Abstract
The synthesis of ribosomal RNA (rRNA) is a tightly regulated central process in all cells. In bacteria efficient expression of all seven rRNA operons relies on the suppression of termination signals (antitermination) and the proper maturation of the synthesized rRNA. These processes depend on N-utilization substance (Nus) factors A, B, E and G, as well as ribosomal protein S4 and inositol monophosphatase SuhB, but their structural basis is only poorly understood. Combining nuclear magnetic resonance spectroscopy and biochemical approaches we show that Escherichia coli SuhB can be integrated into a Nus factor-, and optionally S4-, containing antitermination complex halted at a ribosomal antitermination signal. We further demonstrate that SuhB specifically binds to the acidic repeat 2 (AR2) domain of the multi-domain protein NusA, an interaction that may be involved in antitermination or posttranscriptional processes. Moreover, we show that SuhB interacts with RNA and weakly associates with RNA polymerase (RNAP). We finally present evidence that SuhB, the C-terminal domain of the RNAP α-subunit, and the N-terminal domain of NusG share binding sites on NusA-AR2 and that all three can release autoinhibition of NusA, indicating that NusA-AR2 serves as versatile recruitment platform for various factors in transcription regulation.
INTRODUCTION
Transcription, the first step in gene expression, is tightly controlled by a multitude of transcriptional regulators at all steps of the transcription cycle, i.e. initiation, elongation, and termination, with RNA polymerase (RNAP) being responsible for RNA synthesis. Under certain circumstances termination signals are suppressed by a process called antitermination (AT), a ubiquitous mechanism to regulate the expression of viral genes in bacteria, bacterial genes, and possibly, genes of archaea and eukaryotes. AT mechanisms were first discovered in context with bacteriophage λ, where they are essential for the phage's life cycle as they control the expression of early and late genes (1). Phage protein λN combines with Escherichia coli host proteins, the N-utilization substance (Nus) factors A, B, E, and G, as well as specific RNA sequences, nutL and nutR, that contain two protein binding elements, boxA and boxB, and the elongation complex (EC) (2). In the resulting λN- and Nus factor containing antitermination complex (TAC) RNAP is modulated to suppress termination signals (3–7). Nus factors are not only involved in the life cycle of bacteriophages, but are required for the correct expression of genes in bacteria. NusG is member of the only universally conserved family of transcription factors (8) and, in E. coli, consists of an N- and a C-terminal domain (NTD, CTD) connected by a flexible linker (9). NusG-NTD binds to RNAP to increase its processivity, NusG-CTD serves as interaction platform for various binding partners, e.g. it interacts with ribosomal protein S10 to couple transcription and translation or with termination factor Rho to stimulate Rho-dependent termination (10,11). During AT NusG-CTD binds to NusE, which is identical to S10, and anchors the NusB:NusE:boxA complex to the RNAP (12,13). Highly conserved NusA is composed of six domains in E. coli and involved in AT processes (14,15), RNA folding (16), pausing and intrinsic as well as Rho-dependent termination (17). NusA-NTD binds to the β flap tip helix, which forms the mouth of the RNA exit channel, and modulates pausing and termination (5,7,18). The following three domains, S1 and K homology (KH) 1 and KH2, form a compact RNA binding region called NusA-SKK (19). In E. coli and other γ-proteobacteria, NusA-SKK is followed by two acidic repeat (AR) domains, AR1 and AR2 (20). NusA-AR1 binds λN during λN-dependent AT (21–23). In free NusA NusA-AR2 interacts with NusA-KH1, autoinhibiting NusA by preventing RNA binding. This autoinhibition can be released by interaction of NusA-AR2 with the CTD of the α subunit of RNAP (αCTD) (24–26). Moreover, NusA-AR2 can bind to NusG-NTD, an interaction that might be involved in NusG recruitment (27).
In many bacteria, Nus factors are also required for the correct expression of ribosomal RNA (rRNA) genes (6,28). rRNA and transfer RNA (tRNA) make up more than 95% of the total RNA in a bacterial cell and ribosome biosynthesis consumes a major fraction of the energy of the cell (29). Thus, synthesis, cleavage, and maturation of rRNAs as well as their assembly with ribosomal proteins need to be tightly controlled. E. coli harbors seven rRNA operons, each comprising the genes coding for 16S, 23S and 5S rRNA. All operons have a leader sequence upstream of the 16S gene and a spacer element between the 16S and the 23S genes. Similar to λN-dependent AT sequences, E. coli rRNA operons contain boxA and boxB elements, both in the leader and in the spacer regions, but in reversed order (30–34). The ribosomal (rrn) boxA element is essential for rrn AT, highly conserved in all seven rrn operons in E. coli (consensus sequence: UGCUCUUUA), and differs only slightly from λ boxA (33). The boxA element is specifically recognized by the NusB:NusE heterodimer (35), with NusE being the active AT component while NusB acts as loading factor for NusE (13,36). boxB is neither a consensus sequence nor required for rrn AT, but has the potential to form an RNA hairpin loop structure (33,34). Additionally, a linear, less well characterized boxC element consisting of an alternating GU sequence can be found 3′ of boxA in the rrn leader region or following boxB in lambdoid nut sites (33,34). Similar to boxB, boxC is not necessary for rrn AT (33), but a boxC element can also be found in other species such as Mycobacterium tuberculosis, where it is part of a binding site that is recognized by NusA (37).
In bacteria, long non-coding RNAs are target for Rho-dependent termination. Thus, RNAP is modified at boxA elements of the leader and the spacer region of rRNA operons to suppress Rho-dependent termination via assembly of an rrn TAC that comprises at least NusA, NusB, NusE and NusG (6,38,39). Complete rrn AT cannot be achieved by using purified Nus factors in vitro (6), but cell extract is able to stimulate rrn AT, and ribosomal protein S4 has been identified as key player in rrn AT (40). Moreover, inositol monophosphatase SuhB (41) and translation-associated heat shock protein YbeY (42) are suggested to contribute to correct rRNA biosynthesis. Although AT has long been the accepted role for Nus factors in rRNA synthesis, recent studies suggest that the major role of the Nus factor- and SuhB-modified EC in rRNA operons is to act as RNA chaperone and to co-ordinate transcription with folding of the 16S rRNA and processing by RNase III, ensuring the proper production of 30S subunits (43). Thus, it is unclear if the predominant role of SuhB and Nus factors is to stimulate rrn AT or to function post-transcriptionally or if they are essential for both activities as these are not mutually exclusive. It is proposed that SuhB associates directly with RNAP (44), maybe in a NusB-dependent manner (41), and promotes RNA loop formation between elongating RNAP and the boxA-bound NusB:NusE complex (41). To date, the structural basis for proper rRNA synthesis ensured by Nus and other factors remains elusive.
Here, we combined biochemical approaches with solution-state nuclear magnetic resonance (NMR) spectroscopy to demonstrate that SuhB can indeed be part of a Nus factor-, and optionally S4-, containing EC halted at an rrn AT site. Although it is not yet clear if the resulting complex is involved in AT or acts posttranscriptionally, we will refer to it as rrn-TAC. We show that SuhB specifically interacts with NusA-AR2, but that it also binds to RNA as well as to RNAP. Moreover, our analyses reveal that SuhB, RNAP αCTD and NusG-NTD share binding sites on NusA-AR2, and that all three interactions release autoinhibition of NusA, confirming NusA as central regulator in transcription.
MATERIALS AND METHODS
Cloning and mutagenesis
The genes encoding SuhB and S4 were amplified from chromosomal E. coli DNA by polymerase chain reaction using the primers listed in Supplementary Table S1. Each gene was cloned into a pETGB1a expression vector (provided by Gunter Stier, EMBL Heidelberg, Germany) via BamHI and NcoI restriction sites resulting in the recombinant plasmids pETGB1a_suhb and pETGB1a_s4, respectively. The recombinant target proteins carry a hexa-histidine tag followed by the B1 domain of streptococcal protein G (GB1) and a Tobacco Etch Virus (TEV) cleavage site at their N-termini.
The gene coding for NusA lacking the AR2 domain (amino acids 1–426, NusAΔAR2) was amplified from pTKK19_NusA(1–495) (27) with primers listed in Supplementary Table S1. The gene was cloned into a modified vector pET19b via NdeI and XhoI restriction sites (pET19bmod_nusAΔAR2) so that the resulting fusion protein harbors a hexa-histidine tag followed by a TEV protease cleavage site at its N-terminus.
The exchange of D443 in NusA-AR2 by a cysteine was done according to the QuickChange Site-directed Mutagenesis protocol from Stratagene (Stratagene, La Jolla, USA) using pET19b_nusA-AR2 (24) as template and primers AR2-D443C-FW and AR2-D443C-RV (Supplementary Table S1), resulting in pET19b_nusA-AR2(D443C).
Gene expression and protein purification
Full length NusA was produced as described (27), as was NusA-NTD (45), NusA-SKK (46), NusA-AR1-AR2 (20), NusA-AR2 (27), NusB (12), NusEΔ/B (12), NusG (47), NusG-NTD (47), RNAP (48), α-, β-, β′-, ω-subunit (45) and RNAP α-CTD (20). The production and purification of NusA-AR2D443C were the same as for NusA-AR2.
s4 was expressed in E. coli BL21 (λ DE3) (Novagen, Madison, USA) harboring the plasmid pETGB1a_s4. Lysogeny broth (LB) medium supplemented with 30 μg/ml kanamycin was inoculated with a preculture to an optical density at 600 nm (OD600) of 0.2 and grown at 37°C. At an OD600 of 0.7 expression was induced by addition of 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). Cells were harvested 4 h after induction by centrifugation (6 000 × g, 10 min, 4°C), resuspended in buffer S4-A (50 mM tris(hydroxymethyl)aminomethane (Tris)/HCl, pH 7.5, 500 mM NaCl, 10 mM imidazole, 10% (v/v) glycerol, 2 mM Dithiothreitol (DTT)), and lysed by a microfluidizer (Microfluidics, Newton, USA). Upon centrifugation (25 000 × g, 30 min, 4°C) the supernatant was applied to a 5 ml HisTrap HP chelating column (GE Healthcare, Chalfont St Giles, UK), which was subsequently washed with buffer S4-A. Elution was carried out via a step gradient with increasing imidazole concentrations (10 mM–1 M in buffer S4-A). Fractions containing His6-Gb1-S4 were combined and cleaved during overnight dialysis against buffer S4-B (50 mM Tris/HCl, pH 7.5, 400 mM NaCl, 10 mM imidazole, 10% (v/v) glycerol, 2 mM DTT; molecular weight cut-off (MWCO) 3 500 Da) at 4°C by TEV protease. The dialysate was applied to a 5 ml HisTrap HP chelating column connected to a 5 ml Heparin HP column (GE Healthcare, Munich, Germany). After washing with buffer S4-A, the HisTrap HP chelating column was removed and the Heparin HP column was eluted with a constant gradient from 400 mM to 1 M NaCl in buffer S4-A. Fractions containing pure S4 were combined, dialysed against 50 mM 3-(N-morpholino)propanesulfonic acid (MOPS) buffer, pH 7.0, 300 mM NaCl, 150 mM d-glucose, 2 mM DTT (MWCO 3 500 Da) at 4°C, concentrated by ultrafiltration (MWCO 3 000 Da), shock frozen in liquid nitrogen, and subsequently stored at −80°C.
Expression of suhb was carried out in E. coli Rosetta (λ DE3) plysS (Novagen, Madison, USA) containing pETGB1a_suhb. The procedure was the same as for s4, except that LB medium was supplemented with 34 μg/ml chloramphenicol in addition to 30 μg/ml kanamycin. Furthermore, 0.5 mM IPTG was used for induction and cells were resuspended in buffer SuhB-A (50 mM Tris/HCl, pH 7.5, 150 mM NaCl, 0.5 mM phenylmethane sulfonyl fluoride (PMSF)). Cell lysis and protein purification were analogous to the procedures described for S4, with buffers as follows. Elution of the first Ni affinity chromatography was performed with a step gradient from 0 to 500 mM imidazole in buffer SuhB-A. Subsequent dialysis and cleavage by TEV protease was carried out in buffer SuhB-B (50 mM Tris/HCl, pH 7.5, 150 mM NaCl). The target protein was eluted from the Heparin HP column via a constant gradient with increasing NaCl concentration (150 mM–1 M NaCl in buffer SuhB-B). Pure SuhB was finally dialysed against 50 mM Tris/HCl, pH 7.5, 500 mM NaCl, 10% (v/v) glycerol, 2 mM DTT (MWCO 10 000 Da). Analytical gel filtration on a Superdex 75 10/300 GL column (GE Healthcare, Munich, Germany) showed that the protein existed as monomer in solution.
NusAΔAR2 was produced in E. coli BL21 (λ DE3) harboring the plasmid pET19bmod_nusAΔAR2. The expression procedure resembled the expression of the s4 gene, with exception that the LB medium was supplemented with 100 μg/ml ampicillin. For purification, cells were resuspended in buffer NusAΔAR2-A (20 mM Tris/HCl, pH 7.9, 500 mM NaCl, 5 mM imidazole, 1 mM β-mercaptoethanol (β-ME)). Cell lysis and the first step of protein purification (Ni affinity chromatography) were analogous to the procedures described for S4, with a step gradient of 5 mM–1 M imidazole in buffer NusAΔAR2-A. After cleavage by TEV protease during overnight dialysis at 4°C against buffer NusAΔAR2-B (20 mM Tris/HCl, pH 8.0, 1 mM β-ME) (MWCO 10 000 Da) the protein solution was applied to a HisTrap HP chelating column coupled to a QXL column (GE Healthcare, Chalfont St Giles, UK). The target protein was eluted from the QXL column via a constant gradient with increasing NaCl concentration (150 mM–1 M NaCl in buffer NusAΔAR2-B). Pure NusAΔAR2 was dialysed against 50 mM MOPS, pH 7.0, 300 mM NaCl, 150 mM d-glucose, 2 mM DTT (MWCO 3 500 Da) at 4°C, concentrated by ultrafiltration (MWCO 10 000 Da), shock frozen in with liquid nitrogen and stored at −80°C.
Quality control of recombinant proteins
The purity was checked by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), the absence of nucleic acids by recording UV/visible spectra on a Nanodrop ND-100 spectrometer (PEQLAB, Erlangen, Germany) from 220 to 600 nm and calculating the ratio of the absorbance at 260 and 280 nm. Concentrations were determined by measuring the absorbance at 280 nm in a 10 mm quartz cuvette (Hellma, Müllheim, Germany) on a Biospectrometer basic (Eppendorf, Hamburg, Germany). The identitiy of SuhB and S4, respectively, was checked by peptide mass finger printing (Department of Biochemistry, University of Bayreuth, Germany). Homogeneity was ensured by analytical gel filtration on a Superdex 75 or a Superdex 200 10/300 GL column (GE Healthcare, Munich, Germany). The folding state was assessed by circular dichroism (CD) spectroscopy (1 mm quartz cuvette; J-1100, JASCO, Pfungstadt, Germany) as well as one dimensional (1D) 1H-NMR spectra in the case of S4 and SuhB.
RNA synthesis and purification
rrnG RNA was produced via T7 RNAP-based in vitro transcription using T7 RNAP variant P226K (T7 RNAPP226K) to enhance transcription efficiency. The rrnG DNA template was chemically synthesized (metabion, Planegg, Germany) and contained a T7 promoter, a GGG sequence at the transcription start site to enhance transcription efficiency, and the gene coding for rrnG RNA comprising boxA, boxB, and boxC (Supplementary Table S1). Transcription reactions were performed in 40 mM Tris/HCl, pH 8.1, 5 mM DTT, 1 mM spermidine, 28 mM MgCl2, 8% (w/v) polyethylene glycol (PEG) 8 000, 0,01% (v/v) Triton X-100 containing 100 μM rrnG DNA template, 4 mM NTPs (ATP, GTP, UTP, CTP) and 100 μg/ml T7 RNAPP226K at 37°C for 4 h. Afterward, the reaction was stopped via addition of ethylenediaminetetraacetic acid (EDTA) to a final concentration of 66.8 mM and an ethanol precipitation was performed. The rrnG RNA was purified via a preparative 20% acrylamide/8 M urea PAGE and subsequent electro elution. The RNA was dialyzed against water, concentrated, lyophilized and stored at −80°C.
Isotopic labeling of proteins
15N-labeled proteins were produced by growing E. coli cells in M9 medium (49,50) containing (15NH4)2SO4 (CortecNet, Voisins-Le-Bretonneux, France). For the production of perdeuterated proteins, E. coli cells were grown in M9 medium (49,50) prepared with increasing amounts of D2O (25% (v/v), 50% (v/v), 99.9% (v/v) D2O; Eurisotop, Saint-Aubin, France) with d7-glucose as carbon source. Site-specific [1H,13C]-labeling of Ile, Leu and Val methyl groups ([I,L,V]-labeling) in perdeuterated proteins was performed according to published protocols (51), i.e. expression was carried out as described for the production of perdeuterated proteins, but 60 mg/l 2-keto-3-d3–4-13C-butyrate and 100 mg/l 2-keto-3-methyl-d3–3-d1–4-13C-butyrate (both from Eurisotop, St. Aubin Cedex, France) were added 1 h prior to induction. Expression and purification were as described for the production of unlabeled proteins.
NMR experiments
NMR experiments were carried out on Bruker Avance 700 MHz, Bruker Ascend Aeon 900 MHz and Bruker Ascend Aeon 1 000 MHz spectrometers, equipped with cryogenically cooled inverse triple resonance probes. Data was processed using in-house routines. Two dimensional (2D)/three dimensional (3D) spectra were visualized and analysed by NMRViewJ (One Moon Scientific, Inc., Westfield, NJ, USA), 1D spectra by MatLab (The MathWorks, Inc., Version 7.1.0.183). Assignments for the backbone amide resonances of NusA-AR2 (20), NusA-AR1-AR2(20), NusA-SKK (24) and αCTD (24) were taken from previous studies, the assignment of methyl group signals in full length NusA were adapted from the assignments of individual domains (20,24,52).
All spectra were recorded at 298 K with an initial sample volume of 250 μl, if not stated otherwise. Proteins were in 50 mM MOPS, pH 7.0, 300 mM NaCl, 150 mM d-glucose, 10 mM MgCl2, 2 mM DTT, which was prepared with either H2O for 15N- and 2H,15N-labeled samples or 99.9% D2O for [I,L,V]-labeled proteins. 1D spectra and signal intensities of 2D spectra were normalized by protein concentration, number of scans, and length of the 90° proton pulse; the receiver gain was kept constant.
The SuhB binding surface of NusA-AR2 was determined by analyzing the signal intensity in all titration steps of 2D spectra quantitatively. For each titration step the ratios of remaining signal intensities and signal intensities in the spectrum of the free, labeled protein were calculated, yielding relative signal intensities. The mean value of all relative signal intensities in each titration step was determined and thresholds were defined at 1 and 1.5σ of the mean value. Residues with relative signal intensities below these thresholds were classified as moderately or strongly affected, respectively.
Chemical shift changes in methyl-transverse relaxation-optimized spectroscopy (TROSY)-based interaction studies were in the fast regime on the chemical shift timescale and thus analysed by calculating the normalized chemical shift perturbation (Δδnorm) according to equation (1) for [1H,13C] correlation spectra.
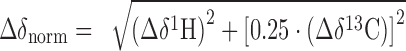 |
(1) |
where Δδ is the resonance frequency difference in ppm.
Leu and Val residues were considered as affected if at least one of the two signals showed a Δδnorm ≥0.04 ppm and only unambiguously assigned signals were used in the analysis. Affected residues were mapped on the 3D structure of NusA with the complete amino acids being highlighted instead of only the methyl group to improve visualization.
Analytical size exclusion chromatography
Analytical size exclusion chromatography on a Superdex 200 Increase 10/300 GL column (GE Helathcare, Munich, Germany) was used to study interactions. Stock solutions of proteins and/or nucleic acids were combined to yield mixtures of equimolar concentration (50 μM of each component; in mixtures containing TACs each component was present at 21 μM). The sequence of the random RNA is given in Supplementary Table S1. 100 μl of the mixtures were incubated for 15 min at room temperature, loaded on the column and chromatographed at 0.3 ml/min at room temperature. 0.5 ml fractions were collected and analyzed by SDS-PAGE (17% PA gels) after trichloracetic acid precipitation or urea PAGE (20% PA gels, 8 M urea) to reveal protein and nucleic acid contents, respectively. The identities of S4 and SuhB were checked by peptide mass finger printing (Department of Biochemistry, University of Bayreuth, Germany). Proteins were in 50 mM Na phosphate buffer, pH 7.0, 75 mM NaCl, 150 mM d-glucose, 2 mM DTT. Quantitative analysis of the PA gels was carried out using the Image Lab Software (Bio-Rad Laboratories, Version 5.2.1).
rrnG-TAC assembly
Assembly of the rrnG-TAC and design of the nucleic acids were based on published methods (53). First, an RNA:DNA-hybrid was formed from the template (T) DNA and the RNA (Supplementary Table S1). The stock solution of T DNA (1 mM and in H2O) was diluted with TAC buffer (50 mM Na phosphate buffer, pH 7.0, 75 mM NaCl, 150 mM d-glucose, 2 mM DTT) by 1:1 and mixed with RNA at an equimolar ratio. The mixture was incubated for 1 min at 95°C, then for 10 min at 70°C, and finally cooled to room temperature within 15 min. RNAP (typically at 50–100 μM) was added at 1.2 molar excess over the hybrid, followed by 10 min incubation at room temperature. Finally, the non-template (NT) DNA strand (Supplementary Table S1; 1 mM stock solution in H2O; diluted 1:2 with TAC buffer) was added at a molar ratio of 1:1.2:3 (T-DNA/RNA-hybrid:RNAP:NT-DNA) and incubated for 10 min at 37°C.
Fluorescence anisotropy
For fluorescence anisotropy measurements, NusA-AR2D443C was site-specifically labeled with fluorescein-5-maleimide (ThermoFisher Scientific, Waltham, USA) according to the manufacturer's protocol. In brief, 25 μM NusA-AR2D443C were incubated with 750 μM fluorescein-5-maleimide at 4°C overnight (labeling buffer: 20 mM Na phosphate, pH 7.0, 150 mM NaCl). The solution was then applied to a PD MiniTrap Sephadex G-25 gravity column (GE Healthcare, Munich, Germany) equilibrated with fluorescence buffer (50 mM MOPS, pH 7.0, 300 mM NaCl, 150 mM glucose, 5 mM DTT) and eluted with fluorescence buffer to remove non-reacted fluorescein-5-maleimide and to exchange labeling buffer by fluorescence buffer. Finally, the degree of labeling and the protein concentration were determined by UV/vis spectroscopy on a Nanodrop ND-1000 spectrometer (PEQLAB, Erlangen, Germany).
For the titration of NusA-AR2D443C with SuhB individual 100 μl samples were prepared for each titration step, each containing 25 nM labeled NusA-AR2D443C and increasing concentrations of SuhB up to 600 μM. Measurements were done in black, sterile 96-well microtiter plates (Brand, Wertheim, Germany) at 25°C on a Synergy 2 microplate reader (BioTek, Winooski, USA). Both proteins were in fluorescence buffer. Four independent measurements were carried out per titration step and the anisotropy values were averaged. The mean values were plotted against the SuhB concentration. The data was fitted to a two-component binding equation describing the binding equilibrium of a 1:1 binding system to determine the KD value (54).
Docking
The complex of NusA-AR2 and SuhB was modeled with the HDOCK server (55) using data from the [1H,15N]-heteronuclear single quantum coherence (HSQC) titration of 15N-NusA-AR1-AR2 with SuhB as restraints. Thus, residues 461, 464, 465, 483, 486, 490 and 491 were defined as binding site residues. Model 1 of the NMR ensemble of NusA-AR2 (PDB ID: 1WCN) and chain A of the crystal structure of SuhB (PDB ID: 2QFL) were used as input.
Programs
The PyMOL Molecular Graphics System (Version 1.7, Schrödinger, LLC.) was used for the graphical representation of protein structures.
RESULTS
SuhB integrates into a Nus factor-containing rrn-TAC
SuhB is suggested to be part of the rRNA biosynthesis machinery, and thus we probed SuhB interactions in a TAC that is halted at an rrn AT site and that harbors all Nus factors by analytical gel filtration using a Superdex 200 column. We produced RNAP, NusA, NusE:NusB, NusG and SuhB recombinantly in E. coli and generated an rrnG RNA (68 nucleotides) containing linear boxA and boxC elements as well as a boxB hairpin (Supplementary Figure S1A) by in vitro transcription. We used a NusE variant lacking the ribosome binding loop (NusEΔ) in complex with NusB to increase its stability (13) in all interaction studies. SuhB existed as monomer in solution. To check the integrity of the rrnG RNA we tested its ability to bind to a NusEΔ:NusB heterodimer. The NusEΔ:NusB complex and rrnG RNA were mixed in equimolar concentration and NusEΔ:NusB coeluted with the RNA (Supplementary Figure S1B), in agreement with previous findings (12,13,35). Next we assembled a Nus factor- and SuhB-containing TAC resting at the rrnG AT site (rrnG-TAC). In brief, an EC was assembled using a nucleic acid scaffold (Supplementary Figure S1A, for details see Materials and Methods) that was subsequently incubated with all Nus factors and SuhB. All transcription factors coeluted with RNAP and the rrnG RNA (Figure 1A), demonstrating that all components form a complex. However, quantitative analysis revealed that, in contrast to NusA, NusEΔ:B, and NusG, only ∼20% of SuhB coeluted with the rrnG-TAC, indicating that the affinity of SuhB for the rrnG-TAC is lower than that of the Nus factors. To identify relevant interactions of SuhB in this complex we tested binary systems by mixing SuhB with NusA, NusEΔ:NusB, NusG, RNAP, or RNA in a 1:1 molar ratio (Figure 1B and Supplementary Figure S1C–F). Among the Nus factors SuhB coeluted only with NusA, indicating a direct interaction between them (Figure 1B). Using a NusA variant lacking the AR2 domain (NusAΔAR2) shows that the SuhB:NusA interaction depends on the presence of AR2 as NusAΔAR2 and SuhB eluted separately (Figure 1C). However, a SuhB- and Nus factor-containing rrnG-TAC could still form when NusAΔAR2 was used instead of full length NusA (Figure 1D), suggesting that the SuhB:NusA-AR2 interaction is dispensable for SuhB integration if all components are present simultaneously. SuhB also coeluted with rrnG RNA (Figure 1E) indicating a direct SuhB:rrnG interaction, in agreement with the suggestion that SuhB functions as RNA chaperone and supports RNA loop formation during RNA maturation (41). To test if SuhB exhibits specificity toward rrnG RNA a random RNA was analysed. The elution profiles of SuhB and the random RNA overlapped when run separately, but they were not altered at all when the 1:1 mixture was applied to the column (Supplementary Figure S1E), indicating that SuhB might specifically recognize rrnG RNA. This finding should, however, be corroborated by an orthogonal technique. Finally, no stable SuhB:RNAP complex could be detected (Supplementary Figure S1F).
Figure 1.
Interaction studies. Analyses of analytical size exclusion chromatography runs by Coomassie-stained SDS-PAGE (proteins, in color) and Toluidine blue-stained urea-PAGE (RNA, in grayscale), monitoring interactions between SuhB, NusA, NusEΔ/B, NusG and the rrnG-TAC (A), SuhB and NusA (B), SuhB and NusAΔAR2 (C), SuhB, NusAΔAR2, NusEΔ/B, NusG and the rrnG-TAC (D), SuhB and rrnG RNA (E), and SuhB, S4, NusA, NusEΔ/B, NusG and the rrnG-TAC (F). Mixtures loaded for each run are indicated in the boxes above the gels. The left lane shows the marker (SDS-PAGE standard low range, BioRad). Molecular weights (MW) are indicated on the left, bands are identified on the right or between two gels, and complexes formed are given below the gels. Gels are representative examples of at least two repetitions.
Ribosomal protein S4 also contributes to rrn AT (40) and, indeed, it coeluted with RNAP, rrnG RNA, all Nus factors, and SuhB when all components were present in equimolar concentration (Figure 1F), indicating integration of S4 into a Nus factor- and SuhB-modified rrnG-TAC. As observed for SuhB, only a fraction of S4 (∼25%) coeluted with the rrnG-TAC, suggesting that its affinity for the rrnG-TAC is also lower than that of the Nus factors. A direct interaction between SuhB and S4 could not be detected (Supplementary Figure S1G).
SuhB binds directly to NusA-AR2
We next aimed to corroborate the finding that SuhB directly interacts with NusA-AR2 by solution-state NMR spectroscopy. The addition of full length NusA to 15N-labeled SuhB led to a significant decrease of SuhB signal intensity in the 1D [1H,15N]-heteronuclear single quantum coherence (HSQC) spectrum (Supplementary Figure S2A). The high transversal relaxation rate of the spins of NusA (54.9 kDa) strongly affects magnetization transfer efficiency upon binding, which results in line broadening and thus ultimately in a decrease of signal intensity. Consequently, the loss of 15N-SuhB signal intensity suggests a direct SuhB:NusA interaction. Titrations of 15N-NusEΔ:NusB, 15N-NusB and 15N-NusG with SuhB demonstrated no interaction (Supplementary Figure S2B–D), all in agreement with our gel filtration data. To identify the NusA domain that binds to SuhB, 15N-NusA-NTD, 15N-NusA-SKK and 15N-NusA-AR1-AR2 were titrated with SuhB and 2D [1H,15N]-HSQC spectra were recorded after each step (Figure 2A, B and Supplementary Figure S2E,F). In the presence of SuhB significant changes were only observable in the spectrum of 15N-NusA-AR1-AR2. The chemical shift perturbations were small, but the intensity of several signals decreased dramatically, indicating intermediate chemical exchange on the NMR time scale. Plotting the relative signal intensity of 15N-NusA-AR1-AR2 in the presence of 0.5 equivalents of SuhB against the NusA-AR1-AR2 amino acid position clearly shows that only the intensity of signals corresponding to the NusA-AR2 domain were affected (Figure 2C), suggesting that SuhB specifically recognizes and interacts with NusA-AR2, consistent with the results of the analytical gel filtration.
Figure 2.
SuhB interacts with NusA-AR2. (A and B) 2D [1H,15N]-HSQC spectra of the titration of 125 μM 15N-NusA-AR1-AR2 with SuhB. Spectra corresponding to molar ratios 1:0, 1:0.5, 1:1, 1:2 are in black, orange, cyan and red, respectively (stock concentration of SuhB: 471 μM). The dashed box in (A) indicates the section as in (B). Selected signals are labeled. (C) HSQC-derived relative intensity of NusA-AR1-AR2 in the presence of 0.5 equivalents of SuhB. Orange and red lines indicate thresholds for moderately (1 σ of average relative signal intensity) and strongly (1.5 σ of average relative signal intensity) affected signals, respectively. Disappearing signals are indicated by red dots, crosses mark not assigned residues. (D) Model of the SuhB:NusA-AR2 complex. The model was generated with HDOCK using the chemical shift perturbations of the [1H,15N]-HSQC titration as restraints for NusA-AR2. The model with the lowest HDOCK score is depicted. SuhB (PDB ID: 2QFL), blue, and NusA-AR2 (PDB ID: 1WCN), gray, are in ribbon representation. Affected residues from (C) are mapped on NusA-AR2 (moderately affected residues, orange; strongly affected residues, red). Termini and secondary structure elements are labeled. Panels show the surface representations of SuhB colored according to its electrostatic surface potential (left) and NusA-AR2 colored as in the complex (right).
Mapping of the affected residues on the structure of NusA-AR2 reveals that they form a continuous patch at the C-terminal part of NusA-AR2, involving helices α3, α4, α5 and the loops connecting these helices (Figure 2D). The electrostatic surface potential of NusA-AR2 is mostly negative, whereas SuhB exhibits an extensive positively charged area, a large negatively charged region, and a mostly hydrophobic patch, the latter constituting the dimerization interface (Supplementary Figure S3A, B). Based on the affected residues in NusA-AR2 a docking model was generated without conformational rearrangements. In the lowest energy model NusA-AR2 binds to the positively charged area of SuhB via helices α3 and α5, positioning NusA-AR2 opposite the dimerization interface of SuhB (Figure 2D). NusA-AR2 residues W490 and F491 located in helix α5 pack against the hydrophobic, C-terminal part of SuhB helix α6.
The KD of the SuhB:NusA-AR2 interaction was determined by fluorescence anisotropy measurements. For this, D443 in the isolated NusA-AR2 domain, which is located in helix α2 and thus opposite the SuhB binding site, was exchanged by a Cys residue, resulting in NusA-AR2D443C. This NusA-AR2 variant was site-specifically labeled with fluorescein-5-maleimide and titrated with SuhB (Supplementary Figure S3C), yielding a KD of 83 ± 4 μM. This rather low affinity may be a result of the high salt concentration that had to be present in the buffer to avoid precipitation of SuhB in the stock solution.
SuhB, NusG-NTD and αCTD share binding sites on NusA-AR2
In free NusA the AR2 domain binds to the KH1 domain of the SKK motif, preventing RNA binding by NusA-SKK and rendering NusA autoinhibited (24–26). This autoinhibition can be relieved by the αCTD of RNAP as NusA-SKK and αCTD share binding sites on NusA-AR2 (24). NusA-AR2, however, can also bind to NusG-NTD, an interaction that might be involved in the regulation of Rho-dependent termination or in the recruitment of NusG (27). The binding sites for both NusG-NTD and αCTD on NusA-AR2 involve the C-terminal helix α5 (Supplementary Figure S4). Interestingly, the SuhB binding site of NusA-AR2 also overlaps with the binding sites for αCTD and NusG-NTD (Supplementary Figure S4). All interactions are specific for NusA-AR2 and involve helix α5 of NusA-AR2 with residues W490 and F491 being key determinants (Figure 2C, (24,27)).
To test if interactions of NusA-AR2 with SuhB and αCTD are competitive, we carried out 2D [1H,15N]-HSQC-based displacement experiments with spectra being recorded after each titration step. First, NusA-AR2 was added in equimolar concentration to 15N-αCTD, resulting in chemical shift changes corresponding to 15N-αCTD:NusA-AR2 complex formation (Figure 3A,B), in agreement with previous findings (24). Subsequent titration with SuhB reversed the chemical shift changes (Figure 3A, B), confirming that SuhB displaces the αCTD from NusA-AR2. A reverse displacement experiment was performed (Figure 3C, D) where NusA-AR2 was added to 2H,15N-SuhB in a 1:1 molar ratio, leading to chemical shift perturbations indicating complex formation. Addition of the αCTD reversed those changes, demonstrating the displacement of SuhB from NusA-AR2. The mutual displacement of SuhB and αCTD from NusA-AR2 indicates similar affinities for the SuhB:NusA-AR2 and the αCTD:NusA-AR2 interactions, suggesting that both are physiologically relevant.
Figure 3.
The αCTD and SuhB share binding sites on NusA-AR2. (A and B) [1H,15N]-HSQC displacement of NusA-AR2 from 15N-αCTD by SuhB. 15N-αCTD:NusA-AR2:SuhB = 1:0:0, black; = 1:1:0, orange; = 1:1:1, cyan; = 1:1:2, purple; = 1:1:5, yellow; = 1:1:10, red. Initial concentration of 15N-αCTD: 200 μM; stock concentrations of NusA-AR2 and SuhB were 941 μM and 597 μM, respectively. The dashed box in (A) indicates the section as in (B). In (B), selected signals are labeled and arrows show the changes in their chemical shifts upon addition of NusA-AR2 (orange) and SuhB (red). (C and D) [1H,15N]-BEST-TROSY displacement of NusA-AR2 from 2H,15N-SuhB by αCTD. 2H,15N-SuhB:NusA-AR2:αCTD = 1:0:0, black; = 1:1:0, orange; = 1:1:1, cyan; = 1:1:2, purple; = 1:1:5, red. Initial concentration of 2H,15N-SuhB: 250 μM; stock concentrations of NusA-AR2 and αCTD were 759 and 793 μM, respectively. The dashed box in (C) indicates the section as in (D). In (D), arrows show the changes in the chemical shifts of selected signals upon addition of NusA-AR2 (orange) and αCTD (red).
SuhB, NusG-NTD and αCTD can release autoinhibition of NusA
The αCTD is able to release autoinhibition of NusA by removing NusA-AR2 from NusA-SKK (24–26). Thus, we probed if NusG-NTD and SuhB have the same capability by NMR-based displacement experiments. We first titrated 2H,15N-labeled NusA-SKK with NusA-AR2 and recorded [1H,15N]-band-selective excitation short-transient transverse relaxation-optimized spectroscopy (BEST-TROSY) spectra after each titration step to determine the chemical shift perturbations caused by binding of NusA-AR2 to the NusA-SKK domain (Figure 4A), which were in agreement with previous data (24). Addition of NusG-NTD to the preformed 2H,15N-NusA-SKK:NusA-AR2 complex led to partial reversal of all shifts, indicating displacement of NusA-AR2 from NusA-SKK (Figure 4B). Similarly, SuhB was able to displace NusA-AR2 from NusA-SKK (Figure 4C).
Figure 4.
NusG-NTD and SuhB displace NusA-AR2 from NusA-SKK. (A) Interaction of NusA-SKK with NusA-AR2. [1H,15N]-BEST-TROSY spectra of the titration of 175 μM 2H,15N-NusA-SKK with NusA-AR2. NusA-AR2 was added in a molar ratio of 1:0, black; 1:0.5, orange; 1:1, cyan; 1:2, purple; 1:3, yellow; 1:5, red. The dashed box in the top panel indicates the section shown in the bottom panel. Selected signals are labeled and the changes in their chemical shifts are indicated by arrows. (B) [1H,15N]-BEST-TROSY displacement experiment of NusA-AR2 from 2H,15N-NusA-SKK by NusG-NTD. 2H,15N-NusA-SKK: NusA-AR2:NusG-NTD = 1:0, black; = 1:5, orange; = 1:5:1, cyan; = 1:5:2, purple; = 1:5:5, yellow; = 1:5:10, red. Initial concentration of 2H,15N-NusA-SKK: 200 μM; stock concentrations of NusA-AR2 and NusG-NTD were 941 and 1098 μM, respectively. The dashed box in the top panel indicates the section as in the bottom panel. Selected signals are labeled and arrows show the changes in their chemical shifts upon addition of NusA-AR2 (orange) and NusG-NTD (red). (C) [1H,15N]-BEST-TROSY displacement experiment of NusA-AR2 from 2H,15N-NusA-SKK by SuhB. 2H,15N-NusA-SKK: NusA-AR2:SuhB = 1:0, black; = 1:5, orange; = 1:5:1, cyan; = 1:5:2, purple; = 1:5:5, yellow; = 1:5:10, red. Initial concentration of 2H,15N-NusA-SKK: 200 μM; stock concentrations of NusA-AR2 and SuhB were 969 μM and 562 μM, respectively. The dashed box in the top panel indicates the section as in the bottom panel. Selected signals are labeled and arrows show the changes in their chemical shifts upon addition of NusA-AR2 (orange) and SuhB (red).
The interaction probability of NusA-AR2 with NusA-SKK may, however, be higher in full length NusA as the domains are covalently connected resulting in a higher local concentration. Thus, we repeated the displacement experiments in the context of the full length protein. Due to the high molecular mass of the system we used 1H,13C-labeled methyl groups of Ile, Leu, and Val residues in a perdeuterated background as NMR-active probes ([I,L,V]-NusA). SuhB was titrated to [I,L,V]-NusA and methyl-TROSY spectra were recorded after each titration step (Figure 5A,B). Normalized changes of the chemical shifts were calculated to identify affected residues (Supplementary Table S2), which were then mapped on the three dimensional structure of NusA (Figure 5C). Affected residues can only be found in the C-terminal region of NusA-AR2 and the β-sheet of the NusA-KH1 subdomain, indicating that SuhB is able to release NusA autoinhibition as KH1 is the binding site for the AR2 domain in autoinhibted NusA (24) and the C-terminal region of NusA-AR2 is the binding site for SuhB (see Figure 2D). This finding corroborates our gel filtration data as SuhB is able to interact with NusA even in the absence of any other factors (Figure 1B). Similarly, NusG-NTD (Supplementary Table S3 and Supplementary Figure S5) and the αCTD (Supplementary Table S4 and Supplementary Figure S6) can release the autoinhibition of NusA. However, in the presence of the αCTD, several residues in NusA-NTD are also strongly affected and form a patch opposite the β flap tip helix binding site (Supplementary Figure S6C), indicating that the αCTD can also bind to NusA-NTD, in agreement with a cryo electron microscopy structure of NusA bound to a paused EC where one αCTD is bound to NusA-AR2 and the other αCTD interacts with NusA-NTD (18).
Figure 5.
SuhB releases autoinhibition of NusA. (A and B) [1H,13C]-methyl-TROSY spectra of the titration of 25 μM [I,L,V]-NusA with SuhB. SuhB was added in a molar ratio of 1:0, black; 1:0.5, orange; 1:1, cyan; 1:2, purple; 1:3, yellow; 1:5, red (concentration of SuhB stock: 343 μM). The boxed region in (A) indicates the section as in (B). Selected signals are labeled. (C) Structure of NusA in ribbon representation. Linker regions are indicated by dashed lines. Ile, Leu, and Val residues with assigned methyl groups are shown as sticks with carbon atoms of the methyl groups as black spheres. If a methyl group was affected upon addition of SuhB (Δδnorm ≥ 0.04 ppm) the whole residue was colored in red (see also Supplementary Table S2). PDB IDs: NusA-NTD: 2KWP, SKK-AR1: 5LM9, NusA-AR2: 1WCN.
SuhB interacts weakly with RNAP
SuhB has been reported to associate with RNAP (44) and the EC (41,56), but SuhB did not coelute with RNAP alone in our gel filtration experiments (Supplementary Figure S1F). Nevertheless, we tested if SuhB can bind to RNAP by solution-state NMR spectroscopy. We recorded 1D [1H,15N]-HSQC spectra of 15N-SuhB in the absence and presence of RNAP (Supplementary Figure S7A). Addition of RNAP led to a significant decrease of 15N-SuhB signal intensity due to enhanced magnetization relaxation upon 15N-SuhB:RNAP complex formation. As binding of SuhB to RNAP was not observable in the gel filtration experiments we conclude that this interaction is only weak and transient and may be not detectable in non-equilibrium methods. To identify the interacting RNAP subunit we added individual RNAP subunits to 15N-labeled SuhB and recorded 1D or 2D [1H,15N]-HSQC spectra (Supplementary Figure S7B–E). The spectrum of 15N-SuhB was only significantly affected in the presence of the β or the β’ subunit, suggesting that SuhB might interact with either or both simultaneously.
DISCUSSION
Nus factors and their role in the proper expression of rRNA have been studied for a long time. rrn AT is a documented activity of an rrn-TAC that comprises minimally RNAP, Nus factors and nucleic acids (6). Highly efficient rrn AT, however, cannot be achieved by this ‘minimal’ rrn-TAC, but rrn AT is strongly stimulated in the presence of cell extract (6), indicating that additional factors are involved. There is, for example, solid evidence that ribosomal protein S4 plays a central role in rrn AT (40). Despite some functional similarities of λN-dependent and rrn AT (38,57), no central unstructured component such as λN has yet been identified in rrn AT. Inositol monophosphatase SuhB and translation-associated heat shock protein YbeY are also involved in rRNA biosynthesis (41,42).
Here, we show that SuhB can form a complex with a Nus factor-containing TAC halted at an rrnG AT site with the RNA harboring boxA, boxB and boxC motifs (Figure 1A). This finding is in agreement with in vivo experiments showing that a boxA element and Nus factors recruit SuhB, which then remains associated with RNAP throughout transcription of rRNA (41). S4 is involved in rrn AT (40), and we demonstrate that SuhB can also be integrated into a Nus factor- and S4-containing rrnG-TAC, but S4 interactions within this complex remain elusive (Figure 1F). We studied the interactions of SuhB with all components in this complex and found that SuhB directly interacts with the AR2 domain of NusA, but highly likely with no other Nus factor or S4 (Figures 1 and 2 and Supplementary Figures S1 and S2). E. coli SuhB has an inositol monophosphatase activity, which it exerts—unlike other inositol monophosphatases—as monomer, although it can occur in a monomer-oligomer equilibrium in solution (58). In our experiments SuhB exists as monomer, indicating that SuhB performs its function as transcription factor in its monomeric state. As there is no requirement for an inositol monophosphatase activity in E. coli (59), our findings support the proposal that widely conserved SuhB has two unrelated functions with gene regulation being the primary one (41). The SuhB:NusA-AR2 interaction is dispensable for SuhB integration into a Nus factor containing rrnG-TAC if all components are present simultaneously (Figure 1D). However, SuhB not only interacts with NusA-AR2, but it also binds to rrnG RNA, in agreement with the finding that it may act as RNA chaperone to support the formation of RNA loops formation during RNA maturation (41). Furthermore, NMR experiments indicated that SuhB can also bind to RNAP (Supplementary Figure S7), although this interaction is weak and transient as it was not observable via gel filtration (Supplementary Figure S1F). It has been reported previously that E. coli SuhB associates with elongating RNAP (41) and binds to σ factor-containing RNAP (44) and that SuhB from Pseudomonas aeroguinosa associates with RNAP in vivo (56), but nevertheless it remains to be determined if the SuhB:RNAP interaction detected via NMR spectroscopy is specific and of physiological relevance.
Taking together all findings we suggest several functions for the SuhB:NusA-AR2 interaction during rrn AT (Figure 6). First, it may be involved in the assembly of the rrn-TAC (Figure 6A). Although SuhB can be integrated into a Nus factor-modified rrn-TAC in the absence of the AR2 domain (Figure 1D), the SuhB:NusA-AR2 interaction might, nevertheless, contribute to SuhB recruitment if not all components are present at the same time, but if the rrn-TAC is assembled in a stepwise manner, as known from other TACs. The λN-dependent TAC, for example, is assembled cooperatively and involves binary interactions that are occasionally not stable when studied in isolation (5,7). However, as SuhB binds to rrnG RNA we hypothesize that the RNA:SuhB interaction is the key player in SuhB recruitment, in agreement with reports that SuhB is incorporated in a NusB/boxA-dependent manner (41). No direct interaction between SuhB and NusB could be detected in vitro (Supplementary Figures S1C and S2B), implying that this dependence is most likely indirect. Alternatively, the SuhB:NusA-AR2 interaction could promote the recruitment of other components of the rrn-TAC during the stepwise assembly of the complex, e.g. the NusB:NusE heterodimer. The NusB:NusE complex binds specifically to boxA (12,35), whereas the regions upstream and downstream of boxA constitute a binding site for NusA-SKK (46). In a λN-dependent TAC NusA-SKK and NusB:NusE can bind simultaneously to RNA (5,7) as the order of boxA and boxB is reversed as compared to rRNA operons (33). NusA is recruited early during transcription (60) and could thus hinder the access of NusB:NusE to boxA via two ways: (i) NusA could bind in its autoinhbited state to RNAP via NusA-NTD, sterically blocking access to boxA, or (ii) upon release of autoinhibition by the αCTD, NusA-SKK could bind to the regions flanking boxA, masking the NusB:NusE binding site. SuhB binds to rrnG RNA (Figure 1E), interacts with NusA-AR2 (Figure 2) and can even release autoinhibition of NusA (Figure 5), and may consequently alter the RNA binding properties of NusA. Thus, the SuhB:NusA-AR2 interaction might affect the overall conformation of NusA or directly the NusA-SKK:RNA interaction in a way that removes NusA from boxA, allowing recruitment of NusB:NusE. Finally, the SuhB:NusA-AR2 interaction may additionally stabilize the rrn-TAC during processive AT. If the SuhB:NusA-AR2 interaction remains stable throughout complete transcription of rRNA and if SuhB establishes specific contacts with RNAP remains to be determined.
Figure 6.
Scheme of the possible functions of the SuhB:NusA-AR2 interaction during rrn AT. The SuhB:NusA-AR2 interaction may be involved in the assembly of the SuhB- and Nus factor-containing rrn-TAC (A) or in suppressing Rho-dependent termination by repositioning of NusA (B). SuhB (blue) is depicted in surface, NusA-AR2 (green) in ribbon representation. Important binding sites on RNAP are highlighted in ribbon representation. The solid arrow in (A) illustrates the αCTD:NusA-AR2 interaction and thus the competition of the αCTD and SuhB for NusA-AR2, the dashed arrow in (B) indicates the putative SuhB:RNAP interaction. β-FTH: β flap tip helix, β'-CH: β' clamp helices. PDB IDs: SuhB: 2QFL, NusA-AR2: 1WCN.
Second, the SuhB:NusA-AR2 interaction could modify RNAP into a termination-resistant state (Figure 6B) via different ways: (i) SuhB could affect the positioning of NusA on RNAP. During elongation NusA-NTD interacts with the β flap tip helix at the RNA exit channel while it is concomitantly bound by one of the αCTDs (18). The second αCTD interacts with NusA-AR2, tethering NusA to the RNAP in a defined conformation with limited flexibility (18,46). As indicated by the displacement experiments the affinity of the SuhB:NusA-AR2 and the NusA-AR2: αCTD interaction is similar (Figure 3), and the concentration of SuhB is approximately half the concentration of RNAP molecules (61). Thus, SuhB might displace the αCTD from NusA-AR2 and consequently change the conformation and consequently influence the positioning of NusA, a principle reminiscent of λN-dependent AT (5,7). How this rearrangement could contribute to suppressing Rho-dependent termination remains elusive, although one may speculate that NusA blocks accession of Rho to its recruitment site on the RNA. (ii) SuhB itself could prevent recruitment of Rho by interacting with RNA and thus sterically blocking the Rho binding site of the RNA.
A recent study showed that Nus factors and SuhB can act as RNA chaperones and suggested that this function may be their major role at rRNA operons, ensuring the proper folding of 16S RNA and thus being critical for the biogenesis of the 30S subunit (43). Our results demonstrate that SuhB can be integrated into a Nus factor-containing rrn-TAC, a complex that may be involved in rrn AT as well as in post-transcriptional processes in rRNA biosynthesis. If the SuhB:NusA-AR2 interaction is required for both RNA chaperone and AT activity or just one of them needs to be determined as does the question if further factors are involved in rrn AT. More general, as boxA elements, which recruit NusB:NusE, are widespread, the regulation by Nus factors and SuhB may not be limited to rRNA operons, but may play a role in the expression of many genes in diverse bacterial species, including autoregulation of Nus factors (62).
NusA-AR1 and NusA-AR2 share a high sequence identity of 31.5% and exhibit nearly identical structures (root mean square deviation of main chain atoms: 1.2 Å) with a very similar electrostatic potential surface (20). Nevertheless, SuhB exclusively recognizes NusA-AR2 (Figure 2). This binding specificity can probably be attributed to the presence of two residues in the C-terminal helix α5 of NusA-AR2, W490 and F491. In NusA-AR1, a Leu (L414) and an Ala (A415) residue can be found at corresponding positions. NusG-NTD and the αCTD also specifically bind to NusA-AR2 (24,27), while λN only interacts with NusA-AR1 (20,22). The interaction surfaces on NusA-AR2 with αCTD, NusG-NTD, and SuhB overlap and all involve helix α5 (Supplementary Figure S4), in particular residues W490 and F491. The NusA-AR2 domain constitutes the very C-terminus of NusA and all domains are connected via linkers conferring NusA high intramolecular flexibility. When attached to RNAP via NusA-NTD and to RNA via NusA-SKK, the AR domains can still move virtually independently. This together with the high specificity suggests that NusA-AR2 may serve as versatile and efficient recruitment platform for various transcription factors in E. coli and other γ-proteobacteria. Upon recruitment, these factors may then be made available to interactions with other factors or RNAP to be integrated into regulatory transcription complexes. Moreover, NusA-AR2 shares some features with the acidic activation domains (ADs) found in eukaryotic transcription activators. Similar to NusA, these transcription factors usually exhibit a modular structure, comprising at least one DNA binding domain and one AD, the latter being essential for the interaction with other transcription factors, RNAP or coactivators (63). Generally, ADs show no apparent sequence conservation and are usually intrinsically disordered regions (IDRs) in the absence of binding partners, but, like NusA-AR2, they interact with structurally diverse binding partners (64). ADs are classified according to the preponderance of certain amino acids with the so called acidic ADs containing clusters of negatively charged amino acids (‘acidic blobs’) and consequently a negative net charge (65,66). Although acidic ADs are enriched in acidic amino acids their interaction with binding partners does usually not rely on these residues (67). Instead, specific recognition is mediated via short motifs of bulky hydrophobic residues in the IDRs that are occasionally presented on one side of one or two α-helices which form in the presence of a binding partner (68–71). This recognition mode is reminiscent of NusA-AR2 where W490 and F491, which are located on one side of helix α5 in the mainly negatively charged domain, are the key determinants for the interaction with SuhB, αCTD and NusG-NTD.
Autoinhibition is an important regulatory mechanism that is widespread in nature (72). Intramolecular interactions between different regions of a polypeptide that may even be coupled to conformational changes inhibit the function of at least one of these regions. This negative regulation limits activation of the protein/enzyme to certain physiological conditions and alters the activity of proteins involved in diverse cellular processes, ranging from transcription factors (73) to E3 ubiquitin ligases (74) and protein kinases (75). In NusA the AR2 domain binds to the KH1 domain of the RNA binding SKK motif, preventing RNA binding and thus autoinhibiting NusA to allow context-dependent RNA binding (24,25). NusA autoinhibition can be released by the αCTD (24), NusG-NTD (27) and SuhB that all bind to NusA-AR2 (Figure 5, Supplementary Figures S5 and S6). The ability of NusA-AR2 to specifically recognize various other transcription factors and the fact that these interactions can activate NusA underline the central role of NusA in transcription regulation.
DATA AVAILABILITY
We generated a model of the SuhB:NusA-AR2 complex. Coordinates of SuhB and NusA-AR2 are available in the PDB (2QFL, 1WCN). The coordinates of the best model are provided as Supplementary Data.
Resonance assignments of NusA-AR2, NusA-AR1-AR2, NusA-SKK, NusA-NTD and αCTD were taken from previous studies as indicated in the manuscript.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Paul Rösch for helpful discussions, Ramona Heissmann, Ulrike Persau and Andrea Hager for excellent technical assistance, and the Northern Bavarian NMR Centre (NBNZ) for access to the NMR spectrometers.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Research Foundation [Ro617/21-1 to P.R.]. Funding for open access charge: German Research Foundation and the University of Bayreuth in the funding program Open Access Publishing.
Conflict of interest statement. None declared.
REFERENCES
- 1. Gottesman M.E., Weisberg R.A.. Little lambda, who made thee. Microbiol. Mol. Biol. Rev. 2004; 68:796–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Roberts J.W. RNA and protein elements of E. coli and lambda transcription antitermination complexes. Cell. 1993; 72:653–655. [DOI] [PubMed] [Google Scholar]
- 3. de Crombrugghe B., Mudryj M., DiLauro R., Gottesman M.. Specificity of the bacteriophage lambda N gene product (pN): nut sequences are necessary and sufficient for antitermination by pN. Cell. 1979; 18:1145–1151. [DOI] [PubMed] [Google Scholar]
- 4. Olson E.R., Flamm E.L., Friedman D.I.. Analysis of nutR: a region of phage lambda required for antitermination of transcription. Cell. 1982; 31:61–70. [DOI] [PubMed] [Google Scholar]
- 5. Said N., Krupp F., Anedchenko E., Santos K.F., Dybkov O., Huang Y.-H., Lee C.-T., Loll B., Behrmann E., Bürger J. et al.. Structural basis for λN-dependent processive transcription antitermination. Nat. Microbiol. 2017; 2:17062. [DOI] [PubMed] [Google Scholar]
- 6. Squires C.L., Greenblatt J., Li J., Condon C., Squires C.L.. Ribosomal RNA antitermination in vitro: requirement for Nus factors and one or more unidentified cellular components. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:970–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Krupp F., Said N., Huang Y.-H., Loll B., Bürger J., Mielke T., Spahn C.M.T., Wahl M.C.. Structural basis for the action of an all-purpose transcription anti-termination factor. Mol. Cell. 2019; 74:143–157. [DOI] [PubMed] [Google Scholar]
- 8. Werner F. A nexus for gene expression-molecular mechanisms of Spt5 and NusG in the three domains of life. J. Mol. Biol. 2012; 417:13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mooney R.A., Schweimer K., Rösch P., Gottesman M., Landick R.. Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. J. Mol. Biol. 2009; 391:341–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burmann B.M., Schweimer K., Luo X., Wahl M.C., Stitt B.L., Gottesman M.E., Rösch P.. A NusE:NusG complex links transcription and translation. Science. 2010; 328:501–504. [DOI] [PubMed] [Google Scholar]
- 11. Mitra P., Ghosh G., Hafeezunnisa M., Sen R.. Rho protein: roles and mechanisms. Annu. Rev. Microbiol. 2017; 71:687–709. [DOI] [PubMed] [Google Scholar]
- 12. Burmann B.M., Luo X., Rösch P., Wahl M.C., Gottesman M.E.. Fine tuning of the E. coli NusB:NusE complex affinity to BoxA RNA is required for processive antitermination. Nucleic Acids Res. 2010; 38:314–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Luo X., Hsiao H.-H., Bubunenko M., Weber G., Court D.L., Gottesman M.E., Urlaub H., Wahl M.C.. Structural and functional analysis of the E. coli NusB-S10 transcription antitermination complex. Mol. Cell. 2008; 32:791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Friedman D.I., Baron L.S.. Genetic characterization of a bacterial locus involved in the activity of the N function of phage lambda. Virology. 1974; 58:141–148. [DOI] [PubMed] [Google Scholar]
- 15. Vogel U., Jensen K.F.. NusA is required for ribosomal antitermination and for modulation of the transcription elongation rate of both antiterminated RNA and mRNA. J. Biol. Chem. 1997; 272:12265–12271. [DOI] [PubMed] [Google Scholar]
- 16. Pan T., Artsimovitch I., Fang X.W., Landick R., Sosnick T.R.. Folding of a large ribozyme during transcription and the effect of the elongation factor NusA. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:9545–9550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Artsimovitch I., Landick R.. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:7090–7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guo X., Myasnikov A.G., Chen J., Crucifix C., Papai G., Takacs M., Schultz P., Weixlbaumer A.. Structural basis for nusa stabilized transcriptional pausing. Mol. Cell. 2018; 69:816–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Worbs M., Bourenkov G.P., Bartunik H.D., Huber R., Wahl M.C.. An extended RNA binding surface through arrayed S1 and KH domains in transcription factor NusA. Mol. Cell. 2001; 7:1177–1189. [DOI] [PubMed] [Google Scholar]
- 20. Eisenmann A., Schwarz S., Prasch S., Schweimer K., Rösch P.. The E. coli NusA carboxy-terminal domains are structurally similar and show specific RNAP- and lambdaN interaction. Protein Sci. 2005; 14:2018–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mah T.F., Li J., Davidson A.R., Greenblatt J.. Functional importance of regions in Escherichia coli elongation factor NusA that interact with RNA polymerase, the bacteriophage lambda N protein and RNA. Mol. Microbiol. 1999; 34:523–537. [DOI] [PubMed] [Google Scholar]
- 22. Prasch S., Schwarz S., Eisenmann A., Wöhrl B.M., Schweimer K., Rösch P.. Interaction of the intrinsically unstructured phage lambda N Protein with Escherichia coli NusA. Biochemistry. 2006; 45:4542–4549. [DOI] [PubMed] [Google Scholar]
- 23. Bonin I., Mühlberger R., Bourenkov G.P., Huber R., Bacher A., Richter G., Wahl M.C.. Structural basis for the interaction of Escherichia coli NusA with protein N of phage lambda. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:13762–13767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schweimer K., Prasch S., Sujatha P.S., Bubunenko M., Gottesman M.E., Rösch P.. NusA interaction with the α subunit of E. coli RNA polymerase is via the UP element site and releases autoinhibition. Structure. 2011; 19:945–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mah T.F., Kuznedelov K., Mushegian A., Severinov K., Greenblatt J.. The alpha subunit of E. coli RNA polymerase activates RNA binding by NusA. Genes Dev. 2000; 14:2664–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu K., Zhang Y., Severinov K., Das A., Hanna M.M.. Role of Escherichia coli RNA polymerase alpha subunit in modulation of pausing, termination and anti-termination by the transcription elongation factor NusA. EMBO J. 1996; 15:150–161. [PMC free article] [PubMed] [Google Scholar]
- 27. Strauß M., Vitiello C., Schweimer K., Gottesman M., Rösch P., Knauer S.H.. Transcription is regulated by NusA:NusG interaction. Nucleic Acids Res. 2016; 44:5971–5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Arnvig K.B., Zeng S., Quan S., Papageorge A., Zhang N., Villapakkam A.C., Squires C.L.. Evolutionary comparison of ribosomal operon antitermination function. J. Bacteriol. 2008; 190:7251–7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bremer H., Dennis P.P.. Modulation of chemical composition and other parameters of the cell by growth rate. Escherichia coli and Salmonella: Cellular and Molecular Biology. 1996; 2:Washington DC: ASM Press; 1553–1569. [Google Scholar]
- 30. Li S.C., Squires C.L., Squires C.. Antitermination of E. coli rRNA transcription is caused by a control region segment containing lambda nut-like sequences. Cell. 1984; 38:851–860. [DOI] [PubMed] [Google Scholar]
- 31. Pfeiffer T., Hartmann R.K.. Role of the spacer boxA of Escherichia coli ribosomal RNA operons in efficient 23 S rRNA synthesis in vivo. J. Mol. Biol. 1997; 265:385–393. [DOI] [PubMed] [Google Scholar]
- 32. Heinrich T., Condon C., Pfeiffer T., Hartmann R.K.. Point mutations in the leader boxA of a plasmid-encoded Escherichia coli rrnB operon cause defective antitermination in vivo. J. Bacteriol. 1995; 177:3793–3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Berg K.L., Squires C., Squires C.L.. Ribosomal RNA operon anti-termination. Function of leader and spacer region box B-box A sequences and their conservation in diverse micro-organisms. J. Mol. Biol. 1989; 209:345–358. [DOI] [PubMed] [Google Scholar]
- 34. Friedman D.I., Gottesman M.E.. Hendrix RW, Roberts JW, Stahl F, Weisberg RA. Lytic mode of lambda development. Lambda II. 1983; 13:NY: Cold Spring Harbor Laboratory Press; 21–51. [Google Scholar]
- 35. Greive S.J., Lins A.F., von Hippel P.H.. Assembly of an RNA-protein complex. Binding of NusB and NusE (S10) proteins to boxA RNA nucleates the formation of the antitermination complex involved in controlling rRNA transcription in Escherichia coli. J. Biol. Chem. 2005; 280:36397–36408. [DOI] [PubMed] [Google Scholar]
- 36. DeVito J., Das A.. Control of transcription processivity in phage lambda: Nus factors strengthen the termination-resistant state of RNA polymerase induced by N antiterminator. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:8660–8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Beuth B., Pennell S., Arnvig K.B., Martin S.R., Taylor I.A.. Structure of a Mycobacterium tuberculosis NusA-RNA complex. EMBO J. 2005; 24:3576–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li J., Horwitz R., McCracken S., Greenblatt J.. NusG, a new Escherichia coli elongation factor involved in transcriptional antitermination by the N protein of phage lambda. J. Biol. Chem. 1992; 267:6012–6019. [PubMed] [Google Scholar]
- 39. Condon C., Squires C., Squires C.L.. Control of rRNA transcription in Escherichia coli. Microbiol. Rev. 1995; 59:623–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Torres M., Condon C., Balada J.M., Squires C., Squires C.L.. Ribosomal protein S4 is a transcription factor with properties remarkably similar to NusA, a protein involved in both non-ribosomal and ribosomal RNA antitermination. EMBO J. 2001; 20:3811–3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Singh N., Bubunenko M., Smith C., Abbott D.M., Stringer A.M., Shi R., Court D.L., Wade J.T.. SuhB associates with nus factors to facilitate 30s ribosome biogenesis in escherichia coli. MBio. 2016; 7:e00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Grinwald M., Ron E.Z.. The Escherichia coli translation-associated heat shock protein YbeY is involved in rRNA transcription antitermination. PLoS ONE. 2013; 8:e62297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bubunenko M., Court D.L., Al Refaii A., Saxena S., Korepanov A., Friedman D.I., Gottesman M.E., Alix J.-H.. Nus transcription elongation factors and RNase III modulate small ribosome subunit biogenesis in Escherichia coli. Mol. Microbiol. 2013; 87:382–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang Y., Stieglitz K.A., Bubunenko M., Court D.L., Stec B., Roberts M.F.. The structure of the R184A mutant of the inositol monophosphatase encoded by suhB and implications for its functional interactions in Escherichia coli. J. Biol. Chem. 2007; 282:26989–26996. [DOI] [PubMed] [Google Scholar]
- 45. Drögemüller J., Strauß M., Schweimer K., Wöhrl B.M., Knauer S.H., Rösch P.. Exploring RNA polymerase regulation by NMR spectroscopy. Sci. Rep. 2015; 5:10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Prasch S., Jurk M., Washburn R.S., Gottesman M.E., Wöhrl B.M., Rösch P.. RNA-binding specificity of E. coli NusA. Nucleic Acids Res. 2009; 37:4736–4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Burmann B.M., Scheckenhofer U., Schweimer K., Rösch P.. Domain interactions of the transcription-translation coupling factor Escherichia coli NusG are intermolecular and transient. Biochem. J. 2011; 435:783–789. [DOI] [PubMed] [Google Scholar]
- 48. Zuber P.K., Artsimovitch I., NandyMazumdar M., Liu Z., Nedialkov Y., Schweimer K., Rösch P., Knauer S.H.. The universally-conserved transcription factor RfaH is recruited to a hairpin structure of the non-template DNA strand. Elife. 2018; 7:e36349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Meyer O., Schlegel H.G.. Biology of aerobic carbon monoxide-oxidizing bacteria. Annu. Rev. Microbiol. 1983; 37:277–310. [DOI] [PubMed] [Google Scholar]
- 50. Sambrook J., Russel D.W.. Molecular Cloning: A Laboratory Manual Third Edition. 2001; NY: Cold Spring Harbor Press. [Google Scholar]
- 51. Sprangers R., Kay L.E.. Quantitative dynamics and binding studies of the 20S proteasome by NMR. Nature. 2007; 445:618–622. [DOI] [PubMed] [Google Scholar]
- 52. Drögemüller J., Strauß M., Schweimer K., Jurk M., Rösch P., Knauer S.H.. Determination of RNA polymerase binding surfaces of transcription factors by NMR spectroscopy. Sci. Rep. 2015; 5:16428–16441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Artsimovitch I., Landick R.. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell. 2002; 109:193–203. [DOI] [PubMed] [Google Scholar]
- 54. Hartl M.J., Kretzschmar B., Frohn A., Nowrouzi A., Rethwilm A., Wöhrl B.M.. AZT resistance of simian foamy virus reverse transcriptase is based on the excision of AZTMP in the presence of ATP. Nucleic Acids Res. 2008; 36:1009–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yan Y., Zhang D., Zhou P., Li B., Huang S.-Y.. HDOCK: a web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017; 45:W365–W373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shi J., Jin Y., Bian T., Li K., Sun Z., Cheng Z., Jin S., Wu W.. SuhB is a novel ribosome associated protein that regulates expression of MexXY by modulating ribosome stalling in Pseudomonas aeruginosa. Mol. Microbiol. 2015; 98:370–383. [DOI] [PubMed] [Google Scholar]
- 57. Torres M., Balada J.-M., Zellars M., Squires C., Squires C.L.. In vivo effect of NusB and NusG on rRNA transcription antitermination. J. Bacteriol. 2004; 186:1304–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chen L., Roberts M.F.. Overexpression, purification, and analysis of complementation behavior of E. coli SuhB protein: comparison with bacterial and archaeal inositol monophosphatases. Biochemistry. 2000; 39:4145–4153. [DOI] [PubMed] [Google Scholar]
- 59. Kozloff L.M., Turner M.A., Arellano F., Lute M.. Phosphatidylinositol, a phospholipid of ice-nucleating bacteria. J. Bacteriol. 1991; 173:2053–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mooney R.A., Davis S.E., Peters J.M., Rowland J.L., Ansari A.Z., Landick R.. Regulator trafficking on bacterial transcription units in vivo. Mol. Cell. 2009; 33:97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Schmidt A., Kochanowski K., Vedelaar S., Ahrné E., Volkmer B., Callipo L., Knoops K., Bauer M., Aebersold R., Heinemann M.. The quantitative and condition-dependent Escherichia coli proteome. Nat. Biotechnol. 2016; 34:104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Baniulyte G., Singh N., Benoit C., Johnson R., Ferguson R., Paramo M., Stringer A.M., Scott A., Lapierre P., Wade J.T.. Identification of regulatory targets for the bacterial Nus factor complex. Nat. Commun. 2017; 8:2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988; 335:683–689. [DOI] [PubMed] [Google Scholar]
- 64. Martchenko M., Levitin A., Whiteway M.. Transcriptional activation domains of the Candida albicans Gcn4p and Gal4p homologs. Eukaryotic Cell. 2007; 6:291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ma J., Ptashne M.. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987; 48:847–853. [DOI] [PubMed] [Google Scholar]
- 66. Hope I.A., Struhl K.. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986; 46:885–894. [DOI] [PubMed] [Google Scholar]
- 67. Brzovic P.S., Heikaus C.C., Kisselev L., Vernon R., Herbig E., Pacheco D., Warfield L., Littlefield P., Baker D., Klevit R.E. et al.. The acidic transcription activator Gcn4 binds the mediator subunit Gal11/Med15 using a simple protein interface forming a fuzzy complex. Mol. Cell. 2011; 44:942–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Uesugi M., Nyanguile O., Lu H., Levine A.J., Verdine G.L.. Induced alpha helix in the VP16 activation domain upon binding to a human TAF. Science. 1997; 277:1310–1313. [DOI] [PubMed] [Google Scholar]
- 69. Radhakrishnan I., Pérez-Alvarado G.C., Parker D., Dyson H.J., Montminy M.R., Wright P.E.. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell. 1997; 91:741–752. [DOI] [PubMed] [Google Scholar]
- 70. Drysdale C.M., Dueñas E., Jackson B.M., Reusser U., Braus G.H., Hinnebusch A.G.. The transcriptional activator GCN4 contains multiple activation domains that are critically dependent on hydrophobic amino acids. Mol. Cell Biol. 1995; 15:1220–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cress W.D., Triezenberg S.J.. Critical structural elements of the VP16 transcriptional activation domain. Science. 1991; 251:87–90. [DOI] [PubMed] [Google Scholar]
- 72. Pufall M.A., Graves B.J.. Autoinhibitory domains: modular effectors of cellular regulation. Annu. Rev. Cell Dev. Biol. 2002; 18:421–462. [DOI] [PubMed] [Google Scholar]
- 73. Burmann B.M., Knauer S.H., Sevostyanova A., Schweimer K., Mooney R.A., Landick R., Artsimovitch I., Rösch P.. An α helix to β barrel domain switch transforms the transcription factor RfaH into a translation factor. Cell. 2012; 150:291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Buetow L., Huang D.T.. Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2016; 17:626–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Au-Yeung B.B., Shah N.H., Shen L., Weiss A.. ZAP-70 in signaling, biology, and disease. Annu. Rev. Immunol. 2018; 36:127–156. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We generated a model of the SuhB:NusA-AR2 complex. Coordinates of SuhB and NusA-AR2 are available in the PDB (2QFL, 1WCN). The coordinates of the best model are provided as Supplementary Data.
Resonance assignments of NusA-AR2, NusA-AR1-AR2, NusA-SKK, NusA-NTD and αCTD were taken from previous studies as indicated in the manuscript.








