Abstract
The present study was performed to clarify the significance of DNA methylation alterations during endometrial carcinogenesis. Genome-wide DNA methylation analysis and targeted sequencing of tumor-related genes were performed using the Infinium MethylationEPIC BeadChip and the Ion AmpliSeq Cancer Hotspot Panel v2, respectively, for 31 samples of normal control endometrial tissue from patients without endometrial cancer and 81 samples of endometrial cancer tissue. Principal component analysis revealed that tumor samples had a DNA methylation profile distinct from that of control samples. Gene Ontology enrichment analysis revealed significant differences of DNA methylation at 1034 CpG sites between early-onset endometrioid endometrial cancer (EE) tissue (patients aged ≤40 years) and late-onset endometrioid endometrial cancer (LE) tissue, which were accumulated among ‘transcriptional factors’. Mutations of the CTNNB1 gene or DNA methylation alterations of genes participating in Wnt signaling were frequent in EEs, whereas genetic and epigenetic alterations of fibroblast growth factor signaling genes were observed in LEs. Unsupervised hierarchical clustering grouped EE samples in Cluster EA (n = 22) and samples in Cluster EB (n = 12). Clinicopathologically less aggressive tumors tended to be accumulated in Cluster EB, and DNA methylation levels of 18 genes including HOXA9, HOXD10 and SOX11 were associated with differences in such aggressiveness between the two clusters. We identified 11 marker CpG sites that discriminated EB samples from EA samples with 100% sensitivity and specificity. These data indicate that genetically and epigenetically different pathways may participate in the development of EEs and LEs, and that DNA methylation profiling may help predict tumors that are less aggressive and amenable to fertility preservation treatment.
Genome-wide analysis using 112 samples of endometrial tissue has revealed that genetically and epigenetically different pathways may participate in early- and late-onset endometrial carcinogenesis. DNA methylation profiling may predict tumors that are less aggressive and amenable to fertility preservation treatment.
Introduction
Endometrial cancer is one of the most common gynecological malignancies in the developed countries, with ~320 000 new cases diagnosed annually worldwide (1). Although it is typically a disease of postmenopausal women, ~2–14% of cases occur in women aged 40 years or less (2–4), and in fact recently endometrial cancer in younger patients has shown a marked increase (4). The standard treatment for endometrial cancer is hysterectomy and bilateral salpingo-oophorectomy with or without lymph node dissection, but for younger patients fertility-sparing treatment that avoids surgical menopause needs to be considered. Currently, conservative treatment with hormones such as medroxyprogesterone acetate is used for early-stage early-onset endometrial cancer without myometrial invasion or extrauterine spread (5). However, we have reported previously that the recurrence rate after medroxyprogesterone acetate therapy is 63.2% and the resulting pregnancy rate lower than would be desired (6). In order to improve the outcome of medical treatment for patients with endometrial cancer, there is a need to clarify the molecular basis of endometrial carcinogenesis, especially in young patients.
It is well known that not only genomic but also epigenomic alterations participate in carcinogenesis in various human organs (7,8). DNA methylation alterations are one of the most important epigenomic changes resulting in chromosomal instability and aberrant expression of tumor-related genes (9–11). We and other groups have shown that DNA methylation alterations are frequently associated with tumor aggressiveness and poorer patient outcome, and can be used clinically as prognostic markers in cancers of various organs (12–17). Recently single-CpG resolution genome-wide DNA methylation screening using the Infinium assay has been introduced for analysis of human tissue specimens. However, none of previous studies of endometrial cancer based on the Infinium assay (18–22), including the study by The Cancer Genome Atlas (TCGA) (23), focused on early-onset endometrial cancer.
In this study, to further clarify the significance of DNA methylation alterations during endometrial carcinogenesis and their correlation with genetic abnormalities, we performed genome-wide DNA methylation (methylome) analysis using the high-density EPIC BeadChip on 31 samples of normal control endometrial tissue from patients without endometrial cancer and 81 samples of endometrial cancer tissue including 35 samples of early-onset endometrial cancer.
Materials and methods
Patients and tissue samples
The 81 samples of cancerous tissue were obtained from 81 patients with primary endometrial cancer who underwent hysterectomy at Keio University Hospital between October 2005 and August 2016. Among the patients, 35 were aged 40 years or less (early-onset endometrial cancer) and 46 were older than 40 years (late-onset endometrial cancer). None of these patients had received any preoperative treatment. Supplementary Table 1A, available at Carcinogenesis Online, gives details of the ages and clinicopathological backgrounds of these patients. Histological diagnosis and grading were based on the 2003 World Health Organization classification (24) and the Tumor-Node-Metastasis classification (25). The clinical stage of the disease was based on the 2008 revised International Federation of Gynecology and Obstetrics classification (26). Recurrence was diagnosed by clinicians on the basis of physical examination and imaging modalities such as computed tomography and positron emission tomography.
For comparison, 31 samples of normal control endometrial tissue were obtained from materials that had been surgically resected from patients without endometrial cancer (Supplementary Table 1B, available at Carcinogenesis Online): 17, 6, 6 and 2 patients with uterine leiomyoma, ovarian cancer, cervical cancer and lobular endocervical glandular hyperplasia, respectively.
Tissue specimens were frozen immediately after surgery and preserved in the Keio Women’s Health Biobank in accordance with the Japanese Society of Pathology Guidelines for the handling of pathological tissue samples for genomic research (27). This study was approved by the ethics committee of Keio University Hospital and was performed in accordance with the Declaration of Helsinki. All patients included in this study provided written informed consent for use of their materials.
Infinium analysis
High-molecular-weight DNA was extracted from fresh frozen tissue samples using phenol–chloroform, followed by dialysis. DNA methylation status at 866 895 CpG loci was examined at single-CpG resolution using the Infinium MethylationEPIC BeadChip (Illumina, San Diego, CA) (28). Thereafter, 500 ng aliquots of DNA were subjected to bisulfite conversion using an EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, CA). After hybridization, the specifically hybridized DNA was fluorescence-labeled by a single-base extension reaction and detected using an iScan Reader (Illumina) in accordance with the manufacturer’s protocol. The data were then assembled using GenomeStudio Methylation software (Illumina). At each CpG site, the ratio of the fluorescence signal was measured using a methylated probe relative to the sum of the methylated and unmethylated probes, i.e. the so-called β-value, which ranges from 0.00 to 1.00 reflecting the methylation level of an individual CpG site.
In this assay, the call proportions (P < 0.01 for detection of signals above the background) for 801 probes on autosomes in all of the 112 tissue samples were <90%. As such a low proportion may have been attributable to polymorphism at the probe CpG sites, these probes were excluded from the present assay. In addition, to avoid any gender-specific methylation bias, all 19 681 probes on chromosomes X and Y were excluded, leaving a final total of 846 413 autosomal CpG sites. Correlations between levels of DNA methylation and messenger RNA expression were examined using the dataset for endometrial cancer deposited in the TCGA database (https://tcga-data.nci.nih.gov/docs/publications/ucec_2013/) (23).
Targeted sequencing analysis of 50 tumor-related genes
Targeted sequencing of 50 tumor-related genes was performed in 81 endometrial cancer tissue samples. Library preparation was performed using an Ion AmpliSeq Library Kit 2.0-96LV and Ion AmpliSeq Cancer Hotspot Panel v2 in accordance with the manufacturer’s protocol (Thermo Fisher Scientific, Waltham, MA). The panel is designed to amplify 207 amplicons covering 2849 mutations of 50 oncogenes and tumor suppressor genes deposited in the Catalogue of Somatic Mutations in Cancer (COSMIC) database (https://cancer.sanger.ac.uk) (Supplementary Table 2, available at Carcinogenesis Online). After library preparation, each amplicon library was quantified using an Ion Library Quantitation Kit on the 7500 Fast Real-Time PCR System (Thermo Fisher Scientific) using the relative standard curve method. Next, emulsion PCR and Ion SphereTM particle enrichment were carried out using the Ion OneTouchTM 2 system and Ion OneTouchTM enrichment system (Thermo Fisher Scientific). Purified Ion SphereTM particles were loaded on an Ion 316 or 318 Chip and sequenced using the Ion PGMTM sequencer (Thermo Fisher Scientific). Data were analyzed using Torrent Suite Software v5.2.2 and Ion Reporter Software v5.2–5.4 (Thermo Fisher Scientific). Detected variants with quality scores of <20 and allele frequencies of <2.0% were eliminated in accordance with the manufacturer’s instructions. We filtered out single-nucleotide polymorphisms using the databases of the 1000 Genomes project (http://www.internationalgenome.org/) and 5000 exomes project (http://evs.gs.washington.edu/EVS/). Effects of amino acid substitutions on protein function due to single-nucleotide non-synonymous mutations were estimated using the Sorting Intolerant from Tolerant (SIFT) protocol (29) (http://sift.jcvi.org). Copy number analyses were performed using the baseline of five normal human kidney tissue samples without kidney cancer, and confidence levels of <10 were eliminated in accordance with the manufacturer’s instructions. The incidence of genetic aberrations in the present cohort was compared against the dataset for endometrial cancer deposited in the TCGA database (https://tcga-data.nci.nih.gov/docs/publications/ucec_2013/) (23).
GO enrichment analysis and pathway analysis
In order to reveal the function of proteins encoded by genes showing DNA methylation alterations and gene mutations, and to reveal the biological processes in which such proteins participate, Gene Ontology (GO) enrichment analysis and pathway analysis were conducted using the GeneGO MetacoreTM software package, version 6.7 (Thomson Reuters, New York, NY).
Statistics
Infinium probes showing significant differences in DNA methylation levels between sample groups were defined by Welch’s t-test and adjusted by Bonferroni correction. The DNA methylation profiles of the 112 tissue samples were analyzed using principal component analysis. Differences in the incidences of genetic aberrations between sample groups were examined using Fisher’s exact test. Unsupervised hierarchical clustering based on DNA methylation levels (Euclidean distance, Ward method and Canberra distance, complete linkage method) was performed. The associations between clinicopathological variables and DNA methylation alterations were evaluated using Fisher’s exact test, Mann–Whitney U test or Welch’s t-test. The receiver operating characteristic (ROC) curve was generated, and the Youden index (30) for each probe was used as a cutoff value for examining distinctions between the sample groups. Programming language R and IBM SPSS statistics 20.0 were used to analyze the data.
Results
DNA methylation alterations during endometrial carcinogenesis
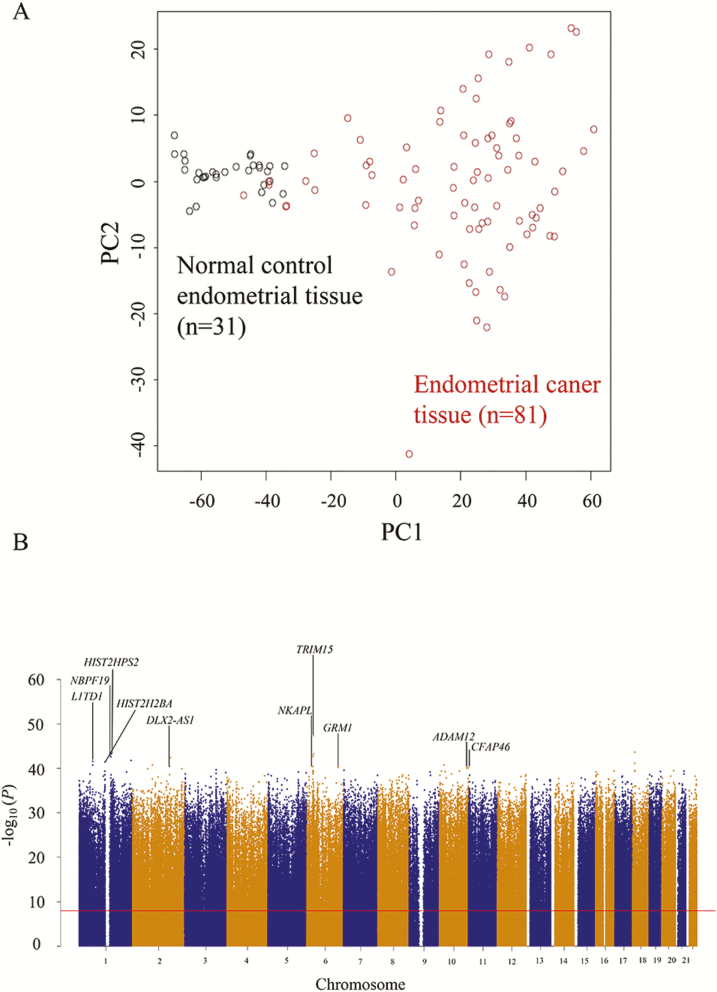
First, to obtain a general overview of endometrial cancers, we identified 58 958 probes that were aberrantly methylated in 81 endometrial cancer tissue samples in comparison with the 31 normal control endometrial tissue samples (Welch’s t-test, adjusted Bonferroni correction [α = 1.18 × 10−8] and ∆βendometrial cancer tissue − normal control endometrial tissue value of >0.25 or < −0.25), indicating that DNA methylation alterations had occurred during endometrial carcinogenesis. Principal component analysis using the DNA methylation levels of these 58 958 probes revealed that endometrial cancer tissue samples had a DNA methylation profile that differed distinctly from that of normal control endometrial tissue samples (Figure 1A). The leading 10 genes, i.e. TRIM15, HIST2H3PS2, NBPF19, L1TD1, HIST2H2BA, NKAPL, DLX2-AS1, GRM1, ADAM12 and CFAP46, showing significant differences in DNA methylation levels between endometrial cancer tissue and normal control endometrial tissue are labeled in the Manhattan plot (Figure 1B). To the best of our knowledge, no previous English-language articles have yet reported correlations between these 10 genes and endometrial cancers; this study may have revealed novel genes potentially associated with endometrial carcinogenesis, although the expression levels and functions of these genes will need to be further examined in connection with endometrial cancers.
Figure 1.
DNA methylation profiles of normal control endometrial tissue (n = 31) and endometrial cancer tissue (n = 81). (A) Principal component analysis was performed using the 58 958 probes showing significant differences in DNA methylation levels between normal control endometrial tissue and endometrial cancer tissue samples (Welch’s t-test, adjusted Bonferroni correction [α = 1.18 × 10−8] and ∆βendometrial cancer tissue − normal control endometrial tissue value of >0.25 or < −0.25). (B) Manhattan plot constructed using all 846 413 probes. The Bonferroni corrected P value (1.18 × 10−8) is indicated by the red line. The leading 10 genes, i.e. TRIM15, HIST2H3PS2, NBPF19, L1TD1, HIST2H2BA, NKAPL, DLX2-AS1, GRM1, ADAM12 and CFAP46, showing significant differences in DNA methylation levels between normal control endometrial tissue and endometrial cancer tissue samples (Δβendometrial cancer tissue − normal control endometrial tissue value of >0.25 or < −0.25) are labeled. N/A, not annotated (designed for the intergenic regions).
Somatic mutations and copy number alterations in endometrial cancer
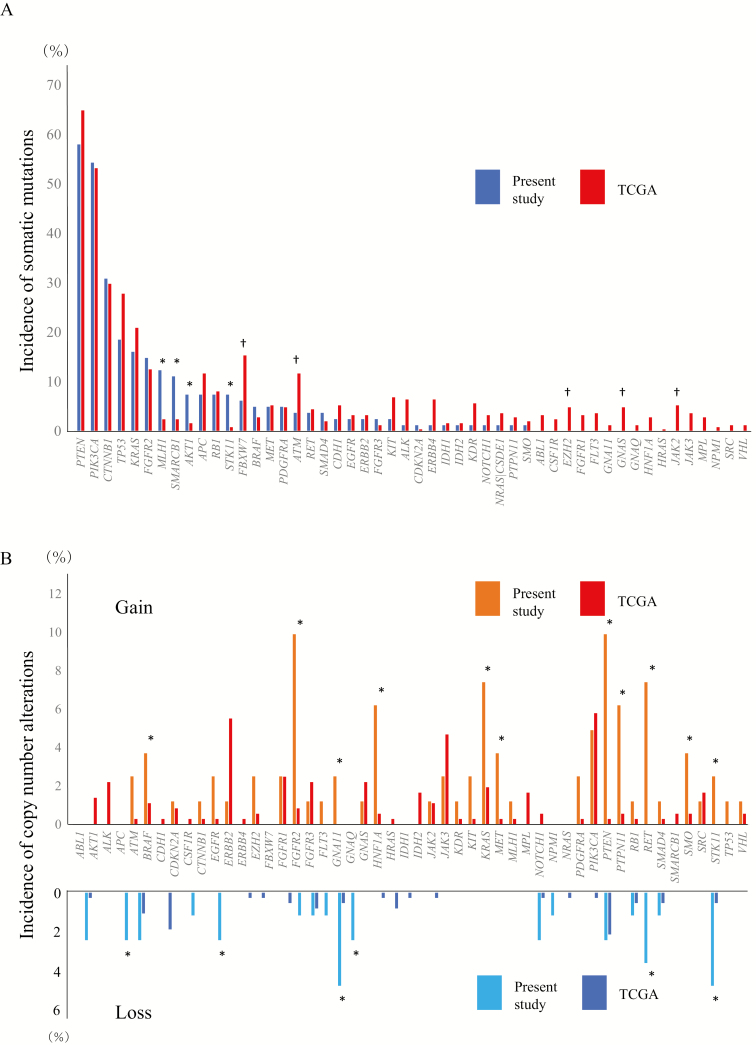
Five hundred and thirty-five somatic mutations were detected in 81 endometrial cancer tissue samples (6.6 ± 2.9 mutations per sample): 230 synonymous and 305 non-synonymous. The non-synonymous mutations consisted of 242 missense mutations (79%), 44 nonsense mutations (14%), 14 frameshift deletions (5%), 4 frameshift insertions (1%) and 1 frameshift block substitution (0.3%). Genes showing the highest incidences of somatic mutations included PTEN in 47 patients (58%), PIK3CA in 44 patients (54%), CTNNB1 in 25 patients (31%) and TP53 in 15 patients (19%). When compared with data for endometrial cancer deposited in the TCGA database (https://tcga-data.nci.nih.gov/docs/publications/ucec_2013/) (23), the incidences of somatic mutations of the MLH1 (P = 1.08 × 10−3), SMARCB1 (P = 3.01 × 10−3), AKT1 (P = 0.017) and STK11 (P = 3.47 × 10−3) genes were higher, and the incidences of those of the FBXW7 (P = 0.037), ATM (P = 0.049), EZH2 (P = 0.043), GNAS (P = 0.043) and JAK2 (P = 0.044) genes were lower in our cohort, whereas significant differences were not found in the remaining 41 genes (Figure 2A). The incidences of copy number alterations of all the 50 genes examined were lower than 10%, and this low incidence was again in accordance with the data deposited in the TCGA database (https://tcga-data.nci.nih.gov/docs/publications/ucec_2013/) (23) (Figure 2B).
Figure 2.
The incidence of somatic mutations (A) and copy number alterations (gain and loss) (B) of the 50 examined tumor-related genes in endometrial cancer tissue in the present cohort (n = 81) and data deposited in TCGA database (https://tcga-data.nci.nih.gov/docs/publications/ucec_2013/) (n = 248) (23). Genes showing significantly higher or lower incidence (P < 0.05) in the present cohort than in the TCGA database are indicated by * and †, respectively.
Differences in DNA methylation profile between early- and late-onset endometrioid endometrial cancers
In order to avoid the bias caused by differences in histological subtypes, we then focused on the major subtype, i.e. endometrioid endometrial cancer. We again identified 63 033 probes that showed significant differences in DNA methylation levels between the 31 normal control endometrial tissue samples and 74 endometrioid endometrial cancer tissue samples (Welch’s t-test, adjusted Bonferroni correction [α = 1.18 × 10−8] and ∆βendometrioid endometrial cancer tissue − normal control endometrial tissue value of >0.25 or < −0.25).
Next, we focused on differences between early-onset endometrioid endometrial cancer (EE, patients aged ≤40 years) and late-onset endometrioid endometrial cancer (LE, patients aged >40 years). Clinicopathological parameters of early- and late-onset cases are summarized in Supplementary Table 3, available at Carcinogenesis Online. Among the 63 033 probes, 1034 showed significant differences in DNA methylation levels between the 34 samples of EE tissue and 40 samples of LE tissue (Welch’s t-test P < 0.01; ∆βLE − EE value >0.25 or < −0.25; Supplementary Table 4, available at Carcinogenesis Online). As several patients (n = 11) aged <50 years were included in the LE group, a relatively small number of CpG sites showed significant differences in DNA methylation levels between EE and LE samples. All 1034 probes again showed significant differences in DNA methylation levels between LE samples and normal control endometrial tissue samples (Welch’s t-test P < 0.01; ∆βLE − normal control endometrial tissue value >0.25 or <−0.25), whereas only 102 of 1034 probes showed significant differences in DNA methylation levels between EE samples and normal control endometrial tissue samples (Welch’s t-test P < 0.01; ∆βEE − normal control endometrial tissue value >0.25 or < −0.25). Among these 102 probes, the DNA methylation levels of 101 probes were higher in EE samples than in normal control endometrial tissue samples and even higher in LE samples.
Three hundred and seventy-one genes, for which the 1034 probes showing significant differences of DNA methylation levels between EE samples and LE samples were designed, were evaluated for protein function by enrichment analysis using the MetaCore software and compared with the protein function distribution of genes within the GeneGo database. These genes were clearly overrepresented by ‘transcriptional factors’ (P = 1.67 × 10−32), being 6.004 times more abundant than expected. Indeed 66 of 371 genes were categorized as transcription factors by MetaCore software (shown by asterisks in Supplementary Table 4, available at Carcinogenesis Online). The top 20 statistically significant GO molecular functions in which the 371 genes participated are listed in Table 1. Fifteen of the top 20 functions were related to DNA binding or transcriptional regulation, and are shown by asterisks, and transcription factors included in the top 20 GO molecular functions are underlined in Table I. All 189 statistically significant GO molecular functions (P < 0.05) are listed in Supplementary Table 5, available at Carcinogenesis Online.
Table 1.
Top 20 statistically significant GO molecular functions revealed by MetaCore software analysis using the 371 genes, for which the 1034 probes showing differences in DNA methylation levels between samples of early-onset endometrioid endometrial cancer tissue (patients aged <40 years) and late-onset endometrioid endometrial cancer tissue were designed (listed in Supplementary Table 4, available at Carcinogenesis Online)
| Molecular functions | P value | Included genes showing differences in DNA methylation levels |
|---|---|---|
| Sequence-specific DNA bindinga | 2.05 × 10−19 | ASCL1, ASCL2, ASCL4, BARHL2, DMARTA2, DRGX, EN1, EVX1, EVX2, FEZF2, FLI1, FOXD3, FOXE1, FOXI2, GATA4, GCM2, HAND2, HIC1, IRF4, LHX1, LHX4, LHX5, MNX1, MSC, MKX1-1, MYOD1, NKX2-6, NKX6-2, STN1, ONECUT2, OTP, OTX1, PAX3, PAX5, PAX6, PAX7, PHOX2A, PITX2, POU3F3, POU4F2, PRDM14, PROX1, RAX, SALL3, SATB2, SIX6, SOX1, SOX11, SOX14, SOX2, TBXT, TBX15, TBX18, TBX5, TLX2, TP73, VSX1, ZFHX4, ZIC5, ZNF516 |
| Transcription factor activity, sequence-specific DNA bindinga | 2.39 × 10−19 | ASCL1, ASCL2, ASCL4, BARHL2, BHLHE22, BHLHE23, BNC1, DMRTA2, DRGX, DBX1, EN1, EVX1, EVX2, FEZF2, FLI1, FOXD3, FOXE1, FOXI2, SKOR1, GATA4, GCM2, GFI1, HAND2, HIC1, IRF4, LHX1, LHX4, LHX5, MNX1, MSC, MKX1-1, MYOD1, NKX2-6, NKX6-2, OLIG2, ONECUT1, ONECUT2, OTP, OTX1, PAX3, PAX5, PAX6, PAX7, PHOX2A, PITX2, POU3F3, POU4F2, POU6F2, PRDM14, PROX1, RAX, SALL3, SATB2, SIM1, SIX6, SOX1, SOX11, SOX14, SOX2, TBXT, TBX15, TBX18, TBX5, TLX2, TP73, VSX1, ZFHX4, ZIC5, ZNF132, ZNF229, ZNF334, ZNF516, ZNF667, ZSCAN1, ZSCAN23 |
| Nucleic acid binding transcription factor activitya | 2.48 × 10−19 | ASCL1, ASCL2, ASCL4, BARHL2, BHLHE22, BHLHE23, BNC1, DMRTA2, DRGX, DBX1, EN1, EVX1, EVX2, FEZF2, FLI1, FOXD3, FOXE1, FOXI2, SKOR1, GATA4, GCM2, GFI1, HAND2, HIC1, IRF4, LHX1, LHX4, LHX5, MNX1, MSC, MKX1-1, MYOD1, NKX2-6, NKX6-2, OLIG2, ONECUT1, ONECUT2, OTP, OTX1, PAX3, PAX5, PAX6, PAX7, PHOX2A, PITX2, POU3F3, POU4F2, POU6F2, PRDM14, PROX1, RAX, SALL3, SATB2, SIM1, SIX6, SOX1, SOX11, SOX14, SOX2, TBXT, TBX15, TBX18, TBX5, TLX2, TP73, VSX1, ZFHX4, ZIC5, ZNF132, ZNF229, ZNF334, ZNF516, ZNF667, ZSCAN1, ZSCAN23 |
| RNA polymerase II transcription factor activity, sequence-specific DNA bindinga | 3.14 × 10−16 | ASCL1, ASCL2, ASCL4, BARHL2, BHLHE22, BHLHE23, BNC1, DMRTA2, DRGX, DBX1, EN1, EVX1, EVX2, FEZF2, FLI1, FOXD3, FOXE1, FOXI2, SKOR1, GATA4, GCM2, GFI1, HAND2, HIC1, IRF4, LHX1, LHX4, LHX5, MNX1, MSC, MKX1-1, MYOD1, NKX2-6, NKX6-2, OLIG2, ONECUT1, ONECUT2, OTP, OTX1, PAX3, PAX5, PAX6, PAX7, PHOX2A, PITX2, POU3F3, POU4F2, POU6F2, PRDM14, PROX1, RAX, SALL3, SATB2, SIM1, SIX6, SOX1, SOX11, SOX14, SOX2, TBXT, TBX15, TBX18, TBX5, TLX2, TP73, VSX1, ZFHX4, ZIC5, ZNF132, ZNF229, ZNF334, ZNF516, ZNF667, ZSCAN1, ZSCAN23 |
| DNA bindinga | 2.05 × 10−10 | ASCL1, ASCL2, ASCL4, BARHL2, BHLHE22, BHLHE23, BNC1, DMRTA2, DRGX, DBX1, EN1, EVX1, EVX2, FEZF2, FLI1, FOXD3, FOXE1, FOXI2, SKOR1, GATA4, GCM2, GFI1, HAND2, HIC1, IRF4, LHX1, LHX4, LHX5, MNX1, MSC, MKX1-1, MYOD1, NKX2-6, NKX6-2, OLIG2, ONECUT1, ONECUT2, OTP, OTX1, PAX3, PAX5, PAX6, PAX7, PHOX2A, PITX2, POU3F3, POU4F2, POU6F2, PRDM14, PROX1, RAX, SALL3, SATB2, SIM1, SIX6, SOX1, SOX11, SOX14, SOX2, TBXT, TBX15, TBX18, TBX5, TLX2, TP73, VSX1, ZFHX4, ZIC5, ZNF132, ZNF229, ZNF334, ZNF516, ZNF667, ZSCAN1, ZSCAN23 |
| Transcription regulatory region sequence-specific DNA bindinga | 2.15 × 10−10 | ASCL1, ASCL2, ASCL4, BARHL2, BHLHE22, EN1, FEZF2, FLI1, FOXD3, GATA4, HAND2, IRF4, MNX1, MSC, MKX1-1, MYOD1, NKX6-2, ONECUT2, OTX1, PAX5, PAX6, PHOX2A, PITX2, POU4F2, PRDM14, PROX1, RAX, SATB2, SOX1, SOX11, SOX2, TBXT, TBX15, TBX18, TBX5, ZIC5, ZNF516 |
| Transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific bindinga | 2.81 × 10−10 | ASCL1, ASCL2, BARHL2, EN1, FLI1, GCM2, GIF1, HAND2, IRF4, MYOD1, NKX6-2, ONECUT1, ONECUT2, OTX1, PAX5, PAX6, PHOX2A, PITX2, POU4F2, PROX1, RAX, SATB2, SOX1, SOX11, SOX2, TBX15, TBX18, TP73 |
| Neurokinin receptor binding | 5.08 × 10−10 | TAC1 |
| Substance P receptor binding | 5.08 × 10−10 | TAC1 |
| Sequence-specific double-stranded DNA bindinga | 7.65 × 10−10 | ASCL1, ASCL2, ASCL4, BARHL2, EN1, FEZF2, FLI1, FOXD3, GATA4, HAND2, IRF4, LHX1, MSC, MKX1-1, MYOD1, NKX6-2, ONECUT2, OTX1, PAX5, PAX6, PHOX2A, PITX2, POU4F2, PRDM14, PROX1, RAX, SATB2, SOX1, SOX11, SOX2, TBXT, TBX15, TBX18, TBX5, TP73, ZIC5, ZNF516 |
| Transcription regulatory region DNA bindinga | 9.47 × 10−10 | ASCL1, ASCL2, ASCL4, BARHL2, BHLHE22, EN1, FEZF2, FLI1, FOXD3, GATA4, GFI1, HAND2, IRF4, LHX1, MSC, MKX1-1, MYOD1, NKX6-2, ONECUT2, OTX1, PAX5, PAX6, PHOX2A, PITX2, POU4F2, PRDM14, PROX1, RAX, SATB2, SOX1, SOX11, SOX2, TBXT, TBX15, TBX18, TBX5, TP73, ZIC5, ZNF516 |
| Regulatory region DNA bindinga | 1.01 × 10−9 | ASCL1, ASCL2, ASCL4, BARHL2, BHLHE22, EN1, FEZF2, FLI1, FOXD3, GATA4, GFI1, HAND2, IRF4, LHX1, MSC, MKX1-1, MYOD1, NKX6-2, ONECUT2, OTX1, PAX5, PAX6, PHOX2A, PITX2, POU4F2, PRDM14, PROX1, RAX, SATB2, SOX1, SOX11, SOX2, TBXT, TBX15, TBX18, TBX5, TP73, ZIC5, ZNF516 |
| Regulatory region nucleic acid bindinga | 1.13 × 10−9 | ASCL1, ASCL2, ASCL4, BARHL2, BHLHE22, EN1, FEZF2, FLI1, FOXD3, GATA4, GFI1, HAND2, IRF4, LHX1, MSC, MKX1-1, MYOD1, NKX6-2, ONECUT2, OTX1, PAX5, PAX6, PHOX2A, PITX2, POU4F2, PRDM14, PROX1, RAX, SATB2, SOX1, SOX11, SOX2, TBXT, TBX15, TBX18, TBX5, TP73, ZIC5, ZNF516 |
| Neuropeptide receptor activity | 3.30 × 10−9 | GALR1, QRFPR, GPR139, NPFFR2, NPY2R, NPY5R, PROKR2, SSTR1, SSTR4, SORCS3 |
| Gated channel activity | 6.48 × 10−9 | CACNG7, CNGA3, GABRA1, GABRA2, GABRB1, GLRA3, GRIA4, KCNAB1, KCNH8, KCNK9, KCNQ5, KCNA6, KCNJ3, KCNA1, KCNA3, KCNA4, KCNV1, GRIN3A, CACNA1A, RYR2, KCNT2, TTYH1, CHRNA4 |
| Double-stranded DNA bindinga | 7.49 × 10−9 | ASCL1, ASCL2, ASCL4, BARHL2, EN1, FEZF2, FLI1, FOXD3, FOXE1, GATA4, HAND2, IRF4, LHX1, MSC, MKX1-1, MYOD1, NKX6-2, ONECUT2, OTX1, PAX5, PAX6, PHOX2A, PITX2, POU4F2, PRDM14, PROX1, RAX, SATB2, SOX1, SOX11, SOX2, TBXT, TBX15, TBX18, TBX5, TP73, ZIC5, ZNF516 |
| RNA polymerase II regulatory region sequence-specific DNA bindinga | 9.49 × 10−9 | ASCL1, ASCL2, ASCL4, BARHL2, EN1, FEZF2, FLI1, FOXD3, GATA4, HAND2, IRF4, MSC, MYOD1, NKX6-2, ONECUT2, OTX1, PAX5, PAX6, PHOX2A, PITX2, POU4F2, PRDM14, PROX1, RAX, SATB2, SOX1, SOX11, TBXT, TBX15, TBX18, TBX5, TP73, ZIC5 |
| Voltage-gated potassium channel activity | 1.06 × 10−8 | CNGA3, KCNAB1, KCNH8, KCNK9, KCNQ5, KCNA6, KCNJ3, KCNA1, KCNA3, KCNA4, KCNV1, KCNT2 |
| RNA polymerase II regulatory region DNA bindinga | 1.19 × 10−8 | ASCL1, ASCL2, ASCL4, BARHL2, EN1, FEZF2, FLI1, FOXD3, GATA4, HAND2, IRF4, MSC, MYOD1, NKX6-2, ONECUT2, OTX1, PAX5, PAX6, PHOX2A, PITX2, POU4F2, PRDM14, PROX1, RAX, SATB2, SOX1, SOX11, TBXT, TBX15, TBX18, TBX5, TP73, ZIC5 |
| Transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific bindinga | 1.24 × 10−8 | BARHL2, FEZF2, FLI1, GATA4, GCM2, HAND2, IRF4, LHX4, MYOD1, ONECUT1, ONECUT2, OTX1, PAX5, PAX6, PHOX2A, PITX2, POU4F2, RAX, SATB2, SIX6, SOX1, SOX11, SOX2, TBX5, TLX2,, TP73 |
aGO molecular functions involved in DNA binding or transcriptional regulation. Genes for which the protein class is a transcription factor are indicated by underlining.
Differences in genetic aberrations between EE and LE
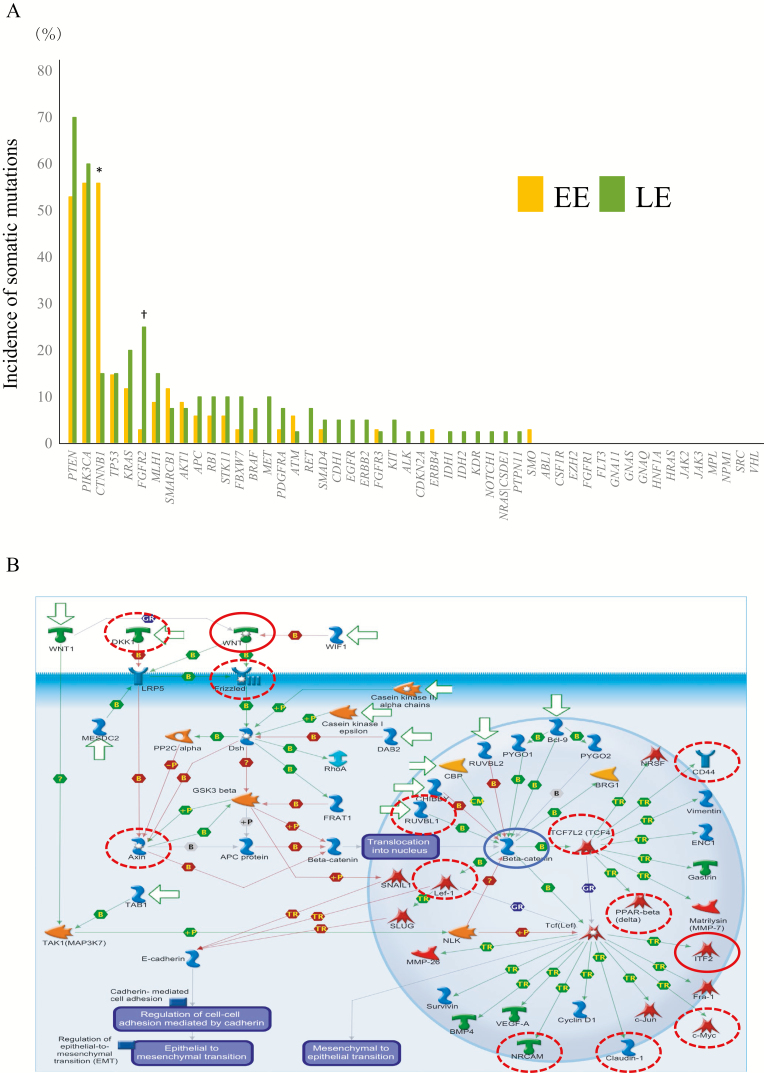
Two hundred and one somatic mutations (90 synonymous and 111 non-synonymous) were detected in 34 samples of EE (5.9 ± 2.2 mutations per sample), whereas 300 mutations (119 synonymous and 181 non-synonymous) were detected in 40 samples of LE (7.5 ± 3.3 mutations per sample). The incidence of somatic mutation of the CTNNB1 gene was significantly higher in EE samples than in LE samples (P = 4.26 × 10−4), and the incidence of that of the FGFR2 gene was significantly higher in LE samples than in EE samples (P = 8.77 × 10−3), whereas significant differences were not found in the remaining 48 genes (Figure 3A). Average SIFT scores for the CTNNB1 and FGFR2 genes were both 0 in EE samples and LE samples, respectively, indicating that these somatic mutations impair protein function (a SIFT score of <0.05 means ‘damaging’ (29)).
Figure 3.
Differences in genetic and epigenetic states between EE (patients aged ≤40 years) and LE. (A) The incidence of somatic mutations of the 50 examined tumor-related genes in 34 samples of EE and 40 samples of LE. Genes showing a significantly higher incidence (P < 0.05) of somatic mutations in EE samples than in LE samples and genes showing a significantly higher incidence of somatic mutations in LE samples than in EE samples are indicated by * and †, respectively. (B) The pathway ‘Development WNT signaling pathway’ (P = 3.45 × 10−8) illustrated schematically using MetaCore software. Genes showing DNA hypermethylation in 19 EE samples with somatic mutations of the CTNNB1 gene (CTNNB1-M) relative to 15 EE samples without such mutations (CTNNB1-W) (Welch’s t-test P < 0.01 and ΔβCTNNB1-M − CTNNB1-W value of >0.15) are indicated by red circles. Genes showing DNA hypomethylation in CTNNB1-M samples relative to CTNNB1-W samples (Welch’s t-test P < 0.01 and ΔβCTNNB1-M − CTNNB1-W value of < −0.15) are indicated by dotted red circles. The CTNNB1 gene is indicated by a blue circle.
Correlation between DNA methylation alterations and genetic aberrations in EE and LE
In order to clarify the correlation between DNA methylation alterations and genetic aberrations in EE samples, we identified 2908 probes that showed significant differences in DNA methylation levels between 19 EE samples with somatic mutation of the CTNNB1 gene (tissue samples of early-onset endometrioid endometrial cancer with somatic mutation of the CTNNB1 gene, CTNNB1-M) and 15 EE samples without it (tissue samples of early-onset endometrioid endometrial cancer without somatic mutation of the CTNNB1 gene, CTNNB1-W) (Welch’s t-test P < 0.01; ∆βCTNNB1-M − CTNNB1-W value >0.15 or < −0.15; Supplementary Table 6, available at Carcinogenesis Online). MetaCore pathway analysis revealed that 1419 genes for which these 2908 probes were designed were significantly accumulated in the Wnt signaling pathway (P = 3.45 × 10−8; Figure 3B). We identified 1133 probes that showed significant differences in DNA methylation levels between 10 LE samples with somatic mutation of the FGFR2 gene (tissue samples of late-onset endometrioid endometrial cancer with somatic mutation of the FGFR2 gene, FGFR2-M) and 30 LE samples without it (tissue samples of late-onset endometrioid endometrial cancer without somatic mutation of the FGFR2 gene, FGFR2-W) (Welch’s t-test P < 0.01; ∆β FGFR2-M − FGFR2-W value >0.15 or < −0.15; Supplementary Table 7, available at Carcinogenesis Online). Three of the 567 genes for which these 1133 probes were designed were included in the fibroblast growth factor signaling pathway according to the MetaCore software (Supplementary Figure 1, available at Carcinogenesis Online).
Epigenetic clustering of endometrioid endometrial cancer based on DNA methylation profile
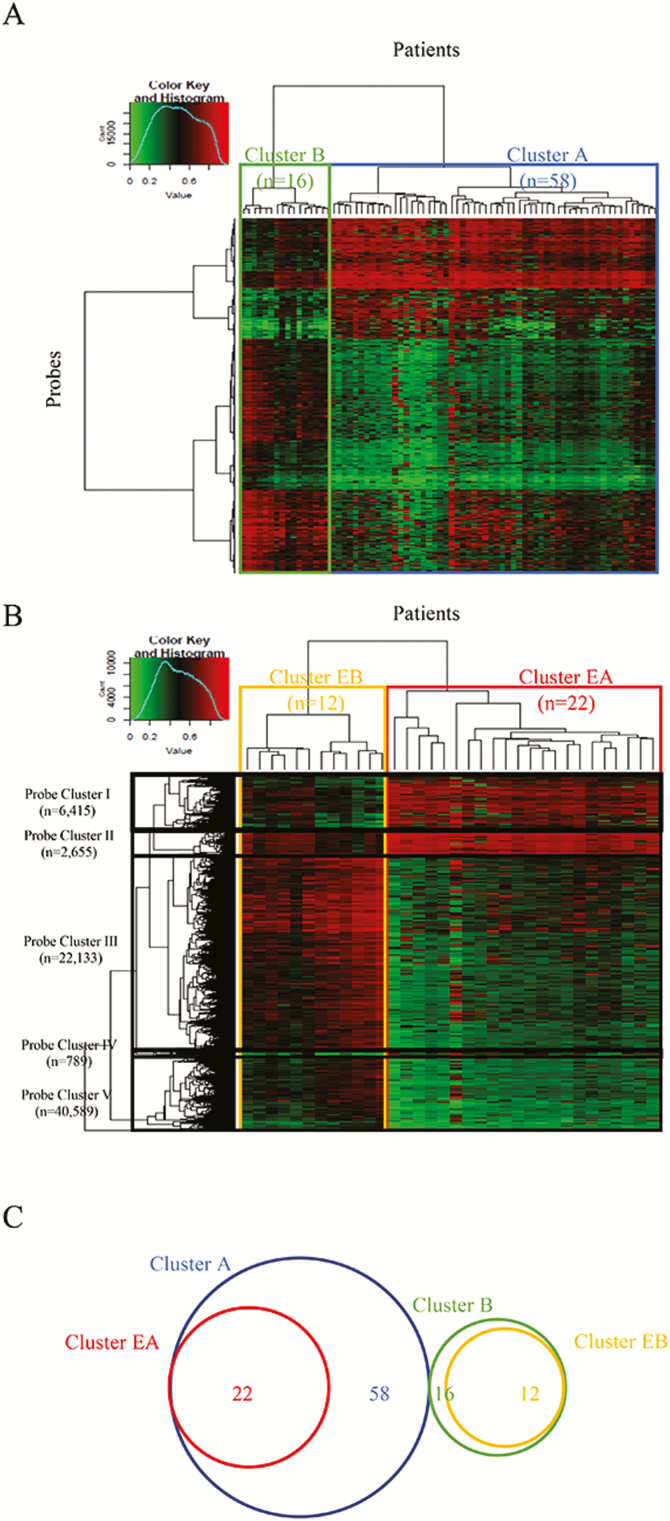
Unsupervised hierarchical clustering using 63 033 probes showing significant differences in DNA methylation levels between the 31 normal control endometrial tissue samples and 74 endometrioid endometrial cancer tissue samples subclustered the patients with cancer into Cluster A (n = 58) and Cluster B (n = 16) (Figure 4A). The clinicopathological parameters of the patients in these clusters are summarized in Supplementary Table 8A, available at Carcinogenesis Online. Patients in Cluster A were significantly older, showed significantly frequent lymphovascular invasion and tended to be more frequently diagnosed as histological grade 3 than those in Cluster B.
Figure 4.
Epigenetic clustering of endometrioid endometrial cancer. (A) Unsupervised hierarchical clustering of endometrioid endometrial cancer tissue samples using DNA methylation levels on 63 033 probes showing significant differences in DNA methylation levels between 31 samples of normal control endometrial tissue and 74 samples of endometrioid endometrial cancer tissue (Welch’s t-test, adjusted Bonferroni correction [α = 1.18 × 10−8] and Δβendometrioid endometrial cancer tissue − normal control endometrial tissue value of >0.25 or < −0.25). On the basis on DNA methylation status, 74 patients were subclustered into Cluster A (n = 58) and Cluster B (n = 16). Correlations between this epigenetic clustering and clinicopathological parameters are summarized in Supplementary Table 8A, available at Carcinogenesis Online. (B) Unsupervised hierarchical clustering of EE samples using DNA methylation levels on the 40 589 probes showing significant differences in DNA methylation between 31 normal control endometrial tissue samples and 34 EE samples (Welch’s t-test, adjusted Bonferroni correction [α = 1.18 × 10−8] and ΔβEE − normal control endometrial tissue value of >0.25 or < −0.25). On the basis on DNA methylation status, the 34 EE patients were subclustered into Cluster EA (n = 22) and Cluster EB (n = 12). Correlations between this epigenetic clustering and clinicopathological parameters of the patients are summarized in Supplementary Table 8B, available at Carcinogenesis Online. Probe Clusters I–V are shown on the left side of the heatmap. (C) Venn diagram showing the relationship between Clusters A and B of endometrioid endometrial cancer tissue samples and Clusters EA and EB of EE samples. All samples belonging to Cluster EA are included in Cluster A, whereas all samples belonging to Cluster EB are included in Cluster B without exception.
Kaplan–Meier survival curves of patients belonging to Clusters A and B were plotted for a period ranging from 263 to 4034 days (median, 1605 days). Although the cancer-free and overall patient survival rates tended to be lower in Cluster A than in Cluster B, such differences did not reach statistically significant levels (P = 0.24 and 0.53, respectively, log-rank test), probably due to the small number of deaths or recurrent cases (Supplementary Figure 2, available at Carcinogenesis Online). Moreover, all recurrence-positive (n = 4) and disease-specific death-positive (n = 1) cases were included in Cluster A (Supplementary Table 8A, available at Carcinogenesis Online).
Unsupervised hierarchical clustering using 40 589 probes showing significant differences in DNA methylation levels between the 31 normal control endometrial tissue samples and 34 EE samples (Welch’s t-test, adjusted Bonferroni correction [α = 1.18 × 10−8]; ΔβEE − normal control endometrial tissue value >0.25 or < −0.25) subclustered 34 patients with EE into samples in Cluster EA (n = 22) and samples in Cluster EB (n = 12) (Figure 4B). The clinicopathological parameters of the patients in these clusters are summarized in Supplementary Table 8B, available at Carcinogenesis Online. Patients in Cluster EA tended to show more frequent lymphovascular invasion and to be more frequently diagnosed as histological grade 3 than those in Cluster EB.
The cancer-free survival rate of patients belonging to Cluster EA tended to be lower than that of patients in Cluster EB, although the difference did not reach a statistically significant level (P = 0.37, log-rank test), probably due to the small number of recurrent cases (Supplementary Figure 2, available at Carcinogenesis Online). Only one recurrence case was included in Cluster EA (Supplementary Table 8B, available at Carcinogenesis Online). As the overall survival rate was 100% in both Clusters EA and EB, the log-rank test was not performed. Moreover, all patients belonging to Cluster EA were included in Cluster A and all patients belonging to Cluster EB were included in Cluster B without exception (Figure 4C).
As seen in Figure 4B, Probe Clusters I and III showed obvious differences in DNA methylation levels between Clusters EA and EB. Furthermore, the ratio of probes located in CpG islands, CpG island shores and CpG island shelves that were important for transcription regulation to all probes belonging to Probe Cluster I was significantly higher than that of probes in Probe Cluster III. We then focused on Probe Cluster I, in which 927 of the 6415 probes were designed from the transcription start site to 1500 bp upstream of the transcription start site of the 470 genes. One hundred and one (Supplementary Table 9A, available at Carcinogenesis Online) of the 470 genes showed a significant (P < 0.05) inverse correlation (r < −0.2) between DNA methylation levels and the messenger RNA expression levels in the TCGA database (https://tcga-data.nci.nih.gov/docs/publications/ucec_2013/) (23). The 101 genes were also evaluated for protein function by enrichment analysis using the MetaCore software and compared with the protein function distribution of genes within the GeneGo database (Supplementary Table 9B, available at Carcinogenesis Online). Fifty-nine statistically significant GO molecular functions in which the 101 genes participated are listed in Supplementary Table 9C, available at Carcinogenesis Online. DNA methylation levels of 18 of the 101 genes, including HOXA9, HOXD10 and SOX11, were significantly correlated with a higher incidence of lymphovascular invasion of EE (Supplementary Table 10, available at Carcinogenesis Online).
Identification of marker CpG sites for distinguishing Cluster EB from Cluster EA
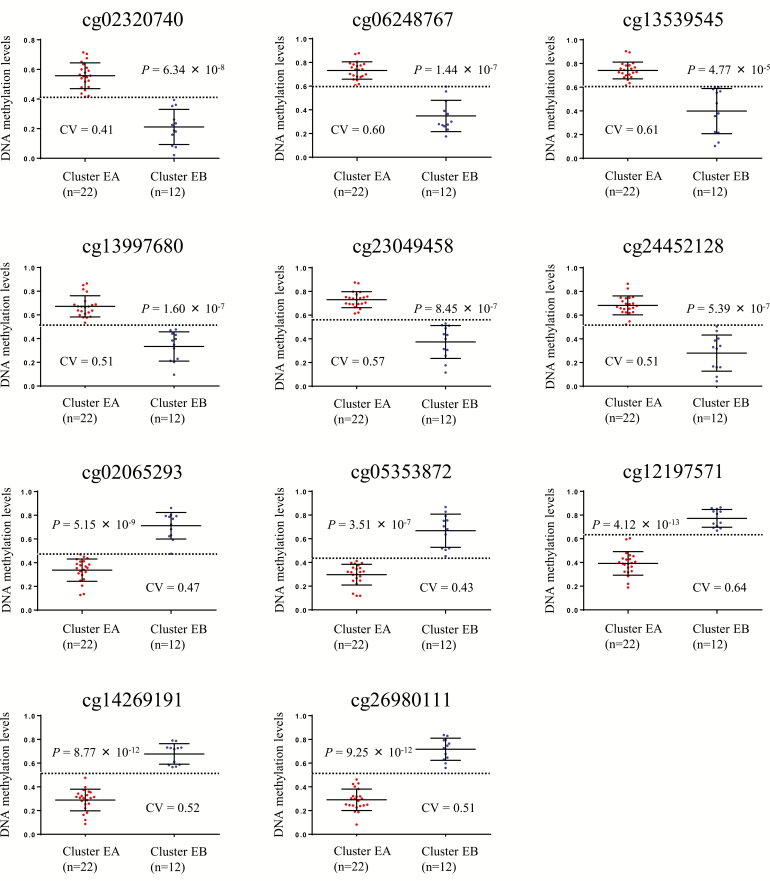
In order to identify diagnostic markers capable of distinguishing the less aggressive Cluster EB from Cluster EA, ROC analysis was performed using the top 100 probes showing the largest differences in DNA methylation levels between EA and EB samples, and the corresponding area under curve (AUC) values were calculated. Among the 100 probes, 11 showed AUC values of 1 (Supplementary Table 11, available at Carcinogenesis Online). The Youden index was used as a cutoff value for each of the 11 probes. As shown by the scattergrams in Figure 5, the use of such cutoff values was able to discriminate EB samples from EA samples with 100% sensitivity and specificity. The DNA methylation levels of these 11 candidate marker CpG sites were successfully verified using another quantification method, MassARRAY: significant correlations between DNA methylation levels demonstrated by Infinium assay and those by MassARRAY were confirmed (Supplementary Figure 3, available at Carcinogenesis Online).
Figure 5.
Scattergrams of DNA methylation levels for all 11 probes showing an area under the curve value of 1 in receiver operating characteristic curve analysis for discrimination of EB samples (n = 12) from EA samples (n = 22). P values by Welch’s t-test for each probe are shown in each panel. Cutoff values (CVs) are shown by a dotted line in each panel. Using each probe and its CV, EB samples were discriminated from EA samples with 100% sensitivity and specificity.
Discussion
We focused on the differences between early- and late-onset cancers of the major histological subtype, i.e. endometrioid endometrial cancer. The incidence of somatic mutations of the CTNNB1 gene was significantly higher in EE samples than in LE samples, whereas somatic mutations of the FGFR2 gene were more frequent in LE samples than in EE samples. Moreover, DNA methylation alterations and genetic aberrations may cooperatively activate the Wnt signaling pathway (31) during the development of EE. Although an inverse association between immunohistochemically detected nuclear accumulation of β-catenin and the age of patients with endometrial cancer has been reported previously (32), and mutations of the CTNNB1 gene have been shown to be accumulated in a subset of endometrioid endometrial cancers arising in young and obese patients (33), this study has comprehensively revealed epigenetic and genetic alterations of genes participating in Wnt signaling in EE for the first time. Similarly, DNA methylation alterations and gene aberrations may cooperatively participate in the fibroblast growth factor signaling pathway in late-onset endometrioid endometrial carcinogenesis. Although activating mutation of the FGFR2 gene has been considered a therapeutic target for endometrioid endometrial cancer (34,35), its correlation with late-onset carcinogenesis has never been reported previously. Thus, the pathways contributing to the development of early- and late-onset endometrial cancer appear to partially differ.
With regard to the genome-wide DNA methylation profile, all 1034 probes showing significant differences in DNA methylation levels between EE and LE samples again showed significant differences between LE and normal control endometrial tissue samples. On the other hand, only 102 probes of the 1034 probes showed significant differences in DNA methylation levels between EE and normal control endometrial tissue samples, indicating that DNA methylation alterations for the majority of the 1034 probes occurred specifically in LE samples. GO enrichment analysis indicated that 371 genes, for which the 1034 probes were designed, were clearly overrepresented by ‘transcriptional factors’ and were accumulated in signaling pathways participating in transcriptional regulation. The late-onset-specific DNA methylation profile may modify the clinicopathological features of LE samples as a result of the differences in the gene expression profiles. In fact, myometrial invasion, one of the most important prognostic factors of endometrioid endometrial cancer (2), was more frequent in LE than in EE samples.
Epigenetic clustering based on DNA methylation profiles showed that endometrioid endometrial cancers belonging to Cluster A were clinicopathologically more aggressive than those belonging to Cluster B. This tendency was also evident in patients belonging to Cluster EA, and all patients in Cluster EA were included in Cluster A without exception. The 101 hallmark genes for Cluster EA may drive the development and progression of different tumor subtypes. For example, the DNA methylation levels of 18 genes, including HOXA9, HOXD10 and SOX11, were significantly correlated with more frequent lymphovascular tumor invasion. DNA hypermethylation of the HOXA9 homeobox gene is reportedly associated with a higher grade of serous ovarian cancer (36), recurrence of bladder cancer (37) and non-small cell lung cancer (38) and mortality in non-infant patients with neuroblastoma (39). Decreased expression of the HOXD10 homeobox gene has been reported in prostate (40), breast (41), thyroid (42), colorectal (43), ovarian (44) and endometrial (45) cancer. Promoter methylation of the SOX11 gene has reportedly been associated with cell growth, invasion or poor prognosis of gastric cancer (46), hematopoietic malignancies (47) and nasopharyngeal carcinoma (48). It is feasible that DNA methylation alterations of these genes participate in determining the clinicopathological characteristics of Cluster EA. These data indicate that clinicopathological features are strongly defined on the basis of DNA methylation profiles, even in EE.
ROC analysis identified markers at CpG sites that were able to distinguish epigenetic Cluster EB from Cluster EA: 11 CpG sites showed AUC values of 1 for such discrimination, and Cluster EB of this cohort was diagnosed using the 11 marker CpG sites with both a sensitivity and specificity of 100%. Although some of the 11 marker CpGs were located in CpG islands around the transcription start sites, an inverse correlation between levels of DNA methylation and messenger RNA expression was not confirmed in the data for endometrial cancer deposited in the TCGA database (https://tcga-data.nci.nih.gov/docs/publications/ucec_2013/) (23). Therefore, DNA methylation alterations of such genes may not result in alterations of expression, and such genes may not be potential therapeutic targets. Instead, as the present data have indicated that the clinicopathological features of endometrial cancers may be determined by their DNA methylation profiles, these 11 markers may be able to reproducibly identify less aggressive cancers, such as those belonging to Cluster EB. Although a validation study will, of course, be needed, DNA methylation diagnostics of biopsy specimens using appropriate marker CpG sites, such as these 11 CpG sites, may help to indicate the feasibility of fertility preservation therapy for patients with EE.
As all samples belonging to Clusters EA and EB were included in Clusters A and B, respectively, it is not surprising that the 11 CpG sites showed excellent AUCs even in ROC analysis for discrimination of the more aggressive Cluster A from the less aggressive Cluster B (Supplementary Table 12, available at Carcinogenesis Online): The 11 CpG sites may also become good markers for prognostication of all age groups of patients with endometrioid endometrial cancers. Moreover, the DNA methylation levels of the 11 CpG sites were not significantly correlated with clinicopathological parameters reflecting tumor aggressiveness, i.e. histological grade and stage, or patient outcome, in cancers of the lung, stomach, breast, colon and ovary deposited in the TCGA database (data not shown), suggesting that the prognostic potential of the 11 CpG sites may be specific to endometrioid endometrial cancers.
In summary, genome-wide DNA methylation analysis using EPIC array and targeted sequencing of tumor-related genes for 112 endometrial tissue samples have indicated that genetically and epigenetically different pathways may participate in the development of early- and late-onset endometrial cancers. As DNA methylation profiles may determine the clinicopathological features of endometrial cancers, such profiling may predict tumors that are less aggressive and amenable to fertility preservation treatment.
Funding
The Project for Utilizing Glycans in the Development of Innovative Drug Discovery Technologies (17ae0101020h0002); Expedite Effective Drug Discovery by Government, Academia and Private partnership (17ak0101043s0603) from the Japan Agency for Medical Research and Development (AMED) and also KAKENHI (16H02472 and 16K08720) from the Japan Society for the Promotion of Science.
Conflict of Interest Statement: None declared.
Supplementary Material
Glossary
Abbreviations
- CTNNB1-M
tissue samples of early-onset endometrioid endometrial cancer with somatic mutation of the CTNNB1 gene
- CTNNB1-W
tissue samples of early-onset endometrioid endometrial cancer without somatic mutation of the CTNNB1 gene
- EE
early-onset endometrioid endometrial cancer
- FGFR2-M
tissue samples of late-onset endometrioid endometrial cancer with somatic mutation of the FGFR2 gene
- FGFR2-W
tissue samples of late-onset endometrioid endometrial cancer without somatic mutation of the FGFR2 gene
- GO
Gene Ontology
- LE
late-onset endometrioid endometrial cancer
- ROC
receiver operating characteristic
- TCGA
The Cancer Genome Atlas
References
- 1. Ferlay J., et al. (2015) Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer, 136, E359–E386. [DOI] [PubMed] [Google Scholar]
- 2. Yamagami W., et al. (2015) Annual report of the Committee on Gynecologic Oncology, the Japan Society of Obstetrics and Gynecology. J. Obstet. Gynaecol. Res., 41, 167–177. [DOI] [PubMed] [Google Scholar]
- 3. Ota T., et al. (2005) Clinicopathologic study of uterine endometrial carcinoma in young women aged 40 years and younger. Int. J. Gynecol. Cancer, 15, 657–662. [DOI] [PubMed] [Google Scholar]
- 4. Duska L.R., et al. (2001) Endometrial cancer in women 40 years old or younger. Gynecol. Oncol., 83, 388–393. [DOI] [PubMed] [Google Scholar]
- 5. Laurelli G., et al. (2011) Conservative treatment of early endometrial cancer: preliminary results of a pilot study. Gynecol. Oncol., 120, 43–46. [DOI] [PubMed] [Google Scholar]
- 6. Yamagami W., et al. (2018) Is repeated high-dose medroxyprogesterone acetate (MPA) therapy permissible for patients with early stage endometrial cancer or atypical endometrial hyperplasia who desire preserving fertility? J. Gynecol. Oncol., 29, e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jones P.A., et al. (2016) Targeting the cancer epigenome for therapy. Nat. Rev. Genet., 17, 630–641. [DOI] [PubMed] [Google Scholar]
- 8. Baylin S.B., et al. (2016) Epigenetic determinants of cancer. Cold Spring Harb. Perspect Biol., 8, a019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ahuja N., et al. (2016) Epigenetic therapeutics: a new weapon in the war against cancer. Annu. Rev. Med., 67, 73–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kanai Y. (2010) Genome-wide DNA methylation profiles in precancerous conditions and cancers. Cancer Sci., 101, 36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arai E., et al. (2010) DNA methylation profiles in precancerous tissue and cancers: carcinogenetic risk estimation and prognostication based on DNA methylation status. Epigenomics, 2, 467–481. [DOI] [PubMed] [Google Scholar]
- 12. Soto J., et al. (2016) The impact of next-generation sequencing on the DNA methylation-based translational cancer research. Transl. Res., 169, 1–18.e1. [DOI] [PubMed] [Google Scholar]
- 13. Robles A.I., et al. (2015) An Integrated Prognostic Classifier for Stage I Lung Adenocarcinoma based on mRNA, microRNA, and DNA Methylation Biomarkers. J. Thorac. Oncol., 10, 1037–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yamanoi K., et al. (2015) Epigenetic clustering of gastric carcinomas based on DNA methylation profiles at the precancerous stage: its correlation with tumor aggressiveness and patient outcome. Carcinogenesis, 36, 509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tian Y., et al. (2014) Prognostication of patients with clear cell renal cell carcinomas based on quantification of DNA methylation levels of CpG island methylator phenotype marker genes. BMC Cancer, 14, 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sato T., et al. (2013) DNA methylation profiles at precancerous stages associated with recurrence of lung adenocarcinoma. PLoS One, 8, e59444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Arai E., et al. (2012) Single-CpG-resolution methylome analysis identifies clinicopathologically aggressive CpG island methylator phenotype clear cell renal cell carcinomas. Carcinogenesis, 33, 1487–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Trimarchi M.P., et al. (2017) Identification of endometrial cancer methylation features using combined methylation analysis methods. PLoS One, 12, e0173242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Farkas S.A., et al. (2017) Epigenetic changes as prognostic predictors in endometrial carcinomas. Epigenetics, 12, 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang B., et al. (2014) Comparative DNA methylome analysis of endometrial carcinoma reveals complex and distinct deregulation of cancer promoters and enhancers. BMC Genomics, 15, 868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jiao Y., et al. (2014) A systems-level integrative framework for genome-wide DNA methylation and gene expression data identifies differential gene expression modules under epigenetic control. Bioinformatics, 30, 2360–2366. [DOI] [PubMed] [Google Scholar]
- 22. Sánchez-Vega F., et al. (2013) Recurrent patterns of DNA methylation in the ZNF154, CASP8, and VHL promoters across a wide spectrum of human solid epithelial tumors and cancer cell lines. Epigenetics, 8, 1355–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Levine D.A.; The Cancer Genome Atlas Research Network (2013) Integrated genomic characterization of endometrial carcinoma. Nature, 497, 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Silverberg S.G., et al. (2003) Epithelial tumours and related lesions. In: Tavassoli F.A. and Devilee P (eds) World Health Organization Classification of Tumours: Pathology and Genetics—Tumours of the Breast and Female Genital Organs. IARC Press, Lyon, France, pp. 217–232. [Google Scholar]
- 25. Sobin L.H., et al. ; International Union Against Cancer (UICC). (2009) TNM Classification of Malignant Tumours. 7th edn. Wiley-Liss, New York, NY. [Google Scholar]
- 26. Pecorelli S. (2009) Revised FIGO staging for carcinoma of the vulva, cervix, and endometrium. Int. J. Gynaecol. Obstet., 105, 103–104. [DOI] [PubMed] [Google Scholar]
- 27. Kanai Y., et al. (2018) The Japanese Society of Pathology Guidelines on the handling of pathological tissue samples for genomic research: standard operating procedures based on empirical analyses. Pathol. Int., 68, 63–90. [DOI] [PubMed] [Google Scholar]
- 28. Bibikova M., et al. (2009) Genome-wide DNA methylation profiling using Infinium® assay. Epigenomics, 1, 177–200. [DOI] [PubMed] [Google Scholar]
- 29. Ng P.C., et al. (2002) Accounting for human polymorphisms predicted to affect protein function. Genome Res., 12, 436–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fan J., et al. (2006) Understanding receiver operating characteristic (ROC) curves. CJEM, 8, 19–20. [DOI] [PubMed] [Google Scholar]
- 31. Yuexin L., et al. (2014) Clinical significance of CTNNB1 mutation and Wnt pathway activation in endometrioid endometrial carcinoma. J. Natl. Cancer Inst., 106, dju245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Peiró G., et al. (2013) Association of mammalian target of rapamycin with aggressive type II endometrial carcinomas and poor outcome: a potential target treatment. Hum. Pathol., 44, 218–225. [DOI] [PubMed] [Google Scholar]
- 33. Klaus A., et al. (2008) Wnt signalling and its impact on development and cancer. Nat. Rev. Cancer, 8, 387–398. [DOI] [PubMed] [Google Scholar]
- 34. Pollock P.M., et al. (2007) Frequent activating FGFR2 mutations in endometrial carcinomas parallel germline mutations associated with craniosynostosis and skeletal dysplasia syndromes. Oncogene, 26, 7158–7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Byron S.A., et al. (2009) FGFR2 as a molecular target in endometrial cancer. Future Oncol., 5, 27–32. [DOI] [PubMed] [Google Scholar]
- 36. Montavon C., et al. (2012) Prognostic and diagnostic significance of DNA methylation patterns in high grade serous ovarian cancer. Gynecol. Oncol., 124, 582–588. [DOI] [PubMed] [Google Scholar]
- 37. Reinert T., et al. (2012) Diagnosis of bladder cancer recurrence based on urinary levels of EOMES, HOXA9, POU4F2, TWIST1, VIM, and ZNF154 hypermethylation. PLoS One, 7, e46297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hwang J.A., et al. (2015) HOXA9 inhibits migration of lung cancer cells and its hypermethylation is associated with recurrence in non-small cell lung cancer. Mol Carcin., 54, E72–80 [DOI] [PubMed] [Google Scholar]
- 39. Alaminos M., et al. (2004) Clustering of gene hypermethylation associated with clinical risk groups in neuroblastoma. J. Natl. Cancer Inst., 96, 1208–1219. [DOI] [PubMed] [Google Scholar]
- 40. Mo R.J., et al. (2017) Decreased HoxD10 expression promotes a proliferative and aggressive phenotype in prostate cancer. Curr. Mol. Med., 17, 70–78. [DOI] [PubMed] [Google Scholar]
- 41. Vardhini N.V., et al. (2014) HOXD10 expression in human breast cancer. Tumour Biol., 35, 10855–10860. [DOI] [PubMed] [Google Scholar]
- 42. Cao Y.M., et al. (2018) Aberrant hypermethylation of the HOXD10 gene in papillary thyroid cancer with BRAFV600E mutation. Oncol. Rep., 39, 338–348. [DOI] [PubMed] [Google Scholar]
- 43. Wang Y., et al. (2016) miR-10b promotes invasion by targeting HOXD10 in colorectal cancer. Oncol. Lett., 12, 488–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nakayama I., et al. (2013) Loss of HOXD10 expression induced by upregulation of miR-10b accelerates the migration and invasion activities of ovarian cancer cells. Int. J. Oncol., 43, 63–71. [DOI] [PubMed] [Google Scholar]
- 45. Osborne J., et al. (1998) Expression of HOXD10 gene in normal endometrium and endometrial adenocarcinoma. J. Soc. Gynecol. Investig., 5, 277–280. [DOI] [PubMed] [Google Scholar]
- 46. Hu X., et al. (2015) Aberrant SOX11 promoter methylation is associated with poor prognosis in gastric cancer. Cell Oncol., 38, 183–194. [DOI] [PubMed] [Google Scholar]
- 47. Gustavsson E., et al. (2010) SOX11 expression correlates to promoter methylation and regulates tumor growth in hematopoietic malignancies. Mol Cancer, 9, 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang S., et al. (2013) Promoter methylation status of the tumor suppressor gene SOX11 is associated with cell growth and invasion in nasopharyngeal carcinoma. Cancer Cell Int., 13, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.