ABSTRACT
Influenza A virus can evade host innate immune response that is involved in several viral proteins with complicated mechanisms. To date, how influenza A M2 protein modulates the host innate immunity remains unclear. Herein, we showed that M2 protein colocalized and interacted with MAVS (mitochondrial antiviral signaling protein) on mitochondria, and positively regulated MAVS-mediated innate immunity. Further studies revealed that M2 induced reactive oxygen species (ROS) production that was required for activation of macroautophagy/autophagy and enhancement of MAVS signaling pathway. Importantly, the proton channel activity of M2 protein was demonstrated to be essential for ROS production and antagonizing the autophagy pathway to control MAVS aggregation, thereby enhancing MAVS signal activity. In conclusion, our studies provided novel insights into mechanisms of M2 protein in modulating host antiviral immunity and uncovered a new mechanism into biology and pathogenicity of influenza A virus.
Abbreviations: AKT/PKB: AKT serine/threonine kinase; Apo: apocynin; ATG5: autophagy related 5; BAPTA-AM: 1,2-Bis(2-aminophenoxy) ethane-N,N,N′,N′-tetraacetic acid tetrakis; BECN1: beclin 1; CARD: caspase recruitment domain; CCCP: carbonyl cyanide m-chlorophenylhydrazone; CQ: chloroquine; DCF: dichlorodihyd-rofluorescein; DPI: diphenyleneiodonium; DDX58: DExD/H-box helicase 58; eGFP: enhanced green fluorescent protein; EGTA: ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid; ER: endoplasmic reticulum; hpi: hours post infection; IAV: influenza A virus; IFN: interferon; IP: immunoprecipitation; IRF3: interferon regulatory factor 3; ISRE: IFN-stimulated response elements; LIR: LC3-interacting region; MAP1LC3B/LC3B: microtubule associated protein 1 light chain 3 beta; MAVS: mitochondrial antiviral signaling protein; MMP: mitochondrial membrane potential; MOI, multiplicity of infection; mRFP: monomeric red fluorescent protein; MTOR: mechanistic target of rapamycin kinase; NC: negative control; NFKB/NF-κB: nuclear factor kappa B; PI3K: class I phosphoinositide 3-kinase; RLR: RIG-I-like-receptor; ROS: reactive oxygen species; SEV: sendai virus; TM: transmembrane; TMRM: tetramethylrhodamine methylester; VSV: vesicular stomatitis virus
KEYWORDS: Autophagy, influenza M2 protein, innate immunity, ion channel activity, MAVS aggregates
Introduction
Influenza A virus (IAV) causes significant morbidity and mortality in humans and animals worldwide. IAV infection induces rapidly host innate immune response via a variety of regulatory mechanisms [1], which is essential for pathogen clearance and host survival. However, excessive inflammatory response defined as ‘cytokine storm’ leads to host immunopathology. Influenza A M2 protein, one of the best characterized viroporin to date, plays a well-established biological role in viral pathogenesis [2]. However, the detailed mechanisms remain not completely understood. It is a homotetrameric integral membrane protein, with each chain of the mature protein containing 97 amino acid residues, comprising a 24-residue N-terminal ectodomain, a single 19-residue transmembrane (TM) domain, and a 54-residue cytoplasmic tail [3]. M2 protein possesses a proton-selective ion channel activity, which is the target of antiviral drug (amantadine) and involved in both viral entry and synthesis of new viruses during viral assembly and budding [4]. Histidine residue at position 37 in its TM domain is associated with proton channel selective activity and conduction, which can be abolished when His37 is replaced by glycine, alanine, glutamic acid, serine or threonine [5,6]. The M2 proton channel activity is required for the activation of NLRP3 (NLR family pyrin domain containing 3) inflammasome in primed macrophages and dendritic cells [7]. It has been documented that M2 proton channel activity contributes to the block of autophagosomes-lysosomes fusion, and that the effect of M2 on autophagy arrest can be inhibited by amantadine [8]. Other studies have also demonstrated that the influenza virus infection can trigger autophagy initiation as well as inhibit autophagosome maturation, and M2 is involved in initiating the formation of autophagosomes in infected cells [9–12]. However, the underlying pathway of how M2 initiates the autophagy process remains not completely understood. M2 is expressed in the endoplasmic reticulum (ER), Golgi and then at the plasma membrane in a sequential manner in the infected cells [7]. However, whether M2 protein could anchor to other organelles remains unknown.
Multiple factors such as nutritional deprivation, growth factor depletion, hypoxia, proteins aggregates, damaged organelles, DNA damage and infection could initiate an autophagic response [13], and in turn the aggregated proteins such as MAVS aggregates could be degraded and cleared in the autophagic process [14]. ROS [15,16] and intracellular Ca2+ [15,17] can serve as signaling molecules in autophagy, regulating inflammation and innate immunity. Autophagy modulates the RIG-I-like-receptor (RLR) signaling in two ways. First, mitochondria accumulate within the cell in the absence of autophagy, leading to the building up of MAVS, a key signaling protein for RLRs. Second, damaged mitochondria that are not degraded in the absence of autophagy provide a source of ROS, which amplifies the RLR signaling [18]. Recent studies demonstrate that autophagy negatively regulates the stability of MAVS or MAVS aggregates that are not only functional but also potently propagate downstream signaling via directly binding of ATG5 [19,20] and MAP1LC3B/LC3B [21,22] to MAVS through a conserved classical LC3-interacting region (LIR) motif in the caspase recruitment domain (CARD) of MAVS, thereby attenuating the MAVS-mediated antiviral signaling. As a viropinrin, whether and how M2 protein is involved in modulating host innate immunity remains unclear.
In this study, we provided the first evidence that M2 was able to anchor to mitochondria, promoted the mitochondria fusion and increased the mitochondria number. Furthermore, we showed that M2 triggered extracellular Ca2+ influx-dependent ROS production, and subsequently led to the activation of ATG5 and inhibition of AKT/PKB and MTOR (mammalian target of rapamycin kinase) activity through class I phosphoinositide 3-kinase (PI3K)-AKT-MTOR signaling pathway, ultimately triggering the activation of autophagy. In addition, we revealed that M2 antagonized the autophagy and interacted with MAVS, resulting in reducing the MAVS-ATG5 and MAVS-LC3B complexes formation, thus increasing MAVS aggregations that induce amplification of RLR signaling. Finally, we demonstrated that the proton channel activity of M2 protein was crucial to enhance activation of host innate immunity.
Results
M2 induces autophagy through ATG5 and PI3K-AKT-MTOR pathway
Autophagy begins with the activation of class III phosphatidylinositol 3-kinase (PtdIns3K) complex, of which, BECN1/Beclin1 not only participates in autophagosome formation, but also interacts with different protein complexes regulating the autophagosome-lysosome fusion [12]. MTOR, a well-characterized suppressor in autophagic pathway, is important in autophagy processing, and AKT has been shown to contribute to positive regulation of MTOR [23]. The ATG12–ATG5 complexes are key regulators that are required to activate autophagy, and are involved in one of the two ubiquitin-like conjugation systems mediating vesicle elongation during the autophagosome formation process [24]. We showed that both the H5N1 A/duck/Hubei/Hangmei01/2006 (H5N1/HM) virus and its M2 protein were able to induce autophagosome accumulation, evidenced by the results of both western blot (Figure 1(a), Figure S1(a and d)) and immunofluorescence analysis (Figure S1(b and c)), which was in agreement with the findings as reported previously [25]. Influenza virus may trigger autophagy initiation as well as block the autophagosome fusion with lysosome [9–12]. To explore whether M2 could mediate autophagy initiation, we knocked down the autophagy genes ATG5 and BECN1, and detected the LC3B lipidation in the M2-expressing cells that were treated with or without chloroquine (CQ, an inhibitor blocking the fusion of autophagosome with lysosome), respectively. As shown in Figure S1(e and f), both ATG5 and BECN1 were efficiently knocked down, and M2-mediated increase of LC3B-II expression was significantly suppressed when silenced ATG5 or BECN1 in the absence of CQ, suggesting that M2 was able to trigger the initiation of autophagy by ATG5 and BECN1. Because CQ can activate a non-canonical autophagy response [26], when the cells were exposed to CQ, M2-induced LC3B-II increase was dramatically enhanced in ATG5-, BECN1- or NC (negative control)-silencing groups compared with cells treated without CQ, which was inconsistent with the findings of a previous study [25]. Intriguingly, LC3B-II expression in ATG5-silencing cells treated with CQ was still much lower than the NC-silencing cells treated with CQ, whereas the amount of LC3B-II induced by M2 was comparable in BECN1 and NC-silencing cells that were treated with CQ. These results indicated that M2 possibly blocked the autolysosome formation, in which process BECN1 played a crucial role. This was consistent with the results of a previous study that showed the first 60 amino acids of M2 enable binding to BECN1 and are sufficient for inhibition of autophagic influx [25]. To confirm that M2 blocks the fusion of autophagosomes with lysosomes, we used a tandem reporter construct, mRFP-GFP-LC3; the green fluorescent protein (GFP) of this tandem autophagosome reporter is sensitive and attenuated in an acidic pH environment by lysosomal degradation, whereas the red fluorescent protein (mRFP) is not; therefore, the fusion of autophagosomes with lysosomes will result in the loss of yellow fluorescence and only appearance of the red fluorescence of mRFP. In either the H5N1/HM virus-infected (Figure S1(g), Middle) or M2-transfected (Figure S1(g), Down) cells, the LC3 positive autophagic vacuoles were yellow, suggesting impaired autophagosome fusion with lysosomes. Moreover, both M2 mutant (M2H37G, expresses equally to the WT M2), which abolished the proton channel activity and amantadine (the proton channel activity inhibitor) exhibited significant depression in LC3B-II expression (Figure S1(d)). Similarly, there was a substantial decrease in the number of GFP-LC3 puncta visualized in H5N1/HM-infected cells treated with amantadine (Figure S1(b)) and M2H37G-transfected cells (Figure S1(c)). These findings were consistent with the results of a recent study that showed proton channel activity of M2 contributes to the autophagy arrest [8].
Figure 1.
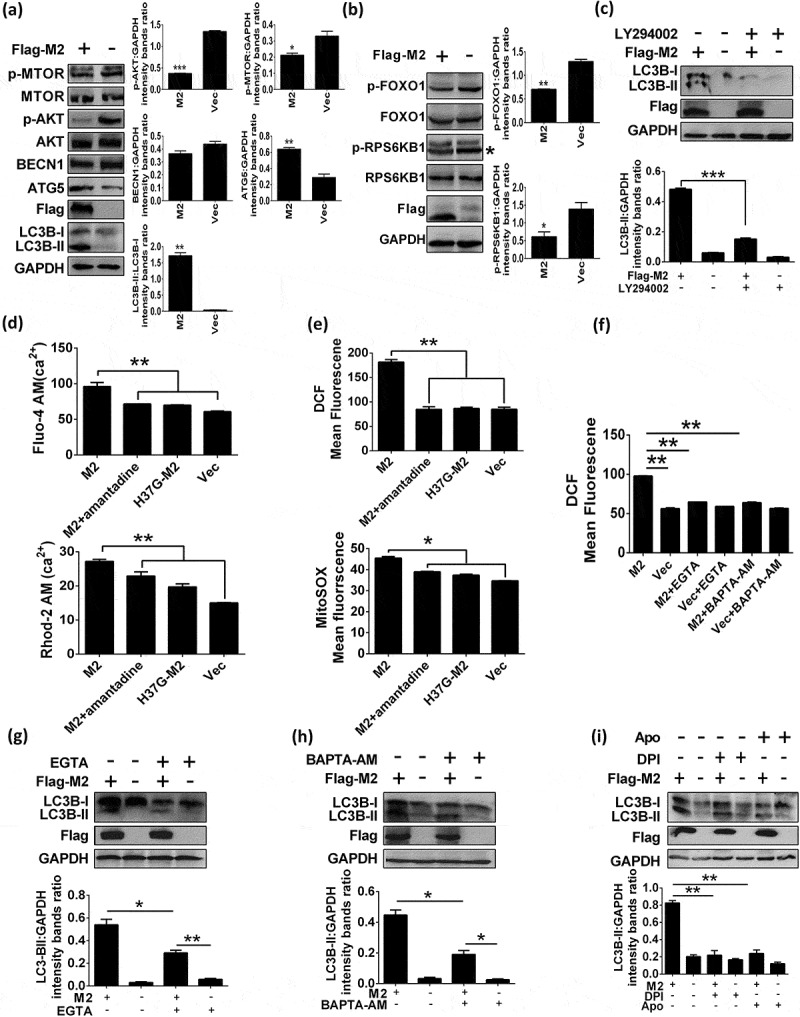
Influenza M2 protein induces autophagy through ATG5 and PI3K-AKT-MTOR pathway and cellular responses. (a and b) HEK 293T cells were transfected with indicated plasmids for 24 h, cells lysates were analyzed by western blot. * represents the indicated protein. (c) HEK 293T cells were pretreated with 10 μM LY294002 for 6 h, and then transfected with Flag-M2 for another 24 h. Cells lysates were evaluated by western blot. (d) HEK 293T cells were transfected with indicated plasmids for 12 h and treated with 5 μM amantadine. The Fluo-4 AM (Up) and Rhod-2 AM (Down) fluorescence was tested by BD FACSCalibur system after 12 h treatment. (e) HEK 293T cells were treated as in (d). Mean DCF (Up) and MitoSOX (Down) fluorescence was determined via flow cytometry. (f) HEK 293T cells were transfected with Flag-M2 and treated with 0.4 mM EGTA or 16 μM BAPTA-AM, respectively. DCF fluorescence was analyzed as (e). (g-i) Flag-M2-transfected HEK 293T cells were treated with 0.4 mM EGTA (g), or 16 μM BAPTA-AM (h), or 3 μM DPI (i), or 0.1 mM Apo (i) for 24 h. Cell lysates were analyzed by western blot. Error bars, mean ± SD of 3 experiments (*p < 0.05; **p < 0.01; ***p < 0.001).
However, which signaling pathway (s) used by M2 to initiate formation of autophagosome remains unclear. Thus, we sought to explore whether M2-induced autophagy depended on ATG5, the PtdIns3K complex containing BECN1 or the PI3K-AKT-MTOR signaling pathway. As shown in Figure 1(a and b), M2 expression resulted in a significant reduction of AKT (Ser473) phosphorylation and of MTOR (Ser2448) phosphorylation as well as one of their respective downstream targets of AKT [27], FOXO1 (Ser256) phosphorylation and RPS6KB1 (Thr389) phosphorylation (a downstream effector of MTOR signaling [27]), although the intensity of whole AKT, MTOR, FOXO1 and RPS6KB1 were comparable in both WT M2- and control-transfected HEK 293T cells. Meanwhile, ATG5 was substantially upregulated but BECN1 was moderately downregulated. Furthermore, LY294002, a known chemical PtdIns3K inhibitor [28], remarkably decreased the M2-induced LC3B-II expression (Figure 1(c)). Collectively, these results indicated that M2-induced autophagy in HEK 293T cells was by activating ATG5 and inhibiting AKT and MTOR activity through the PI3K-AKT-MTOR signaling pathway.
M2-induced elevation of Ca2+ and ROS production are essential for M2-triggered autophagosome formation
Viruses (poliovirus, rhinovirus, coxsackievirus, HBV and EMCV) have been identified as pathogens that encode viroporins, which mediate increase of [Ca2+]i, thereby activating a calcium-dependent signaling pathway to initiate autophagy [29,30]. To investigate whether M2-induced autophagy is due to the Ca2+ concentration alteration, we first evaluated Ca2+ dynamics including intracellular [Ca2+]i and mitochondrial [Ca2+]m affected by M2, respectively. M2 expression resulted in dramatic elevations of [Ca2+]i and [Ca2+]m, whereas both amantadine and M2H37G blocked these effects (Figure 1(d)), indicating that M2 altered the Ca2+ dynamics that were proton channel activity-dependent. Increased [Ca2+]m can result in some effects including increasing electron transport, elevating ROS production, and causing the opening of permeability transition pore under some conditions [31]. Influenza M2 has also been reported to trigger apparent elevation of ROS production [32]. To investigate whether M2 is really able to potentiate the ROS production in concert with the elevated levels of Ca2+, impact of M2 on the intracellular and mitochondrial ROS production was determined by utilizing the fluorescent dye DCF-DA and mitochondria-targeting superoxide indicator MitoSOX, respectively. Results showed that the mean DCF and MitoSOX fluorescence were much weaker in cells expressing M2 treated with amantadine or M2H37G protein than those in M2-transfected cells without treatment with amantadine (Figure 1(e)), indicating that M2 induced intense ROS generation, depending on its proton channel activity. Moreover, the H5N1/HM virus-induced elevations of [Ca2+]i, [Ca2+]m and ROS production were also drastically depressed by amantadine in H5N1/HM-infected A549 cells at 24 h post infection (hpi) (Figure S2(a)), indicating that the proton channel activity of M2 contributed to the cellular response changes. Next we determined whether M2 really triggered Ca2+-dependent ROS production, and showed that both EGTA (a chelator of extracellular Ca2+) and BAPTA-AM (a chelator to block cytosolic calcium) significantly reduced the M2-induced ROS generation (Figure 1(f)). These results indicated that M2-induced increase of ROS production depended on the elevation of [Ca2+]i.
We next determined whether M2-induced increase of [Ca2+]i is responsible for induction of autophagy. The results showed that LC3B-II was significantly reduced in cells treated with EGTA when compared to those without treatment with EGTA (Figure 1(g)). Similar results were obtained when BAPTA-AM was applied in analogous assays (Figure 1(h)). Meanwhile, rapamycin (an autophagy inducer)-induced autophagy was also suppressed by EGTA and BAPTA-AM (Figure S2(d and f)), respectively, suggesting that rapamycin-induced autophagy may also be through the enhanced Ca2+ signaling, which was consistent with the results of a previous study [33]. A significant decrease was also observed in the abundance of M2-induced LC3B-II when cells were treated with the NADPH oxidase inhibitor, diphenyleneiodonium (DPI) or apocynin (Apo) (Figure 1(i)). These results indicated that ROS production were essential for M2-induced autophagy. Simultaneously, EGTA and BAPTA-AM markedly lowered the degree of M2-induced increase of [Ca2+]i (Figure S2(b)), and intracellular ROS also exerted a drastic decrease when cells were treated with either DPI or Apo (Figure S2(b)). Noticeably, the concentrations of inhibitors used did not affect both cell viability as determined by MTT detection (data not shown) and M2 expression (Figure 1(g–i)). Accordingly, these data demonstrated that M2-triggered autophagosome formation was a result of M2 viroporin-mediated elevation of cellular Ca2+ concentration and ROS production.
M2 protein anchors to mitochondria and disrupts mitochondrial dynamics
In the infected cells, M2 protein, upon synthesized, is transported to the ER, travels to the Golgi apparatus, and then is sorted at the trans Golgi network (TGN) for transportation to the plasma membrane and assembled into the virus particle [34]. Several viroporins have been reported to be partially colocalized with mitochondria and alter the normal morphology of mitochondrial network [35–40]. To investigate whether M2 protein is able to anchor to mitochondria, we performed immunofluorescence analysis. Results showed that partial of M2 protein colocalized with the ER and Golgi marker in the A549 cells that were infected with H5N1/HM virus (Figure S3(a and b)), as reported by the previous studies [41,42], and partial colocalized with red fluorescent protein RFP-tagged mito (dsRed-mito), a mitochondrial marker protein, in the A549 cells that were transfected with the M2 (Figure 2(a)) or infected with the H5N1/HM virus (Figure 2(b), Figures S4, S5, Movie S1, Model S1 and S2). Western blot analysis of subcellular fractions further demonstrated that M2 protein was substantially enriched in the mitochondrial fraction in both M2-transfected cells (Figure 2(c); Left) and H5N1/HM-infected cells (Figure 2(c); Right). These results suggested that M2 could be transported to mitochondria. To examine the effect of M2 on the mitochondria number, we quantified the amount of mitochondrial DNA present in the cells in relation to genomic DNA by quantitative PCR (Figure 2(d)) and semi-quantitative PCR (Figure S6(a)). Results revealed a 1.5- to 2-fold increase in the total amount of mitochondrial DNA in M2-transfected HEK 293T cells compared to that in control-transfected cells. Moreover, a significant increase was observed in the expression of mitochondrial protein TOMM20 (for outer membrane) and COX4I1 (for inner membrane) in M2-expressing cells (Figure 2(e)). These results suggested that M2 led to increase of mitochondria number. We hypothesized that M2 regulated the level of mitochondria number in the following manners: (i) an increase in the accumulation of damaged mitochondria during the mitochondria fission; (ii) an increase in the number of new-synthetic mitochondria per cell. Next, we further explored whether M2 could affect the mitochondrial dynamics. A dramatic decrease in the levels of mitochondrial fission-related proteins including DNM1L, MFF and FIS1, but a drastic increase in fusion-related OPA1 protein and a moderate increase in fusion-related MFN1, MFN2 proteins were detected in M2-expressing cells when compared to control- and M2H37G-expressing cells (Figure 2(f and g)), indicating that M2 facilitated mitochondria fusion and exhibited an ion channel activity-dependent manner. Importantly, PRKN/PARK2, an E3 ubiquitin ligase which ubiquitinates itself and its mitochondrial substrates, MFN1, MFN2 and VDAC [43,44], significantly declined in the M2-expressing cells; in contrast, M2H37G reversed this effect (Figure 2(f and g)). PPARGC1A, a transcriptional coactivator required for the transcription of nuclear-encoded mitochondrial genes and a master regulator of mitochondrial biogenesis in health and disease [45,46], was significantly increased in both its mRNA level (Figure S6(b)) and protein level (Figure S6(c)) in M2-transfected cells, depending on its proton channel activity. Strikingly, the mRNA levels of PPARGC1A downstream target genes, directly involved in mitochondrial biogenesis (ATP5F1B, MT-CO2, NRF1, NFE2L2/NRF2, TFAM), resulted to be generally unchanged or significantly increased (Figure S6(b)). Accordingly, these data indicated that M2 protein could anchor to mitochondria, facilitate mitochondrial fusion, increase mitochondrial number by increasing the mitochondria new-biogenesis, and exert a proton channel activity-dependent manner.
Figure 2.

Influenza virus M2 anchors to mitochondria and disrupts the mitochondrial dynamics. (a) A549 cells were co-transfected with indicated plasmids for 24 h and analyzed for the colocalization of M2 and dsRed-mito. Scale bar: 10 μm. It was the representative of 20 cells. (b) A549 cells were transfected with dsRed-mito, and then infected with the H5N1/HM virus for 24 h and analyzed for the colocalization of M2 with dsRed-mito. Scale bar: 10 μm. It was the representative of 20 cells. (c) HEK 293T cells were transfected with Flag-M2 (left) or A549 cells were infected with the H5N1/HM virus (Right), respectively. Subcellular fractions were purified for western blot analysis (Fractions: Cyto, purified cytosolic; Mito, purified mitochondria. Organelle markers: TOMM20, mitochondria; GAPDH, cytoplasm). (d) HEK 293T cells were transfected with Flag-M2 for 24 h. Mitochondrial DNA copy number was measured by quantitative PCR and normalized to nuclear DNA levels in a ratio of mtDNA MT-COI over RNA18S rDNA. Relative mitochondrial DNA copy numbers were depicted. (e) HEK 293T cells were transfected with Flag-M2 for 24 h and analyzed by western blot. (f and g) HEK 293T cells were transfected with Flag-M2 and Flag-M2H37G, respectively. Cell lysates were evaluated by western blot. Error bars, mean ± SD of 3 independent experiments (*p < 0.05; **p < 0.01; ***p < 0.001).
M2 protein potentiates the innate immune response
We have shown that M2 could anchor to mitochondria, which serve as platforms for innate immunity, therefore we further examined whether M2 is involved in regulating the innate immunity. RT-PCR analysis of cellular mRNAs in response to SEV stimulation showed that M2 facilitated the production of IFNB, CXCL8 and CCL5 (Figure 3(a)). M2 also dramatically enhanced SEV-induced activation of IFNB (Figure 3(b)), NFKB/NF-κB (Figure 3(c)) and ISRE (IFN-stimulated response elements) (Figure 3(d)) promoters. IRF3 and NFKB are the essential transcription factors for type I IFN production and cytokine synthesis, and their phosphorylation and nuclear translocation are the hallmarks of IRF3 and NFKB activation [47,48]. We next investigated whether M2 is able to activate these signals. Results showed that SEV infection markedly potentiated IRF3 (Figure 3(e)) and RELA/p65 (Figure 3(f)) phosphorylation in the M2-expressing cells in contrast to the control cells. Overexpression of M2 also significantly increased the Pro-IL1B expression upon SEV infection (Figure 3(f)), which was in agreement with the results of a previous study [7]. These results suggested that M2 potentiated the SEV-induced innate immune response.
Figure 3.
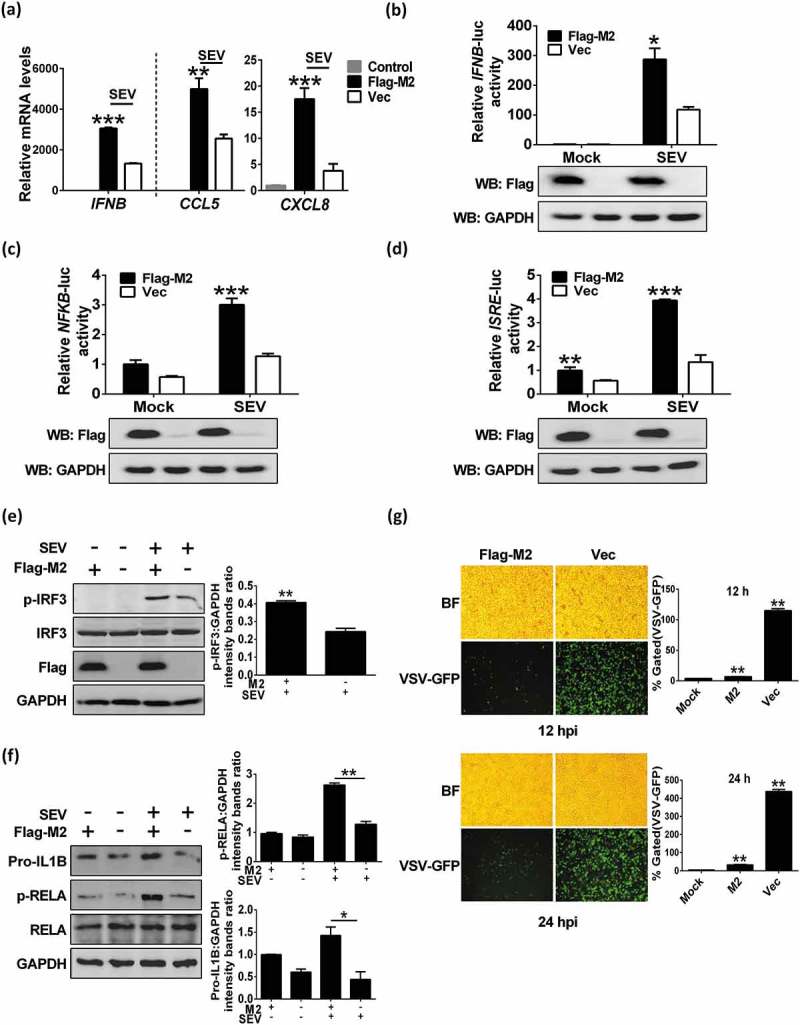
M2 protein potentiates innate immune response. (a) HEK 293T cells were transfected with Flag-M2 for 24 h and infected with SEV. The levels of IFNB, CXCL8 and CCL5 mRNAs were measured by RT-PCR at 12 hpi. (b-d) HEK 293T cells were transfected with indicated plasmids for 24 h, and then infected with SEV. Luciferase assays were performed at 12 hpi. Expression of M2 protein was confirmed by western blot. (e and f) HEK 293T cells (e) or A549 cells (f) were transfected with Flag-M2, then cells were further infected with SEV at 24 h post transfection. After 12 hpi, cells were harvested and analyzed by western blot. (g) HEK 293T cells were transfected with Flag-M2 for 24 h, and then were infected with VSV-eGFP (MOI = 0.1) for 12 h or 24 h. Replication of VSV was analyzed by microscopy and flow cytometry (BF, bright field). The presented image is a representative result from 3 independent experiments. Error bars, mean ± SD of 3 independent experiments (*p < 0.05; **p < 0.01; ***p < 0.001).
Because M2 enhanced virus-triggered induction of IFNB, we next determined whether M2 plays a role in the cellular antiviral response. Results showed that M2 expression exhibited strong inhibition of a recombinant vesicular stomatitis virus (VSV-eGFP) replication at both 12 hpi and 24 hpi (Figure 3(g)) as well as the recombinant Newcastle disease virus (NDV-eGFP) replication. As shown in Figure S7(a), overexpression of M2 protein extremely inhibited NDV replication, as evidenced by the results of fluorescence microscopy (up) and diminished GFP expression (down). Collectively, these data indicated that M2 was involved in cellular antiviral responses.
We have demonstrated that M2 facilitated mitochondria fusion whereas the M2H37G hindered it, we hypothesized that the M2 ion channel activity is crucial for M2-enhanced activation of innate immunity. As shown in Figure S7(b-d), cells overexpressing M2 and treated with amantadine or overexpressing M2H37G markedly attenuated SEV-induced activation of IFNB, NFKB, ISRE promoter activities when compared to those overexpressing M2 without treatment with amantadine. Simultaneously, further results indicated that the elevation of transcription levels of IFNB, CXCL8 and CCL5 potentiated by M2 under SEV infection were significantly weakened by treating with amantadine or overexpressing M2H37G (Figure S7(e)). Furthermore, overexpressing M2 and treatment with amantadine or overexpressing M2H37G compromised the inhibition ability of M2 to VSV replication (Figure S7(f)). Interestingly, the M2H37G protein was not detected in the mitochondrial fraction via western blot analysis (Figure S7(g)), suggesting that mitochondrial-localization was important for M2 in antiviral innate immunity. Collectively, these data suggested that the proton channel activity was crucial for M2-enhanced activation of innate immunity.
MAVS is the potential target of M2 protein
SEV is a strong inducer of the RLRs-mediated IFN signaling pathway, and MAVS is an adaptor protein of DDX58/RIG-I [49]. Next, we examined the role of M2 in MAVS-mediated antiviral signaling. Overexpression of M2 dramatically potentiated DDX58- or MAVS-induced transcription of endogenous IFNB, CXCL8 and CCL5 genes (Figure 4(a)). Consistently, DDX58- and MAVS-activated IFNB and NFKB promoter activities were potently enhanced by M2 (Figure 4(b and c)). Importantly, M2 expression dramatically increased SEV-activated MAVS level (Figure S7(h)). To explore whether MAVS is required for M2-enhanced innate signaling, we tested the effect of MAVS knockdown on M2-enhanced innate immune signaling with DDX58 stimulation, and showed that the knockdown of MAVS significantly lowered DDX58-stimulated IFNB promoter activity when compared with the control (Figure S7(i)). These data together suggested that M2 positively regulated the MAVS-mediated innate immune response. MAVS contains a C-terminal TM domain that is essential for antiviral signaling pathway, localization of MAVS to mitochondrial outer membrane and MAVS self-association, and an N-terminal CARD that is required for binding to downstream effector [19,50]. Because M2 could anchor to mitochondria, we hypothesized that M2 directly interacts with MAVS. As expected, results of coimmunoprecipitation assays demonstrated that M2 did physically interact with HA-MAVS (Figure 4(d)) and Flag-MAVS (Figure 4(e)). Very importantly, M2 also coimmunoprecipitated with endogenous MAVS in H5N1/HM-infected A549 cells (Figure 4(f)). Coincidently, results of immunofluorescence analysis revealed that very large MAVS aggregates were formed, a significant portion of which was colocalized with M2 at the mitochondria when cells coexpressed M2 and MAVS; in contrast, MAVS was uniformly distributed in the cytoplasm without M2 expression (Figure 4(g)). Taken together, our findings indicated that M2 physically interacted with MAVS, and their interaction occurred at the mitochondria.
Figure 4.
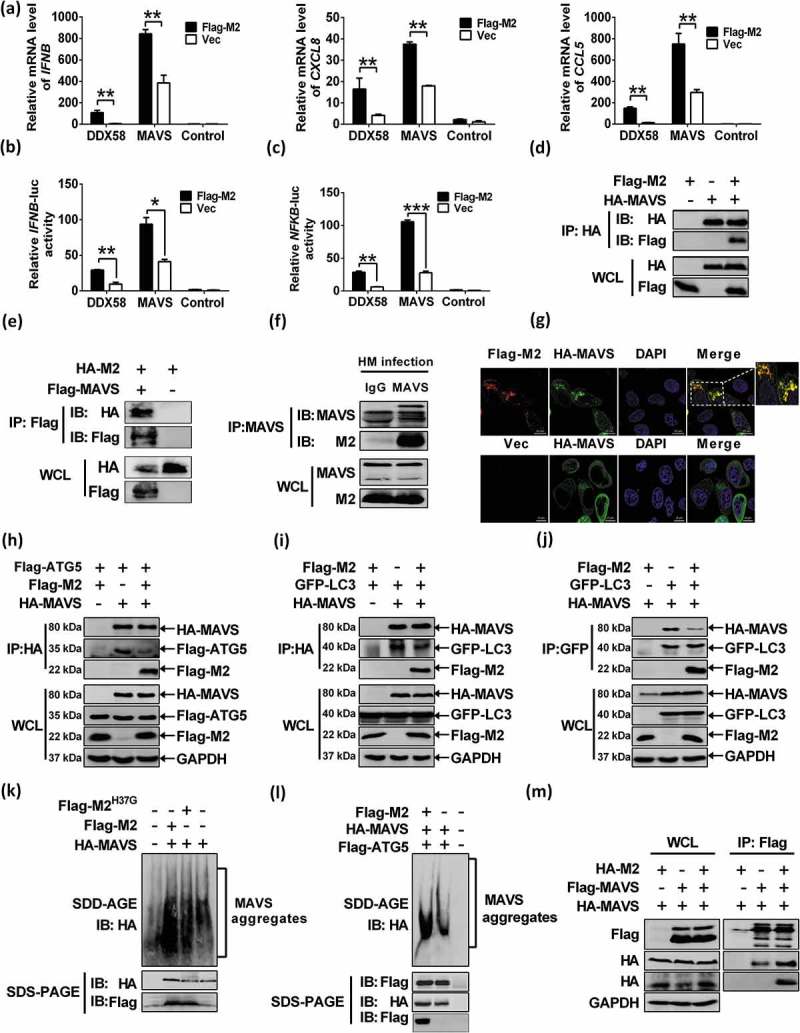
MAVS is the potential target of M2 protein. (a) HEK 293T cells were transfected with indicated plasmids. RNA was prepared for IFNB, CXCL8 and CCL5 detection by RT-PCR. (b and c) HEK 293T cells were transfected with indicated plasmids. Luciferase assays were performed at 24 h post transfection. (d) HEK 293T cells were transfected with plasmids encoding Flag-M2 and HA-MAVS. Cell lysates were subjected to IP. (e) HEK 293T cells were transfected with HA-M2 and Flag-MAVS and cell lysates were subjected to the IP. (f) Lysates of H5N1/HM-infected A549 cells were prepared, and immunoprecipitated with the anti-MAVS antibody or control IgG. (g) HeLa cells were transfected with Flag-M2 and HA-MAVS for 24 h, and then analyzed for the colocalization between M2 and MAVS. (h) HEK 293T cells were transfected with Flag-ATG5 and HA-MAVS or control vector together with Flag-M2. Cell lysates were subjected to IP. (i and j) HEK 293T cells were transfected with indicated plasmids. Cell lysates were immunoprecipitated with either anti-HA (i) or anti-GFP (j) antibody. (k and l) HEK 293T cells were transfected with indicated constructs for 24 h, cells lysates were analyzed by SDD-AGE and SDS-PAGE assays. (m) HEK 293T cells were transfected with indicated plasmids. Cell lysates were subjected to the IP. Error bars, mean ± SD of 3 independent experiments (*p < 0.05; **p < 0.01; ***p < 0.001).
Viral infection efficiently induces MAVS conformational switch that leads to formation of very large MAVS aggregates, which potently activate IRF3 and propagate the antiviral signaling [51]. ATG5 can interact with MAVS and function as a negative regulator in MAVS signaling by controlling MAVS aggregation [19,20]. We have shown that M2 interacted with MAVS, we next investigated whether M2 is an alternative substrate of ATG5, which will impair the binding of ATG5 to MAVS. Results of the coimmunoprecipitation assays revealed that M2 expression markedly decreased the binding of ATG5 to MAVS (Figure 4(h)). Furthermore, results of luciferase assays indicated that the suppressed effect on IFNB promoter activity by ATG5 was reversed when the cells overexpressed with M2 under MAVS stimulation (Figure S7(j)). These results supported that M2 sequestered MAVS-ATG5 interaction. Recent studies have shown that MAVS induces formation of LC3B puncta and interacts with LC3B to suppress MAVS-mediated innate immunity [21,22]. Therefore, we determined whether M2 is able to affect the interaction of MAVS with LC3B, and showed that M2 transfection decreased either the binding of LC3B to MAVS (Figure 4(i)), or MAVS to LC3B (Figure 4(j)). These results suggested that M2 negatively regulated the MAVS-ATG5 and MAVS-LC3B complexes formation, thereby enhancing the MAVS-mediated innate immunity.
To further determine whether M2 modulated MAVS signaling truly by controlling MAVS aggregation, we employed the immunostaining assay and semi-denaturing detergent agarose gel electrophoresis (SDD-AGE) analysis as described previously [51]. Results showed that the MAVS aggregates in cells transfected with control vector, M2 or M2H37G and MAVS were detected. As shown in SDD-AGE assays (Figure 4(k)), enhancement of MAVS aggregations was observed in cells coexpressing MAVS and M2 in contrast to those transfected with either the control vector or the M2H37G. Consistent with the above results, M2 apparently reversed the reduction of MAVS aggregations that were negatively regulated by ATG5 (Figure 4(l)). Our further studies showed that M2 was able to increase the MAVS self-association (Figure 4(m)), which has been shown to mediate antiviral innate immune signaling [50]. Taken together, these data suggested that MAVS was the potential target of M2, and M2 strengthened the MAVS-mediated antiviral signaling by increasing MAVS aggregates formation.
The his37 of M2 and the integrity of MAVS are required for the interaction of M2 with MAVS
Both IAV M2 and MAVS interact with LC3B depending on their respective conserved LIR motif in the cytoplasmic tail and the CARD domain [21,22,52]. We next sought to determine whether M2 bound MAVS also via its LIR domain. Unexpectedly, results showed that all mutants in the M2 LIR motif (M2F91S, M2I94S and M2ΔLIR) did not impair the interactions of MAVS with M2 (Figure 5(a)), indicating that the M2 LIR motif was not critical for interaction with MAVS. As overexpression of MAVS could activate autophagy [19], we hypothesized that the ability of M2 binding MAVS is possibly related to M2-induced autophagy level. Since M2H37G failed to induce autophagy, we further investigated if M2H37G can bind MAVS. Results showed that M2H37G failed to interact with MAVS (Figure 5(b and c)). Simultaneously, M2H37G did not affect the interaction between MAVS with either ATG5 or LC3B (Figure 5(d–f)). Collectively, these data indicated that the His37 of M2 was essential for interaction with MAVS and sequestering interaction of MAVS with either ATG5 or LC3B.
Figure 5.

M2 His37 and the integrity of MAVS are required for the interaction between M2 and MAVS. (a) HEK 293T cells were transfected with Flag-MAVS alone or cotransfected with HA-M2-mutants (F91S, I94S and ΔLIR). Cell lysates were subjected to IP and further analyzed by western blot. (b) HEK 293T cells were transfected with Flag-M2 or Flag-M2H37G alone or along with HA-MAVS, respectively. Cell lysates were immunoprecipitated with anti-Flag and analyzed by western blot. (c) HEK 293T cells were transfected with HA-MAVS or both HA-MAVS and Flag-M2H37G. Cell lysates were immunoprecipitated with anti-HA antibody and then analyzed by western blot. (d-f) HEK 293T cells were transfected with indicated plasmids for 24 h, cell lysates were subjected to IP with respective antibodies. (g-i) HEK 293T cells were cotransfected with each HA-MAVS-mutant (Y9A, I12A, ΔLIR, 1N, 1C, ΔTM and ΔCARD) with Flag-M2. Cell lysates were immunoprecipitated with anti-Flag antibody (g) or anti-HA antibody (h and i), and then analyzed by western blot.
We next explored whether MAVS bound to M2 in a LIR-dependent manner. Results showed that the interactions were detected between M2 with each MAVS LIR mutant (MAVSY9A, MAVSI12A or MAVSΔLIR) (Figure 5(g and h)) that was able to activate MAVS signaling (data not shown). These results indicated that the LIR motif was not essential for MAVS binding to M2. However, other MAVS mutants, which include mutants that only contains N-terminal domain (MAVS-1N) or C-terminal domain (MAVS-1C), or lacking either the N-terminal CARD-like domain (MAVS-ΔCARD) or the C-terminal TM domain (MAVS-ΔTM) constructed as described previously [53,54], and were unable to active MAVS signaling and induce autophagy (data not shown), failed to bind to M2 (Figure 5(i)). These results indicated that the integrity of MAVS was required for MAVS binding to M2 and that M2 modulated antiviral signaling through its interaction with MAVS.
M2 enhances MAVS signaling via increasing ROS production
Ca2+ [55] and ROS [56–58] are closely associated with the innate immune system. Next we determined whether M2-enhanced innate antiviral response is modulated by Ca2+ and ROS. Results showed that the promoter activities of IFNB (Figure 6(a, c) and S8(a-d)) and NFKB (Figure 6(b and d)), enhanced by M2 overexpression under SEV infection, were effectively inhibited by EGTA, BAPTA-AM, DPI and Apo in a dose-dependent manner. Furthermore, results of RT-PCR assays also revealed that treatment with EGTA, BAPTA-AM, DPI or Apo resulted in obvious decrease of the mRNA of IFNB (Figure S8(e–h)). Altogether, these results suggested that M2-enhanced innate immunity required the production of Ca2+ from extracellular and of ROS by the NADPH oxidase.
Figure 6.

M2 protein enhances MAVS signaling through increasing ROS production. (a-d) HEK 293T cells were transfected with indicated plasmids. Cells were then pretreated with EGTA (a and b), BAPTA-AM (a and b), DPI (c and d), Apo (c and d) prior to mock-infection or infection with SEV, respectively. Luciferase assays were performed at 12 hpi. (e) HEK 293T cells were transfected with indicated plasmids for 24 h, FACS analysis were performed to determine cellular ROS production. (f) HEK 293T cells were pretreated with Apo for 4 h, and transfected with indicated plasmids. Cells were lysed to measure the IFNB induction at 24 h post transfection. (g) HEK 293T cells were pretreated with Apo for 4 h, and transfected with indicated plasmids for 24 h, cells lysates were analyzed by either SDD-AGE or SDS-PAGE assays. Error bars, mean ± SD of 3 independent experiments (*p < 0.05; **p < 0.01; ***p < 0.001).
Given that M2 enhanced MAVS signaling activity and accordingly increased cellular ROS levels (Figure 6(e)), we tried to determine whether the increase of MAVS signaling by M2 is attributed to the increase of ROS levels. As expected, Apo treatment was sufficient to suppress the apparent increase in the levels of MAVS signaling activity by overexpressing M2 in MAVS-stimulating cells (Figure 6(f)). Intriguingly, Apo significantly inhibited the M2-enhanced MAVS aggregations (Figure 6(g)). These results indicated that M2 positively regulated MAVS signaling through increasing ROS production as M2 was essential for ROS production and physically interacted with MAVS.
M2 antagonizes the autophagy process to enhance the innate immune response
Autophagy plays a vital role in innate and adaptive immune responses [59,60]. MAVS acts as a potential autophagy receptor to mediate mitochondrial turnover upon excessive activation of RLR signaling [22]. Our results have demonstrated that M2 was able to not only induce autophagy but also enhance the MAVS signaling activity, we next investigated whether there is a link between M2-induced autophagy process and M2-enhanced MAVS-mediated innate immunity. First, the effect of M2-enhanced MAVS activation was detected when the autophagy level was disrupted by knocking down either ATG5 or BECN1. Results showed that ATG5 and BECN1 were efficiently knocked down (Figure 7(a and b)). Furthermore, knocking down either ATG5 or BECN1 downregulated the LC3B-II level but upregulated exogenous MAVS expression and the MAVS aggregations when the cells were transfected with M2 under MAVS stimulation (Figure 7(a and b)), suggesting that M2 accelerated the MAVS activation when autophagy initiation was depressed. Additionally, M2-enhanced IFNB promoter activity further potentiated either in the ATG5 or BECN1-silencing cells, in contrast to the NC-silencing cells with either MAVS (Figure 7(c and e)) or SEV stimulation (Figure 7(d)). Taken together, these data indicated that M2 antagonized the autophagy process to enhance the innate immune response, but the host also negatively regulated the M2-enhanced innate immune responses by inducing the autophagy process.
Figure 7.

M2 antagonizes the autophagy process to enhance the innate immune response. (a) After transfection with siATG5 for 6 h, the HEK 293T cells were further transfected with indicated plasmids for 24 h. The cells lysates were collected and analyzed by either SDD-AGE or SDS-PAGE assays. (b) HEK 293T cells were transfected with siBECN1 for 6 h, cells were further transfected with indicated plasmids for 24 h, and cells lysates were analyzed by either SDD-AGE or SDS-PAGE assays. (c) HEK 293T cells were transfected with siATG5 for 6 h, the cells were further transfected with indicated plasmids for 24 h, and then luciferase assays were performed. (d) HEK 293T cells were transfected with siATG5 for 6 h, the cells were further transfected with indicated plasmids for 24 h, and then mock-infected or infected with SEV for 12 h. The luciferase assays were performed. (e) HEK 293T cells were transfected with siBECN1 for 6 h, cells were further transfected with indicated plasmids. At 24 h post transfection, luciferase assays were performed. Error bars, mean ± SD of 3 independent experiments (*p < 0.05; **p < 0.01; ***p < 0.001).
Discussion
Mitochondria function as platforms for innate antiviral signal transduction in mammals through MAVS [19] and activation of MAVS can change morphology of mitochondrion [19,51]. The mitochondrial dynamics, the physical properties of mitochondria and the resulting changes in MAVS distribution and aggregates play important roles in modulating the innate immune response. Mitochondrial elongation and fusion facilitate the MAVS aggregates into active complexes, specifically for enhancing the MAVS-mediated signaling capacity [50,51,61–63], whereas mitochondrial fragmentation and fission attenuate the RLR signaling [64]. Several viroporins have been reported to be partially colocalized with mitochondria and alter the normal morphology of mitochondrial network [35–37,39,40]. In this study, we demonstrated that IAV M2 protein was able to anchor to mitochondria in both the virus-infected and M2-overexpressing cells. Based on our knowledge, it was the first time. Furthermore, we showed that M2 promoted mitochondria fusion via downregulating expression of PRKN, thereby increasing the expression of mitochondrial fusion proteins, exerting an ion channel activity-dependent effect. Therefore, M2 accelerated the MAVS self-association to facilitate the MAVS aggregates formation. In addition, M2 increased the mitochondria number by regulating mitochondrial new-biosynthesis that was dependent on its proton channel activity, thereby positively modulating the increase of MAVS aggregates, which further promoted the MAVS-mediated innate antiviral immunity. Some signals such as death signals or calcium signaling has been reported to affect mitochondrial dynamics [62]. It is possibly that M2-triggered increase in the level of Ca2+ promotes the mitochondrial fusion. However, the underlying mechanism remains obscure, and needs to be further investigated. Previous studies have reported that ER-mitochondria contacts are increased at the mitochondria-associated membrane (MAM) upon infection-induced mitochondrial elongation and fusion, and that the MAM functions as a crucial subcellular domain in generation of type I IFNs and cytokines [62,65]. We have shown that part of M2 protein could localize to the ER (Figure S3(a)), it is likely that the mitochondrial-targeting M2 protein translocates from the ER to perform its function to enhance the innate immune response. However, whether the mitochondrial-targeting M2 protein is a dynamic process and the relation between the mitochondria- and ER-targeting M2 proteins in regulating the MAVS signaling, are still unclear and need to be further explored. Moreover, although we demonstrated that M2 was able to localize to the mitochondria, the sub-mitochondrial localization of the M2 protein and the detailed mechanisms remain not completely clear and need to be further investigated. Mitochondrial membrane potential (MMP) is required for MAVS-mediated antiviral signaling [66], we also analyzed the MMP affected by M2 via MMP-sensitive dye (tetramethylrhodamine methylester [TMRM]) staining, and showed that CCCP (carbonyl cyanide m-chlorophenylhydrazone), a protonophore that dissipates MMP, dramatically collapsed the MMP, whereas M2 slightly increased the MMP, depending on its ion channel activity (Figure S2(c)), which was in line with M2-mediated mitochondrial fusion and enhancement of MAVS signaling.
Previous reports have alleged that ROS [67] and Ca2+ [68] act as signaling molecules in intracellular autophagy process. Our studies further demonstrated that M2-mediated increase of [Ca2+]i from extracellular ion entering into the cytoplasm resulted in ROS production that directly or indirectly activated ATG5 and inhibited AKT and MTOR activity through PI3K-AKT-MTOR signaling pathway, thereby triggering autophagy initiation, indicating that M2 was able to trigger autophagy initiation as well as inhibit autophagosome maturation. Recent studies have shown that IAV PR8 strain (H1N1) infection [69] as well as CQ [26] can activate a non-canonical autophagy response, different results obtained by our studies were possibly attributed to a different virus strain used, as a previous study has reported that the H5N1 virus but not the H1N1 virus infection induces an AKT-TSC2-MTOR signaling pathway-dependent autophagic process [70]. Activation of MAVS has been shown to be involved in the positive modulation of ROS production and autophagy [19] and autophagy controls DDX58-MAVS signaling by repressing ROS production [18,57]. In the present study, our results indicated that M2 served as another factor besides PB1-F2 [71] leading to excessive Ca2+ flux-dependent ROS generation for the pathogenicity of IAV, and that the proton channel activity played an indispensable role in this effect. Moreover, M2-triggered increase of ROS production regulated the MAVS aggregations, thus upregulated MAVS-mediated innate immune response. Autophagy suppresses MAVS-mediated antiviral signaling through also regulating the MAVS aggregates and stability via the direct binding of ATG5 [19,20] and LC3B [21,22] to MAVS. Noticeably, our results demonstrated that the His37 of M2 was required for M2 to anchor to mitochondria, colocalize and physically interact with MAVS, and sequester the ATG5-MAVS and LC3B-MAVS interactions. Therefore, it acted as an opposite regulator as did ATG5 and LC3B by redistributing MAVS to aggregates observed upon activation of RLR signaling, thereby functioning as a positive regulator in MAVS-mediated antiviral innate immunity. So, M2-mediated increase of ATG5 and LC3B protein were re-involved in the autophagosome formation, in consistent with results that ATG5 played a critical role in the initial process of M2-induced autophagy. Both M2-induced autophagy and enhanced innate antiviral response were depressed by the Ca2+ and ROS inhibitors, which was most likely due to a lower inhibitory effect of autophagy on the M2-enhcanced innate antiviral response than the enhanced effect of ROS on M2-enhanced innate immune response. The underlying mechanisms need to be further determined in future studies. Both M2 and MAVS were able to localize to the mitochondria, but M2H37G failed to localize to the mitochondria (Figure S7(g)), we speculated that the ability of M2 binding MAVS was possibly related to its mitochondrial-targeting. The detailed mechanisms on how M2 translocated to the mitochondria while M2H37G failed to, need to be further investigated. It has been well established that the late stage of autophagy, the process of autolysosome formation, is involved in the removal of the aggregated proteins such as MAVS aggregates by the lysosome enzyme [14]. However, our current and other previous studies [8] showed that the proton channel activity of M2 was able to block the autolysosome formation, thereby reducing the elimination of excessive ROS and the MAVS aggregates, which in turn led to the amplification of MAVS signaling in the regulation of innate immune response. These results suggested that the M2 proton channel activity was the prerequisite for M2-enhanced innate immunity. Based on this, we tried to assure that the M2 proton channel activity is responsible for M2-enhanced innate immunity in infection, but we failed to rescue the recombinant IAV containing M2H37G mutation. Gannagé et al [25] recued the M2-deficient recombinant influenza A/PR8/34 virus (a single round of replications in cell culture), but it might be due to a different virus strain used. Noticeably, we also showed that M2 from several other subtypes of IAV (PR8 H1N1, avian H9N2, swine H1N1 and avian H7N9) could also enhance the MAVS signaling (data not shown). However, overactivation of MAVS signaling could lead to detrimental levels of inflammation or other immunopathological consequences that are harmful to the host. IAV infection induces rapidly host innate immunity, whereas a dysregulation of and an excessively exaggerated immune response ‘cytokine storm’ can cause severe pathogenicity that often directly contributes to morbidity and mortality of IAV infection. However, the link between the direct effect of M2 on the immune signaling and consequent damage to the cells is still unclear and needs to be investigated in future studies.
Therefore, our study unveiled a novel mechanism used by M2 to antagonize the autophagy pathway in order to feedforward modulate MAVS-mediated antiviral signaling and provided novel insight into biology and pathogenicity of IAV. Moreover, improved understanding of M2 protein-enhanced MAVS signaling may facilitate development of novel antiviral therapeutics for the prevention and control of influenza virus infection. The proposed underlying mechanism used by influenza A M2 protein to regulate host innate immune response was shown in Figure 8.
Figure 8.

Schematic diagram of proposed mechanisms underlying the regulation of innate immune response by influenza virus H5N1/HM M2 protein. Proton channel activity of M2 protein was essential for the increase of Ca2+ entry into cytoplasm from extracellular with a subsequent elevation in ROS production. On one hand, the increase level of ROS directly or indirectly led to the activation of ATG5 and inhibition of AKT and MTOR activity through the PI3K-AKT-MTOR pathway, thereby triggering M2-induced autophagy process. One the other hand, M2 protein anchored to the mitochondria, increased the mitochondrial number and accelerated the mitochondria fusion, which resulted in increasing ROS-dependent MAVS aggregations. The MAVS aggregates can be packed into the autophagosome and be degraded by the lysosomal enzyme in the process of autolysosome formation. LC3B and ATG5 interact with MAVS in this process, thereby attenuating MAVS-mediated antiviral signaling pathway. However, M2 physically interacted with MAVS and sequestered the MAVS-ATG5 and MAVS-LC3B complexes formation, thus leading to the release of MAVS aggregates from the ATG5-MAVS and LC3B-MAVS complexes. Furthermore, M2 protein inhibited the autolysosome formation, which suppressed the MAVS aggregates degradation. The released MAVS aggregates from ATG5-MAVS and LC3B-MAVS complexes in turn participated in the MAVS-mediated innate immune response, which resulted in subsequent amplification of IFN and inflammatory cytokine signaling by RLR signaling.
Materials and methods
Cells and viruses
Human lung epithelial cell line A549, human embryonic kidney 293T and HeLa cells were purchased from ATCC (CCL-185™, CRL-3216™ and CCL-2™). A549 cells were maintained in HAM’S/F-12 (HyClone, SH30026.01) medium; HEK 293T and HeLa cells were maintained in RPMI 1640 (HyClone, SH30809.01) medium. They were supplemented with 10% heat-inactivated fetal bovine serum (FBS; PAN biotech, P30-3302), and incubated in a 37°C humidified incubator with 5% CO2. A/duck/Hubei/Hangmei01/2006(H5N1) (H5N1/HM) was isolated from duck brain tissues and stored in our laboratory [72]. A549 cells were infected with H5N1/HM at multiplicity of infection (MOI) of 0.1. After 1 h adsorption, cells were washed once with warm phosphate-buffered saline (PBS (PH 7.2); HyClone, SH30256.01) and then incubated in HAM’S/F-12 containing 1% FBS at 37°C. After 24 h post infection (hpi), cells were subjected to either BD FACSCaliburTM Flow Cytometer (BD Biosciences, San Jose, CA 95131 USA) or immunofluorescence assays. All experiments with the H5N1/HM were performed in the Biosafety Level 3 laboratory at Huazhong Agricultural University. The recombinant vesicular stomatitis virus encoding enhanced green fluorescence protein (VSV-eGFP) was a gift from Harbin Veterinary Research Institute and eGFP expression levels directly reflected the degree of VSV replication. eGFP expression was visualized by fluorescence microscopy and analyzed by FACS. NDV was conserved by the State Key Laboratory of Agricultural Microbiology of China, and Sendai virus (SEV) was kindly provided by Professor Zhengfan Jiang (Peking University, China).
Chemicals and reagents
Amantadine (Sigma-Aldrich, A1260), apocynin (Apo; Sigma-Aldrich, A10809), 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid tetrakis (BAPTA-AM; Sigma-Aldrich, A1076), carbonyl cyanide 3-chlorophenylhydrazone (CCCP; Sigma-Aldrich, C2759), chloroquine (CQ; Sigma-Aldrich, C6628), dimethyl sulphoxide (DMSO; Sigma-Aldrich, D4540), diphenyleneiodonium chloride (DPI; Sigma-Aldrich, D2926), ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid (EGTA; Sigma-Aldrich, E0396), LY294002 (Sigma-Aldrich, L9908), rapamycin (Sigma-Aldrich, 553,210), oxidant-sensitive dye 5-(and-6)-chloromethyl-2ʹ,7ʹ-dichlorodihyd-rofluorescein diacetate (CM-H2DCF-DA; Invitrogen, C6827) and MitoSOX Red mitochondrial superoxide indicators (Invitrogen, M36008) incubated cells at the corresponding concentrations.
Antibodies
The antibodies used and their sources are as follows: rabbit anti-AKT (Cell Signaling Technology, 4691S), rabbit anti-p-AKT-S473 (Cell Signaling Technology, 4060S), rabbit anti-COX4I1 (Cell Signaling Technology, 4844S), rabbit anti-IRF3 (Cell Signaling Technology, 11904T), rabbit anti-NFKB RELA/p65 (Cell Signaling Technology, 8242S), rabbit anti-p-NFKB RELA/p65-S536 (Cell Signaling Technology, 3033S), rabbit anti-BECN1 (ABclonal Biotechnology, A7353), rabbit anti-MFN1 (ABclonal Biotechnology, A9880), rabbit anti-MFN2 (ABclonal Biotechnology, A5750), rabbit anti-DNM1 (ABclonal Biotechnology, A2586), rabbit anti-MFF (ABclonal Biotechnology, A4874), rabbit anti-FIS1 (ABclonal Biotechnology, A5821), rabbit anti-PPARGC1A (ABclonal Biotechnology, A12348), rabbit anti-PRKN (ABclonal Biotechnology, A0968), rabbit anti-RPS6KB1 (ABclonal Biotechnology, A2190), rabbit anti-p-RPS6KB1-T389 (ABclonal Biotechnology, AP0564), rabbit anti-FOXO1 (ABclonal Biotechnology, A13862), rabbit anti-p-FOXO1-S256 (ABclonal Biotechnology, AP0172), rabbit anti-MTOR (ABclonal Biotechnology, A11345), rabbit anti-p-MTOR-S2448 (ABclonal Biotechnology, AP0094), rabbit anti-p-IRF3-S386 (ABclonal Biotechnology, AP0091), rabbit anti-IL1B (ABclonal Biotechnology, A1112), rabbit anti-control IgG (ABclonal Biotechnology, AC005), mouse anti-HSP90B1 (Proteintech, 60012–1-1 g), rabbit anti-MAVS (Proteintech, 14341–1-AP), rabbit anti-TOMM20 (Proteintech, 11802–1-AP), rabbit anti-IAV M2 and NP (Gene Tex, GTX125951 and GTX30852), rabbit anti-Golgi Marker-AE-6 (Santa Cruz Biotechnology, sc58770), rabbit anti-MAP1LC3B/LC3B (Novus Biologicals, NB100-2220), mouse anti-OPA1 (BD biosciences, 612607), mouse anti-ATG5 (Arigo biolaboratories, ARG54822), mouse anti-GAPDH (California Bioscience, CB100127), mouse anti-GFP-tag (CWBIO, CW0258A), mouse anti-HA-tag (MBL, M180-3), and mouse anti-Flag M2 (Sigma-Aldrich, F1804) antibodies were incubated overnight at 4°C. Horseradish peroxidase (HRP)-conjugated anti-mouse (Beijing Biodragon Inmmunotechnologies, BF03001) and anti-rabbit (Beijing Biodragon Inmmunotechnologies, BF03008) secondary antibodies were incubated 1 h at room temperature. FITC-goat anti-mouse (Sungene Biotech, GM200G-02C), FITC-goat anti-rabbit (Sungene Biotech, GR200G-02C), 594-conjugated goat anti-mouse (Sungene Biotech, GM200G-43C), 594-conjugated goat anti-rabbit (Sungene Biotech, GR200G-43C) or 405-goat anti-mouse (Sungene Biotech, GM200G-46C) secondary antibodies were incubated 1 h at room temperature.
Plasmids and siRNA oligonucleotides
Full-length cDNA encoding H5N1/HM M2 and human LC3B proteins were amplified by reverse transcription (RT)-PCR from total RNA extracted from A549 cells infected with H5N1/HM using specific primers (available upon request) and cloned into p3xFlag-CMV (Flag-M2), HA-pCAGGS (HA-M2) and pEGFP-N1 (GFP-LC3). The 3 empty vectors [p3xFlag-CMV (Sigma, E7908), HA-pCAGGS (YouBia, VT1076) and pEGFP-N1 (YouBia, VT1110)] were stored in our laboratory. DDX58, MAVS expression plasmids and 3 luciferase (Luc) reporter plasmids [IFNB promoter (IFNB-luc), NFKB promoter (NFKB-luc) and ISRE promoter (ISRE-luc)], were kindly provided by Professor Zhengfan Jiang (Key Laboratory of Cell Proliferation and Differentiation of the Ministry of Education, School of Life Sciences, Peking University, Beijing, China). A Renilla control plasmid (pGL4.75 hRluc/CMV; Promega, E6931) was used as a control. The construct encoding a mitochondrial-targeting signal fused to RFP gene (dsRed-mito) was stored in our laboratory. The tandem fluorescent monomeric red fluorescent protein (mRFP)-GFP-LC3 (ptfLC3) construct [73] was kindly provided by Dr. Chan Ding (Shanghai Veterinary Research Institute, China). The M2 mutants (H37G, F91S, I94S and ΔLIR) and MAVS mutants (Y9A, I12A, ΔLIR, 1N, 1C, ΔTM and ΔCARD) were generated by using A QuickChange XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) and overlap cloning techniques. RNAi oligonucleotides 5′-GATTCATGGAATTGAGCCA-3′ were used for the ATG5, and 5′-CAGUUUGGCACAAUCAAUA-3′ were used for the BECN1 siRNA experiments.
Real-time quantitative PCR (RT-qPCR)
To determine the effect of M2 protein on the expressions of IFNB, CXCL8 and CCL5, HEK 293T cells seeded in 24-well plates were transfected with the empty vector, or H5N1/HM WT M2 (Flag-M2), or mutant- M2H37G (Flag-M2H37G) plasmid. After 24 h post transfection, cells were mock-infected or infected with SEV for 12 h. To determine the effect of M2 protein on the RLR signaling pathway, plasmids encoding DDX58 or MAVS were co-transfected with Flag-M2 or Flag-M2H37G for 24 h. Total RNA was extracted using TRIzol (Invitrogen, 15596018) according to the manufacturer’s instructions and 1 μg of RNA was reversely transcribed. Gene expression was determined by SYBR Green-based RT-PCR using an ABI ViiA 7 PCR system (Apllied Biosystems, Foster City, CA, USA). Each gene expression was normalized to GAPDH. The primers used in this study were available upon request.
To analyze the expression levels of mitochondrial DNA, total cellular DNA was extracted from the HEK 293T cells transfected with the indicated plasmids using Easy Pure Genomic DNA kit (Transgen Biotech, EE101-01) and subsequently RT-PCR was conducted. Human mtDNA MT-COI primers and the RNA18S rDNA primers used were adapted based on a previous publication [74].
Western blot
For the SDS-PAGE assay, protein samples were prepared by using cell lysis buffer for Western Blotting and IP (Beyotime, P0013) or mammalian cell lysis buffer (CWBIO, CW0889) containing EDTA-free protease inhibitor cocktail (Roche, 04693132001). After SDS-PAGE separation, samples were transferred to nitrocellulose membrane (GE Healthcare Life sciences, 10600001 and 10600002), which was blocked in Tris-buffered saline with Tween (TBST; Sigma, SRE0031) containing 2% bovine serum albumin (BSA; Sigma, A1933), then reacted with the corresponding primary antibodies, followed by HRP-conjugated secondary antibodies, and signals were detected by using a WesternBrightTM ECL detection kit (advansta, K-12045-D50) in an ECL detection system (Amersham Biosciences, Piscataway, NJ, USA). All bands of western blots were detected within the linear range. The semidenaturing detergent agarose gel electrophoresis (SDD-AGE) was carried out as described previously [51]. Briefly, HEK 293T cells transfected with indicated plasmids were homogenized in a 1 × sample buffer without β-mercaptoethanol [1 × TBE (Wuhan Servicebio Technology Co., Ltd, G3002), 10% glycerin (Sigma, G2289), 2% SDS (Sigma, L3771), 0.0025% bromophenol blue (Sigma, 115–39-9) and Roche EDTA-free protease inhibitor cocktail] by repeated Dounce homogenization. Protein samples without boiling were resuspended and loaded onto a vertical 1.5% agarose gel in the running buffer (1 × TBE and 0.1% SDS) for 1 h with a constant voltage of 100 V at 4°C. Immunoblotting was carried out by standard procedures.
Luciferase reporter assay
HEK 293T cells grown in 24-well plates were cotransfected with 0.25 μg/well of reporter plasmids IFNB-luc, NFKB-luc or ISRE-luc, respectively, 0.01 μg/well of plasmid pGL4.75 (hRluc/CMV), 0.25 μg/well DDX58 or MAVS, respectively, and 0.5 μg/well of Flag-M2, Flag-M2H37G expression plasmid or an empty vector for 24 h. When necessary, DDX58 and MAVS transfection were replaced by infecting with SEV to stimulate cells at 24 h post transfection. Cells were lysed at 12 hpi, and firefly luciferase and Renilla luciferase activities were determined with the Dual-Luciferase reporter assay system (Promega, E1910) according to the manufacturer’s protocol. Data were presented as relative firefly luciferase activities normalized to Renilla luciferase activities and were representative of 3 independent experiments.
Immunofluorescence and confocal microscopy
A549 cells or HeLa cells were plated on coverslips in 12-well plates, and infected with H5N1/HM (or transfected with indicated plasmids). At 24 h post infection (or post transfection), cells were fixed with 4% paraformaldehyde for 15 min, permeabilized with 0.1% Triton X-100 (Sigma-Aldrich, X100) in PBS for 10 min, and blocked with 1% BSA for 1 h at room temperature, and then incubated with corresponding antibodies at 4°C overnight. Cells were then incubated in Alexa Fluor FITC- and 594-conjugated goat anti-mouse or anti-rabbit secondary antibodies described above for 2 h. Nuclei were stained with DAPI (Invitrogen, 00–4959-52) for 10 min. All cells were washed with PBS 5 times after each step and were imaged by confocal microscopy (Carl Zeiss LSM 880 Confocal Microscope and ZEN 2.3 LITE software) or super-resolution techniques (NIKON N-SIM).
Detection of Ca2+ accumulation
The culture medium of either the transfected or infected cells was changed to HBSS (BasalMedia, B430KJ) that did not include phenol red. 5 μM Fluo-4 AM (Invitrogen, F23917) or 10 μM Rhod-2 AM (AAT Bioquest, 21,060) in HBSS was added as an indicator of Ca2+ in the cytosol [Ca2+]i or mitochondrial [Ca2+]m, respectively. After incubation at 37°C for 30 min, cells were washed twice with HBSS. Fluorescence was analyzed via the BD FACSCaliburTM Flow Cytometer System.
ROS detection
The transfected HEK 293T cells, the mock infected and infected A549 cells with different treatment were disposed and ROS was detected as described previously [75].
Mitochondrial membrane potential (MMP) detection
The culture medium of the transfected cells were changed to RPMI 1640 and were supplemented with 60 nM TMRM (AAT Bioquest, 22221). After incubation for 30 min at 37°C, fluorescence was determined by using the BD FACSCalibur system.
Subcellular fractionation
Pure mitochondrial fractionation was isolated by utilizing the Mitochondria/Cytosol Fractionation Kit (Millipore, MT1000) according to the manufacturer’s instructions. Briefly, HEK 293T cells and A549 cells were plated onto a 15-cm dish, the HEK 293T cells were transfected with 20 μg of Flag-M2 or Flag-M2H37G; and A549 cells were infected with H5N1/HM at 0.1 MOI for 24 h, respectively. Cells were washed with ice-cold PBS twice and then detached with a cell scraper. Subsequently, the cell pellets were collected after centrifugation and resuspended with 800 μl of Isotonic Mitochondrial Buffer with protease inhibitor cocktail at 1:100 dilutions. Cells were homogenized with a 1 ml Dounce homogenizer via 40 strokes on ice and were centrifuged at 600 × g for 10 min. The supernatants were recentrifuged at 10,000 × g for 30 min. All centrifugation was performed at 4°C. The mitochondrial fraction in the pellets was resuspended with 40 μl of the Mitochondrial Lysis Buffer containing protease inhibitors. The supernatants were considered as the cytosolic fraction. Proteins from each fraction were analyzed by western blot using the indicated antibodies.
Statistical analyses
Data were expressed as means ± standard errors of the means (SEM). Statistical analysis was performed by paired two-tailed Student’s t test. P value equal or lower to 0.05 was considered significant (*p < 0.05, **p < 0.01, ***p < 0.001).
Funding Statement
This work was supported by National Key Research and Development Program (2016YFD0500205 & 2016YFC1200201), Fundamental Research Funds for the Central Universities (2662017PY029), National Natural Science Foundation of China (NO. 31761133005 & 31572545 & 31772752), the Outstanding Youth Science Foundation of Hubei Province (2016CFA056).
Acknowledgments
We thank Ms. Xiao Xiao for her critically proof reading the manuscript, professor Chan Ding from Shanghai Veterinary Research Institute providing the tandem fluorescent monomeric red fluorescent protein (mRFP)-GFP-LC3 (ptfLC3) construct and professor Zhengfan Jiang from Peking University for providing 3 luciferase reporter constructs including IFNB promoter (IFNB-luc), NFKB promoter (NFKB-luc) and ISRE promoter (ISRE-luc) as well as the Sendai virus. This study was supported by National Key Research and Development Program (2016YFD0500205 & 2016YFC1200201), Fundamental Research Funds for the Central Universities (2662017PY029), National Natural Science Foundation of China (NO. 31761133005 & 31572545 & 31772752), the Outstanding Youth Science Foundation of Hubei Province (2016CFA056).
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental material
Supplemental data for this article can be accessed here.
References
- [1].Chen C, Chi X, Bai Q, et al. Mechanisms underlying interferon-mediated host innate immunity during influenza A virus infection. Sheng Wu Gong Cheng Xue Bao. 2015;31(12):1671. [PubMed] [Google Scholar]
- [2].To J, Surya W, Torres J.. Targeting the channel activity of viroporins. Adv Protein Chem Struct. 2016;104:307–355. PubMed PMID: WOS:000381638000009; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mould JA, Li H-C, Dudlak CS, et al. Mechanism for proton conduction of the M2 ion channel of influenza A virus. J Biol Chem. 2000. March 24;275(12):8592–8599. PubMed PMID: BCI:BCI200000349462. [DOI] [PubMed] [Google Scholar]
- [4].Chen BJ, Leser GP, Jackson D, et al. The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. J Virol. 2008. October;82(20):10059–10070. PubMed PMID: WOS:000260109100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Pinto LH, Lamb RA.. Controlling influenza virus replication by inhibiting its proton channel. Mol Biosyst. 2007. January;3(1):18–23. PubMed PMID: WOS:000243429900013; English. [DOI] [PubMed] [Google Scholar]
- [6].Stouffer AL, Acharya R, Salom D, et al. Structural basis for the function and inhibition of an influenza virus proton channel. Nature. 2008. January 31;451(7178):596–599. PubMed PMID: 18235504; PubMed Central PMCID: PMC3889492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ichinohe T, Pang IK, Iwasaki A. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat Immunol. 2010. May;11(5):404–U61. PubMed PMID: WOS:000276852200009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ren Y, Li C, Feng L, et al. Proton channel activity of influenza A virus matrix protein 2 contributes to autophagy arrest. J Virol. 2016. January;90(1):591–598. PubMed PMID: WOS:000366899000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zhirnov OP, Klenk HD. Influenza A virus proteins NS1 and hemagglutinin along with M2 are involved in stimulation of autophagy in infected cells. J Virol. 2013. December;87(24):13107–13114. PubMed PMID: WOS:000327443300003; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhou Z, Jiang XJ, Liu D, et al. Autophagy is involved in influenza A virus replication. Autophagy. 2009. April 1;5(3):321–328. PubMed PMID: WOS:000265626300007; English. [DOI] [PubMed] [Google Scholar]
- [11].Liu G, Zhong MG, Guo CW, et al. Autophagy is involved in regulating influenza A virus RNA and protein synthesis associated with both modulation of Hsp90 induction and mTOR/p70S6K signaling pathway. Int J Biochem Cell B. 2016. March;72:100–108. PubMed PMID: WOS:000371844000010; English. [DOI] [PubMed] [Google Scholar]
- [12].Rossman JS, Lamb RA. Autophagy, apoptosis, and the influenza virus M2 protein. Cell Host Microbe. 2009. October 22;6(4):299–300. PubMed PMID: 19837369. [DOI] [PubMed] [Google Scholar]
- [13].Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010. October 22;40(2):280–293. PubMed PMID: WOS:000284028100010; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bjrky G, Lamark T, Brech A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171(4):603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Filippi-Chiela EC, Viegas MS, Thome MP, et al. Modulation of autophagy by calcium signalosome in human disease. Mol Pharmacol. 2016. September 1;90(3):371–384. PubMed PMID: WOS:000388444000023; English. [DOI] [PubMed] [Google Scholar]
- [16].Scherz-Shouval R, Shvets E, Elazar I. Oxidation as a post-translational modification that regulates autophagy. Autophagy. 2007. Jul–Aug;3(4):371–373. PubMed PMID: WOS:000247554100015; English. [DOI] [PubMed] [Google Scholar]
- [17].Mogensen TH, Melchjorsen J, Hollsberg P, et al. Activation of NF-kappa B in virus-infected macrophages is dependent on mitochondrial oxidative stress and intracellular calcium: downstream involvement of the kinases TGF-beta-activated kinase 1, mitogen-activated kinase/extracellular signal-regulated kinase kinase 1, and I kappa B kinase. J Iimmunol. 2003. June 15;170(12):6224–6233. PubMed PMID: WOS:000183411100051 [DOI] [PubMed] [Google Scholar]
- [18].Tal MC, Iwasaki A. Autophagic control of RLR signaling. Autophagy. 2009. July 1;5(5):749–750. PubMed PMID: WOS:000268205300030; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhao YY, Sun XF, Nie XL, et al. COX5B regulates MAVS-mediated antiviral signaling through interaction with ATG5 and repressing ROS production. Plos Pathog. 2012. December;8(12). DOI: 10.1371/journal.ppat.1003086 PubMed PMID: WOS:000312907100038; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Jounai N, Takeshita F, Kobiyama K, et al. The Atg5-Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci U S A. 2007. August 28;104(35):14050–14055. PubMed PMID: WOS:000249187500036; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Cheng JB, Liao YJ, Xiao L, et al. Autophagy regulates MAVS signaling activation in a phosphorylation-dependent manner in microglia. Cell Death Differ. 2017. February;24(2):276–287. PubMed PMID: WOS:000395789500010; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sun X, Sun L, Zhao Y, et al. MAVS maintains mitochondrial homeostasis via autophagy. Cell Discov. 2016;2:16024 PubMed PMID: 27551434; PubMed Central PMCID: PMC4986202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhang LF, Wu JH, Ling MT, et al. The role of the PI3K/Akt/mTOR signalling pathway in human cancers induced by infection with human papillomaviruses. Mol Cancer. 2015. April 17:14 DOI: 10.1186/s12943-015-0361-x PubMed PMID: WOS:000369749300001; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Romanov J, Walczak M, Ibiricu I, et al. Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. Embo J. 2012. November 14;31(22):4304–4317. PubMed PMID: WOS:000311157500007; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gannage M, Dormann D, Albrecht R, et al. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe. 2009. October 22;6(4):367–380. PubMed PMID: WOS:000271227700012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Jacquin E, Leclercmercier S, Judon C, et al. Pharmacological modulators of autophagy activate a parallel noncanonical pathway driving unconventional LC3 lipidation. Autophagy. 2017;13(5):854–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kuo PL, Hsu YL, Cho CY. Plumbagin induces G2-M arrest and autophagy by inhibiting the AKT/mammalian target of rapamycin pathway in breast cancer cells. Mol Cancer Ther. 2006. December;5(12):3209–3221. PubMed PMID: 17172425. [DOI] [PubMed] [Google Scholar]
- [28].Jiang H, Xiao J, Kang B, et al. PI3K/SGK1/GSK3Î2 signaling pathway is involved in inhibition of autophagy in neonatal rat cardiomyocytes exposed to hypoxia/reoxygenation by hydrogen sulfide. Exp Cell Res. 2015;345(2):134–140. [DOI] [PubMed] [Google Scholar]
- [29].Crawford SE, Hyser JM, Utama B, et al. Autophagy hijacked through viroporin-activated calcium/calmodulin-dependent kinase kinase-beta signaling is required for rotavirus replication. Proc Natl Acad Sci U S A. 2012. December 11;109(50):E3405–E3413. PubMed PMID: WOS:000312605600008; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ito M, Yanagi Y, Ichinohe T. Encephalomyocarditis virus viroporin 2B activates NLRP3 inflammasome. Plos Pathog. 2012. August;8(8). DOI: 10.1371/journal.ppat.1002857 PubMed PMID: WOS:000308558000034; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Li YC, Boehning DF, Qian T, et al. Hepatitis C virus core protein increases mitochondrial ROS production by stimulation of Ca2+ uniporter activity. Faseb J. 2007. August;21(10):2474–2485. PubMed PMID: WOS:000248454400022; English. [DOI] [PubMed] [Google Scholar]
- [32].Lazrak A, Iles KE, Liu G, et al. Influenza virus M2 protein inhibits epithelial sodium channels by increasing reactive oxygen species. Faseb J. 2009. November;23(11):3829–3842. PubMed PMID: 19596899; PubMed Central PMCID: PMC2775009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Sun F, Xu X, Wang X, et al. Regulation of autophagy by Ca(2). Tumour Biol. 2016. November 18 DOI: 10.1007/s13277-016-5353-y PubMed PMID: 27864685; PubMed Central PMCID: PMC5250648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhu P, Liang L, Shao X, et al. Host cellular protein TRAPPC6A delta interacts with influenza A virus M2 protein and regulates viral propagation by modulating M2 trafficking. J Virol. 2017. January;91(1). DOI: 10.1128/jvi.01757-16 PubMed PMID: WOS:000393189200025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Scrima R, Piccoli C, Moradpour D, et al. Targeting Endoplasmic Reticulum and/or Mitochondrial Ca(2+) Fluxes as Therapeutic Strategy for HCV Infection. Front Chem. 2018;6:73 PubMed PMID: 29619366; PubMed Central PMCID: PMC5871704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Madan V, Sánchezmartínez S, Carrasco L, et al. A peptide based on the pore-forming domain of pro-apoptotic poliovirus 2B viroporin targets mitochondria. Biochim Biophys Acta. 2010;1798(1):52. [DOI] [PubMed] [Google Scholar]
- [37].Nomura-Takigawa Y, Nagano-Fujii M, Deng L, et al. Non-structural protein 4A of Hepatitis C virus accumulates on mitochondria and renders the cells prone to undergoing mitochondria-mediated apoptosis. J Gen Virol. 2006. July;87(Pt 7):1935–1945. PubMed PMID: 16760395. [DOI] [PubMed] [Google Scholar]
- [38].Griffin SD, Harvey R, Clarke DS, et al. A conserved basic loop in hepatitis C virus p7 protein is required for amantadine-sensitive ion channel activity in mammalian cells but is dispensable for localization to mitochondria. J Gen Virol. 2004;85(Pt 2):451. [DOI] [PubMed] [Google Scholar]
- [39].Madan V, Castello A, Carrasco L. Viroporins from RNA viruses induce caspase-dependent apoptosis. Cell Microbiol. 2008. February;10(2):437–451. PubMed PMID: WOS:000252205900016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].D’Agostino DM, Bernardi P, Chieco-Bianchi L, et al. Mitochondria as functional targets of proteins coded by human tumor viruses. Adv Cancer Res. 2005;94:87–142. PubMed PMID: WOS:000231116900003; English. [DOI] [PubMed] [Google Scholar]
- [41].Henkel JR, Apodaca G, Altschuler Y, et al. Selective perturbation of apical membrane traffic by expression of influenza M2, an acid-activated ion channel, in polarized madin-darby canine kidney cells. Mol Biol Cell. 1998;9(9):2477–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Henkel JR, Gibson GA, Poland PA, et al. Influenza M2 proton channel activity selectively inhibits trans-Golgi network release of apical membrane and secreted proteins in polarized Madin-Darby canine kidney cells. J Cell Biol. 2000. February 7;148(3):495–504. PubMed PMID: 10662775; PubMed Central PMCID: PMC2174804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ni HM, Williams JA, Ding WX. Mitochondrial dynamics and mitochondrial quality control. Redox Biol. 2015. April;4:6–13. PubMed PMID: WOS:000350813800002; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Gegg ME, Cooper JM, Chau K-Y, et al. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy (vol 19, pg 4861, 2010). Hum Mol Genet. 2013. April 15;22(8):1697 PubMed PMID: WOS:000316965400019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Pacelli C, Rasmo DD, Signorile A, et al. Mitochondrial defect and PGC-1 alpha dysfunction in parkin-associated familial Parkinson’s disease. FEBS J. 2011. June;278:270 PubMed PMID: WOS:000292333102078. [DOI] [PubMed] [Google Scholar]
- [46].De Luca A, Fiorillo M, Peiris-Pages M, et al. Mitochondrial biogenesis is required for the anchorage-independent survival and propagation of stem-like cancer cells. Oncotarget. 2015. June 20;6(17):14777–14795. PubMed PMID: WOS:000359010700012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ramos HJ, Gale M. RIG-I like receptors and their signaling crosstalk in the regulation of antiviral immunity. Curr Opin Virol. 2011. September;1(3):167–176. PubMed PMID: WOS:000312112100003; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Mitchell S, Vargas J, Hoffmann A. Signaling via the NF kappa B system. Wiley Interdiscip Rev Syst Biol Med. 2016. May–Jun;8(3):227–241. PubMed PMID: WOS:000374680600003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Meylan E, Curran J, Hofmann K, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005. October 20;437(7062):1167–1172. PubMed PMID: WOS:000232660500047; English. [DOI] [PubMed] [Google Scholar]
- [50].Tang ED, Wang CY. MAVS self-association mediates antiviral innate immune signaling. J Virol. 2009. April 15;83(8):3420–3428. PubMed PMID: WOS:000264327300001; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hou F, Sun L, Zheng H, et al. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011. August 05;146(3):448–461. PubMed PMID: 21782231; PubMed Central PMCID: PMC3179916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Beale R, Wise H, Stuart A, et al. A LC3-interacting motif in the influenza A virus M2 protein is required to subvert autophagy and maintain virion stability. Cell Host Microbe. 2014. February 12;15(2):239–247. PubMed PMID: WOS:000331475900013; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kawai T, Takahashi K, Sato S, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005. October;6(10):981–988. DOI: 10.1038/ni1243 PubMed PMID: WOS:000232027200016; English. [DOI] [PubMed] [Google Scholar]
- [54].Seth RB, Sun LJ, Ea CK, et al. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappa B and IRF3. Cell. 2005. September 9;122(5):669–682. PubMed PMID: WOS:000231844300008. [DOI] [PubMed] [Google Scholar]
- [55].Tang SQ, Chen TY, Yang MJ, et al. Extracellular calcium elicits feedforward regulation of the Toll-like receptor-triggered innate immune response. Cell Mol Immunol. 2017. February;14(2):180–191. PubMed PMID: WOS:000394350700003; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Soucy-Faulkner A, Mukawera E, Fink K, et al. Requirement of NOX2 and reactive oxygen species for efficient RIG-I-mediated antiviral response through regulation of MAVS expression. Plos Pathog. 2010. June;6(6). DOI: 10.1371/journal.ppat.1000930 PubMed PMID: WOS:000279806300008; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Tal MC, Sasai M, Lee HK, et al. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc Natl Acad Sci U S A. 2009. February 24;106(8):2770–2775. PubMed PMID: WOS:000263652900055; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Gonzalez-Dosal R, Horan KA, Rahbek SH, et al. HSV infection induces production of ROS, which potentiate signaling from pattern recognition receptors: role for S-glutathionylation of TRAF3 and 6. Plos Pathog. 2011. September;7(9). DOI: 10.1371/journal.ppat.1002250 PubMed PMID: WOS:000295409000050; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Nakahira K, Cloonan SM, Mizumura K, et al. Autophagy: a crucial moderator of redox balance, inflammation, and apoptosis in lung disease. Antioxid Redox Signal. 2014. January 20;20(3):474–494. PubMed PMID: 23879400; PubMed Central PMCID: PMC3894710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Puleston DJ, Simon AK. Autophagy in the immune system. Immunology. 2014. January;141(1):1–8. PubMed PMID: 23991647; PubMed Central PMCID: PMC3893844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Fujita T. Virus-infection or 5 ‘ ppp-RNA activates antiviral signal through redistribution of IPS-1 mediated by MFN1. Cytokine. 2010. Oct–Nov;52(1–2):42 PubMed PMID: WOS:000282418400155; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Castanier C, Garcin D, Vazquez A, et al. Mitochondrial dynamics regulate the RIG-I-like receptor antiviral pathway. EMBO Rep. 2010. February;11(2):133–138. PubMed PMID: WOS:000274196600015; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Jacobs JL, Coyne CB. Mechanisms of MAVS regulation at the mitochondrial membrane. J Mol Biol. 2013. December 13;425(24):5009–5019. PubMed PMID: WOS:000328808500011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Yoshizumi T, Ichinohe T, Sasaki O, et al. Influenza A virus protein PB1-F2 translocates into mitochondria via Tom40 channels and impairs innate immunity. Nat Commun. 2014. August 5 DOI: 10.1038/Ncomms5713 PubMed PMID: WOS:000341079100002; English. [DOI] [PubMed] [Google Scholar]
- [65].Horner SM, Liu HM, Park HS, et al. Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc Natl Acad Sci U S A. 2011. August 30;108(35):14590–14595. PubMed PMID: 21844353; PubMed Central PMCID: PMC3167523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Koshiba T, Yasukawa K, Yanagi Y, et al. Mitochondrial membrane potential is required for MAVS-mediated antiviral signaling. Sci Signal. 2011. February 1;4(158). DOI: 10.1126/scisignal.2001147 PubMed PMID: WOS:000286801000002; English. [DOI] [PubMed] [Google Scholar]
- [67].Huang J, Lam GY, Brumell JH. Autophagy signaling through reactive oxygen species. Antioxid Redox Sign. 2011. June;14(11):2215–2231. PubMed PMID: WOS:000290559600014; English. [DOI] [PubMed] [Google Scholar]
- [68].Zhang F, Xing S, Li Z. Antagonistic effects of lycopene on cadmium-induced hippocampal dysfunctions in autophagy, calcium homeostatis and redox. Oncotarget. 2017. May 29 DOI: 10.18632/oncotarget.18249 PubMed PMID: MEDLINE:28615536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Fletcher K, Ulferts R, Jacquin E, et al. The WD40 domain of ATG16L1 is required for its non-canonical role in lipidation of LC3 at single membranes. Embo J. 2018. February 15;37(4). DOI: 10.15252/embj.201797840 PubMed PMID: WOS:000425150500006; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Sun Y, Li C, Shu Y, et al. Inhibition of autophagy ameliorates acute lung injury caused by avian influenza A H5N1 infection. Sci Signal. 2012;5(212):ra16. [DOI] [PubMed] [Google Scholar]
- [71].Shin N, Pyo CW, Jung KI, et al. Influenza A virus PB1-F2 is involved in regulation of cellular redox state in alveolar epithelial cells. Biochem Biophys Res Commun. 2015. April 17;459(4):699–705. PubMed PMID: WOS:000352922200023; English. [DOI] [PubMed] [Google Scholar]
- [72].Zou W, Yu ZJ, Zhou HB, et al. Genetic characterization of an H5N1 avian influenza virus with neurovirulence in ducks. Virus Genes. 2009. April;38(2):263–268. PubMed PMID: WOS:000263900500009; English. [DOI] [PubMed] [Google Scholar]
- [73].Sun YJ, Yu SQ, Ding N, et al. Autophagy benefits the replication of newcastle disease virus in chicken cells and tissues. J Virol. 2014. January;88(1):525–537. PubMed PMID: WOS:000329194600048; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Bai RK, Perng CL, Hsu CH, et al. Quantitative PCR analysis of mitochondrial DNA content in patients with mitochondrial disease. Ann Ny Acad Sci. 2004;1011:304–309. PubMed PMID: WOS:000221896100029; English. [DOI] [PubMed] [Google Scholar]
- [75].Lin X, Wang RF, Zou W, et al. The influenza virus H5N1 infection can induce ROS production for viral replication and host cell death in A549 cells modulated by human Cu/Zn superoxide dismutase (SOD1) overexpression. Viruses-Basel. 2016. January;8(1). DOI: 10.3390/V8010013 PubMed PMID: WOS:000373915000001; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


