Abstract
Soil salinization is a major constraint of agriculture in semiarid ecosystems. In this study soil microcosms were applied to evaluate the impact of a lower- and higher-level salinization treatment of a pristine scrubland soil on the abundance of Bacteria, Archaea, and Fungi, and on prokaryotic diversity in bare soil and the rhizosphere of wheat assessed by qPCR and high-throughput sequencing of 16S rRNA gene amplicons. Furthermore, the impact of soil straw amendment as a salt-stress alleviation strategy was studied. While the low-level salinity stimulated plant growth, the seedlings did not survive under the higher-level salinity unless the soil was amended with straw. Without the straw amendment, salinization had only minor effects on the microbial community in bare soil. On the other hand, it decreased prokaryotic diversity in the rhizosphere of wheat, but the straw amendment was effective in mitigating this effect. The straw however, was not a significant nutrient source for the rhizosphere microbiota but more likely acted indirectly by ameliorating the salinity stress on the plant. Members of Proteobacteria, Actinobacteria, and Firmicutes were abundant among the bacteria that reacted to soil salinization and the straw amendment but showed inconsistent responses indicating the large physiological diversity within these phyla.
Subject terms: Soil microbiology, Agroecology
Introduction
In semiarid ecosystems, the extension of agricultural land area for cultivation of crops has caused the replacement of the original vegetation, typically scrublands, and thereby a dramatic loss of floral and faunal biodiversity1,2. Converting scrublands to croplands strongly effects soil properties introducing frequent irrigation and mixing by tillage to previously water-limited, stratified systems with low nutrient input3,4. Under agricultural management, the land area also receives fertilizers, pesticides, and organic matter from crop residues. It has been shown that conversion from scrubland to cropland causes a loss of soil organic carbon and phosphate but an increase in soil salinity5. While the lost phosphate can be substituted by fertilization, the loss of carbon and the increasing salinity can impede agricultural productivity, especially by low quality irrigation water and poorly drained soils6.
Soil salinization is one of the major constraints for agriculture in semiarid ecosystems and threatens food security on a global scale with affected areas spreading annually and predicted to reach 50% of arable land by 20507,8. Economically important crops show significant decrease in yield above a salinity threshold of 1 to 8 dS m−1 depending on the species, with wheat and barley being more tolerant than maize and most vegetables9. Plants respond to salinity stress by osmotic adjustment accumulating solutes to balance the osmotic pressure10. This, however, has a substantial energy demand that restricts plant growth11. In addition, the accumulation of sodium ions in the leaves has a direct inhibitory effect on photosynthesis12. There are also below-ground responses: reduced primary root growth but increased lateral root development has been found in durum wheat and Arabidopsis thaliana under salt stress13,14. When facing salinity stress, a plant may benefit from the activity of the microbial community colonizing its rhizosphere15,16. For instance, the bacterial production of extracellular polymeric substances (EPS) can aid the plants in resisting drought and salinity17,18. Beyond the direct adverse effects of salinity on plants, it also destabilizes soil structure through replacing bivalent cations like calcium (Ca2+) by sodium (Na+) on the cation exchange sites19. As a consequence, salinized soils do not only lose soil organic carbon but also their structural stability further decreasing soil fertility.
Strategies to maintain agricultural productivity and to alleviate the salinity stress include the development of salt-tolerant cultivars20,21 and the implementation of alternative cropping systems22 and specific management practices, e.g., drip-irrigation or soil drainage23,24. Salinity stress can also be reduced by amending soils with organic substances like manure, composts, or straw25–28. The introduction of such particulate organic materials can have several beneficial effects including improvement of soil structure and promotion of growth of microbial biomass that provides EPS and enzymatic activities which facilitate soil aggregate formation and the stabilization of organic carbon29.
Considering the importance of microbiologically mediated activities to support plant growth and improve soil structure in salinity affected soils, our objective in this study was to analyze how salinity affects microbial abundance and prokaryotic diversity in bare soil and in the rhizosphere of wheat in soil from a semiarid ecosystem, and how amending the soil with straw modifies these effects. More specifically, we conducted a two phase experiment under greenhouse conditions with scrubland soil from a semiarid ecosystem without any history of agricultural use which was either amended with straw or not before the experiment: During the first phase, the soil received either non-saline water, or water with low salinity or high salinity allowing evaporation but excluding drainage to cause the accumulation of salts as it is common for semiarid ecosystems24. Subsequently, wheat seedlings were planted into the soil and grown for 7 weeks to obtain rhizosphere samples.
We hypothesized that (1) the addition of saline water transiently increases microbial abundance in bare soil due to the mobilization of carbon but, as a selective factor, reduces prokaryotic diversity in both the bare soil and in the rhizosphere. Furthermore, we expected that (2) straw amendment provides an additional carbon and energy source resulting in increased microbial abundance and ameliorating the adverse effect of salinity on soil prokaryotic diversity. The microbial communities in the bare soil and rhizosphere samples were characterized by high-throughput sequencing of 16S rRNA gene amplicons to assess the diversity of Bacteria and Archaea, and by qPCR of 16S rRNA genes as an estimate of the abundances of Bacteria and Archaea, and ITS sequences to estimate fungal population sizes.
Materials and Methods
Soil
The soil was a Xerosol collected from a scrubland located in the La Paz-El Carrizal basin, 12 km west of the city of La Paz, Baja California, Mexico on November 13, 2012. The sampling site was within a protected area without any previous history of agricultural use (GPS position: 24°07′43″N, 110°26′03″W). Details about the vegetation and soil properties have been described elsewhere30,31. Briefly, the soil had pH 7.9, electrical conductivity (EC1:5) 0.41 dS m−1, total carbon 3.0 mg C g−1 soil dry weight (d.w.), total nitrogen (Nt) 0.7 mg N g−1 d.w., and water holding capacity (WHC) 27% (w/w) g−1 d.w. After sampling, the soil was sieved (mesh size 2 mm), air-dried, and kept for 15 weeks at room temperature before the experiment.
Experimental setup
The experiment consisted of two phases. In the first phase a salinization treatment was performed: soil with or without straw amendment was irrigated with non-saline water or water with low or high salinity to reach three different levels of soil salinity (control, low, high). In the second phase, wheat seedlings were grown in the soil to obtain rhizosphere samples. At the start of phase 1, glass beakers (0.2 L) were filled with 100 g soil or soil amended with straw. The straw was a mixture of equal amounts of dried whole plant material from maize “Simao”, sorghum “Tarzan”, and summer wheat “Ethos” ground to a particle size of 1 mm. The material contained 48% (w/w) carbon and 0.8% (w/w) nitrogen, and was mixed to the soil to reach 2% (w/w). The salinization treatment was achieved by irrigating the soil with water with different levels of salinity: the controls received sterile distilled water (0 dS m−1), in the low salinity treatment sterile distilled water with 1.125 g NaCl L−1 (1.9 dS m−1) was used, while in the high salinity treatment sterile distilled water with 7 g NaCl L−1 (10.8 dS m−1) was applied. Each combination of straw amendment and salinity treatment was replicated four times resulting in 24 glass beakers with soil in total. The soil was irrigated according to the salinization treatment once a week for eight weeks maintaining 50% WHC. For an additional three weeks, all beakers were irrigated with non-saline sterile distilled water to allow the equilibration of the soil conditions. The soil was then removed from each beaker, sieved (2 mm), and divided into two parts: 60 g was filled back into the beaker while 40 g was collected and analyzed as phase 1 bare soil sample. An aliquot of 2 g from each sample was frozen and kept at −80 °C until DNA extraction. The remaining 38 g of each sample was air-dried and used for chemical analyses. In phase 2, three seedlings of the wheat cultivar Opata, germinated on wet filter paper four days before, were planted into the remaining 60 g of soil in each beaker. The plants were watered 2–3 times a week according to their needs with non-saline water and fertilized weekly with 0.06 mg g−1 soil d.w. Wuxal Basis Suspension (Schering AG, Düsseldorf-Heerdt, Germany). The phase 2 rhizosphere samples were collected after 7 weeks of plant growth. Plants were uprooted and loosely adhering soil was removed from the roots by shaking. Roots of the three individual plants per beaker were combined and washed in 30 ml sterile saline (0.85% NaCl) for 30 min at 4 °C in an orbital shaker (Model 3040, GFL, Burgwedel, Germany) with 10 rpm. The roots were then removed and the cells were collected from the solution by centrifugation at 4,100 × g for 30 min at 4 °C. Pellets were stored at −80 °C until DNA extraction. The remaining soil (phase 2 bulk soil) was sieved (2 mm) and air-dried for chemical analyses. The roots and shoots were oven-dried and weighed to measure plant biomass. The seedlings did not survive in the soil from the high salinity treatment without straw amendment. Additionally, the plants died in one beaker from the control treatment without straw amendment, hence only three replicates were sampled from this treatment combination.
Soil chemical and physical analyses
The electrical conductivity (EC) as an indicator for salinity was measured as dS m−1 in a 1:5 soil-to-water ratio mixture after 1 h of shaking on an orbital shaker32. Soil pH was determined in 0.01 M CaCl2 using a soil-to-solution ratio of 1:2 (w/v). Total carbon (Ct [%]) and nitrogen (Nt [%]) were measured via dry combustion using an elemental analyzer (LECO TruMac, Elementar, Germany). The water holding capacity (WHC; % H2O g−1 w/w) was measured gravimetrically after allowing thoroughly wetted soils to drain on a sand bed for 3 h. Details of the methods can be found elsewhere33.
DNA extraction
DNA was extracted using the FastDNA SPIN kit for soil (MP Biomedicals, Illkirch, France). The extraction included two bead beating steps of 45 s at 6.5 m s−1 on a FastPrep-24 system (MP Biomedicals) and one additional washing step of the binding matrix with 1 ml 5.5 M guanidine thiocyanate (Carl Roth, Karlsruhe, Germany). DNA extraction failed from a phase 2 rhizosphere sample from the control treatment with straw amendment; therefore, there were only three replicates from this treatment combination.
Quantification of the bacterial, archaeal, and fungal communities
Population sizes of the bacterial and archaeal communities were determined by quantitative real-time PCR applying the Maxima Probe qPCR ROX Master Mix (Thermo Fisher Scientific, Epsom, UK) with 0.5 mM of each of the primers and 0.2 mM of the FAM-labeled TaqMan probe. Fungi were quantified with the Maxima SYBRGreen/ROX qPCR Master Mix (Thermo Fisher Scientific) and 0.5 mM of each primer. The amplification was followed by a melt curve analysis. A total of 2 µl of template DNA diluted 50-fold in TE-buffer (10 mM Tris, 1 mM EDTA, pH 8) were used in each 20 µl reaction. All reactions were performed in duplicates in a StepOnePlus Real Time PCR System (Life Technologies GmbH, Darmstadt, Germany). Standard curves were obtained from 10-fold dilutions of the pGEM-T vector (Promega, Mannheim, Germany) containing the 16S rRNA gene of Bacillus subtilis, or Methanobacterium oryza, or the ITS of Fusarium culmorum. PCR conditions, primer and probe sequences with references are listed in the Table S1. The average PCR efficiency was 98% for Bacteria and Archaea and 85% for Fungi with R2 > 0.99.
Illumina PCR-amplicon sequencing and data processing
The V4 region of the 16S rRNA gene was amplified with primers S-D-Arch-0519-a-S-15 (5′-CAGCMGCCGCGGTAA-3′) and S-D-Bact-0785-a-A-21 (5′-GACTACHVGGGTATCTAATCC-3′)34 and sequenced according to the protocol of Kozich et al.35. Primer sequences are listed in the Table S2. Two PCR amplifications were carried out from each DNA extract with the FastStart High Fidelity PCR System (Roche Diagnostics, Mannheim, Germany) in 50 µl final volume. Each reaction contained 1 µl template DNA, 0.4 µM of each primer, 200 µM of each dNTP, 5% dimethyl sulfoxide and 2.5 U FastStart High Fidelity Enzyme Blend in a reaction buffer with 1.8 mM MgCl2. The temperature program of the reactions was initial denaturation at 95 °C for 2 min followed by 35 cycles of 95 °C for 30 s, 50 °C for 30 s, and 72 °C for 1 min and ended with an extension step at 72 °C for 5 min. Products of the two reactions from the same sample were pooled and purified with HiYield PCR Clean-up & Gel-Extraction kit (SLG) followed by quantification with Quant-iT PicoGreen dsDNA assay (Invitrogen, Darmstadt, Germany). Equimolar amounts of the purified PCR products were pooled and sent to StarSEQ (Mainz, Germany) for 250 bp paired-end sequencing on a MiSeq instrument. The sequence data was processed with dada2 version 1.6.036 in R 3.4.1 (www.r-project.org). Forward and reverse reads were truncated at positions 235 and 150, respectively, or at any position with a quality score of two. Reads with ambiguous bases or over two expected errors were discarded. Error models were constructed from one million randomly selected reads. The reads were binned into sequence variants (SVs) based on the error models using the pool option. Forward and reverse sequences were merged, and then chimeric sequences were identified with the removeBimeraDenovo function and removed. SVs were classified based on the SILVA reference release 12837 accepting only results with at least 50% bootstrap support. SVs classified as chloroplast or mitochondrial sequences were deleted from the dataset. All sequences were deposited at the European Nucleotide Archive under the accession number PRJEB30355.
Statistical analyses
Plant biomass was compared between the treatment combinations with Tukey-Kramer tests in JMP 13.0.0 (SAS Institute, Cary, NC). The qPCR results were subjected to log10 transformation and analyzed in JMP 13.0.0. Copy numbers in the different salinity treatments were compared with Tukey-Kramer tests in case of the phase 1 bare soil samples with and without straw and the phase 2 rhizosphere samples with straw. In case of the phase 2 rhizosphere samples without straw amendment, t-tests were used to compare the control and low salinity treatment. The effect of the straw amendment under the different salinity levels was assessed with t-tests. SV richness was estimated from the sequencing data using objective Bayes procedure based on negative binomial models38 implemented in the breakaway R package version 3.039. Simpson diversity was estimated by bootstrapping with 500 iterations using the ‘resample_estimate’ function of the breakaway package. The mean SV richness and Simpson diversity estimates and their standard errors were used in the ‘betta’ function40 to test for significant differences between the control and the low or high salinity treatments, or between the samples with and without straw amendment from the same salinity treatment. For preparing ordination plots, SVs that didn’t have at least 0.1% average relative abundance in the samples included in the ordination were removed from the dataset to decrease sparsity. Remaining zeroes were then replaced with the count zero multiplicative method implemented in the zCompositions R package version 1.1.141 and the data was subjected to centered log-ratio (CLR) transformation to remove compositional effects and correct for differences in sequencing depth42. Principal component analysis (PCA) was performed in R with the vegan package version 2.4.643. Variation partitioning from the vegan package was employed on the CLR-transformed data matrices used in PCA to assess the proportion of the variation in the data explained by the salinity treatment and the straw amendment. ALDEx244 from the R package version 1.6.0 was used to identify SVs differentially abundant between the control and the low or high salinity treatments, or between the samples with and without straw amendment from the same salinity treatment. Only SVs with at least 0.1% average relative abundance were included in these analyses. In the ALDEx2 results, significance was assessed based on Welch’s t-test with Benjamini and Hochberg’s correction45 to maintain a 10% false discovery rate. Figures summarizing the results were prepared in Cytoscape 3.4.0 (www.cytoscape.org).
Results
Effect of salinization and straw amendment on soil chemical parameters and plant growth
The low and high salinization treatments during phase 1 increased the electric conductivity in the soil, as intended, but did not alter the pH (Table 1). The straw amended soils had a higher C:N ratio and were slightly more neutral in pH. Theoretically, the straw amendment should have increased the total soil C by 0.96% (w/w) but the difference between the soil with and without straw at the end of phase 1 was lower, possibly due to microbial mineralization during the 11 weeks of incubation. In the bulk soil at the end of phase 2, the EC was slightly lower than in the bare soil at the end of phase 1. Nevertheless, the differences between the control, low, and high salinity treatments were still large, indicating that the salinity stress persisted during the cultivation of wheat.
Table 1.
Soil chemical parameters (average ± SD) in phase 1 bare soil and in phase 2 bulk soil in the different salinity treatments.
| Water treatment | Total soil C (%, w/w) | Total soil N (%, w/w) | C:N ratio | pH | EC1:5 [dS m−1] |
|---|---|---|---|---|---|
| Phase 1: bare soil after the salinization treatment | |||||
| Soil without straw | |||||
| Control | 0.28 ± 0.02 | 0.033 ± 0.005 | 8.5 ± 1.5 | 7.8 ± 0.3 | 0.076 ± 0.004 |
| Low salinity | 0.29 ± 0.01 | 0.037 ± 0.003 | 8.1 ± 1.0 | 7.7 ± 0.0 | 0.348 ± 0.018 |
| High salinity | 0.29 ± 0.01 | 0.035 ± 0.004 | 8.5 ± 1.1 | 7.6 ± 0.3 | 1.728 ± 0.066 |
| Soil amended with straw | |||||
| Control | 0.98 ± 0.03 | 0.059 ± 0.005 | 16.7 ± 1.5 | 7.5 ± 0.1 | 0.179 ± 0.003 |
| Low salinity | 0.96 ± 0.09 | 0.056 ± 0.006 | 17.3 ± 2.5 | 7.5 ± 0.0 | 0.465 ± 0.018 |
| High salinity | 1.12 ± 0.13 | 0.054 ± 0.009 | 21.2 ± 3.9 | 7.5 ± 0.0 | 1.678 ± 0.072 |
| Phase 2: bulk soil after plant growth | |||||
| Soil without straw | |||||
| Control | 0.31 ± 0.01 | 0.034 ± 0.005 | 9.0 ± 0.8 | 7.6 ± 0.0 | 0.147 ± 0.032 |
| Low salinity | 0.31 ± 0.01 | 0.029 ± 0.005 | 10.8 ± 1.8 | 7.6 ± 0.0 | 0.280 ± 0.061 |
| High salinity1 | 0.31 ± 0.01 | 0.025 ± 0.004 | 12.6 ± 1.9 | 7.7 ± 0.0 | 1.612 ± 0.206 |
| Soil amended with straw | |||||
| Control | 0.83 ± 0.04 | 0.045 ± 0.006 | 18.4 ± 1.7 | 7.3 ± 0.0 | 0.153 ± 0.006 |
| Low salinity | 0.99 ± 0.03 | 0.043 ± 0.004 | 23.2 ± 2.0 | 7.4 ± 0.0 | 0.347 ± 0.036 |
| High salinity | 1.07 ± 0.11 | 0.047 ± 0.005 | 22.9 ± 2.3 | 7.4 ± 0.0 | 1.153 ± 0.076 |
The low salinity level stimulated, regardless whether the soil had been amended with straw, the shoot growth of the wheat plants. Under high salinity, however, the seedlings did not survive in the soil without straw. In the straw amended soil the shoot biomass was not significantly different under control and high salinity conditions (Table S3). Root growth was strongly stimulated by the straw amendment under the non-saline control conditions. Low salinity had no significant effect on the root mass but it was significantly reduced under high salinity.
Abundances of microbial groups based on qPCR
In the bare soil from phase 1, the straw amendment caused a strong increase in bacterial and fungal abundance regardless of the level of salinity (p < 0.001) but slightly decreased the abundance of Archaea (p < 0.005) as indicated by qPCR (Table 2A). Without the straw amendment, there was a small but significant increase in soil bacterial and archaeal abundance in the high salinity treatment compared to the control, but no significant difference was found between the control and the low salinity treatment. In the straw amended soil, the salinity treatment did not affect bacterial abundance, while archaeal rRNA gene numbers were slightly higher under high salinity than in the control. Soil salinity had no significant effect on fungal abundance in the bare soil regardless of the straw amendment.
Table 2.
16S rRNA gene or ITS sequence numbers (log10 average ± SD) of microbial groups per g soil (dry weight) in bare soil after phase 1 and per g root (fresh weight) in the rhizosphere of wheat from phase 2.
| Treatments | Control | Low salinity | High salinity |
|---|---|---|---|
| A. Phase 1: bare soil | |||
| Soil without straw | |||
| Bacteria | 9.38 ± 0.04 B | 9.42 ± 0.05 AB | 9.47 ± 0.02 A |
| Archaea | 8.08 ± 0.05 B | 8.11 ± 0.05 AB | 8.19 ± 0.03 A |
| Fungi | 6.91 ± 0.09 A | 7.03 ± 0.19 A | 6.96 ± 0.04 A |
| Soil amended with straw | |||
| Bacteria | 10.30 ± 0.02 A | 10.31 ± 0.03 A | 10.23 ± 0.02 A |
| Archaea | 7.84 ± 0.03 B | 7.91 ± 0.05 AB | 7.93 ± 0.04 A |
| Fungi | 9.13 ± 0.13 A | 9.04 ± 0.10 A | 9.18 ± 0.10 A |
| B. Phase 2: rhizosphere soil. | |||
| Soil without straw | |||
| Bacteria | 10.42 ± 0.05* | 10.09 ± 0.08* | No plant growth |
| Archaea | 7.99 ± 0.14 | 8.08 ± 0.13 | |
| Fungi | 9.03 ± 0.24 | 9.63 ± 0.40 | |
| Soil amended with straw | |||
| Bacteria | 10.44 ± 0.10 A | 10.58 ± 0.09 A | 10.62 ± 0.10 A |
| Archaea | 7.63 ± 0.14 B | 7.92 ± 0.20 AB | 8.06 ± 0.13 A |
| Fungi | 9.44 ± 0.13 A | 9.41 ± 0.10 A | 9.12 ± 0.28 A |
Different capital letters indicate significant (P < 0.05) differences within the same line of the table based on Tukey-Kramer tests. *Indicates significant (P < 0.05) differences based on t-tests.
In the rhizosphere (phase 2), the straw amendment had only minor effects on microbial abundance: Under non-saline conditions it decreased archaeal rRNA gene numbers (p = 0.03), and under low salinity it increased bacterial abundance (p = 0.002). Soil salinity caused a decrease in bacterial abundance without the straw amendment but had no such effect in the rhizosphere of plants grown in the straw amended soil (Table 2B). Archaea, on the other hand, became more abundant under high salinity with the straw amendment compared to the control. Fungal abundance in the rhizosphere was neither affected by the straw amendment nor the salinization.
Microbial richness and diversity in the bare soil and wheat rhizosphere
From the bare soil samples (phase 1), 1,729,506 high-quality sequences (47,219 to 95,177 sequences per sample) were obtained by sequencing of the prokaryotic 16S rRNA gene amplicons. In each sample 96.9 to 99.8% of the sequences were assigned to the following ten phyla: Firmicutes (23.4–46.7%), Proteobacteria (13.3–43.7%), Actinobacteria (8.0–38.1%), Thaumarchaeota (0.2–7.1%), Gemmatimonadetes (0.5–5.6%), Chloroflexi (0.7–4.1%), Bacteroidetes (0.3–3.7%), Acidobacteria (0.1–3.3%), Planctomycetes (0.3–2.2%), and Verrucomicrobia (0.0–4.0%) (Table S4). The five most abundant classes were Bacilli (23.2 to 46.7%), Alphaproteobacteria (4.9 to 35.9%), Gammaproteobacteria (1.4 to 6.7%), Thermoleophilia (1.3 to 20.3%), and Actinobacteria (5.0 to 16.3%). The straw amendment significantly decreased the richness and the diversity of the bacterial and archaeal community in the bare soil (P < 0.001) (Figs 1A and S1A). High salinity had a negative effect on richness (p < 0.05) which was stronger with the straw amendment than without it. The salinity treatment, however, did not have a significant effect on diversity according to the Simpson index.
Figure 1.
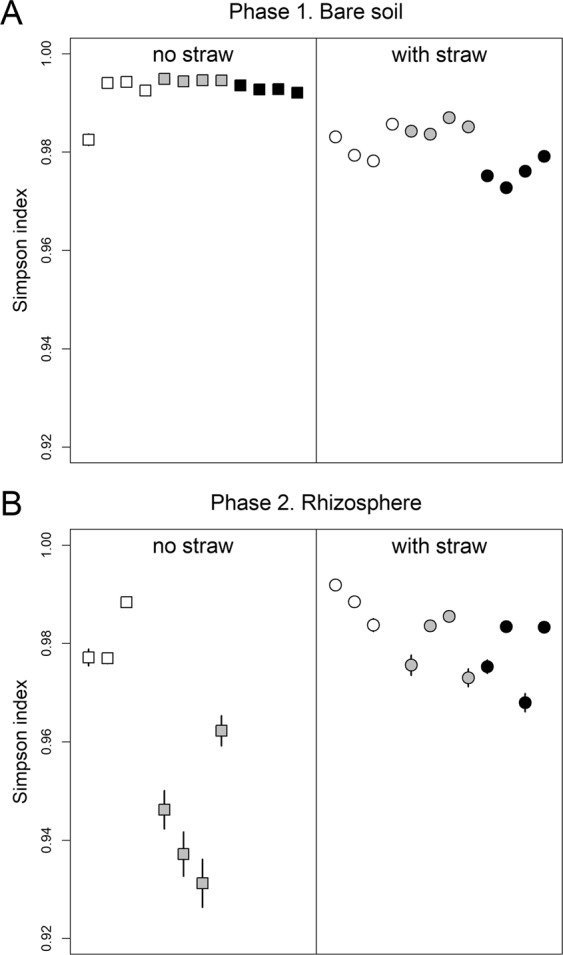
Estimated Simpson diversity index of the bacterial and archaeal community in (A) the bare soil from phase 1 and (B) in the rhizosphere from phase 2. Circles and squares indicate the mean estimates for each sample, lines the standard errors. Control: empty symbols; low salinity: grey symbols; high salinity: black symbols.
From the rhizosphere samples (phase 2), a total of 1,010,317 high-quality sequences (21,521 to 93,126 sequences per sample) were retrieved. In each sample 97.5 to 99.6% of the sequences were from the ten most dominant phyla: Proteobacteria (36.8–56.2%), Firmicutes (1.3–30.0%), Actinobacteria (7.0–20.3%), Verrucomicrobia (1.9–27.6%), Bacteroidetes (2.7–14.7%), Gemmatimonadetes (0.7–12.7%), Chloroflexi (0.4–4.8%), Planctomycetes (1.1–3.5%), Acidobacteria (0.0–3.9%), and Thaumarchaeota (0.1–3.8%) (Table S4). The five most abundant classes in the rhizosphere were Alphaproteobacteria (19.1 to 43.8%), Gammaproteobacteria (4.1 to 25.7%), Bacilli (1.2 to 26.7%), Verrucomicrobiae (0.2 to 25.2%), and Actinobacteria (4.5 to 16.3%). The straw amendment significantly decreased (p < 0.001) the richness of the bacterial and archaeal community in the rhizosphere (Fig. S1B). However, it mitigated the negative effect of salinity on Simpson diversity: both the low and the high salinity treatments had a significant negative effect on diversity (p < 0.004) but it was stronger in the rhizosphere of plants grown in the soil without straw added (Fig. 1B).
Responses of the microbial community structure to salinity and straw amendment
The effects of soil salinization and straw amendment on the structure of the bacterial and archaeal communities were quantified and compared with variation partitioning and the differences in community structure between the samples were visualized by PCA. Among the bare soil samples from phase 1, the straw amendment had a large effect on the community structure explaining the majority of the variation in the data (Fig. 2A). Comparably, the effect of soil salinization was 9-fold smaller. The control and the low salinity samples don’t separate strongly on the PCA plot (Fig. 3A). The effect of the high salinity treatment was larger and especially pronounced in the straw amended soil.
Figure 2.
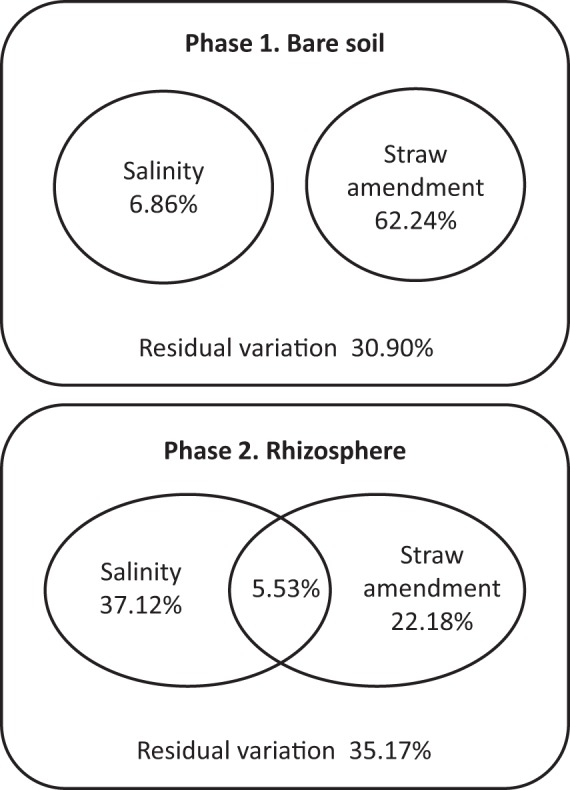
Variation partitioning results. The percentages are the proportion of the variation in the sequencing results which could be explained by salinity or straw amendment. The shared partitions indicate the proportion of variation explained by both factors.
Figure 3.

Principal component analysis plots of phase 1 bare soil (A) and phase 2 wheat rhizosphere samples (B). Circles indicate samples from the straw amendment, squares indicate samples without straw. Control: empty symbols; low salinity: grey symbols; high salinity: black symbols.
In the rhizosphere samples from phase 2, salinity had a larger influence on the bacterial and archaeal community structure, explaining 42.7% of the variation in the data, than the straw amendment which could account for 27.7% (Fig. 2B). In contrast to the bare soil, soil salinization already at a low level caused a change in community structure in the rhizosphere as indicated by the clear separation between the control and low salinity samples on the PCA plot. The high-level salinity, which could only be tested in straw-amended soil, had an even stronger influence on the community structure (Fig. 3B).
SVs responding to soil salinization
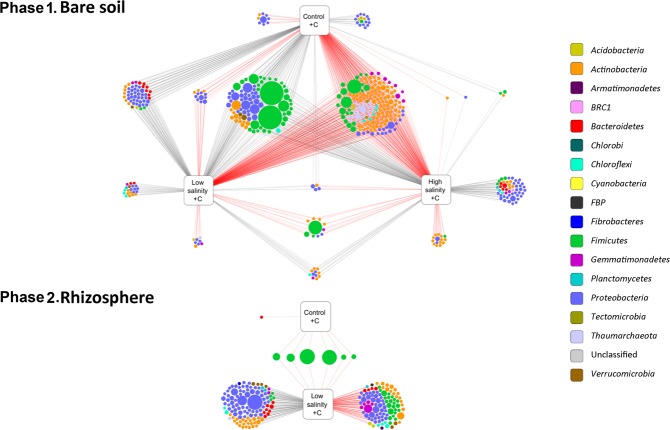
In bare soil from phase 1 without the straw amendment, only six SVs showed a significant change in relative abundance in response to the low salinity treatment: Four SVs, two classified as Proteobacteria and two as Gemmatimonadetes, increased while two from Proteobacteria decreased (Fig. 4, upper panel). In contrast, a total of 44 SVs responded to the high salinity treatment (16 increased, 28 decreased). Most of them were classified as Actinobacteria and Proteobacteria. There was no apparent link between the taxonomic classification of these SVs and the direction of their response to salinity. All SVs that were found to be significantly differentially abundant between treatments are listed in supplementary file 1.
Figure 4.
SVs responding to soil salinity. Circles represent SVs colored according to their phylum-level classification and sized by their average relative abundance. Lines connect SVs to treatments in which their abundance changed significantly compared to the control with the corresponding carbon treatment. Black lines indicate significant increase, red lines significant decrease.
In straw amended bare soil, nine SVs, all of them belonging to Proteobacteria, responded significantly to the low salinity treatment while in the high salinity treatment the relative abundance of 67 SVs increased and 115 SVs decreased significantly compared to the control (Fig. 4, upper panel). The 67 SVs represented eight phyla, the most dominants being Proteobacteria and Actinobacteria. The 115 SVs that responded negatively to high salinity were members of nine phyla including Proteobacteria, Actinobacteria and Firmicutes. The most abundant among these were two SVs from family Bacillaceae (Firmicutes) with 2.24% and 1.54% average relative abundance.
In the phase 2 rhizosphere samples, no SV with a relative abundance larger than 0.1% was found to have a significant response to the low salinity treatment, regardless whether the soil was previously amended with straw or not. However, in the straw-amended soils, 299 SVs showed a significant response to the high salinity treatment: 144 SVs increased and 155 SVs decreased in relative abundance (Fig. 4, lower panel). These SVs covered a wide range of taxonomic diversity representing 13 phyla with a large overlap in the classification of the SVs that responded positively and the ones that responded negatively. A strong difference, however, was found among Firmicutes which, except for a single SV, were only present among the SVs that increased in relative abundance under high salinity. The two most dominants of these SVs were from Planococcaceae with 2.24% and 2.17% average relative abundances. The others belonged to Planococcaceae, Bacillaceae, and Paenibacillaceae. In contrast, Acidobacteria were only present among the SVs that decreased in relative abundance under high salinity with 12 SVs, most of them classified into Blastocatellaceae and subgroups 6 and 10.
SVs responding to the straw amendment
In the bare soil from phase 1, the number of SVs that significantly responded to the straw amendment (468 SVs) was much higher than the number of SVs affected by salinity (219 SVs). The majority, 251 out of 468, of the SVs that changed in relative abundance in response to the straw amendment did so independent of the level of salinity: 79 of them increased while 172 decreased (Fig. 5, upper panel). Most of the 79 SVs that showed significant increase were classified as Bacillales (Firmicutes), Alphaproteobacteria (Proteobacteria), and Actinobacteria. The most dominants of them were two SVs from Planococcaceae (Firmicutes) with 3.93% and 3.90% relative abundances. In comparison, the 172 SVs that responded negatively to the straw amendment under all salinity conditions encompassed a larger taxonomic diversity with members of eight phyla. Actinobacteria was dominant in this group with numerous SVs classified as Rubrobacter, Solirubrobacter, and Gaiellales. Other taxa represented by several SVs in this group included Thaumarchaeota, Deltaproteobacteria, Gemmatimonadetes, and Bacillaceae and Paenibacillaceae (Firmicutes). Only a small number of SVs showed a significant response to the straw amendment in just the control (26 SVs) or just the low salinity (29 SVs) treatments. In contrast, 19.5% of the SVs that significantly changed their relative abundance under high salinity due to the straw amendment were not found to respond significantly in the control and low salinity treatments. Most of these SVs were classified as Alphaproteobacteria and Actinobacteria.
Figure 5.
SVs responding to the straw amendment. Circles represent SVs colored according to their phylum-level classification and sized by their average relative abundance. Lines connect SVs to treatments in which their abundance changed significantly compared to the corresponding salinity treatment without the carbon amendment. Black lines indicate significant increase, red lines significant decrease.
In the rhizosphere samples from phase 2, only seven SVs were found to respond significantly to the straw amendment under the non-saline control conditions (Fig. 5, lower panel). Except for one, these SVs also increased in relative abundance due to the straw addition under low salinity. They were classified as Bacillales (Firmicutes) and included the two abundant Planococcaceae SVs that also responded to the straw amendment, independent of the level of salinity, in the bare soil in phase 1. In contrast to the non-saline control conditions, 217 SVs reacted to the straw amendment in the rhizosphere under low salinity, 106 of them with an increase and 111 with a decrease in relative abundance. Firmicutes, Proteobacteria, and Actinobacteria were dominant among the SVs that increased, and Proteobacteria and Actinobacteria among the ones that showed a negative response.
Discussion
The experimental design of this study allowed distinguishing the effect of a low-level of soil salinization which was stimulatory to the shoot growth of wheat plants, from a high-level salinity which inhibited plant growth unless straw was added to the soil. This demonstrates the efficiency of straw amendment in ameliorating salinity stress on wheat. Under non-saline conditions, root growth was strongly stimulated by the straw amendment likely due to the additional nitrogen and other minerals provided by the straw. Furthermore, the 1 mm-sized straw particles could have altered the soil structure thereby facilitating root growth. Additions of organic particles to soils have been shown to affect root morphology and biomass46. The straw amendment could promote plant growth by counteracting soil compaction which is an important factor in the adverse effects on wheat in salt affected soils47.
The soil used in this study had not experienced previous treatments with saline water, and we hypothesized that the addition of saline water to the bare soil would therefore mobilize soil organic carbon and other nutrients19,48 which would support microbial growth. Such growth would result in higher microbial abundance but concomitantly decrease microbial diversity due to enrichment of a few community members. Salinization did increase the abundance of Bacteria and Archaea in bare soil. However, without straw amendments this effect was found to be small and only detectable under high-level salinity. Fungal abundance was unaffected by soil salinization. Considering the relatively low C content of the soil used in this study, the quantity of this mobilized carbon may have been too small to support strong growth, or this effect was only transient and mostly dissipated over the three weeks equilibration period between the end of the salinization treatment and the sampling. The lack of a strong response in diversity or community structure to salinization in the bare soil was therefore presumably due to the low level of microbial activity. Accordingly, providing available carbon in the form of the straw amendment enhanced the effect of salinity on the richness and structure of the bacterial and archaeal community, largely increasing the number of SVs that changed in relative abundance in response to high-level salinity. Based on the qPCR data, Bacteria and especially Fungi were the main beneficiaries of the straw additions, but the abundance of Archaea significantly declined, indicating that they played a negligible role in straw decomposition. In accordance, the relative abundance of SVs affiliated to Thaumarchaeota decreased in relative abundance in response to straw additions. Most of these archaeal SV were affiliated with Nitrososphaera, which are suspected to perform autotrophic nitrification in soil49. Thus, they are probably less competitive when organic carbon, as supplied by straw, becomes available. SVs representing Gemmatimonadetes also exclusively declined in response to the straw amendment. The addition of wheat straw to soil reduced Gemmatimonadetes in other studies as well50.
In the rhizosphere, our hypothesis that salinity would decrease microbial diversity was found to be true even under the low-level salinity treatment. Salinity also had a much stronger influence on the bacterial and archaeal community structure in the rhizosphere than in the bare soil. As opposed to the bare soil, the rhizosphere is a hot spot of microbial activity where carbon is available from plant root exudates51,52. In this sense, it is more similar to the bare soil with straw amendment where the effect of salinization was also more pronounced. Additionally, physiological changes induced by the salinity stress in the plant13,14 can contribute to the microbial responses in the rhizosphere.
A striking difference was found in the response of Firmicutes SVs to the high-level salinization between bare soil and rhizosphere. In bare soil, they were dominant among the SVs that declined in relative abundance, while in the rhizosphere numerous Firmicutes SVs increased. The phylum Firmicutes thus contains taxa with contrasting survival capacities: one group of SVs represented mainly by Tumebacillus and Bacillaceae that appeared to grow on the nutrients supplied by straw but were sensitive to salinity, while other SVs, mainly members of Planococcaceae but also Paenibacillaceae and Bacillaceae, were abundant in the rhizosphere and tolerant to salinity. All cultivable members of the Planococcaceae isolated from a wide range of relatively extreme environments but not from rhizosphere and semiarid soil, are halotolerant53. Salt-tolerance is also a relatively common property of many bacilli. The fact that members of Bacillaceae occurred in both groups (bare soil – salt-sensitive; rhizosphere – salt-resistant) underlines the high diversity of this taxon, which becomes evident when considering their wide ecological range54,55.
Interestingly, the straw amendment enhanced the effect of soil salinization on bacterial and archaeal richness and community structure in the bare soil, but it mitigated the salinity effect on diversity in the rhizosphere. The former can be explained by the nutrients delivered by the straw inducing microbial growth in the bare soil rendering the community more susceptible to the selective pressure of salinity compared to the less active community in the soil without the straw amendment. In the rhizosphere, on the other hand, carbon is available from root exudates and other rhizodeposits56, and, as shown by the qPCR results, the nutrients from the straw amendment had only minor impact on microbial growth. Therefore, the straw likely decreased the effect of salinity on the rhizosphere microbiota indirectly, by mitigating the salinity stress on the plant.
The results of this study demonstrate the importance of soil microhabitats with different conditions, i.e. the rhizosphere or the presence of straw particles, in modulating the response of soil microbial communities to salinization in semi-arid soils. Contrasting responses of the microbial community were detected. Soil salinization had little or no detectable effect on microbial abundance and prokaryotic diversity in the soil when no external carbon source was provided. On the other hand, it decreased prokaryotic diversity in the rhizosphere of wheat grown in the soil. Straw amendment mitigated this effect. However, it was not a significant nutrient source for the microbial communities in the rhizosphere but more likely acted indirectly by ameliorating the salinity stress on the plant. Diverse responses were found within Proteobacteria, Actinobacteria, and Firmicutes to salinity and the straw amendment indicating the large physiological versatility within these highly diversified phyla.
Supplementary information
Acknowledgements
We thank Britta Müller, Jana Usarek, and Karin Trescher for the excellent technical assistance. The study was financially supported by the German Federal Ministry for Education and Research BMBF (project number 01DN12067), the National Basic Research Program of China (2015CB150501), National Natural Science Foundation of China (41530857), and CONACYT-DLR (B330/252/11).
Author Contributions
Christoph Tebbe, Kornelia Smalla and Thelma Castellanos created the idea of this project. Christoph Tebbe and Astrid Näther designed the experiment. Christoph Tebbe, Thelma Castellanos and Angel Carrillo selected the sampling sites and collected the soils. Astrid Näther carried out the laboratory work with support from Angel Carrillo. Márton Szoboszlay and Bei Liu analyzed the data gathered in this project in consultation with Christoph Tebbe. Márton Szoboszlay and Christoph Tebbe wrote the manuscript. Zhongjun Jia, Kornelia Smalla, and Thelma Castellanos provided comments and consultation.
Data Availability
Sequencing data generated in this study is available from the European Nucleotide Archive under the accession number PRJEB30355.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Márton Szoboszlay and Astrid Näther contributed equally.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-46070-6.
References
- 1.Foley JA, et al. Global consequences of land use. Science. 2005;309:570–574. doi: 10.1126/science.1111772. [DOI] [PubMed] [Google Scholar]
- 2.Verges FAR, Velazquez A, Bocco G, Espejel I. Multi-scale land cover dynamics of semiarid scrubland in Baja California, Mexico. Regional Environmental Change. 2014;14:1315–1328. doi: 10.1007/s10113-013-0574-8. [DOI] [Google Scholar]
- 3.Belnap J. The world at your feet: Desert biological soil crusts. Frontiers in Ecology and the Environment. 2003;1:181–189. doi: 10.1890/1540-9295(2003)001[0181:TWAYFD]2.0.CO;2. [DOI] [Google Scholar]
- 4.Bowker MA, Maestre FT, Escolar C. Biological crusts as a model system for examining the biodiversity-ecosystem function relationship in soils. Soil Biology & Biochemistry. 2010;42:405–417. doi: 10.1016/j.soilbio.2009.10.025. [DOI] [Google Scholar]
- 5.Ding Guo-Chun, Piceno Yvette M., Heuer Holger, Weinert Nicole, Dohrmann Anja B., Carrillo Angel, Andersen Gary L., Castellanos Thelma, Tebbe Christoph C., Smalla Kornelia. Changes of Soil Bacterial Diversity as a Consequence of Agricultural Land Use in a Semi-Arid Ecosystem. PLoS ONE. 2013;8(3):e59497. doi: 10.1371/journal.pone.0059497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Judkins G, Myint S. Spatial variation of soil salinity in the Mexicali Valley, Mexico: Application of a practical method for agricultural monitoring. Environmental Management. 2012;50:478–489. doi: 10.1007/s00267-012-9889-3. [DOI] [PubMed] [Google Scholar]
- 7.Butcher K, Wick AF, DeSutter T, Chatterjee A, Harmon J. Soil salinity: A threat to global food security. Agronomy Journal. 2016;108:2189–2200. doi: 10.2134/agronj2016.06.0368. [DOI] [Google Scholar]
- 8.Shrivastava P, Kumar R. Soil salinity: A serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi Journal of Biological Sciences. 2015;22:123–131. doi: 10.1016/j.sjbs.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maas, E. V. In Agricultural salinity assessment and management Vol. 71 Manuals and Reports on Engineering (ed. Tanji, K. K.) 262–304 (American Society of Civil Engineering, 1990).
- 10.Chinnusamy V, Jagendorf A, Zhu JK. Understanding and improving salt tolerance in plants. Crop Sci. 2005;45:437–448. doi: 10.2135/cropsci2005.0437. [DOI] [Google Scholar]
- 11.Munns R, Gilliham M. Salinity tolerance of crops - what is the cost? New Phytologist. 2015;208:668–673. doi: 10.1111/nph.13519. [DOI] [PubMed] [Google Scholar]
- 12.Wang M, Xia GM. The landscape of molecular mechanisms for salt tolerance in wheat. Crop Journal. 2018;6:42–47. doi: 10.1016/j.cj.2017.09.002. [DOI] [Google Scholar]
- 13.Jung, J. K. H. & McCouch, S. Getting to the roots of it: genetic and hormonal control of root architecture. Frontiers in Plant Science4, 10.3389/fpls.2013.00186 (2013). [DOI] [PMC free article] [PubMed]
- 14.Rahnama A, Munns R, Poustini K, Watt M. A screening method to identify genetic variation in root growth response to a salinity gradient. Journal of Experimental Botany. 2011;62:69–77. doi: 10.1093/jxb/erq359. [DOI] [PubMed] [Google Scholar]
- 15.Qin Y, Druzhinina IS, Pan XY, Yuan ZL. Microbially mediated plant salt tolerance and microbiome-based solutions for saline agriculture. Biotechnology Advances. 2016;34:1245–1259. doi: 10.1016/j.biotechadv.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 16.Vimal SR, Singh JS, Arora NK, Singh S. Soil-plant-microbe interactions in stressed agriculture management: A Review. Pedosphere. 2017;27:177–192. doi: 10.1016/S1002-0160(17)60309-6. [DOI] [Google Scholar]
- 17.Costa, O. Y. A., Raaijmakers, J. M. & Kuramae, E. E. Microbial extracellular polymeric substances: Ecological function and impact on soil aggregation. Frontiers in Microbiology9, 10.3389/fmicb.2018.01636 (2018). [DOI] [PMC free article] [PubMed]
- 18.Upadhyay SK, Singh JS, Singh DP. Exopolysaccharide-producing plant growth-promoting rhizobacteria under salinity condition. Pedosphere. 2011;21:214–222. doi: 10.1016/S1002-0160(11)60120-3. [DOI] [Google Scholar]
- 19.Setia R, Rengasamy P, Marschner P. Effect of exchangeable cation concentration on sorption and desorption of dissolved organic carbon in saline soils. Science of the Total Environment. 2013;465:226–232. doi: 10.1016/j.scitotenv.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 20.Wang WX, Vinocur B, Altman A. Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta. 2003;218:1–14. doi: 10.1007/s00425-003-1105-5. [DOI] [PubMed] [Google Scholar]
- 21.Fita, A., Rodriguez-Burruezo, A., Boscaiu, M., Prohens, J. & Vicente, O. Breeding and domesticating crops adapted to drought and salinity: A new paradigm for increasing food production. Frontiers in Plant Science6, 10.3389/fpls.2015.00978 (2015). [DOI] [PMC free article] [PubMed]
- 22.Jacobsen SE, Jensen CR, Liu F. Improving crop production in the arid Mediterranean climate. Field Crops Research. 2012;128:34–47. doi: 10.1016/j.fcr.2011.12.001. [DOI] [Google Scholar]
- 23.Plaut Z, Edelstein M, Ben-Hur M. Overcoming salinity barriers to crop production using traditional methods. Critical Reviews in Plant Sciences. 2013;32:250–291. doi: 10.1080/07352689.2012.752236. [DOI] [Google Scholar]
- 24.Sakadevan, K. & Nguyen, M. L. In Advances in Agronomy, Vol 109 Vol. 109 Advances in Agronomy (ed. Sparks, D. L.) 55–74 (2010).
- 25.Diacono M, Montemurro F. Effectiveness of organic wastes as fertilizers and amendments in salt-affected soils. Agriculture. 2015;5:221–230. doi: 10.3390/agriculture5020221. [DOI] [Google Scholar]
- 26.Li XG, Li FM, Cui ZJ, Rengel Z. Decomposition of maize straw in saline soil. Biology and Fertility of Soils. 2006;42:366–370. doi: 10.1007/s00374-005-0042-9. [DOI] [Google Scholar]
- 27.Li JG, et al. Soil salinization research in China: Advances and prospects. Journal of Geographical Sciences. 2014;24:943–960. doi: 10.1007/s11442-014-1130-2. [DOI] [Google Scholar]
- 28.Wahid A, Akhtar S, Ali I, Rasul E. Amelioration of saline-sodic soils with organic matter and their use for wheat growth. Communications in Soil Science and Plant Analysis. 1998;29:2307–2318. doi: 10.1080/00103629809370112. [DOI] [Google Scholar]
- 29.Wichern J, Wichern F, Joergensen RG. Impact of salinity on soil microbial communities and the decomposition of maize in acidic soils. Geoderma. 2006;137:100–108. doi: 10.1016/j.geoderma.2006.08.001. [DOI] [Google Scholar]
- 30.Arriaga L, Maya Y. Spatial variability in decomposition rates in a desert scrub of Northwestern Mexico. Plant Ecology. 2007;189:213–225. doi: 10.1007/s11258-006-9178-4. [DOI] [Google Scholar]
- 31.Maya Y, Arriaga L. Litterfall and phenological patterns of the dominant overstorey species of a desert scrub community in north-western Mexico. Journal of Arid Environments. 1996;34:23–35. doi: 10.1006/jare.1996.0090. [DOI] [Google Scholar]
- 32.Rhoades, J. D. In Advances in Agronomy, Vol 49 Vol. 49 Advances in Agronomy (ed. Sparks, D. L.) 201–251 (1993).
- 33.Staff, S. S. In Soil Survey Investigations Report Vol. 42 (U.S. Department of Agriculture, Natural Resources Conservation Service, 2014).
- 34.Klindworth A, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:11. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microb. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Callahan BJ, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–3. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pruesse E, et al. Nucleic Acids Res. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB; pp. 7188–7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barger K, Bunge J. Objective Bayesian estimation for the number of species. Bayesian Anal. 2010;5:765–785. doi: 10.1214/10-BA527. [DOI] [Google Scholar]
- 39.Willis A, Bunge J. Estimating diversity via frequency ratios. Biometrics. 2015;71:1042–1049. doi: 10.1111/biom.12332. [DOI] [PubMed] [Google Scholar]
- 40.Willis A, Bunge J, Whitman T. Improved detection of changes in species richness in high diversity microbial communities. J. R. Stat. Soc. Ser. C-Appl. Stat. 2017;66:963–977. doi: 10.1111/rssc.12206. [DOI] [Google Scholar]
- 41.Palarea-Albaladejo J, Martin-Fernandez JA. zCompositions - R Package for multivariate imputation of left-censored data under a compositional approach. Chemometrics Intell. Lab. Syst. 2015;143:85–96. doi: 10.1016/j.chemolab.2015.02.019. [DOI] [Google Scholar]
- 42.Gloor GB, Wu JR, Pawlowsky-Glahn V, Egozcue JJ. It’s all relative: analyzing microbiome data as compositions. Ann. Epidemiol. 2016;26:322–329. doi: 10.1016/j.annepidem.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 43.Oksanen, J. et al. vegan: Community Ecology Package. Ordination methods, diversity analysis and other functions for community and vegetation ecologists. Version 2.4-6, https://CRAN.R-project.org/package=vegan. (2018).
- 44.Fernandes AD, et al. Unifying the analysis of high-throughput sequencing datasets: characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis. Microbiome. 2014;2:13. doi: 10.1186/2049-2618-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benjamini Y, Hochberg Y. Controlling false discovery rate - A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B-Methodol. 1995;57:289–300. [Google Scholar]
- 46.Olmo M, Villar R, Salazar P, Alburquerque JA. Changes in soil nutrient availability explain biochar’s impact on wheat root development. Plant and Soil. 2016;399:333–343. doi: 10.1007/s11104-015-2700-5. [DOI] [Google Scholar]
- 47.Saqib M, Akhtar J, Qureshi RH. Pot study on wheat growth in saline and waterlogged compacted soil I. Grain yield and yield components. Soil & Tillage Research. 2004;77:169–177. doi: 10.1016/j.still.2003.12.004. [DOI] [Google Scholar]
- 48.Daliakopoulos IN, et al. The threat of soil salinity: A European scale review. Science of the Total Environment. 2016;573:727–739. doi: 10.1016/j.scitotenv.2016.08.177. [DOI] [PubMed] [Google Scholar]
- 49.Offre, P., Spang, A. & Schleper, C. Archaea in biogeochemical cyccles, in Annual Review of Microbiology, Vol 67 (ed. Gottesman, S.) 437–457 (2013). [DOI] [PubMed]
- 50.Li CH, Yan K, Tang LS, Jia ZJ, Li Y. Change in deep soil microbial communities due to long-term fertilization. Soil Biology & Biochemistry. 2014;75:264–272. doi: 10.1016/j.soilbio.2014.04.023. [DOI] [Google Scholar]
- 51.Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM. The role of root exudates in rhizosphere interations with plants and other organisms. Annu Rev Plant Biol. 2006;57:233–266. doi: 10.1146/annurev.arplant.57.032905.105159. [DOI] [PubMed] [Google Scholar]
- 52.Kuzyakov Y, Xu XL. Competition between roots and microorganisms for nitrogen: mechanisms and ecological relevance. New Phytologist. 2013;198:656–669. doi: 10.1111/nph.12235. [DOI] [PubMed] [Google Scholar]
- 53.See-Too WS, et al. Planococcus versutus sp nov., isolated from soil. International Journal of Systematic and Evolutionary Microbiology. 2017;67:944–950. doi: 10.1099/ijsem.0.001721. [DOI] [PubMed] [Google Scholar]
- 54.Ceuppens S, Boon N, Uyttendaele M. Diversity of Bacillus cereus group strains is reflected in their broad range of pathogenicity and diverse ecological lifestyles. FEMS Microbiology Ecology. 2013;84:433–450. doi: 10.1111/1574-6941.12110. [DOI] [PubMed] [Google Scholar]
- 55.Earl AM, Losick R, Kolter R. Ecology and genomics of Bacillus subtilis. Trends in Microbiology. 2008;16:269–275. doi: 10.1016/j.tim.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dennis PG, Miller AJ, Hirsch PR. Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiology Ecology. 2010;72:313–327. doi: 10.1111/j.1574-6941.2010.00860.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data generated in this study is available from the European Nucleotide Archive under the accession number PRJEB30355.