A new transcriptome-wide method produces reliable m6A maps at single-base resolution and reveals site conservation in mammals.
Abstract
N6-methyladenosine (m6A) is one of the most abundant messenger RNA modifications in eukaryotes involved in various pivotal processes of RNA metabolism. The most popular high-throughput m6A identification method depends on the anti-m6A antibody but suffers from poor reproducibility and limited resolution. Exact location information is of great value for understanding the dynamics, machinery, and functions of m6A. Here, we developed a precise and high-throughput antibody-independent m6A identification method based on the m6A-sensitive RNA endoribonuclease recognizing ACA motif (m6A-sensitive RNA-Endoribonuclease–Facilitated sequencing or m6A-REF-seq). Whole-transcriptomic, single-base m6A maps generated by m6A-REF-seq quantitatively displayed an explicit distribution pattern with enrichment near stop codons. We used independent methods to validate methylation status and abundance of individual m6A sites, confirming the high reliability and accuracy of m6A-REF-seq. We applied this method on five tissues from human, mouse, and rat, showing that m6A sites are conserved with single-nucleotide specificity and tend to cluster among species.
INTRODUCTION
Among more than 100 RNA chemical modifications, N6-methyladenosine (m6A) is one of the most abundant forms in eukaryotic mRNA, accounting for 0.1 to 0.4% of all adenosine (1). The m6A modification influences various stages of mRNA metabolism and the biogenesis of long noncoding RNA and microRNA, with effects across diverse biological processes including neuronal development, cell fate transition, immune response, DNA damage response, and tumorigenesis (2–10). Most m6A sites were found in conserved motif DRACH (D=G/A/U, R=G/A, H=A/U/C) (1), with an estimated methylation ratio ranging from 6 to 80% (11). Previous whole-transcriptome m6A maps have suggested that m6A modifications are enriched around stop codon, implying the functional importance of distribution pattern for m6A (12–14).
The chemical properties of m6A are similar to adenosine, making it difficult to discriminate by chemical reactions (15). The recently developed high-sensitivity liquid chromatography–tandem mass spectrometry and blotting methods relying on antibodies were widely used to quantify the overall m6A level. To detect individual sites, methyl-sensitive ligase has been applied to confirm the methylation status of specific adenosines (16, 17), while the method called SCARLET (site-specific cleavage and radioactive-labeling followed by ligation-assisted extraction and thin-layer chromatography) can quantify the methylation level of individual m6A site (11). Other methods were developed to identify m6A in single-base resolution during reverse transcription, taking advantage of m6A-sensitive reverse transcriptase (18, 19), chemoenzymatic substitution of the N6-methyl group (20), or selective dTTP (deoxythymidine triphosphate) analog such as 4SedTTP (21). Along with the rapid progress of single-molecule sequencing technologies, Oxford Nanopore Technologies sequencing platform is able to detect modifications on model RNA (22), although the systematic error prevents the practical application in biological samples. Nevertheless, comprehensive interrogating of m6A at the transcriptome level is pivotal to reveal the biological importance of this mRNA modification. Methylated RNA immunoprecipitation and sequencing (MeRIP-seq or m6A-seq) has been widely used to profile m6A, identifying approximate region with m6A in ~100 nucleotide (nt) length, while the exact location of individual m6A site remains undetermined (12, 13). Many refined methods have been developed to improve resolution, such as PA-m6A-seq, miCLIP, and m6A-CLIP (23–26). However, all of these methods are dependent on m6A-specific antibodies, suffering from poor reproducibility and complicated process. In addition, affinity variation and batch effects of antibodies make it difficult to quantify the methylation level (27). Therefore, a convenient and single-base resolution method is still needed for whole-transcriptome m6A identification and quantification, advancing the comprehension of m6A for its dynamics and cellular functions in posttranscriptomic regulation.
Several DNA endonucleases belonging to the restriction-modification system have demonstrated sensitivity to DNA methylation. For instance, Dpn I specifically cleaves methylated sites, while Dpn II is blocked by the same modified sequence. This feature has been adopted in genome-wide studies to detect the DNA 6mA modification in single-base resolution (6mA-RE-seq/DA-6mA-seq) (28). For RNA endoribonucleases, a large group of sequence-specific enzymes belonging to the bacterial type II toxin-antitoxin system has been found with determined cleavage motifs. Recently, an Escherichia coli toxin and RNA endoribonuclease, MazF, was reported to be sensitive to m6A modification within ACA motif, specifically cleaving the unmethylated ACA motif, leaving methylated (m6A)CA motifs intact (29). By screening the endoribonuclease pool, we identified ChpBK as another m6A-sensitive endoribonuclease, which can discriminate m6A-modified motifs from unmodified sequences. Taking advantage of the m6A-sensitive endoribonucleases, we developed m6A-sensitive RNA-endoribonuclease–facilitated sequencing method or m6A–REF-seq, which can identify transcriptomic m6A sites at specific motifs and quantify the methylation level in single-base resolution. To validate the m6A sites identified by this method, we used a ligation-based method testing individual sites and confirmed the high-reliability results. Further analysis revealed distribution pattern of m6A in single-nucleotide resolution. Last, we applied m6A-REF-seq to five tissues from human, mouse, and rat, revealing the conservation of m6A at both single-base and regional levels across diverse tissues and species.
RESULTS
Identification and quantification of m6A by endoribonuclease
The application of methylation-sensitive DNA endonuclease in genome-wide 6mA identification inspired us to inspect the possibility of finding similar tools for m6A determination (28). To discover endoribonuclease with m6A sensitivity, we expressed and screened the candidate enzymes by testing the cleavage capacity to synthetic RNA oligonucleotides (RNA oligos) with or without m6A. Two enzymes, MazF and ChpBK, both belonging to the bacterial toxin-antitoxin system, were able to distinguish m6A from unmethylated A. MazF recognized and cleaved the motif sequence ACA from the 5′ side of first A, leaving the methylated (m6A)CA motif intact (Fig. 1A). The cleavage activity of ChpBK was also blocked by m6A at the recognition motif UAC (Fig. 1B). Synthetic RNA oligos with different m6A/A ratios were used to simulate the partially methylated sites on mRNA. The cleavage assay demonstrated that the fraction of digested oligo versus intact part was proportional to the methylation ratio, implying that MazF has the potential to quantify the methylation level (Fig. 1, C and D, and fig. S1).
Fig. 1. m6A identification method based on m6A sensitive RNA endoribonuclease.
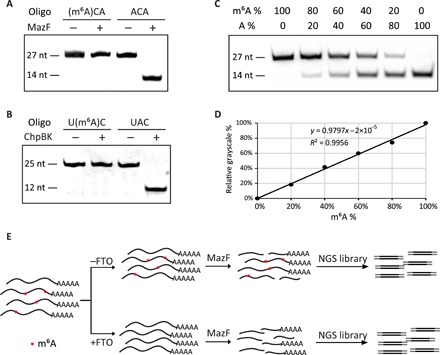
(A) Validation for the methylation sensitivity of endoribonuclease MazF by synthetic m6A-containing RNA oligonucleotide. (B) Validation for the methylation sensitivity of endoribonuclease ChpBK. (C) Various ratios of m6A-containing oligo mixed with unmethylated oligo digested by endoribonuclease. The m6A/A ratios of synthetic oligo are indicated to imitate practical RNAs with different m6A ratios. (D) Relative grayscale of proportionally mixed RNA oligo digested by endoribonuclease. (E) The schematic diagram of m6A-REF-seq.
The high sensitivity and specificity of selected endoribonuclease that enzymatically distinguished m6A urged us to develop a new high-throughput method to identify single-base m6A at the transcriptome level. Since the MazF recognition motif ACA is more prevalent than that of ChpBK among known m6A sites, we chose MazF in the following studies. In general, mRNA from human embryonic kidney (HEK) 293T cells was digested into fragments by MazF. After end repair and purification, the mRNA fragments were ligated to next-generation sequencing (NGS) adaptors and amplified by polymerase chain reaction (PCR). To prevent potential false positives caused by unknown factors, we also included a parallel sample batch demethylated by fat mass and obesity-associated protein (FTO) in advance as the negative control (fig. S2). We call this method m6A-REF-seq, which stands for m6A-sensitive RNA-Endoribonuclease-Facilitated sequencing (Fig. 1E).
Transcriptome-wide identification of m6A sites in single-base resolution
We developed a pipeline to analyze the high-throughput sequencing data generated by m6A-REF-seq (Fig. 2A). As we anticipated, most reads contained the ACA at the 5′ terminus, in line with the effect of MazF treatment (fig. S3). The intact ACA motif sequence observed internally within an RNA fragment was supposed to be methylated. We verified this using the known m6A site with ACA motif on the 18S ribosomal RNA (rRNA) as an example. The unmethylated rRNA fragments were cleaved on the ACA motif, and the methylated fragments were retained (Fig. 2B). Next, we scanned each ACA motif within the transcriptome and interrogated the methylation sites by calculating the reads harboring intact internal ACA motifs. To further eliminate the potential false positive caused by RNA structure, we predicted the probability of each RNA fragment forming secondary structure according to minimum free energy (30) and removed the candidate sites tending to reside in double-stranded regions. We performed three biological replicates and retained the recurrent candidate sites detected in at least two replicates (Fig. 2C and table S1). For each site, the ratio of sequence reads with internal ACA motif versus reads split at the end represents the relative methylation ratio. We compared the methylation ratio of each candidate site in MazF treatment to the sample treated with FTO in advance, which was considered as demethylated RNA. Most of m6A sites had marked reductions in methylation level with FTO treatment (Fig. 2D). Only candidate sites with a substantial decrease of methylation ratio in FTO treatment were considered as accurate m6A sites. In total, we identified 4260 high-confidence m6A sites for further studies.
Fig. 2. Transcriptome-wide distribution of m6A revealed by m6A-REF-seq.
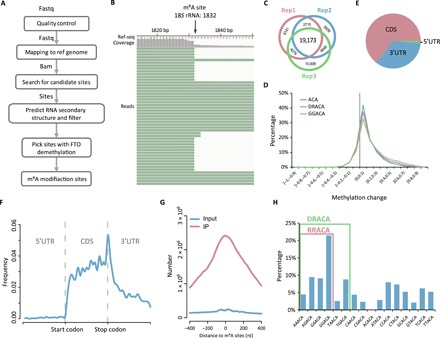
(A) The analysis pipeline of m6A-REF-seq data. (B) The snapshot of sequencing reads shows a known m6A site in 18S rRNA. (C) Overlap sites among three replicates after removing RNA secondary structure. (D) The overall shift of methylation ratio after FTO treatment. The value of methylation change indicates the methylation ratio of each m6A site in MazF minus that after FTO treatment. (E) Transcriptome-wide distribution of m6A. Pie chart shows the percentages of m6A sites located within CDS, 5′UTR, and 3′UTR. (F) Single-base m6A sites from m6A-REF-seq show a typical transcriptome-wide distribution pattern of m6A. (G) Overlap of m6A sites to the m6A peaks identified by antibody-based method. (H) The proportion of motifs containing m6A sites. The red square includes the RRACA motif and the green square includes the DRACA motif.
About 65% of these sites were located in the coding sequence (CDS) region, and others were in untranslated regions (UTRs) (Fig. 2E). The transcriptome-wide distribution showed strong enrichment surrounding the stop codon (Fig. 2F), in line with the typical pattern found by antibody-dependent methods such as MeRIP-seq or m6A-seq. To compare the m6A location with MeRIP-seq in detail, we downloaded the profiling data deposited in Gene Expression Omnibus (GEO) database (GSE29714) (12) and calculated the cumulated reads distribution of input and IP (immunoprecipitation) samples within the range of −400 to 400 nt flanking to every m6A site identified by m6A-REF-seq. The reads from IP samples were markedly concentrated in this region, with a peak summit at almost the exact position of individual m6A sites; whereas reads from input sample did not show any enrichment, implying that the m6A sites identified by m6A-REF-seq coincided well with the m6A peaks reported by MeRIP-seq (Fig. 2G). By expanding the context sequences flanking m6A sites, we found that GGACA was the most enriched consensus motif. More generally, the consensus motifs DRACA and RRACA were overrepresented, accounting for 56 and 45% of all identified m6A sites, respectively (Fig. 2H). These results were in concordance with previous studies, in which DRACH/RRACH was the most preferred m6A motif in native mRNA (12, 13, 24).
High reliability of individual m6A sites identified by m6A-REF-seq
To show the reliability of m6A-REF-seq as a new method, we sought another independent principle to validate individual m6A sites. The T3 ligase was reported to be sensitive to m6A sites during the ligation reaction, which had been applied practically to determine m6A sites in mRNA (17). We adopted this method and designed the probe L (left) and probe R (right) based on the sequence flanking the exact site (table S2) and then used the T3 ligase to concatenate the two probes to an integrated template, which could be amplified by PCR.According to previous result, the ligation efficiency at m6A site was significantly suppressed comparing to that at unmethylated A sites (17), and PCR amplification magnified the difference of ligation efficiency (fig. S4). Thus, the amount of PCR products could be assessed on gel to represent the ligation efficiency, implying the methylation status of the interrogated site (Fig. 3A). We also applied the same protocol on FTO-demethylated mRNA as negative control. In total, we picked nine m6A sites for validation, including eight m6A sites and one unmethylated site. Six of eight m6A sites were validated by the ligase-based method, while the unmethylated site was also confirmed (Fig. 3B and fig. S5). To the best of our knowledge, this is the first batch of individual m6A sites confirmed by two independent methods. One validated m6A site (chr1: 29070177; Fig. 3B) is located at 343 nt upstream from the stop codon of m6A reader protein YTHDF2, suggesting a self-regulating feedback loop mediated by RNA modification.
Fig. 3. Single-base validation using ligation-amplification method and qPCR.
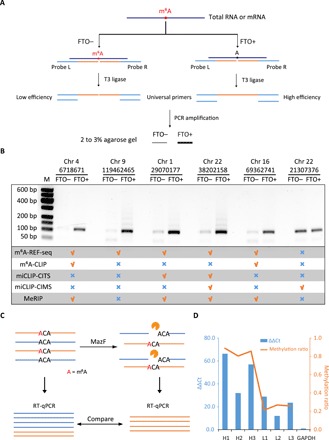
(A) Schematic diagram of ligation-amplification method for single-base m6A validation. (B) Validation results of six individual sites. Five of the sites are validated to be m6A sites, whereas the other one is confirmed to be not modified. Data for m6A-CLIP, miCLIP-CITS, miCLIP-CIMS, and MeRIP-seq are downloaded from published literatures. (C) Schematic diagram of qPCR to quantify the methylation level of a specific m6A site. The undigested mRNA sample is treated as control. (D) qPCR results and methylation ratios of six m6A sites. H1 to H3 represent the highly methylated sites (>0.75), while L1 to L3 represent the weakly methylated sites (<0.35). The left y axis represents the ΔΔCt values, and the right y axis represents the methylation ratio determined by m6A-REF-seq. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Two other antibody-dependent methods, m6A-CLIP and miCLIP, had reported comprehensive m6A maps at transcriptome level. We checked whether the six individual m6A sites could be identified by these two methods. Peaks from MeRIP-seq data were also included as reference. Of the six sites, one of them was not reported in either previous method. The negative control site that supposed to be unmethylated was reported by miCLIP as an m6A site but not by other methods (Fig. 3B). We suspect that differences in samples, as well as the dynamic nature of m6A methylation, contribute to these inconsistencies and potential false positives. Nevertheless, these results underline the need of independent methods to confirm authentic m6A sites for downstream analysis, especially considering m6A as a highly dynamic RNA modification.
Quantification of m6A abundance by m6A-REF-seq
m6A-REF-seq not only determines the methylation status but also can be used to quantify the methylation level of each m6A site by calculating the ratio of sequence reads with internal ACA versus reads split at the motif. To verify this, we designed a digestion-quantification assay that used reverse transcription quantitative PCR (RT-qPCR) to evaluate the methylation level of identified m6A sites. First, mRNA was treated by MazF. The unmethylated sites located in ACA motif were cleaved, leaving intact methylated sites. After reverse transcription, complementary DNA (cDNA) spanning the intended m6A site was amplified by PCR to estimate the relative abundance. The same amount of mRNA without MazF digestion was amplified simultaneously as a control to compare with the digested sample (Fig. 3C). We picked six m6A sites including three sites with high methylation level and three sites with low methylation level determined by m6A-REF-seq (table S3). The ΔΔCt values calculated from RT-qPCR results were comparable to the average methylation ratio determined by m6A-REF-seq (Fig. 3D and fig. S6), demonstrating that m6A-REF-seq can principally quantify the methylation fraction of m6A sites.
Characterizing m6A distribution pattern in single-nucleotide resolution
Previous studies demonstrated that one of the most remarkable features of m6A was the distribution enrichment surrounding stop codon (12, 13). However, limited by the resolution of antibody-based methods, it was hard to know the precise position relative to the exact stop codon site, which divides coding regions and 3′UTRs. The single-nucleotide m6A identification method largely increased the resolution of m6A map. Our results showed that this stop codon–concentrated pattern could be reproduced by using single-nucleotide m6A sites, without any preference to coding regions or 3′UTRs (Fig. 4A). Meanwhile, it was worth noting that consensus motifs DRACA and DRACH were also enriched near the stop codon, suggesting that sequence motif might be one factor to shape this pattern (Fig. 4A).
Fig. 4. Single-base method reveals high resolution features of m6A.
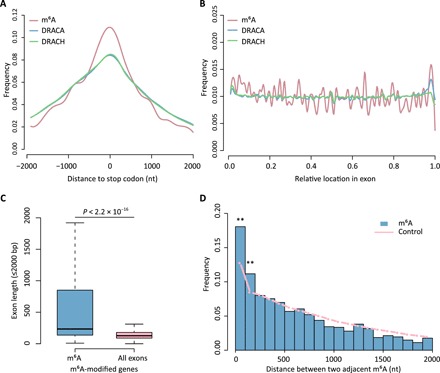
(A) The distance of individual m6A sites to the stop codons. DRACA and DRACH background motifs are extracted from the transcriptome. (B) The relative position of m6A sites within exons. (C) The length distribution of m6A-containing exons versus all exons in m6A-modified genes. (D) The distances between two m6A sites are significantly enriched within 200 bp regions compared to random sampling (Fisher’s exact test, **P < 0.01).
m6A had been proposed to be involved in various key regulatory processes of RNA metabolisms, including splicing (31). We calculated the relative distances of m6A sites to intron-exon boundaries in each exon and plotted in Fig. 4B. The result showed m6A distributed evenly along exon regions, with slight depletion toward the 3′ end. By mapping single m6A sites to exons, we compared the lengths of exons containing m6A modifications versus all exons. The lengths of m6A-containing exons were significantly greater than all exons (P < 2.2 × 10−16, Wilcoxon test; Fig. 4C), in line with previous report that m6A modification usually occurred within long exons (13).
Some indirect evidence showed that m6A tended to locate in short regions, suggesting a specialized working model of m6A as clusters. However, antibody-based methods could barely divide multiple sites from one m6A peak to demonstrate clustering. Using the single-nucleotide m6A positions identified by m6A-REF-seq, we calculated the distances between each two adjacent m6A sites and compared to the control dataset, which contained the same number of sites randomly sampled from transcriptome 1000 times. Our results showed that two adjacent m6A sites were statistically prone to aggregate within 200-nt regions (P = 6.6 × 10−5 and P = 0.0065, Fisher’s exact test), supporting the hypothesis that m6A functions as clusters of modification (Fig. 4D).
Conservation of m6A sites in mammals
We next applied m6A-REF-seq to identify m6A modifications in different tissues of human, mouse, and rat and identified more than 3500 individual m6A sites in each sample (tables S4 and S5). The transcriptomic distribution displayed similar patterns among tissues in three species (Fig. 5A and figs. S7 to S9). To examine the conservation of m6A at the individual gene level, we first collected 13,117 one-to-one orthologs in three species. By mapping the m6A sites to these ortholog genes in each type of tissues, we identified 2439 human genes, 3596 mouse genes, and 1343 rat genes in the brain, among which 571 of them were shared in three species. The numbers of shared genes were 536 and 262 in the kidney and the liver, respectively (figs. S8 and S9). The ortholog genes modified by m6A in all three species were significantly more than expected even after adjusting for gene expression (Fig. 5B, fig. S10, and table S6), indicating a well-conserved distribution of m6A at the gene level. Similarly, previous studies have reported that the count of shared m6A peaks between human and mouse were significantly higher than expected, proposing the conservation and important functions of m6A modification (13).
Fig. 5. Conservation of m6A in mammals.
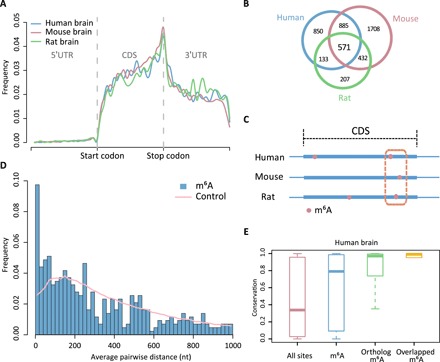
(A) Metagene plots of m6A in the brain of human, mouse, and rat. (B) Shared m6A-modified genes among three species. (C) Diagram showing the m6A sites conserved in the corresponding short regions from different species. (D) Frequency of distances for pairwise m6A in brain. Randomly picked ACA motifs are assigned for the same analysis as control. (E) Conservation scores of all m6A sites, methylation sites in ortholog genes, and conserved m6A sites are compared to that for all A sites in ACA motifs (P < 9 × 10−10, Wilcoxon test).
Since m6A-REF-seq identified high-confidence sites, we were able to investigate the conservation of m6A at single-nucleotide level. Previous work analyzed the evolutionary conservation of yeast and human m6A sites in protein-coding regions and concluded that m6A were overall not conserved in yeasts and only slightly conserved in mammals (32). However, the m6A sites used in this work were predicted based on the consensus motif DRACH, which could contain underestimated false positives. Therefore, we mapped the single-nucleotide m6A sites identified by m6A-REF-seq to orthologous genes in three species (table S6). By using similar method (32), we tested whether m6A site were more conserved than unmethylated A. We randomly sampled the human unmethylated A sites as the same number of m6A sites in each orthologous gene and tested whether the corresponding sites in mouse and rat remaining to be base A (fig. S11). A total of 1854 to 3722 m6A sites were compared to the same number of unmethylated A sites from 1286 to 2306 genes across three tissues. The results indicated that m6A sites were more conserved than unmethylated A sites in brain, kidney, and liver tissues for all three codon positions (P = 0.002, 0.021, and 0.075, respectively; figs. S11 to S13), confirming the m6A conservation at single-nucleotide level.
Furthermore, we tested whether the nonpairwise m6A sites tended to cluster in narrow corresponding regions among species. The shortest distances between pairwise m6A sites in ortholog genes were averaged (Fig. 5C). Random sampling of the same number of ACA motifs were performed for 1000 times as control. The average distances between pairwise m6A sites were shorter than the control (Fig. 5D, and figs. S8 and S9), indicating that, although some m6A sites were not conserved at single-nucleotide level, they tended to cluster within conserved short regions across multiple species.
More extensively, we computed the PhastCons scores representing the conservation of each nucleotide among multiple species (33). Identified m6A sites in the brain were attributed and classified into sites conserved in three species or sites residing in ortholog genes. As expected, conserved m6A sites at single-base level and sites conserved at gene level had highest conservation scores (medians 0.991 and 0.970, respectively). The scores of overall m6A sites were also distinctively higher than that of all A sites in ACA motifs with the medians 0.791 and 0.338, respectively (P < 9 × 10−10, Wilcoxon test; Fig. 5E). Similar results hold for other tissues in human, mouse, and rat (figs. S8, S9, and S14).
DISCUSSION
Antibody-dependent pull-down methods are widely used to profile transcriptomic m6A, the most prevalent modification in mRNA. An antibody-independent, easy-to-use method is strongly desired to cross-validate the known m6A sites and study the dynamic functions in single-nucleotide resolution. We developed m6A-REF-seq, an antibody-independent, high-throughput, and single-base m6A detection method based on m6A-sensitive RNA endoribonuclease. High sensitivity and specificity of endoribonuclease that we found to discriminate m6A modification, as well as stringent data process by removing potential RNA secondary structure and the introducing of FTO demethylation control, ensured reliability of m6A sites identified by this method. Transcriptome-wide m6A maps in human HEK293T cell and mammalian tissues displayed similar distribution pattern to previous antibody-based pull-down methods. To validate the m6A sites identified by m6A-REF-seq, we introduced another independent method, which used T3 ligase to differentiate individual m6A or A at template RNA. The high consistency between different methods demonstrated the efficiency of m6A-REF-seq with high confidence. We also applied qPCR to show that m6A-REF-seq could quantify the methylation level of individual m6A site.
The RNA endoribonuclease MazF has high sensitivity and specificity for RNA substrates. It works very well on extremely limited substrate, as little as nanograms or even picograms. Rapid and simplified experiment design, without the need for antibody-enrichment step, largely reduces the requirement of starting RNA amount and sample preparation time. Therefore, this method could be applied to the studies for rare biological materials such as samples from pathological tissues or early embryos. In addition, m6A-REF-seq offers the potential to capture the subtle changes of m6A during metabolic processes, advancing the dynamic studies of this posttranscriptional RNA modification in different life stages.
By screening RNA endoribonucleases, we found two m6A-sensitive enzymes from the bacterial type II toxin-antitoxin system, which recognized specific motifs “ACA” and “UAC,” respectively. To cover all the RRACH motifs in the transcriptome, we need new enzyme(s) capable of recognizing more universal sequence motifs while retaining the m6A sensitivity. On the basis of our preliminary screening, we speculated that the m6A sensitivity might be a general feature of endoribonucleases belonging to type II TA system. Further exploring in the endoribonucleases pool would be helpful to expand the toolbox for RNA modification studies. Another strategy is to transform the existing enzyme to adapt for new recognition sequences while retaining the methylation sensitivity. We studied the structure of MazF protein and mutated the 56th lysine to alanine. The MazF-K56A mutant showed irregular recognition sequences not meeting our anticipation, although the methyl sensitivity was retained (fig. S15). On the other hand, directed evolution would be a promising way to expand the recognition sequence scope of MazF or other related proteins, as long as dedicated screening system was created.
We applied m6A-REF-seq to five different tissues from human, mouse, and rat. The single-base m6A maps displayed a distribution pattern similar to previous findings that m6A tended to enrich surrounding stop codons. This unique pattern seemed to be conserved among different species. However, because of limited resolution, previous methods could not tell to what extent the modification is conserved. In this study, we mapped the m6A sites to orthologous genes in these three species and compared the conservation score at single-nucleotide level. We found that significantly more m6A-modified genes were shared among species than expected, and a large group of m6A sites could be aligned to the syntenic regions, showing that the m6A modification was conserved at both the gene and individual base level. The well-conserved feature restated the biological importance of m6A, implying other uncovered roles of this RNA modification.
Together, m6A-REF-seq provides a new perspective for single-base m6A identification at the transcriptome level. The new principle eliminates the need for m6A-specific antibodies, making it unique and amenable for future applications in small and precious samples. The features of this efficient and convenient method will largely expand the scope of m6A studies.
MATERIALS AND METHODS
RNA materials
The RNA oligos synthesized from Takara Bio Inc. and Sangon Biotech were as follows: RNA-m6ACA (5′-UUGGUUUUUUUUGG(m6A)CAUGUAUAUAGU-3′), RNA-ACA (5′-UUGGUUUUUUUUGGACAUGUAUAUAGU-3′), RB1-Um6AC (5′-GUUGUGUGAUAU(m6A)CAUAUGGUGGUG-3′), and RB1-UAC (5′ GUUGUGUGAUAUACAUAUGGUGGUG-3′).
HEK293T cell line was cultured with Dulbecco’s modified Eagle’s medium (DMEM; Corning) supplemented with 10% (v/v) fetal bovine serum (FBS; Gibco) at 37°C in the presence of 5% CO2 and tested to be mycoplasma negative. Total RNA from HEK293T cells was extracted using TRIzol reagent (Thermo Fisher Scientific), and the mRNA was purified using the Dynabeads mRNA Purification Kit (Thermo Fisher Scientific, catalog no. 61006). The mRNA of human tissues (brain, liver, and kidney), mouse tissues (brain, liver, heart, testis, and kidney), and rat tissues (brain, liver, and kidney) were purchased from Takara Bio Inc.
RNA endoribonuclease validation for MazF
RNA oligos (10 pmol) were incubated with 2.5 U of MazF (mRNA intereferase MazF, Takara Bio Inc., code no. 2415A) in the 20-μl reaction mixture of MazF buffer [40 mM sodium phosphate (pH 7.5) and 0.01% Tween 20] at 37°C for 30 min. The RNA oligos were mixed with the m6A percentage of 0, 20, 40, 60, 80, and 100%, and the oligo mixtures were incubated with 2.5 U of MazF in the 20-μl reaction mixtures. The samples were loaded into 15% urea polyacrylamide gel and electrophoresed in 0.5× tris-boric acid–EDTA (TBE) buffer.
The FTO demethylation reaction was conducted in the reaction mixture that contained 10 pmol of m6A RNA oligo, 0 to 10 μg of FTO demethylase, 283 μM (NH4)2Fe(SO4)2∙6H2O, 300 μM α-ketoglutarate (α-KG), 2 mM l-ascorbic acid, 20 U of RNase inhibitor (Takara Bio Inc., code no. 2313A), and 50 mM tris-HCl buffer (pH 7.5). The reaction was stopped by 40 mM EDTA after 3 hours of incubation at room temperature. The MazF cleavage reaction was performed to test the efficiency of FTO demethylation.
Dot-blot assay of FTO demethylation
m6A FTO demethylase with different concentrations (0, 0.25, and 2.5 μg) was used in the dot-blot assay to test the demethylation of RNA. The total RNA was incubated with FTO, 283 μM (NH4)2Fe(SO4)2∙6H2O, 300 μM α-KG, and 2 mM l-ascorbic acid for 3 hours at room temperature. After purification by an RNA Clean & Concentrator-5 kit (Zymo Research), the concentration of total RNA was determined by the Qubit RNA HS Assay Kit (Thermo Fisher Scientific) and diluted to 10-μl volume with 500 ng. Spotting and thermal cross-linking were carried out on a metal base plate preheated to 70°C. After thermal cross-linking for 15 min, ultraviolet cross-linking was conducted twice with 2000 kJ. Then, 3% bovine serum albumin (BSA) was used to block at room temperature for 1 hour. The anti-m6A antibody (Synaptic Systems, catalog no. 202003) was diluted to 1:2000 and incubated with the nylon membrane for 2 hours. The nylon membrane was washed for three times with 1% tris-buffered saline with Tween 20 (TBST) and then incubated with the 1:5000 diluted anti-rabbit antibody for 1 hour. The membrane was washed for five times with 1% TBST. After draining off the water, the chemiluminescence detection signal was performed using the enhanced chemiluminesence (ECL) system. The membrane was washed twice with 1% TBST. After draining off the water, it was dyed with methylene blue solution for 8 min and then washed with 1% TBST. The signal was detected by a white light system.
Expression and purification of recombinant ChpBK protein
To express recombinant ChpBK protein [National Center for Biotechnology Information (NCBI reference sequence: NP_418646.1], E. coli BL21(DE3) strain carrying PET-28a-chpBK plasmid was grown in lysogeny broth (LB) medium (Sangon Biotech, A507002) supplemented with Kan (10 μg/ml) at 37°C and 200 rpm until the optical density at 600 nm (OD600) reach to 0.5. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.4 mM, and cell growth resumed for 2 hours under the same conditions. Harvested cell pellets were lysed by sonication in 30 ml of buffer A [10 mM bis-tris (pH 7.0) and 30 mM imidazole] and centrifuged for 20 min at 16,000g and 4°C. The soluble fraction was incubated on ice for 30 min in the presence of DNase and RNase (10 g/ml; Thermo Fisher Scientific). The His-ChpBK fusion protein was purified from clarified lysates using Ni-NTA (nickel-nitrilotriacetic acid) beads (Invitrogen). Protein purity was determined by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) analysis, and samples were quantified using the Bio-Rad protein assay and stored at −80°C.
Expression and purification of MazF-K56A in E. coli
The 56th lysine of MazF protein was mutated to alanine. The protein sequence was shown below, where the underlined bold “A” indicates the mutated amino acid: MazF-K56A (MVSRYVPDMGDLIWVDFDPTKGSEQAGHRPAVVLSPFMYNNKTGMCLCVPCTTQSAGYPFEVVLSGQERDGVALADQVKSIAWRARGATKKGTVAPEELQLIKAKINVLIG).
The plasmid DNA pCOLD-MazF was transformed into E. coli Rossetta strain, and a single colony was picked by sterilized loop and inoculated into a 50-ml culture (containing ampicillin) in sterile culture flask. E. coli was cultured at 37°C until OD600 reached 0.6 to 0.8. IPTG (1 mM) was added to induce protein expression, and cells grew overnight at 18°C. Cells were harvested by centrifugation and suspended in 200 ml of lysis buffer [50 mM tris (pH 8.0), 350 mM NaCl, and 10 mM imidazole]. Cell pellets were lysed by sonication, and the supernatant was collected by centrifugation and applied to nickel affinity purification using HisTrap HP. The column was washed by lysis buffer to remove nonspecific binding protein, and target protein was eluted with linear gradient of elution buffer [50 mM tris (pH 8.0), 350 mM NaCl, and 500 mM imidazole]. We concentrated the protein and changed protein storage buffer to ion-exchange binding buffer [20 mM Hepes and 50 mM NaCl (pH 7.5)] by HiTrap Desalting column. Tobacco etch virus (TEV) protease was used to remove the His-tag, and target protein was loaded to HiTrap HP Q column. Target protein was eluted by linear gradient of ion-exchange elution buffer [20 mM Hepes and 1 M NaCl (pH 7.5)]. The concentrated protein was analyzed by SDS-PAGE.
RNA endoribonuclease validations for ChpBK and MazF-K56A
The digestion reaction of ChpBK was conducted in a 20-μl reaction mixture with 10 pmol of RNA oligo (RB1), 1 μg of ChpBK, 20 U of RNase inhibitor, and 10 mM tris-HCl buffer (pH 7.5). The mixture was incubated at 37°C for 30 min. The digestion reaction of ChpBK with different m6A percentage was conducted as described above. The MazF-K56A cleavage reaction was performed in a 20-μl reaction mixture with 10 pmol of RNA oligo, 2 μg of MazF-K56A, and 1× MazF buffer [40 mM sodium phosphate (pH 7.5) and 0.01% Tween 20] at 37°C for 30 min. All samples were loaded on the 15% urea polyacrylamide gel and electrophoresed in 0.5× TBE buffer.
Library construction for NGS
mRNA was first heated at 85°C for 5 min and chilled on ice for 2 min to denature RNA secondary structure. The FTO demethylation reaction was conducted before MazF reaction in a 20-μl reaction mixture containing 100 to 200 ng mRNA, 2.5 μg of FTO demethylase, 283 μM (NH4)2Fe(SO4)2∙6H2O, 300 μM α-KG, 2 mM l-ascorbic acid, 20 U of RNase inhibitor, and 50 mM tris-HCl buffer (pH 7.5). After incubating at room temperature for 3 hours, the reaction was stopped by adding 40 mM EDTA. To prevent the influence of FTO buffer, the MazF treatment was supplemented with the same reaction mixture except FTO protein. The MazF and FTO-MazF samples were heated at 85°C for 5 min, chilled on ice for 2 min, and then incubated with 10 U of MazF in the MazF reaction buffer at 37°C for 30 min. The fragmented mRNA was purified by an RNA Clean & Concentrator-5 kit (Zymo Research) and eluted into 20 μl of RNase-free water. The eluted RNA fragment was end repaired by the T4 polynucleotide kinase (T4PNK; Vazyme Biotech) in 50-μl reaction mixture with 1× T4PNK buffer and incubated at 37°C for 30 min. After purification by an RNA Clean & Concentrator-5 kit (Zymo Research), the concentration of fragmented mRNA was measured by the Qubit RNA HS Assay Kit (Thermo Fisher Scientific). The NGS library was constructed by using the VAHTS Small RNA Library Prep Kit (Vazyme Biotech, NR801) with 10 ng of fragmented mRNA and amplified by PCR for 15 cycles. Libraries were loaded on the 8% urea polyacrylamide gel and electrophoresed in 0.5× TBE buffer, and purified libraries were sequenced on the Illumina HiSeq X10 platform. Three replicates have been conducted for mRNA of HEK293T cell line. The same protocol was also conducted for mammalian tissues.
NGS data analyses
First, quality control was performed on raw sequencing reads by the open software FastQC (www.bioinformatics.babraham.ac.uk/projects/fastqc/). Then, adaptors were filtered by cutadapt (34) with at least 15 nt remaining length of paired-end reads. The clean reads were mapped to the reference genome (hg19) using HISAT2 (35) with default parameters, except reporting only one result. Integrative Genomics Viewer was used for data visualization (36). We scanned the motif ACA (and the reverse complementary motif TGT) on the reference genome and mapping result file, respectively, counting the number of ACA/TGT at the internal or terminal of a sequence read. As short sequence reads with lengths less than 15 nt were filtered, only ACA/TGT with the distance from the end of a read more than 15 nt was treated as “internal” of a read. Each ACA/TGT motif on the reference genome, which has more than 10 reads supporting the motif located internal of reads, was treated as a candidate m6A site. The ratio of sequence reads with internal ACA versus reads split at the end represents the relative methylation ratio of each site. Candidate sites with repeated or continuous ACA motifs were removed.
As the RNA secondary structure could affect the reaction efficiency, we predicted the probability of each RNA fragment forming secondary structure and removed the candidate sites tending to reside in double-stranded regions. The RNAfold program of ViennaRNA package (30) was used to predict intramolecular secondary structure of each read, which supported the candidate sites. The condensed representation of the pair probabilities of each nucleotide was parsed according to the tutorials of ViennaRNA package. The pairing probability value of NNACA motif was calculated, and candidate sites with high pairing probability were discarded. The FTO demethylation treatment was also treated as a negative control. Candidate sites with at least 10% methylation ratio decrease were remained as m6A sites. The metagene plot of m6A sites, which was described the relative location of each site on the 3′UTR, CDS, and 5′UTR of mRNA, was calculated and plotted as well. The raw reads of MeRIP-seq datasets were downloaded from Meyer et al. (12) (GSE29714) and mapped to human (hg19) reference genome using HISAT2. The reads within the range of −400 to 400 nt flanking to every m6A site were plotted. As the sequencing data were single end, we extended the reads to 200 nt to simulate the IP peak.
Single-base validation using ligase-based method
Refined single-base T3 ligase–based method (17) was used to validate m6A sites identified by m6A-REF-seq. The FTO demethylation sample was used as negative control (i.e., the FTO+ treatment in Fig. 3A). DNA probe L and probe R were designed to match the flanking sequences of intended site. Probe L was modified with a phosphate at 5′ terminal. Probe R was modified with two ribonucleotides at 3′ end. The ligation reaction mixture A consisted of 20 nM probe L, 20 nM probe R, 1× T3 ligation buffer [New England Biolabs (NEB)], and 300 ng of total RNA with or without FTO treatment. The ligation reaction mixture B consisted of appropriate amount of T3 DNA ligase (NEB) with ligation buffer. Mixture A was heated at 85°C for 3 min and then incubated at 35°C for 10 min, and then the ligation reaction mixture B was added to the final volume of 10 μl. This mixture was incubated at room temperature for ligation reaction for 8 to 10 min and chilled on ice immediately. The volume of 1 μl of ligation product was used to PCR amplified for 24 cycles in the 2× Taq mix (Vazyme Biotech). The samples were loaded into the 2 to 3% agarose gel and electrophoresed in 0.5× tris-acetate-EDTA (TAE) buffer to detect the signal. The previous m6A-CLIP and miCLIP datasets were downloaded from Ke et al. (25) and Linder et al. (24), respectively.
qPCR validation for m6A quantification
The MazF treatment was performed in 10-μl volume with 500 ng of mRNA and 10 U of MazF at 37°C for 30 min. The digested mRNA and nondigested mRNA were subjected to reverse transcription using 5× TransScript All-in-One First-Strand cDNA Synthesis SuperMix for qPCR (One-Step gDNA Removal, TransGen Biotech, AT341-02). TB Green Premix Ex Taq II (Tli RNaseH Plus, Takara Bio Inc., code no. RR820A) was used on BIO-CFX96 Touch.
Characterizing m6A distribution pattern in single-base resolution
Consensus motifs DRACA and DRACH were scanned on the reference genome, and the distances of A sites in the two motifs to stop codons were calculated and plotted as well. The relative position in exon for m6A, regular A sites in DRACA, and DRACH was computed. To estimate the distance between two adjacent m6A sites, only genes with more than two m6A sites were analyzed. We randomly sampled the same number of ACA motifs as the number of observed m6A and repeated this step for 1000 times as control. The distances between adjacent A sites in control were calculated. Fisher’s exact test was used to test the significance of first two bins.
Conservation of m6A in the same mammal tissues
m6A-REF-seq protocol was used to identify m6A sites in the different tissues of human (brain, liver, and kidney), mouse (brain, liver, heart, testis, and kidney), and rat (brain, liver, and kidney). The one-to-one orthologous genes in pairwise species of human (GRCh37.p13), mouse (GRCm38.p6), and rat (Rnor_6.0) were downloaded from Ensembl (Ensembl genes 94). A total of 13,117 one-to-one orthologous genes among three species were retained for downstream analyses. Protein sequences of 13,117 orthologs in human, mouse, and rat were aligned by MUSCLE (37) and then converted to corresponding coding sequence alignments using PAL2NAL (38). The shared m6A-modified genes and share m6A sites among species were calculated and tested for significances based on the well-aligned orthologous coding-region sequences as mentioned above. The average pairwise distances among closely m6A sites in three species were calculated and compared with random control sample. The control sample was randomly picked for the same number of ACA motifs as the number of observed m6A for 1000 times. Analyses of conservation score were conducted by using PhastCons scores of human (hg19), mouse (mm10), and rat (rn6) downloaded from University of California, Santa Cruz (UCSC).
Supplementary Material
Acknowledgments
Funding: This work was supported by the Ministry of Science and Technology of China (National Science and Technology Major Project, grant no. 2018YFA0109100), National Science Foundation of China (grant nos. 91753129 and 31870808), and Natural Science Foundation of Guangdong Province (grant nos. 2018B030306044). Author contributions: Zhang Zhang, C.H., and G.Z.L. conceived the project. Zhang Zhang, L.-Q.C., and Y.-L.Z. conducted the experiments. Zhang Zhang analyzed the NGS data. Zijie Zhang, I.A.R., C.-G.Y., J.R., and W.X. provided the supporting materials and ideas. Zhang Zhang, L.-Q.C., C.H., and G.-Z.L. wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript. Competing interests: C.H. is a scientific founder and a member of the scientific advisory board of Accent Therapeutics, Inc. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. Data were deposited in NCBI’s GEO under accession number GSE125240.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/7/eaax0250/DC1
Fig. S1. Quantitative demonstration of various fractions of m6A-containing oligo mixed with unmethylated oligo digested by ChpBK.
Fig. S2. FTO demethylate assay.
Fig. S3. The base composition of mRNA products cleaved by MazF.
Fig. S4. The single-site validation for 18S rRNA control site and m6A site.
Fig. S5. The single-site validation of eight m6A sites and one unmethylated site.
Fig. S6. Quantitative PCR results of six m6A sites.
Fig. S7. Metagene plots of m6A in mouse heart and mouse testis.
Fig. S8. Conservation of m6A in mammalian kidney.
Fig. S9. Conservation of m6A in mammalian liver.
Fig. S10. Gene expression and the expected number of shared m6A-modified genes.
Fig. S11. Evolutionary conservation between methylated and unmethylated A sites in the same genes of brain tissue.
Fig. S12. Evolutionary conservation between methylated and unmethylated A sites in the same genes of kidney tissue.
Fig. S13. Evolutionary conservation between methylated and unmethylated A sites in the same genes of liver tissue.
Fig. S14. Conservation scores for brain.
Fig. S15. Validation for the methylation sensitivity of mutated MazF-K56A.
Table S1. Basic information of sequencing data for human HEK293T cell line.
Table S2. Designed probes and universal primers for T3 ligase–based validation.
Table S3. Designed primers for Quantitative PCR validation.
Table S4. Basic information of sequencing data for mammalian tissues.
Table S5. m6A sites identified by m6A-REF-seq for mammalian tissues.
Table S6. Shared m6A-modified genes and m6A sites between pairwise species and the significance.
REFERENCES AND NOTES
- 1.Fu Y., Dominissini D., Rechavi G., He C., Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 15, 293–306 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Barbieri I., Tzelepis K., Pandolfini L., Shi J., Millán-Zambrano G., Robson S. C., Aspris D., Migliori V., Bannister A. J., Han N., De Braekeleer E., Ponstingl H., Hendrick A., Vakoc C. R., Vassiliou G. S., Kouzarides T., Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature 552, 126–131 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batista P. J., Molinie B., Wang J., Qu K., Zhang J., Li L., Bouley D. M., Lujan E., Haddad B., Daneshvar K., Carter A. C., Flynn R. A., Zhou C., Lim K.-S., Dedon P., Wernig M., Mullen A. C., Xing Y., Giallourakis C. C., Chang H. Y., m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15, 707–719 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gokhale N. S., McIntyre A. B. R., McFadden M. J., Roder A. E., Kennedy E. M., Gandara J. A., Hopcraft S. E., Quicke K. M., Vazquez C., Willer J., Ilkayeva O. R., Law B. A., Holley C. L., Garcia-Blanco M. A., Evans M. J., Suthar M. S., Bradrick S. S., Mason C. E., Horne S. M., N6-Methyladenosine in Flaviviridae Viral RNA Genomes Regulates Infection. Cell Host Microbe 20, 654–665 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lence T., Akhtar J., Bayer M., Schmid K., Spindler L., Ho C. H., Kreim N., Andrade-Navarro M. A., Poeck B., Helm M., Roignant J.-Y., m6A modulates neuronal functions and sex determination in Drosophila. Nature 540, 242–247 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Patil D. P., Chen C.-K., Pickering B. F., Chow A., Jackson C., Guttman M., Jaffrey S. R., m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369–373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiang Y., Laurent B., Hsu C.-H., Nachtergaele S., Lu Z., Sheng W., Xu C., Chen H., Ouyang J., Wang S., Ling D., Hsu P.-H., Zou L., Jambhekar A., He C., Shi Y., RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature 543, 573–576 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y., Li Y., Yue M., Wang J., Kumar S., Wechsler-Reya R. J., Zhang Z., Ogawa Y., Kellis M., Duester G., Zhao J. C., N6-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat. Neurosci. 21, 195–206 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winkler R., Gillis E., Lasman L., Safra M., Geula S., Soyris C., Nachshon A., Tai-Schmiedel J., Friedman N., Le-Trilling V. T. K., Trilling M., Mandelboim M., Hanna J. H., Schwartz S., Stern-Ginossar N., m6A modification controls the innate immune response to infection by targeting type I interferons. Nat. Immunol. 20, 173–182 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Vu L. P., Pickering B. F., Cheng Y., Zaccara S., Nguyen D., Minuesa G., Chou T., Chow A., Saletore Y., MacKay M., Schulman J., Famulare C., Patel M., Klimek V. M., Garrett-Bakelman F. E., Melnick A., Carroll M., Mason C. E., Jaffrey S. R., Kharas M. G., The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 23, 1369–1376 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu N., Parisien M., Dai Q., Zheng G., He C., Pan T., Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA 19, 1848–1856 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer K. D., Saletore Y., Zumbo P., Elemento O., Mason C. E., Jaffrey S. R., Comprehensive analysis of mRNA methylation reveals enrichment in 3’UTRs and near stop codons. Cell 149, 1635–1646 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M., Sorek R., Rechavi G., Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Schwartz S., Agarwala S. D., Mumbach M. R., Jovanovic M., Mertins P., Shishkin A., Tabach Y., Mikkelsen T. S., Satija R., Ruvkun G., Carr S. A., Lander E. S., Fink G. R., Regev A., High-Resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell 155, 1409–1421 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yue Y., Liu J., He C., RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 29, 1343–1355 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao Y., Wang Y., Tang Q., Wei L., Zhang X., Jia G., An elongation and ligation-based qPCR amplification method for the radiolabeling-free detection of locus-specific N6-methyladenosine modifications. Angew. Chem. 57, 15995–16000 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Liu W., Yan J., Zhang Z., Pian H., Liu C., Li Z., Identification of a selective DNA ligase for accurate recognition and ultrasensitive quantification of N6-methyladenosine in RNA at one-nucleotide resolution. Chem. Sci. 9, 3354–3359 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harcourt E. M., Ehrenschwender T., Batista P. J., Chang H. Y., Kool E. T., Identification of a selective polymerase enables detection of N6-methyladenosine in RNA. J. Am. Chem. Soc. 135, 19079–19082 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang S., Wang J., Zhang X., Fu B., Song Y., Ma P., Gu K., Zhou X., Zhang X., Tian T., Zhou X., N6-Methyladenine hinders RNA- and DNA-directed DNA synthesis: Application in human rRNA methylation analysis of clinical specimens. Chem. Sci. 7, 1440–1446 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shu X., Dai Q., Wu T., Bothwell I. R., Yue Y., Zhang Z., Cao J., Fei Q., Luo M., He C., Liu J., N6-Allyladenosine: A New Small Molecule for RNA Labeling Identified by Mutation Assay. J. Am. Chem. Soc. 139, 17213–17216 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong T., Yuan Y., Chen Z., Xi K., Wang T., Xie Y., He Z., Su H., Zhou Y., Tan Z.-J., Weng X., Zhou X., Precise antibody-independent m6A identification via 4SedTTP-involved and FTO-assisted strategy at single-nucleotide resolution. J. Am. Chem. Soc. 140, 5886–5889 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Garalde D. R., Snell E. A., Jachimowicz D., Sipos B., Lloyd J. H., Bruce M., Pantic N., Admassu T., James P., Warland A., Jordan M., Ciccone J., Serra S., Keenan J., Martin S., McNeill L., Wallace E. J., Jayasinghe L., Wright C., Blasco J., Young S., Brocklebank D., Juul S., Clarke J., Heron A. J., Turner D. J., Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods 15, 201–206 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Chen K., Lu Z., Wang X., Fu Y., Luo G.-Z., Liu N., Han D., Dominissini D., Dai Q., Pan T., He C., High-resolution N6-methyladenosine (m6A) map using photo-crosslinking-assisted m6A sequencing. Angew. Chem. Int. Ed. 54, 1587–1590 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linder B., Grozhik A. V., Olarerin-George A. O., Meydan C., Mason C. E., Jaffrey S. R., Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12, 767–772 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ke S., Alemu E. A., Mertens C., Gantman E. C., Fak J. J., Mele A., Haripal B., Zucker-Scharff I., Moore M. J., Park C. Y., Vågbø C. B., Kusśnierczyk A., Klungland A., Darnell J. E. Jr., Darnell R. B., A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev. 29, 2037–2053 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X., Xiong X., Yi C., Epitranscriptome sequencing technologies: Decoding RNA modifications. Nat. Methods 14, 23–31 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Molinie B., Wang J., Lim K. S., Hillebrand R., Van Wittenberghe N., Howard B. D., Daneshvar K., Mullen A. C., Dedon P., Xing Y., Giallourakis C. C., M6A-LAIC-seq reveals the census and complexity of the m6A epitranscriptome. Nat. Methods 13, 692–698 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo G.-Z., Wang F., Weng X., Chen K., Hao Z., Yu M., Deng X., Liu J., He C., Characterization of eukaryotic DNA N6-methyladenine by a highly sensitive restriction enzyme-assisted sequencing. Nat. Commun. 7, 11301 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imanishi M., Tsuji S., Suda A., Futaki S., Detection of N6-methyladenosine based on the methyl-sensitivity of MazF RNA endonuclease. Chem. Commun. 53, 12930–12933 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Lorenz R., Bernhart S. H., zu Siederdissen C. H., Tafer H., Flamm C., Stadler P. F., Hofacker I. L., ViennaRNA Package 2.0. Algorithms Mol. Biol. 6, 26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao W., Adhikari S., Dahal U., Chen Y.-S., Hao Y.-J., Sun B.-F., Sun H.-Y., Li A., Ping X.-L., Lai W.-Y., Wang X., Ma H.-L., Huang C.-M., Yang Y., Huang N., Jiang G.-B., Wang H.-L., Zhou Q., Wang X.-J., Zhao Y.-L., Yang Y.-G., Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61, 507–519 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Liu Z., Zhang J., Most m6A RNA modifications in protein-coding regions are evolutionary unconserved and likely nonfunctional. Mol. Biol. Evol. 35, 666–675 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siepel A., Bejerano G., Pedersen J. S., Hinrichs A. S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L. W., Richards S., Weinstock G. M., Wilson R. K., Gibbs R. A., Kent W. J., Miller W., Haussler D., Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–1050 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2013). [Google Scholar]
- 35.Kim D., Langmead B., Salzberg S. L., HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thorvaldsdóttir H., Robinson J. T., Mesirov J. P., Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suyama M., Torrents D., Bork P., PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34, W609–W612 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/7/eaax0250/DC1
Fig. S1. Quantitative demonstration of various fractions of m6A-containing oligo mixed with unmethylated oligo digested by ChpBK.
Fig. S2. FTO demethylate assay.
Fig. S3. The base composition of mRNA products cleaved by MazF.
Fig. S4. The single-site validation for 18S rRNA control site and m6A site.
Fig. S5. The single-site validation of eight m6A sites and one unmethylated site.
Fig. S6. Quantitative PCR results of six m6A sites.
Fig. S7. Metagene plots of m6A in mouse heart and mouse testis.
Fig. S8. Conservation of m6A in mammalian kidney.
Fig. S9. Conservation of m6A in mammalian liver.
Fig. S10. Gene expression and the expected number of shared m6A-modified genes.
Fig. S11. Evolutionary conservation between methylated and unmethylated A sites in the same genes of brain tissue.
Fig. S12. Evolutionary conservation between methylated and unmethylated A sites in the same genes of kidney tissue.
Fig. S13. Evolutionary conservation between methylated and unmethylated A sites in the same genes of liver tissue.
Fig. S14. Conservation scores for brain.
Fig. S15. Validation for the methylation sensitivity of mutated MazF-K56A.
Table S1. Basic information of sequencing data for human HEK293T cell line.
Table S2. Designed probes and universal primers for T3 ligase–based validation.
Table S3. Designed primers for Quantitative PCR validation.
Table S4. Basic information of sequencing data for mammalian tissues.
Table S5. m6A sites identified by m6A-REF-seq for mammalian tissues.
Table S6. Shared m6A-modified genes and m6A sites between pairwise species and the significance.


