Abstract
To analyze the effect of gravity on the structure of germinal tissues, we examined tissues of the testes and duct deferens of mice that were exposed to space flight conditions for 21–24 days (experiment Rodent Research-4, SpaceX-10 mission, February 2017, USA). We evaluated the levels of cytoskeletal proteins, sperm-specific proteins, and epigenetic events; in particular, we evaluated levels of 5-hydroxymethylcytosine and of enzymes that regulate DNA methylation/demethylation. We did not detect changes in the levels of cytoskeletal proteins, sperm-specific proteins, DNA-methylases, DNA demethylases, DNA acetylases, or histone deacetylases. However, there were changes at the gene expression level. In particular, there was an increase in the demethylase Tet2 and a decrease in the histone deacetylase Hdac1. These gene expression changes may be of key importance during the early period of readaptation since they could lead to an increase in the expression of target genes.
Subject terms: Cytoskeleton, Epigenetic memory
Introduction
The role of gravity in the early development of mammals, particularly during prenatal development, remains unclear despite a number of previous studies1–8.
It is known that the exposure of female rats to zero gravity in the second half of pregnancy does not cause significant changes in embryos3,6, which may be due to the damping of the external mechanical field by the amniotic fluid. On the other hand, the preimplantation development and early stages of gastrulation may be more sensitive to changes in external mechanical stress. For example, pregnancy was not achieved in mice during space flight conditions, which researchers attributed to the lack of implantation and, as a result, the abortion of the preimplantation embryos7.
In addition, there is every reason to believe that germ cells can change their structure in microgravity conditions. For male germ cells, it is known that the speed of the movement of the mouse spermatozoa decreases after a 7-day antiorthostatic hanging, which simulates the effects of weightlessness9; although, for the sperm of sea urchin in space flight (STS-81, STS-84), FP130 protein phosphorylation, which is associated with the activation of motility, occurred 3–4 times faster than in conditions of 1 g10. Additionally, the expression of genes encoding pro-apoptotic proteins increased11, the differentiation potential of progenitor cells changed12, and the efficiency of spermatogenesis decreased13,14. However, the causes of such changes in sperm are not at all clear.
Our previous data suggested that after a 30-day antiorthostatic suspension, changes in the patterns of cytoskeletal proteins in mouse spermatozoa and testes were not observed, although the relative mRNA levels of the corresponding genes changed15. It is known that the external mechanical field plays an important role in establishing the patterns of expression of various genes, primarily those encoding cytoskeletal proteins and the associated signaling cascades, in different cell types, including stem cells16–22. However, how an external signal that the mechanical field has changed is transduced and leads to a change in gene expression remains completely unclear.
In higher mammals, there are a large number of methods for regulating gene expression, such as various posttranslational modifications of histones, chromatin remodeling, RNA interference, and DNA methylation, but most of these processes have hardly been studied under microgravity conditions. Few and partially contradictory data are associated with the determination of the methylation levels in different cell types in conditions of microgravity: hypermethylation took place in rice germs under space flight conditions23, hypomethylation was observed in human T lymphocytes24, and in lymphoblastoid cells, there was a change in the methylation level and in the hydroxymethylation level25. The data from our previous study showed that in space flight conditions, the total level of DNA methylation in the heart and lung tissues of mice increased by 21% and 32%, respectively, which correlated with changes in gene expression levels26.
Therefore, the purpose of our work was to evaluate the levels of the cytoskeletal proteins and the mRNA levels of the genes that encode them in the testes and duct deferens of mice whose tissues were fixed in space flight conditions. In addition, we sought to evaluate epigenetic events, in particular, the levels of 5-hydroxymethylcytosine and of the enzymes that regulate DNA methylation/demethylation.
Results
The levels of the cytoskeletal proteins and the corresponding mRNA
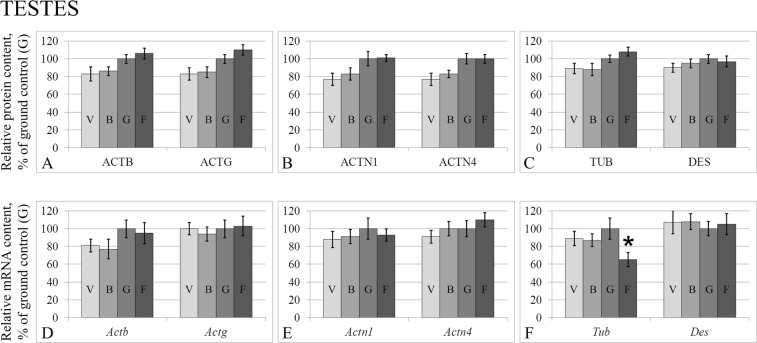
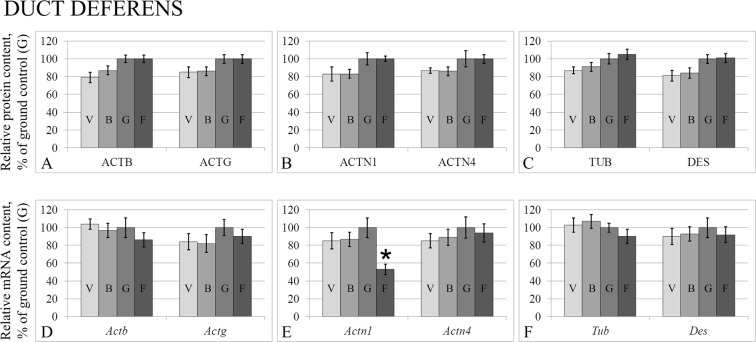
In the testes (Fig. 1) and the duct deferens (Fig. 2), no changes in the relative levels of beta-actin, gamma-actin, alpha-actinin-1, alpha-actinin-4, beta-tubulin and desmin were observed. There were also no changes in the relative levels of the corresponding mRNA, except for tubulin in the testes (Fig. 1F), where the mRNA content of the flight group was lower than that of the control group by 35% (p < 0.05) and alpha-actinin-1 in the duct deferens (Fig. 2E) in group F, where the Actn1 mRNA level was 47% lower than that of the control group (p < 0.05).
Figure 1.
Relative protein contents of cytoskeletal proteins in the testis tissue (A–C) and according mRNA levels (D–F). ACTB – Beta-actin. ACTG – Gamma-actin. ACTN1 – Alpha-actinin-1. ACTN4 – Alpha-actinin-4. TUB – Beta-tubulin. DES – desmin. V – vivarium control, B – basal control, G – ground control, F – flight group. *p < 0.05 in comparison with the G group.
Figure 2.
Relative protein contents of cytoskeletal proteins in the duct deferens tissue (A–C) and according mRNA levels (D–F). ACTB – Beta-actin. ACTG – Gamma-actin. ACTN1 – Alpha-actinin-1. ACTN4 – Alpha-actinin-4. TUB – Beta-tubulin. DES – desmin. V – vivarium control, B – basal control, G – ground control, F – flight group. *p < 0.05 in comparison with the G group.
Levels of proteins that are specific to different stages of spermatogenesis and their corresponding mRNA levels
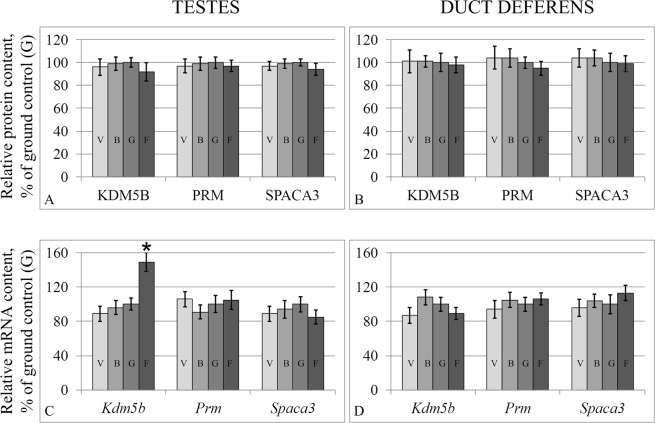
Similar to cytoskeletal proteins, the levels of proteins that are specific to early spermatogonia (KDM5B), spermatids (PRM1) and mature spermatozoa (SPACA3) did not differ in any of the study groups (Fig. 3), both in the testes and in the duct deferens. The mRNA levels were also the similar, with the exception of the Kdm5B mRNA level, which in the flight group in the testes (Fig. 3C) was higher than that of the control by 49% (p < 0.05).
Figure 3.
Relative protein contents of sperm-specific proteins in the testis (A) and duct deferens tissues (B) and according mRNA levels (C – testes, D – duct deferens). KDM5B (Jarid1B) – specific for spermatogonia; PRM (protamine) – specific for spermatids; Spaca 3 – specific for mature sperm. V – vivarium control, B – basal control, G – ground control, F – flight group. *p < 0.05 in comparison with the G group.
The relative levels of 5-hydroxymethylcytosine
The levels of 5-hydroxymethylcytosine did not differ in the study groups, both in the testes and in the duct deferens (Fig. 4).
Figure 4.
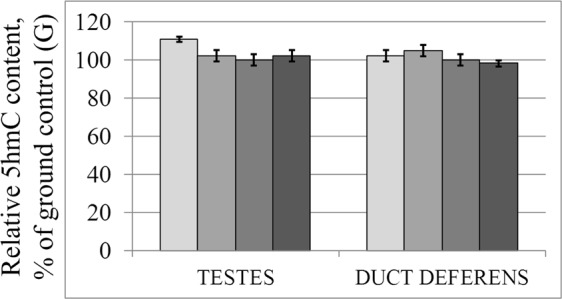
Relative 5-hydroxymethycytosine level in the testis and duct deferens tissues. V – vivarium control, B – basal control, G – ground control, F – flight group.
Methylases/demethylases and acetylases/histone deacetylases and their mRNA levels
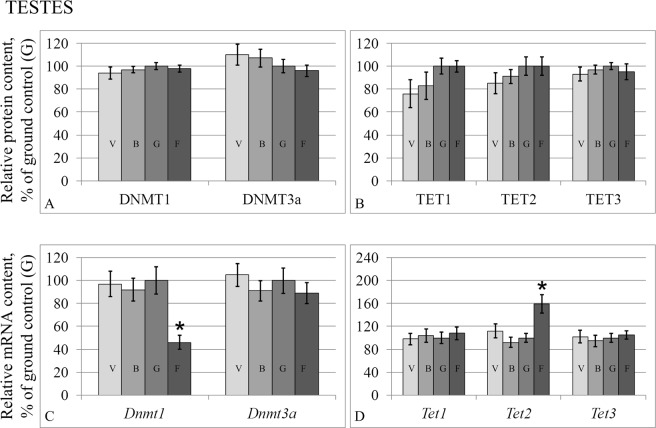
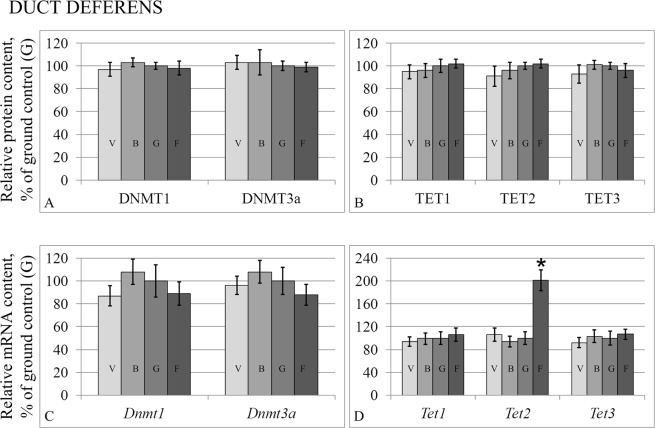
The levels of the S-phase methylase DNMT1 did not change in the testes (Fig. 5A) or in the duct deferens (Fig. 6A); however, the expression level of the corresponding gene in the testes (Fig. 5C) of group F was lower than that of group G by 54% (p < 0.05). At the same time, there was no change in the protein or mRNA levels of the de novo methylase DNMT3a either in the testes or in the duct deferens.
Figure 5.
Relative protein contents of methylation/demethylation fermentas in the testis tissue (A,B) and according mRNA levels (C,D). DNMT1 – S-phase methylase DNMT3a – de novo methylase. TET 1,2,3 – demethylases of the TET-family. V – vivarium control, B – basal control, G – ground control, F – flight group. *p < 0.05 in comparison with the G group.
Figure 6.
Relative protein contents of methylation/demethylation fermentas in the duct deferens tissue (A,B) and according mRNA levels (C,D). DNMT1 – S-phase methylase DNMT3a – de novo methylase. TET 1,2,3 – demethylases of the TET-family. V – vivarium control, B – basal control, G – ground control, F – flight group. *p < 0.05 in comparison with the G group.
At the same time, the relative protein and mRNA levels of the TET family demethylases of group F in the testes (Fig. 5B) and duct deferens (Fig. 6B) did not change during flight, except for TET2 mRNA. In the flight group, the relative TET2 mRNA level increased in the testes (Fig. 5D) by 59% (p < 0.05) and in the duct deferens (Fig. 6D) by 101% (p < 0.05).
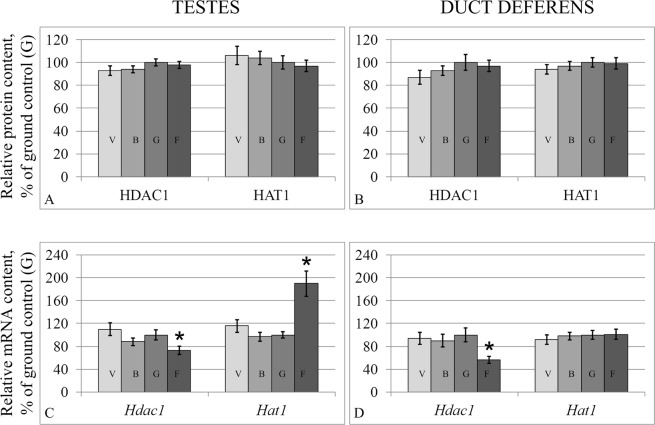
After flight, the level of HDAC1 in the testes (Fig. 7A) and the duct deferens did not change (Fig. 7B), although the relative level of the mRNA decreased by 27% (p < 0.05) (Fig. 7C) and 44% (p < 0.05) (Fig. 7D), respectively.
Figure 7.
Relative protein contents of acetylation/deacetylation fermentas in the testis (A) and duct deferens tissues (B) and according mRNA levels (C,D). HAT1 – histone acetylase. HDAC1 – histone deacetylase V – vivarium control, B – basal control, G – ground control, F – flight group. *p < 0.05 in comparison with the G group.
The level of the histone acetylase HAT1 did not change in the study groups (Fig. 7A,B); however, the relative level of the corresponding mRNA increased after flight in the testes (Fig. 7C) by 90% (p < 0.05), although in the duct deferens it remained unchanged.
Discussion
Studies of the influence of weightlessness on the structure and function of male germ cells not only are applied research but also are of fundamental importance to the understanding of the evolutionary aspects of ontogenesis. However, on Earth, all such studies have limitations that are inherent to all modeling experiments. Therefore, the data from tissue studies that were recorded directly in space flight conditions are of particular interest.
In this study, we had the opportunity to study the duct deferens and the testes of mice that were in zero gravity for approximately 23 days and were euthanized there.
The results suggested that there was no change in the levels of the studied cytoskeletal proteins (beta- and gamma-actin, alpha-actinin 1 and 4, beta-tubulin and desmin) in the flight group, although there was a decrease in Actn1 mRNA in the duct deferens and in beta-tubulin in the testes. Interestingly, in the heart and lungs after a 34–37-day flight (in the Rodent Research-1 experiment that also performed tissue fixation under weightlessness), there were practically no changes in the levels of cytoskeletal proteins, although there were significantly more changes in the mRNA levels26. The latter may be because in the early stages of space flight, the heart and lungs are subjected to an increase of external mechanical stress due to a volume shift in the cranial direction, unlike the testes and the duct deferens. However, it should be noted that testes tissues have high levels of cellular heterogeneity, which does not allow for the identification of the contributions of individual cell types to the results that we obtained. Therefore, we decided to examine the relative content of proteins and mRNA which expression correlates with different degrees of sperm maturity, both in the duct deferens and in the testes.
KDM5B (a histone demethylase that demethylates ‘Lys-4’ of histone H3) was chosen as a marker for early spermatogonia, since their differentiation into spermatocytes requires inactivation of this gene27. In the duct deferens, we did not observe changes in the protein levels and mRNA levels of the encoding gene, Kdm5B. However, in the testes, despite the absence of a change in the protein level, the mRNA level of this gene increased in the flight group, which may indicate a decrease in the differentiation potential and, accordingly, in the mature forms of sperm. We observed a similar effect after a 30-day antiorthostatic suspension, where a decrease in the number of the mature forms of sperm correlated with an increase in the mRNA level of the Kdm5B gene15. However, in this case, we did not note changes in the levels of protamines, the expression of which is highest in spermatids28, as well as sperm lysozyme-like protein 1 (SLLP1, Spaca3), which is localized to the acrosome of mature spermatozoa29. Perhaps this is because more mature forms of sperm are localized in the caudal epididymis, which was not available for analysis.
Thus, despite the absence of changes in the levels of cytoskeletal and sperm-specific proteins, changes in the expression of some proteins were observed.
On the one hand, this situation correlates with our previous data after Rodent Research-1 mission26 and may indicate a change in the efficiency of translation and/or proteolysis. Earlier, we proposed the role of actin-binding proteins in the cell mechanosensitivity at early stages of microgravity30. One of them, alpha-actinin-1 interacts with phospholipase D in the cortical cytoskeleton, and it may regulate efficiency of translation31.
On the other hand, the question remains: what caused the changes in the observed gene expression? The reasons for the changes in expression could be associated with a wide range of factors, such as histone modifications, but for higher mammals, it is most likely because of a change in the DNA methylation levels of CpG-islands in the promoter regions of genes.
In this study, we attempted to analyze the total DNA methylation level by restriction analysis and, accordingly, CG-islands in the promoter regions of the cytoskeletal genes were studied; however, the isolated genomic DNA was not of a high enough quality to conduct such an analysis.
Nevertheless, we were able to evaluate the level of 5-hydroxymethylcytosine (5hmC), which is an intermediate product between the fully methylated and fully demethylated states32. There were no changes in the 5-hydroxymethylcytosine levels in the testes or duct deferens. It should be noted that in the ovaries of mice after 23 days of antiorthostatic suspension, there were also no changes in the 5hmC levels, although we noted a change in the expression of the gene encoding one of the TET-family demethylases, TET2. Therefore, in this work, we also decided to examine the content and expression of genes encoding key methylases/demethylases.
The protein content of the S-phase methylase DNMT1, de novo methylase DNMT3a, and demethylases of the TET family (TET1, TET2, and TET3) of the flight group both in the testes and duct deferens remained at the same level as the control group. However, the gene expression changed in some cases. For example, the expression of the Dnmt1 gene decreased in the testes, and the expression of Tet2 increased both in the testes and in the duct deferens, and in the previous experiment that examined the ovaries of mice after antiorthostatic suspension33. Since the DNMT1 methylase and proteins of the TET family are part of a complex that interacts with DNA34, it can be assumed that a decrease in the expression of one of the participants of the complex could lead to a decrease in the number of active complexes and, as a consequence, to a decrease in the DNA demethylation level. On the other hand, the acetylation of TET leads to an increase in its demethylation activity, and the deacetylation of HDAC1/2 proteins leads to the inhibition of their activity involving demethylases34. Interestingly, as previously observed33, an increase in Tet2 expression in the flight group correlated with a decrease in Hdac1 deacetylase expression. At the same time, a decrease in Dnmt1 methylase expression was accompanied by an increase in Hat1 histone acetylase expression in the testes of mice of the flight group.
In summary, the expression of the Hdac1 deacetylase gene was reduced in the duct deferens, while the mRNA levels of the acetylase Hat1 remained constant; and in the testes, while the expression of the deacetylase was reduced, the expression of the acetylase was increased. Histone acetylation can regulate the efficiency of transcriptional activation both by directly changing the charge35 and causing chromatin remodeling and by the recruitment of transcriptional activators36–38. Therefore, in a case where the expression level of the acetylases/deacetylases changed at the translational level, we could expect to see a greater change in the expression of other genes, such as those encoding cytoskeletal proteins.
Conclusions
In general, in this study of the duct deferens and testes tissues of mice that were euthanized under weightless conditions, we did not detect changes in the levels of cytoskeletal proteins, sperm-specific proteins, DNA methylases, DNA demethylases, DNA acetylases and histone deacetylases. This may indicate that during 3 weeks of space flight, an adaptive protein profile formed in the germinative tissues of male mice.
However, we did observe changes in the gene expression, especially of Tet2 demethylase (an increase) and of the histone deacetylase Hdac1 (a decrease). An increase of mechanical stress, such as the increase that is associated with a return to Earth’s gravity conditions, can trigger the realization of this pattern at the protein level and subsequent changes of cells structure and functions. That is why further detailed research is required to develop effective strategies to protect human health during the early period of readaptation to varying gravity after spending a long period under weightless conditions.
Materials and Methods
Experimental design
In the “Rodent Research-4” (RR-4) experiment, C57BL/6J mice obtained from the Jackson Laboratory (Jackson Laboratory, Bar Harbor, ME) that were 9–12 weeks old at the time of launch were used. The animals were placed in a transport container on February 18, 2017. For four days, the transport container remained in the capsule of the Dragon SV on the launch pad. The SpaceX-10 spacecraft was launched on February 23, 2017. The animals were transferred from the transport container into an animal habitat unit onboard the ISS on February 25, 2017. During flight conditions, the animals received food and water ad libitum. All description of transport container and habitat with animal welfare were provided by Ronca A.E. et al.39.
The animals in the flight group (F) were kept in microgravity for 21–24 days, after which they were euthanized. Euthanasia was made by cardiac puncture and blood collection with pre-made anesthesia. Then, the carcasses were wrapped in two layers of foil and placed in plastic bags (so that each carcass was completely surrounded by ice), which were inserted into piles for storage at low temperature. The period between euthanasia and freezing of the carcass was approximately 2 minutes. The carcasses were stored in the MELFI freezer until returning to Earth.
The animals in the basal control group (B) were euthanized in normal laboratory conditions shortly after the launch of SpaceX-10. Animals of the vivarium control group (V) were kept in standard vivarium conditions and received water and food ad libitum. The animals in the ground control group (G) were euthanized in the laboratory after staying in microgravity conditions while being recorded with a camera (at the John F. Kennedy Space Center, USA), simulating the same environmental conditions as on the ISS.
The schedule for working with the animals of groups B, V and G corresponded to the schedule for working with group F.
All animal procedures were approved by the Ames Institutional Animal Care and Use Committee (Ames, USA).
Organs were isolated from the frozen carcasses on Earth. The right testis (number of animals: B - 10, V - 10, G - 10, F - 6) and duct deferens (number of animals in the groups: B - 10, V - 10, G - 10, F - 6) were frozen in liquid nitrogen for determination of protein content; the left testis (number of animals: B - 10, V - 10, G - 10, F - 8) and duct deferens (number of animals: B - 10, V - 10, G - 10, F - 8) were placed in the RNAlater Stabilization Solution (Qiagen, Germany) for future work with nucleic acids. For each group of animals, we examined the biomaterial of a minimum of 6 animals.
The samples, in accordance with the NASA-Roskosmos protocol “Utilization Sharing Plan on-board ISS” (signed on July 18, 2013), were delivered to Russia on dry ice without defrosting, which was ensured by the use of temperature sensors.
Determination of protein content by western blot
Isolation of total protein from the frozen tissues and denaturing electrophoresis were performed according to the Laemmle method using a Bio-Rad system (USA). The protein concentration of each sample was measured, and the same amount of protein was loaded into each well. Transfer to the nitrocellulose membrane was performed according to Towbin et al.40.
To identify each protein, specific antibodies were used at the dilution recommended by the respective manufacturer. Specific antibodies raised in mice were used to examine the cytoskeletal proteins: beta-actin (at a dilution of 1: 500), gamma-actin (1: 200), alpha-actinin-1 (1: 200), alpha-actinin-4 (1: 200), beta-tubulin (1: 200), and desmin (1: 200) (all from Santa Cruz Biotechnology, Inc., USA). Specific antibodies raised in rabbits for the listed proteins were used at the following dilutions: S-phase methylase DNMT1 (1: 1000), de novo methylase DNMT3a (1: 2000), 5-methylcytosine hydroxylase (demethylase) TET1 (2 µg/ml), TET2 (1: 1000), TET3 (1: 1000), histone acetylase KAT1/HAT1 (1: 1000), and histone deacetylase HDAC1 (1: 1000) (Abcam, UK); JARID1B (KDM5B) (1: 100) (Bioss Antibodies, USA); protamine (1: 250) (Sigma, Germany); and SPACA3 (1: 1000) (OriGene, USA). Biotinylated goat antibodies against mouse IgG (1:20,000) (Sigma, Germany) and rabbit IgG (1:10,000) (Jackson ImmunoResearch Lab, Inc., USA) were used as secondary antibodies. Further, all membranes were treated with a solution of streptavidin conjugated with horseradish peroxidase (Sigma, Germany) at a dilution of 1:10,000. Protein bands were detected using 3,3′-diaminobenzidine (Merck, USA), and the data were analyzed using ImageJ.
Determination of the mRNA relative content by quantitative PCR
Total RNA from tissues, stained in the RNAlater Stabilization Solution, was isolated using the RNeasy Micro Kit (Qiagen, Germany) according to the manufacturer’s instructions. Reverse transcription was performed using d (T)15 as a primer and 500 ng of RNA. The relative mRNA content of the studied genes was estimated using real-time PCR with primers selected by the Primer3Plus program (Table 1), the results were processed using the 2(−Delta Delta CT) method41.
Table 1.
Primer sequences and product sizes.
| Gene | Primer sequence, forward/reverse (5′…3′) | Product size, bp |
|---|---|---|
| Actb (beta-actin) | gctgcgttttacaccctttc/gtttgctccaaccaactgct | 218 |
| Actg (gamma-actin) | ctggtggatctctgtgagca/tcaggagggaagaaaccaga | 184 |
| Actn1 (alpha-actinin 1) | ggtcagcagcaacctcctc/tctttctccaccttctctcca | 167 |
| Actn4 (alpha-actinin 4) | accctgaacagactcccttg/gatcgacaagcctccatctc | 168 |
| Tubb2b (beta-tubulin 2B) | ggcagcaagaagctaacgag/cgaacacgaagttgtctggc | 302 |
| Des (desmin) | gtgaagatggccttggatgt/cgggtctcaatggtcttgat | 182 |
| Kdm5b(lysine (K)-specific demethylase 5B, JARID1B) | agtggctttcctgttcgaga/aagcacatgcccacatacaa | 173 |
| Prm1(protamine 1) | atggccagataccgatgct/cgagatgctcttgaagtctgg | 231 |
| Spaca3(sperm acrosome associated 3, SLLP1) | caaggccaaggtcttcagtc/tcagcttcatgatccacagc | 150 |
| Dnmt1 (S-phase methylation) | ccggaaactcacttggacga/tttggcagctggatctctgg | 90 |
| Dnmt3A (de novo methylation) | agagcgctttgactccacat/ggaccaggaaaaacaaacga | 150 |
| Tet1 (cytosine demethylase) | gtgtgggtcgatggctctat/cttattcccaccaccgctaa | 208 |
| Tet2 (cytosine demethylase) | gttctcaacgagcaggaagg/tgagatgcggtactctgcac | 185 |
| Tet3(cytosine demethylase) | ttctatccgggaactcatgg/ccaggccaggatcaagataa | 226 |
| Hat1 (histone aminotransferase 1) | agagtgccgtggagaagaaa/tttcatcatccccaaagagc | 150 |
| Hdac1 (histone deacetylase 1,2,3,4,6,9) | ccatgaagcctcaccgaat/caaacaccggacagtcctca | 226 |
Determination of the 5-hydroxymethylcytosine (5hmC) content in DNA by the dot blot method
To determine the methylation levels, total DNA was isolated from the tissues, stained in the RNAlater Stabilization Solution, using a DNA extraction kit (Sintol, Russia) based on the phenol/chloroform method according to the manufacturer’s instructions. The isolated DNA was applied to a nitrocellulose membrane, for the preliminarily measurement of the concentration and for denaturing (+95 °С for 5 min and then +4 °С for 3 min), at three dilutions −1 μg, 500 ng, and 200 ng. The membranes were air-dried, welded to the membrane using ultraviolet light and incubated in 4% skim milk overnight at +4 °C.
To evaluate the 5hmC content, specific antibodies raised in rabbits (Abcam, United Kingdom) were used at a dilution of 0.5 μg/ml in accordance with the manufacturer’s instructions. Biotinylated goat anti-rabbit IgG antibodies (Jackson ImmunoResearch Lab., Inc., USA) were used as secondary antibodies at a dilution of 1:10,000. Then, the membranes were treated with a streptavidin solution that was conjugated with horseradish peroxidase (Sigma, Germany) at a dilution of 1:10,000. Dots were detected using 3,3′-diaminobenzidine (Merck, USA). ImageJ was used to analyze the data.
Statistical analysis
The results obtained were analyzed by ANOVA, using a post hoc t-test with significance level p < 0.05 to assess the significance of the differences between the groups. The data are presented as M ± SE, where M is the arithmetic average and SE is the error of the mean.
All methods were performed in accordance with the relevant guidelines and regulations.
Acknowledgements
The authors thank all colleagues from the NASA Ames Research Center and the Johnson Research Center who provided us the opportunity to participate in this project, especially Nicole Rayl, Jennifer Buchli, Julie Robinson, Pete Hasbrook and all astronauts participating in the “Rodent Research” project. We are very grateful to Galina Tverskaya for her support and help during the organization of our cooperative research. The authors thank Russian Cosmonauts Andrey Borisenko and Oleg Novitskiy for their participation in the “Rodent Research” project. The tissue sharing was conducted in accordance with the NASA-Roscosmos protocol “Utilization Sharing Plan on-board ISS” (signed July 18, 2013). This work was financially supported by the program for fundamental research SSC RF – IBMP RAS; program “Postgenomic technologies and perspective solutions in the biomedicine” of the RAS Presidium; Russian Academic Excellence Project 5–100.
Author Contributions
Irina V. Ogneva – conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, resources, software, supervision, validation, visualization, writing. Maria A. Usik – formal analysis, investigation, methodology, visualization, writing. Sergey S. Loktev – formal analysis, investigation, visualization. Yuliya S. Zhdankina – investigation, methodology, validation. Nikolay S. Biryukov – investigation, methodology, validation. Oleg I. Orlov – project administration, resources. Vladimir N. Sychev – project administration, resources, writing.
Data Availability
All data generated or analyzed during this study are included in this article.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ontogenesis of mammals in microgravity. NASA TM – 103978 (1993).
- 2.Fritzsch B, Bruce LL. Utricular and saccular projections of fetal rats raised in normal gravity and microgravity. ASGSB Bull. 1995;9:97. [Google Scholar]
- 3.Serova LV, Natochkin IV, Nosovskii AM, Shakhmatova EI, Fast T. Effect of weightlessness on the mother-fetus system (results of embryological experiment NIH-R1 abroad the “Space Shuttle”. Aviakosm Ekolog Med. 1996;30(6):4–8. [PubMed] [Google Scholar]
- 4.Savel’ev SV, Serova LV, Besova NV, Nosovskii AM. Effect of weightlessness on rats endocrine system development. Aviakosm Ekolog Med. 1998;32(2):31–36. [PubMed] [Google Scholar]
- 5.Schenker E., Forkheim K. Mammalian mice embryo early development in weightlessness environment on STS 80 space flight. Israel Aerospace Medicine Institute Report (1998).
- 6.Boonstra, J. Growth factor-induced signal transduction in adherent mammalian cells is sensitive to gravity. FASEB J. 13 Suppl: S35 42 (1999). [DOI] [PubMed]
- 7.Kojima Y, et al. Effects of simulated microgravity on mammalian fertilization and preimplantation embryonic development in vitro. Fertil Steril. 2000;74(6):1142–1147. doi: 10.1016/S0015-0282(00)01583-1. [DOI] [PubMed] [Google Scholar]
- 8.Serova LV. Microgravity and development of the mammals: problems results prospects. Aviakosm Ekolog Med. 2001;35(2):32–35. [PubMed] [Google Scholar]
- 9.Kamiya H, et al. Effect of simulated microgravity on testosterone and sperm motility in mice. J Androl. 2003;24(6):885–890. doi: 10.1002/j.1939-4640.2003.tb03140.x. [DOI] [PubMed] [Google Scholar]
- 10.Tash JS, Bracho GE. Microgravity alters protein phosphorylation changes during initiation of sea urchin sperm motility. FASEB J. 1999;13(Suppl):S43–54. doi: 10.1096/fasebj.13.9001.s43. [DOI] [PubMed] [Google Scholar]
- 11.Li HY, et al. Simulated microgravity conditions and carbon ion irradiation induce spermatogenic cell apoptosis and sperm DNA damage. Biomed Environ Sci. 2013;26(9):726–734. doi: 10.3967/0895-3988.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 12.Tash JS, Johnson DC, Enders GC. Long-term (6wk) hindlimb suspension inhibits spermatogenesis in adult male rats. J Appl Physiol. 2002;92:1191–1198. doi: 10.1152/japplphysiol.00931.2001. [DOI] [PubMed] [Google Scholar]
- 13.Pellegrini M, et al. Microgravity promotes differentiation and meiotic entry of postnatal mouse male germ cells. PLoS One. 2010;5(2):e9064. doi: 10.1371/journal.pone.0009064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, et al. Mouse undifferentiated spermatogonial stem cells cultured as aggregates under simulated microgravity. Andrologia. 2014;46(9):1013–1021. doi: 10.1111/and.12189. [DOI] [PubMed] [Google Scholar]
- 15.Usik MA, Ogneva IV. Cytoskeleton structure in mouse sperm and testes after 30 days of hindlimb unloading and 12 hours of recovery. Cell Physiol Biochem. 2018;51:375–392. doi: 10.1159/000495235. [DOI] [PubMed] [Google Scholar]
- 16.Zayzafoon M, Gathings WE, McDonald JM. Modeled microgravity inhibits osteogenic differentiation of human mesenchymal stem cells and increases adipogenesis. Endocrinology. 2004;145(5):2421–2432. doi: 10.1210/en.2003-1156. [DOI] [PubMed] [Google Scholar]
- 17.Meyers VE, Zayzafoon M, Gonda SR, Gathings WE, McDonald JM. Modeled microgravity disrupts collagen I/integrin signaling during osteoblastic differentiation of human mesenchymal stem cells. J Cell Biochem. 2004;93(4):697–707. doi: 10.1002/jcb.20229. [DOI] [PubMed] [Google Scholar]
- 18.Meyers VE, Zayzafoon M, Douglas JT, McDonald JM. RhoA and cytoskeletal disruption mediate reduced osteoblastogenesis and enhanced adipogenesis of human mesenchymal stem cells in modeled microgravity. J Bone Miner Res. 2005;20(10):1858–1866. doi: 10.1359/JBMR.050611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dai ZQ, Wang R, Ling SK, Wan YM, Li YH. Simulated microgravity inhibits the proliferation and osteogenesis of rat bone marrow mesenchymal stem cells. Cell Prolif. 2007;40(5):671–684. doi: 10.1111/j.1365-2184.2007.00461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel MJ, et al. Identification of mechanosensitive genes in osteoblasts by comparative microarray studies using the rotating wall vessel and the random positioning machine. J Cell Biochem. 2007;101(3):587–599. doi: 10.1002/jcb.21218. [DOI] [PubMed] [Google Scholar]
- 21.Gershovich PM, Gershovich JG, Buravkova LB. Simulated microgravity alters actin cytoskeleton and integrin-mediated focal adhesions of cultured human mesenchymal stromal cells. J Grav Physiol. 2008;15(1):203–204. [Google Scholar]
- 22.Pan Z, et al. Effects of hindlimb unloading on ex vivo growth and osteogenic/adipogenic potentials of bone marrow-derived mesenchymal stem cells in rats. Stem Cells Dev. 2008;17(4):795–804. doi: 10.1089/scd.2007.0254. [DOI] [PubMed] [Google Scholar]
- 23.Ou X, et al. Spaceflight induces both transient and heritable alterations in DNA methylation and gene expression in rice (Oryza sativa L.) Mutat Res. 2009;662(1-2):44–53. doi: 10.1016/j.mrfmmm.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 24.Singh KP, Kumari R, Dumond JW. Simulated microgravity-induced epigenetic changes in human lymphocytes. J Cell Biochem. 2010;111(1):123–129. doi: 10.1002/jcb.22674. [DOI] [PubMed] [Google Scholar]
- 25.Chowdhury B, et al. A Study of Alterations in DNA Epigenetic Modifications (5mC and 5hmC) and Gene Expression Influence by Simulated Microgravity in Human Lymphoblastoid Cells. PLoS One. 2016;11(1):e0147514. doi: 10.1371/journal.pone.0147514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogneva IV, Loktev SS, Sychev VN. Cytoskeleton structure and total methylation of mouse cardiac and lung tissue during space flight. PLoS One. 2018;13(5):e0192643. doi: 10.1371/journal.pone.0192643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat. Rev. Cancer. 2005;5(8):615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 28.Depa-Martynow M, Kempisty B, Lianeri M, Jagodzinski PP, Jedrzejczak P. Association between fertilin beta, protamines 1 and 2 and spermatid-specific linker histone H1-like protein mRNA levels, fertilization ability of human spermatozoa, and quality of preimplantation embryos. Folia Histochem Cytobiol. 2007;45(Suppl 1):S79–85. [PubMed] [Google Scholar]
- 29.Lea, I. A., Widgren, E. E. & O’Rand, M. G. Association of sperm protein 17 with A-kinase anchoring protein 3 in flagella. Reprod Biol Endocrinol. 2: 57, 7 pages (2004) [DOI] [PMC free article] [PubMed]
- 30.Ogneva IV, Biryukov NS, Leinsoo TA, Larina IM. Possible role of non-muscle alpha-actinins in muscle cell mechanosensitivity. PLoS One. 2014;9:e96395. doi: 10.1371/journal.pone.0096395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park JB, et al. Cardiac phospholipase D2 localizes to sarcolemmal membranes and is inhibited by alpha-actinin in an ADP-ribosylation factor-reversible manner. J Biol Chem. 2000;275:21295–21301. doi: 10.1074/jbc.M002463200. [DOI] [PubMed] [Google Scholar]
- 32.Guo JU, Su Y, Zhong C, Ming GL, Song H. Emerging roles of TET proteins and 5-hydroxymethylcytosines in active DNA demethylation and beyond. Cell Cycle. 2011;10(16):2662–2668. doi: 10.4161/cc.10.16.17093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Usik, M. A. Ogneva IV. The regulation of the DNA methylation in the ovaries of mice under 23-days antiorthostatic suspension. Front. Physiol. Conference Abstract: 39th ISGP Meeting & ESA Life Sciences Meeting (2018).
- 34.Zhang YW, et al. Acetylation enhances TET2 function in protecting against abnormal DNA methylation during oxidative stress. Molecular Cell. 2017;6:323–335. doi: 10.1016/j.molcel.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vettese-Dadey M, et al. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 1996;15:2508–2518. doi: 10.1002/j.1460-2075.1996.tb00608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dhalluin C, et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 37.Hassan AH, et al. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111:369–379. doi: 10.1016/S0092-8674(02)01005-X. [DOI] [PubMed] [Google Scholar]
- 38.Shogren-Knaak M, et al. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 39.Ronca AE, et al. Behavior of mice abroad the International Space Station. Sci Rep. 2019;9:4717. doi: 10.1038/s41598-019-40789-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Towbin H, Staehlin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some application. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;2:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this article.