Abstract
Protein-DNA interactions are critical to the regulation of gene expression, but it remains challenging to define how cell-to-cell heterogeneity in protein-DNA binding influences gene expression variability. Here we report a method for the simultaneous quantification of protein-DNA contacts by combining single-cell DNA adenine methyltransferase identification (DamID) with mRNA sequencing of the same cell (scDam&T-seq). We apply scDam&T-seq to reveal how genome-lamina contacts or chromatin accessibility correlate with gene expression in individual cells. Furthermore, we provide single-cell genome-wide interaction data on a Polycomb-group protein, RING1B, and the associated transcriptome. Our results show that scDam&T-seq is sensitive enough to distinguish mouse embryonic stem cells cultured under different conditions and their different chromatin landscapes. Our method will enable analysis of protein-mediated mechanisms that regulate cell type-specific transcriptional programs in heterogeneous tissues.
Recent advances in measuring genome architecture (Hi-C, DamID)1–4, chromatin accessibility (ATAC-seq and DNaseI-seq)5–7, various DNA modifications8–13 and histone post-translational modifications (ChIP-seq)14 in single cells have enabled characterization of cell-to-cell heterogeneity in gene regulation. More recently, multi-omics methods to study single-cell associations between genomic or epigenetic variations and transcriptional heterogeneity15–19 have allowed researchers to link upstream regulatory elements to transcriptional output from the same cell. At all gene-regulatory levels, protein-DNA interactions play a critical role in determining transcriptional outcomes, however, no method exists to obtain combined measurements of protein-DNA contacts and transcriptomes in single cells. We have therefore developed scDam&T-seq, a multi-omics method that harnesses DamID to map genomic protein localizations together with mRNA-sequencing from the same cell.
The DamID technology involves expression of a protein of interest tethered to E. coli DNA adenine methyltransferase (Dam)20. This enables detection of protein-DNA interactions through exclusive adenine methylation at GATC motifs. In vivo expression of the DamID-constructs requires transient or stable expression at low to moderate levels21. An important distinction between DamID and ChIP is the cumulative nature of the adenine methylation in living cells, allowing interactions to be measured over varying time windows. This property can be exploited to uncover protein-DNA contact histories22. For single-cell applications, a major advantage of DamID is the minimal sample handling which reduces biological losses and enables amplifications of different molecules in the same reaction mixture. To make DamID compatible with transcriptomics, we adapted the method for linear amplification, which allows simultaneous processing of DamID and mRNA by in vitro transcription without nucleotide separation.
As a proof-of-principle, we first benchmarked scDam&T-seq to the previously reported single-cell DamID (scDamID) method. Single KBM7 cells expressing either untethered Dam or Dam-LMNB1 were sorted into 384-well plates by FACS as previously described2. For scDam&T-seq, poly-adenylated mRNA is reverse transcribed into cDNA followed by second strand synthesis to create double-stranded cDNA molecules (Fig. 1a and methods). Next, the DamID-labelled DNA is digested with the restriction enzyme DpnI, followed by adapter ligation to digested gDNA (Fig. 1a), cells are pooled, and cDNA and ligated gDNA molecules are simultaneously amplified by in vitro transcription. Finally, the amplified RNA molecules are processed into Illumina libraries, as described previously23 (Fig. 1a and methods).
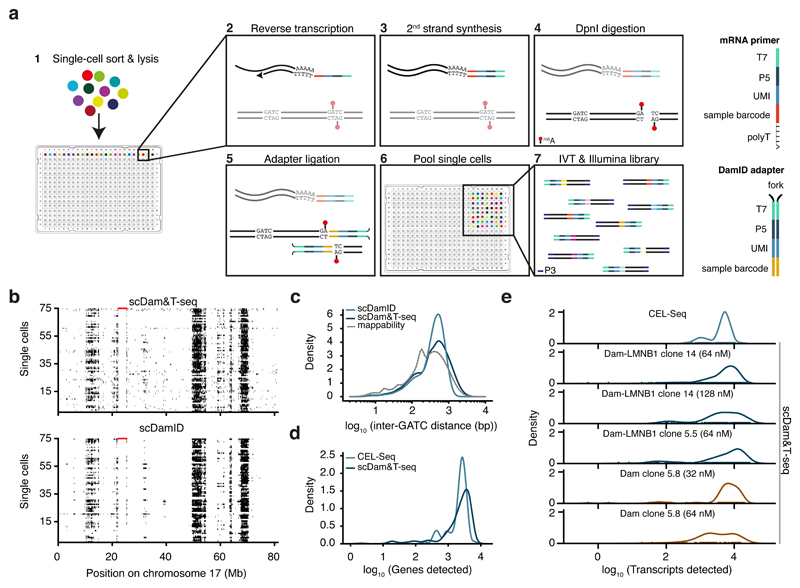
Figure 1. Quantitative comparison of scDamID, CEL-Seq and scDam&T-seq applied to KBM7 cells.
a) Schematic overview of scDam&T-seq. b) Binarized OE values (black: OE >= 1) of Dam-LMNB1 signal on chromosome 17, measured with scDam&T-seq and scDamID2 in 75 single cells with highest sequencing depth. Each row represents a single cell; each column a 100-kb bin along the genome. Unmappable genomic regions are indicated in red along the top of the track. c) Distribution of inter-GATC distances of mappable GATC fragments genome-wide (dotted line), and observed in experimental data with scDamID and scDam&T-seq for Dam-LMNB1. d) Distributions of the number of unique genes detected using CEL-Seq2 and scDam&T-seq on the same Dam-LMNB1 clone. e) Distribution of the number of unique transcripts detected by CEL-Seq (top) and scDam&T-seq for Dam and Dam-LMNB1 clones with varying DamID adapter concentrations.
The crucial modification compared to the original scDamID protocol is the linear amplification of the m6A-marked genome. The advantages of linear amplification include (1) compatibility with mRNA sequencing, (2) unbiased genomic recovery due to the amplification of single ligation events, (3) a >100-fold increase in throughput due to combined sample amplification and library preparation and (4) a resulting substantial cost reduction. Additional improvements of scDam&T-seq involve the inclusion of unique molecule identifiers (UMI) for both gDNA- and mRNA-derived reads and the use of liquid-handling robots to increase throughput and obtain more consistent sample quality (Fig. 1a and methods).
We qualitatively and quantitatively compared scDam&T-seq to previously published scDamID data in KBM7 cells2. As illustrated for chromosome 17, observed over expected (OE) scores2 captured the same LADs and cell-to-cell heterogeneity in genome-nuclear lamina (NL) interactions as previously described (Fig. 1b and Supplementary Fig. 1a). This is also illustrated by the high concordance (r = 0.97) in the contact frequencies (CFs), i.e. the fraction of cells in contact (OE >= 1) with the NL for 100-kb genomic windows (Supplementary Fig. 1b). In addition, scDam&T-seq and scDamID are similarly enriched on LADs in HT1080 cells24 (Supplementary Fig. 1c) and run-length analysis show similar prevalence of long stretches of genome-NL contacts in single cells (Supplementary Fig. 1d). Finally, comparison of auto-correlation of in silico population samples show similar underlying genomic structures, with Dam-LMNB1 measuring larger structures than untethered Dam, as indicated by the lower rate of auto-correlation decay (Supplementary Fig. 1e). Altogether these results show that scDam&T-seq successfully capturers the distribution and variability of genome-NL interactions in single cells. The median scDam&T-seq complexity of 42,192 unique DamID reads per cell, is ~4-fold reduced compared to scDamID (Supplementary Fig. 1f). This difference may be attributed to greater sequencing depth in combination with selection and manual library preparation of single cells with the highest methylation levels for scDamID, as opposed to unbiased high-throughput preparation of scDam&T-seq libraries (Supplementary Fig. 1f). Besides increased throughput, linear amplification of the DamID-products reduced the loss of reads resulting from incorrect adapter sequences (Supplementary Fig. 1g) and a more accurate genome-wide distribution of GATC fragments (Fig. 1c).
Next, we benchmarked the transcriptomic measurements from scDam&T-seq to previously obtained CEL-Seq data for KBM7 cells2. Both methods detected the expression of comparable number of genes (Median: CEL-Seq = 2508.5, scDam&T-seq = 2282.5) (Fig. 1d), and unique transcripts (Median: CEL-Seq = 4920, scDam&T-seq = 4009.5) (Supplementary Fig. 2a). Transcriptomes measured by scDam&T-seq and CEL-Seq show a high degree of correlation (Supplementary Fig. 2b left panel) and display comparable single-cell variations indicated by the fraction of cells with detected genes (Supplementary Fig. 2c left panel), as well as by the relationship between mean gene expression and the coefficient of variation (Supplementary Fig. 2d). These correlations are similar when comparing independent scDam&T-seq libraries (Supplementary Fig 2b and 2c right panels). We observe batch effects between clones, libraries and methods (Supplementary Fig. 2e). Principle component analysis to quantify batch effects in CEL-Seq and scDam&T-seq libraries showed that 16% of the total variance in transcriptional profiles can be attributed to differences between methods (scDam&T-seq and CEL-Seq) while for reference 9.7% is explained by clonal origin (Dam versus Dam-LMNB1) and 2.2% can be ascribed to differences between libraries (see methods for details). Lastly, the overall efficiency and characteristics of mRNA detection is very similar to CEL-Seq (Fig. 1e and Supplementary Fig. 2f and 2g), yet appear to reduce with increasing gDNA adapter concentrations (Fig. 1e). However, no correlations were found between the DamID and mRNA detection efficiencies within each condition (Supplementary Fig. 2h). Since lowering the double-stranded adapter concentrations does not affect DamID complexity (Supplementary Fig. 1f), mRNA detection may be further improved with reduced double-stranded adapter concentrations. In conclusion, scDam&T-seq produces single-cell data that are qualitatively and quantitatively comparable to its uncombined counterparts.
We also established scDam&T-seq in hybrid (129/Sv:CAST/EiJ) mESCs25 with auxin-inducible conditional DamID expression26 (Supplementary Fig. 3a). The median complexity of the scDam&T-seq libraries in mESCs is comparable to KBM7 cells (Supplementary Table 1) and strong overlap of DamID signal between the Dam-LMNB1 expressing mESCs and published Dam-LMNB1 bulk data27 validates the applicability of scDam&T-seq to different cell types (Supplementary Fig. 3b).
The untethered Dam enzyme was previously reported to accurately mark accessible chromatin28 we therefore wished to test the applicability of scDam&T-seq to quantify DNA accessibility and transcriptomes in single cells. We first quantified the levels of Dam methylation at transcription start sites (TSSs) and observed a sharp peak of Dam-signal that scaled in accordance with increasing gene expression levels (Fig. 2a). Similar experiments with AluI digestions did not show signatures of accessibility around TSSs of actively transcribed genes (Fig. 2b), indicating that the observed Dam accessibility patterns are the result of in vivo Dam methylation at accessible regions of the genome, and not restriction enzyme accessibility. We also observed strong Dam enrichment at active enhancers (Fig. 2c). Nucleosomes are regularly spaced around genomic elements like CTCF sites, which is a feature also observed in the scDam&T-seq data obtained with untethered Dam (Fig. 2d). The observed periodicity of 174 bp is in agreement with the reported spacing of nucleosomes in human cells29–30 (Supplementary Fig. 4a). Remarkably, the same periodicity is also apparent in single-cell samples (Fig. 2e), indicating that Dam can serve to determine nucleosome positioning in vivo in single cells.
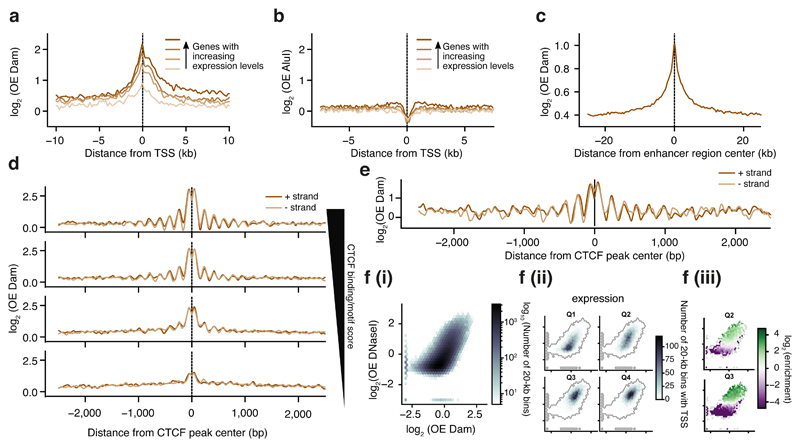
Figure 2. Untethered Dam marks accessible chromatin in single cells.
a) Log-transformed OE-values (log2OE) of Dam signal from an in silico population sample on TSS of genes grouped into four equal-sized categories with increasing expression levels (ordered light to dark). b) log2OE values obtained from AluI-derived fragments for identical TSSs as in (a). c) log2OE values of Dam signal from an in silico population sample at active enhancers (see methods for more details defining active enhancers). d) log2OE values of Dam signal from an in silico population sample at CTCF sites, stratified in four regimes of increasing CTCF binding activity (see methods for details on stratification). e) Example of log2OE Dam signal of a single-cell sample at CTCF sites with the highest CTCF binding activity. f) Relation between DNaseI (y-axis) and in silico population Dam data (x-axis): (i) Density of genomic 20-kb bins. (ii) Density of 20-kb bins with (one or more) TSSs of a gene, stratified in four gene expression quartiles from lowest (Q1) to highest (Q4) expression. (iii) Significant enrichment (red) and depletion (blue) of transcribed 20-kb regions for the two expression quantiles (Q2 and Q3). Points in the plot with fewer than 10 20-kb bins were kept gray, as well as (statistically) insignificant enrichments/depletions (see methods).
scDam&T data correlate strongly for open chromatin with DNaseI, but less at relatively condensed chromatin, where Dam distinguishes between a larger range of chromatin accessibilities (Fig. 2f (i)). This increased sensitivity is functionally related to genes with low expression levels. Stratifying genes into four expression quantiles, shows a strong depletion of DNaseI marked regions of the second expression quantile as opposed to moderate Dam signal for the same genomic regions (Fig. 2f (ii) and (iii)). This increased sensitivity of Dam in measuring lowly transcribed gene regions may be attributed to the ability of Dam to mark gene-units encompassing both active gene promoters (marked by H3K4me3) and gene bodies (marked by H3K36me3) (Supplementary Fig. 4b), whereas DNaseI has been reported to primarily detect active promoters31. Finally, we compared scDam&T-seq in mESCs cells to scNMT-seq: a method for single-cell detection of 5-methylcytosine (5mC), chromatin accessibility and mRNA19. scDam&T-seq and scNMT-seq display similar nucleosome positioning characteristics at DNaseI hypersensitivity sites (DHSs), with a 30-fold shallower sequencing depth for scDam&T-seq (Supplementary Fig. 4c). The numbers of detected genes are also very similar between methods at comparable sequencing depths (Supplementary Fig. 4d). scDam&T-seq therefore provides data quality similar to scNMT-seq, yet at greatly reduced sequencing depth.
We next determined the single-cell associations of genome-NL contacts or chromatin accessibility with gene expression in mESCs. First, the single-cell DamID profiles were converted into binary contact maps as previously described2 (Fig. 3a, step 1). For the untethered Dam enzyme, regions of high contact frequency (CF) indicate transcriptional active open chromatin configurations, while high CF regions for Dam-LMNB1 indicate association with the nuclear lamina (NL) and therefore a repressed chromatin state. Previously in KBM7 cells, the frequency with which genomic regions associate with the NL was shown to inversely correlate with gene activities2. Indeed, in mESCs, we observe that mean expression levels gradually drop with increased genome-NL CFs (Fig. 3b, left). In contrast and as expected, increased Dam CFs positively correlate with mean gene expression levels (Fig. 3b, right). To investigate the impact of genome-NL contacts and chromatin accessibility on gene expression in single cells, we determined the log fold-change in expression (log2FC) in cells showing contact and no-contact states per genomic bin (Fig. 3a, steps 2 and 3). Intriguingly, a genome-wide negative association between genome contact and expression was observed for Dam-LMNB1, and a positive association for the untethered Dam (Fig. 3c). Thus, cell-to-cell variations in genome-NL contacts impact on gene expression; regions are more likely to be active in those cells where they are detached from the NL. The positive association between log2FC in expression and contact with Dam indicates that, between single cells, a genomic region is more likely active when in an open chromatin state. These single-cell associations are largely independent of mean expression levels and expression variance (Supplementary Fig. 5a-d). Interestingly, the negative relationship between genome-NL contact and gene expression is only observed for genomic regions that infrequently associate with the NL (Fig. 3d left panel), while genes residing within medium to high open chromatin are transcriptionally most sensitive to changes in chromatin accessibility (Fig 3d. right panel). The small effect size between the associations of Dam and Dam-LMNB1 with transcription could be resulting from the limited time resolution of these experiments (12 hours) and/or the effect of the relatively large 100-kb bins. A cell line with elevated Dam-expression levels combined with more rapid inducibility may improve this. These data suggest that genomic regions that typically reside in the nucleoplasm are most sensitive to occasional NL association, and that genes respond differently to changes in accessibility depending on their chromatin contexts. Interestingly, the LADs in the low CF range are relatively depleted of constitutive chromatin marked by H3K9me3, and enriched for the facultative heterochromatin modification H3K27me3 (Supplementary Fig. 5e, top). Consistently, the chromatin state of the low CF regions is enriched for cell-type specific (facultative) fLADs, as opposed to cell-type invariant (constitutive) cLADs (Supplementary Fig. 5f, top). The opposite patterns can be observed for the Dam contact regions (Supplementary Fig. 5e and 5f, bottom). Collectively, these observations suggest that fLADs are more susceptible to dissociation from the NL and subsequent transcriptional activation compared to the H3K9me3-enriched cLADs.
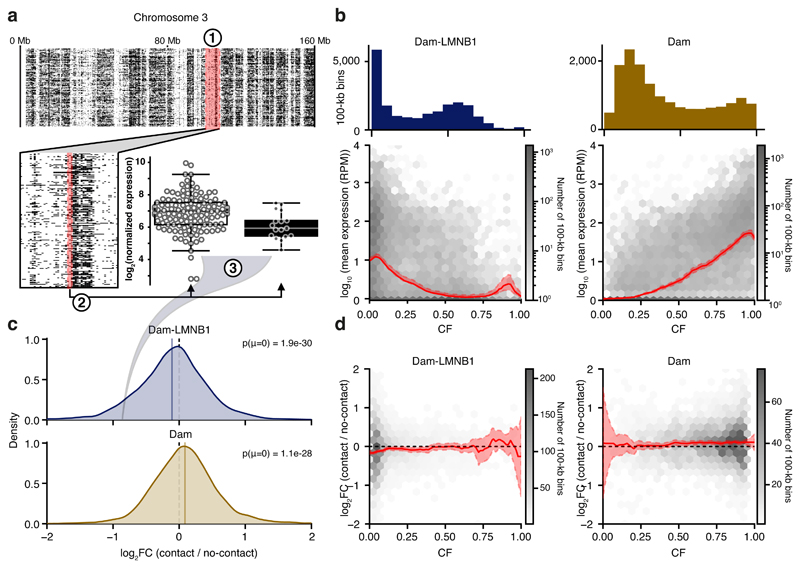
Figure 3. Parallel transcriptomic and DamID measurements link transcriptional dependencies to heterogeneity in DamID contacts.
a) Schematic of analysis to determine the log2 fold-change (log2FC) in transcription between contact and no-contact states. (1) Per genomic bin (of 100 kb), the single-cell samples are binarized into two groups, having either high (OE >= 1, black) or low (OE < 1, white) DamID signal, corresponding to a DamID contact and no contact, respectively. (2) The expression of the two groups of samples in that genomic bin is computed, and (3) a group-wise log2FC in expression is calculated. The example shows one bin on chromosome 3 where mESC Dam-LMNB1 contact is associated with a decrease in expression of about 2-fold (-1 log2FC) compared to no Dam-LMNB1 contact. The example bin displays NL-contacts in 17 out of 143 single cells and log2FC is determined based on the expression of genes in the 100-kb bin (containing 2 expressed genes). Box plots indicate the 25th and 75th percentile (box), median (line) and 1.5 times the inter-quartile range (IQR) past the 25th and 75th percentiles (whiskers). Data points are overlaid as circles. n = 126 and n = 17, in left and right box, respectively. b) Relation between expression (y-axis) and contact frequency (CF) (x-axis) defined as fraction of cells that show high DamID signals (OE >= 1) across 100-kb genomic bins. Dam-LMNB1 (left) and Dam (right) are shown, as well as the genome-wide distribution of CF values across mappable bins (histogram on top). The solid line indicates the mean, shaded area indicates 1.96 times the standard deviation around the mean. c) Distribution of expression log2FC values with Dam-LMNB1 (top) and Dam (bottom), genome-wide across 100-kb bins. Note that only 100-kb bins with at least 3 single-cell samples in both groups, and having expression in at least 20% of the single-cell samples were included in the analysis. P-values of a two-sided one-sample t-test are indicated. d) Relation between expression log2FC values and CF, for Dam-LMNB1 (left) and Dam (right). The red shadings indicate 95% confidence intervals. The solid line indicates the mean, shaded area indicates 1.96 times the standard deviation around the mean.
We next investigated how DNA accessibility relates to gene expression at an allelic resolution. First, to account for potential allelic copy number variations (CNVs) that would introduce biases in our analysis, we performed single-cell reduced-representation whole genome sequencing by substituting DpnI with AluI in the scDam&T-seq protocol (Supplementary Fig. 6a). Chromosomes 5, 8 and 12 were found frequently (partially) duplicated or lost and were excluded from our analyses (Supplementary Fig. 6a). For the Dam data, approximately 45% of reads could be attributed to either allele and the same CNVs were apparent in the resulting allelic single-cell chromatin accessibility tracks (Supplementary Fig. 6b). Surprisingly, we also detected a small fraction of cells that displayed a reverse DNA accessibility bias on chromosome 12, and a corresponding allelic bias in transcription for one cell (Supplementary Figure 6c). After excluding chromosomes with frequent CNVs as well as samples showing a CNV on any other chromosome, we found a positive allelic single-cell association between chromatin accessibility and transcription (Supplementary Fig. 6d). Therefore, scDam&T-seq can be employed to investigate single-cell allelic relationships between expression and chromatin states.
Next, we established scDam&T-seq as an in silico cell sorting strategy to identify and group cell types based on their transcriptomes and uncover the underlying cell type-specific gene regulatory landscapes from DamID data. We first performed a scDam&T-seq proof-of-principle experiment on mESCs cultured under 2i or serum conditions. scDam&T-seq derived transcriptomics separated into two distinct clusters based on independent-component analysis (Fig. 4a). Expression analysis showed signature genes differentially expressed between the two conditions (Supplementary Fig. 7a). DNA accessibility profiles generated from the two in silico transcriptome clusters showed differential accessibility patterns on a genome-wide scale. PEG10, a gene strongly upregulated under serum conditions, showed increased accessibility at the TSS and along the gene body (Fig. 4b). Interestingly, this increased accessibility stretches beyond the PEG10 gene locus, encompassing a large topologically associating domain (TAD). Genome-wide TAD analysis reveals that global changes in chromatin accessibility between 2i and serum conditions occur more within TAD domains than for randomized domains of the same size (Supplementary Fig.7b). Thus, chromatin relaxation of the TAD that encompasses PEG10 in serum conditions is illustrative of a broader phenomenon occurring within the genome-wide TAD framework. At the gene level, differential up-regulation in either 2i or serum conditions is also associated with increased DNA accessibility (Fig. 4c and Supplementary Fig. 7c). Interestingly, the increased accessibility at the TSS extends into the gene body (Supplementary Fig. 7d). The same increased accessibility is also observed in single cells for the top 5 differentially expressed genes between conditions (Supplementary Fig. 7e). Together, these results demonstrate that scDam&T-seq can be used to effectively generate cell type-specific DNA accessibility profiles from heterogeneous mixtures of cells, based on in silico identification and grouping of cell types.
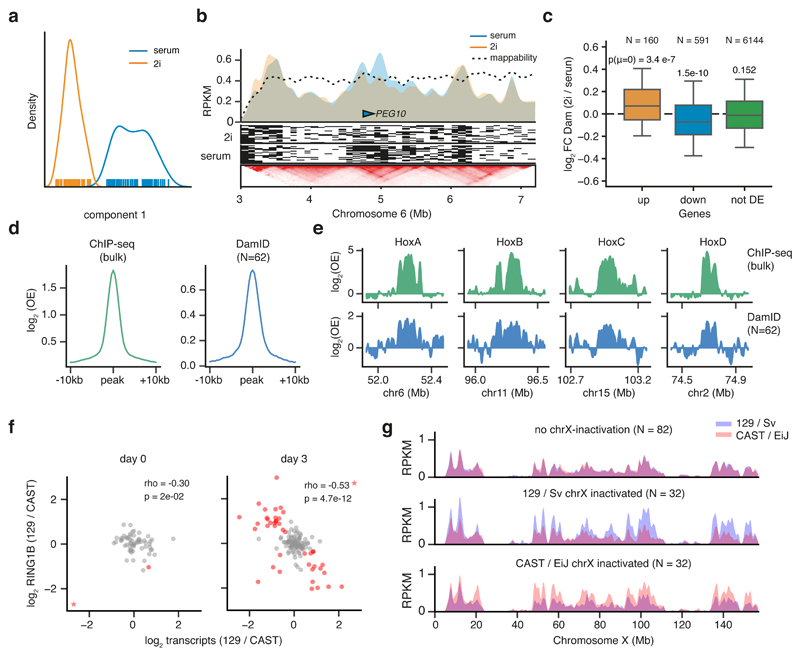
Figure 4. scDam&T-seq enables in silico cell sorting and reconstruction of corresponding cell type specific gene regulatory landscapes.
a) Independent component analysis (ICA) on Dam-expressing mESCs cultured in 2i (orange) or serum (blue) conditions. b) DNA accessibility profiles in 2i and serum conditions of in silico populations (top track) and single cells (signal binarized to high (OE >= 1) as black, and low (OE < 1) as white). The lower panel shows mESC HiC data34 at the same locus, displayed with the 3D genome browser35. c) Fold-change in Dam signal (RPM) between 2i and serum conditions for genes that show statistically significant up-regulation (orange), down-downregulation (blue) or are unaffected (green) in 2i conditions compared to serum. Box plots indicate the 25th and 75th percentile (box), median (line) and 1.5 times the inter-quartile range (IQR) past the 25th and 75th percentiles (whiskers). P-values indicate the result of a two-sided t-test against a mean of 0. n = 158, 577 and 6056 genes, in boxes from left-to-right, respectively. d) Average log2OE signal over all RING1B ChIP-seq peaks obtained with ChIP-seq (left) and scDam&T-seq (right) in 2-kb bins. e) Signal (log2OE) over the four HOX gene clusters for RING1B ChIP-seq and RING1B DamID. In (d) and (e), population ChIP-seq data was normalized for the corresponding input control; RING1B DamID data represents an in silico population of 62 single cells and was normalized with an in silico population Dam sample. f) Relationship between allelic bias in transcription and DamID on chromosome X. Spearman’s rho and p-values (two-sided test, determined by bootstrap) are indicated. Cells are indicated in red when both the transcriptional and DamID allelic biases deviated more than expected based on the somatic chromosomes (see methods). Cells marked as a star fell outside the shown data range; the cell marked as a star in the serum condition is suspected of having lost one chromosome X allele and was excluded from the Spearman correlation. g) Average allelic DamID profiles for cells that had a transcriptional bias on chromosome X towards neither allele (top), towards 129/Sv (middle), or towards CAST/EiJ (bottom) for chromosome X.
Finally, to further test the in silico sorting strategy to profile gene regulatory landscapes, we chose the Polycomb-repressive-complex 1 (PRC1) subunit RING1B (RNF2), which is responsible for the ubiquitination of histone H2AK11932. Because of the role of PRC1 and 2 complexes in the regulation of X chromosome inactivation, we tested whether scDam&T-seq can be employed to identify the randomly inactivated allele in combination with RING1B occupancy in single cells. In undifferentiated mESCs, the cumulative single-cell RING1B scDam&T-seq data is strongly enriched over RING1B binding sites detected by ChIP-seq (Fig. 4d). Similarly, the patterns of enrichment on HOX genes are very comparable (Fig. 4e) and genome-wide scDam&T-seq and ChIP-seq correlate well (Supplementary Fig. 7f). At day 3 of differentiation, random X inactivation is apparent in a fraction of single cells based on the ratio of allelic expression on chromosome X, a pattern which is not observed for autosomal transcripts (Supplementary Fig. 7g). The allelic bias in transcription correlates with increased RING1B levels on the transcriptionally repressed allele (Fig. 4f and 4g), a pattern that is not observed for autosomes of the same cells (Supplementary Fig. 7h). The observed increased levels of RING1B on the inactive X chromosome are consistent with the identification of H2AK119 ubiquitination as one of the earliest events during X inactivation33(Supplementary Fig. 7i). These results demonstrate that scDam&T-seq can be employed to systematically dissect the regulatory mechanisms underlying X chromosome inactivation in single cells.
In summary, scDam&T-seq allows simultaneous quantifications of DNA-protein interactions and transcription from single cells. We have shown that scDam&T-seq enables measuring the impact of spatial genome organization and chromatin states on gene expression and it can be applied to sort cell types in silico and obtain their associated gene regulatory landscapes. Applied to dynamic biological processes, scDam&T-seq should prove especially powerful to identify protein-mediated mechanisms that regulate cell type-specific transcriptional programs in dynamic processes and heterogeneous tissues.
Online methods
Cell culture
Haploid KBM7 cells were cultured in suspension in IMDM (Gibco) supplemented with 10% Fetal Bovine Serum (FBS; Sigma) and 1% Pen/Strep (Gibco). The same Shield1-inducible Dam-LMNB1 and Dam stable clonal KBM7 cell lines were used as in 2. Cells were split every 3 days. F1 hybrid 129/Sv:Cast/EiJ mouse embryonic stem cells (mESCs)25 were cultured on irradiated primary mouse embryonic fibroblasts (MEFs), in ES cell culture media; G-MEM (Gibco) supplemented with 10% FBS (Sigma), 1% Pen/Strep (Gibco), 1x GlutaMAX (Gibco), 1x non-essential amino acids (Gibco), 1x sodium pyruvate (Gibco), 0.1 mM β-mercaptoethanol (Sigma) and 103 U/mL ESGROmLIF (EMD Millipore, ESG1107). Cells were split every 3 days. Expression of constructs was suppressed by addition of 1 mM indole-3-acetic acid (IAA; Sigma, I5148). 2i F1 hybrid 129/Sv:Cast/EiJ mESCs were cultured for 2 weeks on primary MEFs in 2i ES cell culture media; 48% DMEM/F12 (Gibco) and 48% Neurobasal (Gibco), supplemented with 1x N2 (Gibco), 1x B27 supplement (Gibco), 1x non-essential amino acids (Gibco), 1% Pen/Strep (Gibco), 0.1 mM β-mercaptoethanol (Sigma), 0.5% BSA (Sigma), 1 μM PD0325901 (Axon Medchem, 1408), 3 μM CHIR99021 (Axon Medchem, 1386) and 103 U/mL ESGROmLIF (EMD Milipore). Cells were split every 3 days. Expression of constructs was suppressed by addition of 1mM IAA (Sigma). The stable mESC clones were differentiated by culturing them on gelatin-coated 6 well plates after MEF depletion, in monolayer differentiation media; IMDM (Gibco) supplemented with 15% FBS (Sigma), 1% Pen/Strep (Gibco), 1x GlutaMAX (Gibco), 1x non-essential amino acids (Gibco), 50 μg/mL ascorbic acid (Sigma, A4544) and 37.8 μL/L monothioglycerol (Sigma, M1753). Expression of constructs was suppressed by addition of 1 mM IAA (Sigma). After MEF depletion, one confluent 6 well of mESCs was split 1:15 on 6 gelatin-coated wells of a 6-well plate in differentiation media for 3 days. The medium was changed every other day.
Generating cell lines
Stable clonal Dam and Dam-LMNB1 F1 hybrid mESC lines were created by co-transfection of the EF1alpha-Tir1-IRES-neo and hPGK-AID-Dam-mLMNB1 or hPGK-AID-Dam plasmids in a ratio of 1:5. Cells were trypsinized and 0.5 x 106 cells were plated directly with Effectene transfection mixture (Qiagen) in 60% Buffalo Rat Liver (BRL)-conditioned medium; 120 mL BRL medium (in-house production), 80 mL G-MEM (Gibco) supplemented with 10% FBS (Sigma), 1% Pen/Strep (Gibco), 1x GlutaMAX (Gibco), 1x non-essential amino acids (Gibco), 1x sodium pyruvate (Gibco), 0.1 mM β-mercaptoethanol (Sigma) and 103 U/mL ESGROmLIF (EMD Millipore) on gelatin-coated wells of a 6-well plate. The transfection was according to the kit protocol. Cells were selected for 10 days with 250 μg/mL G418 (Thermofischer) and selection of the clones was based on methylation levels, determined by DpnII-qPCR assays as previously described2. To reduce the background methylation levels in the presence of 1 mM IAA (Sigma), we transduced the selected clones of both AID-Dam-LMNB1 and Dam-only with extra hPGK-Tir1-puro lentivirus followed by selection with 0.8 μg/mL puromycin. Positive clones were screened for IAA induction in the presence and absence of IAA by DpnII-qPCR assays and DamID PCR products as previously described2. Stable clonal AID-Dam-RING1B F1 hybrid mESCs were created by lentiviral co-transduction of pCCL-EF1α-Tir1-IRES-puroR and pCCL-hPGK-HA-AID-Dam-RING1B virus in a 4:1 ratio, after which the cells were selected for 10 days on gelatin-coated 10 cm dishes in BRL-conditioned medium containing 0.8 μg/mL puromycin (Sigma) and 0.5 mM IAA (Sigma). Individual puromycin resistant colonies were tested for the presence of the constructs by PCR using primers fw-ttcaacaaaagccaggatcc and rev-gacagcggtgcataaggcgg. Positive clones were furthermore screened for their level of induction upon IAA removal by DamID PCR products.
DamID induction
Expression of Dam-LMNB1 and Dam constructs was induced in the KBM7 cells with 0.5 nM Shield1 (Glixx laboratories, 02939) 15 hours prior to harvesting as described previously2. Expression of Dam-LMNB1 or Dam constructs was induced in the F1 mESCs by IAA washout with PBS (in-house production) 12 hours prior to harvesting. Based on the growth curve of cells counted at time points 12, 24, 30, 36, 42, 48, 54, 60, 72 and 84 hours after plating, the generation time of both the Dam-LMNB1 and Dam cell lines was estimated at 12 hours (data not shown). Considering that 55% of the cells are in G1 and early S phase, the estimated time these cells reside in G1 and early S phase is 6.75 hours.
Cell harvesting and sorting
KBM7 cells were harvested in PBS (in-house production), stained with 0.5 μg/mL DAPI (Sigma) for live/dead selection. Single cells were sorted based on small forward and side-scatter values (30% of total population) and selected for double positive Fucci profile as described previously2, 36. F1 mESCs expressing Dam-LMNB1, Dam or Dam-RING1B were collected in plain or 2i ES cell culture media and stained with 30 μg/mL Hoechst 34580 (Sigma, 63493) for 45 minutes at 37 °C. mESC singlets were sorted based on forward and side-scatter properties, and in mid-S phase of the cell cycle based on DNA content histogram. Differentiated F1 mESCs expressing Dam-RING1B were collected in differentiation media and stained with 30 μg/mL Hoechst 34580 for 45 minutes at 37 °C. The same cells were stained with 1 μg/mL propidium iodide (Sigma) for live/dead selection. Differentiated mESCs singlets were sorted based on forward and side-scatter properties, and in G1, S and G2/M phase of the cell cycle based on DNA content histogram. One cell was sorted per well of 384-well plates (Biorad, HSP3801) using the BD FACSJazz cell sorter. Wells contained 4 μL mineral oil (Sigma) and 100 nL of 15 ng/μL unique CEL-Seq2 primer23.
scDam&T-seq. Robotic preparation
4 μL mineral oil was dispensed manually into each well of a 384-well plate using a multichannel pipet. 100 nL of unique CEL-Seq primer was dispensed per well using the mosquito HTS robot (TTP Labtech). The NanodropII robot (BioNex) was used for all subsequent dispensing steps at 12 p.s.i. pressure. After sorting, 100 nL lysis mix was added (0.8 U RNase inhibitor (Clontech, 2313A), 0.07 % Igepal, 1 mM dNTPs, 1:500,000 ERCC RNA spike-in mix (Ambion, 4456740)). Each single cell was lysed at 65 °C for 5 minutes and 150 nL reverse transcription mix was added (1x First Strand Buffer (Invitrogen, 18064-014), 10 mM DTT (Invitrogen, 18064-014), 2 U RNaseOUT Recombinant Ribonuclease Inhibitor (Invitrogen, 10777019), 10 U SuperscriptII (Invitrogen, 18064014)) and the plate was incubated at 42 °C for 1 hour, 4 °C for 5 minutes and 70 °C for 10 minutes. Next, 1.92 μL of second strand synthesis mix was added (1x second strand buffer (Invitrogen, 10812014), 192 μM dNTPs, 0.006 U E. coli DNA ligase (Invitrogen, 18052019), 0.013 U RNAseH (Invitrogen, 18021071)) and the plate was incubated at 16 °C for 2 hours. 500 nL of protease mix was added (1x NEB CutSmart buffer, 1.21 mg/mL ProteinaseK (Roche, 000000003115836001)) and the plate was incubated at 50 °C for 10 hours and 80 °C for 20 minutes. Next, 230 nL DpnI mix was added (1x NEB CutSmart buffer, 0.2 U NEB DpnI) and the plate was incubated at 37 °C for 4 hours and 80 °C for 20 minutes. Finally, 50 nL of DamID2 adapters were dispensed (final concentrations varied between 32 and 128 nM), together with 450 nL of ligation mix (1x T4 Ligase buffer (Roche, 10799009001), 0.14 U T4 Ligase (Roche, 10799009001)) and the plate was incubated at 16 °C for 12 hours and 65 °C for 10 minutes. Contents of all wells with different primers and adapters was pooled and incubated with 0.8 volume magnetic beads (CleanNA, CPCR-0050) diluted 1:4 or 1:8 with bead binding buffer (20% PEG8000, 2.5M NaCl) for 10 minutes, washed twice with 80% ethanol and resuspended in 7 μL nuclease-free water before in vitro transcription at 37 °C for 14 hours using the MEGAScript T7 kit (Invitrogen, AM1334). Library preparation was done as described in the CEL-Seq protocol with minor adjustments23. Amplified RNA (aRNA) was cleaned and size-selected by incubating with 0.8 volume magnetic beads (CleanNA) for 10 min, washed twice with 80% ethanol and resuspended in 22 μL nuclease-free water, and fragmented at 94 °C for 2 minutes in 0.2 volume fragmentation buffer (200 mM Tris-acetate pH 8.1, 500 mM KOAc, 150 mM MgOAc). Fragmentation was stopped by addition of 0.1 volume fragmentation STOP buffer (0.5 M EDTA pH8) and quenched on ice. Fragmented aRNA was incubated with 0.8 volume magnetic beads (CleanNA) for 10 minutes, washed twice with 80% ethanol and resuspended in 12 μL nuclease-free water. Thereafter, library preparation was done as previously described23 using 5 μL of aRNA and PCR cycles varied between 8 and 10. Libraries were run on the Illumina NextSeq platform with high output 75 bp paired-end sequencing.
DamID adapters
The adapter was designed (5’ to 3’) with a 4 nt fork, a T7 promoter, the 5’ Illumina adapter (as used in the Illumina small RNA kit), a 3 nt UMI (unique molecular identifier), an 8 nt unique barcode followed by CA. The Dam-RING1B mESCs were processed with different adapters. These contained a 6 nt fork, a 6 nt unique barcode followed by GA. The barcodes were designed with a hamming distance of at least two between them. Bottom sequences contained a phosphorylation site at the 5’ end. Adapters were produced as standard desalted oligos. Top and bottom sequences were annealed at a 1:1 volume ratio in annealing buffer (10 mM Tris pH 7.5–8.0, 50 mM NaCl, 1 mM EDTA) by immersing tubes in boiling water, then allowing to cool to room temperature. The oligo sequences can be found in Supplementary Table 2.
CEL-Seq primers
The RT primer was designed according to the Yanai protocol23 with an anchored polyT, an 8 nt unique barcode, a 6 nt UMI (unique molecular identifier), the 5’ Illumina adapter (as used in the Illumina small RNA kit) and a T7 promoter. The barcodes were designed with a hamming distance of at least two between them. Primers are desalted at the lowest possible scale, stock solution 1 μg/μL. The oligo sequences can be found in Supplementary Table 3.
Raw data preprocessing
First mates in the raw read pairs (i.e. "R1" or "read1") conform to a layout of either: 5'-[3 nt UMI][8 nt barcode]CA[gDNA]-3' in the case of gDNA (DamID and AluI restriction) reads, or 5'-[6 nt UMI][8 nt barcode][unalignable sequence]-3'in the case of transcriptomic reads. In the case of transcriptomic reads, the second mate in the read pair contains mRNA sequence. Raw reads were processed by demultiplexing on barcodes (simultaneously using the DamID and transcriptomic barcodes), allowing no mismatches. The UMI sequences were extracted and stored alongside the names of the reads for downstream processing.
Sequence alignments
After demultiplexing of the read pairs using the first mate and removal of the UMI and barcode sequences, the reads were aligned. In the case of gDNA-derived reads, a 'GA' dinucleotide was prepended to the sequences of read1 ('AG' in the case of AluI), and the gDNA sequence of read1 was then aligned to a reference genome using bowtie2 (v.2.3.2) using parameters --seed 42 --very-sensitive -N 1. For transcriptome-derived reads, read2 was aligned using tophat2 (v2.1.1) using parameters --segment-length 22 --read-mismatches 4 --read-edit-dist 4 --min-anchor 6 --min-intron-length 25 --max-intron-length 25000 --no-novel-juncs --no-novel-indels --no-coverage-search --b2-very-sensitive --b2-N 1 --b2-gbar 200 and using transcriptome-guiding (options --GTF and --transcriptome-index). Human data was aligned to hg19 (GRCh37) including the mitochondrial genome, the sex chromosomes and unassembled contigs. Transcriptomic reads were aligned using transcript models from GENCODE (v26) (www.gencodegenes.org/releases/grch37_mapped_releases.html). mESC data was aligned to reference genomes generated by imputing 129S1/SvImJ and CAST/EiJ SNPs obtained from the Sanger Mouse Genomes project37 onto the mm10 reference genome. The mitochondrial genome, sex chromosome and unassembled contigs were included during the alignments. Transcriptomic reads were aligned using a GTF file with transcript annotations obtained from ENSEMBL (release 89) (ftp://ftp.ensembl.org/pub/release-89/gtf/mus_musculus/Mus_musculus.GRCm38.89.gtf.gz). Both human and mouse transcriptome references were supplemented with ERCC mRNA spike-in sequences (https://assets.thermofisher.com/TFS-Assets/LSG/manuals/cms_095047.txt). For both genomic and transcriptomic data, reads that yielded an alignment with mapping quality (BAM field 'MAPQ') lower than 10 were discarded, as well as reads aligning to the mitochondrial genome or unassembled contigs. For the genomic data, reads not aligning exactly at the expected position (5' of the motif, either GATC in the case of DpnI restriction, or AGCT in the case of AluI restriction) were discarded. For the transcriptomic data, reads not aligning to an exon of a single gene (unambiguously) were discarded. The mESC reads were assigned to the 129S1/SvImJ or CAST/EiJ genotype by aligning reads to both references. Reads that aligned with lower edit-distance (SAM tag 'NM') or higher alignment-score (SAM tag 'AS') in case of equal edit-distance to one of the genotypes were assigned to that genotype. Reads aligning with equal edit-distance and alignment-score to both genotypes were considered of 'ambiguous' genotype. For analyses comparing allelic signals, counts with 'ambiguous' genotype were discarded (Fig. 4f-g, Supplementary Fig. 6, 7g-h). For all other figures concerning mESC data, UMI-unique data of the two alleles was summed together with the ambiguously assigned data.
PCR duplicate filtering
For the genomic data (DamID and AluI-WGS), the number of reads per motif, strand and UMI were counted. Read counts were collapsed using the UMIs (i.e. multiple reads with the same UMI count as 1) after an iterative filtering step where the most abundant UMI causes every other UMI sequence with a Hamming-distance of 1 to be filtered out. E.g., observing the three UMIs 'AAA', 'GCG' and 'AAT' in decreasing order would count as 2 unique events (with UMIs 'AAA' and 'GCG', since 'AAT' is within 1 Hamming distance from 'AAA'). The number of observed unique UMIs was taken as the number of unique methylation events (for DamID) or unique transcripts (for the transcriptomics). For the data from KBM7 (a near-complete haploid cell line) at most 1 unique event per GATC position and strand was kept. For the mESC data at most 1 unique event per GATC position, strand and allele was kept, or 2 unique events, in the case of 'ambiguous' allelic assignment.
Filtering of samples
We observed that the number of unique methylation events and unique transcripts per single-cell sample followed a bimodal distribution in most datasets. To discard samples that that clearly failed, we applied the following cutoffs: only single-cell samples with at least 103.7 unique DamID events and at least 103 unique transcripts were taken into consideration for the analyses. These cutoffs were applied jointly for all analyses, regardless of whether genomic and/or transcriptomic signals were used. These numbers were established on our earliest (human and mouse) datasets, by fitting a two component gaussian mixture model to the observed unique counts (with all samples across the datasets).
Normalization of expression values
UMI-unique transcript counts per gene were further normalized using scran38–39. We used computeSumFactors with reduced sizes parameter where our sample sizes were too small for default parameters, and using only genes expressed in at least 1% of all samples, and other parameters left to their default values. Expression values were then converted to log-transformed counts per million (TPM, transcripts per million reads) using logcounts.
Binning and calculation of OE values
DamID and WGS data was binned using consecutive non-overlapping 100-kb bins. For analyses at TSS, enhancer and CTCF sites, data was binned with high resolution (a binsize of 10 bp was used). In order to calculate observed-over-expected (OE) values, the mappability of each motif (GATC or AGCT) was determined by generating 65 nt. long sequences (in both orientations) from the reference genome(s) and aligning and processing them identically to the data. By binning the in silico generated reads, the maximum amount of mappable unique events per bin was determined.
OE values were calculated using
where O is the number of observed unique methylation events per bin, E is the number of mappable unique events per bin, ψ is the pseudocount (1, unless otherwise stated), TO and TE are the total number of unique methylation events observed and mappable, respectively in the sample and B is the number of bins. For analyses across multiple windows, e.g. windows around TSSs or CTCF sites, O and E were summed across the windows, prior to calculation of the OE values.
For the definition of "contact", regions with OE values >= 1 were considered as "in-contact". For further details and justification, see2, Extended Experimental Procedures and Supplementary Fig. 2a in particular. Contact frequency was defined as the fraction of samples (passing cutoffs) showing "contact" (OE >= 1), an is expressed as fraction in [0, 1] per genomic bin.
Comparison scDam&T-seq to Kind Cell 2015 data
For the comparisons with individual measurements of scDamID and single-cell transcriptomics (CEL-Seq)2 with scDam&T-seq (Fig. 1) the scDam&T-seq data was made comparable to the published data by truncating the reads at the 3' end such that gDNA and mRNA sequence lengths were identical to the published data, which was sequenced with shorter reads. Furthermore, UMIs were completely left out of consideration for the DamID measurements. For the transcriptional measurements, the UMIs were truncated to 4 nt to make the data comparable to the published CEL-Seq data.
Signal of scDam&T-seq LMNB1 data on microarray-defined LADs
Comparisons of LMNB1 data obtained with scDam&T-seq to independently identified LADs (Supplementary Fig. 1c for human data and Supplementary Fig. 3b for mouse data) were made using published HT108040 and mESC27 data. We used the LAD coordinates available from the supplementary materials. We remapped LAD coordinates using liftOver (from mm9 to mm10 and from hg18 to hg19, for mouse and human data, respectively), and discarded LADs that spanned less than 500 kb after the liftOver procedure.
Run length analysis
Run length analysis was done as described in 2 with the exception that we did not remove bins from the analysis with a CF of zero. Random shuffling with preservation of marginal distributions was done as described previously4.
Autocorrelation analysis
Autocorrelation of raw signals was analyzed with a maximum resolution limited by a bin size of 100 bp. In silico population profiles were generated for each indicated condition and downsampled to 50 times the DamID methylation count cutoff of 103.7. Only chromosomes larger than 100 Mb were considered in the analysis, as autocorrelation of large distances cannot be measured on shorter chromosomes. Furthermore, sex chromosomes were discarded. We used an FFT approach to determine the statistical autocorrelation of signal at each chromosome, then summed the autocorrelation profiles to arrive at the genome-wide autocorrelation profiles.
Assessment of technical batch effects on variance in transcriptomics data
Principal component analysis (PCA) on the transcriptome data shows that batch effects always appear in the first, or first few principal components. This is unsurprising, since these single-cell samples are biologically homogeneous (ie. clonal cells, FACS-sorted in the same cell phase). To assess to which degree technical effects influence variance in the transcriptomics data, we employed an approach analogous to Bushel 2008 (pvca: Principal Variance Component Analysis (PVCA). R package version 1.22.0),41 with the exception that we fitted simple ordinary least-squares models (with one factor) rather than mixed linear models. Weighing the coefficient of determination for batch effect of each principal component with variance explained by the principal component a total of 16% of data variance can be explained by method, between scDam&T-seq and CEL-Seq (Supplementary Fig. 2d). For reference, 2.2% of total data variance can be explained by batch when contrasting two scDam&T-seq libraries and 9.7% of total variance in expression data can be explained by clonal origin when contrasting Dam-LMNB1 and Dam transcriptomes measured by scDam&T-seq. Finally, we also used ComBat42 to estimate the amount of data variance explained by these technical variables, by comparing the amount of data variance before and after removing "batch effects". We obtained similar ratios of variance explained but in general observe lower amounts of total data variance explained by batch (8.9% explained when using CEL-Seq vs scDam&T-seq as batch, 3.6% by clonal origin, 3.0% when contrasting two Dam-LMNB1 batches).
Using PCA on our mESC 2i vs serum transcriptomics data showed a high degree of separation between 2i and serum samples on the first principal component, but also strong association with sample depths (despite using best practices to normalize our single-cell transcriptomics data). We therefore employed a 2-component independent component analysis (ICA) to deconvolve sample depth effects from the 2i/serum effects on the (normalized) transcriptomics data. The ICA separating 2i and serum samples is shown in Fig. 4a.
TSS, CTCF and enhancer locations
For the analyses at TSSs, one isoform per gene was chosen from the gene annotations, by preferentially taking isoforms that carry the GENCODE "basic" tag, have a valid, annotated CDS (start and stop codon, and CDS length being a multiple of 3 nt), with ties broken by the isoform with longest CDS, and shortest gene length (distance from 5' nucleotide of first exon to 3' nucleotide of last exon). As TSS, the most 5' position of the first exon was taken. CTCF sites were obtained by integrating ENCODE ChIP-seq data (wgEncodeRegTfbsCellsV3, K562 CTCF ChIP-seq tracks) with CTCF motif sites (factorbookMotifPos obtained via the UCSC genome browser (http://genome.ucsc.edu)43. Only CTCF ChIP-seq peaks that contained a CTCF binding motif with score of at least 1.0 within 500 bp. of the center of the ChIP-seq peak were considered. The ChIP-seq peaks were subdivided by ChIP-seq binding score (reported in the ENCODE processed data file), and the group of peaks with maximum score (of 1,000) was subdivided into three groups by the motif score, such that 4 approximately equal-sized groups of CTCF-bound loci were obtained. Enhancer locations were given by the ENCODE HMM chromatin segmentation for K562 cells44. The centers of segments annotated as "4/Strong enhancer" and "5/Strong enhancer" were used in our analysis.
H3K4me3, H3K36me3, RING1B and DNase data (external datasets)
H3K4me3 ChIP-seq, H3K36me3 ChIP-seq and DNase data was obtained from ENCODE (sample IDs GSM788087, GSM733714 and GSE90334_ENCFF038VUM, respectively) as processed bigWig files. In order to calculate OE values for these datasets, whole-genome mappability as determined by the ENCODE project was used (wgEncodeCrgMapabilityAlign36mer). RING1B ChIP-seq data and corresponding input control were obtained from the Gene Expression Omnibus (GEO) (GSM2393579, GSM2393592) and aligned to the GRCm28 mouse reference index with bowtie2 (v2.3.3.1) using parameters --seed 42 --very-sensitive -N 1. Genome-wide coverage was obtained with bamCoverage from the DeepTools toolkit (v3.1.2) using parameters --ignoreDuplicates --minMappingQuality 10. ChIP-seq domains were called with the callpeak tool of MACS2 (v 2.1.1.20160309) using parameters --keep-dup 1 --seed 42 --broad --broad-cutoff 0.005.
Comparison DNase and scDam&T-seq Dam stratified by expression
For Fig. 2f, we used an independent microarray expression dataset (GSE56465, only the haploid KBM7 samples). Microarray probes which had no gene ID assigned to them were discarded. For gene IDs with multiple assigned probes, the median value was taken. Only gene IDs present in GENCODE v26 were used in our analysis. We stratified all genes with at least 1 expression datum (microarray probe) into four expression quantiles. Fig. 2f(ii) shows the density of TSSs of genes with the indicated expression quantiles, according to the scDam&T-seq Dam and DNase OE value of the 20 kb-bin in which those TSS lie. To determine whether a point in the scDam&T-seq-DNase space was enriched for 20-kb bins contained a TSS of the indicated expression quantile, we used the "Significant fold-change" approach, outlined in45. Briefly, a normal-approximation using the expected value np, with p = Ng/(4N) where n is the number of 20-kb bins with given scDam&T-seq Dam and DNase value, Ng is the total number of 20-kb bins with a TSS and is the total number of (mappable) 20-kb bins), and a variance of n⋆p(1 − p), where n⋆ is max(25,n) is used to define a confidence interval (we used a critical value of α = 20%) to determine whether the actual number of observed 20-kb bins with a TSS of gene in the quantile constitutes enrichment or depletion.
Comparison of scDam&T-seq to scNMTseq
Trancriptomics data from scDam&T-seq (mESC serum) and scNMTseq were downsampled to 1.5105 raw reads per single cell. Single-cell samples with fewer reads were left out of the transcriptomics comparison. The detected number of genes per cell for both methods is shown in Supplementary Fig 4d. GpC accessibility data from scNMTseq was obtained from the processed data of GSE109262.
logFC between contact/no contact groups of samples
logFCs between single-cell samples that showed contact and those that showed no contact (see Fig. 3a) were computed as follows:
In 100-kb bins across the genome the logFC in gene expression was calculated between samples that have a DamID OE value ≥ 1 vs. samples that have a DamID OE value lower than 1. The expression per bin was determined by the sum over all genes that have their TSS in that bin. Genomic bins that were considered unmappable (fewer than 2 GATCs per kb) were excluded, as well as bins where either group of samples (high OE, low OE) contained fewer than 3 samples, or where fewer than 7.5% of all samples showed any expression. Finally, an additional cutoff on samples was used (besides the manuscript-wide cutoffs on DamID event and transcript counts) to exclude samples with anomalous genome-wide DamID patterns (judging by their high-OE bins). The distributions of total fraction of high-OE bins across the genome (bins meeting the mappability and expression cutoffs described above) over all samples (for Dam-LMNB1 and Dam separately) was modeled as a Gaussian mixture with k = 1,2…5 Gaussian components with independent means and variances. Using a 25-fold randomized 50% split of samples, we fitted the Gaussian mixture on one half, and measured the goodness of fit using the other half (using the Akaike information criterion, AIC, which penalizes goodness-of-fit for the number of model parameters). We took the mean of each cross-validation and repeated this process 10 times, for each k. We then took the number of Gaussian components k that minimized the mean AIC, which was 2 for both Dam-LMNB1 and untethered Dam. Samples assigned to the Gaussian component with the majority of samples, with probability of at least 67%, were used further in the analysis of logFC in expression.
Rolling mean and standard deviations as function of CF
In Fig. 3 and related supplementary figures, a rolling mean is shown together with confidence interval for the mean. To obtain these measurements we calculated the mean and standard deviations of the metric on the y-axis for each point on the x-axis using a local linear regression approach where data points are weighted according to an exponential decay, i.e. exp(−d/τ). Here d is the distance between the point at the x-axis where the mean is being determined and the data point, and τ is a "decay factor" (or effective radius). For regressions against CF (Fig. 3b, Fig. 3d and Supplementary Fig. 5a) a radius of 0.025 [CF units] was used. The shadings indicate a 95% confidence interval for the means and are determined by 1.96 times the standard deviations, measured using the same exponentially weighted approach as the means.
Variance-to-mean ratios
In our expression data we observed a variance-to-mean ratio (VMR) that increased with increasing mean expression, indicative of overdispersion (wrt. Poisson-distributed counts). We de-trended the VMR from the (log-normalized) mean expression using local linear regression with exponentially decaying weights (see the above paragraph). Supplementary Fig. 5b shows this "de-trended" VMR on the x-axis. Note that, since the logFC between high-OE and low-OE samples is largely independent on mean expression (see Supplementary Fig 5a), raw VMR values show very similar results. The rolling mean and confidence interval in Supplementary Fig. 5b uses local linear regression with a radius of 0.25 [log10(VMR) units].
Relationship between TAD structure and differential accessibility in 2i versus serum
TADs were obtained from34 and converted to a 100 kb resolution. Specifically, TAD boundaries were taken to be the midpoint between TADs and rounded to the nearest 100-kb point. The variance in log2FC serum/2i accessibility (DamID) data in 100-kb bins within each TAD was calculated for all TADs that contained at least three 100-kb bins with at least 2 mappable GATC motifs per kb. Subsequently, the order of the TADs was randomized per chromosome and the new TAD coordinates were used to calculate a control variance distribution. This process was repeated 50 times. P-values between the distributions corresponding to the original and randomized TAD structure were calculated using a two-sided Mann-Whitney U test with continuity correction.
Testing for differential gene expression between 2i and serum in mESCs
To determine genes differentially expressed between 2i and serum conditions, we employed egdeR46, using the exactTest function) with sample totals determined by scran (computeSumFactors) rather than edgeR's internal sample normalization routines. Panels in Fig4 consider genes with a false discovery rate (FDR) smaller than 5% and an absolute logFC greater than 2.0 as either up- or down-regulated. For Fig. 4c, genes with absolute logFC smaller than 1.3 and unadjusted p-value greater than 0.5 were considered as "not differentially expressed". For Supplementary Fig. 7c, where all genes (regardless of statistically significant differential expression) are shown, we removed lowly expressed genes by setting a threshold such that 95% of the differentially expressed genes meet that threshold.
Detecting chrX allelic biases in DamID and transcription during differentiation
Allelic coverage in undifferentiated mESCs indicated a CAST/EiJ duplication of the final ~20 Mb of chromosome X. The analyses described below therefore include only the first 150 Mb of chromosome X. In order to detect allelic biases on chromosome X in DamID and transcription data, the log2 fold-change of 129/Sv over CAST/EiJ was calculated for the total number of DamID counts and transcripts on chrX (with a pseudocount of 1). Subsequently, allelic DamID and transcripts counts on the somatic chromosomes were subsampled such that the combined depth of both alleles corresponded to that of chromosome X. The allelic counts were then used to calculate log2 fold-change values. One cell in the serum condition showed high CAST/EiJ DamID counts (134) and transcript number (47), while showing no data for 129/Sv (0 counts, 0 transcripts). No such discrepancy was seen for the somatic chromosomes, suggesting that this cell lost its maternal chromosome X. Therefore, the cell was excluded in the calculation of Spearman's correlation coefficient. For differentiation day 3, cells that had a transcriptional chrX allelic bias that exceeded the mean +/- 1 s.d. of the somatic chromosome allelic biases were marked as having 129/Sv or CAST/EiJ X-inactivation, while the remaining cells were labelled as not showing X-inactivation. For the cells in these three categories, the average RPKM values on chrX and chr6 were calculated for the two alleles.
By figure details on the statistics can be found in Supplementary Table 4.
Reporting Summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Supplementary Material
Acknowledgements
We would like to thank the members of the Kind, Dey and van Oudenaarden labs for their comments on the manuscript and J Gribnau (Erasmus UMC) for kindly providing the 129/Sv:CAST/EiJ mESCs and for advice on differentiation. We would like to thank B de Barbanson and J Yeung for suggestions regarding computational analyses and statistics, Reinier van der Linden for FACS and M Muraro and L Kester for input on the scDam&T-seq technique. We acknowledge support from the Center for Scientific Computing at UCSB: an NSF MRSEC (DMR-1720256) and NSF CNS-1725797. This work was funded by a European Research Council Starting grant (ERC-StG 678423-EpiID) and a Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO) Open (824.15.019) and ALW/VENI grant (016.181.013). The Oncode Institute is supported by KWF Dutch Cancer Society.
Footnotes
Data availability statement. The sequencing data from this study are available from the Gene Expression Omnibus, accession number GSE108639 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE108639).
Code availability statement. All computational codes used for this study are available upon request.
Author Contributions
K.R., S.S.D. and J.K. designed the study and wrote the manuscript. S.S.D. developed the method. C.M.M. optimized the method and performed all the experiments unless stated otherwise. S.S.d.V. and K.L.d.L. assisted with experiments. S.S.d.V. created the mESC lines. K.R. performed all the analysis except when stated otherwise. F.R. performed cloning and all analyses pertaining the RING1B data. A.C. performed the analysis of Supplementary Fig. 4d. and exploratory analyses together with S.S.D. D.M. provided input during initial technology development. J.K. and S.S.D conceived and supervised the study.
Competing interest statement. The authors declare that they have no competing financial interests.
References
- 1.Nagano T, et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502:59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kind J, et al. Genome-wide maps of nuclear lamina interactions in single human cells. Cell. 2015;163:134–147. doi: 10.1016/j.cell.2015.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flyamer IM, et al. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature. 2017;544:110–114. doi: 10.1038/nature21711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stevens TJ, et al. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature. 2017;544:59–64. doi: 10.1038/nature21429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cusanovich DA, et al. Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 2015;348:910–914. doi: 10.1126/science.aab1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buenrostro JD, et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523:486–490. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin W, et al. Genome-wide detection of DNase I hypersensitive sites in single cells and FFPE tissue samples. Nature. 2015;528:142–146. doi: 10.1038/nature15740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo H, et al. Single-cell methylome landscapes of mouse embryonic stem cells and early embryos analyzed using reduced representation bisulfite sequencing. Genome research. 2013;23:2126–2135. doi: 10.1101/gr.161679.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smallwood SA, et al. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nature methods. 2014;11:817–820. doi: 10.1038/nmeth.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farlik M, et al. Single-cell DNA methylome sequencing and bioinformatic inference of epigenomic cell-state dynamics. Cell reports. 2015;10:1386–1397. doi: 10.1016/j.celrep.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mooijman D, et al. Single-cell 5hmC sequencing reveals chromosome-wide cell-to-cell variability and enables lineage reconstruction. Nature biotechnology. 2016;34:852–856. doi: 10.1038/nbt.3598. [DOI] [PubMed] [Google Scholar]
- 12.Zhu C, et al. Single-Cell 5-Formylcytosine Landscapes of Mammalian Early Embryos and ESCs at Single-Base Resolution. Cell Stem Cell. 2017;20:720–731. doi: 10.1016/j.stem.2017.02.013. [DOI] [PubMed] [Google Scholar]
- 13.Wu X, et al. Simultaneous mapping of active DNA demethylation and sister chromatid exchange in single cells. Genes & Development. 2017;31:511–523. doi: 10.1101/gad.294843.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rotem A, et al. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nature biotechnology. 2015;33:1165–1172. doi: 10.1038/nbt.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dey S, et al. Integrated genome and transcriptome sequencing of the same cell. Nature biotechnology. 2015;33:285–289. doi: 10.1038/nbt.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macaulay IC, et al. G&T-seq: parallel sequencing of single-cell genomes and transcriptomes. Nature methods. 2015;12:519–522. doi: 10.1038/nmeth.3370. [DOI] [PubMed] [Google Scholar]
- 17.Hou Y, et al. Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Research. 2016;26:304–319. doi: 10.1038/cr.2016.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Angermueller C, et al. Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nature methods. 2016;13:229–232. doi: 10.1038/nmeth.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark SJ, et al. scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nature communications. 2018;9:781. doi: 10.1038/s41467-018-03149-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steensel van B, et al. Chromatin profiling using targeted DNA adenine methyltransferase. Nature genetics. 2001;27:304–308. doi: 10.1038/85871. [DOI] [PubMed] [Google Scholar]
- 21.Vogel MJ, et al. Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nature protocols. 2007;2:1467–1478. doi: 10.1038/nprot.2007.148. [DOI] [PubMed] [Google Scholar]
- 22.Kind J, et al. Single-cell dynamics of genome-nuclear lamina interactions. Cell. 2013;153:178–192. doi: 10.1016/j.cell.2013.02.028. [DOI] [PubMed] [Google Scholar]
- 23.Hashimshony T, et al. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq. Genome biology. 2016;17:77. doi: 10.1186/s13059-016-0938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meuleman W, et al. Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Research. 2013;23:270–281. doi: 10.1101/gr.141028.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monkhorst K, et al. X inactivation counting and choice is a stochastic process: evidence for involvement of an X-linked activator. Cell. 2008;132:410–421. doi: 10.1016/j.cell.2007.12.036. [DOI] [PubMed] [Google Scholar]
- 26.Nishimura K, et al. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nature methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 27.Peric-Hupkes D, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Molecular cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aughey GN, et al. CATaDa reveals global remodelling of chromatin accessibility during stem cell differentiation in vivo. Elife. 2018;7 doi: 10.7554/eLife.32341. pii: e32341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schones DE, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valouev A, et al. Determinants of nucleosome organization in primary human cells. Nature. 2011;474:516–520. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boyle AP, et al. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 33.Zylicz JJ, et al. The Implication of Early Chromatin Changes in X Chromosome Inactivation. Cell. 2019;176:182–197. doi: 10.1016/j.cell.2018.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonev B, et al. Multiscale 3D Genome Rewiring during Mouse Neural Development. Cell. 2017;171:557–572.e524. doi: 10.1016/j.cell.2017.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, et al. The 3D Genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biology. 2018;19:151. doi: 10.1186/s13059-018-1519-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakaue-Sawano A, et al. Visualizing spatiotemporal dynamics of multicellular cell cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 37.Keane TM, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–94. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lun AT, et al. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 2016;17:75. doi: 10.1186/s13059-016-0947-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lun AT, et al. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res. 2016;5:2122. doi: 10.12688/f1000research.9501.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meuleman W, et al. Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Research. 2013;23:270–280. doi: 10.1101/gr.141028.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boedigheimer MJ, et al. Sources of variation in baseline gene expression levels from toxicogenomics study control animals across multiple laboratories. BMC Genomics. 2008;9:285. doi: 10.1186/1471-2164-9-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson WE, et al. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2006;1:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 43.Kent WJ, et al. The human genome browser at UCSC. Genome Research. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Knijnenburg TA, et al. Multiscale representation of genomic signals. Nature Methods. 2014;11:689–694. doi: 10.1038/nmeth.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson MD, et al. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.