Abstract
Mammals have coevolved with a large community of symbiotic, commensal, and some potentially pathogenic microbes. The trillions of bacteria and hundreds of species in our guts form a relatively stable community that resists invasion by outsiders, including pathogens. This powerful protective force is referred to as colonization resistance. We discuss the variety of proposed or demonstrated mechanisms that can mediate colonization resistance and some potential ways to manipulate them for improved human health. Instances in which certain bacterial pathogens can overcome colonization resistance are also discussed.
The bacteria and other microbes inhabiting the animal gut (the gut microbiota) provide major beneficial functions to the host. These include nutrition (breakdown of indigestible polysaccharides and production of vitamins) and protection against microbes that could harm the host. The idea that certain good bacteria could compete with harmful ones was first proposed by Élie Metchnikoff in the early 20th Century. With the advent of antibiotics and germ-free animals, it was demonstrated that the resident bacteria in the gut play a large role in preventing pathogens from colonizing and causing disease, in animals and humans.1 This phenomenon was later coined colonization resistance.2 In addition to defense against strict pathogens, the same concept applies to control of indigenous but potentially dangerous pathobionts, as well as exclusion of innocuous foreign species, such as probiotics. There are a wide variety of mechanisms now known to participate in colonization resistance. Many involve direct interactions between bacterial cells, whereas others act by modulating host physiology, especially the immune system. In addition, host genetics, diet, and antibiotic use can modify the composition and function of the microbiota and, thus, affect colonization resistance. In the clinic, fecal microbiota transplantation is used effectively to treat antibiotic-induced diarrhea caused by the overgrowth of Clostridium difficile.3 We will discuss the history and new developments in understanding this phenomenon as it relates to enteric pathogens, probiotics, prebiotics, and improving human health.
Direct Mechanisms
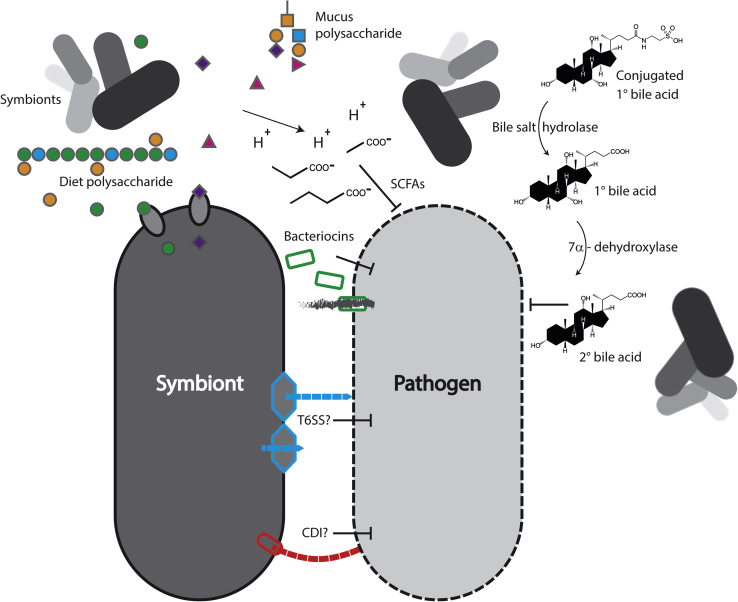
Bacteria can reach densities approaching 1011 cells/g in the mammalian large intestine4 and are in constant competition with each other for survival. Nutrients, including carbon, nitrogen, and energy sources, as well as other essential molecules can be limited, especially in the large intestine. As a consequence of this highly competitive environment, bacteria have evolved ways to suppress or kill each other. These direct mechanisms of resistance are summarized in Figure 1.
Figure 1.
Direct mechanisms. Anaerobic symbionts (top left) digest host mucus and dietary polysaccharides, releasing monosaccharides for another symbiont to take up, excreting short-chain fatty acids (SCFAs), and acidifying the environment, which suppresses growth of a pathogen. Bacteria deconjugate bile acids (top right), and others produce 7α-dehydroxylases that convert them to secondary bile acids, which also inhibit the pathogen. A symbiont produces bacteriocins (green), which form pores in the pathogen, allowing leakage of cellular contents (gray). Symbionts could also theoretically target pathogens with contact-dependent mechanisms, like the type 6 secretion system (T6SS; blue) or contact-dependent inhibition (CDI; red).
Nutritional Competition
Monosaccharides, disaccharides, and most proteins ingested by mice or humans are digested and absorbed in the small intestine. This leaves only complex plant polysaccharides and host-secreted mucus as food sources for the dense community of bacteria in the large intestine. Metabolizing these polysaccharides requires specific enzymes. Some bacteria that predominate in the large intestine, such as Bacteroides species, possess a vast array of genes to harvest sugars from host and diet polysaccharides.5 Others, like the often pathogenic Enterobacteriaceae, can generally only use simpler sugars and amino acids for their carbon, nitrogen, and energy needs; and they are found at low levels in the healthy gut. When resident bacteria are acutely killed with an antibiotic, there is a surplus of free monosaccharides released from host glycans, such as sialic acid and fucose, which are taken advantage of by some pathogens, like Salmonella enterica serovar Typhimurium (S. typhimurium).6 Increased sugars and free amino acids available after antibiotic treatment may also be exploited by C. difficile, a major human pathogen that causes pseudomembranous colitis.6, 7, 8, 9 Constant scavenging of all available nutrients in the normal gut may, therefore, prevent invasion and maintain a stable beneficial community. On the basis of in vitro experiments, Freter et al proposed that the steady-state abundance of different taxa in the gut was determined by “one or a few nutritional substrates which a given strain can utilize most efficiently.”10,pp.676 This nutrient niche hypothesis is supported by in vitro and in vivo experiments.11, 12, 13 When a single host-produced sugar, fucose, is removed from the mouse gut, the microbiota does change in composition,14 but beyond that, the microbiota's ability to suppress pathogens and pathobionts is altered.15, 16, 17 On the other hand, removal of a host-supplied sugar will also make it unavailable to pathogens or pathobionts, even when nutrient scavenging is disrupted, and therefore can improve resistance (as in the case of sialic acid).18 Interestingly, transgenic expression in mice of a sugar structure not normally found in their stomach was also able to affect the phenotype of the pathobiont Helicobacter pylori.19 The host sugar structures (glycosylation) in the gut can be modified by a variety of signals, including microbial colonization,20 immune cell activation,15, 17 and various chemicals,21 so manipulation of the host-produced nutrients in the gut could be a way to alter the microbiota composition or function.
Diet can have a rapid and profound impact on microbiota composition and function and, consequently, colonization resistance.22 Complex plant polysaccharides (fiber) in the diet are a major food source for the anaerobic bacteria that dominate the lower gastrointestinal tract. Removing fermentable polysaccharides from the diet causes a shift in overall community structure and, over time, can lead to permanent loss of species from the gut.23 Loss of protective species or hampering of their normal functions in the absence of their preferred food sources could lead to reduced colonization resistance. For example, when deprived of diet polysaccharides, bacteria switch feeding preferences toward mucus glycans and proteins, degrading them, reducing the effectiveness of this protective barrier, and increasing the damage caused by Citrobacter rodentium.24 Feeding a polysaccharide-free diet also allowed easier invasion of C. difficile, which could be rescued by supplementation with inulin, a fructose-based polysaccharide found in many plants.25 Rice bran added to an otherwise low-fiber diet reduced S. typhimurium colonization.26 However, another study of several purified polysaccharides found that most did not reduce S. typhimurium loads and, in fact, some had the opposite effect.27 It is important to take into account the exact source and structure of polysaccharides (of which there are an almost infinite variety), the dietary context, and the preexisting microbiota and its capacity to respond to a given substrate. Other plant-derived compounds in the diet, besides polysaccharides, could also have myriad effects on both direct and indirect colonization resistance.28 Western diets, which are higher in fat and simple sugars but low in complex polysaccharides, favor the expansion of endogenous Proteobacteria, such as Escherichia coli, and allow easier introduction of a human pathobiont associated with inflammatory bowel disease, adherent-invasive E. coli.29 As discussed later, high-fat diets can also alter the microbiota and colonization resistance via bile acid production, as well as having indirect effects on the immune system. Low-protein diets, on the contrary, can enhance protection against C. difficile, possibly by reducing the free amino acids available to it.7, 30 Of course, malnutrition will have many detrimental effects on host functions, but permanent changes to the microbiota are also apparent in malnourished children.31 This could mean that there are additional microbiota-intrinsic defects in colonization resistance in these already vulnerable individuals. Because diet can have innumerable effects on host physiology, separating the indirect effects from direct influences on the microbiota is not a simple task.
Prebiotics are dietary nutrients, typically polysaccharides, used to target subsets of the indigenous microbiota and bolster their beneficial functions. Identifying prebiotics, such as inulin,25 that specifically enhance colonization resistance and the mechanisms behind this could have immense clinical benefits. Nutrient niches can even be artificially generated in the gut via diet. Two recent studies demonstrated that, by introducing a specific polysaccharide in the diet (porphyran from seaweed) along with a bacterial strain uniquely capable of using it, the strain could be stably inserted into the gut community.32, 33
If nutritional competition is responsible for steady-state microbiota structure and exclusion of outsiders, are all functional niches constantly filled in a healthy gut? This may not always be the case in humans. In one study, subjects were fed a probiotic, Bifidobacterium longum, and then examined for persistence of the strain in their stool. Although probiotics generally do not stably colonize in humans, surprisingly approximately a third of the subjects maintained detectable levels of the strain up to 200 days later.34 Before treatment, the persisters generally had lower amounts of endogenous B. longum in their gut and/or lower amounts of B. longum genes, including some related to carbohydrate use. This suggests that, for unknown reasons, some individuals had an open functional niche that the strain was able to fill. Another recent study gave subjects a mix of 11 probiotics and also found individuals who had persistence of some strains in the large intestinal mucus layer.35 Persistence seemed to be controlled by differences in their microbiota because the result could be recapitulated by transfer to mice, but the underlying mechanisms that control persistence of probiotics are not yet clear.
Overall, nutritional competition seems to be a powerful force for excluding nonnative bacteria from the gut ecosystem, including beneficial probiotics. Fortunately, the nutritional landscape in the gut can be easily modified via diet. Prebiotics, such as plant polysaccharides, may be used to target specific endogenous beneficial species and enhance their functions. Probiotic strains, delivered along with prebiotics that they can use (a combination known as a synbiotic), could facilitate introduction of beneficial strains to the existing gut community.
Bactericidal/Bacteriostatic Mechanisms
Perhaps because competition for growth substrates is so intense, gut bacteria have also developed many ways of suppressing or killing competitors. Indeed, dominant growth suppression may be more important than nutrient limitation for colonization resistance in some cases.36 Bacteriocins, for instance, are a large, heterogeneous group of peptides produced by bacteria, with diverse bacteriostatic or bactericidal activities.37 Bacteriocin production is common in bacteria used as probiotics, fermented foods, and the gut microbiota, and many have been developed as possible replacements for traditional antibiotics. How important is production of antibacterial molecules for competition in the gut? Production or susceptibility to colicins (a family of bacteriocins) affected E. coli competition in antibiotic-treated mouse models,38, 39 but there is also evidence that they mediate ongoing competition among E. coli strains in the natural microbiota as well.40 A bacteriocin of the common gut resident Enterococcus faecalis gave it a competitive advantage against other Enterococci in mice and allowed it to repel a related pathogenic species, vancomycin-resistant Enterococcus faecium.41 Bacteroides species, which are highly abundant in mouse and human gut, produce many bactericidal or bacteriostatic secreted proteins as well, some of which have been shown to mediate intraspecies competition in vivo.42
Because of their relative ease of discovery and therapeutic utility, most of the antibacterial factors known in the gut are secreted, soluble compounds. However, mechanisms of suppression that require direct cell-cell contact are increasingly being appreciated as well. The contact-dependent inhibition system was discovered in E. coli,43 and homologous genes are distributed throughout the Proteobacteria and possibly other phyla as well.44 The contact-dependent inhibition consists of two genes that compose a two-partner secretion system (in the type 5 secretion system family). They assemble a long filament that extends out from the cell. This binds target cells via a specific receptor and delivers the inhibitory activity found in the filament protein's effector domain. This effector domain is variable between strains and interchangeable, and effectors have been found with DNase, RNase, and pore-forming activities. A cognate immunity protein is encoded along with the system, to inactivate the effector and prevent self-inhibition.
The type 6 secretion system (T6SS) is another recently recognized player in contact-dependent competition in the gut. The T6SS was originally identified in several Gram-negative species as a secretion system that was involved in interactions with eukaryotic cells.45, 46, 47, 48 Subsequently, this was extended to interbacterial action as well49 and, specifically, intraspecies killing.50 The T6SS system works by spearing nearby cells and delivering effectors into their cytoplasm. These effectors can degrade nucleotides, cell walls, or membranes, or they have other activities. Like the contact-dependent inhibition, the effectors are often found with a cognate immunity protein. The T6SS genes are widespread in Gram-negative bacteria, especially Proteobacteria, and a related but distinct family exists in the Bacteroidales.51, 52 More important, T6SS effector and immunity genes may contribute to ongoing competition among the abundant Bacteroides species in the mouse and human gut.53, 54, 55 Some pathogens can also use their T6SS to kill resident bacteria and enable their invasion of the gut community.56, 57 Another secretion system, called Esx or type 7, can mediate intraspecies and interspecies killing by Gram-positive bacteria.58, 59 Notably, at least one toxin family secreted by this system is abundant among the Firmicutes, such as Clostridia and Bacilli, in the human microbiome.59
Contact-dependent systems of growth inhibition and killing are increasingly being discovered in the gut microbiota and could likely participate in resistance to pathogens. The modularity of effector/immunity genes common to these systems may make them amenable to engineering. These contact-dependent mechanisms also highlight the value of studying the microscopic structure and spatial relationships in the gut community.60 However, the overall role of bactericidal/bacteriostatic mechanisms in mediating colonization resistance against pathogens, pathobionts, and probiotics remains poorly understood.
Metabolites and Chemical Transformations
Bacteria also produce metabolic by-products or modify compounds in the gut that can affect their growth. Short-chain fatty acids (SCFAs) are a major product of bacterial fermentation in the large intestine, especially downstream of polysaccharide digestion, and they can have growth-inhibiting effects on pathogenic bacteria, such as E. coli, S. typhimurium, and C. difficile.61, 62, 63 Caused, in part, by SCFA excretion, microbial metabolism generally lowers pH in the gut, especially in the large intestine.64 Oxygen is normally scarce in the large intestine lumen, which has been attributed to bacterial and host metabolism, the latter driven by epithelial catabolism of the SCFA butyrate.65, 66 Dysbiosis, a general deviation in community structure associated with disease and breakdown of colonization resistance, frequently coincides with increased oxygen availability, expansion of facultative anaerobes, such as pathogenic Proteobacteria,67 and reduction in butyrate and butyrate producers. Thus, prebiotic polysaccharides that enhance SCFA production could improve colonization resistance in several possible ways.
Bile acids are produced in the liver from cholesterol, stored in the gallbladder, and secreted into the duodenum after eating. Their amphipathic qualities help dissolve fat and fat-soluble vitamins for absorption, but they are antibacterial to varying degrees as well. The primary bile acids in humans are cholic acid and chenodeoxycholic acid, which are synthesized in the liver and conjugated to taurine or glycine. Once in the gut, bile salt hydrolases, produced by many different bacterial taxa, deconjugate them from the amino acid, possibly to reduce their toxicity or to obtain the taurine or glycine itself. Once deconjugated, the primary bile acids can be converted into a variety of secondary bile acids by enzymes produced by rare bacterial species in the gut.68 7α-Dehydroxylation, for example, can convert cholic acid to deoxycholic acid and chenodeoxycholic acid to lithocholic acid. Both of these secondary bile acids can suppress growth of C. difficile.69 At the same time, the conjugated primary bile acid taurocholic acid promotes C. difficile spore germination.70 Thus, microbial transformation of bile acids may partially explain susceptibility to C. difficile.9, 69, 71 Bile acids are also modulated by diet: excessive taurocholic acid, induced by a high-fat diet, promoted expansion of a pathobiont, Bilophila wadsworthia, leading to worsened colitis in that model.72 Although impacts on fat absorption and metabolism must be considered, modulating the levels of bile acids via diet, specific bacteria,69, 73 or drugs could be a means to enhance colonization resistance.
Indirect Mechanisms and Modulation of Host Physiology
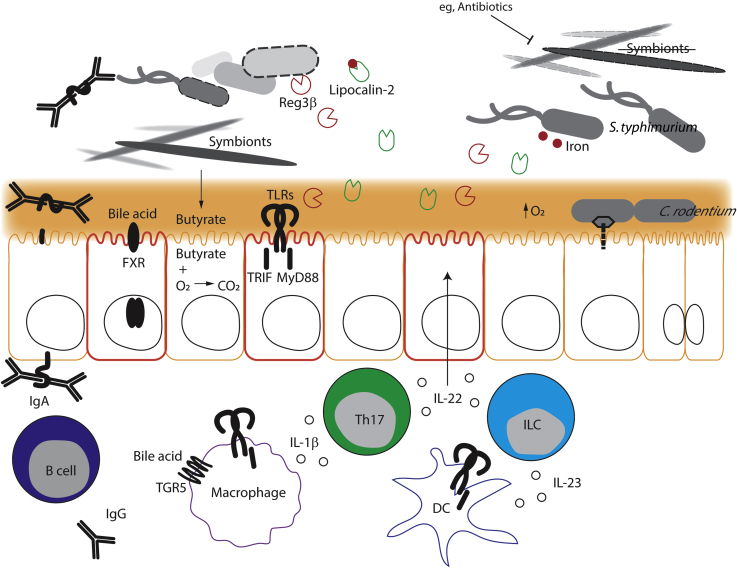
Although the fierce ongoing competition between bacteria in the gut is largely responsible for maintaining a healthy, beneficial, and stable community and effectively repelling invaders, the host organism also has a role to play. The host can shape the structure and activity of the microbiota, thereby affecting its functions, including its level of colonization resistance. At the same time, the host immune system and other aspects of its biology are heavily influenced by the microbiota (Figure 2).
Figure 2.
Indirect mechanisms. Dimeric IgA, produced by a B cell in the lamina propria, is transcytosed by the poly-Ig receptor into the lumen, where it binds a bacterium's flagella. Farnesoid X receptor (FXR) and TGR5 on epithelial cells and macrophages up-regulate defenses or modulate inflammation, respectively, in response to bile acids. The short-chain fatty acid butyrate, produced by anaerobic Clostridia, promotes oxygen respiration in an epithelial cell, reducing the oxygen concentration at the epithelial surface. Toll-like receptors (TLRs) on epithelial cells, macrophages, and dendritic cells (DCs) can sense microbial molecules and signal through myeloid differentiation primary response 88 (MyD88) or TIR-domain-containing adapter-inducing interferon-β (TRIF) adaptors. Macrophages make IL-1β and DCs make IL-23 cytokines in response to TLR stimulation, which induces type 17 helper T cells (Th17s) and innate-like lymphocytes (ILCs) to secrete IL-22. This acts on epithelial cells, causing them to produce regenerating islet-derived protein 3β (Reg3β) (red) and lipocalin-2 (green), which attack a pathogen in the lumen and sequester iron (red circles) from it, respectively. On the right side, protective symbionts, like Clostridia, have been depleted (eg, by antibiotics), resulting in increased oxygen in the lumen. Salmonella enterica serovar Typhimurium (S. typhimurium) is resistant to Reg3β and can capture iron from lipocalin-2. Meanwhile Citrobacter rodentium uses its type 3 secretion system to inject effectors into an epithelial cell and cause hyperplasia, further increasing oxygen levels and supporting its replication.
Innate Defense Mechanisms
Factors that can prevent or promote growth of microbes and, thus, select which bacteria can colonize mucosal surfaces, like the gut, are evidently ancient. The hydra, for example, an extremely simple type of aquatic animal, nonetheless selects and maintains a species-specific group of bacterial symbionts. This is accomplished, in part, through production of different antibacterial compounds.74 Transferring microbiota between more complex animals (eg, zebra fish and mice) has confirmed that the host significantly shapes the bacterial community in the gut.75 Many mammalian genes are regulated by microbial colonization, including antibacterial factors.76 Some of these genes are regulated through sensing pathways of the innate immune system: toll-like receptors (TLRs), which signal through the adaptor myeloid differentiation primary response 88 (MyD88) or TIR-domain-containing adapter–inducing interferon-β (TRIF), or nucleotide-binding oligomerization domain–containing protein (NOD)-like receptors, which sense microbial molecules in the cytosol and assemble inflammasomes.77 These receptors sense conserved microbial molecules, such as cell wall or outer membrane components, which are present in both symbionts and pathogens. TLRs and NOD-like receptors, as well as other microbial pattern sensors, can be expressed by epithelial cells and hematopoietic cells, such as macrophages and dendritic cells. Steady-state sensing of the microbiota through TLRs does occur and regulates many genes in the gut as the microbiota develops.78 Although many of these are antimicrobial genes, the impact of TLR signaling on the steady-state composition of the microbiota seems to be minimal in well-controlled experiments.78 However, the system can be used to artificially boost resistance. Systemic injection of TLR ligands, for instance, activates a signaling pathway that culminates in production of antimicrobial peptides and protects against vancomycin-resistant E. faecium.79 Dendritic cells in the intestinal lamina propria are activated through their TLRs to produce IL-23. This causes production of another cytokine, IL-22, which is crucial for epithelial defense and repair in the gut,80 and can be produced by innate-like lymphocytes or type 17 helper T cells. The IL-22 induces antimicrobial peptides, particularly regenerating islet-derived protein 3β (Reg3β) and Reg3γ, in epithelial cells, resulting in reduced lumen colonization and invasion of vancomycin-resistant E. faecium.79 However, no protective role for MyD88/TRIF signaling or IL-22 was observed in colonization resistance against S. typhimurium.81 In fact, effectors induced by IL-22 may give S. typhimurium an advantage over other bacteria in the gut.82 Further studies are needed to understand the physiological role of IL-22 and antimicrobial peptides in the regulation of pathogen colonization resistance.
Bacterial metabolites can also have a multitude of effects on host physiology and, especially, immune system function through many different signaling pathways.83 SCFAs, products of polysaccharide digestion, can affect both innate inflammation as well as B- and T-cell differentiation. Bile acids that have been modified by bacteria can activate antimicrobial genes in the ileum via the farnesoid X receptor or modulate inflammation in immune cells through another receptor, TGR5.83 The aryl hydrocarbon receptor senses microbial and diet-derived molecules and can regulate epithelial, dendritic cell, T-cell, and innate-like lymphocyte function.83 These are only a few known examples of the large variety of microbiota-derived metabolites that can potentially affect the host animal.
B Cells, Igs, and Adaptive Immunity
The adaptive immune system may add another layer of specificity to control of the microbiota. Humans secrete several grams of IgA daily into the gut. Most of the bacterial cells in the small intestine are coated with IgA, with less coating in the large intestine and feces.84, 85 IgA production is driven by the microbiota, being lower overall in germ-free mice but induced by introduction of certain bacteria or bacterial products.86, 87 Most of the secreted IgA in the steady state is produced by B cells independently of T-cell help and is relatively low affinity but polyreactive, in that a single antibody can bind to more than one bacterial species or even phylum.84, 88, 89, 90 This microbiota-stimulated, secreted IgA could theoretically mediate colonization resistance in two ways: by direct binding to a pathogen/pathobiont or by modulating the composition or function of the resident microbiota.
Indeed, IgA may preferentially target dangerous bacteria in the normal gut community (ie, pathobionts) because transferring the IgA-coated population from humans with inflammatory bowel disease to mice worsened disease in a colitis model.91 IgA also targets flagellar proteins in the normal microbiota, down-regulating their expression and reducing motility/invasion.92 These antibodies can cross-react with pathogens, such as S. typhimurium, and so serve as a preformed line of defense. Indeed, the amount of preexisting IgA that bound S. typhimurium differed between two common inbred mouse strains, and higher amounts correlated with better survival after oral infection, suggesting that this preexisting pathogen-reactive antibody could support colonization resistance.93
Although IgA is actively transported into the gut lumen, IgG antibodies do not normally cross the barrier unless it is damaged. However, there is IgG in the serum that reacts against gut bacteria.94, 95 This antibody may be stimulated by and help to control the normal low-level translocation of bacteria from the gut. If the epithelial barrier is breached by a pathogen or damaged, the premade IgG is ready to help control infection.94
How large a role secreted IgA plays in shaping the normal gut microbiota remains controversial. Mice that lack secreted IgA for various reasons have been reported to have widely varying amounts of dysbiosis.96, 97 Similarly, mice that lack the polymeric Ig receptor and cannot transport IgA or IgM into the gut lumen were found to be either more or less susceptible to S. typhimurium infection than wild-type mice.98, 99 The reason for these conflicting results is unclear, but they are likely explained by underlying microbiota differences.
Aside from changing community composition, could IgA that coats symbiotic bacteria modulate their protective function? IgA against the symbiont Bacteroides thetaiotaomicron down-regulated its expression of the target epitope and reduced its fitness in the gut.100 Even binding of a nonspecific IgA, likely via glycosylation on the constant region, can modulate B. thetaiotaomicron gene expression in the gut.101 Although antibody binding is usually thought of as having an exclusionary function, IgA surprisingly promotes the survival of some symbionts in humans and mice, perhaps by retaining them in a favorable physical niche.102, 103
Selective IgA deficiency in humans is relatively common (approximately 0.1% to 1%), and although it often goes unnoticed, it does increase the overall risk of infection (especially respiratory and gastrointestinal tract) and autoimmunity, including celiac disease and inflammatory bowel disease,104, 105 which are tentatively linked to pathogen or pathobiont infections, respectively. In the absence of IgA, IgM can also be secreted in comparable quantities and bind to similar bacterial populations but may not compensate fully.84, 103
Although mostly dispensable for steady-state IgA, T cells can affect colonization resistance via cytokine production. Type 17 helper T cells, for instance, are produced in response to some resident bacteria, including segmented filamentous bacteria, a unique species that attaches directly to intestinal epithelial cells.106 IL-1β production by macrophages, induced by the microbiota via TLRs, stimulates type 17 helper T-cell development.107 The type 17 helper T cells produce IL-22, which induces antibacterial and tissue repair genes in the epithelium and results in improved resistance to C. rodentium.106
Indirect mechanisms of colonization resistance, which are induced in the host by the microbiota, are exceedingly difficult to dissect away from the influence of the bacteria directly. Although it is clear that the host organism has some ability to shape its microbiota, through primitive innate mechanisms as well as more specific adaptive ones, it seems that we are largely bystanders when it comes to colonization resistance.
Pathogen Evasion of Colonization Resistance
Colonization resistance is generally effective: for example, large doses (≥1011 is not uncommon) of different probiotics are regularly ingested by humans and usually do not establish in the gut. The dose of S. typhimurium needed to successfully infect mice decreases by many orders of magnitude after treatment with an antibiotic.1 Clostridium difficile colitis in humans is also strongly associated with prior antibiotic use.108 Despite this, some pathogens seem to have developed ways to partially circumvent colonization resistance. One is C. rodentium, a mouse-specific pathogen that infects the large intestine, causes colonic hyperplasia, and is then cleared within 3 weeks (in most mouse strains). The dose required for a successful infection with C. rodentium is fairly low when spreading naturally between mice.109 Most important, once an infection is established, the levels of C. rodentium shed into the large intestine lumen and feces quickly increase to 109/g, much higher than any endogenous Proteobacteria. How does it achieve this?
Citrobacter rodentium is similar to the human diarrhea-causing pathogens enteropathogenic and enterohemorrhagic E. coli. All three pathogens exhibit a similar specialized lifestyle in the gut: direct attachment to the epithelium and formation of lesions (giving them the name attaching/effacing pathogens). This attaching/effacing behavior is made possible by a cluster of virulence genes that the strains share, called the locus of enterocyte effacement. The locus of enterocyte effacement comprises a type 3 secretion system and associated effectors, as well as an essential surface protein, intimin. The ability of C. rodentium to live and replicate at the epithelial surface likely lets it avoid competition with most bacteria in the large intestine lumen and, therefore, the main force of colonization resistance.
The epithelial cell surface is already higher in oxygen than the lumen, which excludes the obligate anaerobes that make up most of the microbiota. Citrobacter rodentium further increases oxygenation by triggering epithelial hyperplasia via type 3 secretion system–injected effectors.110 Although it does not possess enzymes to digest mucus glycans, there may be other unique nutrient sources in this epithelial niche that C. rodentium exploits. Once the host initiates an IgG antibody response against locus of enterocyte effacement virulence factors, including surface intimin, opsonized virulent C. rodentium are removed by luminal neutrophils while avirulent bacteria are forced away from the protected niche at the epithelial surface and into the lumen. Without the advantage of their virulence factors, they are unable to compete with the microbiota and are cleared from the gut.111 This clearance is probably caused, in part, by nutrient competition with other bacteria in the lumen.111
Salmonella typhimurium exhibits a different lifestyle in mice than attaching/effacing pathogens. Unlike the diarrheal infection it causes in humans, S. typhimurium in mice is largely focused on invasion across the epithelium and into systemic organs, causing a typhoid-like disease without intestinal inflammation. However, if colonization resistance is ablated with antibiotics, it deploys several strategies to compete with the microbiota in the gut. Like C. rodentium, it uses type 3 secretion systems to target host epithelial and immune cells and induces inflammation and oxygenation of the cecum. This results in new energy and nutrient sources that it can exploit better than resident microbes.66, 112, 113 It has also evolved resistance to some host antimicrobial factors (eg, Reg3β114 and lipocalin-2).115 Similar to C. rodentium, its eventual clearance from the gut requires a fully functional microbiota.116 Whether humans exhibit a different disease from S. typhimurium because of differences in basic physiology or the microbiota is still unclear, although interestingly the microbiota from a healthy human did not protect mice as effectively as their native bacteria.117
Conclusions and Future Perspectives
Colonization resistance is a powerful phenomenon that arises from the extremely complex interactions of thousands of strains of bacteria in the gut with each other and with the host animal. Understanding the mechanisms of colonization resistance (Figures 1 and 2) is crucial to preventing and treating human disease. Approximately half a million children die each year from diarrheal diseases, for example. The infant microbiota is not fully developed and is intrinsically defective in colonization resistance ability.81 Maturation of the microbiota to a protective adult state in mice occurs around the time that solid food is starting to be introduced. This suggests that dietary interventions could be a way to boost the development of colonization resistance in infants. Synbiotic combinations of protective bacteria and prebiotics that support them could be even more powerful, if safe and effective strains can be isolated. Infants also have immature immune systems, and so they are defective in indirect mechanisms of colonization resistance, such as antibody production. This gap is filled by maternal antibodies transferred across the placenta and in breast milk but could be enhanced by maternal immunization.
The arsenal of weapons that bacteria use to compete with each other has historically been and continues to be a source of useful compounds. Discovering new types of antibacterials is especially important now as resistance to common antibiotics continues to increase, and the adverse effects of broad-spectrum antibiotics on the microbiota are increasingly appreciated. Discovering antibacterials from the gut microbiome that are more specific and with fewer adverse effects is another advantage of understanding colonization resistance.
Transfers of the total microbiota (fecal microbiota transplantation) are more effective than antibiotics at treating C. difficile infection,3 and they are being explored for other diseases as well. This presents an interesting opportunity to try to predict and understand why certain species from the donor can successfully colonize the recipient.118 Fecal microbiota transplantations could reveal mechanisms controlling normal microbiota stability and exclusion of nonnative strains.
Colonization resistance is not only a fascinating and complex phenomenon that incorporates aspects of ecology, microbiology, biochemistry, and immunology, but is also incredibly relevant to promoting human health and treating a wide range of diseases, from infection to autoimmunity to metabolic disorders.
Footnotes
Supported by NIH grants F32DK118800 (J.M.P.), DK091191 (G.N.), and DK095782 (G.N.).
Disclosures: None declared.
The Rous-Whipple Award is given by the American Society for Investigative Pathology (ASIP) to a senior pathologist with a distinguished career in experimental pathology research and continued productivity at the time of the award. Gabriel Núñez, M.D., recipient of the 2019 ASIP Rous-Whipple Award, delivered a lecture entitled “Role of the Microbiota in Host Defense and Inflammatory Disease” on October 21, 2018, at the Pathobiology for Investigators, Students, and Academicians (PISA) 2018 Annual Meeting in Ann Arbor, MI.
References
- 1.Bohnhoff M., Drake B.L., Miller C.P. Effect of streptomycin on susceptibility of intestinal tract to experimental Salmonella infection. Proc Soc Exp Biol Med. 1954;86:132–137. doi: 10.3181/00379727-86-21030. [DOI] [PubMed] [Google Scholar]; Bohnhoff M, Drake BL, Miller CP: Effect of streptomycin on susceptibility of intestinal tract to experimental Salmonella infection. Proc Soc Exp Biol Med 1954, 86:132-137. [DOI] [PubMed]
- 2.van der Waaij D., Berghuis-de Vries J.M., Lekkerkerk L-v. Colonization resistance of the digestive tract in conventional and antibiotic-treated mice. J Hyg. 1971;69:405–411. doi: 10.1017/s0022172400021653. [DOI] [PMC free article] [PubMed] [Google Scholar]; van der Waaij D, Berghuis-de Vries JM, Lekkerkerk L-v: Colonization resistance of the digestive tract in conventional and antibiotic-treated mice. J Hyg 1971, 69:405-411. [DOI] [PMC free article] [PubMed]
- 3.van Nood E., Vrieze A., Nieuwdorp M., Fuentes S., Zoetendal E.G., de Vos W.M., Visser C.E., Kuijper E.J., Bartelsman J.F., Tijssen J.G., Speelman P., Dijkgraaf M.G., Keller J.J. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]; van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JF, Tijssen JG, Speelman P, Dijkgraaf MG, Keller JJ: Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med 2013, 368:407-415. [DOI] [PubMed]
- 4.Sender R., Fuchs S., Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14:e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sender R, Fuchs S, Milo R: Revised estimates for the number of human and bacteria cells in the body. PLoS Biol 2016, 14:e1002533. [DOI] [PMC free article] [PubMed]
- 5.Martens E.C., Chiang H.C., Gordon J.I. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; Martens EC, Chiang HC, Gordon JI: Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 2008, 4:447-457. [DOI] [PMC free article] [PubMed]
- 6.Ng K.M., Ferreyra J.A., Higginbottom S.K., Lynch J.B., Kashyap P.C., Gopinath S., Naidu N., Choudhury B., Weimer B.C., Monack D.M., Sonnenburg J.L. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, Sonnenburg JL: Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 2013, 502:96-99. [DOI] [PMC free article] [PubMed]
- 7.Battaglioli E.J., Hale V.L., Chen J., Jeraldo P., Ruiz-Mojica C., Schmidt B.A., Rekdal V.M., Till L.M., Huq L., Smits S.A., Moor W.J., Jones-Hall Y., Smyrk T., Khanna S., Pardi D.S., Grover M., Patel R., Chia N., Nelson H., Sonnenburg J.L., Farrugia G., Kashyap P.C. Clostridioides difficile uses amino acids associated with gut microbial dysbiosis in a subset of patients with diarrhea. Sci Transl Med. 2018;10:eaam7019. doi: 10.1126/scitranslmed.aam7019. [DOI] [PMC free article] [PubMed] [Google Scholar]; Battaglioli EJ, Hale VL, Chen J, Jeraldo P, Ruiz-Mojica C, Schmidt BA, Rekdal VM, Till LM, Huq L, Smits SA, Moor WJ, Jones-Hall Y, Smyrk T, Khanna S, Pardi DS, Grover M, Patel R, Chia N, Nelson H, Sonnenburg JL, Farrugia G, Kashyap PC: Clostridioides difficile uses amino acids associated with gut microbial dysbiosis in a subset of patients with diarrhea. Sci Transl Med 2018, 10:eaam7019. [DOI] [PMC free article] [PubMed]
- 8.Jenior M.L., Leslie J.L., Young V.B., Schloss P.D. Clostridium difficile colonizes alternative nutrient niches during infection across distinct murine gut microbiomes. mSystems. 2017;2:e00063-17. doi: 10.1128/mSystems.00063-17. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jenior ML, Leslie JL, Young VB, Schloss PD: Clostridium difficile colonizes alternative nutrient niches during infection across distinct murine gut microbiomes. mSystems 2017, 2:e00063-17. [DOI] [PMC free article] [PubMed]
- 9.Theriot C.M., Koenigsknecht M.J., Carlson P.E., Jr., Hatton G.E., Nelson A.M., Li B., Huffnagle G.B., Z Li J., Young V.B. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun. 2014;5:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]; Theriot CM, Koenigsknecht MJ, Carlson PE, Jr., Hatton GE, Nelson AM, Li B, Huffnagle GB, J ZL, Young VB: Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun 2014, 5:3114. [DOI] [PMC free article] [PubMed]
- 10.Freter R., Brickner H., Botney M., Cleven D., Aranki A. Mechanisms that control bacterial populations in continuous-flow culture models of mouse large intestinal flora. Infect Immun. 1983;39:676–685. doi: 10.1128/iai.39.2.676-685.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]; Freter R, Brickner H, Botney M, Cleven D, Aranki A: Mechanisms that control bacterial populations in continuous-flow culture models of mouse large intestinal flora. Infect Immun 1983, 39:676-685. [DOI] [PMC free article] [PubMed]
- 11.Guiot H.F. Role of competition for substrate in bacterial antagonism in the gut. Infect Immun. 1982;38:887–892. doi: 10.1128/iai.38.3.887-892.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]; Guiot HF: Role of competition for substrate in bacterial antagonism in the gut. Infect Immun 1982, 38:887-892. [DOI] [PMC free article] [PubMed]
- 12.Sweeney N.J., Klemm P., McCormick B.A., Moller-Nielsen E., Utley M., Schembri M.A., Laux D.C., Cohen P.S. The Escherichia coli K-12 gntP gene allows E. coli F-18 to occupy a distinct nutritional niche in the streptomycin-treated mouse large intestine. Infect Immun. 1996;64:3497–3503. doi: 10.1128/iai.64.9.3497-3503.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sweeney NJ, Klemm P, McCormick BA, Moller-Nielsen E, Utley M, Schembri MA, Laux DC, Cohen PS: The Escherichia coli K-12 gntP gene allows E. coli F-18 to occupy a distinct nutritional niche in the streptomycin-treated mouse large intestine. Infect Immun 1996, 64:3497-3503. [DOI] [PMC free article] [PubMed]
- 13.Wilson K.H., Perini F. Role of competition for nutrients in suppression of Clostridium difficile by the colonic microflora. Infect Immun. 1988;56:2610–2614. doi: 10.1128/iai.56.10.2610-2614.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wilson KH, Perini F: Role of competition for nutrients in suppression of Clostridium difficile by the colonic microflora. Infect Immun 1988, 56:2610-2614. [DOI] [PMC free article] [PubMed]
- 14.Kashyap P.C., Marcobal A., Ursell L.K., Smits S.A., Sonnenburg E.D., Costello E.K., Higginbottom S.K., Domino S.E., Holmes S.P., Relman D.A., Knight R., Gordon J.I., Sonnenburg J.L. Genetically dictated change in host mucus carbohydrate landscape exerts a diet-dependent effect on the gut microbiota. Proc Natl Acad Sci U S A. 2013;110:17059–17064. doi: 10.1073/pnas.1306070110. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kashyap PC, Marcobal A, Ursell LK, Smits SA, Sonnenburg ED, Costello EK, Higginbottom SK, Domino SE, Holmes SP, Relman DA, Knight R, Gordon JI, Sonnenburg JL: Genetically dictated change in host mucus carbohydrate landscape exerts a diet-dependent effect on the gut microbiota. Proc Natl Acad Sci U S A 2013, 110:17059-17064. [DOI] [PMC free article] [PubMed]
- 15.Pickard J.M., Maurice C.F., Kinnebrew M.A., Abt M.C., Schenten D., Golovkina T.V., Bogatyrev S.R., Ismagilov R.F., Pamer E.G., Turnbaugh P.J., Chervonsky A.V. Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature. 2014;514:638–641. doi: 10.1038/nature13823. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pickard JM, Maurice CF, Kinnebrew MA, Abt MC, Schenten D, Golovkina TV, Bogatyrev SR, Ismagilov RF, Pamer EG, Turnbaugh PJ, Chervonsky AV: Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature 2014, 514:638-641. [DOI] [PMC free article] [PubMed]
- 16.Pham T.A., Clare S., Goulding D., Arasteh J.M., Stares M.D., Browne H.P., Keane J.A., Page A.J., Kumasaka N., Kane L., Mottram L., Harcourt K., Hale C., Arends M.J., Gaffney D.J., Sanger Mouse Genetics Project. Dougan G., Lawley T.D. Epithelial IL-22RA1-mediated fucosylation promotes intestinal colonization resistance to an opportunistic pathogen. Cell Host Microbe. 2014;16:504–516. doi: 10.1016/j.chom.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pham TA, Clare S, Goulding D, Arasteh JM, Stares MD, Browne HP, Keane JA, Page AJ, Kumasaka N, Kane L, Mottram L, Harcourt K, Hale C, Arends MJ, Gaffney DJ, Sanger Mouse Genetics Project, Dougan G, Lawley TD: Epithelial IL-22RA1-mediated fucosylation promotes intestinal colonization resistance to an opportunistic pathogen. Cell Host Microbe 2014, 16:504-516. [DOI] [PMC free article] [PubMed]
- 17.Goto Y., Obata T., Kunisawa J., Sato S., Ivanov I.I., Lamichhane A., Takeyama N., Kamioka M., Sakamoto M., Matsuki T., Setoyama H., Imaoka A., Uematsu S., Akira S., Domino S.E., Kulig P., Becher B., Renauld J.C., Sasakawa C., Umesaki Y., Benno Y., Kiyono H. Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science. 2014;345:1254009. doi: 10.1126/science.1254009. [DOI] [PMC free article] [PubMed] [Google Scholar]; Goto Y, Obata T, Kunisawa J, Sato S, Ivanov, II, Lamichhane A, Takeyama N, Kamioka M, Sakamoto M, Matsuki T, Setoyama H, Imaoka A, Uematsu S, Akira S, Domino SE, Kulig P, Becher B, Renauld JC, Sasakawa C, Umesaki Y, Benno Y, Kiyono H: Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science 2014, 345:1254009. [DOI] [PMC free article] [PubMed]
- 18.Huang Y.L., Chassard C., Hausmann M., von Itzstein M., Hennet T. Sialic acid catabolism drives intestinal inflammation and microbial dysbiosis in mice. Nat Commun. 2015;6:8141. doi: 10.1038/ncomms9141. [DOI] [PMC free article] [PubMed] [Google Scholar]; Huang YL, Chassard C, Hausmann M, von Itzstein M, Hennet T: Sialic acid catabolism drives intestinal inflammation and microbial dysbiosis in mice. Nat Commun 2015, 6:8141. [DOI] [PMC free article] [PubMed]
- 19.Pohl M.A., Romero-Gallo J., Guruge J.L., Tse D.B., Gordon J.I., Blaser M.J. Host-dependent Lewis (Le) antigen expression in Helicobacter pylori cells recovered from Leb-transgenic mice. J Exp Med. 2009;206:3061–3072. doi: 10.1084/jem.20090683. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pohl MA, Romero-Gallo J, Guruge JL, Tse DB, Gordon JI, Blaser MJ: Host-dependent Lewis (Le) antigen expression in Helicobacter pylori cells recovered from Leb-transgenic mice. J Exp Med 2009, 206:3061-3072. [DOI] [PMC free article] [PubMed]
- 20.Bry L., Falk P.G., Midtvedt T., Gordon J.I. A model of host-microbial interactions in an open mammalian ecosystem. Science. 1996;273:1380–1383. doi: 10.1126/science.273.5280.1380. [DOI] [PubMed] [Google Scholar]; Bry L, Falk PG, Midtvedt T, Gordon JI: A model of host-microbial interactions in an open mammalian ecosystem. Science 1996, 273:1380-1383. [DOI] [PubMed]
- 21.Terahara K., Nochi T., Yoshida M., Takahashi Y., Goto Y., Hatai H., Kurokawa S., Jang M.H., Kweon M.N., Domino S.E., Hiroi T., Yuki Y., Tsunetsugu-Yokota Y., Kobayashi K., Kiyono H. Distinct fucosylation of M cells and epithelial cells by Fut1 and Fut2, respectively, in response to intestinal environmental stress. Biochem Biophys Res Commun. 2011;404:822–828. doi: 10.1016/j.bbrc.2010.12.067. [DOI] [PubMed] [Google Scholar]; Terahara K, Nochi T, Yoshida M, Takahashi Y, Goto Y, Hatai H, Kurokawa S, Jang MH, Kweon MN, Domino SE, Hiroi T, Yuki Y, Tsunetsugu-Yokota Y, Kobayashi K, Kiyono H: Distinct fucosylation of M cells and epithelial cells by Fut1 and Fut2, respectively, in response to intestinal environmental stress. Biochem Biophys Res Commun 2011, 404:822-828. [DOI] [PubMed]
- 22.Turnbaugh P.J., Ridaura V.K., Faith J.J., Rey F.E., Knight R., Gordon J.I. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]; Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI: The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med 2009, 1:6ra14. [DOI] [PMC free article] [PubMed]
- 23.Sonnenburg E.D., Smits S.A., Tikhonov M., Higginbottom S.K., Wingreen N.S., Sonnenburg J.L. Diet-induced extinctions in the gut microbiota compound over generations. Nature. 2016;529:212–215. doi: 10.1038/nature16504. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, Sonnenburg JL: Diet-induced extinctions in the gut microbiota compound over generations. Nature 2016, 529:212-215. [DOI] [PMC free article] [PubMed]
- 24.Desai M.S., Seekatz A.M., Koropatkin N.M., Kamada N., Hickey C.A., Wolter M., Pudlo N.A., Kitamoto S., Terrapon N., Muller A., Young V.B., Henrissat B., Wilmes P., Stappenbeck T.S., Nunez G., Martens E.C. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell. 2016;167:1339–1353.e21. doi: 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]; Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, Young VB, Henrissat B, Wilmes P, Stappenbeck TS, Nunez G, Martens EC: A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 2016, 167:1339-1353 e21. [DOI] [PMC free article] [PubMed]
- 25.Hryckowian A.J., Van Treuren W., Smits S.A., Davis N.M., Gardner J.O., Bouley D.M., Sonnenburg J.L. Microbiota-accessible carbohydrates suppress Clostridium difficile infection in a murine model. Nat Microbiol. 2018;3:662–669. doi: 10.1038/s41564-018-0150-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hryckowian AJ, Van Treuren W, Smits SA, Davis NM, Gardner JO, Bouley DM, Sonnenburg JL: Microbiota-accessible carbohydrates suppress Clostridium difficile infection in a murine model. Nat Microbiol 2018, 3:662-669. [DOI] [PMC free article] [PubMed]
- 26.Kumar A., Henderson A., Forster G.M., Goodyear A.W., Weir T.L., Leach J.E., Dow S.W., Ryan E.P. Dietary rice bran promotes resistance to Salmonella enterica serovar Typhimurium colonization in mice. BMC Microbiol. 2012;12:71. doi: 10.1186/1471-2180-12-71. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kumar A, Henderson A, Forster GM, Goodyear AW, Weir TL, Leach JE, Dow SW, Ryan EP: Dietary rice bran promotes resistance to Salmonella enterica serovar Typhimurium colonization in mice. BMC Microbiol 2012, 12:71. [DOI] [PMC free article] [PubMed]
- 27.Petersen A., Heegaard P.M., Pedersen A.L., Andersen J.B., Sorensen R.B., Frokiaer H., Lahtinen S.J., Ouwehand A.C., Poulsen M., Licht T.R. Some putative prebiotics increase the severity of Salmonella enterica serovar Typhimurium infection in mice. BMC Microbiol. 2009;9:245. doi: 10.1186/1471-2180-9-245. [DOI] [PMC free article] [PubMed] [Google Scholar]; Petersen A, Heegaard PM, Pedersen AL, Andersen JB, Sorensen RB, Frokiaer H, Lahtinen SJ, Ouwehand AC, Poulsen M, Licht TR: Some putative prebiotics increase the severity of Salmonella enterica serovar Typhimurium infection in mice. BMC Microbiol 2009, 9:245. [DOI] [PMC free article] [PubMed]
- 28.Wlodarska M., Willing B.P., Bravo D.M., Finlay B.B. Phytonutrient diet supplementation promotes beneficial Clostridia species and intestinal mucus secretion resulting in protection against enteric infection. Sci Rep. 2015;5:9253. doi: 10.1038/srep09253. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wlodarska M, Willing BP, Bravo DM, Finlay BB: Phytonutrient diet supplementation promotes beneficial Clostridia species and intestinal mucus secretion resulting in protection against enteric infection. Sci Rep 2015, 5:9253. [DOI] [PMC free article] [PubMed]
- 29.Agus A., Denizot J., Thevenot J., Martinez-Medina M., Massier S., Sauvanet P., Bernalier-Donadille A., Denis S., Hofman P., Bonnet R., Billard E., Barnich N. Western diet induces a shift in microbiota composition enhancing susceptibility to adherent-invasive E. coli infection and intestinal inflammation. Sci Rep. 2016;6:19032. doi: 10.1038/srep19032. [DOI] [PMC free article] [PubMed] [Google Scholar]; Agus A, Denizot J, Thevenot J, Martinez-Medina M, Massier S, Sauvanet P, Bernalier-Donadille A, Denis S, Hofman P, Bonnet R, Billard E, Barnich N: Western diet induces a shift in microbiota composition enhancing susceptibility to Adherent-Invasive E. coli infection and intestinal inflammation. Sci Rep 2016, 6:19032. [DOI] [PMC free article] [PubMed]
- 30.Moore J.H., Pinheiro C.C., Zaenker E.I., Bolick D.T., Kolling G.L., van Opstal E., Noronha F.J., De Medeiros P.H., Rodriguez R.S., Lima A.A., Guerrant R.L., Warren C.A. Defined nutrient diets alter susceptibility to Clostridium difficile associated disease in a murine model. PLoS One. 2015;10:e0131829. doi: 10.1371/journal.pone.0131829. [DOI] [PMC free article] [PubMed] [Google Scholar]; Moore JH, Pinheiro CC, Zaenker EI, Bolick DT, Kolling GL, van Opstal E, Noronha FJ, De Medeiros PH, Rodriguez RS, Lima AA, Guerrant RL, Warren CA: Defined nutrient diets alter susceptibility to clostridium difficile associated disease in a murine model. PLoS One 2015, 10:e0131829. [DOI] [PMC free article] [PubMed]
- 31.Subramanian S., Huq S., Yatsunenko T., Haque R., Mahfuz M., Alam M.A., Benezra A., DeStefano J., Meier M.F., Muegge B.D., Barratt M.J., VanArendonk L.G., Zhang Q., Province M.A., Petri W.A., Jr., Ahmed T., Gordon J.I. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014;510:417–421. doi: 10.1038/nature13421. [DOI] [PMC free article] [PubMed] [Google Scholar]; Subramanian S, Huq S, Yatsunenko T, Haque R, Mahfuz M, Alam MA, Benezra A, DeStefano J, Meier MF, Muegge BD, Barratt MJ, VanArendonk LG, Zhang Q, Province MA, Petri WA, Jr., Ahmed T, Gordon JI: Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 2014, 510:417-421. [DOI] [PMC free article] [PubMed]
- 32.Shepherd E.S., DeLoache W.C., Pruss K.M., Whitaker W.R., Sonnenburg J.L. An exclusive metabolic niche enables strain engraftment in the gut microbiota. Nature. 2018;557:434–438. doi: 10.1038/s41586-018-0092-4. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shepherd ES, DeLoache WC, Pruss KM, Whitaker WR, Sonnenburg JL: An exclusive metabolic niche enables strain engraftment in the gut microbiota. Nature 2018, 557:434-438. [DOI] [PMC free article] [PubMed]
- 33.Kearney S.M., Gibbons S.M., Erdman S.E., Alm E.J. Orthogonal dietary niche enables reversible engraftment of a gut bacterial commensal. Cell Rep. 2018;24:1842–1851. doi: 10.1016/j.celrep.2018.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kearney SM, Gibbons SM, Erdman SE, Alm EJ: Orthogonal dietary niche enables reversible engraftment of a gut bacterial commensal. Cell Rep 2018, 24:1842-1851. [DOI] [PMC free article] [PubMed]
- 34.Maldonado-Gomez M.X., Martinez I., Bottacini F., O'Callaghan A., Ventura M., van Sinderen D., Hillmann B., Vangay P., Knights D., Hutkins R.W., Walter J. Stable engraftment of Bifidobacterium longum AH1206 in the human gut depends on individualized features of the resident microbiome. Cell Host Microbe. 2016;20:515–526. doi: 10.1016/j.chom.2016.09.001. [DOI] [PubMed] [Google Scholar]; Maldonado-Gomez MX, Martinez I, Bottacini F, O'Callaghan A, Ventura M, van Sinderen D, Hillmann B, Vangay P, Knights D, Hutkins RW, Walter J: Stable engraftment of Bifidobacterium longum AH1206 in the human gut depends on individualized features of the resident microbiome. Cell Host Microbe 2016, 20:515-526. [DOI] [PubMed]
- 35.Zmora N., Zilberman-Schapira G., Suez J., Mor U., Dori-Bachash M., Bashiardes S., Kotler E., Zur M., Regev-Lehavi D., Brik R.B., Federici S., Cohen Y., Linevsky R., Rothschild D., Moor A.E., Ben-Moshe S., Harmelin A., Itzkovitz S., Maharshak N., Shibolet O., Shapiro H., Pevsner-Fischer M., Sharon I., Halpern Z., Segal E., Elinav E. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell. 2018;174:1388–1405.e21. doi: 10.1016/j.cell.2018.08.041. [DOI] [PubMed] [Google Scholar]; Zmora N, Zilberman-Schapira G, Suez J, Mor U, Dori-Bachash M, Bashiardes S, Kotler E, Zur M, Regev-Lehavi D, Brik RB, Federici S, Cohen Y, Linevsky R, Rothschild D, Moor AE, Ben-Moshe S, Harmelin A, Itzkovitz S, Maharshak N, Shibolet O, Shapiro H, Pevsner-Fischer M, Sharon I, Halpern Z, Segal E, Elinav E: Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell 2018, 174:1388-1405 e21. [DOI] [PubMed]
- 36.Pultz N.J., Stiefel U., Subramanyan S., Helfand M.S., Donskey C.J. Mechanisms by which anaerobic microbiota inhibit the establishment in mice of intestinal colonization by vancomycin-resistant Enterococcus. J Infect Dis. 2005;191:949–956. doi: 10.1086/428090. [DOI] [PubMed] [Google Scholar]; Pultz NJ, Stiefel U, Subramanyan S, Helfand MS, Donskey CJ: Mechanisms by which anaerobic microbiota inhibit the establishment in mice of intestinal colonization by vancomycin-resistant Enterococcus. J Infect Dis 2005, 191:949-956. [DOI] [PubMed]
- 37.Cotter P.D., Ross R.P., Hill C. Bacteriocins: a viable alternative to antibiotics? Nat Rev Microbiol. 2013;11:95–105. doi: 10.1038/nrmicro2937. [DOI] [PubMed] [Google Scholar]; Cotter PD, Ross RP, Hill C: Bacteriocins - a viable alternative to antibiotics? Nat Rev Microbiol 2013, 11:95-105. [DOI] [PubMed]
- 38.Gillor O., Giladi I., Riley M.A. Persistence of colicinogenic Escherichia coli in the mouse gastrointestinal tract. BMC Microbiol. 2009;9:165. doi: 10.1186/1471-2180-9-165. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gillor O, Giladi I, Riley MA: Persistence of colicinogenic Escherichia coli in the mouse gastrointestinal tract. BMC Microbiol 2009, 9:165. [DOI] [PMC free article] [PubMed]
- 39.Kirkup B.C., Riley M.A. Antibiotic-mediated antagonism leads to a bacterial game of rock-paper-scissors in vivo. Nature. 2004;428:412–414. doi: 10.1038/nature02429. [DOI] [PubMed] [Google Scholar]; Kirkup BC, Riley MA: Antibiotic-mediated antagonism leads to a bacterial game of rock-paper-scissors in vivo. Nature 2004, 428:412-414. [DOI] [PubMed]
- 40.Gordon D.M., Riley M.A., Pinou T. Temporal changes in the frequency of colicinogeny in Escherichia coli from house mice. Microbiology. 1998;144:2233–2240. doi: 10.1099/00221287-144-8-2233. [DOI] [PubMed] [Google Scholar]; Gordon DM, Riley MA, Pinou T: Temporal changes in the frequency of colicinogeny in Escherichia coli from house mice. Microbiology 1998, 144 (Pt 8):2233-2240. [DOI] [PubMed]
- 41.Kommineni S., Bretl D.J., Lam V., Chakraborty R., Hayward M., Simpson P., Cao Y., Bousounis P., Kristich C.J., Salzman N.H. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature. 2015;526:719–722. doi: 10.1038/nature15524. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kommineni S, Bretl DJ, Lam V, Chakraborty R, Hayward M, Simpson P, Cao Y, Bousounis P, Kristich CJ, Salzman NH: Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature 2015, 526:719-722. [DOI] [PMC free article] [PubMed]
- 42.Roelofs K.G., Coyne M.J., Gentyala R.R., Chatzidaki-Livanis M., Comstock L.E. Bacteroidales secreted antimicrobial proteins target surface molecules necessary for gut colonization and mediate competition in vivo. MBio. 2016;7:e01055-16. doi: 10.1128/mBio.01055-16. [DOI] [PMC free article] [PubMed] [Google Scholar]; Roelofs KG, Coyne MJ, Gentyala RR, Chatzidaki-Livanis M, Comstock LE: Bacteroidales Secreted Antimicrobial Proteins Target Surface Molecules Necessary for Gut Colonization and Mediate Competition In Vivo. MBio 2016, 7:e01055-16. [DOI] [PMC free article] [PubMed]
- 43.Aoki S.K., Pamma R., Hernday A.D., Bickham J.E., Braaten B.A., Low D.A. Contact-dependent inhibition of growth in Escherichia coli. Science. 2005;309:1245–1248. doi: 10.1126/science.1115109. [DOI] [PubMed] [Google Scholar]; Aoki SK, Pamma R, Hernday AD, Bickham JE, Braaten BA, Low DA: Contact-dependent inhibition of growth in Escherichia coli. Science 2005, 309:1245-1248. [DOI] [PubMed]
- 44.Aoki S.K., Diner E.J., de Roodenbeke C.T., Burgess B.R., Poole S.J., Braaten B.A., Jones A.M., Webb J.S., Hayes C.S., Cotter P.A., Low D.A. A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature. 2010;468:439–442. doi: 10.1038/nature09490. [DOI] [PMC free article] [PubMed] [Google Scholar]; Aoki SK, Diner EJ, de Roodenbeke CT, Burgess BR, Poole SJ, Braaten BA, Jones AM, Webb JS, Hayes CS, Cotter PA, Low DA: A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature 2010, 468:439-442. [DOI] [PMC free article] [PubMed]
- 45.Pukatzki S., Ma A.T., Sturtevant D., Krastins B., Sarracino D., Nelson W.C., Heidelberg J.F., Mekalanos J.J. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci U S A. 2006;103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ: Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci U S A 2006, 103:1528-1533. [DOI] [PMC free article] [PubMed]
- 46.Bladergroen M.R., Badelt K., Spaink H.P. Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol Plant Microbe Interact. 2003;16:53–64. doi: 10.1094/MPMI.2003.16.1.53. [DOI] [PubMed] [Google Scholar]; Bladergroen MR, Badelt K, Spaink HP: Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol Plant Microbe Interact 2003, 16:53-64. [DOI] [PubMed]
- 47.Rao P.S., Yamada Y., Tan Y.P., Leung K.Y. Use of proteomics to identify novel virulence determinants that are required for Edwardsiella tarda pathogenesis. Mol Microbiol. 2004;53:573–586. doi: 10.1111/j.1365-2958.2004.04123.x. [DOI] [PubMed] [Google Scholar]; Rao PS, Yamada Y, Tan YP, Leung KY: Use of proteomics to identify novel virulence determinants that are required for Edwardsiella tarda pathogenesis. Mol Microbiol 2004, 53:573-586. [DOI] [PubMed]
- 48.Zheng J., Leung K.Y. Dissection of a type VI secretion system in Edwardsiella tarda. Mol Microbiol. 2007;66:1192–1206. doi: 10.1111/j.1365-2958.2007.05993.x. [DOI] [PubMed] [Google Scholar]; Zheng J, Leung KY: Dissection of a type VI secretion system in Edwardsiella tarda. Mol Microbiol 2007, 66:1192-1206. [DOI] [PubMed]
- 49.Hood R.D., Singh P., Hsu F., Guvener T., Carl M.A., Trinidad R.R., Silverman J.M., Ohlson B.B., Hicks K.G., Plemel R.L., Li M., Schwarz S., Wang W.Y., Merz A.J., Goodlett D.R., Mougous J.D. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe. 2010;7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hood RD, Singh P, Hsu F, Guvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, Li M, Schwarz S, Wang WY, Merz AJ, Goodlett DR, Mougous JD: A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 2010, 7:25-37. [DOI] [PMC free article] [PubMed]
- 50.Russell A.B., Hood R.D., Bui N.K., LeRoux M., Vollmer W., Mougous J.D. Type VI secretion delivers bacteriolytic effectors to target cells. Nature. 2011;475:343–347. doi: 10.1038/nature10244. [DOI] [PMC free article] [PubMed] [Google Scholar]; Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD: Type VI secretion delivers bacteriolytic effectors to target cells. Nature 2011, 475:343-347. [DOI] [PMC free article] [PubMed]
- 51.Russell A.B., Wexler A.G., Harding B.N., Whitney J.C., Bohn A.J., Goo Y.A., Tran B.Q., Barry N.A., Zheng H., Peterson S.B., Chou S., Gonen T., Goodlett D.R., Goodman A.L., Mougous J.D. A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe. 2014;16:227–236. doi: 10.1016/j.chom.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; Russell AB, Wexler AG, Harding BN, Whitney JC, Bohn AJ, Goo YA, Tran BQ, Barry NA, Zheng H, Peterson SB, Chou S, Gonen T, Goodlett DR, Goodman AL, Mougous JD: A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe 2014, 16:227-236. [DOI] [PMC free article] [PubMed]
- 52.Chatzidaki-Livanis M., Geva-Zatorsky N., Comstock L.E. Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. Proc Natl Acad Sci U S A. 2016;113:3627–3632. doi: 10.1073/pnas.1522510113. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chatzidaki-Livanis M, Geva-Zatorsky N, Comstock LE: Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. Proc Natl Acad Sci U S A 2016, 113:3627-3632. [DOI] [PMC free article] [PubMed]
- 53.Wexler A.G., Bao Y., Whitney J.C., Bobay L.M., Xavier J.B., Schofield W.B., Barry N.A., Russell A.B., Tran B.Q., Goo Y.A., Goodlett D.R., Ochman H., Mougous J.D., Goodman A.L. Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc Natl Acad Sci U S A. 2016;113:3639–3644. doi: 10.1073/pnas.1525637113. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wexler AG, Bao Y, Whitney JC, Bobay LM, Xavier JB, Schofield WB, Barry NA, Russell AB, Tran BQ, Goo YA, Goodlett DR, Ochman H, Mougous JD, Goodman AL: Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc Natl Acad Sci U S A 2016, 113:3639-3644. [DOI] [PMC free article] [PubMed]
- 54.Hecht A.L., Casterline B.W., Earley Z.M., Goo Y.A., Goodlett D.R., Bubeck Wardenburg J. Strain competition restricts colonization of an enteric pathogen and prevents colitis. EMBO Rep. 2016;17:1281–1291. doi: 10.15252/embr.201642282. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hecht AL, Casterline BW, Earley ZM, Goo YA, Goodlett DR, Bubeck Wardenburg J: Strain competition restricts colonization of an enteric pathogen and prevents colitis. EMBO Rep 2016, 17:1281-1291. [DOI] [PMC free article] [PubMed]
- 55.Verster A.J., Ross B.D., Radey M.C., Bao Y., Goodman A.L., Mougous J.D., Borenstein E. The landscape of type VI secretion across human gut microbiomes reveals its role in community composition. Cell Host Microbe. 2017;22:411–419.e4. doi: 10.1016/j.chom.2017.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]; Verster AJ, Ross BD, Radey MC, Bao Y, Goodman AL, Mougous JD, Borenstein E: The Landscape of Type VI Secretion across Human Gut Microbiomes Reveals Its Role in Community Composition. Cell Host Microbe 2017, 22:411-419 e4. [DOI] [PMC free article] [PubMed]
- 56.Anderson M.C., Vonaesch P., Saffarian A., Marteyn B.S., Sansonetti P.J. Shigella sonnei encodes a functional T6SS used for interbacterial competition and niche occupancy. Cell Host Microbe. 2017;21:769–776.e3. doi: 10.1016/j.chom.2017.05.004. [DOI] [PubMed] [Google Scholar]; Anderson MC, Vonaesch P, Saffarian A, Marteyn BS, Sansonetti PJ: Shigella sonnei Encodes a Functional T6SS Used for Interbacterial Competition and Niche Occupancy. Cell Host Microbe 2017, 21:769-776 e3. [DOI] [PubMed]
- 57.Sana T.G., Flaugnatti N., Lugo K.A., Lam L.H., Jacobson A., Baylot V., Durand E., Journet L., Cascales E., Monack D.M. Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc Natl Acad Sci U S A. 2016;113:E5044–E5051. doi: 10.1073/pnas.1608858113. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sana TG, Flaugnatti N, Lugo KA, Lam LH, Jacobson A, Baylot V, Durand E, Journet L, Cascales E, Monack DM: Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc Natl Acad Sci U S A 2016, 113:E5044-E5051. [DOI] [PMC free article] [PubMed]
- 58.Cao Z., Casabona M.G., Kneuper H., Chalmers J.D., Palmer T. The type VII secretion system of Staphylococcus aureus secretes a nuclease toxin that targets competitor bacteria. Nat Microbiol. 2016;2:16183. doi: 10.1038/nmicrobiol.2016.183. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cao Z, Casabona MG, Kneuper H, Chalmers JD, Palmer T: The type VII secretion system of Staphylococcus aureus secretes a nuclease toxin that targets competitor bacteria. Nat Microbiol 2016, 2:16183. [DOI] [PMC free article] [PubMed]
- 59.Whitney J.C., Peterson S.B., Kim J., Pazos M., Verster A.J., Radey M.C., Kulasekara H.D., Ching M.Q., Bullen N.P., Bryant D., Goo Y.A., Surette M.G., Borenstein E., Vollmer W., Mougous J.D. A broadly distributed toxin family mediates contact-dependent antagonism between gram-positive bacteria. Elife. 2017;6:e26938. doi: 10.7554/eLife.26938. [DOI] [PMC free article] [PubMed] [Google Scholar]; Whitney JC, Peterson SB, Kim J, Pazos M, Verster AJ, Radey MC, Kulasekara HD, Ching MQ, Bullen NP, Bryant D, Goo YA, Surette MG, Borenstein E, Vollmer W, Mougous JD: A broadly distributed toxin family mediates contact-dependent antagonism between gram-positive bacteria. Elife 2017, 6:e26938. [DOI] [PMC free article] [PubMed]
- 60.Mark Welch J.L., Hasegawa Y., McNulty N.P., Gordon J.I., Borisy G.G. Spatial organization of a model 15-member human gut microbiota established in gnotobiotic mice. Proc Natl Acad Sci U S A. 2017;114:E9105–E9114. doi: 10.1073/pnas.1711596114. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mark Welch JL, Hasegawa Y, McNulty NP, Gordon JI, Borisy GG: Spatial organization of a model 15-member human gut microbiota established in gnotobiotic mice. Proc Natl Acad Sci U S A 2017, 114:E9105-E9114. [DOI] [PMC free article] [PubMed]
- 61.Bohnhoff M., Miller C.P., Martin W.R. Resistance of the mouse's intestinal tract to experimental salmonella infection, I: factors which interfere with the initiation of infection by oral inoculation. J Exp Med. 1964;120:805–816. doi: 10.1084/jem.120.5.805. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bohnhoff M, Miller CP, Martin WR: Resistance of the mouse's intestinal tract to experimental salmonella infection. I. Factors which interfere with the initiation of infection by oral inoculation. J Exp Med 1964, 120:805-816. [DOI] [PMC free article] [PubMed]
- 62.Rolfe R.D. Role of volatile fatty acids in colonization resistance to Clostridium difficile. Infect Immun. 1984;45:185–191. doi: 10.1128/iai.45.1.185-191.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]; Rolfe RD: Role of volatile fatty acids in colonization resistance to Clostridium difficile. Infect Immun 1984, 45:185-191. [DOI] [PMC free article] [PubMed]
- 63.Jacobson A., Lam L., Rajendram M., Tamburini F., Honeycutt J., Pham T., Van Treuren W., Pruss K., Stabler S.R., Lugo K., Bouley D.M., Vilches-Moure J.G., Smith M., Sonnenburg J.L., Bhatt A.S., Huang K.C., Monack D. A gut commensal-produced metabolite mediates colonization resistance to Salmonella infection. Cell Host Microbe. 2018;24:296–307.e7. doi: 10.1016/j.chom.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jacobson A, Lam L, Rajendram M, Tamburini F, Honeycutt J, Pham T, Van Treuren W, Pruss K, Stabler SR, Lugo K, Bouley DM, Vilches-Moure JG, Smith M, Sonnenburg JL, Bhatt AS, Huang KC, Monack D: A Gut Commensal-Produced Metabolite Mediates Colonization Resistance to Salmonella Infection. Cell Host Microbe 2018, 24:296-307 e7. [DOI] [PMC free article] [PubMed]
- 64.Maier B.R., Onderdonk A.B., Baskett R.C., Hentges D.J. Shigella, indigenous flora interactions in mice. Am J Clin Nutr. 1972;25:1433–1440. doi: 10.1093/ajcn/25.12.1433. [DOI] [PubMed] [Google Scholar]; Maier BR, Onderdonk AB, Baskett RC, Hentges DJ: Shigella, indigenous flora interactions in mice. Am J Clin Nutr 1972, 25:1433-1440. [DOI] [PubMed]
- 65.Kelly C.J., Zheng L., Campbell E.L., Saeedi B., Scholz C.C., Bayless A.J., Wilson K.E., Glover L.E., Kominsky D.J., Magnuson A., Weir T.L., Ehrentraut S.F., Pickel C., Kuhn K.A., Lanis J.M., Nguyen V., Taylor C.T., Colgan S.P. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe. 2015;17:662–671. doi: 10.1016/j.chom.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kelly CJ, Zheng L, Campbell EL, Saeedi B, Scholz CC, Bayless AJ, Wilson KE, Glover LE, Kominsky DJ, Magnuson A, Weir TL, Ehrentraut SF, Pickel C, Kuhn KA, Lanis JM, Nguyen V, Taylor CT, Colgan SP: Crosstalk between Microbiota-Derived Short-Chain Fatty Acids and Intestinal Epithelial HIF Augments Tissue Barrier Function. Cell Host Microbe 2015, 17:662-671. [DOI] [PMC free article] [PubMed]
- 66.Rivera-Chavez F., Zhang L.F., Faber F., Lopez C.A., Byndloss M.X., Olsan E.E., Xu G., Velazquez E.M., Lebrilla C.B., Winter S.E., Baumler A.J. Depletion of butyrate-producing Clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host Microbe. 2016;19:443–454. doi: 10.1016/j.chom.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; Rivera-Chavez F, Zhang LF, Faber F, Lopez CA, Byndloss MX, Olsan EE, Xu G, Velazquez EM, Lebrilla CB, Winter SE, Baumler AJ: Depletion of Butyrate-Producing Clostridia from the Gut Microbiota Drives an Aerobic Luminal Expansion of Salmonella. Cell Host Microbe 2016, 19:443-454. [DOI] [PMC free article] [PubMed]
- 67.Litvak Y., Byndloss M.X., Tsolis R.M., Baumler A.J. Dysbiotic Proteobacteria expansion: a microbial signature of epithelial dysfunction. Curr Opin Microbiol. 2017;39:1–6. doi: 10.1016/j.mib.2017.07.003. [DOI] [PubMed] [Google Scholar]; Litvak Y, Byndloss MX, Tsolis RM, Baumler AJ: Dysbiotic Proteobacteria expansion: a microbial signature of epithelial dysfunction. Curr Opin Microbiol 2017, 39:1-6. [DOI] [PubMed]
- 68.Ridlon J.M., Kang D.J., Hylemon P.B. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]; Ridlon JM, Kang DJ, Hylemon PB: Bile salt biotransformations by human intestinal bacteria. J Lipid Res 2006, 47:241-259. [DOI] [PubMed]
- 69.Buffie C.G., Bucci V., Stein R.R., McKenney P.T., Ling L., Gobourne A., No D., Liu H., Kinnebrew M., Viale A., Littmann E., van den Brink M.R., Jenq R.R., Taur Y., Sander C., Cross J.R., Toussaint N.C., Xavier J.B., Pamer E.G. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]; Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, Littmann E, van den Brink MR, Jenq RR, Taur Y, Sander C, Cross JR, Toussaint NC, Xavier JB, Pamer EG: Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 2015, 517:205-208. [DOI] [PMC free article] [PubMed]
- 70.Wilson K.H. Efficiency of various bile salt preparations for stimulation of Clostridium difficile spore germination. J Clin Microbiol. 1983;18:1017–1019. doi: 10.1128/jcm.18.4.1017-1019.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wilson KH: Efficiency of various bile salt preparations for stimulation of Clostridium difficile spore germination. J Clin Microbiol 1983, 18:1017-1019. [DOI] [PMC free article] [PubMed]
- 71.Theriot C.M., Bowman A.A., Young V.B. Antibiotic-induced alterations of the gut microbiota alter secondary bile acid production and allow for Clostridium difficile spore germination and outgrowth in the large intestine. mSphere. 2016;1:e00045-15. doi: 10.1128/mSphere.00045-15. [DOI] [PMC free article] [PubMed] [Google Scholar]; Theriot CM, Bowman AA, Young VB: Antibiotic-Induced Alterations of the Gut Microbiota Alter Secondary Bile Acid Production and Allow for Clostridium difficile Spore Germination and Outgrowth in the Large Intestine. mSphere 2016, 1:e00045-15. [DOI] [PMC free article] [PubMed]
- 72.Devkota S., Wang Y., Musch M.W., Leone V., Fehlner-Peach H., Nadimpalli A., Antonopoulos D.A., Jabri B., Chang E.B. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature. 2012;487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]; Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, Antonopoulos DA, Jabri B, Chang EB: Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature 2012, 487:104-108. [DOI] [PMC free article] [PubMed]
- 73.Yao L., Seaton S.C., Ndousse-Fetter S., Adhikari A.A., DiBenedetto N., Mina A.I., Banks A.S., Bry L., Devlin A.S. A selective gut bacterial bile salt hydrolase alters host metabolism. Elife. 2018;7:e37182. doi: 10.7554/eLife.37182. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yao L, Seaton SC, Ndousse-Fetter S, Adhikari AA, DiBenedetto N, Mina AI, Banks AS, Bry L, Devlin AS: A selective gut bacterial bile salt hydrolase alters host metabolism. Elife 2018, 7:e37182. [DOI] [PMC free article] [PubMed]
- 74.Franzenburg S., Walter J., Kunzel S., Wang J., Baines J.F., Bosch T.C., Fraune S. Distinct antimicrobial peptide expression determines host species-specific bacterial associations. Proc Natl Acad Sci U S A. 2013;110:E3730–E3738. doi: 10.1073/pnas.1304960110. [DOI] [PMC free article] [PubMed] [Google Scholar]; Franzenburg S, Walter J, Kunzel S, Wang J, Baines JF, Bosch TC, Fraune S: Distinct antimicrobial peptide expression determines host species-specific bacterial associations. Proc Natl Acad Sci U S A 2013, 110:E3730-E3738. [DOI] [PMC free article] [PubMed]
- 75.Rawls J.F., Mahowald M.A., Ley R.E., Gordon J.I. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. 2006;127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]; Rawls JF, Mahowald MA, Ley RE, Gordon JI: Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell 2006, 127:423-433. [DOI] [PMC free article] [PubMed]
- 76.Larsson E., Tremaroli V., Lee Y.S., Koren O., Nookaew I., Fricker A., Nielsen J., Ley R.E., Backhed F. Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88. Gut. 2012;61:1124–1131. doi: 10.1136/gutjnl-2011-301104. [DOI] [PMC free article] [PubMed] [Google Scholar]; Larsson E, Tremaroli V, Lee YS, Koren O, Nookaew I, Fricker A, Nielsen J, Ley RE, Backhed F: Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88. Gut 2012, 61:1124-1131. [DOI] [PMC free article] [PubMed]
- 77.Brubaker S.W., Bonham K.S., Zanoni I., Kagan J.C. Innate immune pattern recognition: a cell biological perspective. Annu Rev Immunol. 2015;33:257–290. doi: 10.1146/annurev-immunol-032414-112240. [DOI] [PMC free article] [PubMed] [Google Scholar]; Brubaker SW, Bonham KS, Zanoni I, Kagan JC: Innate immune pattern recognition: a cell biological perspective. Annu Rev Immunol 2015, 33:257-290. [DOI] [PMC free article] [PubMed]
- 78.Rakoff-Nahoum S., Kong Y., Kleinstein S.H., Subramanian S., Ahern P.P., Gordon J.I., Medzhitov R. Analysis of gene-environment interactions in postnatal development of the mammalian intestine. Proc Natl Acad Sci U S A. 2015;112:1929–1936. doi: 10.1073/pnas.1424886112. [DOI] [PMC free article] [PubMed] [Google Scholar]; Rakoff-Nahoum S, Kong Y, Kleinstein SH, Subramanian S, Ahern PP, Gordon JI, Medzhitov R: Analysis of gene-environment interactions in postnatal development of the mammalian intestine. Proc Natl Acad Sci U S A 2015, 112:1929-1936. [DOI] [PMC free article] [PubMed]
- 79.Kinnebrew M.A., Ubeda C., Zenewicz L.A., Smith N., Flavell R.A., Pamer E.G. Bacterial flagellin stimulates toll-like receptor 5-dependent defense against vancomycin-resistant Enterococcus infection. J Infect Dis. 2010;201:534–543. doi: 10.1086/650203. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kinnebrew MA, Ubeda C, Zenewicz LA, Smith N, Flavell RA, Pamer EG: Bacterial flagellin stimulates Toll-like receptor 5-dependent defense against vancomycin-resistant Enterococcus infection. J Infect Dis 2010, 201:534-543. [DOI] [PMC free article] [PubMed]
- 80.Zheng Y., Valdez P.A., Danilenko D.M., Hu Y., Sa S.M., Gong Q., Abbas A.R., Modrusan Z., Ghilardi N., de Sauvage F.J., Ouyang W. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]; Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, Ouyang W: Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med 2008, 14:282-289. [DOI] [PubMed]
- 81.Kim Y.G., Sakamoto K., Seo S.U., Pickard J.M., Gillilland M.G., 3rd, Pudlo N.A., Hoostal M., Li X., Wang T.D., Feehley T., Stefka A.T., Schmidt T.M., Martens E.C., Fukuda S., Inohara N., Nagler C.R., Nunez G. Neonatal acquisition of Clostridia species protects against colonization by bacterial pathogens. Science. 2017;356:315–319. doi: 10.1126/science.aag2029. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kim YG, Sakamoto K, Seo SU, Pickard JM, Gillilland MG, 3rd, Pudlo NA, Hoostal M, Li X, Wang TD, Feehley T, Stefka AT, Schmidt TM, Martens EC, Fukuda S, Inohara N, Nagler CR, Nunez G: Neonatal acquisition of Clostridia species protects against colonization by bacterial pathogens. Science 2017, 356:315-319. [DOI] [PMC free article] [PubMed]
- 82.Behnsen J., Jellbauer S., Wong C.P., Edwards R.A., George M.D., Ouyang W., Raffatellu M. The cytokine IL-22 promotes pathogen colonization by suppressing related commensal bacteria. Immunity. 2014;40:262–273. doi: 10.1016/j.immuni.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; Behnsen J, Jellbauer S, Wong CP, Edwards RA, George MD, Ouyang W, Raffatellu M: The cytokine IL-22 promotes pathogen colonization by suppressing related commensal bacteria. Immunity 2014, 40:262-273. [DOI] [PMC free article] [PubMed]
- 83.Kim C.H. Immune regulation by microbiome metabolites. Immunology. 2018;154:220–229. doi: 10.1111/imm.12930. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kim CH: Immune regulation by microbiome metabolites. Immunology 2018, 154:220-229. [DOI] [PMC free article] [PubMed]
- 84.Bunker J.J., Flynn T.M., Koval J.C., Shaw D.G., Meisel M., McDonald B.D., Ishizuka I.E., Dent A.L., Wilson P.C., Jabri B., Antonopoulos D.A., Bendelac A. Innate and adaptive humoral responses coat distinct commensal bacteria with immunoglobulin A. Immunity. 2015;43:541–553. doi: 10.1016/j.immuni.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bunker JJ, Flynn TM, Koval JC, Shaw DG, Meisel M, McDonald BD, Ishizuka IE, Dent AL, Wilson PC, Jabri B, Antonopoulos DA, Bendelac A: Innate and adaptive humoral responses coat distinct commensal bacteria with immunoglobulin A. Immunity 2015, 43:541-553. [DOI] [PMC free article] [PubMed]
- 85.van der Waaij L.A., Limburg P.C., Mesander G., van der Waaij D. In vivo IgA coating of anaerobic bacteria in human faeces. Gut. 1996;38:348–354. doi: 10.1136/gut.38.3.348. [DOI] [PMC free article] [PubMed] [Google Scholar]; van der Waaij LA, Limburg PC, Mesander G, van der Waaij D: In vivo IgA coating of anaerobic bacteria in human faeces. Gut 1996, 38:348-354. [DOI] [PMC free article] [PubMed]
- 86.Moreau M.C., Ducluzeau R., Guy-Grand D., Muller M.C. Increase in the population of duodenal immunoglobulin A plasmocytes in axenic mice associated with different living or dead bacterial strains of intestinal origin. Infect Immun. 1978;21:532–539. doi: 10.1128/iai.21.2.532-539.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]; Moreau MC, Ducluzeau R, Guy-Grand D, Muller MC: Increase in the population of duodenal immunoglobulin A plasmocytes in axenic mice associated with different living or dead bacterial strains of intestinal origin. Infect Immun 1978, 21:532-539. [DOI] [PMC free article] [PubMed]
- 87.Hapfelmeier S., Lawson M.A., Slack E., Kirundi J.K., Stoel M., Heikenwalder M., Cahenzli J., Velykoredko Y., Balmer M.L., Endt K., Geuking M.B., Curtiss R., 3rd, McCoy K.D., Macpherson A.J. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science. 2010;328:1705–1709. doi: 10.1126/science.1188454. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hapfelmeier S, Lawson MA, Slack E, Kirundi JK, Stoel M, Heikenwalder M, Cahenzli J, Velykoredko Y, Balmer ML, Endt K, Geuking MB, Curtiss R, 3rd, McCoy KD, Macpherson AJ: Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science 2010, 328:1705-1709. [DOI] [PMC free article] [PubMed]
- 88.Macpherson A.J., Gatto D., Sainsbury E., Harriman G.R., Hengartner H., Zinkernagel R.M. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science. 2000;288:2222–2226. doi: 10.1126/science.288.5474.2222. [DOI] [PubMed] [Google Scholar]; Macpherson AJ, Gatto D, Sainsbury E, Harriman GR, Hengartner H, Zinkernagel RM: A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science 2000, 288:2222-2226. [DOI] [PubMed]
- 89.Bunker J.J., Erickson S.A., Flynn T.M., Henry C., Koval J.C., Meisel M., Jabri B., Antonopoulos D.A., Wilson P.C., Bendelac A. Natural polyreactive IgA antibodies coat the intestinal microbiota. Science. 2017;358:eaan6619. doi: 10.1126/science.aan6619. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bunker JJ, Erickson SA, Flynn TM, Henry C, Koval JC, Meisel M, Jabri B, Antonopoulos DA, Wilson PC, Bendelac A: Natural polyreactive IgA antibodies coat the intestinal microbiota. Science 2017, 358. [DOI] [PMC free article] [PubMed]
- 90.Okai S., Usui F., Yokota S., Hori I.Y., Hasegawa M., Nakamura T., Kurosawa M., Okada S., Yamamoto K., Nishiyama E., Mori H., Yamada T., Kurokawa K., Matsumoto S., Nanno M., Naito T., Watanabe Y., Kato T., Miyauchi E., Ohno H., Shinkura R. High-affinity monoclonal IgA regulates gut microbiota and prevents colitis in mice. Nat Microbiol. 2016;1:16103. doi: 10.1038/nmicrobiol.2016.103. [DOI] [PubMed] [Google Scholar]; Okai S, Usui F, Yokota S, Hori IY, Hasegawa M, Nakamura T, Kurosawa M, Okada S, Yamamoto K, Nishiyama E, Mori H, Yamada T, Kurokawa K, Matsumoto S, Nanno M, Naito T, Watanabe Y, Kato T, Miyauchi E, Ohno H, Shinkura R: High-affinity monoclonal IgA regulates gut microbiota and prevents colitis in mice. Nat Microbiol 2016, 1:16103. [DOI] [PubMed]
- 91.Palm N.W., de Zoete M.R., Cullen T.W., Barry N.A., Stefanowski J., Hao L., Degnan P.H., Hu J., Peter I., Zhang W., Ruggiero E., Cho J.H., Goodman A.L., Flavell R.A. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158:1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, Degnan PH, Hu J, Peter I, Zhang W, Ruggiero E, Cho JH, Goodman AL, Flavell RA: Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 2014, 158:1000-1010. [DOI] [PMC free article] [PubMed]
- 92.Cullender T.C., Chassaing B., Janzon A., Kumar K., Muller C.E., Werner J.J., Angenent L.T., Bell M.E., Hay A.G., Peterson D.A., Walter J., Vijay-Kumar M., Gewirtz A.T., Ley R.E. Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell Host Microbe. 2013;14:571–581. doi: 10.1016/j.chom.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cullender TC, Chassaing B, Janzon A, Kumar K, Muller CE, Werner JJ, Angenent LT, Bell ME, Hay AG, Peterson DA, Walter J, Vijay-Kumar M, Gewirtz AT, Ley RE: Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell Host Microbe 2013, 14:571-581. [DOI] [PMC free article] [PubMed]
- 93.Fransen F., Zagato E., Mazzini E., Fosso B., Manzari C., El Aidy S., Chiavelli A., D'Erchia A.M., Sethi M.K., Pabst O., Marzano M., Moretti S., Romani L., Penna G., Pesole G., Rescigno M. BALB/c and C57BL/6 mice differ in polyreactive IgA abundance, which impacts the generation of antigen-specific IgA and microbiota diversity. Immunity. 2015;43:527–540. doi: 10.1016/j.immuni.2015.08.011. [DOI] [PubMed] [Google Scholar]; Fransen F, Zagato E, Mazzini E, Fosso B, Manzari C, El Aidy S, Chiavelli A, D'Erchia AM, Sethi MK, Pabst O, Marzano M, Moretti S, Romani L, Penna G, Pesole G, Rescigno M: BALB/c and C57BL/6 mice differ in polyreactive IgA abundance, which Impacts the generation of antigen-specific IgA and microbiota diversity. Immunity 2015, 43:527-540. [DOI] [PubMed]
- 94.Zeng M.Y., Cisalpino D., Varadarajan S., Hellman J., Warren H.S., Cascalho M., Inohara N., Nunez G. Gut microbiota-induced immunoglobulin G controls systemic infection by symbiotic bacteria and pathogens. Immunity. 2016;44:647–658. doi: 10.1016/j.immuni.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zeng MY, Cisalpino D, Varadarajan S, Hellman J, Warren HS, Cascalho M, Inohara N, Nunez G: Gut microbiota-induced immunoglobulin G controls systemic infection by symbiotic bacteria and pathogens. Immunity 2016, 44:647-658. [DOI] [PMC free article] [PubMed]
- 95.Koch M.A., Reiner G.L., Lugo K.A., Kreuk L.S., Stanbery A.G., Ansaldo E., Seher T.D., Ludington W.B., Barton G.M. Maternal IgG and IgA antibodies dampen mucosal T helper cell responses in early life. Cell. 2016;165:827–841. doi: 10.1016/j.cell.2016.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]; Koch MA, Reiner GL, Lugo KA, Kreuk LS, Stanbery AG, Ansaldo E, Seher TD, Ludington WB, Barton GM: Maternal IgG and IgA antibodies dampen mucosal T helper cell responses in early life. Cell 2016, 165:827-841. [DOI] [PMC free article] [PubMed]
- 96.Suzuki K., Meek B., Doi Y., Muramatsu M., Chiba T., Honjo T., Fagarasan S. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc Natl Acad Sci U S A. 2004;101:1981–1986. doi: 10.1073/pnas.0307317101. [DOI] [PMC free article] [PubMed] [Google Scholar]; Suzuki K, Meek B, Doi Y, Muramatsu M, Chiba T, Honjo T, Fagarasan S: Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc Natl Acad Sci U S A 2004, 101:1981-1986. [DOI] [PMC free article] [PubMed]
- 97.Sait L., Galic M., Strugnell R.A., Janssen P.H. Secretory antibodies do not affect the composition of the bacterial microbiota in the terminal ileum of 10-week-old mice. Appl Environ Microbiol. 2003;69:2100–2109. doi: 10.1128/AEM.69.4.2100-2109.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sait L, Galic M, Strugnell RA, Janssen PH: Secretory antibodies do not affect the composition of the bacterial microbiota in the terminal ileum of 10-week-old mice. Appl Environ Microbiol 2003, 69:2100-2109. [DOI] [PMC free article] [PubMed]
- 98.Betz K.J., Maier E.A., Amarachintha S., Wu D., Karmele E.P., Kinder J.M., Steinbrecher K.A., McNeal M.M., Luzader D.H., Hogan S.P., Moore S.R. Enhanced survival following oral and systemic Salmonella enterica serovar Typhimurium infection in polymeric immunoglobulin receptor knockout mice. PLoS One. 2018;13:e0198434. doi: 10.1371/journal.pone.0198434. [DOI] [PMC free article] [PubMed] [Google Scholar]; Betz KJ, Maier EA, Amarachintha S, Wu D, Karmele EP, Kinder JM, Steinbrecher KA, McNeal MM, Luzader DH, Hogan SP, Moore SR: Enhanced survival following oral and systemic Salmonella enterica serovar Typhimurium infection in polymeric immunoglobulin receptor knockout mice. PLoS One 2018, 13:e0198434. [DOI] [PMC free article] [PubMed]
- 99.Wijburg O.L., Uren T.K., Simpfendorfer K., Johansen F.E., Brandtzaeg P., Strugnell R.A. Innate secretory antibodies protect against natural Salmonella typhimurium infection. J Exp Med. 2006;203:21–26. doi: 10.1084/jem.20052093. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wijburg OL, Uren TK, Simpfendorfer K, Johansen FE, Brandtzaeg P, Strugnell RA: Innate secretory antibodies protect against natural Salmonella typhimurium infection. J Exp Med 2006, 203:21-26. [DOI] [PMC free article] [PubMed]
- 100.Peterson D.A., Planer J.D., Guruge J.L., Xue L., Downey-Virgin W., Goodman A.L., Seedorf H., Gordon J.I. Characterizing the interactions between a naturally primed immunoglobulin A and its conserved Bacteroides thetaiotaomicron species-specific epitope in gnotobiotic mice. J Biol Chem. 2015;290:12630–12649. doi: 10.1074/jbc.M114.633800. [DOI] [PMC free article] [PubMed] [Google Scholar]; Peterson DA, Planer JD, Guruge JL, Xue L, Downey-Virgin W, Goodman AL, Seedorf H, Gordon JI: Characterizing the interactions between a naturally primed immunoglobulin A and its conserved Bacteroides thetaiotaomicron species-specific epitope in gnotobiotic mice. J Biol Chem 2015, 290:12630-12649. [DOI] [PMC free article] [PubMed]
- 101.Nakajima A., Vogelzang A., Maruya M., Miyajima M., Murata M., Son A., Kuwahara T., Tsuruyama T., Yamada S., Matsuura M., Nakase H., Peterson D.A., Fagarasan S., Suzuki K. IgA regulates the composition and metabolic function of gut microbiota by promoting symbiosis between bacteria. J Exp Med. 2018;215:2019–2034. doi: 10.1084/jem.20180427. [DOI] [PMC free article] [PubMed] [Google Scholar]; Nakajima A, Vogelzang A, Maruya M, Miyajima M, Murata M, Son A, Kuwahara T, Tsuruyama T, Yamada S, Matsuura M, Nakase H, Peterson DA, Fagarasan S, Suzuki K: IgA regulates the composition and metabolic function of gut microbiota by promoting symbiosis between bacteria. J Exp Med 2018, 215:2019-2034. [DOI] [PMC free article] [PubMed]
- 102.Donaldson G.P., Ladinsky M.S., Yu K.B., Sanders J.G., Yoo B.B., Chou W.C., Conner M.E., Earl A.M., Knight R., Bjorkman P.J., Mazmanian S.K. Gut microbiota utilize immunoglobulin A for mucosal colonization. Science. 2018;360:795–800. doi: 10.1126/science.aaq0926. [DOI] [PMC free article] [PubMed] [Google Scholar]; Donaldson GP, Ladinsky MS, Yu KB, Sanders JG, Yoo BB, Chou WC, Conner ME, Earl AM, Knight R, Bjorkman PJ, Mazmanian SK: Gut microbiota utilize immunoglobulin A for mucosal colonization. Science 2018, 360:795-800. [DOI] [PMC free article] [PubMed]
- 103.Fadlallah J., El Kafsi H., Sterlin D., Juste C., Parizot C., Dorgham K., Autaa G., Gouas D., Almeida M., Lepage P., Pons N., Le Chatelier E., Levenez F., Kennedy S., Galleron N., de Barros J.P., Malphettes M., Galicier L., Boutboul D., Mathian A., Miyara M., Oksenhendler E., Amoura Z., Dore J., Fieschi C., Ehrlich S.D., Larsen M., Gorochov G. Microbial ecology perturbation in human IgA deficiency. Sci Transl Med. 2018;10:eaan1217. doi: 10.1126/scitranslmed.aan1217. [DOI] [PubMed] [Google Scholar]; Fadlallah J, El Kafsi H, Sterlin D, Juste C, Parizot C, Dorgham K, Autaa G, Gouas D, Almeida M, Lepage P, Pons N, Le Chatelier E, Levenez F, Kennedy S, Galleron N, de Barros JP, Malphettes M, Galicier L, Boutboul D, Mathian A, Miyara M, Oksenhendler E, Amoura Z, Dore J, Fieschi C, Ehrlich SD, Larsen M, Gorochov G: Microbial ecology perturbation in human IgA deficiency. Sci Transl Med 2018, 10:eaan1217. [DOI] [PubMed]
- 104.Ludvigsson J.F., Neovius M., Hammarstrom L. Risk of infections among 2100 individuals with IgA deficiency: a nationwide cohort study. J Clin Immunol. 2016;36:134–140. doi: 10.1007/s10875-015-0230-9. [DOI] [PubMed] [Google Scholar]; Ludvigsson JF, Neovius M, Hammarstrom L: Risk of Infections Among 2100 Individuals with IgA Deficiency: a Nationwide Cohort Study. J Clin Immunol 2016, 36:134-140. [DOI] [PubMed]
- 105.Ludvigsson J.F., Neovius M., Hammarstrom L. Association between IgA deficiency & other autoimmune conditions: a population-based matched cohort study. J Clin Immunol. 2014;34:444–451. doi: 10.1007/s10875-014-0009-4. [DOI] [PubMed] [Google Scholar]; Ludvigsson JF, Neovius M, Hammarstrom L: Association between IgA deficiency & other autoimmune conditions: a population-based matched cohort study. J Clin Immunol 2014, 34:444-451. [DOI] [PubMed]
- 106.Ivanov, Atarashi K., Manel N., Brodie E.L., Shima T., Karaoz U., Wei D., Goldfarb K.C., Santee C.A., Lynch S.V., Tanoue T., Imaoka A., Itoh K., Takeda K., Umesaki Y., Honda K., Littman D.R. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ivanov, II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR: Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009, 139:485-498. [DOI] [PMC free article] [PubMed]
- 107.Shaw M.H., Kamada N., Kim Y.G., Nunez G. Microbiota-induced IL-1beta, but not IL-6, is critical for the development of steady-state TH17 cells in the intestine. J Exp Med. 2012;209:251–258. doi: 10.1084/jem.20111703. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shaw MH, Kamada N, Kim YG, Nunez G: Microbiota-induced IL-1beta, but not IL-6, is critical for the development of steady-state TH17 cells in the intestine. J Exp Med 2012, 209:251-258. [DOI] [PMC free article] [PubMed]
- 108.Tedesco F.J., Barton R.W., Alpers D.H. Clindamycin-associated colitis: a prospective study. Ann Intern Med. 1974;81:429–433. doi: 10.7326/0003-4819-81-4-429. [DOI] [PubMed] [Google Scholar]; Tedesco FJ, Barton RW, Alpers DH: Clindamycin-associated colitis. A prospective study. Ann Intern Med 1974, 81:429-433. [DOI] [PubMed]
- 109.Wiles S., Dougan G., Frankel G. Emergence of a “hyperinfectious” bacterial state after passage of Citrobacter rodentium through the host gastrointestinal tract. Cell Microbiol. 2005;7:1163–1172. doi: 10.1111/j.1462-5822.2005.00544.x. [DOI] [PubMed] [Google Scholar]; Wiles S, Dougan G, Frankel G: Emergence of a 'hyperinfectious' bacterial state after passage of Citrobacter rodentium through the host gastrointestinal tract. Cell Microbiol 2005, 7:1163-1172. [DOI] [PubMed]
- 110.Lopez C.A., Miller B.M., Rivera-Chavez F., Velazquez E.M., Byndloss M.X., Chavez-Arroyo A., Lokken K.L., Tsolis R.M., Winter S.E., Baumler A.J. Virulence factors enhance Citrobacter rodentium expansion through aerobic respiration. Science. 2016;353:1249–1253. doi: 10.1126/science.aag3042. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lopez CA, Miller BM, Rivera-Chavez F, Velazquez EM, Byndloss MX, Chavez-Arroyo A, Lokken KL, Tsolis RM, Winter SE, Baumler AJ: Virulence factors enhance Citrobacter rodentium expansion through aerobic respiration. Science 2016, 353:1249-1253. [DOI] [PMC free article] [PubMed]
- 111.Kamada N., Kim Y.G., Sham H.P., Vallance B.A., Puente J.L., Martens E.C., Nunez G. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science. 2012;336:1325–1329. doi: 10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kamada N, Kim YG, Sham HP, Vallance BA, Puente JL, Martens EC, Nunez G: Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science 2012, 336:1325-1329. [DOI] [PMC free article] [PubMed]
- 112.Winter S.E., Thiennimitr P., Winter M.G., Butler B.P., Huseby D.L., Crawford R.W., Russell J.M., Bevins C.L., Adams L.G., Tsolis R.M., Roth J.R., Baumler A.J. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]; Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, Baumler AJ: Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 2010, 467:426-429. [DOI] [PMC free article] [PubMed]
- 113.Faber F., Tran L., Byndloss M.X., Lopez C.A., Velazquez E.M., Kerrinnes T., Nuccio S.P., Wangdi T., Fiehn O., Tsolis R.M., Baumler A.J. Host-mediated sugar oxidation promotes post-antibiotic pathogen expansion. Nature. 2016;534:697–699. doi: 10.1038/nature18597. [DOI] [PMC free article] [PubMed] [Google Scholar]; Faber F, Tran L, Byndloss MX, Lopez CA, Velazquez EM, Kerrinnes T, Nuccio SP, Wangdi T, Fiehn O, Tsolis RM, Baumler AJ: Host-mediated sugar oxidation promotes post-antibiotic pathogen expansion. Nature 2016, 534:697-699. [DOI] [PMC free article] [PubMed]
- 114.Stelter C., Kappeli R., Konig C., Krah A., Hardt W.D., Stecher B., Bumann D. Salmonella-induced mucosal lectin RegIIIbeta kills competing gut microbiota. PLoS One. 2011;6:e20749. doi: 10.1371/journal.pone.0020749. [DOI] [PMC free article] [PubMed] [Google Scholar]; Stelter C, Kappeli R, Konig C, Krah A, Hardt WD, Stecher B, Bumann D: Salmonella-induced mucosal lectin RegIIIbeta kills competing gut microbiota. PLoS One 2011, 6:e20749. [DOI] [PMC free article] [PubMed]
- 115.Raffatellu M., George M.D., Akiyama Y., Hornsby M.J., Nuccio S.P., Paixao T.A., Butler B.P., Chu H., Santos R.L., Berger T., Mak T.W., Tsolis R.M., Bevins C.L., Solnick J.V., Dandekar S., Baumler A.J. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe. 2009;5:476–486. doi: 10.1016/j.chom.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; Raffatellu M, George MD, Akiyama Y, Hornsby MJ, Nuccio SP, Paixao TA, Butler BP, Chu H, Santos RL, Berger T, Mak TW, Tsolis RM, Bevins CL, Solnick JV, Dandekar S, Baumler AJ: Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe 2009, 5:476-486. [DOI] [PMC free article] [PubMed]
- 116.Endt K., Stecher B., Chaffron S., Slack E., Tchitchek N., Benecke A., Van Maele L., Sirard J.C., Mueller A.J., Heikenwalder M., Macpherson A.J., Strugnell R., von Mering C., Hardt W.D. The microbiota mediates pathogen clearance from the gut lumen after non-typhoidal Salmonella diarrhea. PLoS Pathog. 2010;6:e1001097. doi: 10.1371/journal.ppat.1001097. [DOI] [PMC free article] [PubMed] [Google Scholar]; Endt K, Stecher B, Chaffron S, Slack E, Tchitchek N, Benecke A, Van Maele L, Sirard JC, Mueller AJ, Heikenwalder M, Macpherson AJ, Strugnell R, von Mering C, Hardt WD: The microbiota mediates pathogen clearance from the gut lumen after non-typhoidal Salmonella diarrhea. PLoS Pathog 2010, 6:e1001097. [DOI] [PMC free article] [PubMed]
- 117.Chung H., Pamp S.J., Hill J.A., Surana N.K., Edelman S.M., Troy E.B., Reading N.C., Villablanca E.J., Wang S., Mora J.R., Umesaki Y., Mathis D., Benoist C., Relman D.A., Kasper D.L. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, Reading NC, Villablanca EJ, Wang S, Mora JR, Umesaki Y, Mathis D, Benoist C, Relman DA, Kasper DL: Gut immune maturation depends on colonization with a host-specific microbiota. Cell 2012, 149:1578-1593. [DOI] [PMC free article] [PubMed]
- 118.Smillie C.S., Sauk J., Gevers D., Friedman J., Sung J., Youngster I., Hohmann E.L., Staley C., Khoruts A., Sadowsky M.J., Allegretti J.R., Smith M.B., Xavier R.J., Alm E.J. Strain tracking reveals the determinants of bacterial engraftment in the human gut following fecal microbiota transplantation. Cell Host Microbe. 2018;23:229–240.e5. doi: 10.1016/j.chom.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; Smillie CS, Sauk J, Gevers D, Friedman J, Sung J, Youngster I, Hohmann EL, Staley C, Khoruts A, Sadowsky MJ, Allegretti JR, Smith MB, Xavier RJ, Alm EJ: Strain tracking reveals the determinants of bacterial engraftment in the human gut following fecal microbiota transplantation. Cell Host Microbe 2018, 23:229-240 e5. [DOI] [PMC free article] [PubMed]