SUMMARY
In the developing brain, cell surface proteins play crucial roles, but their protein-protein interaction network remains largely unknown. A proteomic screen identified 200 interactions, 89 of which were not previously published. Among these interactions, we find that the IgLONs, a family of five cell surface neuronal proteins implicated in various human disorders, interact as homo- and heterodimers. We reveal their interaction patterns and report the dimeric crystal structures of Neurotrimin (NTRI), IgLON5 and the neuronal growth regulator 1 (NEGR1)/IgLON5 complex. We show that IgLONs maintain an extended conformation and that their dimerization occurs through the first Ig domain of each monomer and is Ca2+-independent. Cell aggregation shows that NTRI and NEGR1 homo- and heterodimerize in trans. Taken together, we report 89 unpublished cell surface ligand-receptor pairs and describe structural models of trans interactions of IgLONs showing that their structures are compatible with a model of interaction across the synaptic cleft.
Keywords: Ligand-receptor pair, ELISA, IgLON, Protein crystallography, SAXS
Graphical Abstract
In Brief
Many aspects of synapse formation, specification, and maturation rely on interactions among a rich repertoire of cell surface glycoproteins with adhesive and repulsive properties. Although the identity of these proteins is known, their network of interactions remains largely untapped. Ranaivoson et al. have identified a number of protein-protein interactions and have determined the structures of three members of the IgLONs, a family of five proteins of the immunoglobulin superfamily that has recently been implicated in a wide range of human disease.
INTRODUCTION
The specification of each synaptic contact and the precise and dynamic balance between excitatory and inhibitory connections are enabled by a rich repertoire of cell adhesion and signaling molecules with diverse structural and functional identities (de Wit and Ghosh, 2016). The composition of the cell surface proteome has been analyzed in detail in the past decade by numerous groups (Bausch-Fluck et al., 2015; Butko et al., 2013), and the extracellular domains (ectodomains) of these proteins are composed of a variety of individually-folded domains, which can independently interact with different domains in other proteins. Thus, it is likely that the global network of potential interactions between neuronal ligand-receptor pairs is currently underestimated. In the past decade, taking advantage of variations of the classical enzyme-linked immunosorbent assay (ELISA), several groups have tackled this problem and have reported the discovery of new ligand-receptor pairs using proteins from Drosophila, zebrafish, and a variety of mammalian cell surface proteins (Bushell et al., 2008; Ozkan et al., 2013; Sollner and Wright, 2009; Wojtowicz et al., 2007). However, a complete interactome study of these mammalian central nervous system proteins remains to be performed.
The immunoglobulin superfamily (IgSF) is a large group of diverse cell surface proteins that includes the IgLONs (Tan et al., 2017). The five-member IgLON family comprises OPCML (opioid-binding cell adhesion molecule, IgLON1), NTRI (Neurotrimin, IgLON2), LSAMP (limbic system–associated membrane protein, IgLON3), NEGR1 (neuronal growth regulator 1, IgLON4), and IgLON5 (IgLON family member 5), all of which are highly expressed in the rat CNS (Struyk et al., 1995). OPCML, NTRI, LSAMP, and NEGR1 have been shown to accumulate pre- and post-synaptically in different brain areas such as the hippocampus and the cerebral cortex (Miyata et al., 2003; Zacco et al., 1990). Cortical neurons grown on a substrate of recombinant OPCML or LSAMP demonstrate a dose dependent increase in neurite outgrowth (Gil et al., 1998; Sanz et al., 2015). In addition, LSAMP was identified as a negative regulator of myelination (Sharma et al., 2015). Some of the IgLON proteins have been described to form homophilic and heterophilic complexes on the cell surface or with other cells, likely modulating their functions such as adhesion and neurite outgrowth and may have axonal fasciculation and synaptogenic functions (Gil et al., 1998; Lodge et al., 2000). The specificity of the interactions among IgLON proteins, however, has never been systematically investigated.
In humans, genetic abnormalities of individual IgLONs have been implicated in a variety of disorders. Four single-nucleotide polymorphisms (SNPs) in intron 1 of the NTRI gene and one SNP in intron 1 of the OPCML gene have been found to be associated with late-onset Alzheimer’s disease (Liu et al., 2007), while modulation of OPCML expression is associated with cancer progression (Cui et al., 2008). Genetic variants in the NTRI gene have also been implicated in childhood aggressiveness in children with attention deficit/hyperactivity disorder (Brevik et al., 2016) and developmental delay (Minhas et al., 2013), whereas single-nucleotide polymorphisms of LSAMP have been associated with schizophrenia and depression (Karis et al., 2018; Koido et al., 2014). Interstitial microdeletions at chromosome 1p31.1 involving only NEGR1 were linked to learning and behavioral problems, hypotonia, hypermobility, scoliosis and aortic root dilation (Genovese et al., 2015). NEGR1 was also identified as a candidate gene involved in dyslexia (Veerappa et al., 2013) and severe obesity (Zandona et al., 2017). More recently, anti-IgLON5 antibodies have been associated with sleep disorder, cognitive dysfunction, gait abnormalities and other neurological symptoms (Gaig et al., 2017; Gelpi et al., 2016).
Structurally, IgLONs are ~340 amino acids long and tethered to the cell membrane by a glycosylphosphatidylinositol (GPI)-anchor. IgLONs possess three C2-type Ig-like domains (Ig1–3), with one conserved disulfide bond in each Ig domain. Interestingly, IgLON5 and NTRI have one additional nonconserved Cys within the first 90 amino acids (C45 and C83 respectively), suggesting that they may form covalent dimers (Struyk et al., 1995). IgLONs contain between 5 and 7 putative N-linked glycosylation sites, three of which are conserved across all five genes (NTRI Asp70, Asp292, and Asp305).
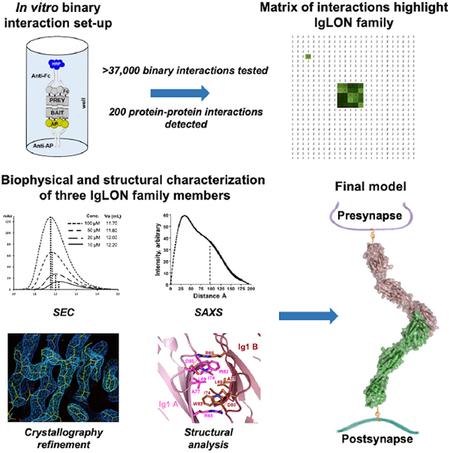
In this work, using an ELISA-based ligand-receptor identification protocol adapted for 384-well plates and a selected set of 207 human and mouse neuronal cell surface proteins, we have screened 37,467 binary pairs and have identified 200 protein-protein interactions. Of these, 89 were previously unpublished. The purpose of this screen was to deorphanize known neuronal receptors and to identify other ligand-receptor pairs that may play important roles in neuronal and brain development. Among these interactions, we have characterized multiple homo- and heterophilic interactions among members of the IgLON family of proteins and we have solved the crystal structures of NTRI, IgLON5, and the complex between NEGR1 and IgLON5. These structures show that IgLONs homo- and heterodimerize with high affinity through the first Ig domain and can interact in trans across different cells and/or across the synaptic cleft.
RESULTS
Pairwise interactions between the ectodomains of cell surface neuronal proteins were performed utilizing a modified ELISA-based binding assay (Figure 1, top panel) based on a previously described high-throughput protein-protein interaction screen (Visser et al., 2015; Wojtowicz et al., 2007). Modifications were aimed at increasing assay throughput, while improving precision and consistency and reducing costs, by using robotic liquid handlers capable of 384-well format plates.
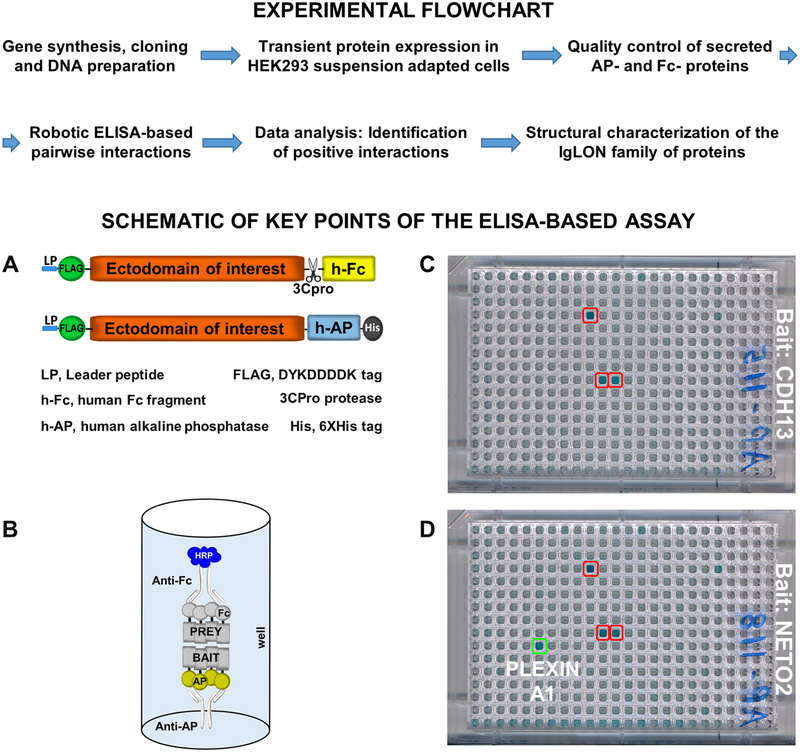
Figure 1 – Flowchart of the ELISA-based ligand receptor assay –
Top panel – Flowchart of the main steps composing the experimental methodology of the ELISA-based assay. Bottom panel: A – Schematic diagram of the vectors used in the assay and their relevant features. B – Schematic of the ELISA setup to show the orientation of the bait and prey. HRP develops a blue color. C – Typical results of a negative 384-well plate. Red boxed wells are false positives (present in every plate of the same batch of one experiment, compare with D). D – 384-well plate containing a potential positive interaction (green box, the interaction appears only in one plate).
Gene selection, production, and protein quality control –
A library of 207 human cell surface neuronal genes selected among families of well-known synaptic adhesion molecules (e.g. neurexins, neuroligins, etc.), axon guidance cues and receptors (netrins, semaphorins, etc.), and other proteins for which little information is available (e.g. IgLONs, Cochlin, etc.) (Figure 2a) was cloned. The topologies of the proteins that we have included in this screen fall into five categories: type I transmembrane (117/207; 56.5%), secreted (54/207; 26.1%), GPI-linked (18/207; 8.7%), N-terminal ectodomain of multipass (GPCR) transmembrane proteins (17/207; 8.2%), and type II transmembrane (1/205; 0.5%) (Figure 2b). For all of these genes, the coding region of the mature extracellular region (i.e. no leader peptide) was cloned into expression vectors at the N-termini of either the human alkaline phosphatase (AP) or the Fc region of human IgG1 (Fc). These recombinant AP- or Fc-tagged proteins were secreted into the media after transient transfection, and used in the ELISA-based screen without further purification (Figure 1a, b). Most proteins expressed at levels sufficient for downstream protein-protein interaction studies. Of 207 constructs, 22 (~10.6%) failed to express protein at levels detectable by Western blot (Figures 2c, S5) but they were still included in the screen because the amount of protein needed to obtain a positive signal is small. Some of the control proteins, not detected by Western blot, still provided positive interactions with known partners (Figure S5 legend). These findings are consistent with other studies that have established that low expression levels do not necessarily preclude the detection of protein interaction partners (Ozkan et al., 2013; Visser et al., 2015).
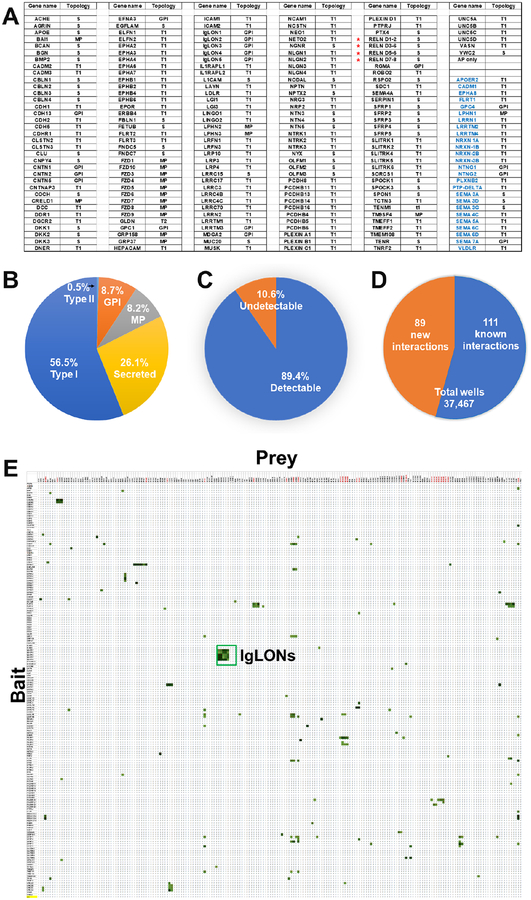
Figure 2 – List of genes used in this study and interaction results –
A – The list contains information about the topology of each protein. Black font indicates genes that have been used as both Fc- and AP fusions, Blue font indicates genes that have been used as Fc-fusions only as positive interactions. In the topology column: S, secreted; MP, multipass membrane; T1, type I protein; GPI, GPI anchor protein. * The RELN gene encodes for Reelin, which is composed by 8 Reelin repeats. Because of its size and intrinsic cleavage, we have cloned this gene into four fragments. B – Pie chart showing the topology distribution of the proteins. C – Graph indicating the number of Fc-fusion detected by western blot expression, shown in Figure S5. D – Pie chart diagram of the number of wells used to identify the 200 interactions and their breakdown as “new” and “known.” E – The matrix of data for 181 × 207 pairwise interactions. Rows contain AP-fusion baits, including the AP-only construct as negative control, whereas columns contain 207 Fc-fusion preys. The green color scale represents values from <2 (White) to 20 (dark green). The largest cluster boxed in green represents all IgLON interactions. Red preys were used as positive control Fc proteins. Source data file attached as supplemental excel file.
ELISA-based ligand-receptor screen –
The screen was designed to test 181 genes in AP- and Fc-tagged formats with all protein ectodomains tested in both orientations (i.e. AP-X vs Fc-Y and AP-Y vs Fc-X) resulting in 181×181 = 32,761 experimental points and testing of 16,381 unique pairwise potential interactions. In addition, we included 26 Fc-tagged proteins as positive control interactions (Figure 2a), for a total of 181×207 = 37,467 experimental points (Figure 2d). Each pair of potentially interacting ectodomains was tested in a reciprocal orientation (Figure 2e), because, in addition to test their potential for homodimerization, we and others have observed instances in which a ligand-receptor interaction can be detected in one orientation but not the other (Ozkan et al., 2013; Visser et al., 2015). In our experiments, a small group of Fc prey protein fusions consistently behaved as false positives as their wells displayed OD650 readings of >5-fold over the background reading (FOB) in virtually every plate, including the AP-only bait used as a control (Figure 1c). Analysis of the amino-acid sequences of some of these false-positive preys revealed that they have long stretches of basic residues or high isoelectric points (pI) (e.g. TMEM108, pI: 8.7; SFRP1, pI: 9.1). As the blocking agent (casein) has a pI of ~5.1 we presume that non-specific electrostatic attraction causes these false positive interactions. Therefore, true positive interactions were defined as those wells that exhibited an OD650 >2.5 FOB and that were only present in a single plate run within a batch of baits tested (typically 15 to 20 baits), during a given experiment (as opposed to non-specific binders that developed in every plate) (Figure 1d). The combination of 2.5 FOB and being unique to a single bait (we typically ran one bait per plate) increases the chances to identify low affinity interactions. Using the criteria outlined above, we identified 200 interacting pairs, 89 of which (Table S1), to the best of our knowledge, have not been reported in the current scientific literature (Table S5 for a list of known ligand-receptor pairs).
Interactions identified by the screen –
Overall, the screen confirmed numerous known ligand-receptor pairs such as the interactions between Reelin and both known receptors, VLDLR and ApoER2 (D’Arcangelo et al., 1995), between Neuroligin 1, 3, and 4 and the three b isoforms of Neurexin (Comoletti et al., 2006), etc. (Figure 2e, Table S1). Figure 2e and the attached data source file contain the entire dataset of positive (green cells) and negative (white cells) interactions. The positive interactions are then reported in alphabetical order for bait and the FOB values in Table S1. Importantly, the screen identified several pairs that could not be inferred by homology with known interactions. Among the newly identified interactions, the majority are 1:1 interactions or pairs (i.e. CLSTN2 only binds to VLDLR). Interestingly, we have also identified numerous interactions where the bait (i.e. SFRP1-AP fusion) binds to multiple and structurally diverse preys. As expected, some of these ligands belong to the same family of proteins. For example, NTN4 appears to interact with LRRTM1, LRRTM2 and LRRTM4; NTN3 interacts with UNC5B and UNC5C, among others. Among the numerous ligand-receptor pairs identified, the IgLON family of proteins, showed a strong, consistent pattern of interaction within the family (Figure 2e). All five IgLONs interact specifically with themselves, and each of them also binds to the other four members of the family. Owing to the importance of this family of proteins in brain development and disease, we sought to further characterize the interactions among family members and to characterize the structural basis of their molecular recognition.
The IgLON family of proteins participate in homo- and heterophilic interactions –
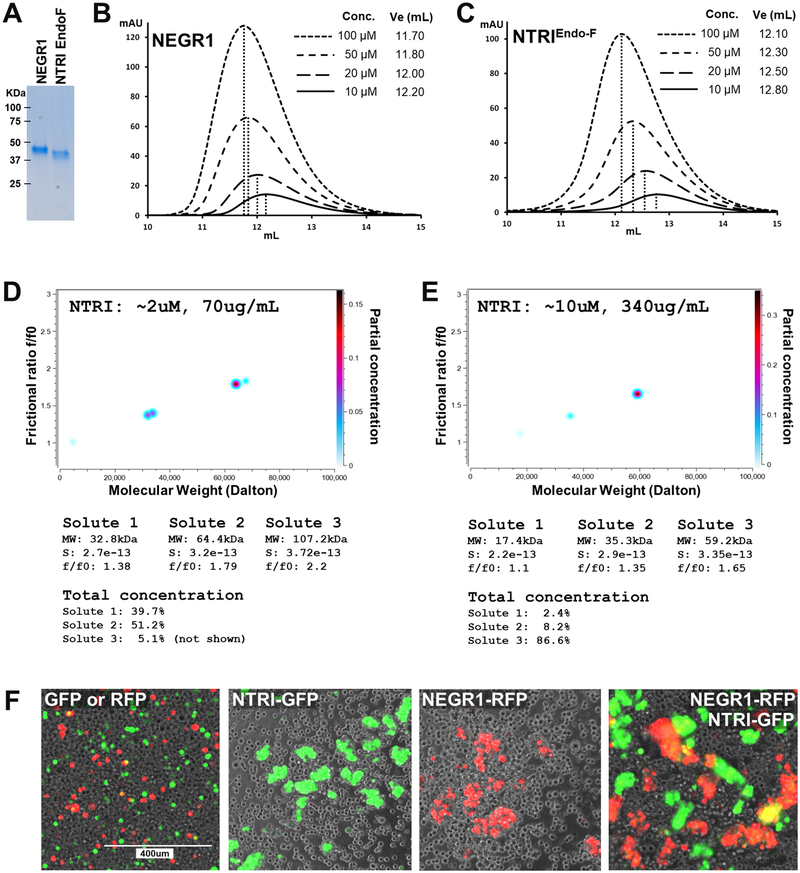
IgLONs share high sequence similarities within the family, with ~27% amino-acid identity when comparing all five members, and between ~42% and ~77% sequence identity by pairwise comparisons between individual members (Table S2). As all IgLONs appear to homo- and heterodimerize, we used size exclusion chromatography (SEC) to inspect their oligomerization, using NEGR1 and NTRI as representative IgLONs (Figure 3a–c). Four separate concentrations of purified NEGR1 (no Fc or AP tags) show that the retention volume of the protein progressively decreases from 12.20 mL to 11.70 mL, indicating that the protein undergoes a concentration-dependent oligomerization. To rule out a potential modulatory effect of the N-linked glycosylation in the dimerization, NTRI was also treated in non-denaturing conditions with endoglycosidase F1 (Endo-F1) prior to SEC. Endo-F1 treated NTRI injected at the same concentrations as NEGR1 eluted at volumes ranging from 12.80 mL to 12.10 mL, indicating that the oligomerization is common among the IgLONs and it is not affected by glycosylation and its processing.
Figure 3 – Solution characterization of NTRI –
A – Coomassie blue staining of purified NEGR1 and NTRI used for SEC experiment. EndoF indicates that the purified protein was treated with Endo F1 under native conditions, causing faster migration in SDS-PAGE. B, C – Four different concentrations of purified NEGR1 and NTRI were subjected to SEC. Higher concentrations of the protein produce a leftward shift in the elution profile. In each graph, the inset contains the concentration of the injected protein and the elution volume. D, E – Sedimentation velocity analysis of two concentrations of purified NTRI, two-dimensional spectrum/Monte Carlo analyses: D shows the presence of two main species (total concentration ~51% and ~40%) of the appropriate MW; E shows that at a higher concentration, one main species of ~59kDa (total concentration ~87%) becomes prevalent; see numerical tables below D and E images. F – Cell aggregation assay of four different conditions as shown in the labels. Three independent experiments were performed, and representative images are shown.
Analytical ultracentrifugation (AUC) –
To ascertain the precise hydrodynamic and oligomerization behavior of purified NTRI, we resorted to analytical ultracentrifugation. Sedimentation velocity experiments indicate that purified NTRI is essentially in a monomer-dimer equilibrium. At a concentration of ~0.07 mg/mL (2.0 μM) NTRI appears as ~40% monomer and ~50% dimer, whereas at ~0.34 mg/mL (10 μM) it is ~87% dimeric (Figure 3d, e). Moreover, the f/f0 ratio increases from ~1.4 to ~1.7 with increasing protein concentration, indicating that monomeric NTRI is dimensionally asymmetric in solution and that the anisotropy increases with dimerization. Finally, Monte Carlo simulations indicated that the dissociation constant KD of the homodimer is ~110 nM (95% confidence interval: 51 to 170 nM) (Table S3). As these experiments were performed in phosphate buffer (PBS), they clearly indicate that the homodimerization of NTRI does not require Ca2+.
Cell aggregation assays –
To understand whether homo- and/or heterodimerization enable IgLONs to interact in trans between different cells or across the synapse, we took advantage of a cell aggregation assay. To tether the ectodomains of these proteins to the cell membrane, the extracellular domains of NTRI and NEGR1 were cloned in frame with the transmembrane and intracellular domains of CASPR2 and were co-transfected with either EGFP or RFP. Four days after transfection, an equal number of transfected cells were mixed in the following groups: EGFP alone mixed with RFP alone as negative control, either NTRI/EGFP or NEGR1/RFP (to test for homophilic trans-interaction), and NTRI/EGFP mixed with NEGR1/RFP (to test for heterophilic trans-interaction). After an hour of incubation, both homophilic and heterophilic trans-interactions were robustly observed as cell aggregates, whereas cells expressing EGFP or RFP alone did not form aggregates larger than 2 cells (i.e. dividing cells) (Figure 3f). This demonstrates that both NTRI and NEGR1 can mediate transcellular interactions by either homophilic or heterophilic protein-protein interactions.
Structural characterization of NTRI –
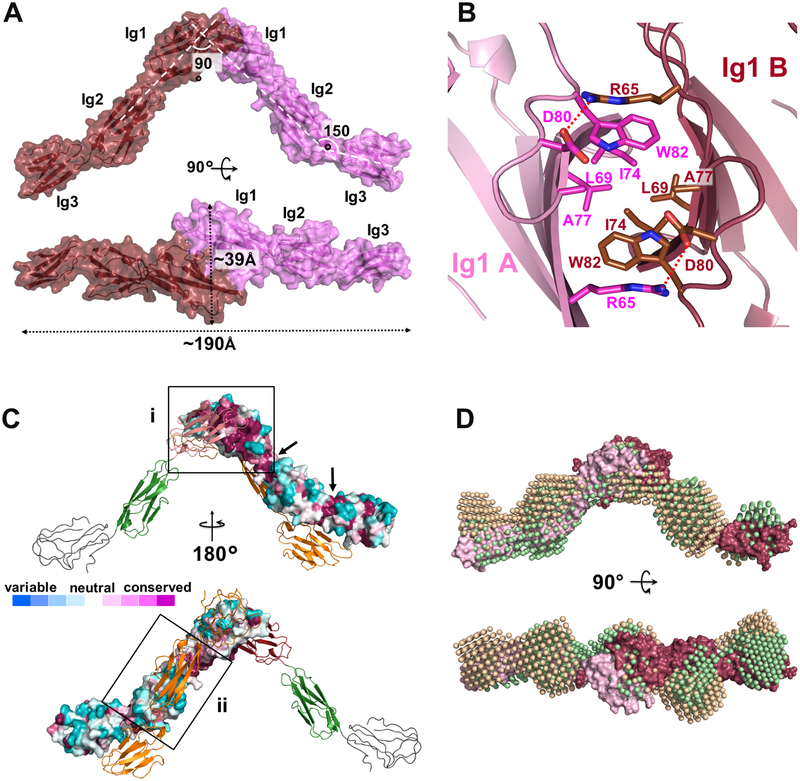
The mature extracellular domain of FLAG-NTRI was crystallized in space group P212121, as a homodimer, at a resolution of 3.45 Å (Figure 4; PDB ID: 6DLF). The structure was modeled between Ala41 and Leu310, with the exception of residues E227AKGTGVPVGQKG239, residues D258DKRLIEGKKGV269, and residues F282FNVSEHDY290. The final Rwork and Rfree values were 27.35 % and 30.45 %, respectively (Table 1). The structure of each NTRI protomer shows the first two Ig domains in a linear arrangement, with Ig3 positioned at a ~150° angle (Figure 4a), similar to the structure of other Ig-domain proteins such as Nectin-1 (Zhang et al., 2011). Missing electron density and significantly higher B-factors suggest a mobile Ig3 domain. The maximum dimension of the NTRI protomer is ~120 Å and the boundaries of each Ig domains are reported in the sequence alignment of Figure S1. Of the six putative N-linked glycosylation sites (N44, N70, N152, N284, N292, and N305) present in NTRI, only N44, N70 and N152 could be identified and were partially built in the final model, with the exclusion of N44 in one of the protomers. The extra Cys at position 83 remains unpaired and it does not form an intra- or inter-molecular disulfide bond as previously speculated (Struyk et al., 1995).
Figure 4 – Three-dimensional structure of NTRI –
A – “Side” and “top” views of the surface rendering of the crystal structure of the NTRI homodimer. The three Ig domains belonging to each monomer are visible through the semitransparent surface. Dotted double arrows show measurements in Å. B – Top view of the Ig1-Ig1 interface of NTRI showing the hydrophobic residues in the center of the image and salt bridge between the two protomers (D80 and R65). C – Surface conservation map to identify the biologically-relevant dimer – Top, the biological dimeric interface (i) is identified inside the square where most of the residues are conserved (purple). Arrows indicate conserved junctions between Ig domains. Bottom, surface of the artefactual crystal dimer (ii) contains mostly variable residues (white to blue). D – Overlay of two distinct SAXS models obtained by ab initio reconstructions (green and beige bead representation) with the NTRI crystal structure (red and pink, surface representation) to highlight the similarity between model pairs.
Table 1 –
Crystallographic data collection and refinement statistics
| IgLON5 + NEGR1 |
IgLON5 + IgLON5 |
NTRI + NTRI |
|
|---|---|---|---|
| Data Collection | |||
| Space Group | C 2 | / 222 | P 212121 |
| Cell Dimensions | |||
| a, b, c (Å) | 120.34 305.80 60.03 | 119.21 138.23 160.84 | 76.52 102.0 134.03 |
| α, β, γ (°) | 90 100.777 90 | 90 90 90 | 90 90 90 |
| Resolution (Å) | 48.60–3.30 (3.50–3.30) | 48.90–4.0 (4.23–4.0) | 47.67–3.45 (3.65–3.45) |
| Rsym (%) | 13.4 (129.8) | 24.3 (239.2) | 9.7 (95.1) |
| </>/<σ/> | 6.70 (0.83) | 7.12 (0.82) | 12.06 (1.44) |
| CC1/2 | 99.6 (39.7) | 99.5 (33.6) | 99.8 (75.8) |
| Completeness (%) | 99.0 (96.5) | 99.4 (97.9) | 99.7 (99.0) |
| Redundancy | 3.5 (3.4) | 6.6 (6.4) | 6.3 (6.9) |
| Refinement | |||
| Resolution (Å) | 48.60–3.30 (3.413.30) | 48.90–4.0 (4.18–4.0) | 47.67–3.45 (3.71–3.45) |
| Reflections | 31534 | 11530 | 14353 |
| Rwork (%) | 24.49 | 26.02 | 27.35 |
| Rfree (%)* | 28.59 | 30.78 | 30.45 |
| Number of atoms | |||
| Protein | 7596 | 3905 | 3384 |
| Ligands/Glycans | 260 | 126 | 84 |
| Average B-factors (Å2) | |||
| All | 124.1 | 226.17 | 159.73 |
| Protein | 123.3 | 225.82 | 158.99 |
| Ligands/Glycans | 145.5 | 236.82 | 189.65 |
| R.m.s deviations from ideality | |||
| Bond Lengths (Å) | 0.003 | 0.003 | 0.003 |
| Bond Angles (°) | 0.569 | 0.634 | 0.758 |
| Geometry | |||
| Ramachandran | |||
| Outliers (%) | 0.0 | 0.0 | 0.0 |
| Allowed (%) | 4.03 | 3.90 | 1.54 |
| Favored (%) | 95.97 | 96.10 | 98.46 |
| Rotamer outliers (%) | 2.0 | 1.04 | 0.0 |
| Cbeta outliers (%) | 0.0 | 0.0 | 0.0 |
| All-atom clashscore | 9.05 | 9.13 | 6.68 |
5% of reflections was not used during refinement as cross validation.
NTRI homodimer –
The asymmetric unit contains one homodimer bound in a “head to head” fashion through a common interfacing surface of the two Ig1 domains (775.5 Å2 of buried surface area), orienting the two monomers at a ~90° angle to each other (Figure 4a). Central to this interface, residues L69, I74, A77 and W82 of one chain form a hydrophobic cluster with the same residues on the other protomer of the dimer. This central hydrophobic patch is bordered at each side by a putative salt bridge formed by R65 and D80 (Figure 4b). Interestingly, the modality of interaction of each protomer brings the total dimension of the dimer to ~190 Å, which approximately matches the size of the cleft of excitatory synapses, which measure ~200 to 220 Å (Perez de Arce et al., 2015). The conserved glycan at N70 is in an invagination formed between the two interacting Ig1 domains. This suggests a structural role for this glycosylation site, possibly important for aligning Ig1 domains relative to each other (Figure S2). Despite Endo F1 treatment prior to crystallization, two sugar moieties could be built at N70 in one of the two protomers, further indicating low accessibility to the glycan moiety. Interestingly, in our crystal packing, a second relatively large inter-molecular contact (708.9 Å2 of buried surface area) occurs with a neighboring symmetric protomer, where the two molecules contact each other via their Ig2 domains in an anti-parallel orientation (Figure 4c). This second contact is likely to be a crystal packing artifact, as strongly suggested by a conservation analysis of surface residues using 150 Ig-like unique sequences (Ashkenazy et al., 2010). Our residue-conservation analysis also revealed that both Ig1-Ig2 and Ig2-Ig3 junctions correspond to highly conserved regions, indicating that the rigidity or the flexibility across all three domains is functionally important and evolutionarily preserved (Figure 4c, arrows).
Solution scattering of NTRI –
To confirm the crystallographic model of the NTRI dimer in solution, small angle X-ray scattering (SAXS), was used. Guinier plots of the SAXS data are linear (Figure S3a, inset) as expected for monodisperse samples of excellent quality (Trewhella et al., 2017), and the radius of gyration (Rg, 51.2 Å) is indicative of an elongated dimer. Consistent with SEC and AUC data, the calculated MW from two different concentrations (0.7 mg/mL and 0.39 mg/mL) is consistent with a monomer-dimer equilibrium (Figure S3c). P(r) analysis also indicates that the maximum linear dimension (Dmax) value of NTRI is ~190 Å (Figure S3b), which is virtually identical to what was calculated from the crystal structure. The secondary peak of the P(r), centered at ~100 Å likely corresponds to the two interacting Ig1 domains, further validating the dimerization organization of NTRI. Taken together, these solution scattering measurements support the crystallography data and demonstrate that this protein preparation is suitable for further structural analysis using the SAXS data. To obtain an assessment of the three-dimensional shape of the NTRI dimer, we used an ab initio approach implemented in DAMMIF (ATSAS software package version 2.8.3) (Franke et al., 2017). Most NTRI models appear to be elongated and loosely V-shaped (average X2 of the fit of 15 models was 1.033 ±0.007), similar to the crystal structure of the dimer, including the position of each Ig domain (Figure 4a, d). These experiments, in addition to the surface conservation analysis, rule out the anti-parallel Ig2-Ig2 interaction (Figure 4c) and confirm that the crystallographic structure is also present in solution, and likely represents the biologically-relevant dimer.
Structural characterization of the NEGR1/IgLON5 heterodimer and the IgLON5 homodimer –
To further understand the structures and interactions within the IgLON family of proteins, we determined the crystal structure of the NEGR1/IgLON5 complex (PDB ID: 6DLD) at 3.3 Å resolution. This heterodimer crystallized in space group C121, with two heterodimers in the asymmetric unit. IgLON5 was modeled with all residues between residues Ser30 and Glu313, and NEGR1 was built between residues Ala37 and Leu312, except for residues V235TPGRSG241. Similar to Cys83 in NTRI, the extra Cys at position 45 in IgLON5 remains unpaired and does not participate in intra- or intermolecular disulfide bonding. The complex, whose two interacting proteins could be unequivocally distinguished from each other due to the presence of distinct glycosylation sites and characteristic side chain densities, yielded Rwork and Rfree scores of 24.49 % and 28.59 % respectively (Table 1). Using a different set of crystals, the extracellular domain of IgLON5 alone was crystallized in space group I222, with one homodimer in the asymmetric unit, and the structure was solved at 4.0 Å resolution (PDB ID: 6DLE). Due to a poor definition of the electron density maps, the third domain was positioned using non-crystallographic symmetry refinement. The final model gave Rwork and Rfree values of 26.02 % and 30.78 % (Table 1).
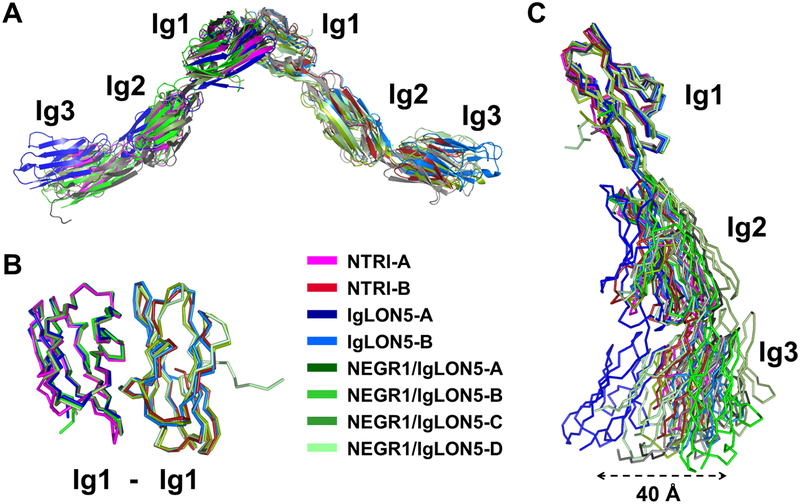
To measure the similarity between the Ig1 dimerization domains, the two interacting Ig1 domains of the NEGR1/IgLON5 heterodimer were superimposed on to the NTRI equivalent fragments, resulting in RMSD values of ~0.8 Å for 153 and 160 Cα atoms, respectively (Table S4, Figure 5b), indicating a high structural identity of dimeric Ig1 across these structures. Furthermore, the NTRI interface forming the central hydrophobic cluster is structurally conserved in the NEGR1/IgLON5 heterodimer and the IgLON5 homodimer, with contributions from L72, I77, A80 and W85 in NEGR1 and L66, I71, A74 and W79 in IgLON5. Similarly, a putative salt bridge at the edge of the interface is also observed (i.e. K68 and D83 in NEGR1, and R62 and D77 in IgLON5) (Figure S4). Superimpositions between entire structures (Figure 5a) exhibit much larger RMSD values (3–7 Å), indicating that the IgLON dimers accommodate some degree of flexibility (Table S4). This flexibility is particularly apparent when superimposing the Ig1 domains of all the available monomers (Figure 5c), suggesting that the inter-domain flexibility across Ig2 and Ig3 domains allows IgLON protomers to adapt to their inter-cellular environment and more flexibility to interact in trans or in cis.
Figure 5 – Superimposed IgLON dimers –
A – Overall superimposition of all the IgLON dimeric crystal structures, in cartoon representation. B – Cα-traces of superimposed Ig1-Ig1 dimeric assemblies as observed in the different crystals. C – Superimposition of Ig1 domains of all the monomers, illustrating an overall flexibility of the IgLONs, to highlight their structural and architectural similarities.
Fragment binding –
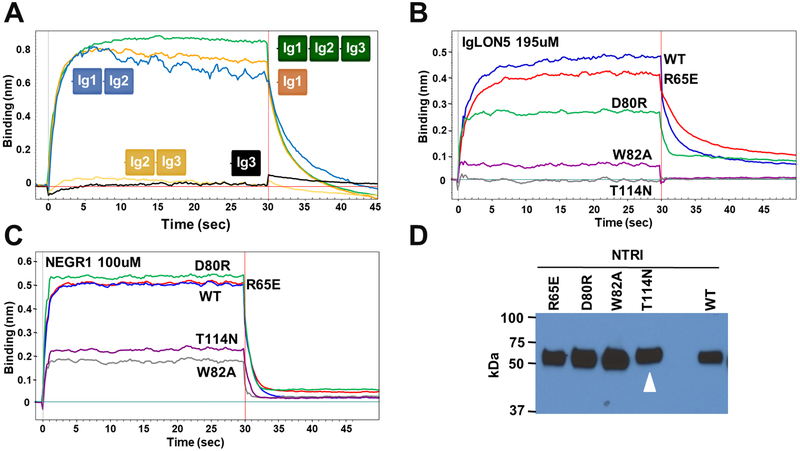
To confirm that the Ig1 domain is necessary and sufficient for the interaction, we engineered deletion constructs containing combinations of only one or two Ig domains and we used Bio-Layer Interferometry (BLI) to assess the interaction of NTRI with purified NEGR1. As expected, these experiments reveal that the Ig1 domain alone is sufficient and necessary for the NTRI/NEGR1 interaction (Figure 6a). To further test whether the dimeric structure through Ig1 is the mode of interaction, we engineered the following mutations in NTRI: R65E, D80R, W82A, and T114N (which introduces a new N-linked glycosylation site) and tested the binding with both NEGR1 and IgLON5 (Figure 6b–d). Consistent with the analysis of the NTRI crystal structure, W82A and T114N disrupt the interaction for both NEGR1 and IgLON5, whereas the other two mutants R65E, D80R interfere with the association to a lesser degree. Finally, a set of mutations homologous to those previously described in NTRI was made in IgLON5 (not shown), and binding data showed that disruption of binding between the proteins is most severe when mutating the hydrophobic residues outlined above, supporting our assignment of the relevant biological interface. Together, these experiments indicate that the physiological interacting surface is made by the dimerization of the Ig1 domains and show the role of some of these amino acids in the interaction, supporting our interpretation of the crystal structures.
Figure 6 – Determination of the associating domains between IgLON protomers –
A – BLI curves of purified NEGR1 (100 mM) interacting with NTRI-Fc or its shorter constructs immobilized on a sensor tip, highlighting the associating deletion constructs (Ig1-Fc, Ig1–2-Fc). B – IgLON5 (195 μM) was used to test the binding of four NTRI-Fc mutants. C – The same NTRI mutants were tested by using 100 μM of purified NEGR1. D – Relative expression of the NTRI-Fc mutants in the cell culture medium. As expected, the introduction of an N-linked glycosylation site (T114N) causes a slightly slower migration of the mutant (white arrowhead).
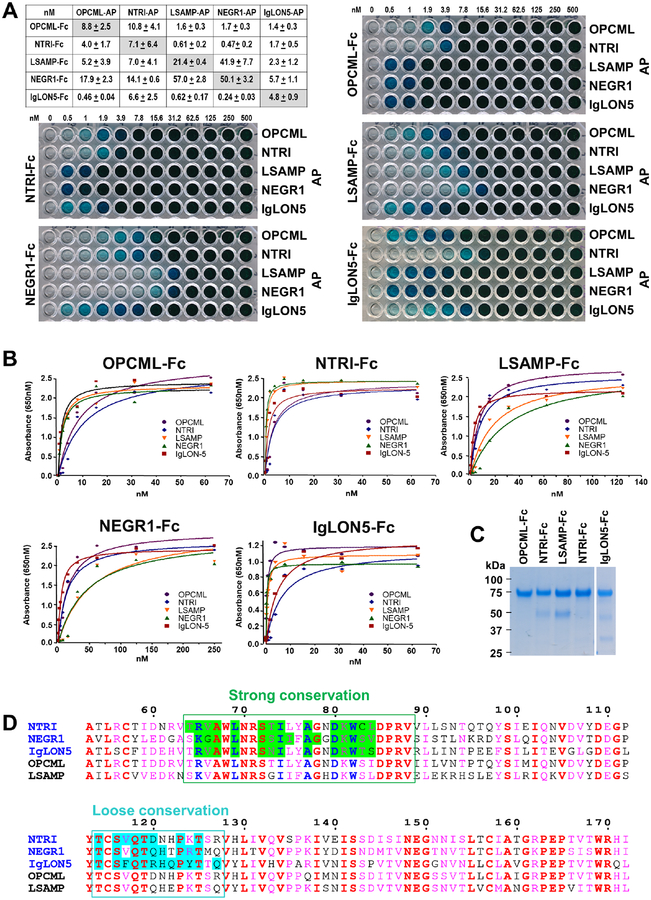
Relative affinity measurements of IgLONs using titration ELISA –
AUC measurements indicated that the dissociation constants of the NTRI homodimer is ~110 nM (Table S3). To compare binding affinities among all IgLONs we used a similar ELISA-based approach that enabled us to determine their interactions. In this experiment, we utilized a fixed concentration of AP-IgLONs to coat each of 12 wells and decreasing concentrations of each purified IgLON-Fc, from 500 nM to 0.5 nM in two-fold dilutions and buffer only (0 nM). In all experiments, we observed concentration-dependent, saturable binding curves, indicative of specific interactions with apparent KDs in the low to sub-nM range (Figure 7a, b). These apparent affinities are useful to compare and rank the order of the interactions among different combinations of IgLON homo- and heterodimers (Figure 7a). We should note however, that the observed binding constants are likely to be significantly overestimated because the purified Fc-IgLONs used in these experiments are tetramerized by the anti-Fc
Figure 7 – ELISA apparent affinity of all IgLONs –
A – Table of apparent affinities among all IgLON combinations and five representative titrations used to calculate these affinities. Numbers on top of the plates (0 to 500) indicate each FLAG purified IgLON-Fc concentration (nM) used in the specific column. B – Titration binding curves used to calculate the affinities reported in A. For clarity, only the first part of the curve is shown, typically to 70 nM to 250 nM concentrations. C – Coomassie blue staining of the five IgLON-Fc purified proteins used as prey. D – Alignment of the five human IgLON sequences at the Ig1-Ig1 interface region (numbering using NTRI sequence). Conserved residues are in red and group similarities are in pink. Using the structures determined in this study (sequence names in blue), interface-buried residues were computed using PDBePISA and boxed. They fall into two categories: a strongly conserved area (green) that include the key interface residues described in Figure 4b (blue font) and a loosely conserved area (cyan). The sequence alignment was done using ESPript 3 (Robert and Gouet, 2014).
antibody, which provides an avidity effect on the IgLON-IgLON interactions. Our observations are consistent with similar titration experiments using FLRT3 and UNC5D in an identical strategy (Visser et al., 2015) and to previous experiments using similar technologies (AVEXIS and ECIA), in which it was shown that pentamerization of the prey protein could improve the sensitivity of detection over monomeric proteins by over 250-fold (Bushell et al., 2008; Ozkan et al., 2013). Examination of the surface residues buried at the Ig1-Ig1 interfaces of the structures presented here and the key residues responsible for the NTRI dimerization (Figure 4b) are all strongly conserved among the five IgLONs. There is also a second region in the binding interface (114–127 in NTRI) which is more loosely conserved (Figure 7d) and may be responsible for interaction stabilization, which may explain the observed variation in apparent affinities.
DISCUSSION
Cell surface glycoproteins (surfaceome) are key elements in the specification of each synaptic contact in the vertebrate brain. Although these proteins have diverse structural and functional identities, they work in concert during the development and stabilization of synaptic connections (de Wit and Ghosh, 2016). Although the composition of the mammalian surfaceome is known in some detail, the understanding of the function and interaction network of its receptors is in its infancy.
By using a partial library of human and mouse cell surface neuronal proteins, we have identified many unpublished ligand-receptor pairs, and we have characterized the binding and three-dimensional structures of three IgLON protein family members. The ELISA-based approach we have used is based on a similar protocol described by Wojtowicz (Wojtowicz et al., 2007) and Visser (Visser et al., 2015), but adapted to a 384-well format (Ozgul et al., 2019). The method has been extensively validated, and advantages and disadvantages have been discussed in depth elsewhere (Bushell et al., 2008; Ozkan et al., 2013; Visser et al., 2015; Wojtowicz et al., 2007). An additional limitation of this approach relates to the cloning boundaries chosen when synthetic genes are designed. All of our genes are cloned in frame with the prolactin leader peptide, however, it’s possible that cleavage of the natural signal peptide may be important to generate a mature functional protein. For example, ELFN1 interaction with mGluRs appears to be dependent on the presence of the first amino acid after cleavage of the endogenous leader peptide (Dunn et al., 2018) and the presence of the FNIII domain. Probably the major limitation of this protein-protein interaction assay relates to the completeness of the library, whereby pairs of proteins can be identified only if they are present in the library. Nevertheless, even using a limited library of genes we have identified 200 ligand-receptors pairs, the majority of which were already known and serve as important built-in controls that reflect the sensitivity and robustness of our screen. Because all positive interactions have been identified only by ELISA, it will be important for other interested research groups to independently validate them using unrelated biochemical or biophysical methods. This aspect also highlights the bias of our gene selection toward well studied families of proteins, as does the low rate of false negatives, estimated by the addition of 26 Fc proteins with known interactors. Furthermore, as we detail elsewhere (Ozgul et al., 2019), our ELISA assay identifies interactions with affinities of ~10–20 μM or tighter. Hence, we suspect that some published interactions will not be identified in our screen if the affinity of the interaction is weaker than ~10–20 μM.
Interestingly, we identified several receptors (bait) that bind to more than one ligand (prey) such as SFRP1, LRRTMs, NTNs, Cochlin, etc. While this promiscuity has been identified previously with the panoply of proteins directly interacting with the Neurexins (Boucard et al., 2012; de Wit et al., 2009; Geppert et al., 1998; Ichtchenko et al., 1995; Ko et al., 2009; Matsuda et al., 2016; Sterky et al., 2017; Sugita et al., 2001; Uemura et al., 2010; Zhang et al., 2010), we expanded the repertoire of proteins that bind multiple, structurally diverse, ligands. Although these unpublished protein-protein interactions need independent validation, our high throughput screening results suggest that “hub” proteins, intended as receptors that bind to multiple ligands, are more common among synaptic proteins than previously appreciated.
The IgLON protein family was of particular interest because we found that each IgLON binds to all four other members and because all five genes have been linked to human diseases. Although interactions among certain members of the IgLON family were previously described (Gil et al., 1998), the specificity of interaction within the family, to the best of our knowledge, has never been reported. While the architecture of the IgLONs was not known at the time, Eagleson and co-workers used domain deletion to identify neurite outgrowth promoting activity of LSAMP to its first Ig domain (Eagleson et al., 2003). Our study indicates that all IgLONs are constitutive homodimers interacting through their first Ig domains enabling them to bind across cells or across the synaptic cleft with high affinity and in a Ca2+-independent manner. Owing to their high affinity of interaction and their overall inter-domain flexibility, cis interactions are still possible, especially in the absence of other IgLON protomers in the vicinity. Because the affinity of the homodimer interaction is, on average, weaker than that of the heterodimer (Figure 7a), heterodimerization and trans interaction may remain dominant in vivo and in vitro, as revealed by the fact that we were able to crystalize the NEGR1/IgLON5 complex. Similarly, a steady-state experiment like the ELISA assay, will allow the formation of both homo- and hetero-complexes if their affinities are similar. Considering the binding affinities measured by titration ELISA are likely to be ~2 orders of magnitude overestimated because of the avidity effect of the capture antibodies, they are consistent with the dissociation constant determined by solution AUC measurements (KD =~110 nM). From a structural perspective, this high affinity derives from the complementary shape and size of the interface and from its highly hydrophobic nature (Figure 4b).
The dimeric architecture determined by X-ray crystallography was confirmed in solution by three independent technologies: SEC, AUC, and SAXS. SEC and AUC identify a monomer-dimer equilibrium and variations of the f/f0 values indicate that the dimer has a greater anisotropy than the monomer. SAXS parameters and ab initio modeling clearly resolve NTRI as an elongated V-shaped structure of ~190 Å long, and these structural models precisely overlap with all three crystal structures. As expected from their high sequence identities, pairwise superimpositions of homo- or heterodimeric Ig1-Ig1 dimers in all crystal forms presented in this study have backbone RMSD values less than 1 Å, indicating that the architecture of the Ig1-Ig1 dimeric association is rigorously preserved across the IgLON family (Figure 5b). However, superimposition of the entire structures yields higher RMSD values, indicating that, as a whole, IgLONs have, some degree of inter-domain flexibility, which appears to be evolutionarily conserved and important for their biological function.
Functionally, because IgLONs are GPI anchored proteins, they can act as structural elements that keep pre- and post-synaptic sides together and aligned; however, they lack intracellular signaling capabilities. Interestingly, a recent report indicates that at least NTRI and NEGR1 are specifically shed from the cell surface (Sanz et al., 2015). Ectodomain shedding can interrupt a signal and/or generate a new biologically active fragment, which can diffuse in nearby spaces and bind to other unknown receptors. A recent report shows that NEGR1 promotes neuronal arborization in Tyrosine-protein kinase Fibroblast growth factor receptor 2 (FGFR2)- and ERK1/2-dependent manners (Szczurkowska et al., 2018), suggesting that FGFR2 may be a receptor for NEGR1. Other receptors for other IgLONs remain to be discovered.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Davide Comoletti (comoleda@rwjms.rutgers.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
HEK293 Cell Culture
HEK293S GNT1-cells (from ATCC) were cultured in DMEM with 5% fetal bovine serum (FBS) at 37°C, 5% CO2. For stable cell line protein production for crystallography experiments, HEK293S GNT1-cells were cultured in DMEM containing 5% FBS supplemented with 500 μg/mL of G418 at 37°C, 5% CO2. HEK293F suspension adapted cells (from Thermo Fisher Scientific) were cultured in FreeStyle™ 293 Expression Medium at 37°C, 5% CO2 and used for ELISA experiments.
METHOD DETAILS
Molecular biology –
Amino acid sequences from a gene target list were derived from the appropriate human or mouse entry in the UNIPROT data base (Uniprot.org). Cloning boundaries were chosen between the first amino acid after the signal peptide and the last amino acid before the transmembrane domain, the ω site for GPI-linked proteins, or the very last amino acid of secreted proteins. Genes were custom synthesized by Gen9 (Cambridge, MA) and fully sequenced, and the DNA sequence of each ecto-domain was cloned in frame as C-terminal Fc or AP-fusion proteins using 5’ NotI and 3’ XbaI sites into modified pCMV6-XL4 expression vectors. Each expression vector was verified by DNA sequencing. Between the ectodomain of the protein of interest and the Fc domain, there is a 3CPro cleavage site (LEVLFQ/GP) that can be used to cleave the protein for purification purposes. The AP construct differs from the Fc construct as it lacks the 3CPro cleavage site, contains AP in place of the Fc and also carries a His6 tag at its C-terminus, in frame with the AP. The fact that AP is enzymatically active provides a simple way to test expression and measure accurate protein concentration. Both constructs carry an N-terminal DYKDDDDK tag inserted after the prolactin leader peptide (MDSKGSSQKGSRLLLLLVVSNLLLCQGVVSTPVV). A few of the genes were obtained from collaborators and although these constructs do not have the prolactin leader peptide and the DYKDDDDK tag, they all maintain the Fc and AP moieties necessary for the ELISA assay.
Protein expression –
For the ELISA-based assay, proteins were expressed by HEK293F suspension adapted cells obtained from ThermoFisher Scientific. These cells were cultured in FreeStyle™ 293 Expression Medium and transfection was achieved by mixing 2μg of cDNA with 6μg of polyethylenimine (PEI) and adding the mixture to one million cells. Conditioned media was harvested 4 days after transfection and kept at 4°C for up to one week. Fc constructs were routinely characterized by SDS-PAGE to ensure protein integrity and confirm the MW of the sample. Fc fusion proteins were quantified by Western blot using a known quantity of Fc-only protein. AP fusion proteins are quantified using calf intestinal alkaline phosphatase (CIP, New England Biolabs) activity. Briefly, the activity of 5 μL of conditioned medium is compared with the activity of 1 μL of CIP at room temperature after 1 minute in 50 μL of reaction in FreeStyle. All of the AP-fusion proteins expressed at levels above 1 U/μL of CIP (where 1 U=10 pg of purified CIP).
ELISA protocol –
An ELISA-based assay was used to test the binding between ectodomain-Fc and ectodomain-AP fusions. Each data point was generated once, but each ectodomain was tested in both orientations as detailed on page 5. 15 μl of a solution at 3 μg/mL of mouse anti-AP (IgGAb-1 clone 8B6.18 Thermo Fisher Scientific; Waltham, MA) in 1× PBS was added to each well of 384-well plates (20 to 25 plates each experiment) using an automated multichannel pipette (Viaflo Assist, Integra), sealed and incubated overnight at 4°. The following day, plates were was hed, and 1% casein was added as a blocking agent, which was removed after 1 hr at room temperature using an automated microplate washer (HydroSpeed, Tecan). Subsequently, to each well, 9 μl of ecto-AP conditioned medium containing 2 μL of monoclonal mouse anti-human IgG1-HRP (2 μg/ml; Serotec; Raleigh, NC) was added using an automated plate copier (Viaflo96, Integra) along with 7 μl of ecto-Fc culture medium. Plates were sealed and incubated for 4 hr at room temperature in the dark.
Plates were subsequently washed, and 15 μL 1-Step Ultra TMB-ELISA HRP substrate was added using an automated multichannel pipette (Viaflo Assist, Integra); after 1 hr incubation at room temperature, the absorbance at 650 nm was recorded with a microplate Spectramax i3 plate reader (Molecular Devices). Finally, plates were scanned to obtain matching images of the 650 nm reading. Positive controls (known interactors like bNRXN/NLGN1, and FLRT3/LPHN3) were used to gauge the sensitivity of the assay in one plate and negative controls (wells where the AP antibody was omitted or the Fc-only sample was added) were used to obtain background values to quantify the positive reactions (for 10 random plates, the mean background Abs650 = 0.11 ± 0.074). Interacting proteins were identified as those pairs that had absorbance readings at 650 (Abs650) >2.5-fold binding above background (FOB) levels and were not present in more than one plate (bait) (Ozgul et al., In press).
Biolayer Interferometry (BLI) analysis –
BLI binding experiments were conducted in technical triplicate using a BLItz instrument (ForteBio, Menlo Park, CA) at room temperature. Anti-human Fc capture Biosensors were pre-wetted for 10 min in 300 μl of 10 mM Hepes, pH 7.4, 150 mM NaCl, 5 mM CaCl2, 5 mM MgCl2, and 1% (w/v) bovine serum albumin (BSA) prior to use. Subsequently, the sensor tips were incubated for 10 min with conditioned medium of HEK293 cells transiently transfected with the appropriate IgLON-Fc, to capture the expressed protein. The binding reaction occurred under agitation in a 4 μl drop containing various concentrations of purified proteins. Both association and dissociation were allowed to occur for 30 s. Nonspecific binding and instrument noise were subtracted by using a sensor tip saturated with Fc fragment alone.
Protein expression and purification for crystallography –
Cell culture and transfection –
HEK293S GnTI− cells were obtained from American Type Culture Collection (ATCC CRL-3022). These cells lack N-acetylglucosaminyltransferase I (GnTI) activity, and consequently glycosylation remains restricted to a homogeneous seven-residue oligosaccharide (Reeves et al., 2002), thus improving the chance of obtaining diffracting crystals. These cells were grown in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 5% fetal bovine serum (FBS), maintained in a humidified incubator at 5% CO2 and 95% air. Stable cells lines were made for each construct by transfecting each cDNA using the Calcium Phosphate method. Cells were selected in DMEM with 5% FBS supplemented with 500 μg/mL of G418 (Geneticin, Sigma) (Comoletti et al., 2003). Stable lines underwent a round of clonal selection using Pyrex cloning rings (Corning) before final amplification, freezing of the highest expressing clones, and large-scale production.
Expression and purification –
Proteins were expressed as secreted entities in the cell culture medium. The stable cell lines expressing the ectodomain of the IgLON-Fc constructs were maintained at 37°C and 5% CO2 in DMEM containing 2–5% FBS. Proteins were affinity purified using Protein-A Sepharose 4 fast flow resin (GE Healthcare). Saturated resin was washed extensively (50mM Tris HCl pH 7.4 and 450mM NaCl), equilibrated with 50 mM Tris HCl pH 7.4, 150 mM NaCl and 1 mM DTT, and subsequently cleaved with 3C protease (10 μg/mL of enzyme in 50 mM Tris HCl pH 7.4 and 150 mM NaCl, and 1 mM DTT) to remove the Fc fragment and allow elution of the ectodomain. Eluted protein was concentrated to 2 mL with Vivaspin concentrators (Sartorius-Stedim) and further purified by size exclusion chromatography using a HiLoad 16/600 Superdex 200 PG (GE Healthcare). The purified preparation underwent further de-glycosylation by an overnight incubation with a GST-tagged Endoglycosidase F1 (EndoF1) in a 1:20 (w:w, enzyme:protein) ratio, at 4°C. Endo-F1 was subsequently removed on a GSTrap™ FF column (GE Healthcare), and the protein samples were further concentrated to between 3.4 and 7.2 mg/ml. For analytical size exclusion chromatography, 100 μL samples (between 10 and 100 μM) were loaded onto a Superdex 200 10/300 GL column (GE Healthcare) equilibrated in 150 mM NaCl, 10 mM Hepes at pH 7.4. Protein was used immediately or aliquoted and flash-frozen in liquid nitrogen and stored at −80°C until needed.
Crystallization and diffraction data collection –
The FLAG-NTRI-Fc construct was purified as detailed above. NTRI crystal agglomerates appeared after about three weeks in vapor-diffusion hanging drops at 20°C, mixing 1 μL protein solution with 1 μL of 1.2 M Na,K-Tartrate, 0.1 M Tris-HCl at pH 8.5. One such agglomerate was used to prepare microseeds using a Seed Bead (Hampton Research) and single crystals grew to their maximal size at 20°C after 3–4 months using 0.9 M Na,K-Tartrate, 0.1 M Tris-HCl at pH 8.5 as crystallization solution. The single crystals (~400×400×400 μm in size) were harvested with cryo-loops, soaked with 1.0 M Na,K-Tartrate, 0.1 M TRIS-HCl at pH 8.5, supplemented with 20% (v/v) glycerol for their cryo-protection and flash-cooled directly in liquid nitrogen before diffraction data collection. Purified IgLON5 and NEGR1 were mixed at a 1:1 molar ratio and single crystals were grown at 20°C to full size in 1.2 M MgS O4, 0.1 MES-NaOH at pH 6.5 with 20% (v/v) glycerol as cryo-protectant. IgLON5 crystals were grown to full size at 20°C in 0.6 M Na,K-Tartrate, 0.1M BTP at pH 7.0, with 20% (v/v) glycerol as cryo-protectant.
Diffraction data sets were collected at Cornell High Energy Synchrotron Source (CHESS) beamline F1. Complete data sets were collected from individual crystals under a cryogenic stream at 100K and diffraction data were processed with XDS (Kabsch, 2010). The NTRI dataset was scaled using AIMLESS (Evans and Murshudov, 2013), while the IgLON5 homodimer and IgLON5:NEGR1 complex were scaled using the XDS package. Datasets for NTRI, IgLON5 and the IgLON5:NEGR1 complex with best overall quality (collected to resolutions of 3.45 Å, 4.0 Å and 3.3 Å, respectively) were used to solve the corresponding X-ray crystal structures.
Structure solution and refinement –
The NTRI structure solution was found by molecular replacement with MoRDa (Vagin and Lebedev, 2015) via the CCP4 online server (Winn et al., 2011), using the Drosophila melanogaster DIP-alpha Domain 1+2 (PDB, 5EO9 chain B) as a search model (30.0% identity and 61.8% similarity). It consisted of a partial model made of two monomers of the two first Ig domains (Ig1 and Ig2) of NTRI in a ‘head-to-tail’ arrangement. The placed Ig domains in NTRI were then used as search models to solve the structure of the IgLON5:NEGR1 complex by molecular replacement using Phaser (McCoy et al., 2007). Ig2 of IgLON5 was used as a search model for the IgLON5’s Ig3 in Phaser, and the complex was then modeled manually using COOT (Emsley et al., 2010) and refined using Phenix (Afonine et al., 2012). With all domains placed in the IgLON5:NEGR1 model, knowledge of its architecture was used to manually place the main beta strands of NTRI’s third domain in the relatively weak density available using COOT and the model was further refined using Phenix. Using the IgLON5 molecule in the complex structure as a search model, Phaser was then used to place the individual Ig domains in the IgLON5 homodimer electron density, which was further refined using Phenix. In each model, amino-acid residues were excluded where there was not sufficient electron density to unambiguously place them, particularly in the third domain, which displayed comparatively poor electron density compared to Ig1 and Ig2 domains. Further refinement and model building in iterative cycles led us to remove several poorly defined residues. However, the poorly-defined Ig3 domain in monomer A was kept in the final model of IgLON5, as its position was restrained by non-crystallographic symmetry.
Small angle X-ray scattering –
NTRI was purified and buffer exchanged in phosphate buffer saline (PBS) at pH 7.4 by size exclusion chromatography, and the fraction corresponding to the main peak was collected without further concentration. The NTRI fraction and PBS from nearby fractions (as blank, to record background signal) were used to collect SAXS data using a synchrotron beam line (G1, CHESS, Cornell). Twenty exposures of one second each at 20°C were taken, and, after ensuring that no radiation damage was detected, data reduction (merging, averaging and buffer subtraction) and initial assessment of the data quality (e.g. Guinier plot, and Kratky plot) were done with BioXTAS RAW (Hopkins et al., 2017). Subsequent data analysis was carried out with ATSAS 2.8.3 software package (Franke et al., 2017). In particular, the probable inter-atomic distance distribution of vector lengths, P(r), was calculated using the indirect Fourier transform method of Svergun as implemented in the program Primus. DAMMIF (Franke et al., 2017) ab initio bead modeling was performed against NTRI SAXS data and provided 15 independent models which were spatially aligned and averaged by DAMAVER and DAMFILT (Franke et al., 2017). Final alignment with the crystal structure of NTRI was done manually in PyMol.
Analytical ultracentrifugation –
The experiments were performed in a Beckman Optima XL-I analytical ultracentrifuge at the Center for Analytical Ultracentrifugation of Macromolecular Assemblies at the University of Texas Health Science Center at San Antonio. NTRI samples were measured at 0.07 mg/mL (~0.67 OD at 230nm; ~2.0 μM) and at ~0.34 mg/mL (~0.8 OD at 280 nm; 10 μM) in PBS at pH 7.4. All samples were run at 35,000 rpm, and scanned at 230 nm and 281 nm, and data were acquired in intensity mode. Data were analyzed with UltraScan-III v.3.1, release 1964. Experimental data were collected at 20°C using 1.2 cm epon 2-channel centerpieces (Beckman-Coulter), using an An60Ti rotor. The partial specific volume was estimated by UltraScan from protein sequence analogous to methods outlined by Laue et al. (Laue et al., 1992) and found to be 0.726 mL/g.
AUC data analysis:
Sedimentation and diffusion transport in the ultracentrifugation cell is described by the Lamm equation, which can be solved using adaptive finite element methods (Cao and Demeler, 2005, 2008). Two-dimensional spectrum analysis (2DSA) and whole boundary data obtained in sedimentation velocity experiments are fitted by linear combinations of such solutions using advanced optimization routines (Gorbet et al., 2014). Sedimentation velocity data were analyzed according to methods described by Demeler (Demeler, 2010). Optimization was performed by 2DSA with simultaneous removal of time- and radially-invariant noise contributions (Schuck and Demeler, 1999). Diffusion-corrected integral sedimentation coefficient distributions were obtained from the enhanced van Holde – Weischet analysis (Demeler and van Holde, 2004). Molecular weights and frictional ratios were determined with the parametrically constrained spectrum analysis (Gorbet et al., 2014). A reversibly self-associating monomer-dimer model was fitted to the sedimentation velocity data using the genetic algorithm-Monte Carlo analysis (Demeler et al., 2010). During the fit the known molecular weight (which included the six N-linked glycosylation sites), was fixed, and the partial specific volume, frictional ratios, KD and koff rate were floated. All calculations were performed on the Stampede and Lonestar cluster at the Texas Advanced Computing Center at the University of Texas at Austin.
Cell aggregation assay –
The extracellular domains of NTRI and NEGR1 were cloned using 5’ NotI and 3’ XbaI sites into a pcDNA3.1 plasmid in frame with the transmembrane and intracellular domains of CASPR2, which also provides the leader peptide. HEK293F suspension cells (0.8 mL at 1 million cell/mL) were co-transfected with 2 mg of NTRI-C2 and pGFP or NEGR1-C2 and pRFP plasmids at a 1:1 ratio using PEI reagent according to manufacturer`s protocol. Four days post transfection equal amounts of cells were mixed in a volume of 120 mL and let to incubate for 0, 30, 60 min, and occasionally swirled. After each time point, 30 mL drops were used to take images with a Zeiss LSM 700 confocal microscope using three channels for every picture: DIC, GFP (488 nm) and RFP (647nm).
QUANTIFICATION AND STATISTICAL ANALYSIS
See table 1 for crystallography methods and statistical analysis.
DATA AND SOFTWARE AVAILABILITY
The atomic coordinates of NTRI and IgLON5 homodimers and the NEGR1/IgLON5 heterodimer have been deposited in the Protein Data Bank under ID codes 6DLF, 6DLE and 6DLD, respectively.
Supplementary Material
Highlights.
A proteomic screen found 200 neuronal cell surface protein-protein interactions
89 of these interactions appeared to be previously unpublished
The five interacting IgLON proteins were identified and further characterized
Ranaivoson et al. present the structures of NTRI, IgLON5 and the NEGR1/IgLON5 complex
ACKNOWLEDGEMENTS
We would like to thank Woj Wojtowicz for generously providing some of the plasmids used here and for priceless advice during the development of the assay; Dr Anne Houdusse for providing part of her infrastructures at the Institut Curie (France) to F.M.R for the completion of the study.
This work was funded in part by the RWJ Foundation grant #74260, NJDOH CBIR16PIL035, National Science Foundation grant IOS-1755189 to DC and MCB-1450895 to DC & GTM, NIH grant R01 NS097161 to EÖ, AUC analysis was supported by NIH grant GM120600 and NSF grant NSF-ACI-1339649 (to BD). Supercomputer calculations were performed on Comet at the San Diego Supercomputing Center (support through NSF/XSEDE grant TG-MCB070039N to BD) and on Lonestar-5 at the Texas Advanced Computing Center (supported through UT grant TG457201 to BD). LST was supported by the NIGMS under T32 GM008339.
This work is based upon research conducted at the Cornell High Energy Synchrotron Source (CHESS), which is supported by the National Science Foundation under award DMR-1332208, using the Macromolecular Diffraction at CHESS (MacCHESS) facility, which is supported by award GM-103485 from the National Institute of General Medical Sciences, National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
G.T.M. is a founder of Nexomics Biosciences, Inc.
REFERENCES
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, and Adams PD (2012). Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68, 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H, Erez E, Martz E, Pupko T, and Ben-Tal N (2010). ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res 38, W529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bausch-Fluck D, Hofmann A, Bock T, Frei AP, Cerciello F, Jacobs A, Moest H, Omasits U, Gundry RL, Yoon C, et al. (2015). A mass spectrometric-derived cell surface protein atlas. PLoS One 10, e0121314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucard AA, Ko J, and Sudhof TC (2012). High affinity neurexin binding to cell adhesion G-protein-coupled receptor CIRL1/latrophilin-1 produces an intercellular adhesion complex. J Biol Chem 287, 9399–9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brevik EJ, van Donkelaar MM, Weber H, Sanchez-Mora C, Jacob C, Rivero O, Kittel-Schneider S, Garcia-Martinez I, Aebi M, van Hulzen K, et al. (2016). Genome-wide analyses of aggressiveness in attention-deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet 171, 733–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushell KM, Sollner C, Schuster-Boeckler B, Bateman A, and Wright GJ (2008). Large-scale screening for novel low-affinity extracellular protein interactions. Genome Res 18, 622–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butko MT, Savas JN, Friedman B, Delahunty C, Ebner F, Yates JR 3rd, and Tsien RY (2013). In vivo quantitative proteomics of somatosensory cortical synapses shows which protein levels are modulated by sensory deprivation. Proc Natl Acad Sci U S A 110, E726–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W, and Demeler B (2005). Modeling analytical ultracentrifugation experiments with an adaptive space-time finite element solution of the Lamm equation. Biophys J 89, 1589–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W, and Demeler B (2008). Modeling analytical ultracentrifugation experiments with an adaptive space-time finite element solution for multicomponent reacting systems. Biophys J 95, 54–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo RA, Ozkan E, Menon KP, Nagarkar-Jaiswal S, Lee PT, Jeon M, Birnbaum ME, Bellen HJ, Garcia KC, and Zinn K (2015). Control of Synaptic Connectivity by a Network of Drosophila IgSF Cell Surface Proteins. Cell 163, 1770–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comoletti D, Flynn R, Jennings LL, Chubykin A, Matsumura T, Hasegawa H, Sudhof TC, and Taylor P (2003). Characterization of the interaction of a recombinant soluble neuroligin-1 with neurexin-1beta. J Biol Chem 278, 50497–50505. [DOI] [PubMed] [Google Scholar]
- Comoletti D, Flynn RE, Boucard AA, Demeler B, Schirf V, Shi J, Jennings LL, Newlin HR, Sudhof TC, and Taylor P (2006). Gene selection, alternative splicing, and post-translational processing regulate neuroligin selectivity for beta-neurexins. Biochemistry 45, 12816–12827. [DOI] [PubMed] [Google Scholar]
- Cui Y, Ying Y, van Hasselt A, Ng KM, Yu J, Zhang Q, Jin J, Liu D, Rhim JS, Rha SY, et al. (2008). OPCML is a broad tumor suppressor for multiple carcinomas and lymphomas with frequently epigenetic inactivation. PLoS One 3, e2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Arcangelo G, Homayouni R, Keshvara L, Rice DS, Sheldon M, and Curran T (1999). Reelin is a ligand for lipoprotein receptors. Neuron 24, 471–479. [DOI] [PubMed] [Google Scholar]
- D’Arcangelo G, Miao GG, Chen SC, Soares HD, Morgan JI, and Curran T (1995). A protein related to extracellular matrix proteins deleted in the mouse mutant reeler. Nature 374, 719–723. [DOI] [PubMed] [Google Scholar]
- de Wit J, and Ghosh A (2016). Specification of synaptic connectivity by cell surface interactions. Nat Rev Neurosci 17, 22–35. [DOI] [PubMed] [Google Scholar]
- de Wit J, Sylwestrak E, O’Sullivan ML, Otto S, Tiglio K, Savas JN, Yates JR 3rd, Comoletti D, Taylor P, and Ghosh A (2009). LRRTM2 interacts with Neurexin1 and regulates excitatory synapse formation. Neuron 64, 799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeler B (2010). Methods for the design and analysis of sedimentation velocity and sedimentation equilibrium experiments with proteins Curr Protoc Protein Sci Chapter 7, Unit 7 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeler B, Brookes E, Wang R, Schirf V, and Kim CA (2010). Characterization of reversible associations by sedimentation velocity with UltraScan. Macromol Biosci 10, 775–782. [DOI] [PubMed] [Google Scholar]
- Demeler B, and van Holde KE (2004). Sedimentation velocity analysis of highly heterogeneous systems. Anal Biochem 335, 279–288. [DOI] [PubMed] [Google Scholar]
- Dunn HA, Patil DN, Cao Y, Orlandi C, and Martemyanov KA (2018). Synaptic adhesion protein ELFN1 is a selective allosteric modulator of group III metabotropic glutamate receptors in trans. Proc Natl Acad Sci U S A 115, 5022–5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eagleson KL, Pimenta AF, Burns MM, Fairfull LD, Cornuet PK, Zhang L, and Levitt P (2003). Distinct domains of the limbic system-associated membrane protein (LAMP) mediate discrete effects on neurite outgrowth. Mol Cell Neurosci 24, 725–740. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans PR, and Murshudov GN (2013). How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr 69, 1204–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke D, Petoukhov MV, Konarev PV, Panjkovich A, Tuukkanen A, Mertens HDT, Kikhney AG, Hajizadeh NR, Franklin JM, Jeffries CM, et al. (2017). ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J Appl Crystallogr 50, 1212–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaig C, Graus F, Compta Y, Hogl B, Bataller L, Bruggemann N, Giordana C, Heidbreder A, Kotschet K, Lewerenz J, et al. (2017). Clinical manifestations of the anti-IgLON5 disease. Neurology 88, 1736–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelpi E, Hoftberger R, Graus F, Ling H, Holton JL, Dawson T, Popovic M, Pretnar-Oblak J, Hogl B, Schmutzhard E, et al. (2016). Neuropathological criteria of anti-IgLON5-related tauopathy. Acta Neuropathol 132, 531–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese A, Cox DM, and Butler MG (2015). Partial Deletion of Chromosome 1p31.1 Including only the Neuronal Growth Regulator 1 Gene in Two Siblings. J Pediatr Genet 4, 23–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geppert M, Khvotchev M, Krasnoperov V, Goda Y, Missler M, Hammer RE, Ichtchenko K, Petrenko AG, and Sudhof TC (1998). Neurexin I alpha is a major alpha-latrotoxin receptor that cooperates in alpha-latrotoxin action. J Biol Chem 273, 1705–1710. [DOI] [PubMed] [Google Scholar]
- Gil OD, Zanazzi G, Struyk AF, and Salzer JL (1998). Neurotrimin mediates bifunctional effects on neurite outgrowth via homophilic and heterophilic interactions. J Neurosci 18, 9312–9325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil OD, Zhang L, Chen S, Ren YQ, Pimenta A, Zanazzi G, Hillman D, Levitt P, and Salzer JL (2002). Complementary expression and heterophilic interactions between IgLON family members neurotrimin and LAMP. J Neurobiol 51, 190–204. [DOI] [PubMed] [Google Scholar]
- Gorbet G, Devlin T, Hernandez Uribe B.I., Demeler AK, Lindsey ZL, Ganji S, Breton S, Weise-Cross L, Lafer EM, Brookes EH, et al. (2014). A parametrically constrained optimization method for fitting sedimentation velocity experiments. Biophys J 106, 1741–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins JB, Gillilan RE, and Skou S (2017). BioXTAS RAW: improvements to a free open-source program for small-angle X-ray scattering data reduction and analysis. J Appl Crystallogr 50, 1545–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichtchenko K, Hata Y, Nguyen T, Ullrich B, Missler M, Moomaw C, and Sudhof TC (1995). Neuroligin 1: a splice site-specific ligand for beta-neurexins. Cell 81, 435–443. [DOI] [PubMed] [Google Scholar]
- Kabsch W (2010). Xds. Acta Crystallogr D Biol Crystallogr 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karis K, Eskla KL, Kaare M, Taht K, Tuusov J, Visnapuu T, Innos J, Jayaram M, Timmusk T, Weickert CS, et al. (2018). Altered Expression Profile of IgLON Family of Neural Cell Adhesion Molecules in the Dorsolateral Prefrontal Cortex of Schizophrenic Patients. Front Mol Neurosci 11, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J, Fuccillo MV, Malenka RC, and Sudhof TC (2009). LRRTM2 functions as a neurexin ligand in promoting excitatory synapse formation. Neuron 64, 791–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koido K, Janno S, Traks T, Parksepp M, Ljubajev U, Veiksaar P, Must A, Shlik J, Vasar V, and Vasar E (2014). Associations between polymorphisms of LSAMP gene and schizophrenia. Psychiatry Res 215, 797–798. [DOI] [PubMed] [Google Scholar]
- Laue T, Shah BD, Ridgeway T, and Pelletier SL, (1992). Computer-aided interpretation of sedimentation data for proteins. [Google Scholar]
- Lee K, Kim Y, Lee SJ, Qiang Y, Lee D, Lee HW, Kim H, Je HS, Sudhof TC, and Ko J (2013). MDGAs interact selectively with neuroligin-2 but not other neuroligins to regulate inhibitory synapse development. Proc Natl Acad Sci U S A 110, 336–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Arias-Vasquez A, Sleegers K, Aulchenko YS, Kayser M, Sanchez-Juan P, Feng BJ, Bertoli-Avella AM, van Swieten J, Axenovich TI, et al. (2007). A genomewide screen for late-onset Alzheimer disease in a genetically isolated Dutch population. Am J Hum Genet 81, 17–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodge AP, Howard MR, McNamee CJ, and Moss DJ (2000). Co-localisation, heterophilic interactions and regulated expression of IgLON family proteins in the chick nervous system. Brain Res Mol Brain Res 82, 84–94. [DOI] [PubMed] [Google Scholar]
- Matsuda K, Budisantoso T, Mitakidis N, Sugaya Y, Miura E, Kakegawa W, Yamasaki M, Konno K, Uchigashima M, Abe M, et al. (2016). Transsynaptic Modulation of Kainate Receptor Functions by C1q-like Proteins. Neuron 90, 752–767. [DOI] [PubMed] [Google Scholar]
- Matsukawa H, Akiyoshi-Nishimura S, Zhang Q, Lujan R, Yamaguchi K, Goto H, Yaguchi K, Hashikawa T, Sano C, Shigemoto R, et al. (2014). Netrin-G/NGL complexes encode functional synaptic diversification. J Neurosci 34, 15779–15792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007). Phaser crystallographic software. J Appl Crystallogr 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minhas HM, Pescosolido MF, Schwede M, Piasecka J, Gaitanis J, Tantravahi U, and Morrow EM (2013). An unbalanced translocation involving loss of 10q26.2 and gain of 11q25 in a pedigree with autism spectrum disorder and cerebellar juvenile pilocytic astrocytoma. Am J Med Genet A 161A, 787–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata S, Matsumoto N, Taguchi K, Akagi A, Iino T, Funatsu N, and Maekawa S (2003). Biochemical and ultrastructural analyses of IgLON cell adhesion molecules, Kilon and OBCAM in the rat brain. Neuroscience 117, 645–658. [DOI] [PubMed] [Google Scholar]
- Murai KK, and Pasquale EB (2003). ‘Eph’ective signaling: forward, reverse and crosstalk. J Cell Sci 116, 2823–2832. [DOI] [PubMed] [Google Scholar]
- O’Sullivan ML, de Wit J, Savas JN, Comoletti D, Otto-Hitt S, Yates JR 3rd, and Ghosh A (2012). FLRT proteins are endogenous latrophilin ligands and regulate excitatory synapse development. Neuron 73, 903–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozgul S, von Daake S, Kakehi S, Sereni D, Denissova N, Hanlon C, Huang YJ, Everett JK, Yin C, Montelione GT, et al. (2019). An ELISA-Based Screening Platform for Ligand-Receptor Discovery. Methods Enzymol 615, 453–475. [DOI] [PubMed] [Google Scholar]
- Ozgul S, von Daake S, Kakehi S, Sereni D, Dennisova N, Hanlon C, Huang YJ, Everett JK, Yin C, Montelione GT, et al. (In press). An ELISA-Based Screening Platform for Ligand-Receptor Discovery. Methods in Enzymology. [DOI] [PubMed] [Google Scholar]
- Ozkan E, Carrillo RA, Eastman CL, Weiszmann R, Waghray D, Johnson KG, Zinn K, Celniker SE, and Garcia KC (2013). An extracellular interactome of immunoglobulin and LRR proteins reveals receptor-ligand networks. Cell 154, 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez de Arce K, Schrod N, Metzbower SWR, Allgeyer E, Kong GK, Tang AH, Krupp AJ, Stein V, Liu X, Bewersdorf J, et al. (2015). Topographic Mapping of the Synaptic Cleft into Adhesive Nanodomains. Neuron 88, 1165–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves PJ, Callewaert N, Contreras R, and Khorana HG (2002). Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci U S A 99, 13419–13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert X, and Gouet P (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42, W320–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Marrero EN, Vincelli G, Jeffries CM, Shaikh TR, Pakos IS, Ranaivoson FM, von Daake S, Demeler B, De Jaco A, Perkins G, et al. (2016). Structural Characterization of the Extracellular Domain of CASPR2 and Insights into Its Association with the Novel Ligand Contactin1. J Biol Chem 291, 5788–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz R, Ferraro GB, and Fournier AE (2015). IgLON cell adhesion molecules are shed from the cell surface of cortical neurons to promote neuronal growth. J Biol Chem 290, 4330–4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P, and Demeler B (1999). Direct sedimentation analysis of interference optical data in analytical ultracentrifugation. Biophys J 76, 2288–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiradake E, del Toro D, Nagel D, Cop F, Hartl R, Ruff T, Seyit-Bremer G, Harlos K, Border EC, Acker-Palmer A, et al. (2014). FLRT structure: balancing repulsion and cell adhesion in cortical and vascular development. Neuron 84, 370–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K, Schmitt S, Bergner CG, Tyanova S, Kannaiyan N, Manrique-Hoyos N, Kongi K, Cantuti L, Hanisch UK, Philips MA, et al. (2015). Cell type- and brain region-resolved mouse brain proteome. Nat Neurosci 18, 1819–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JP, Lelianova VG, Ermolyuk YS, Vysokov N, Hitchen PG, Berninghausen O, Rahman MA, Zangrandi A, Fidalgo S, Tonevitsky AG, et al. (2011). Latrophilin 1 and its endogenous ligand Lasso/teneurin-2 form a high-affinity transsynaptic receptor pair with signaling capabilities. Proc Natl Acad Sci U S A 108, 12113–12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner C, and Wright GJ (2009). A cell surface interaction network of neural leucine-rich repeat receptors. Genome Biol 10, R99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterky FH, Trotter JH, Lee SJ, Recktenwald CV, Du X, Zhou B, Zhou P, Schwenk J, Fakler B, and Sudhof TC (2017). Carbonic anhydrase-related protein CA10 is an evolutionarily conserved pan-neurexin ligand. Proc Natl Acad Sci U S A 114, E1253–E1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struyk AF, Canoll PD, Wolfgang MJ, Rosen CL, D’Eustachio P, and Salzer JL (1995). Cloning of neurotrimin defines a new subfamily of differentially expressed neural cell adhesion molecules. J Neurosci 15, 2141–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita S, Saito F, Tang J, Satz J, Campbell K, and Sudhof TC (2001). A stoichiometric complex of neurexins and dystroglycan in brain. J Cell Biol 154, 435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczurkowska J, Pischedda F, Pinto B, Manago F, Haas CA, Summa M, Bertorelli R, Papaleo F, Schafer MK, Piccoli G, et al. (2018). NEGR1 and FGFR2 cooperatively regulate cortical development and core behaviours related to autism disorders in mice. Brain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Oida K, Ookubo M, Suzuki J, Kohno M, Murase T, Yamamoto T, and Nakai T (1996). Very low density lipoprotein receptor binds apolipoprotein E2/2 as well as apolipoprotein E3/3. FEBS Lett 386, 197–200. [DOI] [PubMed] [Google Scholar]
- Tan RPA, Leshchyns’ka I, and Sytnyk V (2017). Glycosylphosphatidylinositol-Anchored Immunoglobulin Superfamily Cell Adhesion Molecules and Their Role in Neuronal Development and Synapse Regulation. Front Mol Neurosci 10, 378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas LA, Akins MR, and Biederer T (2008). Expression and adhesion profiles of SynCAM molecules indicate distinct neuronal functions. J Comp Neurol 510, 47–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewhella J, Duff AP, Durand D, Gabel F, Guss JM, Hendrickson WA, Hura GL, Jacques DA, Kirby NM, Kwan AH, et al. (2017). 2017 publication guidelines for structural modelling of small-angle scattering data from biomolecules in solution: an update. Acta Crystallogr D Struct Biol 73, 710–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Lee SJ, Yasumura M, Takeuchi T, Yoshida T, Ra M, Taguchi R, Sakimura K, and Mishina M (2010). Trans-synaptic interaction of GluRdelta2 and Neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell 141, 1068–1079. [DOI] [PubMed] [Google Scholar]
- Vagin A, and Lebedev A (2015). MoRDa, an automatic molecular replacement pipeline, Vol 71. [Google Scholar]
- Veerappa AM, Saldanha M, Padakannaya P, and Ramachandra NB (2013). Family-based genome-wide copy number scan identifies five new genes of dyslexia involved in dendritic spinal plasticity. J Hum Genet 58, 539–547. [DOI] [PubMed] [Google Scholar]
- Visser JJ, Cheng Y, Perry SC, Chastain AB, Parsa B, Masri SS, Ray TA, Kay JN, and Wojtowicz WM (2015). An extracellular biochemical screen reveals that FLRTs and Unc5s mediate neuronal subtype recognition in the retina. Elife 4, e08149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P, Pattarini R, Rong Y, Guo H, Bansal PK, Kusnoor SV, Deutch AY, Parris J, and Morgan JI (2012). The Cbln family of proteins interact with multiple signaling pathways. J Neurochem 121, 717–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson NH, and Key B (2006). Neogenin interacts with RGMa and netrin-1 to guide axons within the embryonic vertebrate forebrain. Dev Biol 296, 485–498. [DOI] [PubMed] [Google Scholar]
- Winberg ML, Noordermeer JN, Tamagnone L, Comoglio PM, Spriggs MK, Tessier-Lavigne M, and Goodman CS (1998). Plexin A is a neuronal semaphorin receptor that controls axon guidance. Cell 95, 903–916. [DOI] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. (2011). Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtowicz WM, Wu W, Andre I, Qian B, Baker D, and Zipursky SL (2007). A vast repertoire of Dscam binding specificities arises from modular interactions of variable Ig domains. Cell 130, 1134–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi S, Hampel F, Hata K, del Toro D, Schwark M, Kvachnina E, Bastmeyer M, Yamashita T, Tarabykin V, Klein R, et al. (2011). FLRT2 and FLRT3 act as repulsive guidance cues for Unc5-positive neurons. The EMBO Journal 30, 2920–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacco A, Cooper V, Chantler PD, Fisher-Hyland S, Horton HL, and Levitt P (1990). Isolation, biochemical characterization and ultrastructural analysis of the limbic system-associated membrane protein (LAMP), a protein expressed by neurons comprising functional neural circuits. J Neurosci 10, 73–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandona MR, Sangalli CN, Campagnolo PD, Vitolo MR, Almeida S, and Mattevi VS (2017). Validation of obesity susceptibility loci identified by genome-wide association studies in early childhood in South Brazilian children. Pediatr Obes 12, 85–92. [DOI] [PubMed] [Google Scholar]
- Zhang C, Atasoy D, Arac D, Yang X, Fucillo MV, Robison AJ, Ko J, Brunger AT, and Sudhof TC (2010). Neurexins physically and functionally interact with GABA(A) receptors. Neuron 66, 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Sliwkowski MX, Mark M, Frantz G, Akita R, Sun Y, Hillan K, Crowley C, Brush J, and Godowski PJ (1997). Neuregulin-3 (NRG3): a novel neural tissue-enriched protein that binds and activates ErbB4. Proc Natl Acad Sci U S A 94, 9562–9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Yan J, Lu G, Guo Z, Fan Z, Wang J, Shi Y, Qi J, and Gao GF (2011). Binding of herpes simplex virus glycoprotein D to nectin-1 exploits host cell adhesion. Nat Commun 2, 577. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates of NTRI and IgLON5 homodimers and the NEGR1/IgLON5 heterodimer have been deposited in the Protein Data Bank under ID codes 6DLF, 6DLE and 6DLD, respectively.