Condensins are evolutionarily conserved protein complexes that mediate chromosome compaction during cell division and have been implicated in gene regulation during interphase. Here, Street et al. analyzed the gene regulatory role of an X-specific...
Keywords: chromatin, gene expression, gene regulation, X chromosome, transcription, histone modifications, dosage compensation, condensin, C. elegans
Abstract
Condensins are evolutionarily conserved protein complexes that are required for chromosome segregation during cell division and genome organization during interphase. In Caenorhabditis elegans, a specialized condensin, which forms the core of the dosage compensation complex (DCC), binds to and represses X chromosome transcription. Here, we analyzed DCC localization and the effect of DCC depletion on histone modifications, transcription factor binding, and gene expression using chromatin immunoprecipitation sequencing and mRNA sequencing. Across the X, the DCC accumulates at accessible gene regulatory sites in active chromatin and not heterochromatin. The DCC is required for reducing the levels of activating histone modifications, including H3K4me3 and H3K27ac, but not repressive modification H3K9me3. In X-to-autosome fusion chromosomes, DCC spreading into the autosomal sequences locally reduces gene expression, thus establishing a direct link between DCC binding and repression. Together, our results indicate that DCC-mediated transcription repression is associated with a reduction in the activity of X chromosomal gene regulatory elements.
REGULATION of chromosome structure is essential for the establishment and maintenance of accurate gene expression. A key regulator of chromosome structure across all organisms is condensin, a multi-subunit protein complex that belongs to the family of structural maintenance of chromosomes (SMC) complexes (Hirano 2006; van Ruiten and Rowland 2018). Condensins are required for chromosome condensation and segregation in all eukaryotes (Hirano 2016). Condensins are also important for genome organization and have been implicated in gene regulation during interphase (Paul et al. 2018). Genome-wide binding experiments indicate that condensins bind to a subset of gene regulatory elements including promoters, enhancers, tRNA genes, and topologically associated domain (TAD) boundaries (Jeppsson et al. 2014). However, the link between condensin binding at these sites and its function in gene regulation remains unknown.
Here, we addressed the link between condensin and transcription by using a clear paradigm for the gene regulatory function of condensins, the Caenorhabditis elegans dosage compensation complex (DCC). Like most metazoans, C. elegans contain two types of condensins (I and II) that partially differ in their subunit composition, chromosomal binding, and function (Csankovszki et al. 2009). In addition to the canonical condensins, C. elegans contain a third condensin, condensin IDC (hereafter DC), that differs from condensin I by a single SMC-4 variant, DPY-27 (Csankovszki et al. 2009). Condensin DC interacts with additional subunits necessary for DCC binding and function (Figure 1A) [reviewed in Albritton and Ercan (2018)]. The DCC specifically binds to both hermaphrodite X chromosomes and represses each by one-half to equalize X chromosomal transcript levels between XX hermaphrodites and XO males (Jans et al. 2009; Kruesi et al. 2013; Kramer et al. 2015, 2016).
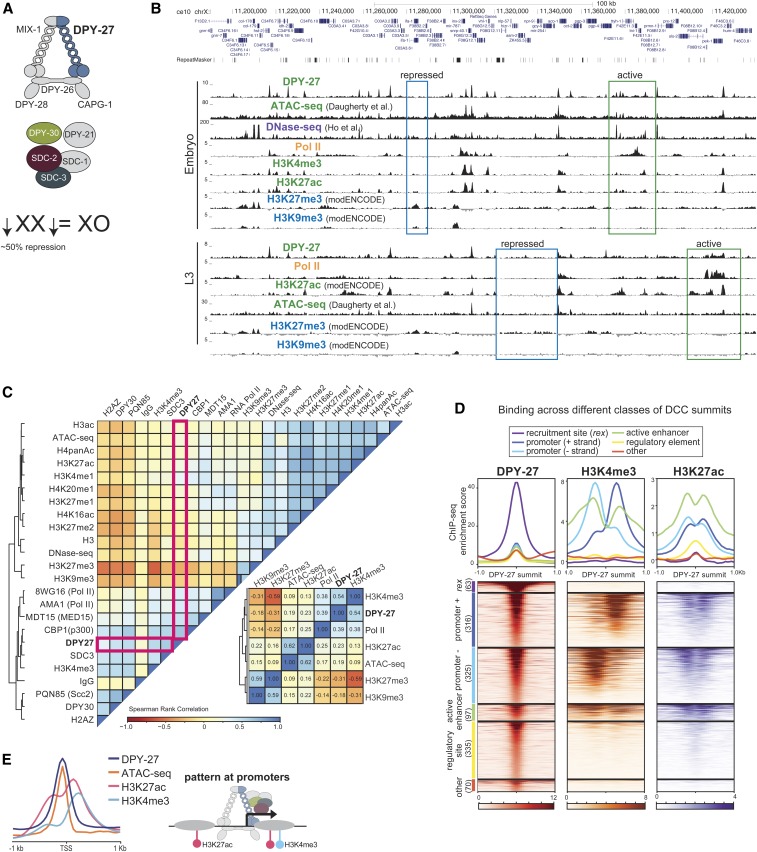
Figure 1.
DCC binding correlates with active chromatin marks at gene regulatory elements. (A) The C. elegans DCC contains a specialized condensin complex (condensin DC) that is distinguished from canonical condensin I by a single SMC-4 variant, DPY-27. The noncondensin DCC subunits SDC-2, SDC-3, and DPY-30 interact with condensin DC, and are required for its recruitment to the X chromosomes. DPY-21 is a histone demethylase that converts H4K20me2 to H4K20me1. DCC binds to and represses X chromosomes in hermaphrodites by approximately twofold. (B) ChIP-seq, DNase-seq (Ho et al. 2017), and ATAC-seq (Daugherty et al. 2017) profiles at a representative 250-kb region of the X chromosome in embryos and L3 larval stage worms. Example active and repressed chromatin regions are labeled in green and blue, respectively. DPY-27 (DCC) binding overlaps with Pol II binding, active chromatin marks, and accessible regions (ATAC-seq). (C) Spearman’s rank correlation values are shown for average ChIP-seq scores of histone modifications, and ATAC-seq and DNase-seq signals within 1-kb contiguous windows across the X chromosome in wild-type embryos. Zoomed-in plot highlights that DCC (DPY-27) binding positively correlates more with promoter marks (H3K4me3) and Pol II, with active enhancers (H3K27ac), and regulatory regions (ATAC-seq), and negatively correlates with repressive marks (H3K27me3 and H3K9me3). (D) The DPY-27 ChIP-seq peak summit coordinates were categorized according to their overlap with recruitment elements on the X chromosome [rex sites defined in Albritton et al. (2017)], promoters (+ and − strand) [within 250 bp of a GRO-seq (Kruesi et al. 2013) or 500 bp of a WormBase-defined TSS], active enhancers (overlapping a H3K27ac peak that is not a promoter), regulatory elements (overlapping an ATAC-seq or DNase-seq peak, and not a promoter or active enhancer), and other, unknown categories. DPY-27 (DCC), H3K4me3, and H3K27ac wild-type embryo ChIP-seq patterns are plotted across the DPY-27 ChIP-seq peak summits belonging to each category. (E) DCC, H3K4me3, and H3K27ac ChIP-seq and ATAC-seq signals are plotted across X chromosome TSSs [defined by GRO-seq (Kruesi et al. 2013)]; DCC signal coincides with the accessibility peak at promoters. ATAC-seq, assay for transposase-accessible chromatin using sequencing; ChIP-seq, chromatin immunoprecipitation sequencing; chr, chromosome; DC, condensin IDC; DCC, dosage compensation complex; DNase-seq, DNAse I hypersensitive site sequencing; GRO-seq, global run-on sequencing; modENCODE, Model Organism ENCyclopedia Of DNA Elements; Pol II, RNA polymerase II; SMC, structural maintenance of chromosomes; TSS, transcription start site.
The current model for DCC binding to the X chromosomes involves two steps: recruitment and spreading (Csankovszki et al. 2004). Recruitment is mediated in a hierarchical manner, whereby the DCC enters the X at a small number of strong recruitment sites, which are fully distinguished from the autosomes by the presence of multiple copies of a 12-bp recruitment motif (McDonel et al. 2006; Ercan et al. 2007; Jans et al. 2009) within high-occupancy transcription factor target (HOT) sites (Albritton et al. 2017). The stronger and weaker recruitment sites are thought to cooperate over long distances to robustly recruit the DCC to the X (Albritton et al. 2017). Unlike recruitment, spreading is an X-sequence-independent process and can occur on DNA physically attached to the X (Ercan et al. 2009). An estimated 50–100 recruitment sites separated by 0.1–1-Mb distances support binding of the DCC across the ∼17-Mb X chromosome (Jans et al. 2009; Kranz et al. 2013; Albritton et al. 2017).
Global run-on sequencing (GRO-seq) (Kruesi et al. 2013) and chromatin immunoprecipitation sequencing (ChIP-seq) (Kramer et al. 2015) analyses have shown that the DCC is required to reduce RNA polymerase II (RNA Pol II) binding at X chromosomal promoters. DCC-mediated repression appears to be chromosome-wide, with no large groups of genes escaping from dosage compensation (Kramer et al. 2015, 2016). Previous work has highlighted multiple roles for the DCC in the regulation of X chromosome structure; the DCC is required for the ∼40% compaction of the X chromosome compared to autosomes (Lau et al. 2014), the regulation of subnuclear localization of the X chromosomes (Sharma et al. 2014; Snyder et al. 2016), and the regulation of TADs on the X (Crane et al. 2015; Brejc et al. 2017). The DCC was also shown to increase and decrease the levels of H4K20me1 and H4K16ac, respectively, on the X chromosomes (Vielle et al. 2012; Wells et al. 2012). Reduction of H4K16ac on the X occurs downstream of H4K20me1 enrichment, and requires the deacetylase SIR-2.1 (Wells et al. 2012). H4K20me1 enrichment on the X is mediated by the H4K20me2 demethylase DPY-21, which physically interacts with the condensin core of the DCC (Brejc et al. 2017). How increased H4K20me1 and decreased H4K16ac mechanistically contribute to X chromosome repression is unclear (Kramer et al. 2015). In addition, previous studies did not address if the DCC regulates the level or the distribution of other histone modifications, such as H3K4me3 and H3K27ac, which are tightly linked to transcription regulation.
To address this question, we analyzed the distribution of several histone modifications in wild-type, DCC mutant, and DCC-depleted conditions, as well as in X-to-autosome fusion strains in which the DCC ectopically spreads into autosomal sequences (Ercan et al. 2009). In wild-type embryos and L3 larval animals, DCC-binding sites coincide with accessible putative gene regulatory elements marked by ATAC-seq (assay for transposase-accessible chromatin using sequencing) (Daugherty et al. 2017). In DCC mutant (dpy-21 null) or DCC-depleted [dpy-27 RNAi (RNA interference)] embryos, the levels of repressive histone modifications, including H3K9me3, remain unchanged while levels of active histone modifications, including H3K4me3 and H3K27ac, increase at X chromosomal promoters compared to autosomal ones. Further linking DCC binding to the regulation of active histone marks and gene expression, in X;V fusion chromosomes, ectopic spreading of the DCC into an autosomal sequence locally reduces both gene expression and H3K4me3. We also found that DCC depletion does not affect binding of PHA-4 transcription factor, the cohesin loader PQN-85 (Scc2 homolog), nor the putative H3K27 acetylase CBP-1, thus ruling out a model in which DCC indiscriminately reduces binding of proteins to the X chromosomes, as measured by ChIP-seq. Taken together, our results are consistent with a model in which the DCC fine-tunes transcription across the X through targeting and modulating the activity of gene regulatory elements by directly or indirectly reducing the levels of specific active histone modifications.
Materials and Methods
Worm strains and growth
Mixed developmental-stage embryos (wild-type N2) were isolated from gravid adults by bleaching. Mutant strains used in this study were CB428 (dpy-21(e428) V), OP37 (wgIs37 [pha-4::TY1::EGFP::3xFLAG + unc-119(+)]), YPT41 (X;II) and YPT47 (also known as 15eh#1, X;V) (Lowden et al. 2008), and GW638 (met-2(n4256) set-25(n5021) III) (Towbin et al. 2012). For ChIP samples, embryos or larvae were incubated in 2% formaldehyde for 30 min. Synchronized L3 worms were isolated by growing starved L1s for 24 hr at 22°. L1–L3 worms were isolated from asynchronous plates by passing larvae through a 20-μm filter, which the embryos and larvae with the expanded germline were not capable of flowing through. Large-scale RNAi knockdown for ChIP and RNA sequencing (RNA-seq) analyses was performed as described previously (Kranz et al. 2013). Briefly, bacteria with RNAi-inducing plasmids were grown in liquid and concentrated 130-fold to seed 6 × 10 cm plates. Synchronized N2 L1s were plated on RNAi plates and grown at 20° for 4 days to obtain gravid adults. Knockdown was verified by western blot analysis of DPY-27 compared to control (vector only) RNAi. In previous work, we found that knockdown of DPY-27 in embryos isolated from mothers that were fed RNAi bacteria was more efficient than knockdown in L3s isolated after feeding L1s for 1 day (Kramer et al. 2015), thus RNAi experiments were performed in embryos.
Antibodies and ChIP
Experiments were from at least two biological replicates with matching input samples as reference (Supplemental Material, File S1). ChIP-seq (Kranz et al. 2013) and mRNA-seq (Albritton et al. 2014) experiments were performed as previously described. Information on antibodies used in this study is given in File S1. Two new antibodies were used. The MDT-15 antibody was validated by western blot analysis upon RNAi knockdown and immunoprecipitation (Figure S7). The CBP-1 antibody did not show a measurable signal on western blot hybridization and immunofluorescence assays, but showed the expected ChIP-seq pattern overlapping with H3K27ac (Figure S1) and immunoprecipitated CBP-1 specifically, as analyzed by mass spectrometry (MS) (File S1). Briefly, whole-embryo extract was prepared by douncing and sonicating embryos (5 min, 30 sec on and 30 sec off in a Bioruptor) in lysis buffer (40 mM HEPES pH 7.5, 10% glycerol, 150 mM NaCl, 1 mM EDTA, and 0.5% NP-40) complemented with protease inhibitors. After spinning insoluble material at 17,000 × g for 15 min, 2 mg of protein were incubated overnight with 5 μg of rabbit polyclonal CBP-1 antibody and IgG as negative control, collected on protein A sepharose beads, washed five times using immunoprecipitation buffer (50 mM HEPES-KOH pH 7.6, 1 mM EDTA, and 150 mM NaCl), and subjected to trypsin digestion and MS by the New York University Medical School Proteomics Facility on an Orbitrap Fusion Lumos. Tandem MS spectra were searched against a Uniprot C. elegans database using Proteome Discoverer 1.4 (File S1).
ChIP-seq data processing
Single-end sequencing was performed by Illumina Genome Analyzer IIx, HiSeq-2000, HiSeq-2500, HiSeq-4000, or NextSeq 500. The raw and processed data are provided at the Gene Expression Omnibus database (GEO, http://www.ncbi.nlm.nih.gov/geo) under accession number GSE122639. ChIP data processing and peak finding was performed as described previously (Kranz et al. 2013). Briefly, 50–75-bp single-end reads were aligned to the C. elegans genome version WS220 using bowtie version 1.2.0 (Langmead et al. 2009), allowing two mismatches in the seed, returning the best alignment, and restricting multiple alignments to four sites in the genome. Mapped reads from ChIP and input were used to call peaks, and obtain read coverage per base using MACS version 1.4.3 (Zhang et al. 2008) with default parameters. ChIP scores per base were obtained by normalizing to the median coverage and subtracting the input coverage. To obtain summits for binding profiles that were a combination of focused and broad patterns, large peaks were split using PeakSplitter version 1.0 (Salmon-Divon et al. 2010) with a minimum height cut-off of 4 and a separation float of 0.86. The replicates, numbers of reads, and access information for the data sets are provided in File S1.
ChIP-seq data analysis
Data were visualized using University of California, Santa Cruz genome browser, ce10 (http://genome.ucsc.edu/). Heatmaps of ChIP enrichment across WS220- and GRO-seq-defined transcription start sites (TSSs) (Kruesi et al. 2013) were produced using Deeptools (Ramírez et al. 2014) with default parameters in Galaxy (doi: 10.1093/nar/gkw343). Changes in ChIP binding scores across TSS and peak summits were calculated by standardizing average ChIP scores within a 1-kb window centering at the TSS, or a 200-bp window centered at the summit through calculating = {log2 (mutant / wild-type) − mean [log2 (mutant / wild-type)]} / SD [log2 (mutant / wild-type)]. Box plots were produced in R using ggplot2 (https://ggplot2.tidyverse.org/). The whiskers extend from the hinge to the largest value no further than ± 1.5 IQR [interquartile range (distance between the first and third quartiles)] from the hinge. Outliers were not plotted. The notch shows the 95% C.I. of the median [median ± 1.58 * IQR / square root (sqrt) (n)]. Data analysis scripts are available at the Ercan laboratory pages on GitHub: https://github.com/ercanlab/street_et_al_2019/.
mRNA-seq data processing and analysis
Single-end sequencing was performed by Illumina HiSeq-2000. mRNA-seq data processing was performed as described previously (Albritton et al. 2017). Briefly, 50-bp single-end reads were aligned to the C. elegans genome version WS220 using TopHat version 2.1.1 (Trapnell et al. 2012), using default parameters. Count data were calculated using HTSeq version 0.6.1 (Anders et al. 2015) and normalized using the R package DESeq2 (Anders and Huber 2010). The resulting mRNA levels and expression ratios are provided in File S1.
Data availability statement
All data sets used in this publication, including processed files, can be found on the GEO under accession number GSE122639. Individual sample indexes can be found in File S1. Scripts and code used for analysis of this data are deposited at https://github.com/ercanlab. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.8160218.
Results
The DCC is preferentially enriched at active gene regulatory elements on the X
To understand the DCC’s effect on histone modifications, we first compared the genomic distribution of the DCC to marks of active and repressed chromatin, in embryos and L3 larval stages (Figure 1B). A combination of new and published ChIP-seq data, including those from the Model Organism ENCyclopedia Of DNA Elements (Gerstein et al. 2010), and published accessibility data for ATAC-seq (Daugherty et al. 2017) and DNAse I hypersensitive site sequencing (DNase-seq) (Ho et al. 2017), were used. New data were produced from at least two biological replicates that correlated based on visual examination of genome browser tracks (Figure S1). Summary and access information on all data sets is provided in File S1.
Genome browser analysis of DCC binding revealed a correlation with active chromatin (Figure 1B), as previously noted (Ercan et al. 2007; Jans et al. 2009). We used data from more recent studies to refine the comparisons. DCC-binding sites coincide with ATAC-seq peaks at promoters and enhancers containing RNA Pol II, H3K4me3, and H3K27ac (Figure 1B) (Daugherty et al. 2017). Conversely, marks of repressive chromatin, including H3K27me3 and H3K9me3, do not coincide with DCC binding (Figure 1B). Comparison of additional DCC subunits DPY-30 and SDC-3, histone modifications, and proteins including the transcription factor PHA-4, the cohesin loader subunit PQN-85 (Scc2 homolog), the putative H3K27 acetylase CBP-1 (p300 homolog), and the mediator subunit MDT-15 (Med15 homolog) support the conclusion that DCC binding coincides with gene regulatory sites (Figure S2). To further refine which proteins the DCC best correlates with, we plotted the Spearman’s rank correlation of average ChIP-seq enrichment within 1-kb contiguous windows across the X chromosome (Figure 1C). DCC subunit DPY-27 binding correlates best with H3K4me3, RNA Pol II, MDT-15, and CBP-1.
The complex pattern of the DCC ChIP-seq profile suggests different modes of binding
The DCC has a complex pattern of binding as measured by ChIP-seq, including somewhat uniform baseline enrichment across the X, peaks of different heights at promoters, enhancers, and within genes, and strong enrichment at the recruitment sites (Ercan et al. 2007; Albritton et al. 2017). To further categorize the sites of DCC enrichment, we focused on the top 50% of peaks sorted by their ChIP-seq score at the summit, mostly eliminating peak calls due to baseline DCC binding (Figure S3). Next, we categorized the DCC peaks as those located at recruitment sites (Albritton et al. 2017), promoters [within 250 bp of a GRO-seq or 500 bp of a WormBase-defined TSS (Kruesi et al. 2013)], active enhancers (overlapping a H3K27ac peak that is not a promoter), gene regulatory elements (overlapping an ATAC-seq or DNase-seq peak, and not a promoter or active enhancer), and unknown categories. We then plotted DCC, H3K4me3, and H3K27ac enrichment patterns across the DCC summits in each category (Figure 1D). This analysis revealed that the majority of DCC-binding peaks occur at active promoters and enhancers, and that the strength of DCC binding correlates with the activity of the gene regulatory site, as measured by H3K4me3 and H3K27ac enrichment. DCC binding at promoters coincides with chromatin accessibility, and surrounding H3K27ac and H3K4me3 enrichment at the +1 nucleosome (Figure 1E). Approximately one-third of DCC-binding sites show little H3K4me3 and H3K27ac enrichment (Figure 1D), suggesting that DCC binding is not restricted to elements marked by these modifications.
DCC binding at promoters partially correlates with their transcriptional activity
To understand the different modes of DCC binding, we next scrutinized the level of correlation between DCC binding and transcription, as shown by ChIP-chip analysis comparing DPY-27 and RNA Pol II in embryos and L4/young adults (Ercan et al. 2009). Similarly, DCC and RNA Pol II binding [as measured by ChIP-seq using an 8WG16 antibody recognizing the unmodified C-terminal of AMA-1 (large subunit)] show DCC enrichment changes at genes differentially bound by RNA Pol II in embryos and L3s (Figure 2A). Supporting the conclusion that DCC correlates with active transcription, DCC binding at promoters is higher at genes that are being transcribed, compared to silent genes and genes whose mRNAs were maternally deposited in embryos (Figure 2B).
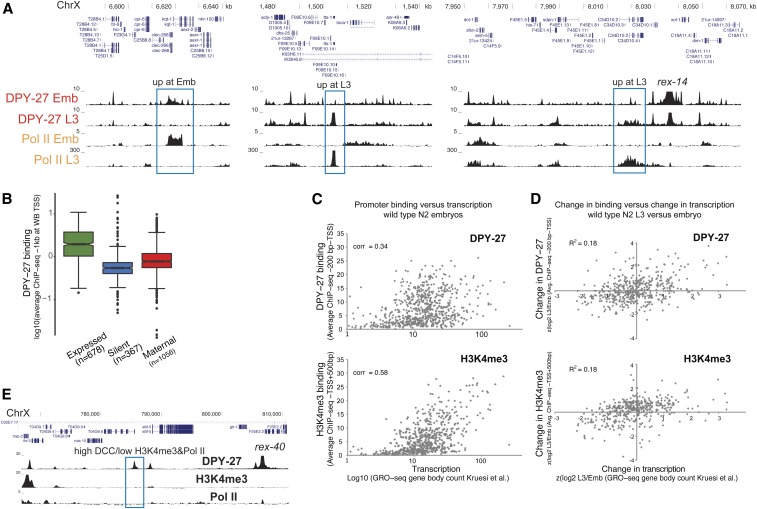
Figure 2.
DCC enrichment at promoters partially correlates with transcriptional activity. (A) DPY-27 ChIP-seq binding across example X chromosomal regions with differential transcription, as shown by Pol II ChIP-seq in embryos vs. L3s. (B) Average DPY-27 ChIP-seq score at 1-kb windows centering around the X chromosomal WB-defined TSS sites were plotted. Genes were categorized as expressed [N2 embryos FPKM > 1 (Kramer et al. 2015) and detected in GRO-seq (Kruesi et al. 2013)], silent (FPKM = 0 and not detected in GRO-seq), and maternally loaded (FPKM > 1 and not detected in GRO-seq). (C) Average DPY-27 and H3K4me3 ChIP-seq scores at proximal promoters [200 bp downstream to – TSS defined by Kruesi et al. (2013)] were plotted on the y-axis, and transcription levels of genes [GRO-seq counts at corresponding gene bodies (Kruesi et al. 2013)] were plotted on the x-axis. Spearman’s rank correlation coefficients are shown on the top left of each plot. (D) Changes in DPY-27 binding at promoters on the y-axis [z score of log2 L3/embryo ratio of average ChIP-seq score within proximal promoters as in (C)] were compared to changes in transcription on the x-axis [z score of log2 L3/embryo of transcription level as in (C)] in L3 vs. embryos. Changes in DPY-27 and H3K4me3 partially correlate with the change in transcription at individual promoters. (E) University of California, Santa Cruz browser view of DPY-27, H3K4me3, and Pol II ChIP-seq signals across a 40-kb region containing a recruitment site. The DCC-binding peak highlighted with a blue rectangle shows low Pol II and H3K4me3, suggesting that DCC enrichment and transcriptional activity at promoters can be uncoupled. ChIP-seq, chromatin immunoprecipitation sequencing; Chr, chromosome; DCC, dosage compensation complex; FPKM, fragments per kilobase of transcript per million mapped reads; GRO-seq, global run-on sequencing; Pol II, RNA polymerase II; TSS, transcription start site; WB, WormBase.
However, the level of positive correlation between the DCC and transcription [as measured by GRO-seq (Kruesi et al. 2013), Spearman’s rank correlation of 0.34] is less than that observed between H3K4me3 and transcription (0.58), suggesting that the link between DCC and RNA Pol II binding is weaker than that of H4K3me3 (Figure 2C). To evaluate how the DCC and H3K4me3 are tuned to transcription at individual promoters, we plotted the change in DCC or H3K4me3 levels vs. the change in transcription between embryos and L3s (Figure 2D). While there is a slight positive correlation, both DCC and H3K4me3 do not perfectly follow transcription changes at individual promoters. Furthermore, at and near recruitment sites, we found sites with high DCC, and low H3K4me3 and RNA Pol II, levels (Figure 2E). These results suggest that while DCC binding generally correlates with transcriptional activity, the two are not strictly coupled.
The DCC reduces the levels of active histone modifications on the X
To determine the DCC’s effect on gene regulatory elements, we analyzed several histone modifications associated with active and repressed chromatin upon DCC knockdown (dpy-27 RNAi) and mutation (dpy-21(e428) V) in embryos. Since DCC represses transcription by approximately twofold, we expected and observed subtle changes. To quantify such subtle changes, we used the autosomes as an internal control for ChIP efficiency and calculated the standardized ratio of ChIP enrichment in mutant vs. wild-type worms. This approach detected previously described X-specific changes upon DCC mutation and knockdown, including H4K20me1, H4K16ac (Vielle et al. 2012; Wells et al. 2012) (Figure 3A), and RNA Pol II (Pferdehirt et al. 2011; Kramer et al. 2015) (Figure 3B). The levels of histone H3 and negative control IgG at promoters do not show a significant change (Figure 3C), ruling out a nonspecific effect on nucleosome occupancy. We then applied the same analyses to additional histone modifications and found a DCC-dependent reduction in the X chromosomal levels of H3K4me3 (Figure 3D), H3K27ac (Figure 3E), and H4pan-ac (K5,8,12,16), but not H3ac (Figure 3F), suggesting that DCC activity correlates with a reduction in specific active histone modifications at X chromosomal promoters.
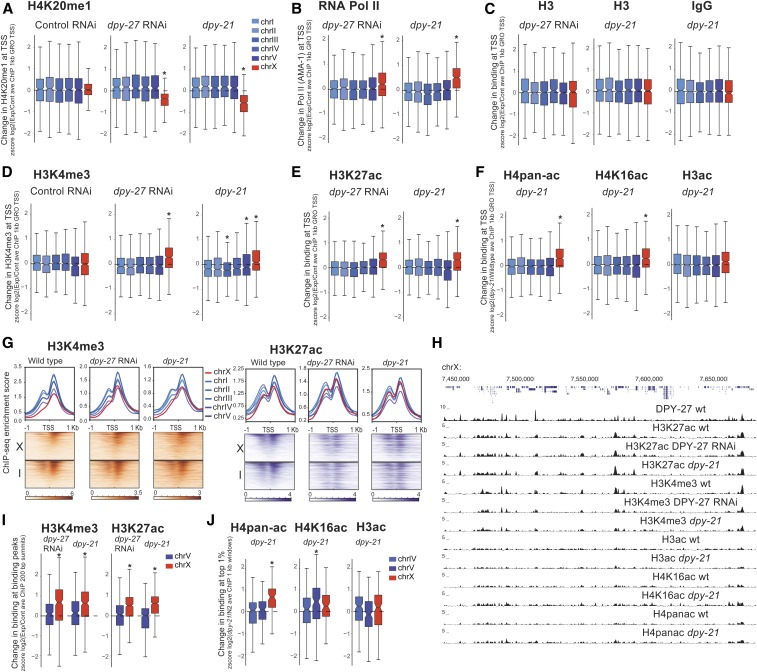
Figure 3.
DCC is required for the reduction of active histone modifications on the X chromosome. Changes in levels of histone modifications upon DCC defects in embryos are plotted. Average ChIP enrichment within 1-kb windows centered at the GRO-seq-defined TSSs (Kruesi et al. 2013) was calculated in wild-type (N2), DCC mutant (dpy-21), and DCC-depleted (dpy-27 RNAi) embryos. Change in the level of each histone modification was measured by standardizing (z score) the log2 ratio of experimental to control ChIP-seq scores. Values from each chromosome were tested against all the other autosomes using a two-tailed Student’s t-test, and resulting P-values that were 0.001 were marked with an asterisk. This analysis captured the expected changes in H4K20me1 (A) and RNA Pol II (B) at X chromosomal promoters upon DCC defect. (C) Neither H3 nor IgG negative control ChIP-seq data showed a comparable difference in the dpy-21 mutant, and H3 in dpy-27 RNAi suggesting that nucleosome levels were not significantly affected. (D–F) Same analysis of different histone modifications associated with active transcription. Note that H4pan-ac antibody also recognizes H4K16ac, thus changes may be due to this modification. (G) ChIP-seq enrichment for H3K4me3 and H3K27ac in wild-type, dpy-21 mutant, and dpy-27 RNAi knockdown embryos was plotted across the GRO-seq-defined TSSs (Kruesi et al. 2013) on chromosomes X and I. The level of enrichment is ordered in a descending manner using maximum coverage in wild-type worms. Mutant data were plotted in the same order. (H) Genome browser view of ChIP-seq profiles in wild-type, mutant, and knockdown embryos over a 250-kb representative region of the X chromosome. The pattern of enrichment in wild-type worms and mutants is largely similar. (I) As in (A), but change in binding at the wild-type peak summit, rather than a TSS. Standardized (z score) log2 ratio of mutant/wild-type ChIP-seq score within a 200-bp window centering at the summit of peaks in wild-type embryos. (J) Similar analysis as in (I), but change in top 1% of 1-kb windows ordered by average ChIP-seq in wild-type embryos. ave, average; ChIP-seq, chromatin immunoprecipitation sequencing; chr, chromosome; DCC, dosage compensation complex; GRO-seq, global run-on sequencing; RNAi, RNA interference; RNA Pol II, RNA polymerase II; TSS, transcription start site.
DCC depletion caused an increase in active histone modifications at their canonical locations rather than changing their distribution (Figure 3H). H3K4me3 and H3K27ac enrichment across the TSSs in wild-type, dpy-21 mutant, and dpy-27 RNAi conditions showed small differences, but generally, sites with high enrichment in the wild-type worms were still highly enriched in the mutant conditions (Figure 3G). Furthermore, Spearman’s rank correlation values, between wild-type and mutant H3K4me3 and H3K27ac enrichment within 1-kb contiguous windows, were similar on the autosomes and the X (H3K4me3 N2-CB428 on X: 0.59 and on autosomes: 0.57–0.65; H3K4me3 control-dpy-27 RNAi on X: 0.55 and on autosomes: 0.52–0.65; H3K27ac N2-CB428 on X: 0.79 and on autosomes: 0.79–0.85; and H3K27ac N2- dpy-27 RNAi on X: 0.85, and on autosomes: 0.84–0.89), indicating a lack of X-specific change in the distribution of histone modifications upon DCC defects. Collectively, these results suggest that DCC depletion did not eliminate or create new sites of enrichment on the X, but increased the level of H3K4me3 and H3K27ac at their canonical locations.
Since the distribution of modifications remains similar in the DCC knockdown embryos, we analyzed the level of change at their canonical sites (ChIP-seq peaks in wild-type). The levels of H3K4me3 and H3K27ac within 200 bp of their canonical binding summits increase specifically on the X upon DCC defects (Figure 3I). Peak calling on lower and broader ChIP-seq patterns observed for H4pan-ac, H4K16ac, and H3ac was difficult; therefore, to analyze their binding, we took the top 1% of 1-kb windows based on wild-type ChIP enrichment. In the dpy-21 mutant, the level of H4pan-ac increases specifically on the X, but H4K16ac and H3ac do not (Figure 3J). Greater variability in the dpy-21 mutant compared to dpy-27 RNAi may be due to additional dpy-21 activity outside dosage compensation (Kramer et al. 2015; Brejc et al. 2017). For H4K16ac, an X-specific effect at the TSS (Figure 3F), but not the top 1% of the H4K16ac sites (Figure 3J), suggests spatial specificity for DCC-mediated reduction of H4K16ac, possibly by SIR-2.1, which has been shown to be required for H4K16 deacetylation on the X chromosome (Wells et al. 2012).
Since DCC reduces RNA Pol II binding and active histone modifications on the X chromosome (Kruesi et al. 2013; Kramer et al. 2015), we asked if the DCC-dependent decrease in Pol II binding correlates with a decrease in histone modifications at individual promoters. RNA Pol II and DCC binding positively correlate with the levels of active histone modifications at promoters in both wild-type and dpy-21 mutant embryos (Figure S4A). At individual promoters on the X, changes in RNA Pol II binding did not correlate well with changes in active histone modifications (Figure S4B). Nevertheless, RNA Pol II and H3K4me3 ChIP-seq ratios in dpy-21 mutant vs. wild-type worms were positively correlated (0.21 for the AMA-1 antibody). Interestingly, DPY-27 and H3K27ac in dpy-21 vs. wild-type worms negatively correlated specifically on the X (Figure S4B, Spearman’s rank correlation of −0.32 on X chromosome and 0.04 on autosomes), supporting the idea that the DCC binding is linked to a reduction in H3K27ac on the X chromosomes.
The DCC does not affect repressive histone modifications
To test if the DCC affects histone modifications indiscriminately, we performed ChIP-seq analysis of H3K4me1, H3K27me1, H3K27me2, and H3K9me3 (Figure 4A). H3K4me1, H3K27me1, and H3K27me2 did not yield strong signals, precluding clear conclusions (Figure S1). Nevertheless, changes in these modifications were neither X-specific nor consistent between dpy-27 RNAi and dpy-21 mutant worms (Figure S5A). We observed no difference in H3K9me3 distribution on the X chromosomes between control and dpy-27 RNAi-treated embryos (Figure 4A). H3K9me3 levels showed higher variability in control and dpy-27 RNAi conditions, but the difference was not restricted to the X chromosome, suggesting that RNAi treatment affects H3K9me3 across the genome (Figure 4B). We also considered the opposite, and tested if H3K9me3 affects DCC localization by using a strain in which H3K9 methylation is eliminated (Towbin et al. 2012). In the absence of H3K9me3, the RNA Pol II binding pattern was similar to that of wild-type worms (Figure 4C), consistent with the lack of an overt effect on growth in laboratory conditions (Towbin et al. 2012). The reason for a general reduction in ChIP scores in the mutant is unclear. Regardless, RNA Pol II binding is not specifically different on the X chromosome (Figure 4D), consistent with there being no strong link between H3K9me3 and the DCC. Similarly, SDC-3 (a DCC subunit required for DPY-27 recruitment to the X) and CAPG-1 (HEAT domain subunit of condensin DC) binding profiles were similar between the wild-type and mutant (Figure 4C). Furthermore, CAPG-1 peaks in the H3K9me3 mutant largely overlapped with those of the wild-type (Figure 4E) and genomic sites with high H3K9me3 enrichment did not coincide with new SDC-3 sites (Figure 4F). Collectively, these results suggest that the DCC does not regulate and is not regulated by the heterochromatin mark H3K9me3.
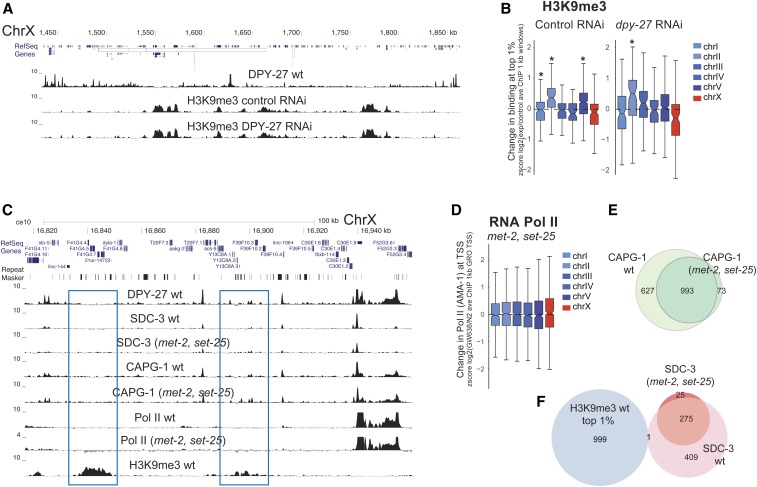
Figure 4.
DCC does not affect the levels of repressive histone marks. (A) ChIP-seq profile of H3K9me3 in control and DPY27 RNAi embryos along a representative region of chromosome X exemplifies no significant change in H3K9me3 upon DCC knockdown. (B) Distribution of standardized (z score) log2 dpy-27 RNAi/N2 ratios of H3K9me3 ChIP-seq average at the top 1% most enriched 1-kb windows. Values from each chromosome were tested against all the other autosomes using a two-tailed Student’s t-test, and resulting P-values that were 0.001 were marked with an asterisk. (C) ChIP-seq profiles of DPY-27, DCC subunits (SDC-3 and CAPG-1), Pol II, and H3K9me3 in wild-type embryos and the H3K9me3 null mutant (GW638, met-2, and set-25) across a representative region of the X chromosome. DCC and Pol II binding profiles remained similar in the met-2, set-25 mutant, including in regions enriched in H3K9me3 in wild-type worms (blue rectangle). (D) Pol II binding in the met-2, set-25 H3K9me3 null mutant (GW638) compared to N2 wild-type worms showed no specific effect on X chromosome expression. Distribution of standardized (z score) log2 mutant/N2 ratio of Pol II ChIP-seq within 1-kb windows centered at the GRO-seq-defined TSSs (Kruesi et al. 2013). (E) ChIP-seq peak overlap of DCC subunit CAPG-1 between wild-type and H3K9me3 null mutant. (F) ChIP-seq peak overlap between SDC-3 and top 1% H3K9me3 enriched 1-kb windows. ave, average; ChIP-seq, chromatin immunoprecipitation sequencing; Chr, chromosome; DCC, dosage compensation complex; GRO-seq, global run-on sequencing; RNAi, RNA interference; Pol II, RNA polymerase II; TSS, transcription start site.
DCC spreading into autosomal loci in X;A fusion chromosomes represses gene expression
To determine if DCC spreading reduces gene expression and histone modification levels locally, we analyzed strains containing X-to-autosome fusion (X;A) chromosomes. Previous work showed that the DCC spreads into the autosomal regions of X;V, X:II, and X:I fusion chromosomes, and that this spreading is linear and reduces with distance from the X (Ercan et al. 2009). Ectopic DCC binding leads to increased H4K20me1 in the autosomal region of spreading (Vielle et al. 2012). An earlier microarray analysis in embryos did not detect a significant difference in gene expression in the fusion strains (Ercan et al. 2009). Subsequent experiments suggested that dosage compensation starts in embryogenesis but is not complete until larval stages (Kramer et al. 2015), therefore we repeated the experiment using mRNA-seq in larvae.
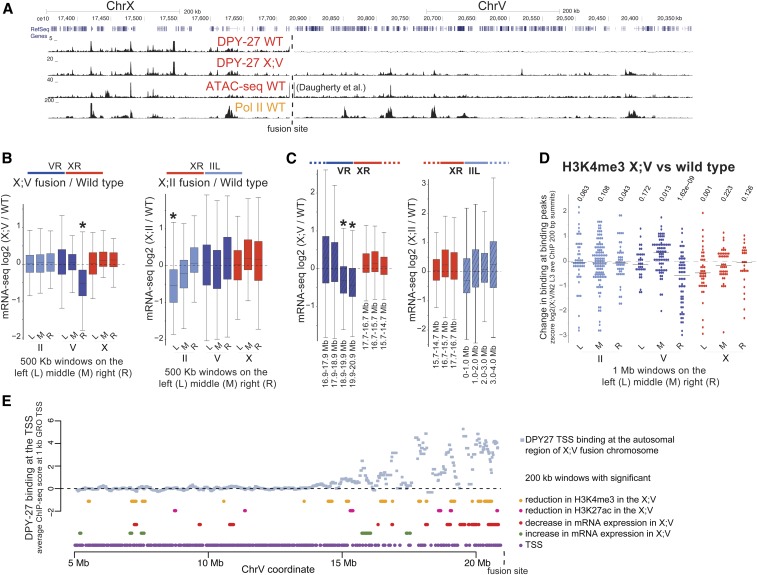
DPY-27 ChIP-seq analysis verified that the DCC binds to the autosomal region of spreading in the X;V fusion chromosomes in larvae (Figure 5A). To test if gene expression specifically changed in the region of spreading, we first took three 0.5-Mb windows at the middle, left- and right-most ends of each chromosome, and plotted the ratio of mRNA-seq levels for genes within each window. Average gene expression was significantly and specifically reduced at the side of X fusion, which was the right-most end of chromosome V in the X;V strain and the left-most end of chromosome II in the X;II strain (Figure 5B). The level of repression reduces with distance from the fusion site (Figure 5C) and is proportional to the level of spreading demonstrated for each fusion (Ercan et al. 2009). These results indicate that DCC spreading into the autosomal regions of the X;V and X;II fusion strains results in repression.
Figure 5.
DCC spreading into X;A fusion chromosomes reduces gene expression. (A) DPY-27 (DCC) ChIP-seq profile in the wild-type and X;V fusion chromosome-containing strains in L3 larvae. The spreading profile of the DCC in the autosomal region of the fusion chromosome is similar to that on the X, as indicated by the Pol II ChIP-seq and ATAC-seq signal in wild-type worms. (B) mRNA-seq analyses in wild-type embryos and strains containing X;V and X;II fusion chromosomes. DESeq log2 expression ratios were calculated and plotted for genes located within the middle, and the left- and right-most 500-kb windows of chromosomes II, V, and X. * P-value 0.001 (two-tailed Fisher’s test for each window against the rest of the windows across the genome). The schematics above the boxplots show which chromosome arms are fused. For X;II, the right end of X was fused to the left end of chromosome II, and for X;V, the right end of X was fused to the right end of chromosome V (Lowden et al. 2008). (C) Similar to (B), but expression ratios were plotted for genes within 1-Mb windows stepping out from the fusion site. The amount of repression decreases as a function of distance from the fusion border, following the pattern of DCC spreading (Ercan et al. 2009). (D) Change in H3K4me3 levels in X:V fusion chromosome compared to wild-type at the middle, left- and right-most 1-Mb windows of chromosomes II, V, and X. Standardized (z score) log2 X;V/wild-type ratios of ChIP-seq score within 200-bp H3K4me3 peak summits were plotted. H3K4me3 slightly but significantly decreases in the DCC spreading region (two-tailed Student’s t-test comparing ratios of each 1-Mb window against the rest across the genome). (E) Average DPY-27 ChIP-seq scores for 1-kb windows centering at GRO-seq-defined TSSs (Kruesi et al. 2013). Changes in expression and histone modifications were calculated by a moving average analysis using a 200-kb window with a 20-kb step size. For each 200-kb window, ChIP-seq and mRNA-seq ratios in X;V/wild-type were compared to the rest of the chromosome, and a P-value statistic was generated through a Student’s t-test. In this analysis, rather than asking if there is a significant change for each gene (as in DEseq), we asked whether the values in each window are higher or lower than the values observed for the rest of the windows along the chromosome. Windows with a P-value 0.01 are clustered toward the region of spreading. ATAC-seq, assay for transposase-accessible chromatin using sequencing; ChIP-seq, chromatin immunoprecipitation sequencing; Chr, chromosome; DCC, dosage compensation complex; GRO-seq, global run-on sequencing; L, left; M, middle; mRNA-seq, mRNA sequencing; R, right; TSS, transcription start site.
DCC spreading into autosomal loci leads to H3K4me3 reduction
In the X;V fusion strain, DCC spreads further (Ercan et al. 2009) and causes stronger repression (Figure 5C). Thus, we assayed the change in H3K4me3 and H3K27ac levels in the X;V fusion chromosomes. Since the expected change was small, we focused the analysis on where the signal was highest by taking a standardized ratio of ChIP-seq enrichment in X;V vs. wild-type at 200 bp around canonical peak summits. Despite high variability, there was a significant reduction in average H3K4me3 levels within the spreading domain of the X;V fusion chromosomes compared to wild-type (Figure 5D). For H3K27ac, there was higher variability across chromosomes, yet a slight reduction within the autosomal region of spreading was also observed (Figure S6A).
To be able to analyze DCC spreading with respect to the subtle changes in gene expression and histone modification levels, we used a sliding window analysis with 200-kb windows and 20-kb steps. For each window, we performed a Student’s t-test asking whether the ChIP-seq or mRNA-seq ratio within each window was significantly increased or decreased compared to the rest of the windows across chromosome V. Windows with P-values < 0.01 were plotted under the DPY-27 ChIP-seq enrichment in the X;V fusion chromosome (Figure 5E). Although noisy, the levels of gene expression, H3K4me3, and H3K27ac were slightly reduced in windows close to the fused end of chromosome V (Figure 5E). The lack of a similar pattern on other chromosomes (Figure S6B) supports the conclusion that DCC spreading into the autosomal region of the fused chromosome reduces active histone modifications and represses transcription.
DCC depletion does not significantly alter binding of PHA-4, CBP-1, and PQN-85
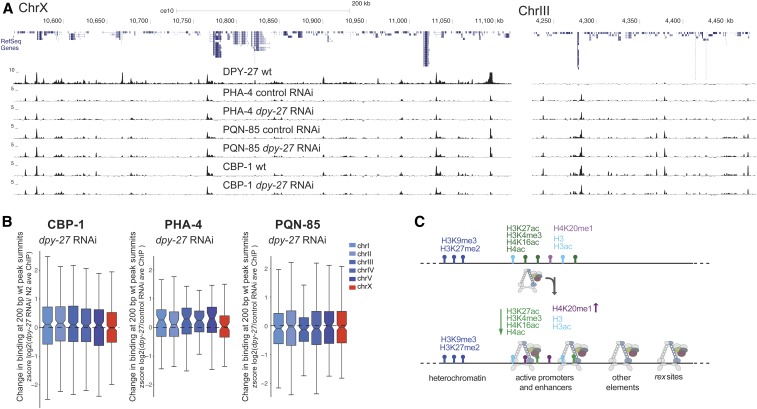
To test if the DCC represses transcription by reducing binding of all proteins to the X chromosomes, we performed ChIP-seq analysis of the transcription factor PHA-4, the putative H3K27 acetylase CBP-1 (p300 homolog), and the cohesin loader subunit PQN-85 (Scc2 homolog) (Figure 6A). We found no X-specific difference in the binding of these proteins as measured by ChIP-seq upon dpy-27 RNAi knockdown (Figure 6B), suggesting that the DCC does not indiscriminately displace proteins from the X.
Figure 6.
DCC knockdown does not indiscriminately reduce protein binding as measured by ChIP-seq. (A) ChIP-seq profiles of DPY-27 (condensin IDC subunit), PHA-4 (FOXA transcription factor), PQN-85 (Saccharomyces cerevisiae Scc2p homolog), and CBP-1 (putative H3K27 acetyltransferase) in representative regions on chromosomes X and III. (B) Analysis as in Figure 3I, plotting changes in protein binding across 200-bp wt peak summits. CBP-1, PQN-85, and PHA-4 levels on the X chromosomes did not change significantly upon DCC knockdown. (C) Summary of DCC binding and regulation of histone modifications on the X chromosomes. DCC-binding sites coincide with gene regulatory elements marked by accessible chromatin on the X. The majority of these elements also contain histone modifications associated with active transcription. The remaining include recruitment elements and sites that do not contain the analyzed histone modifications. DCC activity correlates with X-specific changes in the level of specific histone modifications (denoted by up and down arrows). ave, average; ChIP-seq, chromatin immunoprecipitation sequencing; Chr, chromosome; DCC, dosage compensation complex; RNAi, RNA interference; wt, wild-type.
Discussion
Our analysis of DCC distribution with respect to various chromatin marks reflects multiple modes of binding, including a baseline distribution, strong enrichment at the recruitment sites, and peaks of DCC enrichment at gene regulatory elements, partially correlating with transcription. Our results suggest that the DCC is required to reduce the levels of histone modifications that are associated with active transcription, including H3K4me3 and H3K27ac, but does not regulate, nor is it regulated by, the heterochromatin-associated histone modification H3K9me3. GRO-seq and ChIP-seq analysis of transcription in DCC mutants showed that the DCC reduces RNA Pol II binding to X chromosomal promoters (Pferdehirt et al. 2011; Kruesi et al. 2013; Kramer et al. 2015). Collectively, these results suggest a model in which DCC binding is directly or indirectly linked to a reduction in the activity of X chromosomal gene regulatory elements (Figure 6C).
The first question this model raises is how does the DCC target active gene regulatory elements? Accumulation at active promoters and enhancers is a conserved feature of condensins in C. elegans, Drosophila melanogaster, chicken, mouse, and human cells (Jeppsson et al. 2014). Condensins bind chromosomes by entrapping and/or encircling DNA through multiple interactions mediated by their ring structures (Cuylen et al. 2013; Kschonsak et al. 2017). One mechanism by which condensins may target gene regulatory elements is through binding to accessible DNA in vivo, which tends to coincide with active promoters and enhancers. Another possibility is through specific recruitment by transcription factors. In yeast and mammals, condensins are recruited to tRNA gene promoters and extra transcription factor IIIC (TFIIIC) sites by interacting with TFIIIC (D’Ambrosio et al. 2008; Haeusler et al. 2008; Iwasaki et al. 2010; Kranz et al. 2013; Van Bortle et al. 2014; Yuen et al. 2017), TATA-binding protein (TBP) (Iwasaki et al. 2015), and sequence-specific transcription factors (Kim et al. 2016) . In C. elegans, the strong DCC recruitment elements are HOT sites that are bound by multiple transcription factors (Albritton et al. 2017). Binding to accessible DNA and recruitment by specific transcription factors are not mutually exclusive mechanisms (Robellet et al. 2017). Indeed, condensin DC binding through both DNA accessibility and specific recruiter proteins may result in the complicated pattern of DCC distribution that we observe in vivo.
The second question that our model raises is how are specific histone modifications regulated by the DCC? Our work suggests that the DCC does not indiscriminately reduce binding of proteins to the X chromosome. The DCC may recruit specific histone deacetylases, e.g., sir-2.1, which is required to reduce H4K16ac on the X (Wells et al. 2012). The observation that the DCC reduces H3K27ac but not CBP-1 binding suggests that, similar to H4K16ac, H3K27ac reduction may also depend on a deacetylase. It is also possible that the DCC regulates the binding of specific histone modifying complexes. For instance, physical interaction of the DCC with a subunit of a chromatin-modifying complex may serve as a barrier. Supporting this idea, DPY-30, an essential subunit of the MLL/COMPASS (mixed lineage leukemia/complex of proteins associated with Set1) complex, physically interacts with the DCC (Pferdehirt et al. 2011). Intriguingly, a recent proteomic analysis found that mitotic chromosomes disproportionately lose chromatin-modifying complexes associated with euchromatin and not heterochromatin (Ginno et al. 2018). Furthermore, the level of displacement differed for different histone acetylases (Ginno et al. 2018). It is possible that reduced acetylation is connected to mitotic transcriptional repression, which is thought to be important for chromosome segregation (Sutani et al. 2015). Therefore, DCC-mediated transcriptional repression may have evolved from a conserved condensin role in regulating specific chromatin-modifying complexes in the formation of mitotic chromosomes.
The third question is how do histone modifications regulate RNA Pol II binding to X chromosomal promoters? H3K4me3 and H3K27ac are particularly instructive in models that predict gene expression from histone modifications (Gerstein et al. 2010; Karlic et al. 2010; Zhang and Zhang 2011). We also observed a strong correlation between RNA Pol II, H3K4me3, and H3K27ac. At individual promoters, the differential binding of RNA Pol II and histone modifications upon DCC defect was less correlated, perhaps due to insufficient sensitivity of the ChIP-seq assay in C. elegans embryos, and/or a complex quantitative relationship between Pol II recruitment and histone modifications at a given promoter (Pérez-Lluch et al. 2015). While it remains unclear if and how much H3K4me3 activates transcription directly (Howe et al. 2017), it has been shown that H3K4me3 interacts with specific transcriptional activators (Howe et al. 2017), and ectopic recruitment of H3K4me3 activates and maintains transcription (Cano-Rodriguez et al. 2016). H3K27ac is also associated with transcription activation, presumably by controlling transcription factor binding and RNA Pol II release from promoters (Stasevich et al. 2014). In C. elegans, a small proportion of genes show promoter pausing (Maxwell et al. 2014), thus H3K27ac may instead regulate dynamics of activator binding upstream of RNA Pol II recruitment. Recent work using histone mutants in D. melanogaster suggests that H3K27ac is not required for transcription (McKay et al. 2015; Leatham-Jensen et al. 2019); thus, future work is required to determine how instructive H3K27ac is for transcriptional activation in different systems.
The evolution of diverse dosage compensation strategies reveals how different transcriptional regulatory mechanisms can be coopted to regulate large domains within the genome. The DCC belongs to the deeply conserved family of SMC complexes, which are involved in genome organization and gene regulation across species (Hirano 2006; Dowen and Young 2014; Rowley and Corces 2018). Here, we have shown that the DCC targets gene regulatory elements and that its binding correlates with changes in the level of active histone modifications rather than their distribution, suggesting that C. elegans dosage compensation evolved to control transcriptional output without interfering with the underlying transcriptional program. A similar condensin-mediated tuning of histone modifications on mitotic chromosomes may be important for the proper inheritance of transcriptional programs after cell division. Whether the changes in histone modifications are a cause or consequence of transcriptional repression is an important open question. Understanding how the DCC directly or indirectly modulates histone modifications, and the transcriptional activity of gene regulatory elements, will help reveal mechanisms by which condensin-mediated organization of mitotic chromosomes affects gene regulation across cell division.
Acknowledgments
We thank Susan Gasser for providing the GW638 strain; Dominic Balcon and Jacob Carmichael for help with growing worms; the New York University (NYU) Center for Genomics and Systems Biology, University of North Carolina, and the Max Delbrück Center for Molecular Medicine, Berlin high-throughput sequencing facilities for sequencing; and Gyorgyi Csankovszki for providing the H4K16 antibody. Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health (NIH) under grant number R01 GM-107293. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD-010440). The mass spectrometry was funded in part by an NYU School of Medicine Laura and Isaac Perlmutter Cancer Center grant from the National Cancer Institute (P30 CA-016087), and a shared instrumentation grant for an Orbitrap Fusion Lumos (1S10 OD-010582-01A1).
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.8160218.
Communicating editor: M. Kuroda
Literature Cited
- Albritton S. E., Ercan S., 2018. Caenorhabditis elegans dosage compensation: insights into condensin-mediated gene regulation. Trends Genet. 34: 41–53. 10.1016/j.tig.2017.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albritton S. E., Kranz A. L., Rao P., Kramer M., Dieterich C., et al. , 2014. Sex-biased gene expression and evolution of the x chromosome in nematodes. Genetics 197: 865–883. 10.1534/genetics.114.163311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albritton S. E., Kranz A. L., Winterkorn L. H., Street L. A., Ercan S., 2017. Cooperation between a hierarchical set of recruitment sites targets the X chromosome for dosage compensation. Elife 6: e23645 . 10.7554/eLife.23645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Huber W., 2010. Differential expression analysis for sequence count data. Genome Biol. 11: R106 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P. T., Huber W., 2015. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brejc K., Bian Q., Uzawa S., Wheeler B. S., Anderson E. C., et al. , 2017. Dynamic control of X chromosome conformation and repression by a histone H4K20 demethylase. Cell 171: 85–102.e23. 10.1016/j.cell.2017.07.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano-Rodriguez D., Gjaltema R. A., Jilderda L. J., Jellema P., Dokter-Fokkens J., et al. , 2016. Writing of H3K4Me3 overcomes epigenetic silencing in a sustained but context-dependent manner. Nat. Commun. 7: 12284 10.1038/ncomms12284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane E., Bian Q., McCord R. P., Lajoie B. R., Wheeler B. S., et al. , 2015. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature 523: 240–244. 10.1038/nature14450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csankovszki G., McDonel P., Meyer B. J., 2004. Recruitment and spreading of the C. elegans dosage compensation complex along X chromosomes. Science 303: 1182–1185. 10.1126/science.1092938 [DOI] [PubMed] [Google Scholar]
- Csankovszki G., Collette K., Spahl K., Carey J., Snyder M., et al. , 2009. Three distinct condensin complexes control C. elegans chromosome dynamics. Curr. Biol. 19: 9–19. 10.1016/j.cub.2008.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuylen S., Metz J., Hruby A., Haering C. H., 2013. Entrapment of chromosomes by condensin rings prevents their breakage during cytokinesis. Dev. Cell 27: 469–478. 10.1016/j.devcel.2013.10.018 [DOI] [PubMed] [Google Scholar]
- D’Ambrosio C., Schmidt C. K., Katou Y., Kelly G., Itoh T., et al. , 2008. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 22: 2215–2227. 10.1101/gad.1675708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty A. C., Yeo R. W., Buenrostro J. D., Greenleaf W. J., Kundaje A., et al. , 2017. Chromatin accessibility dynamics reveal novel functional enhancers in C. elegans. Genome Res. 27: 2096–2107. 10.1101/gr.226233.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen J. M., Young R. A., 2014. SMC complexes link gene expression and genome architecture. Curr. Opin. Genet. Dev. 25: 131–137. 10.1016/j.gde.2013.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercan S., Giresi P. G., Whittle C. M., Zhang X., Green R. D., et al. , 2007. X chromosome repression by localization of the C. elegans dosage compensation machinery to sites of transcription initiation. Nat. Genet. 39: 403–408. 10.1038/ng1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercan S., Dick L. L., Lieb J. D., 2009. The C. elegans dosage compensation complex propagates dynamically and independently of X chromosome sequence. Curr. Biol. 19: 1777–1787. 10.1016/j.cub.2009.09.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein M. B., Lu Z. J., Van Nostrand E. L., Cheng C., Arshinoff B. I., et al. , 2010. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330: 1775–1787. 10.1126/science.1196914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginno P. A., Burger L., Seebacher J., Iesmantavicius V., Schubeler D., 2018. Cell cycle-resolved chromatin proteomics reveals the extent of mitotic preservation of the genomic regulatory landscape. Nat. Commun. 9: 4048 10.1038/s41467-018-06007-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler R. A., Pratt-Hyatt M., Good P. D., Gipson T. A., Engelke D. R., 2008. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev. 22: 2204–2214. 10.1101/gad.1675908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., 2006. At the heart of the chromosome: SMC proteins in action. Nat. Rev. Mol. Cell Biol. 7: 311–322. 10.1038/nrm1909 [DOI] [PubMed] [Google Scholar]
- Hirano T., 2016. Condensin-based chromosome organization from bacteria to vertebrates. Cell 164: 847–857. 10.1016/j.cell.2016.01.033 [DOI] [PubMed] [Google Scholar]
- Ho M. C. W., Quintero-Cadena P., Sternberg P. W., 2017. Genome-wide discovery of active regulatory elements and transcription factor footprints in Caenorhabditis elegans using DNase-seq. Genome Res. 27: 2108–2119. 10.1101/gr.223735.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe F. S., Fischl H., Murray S. C., Mellor J., 2017. Is H3K4me3 instructive for transcription activation? Bioessays 39: 1–12. 10.1002/bies.201600095 [DOI] [PubMed] [Google Scholar]
- Iwasaki O., Tanaka A., Tanizawa H., Grewal S. I., Noma K., 2010. Centromeric localization of dispersed Pol III genes in fission yeast. Mol. Biol. Cell 21: 254–265. 10.1091/mbc.e09-09-0790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki O., Tanizawa H., Kim K. D., Yokoyama Y., Corcoran C. J., et al. , 2015. Interaction between TBP and condensin drives the organization and faithful segregation of mitotic chromosomes. Mol. Cell 59: 755–767. 10.1016/j.molcel.2015.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans J., Gladden J. M., Ralston E. J., Pickle C. S., Michel A. H., et al. , 2009. A condensin-like dosage compensation complex acts at a distance to control expression throughout the genome. Genes Dev. 23: 602–618. 10.1101/gad.1751109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppsson K., Kanno T., Shirahige K., Sjogren C., 2014. The maintenance of chromosome structure: positioning and functioning of SMC complexes. Nat. Rev. Mol. Cell Biol. 15: 601–614. 10.1038/nrm3857 [DOI] [PubMed] [Google Scholar]
- Karlic R., Chung H. R., Lasserre J., Vlahovicek K., Vingron M., 2010. Histone modification levels are predictive for gene expression. Proc. Natl. Acad. Sci. USA 107: 2926–2931. 10.1073/pnas.0909344107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. D., Tanizawa H., Iwasaki O., Noma K. I., 2016. Transcription factors mediate condensin recruitment and global chromosomal organization in fission yeast. Nat. Genet. 48: 1242–1252. 10.1038/ng.3647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer M., Kranz A. L., Su A., Winterkorn L. H., Albritton S. E., et al. , 2015. Developmental dynamics of X–chromosome dosage compensation by the DCC and H4K20me1 in C. elegans. PLoS Genet. 11: e1005698 [corrigenda: PLoS Genet. 12: e1005899 (2016)]. 10.1371/journal.pgen.1005698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer M., Rao P., Ercan S., 2016. Untangling the contributions of sex-specific gene regulation and X–chromosome dosage to sex-biased gene expression in Caenorhabditis elegans. Genetics 204: 355–369. 10.1534/genetics.116.190298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz A. L., Jiao C. Y., Winterkorn L. H., Albritton S. E., Kramer M., et al. , 2013. Genome-wide analysis of condensin binding in Caenorhabditis elegans. Genome Biol. 14: R112 10.1186/gb-2013-14-10-r112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruesi W. S., Core L. J., Waters C. T., Lis J. T., Meyer B. J., 2013. Condensin controls recruitment of RNA polymerase II to achieve nematode X-chromosome dosage compensation. Elife 2: e00808 10.7554/eLife.00808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kschonsak M., Merkel F., Bisht S., Metz J., Rybin V., et al. , 2017. Structural basis for a safety-belt mechanism that anchors condensin to chromosomes. Cell 171: 588–600.e24. 10.1016/j.cell.2017.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L., 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau A. C., Nabeshima K., Csankovszki G., 2014. The C. elegans dosage compensation complex mediates interphase X chromosome compaction. Epigenetics Chromatin 7: 31 10.1186/1756-8935-7-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leatham-Jensen M., Uyehara C. M., Strahl B. D., Matera A. G., Duronio R. J., et al. , 2019. Lysine 27 of replication-independent histone H3.3 is required for Polycomb target gene silencing but not for gene activation. PLoS Genet. 15: e1007932 10.1371/journal.pgen.1007932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowden M. R., Meier B., Lee T. W., Hall J., Ahmed S., 2008. End joining at Caenorhabditis elegans telomeres. Genetics 180: 741–754. 10.1534/genetics.108.089920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell C. S., Kruesi W. S., Core L. J., Kurhanewicz N., Waters C. T., et al. , 2014. Pol II docking and pausing at growth and stress genes in C. elegans. Cell Rep. 6: 455–466. 10.1016/j.celrep.2014.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonel P., Jans J., Peterson B. K., Meyer B. J., 2006. Clustered DNA motifs mark X chromosomes for repression by a dosage compensation complex. Nature 444: 614–618. 10.1038/nature05338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay D. J., Klusza S., Penke T. J., Meers M. P., Curry K. P., et al. , 2015. Interrogating the function of metazoan histones using engineered gene clusters. Dev. Cell 32: 373–386. 10.1016/j.devcel.2014.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul M. R., Hochwagen A., Ercan S., 2019. Condensin action and compaction. Curr. Genet. 65: 407–415. 10.1007/s00294-018-0899-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Lluch S., Blanco E., Tilgner H., Curado J., Ruiz-Romero M., et al. , 2015. Absence of canonical marks of active chromatin in developmentally regulated genes. Nat. Genet. 47: 1158–1167. 10.1038/ng.3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pferdehirt R. R., Kruesi W. S., Meyer B. J., 2011. An MLL/COMPASS subunit functions in the C. elegans dosage compensation complex to target X chromosomes for transcriptional regulation of gene expression. Genes Dev. 25: 499–515. 10.1101/gad.2016011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F., Dündar F., Diehl S., Grüning B. A., Manke T., 2014. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42: W187–W191. 10.1093/nar/gku365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robellet X., Vanoosthuyse V., Bernard P., 2017. The loading of condensin in the context of chromatin. Curr. Genet. 63: 577–589. 10.1007/s00294-016-0669-0 [DOI] [PubMed] [Google Scholar]
- Rowley M. J., Corces V. G., 2018. Organizational principles of 3D genome architecture. Nat. Rev. Genet. 19: 789–800. 10.1038/s41576-018-0060-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon-Divon M., Dvinge H., Tammoja K., Bertone P., 2010. PeakAnalyzer: genome-wide annotation of chromatin binding and modification loci. BMC Bioinformatics 11: 415 10.1186/1471-2105-11-415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R., Jost D., Kind J., Gomez-Saldivar G., van Steensel B., et al. , 2014. Differential spatial and structural organization of the X chromosome underlies dosage compensation in C. elegans. Genes Dev. 28: 2591–2596. 10.1101/gad.248864.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M. J., Lau A. C., Brouhard E. A., Davis M. B., Jiang J., et al. , 2016. Anchoring of heterochromatin to the nuclear lamina reinforces dosage compensation-mediated gene repression. PLoS Genet. 12: e1006341 10.1371/journal.pgen.1006341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasevich T. J., Hayashi-Takanaka Y., Sato Y., Maehara K., Ohkawa Y., et al. , 2014. Regulation of RNA polymerase II activation by histone acetylation in single living cells. Nature 516: 272–275. 10.1038/nature13714 [DOI] [PubMed] [Google Scholar]
- Sutani T., Sakata T., Nakato R., Masuda K., Ishibashi M., et al. , 2015. Condensin targets and reduces unwound DNA structures associated with transcription in mitotic chromosome condensation. Nat. Commun. 6: 7815 10.1038/ncomms8815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin B. D., Gonzalez-Aguilera C., Sack R., Gaidatzis D., Kalck V., et al. , 2012. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell 150: 934–947. 10.1016/j.cell.2012.06.051 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., et al. , 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578 [corrigenda: Nat. Protoc. 9: 2513 (2014)]. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bortle K., Nichols M. H., Li L., Ong C. T., Takenaka N., et al. , 2014. Insulator function and topological domain border strength scale with architectural protein occupancy. Genome Biol. 15: R82 10.1186/gb-2014-15-5-r82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ruiten M. S., Rowland B. D., 2018. SMC complexes: universal DNA looping machines with distinct regulators. Trends Genet. 34: 477–487. 10.1016/j.tig.2018.03.003 [DOI] [PubMed] [Google Scholar]
- Vielle A., Lang J., Dong Y., Ercan S., Kotwaliwale C., et al. , 2012. H4K20me1 contributes to downregulation of X-linked genes for C. elegans dosage compensation. PLoS Genet. 8: e1002933 10.1371/journal.pgen.1002933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells M. B., Snyder M. J., Custer L. M., Csankovszki G., 2012. Caenorhabditis elegans dosage compensation regulates histone H4 chromatin state on X chromosomes. Mol. Cell. Biol. 32: 1710–1719. 10.1128/MCB.06546-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen K. C., Slaughter B. D., Gerton J. L., 2017. Condensin II is anchored by TFIIIC and H3K4me3 in the mammalian genome and supports the expression of active dense gene clusters. Sci. Adv. 3: e1700191 10.1126/sciadv.1700191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., et al. , 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Zhang M. Q., 2011. Histone modification profiles are predictive for tissue/cell-type specific expression of both protein-coding and microRNA genes. BMC Bioinformatics 12: 155 10.1186/1471-2105-12-155 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data sets used in this publication, including processed files, can be found on the GEO under accession number GSE122639. Individual sample indexes can be found in File S1. Scripts and code used for analysis of this data are deposited at https://github.com/ercanlab. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.8160218.