Abstract
Signaling pathways can regulate biological responses by the transcriptional regulation of target genes. In yeast, multiple signaling pathways control filamentous growth, a morphogenetic response that occurs in many species including fungal pathogens. Here, we examine the role of signaling pathways that control filamentous growth in regulating adhesion-dependent surface responses, including mat formation and colony patterning. Expression profiling and mutant phenotype analysis showed that the major pathways that regulate filamentous growth [filamentous growth MAPK (fMAPK), RAS, retrograde (RTG), RIM101, RPD3, ELP, SNF1, and PHO85] also regulated mat formation and colony patterning. The chromatin remodeling complex, SAGA, also regulated these responses. We also show that the RAS and RTG pathways coregulated a common set of target genes, and that SAGA regulated target genes known to be controlled by the fMAPK, RAS, and RTG pathways. Analysis of surface growth-specific targets identified genes that respond to low oxygen, high temperature, and desiccation stresses. We also explore the question of why cells make adhesive contacts in colonies. Cell adhesion contacts mediated by the coregulated target and adhesion molecule, Flo11p, deterred entry into colonies by macroscopic predators and impacted colony temperature regulation. The identification of new regulators (e.g., SAGA), and targets of surface growth in yeast may provide insights into fungal pathogenesis in settings where surface growth and adhesion contributes to virulence.
Keywords: MAPK pathways, signaling networks, expression profiling, fungal pathogenesis, microbial predator–prey relationships, pseudohyphal growth, invasive growth, biofilm, mat, complex colony, temperature control
FUNGAL microorganisms exhibit a range of nutrient-related responses. Under certain conditions, fungal cells can differentiate into filamentous or hyphal cells that can expand across, and/or penetrate into, new environments (Soll and Daniels 2016). Many fungal species can also grow in communities of biofilms or mats, which are composed of interconnected cells that attach to each other and to surfaces. One property of mats is the formation of highly organized patterns that result from adhesive contacts between cells. In pathogens, filamentous growth (Lo et al. 1997) and biofilm formation (Desai et al. 2014) are critical determinants of virulence. For example, cells can adhere to medical devices and grow in dense mats that are resistant to antifungal medicines (Chandra et al. 2001; Sudbery et al. 2004; Kumamoto 2005; Ramage et al. 2005; Nett and Andes 2015).
The budding yeast Saccharomyces cerevisiae is a unicellular fungal microbe, and a convenient model for studying nutrient-regulated foraging responses like filamentous growth and mat formation. These responses are best studied in “wild” strain backgrounds (such as Σ1278b) as the responses have been lost in certain laboratory strains due to genetic manipulation (Liu et al. 1996; Dowell et al. 2010; Chin et al. 2012). During filamentous growth, yeast cells differentiate into elongated and polarized filaments that remain connected in pseudohyphae (Gimeno et al. 1992; Cullen and Sprague 2012).
At least 600 genes have been identified by genetic screens (Lorenz and Heitman 1998; Palecek et al. 2000) and genome-wide studies (Jin et al. 2008; Xu et al. 2010; Ryan et al. 2012) that play some role in filamentous growth. A subset of these genes encode signaling pathway components that include at least four major nutrient-sensing pathways [filamentous growth MAPK (fMAPK), RAS, TOR (target of rapamycin), and SNF1], as well as pathways that regulate the response to pH (RIM101), phosphate utilization (PHO85), and mitochondrial stress [the retrograde mitochondria-to-nucleus (RTG) pathway]. In addition, proteins that control the epigenetic modification of histones to alter gene expression have also been implicated in the regulation of filamentous growth (Rpd3p).
Many of the pathways that regulate filamentous growth are functionally connected through their ability to coregulate common target genes (Figure 1A). In some cases, this occurs at the level of transcription. In a pioneering study, it was shown that many of the transcription factors that control filamentous growth control each other’s expression (Borneman et al. 2006). Transcription factors can also converge at common promoter elements. One example is the gene encoding the major cell adhesion molecule in yeast, Flo11p (Kraushaar et al. 2015; Chan et al. 2016). The FLO11 gene contains one of the largest and most highly regulated promoters in the yeast genome, and functions as a hub where multiple transcription factors and chromatin remodeling enzymes bind (Robertson and Fink 1998; Rupp et al. 1999; Palecek et al. 2000; Pan and Heitman 2000; Kuchin et al. 2002; van Dyk et al. 2005; Barrales et al. 2008). Signaling pathways that control filamentous growth can also regulate each other’s activity. The classic example comes from the discovery that the RAS pathway can regulate the activity of the fMAPK pathway (Mösch et al. 1996). It is now clear that many pathways regulate the activity of the fMAPK pathway (Chavel et al. 2010, 2014). One way this may occur is through the protein kinases of the major regulatory pathways, which can regulate each other’s localization and activity (Bharucha et al. 2008).
Figure 1.
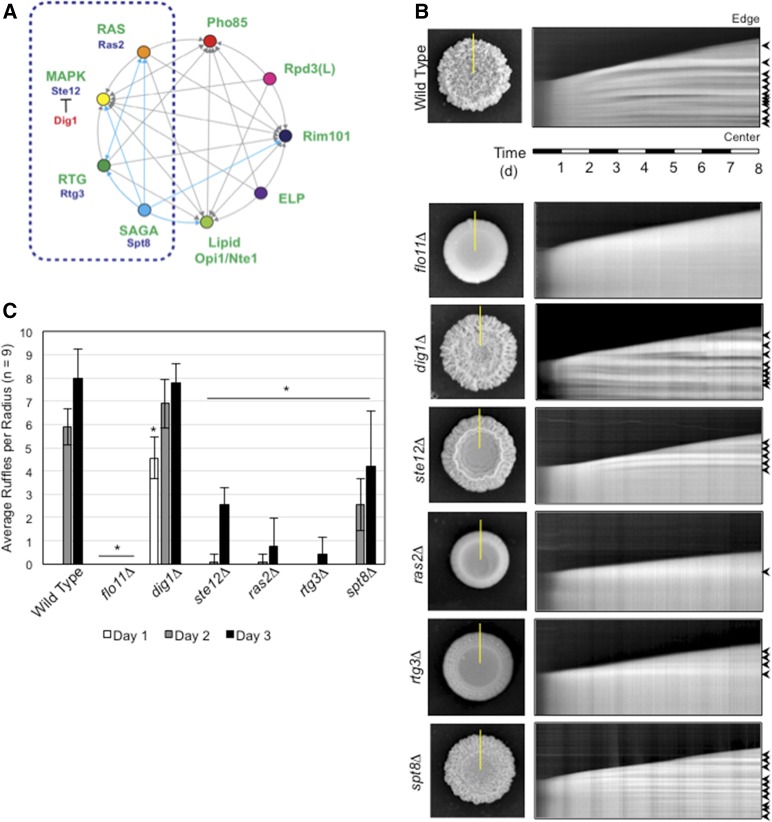
Functionally interconnected signaling pathways regulate mat expansion and colony pattern formation over time. (A) Circles represent signaling pathways or regulatory complexes that control filamentous growth. Arrows refer to target genes coregulated by pathways, as adapted from Chavel et al. (2014). Cyan arrows refer to functional connections identified in this study. The dashed blue box marks the part of the network on which the study is focused. (B) Time-course experiment of colony pattern formation in wild-type cells and the indicated mutants. Colony expansion was examined over an 8-day time period (Videos S1–S7). In the left panels, colonies at the 8-day time period are shown. On the right are kymographs that show colony expansion over time. The yellow lines in the left panels refer to the region selected for kymograph analysis. The panels on the right show colony patterns that developed over time in the region selected by the yellow line, where the bottom of the figure represents the colony center and the top represents the colony perimeter (edge). Arrows mark features that are characteristic of ruffle formation, which is quantitated in (C). (C) Bar graph showing the average number of ruffles in wild-type and the indicated mutant colonies at t = 1 day (white), 2 days (gray), and 3 days (black). Error bars represent SD among nine radii from three biological replicates. * P < 0.05 between the mutant and wild-type for the same time point.
Yeast can also undergo mat formation (Reynolds and Fink 2001; Bojsen et al. 2012), where colonies expand radially across surfaces and form ruffled patterns (Granek and Magwene 2010; Granek et al. 2011; Tam et al. 2018). Mats form wheel-spoke patterns in low-percentage agar media [0.3% agar (Reynolds and Fink 2001)], and wrinkled or ruffled colonies on high-percentage agar media [1–4% (Granek and Magwene 2010; Karunanithi et al. 2012)]. Mat expansion and patterning require contacts between cells that are also mediated by Flo11p. Regulators of mat formation have been identified by direct approaches (Reynolds 2006; Sarode et al. 2011; Váchová et al. 2011), genome-wide deletion screens (Ryan et al. 2012; Voordeckers et al. 2012; Scherz et al. 2014), overexpression screens (Cromie et al. 2017), and comparative expression profiling (Traven et al. 2012; Maršíková et al. 2017). Many of the molecular pathways that control mat formation regulate FLO11 expression. In addition, several regulators of mat formation, like the cell wall integrity sensor Wsc1p (Sarode et al. 2014) and vacuolar protein-sorting genes (Sarode et al. 2011), appear to regulate mat formation by mechanisms that do not involve Flo11p. Despite the identification of many proteins and pathways that regulate mat formation, the benefits that cells derive from forming these complex patterns remain unclear. Furthermore, the stresses encountered during colonial surface growth remain relatively unexplored.
As seen in other fungi, filamentous growth and mat formation are related responses in yeast. Both responses require Flo11p (Lo and Dranginis 1998; Reynolds and Fink 2001), and a corresponding set of regulatory proteins and pathways that control FLO11 expression (Ryan et al. 2012). These include TOR (Cutler et al. 2001; Bojsen et al. 2016), Rim101 (Li and Mitchell 1997; Sarode et al. 2011; Voordeckers et al. 2012), fMAPK (Roberts and Fink 1994; Reynolds and Fink 2001; Granek and Magwene 2010), and RAS (Gimeno et al. 1992; Granek and Magwene 2010; Zara et al. 2011; Ryan et al. 2012; Bojsen et al. 2016). The tRNA modification complex elongator (ELP) regulates all three responses (Abdullah and Cullen 2009). It has also been shown that under nutrient-limiting conditions, mats are composed of filamentous cells (Karunanithi et al. 2012). Given the abovementioned connections between filamentous growth and mat formation, we sought to further define how the network of signaling pathways that control filamentous growth might regulate adhesion-dependent surface growth.
To learn more about the regulation of surface growth in yeast, adhesion-dependent surface responses were examined from several perspectives. In one set of experiments, we developed a method for recording colony pattern formation over time by kymograph analysis, which allowed us to confirm that the major signaling pathways that control filamentous growth also regulate mat formation. A new regulator of filamentous growth, the chromatin remodeling complex SAGA, was also uncovered. Comparative expression profiling identified a large number of target genes as well as new regulatory connections between pathways. By seeking to understand why cells form adhesive connections during surface growth, we identified a role for Flo11p in protection from macroscopic predators. We also showed that colony ruffling aids in temperature regulation. Our findings broaden the role of filamentation regulatory pathways to include the regulation of adhesion-dependent surface responses. Our findings may be relevant to studies of fungal pathogens that must tolerate growth on surfaces to become virulent.
Materials and Methods
Media and growth conditions
Yeast strains were grown and manipulated by standard methods (Sambrook et al. 1989; Rose et al. 1990). For most experiments, colonies were grown on standard YEPD (2%) or YEP-galactose (Gal) semisolid agar media (Cullen 2015a). Some experiments were performed on low-agar (0.3%) media. Glutamate sensitivity experiments used synthetic agar (dextrose, 2%; yeast nitrogen base, 6.7 g/liter; uracil, 20 mg/liter; His, 20 mg/liter; Leu, 120 mg/liter; adenine, 20 mg/liter; Lys, 60 mg/liter; Arg, 20 mg/liter; Trp, 20 mg/liter; Tyr, 30 mg/liter; Thr, 200 mg/liter; Met, 20 mg/liter; and Phe, 50 mg/liter), minimal agar (dextrose, 2%; yeast nitrogen base, 6.7 g/liter; and uracil, 20 mg/liter), and minimal + Glu (dextrose, 2%; yeast nitrogen base, 6.7 g/liter; uracil, 20 mg/liter; and Glu 20 mg/liter) media. To generate low-oxygen (5–15% oxygen) and anaerobic conditions, agar plates were incubated in GasPAK EZ Campy Pouch System (BD 260685; Becton, Dickinson and Company, Franklin Lakes, NJ) or BD GasPAK EZ Anaerobe Pouch System (BD 260683; Becton, Dickinson and Company) bags. To maintain consistent moisture levels, plates were poured and left unwrapped for 3 days at 22° to allow the evaporation of excess moisture. To generate media with reduced moisture levels, YEPD plates were left unwrapped for 10 days at 22°.
Strains and genetic manipulations
Yeast strains are listed in Table 1. Gene deletions were constructed using auxotrophic markers amplified by PCR and introduced into yeast by lithium acetate transformation by standard methods as described (Gietz and Schiest 2007). To generate the spt8Δ mutant, homologous recombination at the SPT8 locus was performed using the pKlURA3 (PC5225) cassette as a template. For some experiments, yeast strains were used from an ordered knockout collection (Ryan et al. 2012). Caenorhabditis elegans strains used in this study include N2 Bristol wild-type and KP4 glr-1(n2461) (Kaplan and Horvitz 1993). Strains were maintained at 20° under standard conditions on nematode growth media (NGM) agar plates seeded with OP50 Escherichia coli bacteria (Brenner 1974).
Table 1. Yeast strains used in the study.
| Straina | Description | Reference |
|---|---|---|
| PJ69-4Ab | MATa trpl-901 leu2-3,112 ura3-52 his3-200 ga14Δ ga18OΔ LYS2::GALl-HIS3 GAL2-ADE2 met2::GAL7-lacZ | James et al. (1996) |
| PC313 | MATa ura3-52 | Liu et al. (1993) |
| PC538 | MATa ura3-52 ste4 FUS1-lacZ FUS1-HIS3 | Cullen et al. (2004) |
| PC549 | MATa ura3-52 ste4 FUS1-lacZ FUS1-HIS3 ste20::URA3 | Cullen and Sprague (2000) |
| PC555 | MATa ura3-52 ste4 FUS1-lacZ FUS1-HIS3 hsl7::URA3 | Cullen and Sprague (2000) |
| PC562 | MATa ura3-52 ste4 FUS1-lacZ FUS1-HIS3 ras2::URA3 | Cullen and Sprague (2002) |
| PC611 | MATa ura3-52 ste4 FUS1-lacZ FUS1-HIS3 ste11::URA3 | Cullen and Sprague (2002) |
| PC999 | MATa ura3-52 ste4 FUS1-lacZ FUS1-HIS3 MSB2-HA | Chavel et al. (2010) |
| PC1029 | MATa ura3-52 ste4 FUS1-lacZ FUS1-HIS3 flo11::KanMX6 | Karunanithi et al. (2010) |
| PC1079 | MATa ura3-52 ste4 FUS1-lacZ FUS1-HIS3 ste12::URA3 | Karunanithi et al. (2010) |
| PC2523 | MATa ura3-52 ste4 FUS1-lacZ -::NAT FUS1-HIS3 flo8::HYG | Chavel et al. (2010) |
| PC2712 | MATa ura3-52 ste4 FUS1-lacZ FUS1-HIS3 GAL-FLO11::KanMX6 | Karunanithi et al. (2010) |
| PC3030 | MATa ura3-52 ste4 FUS1-lacZ FUS1-HIS3 MSB2-HA sin3::NAT | Chavel et al. (2010) |
| PC3039 | MATa ura3-52 ste4 FUS1-lacZ FUS1-HIS3 MSB2-HA dig1::NAT | Chavel et al. (2010) |
| PC3431 | MATa ura3-52 ste4 FUS1-lacZ -::NAT FUS1-HIS3 MSB2-HA sfl1::KlURA3 | Chavel et al. (2010) |
| PC3642 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 MSB2-HA rtg3::NAT | Chavel et al. (2010) |
| PC3652 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 MSB2-HA rtg2::NAT | Chavel et al. (2010) |
| PC3695 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 MSB2-HA rtg1::NAT | Chavel et al. (2014) |
| PC4005 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 gcn5::KlURA3 | This study |
| PC3688 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 opi1::NAT | Chavel et al. (2010) |
| PC4008 | MATa ura3-52 ste4 FUS1-lacZ FUS1-HIS3 spt8::KlURA3 | This study |
| PC5090 | MATa ura3-52 ste4 FUS1-lacZ FUS1-HIS3 nte1::NAT | Chavel et al. (2014) |
| PC5119 | MATa ura3-52 ste4 FUS1-lacZ FUS1-HIS3 pho80::NAT | This study |
| PC6733 | MATa ura3-52 ste4 FUS1-lacZ FUS1-HIS3 ura3-52::pTEF2-mCherry URA3 his5::NAT mCherry::GFPγ-HIS | Chow et al. (2019) |
| PC6016 | MATa can1Δ::Ste2pr-spHIS5 lyp1Δ::Ste3pr-LEU2 his3::hisG leu2Δ0 ura3Δ0 | Ryan et al. (2012) |
| CB13A9c | MATa can1Δ::Ste2pr-spHIS5 lyp1Δ::Ste3pr-LEU2 his3::hisG leu2Δ0 ura3Δ0 dan1Δ | Ryan et al. (2012) |
| CB52G2 | MATa can1Δ::Ste2pr-spHIS5 lyp1Δ::Ste3pr-LEU2 his3::hisG leu2Δ0 ura3Δ0 sip18Δ | Ryan et al. (2012) |
| CB53F12 | MATa can1Δ::Ste2pr-spHIS5 lyp1Δ::Ste3pr-LEU2 his3::hisG leu2Δ0 ura3Δ0 sno4Δ | Ryan et al. (2012) |
| CB60E1 | MATa can1Δ::Ste2pr-spHIS5 lyp1Δ::Ste3pr-LEU2 his3::hisG leu2Δ0 ura3Δ0 gre1Δ | Ryan et al. (2012) |
| CB29E8 | MATa can1Δ::Ste2pr-spHIS5 lyp1Δ::Ste3pr-LEU2 his3::hisG leu2Δ0 ura3Δ0 hsp26Δ | Ryan et al. (2012) |
Unless indicated, strains are derived from the Σ1278b strain background.
Strain from M. Johnston’s laboratory.
Strains from an ordered deletion collection labeled with CB followed by the plate number and location.
Assays for mat formation and filamentous growth
Assays for mat formation were performed as described (Reynolds and Fink 2001; Karunanithi et al. 2012). The plate-washing assay was performed as described (Cullen 2015b).
Microscopy
Mats were examined by bright-field microscopy using an Axioplan 2 fluorescent microscope (Zeiss [Carl Zeiss], Thornwood, NY) with 5×, 10×, 20×, 40×, and 100× PLAN-APOCHROMAT 100×/1.4 (oil) (N.A. 0.17) objectives. Digital images were obtained with an Axiocam MRm camera (Zeiss). Axiovision 4.4 software (Zeiss) was used for image acquisition. Digital images were imported into ImageJ (https://imagej.nih.gov/ij/) in 8-bit format.
Comparative RNA sequencing analysis
To compare the transcriptional response of wild-type cells (PC538) and the ste12Δ (PC1079), dig1Δ (PC3039), rtg3Δ (PC3642), spt8Δ (PC4008), and ras2Δ (PC562) mutants, cells were concentrated (OD A600 = 20) and spotted in 10-μl aliquots onto YEP-Gal (2% agar) for 24 hr. Cells were spotted as six colonies per plate, equidistant to each other and the plate center. All six colonies were harvested for each trial and three separate trials were compared for each strain. The entire colony surface was scraped into 500 μl of distilled water, harvested by centrifugation, washed, and stored at −80°. RNA was harvested by hot-acid phenol–chloroform extraction as described (Adhikari and Cullen 2014). Samples were further purified using a QIAGEN RNeasy Mini Kit (catalog number 74104; QIAGEN, Valencia, CA). RNA concentration and purity was measured via NanoDrop (NanoDrop 2000C; Thermo Fisher Scientific, Waltham, MA). RNA stability was determined by running the sample on an agarose gel.
RNA sequencing (RNA-seq) was performed as described previously (Adhikari and Cullen 2014) by sequencing RNA prepared from three separate cultures. RNA-seq libraries were prepared from total RNA using the TruSeq RNA Sample Prep Kit (Illumina, San Diego, CA). Library size distributions were validated using an Agilent 2200 TapeStation (Agilent Technologies, Santa Clara, CA). Additional library quality control, blending of pooled indexed libraries, and cluster optimization was performed using Invitrogen’s Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA). RNA-seq libraries were pooled (21-plex) and clustered onto a flow cell lane using an Illumina cBot. Sequencing was performed using an Illumina HiSeq 2500 in Rapid Mode employing a paired-end, 50-base read length (PE50) sequencing strategy.
Image analysis and base calling were performed using Illumina’s Real Time Analysis v1.18 software, followed by “demultiplexing” of indexed reads and generation of FASTQ files, using Illumina’s bcl2fastq Conversion Software v1.8.2 (http://support.illumina.com/downloads/bcl2fastq_conversion_software_184.html). For analysis of the RNA-seq data, reads of low quality were filtered out prior to alignment to the reference genome (S. cerevisiae assembly R64-1-1, Ensembl release 75) using TopHat v2.0.9 (Trapnell et al. 2009). Counts were generated from TopHat alignments for each gene using the Python package HTSeq v0.6.1 (Anders et al. 2015). Genes with low counts across all samples were removed.
For comparisons of mutant to wild-type colony samples, differentially expressed genes were identified using the Bioconductor package DESeq2 (Love et al. 2014) with the apeglm package to estimate t-prior shrinkage (Zhu et al. 2018). We employed the IHW package to weight hypotheses and optimize power (Ignatiadis et al. 2016). Differential expression was defined as |log2 (ratio) | ≥ 0.585 (± 1.5 fold) with P < 0.01. Genes were categorized as upregulated [logFC (fold change) > 0.585, P-value < 0.01], down-regulated (logFC < −0.585, P-value < 0.01), or insignificant (logFC < −0.585 or logFC > 0.585, P-value > 0.01). Differentially expressed pathway-specific genes were visualized in principal component analysis (PCA), volcano plot, and Venn diagram figures using ggplot2. Pathway-specific gene expression was visualized using the Kyoto Encyclopedia of Genes and Genomes (Kanehisa and Goto 2000), and the Bioconductor package pathview (Luo and Brouwer 2013). Classification of targets was based on gene ontology (GO) terms and descriptions in the Saccharomyces Genome Database (http://www.yeastgenome.org). For all other comparisons, edgeR v3.18.1 (Robinson et al. 2010) was used. A false discovery rate (FDR) method was employed to correct for multiple testing (Reiner et al. 2003). Differential expression was defined as |log2 (ratio) | ≥ 0.585 (± 1.5 fold) with P-value < 0.01.
GO term analysis
GO term analysis (Ashburner et al. 2000) was performed using the GO enRIchment analysis and visuaLizAtion tool (GOrilla) (Eden et al. 2007, 2009; Mi et al. 2017) using the two unranked lists mode. The Gorilla database had been last updated January 12, 2019. The background list was all of the ORFs identified during the analysis. The target list for genes regulated by all five regulators was identified as genes whose |log2FC| > 0.585 and P-value < 0.01 in all mutant sets. The target list for genes regulated by both RAS and RTG was identified as genes |log2FC| > 0.585 and P-value < 0.01 in only the ras2Δ and rtg3Δ sets. Enriched GO terms were identified by having P < 10−3 and FDR < 0.05. Values for background and target sets, and enriched GO terms, are in Supplemental Material, Table S2.
Quantitative PCR analysis
Differential gene expression was confirmed by quantitative (q) real-time PCR analysis as described previously (Chavel et al. 2014). cDNA libraries from RNA samples were generated using iScript Reverse Transcriptase Supermix (catalog number 1708840; Bio-Rad, Hercules, CA). qPCR was performed using iTaq Universal SYBR Green Supermix (catalog number 1725120; Bio-Rad) on the Bio-Rad CFX384 Real-Time System. Primers were ordered from Sigma ([Sigma Chemical], St. Louis, MO) and are listed in Table S3. FCs in expression were determined by calculating ΔΔCt (Livak and Schmittgen 2001) using ACT1 mRNA as the housekeeping gene for each sample. RNA was prepared from at least three samples and the average of at least three biological replicates was recorded. Statistical significance was determined by the Student’s t-test.
Time-lapse photography
Cells were grown on YEP-Gal semisolid agar media at 22° for 8 days. Photographs were taken using a Nikon D3000 (Nikon, Garden City, NY) digital camera at 30-min intervals using automatic exposure without flash. Graph paper (0.25 cm) was glued to the plate bottom for scaling and later image stabilization; these were cropped out in the final images. Images were imported into ImageJ as an image stack for image stabilization using Kang Li’s image stabilizer plug-in (http://www.cs.cmu.edu/∼kangli/code/Image_Stabilizer.html). Stabilized image series were saved in video format. Kymographs were generated using the reslice tool in ImageJ (https://imagej.nih.gov/ij/).
Ruffles were identified as light bands flanked by dark bands. The number of ruffles per time point was counted manually at 1-day intervals for each kymograph. Statistical analysis was carried out for each time point using an unpaired Student’s t-test.
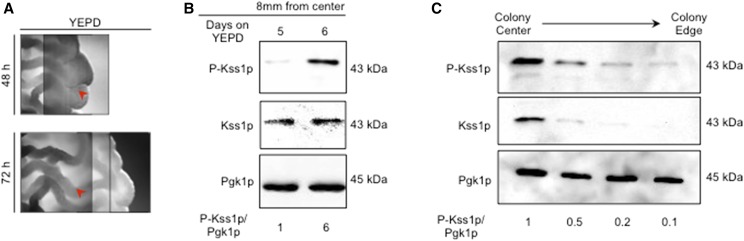
Evaluation of phosphorylated Kss1 levels
Samples were harvested at specific distances from the colony perimeter at t = 0. To determine P-Kss1p levels at the growing edge of a colony, the outermost millimeters of representative colonies were harvested at 1, 2, 3, and 4 days. To determine the changes in P-Kss1p levels before and after the colony ruffles, samples were harvested from regions of the colony 2 mm from the starting colony edge from representative colonies before and after ruffling.
Samples were evaluated by SDS-PAGE analysis. Immunoblots were performed as described (Cullen 2015a). P-Kss1p was detected with phospho-p44/42 primary antibodies (catalog number 4370S; Cell Signaling Technology, Danvers, MA) and anti-rabbit HRP secondary antibodies (catalog number 111-035-144; Jackson ImmunoResearch, West Grove, PA). Total Kss1p was detected by anti-Kss1p antibodies (SC-6775-R; Santa Cruz Biotechnology, Dallas, TX) and with anti-rabbit HRP secondary antibodies (catalog number 111-035-144; Jackson ImmunoResearch). Loading control Pgk1p was identified by anti-Pgk1p primary antibodies (catalog number 459250; Invitrogen) and with anti-mouse HRP secondary antibodies (catalog number 1706516; Bio-Rad).
Effect of S. cerevisiae cell–cell adhesion on pharyngeal uptake by C. elegans
Wild-type yeast cells expressing GFP (PC6733) were grown in synthetic or YEPD liquid media at 30° for 16 hr. Cells were washed twice in M9 buffer (KH2PO4, 3 g/liter; Na2HPO4, 6 g/liter; NaCl, 5 g/liter; and 1 mM MgSO4). As previously described (Bois et al. 2013), adult C. elegans were transferred into suspensions of S. cerevisiae. After 45 min, C. elegans were removed and mounted onto 2% agarose pads, and immobilized with 10 mM sodium azide. Slides were examined with an Axioplan 2 fluorescent microscope (Zeiss) at 40×. Individual S. cerevisiae cells were counted. Independent replicates were performed on three separate days.
Experiments involving yeast colonies exposed to entry by C. elegans
Yeast cells were grown in YEPD liquid media at 30° for 16 hr. For interactions between C. elegans and S. cerevisiae colonies, 200-μl aliquots of cells were dispensed onto NGM (Wood 1988) agar media and grown as colonies at 22° for 72 hr. Under this condition, yeast colonies formed Flo11p-dependent patterns. OP50 E. coli (Brenner 1974) were taken from stock cultures, and 200 μl aliquots were dispensed onto NGM agar media and grown at 22° for 3 days.
To examine colony penetration, adult and fourth-stage larva (L4) C. elegans were transferred from stock OP50 plates to experimental plates around the yeast or E. coli. Mat penetration times were determined by measuring the time from a worm nose first contacting the colony to the tail fully entering the mat, up to 100 sec. The number of stalls and reversals was determined by counting the number of incidents when a worm’s tail would stop forward movement or reverse. The percent of time moving forward was determined by recording the amount of time that a worm was moving forward divided by the total time required to penetrate a mat. Independent replicates were performed on at least three separate days.
To examine the pharyngeal uptake of S. cerevisiae by C. elegans, wild-type (PC538) and flo11Δ (PC1029) yeast were transformed with a plasmid containing GFP-2xPH (PC2560) [CS189 (Stefan et al. 2005) provided by the Emr laboratory (Cornell University)]. Cells were grown in YEPD liquid media at 30° for 16 hr and 200-μl aliquots were spotted onto NGM agar media. Colonies were grown at 22° for 3 days. Worms were transferred directly into colonies and left for 45 min before mounting and imaging. Independent replicates were performed on three separate days.
Videos were captured using an AmScope MD35 camera and AmScope image capture software. The camera was inserted into the eyepiece of a Zeiss SteREO Discovery V8. Dot plots for worm entry times were generated in ggplot2 (Wickham 2016). Statistical analysis was performed using the Student’s t-test.
Infrared imaging
Infrared images were taken using FLIR A325sc (FLIR Systems, Wilsonville, OR) and captured using FLIR ResearchIRMax4 (FLIR Systems), provided by the Sustainable Manufacturing And Robotic Technology center at the University at Buffalo. Average and coolest temperatures for each colony were measured using FLIR ResearchIRMax4 (FLIR Systems).
An insulated housing unit was built to mount the thermal imaging camera above a stage for agar plates. Yeast cells were grown in YEPD liquid media at 30° for 16 hr. Next, 10-μl aliquots were spotted onto YEP-Gal agar media. Colonies were grown for 72 hr at 30°. For imaging, plates were transferred from the incubator to the mount in a photographing area with an ambient temperature of 22°. Plate lids were removed to allow imaging. Images were taken immediately after removing the lid.
Data availability
All strains are available upon request. The Gene Expression Omnibus (GEO) accession number for raw sequencing data is GSE115657. A comparison with previously published expression profiling data sets for cells grown in liquid culture has been described [GEO accession number GSE61783 (Adhikari and Cullen 2014)]. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.8066615.
Results
A signaling network regulates adhesion-dependent surface responses in yeast
Multiple signal transduction pathways regulate filamentous growth (Figure 1A) (Chavel et al. 2010, 2014). A large number of genes that control invasive growth, pseudohyphal formation, and mat formation show significant overlap based on a genome-wide analysis using deletion mutants in the filamentous strain background (Ryan et al. 2012). Thus, we examined and compared how the signaling network that regulates filamentous growth impacts adhesion-dependent surface responses in yeast. We sought to directly compare key pathways in the network (MAPK and RAS) to less well-characterized pathways (RTG) and to a newly identified chromatin remodeling complex (SAGA). All of the pathways were studied in reference to a key target adhesion molecule, Flo11p.
Some strains (e.g., Σ1278b) exhibit ruffled colony morphology when grown on surfaces. This phenotype is dependent on cell adhesion contacts mediated by Flo11p (Reynolds and Fink 2001; Karunanithi et al. 2012). To evaluate colony pattern formation, we performed a time-course experiment followed by kymograph analysis, which can reveal changes in local features over time (Kaksonen et al. 2003). Wild-type cells were spotted on 2% agar media (YEP-Gal), and ruffle formation was examined by photographing colonies at 30-min intervals over an 8-day period (Video S1). In a series of experiments, kymographs were generated along multiple radii of expanding colonies to measure the extent of ruffle formation [Figure 1B, wild-type (PC538), yellow line corresponds to one kymograph at right]. Ruffles were identified as light horizontal bands flanked by dark bands and could be counted manually by this method (Figure 1B, wild-type, black arrows). Measuring the banding pattern from separate sections of the colony allowed us to assess the extent of ruffle formation over time (Figure 1C, wild-type). As has been previously demonstrated (Granek and Magwene 2010), colonies lacking the adhesion molecule Flo11p did not form ruffles (Video S2), which was also evident by kymograph analysis [Figure 1, B and C and Figure S1, flo11Δ (PC1029)]. Therefore, kymograph analysis permitted the numerical assessment of pattern formation in yeast colonies. In principle, kymograph analysis may allow quantification of colony pattern formation in other microbes.
Kymograph analysis was next applied to mutants lacking key components of the major signaling pathways that regulate filamentous growth. As expected (Roberts and Fink 1994; Rupp et al. 1999; Reynolds and Fink 2001; Granek and Magwene 2010), the fMAPK pathway was required for colony ruffling. Specifically, kymograph analysis showed that colonies lacking a negative regulator of the fMAPK pathway, Dig1p (Cook et al. 1996; Tedford et al. 1997; Bardwell et al. 1998; Olson et al. 2000; Breitkreutz et al. 2003; Kusari et al. 2004; Chou et al. 2006; van der Felden et al. 2014), formed more ruffles than wild-type after 1 day of growth [Figure 1, B and C, dig1Δ (PC3039), Figure S1, and Video S3], though this difference was less apparent over longer time periods. Conversely, loss of the fMAPK transcription factor, Ste12p, led to a defect in colony ruffling at the examined time points [Figure 1, B and C, ste12Δ (PC1079), Figure S1, and Video S4]. The fMAPK pathway was also required for mat formation on 0.3% agar (Figure S1; ste12Δ and dig1Δ; Figure S2 provides examples of other fMAPK regulators; ste11Δ and ste20Δ). Also as expected (Gimeno et al. 1992; Rupp et al. 1999; Granek and Magwene 2010; Zara et al. 2011; Ryan et al. 2012), the RAS pathway was required for colony ruffling [Figure 1, B and C, ras2Δ (PC562) and Video S5] and mat formation (Figures S1 and S2). Thus, kymograph analysis can be used to evaluate the roles of signaling pathways in regulating aspects of mat growth and colony pattern formation.
We next tested whether other pathways that regulate filamentous growth also regulated adhesion-dependent colonial responses. The mitochondria-to-nucleus RTG pathway regulates invasive growth (Chavel et al. 2010; González et al. 2017). We found that the RTG pathway was also required for colony ruffling [Figure 1, B and C, rtg3Δ (PC3642), Figure S3, and Video S6] and mat formation on 0.3% agar (Figures S1 and S2).
We next tested the role of other proteins that control filamentous growth. Pho85p regulates filamentous growth (Chavel et al. 2010) and was also required for mat formation (Figure S1 and S2). Rpd3p (Chavel et al. 2010; Voordeckers et al. 2012), lipid regulators Opi1p and Nte1p (Chavel et al. 2014), and Snf1p (Cullen and Sprague 2000; Voordeckers et al. 2012) were also required for mat formation (Figures S1 and S2). Based on these results, we conclude that many of the major signaling pathways that regulate filamentous growth also regulate mat formation and colony patterning.
We hypothesized that the chromatin remodeling complex SAGA (Koutelou et al. 2010) might also regulate adhesion-dependent surface growth. The hypothesis was based on the fact that a component of SAGA, Gcn5p (Georgakopoulos and Thireos 1992; Sterner and Berger 2000), is required for filamentous growth (Chavel et al. 2014). Moreover, SAGA components have previously been shown to control aspects of colonial patterning (Voordeckers et al. 2012). Spt8p, a SAGA component (Winston et al. 1987), also regulates filamentous growth. Kymograph analysis of the spt8Δ mutant showed a defect in ruffle formation at 2 and 3 days [Figure 1, B and C, spt8Δ (PC4008), Figure S3, and Video S7]. The defect was subtle by kymograph analysis but obvious in the time-lapse analysis. The spt8Δ mutant was also defective for colony ruffling and mat pattern formation (Figure S1). Therefore, we include SAGA as a regulator of filamentous surface responses in yeast (Figure 1A, cyan).
Expression profiling of colony surface growth identifies new targets
To explore how filamentation signaling pathways might regulate surface growth, comparative RNA-seq was performed in several mutants that disrupt the main pathways that regulate filamentous growth (dashed blue box in Figure 1A, pathway diagrams can be found in Figure S4, A–D). RNA was prepared from wild-type and mutant colonies with the following genotypes: ste12Δ (fMAPK), dig1Δ (fMAPK), ras2Δ (RAS), rtg3Δ (RTG), and spt8Δ (SAGA). RNA was prepared from colonies under conditions that favored pattern formation (YEP-Gal). Each mutant showed the expected colony patterning. Specifically, the ras2Δ (PC562), rtg3Δ (PC3642), ste12Δ (PC1079), and spt8Δ (PC4008) mutants were less ruffled than wild-type colonies (PC538), and the dig1Δ (PC3039) mutant was more ruffled than wild-type colonies (Figure S4E). After sequencing, PCA of the RNA-seq data showed close clustering of strain replicates, while strains differentiated into their replicate clusters (Table S1).
Comparative RNA-seq analysis between the wild-type colonies and each mutant was performed with the DESeq2 package in R (Love et al. 2014). Differential gene expression was defined by |log2FC| > 0.585 and P-value < 0.01. By this method, 1833 genes were differentially expressed in at least one of the mutants tested (Figure S4F) and represented 29% of the ORFs in the genome (Lin et al. 2013). The annotated data set can be found in Table S1. To identify the most differentially regulated targets between the mutant and wild-type colonies, volcano plots showing all differences in gene expression were generated for each mutant. Individual genes were distributed by change in expression (x-axis, log2FC) and significance of change in expression [y-axis, −log10(P-value)] (Figure 2A). Many of the genes whose expression is known to be induced during filamentous growth were uncovered during the initial analyses (Figure 2A and Table S1). These included targets of the fMAPK pathway: FLO11 (Rupp et al. 1999) (Figure 2A, dig1Δ and ste12Δ; Table S1, dig1Δ and ste12Δ), YLR042C (Roberts et al. 2000) (Figure 2A, ste12Δ; Table S1, dig1Δ and ste12Δ), CLN1 (Madhani et al. 1999) (Table S1, dig1Δ and ste12Δ), PGU1 (Madhani et al. 1999; Roberts et al. 2000) (Figure 2A, dig1Δ and ste12Δ; Table S1, dig1Δ and ste12Δ), SVS1 (Roberts et al. 2000) (Figure 2A, dig1Δ and ste12Δ; Table S1, dig1Δ and ste12Δ), KSS1 (Table S1, dig1Δ and ste12Δ) (Roberts et al. 2000), and MSB2 (Cullen et al. 2004) (Table S1, dig1Δ and ste12Δ). The abovementioned fMAPK pathway targets YLR042C, PGU1 (Roberts et al. 2000), FLO11 (Rupp et al. 1999) are also Ras2p-dependent (Table S1, ras2Δ), potentially through its regulation of the fMAPK pathway (Mösch et al. 1996, 1999; Chavel et al. 2010). Similarly, targets of the RTG pathway were identified in the wild-type-rtg3Δ data set: CIT2, CIT1, IDH1, IDH2 (Liu and Butow 1999) (Figure 2A, rtg3Δ; Table S1, rtg3Δ), and DLD3 (Liu and Butow 2006) (Table S1, rtg3Δ). Genes regulated by SAGA were identified in the wild-type-spt8Δ data set [ADH1, ARG1, BDF2, CTT1, FBA1, GRE2, PGK1, TDH3, and PHO84 (Basehoar et al. 2004; Huisinga and Pugh 2004)] (Table S1, spt8Δ).
Figure 2.
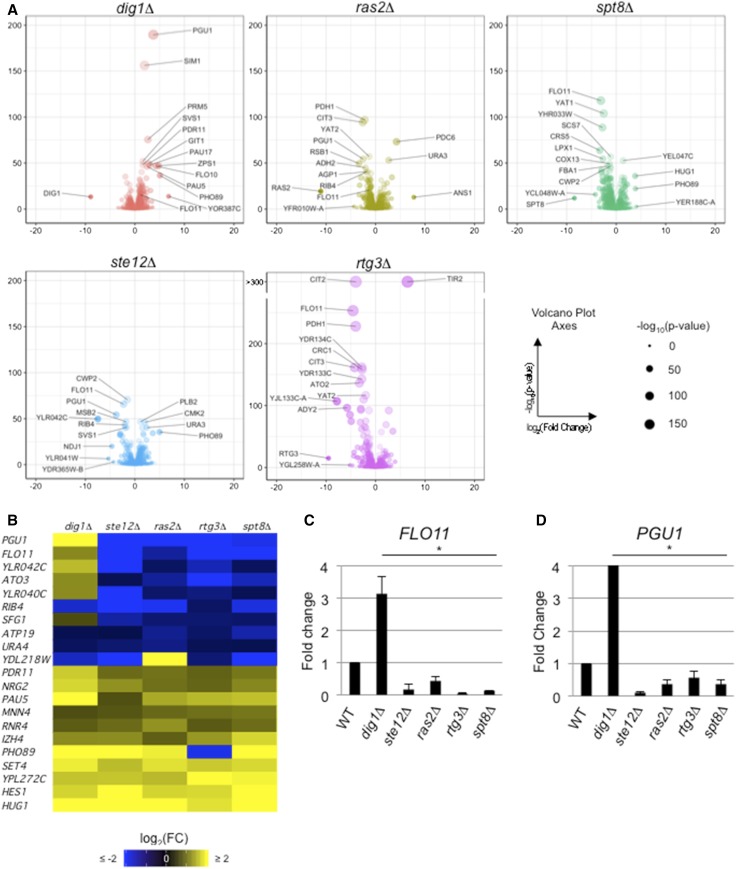
Comparative RNA-seq analysis identifies common and unique targets of signaling pathways that regulate - adhesion-dependent surface growth growth. (A) Volcano plots showing comparative RNA-seq between the indicated mutant and WT. Dot size is −log10(P-value). Volcano plot x-axis is log2(FC); y-axis is −log10(P-value). Labeled dots are 10 targets with lowest P-value or five targets with highest |fold change|. FLO11 has also been labeled on dig1Δ and ras2Δ plots. CIT2 and TIR2 have calculated P-values < 10−300 in the rtg3Δ mutant, as reflected in the break in the y-axis. (B) Heat map showing targets with |log2(FC)| > 0.585 and P-value < 10−3. (C) Bar graph showing fold change in FLO11 mRNA levels, normalized to ACT1 with WT values set to 1, in the indicated mutants by qPCR analysis by the ΔΔCt quantitation method. The experiment was performed in triplicate and error bars represent the SD between experiments. * P < 0.05 for all differences compared to WT. (D) Bar graph showing fold change in PGU1 mRNA levels. See (C) for details. log2(FC), log2(Fold Change); qPCR, quantitative PCR; RNA-seq, RNA sequencing; WT, wild-type.
Confirmation of other targets by qPCR analysis has been performed in related studies including: targets of fMAPK related to the fungal cell wall, OCH1, PRY2, FLO10, and TIP1 (Chow et al. 2018); SUC2 and YLR042C; and GIC2 (Aditi Prabhakar, personal communication). Therefore, comparative RNA-seq analysis identified many genes expected based on previous or parallel studies.
One question we sought to address was the extent of target gene overlap among the signaling pathways that regulate filamentous growth. We found that while most genes were regulated by one pathway (Figure S4G, 742/1833 or 59%), coregulated targets were seen in almost every combination of regulators, including targets coregulated by all five regulators (Figure S4F). This result also demonstrated that the SAGA complex coregulated targets with the fMAPK, RAS, and RTG pathways (Figure S4F), which further validates it as a major regulator of filamentous growth (Figure 1A, cyan arrows). Additionally, Spt8p regulated a target of the RIM101 pathway, NRG1 (Chavel et al. 2014) (Figure 1A, cyan arrow), and a target of lipid biosynthesis, INO1 (Chavel et al. 2014) (Figure 1A, cyan arrow). SAGA did not coregulate targets of the PHO85, RPD3(L), or ELP pathways. Taken together, these data suggest that one function of this network of signaling pathways is to regulate a large number of genes (the more nodes in the network, the greater the number of differentially expressed genes); a second function is to coregulate key targets to amplify target gene expression.
One possible explanation for the regulatory overlap is cross feedback among the pathways. To explore this aspect of signaling network connectivity, we examined whether genes encoding pathway components were themselves targets of other pathways that regulate filamentous growth. The RTG pathway regulated the expression of components of fMAPK (MSB2), RAS (TPK2), and SAGA (SFG73). fMAPK regulated genes encoding components of its own pathway (MSB2, TEC1, and KSS1) and the RAS pathway (BCY1). The RAS pathway regulated one of its own effectors (TPK1) and the fMAPK pathway (TEC1). SAGA regulated the fMAPK pathway (MSB2 and TEC1) and itself (SUS1) (Figure S5, A and B). These results support and extend a previous study, which showed that the activity of the fMAPK is subject to regulation by other filamentation regulatory pathways (Chavel et al. 2010). Interestingly, not all of the interactions would be expected to result in positive feedback. For example, the rtg3Δ mutant showed a ∼1.9-fold increase in TPK2 gene expression (Table S1), which is a PKA subunit and component of the RAS pathway. These data indicate that coregulation of targets may be the result of feedback among the pathways that regulate filamentous growth.
To identify functionally relevant targets whose expression was amplified by multiple pathways that regulate filamentous growth, we focused on genes that were differentially expressed in all regulatory mutants (Figure 2B and Table S2, all mutants). GO term analysis (Ashburner et al. 2000) of the coregulated genes showed significant enrichment in cell adhesion, cell wall constituents, and fungal cell wall proteins (Table S2, all mutants). One of the targets that was coregulated by all five regulators, and that was included in many of the above enriched GO terms, was the gene that encodes the major cell adhesion molecule Flo11p (Figure 2B and Table S2, all mutants). FLO11 is known to be regulated by a large number of proteins and pathways (Rupp et al. 1999; Barrales et al. 2008). We confirmed that fMAPK, RAS, RTG, and Spt8p controlled FLO11 expression by qPCR (Figure 2C). These data fit with the key role that Flo11p plays in regulating adhesion-dependent responses in yeast. Another target regulated by all five mutants, which also had the largest net change in regulation, was PGU1, the gene that encodes a secreted pectinase that is also expressed during filamentous growth (Madhani et al. 1999; Cullen 2015a). This result was confirmed by qPCR analysis (Figure 2D). Taken together, our results are consistent with the idea that multiple signaling pathways and protein complexes coordinately regulate target gene expression during colonial surface growth (Figure 2A). One function of the network is to control the expression of FLO11, a major regulator of adhesion-dependent responses in yeast (Figure 2, B and C).
A regulatory connection between the RAS and RTG pathways
In an analysis of highly significant changes in expression in the ras2Δ and rtg3Δ sets (P < 10−12) (Figure S6), 41 targets were in common and 93% of these targets (39/41) showed the same regulation (both up or down) (Figure S6). For example, citrate synthase, CIT3, was among the significant, differentially expressed genes in both ras2Δ and rtg3Δ (Figure 2A). Other citrate synthases, CIT1 and CIT2, were also coregulated by both the ras2Δ and rtg3Δ mutants (Figure 3A and Table S1). We confirmed that RAS and RTG coregulated CIT2 and CIT3 by qPCR analysis (Figure 3, B and C). Because of the extent of coregulation, especially the coregulation of all citrate synthase isoforms (Graybill et al. 2007), the full set of RAS and RTG coregulated targets was examined further.
Figure 3.
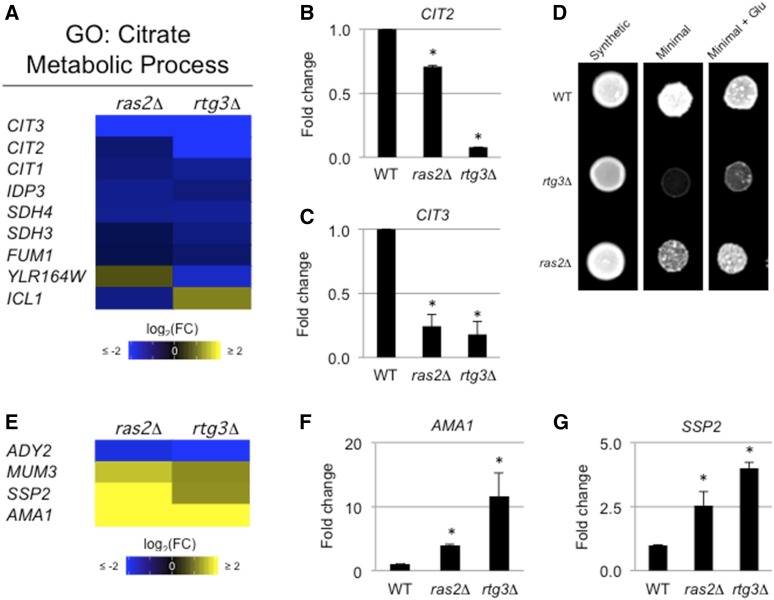
Common and unique target genes of the RAS and RTG pathways. (A) Heat map of targets coregulated by the RAS and RTG pathways with the citrate metabolic process GO term. (B and C) Bar graph showing fold change in (B) CIT2 and (C) CIT3 mRNAs in ras2Δ and rtg3Δ relative to WT. See Figure 2C for details. (D) Strains spotted onto synthetic media, minimal media, or minimal media with glutamate. Cells were grown for 48 hr. (E) Heat map of targets coregulated by RAS and RTG associated with sporulation. (F and G) Bar graphs showing fold change in (F) AMA1 and (G) SSP2 mRNAs in ras2Δ and rtg3Δ relative to WT. See Figure 2C for details. Glu, glutamate; GO, gene ontology; log2(FC), log2(Fold Change); WT, wild-type.
As expected, the RTG pathway regulated mitochondrial gene targets (Table S1). Many of the genes were also regulated by the RAS pathway (Table S1; PDH1, YAT1, and YAT2) (Schmalix and Bandlow 1993; Epstein et al. 2001; Swiegers et al. 2001). Moreover, several hallmark RTG targets were also regulated by the RAS pathway (Table S1; CIT1, CIT2, CIT3, ATO2, and ATO3) (Suissa et al. 1984; Kim et al. 1986). In addition to coregulation of cell wall components (i.e., FLO11), GO term analysis identified significant enrichment of the citrate metabolic process by the overlapping RAS and RTG targets (Figure 3A and Table S2), and the citric acid cycle (Figure S7). Because RAS and RTG coregulated genes that are involved in the citric acid cycle, we hypothesized that RAS might be required to support that mitochondrial role. The mitochondria are critical for glutamate biosynthesis (Liu and Butow 1999; Magasanik and Kaiser 2002) and RTG is required for growth in medium lacking glutamate [Figure 3D, rtg3Δ (Liao and Butow 1993; Small et al. 1995; Chavel et al. 2014)]. More specifically, a loss of citrate synthases results in glutamate auxotrophy in S. cerevisiae (Kim et al. 1986), so we hypothesized that a RAS pathway mutant would be sensitive to limiting glutamate. Like RTG, the RAS pathway was also required for growth in this condition (Figure 3D, ras2Δ). The growth defects of cells lacking an intact RAS or RTG pathway were bypassed by the addition of glutamate (Figure 3D). Therefore, the RAS pathway plays a role in this mitochondrial process.
Similarly, the RAS pathway regulated genes that are required for sporulation (Table S1) and a subset of these genes were coregulated by the RTG pathway [Figure 3E, ADY2 (Rabitsch et al. 2001), AMA1 (Coluccio et al. 2004), SSP2 (Sarkar et al. 2002), and MUM3 (Engebrecht et al. 1998)]. Several of the genes were confirmed by qPCR analysis (Figure 3, F and G). Therefore, the RAS and RTG pathways coregulate functionally related genes and may be more functionally connected than had been previously thought.
Nonreciprocal target genes are regulated by opposing transcriptional regulators of the fMAPK pathway
Ste12p is one of the transcription factors that regulates the fMAPK pathway (Liu et al. 1993). The transcriptional repressor Dig1p inhibits Ste12p-dependent transcription (Cook et al. 1996; Tedford et al. 1997; Bardwell et al. 1998; Olson et al. 2000; Breitkreutz et al. 2003; Kusari et al. 2004; Chou et al. 2006; van der Felden et al. 2014). Although the transcriptional outputs of the pathway can be complex resulting in nonuniform target gene regulation (Breitkreutz et al. 2003), Ste12p and Dig1p were expected to regulate a common set of targets in a reciprocal manner (Figure 4A). We unexpectedly found that the pattern of gene regulation by the two proteins could not be solely explained by reciprocal regulation. In an analysis of the most significant changes in expression in the dig1Δ and ste12Δ sets (P < 10−12) (Figure S8), 20 targets were found to be in common and only 50% (10/20) were reciprocally regulated (up in dig1Δ and down in ste12Δ) (Figure S8). As expected, Ste12p and Dig1p showed reciprocal regulation of a subset of target genes (Figure S8; FLO11/MUC1, PGU1, YLR042C, and BAR1). However, more genes were independently regulated only by Dig1p or only by Ste12p (Figure S8).
Figure 4.
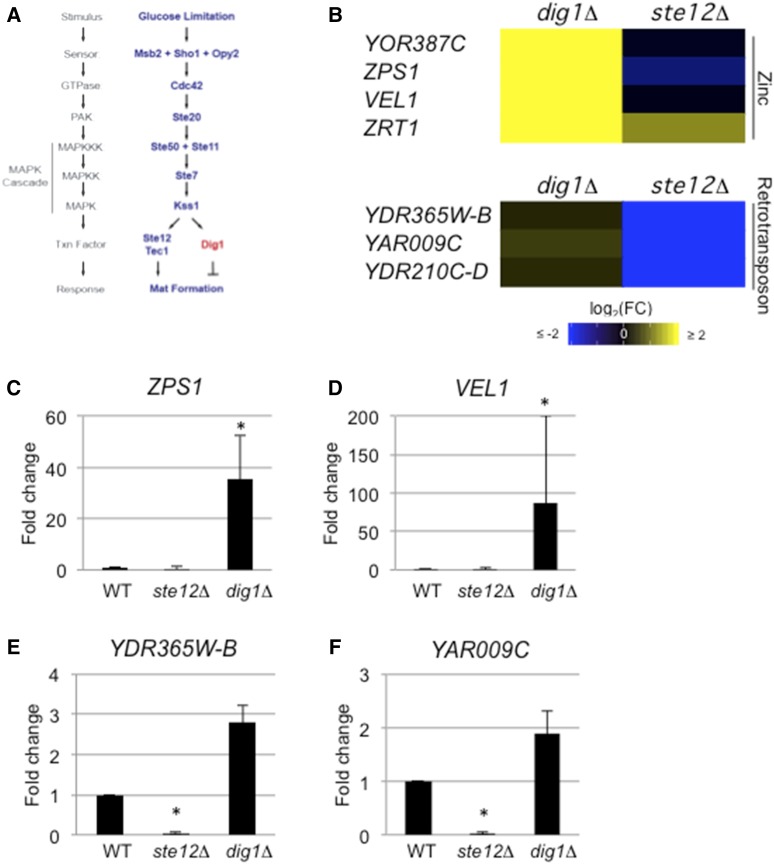
Common and unique genes regulated by Ste12p and Dig1p. (A) Model of the fMAPK pathway showing Ste12p and Dig1p. (B) Heat map of indicated targets regulated in the dig1Δ or ste12Δ mutants. (C–F) Bar graphs showing fold change in target mRNA levels for (C) ZPS1, (D) VEL1, (E) YDR365W-B, and (F) YAR009C. See Figure 2C for details. fMAPK, fungal MAPK; log2(FC), log2(Fold Change); WT, wild-type.
For example, the target with the largest change in expression identified in the dig1Δ volcano plot was YOR387C (> 111-fold increase) (Figure 2A and Table S1). Although YOR387C lacks associated GO terms, a literature search indicated that YOR387C is induced in zinc-limiting conditions (Higgins et al. 2003; Wu et al. 2008). Although YOR387C and its paralog VEL1 (Higgins et al. 2003) were significantly regulated in the dig1Δ mutant, neither gene was found to be regulated in the ste12Δ mutant (Figure 4B and Table S1). Additionally, only the dig1Δ mutant showed a change in expression of ZAP1 (Table S1), a major transcription factor induced by limiting zinc (Zhao and Eide 1997). Interestingly, the ability of cells to take up trace zinc was determined to be critical for flocculation (Yuan 2000), which is a closely related response to filamentous growth and mat formation. A subset of other upregulated targets in the dig1Δ mutant were genes induced under low-zinc conditions [Figure 4B, ZPS1 (Lyons et al. 2000)] as well as a zinc transporter [ZRT1 (Zhao and Eide 1996)]. These genes were not strongly downregulated in the ste12Δ mutant. The expression profiles of ZPS1 and VEL1 were confirmed by qPCR analysis (Figure 4, C and D).
Likewise, the ste12Δ mutant showed downregulation of some target genes that were not altered in the dig1Δ mutant. One of the targets identified in the ste12Δ volcano plot was the retrotransposon YDR365W-B (Kim et al. 1998). Other retrotransposons—YDR365W-B, YOL103W-B, YAR009C, and YDR210C-D (Kim et al. 1998)—were also regulated by Ste12p but less so by Dig1p (Figure 4B). The expression profiles of YDR365W-B and YAR009C were confirmed by qPCR analysis (Figure 4, E and F). Thus, key transcriptional regulators of the fMAPK pathway operate through mechanisms that do not only involve reciprocal regulation between the two proteins.
Changes in colony patterning correspond to changes in MAPK pathway activity
We also examined the development of colony pattern formation under different conditions and over time. Colonies showed different ruffling patterns under different conditions. For example, growth in preferred carbon sources such as glucose media (YEPD) resulted in colony perimeters that were smooth compared to a ruffled colony interior (Figure 5A, adjacent photographs of a colony are shown at different focal planes on the z-axis). Such differences were not observed in more uniformly ruffled colonies grown on a nonpreferred carbon source (YEP-Gal) (Video S1). Over time, the smooth regions of the colony became ruffled (Figure 5A, red arrow marks the same location in the colony).
Figure 5.
Different ruffling patterns in colonies correspond to different levels of fMAPK pathway activity. (A) Bright-field microscopy of a wild-type colony grown in YEPD for 48 and 72 hr. Red arrow indicates same location on a mat at 48 hr when the region is smooth and 72 hr when the region has become ruffled. Different focal planes of the same colony are shown. (B) Immunoblot analysis using p42/p44 antibodies to detect P-Kss1p levels from extracts prepared from cells before and after ruffling. Immunoblots were also performed using antibodies to Kss1p and Pgk1p as loading controls. (C) Immunoblot analysis using p42/p44 antibodies to detect phosphorylated Kss1p at different parts of a colony. Immunoblots were also performed using antibodies to Kss1p and Pgk1p as loading controls. fMAPK, filamentous growth MAPK; YEPD, yeast extract, peptone, and dextrose.
One explanation for differences in colony pattern formation might be that different parts of the colony are exposed to different environments. The fMAPK pathway, which is sensitive to nutrient levels, may induce different responses in different parts of the colony. To test this possibility, samples from regions of the colony were collected and examined for fMAPK activity by phosphorylation of the MAP kinase Kss1p (Cullen 2015a). Samples collected from smooth colony perimeters showed low P∼Kss1p levels (day 5, smooth). Samples collected from the same physical location that had become ruffled (day 6, ruffled) showed high P∼Kss1p levels (Figure 5B). This result indicates that as cells in colonies consume nutrients and subsequently starve, the fMAPK pathway is activated to promote ruffle formation. In line with this possibility, colony interiors showed higher levels of P∼Kss1p compared with colony perimeters (Figure 5C). We note that in this experiment, the levels of total Kss1p protein were also lower at colony perimeters. KSS1 is a target of the fMAPK pathway, and the regulation of total Kss1p may be a way that fMAPK regulates its activity through positive feedback (Roberts et al. 2000). Therefore, signaling pathways may exhibit different activities, and induce target genes to different levels, in a manner that is influenced by nutrient levels (and time) to control adhesion-dependent surface growth.
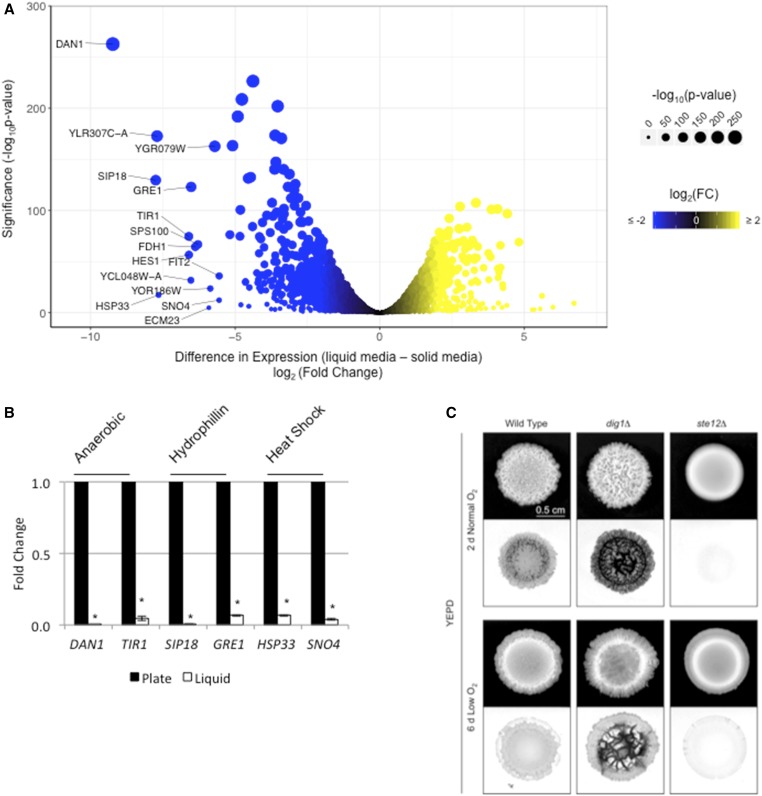
Genes that respond to oxygen, desiccation, and temperature stress were induced during colony surface growth
Growth on surfaces presents unique challenges compared to the uniform growth in liquid cultures. To further explore the response to colonial growth on surfaces, expression profiling data sets were compared between cells grown in liquid (Adhikari and Cullen 2014) and cells grown on an agar surface (this study). Growth on surfaces caused the differential expression of 3267 genes when compared to growth in liquid (Table S1, wild-type liquid–solid, |log2FC| > 0.585 and FDR < 0.05). This represents ∼54% of the yeast genome [6091 genes (Lin et al. 2013)]. Functional classification of the top 15 induced targets (Figure 6A, >25-fold increase in expression) during surface growth included genes induced during anaerobic stress [DAN1 (Sertil et al. 1997) and TIR1 (Cohen et al. 2001), Figure 6A], desiccation/dehydration [SIP18 (Miralles and Serrano 1995) and GRE1 (Garay-Arroyo and Covarrubias 1999), Figure 6A], and elevated temperature [HSP33 (Wilson et al. 2004) and SNO4 (Samanta and Liang 2003), Figure 6A]. An expanded list of the top 50 (>15-fold increase in expression) induced targets included genes that regulate metal homeostasis, RNA processing, and the metabolism of lipids and nonfermentable carbon sources, as well as genes of unknown function (Figure S9).
Figure 6.
Target genes induced during surface growth correspond to the response to specific stresses. (A) Volcano plot showing the difference in expression and significance of all targets between growth in liquid media vs. solid media. x-axis is log2(Fold Change); y-axis is −log10(P-value). Labeled targets are the top 15 induced targets during surface growth. (B) Relative fold change of the indicated gene was determined by qPCR analysis. Expression was normalized to ACT1 in the indicated mutants, as determined by qPCR analysis. The experiment was performed in triplicate and error bars represent the standard difference between experiments. * P < 0.05 for differences compared to wild-type. (C) Wild-type cells and the indicated mutants were spotted on YEPD 2% agar media in normal and low-oxygen conditions. Colonies were grown for the indicated times to compare colonies of comparable sizes before photographing (top panels), washing, and rephotographing colonies (bottom panels) to determine colony patterns and invasive growth. Inverted images of the invasive scars are shown. log2(FC), log2(Fold Change); qPCR, quantitative PCR; YEPD, yeast extract, peptone, and dextrose.
The most differentially induced target during surface growth was the gene that encodes the anaerobic-responsive cell wall mannoprotein Dan1p (Mrsa et al. 1999) (Figure 6A). qPCR analysis showed that DAN1 expression was induced during surface growth (Figure 6B). Examination of the dan1Δ mutant from an ordered gene deletion collection (Ryan et al. 2012) showed a defect in colony ruffling (Figure S10, dan1Δ). Thus, Dan1p may impact colony patterning in response to stresses associated with surface growth. In addition to DAN1, the four members of the Tir family of anaerobic-responsive cell wall mannoproteins (Cohen et al. 2001) also showed transcriptional induction during surface growth (Figure 6A and Table S1, wild-type liquid–solid), which was verified in a study on the regulation of the cell wall (Chow et al. 2018). Therefore, anaerobic-specific changes to the cell wall also occur during surface growth in yeast.
Because targets that encode anaerobic-responsive proteins were among the top induced targets during colony surface growth [DAN1 and the TIR genes (Figure 6A)], the impact of oxygen in colony ruffling and invasive growth was examined. S. cerevisiae undergoes aerobic alcohol fermentation by what is known as the Crabtree effect (Crabtree 1929). In yeast, this includes the repression of genes involved in aerobic respiration in the presence of glucose during exponential growth (De Deken 1966). Studies that explore this phenomenon in yeast have done so in liquid culture (De Deken 1966; Hagman et al. 2014), but the role of oxygen in regulating aspects of colonial growth has not been explored. To determine the role of oxygen on colony pattern formation, wild-type and dig1Δ colonies grown in normal atmospheric oxygen (20% oxygen) were compared to colonies grown in low oxygen (5–15% oxygen). Comparing the colonies at similar sizes (2-day normal oxygen and 6-day low oxygen) showed that both were less ruffled in low oxygen (Figure 6C). Wild-type and dig1Δ colonies showed reduced invasive growth in low oxygen (Figure 6C). Additionally, wild-type, dig1Δ, and ste12Δ colonies grown in limiting oxygen (5–15% oxygen or 0% oxygen) grew more slowly than in normal oxygen (20% oxygen) (Figure S11). Slower growth was not bypassed with excess glucose (Figure S11, 8% Glu) as might be predicted by the Crabtree effect in yeast. Therefore, oxygen levels impact colony patterning and invasive growth.
Another class of genes induced during surface growth has been described as regulated during desiccation. These included the gene that encodes the hydrophilin Sip18p (Miralles and Serrano 1995; Dang and Hincha 2011) (Figure 6A). A second hydrophilin, Gre1p, (Garay-Arroyo and Covarrubias 1999) was also among the top 10 induced targets (Figure 6A). Four of six hydrophilins and putative hydrophilins were induced during surface growth (Table S1, wild-type liquid–solid). Hydrophilins such as Sip18p allow for survival under desiccation stress and during the dehydration/rehydration process (Rodríguez-Porrata et al. 2012). The induction of hydrophilin-encoding genes SIP18 and GRE1 during surface growth was confirmed by qPCR analysis (Figure 6B). By testing mutants from an ordered deletion collection (Ryan et al. 2012), we found that the loss of either hydrophilin, SIP18 or GRE1 (Figure S10A; gre1Δ and sip18Δ), increased mat ruffling and hyper-invasive growth, indicating that intracellular hydration may be a trigger for both invasive growth and colony ruffling (Figure S10B; gre1Δ and sip18Δ). Therefore, cells in mats may experience desiccation stress, and genes induced under this condition can impact colony patterning.
Genes encoding the heat-shock proteins Hsp33p (fourth most-induced target), Sno4p, Hsp26p, and Hsp12p were also induced during mat growth. Additionally, as shown below, a ruffled colony showed a temperature differential of 0.8° between its warmest and coolest regions (see Figure 7D, below). Modest differences in temperature have previously been shown to effect metabolism in S. cerevisiae (Jones and Hough 1970). Spg4p, described as being essential for growth at high temperature (Martinez et al. 2004), was also among the top targets. Using an ordered deletion collection, we explored the roles of SNO4 and HSP26 (Bentley et al. 1992) (Figure S10A; sno4Δ and hsp26Δ), as the hsp33Δ mutant was not present in the deletion collection. Deletion of either of these genes resulted in smoother colonies.
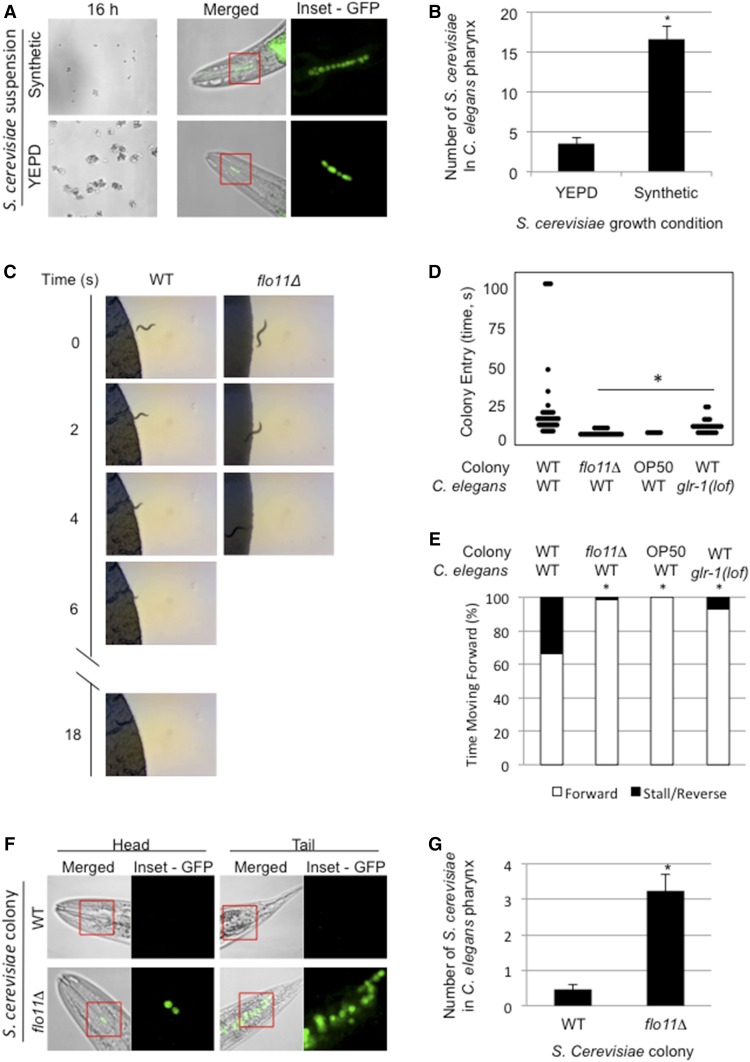
Figure 7.
Impact of Flo11p on predator deterrence in yeast colonies. (A) Suspensions of GFP-labeled WT cells (PC6733) grown for 16 hr in synthetic or YEPD media (16 hr) were examined. Merged fluorescent and brightfield images of heads of mounted C. elegans showing fluorescent S. cerevisiae inside the animal pharynx after 45 min feeding (merged). Fluorescent images of individual S. cerevisiae inside the animal pharynx (inset = GFP). (B) Bar graph showing difference in average number of S. cerevisiae cells in C. elegans pharynx between YEPD- and synthetic-grown suspensions. Error bars are SE. * P < 0.05. (C) Selected images from videos showing WT C. elegans entering WT or flo11Δ yeast colonies. A set of representative videos are available in the supplement (Videos S8–S15). (D) Dot plot showing the time required for worms to enter the indicated colonies. WT (N2) or glr-1(lof) C. elegans worms were placed on plates containing WT or flo11Δ yeast, or the E. coli food strain OP50. (E) Stacked bar graphs comparing C. elegans forward movement to stalls and/or reversals in the indicated yeast or E. coli colonies. (F) Three-day-old colonies of WT (PC538) and flo11Δ (PC1029) cells expressing-plasmid borne GFP (PC2560). C. elegans transferred into the colony for 45 min before mounting. Merged fluorescent images of head and tail regions of C. elegans showing fluorescent S. cerevisiae inside the animal (merged). Fluorescent images of individual S. cerevisiae inside the animal (inset = GFP). (G) Bar graph showing difference in average number of S. cerevisiae cells in the C. elegans pharynx between WT and flo11Δ colonies. Error bars are SE. * P < 0.05. WT, wild-type; YEPD, yeast extract, peptone, and dextrose.
None of the mutants tested were necessary for growth in low-oxygen (5–15% O2), high-temperature (37°), or desiccated conditions (Figure S11). This may be a result of functional redundancy among the major induced targets. Taken together, oxygen, temperature, and desiccation stresses impact colonial growth in yeast. Genes induced during surface growth impact colony patterning that may aid in the response to these stresses.
Flo11p-dependent adhesion protects cells in colonies from a nematode predator
Flo11p is an established regulator of filamentous growth, and its role in maintaining contacts between cells to form filaments has obvious benefits during surface penetration (e.g., invasive growth) into new environments. However, the broader functional implications of Flo11p-dependent adhesion between cells on the colony surface are not clear. The interactions among yeast cells mediated by a related flocculin, Flo1p, can provide protection from toxins and antibiotics (Smukalla et al. 2008), and cell adhesion has more generally been postulated to provide protection to individual cells of a colony (Granek and Magwene 2010). In bacterial communities, the growth of cells in mats can deter predation by other microbes and multicellular predators, like nematodes (Darby et al. 2002). To explore whether cell adhesion contacts might similarly protect fungal cells from predation, we developed a mock predator–prey assay between S. cerevisiae and the nematode C. elegans.
C. elegans is a free-living nematode that is commonly used in research laboratories (Brenner 1974), and has been used to study the effects of C. albicans and S. cerevisiae accumulation in the gut (Jain et al. 2009; Bois et al. 2013). C. elegans feed on bacteria and other microbes including yeasts (Félix and Braendle 2010). The pharynx, a neuromuscular pump, contracts and relaxes rhythmically to draw in liquid and suspended particles to grind them up before transporting them to the intestine. Although the pharynx pumps continuously, previous studies have shown that microbe size is a determining factor in C. elegans feeding; large food is often excluded during this grazing behavior (Avery and Shtonda 2003; Shtonda and Avery 2006; Fang-Yen et al. 2009). One function of cell adhesion molecules is to allow cells to adhere to each other, leading to large colonies. To observe the impact of such adhesion, wild-type S. cerevisiae cells were grown in conditions that either permit cell–cell adherence (Figure 7A, YEPD) or suppress adherence (Figure 7A, synthetic). C. elegans were transferred into suspensions of S. cerevisiae as per established methods (Bois et al. 2013). We found that less-adherent S. cerevisiae (Figure 7A-B, synthetic) were taken into the pharynx of C. elegans in greater numbers than S. cerevisiae that adhered to each other (Figure 7A-B, YEPD). This finding is consistent with the idea that cell–cell adhesion protects cells from ingestion by C. elegans.
We next assessed how yeast cells growing in adhesive colonies might impact the feeding behavior of C. elegans. To test this possibility, the time required for worms to penetrate yeast colonies from nose to tail was measured for ≤ 100 sec (Figure 7, C and D and Videos S8–S15). Worm stalling and reversing out of the colony was also examined (Figure 7E). After 3 days of colony growth, a wild-type yeast strain that formed normal, ruffled colonies slowed C. elegans as it attempted to penetrate the colony (Figure 7, C–E, and Videos S8 and S9). In some cases, worms abandoned entering the colony entirely. In contrast, a flo11Δ colony, which fails to form ruffled colonies, did not deter worms from entering (Figure 7, C–E, and Videos S10 and S11). flo11Δ colonies were equally vulnerable to nematode entry as a colony of OP50, an E. coli laboratory food strain for C. elegans (Brenner 1974) (Figure 7, D and E, and Videos S12 and S13). Additionally, C. elegans transferred directly into flo11Δ S. cerevisiae colonies ingested more cells than C. elegans transferred into wild-type S. cerevisiae colonies (Figure 7, F and G). It is important to note here that our results outline an association between a lack of physical barrier and a higher rate of being consumed by C. elegans. As such, we expect any mutant that lacks the physical barrier conferred by adhesive colonies would be similarly vulnerable.
C. elegans might be excluded from wild-type colonies due to the physical barrier encountered by wild-type colonies expressing FLO11. Additionally, exclusion might also result from a change in a worm’s sensory response to the colony surface. Wild-type C. elegans halt their forward locomotion and initiate backward movement in response to a light touch to their anterior-most tip (nose). This avoidance of touch to the worm’s nose has been termed the nose-touch response (Kaplan and Horvitz 1993), a behavior involving the ASH polymodal nociceptive sensory neurons (with a small contribution from the FLP and OLQ sensory neurons) (Kaplan and Horvitz 1993). The glutamate-gated ion channel GLR-1 functions in the downstream command interneurons and is required for the nose-touch response (Hart et al. 1995; Maricq et al. 1995). To test the possibility that C. elegans do not effectively enter yeast colonies due to the activation of this mechanosensory avoidance response, experiments were performed with C. elegans glr-1(n2461) loss-of-function (lof) mutant animals, which are defective for nose touch, exposed to a wild-type yeast colony. The glr-1(lof) mutant animals penetrated colonies faster than wild-type worms, though this was still slower than wild-type worms penetrating flo11Δ yeast colonies (Figure 7D, and Videos S14 and S15). In addition, the glr-1(lof) mutant animals had fewer stalls and reversals upon initial colony entry than wild-type C. elegans (Figure 7E). Taken together, our data reveal a clear association between the formation of adhesive contacts between yeast cells in surface-growing colonies and predation in a laboratory setting, both by becoming too large to eat and also by forming a physical barrier. Further experiments are needed to establish if protection against macroscopic predators is actually an evolutionary driving force for this phenotype.
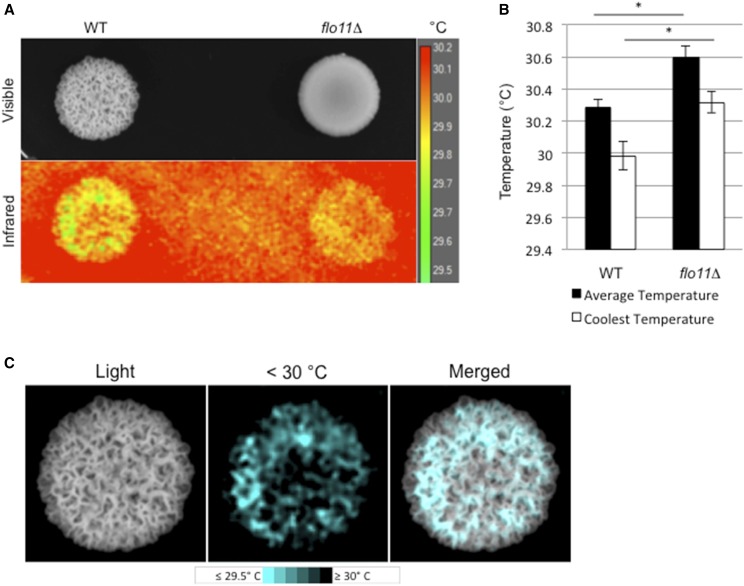
Flo11p-dependent ruffles impact colonial heat dissipation
Colony ruffling increases the surface-to-volume ratio and may provide benefits such as efficient thermoregulation, or other such adaptations to the environment (Palková and Váchová 2006). We also observed that heat-shock response proteins were among the most induced surface-growth targets (Figure 6A, HSP33 and SNO4). To test whether ruffles made by Flo11p-dependent adhesion impact the temperatures of colonies, ruffled wild-type colonies and smooth flo11Δ colonies were examined by infrared imaging (Figure 8A). Thermal imaging showed that the average temperature of a ruffled colony was 0.31° Cooler than the average temperature of a smooth colony (Figure 8B, black bars, P < 0.05, n = 6). The average coolest region of a ruffled colony was 0.33° cooler than the average coolest region of a smooth colony (Figure 8B, white bars, P < 0.05, n = 6). Additionally, the coolest regions of the ruffled colonies (Figure 8C, <30°) were the ruffles themselves (Figure 8C, merged). In a study of cell wall stresses, flo11Δ cells were also shown to have modest temperature sensitivity at 37° (Chow et al. 2018). In summary, Flo11p-dependent ruffling might aid thermoregulation in yeast colonies.
Figure 8.
Effect of cell–cell adhesion on thermoregulation. (A) Visible and infrared images of WT and flo11Δ colonies. Colonies were incubated at 30° and removed from the incubator. The image was taken at < 30 sec after removal of lid. Scale temperature in degrees Celsius. (B) Difference in average and coolest recorded temperatures between WT and flo11Δ. * P < 0.05, n = 6. (C) Visible light (Light) and thermal images of cool colony regions (< 30°) of the WT colony from (A). “Merged” shows the overlapping of cool regions with colony ruffles. WT, wild-type.
Discussion
Here, we explored the role of signaling pathways that regulate filamentous growth in mediating adhesion-based surface responses, including mat formation and colony patterning. Fungal cells commonly grow on surfaces, which poses unique challenges due to the heterogeneity of the nutrients within different parts of the colony and direct exposure to environmental conditions. Understanding the responses to growth on surfaces is important because fungal pathogens exhibit mat growth and form invasive filaments (e.g., hyphae) on the surface of the host, and on inert surfaces during early steps in host colonization.
Analysis of signaling pathways that coregulate adhesion-dependent surface responses
Intracellular signaling pathways can operate in functionally interconnected networks (Levchenko 2003). How multiple pathways operate in a coordinated manner to achieve morphogenetic responses with high fidelity remains a mystery. By examining the major signaling pathways that regulate filamentous growth in yeast, we have identified new roles for the filamentation network in regulating colony patterning and mat growth. Thus, the filamentation regulatory pathways may have a general function in regulating the growth of cells in surface communities.
We also have identified the chromatin remodeling complex, SAGA, to be a regulator of filamentous growth. SAGA and the previously characterized Rpd3p pathway (Bernstein et al. 2000; Chavel et al. 2010) are involved in the epigenetic modification of chromatin. SAGA is an evolutionarily conserved chromatin remodeling complex and a member of the histone acetyltransferase family of proteins (Wang and Dent 2014). SAGA’s functions are diverse and include changes in transcription that result in cell differentiation (Wang and Dent 2014; Hirsch et al. 2015). In yeast, SAGA controls the expression of a specific set of growth-promoting genes (Bruzzone et al. 2018). In fission yeast, TORC1 and TORC2 converge to regulate SAGA in response to nutrient availability (Laboucarié et al. 2017). Moreover, components of SAGA have recently been shown to impact the virulence of Fusarium (Gao et al. 2014).
Expression profiling was performed to evaluate the roles for a subset of pathways that regulate adhesion-dependent surface growth. We identified an unexpectedly large number of differentially expressed genes. Many of the genes were regulated by a single pathway. However, as might be expected of a dense network of functionally connected pathways, each pathway impacted every other pathway by regulating overlapping targets as well as genes encoding pathway components. A key example of this overlap, even in the expanded set of pathways, was the gene encoding the adhesion molecule Flo11p. This may be expected given the critical roles that Flo11p plays in regulating filamentous growth (Rupp et al. 1999), mat formation (Reynolds and Fink 2001), and colonial patterning (Granek and Magwene 2010). Moreover, the FLO11 gene is a hub where many signaling pathways and transcription factors converge (Rupp et al. 1999). The other gene was PGU1, which encodes a secreted plant cell wall-degrading enzyme. Given that one surface that budding yeast commonly encounter is the surface of plants, especially rotting fruit, it may not be surprising that yeast induce this gene when undergoing surface growth. Transcriptional induction of the PGU1 gene may also result from the growth of cells on agar, which is a potential substrate, and may be independently regulated. Therefore, one function of the filamentation regulatory network may be to coordinately regulate target genes that are needed to respond to the challenges of growing on surfaces.
We also uncovered new connections between signaling pathways that regulate adhesion-dependent surface growth. One connection was between the RAS and RTG pathways, which coregulate a substantial number of target genes. The coregulated targets support a function for the RAS pathway in mitochondrial control. The RAS pathway is a global nutrient-sensing pathway in yeast and other organisms (Zaman et al. 2008). The fact that RAS coregulates targets of the RTG pathway supports the idea that RAS plays a critical role in the response to mitochondrial stress. Our results are consistent with previous observations that connect RAS to the overall regulation of the mitochondria. RAS is required for growth on nonfermentable carbon sources, mitochondrial enzyme content (Dejean et al. 2002), and citrate synthase activity (Swiegers et al. 2006; Chavel et al. 2014). Interestingly, components of the RAS pathway, including the GTPase activating protein Ira1p and adenylate cyclase Cyr1p, associate with mitochondrial membranes, which might impact RAS pathway function or activity at this site (Belotti et al. 2012). Moreover, RAS has been implicated in working with RTG to promote longevity in yeast (Kirchman et al. 1999). Thus, the functional interaction between the two pathways to regulate aspects of mitochondrial health might impact the cell’s overall life span. Further experiments will be required to determine how the RAS and RTG pathways coordinate the response to mitochondrial problems. Though the nature of this relationship is unclear, subsequent studies may better characterize the mechanism by which each pathway regulates the other’s hallmark targets.
We also show that Dig1p and Ste12p, which are commonly thought to reciprocally regulate the same set of target genes, actually regulate a partially nonoverlapping set of targets. One that stood out in the Dig1p-regulated set were genes that play a role in zinc uptake and metabolism. This observation is consistent with a previous set of findings that show unique expression profiles and binding sites for this activator–repressor pair (Breitkreutz et al. 2003; Zeitlinger et al. 2003).
Yeast respond to surface growth by expressing a subset of stress-response genes
We also explored the hypothesis that the activity of signaling pathways may change in different parts of a colony over time. We found this to be the case: the activity of the fMAPK pathway was different in different parts of the colony and in different aged colonies. This finding has broad implications in understanding how target genes might be induced during surface growth. For example, it might be expected that other time points or harvesting from certain regions of the colony might result in different expression profiles. The comparative RNA-seq analysis performed here may be an oversimplification of the actual gene expression changes that occur within a colony, which may vary in different parts of a colony and over time.
In the study, we also examined genes that were differentially expressed during surface growth as compared to growth in liquid culture. The genes that were differentially expressed tell us about the challenges of growing in the unique environment of surface growth. Several of the differentially expressed genes identified fit with what one expects cells to encounter when growing on surfaces. One might expect that cells experience anaerobic stress within colonies compared to when growing in liquid conditions, where they are uniformly aerated by shaking. Likewise, cells in colony exteriors would be expected to be vulnerable to desiccation. Interestingly, we show that colony growth patterns change in response to these stresses. Specifically, differentially regulated genes impact colony patterning in response to stress.
One prominent class of differentially expressed genes regulates the heat-shock response. This discovery was at first perplexing, given that cells were grown in liquid and on plates at the same temperature. However, thermal imaging verified that colonies do not have a uniform temperature. This may result from the generation of heat due to metabolic activities of cells within the colony, which may lead to temperature differences in different parts of the colony. We also show that ruffles dissipate heat more efficiently than other parts of the colony. The increased surface area of ruffles may promote heat dissipation. Therefore, one function of colony pattern formation might be to efficiently dissipate heat that results from cellular metabolic activities.
Adhesive contacts between yeast cells may offer protection from macroscopic predators
Yeast and other microbes produce colonies that have a ruffled appearance due to contacts between cells. One established reason for cooperation among individuals in microbial populations is protection (West et al. 2007). For example, cells in the exterior of flocs protect cells in the interior from toxins and antibiotics (Smukalla et al. 2008). Microbial cells are also subject to predation by macroscopic predators. Macroscopic predators, like nematodes, graze on microbial communities. Predator–prey interactions have been studied in bacterial populations, which have developed diverse strategies to evade macroscopic predators. Depending on the species, bacteria have been shown to evade predators by the formation of mats (Darby et al. 2002; DePas et al. 2014; Nandi et al. 2016), by the secretion of proteases (Vaitkevicius et al. 2006) and exopolysaccharides that interfere with the predator’s responses (Begun et al. 2007), and by phase variation, in which bacterial cells express different classes of cell surface proteins to evade detection (Dahl et al. 2011).
Although predator–prey studies are less-well characterized in fungal species, yeast and other fungal microorganisms have ecologically relevant interactions with animals in the wild, including grazing by wasps (Stefanini et al. 2016), flies (Fischer et al. 2017), and other invertebrates. Using two well-characterized laboratory genetic models, we provide evidence that adhesive contacts between yeast cells may deter efficient predation by nematodes. While individual cells in suspension were vulnerable to being ingested, cells that adhered to each other and formed clumps were too large to be eaten. Once cells settled out of suspension and grew into a colony, adhesive colonies formed a physical barrier to deter penetration by worms and also trigger the “nose-touch” mechanosensory behavioral avoidance response. Although the predator–prey model set up in the laboratory may or may not reflect a true ecological relationship between the two species, experiments using model organisms can provide insights into encounters that may occur in the wild. Our data provide an empirical framework to test the hypothesis that the physical interaction among yeast cells in communities can protect individuals from macroscopic predation in the wild.
In conclusion, we have explored the roles of filamentous growth regulatory pathways in regulating multiple adhesion-dependent surface responses in yeast. Dissection of the regulatory pathways by comparative RNA-seq and mutant phenotype analysis revealed new components and targets, which emphasize the common and unique roles that pathways play in coordinately regulating colonial surface responses in this organism. Stress-responsive genes that respond to challenges associated with surface growth were also characterized. The signaling components, target genes, and stress-responsive genes identified here might be common among other fungi, including pathogens. Our results may be particularly relevant to early steps in fungal virulence, where cells must attach to and penetrate surfaces to colonize the host.
Acknowledgments
Clark Driscoll and Aditi Chaubey helped with experiments involving C. elegans. Thanks to laboratory members for comments on the article. The work was supported by grants from the National Institutes of Health (GM-098629 to P.J.C. and GM-015758 to D.M.F.).
Author contributions: J.C. designed and performed experiments and wrote the paper. I.S. analyzed the data. S.J. performed experiments and analyzed the data. O.M. performed experiments. O.G. analyzed the data. D.M.F. designed experiments. A.K. analyzed the data. P.J.C. designed experiments and wrote the paper. The authors have no competing interests in the study.
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25386/genetics.8066615.
Communicating editor: M. Freitag
Literature Cited
- Abdullah U., Cullen P. J., 2009. The tRNA modification complex elongator regulates the Cdc42-dependent mitogen-activated protein kinase pathway that controls filamentous growth in yeast. Eukaryot. Cell 8: 1362–1372. 10.1128/EC.00015-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari H., Cullen P. J., 2014. Metabolic respiration induces AMPK- and Ire1p-dependent activation of the p38-type HOG MAPK pathway. PLoS Genet. 10: e1004734 10.1371/journal.pgen.1004734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P. T., Huber W., 2015. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., et al. , 2000. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat. Genet. 25: 25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L., Shtonda B. B., 2003. Food transport in the C. elegans pharynx. J. Exp. Biol. 206: 2441–2457. 10.1242/jeb.00433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell L., Cook J. G., Zhu-Shimoni J. X., Voora D., Thorner J., 1998. Differential regulation of transcription: repression by unactivated mitogen-activated protein kinase Kss1 requires the Dig1 and Dig2 proteins. Proc. Natl. Acad. Sci. USA 95: 15400–15405. 10.1073/pnas.95.26.15400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrales R. R., Jimenez J., Ibeas J. I., 2008. Identification of novel activation mechanisms for FLO11 regulation in Saccharomyces cerevisiae. Genetics 178: 145–156. 10.1534/genetics.107.081315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basehoar A. D., Zanton S. J., Pugh B. F., 2004. Identification and distinct regulation of yeast TATA box-containing genes. Cell 116: 699–709. 10.1016/S0092-8674(04)00205-3 [DOI] [PubMed] [Google Scholar]
- Begun J., Gaiani J. M., Rohde H., Mack D., Calderwood S. B., et al. , 2007. Staphylococcal biofilm exopolysaccharide protects against Caenorhabditis elegans immune defenses. PLoS Pathog. 3: e57 10.1371/journal.ppat.0030057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belotti F., Tisi R., Paiardi C., Rigamonti M., Groppi S., et al. , 2012. Localization of Ras signaling complex in budding yeast. Biochim. Biophys. Acta 1823: 1208–1216. 10.1016/j.bbamcr.2012.04.016 [DOI] [PubMed] [Google Scholar]
- Bentley N. J., Fitch I. T., Tuite M. F., 1992. The small heat-shock protein Hsp26 of Saccharomyces cerevisiae assembles into a high molecular weight aggregate. Yeast 8: 95–106. 10.1002/yea.320080204 [DOI] [PubMed] [Google Scholar]
- Bernstein B. E., Tong J. K., Schreiber S. L., 2000. Genomewide studies of histone deacetylase function in yeast. Proc. Natl. Acad. Sci. USA 97: 13708–13713 [corrigenda: Proc. Natl. Acad. Sci. USA 98: 5368 (2001)]. 10.1073/pnas.250477697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharucha N., Ma J., Dobry C. J., Lawson S. K., Yang Z., et al. , 2008. Analysis of the yeast kinome reveals a network of regulated protein localization during filamentous growth. Mol. Biol. Cell 19: 2708–2717. 10.1091/mbc.e07-11-1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bois M., Singh S., Samlalsingh A., Lipke P. N., Garcia M. C., 2013. Does Candida albicans Als5p amyloid play a role in commensalism in Caenorhabditis elegans? Eukaryot. Cell 12: 703–711. 10.1128/EC.00020-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojsen R. K., Andersen K. S., Regenberg B., 2012. Saccharomyces cerevisiae–a model to uncover molecular mechanisms for yeast biofilm biology. FEMS Immunol. Med. Microbiol. 65: 169–182. 10.1111/j.1574-695X.2012.00943.x [DOI] [PubMed] [Google Scholar]
- Bojsen R., Regenberg B., Gresham D., Folkesson A., 2016. A common mechanism involving the TORC1 pathway can lead to amphotericin B-persistence in biofilm and planktonic Saccharomyces cerevisiae populations. Sci. Rep. 6: 21874 10.1038/srep21874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman A. R., Leigh-Bell J. A., Yu H., Bertone P., Gerstein M., et al. , 2006. Target hub proteins serve as master regulators of development in yeast. Genes Dev. 20: 435–448. 10.1101/gad.1389306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreutz A., Boucher L., Breitkreutz B. J., Sultan M., Jurisica I., et al. , 2003. Phenotypic and transcriptional plasticity directed by a yeast mitogen-activated protein kinase network. Genetics 165: 997–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruzzone M. J., Grunberg S., Kubik S., Zentner G. E., Shore D., 2018. Distinct patterns of histone acetyltransferase and Mediator deployment at yeast protein-coding genes. Genes Dev. 32: 1252–1265. 10.1101/gad.312173.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C. X., El-Kirat-Chatel S., Joseph I. G., Jackson D. N., Ramsook C. B., et al. , 2016. Force sensitivity in Saccharomyces cerevisiae flocculins. mSphere 1: e00128-16. 10.1128/mSphere.00128-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra J., Kuhn D. M., Mukherjee P. K., Hoyer L. L., McCormick T., et al. , 2001. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J. Bacteriol. 183: 5385–5394. 10.1128/JB.183.18.5385-5394.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavel C. A., Dionne H. M., Birkaya B., Joshi J., Cullen P. J., 2010. Multiple signals converge on a differentiation MAPK pathway. PLoS Genet. 6: e1000883 10.1371/journal.pgen.1000883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavel C. A., Caccamise L. M., Li B., Cullen P. J., 2014. Global regulation of a differentiation MAPK pathway in yeast. Genetics 198: 1309–1328. 10.1534/genetics.114.168252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin B. L., Ryan O., Lewitter F., Boone C., Fink G. R., 2012. Genetic variation in Saccharomyces cerevisiae: circuit diversification in a signal transduction network. Genetics 192: 1523–1532. 10.1534/genetics.112.145573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S., Lane S., Liu H., 2006. Regulation of mating and filamentation genes by two distinct Ste12 complexes in Saccharomyces cerevisiae. Mol. Cell. Biol. 26: 4794–4805. 10.1128/MCB.02053-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow J., Notaro M., Prabhakar A., Free S. J., Cullen P. J., 2018. Impact of fungal MAPK pathway targets on the cell wall. J. Fungi (Basel) 4: E93. 10.3390/jof4030093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow J., Dionne H. M., Prabhakar A., Mehrotra A., Somboonthum J., et al. , 2019. Aggregate filamentous growth responses in yeast. mSphere 4: e00702-18. 10.1128/mSphere.00702-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen B. D., Sertil O., Abramova N. E., Davies K. J., Lowry C. V., 2001. Induction and repression of DAN1 and the family of anaerobic mannoprotein genes in Saccharomyces cerevisiae occurs through a complex array of regulatory sites. Nucleic Acids Res. 29: 799–808. 10.1093/nar/29.3.799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coluccio A., Bogengruber E., Conrad M. N., Dresser M. E., Briza P., et al. , 2004. Morphogenetic pathway of spore wall assembly in Saccharomyces cerevisiae. Eukaryot. Cell 3: 1464–1475. 10.1128/EC.3.6.1464-1475.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook J. G., Bardwell L., Kron S. J., Thorner J., 1996. Two novel targets of the MAP kinase Kss1 are negative regulators of invasive growth in the yeast Saccharomyces cerevisiae. Genes Dev. 10: 2831–2848. 10.1101/gad.10.22.2831 [DOI] [PubMed] [Google Scholar]
- Crabtree H. G., 1929. Observations on the carbohydrate metabolism of tumours. Biochem. J. 23: 536–545. 10.1042/bj0230536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie G. A., Tan Z., Hays M., Sirr A., Jeffery E. W., et al. , 2017. Transcriptional profiling of biofilm regulators identified by an overexpression screen in Saccharomyces cerevisiae. G3 (Bethesda) 7: 2845–2854. 10.1534/g3.117.042440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen P. J., 2015a Evaluating the activity of the filamentous growth mitogen-activated protein kinase pathway in yeast. Cold Spring Harb. Protoc. 2015: 276–283. 10.1101/pdb.prot085092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen P. J., 2015b The plate-washing assay: a simple test for filamentous growth in budding yeast. Cold Spring Harb. Protoc. 2015: 168–171. 10.1101/pdb.prot085068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen P. J., Sprague G. F., Jr., 2000. Glucose depletion causes haploid invasive growth in yeast. Proc. Natl. Acad. Sci. USA 97: 13619–13624. 10.1073/pnas.240345197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen P. J., Sprague G. F., Jr., 2002. The roles of bud-site-selection proteins during haploid invasive growth in yeast. Mol. Biol. Cell 13: 2990–3004. 10.1091/mbc.e02-03-0151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen P. J., Sprague G. F., Jr., 2012. The regulation of filamentous growth in yeast. Genetics 190: 23–49. 10.1534/genetics.111.127456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen P. J., Sabbagh W., Jr., Graham E., Irick M. M., van Olden E. K., et al. , 2004. A signaling mucin at the head of the Cdc42- and MAPK-dependent filamentous growth pathway in yeast. Genes Dev. 18: 1695–1708. 10.1101/gad.1178604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler N. S., Pan X., Heitman J., Cardenas M. E., 2001. The TOR signal transduction cascade controls cellular differentiation in response to nutrients. Mol. Biol. Cell 12: 4103–4113. 10.1091/mbc.12.12.4103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl J. L., Ulrich C. H., Kroft T. L., 2011. Role of phase variation in the resistance of Myxococcus xanthus fruiting bodies to Caenorhabditis elegans predation. J. Bacteriol. 193: 5081–5089. 10.1128/JB.05383-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang N. X., Hincha D. K., 2011. Identification of two hydrophilins that contribute to the desiccation and freezing tolerance of yeast (Saccharomyces cerevisiae) cells. Cryobiology 62: 188–193. 10.1016/j.cryobiol.2011.03.002 [DOI] [PubMed] [Google Scholar]
- Darby C., Hsu J. W., Ghori N., Falkow S., 2002. Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature 417: 243–244. 10.1038/417243a [DOI] [PubMed] [Google Scholar]
- De Deken R. H., 1966. The Crabtree effects and its relation to the petite mutation. J. Gen. Microbiol. 44: 157–165. 10.1099/00221287-44-2-157 [DOI] [PubMed] [Google Scholar]
- Dejean L., Beauvoit B., Bunoust O., Guerin B., Rigoulet M., 2002. Activation of Ras cascade increases the mitochondrial enzyme content of respiratory competent yeast. Biochem. Biophys. Res. Commun. 293: 1383–1388. 10.1016/S0006-291X(02)00391-1 [DOI] [PubMed] [Google Scholar]
- DePas W. H., Syed A. K., Sifuentes M., Lee J. S., Warshaw D., et al. , 2014. Biofilm formation protects Escherichia coli against killing by Caenorhabditis elegans and Myxococcus xanthus. Appl. Environ. Microbiol. 80: 7079–7087. 10.1128/AEM.02464-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai J. V., Mitchell A. P., Andes D. R., 2014. Fungal biofilms, drug resistance, and recurrent infection. Cold Spring Harb. Perspect. Med. 4: a019729. 10.1101/cshperspect.a019729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell R. D., Ryan O., Jansen A., Cheung D., Agarwala S., et al. , 2010. Genotype to phenotype: a complex problem. Science 328: 469 10.1126/science.1189015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E., Lipson D., Yogev S., Yakhini Z., 2007. Discovering motifs in ranked lists of DNA sequences. PLoS Comput. Biol. 3: e39 10.1371/journal.pcbi.0030039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z., 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48 10.1186/1471-2105-10-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engebrecht J., Masse S., Davis L., Rose K., Kessel T., 1998. Yeast meiotic mutants proficient for the induction of ectopic recombination. Genetics 148: 581–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein C. B., Waddle J. A., Hale W., Dave V., Thornton J., et al. , 2001. Genome-wide responses to mitochondrial dysfunction. Mol. Biol. Cell 12: 297–308. 10.1091/mbc.12.2.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang-Yen C., Avery L., Samuel A. D., 2009. Two size-selective mechanisms specifically trap bacteria-sized food particles in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 106: 20093–20096. 10.1073/pnas.0904036106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix M. A., Braendle C., 2010. The natural history of Caenorhabditis elegans. Curr. Biol. 20: R965–R969. 10.1016/j.cub.2010.09.050 [DOI] [PubMed] [Google Scholar]
- Fischer C. N., Trautman E. P., Crawford J. M., Stabb E. V., Handelsman J., et al. , 2017. Metabolite exchange between microbiome members produces compounds that influence Drosophila behavior. Elife 6: e18855. 10.7554/eLife.18855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao T., Zheng Z., Hou Y., Zhou M., 2014. Transcription factors spt3 and spt8 are associated with conidiation, mycelium growth, and pathogenicity in Fusarium graminearum. FEMS Microbiol. Lett. 351: 42–50. 10.1111/1574-6968.12350 [DOI] [PubMed] [Google Scholar]
- Garay-Arroyo A., Covarrubias A. A., 1999. Three genes whose expression is induced by stress in Saccharomyces cerevisiae. Yeast 15: 879–892. [DOI] [PubMed] [Google Scholar]
- Georgakopoulos T., Thireos G., 1992. Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription. EMBO J. 11: 4145–4152. 10.1002/j.1460-2075.1992.tb05507.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R. D., Schiest R. H., 2007. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2: 31–34. 10.1038/nprot.2007.13 [DOI] [PubMed] [Google Scholar]
- Gimeno C. J., Ljungdahl P. O., Styles C. A., Fink G. R., 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68: 1077–1090. 10.1016/0092-8674(92)90079-R [DOI] [PubMed] [Google Scholar]
- González B., Mas A., Beltran G., Cullen P. J., Torija M. J., 2017. Role of mitochondrial retrograde pathway in regulating ethanol-inducible filamentous growth in yeast. Front. Physiol. 8: 148 10.3389/fphys.2017.00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granek J. A., Magwene P. M., 2010. Environmental and genetic determinants of colony morphology in yeast. PLoS Genet. 6: e1000823 10.1371/journal.pgen.1000823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granek J. A., Kayikci O., Magwene P. M., 2011. Pleiotropic signaling pathways orchestrate yeast development. Curr. Opin. Microbiol. 14: 676–681. 10.1016/j.mib.2011.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graybill E. R., Rouhier M. F., Kirby C. E., Hawes J. W., 2007. Functional comparison of citrate synthase isoforms from S. cerevisiae. Arch. Biochem. Biophys. 465: 26–37. 10.1016/j.abb.2007.04.039 [DOI] [PubMed] [Google Scholar]
- Hagman A., Sall T., Piskur J., 2014. Analysis of the yeast short-term Crabtree effect and its origin. FEBS J. 281: 4805–4814. 10.1111/febs.13019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart A. C., Sims S., Kaplan J. M., 1995. Synaptic code for sensory modalities revealed by C. elegans GLR-1 glutamate receptor. Nature 378: 82–85. 10.1038/378082a0 [DOI] [PubMed] [Google Scholar]
- Higgins V. J., Rogers P. J., Dawes I. W., 2003. Application of genome-wide expression analysis to identify molecular markers useful in monitoring industrial fermentations. Appl. Environ. Microbiol. 69: 7535–7540. 10.1128/AEM.69.12.7535-7540.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch C. L., Coban Akdemir Z., Wang L., Jayakumaran G., Trcka D., et al. , 2015. Myc and SAGA rewire an alternative splicing network during early somatic cell reprogramming. Genes Dev. 29: 803–816. 10.1101/gad.255109.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisinga K. L., Pugh B. F., 2004. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell 13: 573–585. 10.1016/S1097-2765(04)00087-5 [DOI] [PubMed] [Google Scholar]
- Ignatiadis N., Klaus B., Zaugg J. B., Huber W., 2016. Data-driven hypothesis weighting increases detection power in genome-scale multiple testing. Nat. Methods 13: 577–580. 10.1038/nmeth.3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain C., Yun M., Politz S. M., Rao R. P., 2009. A pathogenesis assay using Saccharomyces cerevisiae and Caenorhabditis elegans reveals novel roles for yeast AP-1, Yap1, and host dual oxidase BLI-3 in fungal pathogenesis. Eukaryot. Cell 8: 1218–1227. 10.1128/EC.00367-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E. A., 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin R., Dobry C. J., McCown P. J., Kumar A., 2008. Large-scale analysis of yeast filamentous growth by systematic gene disruption and overexpression. Mol. Biol. Cell 19: 284–296. 10.1091/mbc.e07-05-0519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R. C., Hough J. S., 1970. The effect of temperature on the metabolism of baker’s yeast growing on continuous culture. J. Gen. Microbiol. 60: 107–116. 10.1099/00221287-60-1-107 [DOI] [PubMed] [Google Scholar]
- Kaksonen M., Sun Y., Drubin D. G., 2003. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell 115: 475–487. 10.1016/S0092-8674(03)00883-3 [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28: 27–30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan J. M., Horvitz H. R., 1993. A dual mechanosensory and chemosensory neuron in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 90: 2227–2231. 10.1073/pnas.90.6.2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunanithi S., Vadaie N., Chavel C. A., Birkaya B., Joshi J., et al. , 2010. Shedding of the mucin-like flocculin Flo11p reveals a new aspect of fungal adhesion regulation. Curr. Biol. 20: 1389–1395. 10.1016/j.cub.2010.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunanithi S., Joshi J., Chavel C., Birkaya B., Grell L., et al. , 2012. Regulation of mat responses by a differentiation MAPK pathway in Saccharomyces cerevisiae. PLoS One 7: e32294 10.1371/journal.pone.0032294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. M., Vanguri S., Boeke J. D., Gabriel A., Voytas D. F., 1998. Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 8: 464–478. 10.1101/gr.8.5.464 [DOI] [PubMed] [Google Scholar]
- Kim K. S., Rosenkrantz M. S., Guarente L., 1986. Saccharomyces cerevisiae contains two functional citrate synthase genes. Mol. Cell. Biol. 6: 1936–1942. 10.1128/MCB.6.6.1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchman P. A., Kim S., Lai C. Y., Jazwinski S. M., 1999. Interorganelle signaling is a determinant of longevity in Saccharomyces cerevisiae. Genetics 152: 179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutelou E., Hirsch C. L., Dent S. Y., 2010. Multiple faces of the SAGA complex. Curr. Opin. Cell Biol. 22: 374–382. 10.1016/j.ceb.2010.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraushaar T., Bruckner S., Veelders M., Rhinow D., Schreiner F., et al. , 2015. Interactions by the fungal Flo11 adhesin depend on a fibronectin type III-like adhesin domain girdled by aromatic bands. Structure 23: 1005–1017. 10.1016/j.str.2015.03.021 [DOI] [PubMed] [Google Scholar]
- Kuchin S., Vyas V. K., Carlson M., 2002. Snf1 protein kinase and the repressors Nrg1 and Nrg2 regulate FLO11, haploid invasive growth, and diploid pseudohyphal differentiation. Mol. Cell. Biol. 22: 3994–4000. 10.1128/MCB.22.12.3994-4000.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto C. A., 2005. A contact-activated kinase signals Candida albicans invasive growth and biofilm development. Proc. Natl. Acad. Sci. USA 102: 5576–5581. 10.1073/pnas.0407097102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusari A. B., Molina D. M., Sabbagh W., Jr., Lau C. S., Bardwell L., 2004. A conserved protein interaction network involving the yeast MAP kinases Fus3 and Kss1. J. Cell Biol. 164: 267–277. 10.1083/jcb.200310021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laboucarié T., Detilleux D., Rodriguez-Mias R. A., Faux C., Romeo Y., et al. , 2017. TORC1 and TORC2 converge to regulate the SAGA co-activator in response to nutrient availability. EMBO Rep. 18: 2197–2218. 10.15252/embr.201744942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levchenko A., 2003. Dynamical and integrative cell signaling: challenges for the new biology. Biotechnol. Bioeng. 84: 773–782. 10.1002/bit.10854 [DOI] [PubMed] [Google Scholar]
- Li W., Mitchell A. P., 1997. Proteolytic activation of Rim1p, a positive regulator of yeast sporulation and invasive growth. Genetics 145: 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X., Butow R. A., 1993. RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell 72: 61–71. 10.1016/0092-8674(93)90050-Z [DOI] [PubMed] [Google Scholar]
- Lin D., Yin X., Wang X., Zhou P., Guo F. B., 2013. Re-annotation of protein-coding genes in the genome of saccharomyces cerevisiae based on support vector machines. PLoS One 8: e64477 10.1371/journal.pone.0064477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Styles C. A., Fink G. R., 1993. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science 262: 1741–1744. 10.1126/science.8259520 [DOI] [PubMed] [Google Scholar]
- Liu H., Styles C. A., Fink G. R., 1996. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics 144: 967–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Butow R. A., 1999. A transcriptional switch in the expression of yeast tricarboxylic acid cycle genes in response to a reduction or loss of respiratory function. Mol. Cell. Biol. 19: 6720–6728. 10.1128/MCB.19.10.6720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Butow R. A., 2006. Mitochondrial retrograde signaling. Annu. Rev. Genet. 40: 159–185. 10.1146/annurev.genet.40.110405.090613 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D., 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Method. Methods 25: 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lo H. J., Kohler J. R., DiDomenico B., Loebenberg D., Cacciapuoti A., et al. , 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90: 939–949. 10.1016/S0092-8674(00)80358-X [DOI] [PubMed] [Google Scholar]
- Lo W. S., Dranginis A. M., 1998. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol. Biol. Cell 9: 161–171. 10.1091/mbc.9.1.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M. C., Heitman J., 1998. Regulators of pseudohyphal differentiation in Saccharomyces cerevisiae identified through multicopy suppressor analysis in ammonium permease mutant strains. Genetics 150: 1443–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S., 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W., Brouwer C., 2013. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 29: 1830–1831. 10.1093/bioinformatics/btt285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons T. J., Gasch A. P., Gaither L. A., Botstein D., Brown P. O., et al. , 2000. Genome-wide characterization of the Zap1p zinc-responsive regulon in yeast. Proc. Natl. Acad. Sci. USA 97: 7957–7962. 10.1073/pnas.97.14.7957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani H. D., Galitski T., Lander E. S., Fink G. R., 1999. Effectors of a developmental mitogen-activated protein kinase cascade revealed by expression signatures of signaling mutants. Proc. Natl. Acad. Sci. USA 96: 12530–12535. 10.1073/pnas.96.22.12530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magasanik B., Kaiser C. A., 2002. Nitrogen regulation in Saccharomyces cerevisiae. Gene 290: 1–18. 10.1016/S0378-1119(02)00558-9 [DOI] [PubMed] [Google Scholar]
- Maricq A. V., Peckol E., Driscoll M., Bargmann C. I., 1995. Mechanosensory signalling in C. elegans mediated by the GLR-1 glutamate receptor. Nature 378: 78–81 [corrigenda: Nature 379: 749 (1996)]. 10.1038/378078a0 [DOI] [PubMed] [Google Scholar]
- Maršíková J., Wilkinson D., Hlaváček O., Gilfillan G. D., Mizeranschi A., et al. , 2017. Metabolic differentiation of surface and invasive cells of yeast colony biofilms revealed by gene expression profiling. BMC Genomics 18: 814 10.1186/s12864-017-4214-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez M. J., Roy S., Archuletta A. B., Wentzell P. D., Anna-Arriola S. S., et al. , 2004. Genomic analysis of stationary-phase and exit in Saccharomyces cerevisiae: gene expression and identification of novel essential genes. Mol. Biol. Cell 15: 5295–5305. 10.1091/mbc.e03-11-0856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Huang X., Muruganujan A., Tang H., Mills C., et al. , 2017. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 45: D183–D189. https://academic.oup.com/nar/article/45/D1/D183/2605815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralles V. J., Serrano R., 1995. A genomic locus in Saccharomyces cerevisiae with four genes up-regulated by osmotic stress. Mol. Microbiol. 17: 653–662. 10.1111/j.1365-2958.1995.mmi_17040653.x [DOI] [PubMed] [Google Scholar]
- Mösch H. U., Roberts R. L., Fink G. R., 1996. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93: 5352–5356. 10.1073/pnas.93.11.5352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mösch H. U., Kübler E., Krappmann S., Fink G. R., Braus G. H., 1999. Crosstalk between the Ras2p-controlled mitogen-activated protein kinase and cAMP pathways during invasive growth of Saccharomyces cerevisiae. Mol. Biol. Cell 10: 1325–1335. 10.1091/mbc.10.5.1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrsa V., Ecker M., Strahl-Bolsinger S., Nimtz M., Lehle L., et al. , 1999. Deletion of new covalently linked cell wall glycoproteins alters the electrophoretic mobility of phosphorylated wall components of Saccharomyces cerevisiae. J. Bacteriol. 181: 3076–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi M., Berry C., Brassinga A. K., Belmonte M. F., Fernando W. G., et al. , 2016. Pseudomonas brassicacearum strain DF41 kills Caenorhabditis elegans through biofilm-dependent and biofilm-independent mechanisms. Appl. Environ. Microbiol. 82: 6889–6898. 10.1128/AEM.02199-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett, J. E., and D. R. Andes, 2015 Fungal biofilms: in vivo models for discovery of anti-biofilm drugs. Microbiol. Spectr. 3: MB-0008-2014. 10.1128/microbiolspec.MB-0008-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson K. A., Nelson C., Tai G., Hung W., Yong C., et al. , 2000. Two regulators of Ste12p inhibit pheromone-responsive transcription by separate mechanisms. Mol. Cell. Biol. 20: 4199–4209. 10.1128/MCB.20.12.4199-4209.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palecek S. P., Parikh A. S., Kron S. J., 2000. Genetic analysis reveals that FLO11 upregulation and cell polarization independently regulate invasive growth in Saccharomyces cerevisiae. Genetics 156: 1005–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palková Z., Váchová L., 2006. Life within a community: benefit to yeast long-term survival. FEMS Microbiol. Rev. 30: 806–824. 10.1111/j.1574-6976.2006.00034.x [DOI] [PubMed] [Google Scholar]
- Pan X., Heitman J., 2000. Sok2 regulates yeast pseudohyphal differentiation via a transcription factor cascade that regulates cell-cell adhesion. Mol. Cell. Biol. 20: 8364–8372. 10.1128/MCB.20.22.8364-8372.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabitsch K. P., Toth A., Galova M., Schleiffer A., Schaffner G., et al. , 2001. A screen for genes required for meiosis and spore formation based on whole-genome expression. Curr. Biol. 11: 1001–1009. 10.1016/S0960-9822(01)00274-3 [DOI] [PubMed] [Google Scholar]
- Ramage G., Saville S. P., Thomas D. P., Lopez-Ribot J. L., 2005. Candida biofilms: an update. Eukaryot. Cell 4: 633–638. 10.1128/EC.4.4.633-638.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A., Yekutieli D., Benjamini Y., 2003. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19: 368–375. 10.1093/bioinformatics/btf877 [DOI] [PubMed] [Google Scholar]
- Reynolds T. B., 2006. The Opi1p transcription factor affects expression of FLO11, mat formation, and invasive growth in Saccharomyces cerevisiae. Eukaryot. Cell 5: 1266–1275. 10.1128/EC.00022-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds T. B., Fink G. R., 2001. Bakers’ yeast, a model for fungal biofilm formation. Science 291: 878–881. 10.1126/science.291.5505.878 [DOI] [PubMed] [Google Scholar]
- Roberts C. J., Nelson B., Marton M. J., Stoughton R., Meyer M. R., et al. , 2000. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science 287: 873–880. 10.1126/science.287.5454.873 [DOI] [PubMed] [Google Scholar]
- Roberts R. L., Fink G. R., 1994. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 8: 2974–2985. 10.1101/gad.8.24.2974 [DOI] [PubMed] [Google Scholar]
- Robertson L. S., Fink G. R., 1998. The three yeast A kinases have specific signaling functions in pseudohyphal growth. Proc. Natl. Acad. Sci. USA 95: 13783–13787. 10.1073/pnas.95.23.13783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K., 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Porrata B., Carmona-Gutierrez D., Reisenbichler A., Bauer M., Lopez G., et al. , 2012. Sip18 hydrophilin prevents yeast cell death during desiccation stress. J. Appl. Microbiol. 112: 512–525. 10.1111/j.1365-2672.2011.05219.x [DOI] [PubMed] [Google Scholar]
- Rose M. D., Winston F., Heieter P., 1990. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Rupp S., Summers E., Lo H. J., Madhani H., Fink G., 1999. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 18: 1257–1269. 10.1093/emboj/18.5.1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan O., Shapiro R. S., Kurat C. F., Mayhew D., Baryshnikova A., et al. , 2012. Global gene deletion analysis exploring yeast filamentous growth. Science 337: 1353–1356. 10.1126/science.1224339 [DOI] [PubMed] [Google Scholar]
- Samanta M. P., Liang S., 2003. Predicting protein functions from redundancies in large-scale protein interaction networks. Proc. Natl. Acad. Sci. USA 100: 12579–12583. 10.1073/pnas.2132527100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T., 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sarkar P. K., Florczyk M. A., McDonough K. A., Nag D. K., 2002. SSP2, a sporulation-specific gene necessary for outer spore wall assembly in the yeast Saccharomyces cerevisiae. Mol. Genet. Genomics 267: 348–358. 10.1007/s00438-002-0666-5 [DOI] [PubMed] [Google Scholar]
- Sarode N., Miracle B., Peng X., Ryan O., Reynolds T. B., 2011. Vacuolar protein sorting genes regulate mat formation in Saccharomyces cerevisiae by Flo11p-dependent and -independent mechanisms. Eukaryot. Cell 10: 1516–1526. 10.1128/EC.05078-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarode N., Davis S. E., Tams R. N., Reynolds T. B., 2014. The Wsc1p cell wall signaling protein controls biofilm (Mat) formation independently of Flo11p in Saccharomyces cerevisiae. G3 (Bethesda) 4: 199–207. 10.1534/g3.113.006361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherz K., Andersen R., Bojsen L., Gro, Rejkjær, et al. , 2014. Genetic basis for Saccharomyces cerevisiae biofilm in liquid medium. G3 (Bethesda) 4: 1671–1680. 10.1534/g3.114.010892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalix W., Bandlow W., 1993. The ethanol-inducible YAT1 gene from yeast encodes a presumptive mitochondrial outer carnitine acetyltransferase. J. Biol. Chem. 268: 27428–27439. [PubMed] [Google Scholar]
- Sertil O., Cohen B. D., Davies K. J., Lowry C. V., 1997. The DAN1 gene of S. cerevisiae is regulated in parallel with the hypoxic genes, but by a different mechanism. Gene 192: 199–205. 10.1016/S0378-1119(97)00028-0 [DOI] [PubMed] [Google Scholar]
- Shtonda B. B., Avery L., 2006. Dietary choice behavior in Caenorhabditis elegans. J. Exp. Biol. 209: 89–102. 10.1242/jeb.01955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small W. C., Brodeur R. D., Sandor A., Fedorova N., Li G., et al. , 1995. Enzymatic and metabolic studies on retrograde regulation mutants of yeast. Biochemistry 34: 5569–5576. 10.1021/bi00016a031 [DOI] [PubMed] [Google Scholar]
- Smukalla S., Caldara M., Pochet N., Beauvais A., Guadagnini S., et al. , 2008. FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast. Cell 135: 726–737. 10.1016/j.cell.2008.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll D. R., Daniels K. J., 2016. Plasticity of Candida albicans biofilms. Microbiol. Mol. Biol. Rev. 80: 565–595. 10.1128/MMBR.00068-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan C. J., Padilla S. M., Audhya A., Emr S. D., 2005. The phosphoinositide phosphatase Sjl2 is recruited to cortical actin patches in the control of vesicle formation and fission during endocytosis. Mol. Cell. Biol. 25: 2910–2923. 10.1128/MCB.25.8.2910-2923.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanini I., Dapporto L., Berna L., Polsinelli M., Turillazzi S., et al. , 2016. Social wasps are a Saccharomyces mating nest. Proc. Natl. Acad. Sci. USA 113: 2247–2251. 10.1073/pnas.1516453113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner D. E., Berger S. L., 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64: 435–459. 10.1128/MMBR.64.2.435-459.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudbery P., Gow N., Berman J., 2004. The distinct morphogenic states of Candida albicans. Trends Microbiol. 12: 317–324. 10.1016/j.tim.2004.05.008 [DOI] [PubMed] [Google Scholar]
- Suissa M., Suda K., Schatz G., 1984. Isolation of the nuclear yeast genes for citrate synthase and fifteen other mitochondrial proteins by a new screening method. EMBO J. 3: 1773–1781. 10.1002/j.1460-2075.1984.tb02045.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiegers J. H., Dippenaar N., Pretorius I. S., Bauer F. F., 2001. Carnitine-dependent metabolic activities in Saccharomyces cerevisiae: three carnitine acetyltransferases are essential in a carnitine-dependent strain. Yeast 18: 585–595. 10.1002/yea.712 [DOI] [PubMed] [Google Scholar]
- Swiegers J. H., Pretorius I. S., Bauer F. F., 2006. Regulation of respiratory growth by Ras: the glyoxylate cycle mutant, cit2Delta, is suppressed by RAS2. Curr. Genet. 50: 161–171. 10.1007/s00294-006-0084-z [DOI] [PubMed] [Google Scholar]
- Tam A., Green J. E. F., Balasuriya S., Tek E. L., Gardner J. M., et al. , 2018. Nutrient-limited growth with non-linear cell diffusion as a mechanism for floral pattern formation in yeast biofilms. J. Theor. Biol. 448: 122–141. 10.1016/j.jtbi.2018.04.004 [DOI] [PubMed] [Google Scholar]
- Tedford K., Kim S., Sa D., Stevens K., Tyers M., 1997. Regulation of the mating pheromone and invasive growth responses in yeast by two MAP kinase substrates. Curr. Biol. 7: 228–238. 10.1016/S0960-9822(06)00118-7 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S. L., 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traven A., Janicke A., Harrison P., Swaminathan A., Seemann T., et al. , 2012. Transcriptional profiling of a yeast colony provides new insight into the heterogeneity of multicellular fungal communities. PLoS One 7: e46243 10.1371/journal.pone.0046243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Váchová L., Stovícek V., Hlavácek O., Chernyavskiy O., Stĕpánek L., et al. , 2011. Flo11p, drug efflux pumps, and the extracellular matrix cooperate to form biofilm yeast colonies. J. Cell Biol. 194: 679–687. 10.1083/jcb.201103129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaitkevicius K., Lindmark B., Ou G., Song T., Toma C., et al. , 2006. A Vibrio cholerae protease needed for killing of Caenorhabditis elegans has a role in protection from natural predator grazing. Proc. Natl. Acad. Sci. USA 103: 9280–9285. 10.1073/pnas.0601754103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Felden J., Weisser S., Bruckner S., Lenz P., Mosch H. U., 2014. The transcription factors Tec1 and Ste12 interact with coregulators Msa1 and Msa2 to activate adhesion and multicellular development. Mol. Cell. Biol. 34: 2283–2293. 10.1128/MCB.01599-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dyk D., Pretorius I. S., Bauer F. F., 2005. Mss11p is a central element of the regulatory network that controls FLO11 expression and invasive growth in Saccharomyces cerevisiae. Genetics 169: 91–106. 10.1534/genetics.104.033704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voordeckers K., De Maeyer D., van der Zande E., Vinces M. D., Meert W., et al. , 2012. Identification of a complex genetic network underlying Saccharomyces cerevisiae colony morphology. Mol. Microbiol. 86: 225–239. 10.1111/j.1365-2958.2012.08192.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Dent S. Y., 2014. Functions of SAGA in development and disease. Epigenomics 6: 329–339. 10.2217/epi.14.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S. A., Griffin A. S., Gardner A., 2007. Evolutionary explanations for cooperation. Curr. Biol. 17: R661–R672. 10.1016/j.cub.2007.06.004 [DOI] [PubMed] [Google Scholar]
- Wickham H., 2016. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag, New York. [Google Scholar]
- Wilson M. A., St Amour C. V., Collins J. L., Ringe D., Petsko G. A., 2004. The 1.8-A resolution crystal structure of YDR533Cp from Saccharomyces cerevisiae: a member of the DJ-1/ThiJ/PfpI superfamily. Proc. Natl. Acad. Sci. USA 101: 1531–1536. 10.1073/pnas.0308089100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F., Dollard C., Malone E. A., Clare J., Kapakos J. G., et al. , 1987. Three genes are required for trans-activation of Ty transcription in yeast. Genetics 115: 649–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood W. B., 1988. The Nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. [Google Scholar]
- Wu C. Y., Bird A. J., Chung L. M., Newton M. A., Winge D. R., et al. , 2008. Differential control of Zap1-regulated genes in response to zinc deficiency in Saccharomyces cerevisiae. BMC Genomics 9: 370 10.1186/1471-2164-9-370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Shively C. A., Jin R., Eckwahl M. J., Dobry C. J., et al. , 2010. A profile of differentially abundant proteins at the yeast cell periphery during pseudohyphal growth. J. Biol. Chem. 285: 15476–15488. 10.1074/jbc.M110.114926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan D. S., 2000. Zinc-regulated genes in Saccharomyces cerevisiae revealed by transposon tagging. Genetics 156: 45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman S., Lippman S. I., Zhao X., Broach J. R., 2008. How Saccharomyces responds to nutrients. Annu. Rev. Genet. 42: 27–81. 10.1146/annurev.genet.41.110306.130206 [DOI] [PubMed] [Google Scholar]
- Zara G., Budroni M., Mannazzu I., Zara S., 2011. Air-liquid biofilm formation is dependent on ammonium depletion in a Saccharomyces cerevisiae flor strain. Yeast 28: 809–814. 10.1002/yea.1907 [DOI] [PubMed] [Google Scholar]
- Zeitlinger J., Simon I., Harbison C. T., Hannett N. M., Volkert T. L., et al. , 2003. Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell 113: 395–404. 10.1016/S0092-8674(03)00301-5 [DOI] [PubMed] [Google Scholar]
- Zhao H., Eide D., 1996. The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc. Natl. Acad. Sci. USA 93: 2454–2458. 10.1073/pnas.93.6.2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Eide D. J., 1997. Zap1p, a metalloregulatory protein involved in zinc-responsive transcriptional regulation in Saccharomyces cerevisiae. Mol. Cell. Biol. 17: 5044–5052. 10.1128/MCB.17.9.5044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu A., Ibrahim J. G., Love M. I., 2018. Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics. 10.1093/bioinformatics/bty895 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All strains are available upon request. The Gene Expression Omnibus (GEO) accession number for raw sequencing data is GSE115657. A comparison with previously published expression profiling data sets for cells grown in liquid culture has been described [GEO accession number GSE61783 (Adhikari and Cullen 2014)]. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.8066615.