Abstract
Despite the success of antiretroviral therapy (ART), eradication of HIV-1 from brain reservoirs remains elusive. HIV-1 brain reservoirs include perivascular macrophages that are behind the blood-brain barrier and difficult to access by ART. Macrophages express transcription factor FOXO3a and the TNF superfamily cytokine TRAIL, which are known to target HIV-1-infected macrophages for viral inhibition. ONC201 is a novel and potent FOXO3a activator capable of inducing TRAIL. It can cross the blood-brain barrier, and has shown antitumor effects in clinical trials. We hypothesized that activation of FOXO3a/TRAIL by ONC201 will inhibit HIV-1 replication in macrophages. Using primary human monocyte-derived macrophages, we demonstrated that ONC201 dose-dependently decreased replication levels of both HIV-1 laboratory strain and primary strains as determined by HIV-1 reverse transcriptase activity assay. Consistent with data on HIV-1 replication, ONC201 also reduced intracellular and extracellular p24, viral RNA, and integrated HIV-1 DNA in infected macrophages. Blocking TRAIL or knockdown of FOXO3a with siRNA reversed ONC201-mediated HIV-1 suppression, suggesting that ONC201 inhibits HIV-1 through FOXO3a and TRAIL. The anti-HIV-1 effect of ONC201 was further validated in vivo in NOD/scid-IL-2Rgcnull mice. After intracranial injection of HIV-1-infected macrophages into the basal ganglia, we treated the mice daily with ONC201 through intraperitoneal injection for six days. ONC201 significantly decreased p24 levels in both the macrophages and the brain tissues, suggesting that ONC201 suppresses HIV-1 in vivo. Therefore, ONC201 can be a promising drug candidate to combat persistent HIV-1 infection in the brain.
Keywords: HIV-1, reservoir, ONC201, macrophages, FOXO3a, TRAIL
Introduction
HIV continues to be a major global public health issue affecting more than 1.1 million people in the USA and 36.7 million people worldwide (Deeks et al., 2017; Hall et al., 2008). Although application of antiretroviral therapy (ART) dramatically reduces the rate of HIV-1 replication in infected patients, it is not able to eradicate HIV-1 from cellular reservoirs in the lymphatic system, gut, and central nervous system (CNS). Although many types of cells can be infected by HIV-1, cells that form HIV reservoirs are typically long-lived, including memory CD4+ T cells (Chomont et al., 2009), brain perivascular macrophages, and microglia (Pierson et al., 2000). In the CNS, macrophages and microglia are the main cell types that can be productively infected by HIV-1. The persistent infection of these cells often causes complications such as HIV-1 associated neurocognitive disorders (HAND). Because many ART drugs are not able to cross the blood-brain barrier (BBB) and reach therapeutic concentrations within these CNS cell types, they can produce replication-competent HIV-1, which may spread to other tissues and fuel the emergence of drug-resistant viral strains. Thus, strategies targeting these CNS reservoir cells for HIV-1 eradication are urgently needed.
Vulnerabilities of HIV-1-infected brain macrophages and microglia include the FOXO3a pathway and its transcriptional target TRAIL. FOXO3a is a conserved transcription factor downstream of the PI3K/Akt-1 pathway (Kaestner et al., 2000; Kaufmann and Knochel, 1996). In the absence of environmental signals or growth factors, FOXO3a is in an unphosphorylated state and remains transcriptionally active. Growth factors activate the PI3K/Akt-1 signaling pathway, phosphorylating FOXO3a and abrogating its transcriptional activity (Brunet et al., 2001). FOXO3a is predominantly expressed in peripheral lymphoid organs but the specific immunomodulatory role of FOXO3a remains largely undefined (Lin et al., 2004). In memory CD4+ T cells under HIV-1 infection, multiple signals activate IκB kinase, which phosphorylates FOXO3a, decreasing its ability to induce expression of various pro-apoptotic genes (van Grevenynghe et al., 2008). Our previous publications have demonstrated that FOXO3a targets HIV-1-infected macrophages for apoptosis (Cui et al., 2008). Furthermore, we have determined that FOXO3a is phosphorylated (inactive) during neuroinflammation and expression of FOXO3a inhibits reactive astrogliosis in a neuroinflammatory mouse model (Cui et al., 2011). Intriguingly, the transcriptional targets of FOXO3a include TRAIL, which is a member of the TNF superfamily and is also known as TNF Superfamily Member 10 (Pitti et al., 1996; Wiley et al., 1995). TRAIL interacts with at least five unique receptors. TRAIL receptor 1 (R1) and receptor 2 (R2) have death domains and induce cellular apoptosis following ligand binding (Pan et al., 1997; Seol et al., 2001; Walczak et al., 1997). TRAIL-R3, TRAIL-R4, and the fifth receptor named osteoprotegerin do not possess death domains and instead act as decoy receptors (Degli-Esposti et al., 1997; Emery et al., 1998; Pan et al., 1998). Originally thought to target only tumor cells (Pitti et al., 1996; Wiley et al., 1995), TRAIL has been shown to reduce viral burden in HIV-1-infected macrophage and resting memory CD4+ T cells (Huang et al., 2006; Lum et al., 2001b). While its receptors are expressed in macrophage and microglia, TRAIL is expressed in much lower levels in the CNS compared with the lymphoid tissues (Dorr et al., 2002; Huang et al., 2009; Peng et al., 2005; Ryan et al., 2004). The deficiency of FOXO3a signaling during HIV-1 infection and lack of TRAIL expression in the CNS may inadvertently facilitate the forming of HIV-1 brain reservoirs. Therefore, targeting the FOXO3a-TRAIL pathway is a novel strategy to combat HIV reservoirs within the CNS.
Targeting the FOXO3a-TRAIL pathway has been difficult due to a lack of drug candidates. However, recent drug development provides ONC201 as a potent and stable small molecule that can activate FOXO3a and transcriptionally induce TRAIL expression (Allen et al., 2013; Allen et al., 2015; Kline et al., 2016). ONC201 is orally active and can cross the BBB (Allen et al., 2013). It has shown an efficacious antitumor effect and is currently being tested in clinical trials as a promising anticancer agent (Allen et al., 2016). In this study, we have investigated the anti-HIV-1 activity of ONC201. We demonstrate that ONC201 inhibits the HIV-1 infection both in vitro in monocyte-derived macrophages and in vivo in NOD/scid-IL-2Rγcnull (NSG) mouse brains engrafted with HIV-1-infected macrophages. This newly found activity of ONC201 suggests that it can serve as a drug candidate to combat persistent HIV-1 infection in the CNS.
Materials and Methods
Ethics statement and mice
Primary monocyte-derived macrophages (MDM) and peripheral blood lymphocytes (PBL) were used in full compliance with the University of Nebraska Medical Center and National Institutes of Health ethical guidelines, with the Institutional Review Board (IRB) #: 162-93-FB. For animal experiments, all mice were housed in the Comparative Medicine Animal Facilities at the University of Nebraska Medical Center. All procedures were conducted in accordance with the protocol (15-040-06) approved by the Institutional Animal Care and Use Committee at the University of Nebraska Medical Center. Fourteen four-week old male NOD.Cg-Prkdcscidll 2rgtm1Wjl/SzJ mice were purchased from Jackson Laboratories and HIV-1-infected human MDM (5× 105 cells, 3 μl) was injected intracranially (IC) into the basal ganglion of the mice. The coordinates for inoculation is: 0.5 mm posterior to bregma, 3.5 mm lateral and a depth of 3.6 mm. ONC201 (MedKoo Bioscience, Morrisville, NC) was administrated daily through intraperitoneal (IP) injection at 50 mg/kg. Animals were sacrificed seven days after the IC injection.
Isolation and culture of primary MDM and PBL
Human monocytes and PBL were isolated by counter-current centrifugal elutriation from peripheral blood mononuclear cells obtained through leukopheresis from healthy donors (Gendelman et al., 1988). Primary monocytes were cultured as adherent monolayers at a density of 1.1 × 106 cells/well in 24-well plates or 0.25 × 106 cells/well in 96-well plates and differentiated for seven days in differentiation medium consisting of Dulbecco’s Modified Eagle’s Medium (DMEM) (Sigma Chemical Co., St. Louis, MO) supplemented with 10% human serum, 50 μg/ml gentamicin (GIBCO Invitrogen Corp.), 10 μg/ml ciprofloxacin (Thermo Fisher Scientific, Waltham, MA), and 1000 U/ml recombinant human macrophage-colony stimulating factor (M-CSF). After seven days in differentiation, the cells became MDM and M-CSF was removed since the culture can produce M-CSF through autocrine secretion. PBL were stimulated in phorbol 12-myristate 13-acetate (PMA, 20 ng/mL, Sigma #p8139) and ionomycin (1 mM, Sigma #10634) for 5 hours in a T75 flask before being reseeded on 96-well-plates and cultured in RPMI-1640 medium (Gibco, Gaithersburg, MD) supplemented with 10% fetal bovine serum, 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mM L-glutamine, and 100 U/ml IL-2 (R&D Systems, Minneapolis, MN).
HIV-1 Infection of MDM and ONC201 treatment
MDM were infected with HIV-1ADA, a macrophage-tropic strain of HIV-1, at a multiplicity of infection (MOI) of 0.1 virus/target cell (Gendelman et al., 1988). Briefly, viral stocks were diluted in MDM medium (same with the differentiation medium but without M-CSF) for overnight incubation with MDM. On the second day, the medium was removed and substituted with MDM medium and either DMSO (Sigma), ONC201, or ONC201 isomer (MedKoo Bioscience). For selected experiments, a recombinant human TRAIL R2/Fc Chimera Protein (R&D) was added to the cultures along with ONC201. The cultures were re-treated with the same drugs in fresh MDM medium every two days. At four days after infection, the cultures were treated with the same drugs in 0.5 ml/well fresh serum-free Neurobasal™ medium (Thermo Fisher Scientific) for 24 hours before all samples were collected.
Assessment of cell viability
MDM, glioblastoma (ATCC, CRL-1620), and PBL viability was determined by a colorimetric CellTiter 96® AQueous One Solution Assay (Promega, Madison, WI) based on the manufacturer’s instruction. Assays were performed by adding 20 μl of the CellTiter 96® AQueous One Solution Reagent, which contained a tetrazolium compound[3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt; MTS] and an electron coupling reagent (phenazine ethosulfate; PES), directly to each of 100 μl MDM medium in 96-well plates. After two-hour incubation at 37°C in a humidified 5% CO2 atmosphere, the absorbance of each well was recorded at 490 nm using a 96-well plate reader (BioTek, Winooski, VT).
HIV-1 reverse transcriptase (RTase) activity assay
HIV-1 replication levels were determined by HIV-1 RTase activity assay as described previously (Gendelman et al., 1988). RT activity was determined in triplicate samples of cell culture fluids. For this assay, 10 μl of supernatant was incubated in a reaction mixture of 0.05% Nonidet P-40, 10 μg of poly(A)/ml, 0.25 μg of oligo(dT)/ml, 5 mM DTT, 150 mM KCl, 15 mM MgCl2, and [3H]TTP in Tris-HCl buffer (pH 7.9) for 24-h at 37 °C. Radiolabeled nucleotides were precipitated with ice-cold 10% trichloroacetic acid on UniFilter-96 GF/C (FilterMate Harveser, Waltham, MA) in an automatic cell harvester (FilterMate Harveser, Waltham, MA) and washed with 95% ethanol. Radioactivity was determined by liquid scintillation spectroscopy.
Western blots
MDM or tissues of injection sites were homogenized with a PowerGen Homogenizer (Thermo Fisher Scientific) and then lysed by M-PER™ Mammalian Protein Extraction Reagent (Thermo Fisher Scientific, Waltham, MA). Protein concentrations were determined by Bradford protein assay (Bio-Rad, Hercules, CA). Cell lysates were subjected to SDS-PAGE to separate proteins and electrophoretically transferred to Polyvinylidene fluoride membranes (Bio-Rad, Hercules, CA). Membranes were incubated overnight at 4°C with polyclonal antibodies for FOXO3a (Cat#, 75D8, Cell Signaling Technology, Danvers, MA), phosphorylated FOXO3a (Cat#, 9466s, Cell Signaling Technology, Danvers, MA), HIV-1 p24 (Ref#, M0857, DAKO Corp, Carpinteria, CA), human CD68 (Ref#, M0876, DAKO Corp, Carpinteria, CA), caspase-3 (Cat#, 9662, Cell Signaling Technology), and β-actin (Sigma-Aldrich, St. Louis, MO), followed by horseradish peroxidase-linked secondary anti-rabbit or anti-mouse secondary antibodies (Cell signaling Technology, Danvers, MA). Antigen-antibody complexes were visualized by Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific). For quantification of the data, films were scanned with a CanonScan 9950 F scanner (Canon Inc., Tokyo, Japan) and images were analyzed using the public domain NIH Image J program (developed at the U.S. National Institutes of Health and available on the internet at http://rsb.info.nih.gov/nih-image/).
RNA and DNA extraction and real time RT-PCR analysis of HIV-1 gag
Total RNA was isolated with TRIzol (Thermo Fisher Scientific) and RNeasy Mini Kit according to the manufacturer’s protocol (Qiagen Inc., Valencia, CA). Following RNA extraction, DNA was isolated from the remaining organic phase using the DNeasy Blood and Animal Spin-Column kit according to manufacturer’s instructions (Qiagen). Assays-on-demand primers for TRAIL (ID#, Hs00921974_m1), FOXO3a (ID#, Hs00818121_m1), 18S rRNA (Ref#, 4352930#) and GAPDH (ID#, Hs01922876_u1) were purchased from Thermo Fisher Scientific. For quantification of HIV-1 gag, primers and probe were purchased from Thermo Fisher Scientific with the following sequences: forward, 5’ ACA TCA AGC CAT GCA AAT-3’; reverse, 5’-ATC TGG CCT GGT GCA ATA GG-3’; probe, 5’-FAM-CAT CAA TGA GGA AGC TGC AGA ATG GGA TAG A-TAMRA-3’. Real-time reverse-transcription polymerase chain reaction (RT-PCR) was performed in a volume of 10 μl using the one-step quantitative TaqMan Real-time RT-PCR system (Applied Biosystems Inc.). For analysis of HIV-1 gag DNA, real-time PCR was carried out directly on the DNA samples using the TaqMan Universal Master Mix and a StepOne Plus Real-Time PCR System (Thermo Fisher Scientific). Relative FOXO3a, TRAIL, CD4, CCR5 and HIV-1 gag levels were determined and standardized with GAPDH or 18S rRNA internal control using comparative ΔΔCT method. All primers used in the study were tested for amplification efficiencies and the results were similar.
Analysis of HIV-1 proviral DNA integration
Two-step nested PCR assays were used for quantitative HIV-1 DNA integration analysis as previously described (Friedrich et al., 2010; Iordanskiy et al., 2010). The first round was performed in a 25 μl reaction mix to pre-amplify the genomic DNA from infected macrophages. The following primers were used: Alu forward, 5’-GCC TCC CAA AGT GCT GGG ATT ACA G-3’; and HIV-1 gag reverse, 5’-GCT CTC GCA CCC ATC TCT CTC C-3’. The PCR reaction contained 10x EX Taq Buffer, dNTP mix (2.5 mM), EX Taq (all from TaKaRa Bio Inc., Otsu, Shiga, Japan), ddH2O, 100 nM Alu forward primer, 600 nM gag reverse primer, and 100 ng of genomic DNA. The DNA Engine Peltier Thermal Cycler (Bio Rad, Hercules, CA) was programmed to perform a 2-minute hot start at 94°C, followed by 30 steps of denaturation at 93°C for 30 second, annealing at 50°C for 1 minute, and extension at 70°C for two minutes. The second round real-time quantitative PCR was performed by using 5.6 μl of the reaction mix from the pre-amplification step. The amplified DNA was standardized with an internal GAPDH control. The sequences of the primers and probe were as follows: LTR forward, 5’-GCC TCA ATA AAG CTT GCC TTG A-3’; LTR reverse, 5’ -TCC ACA CTG ACT AAA AGG GTC TGA-3’; probe: 5’-FAM-GCG AGT GCC CGT CTG TTG TGT GAC TCT GGT AAC TAG CTC GC-BHQ-3’.
ELISA
Supernatants were collected and protein levels of HIV-1 p24, TRAIL, RANTES (CCL5), MIP-1α (CCL3), and IP-10 (CXCL10) were analyzed by in-house ELISAs (paired antibodies, R&D Systems, Minneapolis, MN) as previously described (Erichsen et al., 2003) and per manufacturer’s instructions.
siRNA transfection
Predesigned siRNA duplexes targeting FOXO3a (ID: 144672) and a negative control siRNA (catalog no. AM4611) were purchased from Thermo Fisher Scientific. At 72 h after HIV-1 infection, cells were transfected with 100 nM siRNA duplexes for 4 h in DMEM culture medium without serum, in the presence of siIMPORTER reagent (EMD Millipore Corporation, Billerica, MA) according to the manufacturer’s instructions. At 72 h post-transfection, cells were harvested and total RNA was extracted for the detection of FOXO3a and TRAIL levels by real-time RT-PCR.
Free-floating immunohistochemistry and image analyses
Animals were euthanized under deep anesthesia and perfused with phosphate-buffered saline (PBS) and then with 4% paraformaldehyde (PFA) in PBS. The brains were removed and immersed in freshly depolymerized 4% PFA in PBS for 48 h and then cryoprotected by 30% sucrose for 48 h. The fixed, cryoprotected brains were frozen and sectioned in the coronal plane at 30 microns using a Cryostat (Leica Microsystems Inc., Bannockburn, IL), with sections collected serially in PBS as previously described (Zhu et al., 2012). Brain sections were then incubated overnight at 4 °C with primary antibodies, followed by secondary antibodies (Molecular Probes, Eugene, OR, 1: 1000) for 1 h at 25 °C. Primary antibodies included P24 (DAKO, 1:1000) and human CD68 (DAKO, 1: 1000). All antibodies were diluted in 5% goat serum in PBS. Cells were counterstained with DAPI (Sigma-Aldrich, 1:2000) to identify the nuclei. Images were taken using a Zeiss Meta 710 confocal microscope (Carl Zeiss Microimaging, LLC) (20× object, tile scan 4 × 4 mode). Section images from eight mice were imported into Image-ProPlus, version 7.0 (Media Cybernetics, Silver Spring, MD) for quantification of p24/DAPI and CD68/DAPI double positive staining. The assessors were blinded during image acquisition and quantification.
Statistical analysis
Data are expressed as means ± SD unless otherwise specified. Statistical analysis was performed by using one-way ANOVA, followed by the Tukey’s-post-test for paired observations, or two-way ANOVA when two independent variables are considered. The two-tailed Student’s t test was used to compare two groups. Significance was determined by a p value < 0.05. All experiments were performed with cells from at least three donors to account for any donor-specific differences. Assays were performed at least three times in triplicate or quadruplicate within each assay.
Results
ONC201 inhibits HIV-1 infection of macrophages in vitro
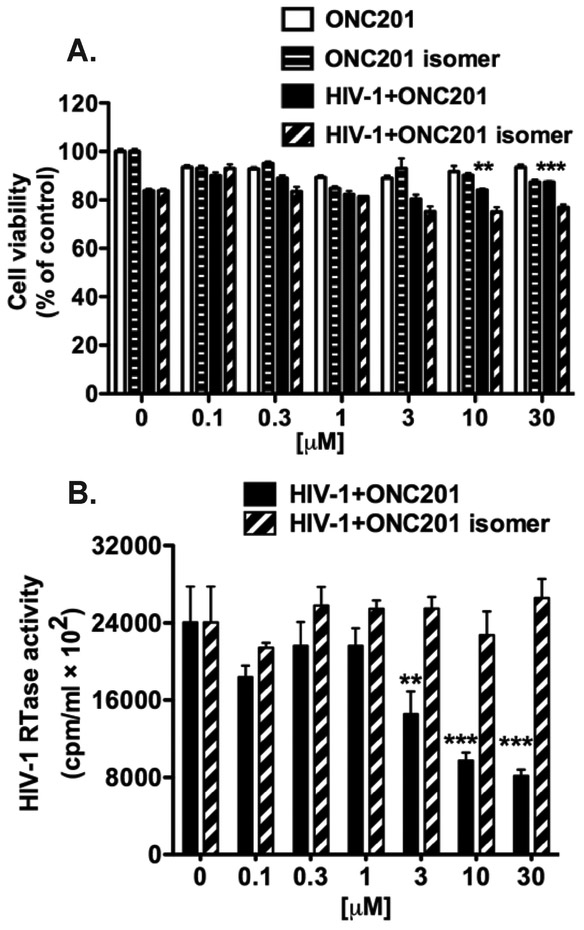
Because ONC201 was originally identified as a TRAIL-inducing anti-tumor agent (Allen et al., 2013), we first sought to confirm its activity in tumor cells. We treated A-172 human glioblastoma cells with doses of ONC201 or its isomer ranging from 0.03 to 30 μM for 72-h and then determined the cell viability through an MTS assay (Supplemental Fig. 1A). Treatment with ONC201, but not ONC201 isomer, induced dose-dependent cytotoxicity in glioblastoma cells, confirming the bioactivity of ONC201. Next, we performed cytotoxicity evaluation of ONC201 on relevant HIV-1 susceptible cells, including primary MDM and PBL. We treated MDM and PBL with either ONC201 or its isomer for five days at concentrations ranging from 0.03 to 30 μM. Treatment with either ONC201 or its isomer did not significantly change cell viability of MDM or PBL at all concentrations tested, as measured by MTS assays, compared with those in the vehicle control DMSO group (0 mM uninfected MDM, Fig. 1A and Supplemental Fig. 1B). Therefore, ONC201 appears to have specific cytotoxicity to tumor cells but not primary human MDM and PBL. In MDM infected with laboratory strain HIV-1ADA, ONC201 treatment significantly increased the cell viability, compared to isomer treatment of the same dosage (Fig. 1A), which is possibly due to the significant decrease of HIV-1 replication at higher doses (10 - 30 μM) of ONC201 treatment (Fig. 1B). The HIV-1 RTase activity assay determines the levels of reverse transcriptase activity in cultural supernatants and is a semi-quantitative measurement of HIV-1 replication levels (Gorantla et al., 2005). Therefore, ONC201 is safe to uninfected human MDM, even at high concentrations, and it has specific antiviral activity against HIV-1 in human MDM.
Figure 1. ONC201 has an antiviral effect on macrophages.
A) Human MDM were plated in 96-well plates and infected with HIV-1 for 24 hours before incubation with doses of ONC201 or its isomer ranging from 0.03 to 30 μM. Uninfected MDM were treated with the same ONC201 and isomer dosages in parallel for comparison. At 5-day post ONC201 and isomer treatments, cell viability was determined by the MTS assays. Data were analyzed by two-way ANOVA, and results shown are the means ± SD of 3 experiments. B) At the same experimental end point as the MTS assays, supernatants were removed and the replication levels of HIV-1 were monitored by RTase activity assay. Results shown are from representative experiments performed with three different donors. Data were analyzed by two-way ANOVA: ** denotes p < 0.01, *** denotes p < 0.001, compared to the HIV-1+ONC201 isomer group of the same treatment dosage.
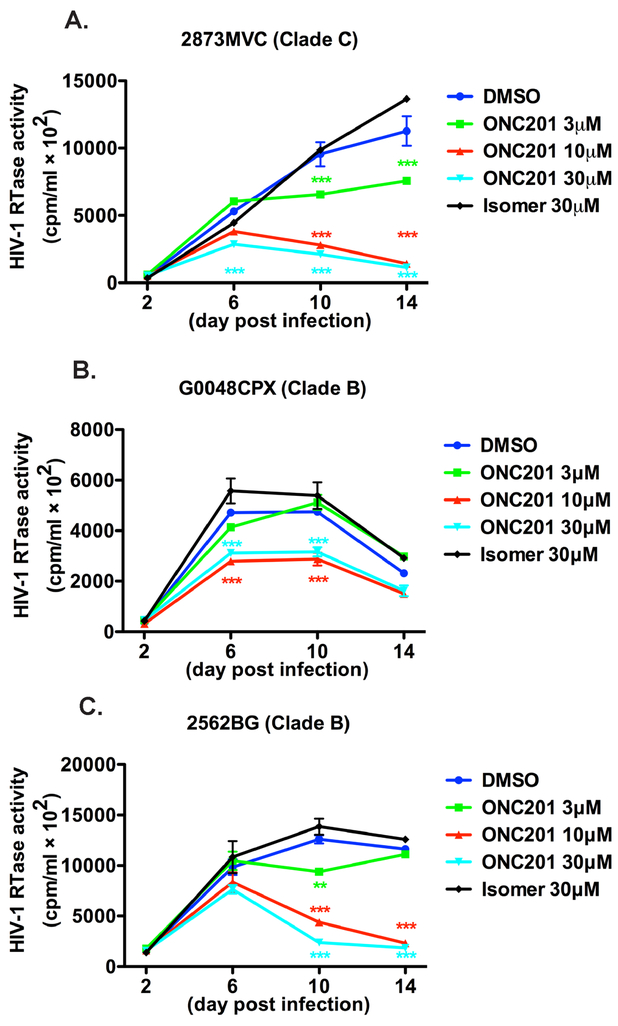
To confirm the antiviral effect of ONC201 on different strains of HIV-1 infection, we infected MDM with two primary HIV-1 Clade B strains (G0048CPX and 2562BG) and one primary HIV Clade C strain (2873MVC), which we characterized previously (Constantino et al., 2011). These HIV strains displayed different replication kinetics and the infection peaked at different times after viral inoculation (Fig. 2). However, for all the strains, ONC201 treatment reduced the viral replication in a dose-dependent manner, which is consistent with the results in HIV-1ADA. Before the peak infection, ONC201 treatment achieved higher degree of viral inhibition compared with those in earlier time points (Fig. 2), suggesting that extending the treatment time has a positive effect on viral inhibition.
Figure 2. ONC201 has an antiviral effect against different primary HIV strains.
A-C) Human MDM were plated in 96-well plates and infected with different primary HIV strains, including 2873MVC (HIV-1 Clade C strain, A), G0048CPX (HIV Clade B strain, B) and 2562BG (HIV Clade B strain, C) for 24 hours before incubation with doses of ONC201 or its isomer ranging from 3 to 30 μM. At 2-, 6-, 10-, and 14-day post ONC201 and isomer treatments, supernatants were collected and the replication levels of HIV-1 were monitored by RTase activity assay. Values represent SEM of biological replicates. Data were analyzed by two-way ANOVA: ** denotes p < 0.01, *** denotes p < 0.001, compared to the HIV-1+ONC201 isomer group of the same treatment dosage.
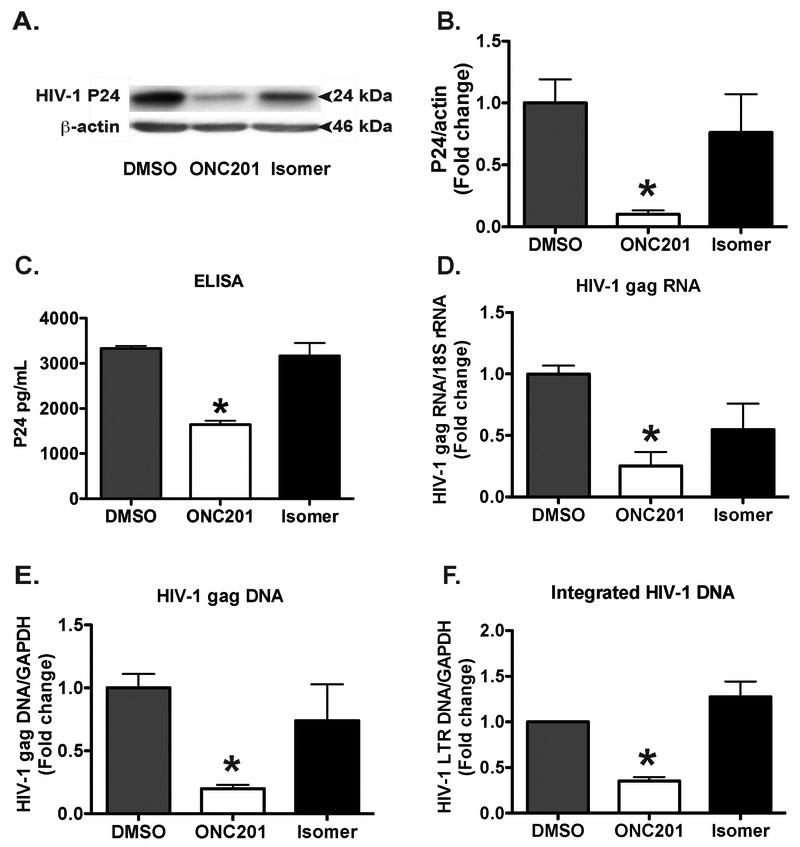
To further characterize the antiviral effect of ONC201 in MDM, we determined both intracellular and extracellular levels of HIV-1 capsid protein p24 through Western blots and ELISA, respectively. Treatment with ONC201 significantly decreased p24 levels in the infected MDM (Fig. 3A-C). Consistent with the p24 data, treatment with ONC201 significantly decreased the levels of HIV-1 gag RNA and DNA when compared with those in the isomer or vehicle control DMSO groups (Fig. 3D and E). Furthermore, we determined the HIV-1 integration through a two-step Alu-based nested PCR method and found that treatment with ONC201 significantly decreased the level of integrated HIV-1 DNA in MDM when compared with those in isomer or DMSO groups (Fig. 3F). Together, ONC201 appears to reduce multiple viral intermediates and products in HIV-1 lifecycle, suggesting it is an effective agent to limit HIV-1 replication in human macrophages.
Figure 3. ONC201 inhibits HIV-1 p24, gag RNA, DNA, and integrated LTR DNA in macrophages.
Human MDM were plated in 24-well plates in triplicate and then infected with HIV-1ADA for 24 hours before incubation with ONC201 or ONC201 isomer at 30 μM for 5 days. A, B) Cell lysates were collected and subjected to SDS-PAGE and immunoblotting for HIV-1 p24. Actin was used as the loading control. Densitometric quantifications of p24 in MDM were presented as a ratio to actin and normalized as fold changes to the vehicle control DMSO group. C) Supernatants were collected and subjected to ELISA for HIV-1 p24. Values represent means ± SEM of three biological replicates. D-F) RNA (D) and DNA (E, F) were isolated from the samples. HIV-1 gag was detected through quantitative real time RT-PCR and real time PCR, respectively. Relative HIV-1 gag and LTR DNA levels were determined and standardized with 18S rRNA or GAPDH internal control. Values represent means ± SEM of three biological replicates. ANOVA analysis: * denotes p < 0.05 compared to the vehicle control DMSO group.
The HIV-1 replication in MDM is productive and cytopathic. To determine whether ONC201 is effective in latently HIV-1 infected cells, we used the pro-monocytic U1 cells (NIH AIDS Reagent Program), which carries latently infected HIV-1 genome that can be stimulated to produce virus (Folks et al., 1987). We treated U1 cells with ONC201 or its isomer in the presence of TNF, a cytokine known to up-regulate viral expression in the cell type, and measured the viral production by RTase activity assay and p24 ELISA. In both assays, ONC201 treatment decreased the level of HIV-1 virus in the culture supernatant in a dose-dependent manner (Supplemental Fig. 2A and B). Notably, the inhibitory effect of ONC201 was seen at concentrations as low as 0.3 μM, suggesting a higher potency of ONC201 against HIV-1 induction from latency compared with the viral inhibition in primary macrophages.
ONC201 has an anti-HIV-1 effect on macrophages in vivo
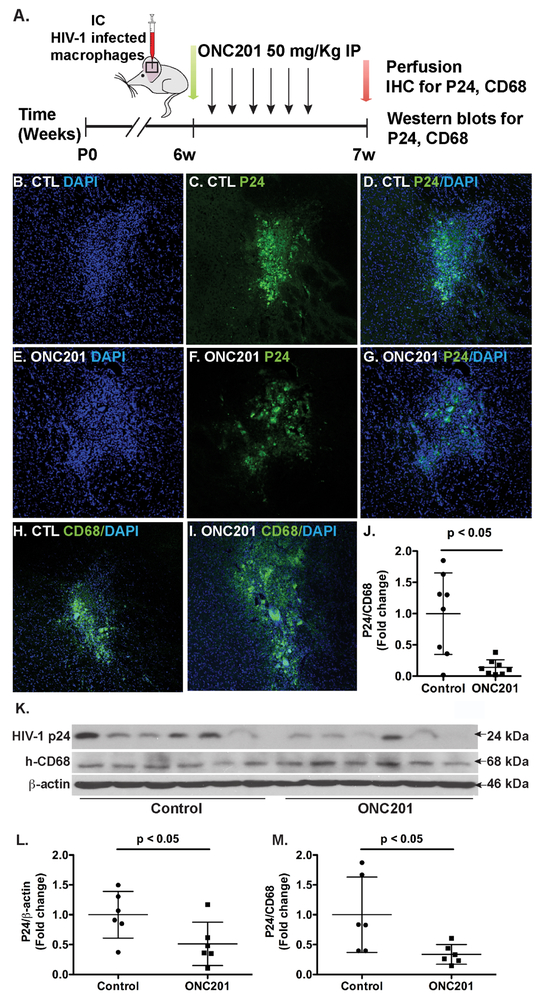
To further determine whether ONC201 has an anti-HIV-1 effect in vivo, we used an established HIV-1 infected NOD/SCID/IL2Rγcnull (NSG) model (Chen et al., 2015; Wu et al., 2009; Ye et al., 2013). HIV-1 infected MDM were intracranially injected into basal ganglion of the NSG immunocompromised mice. At 24-h post MDM engraftment, the mice were administrated daily with either ONC201 (50 mg/kg) or vehicle control via IP for six days. At 7-day post MDM engraftment, the mice were sacrificed for HIV-1 detection (Fig. 4A). To visualize the HIV-1-infected cells in basal ganglion, we first performed immunohistochemistry in serial para-coronal brain slices encompassing the injection track with DAPI (Fig. 4B and E) and p24 (Fig. 4C and F). Both DAPI and p24 clearly marked the injection tracks and their signals were in close proximity to each other in the injection tracks (Fig. 4D and G). To confirm that the p24-positive cells are CD68-positve macrophages, we performed CD68 staining. Because CD68 and p24 antibodies are from the same species (mouse) and alternative antibodies from other species worked poorly in the co-immunostaining (data not shown), we individually labeled the injection site with either CD68 or p24 along with DAPI. CD68 staining revealed a substantial number of macrophages remained in the injection track in both DMSO and ONC201 treatment groups (Fig. 4H and I). We quantified the p24-positive and CD68-positive cells in consecutive slides through a previously established stereological method (Tian et al., 2015; Wang et al., 2017). As expected, we observed that ONC201 administration significantly decreased p24-positive cells as a percentage to total CD68-positive cells (Fig. 4J), compared with those in the vehicle control group. These data suggest that ONC201 reduces the number of HIV-1-infected macrophages in the in vivo NSG mouse model. In order to better quantitatively determine the levels of HIV-1 inhibition, we homogenized the tissues that contained injection sites and subjected the tissue lysates to Western blots (Fig. 4K). The quantification data revealed that p24 levels significantly decreased in the ONC201 group compared with the control group (Fig. 4L). In contrast, CD68 levels were comparable in all of the brain lysates. As a result, the ratio of p24 to CD68 significantly decreased after ONC201 treatment (Fig. 4M), which is consistent with the p24 and CD68 staining data. Together, these data in HIV-1-infected NSG mice demonstrate that ONC201 is able to inhibit HIV-1 replication in macrophages in vivo.
Figure 4. ONC201 reduces HIV-1 infection levels in NSG mouse brains engrafted with infected human macrophages.
HIV-1-infected human macrophages were intracranially injected into the basal ganglia of NOD/scid-IL-2Rgcnull (NSG) mice. Mice were administrated daily with ONC201 (50 mg/kg) or solvent control DMSO through intraperitoneal injections. A) Timeline of experimental procedures and sample collection in the HIV-1-infected NSG mouse model. B-J) Immunostaining of p24 and CD68 in the injection sites of mouse brains: co-immunostaining of p24 and DAPI (B-G); co-immunostaining of CD68 and DAPI (H-I); quantification of the p24/CD68 ratio per area (J). K-M) Brain tissues that contained the injection sites were homogenized for detection of HIV-1 p24 and human CD68 in Western blots. Data were evaluated statistically by an unpaired Student’s t-test, n = 6 per treatment group.
ONC201 activates FOXO3a and induces TRAIL expression in HIV-1 infected macrophages
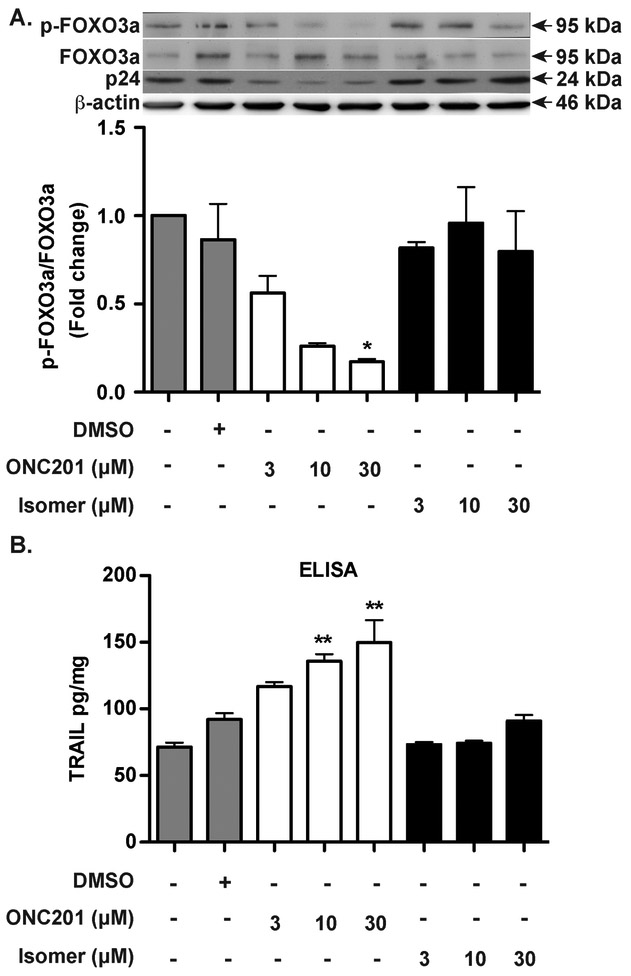
After showing the antiviral effect of ONC201 in vitro and in vivo, we investigated the underlying mechanism of viral inhibition by focusing on the FOXO3a-TRAIL pathway. There are two forms of FOXO3a: the phosphorylated form is inactive and kept in the cytoplasm; once dephosphorylated, it moves to the nucleus and becomes active, inducing pro-apoptotic genes. To assess the status of FOXO3a in HIV-1 infected macrophages after ONC201 treatment, we measured the levels of total FOXO3a, phosphorylated FOXO3a, and β-actin by Western blots. We first confirmed that HIV-1 p24 was indeed reduced by the ONC201 treatment (Fig. 5A). Analysis of FOXO3a phosphorylation revealed that the phosphorylation of FOXO3a, as determined by the ratio of phosphorylated FOXO3a over total FOXO3a, was decreased in a dose-dependent manner after ONC201 treatment compared to the vehicle control DMSO group (Fig. 5A). In contrast, treatment with ONC201 isomer did not significantly change the ratio of phosphor-FOXO3a to total FOXO3a in HIV-1-infected macrophages. This result indicates that FOXO3a is indeed dephosphorylated and activated by ONC201 in MDM. Next, we determined whether ONC201 treatment increases TRAIL expression by measuring soluble TRAIL secretion in the culture supernatants. We found that ONC201 treatment increased TRAIL levels in a dose-dependent manner (Fig. 5B). In contrast, treatment with ONC201 isomer did not significantly change the TRAIL levels in HIV-1-infected macrophages. Because HIV-1 infection is known to decrease FOXO3a phosphorylation and increase TRAIL production in macrophages (Cui et al., 2008; Huang et al., 2009), the changes of FOXO3a phosphorylation and TRAIL production after ONC201 treatment could be due to a change in HIV-1 infection levels. To determine the direct effect of ONC201 on FOXO3a phosphorylation and TRAIL production, we treated uninfected MDM with ONC201 and found that ONC201 decreased FOXO3a phosphorylation (Supplemental Fig. 3A) and increased soluble TRAIL levels (Supplemental Fig. 3B) similar to the data in HIV-1-infected macrophages (Fig. 5). Together, these data confirm that ONC201 activates FOXO3a and induces the downstream expression of TRAIL in HIV-1 infected macrophages.
Figure 5. ONC201 activates FOXO3a and induces TRAIL expression in HIV-1 infected macrophages.
Human MDM were plated in 24-well plates and then infected with HIV-1ADA for 24 hours before incubation with ONC201 or ONC201 isomer at 3, 10, or 30 μM for five days. A) Cell lysates were collected and subsequently subjected to SDS-PAGE and immunoblotting for the detection of phospho-FOXO3a, total FOXO3a, and p24. Actin was used as the loading control. Densitometric quantifications of phospho-FOXO3a in MDM were presented as a ratio to total FOXO3a and normalized as fold changes to the vehicle control DMSO group. B) Soluble TRAIL concentrations after ONC201 or ONC201 isomer treatment were determined from cell culture supernatants using ELISA. Data were normalized with total cellular protein concentrations. Results shown are from representative experiments performed with three different donors. ANOVA analysis: * denotes p <0.05, ** denotes p < 0.01, compared to the vehicle control DMSO group.
The antiviral effect of ONC201 in HIV-1 infected macrophage is dependent on FOXO3a and TRAIL expressions.
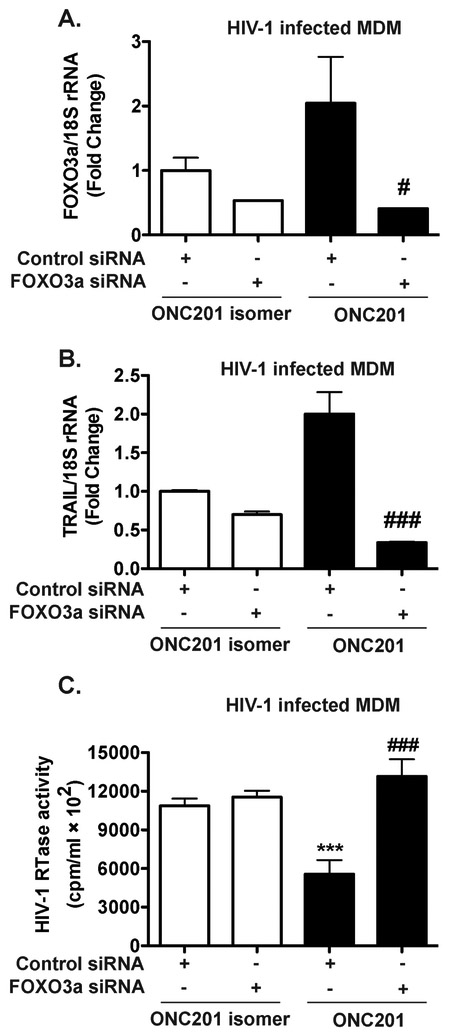
Because downstream targets of ONC201 include TRAIL, which belongs to the TNF superfamily of cytokines, one plausible mechanism for HIV-1 inhibition by ONC201 is through ONC201-induction of cytokines that interfere with the HIV-1 lifecycle. RANTES (CCL5) and MIP-1α (CCL3) are both chemokines that bind to CCR5, which function as a primary co-receptor for HIV-1 entry in MDM (Cocchi et al., 1995; Cocchi et al., 2000). IP-10 (CXCL10) plays a role in chemotaxis of immune cells. We tested the CCL5, CCL3, and CXCL10 levels after ONC201 treatment and found that ONC201 did not significantly increase the levels of these chemokines in the supernatants (Supplemental Fig. 4), suggesting that ONC201 is unlikely to inhibit HIV-1 replication through inducing HIV-1 suppressive chemokines in MDM. To determine if the antiviral effect of ONC201 is mediated by FOXO3a, knockdown of FOXO3a was performed by siRNA targeting FOXO3a in HIV-1-infected MDM. A non-targeting control siRNA was used as the control. We assayed FOXO3a and TRAIL expression at 72-h after siRNA transfection and found that FOXO3a-targeting siRNA reduced the mRNA levels of FOXO3a by 83% (Fig. 6A) and protein levels of FOXO3a by 65% (Supplemental Fig. 5A, B). Similarly, TRAIL expression was downregulated by 85% in HIV-1 infected macrophages (Fig. 6B), suggesting that the siRNA is effective in reducing FOXO3a and its downstream gene transcription. ONC201 significantly decreased the HIV-1 replication levels, as measured by HIV-1 RTase activity assay, compared with those in the isomer treatment group (Fig. 6C). However, FOXO3a knockdown reversed HIV-1 replication to the same level as those in the isomer treated group (Fig. 6C). Consistent with the RTase data, FOXO3a knockdown increased the intracellular p24 level in the ONC201-treated HIV-1 infected MDM (Supplemental Fig. 5C). Together, these data suggest that FOXO3a is required for the antiviral effects of ONC201.
Figure 6. Antiviral effect of ONC201 is dependent upon FOXO3a in HIV-1-infected macrophages.
Human MDM were plated in 24-well plates and then infected with HIV-1ADA for 24 hours before incubation with ONC201 or ONC201 isomer at 30 μM for five days. At two days after HIV-1 infection, MDM were transfected with either non-targeting control siRNA or FOXO3a siRNA. A-B) At the experimental end point, FOXO3a and TRAIL mRNA were detected in total cellular RNA through real time RT-PCR. Data were normalized to 18S rRNA and presented as fold change compared to control siRNA group with isomer treatment. C) HIV-1 infection levels were determined by the RTase activity assay. Values represent means ± SEM of three biological replicates. ANOVA analysis: *** denotes p < 0.001, compared to the control siRNA with isomer treatment group; # denotes p < 0.05, ### denotes p < 0.001, compared to the control siRNA with ONC201 treatment group.
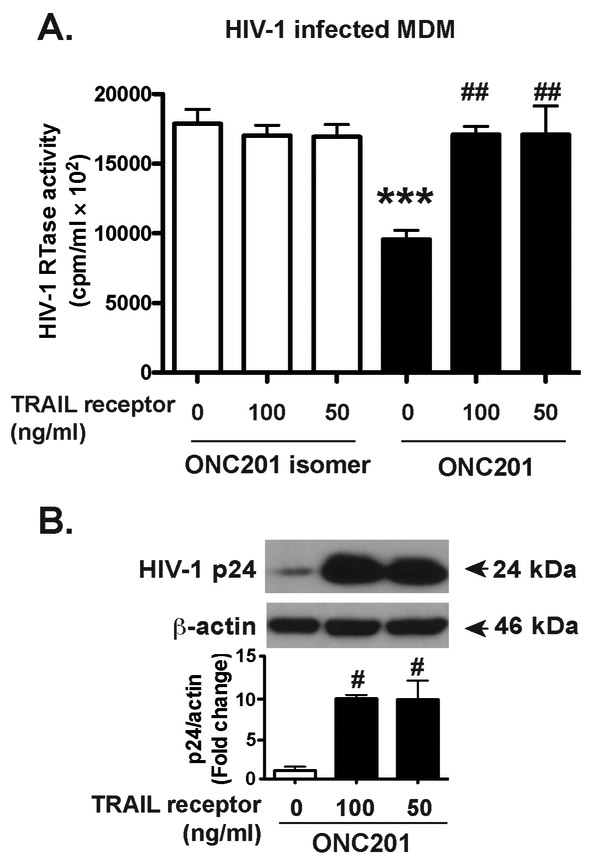
To further determine if TRAIL mediates the antiviral effect of ONC201, we used 50 ng/ml or 100 ng/ml of the soluble TRAIL receptor to act as decoys blocking the activities of both soluble and membrane-associated TRAIL in macrophages. This soluble TRAIL receptor is effective in blocking TRAIL-mediated HIV-1 inhibition in macrophages (Supplemental Fig. 6). In ONC201-treated MDM, we found that treatment of soluble TRAIL receptors reversed the HIV-1 inhibition to the same level as those in the isomer-treated group (Fig. 7A). Consistent with the RTase activity data, treatment with soluble TRAIL receptors also significantly increased intracellular HIV-1 p24 levels (Fig. 7B), suggesting that the antiviral effect of ONC201 in HIV-1 infected macrophages is dependent on TRAIL expressions.
Figure 7. Antiviral effect of ONC201 in HIV-1 infected macrophages is dependent on TRAIL expression.
HIV-1-infected MDM were incubated with ONC201 or ONC201 isomer at 30 μM, along with 100ng/ml, 50ng/ml soluble TRAIL receptor, or 100ng/ml soluble TNF receptor as a control for 5 days. A) HIV-1 infection levels were determined by RTase activity assay. B) Cell lysates were subjected to SDS-PAGE and immunoblotting for HIV-1 p24. Actin was used as the loading control. Densitometric quantifications of p24 in MDM were presented as a ratio to actin and normalized as fold changes to the control group. ANOVA analysis: *** denotes p < 0.001, compared to the control group with isomer treatment; # denotes p < 0.05, ## denotes p < 0.01, compared to the control group with ONC201 treatment.
ONC201 pretreatment increases its efficacies against HIV-1 infection in macrophages
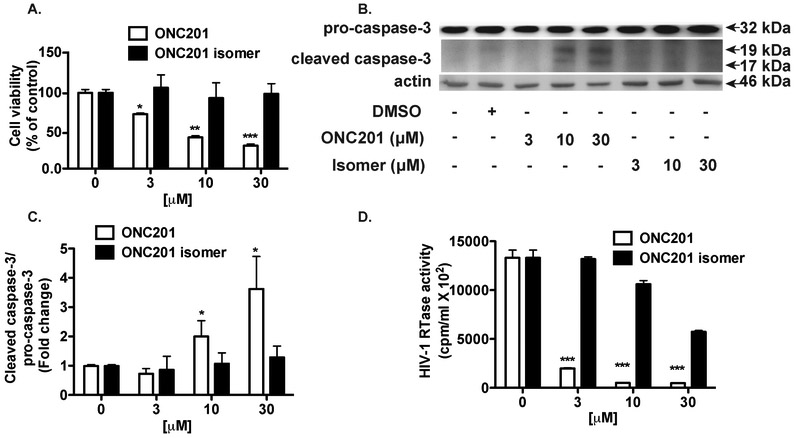
In cancer cells, activation of the FOXO3a-TRAIL pathway by ONC201 leads to cell death and apoptosis, while in HIV-1 infected MDM, treatment of human recombinant TRAIL induces cell death and apoptosis (Allen et al., 2013; Huang et al., 2006). In our data, ONC201 treatment in HIV-1-infected MDM did not significantly impact MDM viability (Fig. 1A). This is likely due to the unique action of ONC201, which reportedly requires at least 72 hours of downstream signaling to induce TRAIL production (Allen et al., 2013). The lagged production of TRAIL by ONC201 during viral propagation in MDM likely undermined the effectiveness of antiviral activities by ONC201. To ensure full TRAIL production, we pretreated MDM with ONC201 for five days before the HIV-1 infection. ONC201 and isomer treatments were continuous both during and after HIV-1 infection. At three-day post infection, we found that ONC201 pretreatment reduced cell viability in a dose-dependent manner (Fig. 8A). Furthermore, cell death of HIV-1 infected macrophages was accompanied by significant increase of caspase cleavage (Fig. 8B, C), indicating that ONC201 pretreatment induces apoptosis in HIV-1 infected macrophages. As expected, analysis of HIV-1 replication by RTase activity assay showed that the ONC201 pretreatment dose-dependently lowered viral levels in the cell culture (Fig. 8D). At 30 μM, ONC201 pretreatment dramatically reduced HIV-1 replication by 96%, a much higher efficacy compared with those in ONC201 treatment post infection. Taken together, these data demonstrate that ONC201 pretreatment increases its efficacies against HIV-1 infection in macrophages and the pretreatment is associated with significant apoptosis in infected macrophages.
Figure 8. ONC201 pretreatment increases its efficacies against HIV-1 infection in macrophages.
Human MDM were plated in 96-well plates and incubated with doses of ONC201 or its isomer ranging from 3 to 30 μM for 5 days before infection with HIV-1. ONC201 and isomer treatments continued during and after the infection. A) At three-day post infection, cell viability was determined by the MTS assays. B) Cell lysates were collected and subsequently subjected to SDS-PAGE and immunoblotting for the detection of cleaved caspase-3 and pro-caspase-3. Actin was used as the loading control. C) Densitometric quantifications of cleaved caspase-3 in MDM were presented as a ratio to pro-caspase-3 and normalized as fold changes to the vehicle control DMSO group. D) Supernatants were collected and the replication levels of HIV-1 were monitored by RTase activity assay. Results shown are from representative experiments performed with three different donors. Data were analyzed by two-way ANOVA: * denotes p < 0.05, ** denotes p < 0.01, *** denotes p < 0.001, compared to the vehicle control DMSO group.
Discussion
Targeting of long-term HIV persistence with novel approaches is essential to combat AIDS in the post-cART era (Carpenter et al., 2000; Krebs et al., 2000; McArthur et al., 1999). In the present study, a novel small molecule ONC201 was tested for its potential antiviral effect on the HIV-infected macrophages. We demonstrated that ONC201 has little toxicity in uninfected human macrophages and exert an anti-HIV-1 effect on infected macrophages in vitro. In a murine HIV Encephalitis model, ONC201 treatment significantly reduced HIV-1 levels in engrafted macrophages, indicating the antiviral effect of ONC201 in the brain reservoir cell type in vivo. We further identified that the antiviral effect of ONC201 is mediated by the FOXO3a/TRAIL pathway, as ONC201 activated FOXO3a and induced TRAIL expression in HIV-1-infected macrophages, and knockdown of FOXO3a or blocking TRAIL abrogated the antiviral activity of ONC201.
Although variants of recombinant TRAIL and the longer-lived TRAIL receptor agonist antibodies have been developed and applied in clinical trials (Camidge et al., 2010; Greco et al., 2008; Leong et al., 2009; Mom et al., 2009; Plummer et al., 2007; Tolcher et al., 2007; Trarbach et al., 2010), those protein-based therapeutics are costly and therapeutic concentrations remain difficult to maintain. Therefore, increasing endogenous TRAIL protein production via pharmacological means is a more practical approach. ONC201 was originally screened from the National Cancer Institute (NCI) Diversity Set II as one of the small molecules capable of up-regulating endogenous TRAIL gene transcription (Allen et al., 2015). There are other potent TRAIL-inducing compounds such as TIC9 reported from the screening. However, ONC201 seems to uniquely potentiates tumor cell death while sparing normal cells and lacks genotoxicity in normal fibroblasts, which could be the result of a relatively milder induction of death receptor 5 compared with those inducted by TIC9 (Allen et al., 2015). This characteristic of ONC201 allows it to specifically target HIV-1 infected macrophages and be further considered for clinical applications.
ONC201 has been well-known in cancer research for its capacity to induce sustained TRAIL upregulation and apoptosis in tumor cells both in vitro and in vivo (Allen et al., 2013; Allen et al., 2015). Our study for the first time demonstrates that ONC201 inhibits HIV-1 replication in macrophages both in vitro and in vivo. The evidence for viral inhibition after ONC201 treatment is multifold. First, the HIV-1 RTase activity assay is a relatively accurate measurement of HIV-1 virion production in the supernatants (Gorantla et al., 2005). Notably, ONC201 decreases HIV-1 RTase activities in MDM through FOXO3a and TRAIL but likely not through a direct inhibition of HIV-1 RTase activities since ONC201 is not able to suppress HIV-1 when directly incubated with HIV-1 virions (Supplemental Fig. 7). Second, two-step Alu-based nested PCR determines the level of HIV-1 integration by measuring HIV-1 LTR DNA as the proviral DNA integrated in the host genome (Brady et al., 2013). ONC201 is able to lower the levels of HIV-1 integration into the genome. Third, ONC201 also decreases intracellular/extracellular HIV-1 p24, GAG RNA, and GAG DNA in HIV-1-infected MDM. Despite its specific effect on HIV-1 replication, at the current working concentration (30 μM), ONC201 is not able to completely eradicate HIV-1 from the cultures. This could be due to the expression of TRAILshort in MDM that confers TRAIL resistance upon ONC201 treatment (Nie et al., 2018), or lack of optimal trimeric TRAIL conformation that is most potent for the antiviral effect (Mongkolsapaya et al., 1999). Thus, other ART drugs may be required along with ONC201 for optimal HIV-1 inhibition.
As a small molecule, ONC201 shows an outstanding capability of BBB penetration, which makes it particularly attractive for combating CNS viral infection. For brain tumors such as glioblastoma, ONC201 penetrates the intact BBB with five times higher concentrations in brain tissue relative to plasma in rodents (Allen et al., 2013). Therefore, after a successful dose escalation phase I trial (Stein et al., 2015), many ongoing clinical trials (clinicaltrials.gov) focus on brain tumors. In the present study, ONC201 is injected through IP into the NSG mice and its antiviral effect against the HIV-1 infected macrophages in the brains indicate that it is able to cross the BBB and achieve the therapeutic outcome in the case of CNS HIV-1 infection.
Our studies identify that the antiviral effect of ONC201 in HIV-1 infected macrophage is dependent on FOXO3a and TRAIL expressions. However, the exact molecular interactions between ONC201 and FOXO3a remain to be determined. Besides being a FOXO3a activator, ONC201 has also been proposed as a selective dopamine receptor 2 (DRD2) antagonist (Arrillaga-Romany et al., 2017; Kline et al., 2018; Stein et al., 2017). Previous studies have shown ONC201 inactivates both Akt-1 and ERK1/2 to activate FOXO3a and induce the transcription of TRAIL in cancer cell lines (Allen et al., 2013). We observed that the phosphorylation of FOXO3a is decreased after ONC201 treatment in the HIV-1 infected MDM, indicating ONC201 targets the same signaling pathway in cancer cells. However, ONC201 is also known to induce endoplasmic reticulum (ER) stress-related or integrated stress response (ISR)-related genes, such as ATF4, CHOP, GADD34, and TRIB3 (Endo Greer and Lipkowitz, 2016). Whether these genes work in tandem with FOXO3a/TRAIL pathway or not warrants future investigations.
The exact mechanism by which ONC201-activated TRAIL signaling reduces HIV-1 replication remains an active research question. One potential mechanism is that TRAIL-mediated apoptosis clears the HIV-1 infected cells while spares the normal cells. This is supported by previous reports demonstrating that leucine-zipper TRAIL, agonistic anti-TRAIL receptor antibodies, and activated NK cell-derived TRAIL induce apoptosis in MDM and peripheral blood lymphocytes and consequently reduce viral burden in HIV-infected individuals (Lum et al., 2001b; Lum et al., 2004; Shepard et al., 2008). Another potential mechanism of TRAIL is that it may exert its antiviral effect though apoptosis-independent mechanisms. TRAIL signaling may change the status of immune cells by interfering with the activation of NF-κB, PKB/Akt and MAPKs, which are relevant factors for HIV-1 replication (Falschlehner et al., 2007). Our current data support the apoptosis theory as a mechanism of action for ONC201. In HIV-1 infected macrophages with ONC201 post-infection treatment, ONC201-mediated TRAIL production inhibits viral replication. However, viral inhibition could also work to negate the effect of HIV-1-induced cell death as seen in Fig. 1A. In HIV-1 infected macrophages with ONC201 pre-infection treatment, ONC201 dramatically lowered HIV-1 replication levels, which is accompanied by lower cell viabilities and increased cleavage of caspase-3. The ONC201-mediated cytotoxicity in HIV-1 infected MDM raises the question of the safety and potential side effects of ONC201. However, in uninfected MDM and PBL, ONC201 treatment does not cause cytotoxicity, possibly due to the reportedly lower expression levels of death receptors on these cells compared to the infected cells (Lum et al., 2001a). Therefore, these data suggest that while cautions need to be considered, ONC201 is relatively safe to normal cells and will be useful in future antiviral therapies.
Immune-activating agents, such as interleukin 2 and anti-CD3 antibody, or for epigenetic modulation, such as Histone deacetylases inhibitors, often failed to purge the virus in vivo (Archin et al., 2012; Chun et al., 1999; Elliott et al., 2014; Prins et al., 1999; Rasmussen et al., 2014). Therefore, new therapeutic agents are warranted to combat persistent HIV-1 infection. Our study demonstrates for the first time that ONC201, a FOXO3a activator and TRAIL inducer, inhibits HIV-1 replication in macrophages both in vitro and in vivo. Notably, ONC201 is the founding member of the imipridone class of compounds that share a unique imidazo pharmacophore. New compounds continue to be developed in this class (Lev et al., 2017; Wagner et al., 2017). These new group of compounds provide us with more drug candidates to modulate immunity and fight viral infections.
Supplementary Material
Highlights.
ONC201 has little toxicity in uninfected human macrophages and exerts anti-HIV-1 effects on infected macrophages in vitro.
In a murine HIV Encephalitis model, ONC201 treatment significantly reduced HIV-1 levels in transplanted macrophages.
The antiviral effect of ONC201 is mediated by the FOXO3a/TRAIL pathway.
Acknowledgements
The following reagent was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: HIV-1 infected U937 Cells (U1) from Dr. Thomas Folks. We kindly thank Li Wu, Na Ly, and Myhanh Che for providing technical support; Justin Peer and Lenal Bottoms for proofreading the manuscript; Julie Ditter, Johna Belling, and Robin Taylor for providing outstanding administrative and secretarial support.
Abbreviations:
- ART
antiretroviral therapy
- CNS
central nervous system
- HAND
HIV-1 associated neurocognitive disorders
- BBB
blood brain barrier
- MDM
monocyte-derived macrophages
- RTase
reverse transcriptase
- ERK1/2
extracellular signal-regulated kinase 1/2
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This work was supported in part by research grants by the National Basic Research Program of China (973 Program Grant No. 2014CB965001 to JCZ), the State Key Program of the National Natural Science Foundation of China (#81830037 to JCZ), the Joint Research Fund for Overseas Chinese, Hong Kong and Macao Young Scientists of the National Natural Science Foundation of China (#81329002 to JCZ), the National Institutes of Health: R01 NS097195 (JCZ), R24 OD018546 (LP, SG), R03 NS094071 (YH), 2P30MH062261-Developmental (YH). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflicts of Interests/Financial Disclosures: NONE
References
- Allen JE, Kline CL, Prabhu VV, Wagner J, Ishizawa J, Madhukar N, Lev A, Baumeister M, Zhou L, Lulla A, Stogniew M, Schalop L, Benes C, Kaufman HL, Pottorf RS, Nallaganchu BR, Olson GL, Al-Mulla F, Duvic M, Wu GS, Dicker DT, Talekar MK, Lim B, Elemento O, Oster W, Bertino J, Flaherty K, Wang ML, Borthakur G, Andreeff M, Stein M, El-Deiry WS, 2016. Discovery and clinical introduction of first-in-class imipridone ONC201. Oncotarget 7, 74380–74392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JE, Krigsfeld G, Mayes PA, Patel L, Dicker DT, Patel AS, Dolloff NG, Messaris E, Scata KA, Wang W, Zhou JY, Wu GS, El-Deiry WS, 2013. Dual inactivation of Akt and ERK by TIC10 signals Foxo3a nuclear translocation, TRAIL gene induction, and potent antitumor effects. Sci Transl Med 5, 171ra117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JE, Krigsfeld G, Patel L, Mayes PA, Dicker DT, Wu GS, El-Deiry WS, 2015. Identification of TRAIL-inducing compounds highlights small molecule ONC201/TIC10 as a unique anti-cancer agent that activates the TRAIL pathway. Mol Cancer 14, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, Parker DC, Anderson EM, Kearney MF, Strain MC, Richman DD, Hudgens MG, Bosch RJ, Coffin JM, Eron JJ, Hazuda DJ, Margolis DM, 2012. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 487, 482–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrillaga-Romany I, Chi AS, Allen JE, Oster W, Wen PY, Batchelor TT, 2017. A phase 2 study of the first imipridone ONC201, a selective DRD2 antagonist for oncology, administered every three weeks in recurrent glioblastoma. Oncotarget 8, 79298–79304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady T, Kelly BJ, Male F, Roth S, Bailey A, Malani N, Gijsbers R, O'Doherty U, Bushman FD, 2013. Quantitation of HIV DNA integration: effects of differential integration site distributions on Alu-PCR assays. J Virol Methods 189, 53–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Park J, Tran H, Hu LS, Hemmings BA, Greenberg ME, 2001. Protein kinase SGK mediates survival signals by phosphorylating the forkhead transcription factor FKHRL1 (FOXO3a). Mol Cell Biol 21, 952–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camidge DR, Herbst RS, Gordon MS, Eckhardt SG, Kurzrock R, Durbin B, Ing J, Tohnya TM, Sager J, Ashkenazi A, Bray G, Mendelson D, 2010. A phase I safety and pharmacokinetic study of the death receptor 5 agonistic antibody PRO95780 in patients with advanced malignancies. Clin Cancer Res 16, 1256–1263. [DOI] [PubMed] [Google Scholar]
- Carpenter CC, Cooper DA, Fischl MA, Gatell JM, Gazzard BG, Hammer SM, Hirsch MS, Jacobsen DM, Katzenstein DA, Montaner JS, Richman DD, Saag MS, Schechter M, Schooley RT, Thompson MA, Vella S, Yeni PG, Volberding PA, 2000. Antiretroviral therapy in adults: updated recommendations of the International AIDS Society-USA Panel. Jama 283, 381–390. [DOI] [PubMed] [Google Scholar]
- Chen Q, Zhang M, Li Y, Xu D, Wang Y, Song A, Zhu B, Huang Y, Zheng JC, 2015. CXCR7 Mediates Neural Progenitor Cells Migration to CXCL12 Independent of CXCR4. Stem Cells 33, 2574–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel MR, Ghattas G, Brenchley JM, Schacker TW, Hill BJ, Douek DC, Routy JP, Haddad EK, Sekaly RP, 2009. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 15, 893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun TW, Davey RT Jr., Engel D, Lane HC, Fauci AS, 1999. Re-emergence of HIV after stopping therapy. Nature 401, 874–875. [DOI] [PubMed] [Google Scholar]
- Cocchi F, DeVico AL, Garzino-Demo A, Arya SK, Gallo RC, Lusso P, 1995. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science 270, 1811–1815. [DOI] [PubMed] [Google Scholar]
- Cocchi F, DeVico AL, Yarchoan R, Redfield R, Cleghorn F, Blattner WA, Garzino-Demo A, Colombini-Hatch S, Margolis D, Gallo RC, 2000. Higher macrophage inflammatory protein (MIP)-1alpha and MIP-1beta levels from CD8+ T cells are associated with asymptomatic HIV-1 infection. Proceedings of the National Academy of Sciences of the United States of America 97, 13812–13817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantino AA, Huang Y, Zhang H, Wood C, Zheng JC, 2011. HIV-1 Clade B and C Isolates Exhibit Differential Replication: Relevance to Macrophage-Mediated Neurotoxicity. Neurotoxicity research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M, Huang Y, Tian C, Zhao Y, Zheng J, 2011. FOXO3a inhibits TNF-α- and IL-1β-induced astrocyte proliferation:Implication for reactive astrogliosis. Glia 59, 641–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M, Huang Y, Zhao Y, Zheng J, 2008. Transcription factor FOXO3a mediates apoptosis in HIV-1-infected macrophages. J Immunol 180, 898–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks SG, Lewin SR, Bekker LG, 2017. The end of HIV: Still a very long way to go, but progress continues. PLoS Med 14, e1002466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degli-Esposti MA, Smolak PJ, Walczak H, Waugh J, Huang CP, DuBose RF, Goodwin RG, Smith CA, 1997. Cloning and characterization of TRAIL-R3, a novel member of the emerging TRAIL receptor family. J Exp Med 186, 1165–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorr J, Bechmann I, Waiczies S, Aktas O, Walczak H, Krammer PH, Nitsch R, Zipp F, 2002. Lack of tumor necrosis factor-related apoptosis-inducing ligand but presence of its receptors in the human brain. The Journal of neuroscience : the official journal of the Society for Neuroscience 22, RC209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott JH, Wightman F, Solomon A, Ghneim K, Ahlers J, Cameron MJ, Smith MZ, Spelman T, McMahon J, Velayudham P, Brown G, Roney J, Watson J, Prince MH, Hoy JF, Chomont N, Fromentin R, Procopio FA, Zeidan J, Palmer S, Odevall L, Johnstone RW, Martin BP, Sinclair E, Deeks SG, Hazuda DJ, Cameron PU, Sekaly RP, Lewin SR, 2014. Activation of HIV transcription with short-course vorinostat in HIV-infected patients on suppressive antiretroviral therapy. PLoS Pathog 10, e1004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery JG, McDonnell P, Burke MB, Deen KC, Lyn S, Silverman C, Dul E, Appelbaum ER, Eichman C, DiPrinzio R, Dodds RA, James IE, Rosenberg M, Lee JC, Young PR, 1998. Osteoprotegerin is a receptor for the cytotoxic ligand TRAIL. J Biol Chem 273, 14363–14367. [DOI] [PubMed] [Google Scholar]
- Endo Greer Y, Lipkowitz S, 2016. ONC201: Stressing tumors to death. Sci Signal 9, fs1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erichsen D, Lopez AL, Peng H, Niemann D, Williams C, Bauer M, Morgello S, Cotter RL, Ryan LA, Ghorpade A, Gendelman HE, Zheng J, 2003. Neuronal injury regulates fractalkine: relevance for HIV-1 associated dementia. J Neuroimmunol 138, 144–155. [DOI] [PubMed] [Google Scholar]
- Falschlehner C, Emmerich CH, Gerlach B, Walczak H, 2007. TRAIL signalling: decisions between life and death. Int J Biochem Cell Biol 39, 1462–1475. [DOI] [PubMed] [Google Scholar]
- Folks TM, Justement J, Kinter A, Dinarello CA, Fauci AS, 1987. Cytokine-induced expression of HIV-1 in a chronically infected promonocyte cell line. Science 238, 800–802. [DOI] [PubMed] [Google Scholar]
- Friedrich B, Li G, Dziuba N, Ferguson MR, 2010. Quantitative PCR used to assess HIV-1 integration and 2-LTR circle formation in human macrophages, peripheral blood lymphocytes and a CD4+ cell line. Virol J 7, 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendelman HE, Orenstein JM, Martin MA, Ferrua C, Mitra R, Phipps T, Wahl LA, Lane HC, Fauci AS, Burke DS, 1988. Efficient isolation and propagation of human immunodeficiency virus on recombinant colony-stimulating factor 1-treated monocytes. J Exp Med 167, 1428–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorantla S, Che M, Gendelman HE, 2005. Isolation, propagation, and HIV-1 infection of monocyte-derived macrophages and recovery of virus from brain and cerebrospinal fluid. Methods Mol Biol 304, 35–48. [DOI] [PubMed] [Google Scholar]
- Greco FA, Bonomi P, Crawford J, Kelly K, Oh Y, Halpern W, Lo L, Gallant G, Klein J, 2008. Phase 2 study of mapatumumab, a fully human agonistic monoclonal antibody which targets and activates the TRAIL receptor-1, in patients with advanced non-small cell lung cancer. Lung Cancer 61, 82–90. [DOI] [PubMed] [Google Scholar]
- Hall HI, Song R, Rhodes P, Prejean J, An Q, Lee LM, Karon J, Brookmeyer R, Kaplan EH, McKenna MT, Janssen RS, Group HIVIS, 2008. Estimation of HIV incidence in the United States. JAMA 300, 520–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Erdmann N, Peng H, Herek S, Davis JS, Luo X, Ikezu T, Zheng J, 2006. TRAIL-mediated apoptosis in HIV-1-infected macrophages is dependent on the inhibition of Akt-1 phosphorylation. J Immunol 177, 2304–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Walstrom A, Zhang L, Zhao Y, Cui M, Ye L, Zheng JC, 2009. Type I interferons and interferon regulatory factors regulate TNF-related apoptosis-inducing ligand (TRAIL) in HIV-1-infected macrophages. PloS one 4, e5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iordanskiy S, Waltke M, Feng Y, Wood C, 2010. Subtype-associated differences in HIV-1 reverse transcription affect the viral replication. Retrovirology 7, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaestner KH, Knochel W, Martinez DE, 2000. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev 14, 142–146. [PubMed] [Google Scholar]
- Kaufmann E, Knochel W, 1996. Five years on the wings of fork head. Mech Dev 57, 3–20. [DOI] [PubMed] [Google Scholar]
- Kline CL, Van den Heuvel AP, Allen JE, Prabhu VV, Dicker DT, El-Deiry WS, 2016. ONC201 kills solid tumor cells by triggering an integrated stress response dependent on ATF4 activation by specific eIF2alpha kinases. Sci Signal 9, ra18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline CLB, Ralff MD, Lulla AR, Wagner JM, Abbosh PH, Dicker DT, Allen JE, El-Deiry WS, 2018. Role of Dopamine Receptors in the Anticancer Activity of ONC201. Neoplasia 20, 80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs FC, Ross H, McAllister J, Wigdahl B, 2000. HIV-1-associated central nervous system dysfunction. Adv Pharmacol 49, 315–385. [DOI] [PubMed] [Google Scholar]
- Leong S, Cohen RB, Gustafson DL, Langer CJ, Camidge DR, Padavic K, Gore L, Smith M, Chow LQ, von Mehren M, O'Bryant C, Hariharan S, Diab S, Fox NL, Miceli R, Eckhardt SG, 2009. Mapatumumab, an antibody targeting TRAIL-R1, in combination with paclitaxel and carboplatin in patients with advanced solid malignancies: results of a phase I and pharmacokinetic study. J Clin Oncol 27, 4413–4421. [DOI] [PubMed] [Google Scholar]
- Lev A, Lulla AR, Wagner J, Ralff MD, Kiehl JB, Zhou Y, Benes CH, Prabhu VV, Oster W, Astsaturov I, Dicker DT, El-Deiry WS, 2017. Anti-pancreatic cancer activity of ONC212 involves the unfolded protein response (UPR) and is reduced by IGF1-R and GRP78/BIP. Oncotarget 8, 81776–81793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Hron JD, Peng SL, 2004. Regulation of NF-kappaB, Th activation, and autoinflammation by the forkhead transcription factor Foxo3a. Immunity 21, 203–213. [DOI] [PubMed] [Google Scholar]
- Lum JJ, Pilon AA, Sanchez-Dardon J, Phenix BN, Kim JE, Mihowich J, Jamison K, Hawley-Foss N, Lynch DH, Badley AD, 2001a. Induction of cell death in human immunodeficiency virus-infected macrophages and resting memory CD4 T cells by TRAIL/Apo2l. J Virol 75, 11128–11136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum JJ, Pilon AA, Sanchez-Dardon J, Phenix BN, Kim JE, Mihowich J, Jamison K, Hawley-Foss N, Lynch DH, Badley AD, 2001b. induction of cell death in human immunodeficiency virus-infected macrophages and resting memory CD4 T cells by TRAIL/Apo2L. Journal of virology 75, 11128–11136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum JJ, Schnepple DJ, Nie Z, Sanchez-Dardon J, Mbisa GL, Mihowich J, Hawley N, Narayan S, Kim JE, Lynch DH, Badley AD, 2004. Differential effects of interleukin-7 and interleukin-15 on NK cell anti-human immunodeficiency virus activity. J Virol 78, 6033–6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur JC, Sacktor N, Selnes O, 1999. Human immunodeficiency virus-associated dementia. Semin Neurol 19, 129–150. [DOI] [PubMed] [Google Scholar]
- Mom CH, Verweij J, Oldenhuis CN, Gietema JA, Fox NL, Miceli R, Eskens FA, Loos WJ, de Vries EG, Sleijfer S, 2009. Mapatumumab, a fully human agonistic monoclonal antibody that targets TRAIL-R1, in combination with gemcitabine and cisplatin: a phase I study. Clin Cancer Res 15, 5584–5590. [DOI] [PubMed] [Google Scholar]
- Mongkolsapaya J, Grimes JM, Chen N, Xu XN, Stuart DI, Jones EY, Screaton GR, 1999. Structure of the TRAIL-DR5 complex reveals mechanisms conferring specificity in apoptotic initiation. Nat Struct Biol 6, 1048–1053. [DOI] [PubMed] [Google Scholar]
- Nie Z, Aboulnasr F, Natesampillai S, Burke SP, Krogman A, Bren GD, Chung TDY, Anderson JR, Smart MK, Katzmann DJ, Rajagopalan G, Cummins NW, Badley AD, 2018. Both HIV-Infected and Uninfected Cells Express TRAILshort, Which Confers TRAIL Resistance upon Bystander Cells within the Microenvironment. J Immunol 200, 1110–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G, Ni J, Yu G-L, Wei Y-F, Dixit VM, 1998. TRUNDD a new member of the TRAIL receptor family that antagonizes TRAIL signalling. FEBS Letters 424, 41–45. [DOI] [PubMed] [Google Scholar]
- Pan G, O'Rourke K, Chinnaiyan AM, Gentz R, Ebner R, Ni J, Dixit VM, 1997. The receptor for the cytotoxic ligand TRAIL. Science 276, 111–113. [DOI] [PubMed] [Google Scholar]
- Peng H, Huang Y, Duan Z, Erdmann N, Xu D, Herek S, Zheng J, 2005. Cellular IAP1 regulates TRAIL-induced apoptosis in human fetal cortical neural progenitor cells. J Neurosci Res 82, 295–305. [DOI] [PubMed] [Google Scholar]
- Pierson T, McArthur J, Siliciano RF, 2000. Reservoirs for HIV-1: mechanisms for viral persistence in the presence of antiviral immune responses and antiretroviral therapy. Annu Rev Immunol 18, 665–708. [DOI] [PubMed] [Google Scholar]
- Pitti RM, Marsters SA, Ruppert S, Donahue CJ, Moore A, Ashkenazi A, 1996. Induction of apoptosis by Apo-2 ligand, a new member of the tumor necrosis factor cytokine family. J Biol Chem 271, 12687–12690. [DOI] [PubMed] [Google Scholar]
- Plummer R, Attard G, Pacey S, Li L, Razak A, Perrett R, Barrett M, Judson I, Kaye S, Fox NL, Halpern W, Corey A, Calvert H, de Bono J, 2007. Phase 1 and pharmacokinetic study of lexatumumab in patients with advanced cancers. Clin Cancer Res 13, 6187–6194. [DOI] [PubMed] [Google Scholar]
- Prins JM, Jurriaans S, van Praag RM, Blaak H, van Rij R, Schellekens PT, ten Berge IJ, Yong SL, Fox CH, Roos MT, de Wolf F, Goudsmit J, Schuitemaker H, Lange JM, 1999. Immuno-activation with anti-CD3 and recombinant human IL-2 in HIV-1-infected patients on potent antiretroviral therapy. AIDS 13, 2405–2410. [DOI] [PubMed] [Google Scholar]
- Rasmussen TA, Tolstrup M, Brinkmann CR, Olesen R, Erikstrup C, Solomon A, Winckelmann A, Palmer S, Dinarello C, Buzon M, Lichterfeld M, Lewin SR, Ostergaard L, Sogaard OS, 2014. Panobinostat, a histone deacetylase inhibitor, for latent-virus reactivation in HIV-infected patients on suppressive antiretroviral therapy: a phase 1/2, single group, clinical trial. Lancet HIV 1, e13–21. [DOI] [PubMed] [Google Scholar]
- Ryan LA, Peng H, Erichsen DA, Huang Y, Persidsky Y, Zhou Y, Gendelman HE, Zheng J, 2004. TNF-Related Apoptosis-Inducing Ligand Mediates Human Neuronal Apoptosis: Links to HIV-1 Associated Dementia. J. Neuroimmunol 148, 127–139. [DOI] [PubMed] [Google Scholar]
- Seol DW, Li J, Seol MH, Park SY, Talanian RV, Billiar TR, 2001. Signaling events triggered by tumor necrosis factor-related apoptosis- inducing ligand (TRAIL): caspase-8 is required for TRAIL-induced apoptosis. Cancer Res 61, 1138–1143. [PubMed] [Google Scholar]
- Shepard BD, De Forni D, McNamara DR, Foli A, Rizza SA, Abraham RS, Knutson K, Wettstein PJ, Lori F, Badley AD, 2008. Beneficial effect of TRAIL on HIV burden, without detectable immune consequences. PLoS One 3, e3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein MN, Bertino JR, Kaufman HL, Mayer T, Moss R, Silk A, Chan N, Malhotra J, Rodriguez L, Aisner J, Aiken RD, Haffty BG, DiPaola RS, Saunders T, Zloza A, Damare S, Beckett Y, Yu B, Najmi S, Gabel C, Dickerson S, Zheng L, El-Deiry WS, Allen JE, Stogniew M, Oster W, Mehnert JM, 2017. First-in-Human Clinical Trial of Oral ONC201 in Patients with Refractory Solid Tumors. Clin Cancer Res 23, 4163–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein MN, Mayer TM, Moss RA, Silk AW, Chan N, Haffty BG, DiPaola RS, Beckett Y, Bentlyewski E, Zheng L, Fang B, Allen J, Mehnert JM, 2015. First-in-human dose escalation study of oral ONC201 in advanced solid tumors. Journal of Clinical Oncology 33, TPS2623–TPS2623. [Google Scholar]
- Tian C, Li Y, Huang Y, Wang Y, Chen D, Liu J, Deng X, Sun L, Anderson K, Qi X, Li Y, Mosley RL, Chen X, Huang J, Zheng JC, 2015. Selective Generation of Dopaminergic Precursors from Mouse Fibroblasts by Direct Lineage Conversion. Sci Rep 5, 12622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolcher AW, Mita M, Meropol NJ, von Mehren M, Patnaik A, Padavic K, Hill M, Mays T, McCoy T, Fox NL, Halpern W, Corey A, Cohen RB, 2007. Phase I pharmacokinetic and biologic correlative study of mapatumumab, a fully human monoclonal antibody with agonist activity to tumor necrosis factor-related apoptosis-inducing ligand receptor-1. J Clin Oncol 25, 1390–1395. [DOI] [PubMed] [Google Scholar]
- Trarbach T, Moehler M, Heinemann V, Kohne CH, Przyborek M, Schulz C, Sneller V, Gallant G, Kanzler S, 2010. Phase II trial of mapatumumab, a fully human agonistic monoclonal antibody that targets and activates the tumour necrosis factor apoptosis-inducing ligand receptor-1 (TRAIL-R1), in patients with refractory colorectal cancer. Br J Cancer 102, 506–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Grevenynghe J, Procopio FA, He Z, Chomont N, Riou C, Zhang Y, Gimmig S, Boucher G, Wilkinson P, Shi Y, Yassine-Diab B, Said EA, Trautmann L, El Far M, Balderas RS, Boulassel MR, Routy JP, Haddad EK, Sekaly RP, 2008. Transcription factor FOXO3a controls the persistence of memory CD4(+) T cells during HIV infection. Nat Med 14, 266–274. [DOI] [PubMed] [Google Scholar]
- Wagner J, Kline CL, Ralff MD, Lev A, Lulla A, Zhou L, Olson GL, Nallaganchu BR, Benes CH, Allen JE, Prabhu VV, Stogniew M, Oster W, El-Deiry WS, 2017. Preclinical evaluation of the imipridone family, analogs of clinical stage anti-cancer small molecule ONC201, reveals potent anti-cancer effects of ONC212. Cell Cycle 16, 1790–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak H, Degli-Esposti MA, Johnson RS, Smolak PJ, Waugh JY, Boiani N, Timour MS, Gerhart MJ, Schooley KA, Smith CA, Goodwin RG, Rauch CT, 1997. TRAIL-R2: a novel apoptosis-mediating receptor for TRAIL. Embo J 16, 5386–5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li Y, Zhao R, Wu B, Lanoha B, Tong Z, Peer J, Liu J, Xiong H, Huang Y, Zheng J, 2017. Glutaminase C overexpression in the brain induces learning deficits, synaptic dysfunctions, and neuroinflammation in mice. Brain Behav Immun 66, 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley SR, Schooley K, Smolak PJ, Din WS, Huang CP, Nicholl JK, Sutherland GR, Smith TD, Rauch C, Smith CA, et al. , 1995. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity 3, 673–682. [DOI] [PubMed] [Google Scholar]
- Wu Y, Peng H, Cui M, Whitney NP, Huang Y, Zheng JC, 2009. CXCL12 increases human neural progenitor cell proliferation through Akt-1/FOXO3a signaling pathway. Journal of neurochemistry 109, 1157–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Huang Y, Zhao L, Li Y, Sun L, Zhou Y, Qian G, Zheng JC, 2013. IL-1beta and TNF-alpha induce neurotoxicity through glutamate production: a potential role for neuronal glutaminase. Journal of neurochemistry 125, 897–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Xu D, Deng X, Chen Q, Huang Y, Peng H, Li Y, Jia B, Thoreson WB, Ding W, Ding J, Zhao L, Wang Y, Wavrin KL, Duan S, Zheng J, 2012. CXCL12 enhances human neural progenitor cell survival through a CXCR7- and CXCR4-mediated endocytotic signaling pathway. Stem Cells 30, 2571–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.