Summary
Neurofibromatosis type 1 (NF1) is a neurodevelopmental disorder in which affected children and adults are at a higher risk for sleep disorders. In an effort to identify potential sleep disturbances in a small-animal model, we leveraged a previously reported Nf1 conditional knockout (Nf1CKO) mouse strain. In contrast to Nf1 mutant flies, the distribution of vigilance states was intact in Nf1CKO mice. However, Nf1CKO mice exhibited increased non-REM sleep (NREM)-to-wake and wake-to-NREM transitions. This sleep disruption was accompanied by decreased bout durations during awake and NREM sleep states under both light and dark conditions. Moreover, Nf1CKO mice have higher percentage delta power during awake and NREM sleep states under all light conditions. Taken together, Nf1CKO mice phenocopy some of the sleep disturbances observed in NF1 patients, and provide a tractable platform to explore the molecular mechanisms governing sleep abnormalities in NF1.
Keywords: NREM Sleep, Sleep fragmentation, Sleep disturbance, Encephalopathy
Sleep disorders are commonly reported in patients with the Neurofibromatosis type 1 (NF1) neurogenetic syndrome, caused by germline mutations in the NF1 gene. In these studies, parasomnias, difficulties initiating sleep, early morning awakenings, and excessive sleep/wake transitions were mostly frequently described in children and adults with NF1 (Leschziner et al., 2013, Licis et al., 2013). Of these sleep-related problems, difficulties with sleep initiation and maintenance, as well as sleep/wake transitions, predominated in both males and females with NF1. Since individuals with NF1 are prone to executive function, learning, and social perception problems, disruptions in sleep can exacerbate these cognitive and behavioral issues.
To understand the basis for sleep disturbances in people with NF1, researchers have studied Nf1 mutant flies engineered to lack Nf1 protein (neurofibromin) expression. In these studies, altered circadian clock rhythms reflect loss of neurofibromin-negative regulation of ERK activity (Williams et al., 2001). In addition, Nf1 mutant flies exhibit a sexually dimorphic short sleep phenotype, observed only in male Nf1-null flies (Bai and Sehgal, 2015). Interestingly, the severe sleep fragmentation in Nf1-mutant flies could be genetically separated from the Nf1 circadian rhythm defect.
In an effort to establish a mammalian model for NF1 sleep abnormalities, we employed a previously-published Nf1 conditional knockout strain (Nf1CKO) in which a germline Nf1 null mutation in all cells is coupled with somatic Nf1 loss in GFAP-expressing neuroglial progenitor cells in the E15.5 developing brain following Cre recombination. Using this platform, we demonstrate that Nf1-mutant mice have increased sleep fragmentation and delta power.
Methods
Mice
All experiments were performed in 6-month-old Nf1flox/mut; GFAP-Cre mice (Nf1CKO mice; n= 10 males, n=16 females; total n=26) and littermate control Nf1flox/flox mice (CTL mice; n= 10 males, n=15 females; total n=25) from 14 litters maintained on an inbred C57BL/6J background, back-crossed more than 20 times prior to the experiments. The mice had ad libitum access to 53WU irradiated PicoLab Rodent Diet 20 (LabDiet) and water. Mice were bred and housed according to an approved Animal Studies protocol at the Washington University School of Medicine.
Electroencephalography (EEG) recording and analysis
EEG screw electrodes were secured at (bregma) anterior +0.8mm, lateral 0.5mm, over the parietal cortex (posterior −2.5mm, lateral +/− 1.5) and a ground screw was placed over the cerebellum (posterior −6.4mm, lateral +/− 0.5). The exposed ends of two wire electrodes were inserted in the neck muscle for nuchal electromyography (EMG) recordings. Bilateral cortical EEG signals were acquired in freely moving mice using a referential montage with XLTEK amplifiers and acquisition software. Signals were amplified at 10,000× with high-pass (0.5Hz) and low-pass (100Hz) filters using LabChart software (ADInstruments). EMG signals were high-pass (10Hz) and low-pass (300Hz) filtered. EEG and EMG signals were digitized at 500Hz and collected in 24-hour sessions. Vigilance states were manually scored (Satoh et al., 2013) using a combination of the EEG, EMG and respective spectral power representations in 10-sec epochs, with data averaged for each hour during the 12-hour light/dark cycle. We defined a transition as a shift in vigilance state occurring at a time point greater than half of the total epoch duration (>5 sec). Bouts were calculated based on the time spent in each vigilance state before transitioning to the next state, and are displayed as mean durations for the light/dark phase. The percentage of total power in delta range (1–4Hz) was calculated using fast Fourier transforms in LabChart with a 512-bin size and a Hann data window with spectral data extracted from each vigilance epoch.
Statistics
Statistical analyses were performed using SigmaStat 3.5 and GraphPad Prism 5.03. Quantitative differences between Nf1CKO and control mice were analyzed by Student’s t-test for single time points, one-way or two-way ANOVA for repeated measures with Bonferroni post-test comparisons. Statistical significance was defined as p<0.05.
Results and Discussion
Based on previous studies in Drosophila demonstrating that Nf1 loss impairs circadian rhythms and sleep (Williams et al., 2001, Bai et al., 2018), we leveraged a previously reported conditional knockout Nf1 mouse strain (Nf1CKO). While patients with NF1 harbor only one mutation in the NF1 gene in neurons, we chose to employ Nf1 mutant mice with bi-allelic Nf1 loss in neurons, astrocytes and neuroglial precursors for several reasons. First, this Nf1 CKO strain exhibits reproducible and quantitative NF1-related cognitive anomalies, as previously demonstrated by behavioral studies measuring attention (Brown et al., 2010) and learning deficits (Diggs-Andrews et al., 2013, Cui et al., 2008), unlike the less reproducible phenotypes exhibited by heterozygous animals in our hands (Brown JA, unpublished results). Second, sleep or circadian rhythm defects were only observed in flies completely lacking Nf1 gene expression (Williams et al., 2001, Bai and Sehgal, 2015). Third, previous studies using Nf1+/− mice revealed no statically significant abnormalities in circadian rhythms (Weiss et al., 2017). Since Nf1-null mice are not viable, we employed the Nf1CKO strain to perform a pilot study comparing vigilance states over a 24-hr period.
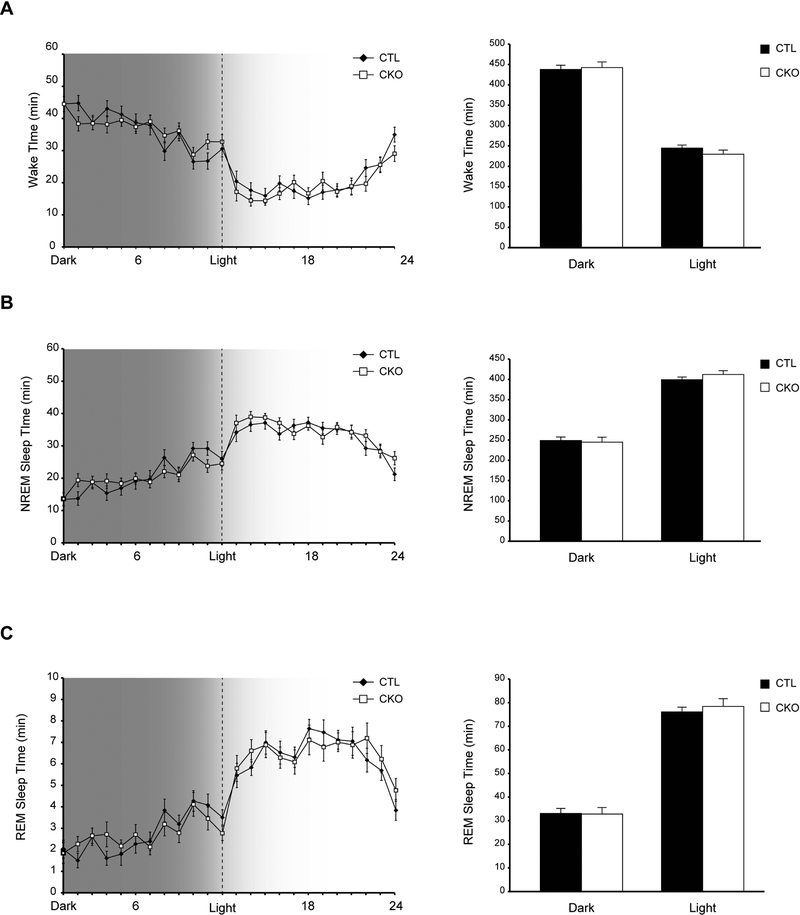
Following a two-week acclimation in 12-hr light/dark cycles, we first assessed Nf1CKO and control (CTL) mouse vigilance state patterns by electroencephalographic (EEG) and electromyographic (EMG) recordings, and analyzed the time spent by individual animals in three vigilance states: awake (wake), non-REM (NREM) and REM sleep (Figure 1). Nf1CKO mice spent similar total amounts of time in wake, NREM and REM-sleep, both in light and dark conditions, relative to controls. These data suggest that, unlike the disrupted circadian system in Drosophila, the daily distribution of vigilance states is intact in Nf1CKO mutant mice. The apparent differences between the two animal models could reflect differences in genetic engineering, specifically the coupling of Nf1 heterozygosity with Nf1 loss in brain cells in Nf1CKO mice. Alternatively, the molecular mechanisms regulating circadian rhythms and sleep may be different between diurnal and nocturnal animals (Vosko et al., 2009). Future studies will be required to clarify the etiologies for the observed differences between these two model systems.
Figure 1.
The daily distribution of vigilance states is intact in Nf1CKO mice A-C Graphs indicating the time spent (A) awake, (B) in non-REM (NREM) sleep or (C) in REM sleep by Nf1 conditional knockout (Nf1CKO; CKO) or wild-type control (CTL) mice over a 24-hour period with 12 hour dark/light cycles. Data are represented as means ± SEM. n.s., not significant
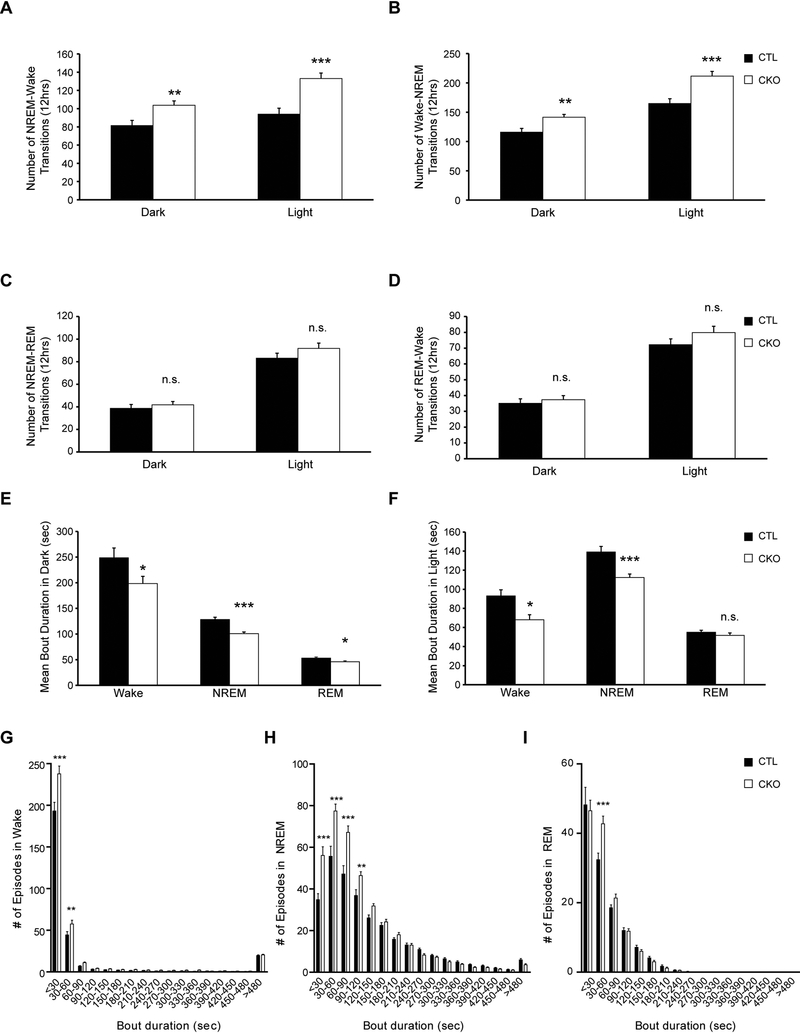
Second, we examined sleep/wake patterns by analyzing vigilance state transitions in Nf1CKO and control mice (Figure 2). Independent of light conditions, Nf1CKO mice had more NREM-wake (Figure 2A) and wake-NREM transitions than control animals over 12 hours (Figure 2B), but similar NREM-REM and REM-wake transitions (Figure 2C–D). Moreover, Nf1CKO mice had shorter mean awake and NREM sleep bout durations relative to controls, both in the light and the dark, but only displayed shorter mean REM sleep bouts in the dark (Figure 2E–F).
Figure 2.
Nf1CKO mice exhibit increased NREM sleep fragmentation relative to littermate controls A-B. Nf1CKO mice (CKO) display an increased number of both (A) NREM-wake and (B) wake-NREM transitions compared to wild type controls (CTL). C-D. Nf1CKO mice exhibit similar numbers of (C) NREM-REM and (D) REM-wake transitions relative to controls. E-F Nf1CKO mice display decreased wake and NREM bout durations (E) in the dark and (F) in the light relative to controls. (G-I) Distributions of bout durations, binned in 30-second periods, illustrate that Nf1CKO mice display an increased number of short awake (G), NREM (H) and REM (I) bouts compared to controls. Data are represented as means ± SEM. *p<0.05; **p<0.01 ***p<0.001; n.s., not significant
Third, analysis of sleep bout durations revealed a disruption in the bout distribution of wake and sleep in Nf1CKO mice (Figure 2G–I). As such, Nf1-mutant mice displayed an increase in the number of short wake bouts (<60 sec; Figure 2G) and displayed less consolidated sleep with an increase in <120-sec NREM bouts (Figure 2H). Analysis of REM sleep showed small differences in overall bout distribution between genotypes, with more 30–60sec REM bouts in Nf1CKO mice (Figure 2I). Taken together, the finding of increased sleep fragmentation parallels the sleep deficit (sleep maintenance) observed in patients with NF1 (Licis et al., 2013), and suggests that this model could be employed to discern the molecular basis for this defect in mammals. For example, in other experimental systems, sleep fragmentation induces cognitive deficits by regulating NADPH oxidase (Nair et al., 2011), as well as negatively impacts on spatial learning and synaptic plasticity (Wallace et al., 2015), raising the intriguing possibility that sleep fragmentation in patients with NF1 could exacerbate their co-existing learning and memory problems.
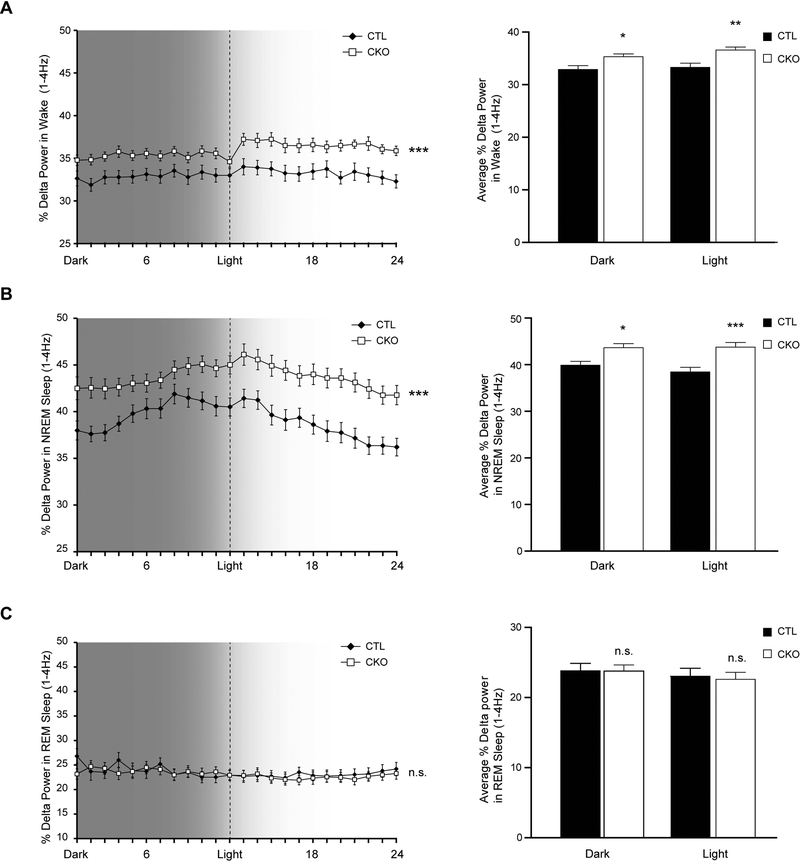
Finally, as sleep fragmentation can be indicative of global brain dysfunction (encephalopathy) (McCoy and Strecker, 2011), we measured spectral EEG power including slow wave delta power and higher frequency theta power in all three vigilance states. While there was no difference in the average percentage theta power between genotypes (4–8.5Hz; data not shown), Nf1CKO mice exhibited increased delta power (1–4Hz) compared to controls, both in awake (Figure 3A) and in NREM sleep (Figure 3B), but not during REM sleep (Figure 3C) over 24 hours of recording. The increased delta power in Nf1CKO mice, could signify hypersomnolence in Nf1CKO mice, in contrast to patients who lack excessive somnolence (Licis et al., 2013). Alternatively, increased delta power is often interpreted to reflect slower overall background EEG activity, which can also influence global cognitive function (Astill et al., 2012), further compounding the co-morbid learning and attention deficits found in people with NF1 (Knyazev, 2012).
Figure 3.
Nf1CKO mice display increased delta power in awake and NREM sleep states compared to control mice A-B. Nf1CKO mice (CKO) have elevated delta power (1–4Hz) during (A) wake and (B) NREM sleep, but not during (C) REM sleep in both light and dark periods. The panels at the right illustrate the % Delta power during the 12 hours of dark and 12 hours of light. Data are represented as means ± SEM. *p<0.05, **p<0.01, ***p<0.0001; n.s., not significant
In conclusion, this study demonstrates, for the first time, that Nf1 mutant mice exhibit reproducible sleep disturbances similar to children and adults with NF1. As sleep abnormalities represent a common morbidity in patients with this condition, future studies employing this and other mammalian preclinical platforms will be critical for addressing circadian rhythm patterns and other long-term sleep issues, elucidating the molecular mechanisms governing sleep disturbances, as well as for evaluating potential therapeutic agents that can improve sleep and life quality in this at risk population.
Supplementary Material
Acknowledgements
This work was funded by an NIH grant (U54 HD087011) to the Washington University Intellectual and Developmental Disabilities Center (MW) and a Research Program Award from the NINDS (1-R35-NS097211–01) to DHG.
Footnotes
Conflict of Interests. The authors declare no conflict of interests.
References
- Astill RG, Van Der Heijden KB, Van Ijzendoorn MH and Van Someren EJ Sleep, cognition, and behavioral problems in school-age children: a century of research meta-analyzed. Psychol Bull, 2012, 138: 1109–38. [DOI] [PubMed] [Google Scholar]
- Bai L, Lee Y, Hsu CT et al. A Conserved Circadian Function for the Neurofibromatosis 1 Gene. Cell Rep, 2018, 22: 3416–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L and Sehgal A Anaplastic Lymphoma Kinase Acts in the Drosophila Mushroom Body to Negatively Regulate Sleep. PLoS Genet, 2015, 11: e1005611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JA, Emnett RJ, White CR et al. Reduced striatal dopamine underlies the attention system dysfunction in neurofibromatosis-1 mutant mice. Hum Mol Genet, 2010, 19: 4515–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa RM, Federov NB, Kogan JH et al. Mechanism for the learning deficits in a mouse model of neurofibromatosis type 1. Nature, 2002, 415: 526–30. [DOI] [PubMed] [Google Scholar]
- Cui Y, Costa RM, Murphy GG et al. Neurofibromin regulation of ERK signaling modulates GABA release and learning. Cell, 2008, 135: 549–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggs-Andrews KA, Tokuda K, Izumi Y, Zorumski CF, Wozniak DF and Gutmann DH Dopamine deficiency underlies learning deficits in neurofibromatosis-1 mice. Ann Neurol, 2013, 73: 309–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knyazev GG EEG delta oscillations as a correlate of basic homeostatic and motivational processes. Neurosci Biobehav Rev, 2012, 36: 677–95. [DOI] [PubMed] [Google Scholar]
- Leschziner GD, Golding JF and Ferner RE Sleep disturbance as part of the neurofibromatosis type 1 phenotype in adults. Am J Med Genet A, 2013, 161A: 1319–22. [DOI] [PubMed] [Google Scholar]
- Licis AK, Vallorani A, Gao F et al. Prevalence of Sleep Disturbances in Children With Neurofibromatosis Type 1. J Child Neurol, 2013, 28: 1400–05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mccoy JG and Strecker RE The cognitive cost of sleep lost. Neurobiol Learn Mem, 2011, 96: 564–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair D, Zhang SX, Ramesh V et al. Sleep fragmentation induces cognitive deficits via nicotinamide adenine dinucleotide phosphate oxidase-dependent pathways in mouse. Am J Respir Crit Care Med, 2011, 184: 1305–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh A, Brace CS, Rensing N et al. Sirt1 extends life span and delays aging in mice through the regulation of Nk2 homeobox 1 in the DMH and LH. Cell Metab, 2013, 18: 416–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosko AM, Hagenauer MH, Hummer DL and Lee TM Period gene expression in the diurnal degu (Octodon degus) differs from the nocturnal laboratory rat (Rattus norvegicus). Am J Physiol Regul Integr Comp Physiol, 2009, 296: R353–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace E, Kim DY, Kim KM et al. Differential effects of duration of sleep fragmentation on spatial learning and synaptic plasticity in pubertal mice. Brain Res, 2015, 1615: 116–28. [DOI] [PubMed] [Google Scholar]
- Weiss JB, Weber SJ, Torres ERS, Marzulla T and Raber J Genetic inhibition of Anaplastic Lymphoma Kinase rescues cognitive impairments in Neurofibromatosis 1 mutant mice. Behav Brain Res, 2017, 321: 148–56. [DOI] [PubMed] [Google Scholar]
- Williams JA, Su HS, Bernards A, Field J and Sehgal A A circadian output in Drosophila mediated by neurofibromatosis-1 and Ras/MAPK. Science, 2001, 293: 2251–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.