Abstract
Background
Salinity is a major abiotic stress that limits the growth, productivity, and geographical distribution of plants. A comparative proteomics and gene expression analysis was performed to better understand salinity tolerance mechanisms in chickpea.
Results
Ten days of NaCl treatments resulted in the differential expression of 364 reproducible spots in seedlings of two contrasting chickpea genotypes, Flip 97-43c (salt tolerant, T1) and Flip 97-196c (salt susceptible, S1). Notably, after 3 days of salinity, 80% of the identified proteins in T1 were upregulated, while only 41% in S2 had higher expression than the controls. The proteins were classified into eight functional categories, and three groups of co-expression profile. The second co-expressed group of proteins had higher and/or stable expression in T1, relative to S2, suggesting coordinated regulation and the importance of some processes involved in salinity acclimation. This group was mainly enriched in proteins associated with photosynthesis (39%; viz. chlorophyll a-b binding protein, oxygen-evolving enhancer protein, ATP synthase, RuBisCO subunits, carbonic anhydrase, and fructose-bisphosphate aldolase), stress responsiveness (21%; viz. heat shock 70 kDa protein, 20 kDa chaperonin, LEA-2 and ascorbate peroxidase), and protein synthesis and degradation (14%; viz. zinc metalloprotease FTSH 2 and elongation factor Tu). Thus, the levels and/or early and late responses in the activation of targeted proteins explained the variation in salinity tolerance between genotypes. Furthermore, T1 recorded more correlations between the targeted transcripts and their corresponding protein expression profiles than S2.
Conclusions
This study provides insight into the proteomic basis of a salt-tolerance mechanism in chickpea, and offers unexpected and poorly understood molecular resources as reliable starting points for further dissection.
Electronic supplementary material
The online version of this article (10.1186/s12870-019-1793-z) contains supplementary material, which is available to authorized users.
Keywords: Chickpea, Differentially expressed proteins, Metabolic pathway, Salt-tolerance mechanism
Background
Salinity stress is one of the most significant constraints to plant growth and productivity as plants are unable to move or escape from this stress [1]. The metabolic imbalances triggered by osmotic stress, ion toxicity, and nutritional deficiency due to salinity can cause oxidative stress [2]. Plants resort to adaptive strategies to cope with salinity, including the accumulation of compatible osmolytes and proteins involved in stress tolerance. Increasing evidence has revealed that the modification of protein synthesis or degradation at both quantitative and qualitative levels is an essential metabolic process that critically influences salinity or dehydration tolerance [3, 4].
Numerous studies have offered insight into plant adaptive mechanisms for high productivity and stress tolerance. High throughput genetic screening techniques have identified significant cellular and molecular responses to various stresses as part of the complex gene network [5]. However, a clearer picture is needed of the tolerance mechanisms in plants. Recent studies have focused on high throughput proteomics analyses to look beyond transcriptomics, as various gene products are subject to post-translation modifications [6]. These studies have characterized the molecular basis for salinity tolerance at the proteome level, in crops such as rice [7], wheat [8], and Medicago truncatula [9]. While proteome research is quite advanced in these plants, there are no specific reports of this kind on chickpea (Cicer arietinum L.).
Chickpea is the third most important pulse legume and is grown for its rich protein grains. It is an excellent crop for natural resource management due to its symbiotic relationship with both nitrogen-fixing bacteria and arbuscular mycorrhizal fungi [10]. However, chickpea growth and development are limited by moderate to severe salinity stress [11].
The major mechanisms involved in plant adaptations to salinity are regulated by modifications in gene and protein expression [12]. However, proteomics analyses of chickpea responses to abiotic stress have been mainly assessed under dehydration at different subcellular levels—including nuclear [13, 14], membrane [15], and extracellular matrix (ECM) proteome [16]—in the leaves of 3-week-old seedlings. Recently, proteome changes in response to heat stress were studied in the leaves of two chickpea genotypes [17]. Based on these studies, the tolerance of chickpea to heat and dehydration stresses was attributed to altered expression of numerous functional proteins, especially those involved in cell defense and rescue, photosynthesis and energy metabolism, redox homeostasis, and signal transduction.
Despite the importance of chickpea and its salt sensitivity, there have been no large-scale proteomics studies undertaken in this area. The salinity response in plants is a complex phenomenon, where the exact structural and functional adjustments caused by this stress are poorly understood. Plants under saline stress have evolved sophisticated mechanisms, including specific ion uptake/exclusion, toxic ion compartmentalization, synthesis of appropriate products, increases in antioxidative enzymes, regulation of photosynthetic and energy metabolism, hormonal adjustments, and cell structure alterations [6]. The principal features of salt tolerance at the proteomics level in pea [18], soybean [19], and maize [20] are photosynthesis, proteolysis, protein biosynthesis, osmotic homeostasis, defiance, and stress-related proteins. The key proteins contributing to salt tolerance responses in these studies were ribulose-1, 5-bisphosphate carboxylase (Rubisco), heat shock proteins (HSPs), late embryogenesis abundant (LEA) proteins, antioxidative enzymes, glucose-6-phosphate 1-dehydrogenase, glycerate dehydrogenase, NADPH-producing dehydrogenase, glutamate synthase, and glutamine synthetase.
This study aimed to isolate novel salt-stress-responsive proteins regulated in chickpea leaves. The 2-DE along with LC-MS/MS analyses were used to characterize 65 differential expressed proteins (DEPs), which were classified based on their putative functions. We provide evidence for the expression of novel proteins that have never been associated with chickpea under abiotic stress or plant response to salinity. Supplementary information from the gene expression analysis was derived to better understand the molecular mechanisms of the salinity response. Overall, this study offers new insights into the molecular mechanisms of salt tolerance underlying responsive proteins and genes in chickpea genotypes.
Results
2-DE maps and identification of DEPs
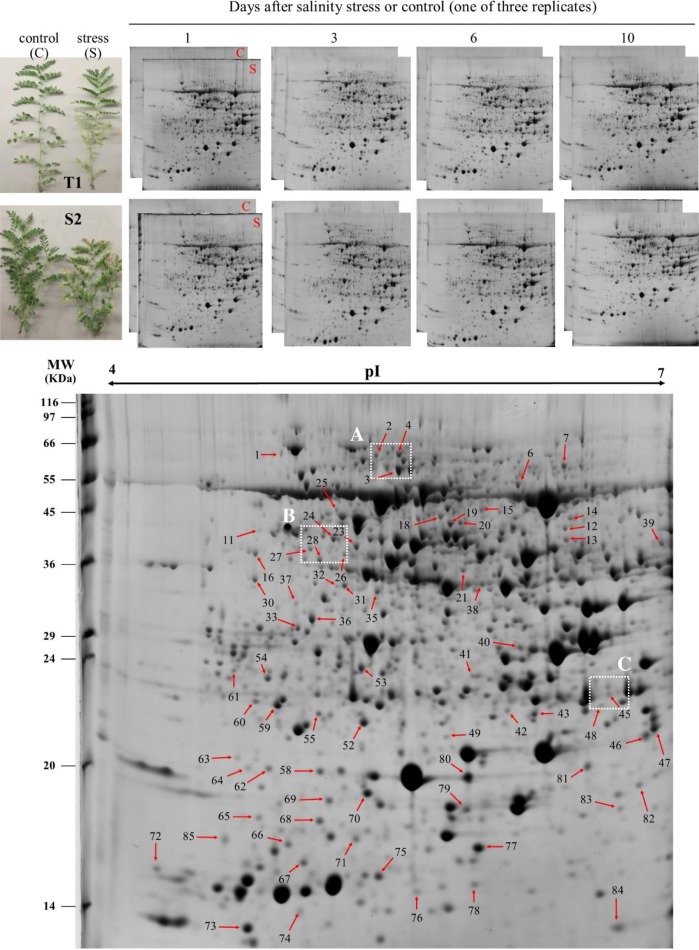
The 2-DE map analyses of the complete leaf of T1 and S2 seedlings exposed to 0 and 100 mM NaCl for 10 days revealed at least 1400 reproducible protein spots that were present in both genotypes and all 2-DE gels (Additional file 1: Figure S1 and Additional file 2: Figure S2). Representative 2-DE gels of the proteome of two chickpea genotypes under saline and control conditions for 10 days is presented in Fig. 1; high-resolution protein spots were obtained in the 4–7 (linear) pI and 6.5–200 kDa Mr range. Of these, 364 spots were classified as DEPs in at least one combination of genotype (T1 and S2) × time (1, 3, 6 and 10 DAT) in the salinity treatment (100 mM), relative to the respective control (0 mM). As a result, 65 DEPs were selected based on quantitative data (more than two fold-changes) from the Image Master analysis and visual inspection of 2-DE gels (classified as ‘high-quality’ spots with differential expression patterns during 10 days of stress). Nine spots in three typical gel regions are represented in zoom images in Fig. 2, displaying differential expression of some proteins. The LC-MS/MS analysis positively identified just over 98% of selected proteins (64 spots) with significant (p ≤ 0.05) hits, with an average MASCOT score of ~ 500, representing a very high rate of success identification. More than 80% of identified proteins were hit to legume species, 47 of which were matched to protein isoforms of C. arietinum species (Table 3). The MASCOT score, coverage percent, theoretical/experimental pI, and Mr are also summarized in Table 3.
Fig. 1.
Representative 2-DE gel electrophoresis maps of leaf proteins for two chickpea genotypes after different days of salinity stress and the control. Three boxed areas (A–C) marked with dotted lines represent the zoomed in gel sections in Fig. 2. The numbers correspond with the spot ID, mentioned in Table 3
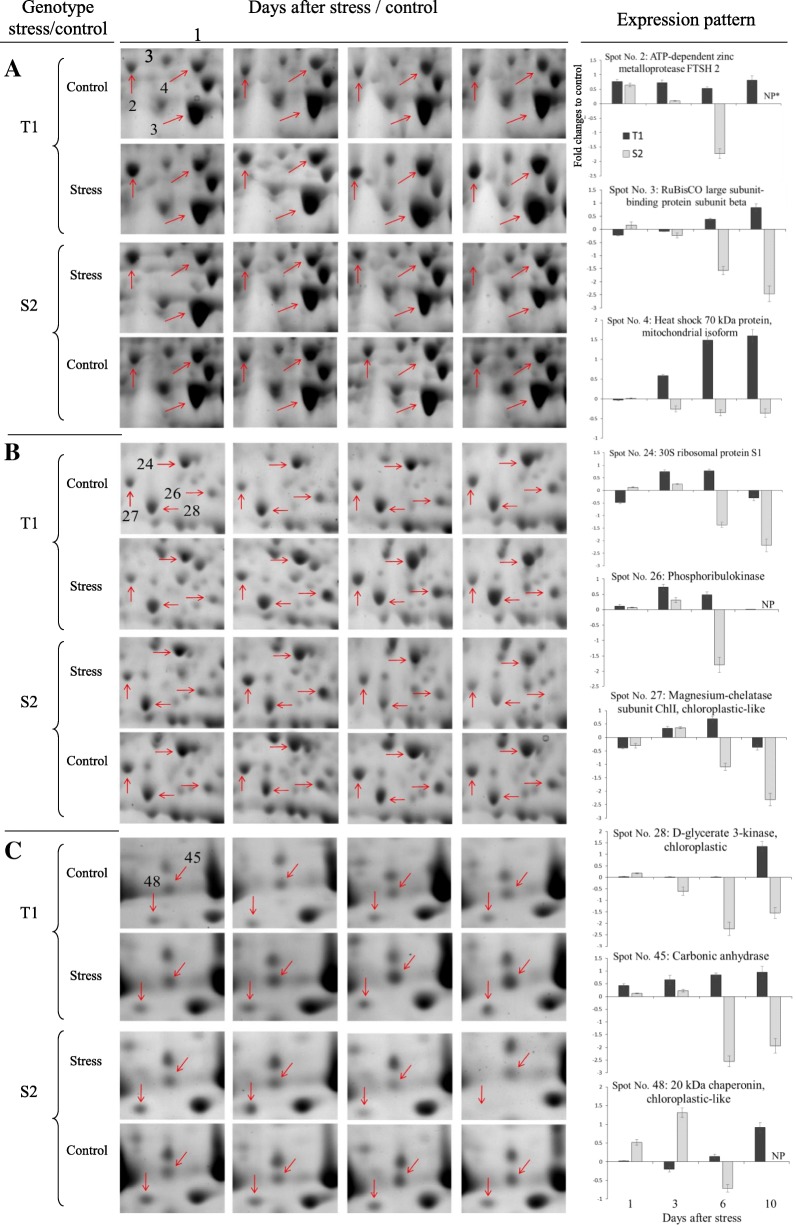
Fig. 2.
The relative expression patterns of nine representative protein spots in two chickpea genotypes under 0 and 100 mM NaCl. Boxed areas (a–c) are zoomed in gel sections, corresponding to the dotted line boxes in Fig. 1. The expression patterns in terms of fold-changes to control are presents on the right. NP* represents not present or absent spot
Table 3.
Details of the identified 2-DE-resolved exclusive DEPs by LC MS/MS in the leaves of tolerant (Flip 97-43c; black columns) versus susceptible (Flip 97-196c; gray columns) chickpea seedlings treated with 100 mM NaCl stress for 1, 3, 6, and 10 days. The key potential proteins contributing to the differential response of the two genotypes to salinity are indicated by * next to their spot numbers
| Spot no. | Identified protein | Ac. No. a | Species | MASCOT score | % Coverage (matches) b | TMr/ EMr c | TpI/ EpI d | Time kinetics (DAT)e | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 3 | 6 | 10 | ||||||||
| 1. Photosynthesis and bioenergy | |||||||||||
| 3* | RuBisCO large subunit-binding protein subunit beta | XP_012567814.1 | Cicer arietinum | 1137 | 44 (93) | 62.9/59.2 | 5.85/5.53 |
|
|||
| 70 | RuBisCO large subunit, partial (chloroplast) 2 | ALB07273.1 | Acanthus ebracteatus | 71 | 32 (15) | 14.4/17.5 | 9.39/5.37 |
|
|||
| 84 | Ribulose bisphosphate carboxylase small chain | Q9ZP07 | Cicer arietinum | 270 | 29 (64) | 20.4/9.25 | 9.03/6.72 |
|
|||
| 18 | ATP synthase beta subunit, partial (chloroplast) | AAK72753.1 | Cucurbita pepo | 925 | 38 (128) | 51.9/45.2 | 5.04/5.76 |
|
|||
| 33 | ATP synthase subunit beta, chloroplastic | B5LMK9 | Cicer arietinum | 585 | 39 (58) | 52.9/28.7 | 5.16/4.99 |
|
|||
| 53* | ATP synthase subunit alpha | A0A023HPK1 | Trifolium aureum | 3840 | 27 (149) | 55.8/24.1 | 5.16/5.33 |
|
|||
| 49* | psbP domain-containing protein 1, chloroplastic | XP_004494530.1 | Cicer arietinum | 643 | 65 (31) | 28.6/20.5 | 8.89/5.79 |
|
|||
| 52* | Chlorophyll a-b binding protein 3, chloroplastic | XP_004491629.1 | Cicer arietinum | 244 | 25 (55) | 29.4/21.3 | 3.32/5.34 |
|
|||
| 64* | Oxygen-evolving enhancer protein 1, chloroplastic | XP_004509219.1 | Cicer arietinum | 214 | 35 (37) | 34.9/19.0 | 6.24/4.72 |
|
|||
| 79 | Oxygen-evolving enhancer protein 2, chloroplastic | XP_004499534.1 | Cicer arietinum | 537 | 32 (60) | 28.7/16.5 | 6.90/5.90 |
|
|||
| 74 | Photosystem I reaction center subunit II, chloroplastic-like | XP_004511355.1 | Cicer arietinum | 625 | 34 (37) | 23.7/12.5 | 9.66/5.00 |
|
|||
| 68 | 2Fe-2S iron-sulfur cluster-binding domain protein | G7IUN0 | Medicago truncatula | 70 | 8 (1) | 21.5/15.7 | 6.82/5.11 |
|
|||
| 69 | Cytochrome b6-f complex iron-sulfur subunit | A0A067FP73 | Gossypium raimondii | 61 | 13 (4) | 32.6/16.7 | 8.48/5.16 |
|
|||
| 27 | Magnesium-chelatase subunit ChlI, chloroplastic-like | XP_004485870.1 | Cicer arietinum | 244 | 34 (32) | 45.4/39.5 | 5.32/5.06 |
|
|||
| 26 | Phosphoribulokinase, chloroplastic | XP_004512451.1 | Cicer arietinum | 369 | 30 (28) | 45.5/38.8 | 6.41/5.24 |
|
|||
| 45* | Carbonic anhydrase, chloroplastic isoform X2 | XP_004489275.1 | Cicer arietinum | 409 | 19 (36) | 35.8/22.3 | 6.61/6.66 |
|
|||
| 38* | Fructose-bisphosphate aldolase 1, chloroplastic | XP_004507507.1 | Cicer arietinum | 1376 | 21 (68) | 43.2/33.5 | 6.28/5.98 |
|
|||
| 63 | Phosphoglycerate kinase, cytosolic-like | XP_004488763.1 | Trifolium aureum | 1452 | 35 (121) | 49.8/19.2 | 7.79/4.66 |
|
|||
| 78 | Transketolase, chloroplastic | XP_012570278.1 | Cicer arietinum | 605 | 18 (25) | 79.9/13.5 | 6.00/5.95 |
|
|||
| 2. Photorespiration | |||||||||||
| 7 | Glycine dehydrogenase/decarboxylating, mitochondrial | XP_004498896.2 | Cicer arietinum | 653 | 26 (42) | 115.3/60.2 | 7.99/6.43 |
|
|||
| 21 | Glutamate--glyoxylate aminotransferase | XP_004489787.1 | Cicer arietinum | 359 | 33 (28) | 53.3/35.7 | 5.83/5.88 |
|
|||
| 28* | D-glycerate 3-kinase, chloroplastic | XP_004486714.1 | Cicer arietinum | 116 | 19 (12) | 46.4/37.2 | 5.39/5.11 |
|
|||
| 3. Stress responsive | |||||||||||
| 1 | 70 kDa heat shock protein | Q1SKX2 | Medicago truncatula | 153 | 11 (12) | 75.7/64.7 | 5.19/4.96 |
|
|||
| 4* | Heat shock 70 kDa protein, mitochondrial isoform | XP_012572445.1 | Cicer arietinum | 833 | 40 (67) | 72.1/64.7 | 5.70/5.54 |
|
|||
| 11 | Heat shock protein 70 kDa | A0A0A1HAD2 | Chrysanthemum morifolium | 265 | 13 (55) | 70.8/42.2 | 5.12/4.86 |
|
|||
| 58 | Heat shock 22 kDa protein, mitochondrial isoform X1 | XP_004506342.1 | Cicer arietinum | 249 | 25 (22) | 25.1/18.75 | 6.78/5.11 |
|
|||
| 81 | Cold shock protein | A0A088FZS5 | Cicer arietinum | 1497 | 31 (51) | 19.2/18.5 | 6.29/6.55 |
|
|||
| 48* | 20 kDa chaperonin, chloroplastic-like | XP_004508023.1 | Cicer arietinum | 413 | 50 (17) | 26.7/21.5 | 9.05/6.63 |
|
|||
| 30* | uncharacterized protein, Homologues with LEA-2 | XP_004513216.1 | Cicer arietinum | 400 | 59 (49) | 34.3/35.0 | 4.63/ 4.76 |
|
|||
| 31 | Thiamine thiazole synthase, chloroplastic | I3TAU0 | Lotus japonicus | 133 | 17 (12) | 37.7/33.2 | 5.02/5.24 |
|
|||
| 40 | Xanthoxin dehydrogenase | XP_004499827.1 | Cicer arietinum | 573 | 41 (49) | 29.6/28.3 | 5.89/ 6.19 |
|
|||
| 42* | Glutathione s-transferase | A0A0X9LEN0 | Cicer arietinum | 270 | 34 (38) | 25.6/21.8 | 6.04/6.12 |
|
|||
| 41* | L-ascorbate peroxidase, cytosolic | XP_004505943.1 | Cicer arietinum | 243 | 37 (49) | 27.1/23.2 | 5.65/ 5.94 |
|
|||
| 55 | L-ascorbate peroxidase, cytosolic | XP_004505943.1 | Cicer arietinum | 501 | 81 (67) | 27.1/21.5 | 5.65/5.10 |
|
|||
| 77 | Superoxide dismutase [Cu-Zn] | A0A0V0HK97 | Solanum chacoense | 1057 | 15 (72) | 22.4/14.5 | 6.08/5.96 |
|
|||
| 83 | Apolipoprotein D-like | XP_004507714.1 | Cicer arietinum | 128 | 35 (29) | 21.3/16.5 | 6.84/6.73 |
|
|||
| 4. Protein synthesis and degradation | |||||||||||
| 2* | ATP-dependent zinc metalloprotease FTSH 2 | XP_004504668.1 | Cicer arietinum | 945 | 36 (74) | 74.6/62.7 | 5.60/5.34 |
|
|||
| 6 | Probable mitochondrial-processing peptidase subunit beta | XP_004492264.1 | Cicer arietinum | 520 | 41 (56) | 59.4/54.5 | 6.22/6.18 |
|
|||
| 60 | Metacaspase-4-like | XP_004510001.1 | Cicer arietinum | 158 | 10 (14) | 45.2/22.0 | 5.08/4.76 |
|
|||
| 25 | 26S protease regulatory subunit 6A homolog | XP_004495708.1 | Cicer arietinum | 382 | 50 (29) | 47.4/47.0 | 4.98/5.20 |
|
|||
| 43 | Proteasome subunit beta type | A0A151TBQ9 | Cajanus cajan | 68 | 21 (10) | 24.6/21.5 | 6.89/6.27 |
|
|||
| 13 | Elongation factor Tu, mitochondrial | XP_004493639.2 | Cicer arietinum | 272 | 30 (54) | 49.1/40.2 | 6.58/6.44 |
|
|||
| 20 | Elongation factor Tu, chloroplastic | XP_004501869.1 | Cicer arietinum | 388 | 30 (33) | 53.5/44.0 | 6.25/5.87 |
|
|||
| 23* | Elongation factor Tu, chloroplastic | XP_004501869.1 | Cicer arietinum | 562 | 29 (52) | 53.5/39.2 | 6.25/5.31 |
|
|||
| 24 | 30S ribosomal protein S1, chloroplastic | XP_004486520.1 | Cicer arietinum | 721 | 64 (48) | 44.5/40.7 | 5.33/5.18 |
|
|||
| 72 | 60S acidic ribosomal protein P3-like | XP_004504130.1 | Cicer arietinum | 420 | 38 (30) | 12.1/14.2 | 4.20/4.23 |
|
|||
| 85 | 50S ribosomal protein L9, chloroplastic | XP_004506080.1 | Cicer arietinum | 27 | 9 (3) | 22.4/15.7 | 9.78/4.6 |
|
|||
| 16* | peptidyl-prolyl cis-trans isomerase CYP38, chloroplastic | XP_004489294.1 | Cicer arietinum | 628 | 43 (68) | 50.1/38.5 | 5.12/4.76 |
|
|||
| 5. Amino acid and nitrogen metabolism | |||||||||||
| 19 | S-adenosylmethionine synthase | C3TS15 | Cicer arietinum | 462 | 50 (61) | 43.3/43.7 | 5.50/5.81 |
|
|||
| 35 | Glutamine synthetase leaf isozyme, chloroplastic | Q9XQ94 | Medicago sativa | 65 | 14 (30) | 47.1/33.0 | 6.29/5.42 |
|
|||
| 39* | Glycerate dehydrogenase | XP_004497175.1 | Cicer arietinum | 963 | 36 (80) | 42.2/39.2 | 6.62/6.95 |
|
|||
| 46 | Hydroxyacylglutathione hydrolase 2, mitochondrial-like isoform X | XP_004486896.1 | Cicer arietinum | 20 | 5 (2) | 36.7/21.3 | 9.03/6.87 |
|
|||
| 6. Other metabolisms (TCA, PP, and purine) | |||||||||||
| 12 | Isocitrate dehydrogenase [NADP] | G7KFV7 | Medicago truncatula | 137 | 14 (39) | 45.9/42.7 | 5.99/6.43 |
|
|||
| 15 | 6-phosphogluconate dehydrogenase, decarboxylating 3 | XP_004491970.2 | Cicer arietinum | 524 | 43 (77) | 53.6/47.2 | 5.88/5.99 |
|
|||
| 36 | Phosphoribosylformylglycinamidine cyclo-ligase, chloroplastic/mitochondrial-like | XP_004500242.1 | Cicer arietinum | 131 | 18 (14) | 41.1/29.7 | 5.65/5.06 |
|
|||
| 7. Signaling | |||||||||||
| 47* | Auxin-binding protein ABP19a-like | XP_004513480.1 | Cicer arietinum | 569 | 28 (46) | 21.9/20.7 | 6.95/6.91 |
|
|||
| 80* | Low molecular weight phosphotyrosine protein phosphatase | XP_004506147.1 | Cicer arietinum | 287 | 12 (12) | 27.1/17.7 | 7.62/5.9 |
|
|||
| 8. Gene transcription and replication | |||||||||||
| 82 | Nascent polypeptide-associated complex subunit beta | B7FMW7 | Medicago truncatula | 327 | 42 (24) | 17.5/17.5 | 6.75/6.83 |
|
|||
| 14 | Polyadenylate-binding protein RBP45B isoform X2 | XP_012573405.1 | Cicer arietinum | 277 | 16 (14) | 34.8/44.7 | 6.31/6.46 |
|
|||
| 37* | 33 kDa ribonucleoprotein, chloroplastic | A0A0B2SU82 | Glycine soja | 66 | 3 (2) | 30.4/31.5 | 8.67/4.98 |
|
|||
| 54* | 29 kDa ribonucleoprotein A, chloroplastic | XP_004497514.1 | Cicer arietinum | 110 | 16 (5) | 30.7/23.2 | 5.36/4.83 |
|
|||
| 65 | LOC101493535 isoform X1, homologous with smad/FHA domain protein | XP_004502003.1 | Cicer arietinum | 1469 | 35 (93) | 24.3/16.5 | 5.69/4.78 |
|
|||
| 71 | Histone H2B | A0A0K9PL95 | Zostera marina | 109 | 20 (18) | 15.5/15.3 | 10.05/5.3 |
|
|||
| 75 | Glycine-rich RNA-binding protein-like | XP_004507449.1 | Cicer arietinum | 252 | 22 (27) | 16.1/13.5 | 6.32/5.43 |
|
|||
aAccession numbers correspond to NCBI or Uniprot entries
bPercentage of the protein sequence covered by matching peptides and number of matched peptides in the database
cTheoretical molecular weight (Mr) and pI, predicted from MS/MS analysis
dExperimental molecular weight (Mr) and pI were estimated using standard protein markers and automatic assignment by image analyzer (Image Master) software
eProtein expression profile of T1 (dark columns) and S2 (light columns) genotype relative to control, shown as horizontal axis
In several cases, some redundancy was observed, where the same protein resolved into multiple spots in the same gel, e.g., RuBisCO, ATP synthase, heat shock proteins (HSPs), L-ascorbate peroxidase, and elongation factor Tu. These relative DEPs recorded some differences in pI, Mr, or both. This could be due to their probable post-translational modification(s) (including phosphorylation, de-amidation, acetylation, glycation, and glutathionylation), multiple isoforms, or in some cases these proteins could be degraded products, possibly from extraction, as evident in the gel-based size [3].
Generally, downregulation was the dominant trend for protein expression in chickpea in response to salinity (Table 1). Almost half of the 512 spots (two genotypes × four sampling times × 64 DEPs) were downregulated, especially S2 at 6 and 10 DAT. However, T1 at 3 and 6 DAT recorded the most spots with upregulation. Sixty-four DEPs, identified with a significant match, were considered for further functional clustering and expression profile across genotype and time of salinity.
Table 1.
Comparison of salt-responsive DEPs in the leaves of two contrasting chickpea genotypes (T1 and S2) during 1, 3, 5, and 10 days of 100 mM NaCl stress
| Ex. pattern | Genotype | Days after salinity treatment | Total | ||||
|---|---|---|---|---|---|---|---|
| 1 | 3 | 6 | 10 | ||||
| * Up | T1 | 24 | 51 | 35 | 21 | 131 | 189 |
| S2 | 24 | 26 | 2 | 6 | 58 | ||
| ** Un | T1 | 12 | 6 | 17 | 2 | 37 | 76 |
| S2 | 18 | 16 | 2 | 3 | 39 | ||
| *** Down | T1 | 28 | 7 | 12 | 41 | 88 | 247 |
| S2 | 22 | 22 | 60 | 55 | 159 | ||
*Up, up-regulated DEPs under salinity stress; **Un, DEPs showing no significant (p ≤ 0.05) response to salinity in at least one time point; *** Down, down-regulated DEPs under salinity
Functional classification of DEPs
The 64 identified protein spots were grouped into eight functional categories (Fig. 3a and Table 3) according to their assignment of putative biological function(s) by searching InterPro or associated literature, with an emphasis on the features related to salinity or dehydration responses in seedlings of chickpea, pulses, or other model plants such as Arabidopsis.
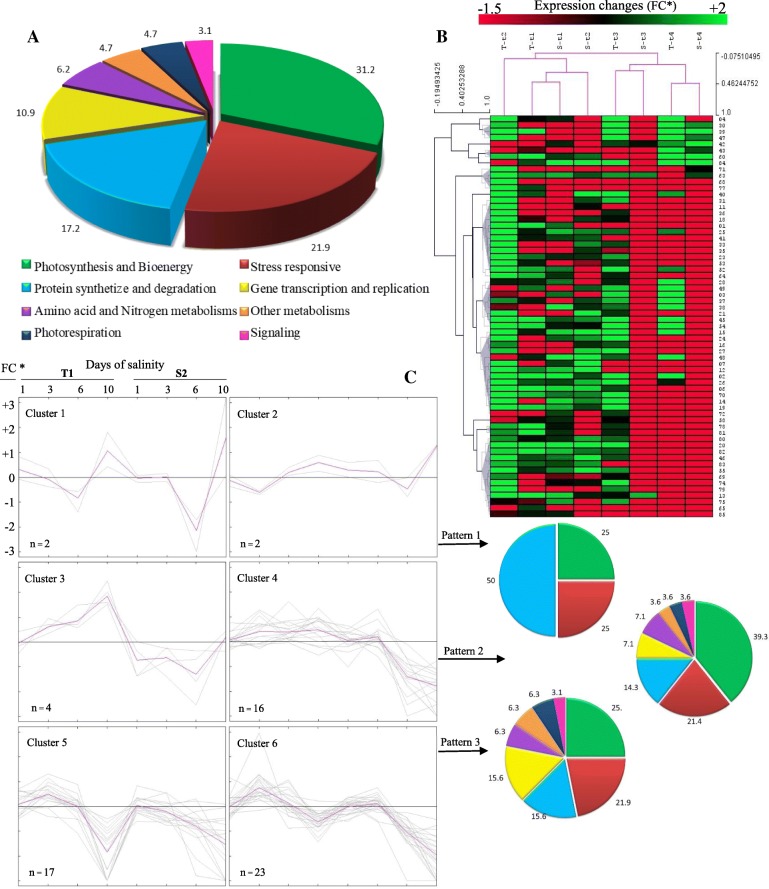
Fig. 3.
a An outline of functional classification for 64 differentially expressed proteins identified in the seedling leaves of two chickpea genotypes under salinity. This classification is according to the assignment of protein putative function(s) by searching in UniProt, NCBI, and associated literature; b Self-organizing tree algorithm (SOTA) clustering analysis of expression profiles for identified proteins under salinity for 1, 3, 5 and 10 days (t1, t2, t3, and t4, respectively); c Proteins were classified into six clusters and three patterns, based on their expression profiles, listed in Additional file 3: Table S1. The gray lines represent the expression profile of each protein, while the mean expression profile is marked in pink for each cluster. The expression change is based on fold-changes over respective controls (FC *)
The functional analysis (Fig. 3a) indicated that ‘photosynthesis and bioenergy’ represented some 31% of the total protein spots, with ~ 22% related to ‘stress responsiveness’, which reflects the role of energy production and defense strategies within the two genotypes. Other categories included ‘protein synthesis and degradation’ (17.2%), followed by ‘gene transcription and replication’ (10.9%), ‘amino acid and nitrogen metabolism’ (6.2%), ‘photorespiration’ (4.7%), ‘signaling’ (3.1%), and ‘other metabolisms’ (4.7%). Furthermore, DEPs were classified based on their expression patterns (Fig. 3c and Additional file 3: Table S1).
Dynamics of DEP networks
To summarize the comprehensive overview of dynamic profiles and coordinated regulation of DEPs, SOTA analysis was undertaken using fold-expression-changes data relative to the control. The analysis yielded six distinct expression clusters based on similar expression profiles for both genotypes during 10 days of salinity (Fig. 3c). The most abundant group appeared in Clusters 4, 5 and 6 only, containing proteins involved in photosynthesis and bioenergy (18 spots), stress responsiveness (11 proteins), protein synthesis and degradation (9 proteins), and gene transcription and replication (7 proteins) (Additional file 3: Table S1). Three major patterns of expression were detected (Fig. 3c and Additional file 3: Table S1):
Clusters 1 and 2 had an initial downregulation of DEPs, which were upregulated during later stages, especially at 10 DAT. These proteins were mainly involved in protein synthesis and degradation (50%).
Clusters 3 and 4 had higher expression in T1, compared with little or no change in S2, relative to their respective controls. These groups were enriched in proteins involved in photosynthesis and bioenergy (39%), stress responsiveness (21%), and protein synthesis and degradation (14%). The upregulation and/or stable expression of these DEPs may be responsible for the higher salinity tolerance in T1 than S2.
Clusters 5 and 6 had upregulation of DEPs in the early days of stress (3 DAT), especially in T1, which were downregulated or not expressed at later stages (10 DAT), particularly in S2. This cluster is enriched in proteins associated with photosynthesis and bioenergy (25%), stress responsiveness (22%), and protein synthesis and degradation (16%).
Figure 3b shows the results of the further analysis of SOTA to cluster the overall response of both genotypes (T1 and S2) based on their expression profile for all 64 DEPs over four time-points (t1 to t4). Both genotypes behaved similarly and clustered together at all time-points, except for 3 DAT (t2) where large differences in protein expression level were observed between the genotypes, with T1 upregulated in 51 protein spots compared with 22 in S2. In terms of the overall expression pattern of proteins across all time-points, two main clusters were formed (Fig. 3b):
Cluster 1 had four members with upregulation at early stages of salinity (t1 and t2) in both genotypes, with the highest expression being 80% DEPs in T-t2 (T1 genotype at 3 DAT as third time-point of stress).
Cluster 2 showed downregulation of DEPs as salinity progressed, with more than 95% DEPs downregulated in S-t3, followed by 86% in S-t4 (both sensitive genotype); less downregulation occurred at the corresponding time-points in the tolerant genotype.
The critical difference between the two genotypes could be related to the 14 proteins that were upregulated in T1 at 10 DAT (T-t4). These DEPs could be responsible for the higher salinity tolerance in T1 over S2, specifically at later stages in the salinity treatment.
Generally, the cellular defense mechanism seems to depend on the progress of salinity. Therefore, both early and late responses could be seen. The fact that the studied critical proteins were identified as salinity-responsive cases gives the first functional annotation to their genes and possible role in salinity response, so further investigation was performed as a complementary and conforming experiment at the transcript level.
Comparison between proteins and transcripts
Six representative leaf DEPs were used to further characterize patterns of gene expression by RT-PCR (Table 2). The choice of these genes was based on their possible biological functions during salinity stress, and their differential proteomics-derived patterns between two genotypes with emphasis on upregulation in T1 and downregulation in S2 (Clusters 3 and 4). The expression of transcripts was calculated as fold-changes relative to their respective controls (Fig. 4 and Additional file 5: Figure S4). Trends were compared to respective protein profiles using Pearson correlation analysis (Fig. 4).
Table 2.
Selected genes for transcript profiling based on biological function and expression profiles of corresponding proteins
| Gene | Biological function | Spot no. /cluster | Primer sequence (5′ – 3′) | PCR product size (bp) | GeneBank accession no. |
|---|---|---|---|---|---|
| Actin | Reference gene | – |
F:TGTTCCCCGGAATTGCTGATAGAATGAGC R:TTGGAAAGTGCTGAGAGATGCCAAAATGGAG |
146 | AJ012685 |
| Carbonic anhydrase | Photosynthesis | 45 / 4 |
F: TTGAAGTGAAGGAAACC R: AGAAGAAATGGGAAAGG |
195 | LOC101498889 |
| Glycerate dehydrogenase | Amino acid & nitrogen metabolism | 39 / 3 |
F: GAAAGACTCGCCAAGA R: GGGCTCATCCTCAAAC |
141 | LOC101496862 |
| Heat shock 70 kDa protein | Stress responsive | 4 / 3 |
F: CTTGATGTAACGCCACTT R: GTCCACCAGATGACCTAATA |
346 | LOC101502051 |
| L-ascorbate peroxidase | Stress responsive | 55 / 5 |
F: AAATCTTACCCAACCGTCA R: CAACAACACCACCCAACT |
304 | LOC101497640 |
| ATP-dependent zinc metalloprotease FTSH2 | Protein synthesis and degradation | 2 / 4 |
F: AAAGAACAACCGTGAAGCAA R: CGGCGACTGGTAATGGA |
152 | LOC101512859 |
| 6-phosphogluconate dehydrogenase | Other metabolisms: PP | 15 / 4 |
F: TGACAGCAAGGCAAACAACTC R: CAGGCATACGACAGAAACCC |
112 | LOC101509898 |
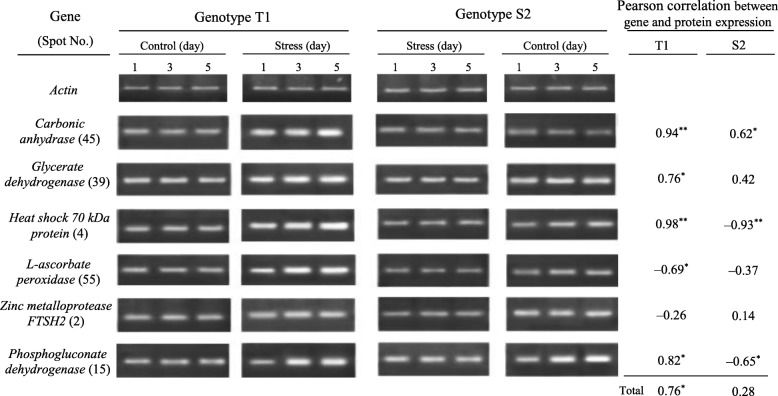
Fig. 4.
Expression profile of genes encoding six selected DEPs in salt-stressed chickpea genotypes. Transcript level was determined by RT-PCR after normalization to the actin gene at three time-points: 1, 3, and 5 days of salt stress relative to the control using three individual replicates. Correlation of gene expression with corresponding protein abundance using Pearson’s correlation analyses was calculated (* p ≤ 0.05, ** p ≤ 0.01)
There was a significant (p ≤ 0.05) correlation between protein abundance and RT-PCR signal for five genes (45, 39, 4, 55, and 15) in salt-stressed T1 and three genes (4, 15, and 45) in salt-stressed S2. Two proteins in T1 (55 and 2) and three proteins in S2 (4, 55, and 15) behaved against their mRNA changes during salinity stress. The calculated Pearson’s correlation was 0.76 and 0.28 in T1 and S2, respectively, which implies that the correlation between transcripts and proteins was significantly (p ≤ 0.05) higher in T1 than S2.
Discussion
Plant response to salinity is a complex process that requires changes in gene and protein expression profiles. This study aimed to better understand salinity tolerance mechanisms in chickpea. The data revealed significant differences in protein and gene expression patterns between two contrasting chickpea genotypes (T1 and S2) under salinity stress (100 mM NaCl) for 10 days. This is the first comprehensive proteome analysis of chickpea under saline conditions.
Role of photosynthesis and bioenergy in salt tolerance
Salinity changes the integrity and functionality of chloroplasts, which in turn impacts cell function as a whole [21]. In the present study, salinity had the most significant effect on the abundance of the functional group of proteins related to light-harvesting complexes, electron transfer pathway, and photosynthetic Calvin-Benson Cycle. The enzymes in this group provide substrates for the synthesis of ATP, NADPH, sucrose, starch, and proteins (Fig. 5), which improve plant biomass, yield, and resistance to stresses [22].
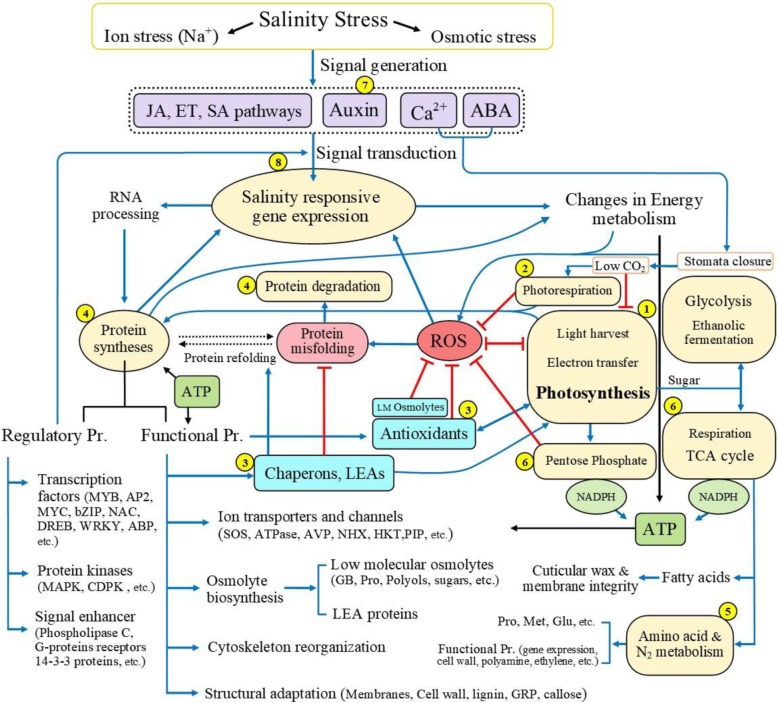
Fig. 5.
Schematic outline of the salinity-induced response pathway based on proteome, gene expression, and physiological changes in chickpea leaves. Numbers in yellow circles are corresponding to the protein(s) mentioned in Table 2. Blue arrows indicate induction or increase; red arrows indicate repression or decrease, and dotted arrows indicate possible or reversible alterations. Salinity activates several signaling cascades, regulates gene expression, and promotes regulatory and functional protein biosynthesis, in which the main role of ROS, antioxidants, and chaperones under saline condition is presented. Tolerant seedlings inhibited or decreased major metabolic pathways (i.e., photosynthesis, photorespiration, TCA cycle, and amino acid metabolism) less than susceptible seedlings to provide more energy and other compatible metabolites. Importantly, salt tolerance relies on higher transporter activities to remove H2O2, Na+, etc., and enhance osmotic regulation, and membrane and cell wall re-modulation to cope with the stress
Five identified DEPs (49, 52, 74, 68, and 69) participated in the light-harvesting reaction and photosystems. Spots 49 and 52 were identified as the psbP domain-containing protein 1 (PPD1) and chlorophyll a-b binding protein 3 (CAB) were upregulated in T1, more so at 10 and 3 DAT, respectively, but downregulated in S2 at the corresponding time-points (Table 3). The increased CAB and PPD1 in T1, which clustered together (Fig. 2c and Additional file 3: Table S1, Cluster 4), transferred more excitation energy to the reaction center, where the accumulated plastocyanin can donate more electrons to photosystem I (PSI) to reduce NADP+ to NADPH [23]. Moreover, PPD1, a nuclear-encoded and thylakoid luminal protein, is an essential factor for PSI [24] and PSII [25] assembly and activity. The initial upregulation (0–12 h) and subsequent downregulation (144 h) of CAB and PPD1 in response to salinity have also been recorded in soybean genotypes [23].
Two protein spots were identified as chloroplastic oxygen-evolving enhancer protein (OEE) 1 (spot 64) and 2 (spot 79). The upregulation of both OEEs in T1, relative to S2, mirror greater efficiency in water-splitting and PSII core assembly/stability [25] in the first 6 days of stress (Table 3). Salt-stress treatments have paradoxical impacts on OEE; for instance, salinity causes its upregulation in Triticum aestivum [26] and halophyte Halogeton glomeratus [27], but downregulation in Spinacia oleracea [25].
Three DEPs were identified as subunits of ATP synthase—alpha (spot 53) and beta (spots 18 and 33)—which participate in photosynthesis. These proteins (Fig. 3c, Cluster 4) were markedly (p ≤ 0.05) upregulated at 3 DAT in T1, but significantly (p ≤ 0.05) downregulated at later stages of salt stress (6 and 10 DAT) in the seedling leaves of S2. These results suggest that energy synthesis is inhibited in salt-stressed S2 seedlings at late-stress stages, and the synthesis of ATP correlates well with light-harvesting and electron transfer proteins. ATP-dependent synthase/protease could have chaperone-like activity [28], which is a crucial coping strategy for plants under salt stress [29]. In addition, the increased abundance of this complex in T1 under salt stress has an indirect role in the translocation of excess Na+ and Cl− ions into the vacuole [30]. The increased ATP supply in T1 seems to meet the increased stress-related energy demand to increase Calvin cycle efficiency. Consistent with our results, the higher expression of this enzyme in tolerant than susceptible genotypes under salinity or dehydration stress has been reported in barley [27] and common bean [31]. In contrast, ATP synthase expression was significantly downregulated in a tolerant cowpea cultivar, but upregulated in a sensitive one, after salinity stress [32].
Eight proteins were engaged in carbon assimilation: carbonic anhydrase (CA, spot 45), phosphoribulokinase (PRK, spot 26), ribulose 1,5-bisphosphate carboxylase/oxygenase (RuBisCO) large (spots 3 and 70) and small (spot 84) subunits, phosphoglycerate kinase (PGK, spot 63), fructose-bisphosphate aldolase (spot 38), and transketolase (TKT, spot 78). Carbonic anhydrase provides inorganic carbon to improve photosynthetic efficiency and cell resistance to cytotoxic concentrations of H2O2 under salinity [33], which increased in T1 but decreased in S2 at the late salinity stages (Table 3 and Fig. 2c). Phosphoribulokinase (Fig. 2b) provides an immediate CO2 acceptor and is considered the regulatory enzyme of the Calvin cycle [34]. The dissolved CO2 is fixed by RuBisCO to produce 3-phosphoglycerates [27]. RuBisCO subunits increased in T1, but the large subunits decreased in S2 (Table 3 and Fig. 2a), which implies higher sensitivity of this subunit to salinity. Transketolase is involved in the regeneration phase of the Calvin cycle and the pentose phosphate pathway, thus influencing plant productivity [35]. This enzyme was upregulated in T1 during the early salt-stress stage, which decreased in the later stages in both genotypes. This suggests that the Calvin cycle in chickpea can be slowed by salinity, and photosynthetic enzymes in S2 are more severely and rapidly impaired than in T1. Similar impairments in carbon assimilation have been reported in proteomic studies in salt-stressed soybean genotypes [23] and H2O2-stressed rice seedlings [36].
In addition, spot 27 was identified as magnesium chelatase, the first committed enzyme in the chlorophyll biosynthesis pathway [37]. This finding is consistent with our previous studies where salt-stressed T1 recorded higher photosynthesis [38] and biomass [39] than salt-stressed S2. These results suggest that Mg-chelatase is a salt-tolerance factor in T1 by affecting photosynthesis and biomass. The up- and down-regulation of Mg-chelatase after salinity stress has also been reported in barley [40].
In general, the above results suggest that the salinity-tolerant chickpea genotype (T1) withstands salinity through the upregulation of proteins related to light-dependent reactions and the electron transfer pathway (particularly PPD1, CAB, OEE, and ATP synthase). These processes, in turn, provide sufficient capacity and energy equivalents required for the Calvin cycle [22, 23].
Stress protection by photorespiration
Photorespiration (C2 cycle) results from the oxygenase reaction catalyzed by RuBisCO to produce CO2 and NH3, which wastes ATP and reduction equivalents. Due to this inadequacy, photorespiration may act as an energy sink to inhibit further reductions in the photosynthetic electron transport chain and photoinhibition, which are crucial for plants under stress conditions [41]. Besides, photorespiration generates other metabolites such as serine and glycine for the synthesis of glutathione [42]. Glutathione is an antioxidative factor in plants [42]; accordingly, photorespiration can be involved in stress protection (Fig. 5).
Three DEPs were involved in photorespiration: glycine dehydrogenase (GDH, spot 7), glutamate glyoxylate aminotransferase (GGAT, spot 21), and D-glycerate 3-kinase (GLYK, spot 28). A differential expression pattern was recorded for these DEPs in the two genotypes, which were upregulated at 3 DAT (GDH and GGAT) and 10 DAT (GLYK) in T1 and downregulated at a later stage of stress in S2. Similarly, GDH declined in a drought-sensitive cultivar of C. dactylon but increased or remained stable in a tolerant cultivar [43].
Glutamate glyoxylate aminotransferase plays a central role in the biosynthesis and metabolism of nitrogen and certain amino acids, including Gln, Glu, Ser, and Gly. This enzyme catalyzes the phosphorylation of glycerate as the concluding reaction of the C2 cycle in chloroplasts [41, 44]. Despite the critical roles of GGAT and GLYK, studies investigating stress responses rarely focus on them. Recently, GGAT was upregulated in faba bean under drought stress and recognized as an important factor in drought tolerance [45]. The elevated abundance of C2-cycle-related proteins in T1 under salinity appears to be an essential process in the control of amino acid and glutathione biosynthesis and other metabolisms.
Defense and detoxification under salinity
Fourteen DEPs were recognized as stress-responsive and implicated in detoxification and defense mechanisms such as ROS production, control of oxidative damage, protein stabilization, and hyperosmotic stress.
Heat shock proteins (HSPs) and late embryogenesis abundant (LEA) proteins
The HSP family are high-temperature-inducible chaperones that regulate normal plant growth processes by helping with protein folding and preventing protein aggregation. Three HSP 70 molecular chaperones (spots 1, 11 and 4) and one 20 kDa chaperonin (spot 48) were significantly upregulated in T1, especially at 3 and 10 DAT, respectively (Table 3). However, mitochondrial isoforms (HSP 70, spot 4 and HSP 22, spot 58) progressively increased (spot 4) or decreased (spot 58) during the stress. The higher abundance of chaperones and chaperonin in T1 implies that protein refolding and stabilization might be enhanced to cope with salinity.
In addition, the LEA (spot 30) proteins usually involved in salt tolerance, known as high-molecular osmolytes, function to protect the steady structure of proteins, chlorophyll, membranes, and cells [46]. Upregulation of LEA proteins in T1 could be involved in the process of adaptation to saline conditions (Fig. 5).
Antioxidant and detoxifying enzymes
As illustrated in Fig. 5, salinity perturbs water uptake leading to stomatal closure in response to ABA and Ca2+, which reduces the entrance of CO2, resulting in a net reduction of photosynthesis and subsequent accumulation of ROS with oxidative stress [47, 48]. Beside chloroplasts, ROS are produced in mitochondria, peroxisomes, apoplasts, and their membranes, and involved in aerobic metabolism and active electron transport [35]. In this study, three defense-related enzymes were significantly regulated by salinity. The Cu/Zn superoxide dismutase (spot 77, SOD) and ascorbate peroxidase (spots 41 and 55, APX) were classified in Cluster 5 (Fig. 3c and Additional file 3: Table S1), and upregulated in T1 at 3 DAT and significantly (p ≤ 0.05) downregulated at later stages of stress. Glutathione S-transferase (spot 42, GST) had the reverse pattern under salinity and was grouped in Cluster 2. Superoxide dismutase acts as the first line of defense by transforming superoxide into H2O2, and any excess is removed by the activities of APX [49]. The GST represent a major group of detoxification enzymes in the GPX/GST pathway, which removes H2O2 [6].
Other defensive proteins
Spots 31 and 40 were identified as thiamine thiazole synthase and xanthoxin dehydrogenase, which are involved in the biosynthesis of thiamine [50] and ABA [51], respectively. Thiamine (B1 vitamin) may be involved in DNA damage tolerance in plant cells under oxidative stress [50]. In addition to ABA biosynthesis and regulation, xanthoxin dehydrogenase participates in proline biosynthesis and the sugar-mediated signaling pathway [52]. The elevated levels of these enzymes in T1 at 3 and 6 DAT can protect seedlings against the deleterious effects of ROS.
The cold-shock domain protein (spot 81) may act as an RNA-chaperone in the regulation of translation [53]. It has been induced in the leaves of rice [54] and roots of wheat [55] under salt stress. It is possible that the higher expression of the cold-shock protein in T1 than S2 in the early stages of salinity helps in the translation process by removing secondary structures of mRNA and regulating gene expression by dsDNA interaction.
Apolipoprotein D (ApoD, spot 83), expressed in T1 more than S2 (Table 3), is a small plasma membrane-associated protein known as lipocalin. In Arabidopsis, ApoD has protective functions against oxidative stresses induced by freezing, light, and paraquat [56]. Apolipoprotein D binds and scavenges peroxidated lipids, which helps to maintain membrane integrity. Taken together, the elevated levels of these scavengers, especially in T1 under salinity, are likely strategies for chickpea to cope with the deleterious effects of ROS.
Protein metabolism under salinity
Proteins associated with metabolism accounted for nearly 17% of DEPs in stressed seedlings (Fig. 3a), and were divided into two functional groups: protein biogenesis and protein degradation. The first group consisted of seven identities including three elongation factor Tu (EF, mitochondrial: spot 13 and chloroplastic: spots 20 and 23), three ribosomal proteins (RP, 30S: spot 24; 50S: spot 85 and 60S: spot 72), and one peptidyl-prolyl cis-trans isomerase (PPIase, spot 16). In response to salt stress, the EFs were significantly upregulated at 1 and 3 DAT, especially in T1, and then sharply downregulated at 10 DAT in both genotypes, particularly S2 (Table 3). Elongation factors contribute to the initiation and elongation of newly growing peptide chains [36], which may explain the enhanced biosynthesis or repair of salt-stressed proteins. In confirming this, 30S RP and PPIase were upregulated at 3 DAT, especially in T1, which indicates T1’s greater capacity for protein biogenesis, folding and stability. A decreased relative abundance of synthetic proteins at later stages of salinity, especially in S2, indicate further suppression of protein biosynthesis in salt-stressed S2. Similar data have been reported in salt-susceptible genotypes of common bean [31] and Brassica napus [57] seedlings.
Salinity usually causes protein damage or misfolding, mainly due to ROS accumulation (Fig. 5). The replacement of dysfunctional proteins with newly activated ones is vital during the stress [58]. In this investigation, the initial upregulation of ATP-dependent zinc metalloprotease FTSH 2 (spot 2) and 26S protease regulatory (spot 25) and later increases in metacaspase (spot 60) and proteasome (spot 43), especially in T1 (Table 3), may support this mechanism. The continuous upregulation of FTSH (spot 2; Fig. 2a) in T1, an ATP-dependent metalloprotease, reflects the enhanced replacement of the D1 core component of PSII during salinity [59]. In addition to protease activity, FTSH is a molecular chaperone [60]. Therefore, under salinity FTSH could help to maintain quality control of certain membrane and cytoplasmic proteins, which in turn, could be a salt-tolerance factor in chickpea. However, the observed differential regulation of distinct components in the protein biogenesis and degradation machinery suggests that a complicated mechanism is involved in controlling protein metabolism under salinity, which depends on time and genotype.
Amino acid and nitrogen metabolism
Our analysis revealed changes in four identities related to the accumulation of organic solutes and nitrogen compounds under salinity (Table 2). The S-adenosylmethionine synthase (SAM-S, spot 19) is a donor of the methyl group in the methylation reactions of proteins, nucleic acids, polysaccharides, and fatty acids, and a precursor for ethylene and polyamine biosynthesis. This enzyme was upregulated in T1 up to 6 DAT but then significantly declined, especially in S2. Higher expression of the SAM-S encoding gene in Suaeda salsa [61] and its protein in soybean leaves [23] increased salinity tolerance.
The higher abundance of glutamine synthetase (GS, spot 35) in T1 than S2 produced more glutamate, causing re-assimilation of the extra ammonia released during salinity, and ensuring the production of nitrogenous compounds related to the stress, which can enhance photorespiration to improve stress resistance [45]. Glycine dehydrogenase (GLDC; spot 39) is crucial for the biosynthesis of glycine betaine and facilitates osmotic adjustment [62]. The observed upregulation of GS2 and GLDC in T1 resulted in the accumulation of compatible organic solutes in leaves, which could be linked to salt tolerance. Several studies have also shown that salt stress upregulates GS and GLDC [27].
Modifications in stress signaling and gene regulation
The signal transduction pathways can regulate gene expression, which leads to the expression of responsive proteins, specifically in the case of abiotic stresses. Spot 47 was characterized as an auxin-binding protein (ABP19a), which notably participates in signal transduction in the presence of abiotic stresses. It functions as an auxin receptor and has an essential role in many developmental processes [63]. The progressively increasing levels of ABP19 over time in T1, relative to S2, suggest an important role of auxin in salinity stress responses. Upregulation of some members of the ABP family has occurred under abiotic stress in maize [23]. To our knowledge, little information is available on ABP members and their roles in response to salinity.
An initial upregulation followed by later downregulation in the expression of the nascent polypeptide-associated complex subunit beta (NAC, spot 82) was observed in both genotypes. This transcriptional reprogramming factor associated with plant stress responses can bind to ribosome-associated nascent polypeptide chains to regulate its sorting and translocation [64]. In response to salt stress, there are reports of NAC upregulated in tomato [65] and downregulated in rice roots [66].
Two ribonucleoproteins (RNP, 33 kDa: spot 37 and 29 kDa: spot 54) that appeared in Cluster 4 (Fig. 3c and Additional file 3: Table S1) are involved in the 3-end processing of chloroplast mRNAs, and their upregulation in T1 may be related to the translation of defense-related genes in the chloroplast [67]. Another RNA processing DEP was a glycine-rich RNA-binding protein (spot 75), which decreased in both genotypes at 10 DAT.
Other metabolisms
Other proteins associated with the TCA cycle (isocitrate dehydrogenase, IDH, spot 12), oxidative pentose phosphate pathway (6-phosphogluconate dehydrogenase, 6PGDH, spot 15), and purine metabolism (phosphoribosylformylglycinamidine cyclo-ligase, spot 36) were initially upregulated, especially in T1, and then downregulated at 6 and 10 DAT, more so in S2.
Isocitrate dehydrogenase provides NADPH for plants to cope with oxidative stress, and supplies 2-oxoglutarate, which is involved in the glutamine–glutamate synthase cycle. It is crucial, especially in carbon-limiting conditions that experience salinity stress [68]. Similarly, an increase in IDH abundance and the citrate cycle to generate more energy to combat salinity has been reported in tolerant genotypes of rice [69] and wheat [55]. Taken together, an increase in both IDH and 6PGDH abundance in T1 may have accumulated NADPH to supply the necessary energy for salt tolerance.
Transcriptional investigation
To correlate the levels of identified DEPs with their gene expression patterns, an RT-PCR assay was used to analyze mRNA levels in six corresponding genes (Table 2; Fig. 4; Additional file 5: Figure S4). Five transcripts in T1 (carbonic anhydrase, glycerate dehydrogenase, HSP 70 kDa, ascorbate peroxidase, and phosphogluconate dehydrogenase) and two transcripts in S2 (carbonic anhydrase and HSP 70 kDa) recorded similar trends in their protein profiles. This consistency suggests that these proteins are initially regulated at the transcriptional level, do not misfold or dysfunction after salt treatment, and induce related signal transduction pathways to resist salinity stress [70].
One gene in T1 (metalloprotease) and four in S2 (glycerate dehydrogenase, ascorbate peroxidase, metalloprotease, and phosphogluconate dehydrogenase) displayed different or reversed trends between mRNA and protein levels (Fig. 4; Additional file 5: Figure S4). This inconsistency might be due to the complex mechanisms of protein expression regulations [71] and the presence of multigene families [72]. On the other hand, the parallel and independent changes between protein and mRNA abundance for these genotypes might reflect the complex, mediated regulatory mechanisms of plants in response to salinity [20, 22, 70]. Moreover, correlations between the transcriptome and proteome are not always straightforward and vary due to species, growth stage, and environmental conditions.
Conclusions
Comparative proteomic analyses using two contrasting chickpea genotypes under control and salt-stressed conditions provided the basis for revealing salt-tolerance mechanisms. Based on our current and previous observations, as well as other reports, a model elucidating the role of proteins and the mechanisms underlying salinity tolerance is depicted in Fig. 5. Salinity stress can be characterized by enhanced ion toxicity and decreased osmotic potential in plants. In response, plants induce signaling events in the plasma membrane that change gene expression and reduce water release due to stomatal closure. The differential salinity response in T1 and S2 could be related to the reprogramming of numerous DEP expression patterns that induce changes in energy metabolism, including photosynthesis, stress-responsive proteins, protein processes, signaling, and energy metabolism. These salinity-tolerance-associated proteins could be key factors that regulate these pathways, including chlorophyll a-b binding protein, oxygen-evolving enhancer protein, ATP synthase, carbonic anhydrase, RuBisCO, HSP and LEA families, ascorbate peroxidase, elongation factor Tu, auxin-binding protein, and ribonucleoproteins. These findings highlight the significance of photosynthesis-related and stress-responsive proteins in the adaptation of chickpea to salinity stress.
Furthermore, hierarchical clustering data revealed key proteins, especially in Clusters 3 and 4, involved in a dynamic network for salt tolerance in chickpea. The higher correlation between some transcripts and their functional proteins, observed in T1, may be important in salt tolerance, and suggests that these proteins are regulated at the transcriptional level in the tolerant genotype and/or less degraded under saline conditions, relative to the sensitive genotype. The proteomic analyses revealed some novel and unexpected proteins not yet reported in chickpea or in relation to salinity.
Methods
Plant materials and salinity treatments
In our previous studies, screening of chickpea genotypes for salinity tolerance revealed Flip 97-43c (T1) and Flip 97-196c (S2) as relatively tolerant and susceptible genotypes, respectively [39, 73]. Seeds of these two genotypes were provided by the Research Center for Plant Sciences, Ferdowsi University of Mashhad, Iran. In the current study, the seeds were sterilized with 3% (w/v) sodium hypochlorite and 70% ethanol, rinsed five times with sterilized distilled water and germinated on wet, sterile filter papers in the dark at 25 ± 2 °C for 48 h. Three uniformly germinated seeds were transferred to 3 l pots containing a mixture of field soil and sand (2:1, w/w), and later thinned to two uniform seedlings per pot. The experiment was conducted in a greenhouse (16/8 h, light/dark cycle; 28 °C/18 °C, day/night). Every 3 days at 16:00, each pot was irrigated to maintain the soil moisture content at ~ 70%. Twenty-one-day-old seedlings of each genotype were subjected to 0 and 100 mM NaCl (Sigma, USA) that had been dissolved in distilled water. The pots were arranged in a completely randomized design with three replicates. Fresh leaves were collected after 1, 3, 6, and 10 d for proteomics and 1, 3 and 5 d for gene expression experiments to extract protein and mRNA, respectively, then frozen in liquid nitrogen and stored at − 80 °C. The choice of NaCl concentration and stress time-points was based on our previous studies, and other proteomics and gene expression analysis reports [6, 73].
Protein extraction and concentration assay
Total protein extraction was performed using the method of Goggin [74]. Briefly, 1 g of leaves was ground to a fine powder in liquid nitrogen, and placed in two 2 ml tubes with two volumes of extraction buffer containing 7 M urea, 2 M thiourea, 2% (v/v) Triton X-100, 20 mM DTT, and 4% (w/v) CHAPS. After 20 min incubation on ice with gentle rocking, the tubes were centrifuged at 12,000 g for 10 min. For purification, the supernatant was precipitated in 9 ml chilled acetone (− 80 °C), incubated for 1 h at − 80 °C, then centrifuged at 14,000 g for 30 min. The green supernatant was removed. The pellet was washed with 5 ml chilled acetone, dried at room temperature, and resuspended in minimal IEF buffer containing 7 M urea, 2 M thiourea, 4% (w/v) CHAPS, 60 mM DTT, 2% (v/v) immobilized pH gradient (IPG) buffer (pH 4–7) for 10 min with gentle rocking. All centrifugations were carried out at 2 °C. The protein concentration in the supernatant was determined by Bradford assay using crystalline BSA as standard [75].
Two-dimensional polyacrylamide gel electrophoresis (2D-PAGE)
2D-PAGE of protein samples was carried out according to the method of O’Farrell [76]. In brief, IPG strips (pH 4–7 linear, 17 cm, BioRad) were loaded with 350 μl 2-DE rehydration buffer containing 500 μg protein and, after 1 h, covered with 2 ml mineral oil. The passive rehydration step was performed at room temperature for 16 h, then IEF was carried out with a PROTEAN IEF Cell system (BioRad, USA) under the following conditions: 1 h at 300 V, gradient from 300 V to 6000 V over 2 h, and hold at 6000 V for 6 h (total 40,000 kVh). The current was 50 μA per strip, and the temperature was maintained at 20 °C. The focused IPG strips were reduced in 10 ml equilibrium buffer (6 M urea, 50 mM Tris pH 8.8, 30% [v/v] glycerol, 2% [w/v] SDS and 0.002% [w/v] bromophenol blue) with gentle shaking containing 65 mM DTT (Sigma, USA) for 15 min. Then, the strips were placed in the same buffer for alkylation containing 135 mM iodoacetamide (Sigma, USA) for 15 min, followed by brief washing in 1× SDS–Tris–glycine running buffer, as described by Chivasa et al. [77].
For second dimension separation of proteins, the developed IPG strips were applied to SDS-PAGE gels containing 12.5% (w/v) polyacrylamide (Sigma, USA) using a PROTEAN II Multi Cell (BioRad, USA), with protein markers (Fermentas, Waltham, MA, USA) loaded on the left side and overlaid with agarose solution. The electrophoresis was run at 15 mA per gel for 30 min, subsequently increased to 30 mA/gel for 8 h. Following SDS-PAGE, gels were washed in ddH2O and stained with 0.12% (w/v) Coomassie Brilliant Blue G-250 solution followed by destaining in 1% (v/v) acetic acid as described by Candiano et al. [78].
Image acquisition and statistical analysis
The gels were scanned using a GS-800 calibrated densitometer (BioRad) at 600 dpi resolution. Spot detection, spot measurement, background subtraction, and spot matching were conducted using Image Master 2D Platinum (ver. 6.01). Following automatic spot detection and matching, all gel images were double-checked manually. To normalize spot volumes, the quantitative amount of protein spots was expressed as percent volumes (percentage of total volume in all spots in the gel) to overcome experimental errors introduced due to differential staining. The molecular mass (Mr) and isoelectric pH (pI) of proteins were calculated on digitized gels using standard protein markers (Sigma, USA) and interpolation of values on the IPGs, respectively.
Significant differences between means were calculated by SPSS software (ver. 24) using one-way analysis of variance and Duncan’s multiple range test. Spots that were consistently present in all three replicates gels and recorded statistically significant (p ≤ 0.05) changes between time-points or relative to their respective control were accepted for further analysis. These spots were then filtered, according to at least two fold-changes in the expression. The data were expressed as fold-changes that were calculated by an increase (+) or decrease (−) of percent volume in each protein spot at each time-point relative to the same protein spot in the control.
Protein identification and database searching
Selected protein spots were excised from 2-DE gels and digested with trypsin, and the peptides were extracted based on standard techniques [79] at Proteomics International, Nedlands, Western Australia. Protein spots were identified using liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) with an Agilent 1260 Infinity HPLC system (Agilent) coupled to an Agilent 1260 Chipcube Nanospray interface (Agilent) on an Agilent 6540 mass spectrometer (Agilent). Peptides were loaded onto a ProtID-Chip-150 C18 column (Agilent) and separated with a linear gradient of water/acetonitrile/0.1% formic acid (v/v). To identify proteins of interest, the raw spectra were submitted to the MSPnr100 database search using MASCOT sequence matching software (Matrix Science, London, UK; http://www.matrixscience.com). Database interrogation parameters for LC-MS/MS analysis on the Agilent 6540 mass spectrometer were as follows: taxonomy (Viridiplantae: green plants), variable modifications (carbamidomethyl, oxidation of methionine residues), mass values (monoisotopic), protein mass (unrestricted), peptide mass tolerance (±0.2 Da), MS/MS tolerance (± 0.2 Da), peptide charge state (2+, 3+ and 4+), enzyme (trypsin), and maximum missed cleavages (1). Only significant hits, as defined by the MASCOT probability analysis (p ≤ 0.05) were accepted.
Functional classification and clustering analysis
The identified proteins were grouped into gene ontology categories and mapped based on their putative functional and likely metabolic roles by investigations in Blast2GO software and databases as Kyoto Encyclopedia of Genes and Genomes (https://www.genome.jp), NCBI and UniProt [80]. Furthermore, literature reviews were used, if available, to confirm biological processes.
The co-expression pattern of the identified proteins was specified using a self-organizing tree algorithm (SOTA) hierarchical clustering. The changes in fold expression values across four time-points (t1–t4) were log transformed, and clustering was undertaken with Pearson correlation as distance with 10 cycles and a maximum cell diversity of 0.8, using Multi-Experimental Viewer (The Institute for Genomic Research) version 4.5.1 [81].
RNA extraction, cDNA synthesis, and semi-quantitative RT-PCR
Total RNA was extracted from the leaves in three biological replicates at three time-points (1, 3 and 5 days after salt treatment [DAT]) using RNeasy Plant Mini Kit (Qiagen, Germany), and treated with RNase-Free DNase Set (Qiagen, Germany) for further purification, in accordance with the manufacturer’s instructions. The extracted RNA integrity and concentrations were assessed by gel electrophoresis (1.5% (w/v) agarose) and a NanoDrop spectrophotometer (Thermo NanoDrop 2000 UV-vis). The cDNAs were generated from each sample using gene-specific primers (Table 2) by Maxima First Strand cDNA Synthesis kit (Fermentas, Waltham, MA, USA) as follows: 6 μl of DEPC water, 1 μl of 100 ng/μl RNA, 2 μl of 5 × Reaction Mix, and 1 μl of Max Enzyme. The mixture was incubated for 10 min at 25 °C, 15 min at 50 °C, and 5 min at 85 °C.
The PCR reactions were performed in a 25 μl reaction volume containing 1–2 μl of the cDNA as template, 1 μM of each primer, 200 μM of dNTP, 1 unit of Taq polymerase enzyme, and 1 × PCR buffer with 1.5 mM MgCl2. A pre-test was conducted with 25, 28, 31, and 34 PCR cycles to determine the optimal number of cycles for amplification of selected genes. The amplification at cycle 28 was in the exponential range and had not reached a plateau yet (Additional file 4: Figure S3). The RT-PCR conditions consisted of an initial incubation of 95 °C for 7 min, followed by 28 cycles of 95 °C for 20 s, annealing at 50–60 °C for 25 s, extension at 72 °C for 30 s, ending with a 7-min incubation at 72 °C.
To design RT-PCR primers, the sequence of each differentially expressed protein (DEP) was first used as a tBLASTn search term against chickpea expressed sequence tags (ESTs). The best aligned EST, along with its corresponding UniGene, were selected; all ESTs clustering with this UniGene were electronically assembled, and the assembled sequence with emphasis on the part of the sequence which contained the identified peptides, was used for primer design using Primer Premier 5. The primer sequences and other features for the RT-PCR assays are in Table 2.
Additional files
Figure S1. Leaf proteome of T1, a salt-tolerant chickpea genotype after (A) 1, (B) 3, (C) 6, and (A) 10 days of 100 mM NaCl stress. An equal amount (500 μg) of protein from all samples was resolved by 2-DE. The experiment was performed in three replicates and the gels of unstressed seedlings (control) are not shown. (DOCX 387 kb)
Figure S2. Leaf proteome of S2, a salt-susceptible chickpea genotype after (A) 1, (B) 3, (C) 6, and (A) 10 days of 100 mM NaCl stress. An equal amount (500 μg) of protein from all samples was resolved by 2-DE. The experiment was performed in three replicates and the gels of unstressed seedlings (control) are not shown. (DOCX 5332 kb)
Table S1. Classification of different proteins represented in salinity-stressed chickpea seedlings compared to their respective controls based on their expression patterns. (DOCX 20 kb)
Figure S3. Determination of the optimal number of PCR cycles for selected gene amplification. (DOCX 105 kb)
Figure S4. The effects of 100 mM salt stress for 1, 3, and 5 days on mRNA expressions, and 1, 3, 6, and 10 days of protein changes in abundance of (A) carbonic anhydrase, (B) glycerate dehydrogenase, (C) heat shock 70 kDa protein, (D) L-ascorbate peroxidase, (E) zinc metalloprotease FTSH2, and (F) phosphogluconate dehydrogenase in the seedling leaves of chickpea genotypes T1 and S2. Transcript levels were determined by RT-PCR, using the chickpea actin gene as a control for normalization, and expressed as fold changes (increase or decrease) relative to the respective control. Data represents the mean of three biological replicates and the vertical bars indicate ±SE. (DOCX 287 kb)
Acknowledgments
We are grateful to the Institute of Agriculture, UWA for its partial financial and technical support. We also thank Research Center for Plant Sciences and The Research Institute of Biotechnology, FUM.
Funding
This work was financially supported by the grants from Ferdowsi University of Mashhad (FUM) and the University of Western Australia (UWA). The funders had no role in the study design, data analysis and interpretation, and manuscript writing, but just provided the financial.
Availability of data and materials
All the data generated or analyzed during this study are included in this published article.
Abbreviations
- 2-DE
Two-dimensional electrophoreses
- 6PGDH
6-phosphogluconate dehydrogenase
- ABP
Auxin-binding protein
- ApoD
Apolipoprotein D
- APX
Ascorbate peroxidase
- CA
Carbonic anhydrase
- CAB
Chlorophyll a-b binding protein 3
- DAT
Days after salt treatment
- DEPs
Differentially expressed proteins
- ECM
Extracellular matrix
- EF
Elongation factor Tu
- FBPA
Fructose-bisphosphate aldolase
- GDH
Glycine dehydrogenase
- GGAT
Glutamate glyoxylate aminotransferase
- GLDC
Glycine dehydrogenase
- GLYKD
Glycerate 3-kinase
- GS
Glutamine synthetase
- GST
S-transferase
- HSPs
Heat shock proteins
- IDH
Isocitrate dehydrogenase
- LC-MS/MS
Liquid chromatography–mass spectrometry
- LEA
Late embryogenesis abundant
- MAPK
Mitogen-activated protein kinase
- Mr
Molecular mass
- NAC
Nascent polypeptide-associated complex subunit beta
- OEE
Oxygen-evolving enhancer protein
- PGK
Phosphoglycerate kinase
- pI
Isoelectric pH
- PPD1
PsbP domain-containing protein 1
- PPIase
Peptidyl-prolyl cis-trans isomerase
- PRK
Phosphoribulokinase
- PS
Photosystem
- PTM
Post-translational modification
- RNP
Ribonucleoproteins
- ROS
Reactive oxygen species
- RP
Ribosomal protein
- RuBisCO
Ribulose 1,5-bisphosphate carboxylase/oxygenase
- SAM-S
S-adenosylmethionine synthase
- SOD
Cu/Zn superoxide dismutase
- SOTA
Self-organizing tree algorithm
- TKT
Transketolase
Authors’ contributions
MA conceived and designed the experiments, carried out all experimental work, analyzed the data, and wrote the manuscript. SV assisted in the experimental design, methodology, interpretation of data and editing the manuscript. SM, and AB participated in project design, and co-supervised the experimental work. KS helped to perform mass spectrometric analyses and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gupta B, Sengupta A, Saha J, Gupta K. Plant abiotic stress:‘omics’ approach. J Plant Biochem Physiol. 2013;1(3):108–109. doi: 10.4172/2329-9029.1000e108. [DOI] [Google Scholar]
- 2.Ashraf M, Harris P. Potential biochemical indicators of salinity tolerance in plants. Plant Sci. 2004;166(1):3–16. doi: 10.1016/j.plantsci.2003.10.024. [DOI] [Google Scholar]
- 3.Subba P, Kumar R, Gayali S, Shekhar S, Parveen S, Pandey A, Datta A, Chakraborty S, Chakraborty N. Characterisation of the nuclear proteome of a dehydration-sensitive cultivar of chickpea and comparative proteomic analysis with a tolerant cultivar. Proteomics. 2013;13(12–13):1973–1992. doi: 10.1002/pmic.201200380. [DOI] [PubMed] [Google Scholar]
- 4.Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, Satou M, Akiyama K, Taji T, Yamaguchi-Shinozaki K, Carninci P, Kawai J, Hayashizaki Y, Shinozaki K. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 2002;31(3):279–292. doi: 10.1046/j.1365-313X.2002.01359.x. [DOI] [PubMed] [Google Scholar]
- 5.Hasegawa PM, Bressan RA, Zhu J-K, Bohnert HJ. Plant cellular and molecular responses to high salinity. Annu Rev Plant Biol. 2000;51(1):463–499. doi: 10.1146/annurev.arplant.51.1.463. [DOI] [PubMed] [Google Scholar]
- 6.Zhang H, Han B, Wang T, Chen S, Li H, Zhang Y, Dai S. Mechanisms of plant salt response: insights from proteomics. J Proteome Res. 2011;11(1):49–67. doi: 10.1021/pr200861w. [DOI] [PubMed] [Google Scholar]
- 7.Xu J, Lan H, Fang H, Huang X, Zhang H, Huang J. Quantitative proteomic analysis of the rice (Oryza sativa L.) salt response. PLoS ONE. 2015;10(3):e0120978. doi: 10.1371/journal.pone.0120978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo G, Ge P, Ma C, Li X, Lv D, Wang S, Ma W, Yan Y. Comparative proteomic analysis of salt response proteins in seedling roots of two wheat varieties. J Proteome. 2012;75(6):1867–1885. doi: 10.1016/j.jprot.2011.12.032. [DOI] [PubMed] [Google Scholar]
- 9.Long R, Li M, Zhang T, Kang J, Sun Y, Cong L, Gao Y, Liu F, Yang Q. Comparative proteomic analysis reveals differential root proteins in Medicago sativa and Medicago truncatula in response to salt stress. Front Plant Sci. 2016;7:424. doi: 10.3389/fpls.2016.00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barker David G., Bianchi Sylvie, Blondon François, Dattée Yvette, Duc Gérard, Essad Sadi, Flament Pascal, Gallusci Philippe, Génier Gérard, Guy Pierre, Muel Xavier, Tourneur Jacques, Dénarié Jean, Huguet Thierry. Medicago truncatula, a model plant for studying the molecular genetics of theRhizobium-legume symbiosis. Plant Molecular Biology Reporter. 1990;8(1):40–49. doi: 10.1007/BF02668879. [DOI] [Google Scholar]
- 11.Toker C, Lluch C, Tejera N, Serraj R, Siddique K. Abiotic stresses. In: Yadav SS, Redden RJ, Chen W, Sharma B, editors. Chickpea breeding and management. Wallingford: Cabi Publishing; 2007. pp. 474–496. [Google Scholar]
- 12.Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- 13.Pandey A, Choudhary MK, Bhushan D, Chattopadhyay A, Chakraborty S, Datta A, Chakraborty N. The nuclear proteome of chickpea (Cicer arietinum L.) reveals predicted and unexpected proteins. J Proteome Res. 2006;5(12):3301–3311. doi: 10.1021/pr060147a. [DOI] [PubMed] [Google Scholar]
- 14.Pandey A, Chakraborty S, Datta A, Chakraborty N. Proteomics approach to identify dehydration responsive nuclear proteins from chickpea (Cicer arietinum L.) Mol Cell Proteomics. 2008;7(1):88–107. doi: 10.1074/mcp.M700314-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Jaiswal DK, Ray D, Subba P, Mishra P, Gayali S, Datta A, Chakraborty S, Chakraborty N. Proteomic analysis reveals the diversity and complexity of membrane proteins in chickpea (Cicer arietinum L.) Proteome Sci. 2012;10(1):59–75. doi: 10.1186/1477-5956-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhushan D, Pandey A, Choudhary MK, Datta A, Chakraborty S, Chakraborty N. Comparative proteomics analysis of differentially expressed proteins in chickpea extracellular matrix during dehydration stress. Mol Cell Proteomics. 2007;6(11):1868–1884. doi: 10.1074/mcp.M700015-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Parankusam S, Bhatnagar-Mathur P, Sharma KK. Heat responsive proteome changes reveal molecular mechanisms underlying heat tolerance in chickpea. Environ Exp Bot. 2017;141:132–144. doi: 10.1016/j.envexpbot.2017.07.007. [DOI] [Google Scholar]
- 18.Kav NN, Srivastava S, Goonewardene L, Blade SF. Proteome-level changes in the roots of Pisum sativum in response to salinity. Ann Appl Biol. 2004;145(2):217–230. doi: 10.1111/j.1744-7348.2004.tb00378.x. [DOI] [Google Scholar]
- 19.Pi E, Qu L, Hu J, Huang Y, Qiu L, Lu H, Jiang B, Liu C, Peng T, Zhao Y, Wang H, Tsai S-N, Ngai S, Du L. Mechanisms of soybean roots’ tolerances to salinity revealed by proteomic and phosphoproteomic comparisons between two cultivars. Mol Cell Proteomics. 2016;15(1):266–288. doi: 10.1074/mcp.M115.051961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo M, Zhao Y, Wang Y, Shi Z, Zhang P, Zhang Y, Song W, Zhao J. Comparative proteomics of contrasting maize genotypes provides insights into salt-stress tolerance mechanisms. J Proteome Res. 2017;17(1):141–153. doi: 10.1021/acs.jproteome.7b00455. [DOI] [PubMed] [Google Scholar]
- 21.Velikova V, Ghirardo A, Vanzo E, Merl J, Hauck SM. Schnitzler Jr-P. genetic manipulation of isoprene emissions in poplar plants remodels the chloroplast proteome. J Proteome Res. 2014;13(4):2005–2018. doi: 10.1021/pr401124z. [DOI] [PubMed] [Google Scholar]
- 22.Wang L, Liang W, Xing J, Tan F, Chen Y, Huang L, Cheng C-L, Chen W. Dynamics of chloroplast proteome in salt-stressed mangrove Kandelia candel (L.) Druce. J Proteome Res. 2013;12(11):5124–5136. doi: 10.1021/pr4006469. [DOI] [PubMed] [Google Scholar]
- 23.Benešová M, et al. The physiology and proteomics of drought tolerance in maize: early stomatal closure as a cause of lower tolerance to short-term dehydration? PLoS One. 2012;7(6):e38017. doi: 10.1371/journal.pone.0038017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J, Yang H, Lu Q, Wen X, Chen F, Peng L, Zhang L, Lu C. PsbP-domain protein1, a nuclear-encoded thylakoid lumenal protein, is essential for photosystem I assembly in Arabidopsis. Plant Cell. 2012;24(12):4992–5006. doi: 10.1105/tpc.112.106542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bagheri R, Bashir H, Ahmad J, Iqbal M, Qureshi MI. Spinach (Spinacia oleracea L.) modulates its proteome differentially in response to salinity, cadmium and their combination stress. Plant Physiol Biochem. 2015;97:235–245. doi: 10.1016/j.plaphy.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 26.Kamal A, Cho K, Choi J-S, Jin Y, Park C-S, Lee J, Woo S. Patterns of protein expression in water-stressed wheat chloroplasts. Biol Plant. 2013;57(2):305–312. doi: 10.1007/s10535-012-0290-0. [DOI] [Google Scholar]
- 27.Wang J, et al. Physiological and proteomic analyses of salt stress response in the halophyte Halogeton glomeratus. Plant Cell Environ. 2015;38(4):655–669. doi: 10.1111/pce.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki CK, Rep M, van Dijl JM, Suda K, Grivell LA, Schatz G. ATP-dependent proteases that also chaperone protein biogenesis. Trends Biochem Sci. 1997;22(4):118–123. doi: 10.1016/S0968-0004(97)01020-7. [DOI] [PubMed] [Google Scholar]
- 29.Liu CW, Chang TS, Hsu YK, Wang AZ, Yen HC, Wu YP, Wang CS, Lai CC. Comparative proteomic analysis of early salt stress responsive proteins in roots and leaves of rice. Proteomics. 2014;14(15):1759–1775. doi: 10.1002/pmic.201300276. [DOI] [PubMed] [Google Scholar]
- 30.Gao L, Yan X, Li X, Guo G, Hu Y, Ma W, Yan Y. Proteome analysis of wheat leaf under salt stress by two-dimensional difference gel electrophoresis (2D-DIGE) Phytochemistry. 2011;72(10):1180–1191. doi: 10.1016/j.phytochem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 31.Zadražnik T, Hollung K, Egge-Jacobsen W, Meglič V, Šuštar-Vozlič J. Differential proteomic analysis of drought stress response in leaves of common bean (Phaseolus vulgaris L.) J Proteome. 2013;78:254–272. doi: 10.1016/j.jprot.2012.09.021. [DOI] [PubMed] [Google Scholar]
- 32.de Abreu CEB, dos Santos AG, de Oliveira Monteiro-Moreira AC, Costa JH, de Brito LH, Moreno FBMB, Prisco JT, Gomes-Filho E. Proteomic analysis of salt stress and recovery in leaves of Vigna unguiculata cultivars differing in salt tolerance. Plant Cell Rep. 2014;33(8):1289–1306. doi: 10.1007/s00299-014-1616-5. [DOI] [PubMed] [Google Scholar]
- 33.Räisänen SR, Lehenkari P, Tasanen M, Rahkila P, Härkönen PL, Väänänen HK. Carbonic anhydrase III protects cells from hydrogen peroxide-induced apoptosis. FASEB J. 1999;13(3):513–522. doi: 10.1096/fasebj.13.3.513. [DOI] [PubMed] [Google Scholar]
- 34.Graciet E, Lebreton S, Gontero B. Emergence of new regulatory mechanisms in the Benson–Calvin pathway via protein–protein interactions: a glyceraldehyde-3-phosphate dehydrogenase/CP12/phosphoribulokinase complex. J Exp Bot. 2004;55(400):1245–1254. doi: 10.1093/jxb/erh107. [DOI] [PubMed] [Google Scholar]
- 35.Yang S, Li X, Ma Y, Sun X, Yang Y, Yang Y. Proteome response of wild wheat relative Kengyilia thoroldiana to drought stress. Can J Plant Sci. 2014;95(2):237–249. doi: 10.4141/cjps-2014-294. [DOI] [Google Scholar]
- 36.Wan X-Y, Liu J-Y. Comparative proteomics analysis reveals an intimate protein network provoked by hydrogen peroxide stress in rice seedling leaves. Mol Cell Proteomics. 2008;7(8):1469–1488. doi: 10.1074/mcp.M700488-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bandehagh A, Salekdeh GH, Toorchi M, Mohammadi A, Komatsu S. Comparative proteomic analysis of canola leaves under salinity stress. Proteomics. 2011;11(10):1965–1975. doi: 10.1002/pmic.201000564. [DOI] [PubMed] [Google Scholar]
- 38.Arefian M, Vessal S, Shafaroudi SM, Bagheri A. Comparative analysis of the reaction to salinity of different chickpea (Cicer aretinum L.) genotypes: a biochemical, enzymatic and transcriptional study. J Plant Growth Regul. 2017;37(2):391–402. doi: 10.1007/s00344-017-9737-z. [DOI] [Google Scholar]
- 39.Arefian M, Vessal S, Bagheri A. Trend of changes for morphophysiological characteristics of chickpea (Cicer arietinum L.) under salinity stress (NaCl) condition. Iran J Pulse Res. 2014;5(1):117–128. [Google Scholar]
- 40.Rasoulnia A, Bihamta MR, Peyghambari SA, Alizadeh H, Rahnama A. Proteomic response of barley leaves to salinity. Mol Biol Rep. 2011;38(8):5055–5063. doi: 10.1007/s11033-010-0651-8. [DOI] [PubMed] [Google Scholar]
- 41.Wingler A, Lea PJ, Quick WP, Leegood RC. Photorespiration: metabolic pathways and their role in stress protection. Philos Trans R Soc Lond Ser B Biol Sci. 2000;355(1402):1517–1529. doi: 10.1098/rstb.2000.0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noctor G, Arisi A-CM, Jouanin L, Kunert KJ, Rennenberg H, Foyer CH. Glutathione: biosynthesis, metabolism and relationship to stress tolerance explored in transformed plants. J Exp Bot. 1998;49(321):623–647. [Google Scholar]
- 43.Zhao Y, Du H, Wang Z, Huang B. Identification of proteins associated with water-deficit tolerance in C4 perennial grass species, Cynodon dactylon × Cynodon transvaalensis and Cynodon dactylon. Physiol Plant. 2011;141(1):40–55. doi: 10.1111/j.1399-3054.2010.01419.x. [DOI] [PubMed] [Google Scholar]
- 44.Boldt R, Edner C, Kolukisaoglu Ü, Hagemann M, Weckwerth W, Wienkoop S, Morgenthal K, Bauwe H. D-GLYCERATE 3-KINASE, the last unknown enzyme in the photorespiratory cycle in Arabidopsis, belongs to a novel kinase family. Plant Cell. 2005;17(8):2413–2420. doi: 10.1105/tpc.105.033993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li P, Zhang Y, Wu X, Liu Y. Drought stress impact on leaf proteome variations of faba bean (Vicia faba L.) in the Qinghai–Tibet plateau of China. 3 Biotech. 2018;8(2):110–122. doi: 10.1007/s13205-018-1088-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chinnusamy V, Zhu J, Zhu J-K. Salt stress signaling and mechanisms of plant salt tolerance. Genet Eng. 2006;27:141–177. doi: 10.1007/0-387-25856-6_9. [DOI] [PubMed] [Google Scholar]
- 47.Meloni DA, Oliva MA, Martinez CA, Cambraia J. Photosynthesis and activity of superoxide dismutase, peroxidase and glutathione reductase in cotton under salt stress. Environ Exp Bot. 2003;49(1):69–76. doi: 10.1016/S0098-8472(02)00058-8. [DOI] [Google Scholar]
- 48.Cruz de Carvalho MH. Drought stress and reactive oxygen species: production, scavenging and signaling. Plant Signal Behav. 2008;3(3):156–165. doi: 10.4161/psb.3.3.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fadzilla NM, Finch RP, Burdon RH. Salinity, oxidative stress and antioxidant responses in shoot cultures of rice. J Exp Bot. 1997;48(2):325–331. doi: 10.1093/jxb/48.2.325. [DOI] [Google Scholar]
- 50.Tunc-Ozdemir M, Miller G, Song L, Kim J, Sodek A, Koussevitzky S, Misra AN, Mittler R, Shintani D. Thiamin confers enhanced tolerance to oxidative stress in Arabidopsis. Plant Physiol. 2009;151(1):421–432. doi: 10.1104/pp.109.140046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwartz SH, Leon-Kloosterziel KM, Koornneef M, Zeevaart JA. Biochemical characterization of the aba2 and aba3 mutants in Arabidopsis thaliana. Plant Physiol. 1997;114(1):161–166. doi: 10.1104/pp.114.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.León P, Sheen J. Sugar and hormone connections. Trends Plant Sci. 2003;8(3):110–116. doi: 10.1016/S1360-1385(03)00011-6. [DOI] [PubMed] [Google Scholar]
- 53.Karlson D, Nakaminami K, Toyomasu T, Imai R. A cold-regulated nucleic acid-binding protein of winter wheat shares a domain with bacterial cold shock proteins. J Biol Chem. 2002;277(38):35248–35256. doi: 10.1074/jbc.M205774200. [DOI] [PubMed] [Google Scholar]
- 54.Dooki AD, Mayer-Posner FJ, Askari H, Zaiee AA, Salekdeh GH. Proteomic responses of rice young panicles to salinity. Proteomics. 2006;6(24):6498–6507. doi: 10.1002/pmic.200600367. [DOI] [PubMed] [Google Scholar]
- 55.Peng Z, Wang M, Li F, Lv H, Li C, Xia G. A proteomic study of the response to salinity and drought stress in an introgression strain of bread wheat. Mol Cell Proteomics. 2009;8(12):2676–2686. doi: 10.1074/mcp.M900052-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Charron J-BF, Ouellet F, Houde M, Sarhan F. The plant apolipoprotein D ortholog protects Arabidopsis against oxidative stress. BMC Plant Biol. 2008;8(1):86–98. doi: 10.1186/1471-2229-8-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jia H, Shao M, He Y, Guan R, Chu P, Jiang H. Proteome dynamics and physiological responses to short-term salt stress in Brassica napus leaves. PLoS One. 2015;10(12):e0144808. doi: 10.1371/journal.pone.0144808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smalle J, Vierstra RD. The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol. 2004;55:555–590. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- 59.Zörb C, Herbst R, Forreiter C, Schubert S. Short-term effects of salt exposure on the maize chloroplast protein pattern. Proteomics. 2009;9(17):4209–4220. doi: 10.1002/pmic.200800791. [DOI] [PubMed] [Google Scholar]
- 60.Schumann W. FtsH–a single-chain charonin? FEMS Microbiol Rev. 1999;23(1):1–11. doi: 10.1111/j.1574-6976.1999.tb00389.x. [DOI] [PubMed] [Google Scholar]
- 61.Ma X, Wang Z, Qi Y, Zhao Y, Zhang H. Isolation of S-adenosylmethionine synthetase gene from Suaeda salsa and its differential expression under NaCl stress. Acta Bot Sin. 2003;45(11):1359–1365. [Google Scholar]
- 62.Gao F, Zhou Y, Zhu W, Li X, Fan L, Zhang G. Proteomic analysis of cold stress-responsive proteins in Thellungiella rosette leaves. Planta. 2009;230(5):1033–1046. doi: 10.1007/s00425-009-1003-6. [DOI] [PubMed] [Google Scholar]
- 63.Ohmiya A, Tanaka Y. Kadowaki K-i, Hayashi T. cloning of genes encoding auxin-binding proteins (ABP19/20) from peach: significant peptide sequence similarity with germin-like proteins. Plant Cell Physiol. 1998;39(5):492–499. doi: 10.1093/oxfordjournals.pcp.a029396. [DOI] [PubMed] [Google Scholar]
- 64.Nuruzzaman M, Sharoni AM, Kikuchi S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front Microbiol. 2013;4:248. doi: 10.3389/fmicb.2013.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen S, Gollop N, Heuer B. Proteomic analysis of salt-stressed tomato (Solanum lycopersicum) seedlings: effect of genotype and exogenous application of glycinebetaine. J Exp Bot. 2009;60(7):2005–2019. doi: 10.1093/jxb/erp075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yan S, Tang Z, Su W, Sun W. Proteomic analysis of salt stress-responsive proteins in rice root. Proteomics. 2005;5(1):235–244. doi: 10.1002/pmic.200400853. [DOI] [PubMed] [Google Scholar]
- 67.Lv D-W, Subburaj S, Cao M, Yan X, Li X, Appels R, Sun D-F, Ma W, Yan Y-M. Proteome and phosphoproteome characterization reveals new response and defense mechanisms of Brachypodium distachyon leaves under salt stress. Mol Cell Proteomics. 2013;13(2):632–652. doi: 10.1074/mcp.M113.030171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hodges M, Flesch V, Gálvez S, Bismuth E. Higher plant NADP-dependent isocitrate dehydrogenases, ammonium assimilation and NADPH production. Plant Physiol Biochem. 2003;41(6):577–585. doi: 10.1016/S0981-9428(03)00062-7. [DOI] [Google Scholar]
- 69.Li X-J, Yang M-F, Chen H, Qu L-Q, Chen F, Shen S-H. Abscisic acid pretreatment enhances salt tolerance of rice seedlings: proteomic evidence. Biochem Biophys Acta. 2010;1804(4):929–940. doi: 10.1016/j.bbapap.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 70.Meng F, et al. Physiological and proteomic responses to salt stress in chloroplasts of diploid and tetraploid black locust (Robinia pseudoacacia L.) Sci Rep. 2016;6:23098–23113. doi: 10.1038/srep23098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Igwe EI, Essler S, Al-Furoukh N, Dehne N, Brüne B. Hypoxic transcription gene profiles under the modulation of nitric oxide in nuclear run on-microarray and proteomics. BMC Genomics. 2009;10(1):408. doi: 10.1186/1471-2164-10-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu WF, Shi WM. Expression profiling of the 14-3-3 gene family in response to salt stress and potassium and iron deficiencies in young tomato (Solanum lycopersicum) roots: analysis by real-time RT–PCR. Ann Bot. 2006;98(5):965–974. doi: 10.1093/aob/mcl189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arefian M, Malekzadeh S. Physiological and gene expression analysis of extreme chickpea (Cicer arietinum L.) genotypes in response to salinity stress. Acta Physiol Plant. 2015;37(9):1–11. doi: 10.1007/s11738-015-1945-1. [DOI] [Google Scholar]
- 74.Goggin DE, Powles SB, Steadman KJ. Selection for low or high primary dormancy in Lolium rigidum gaud seeds results in constitutive differences in stress protein expression and peroxidase activity. J Exp Bot. 2011;62(3):1037–1047. doi: 10.1093/jxb/erq334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72(1):248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 76.O’Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250(10):4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 77.Chivasa S, Ndimba BK, Simon WJ, Robertson D, Yu XL, Knox JP, Bolwell P, Slabas AR. Proteomic analysis of the Arabidopsis thaliana cell wall. Electrophoresis. 2002;23(11):1754–1765. doi: 10.1002/1522-2683(200206)23:11<1754::AID-ELPS1754>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 78.Candiano G, Bruschi M, Musante L, Santucci L, Ghiggeri GM, Carnemolla B, Orecchia P, Zardi L, Righetti PG. Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis. 2004;25(9):1327–1333. doi: 10.1002/elps.200305844. [DOI] [PubMed] [Google Scholar]
- 79.Bringans S, Eriksen S, Kendrick T, Gopalakrishnakone P, Livk A, Lock R, Lipscombe R. Proteomic analysis of the venom of Heterometrus longimanus (Asian black scorpion) Proteomics. 2008;8(5):1081–1096. doi: 10.1002/pmic.200700948. [DOI] [PubMed] [Google Scholar]
- 80.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 81.Romijn EP, Christis C, Wieffer M, Gouw JW, Fullaondo A, van der Sluijs P, Braakman I, Heck AJ. Expression clustering reveals detailed co-expression patterns of functionally related proteins during B cell differentiation. Mol Cell Proteomics. 2005;4(9):1297–1310. doi: 10.1074/mcp.M500123-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Leaf proteome of T1, a salt-tolerant chickpea genotype after (A) 1, (B) 3, (C) 6, and (A) 10 days of 100 mM NaCl stress. An equal amount (500 μg) of protein from all samples was resolved by 2-DE. The experiment was performed in three replicates and the gels of unstressed seedlings (control) are not shown. (DOCX 387 kb)
Figure S2. Leaf proteome of S2, a salt-susceptible chickpea genotype after (A) 1, (B) 3, (C) 6, and (A) 10 days of 100 mM NaCl stress. An equal amount (500 μg) of protein from all samples was resolved by 2-DE. The experiment was performed in three replicates and the gels of unstressed seedlings (control) are not shown. (DOCX 5332 kb)
Table S1. Classification of different proteins represented in salinity-stressed chickpea seedlings compared to their respective controls based on their expression patterns. (DOCX 20 kb)
Figure S3. Determination of the optimal number of PCR cycles for selected gene amplification. (DOCX 105 kb)
Figure S4. The effects of 100 mM salt stress for 1, 3, and 5 days on mRNA expressions, and 1, 3, 6, and 10 days of protein changes in abundance of (A) carbonic anhydrase, (B) glycerate dehydrogenase, (C) heat shock 70 kDa protein, (D) L-ascorbate peroxidase, (E) zinc metalloprotease FTSH2, and (F) phosphogluconate dehydrogenase in the seedling leaves of chickpea genotypes T1 and S2. Transcript levels were determined by RT-PCR, using the chickpea actin gene as a control for normalization, and expressed as fold changes (increase or decrease) relative to the respective control. Data represents the mean of three biological replicates and the vertical bars indicate ±SE. (DOCX 287 kb)
Data Availability Statement
All the data generated or analyzed during this study are included in this published article.