Abstract
In the past 15 years, cell-based microscopy has evolved its focus from observing cell function to aiming to predict it. In particular—powered by breakthroughs in computer vision, large-scale image analysis and machine learning—high-throughput and high-content microscopy imaging have enabled to uniquely harness single-cell information to systematically discover and annotate genes and regulatory pathways, uncover systems-level interactions and causal links between cellular processes, and begin to clarify and predict causal cellular behaviour and decision making. Here we review these developments, discuss emerging trends in the field, and describe how single-cell ‘omics and single-cell microscopy are imminently in an intersecting trajectory. The marriage of these two fields will make possible an unprecedented understanding of cell and tissue behaviour and function.
Keywords: causal cell behaviour, gene regulatory networks, genome-wide screening, high-content microscopy, high-throughput microscopy, Machine Learning
The unique role of microscopy in illuminating single cell biology
Single cell biology will fundamentally revolutionize biology and healthcare. Building on the arsenal of ‘omics technologies developed in the past 20 years—genomics, transcriptomics, metabolomics, epigenomics, proteomics, etc.—and applying them to comprehensively molecularly profile individual cells in unparalleled detail, this young field aims to: systematically elucidate what brings about cell-to-cell heterogeneity within cell populations and tissues; clarify the link between genotype and phenotype; characterize the broad diversity of existing cellular types, states, transitions and histories observed in the living; provide quantitative/predictive insights on how the pheno-genotypic landscape of cells changes upon physiological or pathological triggers; and ultimately leverage that knowledge to reveal new biology as well as help develop improved cell-based diagnostics and tools for bio/healthcare applications [1–4]. For instance the Human Cell Atlas consortium aims to exploit single-cell technologies to create the most comprehensive reference maps to date of all human cells [5]. Similarly, the use of single-cell ‘omics technologies to track embryonic development cell by cell was highlighted as part of Science Magazine’s 2018 Breakthrough of the Year [6]. Yet for all that single-cell ‘omics can give a deep molecular snapshot of cells and even their lineage relationships and history (for instance using molecular barcoding approaches [7–9]), those approaches fail to fully capture the true organization and spatiotemporal dynamics of cells and tissues as they grow, divide, die, migrate or change their structure/function through time, and hence can infer only indirectly how cells make decisions in space and in time in a cause–effect manner. This is where microscopy, particularly ‘live cell’ fluorescence light microscopy, is unchallenged and unparalleled [10–14]. This essay describes how high-throughput and high-content microscopy imaging make it possible to exploit spatial and dynamical information at the single-cell level to discover genes and pathways, observe and predict cell structure and function, and how it could shed unique light into causal cell function and evolution in ways complementary and inaccessible to other single-cell approaches.
From high-throughput screening to high-throughput and high-content cell biology
The combined use of large-scale (‘high-throughput’), cell-based fluorescence light microscopy with computational image analysis and feature extraction (‘high-content’) was originally published as a way to quantitatively profile at scale the effect of drugs on cells, in the context of pharmacological ‘fixed cell’ end-point assays for drug screening/development where it has been used since extensively [15–18]. However, it quickly became obvious that this could provide a new way to perform cell biological [19] experiments at scale in an unbiased, systematic and quantitative manner (Figure 1). One of the first published studies to show this was a budding yeast (S. cerevisiae) study using 4718 knock-out (KO) gene deletion mutant cell lines to look for genes whose mutation could yield defective cell wall, actin and nuclear DNA morphological phenotypes. That fixed-cell, endpoint assay study identified 2378 gene candidates potentially involved in controlling different aspects of cell morphology and found that similar phenotypes were caused by deletions of functionally related genes [20], providing the first illustration of the potential of this approach to do comprehensive—potentially genome-wide—reverse genetics investigations. In the 10 years that followed, and gradually empowered by the GFP revolution as well as increasingly refined techniques for systematic gene KO or knock-down (KD; RNAi/siRNA and now CRISPR), a plethora of similar studies followed using cell types ranging from yeasts to human cell lines and aiming to carry out genome-wide gene discovery and functional assignment [19,21–29]. For instance, the landmark project MitoCheck used RNA interference to KD ∼21 000 human protein coding genes in live HeLa cells and systematically look for candidate human cell division genes through the genome [30]. By quantitatively and automatically analysing more than 180 000 timelapse epifluorescence microscopy movies of cells stably expressing histone H2B-GFP (>17 000 000 images), MitoCheck identified and validated 572 genes involved in cell division—over half of them never before linked to mitosis regulation – providing the largest genomic catalogue of mitotic genes until then. High-throughput microscopy phenotyping has been used with success to obtain genomic catalogues of—and discover many new genes involved in – countless basic cellular processes including cell division [30], cell migration [31], endocytosis [32,33], Golgi organization [34,35], mitochondrial quality control [36] and structure [37], the microtubule cytoskeleton [38,39] and viral infection [40]. Notably, despite all those seminal works and others, functional discovery and annotation across genomes is still very much a task in progress [41]. In the case of high-throughput microscopy screening, the reasons for this are numerous and include the fact that the translation of quantitative measurements into well-defined annotations in agreed ontologies is particularly challenging [42]. In addition microscopy-based screens have traditionally suffered from poor reproducibility [43] (i.e. a low overlap between gene lists obtained from independent screens looking at a same process) and it has been shown that the identification by microscopy-based screening of candidate genes associated with a given cell function is probabilistic and does not allow in any one screen to identify all possible genes associated with that function [39], a coverage problem that may only be gradually solved through incremental integration of complementary screens in the future.
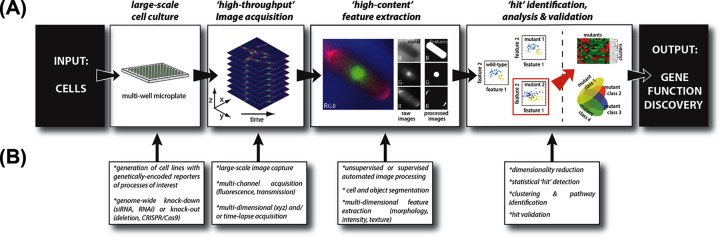
Figure 1. The principle of high-throughput/high-content microscopy screens to identify and study genes’ functions at scale.
(A) Typical high-throughput/high-content microscopy pipeline, where the inputs are cells and cell images, and the output is high-dimensional, single-cell feature data. (B) Implementation steps involved in the context of a single perturbation (KD/KO) genome-wide phenotypic screen, allowing to systematically discover gene functions at scale.
Using computational image analysis and machine learning for deep single-cell phenotyping
Of note, the introduction of high-content microscopy phenotyping was key to enable richer and deeper phenotypic characterization at the single-cell level, thanks to advances in numerical feature extraction, comparison and analysis [11,44–46]. Computed single-cell-based morphological, intensity and texture features can be simple and biologically interpretable, e.g. the number, size, intensity and positions of endocytic vesicles [33] or the length, number and curvature of microtubules [39], or they can be more complex mathematical constructions, like the CHARM feature set [47]. In addition, cell population-level features like cellular position and context [48,49] can be obtained. The data science pipelines needed to extract information from such descriptors is then more involved [4,50] and include comparison of conditions [51,52] and use of machine learning methods [53] (e.g. unsupervised clustering or supervised classification) to both explore cell-to-cell variability and to group cells or conditions according to phenotypic similarity for functional annotation or mechanistic characterization [54]. The publication of open source libraries and software packages performing those tasks and automating them have been fundamental to advances in the field; they are described in more detail elsewhere [45,50].
An important recent development is Deep Learning, a class of machine learning/artificial intelligence (ML/AI) algorithms that can be easier to use and far more efficient than their predecessors, and particularly adapted to images through Convolutional Neural Networks (CNNs) [55]. They have been applied to high-throughput/high-content microscopy and are sure to become a key part of analysis pipelines from now on. They can be used as a feature learning step included in a classical pipeline [56] (for example to better segment/detect cells or subcellular structures that are then analysed with more classical methods), or more thoroughly as an end-to-end trainable phenotyping strategy [57], and increasingly for fluorescent label prediction from label-free samples [58–60]. While the power and usefulness of Deep Learning approaches is clear, their downside is that they need a large amount of annotated data and computing power, and are more often of use in a supervised rather than unsupervised setting. They are also ‘black-boxes’, notoriously hard to interpret or understand with sometimes unexpected failure modes [61,62]. To mitigate those issues, on top of a good understanding of those methods special care has to be taken in evaluating the algorithms used and their results. This includes establishing standard internal controls within the algorithmic pipelines—essentially using good practices for machine learning—and, at the level of whole studies, checking for consistency of biological end results and experimentally testing predictions.
Reconstructing regulatory networks and systems-level cellular wiring
We have so far seen how high-throughput/high-content microscopy can be used to extract rich information at the single-cell level to identify and characterise the function of genes, by using gene KD/KO strategies for example. However genes do not operate independently but as regulatory networks (interaction networks, transcription networks, metabolic or signalling networks) and uncovering those networks is a key goal of functional genomics. One common simplifying assumption to try and estimate network-level information from single condition experiments is to link conditions leading to a similar phenotype [63]. However such phenotypic similarity networks, while shown to be enriched in physical or genetic interactions and to work with some success [64], are typically noisy and hard to interpret.
Revealing regulatory networks by double gene KD/KO
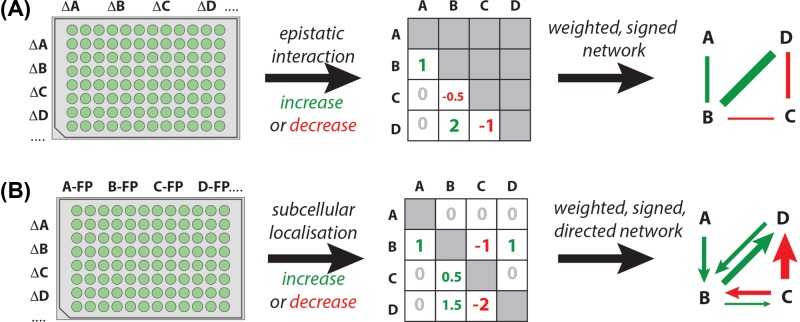
To go further and investigate the existence of a network link between two genes one has to change the experiment so that each condition probed is the combination of two interventions (Figure 2). For instance, Synthetic Genetic Array (SGA) approaches have achieved that by combinatorially crossing large collections of budding yeast KO cell lines—each knocked out for one gene in the genome—among themselves, and imaging and analysing images of the resulting colonies’ size and appearance in the plates to look for double KOs whose combinations have positive or negative (synthetic) epistatic effect on cell fitness [65,66]. The resulting ∼1 million gene–gene interactions (edges) scored in this way can then be used to generate a global genetic interaction network map or ‘wiring diagram’ of a typical S. cerevisiae cell. Similar approaches have been applied to mammalian cells using finer epifluorescence microscopy-based readouts [67–69]. Building on those methods, the combination of high-content microscopy phenotyping with double KO/KD interventions allows looking for more complex epistatic relationships between genes, either by considering each single phenotype independently under a multiplicative assumption [70,71] or by combining them to infer directed interaction networks [67]. Monitoring how the synthetic double KO/KD phenotypes change over time allows mapping how regulatory networks rewire, giving a much more complex picture of regulatory network dynamics [72].
Figure 2. Reconstructing gene/protein networks and systems-level interactions between cellular processes.
Using two interventions, either by double gene KD/KO (A) or by combining gene KD/KO with fluorescent protein (FP) tagging (B), allows the reconstruction of functional interactions between genes/proteins and construction of regulatory networks.
Revealing regulatory networks by combining gene KD/KO and protein localization
Another way to combinatorially probe and reveal edges in regulatory networks is by combining the use of gene KD/KO strategies with fluorescently tagged protein (re)localization, to build a so-called Localisation Interdependency Network (LIN) [73]. According to this approach, if the protein produced by gene B becomes de-localized in cells as a function of KD/KO of gene A, then the localization (and function) of B depends on A, thereby directly revealing a directed edge going from gene/protein A to gene/protein B. When done combinatorially across many genes by high-throughput epifluorescence microscopy imaging this procedure allows the generation of a signed, directed and weighted network connecting those genes without need for directionality inference – thereby overcoming an intrinsic limitation of double gene KD/KO approaches. Technical challenges with the LIN approach include the fact that fluorescently tagging proteins using genetically encoded fluorescent tags (like GFP) often compromises their function, hence careful quality control and validation is required, as well as challenges with quantifying intracellular protein localisation changes and phenotypes. This technique was used with success to investigate interactions between the core ∼40 cell polarity regulators of fission yeast (S. pombe), revealing the most complete picture of the cell polarity network for that cell type to date and discovering 554 pairwise interactions among the polarity regulators (98% of them novel) as well as ‘modular’ interactions between subgroups of regulators in the network. A similar much larger scale approach was used in S. cerevisiae combining SGA and high-throughput/high-content microscopy phenotyping to identify how the entire budding yeast proteome changes over time in response to drugs like rapamycin and hydroxyurea [74]. These approaches, as well as emerging perturbation-free approaches exploiting inherent cellular fluctuations in fluorescently labelled proteins [75,76], are enabling to map information flow in regulatory networks at unprecedented spatial and temporal resolution.
Inferring systems-level interactions and causal links between cellular processes
Another means of deriving biologically meaningful networks from multivariate single-cell data is using Bayesian network inference through a Bayesian graphical model of the probability distribution of the measurements. By computing conditional independencies Bayesian network inference allows the investigation of possible causality relationships between variables. This approach was proposed early on for use in flow cytometry [77], where single cell fluorescence measurements of phosphoproteins can be linked to activity and a signalling network can be inferred. In high-content screening, it was introduced to look at causality relationships between cellular/subcellular features, to allow building a high level system-wide description of the processes under study. Using Bayesian network inference the projects HepatoSys and Endotrack were able for example to identify and predict key differences in the design principles of the endocytosis of Transferrin versus that of Epidermal Growth Factor in human cell lines [33]. Similarly in the ‘multi-process’ phenomics project SYSGRO, which monitored how fission yeast cell shape, microtubule organization and cell cycle progression co-vary simultaneously across a genome-wide collection of mutant cell lines, Bayesian network inference was used to predict directional systems-level functional links between cell shape and microtubule control that could be successfully experimentally validated [39]. It is important to point out that although potentially very powerful such network inference methods are not infallible and the computational predictions derived from them (the topology and directionality of the network) must be experimentaly validated, a step unfortunately too often missing in such studies. In the future methods taking full advantage of high-dimensional, multi-process, multi-parametric single-cell information measured jointly in a cell/cell population [78,79] promise to increasingly provide a goldmine of discovery into how cells work as integrated systems.
Pushing the limits of single-cell high-content imaging: multi-scale, dynamical, functional
High-throughput/high-content microscopy is naturally evolving, as is microscopy as a whole, away from purely cell-level assays and questions towards the two nearest scales, tissues and organs above and single molecules below. In both cases technical obstacles abound but recent works are promising. At the larger scale, beyond the more classical methods extending the study of organoids at higher throughput [80], methods based on microfluidics for the generation of microencapsulated organoids on matrigel beads have been proposed [81]. At the smaller scale, an automated workflow for single-molecule localization microscopy on 96-well plates has been proposed [82], with the main issues being optimising acquisition, processing, analysis and storage to manage the data deluge that those methods generate. Equivalent techniques have also been used to study the nanoscale organization of bacterial cell division [83].
Another important milestone has been achieved by moving high-throughput/high-content microscopy into the realm of measuring 3D protein dynamics in live cells, at scale. Building on its predecessor MitoCheck, the follow on MitoSys project recently used Fluorescence Correlation Spectroscopy (FCS) to build the most resolved ‘mitotic cell atlas’ of the dynamics and potential interaction in 4D (3D + time) of proteins controlling cell division [84]. Focusing on 28 mitotic proteins tagged with eGFP and using supervised machine learning, the authors were furthermore able to assign protein quantities to six organelles (chromosomes, nuclear envelope, kinetochores, spindle, centrosomes and midbody) and determine the timing, stoichiometry and dissociation rates in those organelles for multiple mitotic proteins, demonstrating for example that AURKB kinase and its regulator CDCA8/borealin only partially colocalize at the midbody and that AURKB exhibits most likely an additional localization at the contractile cytokinetic ring consistent with a known cytokinetic function for that kinase. This provides a template for the elucidation of dynamical protein atlases in cells.
Finally, an area where the limits of high-content imaging are also being pushed is the integration of single-cell ‘omics with imaging, enabled by techniques such as mass spectrometry imaging (MSI) combined with microscopy [85–87] and in situ single-cell RNA profiling [88–92]. In the future these approaches are bound to give new and substantial functional depth to the microscopic study of single cell biology.
Sharing and integrating large microscopy phenomics datasets to boost discovery
One characteristic of high-content studies – particularly single-cell based – is the size and complexity of the data and metadata generated, from raw images to segmented single cells to the numerical features computed from those cells and the annotation eventually associated to them. Trying to organise that complexity has been the focus of much work in the community, from the identification of specific/suitable file formats [93,94], to the generation of databases [95,96] across modalities [97] and of interactive data visualisation tools [98]. Not surprisingly, given the size and very high cost of large high-throughput/high-content microscopy projects, initiatives to promote data sharing for re-use and cross analysis are becoming more common [99]. An example worth noting is the Image Data Resource (IDR, https://idr.openmicroscopy.org), the largest community-driven microscopy phenomics dataset resource in the world, which aims to bring together, organise and share with the community reference large-scale microscopy phenomics datasets (original image data, image-derived feature data, annotations and metadata) generated the world over using multiple imaging modalities in common ontologies and databases, so that the community is able to continue the discovery process by mining and integrating the datasets among themselves and with other existing pheno-genomic resources (STRING, GO, etc.) and to promote integration across organisms, biological processes and scales [100]. Although many of the >40 studies currently contained in IDR did not include single-cell data when originally published (particularly the older high-throughput microscopy datasets), an increasing number of the novel studies does. Likewise, many IDR image data sets can be reanalysed at the single-cell level even though the original publications describing them did not. In the future, by enabling the re-analysis and integration of an increasing number of datasets this unique resource promises to provide a precious trove of microscopy phenomics data for single-cell exploration and to help catalyze future biological discovery.
Exploiting single-cell data to predict cell structure-dynamics with artificial intelligence
An important avenue of research brought back to the forefront by recent work is generative models. Generative models aim to generate (i.e. simulate) new typical examples of the data under study to investigate statistical variability, allow a more accurate and intuitive way to describe a particular localisation or enable better and more realistic cellular models [101]. For instance in an earlier study the authors were able to learn conditional generative models of punctuate patterns knowing microtubule localisation, enabling the study of relative positions of organelles with single-cell resolution [102]. Strikingly, very recent work shows that Deep Neural Networks, on top of their use in discrimination and classification seen in earlier Deep Learning applications, can also be fantastic tools as generative models [103] (Figure 3). For example, so-called Generative Adversarial Networks (GANs) have been proposed by several groups as a means to do ‘impossible experiments’, i.e. to complete the data that is available with data that cannot be obtained but can be ‘guessed’ (i.e. predicted) [104,105]. Other uses of GANs include generating data to help in the training of data hungry Deep Neural Networks [106,107] or uses in correlative microscopy by learning an imaging modality from another (for example to generate super-resolution microscopy images from wide-field microscopy images [108] or predict labels from label-free images). We anticipate that in the future GANs and related methods will provide an invaluable tool to allow the enrichment of available datasets with computationally generated but biologically meaningful additions (including e.g. by data augmentation [109,110]) and will enable previously inaccessible studies. Importantly, going forward and as with Deep Learning in general, thorough, unbiased and study-specific evaluation and controls will be crucial to ensure trust and reproducibility. This might become particularly important in the future to prevent and avoid abuse of such powerful methods to generate synthetic ‘fake data’ [111], a recognized threat in the realms of social media, public discourse and politics that has not been contemplated until now in the scientific arena and might gravely corrupt scientific publications and databases if neglected.
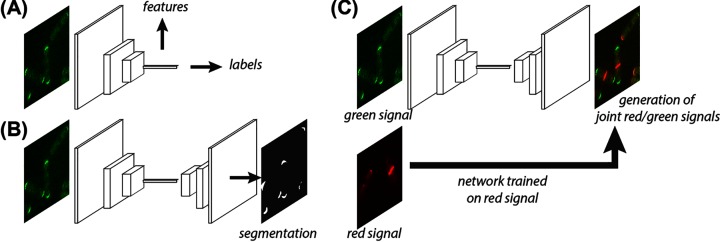
Figure 3. Predicting spatio-temporal cellular structure using Deep Learning.
The three main current uses of Deep Learning are: (A) phenotypically labelled images of whole image fields or single cells are used to learn a classification and/or a set of features for future uses; (B) pixel-level segmentation masks are used to train pixel-to-pixel networks for automated classification; (C) generative networks are given pairs of multi-channel images and trained to generate one channel given the others.
Towards predicting causal cell behaviour and decision making
Lastly, an area where single-cell microscopy phenomics can give unique insights into cell function is clarifying cellular decision-making and how the behaviour of a heterogeneous population of cells evolves through time, such that some cells take one fate and other cells another. While single-cell ‘omics information can enable the inference of some aspects of cell lineage relationships and history, the information is ‘time implicit’ (i.e. the true timing and duration of events monitored are not known) and space agnostic. How cells grow, divide, die or migrate [112,113] or change their structure/function/location/fate through time within a population or tissue, the cell-to-cell temporal noise in those single-cell decisions, the true timing and time scale of those decisions on a cell-by-cell basis, and how all of this impacts on how each cell and the heterogeneous cell population and its progeny as a whole evolve [114] are dynamical informations simply inaccessible to single-cell ‘omics approaches [24,115]. The time and location of cell death events in a tissue clearly exemplify this blind spot of ‘omics single-cell approaches: those informations though crucial for cell and tissue patterning and homeostasis [116,117] just cannot be derived from inferred cell lineages; only real time measurement of those events could inform about them. Time-resolved multi-process microscopy phenotyping and cellular lineaging hence plays a unique and powerful role in making available such dynamical informations [118,119], and as such (although currently not yet very common) will continue to play an indispensable role in allowing to accurately measure and – powered by predictive analytics approaches from e.g. ML/AI – to precisely predict causally the behaviour, function and fate [120] of single-cells, cell populations and tissues (Figure 4). For example continuous multi-generational, time-lapse multi-colour single-cell tracking and lineaging by epifluorescence microscopy of mouse Embryonic Stem Cells (mESCs) expressing the fluorescently labelled pluripotency transcription factor NanogVENUS combined with exact Bayesian inference revealed that in mESCs Nanog autoregulates by weak negative feedback [121], and Deep Neural Networks were able—using brightfield-derived information—to predict mouse hematopoietic stem and progenitor cell (HSPC) lineage choice during differentiation up to three generations before it could be detected by immuno-fluorescence using conventional markers [122]. These examples illustrate the enormous future potential of combining large amounts of biological information (‘biological Big Data’) derived from continuous time-resolved microscopy with predictive analytics not just for the discovery of basic biological mechanisms but also for improving technologies for tissue bioengineering as well as to enable highly precise and predictive diagnostics of clinical samples [123].
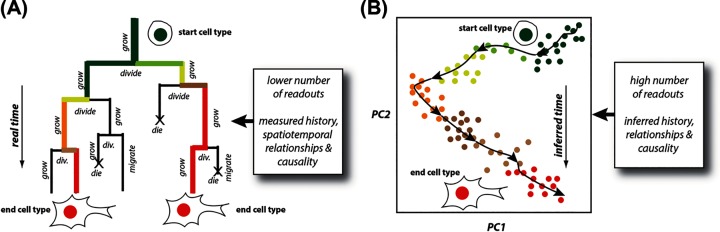
Figure 4. Measuring and predicting causal cell behaviour by time-resolved, continuous single-cell microscopy.
(A) Single-cell microscopy (represented by a multi-generation cell lineage) allows tracking in real time spatiotemporal cell decisions – e.g. how cells grow, divide, die, migrate or change their structure, function, location and fate within a population – and the cell-to-cell heterogeneity of those decisions in ways inaccessible and complementary to (B) single-cell ‘omics (represented by a pseudo-time ordered PCA plot of ‘omics data from a cell population; each cell is 1 dot). As explained in the text this is epitomised by the time and location of cell death events (marked with an ‘X’ in (A)), which though crucial to cell population and tissue evolution are invisible to single-cell ‘omics approaches but visible by single-cell microscopy.
Conclusions
Since its birth 15 years ago high-throughput/high-content microscopy has enabled the systematic identification of genes, pathways and cell biological mechanisms by exploiting single-cell derived rich structural as well as dynamical information about cells, their context and their evolution within cell populations. The recent marriage of single-cell microscopy phenotyping with predictive data analytics powered by ML/AI is dramatically shifting the focus from observing and exploiting cell behaviour to learning how to accurately predict it, with promising basic and biomedical applications. In many ways, single-cell microscopy phenotyping remains for the most part an orthogonal source of biological Big Data separate from ‘omics derived single-cell data, with different tradeoff between spatiotemporal resolution and the amount of readouts being followed. We foresee that in the future substantial effort will be invested in producing a similar marriage between ‘omics single-cell data strategies and high-throughput/high-content microscopy phenomics—currently sparse and mostly challenging [124–126]—and that this will bring about a fundamental step change in our capacity to predict and potentially control cell function. With microscopy the future is bright.
Summary
In the past 15 years, single-cell based microscopy has evolved its focus from observing cell function to aiming to predict it.
This has been possible thanks to breakthroughs in computer vision, large-scale image analysis and machine learning.
In this way high-throughput/high-content microscopy has enabled to discover and annotate genes, pathways and links between processes, and to begin clarifying cell decision making.
In the future, the marriage of single-cell ‘omics and single-cell microscopy will make possible an unprecedented understanding of cell and tissue behaviour and function.
Abbreviations
- GAN
Generative Adversarial Network
- IDR
Image Data Resource
- KD
knock-down
- KO
knock-out
- LIN
Localisation Interdependency Network
- mESC
mouse Embryonic Stem Cell
- ML/AI
machine learning/artificial intelligence
- SGA
Synthetic Genetic Array
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1. (2017) Single-cell biology. Nature https://www.nature.com/collections/gbljnzchgg [Google Scholar]
- 2.Haselgrübler T.et al. (2014) High-throughput, multiparameter analysis of single cells. Anal. Bioanal. Chem. 406, 3279–3296 10.1007/s00216-013-7485-x [DOI] [PubMed] [Google Scholar]
- 3.Perkel J.M. (2015) Single-cell biology: the power of one. Science 350, 696–698 10.1126/science.350.6261.696 [DOI] [Google Scholar]
- 4.Yuan G.C.et al. (2017) Challenges and emerging directions in single-cell analysis. Genome Biol. 18, 84. 10.1186/s13059-017-1218-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Human Cell Atlas, https://www.humancellatlas.org/ [Google Scholar]
- 6. (2018) Breakthrough of the Year 2018, http://vis.sciencemag.org/breakthrough2018/ [Google Scholar]
- 7.Alemany A., Florescu M., Baron C.S., Peterson-Maduro J. and van Oudenaarden A (2018) Whole-organism clone tracing using single-cell sequencing. Nature 556, 108, https://www.nature.com/articles/nature25969-supplementary-information 10.1038/nature25969 [DOI] [PubMed] [Google Scholar]
- 8.McKenna A.et al. (2016) Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science 353, aaf7907. 10.1126/science.aaf7907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagner D.E.et al. (2018) Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360, 981–987 10.1126/science.aar4362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sandmann T. and Boutros M. (2012) Screens, maps & networks: from genome sequences to personalized medicine. Curr. Opin. Genet. Development 22, 36–44 [DOI] [PubMed] [Google Scholar]
- 11.Roukos V. and Misteli T. (2014) Deep imaging: the next frontier in microscopy. Histochem. Cell Biol. 142, 125–131 10.1007/s00418-014-1239-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horwitz R. (2016) Integrated, multi-scale, spatial-temporal cell biology – a next step in the post genomic era. Methods 96, 3–5 10.1016/j.ymeth.2015.09.007 [DOI] [PubMed] [Google Scholar]
- 13.Skylaki S., Hilsenbeck O. and Schroeder T. (2016) Challenges in long-term imaging and quantification of single-cell dynamics. Nat. Biotechnol. 34, 1137. 10.1038/nbt.3713 [DOI] [PubMed] [Google Scholar]
- 14.Del Sol A., Thiesen H.J., Imitola J. and Carazo Salas R.E. (2017) Big-data-driven stem cell science and tissue engineering: vision and unique opportunities. Cell Stem Cell 20, 157–160 10.1016/j.stem.2017.01.006 [DOI] [PubMed] [Google Scholar]
- 15.Korn K. and Krausz E. (2007) Cell-based high-content screening of small-molecule libraries. Curr. Opin. Chem. Biol. 11, 503–510 10.1016/j.cbpa.2007.08.030 [DOI] [PubMed] [Google Scholar]
- 16.Lang P., Yeow K., Nichols A. and Scheer A. (2006) Cellular imaging in drug discovery. Nat. Rev. Drug Discov. 5, 343. 10.1038/nrd2008 [DOI] [PubMed] [Google Scholar]
- 17.Perlman Z.E.et al. (2004) Multidimensional drug profiling by automated microscopy. Science 306, 1194–1198 10.1126/science.1100709 [DOI] [PubMed] [Google Scholar]
- 18.Rausch O. (2006) High content cellular screening. Curr. Opin. Chem. Biol. 10, 316–320 10.1016/j.cbpa.2006.06.004 [DOI] [PubMed] [Google Scholar]
- 19.Mattiazzi Usaj M.et al. (2016) High-content screening for quantitative cell biology. Trends Cell Biol. 26, 598–611 10.1016/j.tcb.2016.03.008 [DOI] [PubMed] [Google Scholar]
- 20.Ohya Y.et al. (2005) High-dimensional and large-scale phenotyping of yeast mutants. PNAS 102, 19015–19020 10.1073/pnas.0509436102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erfle H., Simpson J.C., Bastiaens P.I.H. and Pepperkok R. (2004) siRNA cell arrays for high-content screening microscopy. BioTechniques 37, 454–462 10.2144/04373RT01 [DOI] [PubMed] [Google Scholar]
- 22.Pepperkok R. and Ellenberg J. (2006) High-throughput fluorescence microscopy for systems biology. Nat. Rev. Mol. Cell Biol. 7, 690. 10.1038/nrm1979 [DOI] [PubMed] [Google Scholar]
- 23.Wollman R. and Stuurman N (2007) High throughput microscopy: from raw images to discoveries. J. Cell Sci. 120, 3715–3722 10.1242/jcs.013623 [DOI] [PubMed] [Google Scholar]
- 24.Muzzey D. and Oudenaarden A.V. (2009) Quantitative time-lapse fluorescence microscopy in single cells. Annu. Rev. Cell Dev. Biol. 25, 301–327 10.1146/annurev.cellbio.042308.113408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conrad C. and Gerlich D.W. (2010) Automated microscopy for high-content RNAi screening. J. Cell Biol. 188, 453–461 10.1083/jcb.200910105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boutros M., Heigwer F. and Laufer C. (2015) Microscopy-based high-content screening. Cell 163, 1314–1325 10.1016/j.cell.2015.11.007 [DOI] [PubMed] [Google Scholar]
- 27.Ohya Y., Kimori Y., Okada H. and Ohnuki S. (2015) Single-cell phenomics in budding yeast. Mol. Biol. Cell 26, 3920–3925 10.1091/mbc.E15-07-0466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moreau D. and Gruenberg J. (2016) Automated microscopy and high content screens (phenotypic screens) in academia labs. CHIMIA Int. J. Chem. 70, 878–882 10.2533/chimia.2016.878 [DOI] [PubMed] [Google Scholar]
- 29.Rallis C. and Bähler J. (2016) Cell-based screens and phenomics with fission yeast. Crit. Rev. Biochem. Mol. Biol. 51, 86–95 10.3109/10409238.2015.1103205 [DOI] [PubMed] [Google Scholar]
- 30.Neumann B., Walter T., Hériché J-K., Bulkescher J., Erfle H., Conrad C.. et al. (2010) Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature 464, 721, 10.1038/nature08869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams S.P.et al. (2017) Systematic high-content genome-wide RNAi screens of endothelial cell migration and morphology. Scientific Data 4, 170009, https://www.nature.com/articles/sdata20179-supplementary-information 10.1038/sdata.2017.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pelkmans L.et al. (2005) Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature 436, 78, https://www.nature.com/articles/nature03571-supplementary-information 10.1038/nature03571 [DOI] [PubMed] [Google Scholar]
- 33.Collinet C.et al. (2010) Systems survey of endocytosis by multiparametric image analysis. Nature 464, 243, https://www.nature.com/articles/nature08779-supplementary-information 10.1038/nature08779 [DOI] [PubMed] [Google Scholar]
- 34.Chia J.et al. (2013) RNAi screening reveals a large signaling network controlling the Golgi apparatus in human cells. Mol. Syst. Biol. 9, 677. 10.1038/msb.2013.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galea G. and Simpson J.C. (2013) Methods in Cell Biology (Perez Franck and Stephens David J., eds), vol. 118, pp. 281–295, Academic Press; [DOI] [PubMed] [Google Scholar]
- 36.Hasson S.A.et al. (2013) High-content genome-wide RNAi screens identify regulators of parkin upstream of mitophagy. Nature 504, 291, https://www.nature.com/articles/nature12748-supplementary-information 10.1038/nature12748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tavassoli S., Chao J.T. and Loewen C. (2009) A high-throughput method to globally study the organelle morphology in S. cerevisiae. J. Vis. Exp 25, e1224. 10.3791/1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vizeacoumar F.J.et al. (2010) Integrating high-throughput genetic interaction mapping and high-content screening to explore yeast spindle morphogenesis. J. Cell Biol. 188, 69–81 10.1083/jcb.200909013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Graml V.et al. (2014) A genomic multiprocess survey of machineries that control and link cell shape, microtubule organization, and cell-cycle progression. Dev. Cell 31, 227–239 10.1016/j.devcel.2014.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mercer J.et al. (2012) RNAi screening reveals proteasome- and Cullin3-dependent stages in vaccinia virus infection. Cell Reports 2, 1036–1047 10.1016/j.celrep.2012.09.003 [DOI] [PubMed] [Google Scholar]
- 41.Wood V.et al. (2019) Hidden in plain sight: what remains to be discovered in the eukaryotic proteome? Open Biol. 9, 180241. 10.1098/rsob.180241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Serrano-Solano B., Díaz Ramos A., Hériché J.-K. and Ranea J.A.G. (2017) How can functional annotations be derived from profiles of phenotypic annotations? BMC Bioinformatics 18, 96. 10.1186/s12859-017-1503-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barr A.R. and Bakal C. (2012) A direct look at RNAi screens. Mol. Syst. Biol. 8, 580. 10.1038/msb.2012.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sommer C. and Gerlich D.W. (2013) Machine learning in cell biology – teaching computers to recognize phenotypes. J. Cell Sci. 126, 5529–5539 10.1242/jcs.123604 [DOI] [PubMed] [Google Scholar]
- 45.Chessel A. (2017) An overview of data science uses in bioimage informatics. Methods 115, 110–118 10.1016/j.ymeth.2016.12.014 [DOI] [PubMed] [Google Scholar]
- 46.Smith K.et al. (2018) Phenotypic image analysis software tools for exploring and understanding big image data from cell-based assays. Cell Syst. 6, 636–653 10.1016/j.cels.2018.06.001 [DOI] [PubMed] [Google Scholar]
- 47.Orlov N.et al. (2008) WND-CHARM: multi-purpose image classification using compound image transforms. Pattern Recognit. Lett. 29, 1684–1693 10.1016/j.patrec.2008.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Snijder B.et al. (2009) Population context determines cell-to-cell variability in endocytosis and virus infection. Nature 461, 520, https://www.nature.com/articles/nature08282-supplementary-information 10.1038/nature08282 [DOI] [PubMed] [Google Scholar]
- 49.Snijder B.et al. (2012) Single-cell analysis of population context advances RNAi screening at multiple levels. Mol. Syst. Biol. 8, 579. 10.1038/msb.2012.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Caicedo J.C.et al. (2017) Data-analysis strategies for image-based cell profiling. Nat. Methods 14, 849. 10.1038/nmeth.4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pauwels E.et al. (2012) A probabilistic model for cell population phenotyping using HCS data. PLoS One 7, e42715. 10.1371/journal.pone.0042715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sommer C., Hoefler R., Samwer M. and Gerlich D.W. (2017) A deep learning and novelty detection framework for rapid phenotyping in high-content screening. Mol. Biol. Cell 28, 3428–3436 10.1091/mbc.e17-05-0333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scheeder C., Heigwer F. and Boutros M. (2018) Machine learning and image-based profiling in drug discovery. Curr. Opin. Syst. Biol. 10, 43–52 10.1016/j.coisb.2018.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caicedo J.C., Singh S. and Carpenter A.E. (2016) Applications in image-based profiling of perturbations. Curr. Opin. Biotechnol. 39, 134–142 10.1016/j.copbio.2016.04.003 [DOI] [PubMed] [Google Scholar]
- 55.LeCun Y., Bengio Y. and Hinton G. (2015) Deep learning. Nature 521, 436. 10.1038/nature14539 [DOI] [PubMed] [Google Scholar]
- 56.Dürr O. and Sick B (2016) Single-cell phenotype classification using deep convolutional neural networks. J. Biomol. Screen 21, 998–1003 10.1177/1087057116631284 [DOI] [PubMed] [Google Scholar]
- 57.Kraus O.Z.et al. (2017) Automated analysis of high-content microscopy data with deep learning. Mol. Syst. Biol. 13, 924. 10.15252/msb.20177551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brent R. and Boucheron L (2018) Deep learning to predict microscope images. Nat. Methods 15, 868–870 10.1038/s41592-018-0194-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Christiansen E.M.et al. (2018) In silico labeling: predicting fluorescent labels in unlabeled images. Cell 173, 792.e719–803.e719 10.1016/j.cell.2018.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ounkomol C., Seshamani S., Maleckar M.M., Collman F. and Johnson G.R. (2018) Label-free prediction of three-dimensional fluorescence images from transmitted-light microscopy. Nat. Methods 15, 917–920 10.1038/s41592-018-0111-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nguyen A., Yosinski J. and Clune J. (2015) Deep neural networks are easily fooled: high confidence predictions for unrecognizable images. arXiv:1412.1897 [Google Scholar]
- 62.Su J., Vasconcellos Vargas D. and Kouichi S. (2017) One pixel attack for fooling deep neural networks. arXiv:1710.08864 [Google Scholar]
- 63.Markowetz F. (2010) How to understand the cell by breaking it: network analysis of gene perturbation screens. PLoS Comput. Biol. 6, e1000655. 10.1371/journal.pcbi.1000655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bakal C., Aach J., Church G. and Perrimon N. (2007) Quantitative morphological signatures define local signaling networks regulating cell morphology. Science 316, 1753–1756 10.1126/science.1140324 [DOI] [PubMed] [Google Scholar]
- 65.Costanzo M.et al. (2016) A global genetic interaction network maps a wiring diagram of cellular function. Science 353, aaf1420. 10.1126/science.aaf1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Usaj M.et al. (2017) TheCellMap.org: a web-accessible database for visualizing and mining the global yeast genetic interaction network. G3: Genes|Genomes|Genetics 7, 1539–1549 10.1534/g3.117.040220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Laufer C., Fischer B., Billmann M., Huber W. and Boutros M. (2013) Mapping genetic interactions in human cancer cells with RNAi and multiparametric phenotyping. Nat. Methods 10, 427, https://www.nature.com/articles/nmeth.2436-supplementary-information 10.1038/nmeth.2436 [DOI] [PubMed] [Google Scholar]
- 68.Roguev A.et al. (2013) Quantitative genetic-interaction mapping in mammalian cells. Nat. Methods 10, 432, https://www.nature.com/articles/nmeth.2398-supplementary-information 10.1038/nmeth.2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Billmann M. and Boutros M. (2017) Systematic epistatic mapping of cellular processes. Cell Division 12, 2. 10.1186/s13008-016-0028-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Horn T.et al. (2011) Mapping of signaling networks through synthetic genetic interaction analysis by RNAi. Nat. Methods 8, 341, https://www.nature.com/articles/nmeth.1581-supplementary-information 10.1038/nmeth.1581 [DOI] [PubMed] [Google Scholar]
- 71.Fischer B.et al. (2015) A map of directional genetic interactions in a metazoan cell. eLife 4, e05464. 10.7554/eLife.05464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Heigwer F.et al. (2018) Time-resolved mapping of genetic interactions to model rewiring of signaling pathways. eLife 7, e40174. 10.7554/eLife.40174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dodgson J., Chessel A., Vaggi F., Giordan M., Yamamoto M., Arai K.. et al. (2017) Reconstructing regulatory pathways by systematically mapping protein localization interdependency networks. bioRxiv 116749, 10.1101/116749 [DOI] [Google Scholar]
- 74.Chong Y.T.et al. (2015) Yeast proteome dynamics from single cell imaging and automated analysis. Cell 161, 1413–1424 10.1016/j.cell.2015.04.051 [DOI] [PubMed] [Google Scholar]
- 75.Machacek M.et al. (2009) Coordination of Rho GTPase activities during cell protrusion. Nature 461, 99, https://www.nature.com/articles/nature08242-supplementary-information 10.1038/nature08242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Isogai T. and Danuser G. (2018) Discovery of functional interactions among actin regulators by analysis of image fluctuations in an unperturbed motile cell system. Phil. Trans. R. Soc. B 373, 20170110. 10.1098/rstb.2017.0110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sachs K., Perez O., Pe’er D., Lauffenburger D.A. and Nolan G.P. (2005) Causal protein-signaling networks derived from multiparameter single-cell data. Science 308, 523–529 10.1126/science.1105809 [DOI] [PubMed] [Google Scholar]
- 78.Gut G., Herrmann M.D. and Pelkmans L (2018) Multiplexed protein maps link subcellular organization to cellular states. Science 361, eaar7042. 10.1126/science.aar7042 [DOI] [PubMed] [Google Scholar]
- 79.Valm A.M.et al. (2017) Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature 546, 162, https://www.nature.com/articles/nature22369-supplementary-information 10.1038/nature22369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li L., LaBarbera D.V. (2017) 3D High-content screening of organoids for drug discoveryin Comprehensive medicinal chemistry III,Chackalamannil S, Rotella D., Ward S.(eds.) Elsevier, Amsterdam, The Netherlands [Google Scholar]
- 81.Laperrousaz B.et al. (2018) Direct transfection of clonal organoids in Matrigel microbeads: a promising approach toward organoid-based genetic screens. Nucleic Acids Res. 46, e70–e70 10.1093/nar/gky030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Beghin A.et al. (2017) Localization-based super-resolution imaging meets high-content screening. Nat. Methods 14, 1184, https://www.nature.com/articles/nmeth.4486-supplementary-information 10.1038/nmeth.4486 [DOI] [PubMed] [Google Scholar]
- 83.Holden S.J.et al. (2014) High throughput 3D super-resolution microscopy reveals Caulobacter crescentus in vivo Z-ring organization. Proc. Natl. Acad. Sci. 111, 4566–4571 10.1073/pnas.1313368111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cai Y.et al. (2018) Experimental and computational framework for a dynamic protein atlas of human cell division. Nature 561, 411–415 10.1038/s41586-018-0518-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Buchberger A.R., DeLaney K., Johnson J. and Li L. (2018) Mass spectrometry imaging: a review of emerging advancements and future insights. Anal. Chem. 90, 240–265 10.1021/acs.analchem.7b04733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dong Y., Li B. and Aharoni A. (2016) More than pictures: when MS imaging meets histology. Trends Plant Sci. 21, 686–698 10.1016/j.tplants.2016.04.007 [DOI] [PubMed] [Google Scholar]
- 87.Miura D., Fujimura Y. and Wariishi H (2012) In situ metabolomic mass spectrometry imaging: recent advances and difficulties. J. Proteomics 75, 5052–5060 10.1016/j.jprot.2012.02.011 [DOI] [PubMed] [Google Scholar]
- 88.Eng C.-H.L.et al. (2019) Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH+. Nature 568, 235–239 10.1038/s41586-019-1049-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ke R.et al. (2013) In situ sequencing for RNA analysis in preserved tissue and cells. Nat. Methods 10, 857, https://www.nature.com/articles/nmeth.2563-supplementary-information 10.1038/nmeth.2563 [DOI] [PubMed] [Google Scholar]
- 90.Lee J.H.et al. (2014) Highly multiplexed subcellular RNA sequencing in situ. Science 343, 1360–1363 10.1126/science.1250212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lubeck E., Coskun A.F., Zhiyentayev T., Ahmad M. and Cai L. (2014) Single-cell in situ RNA profiling by sequential hybridization. Nat. Methods 11, 360, https://www.nature.com/articles/nmeth.2892-supplementary-information 10.1038/nmeth.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Strell C.et al. (2019) Placing RNA in context and space – methods for spatially resolved transcriptomics. FEBS J. 286, 1468–1481 10.1111/febs.14435 [DOI] [PubMed] [Google Scholar]
- 93.GitHub – CellH5/cellh5, https://github.com/CellH5/cellh5 [Google Scholar]
- 94.Millard B.L., Niepel M., Menden M.P., Muhlich J.L. and Sorger P.K. (2011) Adaptive informatics for multifactorial and high-content biological data. Nat. Methods 8, 487, https://www.nature.com/articles/nmeth.1600-supplementary-information 10.1038/nmeth.1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Allan C.et al. (2012) OMERO: flexible, model-driven data management for experimental biology. Nat. Methods 9, 245, https://www.nature.com/articles/nmeth.1896-supplementary-information 10.1038/nmeth.1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Marée R.et al. (2016) Collaborative analysis of multi-gigapixel imaging data using cytomine. Bioinformatics 32, 1395–1401 10.1093/bioinformatics/btw013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Carr A.et al. (2012) InterMine: a flexible data warehouse system for the integration and analysis of heterogeneous biological data. Bioinformatics 28, 3163–3165 10.1093/bioinformatics/bts577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Antal B., Chessel A. and Carazo Salas R.E. (2015) Mineotaur: a tool for high-content microscopy screen sharing and visual analytics. Genome Biol. 16, 283. 10.1186/s13059-015-0836-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Molloy J.C. (2011) The open knowledge foundation: open data means better science. PLoS Biol. 9, e1001195. 10.1371/journal.pbio.1001195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Williams E.et al. (2017) Image Data Resource: a bioimage data integration and publication platform. Nat. Methods 14, 775, https://www.nature.com/articles/nmeth.4326-supplementary-information 10.1038/nmeth.4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Murphy R.F. (2016) Building cell models and simulations from microscope images. Methods 96, 33–39 10.1016/j.ymeth.2015.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Johnson G.R., Li J., Shariff A., Rohde G.K. and Murphy R.F. (2015) Automated learning of subcellular variation among punctate protein patterns and a generative model of their relation to microtubules. PLoS Comput. Biol. 11, e1004614. 10.1371/journal.pcbi.1004614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Belthangady C. and Royer L. (2018) Applications, promises, and pitfalls of deep learning for fluorescence image reconstruction. Preprint 10.20944/preprints201812.0137.v1 [DOI] [PubMed] [Google Scholar]
- 104.Johnson G.R., Donovan-Maiye R.M. and Maleckar M.M. (2017) Generative modeling with conditional autoencoders: building an integrated cell. arXiv:1705.00092 [Google Scholar]
- 105.Osokin A., Chessel A., Carazo Salas R.E. and Vaggi F. (2017) GANs for biological image synthesis. arXiv:1708.04692 [Google Scholar]
- 106.Goldsborough P., Pawlowski N., Caicedo J.C., Singh S. and Carpenter A. (2017) CytoGAN: generative modeling of cell images. bioRxiv 227645, 10.1101/227645 [DOI] [Google Scholar]
- 107.Ouyang W., Aristov A., Lelek M., Hao X. and Zimmer C (2018) Deep learning massively accelerates super-resolution localization microscopy. Nat. Biotechnol. 36, 460, https://www.nature.com/articles/nbt.4106-supplementary-information 10.1038/nbt.4106 [DOI] [PubMed] [Google Scholar]
- 108.Wang H.et al. (2019) Deep learning enables cross-modality super-resolution in fluorescence microscopy. Nat. Methods 16, 103–110 10.1038/s41592-018-0239-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Calimeri F., Marzullo A., Stamile C. and Terracina G. (2017) Biomedical Data Augmentation Using Generative Adversarial Neural Networksin Artificial Neural Networks and Machine Learning – ICANN 2017,Lintas A., Rovetta S., Verschure P.F.M.J, Villa A.E.P.(eds.) Springer International Publishing, 10.1007/978-3-319-68612-7 [DOI] [Google Scholar]
- 110.Kiyoiti dos Santos Tanaka F.H. and Aranha C. (2019) Data augmentation using GANs. arXiv [Google Scholar]
- 111.Borel B. (2018) Clicks, lies and videotape. Sci. Am. 319, 38–43 10.1038/scientificamerican1018-38 [DOI] [PubMed] [Google Scholar]
- 112.Keller P.J., Schmidt A.D., Wittbrodt J. and Stelzer E.H.K. (2008) Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science 322, 1065–1069 10.1126/science.1162493 [DOI] [PubMed] [Google Scholar]
- 113.McDole K.et al. (2018) In Toto imaging and reconstruction of post-implantation mouse development at the single-cell level. Cell 175, 859.e833–876.e833 10.1016/j.cell.2018.09.031 [DOI] [PubMed] [Google Scholar]
- 114.Wait E., Winter M., Mankowski W., Cohen A.R. and Temple S. (2016) LEVER: software tools for segmentation, tracking and lineaging of proliferating cells. Bioinformatics 32, 3530–3531 10.1093/bioinformatics/btw406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Snijder B. and Pelkmans L. (2011) Origins of regulated cell-to-cell variability. Nat. Rev. Mol. Cell Biol. 12, 119, https://www.nature.com/articles/nrm3044-supplementary-information 10.1038/nrm3044 [DOI] [PubMed] [Google Scholar]
- 116.Suzanne M. and Steller H. (2013) Shaping organisms with apoptosis. Cell Death Differ. 20, 669. 10.1038/cdd.2013.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Galluzzi L.et al. (2018) Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 25, 486–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schroeder T. (2011) Long-term single-cell imaging of mammalian stem cells. Nat. Methods 8, S30. 10.1038/nmeth.1577 [DOI] [PubMed] [Google Scholar]
- 119.Keller P.J. (2013) Imaging morphogenesis: technological advances and biological insights. Science 340, 1234168. 10.1126/science.1234168 [DOI] [PubMed] [Google Scholar]
- 120.Etzrodt M., Endele M. and Schroeder T (2014) Quantitative single-cell approaches to stem cell research. Cell Stem Cell 15, 546–558 10.1016/j.stem.2014.10.015 [DOI] [PubMed] [Google Scholar]
- 121.Feigelman J.et al. (2016) Analysis of cell lineage trees by exact bayesian inference identifies negative autoregulation of Nanog in mouse embryonic stem cells. Cell Syst. 3, 480e413–490.e413 10.1016/j.cels.2016.11.001 [DOI] [PubMed] [Google Scholar]
- 122.Buggenthin F.et al. (2017) Prospective identification of hematopoietic lineage choice by deep learning. Nat. Methods 14, 403, https://www.nature.com/articles/nmeth.4182-supplementary-information 10.1038/nmeth.4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Orth A., Schaak D. and Schonbrun E. (2017) Microscopy, meet big data. Cell Syst. 4, 260–261 10.1016/j.cels.2017.03.009 [DOI] [PubMed] [Google Scholar]
- 124.Hormoz S.et al. (2016) Inferring cell-state transition dynamics from lineage trees and endpoint single-cell measurements. Cell Syst. 3, 419e418–433.e418 10.1016/j.cels.2016.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Spanjaard B. and Junker J.P. (2017) Methods for lineage tracing on the organism-wide level. Curr. Opin. Cell Biol. 49, 16–21 10.1016/j.ceb.2017.11.004 [DOI] [PubMed] [Google Scholar]
- 126.Yuan J., Sheng J. and Sims P.A. (2018) SCOPE-Seq: a scalable technology for linking live cell imaging and single-cell RNA sequencing. Genome Biol. 19, 227. 10.1186/s13059-018-1607-x [DOI] [PMC free article] [PubMed] [Google Scholar]