 | 1 | 1 | |||
| Guinea | 3 | 3 | 3 | 2 | 2 | ||||
| Concord | 7 | 2 | 2 | 2 | 1 | 1 | |||
| Derby | 51 | 3 | 2 | 1 | 1 | ||||
| Enteritidis | 2,940 | 186 | 2 | 2 | 1 | 1# | |||
| Ituri | 2 | 2 | 2 | 2 | 1 | 1# | |||
| Manhattan | 41 | 2 | 2 | 2 | 1 | 1 | |||
| Montevideo | 405 | 3 | 2 | 2 | 1 | 1 | |||
| Muenster | 27 | 4 | 2 | 2 | 2 | ||||
| Senftenberg | 63 | 5 | 2 | ||||||
| Telelkebir | 14 | 2 | 2 | 2 | 2 | ||||
| I 4,[5],12:-:1,2 | 7 | 3 | 1 | ||||||
| I 4,[5],12:d:- | 4 | 1 | 1 | 1 | 1 | ||||
| IV 44:z4,z23:- | 5 | 1 | 1 | 1 | 1 | ||||
| Agona | 203 | 7 | 1 | ||||||
| Anatum | 111 | 3 | 1 | ||||||
| Apapa | 2 | 1 | 1 | 1 | 1 | ||||
| Berta | 106 | 2 | 1 | 1 | 1# | ||||
| Cannstatt | 3 | 1 | 1 | ||||||
| Give | 49 | 2 | 1 | 1 | 1 | ||||
| Hadar | 107 | 8 | 1 | 1 | 1 | ||||
| Haifa | 1 | 1 | 1 | 1 | 1 | ||||
| Havana | 20 | 1 | 1 | ||||||
| Heidelberg | 465 | 4 | 1 | 1 | 1 | ||||
| Isangi | 1 | 1 | 1 | 1 | 1 | ||||
| Javiana | 973 | 7 | 1 | 1 | 1 | ||||
| Kouka | 1 | 1 | 1 | 1 | 1 | ||||
| Ouakam | 1 | 1 | 1 | 1 | 1 | ||||
| Panama | 83 | 1 | 1 | 1 | 1 | ||||
| Paratyphi B var. L(+) tartrate+ | 218 | 7 | 1 | ||||||
| Reading | 24 | 1 | 1 | 1 | 1 | ||||
| Urbana | 20 | 2 | 1 | 1 | 1 | ||||
| Virchow | 35 | 11 | 1 | 1 | 1 | ||||
| All other serotypes | 3,236 | 57†† | |||||||
| Rough/nonmotile isolates | 84 | 5 | |||||||
| Unknown serotype | 331 | 11 | |||||||
| Total | 15,897 | 489 | 117 | 72 | 4 | 44¶ | 20 | 3¶ | 4# |
Ciprofloxacin nonsusceptibility includes ciprofloxacin resistant (MIC ≥1 μg/mL) and intermediate (MIC 0.12–0.5 μg/mL).
Study phenotype is ciprofloxacin MIC ≥0.25 μg/mL with nalidixic acid susceptibility.
No isolates tested for PMQR genes had qnrC, qnrD, oqxA, oqxB, or qepA detected.
Three Salmonella Litchfield isolates had both qnrB and aac(6’)-Ib-cr genes.
The Salmonella I 4,[5],12:i:- and Berta isolates with no PMQR genes detected had ciprofloxacin MICs ≤0.015 μg/mL upon retesting, while the Enteritidis and Ituri isolates with no PMQR genes detected had gyrA mutations; the Ituri isolate also had a parC mutation.
Includes 13 Infantis, 12 Kentucky, 8 Choleraesuis, 5 Dublin, 2 Albert, 2 Nitra, 2 Oranienburg, 2 Potsdam, 2 Schwarzengrund, 1 I 6,7:r:-, 1 IIIa 50:z4,z23:-, 1 Bredeney, 1 Cubana, 1 Grumpensis, 1 Indiana, 1 London, 1 Oslo, 1 Poona.
WGS or PCR for PMQR genes was performed for 72 (62%) of the 117 study-phenotype isolates and 68 (94%) isolates from 34 serotypes had PMQR genes detected (Table 1). The qnr genes detected were qnrB (44), qnrS (20), and qnrA (4). Three Salmonella Litchfield isolates with qnrB genes also had aac(6’)-Ib-cr genes and were ciprofloxacin resistant (MIC 1 μg/mL). No other isolate had >1 PMQR gene detected and no oqxA, oqxB, qepA, qnrC, or qnrD genes were detected. All 13 Salmonella Newport isolates with the study phenotype had qnrB genes, while all Salmonella Bareilly, Saintpaul, and Corvallis isolates tested had qnrS genes. A mix of different qnr gene types was found among serotype Typhimurium, Litchfield, and Stanley isolates. Four isolates tested had no PMQR genes detected. Two of these isolates had QRDR mutations; an Enteritidis isolate with a Gly81Asp mutation in gyrA was ciprofloxacin intermediate (MIC 0.25 μg/mL), while a Salmonella Ituri isolate with this mutation and a Ser80Arg mutation in parC was ciprofloxacin resistant (MIC 1 μg/mL). No other study-phenotype isolates tested had QRDR mutations detected. Two study-phenotype isolates with no quinolone resistance mechanisms detected had ciprofloxacin MICs ≤0.015 μg/mL upon retesting.
Patient Exposures
Travel information was available for 81 (69%) of the 117 patients with study-phenotype isolates. Of these, 24 (30%) patients had a history of recent international travel before illness began; qnr genes were detected in all isolates from the travelers (Table 2). Among 23 travelers with reported travel destinations, 14 (61%) traveled to Asia, 6 (26%) to Latin America or the Caribbean, and 3 (13%) to Africa. Sixteen serotypes were identified among the 24 traveler isolates; Typhimurium was the most common. Isolates from the 14 travelers to Asia had qnrS (10), qnrA (2), and qnrB (2) genes; the most common serotypes among these isolates were Stanley (3), Typhimurium (3), Corvallis (2), and Montevideo (2). All 6 isolates from patients with travel to Latin America or the Caribbean had qnrB genes and each had a different serotype. Two Salmonella Concord isolates, one with qnrA and another with qnrB, were from infants adopted from Ethiopia.
TABLE 2.
Characteristics of nontyphoidal Salmonella isolates with plasmid-mediated quinolone resistance (PMQR) from patients in the United States with international travel, by travel destination, 2008–2014
| Travel region and destination(s) | Serotype | PMQR gene type | Resistance† | Ciprofloxacin/nalidixic acid MIC (μg/mL) | Specimen source | Patient age in years/sex | Year |
|---|---|---|---|---|---|---|---|
| Asia | |||||||
| Malaysia | Typhimurium | qnrS | SSuT | 0.5/16 | Stool | 28/M | 2009 |
| Malaysia | Typhimurium | qnrS | ACSSuTKanCot | 0.5/8 | Stool | 25/F | 2010 |
| Malaysia | Corvallis | qnrS | None | 0.5/16 | Stool | 53/M | 2009 |
| Cambodia | I 4,[5],12:i:- | qnrS | ACSSuTCxTio | 0.25/8 | Stool | 39/F | 2014 |
| Cambodia | Virchow | qnrS | None | 0.5/16 | Stool | 29/M | 2009 |
| Philippines | Stanley | qnrA | SSuTCot | 0.25/16 | Stool | 36/M | 2012 |
| Philippines | Stanley | qnrA | SuTCot | 0.25/8 | Stool | 61/F | 2013 |
| Thailand | Corvallis | qnrS | None | 0.5/16 | Blood | 56/F | 2012 |
| Thailand | Montevideo | qnrS | SuCot | 0.25/4 | Stool | 26/F | 2013 |
| China, Hong Kong, Vietnam | Stanley | qnrS | ACSuTCot | 0.25/4 | Stool | 30/M | 2014 |
| Indonesia | Typhimurium | qnrS | ASu | 0.5/8 | Stool | 7/NA | 2011 |
| Israel | Hadar | qnrB | ST | 0.5/16 | Stool | <1/M | 2012 |
| Taiwan | Montevideo | qnrB | SuTCot | 0.25/16 | Stool | 4/M | 2011 |
| Vietnam‡ | Panama | qnrS | ACSSuTCipGenCotAzm | 1/16 | Blood | 4/M | 2011 |
| Latin America/Caribbean | |||||||
| Mexico | Braenderup | qnrB | None | 0.5/8 | Stool | 42/M | 2012 |
| Mexico§ | Heidelberg | qnrB | None | 0.5/16 | Stool | 6/M | 2012 |
| Mexico | Muenster | qnrB | CSSuTCot | 0.5/16 | Stool | 20/F | 2010 |
| Mexico | Typhimurium | qnrB | CSSuT | 0.5/16 | Stool | 56/M | 2010 |
| Dominican Republic | Derby | qnrB | None | 0.25/16 | Stool | 44/M | 2009 |
| Dominican Republic | Kouka | qnrB | ASSuTGenCot | 0.25/16 | Stool | 55/F | 2009 |
| Africa | |||||||
| Ethiopia | Concord | qnrA | ASSuTCxTioGenCot | 0.25/8 | Stool | <1/F | 2008 |
| Ethiopia | Concord | qnrB | ACSSuTCxTioGenCot | 0.25/8 | Stool | <1/F | 2008 |
| Egypt | Haifa | qnrB | ASSuTCxGenKanCot | 0.25/8 | Stool | 58/F | 2010 |
| Unknown | |||||||
| Unknown¶ | Javiana | qnrS | None | 0.5/16 | Stool | 32/F | 2010 |
A, ampicillin; Azm, azithromycin; C, chloramphenicol; Cip, ciprofloxacin; Cot, trimethoprim-sulfamethoxazole; Cx, ceftriaxone; Gen, gentamicin; Kan, kanamycin; NA, not available; S, streptomycin; Su, sulfisoxazole; T, tetracycline; Tio, ceftiofur.
13 agents in 8 classes were tested in all years: aminoglycosides (Gen, S); β-lactam/β-lactamase inhibitor combinations (amoxicillin-clavulanic acid); cephems (Cx, Tio, cefoxitin); folate pathway inhibitors (Su, Cot); penicillins (A); phenicols (C); quinolones (Cip, nalidixic acid); tetracyclines (T). The aminoglycosides amikacin and kanamycin were tested through 2010 and 2013, respectively. The macrolide Azm was tested in 2011–2014; Azm resistance was defined as MIC ≥32 μg/mL.
Child played with fish in a fishbowl for several hours while in Vietnam.
Child handled turtles in Mexico.
Patient was a refugee; country of origin is unknown.
Seventeen (71%) of the 24 isolates from travelers had resistance to other agents, including trimethoprim-sulfamethoxazole (50%), ampicillin (38%), and ceftriaxone (17%). Among 12 traveler isolates tested for azithromycin resistance, 1 (8%) was resistant. Twelve (50%) isolates from travelers were MDR and 6 (25%) were resistant to ≥5 antimicrobial classes. The median age of travelers was 29.5 years (mean 30, range <1‒61) and 5 (21%) were ≤5 years of age. Among 23 travelers with sex reported, 12 (52%) were male. Salmonella was isolated from stool in 22 (92%) travelers and blood in 2 (8%). Among 21 travelers with this information, 5 (24%) were hospitalized. No deaths were reported among 16 travelers with health outcome information.
Information about reptile and amphibian exposure was available for 67 (57%) of the 117 patients with study-phenotype isolates; 16 (24%) reported this exposure (Table 3). Isolates from the 16 patients were submitted by health departments in 12 states. Twelve serotypes were identified among these isolates; the most common were Litchfield (4) and Telelkebir (2); the others were Apapa, Give, Guinea, Heidelberg, Manhattan, Ouakam, Saintpaul, Urbana, I 4,[5],12:d:-, and IV 44:z4,z23:−. All isolates from patients with reported reptile or amphibian exposure had a qnrB (12) or qnrS (4) gene. Three Salmonella Litchfield isolates with qnrB genes also had aac(6’)-lb-cr genes and were ciprofloxacin resistant (MIC 1 μg/mL), while another Salmonella Litchfield isolate had only a qnrS gene and was ciprofloxacin intermediate (MIC 0.25 μg/mL). Exposures to lizards (bearded dragons, iguanas, geckos, and chameleons), turtles, frogs, and snakes were reported. Five (31%) patients reported exposure to >1 reptile or amphibian type.
TABLE 3.
Characteristics of nontyphoidal Salmonella isolates with plasmid-mediated quinolone resistance (PMQR) from patients with reptile or amphibian exposure, by animal type, United States, 2008–2014
| Reptile/amphibian type(s) | Serotype | PMQR gene type | Resistance† | Ciprofloxacin/nalidixic acid MIC (μg/mL) | Specimen source | Patient age in years/sex | Year |
|---|---|---|---|---|---|---|---|
| Lizards | |||||||
| Bearded dragon | IV 44:z4,z23:- | qnrB | S | 0.5/16 | Stool | <1/F | 2014 |
| Bearded dragon | Apapa | qnrB | None | 0.5/16 | Stool | 22/M | 2014 |
| Bearded dragon | Telelkebir | qnrB | None | 0.5/16 | Stool | 61/F | 2014 |
| Geckos | Litchfield | qnrB, aac(6’)-Ib-cr | SuTCipCot | 1/16 | Stool | 1/M | 2008 |
| Iguana | Give | qnrB | None | 0.5/16 | Stool | 37/F | 2010 |
| Turtles | |||||||
| Turtles‡ | Heidelberg | qnrB | None | 0.5/16 | Stool | 6/M | 2012 |
| Turtle | Litchfield | qnrB, aac(6’)-Ib-cr | SuTCipCot | 1/16 | Stool | 3/F | 2009 |
| Turtle | Ouakam | qnrB | None | 0.5/16 | Urine | 14/F | 2012 |
| Turtles | Telelkebir | qnrB | None | 0.5/16 | Stool | 60/F | 2011 |
| Frogs | |||||||
| Frog | I 4,[5],12:d:- | qnrS | ASuCot | 0.25/4 | Stool | 3/F | 2010 |
| Frog | Manhattan | qnrS | AT | 0.5/16 | Stool | 4/F | 2012 |
| Multiple | |||||||
| Chameleon, snake | Litchfield | qnrS | TCot | 0.25/8 | Stool | 30/F | 2012 |
| Frogs, turtles | Litchfield | qnrB, aac(6’)-Ib-cr | SuTCipCot | 1/8 | Stool | 48/F | 2013 |
| Geckos, iguana, turtles | Saintpaul | qnrS | TCot | 0.5/8 | Stool | 19/M | 2010 |
| Many types of reptiles§ | Guinea | qnrB | None | 0.25/16 | Urine¶ | 50/F | 2014 |
| Lizard#, snake | Urbana | qnrB | None | 0.5/16 | Urine | 20/F | 2014 |
A, ampicillin; Cip, ciprofloxacin; Cot, trimethoprim-sulfamethoxazole; Su, sulfisoxazole; T, tetracycline.
13 agents in 8 classes were tested in all years: aminoglycosides (gentamicin, streptomycin); β-lactam/β-lactamase inhibitor combinations (amoxicillin-clavulanic acid); cephems (ceftriaxone, ceftiofur, cefoxitin); folate pathway inhibitors (Su, Cot); penicillins (A); phenicols (chloramphenicol); quinolones (Cip, nalidixic acid); and tetracyclines (T). The aminoglycosides amikacin and kanamycin were tested through 2010 and 2013, respectively. The macrolide Azm was tested in 2011–2014; Azm resistance was defined as MIC ≥32 μg/mL.
Child handled turtles in Mexico.
Patient ran a reptile rescue operation and had exposure to many types of reptiles.
Patient reported that she did not have diarrhea.
Type not specified.
Eight (50%) of the 16 isolates from patients with reported reptile or amphibian exposure had additional resistance, including trimethoprim-sulfamethoxazole (38%) and ampicillin (13%). Three (19%) isolates were MDR. Unlike isolates from travelers, none had resistance to ceftriaxone or to >3 antimicrobial classes. The median age of patients with reptile or amphibian exposure was 19.5 years (mean 24, range <1‒61) and 5 (31%) patients were ≤5 years. Most (75%) patients were female and 3 (19%) were hospitalized. Patients had stool (81%) or urine (19%) isolates. No deaths were reported among 13 patients with outcome information.
One study-phenotype Salmonella Newport isolate was from a person who became ill after caring for a foal with a Newport infection. The patient’s isolate had a qnrB gene and resistance to agents (including ampicillin, ceftriaxone, and trimethoprim-sulfamethoxazole) in 7 antimicrobial classes.
Three of the 117 surveillance isolates with the study phenotype were from patients epidemiologically linked to a 102-person multistate Salmonella Newport outbreak associated with tomatoes from a few restaurants and a caterer in 2012. All 3 isolates had qnrB genes, the outbreak PFGE pattern, and resistance to amoxicillin-clavulanic acid, ampicillin, cefoxitin, and ceftriaxone. The outbreak strain was isolated from a tomato sampled at one of the restaurants; it had a qnrB gene. The source of the tomatoes was not identified.
Enhanced Outbreak Isolate Testing
We identified 2 additional foodborne outbreaks with study-phenotype isolates by reviewing antimicrobial susceptibility results for isolates submitted to CDC through enhanced testing of outbreak isolates during 2008‒2014. In 2013, a 43-person Salmonella Newport outbreak was associated with a Chinese restaurant and a 4-person Salmonella Muenchen outbreak was associated with a barbeque restaurant. Salmonella strains isolated from patients associated with both outbreaks had qnrB genes.
DISCUSSION
PMQR is emerging among Salmonella causing human infections in the United States. We detected PMQR genes in 68 Salmonella isolates from humans comprising 34 serotypes. Our analysis indicates that infections with Salmonella harboring PMQR genes were acquired both internationally and domestically. Likely sources of domestically-acquired infections were reptile and amphibian exposure and food.
International travel before illness onset has been reported in 9% of persons with laboratory-confirmed nontyphoidal Salmonella infections in the United States (Kendall et al., 2012). It has also been reported as a source of infection with Salmonella having PMQR, particularly travel to Asia (Hopkins et al., 2007; Hopkins et al., 2008; Murray et al., 2008; Sjölund-Karlsson et al., 2009; Sjölund-Karlsson et al., 2010). In our study, 30% of patients with travel information and study-phenotype isolates traveled abroad, mostly to Asia. Although we cannot confirm that all 24 patients in our study with reported international travel were infected abroad, many of the infections were caused by serotypes that are uncommon in the United States, but common in the destinations visited (CDC, 2017; Johnson et al., 2011). For example, serotypes Corvallis and Stanley are endemic in Asia (Hendriksen et al., 2009a; Hendricksen et al.; 2012; Van et al., 2012), and serotype Concord has been reported in Ethiopian adoptees (Hendriksen et al., 2009b).
We previously reported that a high proportion of nalidixic acid-resistant Salmonella Enteritidis infections in the United States are associated with international travel (O’Donnell et al., 2014). In the present study, we found that Enteriditis was the serotype with the greatest number of ciprofloxacin-nonsusceptible isolates, but notably, 98% (183/186) of these isolates were nalidixic acid resistant and only 1% (2/186) had the study phenotype. This is consistent with a recent Canadian study that found Enteritidis had the greatest number of ciprofloxacin-nonsusceptible isolates, but only 4% (2/51) of ciprofloxacin-nonsusceptible isolates had PMQR genes (Kim et al., 2016). Clonal expansion of isolates with QRDR mutations may contribute to high quinolone-resistance levels in this serotype (Kilmartin et al., 2005).
Among patients with study-phenotype isolates, almost one quarter with available information reported reptile or amphibian exposure. In comparison, a 2006–2007 population survey found that 7.4% of respondents reported such exposure (CDC FoodNet, unpub. data), while a 1996–1997 case-control study attributed 6% of all sporadic Salmonella infections to reptile or amphibian contact (Mermin et al., 2004). All patients in our study who reported reptile or amphibian exposure had isolates with PMQR genes and most of these isolates had serotypes that are reptile associated (Ackman et al., 1995; Editorial Team, 2008; Guerra et al., 2010; Whitten et al., 2015); these included several serotypes (e.g., Apapa, Telelkebir, IV 44:z4,z23:-) that are uncommon in humans (CDC, 2017). Three Salmonella Litchfield isolates from these patients were ciprofloxacin resistant and were the only study isolates tested with >1 PMQR gene detected, consistent with reports of higher-level quinolone resistance in strains carrying 2 or more unrelated PMQR genes (Jacoby et al., 2014; Lin et al., 2015; Rodríguez-Martínez et al., 2016).
PMQR has been reported in Salmonella (Guerra et al., 2010; Veldman et al., 2011) and other Enterobacteriaceae (Ahmed et al., 2007; Cortés-Cortés et al., 2016; Unger et al., 2017) from reptiles outside the United States. Interestingly, a study of Salmonella strains isolated from reptiles in Germany detected both qnrB and aac(6’)-Ib-cr genes in a Salmonella Litchfield turtle isolate that had a ciprofloxacin MIC of 1 μg/mL and resistance to trimethoprim-sulfamethoxazole and tetracycline (Guerra et al., 2010), like 3 Litchfield isolates from patients with reptile exposure in our study. More recently, other researchers in Germany reported identifying a number of resistance genes, including qnrS1, mcr-1, blaCTX-M-55, and mph(A) in MDR Escherichia coli isolates from lizards imported from Vietnam (Unger et al., 2017). Reptiles and amphibians may acquire resistant bacteria from their environment, food or water sources, or other animals, and use of antimicrobial agents in these animals may select for resistant strains. In Louisiana, high-level plasmid-mediated gentamicin resistance was found in Salmonella, E. coli, and other bacteria isolated from pet turtle farms that attempted to eradicate Salmonella using gentamicin (Diaz et al., 2006).
We retrospectively identified PMQR in 3 foodborne salmonellosis outbreaks and 1 sporadic infection that was likely acquired from contact with an ill foal. PMQR genes have been found in Salmonella and other bacteria from a variety of animals (wild and domestic) and foods in many countries (Ahmed et al., 2007; Jacoby et al., 2014; Lin et al., 2015; Rodríguez-Martínez et al., 2016; Veldman et al., 2011; Yanat et al., 2017). In the United States, Salmonella with PMQR were recently identified in imported and domestic foods and in food animals. Scientists at the U.S. Food and Drug Administration (FDA) found Salmonella with PMQR in seafood, spices, and other foods imported from Asia and to a lesser extent, from Latin America (Akiyama & Khan, 2012; Bae et al., 2016)(G.H. Tyson, pers. comm.). NARMS scientists identified qnr genes in 2013–2015 Salmonella isolates from 3 retail pork chop samples and cecal samples from multiple swine, 3 cattle, and 1 turkey (FDA, 2017; Tyson et al., 2017). PMQR was also identified in isolates linked to a 2015 MDR Salmonella I 4,[5],12:i:- outbreak in Wisconsin associated with pork consumption (Elbadawi et al., 2016) and a 2015–2017 multistate, MDR Salmonella Heidelberg outbreak associated with exposure to dairy bull calves (CDC NARMS, unpub. data). Fluoroquinolones are approved for the treatment and control of certain infections in cattle and pigs in the United States, but their use is prohibited in poultry (FDA, n.d.). Extralabel fluoroquinolone use has been prohibited in food animals in the United States since 1997 (FDA, n.d.) and fluoroquinolone drug approvals for chickens and turkeys were withdrawn by FDA in 2001 and 2005 because they caused fluoroquinolone-resistant Campylobacter infections in humans (Karp et al., 2017).
We identified 2 study-phenotype isolates with a rare gyrA mutation (Gly81Asp) reported to confer greater resistance to fluoroquinolones than to nalidixic acid, unlike most QRDR mutations (Cattoir et al., 2006; Hopkins et al., 2005); one of these isolates also had a parC mutation (Ser80Arg). A French study reported the co-occurrence of these 2 mutations in a clinical E. coli isolate from a patient treated with ofloxacin; the isolate had a ciprofloxacin MIC of 1 μg/mL and nalidixic acid susceptibility (Cattoir et al., 2006), similar to a Salmonella Ituri isolate with the same mutations in our study.
Our study has several limitations. We did not capture all isolates with PMQR because we excluded isolates with nalidixic acid resistance and those with a ciprofloxacin MIC ≤0.12 μg/mL, which could harbor PMQR genes. As a result, we could not calculate the prevalence of PMQR among Salmonella isolated from humans and we may have excluded some PMQR-containing isolates with multiple quinolone resistance mechanisms. Because isolates tested by PCR or WGS were not randomly selected, we do not know if the 45 study-phenotype isolates that we did not test had the same high prevalence of PMQR as those tested. We also did not perform susceptibility testing at the same time as WGS and PCR. Plasmid loss likely occurred in 2 isolates with no quinolone-resistance mechanisms detected; they had the study phenotype upon initial but not subsequent testing. Additionally, our study was descriptive and did not include a comparison group. We relied on patient information previously collected by health departments, and exposure and outcome information was incomplete. Information on reptile and amphibian exposures was more complete in later years.
The emergence of PMQR among Salmonella in the United States is of public health concern because it has the potential to spread rapidly, lead to high-level quinolone resistance, and make infections harder to treat (Gay et al., 2006; Jacoby et al., 2014; Robicsek et al., 2006). Moreover, PMQR genes are often located on transferable plasmids with other resistance determinants and use of a single drug can select for MDR strains (Gay et al., 2006; Robicsek et al., 2006). The use of antimicrobial agents is one of the main factors driving resistance (CDC, 2013), therefore, judicious use of quinolones and other antimicrobial agents in humans and animals is critically important for preserving their effectiveness. Following safe food handling practices; consuming food and water from safe sources, particularly while traveling internationally; and following public health recommendations regarding the proper handling of reptiles and other animals will help prevent both susceptible and resistant infections. The expanded use of WGS can facilitate characterization of specific genes, mobile elements, and mutations conferring quinolone resistance among Salmonella from humans, animals, and foods. Comparing molecular data for isolates from various sources and linking them to detailed exposure and antimicrobial use information may help identify factors that contribute to the spread of quinolone resistance and target prevention measures.
Supplementary Material
Impacts.
Plasmid-mediated quinolone resistance (PMQR) is emerging among nontyphoidal Salmonella causing human infections in the United States.
International travel, reptile and amphibian exposure, and food are likely sources of human infection with Salmonella having PMQR.
PMQR may facilitate the spread of quinolone resistance, lead to higher-level quinolone resistance, and make infections harder to treat.
Acknowledgments
We thank state and local health departments for their ongoing participation in NARMS and for collecting and sharing with us public health investigation information for salmonellosis cases and outbreaks. We also thank our colleagues in CDC’s National Outbreak Reporting System Team and Outbreak Response and Prevention Branch for providing outbreak investigation information; colleagues in CDC’s FoodNet Team for providing population survey data; and NARMS colleagues at FDA and the Food Safety and Inspection Service of the U.S. Department of Agriculture for testing and providing information about food and animal isolates. Additionally, we acknowledge FDA; the University of California, Davis; and the 100K Pathogen Genome Project for sequencing a few isolates included in this study.
Footnotes
Conflict of interest
The authors have no conflicts of interest to declare.
Disclaimer: The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Contributor Information
Beth E. Karp, Centers for Disease Control and Prevention, Division of Foodborne, Waterborne, and Environmental Diseases, Atlanta, GA, USA
Davina Campbell, Oak Ridge Institute for Science and Education, Oak Ridge, TN, USA.
Jessica C. Chen, IHRC Inc., Atlanta, GA, USA
Jason P. Folster, Centers for Disease Control and Prevention, Division of Foodborne, Waterborne, and Environmental Diseases, Atlanta, GA, USA
Cindy R. Friedman, Centers for Disease Control and Prevention, Division of Foodborne, Waterborne, and Environmental Diseases, Atlanta, GA, USA
References
- Aarestrup FM, Wiuff C, Mølbak K, & Threlfall EJ (2003). Is it time to change fluoroquinolone breakpoints for Salmonella spp.? Antimicrobial Agents and Chemotherapy, 47, 827–829. 10.1128/AAC.47.2.827-829.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackman DM, Drabkin P, Birkhead G, & Cieslak P (1995). Reptile-associated salmonellosis in New York State. Pediatric Infectious Diseases Journal, 14, 955–959. [DOI] [PubMed] [Google Scholar]
- Ahmed AM, Motoi Y, Sato M, Maruyama A, Watanabe H, Fukumoto Y, & Shimamoto T (2007). Zoo animals as reservoirs of gram-negative bacteria harboring integrons and antimicrobial resistance genes. Applied and Environmental Microbiology, 73, 6686–6690. 10.1128/AEM.01054-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama T, & Khan AA (2012). Isolation and characterization of small qnrS1-carrying plasmids from imported seafood isolates of Salmonella enterica that are highly similar to plasmids of clinical isolates. FEMS Immunology & Medical Microbiology, 64, 429–432. 10.1111/j.1574-695X.2011.00921.x. [DOI] [PubMed] [Google Scholar]
- Albornoz E, Tijet N, De Belder D, Gomez S, Martino F, Corso A,…Petroni A (2017). qnrE1, a member of a new family of plasmid-located quinolone resistance genes, originated from the chromosome of Enterobacter species. Antimicrobial Agents and Chemotherapy, 61, e02555–16. 10.1128/AAC.02555-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae D, Kweon O, & Khan AA (2016). Isolation and characterization of antimicrobial-resistant nontyphoidal Salmonella enterica serovars from imported food products. Journal of Food Protection, 79, 1348–1354. 10.4315/0362-028X.JFP-15-564. [DOI] [PubMed] [Google Scholar]
- Cattoir V, Lesprit P, Lascols C, Denamur E, Legrand P, Soussy CJ, & Cambau E (2006). In vivo selection during ofloxacin therapy of Escherichia coli with combined topoisomerase mutations that confer high resistance to ofloxacin but susceptibility to nalidixic acid. Journal of Antimicrobial Chemotherapy, 58, 1054–1057. 10.1093/jac/dkl361. [DOI] [PubMed] [Google Scholar]
- Cavaco LM, & Aarestrup FM (2009). Evaluation of quinolones for use in detection of determinants of acquired quinolone resistance, including the new transmissible resistance mechanisms qnrA, qnrB, qnrS, and aac(6’)Ib-cr, in Escherichia coli and Salmonella enterica and determinations of wild-type distributions. Journal of Clinical Microbiology, 47, 2751–2758. 10.1128/JCM.00456-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. (2013). Antibiotic resistance threats in the United States, 2013. Atlanta, GA: U.S. Department of Health and Human Services, CDC; http://www.cdc.gov/drugresistance/threat-report-2013/. [Google Scholar]
- Centers for Disease Control and Prevention. (2015). National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): human isolates final report, 2013. Atlanta, GA: U.S. Department of Health and Human Services, CDC; http://www.cdc.gov/narms/reports/index.html. [Google Scholar]
- Centers for Disease Control and Prevention. (2016). National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): human isolates final report, 2014. Atlanta, GA: U.S. Department of Health and Human Services, CDC; http://www.cdc.gov/narms/reports/index.html. [Google Scholar]
- Centers for Disease Control and Prevention. (2017). National enteric disease surveillance: Salmonella annual report, 2014. Atlanta, GA: U.S. Department of Health and Human Services, CDC; https://www.cdc.gov/nationalsurveillance/salmonella-surveillance.html. [Google Scholar]
- Cortés-Cortés G, Lozano-Zarain P, Torres C, Castañeda M, Sánchez GM, Alonso CA,…Rocha-Gracia RC (2016). Detection and molecular characterization of Escherichia coli strains producers of extended-spectrum and CMY-2 type beta-lactamases, isolated from turtles in Mexico. Vector-Borne Zoonotic Diseases, 16, 595–603. 10.1089/vbz.2014.1725. [DOI] [PubMed] [Google Scholar]
- Crump JA, Barrett TJ, Nelson JT, & Angulo FJ (2003). Reevaluating fluoroquinolone breakpoints for Salmonella enterica serotype Typhi and for non-Typhi salmonellae. Clinical Infectious Diseases, 37, 75–81. 10.1086/375602. [DOI] [PubMed] [Google Scholar]
- Díaz MA, Cooper RK, Cloeckaert A, & Siebeling RJ (2006). Plasmid-mediated high-level gentamicin resistance among enteric bacteria isolated from pet turtles in Louisiana. Applied and Environmental Microbiology, 72, 306–312. 10.1128/AEM.72.1.306-312.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Editorial Team, Bertrand S, Rimhanen-Finne R, Weill FX, Rabsch W, Thornton L,…Heck M (2008). Salmonella infections associated with reptiles: the current situation in Europe. Eurosurveillance, 13(24). pii: 18902 http://www.eurosurveillance.org/content/10.2807/ese.13.24.18902-en. [PubMed] [Google Scholar]
- Elbadawi LI, DeSalvo T, Klos R, Monson T, Warshauer D, Folster JP,…Davis JP (2016). Multidrug-resistant Salmonella serovar I 4,[5],12:i:- infections associated with pork consumption - Wisconsin, 2015. In: Abstracts of the 65th Annual Epidemic Intelligence Service (EIS) Conference, May 2–5, 2016 Atlanta, GA: Centers for Disease Control and Prevention. https://www.cdc.gov/eis/conference/archives.html#anchor_1521471026916. [Google Scholar]
- Food and Drug Administration. (n.d.). Extralabel use and antimicrobials. Rockville, MD: U.S. Department of Health and Human Services, FDA; https://www.fda.gov/animalveterinary/safetyhealth/antimicrobialresistance/ucm421527.htm (accessed 23 January 2018). [Google Scholar]
- Food and Drug Administration. (2017). NARMS Now. Rockville, MD: U.S. Department of Health and Human Services, FDA; https://www.fda.gov/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/NationalAntimicrobialResistanceMonitoringSystem/ucm570685.htm (accessed 23 January 2018). [Google Scholar]
- Gay K, Robicsek A, Strahilevitz J, Park CH, Jacoby G, Barrett T,…Hooper DC (2006). Plasmid-mediated quinolone resistance in non-Typhi serotypes of Salmonella enterica. Clinical Infectious Diseases, 46, 297–304. 10.1086/505397. [DOI] [PubMed] [Google Scholar]
- Guerra B, Helmuth R, Thomas K, Beutlich J, Jahn S, & Schroeter A (2010). Plasmid-mediated quinolone resistance determinants in Salmonella spp. isolates from reptiles in Germany. Journal of Antimicrobial Chemotherapy, 65, 2043–2045. 10.1093/jac/dkq242. [DOI] [PubMed] [Google Scholar]
- Hendriksen RS, Bangtrakulnonth A, Pulsrikarn C, Pornruangwong S, Noppornphan G, Emborg HD, & Aarestrup FM (2009a). Risk factors and epidemiology of the ten most common Salmonella serovars from patients in Thailand: 2002–2007. Foodborne Pathogens and Disease, 6, 1009–1019. 10.1089/fpd.2008.0245. [DOI] [PubMed] [Google Scholar]
- Hendriksen RS, Le Hello S, Bortolaia V, Pulsrikarn C, Nielsen EM, Pornruangmong S,…Aarestrup FM (2012). Characterization of isolates of Salmonella enterica serovar Stanley, a serovar endemic to Asia and associated with travel. Journal of Clinical Microbiology. 50, 709–720. 10.1128/JCM.05943-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriksen RS, Mikoleit M, Kornschober C, Rickert RL, Duyne SV, Kjelsø C,…Aarestrup FM (2009b). Emergence of multidrug-resistant Salmonella Concord infections in Europe and the United States in children adopted from Ethiopia, 2003–2007. Pediatric Infectious Diseases Journal, 28, 814–818. 10.1097/INF.0b013e3181a3aeac. [DOI] [PubMed] [Google Scholar]
- Hooper DC, & Jacoby GA. (2015). Mechanisms of drug resistance: quinolone resistance. Annals of the New York Academy of Sciences, 1354, 12–31. 10.1111/nyas.12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins KL, Davies RH, & Threlfall EJ (2005). Mechanisms of quinolone resistance in Escherichia coli and Salmonella: recent developments. International Journal of Antimicrobial Agents, 25, 358–373. 10.1016/j.ijantimicag.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Hopkins KL, Day M, & Threlfall EJ (2008). Plasmid-mediated quinolone resistance in Salmonella enterica, United Kingdom. Emerging Infectious Diseases, 14, 340–342. 10.3201/eid1402.070573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins KL, Wootton L, Day MR, & Threlfall EJ (2007). Plasmid-mediated quinolone resistance determinant qnrS1 found in Salmonella enterica strains isolated in the UK. Journal of Antimicrobial Chemotherapy, 59, 1071–1075. 10.1093/jac/dkm081. [DOI] [PubMed] [Google Scholar]
- Humphries RM, Fang FC, Aarestrup FM, & Hindler JA (2012). In vitro susceptibility testing of fluoroquinolone activity against Salmonella: recent changes to CLSI standards. Clinical Infectious Diseases, 55, 1107–1113. 10.1093/cid/cis600. [DOI] [PubMed] [Google Scholar]
- Hunt M, Mather AE, Sánchez-Busó L, Page AJ, Parkhill J, Keane JA, & Harris SR (2017). ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. bioRxiv. 10.1101/118000. [DOI] [PMC free article] [PubMed]
- Jacoby GA, Strahilevitz J, & Hooper DC (2014). Plasmid-mediated quinolone resistance. Microbiology Spectrum. 2(5): PLAS-0006–2013. 10.1128/microbiolspec.PLAS-0006-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LR, Gould LH, Dunn JR, Berkelman R, & Mahon BE (2011). Salmonella infections associated with international travel: a Foodborne Diseases Active Surveillance Network (FoodNet) study. Foodborne Pathogens and Disease, 8, 1031–1037. 10.1089/fpd.2011.0854. [DOI] [PubMed] [Google Scholar]
- Karp B,E., Tate H, Plumblee JR, Dessai U, Whichard JM, Thacker EL,…McDermott PF (2017). National Antimicrobial Resistance Monitoring System: Two decades of advancing public health through integrated surveillance of antimicrobial resistance. Foodborne Pathogens and Disease, 14, 545–557. 10.1089/fpd.2017.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall ME, Crim S, Fullerton K, Han PV, Cronquist AB, Shiferaw B,…Mahon BE (2012). Travel-associated enteric infections diagnosed after return to the United States, Foodborne Diseases Active Surveillance Network (FoodNet), 2004–2009. Clinical Infectious Diseases, 54 Suppl 5, S480–S487. 10.1093/cid/cis052. [DOI] [PubMed] [Google Scholar]
- Kilmartin D, Morris D, O’Hare C, Corbett-Feeney G, & Cormican M (2005). Clonal expansion may account for high levels of quinolone resistance in Salmonella enterica serovar Enteritidis. Applied and Environmental Microbiology, 71, 2587–2591. 10.1128/AEM.71.5.2587-2591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Han X, Bae J, Chui L, Louie M, Finley R,…Jeon B (2016). Prevalence of plasmid-mediated quinolone resistance (PMQR) genes in non-typhoidal Salmonella strains with resistance and reduced susceptibility to fluoroquinolones from human clinical cases in Alberta, Canada, 2009–13. Journal of Antimicrobial Chemotherapy, 71, 2988–2990. 10.1093/jac/dkw232. [DOI] [PubMed] [Google Scholar]
- Lin D, Chen K, Wai-Chi Chan E, & Chen S (2015). Increasing prevalence of ciprofloxacin-resistant food-borne Salmonella strains harboring multiple PMQR elements but not target gene mutations. Scientific Reports, 5, 14754 10.1038/srep14754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermin J, Hutwagner L, Vugia D, Shallow S, Daily P, Bender J,…Angulo FJ (2004). Reptiles, amphibians, and human Salmonella infection: a population-based, case-control study. Clinical Infectious Diseases, 38 Suppl 3, S253–S261. 10.1086/381594. [DOI] [PubMed] [Google Scholar]
- Murray A, Mather H, Coia JE, & Brown DJ (2008). Plasmid-mediated quinolone resistance in nalidixic-acid-susceptible strains of Salmonella enterica isolated in Scotland. Journal of Antimicrobial Chemotherapy, 62, 1153–1155. 10.1093/jac/dkn340. [DOI] [PubMed] [Google Scholar]
- O’Donnell AT, Vieira AR, Huang JY, Whichard J, Cole D, & Karp BE (2014). Quinolone-resistant Salmonella enterica serotype Enteritidis infections associated with international travel. Clinical Infectious Diseases, 59, e139–e141. 10.1093/cid/ciu505. [DOI] [PubMed] [Google Scholar]
- Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, & Barrett TJ (2006). Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathogens and Disease, 3, 59–67. 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- Robicsek A, Jacoby GA, & Hooper DC (2006). The worldwide emergence of plasmid-mediated quinolone resistance. The Lancet Infectious Diseases, 6, 629–640. 10.1016/S1473-3099(06)70599-0. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Martínez JM, Machuca J, Cano ME, Calvo J, Martínez-Martínez L, & Pascual A (2016). Plasmid-mediated quinolone resistance: Two decades on. Drug Resistance Updates, 29, 13–29. 10.1016/j.drup.2016.09.001. [DOI] [PubMed] [Google Scholar]
- Sjölund-Karlsson M, Folster JP, Pecic G, Joyce K, Medalla F, Rickert R, & Whichard JM (2009). Emergence of plasmid-mediated quinolone resistance among non-Typhi Salmonella enterica isolates from humans in the United States. Antimicrobial Agents and Chemotherapy, 53, 2142–2144. 10.1128/AAC.01288-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjölund-Karlsson M, Howie R, Rickert R, Krueger A, Tran TT, Zhao S,…McDermott PF (2010). Plasmid-mediated quinolone resistance among non-Typhi Salmonella enterica isolates, USA. Emerging Infectious Diseases, 16, 1789–1791. 10.3201/eid1611.100464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strockbine NA, Bopp CA, Fields PI, Kaper JB, & Nataro JP (2015). Escherichia, Shigella, and Salmonella In Jorgensen JH, Pfaller MA, Carroll KC, Funke G, Landry ML, Richter SS, & Warnock DW Manual of clinical microbiology, 11th ed. (pp. 685–713). Washington DC: ASM Press. [Google Scholar]
- Tyson GH, Tate HP, Zhao S, Li C, Dessai U, Simmons M, McDermott PF (2017). Identification of plasmid-mediated quinolone resistance in Salmonella isolated from swine ceca and retail pork chops in the United States. Antimicrobial Agents and Chemotherapy, 22, e01318–17. 10.1128/AAC.01318-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger F, Eisenberg T, Prenger-Berninghoff E, Leidner U, Ludwig ML, Rothe M,… Ewers C (2017). Imported reptiles as a risk factor for the global distribution of Escherichia coli harbouring the colistin resistance gene mcr-1. International Journal of Antimicrobial Agents, 49, 122–123. 10.1016/j.ijantimicag.2016.10.007. [DOI] [PubMed] [Google Scholar]
- Van TT, Nguyen HN, Smooker PM, & Coloe PJ (2012). The antibiotic resistance characteristics of non-typhoidal Salmonella enterica isolated from food-producing animals, retail meat and humans in South East Asia. International Journal of Food Microbiology, 154, 98–106. 10.1016/j.ijfoodmicro.2011.12.032. [DOI] [PubMed] [Google Scholar]
- Veldman K, Cavaco LM, Mevius D, Battisti A, Franco A, Botteldoorn N,…Aarestrup FM (2011) International collaborative study on the occurrence of plasmid-mediated quinolone resistance in Salmonella enterica and Escherichia coli isolated from animals, humans, food and the environment in 13 European countries. Journal of Antimicrobial Chemotherapy, 66, 1278–1286. 10.1093/jac/dkr084. [DOI] [PubMed] [Google Scholar]
- Whitten T, Bender JB, Smith K, Leano F, & Scheftel J (2015). Reptile-associated salmonellosis in Minnesota, 1996–2011. Zoonoses and Public Health, 62, 199–208. 10.1111/zph.12140. [DOI] [PubMed] [Google Scholar]
- World Health Organization. (2017). Critically important antimicrobials for human medicine, 5th revision 2016. http://www.who.int/foodsafety/publications/antimicrobials-fifth/en/.
- Yanat B, Rodríguez-Martínez JM, & Touati A (2017). Plasmid-mediated quinolone resistance in Enterobacteriaceae: a systematic review with a focus on Mediterranean countries. European Journal of Clinical Microbiology & Infectious Diseases, 36, 421–435. 10.1007/s10096-016-2847-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.