Abstract
Australian Genomics is a national collaborative research partnership of more than 80 organizations piloting a whole-of-system approach to integrating genomics into healthcare that is based on federation principles. The aim of Australian Genomics is to assess the application of genomic testing in healthcare at the translational interface between research and clinical delivery, with an emphasis on robust evaluation of outcomes. It encompasses two bodies of work: a research program prospectively providing genomic testing through exemplar clinical projects in rare diseases, cancers, and reproductive carrier screening and interdependent programs for advancing the diagnostic, health informatics, regulatory, ethical, policy, and workforce infrastructure necessary for the integration of genomics into the Australian health system.
Main Text
Genomic sequencing is rapidly transitioning from the research environment into clinical practice, transforming patient diagnosis and management, particularly in rare diseases and cancer. Internationally, governments are investing heavily in genomic medicine;1 genomic data from over 60 million patients are expected to be generated in healthcare globally in the next five years.2 Translational genomic medicine initiatives operate at the interface between research and clinical care.3 Participating patients often gain a direct benefit from access to genomic testing, and translational research contributes to the development of clinical and laboratory expertise, workforce capacity, and the infrastructure necessary to prepare healthcare systems for wider implementation of evidence-based genomic medicine.4
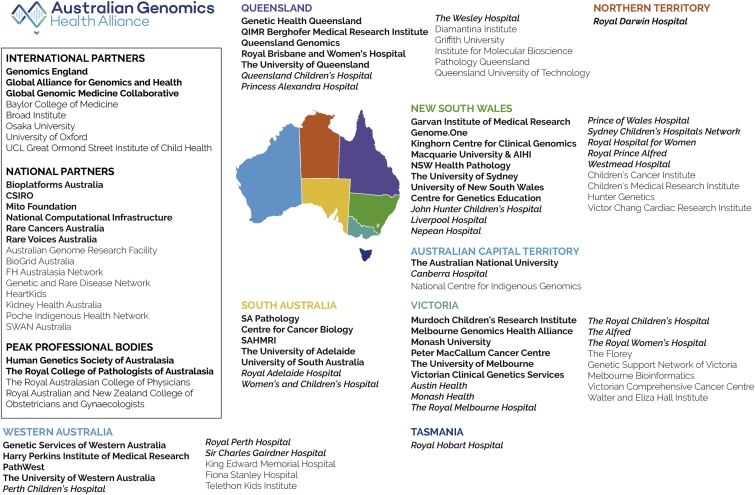
The single-payer nature of some public healthcare systems, notably the National Health Service (NHS) in England, has enabled a centralized approach to the implementation of whole-genome sequencing (WGS) in healthcare, through the establishment of Genomics England, a single national provider of sequencing, bioinformatics, and data storage infrastructure.5 In contrast, the responsibility for public healthcare in Australia is shared between state and federal governments, and hence the integration of clinical genomics requires a coordinated whole-of-nation approach based on a federated system. The Australian Genomics Health Alliance (Australian Genomics) is a collaborative research partnership of more than 80 organizations, including diagnostic laboratories, clinical genetics services, and research and academic institutions in all six states and two territories (Figure 1). After a targeted call by the National Health and Medical Research Council (NHMRC), Australian Genomics was awarded a five-year, AUD$25M (USD$17M) grant in November 2015 to demonstrate the value and practical strategies of implementing genomic medicine into the Australian healthcare system. Australian Genomics has established close working relationships with state-government-funded genomics programs, leveraging an additional ∼AUD$100M (USD$70M). This includes funding from the Victorian state government (Melbourne Genomics Health Alliance),4 the New South Wales state government (Sydney Genomics Collaborative), the Queensland state government (Queensland Genomics Health Alliance), and the Australian Capital Territory government (Canberra Clinical Genomics). Australian Genomics has also secured over AUD$30M (USD$21M) in philanthropic and competitive grant funding. Three years into the NHMRC grant, we review progress toward delivering a collaborative, federated model for the integration of genomics into the Australian healthcare system.
Figure 1.
Australian Genomics: A Collaborative Research Partnership of More Than 80 Organization
Fit for Purpose: Genomic Medicine in the Australian Healthcare System
The Australian state, territory, and federal governments share responsibility for the publicly-funded healthcare system. The federal government has responsibility for the universal public health insurance scheme that supports a range of services (Medicare Benefits Scheme, MBS). Both the federal and respective state or territory governments provide funding for public hospitals. Clinical genetics services are funded by state governments, but there are a limited number of genetic tests, including chromosomal microarray, that are funded federally through MBS. In addition, many Australians choose to have private health insurance, but this does not currently cover the cost of genetic testing.
In 2015, Australian Genomics undertook a landscape analysis to examine the opportunities for and challenges to the integration of genomics into the Australian healthcare system. This identified a number of national disease-specific centers of excellence in research and clinical translation that were already assembling large patient cohorts and performing next-generation-sequencing-based diagnostic testing; undiagnosed patients then progressed into discovery research. This mapping exercise also revealed considerable sequencing and bioinformatics capacity and genomic data analysis expertise, primarily in the research environment, and significant investments in clinical genomics in some but not all states. However, the state-based provision of diagnostic services and the divide between state and federal health funding posed the risks of duplication and inefficient use of resources and the creation of silos of clinical and genomic data, thereby reducing opportunities for collaboration. It also had the potential to create gaps in funding and inequalities in patient access, as well as to perpetuate uncoordinated approaches to diagnostic, ethical, and privacy standards and frameworks.
Design and Delivery of Australian Genomics
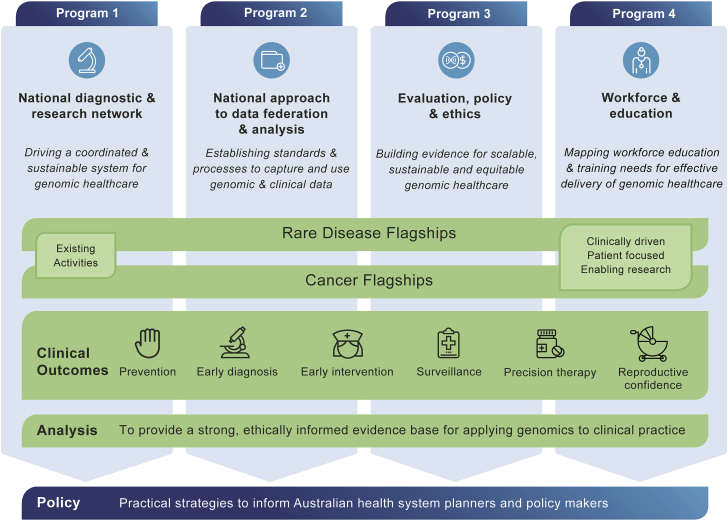
The aim of Australian Genomics is to accelerate and evaluate the application of genomic testing in healthcare and to do so while adopting a collaborative approach based on federation principles. Australian Genomics encompasses two bodies of interrelated work (Figure 2):
-
1.
Prospective recruitment of patients for genomic testing through exemplar rare disease and cancer flagship projects and referral to linked disease-specific research for undiagnosed cases, as well as
-
2.
Interdependent translational research programs to advance the diagnostic, health informatics, regulatory, ethical, policy, and workforce infrastructure necessary for the integration of genomics into the Australian health system.
Figure 2.
The Australian Genomics Model: Genomic Testing
Genomic testing is provided prospectively through rare disease and cancer flagship projects, while four interdependent programs simultaneously address diagnostic, health informatics, ethics, regulatory, and workforce challenges and enable evaluation to inform future policy.
The Australian Genomics approach is based on federation principles: clinical recruitment and genomic testing are accessible in all six states and two territories, and the collaborative networks underpinning the clinical and research activities have national representation and shared leadership across the states.
A National Steering Committee comprised of diverse expertise from across the states provides operational and strategic oversight of Australian Genomics. A National Implementation Committee provides two-way linkage with state and federal government health-system planners and policy makers. Broad national stakeholder engagement is facilitated by three advisory groups: the Independent Advisory Board, the Joint Committee on Digital Health and Genomics, and the Community Advisory Group. Australian Genomics is strongly engaged with the international genomic community as a member of the Global Alliance for Genomics and Health (GA4GH) and the Global Genomic Medicine Collaborative (G2MC), and it has an established partnership with Genomics England. In 2017, Australian Genomics became a GA4GH Driver Project, and it actively participates in the development and implementation of tools, solutions, and policies for global genomic data sharing.
Rare Disease and Cancer Flagship Projects
Central to the Australian Genomics approach are the clinical flagship projects, which prospectively recruit patients for genomic testing in specific disease areas. The flagships build from existing expertise and infrastructure to create national clinical and research networks, as well as assemble cohorts to enable gene discovery, conduct studies into disease mechanisms, and enable access to clinical trials. These cohorts also provide the platform for evaluating genomic testing’s impact, including diagnostic and clinical utility and cost effectiveness, and for evaluating new approaches to service delivery.4 Importantly, the flagships provide experiential learning for the multi-disciplinary working groups implementing and delivering the projects, further building state-based expertise. There are currently 17 rare disease and cancer flagship projects across 32 clinical sites, building capacity and promoting national partnership (Table 1).
Table 1.
Australian Genomics Flagship Projects
| Flagship | Sites | Methodology | Duration | Cohort Size | |
|---|---|---|---|---|---|
| Rare disease |
Neuromuscular disorders | NSW, QLD, SA, VIC, WA | Custom capture panel and RNAseq | 2016–2018 | 120 |
| Mitochondrial disorders (with Mito Foundation) | NSW, QLD, SA,VIC, WA | WES + mtDNA or WGS | 2016–2019 | 150 | |
| Neurodevelopmental Disabilities: epileptic encephalopathy | NSW, QLD, SA,TAS, VIC, WA | WES | 2016–2018 | 105 | |
| Neurodevelopmental disabilities: brain malformations | NSW, QLD, SA, VIC, WA, NT | WES | 2016–2018 | 100 | |
| Neurodevelopmental disabilities: Leukodystrophies | NSW, QLD, SA, VIC, WA, NT | WES | 2017–2019 | 50 | |
| Neurodevelopmental disabilities: intellectual disabilities | NSW, QLD, SA, VIC, WA | WES or WGS | 2016–2019 | 100 (trios) | |
| Renal genetics (with Kidgen and Melbourne Genomics Health Alliance) | NSW, QLD, SA,VIC, WA | WES or Panel | 2016–2018 | 365 | |
|
Genetic immunology |
ACT, NSW, SA,VIC, WA |
WES or WGS |
2016–2018 |
110 (trios) |
|
| ChILDRANZ interstitial lung disease | NSW, QLD, SA, VIC, WA, TAS, NT | WES | 2018–2020 | 120 (trios) | |
| Acute care genomics | NSW, QLD, SA, VIC | WES or WGS | 2018–2020 | 250 (trios) | |
| Cardiovascular genetic disorders | NSW, QLD, SA, VIC, WA | WGS | 2018–2022 | 600 | |
| HIDDEN renal genetic disorders | NSW, QLD, SA, VIC, WA, NT | WGS | 2018–2020 | 200 | |
| Cancer |
Acute lymphoblastic leukemia | NSW, NT, QLD, SA, VIC, WA | RNA Seq | 2016–2020 | 300 |
| Somatic cancer (with Melbourne Genomics Health Alliance) | NSW, QLD, VIC, WA | Panel | 2016–2018 | 400 | |
| Germline cancer - pediatric, adolescent, young adults (with NSW Cancer Genomics) | NSW | WGS | 2016–2020 | 1,400 | |
| Hereditary cancer syndromes (with ICCon) | NSW, QLD, SA, VIC, WA | WGS | 2016–2020 | 190 | |
| Lung cancer diagnosis (with CCQ, Cancer Australia) | QLD, NSW, VIC, SA | WGS or WES | 2018–2020 | 150 | |
|
SUPER WGS (with Cancer Australia and VCA) |
NSW, VIC, NT, ACT |
WGS |
2018–2020 |
100 |
|
| GHFM | Mackenzie’s Mission | NSW, VIC, WA; QLD, NT, SA, ACT, TAS | Panel or WES | 2019–2022 | 10,000 (couples) |
| Total recruitment: 4,810 (NHMRC) 20,000 (GHFM) | |||||
Abbreviations are as follows: NSW = New South Wales, VIC = Victoria, WA = Western Australia, QLD = Queensland, NT = Northern Territory, SA = South Australia, ACT = Australian Capital Territory, TAS = Tasmania, WES = whole-exome sequencing, WGS = whole-genome sequencing, NHMRC = National Health and Medical Research Council, and GHFM = Genomics Health Futures Mission.
The initial flagship projects were aligned with existing disease-specific national research programs. Subsequent projects were selected via a competitive approach and independent assessment on the basis of criteria including the expected clinical and economic impact of genomic testing in the chosen cohort, project feasibility, implementation capacity, and the promotion and leveraging of national collaboration. Where feasible, a hybrid implementation-effectiveness study design has been adopted, whereby the implementation of genomic testing is studied simultaneously with the investigation of the outcomes of testing.4, 6
National patient recruitment ensures that clinical, diagnostic, and research pathways are developed within state-specific infrastructures. However, it also highlights some of the difficulties inherent in building a national approach in the Australian context. For example, the ethics and governance approval process across 32 recruitment sites in every Australian state and territory (HREC/16/MH/251) took three years at a cost of >AUD$1M (USD$0.7M) (unpublished data). Now that this is in place, the rollout of additional flagships is much more efficient, taking only three months from the submission of an ethics amendment to the first participant recruitment. Similarly, applying for and accessing both federal and individual state-based health datasets for evaluation purposes requires significant investment.
The flagship projects are nationally coordinated and supported by a distributed network of Australian Genomics project officers and genetic counselors. Approximately one full-time equivalent genetic counselor is employed per 100 patients recruited/annum, simultaneously serving to build genomics capacity and capability in the Australian genetic counselor workforce. Patients are recruited prospectively through the 32 participating clinical services on the basis of defined patient selection criteria. By using consensus expert opinion, each flagship working group has developed a minimal optimal clinical dataset to be collected at enrolment to enable effective genomic data analysis. Data are captured in the centralized Australian Genomics study REDCap database,7 and this has been standardized through a REDCap plugin developed as part of the program. This accesses terminology value sets from the Australian Genomics Clinical Terminology Server. The terminology server supports SNOMED CT (Systematized Nomenclature of Medicine -- Clinical Terms),8 the Human Phenotype Ontology,9 and other related vocabularies such as Online Mendelian Inheritance in Man (OMIM). Sequencing modalities that are being compared between different flagships include whole-genome sequencing, whole-exome sequencing, mitochondrial genome sequencing, RNA sequencing, and large capture panels. The majority of rare disease testing uses a singleton (rather than a trio) approach to optimize resource use. Australian Genomics adopts a decentralized approach to clinical genomic testing; DNA extraction, sequencing, bioinformatics analysis, data interpretation, and reporting occur through participating state-based accredited diagnostic laboratories, reflecting the decentralized nature of our healthcare system. There is a high degree of integration between local clinical and laboratory teams during the process of data analysis and interpretation, and variant data from genomic tests are reviewed at multidisciplinary meetings prior to reporting. Clinical genomic reports are issued directly to requesting physicians, and diagnostic outcomes and the clinical utility of results are captured centrally. The consent process provides participants with the opportunity to store their data and participate in further research; this has been done with the intention of contributing enriched patient cohorts to gene discovery and striving to resolve diagnoses for more families in the future.
Australian Genomics Programs
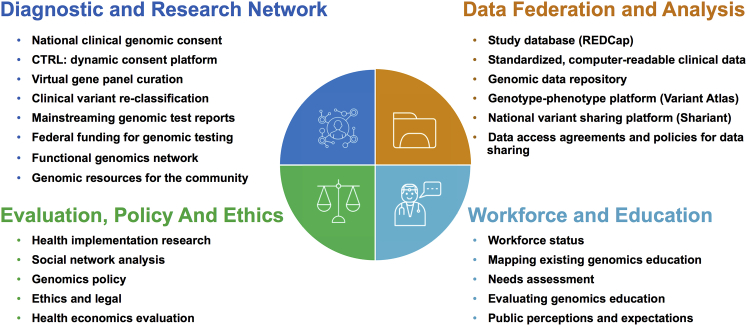
The four Australian Genomics programs intersect with the flagship projects and bring together expertise from a diverse group of clinicians, researchers, diagnostic scientists, bioinformaticians, health informaticians, implementation scientists, lawyers, bioethicists, health economists, policy analysts, educators, and patient groups to work on a range of sub-projects to address specific barriers and/or develop resources, tools, and platforms (Figure 3 and Table S1). There are 50 working groups, comprising over 400 member investigators, currently active within Australian Genomics.
Figure 3.
Australian Genomics Programs and Sub-projects. Additional information is available in Table S1.
Although clinical genomic testing remains the responsibility of accredited diagnostic laboratories, the National Diagnostic and Research Network (Program 1) is examining current practices and building a nationally consistent approach to consent, genomic data analysis, variant re-classification, and reporting, as well as developing patient information resources. The incorporation of functional genomic assays into diagnostic pipelines will be facilitated through the related Australian Functional Genomics Network, which links clinicians with researchers and will in the future provide seed funding for pilot projects. Australian Genomics is also leading applications to the Medical Services Advisory Committee to recommend funding for genomic testing at the federal government level. An initial application for genomic testing for pediatric patients with syndromic and non-syndromic intellectual disability, made on the basis of evidence generated in Australian cohorts,10, 11, 12, 13 has recently been recommended for funding. This will serve as a model for future applications as evidence is progressively gathered in other patient groups.
The decentralized approach to genomic test delivery in Australia poses a significant risk of creating silos of clinical, phenotypic, and genomic data at individual service or state level, and this will hinder the realization of the full benefits of genomics in healthcare.14 Although initial data storage is occurring with on-premises computing infrastructure at the laboratory and research institute level, the key priority for the National Approach to Data Federation and Analysis program (Program 2) is to establish cloud-based, scalable, shared, and standardized data repositories for the clinical, phenotypic, and genomic information generated through the flagships, compliant with Global Alliance for Genomics and Health (GA4GH) standards (Figure S1). All data are stored in compliance with local clinical, laboratory, and research guidelines, and we have undertaken data breach testing as part of our role as a GA4GH Driver Project. The solutions we are piloting will inform the implementation of genomics in Australia’s digital health systems—from hospital electronic medical records to the national My Health Record.15 Australian Genomics has developed guidelines and tools to enable Australians to participate in data sharing for research purposes if they so choose (Figure 3 and Table S1). Consent is structured in compliance with the GA4GH framework for responsible data sharing16 and consent clauses. Under the terms of the original NHMRC grant, our principal focus is to enable data sharing for healthcare and academic research. Private entities may be granted access to data, provided the proposed use and ethics approval match the data permissions from the original consent.
The Evaluation, Policy, and Ethics program (Program 3) applies an interdisciplinary research model to bring together expertise in implementation science, health economics, bioethics, and law to inform genomics health policy and clinical practice. Modeling and evaluating health economic data collected through the clinical flagship projects builds evidence for the value of health and non-health impacts of genomic medicine. The program also employs implementation science methods and theories to understand and promote the uptake of genomic medicine into routine healthcare in clinical, organizational, and policy contexts.17 A social network analysis has demonstrated collaboration within and external to Australian Genomics is accelerating at unprecedented levels.18 Applied ethical and legal research is undertaken across a variety of aspects, including genomic data sharing, ownership of genetic information, clinical and research consent, insurance discrimination, and rapid genomic testing in acute pediatrics.19
The Genomics Workforce and Education program (Program 4) provides evidence to inform the education and training of those whose professional roles will be impacted by clinical genomics. Broadly, this program is mapping the current landscape of education delivery in genomics, identifying the genomic education needs and preferences of genetic and non-genetic health professionals,20, 21, 22 and developing tools to support the development and evaluation of effective education programs. A combination of qualitative and quantitative methods are examining perspectives of a wide range of stakeholders, including health professionals, educators, patients, the community, scientists, and policy makers. Tools have been developed to support the development of evidence-based genomic education (a “program logic model”) and evaluation (“evaluation framework”). A draft program logic model was reviewed by Australian and international experts and is now being piloted in the UK, USA, Canada, and Australia. Australian Genomics also founded the Genomics Education Network of Australasia (GENA) in mid-2018 to foster a community of practice and share experiences, tools, and exemplars of health-professional genomic education and evaluation.
National Policy Impact
Australian Genomics is working within a climate of dynamic policy development and new funding initiatives for genomics in Australia. In March 2016, the Australian Health Ministers’ Advisory Council (AHMAC) directed the development of a whole-of-government framework to guide and harmonize state and federal government policies to integrate genomics into Australian healthcare. The National Health Genomics Policy Framework23 draws on a national, public, and targeted stakeholder consultation and was endorsed by the Council of Australian Governments (COAG) Health Council in November 2017. An implementation plan was subsequently published in late 2018.24 These, together with the recently published Future of Precision Medicine in Australia report by the Australian Council of Learned Academies25 and the Australia 2030: Prosperity through Innovation by Innovation and Science Australia,26 lay out a vision for the integration of genomics and precision medicine into the Australian healthcare system.
As well as setting out a new policy framework, the Australian federal government committed further funding into genomics research through the Medical Research Futures Fund in 2018. The Genomics Health Futures Mission (GHFM) constitutes a substantial investment of AUD$500M (USD$348M) over ten years. This mission builds upon the targeted genomic evaluations in rare diseases and cancers to expand both the scale and scope of current genomic translational research; it aims to transform the lives of more than 200,000 Australians. It is envisaged that the long-term outcome of the GHFM will be enabling the seamless transition of genomics research into clinical practice as the evidence for each clinical indication becomes available.
Australian Genomics is delivering two of the initial projects of the GHFM. Mackenzie’s Mission, an AUD$20M (USD$14M) reproductive carrier-screening project, will provide information to inform reproductive choices in 10,000 couples on the basis of variants in around 500 genes associated with severe and often fatal genetic conditions affecting children. The GHFM has also supported an AUD$6M (USD$4M) expansion of the Australian Genomics Cardiovascular Genetic Disorders flagship project, which will now have the capacity to provide sequencing to 600 families living with congenital heart disease, cardiomyopathies, and arrhythmias. This flagship will also establish a network that interfaces between clinical care for patients and families with cardiovascular genetic disorders, research genomic analysis, and functional genomics.
Future Directions
Australian Genomics builds upon extensive state and federal genomic research and infrastructural investment, estimated at AUD$250M (USD$174M) in the past six years. Of this, direct co-investment into the Australian Genomics program is estimated at over AUD$40M (USD$28M). Over the past three years, Australian Genomics has grown to include over 400 investigators, established 50 working groups, promoted the creation of extensive policy, clinical, researcher, and patient networks, increased inter-disciplinary and inter-state interactions, and enabled the prospective recruitment, phenotyping, and genomic testing of rare disease and cancer patient cohorts. Now that the scaffolding infrastructure has been established, this core research capability can be maintained at ∼AUD$4M (USD$2.8M) per annum.
Australian Genomics has built a collaborative, federated, national learning network that can adapt to change and maintain a continuous virtuous cycle between research and clinical practice in genomics, with an emphasis on robust evaluation of outcomes. It has by no means been easy to get to this point, and yet the hard part of the journey is still to come: embedding genomics into routine, mainstream clinical practice when and where it is needed.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
The Australian Genomics Health Alliance (Australian Genomics) project is funded by a National Health and Medical Research Council (NHMRC) Targeted Call for Research grant (GNT1113531). Mackenzie’s Mission and the Australian Genomics Cardiovascular Genetics Disorders flagship are funded by the Genomics Health Futures Mission, which is part of the Australian government’s Medical Research Futures Fund. The research conducted at the Murdoch Children’s Research Institute was supported by the Victorian government’s Operational Infrastructure Support Program.
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.06.003.
Web Resources
Australian Genomics, https://www.australiangenomics.org.au
Australian Genomics Clinical Terminology Server, https://genomics.ontoserver.csiro.au
Australian Functional Genomics Network, https://www.functionalgenomics.org.au
Global Alliance for Genomics and Health, https://www.ga4gh.org
Global Genomic Medicine Collaborative (G2MC), https://g2mc.org
Online Mendelian Inheritance in Man (OMIM), https://www.ncbi.nlm.nih.gov/omim
Human Phenotype Ontology, https://hpo.jax.org/app/
Supplemental Data
References
- 1.Stark Z., Dolman L., Manolio T.A., Ozenberger B., Hill S.L., Caulfied M.J., Levy Y., Glazer D., Wilson J., Lawler M. Integrating genomics into healthcare: A global responsibility. Am. J. Hum. Genet. 2019;104:13–20. doi: 10.1016/j.ajhg.2018.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; Stark, Z., Dolman, L., Manolio, T.A., Ozenberger, B., Hill, S.L., Caulfied, M.J., Levy, Y., Glazer, D., Wilson, J., Lawler, M., et al. (2019). Integrating genomics into healthcare: A global responsibility. Am. J. Hum. Genet. 104, 13-20. [DOI] [PMC free article] [PubMed]
- 2.Birney E., Vamathevan J., Goodhand P. Genomics in healthcare: GA4GH looks to 2022. bioRxiv. 2017 [Google Scholar]; Birney, E.V.J., and Goodhand, P. (2017). Genomics in healthcare: GA4GH looks to 2022. bioRxiv. 10.1101/203554.
- 3.Wolf S.M., Amendola L.M., Berg J.S., Chung W.K., Clayton E.W., Green R.C., Harris-Wai J., Henderson G.E., Jarvik G.P., Koenig B.A. Navigating the research-clinical interface in genomic medicine: Analysis from the CSER Consortium. Genet. Med. 2018;20:545–553. doi: 10.1038/gim.2017.137. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wolf, S.M., Amendola, L.M., Berg, J.S., Chung, W.K., Clayton, E.W., Green, R.C., Harris-Wai, J., Henderson, G.E., Jarvik, G.P., Koenig, B.A., et al. (2018). Navigating the research-clinical interface in genomic medicine: Analysis from the CSER Consortium. Genet. Med. 20, 545-553. [DOI] [PMC free article] [PubMed]
- 4.Gaff C.L., Winship I.M., Forrest S.M., Hansen D.P., Clark J., Waring P.M., South M., Sinclair A.H. Preparing for genomic medicine: A real world demonstration of health system change. NPJ Genom Med. 2017;2:16. doi: 10.1038/s41525-017-0017-4. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gaff, C.L., M Winship, I., M Forrest, S., P Hansen, D., Clark, J., M Waring, P., South, M., and H Sinclair, A. (2017). Preparing for genomic medicine: A real world demonstration of health system change. NPJ Genom Med 2, 16. [DOI] [PMC free article] [PubMed]
- 5.Turnbull C., Scott R.H., Thomas E., Jones L., Murugaesu N., Pretty F.B., Halai D., Baple E., Craig C., Hamblin A., 100 000 Genomes Project The 100 000 Genomes Project: Bringing whole genome sequencing to the NHS. BMJ. 2018;361:k1687. doi: 10.1136/bmj.k1687. [DOI] [PubMed] [Google Scholar]; Turnbull, C., Scott, R.H., Thomas, E., Jones, L., Murugaesu, N., Pretty, F.B., Halai, D., Baple, E., Craig, C., Hamblin, A., et al.; 100 000 Genomes Project (2018). The 100 000 Genomes Project: Bringing whole genome sequencing to the NHS. BMJ 361, k1687. [DOI] [PubMed]
- 6.Curran G.M., Bauer M., Mittman B., Pyne J.M., Stetler C. Effectiveness-implementation hybrid designs: Combining elements of clinical effectiveness and implementation research to enhance public health impact. Med. Care. 2012;50:217–226. doi: 10.1097/MLR.0b013e3182408812. [DOI] [PMC free article] [PubMed] [Google Scholar]; Curran, G.M., Bauer, M., Mittman, B., Pyne, J.M., and Stetler, C. (2012). Effectiveness-implementation hybrid designs: Combining elements of clinical effectiveness and implementation research to enhance public health impact. Med. Care 50, 217-226. [DOI] [PMC free article] [PubMed]
- 7.Harris P.A., Taylor R., Thielke R., Payne J., Gonzalez N., Conde J.G. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inform. 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]; Harris, P.A., Taylor, R., Thielke, R., Payne, J., Gonzalez, N., and Conde, J.G. (2009). Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inform. 42, 377-381. [DOI] [PMC free article] [PubMed]
- 8.Donnelly K. SNOMED-CT: The advanced terminology and coding system for eHealth. Stud. Health Technol. Inform. 2006;121:279–290. [PubMed] [Google Scholar]; Donnelly, K. (2006). SNOMED-CT: The advanced terminology and coding system for eHealth. Stud. Health Technol. Inform. 121, 279-290. [PubMed]
- 9.Robinson P.N., Köhler S., Bauer S., Seelow D., Horn D., Mundlos S. The Human Phenotype Ontology: A tool for annotating and analyzing human hereditary disease. Am. J. Hum. Genet. 2008;83:610–615. doi: 10.1016/j.ajhg.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]; Robinson, P.N., Kohler, S., Bauer, S., Seelow, D., Horn, D., and Mundlos, S. (2008). The Human Phenotype Ontology: A tool for annotating and analyzing human hereditary disease. Am. J. Hum. Genet. 83, 610-615. [DOI] [PMC free article] [PubMed]
- 10.Stark Z., Schofield D., Alam K., Wilson W., Mupfeki N., Macciocca I., Shrestha R., White S.M., Gaff C. Prospective comparison of the cost-effectiveness of clinical whole-exome sequencing with that of usual care overwhelmingly supports early use and reimbursement. Genet. Med. 2017;19:867–874. doi: 10.1038/gim.2016.221. [DOI] [PubMed] [Google Scholar]; Stark, Z., Schofield, D., Alam, K., Wilson, W., Mupfeki, N., Macciocca, I., Shrestha, R., White, S.M., and Gaff, C. (2017). Prospective comparison of the cost-effectiveness of clinical whole-exome sequencing with that of usual care overwhelmingly supports early use and reimbursement. Genet. Med. 19, 867-874. [DOI] [PubMed]
- 11.Stark Z., Schofield D., Martyn M., Rynehart L., Shrestha R., Alam K., Lunke S., Tan T.Y., Gaff C.L., White S.M. Does genomic sequencing early in the diagnostic trajectory make a difference? A follow-up study of clinical outcomes and cost-effectiveness. Genet. Med. 2019;21:173–180. doi: 10.1038/s41436-018-0006-8. [DOI] [PubMed] [Google Scholar]; Stark, Z., Schofield, D., Martyn, M., Rynehart, L., Shrestha, R., Alam, K., Lunke, S., Tan, T.Y., Gaff, C.L., and White, S.M. (2019). Does genomic sequencing early in the diagnostic trajectory make a difference? A follow-up study of clinical outcomes and cost-effectiveness. Genet. Med. 21, 173-180. [DOI] [PubMed]
- 12.Stark Z., Tan T.Y., Chong B., Brett G.R., Yap P., Walsh M., Yeung A., Peters H., Mordaunt D., Cowie S., Melbourne Genomics Health Alliance A prospective evaluation of whole-exome sequencing as a first-tier molecular test in infants with suspected monogenic disorders. Genet. Med. 2016;18:1090–1096. doi: 10.1038/gim.2016.1. [DOI] [PubMed] [Google Scholar]; Stark, Z., Tan, T.Y., Chong, B., Brett, G.R., Yap, P., Walsh, M., Yeung, A., Peters, H., Mordaunt, D., Cowie, S., et al.; Melbourne Genomics Health Alliance (2016). A prospective evaluation of whole-exome sequencing as a first-tier molecular test in infants with suspected monogenic disorders. Genet. Med. 18, 1090-1096. [DOI] [PubMed]
- 13.Tan T.Y., Dillon O.J., Stark Z., Schofield D., Alam K., Shrestha R., Chong B., Phelan D., Brett G.R., Creed E. Diagnostic impact and cost-effectiveness of whole-exome sequencing for ambulant children with suspected monogenic conditions. JAMA Pediatr. 2017;171:855–862. doi: 10.1001/jamapediatrics.2017.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tan, T.Y., Dillon, O.J., Stark, Z., Schofield, D., Alam, K., Shrestha, R., Chong, B., Phelan, D., Brett, G.R., Creed, E., et al. (2017). Diagnostic impact and cost-effectiveness of whole-exome sequencing for ambulant children with suspected monogenic conditions. JAMA Pediatr. 171, 855-862. [DOI] [PMC free article] [PubMed]
- 14.Global Alliance for Genomics and Health. A federated ecosystem for sharing genomic, clinical data. Science. 2016;352:1278–1280. doi: 10.1126/science.aaf6162. [DOI] [PubMed] [Google Scholar]; Global Alliance for Genomics and Health (2016). GENOMICS. A federated ecosystem for sharing genomic, clinical data. Science 352, 1278-1280. [DOI] [PubMed]
- 15.Hansen D.P., Dinger M.E., Hofmann O., Thorne N., Boughtwood T.F. Preparing Australia for genomic medicine: Data, computing and digital health. Med. J. Aust. 2019;210(Suppl 6):S30–S32. doi: 10.5694/mja2.50032. [DOI] [PubMed] [Google Scholar]; Hansen, D.P., Dinger, M.E., Hofmann, O., Thorne, N., and Boughtwood, T.F. (2019). Preparing Australia for genomic medicine: Data, computing and digital health. Med. J. Aust. 210 (Suppl 6), S30-S32. [DOI] [PubMed]
- 16.Knoppers B.M. Framework for responsible sharing of genomic and health-related data. HUGO J. 2014;8:3. doi: 10.1186/s11568-014-0003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]; Knoppers, B.M. (2014). Framework for responsible sharing of genomic and health-related data. HUGO J. 8, 3. [DOI] [PMC free article] [PubMed]
- 17.Taylor N., Best S., Martyn M., Long J.C., North K.N., Braithwaite J., Gaff C. A transformative translational change programme to introduce genomics into healthcare: A complexity and implementation science study protocol. BMJ Open. 2019;9:e024681. doi: 10.1136/bmjopen-2018-024681. [DOI] [PMC free article] [PubMed] [Google Scholar]; Taylor, N., Best, S., Martyn, M., Long, J.C., North, K.N., Braithwaite, J., and Gaff, C. (2019). A transformative translational change programme to introduce genomics into healthcare: A complexity and implementation science study protocol. BMJ Open 9, e024681. [DOI] [PMC free article] [PubMed]
- 18.Long J.C., Pomare C., Best S., Boughtwood T., North K., Ellis L.A., Churruca K., Braithwaite J. Building a learning community of Australian clinical genomics: A social network study of the Australian Genomic Health Alliance. BMC Med. 2019;17:44. doi: 10.1186/s12916-019-1274-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; Long, J.C., Pomare, C., Best, S., Boughtwood, T., North, K., Ellis, L.A., Churruca, K., and Braithwaite, J. (2019). Building a learning community of Australian clinical genomics: A social network study of the Australian Genomic Health Alliance. BMC Med. 17, 44. [DOI] [PMC free article] [PubMed]
- 19.Gyngell C., Newson A.J., Wilkinson D., Stark Z., Savulescu J. Rapid challenges: Ethics and genomic neonatal intensive care. Pediatrics. 2019;143(Suppl 1):S14–S21. doi: 10.1542/peds.2018-1099D. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gyngell, C., Newson, A.J., Wilkinson, D., Stark, Z., and Savulescu, J. (2019). Rapid challenges: Ethics and genomic neonatal intensive care. Pediatrics 143 (Suppl 1), S14-S21. [DOI] [PMC free article] [PubMed]
- 20.Janinski M., McClaren B., Nisselle A., Dunlop K., Prichard Z., Terrill B., Metcalfe S. Perspectives of education providers on education and training needs of non-genetic health professionals. 2018. https://www.australiangenomics.org.au/resources/publications/reports/; Janinski, M., McClaren, B., Nisselle, A., Dunlop, K., Prichard, Z., Terrill, B., and Metcalfe, S. (2018). Perspectives of education providers on education and training needs of non-genetic health professionals. https://www.australiangenomics.org.au/resources/publications/reports/
- 21.McClaren B., Nisselle A., Prichard Z., Dunlop K., Terrill B., Gaff C., Metcalfe S. Mapping existing education and training for the Australian clinical genomics workforce. 2018. https://www.australiangenomics.org.au/resources/publications/reports/; McClaren, B., Nisselle, A., Prichard, Z., Dunlop, K., Terril, B., Gaff, C., and Metcalfe, S. (2018). Mapping existing education and training for the Australian clinical genomics workforce. https://www.australiangenomics.org.au/resources/publications/reports/
- 22.Nisselle A., Macciocca I., McKenzie F., Vuong H., Dunlop K., McClaren B., Metcalfe S., Gaff C., Australian Genomics Workforce & Education Working Group Readiness of clinical genetic healthcare professionals to provide genomic medicine: An Australian census. J. Genet. Couns. 2019;28:367–377. doi: 10.1002/jgc4.1101. [DOI] [PubMed] [Google Scholar]; Nisselle, A., Macciocca, I., McKenzie, F., Vuong, H., Dunlop, K., McClaren, B., Metcalfe, S., and Gaff, C.; Australian Genomics Workforce & Education Working Group (2019). Readiness of clinical genetic healthcare professionals to provide genomic medicine: An Australian census. J. Genet. Couns. 28, 367-377. [DOI] [PubMed]
- 23.Australian Government, Department of Health National Health Genomics Policy Framework. 2017. http://www.health.gov.au/internet/main/publishing.nsf/Content/national-health-genomics-policy-framework-2018-2021; Australian Government, Department of Health (2017). National Health Genomics Policy Framework. http://www.health.gov.au/internet/main/publishing.nsf/Content/national-health-genomics-policy-framework-2018-2021
- 24.Australian Government, Department of Health National Health Genomics Policy Framework Implementation Plan. 2018. https://www.coaghealthcouncil.gov.au/Portals/0/Genomics%20Policy%20Framework%20Implementation.pdf; Australian Government, Department of Health (2018). National Health Genomics Policy Framework Implementation Plan. https://www.coaghealthcouncil.gov.au/Portals/0/Genomics%20Policy%20Framework%20Implementation.pdf
- 25.Australian Council of Learned Academies The Future of Precision Medicine in Australia. 2018. https://acola.org.au/wp/wp-content/uploads/PMED-20180220.pdf; Australian Council of Learned Academies (2018). The Future of Precision Medicine in Australia. https://acola.org.au/wp/wp-content/uploads/PMED-20180220.pdf
- 26.Innovation and Science Australia Australia 2030: Prosperity through Innovation. 2018. https://industry.gov.au/Innovation-and-Science-Australia/Australia-2030/Pages/default.aspx; Innovation and Science Australia (2018). Australia 2030: Prosperity through Innovation. https://industry.gov.au/Innovation-and-Science-Australia/Australia-2030/Pages/default.aspx
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.