Oxygen is used as a substrate to derive energy by the bacterial pathogen Staphylococcus aureus during infection; however, S. aureus can also grow fermentatively in the absence of oxygen. To successfully cause infection, S. aureus must tailor its metabolism to take advantage of respiratory activity. Different proteins are required for growth in the presence or absence of oxygen; therefore, when cells transition between these conditions, several proteins would be expected to become unnecessary. In this report, we show that regulated proteolysis is used to modulate energy metabolism in S. aureus. We report that the ClpCP protein complex is involved in specifically modulating aerobic respiratory growth but is dispensable for fermentative growth.
KEYWORDS: ClpC, ClpP, SrrAB, Staphylococcus aureus, TrfA, fermentation, metabolism, respiration
ABSTRACT
The staphylococcal respiratory regulator (SrrAB) modulates energy metabolism in Staphylococcus aureus. Studies have suggested that regulated protein catabolism facilitates energy homeostasis. Regulated proteolysis in S. aureus is achieved through protein complexes composed of a peptidase (ClpQ or ClpP) in association with an AAA+ family ATPase (typically, ClpC or ClpX). In the present report, we tested the hypothesis that SrrAB regulates a Clp complex to facilitate energy homeostasis in S. aureus. Strains deficient in one or more Clp complexes were attenuated for growth in the presence of puromycin, which causes enrichment of misfolded proteins. A ΔsrrAB strain had increased sensitivity to puromycin. Epistasis experiments suggested that the puromycin sensitivity phenotype of the ΔsrrAB strain was a result of decreased ClpC activity. Consistent with this, transcriptional activity of clpC was decreased in the ΔsrrAB mutant, and overexpression of clpC suppressed the puromycin sensitivity of the ΔsrrAB strain. We also found that ClpC positively influenced respiration and that it did so upon association with ClpP. In contrast, ClpC limited fermentative growth, while ClpP was required for optimal fermentative growth. Metabolomics studies demonstrated that intracellular metabolic profiles of the ΔclpC and ΔsrrAB mutants were distinct from those of the wild-type strain, supporting the notion that both ClpC and SrrAB affect central metabolism. We propose a model wherein SrrAB regulates energy homeostasis, in part, via modulation of regulated proteolysis.
IMPORTANCE Oxygen is used as a substrate to derive energy by the bacterial pathogen Staphylococcus aureus during infection; however, S. aureus can also grow fermentatively in the absence of oxygen. To successfully cause infection, S. aureus must tailor its metabolism to take advantage of respiratory activity. Different proteins are required for growth in the presence or absence of oxygen; therefore, when cells transition between these conditions, several proteins would be expected to become unnecessary. In this report, we show that regulated proteolysis is used to modulate energy metabolism in S. aureus. We report that the ClpCP protein complex is involved in specifically modulating aerobic respiratory growth but is dispensable for fermentative growth.
INTRODUCTION
Staphylococcus aureus is a commensal bacterium that colonizes between 20% and 50% of the healthy human population (1–4). Typically, S. aureus does not cause the human carrier harm; however, it is capable of causing both invasive and noninvasive infections (5–7). Historically, S. aureus infections were acquired within a hospital environment (8), but the onset or occurrence of S. aureus infections is increasing in community settings (5, 9).
S. aureus is a facultative anaerobe. It can derive energy for growth using either fermentative or respiratory pathways (10). Oxygen concentrations vary within healthy human tissues (between ∼1.5% and 19.7%), and in infected tissues, they are estimated to be less than 1% (11–13). In the context of S. aureus infections, the concentration of oxygen at the site of infection progressively decreases as an infection proceeds (14). Consequently, the ability of S. aureus to fine-tune respiratory or fermentative metabolism with respect to the oxygen concentration is likely to be crucial for achieving optimum energy production and utilization. Consistent with this idea, the pathogenesis of strains with an impaired ability to respire or ferment is attenuated (15–17).
Bacteria use a variety of regulatory mechanisms to facilitate adaptation toward changes in their environments. Regulation by means of proteolysis is a strategy adopted in processes as diverse as stress response to cell division (18, 19). It has been argued that regulated proteolysis allows cells to efficiently remove proteins that have been rendered superfluous to cellular needs (20). Two rather distinct sets of proteins are required to facilitate fermentative and respiratory growth; therefore, when cells transition between either of the conditions, a number of proteins will be rendered unnecessary. The concentration and availability of a terminal electron acceptor (TEA) constitute one factor that could necessitate reliance of cells upon regulated proteolysis to facilitate energetic homeostasis. Regulated proteolysis is employed to modulate respiratory metabolism in yeast (21, 22). It is unclear whether regulated proteolysis and respiratory/fermentative metabolism are linked in S. aureus.
Proteolysis of S. aureus cytoplasmic proteins is achieved by chaperone-protease complexes (Clp complexes) (19). The complexes are two-component proteases consisting of peptidase and ATPase subunits (19). S. aureus harbors two peptidase subunits: ClpP and ClpQ (19, 23, 24). ClpP is the dominant peptidase under standard laboratory growth conditions (23, 24). S. aureus harbors multiple ATPase subunits that each carry a domain typical of the AAA+ protein superfamily (19). Only a subset of ATPases contain the ClpP recognition tripeptide and can interact with ClpP (25, 26). In S. aureus, ClpC and ClpX are capable of interacting with ClpP, while ClpL and ClpB are thought to not interact with ClpP (19). Apart from interacting with ClpP, the ATPases also interact with a second class of proteins termed cofactor or adaptor proteins. The adaptor proteins facilitate the recognition and targeting of proteins for degradation. Three adaptor proteins have been identified in S. aureus: YjbH (27), TrfA (28), and McsB (29). TrfA, a homolog of MecA in Bacillus subtilis, has been suggested to function as an adaptor for ClpC (30). ClpP-dependent complexes are integral for numerous cellular processes, thereby establishing regulated proteolysis as a global modulator of cellular physiology.
Bacteria utilize two-component regulatory systems (TCRS) to adapt to their surroundings. TCRS allow bacteria to integrate several stimuli into signaling circuits, allowing for a tailored response toward their environment (reviewed in reference 31). Classical TCRS consist of two proteins: a histidine kinase (HK) and a response regulator (RR). The HK is capable of interacting with intracellular and/or extracellular stimuli. The RR is typically cytosolic and may also be a transcription factor with one or more of the following functionalities: it can undergo autophosphorylation, transfer phosphoryl groups to the RR, or remove phosphoryl groups from the RR. Interaction with a signal molecule alters the functionality of the HK, thereby affecting the levels of the phosphoryl group on the RR. In most cases but not all, the levels of phosphoryl groups on the RR at any given point determine whether system output is increased or decreased.
The staphylococcal respiratory regulator (SrrAB) TCRS was identified as a system that modulates the expression of staphylococcal virulence factors when oxygen tension is decreased (32–34). SrrA is a DNA binding RR (33, 35). SrrB is the HK and is membrane spanning (33). Proteomic and microarray studies have established SrrAB as a pleiotropic regulator of energy metabolism (14, 36, 37), and SrrAB positively influences aerobic respiration (14, 35, 36). In the absence of oxygen or upon its limitation, SrrAB positively influences fermentative growth (37). Biofilms are crucial in staphylococcal pathogenesis (6, 38, 39), and SrrAB positively influences the formation of biofilms in low-oxygen environments (36, 40). S. aureus strains lacking SrrAB also display attenuated survival in models of infection (17, 41).
RESULTS
A ΔsrrAB strain has increased sensitivity to puromycin.
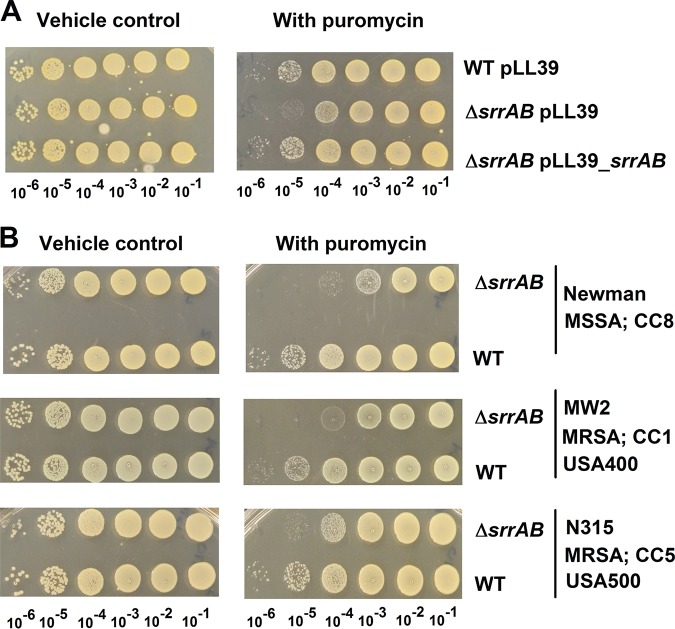
We reasoned that if SrrAB regulates a factor involved in protein turnover, then a ΔsrrAB strain would display altered growth under conditions that cause protein misfolding. Puromycin is a tRNA analog that causes premature termination of protein translation; therefore, puromycin imposes a high demand for the proteolytic machinery (24). The USA300_LAC ΔsrrAB mutant strain displayed a pronounced growth defect in solid and liquid aerobic media supplemented with puromycin but not in its absence (Fig. 1; see also Fig. S1 in the supplemental material). Growth on both solid and liquid media was examined, since regulatory networks in S. aureus can be altered between these two growth environments (42). Serial dilutions of the USA300_LAC (WT) and ΔsrrAB strains were placed upon solid tryptic soy broth (TSB) medium in the presence or absence of puromycin and growth was analyzed. In the presence of puromycin, the ΔsrrAB strain formed colonies that were smaller in size and the number of colonies formed was decreased by ∼10- to 100-fold (Fig. 1), while in the absence of puromycin, the ΔsrrAB strain formed colonies of the same size and frequency as the WT strain. Likewise, in liquid medium, the ΔsrrAB strain displayed an increased generation time, and its growth was inhibited at lower concentrations of puromycin than for the WT (Fig. S1B and C). The return of srrAB genes to the chromosome of the ΔsrrAB strain, in a nonendogenous location, restored puromycin sensitivity to WT levels (Fig. 1 and S1). Puromycin resistance was mildly, but consistently, enhanced in a WT strain carrying srrAB in a multicopy plasmid (Fig. S1D).
FIG 1.
SrrAB is involved in puromycin resistance in diverse isolates of S. aureus. (A) A ΔsrrAB strain is deficient in growth upon solid TSB medium containing puromycin. Growth for the WT with pLL39 or the ΔsrrAB strain with pLL39 or pLL39_srrAB is displayed on solid TSB medium with or without puromycin. (B) Growth for the Newman (JMB 1422), Newman ΔsrrAB (JMB 4751), MW2 (JMB 1324), MW2 ΔsrrAB (JMB 7573), N315 (JMB 7570), and N315 ΔsrrAB (JMB 7574) strains is displayed on solid TSB medium with or without puromycin. MRSA, methicillin-resistant S. aureus; MSSA, methicillin-sensitive S. aureus; CC, clonal-complex type, USA number, the pulsed-field gel electrophoresis type. Photographs are representative of at least three independent experiments. The numbers beneath each photograph denote the serial dilution that cells were removed from before plating.
Regulatory networks can differ between isolates of S. aureus (43, 44). The influence of SrrAB on puromycin resistance was examined in alternate isolates of S. aureus (Newman, MW2, and N315) that differ in their physiologies and in their expression of virulence factors. In the Newman strain, the global virulence regulator SaeS contains a point mutation (SaeS P18) that imparts constitutive kinase activity (45). The growth of strains lacking SrrAB was attenuated on solid medium containing puromycin for each isolate examined (Fig. 1). The data in Fig. 1 and S1 suggested that SrrAB positively influences puromycin resistance in S. aureus.
Genetic evidence suggests that decreased expression of clpC in the ΔsrrAB strain results in sensitivity to puromycin.
We tested the hypothesis that SrrAB controls the activity of a factor that facilitates protein turnover. Strains deficient in the activity of Clp proteolytic complexes have deficient growth in the presence of puromycin (24). Proteomic analyses by Throup et al. identified a putative ATPase, with homology to the Lactococcus lactis protein CAA44207, which had altered abundance in an S. aureus strain lacking SrrAB (37). Basic Local Alignment Search Tool (BLAST) analyses found that ClpL (SAUSA300_2486) and ClpC (SAUSA300_0510) had the highest similarity to CAA44207. We found that a ΔclpC strain had deficient growth on solid medium containing puromycin, whereas a clpL::Tn strain did not (Fig. 2A).
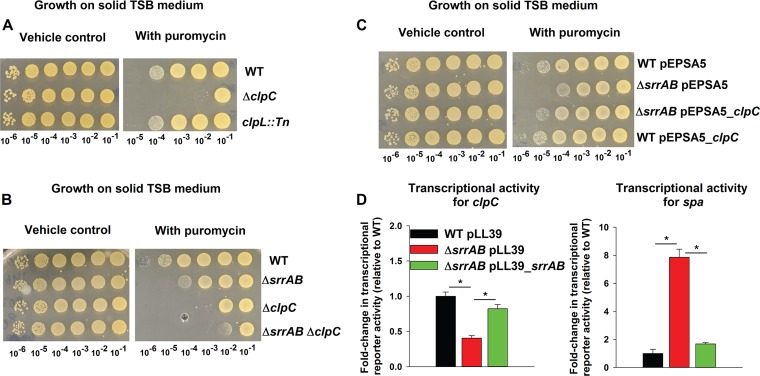
FIG 2.
Genetic evidence suggests that decreased expression of ClpC in a ΔsrrAB strain results in sensitivity to puromycin. (A) ClpC is required for puromycin resistance, while ClpL is dispensable. Growth for the WT (JMB 1100), ΔclpC (JMB 8025), and clpL::Tn (JMB 4850) strains is displayed on solid TSB medium with or without puromycin. (B) The puromycin sensitivity phenotypes of the ΔsrrAB and ΔclpC mutations are not additive. Growth for the WT, ΔsrrAB (JMB 1467), ΔclpC, and ΔsrrAB ΔclpC (JMB 8027) strains is displayed on solid TSB medium with or without puromycin. (C) Overexpression of clpC suppresses the puromycin sensitivity phenotype of the ΔsrrAB strain. The growth profiles of WT and ΔsrrAB strains with either pEPSA5 (empty vector) or pEPSA5_clpC are displayed on solid TSB medium with or without puromycin. (D) Transcriptional activity of clpC is decreased, while that for spa is increased, in a strain lacking SrrAB. Transcriptional activities are displayed for the WT with pLL39 and the ΔsrrAB strain with pLL39 or pLL39_srrAB and simultaneously carrying a multicopy plasmid containing gfp under the transcriptional control of either the clpC or spa promoter. Data in panel D represent the averages from triplicate cultures and error bars represent standard deviations. Photographs shown in panels A, B, and C are representative of at least three independent experiments, and the numbers beneath each photograph denote the serial dilution that cells were removed from before plating. Where indicated, Student t tests (two tailed) were performed. *, P < 0.05.
Epistasis experiments were used to examine potential interactions between srrAB and clpC. The puromycin sensitivities of the ΔsrrAB, ΔclpC, and ΔsrrAB ΔclpC strains were analyzed in liquid medium and on solid medium. The puromycin sensitivity phenotypes associated with the ΔsrrAB and ΔclpC mutations were nonadditive. The size and frequency of colonies formed by the ΔsrrAB ΔclpC double mutant were similar to those observed for the ΔclpC strain (Fig. 2B). We found that the presence of puromycin increased the lag times necessary to initiate outgrowth (see Fig. S2A). The puromycin-dependent growth inhibition levels associated with the ΔsrrAB and ΔclpC mutations were not additive in liquid medium (Fig. S2B). These data suggest that SrrAB may affect puromycin resistance via ClpC.
We introduced clpC, under the transcriptional control of a xylose-inducible promoter (pEPSA5_clpC), into the WT and ΔsrrAB strains and assessed puromycin sensitivities. In the presence of puromycin, the ΔsrrAB strain carrying empty vector formed ∼10-fold fewer colonies than the WT strain carrying an empty vector (Fig. 2C). However, the ΔsrrAB strain carrying pEPSA5_clpC formed a similar number of colonies as the WT carrying an empty vector. The presence of pEPSA5_clpC had no noticeable effect on the WT at the puromycin concentration utilized.
The influence of SrrAB on clpC transcription was examined. A transcriptional reporter was constructed wherein the gene encoding green fluorescent protein (GFP) was placed under the transcriptional control of the clpC promoter. SrrA is a transcriptional repressor of the gene encoding protein A (Spa) (33), and a spa transcriptional reporter was included as a positive control. The transcriptional activities of clpC and spa were decreased and increased, respectively, in the ΔsrrAB strain (Fig. 2D). The transcriptional activities of both genes were restored to near WT levels by the reintroduction of srrAB to the ΔsrrAB strain.
Metabolomics analyses demonstrate the influence of SrrAB and ClpC on metabolism.
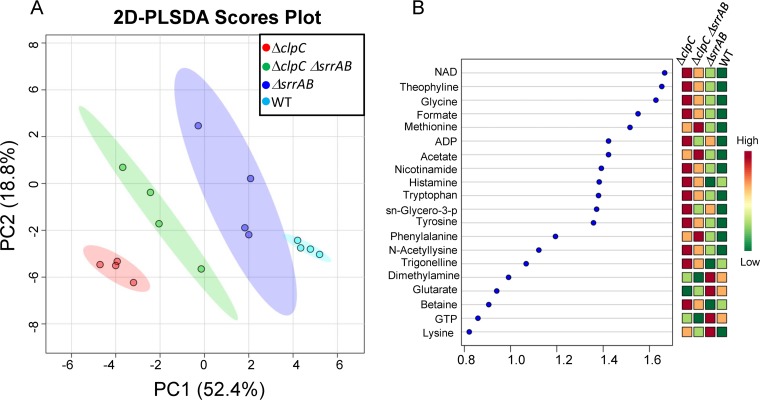
We tested the hypothesis that SrrAB affects energy homeostasis via ClpC. To this end, we examined the effect of SrrAB and ClpC upon cellular energetics using one-dimensional (1D) 1H nuclear magnetic resonance (NMR) for the global profiling of cellular metabolites following aerobic growth, as previously described (46). Intracellular metabolite levels were quantified in the WT, ΔsrrAB, ΔclpC, and ΔsrrAB ΔclpC strains. A previous study found that a strain lacking ClpC, cultured to stationary phase, had altered transcript levels for genes encoding components of the electron transfer chain and oxidative phosphorylation (46). To allow comparisons with previous studies, our analyses were also conducted in stationary phase, following 48 h of growth. Two-dimensional principal-component analysis (2D-PCA) and two-dimensional partial least-squares discriminant (2D PLS-DA) multivariate statistical analyses demonstrated that the WT, ΔsrrAB, ΔclpC, and ΔsrrAB ΔclpC strains exhibit distinct metabolic profiles (Fig. 3A) (relative metabolite concentrations are listed in Table S1). Associated variable importance in projection (VIP) scores resulting from the 2D PLS-DA analysis highlighted the metabolites whose concentration changes contributed the most to the separation (VIP score > 1.0) of the metabolic profiles of the four strains (Fig. 3B). Most notably, the mutant strains displayed higher levels of NAD+ than the WT (with NAD+ concentrations in the following order: ΔclpC > ΔsrrAB ΔclpC > ΔsrrAB > WT). Other interesting metabolite pattern changes included higher levels of formate, niacinamide (ΔclpC > ΔsrrAB ΔclpC > ΔsrrAB > WT), and ADP (ΔclpC > ΔsrrAB > ΔsrrAB ΔclpC > WT) in the mutant strains than in the WT. In addition, GTP levels were lower in the ΔclpC and ΔsrrAB ΔclpC mutants than in the WT.
FIG 3.
2D partial least-square discriminant analysis (2D PLS-DA) score plot and associated VIP scores following aerobic growth for 48 h. (A) 2D PLS-DA score plot separating the WT (JMB 1100), ΔsrrAB (JMB 1467), ΔclpC (JMB 8025), and ΔsrrAB ΔclpC (JMB 8027) S. aureus strains according to their distinct metabolic profiles, with principal component 1 (PC1) and PC2 accounting for ~52% and ~19% of the variance, respectively. (B) Variable importance in projection (VIP) scores ranking the most important metabolites that contributed to the separation of the different strains in the 2D PLS-DA score plot. Metabolites with associated VIP scores of ≥1.0 were considered to be significant contributors to the separation between the metabolic profiles of the different strains. sn-Glycero-3-p, sn-glycero-3-phosphocholine.
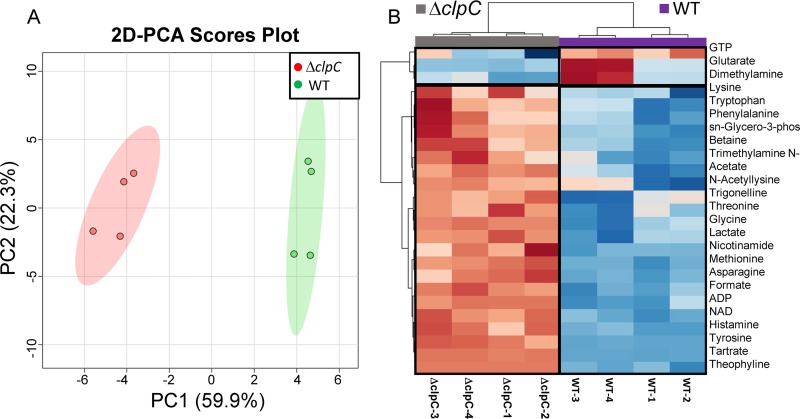
We compared the metabolite profile of each strain to that of the WT. The 2D-PCA plot and heat map representation of metabolites whose levels were significantly altered between ΔclpC and WT are shown in Fig. 4. The 2D-PCA plot separates ΔclpC and WT primarily in the principal component 1 (PC1) dimension, where PC1 accounts for ∼60% of the variance and PC2 accounts for ∼22% of the variance (Fig. 4A). We found that ADP, NAD+, lactate, acetate, and formate levels were higher in the ΔclpC sample group (Fig. 4B), supporting the notion that the energy status of the ΔclpC strain is decreased. Levels of GTP and 2-oxo-glutarate were also lower in the ΔclpC strain. Taken together, these data suggest that the ΔclpC strain is, in all likelihood, exhibiting reduced oxidative phosphorylation and tricarboxylic acid (TCA) cycle activity.
FIG 4.
2D-PCA score plot and corresponding hierarchical clustering analysis (HCA) of the metabolite profiles of ΔclpC and WT strains following aerobic growth for 48 h. (A) 2D-PCA score plot separating the ΔclpC (JMB 8025) and WT (JMB 1100) cell cultures and revealing that the two strains exhibit distinct metabolic profiles, with PC1 and PC2 accounting for ∼60% and ∼22% of the variance, respectively. (B) Heat map visualization of the top 25 metabolites that contributed most significantly to the metabolic profile separation of the ΔclpC mutant from the WT strain, based on Euclidean distance calculated from metabolite abundance and a Ward clustering algorithm. Boxed regions highlight metabolites whose levels were higher (red) or lower (blue) in the ΔclpC than in the WT groups, using a relative metabolite abundance scale of +2 (red) to −2 (blue). The HCA analysis indicates that, within these top 25 metabolites whose levels were significantly altered, a greater proportion of metabolites are found in higher levels in the ΔclpC mutant than in the WT. sn-Glycero-3-phos, sn-glycero-3-phosphocholine; Trimethylamine N-, trimethylamine N-oxide.
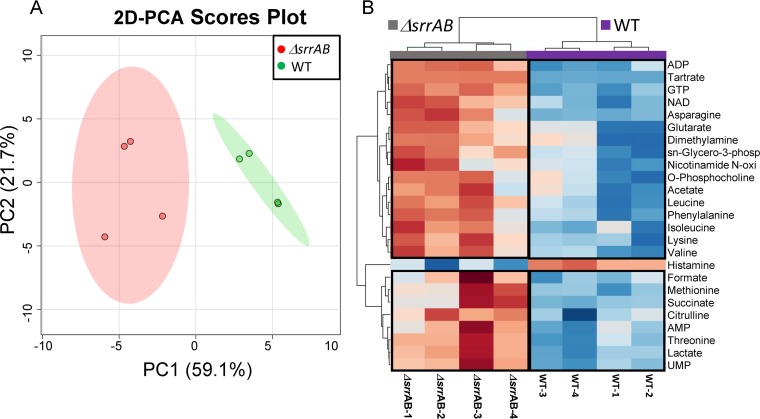
Similarly, the metabolic profile of the ΔsrrAB strain separates from that of WT in the 2D-PCA score plot, primarily in the PC1 dimension, with PC1 and PC2 accounting for 59% and 22%, respectively, of the variance (Fig. 5A). Similarly to the ΔclpC strain, the ADP, NAD+, acetate, formate, and lactate levels were higher in the ΔsrrAB strain (Fig. 5B). Levels of AMP, UMP, succinate, and citrulline were also increased in the ΔsrrAB strain. Importantly, metabolite abundance in the ΔsrrAB ΔclpC strain was nonadditive with respect to the metabolite levels quantified for the individual ΔsrrAB and ΔclpC mutations for ∼65% of the metabolites examined (25/39) (Table 1; see also Fig. S3). Notably, a number of the metabolites that displayed nonadditivity are crucial in the maintenance of cellular redox and energy homeostasis (for example, NAD+, NADP+, GTP, and ADP). Consistent with the NMR analyses, the NAD+/NADH ratios were significantly altered in strains lacking SrrAB or ClpC compared to that in the WT upon reexamination using a biochemical assay (Table 2). Moreover, the magnitude of change observed in the ΔsrrAB ΔclpC double mutant strain was statistically indistinguishable from those in the single ΔsrrAB strains. We do note that the metabolite level patterns were different between the ΔsrrAB, ΔclpC, and the ΔsrrAB ΔclpC strain mutants for ∼30% of the metabolites examined (Table S1). We conclude that while the ΔclpC and ΔsrrAB mutations do not influence central metabolism in exactly the same way, it is likely that a portion of the effect of SrrAB on metabolism is controlled via alterations in ClpC activity.
FIG 5.
2D-PCA score plot and hierarchical clustering analysis (HCA) of the metabolite profiles of the ΔsrrAB and WT strains following aerobic growth for 48 h. (A) The 2D-PCA score plot separates the metabolic profile of ΔsrrAB (JMB 1467) from that of WT (JMB 1100), with PC1 and PC2 accounting for 59% and 22% of the variance, respectively. (B) Heat map visualization of the top 25 metabolites whose level changes contributed the separation between the ΔsrrAB and WT groups. Boxed regions indicate metabolites whose levels were higher (red) or lower (blue) when comparing the ΔsrrAB and WT groups, using a relative metabolite abundance scale of +2 (red) to −2 (blue). Most of the metabolites contributing to the separation between the two groups were in higher relative abundance in the ΔsrrAB strain than in the WT strain. sn-Glycero-3-phosp, sn-glycero-3-phosphocholine; Nicotinamide N-oxi, nicotinamide N-oxide.
TABLE 1.
Relative abundances of select metabolite levels for which the ΔsrrAB and ΔclpC mutations are nonadditivea
| Metabolite | ΔclpC |
ΔsrrAB |
ΔsrrAB ΔclpC |
|||
|---|---|---|---|---|---|---|
| FC | P value | FC | P value | FC | P value | |
| Acetate | 1.6 ± 0.3 | 1.7E−03 | 1.4 ± 0.3 | 3.4E−02 | 1.7 ± 0.4 | 1.3E−03 |
| ADP | 1.9 ± 0.2 | 8.7E−05 | 1.8 ± 0.3 | 4.8E−04 | 1.6 ± 0.4 | 2.1E−03 |
| Asparagine | 1.2 ± 0.1 | 2.1E−04 | 1.5 ± 0.3 | 2.5E−03 | 1.3 ± 0.2 | 1.0E−02 |
| Citrulline | 1.3 ± 0.3 | 1.4E−01 | 1.4 ± 0.3 | 1.4E−02 | 0.7 ± 0.1 | 3.0E−03 |
| Formate | 1.4 ± 0.1 | 4.6E−05 | 1.2 ± 0.2 | 3.5E−02 | 1.2 ± 0.1 | 4.4E−03 |
| Glutarate | 0.5 ± 0.3 | 4.8E−02 | 2.4 ± 2.0 | 1.5E−02 | 0.9 ± 0.6 | 7.4E−01 |
| GTP | 0.8 ± 0.1 | 3.1E−02 | 1.4 ± 0.1 | 2.8E−05 | 0.5 ± 0.0 | 3.6E−06 |
| Lactate | 1.3 ± 0.1 | 7.3E−04 | 1.5 ± 0.3 | 1.6E−03 | 1.2 ± 0.3 | 2.0E−01 |
| Lysine | 1.2 ± 0.1 | 1.5E−02 | 1.3 ± 0.1 | 3.9E−03 | 1.2 ± 0.1 | 5.3E−03 |
| Methionine | 1.9 ± 0.2 | 6.5E−06 | 1.4 ± 0.2 | 1.1E−02 | 2.0 ± 0.4 | 2.7E−03 |
| NAD | 1.9 ± 0.2 | 2.1E−05 | 1.3 ± 0.2 | 1.4E−03 | 1.9 ± 0.2 | 4.2E−04 |
| NADP | 1.2 ± 0.3 | 3.4E−01 | 1.1 ± 0.3 | 4.9E−01 | 1.0 ± 0.2 | 8.5E−01 |
| N-Alpha-acetyllysine | 1.7 ± 0.6 | 3.6E−02 | 1.2 ± 1.0 | 5.8E−01 | 1.4 ± 1.1 | 1.9E−01 |
| Niacinamide | 1.7 ± 0.3 | 1.5E−03 | 1.2 ± 0.3 | 1.2E−01 | 1.6 ± 0.5 | 2.0E−02 |
| Nicotinamide N-oxide | 1.2 ± 0.1 | 1.7E−01 | 1.6 ± 0.7 | 2.9E−02 | 1.2 ± 0.3 | 2.0E−01 |
| O-Phosphocholine | 1.2 ± 0.3 | 1.7E−01 | 1.5 ± 0.5 | 3.0E−02 | 1.6 ± 0.5 | 1.1E−02 |
| Phenylalanine | 1.3 ± 0.1 | 6.4E−03 | 1.3 ± 0.2 | 6.3E−03 | 1.3 ± 0.2 | 5.2E−03 |
| sn-Glycero-3-phosphocholine | 1.6 ± 0.1 | 1.1E−03 | 1.3 ± 0.2 | 7.2E−03 | 1.1 ± 0.2 | 9.3E−02 |
| Succinate | 0.9 ± 0.0 | 2.6E−01 | 2.1 ± 0.9 | 1.7E−02 | 1.2 ± 0.5 | 3.3E−01 |
| Theophylline | 7.5 ± 0.7 | 2.5E−06 | 1.4 ± 0.6 | 3.5E−01 | 7.0 ± 1.9 | 3.2E−06 |
| Threonine | 1.3 ± 0.1 | 3.9E−03 | 1.5 ± 0.3 | 2.7E−03 | 1.2 ± 0.3 | 1.7E−01 |
| Trigonelline | 1.9 ± 0.7 | 1.5E−02 | 1.1 ± 1.3 | 8.9E−01 | 1.5 ± 1.1 | 1.1E−01 |
| Tryptophan | 1.3 ± 0.1 | 9.1E−03 | 1.2 ± 0.2 | 4.5E−02 | 1.3 ± 0.2 | 2.3E−03 |
| Tyrosine | 1.4 ± 0.1 | 5.9E−06 | 1.2 ± 0.2 | 3.6E−02 | 1.3 ± 0.2 | 2.3E−02 |
| UMP | 1.1 ± 0.1 | 1.1E−01 | 1.5 ± 0.3 | 2.7E−03 | 0.9 ± 0.2 | 1.5E−01 |
Data presented as fold changes (FC) and significance (P values) relative to the WT.
TABLE 2.
NAD+/NADH ratio after 48 h of growth
| Strain or genotype | NAD+ | NADH | NAD+/NADH ratioa |
|---|---|---|---|
| WT | 17.02 ± 1.75 | 0.23 ± 0.12 | 99.0 ± 15.3 |
| ΔclpC | 20.78 ± 0.98 | 0.18 ± 0.09 | 144.5 ± 19.4b |
| ΔsrrAB | 20.86 ± 1.13 | 0.34 ± 0.02 | 61.0 ± 3.5b |
| ΔsrrAB ΔclpC | 22.06 ± 1.35 | 0.46 ± 0.20 | 56.2 ± 6.0b |
Metabolite concentrations standardized to pmol per 108 bacteria before determining the ratios.
P < 0.05 versus WT by Students t tests.
ClpCP is required for optimal growth in a medium that only supports respiratory growth.
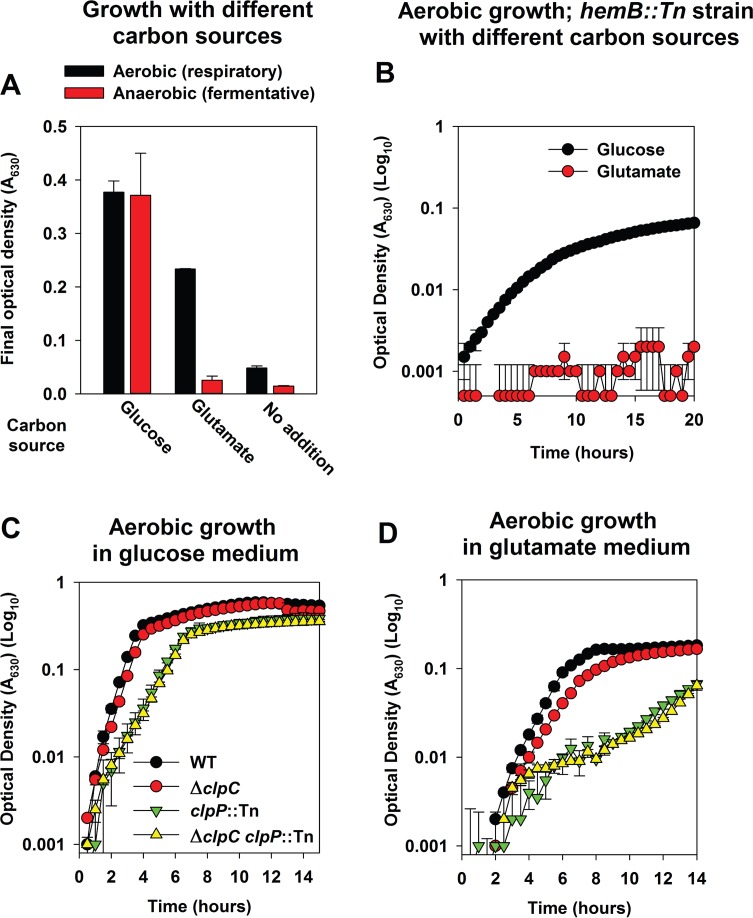
SrrAB regulates aerobic respiration, and the data presented suggested that a ΔclpC strain may be deficient in oxidative phosphorylation and tricarboxylic acid (TCA) cycle activity (35, 36). This prompted us to test the hypothesis that ClpC positively influences respiratory growth. We examined the growth profiles of strains in a medium that supports only respiratory growth. S. aureus is capable of utilizing glucose as well as glutamate as a source of carbon (47, 48). Glutamate feeds directly into the TCA cycle at the α-ketoglutarate entry point, and we reasoned that glutamate would support respiratory growth almost exclusively. Defined medium containing glutamate as a carbon source (DFM-glutamate) did not support fermentative (anaerobic) growth but did support respiratory (aerobic) growth (Fig. 6A). Defined medium with glucose as a carbon source (DFM-glucose) supported both respiratory and fermentative growth (Fig. 6A). Heme is necessary for the function of S. aureus terminal oxidases, and consequently, a heme auxotroph is incapable of respiring. A hemB::Tn strain was capable of aerobic growth in DFM-glucose but not DFM-glutamate medium (Fig. 6B), leading to the conclusion that DFM-glutamate supports only respiratory growth. The ΔclpC strain had a modest, but repeatable, growth defect in DFM-glutamate but not DFM-glucose medium (Fig. 6C and D; black versus red circles).
FIG 6.
The ClpCP complex is required for optimal growth in a medium that only supports respiratory growth. (A) S. aureus can utilize glutamate as a source of carbon aerobically but not fermentatively. Final growth yields are displayed for the WT (JMB 1100) cultured in the presence or absence of oxygen (fermentative growth) and in defined minimal medium containing the canonical 20 amino acids and glucose (DFM-glucose) or glutamate (DFM-glutamate) as a carbon source or in the absence of a carbon source. (B) A heme auxotroph is unable to utilize glutamate as a carbon source during aerobic growth. Growth profiles are displayed for a hemB::Tn (JMB 6037) strain cultured aerobically in either DFM-glucose or DFM-glutamate. (C and D) A strain lacking ClpC or ClpCP is substantially attenuated for aerobic growth upon DFM-glutamate. Growth profiles are displayed for the WT, ΔclpC (JMB 8025), clpP::Tn (JMB 4898), and ΔclpC clpP::Tn (JMB 8029) strains cultured aerobically in either DFM-glucose (C) or DFM-glutamate (D). Data in all panels represent the averages from duplicate cultures and error bars represent standard deviations. Error bars are displayed for all points but may be too small to see on occasion.
ClpC can associate with a peptidase that forms a proteolytic complex. S. aureus harbors the ClpP and ClpQ peptidases. To understand which peptidase ClpC associates with to influence respiratory growth, the puromycin sensitivities of strains lacking ClpC, ClpP, or ClpQ were assessed. Concentrations of puromycin that attenuated growth of a ΔclpC strain also attenuated growth of a clpP::Tn strain, while the growth of a clpQ::Tn strain was unaffected (data not shown). This suggested that ClpC associates with ClpP; therefore, we conducted epistasis experiments to further understand the role of ClpP.
The growth of a ΔclpC clpP::Tn strain was examined relative to that of its parental strains in different media. The phenotypic effects of the ΔclpC and clpP::Tn mutations were nonadditive, and the ΔclpC clpP::Tn strain phenocopied the clpP::Tn strain in both DFM-glucose and DFM-glutamate media (Fig. 6C and D). The growth of the clpP::Tn strain was attenuated to a greater degree than for the ΔclpC strain in both media (Fig. 6C and D).
ClpP is required for optimal fermentative growth while ClpC is dispensable.
SrrAB positively influences fermentative growth (37). A previous study found that the transcription of clpP but not clpC is increased upon a shift to fermentative growth (49). In conjunction with the metabolomics data presented, we reasoned that ClpC was not required for fermentative growth and that SrrAB would not modulate clpC transcription during fermentative growth.
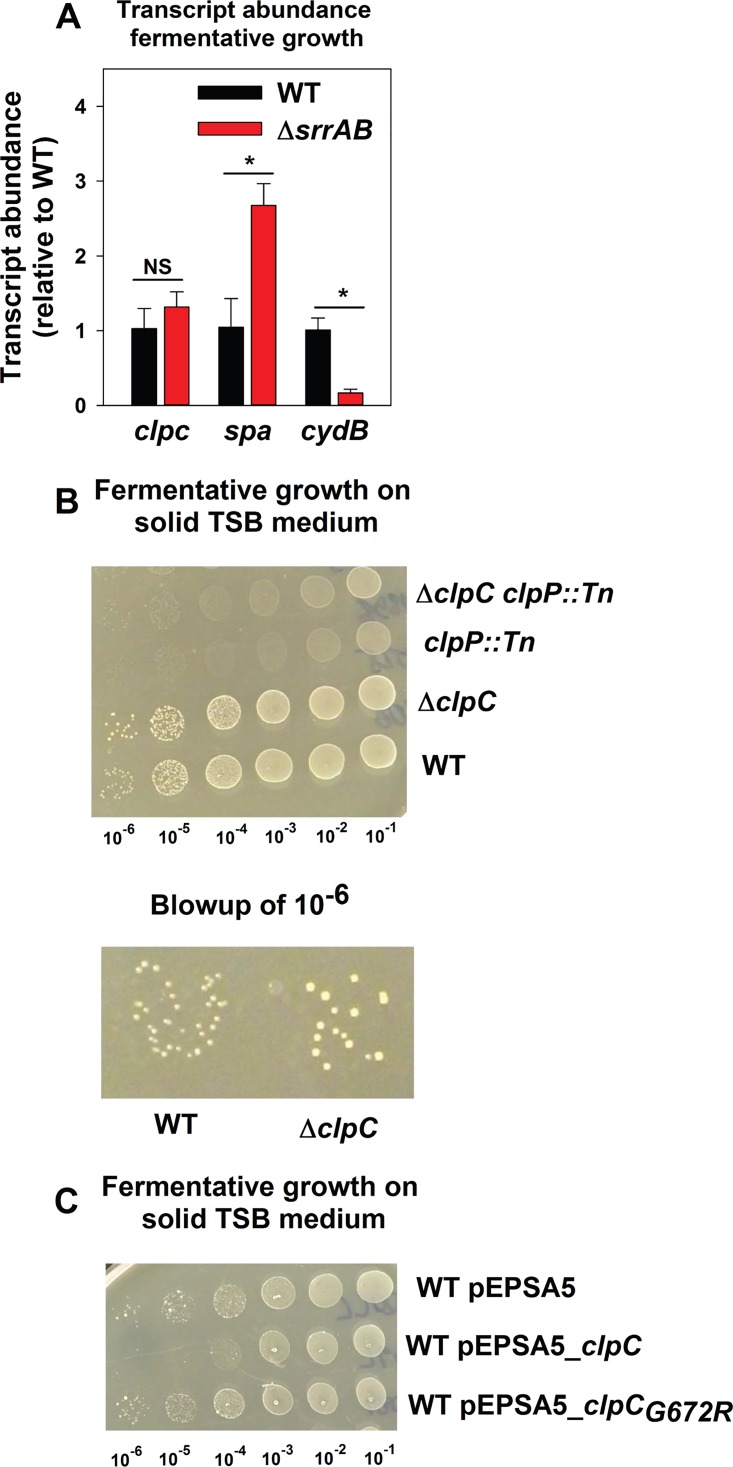
The transcript levels for clpC were examined in the WT and ΔsrrAB strains following fermentative growth. Transcript levels for spa and cydB, which are negatively and positively modulated by SrrAB, were assessed as controls (33, 35). Transcript levels were examined instead of transcriptional activity using the reporter constructs, since the folding of GFP is impaired in the absence of oxygen (50). Transcript levels for spa and cydB were increased and decreased, respectively, in the ΔsrrAB strain (Fig. 7A). However, transcription of clpC was unaltered (Fig. 7A).
FIG 7.
ClpP is required for optimal fermentative growth, while ClpC is dispensable. (A) SrrAB influences transcript levels for spa and cydB but not clpC during fermentative growth. The WT (JMB 1100) and ΔsrrAB (JMB 1467) strains were cultured fermentatively, mRNA was extracted, and the abundances of the clpC, spa, and cydB transcripts were quantified. The data were normalized to 16S rRNA levels and thereafter to levels observed in the WT. (B) ClpP is required for optimal fermentative growth, while ClpC is dispensable. Fermentative growth is displayed for the WT, ΔclpC (JMB 8025), clpP::Tn (JMB 4898), and ΔclpC clpP::Tn (JMB 8029) strains on solid TSB medium. (C) Increased expression of clpC inhibits growth of fermenting S. aureus. Fermentative growth is displayed for the WT strain with pEPSA5 (empty vector), pEPSA5_clpC, or pEPSA5_clpCG672R on solid TSB medium. Data in panel A represent the averages from triplicate cultures and error bars represent standard deviations. Photographs in panels B and C are representative of at least three independent experiments, and the numbers beneath each photograph denote the serial dilution that cells were removed from before plating. Where indicated, Student t tests (two tailed) were performed on the data. *, P < 0.05.
We examined whether ClpC, ClpP, or the ClpCP complex had a role during fermentative growth. Relative to that of the WT, the growth of the clpP::Tn strain was severely attenuated during fermentative culture (Fig. 7B). The ΔclpC strain was more proficient in fermentative growth than the WT and consistently formed bigger colonies (∼65% larger; relative sizes presented in Fig. S4, and representative image presented in Fig. 7B), suggesting ClpC limits fermentative growth. The phenotypic effects of the ΔclpC and clpP::Tn mutations were nonadditive (Fig. 7B).
The glycine at position 672 of ClpC is required for the interaction between ClpC and ClpP (51). The introduction of pEPSA5_clpC but not pEPSA5_clpCG672R inhibited fermentative growth of the WT strain (Fig. 7C). The WT strain carrying pEPSA5_clpC formed ∼100-fold fewer colonies that the WT carrying empty vector (Fig. 7C). The WT strain carrying pESPSA5_clpCG672R behaved similarly to the WT strain carrying empty vector (Fig. 7C). From Fig. 7, we concluded that ClpC is dispensable for fermentative growth, while ClpP is required. Further, dysregulation of ClpC levels during fermentative growth inhibits growth, which is likely due its interference with ClpP function(s).
DISCUSSION
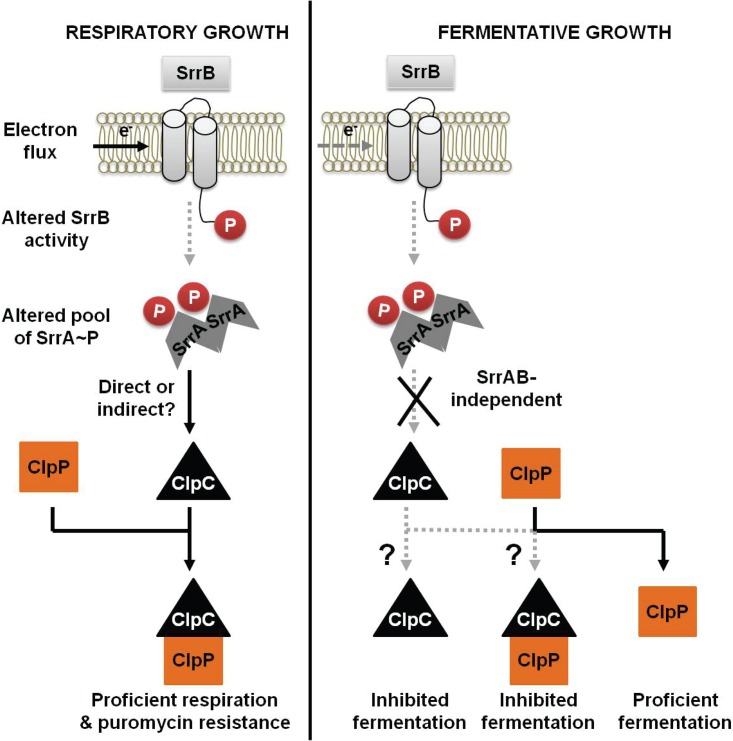
Energy homeostasis is crucial for cellular physiology. SrrAB modulates aerobic respiration as well as fermentation in S. aureus (36, 37). Regulated proteolysis is involved in modulating respiratory metabolism in alternate organisms (18, 21); however, it is unclear whether a similar mechanism exists in S. aureus. We tested the hypothesis that the SrrAB TCRS links energy homeostasis and regulates proteolysis by modulating the levels of a factor involved in protein turnover. Our analyses using a combination of genetics and metabolomics lead us to propose a model wherein SrrAB affects puromycin resistance and energy homeostasis, at least in part, by modulating ClpC levels, which functions in protein turnover (Fig. 8). Consistent with our model, the growth of a ΔsrrAB strain was deficient on media that impose a high demand for the cellular proteolytic machinery. Moreover, the transcriptional activity for clpC was decreased in a strain lacking SrrAB during respiratory growth, and overexpression of clpC suppressed the puromycin sensitivity phenotype of the srrAB mutant. We also note that early studies by Throup et al. (37) suggest that ClpC levels were altered in a srrAB mutant. One interpretation of this is that SrrAB regulates particular oxidative phosphorylation factors in a posttranslational manner. This idea would be consistent with our findings presented herein.
FIG 8.
A working model for the influence of respiration upon SrrAB-dependent regulation of ClpC in S. aureus. Respiratory growth results in increased expression of clpC in an SrrAB-dependent manner. The increased ClpC leads to increased activity of the ClpCP proteolytic machinery and thereby facilitates proficient respiration and resistance toward protein misfolding stress. In contrast, during fermentative growth, SrrAB does not alter clpC expression and ClpP is necessary for proficient growth, while ClpC limits growth. Further experimentation is required to understand whether ClpC limits growth fermentatively solely upon association with ClpP or if it functions via an alternate mechanism.
To understand whether SrrAB modulation of ClpC affects energy homeostasis, we conducted NMR-based metabolomics analyses. Our studies revealed that the ΔsrrAB and ΔclpC mutations both result in altered concentrations of metabolites involved in balancing cellular redox and energy or of amino acids that serve as precursors for the TCA cycle. Importantly metabolite analyses in a ΔsrrAB ΔclpC double mutant strain found that the effects of the ΔclpC and ΔsrrAB mutations are not additive for ∼65% of the metabolites examined (25/39). A number of these metabolites serve as reporters of cellular energy status such as (ADP and NAD+) and respiratory growth (acetate, formate, and lactate). Moreover, the growth of the ΔclpC strain was deficient on a medium that supported only respiratory growth. Taken together, these data are highly suggestive of a role for ClpC in directing energy metabolism. However, we do note that the ΔsrrAB ΔclpC double mutant strain did not completely phenocopy the ΔclpC and ΔsrrAB single mutants. Thus, while our data are supportive of our hypothesis that SrrAB affects energy homeostasis via ClpC, we do not rule out the possibility that mutations in either gene could have direct or indirect (or both) effects on metabolism. Alternatively, the results could arise as an indirect consequence of altering regulated proteolysis, which subsequently results in global changes in the cell.
The growth conditions of our NMR-based metabolomics experiments were selected to allow comparisons with previous studies conducted on ClpC function using transcriptomics and proteomics. Chatterjee et al. found that a strain lacking ClpC had decreased transcript levels for genes encoding NADH dehydrogenases and 2-oxoglutarate ferredoxin oxidoreductase (46). They also found that the protein levels for the ATP synthase subunits were decreased in abundance in a clpC mutant (46). These data align well with our results indicating that a ΔclpC strain alters the NAD/NADH ratio in the cell. A separate study found that a number of proteins that are involved in the synthesis of cofactors, which are essential for respiration, are substrates for ClpC (52). Prominent among these were SufCD, which are required to synthesize iron-sulfur (Fe-S) cofactors (52). S. aureus strains unable to properly process Fe-S proteins into their mature forms display global shifts in carbon flux and metabolic defects (47, 51, 53–55). Consistent with these studies, we found that a clpC mutant had deficient respiratory growth. ClpC acts either independently as a molecular chaperone or in conjunction with the ClpP peptidase. A clpP::Tn strain had attenuated growth on media supporting only respiratory growth, and the phenotypic effects of the ΔclpC and clpP::Tn mutations were not additive. Thus, we concluded that the ClpCP proteolytic complex functions in directing respiratory metabolism. These findings are consistent with previous observations that several proteins involved in energy metabolism may be direct substrates of the ClpCP system (56).
We also examined the roles of ClpC, ClpP, and ClpCP during fermentative growth. ClpP was required for optimal growth during fermentative culture. Interestingly, the ΔclpC strain consistently formed bigger colonies than the WT strain when growing fermentatively but not aerobically. This suggested that the presence of ClpC limits fermentative growth. SrrAB did not affect clpC transcript levels during fermentative growth. Our metabolic profile analyses support this observation, as elevated levels of fermentative products/metabolites were measured in the ΔclpC mutant compared to that in the WT. These data are also consistent with previous findings that the transcription of clpP but not clpC is increased upon transition to fermentative growth (49). Overexpression of clpC but not clpCG627R inhibited fermentative growth of the WT strain. Therefore, we propose that dysregulated levels of ClpC interfere with ClpP function and thereby limit fermentative growth. However, future experiments are necessary to uncover the precise mechanism by which overexpression of clpC decreases fermentative growth.
In summary, this study demonstrates that both SrrAB and ClpC affect energy metabolism. The data presented suggest that SrrAB influences energy metabolism, in part, by modulating ClpCP-directed regulated proteolysis. Metabolomics and physiological analyses establish that ClpCP is involved in altering aerobic respiratory metabolism but is dispensable for fermentative growth.
MATERIALS AND METHODS
Materials.
Restriction enzymes, deoxynucleoside triphosphates, quick DNA ligase kit, and Phusion DNA polymerase were purchased from New England BioLabs. The plasmid miniprep kit, RNAprotect, and the gel extraction kit were purchased from Qiagen. Lysostaphin was purchased from Ambi Products. DNase I was purchased from Ambion. High-Capacity cDNA reverse transcription kits and TRIzol were purchased from Life Technologies. Oligonucleotides were purchased from Integrated DNA Technologies, and sequences are listed in Table S2 in the supplemental material. Tryptic soy broth (TSB) was purchased from MP Biomedicals. Unless specified, all chemicals were purchased from Sigma-Aldrich and were of the highest purity available.
Bacterial growth conditions.
Unless specifically stated otherwise, the S. aureus strains used in this study (Table 3) were constructed in the community-associated USA300 strain LAC that was cured of the native plasmid pUSA03, which confers erythromycin resistance (57). S. aureus strains were cultured at 37°C. For aerobic growth in liquid, cultures were grown with shaking at 200 rpm at a flask/tube headspace-to-culture medium volume ratio of 10 (with the exception of analyses conducted in 96-well microtiter plates). Anaerobic growth was achieved by either incubation with a flask/tube headspace-to-culture medium volume ratio of 0, as described earlier (47, 53), or by incubation in a Coy anaerobic chamber equipped with an oxygen-scavenging catalyst to maintain oxygen levels lower than 1 ppm. For culture in 96-well microtiter plates, each well contained 200 μl total volume (detailed procedure below). Difco BioTek agar was added (15 g liter−1) for solid medium. The staphylococcal defined medium recipe was as described previously and contained 10 mg ml−1 (NH4)2SO4, 45 mg ml−1 KH2PO4, 105 mg ml−1 K2HPO4, 6.42 mg ml−1 NaCl, 2.23 mg ml−1 KCl, 0.5 μg ml−1 nicotinic acid, 0.5 μg ml−1 thiamine, 0.5 μg ml−1 pantothenic acid, 3 ng ml−1 biotin, and 0.25 ng ml−1 of each individual amino acid (47, 53). All components were prepared using distilled and deionized water, but the water added to the medium was only deionized. Glutamate and glucose were added as carbon sources at 44 mM and 22 mM, respectively. When selecting for plasmids or episome insertions, antibiotics were added at the final following concentrations: 150 μg ml−1 ampicillin, 30 μg ml−1 chloramphenicol (Cm), 10 μg ml−1 erythromycin (Erm), and 3 μg ml−1 tetracycline (Tet). For routine plasmid maintenance, media were supplemented with 10 μg ml−1 or 3.3 μg ml−1 of chloramphenicol or erythromycin, respectively.
TABLE 3.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Genetic background | Source and/or reference |
|---|---|---|---|
| Strains | |||
| S. aureus | |||
| JMB 1100 | USA300_LAC (Erm sensitive), MRSA, USA300, CC8 | LAC | 57 |
| JMB 1467 | ΔsrrAB (SAUSA300_1441-42) | LAC | 62 |
| JMB 2047 | ΔsrrAB::tet | LAC | 35 |
| JMB 1103 | Restriction minus, MSSA, CC8 | RN4220 | 63 |
| JMB 1422 | Parent, MSSA, CC8 | Newman | 64 |
| JMB 4751 | ΔsrrAB::tet | Newman | This work |
| JMB 1324 | Parent, MRSA, USA400, CC1 | MW2 | 65 |
| JMB 7573 | ΔsrrAB::tet | MW2 | This work |
| JMB 7570 | Parent, MRSA, USA100, CC5 | N315 | 66 |
| JMB 7574 | ΔsrrAB::tet | N315 | This work |
| JMB 4898 | clpP::Tn(erm) | LAC | BEI Resources, 67 |
| JMB 8025 | ΔclpC::tet | LAC | 51 |
| JMB 8027 | ΔsrrAB ΔclpC::tet | LAC | This work |
| JMB 8029 | ΔclpC::tet clpP::Tn(erm) | LAC | This work |
| JMB 7714 | clpQ::Tn(erm) | LAC | BEI Resources, 67 |
| JMB 4850 | clpL::Tn(erm) | LAC | BEI Resources, 67 |
| JMB 6037 | hemB::Tn(erm) | LAC | BEI Resources, 67 |
| Escherichia coli PX5 | Protein Express | ||
| Saccharomyces cerevisiae FY2 | W. Belden | ||
| Plasmids | |||
| pCM28 | Insertless cloning vector, genetic complementation, multicopy | 62 | |
| pCM11 | Parent vector for construction of transcriptional reporter, multicopy | 68 | |
| pCM11_PclpC | clpC transcriptional reporter | This work | |
| pCM11_Pspa | spa transcriptional reporter | This work | |
| pCM28_srrAB | srrAB complementation, multicopy | 35 | |
| pLL39 | Insertless cloning vector, genetic complementation, episome | 69 | |
| pLL39_srrAB | srrAB complementation, episome | 40 | |
| pEPSA5 | Cloning vector, genetic complementation, xylose-inducible promoter, multicopy | 70 | |
| pEPSA5_clpC | clpC complementation, multicopy | 51 | |
| pEPSA5_clpCG672R | clpC complementation, multicopy | 51 |
(i) Liquid growth analyses.
Strains were cultured overnight in TSB (∼18 h of growth) and subsequently inoculated in minimal medium to a final optical density (OD) of 0.02 (A600) unit. For assessing nutritional requirements in chemically defined medium, the cell pellet was washed twice with phosphate-buffered saline (PBS) prior to inoculation to prevent carryover of rich medium components.
Puromycin sensitivities were examined in TSB medium, and the media were amended with antibiotic at the point of inoculation. The puromycin concentrations ranged between ∼0.5 and 10 μg ml−1. Aerobic growth was monitored using a BioTek 808E visible absorption spectrophotometer equipped with an incubator and set at medium shake speed. For anaerobic growth, the microtiter plate was incubated in an air incubator inside a Coy anaerobic chamber. Where final optical densities are presented, growth was assessed after overnight growth (∼18 h).
(ii) Solid growth analyses.
Strains were cultured overnight in TSB (∼18 h of growth). The strains were serial diluted using 1× PBS, and 5-μl aliquots of the dilutions were spot plated on solid medium. Where mentioned, the TSB solid medium was amended with between 4.5 and 7 μg ml−1 of puromycin. Plates were subsequently incubated at 37°C overnight before photographs were taken. Where mentioned, anaerobic growth was achieved by incubation in an air incubator inside a Coy anaerobic chamber.
(iii) Colony size analyses.
Relative colony sizes were determined using the particle size tool in the ImageJ software. Sizes for at least ten colonies for each strain were determined.
Recombinant DNA and genetic techniques.
Plasmids were passaged through RN4220 and subsequently transduced into the appropriate strains using bacteriophage 80α (58). Mutant strains and plasmids were verified using PCR or by sequencing PCR products or plasmids. DNA sequencing was performed at Genewiz (South Plainfield, NJ).
Creation of plasmids and mutant strains.
pCM11_clpC was created using the clpC hindIII and clpC kpnI primer pair. pCM11_spa was made using the spa Pro for HindIII and spa Pro Rev KpnI primer pair. Digested PCR products were ligated into similar digested vectors.
Transcriptional reporter fusion assay.
Strains cultured overnight in liquid TSB-Erm medium were diluted in fresh liquid TSB-Erm medium to a final OD of 0.1 (A600) and cultured with shaking. At periodic intervals, culture density and fluorescence were assessed as described previously (47, 53). Fluorescence data were normalized with respect to a strain not carrying a GFP-based transcriptional reporter to normalize for background fluorescence values. The resulting data were normalized to the culture OD. Finally, the data were normalized relative to the wild-type (WT) strain, or as specified in the figure legend.
RNA extractions and real-time quantitative PCR.
The abundances of RNAs were determined using a previously described cDNA library (35, 53). Briefly, cells were cultured in capped microcentrifuge tubes at a headspace-to-volume (H/V) ratio of 0. Anaerobic conditions were verified by the addition of resazurin to control tubes. Cultures were grown for 4.5 h. Thereafter, cells were treated with RNAprotect reagent, mRNA was obtained, cDNA libraries were constructed, and real-time quantitative PCR (RT-qPCR) was performed as described earlier (35, 53).
NMR metabolomics.
(i) Bacterial strains and growth conditions. Aerobic growth for all strains was assessed on four biological replicates for each cell group. Overnight cultures diluted 1:1,000 were used to inoculate 25 ml of fresh TSB in a 250-ml flask with 220-rpm agitation at 37°C. Aliquots of 10 ml were collected at 48 h, centrifuged at 5,000 rpm for 5 min, rinsed once with 1 ml of 1× PBS, and centrifuged at 5,000 rpm for 5 min. The supernatant was discarded, and cell pellets were frozen at −80°C until further use. An additional 10 μl of the culture was utilized to determine the CFU, and 5 ml was utilized for NAD+/NADH assays.
(ii) Polar metabolite extraction. Frozen cell pellets were resuspended in 1 ml of a 2:1 methanol-chloroform mixture and transferred to FastPrep lysis B matrix tubes (MP Biomedicals). Cells were lysed using the FastPrep-24 5G instrument and designated S. aureus settings (2 cycles at a speed of 6.0 m/s for 40 s); 300 μl of each layer of a 1:1 aqueous chloroform solution was added to each cell lysate. The tubes were vortexed, placed at −20°C for 20 min, and centrifuged at 14,000 × g for 10 min. 800 μl of the aqueous phase was transferred to microcentrifuge tubes and placed in a SpeedVac (no heat, manual run, volatile solvent) to dry overnight. Samples were resuspended in 600 μl of NMR buffer (0.25 mM 4,4-dimethyl-4-silapentane-1-sulfonic acid [DSS], 0.4 mM imidazole, 25 mM phosphate buffer, 90% H2O–10% D2O) and transferred to 5-mm Bruker NMR tubes.
(iii) NMR experiments. 1D 1H NMR spectra for each sample were obtained at 298 K on a Bruker 600-MHz (1H Larmor frequency) AVANCE III solution NMR spectrometer, equipped with an automatic SampleJet sample loading system as well as a 5-mm triple resonance (1H, 15N, and 13C) liquid helium-cooled TCI probe (cryoprobe) and Topspin software (Bruker version 3.2). 1D 1H experiments were performed using the “zgesgp” Bruker pulse sequence with 256 scans, 1H spectral window of 9,615.38 Hz. Free induction decays (FIDs) were collected in 32K data points, with a dwell time interval of 52 μs amounting to an acquisition time of ∼1.7 s, and using an additional 1-s relaxation recovery delay between spectrum acquisitions. For each sample, the Topspin software (Bruker version 3.2) was used for phasing, baseline correction, and suppression of the water NMR signal using the baseline correction module “qfil” with a filter width (BCFW) of 0.2 ppm. Spectral analysis and metabolite identification and quantitation were performed using Chenomx software (version 8.0) (Chenomx Inc., Edmonton, AB, Canada) (59). For each NMR spectrum, the baseline was further corrected prior to metabolite identification and quantitation using the Chenomx small-molecule spectral database for 600-MHz (1H Larmor frequency) magnetic field strength NMR spectrometers. NMR spectral patterns were fitted for each sample independently, and an internal DSS (0.25 mM) standard was used for metabolite quantitation. Metabolite concentrations were further normalized to viable cell counts (i.e., CFU). Metabolite identification (ID), when ambiguous due to partial 1H NMR signal overlap, was further confirmed by recording 2D 1H-1H total correlation spectroscopy (TOCSY) NMR spectra or by spiking, when available, pure metabolite standards into samples. A total of 42 compounds were identified upon analysis of the 1D 1H NMR spectra, and concentrations reported in millimolar were exported to Excel spreadsheets for further analysis using the MetaboAnalyst software (60).
(iv) Multivariate data analysis. Metabolite concentrations (in mM) for all four biological replicates obtained from the Chenomx software data analysis were normalized to the total number of viable cells as determined by CFU, converted to attomoles/CFU, and saved as a CSV file. These data sets were analyzed, and metabolic profile trends between different sample groups were assessed using multivariate statistical analysis methods, including unsupervised 2D principal-component analysis (2D-PCA) and supervised 2D partial least-squares discriminant (2D PLS-DA) analyses using the multivariate statistical analysis modules of MetaboAnalyst (60). All metabolite concentration data sets were imported into MetaboAnalyst using log transformation and autoscaling settings. Log transformation was used to ensure Gaussian distribution of the data, and autoscaling was used for data scaling. This approach ensures that the variance from the more abundant metabolites does not dominate the variance-covariance matrix of the multivariate statistical analysis and ensures that all changes from metabolites whose concentrations span several orders of magnitude contribute to the analysis, as described in reference 61.
(v) Metabolic pathway analysis/VIP scores. The most important metabolites contributing to the variance were ranked by 2D PLS-DA VIP (variable importance in projection) scores. Metabolites with VIP scores of ≥1.0 were considered the most significant contributors to the separation of the metabolic profiles of the different S. aureus sample groups, as shown in the 2D PLS-DA score plots. VIP scores resulting from the MetaboAnalyst data analysis are shown in Fig. 4B.
(vi) Data interpretation and analyses. Heat maps were generated using the Cluster Analysis module in MetaboAnalyst. Except for displaying the top 25 metabolites using the t test/analysis of variance (ANOVA) setting, default settings were used (Euclidian distance measure and Ward clustering algorithm). During analyses, data were curated using the interquartile range (IQR) rule. The lower and upper outlier bounds were determined by 1.5 IQR subtraction from the first quartile and 1.5 IQR addition to the third quartile, respectively. Data values falling outside these bounds were omitted as outliers.
NAD/NADH analysis.
Total NAD+ and NADH concentrations were determined using 5-ml aliquots of cell culture and standard NAD+/NADH enzymatic quantitation kits (BioVision, K337-100) according to the manufacturer’s protocol and by normalizing resultant data to viable cell counts (i.e., CFU). After 48 h of culture, cells were washed with 1× PBS and lysed via 2 cycles of freeze/thaw in NADH/NAD+ extraction buffer. The resulting supernatant was split into two reactions for each sample, and one was used to measure the concentration of NADH present and the other to measure total NAD+ (NADt) corresponding to [NADH] + [NAD+]. NADH samples were heated to 60°C for 30 min to remove any NAD+ present. NADH and NADt samples were then loaded into 96-well plates, and NAD+ cycling enzyme mix was added. The reactions were allowed to incubate at room temperature for 5 min prior to the addition of developer mix, and the reactions were allowed to sit for 1 h prior to the measurement of optical density at 450 nm. Standard curves were generated using standards of 10 pmol/μl NADH and NAD+ provided by the manufacturer. NAD+/NADH concentration measurements were conducted on four biological replicates for each sample group.
Data availability.
We have deposited all the NMR metabolomics data and associated data processing files into the Metabolomics Workbench database under data accession number ST001174.
Supplementary Material
ACKNOWLEDGMENTS
The Boyd lab is supported by the Charles and Johanna Busch foundation, USDA MRF project NE-1028, and grant 1R01AI139100-01 from the NIAID. A.A.M. was supported by the Douglas Eveleigh fellowship from the Microbial Biology Graduate Program and an Excellence Fellowship from Rutgers University. C.A.E. was supported by a Rutgers Aresty undergraduate research fellowship. The research in the Copié lab was supported by a grant from the Montana University System Research Initiative (51040-MUSRI2015-03). The NMR metabolomics studies were conducted on Montana State University"s (MSU’s) 600-MHz Bruker AVANCE III solution NMR spectrometer. Support for the NMR instruments, console upgrades, and MSU’s NMR Center has been provided by the NIH Shared Instrumentation Grant (SIG) program (grant numbers 1S10RR13878 and 1S10RR026659), the NSF MRI program (NSF grant number DBI-1532078), the Murdock Charitable Trust, and MSU’s Vice President for Research Economic Development’s office.
We thank William Belden for use of his real-time thermocycler.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00188-19.
REFERENCES
- 1.Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, Spratt BG. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc Natl Acad Sci U S A 99:7687–7692. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graham PL III, Lin SX, Larson EL. 2006. A U.S. population-based survey of Staphylococcus aureus colonization. Ann Intern Med 144:318–325. doi: 10.7326/0003-4819-144-5-200603070-00006. [DOI] [PubMed] [Google Scholar]
- 3.Ohara-Nemoto Y, Haraga H, Kimura S, Nemoto TK. 2008. Occurrence of staphylococci in the oral cavities of healthy adults and nasal oral trafficking of the bacteria. J Med Microbiol 57:95–99. doi: 10.1099/jmm.0.47561-0. [DOI] [PubMed] [Google Scholar]
- 4.Zafar U, Johnson LB, Hanna M, Riederer K, Sharma M, Fakih MG, Thirumoorthi MC, Farjo R, Khatib R. 2007. Prevalence of nasal colonization among patients with community-associated methicillin-resistant Staphylococcus aureus infection and their household contacts. Infect Control Hosp Epidemiol 28:966–969. doi: 10.1086/518965. [DOI] [PubMed] [Google Scholar]
- 5.Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, Craig AS, Zell ER, Fosheim GE, McDougal LK, Carey RB, Fridkin SK. 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 6.Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG Jr. 2015. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev 28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williamson DA, Lim A, Thomas MG, Baker MG, Roberts SA, Fraser JD, Ritchie SR. 2013. Incidence, trends and demographics of Staphylococcus aureus infections in Auckland, New Zealand, 2001-2011. BMC Infect Dis 13:569. doi: 10.1186/1471-2334-13-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Said-Salim B, Mathema B, Kreiswirth BN. 2003. Community-acquired methicillin-resistant Staphylococcus aureus: an emerging pathogen. Infect Control Hosp Epidemiol 24:451–455. doi: 10.1086/502231. [DOI] [PubMed] [Google Scholar]
- 9.Tenover FC, McDougal LK, Goering RV, Killgore G, Projan SJ, Patel JB, Dunman PM. 2006. Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. J Clin Microbiol 44:108–118. doi: 10.1128/JCM.44.1.108-118.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mashruwala AA, Gries CM, Scherr TD, Kielian T, Boyd JM. 2017. SaeRS is responsive to cellular respiratory status and regulates fermentative biofilm formation in Staphylococcus aureus. Infect Immun 85:e00157-17. doi: 10.1128/IAI.00157-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnold F, West D, Kumar S. 1987. Wound healing: the effect of macrophage and tumour derived angiogenesis factors on skin graft vascularization. Br J Exp Pathol 68:569–574. [PMC free article] [PubMed] [Google Scholar]
- 12.Carreau A, El Hafny-Rahbi B, Matejuk A, Grillon C, Kieda C. 2011. Why is the partial oxygen pressure of human tissues a crucial parameter? Small molecules and hypoxia. J Cell Mol Med 15:1239–1253. doi: 10.1111/j.1582-4934.2011.01258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vogelberg KH, Konig M. 1993. Hypoxia of diabetic feet with abnormal arterial blood flow. Clin Investig 71:466–470. [DOI] [PubMed] [Google Scholar]
- 14.Wilde AD, Snyder DJ, Putnam NE, Valentino MD, Hammer ND, Lonergan ZR, Hinger SA, Aysanoa EE, Blanchard C, Dunman PM, Wasserman GA, Chen J, Shopsin B, Gilmore MS, Skaar EP, Cassat JE. 2015. Bacterial hypoxic responses revealed as critical determinants of the host-pathogen outcome by TnSeq analysis of Staphylococcus aureus invasive infection. PLoS Pathog 11:e1005341. doi: 10.1371/journal.ppat.1005341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammer ND, Reniere ML, Cassat JE, Zhang Y, Hirsch AO, Indriati Hood M, Skaar EP. 2013. Two heme-dependent terminal oxidases power Staphylococcus aureus organ-specific colonization of the vertebrate host. mBio 4:e00241-13. doi: 10.1128/mBio.00241-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vitko NP, Grosser MR, Khatri D, Lance TR, Richardson AR. 2016. Expanded glucose import capability affords Staphylococcus aureus optimized glycolytic flux during infection. mBio 7:e00296-16. doi: 10.1128/mBio.00296-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richardson AR, Libby SJ, Fang FC. 2008. A nitric oxide-inducible lactate dehydrogenase enables Staphylococcus aureus to resist innate immunity. Science 319:1672–1676. doi: 10.1126/science.1155207. [DOI] [PubMed] [Google Scholar]
- 18.Gottesman S. 1999. Regulation by proteolysis: developmental switches. Curr Opin Microbiol 2:142–147. doi: 10.1016/S1369-5274(99)80025-3. [DOI] [PubMed] [Google Scholar]
- 19.Frees D, Savijoki K, Varmanen P, Ingmer H. 2007. Clp ATPases and ClpP proteolytic complexes regulate vital biological processes in low GC, Gram-positive bacteria. Mol Microbiol 63:1285–1295. doi: 10.1111/j.1365-2958.2007.05598.x. [DOI] [PubMed] [Google Scholar]
- 20.Michalik S, Liebeke M, Zuhlke D, Lalk M, Bernhardt J, Gerth U, Hecker M. 2009. Proteolysis during long-term glucose starvation in Staphylococcus aureus COL. Proteomics 9:4468–4477. doi: 10.1002/pmic.200900168. [DOI] [PubMed] [Google Scholar]
- 21.van Dijl JM, Kutejova E, Suda K, Perecko D, Schatz G, Suzuki CK. 1998. The ATPase and protease domains of yeast mitochondrial Lon: roles in proteolysis and respiration-dependent growth. Proc Natl Acad Sci U S A 95:10584–10589. doi: 10.1073/pnas.95.18.10584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki CK, Suda K, Wang N, Schatz G. 1994. Requirement for the yeast gene LON in intramitochondrial proteolysis and maintenance of respiration. Science 264:891. doi: 10.1126/science.8178144. [DOI] [PubMed] [Google Scholar]
- 23.Frees D, Thomsen LE, Ingmer H. 2005. Staphylococcus aureus ClpYQ plays a minor role in stress survival. Arch Microbiol 183:286–291. doi: 10.1007/s00203-005-0773-x. [DOI] [PubMed] [Google Scholar]
- 24.Frees D, Qazi SN, Hill PJ, Ingmer H. 2003. Alternative roles of ClpX and ClpP in Staphylococcus aureus stress tolerance and virulence. Mol Microbiol 48:1565–1578. doi: 10.1046/j.1365-2958.2003.03524.x. [DOI] [PubMed] [Google Scholar]
- 25.Kim YI, Levchenko I, Fraczkowska K, Woodruff RV, Sauer RT, Baker TA. 2001. Molecular determinants of complex formation between Clp/Hsp100 ATPases and the ClpP peptidase. Nat Struct Biol 8:230–233. doi: 10.1038/84967. [DOI] [PubMed] [Google Scholar]
- 26.Singh SK, Rozycki J, Ortega J, Ishikawa T, Lo J, Steven AC, Maurizi MR. 2001. Functional domains of the ClpA and ClpX molecular chaperones identified by limited proteolysis and deletion analysis. J Biol Chem 276:29420–29429. doi: 10.1074/jbc.M103489200. [DOI] [PubMed] [Google Scholar]
- 27.Gohring N, Fedtke I, Xia G, Jorge AM, Pinho MG, Bertsche U, Peschel A. 2011. New role of the disulfide stress effector YjbH in beta-lactam susceptibility of Staphylococcus aureus. Antimicrob Agents Chemother 55:5452–5458. doi: 10.1128/AAC.00286-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jousselin A, Kelley WL, Barras C, Lew DP, Renzoni A. 2013. The Staphylococcus aureus thiol/oxidative stress global regulator Spx controls trfA, a gene implicated in cell wall antibiotic resistance. Antimicrob Agents Chemother 57:3283–3292. doi: 10.1128/AAC.00220-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elsholz AK, Hempel K, Michalik S, Gronau K, Becher D, Hecker M, Gerth U. 2011. Activity control of the ClpC adaptor McsB in Bacillus subtilis. J Bacteriol 193:3887–3893. doi: 10.1128/JB.00079-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Donegan NP, Marvin JS, Cheung AL. 2014. Role of adaptor TrfA and ClpPC in controlling levels of SsrA-tagged proteins and antitoxins in Staphylococcus aureus. J Bacteriol 196:4140–4151. doi: 10.1128/JB.02222-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu Rev Biochem 69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 32.Pragman AA, Ji Y, Schlievert PM. 2007. Repression of Staphylococcus aureus SrrAB using inducible antisense srrA alters growth and virulence factor transcript levels. Biochemistry 46:314–321. doi: 10.1021/bi0603266. [DOI] [PubMed] [Google Scholar]
- 33.Pragman AA, Yarwood JM, Tripp TJ, Schlievert PM. 2004. Characterization of virulence factor regulation by SrrAB, a two-component system in Staphylococcus aureus. J Bacteriol 186:2430–2438. doi: 10.1128/jb.186.8.2430-2438.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yarwood JM, McCormick JK, Schlievert PM. 2001. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J Bacteriol 183:1113–1123. doi: 10.1128/JB.183.4.1113-1123.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mashruwala AA, Boyd JM. 2017. The Staphylococcus aureus SrrAB regulatory system modulates hydrogen peroxide resistance factors, which imparts protection to aconitase during aerobic growth. PLoS One 12:e0170283. doi: 10.1371/journal.pone.0170283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kinkel TL, Roux CM, Dunman PM, Fang FC. 2013. The Staphylococcus aureus SrrAB two-component system promotes resistance to nitrosative stress and hypoxia. mBio 4:e00696-13. doi: 10.1128/mBio.00696-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Throup JP, Zappacosta F, Lunsford RD, Annan RS, Carr SA, Lonsdale JT, Bryant AP, McDevitt D, Rosenberg M, Burnham MK. 2001. The srhSR gene pair from Staphylococcus aureus: genomic and proteomic approaches to the identification and characterization of gene function. Biochemistry 40:10392–10401. doi: 10.1021/bi0102959. [DOI] [PubMed] [Google Scholar]
- 38.Joo HS, Otto M. 2012. Molecular basis of in vivo biofilm formation by bacterial pathogens. Chem Biol 19:1503–1513. doi: 10.1016/j.chembiol.2012.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otto M. 2008. Staphylococcal biofilms. Curr Top Microbiol Immunol 322:207–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mashruwala AA, Van De Guchte A, Boyd JM. 2017. Impaired respiration elicits SrrAB-dependent programmed cell lysis and biofilm formation in Staphylococcus aureus. Elife 6:e23845. doi: 10.7554/eLife.23845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richardson AR, Dunman PM, Fang FC. 2006. The nitrosative stress response of Staphylococcus aureus is required for resistance to innate immunity. Mol Microbiol 61:927–939. doi: 10.1111/j.1365-2958.2006.05290.x. [DOI] [PubMed] [Google Scholar]
- 42.Bose JL, Daly SM, Hall PR, Bayles KW. 2014. Identification of the Staphylococcus aureus vfrAB operon, a novel virulence factor regulatory locus. Infect Immun 82:1813–1822. doi: 10.1128/IAI.01655-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herbert S, Ziebandt AK, Ohlsen K, Schafer T, Hecker M, Albrecht D, Novick R, Gotz F. 2010. Repair of global regulators in Staphylococcus aureus 8325 and comparative analysis with other clinical isolates. Infect Immun 78:2877–2889. doi: 10.1128/IAI.00088-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Memmi G, Nair DR, Cheung A. 2012. Role of ArlRS in autolysis in methicillin-sensitive and methicillin-resistant Staphylococcus aureus strains. J Bacteriol 194:759–767. doi: 10.1128/JB.06261-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mainiero M, Goerke C, Geiger T, Gonser C, Herbert S, Wolz C. 2010. Differential target gene activation by the Staphylococcus aureus two-component system SaeRS. J Bacteriol 192:613–623. doi: 10.1128/JB.01242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chatterjee I, Schmitt S, Batzilla CF, Engelmann S, Keller A, Ring MW, Kautenburger R, Ziebuhr W, Hecker M, Preissner KT, Bischoff M, Proctor RA, Beck HP, Lenhof HP, Somerville GA, Herrmann M. 2009. Staphylococcus aureus ClpC ATPase is a late growth phase effector of metabolism and persistence. Proteomics 9:1152–1176. doi: 10.1002/pmic.200800586. [DOI] [PubMed] [Google Scholar]
- 47.Mashruwala AA, Bhatt S, Poudel S, Boyd ES, Boyd JM. 2016. The DUF59 containing protein SufT is involved in the maturation of iron-sulfur (FeS) proteins during conditions of high FeS cofactor demand in Staphylococcus aureus. PLoS Genet 12:e1006233. doi: 10.1371/journal.pgen.1006233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller RD, Fung DY. 1973. Amino acid requirements for the production of enterotoxin B by Staphylococcus aureus S-6 in a chemically defined medium. Appl Microbiol 25:800–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fuchs S, Pane-Farre J, Kohler C, Hecker M, Engelmann S. 2007. Anaerobic gene expression in Staphylococcus aureus. J Bacteriol 189:4275–4289. doi: 10.1128/JB.00081-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cubitt AB, Heim R, Adams SR, Boyd AE, Gross LA, Tsien RY. 1995. Understanding, improving and using green fluorescent proteins. Trends Biochem Sci 20:448–455. doi: 10.1016/S0968-0004(00)89099-4. [DOI] [PubMed] [Google Scholar]
- 51.Mashruwala AA, Roberts CA, Bhatt S, May KL, Carroll RK, Shaw LN, Boyd JM. 2016. Staphylococcus aureus SufT: an essential iron-sulphur cluster assembly factor in cells experiencing a high-demand for lipoic acid. Mol Microbiol 102:1099–1119. doi: 10.1111/mmi.13539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Graham JW, Lei MG, Lee CY. 2013. Trapping and identification of cellular substrates of the Staphylococcus aureus ClpC chaperone. J Bacteriol 195:4506–4516. doi: 10.1128/JB.00758-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mashruwala AA, Pang YY, Rosario-Cruz Z, Chahal HK, Benson MA, Mike LA, Skaar EP, Torres VJ, Nauseef WM, Boyd JM. 2015. Nfu facilitates the maturation of iron-sulfur proteins and participates in virulence in Staphylococcus aureus. Mol Microbiol 95:383–409. doi: 10.1111/mmi.12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roberts CA, Al-Tameemi HM, Mashruwala AA, Rosario-Cruz Z, Chauhan U, Sause WE, Torres VJ, Belden WJ, Boyd JM. 2017. The Suf iron-sulfur cluster biosynthetic system is essential in Staphylococcus aureus, and decreased Suf function results in global metabolic defects and reduced survival in human neutrophils. Infect Immun 85:e00100-17. doi: 10.1128/IAI.00100-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rosario-Cruz Z, Boyd JM. 2016. Physiological roles of bacillithiol in intracellular metal processing. Curr Genet 62:59–65. doi: 10.1007/s00294-015-0511-0. [DOI] [PubMed] [Google Scholar]
- 56.Feng J, Michalik S, Varming AN, Andersen JH, Albrecht D, Jelsbak L, Krieger S, Ohlsen K, Hecker M, Gerth U, Ingmer H, Frees D. 2013. Trapping and proteomic identification of cellular substrates of the ClpP protease in Staphylococcus aureus. J Proteome Res 12:547–558. doi: 10.1021/pr300394r. [DOI] [PubMed] [Google Scholar]
- 57.Boles BR, Thoendel M, Roth AJ, Horswill AR. 2010. Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS One 5:e10146. doi: 10.1371/journal.pone.0010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Novick RP. 1991. Genetic systems in staphylococci. Methods Enzymol 204:587–636. doi: 10.1016/0076-6879(91)04029-N. [DOI] [PubMed] [Google Scholar]
- 59.Mercier P, Lewis MJ, Chang D, Baker D, Wishart DS. 2011. Towards automatic metabolomic profiling of high-resolution one-dimensional proton NMR spectra. J Biomol NMR 49:307–323. doi: 10.1007/s10858-011-9480-x. [DOI] [PubMed] [Google Scholar]
- 60.Xia J, Wishart DS. 2011. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat Protoc 6:743–760. doi: 10.1038/nprot.2011.319. [DOI] [PubMed] [Google Scholar]
- 61.Xia J, Wishart DS. 2016. Using MetaboAnalyst 3.0 for comprehensive metabolomics data analysis. Curr Protoc Bioinformatics 55:14.10.1–14.10.91. doi: 10.1002/cpbi.11. [DOI] [PubMed] [Google Scholar]
- 62.Pang YY, Schwartz J, Bloomberg S, Boyd JM, Horswill AR, Nauseef WM. 2014. Methionine sulfoxide reductases protect against oxidative stress in Staphylococcus aureus encountering exogenous oxidants and human neutrophils. J Innate Immun 6:353–364. doi: 10.1159/000355915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kreiswirth BN, Lofdahl S, Betley MJ, O'Reilly M, Schlievert PM, Bergdoll MS, Novick RP. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 64.Duthie ES, Lorenz LL. 1952. Staphylococcal coagulase; mode of action and antigenicity. J Gen Microbiol 6:95–107. doi: 10.1099/00221287-6-1-2-95. [DOI] [PubMed] [Google Scholar]
- 65.Anonymous. 1999. From the Centers for Disease Control and Prevention. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus—Minnesota and North Dakota, 1997-1999. JAMA 282:1123–1125. [PubMed] [Google Scholar]
- 66.Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, Lian J, Ito T, Kanamori M, Matsumaru H, Maruyama A, Murakami H, Hosoyama A, Mizutani-Ui Y, Takahashi NK, Sawano T, Inoue R, Kaito C, Sekimizu K, Hirakawa H, Kuhara S, Goto S, Yabuzaki J, Kanehisa M, Yamashita A, Oshima K, Furuya K, Yoshino C, Shiba T, Hattori M, Ogasawara N, Hayashi H, Hiramatsu K. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225–1240. doi: 10.1016/S0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 67.Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW. 2013. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4:e00537. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Malone CL, Boles BR, Lauderdale KJ, Thoendel M, Kavanaugh JS, Horswill AR. 2009. Fluorescent reporters for Staphylococcus aureus. J Microbiol Methods 77:251–260. doi: 10.1016/j.mimet.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Luong TT, Lee CY. 2007. Improved single-copy integration vectors for Staphylococcus aureus. J Microbiol Methods 70:186–190. doi: 10.1016/j.mimet.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Forsyth RA, Haselbeck RJ, Ohlsen KL, Yamamoto RT, Xu H, Trawick JD, Wall D, Wang LS, Brown-Driver V, Froelich JM, Kedar GC, King P, McCarthy M, Malone C, Misiner B, Robbins D, Tan ZH, Zhu ZY, Carr G, Mosca DA, Zamudio C, Foulkes JG, Zyskind JW. 2002. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol Microbiol 43:1387–1400. doi: 10.1046/j.1365-2958.2002.02832.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We have deposited all the NMR metabolomics data and associated data processing files into the Metabolomics Workbench database under data accession number ST001174.