 A novel water soluble, phosphate-based enrichable, retro-Michael addition-driven MS-cleavable and multi-targeting cross-linker was developed.
A novel water soluble, phosphate-based enrichable, retro-Michael addition-driven MS-cleavable and multi-targeting cross-linker was developed.
Abstract
Chemical cross-linking mass spectrometry (XL-MS) is a powerful technology for obtaining protein structural information and studying protein–protein interactions. We report phospho-bisvinylsulfone (pBVS) as a novel water-soluble, MS-cleavable, phosphate-based enrichable and multi-targeting cross-linker. In this approach, the fragmentation of pBVS cross-linked peptides occurs in situ through retro-Michael addition. The phosphate group is successfully used as a small affinity tag to isolate cross-linked peptides from the highly abundant non-cross-linked peptides. In addition, the linker targets multiple types of amino acid residues, including cysteine, lysine and histidine. This method was applied to cross-link bovine serum albumin (BSA), myoglobin and Lbcpf1 demonstrating the ability to yield accurate and abundant information to facilitate protein structure elucidation.
Introduction
Chemical cross-linking-mass spectrometry (XL-MS) is emerging as a powerful tool to study protein–protein interactions at the proteome level and derive tertiary structural information about proteins and protein complexes.1,2 With the increasing contributions of this technology to structural biology,3–5 optimized XL-MS methods have been constantly developed and refined.6–8 However, some technical challenges remain. The first challenge is the interpretation of cross-linking spectra on a large scale due to the exponential growth of the search space and poor spectral quality.9–11 To address this limitation, MS-cleavable cross-linking reagents such as PIR,12 DSSO,13 DSBU, BuUrBu6,14–16 and DAU17 have been developed. A short, MS-cleavable linker, CDI, has also been reported recently.18 The methods above can simplify the interpretation workflow and convert the computational complexity from O (n2) to O (n), consequently generating far fewer false positive hits. The second challenge is that most protein–protein interactions are weak or transient, and non-cross-linked peptides are present in a large excess compared to cross-linked peptides. Due to the stochastic nature of MS experiments, most cross-linked peptides are not observable by MS.10,19 To remove these non-cross-linked peptides, an affinity tag was incorporated into the cross-linking reagent to selectively enrich cross-linked peptide pairs.20,21 For example, cross-linking reagents containing biotin as an affinity handle have been developed to boost the detection of cross-linked peptide pairs.21–24 Another issue is that most cross-linking reagents target only one certain type of residue. For example, N-hydroxysuccinimide (NHS) esters, which are one of the most widely used cross-linking reagents, mainly react with lysine.1,2,25 The specificity leads to fewer identifiable cross-linked peptide pairs, especially when there is no lysine available for cross-linking. Methods targeting acidic residues (Glu and Asp) have also been developed.26,27 A SuFEx chemical cross-linker that can target different types of amino acid residues was recently reported.28 It is necessary to develop an MS-cleavable, enrichable and multi-targeting cross-linker to study protein–protein interactions and elucidate protein structures.
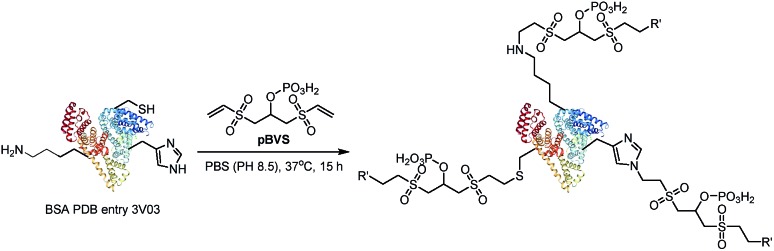
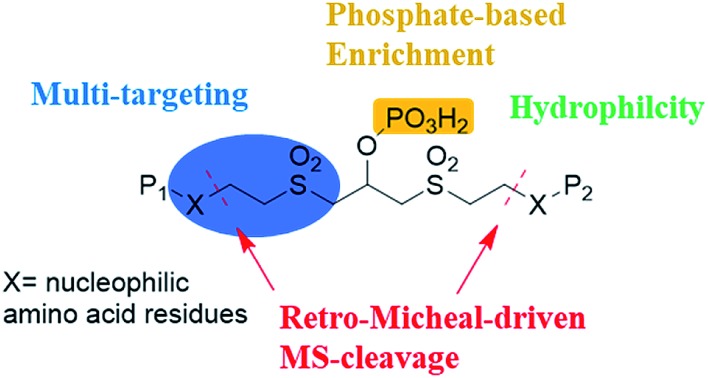
Vinyl sulfones can react with several nucleophilic residues including cysteine, lysine and histidine.29,30 Vinyl sulfone-based Michael addition is an attractive methodology for protein modification,30,31 and we have paid much attention to the development of vinyl sulfones for conjugation with peptides and proteins.32–35 Additionally, it is well-known that a high energy will induce retro-Michael addition, which triggers the preferential cleavage of the labile conjugation bond.36,37 In this study, we intend to employ bisvinylsulfones as a novel class of MS-cleavable cross-linkers. In addition, although biotin is widely used as an affinity tag to enrich cross-linked peptides, the bulky biotin tag will certainly hinder the cross-linking efficiency due to steric effects and therefore result in fewer identifiable cross-linked peptide pairs.38,39 Phosphate-based purification has been widely employed in proteomics analysis due to the affinity of phosphate to TiO2.40 In this study, a phosphate group was introduced into bisvinylsulfones in an attempt to selectively enrich cross-linked peptides using the TiO2 method, and we expect the smaller affinity tag to minimize the steric impact of the cross-linker. Therefore, phospho-bisvinylsulfone (pBVS) was developed as a novel class of MS-cleavable and enrichable cross-linkers (Fig. 1). To the best of our knowledge, this is the first use of a small phosphate group as an affinity tag. Cleaving the cross-linked peptides into two separate peptide chains through retro-Michael addition is also a novel distinct fragmentation mechanism. In addition, the linker demonstrates the ability to react with multiple types of amino acids under physiological conditions.
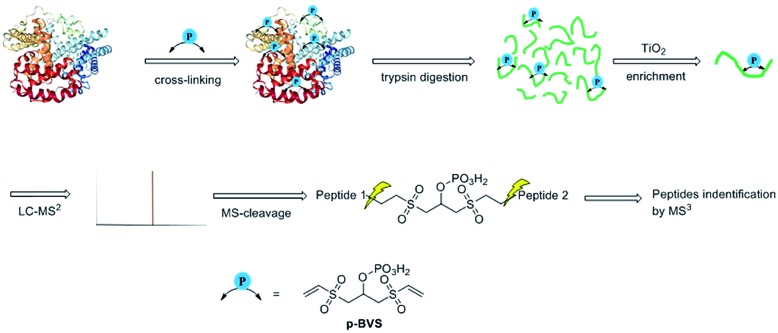
Fig. 1. XL-MS analysis workflow for identifying pBVS cross-linked peptides.
Results and discussion
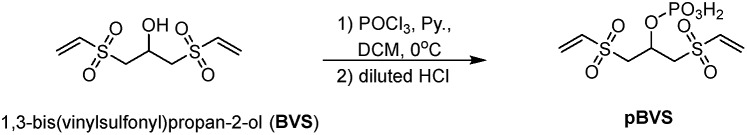
pBVS was obtained from commercially available 1,3-bis(vinylsulfonyl)propan-2-ol (BVS) by one step phosphorylation (Scheme 1). Bovine serum albumin (BSA) was utilized to evaluate the cross-linking efficiency. The preliminary results showed that pBVS was less reactive to amino residues than NHS esters. However, pBVS shows good stability in aqueous solution, whereas NHS is sensitive to hydrolysis. After cross-linking with pBVS, the protein was digested,41 and the resulting cross-linked peptides were enriched with TiO2 (ref. 38) and analysed using collision-induced dissociation (CID)-MS2-MS3. 29 cross-linked peptides were identified, when the reaction was performed for 4 hours at room temperature and pH 7.4. To improve the efficiency of cross-linking, the reaction conditions were optimized with respect to time, temperature and pH value (ESI, Table S1†). 49 cross-linkers were found when the reaction was performed for 15 hours at 37 °C and pH 8.5.
Scheme 1. Synthesis of pBVS.
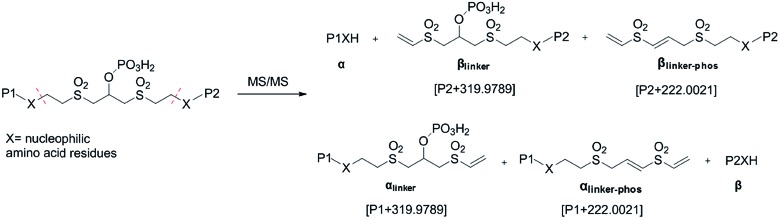
As initially expected, the fragmentation of pBVS cross-linked peptides occurred in situ through retro-Michael addition under low-energy CID conditions. The cross-linked peptide P1–P2 was first fragmented into two pairs: α/βlinker/βlinker-phos and β/αlinker/αlinker-phos (Fig. 2). A mass difference of 319.9789 was expected to be detected under low-energy MS/MS conditions. The αlinker and βlinker ions underwent further cleavage, which generated another mass tag (222.0021). These two mass tags (319.9789 and 222.0021) could be employed to trigger MS3 analysis for the identification of cross-linked peptide sequences.
Fig. 2. Fragmentation products of pBVS-crosslinked peptides.
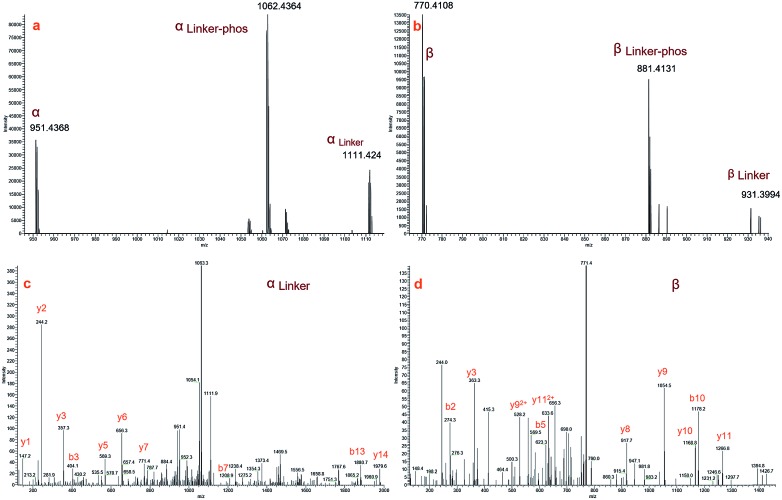
Fig. 3a and b display zoomed MS2 spectra of a cross-linked α–β peptide ion (m/z = 1254.2218, z = 3) in the m/z ranges 940–1120 and 750–940, respectively, in which the two fragment ion pairs αlinker/αlinker-phos/α and βlinker/βlinker-phos/β (z = 2) were detected. The mass difference between the ion αlinker and the ion α is 319.9744, equivalent to the exact mass of one pBVS linker with a mass error of approximately 0.005 Da. The mass difference between αlinker-phos and α is 221.9992, which is equivalent to the exact mass of one pBVS linker-phos with a mass error of approximately 0.003 Da. The αlinker and β ions were further fragmented by higher-energy collisional dissociation (HCD) (Fig. 3c and d). Subsequent database searching identified peptide α as NECFLSHKDDSPDLPK and β as LCVLHEKTPVSEK. In addition, the mass sum of the αlinker ion (1111.424, z = 2) and the β ion (770.4108, z = 2) is 3759.6384 Da, equivalent to the mass of the precursor ion 1254.2218 (z = 3, mass = 3759.642 Da) with an error less than 1 ppm. This finding suggests that the two fragment ions (ion 1111.424 and ion 770.4108) were formed by the cleavage of one labile bond of the precursor ion 1254.2218. We hence concluded that these two pairs of peptides were cross-linked by pBVS. The assignment of the ions in Fig. 3a and b and the detailed interpretation of α/αlinker-phos/αlinker and β/βlinker-phos/βlinker ions are shown in the ESI† (Tables S2–S8, Fig. S1–S6†).
Fig. 3. MS2 spectra of a representive cross-linked peptide pair (α–β, NECFLSHKDDSPDLPK–LCVLHEKTPVSEK): (a) MS2 spectrum of α peptide ions; (b) MS2 spectrum of β peptide ions; (c) MS3 spectrum of αlinker; and (d) MS3 spectrum of β.
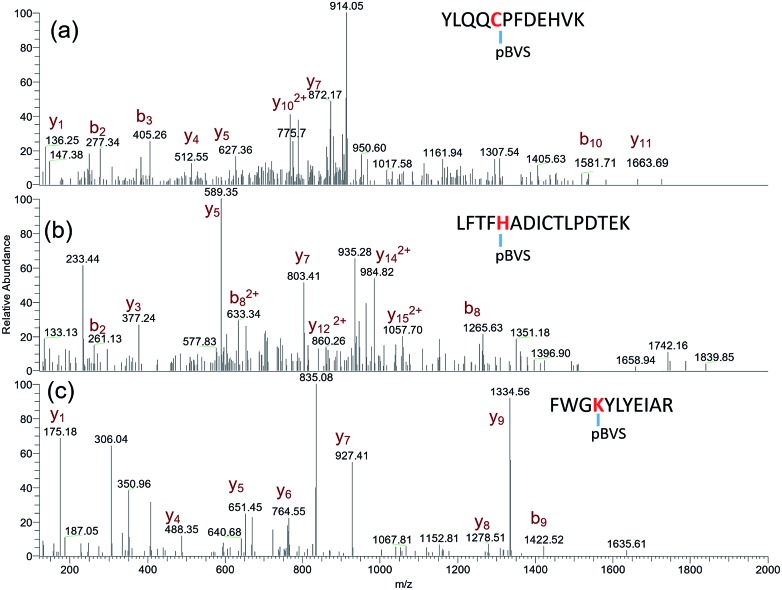
The reactivity of pBVS towards different types of residues was also explored. As expected, pBVS is reactive to nucleophilic amino acid residues, including cysteine, lysine and histidine, via Michael addition (Scheme 2). As shown in Fig. 4a, the peptide YLQQCPFDEHVK contains one linker, and the existence of b5/b6/y7/y8 ions revealed that cross-linking occurred at C5 (Table S9†). The absence of [b5-pBVS] ions and [y12-pBVS] ions suggests that the pBVS linker is not dissociated from the peptide under high-energy collision dissociation (HCD) conditions. For the fragmentation of αlinker/βlinker ions, cleavages occurred along the peptide backbone rather than at the position between the pBVS linker and the peptide, and the feature can be utilized to localize the exact linked sites. Fig. 4b and c display the spectra of ions that were identified as LFTFHADICTLPDTEK and FWGKYLYEIAR, and the observation of a series of MS3 ions revealed that cross-linking occurred at H5 and K4, respectively (Tables S10 and S11†).
Scheme 2. The reaction between pBVS with nucleophilic amino acid residues via Micheal addition.
Fig. 4. pBVS as a multi-targeting cross linker: MS3 spectra of YLQQCPFDEHVK + pBVS (a), LFTFHADICTLPDTEK + pBVS (b), and FWGKYLYEIAR + pBVS (c).
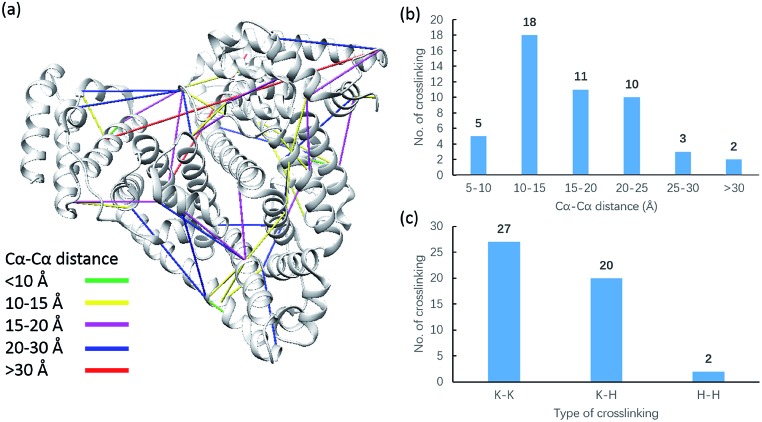
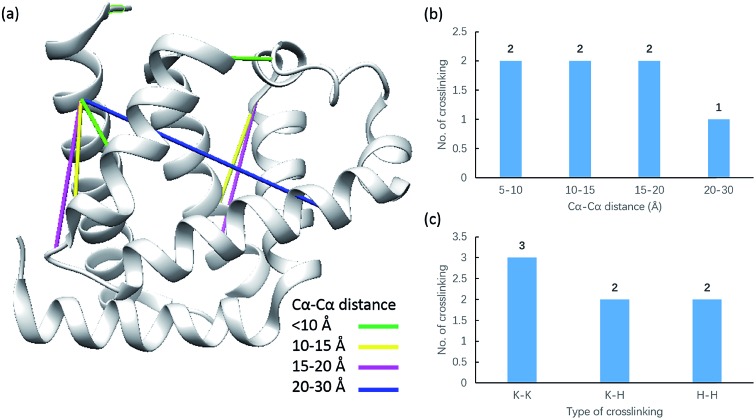
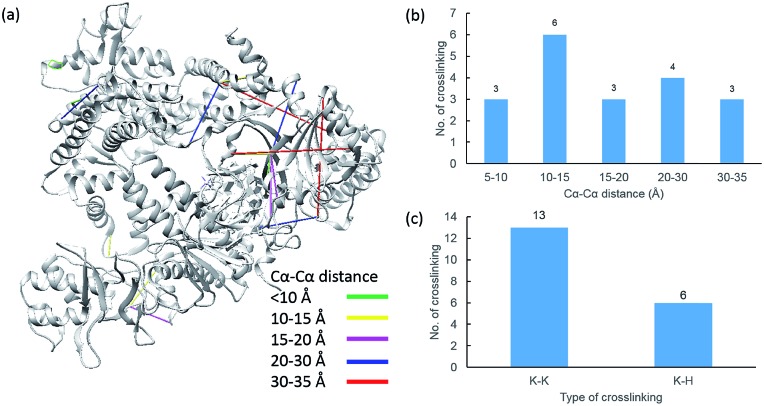
Due to the linker's ability to facilitate both MS spectrum interpretation and sample purification, the identification of cross-linked pairs has been greatly improved compared to the results of previously reported methods.18,28 A total of 49 unique cross-linked peptides, including 27 Lys–Lys, 20 Lys–His and 2 His–His, were detected by LC/MS/MS analysis from BSA (Fig. 5a–c, Table S12†). Unlike existing NHS cross-linkers that target only lysine, our method is able to target histidine in addition to lysine and thus demonstrates the ability to provide more information for protein structure elucidation. We also identified cross-linked peptides without enrichment to demonstrate the effects of the phosphate-based enrichment strategy. Twelve unique cross-linked peptides were detected (Table S13†), which confirmed the usefulness of the new enrichable strategy. To verify our data, the identified cross-linkages were mapped onto the crystal structures of BSA (Protein Data Bank entry: ; 3V03) to determine the Cα–Cα distance of the linked residues. Considering the spacer length of pBVS, which is ∼15 Å, and the distances contributed by amino acid side chains, as well as the structural flexibility of the protein, the distance cut off for two connectable residues is set to 30 Å. Forty seven out of the 49 identified linkages (96%) were calculated to have Cα–Cα distances of less than 30 Å (Fig. 5). The two outliers could be explained by protein flexibility or dimerization. This result is consistent with the structural features of BSA, which indicates that the BSA structure was not disturbed by cross-linking. We also employed pBVS to investigate the structure of myoglobin. Seven cross-linked peptides, including 3 Lys–Lys, 2 Lys–His and 2 His–His, were detected by LC/MS/MS analysis (Fig. 6a–c, Table S14†).
Fig. 5. Assessment of cross linkages with the BSA 3D structure: (a) cross-links mapped onto the crystal structures of BSA (PDB entry 3V03); (b) distribution of the Cα–Cα distance of the identified linkages; and (c) types of cross-linking sites identified from the pBVS cross-linked BSA sample, with numbers indicated on top.
Fig. 6. Assessment of cross linkages with the myoglobin 3D structure: (a) cross-links mapped onto the crystal structures of myoglobin (PDB entry 1dwr); (b) distribution of the Cα–Cα distance of the identified linkages; and (c) types of cross-linking sites identified from the pBVS cross-linked myoglobin sample, with numbers indicated on top.
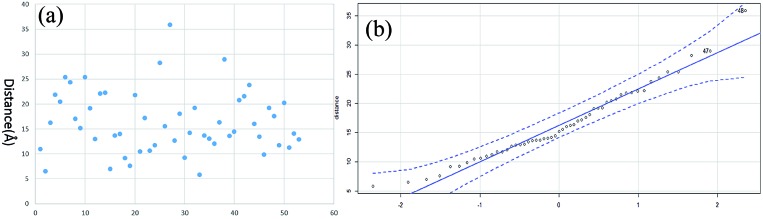
The distance data for all the linkages obtained from BSA and myoglobin except the outlier (60.5 Å) are plotted in Fig. 7a. The median value of the dataset is 15.2 Å, which is equal to the pBVS spacer length (∼15 Å). The QQ plot in Fig. 7b shows that the dataset follows a normal distribution. This suggests that the distance error is mainly due to the flexibility of the amino acid side chains. One shortcoming of XL-MS for protein structure analysis is that the distance constraint is too loose to provide accurate structural information. The results of this study show that the method generates accurate distance information.
Fig. 7. The distribution of all the linkages: (a) Cα–Cα distance and (b) QQ plot.
We further apply the reagent for structural elucidation of the Cpf1 protein from Lachnospiraceae bacteria (Lbcpf1), which is a single RNA-guided endonuclease, classified as a type V CRISPR system.42,43 19 unique linkages, including 13 Lys–Lys and 6 Lys–His, were detected (Fig. 8, Table S15†). When the cross-links were mapped onto the crystal structure (PDB entry: ; 5ID6), we found that 13 out of 19 linkages appeared in helical I (H1) and helical II (H2) domains, and 6 linkages were produced between H1 and its neighbouring OBD domain.43
Fig. 8. Assessment of cross linkages with the Cpf1 3D structure: (a) cross-links mapped onto the crystal structures of Cpf1 (PDB entry 5ID6); (b) distribution of the Cα–Cα distance of the identified linkages; and (c) types of cross-linking sites identified from the pBVS cross-linked Cpf1 sample, with numbers indicated on top.
To evaluate whether pVBS could be used in complex samples, we also applied the reagent to HeLa whole cell lysate. Unfortunately, the preliminary results show that most of the peptides detected were phosphopeptides rather than cross-linked peptides. This suggests that the method is more suitable to study samples in which protein phosphorylation is absent or less abundant.
Conclusions
In this study, a novel MS-cleavable and enrichable cross-linking reagent, phospho-bisvinylsulfone (pBVS), was developed. This stable and water-soluble linker possesses an innovative modular scaffold and targets multiple types of amino acids (i.e., lysine, histidine and cysteine) via Michael addition without any by-product. Under low-CID MS/MS conditions, cross-linked peptides are cleaved into two individual peptide chains through retro-Michael addition, which is a novel fragmentation pattern in XL-MS. A phosphate group as a small affinity tag is successfully used to isolate the cross-linked peptides from highly abundant non-cross-linked peptides. This novel strategy was applied to the cross-linking of the protein BSA, myoglobin and Lbcpf1, and the results demonstrated that pBVS-based linkages provide accurate and abundant information to facilitate protein structure elucidation. We also found one limitation of the method is that phosphopeptides will also be enriched and pBVS will not be suitable for complex samples with abundant phosphopeptides. Future studies will explore other types of vinyl sulfone-based cross-linkers suitable for complex samples.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
We thank Dr Heqiao Zhang for his help in data analysis of BSA cross-links. We also thank Dr Sheng Li, Shangshang Wang, Dr Lixia Zhao and Dr Yan Nie (ShanghaiTech University, SIAIS Protein & Gene Platform) for providing Lbcpf1.
Footnotes
†Electronic supplementary information (ESI) available: Experimental procedures and full spectroscopic data are available. See DOI: 10.1039/c9sc00893d
References
- Yu C., Huang L. Anal. Chem. 2018;90:144–165. doi: 10.1021/acs.analchem.7b04431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinz A. Angew. Chem., Int. Ed. 2018;57:6390–6396. doi: 10.1002/anie.201709559. [DOI] [PubMed] [Google Scholar]
- Limpikirati P., Liu T., Vachet R. W. Methods. 2018;144:79–93. doi: 10.1016/j.ymeth.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobucci C., Gotze M., Ihling C. H., Piotrowski C., Arlt C., Schafer M., Hage C., Schmidt R., Sinz A. Nat. Protoc. 2018;13:2864–2889. doi: 10.1038/s41596-018-0068-8. [DOI] [PubMed] [Google Scholar]
- Sinz A. Anal. Bioanal. Chem. 2017;409:33–44. doi: 10.1007/s00216-016-9941-x. [DOI] [PubMed] [Google Scholar]
- Pan D., Brockmeyer A., Mueller F., Musacchio A., Bange T. Anal. Chem. 2018;90:10990–10999. doi: 10.1021/acs.analchem.8b02593. [DOI] [PubMed] [Google Scholar]
- Horne J. E., Walko M., Calabrese A. N., Levenstein M. A., Brockwell D. J., Kapur N., Wilson A. J., Radford S. E. Angew. Chem., Int. Ed. 2018;57:16688–16692. doi: 10.1002/anie.201809149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez C. B., Block S. A., Yu C., Soohoo S. M., Huszagh A. S., Rychnovsky S. D., Huang L. Anal. Chem. 2018;90:7600–7607. doi: 10.1021/acs.analchem.8b01287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Li N., Yu W. J. Proteome Res. 2017;16:3942–3952. doi: 10.1021/acs.jproteome.7b00338. [DOI] [PubMed] [Google Scholar]
- Liu F., Rijkers D. T., Post H., Heck A. J. Nat. Methods. 2015;12:1179–1184. doi: 10.1038/nmeth.3603. [DOI] [PubMed] [Google Scholar]
- Lu L., Millikin R. J., Solntsev S. K., Rolfs Z., Scalf M., Shortreed M. R., Smith L. M. J. Proteome Res. 2018;17:2370–2376. doi: 10.1021/acs.jproteome.8b00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Munske G. R., Siems W. F., Bruce J. E. Anal. Chem. 2005;77:311–318. doi: 10.1021/ac0488762. [DOI] [PubMed] [Google Scholar]
- Kao A., Chiu C. L., Vellucci D., Yang Y., Patel V. R., Guan S., Randall A., Baldi P., Rychnovsky S. D., Huang L. Mol. Cell. Proteomics. 2011;10:M110.002212. doi: 10.1074/mcp.M110.002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. L., Gotze M., Bartolec T. K., Hart-Smith G., Wilkins M. R. Anal. Chem. 2018;90:9101–9108. doi: 10.1021/acs.analchem.8b01525. [DOI] [PubMed] [Google Scholar]
- Arlt C., Gotze M., Ihling C. H., Hage C., Schafer M., Sinz A. Anal. Chem. 2016;88:7930–7937. doi: 10.1021/acs.analchem.5b04853. [DOI] [PubMed] [Google Scholar]
- Muller M. Q., Dreiocker F., Ihling C. H., Schafer M., Sinz A. Anal. Chem. 2010;82:6958–6968. doi: 10.1021/ac101241t. [DOI] [PubMed] [Google Scholar]
- Iacobucci C., Piotrowski C., Rehkamp A., Ihling C. H., Sinz A. J. Am. Soc. Mass Spectrom. 2019;30:139–148. doi: 10.1007/s13361-018-1952-8. [DOI] [PubMed] [Google Scholar]
- Hage C., Iacobucci C., Rehkamp A., Arlt C., Sinz A. Angew. Chem., Int. Ed. 2017;56:14551–14555. doi: 10.1002/anie.201708273. [DOI] [PubMed] [Google Scholar]
- Hoopmann M. R., Zelter A., Johnson R. S., Riffle M., MacCoss M. J., Davis T. N., Moritz R. L. J. Proteome Res. 2015;14:2190–2198. doi: 10.1021/pr501321h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X., Navare A. T., Chavez J. D., Eng J. K., Schweppe D. K., Bruce J. E. J. Proteome Res. 2017;16:720–727. doi: 10.1021/acs.jproteome.6b00752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Bruce J. E. Mol. BioSyst. 2010;6:939–947. doi: 10.1039/b920876c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holding A. N. Methods. 2015;89:54–63. doi: 10.1016/j.ymeth.2015.06.010. [DOI] [PubMed] [Google Scholar]
- Kaake R. M., Wang X., Burke A., Yu C., Kandur W., Yang Y., Novtisky E. J., Second T., Duan J., Kao A., Guan S., Vellucci D., Rychnovsky S. D., Huang L. Mol. Cell. Proteomics. 2014;13:3533–3543. doi: 10.1074/mcp.M114.042630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alley S. C., Ishmael F. T., Jones A. D., Benkovic S. J. J. Am. Chem. Soc. 2000;122:6126–6127. [Google Scholar]
- Rappsilber J. Eur. J. Org. Chem. 2011;173:530–540. doi: 10.1016/j.jsb.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez C. B., Yu C., Novitsky E. J., Huszagh A. S., Rychnovsky S. D., Huang L. Anal. Chem. 2016;88:8315–8322. doi: 10.1021/acs.analchem.6b02240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner A., Joachimiak L. A., Unverdorben P., Walzthoeni T., Frydman J., Forster F., Aebersold R. Proc. Natl. Acad. Sci. U. S. A. 2014;111:9455–9460. doi: 10.1073/pnas.1320298111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Wu H., Schnier P. D., Liu Y., Liu J., Wang N., DeGrado W. F., Wang L. Proc. Natl. Acad. Sci. U. S. A. 2018;115:11162–11167. doi: 10.1073/pnas.1813574115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koniev O., Wagner A. Chem. Soc. Rev. 2015;44:5495–5551. doi: 10.1039/c5cs00048c. [DOI] [PubMed] [Google Scholar]
- Morales-Sanfrutos J., Lopez-Jaramillo J., Ortega-Munoz M., Megia-Fernandez A., Perez-Balderas F., Hernandez-Mateo F., Santoyo-Gonzalez F. Org. Biomol. Chem. 2010;8:667–675. doi: 10.1039/b920576d. [DOI] [PubMed] [Google Scholar]
- Maruani A., Richards D. A., Chudasama V. Org. Biomol. Chem. 2016;14:6165–6178. doi: 10.1039/c6ob01010e. [DOI] [PubMed] [Google Scholar]
- Chen H., Huang R., Li Z., Zhu W., Chen J., Zhan Y., Jiang B. Org. Biomol. Chem. 2017;15:7339–7345. doi: 10.1039/c7ob01866e. [DOI] [PubMed] [Google Scholar]
- Huang R., Li Z., Ren P., Chen W., Kuang Y., Chen J., Zhan Y., Chen H., Jiang B. J. Struct. Biol. 2018;2018:829–836. [Google Scholar]
- Huang R., Li Z., Sheng Y., Yu J., Wu Y., Zhan Y., Chen H., Jiang B. Org. Lett. 2018;20:6526–6529. doi: 10.1021/acs.orglett.8b02849. [DOI] [PubMed] [Google Scholar]
- Li Z., Huang R., Xu H., Chen J., Zhan Y., Zhou X., Chen H., Jiang B. Org. Lett. 2017;19:4972–4975. doi: 10.1021/acs.orglett.7b02464. [DOI] [PubMed] [Google Scholar]
- Selvi T., Velmathi S. J. Org. Chem. 2018;83:4087–4091. doi: 10.1021/acs.joc.7b03151. [DOI] [PubMed] [Google Scholar]
- Kobetic R., Kazazic S., Kovacevic B., Glasovac Z., Krstulovic L., Bajic M., Zinic B. J. Am. Soc. Mass Spectrom. 2015;26:833–842. doi: 10.1007/s13361-014-1068-8. [DOI] [PubMed] [Google Scholar]
- Petrotchenko E. V., Borchers C. H. Mass Spectrom. Rev. 2010;29:862–876. doi: 10.1002/mas.20293. [DOI] [PubMed] [Google Scholar]
- Chowdhury S. M., Du X. X., Tolic N., Wu S., Moore R. J., Mayer M. U., Smith R. D., Adkins J. N. Anal. Chem. 2009;81:5524–5532. doi: 10.1021/ac900853k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L. R., Zhu Z., Chan K. C., Issaq H. J., Dimitrov D. S., Veenstra T. D. J. Proteome Res. 2007;6:4150–4162. doi: 10.1021/pr070152u. [DOI] [PubMed] [Google Scholar]
- Wisniewski J. R., Zougman A., Nagaraj N., Mann M. Nat. Methods. 2009;6:359–U360. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- Zetsche B., Gootenberg J. S., Abudayyeh O. O., Slaymaker I. M., Makarova K. S., Essletzbichler P., Volz S. E., Joung J., van der Oost J., Regev A., Koonin E. V., Zhang F. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong D., Ren K., Qiu X., Zheng J., Guo M., Guan X., Liu H., Li N., Zhang B., Yang D., Ma C., Wang S., Wu D., Ma Y., Fan S., Wang J., Gao N., Huang Z. Nature. 2016;532:522–526. doi: 10.1038/nature17944. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.