Abstract
Genome-wide association studies (GWASs) have identified approximately 100 colorectal cancer (CRC) risk loci. However, the causal genes in these loci have not been systematically interrogated. We conducted a high-throughput RNA-interference functional screen to identify the genes essential for proliferation in the CRC risk loci of Asian populations. We found that ATF1, located in the 12q13.12 region, functions as an oncogene that facilitates cell proliferation; ATF1 has the most significant effect of the identified genes and promotes CRC xenograft growth by affecting cell apoptosis. Next, by integrating a fine-mapping analysis, a two-stage affected-control study consisting of 6,213 affected individuals and 10,388 controls, and multipronged experiments, we elucidated that two risk variants, dbSNP: rs61926301 and dbSNP: rs7959129, that located in the ATF1 promoter and first intron, respectively, facilitate a promoter-enhancer interaction, mediated by the synergy of SP1 and GATA3, to upregulate ATF1 expression, thus synergistically predisposing to CRC risk (OR = 1.77, 95% CI = 1.42–2.21, p = 3.16 × 10−7; Pmultiplicative-interaction = 1.20 × 10−22; Padditive-interaction = 6.50 × 10−3). Finally, we performed RNA-seq and ChIP-seq assays in CRC cells treated with ATF1 overexpression in order to dissect the target programs of ATF1. Results showed that ATF1 activates a subset of genes, including BRAF, NRAS, MYC, BIRC2, DAAM1, MAML2, STAT1, ID1, and NKD2, related to apoptosis, Wnt, TGF-β, and MAPK pathways, and these effects could cooperatively increase the risk of CRC. These findings reveal the clinical potential of ATF1 in CRC development and illuminate a promoter-enhancer interaction module between the ATF1 regulatory elements dbSNP: rs61926301 and dbSNP: rs7959129, and they bring us closer to understanding the molecular drivers of cancer.
Keywords: high-throughput RNA interference screen, ATF1, enhancer-promoter interaction, apoptosis/ Wnt/TGF-β/MAPK pathway
Introduction
The global cancer burden is rising rapidly due to the aging of the population and the adoption of new, unhealthy lifestyle behaviors.1 In China, colorectal cancer (CRC; MIM: 114500) is the third most common cancer diagnosed in adults and the fifth leading cause of death from cancer.2 Genome-wide association studies (GWASs) have become a powerful tool for uncovering genetic susceptibility factors for complex diseases. To date, approximately 100 CRC GWAS-identified risk loci have been identified,3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14 and they explain a substantial proportion of the genetic heritability of CRC. Efforts have now focused on investigating the functional basis of these associations in order to identify new prevention and therapy targets.
Most GWAS-identified single nucleotide polymorphisms (SNPs) are in noncoding regions and are a long distance from nearby annotated genes. It is believed that causal SNPs are in linkage disequilibrium (LD) with the corresponding tag SNP and are located in regulatory regions that control gene expression through long-range interactions. However, the functional characterization of these causal variants through traditional fine-mapping analysis is a major challenges because there may be multiple potential functional variants that need to be tested.15, 16, 17 Therefore, first identifying the causal genes in these loci might be a better way to elucidate targets for prevention and therapy. Functional genomic screening based on a high-throughput RNA interference (RNAi) or CRISPR-Cas9 interrogation has been suggested to be a powerful tool for unravelling cancer dependency genes and gene interaction networks.18, 19, 20, 21 This approach may also be useful for systematically identifying causal genes in the GWAS loci.
In the present study, we integrated a high-throughput RNAi-based functional interrogation, a large-scale population study, and a series of biochemical experiments in order to elucidate the potential role of genes in the CRC GWAS-identified risk loci. We demonstrated that ATF1 (MIM: 123803), located in the 12q13.12 region, functions as an oncogene by affecting cell apoptosis, and two causal SNPs, located in the ATF1 promoter and first intron, synergistically predispose to CRC risk through a promoter-enhancer interaction mediated by SP1 and GATA3 (MIM: 189906 and MIM: 131320), and these findings will provide important clues for the etiology of CRC.
Material and Methods
A Functional Genomic Screen with a High-Throughput RNAi Interrogation
We selected candidate genes on the basis of CRC GWASs, which identified 15 loci associated with CRC risk (2016.12, Table S1) in Asian (ASN) populations. To select candidate genes in each region for functional screening, we performed fine mapping by extending 1 Mb upstream and downstream of the tag SNPs. After we excluded microRNAs, noncoding RNAs, and pseudogenes on the basis of their functional annotation in the National Center for Biotechnology Information database, we ultimately selected a total of 157 protein-coding genes (Table S2) for a proliferation measurement of CRC cells by a large-scale RNAi interrogation. The siRNA library was provided by ViewSolid Biotech, and the repression efficiencies were guaranteed by the provider. Both p < 0.05 and an n-fold change >1.1 or <0.9 were selected as the threshold of significance.
Integrative Expression Quantitative Trait Locus (eQTL) Analysis and Genotype Imputation
The LD SNPs (r2 ≥ 0.2, ASN) of dbSNP: rs1169571 were downloaded from the Haploreg database. Individual genotypes and ATF1 mRNA expression were downloaded from the TCGA (The Cancer Genome Atlas) data portal. To increase the power for eQTL analysis, we imputed the variants for all CRC samples from TCGA with IMPUTE2, and we used 1000 Genomes Phase 3 as the reference panel. Then, we performed an integrative eQTL analysis between those SNPs and ATF1 mRNA expression by using the TCGA CRC data and adjusting for the effect of copy number variation, CpG methylation levels, population structures (principal components), and clinical parameters (age, sex, and tumor stage) on gene expression. The details of the genotype imputation and principal components calculation can be seen in our previous study.22
We performed a functional annotation for eQTLs with multiple bioinformatic tools, including the Haploreg database, ANNOVAR, rSNPBase, RegulomeDB, and CistromeDB, and this annotation integrated multiple histone modification ChIP-seq peaks, TFs ChIP-seq peaks, and DNase hypersensitive site data. Finally, we selected functional variants with the highest potential in each LD block (r2 ≥ 0.8) for further population and experimental validation.
Cell Lines
HCT116, SW480, LoVo, HCT15, HT115, CoLo205, LS123, and SNU-C1 cell lines were obtained from the China Center for Type Culture Collection and were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (GIBCO) and 1% antibiotics at 37°C in a humidified atmosphere of 5% CO2. All cell lines that we used in this study were authenticated by short tandem repeat profiling (Applied Biosystems) and tested for the absence of mycoplasma contamination (MycoAlert).
Construction of Plasmids
DNA fragments totaling 1,100 bp and surrounding the SNP dbSNP: rs61926301 G or T allele were subcloned into pGL3-Basic vector (Promega). DNA fragments totaling 1,120 bp and surrounding the SNP dbSNP: rs7959129 G or T allele were subcloned into pGL3-Promoter vector (Promega) in both forward and reverse orientations. The full-length cDNAs of SP1, GATA3, and ATF1 were subcloned into pcDNA3.1(+) vector (Invitrogen), respectively. All plasmids were commercially synthesized by Genewiz Biological Technology.
Transient Transfections and Lentiviral Transduction
For transient transfections, we transfected all CRC cell lines with lipofectamine 3000 (Invitrogen). For lentivirus production and transfection, we subcloned the full-length cDNA of ATF1 into pLVX-3FLAG-PGK-Puro vector (pLV-ATF1), and we used empty pLVX-3FLAG-PGK-Puro vector (pLV-EV) as a control. We produced the lentivirus in 293T cells by transfecting pLV-ATF1 or pLV-EV plasmids with X-tremeGENE9 transfection reagent (Roche), and we ultimately used it to infect HCT116 cells. We used the Lenti-XTM concentrator to concentrate the lentiviruses, and we chose puromycin (2 mg/mL) for antibiotic selection. We determined the transfection effect by qRT-PCR and immunoblotting (Figures 2A, 2E, S7B, and S7D).
Figure 2.
ATF1 Promotes Cell Proliferation and Xenograft Tumor Growth in CRC by Affecting Cell Apoptosis
(A–D) The effect of ATF1 overexpression (A) or knockdown (C). The overexpression of ATF1 substantially promotes the rate of CRC cell proliferation in the SW480 and HCT116 cell lines (B), whereas knockdown of ATF1 significantly inhibits the proliferation of SW480 and HCT116 cells (D). Results were shown as the means ± SEM from three experiments, each with six replicates.
(E–H) The effect of ATF1 overexpression by lentiviral transduction (E) or ATF1 knockout by CRISPR-Cas9 (F) on the colony formation ability of CRC cells (G and H). The results present colony formation ability relative to control cells (set to 100%); data were shown as the means ± SEM from three experiments, each with three replicates.
(I and J) The image and growth curve of a xenograft tumor of HCT16 cells; the tumor was treated with ATF1 overexpression (I) or ATF1 knockout (J) in nude mice. Results were shown as the means ± SEM for five mice in each group. All ∗p < 0.05 and ∗∗p < 0.01 values derived from comparison with control cells were calculated via a two-sided Student’s t test.
(K–N) The effect of ATF1 knockdown (K and M) and ATF1 overexpression (L and N) on the apoptosis and cell-cycle process of CRC cells. Data were a representative result from three repeated experiments, each with six replicates. ∗∗p < 0.01, derived from a comparison with controls via a two-sided Student’s t test in the apoptosis analysis and Pearson’s χ2 test in the cell-cycle analysis.
RNA Interference and CRISPR-Cas9-Mediated Genome Editing
siRNA oligonucleotides targeting SP1, GATA3, ATF1, and a nontargeting siRNA control were purchased from RiboBio (Table S6). A modified ATF1 knockout CRC HCT116 cell line was generated by CRISPR-Cas9 technology (Genloci Biotechnologies). Single guide RNAs (sgRNAs) targeting ATF1 were cloned into the pGK1.1-CRISPR-Cas9 vector (cat# GP0134). The sgRNA sequences targeting ATF1 sites are shown in Table S6, and the effects of ATF1 knockdown or knockout were determined via qRT-PCR and immunoblotting (Figures 2C, 2F, S7A, and S7C).
Cell Proliferation Assays
Cells were seeded and transfected in 24-well plates (5 × 104 cells per well). After 24 h, the cells were harvested by trypsin digestion and subsequently seeded in 96-well, flat-bottom plates; each well contained 2,500 cells in 100 μL of cell suspension. After a certain time in culture, cell viability was measured with CCK-8 assays (Dojindo) according to the manufacturer’s recommendations.
Colony-Formation Assays
Cells were seeded into 6-well cell culture plates at a density of 2,000 cells per well. After 10 days, the cells were washed with cold PBS twice, fixed with 3.7% formaldehyde, and were stained with crystal violet. The colony number in each well was counted.
Xenograft Growth of CRC HCT116 Cells in Nude Mice
Female BALB/c nude mice at age 4–5 weeks, purchased from Beijing HuaFuKang Bioscience, were allowed to acclimate to local conditions for 1 week and maintained under a 12-h-dark/12-h-light cycle with food and water provided ad libitum. The mice (five in each group) were subcutaneously injected in the back flank with 0.1 mL of cell suspension containing 1 × 106 CRC HCT116 cells. When a tumor was palpable, it was measured every five days, and its volume was calculated according to the formula: volume = 0.5 × length × width.2 Tumor tissue was fixed with paraformaldehyde, and then it was subjected to hematoxylin and eosin (H&E) staining, Ki67, and ATF1 immunohistochemical analyses. All experimental procedures were performed in accordance with the relevant institutional and national guidelines and approved by the institutional animal care and use committee of Huazhong University of Science and Technology.
Immunohistochemical Analysis
Formalin-fixed, paraffin-embedded xenograft tumor tissue mouse samples were stained with H&E and analyzed under a light microscope. Immunohistochemical staining was applied to further determine the ATF1 or Ki67 expression in the mouse xenograft tumor tissues, which were incubated with primary antibodies against ATF1 or Ki67 (1:50, Abcam, ab181569 and ab15580, respectively) at 4°C overnight and then detected with an ABC kit (Thermo Fisher Scientific).
qRT-PCR and Immunoblotting
For the qRT-PCR assay, total RNA was extracted from cells or tissues with TRIzol reagent (Thermo Fisher Scientific). Reverse transcription was performed with the SuperScript III First-Strand Synthesis System (Invitrogen), and quantitative PCR was performed with Power SYBR Green PCR Master Mix (Applied Biosystems). Target gene expression was normalized to that of GAPDH. All specific primers that were used for qPCR are listed in Table S6. For immunoblot analysis, total protein was harvested with RIPA lysis buffer supplemented with the protease inhibitor PMSF (Beyotime). Proteins were incubated with antibodies against ATF1 (1:1,000, Proteintech, Cat#11946-1-AP), Flag (1:1,000, CST, Cat#14793), or β-actin (1:1,000, Proteintech, Cat# 60008-1-Ig) at 4°C overnight.
Electrophoretic Mobility Shift Assays
Complementary DNA oligonucleotides that were centered on the variant dbSNP: rs61926301 alleles or dbSNP: rs7959129 alleles (Table S6) were commercially synthesized by Takara and labeled with biotin at the 3′ end. Nuclear extracts of cells were obtained with the Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime). Electrophoretic mobility shift assays (EMSAs) were performed with the EMSA/Gel-Shift Kit (Beyotime) according to the manufacturer’s instructions. Additionally, for the competitive binding assay, unlabeled probes were added to the reaction mixtures at a 10-fold or 100-fold excess compared with the labeled probes and incubated for 20 min prior to the addition of labeled probes. For super-shift reactions, 2–3 μg of anti-SP1 or anti-GATA3 antibody (Abcam, ab13370 and ab199428, respectively) was incubated with reaction mixtures for 20 min at room temperature before the addition of labeled DNA probes.
Chromatin Immunoprecipitation Sequencing or qPCR (ChIP-seq or ChIP-qPCR)
ChIP assays were performed with a ChIP assay kit (Cat#10086, Millipore) according to the manufacturer’s instructions. Cells were crosslinked with 1% formaldehyde, and glycine was added to stop fixation. Genomic DNA was extracted from the fixed-chromatin cells and sheared by sonication. Antibodies against SP1 (Abcam, ab13370), GATA3 (Abcam, ab199428), or Flag-ATF1 (CST, Cat#14793) and a nonspecific rabbit IgG (Santa Cruz) were subsequently incubated with the cross-linked protein and DNA overnight for immunoprecipitation with protein A/G magnetic beads. DNA fragments were purified and collected by a Dr.GenTLE Precipitation Carrier kit (Takara). The purified DNA library was sequenced (BerryGenomics) or analyzed by qPCR. The primers that we used for ChIP-qPCR are shown in Table S6.
Dual-Luciferase Reporter Assay
The luciferase reporter assay was performed with a dual-Luciferase Reporter Kit (Promega) according to the manufacturer’s recommendations. Reporter plasmids (PGL3-Basic or PGL3-Promoter) and a constitutively active pRL-SV40 Renilla luciferase plasmid (Promega) were co-transfected via Lipofectamine 3000 (Invitrogen) in SW480 and HCT116 cell lines. For each sample, luciferase activity was determined by normalizing the luminescence value of Renilla luciferase to that of firefly luciferase.
Chromosome Conformation Capture Assays
Chromosome conformation capture (3C) assays were performed as previously described23 in CRC cell lines carrying different genotypes of dbSNP: rs61926301 and dbSNP: rs7959129. Cells were fixed with formaldehyde, which was stopped with glycine, and lysed in lysis buffer (10 mM Tris-HCl [pH 7.5]; 10 mM NaCl; 5 mM MgCl2; 0.1 mM EGTA; and 1× complete protease inhibitor; 11836145001 Roche), then digested with Mbo-I enzyme (New England Biolabs) at 37°C overnight. Ligation was performed with T4 ligase (Thermo Fisher Scientific) at 16°C for 6 h. The cross-linked DNA fragments were extracted by phenol/chloroform and precipitated with ethanol. A bacterial artificial chromosome (BAC) clone that covered the genome segment of the target regions and that was applied to eliminate amplification efficiency differences among different primers was treated with the same procedures. In addition, cell background differences were normalized with GAPDH. Physical interactions among anchor and test primers were measured by qPCR. All 3C-qPCR primers (Table S6) were synthesized by TSINGKE Biological Technology (Wuhan).
RNA Sequencing
Total RNA was extracted with TRIzol reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. RNA sequencing (RNA-seq) libraries were constructed with the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England Biolabs) according to the manufacturer’s recommendations. After cluster generation, the libraries were sequenced, and 125 bp paired-end reads were generated (Novogene). For all cell line studies, samples were done in triplicate.
Study Subjects
Two-stage affected-control studies were conducted to evaluate the associations between eQTLs and CRC risk. The 1,524 CRC-affected individuals and 1,522 cancer-free controls in the discovery stage were recruited from the cancer hospital of the Chinese Academy of Medical Sciences in Beijing, China. The 4,689 affected individuals and 8,866 cancer-free controls in the replication stage were recruited from the Tongji Hospital of Huazhong University of Science and Technology (HUST), Wuhan, China. All cases were histopathologically or cytologically confirmed by at least two local pathologists, and the individuals had not had chemotherapy or radiotherapy before blood collection according to the World Health Organization classification. All controls were cancer-free individuals selected from a community nutritional survey in the same region where the affected individuals were recruited, and they were matched to the affected individuals by gender and age (± 5 years).16 At recruitment, peripheral blood samples and demographic characteristics, including age, gender, smoking status, and drinking status, were obtained from the medical records of these individuals. The detailed definitions of smoking and drinking statuses have been previously described.24 Informed consent was obtained from each subject, and this study was approved by the Chinese Academy of Medical Sciences Cancer Institute and the institutional review board of Tongji Medical College, HUST.
Genotyping and Quality Control
Genomic DNA was extracted from blood samples with the Relax Gene Blood DNA System Kit (Tiangen). SNPs were genotyped with the TaqMan SNP Genotyping system in both stages. Quality control was implemented as follows: (1) affected and control samples were mixed in the plates, and the persons who performed the genotyping assay were unaware of the affected or control status; (2) positive and negative (no DNA) samples were included on every 384-well assay plate; (3) there were 5% duplicate samples that had a concurrence rate of 100% in each 384-well plate; (4) the SNPs had genotyping call rates > 95%; and (5) SNPs that deviated from the Hardy-Weinberg equilibrium (HWE) in controls were excluded.
Association and Interaction Analyses
For association analyses, unconditional logistic regression was employed to estimate odds ratios (ORs) and 95% confidence intervals (CIs) for the associations between candidate SNPs and CRC risk, with adjustments for gender, age group, smoking status, and drinking status. Multiple genetic models, such as allelic, dominant, recessive, and additive genetic models, were applied to assess the genetic susceptibility of variants to CRC. P values < 0.05 were considered statistically significant in SPSS (21.0).
For interaction analyses, gene-gene interaction were evaluated by a bootstrapping test of goodness-of-fit for additive interaction and by an unconditional logistic regression analysis for multiplicative interaction after adjusting for gender, age group, smoking status, and drinking status. P values < 0.05 were considered statistically significant by Stata (11.0) or SPSS (21.0).
Statistical Analyses and Computational Analyses
For statistical analysis, we used a goodness-of-fit χ2 test to assess the Hardy-Weinberg equilibrium for the genotype distribution of each SNP in the controls. Either Pearson’s χ2 test or a two-sided Student’s t test was applied to estimate the significance of the differences in gender, age, age group, smoking status, and drinking status between affected individuals and controls. For the functional assays, in the relevant figures, the figure legends denote the experiments’ statistical details, including the statistical tests used, the numbers of replicates, and the data presentation type. All statistical analyses were performed in R software (3.30) or SPSS software (21.0).
For computational analyses, all the sequencing data were aligned to the human genome (GRCh38/hg37) unless indicated specifically. The quantification and differential expression of RNA-seq genes were analyzed with Cuffdiff tools (v2.1.1), and a false discovery rate (FDR) p value < 0.05 was considered significant. ChIP-seq fastq files were generated with Illumina’s CASAVA software and mapped to the human genome (hg38) with the Bowtie2 tool. ChIP-seq bedgraph files were generated with Bedtools, and ChIP-seq peaks were called with the MACS1.4 software. ChIP binding peaks were intersected with the promoter regions of genes annotated by Gencode v24. The promoter is defined as the region between two kb upstream and 100 bp downstream of any transcription start site of a coding transcript gene.25 We used a paired Student’s t test to test gene expression differences between tumor tissues and matched, adjacent normal tissues. Gene coexpression was tested by Spearman’s correlation, and genes with p < 0.05 and |r| > 0.25 were considered statistically significant.
Results
Functional Genomic Screens Based on High-Throughput RNAi Interrogation Reveal ATF1 to Be an Oncogene in CRC
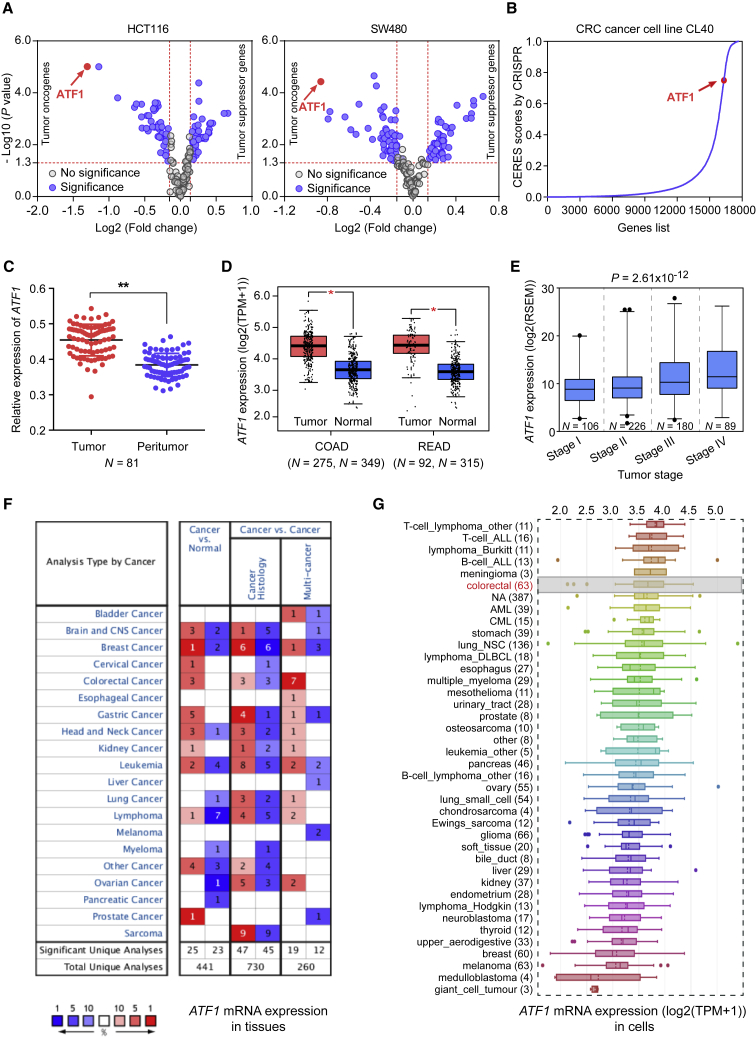
We screened 157 protein-coding genes in Asian CRC GWAS-identified loci (Table S1) for their effects on cancer cell proliferation by using an RNAi-based on-chip approach. A total of 78 genes have a significant effect on cell proliferation in both HCT116 and SW480 cells (Table S2 and Figure 1A). Among these genes, ATF1 has the most significant effect in both cell lines (Figure 1A). The essential role of ATF1 in cell proliferation is also verified in the CRC CL40 cell line from the genome-wide CRISPR-Cas9-based loss-of-function screening data19 (Figure 1B). The mRNA levels of ATF1 are significantly higher in tumors than those in normal tissues from our own CRC patients (Figure 1C) and from the TCGA, GTEX, and Oncomine database data (Figures 1D, 1F, and S1A). Data from the Cancer Cell Line Encyclopedia (CCLE) also suggested that ATF1 is highly expressed in CRC cell lines and that the high expression in these cell lines ranks ahead of that among 1,036 human cancer cell lines (Figure 1G). Moreover, ATF1 amplification also frequently occurs across cancer types (Figure S1B). We also examined the effect of ATF1 expression on the tumor stages of CRC in the TCGA cohort and found that ATF1 is overexpressed in advanced CRC (Figure 1E). Collectively, these data illustrated that the upregulation of ATF1 expression correlates with the development of CRC, suggesting that ATF1 may function as an oncogene in CRC.
Figure 1.
Functional Genomic Screening Reveals that ATF1 Is an Oncogene in CRC
(A) Functional genomic screening based on high-throughput RNAi interrogation was used to identify genes essential for cell proliferation in the CRC risk loci in Asian populations in HCT116 and SW480 cells. Both p < 0.05 and an n- fold change >1.1 or <0.9 were selected as the threshold of significance and calculated by a two-sided Student’s t test.
(B) According to the data of a genome-wide CRISPR-Cas9-based loss-of-function screen, ATF1 is essential for cell growth; higher CERES scores are found in CRC CL40 cells. Higher CERES scores demonstrate an elevated dependency of cell viability on given genes.
(C and D) ATF1 is significantly overexpressed in tumors compared to in the normal tissues from our CRC patients and the TCGA and GTEX datasets. Data were shown as the mean ± SD and all ∗p < 0.05 and ∗∗p < 0.01 values were calculated by a two-sided Student’s t test. Abbreviations are as follows: COAD = colon adenocarcinoma and READ = rectal adenocarcinoma.
(E) ATF1 expression levels were measured in different tumor stages of CRC. Data were presented as the mean ± SD from the TCGA database. P values were calculated by one-way ANOVA.
(F and G) ATF1 expression levels were evaluated in multiple tumor tissue types from the Oncomine database and in 1,036 human cancer cell lines from the CCLE database.
We next examined the effect of ATF1 on cell phenotypes and found that the overexpression of ATF1 in HCT116 and SW480 cells substantially increases the CRC cell proliferation rate (Figures 2A, 2B, and S1C), whereas the knockdown or knockout of ATF1 substantially reduces this effect (Figures 2C, 2D, and S1D). The colony-formation ability of CRC cells is markedly stimulated by the ATF1 overexpression but substantially attenuated by the ATF1 knockout (Figures 2E–H). Furthermore, to assess whether ATF1 is also a tumor oncogene in vivo, we overexpressed and knocked out ATF1 in HCT116 cells and then injected these cells subcutaneously into nude mice. The growth rate of the xenograft overexpressing ATF1 is significantly higher than that of the controls (Figure 2I). In contrast, ATF1 knockout results in a significant reduction in the growth of xenografts compared to in control counterparts (Figure 2J). Consistent with this finding, the levels of Ki67 and ATF1 protein in the xenograft also present similar trends (Figures S2A and S2B).
To test the ATF1 biological mechanism affecting CRC cell proliferation, we further examined the effect of ATF1 on CRC cell apoptosis and cell-cycle progression. Flow cytometry analysis showed that compared with control counterparts, ATF1 overexpression significantly inhibits apoptosis (Figures 2L and S2D), whereas the knockdown of ATF1 results in a substantial increase in the number of apoptotic cells (Figures 2K and S2C). However, the overexpression and knockdown of ATF1 do not have significant effects on the cell cycle of CRC cells (Figures 2M, 2N, S2E, and S2F), demonstrating that the promotive effect of ATF1 on cell proliferation may be mediated through inhibiting apoptosis rather than altering the cell cycle.
Fine Mapping Identifies Two Putative Functional Variants Affecting ATF1 Expression
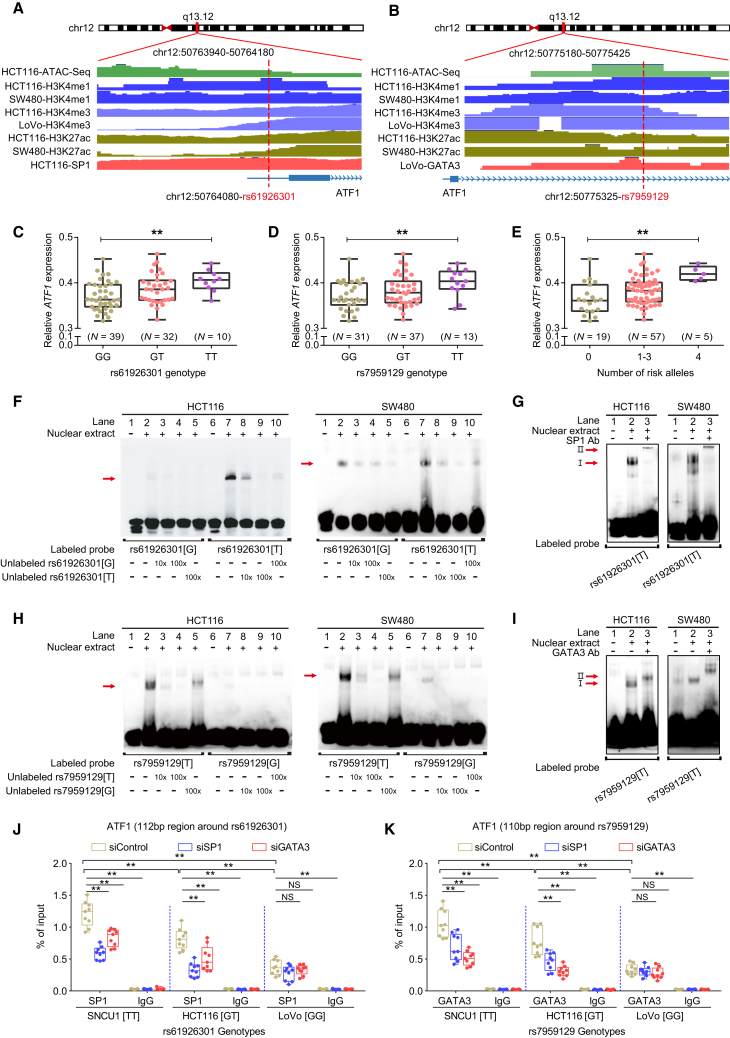
ATF1 is located in the 12q13.12 region; dbSNP: rs11169571 is the tag SNP identified in Chinese populations,12 and dbSNP: rs11169552 is the tag SNP found in European populations.13 Causal variants in this region have not been systematically investigated. Thus, we first performed an eQTL analysis between all SNPs in LD with the tag dbSNP: rs11169571 LD (r2 ≥ 0.2) and ATF1 mRNA expression. The results revealed that two LD blocks (r2 ≥ 0.8) present significant eQTLs with ATF1 expression (Figures S3A–S3C). We then performed a functional annotation for SNPs in these two blocks by using multiple bioinformatic tools, including the Haploreg database, ANNOVAR, rSNPBase, RegulomeDB, and CistromeDB. Variants with the highest potential to be functional in each LD block (r2 ≥ 0.8) were selected as candidate causal variants. We found that the two SNPs, dbSNP: rs61926301 and dbSNP: rs7959129, located in the ATF1 promoter and first intron, respectively, are enriched in active histone modification peaks (H3K4me1, H3K4me3, and H3K27ac) and open chromatin accessibility (ATAC-seq peaks) (Figures 3A and 3B). Finally, we validated the correlation between these two SNPs and ATF1 mRNA expression in our own CRC samples, and the results are in line with the data from the TCGA database, showing that carriers with the dbSNP: rs61926301[TT] or dbSNP: rs7959129[TT] genotype have higher ATF1 expression than those with other genotypes (Figures 3C–3E, S3D, and S3E). Notably, dbSNP: rs61926301 is in independent LD with dbSNP: rs7959129 (r2 = 0.22). Collectively, these results illustrated that these two variants have allele-specific differences that affect ATF1 expression.
Figure 3.
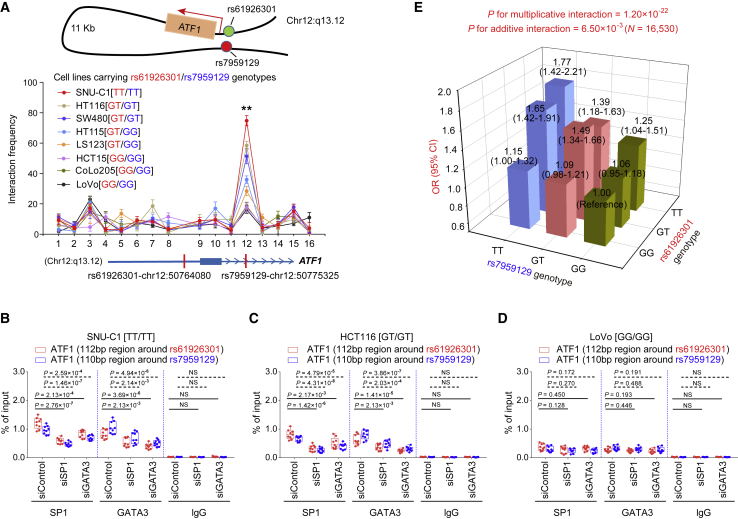
SP1 and GATA3 preferentially bind to the dbSNP: rs61926301[T] and dbSNP: rs7959129[T] alleles at the ATF1 promoter and first intron region, respectively
(A and B) Epigenetic annotation for the region surrounding dbSNP: rs61926301 (D) or dbSNP: rs7959129 (E) in CRC cell lines. Data including ATAC-seq peaks, TF (SP1 or GATA3) peaks, and multiple histone (H3k4me1, H3K4me3, and H3k27ac) modification peaks were obtained from the ENCODE database.
(C–E) eQTL analyses of ATF1 expression with the dbSNP: rs61926301 genotype (A), the dbSNP: rs7959129 genotype (B), and number of both risk alleles from our CRC patient samples. Data were shown as the mean ± SD and all ∗∗p < 0.01 values were calculated by linear regression analysis.
(F–I) EMSAs and SP1 and GATA3 super-shift EMSAs with biotin-labeled probes containing dbSNP: rs61926301 (F and G) or dbSNP: rs7959129 (H and I) in HCT116 and SW480 cells. Arrows indicate allele-specific bands that interact with nuclear protein in the cells. “I” represents the allele-specific binding band. “II” represents the super-shifted band. In addition, 10× and 100× respectively represent 10-fold and 100-fold excess amounts of an unlabeled probe compared with the amount of the labeled probe. “+” and “−” indicate added and not added, respectively.
(J and K) The binding of SP1 and GATA3 to the region surrounding dbSNP: rs61926301 (J) or dbSNP: rs7959129 (K) was measured by ChIP-qPCR assays in the SNU-C1, HCT116, and LoVo cell lines, which carry different genotypes of both SNPs. Data were presented as the mean ± SD from three repeated experiments, each with three replicates. All ∗∗p < 0.01 values were derived from a comparison with controls via a two-sided Student’s t test.
SP1 and GATA3 Preferentially Bind to the Risk Alleles of rs61926301 and rs7959129 at the ATF1 Promoter and First Intron Region, Respectively
Having demonstrated that dbSNP: rs61926301 and dbSNP: rs7959129 are associated with ATF1 mRNA expression, we next sought to elucidate the underlying mechanisms. SNPs in regulatory regions can function by modulating TF binding. To identify which TFs bind to the regions containing these two causal SNPs, we took a multipronged approach. We first used the Cistrome database to predict potential TF motifs by imputing the sequences around each candidate SNP, and it revealed that dbSNP: rs61926301 maps within the binding motif of SP1, whereas dbSNP: rs7959129 maps within the binding motif of GATA3 (Figure S3F). This observation was further supported by ChIP-seq data of SP1 and GATA3 in CRC cell lines (Figures 3A and 3B).
Moreover, we validated the binding of SP1 and GATA3 to these two regions with EMSA assays. The results showed that the dbSNP: rs61926301[T] allele but not the dbSNP: rs61926301[G] allele preferentially binds to nuclear extracts, and the binding signal is gradually attenuated in a dose-dependent manner with the addition of the unlabeled probe containing the dbSNP: rs61926301[T] allele but not the dbSNP: rs61926301[G] allele (Figure 3F). An additional super-shift EMSA showed that the dbSNP: rs61926301[T] allele is significantly enriched for SP1 (Figure 3G). Similarly, the dbSNP: rs7959129[T] allele binds more preferentially to nuclear extracts than does the dbSNP: rs7959129[G] allele, and it is significantly enriched with GATA3 (Figures 3H and 3I). These findings indicated that dbSNP: rs61926301 and dbSNP: rs7959129 have an allele-specific affinity for SP1 and GATA3 binding, respectively, in CRC cell lines. Finally, we further validated this observation in vivo by using ChIP-qPCR assays in three CRC cell lines (SNU-C1[TT/TT], HCT116[GT/GT] and LoVo [GG/GG]) with different dbSNP: rs61926301 or dbSNP: rs7959129 genotypes. We found that a stronger SP1 binding is enriched in the dbSNP: rs61926301 region, whereas a stronger GATA3 binding is enriched in the dbSNP: rs7959129 region in the SNU-C1 and HCT116 cells compared with the LoVo cells (Figures 3J and 3K), suggesting that SP1 and GATA3 preferentially bind to the dbSNP: rs61926301[T] and dbSNP: rs7959129[T] alleles, respectively, in an allele-specific manner. Intriguingly, these binding signals are significantly attenuated when SP1 or GATA3 is knocked down (Figures 3J and 3K), demonstrating that SP1 and GATA3 bind to risk alleles of the two variants in a mutually dependent manner.
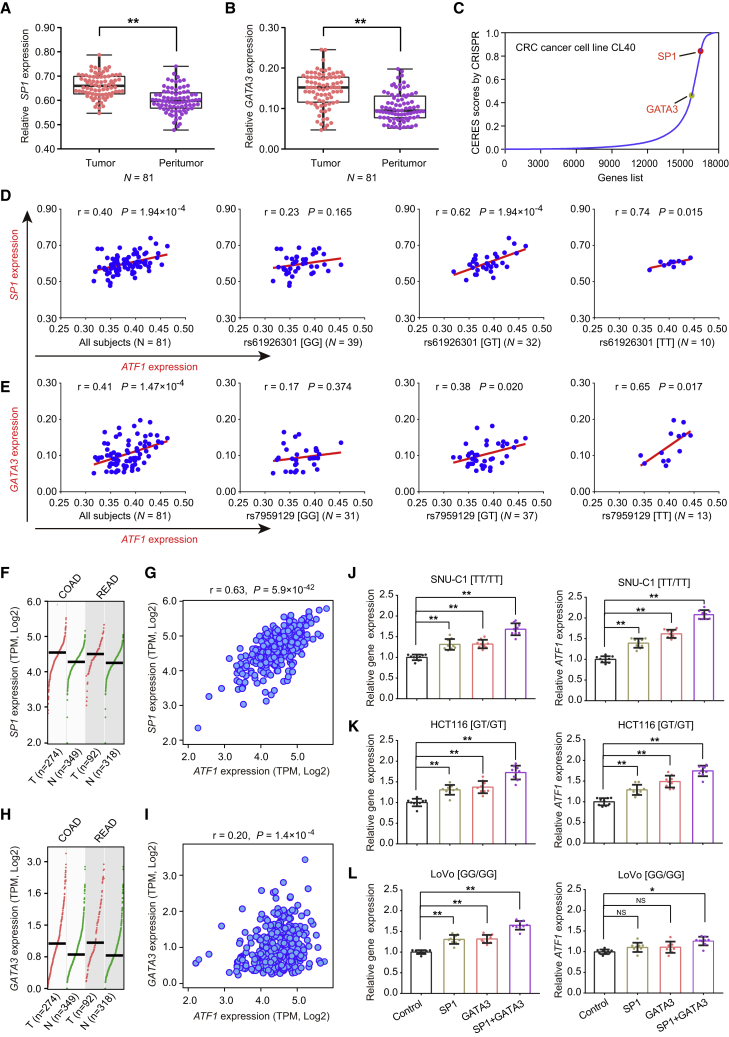
SP1 and GATA3 are well-known transcription factors in tumors.26, 27, 28, 29, 30 In both our CRC-affected patients and the TCGA samples, SP1 and GATA3 are significantly overexpressed in tumor tissues compared with adjacent normal tissues (Figures 4A, 4B, 4F, and 4G). This result is in line with the data from the genome-wide CRISPR-Cas9-based loss-of-function screens of CRC CL40 cells,19 revealing that SP1 and GATA3 are essential for cell viability (Figure 4C). Moreover, positive correlations between the expression of SP1 or GATA3 and ATF1 expression are observed in both our CRC-affected patient cohorts (Figures 4D and 4E) and the TCGA CRC samples (Figures 4H and 4I). Intriguingly, significant correlations occur only in carriers with the dbSNP: rs61926301[T] or dbSNP: rs7959129[T] allele (Figures 4D and 4E). Furthermore, when SP1 or GATA3 is overexpressed in the three cell lines with different genotypes, ATF1 expression is increased in the SNCU-1 and HCT116 cells with the dbSNP: rs61926301[T] and dbSNP: rs7959129[T] alleles (Figures 4J and 4K), but not in the LoVo cells lacking the two alleles (Figure 4L). Analogously, when SP1 or GATA3 is knocked down in these three cell lines, ATF1 expression is decreased concomitantly in the SNCU-1 and HCT116 CRC cells, but not in the LoVo cells (Figures S4A–S4C), further suggesting that the regulatory effect of SP1 and GATA3 on ATF1 expression occurs in an allele-specific manner.
Figure 4.
Transcription Factors SP1 and GATA3 Correlate with ATF1 Expression in an Allele-Specific Manner
(A and B) SP1 (A) and GATA3 (B) are significantly overexpressed in tumors compared with the adjacent normal tissues from our CRC patients. Data were shown as the mean ± SD, and ∗∗p < 0.01 values were calculated with two-sided paired Student’s t test.
(C) SP1 and GATA3 are essential for cell growth; higher CERES scores are found in the CRC CL40 cells from the genome-wide CRISPR-Cas9-based loss-of-function screen data.
(D and E) The correlations of SP1 and GATA3 expression with ATF1 expression were measured in our CRC patients. All P values and r values were calculated with Spearman’s correlation analysis.
(F and G) SP1 (F) and GATA3 (G) are significantly overexpressed in tumors compared to adjacent normal tissues from the TCGA and GTEX datasets. Data were shown as the mean ± SD, and all ∗p < 0.05 values were calculated with a two-sided Student’s t test.
(H–L) The correlations of SP1 and GATA3 expression and ATF1 expression were calculated in TCGA CRC samples (H and I) and CRC cells (SNU-C1 [J], HCT116 [K], and LoVo [L]) with different genotypes of dbSNP: rs61926301 and dbSNP: rs7959129. ∗p < 0.05 and ∗∗p < 0.01; all p and r values in Figures 4H and 4I were calculated by Spearman’s correlation analysis, and all p values in Figures 4J–4L were calculated by a two-sided Student’s t test. Notably, in Figures 4J–4L, the left panels show expression of these genes (SP1, GATA3, and SP1+GATA3) in the x axis, and the right panels show the expression of ATF1.
The Risk Alleles of Two Variants Facilitate a Promoter-Enhancer Interaction Mediated by SP1 and GATA3 to Upregulate ATF1 Expression
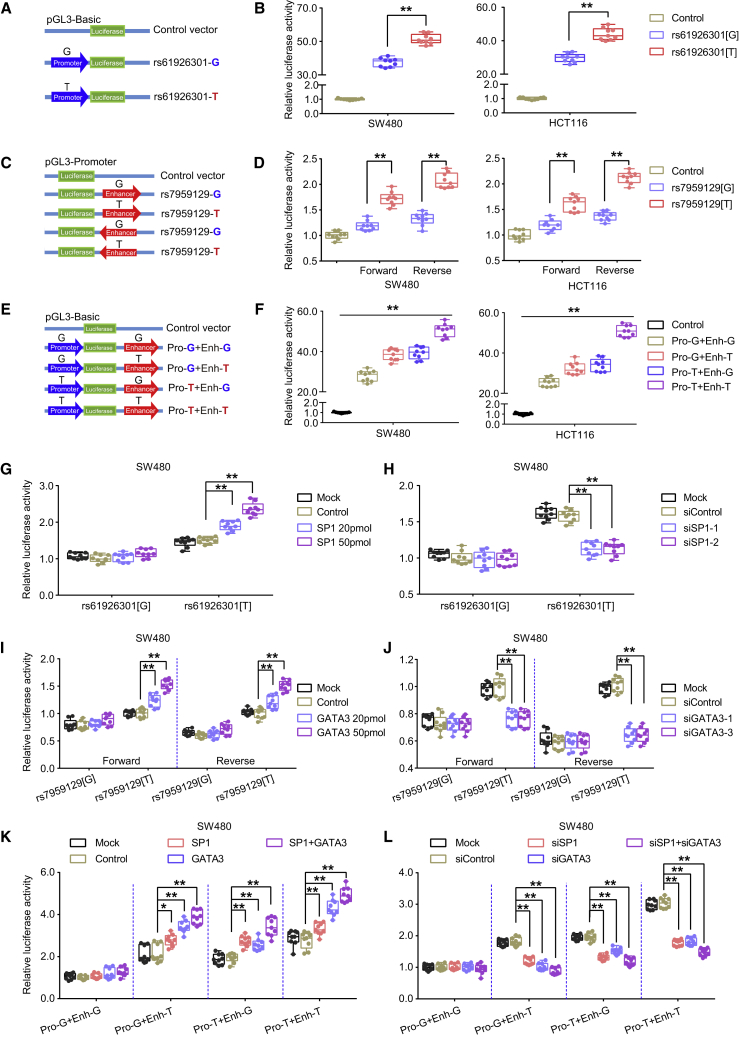
We then tested whether these two SNPs function in a promoter-enhancer manner to affect ATF1 expression. We performed luciferase reporter assays and found that the construct containing the dbSNP: rs61926301[T] allele exhibits higher promoter activity than that containing the dbSNP: rs61926301[G] allele (Figures 5A and 5B). Consistently, the construct containing the dbSNP: rs7959129[T] allele exhibits higher enhancer activity than that containing the dbSNP: rs7959129[G] allele in both forward and reverse orientations (Figures 5C and 5D). In addition, when we overexpressed SP1 or GATA3 in SW480 and HCT116 cells at an increasing dose, the luciferase activity differences between the risk and non-risk alleles of dbSNP: rs61926301 and dbSNP: rs7959129 are enhanced in a dose-dependent manner (Figures 5G, 5I, S4E, and S4G). In contrast, the differences in luciferase activity between both alleles of dbSNP: rs61926301 and dbSNP: rs7959129 are significantly attenuated when SP1 and GATA3, respectively, are knocked down (Figures 5H 5J, S4F, and S4H), suggesting the allele-specific differences of the two variants in ATF1 transcriptional activity are modulated by SP1 and GATA3. Remarkably, we found that the construct containing both the dbSNP: rs61926301[T] and the dbSNP: rs7959129[T] allele presents the highest luciferase activity among all tested constructs (Figures 5E 5F, and S4D), a finding in line with the eQTL results of the synergistic effects of these two variants, showing that carriers with both the dbSNP: rs61926301[TT] and dbSNP: rs7959129[TT] genotypes have the highest ATF1 expression levels among all tested groups (Figure 4C). The synergistic effect is provoked when SP1 or GATA3 is overexpressed, but it is substantially attenuated when SP1 or GATA3 is knocked down in CRC cell lines (Figures 5K, 5L, and S4I–S4L), indicating that the synergistic effect of these two SNPs is mediated by the TFs SP1 and GATA3.
Figure 5.
rs61926301 and rs7959129 Synergistically Facilitate ATF1 Transcriptional Activity Mediated by the Transcription Factors SP1 and GATA3
(A and B) Relative reporter gene activity of the constructs containing the dbSNP: rs61926301[G] or dbSNP: rs61926301[T] allele in CRC SW480 and HCT116 cells.
(C and D) Relative reporter gene activity of the constructs containing the dbSNP: rs7959129[G] or dbSNP: rs7959129[T] allele in both forward and reverse orientations in CRC cell lines.
(E and F) Relative reporter gene activity of the combined constructs containing both dbSNP: rs61926301 and dbSNP: rs7959129 in CRC cell lines.
(G and H) Effect of SP1 overexpression or SP1 knockdown on the relative luciferase activity of constructs containing the dbSNP: rs61926301[G] or dbSNP: rs61926301[T] alleles in SW480 cells.
(I and J) Effect of GATA3 overexpression or GATA3 knockdown on the relative luciferase activity of the constructs containing the dbSNP: rs7959129[G] or dbSNP: rs7959129[T] alleles in SW480 cells.
(K and L) Effect of SP1 and GATA3 overexpression or SP1 and GATA3 knockdown on the relative luciferase activity of the combined constructs containing both dbSNP: rs61926301 and dbSNP: rs7959129 in SW480 cells. All experiments were performed in triplicate, and each had three replicates. Data were shown as the mean ± SD, and all ∗∗p < 0.01 and ∗p < 0.05 values were calculated with a two-sided Student’s t test.
We further experimentally validated the interaction by allele-specific 3C assays in multiple CRC cell lines with different genotypes of these two SNPs. When anchored at the ATF1 promoter containing dbSNP: rs61926301, the region containing dbSNP: rs7959129 shows a stronger interaction with the ATF1 promoter containing dbSNP: rs61926301 than any of the other neighboring Mbo-I cutting sites tested (Figure 6A). Notably, the interaction frequency is more significant in the cell lines carrying both the dbSNP: rs61926301[T] allele and the dbSNP: rs7959129[T] allele (SNU-C1, HCT116, and SW480 cells) than in the other cell lines lacking either the dbSNP: rs61926301[T] allele or the dbSNP: rs7959129[T] allele (HT115, LS123, HCT15, CoLo205, and LoVo cells, Figure 6A). The genotype-specific ChIP-qPCR results also showed that the binding peaks of SP1 not only overlap the region containing dbSNP: rs61926301 but also overlap the region containing dbSNP: rs7959129; they also showed that the binding peaks of GATA3 also overlap these two regions (Figures 6B and 6C). Interestingly, the binding of SP1 and GATA3 to these two regions is more significant in SNU-C1 and HCT116 cells than in LoVo cells (Figures 6B–6D). Moreover, the binding signals are substantially attenuated with the knockdown of either SP1 or GATA3 expression (Figures 6B and 6C), further supporting the conclusion that the synergy of the TFs SP1 and GATA3 facilitates the physical interaction of these two regions in an allele-specific manner.
Figure 6.
Risk Alleles of rs61926301 and rs7959129 Facilitate a Promoter-Enhancer Interaction Mediated by SP1 and GATA3 to Upregulate ATF1 Expression
(A) Enrichment quantification of allele-specific 3C profiles in multiple CRC cell lines with different dbSNP: rs61926301 and dbSNP: rs7959129 genotypes depicts the relative interaction frequencies between the ATF1 promoter region containing dbSNP: rs61926301 as the anchor and representative Mbo-I enzyme cutting sites indicated by a dot plot, including a region containing dbSNP: rs7959129. Data were shown as the mean ± SEM from three independent experiments, and each had three replicates. All ∗∗p < 0.01 values were calculated by a two-sided Student’s t test.
(B–D) SP1 and GATA3 ChIP-qPCR signals of DNA fragments spanning dbSNP: rs61926301 and DNA fragments spanning dbSNP: rs7959129 in SNU-C1 (B), HCT116 (C), and LoVo (D) cells with different genotypes of the two variants. Results were presented as the mean ± SD from three experiments, each with triplicates. All p values were calculated by a two-sided Student’s t test.
(E) Two-stage case-control studies totally consisting of 6,213 cases and 10,388 controls were done to evaluate the interaction between dbSNP: rs61926301 and dbSNP: rs7959129 in both multiplicative and additive interaction models.
The rs61926301 and rs7959129 Variants are Associated with CRC Risk and Exhibit a Significant Interaction
To further strengthen the finding that these two SNPs are associated with CRC risk, we performed a two-stage case-control study, totally consisting of 6,213 affected individuals and 10,388 controls. The demographic characteristics of the study subjects are detailed in Table S3. As shown in Table 1, dbSNP: rs61926301 and dbSNP: rs7959129 confer genetic predisposition to CRC in both stages, after adjusting for gender, age group, smoking status, and drinking status. Furthermore, we combined the results from the discovery and replication stages and found that the dbSNP: rs61926301[TT] and dbSNP: rs7959129[TT] genotypes are associated with an increased risk of CRC and have ORs of 1.18 (95% CI = 1.13–1.24, p = 7.97 × 10−12) and 1.16 (95% CI = 1.11–1.22, p = 1.17 × 10−10), respectively. Intriguingly, significant interactions are found between these two SNPs in both multiplicative (p = 1.20 × 10−22) and additive models (p = 6.50 × 10−3, Figure 6E and Table S4). Compared to the individuals carrying both non-risk genotypes, the carriers heterozygous for both risk genotypes display a stronger genetic predisposition to CRC and have ORs ranging from 1.06 to 1.65, and the carriers homozygous for both risk genotypes present the strongest CRC risk (combined data OR = 1.77, 95% CI = 1.42–2.21, p = 3.16 × 10−7, Table S4).
Table 1.
Association Analyses between Individual SNPs and CRC Risk in the Discovery, Replication, and Combined Samples
| SNP | Genotypes |
Discovery Phase |
Replication Phase |
Combined Study |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Affected/controls | OR (95% CI)† | P value‡ | Affected/controls | OR (95% CI)† | P Value‡ | Affected/controls | OR (95% CI)† | P value‡ | ||
| dbSNP: rs61926301 | GG | 538/649 | 1.00 (reference) | 2,133/4,389 | 1.00 (reference) | 2,671/5,038 | 1.00 (reference) | |||
| GT | 787/712 | 1.30 (1.12–1.52) | 8.05 × 10−4 | 2,003/3,567 | 1.16 (1.08–1.25) | 9.08 × 10−5 | 2,790/4,279 | 1.24 (1.16–1.32) | 6.46 × 10−10 | |
| TT | 177/129 | 1.59 (1.23–2.06) | 4.31 × 10−4 | 553/910 | 1.26 (1.12–1.42) | 1.26 × 10−4 | 730/1,039 | 1.33 (1.19–1.48) | 1.63 × 10−7 | |
| Dominant | 1.35 (1.16–1.56) | 9.06 × 10−5 | 1.18 (1.10–1.27) | 4.01 × 10−6 | 1.25 (1.18–1.34) | 3.33 × 10−12 | ||||
| Recessive | 1.37 (1.07–1.74) | 1.12 × 10−2 | 1.17 (1.05–1.31) | 5.29 × 10−3 | 1.20 (1.08–1.33) | 4.29 × 10−4 | ||||
| Additive | 1.28 (1.14–1.43) | 2.92 × 10−5 | 1.14 (1.08–1.20) | 2.26 × 10−6 | 1.18 (1.13–1.24) | 7.97 × 10−12 | ||||
| dbSNP: rs7959129 | GG | 505/630 | 1.00 (reference) | 1,686/3,583 | 1.00 (reference) | 2,191/4,213 | 1.00 (reference) | |||
| GT | 753/668 | 1.38 (1.18–1.63) | 7.46 × 10−5 | 2,208/3,965 | 1.17 (1.08–1.27) | 7.31 × 10−5 | 2,961/4,633 | 1.21 (1.13–1.30) | 9.03 × 10−8 | |
| TT | 259/212 | 1.49 (1.19–1.85) | 4.49 × 10−4 | 795/1,318 | 1.30 (1.17–1.45) | 1.58 × 10−6 | 1,054/1,530 | 1.32 (1.20–1.46) | 9.25 × 10−9 | |
| Dominant | 1.40 (1.20–1.63) | 1.54 × 10−5 | 1.20 (1.12–1.30) | 1.05 × 10−6 | 1.24 (1.16–1.32) | 3.00 × 10−10 | ||||
| Recessive | 1.24 (1.01–1.51) | 3.94 × 10−2 | 1.19 (1.08–1.31) | 4.83 × 10−4 | 1.19 (1.09–1.30) | 1.02 × 10−4 | ||||
| Additive | 1.25 (1.13–1.39) | 3.28 × 10−5 | 1.15 (1.09–1.21) | 1.72 × 10−7 | 1.16 (1.11–1.22) | 1.17 × 10−10 | ||||
Abbreviations are as follows: OR = odds ratio and CI = confidence interval.
The calculation of ORs and 95% CIs was conducted under assumption that variant alleles were risk alleles.
All p values were calculated with an unconditional logistic regression model after adjusting for gender, age group, smoking status, and drinking status.
ATF1 Activates a Subset of Genes Associated with Apoptosis, Wnt, TGF-β, and MAPK Pathways and Facilitates the Early Onset of CRC
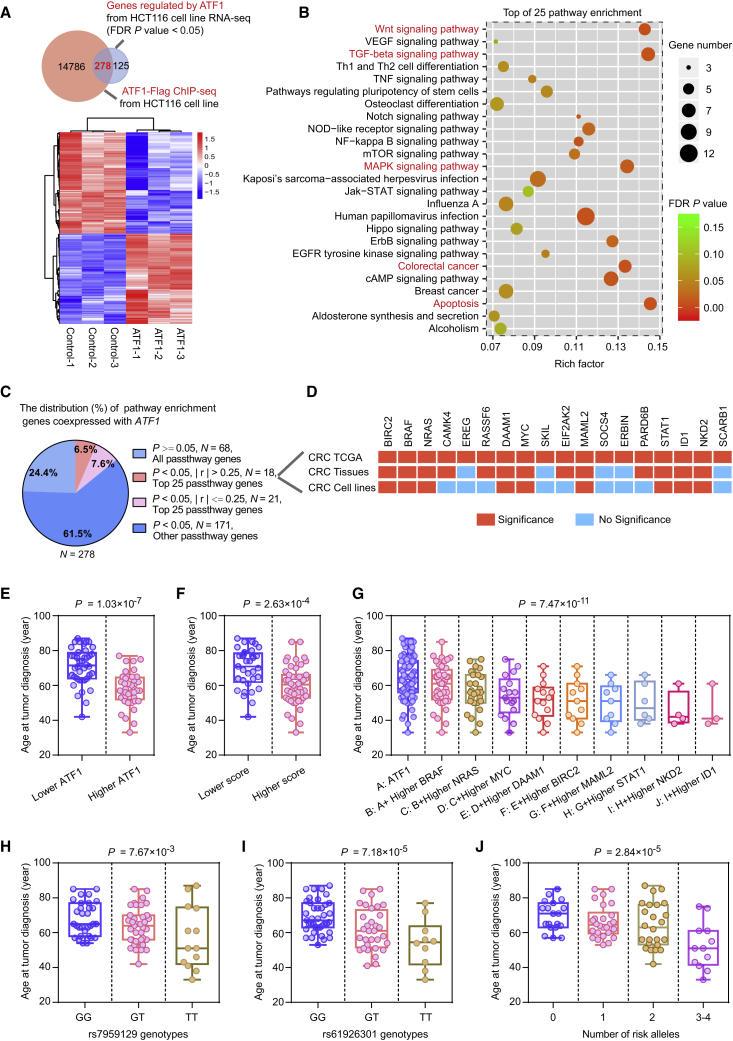
The exact mechanism and downstream transcriptional programs by which ATF1 provokes tumor activity are not well understood. We performed ChIP-seq and RNA-seq in HCT116 cells treated with either pLV-empty vector (control) or pLV-ATF1 (ATF1). Integrating RNA-seq and ChIP-seq data, we identified 278 differentially expressed genes regulated by ATF1 (Figure 7A and Table S5). KEGG pathway analysis of the 278 genes showed that several pathological pathways, including apoptosis, Wnt, TGF-β, and MAPK pathways, that are associated with CRC are significantly enriched (Figure 7B). We then tested the coexpression of ATF1 and its target genes in the TCGA cohort and our own CRC patients, and we validated this coexpression with real-time qPCR in CRC cells treated with either siRNAs targeting ATF1 or pcDNA-ATF1. Ultimately, a total of nine genes including BRAF, NRAS, MYC, BIRC2, DAAM1, MAML2, STAT1, ID1, and NKD2 (MIM: 164757; 164790; 190080; 601712; 606626; 607537; 600555; 600349; and 607852, respectively) are significantly correlated with ATF1 expression in three independent datasets (Figures 7C, 7D, and S5A–S5C), and interestingly, these genes are closely involved in the pathological activation of cell apoptosis, Wnt, TGF-beta, and MAPK pathways (Figure 7B, marked in red). Collectively, these findings illustrated that ATF1 could contribute to CRC tumorigenesis, which might be largely attributed to the pathological activation of these oncogenic pathways.
Figure 7.
ATF1 Activates a Panel of Genes Associated with Apoptosis, Wnt, TGF-β, and MAPK Pathways and Facilitates the Early Onset of CRC
(A) A Venn diagram and heatmap from RNA-seq and ChIP-seq data in HCT116 cells treated with ATF1 overexpression depict the differentially expressed genes regulated by ATF1. An FDR of p < 0.05 was considered statistically significant and was calculated by a two-sided Student’s t test.
(B) Pathway enrichment analysis of 278 ATF1 target genes revealed that the majority of these genes are involved in multiple oncogenic pathways, such as apoptosis, Wnt, TGF-beta, and MAPK pathways (marked in red).
(C) The correlations of ATF1 expression with the expression of its target genes were evaluated in the TCGA CRC samples. The values p < 0.05 and |r| > 0.25 were selected as the threshold of significance and calculated by Pearson’s correlation analysis.
(D) The correlation relationships between ATF1 and the genes in the top 25 pathways of the pathway enrichment analyses in the three independent datasets, including the TCGA cohort, our CRC patient sets, and CRC cell lines. Results were calculated by Pearson’s correlation analysis in the TCGA samples and our CRC patients and by a two-sided Student’s t test in the CRC cell lines. The red and blue diagram indicates significance or non-significance, respectively.
(E–J) The associations between age at CRC diagnosis and ATF1 expression (E), ATF1 scores (F), the synergistic effect of ATF1 and its target genes (G), the dbSNP: rs61926301 and dbSNP: rs7959129 genotypes (H and I), and the number of risk alleles of both SNPs (J) in our CRC patient sets. ATF1 activity scores were calculated on the basis of the expression of ATF1 and a panel of ATF1 target genes. Data were shown as the mean ± SD and all p values in Figures 7E and 7F were calculated by a two-sided Student’s t test, whereas those in Figures 7G–7J were calculated by linear regression analysis.
To further gain insight into the potential roles ATF1 and its target genes play in CRC susceptibility, we evaluated the clinical significance of ATF1 and its regulated genes in our CRC patients and the TCGA cohort. The age at CRC diagnosis is significantly younger in the patients with higher ATF1 expression than those with lower ATF1 expression (Figure 7E). We then devised a representative ATF1 activity score based on the expression levels of ATF1 and its target genes associated with apoptosis, Wnt, TGF-beta, and MAPK pathways. The early onset of CRC occurs more frequently in the patients with higher scores (Figure 7F). Intriguingly, the age at CRC diagnosis gradually decreases as the number of patients with high ATF1 target-gene expression increases (Figures 7G and S6A). Consistent with these findings, the analysis of data from the TCGA CRC samples also presents similar results (Figures S6B and S6C). Additionally, we also found that the carriers with the dbSNP: rs7959129[TT] or dbSNP: rs61926301[TT] genotype have a younger age at CRC diagnosis than carriers with other genotypes (Figures 7H and 7I), and the age at CRC diagnosis is gradually decreased when the number of risk alleles for these two SNPs is increased (Figure 7J). Together, these integrated analyses show that the risk SNPs dbSNP: rs61926301 and dbSNP: rs7959129 and high ATF1 expression are associated with the early onset of CRC, indicating that the synergistic effects of ATF1 and its target genes are beneficial for preventing early-onset CRC.
Discussion
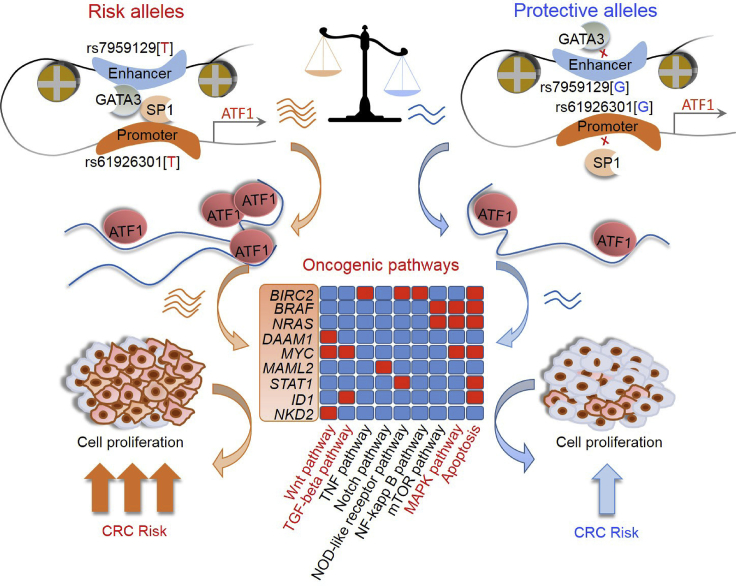
Early twin studies have suggested that genetic factors contribute to ∼35% of CRC risk,31 and GWASs have collectively identified approximately 100 CRC risk loci.3, 5, 6, 7, 8, 9, 11 Despite the need to better understand CRC development, an understanding of the functional mechanisms of these genes or risk SNPs in loci remains largely elusive. Here, through a high-throughput RNAi functional genomic screen, a large-scale population study consisting of 6,213 affected individuals and 10,388 controls, and a series of experiments at the functional level, we demonstrated for the first time that ATF1, which is located in the CRC susceptibility locus 12q13.12, is overexpressed in CRC tumors and activates a panel of genes associated with apoptosis, Wnt, TGF-β and MAPK pathways, which ultimately contribute to cell proliferation and xenograft growth in CRC. At the regulatory level, we illuminated that risk alleles of two risk SNPs, dbSNP: rs61926301 and dbSNP: rs7959129, which are located in the promoter and first intron of ATF1, respectively, facilitate a promoter-enhancer interaction mediated by the TFs SP1 and GATA3 to upregulate ATF1 expression, thus synergistically predisposing to CRC susceptibility (OR = 1.77, 95% CI = 1.42–2.21, p = 3.16 × 10−7; Figure 8).
Figure 8.
Graphical Representation of the Regulation and Function of ATF1 in CRC
Compared with the dbSNP: rs61926301[G] and dbSNP: rs7959129[G] alleles, the CRC risk variant dbSNP: rs61926301[T] and dbSNP: rs7959129[T] alleles increase the binding of TFs SP1 and GATA3 to the promoter and first intron region of ATF1, respectively, facilitating a promoter-enhancer interaction that affects ATF1 expression, and thus synergistically predisposing to CRC risk. Furthermore, ATF1 activates a subset of genes associated with cell apoptosis, Wnt, TGF-β, and MAPK pathways; the genes include BRAF, NRAS, MYC, BIRC2, DAAM1, MAML2, STAT1, ID1, and NKD2, which ultimately contribute to CRC cell growth in vitro and in vivo and result in an increasing risk of CRC.
ATF1 encodes a sequence-specific activating TF containing a bZIP DNA-binding domain, which plays critical roles in driving gene expression programs that are related to growth, survival, and other cellular activities. Previous studies characterized that ATF1 often cooperates with CREB (MIM: 123810) to nucleate constitutive heterochromatin.32, 33 In addition, ATF1 has also been reported to be fused with the Ewing’s sarcoma gene (EWS, MIM: 133450) and to be associated with the development of clear cell sarcoma (MIM: 612219).34, 35 However, the exact mechanism by which ATF1 acts in CRC is not well understood. In the present study, we revealed that ATF1 is overexpressed in CRC tumor tissues compared with their paired normal tissues in two independent cohorts and that ATF1 amplification also frequently occurs across multiple cancer types. Moreover, ATF1 is more highly expressed in advanced stages of CRC. Mechanistically, ATF1 overexpression could provoke cell proliferation and xenograft growth in vitro and in vivo by affecting cell apoptosis, and a reduction in ATF1 expression significantly attenuated this effect. Together, these findings shed light on the important role of ATF1 in the development of CRC. However, noncoding RNAs, which have been suggested to play important roles in cancers, are not involved in this study.
In light of the crucial role of ATF1 in CRC, it is essential to establish the precise regulatory mechanisms of ATF1-enhanced tumor activity. Cell- and tissue-specific gene expression programs in humans are generally controlled by TF binding and gene regulatory elements called enhancers or promoters.36 Enhancer-promoter interaction, which is one of the transcriptional regulation mechanisms, has been illustrated to be a general feature of mammalian gene control and essential for embryonic and adult cell viability and tumor pathological activation.36, 37 In this study, on the basis of allele-specific 3C and ChIP-qPCR results, we found that the risk SNPs dbSNP: rs61926301 and dbSNP: rs7959129, located in the ATF1 promoter and first intron region, respectively, facilitate a promoter-enhancer interaction that modulates the expression of ATF1 and is mediated by SP1 and GATA3, and this finding partly provides important clues as to how these two polymorphisms confer susceptibility to CRC. This finding is in line with the eQTL results showing that ATF1 expression is gradually elevated as the number of risk alleles of both variants increases for these two SNPs. Intriguingly, we also found a significant interaction between these two variants in a multiplicative model (p = 1.20 × 10−22) and in an additive model (p = 6.50 × 10−3) in the risk of CRC.
In addition, our results indicated that the TFs SP1 and GATA3 bind to the SNPs dbSNP: rs61926301 and dbSNP: rs7959129, respectively, to promote ATF1 expression, and there is higher affinity at the risk alleles. Consistently, SP1 is a well-known transcription factor that has been reported to be important in cell growth, differentiation, and the apoptosis of various tumors.26, 27 GATA3 is frequently mutated and functions as a pioneer TF that participates in a cellular reprogramming event in breast cancer (MIM: 114480).29, 30 Here, we showed that both SP1 and GATA3 are overexpressed in tumors compared with counterpart normal tissues, not only in TCGA samples, but also in our CRC-affected patients. Moreover, both of the TFs are essential for CRC cell proliferation according to the data from a CRISPR-Cas9-based loss-of-function screen.19 Notably, the binding of SP1 to the dbSNP: rs61926301[T] allele and that of GATA3 to the dbSNP: rs7959129[T] allele is significantly attenuated by the knockdown of either SP1 or GATA3. Therefore, it is possible that the synergy of these two TFs facilitates the promoter-enhancer interaction of ATF1.
Pathological activation of apoptosis, Wnt, TGF-β, and MAPK pathways is closely involved in the initiation and progression of various tumors, including CRC tumors, by promoting programs, such as cell proliferation, inflammation invasion, and metastasis, that are essential for tumorigenesis.38, 39, 40, 41 Hence, there are intense efforts to look into these pathways for biomarkers and therapeutic targets.40, 41, 42 In this study, by integrating RNA-seq and ChIP-seq data, we identified a subset of genes, including BRAF, NRAS, MYC, BIRC2, DAAM1, MAML2, STAT1, ID1, and NKD2, regulated by ATF1 and significantly enriched in these above pathways. Notably, emerging evidence has indicated that these ATF1 target genes, including MYC, BRAF, NRAS, BIRC2, and ID1, play important roles in cell apoptosis and proliferation in the progression of multiple cancers, including CRC.39, 43, 44 Therefore, these findings suggest that ATF1 can facilitate CRC development, and most of that facilitation is due to the pathological activation of these oncogenic pathways.
In contrast to the decreasing trends in adults 50 years or older, the incidence and mortality rates of CRC are increasing among all age groups between 20 and 49 years.45 According to population-based projections, by 2030, colon and rectal cancer will increase by 90% and 124% among individuals aged 20 to 34 years, and by 28% and 46% among those aged 35 to 49 years.46 Early-onset cancer is a hallmark of cancer predisposition.47 However, the drivers for the increases in the incidence of early-onset CRC have not been well elucidated. Here, we found that the synergy of ATF1 and its target genes related to apoptosis, Wnt, TGF-β, and MAPK pathways is significantly associated with the early onset of CRC in both the TCGA cohort and our CRC-affected patients. Consistently, the risk SNP genotypes are also associated with an increasingly early onset of CRC. Altogether, these findings indicate the potential of ATF1 and its regulatory elements dbSNP: rs7959129 and dbSNP: rs61926301 as risk stratification markers for the management and prevention of CRC.
In summary, through a high-throughput RNAi-based functional interrogation, a large-scale population study, and multipronged experiments, we revealed the mechanism of ATF1 and its regulatory elements dbSNP: rs61926301 and dbSNP: rs7959129 in the development of CRC. These findings not only bring us closer to an understanding of the molecular drivers of cancers, but also highlight the potential use of ATF1 in the prevention of CRC.
Acknowledgments
This work was supported by the National Key Research and Development Plan Program (2016YFC1302702 and 2016YFC1302703), the National Program for Support of Top-notch Young Professionals, the Program for Huazhong University of Science and Technology (HUST) Academic Frontier Youth Team for X.M., the National Natural Science Foundation of China (81502875), and the Fundamental Research Funds for the Central Universities, HUST (2018JYCXJJ013).
Published: June 13, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.05.004.
Accession Numbers
The accession number for the human Flag-ATF1 ChIP-seq data reported in this paper is GEO: GSE130477 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE130477).
Web Resources
cBioPortal, http://cbioportal.org
CistromeDB, http://cistrome.org/db/#/
CRISPR Design Web Tool, http://crispr.mit.edu
Depmap, https://depmap.org/portal
ENCODE, https://www.encodeproject.org/
GTEX, https://gtexportal.org
GWAS NHGRI Catalog, https://www.ebi.ac.uk/gwas/
OMIM, https://omim.org/
Oncomine, https://www.oncomine.org
RegulomeDB, http://regulome.stanford.edu/
rSNPBase, http://rsnp.psych.ac.cn/
TCGA portal, https://portal.gdc.cancer.gov/
Declaration of Interests
The authors declare no competing interests.
Supplemental Data
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]; Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R.L., Torre, L.A., and Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394-424. [DOI] [PubMed]
- 2.Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F., Jemal A., Yu X.Q., He J. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]; Chen, W., Zheng, R., Baade, P.D., Zhang, S., Zeng, H., Bray, F., Jemal, A., Yu, X.Q., and He, J. (2016). Cancer statistics in China, 2015. CA Cancer J. Clin. 66, 115-132. [DOI] [PubMed]
- 3.Tenesa A., Farrington S.M., Prendergast J.G.D., Porteous M.E., Walker M., Haq N., Barnetson R.A., Theodoratou E., Cetnarskyj R., Cartwright N. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat. Genet. 2008;40:631–637. doi: 10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tenesa, A., Farrington, S.M., Prendergast, J.G.D., Porteous, M.E., Walker, M., Haq, N., Barnetson, R.A., Theodoratou, E., Cetnarskyj, R., Cartwright, N., et al. (2008). Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat. Genet. 40, 631-637. [DOI] [PMC free article] [PubMed]
- 4.Broderick P., Carvajal-Carmona L., Pittman A.M., Webb E., Howarth K., Rowan A., Lubbe S., Spain S., Sullivan K., Fielding S., CORGI Consortium A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat. Genet. 2007;39:1315–1317. doi: 10.1038/ng.2007.18. [DOI] [PubMed] [Google Scholar]; Broderick, P., Carvajal-Carmona, L., Pittman, A.M., Webb, E., Howarth, K., Rowan, A., Lubbe, S., Spain, S., Sullivan, K., Fielding, S., et al.; CORGI Consortium (2007). A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat. Genet. 39, 1315-1317. [DOI] [PubMed]
- 5.Houlston R.S., Webb E., Broderick P., Pittman A.M., Di Bernardo M.C., Lubbe S., Chandler I., Vijayakrishnan J., Sullivan K., Penegar S., COGENT Study. Colorectal Cancer Association Study Consortium. CoRGI Consortium Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat. Genet. 2008;40:1426–1435. doi: 10.1038/ng.262. [DOI] [PMC free article] [PubMed] [Google Scholar]; Houlston, R.S., Webb, E., Broderick, P., Pittman, A.M., Di Bernardo, M.C., Lubbe, S., Chandler, I., Vijayakrishnan, J., Sullivan, K., Penegar, S., et al.; COGENT Study; Colorectal Cancer Association Study Consortium; CoRGI Consortium (2008). Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat. Genet. 40, 1426-1435. [DOI] [PMC free article] [PubMed]
- 6.Tomlinson I., Webb E., Carvajal-Carmona L., Broderick P., Kemp Z., Spain S., Penegar S., Chandler I., Gorman M., Wood W., CORGI Consortium A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat. Genet. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]; Tomlinson, I., Webb, E., Carvajal-Carmona, L., Broderick, P., Kemp, Z., Spain, S., Penegar, S., Chandler, I., Gorman, M., Wood, W., et al.; CORGI Consortium (2007). A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat. Genet. 39, 984-988. [DOI] [PubMed]
- 7.Jia W.-H., Zhang B., Matsuo K., Shin A., Xiang Y.-B., Jee S.H., Kim D.-H., Ren Z., Cai Q., Long J., Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO) Colon Cancer Family Registry (CCFR) Genome-wide association analyses in East Asians identify new susceptibility loci for colorectal cancer. Nat. Genet. 2013;45:191–196. doi: 10.1038/ng.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jia, W.-H., Zhang, B., Matsuo, K., Shin, A., Xiang, Y.-B., Jee, S.H., Kim, D.-H., Ren, Z., Cai, Q., Long, J., et al.; Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO); Colon Cancer Family Registry (CCFR) (2013). Genome-wide association analyses in East Asians identify new susceptibility loci for colorectal cancer. Nat. Genet. 45, 191-196. [DOI] [PMC free article] [PubMed]
- 8.Wang M., Gu D., Du M., Xu Z., Zhang S., Zhu L., Lu J., Zhang R., Xing J., Miao X. Common genetic variation in ETV6 is associated with colorectal cancer susceptibility. Nat. Commun. 2016;7:11478. doi: 10.1038/ncomms11478. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang, M., Gu, D., Du, M., Xu, Z., Zhang, S., Zhu, L., Lu, J., Zhang, R., Xing, J., Miao, X., et al. (2016). Common genetic variation in ETV6 is associated with colorectal cancer susceptibility. Nat. Commun. 7, 11478. [DOI] [PMC free article] [PubMed]
- 9.Zeng C., Matsuda K., Jia W.-H., Chang J., Kweon S.-S., Xiang Y.-B., Shin A., Jee S.H., Kim D.-H., Zhang B., Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO) Colorectal Transdisciplinary (CORECT) Study. Colon Cancer Family Registry (CCFR) Identification of susceptibility loci and genes for colorectal cancer risk. Gastroenterology. 2016;150:1633–1645. doi: 10.1053/j.gastro.2016.02.076. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zeng, C., Matsuda, K., Jia, W.-H., Chang, J., Kweon, S.-S., Xiang, Y.-B., Shin, A., Jee, S.H., Kim, D.-H., Zhang, B., et al.; Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO); Colorectal Transdisciplinary (CORECT) Study; Colon Cancer Family Registry (CCFR) (2016). Identification of susceptibility loci and genes for colorectal cancer risk. Gastroenterology 150, 1633-1645. [DOI] [PMC free article] [PubMed]
- 10.Zhang B., Jia W.H., Matsuda K., Kweon S.S., Matsuo K., Xiang Y.B., Shin A., Jee S.H., Kim D.H., Cai Q., Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO) Colorectal Transdisciplinary (CORECT) Study. Colon Cancer Family Registry (CCFR) Large-scale genetic study in East Asians identifies six new loci associated with colorectal cancer risk. Nat. Genet. 2014;46:533–542. doi: 10.1038/ng.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhang, B., Jia, W.H., Matsuda, K., Kweon, S.S., Matsuo, K., Xiang, Y.B., Shin, A., Jee, S.H., Kim, D.H., Cai, Q., et al.; Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO); Colorectal Transdisciplinary (CORECT) Study; Colon Cancer Family Registry (CCFR) (2014). Large-scale genetic study in East Asians identifies six new loci associated with colorectal cancer risk. Nat. Genet. 46, 533-542. [DOI] [PMC free article] [PubMed]
- 11.Zhang B., Jia W.-H., Matsuo K., Shin A., Xiang Y.-B., Matsuda K., Jee S.H., Kim D.-H., Cheah P.Y., Ren Z. Genome-wide association study identifies a new SMAD7 risk variant associated with colorectal cancer risk in East Asians. Int. J. Cancer. 2014;135:948–955. doi: 10.1002/ijc.28733. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhang, B., Jia, W.-H., Matsuo, K., Shin, A., Xiang, Y.-B., Matsuda, K., Jee, S.H., Kim, D.-H., Cheah, P.Y., Ren, Z., et al. (2014). Genome-wide association study identifies a new SMAD7 risk variant associated with colorectal cancer risk in East Asians. Int. J. Cancer 135, 948-955. [DOI] [PMC free article] [PubMed]
- 12.Chang J., Tian J., Yang Y., Zhong R., Li J., Zhai K., Ke J., Lou J., Chen W., Zhu B. A rare missense variant in TCF7L2 associates with colorectal cancer risk by interacting with a GWAS-identified regulatory variant in the MYC enhancer. Cancer Res. 2018;78:5164–5172. doi: 10.1158/0008-5472.CAN-18-0910. [DOI] [PubMed] [Google Scholar]; Chang, J., Tian, J., Yang, Y., Zhong, R., Li, J., Zhai, K., Ke, J., Lou, J., Chen, W., Zhu, B., et al. (2018). A rare missense variant in TCF7L2 associates with colorectal cancer risk by interacting with a GWAS-identified regulatory variant in the MYC enhancer. Cancer Res. 78, 5164-5172. [DOI] [PubMed]
- 13.Peters U., Jiao S., Schumacher F.R., Hutter C.M., Aragaki A.K., Baron J.A., Berndt S.I., Bézieau S., Brenner H., Butterbach K., Colon Cancer Family Registry and the Genetics and Epidemiology of Colorectal Cancer Consortium Identification of genetic susceptibility loci for colorectal tumors in a genome-wide meta-analysis. Gastroenterology. 2013;144:799–807.e24. doi: 10.1053/j.gastro.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]; Peters, U., Jiao, S., Schumacher, F.R., Hutter, C.M., Aragaki, A.K., Baron, J.A., Berndt, S.I., Bezieau, S., Brenner, H., Butterbach, K., et al.; Colon Cancer Family Registry and the Genetics and Epidemiology of Colorectal Cancer Consortium (2013). Identification of genetic susceptibility loci for colorectal tumors in a genome-wide meta-analysis. Gastroenterology 144, 799-807.e24. [DOI] [PMC free article] [PubMed]
- 14.Huyghe J.R., Bien S.A., Harrison T.A., Kang H.M., Chen S., Schmit S.L., Conti D.V., Qu C., Jeon J., Edlund C.K. Discovery of common and rare genetic risk variants for colorectal cancer. Nat. Genet. 2019;51:76–87. doi: 10.1038/s41588-018-0286-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; Huyghe, J.R., Bien, S.A., Harrison, T.A., Kang, H.M., Chen, S., Schmit, S.L., Conti, D.V., Qu, C., Jeon, J., Edlund, C.K., et al. (2019). Discovery of common and rare genetic risk variants for colorectal cancer. Nat. Genet. 51, 76-87. [DOI] [PMC free article] [PubMed]
- 15.Wang H., Haiman C.A., Burnett T., Fortini B.K., Kolonel L.N., Henderson B.E., Signorello L.B., Blot W.J., Keku T.O., Berndt S.I. Fine-mapping of genome-wide association study-identified risk loci for colorectal cancer in African Americans. Hum. Mol. Genet. 2013;22:5048–5055. doi: 10.1093/hmg/ddt337. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang, H., Haiman, C.A., Burnett, T., Fortini, B.K., Kolonel, L.N., Henderson, B.E., Signorello, L.B., Blot, W.J., Keku, T.O., Berndt, S.I., et al. (2013). Fine-mapping of genome-wide association study-identified risk loci for colorectal cancer in African Americans. Hum. Mol. Genet. 22, 5048-5055. [DOI] [PMC free article] [PubMed]
- 16.Gong J., Tian J., Lou J., Wang X., Ke J., Li J., Yang Y., Gong Y., Zhu Y., Zou D. A polymorphic MYC response element in KBTBD11 influences colorectal cancer risk, especially in interaction with an MYC-regulated SNP rs6983267. Ann. Oncol. 2018;29:632–639. doi: 10.1093/annonc/mdx789. [DOI] [PubMed] [Google Scholar]; Gong, J., Tian, J., Lou, J., Wang, X., Ke, J., Li, J., Yang, Y., Gong, Y., Zhu, Y., Zou, D., et al. (2018). A polymorphic MYC response element in KBTBD11 influences colorectal cancer risk, especially in interaction with an MYC-regulated SNP rs6983267. Ann. Oncol. 29, 632-639. [DOI] [PubMed]
- 17.Gong J., Tian J., Lou J., Ke J., Li L., Li J., Yang Y., Gong Y., Zhu Y., Zhang Y. A functional polymorphism in lnc-LAMC2-1:1 confers risk of colorectal cancer by affecting miRNA binding. Carcinogenesis. 2016;37:443–451. doi: 10.1093/carcin/bgw024. [DOI] [PubMed] [Google Scholar]; Gong, J., Tian, J., Lou, J., Ke, J., Li, L., Li, J., Yang, Y., Gong, Y., Zhu, Y., Zhang, Y., et al. (2016). A functional polymorphism in lnc-LAMC2-1:1 confers risk of colorectal cancer by affecting miRNA binding. Carcinogenesis 37, 443-451. [DOI] [PubMed]
- 18.Zhao Y., Wei L., Shao M., Huang X., Chang J., Zheng J., Chu J., Cui Q., Peng L., Luo Y. BRCA1-associated protein increases invasiveness of esophageal squamous cell carcinoma. Gastroenterology. 2017;153:1304–1319.e5. doi: 10.1053/j.gastro.2017.07.042. [DOI] [PubMed] [Google Scholar]; Zhao, Y., Wei, L., Shao, M., Huang, X., Chang, J., Zheng, J., Chu, J., Cui, Q., Peng, L., Luo, Y., et al. (2017). BRCA1-associated protein increases invasiveness of esophageal squamous cell carcinoma. Gastroenterology 153, 1304-1319.e5. [DOI] [PubMed]
- 19.Meyers R.M., Bryan J.G., McFarland J.M., Weir B.A., Sizemore A.E., Xu H., Dharia N.V., Montgomery P.G., Cowley G.S., Pantel S. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 2017;49:1779–1784. doi: 10.1038/ng.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]; Meyers, R.M., Bryan, J.G., McFarland, J.M., Weir, B.A., Sizemore, A.E., Xu, H., Dharia, N.V., Montgomery, P.G., Cowley, G.S., Pantel, S., et al. (2017). Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 49, 1779-1784. [DOI] [PMC free article] [PubMed]
- 20.McDonald E.R., 3rd, de Weck A., Schlabach M.R., Billy E., Mavrakis K.J., Hoffman G.R., Belur D., Castelletti D., Frias E., Gampa K. Project DRIVE: A compendium of cancer dependencies and synthetic lethal relationships uncovered by large-scale, deep RNAi screening. Cell. 2017;170:577–592.e10. doi: 10.1016/j.cell.2017.07.005. [DOI] [PubMed] [Google Scholar]; McDonald, E.R., 3rd, de Weck, A., Schlabach, M.R., Billy, E., Mavrakis, K.J., Hoffman, G.R., Belur, D., Castelletti, D., Frias, E., Gampa, K., et al. (2017). Project DRIVE: A compendium of cancer dependencies and synthetic lethal relationships uncovered by large-scale, deep RNAi screening. Cell 170, 577-592.e10. [DOI] [PubMed]
- 21.Fulco C.P., Munschauer M., Anyoha R., Munson G., Grossman S.R., Perez E.M., Kane M., Cleary B., Lander E.S., Engreitz J.M. Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science. 2016;354:769–773. doi: 10.1126/science.aag2445. [DOI] [PMC free article] [PubMed] [Google Scholar]; Fulco, C.P., Munschauer, M., Anyoha, R., Munson, G., Grossman, S.R., Perez, E.M., Kane, M., Cleary, B., Lander, E.S., and Engreitz, J.M. (2016). Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science 354, 769-773. [DOI] [PMC free article] [PubMed]
- 22.Tian J., Wang Z., Mei S., Yang N., Yang Y., Ke J., Zhu Y., Gong Y., Zou D., Peng X. CancerSplicingQTL: A database for genome-wide identification of splicing QTLs in human cancer. Nucleic Acids Res. 2019;47(D1):D909–D916. doi: 10.1093/nar/gky954. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tian, J., Wang, Z., Mei, S., Yang, N., Yang, Y., Ke, J., Zhu, Y., Gong, Y., Zou, D., Peng, X., et al. (2019). CancerSplicingQTL: A database for genome-wide identification of splicing QTLs in human cancer. Nucleic Acids Res. 47 (D1), D909-D916. [DOI] [PMC free article] [PubMed]
- 23.Hagège H., Klous P., Braem C., Splinter E., Dekker J., Cathala G., de Laat W., Forné T. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nat. Protoc. 2007;2:1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]; Hagege, H., Klous, P., Braem, C., Splinter, E., Dekker, J., Cathala, G., de Laat, W., and Forne, T. (2007). Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat. Protoc. 2, 1722-1733. [DOI] [PubMed]
- 24.Chang J., Zhong R., Tian J., Li J., Zhai K., Ke J., Lou J., Chen W., Zhu B., Shen N. Exome-wide analyses identify low-frequency variant in CYP26B1 and additional coding variants associated with esophageal squamous cell carcinoma. Nat. Genet. 2018;50:338–343. doi: 10.1038/s41588-018-0045-8. [DOI] [PubMed] [Google Scholar]; Chang, J., Zhong, R., Tian, J., Li, J., Zhai, K., Ke, J., Lou, J., Chen, W., Zhu, B., Shen, N., et al. (2018). Exome-wide analyses identify low-frequency variant in CYP26B1 and additional coding variants associated with esophageal squamous cell carcinoma. Nat. Genet. 50, 338-343. [DOI] [PubMed]
- 25.Roe J.S., Hwang C.I., Somerville T.D.D., Milazzo J.P., Lee E.J., Da Silva B., Maiorino L., Tiriac H., Young C.M., Miyabayashi K. Enhancer reprogramming promotes pancreatic cancer metastasis. Cell. 2017;170:875–888.e20. doi: 10.1016/j.cell.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; Roe, J.S., Hwang, C.I., Somerville, T.D.D., Milazzo, J.P., Lee, E.J., Da Silva, B., Maiorino, L., Tiriac, H., Young, C.M., Miyabayashi, K., et al. (2017). Enhancer reprogramming promotes pancreatic cancer metastasis. Cell 170, 875-888.e20. [DOI] [PMC free article] [PubMed]
- 26.Vizcaíno C., Mansilla S., Portugal J. Sp1 transcription factor: A long-standing target in cancer chemotherapy. Pharmacol. Ther. 2015;152:111–124. doi: 10.1016/j.pharmthera.2015.05.008. [DOI] [PubMed] [Google Scholar]; Vizcaino, C., Mansilla, S., and Portugal, J. (2015). Sp1 transcription factor: A long-standing target in cancer chemotherapy. Pharmacol. Ther. 152, 111-124. [DOI] [PubMed]
- 27.Chen X., Zeng K., Xu M., Hu X., Liu X., Xu T., He B., Pan Y., Sun H., Wang S. SP1-induced lncRNA-ZFAS1 contributes to colorectal cancer progression via the miR-150-5p/VEGFA axis. Cell Death Dis. 2018;9:982. doi: 10.1038/s41419-018-0962-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chen, X., Zeng, K., Xu, M., Hu, X., Liu, X., Xu, T., He, B., Pan, Y., Sun, H., and Wang, S. (2018). SP1-induced lncRNA-ZFAS1 contributes to colorectal cancer progression via the miR-150-5p/VEGFA axis. Cell Death Dis. 9, 982. [DOI] [PMC free article] [PubMed]
- 28.Yang Z., He L., Lin K., Zhang Y., Deng A., Liang Y., Li C., Wen T. The KMT1A-GATA3-STAT3 circuit is a novel self-renewal signaling of human bladder cancer stem cells. Clin. Cancer Res. 2017;23:6673–6685. doi: 10.1158/1078-0432.CCR-17-0882. [DOI] [PubMed] [Google Scholar]; Yang, Z., He, L., Lin, K., Zhang, Y., Deng, A., Liang, Y., Li, C., and Wen, T. (2017). The KMT1A-GATA3-STAT3 circuit is a novel self-renewal signaling of human bladder cancer stem cells. Clin. Cancer Res. 23, 6673-6685. [DOI] [PubMed]
- 29.Takaku M., Grimm S.A., Shimbo T., Perera L., Menafra R., Stunnenberg H.G., Archer T.K., Machida S., Kurumizaka H., Wade P.A. GATA3-dependent cellular reprogramming requires activation-domain dependent recruitment of a chromatin remodeler. Genome Biol. 2016;17:36. doi: 10.1186/s13059-016-0897-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; Takaku, M., Grimm, S.A., Shimbo, T., Perera, L., Menafra, R., Stunnenberg, H.G., Archer, T.K., Machida, S., Kurumizaka, H., and Wade, P.A. (2016). GATA3-dependent cellular reprogramming requires activation-domain dependent recruitment of a chromatin remodeler. Genome Biol. 17, 36. [DOI] [PMC free article] [PubMed]
- 30.Takaku M., Grimm S.A., Roberts J.D., Chrysovergis K., Bennett B.D., Myers P., Perera L., Tucker C.J., Perou C.M., Wade P.A. GATA3 zinc finger 2 mutations reprogram the breast cancer transcriptional network. Nat. Commun. 2018;9:1059. doi: 10.1038/s41467-018-03478-4. [DOI] [PMC free article] [PubMed] [Google Scholar]; Takaku, M., Grimm, S.A., Roberts, J.D., Chrysovergis, K., Bennett, B.D., Myers, P., Perera, L., Tucker, C.J., Perou, C.M., and Wade, P.A. (2018). GATA3 zinc finger 2 mutations reprogram the breast cancer transcriptional network. Nat. Commun. 9, 1059. [DOI] [PMC free article] [PubMed]
- 31.Lichtenstein P., Holm N.V., Verkasalo P.K., Iliadou A., Kaprio J., Koskenvuo M., Pukkala E., Skytthe A., Hemminki K. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N. Engl. J. Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]; Lichtenstein, P., Holm, N.V., Verkasalo, P.K., Iliadou, A., Kaprio, J., Koskenvuo, M., Pukkala, E., Skytthe, A., and Hemminki, K. (2000). Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N. Engl. J. Med. 343, 78-85. [DOI] [PubMed]
- 32.Jia S., Noma K., Grewal S.I. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science. 2004;304:1971–1976. doi: 10.1126/science.1099035. [DOI] [PubMed] [Google Scholar]; Jia, S., Noma, K., and Grewal, S.I. (2004). RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science 304, 1971-1976. [DOI] [PubMed]
- 33.Kvietikova I., Wenger R.H., Marti H.H., Gassmann M. The transcription factors ATF-1 and CREB-1 bind constitutively to the hypoxia-inducible factor-1 (HIF-1) DNA recognition site. Nucleic Acids Res. 1995;23:4542–4550. doi: 10.1093/nar/23.22.4542. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kvietikova, I., Wenger, R.H., Marti, H.H., and Gassmann, M. (1995). The transcription factors ATF-1 and CREB-1 bind constitutively to the hypoxia-inducible factor-1 (HIF-1) DNA recognition site. Nucleic Acids Res. 23, 4542-4550. [DOI] [PMC free article] [PubMed]
- 34.Yamada K., Ohno T., Aoki H., Semi K., Watanabe A., Moritake H., Shiozawa S., Kunisada T., Kobayashi Y., Toguchida J. EWS/ATF1 expression induces sarcomas from neural crest-derived cells in mice. J. Clin. Invest. 2013;123:600–610. doi: 10.1172/JCI63572. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yamada, K., Ohno, T., Aoki, H., Semi, K., Watanabe, A., Moritake, H., Shiozawa, S., Kunisada, T., Kobayashi, Y., Toguchida, J., et al. (2013). EWS/ATF1 expression induces sarcomas from neural crest-derived cells in mice. J. Clin. Invest. 123, 600-610. [DOI] [PMC free article] [PubMed]
- 35.Davis I.J., Kim J.J., Ozsolak F., Widlund H.R., Rozenblatt-Rosen O., Granter S.R., Du J., Fletcher J.A., Denny C.T., Lessnick S.L. Oncogenic MITF dysregulation in clear cell sarcoma: Defining the MiT family of human cancers. Cancer Cell. 2006;9:473–484. doi: 10.1016/j.ccr.2006.04.021. [DOI] [PubMed] [Google Scholar]; Davis, I.J., Kim, J.J., Ozsolak, F., Widlund, H.R., Rozenblatt-Rosen, O., Granter, S.R., Du, J., Fletcher, J.A., Denny, C.T., Lessnick, S.L., et al. (2006). Oncogenic MITF dysregulation in clear cell sarcoma: Defining the MiT family of human cancers. Cancer Cell 9, 473-484. [DOI] [PubMed]
- 36.Weintraub A.S., Li C.H., Zamudio A.V., Sigova A.A., Hannett N.M., Day D.S., Abraham B.J., Cohen M.A., Nabet B., Buckley D.L. YY1 is a structural regulator of enhancer-promoter loops. Cell. 2017;171:1573–1588.e1528. doi: 10.1016/j.cell.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; Weintraub, A.S., Li, C.H., Zamudio, A.V., Sigova, A.A., Hannett, N.M., Day, D.S., Abraham, B.J., Cohen, M.A., Nabet, B., Buckley, D.L., et al. (2017). YY1 is a structural regulator of enhancer-promoter loops. Cell 171, 1573-1588.e1528. [DOI] [PMC free article] [PubMed]
- 37.Zhang Y., Wong C.H., Birnbaum R.Y., Li G., Favaro R., Ngan C.Y., Lim J., Tai E., Poh H.M., Wong E. Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature. 2013;504:306–310. doi: 10.1038/nature12716. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhang, Y., Wong, C.H., Birnbaum, R.Y., Li, G., Favaro, R., Ngan, C.Y., Lim, J., Tai, E., Poh, H.M., Wong, E., et al. (2013). Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature 504, 306-310. [DOI] [PMC free article] [PubMed]
- 38.Calon A., Espinet E., Palomo-Ponce S., Tauriello D.V., Iglesias M., Céspedes M.V., Sevillano M., Nadal C., Jung P., Zhang X.H. Dependency of colorectal cancer on a TGF-β-driven program in stromal cells for metastasis initiation. Cancer Cell. 2012;22:571–584. doi: 10.1016/j.ccr.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; Calon, A., Espinet, E., Palomo-Ponce, S., Tauriello, D.V., Iglesias, M., Cespedes, M.V., Sevillano, M., Nadal, C., Jung, P., Zhang, X.H., et al. (2012). Dependency of colorectal cancer on a TGF-β-driven program in stromal cells for metastasis initiation. Cancer Cell 22, 571-584. [DOI] [PMC free article] [PubMed]
- 39.Sanchez-Vega F., Mina M., Armenia J., Chatila W.K., Luna A., La K.C., Dimitriadoy S., Liu D.L., Kantheti H.S., Saghafinia S., Cancer Genome Atlas Research Network oncogenic signaling pathways in The Cancer Genome Atlas. Cell. 2018;173:321–337.e10. doi: 10.1016/j.cell.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sanchez-Vega, F., Mina, M., Armenia, J., Chatila, W.K., Luna, A., La, K.C., Dimitriadoy, S., Liu, D.L., Kantheti, H.S., Saghafinia, S., et al.; Cancer Genome Atlas Research Network (2018). oncogenic signaling pathways in The Cancer Genome Atlas. Cell 173, 321-337.e10. [DOI] [PMC free article] [PubMed]
- 40.Tammela T., Sanchez-Rivera F.J., Cetinbas N.M., Wu K., Joshi N.S., Helenius K., Park Y., Azimi R., Kerper N.R., Wesselhoeft R.A. A Wnt-producing niche drives proliferative potential and progression in lung adenocarcinoma. Nature. 2017;545:355–359. doi: 10.1038/nature22334. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tammela, T., Sanchez-Rivera, F.J., Cetinbas, N.M., Wu, K., Joshi, N.S., Helenius, K., Park, Y., Azimi, R., Kerper, N.R., Wesselhoeft, R.A., et al. (2017). A Wnt-producing niche drives proliferative potential and progression in lung adenocarcinoma. Nature 545, 355-359. [DOI] [PMC free article] [PubMed]
- 41.Corcoran R.B., André T., Atreya C.E., Schellens J.H.M., Yoshino T., Bendell J.C., Hollebecque A., McRee A.J., Siena S., Middleton G. Combined BRAF, EGFR, and MEK inhibition in patients with BRAFV600E-mutant colorectal cancer. Cancer Discov. 2018;8:428–443. doi: 10.1158/2159-8290.CD-17-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]; Corcoran, R.B., Andre, T., Atreya, C.E., Schellens, J.H.M., Yoshino, T., Bendell, J.C., Hollebecque, A., McRee, A.J., Siena, S., Middleton, G., et al. (2018). Combined BRAF, EGFR, and MEK inhibition in patients with BRAFV600E-mutant colorectal cancer. Cancer Discov. 8, 428-443. [DOI] [PMC free article] [PubMed]
- 42.Sullivan R.J., Infante J.R., Janku F., Wong D.J.L., Sosman J.A., Keedy V., Patel M.R., Shapiro G.I., Mier J.W., Tolcher A.W. First-in-class ERK1/2 inhibitor ulixertinib (BVD-523) in patients with MAPK mutant advanced solid tumors: Results of a phase I dose-escalation and expansion study. Cancer Discov. 2018;8:184–195. doi: 10.1158/2159-8290.CD-17-1119. [DOI] [PubMed] [Google Scholar]; Sullivan, R.J., Infante, J.R., Janku, F., Wong, D.J.L., Sosman, J.A., Keedy, V., Patel, M.R., Shapiro, G.I., Mier, J.W., Tolcher, A.W., et al. (2018). First-in-class ERK1/2 inhibitor ulixertinib (BVD-523) in patients with MAPK mutant advanced solid tumors: Results of a phase I dose-escalation and expansion study. Cancer Discov. 8, 184-195. [DOI] [PubMed]
- 43.Pulikkan J.A., Hegde M., Ahmad H.M., Belaghzal H., Illendula A., Yu J., O’Hagan K., Ou J., Muller-Tidow C., Wolfe S.A. CBFβ-SMMHC inhibition triggers apoptosis by disrupting MYC chromatin dynamics in acute myeloid leukemia. Cell. 2018;174:172–186.e21. doi: 10.1016/j.cell.2018.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pulikkan, J.A., Hegde, M., Ahmad, H.M., Belaghzal, H., Illendula, A., Yu, J., O’Hagan, K., Ou, J., Muller-Tidow, C., Wolfe, S.A., et al. (2018). CBFβ-SMMHC inhibition triggers apoptosis by disrupting MYC chromatin dynamics in acute myeloid leukemia. Cell 174, 172-186.e21. [DOI] [PMC free article] [PubMed]
- 44.Phesse T.J., Myant K.B., Cole A.M., Ridgway R.A., Pearson H., Muncan V., van den Brink G.R., Vousden K.H., Sears R., Vassilev L.T. Endogenous c-Myc is essential for p53-induced apoptosis in response to DNA damage in vivo. Cell Death Differ. 2014;21:956–966. doi: 10.1038/cdd.2014.15. [DOI] [PMC free article] [PubMed] [Google Scholar]; Phesse, T.J., Myant, K.B., Cole, A.M., Ridgway, R.A., Pearson, H., Muncan, V., van den Brink, G.R., Vousden, K.H., Sears, R., Vassilev, L.T., et al. (2014). Endogenous c-Myc is essential for p53-induced apoptosis in response to DNA damage in vivo. Cell Death Differ. 21, 956-966. [DOI] [PMC free article] [PubMed]
- 45.Siegel R.L., Miller K.D., Jemal A. Colorectal cancer mortality rates in adults aged 20 to 54 years in the United States, 1970-2014. JAMA. 2017;318:572–574. doi: 10.1001/jama.2017.7630. [DOI] [PMC free article] [PubMed] [Google Scholar]; Siegel, R.L., Miller, K.D., and Jemal, A. (2017). Colorectal cancer mortality rates in adults aged 20 to 54 years in the United States, 1970-2014. JAMA 318, 572-574. [DOI] [PMC free article] [PubMed]
- 46.Bailey C.E., Hu C.Y., You Y.N., Bednarski B.K., Rodriguez-Bigas M.A., Skibber J.M., Cantor S.B., Chang G.J. Increasing disparities in the age-related incidences of colon and rectal cancers in the United States, 1975-2010. JAMA Surg. 2015;150:17–22. doi: 10.1001/jamasurg.2014.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bailey, C.E., Hu, C.Y., You, Y.N., Bednarski, B.K., Rodriguez-Bigas, M.A., Skibber, J.M., Cantor, S.B., and Chang, G.J. (2015). Increasing disparities in the age-related incidences of colon and rectal cancers in the United States, 1975-2010. JAMA Surg. 150, 17-22. [DOI] [PMC free article] [PubMed]
- 47.Pearlman R., Frankel W.L., Swanson B., Zhao W., Yilmaz A., Miller K., Bacher J., Bigley C., Nelsen L., Goodfellow P.J., Ohio Colorectal Cancer Prevention Initiative Study Group Prevalence and spectrum of germline cancer susceptibility gene mutations among patients with early-onset colorectal cancer. JAMA Oncol. 2017;3:464–471. doi: 10.1001/jamaoncol.2016.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pearlman, R., Frankel, W.L., Swanson, B., Zhao, W., Yilmaz, A., Miller, K., Bacher, J., Bigley, C., Nelsen, L., Goodfellow, P.J., et al.; Ohio Colorectal Cancer Prevention Initiative Study Group (2017). Prevalence and spectrum of germline cancer susceptibility gene mutations among patients with early-onset colorectal cancer. JAMA Oncol. 3, 464-471. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.