Abstract
In systemic lupus erythematosus (SLE), defective clearance of apoptotic debris and activation of innate cells result in a chronically activated type 1 interferon (T1 IFN) response, which can be measured in peripheral blood mononuclear cells (PBMCs) of most patients. Metformin, a widely used prescription drug for Type 2 Diabetes, has a therapeutic effect in several mouse models of lupus through mechanisms involving inhibition of oxidative phosphorylation and a decrease in CD4+ T cell activation. Here, we report that in CD4+ T cells from human healthy controls and human SLE patients, metformin inhibits the transcription of interferon stimulated genes (ISGs) after IFNα treatment. Accordingly, metformin inhibited the phosphorylation of pSTAT1 (Y701) and its binding to interferon stimulated response elements (ISRE) that control ISG expression. These effects were independent of AMPK activation or mTORC1 inhibition, but were replicated using inhibitors of the electron transport chain respiratory complexes I, III, and IV. This indicates that mitochondrial respiration is required for ISG expression in CD4+ T cells, and provides a novel mechanism by which metformin may exert a beneficial therapeutic effect in autoimmune diseases.
Introduction
Systemic lupus erythematosus (SLE) is an autoimmune disease of complex etiology. At least half of SLE patients have elevated levels of non-virally induced expression of type 1 interferon (T1 IFN) stimulated genes in peripheral blood mononuclear cells (PBMCs) (1–3). This phenomenon has been termed the “interferon signature of lupus” (4). There are many contributors to the increased amount of T1 IFN in SLE patients, including familial high serum IFNα levels (5). However, a major cause is the chronic inflammation resulting from defective clearing of apoptotic cells, which triggers TLR7 and TLR9 activation as well as FCγR signaling on plasmacytoid dendritic cells (pDC), which subsequently produce T1 IFN (4; 6). B cells from non-autoimmune healthy controls (HC) or SLE patients can also produce IFNα after TLR9 stimulation (7) and IFNβ after TLR3, TLR7, or TLR9 stimulation (8). IFNα signaling impairs the generation of FOXP3hi regulatory T cells (Treg) and increased the frequency of Th1 cells, as suggested by the effect of SLE plasma on healthy PBMCs (9). IFNα also increases T cell chemotaxis and adhesion by inducing LFA-1 and VLA-4 clustering on the cell membrane (10).
Among new therapeutic approaches considered in SLE is a monoclonal antibody against the IFNAR1/2 receptor, Anifrolumab, which passed primary endpoints in a phase II trial (11). Patients stratified into a high IFN signature benefitted more than patients in the low IFN signature group. IFNα kinoid (a modified IFNα2b attached to a carrier protein) used to induce a pan anti-IFNα response, has also been tested, and exhibited a good safety profile (12). These results suggest that T1 IFN is a promising therapeutic target in SLE and warrants further investigation. In mouse models of SLE, the T1 IFN signature is not as prominent as observed in SLE patients, but it does play an important role. Deletion of Ifnar1, one of the genes encoding the IFN receptor dimer, dramatically reduces disease in pristane-induced lupus (13) as well as in NZB mice (14). In many spontaneous models, artificially increasing IFNα levels with virally induced expression or by other methods accelerates the development of autoimmune symptoms (4). In the NZM2410-derived triple congenic model (15), DCs display a strong interferon signature (16) and there is an increased frequency of PDCA1+ pDCs in the marginal zone (17).
We and others have shown that CD4+ T cells from SLE patients are characterized by an increased cellular metabolism, including increased oxidative phosphorylation (OXPHOS), glycolysis and mechanistic target of rapamycin kinase (mTOR) activation (18; 19). Accordingly, we have shown that treatment with a combination of two metabolic inhibitors, metformin and 2-deoxyglucose (2DG) reversed autoimmune pathology in several mouse models of lupus (18; 20). 2DG is a glucose analog that blocks hexokinase function at the first step of glycolysis. Metformin blocks complex I of the electron transport chain (ETC), therefore inhibiting OXPHOS and ATP production (21; 22), leading to AMP-activated protein kinase (AMPK) activation in numerous cell types (23). In cancer cells, it has been demonstrated that reduced proliferation and tumor volume in xenografts was directly related to complex I inhibition (22). Metformin treatment can also lead to suppression of mTORC1 activity (24), a nutrient sensing complex which is increased in T cells from SLE patients (25). Metformin reduced oxygen consumption, activation, and IFNγ production in CD4+ T cells of lupus mice and from HC and SLE patients in vitro, as well as in vivo in lupus mice (18). In a trial as add-on therapy in SLE patients, metformin showed a significant steroid-sparing effect (26). Mechanistically, metformin inhibited IFNα production from pDCs stimulated with either CpG or mitochondrial DNA (26). The IFN signature was not examined in this study, but a significant decrease in IFNα production suggests that metformin would prevent autocrine or paracrine ISG expression. Currently, the mechanisms by which metformin reduced the inflammatory function of lupus CD4+ T cells and pDCs have not been reported.
The goal of this study was to investigate the mechanisms by which metformin affects primary human lymphocytes from SLE patients as well as healthy subjects by transcriptional profiling analysis. We found that metformin inhibited ISG expression in CD4+ T cells but not in CD19+ B cells. Metformin decreased signal transducer and activator of transcription 1 (STAT1) activation and binding to the promoter of ISGs. This effect was independent of AMPK and mTOR activation. Rotenone, antimycin-a, and oligomycin inhibited ISG expression to a similar degree as metformin, indicating a role for the involvement of the ETC in T1 IFN signaling or ISG expression. These results demonstrate a novel role of the ETC in the T1 IFN response in CD4+ T cells, and suggest that it may function as a mechanism by which metformin reduces T1 IFN inflammation in SLE.
Materials and Methods
Recruitment of HC and SLE patients
Peripheral blood samples from five HC and five SLE patients (Table 1, microarray group) were obtained following written consent according to protocols approved by the University of Florida institutional review board (IRB201300125). SLE patients were recruited from outpatients who fulfilled 4 of 11 SLE ACR 1997 criteria and were on regular visits to the UF medical specialties clinic. Samples were collected in heparinized sodium tubes (BD Biosciences) HC subjects were selected to be female and have similar racial distribution as SLE patients (Table 1). CD4+ T cells were isolated using RosetteSep CD4+ T cell isolation cocktail (StemCell) with about 80% post-isolation purity (Supp. Fig. 1A) based on CD4+ flow cytometry evaluation (OKT4, BioLegend), compared with 12% in unfractionated whole blood. Wells were pre-coated with 1 ug/mL anti-CD3ε (UCHT1, BD Biosciences) for 8 h at 37°C in PBS which was removed prior to cell culture. CD4+ T cells were cultured for 24 h (2 × 106 per well) with 1 ug/mL anti-CD28 (L293 BD biosciences), with or without 1.0 mM metformin (Sigma) in complete RPMI 1640 media (Corning) with antibiotic-antimycotic cocktail (Thermo Fisher) and 10% Fetal Bovine Serum (SAFC). Cells were harvested and RNA isolated with RNeasy Mini Kit (Qiagen).
Table 1.
Demographics of SLE patients and HC donors of CD4+ T cells for microarray and qRT-PCR experiments.
| Microarray | qRT-PCR | |||
|---|---|---|---|---|
| SLE | HC | SLE | HC | |
| Female # | 5 | 5 | 5 | 5 |
| Age mean (range) | 60.6 (52 to 70) | 34.8 (23 to 43) | 32.2 (25 to 39) | 32.0 (26 to 38) |
| African American | 3 (60%) | 3 (60%) | 5 (100%) | 5 (100%) |
| Caucasian | 2 (40%) | 2 (40%) | 0 | 0 |
| SLEDAI avg. (range) | 1.8 (from 0–8) | N/A | 3.6 (0 to 8) | N/A |
| Hydroxychloroquine | 5 (100%) | N/A | 4 (80%) | N/A |
| Mycophenolate Mofetil | 2 (40%) | N/A | 4 (80%) | N/A |
| Prednisone | 2 (40%) | N/A | 0 (0%) | N/A |
| Azathioprine | 1 (20%) | N/A | 0 (0%) | N/A |
| Tacrolimus | 0 (0%) | N/A | 1 (20%) | N/A |
CD4+ T cell Metformin Microarray
Global gene expression was analyzed by HTA 2.0 microarray chips (Affymetrix/ Thermo Fisher) with 250 ng input RNA used for hybridization targets. RNA amplification, fragmenting, and labeling was carried out according to manufacturer recommendations. Microarray data was analyzed with Partek Genomics Suite (v6.6-v7.0) software using the robust multi-array average (RMA) normalization procedure. After normalization, probes were averaged to obtain final expression values for each gene. To visualize group separation, principle component analysis (PCA) using all probes was performed. Two-factor ANOVA testing was performed based on SLE or HC status and metformin treatment. Pathway discovery analysis was performed on paired comparisons between metformin-treated and controls both for all samples and separately for HC and SLE samples with the Gene Set Enrichment Analysis program (GSEA, Broad Institute). Data is publically available on the Gene Expression Omnibus, accession number GSE119446. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE119446
QRT-PCR Analysis of Interferon Stimulated Gene Expression
Leukocyte fractions of peripheral blood from healthy donors (HD) were obtained through LifeSouth BloodBank (UF IRB approval IRB201700257). HD were not restricted by sex or race but were limited to individuals between 18 and 65 years old. PBMCs were isolated with gradient centrifugation using Ficoll-Paque (GE Healthcare). CD4+ T cells or CD19+ B cells were isolated with EasySep magnetic isolation following manufacturer provided protocols (StemCell). The purity of CD4+ T cells was typically > 90% and the purity of CD19+ B cells was > 95% (Supp. Fig. 1B and C). CD4+ T cells were cultured for 24 h in wells pre-coated with 1 ug/mL anti-CD3ε and 1ug/mL anti-CD28 stimulation in complete RPMI media, with or without 2 mM metformin. B cells were cultured in complete RPMI media. Cells were treated with 0.4 ng/mL (100 IU) human IFNα2a for the final 2 h or with 10 ng/mL of IL-2 (both from Shenandoah Biotech.) for the final 4 h. In some experiments, the anti-CD3/CD28 stimulated CD4+ T cells were treated with the combination of 2 mM metformin and IFNα for 2 h. RNA was isolated with RNeasy Mini Kit (Qiagen) and used for qRT-PCR with poly-T16 primed reverse transcription using ImProm II Reverse Transcriptase (Promega). Gene expression was quantified using SYBR Green Dye (Bio-RAD) on the BioRad CFX Connect system, using intron-skipping qRT-PCR primers (Supp. Table 1). The PCR thermo-cycling protocol was 30 s at 95°C, 30 s at 60°C, and 30 s at 72°C repeated for 40 cycles. Expression was calculated using the 2ΔCq method with difference in Cq values taken between the housekeeping gene PPIA against the gene of interest.
QRT-PCR-Based Interferon Score
An overall score to represent IFNα stimulation was calculated using a previously published method (27). The baseline expression level was calculated in unstimulated cells using 6 genes (MX1, ISG20, GBP1, IRF7, ISG15, OASL). CD4+ T cells from either anonymous HD or African American HC and SLE patients (Table 1, qRT-PCR group) were treated with 4 conditions: stimulation only (anti-CD3ε/antiCD28), stimulation with metformin, stimulation with IFNα, or stimulation with IFNα and metformin. For the control stimulation only group, the mean and standard deviation was calculated for each gene. Score values for each gene are a z-score calculated by subtracting the control group mean from each sample’s expression value, and then dividing by control group standard deviation. The final interferon score is the average of 6 genes. The same approach was used for experiments with rotenone, antimycin-a, and oligomycin.
Chromatin Immunoprecipitation
CD4+ T cells cultured in complete RPMI (2 × 106 per well) as described previously were stimulated with 0.4 ng/mL IFNα2a for 1 h, then immediately fixed for 10 min in 0.04% formaldehyde and sonicated using a Branson model SFX instrument for an average fragment size of 500 bp. Chromatin immunoprecipitation (CHiP) was carried out using the Millipore-Sigma agarose ChiP kit. DNA-protein complex were bound by antibody overnight using 1:300 dilution of anti-STAT1 (polyclonal, Cell Signaling Technologies) or 1:500 dilution of Rabbit IgG (Thermo Fisher) as control per sample. ChiP DNA was purified with chloroform:phenol:isoamyl alcohol (Sigma) and isopropanol centrifugation. Rabbit IgG or anti-STAT1 antibody precipitated fractions were compared to total input chromatin for each treatment. 1 uL of DNA was used for qPCR. Primers in a genomic area devoid of genes or regulatory features were designed in the USCS genome browser, and used as a non-specific control for baseline signal level. ISRE sites for STAT1:STAT2 consensus binding sequence YAGTTTC(A/T)YTTTYCC were obtained for MX1 (28) and were profiled in silico with the Jaspar database (http://jaspar.binf.ku.dk/) for ISG15. Primer sequences appear in Supp. Table 1.
Intracellular Flow Cytometry
CD4+ T cells were prepared for intracellular staining using the FOXP3/Transcription Factor Staining Buffer Set (Thermo Fisher), according to manufacturer instructions. Briefly, cells were permeabilized overnight and stained for 1 h with antibodies against AMPK (23A3, Cell Signaling), pAMPKα (40H9, Cell Signaling), p-ribosomal protein S6 (D57.2.2E, Cell Signaling), pSTAT5 Y694 (SRBCZX, eBioscience). For AMPK and pAMPKα, unlabeled primary antibodies were detected with secondary goat anti-rabbit H+L AF488 (Invitrogen). Data was acquired on a LSR Fortessa instrument and analyzed with FlowJo V10 (FlowJo LLC).
Western Blot
CD4+ T cells were cultured for 24 h in complete RPMI (2 × 106 per well) with anti-CD3ε/anti-CD28 stimulation with or without 2.0 or 4.0 mM metformin and 0.4 ng/mL IFNα2a stimulation during the final hour. To prepare the protein lysate, cells were pelleted and resuspended in 100 uL of passive lysis buffer (Promega) with protease inhibitor cocktail (Sigma-Aldrich) on ice for 20 min with occasional mixing. Tubes were spun at 10,000 rpm for 15 min to remove cell debris. Supernatant was used for Western blots in 1:1 ratio with 2x Laemmli Sample Buffer (BioRad). Blocking was performed with 5% milk in TBS buffer. Antibodies against AMPK (23A3), p-T172 AMPKα (40H9), STAT1 (polyclonal), p-Y701 STAT1 (58D6), p-rpS6 (polyclonal), LKB1 (27D10), pS428 LKB1 (C67A3), ACC (polyclonal), pS79 ACC (polyclonal), were purchased from Cell Signaling. Goat anti-rabbit IgG conjugated to HRP was used as the secondary antibody (Cell Signaling). Anti-GAPDH–HRP conjugate was used for loading control (FL-335, Santa Cruz Biotechnology). Signal was detected with chemiluminescent substrate (Immobilon, Millipore). For repeat measurements of different proteins on the same membrane, PVDF stripping buffer was applied for 30 min (Research Products International) and the membrane reblocked with 5% milk in TBS afterwards. Western blot results were quantified by densitometric analysis using ImageJ software (NIH, Bethesda).
Extracellular Metabolism Analysis
CD4+ T cells cultured with anti-CD3ε and anti-CD28 stimulation overnight with or without 1.0 mM metformin from 4 HD samples with 2 – 4 technical replicates per donor. 0.2 × 106 cells were analyzed with the Seahorse XF96 instrument (Agilent) using the mitochondrial stress test with addition of 1 μM oligomycin at 16 min, 1.25 μM CCCP at 56 min, and 1 μM Rotenone and Antimycin A at 88 min. Basal oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were calculated as the mean values of the first three readings before the addition of oligomycin.
mtROS and ATP measurement
Measurement of total cellular reactive oxygen species (ROS) was done using mitoSOX Red (Thermo Fisher/Invitrogen) staining of CD4+ T cells after culture at 37C for 30 minutes. Cells were analyzed by flow cytometry (PE channel) immediately after staining without fixation with formaldehyde.
Cellular ATP was measured with a firefly luciferase ATP kit (Thermo Fisher/Invitrogen). CD4+ T cells were lysed in passive lysis buffer (Promega) and incubated with substrate as indicated by the manufacturer protocol. Luminescence was detected using a Lumat LB 9507 device (Berthold Technologies).
Statistical Analysis
GraphPad Prism 7.0 was used for all statistical testing of gene expression and ChiP data. For paired analyses, paired t-tests were used with a t-test ratio correction. Microarray statistics were calculated in Partek Genomics Suite (v6.6–7.0). PCA analysis was performed with probe-summarized data after RMA normalization. Two-factor ANOVA was performed with SLE or HC status and well as metformin treatment as the factors and using the step-up FDR method for multiple test correction. GSEA software calculated p-values for pathway analysis using raw microarray expression values.
Results
Metformin and SLE Status Differently Affect the Human CD4+ T cell Transcriptional Signature
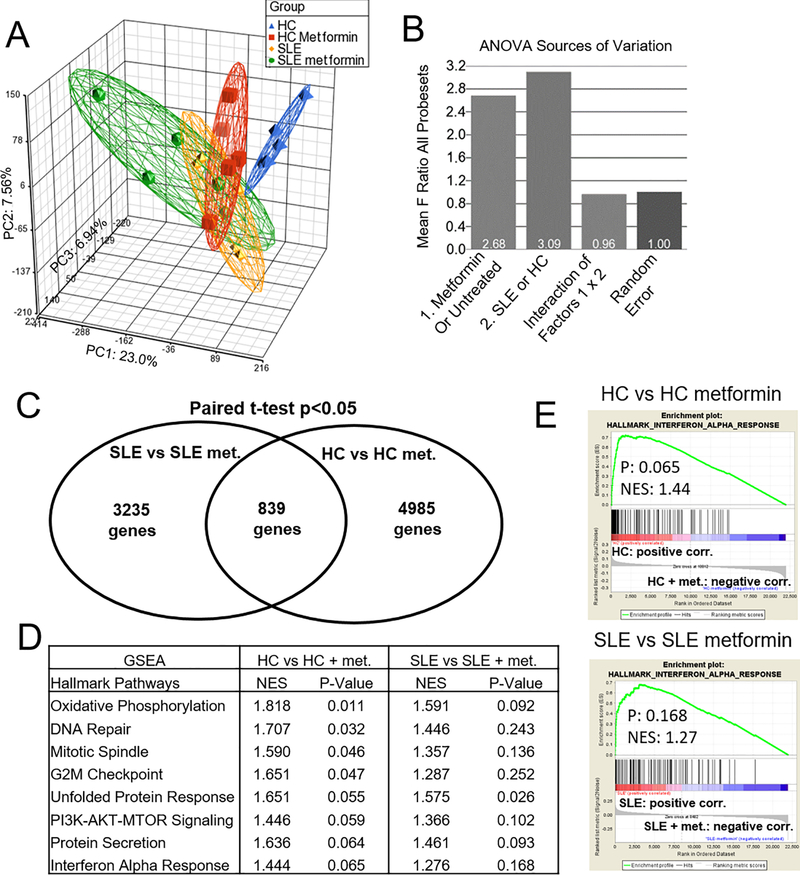
We evaluated the effect of metformin on CD4+ T cells from HC and SLE donors (Table 1) stimulated with anti-CD3ε and anti-CD28 by microarray analysis. PCA showed a distinct transcriptome between untreated SLE and HC CD4+ T cells, as well as an effect of metformin that was more pronounced in HC than in SLE cells (Fig. 1A). Two-factor ANOVA testing was performed based on HC or SLE status and presence of metformin. Analysis of factor contribution to variation showed that metformin treatment status or HC vs. SLE status had a similar contribution to the averaged F ratio, and these factors did not interact with each other significantly, as seen by an average F ratio of interaction less than random error (Fig. 1B). This suggested that metformin exerted an effect on both HC and SLE T cells. HC or SLE samples were compared to matched metformin treated samples by paired t-test to determine overlap of genes different at a p-value of <0.05 (Fig. 1C). 839 transcripts overlapped between the two groups while 3235 were unique to the effect of metformin on SLE cells and 4985 to the effect of metformin on HC cells. Random noise would be expected to result in approximately 3750 genes significant only by chance at an error rate of 0.05. Further statistical analysis was conducted to identify pathways discovered by the microarray for in depth validation.
FIGURE 1.
Metformin and SLE status affect the CD4+ T cell transcriptional signature. (A) PCA of CD4+ T cells from HC, SLE, and their paired metformin-treated samples (N = 5 each). (B) Two-way ANOVA analysis of source of variation using mean square of all genes divided by mean square error. (C) Comparison of genes significant at p<0.05 evaluated by paired t-test for HC vs HC metformin treated or SLE vs SLE metformin treated samples (D) Gene set enrichment analysis (GSEA) of pathways in HC vs. HC metformin-treated and SLE vs. SLE metformin-treated. (E) GSEA plots of the interferon alpha response pathway, HC vs. HC metformin-treated and SLE vs. SLE metformin-treated.
To analyze which pathways were affected by metformin in HC and in SLE CD4+ T cells, Gene Set Enrichment Analysis (GSEA) was performed separately for HC and SLE groups (Fig. 1D). The top differentially expressed pathways were similar in both groups and included metabolic pathways such as oxidative phosphorylation and mTOR signaling. This was consistent with the known functions of metformin and its previously reported effect on gene expression in murine hepatocytes and a human adrenal cell line (29; 30). DNA repair and two cell cycle pathways were also significantly affected by metformin, in agreement with the impaired cancer cell proliferation in the presence of metformin (31). Among the results there was also a group of genes categorized Interferon Alpha Response. Although the pathway p-values did not reach the 0.05 significance threshold, this may be due to a relatively short list of differentially expressed genes in the untreated vs. metformin treated HC group. We decided to pursue this pathway for validation because of the significance of type 1 IFN in lupus. Enrichment plots for both the HC and SLE groups showed positive curves skewing to the untreated group as compared with metformin treatment (Fig. 1E). These results suggested that in addition to metabolic changes, metformin reduced the level of ISG expression in CD4+ T cells.
Metformin Inhibits ISG Expression in CD4+ T Cells but not B cells
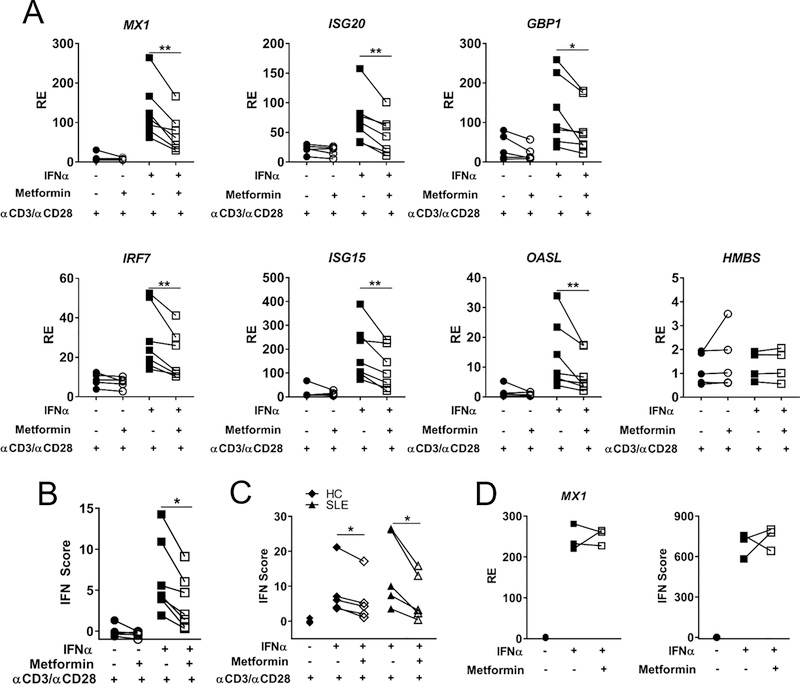
To validate whether metformin reduces ISG expression, CD4+ T cells were isolated from anonymous HD subjects and stimulated with anti-CD3ε/anti-CD28 and IFNα in the presence or absence of metformin. Six ISGs were chosen to represent the IFNα response, based on differently expressed genes in the microarray as well as ISGs known to be elevated in SLE PBMC (32). A range of IFNα concentrations were tested (Supp. Fig. 2A) and 100 IU/mL or 0.4 ng/mL was subsequently selected based on optimal ISG induction. 24 h treatment with metformin reduced the expression of all six ISGs stimulated for 2 h with IFNα as compared with control cells that were stimulated without metformin (Fig. 2A). Similar results were obtained with a 2 h co-treatment with metformin and IFNα (Supp. Fig. 2B), indicating that pre-treatment was not necessary for metformin’s inhibitory effect. The expression of the control gene hydroxymethylbilane synthase (HMBS) was unchanged by metformin or IFNα, indicating the effects on ISG expression were specific to IFNα signaling and that no global transcriptional inhibition was induced by metformin under these conditions. To summarize the degree of T1 IFN activation, an IFN score was calculated from the individual qRT-PCR results using an established method (27). Metformin significantly inhibited the IFN score in CD4+ T cells from HD (Fig. 2B) as well as in CD4+ T cells from African American SLE patients and matched HCs (Table 1 and Fig. 2C). As expected, ISG genes were expressed at significantly higher levels in ex-vivo CD4+ T cells from SLE patients than from HCs (data not shown). The IFN score after anti-CD3/CD28 plus IFNα stimulation as well as the level of inhibition by metformin were however similar between SLE patients and HCs (Fig. 2C). To test if this effect was specific to CD4+ T cells, CD19+ B cells were analyzed for ISG expression following the same conditions of metformin and IFNα treatment (Fig. 2D). Surprisingly, B cells showed no reduction in ISG expression after metformin treatment. This data suggests that metformin inhibits the activation of the T1 IFN signaling pathway in a manner which depends on cell-specific signaling or a metabolic profile specific to CD4+ T cells.
FIGURE 2.
Metformin Inhibits ISG Expression in CD4+ T Cells but not B cells. (A) CD4+ T cells from HD cultured with indicated treatments and analyzed for expression of 6 ISGs and 1 control gene HMBS. (B) IFN score for anonymous healthy donors. (C) IFN score for female African American SLE or HC T cells. (D) Expression of MX1 in CD19+ B cells from anonymous HDs and overall IFN score. Each pair of connected symbols represents an individual HD. Data were analyzed with paired t-tests, * p < 0.05, ** p <0.01. N = 7 anonymous HDs (A, B) or 3 in (D) and N = 5 for SLE and matched HC samples (C).
Metformin Reduces pSTAT1 Activation Independently of AMPK Activation Status
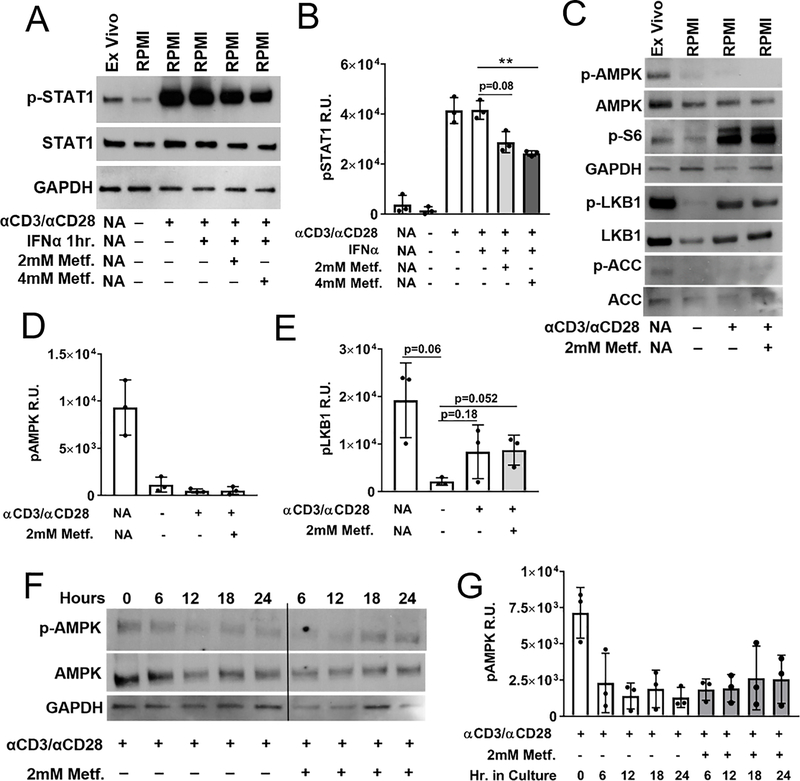
T1 IFN signaling is mediated by ISGF3, which is composed of pSTAT1, pSTAT2, and IRF9. To assess whether metformin inhibits activation of this complex, HD CD4+ T cells were analyzed for p-Y701 STAT1. Western blot analysis showed reduced levels of pSTAT1 in cells treated with metformin before IFNα stimulation (Fig. 3 A, B). This indicated that a reduced formation of the signaling complex downstream of IFNAR activation could be responsible for the decreased ISG expression caused by metformin.
FIGURE 3.
Metformin inhibits pSTAT1 activation independently of AMPK activation. Freshly isolated ex vivo HD CD4+ T cells were compared against 24 h cultured HD CD4+ T cells. Treatment conditions are indicated as + or −, either applied or not applied. (A) Representative Western blot analysis of HD CD4+ T cells for pSTAT1 (Y701), STAT1 or GAPDH as a loading control stimulated with the conditions indicated below. (B) Quantification of three separate Western blot results for pSTAT1 intensity. (C) Representative Western blot analysis of HD CD4+ T cells for pAMPKα (Thr172), AMPK, pLKB1 (S428), LKB1, pACC (S79), ACC, pS6, and GAPDH as a loading control. Quantification of three separate Western blot results for pAMPKα (D) and pLKB1 (E). (F) Representative time course of pS6 and pAMPKα expression with and without metformin with quantitation of three experiments shown in (G). Graphs show means and standard deviations compared with t-tests. * p < 0.05, ** p < 0.01.
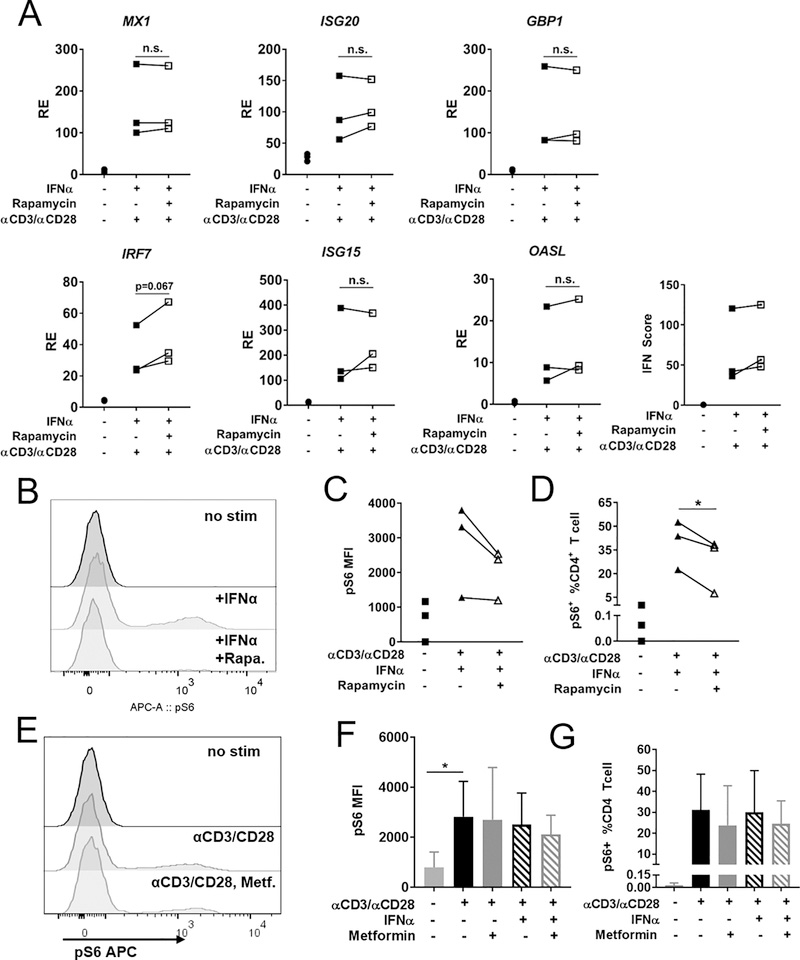
A known outcome of metformin treatment is the activation of serine/threonine kinase 11 (LKB1) kinase, which phosphorylates AMPKα subunit on Thr172 (23). A downstream target of activated AMPK is acetyl-coa carboxylase (ACC), which is considered a specific indicator of AMPK activity (33). Metformin can also repress mTORC1 by phosphorylating TSC2, and this activity has been reported to be independent of AMPK activation (34). PT172 AMPKα was detectable in ex vivo CD4+ T cells immediately following T cell isolation (Fig. 3C), but not after overnight stimulation with anti-CD3ε/anti-CD28, with or without metformin (Fig. 3D). LKB1 phosphorylation was also detectable immediately following T cell isolation, and CD3/CD28 stimulation was required to see activation in vitro. Metformin treatment did not increase pLKB1 (Fig. 3C, E). In agreement with pAMK expression, a weak signal for phosphorylated ACC was observed in ex vivo CD4+ T cells, but not in culture with and without metformin (Fig. 3C), further supporting minimal AMPK activation with or without metformin in our culture conditions. The glucose-rich RPMI media used in our assay probably compensated for the energy loss due to the ETC inhibition by metformin (35). Importantly, this result indicates that metformin inhibits ISG expression independently of AMPK activation. Ribosomal protein S6, a target of mTORC1 activation, was phosphorylated after anti-CD3ε/anti-CD28 stimulation, but was not reduced by metformin (Fig. 3C). This indicated that mTORC1 was not repressed by metformin in our assay conditions, and suggested that the expression of ISGs in CD4+ T cells is independent of mTORC1 activation. To confirm that mTORC1 activity was not required for ISG expression, CD4+ T cells were treated with rapamycin for 24 h and ISG expression was examined after IFNα stimulation (Fig. 4A). This dose of rapamycin (50 nM) decreased the pS6 signal as evaluated by flow cytometry (Fig. 4B–D). In contrast, metformin did not impact pS6 levels in CD4+ T cells (Fig. 4E–G). This further supported that mTORC1 inhibition does not play a role in ISG mRNA expression.
FIGURE 4.
Rapamycin does not reduce ISG expression. The effect of 24 h rapamycin treatment and anti-CD3ε/anti-CD28 stimulation was compared with final 2 h of IFNα stimulation (A) ISG expression in HD CD4+ T cells with or without 50 nM Rapamycin treatment. (B) Representative FACS plots of pS6 in CD4+ T cells in same conditions as described above. (C) Quantitation of pS6 MFI in CD4+ T cells as shown in (B). (D) Quantitation of pS6+ cells as % of total CD4+ T cells shown in (B). (E) Representative FACS plots of pS6 after metformin treatment. (F) Quantitation of pS6 MFI in CD4+ T cells shown in (E). (G) Quantitation of pS6+ cells as % of total CD4+ T cells shown in (E). Each linked symbol represents an individual donor (N = 3), analyzed by paired t-test.
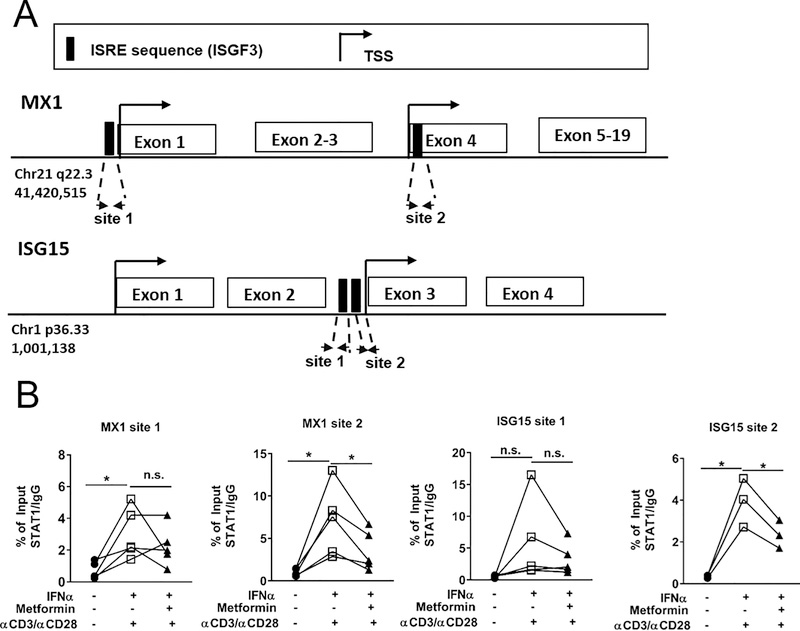
Metformin Prevents ISGF3 Binding to MX1 and ISG15 ISREs
To investigate whether metformin treatment prevented the recruitment and binding of the ISGF3 complex to ISGs, ChiP experiments were carried out on HD CD4+ T cells with an antibody against STAT1, a component of the ISGF3 complex. Two ISRE sites for MX1 (28) and two ISRE sites predicted for ISG15 were examined (Fig. 5A). Metformin significantly decreased STAT1 binding to MX1 and ISG15 sites 2, and a similar trend was observed for MX1 and ISG15 sites 1 (Fig. 5B). Taken together, these data demonstrate that treatment of CD4+ T cells with metformin before IFNα stimulation prevents full signaling through pathways that activate p-Y701 STAT1.
FIGURE 5.
Metformin prevents ISGF3 binding to MX1 and ISG15. (A) Analysis of human MX1 and ISG15 promoters showing putative ISGF3 binding sequences. (B) qPCR of ChiP assays against STAT1 for the MX1 and ISG15 ISRE sites on chromatin isolated from CD4+ T cells stimulated as indicated in legend below. Results shown as enrichment relative to non-specific IgG for each of the 3–5 HDs and analyzed with paired t-tests * p < 0.05, ** p < 0.01.
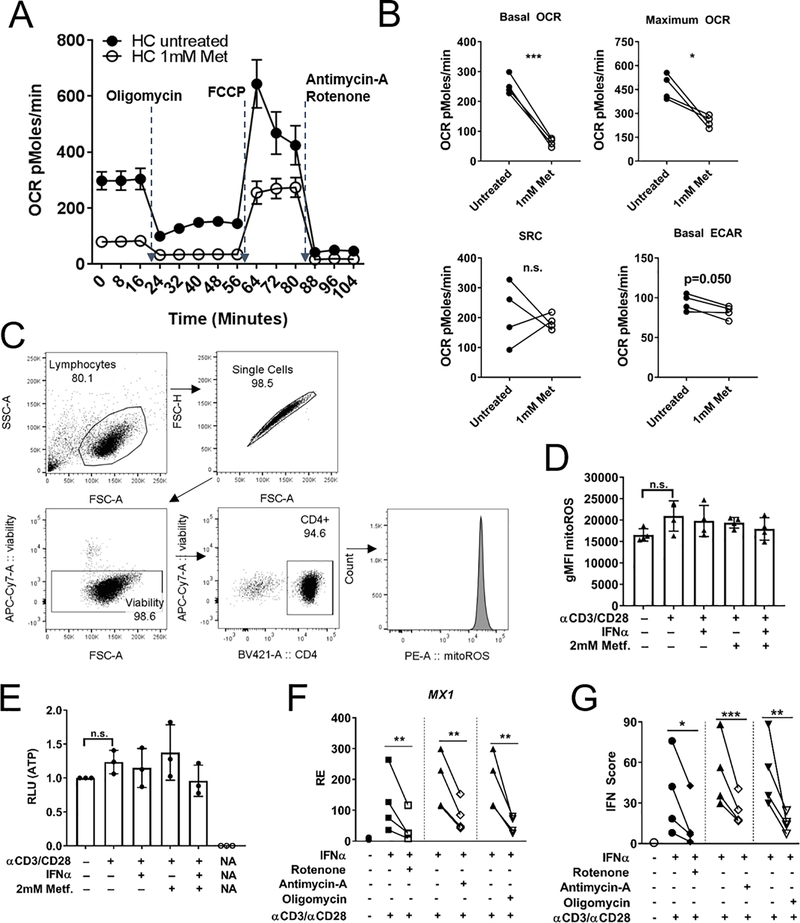
Mitochondrial ETC Inhibitors Reduce ISG expression in CD4+ T cells
To investigate whether inhibition of complex I of the ETC plays a role in reducing ISGF3 signaling during metformin treatment, CD4+ T cell OCR was measured by detection of extracellular metabolism with the Seahorse instrument. Metformin decreased basal and maximal OCR, but the spare respiratory capacity (SRC, the difference between maximal and basal respiration) was unchanged (Fig. 6A, B). These results are consistent with data demonstrating that metformin binds and inhibits complex 1 of the ETC (22). ECAR, a measure of glycolysis, showed a small decrease. The concentration of metformin used in our assays (up to 2 mM) did not affect cell viability (Supp. Fig. 1D). The inhibition of mitochondrial respiration by metformin was not sufficient to decrease mtROS (Fig. 6C and D) and ATP (Fig. 6E) production in HD CD4+ T cells after stimulation with anti-CD3/CD28 and IFNa, although there was a trend. This indicated that cellular energy homeostasis was maintained under the culture conditions.
FIGURE 6.
Mitochondrial ETC inhibitors reduce ISG expression in CD4+ T cells. (A) HD CD4+ T cells stimulated with anti-CD3ε/anti-CD28 with and without metformin analyzed with a mitochondrial stress test based on injecting metabolic inhibitors, oligomycin (1μM) inhibits ATP synthase, FCCP (1.25 μM) uncouples proton gradient, rotenone blocks complex I (1μM), antimycin-A blocks complex III (1μM). Symbols represent means and standard deviation for N = 4. (B) Summary data for OCR, SRC and ECAR for CD4+ T cells shown in (A). (C) Gating strategy for evaluating mtROS by mitoSOX red staining of cultured HD CD4+ T cells. (D) Summary of mtROS levels in CD4+ T cells. (E) ATP measurement in cultured HD CD4+ T cells quantified in luciferase relative light units (RLU). (F) MX1 expression in HD CD4+ T cells stimulated as in (C) and treated with indicated ETC inhibitors (all at 100nM). (G) Interferon score of HD CD4+ T cells treated with indicated ETC inhibitors. Paired t-tests shown for (B, E, F) * p<0.05, ** p<0.01.
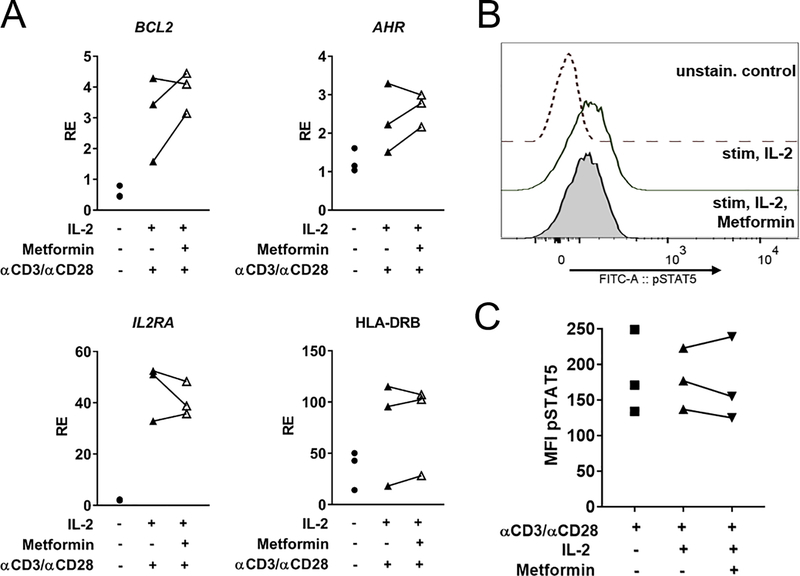
To test whether inhibition of the other ETC complexes would also inhibit IFNα signaling, the metabolic inhibitors rotenone (complex I), antimycin-A (complex III) or oligomycin (ATP synthase/complex V) were used at a concentration of 100 nM to treat HD CD4+ T cells. All three inhibitors significantly decreased MX1 expression and the IFN score (Fig. 6F, G) without affecting cell viability (Supp. Fig. 1D). To test whether this result was the result of lack of global phosphate availability due to decreased ATP production, the expression of a set of IL-2/STAT5 dependent genes (IL2RA, BCL2, AHR, HLA-DRB) (36) was examined after 2 mM metformin treatment in HD CD4+ T cells stimulated with anti-CD3ε/anti-CD28 and IL-2 (Fig. 7A). Metformin did not decrease the expression of these genes, and pSTAT5 (Y694) levels were unchanged as assessed by flow cytometry (Fig. 7B, C). This data supports that the inhibition of STAT1 activation by metformin does not correspond to a global inhibition of STAT activation.
FIGURE 7.
IL-2 target genes are not inhibited by metformin. (A) HD CD4+ T cells stimulated with anti-CD3ε/anti-CD28 with or without metformin for 24 h and treated with recombinant IL-2 for the final 4 h were assessed for the expression of four IL-2/pSTAT5 controlled genes (B) Representative pSTAT5 (Y694) staining in HD CD4+ T cells (C) Flow cytometry data for pSTAT5 (Y694) intensity in HD CD4+ T cells. n=3 for data in (A) and (C).
Discussion
We report that metformin significantly reduced gene expression of IFNα stimulated genes in human CD4+ T cells from both health subjects and SLE patients. Our results were obtained with physiological levels of IFNα2a, at a concentration comparable to elevated levels found in SLE patient serum (37). In this type of activation, cells experience activation through the TCR first, and an interferon signal second, which serves as an inflammatory and anti-apoptotic signal (38). Metformin reduced STAT1 activation in CD4+ T cells and prevented binding of pSTAT1 to MX1 and ISG15 gene promoters. Reduced ISG expression and pSTAT1 activation by IFNα suggests an overall anti-inflammatory effect of metformin.
Very few studies have examined the effects of metformin in the context of a T1 IFN response. Increased T1 IFN signaling has been reported after metformin treatment in cell lines infected with HCV (39). Increased viral clearance was found in a subset of HCV patients treated with pegylated-IFNα with metformin add-on therapy (40). Results obtained in HUVEC cell lines (41) demonstrated a robust decrease in pSTAT1 activation after treatment with a pharmacological activator of AMPK. We confirmed by microarray analysis that metformin activates canonical AMPK associated pathways in activated CD4+ T cells. However, pAMPKα could not be detected in CD4+ T cells after 24 h culture. In these conditions, metformin reduced pSTAT1 activation and ISG expression. In addition, rapamycin treatment of CD4+ T cells had no effect on T1 IFN gene expression, and metformin had no noticeable effect on mTORC1 activation. This further suggests that AMPK activation is not required for metformin to decrease ISG expression, since AMPK represses mTORC1 activation.
The significant decrease in ISG expression after metformin treatment was replicated using three different ETC inhibitors, rotenone, antimycin-a, and oligomycin. Their effectiveness at suppressing ISG expression without significant impact on cell viability suggests oxidative phosphorylation is required for the activation of the T1 IFN response in CD4+ T cells. An increase in oxidative metabolism and fatty acid oxidation has been reported in mouse pDCs stimulated with IFNα (42). Blocking either pyruvate transport into the mitochondria or fatty acid oxidation inhibited the effects of IFNα in increasing oxygen consumption in pDCs, and subsequently their activation (42). In our study, metformin did not change ISG expression in CD19+ human B cells. B cell differentiation and function is regulated by mitochondria (43), but the role of OXPHOS in this process has not been specifically addressed as it has been for T cells (44). It is also unknown whether OXPHOS is elevated in B cells from SLE patients in the same manner as in CD4+ T cells. To determine why there are differences between T cell and B cells, future studies need to examine differences in IFNα response, activation, and specific metabolism pathways in the context of metformin treatment. It would be interesting to determine whether the effect of metformin on ISG expression in autocrine IFNα signaling in pDCs is similar to CD4+ T cells or CD19+ B cells. It has been shown that human pDCs secrete less IFNα when stimulated by mitochondrial DNA or CpG in the presence of metformin, but how this affects their own gene expression is unclear (26).
More studies are needed to elucidate which molecular and cellular mediators are responsible for the effect of metformin on T1 IFN signaling. One candidate pathway is the localization of STAT family members to the mitochondria, where they have non-canonical roles in supporting oxidative metabolism (45). STAT3-deficient cells have reported respiratory deficiencies (46), and treatment of mice with metformin decreased levels of pSTAT3 in T cells (47). There may be a link between the inhibition of Th1 (18) and Th17 (47) polarization by metformin and the inhibition of mitochondrial localization of pSTAT1 and pSTAT3. Our results indicate that metformin did not reduce IL-2 stimulated genes in CD4+ T cells, suggesting that pSTAT5 is not inhibited the same way as pSTAT1 or pSTAT3. There is some evidence that pSTAT5 can also localize to the mitochondria, although the function of this is unclear (48). A preservation of STAT5 signaling coupled with STAT1 and STAT3 inhibition is consistent with metformin facilitating Treg function/differentiation, such as an increased number of circulating Treg cells reported after metformin treatment of multiple sclerosis patients with metabolic syndrome (49).
The therapeutic potential of metformin has been recognized as an add-on therapy for SLE patients, because metformin was capable of reducing the frequency of flares as well as the dose of prednisone in a preliminary trial (26). An expansion of this study is currently in clinical trial (NCT02741960). If metformin reduces IFNα signaling, at least in CD4+ T cells, it can be hypothesized that a greater benefit in clinical parameters would be seen in patients with a high baseline IFN signature, similar to greater benefits in anti-IFNAR1 therapy from the Anifrolumab trial in a subgroup of SLE patients with a high initial interferon signature (11).
Supplementary Material
Key Points.
Metformin affects gene expression in healthy control and SLE patient CD4 T cells.
Metformin reduces activation of type 1 interferon stimulated genes in CD4 T cells.
Inhibition of electron transport chain activity reduces type 1 interferon response.
Acknowledgements
The authors would like to thank Cecilia Lopez for microarray assistance with hybridization and scanning as well as analysis and discussion of data. We also thank Annie Chan from the UF Rheumatology Clinic for recruiting SLE patients and HCs and collecting blood samples. We are grateful to Dr. Dan Perry and Howie Seay for technical advice with T cell isolation and stimulation. Thanks to Caleb Cornaby for proofreading of the manuscript.
This study is supported by grants from the NIH (R01AI045050 and R01 AI128901) to L.M and P01 AI42288 to T.M.B. AAT is supported by 5T32DK104721.
Footnotes
Disclosures
No financial disclosure of conflicts of interest.
References
- 1.Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, Shark KB, Grande WJ, Hughes KM, Kapur V, Gregersen PK, and Behrens TW 2003. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci U S A. 100:2610–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett L, Palucka AK, Arce E, Cantrell V, Borvak J, Banchereau J, and Pascual V 2003. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med. 197:711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han GM, Chen SL, Shen N, Ye S, Bao CD, and Gu YY 2003. Analysis of gene expression profiles in human systemic lupus erythematosus using oligonucleotide microarray. Genes Immun. 4:177–186. [DOI] [PubMed] [Google Scholar]
- 4.Crow MK 2014. Type I interferon in the pathogenesis of lupus. J Immunol. 192:5459–5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niewold TB, Hua J, Lehman TJ, Harley JB, and Crow MK 2007. High serum IFN-alpha activity is a heritable risk factor for systemic lupus erythematosus. Genes Immun. 8:492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bave U, Magnusson M, Eloranta ML, Perers A, Alm GV, and Ronnblom L 2003. Fc gamma RIIa is expressed on natural IFN-alpha-producing cells (plasmacytoid dendritic cells) and is required for the IFN-alpha production induced by apoptotic cells combined with lupus IgG. J Immunol. 171:3296–3302. [DOI] [PubMed] [Google Scholar]
- 7.Ward JM, Ratliff ML, Dozmorov MG, Wiley G, Guthridge JM, Gaffney PM, James JA, and Webb CF 2016. Human effector B lymphocytes express ARID3a and secrete interferon alpha. J Autoimmun. 75:130–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamilton JA, Wu Q, Yang P, Luo B, Liu S, Li J, L. M. A, Sanz I, Chatham WW, Hsu HC, and Mountz JD 2018. Cutting Edge: Intracellular IFN-beta and distinct type I IFN expression patterns in circulating systemic lupus erythematosus B cells. J Immunol. 201:2203–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Golding A, Rosen A, Petri M, Akhter E, and Andrade F 2010. Interferon-alpha regulates the dynamic balance between human activated regulatory and effector T cells: implications for antiviral and autoimmune responses. Immunology. 131:107–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Avraamides G, Ng CY, David R, Gu Y, Fazekasova H, Mirenda V, Foster GR, Runkel L, Lombardi G, and Marelli-Berg FM 2007. IFN-alpha2 induces leukocyte integrin redistribution, increased adhesion, and migration. J Interferon Cytokine Res. 27:291–303. [DOI] [PubMed] [Google Scholar]
- 11.Furie R, Khamashta M, Merrill JT, Werth VP, Kalunian K, Brohawn P, Illei GG, Drappa J, Wang L, and Yoo S 2017. Anifrolumab, an anti-interferon-alpha receptor monoclonal antibody, in moderate-to-severe systemic lupus erythematosus. Arthritis Rheumatol. 69:376–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lauwerys BR, Hachulla E, Spertini F, Lazaro E, Jorgensen C, Mariette X, Haelterman E, Grouard-Vogel G, Fanget B, Dhellin O, Vandepapeliere P, and Houssiau FA 2013. Down-regulation of interferon signature in systemic lupus erythematosus patients by active immunization with interferon alpha-kinoid. Arthritis Rheum. 65:447–456. [DOI] [PubMed] [Google Scholar]
- 13.Nacionales DC, Kelly-Scumpia KM, Lee PY, Weinstein JS, Lyons R, Sobel E, Satoh M, and Reeves WH 2007. Deficiency of the type I interferon receptor protects mice from experimental lupus. Arthritis Rheum. 56:3770–3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santiago-Raber ML, Baccala R, Haraldsson KM, Choubey D, Stewart TA, Kono DH, and Theofilopoulos AN 2003. Type-I interferon receptor deficiency reduces lupus-like disease in NZB mice. J Exp Med. 197:777–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morel L, Yu Y, Blenman KR, Caldwell RA, and Wakeland EK 1996. Production of congenic mouse strains carrying genomic intervals containing SLE-susceptibility genes derived from the SLE-prone NZM2410 strain. Mamm Genome. 7:335–339. [DOI] [PubMed] [Google Scholar]
- 16.Sriram U, Varghese L, Bennett HL, Jog NR, Shivers DK, Ning Y, Behrens EM, Caricchio R, and Gallucci S 2012. Myeloid dendritic cells from B6.NZM Sle1/Sle2/Sle3 lupus-prone mice express an IFN signature that precedes disease onset. J Immunol. 189:80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sang A, Zheng YY, Yin Y, Dozmorov I, Li H, Hsu HC, Mountz JD, and Morel L 2014. Dysregulated cytokine production by dendritic cells modulates B cell responses in the NZM2410 mouse model of lupus. PLoS One. 9:e102151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin Y, Choi SC, Xu Z, Perry DJ, Seay H, Croker BP, Sobel ES, Brusko TM, and Morel L 2015. Normalization of CD4+ T cell metabolism reverses lupus. Sci Transl Med. 7:274ra218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perl A 2015. mTOR activation is a biomarker and a central pathway to autoimmune disorders, cancer, obesity, and aging. Ann N Y Acad Sci. 1346:33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin Y, Choi SC, Xu Z, Zeumer L, Kanda N, Croker BP, and Morel L 2016. Glucose Oxidation is critical for CD4+ T cell activation in a mouse model of systemic lupus erythematosus. J Immunol. 196:80–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mehta MM, and Chandel NS 2015. Targeting metabolism for lupus therapy. Sci Transl Med. 7:274fs275. [DOI] [PubMed] [Google Scholar]
- 22.Wheaton WW, Weinberg SE, Hamanaka RB, Soberanes S, Sullivan LB, Anso E, Glasauer A, Dufour E, Mutlu GM, Budigner GS, and Chandel NS 2014. Metformin inhibits mitochondrial complex I of cancer cells to reduce tumorigenesis. Elife. 3:e02242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He L, and Wondisford FE 2015. Metformin action: concentrations matter. Cell Metab. 21:159–162. [DOI] [PubMed] [Google Scholar]
- 24.Lei Y, Yi Y, Liu Y, Liu X, Keller ET, Qian CN, Zhang J, and Lu Y 2017. Metformin targets multiple signaling pathways in cancer. Chin J Cancer. 36:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kato H, and Perl A 2014. Mechanistic target of rapamycin complex 1 expands Th17 and IL-4+ CD4-CD8- double-negative T cells and contracts regulatory T cells in systemic lupus erythematosus. J Immunol. 192:4134–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang H, Li T, Chen S, Gu Y, and Ye S 2015. neutrophil extracellular trap mitochondrial DNA and its autoantibody in systemic lupus erythematosus and a proof-of-concept trial of metformin. Arthritis Rheumatol. 67:3190–3200. [DOI] [PubMed] [Google Scholar]
- 27.Petri M, Singh S, Tesfasyone H, Dedrick R, Fry K, Lal P, Williams G, Bauer J, Gregersen P, Behrens T, and Baechler E 2009. Longitudinal expression of type I interferon responsive genes in systemic lupus erythematosus. Lupus. 18:980–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ronni T, Matikainen S, Lehtonen A, Palvimo J, Dellis J, Van Eylen F, Goetschy JF, Horisberger M, Content J, and Julkunen I 1998. The proximal interferon-stimulated response elements are essential for interferon responsiveness: a promoter analysis of the antiviral MxA gene. J Interferon Cytokine Res. 18:773–781. [DOI] [PubMed] [Google Scholar]
- 29.Liu ZQ, Song XM, Chen QT, Liu T, Teng JT, Zhou K, and Luo DQ 2016. Effect of metformin on global gene expression in liver of KKAy mice. Pharmacol Rep. 68:1332–1338. [DOI] [PubMed] [Google Scholar]
- 30.Udhane SS, Legeza B, Marti N, Hertig D, Diserens G, Nuoffer JM, Vermathen P, and Fluck CE 2017. Combined transcriptome and metabolome analyses of metformin effects reveal novel links between metabolic networks in steroidogenic systems. Sci Rep. 7:8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hadad SM, Hardie DG, Appleyard V, and Thompson AM 2014. Effects of metformin on breast cancer cell proliferation, the AMPK pathway and the cell cycle. Clin Transl Oncol. 16:746–752. [DOI] [PubMed] [Google Scholar]
- 32.Chiche L, Jourde-Chiche N, Whalen E, Presnell S, Gersuk V, Dang K, Anguiano E, Quinn C, Burtey S, Berland Y, Kaplanski G, Harle JR, Pascual V, and Chaussabel D 2014. Modular transcriptional repertoire analyses of adults with systemic lupus erythematosus reveal distinct type I and type II interferon signatures. Arthritis Rheumatol. 66:1583–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mihaylova MM, and Shaw RJ 2011. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol. 13:1016–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu X, Chhipa RR, Pooya S, Wortman M, Yachyshin S, Chow LM, Kumar A, Zhou X, Sun Y, Quinn B, McPherson C, Warnick RE, Kendler A, Giri S, Poels J, Norga K, Viollet B, Grabowski GA, and Dasgupta B 2014. Discrete mechanisms of mTOR and cell cycle regulation by AMPK agonists independent of AMPK. Proc Natl Acad Sci U S A. 111:E435–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salt IP, Johnson G, Ashcroft SJ, and Hardie DG 1998. AMP-activated protein kinase is activated by low glucose in cell lines derived from pancreatic beta cells, and may regulate insulin release. Biochem J. 335 ( Pt 3):533–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu A, Snowhite I, Vendrame F, Rosenzwajg M, Klatzmann D, Pugliese A, and Malek TR 2015. Selective IL-2 responsiveness of regulatory T cells through multiple intrinsic mechanisms supports the use of low-dose IL-2 therapy in type 1 diabetes. Diabetes. 64:2172–2183. [DOI] [PubMed] [Google Scholar]
- 37.Kim T, Kanayama Y, Negoro N, Okamura M, Takeda T, and Inoue T 1987. Serum levels of interferons in patients with systemic lupus erythematosus. Clin Exp Immunol. 70:562–569. [PMC free article] [PubMed] [Google Scholar]
- 38.Crouse J, Kalinke U, and Oxenius A 2015. Regulation of antiviral T cell responses by type I interferons. Nat Rev Immunol. 15:231–242. [DOI] [PubMed] [Google Scholar]
- 39.Tsai WL, Chang TH, Sun WC, Chan HH, Wu CC, Hsu PI, Cheng JS, and Yu ML 2017. Metformin activates type I interferon signaling against HCV via activation of adenosine monophosphate-activated protein kinase. Oncotarget. 8:91928–91937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Romero-Gomez M, Diago M, Andrade RJ, Calleja JL, Salmeron J, Fernandez-Rodriguez CM, Sola R, Garcia-Samaniego J, Herrerias JM, De la Mata M, Moreno-Otero R, Nunez O, Olveira A, Duran S, and Planas R 2009. Treatment of insulin resistance with metformin in naive genotype 1 chronic hepatitis C patients receiving peginterferon alfa-2a plus ribavirin. Hepatology. 50:1702–1708. [DOI] [PubMed] [Google Scholar]
- 41.Rutherford C, Speirs C, Williams JJ, Ewart MA, Mancini SJ, Hawley SA, Delles C, Viollet B, Costa-Pereira AP, Baillie GS, Salt IP, and Palmer TM 2016. Phosphorylation of Janus kinase 1 (JAK1) by AMP-activated protein kinase (AMPK) links energy sensing to anti-inflammatory signaling. Sci Signal. 9:ra109. [DOI] [PubMed] [Google Scholar]
- 42.Wu D, Sanin DE, Everts B, Chen Q, Qiu J, Buck MD, Patterson A, Smith AM, Chang CH, Liu Z, Artyomov MN, Pearce EL, Cella M, and Pearce EJ 2016. Type 1 interferons induce changes in core metabolism that are critical for immune function. Immunity. 44:1325–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sandoval H, Kodali S, and Wang J 2018. Regulation of B cell fate, survival, and function by mitochondria and autophagy. Mitochondrion. 41:58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Geltink RIK, Kyle RL, and Pearce EL 2018. Unraveling the Complex interplay between T cell metabolism and function. Annu Rev Immunol. 36:461–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meier JA, and Larner AC 2014. Toward a new STATe: the role of STATs in mitochondrial function. Semin Immunol. 26:20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wegrzyn J, Potla R, Chwae YJ, Sepuri NB, Zhang Q, Koeck T, Derecka M, Szczepanek K, Szelag M, Gornicka A, Moh A, Moghaddas S, Chen Q, Bobbili S, Cichy J, Dulak J, Baker DP, Wolfman A, Stuehr D, Hassan MO, Fu XY, Avadhani N, Drake JI, Fawcett P, Lesnefsky EJ, and Larner AC 2009. Function of mitochondrial Stat3 in cellular respiration. Science. 323:793–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee SY, Lee SH, Yang EJ, Kim EK, Kim JK, Shin DY, and Cho ML 2015. Metformin ameliorates inflammatory bowel disease by suppression of the STAT3 signaling pathway and regulation of the between Th17/Treg balance. PLoS One. 10:e0135858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chueh FY, Leong KF, Cronk RJ, Venkitachalam S, Pabich S, and Yu CL 2011. Nuclear localization of pyruvate dehydrogenase complex-E2 (PDC-E2), a mitochondrial enzyme, and its role in signal transducer and activator of transcription 5 (STAT5)-dependent gene transcription. Cell Signal. 23:1170–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Negrotto L, Farez MF, and Correale J 2016. Immunologic Effects of Metformin and Pioglitazone Treatment on Metabolic Syndrome and Multiple Sclerosis. JAMA Neurol. 73:520–528. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.