Abstract
Mitochondria, a nearly ubiquitous feature of eukaryotes, are derived from an ancient symbiosis. Despite billions of years of cooperative coevolution—in what is arguably the most important mutualism in the history of life—the persistence of mitochondrial genomes also creates conditions for genetic conflict with the nucleus. Because mitochondrial genomes are present in numerous copies per cell, they are subject to both within- and among-organism levels of selection. Accordingly, “selfish” genotypes that increase their own proliferation can rise to high frequencies even if they decrease organismal fitness. It has been argued that uniparental (often maternal) inheritance of cytoplasmic genomes evolved to curtail such selfish replication by minimizing within-individual variation and hence, within-individual selection. However, uniparental inheritance creates conditions for cytonuclear conflict over sex determination and sex ratio, as well as conditions for sexual antagonism when mitochondrial variants increase transmission by enhancing maternal fitness but have the side-effect of being harmful to males (i.e., “mother’s curse”). Here, we review recent advances in understanding selfish replication and sexual antagonism in the evolution of mitochondrial genomes and the mechanisms that suppress selfish interactions, drawing parallels and contrasts with other organelles (plastids) and bacterial endosymbionts that arose more recently. Although cytonuclear conflict is widespread across eukaryotes, it can be cryptic due to nuclear suppression, highly variable, and lineage-specific, reflecting the diverse biology of eukaryotes and the varying architectures of their cytoplasmic genomes.
Introduction
Mitochondria are a key feature of eukaryotic life, and their persistence represents one of the most enduring biological unions, as mitochondria are the descendants of ancient bacterial endosymbionts that were acquired prior to the last eukaryotic common ancestor [1]. Remarkably, mitochondria still maintain independent genomes in nearly all eukaryotic lineages. Coevolution between mitochondrial (mt) and nuclear genomes is generally considered to be mutualistic, because precise interactions between mt- and nuclear-encoded gene products are necessary for fundamental cellular functions [2]. The importance and near ubiquity of mitochondria has led to hypotheses that implicate mitonuclear coevolution in nearly all aspects of eukaryotic biology - from speciation to the origins of sex (Box 1) [3-11].
Box 1. The role of mitonuclear conflict in key eukaryotic transitions.
The acquisition of the mt endosymbiont and the resulting establishment of mitonuclear interactions has been suggested as a driving force for the evolution of nearly every feature that distinguishes eukaryotes from prokaryotes. Organismal complexity, genome complexity, speciation processes, sexual reproduction, the presence of two sexes, sexual selection, apoptosis, aging, the sequestered germline, the nuclear membrane, and introns have all been argued to have arisen at least in part due to mitochondria [3-11].
Many of these hypotheses assume mutualistic coevolution between mt and nuclear genomes, but mitonuclear conflict could provide insights into these key transitions as well. For example, genomic and organismal complexity has been hypothesized to have arisen due to increased energy supplied by the mt endosymbiont [2, 11]. However, mechanisms of selfish replication in mt genomes may have selected for novel genes and functions in nuclear genomes to combat selfishness, which would have also increased complexity. Reproductive isolation between populations has been attributed to breaking up coadapted mitonuclear complexes in offspring, resulting in reduced hybrid fitness [5, 40, 192]. However, in angiosperms, CMS is often manifested in hybrids, not because mutualistically coadapted complexes are disrupted, but because selfish mt variants are placed against a naïve nuclear background that lacks the proper counteradaptations. Finally, in a recent hypothesis on sexual selection, male ornaments were proposed to act as a signal of how well mt and nuclear genomes are coadapted to one another, resulting in greater OXPHOS efficiencies and more attractive ornaments with greater coadaptation [193]. Another view might be that male ornaments can only be maintained when selfish mt genomes are brought under control by nuclear responses.
While such hypotheses for mitonuclear conflict in key eukaryotic features are speculative, researchers investigating the implications of mitonuclear interactions would do well to consider the role of antagonistic, as well as mutualistic, coevolution between the genomes.
Despite their longstanding symbiosis, the presence of independently replicating mt genomes in eukaryotes creates the opportunity for selfish conflict between mt and nuclear genes. The term conflict can be used in different ways, but here we mean more than simple genetic incompatibilities that arise due to disruption of coadapted mitonuclear genotypes during hybridization and genetic admixture [5, 12-14]. Rather, we are referring to cases in which opposing selection pressures act on mt and nuclear genomes such that their evolutionary “interests” are at odds with each other [15]. For example, mutations that benefit the transmission of mt genomes but reduce the transmission of nuclear genomes create mitonuclear conflict. Such opposing selection pressures were likely much more prominent early in eukaryotic evolution when mitochondria had larger genomes and greater independence from the nucleus. However, in the billions of years since the origin of mitochondria, the control of nearly all mt functions has been transferred to the nucleus in a process of cytonuclear integration that is sometimes termed “domestication” [16-19].
Mt domestication likely eliminated many sources of mitonuclear conflict, but not all of them. The only lineages that have completely lost their mt genomes lack electron transport systems and survive as parasites on the cellular energy of other eukaryotes [1, 20, 21]. There is active research into the classic question of whether mt genomes have been maintained due to adaptive processes or functional constraints [17, 22-25]. Regardless of the answer, the persistence of mt genomes over billions of years has made mitonuclear conflict an enduring feature of eukaryotic evolution.
Although there has been longstanding interest in mitonuclear conflict and similar selfish interactions involving plastids (e.g., chloroplasts) and other endosymbionts [15, 26-32], recent work across diverse eukaryotes has employed genomic and modeling-based approaches to yield new insights into the molecular mechanisms of mitonuclear conflict and how such conflict is ameliorated. Here, we review this literature under the two broad themes of selfish mt replication and sexual antagonism, while drawing parallels and contrasts with cytonuclear conflict in plastids and endosymbionts.
Mitochondrial genomes as “selfish little circles”
“…evolution might be determined not by which plasmon form furthered the organism in which it occurred, but by which furthered itself most in competition with other plasmon factors…” Grun 1976 [29]
Selection among and within individuals
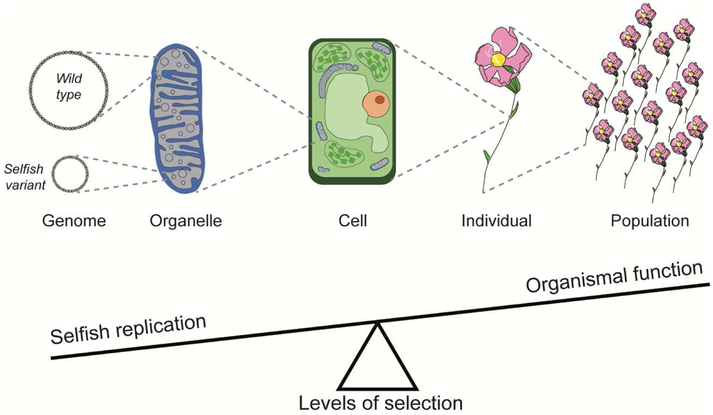
An individual eukaryote may possess up to trillions of cells, each of which may contain hundreds of mitochondria, which in turn may each contain many mt genomes [33, 34] (Fig. 1). Even unicellular eukaryotes that possess only one or a handful of mitochondria can have many copies of the mt genome [35, 36]. Due to their multicopy nature, selection on mt genomes operates both among and within individuals [37-40] (Fig. 1). Consequently, mt genomes that harm the organism can outcompete beneficial mt genomes and rise to high frequencies within an individual because the strength of selection on individual mt genomes for cellular function can be weak compared to the strength of selection to replicate. Haig [30] referred to this as the “tragedy of the cytoplasmic commons” because mt replication benefits the individual genome, while efficient cellular function benefits all the genomic inhabitants of the cell and is therefore a “public good”. In such situations, the self-interest of individuals (mt genomes) can result in the decay of public goods (cellular function) [41].
Fig. 1. The balance of selection among different levels of organization.
Mitochondrial genomes that have a replication advantage can spread within an organism even if they confer deleterious effects on organismal function because selection on mitochondrial genomes acts both among and within individuals.
Mt and nuclear selection pressures will generally remain aligned unless there is variation among mt genomes for selection to act on. Organisms that possess a heterogeneous population of mt genomes are termed heteroplasmic and are commonly observed. For example, even with low sensitivity detection methods, heteroplasmic females possessing mt genomes that differ by ~3% have been found to occur naturally at frequencies of up to 12% in Drosophila individuals [42]. Most individuals will be heteroplasmic at some point in their life cycle as a result of mutations in individual mt genome copies [43-45]. Mt mutation rates are elevated compared to nuclear rates in some eukaryotic lineages [46-49], and all mt variants that are fixed between populations or species began as heteroplasmic variants within an individual. Because multiple copies of the mt genome are also transmitted across generations (albeit often in reduced or “bottlenecked” numbers – see “Nuclear responses to suppress selfish replicators” below), offspring can inherit heteroplasmies [50]. In such heterogenous populations of mt genomes, copies that over-replicate have the potential to spread even if they have harmful effects on organismal fitness. The frequencies of these selfish replicators will be shaped by a balance between the different processes of mutation, multilevel selection, and drift (e.g., due to transmission bottlenecks).
Several elegant studies have experimentally manipulated organismal population sizes to examine selfish mt transmission and illustrate the conflicting levels of selection in systems such as yeast and nematodes [51-53]. Such experiments reduce the efficacy of selection among individuals by lowering the effective population size. However, reducing the number of organisms has no effect on the number of mt genome copies within each organism, so selection remains strong among mt genome copies within individuals. These studies have generally confirmed that reducing organismal selection “tilts the balance” toward the proliferation of selfish mt elements (Fig. 1).
Mechanisms of selfish replication
Several examples and mechanisms of mt selfish replication have been identified or hypothesized. Here, we describe six different processes that may lead to over-replication of otherwise deleterious mt genomes (Table 1). First, sequence variants in mt genomes can directly reduce genome replication time [54]. Smaller mt genomes arising via deletions are expected to replicate faster and spread within an individual, even though deletions of key genes can have major effects on oxidative phosphorylation (OXPHOS) activity and organismal fitness. Mt variants in Caenorhabditis nematodes with large deletions in NAD5 may serve as an example, as they have a replication advantage but compromise organismal energetics [54-56]. Sequence variants that preferentially recruit replication machinery may also accelerate DNA replication and increase in frequency within a variable mt population. For example, petite mutants in baker’s yeast (Saccharomyces cerevisiae) arise spontaneously at high frequencies, often via large deletions in the mt genome. Therefore, growth is slowed due to compromised respiration, resulting in a small or “petite” colony phenotype [57]. Petite mutants also have duplicated ORI sequences in their mt genomes, which are thought to promote replication and may act as an origin of mt replication (but see [58]), giving them an additional replication advantage [59]. Enhanced or expanded origins of replication may also explain evidence in the fruit fly Drosophila melanogaster that larger mt genomes can have a within-individual replication advantage [60]. In Drosophila where a foreign mt genome has been introduced to compete with the native variant, it was shown that the noncoding region of the genome (containing the origin of replication) governs selfish transmission [40]. In facultative aerobes such as yeast, selfish mt variants with defective oxidative phosphorylation can rise to fixation (homoplasmy) when the yeast are in an anaerobic phase, because countervailing selection against them at the individual level is weak. However, in species such as nematodes where respiration is critical, selfish mt variants will rise in frequency (heteroplasmy), but this process will be counterbalanced by selection on organismal function.
Table 1.
Six possible mechanisms of selfish mt replication and six possible nuclear responses.
| Mechanism | Details | Example taxa | Citations |
|---|---|---|---|
| Selfish replication | |||
| 1. Faster replication | Via deletions and preferential recruitment of replication machinery | Humans, nematodes, drosophila, yeast | [40, 51, 53-55, 57, 59, 190, 191] |
| 2. Moral hazard hypothesis | Poor organelles result in increased DNA replication | Nematodes (and simulations) | [67-70] |
| 3. Selfish organelle growth and division | Genome variation causes replication of organelles, not DNA | Plastids in evening primrose? | [72] |
| 4. Epigenetic tagging | Replication and transcription may be mutually exclusive | Mammals, possibly in copepods | [73-75] |
| 5. Organelle inheritance bias | Variants causing organelles to move into germline | None, relevant mechanisms proposed in bivalve molluscs | [76] |
| 6. Warfare | Organelles producing toxins to disrupt competing organelles | None, relevant observations in heteroplasmic mice | [79] |
| Nuclear responses | |||
| 1. Gene transfer | Functional movement of genes from the mt to the nuclear genome | Mt replication – all eukaryotes; dNTP levels – yeast | [1, 81] |
| 2. Organelle selection | Selective mitophagy aided by mt fusion/fission cycles | Characterized most extensively in mammals | [6, 70, 82-84, 88] |
| 3. Cell selection | Mt function in germline selective sieves and apoptosis | Characterized most extensively in mammals | [90-92, 94] |
| 4. Sexual recombination | Sex may counteract the parasitic nature of selfish replication | None; modeling studies contrarily suggest sex evolved after mt control | [4, 32, 98-100] |
| 5. Germline bottlenecks | Reduces heteroplasmy | Mammals; parallels in young endosymbionts | [101, 102, 106, 107] |
| 6. Uniparental inheritance | Reduces heteroplasmy | Fig. 2 | [97, 108] |
The multicopy nature of mt genomes can sometimes be extreme. In one remarkable example, the amoeba endosymbiont Perkinsela has a single mitochondrion and a mt genome that encodes only six proteins; nevertheless, this endosymbiont has so many mitochondrial genome copies that it has more DNA in its single mitochondrion than is present in the nuclear genomes of either the endosymbiont or the host [61, 62]. This raises questions as to whether highly multicopy mt genomes, which are common across eukaryotes, are beneficial for organismal fitness or may have arisen at least in part due to selfish replication.
Functional transfer of mt genes to the nucleus may buffer the effects of mt gene deletions. In this scenario, an intracellular gene transfer duplication occurs such that a mitochondrial gene is present and functional in both the mt genome and the nuclear genome (e.g., [63]). Because the mt and nuclear gene copies are functionally redundant, one copy will likely be lost. Several evolutionary hypotheses have been presented detailing why mt genes are transferred to the nucleus [19, 64, 65]. One possibility is that mt variants in which the transferred gene is deleted might have a replication advantage over intact mt genomes. As such, variants with deletions would spread and preferentially result in the loss of the mt copy and retention of the nuclear copy, completing nuclear gene transfer [66]. Under this hypothesized mechanism, the inherent selfishness of multicopy organelle genomes may actually have acted to strip them of much of the very genetic content that allowed them to function in a selfish fashion to begin with (e.g., mt replication machinery).
A second potential mechanism of selfish replication involves variants that compromise mt function but do not result directly in shorter DNA replication times. One hypothesized cellular response to under-performing mitochondria that harbor such defective genome copies is to increase the replication of all mtDNA within such organelles, including both functional and defective variant copies [15, 67]. The result over time would be a mitochondrial moral hazard that rewards “bad behavior” and produces a large population of poor-performing mt genomes that hitchhike along with upregulated mt DNA replication in their compartments. In other words, the very response of the cell to the defective genome results in the spread of the defective genome. The overall result is a snowball effect in which cellular function continues to decline as more dysfunctional genomes accumulate. Modeling studies provide some evidence for this, with implications for why mt associated diseases caused by heteroplasmy are manifested later in life [68-70]. There is also empirical evidence that a mutant mt haplotype in C. elegans that has a 3.1 kb deletion may proliferate by exploiting the regulatory machinery that maintains the necessary number of wild-type genome copies [67]. These observations provide an alternative (or complementary) interpretation for the role of deletions in selfish over-replication.
Importantly, the potential for spread of selfish mt genome copies within a cell may be shaped by the fact that mitochondria undergo cycles of fusion and fission [71]. In the absence of mt fusion, a selfish mt variant that spreads to fixation within a mitochondrion would still require that mitochondrion to proliferate within the cell in order to spread further. Modeling work suggests that fusion/fission makes it possible for defective mtDNA copies to spread among mitochondria and outcompete wildtype copies within each organelle [70]. This is in contrast to the apparent lack of regular fusion in many plastids and younger endosymbionts, which traps genome copies inside a single organelle or bacterium.
A third mechanism of selfish replication may take place at the organelle level rather than the genome level. Some of the earliest ideas about intracellular competition among organelles were developed based on observations of plastids rather than mitochondria [27, 29]. In particular, in the evening primrose (Oenothera) certain plastid genotypes are known to consistently outcompete others when they are present in a heteroplasmic state. In one recent study, specific variants in plastid-encoded genes were implicated in a mechanism for preferential replication of these “strong” plastids [72]. The authors did not detect differences in genome copy number across plastid types, suggesting that rates of DNA replication were not a primary driver. Instead, variation in the plastid-encoded subunit of the acetyl-CoA carboxylase (accD) was shown to correlate with plastid competitive success and underlie variation in fatty acid production, which the authors hypothesize is rate-determining for plastid membrane growth and plastid division. Although there is no evidence that rapid plastid division is inherently harmful to organismal fitness, these strong plastids are able to spread even when introduced onto genetically incompatible nuclear backgrounds, resulting in loss of photosynthetic function. Thus, they clearly create opportunities for conflicting levels of selection.
Three other mechanisms of selfish replication that have received less attention include mt epigenetic modifications, organelle-level inheritance bias, and “warfare” among organelles. It has been proposed in mammals that mtDNA is epigenetically tagged. Some evidence suggests that tagging can direct the fate of a genome copy to either replication or transcription of mtDNA, but not both [73, 74]. In cytoplasmic hybrids of the marine copepod Tigriopus californicus, mt transcription and DNA copy number are negatively correlated [75], possibly supporting this mechanistic tradeoff. In this context, mt genome copies that are preferentially tagged for replication will spread, but will not be transcribed, and therefore are less likely to be screened for functionality. Meiotic drive, in which certain alleles subvert Mendel’s laws of inheritance and are preferentially transmitted to the next generation, is generally considered only in the context of heterozygous nuclear genes because mitochondria do not undergo meiosis. However, an analogous process of inheritance bias could exist among organelles if particular mitochondria have the ability to move into germline cells during development. Recent evidence in molluscs provides a possible mechanism as mt-encoded genetic products were suggested to affect microtubule motors [76], and cytoskeletal elements play a role in partitioning mitochondria during cell division [77]. Finally, “warfare” could occur among organelles. Many bacteria are characterized by toxin-antitoxin systems, which themselves can be selfish genetic elements [78]. Endosymbionts could encode similar systems to destroy competing endosymbionts, possibly at the expense of organismal function. To our knowledge, no examples of such systems have been described in organelles, but we would predict them to occur more frequently in young endosymbionts that retain larger genomic repertoires. They may also be possible in some lineages where “non-functional” (and putatively selfish) mtDNA is common.
Interestingly, recent studies in heteroplasmic mice and humans found deleterious effects even though individuals that were homoplasmic for either mt variant showed no such effects (i.e., underdominance) [79, 80]. To put it differently, mt genomes that are seemingly fine in isolation can be deleterious in combination. One possible explanation is that active competition between the variants through one or more of the mechanisms described above causes reduced organismal fitness.
Nuclear responses to suppress selfish replicators
Selfish replication in mt genomes is curtailed by several mechanisms, six of which we outline here (Table 1). First is the transfer of the majority of mt genes and control of mt functions to the nucleus during the billions of years since the endosymbiotic origins of eukaryotes [19]. While nuclear gene transfer is often thought to be adaptive because nuclear genes benefit from sexual recombination while mt genes do not [65], there are also neutral reasons why gene transfer should be asymmetrical between the genomes [64], and this remains an active area of research [22, 23]. Ironically, accordingly to the logic described in the previous section, selfish replication in mt genomes may also predispose genes to nuclear transfer [66]. Regardless of the cause, nuclear gene transfer likely provided the nucleus with a novel set of genes that may have played roles in combating selfish mt replication, while stripping mt genomes of most of their weapons for selfish replication. Perhaps most importantly, the machinery that actually controls mtDNA replication is nuclear-encoded in all eukaryotes [1]. The ability to selfishly replicate is presumably more limited in a genome that does not control its own replication. There is also recent evidence that nuclear control over dNTP levels may act to regulate selfish mt genomes in yeast [81].
Secondly, mitochondria may be “screened” to eliminate poor-performing organelles that possess selfish genomes [6, 82-84]. Mitophagy, which was once thought to be largely random, is now known to selectively eliminate underperforming mitochondria, likely as a form of quality control [85-87]. Fusion/fission cycles also act to segregate underperforming organelles with lowered membrane potential from the rest of the mt population [70, 88], allowing them to be selectively eliminated via mitophagy.
Thirdly, the entire cell can be vetted based on the performance of its mitochondria, which may be especially important during germline development [89]. For example, in a 20-week old human female fetus, ~7 million ovarian follicles are present, but only a few hundred will eventually undergo ovulation [90] – amounting to a very effective potential selective sieve during this process (known as atresia). Importantly, recent studies suggest mitochondrial function is especially important during early primordial germ cell development [91, 92] and may also be under selection during later stages of egg development [93]. The central role of mitochondria in apoptosis across cell types also suggests that cells with poor performing mitochondria will be effectively eliminated [94]. Indeed, the role of mitochondria in apoptosis may have initially evolved as a mechanism to regulate poorly functioning and selfish mitochondria.
Fourthly, one advantage of sexual reproduction may be to counteract the physiological consequences of selfish mt genomes. Eukaryotic sex is often viewed as an adaptation to respond to changing environments or parasites [95, 96], and selfish mt replication can be viewed as a form of parasitism [97]. It was recently proposed that sexual recombination provided the genetic variation for early eukaryotes to respond to mt mutations [4], and a logical extension is that sex allowed a more efficient response to selfish mt replication. Several related hypotheses have suggested a role for mitochondria in the evolution of eukaryotic sexual reproduction [98-100]. Interestingly, one modeling study came to an opposite conclusion regarding selfish mt replication. It suggested that selfish endosymbionts may actually spread under sexual reproduction, leading to the conclusion that selfish mitochondria were brought under control before the evolution of sex [32], not as a result of it.
The final two mechanisms of nuclear suppression both act to limit the amount of mt heteroplasmy and, thus, the opportunity for intracellular competition. Germline bottlenecks in females may serve to reduce variation among mt genome copies. Primordial germ cells in human females undergo a massive bottleneck in the number of mt genomes per cell, dropping to 10 or fewer mitochondria and ~200 or fewer mt genome copies during germ line development [101, 102]. Somatic cells induced to form pluripotent stem cells that mimic primordial germ cells also show a large reduction in mtDNA copy number [103, 104]. The mt genomes that survive this bottleneck then undergo a rapid expansion to ~200,000 copies per mature oocyte [105]. Such bottlenecking reduces heteroplasmy in the mature oocyte and embryo, thus restricting the possibility for intracellular competition early in development. Enforcing small inoculum sizes during transmission of younger endosymbionts may serve the same purpose [106, 107]. It is important to note that any selfish mt genomes that survive such bottlenecks could lead to drastic heteroplasmy after subsequent proliferation of mt genomes.
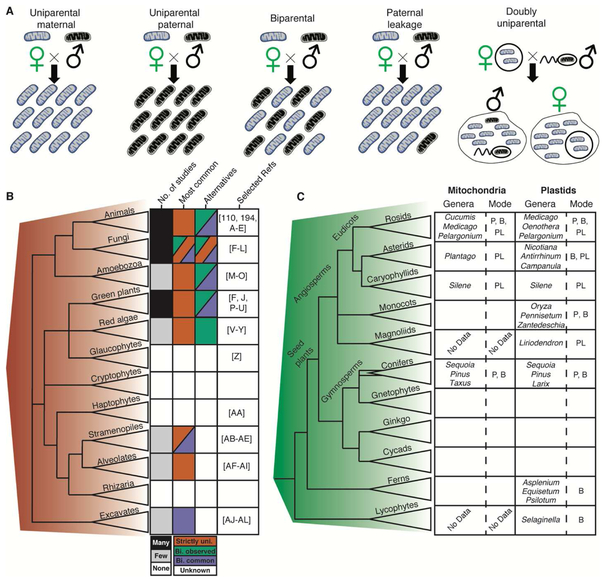
Finally, one of the most widely hypothesized mechanisms to bring selfish mt replication under control is the evolution of uniparental inheritance [27-29, 97]. If mitochondria are transmitted though only a single parent (Fig. 2A), it is much more likely that offspring will receive a homogenous population of mt genomes, reducing the opportunity for intracellular selection. Widespread observations of maternal inheritance in plants and animals have motivated this hypothesis. Uniparental inheritance occurs via many active and often redundant mechanisms that ensure paternal mtDNA is excluded during fertilization or is destroyed shortly thereafter, including prezygotic exclusion/degradation of mitochondria in sperm, prevention of paternal organelles from entering the developing zygote, and degradation of paternal organelles in the zygote [97, 108]. In Drosophila, when a second, deleterious mt genome was introduced to compete with the native genome, the deleterious genome prevailed only when the two genomes were distantly related [40]. This suggests that heteroplasmy arising via mutation within an individual may be relatively harmless compared to the possible divergent heteroplasmies created via biparental inheritance [40]. However, it is important to note that it is an oversimplification to assume that uniparental inheritance of mt genomes is standard. Indeed, for many of the major eukaryotic lineages, there are few or no studies of mt inheritance (Fig. 2B). Moreover, studies of cytoplasmic inheritance in well-sampled groups (e.g., vascular plants) have revealed that it is a strikingly labile trait, with many independent lineages showing evidence of at least episodic biparental inheritance (Fig. 2C). In such taxa, mitochondria may have selfishly escaped the mechanisms that ensure uniparental inheritance. Even in species such as humans, biparental inheritance may be more common than previously appreciated [80, 109, 110].
Fig 2. Variation in mitochondrial inheritance.
(A) Representations of different modes of mitochondrial inheritance, including the doubly uniparental inheritance system in some bivalve molluscs [194, 195]. (B) Summary of studies of mitochondrial inheritance in major eukaryotic groups. (Left) Phylogenetic relationships of major eukaryotic clades as summarized by [1] and [196]. (Right) Grid of four columns (1-4, left to right) displaying information about our knowledge of mitochondrial inheritance in each group. (Col. 1) The number of studies pertaining to mitochondrial inheritance in each group. Few: 1-10 studies; Many: >> 10 studies. (Col. 2) The most common pattern of inheritance in each group based on current literature. Multiple colors per box indicate uncertainty about which pattern is most common. (Col. 3) Alternative patterns observed in the group. Strictly Uni.: Inheritance was found to be strictly uniparental and maternal; Bi. observed: Rare (<1% of individuals) cases of biparental inheritance were observed; Bi. common: biparental inheritance was observed in >1% of individuals. (Col. 4) A non-exhaustive list of references for mitochondrial inheritance in each group. (C) Non-maternal organelle inheritance in vascular plants. (Left) Phylogenetic relationships of major vascular plant clades as summarized by [197]. The double line for conifers represents possible non-monophyly [198]. (Right) Cases of mitochondria and plastid inheritance that depart from strictly maternal transmission including selected examples of genera in which these observations were made. Blank boxes indicate strict maternal inheritance. “No Data” indicates groups in which plastid/mitochondrial transmission in unknown. Modes of inheritance are abbreviated: P, paternal; PL, paternal leakage (similar to Bi. observed in panel B); B, biparental. References used to populate (C) include [27, 97, 117, 197-198, 217, 237-246].
Antagonistic sexual selection due to uniparental inheritance
“In males, cytoplasmic genes in outbreeding species will have no selection on them at all to function properly.” Cosmides and Tooby 1981 [28]
Although uniparental inheritance may have evolved to reduce the spread of “selfish” mt haplotypes, it results in a new arena for conflict between the cytoplasmic and nuclear genomes. Whereas nuclear genes are usually inherited through both sexes, mt genes are often inherited through the maternal lineage (Fig. 2). In these cases, males acquire mitochondria from their mothers, but they do not pass them on to succeeding generations. This uniparental inheritance has two consequences. First, it invokes cytonuclear genetic conflict over sex determination [28, 111] due to active selection on cytoplasmic elements to distort sex determination and sex ratios towards females. Second, because mitochondria are not actively selected to maintain functions in males, variants that reduce male fitness can increase in frequency within populations [28, 112, 113]. We briefly outline these concepts in the next two sections.
Cytonuclear conflict and reproductive manipulation
Mitochondrial variants (as well as plastids and heritable microbes) that increase the number or fitness of females by reducing the number or fitness of males will be selectively favored, and nuclear suppressors of such “rogue” elements will be selected for, resulting in cytonuclear conflict. Among the best examples of this are cytoplasmic male sterility (CMS) in plants [114, 115] and sex ratio distorting cytoplasmic microorganisms in many animals [31, 111].
CMS can result in gynodioecy (i.e., the presence of both hermaphroditic and female individuals) in natural populations of flowering plants and can be revealed through crosses [116]. In these systems, mt genomic variants cause hermaphrodites to become male-sterile (see [114-116] for reviews). At least in some cases, suppression of male function can increase female fitness either through resource reallocation to female functions [117-119], or through improved offspring quality via forced outcrossing [120].
Processes of reproductive manipulation include male-killing (or harming), parthenogenesis, feminization of males, and cytoplasmic incompatibility (CI) – all of which can potentially increase the abundance/fitness of females [31, 111](Fig. 3). All of these processes have been demonstrated in a class of bacterial endosymbionts known as reproductive manipulators – epitomized by Wolbachia, a group of intracellular Alphaproteobacteria found in numerous arthropod and nematode species [31]. Mitochondria may have a more restricted range of mechanisms for reproductive manipulation. They have only been clearly shown to induce male harm/sterility. One recent example from a book louse suggests that certain highly divergent mt variants may cause only daughters to be produced [26]. This could represent a case of mt-induced sex ratio distortion, but the precise mechanism has not been worked out, and there is as yet no conclusive evidence of mitochondrial involvement. To our knowledge, there are no identified cases of mt-mediated parthenogenesis, feminization or CI. Evidence for plastid-mediated reproductive manipulation is even more limited (Fig. 3). Therefore, beyond CMS in plants, clear cases of reproductive manipulation by organelles remain to be established.
Fig. 3. Mechanisms of reproductive manipulation in cytoplasmic genomes.
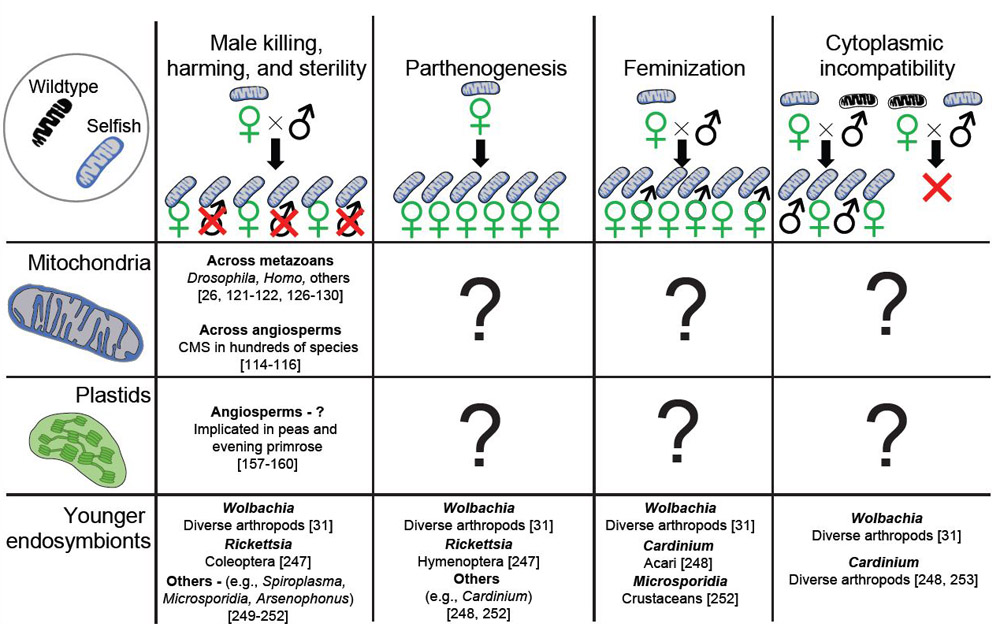
Four mechanisms of reproductive parasitism of arthropods have been described in younger endosymbionts such as Wolbachia: Male killing/harm eliminates or reduces male functions, parthenogenesis results in asexual production of exclusively females, feminization causes males to develop as females, and cytoplasmic incompatibility prevents males with the selfish variant from mating with wildtype females. However, mitochondrial genomes have only been documented to cause male harm/killing, and evidence for plastids causing reproductive manipulation is scarce.
Mother’s curse
Transmitting mt genomes through a single sex or mating type will limit the effects of natural selection to that parent. For example, maternal inheritance results in natural selection being “blind” to mt variation that is male-specific in its effect on phenotype. Mt variants that do not incur costs to females, or which augment female fitness, can therefore spread in a population, even if those variants are deleterious to males. This concept has been expressed in the literature for nearly 40 years [28, 112] and is often referred to as “mother’s curse” [113].
There is an important distinction to make regarding the role of female fitness effects in mother’s curse. In a “weak form” of mother’s curse, a mt mutation with substantial deleterious effects on males may be neutral or nearly neutral in females. In this scenario, the lack of selection on males permits the mt variant to accumulate to appreciable frequencies under mutation-selection balance and potentially to be fixed by genetic drift [112]. One such example is an identified mutation in cytochrome c oxidase 2 (COX2) in Drosophila that impairs male fertility with no apparent harm to female viability or fertility [121]. In contrast, a “strong form” of mother’s curse involves sexually antagonistic mt variants that harm males but have beneficial effects in females and, thus, would actively spread by positive selection. One such example is a CYTB mutation identified in Drosophila that increases in frequency during experimental evolution trials due to increasing female fitness, and in spite of decreasing male fertility [122-124]. Only the strong form of mother’s curse would be considered true selfish conflict, with the extreme example being the spread of a mt variant that entirely eliminates male function but nonetheless spreads because it confers just a slight fitness advantage for females. In contrast, a nuclear-encoded variant with comparable effects on male fitness would only be maintained by selection if it increased female fitness at least two-fold [125].
Individual mt mutations resulting in male-specific harm have been documented in humans [126, 127], fruit flies [121, 122, 128], mice [129], hares [130], and flowering plants [114-116]. Moreover, mt variation was shown to affect the expression of nearly 10% of all nuclear genes in male Drosophila with few effects on gene expression in females [131]. However, in many of these cases it is unclear whether these male-harming variants are beneficial to females (i.e., the strong form of mother’s curse). Recent studies in Drosophila have addressed this shortcoming by measuring both male and female fitness conferred by mt variants and documenting sexually antagonistic effects on trait expression associated with particular mt variants [132], or across whole mtDNA haplotypes [123]. These studies indicate that mt mutations with male-harming effects may have spread due to beneficial female effects (sexual antagonism), not simply due to a lack of selection in males.
Mechanisms of male fitness reduction by mitochondria
The physiological mechanisms underpinning how mt variants can specifically harm male function without impairing female fitness (or even while increasing it) are largely unknown. Two possible mechanisms may act as common explanations shared across multiple lineages. First, sperm are highly active, and sperm motility may be particularly sensitive to ATP production. Mt variants that only slightly compromise mt function may therefore show negative effects in sperm, but not other tissues [15]. Some evidence in humans and Drosophila supports this hypothesis, as certain mt haplotypes confer reduced sperm motility leading to reduced fertility, but no other obvious phenotypes [132, 133]. Second, the number of mtDNA copies in sperm is very limited compared with eggs. As mentioned earlier, mature mammalian oocytes contain ~200,000 mtDNA copies, which must pass through a selective sieve. In contrast, mammalian sperm may contain fewer than 100 mtDNA copies, which are not subject to a selective sieve. The quantity of mt genomes in sperm may therefore be limiting, resulting in mt variants that produce mild phenotypes being detrimental only to sperm function. Detailed quantification of mtDNA copy number in sperm vs. eggs of other eukaryotes can test the generality of this possibility.
Both of these hypotheses assume that sperm or their precursors are dependent on OXPHOS, because while it is possible that mt genomic variants might affect other mt functions, all protein-coding genes in mammalian mtDNA are components of OXPHOS machinery. Contrary to this assumption, there is now general agreement that glycolysis provides the main source of ATP in mammalian sperm while OXPHOS plays a secondary role, if any [134-136]. Therefore, these two mechanisms may serve as null or complementary hypotheses when investigating additional mechanisms.
Although CMS in angiosperms is both widespread and agriculturally important [114], the physiological mechanisms underlying reduced male fitness are still unclear, even though the genetic variation leading to it is well-characterized. In most cases of CMS, the mt gene responsible for CMS (often a chimeric ORF) is expressed in all tissues, whereas phenotypic effects are confined to the anthers [116]. One proposed explanation for these tissue-specific effects is that pollen production is one of the most energetically expensive organismal functions [137, 138], and mt variants that only slightly diminish energetic efficiency might therefore only affect pollen production [139]. However, this explanation has not been explicitly tested. Some evidence in Nicotiana suggests that CMS individuals do show altered mt function but that mt deficiencies may not affect phenotypes overall due to compensating nuclear factors [140]. If this is the case, it is unclear why male phenotypes escape nuclear compensation.
In many CMS species, dysfunction is thought to originate in the tapetum – the specialized tissue surrounding the developing pollen grain [114]. In sunflowers, the death of tapetal cells and meiocytes associated with CMS has been shown to be due to the initiation of a programmed cell death (PCD) pathway [139]. Given the importance of mitochondria in PCD [94] and the precise patterns of PCD necessary during pollen development [141], it is possible that mt-mediated PCD could specifically target developing pollen but few other tissues, possibly by interacting with anther-specific proteases found across angiosperms [139, 142]. Investigating such targeted mechanisms [143] in comparison and contrast with the more general mechanisms outlined above should be a focus of future studies of sexually antagonistic mitonuclear coevolution.
Detailed mechanisms of male-specific harm are beginning to be examined in younger endosymbionts. For example, recent work on Spiroplasma endosymbionts in Drosophila found a single locus in the endosymbiont (Spaid) that causes male-killing by targeting the dosage compensation machinery on the male X chromosome [144]. Recent studies of Wolbachia also identified genes underlying cytoplasmic incompatibility (Fig. 3), with some evidence suggesting similar mechanisms in both lineages [145-147]. Identifying how endosymbionts and mitochondria induce sex-specific phenotypes remains a key arena for future research.
The relatively limited repertoire of organelles as reproductive manipulators
In theory, mt and plastid genomes should benefit from any form of reproductive manipulation that enhances female fitness, so why have many of these mechanisms only been found in younger endosymbionts such as Wolbachia (Fig. 3)? One answer may simply be that similar mechanisms are common in mitochondria and plastids but have largely eluded detection. For example, CMS in plants is often uncovered in crosses between populations or lines, when the sterilizing mitochondria and suppressing nuclear genotypes become decoupled [148]. Other forms of genetic conflict that involve antagonistic coevolution of selfish genetic and elements and suppressors are also often uncovered in interpopulation crosses [149].
Another possibility is that mt and plastid genomes have been domesticated (e.g., genome reduction) to such an extent that the possibilities for reproductive manipulation are limited compared with younger endosymbionts [26]. The prevalence of CMS in angiosperms may serve as an example of how genome architecture and content can dictate which pathways are available. Unlike in animals, angiosperm mt genomes regularly undergo intragenomic recombination, generating structural rearrangements that act as the genetic fuel for reproductive manipulation [114, 115]. In bilaterian animals, the mt genome is more streamlined and may have less mutational fuel (i.e., structural variation) for selection to act on [13]. Testing ideas about why reproductive manipulation is more diverse in younger endosymbionts will require examining cases that represent exceptions to these typical patterns. For example, could reproductive manipulation via mt genomes be more common in nonbilaterian animals, which can possess radically different mt genomes than are typical in bilaterians [48]?
Whereas studies of CMS in angiosperms have provided extensive insight into sexual antagonism, it is less clear if and how mt variation affects reproduction in hermaphroditic animals. Previous studies of mother’s curse in animals have focused solely on species with separate sexes. Hermaphroditic nematodes show mitonuclear epistasis [150, 151], maternal mt inheritance [152], differences in lifespan between males and hermaphrodites, and variation in lifespan due to mitonuclear interactions [153], making them an underutilized study system to investigate mother’s curse [154]. In addition, examining sexual antagonism in organisms with paternal mt transmission would provide a corollary to mother’s curse in which a “father’s curse” would be expected (or by examining isogamous species in which mt transmission is linked to one mating type) [155, 156].
Comparing and contrasting the biology of mitochondria with plastids may help us better understand sexual antagonism. For example, in land plants, plastid genomes tend to be more stable than mt genomes, undergoing fewer rearrangements and thereby limiting the generation of selfish variants. In addition, plastid function may also constrain the possibilities for sexual antagonism compared to mt genomes. The major role of the most abundant type of plastids, chloroplasts, is to perform photosynthesis, which is not critical in male reproductive tissues. Mt respiration plays a more general role for most male functions, including pollen production. It may therefore not be surprising that sexual antagonism has rarely been described in plastid-nuclear interactions, despite uniparental inheritance being common in both mitochondria and plastids (Fig. 2C).Interestingly, the few plastid genes that have been implicated in cytonuclear conflict play roles outside of photosynthesis [157-160]. Such genes are also often implicated as targets of positive selection [161]. In all of these cases, however, the role of plastids in selfish reproductive manipulation remain speculative or incomplete.
Nuclear responses to sexual antagonism
When sexually antagonistic and male-harming mt variants spread to high frequency within a population, it is expected to create strong selection for nuclear responses that counteract these effects. For example, there is a large body of literature describing the nuclear restorer-of-fertility genes that offset CMS caused by mt variants, including evidence of strong positive selection on these nuclear loci that implies an arms-race model of mitonuclear conflict [116, 162-164].
Another potential consequence of sexual antagonism is the evolution of tissue-specific paralogs, which have arisen repeatedly for metazoan nuclear-encoded OXPHOS genes, particularly those in cytochrome c oxidase [165]. These duplicate genes can be highly divergent from each other and often show testis-specific expression. Although optimization in response to tissue-specific metabolic demands may explain the retention of these gene duplicates, one intriguing possibility is that testis-specific paralogs are under positive selection to counteract male-harming mt variants. Some evidence for this hypothesis comes from Drosophila, in which duplicates of mt-targeted genes preferentially show testis-specific expression [166]. These duplicates often relocate far away from the parent gene and are underrepresented on the X chromosome. Duplicates with testis-specific expression are also frequently involved in energy production (e.g., OXPHOS genes), are older than other duplicates in the Drosophila genome, and have higher dN/dS ratios than other gene duplicates (which can indicate positive selection) [166]. On the other hand, in humans, duplicated mt-targeted genes do not exhibit this same enrichment in testis function and are younger than other duplicates [167]. Testis-specific genes in general have also been inferred to be evolving under relaxed selection in humans [168]. An obvious area for future research is to determine why nuclear-encoded OXPHOS duplicates with testis-specific expression evolve under relaxed or positive selection in different lineages, with the latter being expected if their evolution has been shaped by selection for counteracting mt variants that harm male-specific functions.
Many studies investigate the additive effects of mt variants on male function by placing variable mt genomes from different populations on a common nuclear background [123, 131, 132, 169-173]. However, this precludes examining whether nuclear genomes in local populations have responded to selection to counteract male-harming mt variants (i.e., by examining the extent of mt male harm in “home” vs “away” nuclear backgrounds). Recent studies in Drosophila [174-177] and Callosobruchus seed beetles [178, 179] have included multiple nuclear backgrounds as well, with some results indicating sex-specific effects as predicted under mother’s curse and others showing more complicated interactions. It is clear from these studies that environment also plays a role in mediating sex-specific mitonuclear effects (i.e., G × G × E × sex effects). However, these studies are in their infancy and more work is needed to understand the generality of mother’s curse and the frequency of counteracting nuclear mutations.
Summary and future outlook
Although mitochondria are beneficial endosymbionts that have played key roles in shaping eukaryotic evolution (Box 1), the persistence of mt genomes across eukaryotes creates conflicting levels of selection on organismal function and mt genome replication (Fig. 1). Nuclear mechanisms have evolved to counter the spread of selfish mt variants. However, what has been argued to be one of the most widespread countermeasures, uniparental inheritance, also creates the opportunity for sexual antagonism in mt genomes. Here, we have indicated several lines of future research in cytonuclear conflict, including investigating underexplored mechanisms of selfish replication (Table 1), the role for selfish mt genomes during nuclear gene transfer, the prevalence of uniparental inheritance across the majority of eukaryotic diversity (Fig. 2), the reasons why mt and plastid genomes appear to have a limited repertoire for reproductive manipulation (Fig. 3), and the physiological and genetic mechanisms that organelle genomes use to induce sex-specific harm. Importantly, studies investigating selfishness in younger endosymbionts can provide key insights into mitonuclear conflict [106, 180].
Mitonuclear conflict also has applied importance for agriculture and health. CMS in crop species is an important tool for breeding [114, 115, 181-183]. Determining how mt variants cause male-specific harm in CMS could allow for the genetic design of mt genomes that would cause CMS in species of interest, possibly via new mt genomic engineering techniques [184, 185]. Such systems could also be harnessed in metazoans to control pest species [124, 186]. Mitochondrial replacement therapy (MRT) is an emerging germline treatment for mt diseases in which the cytoplasm of an egg from a patient with the disease is replaced by the cytoplasm of a healthy donor, resulting in an offspring that does not inherit the mt disease [187, 188]. However, recent studies have shown that replacement can be imperfect, resulting in a heteroplasmic population of healthy and afflicted mitochondria [189]. Moreover, the mt variant responsible for the disease can rise to fixation after replacement therapy, and drift has been implicated as the primary cause [189]. Determining the role of selfish replication in such dynamics could play a role in improving the efficacy of this therapy and contribute to a broader understanding of the role of selfish genetic conflict in human health.
Acknowledgements
We thank many researchers that provided notes and citations on uniparental inheritance across eukaryotes, Jeff Palmer for providing input on models of gene transfer, and the Kirkpatrick Lab for comments on the manuscript. This work was supported by NIH F32GM116361 to JCH, NSF DGE-1321845 to AMW, NSF MCB 1412260 to DBS, NSF IOS 1456233 to JHW, and ARC FT160100022 and DP170100165 to DKD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Roger AJ, Munoz-Gomez SA, and Kamikawa R (2017). The origin and diversification of mitochondria. Curr. Biol 27, R1177–R1192. [DOI] [PubMed] [Google Scholar]
- 2.Lane N (2014). Bioenergetic constraints on the evolution of complex life. Cold Spring Harb. Perspect. Biol 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hill GE (2015). Mitonuclear ecology. Mol Biol Evol 32, 1917–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Havird JC, Hall MD, and Dowling DK (2015). The evolution of sex: A new hypothesis based on mitochondrial mutational erosion. Bioessays 37, 951–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burton RS, and Barreto FS (2012). A disproportionate role for mtDNA in Dobzhansky-Muller incompatibilities? Mol. Ecol 21, 4942–4957. [DOI] [PubMed] [Google Scholar]
- 6.Radzvilavicius AL, Hadjivasiliou Z, Pomiankowski A, and Lane N (2016). Selection for mitochondrial quality drives evolution of the germline. Plos Biol. 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bendich AJ (2010). Mitochondrial DNA, chloroplast DNA and the origins of development in eukaryotic organisms. Biol. Direct 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hadjivasiliou Z, Pomiankowski A, Seymour RM, and Lane N (2012). Selection for mitonuclear co-adaptation could favour the evolution of two sexes. Proc. R. Soc. Lond. B Biol. Sci 279, 1865–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lane N (2011). Mitonuclear match: Optimizing fitness and fertility over generations drives ageing within generations. Bioessays 33, 860–869. [DOI] [PubMed] [Google Scholar]
- 10.Martin W, and Koonin EV (2006). Introns and the origin of nucleus-cytosol compartmentalization. Nature 440, 41–45. [DOI] [PubMed] [Google Scholar]
- 11.Lane N, and Martin W (2010). The energetics of genome complexity. Nature 467, 929–934. [DOI] [PubMed] [Google Scholar]
- 12.Burton RS, Pereira RJ, and Barreto FS (2013). Cytonuclear genomic interactions and hybrid breakdown. Annu. Rev. Ecol. Evol. Syst 44, 281–302. [Google Scholar]
- 13.Dobler R, Rogell B, Budar F, and Dowling DK (2014). A meta-analysis of the strength and nature of cytoplasmic genetic effects. J. Evol. Biol 27, 2021–2034. [DOI] [PubMed] [Google Scholar]
- 14.Sloan DB, Havird JC, and Sharbrough J (2017). The on-again, off-again relationship between mitochondrial genomes and species boundaries. Mol. Ecol 26, 2212–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burt A, and Trivers R (2006). Selfish mitochondrial DNA. In Genes in conflict: The biology of selfish genetic elements, Burt A and Trivers R, eds. (Cambridge, Massachusetts: The Belknap press of Harvard University Press; ). [Google Scholar]
- 16.Gray MW (2012). Mitochondrial evolution. Cold Spring Harb. Perspect. Biol 4, a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adams KL, and Palmer JD (2003). Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol. Phylogenet. Evol 29, 380–395. [DOI] [PubMed] [Google Scholar]
- 18.Sloan DB, Warren JM, Williams AM, Wu Z, Abdel-Ghany SE, Chicco AJ, and Havird JC (2018). Cytonuclear integration and co-evolution. Nat. Rev. Genet 19, 635–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Timmis JN, Ayliffe MA, Huang CY, and Martin W (2004). Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet 5, 123–135. [DOI] [PubMed] [Google Scholar]
- 20.Hjort K, Goldberg AV, Tsaousis AD, Hirt RP, and Embley TM (2010). Diversity and reductive evolution of mitochondria among microbial eukaryotes. Philos. Trans. R. Soc. Lond. B Biol. Sci 365, 713–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santos HJ, Makiuchi T, and Nozaki T (2018). Reinventing an organelle: The reduced mitochondrion in parasitic protists. Trends Parasitol. 34, 1038–1055. [DOI] [PubMed] [Google Scholar]
- 22.Allen JF (2015). Why chloroplasts and mitochondria retain their own genomes and genetic systems: Colocation for redox regulation of gene expression. Proc. Natl. Acad. Sci. USA 112, 10231–10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bjorkholm P, Harish A, Hagstrom E, Ernst AM, and Andersson SG (2015). Mitochondrial genomes are retained by selective constraints on protein targeting. Proc. Natl. Acad. Sci. USA 112, 10154–10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daley DO, and Whelan J (2005). Why genes persist in organelle genomes. Genome Biol. 6, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Race HL, Herrmann RG, and Martin W (1999). Why have organelles retained genomes? Trends Genet. 15, 364–370. [DOI] [PubMed] [Google Scholar]
- 26.Perlman SJ, Hodson CN, Hamilton PT, Opit GP, and Gowen BE (2015). Maternal transmission, sex ratio distortion, and mitochondria. Proc. Natl. Acad. Sci. USA 112, 10162–10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greiner S, Sobanski J, and Bock R (2015). Why are most organelle genomes transmitted maternally? Bioessays 37, 80–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cosmides LM, and Tooby J (1981). Cytoplasmic inheritance and intragenomic conflict. J. Theor. Biol 89, 83–129. [DOI] [PubMed] [Google Scholar]
- 29.Grun P (1976). Cytoplasmic genetics and evolution, (Columbia University Press; ). [Google Scholar]
- 30.Haig D (2016). Intracellular evolution of mitochondrial DNA (mtDNA) and the tragedy of the cytoplasmic commons. Bioessays 38, 549–555. [DOI] [PubMed] [Google Scholar]
- 31.Werren JH, Baldo L, and Clark ME (2008). Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol 6, 741–751. [DOI] [PubMed] [Google Scholar]
- 32.Radzvilavicius AL, and Blackstone NW (2015). Conflict and cooperation in eukaryogenesis: implications for the timing of endosymbiosis and the evolution of sex. J. Royal Soc. Interface 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bianconi E, Piovesan A, Facchin F, Beraudi A, Casadei R, Frabetti F, Vitale L, Pelleri MC, Tassani S, Piva F, et al. (2013). An estimation of the number of cells in the human body. Ann. Hum. Biol 40, 463–471. [DOI] [PubMed] [Google Scholar]
- 34.Reis RJS, and Goldstein S (1983). Mitochondrial DNA in mortal and immortal human cells - Genome number, integrity, and methylation. J. Biol. Chem 258, 9078–9085. [PubMed] [Google Scholar]
- 35.Blank R, Hauptmann E, and Arnold CG (1980). Variability of mitochondrial population in Chlamydomonas reinhardii. Planta 150, 236–241. [DOI] [PubMed] [Google Scholar]
- 36.Gallaher SD, Fitz-Gibbon ST, Strenkert D, Purvine SO, Pellegrini M, and Merchant SS (2018). High-throughput sequencing of the chloroplast and mitochondrion of Chlamydomonas reinhardtii to generate improved de novo assemblies, analyze expression patterns and transcript speciation, and evaluate diversity among laboratory strains and wild isolates. Plant Journal 93, 545–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rand DM (2001). The units of selection on mitochondrial DNA. Annu. Rev. Ecol. Evol. Syst 32, 415–448. [Google Scholar]
- 38.Birky CW Jr. (1973). On the origin of mitochondrial mutants: Evidence for intracellular selection of mitochondria in the origin of antibiotic-resistant cells in yeast. Genetics 74, 421–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kirk JTO, and Tilney-Bassett RAE (1967). The plastids: their chemistry, structure, growth and inheritance, (London: W. H. Freeman; ). [Google Scholar]
- 40.Ma H, Gutierrez NM, Morey R, Van Dyken C, Kang EJ, Hayama T, Lee Y, Li Y, Tippner-Hedges R, Wolf DP, et al. (2016). Incompatibility between nuclear and mitochondrial genomes contributes to an interspecies reproductive barrier. Cell Metab. 24, 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hardin G (1968). Tragedy of commons. Science 162, 1243–1248. [PubMed] [Google Scholar]
- 42.James AC, and Ballard JWO (2003). Mitochondrial genotype affects fitness in Drosophila simulans. Genetics 164, 187–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Larsson NG (2010). Somatic mitochondrial DNA mutations in mammalian aging In Annual Review of Biochemistry, Vol 79, Volume 79, Kornberg RD, Raetz CRH, Rothman JE and Thorner JW, eds. (Palo Alto: Annual Reviews; ), pp. 683–706. [DOI] [PubMed] [Google Scholar]
- 44.Payne BAI, and Chinnery PF (2015). Mitochondrial dysfunction in aging: Much progress but many unresolved questions. Biochim. Biophys. Acta. - Bioenergetics 1847, 1347–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Payne BAI, Wilson IJ, Yu-Wai-Man P, Coxhead J, Deehan D, Horvath R, Taylor RW, Samuels DC, Santibanez-Koref M, and Chinnery PF (2013). Universal heteroplasmy of human mitochondrial DNA. Hum. Mol. Gen 22, 384–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown WM, George M, and Wilson AC (1979). Rapid evolution of animal mitochondrial-DNA. Proc. Natl. Acad. Sci. USA 76, 1967–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith DR, Hua JM, Lee RW, and Keeling PJ (2012). Relative rates of evolution among the three genetic compartments of the red alga Porphyra differ from those of green plants and do not correlate with genome architecture. Mol. Phylogenet. Evol 65, 339–344. [DOI] [PubMed] [Google Scholar]
- 48.Lavrov DV, and Pett W (2016). Animal mitochondrial DNA as we do not know it: mt genome organization and evolution in nonbilaterian lineages. Genome Biol. Evol 8, 2896–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mower JP, Touzet P, Gummow JS, Delph LF, and Palmer JD (2007). Extensive variation in synonymous substitution rates in mitochondrial genes of seed plants. BMC Evol. Biol 7:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stewart JB, and Chinnery PF (2015). The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nat. Rev. Gen 16, 530–542. [DOI] [PubMed] [Google Scholar]
- 51.Phillips WS, Coleman-Hulbert AL, Weiss ES, Howe DK, Ping S, Wernick RI, Estes S, and Denver DR (2015). Selfish mitochondrial DNA proliferates and diversifies in small, but not large, experimental populations of Caenorhabditis briggsae. Genome Biol. Evol 7, 2023–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jasmin JN, and Zeyl C (2014). Rapid evolution of cheating mitochondrial genomes in small yeast populations. Evolution 68, 269–275. [DOI] [PubMed] [Google Scholar]
- 53.Taylor DR, Zeyl C, and Cooke E (2002). Conflicting levels of selection in the accumulation of mitochondrial defects in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 99, 3690–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clark KA, Howe DK, Gafner K, Kusuma D, Ping S, Estes S, and Denver DR (2012). Selfish little circles: Transmission bias and evolution of large deletion-bearing mitochondrial DNA in Caenorhabditis briggsae nematodes. Plos One 7, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hicks KA, Howe DK, Leung A, Denver DR, and Estes S (2012). In vivo quantification reveals extensive natural variation in mitochondrial form and function in Caenorhabditis briggsae. PLoS One 7, e43837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Howe DK, Baer CF, and Denver DR (2010). High rate of large deletions in Caenorhabditis briggsae mitochondrial genome mutation processes. Genome Biol. Evol 2, 29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen XJ, and Clark-Walker GD (2000). The petite mutation in yeasts: 50 years on. Int. Rev. Cytol 194, 197–238. [DOI] [PubMed] [Google Scholar]
- 58.Chen XJ, and Clark-Walker GD (2018). Unveiling the mystery of mitochondrial DNA replication in yeasts. Mitochondrion 38, 17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.MacAlpine DM, Kolesar J, Okamoto K, Butow RA, and Perlman PS (2001). Replication and preferential inheritance of hypersuppressive petite mitochondrial DNA. Embo Journal 20, 1807–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rand DM (2011). Population genetics of the cytoplasm and the units of selection on mitochondrial DNA in Drosophila melanogaster. Genetica 139, 685–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.David V, Flegontov P, Gerasimov E, Tanifuji G, Hashimi H, Logacheva MD, Maruyama S, Onodera NT, Gray MW, Archibald JM, et al. (2015). Gene loss and error-prone RNA editing in the mitochondrion of Perkinsela, an endosymbiotic kinetoplastid. Mbio 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lukeš J, Wheeler R, Jirsová D, David V, and Archibald JM (2018). Massive mitochondrial DNA content in diplonemid and kinetoplastid protists. IUBMB Life. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adams KL, Song K, Roessler PG, Nugent JM, Doyle JL, Doyle JJ, and Palmer JD (1999). Intracellular gene transfer in action: dual transcription and multiple silencings of nuclear and mitochondrial cox2 genes in legumes. Proc. Natl. Acad. Sci. USA 96, 13863–13868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Doolittle WE (1998). You are what you eat: a gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends Genet. 14, 307–311. [DOI] [PubMed] [Google Scholar]
- 65.Blanchard JL, and Lynch M (2000). Organellar genes - why do they end up in the nucleus? Trends Genet. 16, 315–320. [DOI] [PubMed] [Google Scholar]
- 66.Berg OG, and Kurland CG (2000). Why mitochondrial genes are most often found in nuclei. Mol. Biol. Evo l 17, 951–961. [DOI] [PubMed] [Google Scholar]
- 67.Gitschlag BL, Kirby CS, Samuels DC, Gangula RD, Mallal SA, and Patel MR (2016). Homeostatic responses regulate selfish mitochondrial genome dynamics in C. elegans. Cell Metab. 24, 91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Capps GJ, Samuels DC, and Chinnery PF (2003). A model of the nuclear control of mitochondrial DNA replication. J. Theor. Biol 221, 565–583. [DOI] [PubMed] [Google Scholar]
- 69.Chinnery PF, and Samuels DC (1999). Relaxed replication of mtDNA: A model with implications for the expression of disease. Am. J. Hum. Genet 64, 1158–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tam ZY, Gruber J, HalliweN B, and Gunawan R (2015). Context-dependent role of mitochondrial fusion-fission in clonal expansion of mtDNA mutations. Plos Comput. Biol 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van der Bliek AM, Shen QF, and Kawajiri S (2013). Mechanisms of mitochondrial fission and fusion. Cold Spring Harb. Perspect. Biol 5, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sobanski J, Giavalisco P, Fischer A, Walther D, Schoettler MA, Pellizzer T, Golczyk H, Obata T, Bock R, Sears BB, et al. (2018). Biparental inheritance of chloroplasts is controlled by lipid biosynthesis in evening primroses. bioRxiv 330100, doi: 10.1101/330100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wolf DP, Hayama T, and Mitalipov S (2017). Mitochondrial genome inheritance and replacement in the human germline. Embo Journal 36, 2177–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Milenkovic D, Matic S, Kuhl I, Ruzzenente B, Freyer C, Jemt E, Park CB, Falkenberg M, and Larsson NG (2013). TWINKLE is an essential mitochondrial helicase required for synthesis of nascent D-loop strands and complete mtDNA replication. Hum. Mol. Gen 22, 1983–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ellison CK, and Burton RS (2010). Cytonuclear conflict in interpopulation hybrids: the role of RNA polymerase in mtDNA transcription and replication. J. Evol. Biol 23, 528–538. [DOI] [PubMed] [Google Scholar]
- 76.Pozzi A, Plazzi F, Milani L, Ghiselli F, and Passamonti M (2017). SmithRNAs: Could mitochondria "bend" nuclear regulation? Mol. Biol. Evol 34, 1960–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mishra P, and Chan DC (2014). Mitochondrial dynamics and inheritance during cell division, development and disease. Nat. Rev. Mol. Cell Biol 15, 634–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Van Melderen L, and De Bast MS (2009). Bacterial toxin-antitoxin systems: More than selfish entities? Plos Genet. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sharpley MS, Marciniak C, Eckel-Mahan K, McManus M, Crimi M, Waymire K, Lin CS, Masubuchi S, Friend N, Koike M, et al. (2012). Heteroplasmy of mouse mtDNA is genetically unstable and results in altered behavior and cognition. Cell 151, 333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Luo SY, Valencia CA, Zhang JL, Lee NC, Slone J, Gui BH, Wang XJ, Li Z, Dell S, Brown J, et al. (2018). Biparental inheritance of mitochondrial DNA in humans. Proc. Natl. Acad. Sci. USA 115, 13039–13044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bradshaw E, Yoshida M, and Ling F (2017). Regulation of small mitochondrial DNA replicative advantage by ribonucleotide reductase in Saccharomyces cerevisiae. G3 (Bethesda) 7, 3083–3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stewart JB, Freyer C, Elson JL, and Larsson NG (2008). Purifying selection of mtDNA and its implications for understanding evolution and mitochondrial disease. Nat. Rev. Gen 9, 657–662. [DOI] [PubMed] [Google Scholar]
- 83.Stewart JB, Freyer C, Elson JL, Wredenberg A, Cansu Z, Trifunovic A, and Larsson NG (2008). Strong purifying selection in transmission of mammalian mitochondrial DNA. Plos Biol. 6, 63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fan WW, Waymire KG, Narula N, Li P, Rocher C, Coskun PE, Vannan MA, Narula J, MacGregor GR, and Wallace DC (2008). A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science 319, 958–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Narendra D, Tanaka A, Suen DF, and Youle RJ (2008). Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol 183, 795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, and Youle RJ (2010). PINK1 Is selectively stabilized on impaired mitochondria to activate parkin. Plos Biol. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Youle RJ, and Narendra DP (2011). Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol 12, 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Twig G, Elorza A, Molina AJA, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, et al. (2008). Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. Embo Journal 27, 433–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hill GE, Havird JC, Sloan DB, Burton RS, Greening C, and Dowling DK (2018). Assessing the fitness consequences of mitonuclear interactions in natural populations. Biological Reviews In press. doi: 10.1111/brv.12493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Visser JA, Durlinger ALL, Peters IJJ, van den Heuvel ER, Rose UM, Kramer P, de Jong FH, and Themmen APN (2007). Increased oocyte degeneration and follicular atresia during the estrous cycle in anti-Mullerian hormone null mice. Endocrinology 148, 2301–2308. [DOI] [PubMed] [Google Scholar]
- 91.Hayashi Y, Otsuka K, Ebina M, Igarashi K, Takehara A, Matsumoto M, Kanai A, Soga T, and Matsui Y (2017). Distinct requirements for energy metabolism in mouse primordial germ cells and their reprogramming to embryonic germ cells. Proc. Natl. Acad. Sci. USA 114, 8289–8294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ge HS, Tollner TL, Hu Z, Dai MM, Li XH, Guan HQ, Shan D, Zhang XJ, Lv JQ, Huang CJ, et al. (2012). The importance of mitochondrial metabolic activity and mitochondrial DNA replication during oocyte maturation in vitro on oocyte quality and subsequent embryo developmental competence. Mol. Reprod. Dev 79, 392–401. [DOI] [PubMed] [Google Scholar]
- 93.De Fanti S, Vicario S, Lang M, Simone D, Magli C, Luiselli D, Gianaroli L, and Romeo G (2017). Intra-individual purifying selection on mitochondrial DNA variants during human oogenesis. Hum. Reprod 32, 1100–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Desagher S, and Martinou JC (2000). Mitochondria as the central control point of apoptosis. Trends Cell Biol. 10, 369–377. [DOI] [PubMed] [Google Scholar]
- 95.Hamilton WD, Axelrod R, and Tanese R (1990). Sexual reproduction as an adaptation to resist parasites (a review). Proc. Natl. Acad. Sci. USA 87, 3566–3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lively CM (1987). Evidence from a new-zealand snail for the maintenance of sex by parasitism. Nature 328, 519–521. [Google Scholar]
- 97.Birky CW (1995). Uniparental inheritance of mitochondrial and chloroplast genes - mechanisms and evolution. Proc. Natl. Acad. Sci. USA 92, 11331–11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Garg SG, and Martin WF (2016). Mitochondria, the cell cycle, and the origin of sex via a syncytial eukaryote common ancestor. Genome Biol. Evol 8, 1950–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nedelcu AM (2005). Sex as a response to oxidative stress: stress genes co-opted for sex. Proc. Biol. Sci 272, 1935–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Horandl E, and Speijer D (2018). How oxygen gave rise to eukaryotic sex. Proc. R. Soc. Lond. B Biol. Sci 285, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jenuth JP, Peterson AC, Fu K, and Shoubridge EA (1996). Random genetic drift in the female germline explains the rapid segregation of mammalian mitochondrial DNA. Nat. Gen 14, 146–151. [DOI] [PubMed] [Google Scholar]
- 102.Cree LM, Samuels DC, Lopes S, Rajasimha HK, Wonnapinij P, Mann JR, Dahl HHM, and Chinnery PF (2008). A reduction of mitochondrial DNA molecules during embryogenesis explains the rapid segregation of genotypes. Nat. Gen 40, 249–254. [DOI] [PubMed] [Google Scholar]
- 103.Folmes CDL, Martinez-Fernandez A, Perales-Clemente E, Li X, McDonald A, Oglesbee D, Hrstka SC, Perez-Terzic C, Terzic A, and Nelson TJ (2013). Disease-causing mitochondrial heteroplasmy segregated within induced pluripotent stem cell clones derived from a patient with MELAS. Stem Cells 31, 1298–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Prigione A, Fauler B, Lurz R, Lehrach H, and Adjaye J (2010). The senescence-related mitochondrial/oxidative stress pathway is repressed in human induced pluripotent stem cells. Stem Cells 28, 721–733. [DOI] [PubMed] [Google Scholar]
- 105.Reynier P, May-Panloup P, Chretien MF, Morgan CJ, Jean M, Savagner F, Barriere P, and Malthiery Y (2001). Mitochondrial DNA content affects the fertilizability of human oocytes. Mol. Hum. Reprod 7, 425–429. [DOI] [PubMed] [Google Scholar]
- 106.Bennett GM, and Moran NA (2015). Heritable symbiosis: The advantages and perils of an evolutionary rabbit hole. Proc. Natl. Acad. Sci. USA 112, 10169–10176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rispe C, and Moran NA (2000). Accumulation of deleterious mutations in endosymbionts: Muller's ratchet with two levels of selection. Amer. Nat 156, 425–441. [DOI] [PubMed] [Google Scholar]
- 108.Yu ZS, O'Farrell PH, Yakubovich N, and DeLuca SZ (2017). The mitochondrial DNA polymerase promotes elimination of paternal mitochondrial genomes. Curr. Biol 27, 1033–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pyle A, Hudson G, Wilson IJ, Coxhead J, Smertenko T, Herbert M, Santibanez-Koref M, and Chinnery PF (2015). Extreme-depth re-sequencing of mitochondrial DNA finds no evidence of paternal transmission in humans. Plos Genet. 11, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schwartz M, and Vissing J (2002). Paternal inheritance of mitochondrial DNA. N. Engl. J. Med 347, 576–580. [DOI] [PubMed] [Google Scholar]
- 111.Werren JH, and Beukeboom LW (1998). Sex determination, sex ratios, and genetic conflict. Annu. Rev. Ecol. Evol. Syst 29, 233–261. [Google Scholar]
- 112.Frank SA, and Hurst LD (1996). Mitochondria and male disease. Nature 383, 224. [DOI] [PubMed] [Google Scholar]
- 113.Gemmell NJ, Metcalf VJ, and Allendorf FW (2004). Mother's curse: the effect of mtDNA on individual fitness and population viability. Trends Ecol. Evol 19, 238–244. [DOI] [PubMed] [Google Scholar]
- 114.Schnable PS, and Wise RP (1998). The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci. 3, 175–180. [Google Scholar]
- 115.Kim YJ, and Zhang DB (2018). Molecular control of male fertility for crop hybrid breeding. Trends Plant Sci. 23, 53–65. [DOI] [PubMed] [Google Scholar]
- 116.Budar F, Touzet P, and De Paepe R (2003). The nucleo-mitochondrial conflict in cytoplasmic male sterilities revisited. Genetica 117, 3–16. [DOI] [PubMed] [Google Scholar]
- 117.McCauley DE (2013). Paternal leakage, heteroplasmy, and the evolution of plant mitochondrial genomes. New Phytol. 200, 966–977. [DOI] [PubMed] [Google Scholar]
- 118.McCauley DE, Olson MS, Emery SN, and Taylor DR (2000). Population structure influences sex ratio evolution in a gynodioecious plant. Amer. Nat 155, 814–819. [DOI] [PubMed] [Google Scholar]
- 119.Loussaert D, DeBruin J, San Martin JP, Schussler J, Pape R, Clapp J, Mongar N, Fox T, Albertsen M, Trimnell M, et al. (2017). Genetic male sterility (Ms44) Increases maize grain yield. Crop Sci. 57, 2718–2728. [Google Scholar]
- 120.Lewis D (1941). Male sterility in natural populations of hermaphrodite plants: The equilibrium between females and hermaphrodites to be expected with different types of inheritance. New Phytol. 40, 56–63. [Google Scholar]
- 121.Patel MR, Miriyala GK, Littleton AJ, Yang HK, Trinh K, Young JM, Kennedy SR, Yamashita YM, Pallanck LJ, and Malik HS (2016). A mitochondrial DNA hypomorph of cytochrome oxidase specifically impairs male fertility in Drosophila melanogaster. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Clancy DJ, Hime GR, and Shirras AD (2011). Cytoplasmic male sterility in Drosophila melanogaster associated with a mitochondrial CYTB variant. Heredity 107, 374–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Camus MF, and Dowling DK (2018). Mitochondrial genetic effects on reproductive success: signatures of positive intrasexual, but negative intersexual pleiotropy. Proc. R. Soc. Lond. B Biol. Sci 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wolff JN, Gemmell NJ, Tompkins DM, and Dowling DK (2017). Introduction of a male-harming mitochondrial haplotype via 'Trojan Females' achieves population suppression in fruit flies. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jacobs MS, and Wade MJ (2003). A synthetic review of the theory of gynodioecy. Amer. Nat 161, 837–851. [DOI] [PubMed] [Google Scholar]
- 126.Wallace DC, Singh G, Lott MT, Hodge JA, Schurr TG, Lezza AM, Elsas LJ, and Nikoskelainen EK (1988). Mitochondrial DNA mutation associated with Leber's hereditary optic neuropathy. Science 242, 1427–1430. [DOI] [PubMed] [Google Scholar]
- 127.Milot E, Moreau C, Gagnon A, Cohen AA, Brais B, and Labuda D (2017). Mother's curse neutralizes natural selection against a human genetic disease over three centuries. Nat. Ecol. Evol 1, 1400–1406. [DOI] [PubMed] [Google Scholar]
- 128.Xu H, DeLuca SZ, and O'Farrell PH (2008). Manipulating the metazoan mitochondrial genome with targeted restriction enzymesu. Science 321, 575–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Nakada K, Sato A, Yoshida K, Morita T, Tanaka H, Inoue SI, Yonekawa H, and Hayashi JI (2006). Mitochondria-related male infertility. Proc. Natl. Acad. Sci. USA 103, 15148–15153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Smith S, Turbill C, and Suchentrunk F (2010). Introducing mother's curse: low male fertility associated with an imported mtDNA haplotype in a captive colony of brown hares. Mol. Ecol 19, 36–43. [DOI] [PubMed] [Google Scholar]
- 131.Innocenti P, Morrow EH, and Dowling DK (2011). Experimental evidence supports a sex-specific selective sieve in mitochondrial genome evolution. Science 332, 845–848. [DOI] [PubMed] [Google Scholar]
- 132.Camus MF, Wolf JBW, Morrow EH, and Dowling DK (2015). Single nucleotides in the mtDNA sequence modify mitochondrial molecular function and are associated with sex-specific effects on fertility and aging. Curr. Biol 25, 2717–2722. [DOI] [PubMed] [Google Scholar]
- 133.Ruiz-Pesini E, Lapena AC, Diez-Sanchez C, Perez-Martos A, Montoya J, Alvarez E, Diaz M, Urrieis A, Montoro L, Lopez-Perez MJ, et al. (2000). Human mtDNA haplogroups associated with high or reduced spermatozoa motility. Am. J. Hum. Genet 67, 682–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.du Plessis SS, Agarwal A, Mohanty G, and van der Linde M (2015). Oxidative phosphorylation versus glycolysis: what fuel do spermatozoa use. Asian J. Androl 17, 230–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mukai C, and Okuno M (2004). Glycolysis plays a major role for adenosine triphosphate supplementation in mouse sperm flagellar movement. Biol. Reprod 71, 540–547. [DOI] [PubMed] [Google Scholar]
- 136.Nascimento JM, Shi LZ, Tam J, Chandsawangbhuwana C, Durrant B, Botvinick EL, and Berns MW (2008). Comparison of glycolysis and oxidative phosphorylation as energy sources for mammalian sperm motility, using the combination of fluorescence imaging, laser tweezers, and real-time automated tracking and trapping. J. Cell. Physiol 217, 745–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zabaleta E, Heiser V, Grohmann L, and Brennicke A (1998). Promoters of nuclear-encoded respiratory chain Complex I genes from Arabidopsis thaliana contain a region essential for anther/pollen-specific expression. Plant J. 15, 49–59. [DOI] [PubMed] [Google Scholar]
- 138.Lee SLJ, and Warmke HE (1979). Organelle size and number in fertile and t-cytoplasmic male-sterile corn. Am. J. Bot 66, 141–148. [Google Scholar]
- 139.Balk J, and Leaver CJ (2001). The PET1-CMS mitochondrial mutation in sunflower is associated with premature programmed cell death and cytochrome c release. Plant Cell 13, 1803–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sabar M, De Paepe R, and de Kouchkovsky Y (2000). Complex I impairment, respiratory compensations, and photosynthetic decrease in nuclear and mitochondrial male sterile mutants of Nicotiana sylvestris. Plant Physiol. 124, 1239–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wu HM, and Cheung AY (2000). Programmed cell death in plant reproduction. Plant Mol. Biol 44, 267–281. [DOI] [PubMed] [Google Scholar]
- 142.Walden AR, Walter C, and Gardner RC (1999). Genes expressed in Pinus radiata male cones include homologs to anther-specific and pathogenesis response genes. Plant Physiol. 121, 1103–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Flavell R (1974). A model for the mechanism of cytoplasmic male sterility in plants, with special reference to maize. Plant Sci. Lett 3, 259–263. [Google Scholar]
- 144.Harumoto T, and Lemaitre B (2018). Male-killing toxin in a bacterial symbiont of Drosophila. Nature 557, 252–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Beckmann JF, and Fallon AM (2013). Detection of the Wolbachia protein WPIP0282 in mosquito spermathecae: Implications for cytoplasmic incompatibility. Insect. Biochem. Mol. Biol 43, 867–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Beckmann JF, Ronau JA, and Hochstrasser M (2017). A Wolbachia deubiquitylating enzyme induces cytoplasmic incompatibility. Nat. Microbiol 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.LePage DP, Metcalf JA, Bordenstein SR, On JM, Perlmutter JI, Shropshire JD, Layton EM, Funkhouser-Jones LJ, and Beckmann JF (2017). Prophage WO genes recapitulate and enhance Wolbachia-induced cytoplasmic incompatibility. Nature 543, 243–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Roux F, Mary-Huard T, Barillot E, Wenes E, Botran L, Durand S, Villoutreix R, Martin-Magniette ML, Camilleri C, and Budar F (2016). Cytonuclear interactions affect adaptive traits of the annual plant Arabidopsis thaliana in the field. Proc. Natl. Acad. Sci. USA 113, 3687–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hurst GDD, and Werren JH (2001). The role of selfish genetic elements in eukaryotic evolution. Nat. Rev. Gen 2, 597–606. [DOI] [PubMed] [Google Scholar]
- 150.Ross JA, Koboldt DC, Staisch JE, Chamberlin HM, Gupta BP, Miller RD, Baird SE, and Haag ES (2011). Caenorhabditis briggsae recombinant inbred line genotypes reveal inter-strain incompatibility and the evolution of recombination. Plos Genet. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chang CC, Rodriguez J, and Ross J (2016). Mitochondrial-nuclear epistasis impacts fitness and mitochondrial physiology of interpopulation Caenorhabditis briggsae hybrids. G3 (Bethesda) 6, 209–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wang Y, Zhang Y, Chen LW, Liang Q, Yin XM, Miao L, Kang BH, and Xue D (2016). Kinetics and specificity of paternal mitochondrial elimination in Caenorhabditis elegans. Nat. Commun 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhu ZB, Lu Q, Zeng FF, Wang JJ, and Huang S (2015). Compatibility between mitochondrial and nuclear genomes correlates with the quantitative trait of lifespan in Caenorhabditis elegans. Sci. Rep 5, 17303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ancell H, and Pires-DaSilva A (2017). Sex-specific lifespan and its evolution in nematodes. Semin. Cell Dev. Biol 70, 122–129. [DOI] [PubMed] [Google Scholar]
- 155.Neale DB, Marshall KA, and Sederoff RR (1989). Chloroplast and mitochondrial DNA are paternally inherited in Sequoia sempervirens D. Don Endl. Proc. Natl. Acad. Sci. USA 86, 9347–9349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Nakamura S (2010). Paternal inheritance of mitochondria in Chlamydomonas. J. Plant Res 123, 163–170. [DOI] [PubMed] [Google Scholar]
- 157.Bogdanova VS, Zaytseva OO, Mglinets AV, Shatskaya NV, Kosterin OE, and Vasiliev GV (2015). Nuclear-cytoplasmic conflict in pea (Pisum sativum L.) is associated with nuclear and plastidic candidate genes encoding acetyl-CoA carboxylase subunits. PLoS One 10, e0119835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Stubbe W, and Steiner E (1999). Inactivation of pollen and other effects of genome-plastome incompatibility in Oenothera. Plant Syst. Evol 217, 259–277. [Google Scholar]
- 159.Erixon P, and Oxelman B (2008). Whole-gene positive selection, elevated synonymous substitution rates, duplication, and indel evolution of the chloroplast clpP1 gene. Plos One 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Rockenbach K, Havird JC, Monroe JG, Triant DA, Taylor DR, and Sloan DB (2016). Positive selection in rapidly evolving plastid-nuclear enzyme complexes. Genetics 204, 1507–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bock DG, Andrew RL, and Rieseberg LH (2014). On the adaptive value of cytoplasmic genomes in plants. Mol. Ecol 23, 4899–4911. [DOI] [PubMed] [Google Scholar]
- 162.Fujii S, Bond CS, and Small ID (2011). Selection patterns on restorer-like genes reveal a conflict between nuclear and mitochondrial genomes throughout angiosperm evolution. Proc. Natl. Acad. Sci. USA 108, 1723–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gaborieau L, Brown GG, and Mireau H (2016). The propensity of pentatricopeptide repeat genes to evolve into restorers of cytoplasmic male sterility. Front. Plant Sci 7, 1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Touzet P, and Budar F (2004). Unveiling the molecular arms race between two conflicting genomes in cytoplasmic male sterility? Trends Plant Sci. 9, 568–570. [DOI] [PubMed] [Google Scholar]
- 165.Sinkler CA, Kalpage H, Shay J, Lee I, Malek MH, Grossman LI, and Huttemann M (2017). Tissue-and condition-specific isoforms of mammalian cytochrome c oxidase subunits: from function to human disease. Oxid. Med. Cell. Longev 2017, 1534056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gallach M, Chandrasekaran C, and Betran E (2010). Analyses of nuclearly encoded mitochondrial genes suggest gene duplication as a mechanism for resolving intralocus sexually antagonistic conflict in Drosophila. Genome Biol. Evol 2, 835–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Eslamieh M, Williford A, and Betran E (2017). Few nuclear-encoded mitochondrial gene duplicates contribute to male germline-specific functions in humans. Genome Biol. Evol 9, 2782–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Pierron D, Razafindrazaka H, Rocher C, Letellier T, and Grossman LI (2014). Human testis-specific genes are under relaxed negative selection. Mol. Genet. Genomics 289, 37–45. [DOI] [PubMed] [Google Scholar]
- 169.Camus MF, Clancy DJ, and Dowling DK (2012). Mitochondria, maternal inheritance, and male aging. Curr. Biol 22, 1717–1721. [DOI] [PubMed] [Google Scholar]
- 170.Tourmente M, Hirose M, Ibrahim S, Dowling DK, Tompkins DM, Roldan ERS, and Gemmell NJ (2017). mtDNA polymorphism and metabolic inhibition affect sperm performance in conplastic mice. Reproduction 154, 341–354. [DOI] [PubMed] [Google Scholar]
- 171.Aw WC, Correa CC, Clancy DJ, and Ballard JWO (2011). Mitochondrial DNA variants in Drosophila melanogaster are expressed at the level of the organismal phenotype. Mitochondrion 11, 756–763. [DOI] [PubMed] [Google Scholar]
- 172.Latorre-Pellicer A, Moreno-Loshuertos R, Lechuga-Vieco AV, Sanchez-Cabo F, Torroja C, Acin-Perez R, Calvo E, Aix E, Gonzalez-Guerra A, Logan A, et al. (2016). Mitochondrial and nuclear DNA matching shapes metabolism and healthy ageing. Nature 535, 561–565. [DOI] [PubMed] [Google Scholar]
- 173.Yu X, Gimsa U, Wester-Rosenlof L, Kanitz E, Otten W, Kunz M, and Ibrahim SM (2009). Dissecting the effects of mtDNA variations on complex traits using mouse conplastic strains. Genome Res. 19, 159–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Mossman JA, Tross JG, Li N, Wu ZJ, and Rand DM (2016). Mitochondrial-nuclear interactions mediate sex-specific transcriptional profiles in Drosophila. Genetics 204, 613–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Mossman JA, Biancani LM, Zhu CT, and Rand DM (2016). Mitonuclear epistasis for development time and its modification by diet in Drosophila. Genetics 203, 463–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Mossman JA, Tross JG, Jourjine NA, Li N, Wu ZJ, and Rand DM (2017). Mitonuclear interactions mediate transcriptional responses to hypoxia in Drosophila. Mol. Biol. Evol 34, 447–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Yee WKW, Sutton KL, and Dowling DK (2013). In vivo male fertility is affected by naturally occurring mitochondrial haplotypes. Curr. Biol 23, R55–R56. [DOI] [PubMed] [Google Scholar]
- 178.Immonen E, Collet M, Goenaga J, and Arnqvist G (2016). Direct and indirect genetic effects of sex-specific mitonuclear epistasis on reproductive ageing. Heredity 116, 329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Đorđević M, Stojković B, Savković U, Immonen E, Tucić N, Lazarević J, and Arnqvist G (2017). Sex-specific mitonuclear epistasis and the evolution of mitochondrial bioenergetics, ageing, and life history in seed beetles. Evolution 71, 274–288. [DOI] [PubMed] [Google Scholar]
- 180.Chong RA, and Moran NA (2016). Intraspecific genetic variation in hosts affects regulation of obligate heritable symbionts. Proc. Natl. Acad. Sci. USA 113, 13114–13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Bohra A, Jha UC, Adhimoolam P, Bisht D, and Singh NP (2016). Cytoplasmic male sterility (CMS) in hybrid breeding in field crops. Plant Cell Rep. 35, 967–993. [DOI] [PubMed] [Google Scholar]
- 182.Chen LT, and Liu YG (2014). Male Sterility and Fertility Restoration in Crops In Annual Review of Plant Biology, Vol 65, Volume 65, Merchant SS, ed. (Palo Alto: Annual Reviews; ), pp. 579–606. [DOI] [PubMed] [Google Scholar]
- 183.Saxena RK, Saxena KB, Pazhamala LT, Patel K, Parupalli S, Sameerkumar CV, and Varshney RK (2015). Genomics for greater efficiency in pigeonpea hybrid breeding. Front. Plant Sci 6, 793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Bacman SR, Kauppila JHK, Pereira CV, Nissanka N, Miranda M, Pinto M, Williams SL, Larsson NG, Stewart JB, and Moraes CT (2018). MitoTALEN reduces mutant mtDNA load and restores tRNA. Nat. Med 24, 1696–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Gammage PA, Viscomi C, Simard ML, Costa ASH, Gaude E, Powell CA, Van Haute L, McCann BJ, Rebelo-Guiomar P, Cerutti R, et al. (2018). Genome editing in mitochondria corrects a pathogenic mtDNA mutation in vivo. Nat. Med 24, 1691–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Gemmell NJ, Jalilzadeh A, Didham RK, Soboleva T, and Tompkins DM (2013). The Trojan female technique: a novel, effective and humane approach for pest population control. Proc. R. Soc. Lond. B Biol. Sci 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kang EJ, Wu J, Gutierrez NM, Koski A, Tippner-Hedges R, Agaronyan K, Platero-Luengo A, Martinez-Redondo P, Ma H, Lee Y, et al. (2016). Mitochondrial replacement in human oocytes carrying pathogenic mitochondrial DNA mutations. Nature 540, 270–275. [DOI] [PubMed] [Google Scholar]
- 188.Hyslop LA, Blakeley P, Craven L, Richardson J, Fogarty NME, Fragouli E, Lamb M, Wamaitha SE, Prathalingam N, Zhang Q, et al. (2016). Towards clinical application of pronuclear transfer to prevent mitochondrial DNA disease. Nature 534, 383–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Yamada M, Emmanuele V, Sanchez-Quintero MJ, Sun B, Lallos G, Paull D, Zimmer M, Pagett S, Prosser RW, Sauer MV, et al. (2016). Genetic drift can compromise mitochondrial replacement by nuclear transfer in human oocytes. Cell Stem Cell 18, 749–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Diaz F, Bayona-Bafaluy MP, Rana M, Mora M, Hao H, and Moraes CT (2002). Human mitochondrial DNA with large deletions repopulates organelles faster than full-length genomes under relaxed copy number control. Nucleic Acids Res. 30, 4626–4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Russell OM, Fruh I, Rai PK, Marcellin D, Doll T, Reeve A, Germain M, Bastien J, Rygiel KA, Cerino R, et al. (2018). Preferential amplification of a human mitochondrial DNA deletion in vitro and in vivo. Sci. Rep 8, 1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Hill GE (2016). Mitonuclear coevolution as the genesis of speciation and the mitochondrial DNA barcode gap. Ecol. Evol 6, 5831–5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Hill GE, and Johnson JD (2013). The mitonuclear compatibility hypothesis of sexual selection. Proc. R. Soc. Lond. B Biol. Sci 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zouros E (2013). Biparental inheritance through uniparental transmission: the doubly uniparental inheritance (DUI) of mitochondrial DNA. Evol. Biol 40, 1–31. [Google Scholar]
- 195.Gusman A, Lecomte S, Stewart DT, Passamonti M, and Breton S (2016). Pursuing the quest for better understanding the taxonomic distribution of the system of doubly uniparental inheritance of mtDNA. Peerj 4, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Burki F (2014). The eukaryotic tree of life from a global phylogenomic perspective. Cold Spring Harb. Perspect. Biol 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Davis CC, Xi ZX, and Mathews S (2014). Plastid phylogenomics and green plant phylogeny: almost full circle but not quite there. BMC Biol. 12, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Zhu AD, Fan WS, Adams RP, and Mower JP (2018). Phylogenomic evidence for ancient recombination between plastid genomes of the Cupressus-Juniperus-Xanthocyparis complex (Cupressaceae). BMC Evol. Biol 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Alexander M, Ho SYW, Molak M, Barnett R, Carlborg Ö, Dorshorst B, Honaker C, Besnier F, Wahlberg P, Dobney K, et al. (2015). Mitogenomic analysis of a 50-generation chicken pedigree reveals a rapid rate of mitochondrial evolution and evidence for paternal mtDNA inheritance. Biol. Lett 11, 10–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Zhao X, Li N, Guo W, Hu X, Liu Z, Gong G, Wang A, Feng J, and Wu C (2004). Further evidence for paternal inheritance of mitochondrial DNA in the sheep (Ovis aries). Heredity (Edinb). 93, 399–403. [DOI] [PubMed] [Google Scholar]
- 201.Nunes MDS, Dolezal M, and Schlötterer C (2013). Extensive paternal mtDNA leakage in natural populations of Drosophila melanogaster. Mol. Ecol 22, 2106–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Gyllensten U, Wharton D, Josefsson a, and Wilson a C. (1991). Paternal inheritance of mitochondrial-DNA in mice. Nature 352, 255–257. [DOI] [PubMed] [Google Scholar]
- 203.Kvist L, Martens J, Nazarenko AA, and Orell M (2003). Paternal leakage of mitochondrial DNA in the great tit (Parus major). Mol. Biol. Evol 20, 243–247. [DOI] [PubMed] [Google Scholar]
- 204.Barr CM, Neiman M, and Taylor DR (2005). Inheritance and recombination of mitochondrial genomes in plants, fungi and animals. New. Phytol 168, 39–50. [DOI] [PubMed] [Google Scholar]
- 205.Solieri L (2010). Mitochondrial inheritance in budding yeasts: Towards an integrated understanding. Trends Microbiol. 18, 521–530. [DOI] [PubMed] [Google Scholar]
- 206.Yan Z, Hull CM, Sun S, Heitman J, and Xu J (2007). The mating type-specific homeodomain genes SXI1α and SXI2a coordinately control uniparental mitochondrial inheritance in Cryptococcus neoformans. Curr. Genet 51, 187–195. [DOI] [PubMed] [Google Scholar]
- 207.Yaffe MP (1991). Organelle inheritance in the yeast cell cycle. Trends Cell Biol. 1, 160–164. [DOI] [PubMed] [Google Scholar]
- 208.Xu J (2005). The inheritance of organelle genes and genomes: patterns and mechanisms. Genome 48, 951–958. [DOI] [PubMed] [Google Scholar]
- 209.Basse CW (2014). Mitochondrial inheritance in fungi. Biochim. Biophys. Acta - Bioenerg 1837, 1039–1046. [DOI] [PubMed] [Google Scholar]
- 210.Seitz-Mayr G, Wolf K, and Kaudewitz F (1978). Extrachromosomal inheritance in Schizosaccharomyces pombe. VII. Studies by zygote clone analysis on transmission, segregation, recombination, and uniparental inheritance of mitochondrial markers conferring resistance to antimycin, chloramphenicol, and ery. Mol. Gen. Genet 164, 309–320. [DOI] [PubMed] [Google Scholar]
- 211.Mirfakhrai M, Tanaka Y, and Yanagisawa K (1990). Evidence for mitochondrial DNA polymorphism and uniparental inheritance in the cellular slime mold Polysphondylium pallidum: effect of intraspecies mating on mitochondrial DNA transmission. Genetics 124, 607–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Silliker ME, Liles JL, and Monroe JA (2002). Patterns of mitochondrial inheritance in the myxogastrid Didymium iridis. Mycologia 94, 939–946. [PubMed] [Google Scholar]
- 213.Moriyama Y, and Kawano S (2003). Rapid, selective digestion of mitochondrial DNA in accordance with the matA hierarchy of multiallelic mating types in the mitochondrial inheritance of Physarum polycephalum. Genetics 164, 963–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Reboud X, and Zeyl C (1994). Organelle inheritance in plants. Heredity (Edinb). 72, 132–140. [Google Scholar]
- 215.Corriveau JL, and Coleman AW (2016). Rapid screening method to detect potential biparental inheritance of plastid DNA and results for over 200 angiosperm species. Am. J. Bot 75, 1443–1458. [Google Scholar]
- 216.Whatley JM (1982). Ultrastructure of plastid inheritance: green algae to angiosperms. Biol. Rev 57, 527–569. [Google Scholar]
- 217.Hu Y, Zhang Q, Rao G, and Sodmergen (2008). Occurrence of plastids in the sperm cells of caprifoliaceae: Biparental plastid inheritance in angiosperms is unilaterally derived from maternal inheritance. Plant Cell Physiol. 49, 958–968. [DOI] [PubMed] [Google Scholar]
- 218.Zhang Q, Liu Y, and Sodmergen (2003). Examination of the cytoplasmic DNA in male reproductive cells to determine the potential for cytoplasmic inheritance in 295 angiosperm species. Plant Cell Physiol. 44, 941–951. [DOI] [PubMed] [Google Scholar]
- 219.Hagemann R (2004). The sexual inheritance of plant organelles In: Daniell H, Chase C (eds) Molecular Biology and Biotechnology of Plant Organelles. Springer, Dordrecht [Google Scholar]
- 220.Zuccarello GC, and West JA (2011). Insights into evolution and speciation in the red alga Bostrychia: 15 years of research. Algae 26, 21–32. [Google Scholar]
- 221.Zuccarello GC, West JA, Kamiya M, and King RJ (1999). A rapid method to score plastid haplotypes in red seaweeds and its use in determining parental inheritance of plastids in the red alga Bostrychia (Ceramiales). Hydrobiologia 401, 207–214. [Google Scholar]
- 222.Zuccarello GC, Burger G, West JA, and King RJ (1999). A mitochondrial marker for red algal intraspecific relationships. Mol. Ecol 8, 1443–1447. [DOI] [PubMed] [Google Scholar]
- 223.Choi SJ, Park EJ, Endo H, Kitade Y, and Saga N (2008). Inheritance pattern of chloroplast and mitochondrial genomes in artificial hybrids of Porphyra yezoensis (Rhodophyta). Fish. Sci 74, 822–829. [Google Scholar]
- 224.Chong J, Jackson C, Kim JI, Yoon HS, and Reyes-Prieto A (2014). Molecular markers from different genomic compartments reveal cryptic diversity within glaucophyte species. Mol. Phylogenet. Evol 76, 181–188. [DOI] [PubMed] [Google Scholar]
- 225.Bendif EM, Probert I, Carmichael M, Romac S, Hagino K, and de Vargas C (2014). Genetic delineation between and within the widespread coccolithophore morpho-species Emiliania huxleyi and Gephyrocapsa oceanica (Haptophyta). J. Phycol 50, 140–148. [DOI] [PubMed] [Google Scholar]
- 226.Motomura T, Nagasato C, and Kimura K (2010). Cytoplasmic inheritance of organelles in brown algae. J. Plant Res 123, 185–192. [DOI] [PubMed] [Google Scholar]
- 227.Nagasato C, Motomura T, and Ichimura T (2000). Spindle formation in karyogamy-blocked zygotes of the isogamous brown alga Scytosiphon lomentaria (Scytosiphonales, Phaeophyceae). Eur. J. Phycol 35, 339–347. [Google Scholar]
- 228.Peters AF, Scornet D, Müller DG, Kloareg B, and Cock JM (2004). Inheritance of organelles in artificial hybrids of the isogamous multicellular chromist alga Ectocarpus siliculosus (Phaeophyceae). Eur. J. Phycol 39, 235–242. [Google Scholar]
- 229.Brawley SH, Quatrano RS, and Wetherbee R (1977). Fine-structural studies of the gametes and embryo of Fucus vesiculosus L. (Phaecophyta). III. Cytokinesis and the multicellular embryo. J. Cell Sci 24, 275–294. [DOI] [PubMed] [Google Scholar]
- 230.Hurst LD (1995). Selfish genetic elements and their role in evolution: the evolution of sex and some of what that entails. Philos. Trans. R. Soc. B Biol. Sci 349, 321–332. [DOI] [PubMed] [Google Scholar]
- 231.Nishi M, Hu K, Murray JM, and Roos DS (2008). Organellar dynamics during the cell cycle of Toxoplasma gondii. J. Cell Sci 121, 1559–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Ferguson DJP, Henriquez FL, Kirisits MJ, Muench SP, Prigge ST, Rice DW, Roberts CW, and Mcleod RL (2005). Maternal inheritance and stage-speci c variation of the apicoplast in the intermediate and definitive host. Eukaryot. Cell 4, 814–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Ferguson DJP, Campbell SA, Henriquez FL, Phan L, Mui E, Richards TA, Muench SP, Allary M, Lu JZ, Prigge ST, et al. (2007). Enzymes of type II fatty acid synthesis and apicoplast differentiation and division in Eimeria tenella. Int. J. Parasitol 37, 33–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Ramírez JD, Guhl F, Messenger LA, Lewis MD, Montilla M, Cucunuba Z, Miles MA, and Llewellyn MS (2012). Contemporary cryptic sexuality in Trypanosoma cruzi. Mol. Ecol 21, 4216–4226. [DOI] [PubMed] [Google Scholar]
- 235.Gibson W (2001). Sex and evolution in trypanosomes. Int. J. Parasitol 31, 3–200. [DOI] [PubMed] [Google Scholar]
- 236.Gibson W, and Garside L (1990). Kinetoplast DNA minicircles are inherited from both parents in genetic crosses of Trypanosoma brucei. Parasitol. Res 83, 483–488. [DOI] [PubMed] [Google Scholar]
- 237.Crosby K, and Smith DR (2012). Does the mode of plastid inheritance influence plastid genome architecture? PLoS One 7, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Guzzo F, Baldan B, Bracco F, and Mariani P (1994). Pollen development in Liriodendron tulipifera - some unusual features. Can. J. Bot. Can. Bot 72, 352–358. [Google Scholar]
- 239.Mogensen HL (1996). The hows and whys of cytoplasmic inheritance in seed plants. Am. J. Bot 83, 383–404. [Google Scholar]
- 240.Yao J., Cohen D, and Rowland RE (1994). Plastid DNA inheritance and plastome-genome incompatibility in interspecific hybrids of Zantedeschia (Araceae). Theor. Appl. Genet, 255–260. [DOI] [PubMed] [Google Scholar]
- 241.Snijder RC, Brown FS, and Tuyl Jm. (2007). The role of plastome-genome incompatibility and biparental plastid inheritance in interspecific hybridization in the genus Zantedeschia (Araceae). Floric. Ornam. Biotechnol 1, 150–157. [Google Scholar]
- 242.Barnard-Kubow KB, McCoy MA, and Galloway LF (2017). Biparental chloroplast inheritance leads to rescue from cytonuclear incompatibility. New Phytol. 213, 1466–1476. [DOI] [PubMed] [Google Scholar]
- 243.Levsen N, Bergero R, Charlesworth D, and Wolff K (2016). Frequent, geographically structured heteroplasmy in the mitochondria of a flowering plant, ribwort plantain (Plantago lanceolata). Heredity (Edinb). 117, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.McCauley DE, Sundby AK, Bailey MF, and Welch ME (2007). Inheritance of chloroplast DNA is not strictly maternal in Silene vulgaris (Caryophyllaceae): Evidence from experimental crosses and natural populations. Am. J. Bot 94, 1333–1337. [DOI] [PubMed] [Google Scholar]
- 245.Renzaglia KS, Renzaglia KS, Bernhard DL, Bernhard DL, Garbary DJ, and Garbary DJ (1999). Developmental ultrastructure of the male gamete of Selaginella. Int. J. Plant Sci 160, 14–28. [Google Scholar]
- 246.Zhong ZR, Li N, Qian D, Jin JH, and Chen T (2011). Maternal inheritance of plastids and mitochondria in Cycas L. (Cycadaceae). Mol. Genet. Genomics 286, 411–416. [DOI] [PubMed] [Google Scholar]
- 247.Perlman SJ, Hunter MS, and Zchori-Fein E (2006). The emerging diversity of Rickettsia. Proc. Biol. Sci 273, 2097–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Duron O, Bouchon D, Boutin S, Bellamy L, Zhou L, Engelstädter J, and Hurst GD (2008). The diversity of reproductive parasites among arthropods: Wolbachia do not walk alone. BMC Biol. 6, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Andreadis TG (1985). Experimental transmission of a microsporidian pathogen from mosquitoes to an alternate copepod host. Proc. Natl. Acad. Sci. USA 82, 5574–5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Werren JH, Skinner SW, and Huger AM (1986). Male-killing bacteria in a parasitic wasp. Science 231, 990–992. [DOI] [PubMed] [Google Scholar]
- 251.Hurst GD, and Jiggins FM (2000). Male-killing bacteria in insects: mechanisms, incidence, and implications. Emerg. Infect. Dis 6, 329–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Bandi C, Dunn AM, Hurst GD, and Rigaud T (2001). Inherited microorganisms, sex-specific virulence and reproductive parasitism. Trends Parasitol. 17, 88–94. [DOI] [PubMed] [Google Scholar]
- 253.Nguyen DT, Morrow JL, Spooner-Hart RN, and Riegler M (2017) Independent cytoplasmic incompatibility induced by Cardinium and Wolbachia maintains endosymbiont coinfections in haplodiploid thrips populations. Evolution 71, 995–1008. [DOI] [PubMed] [Google Scholar]