Abstract
Triple-negative breast cancer (TNBC) is an aggressive disease with a short median time from relapse to death. The increased aggressiveness, drug resistance, disease relapse, and metastasis are associated with the presence of stem cells within tumors. Several stem cell markers, such as CD24, CD44, CD133, ALDH1, and ABCG2, have been reported, but their roles in breast cancer tumorigenesis remain unclear. Herein, we apply RNA nanotechnology to deliver anti-microRNA (miRNA) for TNBC therapy. The thermodynamically and chemically stable three-way junction (3WJ) motif was utilized as the scaffold to carry an RNA aptamer binding to CD133 receptor and a locked nuclei acid (LNA) sequence for miRNA21 inhibition. Binding assays revealed the specific uptake of the nanoparticles to breast cancer stem cells (BCSCs) and TNBC cells. Functional assays showed that cancer cell migration was reduced, miR21 expression was inhibited, and downstream tumor suppressor PTEN and PDCD4 expressions were upregulated. In vitro and in vivo studies revealed that these therapeutic RNA nanoparticles did not induce cytokine secretion. Systemic injection of these RNA nanoparticles in animal trial demonstrated high specificity in TNBC tumor targeting and high efficacy for tumor growth inhibition. These results revealed the clinical translation potential of these RNA nanoparticles for TNBC therapy.
Keywords: RNA nanotechnology, RNA nanoparticles, anti-miRNA, CD133 aptamer, triple-negative breast cancer
Yin et al. utilized a stable three-way junction (3WJ) motif as a scaffold, further functionalized with CD133 aptamer, anti-miR21, and Alexa647 fluorophore, for triple-negative breast cancer targeting and therapy. The multifunctional RNA nanoparticles were capable of inhibiting tumor growth in a mouse model with undetectable toxicity.
Introduction
Triple-negative breast cancer, which accounts for 15%–20% of all breast cancer cases, is among the most aggressive breast cancers and diagnosed more frequently in young African-American women, Hispanic women, and BRCA1 mutation carriers.1 This breast cancer subtype is characterized by negative expression of estrogen receptor, progesterone receptor, and HER-2; therefore, patients with TNBC do not benefit from hormonal or trastuzumab-based therapy.2 This type of breast cancer is tentative to metastasis to brain, bone, and lungs, which is associated with poor prognosis and short survival.3, 4 Currently, TNBC patients usually receive pre-operative neoadjuvant treatment involving the administration of chemotherapeutic drug before surgery. However, the outcome for the majority who still have residual disease after treatment is relatively poor. About 60% of TNBC patients have tumors that are highly resistant to chemotherapy.5, 6 Due to the dismal diagnosis and low efficiency of traditional treatment for TNBC patients, new targeted therapeutic strategies are critically desired.
RNAi emerged in the past decade as a novel and promising gene therapy approach because it can regulate a broad set of genes efficiently and simultaneously.7, 8, 9 There are several types of RNAi agents, including small interfering RNA (siRNA), microRNA (miRNA) mimics, and anti-oncogenic miRNAs, under investigation, showing great potential in cancer treatment.10, 11, 12, 13 However, very few RNAi therapeutic agents are approved by the FDA or even enter clinical trial in phase II or III.14, 15 The poor physicochemical properties of naked RNAi agent result in quick degradation and elimination in blood circulation, thus lowering the bioavailability significantly. Nanotechnology is one of the effective strategies to improve the therapeutic efficacy of RNAi agents by protecting RNAs from quick degradation, targeting tumors efficiently, and prolonging the circulation period.16, 17, 18
RNA nanotechnology has been well developed in recent years as a delivery platform.19, 20, 21, 22 The structural flexibility and versatility make it extraordinarily attractive in biomedical application.23, 24, 25, 26 A variety of RNA architectures can be fabricated via bottom-up self-assembly, such as RNA polygons,27, 28 RNA polyhedrons,29, 30, 31 RNA dendrimers,32 and RNA micelles.33, 34 Furthermore, the approach of modular design and construction enables the RNA scaffold to incorporate functional modules. A three-way junction (3WJ) motif from the packaging RNA (pRNA) of the phi29 DNA packaging motor has been reported by our lab.20 The pRNA-3WJ can be designed and constructed to include therapeutic agents (siRNA, miRNA, chemotherapeutic drugs), targeting ligands (chemical moieties or RNA aptamers), and imaging agents (fluorophores, radioactive molecules) while retaining authentic folding properties.35, 36, 37, 38, 39 In addition, chemical stability has been drastically improved after 2′-fluoro (2′F) modification on the ribose.20, 40, 41 Due to the high thermostability, the three strands of the 3WJ motif can be assembled cohesively and remain intact at ultra-low concentration. No intrinsic toxicity of pRNA-3WJ and no histological damage have been detected after systemic injection into a mouse model.33, 35, 42
With the aim of improving bioavailability of therapeutics and realizing targeted therapy for TNBC, multifunctional RNA nanoparticles are constructed on the basis of the pRNA-3WJ scaffold to deliver anti-miR21. The 2′F-modified scaffold provides a stable platform for in vivo application, while anti-miR21 is modified by a locked nuclei acid (LNA) to enhance binding affinity to oncogenic miR21.36, 37, 43, 44 To further improve targeting specificity, an RNA aptamer against CD133 was incorporated.45, 46 CD133, also known as Prominin-1, is a membrane glycoprotein that has gained its prominence in the cancer research field due to its reported role as a cancer stem cell marker.47, 48, 49 It is commonly believed that CD133 is one of the markers for cancer stem cells, including breast cancer stem cells (BCSCs). It is not only one of the best markers to characterize cancer stem cells but also positively expressed in TNBC patients. It is reported that CD133 showed positive expression in TNBC patients by immunohistochemistry and clinicopathology analysis.50 The expression level corresponded to tumor size, clinical stage, and lymphatic metastasis. Herein, we found CD133 is also expressed on the TNBC MDA-MB-231 cell surface, which can be a potential specific targeting receptor. The RNA nanoparticles have been designed, constructed, and characterized, and they were further applied to TNBC treatment in vitro and in vivo. Tumor-targeting affinity, therapeutic efficacy, as well as gene regulation outcomes have been comprehensively evaluated in our well-established orthotropic model.
Results
Design, Construction, and Characterization of RNA Nanoparticles Carrying Anti-miR21 and the CD133 Aptamer
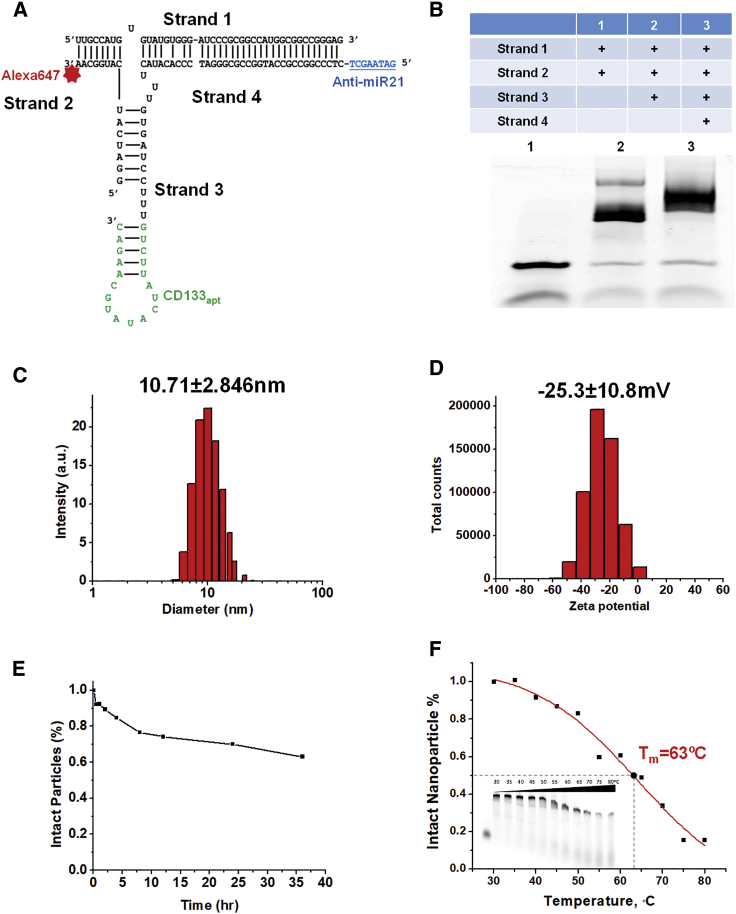
We have previously reported a 3WJ scaffold composed of three short strands, derived from pRNA of bacteriophage phi29 DNA-packaging motor. The unique branched structure of pRNA-3WJ enables the incorporation of different functional modules, including anti-miR21 as a therapeutic agent, CD133 aptamer as a target ligand, and Alexa 647 fluorophore as an imaging molecule (Figure 1A); 8 nt LNA-modified anti-miR21 is included by extension sequence. CD133 RNA aptamer screened by SELEX was capable of binding to the extracellular domain of the CD133 protein.45 The aptamer was attached to another branch of the 3WJ connected by three U bases to provide flexibility for the folding. The NIR fluorophore Alexa 647 was conjugated to the third branch for image-guided nanoparticle tracking. Upon mixing four strands at equal molar concentrations and running through an annealing program in a thermocycler, RNA nanoparticles were produced via a one-pot assembly. 3WJ/CD133apt/anti-miR21 nanoparticles revealed highly efficient stepwise assembly in gel electrophoresis (Figure 1B). Dynamic light scattering (DLS) determined that the average hydrodynamic diameter of 3WJ/CD133apt/anti-miR21 nanoparticles was about 10.71 ± 2.846 nm (Figure 1C). It is within the favorable range of size scale that can facilitate the EPR (enhanced permeability and retention) effect while avoiding rapid renal clearance. The zeta potential was determined to be −25.3 ± 10.8 mV (Figure 1D). The anionic property of RNA nanoparticles can minimize non-specific entry into normal cells, since the cell membrane is also negatively charged.
Figure.1.
Design, Construction, and Characterization of RNA Nanoparticles Carrying Anti-miR21 and the CD133 Aptamer
(A) 2D structure of 3WJ/CD133apt/anti-miR21 (blue underline sequence is 8 nt LNA-modified anti-miR21; green sequence is 19 nt CD133 aptamer; red star is Alexa647). (B) Stepwise assembly of 3WJ/CD133apt/anti-miR21 nanoparticles assayed by 12% TBE native PAGE. (C) Size distribution of 3WJ/CD133apt/anti-miR21 nanoparticles. (D) Zeta-potential of 3WJ/CD133apt/anti-miR21 nanoparticles. (E) Serum stability curve of the nanoparticles. (F) TGGE study for Tm estimation.
To improve chemical stability, 2′F modification was introduced into C and U bases by replacing the 2′-OH group with the F molecule. This replacement can reduce vulnerability of normal 2′-OH RNA attacked by RNase. The complete nanoparticles were incubated with 50% fetal bovine serum (FBS) for different time points and assessed by 12% native PAGE. The gel band intensity was quantified by ImageJ software, revealing that more than 50% of nanoparticles remained intact even at 36 h post-incubation (Figure 1E). Furthermore, temperature gradient gel electrophoresis (TGGE) was utilized to study the thermodynamic stability of RNA nanoparticles. 3WJ/CD133apt/anti-miR21 nanoparticles labeled with Alexa647 were loaded onto multiple wells of 12% native PAGE with a temperature gradient from 30°C to 80°C. The melting temperature (Tm), which represents the temperature at which 50% of nanoparticles are dissociated, was estimated by ImageJ quantification of the gel band intensity. It was found that the Tm of 3WJ/CD133apt/anti-miR21 nanoparticles was about 63°C (Figure 1F), which is much higher than the physiological temperature. These results indicated that 3WJ/CD133apt/anti-miR21 nanoparticles displayed high stability with little degradation or dissociation in the tested condition. The stable property is beneficial for in vivo circulation, distribution, and function.
Binding and Internalization of RNA Nanoparticles Mediated by the CD133 Aptamer
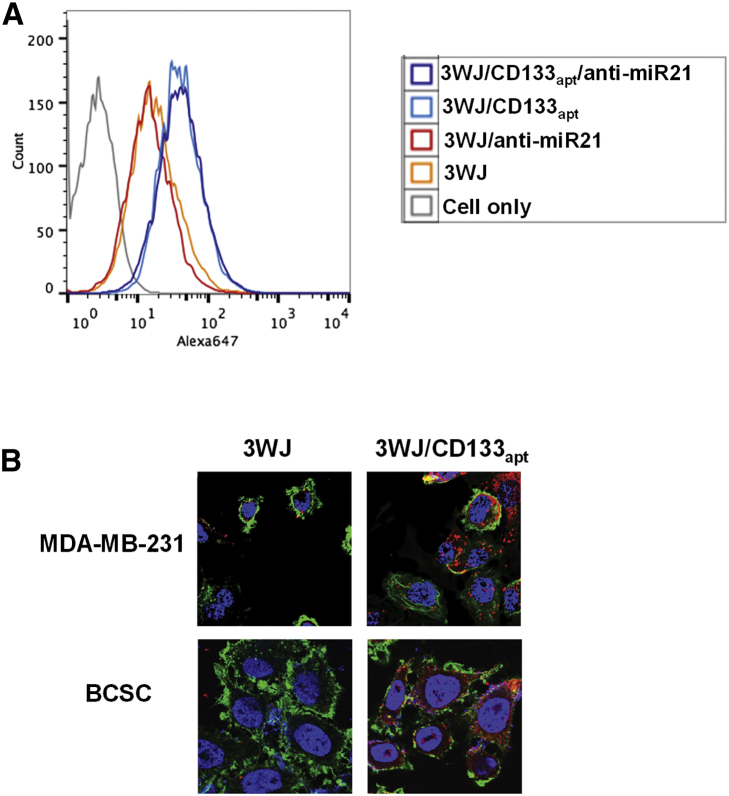
Specificity is one of the key considerations in the drug delivery field. Targeted delivery can not only improve therapy outcome but also reduce normal organ and tissue accumulation, thus lowering side effects. In this study, we incorporated CD133 RNA aptamer, which can form a defined 3D structure to bind to the CD133 marker highly expressed on the surface of BCSCs as well as TNBC. To test the nanoparticle-binding capability, Alexa 647-labeled 3WJ/CD133apt, 3WJ/CD133apt/anti-miR21, 3WJ, and 3WJ/anti-miR21 nanoparticles as control groups were incubated with MDA-MB-231 cells before being assayed by flow cytometry. Compared with control groups, 3WJ/CD133apt and 3WJ/CD133apt/anti-miR21 nanoparticles exhibited higher binding efficiency on MDA-MB-231 cells (Figure 2A), suggesting that CD133 aptamer retained its authentic folding and mediated specific targeting when incorporated into the 3WJ scaffold.
Figure 2.
Binding and Internalization of RNA Nanoparticles Mediated by the CD133 Aptamer
(A) Cell-binding assay by flow cytometry. (B) RNA nanoparticles’ internalization profile in TNBC MDA-MB-231 cells and BCSCs by confocal microscopy (blue, nuclei; green, cytoplasm; and red, RNA nanoparticles).
To evaluate the specificity, the level of CD133 marker expression on MDA-MB-231 cell surface was assessed. HEK293T cell, a CD133-negative cell line, was used as control group. 73.9% of the population of the MDA-MB-231 cells was CD133 positive (Figure S1A). In comparison, HEK293T as negative cells only showed 9% CD133 positive. In addition, the binding affinity of 3WJ/CD133apt to MDA-MB-231 cells (52.3%) was obviously higher than that to HEK293T cells (10.8%) (Figure S1B). Cells with higher CD133 expression exhibited higher 3WJ/CD133apt-binding affinity, suggesting the specificity of the CD133 aptamer.
Furthermore, confocal microscope images were taken to study whether CD133 aptamer-decorated RNA nanoparticles can enter the TNBC MDA-MB-231 cells as well as BCSCs. The images (Figure 2B) demonstrated that the fluorescence of 3WJ/CD133apt nanoparticles is strongly overlapped with the cell cytosol. The CD133 aptamer facilitates more nanoparticles to be internalized into both cell lines. The enhanced internalization profile provides a foundation for 3WJ/CD133apt/anti-miR21 to be applied in TNBC targeting and therapy.
Delivery of Anti-miR21 by the CD133 Aptamer in Cell Culture
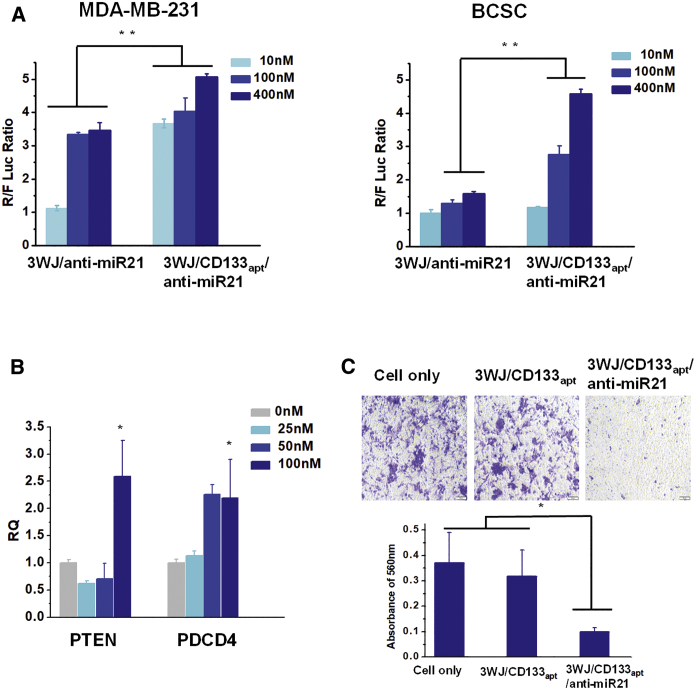
Delivery of anti-miR21 was studied by a very sensitive dual-luciferase assay, as we reported previously.36, 37, 44 A luciferase reporter vector containing the mature miR21 target sequence in the Renilla luciferase 3′ UTR region together with a Firefly luciferase as an internal control was first transfected to cells. The anti-miR21 was able to bind to intracellular miR21 in competition with the miR21 target sequence at the 3′ UTR region of the Renilla luciferase gene, thus restoring the expression of the Renilla luciferase. Therefore, a higher Renilla to Firefly luciferase (R:F luc) ratio is correlated with a higher delivery efficiency of anti-miR21 to inhibit miR21 function. The results (Figure 3A) demonstrated that the delivery of anti-miR21 by CD133 aptamer showed a higher R:F luc ratio on both TNBC MDA-MB-231 cells and BCSCs in a dose-dependent manner, indicating that the CD133 aptamer mediated favorable delivery of anti-miR21 and, subsequently, inhibited the activity of endogenously oncogenic miR21.
Figure 3.
Therapeutic Effects of 3WJ/CD133apt/Anti-miR21 Nanoparticles In Vitro
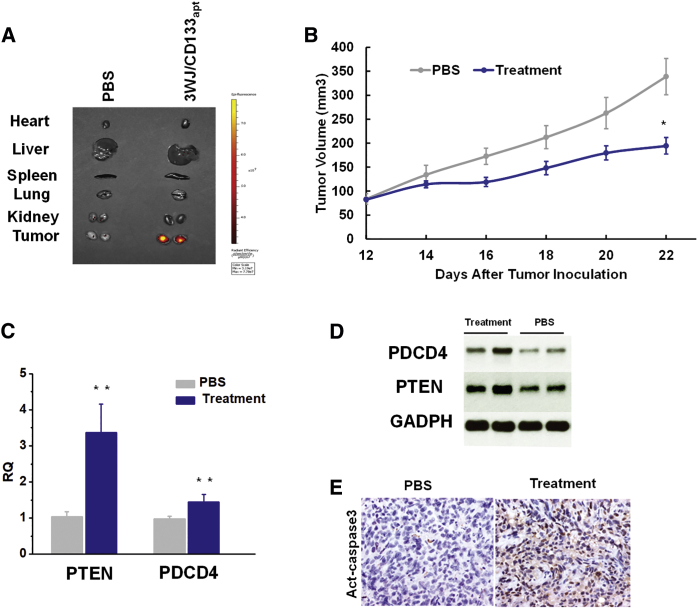
(A) Dual-luciferase assay showing delivery of anti-miR21 to TNBC MDA-MB-231 cells and BCSCs. (B) qRT-PCR demonstrating downstream gene regulation. (C) Cell migration study (top: representative images taken by inverted microscopy; bottom: crystal violet quantification by plate reader). In all plots, data are represented as mean ± SD. *p < 0.05, **p < 0.01.
The delivery efficacy of anti-miR21 was further evaluated by measuring the expression of miR21-targeted downstream gene. It has been reported that the high expression of oncogenic miR21 is associated with cancer progression and metastasis by multiple cellular regulation pathways.51, 52 Several miR21-targeted tumor suppressor genes have been identified, including PTEN and PDCD4.53, 54, 55 Upregulation of PTEN and PDCD4 can induce cell apoptosis and inhibit cell proliferation.56, 57, 58 In our study, we found obvious upregulation of PTEN and PDCD4 after treatment with 3WJ/CD133apt/anti-miR21 nanoparticles, assayed by both qPCR (Figure 3B) and western blot (Figure S1C).
To further evaluate the therapeutic effects, a cell migration study was performed. The invasion and migration capabilities of tumor cells are main contributors to cancer progression and recurrence. In our study, we evaluated TNBC cell migration properties using the Boyden chamber assay. The assay system includes a hollow plastic chamber with a porous membrane, which can be suspended over a larger well containing the cell culture medium. Cells re-suspended in FBS free medium were placed inside the chamber and allowed to migrate to the other side of the membrane. The higher the migration cell number is, the more invasive they are. Treatment with 3WJ/CD133apt/anti-miR21 nanoparticles reduced the invasive property of MDA-MB-231 cells from the analysis of the representative images taken by inverted microscopy (Figure 3C). Less crystal staining was found comparing to the other control groups. Quantitative analysis of the violet crystal revealed that the change was statistically significant (Figure 3C). The 3WJ/CD133apt without anti-miR21 did not alter the migration property, suggesting that this RNA nano-platform itself did not cause toxicity.
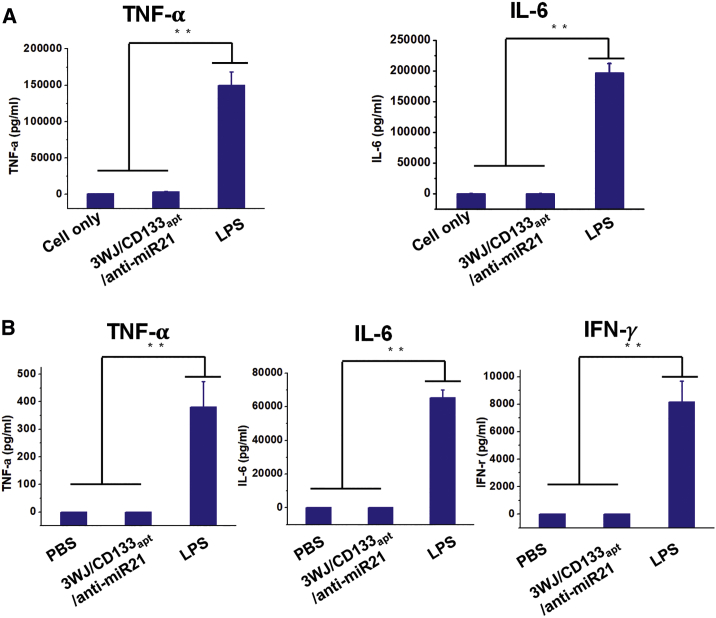
In Vitro and In Vivo Cytokine Induction Studies of RNA Nanoparticles
Immunogenicity is one of the concerns in the use of nanoparticles fabricated with biomaterials. To evaluate immunogenicity of RNA nanoparticles, production of pro-inflammatory cytokines, including TNF-α (tumor necrosis factor α), IL-6 (interleukin-6), and IFN-γ (interferon-γ), was investigated in vitro and in vivo. TNF-α is a cytokine involved in acute phase reaction and systemic inflammation.59 IL-6 is an interleukin that is secreted to stimulate immune response during infection.60 IFN-γ is a subtype of type I interferon involved in pro-inflammatory reaction, released in response to the presence of viral pathogens.61 The results (Figure 4A) showed that 3WJ/CD133apt/anti-miR21 at a therapeutic concentration (1 μM) induced neither TNF-α nor IL-6 production compared to LPS (lipopolysaccharide) positive control while incubating with mouse macrophage-like RAW 264.7 cells in vitro. Furthermore, all three cytokines were undetectable compared to LPS positive control after being systemically injected into CD-1 mice (Figure 4B). The low induction of cytokine suggests that RNA as biomaterial is biocompatible and relatively safe.
Figure 4.
Immunogenicity Study of RNA Nanoparticles In Vitro and In Vivo
(A) TNF-α and IL-6 induction in RAW264.7 cells after treatment. (B) TNF-α, IL-6, and IFN-γ levels in CD-1 mouse serum after treatment. In all plots, data are represented as mean ± SD. **p < 0.01.
Bio-Distribution, Tumor Inhibition, and Gene Regulation Studies in an Animal Trial
To study the TNBC-targeting capacity in vivo, Alexa 647-labeled RNA nanoparticles were systemically administered via the tail vein into the mice bearing TNBC xenografts. Ex vivo images of the main organs (heart, liver, spleen, lung, and kidney) and tumors were taken at 8 h post-injection. Compared with PBS control, 3WJ/CD133apt nanoparticles exhibited a strong fluorescence signal in tumor with little or no signal in other healthy organs (Figure 5A). It proved that RNA nanoparticles were cleared from main organs at 8 h but still accumulated favorably in the tumor.
Figure 5.
Evaluation of Targeting and Therapeutic Effects of 3WJ/CD133apt/Anti-miR21 Nanoparticles in an Animal Trial
(A) Fluorescence image showing RNA nanoparticle distribution in organs at 8 h post-injection. (B) TNBC tumor growth curve over the course of 5 injections. Data are represented as mean ± SEM. *p < 0.05. (C) qRT-PCR assay to study downstream gene expression level at the mRNA level. Data are represented as mean ± SD. **p < 0.01. (D) Western blot assay to study downstream gene expression at the protein level. (E) Immunohistochemistry assay demonstrating Act-caspase3 level in tumor.
The therapeutic effect of 3WJ/CD133apt/anti-miR21 nanoparticles was validated by a tumor inhibition study in an orthotropic TNBC model in nude mice. Treatment with nanoparticles significantly inhibited tumor growth in the MDA-MB-231 xenograft model compared with the control group (Figure 5B). To examine whether anti-miR21 is functionally delivered into tumor tissue, expressions of PTEN and PDCD4 in the harvested tumor samples were assayed by both qRT-PCR at the mRNA level and western blot at the protein level (Figures 5C and 5D). The data revealed that the 3WJ/CD133apt/anti-miR21 nanoparticle treatment upregulated PTEN and PDCD4 expressions in tumor tissues compared to PBS control. Furthermore, the treatment with RNA nanoparticles enhanced cell apoptosis, as indicated by increased active caspase3 (Act-caspase3) level in histological analysis (Figure 5E).
Discussion
RNAi has been extensively studied as a potential cancer treatment strategy. However, several challenges, including rapid degradation in vivo, quick elimination from kidney, non-specific binding and gene regulation, still hindered its clinical translation. We apply RNA nanotechnology to overcome these hurdles. It has been well studied that the stability of the 2′F-modified pRNA-3WJ motif was improved significantly while retaining authentic function. In addition, the three strands of the 3WJ can be self-assembled efficiently upon mix-up at room temperature. The multivalent property allows for a variety of functional module incorporation, including therapeutics, targeting ligands, and imaging modules. Therefore, this pRNA-3WJ motif is an ideal building block for drug delivery platform manipulation.
Herein, we explored the delivery of anti-miR21 by our RNA nanoparticles to treat TNBC, which is highly aggressive with a low survival rate. The ultra-stable pRNA-3WJ motif was utilized as a core scaffold to deliver 8 nt LNA-modified anti-miR21. An RNA aptamer generated via SELEX was introduced to the branch of pRNA-3WJ for specific targeting of CD133. CD133 is an important cell surface marker in cancer stem cells of various solid tumors. In this paper, we focused on using the CD133 aptamer to target mainly TNBC cells as well as BCSCs. The multifunctional RNA nanoparticles were first designed, assembled, and characterized by gel electrophoresis, DLS, serum stability, and TGGE. The results demonstrated a highly efficient assembly, defined size and zeta-potential, and high chemical and thermodynamic stability. To study the targeting capability and specificity, flow cytometry and confocal microscopy were used for comparison between RNA nanoparticles with or without the CD133 aptamer. Within the CD133 aptamer, RNA nanoparticles displayed not only higher binding affinity but also more internalization into breast cancer cells.
Upon confirmation of favorable targeting ability, experiments were expanded to study the delivery of anti-miR21. miR21 is an overexpressed oncogenic gene in TNBC and is attributed to the downregulation of tumor suppressors, such as PTEN and PDCD4. Herein, 8 nt anti-miR21 complimentary to the seed region of miR21 was used to inhibit the function of miR21. LNA modification further made the anti-miR21 a stronger competitor for miR21 binding and suppression. In vitro and in vivo studies demonstrated the effective miR21 knockdown, further upregulating PTEN and PDCD4 expression and inducing cell apoptosis. In an orthotropic TNBC mouse model, the treatment with 3WJ/CD133apt/anti-miR21 resulted in favorable tumor targeting and effective tumor suppression. The RNA nanoparticles displayed high specificity for tumor targeting due to the following reasons. First, the RNA nanoparticles are negatively charged, which can minimize non-specific cell or organ uptake since the cell membrane is also negatively charged. Second, they are larger than the 10-nm cutoff for rapid renal excretion yet small enough to enter cells via endocytosis while avoiding entrapment by liver Kupffer cells and lung, liver, or spleen macrophages. Also, nanometer-scale nanoparticles can penetrate to tumors by the EPR effect, due to the unique leaky vasculature system of a tumor. Finally, CD133 is the cell receptor of TNBC cells with an endocytosis effect. Incorporation of CD133 aptamer further facilitates RNA nanoparticles to recognize the CD133 marker that is highly expressed in TNBC and achieves specific targeting.
To address the safety issue, immunogenicity of RNA nanoparticles was studied by cytokine induction in vitro and in vivo. Consistent with our previous report, the 3WJ-based nanoparticles did not show the induction of serval pro-inflammatory-associated cytokines, including TNF-α, IL-6, and IFN-γ. We have recently shown that the immune response of RNA nanoparticles is shape, size, structure, and sequence dependent, and thus it is tunable.25 With careful control, we have been able to produce RNA nanoparticles with low or no immune response. Several other factors also account for the low immunogenicity profile of RNA nanoparticles. First, RNA nanoparticles are composed of ribonucleotides, of which the chemical nature is identical to body RNA molecules as important components of living organisms. Thus, RNA nanoparticles are more biocompatible with a lower tendency for being recognized as invaders by the immune system such as macrophages. Second, it has been reported that some physicochemical parameters, such as surface charge, play a role in the immunorecognition of nanoparticles.62 Cationic nanomaterials have a higher tendency to induce immune-activation than anionic nanomaterials. Thus, the polyanionic nature of RNA nanoparticles minimizes their non-specific interaction with the immune system. Third, the small sizes of RNA nanoparticles are advantageous for not being recognized by the bulky opsonins in the complement cascade, one of the key parts in the immune system, due to the inadequate accommodation on particle surface. Thus, they are less sensitive to phagocytosis.
Taken together, our studies provide further evidence for the feasibility of using RNA nanoparticles for therapeutic delivery, and facilitate this platform to enter into clinical trial.
Materials and Methods
Design, Construction, and Characterization of RNA Nanoparticles Carrying Anti-miR21 and the CD133 Aptamer
The 3WJ/CD133apt/anti-miR21 is composed of four strands (strands 1–4), prepared by RNA synthesizer using solid-phase synthesis. The sequences are described as follows:
Strand 1: 5′-uuG ccA uGu GuA uGu GGG Auc ccG cGG ccA uGG cGG ccG GGA G-3′
Strand 2: 5′-GGA ucA Auc AuG GcA A(C6-NH) (Alexa 647)-3′
Strand 3: 5′-ccc AcA uAc uuu Guu GAu ccu uuc AGA AcG uAu Acu Auu CuG-3′ (underlined sequence is 19 nt CD133apt)
Strand 4: 5′-+G+A+T+A+A+G+C+T CTC CCG GCC GCC ATG GCC GCG GGA T-3′ (underlined sequence is 8 nt LNA-modified anti-miR21 seed region).
2′F modification was made on the cytosine(C) and uracil(U) bases shown in lowercase to improve chemical stability of RNA, as we reported before.63, 64 3WJ/CD133apt/anti-miR21 can be self-assembled upon mixing four strands at the same molar concentrations in TES (10 mM Tris [pH 7.5–8.0], 50 mM NaCl, and 1 mM EDTA) or PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4 [pH 7.4]) buffer, heated to 90°C for 5 min and slowly cooled to 37°C. After the annealing program, 3WJ/CD133apt/anti-miR21 nanoparticles were loaded onto 12% native PAGE in TBE buffer (89 mM Tris-borate and 2 mM EDTA), stained by ethidium bromide (EB), and visualized by Typhoon FLA 7000 (GE Healthcare).
The apparent hydrodynamic diameter and z-potential of assembled 3WJ/CD133apt/anti-miR21 (1 μM in PBS buffer) nanoparticles were measured by a Zetasizer nano-ZS (Malvern Instruments) at 25°C.
To study the chemical stability, 3WJ/CD133apt/anti-miR21 nanoparticles were incubated in 50% FBS at 37°C for different times and assayed by 12% native PAGE in TBE buffer. The intact nanoparticles were quantified by intensity of gel band using ImageJ. A TGGE system (Biometra, Germany) was used to study thermodynamic stability. The assembled nanoparticles were loaded to 12% native PAGE in TBM buffer (89 mM Tris, 200 mM boric acid, and 2.5 mM MgCl2) and allowed to run at 100 V for 5 min. After running into the gel well, the gel was transferred to the TGGE apparatus with a temperature gradient from 30°C to 80°C for 60 min. Next, the gel was stained by EB, visualized by the Typhoon FLA 7000, and analyzed by ImageJ to decide the Tm.
Cell Culture
The human TNBC cell line MDA-MB-231 (American Type Culture Collection [ATCC]) cells were cultured in DMEM/F12 (1:1) medium (Thermo Scientific) containing 10% FBS. Human BCSCs (Celprogen Stem Cell Research & Therapeutics), which have been characterized as CD133-positive stem cells, were cultured in complete culture media with serum.
Macrophage RAW 264.7 and HEK293T cells were cultured in DMEM (Life Technologies) containing 10% FBS. All the cells cultured in flasks were put in a 37°C incubator under 5% CO2 and a humidified atmosphere.
Binding and Internalization of RNA Nanoparticles Mediated by the CD133 Aptamer
For the cell-binding assay, 100 nM Alexa 647-labeled 3WJ, 3WJ/anti-miR21, 3WJ/CD133apt, and 3WJ/CD133apt/anti-miR21 nanoparticles were incubated with MDA-MB-231 cells at 37°C for 1 h. After washing with PBS buffer twice, the cells were re-suspended in PBS buffer for analysis. Other binding studies for 3WJ or 3WJ/CD133apt to MDA-MB-231 or HEK293T cells follow the same procedure as described above.
To quantify CD133 expression on the cell surface, rabbit anti-CD133 polyclonal antibody (Proteintec) was incubated with MDA-MB231 and HEK293 cells for 1 h on ice. After washing by 0.1% BSA in PBS, the second antibody goat anti-rabbit immunoglobulin G (IgG) heavy and light chains (H&Ls) (Alexa Fluor 647) was incubated with cells for 40 min on ice. The cells were washed thoroughly before analysis. Flow cytometry was performed by FACSCalibur (Becton Dickinson) of The Ohio State University Comprehensive Cancer Center (OSUCCC) Analytical Cytometry Shared Resource (ACSR).
For the cell internalization study, confocal microscopy was used to detect fluorophore-labeled nanoparticles intracellularly. MDA-MB-231 cells and BCSCs were seeded on glass slides in a 24-well plate overnight. On the next day, 100 nM Alexa 647-labeled 3WJ/CD133apt and 3WJ nanoparticles were incubated with the MDA-MB-231 cells, and 200 nM nanoparticles were incubated with BCSCs at 37°C for 1 h. Followed by the incubation process and wash step, the cells were fixed by 4% paraformaldehyde (PFA) for 30 min and washed with PBS. To stain the cytoskeleton, 0.1% Triton X-100 (Sigma) in PBS was used to treat the fixed cells for 5 min to increase cell permeability, followed by Alexa Fluor 488 Phalloidin (Invitrogen) staining for 25 min. The glass slides with cells were then mounted with Prolong Gold antifade reagent with DAPI (Life Technologies) to stain the nucleus. The images of nanoparticle uptake to cells were captured by the FluoView FV1000-Filter Confocal Microscope System (Olympus).
Dual-Luciferase Assay to Study the Delivery of Anti-miR21 In Vitro
MDA-MB-231 cells and BCSCs were seeded on 24-well plates at 70% confluence in DMEM/F12 medium with 10% FBS and Complete culture media with serum, respectively. On the second day, 150 ng psi-Check2 plasmid (Promega) containing miR21-binding sequences at the 3′ UTR region of Renilla luciferase gene together with Firefly luciferase gene as internal control was transfected to cells by Lipofectamine 2000 (Life Technologies). At 4 h post-transfection, the medium was discarded and replaced with complete medium for another 2-h incubation. 3WJ/CD133apt/anti-miR21 and the control nanoparticles at final concentrations of 10, 100, and 400 nM were then incubated with cells at 37°C overnight. A dual-luciferase assay kit (Promega) was used to evaluate the inhibition of miR21 function effects, following the manufacturer’s instruction.
qRT-PCR Assay to Study Downstream Gene Expression In Vitro
The TaqMan Gene Expression Assay (Life Technologies) was performed to study miR21 downstream gene expression, including PTEN and PDCD4. 3WJ/CD133apt/anti-miR21 nanoparticles were incubated with MDA-MB-231 cells at final concentrations of 0, 25, 50, and 100 nM for 72 h. Afterward, the total RNA was extracted by TRIzol reagent (Life Technologies), purified by ethanol precipitation, and quantified by Nanodrop (Bio-Tek). The SuperScript III First-Strand Synthesis System (Life Technologies) was used to synthesize the first cDNA strand from total RNA (1 μg). The Taqman Assay was used for RT-PCR. Primers and probe set for human PTEN, PDCD4, and GADPH (housekeeping gene) were purchased from Life Technologies. PCR was performed on StepOne/StepOnePlus systems (Applied Biosystems). The data were analyzed by the ΔΔCT method.
Western Blot Assay to Study Downstream Gene Expression In Vitro
400 nM nanoparticles were incubated with MDA-MB-231 cells in triplet for 72 h. 20 μg protein quantified by BCA Protein Assay Kit (Pierce) run in 12% SDS-PAGE was transferred to polyvinylidene fluoride (PVDF) membrane (Bio-Rad) and then blocked in 5% fat-free milk at room temperature for 2 h. Afterward, the membrane was incubated overnight with primary antibody (Rabbit-PTEN, 1:1,000; Rabbit-PDCD4, 1:1,000; and Mouse-GADPH, 1:10,000) at 4°C. After washing 15 min × 3 times using TBS with Tween 20 (TBST), the membrane was then incubated with secondary antibody (goat polyclonal antibody [pAb] to rabbit IgG and goat pAb to mouse IgG: 1:20,000) at room temperature for 40 min, followed by the same washing steps. Membranes were then incubated with enhanced chemiluminescence (ECL) substrate (Bio-Rad), exposed to Amersham HyperfilmTM (GE Healthcare). The film was developed by TiBA Processor 2000A Series.
Cell Migration Study
MDA-MB-231 cells were seeded in a 6-well plate at 70% confluence 1 day prior to the treatment. On the next day, 3WJ/CD133apt/anti-miR21 and 3WJ/CD133apt nanoparticles were added to the well at a final concentration of 250 nM in triplet. After 24-h treatment, the cells were trypsinized and suspended in FBS-free DMEM/F12(1:1) medium. 50,000 cells in 250 μL DMEM/F12(1:1) medium were put in the upper well of a Boyden chamber (Sigma), which was soaked in the bottom chamber with 700 μL DMEM/F12(1:1) + 10% FBS medium in a 24-well plate. The plate was swirled gently and incubated at 37°C for another 24 h. After the incubation, medium from both upper and bottom chambers was taken out, and the membrane was fixed by 4% PFA for 30 min. Crystal violet (0.05% in 25% methanol) was used to stain the cells on the upper chamber membrane, then washed by PBS. The cells on the inner wall of the upper chamber were wiped out with a cotton tip. Images were taken for crystal violet-stained cells of the lower membrane of the upper chamber by Zeiss Axiovert 200 Inverted microscopy. To quantify the result, the stains were dissolved in 10% acetic acid and absorbance at 560 nm was recorded.
Evaluation of Cytokine Induction of RNA Nanoparticles
For the in vitro pro-inflammatory cytokine induction study, 2.5 × 105 RAW 264.7 cells/well were seeded in a 24-well plate and cultured overnight. On the next day, 1 μM 3WJ/CD133apt/anti-miR21 and LPS were diluted in DMEM and added to cells for incubation in triplet. After 16-h incubation, cell culture supernatants were collected. TNF-α and IL-6 in the collected supernatants were examined by using Mouse ELISA MAX Deluxe sets (BioLegend, San Diego, CA, USA), following the manufacturer’s instructions.
For in vivo evaluation of pro-inflammatory cytokine induction, 3WJ/CD133apt/anti-miR21 (1 μM), LPS (10 μg/mouse), and Dulbecco’s PBS (DPBS) were injected into 4- to 5-week-old female CD-1 mice (n = 3) intravenously via tail vein. At 3.5 h post-injection, blood samples were collected from mice by cardiac puncture and centrifuged at 12,800 × g for 10 min to extract serum. The levels of induced TNF-α, IL-6, and IFN-γ in serum were measured by ELISA as described above, following the manufacturer’s instructions.
Animal Models
All protocols involving animals were performed under the supervision of the University of Kentucky and The Ohio State University Institutional Animal Care and Use Committee (IACUC). To generate the TNBC xenograft model, 6-week-old female NCrnu/nu mice were purchased from Taconic. Orthotropic tumor xenografts were established by injecting 2 × 106 MDA-MB-231 cells/100 μL PBS into the fourth mammary fat pad of nude mice. For the immunogenicity study, 4- to 5-week-old female CD-1 mice purchased from Taconic were used.
In Vivo Bio-Distribution Study of RNA Nanoparticles in a TNBC Xenograft
To investigate the bio-distribution profile of RNA nanoparticles, 100 μL 20 μM Alexa 647-labeled 3WJ/CD133apt nanoparticles were injected into mice with breast TNBC xenograft by intravenous tail vein injection. PBS was injected into mice as a fluorescence negative control. After 8-h injection, the mice were sacrificed by CO2 inhalation, followed by cervical dislocation. Major organs, including heart, lungs, liver, spleen, and kidneys, together with tumors were harvested and subjected to fluorescence imaging via an IVIS Spectrum station (Xenogeny, Caliper Life Sciences), with excitation at 640 nm and emission at 680 nm for bio-distribution assessment.
In Vivo Tumor Inhibition Study by the Functional RNA Nanoparticles
When the tumor volume reached about 80 mm3 (12 days after tumor cell implantation), the mice were randomly divided into two groups (n = 6). The mice of the therapeutic group were injected with 3WJ/CD133apt/anti-miR21 nanoparticles and mice in the control group were injected with PBS. A total of 5 injections (RNA dose 5 mg/kg and LNA dose 0.26 mg/kg) was performed every other day. The tumor volume ((length * width2)/2) was measured and recorded accordingly. The day after the last dose injection, the mice were sacrificed and tumors were harvested for the target gene and protein assays.
To assess the expression level of miR21-targeted PTEN and PDCD4 at both mRNA and protein levels, the harvested tumors were transferred to a clean centrifuge tube and homogenized completely by homogenizer. For the qRT-PCR assay, the total RNA of tumor tissues was extracted by TRIzol (Life Technologies). PTEN and PDCD4 levels were further quantified using a Taqman Assay (Life Technologies) system as described before. For western blot assay, tumor tissues were lysed in radioimmunoprecipitation assay (RIPA) buffer with protease inhibitor cocktail. 15 μg protein quantified by a BCA Protein Assay Kit (Pierce) was loaded and run in 12% SDS-PAGE, and it was assayed using the same protocol as in the in vitro western blot (WB) study, as described before.
The therapeutic effects were further evaluated by histological analysis of active caspase3 activity in tumor tissues. Harvested tumors were sectioned; deparaffined by incubating with xylene (10 min × 3 times); and hydrated from 100% ethanol, 95% ethanol, 85% ethanol, and 70% ethanol to PBS buffer. Slides were then incubated with 3% H2O2 for 20 min to block endogenous peroxidase. For antigen retrieval, the slides were steamed in 10 mM citrate sodium buffer (pH 6.0) for 30 min. 5% goat serum and an Avidin/Biotin Blocking Kit (Vector Laboratories) was used for blocking. The slides were incubated with primary antibody (active caspase-3, Millipore, 1:100) at 4°C overnight, and then incubated with biotinylated goat anti-rabbit IgG at room temperature for 60 min. The conjugated antibody was detected by diaminobenzidine (DAB). All slides were counterstained with hematoxylin and images were taken by a Nikon microscope.
Author Contributions
D.S. and P.G. conceived, designed, and led the project in collaboration with R.X. H.Y. designed and performed the experiments. G.X. contributed to animal experiments and S.G. contributed to immunogenicity studies. C.X. contributed to dye conjugation of RNA. H.Y. wrote the manuscript with input from D.S. and P.G. and help from all authors.
Conflicts of Interest
P.G. is the consultant of Oxford Nanopore Technologies and Nanobio Delivery Pharmaceutical Co. Ltd. He is the cofounder of Shenzhen P&Z Bio-medical Co. Ltd. and its subsidiary US P&Z Biological Technology LLC, as well as ExonanoRNA, LLC and its subsidiary Foshan Wei-Na Biomed, Ltd.
Acknowledgments
The research was supported by DOD awards (W81XWH-15-1-0052 to D.S. and R.X.) as well as NIH grants (R01EB019036, U01CA151648, and U01CA207946 to P.G., R21CA209045 to R.X. and D.S., R01CA207772, and R01CA215095 to R.X.). P.G.’s Sylvan G. Frank Endowed Chair position in Pharmaceutics and Drug Delivery is funded by the CM Chen Foundation.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2019.04.018.
Contributor Information
Peixuan Guo, Email: guo.1091@osu.edu.
Dan Shu, Email: shu.135@osu.edu.
Supplemental Information
References
- 1.Bauer K.R., Brown M., Cress R.D., Parise C.A., Caggiano V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer. 2007;109:1721–1728. doi: 10.1002/cncr.22618. [DOI] [PubMed] [Google Scholar]; Bauer, K.R., Brown, M., Cress, R.D., Parise, C.A., and Caggiano, V. (2007). Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer 109, 1721-1728. [DOI] [PubMed]
- 2.Foulkes W.D., Smith I.E., Reis-Filho J.S. Triple-negative breast cancer. N. Engl. J. Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]; Foulkes, W.D., Smith, I.E., and Reis-Filho, J.S. (2010). Triple-negative breast cancer. N. Engl. J. Med. 363, 1938-1948. [DOI] [PubMed]
- 3.Fadare O., Tavassoli F.A. Clinical and pathologic aspects of basal-like breast cancers. Nat. Clin. Pract. Oncol. 2008;5:149–159. doi: 10.1038/ncponc1038. [DOI] [PubMed] [Google Scholar]; Fadare, O., and Tavassoli, F.A. (2008). Clinical and pathologic aspects of basal-like breast cancers. Nat. Clin. Pract. Oncol. 5, 149-159. [DOI] [PubMed]
- 4.Basho R.K., Yam C., Gilcrease M., Murthy R.K., Helgason T., Karp D.D., Meric-Bernstam F., Hess K.R., Valero V., Albarracin C. Comparative Effectiveness of an mTOR-Based Systemic Therapy Regimen in Advanced, Metaplastic and Nonmetaplastic Triple-Negative Breast Cancer. Oncologist. 2018;23:1300–1309. doi: 10.1634/theoncologist.2017-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]; Basho, R.K., Yam, C., Gilcrease, M., Murthy, R.K., Helgason, T., Karp, D.D., Meric-Bernstam, F., Hess, K.R., Valero, V., Albarracin, C., et al. (2018). Comparative Effectiveness of an mTOR-Based Systemic Therapy Regimen in Advanced, Metaplastic and Nonmetaplastic Triple-Negative Breast Cancer. Oncologist 23, 1300-1309. [DOI] [PMC free article] [PubMed]
- 5.Miller-Kleinhenz J.M., Bozeman E.N., Yang L. Targeted nanoparticles for image-guided treatment of triple-negative breast cancer: clinical significance and technological advances. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2015;7:797–816. doi: 10.1002/wnan.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]; Miller-Kleinhenz, J.M., Bozeman, E.N., and Yang, L. (2015). Targeted nanoparticles for image-guided treatment of triple-negative breast cancer: clinical significance and technological advances. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 7, 797-816. [DOI] [PMC free article] [PubMed]
- 6.Isakoff S.J. Triple-negative breast cancer: role of specific chemotherapy agents. Cancer J. 2010;16:53–61. doi: 10.1097/PPO.0b013e3181d24ff7. [DOI] [PMC free article] [PubMed] [Google Scholar]; Isakoff, S.J. (2010). Triple-negative breast cancer: role of specific chemotherapy agents. Cancer J. 16, 53-61. [DOI] [PMC free article] [PubMed]
- 7.Kim D.H., Rossi J.J. Overview of gene silencing by RNA interference. Curr. Protoc. Nucleic Acid Chem. 2009;Chapter 16:Unit 16.1. doi: 10.1002/0471142700.nc1601s36. [DOI] [PubMed] [Google Scholar]; Kim, D.H., and Rossi, J.J. (2009). Overview of gene silencing by RNA interference. Curr. Protoc. Nucleic Acid Chem. Chapter 16, Unit 16.1. [DOI] [PubMed]
- 8.Kim D.H., Rossi J.J. Strategies for silencing human disease using RNA interference. Nat. Rev. Genet. 2007;8:173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]; Kim, D.H., and Rossi, J.J. (2007). Strategies for silencing human disease using RNA interference. Nat. Rev. Genet. 8, 173-184. [DOI] [PubMed]
- 9.de Fougerolles A., Vornlocher H.P., Maraganore J., Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat. Rev. Drug Discov. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]; de Fougerolles, A., Vornlocher, H.P., Maraganore, J., and Lieberman, J. (2007). Interfering with disease: a progress report on siRNA-based therapeutics. Nat. Rev. Drug Discov. 6, 443-453. [DOI] [PMC free article] [PubMed]
- 10.Devi G.R. siRNA-based approaches in cancer therapy. Cancer Gene Ther. 2006;13:819–829. doi: 10.1038/sj.cgt.7700931. [DOI] [PubMed] [Google Scholar]; Devi, G.R. (2006). siRNA-based approaches in cancer therapy. Cancer Gene Ther. 13, 819-829. [DOI] [PubMed]
- 11.Chakraborty C., Sharma A.R., Sharma G., Doss C.G.P., Lee S.S. Therapeutic miRNA and siRNA: Moving from Bench to Clinic as Next Generation Medicine. Mol. Ther. Nucleic Acids. 2017;8:132–143. doi: 10.1016/j.omtn.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chakraborty, C., Sharma, A.R., Sharma, G., Doss, C.G.P., and Lee, S.S. (2017). Therapeutic miRNA and siRNA: Moving from Bench to Clinic as Next Generation Medicine. Mol. Ther. Nucleic Acids 8, 132-143. [DOI] [PMC free article] [PubMed]
- 12.Hayes J., Peruzzi P.P., Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol. Med. 2014;20:460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]; Hayes, J., Peruzzi, P.P., and Lawler, S. (2014). MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol. Med. 20, 460-469. [DOI] [PubMed]
- 13.Van Roosbroeck K., Fanini F., Setoyama T., Ivan C., Rodriguez-Aguayo C., Fuentes-Mattei E., Xiao L., Vannini I., Redis R.S., D’Abundo L. Combining Anti-Mir-155 with Chemotherapy for the Treatment of Lung Cancers. Clin. Cancer Res. 2017;23:2891–2904. doi: 10.1158/1078-0432.CCR-16-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]; Van Roosbroeck, K., Fanini, F., Setoyama, T., Ivan, C., Rodriguez-Aguayo, C., Fuentes-Mattei, E., Xiao, L., Vannini, I., Redis, R.S., D’Abundo, L., et al. (2017). Combining Anti-Mir-155 with Chemotherapy for the Treatment of Lung Cancers. Clin. Cancer Res. 23, 2891-2904. [DOI] [PMC free article] [PubMed]
- 14.Wittrup A., Lieberman J. Knocking down disease: a progress report on siRNA therapeutics. Nat. Rev. Genet. 2015;16:543–552. doi: 10.1038/nrg3978. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wittrup, A., and Lieberman, J. (2015). Knocking down disease: a progress report on siRNA therapeutics. Nat. Rev. Genet. 16, 543-552. [DOI] [PMC free article] [PubMed]
- 15.Bobbin M.L., Rossi J.J. RNA Interference (RNAi)-Based Therapeutics: Delivering on the Promise? Annu. Rev. Pharmacol. Toxicol. 2016;56:103–122. doi: 10.1146/annurev-pharmtox-010715-103633. [DOI] [PubMed] [Google Scholar]; Bobbin, M.L., and Rossi, J.J. (2016). RNA Interference (RNAi)-Based Therapeutics: Delivering on the Promise? Annu. Rev. Pharmacol. Toxicol. 56, 103-122. [DOI] [PubMed]
- 16.Babar I.A., Cheng C.J., Booth C.J., Liang X., Weidhaas J.B., Saltzman W.M., Slack F.J. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc. Natl. Acad. Sci. USA. 2012;109:E1695–E1704. doi: 10.1073/pnas.1201516109. [DOI] [PMC free article] [PubMed] [Google Scholar]; Babar, I.A., Cheng, C.J., Booth, C.J., Liang, X., Weidhaas, J.B., Saltzman, W.M., and Slack, F.J. (2012). Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc. Natl. Acad. Sci. USA 109, E1695-E1704. [DOI] [PMC free article] [PubMed]
- 17.Draz M.S., Fang B.A., Zhang P., Hu Z., Gu S., Weng K.C., Gray J.W., Chen F.F. Nanoparticle-mediated systemic delivery of siRNA for treatment of cancers and viral infections. Theranostics. 2014;4:872–892. doi: 10.7150/thno.9404. [DOI] [PMC free article] [PubMed] [Google Scholar]; Draz, M.S., Fang, B.A., Zhang, P., Hu, Z., Gu, S., Weng, K.C., Gray, J.W., and Chen, F.F. (2014). Nanoparticle-mediated systemic delivery of siRNA for treatment of cancers and viral infections. Theranostics 4, 872-892. [DOI] [PMC free article] [PubMed]
- 18.Gomes-da-Silva L.C., Fonseca N.A., Moura V., Pedroso de Lima M.C., Simões S., Moreira J.N. Lipid-based nanoparticles for siRNA delivery in cancer therapy: paradigms and challenges. Acc. Chem. Res. 2012;45:1163–1171. doi: 10.1021/ar300048p. [DOI] [PubMed] [Google Scholar]; Gomes-da-Silva, L.C., Fonseca, N.A., Moura, V., Pedroso de Lima, M.C., Simoes, S., and Moreira, J.N. (2012). Lipid-based nanoparticles for siRNA delivery in cancer therapy: paradigms and challenges. Acc. Chem. Res. 45, 1163-1171. [DOI] [PubMed]
- 19.Guo P. The emerging field of RNA nanotechnology. Nat. Nanotechnol. 2010;5:833–842. doi: 10.1038/nnano.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]; Guo, P. (2010). The emerging field of RNA nanotechnology. Nat. Nanotechnol. 5, 833-842. [DOI] [PMC free article] [PubMed]
- 20.Shu D., Shu Y., Haque F., Abdelmawla S., Guo P. Thermodynamically stable RNA three-way junction for constructing multifunctional nanoparticles for delivery of therapeutics. Nat. Nanotechnol. 2011;6:658–667. doi: 10.1038/nnano.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shu, D., Shu, Y., Haque, F., Abdelmawla, S., and Guo, P. (2011). Thermodynamically stable RNA three-way junction for constructing multifunctional nanoparticles for delivery of therapeutics. Nat. Nanotechnol. 6, 658-667. [DOI] [PMC free article] [PubMed]
- 21.Afonin K.A., Bindewald E., Yaghoubian A.J., Voss N., Jacovetty E., Shapiro B.A., Jaeger L. In vitro assembly of cubic RNA-based scaffolds designed in silico. Nat. Nanotechnol. 2010;5:676–682. doi: 10.1038/nnano.2010.160. [DOI] [PMC free article] [PubMed] [Google Scholar]; Afonin, K.A., Bindewald, E., Yaghoubian, A.J., Voss, N., Jacovetty, E., Shapiro, B.A., and Jaeger, L. (2010). In vitro assembly of cubic RNA-based scaffolds designed in silico. Nat. Nanotechnol. 5, 676-682. [DOI] [PMC free article] [PubMed]
- 22.Afonin K.A., Viard M., Koyfman A.Y., Martins A.N., Kasprzak W.K., Panigaj M., Desai R., Santhanam A., Grabow W.W., Jaeger L. Multifunctional RNA nanoparticles. Nano Lett. 2014;14:5662–5671. doi: 10.1021/nl502385k. [DOI] [PMC free article] [PubMed] [Google Scholar]; Afonin, K.A., Viard, M., Koyfman, A.Y., Martins, A.N., Kasprzak, W.K., Panigaj, M., Desai, R., Santhanam, A., Grabow, W.W., Jaeger, L., et al. (2014). Multifunctional RNA nanoparticles. Nano Lett. 14, 5662-5671. [DOI] [PMC free article] [PubMed]
- 23.Khisamutdinov E.F., Jasinski D.L., Guo P. RNA as a boiling-resistant anionic polymer material to build robust structures with defined shape and stoichiometry. ACS Nano. 2014;8:4771–4781. doi: 10.1021/nn5006254. [DOI] [PMC free article] [PubMed] [Google Scholar]; Khisamutdinov, E.F., Jasinski, D.L., and Guo, P. (2014). RNA as a boiling-resistant anionic polymer material to build robust structures with defined shape and stoichiometry. ACS Nano 8, 4771-4781. [DOI] [PMC free article] [PubMed]
- 24.Jaeger L., Westhof E., Leontis N.B. TectoRNA: modular assembly units for the construction of RNA nano-objects. Nucleic Acids Res. 2001;29:455–463. doi: 10.1093/nar/29.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jaeger, L., Westhof, E., and Leontis, N.B. (2001). TectoRNA: modular assembly units for the construction of RNA nano-objects. Nucleic Acids Res. 29, 455-463. [DOI] [PMC free article] [PubMed]
- 25.Guo S., Li H., Ma M., Fu J., Dong Y., Guo P. Size, Shape, and Sequence-Dependent Immunogenicity of RNA Nanoparticles. Mol. Ther. Nucleic Acids. 2017;9:399–408. doi: 10.1016/j.omtn.2017.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]; Guo, S., Li, H., Ma, M., Fu, J., Dong, Y., and Guo, P. (2017). Size, Shape, and Sequence-Dependent Immunogenicity of RNA Nanoparticles. Mol. Ther. Nucleic Acids 9, 399-408. [DOI] [PMC free article] [PubMed]
- 26.Li H., Lee T., Dziubla T., Pi F., Guo S., Xu J., Li C., Haque F., Liang X.J., Guo P. RNA as a stable polymer to build controllable and defined nanostructures for material and biomedical applications. Nano Today. 2015;10:631–655. doi: 10.1016/j.nantod.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; Li, H., Lee, T., Dziubla, T., Pi, F., Guo, S., Xu, J., Li, C., Haque, F., Liang, X.J., and Guo, P. (2015). RNA as a stable polymer to build controllable and defined nanostructures for material and biomedical applications. Nano Today 10, 631-655. [DOI] [PMC free article] [PubMed]
- 27.Jasinski D.L., Khisamutdinov E.F., Lyubchenko Y.L., Guo P. Physicochemically tunable polyfunctionalized RNA square architecture with fluorogenic and ribozymatic properties. ACS Nano. 2014;8:7620–7629. doi: 10.1021/nn502160s. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jasinski, D.L., Khisamutdinov, E.F., Lyubchenko, Y.L., and Guo, P. (2014). Physicochemically tunable polyfunctionalized RNA square architecture with fluorogenic and ribozymatic properties. ACS Nano 8, 7620-7629. [DOI] [PMC free article] [PubMed]
- 28.Ke W., Hong E., Saito R.F., Rangel M.C., Wang J., Viard M., Richardson M., Khisamutdinov E.F., Panigaj M., Dokholyan N.V. RNA-DNA fibers and polygons with controlled immunorecognition activate RNAi, FRET and transcriptional regulation of NF-κB in human cells. Nucleic Acids Res. 2019;47:1350–1361. doi: 10.1093/nar/gky1215. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ke, W., Hong, E., Saito, R.F., Rangel, M.C., Wang, J., Viard, M., Richardson, M., Khisamutdinov, E.F., Panigaj, M., Dokholyan, N.V., et al. (2019). RNA-DNA fibers and polygons with controlled immunorecognition activate RNAi, FRET and transcriptional regulation of NF-κB in human cells. Nucleic Acids Res. 47, 1350-1361. [DOI] [PMC free article] [PubMed]
- 29.Khisamutdinov E.F., Jasinski D.L., Li H., Zhang K., Chiu W., Guo P. Fabrication of RNA 3D Nanoprisms for Loading and Protection of Small RNAs and Model Drugs. Adv. Mater. 2016;28:10079–10087. doi: 10.1002/adma.201603180. [DOI] [PMC free article] [PubMed] [Google Scholar]; Khisamutdinov, E.F., Jasinski, D.L., Li, H., Zhang, K., Chiu, W., and Guo, P. (2016). Fabrication of RNA 3D Nanoprisms for Loading and Protection of Small RNAs and Model Drugs. Adv. Mater. 28, 10079-10087. [DOI] [PMC free article] [PubMed]
- 30.Li H., Zhang K., Pi F., Guo S., Shlyakhtenko L., Chiu W., Shu D., Guo P. Controllable Self-Assembly of RNA Tetrahedrons with Precise Shape and Size for Cancer Targeting. Adv. Mater. 2016;28:7501–7507. doi: 10.1002/adma.201601976. [DOI] [PMC free article] [PubMed] [Google Scholar]; Li, H., Zhang, K., Pi, F., Guo, S., Shlyakhtenko, L., Chiu, W., Shu, D., and Guo, P. (2016). Controllable Self-Assembly of RNA Tetrahedrons with Precise Shape and Size for Cancer Targeting. Adv. Mater. 28, 7501-7507. [DOI] [PMC free article] [PubMed]
- 31.Xu C., Li H., Zhang K., Binzel D.W., Yin H., Chiu W., Guo P. Photo-controlled release of paclitaxel and model drugs from RNA pyramids. Nano Res. 2019;12:41–48. doi: 10.1007/s12274-018-2174-x. [DOI] [PMC free article] [PubMed] [Google Scholar]; Xu, C., Li, H., Zhang, K., Binzel, D.W., Yin, H., Chiu, W., and Guo, P. (2019). Photo-controlled release of paclitaxel and model drugs from RNA pyramids. Nano Res. 12, 41-48. [DOI] [PMC free article] [PubMed]
- 32.Sharma A., Haque F., Pi F., Shlyakhtenko L.S., Evers B.M., Guo P. Controllable self-assembly of RNA dendrimers. Nanomedicine (Lond.) 2016;12:835–844. doi: 10.1016/j.nano.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sharma, A., Haque, F., Pi, F., Shlyakhtenko, L.S., Evers, B.M., and Guo, P. (2016). Controllable self-assembly of RNA dendrimers. Nanomedicine (Lond.) 12, 835-844. [DOI] [PMC free article] [PubMed]
- 33.Shu Y., Yin H., Rajabi M., Li H., Vieweger M., Guo S., Shu D., Guo P. RNA-based micelles: A novel platform for paclitaxel loading and delivery. J. Control. Release. 2018;276:17–29. doi: 10.1016/j.jconrel.2018.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shu, Y., Yin, H., Rajabi, M., Li, H., Vieweger, M., Guo, S., Shu, D., and Guo, P. (2018). RNA-based micelles: A novel platform for paclitaxel loading and delivery. J. Control. Release 276, 17-29. [DOI] [PMC free article] [PubMed]
- 34.Yin H., Wang H., Li Z., Shu D., Guo P. RNA Micelles for the Systemic Delivery of Anti-miRNA for Cancer Targeting and Inhibition without Ligand. ACS Nano. 2019;13:706–717. doi: 10.1021/acsnano.8b07948. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yin, H., Wang, H., Li, Z., Shu, D., and Guo, P. (2019). RNA Micelles for the Systemic Delivery of Anti-miRNA for Cancer Targeting and Inhibition without Ligand. ACS Nano 13, 706-717. [DOI] [PMC free article] [PubMed]
- 35.Cui D., Zhang C., Liu B., Shu Y., Du T., Shu D., Wang K., Dai F., Liu Y., Li C. Regression of gastric cancer by systemic injection of RNA nanoparticles carrying both ligand and siRNA. Sci. Rep. 2015;5:10726. doi: 10.1038/srep10726. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cui, D., Zhang, C., Liu, B., Shu, Y., Du, T., Shu, D., Wang, K., Dai, F., Liu, Y., Li, C., et al. (2015). Regression of gastric cancer by systemic injection of RNA nanoparticles carrying both ligand and siRNA. Sci. Rep. 5, 10726. [DOI] [PMC free article] [PubMed]
- 36.Shu D., Li H., Shu Y., Xiong G., Carson W.E., 3rd, Haque F., Xu R., Guo P. Systemic delivery of anti-miRNA for suppression of triple negative breast cancer utilizing RNA nanotechnology. ACS Nano. 2015;9:9731–9740. doi: 10.1021/acsnano.5b02471. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shu, D., Li, H., Shu, Y., Xiong, G., Carson, W.E., 3rd, Haque, F., Xu, R., and Guo, P. (2015). Systemic delivery of anti-miRNA for suppression of triple negative breast cancer utilizing RNA nanotechnology. ACS Nano 9, 9731-9740. [DOI] [PMC free article] [PubMed]
- 37.Binzel D.W., Shu Y., Li H., Sun M., Zhang Q., Shu D., Guo B., Guo P. Specific Delivery of MiRNA for High Efficient Inhibition of Prostate Cancer by RNA Nanotechnology. Mol. Ther. 2016;24:1267–1277. doi: 10.1038/mt.2016.85. [DOI] [PMC free article] [PubMed] [Google Scholar]; Binzel, D.W., Shu, Y., Li, H., Sun, M., Zhang, Q., Shu, D., Guo, B., and Guo, P. (2016). Specific Delivery of MiRNA for High Efficient Inhibition of Prostate Cancer by RNA Nanotechnology. Mol. Ther. 24, 1267-1277. [DOI] [PMC free article] [PubMed]
- 38.Pi F., Zhang H., Li H., Thiviyanathan V., Gorenstein D.G., Sood A.K., Guo P. RNA nanoparticles harboring annexin A2 aptamer can target ovarian cancer for tumor-specific doxorubicin delivery. Nanomedicine (Lond.) 2017;13:1183–1193. doi: 10.1016/j.nano.2016.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pi, F., Zhang, H., Li, H., Thiviyanathan, V., Gorenstein, D.G., Sood, A.K., and Guo, P. (2017). RNA nanoparticles harboring annexin A2 aptamer can target ovarian cancer for tumor-specific doxorubicin delivery. Nanomedicine (Lond.) 13, 1183-1193. [DOI] [PMC free article] [PubMed]
- 39.Jasinski D.L., Yin H., Li Z., Guo P. Hydrophobic Effect from Conjugated Chemicals or Drugs on In Vivo Biodistribution of RNA Nanoparticles. Hum. Gene Ther. 2018;29:77–86. doi: 10.1089/hum.2017.054. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jasinski, D.L., Yin, H., Li, Z., and Guo, P. (2018). Hydrophobic Effect from Conjugated Chemicals or Drugs on In Vivo Biodistribution of RNA Nanoparticles. Hum. Gene Ther. 29, 77-86 [DOI] [PMC free article] [PubMed]
- 40.Layzer J.M., McCaffrey A.P., Tanner A.K., Huang Z., Kay M.A., Sullenger B.A. In vivo activity of nuclease-resistant siRNAs. RNA. 2004;10:766–771. doi: 10.1261/rna.5239604. [DOI] [PMC free article] [PubMed] [Google Scholar]; Layzer, J.M., McCaffrey, A.P., Tanner, A.K., Huang, Z., Kay, M.A., and Sullenger, B.A. (2004). In vivo activity of nuclease-resistant siRNAs. RNA 10, 766-771. [DOI] [PMC free article] [PubMed]
- 41.Manoharan M., Akinc A., Pandey R.K., Qin J., Hadwiger P., John M., Mills K., Charisse K., Maier M.A., Nechev L. Unique gene-silencing and structural properties of 2′-fluoro-modified siRNAs. Angew. Chem. Int. Ed. Engl. 2011;50:2284–2288. doi: 10.1002/anie.201006519. [DOI] [PMC free article] [PubMed] [Google Scholar]; Manoharan, M., Akinc, A., Pandey, R.K., Qin, J., Hadwiger, P., John, M., Mills, K., Charisse, K., Maier, M.A., Nechev, L., et al. (2011). Unique gene-silencing and structural properties of 2′-fluoro-modified siRNAs. Angew. Chem. Int. Ed. Engl. 50, 2284-2288. [DOI] [PMC free article] [PubMed]
- 42.Zhang Y., Leonard M., Shu Y., Yang Y., Shu D., Guo P., Zhang X. Overcoming Tamoxifen Resistance of Human Breast Cancer by Targeted Gene Silencing Using Multifunctional pRNA Nanoparticles. ACS Nano. 2017;11:335–346. doi: 10.1021/acsnano.6b05910. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhang, Y., Leonard, M., Shu, Y., Yang, Y., Shu, D., Guo, P., and Zhang, X. (2017). Overcoming Tamoxifen Resistance of Human Breast Cancer by Targeted Gene Silencing Using Multifunctional pRNA Nanoparticles. ACS Nano 11, 335-346. [DOI] [PMC free article] [PubMed]
- 43.Obad S., dos Santos C.O., Petri A., Heidenblad M., Broom O., Ruse C., Fu C., Lindow M., Stenvang J., Straarup E.M. Silencing of microRNA families by seed-targeting tiny LNAs. Nat. Genet. 2011;43:371–378. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]; Obad, S., dos Santos, C.O., Petri, A., Heidenblad, M., Broom, O., Ruse, C., Fu, C., Lindow, M., Stenvang, J., Straarup, E.M., et al. (2011). Silencing of microRNA families by seed-targeting tiny LNAs. Nat. Genet. 43, 371-378. [DOI] [PMC free article] [PubMed]
- 44.Lee T.J., Yoo J.Y., Shu D., Li H., Zhang J., Yu J.G., Jaime-Ramirez A.C., Acunzo M., Romano G., Cui R. RNA Nanoparticle-Based Targeted Therapy for Glioblastoma through Inhibition of Oncogenic miR-21. Mol. Ther. 2017;25:1544–1555. doi: 10.1016/j.ymthe.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lee, T.J., Yoo, J.Y., Shu, D., Li, H., Zhang, J., Yu, J.G., Jaime-Ramirez, A.C., Acunzo, M., Romano, G., Cui, R., et al. (2017) RNA Nanoparticle-Based Targeted Therapy for Glioblastoma through Inhibition of Oncogenic miR-21. Mol. Ther. 25, 1544-1555. [DOI] [PMC free article] [PubMed]
- 45.Shigdar S., Qiao L., Zhou S.F., Xiang D., Wang T., Li Y., Lim L.Y., Kong L., Li L., Duan W. RNA aptamers targeting cancer stem cell marker CD133. Cancer Lett. 2013;330:84–95. doi: 10.1016/j.canlet.2012.11.032. [DOI] [PubMed] [Google Scholar]; Shigdar, S., Qiao, L., Zhou, S.F., Xiang, D., Wang, T., Li, Y., Lim, L.Y., Kong, L., Li, L., and Duan, W. (2013). RNA aptamers targeting cancer stem cell marker CD133. Cancer Lett. 330, 84-95. [DOI] [PubMed]
- 46.Guo X., Zhu X., Gao J., Liu D., Dong C., Jin X. PLGA nanoparticles with CD133 aptamers for targeted delivery and sustained release of propranolol to hemangioma. Nanomedicine (Lond.) 2017;12:2611–2624. doi: 10.2217/nnm-2017-0130. [DOI] [PubMed] [Google Scholar]; Guo, X., Zhu, X., Gao, J., Liu, D., Dong, C., and Jin, X. (2017). PLGA nanoparticles with CD133 aptamers for targeted delivery and sustained release of propranolol to hemangioma. Nanomedicine (Lond.) 12, 2611-2624. [DOI] [PubMed]
- 47.Mizrak D., Brittan M., Alison M. CD133: molecule of the moment. J. Pathol. 2008;214:3–9. doi: 10.1002/path.2283. [DOI] [PubMed] [Google Scholar]; Mizrak, D., Brittan, M., and Alison, M. (2008). CD133: molecule of the moment. J. Pathol. 214, 3-9. [DOI] [PubMed]
- 48.Li Z. CD133: a stem cell biomarker and beyond. Exp. Hematol. Oncol. 2013;2:17. doi: 10.1186/2162-3619-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]; Li, Z. (2013). CD133: a stem cell biomarker and beyond. Exp. Hematol. Oncol. 2, 17. [DOI] [PMC free article] [PubMed]
- 49.Kazama S., Kishikawa J., Kiyomatsu T., Kawai K., Nozawa H., Ishihara S., Watanabe T. Expression of the stem cell marker CD133 is related to tumor development in colorectal carcinogenesis. Asian J. Surg. 2018;41:274–278. doi: 10.1016/j.asjsur.2016.12.002. [DOI] [PubMed] [Google Scholar]; Kazama, S., Kishikawa, J., Kiyomatsu, T., Kawai, K., Nozawa, H., Ishihara, S., and Watanabe, T. (2018). Expression of the stem cell marker CD133 is related to tumor development in colorectal carcinogenesis. Asian J. Surg. 41, 274-278. [DOI] [PubMed]
- 50.Han M., Guo L., Zhang Y., Huang B., Chen A., Chen W., Liu X., Sun S., Wang K., Liu A., Li X. Clinicopathological and Prognostic Significance of CD133 in Glioma Patients: A Meta-Analysis. Mol. Neurobiol. 2016;53:720–727. doi: 10.1007/s12035-014-9018-9. [DOI] [PubMed] [Google Scholar]; Han, M., Guo, L., Zhang, Y., Huang, B., Chen, A., Chen, W., Liu, X., Sun, S., Wang, K., Liu, A., and Li, X. (2016). Clinicopathological and Prognostic Significance of CD133 in Glioma Patients: A Meta-Analysis. Mol. Neurobiol. 53, 720-727. [DOI] [PubMed]
- 51.Pfeffer S.R., Yang C.H., Pfeffer L.M. The Role of miR-21 in Cancer. Drug Dev. Res. 2015;76:270–277. doi: 10.1002/ddr.21257. [DOI] [PubMed] [Google Scholar]; Pfeffer, S.R., Yang, C.H., and Pfeffer, L.M. (2015). The Role of miR-21 in Cancer. Drug Dev. Res. 76, 270-277. [DOI] [PubMed]
- 52.Feng Y.H., Tsao C.J. Emerging role of microRNA-21 in cancer. Biomed. Rep. 2016;5:395–402. doi: 10.3892/br.2016.747. [DOI] [PMC free article] [PubMed] [Google Scholar]; Feng, Y.H., and Tsao, C.J. (2016). Emerging role of microRNA-21 in cancer. Biomed. Rep. 5, 395-402. [DOI] [PMC free article] [PubMed]
- 53.Meng F., Henson R., Wehbe-Janek H., Ghoshal K., Jacob S.T., Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]; Meng, F., Henson, R., Wehbe-Janek, H., Ghoshal, K., Jacob, S.T., and Patel, T. (2007). MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 133, 647-658. [DOI] [PMC free article] [PubMed]
- 54.Frankel L.B., Christoffersen N.R., Jacobsen A., Lindow M., Krogh A., Lund A.H. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J. Biol. Chem. 2008;283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]; Frankel, L.B., Christoffersen, N.R., Jacobsen, A., Lindow, M., Krogh, A., and Lund, A.H. (2008). Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J. Biol. Chem. 283, 1026-1033. [DOI] [PubMed]
- 55.Qi L., Bart J., Tan L.P., Platteel I., Sluis Tv., Huitema S., Harms G., Fu L., Hollema H., Berg Av. Expression of miR-21 and its targets (PTEN, PDCD4, TM1) in flat epithelial atypia of the breast in relation to ductal carcinoma in situ and invasive carcinoma. BMC Cancer. 2009;9:163. doi: 10.1186/1471-2407-9-163. [DOI] [PMC free article] [PubMed] [Google Scholar]; Qi, L., Bart, J., Tan, L.P., Platteel, I., Sluis, Tv., Huitema, S., Harms, G., Fu, L., Hollema, H., and Berg, Av. (2009). Expression of miR-21 and its targets (PTEN, PDCD4, TM1) in flat epithelial atypia of the breast in relation to ductal carcinoma in situ and invasive carcinoma. BMC Cancer 9, 163. [DOI] [PMC free article] [PubMed]
- 56.Goberdhan D.C., Wilson C. PTEN: tumour suppressor, multifunctional growth regulator and more. Hum. Mol. Genet. 2003;12:R239–R248. doi: 10.1093/hmg/ddg288. [DOI] [PubMed] [Google Scholar]; Goberdhan, D.C., and Wilson, C. (2003). PTEN: tumour suppressor, multifunctional growth regulator and more. Hum. Mol. Genet. 12, R239-R248. [DOI] [PubMed]
- 57.Chalhoub N., Baker S.J. PTEN and the PI3-kinase pathway in cancer. Annu. Rev. Pathol. 2009;4:127–150. doi: 10.1146/annurev.pathol.4.110807.092311. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chalhoub, N., and Baker, S.J. (2009). PTEN and the PI3-kinase pathway in cancer. Annu. Rev. Pathol. 4, 127-150. [DOI] [PMC free article] [PubMed]
- 58.Lankat-Buttgereit B., Göke R. The tumour suppressor Pdcd4: recent advances in the elucidation of function and regulation. Biol. Cell. 2009;101:309–317. doi: 10.1042/BC20080191. [DOI] [PubMed] [Google Scholar]; Lankat-Buttgereit, B., and Goke, R. (2009). The tumour suppressor Pdcd4: recent advances in the elucidation of function and regulation. Biol. Cell 101, 309-317. [DOI] [PubMed]
- 59.Jaffer U., Wade R.G., Gourlay T. Cytokines in the systemic inflammatory response syndrome: a review. HSR Proc. Intensive Care Cardiovasc. Anesth. 2010;2:161–175. [PMC free article] [PubMed] [Google Scholar]; Jaffer, U., Wade, R.G., and Gourlay, T. (2010). Cytokines in the systemic inflammatory response syndrome: a review. HSR Proc. Intensive Care Cardiovasc. Anesth. 2, 161-175. [PMC free article] [PubMed]
- 60.Tanaka T., Narazaki M., Kishimoto T. IL-6 in inflammation, immunity, and disease. Cold Spring Harb. Perspect. Biol. 2014;6:a016295. doi: 10.1101/cshperspect.a016295. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tanaka, T., Narazaki, M., and Kishimoto, T. (2014). IL-6 in inflammation, immunity, and disease. Cold Spring Harb. Perspect. Biol. 6, a016295. [DOI] [PMC free article] [PubMed]
- 61.Sharif M.N., Tassiulas I., Hu Y., Mecklenbräuker I., Tarakhovsky A., Ivashkiv L.B. IFN-alpha priming results in a gain of proinflammatory function by IL-10: implications for systemic lupus erythematosus pathogenesis. J. Immunol. 2004;172:6476–6481. doi: 10.4049/jimmunol.172.10.6476. [DOI] [PubMed] [Google Scholar]; Sharif, M.N., Tassiulas, I., Hu, Y., Mecklenbrauker, I., Tarakhovsky, A., and Ivashkiv, L.B. (2004). IFN-alpha priming results in a gain of proinflammatory function by IL-10: implications for systemic lupus erythematosus pathogenesis. J. Immunol. 172, 6476-6481. [DOI] [PubMed]
- 62.Merkel O.M., Urbanics R., Bedocs P., Rozsnyay Z., Rosivall L., Toth M., Kissel T., Szebeni J. In vitro and in vivo complement activation and related anaphylactic effects associated with polyethylenimine and polyethylenimine-graft-poly(ethylene glycol) block copolymers. Biomaterials. 2011;32:4936–4942. doi: 10.1016/j.biomaterials.2011.03.035. [DOI] [PubMed] [Google Scholar]; Merkel, O.M., Urbanics, R., Bedocs, P., Rozsnyay, Z., Rosivall, L., Toth, M., Kissel, T., and Szebeni, J. (2011). In vitro and in vivo complement activation and related anaphylactic effects associated with polyethylenimine and polyethylenimine-graft-poly(ethylene glycol) block copolymers. Biomaterials 32, 4936-4942. [DOI] [PubMed]
- 63.Behlke M.A. Chemical modification of siRNAs for in vivo use. Oligonucleotides. 2008;18:305–319. doi: 10.1089/oli.2008.0164. [DOI] [PubMed] [Google Scholar]; Behlke, M.A. (2008). Chemical modification of siRNAs for in vivo use. Oligonucleotides 18, 305-319. [DOI] [PubMed]
- 64.Piao X., Wang H., Binzel D.W., Guo P. Assessment and comparison of thermal stability of phosphorothioate-DNA, DNA, RNA, 2′-F RNA, and LNA in the context of Phi29 pRNA 3WJ. RNA. 2018;24:67–76. doi: 10.1261/rna.063057.117. [DOI] [PMC free article] [PubMed] [Google Scholar]; Piao, X., Wang, H., Binzel, D.W., and Guo, P. (2018). Assessment and comparison of thermal stability of phosphorothioate-DNA, DNA, RNA, 2′-F RNA, and LNA in the context of Phi29 pRNA 3WJ. RNA 24, 67-76. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.