Alterations in metabolite levels provide fast and direct means to regulate enzymatic reactions and, therefore, metabolic pathways. This study documents, for the first time, the metabolic changes that occur in Mycobacterium smegmatis as a response to three stresses, namely, acidic stress, oxidative stress, and nutrient starvation. These stresses are also faced by intracellular mycobacteria during infection and therefore may be extended to frame therapeutic interventions for pathogenic mycobacteria. In addition to the purine-pyrimidine, amino acid, nicotinate-nicotinamide, and energy metabolism pathways that were found to be affected in response to different stresses, a novel putative methylamine biosynthesis pathway was identified to be present in Mycobacterium smegmatis.
KEYWORDS: mycobacteria, metabolomics, methylated amines, osmolyte, stress response, trimethylamine dehydrogenase
ABSTRACT
Mycobacterium smegmatis, the saprophytic soil mycobacterium, is routinely used as a surrogate system to study the human pathogen Mycobacterium tuberculosis. It has also been reported as an opportunistic pathogen in immunocompromised hosts. In addition, it can exist in several ecological setups, thereby suggesting its capacity to adapt to a variety of environmental cues. In this study, we employed untargeted proton nuclear magnetic resonance (1H-NMR)-based metabolomics to identify metabolites and metabolic pathways critical for early adaptive responses to acidic stress, oxidative stress, and nutrient starvation in Mycobacterium smegmatis. We identified 31, 20, and 46 metabolites that showed significant changes in levels in response to acidic, oxidative, and nutrient starvation stresses, respectively. Pathway analyses showed significant perturbations in purine-pyrimidine, amino-acid, nicotinate-nicotinamide, and energy metabolism pathways. Besides these, differential levels of intermediary metabolites involved in α-glucan biosynthesis pathway were observed. We also detected high levels of organic osmolytes, methylamine, and betaine during nutrient starvation and oxidative stress. Further, tracing the differential levels of these osmolytes through computational search tools, gene expression studies (using reverse transcription-PCR [RT-PCR]), and enzyme assays, we detected the presence of a putative pathway of biosynthesis of betaine, methylamine, and dimethylamine previously unreported in Mycobacterium smegmatis.
IMPORTANCE Alterations in metabolite levels provide fast and direct means to regulate enzymatic reactions and, therefore, metabolic pathways. This study documents, for the first time, the metabolic changes that occur in Mycobacterium smegmatis as a response to three stresses, namely, acidic stress, oxidative stress, and nutrient starvation. These stresses are also faced by intracellular mycobacteria during infection and therefore may be extended to frame therapeutic interventions for pathogenic mycobacteria. In addition to the purine-pyrimidine, amino acid, nicotinate-nicotinamide, and energy metabolism pathways that were found to be affected in response to different stresses, a novel putative methylamine biosynthesis pathway was identified to be present in Mycobacterium smegmatis.
INTRODUCTION
The genus Mycobacterium, though more famous for dreaded human pathogens like Mycobacterium tuberculosis, includes almost 200 species of nonpathogenic mycobacteria which form an important constituent of various habitats, including endangered coastal swamps and peat-rich wetlands (1, 2). The hydrophobic cell surface permits the accumulation and persistence of mycobacteria under various environmental conditions and interfaces, including aerosolized droplets (3–7).
Microbes, pathogenic or nonpathogenic, are always under stress due to continually changing habitats (8, 9). Mycobacterium smegmatis, a nonpathogenic saprophyte, has been reported to reside in habitats including soil, water (including drinking water), and air from peat moss processing plants (3, 10). These varied environments pose a spectrum of challenges and growth-limiting conditions, like nutrient deprivation, low pH, and low oxygen.
Despite residing in a completely different habitat, M. smegmatis is used as a model system to study the properties of the human pathogen M. tuberculosis (11, 12). This is primarily due to its avirulent nature and higher growth rate than that of M. tuberculosis, making it easier to handle in laboratory setups. Additionally, it shares several properties with pathogenic mycobacteria, including (i) a thick mycolic acid-rich cell wall, (ii) inducible pH homeostasis, (iii) response to anaerobiosis, and (iv) substrate deprivation (12–15). The presence of antioxidant enzymes, viz., superdioxide dismutase, catalase, and peroxidase in M. smegmatis, which can detoxify reactive oxygen species (ROS) (16), suggests that even in its natural habitat it is prepared to face the challenges of ROS. Lately, M. smegmatis has also been reported to cause opportunistic infections in immunocompromised hosts (17).
Most studies performed to date have attempted to study the adaptation by mycobacteria to different stresses for survival using genomics-, transcriptomics-, and proteomics-based approaches (11, 18–25). Metabolomics, defined as the study of global metabolites in a given biological system at a given time in contrast to the other omics studies, provides insights into the final product of gene transcription. The changes in the metabolome are relatively amplified with respect to the changes in the transcriptome and proteome and thus serve as immediate markers for an early response to any change in an environment (26). Previous studies have attempted to identify distinct metabolic signatures associated with different growth stages and drug responses in M. smegmatis. For instance, distinct metabolic profiles have been reported among various mycobacterial species in different growth stages (27), during stresses such as hypoxia (28), and in M. smegmatis treated with antimycobacterial agents, namely, rifampin, capreomycin (29), and pretomanid (30). These studies identified distinctive metabolic signatures toward growth stages and drug responses in M. smegmatis.
With this background, we hypothesized that a metabolomics approach would help gain new insights into similar or unique responses at metabolic levels in M. smegmatis to different stresses that have not been reported earlier. For this, we employed untargeted proton nuclear magnetic resonance (1H-NMR) spectroscopy-based metabolomics methods to study the responses of M. smegmatis to imbalances in metabolism due to three abiotic stresses, namely, acidic stress given by reducing the pH of the medium, oxidative stress given by the addition of hydrogen peroxide, and nutrient starvation by growing mycobacteria in phosphate-buffered saline (PBS). These three stresses are faced by M. smegmatis both as a soil mycobacterium and as an opportunistic infection inside human phagocytic cells. The study reports, for the first time, a significant increase in the levels of organic osmolytes, methylamine, and betaine as an early adaptive stress response to nutrient starvation and oxidative stress. These observations were then used to trace putative pathways of biosynthesis of these secondary metabolites in M. smegmatis through computational search tools, gene expression studies (using reverse transcription-PCR [RT-PCR]), and enzyme assays.
RESULTS AND DISCUSSION
Differential metabolites associated with stresses.
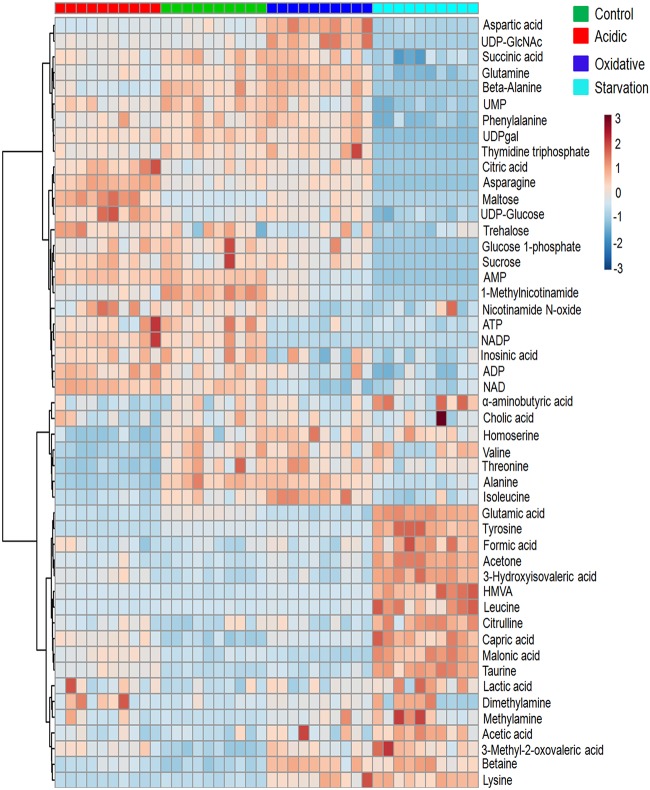
Fifty-six different metabolites, including the standard DSS (4,4-dimethyl-4-silapentane-1-sulfonic acid), were identified and marked on the 1H-NMR spectrum (see Fig. S1a and Table S1 in the supplemental material). Univariate analysis with a minimum cutoff of 1.2-fold change, P value of <0.05, and a false-discovery rate of <0.05 showed 31, 20, and 46 metabolites with significant differential levels in acidic stress, oxidative stress, and nutrient starvation, respectively, compared to the control. Refer to Table S2a to d for comprehensive lists of differential metabolites in response to these stresses and to Materials and Methods for details on the cutoffs applied for analysis of the data. A graphical representation of individual metabolite levels in different stresses is provided as a heat map (Fig. 1), showing the relative concentration of metabolites under the different conditions and across different experiment groups. A noticeable observation was high levels of the osmolytes methylamine, dimethylamine, and betaine in oxidative stress and nutrient starvation compared to the control. Methylamine levels were highest in nutrient starvation (3.45-fold change), while oxidative stress showed 2.85-fold higher methylamine and 2.36-fold higher betaine levels than in the control. Dimethylamine levels increased moderately during all stresses by about 1.39-fold (Table S2).
FIG 1.
Heat map representation of the metabolic changes in M. smegmatis upon exposure to acidic stress, oxidative stress, and nutrient starvation compared to the control. Each row represents a single metabolite detected in the study. Color differences demonstrate the relative concentrations of metabolites across the different conditions and experiment groups. Individual samples (horizontal axis) and metabolites (vertical axis) are separated using hierarchical clustering (Ward’s algorithm), with the dendrogram scaled to represent the distance between each branch (Pearson’s correlation). From each group, 10 biological samples were analyzed. Dark-orange color in a the tile indicates high levels, and dark blue indicates low levels. HMVA, 2-hydroxy-3-methyl valeric acid.
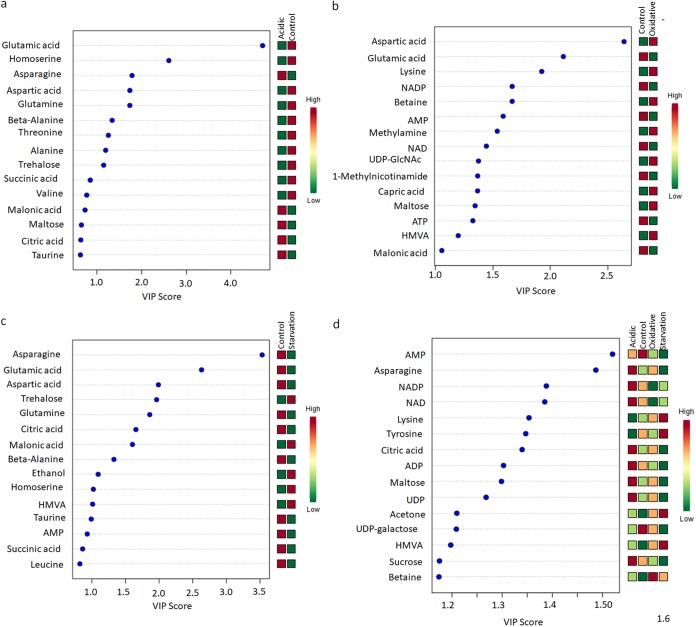
Further, principal-component analysis (PCA) and partial least-squares-discriminate analysis (PLS-DA) were performed to examine the intrinsic variations in the groups and identify a distinct metabolic signature characteristic of the specific stress response (refer to the text and Fig. S2 and S3 for details). Variable importance in projection (VIP) scores obtained from the PLS-DA models were used to identify key metabolites that segregated each stress response from the control. Figure 2 shows the metabolites with the top 15 VIP scores under each stress condition compared to the control. We observed, when all the stresses were compared collectively with the control, that the critical distinguishing metabolites belonged to energy metabolism and amino acid metabolism, while also citing betaine (Fig. 2d). Methylamine appeared as a distinguishable metabolite for oxidative stress but not for nutrient starvation. We also observed that critical metabolites associated with α-glucan biosynthesis (31, 32), namely, trehalose and maltose, were at distinguishable levels, with high VIP scores, during different stresses (Fig. 2).
FIG 2.
Variable importance in projection (VIP) plots with the top 15 discriminating metabolites identified through PLS-DA analyses in descending order of importance. (a) Acidic stress versus the control. (b) Oxidative stress versus the control. (c) Nutrient starvation versus the control. (d) Acidic, oxidative, and nutrient starvation versus the control. The colored boxes on the right indicate the relative concentrations given on a scale of high (red) to low (green) for the corresponding metabolites in each group.
Metabolic pathways perturbed during stresses.
Having determined the differential levels of metabolites during stresses, the data were subjected to metabolic pathway impact analysis to identify the relevant pathways that were perturbed (33). Information related to metabolic pathways with their impact values and corresponding negative natural logs of the P value [−log(P)] is given in Table S4. Table 1 shows the top seven pathways perturbed during stresses. It was interesting to note that although common pathways were perturbed across different stresses, the significance scores [−log(P)] in their pairwise comparisons were found to be different, suggesting differential importance of one pathway over the other in different stresses. In the pairwise comparison of acidic stress versus control, the most significant pathways perturbed based on highest −log(P) were glycine-serine-threonine metabolism and glutamine-glutamate metabolism. In the oxidative stress, nicotinate-nicotinamide metabolism and glycine-serine-threonine metabolism were the principal pathways altered, and, under the nutrient starvation condition, the major pathways perturbed were alanine-aspartate-glutamate metabolism and beta-alanine metabolism. The analyses did not reflect pathways associated with methylated amines despite altered levels of these metabolites during stresses. We presumed that this might have been due to the absence of an annotated pathway of methylated amines in M. smegmatis.
TABLE 1.
Metabolic pathway impact analysis, with the top seven metabolic pathways significantly influenced under acidic stress, oxidative stress, and nutrient starvation compared to the control
| Pathway no. | Pathway impact analysis |
|||||
|---|---|---|---|---|---|---|
| Function by stress type | Total no. of metabolites in pathway | No. of matched metabolites (hits) | −log(P)a | FDR (Benjamini-Hochberg) | Impactb | |
| Acidic stress | ||||||
| 1 | Glycine, serine, and threonine metabolism | 28 | 4 | 20.31 | 1.09E−08 | 0.21 |
| 2 | d-Glutamine and d-glutamate metabolism | 7 | 2 | 18.68 | 3.70E−08 | 0.38 |
| 3 | β-Alanine metabolism | 9 | 2 | 15.17 | 7.28E−07 | 1 |
| 4 | Alanine, aspartate, and glutamate metabolism | 20 | 5 | 15.12 | 7.28E−07 | 0.69 |
| 5 | Pyrimidine metabolism | 37 | 2 | 13.04 | 4.23E−06 | 0.11 |
| 6 | Purine metabolism | 66 | 5 | 12.75 | 5.41E−06 | 0.14 |
| 7 | Nicotinate and nicotinamide metabolism | 13 | 3 | 9.89 | 8.09E−05 | 0.15 |
| Oxidative stress | ||||||
| 1 | Nicotinate and nicotinamide metabolism | 13 | 3 | 21.63 | 8.69E−09 | 0.15 |
| 2 | Glycine, serine, and threonine metabolism | 28 | 4 | 19.08 | 4.63E−08 | 0.21 |
| 3 | d-Glutamine and d-glutamate metabolism | 7 | 2 | 18.39 | 7.41E−08 | 0.38 |
| 4 | Amino sugar and nucleotide sugar metabolism | 38 | 4 | 18.13 | 8.22E−08 | 0.21 |
| 5 | Purine metabolism | 66 | 5 | 16.23 | 3.50E−07 | 0.14 |
| 6 | Alanine, aspartate, and glutamate metabolism | 20 | 5 | 16.23 | 3.50E−07 | 0.69 |
| 7 | β-Alanine metabolism | 9 | 2 | 15.64 | 5.06E−07 | 1 |
| Nutrient starvation stress | ||||||
| 1 | Alanine, aspartate, and glutamate metabolism | 20 | 5 | 34.86 | 7.84E−15 | 0.69 |
| 2 | β-Alanine metabolism | 9 | 2 | 26.77 | 1.13E−11 | 1 |
| 3 | Nicotinate and nicotinamide metabolism | 13 | 3 | 21.8 | 1.22E−09 | 0.15 |
| 4 | Purine metabolism | 66 | 5 | 21.23 | 1.99E−09 | 0.14 |
| 5 | Pyrimidine metabolism | 37 | 2 | 19.33 | 1.02E−08 | 0.11 |
| 6 | Amino sugar and nucleotide sugar metabolism | 38 | 4 | 19.11 | 1.19E−08 | 0.21 |
| 7 | Glycine, serine, and threonine metabolism | 28 | 4 | 18.28 | 2.37E−08 | 0.21 |
−log(P), negative natural log of the P value for each pathway.
Impact, pathway impact value calculated from the pathway topology analysis. Pathways with pathway impact values of ≥0.1, P values of <0.05, and FDRs of <0.05 were considered to be perturbed significantly.
Putative pathway of biosynthesis of methylated amines in M. smegmatis.
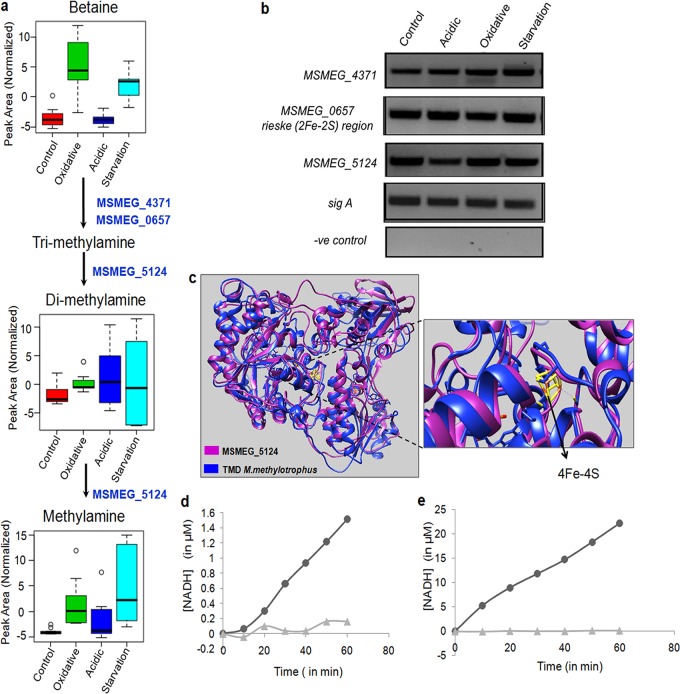
As mentioned above, the metabolomics data showed high intracellular levels of the osmolytes betaine, methylamine, and dimethylamine when M. smegmatis was subjected to stress (Fig. 3a). Besides protecting from ionic imbalances, these osmolytes are also known to play a role in adaptation to various stresses (34–38). While many bacteria, including those of the human microbiota, can produce these methylated amines from carnitine (39), the biosynthesis pathway of methylamines is not reported in M. smegmatis. With no direct supplements of tri-, di-, or methylamine, we asked the question of how these metabolites were produced in M. smegmatis, if the orthologous genes of methylamine biosynthesis pathways were present, and if those genes were expressed under stress conditions. In methylotrophic bacteria, dimethylamine can be produced from trimethylamine by trimethylamine dehydrogenase (TMD) and methylamine from dimethylamine using dimethylamine dehydrogenase (DMD). In certain bacteria, both of these steps are mediated by a single dehydrogenase, which shares similar physical, chemical, spectral, and kinetic properties (40). An open reading frame (ORF) or a gene by this name could not be located within the annotated M. smegmatis genome (https://mycobrowser.epfl.ch/).
FIG 3.
A putative pathway of biosynthesis of methylated amines in M. smegmatis. (a) Putative methylamine pathway showing the identified intermediate metabolites under different stress conditions. The numbers indicate the orthologues of M. smegmatis genes identified in this study that possibly catalyze these reactions similar to those in methylotrophic bacteria (refer to the text for details). (b) RT-PCR of orthologue ORFs of M. smegmatis indicated in panel a under different stress conditions; a negative (−ve) control containing RNA instead of cDNA was used to rule out genomic DNA contamination. (c) Homology modeling of MSMEG_5124 (magenta) using TMD of bacterium W3A1 (blue) with overlapping catalytic sites, as illustrated. (d) The concentration of NADH versus time plot for the reaction with trimethylamine (TMA) as the substrate. The rate curve of the control reaction (MSMEG_5124 plus 2 mM NAD+ only; ▲) is shown along with the rate curve of the reaction (MSMEG_5124 plus 20mM TMA plus 2 mM NAD+; •). (e) A concentration of NADH versus time plot for reaction with dimethylamine (DMA) as the substrate. The rate curve of the control reaction (MSMEG_5124 plus 2mM NAD+ only; ▲) is shown along with the rate curve of the reaction (MSMEG_5124 plus 20mM DMA plus 2 mM NAD+; •).
A protein sequence similarity search using PSI-BLAST was then performed using the protein sequence of TMD from the methylotroph Methylophilus methylotrophus (bacterium W3A1; UniProt accession no. P16099) as the query sequence and the mc2155 genome as the subject. A sequence identity of 26% with a query coverage of 72% were observed with MSMEG_5124 of M. smegmatis. MSMEG_5124 is annotated as 2,4-dienoyl coenzyme A (dienoyl-CoA) reductase (DCR), identified by protein family similarity (https://mycobrowser.epfl.ch/genes/MSMEG_5124).
A literature survey showed that Hubbard et al. had earlier pointed out certain similarities between TMD of Methylophilus methylotrophus and DCR of Escherichia coli (41). These authors solved the crystal structure of Escherichia coli DCR and observed that the domain arrangement and overall polypeptide fold resembled those of TMD. With these clues, MSMEG_5124 was then aligned with E. coli DCR (UniProt accession no. P42593), which showed 100% query coverage and an identity of 54%, suggesting that there is a possibility that although annotated as DCR, MSMEG_5124 also functions as TMD in M. smegmatis. Homology modeling of MSMEG_5124 using TMD of Methylophilus methylotrophus, bacterium W3A1 (Mm-TMD), showed overlapping catalytic sites, as illustrated diagrammatically in Fig. 3c. Cysteine residues for 4Fe-4S in Mm-TMD (CYS315, CYS351, and CYS364) and MSMEG_5124 (CYS335, CYS342, and CYS354) were observed to be conserved. With this supporting information, we checked if this probable ORF was expressed in M. smegmatis under both control and stress conditions, for which RT-PCR of MSMEG_5124 was performed using specific primers (Fig. 3b). The results showed that MSMEG_5124 is expressed in M. smegmatis under all conditions.
With the confirmation of the expression of MSMEG_5124, a putative TMD in M. smegmatis, and evidence of intermediate metabolites, dimethylamine, and methylamine from the metabolomics data, we further cloned and purified MSMEG_5124 to check its possible enzymatic activity. Purified MSMEG_5124 (Fig. S4b) was used for the enzymatic assay, as described in Materials and Methods. The rate plots clearly show that MSMEG_5124 could function as TMD (Fig. 3d) but show higher activity with dimethylamine as the substrate (Fig. 3e), indicating that MSMEG_5124 can utilize both trimethylamine and dimethylamine as substrates, as reported in certain bacteria (40). While it is still required to check if MSMEG_5124 functions as DCR as annotated, these experiments point toward its possible TMD and DMD activities. These observations suggested the presence of a putative pathway of conversion of trimethylamine to dimethylamine and methylamine.
Next, we wanted to know the possible intracellular source of trimethylamine (TMA). Metabolomics data showed that betaine is accumulated during oxidative stress and nutrient starvation. The gene pair carnitine monooxygenase reductase subunit (YeaX) as a complex with an oxygenase component (Rieske [2Fe-2S] [YeaW] region) of E. coli K-12 can use different substrates, namely, carnitine, γ-butyrobetaine, choline, and betaine, to produce trimethylamine, a system which is also reported in many bacteria, including those associated with gut microbiota (42). We next looked for these genes in the genome of M. smegmatis mc2155. With E. coli genes (UniProt accession numbers P76254 and P0ABR7) as references, scanning of mc2155 using PSI-BLAST showed two orthologs, MSMEG_4371 and MSMEG_0657, with similarities to YeaX (38% identity) and YeaW (37% identity), respectively. MSMEG_4371 and MSMEG_0657 are hypothetical ORFs with no experimental evidence of their expression in M. smegmatis. The expression of MSMEG_4371 and MSMEG_0657 in M. smegmatis under both control and stress conditions was checked using RT-PCR (Fig. 3b). Once again, these were observed to be expressed under all conditions. It was noted that the transcript levels of these genes were similar under all stress conditions, although the intermediate metabolites of the pathway were differential in their levels (Fig. 3b). These experiments suggested a putative pathway that could convert betaine into TMA and TMA into methylamines in M. smegmatis. One may therefore speculate that betaine, which accumulated during oxidative stress and nutrient starvation in our metabolomics data, was the possible intracellular source for the synthesis of methylated amines. With the expression of all these hypothetical ORFs from the M. smegmatis genome, confirmation of TMD/DMD activity of MSMEG_5124, and the presence of intermediate metabolites that are catalyzed by these ORFs in our metabolomics data, we propose the existence of a putative pathway of biosynthesis of methylamines in M. smegmatis (Fig. 3a).
Methylamines are reported to be utilized to synthesize formaldehyde and ammonia, which may serve as an advantage to pathogenic mycobacteria, especially during acidic stress (43). Several in vitro biochemical studies have shown that methylamines can provide stability to proteins, a property by which they may function as osmolytes, providing protection from osmotic stress (44). One may expect that a similar feature of methylamine may provide an adaptive advantage during oxidative stress and nutrient starvation. With these observations, one may hypothesize that M. smegmatis lacking gene MSMEG_5124 would be compromised in surviving nutrient starvation and oxidative stress. The importance of this pathway and the ORFs in stress adaptation can be deciphered through knockout/knockdown studies. It would also be interesting to study the homologs of these genes in Mycobacterium tuberculosis, where if found to be essential for survival in oxidative stress and nutrient starvation, they may serve as new drug targets. Our hypothesis is strengthened by recent reports where betaine and trimethylamine N-oxide, which arises from trimethylamine, have been shown to accumulate in tuberculosis granulomas in guinea pigs (45). Betaine also contributes to early steps in the colonization of pathogenic mycobacteria in the host (46).
With recent reports on possible methylotrophy in mycobacteria (47–49), it will be interesting to explore if the mechanisms of protection given by methylamine are similar or different during different stresses. Further studies involving knockout/knockdown of MSMEG_5124 and detailed enzymatic characterization of MSMEG_5124 will shed light on the significance of this ORF in mycobacterial physiology.
Conclusions.
Overall, this study provides the first comprehensive list of differential levels of metabolites in response to three abiotic stresses in M. smegmatis. The comparative metabolic analysis demonstrated that these stresses induce a considerable metabolic shift in M. smegmatis. The metabolic profile shift was most distinct for nutrient starvation compared to acidic stress and oxidative stress. The paper also lists stress-specific metabolites distinguishing the stress response from control, using VIP scores. Pathway analysis using the metabolomics data imparted various degrees of significance to the perturbed pathways. While it was expected that the bacteria would only rewire its energy metabolism in response to stresses, several distinct differences in responses to these three stresses were also noticed. It was noted that while glycine-serine-threonine metabolism was perturbed significantly in both acidic stress and oxidative stress, perturbation in the nicotinate-nicotinamide pathway was more significant in oxidative stress, and alanine-aspartate-glutamate metabolism and beta-alanine metabolism were more significant for the nutrient starvation response. These data point to the similarities and uniqueness in metabolic adaptations by M. smegmatis to these stresses.
Though limited by the number of metabolites identified through 1H-NMR, the strength of the study is the number of replicates used per experimental condition that permitted thorough and meaningful statistical analyses. The baseline information generated through these metabolomics data could be used to trace the presence of an unreported putative methylamine biosynthesis pathway in M. smegmatis, opening new research questions in the field and encouraging further metabolomics studies to reveal additional vital aspects of mycobacterial metabolism not accessible by existing genomic or proteomic approaches. The orthologs of these new pathways can be traced in pathogenic mycobacteria, and their significance may be studied to understand mycobacterial strategies for host invasion.
MATERIALS AND METHODS
Bacterial strains and experimental conditions.
Mycobacterium smegmatis mc2155 (M. smegmatis) was used in the present study. The growth of mycobacteria was achieved as described earlier, with a few modifications (24). M. smegmatis was grown at 37°C with shaking at 180 rpm in 7H9 medium with 0.4% (vol/vol) glycerol and 0.05% (vol/vol) tyloxapol, and supplemented with 10% (vol/vol) oleic acid-albumin-dextrose-catalase (OADC) until its optical density at 600 nm (OD600) reached 0.5 to 0.7. The cultures were harvested, washed with 1× phosphate-buffered saline (PBS), and resuspended in medium mimicking abiotic stress conditions. M. smegmatis was subjected to three stresses, as described earlier (50) and as follows: acidic stress (pH 5.5), oxidative stress (10 mM H2O2) in Sauton’s minimal medium, and nutrient starvation stress in 1× PBS (51, 52). Each stress was given for 4 h. Each of the stress conditions was performed 10 independent times. Each sample was checked for contamination using Ziehl-Neelsen (ZN) staining. Four hours poststress, intracellular metabolites were extracted and subjected to untargeted metabolomics using nuclear magnetic resonance (NMR) spectroscopy.
Extraction of intracellular metabolites.
Intracellular metabolites from M. smegmatis cultures were extracted using a modified version of a previously described extraction method (53). After 4 h of stress, the OD600 was measured, and the cultures were harvested and quenched in liquid nitrogen. Subsequently, the cells were thawed on ice for 5 min and extracted with the methanol-chloroform method. Briefly, 1 ml of methanol-chloroform (2:1) was added, along with 0.1-mm zirconia beads for 10 cycles with an interval of 1 min on ice to lyse the cells. The clear cell lysate was collected by centrifugation at 1,000 rpm for 45 s. To this supernatant, 500 μl of water and 500 μl of chloroform were added and vortexed for 30 s. The suspension was then centrifuged at 12,000 rpm for 30 min. The upper aqueous layer and lower organic phase were collected in 1.5-ml vials. Samples were dried in a vacuum concentrator and resuspended in NMR buffer for analysis.
Sample preparation.
The lyophilized polar samples were reconstituted by dissolving in 580 μl of 100% D2O NMR buffer (20 mM sodium phosphate [pH 7.4] containing 0.4 mM DSS [4,4-dimethyl-4-silapentane-1-sulfonic acid]. The composition for 1 liter of PBS was 0.623 g Na2HPO4·H2O and 4.15 g NaH2PO4·7H2O with the pH adjusted to 7.4. The samples were vortexed for 2 min at room temperature. After centrifugation at 4,000 × g for 2 min, supernatants were transferred to 5-mm NMR tubes for NMR analysis.
NMR spectroscopy analyses and metabolite identification.
All the NMR data were measured at 298 K using a Bruker Avance III HD Ascend NMR spectrometer, equipped with a quad-channel (HCNP-D) cryo-probe and pulsed-field gradients in the x, y, and z directions, operating at a proton frequency of 600.13 MHz. The water suppression pulse sequence noesygppr1d was used to record 1H-NMR spectra, which uses water presaturation and spoiler gradients during the relaxation delay and is of the form RD-G1-90°-t-90°-tm-G2-90°-ACQ, where RD is the relaxation delay of 5 s, t is a short delay typically of ∼3 μs, 90° represents a 90° RF pulse, tm is the mixing time of 100 ms, and ACQ is the data acquisition period. For a given sample, a total of 64 transients were collected into 32,000 data points for each spectrum, with a spectral width of 12 ppm. Pulse width, receiver gain, and water suppression parameters were kept identical among all the 1H experiments recorded for various samples to rule out intensity variations while recording the NMR data. For the 1H-1H total correlation spectroscopy (TOCSY) experiment (mixing time, 80 ms), a total of 2,048 × 1,024 data points with 40 transients per increment in the indirect dimension were recorded, spanning a spectral width of 10 ppm, and free induction decays (FIDs) were weighted by sine-bell function before Fourier transform, in both dimensions.
Spectra were processed and analyzed by using TopSpin (v3.5; Bruker). This includes automatic and manual phase correction, spectra referencing, and baseline correction. The 1H chemical shift dimension was directly referenced to the DSS (4, 4-dimethyl-4-silapentane-1-sulfonic acid) resonance. The distinguishing chemical shifts in the frequency domain that originated from the sets of spectral measurements were allocated to particular metabolites. Metabolite resonances present in the respective 1H-NMR spectrum (see Fig. S1a in the supplemental material) were identified and quantified using the Chenomx NMR suite software (Chenomx, Inc.) by spectral deconvolution approach and confirmed with the Biological Magnetic Resonance Data Bank (BMRB) database (54) and Human Metabolome Database (HMDB) (55). Further confirmation of metabolites was achieved using two-dimensional (2D) total correlation spectroscopy (TOCSY) (Fig. S1b) via the semiautomated software MetaboMiner (56). In all, 40 samples (10 replicates for each of the four conditions) containing ∼9,500 peaks, with an average of ∼237 peaks per sample, were analyzed. All the concentration data such obtained were exported to a spreadsheet file in a matrix format for univariate and multivariate analyses.
Statistical analysis.
To identify differential metabolites of M. smegmatis upon exposure to abiotic stress conditions, univariate analysis (fold change and t test), multivariate analysis, and pathway analysis were performed using the MetaboAnalyst online tool (57). The data matrix file containing metabolomics data under different stresses was uploaded to MetaboAnalyst (https://www.metaboanalyst.ca/).
The data obtained for each condition were normalized against the respective number of cells at the point of metabolite extraction. Each set of data was then preprocessed in the MetaboAnalyst site to achieve normal distribution (i.e., they follow a Gaussian or “normal” distribution), and the methods applied for each condition were briefly tabulated (Table S5). Such normalization is an essential prerequisite for a reliable statistical analysis (57).
A heat map was generated using hierarchical clustering (Ward’s algorithm), with the dendrogram being scaled to represent the distance between each branch (distance measure, Pearson’s correlation). The color differences demonstrate the relative concentrations of metabolites across different conditions and experiment groups. A fold change cutoff of >1.2 or <0.8, P value of ≤0.05, and false-discovery rate (FDR) of ≤0.05 were considered significant (58–61). Variances of the measured values were considered in the statistical analysis. Multivariate analyses, such as PCA and PLS-DA, were performed. The VIP score plot obtained by PLS-DA identified the key metabolites responsible for group segregation. VIP is a weighted sum of squares of the PLS weight, which indicates the importance of the variable to the entire model. While variables with a VIP score of ≥1.0 are generally considered to be statistically significant (58, 61, 62), we considered a VIP score of >1 for identifying distinguishing metabolites in different stresses compared to the control. Pathway analysis was also carried out using MetaboAnalyst tool. This software identifies the most significantly perturbed pathways under specific experimental conditions by utilizing pathway enrichment and topology analysis. The pathway impact is calculated from the pathway-topology analysis (33). We used the default “Global Test” and “Relative Betweenness Centrality” pathway analysis algorithm. We considered the metabolic pathways with pathway impact values of ≥0.1, P value of <0.05, and FDR of <0.05 to be perturbed significantly (20).
Protein structure homology modeling.
The primary amino acid sequence of 2,4-dienoyl-CoA reductase (M. smegmatis strain mc2155) MSMEG_5124 was obtained from Mycobrowser (63, 64) and subjected to domain analysis tools to predict the conserved domains that were present. These include MOTIF (https://www.genome.jp/tools/motif/) and SMART (http://smart.embl-heidelberg.de/). To further confirm the catalytic site of 2,4-dienoyl-CoA reductase (M. smegmatis strain mc2155) MSMEG_5124, we generated a model using SWISS-MODEL (a protein structure homology modeling server) (65) and GENO3D (automatic modeling of three-dimensional protein structure) (66). The predicted protein structure homology was checked by using Chimera version 1.10.2 and the PyMOL viewer.
RNA extraction and semiquantitative RT-PCR.
RNA extraction was performed using the TRIzol method, with minor modifications. After being subjected to stress, the samples were harvested at 3,500 rpm. The resultant pellet was snap-frozen in liquid nitrogen and stored at −80°C until further processing for RNA. The pellet was resuspended in TRIzol reagent (Invitrogen, CA) along with 0.1-mm glass beads and subjected to lysis by bead beating with a pulse on for 1 min and pulse off for 2 min on ice. A final concentration of 200 μg/ml glycogen was added and the samples incubated for 10 min at room temperature (RT). The samples were vortexed vigorously after adding chloroform and incubated at RT for 10 min. The samples were centrifuged at 10,000 rpm and 4°C for 20 min. The upper aqueous layer was collected. To the samples, 0.6 volumes of isopropanol along with 200 μg/ml glycogen were used to precipitate RNA. The pellets were washed with 75% ethanol, air dried at RT, and dissolved in RNase-free water (Qiagen, Hilden, Germany). Any residual DNA contamination was removed by DNase treatment before reverse transcription of the RNA. This DNase-treated RNA was further reverse transcribed with random hexamers as a primer to make cDNA using SuperScript III reverse transcriptase (Invitrogen). RT-PCR was carried out with 1:10 diluted cDNA for 30 cycles with respective primers. The PCR cycling parameters were (i) initial denaturation 95°C for 3 min, (ii) 30 amplification cycles (95°C for 15 s, annealing temperature [as given in Table S6] for 20 s, and 72°C for 20 s), and (iii) final extension 72°C for 10 min. A 1.5% agarose gel was used to visualize the RT-PCR product. All of the experiments were performed more than three times.
Cloning, expression, and purification of MSMEG_5124.
The ORF MSMEG_5124 (2,016 bp) was PCR amplified from the genomic DNA of M. smegmatis (mc2155) using specific primers listed in Table S5 and cloned into pET28a using the BamH1 and HindIII restriction sites. The clone, pET-MS_5124, was confirmed by double digestion (Fig. S4a) and sequencing. E. coli BL-21/plysS was transformed with pET-MS_5124 for overexpression and purification. E. coli BL-21/plysS transformed with pET-MS_5124 was induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 5 h. Cells were harvested and checked for overexpression. The histidine-tagged MSMEG_5124 was purified under native conditions using Talon (Clontech), following the manufacturer’s instructions (Fig. S4b). The protein was eluted by using 150 to 200 mM imidazole. The eluted protein was dialyzed against 50 mM potassium phosphate buffer (pH 7.8) with 5 mM β-mercaptoethanol.
Enzyme assay.
Purified MSMEG_5124 (340 to 350 nM) was used in a 1-ml reaction mixture consisting of reaction buffer (50 mM potassium phosphate [pH 7.8] with 150 mM NaCl), a substrate (either 20 mM trimethylamine [TCI Chemicals] or 20 mM dimethylamine [TCI Chemicals]), and 2 mM NAD+ (MP Biomedicals). The reactions were carried out anaerobically by nitrogen purging. The assay for MSMEG_5124 was carried as per the protocol used by McIntire (40), with slight modifications, where NAD+ was used as an electron acceptor instead of phenazine methosulfate (PMS). NAD+ was used here as an indicator. The conversion of NAD+ to NADH was measured at 340 nm for an hour. Reactions without MSMEG_5124 or substrates were used as controls. A standard plot of known concentrations of NADH was generated to convert the absorbance values to the concentration of NADH generated during the reaction (Fig. S5). The rate curve of the control reaction (MSMEG_5124 and 2 mM NAD+ only) is shown along with the rate curve of reaction (MSMEG_5124 and 2 mM NAD+ with substrates).
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by grants from the Department of Biotechnology (BT/IN/New-Indigo/05/SB/2013 and BT/HRD/NWBA/38/09/2018) to S.B. and a UGC SAP DRS grant to the Department of Biochemistry, School of Life Sciences, University of Hyderabad. A short-term European Molecular Biology Organization fellowship (STF-7009) to A.R. is acknowledged. K.B. was supported by a research fellowship from the Indian Council of Medical Research. Infrastructure support by DBT-CREB, DST-FIST, and DST-PURSE to the Department of Biochemistry and School of Life Sciences, University of Hyderabad, is acknowledged. We acknowledge the High-Field NMR facility at IISER Pune, cofunded by DST-FIST and IISER Pune. S.Y. acknowledges fellowship support from the University Grants Commission.
We are highly grateful to Anant B. Patel (CCMB) for his initial help with NMR and constructive input toward standardizing the NMR protocol. We thank Santosh K. Padhi, Department of Biochemistry, UoH, and his lab for providing reagents and helping in the design, execution, and analysis of the enzyme assay results. We thank Indira Ghosh (JNU, India) for timely suggestions and inputs, S. Dayananda (SLS, UoH), Manolis Matzapetakis (ITQB, Portugal), Ana Maria Varela Coelho (ITQB, Portugal), Pedro Fernandes (IGC, Portugal), Ankita Chatterjee, and Kiran Jakkala for critical input, as well as Krishnaveni Mohareer for help with manuscript preparation.
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00707-18.
REFERENCES
- 1.Falkinham JO., III. 2009. The biology of environmental mycobacteria. Environ Microbiol Rep 1:477–487. doi: 10.1111/j.1758-2229.2009.00054.x. [DOI] [PubMed] [Google Scholar]
- 2.Iivanainen E, Sallantaus T, Katila ML, Martikainen PJ. 1999. Mycobacteria in runoff waters from natural and drained peatlands. J Environ Qual 28:1226–1234. doi: 10.2134/jeq1999.00472425002800040025x. [DOI] [Google Scholar]
- 3.Collins CH, Grange JM, Yates MD. 1984. Mycobacteria in water. J Appl Bacteriol 57:193–211. doi: 10.1111/j.1365-2672.1984.tb01384.x. [DOI] [PubMed] [Google Scholar]
- 4.Parker BC, Ford MA, Gruft H, Falkinham JO III.. 1983. Epidemiology of infection by nontuberculous mycobacteria. IV. Preferential aerosolization of Mycobacterium intracellulare from natural waters. Am Rev Respir Dis 128:652–656. [DOI] [PubMed] [Google Scholar]
- 5.Wendt SL, George KL, Parker BC, Gruft H, Falkinham JO III.. 1980. Epidemiology of infection by nontuberculous Mycobacteria. III. Isolation of potentially pathogenic mycobacteria from aerosols. Am Rev Respir Dis 122:259–263. [DOI] [PubMed] [Google Scholar]
- 6.Santos R, de Carvalho CC, Stevenson A, Grant IR, Hallsworth JE. 2015. Extraordinary solute-stress tolerance contributes to the environmental tenacity of mycobacteria. Environ Microbiol Rep 7:746–764. doi: 10.1111/1758-2229.12306. [DOI] [PubMed] [Google Scholar]
- 7.Yang Y, Richards JP, Gundrum J, Ojha AK. 2018. GlnR activation induces peroxide resistance in mycobacterial biofilms. Front Microbiol 9:1428. doi: 10.3389/fmicb.2018.01428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hallsworth JE. 2018. Stress-free microbes lack vitality. Fungal Biol 122:379–385. doi: 10.1016/j.funbio.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Lovett B, St. Leger RJ. 2015. Stress is the rule rather than the exception for Metarhizium. Curr Genet 61:253–261. doi: 10.1007/s00294-014-0447-9. [DOI] [PubMed] [Google Scholar]
- 10.Grange JM. 1988. Mycobacteria and human disease. Edward Arnold Publishers Ltd, London, United Kingdom. [Google Scholar]
- 11.Roxas BA, Li Q. 2009. Acid stress response of a mycobacterial proteome: insight from a gene ontology analysis. Int J Clin Exp Med 2:309–328. [PMC free article] [PubMed] [Google Scholar]
- 12.O’Toole R, Smeulders MJ, Blokpoel MC, Kay EJ, Lougheed K, Williams HD. 2003. A two-component regulator of universal stress protein expression and adaptation to oxygen starvation in Mycobacterium smegmatis. J Bacteriol 185:1543–1554. doi: 10.1128/JB.185.5.1543-1554.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anes E, Peyron P, Staali L, Jordao L, Gutierrez MG, Kress H, Hagedorn M, Maridonneau-Parini I, Skinner MA, Wildeman AG, Kalamidas SA, Kuehnel M, Griffiths G. 2006. Dynamic life and death interactions between Mycobacterium smegmatis and J774 macrophages. Cell Microbiol 8:939–960. doi: 10.1111/j.1462-5822.2005.00675.x. [DOI] [PubMed] [Google Scholar]
- 14.Gupta S, Chatterji D. 2005. Stress responses in mycobacteria. IUBMB Life 57:149–159. doi: 10.1080/15216540500090611. [DOI] [PubMed] [Google Scholar]
- 15.Röse L, Kaufmann SH, Daugelat S. 2004. Involvement of Mycobacterium smegmatis undecaprenyl phosphokinase in biofilm and smegma formation. Microbes Infect 6:965–971. doi: 10.1016/j.micinf.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 16.Lygren ST, Closs O, Bercouvier H, Wayne LG. 1986. Catalases, peroxidases, and superoxide dismutases in Mycobacterium leprae and other mycobacteria studied by crossed immunoelectrophoresis and polyacrylamide gel electrophoresis. Infect Immun 54:666–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pierre-Audigier C, Jouanguy E, Lamhamedi S, Altare F, Rauzier J, Vincent V, Canioni D, Emile JF, Fischer A, Blanche S, Gaillard JL, Casanova JL. 1997. Fatal disseminated Mycobacterium smegmatis infection in a child with inherited interferon gamma receptor deficiency. Clin Infect Dis 24:982–984. doi: 10.1093/clinids/24.5.982. [DOI] [PubMed] [Google Scholar]
- 18.Li X, Mei H, Chen F, Tang Q, Yu Z, Cao X, Andongma BT, Chou SH, He J. 2017. Transcriptome Landscape of Mycobacterium smegmatis. Front Microbiol 8:2505. doi: 10.3389/fmicb.2017.02505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang R, Prince JT, Marcotte EM. 2005. Mass spectrometry of the M. smegmatis proteome: protein expression levels correlate with function, operons, and codon bias. Genome Res 15:1118–1126. doi: 10.1101/gr.3994105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liew KL, Jee JM, Yap I, Yong PV. 2016. In vitro analysis of metabolites secreted during infection of lung epithelial cells by Cryptococcus neoformans. PLoS One 11:e0153356. doi: 10.1371/journal.pone.0153356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mattow J, Siejak F, Hagens K, Becher D, Albrecht D, Krah A, Schmidt F, Jungblut PR, Kaufmann SH, Schaible UE. 2006. Proteins unique to intraphagosomally grown Mycobacterium tuberculosis. Proteomics 6:2485–2494. doi: 10.1002/pmic.200500547. [DOI] [PubMed] [Google Scholar]
- 22.Rao PK, Li Q. 2009. Protein turnover in mycobacterial proteomics. Molecules 14:3237–3258. doi: 10.3390/molecules14093237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho SH, Goodlett D, Franzblau S. 2006. ICAT-based comparative proteomic analysis of non-replicating persistent Mycobacterium tuberculosis. Tuberculosis (Edinb) 86:445–460. doi: 10.1016/j.tube.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 24.Ganji R, Dhali S, Rizvi A, Rapole S, Banerjee S. 2016. Understanding HIV-Mycobacteria synergism through comparative proteomics of intra-phagosomal mycobacteria during mono- and HIV co-infection. Sci Rep 6:22060. doi: 10.1038/srep22060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ganji R, Dhali S, Rizvi A, Sankati S, Vemula MH, Mahajan G, Rapole S, Banerjee S. 2016. Proteomics approach to understand reduced clearance of mycobacteria and high viral titers during HIV-mycobacteria co-infection. Cell Microbiol 18:355–368. doi: 10.1111/cmi.12516. [DOI] [PubMed] [Google Scholar]
- 26.Wegner A, Meiser J, Weindl D, Hiller K. 2015. How metabolites modulate metabolic flux. Curr Opin Biotechnol 34:16–22. doi: 10.1016/j.copbio.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 27.Drapal M, Perez-Fons L, Wheeler PR, Fraser PD. 2014. The application of metabolite profiling to Mycobacterium spp.: determination of metabolite changes associated with growth. J Microbiol Methods 106:23–32. doi: 10.1016/j.mimet.2014.07.037. [DOI] [PubMed] [Google Scholar]
- 28.Drapal M, Wheeler PR, Fraser PD. 2016. Metabolite analysis of Mycobacterium species under aerobic and hypoxic conditions reveals common metabolic traits. Microbiology 162:1456–1467. doi: 10.1099/mic.0.000325. [DOI] [PubMed] [Google Scholar]
- 29.Man DK, Kanno T, Manzo G, Robertson BD, Lam JKW, Mason AJ. 2018. Rifampin- or capreomycin-induced remodeling of the Mycobacterium smegmatis mycolic acid layer is mitigated in synergistic combinations with cationic antimicrobial peptides. mSphere 3:e00218-18. doi: 10.1128/mSphere.00218-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baptista R, Fazakerley DM, Beckmann M, Baillie L, Mur L. 2018. Untargeted metabolomics reveals a new mode of action of pretomanid (PA-824). Sci Rep 8:5084. doi: 10.1038/s41598-018-23110-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elbein AD, Pastuszak I, Tackett AJ, Wilson T, Pan YT. 2010. Last step in the conversion of trehalose to glycogen: a mycobacterial enzyme that transfers maltose from maltose 1-phosphate to glycogen. J Biol Chem 285:9803–9812. doi: 10.1074/jbc.M109.033944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miah F, Koliwer-Brandl H, Rejzek M, Field RA, Kalscheuer R, Bornemann S. 2013. Flux through trehalose synthase flows from trehalose to the alpha anomer of maltose in mycobacteria. Chem Biol 20:487–493. doi: 10.1016/j.chembiol.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xia J, Wishart DS. 2011. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat Protoc 6:743–760. doi: 10.1038/nprot.2011.319. [DOI] [PubMed] [Google Scholar]
- 34.Whatmore AM, Chudek JA, Reed RH. 1990. The effects of osmotic upshock on the intracellular solute pools of Bacillus subtilis. J Gen Microbiol 136:2527–2535. doi: 10.1099/00221287-136-12-2527. [DOI] [PubMed] [Google Scholar]
- 35.Holmström KO, Welin B, Mandal A, Kristiansdottir I, Teeri TH, Lamark T, Strom AR, Palva ET. 1994. Production of the Escherichia coli betaine-aldehyde dehydrogenase, an enzyme required for the synthesis of the osmoprotectant glycine betaine, in transgenic plants. Plant J 6:749–758. doi: 10.1046/j.1365-313X.1994.6050749.x. [DOI] [PubMed] [Google Scholar]
- 36.Park S, Smith LT, Smith GM. 1995. Role of glycine betaine and related osmolytes in osmotic stress adaptation in Yersinia enterocolitica ATCC 9610. Appl Environ Microbiol 61:4378–4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yancey PH, Blake WR, Conley J. 2002. Unusual organic osmolytes in deep-sea animals: adaptations to hydrostatic pressure and other perturbants. Comp Biochem Physiol A Mol Integr Physiol 133:667–676. doi: 10.1016/S1095-6433(02)00182-4. [DOI] [PubMed] [Google Scholar]
- 38.Burg MB, Ferraris JD. 2008. Intracellular organic osmolytes: function and regulation. J Biol Chem 283:7309–7313. doi: 10.1074/jbc.R700042200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu Y, Jameson E, Crosatti M, Schafer H, Rajakumar K, Bugg TD, Chen Y. 2014. Carnitine metabolism to trimethylamine by an unusual Rieske-type oxygenase from human microbiota. Proc Natl Acad Sci U S A 111:4268–4273. doi: 10.1073/pnas.1316569111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McIntire WS. 1990. Trimethylamine dehydrogenase from bacterium W3A1. Methods Enzymol 188:250–260. doi: 10.1016/0076-6879(90)88042-9. [DOI] [PubMed] [Google Scholar]
- 41.Hubbard PA, Liang X, Schulz H, Kim JJ. 2003. The crystal structure and reaction mechanism of Escherichia coli 2,4-dienoyl-CoA reductase. J Biol Chem 278:37553–37560. doi: 10.1074/jbc.M304642200. [DOI] [PubMed] [Google Scholar]
- 42.Koeth RA, Levison BS, Culley MK, Buffa JA, Wang Z, Gregory JC, Org E, Wu Y, Li L, Smith JD, Tang WHW, DiDonato JA, Lusis AJ, Hazen SL. 2014. γ-Butyrobetaine is a proatherogenic intermediate in gut microbial metabolism of l-carnitine to TMAO. Cell Metab 20:799–812. doi: 10.1016/j.cmet.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim SG, Bae HS, Lee ST. 2001. A novel denitrifying bacterial isolate that degrades trimethylamine both aerobically and anaerobically via two different pathways. Arch Microbiol 176:271–277. doi: 10.1007/s002030100319. [DOI] [PubMed] [Google Scholar]
- 44.Yancey PH, Somero GN. 1979. Counteraction of urea destabilization of protein structure by methylamine osmoregulatory compounds of elasmobranch fishes. Biochem J 183:317–323. doi: 10.1042/bj1830317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Somashekar BS, Amin AG, Rithner CD, Troudt J, Basaraba R, Izzo A, Crick DC, Chatterjee D. 2011. Metabolic profiling of lung granuloma in Mycobacterium tuberculosis infected guinea pigs: ex vivo 1H magic angle spinning NMR studies. J Proteome Res 10:4186–4195. doi: 10.1021/pr2003352. [DOI] [PubMed] [Google Scholar]
- 46.Price CT, Bukka A, Cynamon M, Graham JE. 2008. Glycine betaine uptake by the ProXVWZ ABC transporter contributes to the ability of Mycobacterium tuberculosis to initiate growth in human macrophages. J Bacteriol 190:3955–3961. doi: 10.1128/JB.01476-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boden R, Thomas E, Savani P, Kelly DP, Wood AP. 2008. Novel methylotrophic bacteria isolated from the River Thames (London, UK). Environ Microbiol 10:3225–3236. doi: 10.1111/j.1462-2920.2008.01711.x. [DOI] [PubMed] [Google Scholar]
- 48.McTaggart TL, Beck DA, Setboonsarng U, Shapiro N, Woyke T, Lidstrom ME, Kalyuzhnaya MG, Chistoserdova L. 2015. Genomics of methylotrophy in Gram-positive methylamine-utilizing bacteria. Microorganisms 3:94–112. doi: 10.3390/microorganisms3010094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dubey AA, Wani SR, Jain V. 2018. Methylotrophy in mycobacteria: dissection of the methanol metabolism pathway in Mycobacterium smegmatis. J Bacteriol 200:e00288-18. doi: 10.1128/JB.00288-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Balakrishnan K, Mohareer K, Banerjee S. 2017. Mycobacterium tuberculosis Rv1474c is a TetR-like transcriptional repressor that regulates aconitase, an essential enzyme and RNA-binding protein, in an iron-responsive manner. Tuberculosis (Edinb) 103:71–82. doi: 10.1016/j.tube.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 51.Loebel RO, Shorr E, Richardson HB. 1933. The influence of adverse conditions upon the respiratory metabolism and growth of human tubercle bacilli. J Bacteriol 26:167–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vemula MH, Ganji R, Sivangala R, Jakkala K, Gaddam S, Penmetsa S, Banerjee S. 2016. Mycobacterium tuberculosis zinc metalloprotease-1 elicits tuberculosis-specific humoral immune response independent of mycobacterial load in pulmonary and extra-pulmonary tuberculosis patients. Front Microbiol 7:418. doi: 10.3389/fmicb.2016.00418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tambellini NP, Zaremberg V, Turner RJ, Weljie AM. 2013. Evaluation of extraction protocols for simultaneous polar and non-polar yeast metabolite analysis using multivariate projection methods. Metabolites 3:592–605. doi: 10.3390/metabo3030592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, Schulte CF, Tolmie DE, Kent Wenger R, Yao H, Markley JL. 2008. BioMagResBank. Nucleic Acids Res 36:D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S, Fung C, Nikolai L, Lewis M, Coutouly MA, Forsythe I, Tang P, Shrivastava S, Jeroncic K, Stothard P, Amegbey G, Block D, Hau DD, Wagner J, Miniaci J, Clements M, Gebremedhin M, Guo N, Zhang Y, Duggan GE, Macinnis GD, Weljie AM, Dowlatabadi R, Bamforth F, Clive D, Greiner R, Li L, Marrie T, Sykes BD, Vogel HJ, Querengesser L. 2007. HMDB: the Human Metabolome Database. Nucleic Acids Res 35:D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xia J, Bjorndahl TC, Tang P, Wishart DS. 2008. MetaboMiner–semi-automated identification of metabolites from 2D NMR spectra of complex biofluids. BMC Bioinformatics 9:507. doi: 10.1186/1471-2105-9-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xia J, Wishart DS. 2016. Using MetaboAnalyst 3.0 for comprehensive metabolomics data analysis. Curr Protoc Bioinformatics 55:14.10.1–14.10.91. doi: 10.1002/cpbi.11. [DOI] [PubMed] [Google Scholar]
- 58.Feng Q, Liu Z, Zhong S, Li R, Xia H, Jie Z, Wen B, Chen X, Yan W, Fan Y, Guo Z, Meng N, Chen J, Yu X, Zhang Z, Kristiansen K, Wang J, Xu X, He K, Li G. 2016. Integrated metabolomics and metagenomics analysis of plasma and urine identified microbial metabolites associated with coronary heart disease. Sci Rep 6:22525. doi: 10.1038/srep22525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hou W, Meng X, Zhao A, Zhao W, Pan J, Tang J, Huang Y, Li H, Jia W, Liu F. 2018. Development of multimarker diagnostic models from metabolomics analysis for gestational diabetes mellitus (GDM). Mol Cell Proteomics 17:431–441. doi: 10.1074/mcp.RA117.000121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abdelrahman M, Sawada Y, Nakabayashi R, Sato S, Hirakawa H, El-Sayed M, Hirai MY, Saito K, Yamauchi N, Shigyo M. 2015. Integrating transcriptome and target metabolome variability in doubled haploids of Allium cepa for abiotic stress protection. Mol Breed 35:195. doi: 10.1007/s11032-015-0378-2. [DOI] [Google Scholar]
- 61.Wu H, Chen Y, Li ZG, Liu XH. 2018. Untargeted metabolomics profiles delineate metabolic alterations in mouse plasma during lung carcinoma development using UPLC-QTOF/MS in MSE mode. R Soc Open Sci 5:181143. doi: 10.1098/rsos.181143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ma X, Chi YH, Niu M, Zhu Y, Zhao YL, Chen Z, Wang JB, Zhang CE, Li JY, Wang LF, Gong M, Wei SZ, Chen C, Zhang L, Wu MQ, Xiao XH. 2016. Metabolomics coupled with multivariate data and pathway analysis on potential biomarkers in cholestasis and intervention effect of Paeonia lactiflora Pall. Front Pharmacol 7:14. doi: 10.3389/fphar.2016.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kapopoulou A, Lew JM, Cole ST. 2011. The MycoBrowser portal: a comprehensive and manually annotated resource for mycobacterial genomes. Tuberculosis (Edinb) 91:8–13. doi: 10.1016/j.tube.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 64.Lew JM, Kapopoulou A, Jones LM, Cole ST. 2011. TubercuList–10 years after. Tuberculosis (Edinb) 91:1–7. doi: 10.1016/j.tube.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 65.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L, Schwede T. 2014. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Combet C, Jambon M, Deleage G, Geourjon C. 2002. Geno3D: automatic comparative molecular modelling of protein. Bioinformatics 18:213–214. doi: 10.1093/bioinformatics/18.1.213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.