Abstract
Quorum sensing is a process of bacterial cell-to-cell chemical communication that relies on the production, detection and response to extracellular signalling molecules called autoinducers. Quorum sensing allows groups of bacteria to synchronously alter behaviour in response to changes in the population density and species composition of the vicinal community. Quorum-sensing-mediated communication is now understood to be the norm in the bacterial world. Elegant research has defined quorum-sensing components and their interactions, for the most part, under ideal and highly controlled conditions. Indeed, these seminal studies laid the foundations for the field. In this Review, we highlight new findings concerning how bacteria deploy quorum sensing in realistic scenarios that mimic nature. We focus on how quorums are detected and how quorum sensing controls group behaviours in complex and dynamically changing environments such as multi-species bacterial communities, in the presence of flow, in 3D non-uniform biofilms and in hosts during infection.
Bacteria, once thought capable of only simple processes and single-celled life, are now appreciated for their ability to act collectively in multi-cellular groups1,2. Coordinated behaviours include bioluminescence3,4, virulence factor production5,6, secondary metabolite production7, competence for DNA uptake8,9 and biofilm formation10,11. These processes are futile when under-taken by a single bacterium acting alone. Rather, success requires population-wide coordination of the individual cells. To orchestrate collective behaviours, bacteria use the cell-to-cell communication process called quorum sensing10,12–14. Quorum sensing is mediated by the production, release, accumulation and group-wide detection of extracellular signalling molecules called autoinducers.
Gram-negative quorum-sensing bacteria use small molecules as autoinducers, and two types of cognate receptor detect these autoinducers — cytoplasmic transcription factors or transmembrane two-component histidine sensor kinases (FIG. 1a and FIG. 1b, respectively). In both cases, autoinducer-receptor complexes direct the expression of quorum-sensing-dependent target genes (reviewed previously12). Gram-positive bacteria typically use oligopeptides as autoinducers, and the partner receptors are transmembrane two-component histidine sensor kinases15 (Fig. 1c). Often, autoinducer–receptor complexes activate expression of the gene encoding the autoinducer synthase, which ramps up the extracellular autoinducer concentration as the bacteria enter into quorum-sensing mode16. This feedforward autoinduction loop is thought to synchronize behaviours across the bacterial population.
Fig. 1 |. Quorum-sensing circuits.
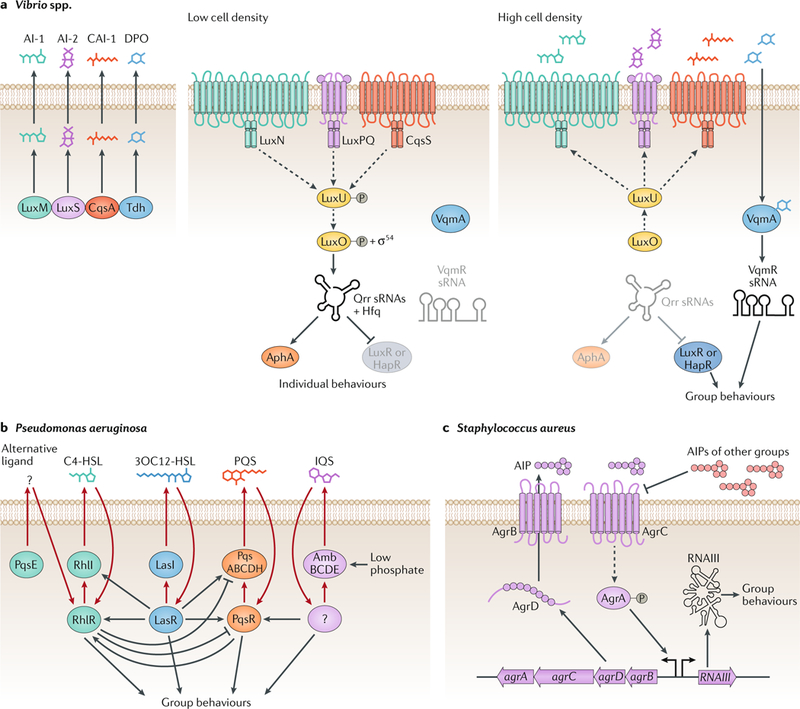
Bacterial quorum sensing relies on networks of autoinducers, autoinducer synthases, partner autoinducer receptors and downstream signal transduction components that convert the information contained in autoinducers into changes in gene expression. a | When Vibrio spp. are at a low cell density, autoinducer levels are low, and their cognate receptors activate a phosphorylation cascade that ultimately results in the activation of the transcription factor AphA, which mediates individual behaviours. By contrast, at high cell density, the synthases LuxM, LuxS, CqsA and Tdh produce high levels of the autoinducers AI-1, AI-2, CAI-1 and DPO, respectively, and the corresponding receptors function as phosphatases. Instead of AphA, LuxR or HapR is produced, which ng loops using LasI and LasR, RhlI, PqsE andmediates group behaviours. b | Pseudomonas aeruginosa employs four interwoven quorum-sensi RhlR, PqsABCDH and PqsR, AmbBCDE and an unknown receptor as the synthases and receptors of the autoinducers 23OC12-HSL, C4-HSL, unknown (PqsE), PQS and IQS, respectively. c | At high cell densities, AgrB from Staphylococcus aureus processes the AgrD precursor peptide and exports the autoinducing peptide AIP, which in turn signals through the AgrC receptor and the downstream transcription factor AgrA. Phosphorylated AgrA induces the production of a regulatory RNA that controls group behaviours. sRNA, small RNA. Dashed lines represent phosphorylation and dephosphorylation. Solid lines represent gene regulation or protein production or small molecule production. Adapted with permission from REF.102, Elsevier.
Bacteria typically integrate information encoded in several quorum-sensing autoinducers into the control of gene expression, which enables intra-species, intragenera and inter-species communication as well as communication with bacteria in the microbiota12 (FIG. 1). Hundreds of traits can be subject to quorum-sensing control in a given bacterial species. In addition to the above autoinduction loop, quorum-sensing circuits frequently harbour several feedback and feedforward regulatory loops that fine tune the response by, for example, altering input–output range and dynamics, reducing noise and committing the cells to the individual or group lifestyle programme17–21. Quorum-sensing circuits can intersect with global regulators (such as the alternative sigma factor RpoN, the RNA-binding proteins Hfq and CsrA and the nucleoid protein Fis) to further refine the control of quorum-sensing-dependent gene expression22–24.
Our current understanding of quorum-sensing mechanisms stems primarily from studying traditional well-mixed pure laboratory cultures. These studies have provided foundational knowledge of the molecular mechanisms underlying quorum sensing in different bacteria. However, bacteria often exist in mixtures of species as well as under non-ideal conditions in which fluctuations occur. Moreover, bacteria form structured surface-bound communities called biofilms25,26. Therefore, in addition to discoveries of new quorum-sensing systems, recent research efforts have focused on defining how quorum sensing plays out in realistic bacterial habitats. In this Review, we concentrate on recent advances in the understanding of autoinducer production and detection under spatially structured and/or fluctuating conditions that mimic natural bacterial niches such as in heterogeneous 3D biofilms, in the presence of fluid flow and within eukaryotic hosts where pathogens encounter the host microbiota.
Quorum sensing in biofilm communities
Bacteria attach to surfaces and, together, build biofilm communities26,27. We now understand that biofilms are a predominant form of bacterial life on Earth and that these sessile communities are relevant in the environment26, medicine25,28 and industry29,30. Biofilm cells are encased in an extracellular matrix composed of polysaccharides, proteins and extracellular DNA31,32. Unlike well-mixed bacterial cultures in liquid, biofilms are heterogeneous and can rearrange over time, raising questions about nutrient acquisition and diffusion33. Moreover, understanding how quorum sensing occurs within the architectural constraints of biofilms is a key question facing the field.
Effects of fluid flow and surface topography on quorum-sensing signalling.
Bacteria form biofilms on diverse surfaces, including soil, river beds, sewage, deep-sea vents and plant and animal tissues26. Natural environments differ from those traditionally used in the laboratory for investigating biofilms by two key features: the presence of irregular surfaces (for example, on rocks, corrugated pipes, intestinal villi, leaves, teeth, and so on) and the presence of fluid flow34. Recent studies striving to mimic natural scenarios have capitalized on advances in microfluidics technologies that enable precise control over surface topography and fluid flow35 (BOX 1).
Box 1 |. Microfluidics technology to investigate bacterial processes under realistic settings that mimic nature.
In recent years, microbiology has been revolutionized by advances in microfluidics technologies that have enabled precise control over physical and chemical conditions for bacterial growth with an unprecedented level of flexibility and quantification. Such technology has allowed experimentalists to mimic natural microbial habitats in the laboratory. Natural features of microbial habitats, such as shear force and nutrient availability, often exhibit dynamics and can be heterogeneously distributed at microbial length scales. By using microfluidics technology coupled with advanced imaging, scientists have begun to successfully investigate how environmental features influence bacterial processes while nonetheless performing controlled experiments to establish causal mechanisms and draw concrete conclusions that are not confounded by the extreme complexity of natural settings. The use of microfluidics for studies of diverse microbial lifestyles has been reviewed in detail elsewhere35,99,100.
Compared with traditional flow cell systems, in which biofilm formation has been studied, microfluidics promote high-throughput experimentation, enabling parallelization coupled with finer control over physical and chemical conditions, and exploration of the influence of geometries of interest on bacterial colonization, gene expression and fitness. For example, a device used to study biofilm streamers was fabricated using soft lithography so that it had corners, a geometry that is not typical of conventional flow cells. In this geometry, which mimics natural surfaces, biofilms of Pseudomonas aeruginosa and Staphylococcus aureus formed 3D streamers that hindered fluid flow and, ultimately, clogged the device34,74,101. These experiments using flow and geometry, rather than straight chambers, allowed the decades-old view concerning how biofilms clog industrial and medical devices to be overturned. Specifically, it was long assumed that biofilms cause clogging from the outside in (that is, biofilms initiate on the walls and grow inward to the centre of the channel). Rather, this experiment showed that biofilms clog from the inside out (that is, biofilms form at the centre of the channel in the flow and they grow outward to the wall of the channel)34. This finding inspired simple theoretical calculations that showed that clogging from the outside in could not occur on timescales relevant to known processes that are prone to clogging.
The ability to exactly control bacterial population density in microfluidics devices down to very few cells has revealed unexpected dynamics of quorum-sensing processes in small populations and confined environments. Another benefit of microfluidics is the ability to segregate bacterial populations using hydrogels or nanoslits while maintaining chemical communication between the isolated populations. This approach is providing insights into the role of spatial heterogeneity during quorum sensing, competition and cooperation in bacterial biofilms40,72.
Despite advances made possible by microfluidics, it is noteworthy that the use of this technology in microbiology is still in its early days and suffers from some limitations: because the fluid volumes are minute, typically less than a microlitre, collection of samples for downstream analyses such as transcriptomics is often difficult; most microfluidics devices are 2D, with few exceptions, and thus do not yet accurately represent natural bacterial habitats; and the range of scales that can be studied in microfluidics devices remains small and is subject to laminar flow, whereas biofilms in nature can develop macroscopic structures and certainly experience turbulent flow. Nonetheless, the use of microfluidics is substantially expanding the scope of possible investigations of bacterial processes that are affected by flow and topography such as quorum sensing and biofilm formation. Microfluidics technology promises to deliver a more comprehensive understanding of bacterial processes in nature.
Bacteria exhibit distinct biofilm formation behaviours with respect to their quorum-sensing states. As examples, Pseudomonas aeruginosa forms biofilms at high cell density (HCD) in response to autoinducer accumulation and detection, whereas Vibrio cholerae and Staphylococcus aureus form biofilms at low cell density (LCD), and autoinducer accumulation and detection repress biofilm formation5,6 (FIG. 1). Irrespective of whether quorum-sensing regulation of biofilm formation is positive or negative, one common theme that has emerged is that the amount of bacterial biomass required to initiate quorum sensing in a particular bacterial population increases with increasing fluid flow rate36–40. Specifically, fluid flow removes autoinducers by advection and, thus, a higher cell density is required to achieve a quorum under flow than in well-mixed liquid cultures. One counter-intuitive result from new studies in this area is that, in bacterial species such as V. cholerae and S. aureus (FIG. 1 ) in which quorum sensing represses biofilm formation, increased biofilm formation occurs under flow compared with under non-flow conditions40 (FIG. 2a,b). Autoinducer removal by flow relieves repression, promoting increased biofilm formation relative to biofilms formed on surfaces lacking flow. Nonetheless, once thick biofilms are established, quorum sensing is activated in the cells residing at the base and interior of the biofilms, presumably because those cells are shielded from autoinducer advection by the neighbouring cells and the deposited extracellular matrix (FIG. 2a,b). Because externally residing cells experience a different flow regime from internally residing cells, cells in distinct regions of the biofilm enact discrete quorum-sensing-controlled gene expression programmes40. Thus, the flow environment drives spatial fate decisions, which enables genetically identical bacteria that exist in close proximity to nonetheless undertake distinct biological functions. We discuss heterogeneity in more depth in the next section, but we note that flow, surface topography and quorum-sensing heterogeneity frequently go hand in hand.
Fig. 2 |. Fluid flow and surface topography influence quorum-sensing dynamics.
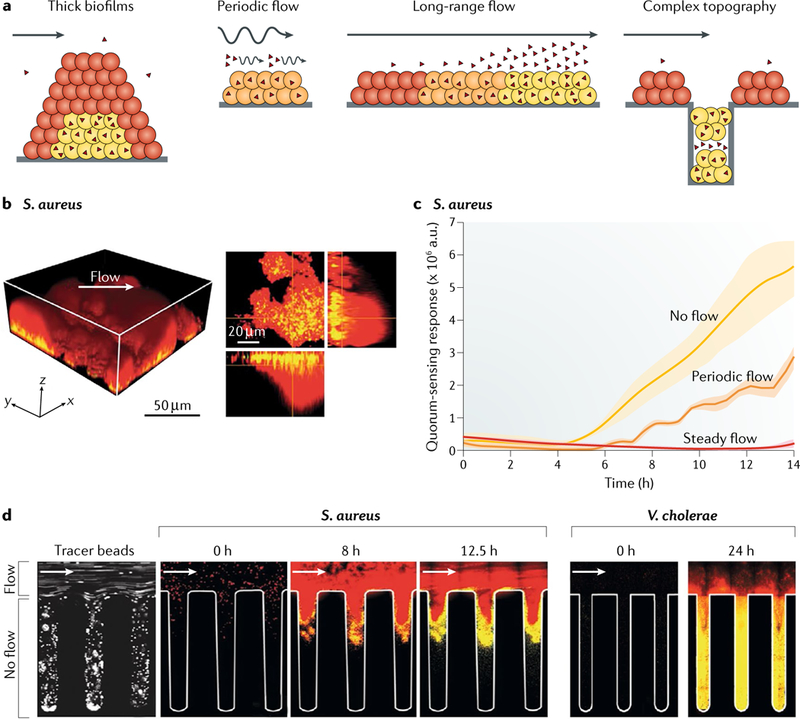
a | Bacterial populations can exhibit heterogeneous quorum-sensing activation patterns under different flow and topography regimes, ranging from quorum-sensing-off cells (red throughout the figure) to partially quorum-sensing-on cells (orange throughout the figure) and fully quorum-sensing-on cells (yellow throughout the figure). Flow (straight arrows for continuous flow and curvy arrows for periodic flow; arrows are pointing in the direction of flow throughout the figure) can wash away endogenously produced autoinducers unless the cells are shielded in a thick biofilm or in crypt-like niches. b | Quorum sensing is activated within thick biofilms of Staphylococcus aureus grown in a microfluidics channel (see Supplementary Movie 1). The left panel shows a 3D view and the right panel shows single optical sections of the x–y plane, 10 μm above the surface–biofilm interface, with z projections shown to the right (x–z plane) and below (y–z plane). The white arrow shows the flow direction. c | Under steady flow, the normalized quorum-sensing output is low in S. aureus compared with no-flow conditions during which autoinducers can accumulate and drive increased quorum-sensing output. Periodic flow leads to quorum-sensing responses that fluctuate between on and off and thus a stepwise increase in quorum-sensing output. d | In the left panel, fluorescent tracer beads flow into a corrugated microfluidics channel with crypt-like cavities, which are shielded from the surface flow and thus trap the beads. Similarly, S. aureus (middle) and Vibrio cholerae (right) growing in the cavities are shielded from flow and, thus, autoinducers can accumulate and turn on quorum sensing (see Supplementary Movie 2). a.u., arbitrary unit. Adapted with permission from REF40, Springer Nature Limited.
Flow, while ubiquitous in living systems, need not be constant. Intermittent flow, which involves transitions between flow and no-flow conditions, or flow and reduced-flow conditions, is common, for example, during rain, intestinal digestion and urination. Under intermittent flow regimes, bacteria in biofilms can fluctuate between two modes: quorum-sensing-on when flow stops and quorum-sensing-off when flow commences, which as described above, track with autoinducer accumulation and advection, respectively40 (FIG. 2c). Evidence of such quorum-sensing transitions comes from analyses of GFP output from the quorum-sensing-activated P3 promoter of S. aureus (Fig. 1c). Over the growth of the biofilm, this quorum-sensing reporter exhibited step-like increases in expression when S. aureus cells experienced periodic flow (FIG. 2c). By contrast, a linear increase in reporter output occurred without flow, and total repression of the reporter occurred under steady flow (FIG. 2c). Thus, intermittent flow can lead to non-uniform quorum-sensing gene expression over time (FIG. 2a). Further studies are required to more comprehensively understand the ramifications of fluctuating flow conditions on quorum sensing, especially in clinical and industrial settings.
In addition to fluid flow, surface topography also influences quorum-sensing dynamics, and as mentioned, often flow and topographical constraints are connected. We provide a few examples here. When bacteria live under flow conditions in a confined geometry, such as in an industrial pipe or in plant phloem, the length of the confined space determines the precise spatial activation of quorum sensing. Experiments using long micro-fluidics channels with physiologically relevant length scales (~0.3 m) showed that quorum sensing was locally repressed near the channel inlet owing to flow-mediated advection of autoinducers, but quorum sensing was highly activated near the outlet where autoinducers, made by cells along the length of the channel, had accumulated40. Thus, in such a regime, quorum-sensing-controlled processes are not carried out uniformly along the length of the confinement. Consistent with this idea, in a long channel, P. aeruginosa exhibited individual behaviours such as motility upstream and quorum-sensing-regulated group behaviours including biofilm formation downstream41. Another study42 also provided insight into how the topography of the growth substrate influences quorum sensing. Using a synthetic cystic fibrosis sputum medium that mimics the cystic fibrosis lung environment with respect to physicochemical properties including viscosity, the authors found that surface topography dictates the spatial range over which successful quorum-sensing signalling can occur. Specifically, biofilm clusters with ~2,000 autoinducer-producing P. aeruginosa cells failed to communicate with other biofilm clusters, whereas communities with >5,000 cells engaged in quorum-sensing signalling with neighbouring clusters that were located hundreds of micrometres away. This observation suggests that, in a viscous environment in which autoinducers are diffusion limited, a higher concentration of autoinducer is required for inter-community communication in P. aeruginosa biofilms.
Another case in which flow and topography combine to drive non-uniform bacterial behaviour involves S. aureus biofilms grown in microfluidics chambers with crevices that mimic intestinal crypts. On the surface outside of the crevices, the bacteria experienced constant flow, and autoinducers were washed away, leading to the repression of quorum sensing40 (FIG. 2a,d). However, bacteria that had colonized the spaces inside the crevices experienced little to no flow and, therefore, those cells transitioned into the quorum-sensing-on mode in response to autoinducer accumulation (FIG. 2a,d). Such localized activation of quorum-sensing signalling facilitated by the coupling of topographical and flow features could increase bacterial colonization of particular niches. Indeed, S. aureus activates the quorum-sensing-dependent production of enterotoxin B only inside of intestinal crypts43,44. The effect of the enterotoxin is to increase the crypt depth. Thus, the very product that quorum-sensing controls is used to rearchitect the space, enabling the cells to escape to a new, shielded niche that more successfully buffers the quorum-sensing programme from flow-mediated perturbation. Similarly, V cholerae activates quorum sensing inside of crevices but not outside of them (FIG. 2d). Specifically, monitoring of a target gene regulated by the quorum-sensing master HCD transcription factor HapR (FIG. 1a) showed that it was expressed inside of crevices where autoinducers accumulated and were detected but not outside of the crevices where flow prevented auto-inducer accumulation40. Perhaps bacteria exploit flow conditions to enable isogenic cells residing in neighbouring but environmentally distinct regions to execute unique quorum-sensing-directed programmes. Presumably, these fine-tuned programmes provide fitness advantages in different locations and/or at different times in the host during infections.
Heterogeneity in quorum sensing.
In contrast to the traditional idea that quorum sensing promotes the synchronous expression of target genes across a bacterial population, recent studies suggest that quorum-sensing-dependent processes can be stochastic: a sub-population of cells can exhibit the quorum-sensing-on mode, whereas the remaining population is in the quorum-sensing-off mode45–50. In most cases, the molecular mechanisms underlying heterogeneity are not yet defined. Although in its early days, this avenue of exploration could lead to increased understanding of how bacteria deploy quorum sensing in natural niches.
Phenotypic heterogeneity exists in the early stages of quorum-sensing-controlled biofilm development in Pseudomonas putida. When the P. putida community is at the microcolony stage, only a subpopulation of cells produces autoinducers45. Curiously, the autoinducer-producing cells do not induce neighbouring isogenic cells to make autoinducers and, therefore, the canonical autoinduction loop is not engaged (FIG. 3a). The authors of this study noted that quorum sensing in P putida activates production of biosurfactants called putisolvins. Stochastic production of putisolvins, which adhere to the surface of the producer cells, caused those cells to disperse, removing them from the community. This feature underpins why neighbouring nonproducer cells did not launch their quorum-sensing cascades and, moreover, had the consequence of delaying overall quorum-sensing induction in the young biofilm. However, in mature biofilms, autoinducers are produced by the entire population, and quorum-sensing signalling becomes homogeneous. The consequence is population-wide production of putisolvins, which leads to the sudden collapse of the biofilm and en masse dispersal of the cells. It is not understood how the transition from dispersal to cross-induction occurs in the population. Another example of quorum-sensing heterogeneity exists in P aeruginosa. When P. aeruginosa cells were confined in small volumes, which enabled the local accumulation of quorum-sensing signals, the major quorum-sensing receptor, LasR (FIG. 1b), activated a target gene–gfp reporter fusion construct when as few as one to three cells were present; however, not all cells in the confined area expressed gfp, suggesting that quorum-sensing initiation was heterogeneous within a clonal population51. Similar observations have been made in Pseudomonas syringae and Xanthomonas campestris46. Furthermore, genetic heterogeneity can occur when quorum-sensing mutants arise in bacterial populations52,53 (discussed in the next section).
Fig. 3 |. Heterogeneity in quorum sensing.
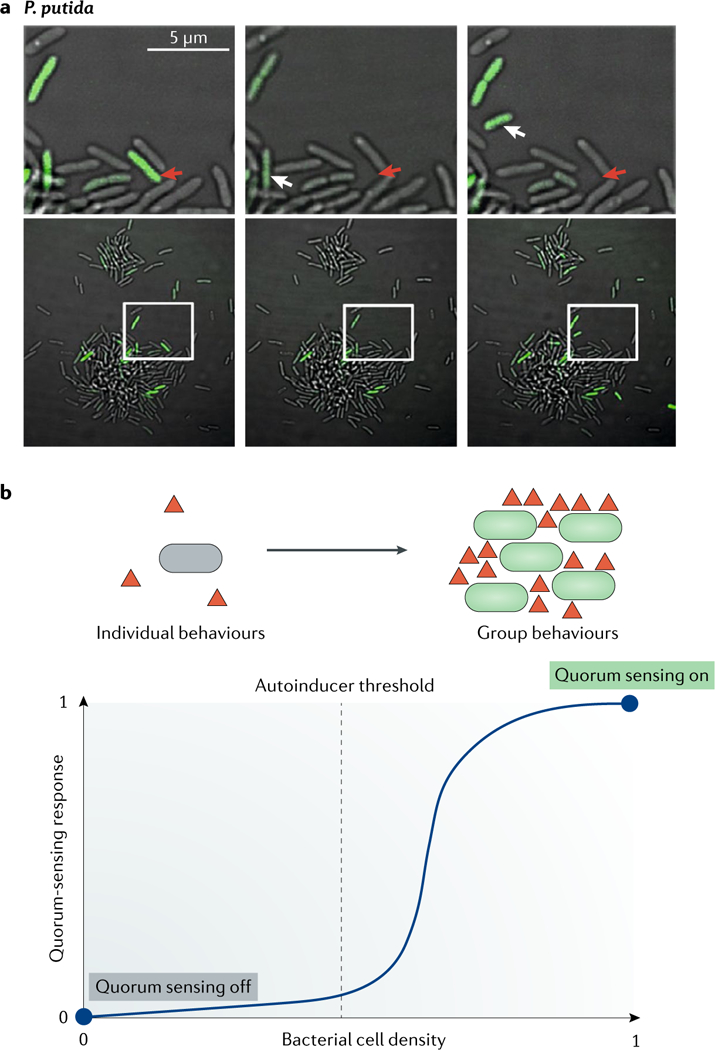
a | Pseudomonas putida can exhibit heterogeneous quorum-sensing responses, in particular, during the early stages of biofilm growth. Only some cells in growing microcolonies produce GFP from a plasmid carrying a quorum-sensing-dependent reporter fusion (lasB–gfp) and the autoinducer receptor. The construct thus reports on individual cell autoinducer production and autoinducer response. Thus, quorum-sensing-regulated putisolvin production occurs only in a subpopulation of cells, and those cells subsequently disperse from the clusters. The upper panel shows a close-up view of the region outlined in the lower panel, and green shows GFP production. The red arrows indicate a cell that leaves the microcolony (top far left; cell absent in middle and right top panels), and the white arrows indicate a cell that moves to the periphery of the microcolony. b | Such heterogeneity can be explained through the concept of quorum sensing as a bistable response function58,60. The dashed line indicates the autoinducer threshold level. The curve shows the quorum-sensing response to different autoinducer (triangles) levels. To achieve bistability, autoinducer production is downregulated in cells that detect it below the threshold value and upregulated in cells that detect it above this threshold. At low cell density, the system is fixed in quorum-sensing-off mode (stable fixed point at 0), and the bacteria exhibit individual behaviours. At high cell density, the system is fixed in quorum-sensing-on mode (stable fixed point at 1), and the bacteria exhibit group behaviours. At intermediate levels (unstable fixed point), transitions between quorum-sensing-on or quorum-sensing-off modes are driven by fluctuations in autoinducer concentration. Part a is reproduced from REF45, CC-BY-4.0.
An emerging theme in this realm is that quorum-sensing heterogeneity is a feature associated with the LCD state of bacterial populations47–51. It is under this condition, when few cells are producing and/or responding to autoinducers, that the population experiences high noise, which, as in other regulatory systems, promotes heterogeneity. Current models to explain phenotypic heterogeneity in autoinducer production typically assume a bistable54 gene regulation programme in which autoinducer synthesis is repressed upon detection of autoinducer concentrations below a critical threshold and autoinducer production is activated when the signal molecules are detected above the critical threshold55–58 (FIG. 3b). In these models, noise at the level of expression of the autoinducer synthase gene causes phenotypic heterogeneity.
Maintaining phenotypic heterogeneity in HCD quorum-sensing populations could allow the bacteria to undertake bet-hedging59 strategies in which, simultaneously, some cells in the population perform individual behaviours whereas others engage in collective activities. Consistent with this idea, modelling efforts suggest that bacteria alter their immediate surroundings by secreting autoinducers and that they respond to their local environment by increasing the rate of autoinducer production, setting up a positive feedback loop that ensures that autoinducers are produced by only a regional subpopulation of cells60. This model proposes that heterogeneity arises from a balance between the fitness advantage gained by the nonproducers who avoid the costly production of autoinducers and the persistence of producers that engage in the autoinduction loop, ultimately allowing separate subpopulations to coexist. Follow-up experimental studies are necessary to test these theoretical models.
The public goods dilemma, cooperation and cheating.
Bacteria frequently secrete extracellular biomolecules to capture nutrients from the environment, hydrolyse solid food sources and construct biofilm communities. Some secreted substances can be used by nonproducing cells and are thus considered to be public goods61. Production of metabolically expensive public goods is often under the control of quorum sensing such that each cell in the population produces its share of the goods, and the community thrives through communal use of the goods62–64. However, exploitation of these goods by nonproducers must be prevented or at least minimized, as conflict over public goods reduces population fitness, and the severity of this conflict appears greater in biofilms than in planktonic populations62,65. Thus, a public goods dilemma exists (FIG. 4a). Several processes, including spatial structure and social policing of the community, are thought to promote cooperation and prevent cheating in bacterial systems that depend on public goods62–70. For example, studies of V cholerae biofilms formed on the solid substrate chitin showed that the public goods dilemma may be solved in two different ways70 (FIG. 4a). Chitin is a solid polymer that must be processed into soluble oligomers or N-acetylglucosamine monomers to be internalized and used as a nutrient by bacteria71. Bacteria secrete chitin-degrading enzymes called chitinases that convert the solid polymer into soluble, digestible units that can be taken up. However, nonproducers can also consume these soluble goods. In thick biofilms, because diffusion out of the biofilm is slow, biofilm-residing cells can fully consume N-acetylglucosamine monomers. Thus, the public goods are privatized, presumably accruing maximum benefit to the producer cells. Indeed, competition experiments show that chitinase producers have a fitness advantage over nonproducer cells in thick bio-films but not in well-mixed liquid cultures70 (FIG. 4b). Second, in biofilms under fluid flow, soluble products of chitin digestion are washed away (advection) and thereby unavailable to nonproducing cells70 (FIG. 4c). In this case, the producing cells also incur a cost because they do not get to consume all of the released nutritious products. However, the producing cells can successfully consume a fraction of the soluble products before they are lost to the flow, presumably owing to the proximity of the chitinase-producing cell to the products of chitin digestion. At least in laboratory setups, this situation provides a competitive advantage to chitinase producers over nonproducers. Both of these mechanisms, thick biofilms and flow-mediated public goods removal, limit the distance over which public-good-producing cells provide goods to neighbours. Thus, both mechanisms primarily benefit the closest cells, which are presumably kin and therefore also producers.
Fig. 4 |. Quorum sensing and the public goods dilemma.
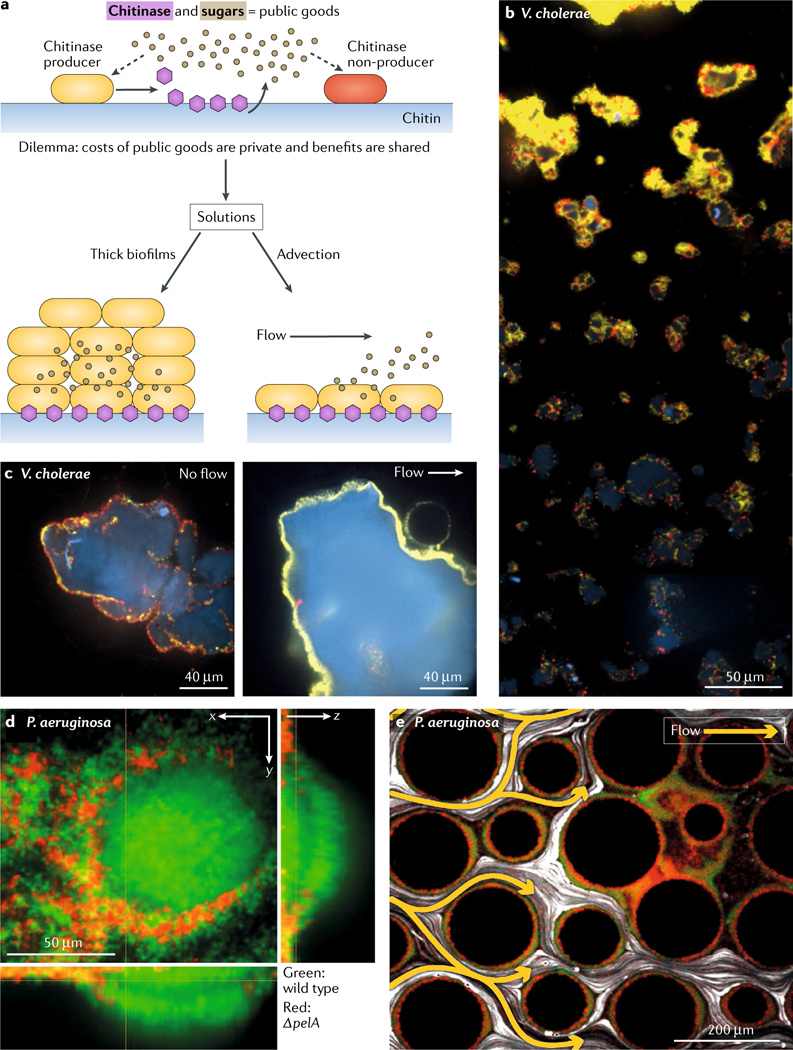
a | Chitin degradation represents a public goods dilemma70. Chitinase producers (yellow in parts a–c) secrete chitinase enzymes (purple hexagons) that degrade the chitin polymer (light blue in parts a–c) into soluble N-acetylglucosamine oligomers (tan circles in part a), which can be imported and catabolized by both chitinase producers and chitinase nonproducers (red in parts a–c). b | In static liquid culture, Vibrio cholerae chitinase producers that compete against chitinase nonproducers on chitin make thick biofilms and outcompete the nonproducers. c | Similarly, chitinase nonproducers fail to accumulate biomass when soluble products of chitin degradation are washed away by flow (right), whereas they can exploit the public good in the absence of flow (left). d | Matrix production confers a competitive advantage to wild-type Pseudomonas aeruginosa (green) over a ΔpelA non-matrix producing mutant (red) in biofilms under flow conditions. The images show that wild-type bacteria contribute to the main biofilm biomass, while the ΔpelA mutant cells are excluded. e | The Pel-deficient P. aeruginosa mutant (red) can occupy locations protected from flow owing to local clogging by wild-type P. aeruginosa (green) biofilm streamers. White lines indicate bead tracks monitoring flow; yellow arrows highlight flow trajectories. Parts a–c are adapted with permission from REF.70, Elsevier. Parts d–e are adapted from REF.72, CC-BY-4.0.
Curiously, under some conditions, spatial structure can also allow wild-type bacteria and cheaters to coexist72. In P aeruginosa, for example, quorum sensing is required for biofilm formation, as the Las quorum-sensing system controls production of the Pel exopolysaccharide, which is a necessary matrix component73. When wild-type P. aeruginosa cells were grown with matrix-nonproducing pelA mutants under flow in straight chambers, matrix producers outcompeted nonproducers because the latter were removed by shear forces72 (FIG. 4d). However, in geometries with topography, wild-type P aeruginosa biofilms deform into 3D streamers34,74 that partially clog flow channels, which locally reduces flow speed. In this situation, the mutant and the wild-type strains could coexist because the non-matrix producers were not washed away and could proliferate using nutrients that slowly entered into the low-flow areas from other areas of the chamber72 (FIG. 4e). Thus, wild-type bacteria modify the dynamics of the environment by forming quorum-sensing-dependent biofilm streamers and thereby allow pelA mutants to survive and coexist.
Autoinducers can also function as public goods and, thus, are prone to exploitation by nonproducing cheaters: P. aeruginosa lasI mutants that lack the LasI5 synthase that produces the autoinducer 3-oxo-dodecanoyl-homoserine lactone (3OC12-HSL) (FIG. 1b) can, nonetheless, respond to 3OC12-HSL produced by wild-type bacteria and, in so doing, outcompete the wild-type population in well-mixed cultures75. When grown on adenosine as the carbon source, however, lasI mutants exhibit a growth defect in monoculture because the LasR receptor that detects and initiates the response to 3OC12-HSL is required to activate expression of nuh, which encodes an intracellular nucleoside hydrolase that is essential for adenosine catabolism. By contrast, in mixed cultures, lasI mutants have a higher relative fitness than wild-type bacteria, as they use the 3OC12-HSL supplied by the wild-type bacteria to activate their cytoplasmic LasR receptor and induce nuh expression, enabling them to consume adenosine. Thus, lasI mutants act as social cheaters. However, increasing the viscosity of the growth medium, which has the consequence of reducing autoinducer diffusion, makes the autoinducers less accessible to nonproducer cells and leads to reduced social cheating by the lasI mutant75.
Another strategy that prevents cheating in situations in which public goods are at stake is social policing66. Mechanistically, quorum-sensing-dependent production of a released public good is tied to the concomitant production of an intracellular private good that is not shared with the community. Studies in P. aeruginosa demonstrate that lasR mutants act as social cheaters when grown with wild-type P aeruginosa on a substrate such as casein that requires the secretion of quorum-sensing-dependent extracellular proteases52. However, such cheating is prevented when the growth medium includes adenosine that, as mentioned above, requires the function of the LasR-activated intracellular enzyme Nuh to metabolize adenosine53. In this context, unlike the lasI mutants, lasR mutants cannot act as cheaters, as both LasR and Nuh are cytoplasmic components and thus private goods that cannot be shared. Similar results have been obtained with the P. aeruginosa RhlR-RhlI system, which controls cyanide production and immunity from cyanide toxicity76,77. Specifically, although cyanide production is costly, wild-type P aeruginosa cyanide-producers are resistant to cyanide, whereas lasR mutant cells are vulnerable because lasR mutants fail to activate expression of the rhlR and rhlI genes encoding the RhlR-RhlI quorum-sensing system77 (FIG. 1b). Thus, lasR cheaters are punished by the cooperating cyanide-producing cells, thereby stabilizing the population. In summary, quorum-sensing-driven co-regulation of two metabolic enzymes, one that serves as a public good and one that serves as a private good, can provide an incentive that reduces social cheating and prevents the collapse of the wild-type population.
Quorum sensing in eukaryotic hosts
Inside hosts, bacteria often exist in mixed-species communities and, therefore, quorum sensing by one species can influence and be influenced by quorum sensing or other activities carried out by neighbouring species. Furthermore, host processes such as the immune response can also influence bacterial quorum sensing and vice versa. Here, we review some recent advances concerning the function of quorum sensing in mixed bacterial communities and how host processes affect quorum-sensing signal transduction during infection.
Quorum sensing and the host-associated microbiota.
Eukaryotes harbour diverse microbial ecosystems that make up the microbiota78,79. Examples include bacterial communities on mammalian skin, in the oral cavity and in the gut. It is estimated that 1013 bacteria reside in the human gut80. Increasing evidence suggests that inter-species and inter-kingdom chemical communication shape the species composition of the gut microbiota81–83. For example, a study investigating the effect of quorum sensing on the gut microbiota following antibiotic-induced dysbiosis in mice reported that AI-2-mediated inter-species communication (FIG. 1) promotes the expansion of Firmicutes over Bacteroidetes84 (FIG. 5a). Specifically, streptomycin treatment of mice caused near complete elimination of Firmicutes, which caused Bacteroidetes to increase in relative abundance and, in so doing, decreased the diversity of the gut microbiota. However, when an engineered Escherichia coli strain overproducing AI-2, a widely used inter-species quorum-sensing autoinducer, was introduced following the antibiotic treatment, a substantial increase in Firmicutes abundance occurred. Interestingly, a greater proportion of Firmicutes species than Bacteroidetes species encode AI-2 quorum-sensing systems, suggesting that, at least in this context, AI-2-mediated communication selectively promotes the growth of AI-2-producing populations.
Fig. 5 |. Quorum sensing and the host microbiota.
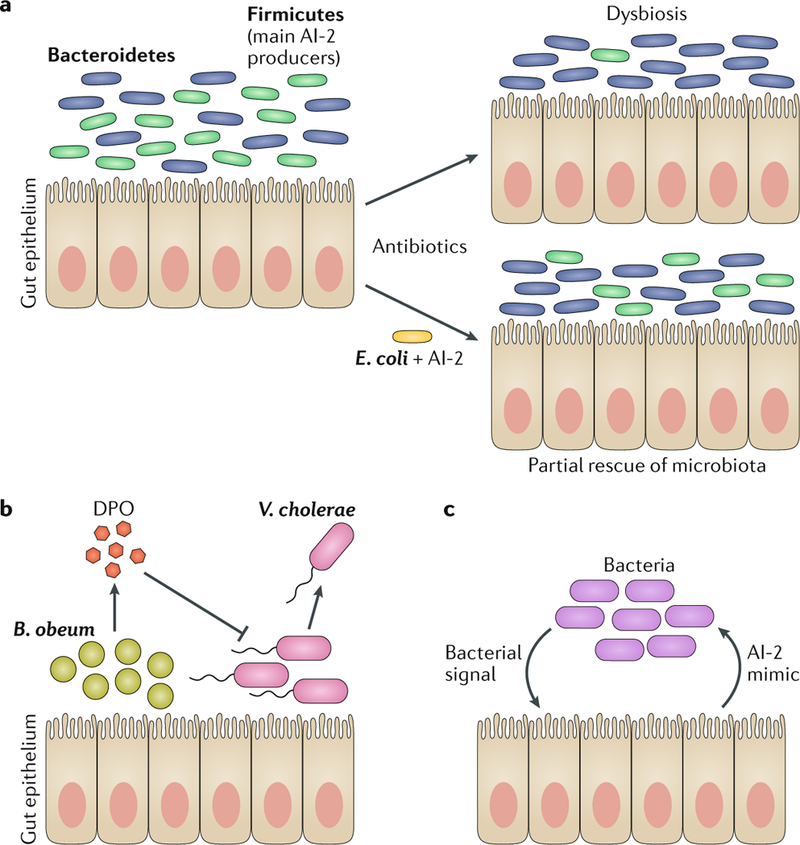
a | Quorum sensing can control the species composition of the gut microbiota. Disruption of the normal microbiota composition by antibiotic treatment leads to a reduction in AI-2-producing bacteria (and AI-2 levels), resulting in dysbiosis. In this instance, members of the Firmicutes phylum (green) are the primary AI-2 producers, and their abundance decreases following antibiotic treatment, while members of the Bacteroidetes phylum (blue) increase in abundance. However, artificially increasing AI-2 levels by introduction of an AI-2 producer (in this case, an engineered strain of Escherichia coli) partially restores the normal gut microbiota composition84. b | The gut commensal bacterium Blautia obeum can produce the DPO autoinducer, and DPO is speculated to inhibit colonization by Vibrio cholerae, possibly providing protection against this pathogen83,85. c | Communication can also occur between mammalian epithelial cells and bacteria. Epithelial cells release an AI-2 mimic in response to bacteria, and this AI-2 mimic is detected by bacterial colonizers via their AI-2 quorum-sensing receptors. Thus, the AI-2 mimic modulates bacterial quorum sensing87.
In the context of pathogenicity, the VqmA-DPO quorum-sensing system of V. cholerae (FIG. 1a) that, at HCD, represses biofilm formation and toxin production and promotes dispersal is postulated to have a key role in V. cholerae transitions between the human host and the aquatic environment85. Surprisingly, in a mouse model of infection, the presence of the gut commensal Blautia obeum limits the severity of V. cholerae infection83. Protection requires that the V. cholerae pathogen possesses VqmA. This finding, coupled with the discovery of DPO as the autoinducer that activates VqmA, suggests that bacteria in the gut microbiota produce DPO, which V. cholerae cells detect via VqmA, and this causes the V. cholerae cells to prematurely disperse from the host (FIG. 5b). However, we note that this interpretation requires experimental validation. In a similar vein, probiotic Bacillus subtilis produces lipopeptides known as fengycins that antagonize the Agr quorum-sensing receptor AgrC (FIG. 1c). The fengycins thereby repress production of Agr-controlled virulence factors and suppress the ability of S. aureus to colonize mice86.
Inter-kingdom communication between bacteria and hosts could also influence colonization. For example, mammalian epithelial cells, but not haematopoietic cells, release an AI-2 mimic in response to interaction with bacteria87 (FIG. 5c). The structure of the AI-2 mimic has not yet been identified. The AI-2 mimic can activate quorum-sensing-dependent regulons in bacteria including in enteric pathogens such as Salmonella enterica subsp. enterica serovar Typhimurium and V. cholerae. Presumably, exploiting the relatively generic inter-species AI-2 autoinducer as the mimic, rather than a species-specific autoinducer, enables the host to interact with a large range of bacterial species present in the gut. Although this remains speculative, perhaps this AI-2 mimic drives wide-spread global changes in gene expression in the gut microbiota.
Host factors influence bacterial quorum sensing.
Microbiota communities that reside on epithelial surfaces are influenced by host factors including innate immune components, mucus composition and diet81,82.Notably, eukaryotes can produce enzymes that quench bacterial quorum-sensing-mediated communication. For example, freshwater hydra88 produce an oxidoreductase that reduces the autoinducer 3OC12-HSL, which is made by the main bacterial colonizer of hydra, Curvibacter sp., to 3OHC12-HSL89 (FIG. 6a). The host-modified 3OHC12-HSL molecule promotes host colonization by Curvibacter sp. However, only the original 3OC12-HSL autoinducer activates a crucial Curvibacter sp. phenotypic switch in which flagellar genes, motility and host dispersal are induced. Thus, hydra, by manipulating the autoinducer, capture Curvibacter sp. Other examples of eukaryotic quorum-quenching mechanisms include production of halogenated furanones by the red algae Delisea pulchra that function as quorum-sensing receptor antagonists90 and mammalian-produced paraoxonases91,92 that function as lactonases that hydrolyse and thereby inactivate homoserine lactone autoinducers (FIG. 6b).
Fig. 6 |. Host factors influence quorum sensing.
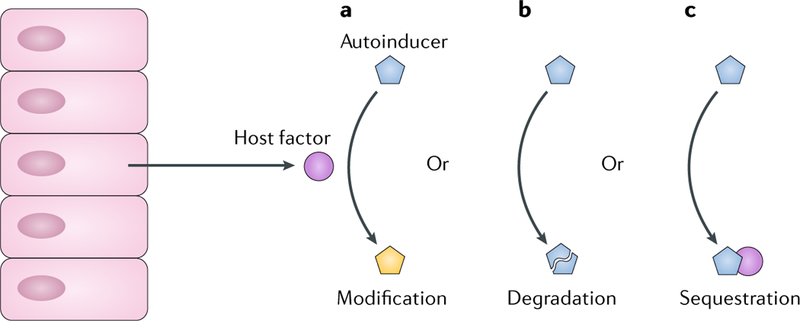
Host-derived enzymes and other proteins can modulate bacterial quorum sensing by altering autoinducer levels through processes including autoinducer modification89 (part a), autoinducer degradation92 (part b) or autoinducer sequestration96,97 (part c). These processes, because they inactivate autoinducers (parts a, b) or make autoinducers unavailable (part c), induce the LCD quorum-sensing state, causing bacteria to enact individual behaviours.
Host factors can also affect quorum-sensing signalling and thereby modulate the outcome of pathogen invasion. For example, chronic wounds are commonly infected with both S. aureus and P. aeruginosa. Curiously, whereas P. aeruginosa readily eliminates S. aureus when co-cultured under standard laboratory conditions, the two species coexist and exhibit synergistic tolerance to antibiotics in chronic wounds93. Quorum-sensing-dependent P aeruginosa exoproducts such as the LasA protease94 and redox active phenazines95 inhibit S. aureus growth in the laboratory co-culture model. However, in the chronic wound, host factors, such as serum albumin, sequester the 3OC12-HSL autoinducer and thereby suppress P aeruginosa LasR-dependent quorum-sensing behaviours96 (FIG. 6c). The consequence is that P. aeruginosa becomes incapable of killing S. aureus, and the two species coexist. Similarly, human apolipoprotein B binds to the S. aureus oligopeptide autoinducer and prevents its interaction with its partner receptor, thus inhibiting S. aureus quorum-sensing-mediated behaviours97 (FIGS 1c, 6c). Likewise, there is evidence from transcriptomic studies that during human infection by P. aeruginosa, quorum sensing is suppressed relative to that in laboratory setups in vitro98. These studies, although preliminary, suggest that host factors have a marked influence on bacterial quorum sensing.
Conclusions
Quorum-sensing-mediated control of bacterial behaviours has a central role in bacterial lifestyle transitions. Environmental features ranging from fluid flow and surface topography to host immune responses and the presence or absence of other bacterial species influence bacterial communication. It is imperative to investigate quorum sensing under complex conditions such as those in biofilms and in the context of the microbiota within eukaryotic hosts for the field to learn how cell-cell communication functions under realistic circumstances and to understand how quorum-sensing-controlled behaviours are deployed outside the laboratory setting. Exciting studies are taking place along these lines, and beyond yielding basic insight, they promise to propel the field forward in efforts to impede quorum sensing in harmful bacteria and promote quorum sensing in beneficial bacteria.
Supplementary Material
Acknowledgements
This work was supported by the Howard Hughes Medical Institute, US National Institutes of Health (NIH) grant 5R37GM065859 and National Science Foundation grant MCB-1713731 (to B.L.B.), as well as by a Life Science Research Foundation Postdoctoral Fellowship through the Gordon and Betty Moore Foundation through grant GBMF2 550.06 and NIH grant 1K99GM129424–01 to S.M.
Footnotes
Competing interests
The authors declare no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information is available for this paper at https://doi.org/10.1038/s41579–019-0186–5.
Phenotypic heterogeneity
Nongenetic variations in traits between individual cells in an isogenic population.
Bet hedging
A strategy that enables diversification of phenotypes within a population with the consequence of reducing the overall risk of death of all the cells in the population. Thus, bet hedging increases fitness under temporally varying conditions.
Social policing
A strategy in which quorum-sensing bacteria link production of costly private goods to production of public goods to punish nonproducers and thereby prevent emergence of social cheaters.
Dysbiosis
A microbial imbalance on or inside a host in which the normal microbiota is disrupted, for example, after treatment with antibiotics.
References
- 1.Bassler BL & Losick R Bacterially speaking. Cell 125, 237–246 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Aguilar C, Vlamakis H, Losick R & Kolter R Thinking about Bacillus subtilis as a multicellular organism. Curr. Opin. Microbiol. 10, 638–643 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engebrecht J, Nealson K & Silverman M Bacterial bioluminescence: Isolation and genetic analysis of functions from Vibrio fischeri. Cell 32, 773–781 (1983). [DOI] [PubMed] [Google Scholar]
- 4.Bassler BL, Wright M & Silverman M Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 13, 273–286 (1994). [DOI] [PubMed] [Google Scholar]
- 5.de Kievit TR & Iglewski BH Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 68, 4839–4849 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bronesky D et al. Staphylococcus aureus RNAIII and its regulon link quorum sensing, stress responses, metabolic adaptation, and regulation of virulence gene expression. Annu. Rev. Microbiol. 70, 299–316 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Barnard AML et al. Quorum sensing, virulence and secondary metabolite production in plant soft-rotting bacteria. Phil. Trans. R. Soc. B Biol. Sci. 362, 1165–1183 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kleerebezem M, Quadri LE, Kuipers OP & de Vos WM Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol. Microbiol. 24, 895–904 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Okada M et al. Structure of the Bacillus Subtilis quorum-sensing peptide pheromone ComX. Nat. Chem. Biol. 1, 23–24 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Miller MB & Bassler BL Quorum sensing in bacteria. Annu. Rev. Microbiol. 55, 165–199 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Hammer BK & Bassler BL Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50, 101–114 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Papenfort K & Bassler BL Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 14, 576–588 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rutherford ST & Bassler BL Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2, a012427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henke JM & Bassler BL Bacterial social engagements. Trends Cell Biol. 14, 648–656 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Novick RP & Geisinger E Quorum sensing in staphylococci. Annu. Rev. Genet. 42, 541–564 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Waters CM & Bassler BL Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21, 319–346 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Svenningsen SL, Tu KC & Bassler BL Gene dosage compensation calibrates four regulatory RNAs to control Vibrio cholerae quorum sensing. EMBO J. 28, 429–439 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lenz DH et al. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118, 69–82 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Ng W-L & Bassler BL Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 43, 197–222 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng L et al. A Qrr noncoding RNA deploys four different regulatory mechanisms to optimize quorum-sensing dynamics. Cell 160, 228–240 (2015).This work shows how regulatory RNAs rely on multiple molecular mechanisms to precisely and dynamically control quorum-sensing behaviours in Vibrio spp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rutherford ST, Kessel, Van JC, Shao Y & Bassler BL. AphA and LuxR / HapR reciprocally control quorum sensing in vibrios. Genes Dev. 25, 397–408 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lenz DH, Miller MB, Zhu J, Kulkarni RV & Bassler BL CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae. Mol. Microbiol. 58, 1186–1202 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Lenz DH & Bassler BL The small nucleoid protein Fis is involved in Vibrio cholerae quorum sensing. Mol. Microbiol. 63, 859–871 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Thompson LS, Webb JS, Rice SA & Kjelleberg S The alternative sigma factor RpoN regulates the quorum sensing gene rhlI in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 220, 187–195 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Donlan RM & Costerton JW Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15, 167–193 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolter R & Greenberg EP The superficial life of microbes. Nature 441, 300–302 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Tolker-Nielsen T Biofilm development. Microbiol. Spectr. 10.1128/microbiolspec.MB-0001-2014 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Rybtke M, Hultqvist LD, Givskov M & Tolker-Nielsen T Pseudomonas aeruginosa biofilm infections: community structure, antimicrobial tolerance and immune response. J. Mol. Biol. 427, 3628–3645 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Escudié R, Cresson R, Delgenés JP & Bernet N Control of start-up and operation of anaerobic biofilm reactors: an overview of 15 years of research. Water Res. 45, 1–10 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Pintelon TRR, Picioreanu C, van Loosdrecht MCM & Johns ML The effect of biofilm permeability on bio-clogging of porous media. Biotechnol. Bioeng. 109, 1031–1042 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Branda SS, Vik Á, Friedman L & Kolter R Biofilms: the matrix revisited. Trends Microbiol. 13, 20–26 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Flemming H & Wingender J The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Stewart PS & Franklin MJ Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 6, 199–210 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Drescher K, Shen Y, Bassler BL & Stone HA Biofilm streamers cause catastrophic disruption of flow with consequences for environmental and medical systems. Proc. Natl Acad. Sci. USA 110, 4345–4350 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rusconi R, Garren M & Stocker R Microfluidics expanding the frontiers of microbial ecology. Annu. Rev. Biophys. 43, 65–91 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim MK et al. Surface-attached molecules control Staphylococcus aureus quorum sensing and biofilm development. Nat. Microbiol. 2, 17080 (2017).This study develops methods to coat surfaces with pro-quorum-sensing and anti-quorum-sensing compounds to manipulate bacterial group behaviours. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kirisits MJ et al. Influence of the hydrodynamic environment on quorum sensing in Pseudomonas aeruginosa biofilms. J. Bacteriol. 189, 8357–8360 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyer A et al. Dynamics of AHL mediated quorum sensing under flow and non-flow conditions. Phys. Biol. 9, 026007 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Purevdorj B, Costerton JW & Stoodley P Influence of hydrodynamics and cell signaling on the structure and behavior of Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 68, 4457–4464 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim MK, Ingremeau F, Zhao A, Bassler BL & Stone HA Local and global consequences of flow on bacterial quorum sensing. Nat. Microbiol. 1, 15005 (2016).This study shows that fluid flow and surface topography influence quorum-sensing outputs in V. cholerae and S. aureus in non-intuitive ways. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siryaporn A, Kim MK, Shen Y, Stone HA & Gitai Z Colonization, competition, and dispersal of pathogens in fluid flow networks. Curr. Biol. 25, 1201–1207 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Darch SE et al. Spatial determinants of quorum signaling in a Pseudomonas aeruginosa infection model. Proc. Natl Acad. Sci. USA 115, 4779–4784 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benjamin MA, Lu J, Donnelly G, Dureja P & McKay DM Changes in murine jejunal morphology evoked by the bacterial superantigen Staphylococcus aureus enterotoxin B are mediated by CD4+T cells. Infect. Immun. 66, 2193–2199 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bronner S, Monteil H & Prévost G Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol. Rev. 28, 183–200 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Cárcamo-Oyarce G, Lumjiaktase P, Kümmerli R & Eberl L Quorum sensing triggers the stochastic escape of individual cells from Pseudomonas putida biofilms. Nat. Commun. 6, 5945 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pradhan BB & Chatterjee S Reversible non-genetic phenotypic heterogeneity in bacterial quorum sensing. Mol. Microbiol. 92, 557–569 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Plener L et al. The phosphorylation flow of the Vibrio harveyi quorum-sensing cascade determines levels of phenotypic heterogeneity in the population. J. Bacteriol. 197, 1747–1756 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grote J, Krysciak D & Streit WR Phenotypic heterogeneity, a phenomenon that may explain why quorum sensing does not always result in truly homogenous cell behavior. Appl. Environ. Microbiol. 81, 5280–5289 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anetzberger C, Pirch T & Jung K Heterogeneity in quorum sensing-regulated bioluminescence of Vibrio harveyi. Mol. Microbiol. 73, 267–277 (2009). [DOI] [PubMed] [Google Scholar]
- 50.Pérez PD & Hagen SJ Heterogeneous response to a quorum-sensing signal in the luminescence of individual Vibrio fischeri. PLOS ONE 5, e15473 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boedicker JQ, Vincent ME & Ismagilov RF Microfluidic confinement of single cells of bacteria in small volumes initiates high-density behavior of quorum sensing and growth and reveals its variability. Angew. Chemie Int. Ed. Engl. 48, 5908–5911 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sandoz KM, Mitzimberg SM & Schuster M Social cheating in Pseudomonas aeruginosa quorum sensing. Proc. Natl Acad. Sci. USA 104, 15876–15881 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dandekar AA, Chugani S & Greenberg EP Bacterial quorum sensing and metabolic incentives to cooperate. Science 338, 264–267 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Veening J-W, Smits WK & Kuipers OP Bistability, epigenetics, and bet-hedging in bacteria. Annu. Rev. Microbiol. 62, 193–210 (2008). [DOI] [PubMed] [Google Scholar]
- 55.Fujimoto K & Sawai S A design principle of group-level decision making in cell populations. PLOS Comput. Biol. 9, e1003110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dockery JD & Keener JP A mathematical model for quorum sensing in Pseudomonas aeruginosa. Bull. Math. Biol. 63, 95–116 (2001). [DOI] [PubMed] [Google Scholar]
- 57.Goryachev AB et al. Transition to quorum sensing in an agrobacterium population: a stochastic model. PLOS Comput. Biol. 1, e37 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pérez-Velázquez J, Gölgeli M & García-Contreras R Mathematical modelling of bacterial quorum sensing: a review. Bull. Math. Biol. 78, 1585–1639 (2016). [DOI] [PubMed] [Google Scholar]
- 59.Veening J-W et al. Bet-hedging and epigenetic inheritance in bacterial cell development. Proc. Natl Acad. Sci. USA 105, 4393–4398 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bauer M, Knebel J, Lechner M, Pickl P & Frey E Ecological feedback in quorum-sensing microbial populations can induce heterogeneous production of autoinducers. eLife 6, e25773 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Griffin AS, West SA & Buckling A Cooperation and competition in pathogenic bacteria. Nature 430, 2–5 (2004). [DOI] [PubMed] [Google Scholar]
- 62.Popat R et al. Quorum-sensing and cheating in bacterial biofilms. Proc. Biol. Sci. 279, 4765–4771 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cordero OX, Ventouras L-A, DeLong EF & Polz MF Public good dynamics drive evolution of iron acquisition strategies in natural bacterioplankton populations. Proc. Natl Acad. Sci. USA 109, 20059–20064 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bruger E & Waters C Sharing the sandbox: evolutionary mechanisms that maintain bacterial cooperation. FJ000Res. 4, 1504 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Darch SE, West SA, Winzer K & Diggle SP Density-dependent fitness benefits in quorum-sensing bacterial populations. Proc. Natl Acad. Sci. USA 109, 8259–8263 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.West SA, Griffin AS, Gardner A & Diggle SP Social evolution theory for microorganisms. Nat. Rev. Microbiol. 4, 597–607 (2006). [DOI] [PubMed] [Google Scholar]
- 67.Oshri RD, Zrihen KS, Shner I, Bendori SO & Eldar A Selection for increased quorum-sensing cooperation in Pseudomonas aeruginosa through the shut-down of a drug resistance pump. ISME J. 12, 2458–2469 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bruger EL & Waters CM Bacterial quorum sensing stabilizes cooperation by optimizing growth strategies. Appl. Environ. Microbiol. 82, 6498–6506 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bruger EL & Waters M Maximizing growth yield and dispersal via quorum sensing promotes cooperation in vibrio bacteria. Appl. Environ. Microbiol. 84, 1–13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Drescher K, Nadell CD, Stone HA, Wingreen NS & Bassler BL Solutions to the public goods dilemma in bacterial biofilms. Curr. Biol. 24, 50–55 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Meibom KL et al. The Vibrio cholerae chitin utilization program. Proc. Natl Acad. Sci. USA 101, 2524–2529 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nadell CD, Ricaurte D, Yan J, Drescher K & Bassler BL Flow environment and matrix structure interact to determine spatial competition in Pseudomonas aeruginosa biofilms. eLife 6, e21855 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sakuragi Y & Kolter R Quorum-sensing regulation of the biofilm matrix genes (pel) of Pseudomonas aeruginosa. J. Bacteriol. 189, 5383–5386 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rusconi R, Lecuyer S, Autrusson N, Guglielmini L & Stone HA Secondary flow as a mechanism for the formation of biofilm streamers. Biophys. J. 100, 1392–1399 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mund A, Diggle SP & Harrison F The fitness of Pseudomonas aeruginosa quorum sensing signal cheats is influenced by the diffusivity of the environment. mBio 8, e00353–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wagner VE, Bushnell D, Passador L, Brooks AI & Iglewski BH. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185, 2080–2095 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang M, Schaefer AL, Dandekar AA & Greenberg EP Quorum sensing and policing of Pseudomonas aeruginosa social cheaters. Proc. Natl Acad. Sci. USA 112, 2187–2191 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McFall-Ngai M et al. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl Acad. Sci. USA 110, 3229–3236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cho I & Blaser MJ The human microbiome: at the interface of health and disease. Nat. Rev. Genet. 13, 260–270 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sender R, Fuchs S & Milo R Revised estimates for the number of human and bacteria cells in the body. PLOS Biol. 14, e1002533 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ley RE et al. Evolution of mammals and their gut microbes. Science 320, 1647–1651 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sommer F & Backhed F The gut microbiota-masters of host development and physiology. Nat. Rev. Microbiol. 11, 227–238 (2013). [DOI] [PubMed] [Google Scholar]
- 83.Hsiao A et al. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature 515, 423–426 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thompson J, Oliveira R, Djukovic A, Ubeda C & Xavier K Manipulation of the quorum sensing signal AI-2 affects the antibiotic-treated gut microbiota. Cell Rep. 10, 1861–1871 (2015). [DOI] [PubMed] [Google Scholar]
- 85.Papenfort K et al. A Vibrio cholerae autoinducer-receptor pair that controls biofilm formation. Nat. Chem. Biol. 13, 551–557 (2017).This manuscript reports a novel Vibrio spp. autoinducer, called DPO, and its receptor, called VqmR, and that the DPO-VqmR complex controls biofilm formation via the action of a regulatory RNA called VqmR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Piewngam P et al. Pathogen elimination by probiotic Bacillus via signalling interference. Nature 562, 532–537 (2018).This study reports that probiotic Bacillus spp. produce fengycin lipopeptides that antagonize S. aureus quorum sensing and inhibit S. aureus colonization of mice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ismail AS, Valastyan JS & Bassler BLA Host-produced autoinducer-2 mimic activates bacterial quorum sensing. Cell Host Microbe 19, 470–480 (2016).This manuscript reports that bacteria respond to an AI-2 mimic produced by human epithelial cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bosch TCG Cnidarian-microbe interactions and the origin of innate immunity in metazoans. Annu. Rev. Microbiol. 67, 499–518 (2013). [DOI] [PubMed] [Google Scholar]
- 89.Pietschke C et al. Host modification of a bacterial quorum-sensing signal induces a phenotypic switch in bacterial symbionts. Proc. Natl Acad. Sci. USA 114, E8488–E8497 (2017).This study demonstrates that Hydra, a genus of metazoans, modify the autoinducers of the primary bacterial colonizer of hydra, Curvibacter spp., and in so doing alter their quorum-sensing behaviours. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Harder T, Campbell AH, Egan S & Steinberg PD Chemical mediation of ternary interactions between marine holobionts and their environment as exemplified by the red alga Delisea pulchra. J. Chem. Ecol. 38, 442–450 (2012). [DOI] [PubMed] [Google Scholar]
- 91.Chun CK, Ozer EA, Welsh MJ, Zabner J & Greenberg EP Inactivation of a Pseudomonas aeruginosa quorum-sensing signal by human airway epithelia. Proc. Natl Acad. Sci. USA 101, 3587–3590 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stoltz DA et al. Drosophila are protected from Pseudomonas aeruginosa lethality by transgenic expression of paraoxonase-1. J. Clin. Invest. 118, 3123–3131 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.DeLeon S et al. Synergistic interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an In vitro wound model. Infect. Immun. 82, 4718–4728 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kessler E, Safrin M, Olson JC & Ohman DE Secreted LasA of Pseudomonas aeruginosa is a staphylolytic protease. J. Biol. Chem. 268, 7503–7508 (1993). [PubMed] [Google Scholar]
- 95.Dietrich LEP, Price-Whelan A, Petersen A, Whiteley M & Newman DK The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol. Microbiol. 61, 1308–1321 (2006). [DOI] [PubMed] [Google Scholar]
- 96.Smith AC et al. Albumin inhibits Pseudomonas aeruginosa quorum sensing and alters polymicrobial interactions. Infect. Immun. 85, e00116–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Peterson MM et al. Apolipoprotein B is an innate barrier against invasive Staphylococcus aureus infection. Cell Host Microbe 4, 555–566 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cornforth DM et al. Pseudomonas aeruginosa transcriptome during human infection. Proc. Natl Acad. Sci. USA 115, E5125–E5134 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yawata Y, Nguyen J, Stocker R & Rusconi R Microfluidic studies of biofilm formation in dynamic environments. J. Bacteriol. 198, 2589–2595 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Norman TM, Lord ND, Paulsson J & Losick R Stochastic switching of cell fate in microbes. Annu. Rev. Microbiol. 69, 381–403 (2015). [DOI] [PubMed] [Google Scholar]
- 101.Kevin Kim M, Drescher K, Shun Pak O, Bassler BL & Stone HA Filaments in curved streamlines: rapid formation of Staphylococcus aureus biofilm streamers. New J. Phys. 16, 065024 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Eickhoff MJ & Bassler BL SnapShot: bacterial quorum sensing. Cell 174, 1328–1328.e1 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


