Abstract
Base editing systems show their power in modeling and correcting the pathogenic mutations of genetic diseases. Previous studies have already demonstrated the editing efficiency of BE3-mediated C-to-T conversion in human embryos. However, the precision and efficiency of a recently developed adenine base editor (ABE), which converts A-to-G editing in human embryos, remain to be addressed. Here we selected reported pathogenic mutations to characterize the ABE in human tripronuclear embryos. We found effective A-to-G editing occurred at the desirable sites using the ABE system. Furthermore, ABE-mediated A-to-G editing in the single blastomere of the edited embryos exhibited high product purity. By deep sequencing and whole-genome sequencing, A or T mutations didn’t increase significantly, and no off-target or insertion or deletion (indel) mutations were detected in these edited embryos, indicating the ABE-mediated base editing in human embryos is precise and controllable. For some sites, since a different editing pattern was obtained from the cells and the embryos targeted with the same single guide RNA (sgRNA), it suggests that ABE-mediated editing might have different specificity in vivo. Taken together, we efficiently generated pathogenic A-to-G mutations in human tripronuclear embryos via ABE-mediated base editing.
Introduction
Next-generation sequencing technology has greatly advanced the discovery of pathogenic mutations in humans. Consequently, the therapeutic approaches have become the focus of modern human genetic research. Gene therapy has been considered as the most desirable strategy for repairing pathogenic mutations in human genetic diseases. Unfortunately, it was very difficult to correct these pathogenic mutations until the arrival of the revolutionary CRISPR/Cas9 technology.1, 2, 3, 4 Nevertheless, it is of great concern that CRISPR targeting is uncontrollable with insertion or deletion (indel) mutations.5 A cytosine base editor (BE3/BE4) opens the potential of gene therapy by precise base editing in different species, including plant6 and mouse,7 and even in humans.8, 9, 10 However, its potential is limited by the fact that CBEs (cytosine base editors) only convert the C to T and possibly induce off-target by using APOBEC.11, 12 A recently developed adenine base editor (ABE) system that converts A to G with the action of an engineered adenine deaminase ecTadA provides us another possible precise tool for gene therapy.13, 14 To demonstrate its feasibility, here we aimed to characterize the effectiveness of the ABE in human embryos.
Results
The ABE System Exhibits Efficient Editing at Desirable Sites in HEK293T Cells
To comprehensively analyze the characteristics of the ABE system in human embryos, we produced the A-to-G mutation in human cells and embryos using ABE7.10. Five genes, TTR,15 ALDOB,16 COL9A2,17 KCNJ11,18 and RPE65,19 which had been reported to have pathogenic A-to-G point mutations, were selected (Figure S1A). We first tested the editing of these genes with the ABE system in HEK293T cells. At 3 days after transfection of ABE7.10 with the corresponding single guide RNAs (sgRNAs) targeting these five genes individually, the genomic DNA sample was extracted from the transfected HEK293T cells, and it was used as the template to amplify the target regions by PCR. The PCR product was subjected to Sanger sequencing.
As expected, double A and double G peaks were observed in the sequencing chromatogram (Figure S1B), indicating the successful A-to-G editing of the targeted sites. Then, we further calculated the base editing efficiency for each target A site based on three independent experiments, by using the EditR software (https://moriaritylab.shinyapps.io/editr_v10/). The results showed that successful base editing was achieved at all targeted sites, but with different editing efficiencies (Figure S1C). The A-to-G conversions were further confirmed for two target sites with the highest editing efficiencies, the A6 of TTR (editing efficiency, 75%) and the A4 of RPE65 (editing efficiency, 71%), by sequencing the PCR product derived from these two genomic DNA samples after the TA cloning (Figure S1D). It is worth noting that, besides the base editing at the desirable sites, the A3 for TTR and the A2 for RPE65 were also edited with an efficiency of about 15% (3 in 20 clones) and 12% (2 in 17 clones), respectively. Taken together, the pathogenic A-to-G mutations of the selected genes had been successfully achieved by the ABE system in HEK293T cells.
The ABE System Efficiently Converts A to G in Discarded Human Tripronuclear Embryos
Then, we tested the ABE in discarded human tripronuclear embryos. First, two reported sgRNAs targeting SITE2 and SITE6,13 each together with mRNA of ABE7.10, were microinjected into the human tripronuclear embryos as described previously.9 3 days later, 12 embryos for each site were obtained for genotyping by Sanger sequencing of the PCR product (Table 1). The editing efficiency was calculated by using EditR (Table S1). The results showed that the A7 of SITE6 was efficiently edited in all embryos. Meanwhile, A3 editing was observed at SITE6 in some embryos (6, 8, and 9). For SITE2, A5 was efficiently edited in all samples, and A2 editing and A8 editing were also detected in several embryos (21 and 22). Notably, editing occurred in most of the SITE2 samples (92%) at an efficiency of more than 80% (Table 1).
Table 1.
Summary of the Used Embryos Edited with ABE
| Serial Number of Injection | Used sgRNA | Edited Embryo |
Blastomere |
|||||
|---|---|---|---|---|---|---|---|---|
| Number | Efficiency >50% |
Efficiency >80% |
Number | 2 Edited Alleles |
3 Edited Alleles |
|||
| Number (%) | Number (%) | Number (%) | Number (%) | |||||
| 1 | SITE6 | 12 | 7 (58) | 3 (25) | NA | |||
| 2 | SITE2 | 12 | 12 (100) | 11 (92) | NA | |||
| 3 | ALDOB | 7 | 0 (0) | 0 (0) | NA | |||
| 4 | COL9A2 | 7 | 6 (86) | 4 (57) | NA | |||
| 5 | KCNJ11 | 7 | 0 (0) | 0 (0) | NA | |||
| 6 | TTR | 12 | 12 (100) | 10 (83) | NA | |||
| 7 | RPE65 | 10 | 6 (60) | 0 (0) | NA | |||
| 8 | TTR + RPE65 | TTR | 10 | 10 (100) | 9 (90) | NA | ||
| RPE65 | 10 | 9 (90) | 5 (50) | NA | ||||
| 9 | TTR | 6 | – | – | 48 | 46 (96) | 37 (77) | |
NA, not applicable.
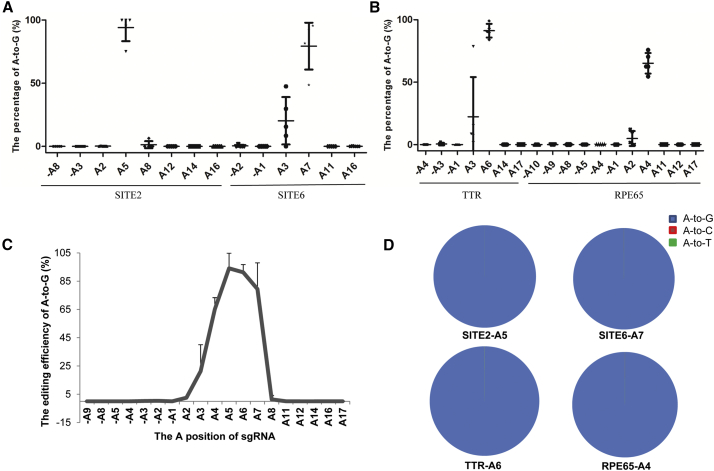
To further test the base conversion efficiency, the DNA samples targeting SITE6 in five embryos (5–7, 9, and 12) and targeting SITE2 in five other embryos (13, 14, and 17–19) were randomly selected for deep sequencing (Table S1). Then, the editing efficiency was measured for all As 10 bp upstream of the sgRNA target region as well as within the sgRNA region. We consistently observed that the A5 of SITE2 and the A7 of SITE6 were edited with the highest efficiencies. In addition, low levels of editing were observed at the A8 and the A2 of SITE2 and the A3 of SITE6 (Figure 1A). No editing occurred at any other sites. Taken together, we achieved efficient A-to-G editing in human embryos with ABE7.10.
Figure 1.
Generation of Specific Pathogenic Point Mutations in Human Tripronuclear Embryos Using the ABE System
(A) Analysis of the ABE-mediated A-to-G editing of the reported sites in human embryos by deep sequencing. Two reported sites (SITE2 and SITE6) were selected to test the A-to-G editing in human tripronuclear embryos. The editing of all A sites 10 bp upstream of the sgRNA and within the sgRNA was analyzed by deep sequencing. Data of the detected embryos described in Table S1 are shown as the mean ± SD (n = 5). (B) Analysis of ABE-mediated A-to-G editing of the novel sites in human embryos by deep sequencing. Two novel sites, TTR and RPE65, were selected for A-to-G editing in human tripronuclear embryos. The editing of all the A sites 10 bp upstream and within the sgRNA was analyzed by deep sequencing. Data of the detected embryos described in Table S1 are shown as the mean ± SD (n = 5). (C) Characterization of the ABE-mediated A-to-G substitution at different positions in human tripronuclear embryos. (D) Analysis of different adenine substitutions of the target sites. The positions with most efficient editing of the 4 target sites were selected for analysis.
Furthermore, the editing at TTR and RPE65 was tested in 12 and 10 respective human tripronuclear embryos, as described above (Table 1). Sanger sequencing of the PCR product showed that the A-to-G conversion occurred in most of the embryos at the pathogenic mutation sites with A6 for TTR and A4 for RPE65, respectively (Table S1). Similarly, some embryos (embryos 47–49, 51, and 53 for TTR and embryos 58, 60, and 64–66 for RPE65) were randomly selected for deep sequencing to further characterize the editing efficiencies and specificities. We consistently observed efficient editing at the A6 of TTR and the A4 of RPE65. Low levels of editing were observed at the A3 of TTR and the A2 of RPE65 (Figure 1B). Based on the deep sequencing results of the above 4 target sites, ABE induces the best base editing efficiencies at the positions 4–7 of the protospacer region in human embryos (Figure 1C). Editing at A3 was also observed, as has been reported before.13 Meanwhile, we also analyzed the fractions of adenine substitutions for the efficiently edited sites of SITE2, SITE6, TTR, and RPE65. Indeed, almost all of the conversions were A to G (Figure 1D), indicating that the ABE system produces highly pure products in human embryos. We also analyzed the indels for TTR and RPE65 and, as expected, no indel was detected (Figure S2).
Sometimes it is necessary to model multiple pathogenic mutations, so we tried to simultaneously edit multiple sites by ABE. Similar to what was described in BE3-mediated multiple C-to-T conversions,9 we simultaneously microinjected two sgRNAs, TTR and RPE65, into 10 embryos (Table S1). After sequencing, the editing efficiencies at the desirable A6 of TTR and A4 for RPE65 were 91% and 70%, respectively (Figure S3A). To further characterize the editing efficiencies of multiple sites in human embryos, five embryos (70, 72, 74, 76, and 77) were randomly selected for deep sequencing. Analysis of the deep sequencing data showed that simultaneous editing by ABE produces similar editing efficiencies and patterns at both TTR and RPE65 (Figure S3B), suggesting that the ABE system could be used to manipulate multiple pathogenic point mutations with high efficiency and accuracy.
The Editing Characteristic in Embryos Is Different from that in 293T Cells for Some Sites
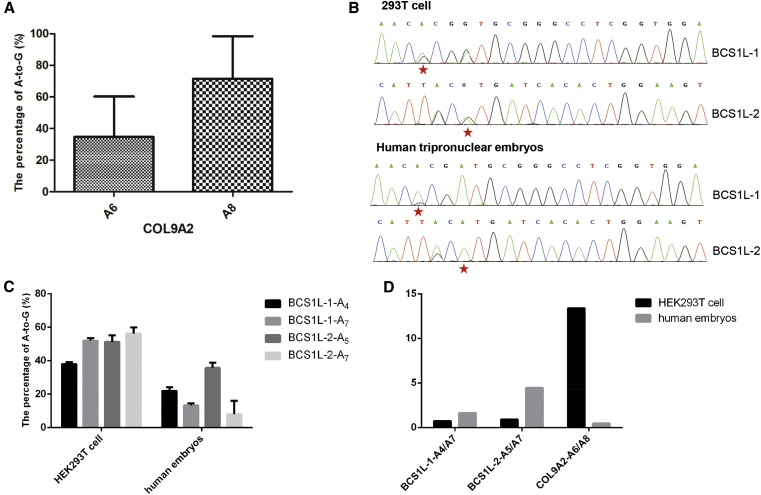
Next, three targets, ALDOB, COL9A2, and KCNJ11, with poor editing in HEK293T cells were tested for the editing efficiencies in human embryos. Each sgRNA together with mRNA of ABE7.10 was microinjected into 7 tripronuclear embryos, respectively (Table 1). We found the sgRNAs of ALDOB and KCNJ11 still gave rise to low base conversion efficiencies in human tripronuclear embryos, similar to those observed in HEK293T cells (Figure S4). However, COL9A2 showed highly efficient editing at A8 in human tripronuclear embryos (Figure 2A), which was hardly detectable in HEK293T cells (Figure S1C), indicating the differences of ABE-mediated base editing between cells and embryos. Moreover, we found 2 other sites (BCS1L-1 and BCS1L-2 sites) using the ABE-NG system that recognized the non-NGG protospacer adjacent motif (PAM) sequences (Figure 2B). The analysis confirmed the differences of ABE-mediated base editing between cells and embryos (Figures 2C and 2D).
Figure 2.
The Editing Characteristic in Embryos Is Different from that in 293T Cells
(A) The editing efficiency of A6 and A8 for the COL9A2 gene in embryos was calculated. Data from three independent experiments are shown as means ± SD. (B) Two target sequences that contained the GGA or AGT PAM were edited using ABE-NG. The representative chromatograms of the Sanger sequencing of target sites from genomic DNA of HEK293T cells (top) and human tripronuclear embryos (bottom) are shown. The red star indicates the pathogenic point. (C) The A4 and A7 sites for the BCS1L-1 gene site were calculated by EditR software via Sanger sequencing of the PCR products derived from the target sites. Data are shown as the mean ± SD (n = 3). The A5 and A7 sites for the BCS1L-2 gene site were calculated with the same method. (D) The ratios of editing efficiency for different points of the three gene sites were calculated using the previous data.
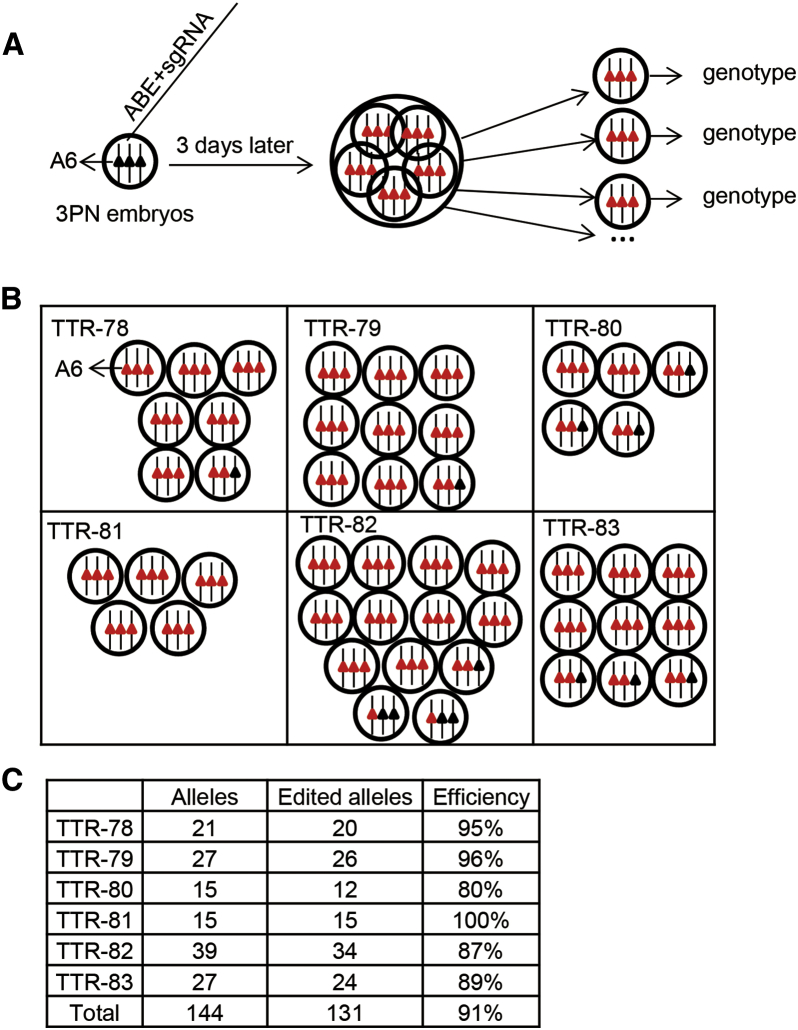
Analysis of Blastomeres for TTR-Edited Embryos
As described above, deep sequencing analysis proves the efficiency and product purity of ABE-mediated base editing in human embryos. This suggests that there is only a low level of other kinds of genotypes in ABE-mediated base editing. To further address this concern, we performed the genotyping analysis for every single blastomere of the edited embryos (Figure 3A). The blastomeres collected from 6 TTR-edited embryos were used for genotyping (Figure 3B). The results showed that the base substitutions occurred at pathogenic A6 with nearly perfect efficiencies for three TTR alleles in every triploid blastomere of the edited embryos (78, 95%; 79, 96%; 80, 80%; 81, 100%; 82, 87%; and 83, 89%) (Figure 3C), indicating the ABE-mediated base editing in human embryos is controllable.
Figure 3.
Analysis of ABE-Mediated Base Editing of the TTR Gene in Blastomeres
(A) Schematic illustration of the experimental procedure for analysis of ABE-mediated base editing in triploid blastomeres. A6 in the target sites was detected. Each string presents one allele in the tripronuclear embryos. The black triangle means adenine and the red triangle means guanine. The genotype was calculated via Sanger sequencing of the target sites. (B) Six embryos for the TTR gene were divided into triploid blastomeres. Each box indicates one embryo. The black triangle means adenine and the red triangle means guanine. (C) Summary of the alleles with the edited A6 site in triploid blastomeres.
Off-Target Analysis by Deep Sequencing and Whole-Genome Sequencing
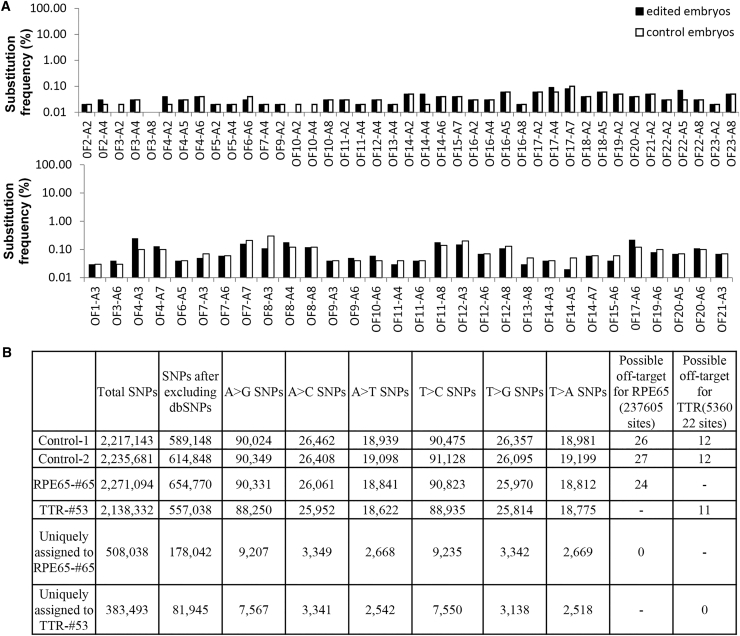
Another concern about genome editing is the off-target base editing.20 To explore the off-target edited sites of ABE in human embryos, the potential off-target sites of the sgRNAs for TTR and RPE65 were predicted by using three different available tools.21, 22, 23 In total, there were 21 predicted potential off-target sites of the TTR sgRNA (Table S2) and 23 off-target sites of the RPE65 sgRNA (Table S3). Then, 10 edited embryos (68–77) were used for the detection of the off-target sites, with 5 embryos injected with the ABE mRNA and the scramble sgRNA as the control. The PCR products of all edited embryos were mixed with an equal amount of DNA for each embryo and subjected to deep sequencing. The results showed that there were no obviously detectable off-target sites (Figure 4A).
Figure 4.
Off-Target Site Analysis
(A) Totals of 23 off-target sites for RPE65 (top) and 21 off-target sites for TTR (bottom) were selected. The off-target frequencies were calculated by deep sequencing. The adenine sites within the positions of 2–8 of sgRNA were examined for off-target frequencies. (B) Whole-genome-wide sequencing analysis was conducted for two control embryos and two edited embryos.
To further demonstrate the fidelity of ABE-mediated base editing in human embryos, we performed whole-genome sequencing (WGS) on genomic DNA isolated from 2 control embryos, TTR-53 and RPE65-65, at the depth of 30–40×. Totals of 237,605 sites for RPE65 and 536,022 sites for TTR, with up to 5-bp mismatches and equal or less than one DNA or RNA bulge in the sgRNA-targeting region, were analyzed. After excluding the false-positive sites depending on The Single Nucleotide Polymorphism Databases (dbSNPs), the mutation frequency of A or T didn’t seem to increase significantly, and no off-target was uniquely assigned to both tested samples (Figure 4B). These results indicated that ABE can mediate precise base editing with high efficiency and fidelity in human embryos.
Discussion
For human genetic diseases, gene therapy has become an ideal alternative strategy to prevent the germline transmission of the founder mutations. To date, there are about 10,000 reported monogenic inherited mutations that may be generated by precise gene-editing technologies. Previously, we have reported the success to precisely convert C to T in human embryos by using a cytosine base editor. However, nearly 50% of pathogenic point mutations were caused by G-to-A conversion instead.13 The newly developed ABE, which converts A to G, provides a potential tool for creating these pathogenic mutations. To test its potential, we performed the ABE-mediated A-to-G editing by microinjection of ABE mRNA together with target sgRNA. As expected, we observed efficient ABE-mediated A-to-G conversion.
Off-target is rare with base editors.5 Indeed, A or T mutations didn’t increase significantly, and no obvious off-target sites were observed in the edited human embryos by ABE based on thorough deep sequencing and whole-genome sequencing analysis, further evidence for BEs as the safe genome editors.24 More interestingly, no other base conversions (A to C or A to T) were induced by ABE, although there was low level of A-to-G conversion at the undesirable sites. This kind of undesirable A-to-G conversion may be prevented by other means, such as the use of different sgRNAs or Cas9. It is worth noting that the ABE-mediated editing showed different editing patterns between HEK293T cells and human embryos for some sites, indicating different specificity of ABE-mediated editing in vivo compared with that in cells. We found 77% (37/48) of the blastomeres harbored the same base conversion at the pathogenic site (Table 1); no other genotype was found, suggesting that ABE-mediated editing is more controllable. Nevertheless, whether a ribonucleoprotein (RNP) system and metaphase II (MII) oocyte injection can further increase the efficiency of base editing need to be tested.25
In summary, we successfully achieved and characterized the ABE-mediated editing in human embryos. The results demonstrated that ABE-mediated base editing was efficient and precise, providing a novel strategy for modeling and correcting the pathogenic G-to-A point mutation.
Materials and Methods
Discarded Human Tripronuclear Zygotes
The study was approved by the Ethics Committee of the Third Affiliated Hospital of Guangzhou Medical University. All of the used tripronuclear zygotes were donated by the patients undergoing in vitro fertilization (IVF) treatment at the Center for Reproductive Medicine of the hospital. The patients signed the informed consent before sample collection.
Plasmid Construction and In Vitro Transcription
The sequence of ABE7.10 was synthesized and cloned into mammal expression plasmid with cytomegalovirus (CMV) or T7 promoter. We replaced the puromycin element of pGL3-U6-sgRNA-phosphoglycerate kinase (PGK)-puromycin (Addgene, 51133) with the GFP sequence to produce the pGL3-U6-sgRNA-PGK-GFP plasmid. The oligos for the used sgRNAs were synthesized and respectively cloned into the pGL3-U6-sgRNA-PGK-GFP vector and pUC57-sgRNA expression vector (Addgene, 51132). All of the plasmids were extracted and quantified with Nanodrop 2000. The pGL3-U6-sgRNA-PGK-GFP plasmids were used to transfect into the HEK293T cells. The pUC57-sgRNA plasmids were used as the template for in vitro transcription. The ABE was transcribed in vitro according to the reported protocol.9 Briefly, the plasmid was digested using BbsI and then was purified using a PCR clean-up kit (Axygene, AP-PCR-250). The linearized plasmid was used as the template to generate the mRNA transcript. The sgRNA was transcribed according to the reported protocol.9 The concentration of the transcribed RNA was measured with Nanodrop 2000, and the RNA samples were stored at −80°C before use for microinjection.
Cell Culture, Transfection, and Identification
HEK293T cells were maintained in DMEM-high glucose (HyClone, SH30243.01) with 10% fetal bovine serum (FBS) (v/v) (Gemini, 900-108). The ABE plasmid was transfected into the HEK293T cells with the corresponding sgRNA expression plasmid by using Lipofectamine 2000 reagent (Life Technologies), according to the manufacturer’s suggested protocols. 3 days later, 50,000 GFP-positive cells were sorted using flow cytometry. The collected cells were lysed in the buffer containing 50 mM KCl, 1.5 mM MgCl2, 10 mM Tris (pH 8.0), 0.5% Nonidet P-40, 0.5% Tween 20, and 100 μg/mL protease K at 65°C for 30 min. Then, the samples were heated at 98°C for 3 min. The target sequences were amplified and sequenced using the primers corresponding to each target gene (Table S4).
Embryo Injection and Culture
The tripronuclear zygotes were picked after 16–18 h of IVF under microscope. The RNA solution was diluted at the concentration of 100 ng/μL for ABE-generated mRNA and 50 ng/μL for sgRNA. The procedure for microinjection was the same as the reported method by using a micromanipulator (Eppendorf, Germany).26 The embryos were kept in embryo culture medium (Vitrolife, Sweden) for 3 days. Then the whole embryos were collected for experiments, or the blastomeres were obtained from these embryos with the acidic Tyrode’s solution (Solarbio, Beijing, China) before use.
Whole-Genome Amplification of Single Embryos
The collected embryos and blastomeres were amplified using Discover-sc Single Cell Kit (Vazyme, N601-01). The product was diluted 100-fold before it was used as the template to amplify the target sites.
Deep Sequencing of On-Target and Off-Target Sites
The off-target sites for TTR and RPE65 were predicted by using three reported softwares (https://zlab.bio/guide-design-resources; https://crispr.cos.uni-heidelberg.de/; http://www.rgenome.net/cas-offinder/)21, 22, 23. The on-target and off-target sites for the edited embryos (Table S1) were amplified using super-fidelity DNA polymerase (Vazyme, p505). The purified PCR products were submitted to sequencing using Hiseq X-10 (2 × 150) platform at CAS-MPG Partner Institute for Computational Biology Omics Core, Shanghai, China. The deep sequencing data were processed using the Burrows-Wheeler Aligner-maximal exact matches (BWA-MEMs) algorithm. The column diagram about the editing efficiency was produced using GraphPad.
Whole-Genome Sequencing for the Off-Target Detection
To comprehensively analyze the off-targets induced by ABE in human embryos, whole-genome sequencing was performed with a depth of 30×–40× using an Illumina HiSeq X Ten (2 × 150 paired end [PE]) at HuaGen Biotech Institute, Shanghai, China. BWA version (v.)0.7.16 was used to map the sequencing data with a human reference genome (GRCh38/hg38). Sequence reads were marked for duplicates using Sambamba v.0.6.7 and realigned using Genome Analysis Toolkit (GATK, v.3.7) IndelRealigner. Variants were identified by GATK HaplotypeCaller, and we evaluated the quality using GATK VariantFiltration. The putative off-target sites were obtained from Cas-OFFinder software, with up to 5 mismatches and equal or less than one DNA or RNA bulge. To figure out the potential off-target sites, A to G was picked out. The common SNPs between the edited and control embryos were discarded.
Data Availability
All the deep sequencing data can be viewed in the National Omics Data Encyclopedia (NODE) (https://www.biosino.org/node/) by pasting the accession (OEP000192) into the text search box or through the URL https://www.biosino.org/node/project/detail/OEP000192. The whole-genome sequencing data have been deposited in the NCBI Sequence Read Archive database (SRA: PRJNA480397).
Author Contributions
J.L., X.H., L.W., and G.L. conceived and designed the project. G.L., X.L., S.H., Y. Zeng, G.Y., Z.L., X.M., and Y. Zhang performed the experiments. X.H., J.L., and L.W. supervised the project. X.H. and G.L. analyzed the data and wrote the paper.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We thank members of the Huang and Liu labs for helpful discussions. We are grateful to Dr. Xiajun Li from ShanghaiTech University for excellent language editing. This work was supported by the grants from the National Natural Science Foundation of China grant (31471400), the Provincial Major Project of Guangdong Provincial Department of Education (2014KZDXM047) and the China Postdoctoral Science Foundation (2017M622659).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.05.021.
Contributor Information
Lisheng Wang, Email: maxwangls@sina.com.
Xingxu Huang, Email: huangxx@shanghaitech.edu.cn.
Jianqiao Liu, Email: liujqssz@gzhmu.edu.cn.
Supplemental Information
References
- 1.Katsanis N. Point: Treating Human Genetic Disease One Base Pair at a Time: The Benefits of Gene Editing. Clin. Chem. 2018;64:486–488. doi: 10.1373/clinchem.2017.278309. [DOI] [PubMed] [Google Scholar]
- 2.Austin C.P., Dawkins H.J.S. Medical research: Next decade’s goals for rare diseases. Nature. 2017;548:158. doi: 10.1038/548158c. [DOI] [PubMed] [Google Scholar]
- 3.Liu Y., Yang Y., Kang X., Lin B., Yu Q., Song B., Gao G., Chen Y., Sun X., Li X. One-Step Biallelic and Scarless Correction of a β-Thalassemia Mutation in Patient-Specific iPSCs without Drug Selection. Mol. Ther. Nucleic Acids. 2017;6:57–67. doi: 10.1016/j.omtn.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kang X., He W., Huang Y., Yu Q., Chen Y., Gao X., Sun X., Fan Y. Introducing precise genetic modifications into human 3PN embryos by CRISPR/Cas-mediated genome editing. J. Assist. Reprod. Genet. 2016;33:581–588. doi: 10.1007/s10815-016-0710-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim D., Lim K., Kim S.T., Yoon S.H., Kim K., Ryu S.M., Kim J.S. Genome-wide target specificities of CRISPR RNA-guided programmable deaminases. Nat. Biotechnol. 2017;35:475–480. doi: 10.1038/nbt.3852. [DOI] [PubMed] [Google Scholar]
- 6.Zong Y., Wang Y., Li C., Zhang R., Chen K., Ran Y., Qiu J.L., Wang D., Gao C. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol. 2017;35:438–440. doi: 10.1038/nbt.3811. [DOI] [PubMed] [Google Scholar]
- 7.Kim K., Ryu S.M., Kim S.T., Baek G., Kim D., Lim K., Chung E., Kim S., Kim J.S. Highly efficient RNA-guided base editing in mouse embryos. Nat. Biotechnol. 2017;35:435–437. doi: 10.1038/nbt.3816. [DOI] [PubMed] [Google Scholar]
- 8.Zhou C., Zhang M., Wei Y., Sun Y., Sun Y., Pan H., Yao N., Zhong W., Li Y., Li W. Highly efficient base editing in human tripronuclear zygotes. Protein Cell. 2017;8:772–775. doi: 10.1007/s13238-017-0459-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li G., Liu Y., Zeng Y., Li J., Wang L., Yang G., Chen D., Shang X., Chen J., Huang X., Liu J. Highly efficient and precise base editing in discarded human tripronuclear embryos. Protein Cell. 2017;8:776–779. doi: 10.1007/s13238-017-0458-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng Y., Li J., Li G., Huang S., Yu W., Zhang Y., Chen D., Chen J., Liu J., Huang X. Correction of the Marfan Syndrome Pathogenic FBN1 Mutation by Base Editing in Human Cells and Heterozygous Embryos. Mol. Ther. 2018;26:2631–2637. doi: 10.1016/j.ymthe.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zuo E., Sun Y., Wei W., Yuan T., Ying W., Sun H., Yuan L., Steinmetz L.M., Li Y., Yang H. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science. 2019;364:289–292. doi: 10.1126/science.aav9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin S., Zong Y., Gao Q., Zhu Z., Wang Y., Qin P., Liang C., Wang D., Qiu J.L., Zhang F., Gao C. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science. 2019;364:292–295. doi: 10.1126/science.aaw7166. [DOI] [PubMed] [Google Scholar]
- 13.Gaudelli N.M., Komor A.C., Rees H.A., Packer M.S., Badran A.H., Bryson D.I., Liu D.R. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsoukas I.G. Commentary: Programmable base editing of A·T to G·C in genomic DNA without DNA cleavage. Front. Genet. 2018;9:21. doi: 10.3389/fgene.2018.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sekijima Y., Campos R.I., Hammarström P., Nilsson K.P., Yoshinaga T., Nagamatsu K., Yazaki M., Kametani F., Ikeda S. Pathological, biochemical, and biophysical characteristics of the transthyretin variant Y114H (p.Y134H) explain its very mild clinical phenotype. J. Peripher. Nerv. Syst. 2015;20:372–379. doi: 10.1111/jns.12143. [DOI] [PubMed] [Google Scholar]
- 16.Esposito G., Imperato M.R., Ieno L., Sorvillo R., Benigno V., Parenti G., Parini R., Vitagliano L., Zagari A., Salvatore F. Hereditary fructose intolerance: functional study of two novel ALDOB natural variants and characterization of a partial gene deletion. Hum. Mutat. 2010;31:1294–1303. doi: 10.1002/humu.21359. [DOI] [PubMed] [Google Scholar]
- 17.Annunen S., Paassilta P., Lohiniva J., Perälä M., Pihlajamaa T., Karppinen J., Tervonen O., Kröger H., Lähde S., Vanharanta H. An allele of COL9A2 associated with intervertebral disc disease. Science. 1999;285:409–412. doi: 10.1126/science.285.5426.409. [DOI] [PubMed] [Google Scholar]
- 18.Gloyn A.L., Reimann F., Girard C., Edghill E.L., Proks P., Pearson E.R., Temple I.K., Mackay D.J., Shield J.P., Freedenberg D. Relapsing diabetes can result from moderately activating mutations in KCNJ11. Hum. Mol. Genet. 2005;14:925–934. doi: 10.1093/hmg/ddi086. [DOI] [PubMed] [Google Scholar]
- 19.Felius J., Thompson D.A., Khan N.W., Bingham E.L., Jamison J.A., Kemp J.A., Sieving P.A. Clinical course and visual function in a family with mutations in the RPE65 gene. Arch. Ophthalmol. 2002;120:55–61. doi: 10.1001/archopht.120.1.55. [DOI] [PubMed] [Google Scholar]
- 20.Kang X.J., Caparas C.I.N., Soh B.S., Fan Y. Addressing challenges in the clinical applications associated with CRISPR/Cas9 technology and ethical questions to prevent its misuse. Protein Cell. 2017;8:791–795. doi: 10.1007/s13238-017-0477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stemmer M., Thumberger T., Del Sol Keyer M., Wittbrodt J., Mateo J.L. CCTop: An Intuitive, Flexible and Reliable CRISPR/Cas9 Target Prediction Tool. PLoS ONE. 2015;10:e0124633. doi: 10.1371/journal.pone.0124633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bae S., Park J., Kim J.S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akcakaya P., Bobbin M.L., Guo J.A., Malagon-Lopez J., Clement K., Garcia S.P., Fellows M.D., Porritt M.J., Firth M.A., Carreras A. In vivo CRISPR editing with no detectable genome-wide off-target mutations. Nature. 2018;561:416–419. doi: 10.1038/s41586-018-0500-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma H., Marti-Gutierrez N., Park S.W., Wu J., Lee Y., Suzuki K., Koski A., Ji D., Hayama T., Ahmed R. Correction of a pathogenic gene mutation in human embryos. Nature. 2017;548:413–419. doi: 10.1038/nature23305. [DOI] [PubMed] [Google Scholar]
- 26.Tang L., Zeng Y., Du H., Gong M., Peng J., Zhang B., Lei M., Zhao F., Wang W., Li X., Liu J. CRISPR/Cas9-mediated gene editing in human zygotes using Cas9 protein. Mol. Genet. Genomics. 2017;292:525–533. doi: 10.1007/s00438-017-1299-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the deep sequencing data can be viewed in the National Omics Data Encyclopedia (NODE) (https://www.biosino.org/node/) by pasting the accession (OEP000192) into the text search box or through the URL https://www.biosino.org/node/project/detail/OEP000192. The whole-genome sequencing data have been deposited in the NCBI Sequence Read Archive database (SRA: PRJNA480397).