Abstract
Cell diversity in multicellular organisms relies on coordination between cell proliferation and the acquisition of cell identity. The equilibrium between these two processes is essential to assure the correct number of determined cells at a given time at a given place. Using genetic approaches and correlative microscopy, we show that Tramtrack-69 (Ttk69, a Broad-complex, Tramtrack and Bric-à-brac - Zinc Finger (BTB-ZF) transcription factor ortholog of the human promyelocytic leukemia zinc finger factor) plays an essential role in controlling this balance. In the Drosophila bristle cell lineage, which produces the external sensory organs composed by a neuron and accessory cells, we show that ttk69 loss-of-function leads to supplementary neural-type cells at the expense of accessory cells. Our data indicate that Ttk69 (1) promotes cell cycle exit of newborn terminal cells by downregulating CycE, the principal cyclin involved in S-phase entry, and (2) regulates cell-fate acquisition and terminal differentiation, by downregulating the expression of hamlet and upregulating that of Suppressor of Hairless, two transcription factors involved in neural-fate acquisition and accessory cell differentiation, respectively. Thus, Ttk69 plays a central role in shaping neural cell lineages by integrating molecular mechanisms that regulate progenitor cell cycle exit and cell-fate commitment.
Keywords: Ttk69, cyclin-E, Hamlet, precursor cells, sensory organs
ORGANISMS are composed of morphologically and functionally distinct cell types. Such cell diversity is generated from a restricted set of precursor cells producing a limited number of differentiated cells. Division of precursor cells gives rise to daughter cells that differentiate and acquire specific fates. The transit from a proliferative to cell cycle-arrested state during this process is tightly regulated and requires changes in transcriptional programs. Disentangling the molecular mechanisms that control the balance between proliferation and differentiation is essential for understanding the formation and maintenance of organisms, as well as human diseases, such as cancer, in which this process is disturbed.
Broad-complex, Tramtrack and Bric-à-brac - Zinc Finger (BTB-ZF) transcription factors are involved in a wide variety of biological processes (Kelly and Daniel 2006). They include Drosophila Broad-complex factors (BR-C), Bric-à-brac (Bab), and several pox virus zinc finger proteins (Chaharbakhshi and Jemc 2016). All possess a protein–protein interaction motif (BTB/POZ) at the N-terminus that allows protein homo- and multimerization, and one or several zinc finger DNA-binding motifs (Bonchuk et al. 2011). These proteins are conserved from Saccharomyces cerevisiae to Homo sapiens, and act as transcriptional repressors or activators, depending on the BTB domain (Siggs and Beutler 2012). The founding BTB-ZF members are all Drosophila transcriptional repressors that regulate processes such as metamorphosis, ovary development, and neurogenesis (Karim et al. 1993; Guo et al. 1995; Sahut-Barnola et al. 1995). In vertebrates, the human BTB-ZF, promyelocytic leukemia zinc finger (PLZF), acts as a tumor suppressor maintaining cell growth inhibition and quiescence by transcriptional repression of the c-myc proto-oncogene (McConnell et al. 2003). Accordingly, plzf loss-of-function has been correlated with prostate and lung cancer (Jin et al. 2017). Moreover, this factor regulates organogenesis by controlling the balance between self-renewal and the differentiation of neural stem cells (Sobieszczuk et al. 2010; Gaber et al. 2013). Overall, BTB-ZF proteins have fundamental and conserved roles during development, controlling cell proliferation and differentiation.
The Drosophila ortholog of PLZF, Tramtrack (Ttk), also plays multiple roles during development, including cell proliferation and cell-fate decisions in the nervous system, intestinal stem cells, photoreceptors, and tracheal cells (Giesen et al. 1997; Lai and Li 1999; Badenhorst 2001; Araujo et al. 2007; Wang et al. 2015; Liu et al. 2016). In particular, Ttk is a key regulator of cell fate in the peripheral nervous system, in which it promotes nonneural instead of neural fates (Guo et al. 1995). Based on initial studies on the even skipped and fushi tarazu (ftz) genes, Ttk is considered to be a transcriptional repressor (Harrison and Travers 1988, 1990; Brown et al. 1991). The ttk locus encodes two proteins, Ttk69 and Ttk88, via alternative splicing (Read and Manley 1992). Both isoforms share a common conserved N-terminal BTB/POZ domain but contain divergent C-terminal zinc finger Cis2His2-like fold group (C2H2)-type domains for DNA binding, conferring specific DNA-binding and probably independent functions for each isoform (Read and Manley 1992). In spite of these differences, Ttk69 appears to have a broader spectrum of functions than Ttk88. For example, Ttk69, but not Ttk88, promotes specific nonneuronal fates such as cone cells during eye development (Lai and Li 1999) and enteroblast cells during intestine development (Wang et al. 2015). Ttk has been also shown to be involved in cell cycle regulation. It has been shown that overexpression of Ttk69, but not Ttk88, causes complete loss of mitosis in the eye disc morphogenetic furrow through the repression of String, the positive regulator of the G2/M transition (Baonza et al. 2002). Similarly, Ttk69 negatively regulates intestinal stem cell proliferation (Wang et al. 2015). Finally, in mechanosensory organs, ttk loss-of-function leads to the complete transformation of sensory cells into neurons (Guo et al. 1995) and the loss of Ttk69 alone also induces cell proliferation (Audibert et al. 2005). To dig into the mechanisms by which Ttk69 controls cell determination and division, we focus on how Ttk69 controls the balance between cell proliferation and the acquisition of cell fate in the bristle system. To this end, we extensively used the ttk1e11 allele, a P-element insertion flanked by a deletion in the ttk locus (Xiong and Montell 1993), which is considered to be a specific null allele for ttk69 (Lai and Li 1999).
The Drosophila external mechanosensory organs, or bristles, are an excellent model system to study the balance between proliferative and determined states of progenitor cells (Fichelson et al. 2005). Each bristle is composed of a shaft and an annular cuticular structure, called the socket, at its base. Bristles are formed during the pupal stage from four specialized cells with a common origin: two outer cells, the socket and shaft cells, and two inner cells, the neuron and the sheath cell (Hartenstein and Posakony 1989). Each cell differs from the other by its size, relative position, and expression of specific markers (Figure 1A and Supplemental Material, Movie S1). They arise from the division of a primary precursor cell (or pI) after a stereotypical sequence of four asymmetric cell divisions (the bristle cell lineage). In the dorsal thorax (notum), pI cells divide to generate a posterior secondary precursor cell (pIIa) and an anterior secondary precursor cell (pIIb). The division of pIIa leads to the formation of the outer cells (the pIIa sublineage), whereas the pIIb cell gives rise to the inner cells (the pIIb sublineage), following two rounds of division. First, pIIb divides to give rise to a glial cell that enters apoptosis shortly after birth and a tertiary precursor cell, pIIIb. Then, pIIIb divides to produce the sheath and the neuron (Gho et al. 1999; Fichelson and Gho 2003). At each of these divisions, the Notch (N) pathway is differentially activated in only one daughter cell. This differential activation ensures the acquisition of different fates by both daughter cells (Guo et al. 1996). As such, the N pathway does not specify particular identities, but its activation triggers different outcomes depending on the cellular context, likely in cooperation with other factors that specify cell fate (Ramat et al. 2016). Only some of these specific factors are known in the bristle lineage. Two are Sequoia (Seq) and Hamlet (Ham), two zinc finger transcription factors, expressed in pIIb sublineage cells, that have a critical role in the acquisition of inner-cell identity (Moore et al. 2002, 2004; Andrews et al. 2009). Indeed, sensory organs (SOs) in ham and seq mutants are composed of external cells only, due to respecification of the inner cells. Moreover, it has also been shown that Seq controls ham expression, indicating that these factors are related in a complex regulatory network of transcription factors (Andrews et al. 2009).
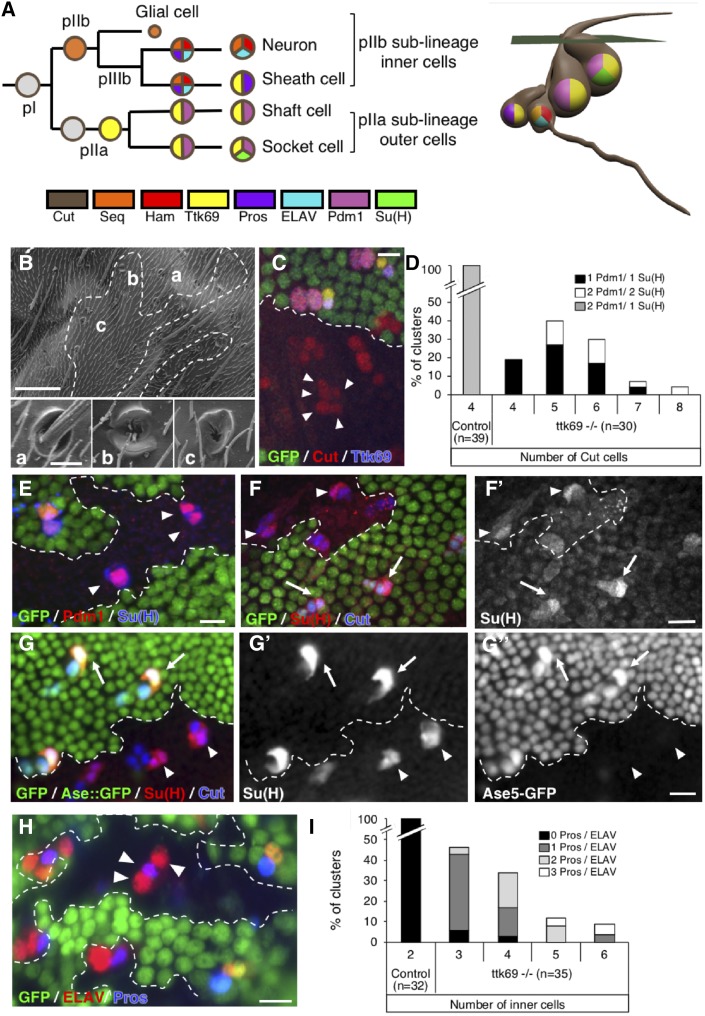
Figure 1.
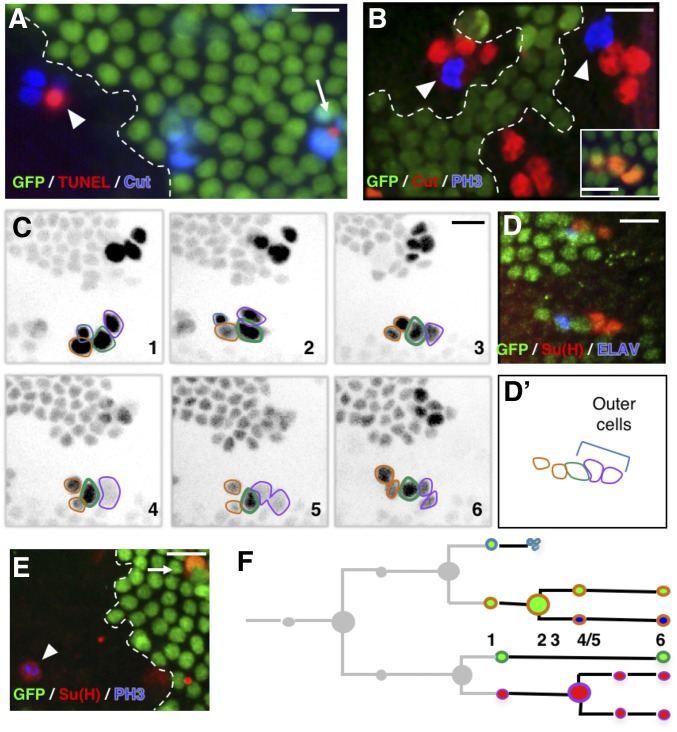
Ttk69 loss-of-function leads to extraterminal cells in SOs. (A) Scheme of the wild-type bristle lineage (left) and of an SO (right). Nuclei are represented by circles and cell markers by specific colors. (B) Scanning electron micrograph of Ttk69 SO at the external level. The Ttk69 clone is outlined by a white dashed line. Control (a) and Ttk69-mutant SOs (b and c). (C and E–H) Ttk69-mutant SO at the cellular level. Ttk69 clones were detected by the absence of GFP and outlined by a white dashed line. Pupae were at 28-hr APF, except in (F) where they were at 23-hr APF. (C) Ttk69-mutant SOs were composed of more than four cells (arrowheads). Sensory cells were identified by Cut (red) and Ttk69 (blue) immunoreactivity. (D) Histogram showing the percentage of SOs harboring four-to-eight cut positive cells in control and mutant SOs. The percentages of SOs with different combinations of Pdm1- and Su(H)-positive cells are shown (black bar, only one Pdm1/Su(H)-positive cell; white bar, two Pdm1/Su(H)-positive cells; and gray bar, 1 Su(H)-positive cell among two Pdm1-positive cells). (E) Outer cells acquired a socket fate. All outer cells [specifically marked by Pdm1 (red)] expressed the socket marker Su(H) (blue) in Ttk69-mutant SOs (arrowheads). (F and G) Autoamplification of Su(H) was impaired in Ttk69-mutant SOs. (F and F’) In 23-hr APF pupae, accumulation of Su(H) protein [F’ and red in (F)] was similar in Ttk69-mutant (arrowheads) and control SOs (arrows). (G–G”) In 28-hr-old APF pupae, Su(H) immunoreactivity [G’ and red in (G)] and Su(H) autoamplification assessed by the ASE5::GFP reporter [G’’ and green in (G)]. Note that Su(H) autoamplification was absent. (H) More than two inner cells (arrowheads) are present in the Ttk69-mutant SO. Inner cells are revealed by ELAV (red) and Pros (blue) immunoreactivity. (I) Histogram showing the percentage of SOs harboring two-to-six pIIb cells in control and mutant SOs. The percentage of clusters with zero (black bars), one (dark gray bars), two (light gray bars), or three (white bars) inner cells positive for both Pros and ELAV immunoreactivity (Pros/ELAV) is indicated. Bar, 50 μm in (B), inset 5 μm; 10 μm in (C and E–H). APF, after pupal formation; SO, sensory organ.
Here, we use the bristle lineage to explore how Ttk69 coordinates terminal cell determination and cell cycle arrest. We show that loss of ttk69 leads to the production of supernumerary progenitor cells and the respecification of cell-fate identity. Notably, we observed a cell transformation in which an outer cell acquired an inner-cell precursor identity; precisely, the pIIa shaft cell adopts a pIIIb cell fate. Since outer and inner cells are cousin cells, this kind of transformation has been previously named cousin–cousin cell transformation (Moore et al. 2004). We identified the CycE gene, encoding the essential cyclin required for entry into S phase, as a Ttk69 downstream gene. In addition, we show that Ttk69 regulates cell-fate acquisition and terminal differentiation by controlling the expression of ham and Suppressor of Hairless (Su(H)), which encodes the transducing transcription factor of N receptor signaling. We propose that Ttk69 is a central node of a transcriptional regulatory network that assures cell lineage completion by controlling the acquisition of terminal cell fates and the arrest of cell proliferation.
Materials and Methods
CycE reporter constructions
To establish transgenic fly strains bearing CycE transcriptional reporters, the eve promoter of the eve.p-LacZ.attB construct (Liberman and Stathopoulos 2009) was replaced with part of the CycE promoter from the 16,4 lacZ construct engineered by H. Richardson (Jones et al. 2000), which contains the full-length CycE promoter region. The 4,6WT fragment covers the Kpn1-Xho1 proximal region of the full-length CycE promoter. All other constructs were derived from the 4,6WT construct, with deletion of the Kpn1-BssHII and Ale1-Nco1 fragments in the ΔAC-lacZ construct, deletion of the BssHII-Ale1 fragment in the ΔB-lacZ construct, and deletion of the Nco1-Xho1 fragment in the ΔD construct. To generate the 4,6m-lacZ promoter bearing mutated Ttk69-binding sites, we used the QuickChange Multi Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA). Experiments were performed following the manufacturer’s instructions. For each Ttk-binding site, specific primers carrying the ACTGC sequence (underlined in the sequence) to replace the canonical AGGAC sequence were generated and are listed from the more proximal to the more distal Ttk-binding sites: 5′-GGCATGTTAAAACTGCTGTTTTAGAACTCAGC-3′; 5′-GCATATGCATGCCACTGCAAAGGAGCCG-3′; 5′-GACGCAGAACAACTGCAGAAGGCGTCG-3′; 5′-GATGTCCCAAAAAGTAGACACTGCTTTAGCTA-3′; 5′-TAAATGTTATCAAACTGCTTGGGGGAGAATTG-3′; 5′-CGACACATAAGCGCACTGCTTTATGGG-3′; 5′-GGGGCACCACTGCATCGAGTATTGAGG-3′; ανδ 5′-CGAGATGCGAACTGCGATTGCAGCAGC-5′. Clones obtained were sequenced and wild-type fragments of the 4,6WT-lacZ construct were replaced after enzymatic digestion by mutated fragments to generate the 4,6m-lacZ construct. Transgenic flies were generated by BestGene (Chino Hills, CA). All constructs were inserted at the same locus, attP40 on chromosome II, using ΦC31 integrase-based tools to avoid expression variations due to genomic environment.
Fly strains
Somatic clones were obtained using the FLP/FRT recombination system (Xu and Rubin 1993). The y, w; FRT82B ttk1e11/CyO^SM5 line [41754 Bloomington Drosophila Stock Center (BDSC)] was crossed with the y, w, Ubx-FLP; FRT82B ubi-nls::GFP line (gift from J. Knoblich) to generate ttk69-null somatic clones. Somatic ham and seq clones were generated using the y, w; FRT40A ham1/CyO^SM5 line (gift from Y. N. Jan) crossed with y, w, Ubx-FLP; FRT40A ubi-nls::GFP and y, w; FRT42D seqA41/CyO^SM5 (gift from H. Bellen) crossed to y, w; UbxFLP, FRT42A ubi-nls::GFP, respectively.
Analysis of the Ttk69 loss-of-function on a ham heterozygous background was obtained using a y, w; FRT40A ham1; FRT82B ttk1e1/CyO^SM5 fly crossed with y, w, UbxFLP; FRT82B ubi-nls::GFP. Analysis of the Ttk69 loss-of-function on a CycE heterozygous background was obtained using a y, w; CycEAR95; FRT82B ttk1e11/CyO^SM5 crossed with y, w, Ubx-FLP; FRT82B ubi-nls::GFP. To study Su(H) autoamplification under Ttk69 loss-of-function conditions, the line y, w; ASE5-GFP; FRT82B ttk1e11/CyO^SM5 was crossed with y, w, Ubx-FLP; FRT82B ubi-nls::GFP. To analyze the CycE promoter, the following CycE transcriptional reporter lines were used: y, w; CycE-4.6-lacZ, y, w; CycE-4,6m-lacZ, y, w; CycE-ΔAC-lacZ, y, w; CycE-ΔB-lacZ, y, w; CycE-ΔD-lacZ, and CycE-D-lacZ corresponding to the 2,9 construct described (and a gift from H. Richardson). These CycE transcriptional reporters were analyzed under Ttk69 loss-of-function conditions in pupae at 28-hr after pupal formation (APF) in lines obtained after crossing y, w, Ubx-FLP; FRT 82B, nls-GFP/TM6 Tb with the following lines: 4,6WT-lacZ; FRT82B ttk1e11/TM6 Tb, ΔCm-lacZ; FRT82B ttk1e11/TM6 Tb, ΔD-lacZ; FRT82B ttk1e11/TM6 Tb, and D-lacZ; FRT82B ttk1e11/TM6 Tb.
The GAL4/UAS (upstream activating sequence) expression system (Brand and Perrimon 1993) was used to express the following UAS constructions in the mechanosensory bristle cell lineage using, as a GAL4 driver, the line neuralizedp72-Gal4 (neurp72) (Bellaïche et al. 2001): UAS-histone H2B::YFP (UAS-H2B::YFP) (Bellaïche et al. 2001), UAS-tramtrack69 (UAS-ttk69) (Badenhorst 2001), UAS-hamlet (UAS-ham), UAS-sequoia (UAS-seq) (gift from H. Bellen), UAS-TrlRNAi (BDSC, UAS-TRiP.GL00699 and UAS-TRiP.HMS02188), UAS-MEP1RNAi [BDSC, UAS-TRiP.GL00319; Vienna Drosophila Resource Center (VDRC), identifier: 24534] and UAS-Mi-2RNAi (VDRC, identifier: 107204). We used the temperature-conditional line UAS-H2B::YFP; neurp72, tub-GAL80ts to overexpress these constructs late during SO formation. Fly crosses were carried out at 18° and pupae were transferred to 30° at 21-hr APF. Pupae were fixed and dissected 7-hr later. Genotypes used in the figures are recapitulated in Table S1.
Immunohistology
Pupal nota were dissected at 17–32-hr APF and processed as previously described (Gho et al. 1996). Primary antibodies were: mouse anti-Cut [#2B10; 1:500; Developmental Studies Hybridoma Bank (DSHB)]; rabbit anti-β-galactosidase (β-Gal) (#55976; 1:500; Cappel); rabbit anti-GFP (#sc-8334; 1:500; Santa Cruz Biotechnology); mouse anti-GFP (No 11 814 460 001, 1:500; Roche), rabbit anti-Pdm1 (gift from T. Préat; École Supérieure de Physique et de Chimie Industrielles, Paris, France; 1:200), rabbit anti-Ttk69 (gift from A. Travers; Medical Research Council, Cambridge, United Kingdom; 1:500), rabbit anti-Ham (gift from Y. N. Jan; Howard Hughes Medical Institute, San Francisco; 1:500), rabbit anti-Seq (gift from H. Bellen; Baylor College of Medicine, Houston; 1:500); rat anti-ELAV (#7E8A10; 1:10; DSHB); mouse anti-ELAV (#9F8A9; 1:100; DSHB); mouse anti-Prospero (Pros) (gift from C. Doe; Institute of Neuroscience, University of Oregon, Eugene; 1:5), rat anti-Su(H) (gift from F. Schweisguth; Institut Pasteur, Paris, France 1:500), and rabbit anti-phospho-Histone H3 (06–510; Upstate; 1:10,000). Alexa 488-conjugated secondary anti-mouse (#A11029), anti-rat (#A11006), and anti-rabbit (#A11034) and Alexa 568-conjugated secondary anti-mouse (#A11031), anti-rat (#A11077), and anti-rabbit (#A11011) antibodies were purchased from Molecular Probes (Eugene, OR) and used at 1:1000. Cy5-conjugated anti-mouse (#715-175-151), anti-rat (#712-175-153), or anti-rabbit (#711-175-152) antibodies were purchased from Jackson Immunoresearch and were used at 1:2000. DNA fragmentation was assayed by terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) and performed as previously described (Fichelson and Gho 2003) (TUNEL kit, Roche). Images were processed with National Institutes of Health (NIH) ImageJ and Photoshop software. All quantification was done using the ImageJ software (NIH).
Quantification and statistical analyses
Quantification of immunostaining was done by calculating the correlated total cell fluorescence (CTCF) with Fiji (Schindelin et al. 2012): CTCF = integrated density – (area of selected cells × mean fluorescence of background). For Figure 1, F and G, Figure 4, D and E, and Figure 5, B and C, to compare different experiments, individual quantifications in control conditions were normalized to the means of similar quantifications in Ttk69-mutant SOs. As such, a ratio of 1 indicated that immunostaining in control and Ttk69-mutant SOs was similar, < 1 that measurements in control conditions were lower than those observed in Ttk69-mutant cells, and conversely when the ratio was > 1 one. To compare the quantification of Ttk69 immunostaining presented in Figure S3 in different experiments under a given condition, individual quantifications of Ttk69 immunostaining were normalized to the mean of quantifications of Su(H) accumulation. In Figure 6, to compare different experiments under a given condition, individual quantifications of immunostaining were normalized to the mean of similar quantifications of ELAV staining observed in neurons.
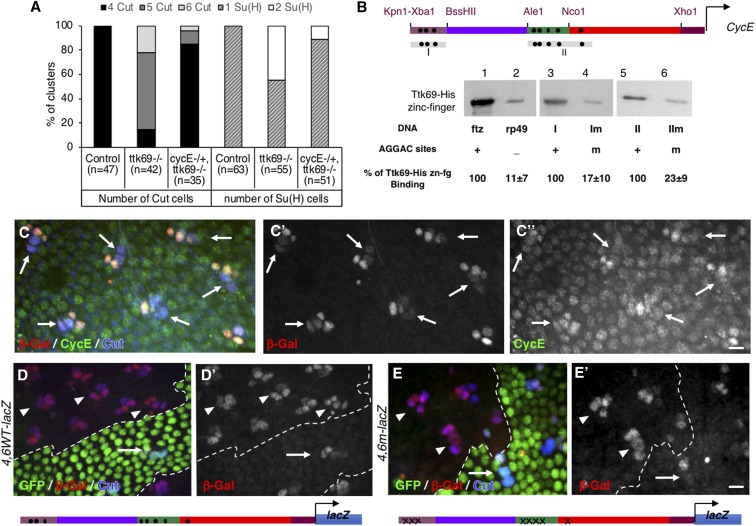
Figure 4.
CycE expression is transcriptionally repressed by Ttk69. (A) The integrity of a Ttk-mutant SO was rescued under CycE heterozygous conditions. Histogram showing the percentage of SO harboring four (black bars) or more (gray bars). Cut-positive cells (left), and one (hatched bars) or two (white bars) Su(H)-positive cells (right), located outside (control) and inside Ttk69 clones in CycE+/+ or CycEAR95/+ heterozygous backgrounds. (B) DNA-mediated Ttk69-His-zinc finger pull-down assay. (Top) Diagram of Ttk69-binding sites (black dots) in the CycE promoter. (Bottom) Magnetic beads were coated with: lane 1, ftz promoter bearing AGGAC-binding sites (positive control); lane 2, rp49 an AGGAC-free promoter (negative control); lanes 3 and 5, I and II regions of the CycE promoter, respectively; and lanes 4 and 6, Im and IIm regions of the CycE promoter, respectively, in which AGGAC-binding sites were replaced by an unrelated ACTGC sequence. (C) The 4.6WT fragment recapitulates endogenous CycE expression in adult bristle sensory cells. Immunostaining of 24-hr APF pupae. Sensory cells in blue (outer cells indicated with arrows). β-Gal in red (shown as a separate channel in the middle C’), CycE in green (shown as a separate channel in C") immunoreactivity. (D and E). Expression pattern of the 4,6WT-lacZ (D and D’) and 4,6m-lacZ (E and E’) CycE transcriptional reporters in control (arrows) and Ttk69-mutant SOs (arrowheads). Ttk69 clones outlined by a white line were detected by the absence of GFP (green). Sensory cells (Cut immunoreactivity, blue) and expression of CycE transcriptional reporters (β-Gal immunoreactivity, red). Bar, 10 μm for (C and D). APF, after pupal formation; β-Gal, β-galactosidase; SO, sensory organ.
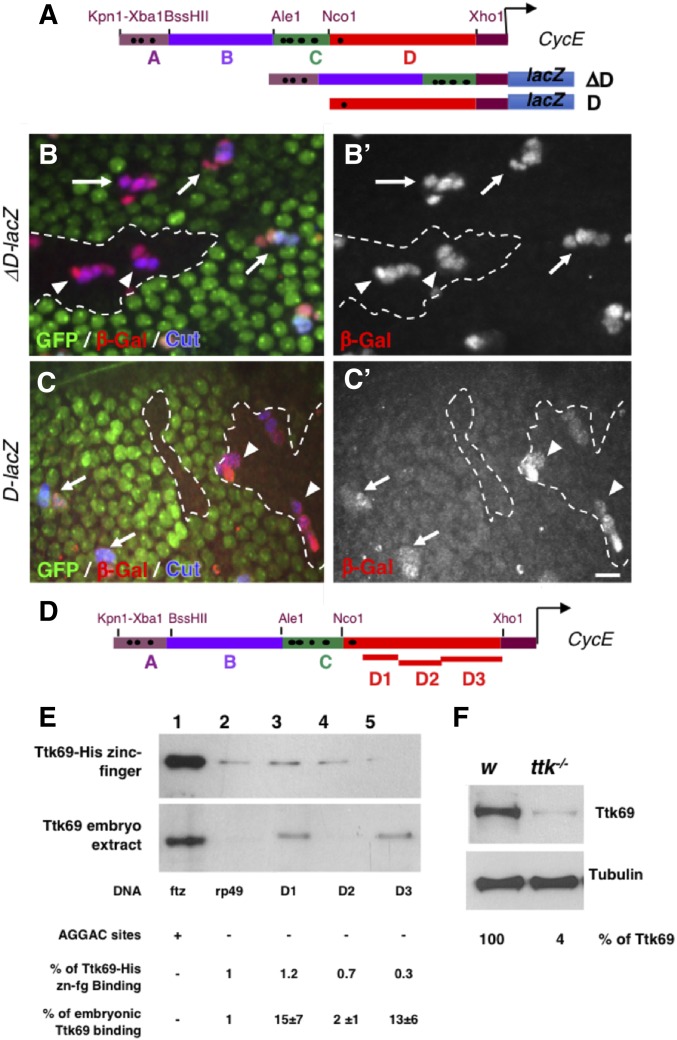
Figure 5.
Ttk69 binds indirectly to the CycE promoter. (A) Diagram of CycE promoter depicting the four specific regions analyzed (A–D). Black dots, canonical AGGAC Ttk-binding sites. ΔD and D CycE transcriptional reporters bearing a deletion of the D region and only the D region, respectively. (B and C) Expression pattern of ΔD-lacZ (B and B’) and D-lacZ (C and C’) in control (arrows) and Ttk69-mutant SOs (arrowheads). Ttk69 clones outlined by a white line were detected by the absence of GFP (green). Sensory cells (Cut immunoreactivity, blue) and expression of CycE transcriptional reporters (β-Gal immunoreactivity, red and bottom panels). (D) As in (A), showing the regions of the D fragment analyzed (D1–D3). (E) DNA-mediated pull-down assay using Ttk69-his-zinc finger (top) and whole Ttk69 protein from an embryo protein extract (bottom). Magnetic beads were coated with: lanes 1 and 2, as in Figure 4B; and lanes 3, 4, and 5 with the D1, D2, and D3 regions, respectively, of the CycE promoter. (F) Detection of Ttk69 protein from a protein extract of one control (white) and one Ttk69-mutant embryo. Tubulin, loading control. Bar, 10 μm for (B and C). β-Gal, β-galactosidase; SO, sensory organ.
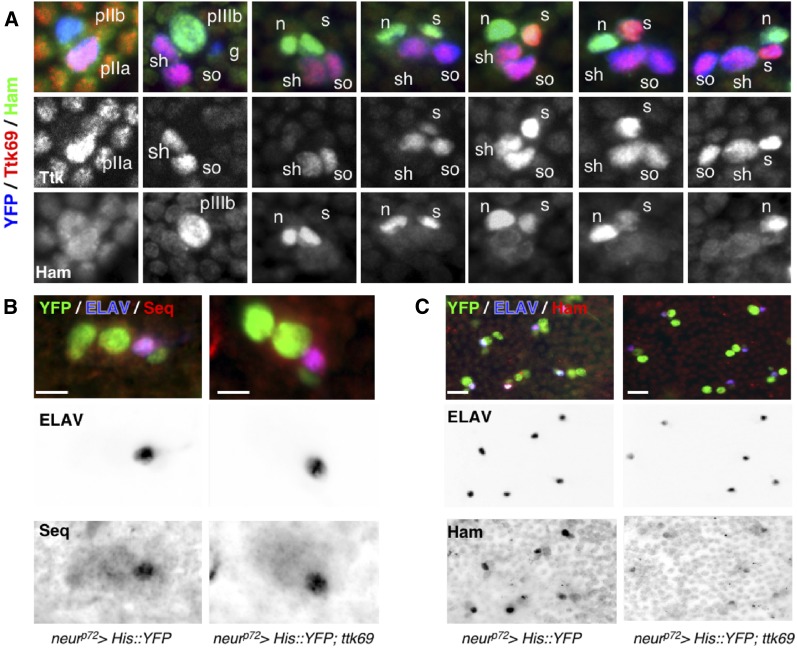
Figure 6.
Ttk69 downregulates hamlet but not sequoia expression. (A) Complementary expression pattern of Ttk69 and Ham proteins in bristle sensory cells. Ttk69 and Ham expression in bristle sensory cells at progressive stages of development from 18–28-hr APF. Sensory cells are shown in blue (YFP staining), Ttk69-positive cells in red (shown as a separate channel in the middle panels), and Ham-positive cells in green (shown as a separate channel in the bottom panels). The sensory cells shown are the precursor cells pIIb, pIIa, and pIIIb, and the terminal cells: glial cells (g), sheath cells (s), neurons (n), shaft cells (sh), and socket cells (so). Ttk69 is first detected in pIIa cells and their daughter cells, and later in the sheath cells. Ham is first detected in pIIIb cells and their daughter cells, before disappearing from the sheath cell when Ttk69 appears in this cell. (B and C) Ttk69 overexpression represses hamlet but not sequoia expression. Analysis of Sequoia (Seq) (B) and Ham (C) protein accumulation after specific expression of Ttk69 in sensory cells. Sensory cells (green); neurons (blue), Ham and Seq (red). ELAV (middle panels), Ham, and Seq channels (right panels) are shown in inverted color. Bar, 5 μm in (A and B), 10 μm in (C). APF, after pupal formation; YFP, yellow fluorescent protein.
Statistical significance was calculated by Mann–Whitney U-test (*** P < 0.001, ns, not significant). Analyses were performed and graphs produced using KaleidaGraph software. Error bars represent SEM.
Time-lapse microscopy
We performed live imaging of SOs in neur-H2B::GFP; FRT82B ttk1e11 pupae following protocols described previously (Simon et al. 2009; Sallé et al. 2012). The neur-H2B::GFP (line 22A4, gift from F. Schweisguth) construct allows SO cells to be followed throughout the progression of the bristle lineage. White pupae were collected and aged until 20-hr APF at 25° in a humid chamber, before dissection and mounting for imaging. Live imaging data were collected using a spinning disk coupled to an Olympus BX-41 microscope (Roper Scientific, 40×, NA 0.75 objective, CoolSnapHQ2 camera). The temperature of the recording chamber was carefully controlled (± 0.1°) using a homemade Peltier device temperature controller fixed to the microscope stage. Systems were driven by Metamorph software (Universal Imaging). Z-stacks of images were acquired every 3 min and assembled using ImageJ software (NIH). At the end of the movies, pupae were dissected and immunostaining was carried out as described previously. Imaged cells were unambiguously identified by their relative position, nuclear size, and order of birth.
Tramtrack69 pull-down
Experiments were performed either using a batch of Escherichia coli-expressed Ttk69::ZF or 20-hr-old white Drosophila embryo protein extracts. Ttk69::ZF was expressed from BL21 (DE3) bacteria transformed with a pET15 vector in which the C-terminal fragment (318–641) of the Ttk69 protein, containing zinc fingers, was cloned in-frame with a histidine tag (gift from A. Travers). Nondenatured E. coli extracts were prepared after a 2-hr induction in 0.1 mM IPTG and embryo extracts were obtained as described by Wodarz (2008) (1 mg devitellinized embryos was always extracted in 5 μl lysis buffer to calibrate extraction). DNA templates were generated by PCR using 5′ biotinylated primers. As controls, a ftz template was obtained using 5′-GGGAGTTGCGCACTTGCTTG-3′ and 5′-GTGCACGCAACGCTGGTGAG-3′ primers, which correspond to the portion of the ftz promoter bearing the canonical AGGAC Ttk69-binding sites, and a RP49 template devoid of this sequence was obtained by PCR using 5′-TGTACTTGGCATCCGCGAG-3′ and 5′-CACCAGCACTTCTCCAACAC-3′. Two CycE templates were obtained using two sets of primers—5′-GCAAGATTATGAATATCTAT-3′ and 5′-GTGTGCGCGCATGCGCAACG-3′, and 5′-GTTGGATTAACCCTTTCTGG-3′ and 5′-AGGATTTAAGTCTCAACTC-3′—to cover fragments I and II, respectively, which correspond to the proximal part of the promoter bearing the canonical Ttk69 AGGAC-binding site. Im and IIm CycE mutated promoters, in which all canonical AGGAC sequences were replaced by a ACTGC sequences, were obtained using the same primers as for fragments I and II of the CycE promoter. The three CycE templates corresponding to the D fragment were obtained using primers 5′-GCTGCCTGCTTGGAGTTGAGAC-3′ and 5′-GGAAGGTCCAAGACGCATGAC-3′ for the D1 fragment, 5′-GTCATGCGTCTTGGACCTTCC-3′ and 5′-TTATGTGCAGATATTGGGCA-3′ for the D2 fragment, and 5′-TTATGTGCAGATATTGGGCA-3′ and 5′-CTCGAGCTGCCAGCGGCTGC-3′ for the D3 fragment. Biotinylated DNA was coupled to streptavidin-coated magnetic beads (M280; Dynal Biotechnology) with 0.1 mg beads per 200 ng DNA overnight at 4°. The beads were washed three times as recommended by the supplier and streptavidin-immobilized DNA saturated for 1 hr in PBS-20% horse serum before incubation for 1 hr with the protein extract in PBS-Triton (0.15%). Protein extracts and beads were prepared separately according to the manufacturer’s instructions. Beads were washed four times with 100 mM NaCl/25 mM NaH2PO4. Extracts were divided equally and added to the beads. At the end of each incubation, the beads were pelleted and precipitated proteins were analyzed by western blot. Ttk69::ZF was revealed using mouse anti-penta-histidine (34660, 1:1000; QIAGEN, Valencia, CA), whereas Ttk69 protein from embryo extracts was revealed using rabbit anti-Ttk69 (1:4000 gift from A. Travers). Specificity of the Ttk69 antibody was tested by analyzing the immunodetection of protein extracts from pools of 10 20-hr-old ttk1e11/TM6 and w1118 embryos. Anti-β tubulin staining (1:10,000; Amersham, Piscataway, NJ) was used as a loading control. Visualization was performed using horseradish peroxidase coupled to anti-mouse or anti-rabbit (1:10,000; Promega, Madison, WI) antibodies coupled to the Super Signal western blotting detection system (Pierce Chemical, Rockford, IL), according to the manufacturer’s instructions.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.8020103.
Results
Ttk69 loss-of-function leads to the formation of SOs with extra inner cells and only one type of outer cell
To precisely determine the involvement of Ttk69 in cell cycle progression and cell-fate determination, we studied somatic clones of ttk1e11, which specifically disrupts Ttk69, hereafter called Ttk69 clones (Lai and Li 1999). SOs inside Ttk69 clones (called Ttk69-mutant SOs) were devoid of the shaft and presented only sockets externally (Figure 1B). At the cellular level, 82% (n = 30) of the mutant organs were composed of more than four cells (up to eight cells) at 28-hr APF (Figure 1C and D, arrowheads). Among the SO cells, one (67%) or two (33%) cells were Pdm1-positive outer cells (n = 30), and in all cases they expressed Su(H), a landmark of socket cells (Figure 1, D and E). These data show that the absence of the shaft structure is associated with the lack of a cell expressing a shaft signature (Pdm1-positive and Su(H)-negative). We observed that in Ttk69-mutant SOs, although the socket was present, it did not have a normal shape (Figure 1B, insets b and c). It was previously shown that normal socket cell differentiation is dependent on autoamplification of the N transcription factor Su(H). Thus, Su(H) is first induced in presumptive socket cells in response to the N pathway and subsequently boosted via its binding to a 3′-enhancer [ASE5, (Barolo et al. 2000; Liu and Posakony 2014)]. Initially, at 23-hr APF, Su(H) expression was similar in control and Ttk69-mutant socket cells (Figure 1, F and F’, quantified in Figure S1A). Later, at 28-hr APF, using an ASE5::GFP reporter to assess Su(H) autoamplification, we failed to detect a GFP signal in Ttk69-mutant socket cells indicating that Su(H) amplification had not occurred (Figure 1, G–G”, compare GFP expression in control socket cells, arrows, with Ttk69-mutant socket cells, arrowhead). This was associated with no further increases in Su(H) levels in Ttk69-mutant socket cells [Figure 1G’, compare Su(H) accumulation in control socket cells, arrows, with Ttk69-mutant socket cells, arrowhead, quantified in Figure S1A]. Since terminal differentiation of socket cells depends on functional Su(H) autoamplification (Barolo et al. 2000), our data showing that Ttk69 impairs Su(H) autoamplification without affecting its initial Su(H) expression indicate that Ttk69 is only required at later stages of socket cell differentiation.
The remaining cells in Ttk69-mutant SOs expressed inner-cell markers. Immunostaining against ELAV and Pros revealed the presence of one-to-four neurons, and one or no sheath cell (Figure 1H). We also observed up to three cells per cluster (n = 35) that were positive for both ELAV and Pros immunostaining (Figure 1, H and I). This suggests the presence of either additional pIIIb precursor cells, or postmitotic cells in which the fate was either completely or not yet well resolved (Ramat et al. 2016). Thus, the Ttk69-mutant lineage is probably not yet completed at 28-hr APF. Overall, these data show that loss-of-function of Ttk69 leads to the formation of SOs composed of extra inner cells with only socket cells as the outer-cell type.
Ttk69 promotes cell cycle arrest and triggers terminal cell-fate identities
Several nonexclusive explanations can account for the presence of SOs with supplementary cells in Ttk69-mutant SOs. One is that the glial cells do not die but divide and produce extra terminal cells. We examined this possibility by studying cell death in Ttk69-mutant SOs. TUNEL assays showed that glial cells undergo apoptosis in Ttk69-mutant SOs at the same time as in control organs located outside the mutant clone (Figure 2A, n = 3). Thus, the supplementary cells do not originate from glial cells that resume proliferation. It is also possible that supplementary cells arise from bristle cells that do not properly exit from the cell cycle and continue to proliferate. We explored this possibility by searching for metaphasic cells using phospho-ser10 histone-3 (PH3) immunoreactivity at 28-hr APF, when control SO cells are already postmitotic. Ttk69-mutant SOs containing four or more cells harbored sensory cells positive for PH3 (Figure 2B, arrowheads, 20% of SOs, n = 25). As these clusters contained the terminal number of cells, these data show that the metaphasic cells were not due to delayed divisions. Thus, these data indicate that Ttk69-mutant SOs harbor supplementary cells due to additional mitoses.
Figure 2.
Extra mitoses in Ttk69-mutant socket cells. (A, B, and E) Ttk69 clones, outlined by a white line, are detected by the absence of GFP (green) in fixed tissues. (A) Apoptosis occurred at the same time in mutant (arrowhead) and control (arrow) SOs. Sensory cells (blue); apoptotic cells (TUNEL staining, red). (B) Extra mitoses [PH3 immunoreactivity (blue), arrowheads] in a Ttk69-mutant SO composed of four cells (red); control cluster in the inset. (C and D) Correlative four-dimensional live imaging and lineage analysis showing an extra division of socket cells. A Ttk69 clone was identified by a lack of GFP expression in epithelial cells and SOs inside the clones were imaged. (C) Representative frames (1–6), depicted in inverted fluorescence, from a time-lapse recording of one Ttk69-mutant SO at 19-hr APF. Glial cell outlined in blue, pIIb and its progeny in orange, the shaft cell in green, and the socket cell and its daughter in purple. Frames 4 and 5, division of the socket cell. Apoptosis of the glial cell between frames 2 and 3. (D and D’) Immunostaining and schematic representation of the same cluster after the time-lapse recording shown in (C). Nonclonal epithelial and sensory cell (GFP expression, green), neuron (ELAV, blue), and socket cell [Su(H), red] immunoreactivity. (E) Cell divisions in full-determined socket cells. PH3 (blue) and Su(H) (red) immunoreactivity was detected in the same cell (arrowhead); control socket cell (arrow). (F) Schematic view of the lineage shown in (C). Cells are encircled using the same color code as in (C) and filled with the same color as in (D). Bar, 10 μm. APF, after pupal formation; SO, sensory organ; TUNEL, terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling.
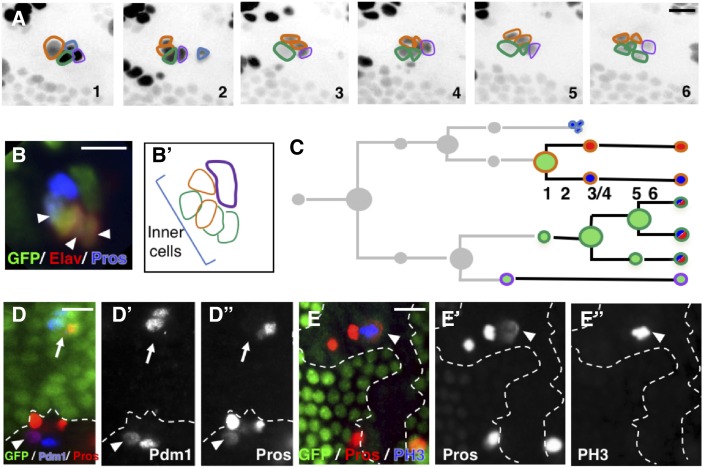
To unambiguously define the origin of the supernumerary cells, we used correlative microscopy that combines live imaging to record the entire pattern of cell divisions in Ttk69-mutant SOs followed by immunolabeling to identify cell identities. During time-lapse recording, sensory cells were identified by the expression of GFP under the control of the neuralized promoter (neur-GFP). At the end of each recording, the imaged notum was fixed and immunolabeled with anti-Su(H), as well as anti-ELAV and anti-Pros to highlight outer and inner cells, respectively. Imaged SOs could be unambiguously recognized within the fixed nota by their relative position with respect to the midline, the position of the macrochaetae, or the rows of microchaetae (Fichelson and Gho 2004). We confirmed the presence of extra cell divisions and revealed an unexpected cell-transformation event. First, there was a supplementary division in a pIIa daughter cell, identified as the future socket cell by its position in the cluster (Figure 2C, panels 4 and 5, and Movie S2). This extra division was symmetric, leading to two Su(H)-positive socket cells (Figure 2, D and D’). We also observed socket cells in mitosis, identified by Su(H) and PH3 immunoreactivity, in fixed material (Figure 2E, arrowhead, n = 3). Thus, future socket cells undergo an extra division in the absence of Ttk69 (Figure 2F). In addition, the anteriorly located pIIa daughter cell, the presumptive shaft cell that normally does not divide, underwent repetitive cell divisions (Figure 3A, panels 3 and 5, and Movie S3). Surprisingly, immunostaining of the resulting clusters showed that cells arising from these extra divisions acquired a neural fate, as they expressed Pros and ELAV (Figure 3, B and B’), two markers expressed in sheath and neuron cells, respectively. This suggests that presumptive shaft cells underwent cousin–cousin cell transformation in which outer cells acquired an inner-cell fate (Figure 3C). Consistent with this possibility, we also observed cells with weak expression of Pdm1 associated with weak expression of Pros in fixed material, suggesting that they were midway through transformation (arrowhead in Figure 3, D–D”). Furthermore, we detected clusters harboring two Pros-positive cells, of which one was dividing (PH3-positive, arrowhead in Figure 3, E–E”). These different lines of evidence led us to conclude that the presumptive shaft cells underwent cell-fate respecification and acquired a pIIIb precursor cell identity. We never observed, by in vivo recordings, cell lineages in which both pIIa daughter cells entered division. This is likely due to the low probability of such cases. We do not favor the possibility that the division of pIIa daughter cells is mutually exclusive as, using fixed material, we observed SOs composed of more than five cells and harboring two socket cells, a situation that required ectopic division of both pIIa daughter cells.
Figure 3.
Presumptive Ttk69-mutant shaft cells undergo cell transformation toward inner precursor cells. (A) Correlative four-dimensional live imaging and lineage analysis showing extra divisions of the presumptive shaft cell. Representative frames (1–6) depicted in inverse fluorescence from a time-lapse recording of one Ttk69-mutant SO at 20-hr APF. Cells outlined as in Figure 2C. Frame 2: apoptosis of the glial cell. Frames 3 and 5: two rounds of division of the presumptive shaft cell. (B and B’) Immunostaining and schematic representation of the same cluster after the time-lapse recording. Sensory cells in green. Inner cells identified by ELAV (red) and Pros (blue) immunoreactivity. (C) Schematic view of the lineage shown in (A). Cells are encircled using the same color code as in (A) and filled with the same colors as in (B). (D and E) Ttk69-mutant clones detected by the absence of GFP (green) are outlined by a white line. (D–D”) Cousin–cousin cell-fate transformation. Arrowhead, mutant cell having both Pdm1 [D’ and blue in (D)] and Pros [D” and red in (D)] immunoreactivities (outer- and inner-cell marker, respectively), situation never observed in control (arrow). (E–E”) Extra mitoses of inner cells revealed by PH3 immunoreactivity [E” and blue in (E)]. Inner cells identified by Pros immunoreactivity [E’ and red in (E)]. Bar, 10 μm. APF, after pupal formation; SO, sensory organ.
The respecification of presumptive shaft cells into inner precursor cells can explain, in part, the observation that Ttk69-mutant sensory clusters harbored up to six neural (ELAV/Pros-positive) cells, showing that extra inner cells originate at the expense of outer cells. Overall, these results suggest that Ttk69 promotes cell-precursor exit, acting both by arresting cell proliferation and triggering terminal cell-fate identity.
Ttk69 induces cell cycle exit via transcriptional repression of CycE expression
Our results suggest that Ttk69 regulates cell proliferation. We have already shown that Ttk69-mutant sensory cells strongly accumulate CycE protein (Audibert et al. 2005). We studied whether supplementary mitosis was related to the accumulation of ectopic CycE by assessing whether a reduction in the dose of CycE could revert the phenotype of supplementary cell divisions observed in Ttk69-mutant SOs. As already described, we observed that most SOs (85%, n = 42) inside Ttk69-mutant clones contained more than the normal four cells (Figure 4A). This dropped to 18% when the clones were induced in a CycEAR95/+ heterozygous background (n = 35). In another set of experiments to study the number of socket cells, 45% of Ttk69-mutant SOs harbored duplicated Su(H)-positive socket cells (n = 55), whereas only 11% did so in the CycEAR95/+ heterozygous background (n = 51). These results show that reducing the dose of CycE is sufficient to markedly reduce the number of supplementary divisions observed in Ttk69-mutant SOs. This strongly suggests that the supplementary mitoses observed in Ttk69-mutant SOs are mainly driven by the increase in CycE levels induced after Ttk69 loss-of-function.
We analyzed the role of Ttk69 in the transcriptional expression of CycE by first testing the capacity of Ttk69 to bind the CycE promoter. It has been previously shown that a 4.6-kb proximal fragment of the whole CycE promoter is able to recapitulate zygotic CycE expression in the embryonic peripheral nervous system (Figure S2A, Jones et al. 2000). We have identified eight AGGAC canonical Ttk-binding sites in this 4.6-kb fragment that are organized in two clusters: three in region I and five in region II (Figure 4B) (Harrison and Travers 1988, 1990). The binding of Ttk69 to these regions was assessed by a DNA-mediated Ttk pull-down assay using the Ttk69 C-terminal domain, containing the C2H2-type zinc finger. The Ttk69 zinc finger domain behaved as expected, since it was efficiently retained on beads coated with DNA fragments bearing canonical Ttk-binding sites, as well as on regions I or II (Figure 4B, lanes 1, 3, and 5). In contrast, we observed low-level retention when beads were coated with DNA free of Ttk-binding sites (rp49, Figure 4B, compare lanes 1 and 2); quantification showed that binding to the rp49 probe was reduced to 11% (n = 5) of that observed using the ftz probe. We also observed low-level retention when the beads were coated with regions I or II in which the Ttk-binding sites were mutated (Im and IIm respectively, Figure 4B, compare lane 3 with 4 and lane 5 with 6). Binding to the Im probe was reduced to 17% (n = 5) of that containing region I and binding to the IIm probe was reduced to 23% (n = 5) of that containing region II. As such, this in vitro approach suggests that the control of CycE expression by Ttk69 relies on the direct binding of this factor to canonical Ttk-binding sites on the CycE promoter. We then tested this possibility in vivo using a lacZ transcriptional reporter strategy. First, we followed lacZ expression under the control of the 4.6WT fragment in bristle sensory cells. We observed endogenous CycE and β-Gal accumulation in inner cells, whereas both were only barely detected in pIIa daughter cells in 24-hr APF pupae (arrows in Figure 4, C–C” and Figure S2B). These data confirm that the expression of the 4.6-kb CycE transgenic reporter mirrors the expression of the endogenous CycE protein (Audibert et al. 2005). Then, we analyzed lacZ expression under the control of a wild-type 4.6 CycE promoter fragment (4.6WT) and a 4.6 CycE fragment in which all eight Ttk69-binding sites were mutated (4.6 m ,in which the AGGAC sequence was replaced by the ACTGC sequence) (Figure 4, D and E). As expected, the expression of the 4.6WT construct was upregulated in Ttk69-mutant SOs. Thus, we observed a particularly high level of β-Gal accumulation in both pIIa daughter cells (Figure 4, D and D’, arrowheads: quantification in Figure S1B). Surprisingly, the expression pattern of the 4.6 m transcriptional reporter, where the eight Ttk69-binding sites had been mutated, was not modified in wild-type SOs (Figure 4, E and E’, arrow and Figure S2, C and C’). Moreover, expression of the 4.6 m-lacZ construct was still upregulated in Ttk69-mutant SOs (Figure 4, E and E’, arrowheads; quantification in Figure S1B). As such, although in vitro experiments showed that Ttk69 binds the CycE promoter, the in vivo results indicated that repression of CycE expression in sensory cells is not mediated through binding to the canonical AGGAC Ttk-binding sites present in the 4.6 fragment.
To accurately define which part of the CycE promoter is required for Ttk69 regulation, we divided the 4.6-kb CycE cis-regulatory fragment into four regions (A–D) and monitored the regulatory activity of constructs with these regions deleted in bristle sensory cells (Figure 5A and Figure S2A). All constructs were inserted at the same locus (using ΦC31 integrase-based tools, except line D-lacZ) to avoid expression variations due to genomic environment. A CycE promoter construct bearing a deletion of region A and C (ΔAC-lacZ), as well as a deletion of region B (ΔB-lacZ), showed an expression pattern similar to that of the 4.6-kb lacZ construct, indicating that the deletion of these regions did not remove the Ttk69 regulatory domain (Figure S2, D, D’, E, and E’). In contrast, deletion of region D (ΔD-lacZ) led to high levels of βGal accumulation in all sensory cells under control conditions (Figure 5, B and B’, arrows and Figure S2, F and F’). Moreover, the expression patterns were similar in Ttk69-mutant SOs and control SOs outside of the clones (Figure 5, B and B’, arrowheads; quantification in Figure S1C). These data show that the cis-regulatory sequence required for Ttk69-mediated downregulation of CycE expression is located in the D-sequence. Next, we analyzed lacZ expression under the control of fragment D (D-lacZ) in wild-type and Ttk69-mutant SOs. Although a low level of β-Gal accumulation was observed with the D-lacZ construct, this construct behaved similarly to the 4.6WT-lacZ construct. β-Gal was barely detected in pIIa control cells (Figure 5, C and C’, arrow and Figure S2, G and G’) while a high level of β-Gal accumulation was observed in mutant SOs (Figure 5, C and C’, arrowhead; quantification in Figure S1C). These data confirm that this region is sufficient to drive Ttk69-mediated CycE regulation. Although the D-fragment harbors a Ttk69-binding site at the more distal location, we do not believe that this binding site plays a role in Ttk69-mediated control of CycE expression since Ttk69 continues to regulate the expression of the reporter when this Ttk69-binding site is mutated (see above and Figure 4, E and E’). As such, these data suggest that Ttk69 may control CycE expression by binding directly to unknown binding sites located in the D-fragment, or indirectly either via another factor that recognizes the D-fragment or via the regulation of the expression of CycE transcriptional regulators.
Ttk69 binds to the CycE promoter independently of canonical sites
To address these possibilities, we divided the D-fragment devoid of the Ttk69-binding site into three pieces (D1–D3) and assessed their roles in Ttk69 binding by performing DNA-mediated Ttk pull-down assays (Figure 5D). As expected, a high and a low amount of Ttk69 precipitated with beads coated with ftz and rp49 probes, respectively (Figure 5E, lanes 1 and 2). When we assessed the binding of the Ttk69 zinc finger domain to the D-fragments, we observed that only a low amount of Ttk69 precipitated (Figure 5E, top panel, lanes 3 to 5). This signal was comparable to the one obtained with beads coated with a fragment devoid of Ttk69-binding sites [Figure 5E, rp49-probe (lane 2), n = 2]. These data indicate that the D-fragments do not retain the Ttk69 zinc finger domain. Next, we tested whether endogenous Ttk69 binds these CycE promoter fragments in association with other partners by performing similar DNA-mediated Ttk pull-down assays, but using embryonic extracts and a specific Ttk69 antibody (Figure 5, E and F). Our data show that endogenous Ttk69 was not retained by the D2 CycE promoter fragment [Figure 5E bottom panel, lane 3; quantification showed that binding to the D2 probe was similar to that observed using the rp49 probe (n = 3)]. Remarkably, it was retained on beads coated with the D1 and D3 fragments [Figure 5E, bottom panel, lanes 2–5, signals using the D1 probe were 15 times and using the D3 probe 13 times that observed for the rp49 probe (n = 3)]. These data show that the binding of Ttk69 to the D1 and D3 CycE promoter fragments required either the native form of Ttk69 or other proteins present in embryonic extracts.
Ttk69 downregulates hamlet expression
We wished to identify Ttk target genes involved in cell fate regulation in SOs. We focused on two candidates, hamlet (ham) and sequoia (seq) (Moore et al. 2004; Andrews et al. 2009), because the phenotype associated with ham and seq loss-of-function, inner-to-outer-cell transformation is similar to that of ttk gain-of-function. In addition, ham and seq genes are expressed in patterns that are complementary to the expression pattern of ttk during the bristle lineage [Figure 1A and Figure 6A, and see Figure S4 in Andrews et al. (2009)]. Thus, we studied the potential epistatic interactions between these three factors.
We first studied ttk expression in SOs when ham and seq were either overexpressed or downregulated. We used a temperature conditional driver to overexpress ham or seq late in development, to avoid potential interference with outer-to-inner-cell transformations induced by seq or ham-overexpression. Under these conditions, we detected no cell-fate transformations, assessed by Su(H) immunoreactivity as a marker of outer-socket fate [Figure S3, A–C, Su(H) panels]. This analysis revealed that Ttk69 expression was unaffected when either ham or seq were overexpressed (Figure S3, A–C, Ttk panels; quantification in Figure S1D). Reciprocally, we failed to observe modifications in the number of Ttk-positive cells early during development in ham or seq mutant SOs (20- and 22-hr APF, Figure S3, D–G). We observed sensory clusters harboring four Ttk69-positive cells only late in the bristle lineage in seq or ham mutant clones (24-hr APF in 12 and 34% of clusters in ham and seq mutant clones, respectively, Figure S3, D–G). These data suggest that supernumerary Ttk cells in ham and seq loss-of-function cells are due to cell transformation induced by the loss-of-function of seq or ham, rather than direct deregulation of ttk69 expression. Thus, we conclude that ttk expression is independent of Ham and Seq.
Next, we analyzed whether the expression of ham or seq is controlled by Ttk69. Thus, we overexpressed Ttk69 in neurons, where it is never detected, and analyzed Seq and Ham protein accumulation. We used a similar strategy as before to overexpress Ttk69 late in development and observed no cell-fate transformation as shown by ELAV immunoreactivity (Figure 6, B and C, ELAV panels). Under these conditions, Seq accumulated at the same level as in the control situation, whereas Ham immunoreactivity was strongly reduced (compare the right panels in Figure 6, B and C for Seq and Ham detection, respectively; quantification in Figure S1, E and F). The observed effects were not due to changes in cell fate, as ELAV expression was unaltered. As such, we conclude that Ttk69 does not affect seq expression, whereas it downregulates ham expression.
Ttk69 maintains nonneural cell fate via repression of hamlet expression
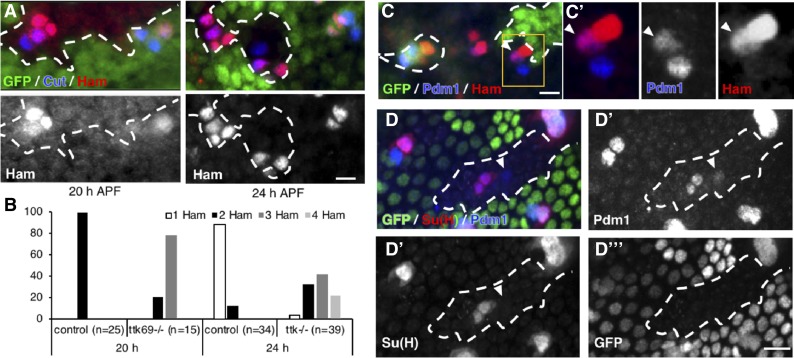
Our results show that Ttk69 downregulates ham expression. This suggests that ham is repressed in Ttk69-expressing cells, in particular in pIIa precursor cells and their progeny. Thus, it is expected that ham would be ectopically expressed in pIIa cells in Ttk69-mutant SOs. Indeed, we observed the presence of three-to-four Ham-positive cells in 50% of Ttk69-mutant SOs analyzed (SOs inside Ttk69 clones) at 24-hr APF, whereas there were no more than two in the control SOs (SOs outside Ttk69 clones) (Figure 7, A and B). However, ectopic expression of ham could be due to the deregulation of ham expression per se or to the cousin–cousin cell transformation already described. We assessed ham expression in Ttk69-mutant SOs at early stages to determine the mechanism behind its ectopic expression. We observed three Ham-positive cells in Ttk69-mutant SOs composed of four cells as early as 20-hr APF, even before the completion of the bristle lineage (Figure 7, A and B). These data show that the ectopic expression of ham in the absence of Ttk69 is an early event during cousin–cousin cell transformation. Moreover, we occasionally observed cells positive for both Ham and Pdm1, a specific marker of pIIa-descendent cells, in Ttk69-mutant sensory clusters (arrowhead in Figure 7, C and C’, n = 4). This signature is consistent with the cells undergoing transformation from outer to inner cells.
Figure 7.
Ttk69 represses hamlet expression to maintain the nonneural cell fate. Ttk69 clones outlined by a white line were detected by the absence of GFP (green). (A) ham expression (red) in Ttk69-mutant SO at 20- and 24-hr APF. Sensory cells in blue. (B) Histogram showing the percentage of SO harboring one (white bars), two (black bars), three (dark gray bars), or four (pale gray bars) Ham-positive cells in control and Ttk69-mutant SOs at 20- and 24-hr APF. (C) Cell transformation of a Pdm1-positive cell to a Ham-expressing cell. Arrowhead, Ttk69-mutant cell having both Pdm1 (blue in C’) and Ham (red in C’) immunoreactivities. Pupae at 22-hr APF. (C’) Higher magnification of the Ttk69-mutant cluster outlined in (C). Merged and separate Pdm1 and Ham channels. (D) Recovery of shaft cells. Arrowhead, a Pdm1-positive, Su(H)-negative Ttk69-mutant cell in a ham heterozygous mutant background. ham+/− pupae at 28-hr APF. (D’–D”’), separate Pdm1, Su(H), and GFP channels. Bar, 10 μm. APF, after pupal formation; SO, sensory organ.
These data suggest that the derepression of ham in pIIa cells in Ttk69-mutants drives cousin–cousin cell transformation, in which pIIa shaft cells adopt a pIIIb cell fate. We tested this possibility by studying whether the reduction of ham expression in Ttk69-mutant SOs could restore shaft identity. We analyzed cell identities in Ttk69-mutant SOs, namely SOs inside Ttk69 clones, in flies that were otherwise ham heterozygous. This analysis was performed in pupae at 28-hr APF, when this cousin–cousin transformation had already taken place in Ttk69-mutant SOs. Under these conditions, in 4 out of 60 Ttk69-mutant SOs analyzed, we observed Pdm1-positive/Su(H) negative cells, a specific sign of shaft cells (Figure 7, D–D’’’, arrowhead). These rescues were significant since the shaft cell type was never observed in Ttk69-mutant SOs under ham normal conditions (see Figure 1D). Attempts to increase the percentage of rescue were aborted since further reduction of ham starts to affect cell fate identities per se. Our results show that a reduction in ham expression can rescue the formation of shaft cells in the absence of Ttk69. Thus, Ttk69 maintains a nonneural cell fate in pIIa daughter cells by inhibiting the adoption of the inner precursor fate via the repression of ham expression.
Discussion
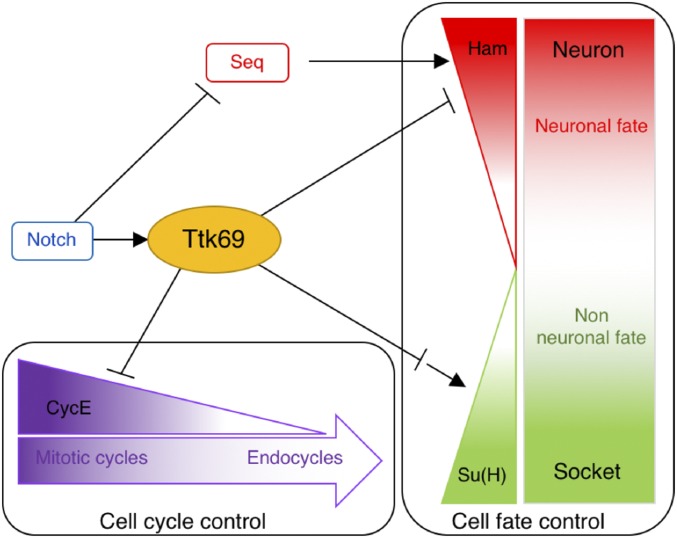
An important goal in developmental biology is to understand the mechanisms by which cell proliferation and cell-fate acquisition are coupled during organogenesis. Here, we show that Ttk69, a member of the evolutionarily conserved BTB-ZF transcription factor family, acts as a link between these two processes. We found that Ttk69 is essential for exiting the proliferative progenitor state and conferring a nonneural fate to the progeny during the formation of mechanosensory bristles. Indeed, using mainly clonal analysis, we show that ectopic cell divisions occur independently of changes in cell fate. In addition, we observed that Ttk69-mutant SOs harbor supplementary terminal cells due to cell transformation that generates extra neural progenitor cells. This was associated with upregulation of CycE, required for S-phase entry, and the ectopic expression of hamlet, a neural determinant. As such, the BTB-ZF transcriptional factor Ttk69 links cell proliferation and cell fate acquisition in the bristle cell lineage (Figure 8).
Figure 8.
Ttk69 as a central node of a transcriptional regulatory network coordinating terminal cell fate acquisition and the arrest of cell proliferation. In response to Notch pathway activation (Guo et al. 1995), Ttk69 (1) downregulates CycE expression inducing transition from a mitotic to endocycle mode of the cell cycle, and (2) downregulates ham and upregulates Su(H) expression. Downregulation of ham expression prevents the acquisition of neural fate induced by the combined action of Seq (Sequoia) and Ham (Moore et al. 2004; Andrews et al. 2009). Upregulation of Su(H) expression allows the terminal differentiation of socket cells.
Ttk69 acts as a dual factor linking cell proliferation and cell fate acquisition
Our results showed that the loss of Ttk69 leads to SOs harboring up to eight terminal cells. Supplementary terminal cells arose from two different mechanisms: an extra division of socket cells and, more importantly, several rounds of extra division due to the respecification of the presumptive shaft cell into a pIIIb precursor cell. It was previously shown that SOs in complete ttk-null pupae are composed of only four neurons (Guo et al. 1995). The fact that no ectopic cells were generated when all Ttk isoforms were absent may reflect different kinetics between cell cycle arrest and cell differentiation. ttk-null cells rapidly acquired an arrested cell cycle and neuronal terminal fate, rather than entering a proliferative precursor state, as after Ttk69 loss-of-function alone. Thus, the specific effects of Ttk69 on the cell cycle were masked in the ttk-null mutant. Use of the Ttk69 loss-of-function mutant made it possible to reveal intermediate cell fates prior to the acquisition of terminal-cell identities. Moreover, the study of mutants that exclusively affected Ttk69 allowed decoupling of the acquisition of cell cycle arrest and cell fate.
The effects of Ttk69 on the core cell cycle machinery were revealed by the ectopic divisions of socket cells observed under Ttk69 loss-of-function conditions. These cells are already committed to acquire a terminal identity, indicating that such ectopic divisions are not associated with changes in cell fate. This shows that Ttk69 impedes cell cycle progression per se. Accordingly, we show that the control of cell cycle progression by Ttk69 involves transcriptional downregulation of CycE expression. Such negative control of CycE expression by Ttk69 appears to be a general effect, as it has also been observed in proliferating glial cells (Badenhorst 2001). Moreover, it has also been shown that Ttk69 represses the expression of string, which encodes the phosphatase (Cdc25) essential for G2/M transition in the imaginal eye disc (Baonza et al. 2002). This suggests that Ttk69 represses cell cycle progression at different phases of the cell cycle. Thus, the induction of ectopic cell divisions in the absence of Ttk69 is probably due to the multiple effects of Ttk69 on cell cycle progression. The diverse targets of Ttk69 in the cell cycle machinery could explain the involvement of this factor in the transition between different modes of the cell cycle (Jordan et al. 2006; Sun et al. 2008). In SOs, pIIa terminal cells underwent endocycles that required fine-tuned control of CycE expression. Indeed, we have already shown that endocycles are abolished in CycE-null mutants, occur at very low CycE levels, and become mitotic cycles at high CycE levels (Simon et al. 2009; Sallé et al. 2012). We show here that Ttk69 is involved in a mechanism that limits CycE levels. Thus, Ttk69 is probably involved in the transition from mitotic cell cycles to endocycles in pIIa terminal cells. Similar transitions between two cycling states associated with two different levels of Ttk69 have been observed in ovary epithelial follicular cells during the transition between endocycles to gene amplification (Jordan et al. 2006; Sun et al. 2008). We propose that Ttk69 contributes to the dampening of CycE levels, allowing cells to transit throughout different modes of the cell cycle.
In addition to cell proliferation, both pIIa daughter cells were differentially affected under Ttk69-mutant conditions, at the level of terminal differentiation for socket cells and of determination for shaft cells. For socket cells, although SOs of adult Ttk69 mutants contain socket cuticular structures, autoamplification of Su(H) expression is impaired leading to misshapen sockets (Barolo et al. 2000). For shaft cells, presumptive shaft cells are respecified and acquire a neural progenitor identity due to the misexpression of ham. Ham is normally expressed in pIIIb precursor cells, and its overexpression induces the formation of SOs bearing supernumerary cells expressing both ELAV and Pros, such as in pIIIb cells [see figure 4 in Moore et al. (2004)]. In addition, ham loss-of-function induces the conversion of terminal inner cells into outer cells, suggesting that Ham is essential for acquisition of the neural-precursor fate (Moore et al. 2004). Moreover, we observed that a reduction of ham levels in Ttk69-mutant clones decreased shaft respecification, in agreement with the fact that Ham is an essential regulator of neural-precursor fate. Finally, we show that Ttk69 repressed ham expression. Overall, these data suggest that respecification of shaft cells is due to the ectopic expression of ham as a consequence of the loss-of-function of Ttk69. ham was also misexpressed in socket cells, but this did not lead to cell transformation. This apparent contradiction may be related to the differential activation of the N pathway between these two sister cells. Indeed, Ttk69 loss-of-function induces a cell fate change in shaft cells, an N-off cell. In contrast, Ttk69 loss-of-function in socket cells, in which the N pathway is activated as soon as the cells are formed (Remaud et al. 2008), impaired only their late differentiation. These results suggest that early activation of the N pathway prevents cell-fate transformation.
We conclude that Ttk69 is required in terminal cells to repress the neural-precursor state by inhibiting both proliferative capacity, and repressing CycE expression and neural fate, by repressing ham. This shows that Ttk69 is a central actor in the coordination between cell cycle arrest and cell-fate acquisition.
Tt69 regulates its downstream genes in several ways
In this study, we identified three genes—CycE, ham, and Su(H)—whose expression was deregulated in Ttk69-mutant clones. Su(H) is positively regulated by Ttk69, as revealed by its downregulation in socket cells in the Ttk69-mutant context. The action of Ttk69 on the Su(H) enhancer may occur in two different ways. Either Ttk69 acts directly as a transcriptional activator or it represses the expression of an unknown factor, as Ttk69 has always been described as a transcriptional repressor. The time required to express this putative relay factor is consistent with the observation that Ttk69 loss-of-function affected only late socket cell differentiation. Furthermore, we showed that such Ttk69-mediated regulation occurs through the Su(H) 3′-end autoregulatory enhancer ASE5. It is interesting to note that the long-lasting, high level of Su(H) expression mediated by the autoregulatory ASE5 loop does not require N pathway signaling (Liu and Posakony 2014). As such, Ttk69 once again affected N-independent processes. Further experiments are required to elucidate how Ttk69 activates the autoamplification of Su(H) expression.
We show here that Ttk69 also regulates CycE. Indeed, we have identified a region in the 4.6-kb CycE promoter fragment where Ttk acts to turn off CycE expression. The analysis of this region revealed an unexpected feature. Indeed, our structure–function analysis failed to identify a region responsible for turning on CycE expression in SO cells. Thus, the reporter was always expressed in these cells in all deletions tested and none of these fragments seemed to be specifically required to turn on CycE expression. One possibility is that the 0.4-kb most proximal region of the CycE promoter (dark pink in Figure 4 and Figure 5) drives CycE expression. Alternatively, there are multiple regions scattered along the 4.6-kb fragments that control CycE expression. Further experiments are required to understand how the 4.6-kb promoter turns on CycE in this system. In contrast, Ttk69-mediated CycE downregulation was driven through a promoter domain (fragment D) independently of the canonical AGGAC Ttk69-binding sites (Brown et al. 1991). Moreover, we showed that the Ttk69 zinc finger domain does not bind subdomains of fragment D (D1 and D3), whereas the native Ttk69 protein, present in a late embryonic extract, does. There are two nonexclusive explanations for this observation. Either an uncharacterized Ttk69 domain outside the zinc finger domain binds directly to the D fragment through a noncanonical binding site or Ttk69 binds indirectly to the CycE promoter via an interaction with trans-acting factors. The first explanation is formally possible, but no DNA-binding domain has been described in the N-terminal portion of the Ttk69 protein. Nevertheless, it is well known that Ttk69 may bind to other noncanonical binding sites. This is the case for the GTCCTG and TTATCCG sequences in the eve and ftz promoters, respectively (Harrison and Travers 1990; Read and Manley 1992). However, we observed that Ttk69 continued to downregulate CycE expression in the absence of these noncanonical sites, making this explanation unlikely. In contrast, several lines of evidence support the second explanation. It is known that the activity of Ttk69 can be influenced by the presence of other DNA-binding factors. Thus, the repressive action of Ttk69 depends on interactions with MEP1 and Mi-2 proteins, which recruit the ATP-dependent chromatin-remodeling complex (the nucleosome remodeling and deacetylase complex) (Reddy et al. 2010). Moreover, it has been shown that while Ttk could bind directly to the eve promoter repressing eve expression (see above), Ttk69 represses eve expression independently of their direct binding to DNA, by interacting with GAGA factors through its BTB/POZ domain. When bound to DNA, GAGA zinc finger factors (Trithorax-like, Trl) activate the transcriptional machinery, but this transcription is inhibited when it is complexed with Ttk69 (Pagans et al. 2004). In bristle sensory cells, RNA interference-mediated loss-of-function of the MEP1, Mi-2, and Trl genes did not affect cell number or cell fates, even when these loss-of-function mutations were analyzed in a sensitized ttk69 heterozygous background (Figure S4). Despite these results, we favor a model in which another factor is required for Ttk69 to bind the D fragment of the CycE promoter. Furthermore, we did not find Ttk binding close to the CycE promoter using genome-wide Ttk-binding profiles from 0–12-hr-old embryos, published by the Model Organism ENCyclopedia Of DNA Elements (modENCODE) project. Moreover, CycE upregulation was not observed in genome-wide expression experiments performed in S2 cells treated with double-stranded RNA directed against Ttk69 (Reddy et al. 2010). These data suggest that Ttk69 does not regulate CycE expression during early embryonic stages. Thus, Ttk-mediated downregulation of CycE expression late in development probably requires cell-specific trans-acting factors.
The involvement of trans-acting factors would explain the diversity of the Ttk response of particular cells at specific developmental stages. Such diversity mediated by trans-acting factors allows Ttk to regulate the expression of a broad spectrum of genes in bristle sensory cells in response to N pathway activation (Figure 8). This is true not only for genes related to cell proliferation, such as CycE, but also those controlling cell fate, such as ham and Su(H). Our data suggest that Ttk69 represses ham in pIIa sublineage cells and activates the Su(H) autoregulatory loop in socket cells. CycE and ham regulation occur earlier in this lineage, and likely involves the binding of Ttk69 to gene promoters as we have shown for CycE. In contrast, Su(H) regulation takes place late in the lineage, implying the probable repression of intermediary relay factors. Trans-acting factors would allow cell-specific responses to Ttk69, whereas intermediary relay factors would allow diversification over time. Thus, since Ttk69 is an N effector, the mechanism of action of this factor increases the spatial and temporal diversity of the N pathway cell response.
Acknowledgments
We thank Y. N. Jan, J. Knoblich, H. Bellen, H. Richardson, and A. Travers for antibodies; the fly community for fly strains; and Heather McLean for critical reading. Funding was provided by the Centre National de la Recherche Scientifique and the Sorbonne Université. A.R. was financed by grants from the Ministère de l’Education Nationale et de la Recherche and the Association pour la Recherche sur le Cancer, France. The funders had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25386/genetics.8020103.
These authors share senior authorship.
Communicating editor: R. Duronio
Literature Cited
- Andrews H. K., Giagtzoglou N., Yamamoto S., Schulze K. L., Bellen H. J., 2009. Sequoia regulates cell fate decisions in the external sensory organs of adult Drosophila. EMBO Rep. 10: 636–641. 10.1038/embor.2009.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo S. J., Cela C., Llimargas M., 2007. Tramtrack regulates different morphogenetic events during Drosophila tracheal development. Development 134: 3665–3676. 10.1242/dev.007328 [DOI] [PubMed] [Google Scholar]
- Audibert A., Simon F., Gho M., 2005. Cell cycle diversity involves differential regulation of Cyclin E activity in the Drosophila bristle cell lineage. Development 132: 2287–2297. 10.1242/dev.01797 [DOI] [PubMed] [Google Scholar]
- Badenhorst P., 2001. Tramtrack controls glial number and identity in the Drosophila embryonic CNS. Development 128: 4093–4101. [DOI] [PubMed] [Google Scholar]
- Baonza A., Murawsky C. M., Travers A. A., Freeman M., 2002. Pointed and Tramtrack69 establish an EGFR-dependent transcriptional switch to regulate mitosis. Nat. Cell Biol. 4: 976–980. 10.1038/ncb887 [DOI] [PubMed] [Google Scholar]
- Barolo S., Walker R. G., Polyanovsky A. D., Freschi G., Keil T., et al. , 2000. A Notch-independent activity of suppressor of hairless is required for normal mechanoreceptor physiology. Cell 103: 957–970. 10.1016/S0092-8674(00)00198-7 [DOI] [PubMed] [Google Scholar]
- Bellaïche Y., Gho M., Kaltschmidt J. A., Brand A. H., Schweisguth F., 2001. Frizzled regulates localization of cell-fate determinants and mitotic spindle rotation during asymmetric cell division. Nat. Cell Biol. 3: 50–57. 10.1038/35050558 [DOI] [PubMed] [Google Scholar]
- Bonchuk A., Denisov S., Georgiev P., Maksimenko O., 2011. Drosophila BTB/POZ domains of “ttk Group” can form multimers and selectively interact with each other. J. Mol. Biol. 412: 423–436. 10.1016/j.jmb.2011.07.052 [DOI] [PubMed] [Google Scholar]
- Brand A. H., Perrimon N., 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Brown J. L., Sonoda S., Ueda H., Scott M. P., Wu C., 1991. Repression of the Drosophila fushi tarazu (ftz) segmentation gene. EMBO J. 10: 665–674. 10.1002/j.1460-2075.1991.tb07995.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaharbakhshi E., Jemc J. C., 2016. Broad-complex, tramtrack, and bric-à-brac (BTB) proteins: critical regulators of development. Genesis 54: 505–518. 10.1002/dvg.22964 [DOI] [PubMed] [Google Scholar]
- Fichelson P., Gho M., 2003. The glial cell undergoes apoptosis in the microchaete lineage of Drosophila. Development 130: 123–133. 10.1242/dev.00198 [DOI] [PubMed] [Google Scholar]
- Fichelson P., Gho M., 2004. Mother-daughter precursor cell fate transformation after Cdc2 down-regulation in the Drosophila bristle lineage. Dev. Biol. 276: 367–377. 10.1016/j.ydbio.2004.08.043 [DOI] [PubMed] [Google Scholar]
- Fichelson P., Audibert A., Simon F., Gho M., 2005. Cell cycle and cell-fate determination in Drosophila neural cell lineages. Trends Genet. TIG 21: 413–420. 10.1016/j.tig.2005.05.010 [DOI] [PubMed] [Google Scholar]
- Gaber Z. B., Butler S. J., Novitch B. G., 2013. PLZF regulates fibroblast growth factor responsiveness and maintenance of neural progenitors. PLoS Biol. 11: e1001676 10.1371/journal.pbio.1001676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gho M., Lecourtois M., Géraud G., Posakony J. W., Schweisguth F., 1996. Subcellular localization of Suppressor of Hairless in Drosophila sense organ cells during Notch signalling. Development 122: 1673–1682. [DOI] [PubMed] [Google Scholar]
- Gho M., Bellaïche Y., Schweisguth F., 1999. Revisiting the Drosophila microchaete lineage: a novel intrinsically asymmetric cell division generates a glial cell. Development 126: 3573–3584. [DOI] [PubMed] [Google Scholar]
- Giesen K., Hummel T., Stollewerk A., Harrison S., Travers A., et al. , 1997. Glial development in the Drosophila CNS requires concomitant activation of glial and repression of neuronal differentiation genes. Development 124: 2307–2316. [DOI] [PubMed] [Google Scholar]
- Guo M., Bier E., Jan L. Y., Jan Y. N., 1995. Tramtrack acts downstream of numb to specify distinct daughter cell fates during asymmetric cell divisions in the Drosophila PNS. Neuron 14: 913–925. 10.1016/0896-6273(95)90330-5 [DOI] [PubMed] [Google Scholar]
- Guo M., Jan L. Y., Jan Y. N., 1996. Control of daughter cell fates during asymmetric division: interaction of Numb and Notch. Neuron 17: 27–41. 10.1016/S0896-6273(00)80278-0 [DOI] [PubMed] [Google Scholar]
- Harrison S. D., Travers A. A., 1988. Identification of the binding sites for potential regulatory proteins in the upstream enhancer element of the Drosophila fushi tarazu gene. Nucleic Acids Res. 16: 11403–11416. 10.1093/nar/16.24.11403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S. D., Travers A. A., 1990. The tramtrack gene encodes a Drosophila finger protein that interacts with the ftz transcriptional regulatory region and shows a novel embryonic expression pattern. EMBO J. 9: 207–216. 10.1002/j.1460-2075.1990.tb08097.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenstein V., Posakony J. W., 1989. Development of adult sensilla on the wing and notum of Drosophila melanogaster. Development 107: 389–405. [DOI] [PubMed] [Google Scholar]
- Jin Y., Nenseth H. Z., Saatcioglu F., 2017. Role of PLZF as a tumor suppressor in prostate cancer. Oncotarget 8: 71317 10.18632/oncotarget.19813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L., Richardson H., Saint R., 2000. Tissue-specific regulation of cyclin E transcription during Drosophila melanogaster embryogenesis. Development 127: 4619–4630. [DOI] [PubMed] [Google Scholar]
- Jordan K. C., Schaeffer V., Fischer K. A., Gray E. E., Ruohola-Baker H., 2006. Notch signaling through tramtrack bypasses the mitosis promoting activity of the JNK pathway in the mitotic-to-endocycle transition of Drosophila follicle cells. BMC Dev. Biol. 6: 16 10.1186/1471-213X-6-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim F. D., Guild G. M., Thummel C. S., 1993. The Drosophila Broad-Complex plays a key role in controlling ecdysone-regulated gene expression at the onset of metamorphosis. Development 118: 977–988. [DOI] [PubMed] [Google Scholar]
- Kelly K. F., Daniel J. M., 2006. POZ for effect – POZ-ZF transcription factors in cancer and development. Trends Cell Biol. 16: 578–587. 10.1016/j.tcb.2006.09.003 [DOI] [PubMed] [Google Scholar]
- Lai Z. C., Li Y., 1999. Tramtrack69 is positively and autonomously required for Drosophila photoreceptor development. Genetics 152: 299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman L. M., Stathopoulos A., 2009. Design flexibility in cis-regulatory control of gene expression: synthetic and comparative evidence. Dev. Biol. 327: 578–589. 10.1016/j.ydbio.2008.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Posakony J. W., 2014. An enhancer composed of interlocking submodules controls transcriptional autoregulation of Suppressor of hairless. Dev. Cell 29: 88–101. 10.1016/j.devcel.2014.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T. M., Lee E. H., Lim B., Shyh-Chang N., 2016. Concise review: balancing stem cell self-renewal and differentiation with PLZF. Stem Cells 34: 277–287. 10.1002/stem.2270 [DOI] [PubMed] [Google Scholar]
- McConnell M. J., Chevallier N., Berkofsky-Fessler W., Giltnane J. M., Malani R. B., et al. , 2003. Growth suppression by acute promyelocytic leukemia-associated protein PLZF is mediated by repression of c-myc expression. Mol. Cell. Biol. 23: 9375–9388. 10.1128/MCB.23.24.9375-9388.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore A. W., Jan L. Y., Jan Y. N., 2002. hamlet, a binary genetic switch between single- and multiple- dendrite neuron morphology. Science 297: 1355–1358. 10.1126/science.1072387 [DOI] [PubMed] [Google Scholar]
- Moore A. W., Roegiers F., Jan L. Y., Jan Y.-N., 2004. Conversion of neurons and glia to external-cell fates in the external sensory organs of Drosophila hamlet mutants by a cousin-cousin cell-type respecification. Genes Dev. 18: 623–628. 10.1101/gad.1170904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagans S., Piñeyro D., Kosoy A., Bernués J., Azorín F., 2004. Repression by TTK69 of GAGA-mediated activation occurs in the absence of TTK69 binding to DNA and solely requires the contribution of the POZ/BTB domain of TTK69. J. Biol. Chem. 279: 9725–9732. 10.1074/jbc.M313200200 [DOI] [PubMed] [Google Scholar]
- Ramat A., Audibert A., Louvet-Vallée S., Simon F., Fichelson P., et al. , 2016. Escargot and Scratch regulate neural commitment by antagonizing Notch activity in Drosophila sensory organs. Development 143: 3024–3034. 10.1242/dev.134387 [DOI] [PubMed] [Google Scholar]
- Read D., Manley J. L., 1992. Alternatively spliced transcripts of the Drosophila tramtrack gene encode zinc finger proteins with distinct DNA binding specificities. EMBO J. 11: 1035–1044. 10.1002/j.1460-2075.1992.tb05142.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy B. A., Bajpe P. K., Bassett A., Moshkin Y. M., Kozhevnikova E., et al. , 2010. Drosophila transcription factor Tramtrack69 binds MEP1 to recruit the chromatin remodeler NuRD. Mol. Cell. Biol. 30: 5234–5244. 10.1128/MCB.00266-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remaud S., Audibert A., Gho M., 2008. S-phase favours notch cell responsiveness in the Drosophila bristle lineage. PLoS One 3: e3646 10.1371/journal.pone.0003646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahut-Barnola I., Godt D., Laski F. A., Couderc J. L., 1995. Drosophila ovary morphogenesis: analysis of terminal filament formation and identification of a gene required for this process. Dev. Biol. 170: 127–135. 10.1006/dbio.1995.1201 [DOI] [PubMed] [Google Scholar]
- Sallé J., Campbell S. D., Gho M., Audibert A., 2012. CycA is involved in the control of endoreplication dynamics in the Drosophila bristle lineage. Development 139: 547–557. 10.1242/dev.069823 [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., et al. , 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9: 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siggs O. M., Beutler B., 2012. The BTB-ZF transcription factors. Cell Cycle 11: 3358–3369. 10.4161/cc.21277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon F., Fichelson P., Gho M., Audibert A., 2009. Notch and Prospero repress proliferation following cyclin E overexpression in the Drosophila bristle lineage. PLoS Genet. 5: e1000594 10.1371/journal.pgen.1000594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobieszczuk D. F., Poliakov A., Xu Q., Wilkinson D. G., 2010. A feedback loop mediated by degradation of an inhibitor is required to initiate neuronal differentiation. Genes Dev. 24: 206–218. 10.1101/gad.554510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Smith L., Armento A., Deng W.-M., 2008. Regulation of the endocycle/gene amplification switch by Notch and ecdysone signaling. J. Cell Biol. 182: 885–896. 10.1083/jcb.200802084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Guo X., Dou K., Chen H., Xi R., 2015. Ttk69 acts as a master repressor of enteroendocrine cell specification in Drosophila intestinal stem cell lineages. Development 142: 3321–3331. 10.1242/dev.123208 [DOI] [PubMed] [Google Scholar]
- Wodarz A., 2008. Extraction and immunoblotting of proteins from embryos. Methods Mol. Biol. 420: 335–345. 10.1007/978-1-59745-583-1_21 [DOI] [PubMed] [Google Scholar]
- Xiong W. C., Montell C., 1993. Tramtrack is a transcriptional repressor required for cell fate determination in the Drosophila eye. Genes Dev. 7: 1085–1096. 10.1101/gad.7.6.1085 [DOI] [PubMed] [Google Scholar]
- Xu T., Rubin G. M., 1993. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 117: 1223–1237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.8020103.