Abstract
Background
Intervertebral disc degeneration (IDD) is associated with low back and neck pain, but the mechanisms underlying its pathogenesis are unclear. In this study, we explored the function of microRNA-149 (miR-149) in inflammatory response mediated by lipopolysaccharide (LPS) in nucleus pulposus (NP) cells.
Material/Methods
Quantitative real-time PCR was used to detect miRNA and mRNA levels, while Western blotting was utilized to determine protein levels. ELISA was used to examine chemokine production. The correlation between miR-149 and MyD88 was assessed by reporter assay. Apoptosis was examined by flow cytometry.
Results
miR-149 expression was significantly decreased after LPS exposure in NP cells. Overexpression of miR-149 reversed LPS-induced inhibition in aggrecan and collagen II expression and attenuated LPS-mediated promotion in the levels of MMP3, ADAMTS4, and inflammatory cytokines. Moreover, we found that miR-149 exerted its function by targeting MyD88 in NP cells.
Conclusions
miR-149 can inhibit the inflammatory response mediated by LPS in NP cells, and might be a potential target for the treatment of IDD.
MeSH Keywords: Inflammation Mediators, MicroRNAs, Myeloid Differentiation Factor 88
Background
Intervertebral disc degeneration (IDD) causes chronic pain in the lower back and neck, but mechanisms underlying its pathogenesis are not fully understood. It has been suggested that maintenance of a homeostatic environment is important for healthy discs. When it is disrupted, metabolic disorders occur, resulting in a loss of matrix protein, increased degradative enzymes, and upregulation of inflammatory cytokines, and subsequent development of IDD [1,2].
Inflammatory cytokines have been shown to be important in the pathophysiological process of IDD [3–5]. For example, IL-1β is upregulated in degenerated discs, which increases the levels of MMPs and ADAMTs, and decreases synthesis of aggrecan and collagen II in NP cells [3]. Similarly, increased production of TNF-α is observed in patients with IDD [6]. TNF-α increases aggrecan degradation and enhances MMP/ADAMT expression, thus contributing to the progression of IDD [7–9]. Additionally, IL-6, IL-10, and IL-21 are also reported to be associated with IDD [5,10]. However, the exact mechanism of inflammation involved in IDD remains unclear.
miRNAs are a class of small noncoding RNAs that are 21–25 nucleotides in length. miRNAs regulate gene expression by targeting specific mRNAs for degradation or translation repression [11]. Inflammatory response regulation is mediated by controlling gene expression in participating immune system and tissue cells. Studies have demonstrated that miRNAs are important in various biological processes, including inflammatory response [12–14]. Moreover, miRNAs have been reported to be implicated in the pathogenesis of IDD. For example, Lv et al. showed that microRNA-146a (miR-146a) promotes IDD by ameliorating inflammation via the TRAF6/NF-κB pathway [15]. miR-194 has been reported to inhibit inflammatory response by TRAF-6 in nucleus pulposus (NP) cells [16]. Similarly, Zhang et al. demonstrated that overexpression of miR-140-5p inhibits LPS-induced human intervertebral disc inflammation and degeneration [17]. miR-149 has been shown to play a critical role in inflammatory response [18,19]; however, whether miR-149 has a function in IDD is not yet known.
Lipopolysaccharide (LPS) has been used as an inflammatory mediator to trigger IDD [16,17]. In this study, we investigated the role of miR-149 in LPS-mediated inflammation in NP cells of intervertebral discs and explored the underlying mechanism.
Material and Methods
Antibodies
Anti-aggrecan (ab36861), anti-Collage II (ab185430), anti-GAPDH (ab9484), anti-ADAMTS4 (ab185722), anti-Bcl2 (ab196495), and anti-MyD88 (ab2064) were obtained from Abcam (Cambridge, MA, USA). Anti-TLR4 (sc-293072) was obtained from Santa Cruz (USA). Antibodies against MMP3 (#14351), capase3 (#9662), caspase9 (#9508), and Bax (#2772), p65 (#3033) and HRP-conjugated second antibodies were purchased from Cell Signaling Technology (Danvers, USA).
Cell culture
Sprague-Dawley rats (6 weeks old) were obtained from the Experimental Animal Center of Shanghai (Shanghai, China). The lumber vertebrae of rats were extracted using a scalpel, and then NP was extracted under a microscope. NP was digested with trypsin for 30 min and then with collagenase II for 3 h. NP cells were filtered with a 100-μm filter and centrifuged at 2000 rpm for 5 min. NP cells were cultured in DMEM medium containing 10% FBS, 100 U/ml penicillin, and 100 mg/ml streptomycin. All animal experiments were approved by the Ethics Committee of the Third Affiliated Hospital of Guangzhou Medical University.
Cell transfection and LPS treatment
MiR-149 mimics were obtained from Genepharma (Shanghai, China). The full-length cDNA of MyD88 was cloned into pcDNA3.1 with a Myc-tag at the C-terminal. Transfection was performed using Liposome 2000 (Invitrogen, USA). LPS (10 μg/ml, Sigma) was used to induce inflammation in NP cells. For co-transfection experiments, NP cells were transfected with miR-149 mimics together with pcDNA 3.1 vector or pcDNA3.1-MyD88 plasmid, and then treated with LPS.
Quantitative real-time PCR (qRT-PCR)
Total RNA was isolated from NP cells with Trizol reagent and transcribed into first-strand cDNA using the PrimeScript RT reagent kit (Takara, China). qRT-PCR was applied to determine mRNA expression by using the SYBR Premix Ex Taq kit (Takara, China). The reaction procedure was as follows: 95°C for 30 s and 40 cycles of 95°C for 30 s, then 60°C for 30 s. GAPDH was utilized as an internal control. For detection of miR-149, reverse transcription and qRT-PCR were performed using the bulge-loop miRNA qPCR primer set (RiboBio, China) according to the manufacturer’s instructions, and a human U6 small nuclear RNA was used for normalization. Gene expression was calculated by 2−ΔΔCt method. Primer sequences are shown in Supplementary Table 1.
ELISA analysis
Levels of inflammatory cytokines were determined by ELISA kits (R&D Systems, USA) following the manufacturer’s instructions.
Western blotting
Total protein was isolated from NP cells. Protein concentration was measured using the BCA method. Protein samples were separated by SDS-PAGE and transferred to polyvinylidene fluoride membranes (Millipore, USA). After blocking with 5% skim milk at room temperature for 1 h, the membrane was incubated with primary antibody at 4°C overnight. After washing with TBS-Tween, the membrane was incubated with HRP-conjugated secondary antibody for 1 h. Protein bands were determined by use of an enhanced chemiluminescence assay kit (Thermo Scientific, USA). The gray value of bands was analyzed using Image J software.
Luciferase assay
Target genes of miR-149 were predicted using miR-Base (http://www.mirbase.org/) and TargetScan Human 7.0 (http://www.targetscan.org/) databases. MyD88 3′UTR and 3′UTR mutants that contain miR-149 mutant binding site were amplified by PCR and then subcloned into pMIR-REPORT vector (Applied Biosystems, USA). The firefly luciferase plasmid and Renilla vector (Promega Corp., Madison, Wisconsin, USA) were co-transfected into NP cells together with miR-149 mimics or negative controls. The luciferase activity was examined by use of a dual-luciferase reporter assay system kit (Promega, USA).
Apoptosis detection
The apoptosis of NP cells was examined with a FITC Annexin V apoptosis Detection kit (BD, USA) according the manufacturer’s protocol. Briefly, after washing twice with PBS and staining with Annexin V- FITC and propidium iodide, cells were analyzed using a flow cytometer (BD Biosciences, USA).
Statistical analysis
All data are shown as mean ±SD. Analysis was performed using SPSS 20.0 software (USA). Statistical comparisons were performed using the t test or one-way ANOVA. A P value of <0.05 was considered statistically significant.
Results
miR-149 regulates ECM-related gene expression
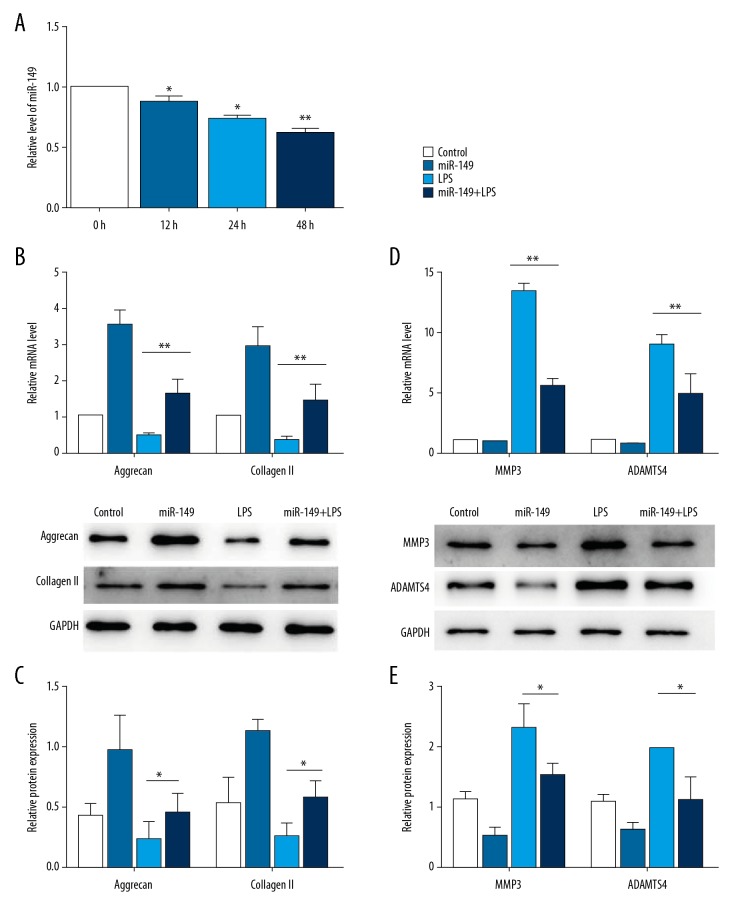
First, we detected whether LPS treatment affected the expression of miR-149. qRT-PCR result showed that miR-149 expression was significantly reduced after LPS treatment for 12 h in NP cells, and the expression levels were further decreased after treatment for 24 h and 48 h (P < 0.05, Figure 1A). It has been reported that LPS exposure decreased aggrecan and collagen expression [16,17]. Consistent with previous results, we found that LPS treatment significantly reduced aggrecan mRNA expression and collagen II mRNA level in NP cells. Interestingly, transfection of miR-149 restored the mRNA expression of aggrecan and collagen II inhibited by LPS (P<0.01, Figure 1B). Similar to the qRT-PCR result, LPS stimulation decreased the protein levels of aggrecan and collagen II as determined by Western blotting, and overexpression of miR-149 abolished the promotional effects of LPS (P<0.05, Figure 1C). Then, we determined the impacts of miR-149 on the protein levels of ECM degradation-related enzymes MMP3 and ADAMTS4. The results showed that LPS significantly induced the mRNA levels of MMP3 and ADAMTS4, and miR-149 transfection inhibited this induction (P<0.05, Figure 1D). Consistently, miR-149 overexpression significantly suppressed the increased protein levels of MMP3 and ADAMTS4 promoted by LPS (P<0.01, Figure 1E).
Figure 1.
Effects of miR-149 on the expression of ECM-related genes. (A) miR-149 expression was detected by qRT-PCR in NP cells after LPS treatment. * P<0.05, ** P<0.01 vs. 0 h. (B, C) Effects of miR-149 on the mRNA levels (B) and protein (C) levels of aggrecan and collagen II in LPS-treated NP cells. * P<0.05, ** P<0.01. (D, E) Effects of miR-149 on the mRNA levels (D) and protein (E) levels of MMP3 and ADAMTS4 in LPS-treated NP cells. * P<0.05, ** P<0.01.
miR-149 inhibits the production of inflammatory cytokines
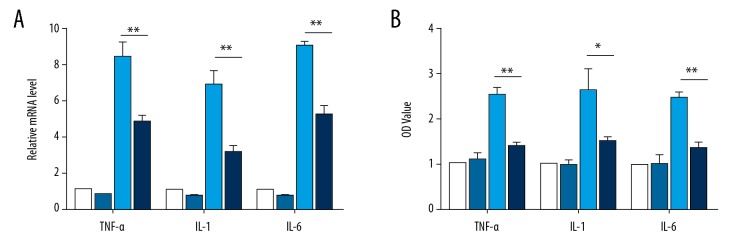
We then examined the effect of miR-149 on the production of inflammatory cytokines in NP cells treated with LPS. LPS exposure markedly increased the mRNA levels of TNF-α, IL-1, and IL-6 as determined by qRT-PCR, whereas transfection with miR-149 suppressed the promotional effects of LPS on these cytokines (P<0.01, Figure 2A). In agreement with this result, ELISA analysis showed the overexpression of miR-149 restrained the enhanced production of TNF-α, IL-1, and IL-6 stimulated by LPS (P<0.05, Figure 2B).
Figure 2.
Effects of miR-149 on the production of LPS-induced inflammatory cytokines. (A) Effects of miR-149 on the mRNA levels of TNF-α, IL-1, and IL-6 in NP cells after LPS stimulation, as determined by qRT-PCR. ** P<0.01. (B) Effects of miR-149 on the production of TNF-α, IL-1, and IL-6 in NP cells after LPS stimulation, as detected by ELISA. * P<0.05, ** P<0.01.
miR-149 represses LPS-induced apoptosis in NP cells
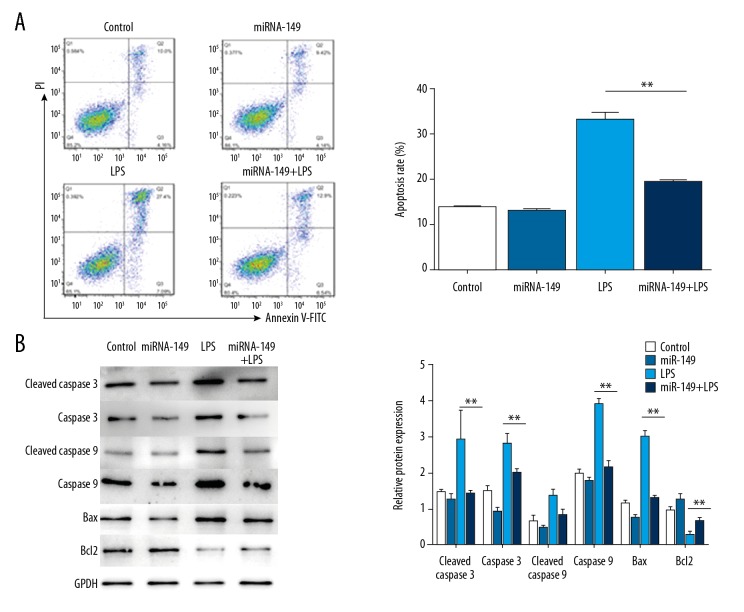
Given the important role of apoptosis in IDD, we examined the impact of miR-149 on LPS-mediated apoptosis in NP cells. Flow cytometry analysis showed that administration of LPS significantly induced apoptosis in NP cells, and transfection of miR-149 suppressed this induction (P<0.01, Figure 3A). In line with this result, Western blotting analysis showed that LPS treatment markedly enhanced the levels of caspase 3, caspase 8, and Bax, but decreased the level of anti-apoptosis protein Bcl2. However, upregulation of miR-149 suppressed the LPS-mediated enhancement in the levels of caspase 3, caspase 8, and Bax, and restored the level of Bcl2 inhibited by LPS (P<0.05, Figure 3B).
Figure 3.
The effect of miR-149 on LPS-triggered apoptosis in NP cells. (A) The effect of miR-149 on LPS-promoted apoptosis in NP cells was determined by flow cytometry. ** P<0.01. (B) The levels of apoptosis-related proteins were analyzed by Western blotting. * P<0.05, ** P<0.01.
miR-149 regulates MyD88 expression
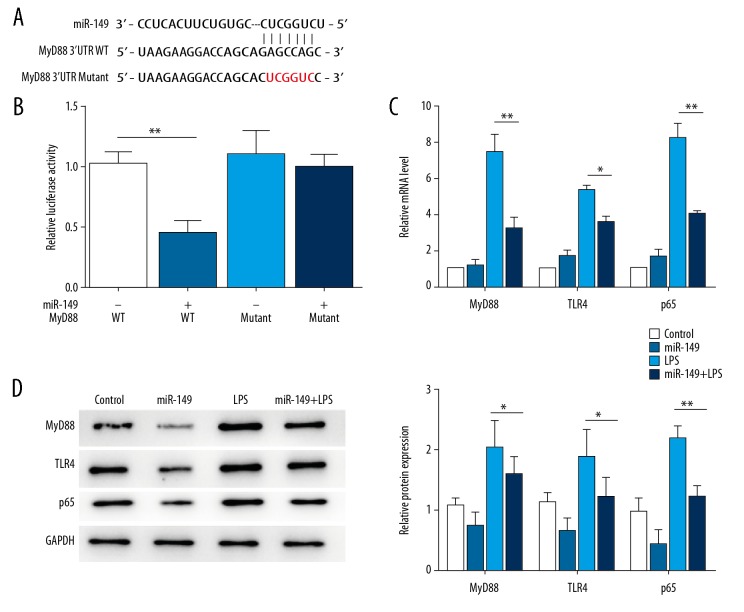
miRNAs usually function via regulating the transcription or degradation of target genes. Through online target prediction, MyD88, which has been reported to play a vital role in IDD [20], was predicted as a potential target of miR-149. To confirm that MyD88 is a target of miR-149, we constructed reporter vectors consisting of the luciferase coding sequence followed by the WT or mutant 3′UTR of MyD88. Cotransfection experiments showed that miR-149 decreased the luciferase activity of MyD88 3′UTR WT, but had no significant effect on MyD88 3′UTR mutant (Figure 4A, 4B). Moreover, in NP cells, LPS treatment significantly enhanced the expression of MyD88, as well as the levels of its receptor TLR4 and downstream effector p65, and these effects of LPS was inhibited by transfection with miR-149 (P<0.01, Figure 4B, 4C).
Figure 4.
MyD88 is a target of miR-149. (A) Sequence of potential binding sites of miR-149 in MyD88 3′-UTR. (B) miR-149 repressed the luciferase activity of MyD88 3′UTR, but had no significant effect on the activity of the mutant. ** P<0.01. (C) Effects of miR-149 on LPS-mediated mRNA expression of MyD88, TLR4, and p65 in NP cells. * P<0.05, ** P<0.01. (D) Effects of miR-149 on LPS-mediated protein expression of MyD88, TLR4, and p65 in NP cells. * P<0.05, ** P<0.01.
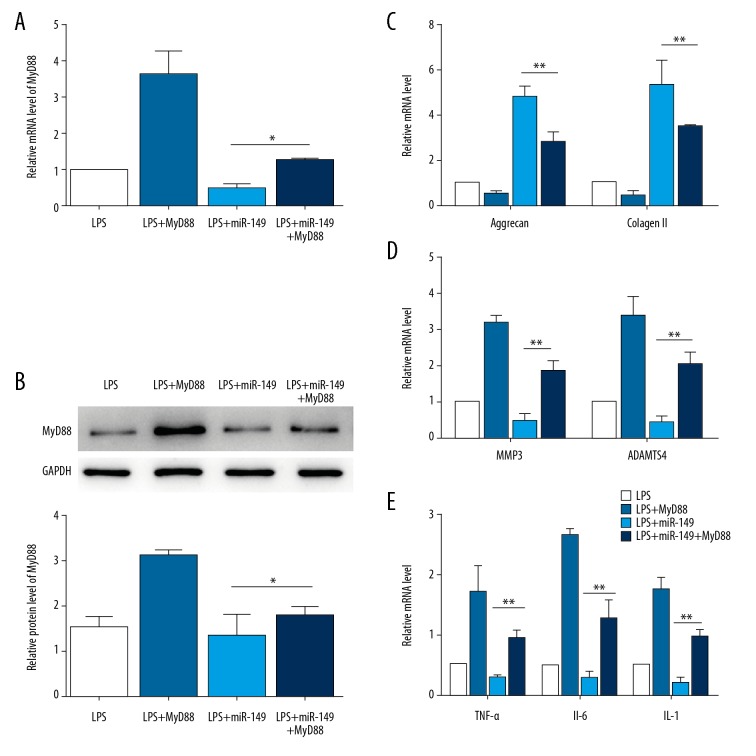
miR-149 regulates LPS-induced ECM degradation and inflammation response via targeting MyD88 in NP cells
Next, we investigated whether miR-149 regulates ECM degradation and inflammation response by targeting MyD88. Overexpression of MyD88 further reduced the expression levels of aggrecan and collagen II decreased by LPS, and enhanced the levels of MMP3, ADAMTS4, TNF-α, IL-1, and IL-6 stimulated by LPS treatment (P<0.01, Figure 5A–5E). While miR-149 abolished LPS-induced inhibition in aggrecan and collagen II expression and suppressed LPS-mediated promotion in the levels of MMP3, ADAMTS4, TNF-α, IL-1, IL-6, overexpression of MyD88 reversed these effects of miR-149 (P<0.01, Figure 5A–5E). Together, these results suggest that miR-149 modulates LPS-induced ECM degradation and inflammation response through MyD88 in NP cells.
Figure 5.
miR-149 inhibits LPS-induced ECM degradation and inflammation response by targeting MyD88. (A) The mRNA expression of MyD88 in LPS-stimulated NP cells transfected with miR-149 and/or MyD88. * P<0.05. (B) The protein expression of MyD88 in LPS-stimulated NP cells transfected with miR-149 and/or MyD88. * P<0.05. (C) MyD88 transfection suppressed the levels of aggrecan and collagen II promoted by miR-149 in LPS-treated NP cells. ** P<0.01. (D) MyD88 transfection restored the levels of MMP3 and ADAMTS4 repressed by miR-149 in LPS-treated NP cells. ** P<0.01. (E) MyD88 transfection rescued the levels of TNFα, IL-1, and IL-6 inhibited by miR-149 in LPS-treated NP cells. ** P<0.01.
Discussion
Over 90% of individuals aged over 50 years have common chronic IDD, but the pathological mechanism of IDD remains largely unknown. In recent years, increasing studies suggest that miRNAs are involved in the progression of IDD. Intriguingly, in this study, we identified that miR-149 has an important function in the process of IDD. We showed that miR-149 expression was decreased in NP cells of intervertebral discs after LPS treatment. Overexpression of miR-149 attenuated LPS-induced ECM degradation, apoptosis, and inflammatory response in NP cells. Our findings imply that miR-149 might be potential target for the treatment of IDD.
Development of IDD involves a multi-step process, in which a major step is degradation of ECM. NP is reported to be important for the maintenance of the biomechanical performance of intervertebral discs, which undergoes ECM changes during IDD [21]. Collagen II and proteoglycan, particularly aggrecan, are the major components of ECM in NP cells and are vital to the normal function of the disc. Loss of collagen II and aggrecan is associated with IDD generation [1]. Here, we found that miR-194 significantly attenuated LPS-induced inhibition in the levels of aggrecan and collagen II. MMPs are primary mediators of ECM degradation, and play a critical role in the destruction of the matrix during IDD [22]. Previous studies have shown that MMP3 and MMP7 are responsible for the degradation of collagen and the core protein of aggrecan [23,24]. In addition, ADAMTS4 is an autocrine factor of the nucleus pulposus cells and is crucial for degrading proteoglycans in IDD [25]. Here, we showed that miR-194 markedly decreased LPS-increased MMP3 and ADAMTS4 expression. Together, our data suggest that miR-194 is critical in LPS-induced ECM degradation.
Accumulating studies have demonstrated that inflammatory cytokines play pivotal roles during the development of IDD. It has been reported that inflammatory cytokines exert their function in IDD via modulating the levels of ECM-degrading enzymes [1]. TNF-α and IL-1β are the 2 most important inflammatory factors. Johnson et al. demonstrated that accumulation of TNF-α and IL-1β in intervertebral discs increases the expression of MMPs [26]. Similarly, Le Maitre et al. reported that activation of IL-1β upregulates MMPs and downregulates collagen II and aggrecan in NP cells [3]. Tian et al. have also shown that treatment with TNF-α and IL-1β significantly increases the expression of ADAMTS4 in NP cells [9]. In this study, we found that overexpression of miR-149 suppressed the production of inflammatory cytokines induced by LPS, including TNF-α, IL-1, and IL-6, indicating that miR-149 has a suppressive effect on LPS-mediated inflammation in NP cells.
To further understand the mechanism underlying the function of miR-149, we searched the potential target of miR-149. We defined that MyD88 is a target of miR-149. MyD88 is an adapter protein that links Toll-like receptors and interleukin-1 receptors with downstream signaling molecules (e.g., NF-kB) and plays an important role in immune response and inflammation response [27]. Previous studies have indicated that MyD88-mediated signaling is critical in the pathogenesis of IDD [20,28]. Consistently, we found that the effects of miR-149 on the levels of aggrecan, collagen II, MMP3, ADAMTS4, TNF-α, IL-1, and IL-6 induced by LPS were reversed by overexpression of MyD88, indicating that miR-149 regulates LPS-induced ECM degradation and inflammation response via targeting MyD88.
Conclusions
miR-149 inhibits the TLR4 signal pathway by targeting MyD88, which suppresses LPS-induced inflammatory responses, leading to decreased ECM degradation and NP cell apoptosis. Therefore, miR-149 may be a possible target for IDD therapy development.
Supplementary Table
Supplementary Table 1.
Primers for qRT-PCR.
| Gene | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| miR-149 | CATCCTTTCTGGCTCCGTGT | GCGTGATTCGTGCTCGTATATC |
| U6 | GCTTCGGCAGCACATATACTAAAAT | CGCTTCACGAATTTGCGTGTCAT |
| Aggrecan | CAGTGAGTTGGACAGTAGTG | CAGATGTTTCTCCACTGACA |
| Collagen II | GGCCCTCAGGGACCTGCCGG | CCAGGGGTACCAGGTTCTCC |
| MMP3 | CCTGGACAAGCAGTTCCAAA | TTCACAATCCTGTAGGAGAT |
| ADAMTS4 | GGAGGCGCCCTTAACTCTGC | GGGCTCCCAGAAGGAGCCTT |
| TNFα | CATCTGCTGGTACCACCAGTT | TGAGCACAGAAAGCATGATC |
| IL-1 | TTGTACAAGGAGAGACAAGC | CAGCTGCAGGGTGGGTGTGC |
| IL-6 | GTGTGA AAGCAGCAAAGAGGC | CTGGAGGTACTCTAGGTATAC |
| MyD88 | GAGATCCGCGAGTTTGAGAC | CTGTTTCTGCTGGTTGCGTA |
| TLR4 | GAGGACTGGGTGAGAAACGA | AGATACACCAACGGCTCTGG |
| p65 | CAAGATCAATGGCAACACGG | CAAGATCAATGGCAACACGG |
| GAPDH | ATGGGAAGCTGGTCATCAAC | GTGGTTCACACCCATCACAA |
Footnotes
Source of support: This research was supported by a grant from the Natural Science Foundation of Guangdong Province (grant no. 2016A030313607)
Conflict of interest
None.
References
- 1.Kepler CK, Ponnappan RK, Tannoury CA, et al. The molecular basis of intervertebral disc degeneration. Spine J. 2013;13:318–30. doi: 10.1016/j.spinee.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 2.Kadow T, Sowa G, Vo N, Kang JD. Molecular basis of intervertebral disc degeneration and herniations: What are the important translational questions? Clin Orthop Relat Res. 2015;473:1903–12. doi: 10.1007/s11999-014-3774-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Le Maitre CL, Freemont AJ, Hoyland JA. The role of interleukin-1 in the pathogenesis of human intervertebral disc degeneration. Arthritis Res Ther. 2005;7:R732–45. doi: 10.1186/ar1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith LJ, Chiaro JA, Nerurkar NL, et al. Nucleus pulposus cells synthesize a functional extracellular matrix and respond to inflammatory cytokine challenge following long-term agarose culture. Eur Cell Mater. 2011;22:291–301. doi: 10.22203/ecm.v022a22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holm S, Mackiewicz Z, Holm AK, et al. Pro-inflammatory, pleiotropic, and anti-inflammatory TNF-αlpha, IL-6, and IL-10 in experimental porcine intervertebral disk degeneration. Vet Pathol. 2009;46:1292–300. doi: 10.1354/vp.07-VP-0179-K-FL. [DOI] [PubMed] [Google Scholar]
- 6.Bachmeier BE, Nerlich AG, Weiler C, et al. Analysis of tissue distribution of TNF-αlpha, TNF-αlpha-receptors, and the activating TNF-αlpha-converting enzyme suggests activation of the TNF-αlpha system in the aging intervertebral disc. Ann NY Acad Sci. 2007;1096:44–54. doi: 10.1196/annals.1397.069. [DOI] [PubMed] [Google Scholar]
- 7.Fujita K, Ando T, Ohba T, et al. Age-related expression of MCP-1 and MMP-3 in mouse intervertebral disc in relation to TWEAK and TNF-αlpha stimulation. J Orthop Res. 2012;30:599–605. doi: 10.1002/jor.21560. [DOI] [PubMed] [Google Scholar]
- 8.Purmessur D, Walter BA, Roughley PJ, et al. A role for TNFalpha in intervertebral disc degeneration: A non-recoverable catabolic shift. Biochem Biophys Res Commun. 2013;433:151–56. doi: 10.1016/j.bbrc.2013.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian Y, Yuan W, Fujita N, et al. Inflammatory cytokines associated with degenerative disc disease control aggrecanase-1 (ADAMTS-4) expression in nucleus pulposus cells through MAPK and NF-kappaB. Am J Pathol. 2013;182:2310–21. doi: 10.1016/j.ajpath.2013.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen B, Liu Y, Zhang Y, et al. IL-21 is positively associated with intervertebral disc degeneration by interaction with TNF-αlpha through the JAK-STAT signaling pathway. Inflammation. 2017;40:612–22. doi: 10.1007/s10753-017-0508-6. [DOI] [PubMed] [Google Scholar]
- 11.Wahid F, Shehzad A, Khan T, Kim YY. MicroRNAs: Synthesis, mechanism, function, and recent clinical trials. Biochim Biophys Acta. 2010;1803:1231–43. doi: 10.1016/j.bbamcr.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 12.Rebane A, Akdis CA. MicroRNAs: Essential players in the regulation of inflammation. J Allergy Clin Immunol. 2013;132:15–26. doi: 10.1016/j.jaci.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 13.Xu D, Song M, Chai C, et al. Exosome-encapsulated miR-6089 regulates inflammatory response via targeting TLR4. J Cell Physiol. 2019;234:1502–11. doi: 10.1002/jcp.27014. [DOI] [PubMed] [Google Scholar]
- 14.O’Connell RM, Rao DS, Baltimore D. microRNA regulation of inflammatory responses. Annu Rev Immunol. 2012;30:295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- 15.Lv F, Huang Y, Lv W, et al. MicroRNA-146a ameliorates inflammation via TRAF6/NF-kappaB pathway in intervertebral disc cells. Med Sci Monit. 2017;23:659–64. doi: 10.12659/MSM.898660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kong L, Sun M, Jiang Z, et al. MicroRNA-194 inhibits lipopolysaccharide-induced inflammatory response in nucleus pulposus cells of the intervertebral disc by targeting TNF receptor-associated factor 6 (TRAF6) Med Sci Monit. 2018;24:3056–67. doi: 10.12659/MSM.907280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Q, Weng Y, Jiang Y, et al. Overexpression of miR-140-5p inhibits lipopolysaccharide-induced human intervertebral disc inflammation and degeneration by downregulating toll-like receptor 4. Oncol Rep. 2018;40:793–802. doi: 10.3892/or.2018.6488. [DOI] [PubMed] [Google Scholar]
- 18.Chen Q, Wu S, Wu Y, et al. MiR-149 suppresses the inflammatory response of chondrocytes in osteoarthritis by down-regulating the activation of TAK1/NF-kappaB. Biomed Pharmacother. 2018;101:763–68. doi: 10.1016/j.biopha.2018.02.133. [DOI] [PubMed] [Google Scholar]
- 19.Zhao Y, Zhao J, Guo X, et al. Long non-coding RNA PVT1, a molecular sponge for miR-149, contributes aberrant metabolic dysfunction and inflammation in IL-1beta-simulated osteoarthritic chondrocytes. Biosci Rep. 2018;38(5) doi: 10.1042/BSR20180576. pii: BSR20180576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qin C, Zhang B, Zhang L, et al. MyD88-dependent Toll-like receptor 4 signal pathway in intervertebral disc degeneration. Exp Ther Med. 2016;12:611–18. doi: 10.3892/etm.2016.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma K, Chen S, Li Z, et al. Mechanisms of endogenous repair failure during intervertebral disc degeneration. Osteoarthritis Cartilage. 2019;27:41–48. doi: 10.1016/j.joca.2018.08.021. [DOI] [PubMed] [Google Scholar]
- 22.Vo NV, Hartman RA, Yurube T, et al. Expression and regulation of metalloproteinases and their inhibitors in intervertebral disc aging and degeneration. Spine J. 2013;13:331–41. doi: 10.1016/j.spinee.2012.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang AC, Hsu SC, Kuo CL, et al. Involvement of matrix metalloproteinases in the inhibition of cell invasion and migration through the inhibition of NF-[kappa]B by the new synthesized ethyl 2-[N-p-chlorobenzyl-(2′-methyl)]anilino-4-oxo-4,5-dihydrofuran-3-carboxylate (JOTO1007) in human cervical cancer Ca ski cells. In Vivo. 2009;23:613–19. [PubMed] [Google Scholar]
- 24.Jing W, Jiang W. MicroRNA-93 regulates collagen loss by targeting MMP3 in human nucleus pulposus cells. Cell Prolif. 2015;48:284–92. doi: 10.1111/cpr.12176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mern DS, Tschugg A, Hartmann S, Thome C. Self-complementary adeno-associated virus serotype 6 mediated knockdown of ADAMTS4 induces long-term and effective enhancement of aggrecan in degenerative human nucleus pulposus cells: A new therapeutic approach for intervertebral disc disorders. PLoS One. 2017;12:e0172181. doi: 10.1371/journal.pone.0172181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson ZI, Schoepflin ZR, Choi H, et al. Disc in flames: Roles of TNF-αlpha and IL-1beta in intervertebral disc degeneration. Eur Cell Mater. 2015;30:104–16. doi: 10.22203/ecm.v030a08. discussion 116–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellman MB, Kim JS, An HS, et al. Toll-like receptor adaptor signaling molecule MyD88 on intervertebral disk homeostasis: In vitro, ex vivo studies. Gene. 2012;505:283–90. doi: 10.1016/j.gene.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L, Chen Q, Wang H, et al. Andrographolide mitigates IL1betainduced human nucleus pulposus cells degeneration through the TLR4/MyD88/NFkappaB signaling pathway. Mol Med Rep. 2018;18:5427–36. doi: 10.3892/mmr.2018.9599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1.
Primers for qRT-PCR.
| Gene | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| miR-149 | CATCCTTTCTGGCTCCGTGT | GCGTGATTCGTGCTCGTATATC |
| U6 | GCTTCGGCAGCACATATACTAAAAT | CGCTTCACGAATTTGCGTGTCAT |
| Aggrecan | CAGTGAGTTGGACAGTAGTG | CAGATGTTTCTCCACTGACA |
| Collagen II | GGCCCTCAGGGACCTGCCGG | CCAGGGGTACCAGGTTCTCC |
| MMP3 | CCTGGACAAGCAGTTCCAAA | TTCACAATCCTGTAGGAGAT |
| ADAMTS4 | GGAGGCGCCCTTAACTCTGC | GGGCTCCCAGAAGGAGCCTT |
| TNFα | CATCTGCTGGTACCACCAGTT | TGAGCACAGAAAGCATGATC |
| IL-1 | TTGTACAAGGAGAGACAAGC | CAGCTGCAGGGTGGGTGTGC |
| IL-6 | GTGTGA AAGCAGCAAAGAGGC | CTGGAGGTACTCTAGGTATAC |
| MyD88 | GAGATCCGCGAGTTTGAGAC | CTGTTTCTGCTGGTTGCGTA |
| TLR4 | GAGGACTGGGTGAGAAACGA | AGATACACCAACGGCTCTGG |
| p65 | CAAGATCAATGGCAACACGG | CAAGATCAATGGCAACACGG |
| GAPDH | ATGGGAAGCTGGTCATCAAC | GTGGTTCACACCCATCACAA |