Abstract
AF4/FMR2 family member 4 (AFF4) is the scaffold protein of the multisubunit super-elongation complex, which plays key roles in the release of RNA polymerase II from promoter-proximal pausing and in the transactivation of HIV-1 transcription. AFF4 consists of an intrinsically disordered N-terminal region that interacts with other super-elongation complex subunits and a C-terminal homology domain (CHD) that is conserved among AF4/FMR2 family proteins, including AFF1, AFF2, AFF3, and AFF4. Here, we solved the X-ray crystal structure of the CHD in human AFF4 (AFF4-CHD) to 2.2 Å resolution and characterized its biochemical properties. The structure disclosed that AFF4-CHD folds into a novel domain that consists of eight helices and is distantly related to tetratrico peptide repeat motifs. Our analyses further revealed that AFF4-CHD mediates the formation of an AFF4 homodimer or an AFF1-AFF4 heterodimer. Results from fluorescence anisotropy experiments suggested that AFF4-CHD interacts with both RNA and DNA in vitro. Furthermore, we identified a surface loop region in AFF4-CHD as a substrate for the P-TEFb kinase cyclin-dependent kinase 9, which triggers release of polymerase II from promoter-proximal pausing sites. In conclusion, the AFF-CHD structure and biochemical analyses reported here reveal the molecular basis for the homo- and heterodimerization of AFF proteins and implicate the AFF4-CHD in nucleic acid interactions. The high conservation of the CHD among several other proteins suggests that our results are also relevant for understanding other CHD-containing proteins and their dimerization behavior.
Keywords: transcription elongation factor, phosphorylation, X-ray crystallography, dimerization, RNA polymerase II, gene regulation, AF4/FMR2 family member 4 (AFF4), C-terminal homology domain (CHD), super elongation complex
Introduction
Transcription of protein-coding genes in eukaryotic cells by RNA polymerase (Pol)2 II is regulated not only during the initiation phase but also during elongation of the RNA chain (1–4). Recent biochemical and structural studies revealed how transcription elongation is regulated via the release of promoter-proximal pausing by the kinase complex P-TEFb (5, 6). P-TEFb exists in several forms in cells, including an inactive 7SK snRNP particle, a complex with BRD4, and a complex with scaffolding and elongation factors within a multisubunit super elongation complex (SEC) (7–9). SEC was shown to play key roles in the transactivation of HIV-1 gene transcription and the release of Pol II that is paused in promoter-proximal regions of genes (10–14).
SEC consists of an AF4/FMR family protein, either AFF1 or AFF4; a Pol II elongation factor, either ELL, ELL2, or ELL3; a partner protein, either EAF1 or EAF2; an ENL family protein, either ENL or AF9; and of P-TEFb, a heterodimer of the kinase CDK9 and a cyclin partner, either CycT1, CycT2a, or CycT2b (15). The subunits ELL and ELL2 associate with Pol II and can increase the catalytical rate of transcription elongation (16). Factors ENL and AF9 are thought to bridge between Pol II and SEC via their interaction with the Pol II–binding elongation factor PAF1 complex (PAF1c) (17).
AFF1 and AFF4 are large proteins that serve as scaffolds for SEC assembly (Fig. 1A). AFF4 interacts with other SEC subunits via different parts of its long, intrinsically disordered N-terminal region. For example, P-TEFb associates with the N-terminal 1–73 residues, ELL2 associates with residues 301–350, and ENL/AF9 associates with residues 761–774 of AFF4 (11, 13, 18–21). Several of these interactions have been characterized structurally over the past two decades (18, 19, 21–23).
Figure 1.
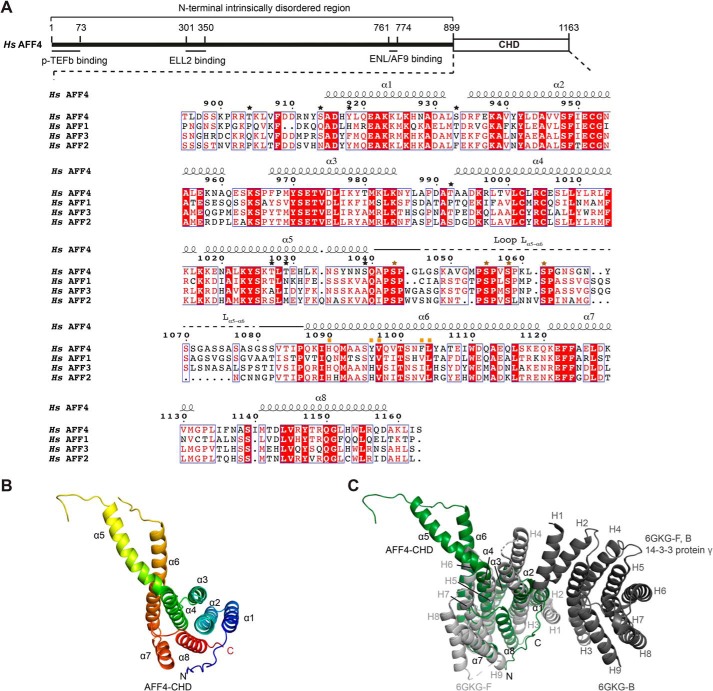
Crystal structure of the AFF4-CHD. A, scheme of the domain organization of AFF4 and the sequence alignment of human AFF4 (NP_055238.1), AFF1 (NP_001160165.1), AFF3 (NP_002276.2), and AFF2 (NP_001162594.1). The interaction regions of AFF4 with P-TEFb, ELL2, and AF9/ENL are shown as lines under the human AFF4 scheme. The alignment was performed using full-length AFF1, AFF2, AFF3, and AFF4 proteins from human, mouse, chicken, and zebrafish and Lilliputian protein from Drosophila using Jalview (muscle with default) (65). The C-terminal sequences of the human AFF proteins are shown here. The C-terminal sequences of the AFF proteins from all above species are shown in Fig. S1. The secondary structure on top of the alignment was assigned based on the crystal structure of AFF4-CHD to the multisequence alignment using ESPript 3 (66). Stars above the sequences indicate residues that are phosphorylated by P-TEFb identified by MS in this study. Black stars show the residues that are not conserved among AFFs. Brown stars show the phosphorylation sites that are conserved among AFFs. The conserved phosphorylation region Lα5–α6 is marked by solid and dashed lines above the sequence. A dashed line indicates the invisible residues in the crystal structure. Mutated residues on the dimerization interface are marked by orange squares. B, crystal structure of AFF4-CHD in one asymmetry unit. The model is colored from N to C using rainbow colors from blue to red. C, superposition of AFF4-CHD monomer with homodimer of 14-3-3 protein γ (Protein Data Bank code 6GKG-F, B (31)). 14-3-3 protein γ is colored in gray, and AFF4-CHD is colored in dark green as figures above. The α-helices of AFF4-CHD are labeled as α1–α8, and the α-helices of 14-3-3 protein γ are labeled as H1–H9. H1, H3, and H4 of the two 14-3-3 protein γ monomers (in light and dark gray) mediate the homodimerization of 14-3-3 protein γ.
In addition to the N-terminal disordered region, AFF4 shares a highly conserved C-terminal homology domain (CHD) with other AF4/FMR family proteins, including AFF1, AFF2, and AFF3. The latter proteins are scaffolds of SEC-L2 and SEC-L3 complexes that contain P-TEFb and AF9/ENL but lack ELL/EAF subunits. The CHD is also found in the Drosophila protein Lilliputian, which is the founding member of the AFF family (10, 24).
The subunits of SEC were shown to be frequent translocation partners in mixed linkage leukemia (MLL). In particular, the fusion of AFF4 to MLL leads to up-regulation of the expression level of MLL target genes (15). It was further shown that MLL–AFF1 and MLL–AFF4 can form a heterodimer via their CHDs in vivo and that heterodimerization correlates with the oncogenic potential of MLL–AFF1 and MLL–AFF4 (25). Furthermore, AFF4 and AFF1 form a heterodimer when co-expressed in vivo (26). Secondary structure predication indicates that the CHD is mainly α-helical and likely to fold into one independent domain (27, 28).
To investigate the structure of the CHD in AFF family proteins and to elucidate the mechanism of CHD dimerization, we solved the X-ray crystal structure of the CHD in human AFF4 (AFF4-CHD) and characterized its biochemical properties by in vitro assays. Our results show that the CHD of AFF4 folds into a helical domain that distantly resembles 14-3-3 proteins and can mediate the formation of an AFF4 homodimer and an AFF1–AFF4 heterodimer. In addition, our fluorescence anisotropy data reveal that the AFF4-CHD interacts with RNA and DNA in vitro. Furthermore, we identified a surface loop region in the AFF4-CHD as a substrate for the P-TEFb kinase. The high conservation of the CHD indicates that our results are relevant for understanding other CHD-containing proteins and their dimerization behavior. Our work contributes to the structural characterization of SEC, which is a key player in gene regulation in eukaryotic cells.
Results
Crystal structure of the AFF4-CHD
To study the structure of the AFF4-CHD, we designed a crystallization construct (residues 899–1163) based on secondary structure prediction and multisequence alignments of AFF proteins from different species (Fig. 1A and Fig. S1). Soluble AFF4-CHD was purified and crystallized as indicated under “Experimental procedures,” and the structure was determined by single-wavelength anomalous diffraction and refined to a resolution of 2.2 Å.
The crystal structure reveals that AFF4-CHD folds into eight α-helices, with α1–α4, α7, and one-half of α6, forming a structure with a concave side, which is filled by the last helix α8 and covered by the N-terminal tail region that folds onto α8 (Fig. 1B). Interestingly, helices α4 and α5 are almost connected as one long helix that bends toward the long helix α6.
AFF4-CHD has a unique fold
To investigate whether the AFF4 dimerization domain is similar to any known functional domains in the Protein Data Bank, we performed 3D structural alignment using the DALI server (29). The results showed that the AFF4-CHD partially aligns with the tetratrico peptide repeat (TPR) motif–containing proteins, such as anaphase-promoting complex subunit 5 (5G05-O) (30), 14-3-3 protein γ (6GKG-F) (31), G-protein-signaling modular 2 (4JHR-B) (32), and 14-3-3 protein β/α (6HEP-B) (33) with Z scores of 9.1, 8.9, 8.9, and 8.8, respectively.
A superposition of each individual structure onto AFF4-CHD was produced with PyMOL (Fig. 1C and Fig. S2, A–C) (29). The root mean square deviations between the structures and AFF4-CHD are 2.7 Å over 452 atoms for 5G05-O, 4.1 Å over 466 atoms for 6GKG-F, 2.2 Å over 424 atoms for 4JHR-B, and 5.4 Å over 468 atoms for 6HEP-B. Based on the superpositions, the arrangement of α1–α4 and half of α6 corresponds to 2.5 TPR motifs. However, AFF4 lacks the conserved amino acids in the featured positions that define the TPR repeats, such that TPRpred could not identify typical TPR motifs in the sequence (34, 35).
Considering that two 14-3-3 proteins stand out in the DALI search, we further compared AFF4-CHD to 14-3-3 proteins (29). The AFF4-CHD lacks the α-helices (N-terminal helix H1 and the two longer helices H3 and H4) that mediate the dimerization of 14-3-3 proteins (Fig. 1C). Instead, the N-terminal residues are unstructured and fold onto α8. This indicates that AFF4-CHD cannot form a similar homodimer as 14-3-3 proteins, which is an important mechanism for regulating protein–protein interactions by 14-3-3 proteins (36). Furthermore, 14-3-3 proteins were shown to bind phosphorylated peptides or other modified peptides via their concave surface formed by helices H3, H5, H7, and H9 (33, 36, 37).
In AFF4-CHD, the C-terminal residues (α7 and α8), which could potentially take the role of H7–H9, adopt a conformation different from H7–H9 in 14-3-3 protein γ (Fig. 1C). When AFF4-CHD is superposed with 14-3-3 protein γ, the loop between α7 and α8 takes the position of H7 in 14-3-3 protein γ; however, α8 folds into the concave surface and blocks the surface, instead of taking the open position as observed for H9 in 14-3-3 protein γ (Fig. 1C). When superposed with 14-3-3 protein β (6HEP-B) in the presence of its ligand in complex with CFTR R-domain peptide pSer753–pSer768, helix α8 of AFF4-CHD clashes with the ligand peptide (Fig. S2C). However, we do not rule out the possibility that α8 and the N-terminal region of AFF4-CHD could change conformation and move away from the ligand-binding concave surface in the presence of natural substrates. In summary, except for a partial and distant similarity to 14-3-3 proteins and TPR motif–containing proteins, AFF4-CHD adopts a unique fold and does not resemble any known structure in the Protein Data Bank.
AFF4-CHD forms homodimers
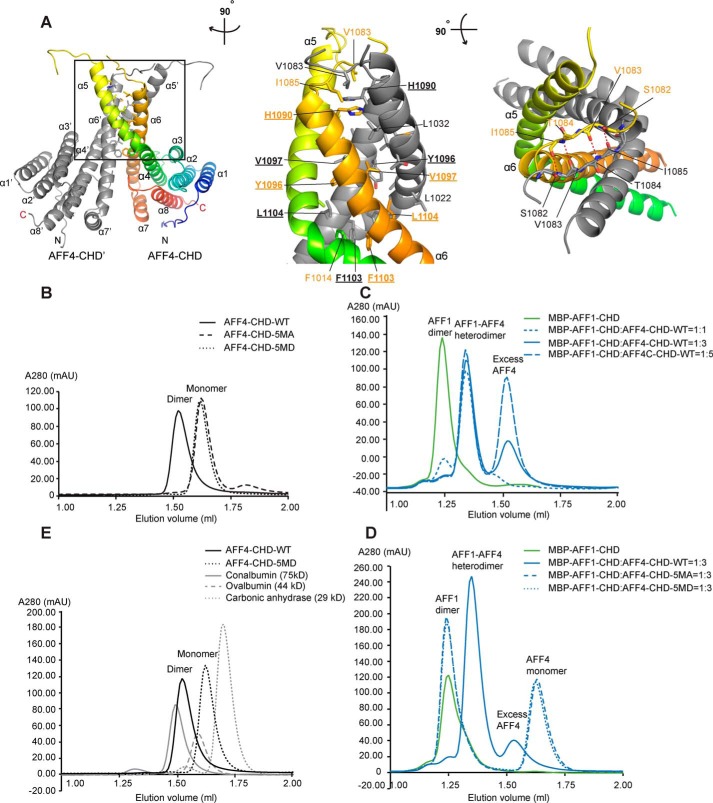
Analysis of the crystal packing showed that AFF4-CHD tightly interacts with a close symmetry mate (AFF4-CHD′) via helices α5 and α6 (α5′ and α6′ for AFF4-C′; Fig. 2A). The interaction involves an interface area of 1655 Å2, as calculated with QtPISA (38, 39).
Figure 2.
AFF4-CHD forms homodimer and can form heterodimer with AFF1-CHD in solution. A, dimerization interface of AFF4-CHD homodimer. The left panel shows the overview of the homodimerization interface. In the middle panel, the model is rotated ∼90° clockwise around the vertical axis relative to the overview in the left panel. In the right panel, the model is rotated ∼90° toward the reader around the horizontal axis relative to the view in the middle panel. The two monomers in the dimer are colored gray (AFF4-CHD′) and rainbow (AFF4-CHD), respectively. Residues that are involved in the interface are labeled in orange (AFF4-CHD) and gray (AFF4-CHD′). The side chains (middle panel) or main chains (right panel) are shown as sticks. Underlined letters and numbers indicate residues that are mutated to alanine or aspartate in the 5MA or 5MD constructs for biochemical studies in panels B–D. In the right panel, red dashed lines indicate hydrogen bonds. B, disruption of the dimerization interface abolished the homodimerization of AFF4-CHD. The state of WT and mutants (5MA and 5MD) of AFF4-CHD are analyzed by analytical gel filtration. The elution profiles of WT and mutants of AFF4-CHD are shown as smoothed lines. The raw data were exported from UNICORN and plotted as smoothed lines in Excel. AFF4-CHD–5MA contains the following mutations: H1090A, Y1096A, V1097A, F1103A, and L1104A. AFF4-CHD–5MD contains mutations H1090D, Y1096D, V1097D, F1103D, and L1104D. The WT, 5MA, and 5MD elution profiles are shown in solid, dashed, and dotted black lines, respectively. C, WT AFF4-CHD forms a heterodimer with MBP–AFF1-CHD. The elution profiles of MBP–AFF1-CHD and premixed MBP–AFF1-CHD/AFF4-CHD are shown as smoothed lines. MBP–AFF1-CHD alone (in green) serves as a control. To form the AFF4-CHD/AFF1-CHD complex, AFF4-CHD and MBP–AFF1-CHD were mixed in different ratios (1:1, 1:3, and 1:5 molar ratios) and incubated overnight at 4 °C. The protein mixtures were sequentially injected onto analytical gel filtration column, and the profiles of the preincubated MBP–AFF1/AFF4 protein at different ratios are presented as solid (1:3) and dashed blue lines (1:1 and 1:5) as indicated in the figure. D, the AFF4-CHD mutants failed to form heterodimers with MBP–AFF1-CHD. The MBP–AFF1-CHD are premixed with AFF4-CHD–WT or AFF4-CHD mutants (5MA and 5MD) in 1:3 ratio and incubated overnight prior injection to analytical gel filtrations. The MBP–AFF1-CHD alone (in green) serves as a control. Solid, dashed, and dotted blue lines indicate the elution profile of MBP–AFF1-CHD premixed WT, 5MA, and 5MD, respectively. E, comparison of the molecular mass of AFF4-CHD–WT and 5MD with size exclusion standards. The elution profiles of AFF4-CHD–WT, AFF4-CHD–5MD, and size exclusion standards including conalbumin, ovalbumin, and carbonic anhydrase are shown as a black solid line, a black dashed line, a gray solid line, and a gray dashed line, respectively.
The helices α5 and α6 and the loop region that connects them (Lα5–α6) form a hydrophobic core and mediate the interaction between the two monomers (Fig. 2A). In addition to the hydrophobic interactions, residues Ile1085 to Ser1082 of loop Lα4–α5 form hydrogen bonds via main chain interaction with the same region of the partner, which further strengthens the interaction (Fig. 2A, right panel).
During protein preparation, we noticed that AFF4-CHD forms homodimers based on the elution volume of the protein during gel filtration (Fig. 2E), which is consistent with the dimerization observed in the crystal structure. Analytical size-exclusion chromatography showed that AFF4-CHD (theoretical molecular mass = 31.9 kDa as a monomer) elutes slightly after conalbumin (75 kDa) and before ovalbumin (44 kDa), and the calculated molecular mass based on the calibration of the column is 64.6 kDa, which indicates that AFF4-CHD forms a dimer in solution (Fig. 2E and Fig. S3C).
To further validate the dimerization interface observed in the crystal structure, we mutated the hydrophobic residues on α6, which play main roles in the dimerization interface, including His1090, Tyr1096, Val1097, Phe1103, and Leu1104, to alanine (5MA) or aspartate (5MD) and compared the elution volume of the WT protein and the mutants on analytical gel filtration (Superdex 200 increase 3.2/300; GE Healthcare). The peak of the WT AFF4-CHD elution peak was located at 1.53 ml, whereas both AFF4-CHD mutants 5MA and 5MD showed an elution peak at 1.62 ml, which indicates that mutation of the hydrophobic residues on the interface abolished homodimerization of AFF4-CHD (Fig. 2, B and E, and Fig. S2D). Static light scattering data confirmed that the molecular mass of AFF4-CHD–WT is 64.38 kDa, and the molecular mass of the dimerization mutant AFF4-CHD–5MD is 32.2 kDa in solution, which supports our dimerization hypothesis (Fig. S3, D and E).
CHDs of AFF4 and AFF1 form a heterodimer
It was reported that AFF4 and AFF1 are able to form a heterodimer in cells and that the CHD was responsible for heterodimerization (15, 26). We therefore investigated whether the CHD of AFF4 and AFF1 also form a heterodimer in vitro and whether our homodimeric structure would be a good model for AFF4-AFF1 heterodimerization. For this purpose, we designed an AFF1-CHD (residues 938–1210) construct based on alignments and on our crystal structure. Considering the similar size of AFF4-CHD and AFF1-CHD, we used an N-terminal His-MBP–tagged AFF1-CHD (MBP–AFF1-CHD), with a theoretical molecular mass of 72.3 kDa for a monomer.
Our analytical size-exclusion chromatography and static light scattering data showed that the molecular mass of MBP–AFF1-CHD is 147 kDa in solution, and the MBP–AFF1-CHD/AFF4-CHD complex is 99 kDa in solution. This indicates that MBP–AFF1-CHD forms a homodimer and that AFF4-CHD was able to replace MBP–AFF1-CHD from the AFF1-CHD homodimer and to form an AFF4-CHD/MBP–AFF1-CHD heterodimer after incubation overnight at 4 °C (Fig. 2C and Fig. S3, F and G). Incubation of MBP–AFF1-CHD with AFF4-CHD indicates that AFF4-CHD replaces most of MBP–AFF1-CHD at 1:1 molar ratio. An excess of AFF4-CHD could eliminate almost all MBP–AFF1-CHD homodimer (Fig. 2C). In contrast, neither of the AFF4-CHD mutants (5MA and 5MD) was able to disrupt the MBP–AFF1-CHD homodimer (Fig. 2D and Fig. S2E). This implies that the dimerization interface observed in the AFF4-CHD homodimer also mediates the interaction between AFF1 and AFF4.
AFF4-CHD interacts with nucleic acids
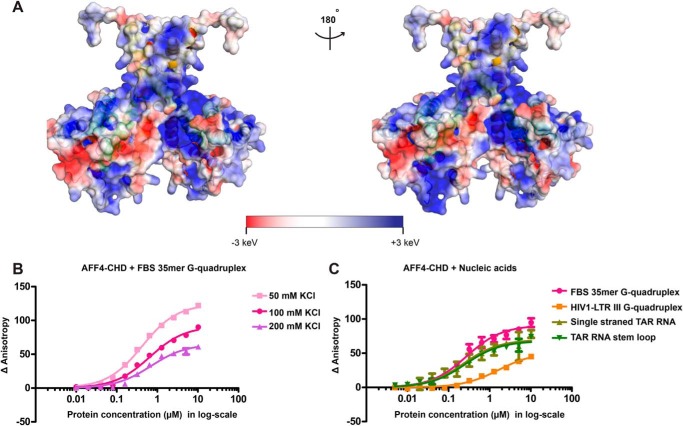
Surface electrostatic potential analysis of our crystal structure revealed several large positively charged patches on AFF4-CHD (Fig. 3A), which implies that AFF4-CHD might be able to interact with nucleic acids. To study the nucleic acid–binding ability of AFF4-CHD, we first tested the binding of a previously reported (24) substrate 5′-FAM–labeled N19-FBS 35mer G-quadruplex RNA under different KCl concentrations by fluorescence anisotropy. The results showed that the variation of KCl concentrations in the assay buffer from 50 to 200 mm only slightly reduced the affinity of the substrate to AFF4-CHD, with a variation of Kd values from 0.422 ± 0.016 to 0.691 ± 0.081 μm (Fig. 3B and Table S2). The standard deviations reported here and in all following fluorescence anisotropy experiments were calculated from triplicated experiments. The sequences of nucleic acids for fluorescence anisotropy are shown in Table S1.
Figure 3.
AFF4-CHD harbors large positively charged surface and associates with nucleic acids. A, electrostatic surface potential generated with APBS software (67, 68). Blue, red, and white represent positive, negative, and neutral residues, respectively, with electrostatic potentials showing from −3 to +3 keV. The right panel shows the back side of the molecule, with 180° rotation relative to the left panel. B, binding of AFF4-CHD to 10 nm fluorescence labeled FBS 35-mer G-quadruplex in buffer containing 50, 100, and 200 mm KCl. C, binding of AFF4-CHD to 10 nm fluorescence labeled FBS 35-mer G-quadruplex, structured and unstructured TAR RNA, and HIV-1 LTR-III G-quadruplex DNA in buffer containing 100 mm KCl. The experiments in B were repeated three times, each time with two pipetting replicates using 10 nm 5′-FAM–labeled RNA and protein concentration from 0.01 to 10 μm. The experiments in C were repeated three times using 10 nm 5′-FAM–labeled RNA and protein concentration from 0.005 to 10 μm. The standard deviation is shown as error bars on the figure. The curves were fitted with quadratic single-site binding equation (see “Experimental procedures”).
We tested the interaction of AFF4-CHD with single-stranded TAR RNA, structured TAR RNA with a stem-loop structure and a G-quadruplex DNA HIV-LTR III, and compared the interaction with N19-FBS 35-mer G-quadruplex RNA in buffer containing 100 mm KCl (Fig. 3C and Table S2). Single-stranded TAR RNA and structured TAR RNA interact with AFF4-CHD with a similar affinity as the N19-FBS RNA (Fig. 3C and Table S2). The affinity for HIV-LTR III RNA is lower with a Kd of 1.647 ± 0.176 μm. Our results indicate that AFF4-CHD binds to RNA regardless of the structure of the RNA (Fig. 3C and Table S2), and the interaction with DNA is weaker (Fig. 3C and Table S2).
AFF4-CHD is phosphorylated by P-TEFb in vitro
AFF4 is a substrate for the P-TEFb kinase CDK9, as revealed by phosphoproteomics (40–43), kinase substrate prediction (GPS 3.0) (44–46), and P-TEFb inhibition (14). To investigate whether AFF4-CHD is phosphorylated by P-TEFb in vitro, we performed phosphorylation assays with purified kinase complex P-TEFb–5aa–TAT, which consists of CDK9 and its partner CycT1 (residues 1–272) fused to HIV-1 protein TAT (residues 1–72) via a GGGSG linker (47). For simplicity we refer to this P-TEFb–5aa–TAT complex as P-TEFb in the following text and in figures.
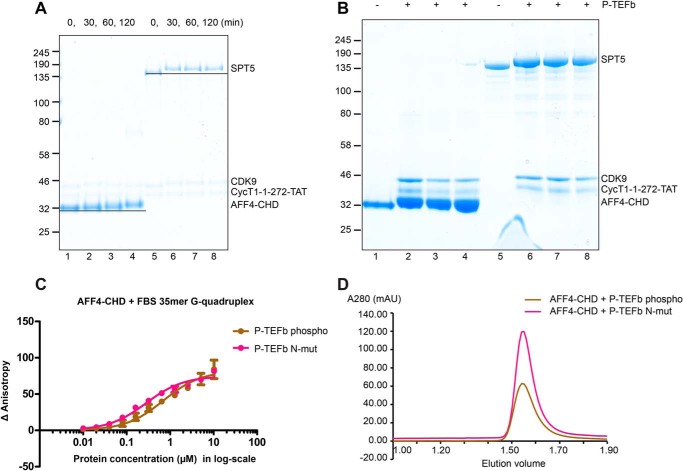
AFF4-CHD was indeed phosphorylated after incubation with P-TEFb and ATP (“Experimental procedures”). Time-course experiments showed that AFF4-CHD slightly shifted upwards in electrophoretic analysis compared with apo-AFF4-CHD, suggesting phosphorylation (Fig. 4A). Mass spectrometric analysis of the phosphorylation product revealed 12 phosphorylated residues that are present in three replicates, Ser914, Ser933, Ser1043, Ser1055, Ser1058, Ser1062, Ser1039, Thr992, Thr904, Thr1027, Thr1029, and Tyr918 (Figs. 1A and 4B). Among the identified phosphorylation sites, Ser1043, Ser1055, Ser1058, and Ser1062 were shown to be phosphorylated in previous phosphoproteomics studies (28, 29, 30, 31). Most of the conserved phosphorylation sites lie in the unstructured loop region Lα5–α6, which contains conserved SP sequences and is mostly invisible in the crystal structure (Fig. 1A and Fig. S1).
Figure 4.
AFF4-CHD is a substrate of P-TEFb kinase. A, time course of the phosphorylation of AFF4-CHD by P-TEFb. AFF4-CHD and SPT4/SPT5 were incubated with P-TEFb over a time course of 2 h, and samples were taken at 0, 30, 60, and 120 min. SPT5 serves as a control to monitor the activity of P-TEFb and demonstrate the shift of protein bands after incubation with P-TEFb in the presence of ATP. B, final products of the phosphorylation assay triplicates for the identification of phosphorylation sites using MS. Lanes 1 and 5 are apo proteins before adding P-TEFb. Lanes 2–4 and 6–8 are the triplicates of the phosphorylation assay after 2 h of incubation. C, binding of AFF4-CHD to 10 nm fluorescence labeled FBS 35-mer G-quadruplex in apo and phosphorylated states. The binding assay was performed after incubating AFF4-CHD with WT P-TEFb kinase or catalytical mutant of P-TEFb kinase in the presence of ATP. The experiments were repeated three times, each time with two pipetting repeats as in Fig. 3C. D, the final products of the three phosphorylation triplicates of the AFF4-CHD incubated with P-TEFb and P-TEFb–N-mut were injected onto the gel filtration column (Superdex 200 increase 3.2/300; GE Healthcare), respectively.
To investigate whether the phosphorylation of AFF4-CHD by P-TEFb affects the RNA binding and homodimerization of AFF4-CHD, we performed fluorescence anisotropy and analytical gel filtration after incubating AFF4-CHD with WT P-TEFb kinase (P-TEFb–WT) or the catalytically dead mutant of P-TEFb (PTEFb-N, D149N) (Fig. S3). Our fluorescence anisotropy results indicate that after phosphorylation by P-TEFb, the affinity of AFF4-CHD to N19-FBS 35-mer sequence is slightly reduced (the Kd changed from 0.274 ± 0.019 to 0.693 ± 0.068 μm) (Fig. 4C and Table S2). Analytical size-exclusion chromatography showed that phosphorylation of AFF4-CHD has no effect on AFF4 homodimerization (Fig. 4D). Taken together, these results indicate that P-TEFb–dependent phosphorylation of AFF4-CHD may slightly modulate nucleic acid interactions but not domain dimerization.
Discussion
Here we report the crystal structure of the CHD of AFF4, the scaffold protein of the SEC. We show that AFF4-CHD folds into a unique structure that distantly resembles 14-3-3 proteins, which can interact with peptides that contain phosphorylated serine residues (29). We further show that AFF4-CHD forms a homodimer in vitro, and mutation of the dimerization interface disrupts both homodimerization and heterodimerization with AFF1-CHD. We further show that AFF4-CHD exhibits positively charged surfaces and can interact with DNA and RNA in vitro. Finally, we show that AFF4-CHD is a substrate of P-TEFb phosphorylation in vitro and identified the phosphorylation sites by MS.
The crystal structure of AFF4-CHD revealed the extensive dimerization interface that includes hydrophobic interactions and hydrogen bonds. Mutagenesis studies show that disruption of the hydrophobic core of the interface was able to abolish both AFF4-CHD homodimer and AFF4-CHD/AFF1-CHD heterodimer in vitro. Sequence alignment indicates that the dimerization interface residues are highly conserved among the CHD of AFF proteins. Thus our results reveal a conserved dimerization mechanism for AFF proteins.
Although our results are consistent with a large body of prior work, there is one discrepancy. Whereas we show that AFF4-CHD can form homodimers when expressed recombinantly, this was not observed by overexpressing AFF4 in vivo (26). It is possible that the protein undergoes additional modifications in vivo, which would prevent homodimerization. It is also possible that in the context of full-length protein, the dimerization interface is shielded by other regions of the protein or by interaction partners such as AFF1.
The heterodimer of AFF1 and AFF4 could be observed in vivo; however, this heterodimer most likely resides outside of the SEC complex, because tandem co-immunoprecipitation experiments showed that AFF1 and AFF4 do not co-exist in the same SEC complex (26). Furthermore, it was shown that the target genes of AFF1 and AFF4 are largely nonoverlapping, with less than 40% of them being regulated by both AFF1 and AFF4 (26). However, how AFF1 and AFF4 may cooperate in regulating the shared target genes remains unclear. In a previous study, it was proposed that during transcription several SEC complexes are recruited to transcribing Pol II, and the recruitment of AFF1-SEC and AFF4-SEC might happen in sequential steps (48). The CHDs might facilitate recruitment of the later SEC to Pol II.
CHD-mediated dimerization is required for the recruitment of intact SEC to MLL targets via the interaction between MLL-fused AFF1 or AFF4 and the full-length AFF4 and AFF1 in the SEC complex, which led to constitutive activation of MLL target genes (49). Indeed, only MLL–AFF4 fusion proteins that include the CHD can induce up-regulation of Hoxa9 transcription and retain oncogenic potential (25, 49). Therefore, our structure of AFF4-CHD might be explored to provide a new target for future therapeutic routes to treat MLL-rearranged leukemias (50).
We further showed that AFF4-CHD harbors large positively charged surface and showed that AFF4-CHD interacts with nucleic acids at micromolar range concentration, with slightly higher affinity to RNA than to DNA substrates. These observations are in line with a previous study showing that the C-terminal region of AFF proteins interacts with N19 RNA. The N19 RNA contains a 35-mer G-quadruplex structure, which was shown to be required for the interaction with AFF proteins (24). However, in our study, we did not observe a preference for N19 35-mer RNA G-quadruplex over other RNAs, maybe because our AFF4-CHD lacks the unstructured region upstream of the CHD, and the N19-FBS-35mer is shorter than the construct used in previous literature (24). An interaction of AFF4-CHD with the G-quadruplex structure of N19 was shown to be important for the splicing of this RNA (24). Identification of natural RNA targets would require suited in vivo approaches such as cross-linking followed by sequencing of the cross-linked RNA regions.
We also showed that P-TEFb phosphorylates AFF4-CHD and identified phosphorylation sites on the domain, among which there are phosphorylation sites that were also observed in vivo (40–42, 51). Although the phosphorylation did not affect dimerization of AFF4-CHD or nucleic acids binding in vitro, it is possible that these modifications might influence the interaction with additional binding partners or regulate the function of AFF4 in cells.
In summary, we provide the structure of the conserved AFF protein CHD and the molecular basis for the homo- and heterodimerization of AFF proteins. In addition, our data implicate the AFF4-CHD in nucleic acid interactions and identified a conserved loop region as a phosphorylation site for P-TEFb. The structure provides a missing building block of the super elongation complex and can be used to design future experiments aimed at dissecting the function of AFF4 and SEC.
Experimental procedures
Preparation of the C-terminal dimerization domain of AFF4 (AFF4-CHD) and AFF1 (AFF1-CHD)
The boundary of the C-terminal domain of AFF4 (AFF4-CHD) and AFF1 (AFF1-CHD) was designed based on the secondary structure prediction using PSIPRED (27, 28). DNA containing the AFF4-CHD (residues 899–1163) was amplified from genomic DNA and was cloned into vector 1B using ligation-independent cloning (MacroLab) (52, 53). This results in an ORF containing an N-terminal hexahistidine tag with a TEV protease cleavage site. Similarly, the AFF1-CHD (residues 938–1210) was cloned into vector 1C (MacroLab) using LIC, which results in an ORF containing an N-terminal hexahistidine-MBP tag with a TEV protease cleavage site. The primers used for cloning are listed in Table S1. The His-AFF4-CHD and His-MBP–tagged AFF1-CHD were expressed overnight at 20 °C in Escherichia coli BL21 (DE3) strain cells (Stratagene), respectively. For selenomethionine labeling, AFF4-CHD was expressed in minimal medium supplemented with selenomethioine (54).
The cells were harvested and resuspended in buffer containing 50 mm HEPES, pH 7.4, 300 mm NaCl, 10% glycerol, 1 mm DTT, and 30 mm imidazole supplemented with protease inhibitors, DNase I and lysozyme and lysed by sonication. Cell lysates were subjected to affinity purification on a nickel column (HisTrap HP; GE Healthcare) and eluted with linear imidazole gradient ranging from 30 to 500 mm. The eluted protein was concentrated and polished by a final gel filtration (Superdex 200 10/300 increase; GE Healthcare). The sample was concentrated to 21 mg/ml and stored at −80 °C in buffer containing 20 mm HEPES, pH 7.5, 300 mm NaCl, and 2 mm DTT. His-MBP–tagged AFF1-CHD was expressed and purified in the same way, concentrated to 12 mg/ml, and stored at −80 °C.
Mutants of AFF4-CHD (5MA and 5MD) were generated by site-directed mutagenesis PCR (QuikChange; Qiagen) using WT AFF4-CHD plasmid as the template. The primers used for mutagenesis are listed in Table S1. The AFF4-CHD 5MA and 5MD were expressed and purified in the same way as the WT protein. Importantly, in the affinity purification step for the AFF4-CHD mutants, only the fractions of the first peak, which is also the main peak, were collected for further purification. The tailing shoulder peak at the end of the imidazole gradient, which roughly contains less than 5% of the whole eluate, might contain mixture of dimer and monomers.
Crystal structure determination
The purified native and selenomethionine-labeled His-AFF4-CHD was crystalized by vapor diffusion using 14–16% PEG8000, 40 mm KH2PO4, and 20% glycerol as reservoir solution. Complete diffraction data at 2.2 Å resolution were obtained for crystals of the selenomethionine-labeled protein on a PILATUS 6M detector at the PXII Beamline of the Swiss Light Source at a temperature of 100 K and processed by XDS and scaled by XSCALE (55). The resolution cutoff was set based on CC½ value (56). Selenium sites were identified with SHELXC and chain-racing and density modification with SHELXD/E (57). The model was manually built in coot using the initial polyalanine model from SHELXC/D/E as a starting model and refined with program PHENIX (58, 59). We further refined the model using coot and PHENIX iteratively. 99.5% of residues were in the favored regions of the Ramachandran plot without any outliers. The stereochemical properties of the structure was verified with MolProbity (60). The refinement statistics are shown in Table 1.
Table 1.
Data collection, refinement, and validation stastics
| Crystal | SeMet |
|---|---|
| Data collection | |
| Space group | I2 (1)2 (1)2 (1) |
| Unit cell dimensions (Å) | 41.53, 79.48, 185.44 |
| Wavelength | 0.97925 |
| Resolution (Å) | 50–2.2 (2.26–2.20) |
| Rsym (%) | 8.5 (173.3%) |
| I / σ (I) | 11.04 (1.11) |
| CC (1/2) | 99.9 (66.3) |
| Completeness (%) | 100.0 (100.0) |
| Redundancy | 6.97 (6.85) |
| Refinement | |
| Resolution (Å) | 46.36–2.20 |
| Unique reflections | 16,066 |
| Rwork/Rfree (%) | 23.4/25.4 |
| No. of atoms | |
| Protein | 1811 |
| Water | 42 |
| Root mean square deviations | |
| Bond lengths | 0.002 |
| Bond angles | 0.460 |
| Ramachandran plot (%) | |
| Favored | 99.54 |
| Allowed | 0.46 |
| Disallowed | 0 |
Analytical gel filtration of AFF4-CHD and MBP–AFF1-CHD
Analytical gel filtration study with purified WT and mutated AFF4-CHD and MBP–AFF1-CHD proteins were performed with ÄKTAmicro FPLC using Superdex 200 3.2/300 increase column (GE Healthcare) in buffer containing 20 mm HEPES, pH 7.4, 300 mm NaCl, 4% glycerol, and 1 mm DTT. For the study of heterodimerization of AFF1 and AFF4, MBP–AFF1-CHD and AFF4-CHD were mix in different molar ratios, 1:0, 1:1, 1:3, and 1:5 in a 50-μl system and incubated overnight at 4 °C. To compare the interaction between MBP–AFF1-CHD and WT and mutants of AFF4-CHD, MBP–AFF1-CHD, and AFF4-CHD, proteins were mixed in 1:3 ratios and incubated at 4 °C overnight. The premixed protein solution was injected onto the gel filtration column, and the elution fractions were collected and analyzed on SDS-PAGE.
To estimate the molecular masses of proteins, the analytical Superdex 200 3.2/300 increase column (GE Healthcare) was calibrated with size exclusion standards ferritin (440 kDa), aldolase (158 kDa), conalbumin (75 kDa), ovalbumin (44 kDa), and carbonic anhydrase (29 kDa) (GE Healthcare and Sigma–Aldrich) at a flow rate of 0.03 ml/min on ÄKTAmicro FPLC (GE Healthcare) in buffer containing 20 mm HEPES, pH 7.4, 300 mm NaCl, 4% glycerol, and 1 mm DTT. The Log molecular mass of each standard was plotted against the elution volume of the very peak of the elution profile using GraphPad Prism. The correlation between molecular mass and elution volume were calculated by fitting the data with the linear regression equation: y = −2.3257x + 8.374, in which y is the Log molecular mass, and x is the elution volume. The molecular masses of AFF4-CHD–WT and AFF4-CHD–5MD were calculated based on the elution volume and the equation.
Static light scattering
The size exclusion chromatogram coupled static light scattering experiments were performed using Superdex 200 10/300 increase column (GE Healthcare) and VISCOTEK 305 TDA detector (Malvern) in buffer containing 20 mm HEPES, pH 7.4, 300 mm NaCl, 4% glycerol, and 1 mm DTT at a flow rate of 0.5 ml/min. The scattering of proteins was measured at 90° (right-angle light scattering) and 7° (low-angle light scattering). The system was first calibrated with BSA. Then samples including AFF4-CHD–WT, AFF4-CHD–5MD, MBP–AFF1-CHD, and preincubated MBP–AFF1-CHD/AFF4-CHD–WT were sequentially measured using the same setup. The data were evaluated with OmniSEC 5.12 software (Malvern) and plotted using GraphPad Prism.
Fluorescence anisotropy
The fluorescence anisotropy experiments were performed using purified AFF4-CHD protein and 5′-FAM–labeled RNA (Table S1) and DNA oligonucleotides in the assay buffer containing 50 mm HEPES, pH 7.4, 1 mm MgCl2, 1 mm EDTA, 1 mm DTT, 10 μg of BSA, 5 μg of yeast tRNA, and 50, 100, or 200 mm KCl. The experimental procedures and buffer condition are modified based on previous literature (24, 61). Briefly, the 5′-FAM–labeled nucleotides were diluted to 75 nm in the assay buffer, and the protein was diluted by a factor of 2 ranging from 40 nm to 40 μm with the assay buffer. 7.5 μl of protein and 4 μl of nucleotides were mixed and incubated on ice for 10 min. 17.5 μl of assay buffer was added to the mixture and incubated for another 20 min at room temperature. The final assay condition contains 10 nm labeled RNA and 5 nm (Fig. 3C) or 10 nm to 10 μm protein in 30 μl of volume. 18 μl of the assay solution was measured with Plate Reader (Infinite M1000pro, TECAN) at an excitation wavelength of 470 ± 5 nm, an emission wavelength of 518 ± 20 nm, a Z position of 26,050 μm, and gain of 80 at 25 °C. Δanisotropy was calculated by subtracting the isotropy of DNA or RNA in buffer alone from the isotropy of the DNA or RNA in the presence of AFF4-CHD. The curves were fitted with quadratic single-site binding equation: Y = Bmax ((([L] + [P] + Kd,app) − Sqrt (([L] + [P] + Kd,app)2 − 4[L][P]))/2[L]) in GraphPad Prism, where Bmax is the maximum specific binding, L is the DNA or RNA concentration, P is the concentration of AFF4-CHD, and Kd,app is the apparent dissociation constant for AFF4-CHD and DNA or RNA.
Phosphorylation assays of AFF4-CHD
The AFF4-CHD was phosphorylated by the kinase complex P-TEFb. To enhance the solubility of P-TEFb, a fusion construct of human CYCT1 (1–272) and HIV-1 TAT (1–72) was made by PCR. A 5-amino acid linker (Gly-Gly-Gly-Ser-Gly) was inserted between CYCT1 and TAT. The resulting gene fusion was inserted into the pSPL vector (62, 63) using the XbaI and StuI restriction sites. Human CDK9 (1–372) bearing an N-terminal His8 tag was cloned into pACEBAC1 as described (5). The pSPL and pACEBAC vectors were combined using Cre recombination (62). Clones were screened to select for single recombination events, which result in a P-TEFb–5aa–TAT construct. The WT P-TEFb–5aa–TAT and the catalytic mutant of P-TEFb–5aa–TAT-N (D149N) were expressed and purified as described in a previous study (5).
The phosphorylation assays of AFF4-CHD were performed in a 20-μl system based on a previous study (5). Briefly, 0.5 μl of AFF4-CHD (675 μm stock) or 0.8 μl of DSIF (48.5 μm) was mixed with 0.5 μl of purified P-TEFb (40 μm stock) and 19 μl of phosphorylation buffer (20 mm HEPES, pH 7.4, 100 mm NaCl, 3 mm MgCl2, 1 mm DTT, 4% glycerol) and incubated for 5 min at 30 °C. 2 μl of sample was taken prior ATP addition as 0 min. 0.4 μl of ATP (50 mm stock) was added to the assay solution and further incubated for a time course of 2 h, and 2 μl of sample was taken at the 30-min, 60-min, and 2-h time points (5). The samples were loaded onto SDS-PAGE and run with MOPS buffer for 42 min at 200 kV (Invitrogen). The time course was repeated three times, and the final products of the triplicates were loaded onto a 10-well SDS-PAGE.
To produce the phosphorylated AFF4-CHD protein for fluorescence anisotropy, 4 μl of AFF4-CHD (675 μm stock) were incubated with either P-TEFb–5aa–TAT or the catalytic mutant of P-TEFb–5aa–TAT (D149N) as described above for 2 h. 0.5 μl of the final product was checked on SDS-PAGE to validate the phosphorylation. The phosphorylated or nonphosphorylated AFF4-CHD was diluted to 40 μm and mixed with nucleic acids as indicated above.
Identification of the phosphorylation sites
As described above, the final products of the three time-course experiments were analyzed on SDS-PAGE. Phosphopeptides obtained from in-gel digest of the protein band were enriched and analyzed as previously described and the MS/MS spectra were searched against the human database with the Andromeda search engine (5, 64). The phosphorylation sites reported in this study were present in all the three replicates with a localization probability higher than 0.99.
Author contributions
Y. C. and P. C. conceptualization; Y. C. data curation; Y. C. and P. C. formal analysis; Y. C. and P. C. validation; Y. C. and P. C. investigation; Y. C. and P. C. visualization; Y. C. and P. C. methodology; Y. C. writing-original draft; Y. C. and P. C. writing-review and editing; P. C. resources; P. C. software; P. C. supervision; P. C. funding acquisition; P. C. project administration.
Supplementary Material
Acknowledgments
We thank Seychelle Vos for providing the plasmids and purification protocol of the P-TEFb–5aa–TAT and P-TEFb–5aa–TAT-D149N and for suggestions on this work, Uwe Plessmann and Sabine Koenig for the MS data, and Hauke Hillen for advice and discussions. We thank Juergen Wawrzinek and Ulrich Steuerwald for managing the in-house X-ray crystallography facility and the Swiss Light Source at the Paul-Scherrer-Institut in Villigen, Switzerland, for data collection.
This work was supported by Deutsche Forschungsgemeinschaft Grants SFB860 and SPP1935, by European Research Council Advanced Investigator Grant TRANSREGULON under Agreement 693023, and by the Volkswagen Foundation (to P. C.). The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (code 6R80) have been deposited in the Protein Data Bank (http://wwpdb.org/).
This article contains Tables S1 and S2 and Figs. S1–S3.
- Pol
- RNA polymerase
- CHD
- C-terminal homology domain
- SEC
- super elongation complex
- TPR
- Tetratrico Peptide Repeat
- CTD
- C-terminal domain
- FAM
- Fluorescein amidite.
References
- 1. Hahn S., and Young E. T. (2011) Transcriptional regulation in Saccharomyces cerevisiae: transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics 189, 705–736 10.1534/genetics.111.127019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li J., and Gilmour D. S. (2011) Promoter proximal pausing and the control of gene expression. Curr. Opin. Genet. Dev. 21, 231–235 10.1016/j.gde.2011.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sainsbury S., Bernecky C., and Cramer P. (2015) Structural basis of transcription initiation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 16, 129–143 10.1038/nrm3952 [DOI] [PubMed] [Google Scholar]
- 4. Jonkers I., and Lis J. T. (2015) Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 16, 167–177 10.1038/nrm3953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vos S. M., Farnung L., Boehning M., Wigge C., Linden A., Urlaub H., and Cramer P. (2018) Structure of activated transcription complex Pol II–DSIF–PAF–SPT6. Nature 560, 607–612 10.1038/s41586-018-0440-4 [DOI] [PubMed] [Google Scholar]
- 6. Vos S. M., Farnung L., Urlaub H., and Cramer P. (2018) Structure of paused transcription complex Pol II–DSIF–NELF. Nature 560, 601–606 10.1038/s41586-018-0442-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. He N., and Zhou Q. (2011) New insights into the control of HIV-1 transcription: when Tat meets the 7SK snRNP and super elongation complex (SEC). J. Neuroimmune Pharmacol. 6, 260–268 10.1007/s11481-011-9267-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lu H., Li Z., Xue Y., Schulze-Gahmen U., Johnson J. R., Krogan N. J., Alber T., and Zhou Q. (2014) AFF1 is a ubiquitous P-TEFb partner to enable Tat extraction of P-TEFb from 7SK snRNP and formation of SECs for HIV transactivation. Proc. Natl. Acad. Sci. U.S.A. 111, E15–E24 10.1073/pnas.1318503111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mück F., Bracharz S., and Marschalek R. (2016) DDX6 transfers P-TEFb kinase to the AF4/AF4N (AFF1) super elongation complex. Am. J. Blood Res. 6, 28–45 [PMC free article] [PubMed] [Google Scholar]
- 10. Luo Z., Lin C., and Shilatifard A. (2012) The super elongation complex (SEC) family in transcriptional control. Nat. Rev. Mol. Cell Biol. 13, 543–547 10.1038/nrm3417 [DOI] [PubMed] [Google Scholar]
- 11. Chou S., Upton H., Bao K., Schulze-Gahmen U., Samelson A. J., He N., Nowak A., Lu H., Krogan N. J., Zhou Q., and Alber T. (2013) HIV-1 Tat recruits transcription elongation factors dispersed along a flexible AFF4 scaffold. Proc. Natl. Acad. Sci. U.S.A. 110, E123–E131 10.1073/pnas.1216971110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hill C. P., and Sundquist W. I. (2013) Building a super elongation complex for HIV. Elife 2, e00577 10.7554/eLife.00577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schulze-Gahmen U., Lu H., Zhou Q., and Alber T. (2014) AFF4 binding to Tat-P-TEFb indirectly stimulates TAR recognition of super elongation complexes at the HIV promoter. Elife 3, e02375 10.7554/eLife.02375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kuzmina A., Krasnopolsky S., and Taube R. (2017) Super elongation complex promotes early HIV transcription and its function is modulated by P-TEFb. Transcription 8, 133–149 10.1080/21541264.2017.1295831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lin C., Smith E. R., Takahashi H., Lai K. C., Martin-Brown S., Florens L., Washburn M. P., Conaway J. W., Conaway R. C., and Shilatifard A. (2010) AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol. Cell 37, 429–437 10.1016/j.molcel.2010.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shilatifard A., Lane W. S., Jackson K. W., Conaway R. C., and Conaway J. W. (1996) An RNA polymerase II elongation factor encoded by the human ELL gene. Science 271, 1873–1876 10.1126/science.271.5257.1873 [DOI] [PubMed] [Google Scholar]
- 17. He N., Chan C. K., Sobhian B., Chou S., Xue Y., Liu M., Alber T., Benkirane M., and Zhou Q. (2011) Human polymerase-associated factor complex (PAFc) connects the super elongation complex (SEC) to RNA polymerase II on chromatin. Proc. Natl. Acad. Sci. U.S.A. 108, E636–E645 10.1073/pnas.1107107108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Qi S., Li Z., Schulze-Gahmen U., Stjepanovic G., Zhou Q., and Hurley J. H. (2017) Structural basis for ELL2 and AFF4 activation of HIV-1 proviral transcription. Nat. Commun 8, 14076 10.1038/ncomms14076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schulze-Gahmen U., and Hurley J. H. (2018) Structural mechanism for HIV-1 TAR loop recognition by Tat and the super elongation complex. Proc. Natl. Acad. Sci. U.S.A. 115, 12973–12978 10.1073/pnas.1806438115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schulze-Gahmen U., Upton H., Birnberg A., Bao K., Chou S., Krogan N. J., Zhou Q., and Alber T. (2013) The AFF4 scaffold binds human P-TEFb adjacent to HIV Tat. Elife 2, e00327 10.7554/eLife.00327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leach B. I., Kuntimaddi A., Schmidt C. R., Cierpicki T., Johnson S. A., and Bushweller J. H. (2013) Leukemia fusion target AF9 is an intrinsically disordered transcriptional regulator that recruits multiple partners via coupled folding and binding. Structure 21, 176–183 10.1016/j.str.2012.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gu J., Babayeva N. D., Suwa Y., Baranovskiy A. G., Price D. H., and Tahirov T. H. (2014) Crystal structure of HIV-1 Tat complexed with human P-TEFb and AFF4. Cell Cycle 13, 1788–1797 10.4161/cc.28756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schulze-Gahmen U., Echeverria I., Stjepanovic G., Bai Y., Lu H., Schneidman-Duhovny D., Doudna J. A., Zhou Q., Sali A., and Hurley J. H. (2016) Insights into HIV-1 proviral transcription from integrative structure and dynamics of the Tat:AFF4:P-TEFb:TAR complex. Elife 5, pii: e15910 10.7554/eLife.15910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Melko M., Douguet D., Bensaid M., Zongaro S., Verheggen C., Gecz J., and Bardoni B. (2011) Functional characterization of the AFF (AF4/FMR2) family of RNA-binding proteins: insights into the molecular pathology of FRAXE intellectual disability. Hum. Mol. Genet. 20, 1873–1885 10.1093/hmg/ddr069 [DOI] [PubMed] [Google Scholar]
- 25. Yokoyama A., Lin M., Naresh A., Kitabayashi I., and Cleary M. L. (2010) A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell 17, 198–212 10.1016/j.ccr.2009.12.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lu H., Li Z., Zhang W., Schulze-Gahmen U., Xue Y., and Zhou Q. (2015) Gene target specificity of the super elongation complex (SEC) family: how HIV-1 Tat employs selected SEC members to activate viral transcription. Nucleic Acids Res. 43, 5868–5879 10.1093/nar/gkv541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jones D. T. (1999) Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202 10.1006/jmbi.1999.3091 [DOI] [PubMed] [Google Scholar]
- 28. Buchan D. W., Minneci F., Nugent T. C., Bryson K., and Jones D. T. (2013) Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 41, W349–W357 10.1093/nar/gkt381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Holm L., and Laakso L. M. (2016) Dali server update. Nucleic Acids Res. 44, W351–W355 10.1093/nar/gkw357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang S., Chang L., Alfieri C., Zhang Z., Yang J., Maslen S., Skehel M., and Barford D. (2016) Molecular mechanism of APC/C activation by mitotic phosphorylation. Nature 533, 260–264 10.1038/nature17973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Smidova A., Alblova M., Kalabova D., Psenakova K., Rosulek M., Herman P., Obsil T., and Obsilova V. (2018) 14-3-3 protein masks the nuclear localization sequence of caspase-2. FEBS J. 285, 4196–4213 10.1111/febs.14670 [DOI] [PubMed] [Google Scholar]
- 32. Pan Z., Zhu J., Shang Y., Wei Z., Jia M., Xia C., Wen W., Wang W., and Zhang M. (2013) An autoinhibited conformation of LGN reveals a distinct interaction mode between GoLoco motifs and TPR motifs. Structure 21, 1007–1017 10.1016/j.str.2013.04.005 [DOI] [PubMed] [Google Scholar]
- 33. Stevers L. M., de Vink P. J., Ottmann C., Huskens J., and Brunsveld L. (2018) A thermodynamic model for multivalency in 14-3-3 protein–protein interactions. J. Am. Chem. Soc. 140, 14498–14510 10.1021/jacs.8b09618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Karpenahalli M. R., Lupas A. N., and Söding J. (2007) TPRpred: a tool for prediction of TPR-, PPR- and SELI-like repeats from protein sequences. BMC Bioinformatics 8, 2 10.1186/1471-2105-8-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zimmermann L., Stephens A., Nam S. Z., Rau D., Kübler J., Lozajic M., Gabler F., Söding J., Lupas A. N., and Alva V. (2018) A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 430, 2237–2243 10.1016/j.jmb.2017.12.007 [DOI] [PubMed] [Google Scholar]
- 36. Yang X., Lee W. H., Sobott F., Papagrigoriou E., Robinson C. V., Grossmann J. G., Sundström M., Doyle D. A., and Elkins J. M. (2006) Structural basis for protein-protein interactions in the 14–3-3 protein family. Proc. Natl. Acad. Sci. U.S.A. 103, 17237–17242 10.1073/pnas.0605779103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Obsil T., and Obsilova V. (2011) Structural basis of 14-3-3 protein functions. Semin. Cell Dev. Biol. 22, 663–672 10.1016/j.semcdb.2011.09.001 [DOI] [PubMed] [Google Scholar]
- 38. Krissinel E., and Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 10.1016/j.jmb.2007.05.022 [DOI] [PubMed] [Google Scholar]
- 39. Krissinel E. (2010) Crystal contacts as nature's docking solutions. J. Comput Chem. 31, 133–143 10.1002/jcc.21303 [DOI] [PubMed] [Google Scholar]
- 40. Daub H., Olsen J. V., Bairlein M., Gnad F., Oppermann F. S., Körner R., Greff Z., Kéri G., Stemmann O., and Mann M. (2008) Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol. Cell 31, 438–448 10.1016/j.molcel.2008.07.007 [DOI] [PubMed] [Google Scholar]
- 41. Oppermann F. S., Gnad F., Olsen J. V., Hornberger R., Greff Z., Kéri G., Mann M., and Daub H. (2009) Large-scale proteomics analysis of the human kinome. Mol. Cell. Proteomics 8, 1751–1764 10.1074/mcp.M800588-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Olsen J. V., Vermeulen M., Santamaria A., Kumar C., Miller M. L., Jensen L. J., Gnad F., Cox J., Jensen T. S., Nigg E. A., Brunak S., and Mann M. (2010) Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3, ra3 [DOI] [PubMed] [Google Scholar]
- 43. Zhou H., Di Palma S., Preisinger C., Peng M., Polat A. N., Heck A. J., and Mohammed S. (2013) Toward a comprehensive characterization of a human cancer cell phosphoproteome. J. Proteome Res. 12, 260–271 10.1021/pr300630k [DOI] [PubMed] [Google Scholar]
- 44. Xue Y., Zhou F., Zhu M., Ahmed K., Chen G., and Yao X. (2005) GPS: a comprehensive www server for phosphorylation sites prediction. Nucleic Acids Res. 33, W184–W187 10.1093/nar/gki393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xue Y., Ren J., Gao X., Jin C., Wen L., and Yao X. (2008) GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol. Cell. Proteomics 7, 1598–1608 10.1074/mcp.M700574-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xue Y., Liu Z., Cao J., Ma Q., Gao X., Wang Q., Jin C., Zhou Y., Wen L., and Ren J. (2011) GPS 2.1: enhanced prediction of kinase-specific phosphorylation sites with an algorithm of motif length selection. Protein Eng. Des. Sel. 24, 255–260 10.1093/protein/gzq094 [DOI] [PubMed] [Google Scholar]
- 47. Czudnochowski N., Bösken C. A., and Geyer M. (2012) Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition. Nat. Commun. 3, 842 10.1038/ncomms1846 [DOI] [PubMed] [Google Scholar]
- 48. Lu X., Zhu X., Li Y., Liu M., Yu B., Wang Y., Rao M., Yang H., Zhou K., Wang Y., Chen Y., Chen M., Zhuang S., Chen L. F., Liu R., and Chen R. (2016) Multiple P-TEFbs cooperatively regulate the release of promoter-proximally paused RNA polymerase II. Nucleic Acids Res. 44, 6853–6867 10.1093/nar/gkw571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Okuda H., Kanai A., Ito S., Matsui H., and Yokoyama A. (2015) AF4 uses the SL1 components of RNAP1 machinery to initiate MLL fusion- and AEP-dependent transcription. Nat. Commun. 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Winters A. C., and Bernt K. M. (2017) MLL-rearranged leukemias: an update on science and clinical approaches. Front. Pediatr. 5, 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dephoure N., Zhou C., Villén J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., and Gygi S. P. (2008) A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 105, 10762–10767 10.1073/pnas.0805139105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Aslanidis C., and de Jong P. J. (1990) Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res. 18, 6069–6074 10.1093/nar/18.20.6069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Aslanidis C., de Jong P. J., and Schmitz G. (1994) Minimal length requirement of the single-stranded tails for ligation-independent cloning (LIC) of PCR products. PCR Methods Appl. 4, 172–177 10.1101/gr.4.3.172 [DOI] [PubMed] [Google Scholar]
- 54. Doublie S. (1997) Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 276, 523–530 10.1016/S0076-6879(97)76075-0 [DOI] [PubMed] [Google Scholar]
- 55. Kabsch W. (2010) XDS. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Karplus P. A., and Diederichs K. (2015) Assessing and maximizing data quality in macromolecular crystallography. Curr. Opin Struct. Biol. 34, 60–68 10.1016/j.sbi.2015.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sheldrick G. M. (2010) Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr. D 66, 479–485 10.1107/S0907444909038360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 60. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vos S. M., Pollmann D., Caizzi L., Hofmann K. B., Rombaut P., Zimniak T., Herzog F., and Cramer P. (2016) Architecture and RNA binding of the human negative elongation factor. Elife 5, pii: e14981 10.7554/eLife.14981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fitzgerald D. J., Berger P., Schaffitzel C., Yamada K., Richmond T. J., and Berger I. (2006) Protein complex expression by using multigene baculoviral vectors. Nat. Methods 3, 1021–1032 10.1038/nmeth983 [DOI] [PubMed] [Google Scholar]
- 63. Berger I., Fitzgerald D. J., and Richmond T. J. (2004) Baculovirus expression system for heterologous multiprotein complexes. Nat. Biotechnol. 22, 1583–1587 10.1038/nbt1036 [DOI] [PubMed] [Google Scholar]
- 64. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. J. Proteome Res. 10, 1794–1805 10.1021/pr101065j [DOI] [PubMed] [Google Scholar]
- 65. Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., and Barton G. J. (2009) Jalview version 2: a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Robert X., and Gouet P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 10.1093/nar/gku316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Holst M. J., and Saied F. (1995) Numerical-solution of the nonlinear Poisson–Boltzmann equation: developing more robust and efficient methods. J. Comput. Chem. 16, 337–364 10.1002/jcc.540160308 [DOI] [Google Scholar]
- 68. Oberoi H., and Allewell N. M. (1993) Multigrid solution of the nonlinear Poisson–Boltzmann equation and calculation of titration curves. Biophys. J. 65, 48–55 10.1016/S0006-3495(93)81032-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.