Significance
Staphylococcus aureus is a pathogen that produces an arsenal of virulence factors to promote infection within different human anatomical sites. To generate the appropriate array of virulence factors, the bacterium utilizes a complex regulatory system. While aspects of this regulatory system have been well-elucidated, still more than 50% of the regulators in S. aureus have not been fully characterized. Here, we demonstrate that a regulator of nucleic acid building block production can moonlight as a direct regulator of virulence genes. This finding exposes a clever tactic employed by S. aureus whereby the bacterium assigns a multidimensional role to a canonical regulator of metabolism to temper its own pathogenesis.
Keywords: MRSA, PurR, purine biosynthesis, pathogenesis, infection
Abstract
The pathogen Staphylococcus aureus colonizes and infects a variety of different sites within the human body. To adapt to these different environments, S. aureus relies on a complex and finely tuned regulatory network. While some of these networks have been well-elucidated, the functions of more than 50% of the transcriptional regulators in S. aureus remain unexplored. Here, we assess the contribution of the LacI family of metabolic regulators to staphylococcal virulence. We found that inactivating the purine biosynthesis regulator purR resulted in a strain that was acutely virulent in bloodstream infection models in mice and in ex vivo models using primary human neutrophils. Remarkably, these enhanced pathogenic traits are independent of purine biosynthesis, as the purR mutant was still highly virulent in the presence of mutations that disrupt PurR’s canonical role. Through the use of transcriptomics coupled with proteomics, we revealed that a number of virulence factors are differentially regulated in the absence of purR. Indeed, we demonstrate that PurR directly binds to the promoters of genes encoding virulence factors and to master regulators of virulence. These results guided us into further ex vivo and in vivo studies, where we discovered that S. aureus toxins drive the death of human phagocytes and mice, whereas the surface adhesin FnbA contributes to the increased bacterial burden observed in the purR mutant. Thus, S. aureus repurposes a metabolic regulator to directly control the expression of virulence factors, and by doing so, tempers its pathogenesis.
The human pathogen Staphylococcus aureus can cause an array of illnesses, ranging from mild skin conditions to life-threatening diseases such as pneumonia or bacteremia (1). Combatting S. aureus infections has become increasingly difficult due to the widespread emergence of methicillin-resistant S. aureus (MRSA) strains that plague both communities and hospitals worldwide (2). The pathogen’s adaptability to different lifestyles—from commensal/skin colonizer to an invasive pathogen that can thrive in different tissues—is due in part to its expansive and dynamic regulatory network (3). While there is a great deal known about how S. aureus activates and/or represses virulence genes under defined laboratory conditions, much remains to be understood about how metabolic fluctuations influence S. aureus pathophysiology in vivo. Metabolic regulation is likely of great importance to S. aureus as it transitions between states of variable energy sources, such as when it moves from a colonization site to an invasive site. Thus, as S. aureus confronts new environments, the bacterium must finely coordinate the expression of virulence factors with the metabolic demands of a particular anatomical compartment.
S. aureus employs a plethora of secreted and surface-associated virulence factors to evoke disease in the host and to evade immune defenses. This diverse arsenal of factors includes cytotoxins, proteases, adhesins, immunomodulators, and autolysins (4). Given the important contribution of these factors to the pathogenesis of the bacterium, it is not surprising that an intricate regulatory pathway controls their expression. S. aureus utilizes a network of dedicated regulators to positively control virulence, which includes the accessory gene regulator (Agr) proteins, S. aureus exoprotein two-component system (SaeRS-TCS), and staphylococcal accessory regulator nucleic acid-binding protein family (Sar). The Agr proteins make up a quorum-sensing system that, when activated, up-regulates the production of toxins and exoenzymes (5). The SaeRS system positively regulates virulence factor production and responds to environmental stimuli such as pH, osmolarity, and host-derived factors (6). The Sar family of transcriptional factors positively regulate the production of virulence factors and modulate the production of the Agr quorum-sensing system (7). The bacterium also negatively regulates toxin production via the repressor of toxins (Rot) (8). Each of these regulatory pathways is connected and complementary, creating a complex network driving virulence factor production.
While the described regulatory pathways used to control virulence are intricate, we only understand a portion of the system’s complexity. S. aureus is predicted to encode 135 transcriptional factors, and less than 50% of them have been fully characterized (9). Moreover, it has been demonstrated that transcriptional factors with roles previously thought to be unrelated to virulence, such as those involved in metabolic homeostasis, are capable of regulating virulence as well. CodY is a transcriptional factor that senses branched-chain amino acids and GTP to regulate related metabolic genes (10). However, the protein also regulates transcripts encoding virulence factors, and thus serves as a multifaceted regulator that can influence both metabolic and virulence outputs (11). Additionally, the catabolite control protein A, CcpA, is a carbon utilization regulator that has been implicated as a positive regulator of virulence, as mutants for ccpA exhibit a reduction in the expression of the Agr effector RNAIII, which results in the down-regulation of virulence factors (12). Given the dual functionality of these metabolic regulators and the vast potential for the large number of uncharacterized regulators, there are likely additional layers of complex virulence regulation than what has been described in the literature.
Here, we aimed to dissect the extent to which members of a family of important metabolic regulators in S. aureus, called the LacI family, influence the severity of staphylococcal pathogenesis. These metabolite-responsive regulators are of great importance to bacteria, as they regulate many key metabolic processes in the cell, including the utilization of glucose, maltose, purines, ribose, and sucrose. LacI family regulators in S. aureus consist of five genes: ccpA, malR, purR, rbsR, and scrR. Through a screen to determine the role LacI transcriptional factors play in MRSA infection, we have revealed that the purine biosynthesis regulator PurR serves as a transcriptional factor that coordinates the expression of genes involved in both metabolism and virulence in S. aureus. Thus, PurR is a critical regulator of staphylococcal pathogenesis.
Results
The LacI Transcriptional Regulator PurR Contributes to Virulence.
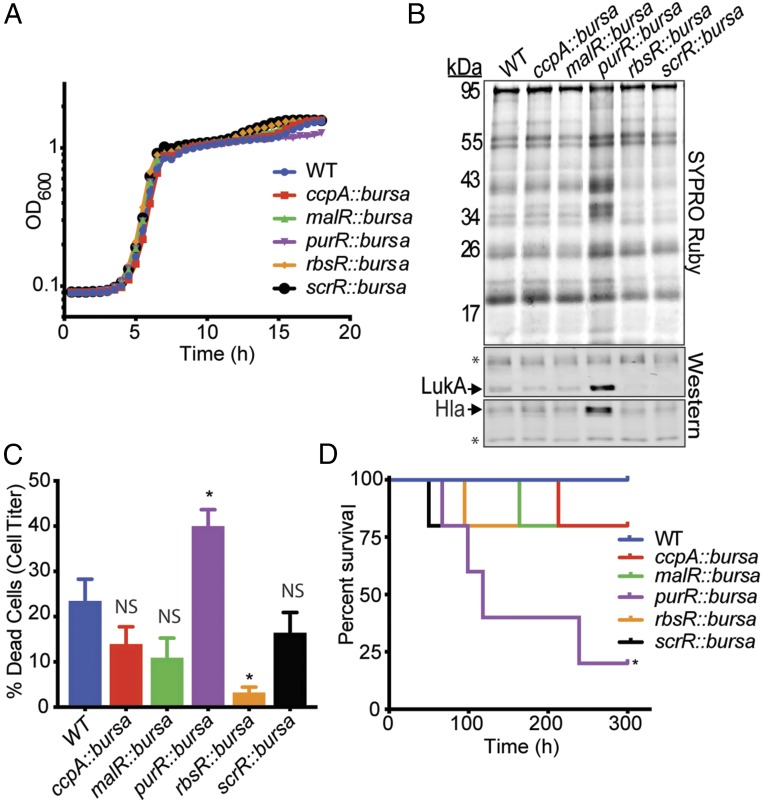
To determine the extent to which LacI transcriptional factors contribute to S. aureus virulence, we conducted a screen to assess virulence using transposon mutants (13) of a representative strain of the epidemic clone in the United States, CA-MRSA USA300 (14), for each LacI transcriptional factor. Before initiating this study, we first determined whether each LacI mutant displayed similar growth in nutrient-rich medium. Upon performing growth curves, we found that each mutant grew with similar kinetics in tryptic soy broth (TSB) and concluded that these media would be appropriate for subsequent experiments comparing the virulence profiles of the mutants (Fig. 1A). It is known that secreted factors play a crucial role in USA300 pathogenesis, as a number of effectors released from the cell support host-cell destruction, immune evasion, and tissue dissemination (4). To assess the makeup of secreted factors (“exoproteins”) released from each of the LacI regulator mutants, we analyzed the supernatants from the strains when grown to exponential phase. Upon performing this qualitative assessment, it was discovered that the mutant for the purine biosynthesis repressor purR produced a greater amount of exoproteins compared with that of the other LacI regulator mutants (Fig. 1B). Moreover, upon examining for the presence of two key USA300 virulence factors produced at exponential phase, leukocidin AB (LukAB) and alpha-toxin (Hla) (15), it was determined that the purR mutant secreted elevated levels of each of these toxins.
Fig. 1.
Phenotypic screen for virulence reveals that purR transposon mutants are hypervirulent. (A) LacI family regulator mutants were assessed for their ability to grow in TSB. Each strain showed similar growth kinetics as determined by OD600 (three replicates per strain). (B) Supernatants from exponentially grown purR::bursa exhibit a greater number of secreted proteins compared with the profiles of the other LacI family regulator mutants. purR::bursa also produced higher levels of the toxins Hla and LukAB. Asterisks indicate loading controls, which are nonspecific bands recognized by the antibodies. (C) Five percent culture filtrates obtained from purR::bursa were more cytotoxic toward primary human PMNs than those of wild-type USA300 or LacI family regulator mutants (n = 5 human donors). Statistical significance was done using a one-way ANOVA and post hoc Holm–Sidak correction of multiple comparisons. Error bars indicate SEM. *P < 0.05. NS, not significant. (D) Mice infected i.v. (2.5 × 107 CFUs) with each LacI mutant or wild-type USA300 succumb to infection with the purR::bursa mutant at a faster rate than that of any other strains tested. Statistical analysis of survival differences between wild type and purR::bursa was performed using the Gehan–Breslow–Wilcoxon test. *P < 0.05.
In accordance with the observation that the purR USA300 mutant produces elevated levels of secreted toxins, we found that supernatants obtained from the mutant were more cytotoxic toward primary human neutrophils (hPMNs) than those of the other LacI regulator mutants and wild-type USA300 (Fig. 1C). This observation is concomitant with an elevated amount of LukAB production, as this secreted factor is responsible for the death of hPMNs (16, 17). To extend the breadth of these findings, we next infected mice i.v. with each of the LacI regulator mutants in a bacteremia model. Intriguingly, we found that mutants for purR acutely enhanced the lethality of USA300 compared to wild-type bacteria and the other LacI regulator mutants (Fig. 1D). Thus, through the assessment of each LacI family regulator’s contribution to virulence, we discovered that the purine biosynthesis regulator PurR represses USA300 virulence traits.
Mutants for purR Cause Rapid Invasive Infection.
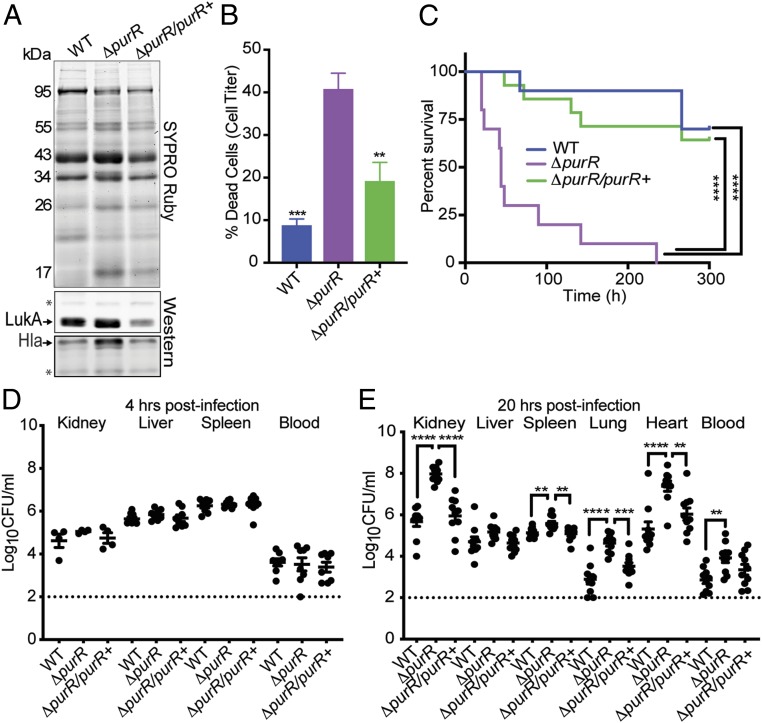
To expand on outcomes generated from our virulence screen (Fig. 1), we created an isogenic mutant for purR (ΔpurR) in USA300 and further assessed its role in pathogenesis. Much like the transposon mutant for purR, we found that the ΔpurR strain secreted a greater amount of virulence factors and that this effect could be complemented with expression of purR on a plasmid (Fig. 2A). As expected, an enhancement in cytotoxicity toward hPMNs was also observed for the isogenic mutant, a phenotype that could be complemented as well (Fig. 2B).
Fig. 2.
Isogenic mutants for purR are highly cytotoxic and cause rapid death. (A) Mutants for purR secrete a greater number of proteins compared with wild-type USA300 and a mutant complemented in trans with purR. Asterisks indicate loading controls, which are nonspecific bands recognized by the antibodies. (B) Five percent culture filtrates obtained from the purR mutant are highly cytotoxic against primary human PMNs compared with wild-type USA300 or the complemented mutant (n = 5 donors). Statistical significance was done using a one-way ANOVA and post hoc Holm–Sidak correction of multiple comparisons. Error bars indicate SEM. **P < 0.01, ***P < 0.001. (C) Mice infected i.v. (2.5 × 107 CFUs) with the purR mutant are acutely susceptible to death compared with wild-type USA300 or the complemented mutant (n = 10 mice per strain). Statistical analysis for survival was performed using the Gehan–Breslow–Wilcoxon test with P values adjusted for multiple comparisons. ****P < 0.0001. (D) Mice administered an i.v. infection (2.5 × 107 CFUs) exhibit no differences in bacterial burden at 4 h postinfection (n = 10 mice per strain; n = 5 kidneys per strain). (E) Mice infected i.v. (2.5 × 107 CFUs) with the purR mutant exhibit significantly higher levels of bacterial burden at 20 h postinfection compared with wild-type S. aureus or the complemented mutant (n = 10 mice per strain). Dotted lines represent the limit of detection in these organs. Statistical significance was done using a one-way ANOVA and post hoc Holm–Sidak correction of multiple comparisons. **P < 0.01, ***P < 0.001, ****P < 0.0001.
To further assess the contribution of PurR in the context of infection in vivo, we complemented the USA300 purR mutant by integrating a single copy of the native gene on the chromosome under the control of its endogenous promoter and performed mouse infections. As with the purR transposon mutant, we found that the ΔpurR isogenic mutant led to acute lethality in a murine model of USA300 bacteremia and that wild-type and complemented bacteria behaved similarly in that they were significantly less lethal (Fig. 2C).
We next sought to determine whether the lethality phenotype witnessed with the purR mutant was associated with increased bacterial burden in mouse tissues commonly infected during S. aureus bacteremia. Due to the acute lethality witnessed in purR mutant infections and our desire to maintain the same bacterial inoculum between infections, we used early time points (4 and 20 h postinfection) to assess bacterial burden. Based on observations that the purR mutant displays a slight growth advantage in minimal media (SI Appendix, Fig. S1), we postulated that the purR mutant might exhibit enhanced replication in nutrient-restricted compartments, such as blood, and other tissues typically infected rapidly after infection (18). However, we found that at 4 h postinfection there were no differences in the number of bacteria recovered from mouse blood or other tissues commonly harboring bacteria at this time point (Fig. 2D). In contrast, at 20 h postinfection, we observed a near pan-organ increase in bacterial burden in mice infected with the USA300 purR mutant, phenotypes that were complemented by expression of purR in trans on the chromosome (Fig. 2E). These data demonstrate that upon infection, the purR mutant exhibits increased bacterial burden in multiple tissues, which is linked with rapid lethality.
The Increased Virulence Exhibited by the purR Mutant Occurs Independent of Purine Production.
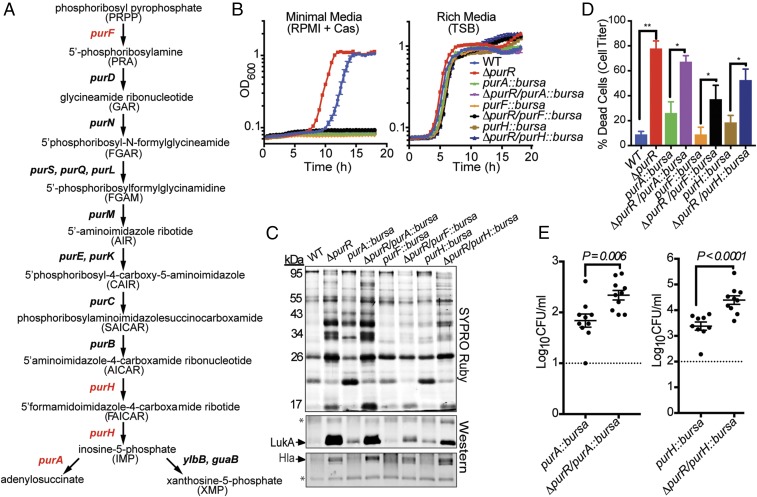
It is well-established that PurR serves as the master negative regulator of de novo purine biosynthesis in bacteria (19, 20). Thus, in the absence of purR there should be overexpression of purine biosynthesis genes and a subsequent increase in purine production. To determine whether the purR mutant’s increased pathogenesis can be attributed to purine overproduction, we generated mutants for purine biosynthesis genes in the USA300 purR mutant background and assessed their contribution to the virulence phenotype (Fig. 3A). It has been established that purine biosynthesis genes are crucial for S. aureus growth in minimal media devoid of purines, as the bacteria cannot overcome the absence of de novo purine biosynthesis (21). Indicative of that, we found that our panel of mutants for purine biosynthesis genes, which included purA, purF, and purH, grew similar to wild-type bacteria in rich media but were incapable of growth in minimal media (Fig. 3B). There was no difference in growth whether the purine biosynthesis mutation was measured alone or in the presence of the purR mutation. Given similar growth kinetics in rich media, we next examined the effects, if any, the purine biosynthesis mutations had on the purR mutant’s hyperproduction of exoproteins. Supernatants from each purine biosynthesis mutant placed in the USA300 purR mutant background retained the increased levels of secreted proteins and exhibited enhanced levels of LukAB and Hla production (Fig. 3C). Concomitant with elevated levels of LukAB, each purine biosynthesis mutant in the purR mutant background showed increased cytotoxicity toward hPMNs compared with its parental strain (Fig. 3D). Collectively, these results support the notion that the increase in cytotoxicity observed by the USA300 purR mutant occurs independent of increased purine production. It is worth noting that purine biosynthesis clearly contributes to the general production of cytotoxins, as each isogenic purine biosynthesis mutant displayed altered secretion and cytotoxic profiles.
Fig. 3.
purR mutant is hypervirulent independent of purine biosynthesis. (A) The purine biosynthesis pathway in S. aureus, as adapted from Mongodin et al. (40). Genes highlighted in red were mutated in our study. (B) Mutants for the purine biosynthesis genes purA, purF, and purH cannot grow in minimal media (RPMI + casamino acids) in either the wild-type or purR mutant background (n = 3 replicates per strain). These mutations do not prevent the strains from growing in rich media (TSB) (n = 3 replicates per strain). (C) Supernatants obtained from purine biosynthesis mutants in the purR mutant background generate elevated levels of secreted proteins, including LukAB and Hla. Asterisks indicate loading controls, which are nonspecific bands recognized by the antibodies. (D) Ten percent culture filtrates obtained from purine biosynthesis mutants in the purR mutant background are more cytotoxic toward primary human PMNs than supernatants obtained from their parental strains (n = 5–8 donors per strain). Statistical analysis was determined using a two-tailed Mann–Whitney U test. Error bars indicate SEM. *P < 0.05, **P < 0.01. (E) Kidneys of mice infected i.v. (2.5 × 107 CFUs) with either ΔpurR/purA::bursa or ΔpurR/purH::bursa for 20 h demonstrate higher bacterial burdens than mice infected with the respective parental strains of these mutants, purA::bursa and purH::bursa (n = 10 mice per strain). Dotted lines represent the limit of detection in these organs. Statistical significance was calculated using a two-tailed t test.
It has been established that S. aureus mutants for purA and purH are highly attenuated in mice (22). To further define if the hyperinfectivity in vivo phenotype observed by the purR mutant could be decoupled from purine biosynthesis, we infected mice in the absence of these key purine biosynthesis enzymes. Despite these debilitating mutations, we observed increased bacterial burden in mice infected with either the purA or purH mutation in the USA300 purR mutant background compared with parental strains (Fig. 3E). These results demonstrate that the purR mutation is able to partially compensate for the effects of mutations in purine biosynthesis genes. Taken together, these ex vivo and in vivo data support the notion that the enhancement in virulence witnessed with the USA300 purR mutant cannot be simply attributed to an overproduction of purines.
Mutants for purR Generate Elevated Virulence Transcripts and Proteins.
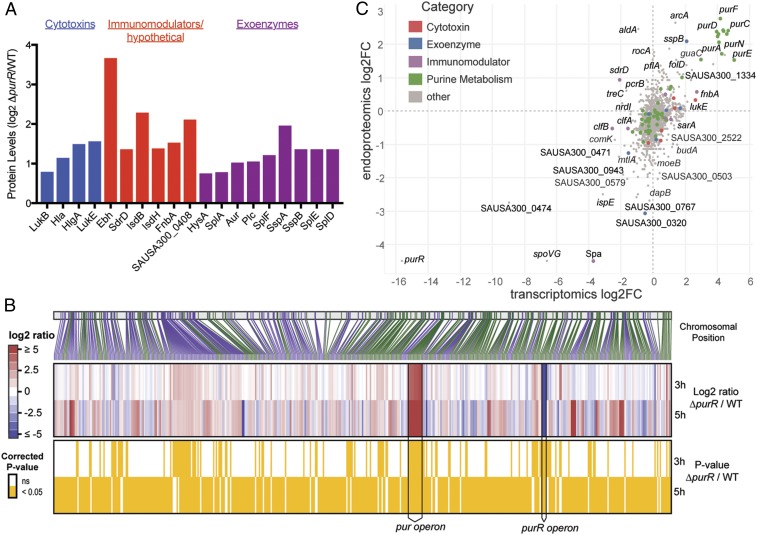
Given our observations that, in a manner independent of purine biosynthesis, inactivation of purR in USA300 results in heightened levels of secreted molecules that make a hypercytotoxic strain, we next sought to identify and quantify the nature of the molecules being secreted by the mutant. To do this, we performed quantitative mass spectrometry analyses on secreted protein fractions from both the purR mutant and wild-type USA300 grown to exponential phase (15). Concomitant with the purR mutant’s increased virulence was an elevation in the production of key staphylococcal factors associated with disease and tissue destruction (Fig. 4A and Dataset S1). Specifically, we witnessed increases in the production of secreted toxins that target and kill human cells, including leukocidins and alpha-toxin (Hla) (23, 24). Additionally, we observed an increase in a number of surface-associated proteins that promote immune evasion, bacterial dissemination, and the acquisition of iron from the host, and an elevation in multiple secreted proteases that have also been shown to support immune evasion (4). To broaden these findings and to understand how PurR may be affecting gene expression at the transcriptional level, we next performed whole-transcriptome RNA-seq analyses. We discovered that disrupting purR results in the differential regulation of 130 genes in USA300 during exponential growth. Among these, 92 are positively regulated and 38 are negatively regulated (Fig. 4B and Dataset S2). As expected, we witnessed the up-regulation in genes within the purine biosynthesis operon, as the repressive activity of PurR had been ablated. In accordance with our proteomics dataset, we also observed a corollary increase in transcripts of gene products associated with virulence (Fig. 4B). Interestingly, we observed a greater than twofold increase in the transcription of the virulence regulator sarA. SarA is a known positive regulator of Hla production and an activator of the Agr system (25–28). A number of the up-regulated genes encoding secreted toxins were also validated with qRT-PCR (SI Appendix, Fig. S2). RNA-seq performed on the purR mutant grown to stationary phase resulted in a significant increase in agrA, agrB, and agrC, which correlates temporally with the preceding activation of sarA. Changes in transcript levels were more profound in general at stationary phase for the purR mutant. This effect can likely be attributed to the differential regulation by PurR at stationary phase and/or secondary outcomes from the regulatory events that occurred during exponential phase (Fig. 4B and SI Appendix, Table S1). Interestingly, when grown to stationary phase, the purR mutant was still acutely lethal to mice during i.v. infection (SI Appendix, Fig. S3). To test the correlation between transcript changes and protein changes in the purR mutant, we integrated the RNA-seq dataset with mass spectrometry done on intracellular proteins (Fig. 4C). This exercise demonstrated corollary changes in both outputs, further bolstering the notion that PurR has a profound effect on gene regulation and the subsequent production of proteins.
Fig. 4.
Mutants for purR transcribe and translate a greater number of virulence effectors. (A) Mutants for purR secrete a greater number of virulence effectors compared with wild-type USA300. Proteins were considered of interest if they contained a signal peptide and displayed absolute fold change >1.5. (B) Heat maps depicting relative fold changes in gene expression between the purR mutant and wild-type USA300 at either exponential or stationary phase. The vertical bar with connecting lines (Top) indicates the gene position on the chromosome. The lines are colored alternating in green or purple, signifying on which strand the gene is encoded. (B, Middle) The expression change in the purR mutant vs. WT, with red signifying an increased expression in the mutant. The yellow panel (Bottom) highlights which genes have expression changes at 3 and/or 5 h. (C) Integration of relative fold changes in gene expression and the changes in intracellular proteins between the purR mutant and wild-type USA300 at exponential phase.
PurR Directly Binds the Promoters of Virulence Genes.
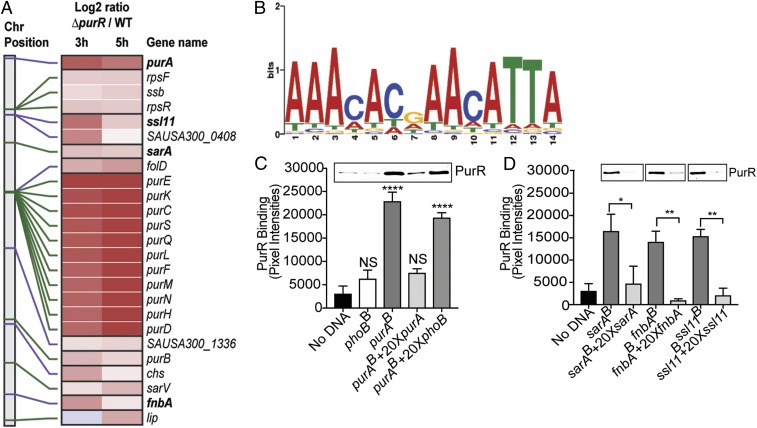
We established that genes involved in virulence are differentially regulated in the absence of purR and that this regulation occurs independent of purine biosynthesis. To examine if PurR is capable of binding to and regulating promoter elements outside of the purine biosynthesis pathway, we performed an in silico search for “PurBoxes” within the USA300 genome using the Bacillus subtilis PurBox (5′-AAACACGAACATTA-3′) consensus sequence as our guide (29). Upon identifying putative transcriptional factor binding sites in the USA300 genome sharing significant homology (P value ≤ 0.0001) with the B. subtilis PurR binding motif using FIMO (Find Individual Motif Occurrences) (30), we found a number of these regions were upstream of single or multigene operons that were up-regulated in the purR mutant (Fig. 5A). Not surprisingly, promoters within the purine biosynthesis pathway had some of the highest matches for homology and also had the greatest increase in gene expression. Interestingly, promoters upstream of nonpurine biosynthesis transcripts that were some of the most highly up-regulated genes in the RNA-seq dataset also contained well-conserved matches for PurR binding sites, such as superantigen-like protein 11 (ssl11) and fibronectin-binding protein A (fnbA). This result bolstered our belief that PurR is capable of regulating loci outside of the purine biosynthesis pathway and that some of these genes either regulate virulence or are themselves virulence factors. Based on these analyses, a consensus S. aureus PurBox was predicted (Fig. 5B). The complete list of regions containing PurBoxes and their P values can be found in Dataset S3.
Fig. 5.
Recombinant PurR binds to virulence gene promoters. (A) The heat map shows motifs recognized by PurR that are positioned upstream of highly expressed genes in the purR mutant. The vertical bar with connecting lines (Left) indicates the gene position on the chromosome. The lines are colored alternating in green or purple, signifying operon groupings. (A, Middle) Expression change in the purR mutant vs. WT, with red signifying an increased expression in the mutant. Gene names are indicated on the far Right. The two Middle panels also have horizontal dividers grouping genes in the same operons. (B) Consensus motif sequence bound by PurR in the S. aureus genome. The height of the letters in the position weight matrix represents information content (in bits) at each position that is related to the degree of certainty of the particular nucleotide at a given position. (C) Recombinant PurR (17 nM) binds to biotinylated purA promoter (800 fmol). This binding activity is not seen with biotinylated phoB promoter (800 fmol), a gene not differentially regulated in the purR mutant and not containing a PurBox. The binding activity of PurR can be competed off with 20× molar excess of nonbiotinylated purA promoter containing the same sequence. XB, biotinylated. The graph includes three biological replicates per binding experiment. Band intensity of PurR was determined using ImageJ software, and statistical significance was calculated using a one-way ANOVA with post hoc Dunnett’s multiple comparisons test. Error bars indicate SEM. ****P < 0.0001. NS, not significant. (D) Recombinant PurR (17 nM) binds to biotinylated promoters of the virulence determinants sarA, ssl11, and fnbA (800 fmol). This activity can be competed off with 20× molar excess of nonbiotinylated promoter containing the same sequence. The graph includes three biological replicates per binding experiment. Band intensity was determined using ImageJ software, and statistical significance was calculated using a one-tailed t test. *P < 0.05, **P < 0.01.
To extend our in silico findings, we performed DNA-binding experiments between purified His-tagged PurR and promoters for virulence factors. We selected our candidate promoters based on their up-regulation in the RNA-seq data, their PurBox homology, and the potential role of their gene product in the purR mutant’s increased virulence. To perform these experiments, equimolar amounts of biotinylated DNA probes were conjugated to streptavidin magnetic beads and then incubated with PurR, and bound PurR was detected via immunoblotting. We found that PurR, as expected, bound the promoter for the purine biosynthesis gene purA, and that this binding could be competed off with a molar excess of nonbiotinylated purA (Fig. 5C). Importantly, the promoter for phoB, a gene that was unchanged based on the RNA-seq data, did not bind to PurR, nor did it compete for PurR binding to the purA promoter DNA. SarA positively regulates both the expression of many virulence factors and virulence factor regulators, including hla and agrBDCA (25–28). sarA was found to be up-regulated in the RNA-seq experiment, and the sarA promoter has high homology for a PurBox (Fig. 5A). In accordance with these observations, we found that PurR binds the sarA promoter and that this binding could be competed off with a molar excess of nonbiotinylated sarA (Fig. 5D). We further extended these findings by demonstrating that PurR also binds to the promoters of fnbA and ssl11 (Fig. 5D). Altogether, these results demonstrate that PurR directly binds to the promoters of virulence genes and other regulators, explaining the observed differential expression of these genes.
Several Virulence Factors Are Responsible for the Enhanced Virulence Observed by the purR Mutant.
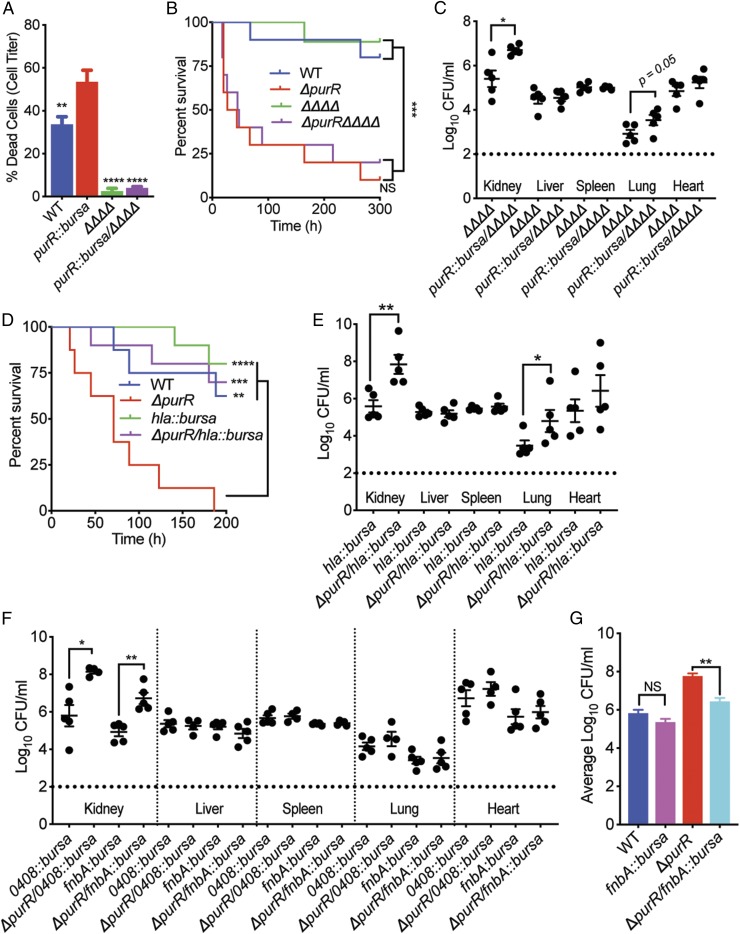
The USA300 purR mutant overproduces an assortment of virulence factors that can contribute to the heightened virulence witnessed in the mouse. Additionally, RNA-seq and PurR–DNA binding experiments demonstrated that PurR regulates sarA, which could further impact the expression of virulence factors. To tease out which factor(s) is responsible for the acute death phenotype observed in the purR mutant infections, we assessed the contribution of the five bicomponent leukocidins (LukAB, LukED, and Panton–Valentine leukocidin, and γ-hemolysins AB and CB) produced by USA300. To do this, we transduced a USA300 strain lacking each leukocidin (ΔΔΔΔ) (31, 32) with the purR::bursa transposon mutant to generate purR::bursa/ΔΔΔΔ. We first examined whether the purR mutant was still hypercytotoxic in the absence of the leukocidins. As expected, loss of the leukocidins ablated the hypercytotoxicity phenotype of the USA300 purR mutant (Fig. 6A). However, upon infecting mice i.v. with purR::bursa/ΔΔΔΔ, we observed no decrease in lethality with respect to the USA300 purR mutation. This result indicated that the acute death phenotype seen with the purR mutant could not be attributed to overproduction of the bicomponent leukocidins (Fig. 6B). Moreover, we found that the leukocidins were not responsible for the enhancement seen in CFUs at 20 h postinfection, as the purR::bursa/ΔΔΔΔ mutant had more bacteria in the kidneys and heart than that of the ΔΔΔΔ mutant (Fig. 6C).
Fig. 6.
purR mutant relies on a set of secreted factors to drive its heightened pathogenesis. (A) PMNs intoxicated with 1.25% supernatants no longer display an elevated susceptibility to supernatants derived from the purR mutant in the absence of the bicomponent pore-forming leukocidins (ΔpurR/ΔΔΔΔ) (n = 3–6 donors). Statistical significance was done using a one-way ANOVA and post hoc Holm–Sidak correction of multiple comparisons. Error bars indicate SEM. **P < 0.01, ****P < 0.0001. (B) Mice infected i.v. (2.5 × 107 CFUs) with the ΔpurR/ΔΔΔΔ mutant do not demonstrate a reduction in susceptibility to death despite the loss of the leukocidins (n = 10 mice per strain). Statistical analysis for survival was performed using the Gehan–Breslow–Wilcoxon test with P values adjusted for multiple comparisons. ***P < 0.001. NS, not significant. (C) Mice infected i.v. (1 × 107 CFUs) with the ΔpurR/ΔΔΔΔ mutant do not demonstrate a reduction in bacterial burden at 20 h postinfection compared with the parental ΔΔΔΔ strain (n = 5 mice per strain). Statistical analysis was examined using a two-tailed Mann–Whitney U test between each infected organ. *P < 0.05. (D) Alpha-toxin drives the purR mutant lethality phenotype in mice. Mice infected i.v. (2.5 × 107 CFUs) with the ΔpurR/hla::bursa mutant are no longer susceptible to death in a purR mutant-dependent manner. Statistical analysis for survival was performed using the Gehan–Breslow–Wilcoxon test with P values adjusted for multiple comparisons. **P < 0.01, ***P < 0.001, ****P < 0.0001. (E) Mice infected i.v. (2.5 × 107 CFUs) with the ΔpurR/hla::bursa mutant do not demonstrate a reduction in bacterial burden at 20 h postinfection compared with the parental hla::bursa strain (n = 5 mice per strain). Statistical analysis was examined using a two-tailed Mann–Whitney U test between each infected organ. *P < 0.05, **P < 0.01. (F) FnbA contributes to the increased bacterial burden phenotype of the purR mutant at 20 h postinfection. Mice infected i.v. (2.5 × 107 CFUs) with mutants for SAUSA300_0408 and fnbA in the purR mutant background were assessed for bacterial burden and only the ΔpurR/fnbA::bursa mutant demonstrated a drop in CFUs within the kidneys (n = 5 mice per strain). Statistical analysis was examined using a two-tailed Mann–Whitney U test between each infected organ. *P < 0.05, **P < 0.01. (G) FnbA significantly contributes to the enhanced bacterial burden phenotype exhibited by the purR mutant. CFU outputs from the kidneys of i.v. infected mice were aggregated and compared (n ≥ 10 mice per strain). Statistical significance was calculated using two-tailed t tests comparing either wild-type USA300 with fnbA::bursa or ΔpurR with ΔpurR/fnbA::bursa. **P < 0.01. NS, not significant.
Hla is a critical virulence factor known to be positively regulated by SarA and we found to be overproduced in the USA300 purR mutant (Figs. 1B and 2A). To determine the role of Hla in the context of the pathogenesis of the purR mutant, we generated a ΔpurR/hla::bursa strain. We found that overproduction of Hla by the purR mutant was indeed responsible for the acute lethality in mice, as deleting hla in the purR mutant background ablated the purR hyperlethality phenotype (Fig. 6D). The overproduction of Hla by the purR mutant likely results in toxinosis during infection of mice, which leads to rapid death (33, 34). To examine whether Hla was also driving the rapid increase in bacterial burden we observe with the mutant following infection, we infected mice i.v. with the ΔpurR/hla::bursa mutant and examined tissues for bacterial burden. As with the leukocidin mutants, Hla was found to not be responsible for driving the increased CFUs in the kidney, as the ΔpurR/hla::bursa strain still had significantly more bacteria than the hla::bursa mutant alone (Fig. 6E).
To understand which staphylococcal factor(s) drives the purR mutant’s enhancement in bacterial burden, we assessed the role of two additional proteins that were significantly up-regulated in both the RNA-seq and proteomic analyses (Fig. 4 A and B) and whose promoters were shown to interact directly with PurR (Fig. 5D). The gene encoding the predicted surface-associated lipoprotein SAUSA300_0408 lies in a two-gene operon with the gene encoding ssl11. These two genes were two of the most significantly up-regulated nonpurine biosynthesis genes in our RNA-seq analyses (Fig. 4B). Because USA300 isolates have a premature stop codon embedded in the ORF of ssl11, we focused on SAUSA300_0408 by transducing the purR mutant with SAUSA300_0408::bursa to generate USA300 ΔpurR/SAUSA300_0408::bursa (ΔpurR/0408::bursa). Another virulence factor that was highly up-regulated in both the RNA-seq and proteomics studies was the fibronectin-binding protein A (fnbA). To test this factor, we transduced the purR mutant with fnbA::bursa to generate USA300 ΔpurR/fnbA::bursa. Upon examining these mutants in the murine bacteremia model of infection, we found that at 20 h postinfection, ΔpurR/fnbA::bursa resulted in a reduction in bacterial burden, while ΔpurR/SAUSA300_0408::bursa did not (Fig. 6F). Albeit the CFU values for ΔpurR/fnbA::bursa were still significantly greater than those of the fnbA::bursa infection alone, consistent with the role of other virulence factors repressed by PurR. To further dissect the role of FnbA in the context of a purR mutant infection, we infected another cohort of mice with either wild-type USA300, ΔpurR, fnbA::bursa mutant, or ΔpurR/fnbA::bursa. Upon aggregating the CFU burden data in the kidneys and comparing the impact of mutating fnbA in either wild-type USA300 or in the purR mutant background, it became clear that mutating fnbA in the purR mutant results in a significant reduction in bacterial burden (Fig. 6G). Together, these data demonstrate that the overproduction of FnbA by the purR mutant, at least in part, facilitates the establishment of infection for USA300 at deeper tissues.
Discussion
In addition to being able to establish infection in essentially every anatomical site within the body, S. aureus is able to persist in a diverse range of environmental settings, including both the community and in the hospital. As such, S. aureus must finely tune virulence factor production to coincide with its continual integration of environmental signals. This study reveals that the canonical regulator of purine biosynthesis in S. aureus and in other bacteria, PurR, moonlights as a direct regulator of virulence in S. aureus. Through our screen of LacI family regulators in USA300, we found that mutants for purR are uniquely hypervirulent and that this phenomenon is due in part to a global increase in virulence factor production. While PurR’s classical role in the cell is to negatively regulate purine production, our data establish that the increase in virulence of cells lacking purR occurs independent of purine biosynthesis. This finding is vital, because a conceivable explanation for the increase in virulence is an unregulated overproduction of purines.
The impact of purine biosynthesis enzymes on the pathogenic fitness of S. aureus has been well-documented (21, 22). It was recently shown that clinical isolates retrieved from cases of persistent MRSA bacteremia expressed higher levels of purine biosynthesis genes and increased production of virulence determinants (35). These data might suggest that purine levels are limiting during the course of bloodstream infections and that MRSA must produce greater amounts of purines de novo, and this coincides with increased production of virulence factors. It would be interesting in future studies to examine levels of circulating purines during infection and assess how PurR activity changes throughout the course of infection. It is very clear that purine biosynthesis genes are crucial during murine infection, as Lan et al. (22) demonstrated that mutations in the purine biosynthesis genes purA and purH prevent S. aureus from infecting the kidneys of mice. Here we show that despite the incredible barrier introduced by disrupting purine biosynthesis, purA and purH mutants in the purR mutant background are still able to infect kidneys at significantly higher levels than their isogenic mutant counterparts. Moreover, purine biosynthesis mutants in the purR mutant background still generate higher amounts of toxins and are more cytotoxic to human phagocytes.
The observation that PurR regulates virulence in addition to purine biosynthesis was bolstered by RNA-seq coupled with proteomic analyses, whereby we observed significant increases in both transcripts encoding toxins and actual secretion of toxin effectors. This dataset validated what was seen experimentally with immunoblots and cytotoxicity studies, in that heightened levels of a subset of leukocidins and alpha-toxin comprised some of the most highly up-regulated genes and protein products. Coinciding with this outcome, upon introducing a purR mutation into a strain lacking leukotoxins (purR::bursa/ΔΔΔΔ), the purR mutant’s cytotoxicity phenotype toward hPMNs was ablated. While leukotoxin overproduction did explain the heightened cytotoxicity toward hPMNs, it did not explain the acute lethality observed in mice. Upon introducing an hla mutation into the purR mutant background (ΔpurR/hla::bursa), we discovered that Hla drives the purR mutant’s acute lethality in mice. Given the high levels of hla/alpha-toxin observed via immunoblot, RNA-seq, and proteomics, perhaps this result is not surprising. Lastly, the overproduction of FnbA by the purR mutant is in part responsible for the increased bacterial burden exhibited by the USA300 purR mutant. Thus, we establish that PurR coordinates the production of an array of virulence factors in S. aureus, which influence the pathogenesis of the organism.
While we had observed phenotypic outcomes that were dependent on the loss of purR, a mechanistic explanation for this phenomenon was not achieved until we performed biochemical studies with purified PurR and potential DNA-binding targets. Using PurR binding sites in B. subtilis as our guide, we performed in silico studies to identify similar sites within the USA300 genome. We found PurBoxes upstream of many genes that were differentially regulated in our RNA-seq studies. To validate these queries, we went on to show that purified PurR does indeed bind to promoters of virulence factors or their regulators. Namely, the promoter for sarA, a gene that was up-regulated in the purR mutant and whose protein product helps drive the expression of virulence factors, was found to be directly recognized by PurR. Additionally, PurR bound to promoters for two of the most highly up-regulated genes in the purR mutant, fnbA and ssl11. Altogether, these findings show that PurR not only regulates genes outside of the purine biosynthesis operon but that some of these genes encode virulence factors.
In the midst of writing this manuscript, Goncheva et al. (36) published a study describing the phenotype of a purR mutant in S. aureus and the role of fnbA in the phenotype. While both studies nicely establish the role of PurR in S. aureus pathogenesis, several major differences distinguish the Goncheva et al. study and the study described herein. Namely, Goncheva et al. performed RNA-seq on exponentially grown S. aureus but observed no transcript changes in the purR mutant aside from purine biosynthesis genes. This surprising result could be due to differences in bacterial growth conditions or protocols for RNA preparation. The group did see changes in fnbA expression using a transcriptional reporter system, indicating that PurR does mediate transcriptional regulation of fnbA. When we coupled RNA-seq with mass spectrometry proteomics, we observed global changes in both transcripts and proteins, many of which were virulence factors. Goncheva et al. not only observed no differences in the secretion profile of the purR mutant compared with wild type but witnessed very little protein secretion overall in either wild-type or mutant bacteria at multiple growth phases. In contrast, we observed increases in overall protein secretion and in toxin levels with the purR mutant, phenotypes that were complemented and that contributed to the observed enhanced cytotoxicity. These differences could be attributed to strain usage, growth conditions, or methods for protein preparation. Goncheva et al. postulated that FnbA drives the acute lethality phenotype seen in mice infected with the purR mutant. However, we found that upon deleting Hla from the purR mutant, the mutant is no longer acutely lethal. Hla has long been shown to induce toxinosis in mice and that overexpression of the toxin leads to acute death (24). Our data, however, agree that FnbA contributes to the heightened level of bacterial burden in tissues observed with the purR mutant, which we hypothesize could in turn lead to increased toxin production and the observed lethal phenotype. Importantly, the current study further dissected the role of PurR by decoupling purine biosynthesis from the purR mutant’s virulence phenotype and demonstrated biochemically that PurR directly binds to the promoters of genes encoding virulence factors to alter their expression. Altogether, these findings establish that PurR moonlights as an important regulator of S. aureus virulence.
The intersection that exists between metabolism and virulence is a growing area of host–pathogen research that has reemerged as a key to understanding infection dynamics. We describe herein a mechanism whereby the regulator of purine biosynthesis, PurR, binds to and regulates the production of virulence factors influencing the pathophysiology of this organism. Examples of metabolic regulators that display dual-functional roles as they pertain to controlling virulence have been previously reported in a number of pathogens. It was recently shown that Citrobacter rodentium, the surrogate in vivo model organism for enterohemorrhagic Escherichia coli, uses an intricate circuit of amino and fatty acid regulators to control the expression of virulence genes in the infected gut (37). Additionally, the intracellular pathogen Listeria monocytogenes uses the branched-chain amino acid sensor CodY to directly control the activity of the master virulence regulator prfA (38). These are just a few examples of multifaceted metabolic regulators, and the work described herein expands on this ever-growing list. Specifically, our findings suggest that S. aureus monitors the concentrations of purines, via PurR, to control its virulence potential. Future studies warrant deciphering the temporal and spatial regulation of virulence by PurR in the host. These studies could form the basis to devise a ligand-based system to lock PurR into a repressive state, thereby disrupting S. aureus purine biosynthesis and the production of critical virulence factors.
Methods
Ethics Statement.
Buffy coats were obtained from anonymous blood donors with informed consent from the New York Blood Center. All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee of NYULMC. All experiments were performed according to NIH guidelines, the Animal Welfare Act, and US federal law.
Bacterial Growth Conditions.
S. aureus strains were routinely grown at 37 °C on tryptic soy agar or in TSB. When appropriate, S. aureus strains were grown in the presence of tetracycline (4 μg/mL), chloramphenicol (10 μg/mL), erythromycin (2.5 μg/mL), or kanamycin (50 μg/mL). E. coli DH5α or BL21 was used for cloning and propagation of plasmids. E. coli culturing was done in Luria–Bertani (LB) broth supplemented with ampicillin (100 μg/mL). Liquid cultures were routinely prepared by diluting overnight cultures at 1:100 and then growing them at 37 °C in 5 mL of growth medium in 15-mL conical tubes. Cultures were incubated at a 45° angle with shaking at 180 rpm.
PurR Motif Discovery.
We used FIMO version 5.0.1 (30) to compute a log-likelihood ratio score for each position in the USA300_FPR3757 genome sequence matching the B. subtilis PurR motif, and applied false discovery rate analysis after estimating a P value and a q value for each position in the given sequence. Motif sequences matching intragenic regions were filtered out, and genes directly located downstream of motif sequences were identified using ANNOVAR (39).
PurR Purification.
The purR locus was cloned into the plasmid pET-41b(+) (EMD Biosciences) to generate a construct containing a 6× histidine tag on the C terminus of PurR. The construct was transformed into E. coli strain BL21 (Agilent Technologies) for purification. Purification protocols were adapted from previously described methods to purify PurR from B. subtilis in E. coli (29). To purify S. aureus PurR, overnight cultures of E. coli harboring pET-41b(+)-purR were diluted 1:20 in LB broth containing 100 μg/mL ampicillin and subcultured for 3 h at 37 °C. Following incubation, the cultures were placed on ice and given 1 mM isopropyl-β-d-thiogalactopyranoside to induce the transcription of purR. Induced cultures were then incubated at 16 °C overnight at 180 rpm for optimal production of PurR. The next morning, cultures were spun down at 10,000 rpm for 15 min and pellets were placed at −80 °C overnight. Following storage at −80 °C, pellets were thawed and resuspended in 40 mL of buffer A1 (pH 8.0, 50 mM NaH2PO4, 500 mM NaCl, 15 mM imidazole, Halt protease inhibitor) and then sonicated six times at 65% amplitude for 20 s. Sonicated fractions were spun down at 10,000 rpm for 30 min and 40 mL soluble fraction was added to 40 mL of buffer A2 (pH 8.0, 50 mM NaH2PO4, 50 mM NaCl, 15 mM imidazole, Halt protease inhibitor). Nickel-nitrilotriacetic acid resin (4 mL) (Qiagen) was then added to the lysate and the mixture was incubated with agitation for 30 min. Following incubation, the sample was applied to a column and washed with 200 mL of buffer A3 (pH 8.0, 50 mM NaH2PO4, 300 mM NaCl, 15 mM imidazole) and 200 mL of buffer B (pH 6.0, 50 mM NaH2PO4, 300 mM NaCl, 15 mM imidazole, 10% glycerol). Purified protein was ultimately eluted with buffer B containing 500 mM imidazole and then dialyzed overnight against 10 mM Hepes (pH 8.0), 50 mM (NH4)2SO4, 300 mM NaCl. Protein samples were then concentrated using Centriprep spin columns (Millipore). To achieve greater purity, protein was separated by size exclusion using the ÄKTA Column Chromatography System (GE Healthcare).
Promoter Pull-Down Assays.
To visualize interactions between PurR and promoters of interest, we generated PCR products for each promoter using oligonucleotides containing biotinylated labels. All PCR products were purified (Qiagen) and then diluted to a concentration of 100 ng/μL. M-280 streptavidin Dynabeads (Invitrogen) that had been washed and resuspended in binding and wash buffer (pH 7.5, 2 M NaCl, 1 mM EDTA, 10 mM Tris) were incubated with 800 fmol of each DNA fragment for 30 min at room temperature on a rotisserie. Following incubation, samples were placed on a magnet and the supernatants were removed. The samples were then washed three times, once with binding and wash buffer and twice with PurR–DNA binding buffer (10 mM Hepes, pH 7.6, 50 mM KCl, 1 mM EDTA, 5 mM MgCl2, 5 mM DTT) (29). The washed DNA-conjugated beads were then resuspended in 200 μL of PurR–DNA binding buffer. Before mixing the beads with protein, PurR protein samples were diluted in PurR–DNA binding buffer and mixed with 5 μg of double-stranded poly(dG:dC) (InvivoGen) per binding reaction; 100 μL of 50 nM PurR protein mixed with poly(dG:dC) was then added to the DNA-conjugated beads and allowed to mix on a shaking platform for 30 min at 30 °C at 550 rpm. In the case of competitions for PurR binding, nonbiotinylated DNA was mixed with the washed DNA-conjugated beads before the addition of PurR. Following incubations with DNA and PurR, beads were again placed on a magnet and supernatants were removed. The beads were then washed twice with PurR–DNA binding buffer. Following the washes, beads were resuspended in 2× SDS sample buffer and boiled for 10 min. Lastly, the samples were placed back on a magnet and the supernatants were collected to be resolved with 12% SDS/PAGE. Following electrophoresis, gels were transferred to a nitrocellulose membrane and then probed with anti-His antibody. Alexa Fluor 680-anti-rabbit antibody was used as a secondary antibody, and the membranes were visualized using the Odyssey Infrared Imaging System (LI-COR Biosciences). Densitometry on the immunoblots was conducted using ImageJ software, version 1.52o (NIH).
Graphical and Statistical Analyses.
Statistical significance was determined using Prism 7.0 (GraphPad Software).
Meetings.
Preliminary data were presented at the Gordon Research Conference on Microbial Toxins and Pathogenicity in July 2018.
Data Availability.
Additional methods and other relevant data are in Datasets S1, S2, and S3 and SI Appendix. Raw and processed mRNA sequence files are accessible at the NCBI Gene Expression Omnibus under accession no. GSE132179. The mass spectrometry data have been deposited in MassIVE ID MSV000083904 and ProteomeXchange ID PXD014162 (41, 42).
Supplementary Material
Acknowledgments
We thank the members of the V.J.T. laboratory for helpful discussions. We also thank Bianca Dunn for help in generating digital images. We thank members of the Genome Technology Center for their assistance in generating the RNA-seq data, as well as other members of the Proteomics Laboratory Core for help with the mass spectrometry experiments. We thank BEI Resources for providing the following strains: NR-47981, NR-47780, NR-47528, NR-46968, NR-47285, and NR-47124. Research reported in this publication was supported in part by the National Institutes of Health, National Institute of Allergy and Infectious Diseases (NIH-NIAID) under Awards R01AI099394 and R01AI105129 (to V.J.T.), R01AI103268 (to B.S. and V.J.T.), and R01AI119145 (to H.v.B.), and NIAID-supported Institutional Research Training Grant on Infectious Disease & Basic Microbiological Mechanisms T32AI007180 (to W.E.S.). The NYU Langone Health Genome Technology Center and the Proteomics Laboratory are partially supported by Cancer Center Support Grant P30CA016087 at the Laura and Isaac Perlmutter Cancer Center. V.J.T. is a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Diseases. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Conflict of interest statement: V.J.T. is an inventor on patents and patent applications filed by New York University which are currently under commercial license to Janssen Biotech, Inc.
This article is a PNAS Direct Submission.
Data deposition: The mRNA sequencing data reported in this paper have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) (accession no. GSE132179). The mass spectrometry data reported in this paper have been deposited in MassIVE ID MSV000083904 and ProteomeXchange ID PXD014162.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1904280116/-/DCSupplemental.
References
- 1.Lowy F. D., Staphylococcus aureus infections. N. Engl. J. Med. 339, 520–532 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Dantes R., et al. ; Emerging Infections Program–Active Bacterial Core Surveillance MRSA Surveillance Investigators , National burden of invasive methicillin-resistant Staphylococcus aureus infections, United States, 2011. JAMA Intern. Med. 173, 1970–1978 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balasubramanian D., Harper L., Shopsin B., Torres V. J., Staphylococcus aureus pathogenesis in diverse host environments. Pathog. Dis. 75, ftx005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thammavongsa V., Kim H. K., Missiakas D., Schneewind O., Staphylococcal manipulation of host immune responses. Nat. Rev. Microbiol. 13, 529–543 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Novick R. P., Geisinger E., Quorum sensing in staphylococci. Annu. Rev. Genet. 42, 541–564 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Liu Q., Yeo W. S., Bae T., The SaeRS two-component system of Staphylococcus aureus. Genes (Basel) 7, E81 (2016).27706107 [Google Scholar]
- 7.Cheung A. L., Nishina K. A., Trotonda M. P., Tamber S., The SarA protein family of Staphylococcus aureus. Int. J. Biochem. Cell Biol. 40, 355–361 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McNamara P. J., Milligan-Monroe K. C., Khalili S., Proctor R. A., Identification, cloning, and initial characterization of rot, a locus encoding a regulator of virulence factor expression in Staphylococcus aureus. J. Bacteriol. 182, 3197–3203 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ibarra J. A., Pérez-Rueda E., Carroll R. K., Shaw L. N., Global analysis of transcriptional regulators in Staphylococcus aureus. BMC Genomics 14, 126 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levdikov V. M., Blagova E., Joseph P., Sonenshein A. L., Wilkinson A. J., The structure of CodY, a GTP- and isoleucine-responsive regulator of stationary phase and virulence in gram-positive bacteria. J. Biol. Chem. 281, 11366–11373 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Majerczyk C. D., et al. , Staphylococcus aureus CodY negatively regulates virulence gene expression. J. Bacteriol. 190, 2257–2265 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seidl K., et al. , Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob. Agents Chemother. 50, 1183–1194 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fey P. D., et al. , A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. MBio 4, e00537-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diep B. A., et al. , Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367, 731–739 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Chapman J. R., et al. , Using quantitative spectrometry to understand the influence of genetics and nutritional perturbations on the virulence potential of Staphylococcus aureus. Mol. Cell. Proteomics 16 (4 suppl. 1), S15–S28 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dumont A. L., et al. , Characterization of a new cytotoxin that contributes to Staphylococcus aureus pathogenesis. Mol. Microbiol. 79, 814–825 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ventura C. L., et al. , Identification of a novel Staphylococcus aureus two-component leukotoxin using cell surface proteomics. PLoS One 5, e11634 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Surewaard B. G., et al. , Identification and treatment of the Staphylococcus aureus reservoir in vivo. J. Exp. Med. 213, 1141–1151 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weng M., Nagy P. L., Zalkin H., Identification of the Bacillus subtilis pur operon repressor. Proc. Natl. Acad. Sci. U.S.A. 92, 7455–7459 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho B. K., et al. , The PurR regulon in Escherichia coli K-12 MG1655. Nucleic Acids Res. 39, 6456–6464 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Connolly J., et al. , Identification of Staphylococcus aureus factors required for pathogenicity and growth in human blood. Infect. Immun. 85, e00337-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lan L., Cheng A., Dunman P. M., Missiakas D., He C., Golden pigment production and virulence gene expression are affected by metabolisms in Staphylococcus aureus. J. Bacteriol. 192, 3068–3077 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alonzo F. III, Torres V. J., The bicomponent pore-forming leucocidins of Staphylococcus aureus. Microbiol. Mol. Biol. Rev. 78, 199–230 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berube B. J., Bubeck Wardenburg J., Staphylococcus aureus α-toxin: Nearly a century of intrigue. Toxins (Basel) 5, 1140–1166 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheung A. L., Koomey J. M., Butler C. A., Projan S. J., Fischetti V. A., Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc. Natl. Acad. Sci. U.S.A. 89, 6462–6466 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheung A. L., Ying P., Regulation of alpha- and beta-hemolysins by the sar locus of Staphylococcus aureus. J. Bacteriol. 176, 580–585 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chien Y., Manna A. C., Cheung A. L., SarA level is a determinant of agr activation in Staphylococcus aureus. Mol. Microbiol. 30, 991–1001 (1998). [DOI] [PubMed] [Google Scholar]
- 28.Zielinska A. K., et al. , Defining the strain-dependent impact of the staphylococcal accessory regulator (sarA) on the alpha-toxin phenotype of Staphylococcus aureus. J. Bacteriol. 193, 2948–2958 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bera A. K., Zhu J., Zalkin H., Smith J. L., Functional dissection of the Bacillus subtilis pur operator site. J. Bacteriol. 185, 4099–4109 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grant C. E., Bailey T. L., Noble W. S., FIMO: Scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boles B. R., Thoendel M., Roth A. J., Horswill A. R., Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS One 5, e10146 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blake K. J., et al. , Staphylococcus aureus produces pain through pore-forming toxins and neuronal TRPV1 that is silenced by QX-314. Nat. Commun. 9, 37 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inoshima I., et al. , A Staphylococcus aureus pore-forming toxin subverts the activity of ADAM10 to cause lethal infection in mice. Nat. Med. 17, 1310–1314 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Powers M. E., Becker R. E., Sailer A., Turner J. R., Bubeck Wardenburg J., Synergistic action of Staphylococcus aureus α-toxin on platelets and myeloid lineage cells contributes to lethal sepsis. Cell Host Microbe 17, 775–787 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li L., et al. , Role of purine biosynthesis in persistent methicillin-resistant Staphylococcus aureus infection. J. Infect. Dis. 218, 1367–1377 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goncheva M. I., et al. , Stress-induced inactivation of the Staphylococcus aureus purine biosynthesis repressor leads to hypervirulence. Nat. Commun. 10, 775 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pifer R., Russell R. M., Kumar A., Curtis M. M., Sperandio V., Redox, amino acid, and fatty acid metabolism intersect with bacterial virulence in the gut. Proc. Natl. Acad. Sci. U.S.A. 115, E10712–E10719 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lobel L., et al. , The metabolic regulator CodY links Listeria monocytogenes metabolism to virulence by directly activating the virulence regulatory gene prfA. Mol. Microbiol. 95, 624–644 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang K., Li M., Hakonarson H., ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mongodin E., et al. , Microarray transcription analysis of clinical Staphylococcus aureus isolates resistant to vancomycin. J. Bacteriol. 185, 4638–4643 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sause W., et al. , RNA-Seq of WT Staphylococcus aureus and purR grown to exponential and stationary phase. NCBI Gene Expression Omnibus. http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE132179. Deposited 4 June 2019.
- 42.Sause W., et al. , The purine biosynthesis regulator PurR moonlights as a virulence regulator in Staphylococcus aureus. MassIVE and proteomeXchange. https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?task=b5fa9a1d3f464bc8b2b83b0492663420 and http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD014162. Deposited 4 June 2019. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Additional methods and other relevant data are in Datasets S1, S2, and S3 and SI Appendix. Raw and processed mRNA sequence files are accessible at the NCBI Gene Expression Omnibus under accession no. GSE132179. The mass spectrometry data have been deposited in MassIVE ID MSV000083904 and ProteomeXchange ID PXD014162 (41, 42).