Zinc is an important transition metal cation and is present as an essential component in many enzymes, such as RNA polymerase. As with other transition metals, zinc is also toxic at higher concentrations so that living cells have to maintain strict control of their zinc homeostasis. Members of the COG0523 family of metal-chaperoning GE3-type GTPases exist in archaea, bacteria, and eucaryotes, including humans, and they may be involved in delivery of zinc to thousands of different proteins. We used a combination of molecular, physiological, and biochemical methods to demonstrate the important but diverse functions of COG0523 proteins in C. metallidurans, which are produced as part of the Zur-controlled zinc starvation response in this bacterium.
KEYWORDS: COG0523, CobW, Cupriavidus, Zur, zinc
ABSTRACT
The Zur regulon is central to zinc homeostasis in the zinc-resistant bacterium Cupriavidus metallidurans. It comprises the transcription regulator Zur, the zinc importer ZupT, and three members of the COG0523 family of metal-chaperoning G3E-type GTPases, annotated as CobW1, CobW2, and CobW3. The operon structures of the zur and cobW1 loci were determined. To analyze the interplay between the Zur regulon components and metal resistance, deletion mutants were constructed from the wild-type strain CH34 and various other strains. The Zur regulon components interacted with the plasmid-encoded and chromosomally encoded metal resistance factors to acquire metals from complexes of EDTA and for homeostasis of and resistance to zinc, nickel, cobalt, and cadmium. The three G3E-type GTPases were characterized in more detail. CobW1 bound only 1 Zn atom per mol of protein with a stability constant slightly above that of 2-carboxy-2′-hydroxy-5′-sulfoformazylbenzene (Zincon) and an additional 0.5 Zn with low affinity. The CobW1 system was necessary to obtain metals from EDTA complexes. The GTPase CobW2 is a zinc storage compound and bound 0.5 to 1.5 Zn atoms tightly and up to 6 more with lower affinity. The presence of MgGTP unfolded the protein partially. CobW3 had no GTPase activity and equilibrated metal import by ZupT with that of the other metal transport systems. It sequestered 8 Zn atoms per mol with decreasing affinity. The three CobWs bound to the metal-dependent protein FolEIB2, which is encoded directly downstream of cobW1. This demonstrated an important contribution of the Zur regulon components to metal homeostasis in C. metallidurans.
IMPORTANCE Zinc is an important transition metal cation and is present as an essential component in many enzymes, such as RNA polymerase. As with other transition metals, zinc is also toxic at higher concentrations so that living cells have to maintain strict control of their zinc homeostasis. Members of the COG0523 family of metal-chaperoning GE3-type GTPases exist in archaea, bacteria, and eucaryotes, including humans, and they may be involved in delivery of zinc to thousands of different proteins. We used a combination of molecular, physiological, and biochemical methods to demonstrate the important but diverse functions of COG0523 proteins in C. metallidurans, which are produced as part of the Zur-controlled zinc starvation response in this bacterium.
INTRODUCTION
The first row of transition metals from Mn to Zn are generally important for life, but the specific contribution of each metal is strongly dependent on the organism in which it is found (1–3). Zinc is needed by all known organisms. Consequently, acquisition of zinc is a major issue for free-living bacteria and also for pathogenic strains and their interaction with their respective hosts (4). Zinc ions are important structural components that keep or fold a polypeptide chain into the desired conformation or serve as a Lewis acid in the catalytic cycle of an enzyme (1). In the metal-resistant betaproteobacterium Cupriavidus metallidurans (5), absence of the zinc importer ZupT (ZIP protein family, TC 2.A.5 [6, 7]) results in a low cellular zinc content and accumulation of a misfolded RpoC (beta-prime subunit of the RNA polymerase) in inclusion bodies (8). RpoC needs a conformational zinc for correct folding, and zinc converts the disordered polypeptide into the compact conformation required to form the RNA polymerase core protein complex (9, 10).
C. metallidurans thrives in environments with high zinc concentrations, e.g., in zinc deserts in Belgium and on gold nuggets in auriferous Australian soils (5, 11–13). The genome of C. metallidurans is composed of a chromosome, a chromid, and two plasmids (14, 15). The most important metal resistance determinants of C. metallidurans are located on the plasmids, e.g., the cobalt-zinc-cadmium resistance determinant czc on plasmid pMOL30 and the cobalt-nickel determinant cnr on plasmid pMOL28 (16). Some genes involved in metal resistance are also located on the chromid (previously designated a second chromosome [15]). The plasmid-free C. metallidurans derivative strain AE104 contains only the chromosome and the chromid and has a metal resistance comparable to that of Escherichia coli (16, 17).
High-level zinc resistance (mid-micromolar to lower-millimolar concentrations) is mediated by the plasmid-encoded double-decameric CzcCBA protein complex of C. metallidurans, which removes periplasmic zinc directly to the outside of the cell (18). The CzcCBA efflux complex is supported by periplasmic metal-binding proteins such as CzcE and CzcJ and by the inner membrane efflux systems CzcP (PIB4-type ATPase, TC 3.A.3) and CzcD (CDF protein, TC 2.A.4), which export surplus ions efficiently from the cytoplasm for further export by CzcCBA (19). At lower zinc concentrations, removal of surplus cytoplasmic zinc by chromosomal or chromid-encoded efflux pumps, the PIB2-type ATPase ZntA or its substitute CadA, suffices (19, 20), although the Czc system is also present in these cells (21–23).
The efflux systems of the inner membrane export surplus ions from the cytoplasm. These were previously imported by ZupT and additionally a battery of at least 9 redundant high-rate, low-specificity importers, which supply a set of metal cations to the cell: (i) the MIT proteins CorA1, CorA2, CorA3, ZntB (TC 1.A.35); (ii) the PiT metal-phosphate importer PitA (TC 2.A.20); (iii) the NiCoT nickel importer HoxN (TC 2.A.52); (iv) the P-type ATPases MgtA and MgtB (TC 3.A.3); and (v) at least one unknown importer (24, 25). These systems import zinc ions as noncognate substrates.
In Escherichia coli, the Zinc uptake regulator Zur, a member of the Fur family of regulators, controls expression of znuABC for high-specificity import of zinc ions (26, 27). In contrast, C. metallidurans does not possess a ZnuABC-type ABC importer (28). Instead, Zur upregulates synthesis of ZupT (28, 29). The ZupT ortholog in E. coli, in contrast, is constitutively expressed (30, 31). ZupT is essential in cells containing a CzcCBA efflux system (8), indicating that transport of zinc from the periplasm to the outside of the cell by CzcCBA and to the inside by ZupT are balanced processes.
C. metallidurans contains, per cell, about 110,000 copies of various zinc-binding proteins (32), many of them with more than one zinc-binding site per protein, summing up to perhaps 200,000 zinc-binding sites in the total proteome of the cell. When cultivated in Tris-buffered mineral salts medium (TMM) containing 200 nM Zn(II) as minor bioelement, C. metallidurans possesses only 70,000 zinc atoms per cell. Under severe starvation conditions generated by absence of the zinc importer ZupT, the bacterium survives with merely 20,000 zinc atoms per cell. C. metallidurans mutants without zinc efflux systems accumulated zinc in lower micromolar concentrations of up to 210,000 Zn atoms per cell (8, 32) and ceased growing at higher concentrations. This indicates the presence of a zinc repository formed by unoccupied zinc-binding sites, which exists in C. metallidurans cells under certain growth conditions, for instance, after cultivation in TMM. At above 210,000 Zn atoms per cell, all zinc-binding sites in the proteome may be occupied so that additional zinc ions start to compete with other metal ions for binding sites or bind to thiol groups in glutathione or proteins, subsequently leading to zinc toxicity.
In addition to zupT (29), Zur controls expression of three more putative operons in C. metallidurans (Fig. 1 and 2): zur-cobW2; cobW3-Rmet_0124, and the operon region Op0317f, which starts with the cobW1 gene (33). There is only one Zur-binding site in each of the zupTp, zurp, and cobW3p promoter regions, allowing synthesis of these proteins in cells grown in non-zinc-amended TMM (33). Two Zur sites are upstream of the gene for the third paralog, CobW1, which is encoded by first gene of operon region Op0317f. Genes for paralogs of important zinc-dependent proteins follow cobW1. For instance, folEIB2 codes for a metal-dependent enzyme involved in folate synthesis and CysS for a cysteinyl-tRNA synthetase. In some CysS paralogs Zn(II) is required for discrimination between cysteine and serine (34, 35). Op0317f is expressed only under conditions of severe zinc starvation (25, 33, 36).
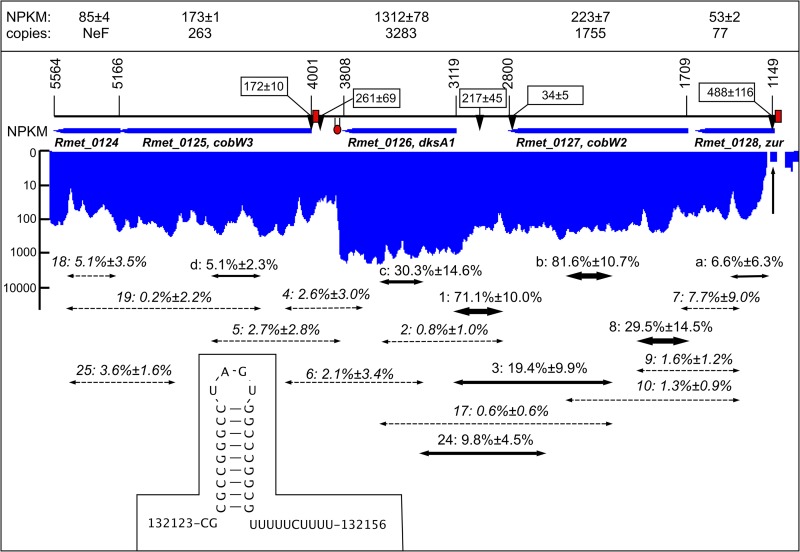
FIG 1.
The zur gene region of C. metallidurans in operon region Op0032r. The zur gene on the chromosome of C. metallidurans is followed by the genes cobW2, dksA1, cobW3, and Rmet_0124 in the same direction of transcription (arbitrary numbering, corresponding to complement bp 134133 to 130825 under accession no. CP000352.1). The respective open reading frames are indicated by blue arrows. The box at the top indicates the NPKM value for each gene (43), which corresponds to the transcriptional activity for each base pair as shown in blue (indicating the complement orientation) below the genes on a logarithmic scale. The “copies” refer to the number of gene products per cell (32). The RNA-Seq data (43) were analyzed in more detail to obtain the transcriptional start sites (TSS) in three independent reads. The mean score and deviation for each TSS is indicated by labeled arrows. The red boxes show the positions of the Zur boxes and the hairpin the Rho-independent terminator downstream of dksA1, which is also shown in the inset. A vertical arrow points to the position of the start codon of zur. The two-headed arrows symbolize the positions of the RT-PCR results (dashed, no visible signal; others, thickness of the arrow indicates the intensity of the band). The numbers above are the names of the primer pairs followed by the mean intensity of the RT-PCR band as a percentage of the intensity of the positive DNA control. The RT-PCR data are listed in Table S1 in the supplemental material.
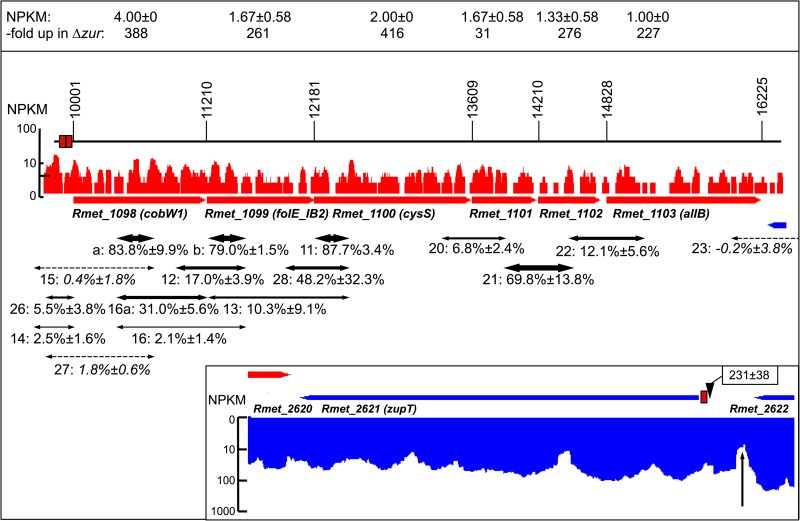
FIG 2.
The cobW1 operon region Op0317f of C. metallidurans and the zupT region. The cobW1 gene on the chromosome of C. metallidurans is followed by the genes Rmet_1099 to Rmet_1103 in the same direction of transcription (arbitrary numbering, corresponding to bp 1194940 to 1201164 under accession no. CP000352.1). The respective open reading frames are indicated by red arrows. The box at the top indicates the NPKM value for each gene (43), which corresponds to the transcriptional activity for each base pair as shown above the genes in red on a logarithmic scale. The genes have a low expression value in C. metallidurans CH34 but are strongly upregulated in the Δzur mutant strain (33), as shown in the upper box. Due to the low activity, transcriptional start sites were not found for this region, but the transcribed regions of the open reading frames were continuously connected by transcripts of 5′ and 3′ untranslated regions (43). Red boxes show the positions of the two Zur boxes. The two-headed arrows symbolize the positions of the RT-PCR results for RNA from the Δzur mutant (dashed, no visible signal; others, thickness of the arrow indicates the intensity of the band). The numbers above are the names of the primer pairs followed by the mean intensity of the RT-PCR band as a percentage of the intensity of the positive DNA control. Together with the continuous coverage with transcripts as measured by RNA-Seq, the RT-PCR data validate the presence of the operon Op0317f_1 (43) from the region of the Zur boxes upstream of cobW1 to the region downstream of Rmet_1103 but not further on (see primer pair 23). The RT-PCR data are also listed in Table S1 in the supplemental material. The inset shows the transcriptional activity of the zupT gene (position complement 2852938 to 2852062 on the chromosome under accession no. CP000352.1, “complement” indicated by the color blue), the position of the Zur box, and the transcriptional start site with the mean value of its score. The vertical arrow indicates a decreasing transcriptional activity of the upstream gene Rmet_2622.
The three CobWs of C. metallidurans belong to the COG0523 group of the G3E family of P-loop, signal recognition-associated SIMIBI class GTPases (37, 38). Besides the COG0523 subfamily, the G3E-GTPases contain other groups of proteins needed to assemble metalloproteins, for instance, UreG and HybB, which facilitate Ni(II) insertion into urease and hydrogenase, respectively (39, 40). Two COG0523 members from E. coli are known. YeiR binds several zinc ions, which stimulates its GTPase activity. Deletion of the gene decreased resistance to EDTA and Cd(II) (41). YjiA has been crystallized. It was shown to prefer Zn(II) over Co(II) and Ni(II), and it bound a maximum of seven Zn(II) per dimer: two in an internal site, one in a bridging site between the protomers, and four in surface sites (42). The comparison of YeiR, YjiA, and the three CobWs (Fig. 3) indicates the following: (i) the presence of a GTP-binding site; (ii) an internal metal-binding site between the Walker motifs, which is absent in CobW3; (iii) an internal histidine-rich loop with a decreasing number of His residues (CobW2 [22 residues] > YeiR [4 residues] > CobW3 [3 residues] = CobW1 [3 residues] > YjiA [1 residue]); and (iv) the presence of a His-rich C terminus exclusively in CobW3 containing 8 additional His residues (Fig. 3), marking CobW3 as an unusual member of the COG0523 group of proteins. All three CobWs of C. metallidurans might bind zinc, other metal ions, and GTP and perform some action driven by GTP hydrolysis. Since their primary sequences differ, they may have different functions in metal homeostasis.
FIG 3.
Alignment of the three CobW proteins from C. metallidurans with other COG0523 members. YciC_Bsu (KIX81565, Bacillus subtilis), YciC_Atu (CUX52153, Agrobacterium tumefaciens), ZigA (ABO13800, Acinetobacter baumannii), YjiA (CQR83726, E. coli), and YeiR (BAA15982, E. coli) were aligned in a multiple alignment with CobW1 (ABF07984, Rmet_1098), CobW2 (ABF07013, Rmet_0127), and CobW3 (ABF07011, Rmet_0125). The Walker motifs and metal-binding sites are indicated. Boxes indicate residues involved in the protomer-bridging zinc site in YjiA (42).
This study investigated the interaction between the Zur regulon components, which are needed to maintain zinc homeostasis under conditions of low and medium zinc availability, and the zinc resistance apparatus in C. metallidurans, which is required at high external zinc concentrations.
RESULTS
Operon structure of the Zur regulon.
The Zur regulon contains genes in three operon regions (33, 36): Op0317f, with cobW1 followed by genes for paralogs of zinc-dependent proteins; Op0032r, with zur, cobW2, dksA1, and cobW3; and Op0734r, with zupT (Fig. 1 and 2). Reverse transcription-PCR (RT-PCR) was used to identify the transcriptional units within zur- and cobW1-containing operon regions but not for zupT, which is most probably a single Zur-controlled gene. RNA was isolated from the plasmid-free strain C. metallidurans AE104 cultivated in Tris-buffered mineral salts medium without addition (TMM) or with 50 μM EDTA and from the Δzur mutant of AE104, strain DN728. The RNA was checked for DNA contamination and its quality verified by RT-PCR experiments using a rpoZ-specific primer pair. The positive control for all RT-PCR experiments was DNA, and the negative control was no added RNA (water control). The signals were normalized to the intensity of the positive DNA control after the intensity of the negative water control had been subtracted. The RT-PCR results were combined with recent whole-transcriptome shotgun sequencing (RNA-Seq) data (43). Moreover, the RNA-Seq data were also used to predict transcriptional start sites.
The algorithm used predicted three operons in Op0032r, i.e., for (i) zur, (ii) cobW2, and (iii) dksA1 and its downstream genes, from the RNA-Seq data on the basis of increasing nucleotide activities per kilobase of exon model per million mapped reads (NPKM) values (43). A detailed analysis (Fig. 1; see Table S1 in the supplemental material) clearly demonstrates common transcripts for zur and cobW2 and a strong transcriptional start site (TSS) in the region of the Zur box upstream of zur but no TSS upstream of cobW2 (Fig. 1; Table S1), together providing evidence for a dicistronic zur-cobW2 operon, Op0032r_1/2, instead of two monocistronic operons, Op0032r_1 and Op0032r_2 (43). However, the NPKM values right at the beginning of the zur gene (vertical arrow in Fig. 1) were between 0 and 1, and of four RT-PCR products spanning this region, one indicated a low transcriptional activity (a, with 6.6% ± 6.3% of the DNA control value) (Fig. 1), while three primer pairs (7, 9, and 10) failed. After the NPKM values increased within zur, RT-PCR yielded positive results (8): the 5′ end of the zur-cobW2 mRNA is unstable, especially at the position of the start codon, which results in a low copy number of 77 Zur proteins per cell compared to 1,755 CobW2 proteins, due to differential mRNA stability (Fig. 1).
The dksA1 gene clearly resides in its own operon, Op0032r_3, with a strong and a low-intensity TSS upstream and a Rho-independent terminator structure downstream. This gene is strongly transcribed, leading to 3,283 DksA1 copies per cell (Fig. 1). RT-PCR indicated transcripts between cobW2 and dksA1 (Fig. 1, 1), from within cobW2 up to the 5′ end of dksA1 (3), or from within cobW2 into dksA1 (24) but not up to the middle of the dksA1 open reading frame (2 and 17). The dksA1 operon Op0032r_3 is not transcribed with cobW2 as part of the Zur regulon. The cobW3 gene downstream of dksA1 possesses its own TSS in the vicinity of the Zur box and is transcribed with an NPKM value of 173, leading to 263 CobW3 copies per cell (Fig. 1). There was no RT-PCR evidence for common transcripts of cobW3 and its downstream gene Rmet_0124, but the distance between the genes is only 10 bp. It is unresolved at this stage if Rmet_0124 is member of the Zur regulon.
Due to a very low transcriptional activity, no RT-PCR products could be obtained for the cobW1 region Op0317f from RNA isolated from strain AE104, and no TSS was identified (Fig. 2). Consequently, RNA was isolated from the AE104 Δzur strain, which constitutively upregulated expression of this region. With RNA from Δzur, RT-PCR indicated continuous transcription from cobW1 to Rmet_1103 but not beyond (Fig. 2). The cobW1 region is indeed organized as one operon, Op0317f_1 (43).
The zupT (complement 2852938 to 2852062 on the chromosome under accession no. CP000352.1) possesses a TSS at position 2852971 in the vicinity of the Zur box (2852963 to 2852941), which is 3 bp away from the start codon of zupT (43). The upstream gene Rmet_2622 shows a declining transcriptional activity at its 3′ end upstream of the zupT TSS (vertical arrow in the inset in Fig. 2), and the downstream gene is on the other DNA strand, so that zupT is probably transcribed as monocistronic mRNA.
Mutants of C. metallidurans.
To study the interaction of the Zur regulon components with metal resistance in C. metallidurans, single, double, and triple mutants were constructed from the C. metallidurans wild-type strain CH34, which contains both plasmids and the full arsenal of efflux systems, and derivatives of the plasmid-free strain AE104. This removed sequentially all components of the zinc homeostasis system of C. metallidurans as putative interaction partners: (i) the plasmid-encoded efflux systems of the periplasm, with Czc being most important; (ii) ZupT; (iii) Zur; (iv) the chromosomally encoded zinc efflux systems ZntA, CadA, DmeF, and FieF in the Δe4 mutant strain of AE104; and (v) ZupT or Zur in the Δe4 strain. As published, ZupT cannot be deleted in strain CH34 because this leads to simultaneous curing of the czc-harboring plasmid pMOL30 (8). The complete cobW1-containing operon Op0317f_1 was removed to eliminate the complete CobW1 system. The gene cobW2 could be interrupted only while cobW3 was inactivated by a marker-free deletion. The names of the single mutants were abbreviated to ΔW1-cluster (ΔcobW1 cluster), ΔW2 (ΔcobW2 disruption), and ΔW3 (ΔcobW3).
Unchallenged cells and metal starvation conditions.
EDTA was added to the medium to simulate metal starvation conditions. As in the plasmid-free strain AE104 (33), expression of cobW1 in CH34 was strongly upregulated by EDTA, and that of cobW2 and cobW3 was upregulated to a minor degree (Table 1). While expression of cobW2 was downregulated at elevated zinc concentrations, that of cobW3 was downregulated at high cobalt concentrations. Although on a low level (Fig. 2), cobW1 was expressed in unchallenged cells. Cadmium had no influence on any cobW expression (Table 1). This assigned CobW1 and CobW2 to zinc and metal starvation, albeit with different roles. CobW3 showed some connection to cobalt homeostasis.
TABLE 1.
Expression of cobW-lacZ reporter gene fusions in wild-type C. metallidurans CH34a
| lacZ fusion | Sp act (U/mg) without addition | Fold upregulationb
with: |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| EDTA |
ZnCl2 |
CoCl2 |
CdCl2 |
||||||
| 50 μM | 1 mM | 10 μM | 0.1 mM | 10 μM | 0.1 mM | 10 μM | 0.1 mM | ||
| cobW1 | 2.1 ± 1.4 | 37 ± 6 | 15 ± 5 | 0.2 ± 0 | ND | 1.0 ± 0 | 1.0 ± 0 | 0.8 ± 0 | 1.0 ± 0.3 |
| cobW2 | 64 ± 20 | 2.6 ± 0.6 | 2.4 ± 0.5 | 0.4 ± 0.1 | 0.1 ± 0 | 0.9 ± 0.2 | 0.9 ± 0.2 | 1.3 ± 0.1 | 1.3 ± 0.1 |
| cobW3 | 123 ± 48 | 1.6 ± 0.5 | 1.5 ± 0.4 | 1.1 ± 0.3 | 0.7 ± 0.2 | 0.4 ± 0.1 | 0.2 ± 0.1 | 0.6 ± 0 | 0.6 ± 0.1 |
Chromosomal lacZ fusions were constructed in wild-type C. metallidurans CH34 by insertion of the lacZ fusion plasmid pECD794-1 downstream of the gene, leaving it functional in case of cobW2 and cobW3. In case of cobW1, lacZ was in the middle, interrupting the gene. All constructions had possible polar effects. Early-exponential-phase cells of these strains were cultivated for 3 h with shaking at 30°C in TMM without or with the indicated additions, and β-galactosidase activity was determined. All values are means ± standard deviations.
Bold, significant upregulation of the specific activity of a fusion compared to that in nonamended medium (D > 1 and ratio > 2-fold; n > 3); bold and italic, significant downregulation; ND, not done.
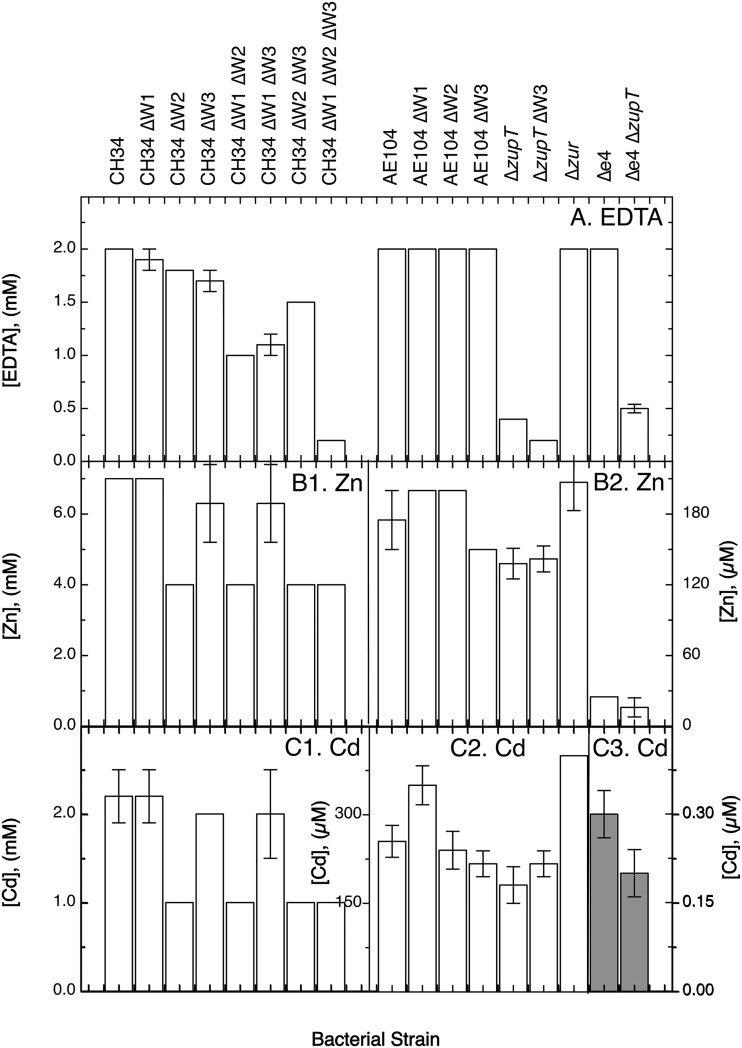
On solid medium, the MIC of EDTA for CH34 mutants decreased slightly in the single mutant strains, with the EDTA resistance levels in the order CH34 > ΔW1-cluster > ΔW2 > ΔW3. Resistance did not decrease in the plasmid-free strain AE104 in a similar manner (Fig. 4). The three CH34 ΔW double mutants were less resistant to EDTA-mediated metal starvation than the single mutants. The ΔW2 ΔW3 double mutant, which still contains the CobW1 system, was more resistant to EDTA than the ΔW1-cluster ΔW2 or ΔW1-cluster ΔW3 mutant. EDTA resistance of the ΔW1-cluster ΔW2 ΔW3 triple mutant dropped sharply to 200 μM EDTA, a resistance level even lower than that in the AE104 Δe4 ΔzupT mutant (Fig. 4). Under metal starvation conditions, the presence of the plasmids in strain CH34 resulted in the need for the three CobWs as compensation, with CobW1 plus the products of the other gene of the cobW1 cluster being more important than the other two CobWs, although these two could functionally substitute for a missing CobW1 system to some degree.
FIG 4.
MICs of C. metallidurans mutant strains. Cells from a first preculture were diluted 20-fold into a second preculture with fresh TMM, cultivated at 30°C with shaking until the stationary phase was reached, diluted 100-fold with fresh TMM, and streaked onto TMM agar plates with increasing concentrations of the test substance. The MIC was determined after incubation at 30°C for 4 days and is the lowest concentration preventing the formation of colonies. The strains are indicated on the top. (A) EDTA; (B) zinc; (C) cadmium. The last two panels in each group have different scales for the y axes, which are additionally highlighted by the gray bars in panel C3.
In liquid TMM, the 50% inhibitory concentration (IC50) of wild-type strain CH34 for EDTA was 6.0 ± 1.1 mM (Table 2), 3-fold the value of the MIC on solid TMM (Fig. 4). In contrast, the MIC values for various metals were higher than the respective IC50 values; e.g., 7.0 versus 2.0 mM for zinc, 8.2 mM versus 1.5 mM for cobalt, and 2.2 mM versus 103 μM for cadmium, respectively (Fig. 4 and Table 2; see Table S2 in the supplemental material). This was probably due to undefinable differences between growth on solid medium and in liquid culture.
TABLE 2.
Zn content and metal resistance in liquid culture of C. metallidurans derivatives
| Bacterial straina | Fold metal zinc content withb
: |
% Znc | % IC50d
|
||||
|---|---|---|---|---|---|---|---|
| No addition | EDTA | Zn | EDTA | ZnCl2 | CdCl2 | ||
| CH34 | 1.00 | 0.98 | 1.86 | 1.00 | 100 | 100 | 100 |
| CH34 ΔW1-cluster | 1.14 | 0.67 | 1.55 | 0.83 | 145 | 105 | 123 |
| CH34 ΔW2 | 1.05 | 0.80 | 2.03 | 1.09 | 60 | 100 | 125 |
| CH34 ΔW3 | 1.22 | 0.73 | 2.19 | 1.18 | 100 | 95 | 103 |
| CH34 ΔW1-cluster ΔW2 | 1.06 | 0.77 | 1.86 | 1.00 | 103 | 85 | 119 |
| CH34 ΔW1-cluster ΔW3 | 1.23 | 0.73 | 1.69 | 0.91 | 142 | 105 | 141 |
| CH34 ΔW2 ΔW3 | 1.17 | 0.92 | 1.92 | 1.03 | 75 | 80 | 44 |
| CH34 ΔW1-cluster ΔW2 ΔW3 | 1.05 | 0.70 | 1.69 | 0.91 | 107 | 65 | 146 |
| AE104 | 1.19 | 0.80 | 2.16 | 1.16 | 53 | 25 | 116 |
| AE104 ΔW1-cluster | 1.19 | 0.81 | 1.50 | 0.81 | 47 | 100 | 51 |
| AE104 ΔW2 | 0.98 | 0.77 | 1.92 | 1.03 | 75 | 120 | 100 |
| AE104 ΔW3 | 0.69 | 0.73 | 2.17 | 1.17 | 128 | 120 | 96 |
| AE104 ΔW1-cluster ΔW3 | 1.08 | 0.73 | 2.16 | 1.16 | 19 | 100 | 91 |
| AE104 ΔW2 ΔW3 | 1.20 | 0.78 | 2.45 | 1.32 | 84 | 100 | 61 |
| ΔzupT | 0.50 | 0.44 | 1.91 | 1.03 | 56 | 27 | 34 |
| ΔzupT ΔW1-cluster | 0.72 | 0.44 | 1.63 | 0.87 | 267 | 115 | 185 |
| ΔzupT ΔW2 | 0.59 | 0.36 | 1.91 | 1.03 | 89 | 110 | 100 |
| ΔzupT ΔW3 | 1.20 | 0.56 | 2.63 | 1.41 | 83 | 110 | 118 |
| ΔzupT ΔW1-cluster ΔW3 | 1.11 | 0.53 | 2.84 | 1.53 | 33 | 96 | 98 |
| ΔzupT ΔW2 ΔW3 | 1.11 | 0.55 | 3.31 | 1.78 | 72 | 100 | 80 |
| Δzur | 1.52 | 1.22 | 2.64 | 1.42 | 56 | 97 | 135 |
| Δzur ΔW1-cluster | 1.38 | 1.17 | 2.23 | 1.20 | 106 | 106 | 103 |
| Δzur ΔW2 | 1.00 | 0.83 | 2.27 | 1.22 | 89 | 90 | 78 |
| Δzur ΔW3 | 1.45 | 1.00 | 2.14 | 1.15 | 72 | 106 | 102 |
| Δzur ΔW1-cluster ΔW2 | 1.03 | 0.97 | 2.42 | 1.30 | 89 | 57 | 84 |
| Δzur ΔW1-cluster ΔW3 | 0.98 | 0.70 | 2.20 | 1.18 | 78 | 92 | 75 |
| Δzur ΔW2 ΔW3 | 1.42 | 0.70 | 2.05 | 1.10 | 100 | 92 | 78 |
| Δe4 | 0.91 | 0.95 | 3.66 | 1.97 | 53 | 1.5 | 18 |
| Δe4 ΔW1-cluster | 1.19 | 0.86 | 2.23 | 1.20 | 88 | 70 | 9.0 |
| Δe4 ΔW3 | 1.20 | 0.72 | 3.09 | 1.66 | 88 | 74 | 7.1 |
| Δe4 ΔW1-cluster ΔW2 | 1.09 | 0.70 | 2.56 | 1.38 | 94 | 70 | ND |
| Δe4 ΔW1-cluster ΔW3 | 0.97 | 0.64 | 2.84 | 1.53 | 100 | 21 | 67 |
| Δe4 ΔW2 ΔW3 | 1.03 | 0.66 | 2.48 | 1.34 | 88 | 71 | 14 |
| Δe4 ΔzupT | 0.42 | 0.67 | 4.13 | 2.22 | 112 | 75 | 105 |
| Δe4 ΔzupT ΔcobW3 | 0.91 | 0.44 | 3.03 | 1.63 | 94 | 74 | 8.1 |
| Δe4 ΔzupT ΔW1-cluster ΔW3 | 1.00 | 0.66 | 2.98 | 1.61 | 100 | 83 | 11 |
| Δe4 ΔzupT ΔW2 ΔW3 | 1.38 | 0.56 | 2.38 | 1.28 | 94 | 72 | 13 |
| Δe4 Δzur | 1.70 | 1.27 | 3.73 | 2.01 | 94 | 62 | 7.1 |
| Δe4 Δzur ΔW2 | 1.38 | 1.20 | 2.91 | 1.56 | 94 | 64 | 7.6 |
The mutants carry a deletion of the complete cobW1 cluster, a deletion of the cobW3 gene, or disruption of cobW2.
The metal content was measured in cells grown in TMM with 100 μM EDTA, 100 μM ZnCl2 (10 μM ZnCl2 in case of the Δe4 mutant), or no addition. The metal content was compared to that of CH34 cells in unamended medium (double-underlined value in shaded cell). If D > 1 (n > 4, deviation bars of the data points do not touch or overlap), a metal content of <67% is in italic and a metal content of >133% is in bold. The 100% value for CH34 cells in nonamended TMM is (64 ± 9) · 103 Zn atoms per cell.
Ratio compared to value for CH34 cells cultivated in the presence of zinc. If D > 1 (n > 4, deviation bars of the data points do not touch or overlap), a metal content of >133% is in bold.
The metal resistance of the mutants was tested in dose-response experiments, and the IC50 (concentration for half-maximal growth inhibition) was calculated. These values were compared as follows: (i) CH34 mutants, including the plasmid-free strain AE104 (double-underlined values), were compared to CH34 wild-type cells (double-underlined values in shaded cells); (ii) AE104 mutants, including ΔzupT, Δzur, and Δe4 (ΔcadA ΔzntA ΔfieF ΔdmeF) mutants (underlined values), were compared to AE104 cells (double-underlined values), and (iii) ΔzupT, Δzur, and Δe4 mutants were compared to these respective parents (underlined values). If D > 1 (n > 3, deviation bars of the data points do not touch or overlap), an IC50 ratio of <67% is in italics and an IC50 ratio of >133% is in bold. The 100% values for CH34 cells in TMM are 6.0 ± 1.13 mM EDTA, 2.0 ± 0.1 mM ZnCl2, 2.6 ± 0.2 mM NiCl2, 1.5 ± 0.2 mM CoCl2, and 103 ± 9 μM CdCl2. ND, not determined.
The IC50 for EDTA dropped with loss of the plasmids in strain AE104 by half to about 3 mM (shaded and double-underlined CH34 values versus double-underlined AE104 values in Table 2) and by a further 50% to about 1.5 mM with loss of zupT, loss of zur, or loss of four zinc efflux systems in strain Δe4 (Table 2, single-underlined values versus double-underlined AE104 values). In the case of ΔzupT, the MIC value was lower than the IC50 value, while in the case of Δzur and Δe4, the MIC value was higher (Fig. 4). This indicated an influence of plasmid-encoded metal-handling systems, metal efflux pumps, ZupT, and Zur on EDTA resistance.
The different growth conditions also influenced whether and how the CobWs were involved in EDTA resistance. While all three were required in colonies of CH34 on solid TMM, CobW3 was not essential in liquid culture (Fig. 4 and Table 2). The CobW1 system and CobW2 had opposing functions, with CobW2 increasing EDTA resistance and the CobW1 system decreasing it. Both effects disappeared in the CH34 ΔW1-cluster ΔW2 mutant and the triple mutant (Table 2). In all CH34 mutants, the cellular zinc content was unchanged in cells grown in nonamended medium and in the presence of 100 μM EDTA (Table 2). The cellular Mg and Fe content was also unchanged under both conditions in strain CH34 derivatives (see Table S3 in the supplemental material), but the nickel content increased and the cobalt content decreased in all CH34 ΔW1-cluster and ΔW2 mutants (see Table S4 in the supplemental material), whereas the ΔW3 mutation had no effect on any metal content in CH34 cells. Together, all three CobWs were needed in strain CH34 with its plasmids on solid medium, the CobW1 system and CobW2 with opposing functions were needed in liquid medium, and the cellular Zn, Fe, and Mg content could always be maintained under mild metal starvation conditions but at the cost of some imbalance in the nickel and cobalt contents, which agrees with the general preferences that C. metallidurans has for its multiple-metal homeostasis (43).
In strain AE104 and its derivatives cultivated in liquid culture, EDTA resistance (IC50) of the ΔW1-cluster mutant dropped by half, which is comparable to the effect of the ΔzupT, the Δzur, and the Δe4 mutations, but it increased again in the ΔzupT ΔW1-cluster double null mutant to 4.75 mM (6 mM × 0.53 × 0.56 × 2.67) (Table 2). There was no further decrease in EDTA resistance in the Δzur or the Δe4 mutant (Table 2). The ΔW3 deletion decreased EDTA resistance again, to about 0.6 mM in the AE104 ΔW1-cluster ΔW3 double mutant and in the AE104 ΔzupT ΔW1-cluster ΔW3 triple mutant (Table 2). The plasmid-free strain AE104 needed cooperation of ZupT and the CobW1 system to acquire metals from EDTA complexes in liquid culture. Deletion of both yielded a CobW3-mediated increase in the ability to acquire metals. Overexpression of the remaining Zur regulon components, at least ZupT and one CobW, or absence of four zinc efflux systems also decreased the EDTA resistance of strain AE104, but cooperation between the CobW1 system, CobW3, and ZupT appeared no longer to be significant.
While the zinc content of AE104 cells did not change with any ΔW deletion, it was, as published previously (8), lower in the ΔzupT mutant cultivated in nonamended TMM (Table 2). A ΔW3 deletion changed this low zinc content back to the level in the AE104 parent cells, even in the ΔzupT ΔW1-cluster and ΔzupT ΔW2 mutant backgrounds (Table 2). On the other hand, deletion of cobW3 increased the cellular zinc content of ΔzupT cells grown in the presence of 100 μM ZnCl2 (Table 2). CobW3 was responsible for the low zinc content of the ΔzupT mutant by decreasing zinc accumulation in the absence of ZupT. This could be managed by decreasing uptake or increasing efflux of the metal cation. The Δe4 ΔzupT mutant displayed a diminished zinc content similar to that of the AE104 ΔzupT mutant, and the Δe4 ΔzupT ΔW3 mutant recovered 91% of the wild-type level. Since the Δe4 mutant contained no known zinc efflux systems, CobW3 should act not upon zinc efflux but upon zinc uptake systems. CobW3 might coordinate metal import by ZupT with that of the other metal uptake systems, which also explains why CobW3 was able to increase EDTA resistance of the AE104 ΔzupT ΔW1-cluster mutant (Table 2).
In TMM, the AE104 Δzur mutant contained 52% more zinc atoms per cell (33) than wild-type AE104 or CH34 (Table 2), representing 33,000 additional zinc atoms. The ΔW2 deletion changed this back to the wild-type level. With 1,800 copies per cell (32) and a 6-fold upregulation in the Δzur mutant (33), binding of only 3 zinc atoms per CobW2 could fully account for the elevated zinc content of the Δzur mutant strain, indicating that CobW2 might be a zinc storage compound. The zinc content of the Δzur ΔW2 ΔW3 mutant increased again (Table 2), in agreement with a role of CobW3 in control of zinc uptake. The Δe4 Δzur ΔW2 mutant contained 20,000 fewer zinc atoms per cell than the Δe4 Δzur mutant, so the efflux systems did not influence zinc storage by CobW2.
Together, the three CobWs may be required here for efficient zinc allocation (CobW1 system), zinc storage (CobW2), or coordination of metal uptake by ZupT with other zinc uptake systems (CobW3), together maintaining cytoplasmic zinc homeostasis in strain CH34. All three could likely be assigned to zinc homeostasis rather than to homeostasis of other metals due to the effect of CobW3 on the low cellular zinc content of the ΔzupT mutant, the effect of CobW2 on the increased zinc content of the Δzur mutant, and the close cooperation of the CobW1 system with ZupT; however, an influence on homeostasis of other metals could not be excluded at this point.
Metal resistance.
Since transition metal cations interfere with each other, metal resistance of strain CH34, of its mutants with altered metal resistance or homeostasis, and of ΔcobW mutants of both types of parent strains was investigated. On solid medium, CobW2 was required for full zinc and cadmium resistance of strain CH34 (Fig. 4). Loss of the plasmids decreases both resistances (16), with the pMOL30-encoded Czc system being responsible (19). CobW2 was no longer required to elevate either resistance in the plasmid-free strain AE104 on solid medium (Fig. 4). Consequently, CobW2 was able to enhance zinc and cadmium resistance only if Czc increased zinc-cadmium resistance from 0.2 mM to 4 mM first (Fig. 4). CobW2 cooperated with Czc. As a zinc storage compound, it could buffer the cellular zinc content to maintain zinc resistance on solid medium (Fig. 4).
In liquid culture, and with up to 100 μM added Zn(II), neither the ΔW2 deletion nor any other ΔW deletion changed the zinc content or influenced zinc resistance, with the exception of a 35% decrease in zinc resistance in the ΔW1-cluster ΔW2 ΔW3 triple mutant of CH34 (Table 2). The zinc content of the CH34 ΔW derivatives remained unchanged, indicating that possible zinc storage by CobW2 was more important on solid medium than in liquid culture. The already-decreased zinc resistance levels in the plasmid-free strain AE104 (IC50 = 500 μM), its ΔzupT mutant (135 μM), or its Δzur mutant (485 μM) were not further decreased in any of the respective ΔW mutant strains obtained, despite the changes in the zinc content of the ΔzupT mutant (Table 2). However, both CobW1 and CobW2 were important in maintaining the level of zinc resistance of the Δe4 mutant at an IC50 of 7.5 μM: without both CobWs, the IC50 dropped to 1.6 μM (Table 2). This decrease did not occur in the respective Δe4 ΔW single mutants, the Δe4 ΔW1-cluster ΔW2 mutant, the Δe4 ΔW2ΔW3 mutant, or the Δe4 ΔzupT mutant, again demonstrating the cooperation of ZupT, the CobW1 system, and CobW3 in maintaining zinc resistance.
The influence of the three CobWs on the cellular metal content and resistance to metals other than zinc is detailed in the results in the supplemental material and summarized here. Resistance to cadmium in liquid culture (IC50 = 0.1 mM [Table 2]) required the four chromosomal efflux systems, most probably CadA and ZntA, and additionally the complete Zur regulon components ZupT plus the three CobWs. The cellular Fe and Mg content almost never changed in the experiments, except for the already published 4-fold increase of the Mg content of Δe4 cells (8), which was decreased again in all of the ΔW mutants (Table S3). In the absence of the four metal efflux systems ZntA, CadA, DmeF, and FieF, all three CobWs were required to maintain a cellular magnesium level of about 10 million atoms per cell.
All CobWs, but especially CobW3, were required for nickel resistance (Table S2). Since the ΔW3 mutant displayed no altered cellular nickel content (Table S4), nickel toxicity was connected not to increased nickel accumulation but rather to enhanced toxicity of the nickel ions present in the cell. As in the case of nickel, the decrease of cobalt resistance in the ΔW mutant strains was not connected to a reciprocal increase in cell-bound cobalt but instead is likely to be mediated by an increased toxicity of the cobalt ions already present in the cell (Tables S2 and S4). This indicated that a main function of the three CobWs might be to shield the cellular zinc homeostasis against the competing ions Co(II), Ni(II), and Cd(II), although a role in cobalt and nickel homeostasis could not be excluded at this stage. CobW1 may accomplish this goal potentially by efficient zinc allocation, and CobW2 may achieve it by buffering and binding zinc.
CobW3 was needed to repress the genes for the MIT-type metal cation importers ZntB and CorA3 in CH34, for a zinc-dependent repression of corA1 in the absence of zupT, and for an EDTA-dependent expression of corA1 in the same strain, but it had only a minor influence on the expression of the genes for metal efflux systems (see Tables S5 and S6 in the supplemental material). This missing repression of corA1 in the ΔzupT strain was paralleled by an increased cellular cobalt and decreased nickel content in cobalt-treated but not in zinc-treated, EDTA-treated, or untreated cells (see Table S7 in the supplemental material). It also resulted in reversion of the decreased zinc content of ΔzupT cells (Table 2) and a decreased cobalt and nickel resistance (results in the supplemental material). Consequently, CobW3 may have some regulatory function on the metal import transportome, more specifically the MIT proteins CorA1, CorA3, and ZntB, and control low-specificity high-rate metal import by these systems. Such a regulatory function may be needed to prevent a short-term overflow of the cytoplasmic metal content mediated by MIT importers during fluctuations of the periplasmic metal content and to allow the cell to adjust the activity of its metal import and export arsenal. This also indicated a major difference between CobW1 and CobW2 on the one hand as zinc delivery and/or storage compounds and CobW3 on the other hand as some kind of regulator.
Purification of the CobWs.
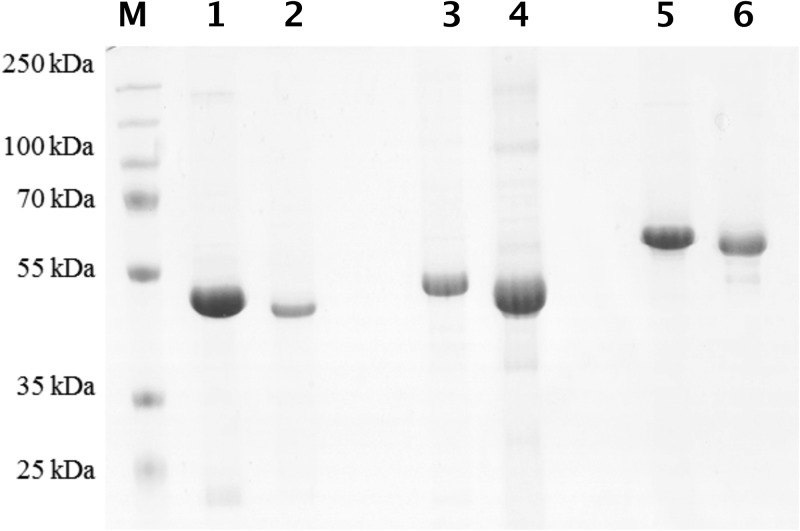
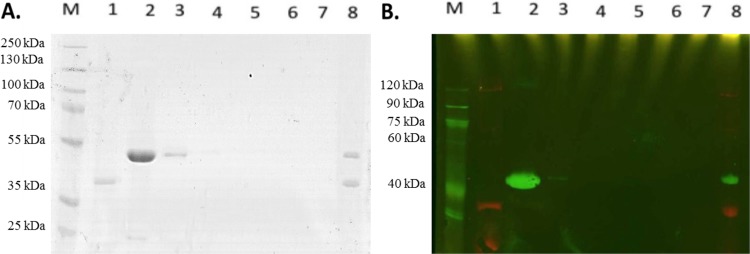
To obtain biochemical evidence for the proposed functions of CobW1, CobW2, and CobW3 as a zinc delivery protein, a zinc storage protein, and a regulator, respectively, the three CobWs were produced in E. coli as His-tagged proteins and purified by Ni-nitrilotriacetic acid (NTA) affinity chromatography. The His tag was removed by a His-tagged version of the Tev protease. A second affinity chromatography step removed unprocessed CobWs and the Tev protease, leaving the respective processed CobW in the flowthrough. A size exclusion chromatography step concluded the purification procedure. A few minor contaminations remained in the preparations (Fig. 5).
FIG 5.
Purified CobW proteins from C. metallidurans. Lanes 1, 3, and 5 show the His-tagged forms of CobW1, CobW2, and CobW3, respectively; lanes 2, 4, and 5 show the corresponding untagged forms. Lane M, size marker. An SDS-polyacrylamide gel, Coomassie blue stain, and 10 μg proteins were used, except for untagged CobW1 (5 μg). The CobW2 and CobW3 preparations contain a few remaining contaminations.
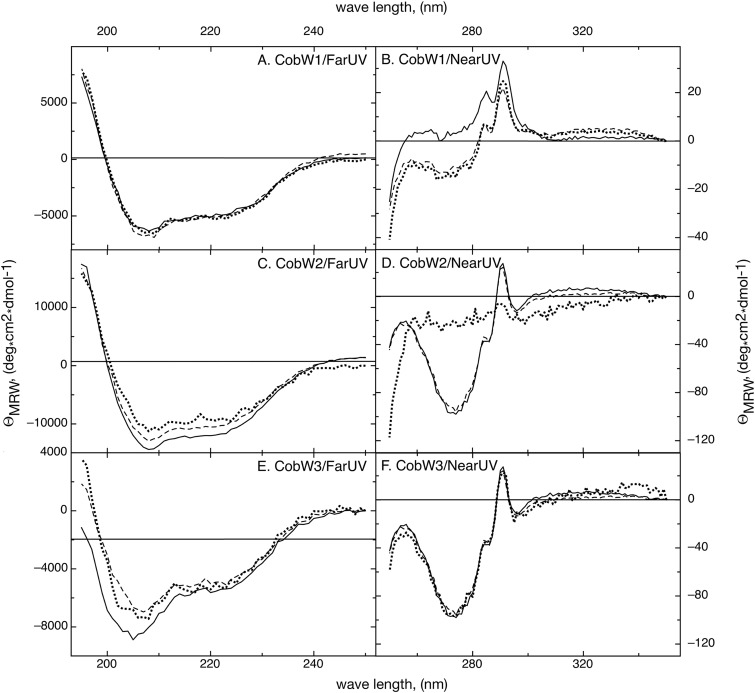
The three CobWs were treated with EDTA to remove metals which had bound to the proteins during production or the purification procedure. Inductively coupled plasma mass spectrometry (ICP-MS) analysis revealed that no metal remained associated with the proteins (Table 3). The conformation of the resulting apo forms of the three CobWs was determined by near- and far-UV-circular dichroism (CD) spectroscopy (Fig. 6) and the alpha-helical and beta-sheet content calculated with the program CDNN (44). The three proteins contained an alpha-helical content between 15% and 17% with around 20% beta sheets, indicating that all three apoproteins were mainly folded.
TABLE 3.
Metal content of the three CobWs from C. metalliduransa
| Protein | Condition | Metal content (mol/mol protein)b
|
||||
|---|---|---|---|---|---|---|
| Zn | Mg | Ni | Co | Cd | ||
| CobW1 | Apo | 0.10 ± 0.02 | 0.01 ± 0.07 | 0.03 ± 0.04 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| ZnCl2 | 2.52 ± 0.67 | 0.03 ± 0.02 | 0.05 ± 0.04 | 0.02 ± 0.04 | 0.00 ± 0.00 | |
| ZnCl2-GTP-MgCl2 | 1.33 ± 0.14 | 0.00 ± 0.00 | 0.16 ± 0.09 | 0.01 ± 0.02 | 0.00 ± 0.00 | |
| Metal mix | 1.07 ± 0.52 | 0.13 ± 0.16 | 1.39 ± 1.24 | 0.98 ± 0.76 | 0.00 ± 0.00 | |
| CobW2 | Apo | 0.20 ± 0.15 | 0.00 ± 0.03 | 0.03 ± 0.04 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| ZnCl2 | 3.65 ± 3.13c | 0.00 ± 0.05 | 0.45 ± 0.37 | 0.03 ± 0.03 | 0.00 ± 0.00 | |
| ZnCl2-GTP-MgCl2 | 6.36 ± 1.30 | 1.43 ± 1.93 | 0.40 ± 0.52 | 0.00 ± 0.00 | 0.00 ± 0.00 | |
| CobW3 | Apo | 0.04 ± 0.04 | 0.00 ± 0.02 | 0.13 ± 0.10 | 0.01 ± 0.01 | 0.00 ± 0.00 |
| ZnCl2 | 7.99 ± 1.21 | 0.26 ± 0.40 | 0.47 ± 0.54 | 0.25 ± 0.39 | 0.00 ± 0.00 | |
| ZnCl2-GTP-MgCl2 | 5.09 ± 1.89 | 2.44 ± 2.37 | 0.15 ± 0.02 | 0.01 ± 0.01 | 0.00 ± 0.00 | |
| Metal mix | 3.95 ± 0.82 | 0.17 ± 0.10 | 2.30 ± 0.52 | 1.19 ± 0.69 | 0.83 ± 0.56 | |
Metals remaining from the purification process were first removed by treatment with EDTA, followed by a desalting step, leading to the apo forms. The apoproteins were metalated with a 10-fold concentration of ZnCl2, of ZnCl2 plus MgCl2 plus GTP, or of a metal mix containing equimolar concentrations of Zn, Ni, Co, and Cd (with each metal in a 10-fold excess), always followed by a desalting step using size exclusion chromatography with a PD10 column. The metal content was determined by ICP-MS.
Metal contents of >0.2 mol/mol are in bold or, in case of high deviations of >50%, in italics. Determinations were 3-fold, or at least 4-fold for the zinc values, and standard deviations are indicated.
Incubation of CobW2 with zinc resulted in two outcomes, each twice: either 0.52 ± 0.09 Zn/CobW2 or 6.79 ± 0.73 Zn/CobW2. The treatment of CobW2 with the metal mix always resulted in denaturation and precipitation of the protein.
FIG 6.
Structural integrity of the purified CobW proteins. Remaining bound metals were removed from the CobW proteins with EDTA (solid lines), and they were subsequently loaded with zinc (dashed lines) or zinc and MgGTP (dotted lines). The conformations of the respective protein preparations were determined using far (A, C, and E)- and near (B, D, and F)-UV-CD spectroscopy. (A and B) CobW1; (C and D) CobW2; (E and F) CobW3. Note the change of the conformation of CobW2 in the presence of zinc and MgGTP (D).
The apo forms of the CobWs were loaded with zinc or with zinc plus MgGTP, and the resulting conformations were compared with those of the apo forms to study a possible influence of these ligands on the secondary and tertiary structures by near-UV-CD and far-UV-CD spectroscopy, respectively (Fig. 6). The secondary structure of CobW1 did not change when these ligands were present (Fig. 6A) despite a zinc content of 2.5 zinc atoms per mol of protein (Table 3). The near-UV-CD spectrum of apo-CobW1 shifted to a higher level on the y axis in the presence of zinc plus MgGTP, while the fine structure of the spectrum did not change (Fig. 6A). The presence of zinc or of zinc plus MgGTP did not change the secondary structure or overall conformation of CobW1 or of CobW3. While the secondary structure of CobW2 did not change in the presence of Zn2+, it was altered when MgGTP was additionally present, indicating a partial unfolding of CobW2 (Fig. 6D).
Binding of zinc to the three CobWs.
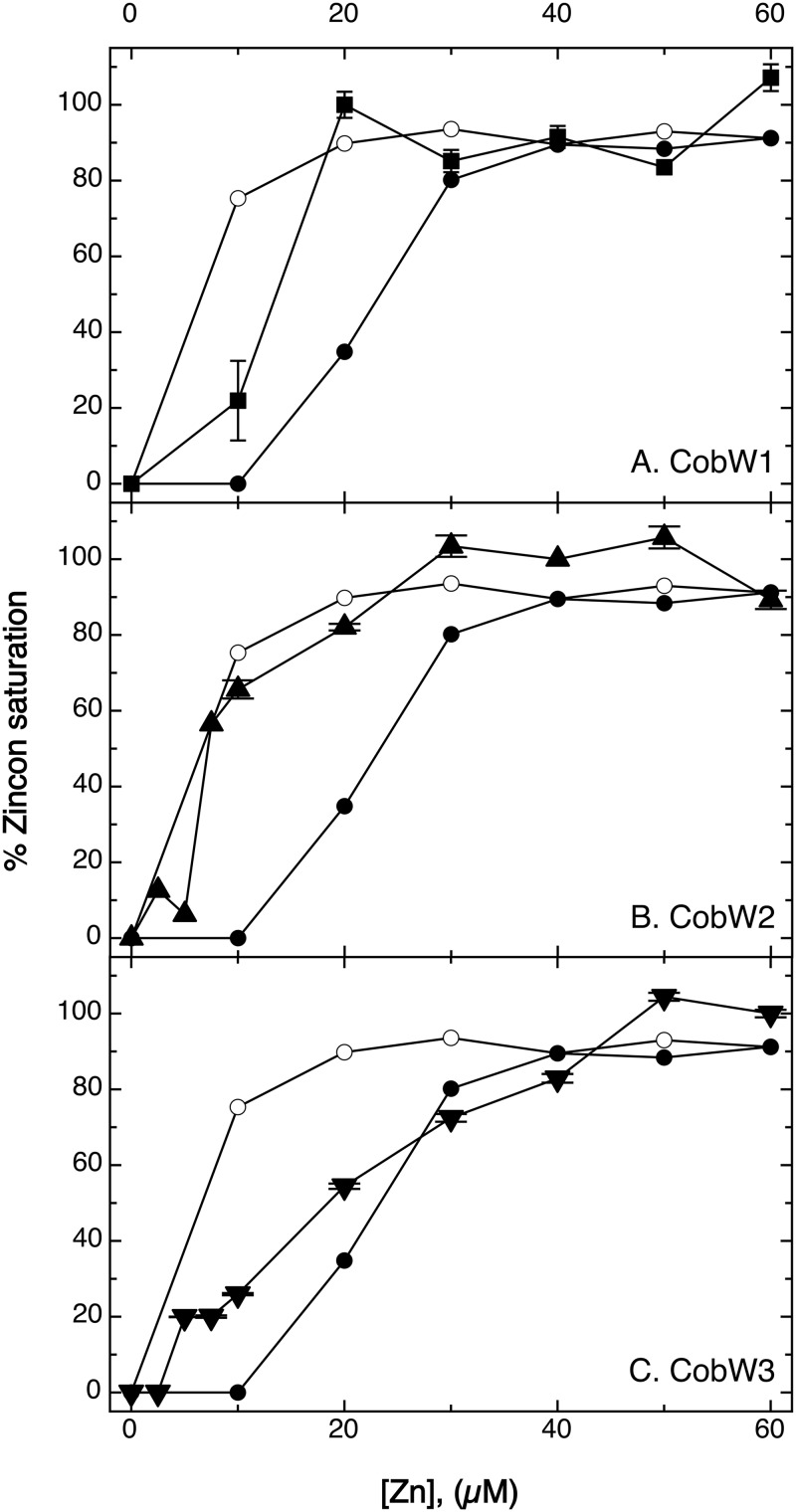
A Zincon (2-carboxy-2′-hydroxy-5′-sulfoformazylbenzene) competition experiment was used to measure binding of zinc to the three CobWs. Zincon has an absorption maximum at 480 nm, which shifts to 620 nm when the metal is bound. Disappearance of the 480 nm peak was used to calculate the percentage of zinc saturation of the Zincon molecule. The dissociation constant of the Zn-Zincon complex is 2.09 · 10−6 M, corresponding to a stability constant of log K1 = 5.89, and no Zn2-Zincon complexes were formed (45). At an equimolar zinc-to-Zincon ratio, Zincon was 75% saturated (Fig. 7). EDTA chelates zinc ions with an apparent stability constant at pH 7 of log K1 = 13.1 (46), outcompeting Zincon. At 10 μM ZnCl2, 10 μM EDTA, and 10 μM Zincon, Zincon was consequently not able to acquire zinc, and the saturation was 0% (Fig. 7). When the zinc concentration was increased to 20 μM, the Zincon saturation was 30%, indicating intermediary binding of zinc by Zincon. Only at 30 μM zinc and above was Zincon more than 75% saturated (Fig. 7).
FIG 7.
Zincon competition experiment. A 10 μM concentration of CobW1 (A) (squares), CobW2 (B) (triangles), or CobW3 (C) (inverted triangles) was mixed with 10 μM Zincon and various concentrations of zinc chloride. The percentage of zinc saturation was calculated from the Zincon absorption at 480 nm (without zinc) and 620 nm (bound zinc). Controls were 10 μM Zincon without protein (open circles) and 10 μM Zincon plus 10 μM EDTA (filled circles). These controls are given in all panels. Results are from triplicates, and standard deviations are shown.
In a mixture of 10 μM (each) apo-CobW1, zinc, and Zincon, Zincon was saturated to 20%, and it was saturated to 100% at 20 μM zinc and above (Fig. 7). The apo forms of the three CobWs were also incubated with a 10-fold excess of zinc, with zinc plus MgGTP, or with a metal mix composed of Zn, Ni, Co, and Cd in a 1:1:1:1 molar ratio, each in a 10-fold excess, and the metal content was determined by ICP-MS (Table 3). Apo-CobW1 contained no metal, so it was fully capable of competing with Zincon for zinc ions. It contained 2.5 Zn atoms per mol of protein in the absence and 1.5 Zn atoms per mol of protein in the presence of MgGTP, and this difference was significant (distance [D] value = 1.47). When treated with the metal mix, CobW1 contained between 0.5 and 1.5 Zn atoms per mol of protein plus 0 to 2.5 Ni atoms and 0 to 2 Co atoms per mol of protein but no Cd, with 1 Zn, 1.5 Ni, and 1 Co being the mean values (Table 3). This demonstrated that CobW1 bound only 1 Zn atom per mol of protein with a stability constant slightly above that of Zincon, an additional 0.5 Zn atom with lower affinity, which could lead to formation of a dimer, and an additional zinc or other cations with very low binding affinity. The decrease in the zinc content in the presence of MgGTP and some residual Mg under these conditions may indicate that transient binding of MgGTP can remove zinc or other ions bound to the low-affinity site(s), although MgGTP did not alter the structure of the protein.
CobW2 blocked excess Zincon binding to zinc at a zinc concentration of 5 μM (Fig. 7). At higher zinc concentrations CobW2 and Zincon competed for zinc, and Zincon was saturated at 30 μM zinc. Incubation of CobW2 with zinc yielded two different outcomes, 0.52 ± 0.09 Zn/monomer in two experiments and 6.79 ± 0.73 Zn/monomer in two other experiments (Table 3). Due to the large internal histidine-rich stretch of CobW2 (Fig. 3), the protein may form different proteoforms, one that binds 1 zinc atom per dimer, and one binding 6 additional Zn ions per monomer. The presence of MgGTP resulted in 6.36 ± 1.30 Zn atoms per monomer plus some Mg, so MgGTP seems to stabilize the zinc-binding proteoform. CobW2 bound 0.5 Zn atom with an affinity comparable to that of Zincon and 6 or even 7 Zn atoms with lower affinity. CobW2 precipitated in the presence of the metal mix.
CobW3 was able to outcompete Zincon at 10 μM zinc (Fig. 7), and the apoprotein had been zinc free (Table 3). Zincon was fully saturated at 50 μM ZnCl2 with intermediary binding between both concentrations. The zinc content of zinc-treated CobW3 was about 8 Zn atoms (6.5 to 9) per mol of protein (Table 3). CobW3 bound one Zn atom per mol of protein with higher affinity than Zincon, a second with a similar affinity leading to 50% saturation, a third and perhaps fourth with low affinity, and an additional 4 Zn atoms per mol with even lower affinity. In the presence of MgGTP, traces of Mg were found in CobW3. A decrease in the zinc content in the presence of MgGTP was not significant (D = 0.94). In the presence of the metal mix, CobW3 bound about 4 zinc ions, plus 2.5 Ni, 1 Co, and 1 Cd CobW3 ions, totaling 8.5 metal ions. Considering the Zincon result, CobW3 bound 4 Zn ions with decreasing affinity and four additional transition metal cations at low-affinity sites.
The three CobWs thus were different. CobW2 bound 0.5 Zn ion tightly, perhaps through a monomer-bridging site and up to 6 more with lower affinity, and the presence of MgGTP may have unfolded the protein partially, which nevertheless stabilized its zinc-binding ability and could present the 6 less firmly bound zinc ions to the outside. This was in agreement with a possible role of CobW2 as a zinc storage and/or zinc-buffering compound. The different affinities of CobW3 for its zinc ions, the inertness of the conformation, and the absence of the GCXCC motif may assign to CobW3 a function altogether different from that of CobW2. CobW1, which contained no histidine-rich stretches, bound 1.5 zinc ion only with considerable affinity, maybe to the GCXCC motif and a monomer-bridging site (Fig. 3), plus one or more additional metal cations to low-affinity sites. MgGTP was able to remove metal from these low-affinity sites.
CobW1 interacts with FolEIB2.
RT-PCR experiments indicated that the Op0317f region was transcribed as an operon (Fig. 2). The gene downstream of cobW1, Rmet_1099, encodes the possible GTP cyclohydrolase paralog FolEIB2, which could be involved in folate biosynthesis. FolEIB2 was heterologously produced in E. coli as a Strep-tagged protein. The resulting protein was undermetalated and contained only 0.27 ± 0.16 Zn atom per mol FolEIB2 instead of 1 Zn per monomer (47).
The Strep-tagged FolEIB2 protein was bound to magnetic beads coated with streptavidin and incubated with His-tagged CobW1 in the presence of zinc and MgGTP. After washing steps, FolEIB2 and CobW1 eluted in the same fraction. Both proteins were identified using multiplex staining of the respective tags (Fig. 8). Omission of zinc resulted in the absence of CobW1 in the elution fraction (see Fig. S1A in the supplemental material), so the interaction of FolEIB2 and CobW1 was zinc dependent; however, only zinc and not the presence of MgGTP was required for binding (Fig. S1B).
FIG 8.
Interaction of FolEIB2 and CobW1. A pulldown assay was performed using magnetic MagStrep XT beads washed three times in buffer W (50 mM Tris-HCl [pH 8], 150 mM NaCl) to bind 10 μg of FolEIB2-Strep-tag for 30 min at 4°C in buffer W additionally containing 500 μM MgCl2, 10 μM GTP, and 10 μM ZnCl2. The supernatant (lanes 1) was removed, and 100 μg CobW1-His tag was added. The mixture was incubated for 3 h at 4°C. Again, the supernatant was removed (lanes 2) and the beads washed with buffer W five times (lanes 3 to 7). Finally, the bound proteins were removed using 2-fold-concentrated SDS sample buffer (lanes 8). (A) Coomassie blue-stained SDS-polyacrylamide gel; (B) multiplex Western blot. The Strep-tag of FolEIB2 was stained using streptavidin-Alexa Fluor conjugate (red), and the His tag of CobW1 was stained with a primary anti-His antibody and a secondary Cy3-labeled anti-mouse antibody (green).
Dimerization of CobW1.
A possible dimerization of apo-CobW1 was analyzed by size exclusion chromatography. The protein preparation contained two peaks corresponding to calculated masses of 73.8 kDa and 54.8 kDa (see Fig. S2A in the supplemental material). Since the predicted size of CobW1 is 44.4 kDa, the two species in the size exclusion chromatogram may represent monomers and dimers of CobW1. This was verified by nondenaturing and SDS-polyacrylamide gel electrophoresis of the separated 73.8-kDa and 54.8-kDa protein species, which were indeed identified as CobW1 dimers (Fig. S2B and C, lane 2) and monomers, respectively (Fig. S2B and C, lane 4). Neither preparation contained zinc (the monomers had 0.01 ± 0.01 Zn per mol of protein and the dimers 0.05 ± 0.03 Zn per mol of protein [n = 3] by ICP-MS]), so zinc was not required for dimer formation. Incubation of the monomers with zinc, MgGTP, or both did not lead to formation of dimers (see Fig. S3 in the supplemental material). CobW1 was able to form monomers and dimers. Zinc, MgGTP, or both did not trigger dimerization of the monomers, but zinc was nevertheless essential for interaction of CobW1 with FolEIB2.
Interaction of the CobWs with other proteins.
Since the cobW2, cobW3, and zur genes are in proximity, CobW3, with its possible regulatory function, may interact with Zur. Another pulldown experiment was performed, this time with Zur as bait. No CobW3 bound to Zur, while interaction of CobW1 and CobW2 with Zur was very weak (see Fig. S4 in the supplemental material). As a control, we demonstrated the binding of CobW2 and CobW3 using FolEIB2 as bait in the presence of zinc (Fig. S1C and D). This suggested that CobW2 and CobW3 may also be able to interact with zinc-dependent proteins, in agreement with their ability to substitute fully, or in part, for a missing CobW1 in CH34 on solid TMM (Fig. 4) and in CH34, AE104, AE104 ΔzupT, and AE104 Δzur in liquid TMM (Table 2), all containing EDTA.
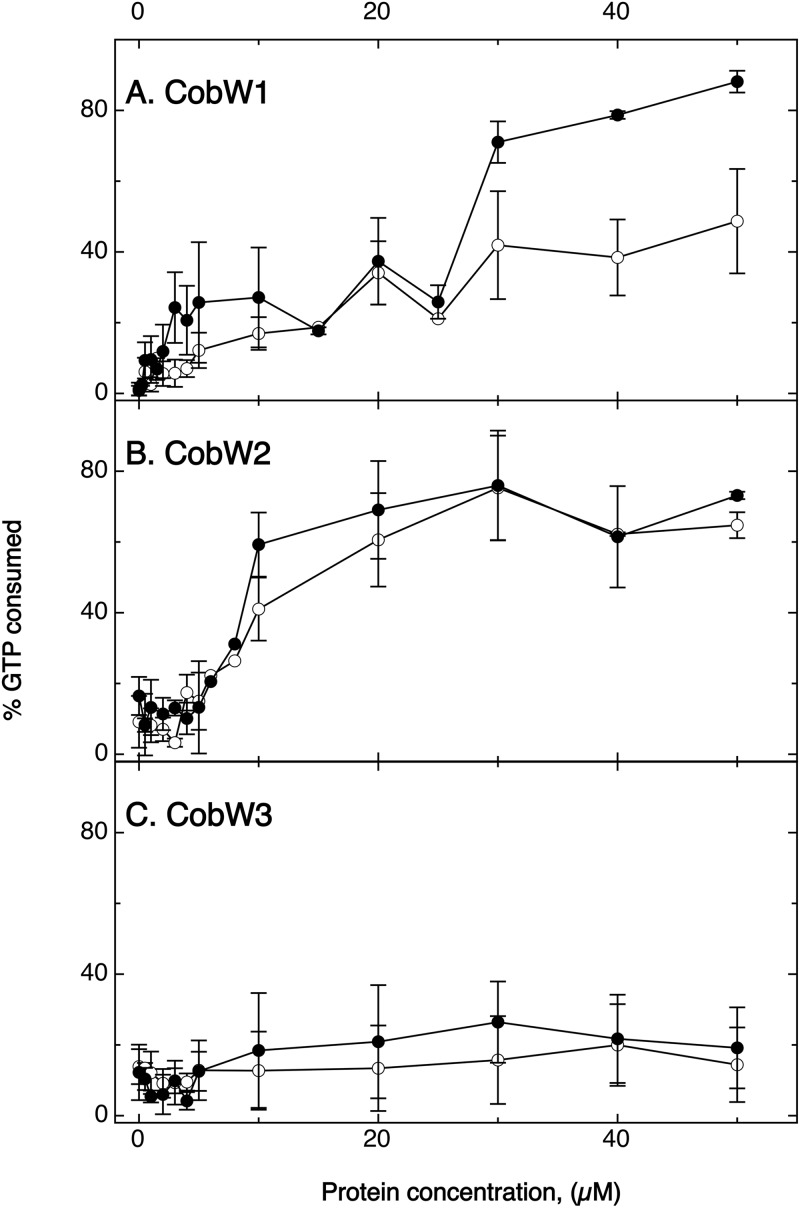
GTPase activity.
The GTPase activity of the three CobWs was measured for the apoproteins in the absence and presence of zinc (Fig. 9). There was no significant consumption of GTP by CobW3 in either the presence or the absence of zinc. The absence of the GCXCC motif was congruent with a metal-activated GTPase activity. CobW3, with its potential function in regulating the presence or activity of metal uptake systems, should be able to titrate zinc and/or other metals by binding them to the His residues at its C terminus, but the protein does not seem to be a metal-stimulated GTPase.
FIG 9.
GTPase activity of the three CobWs. Increasing concentration of CobW1 (A), CobW2 (B), or CobW3 (C) were incubated for 60 min with 5 μM GTP in the presence (closed circles) or absence (open circles) of 50 μM ZnCl2. Residual gamma-phosphate units of the GTP remaining after 60 min were transferred to ADP and the concentration of the resulting ATP measured in a luminescence assay. The percent GTP consumed was plotted against the protein concentration in the assay. Standard deviations indicated (n > 3).
On the other hand, CobW2 cleaved GTP, with 50% of the GTP consumed at 10 μM CobW2, also independent of the presence or absence of zinc (Fig. 9). In summary, the presence of GTP led to a zinc-independent GTPase activity, stabilization of a zinc-binding proteoform (Table 3), and partial unfolding of CobW2 (Fig. 6), so the GTPase activity may present the internal histidine-rich loop of CobW2 to the outside.
Last, CobW1 also exhibited GTPase activity but at higher protein concentrations than CobW2. The presence of Zn stimulated the GTPase activity of CobW1 (Fig. 9), which led not to conformational changes (Fig. 6) but to clearance of zinc bound to a low-affinity site (Table 3) and to binding of CobW1 to FolEIB2 (Fig. 8).
DISCUSSION
Structure-function relationship of members of the G3E family of P-loop GTPases.
The CobW protein subfamily or COG0523 group belongs to the G3E protein family, which is a separate protein family within the SIMIBI protein class of the GTPase superclass of P-loop NTPases (37, 38). Discussing the features of the three CobW proteins from C. metallidurans in the light of the general characteristics should derive constraints for the structure-function relationships and physiological roles of the three CobWs.
All P-loop NTPases possess a common folding motif different from that of other proteins binding nucleoside triphosphates (NTPs), for example, proteins exhibiting a Rossmann fold. In the recurring α-β units of the P-loop NTPases, the β-sheet forms a central core that is surrounded by α helices. The amino acid sequence of the P-loop NTPases contains an N-terminal Walker A motif, GXXXXGK(S/T), in the flexible “P loop” (phosphate binding) between a β strand and an α helix, which binds and positions the triphosphate group of the NTP, and a Walker B motif with a conserved aspartate or glutamate residue (37). The Walker motifs are clearly conserved in the compared CobWs (Fig. 3), firmly assigning to them the function of NTP-binding proteins.
P-loop NTPases are very widespread proteins and contain at least seven monophyletic lineages, such as the protein superclasses RecA/F1F0, AAA+, ABC/PilT, nucleotide kinases, and the GTPases (37). In the latter two superclasses, the β strand that leads to the P loop is adjacent to the Walker B motif (37) (Fig. 3). Many GTPases are molecular switches cycling between a GTP-bound “on state,” which interacts with an effector downstream in an information chain, and a GDP-bound “off state” (38). CobW1 and CobW2, but not CobW3, have GTPase activity (Fig. 9) and are therefore experimentally verified members of the P-loop GTPase superclass, and GTPase action indeed switched CobW1 and CobW2 with respect to their zinc-binding activity or conformation, respectively.
The P-loop GTPases exhibit five common “G” motifs: (i) Walker A (G1), (ii) Walker B (G3), (iii) a switch (G2 or effector region) conserved within the various families of the P-loop GTPases, (iv) the binding motif for the guanine residue [G4, four hydrophobic/apolar amino acids followed by (N/T)(K/Q)XD], and (v) G5, which also interacts with the guanine but is not as strongly conserved (38). The G2 switch motif interacts with the effector molecule and coordinates the Mg(II) ion of the MgGTP complex, so that the interaction with the effector is coupled with MgGTP binding or hydrolysis. The CobW protein YjiA contains a putative metal-binding GCXCC motif in the switch region (48), which is indeed part of its zinc-binding site (42). This region is conserved in all compared CobW proteins except CobW3 (Fig. 3). YjiA contains another zinc-binding site bridging two protomers, which is composed of a glutamate residue 7 positions downstream of the GCXCC motif plus a histidine residue downstream of the Walker B motif of each monomer. This indicates a possible interaction between the three processes (i) zinc-induced dimerization, (ii) binding of zinc to the internal GCXCC metal-binding region, and (iii) GTP binding or hydrolysis. The histidine residue of YjiA downstream of the Walker B motif is not conserved in the other compared CobW-like proteins except perhaps CobW3 (Fig. 3), so zinc-dependent dimerization may not be part of the switching cycle in all members of this group, leaving the interaction between binding of zinc to the internal GCXCC region and GTP binding, exchange, or hydrolysis as a core function of the compared proteins with the exception of CobW3 (Fig. 3).
The P-loop GTPases contain two protein classes, TRAFAC (translation factors, including translational initiation and elongation factors) and SIMIBI (signal recognition particles, e.g., MinD and BioD) (38), which are characterized by β-strand 2 being adjacent and parallel to the β strand containing the Walker B motif (37). As a common functional motif in both classes, the P-loop GTPases assemble with interaction partners including a target protein or protein complex in the GDP-bound “off” state and exchange GDP against GTP when all partners are present. The energy of GTP hydrolysis is subsequently used to release the P-loop GTPase again, often leaving an activated target structure behind, e.g., a ribosome with a loaded aminoacyl-tRNA at the A site or a urease with nickel in its active site. All three CobWs bound to FolEIB2 in the presence of zinc, but MgGTP did not influence binding of CobW1 to FolEIB2. Experimental evidence for a release of CobW1 from FolEIB2 after zinc delivery is lacking at this stage, but it also cannot be excluded that the GTPase activity is needed for release of CobW1 from an unknown target.
One protein family among the SIMIBI class of P-loop GTPases is the G3E (“glutamate E in the G3 motif”) family of proteins with (i) a GXXGXGK(S/T) Walker A motif, (ii) a conserved D residue for binding of the magnesium ion, (iii) another D at the N terminus of the Walker B strand, (iv) a glutamate (E) in the Walker B or G3 motif, and (v) an NKxD G4 motif after β-strand 6 for interaction with a guanine (37, 38). The Walker A motif in the compared CobWs (Fig. 3) is GFLGAGKTT, and the other features are also present with the G4 motif being TKxD in CobW2 and YjiA, so these five proteins are members of the G3E family of GTPases. Another group of factors that insert nickel into the carbon monoxide dehydrogenase and acetyl coenzyme A (acetyl-CoA) synthase are the CooC-like factors (49–51), which belong to the MinD group of the SIMIBI class of proteins (37, 52). Only the asparagine of the G4 motif is retained within the CooC-like proteins, which consequently use ATP instead of GTP. The CooC-like proteins cycle between dimerization, binding to the target protein, and/or metal binding, and ATP hydrolysis finally triggers release from the target protein and metal delivery (51), giving another example for the general mechanism of SIMIBI protein. The strict conservation of the NKxD motif in the compared CobW-like proteins (Fig. 3) indicates that they should interact with GDP/GTP rather than with ATP, which was indeed proven by the GTPase activity of CobW1 and CobW2 (Fig. 9).
The G3E family of GTPases contains four subfamilies, all with members that interact with transition metal cations. A common feature is an intact NKxD, indicating interaction with guanine, and the glutamate in the Walker B signature (37). Members of the G3E subfamily ArgK/MeaB participate in transport of arginine and other cationic amino acids and assembly of B12-dependent methyl-malonyl-CoA mutase (53), perhaps by inserting the cobalt ion. HypB and UreG proteins deliver Ni to hydrogenase and urease, respectively, whereas the exact role of CobW proteins of the COG0523 subfamily is unclear (37).
Other CobW proteins of the COG0523 protein subfamily.
With a few exceptions, members of the CobW or COG0523 subfamily were encoded in operons controlled by the Zur zinc uptake regulator, while the hypB and ureG genes were part of hydrogenase and urease gene clusters, respectively (39). Consequently, a role of the three CobWs of C. metallidurans in zinc homeostasis had been predicted (41). CobW-like proteins have been characterized from several bacteria, i.e., YeiR and YjiA from E. coli (41, 42) YciC from Bacillus subtilis (54) and Agrobacterium tumefaciens (55), and ZigA from Acinetobacter baumannii (56). The Zur regulon of A. tumefaciens encodes in addition to a ZnuACB a second ABC import system (TroABC), the periplasmic zinc-binding protein ZinT, and YciC, which are upregulated under zinc-limited conditions. YciC is important for survival under these conditions but only in a Δtro or ΔzinT deletion mutant (55). Expression control of yciC from B. subtilis is very similar to that of the cobW1 cluster with two Zur-binding sites, which also results in a tight repression when sufficient zinc is available (54). YciC from B. subtilis and CobW1 from C. metallidurans both have a size of 397 amino acyl residues and are 55% identical on the amino acid sequence level, while similarity to the other two CobWs is lower. The YciC proteins are CobW1-like in having no extended histidine stretches in the middle or at the C terminus of the polypeptide chain and no histidine residue that may be involved in zinc-dependent dimerization, but they possess an internal GCXCC motif for possible zinc stimulation of the GTPase activity (Fig. 3).
ZigA from A. baumannii is 77% identical to CobW1, although it contains an additional 18 amino acids (aa) at the N terminus. ZigA also contains a GCXCC metal-binding site between the Walker A and B motifs but no obvious His-rich loop. The protein is needed under metal starvation conditions and binds up to two zinc ions, one with low and one with high affinity. It exhibits a low-level GTPase activity, and GTP hydrolysis may shift ZigA from a dimer to a monomeric state (56), although there is also no conserved histidine residue for zinc-stimulated dimerization (Fig. 3).
Among the CobW-like proteins from E. coli, YjiA is only 29% identical to CobW1 and, with a length of 346 aa, is shorter at the C terminus. YjiA binds Ni(II), Co(II), and Zn(II) ions, but zinc ions outcompete the other two metals. It binds up to 4 Zn ions per monomer, and the GCXCC metal-binding site is involved in binding of one zinc to an internal site adjacent to the GTP-binding region (42). A second Zn ion is bound to a bridging site between the monomers, while the remaining two zinc ions are loosely surface bound by histidine and glutamate residues. While deletion of yjiA does not affect EDTA sensitivity of E. coli, deletion of yeiR decreases resistance to EDTA and to cadmium. YeiR also binds zinc, oligomerizes, and has a low-level GTPase activity (41).
YeiR, YjiA, the two YciCs, ZigA, CobW1, and CobW2 share the conserved GCXCC sequence for the internal zinc-binding site in the switch region between the Walker A and B motifs (shown for proteins in Fig. 3), indicating that for these proteins, but not for CobW3, binding of zinc to this site interferes with GDP/GTP exchange and/or GTP hydrolysis. On the other hand, none of the other proteins possesses the particular histidine residue of YjiA that is involved in zinc-dependent dimerization. Consequently, YeiR, ZigA, the YciCs, and CobW1 may perform similar functions. YjiA is also CobW1-like but possesses the feature of zinc-dependent dimerization. In contrast, CobW2, with its internal histidine stretch, and CobW3, lacking the GCXCC motif, may have unique physiological roles.
CobW1.
CobW1 bound one zinc ion per monomer with a stability constant slightly above that of Zincon, an additional 0.5 Zn ion with low affinity, and a further Zn or other metal cation to sites with even lower metal affinity of up to 1 Zn, 1.5 Ni, and 1 Co (2.5 ions) as mean values in the metal mix or 2.5 Zn ions in the presence of zinc only, with a high deviation of the number of Ni or Co ions bound from the metal mix (Table 3).
The tightly bound single zinc per monomer may be bound to the internal metal-binding “GCXCC” motif between the Walker A, switch, and Walker B motifs, indicating a connection between the bound zinc ion, GTP binding, and/or hydrolysis. In agreement with this, zinc stimulated GTPase activity (Fig. 9). MgGTP decreased the number of zinc ions bound to CobW1. If the GTPase action caused removal of Zn from a high-affinity site, zinc bound to the low-affinity site should be able to enter the high-affinity site and subsequently be removed by the next GTPase-triggered activity. Consequently, the presence of MgGTP should have “cleared” zinc from the low-affinity site or sites, which are also used by Ni and Co, but not Cd, ions (Table 3).
Zinc was also essential for binding of CobW1 to FolEIB2 (see Fig. S1 in the supplemental material), but MgGTP was not required here. Zinc was not necessary to form CobW1 dimers. Amino acid residues required to bind zinc to the monomer-monomer interface, framing the Walker B motif in YjiA (Fig. 3), are less conserved in CobW1. The 0.5 Zn ion bound per monomer with a low affinity and probably to the monomer interface. Ni or Co may even outcompete zinc binding to this site (Table 3). Consequently, formation of dimers was not an important part of any CobW1-mediated activity. This activity was zinc-mediated binding of CobW1 to FolEIB2 and zinc-enhanced GTPase activity leading to removal of less tightly bound zinc ions, or maybe Ni and Co ions. Without Zn, CobW1 catalyzed GTP hydrolysis at a low rate. With increasing availability of zinc, the GTPase activity is stimulated, causing binding to FolEIB2 or other proteins and delivery of Zn, Co, Ni, or other metal cations, which bind to the low-affinity sites of CobW1. FolEIB2 belongs to a family of GTP cyclohydrolases under Zur control, which are metal promiscuous and are able to use Mn(II), Fe(II), Mg(II), Co(II), Zn(II), or Ni(II) in their active center. FolEIA, on the other hand, binds only zinc ions (57). Consequently, CobW1 may deliver other metals under zinc starvation conditions to FolEIB2 and other metal-promiscuous enzymes, saving zinc for yet other enzymes that essentially depend on zinc, for instance, RpoC.
In agreement with this proposal is the fact that the CobW1 cluster is important for EDTA resistance of wild-type strain CH34 on solid medium. In liquid medium, the presence of the CobW1 system decreased EDTA resistance in a CobW2-dependent manner. CobW2, which also bound to FolEIB2, may have mismetalated this and other promiscuous enzymes with zinc, so that zinc was not available for key zinc-dependent proteins, while the promiscuous enzymes acted with lower efficiency, e.g., 14% of the maximum activity in the case of FolEIB from Bacillus subtilis (57). This indicates a close cooperation between CobW1 and CobW2.
CobW2.
CobW2 bound 0.5 zinc ion per monomer tightly and about six more with lower affinity, which could bind to the 22 His, 2 Cys, 8 Asp, and 5 Glu residues within the internal metal-binding site (Fig. 3, positions D-221 to D-267). The tightly bound 0.5 zinc ion could indicate 1 zinc per dimer bound to the monomer-monomer interface or 1 zinc in half of the monomers at the internal metal-binding GCXCC motif between the switch and Walker motifs. There was no influence of zinc on the GTPase activity of the protein, so the zinc-binding site with the highest affinity should be at the monomer-monomer interface.
CobW2 proved to be recalcitrant to in vitro analysis, probably due to the internal histidine-rich stretch, which may be intrinsically disordered. CobW2 precipitated in the presence of the metal mix. When incubation was in the presence of zinc, the experiments had two outcomes, one with 0.5 Zn/CobW2 and one with 6.8 Zn/CobW2, indicating that CobW2 might have two proteoforms, a dimer not able to bind more zinc and a dimer able to bind 6 zinc ions per monomer. The presence of MgGTP and presumably the subsequent GTPase action unfolded the protein partially, thereby stabilizing the zinc-binding proteoform. Consequently, CobW2 may not present zinc to the outside but may rather reside in a “closed” conformation not binding zinc. When an, as-yet-unknown, signal triggers the GTPase activity, the CobW2 dimer switches into an “open” zinc-binding, -presenting, and -buffering form. With 1,800 CobW2 copies per AE104 cell (32) and 6 Zn ions bound per monomer (Table 3), this equates to a buffering capacity of 10,800 zinc ions, and cobW2 expression is also 2-fold upregulated (Table 1) under metal starvation conditions in strain CH34.
CobW2 functions as a zinc buffer and storage compound, which agrees with the data discussed above. It is responsible for the increased zinc content of AE104 Δzur cells, with 21,000 more Zn ions than AE104 (Table 2), and is needed to counterbalance the plasmid-encoded efflux systems, which together mediate full zinc resistance on solid medium (Fig. 4). CobW2-like proteins are widespread, especially in bacteria of the order Burkholderiales. In a database search (September 2018), 3,528 entries were found with the typical internal histidine-rich stretch, albeit with a decreasing number of histidine residues (e.g., only 13 in Cupriavidus necator and 9 in Ralstonia solanacearum), indicating that the zinc-binding capacity of the CobW2 protein in C. metallidurans may have increased during its evolution to counterbalance the increasing efficiency of the Czc system.
CobW3.
CobW3 does not contain an internal GCXCC zinc-binding site between the switch and Walker motifs (Fig. 3), and the presence of zinc and/or MgGTP had no influence on the structure of CobW3. Zinc is not involved in triggering GTP hydrolysis of CobW3, and the protein had barely detectable GTPase activity (Fig. 9). It bound up to 8 zinc atoms per monomer. The presence of a histidine-rich carboxy terminus and the ability to bind the zinc ions with different affinities lead to the hypothesis that CobW3 may titrate the cytoplasmic zinc concentration.
CobW3 was responsible for the low zinc content of the strains AE104 ΔzupT and AE104 Δe4 ΔzupT. This means that CobW3 may decrease zinc uptake by interfering with the minimally nine other zinc uptake systems that C. metallidurans possesses in addition to ZupT (24, 25). While expression of genes for efflux systems is not strongly influenced by CobW3 (see results in the supplemental material) and those encoding uptake systems, with the exception of zupT, are influenced by zinc only at a low level (28, 29), CobW3 may act upon the activity of metal transport systems, e.g., ZntB, CorA1, and CorA3. CobW3-like proteins are also widespread in Burkholderiales, here mostly in Cupriavidus species; however, most of the 2,951 entries found still contained the GCXCC motif, and many have no histidine-rich stretch at the carboxy terminus and retain the amino acid stretch following the switch region (Fig. 3). These proteins resemble CobW1 more than CobW3, so CobW3 may have evolved from CobW1-like proteins by adding histidine residues at the C terminus, changing the GCXCC motif into a non-metal-binding motif, and loss of the complete region in the last step. Increasing the zinc-binding capacity of CobW2 during evolution of C. metallidurans may have resulted in the need for a regulator of the activity of low-specificity, high-rate metal cation importers of the MIT protein family, presumably to prevent an “overflow” of the zinc storage capacity of CobW2 by other metals. Subsequently, ZupT was needed to supply zinc ions in a sufficient number to the cell.
In conclusion, zinc homeostasis in C. metallidurans is composed of three subsystems: a basic transportome, Czc, and the Zur regulon. The basic transportome is composed of the four efflux systems ZntA, CadA, DmeF, and FieF (which are missing in the AE104 Δe4 mutant), at least eight metal uptake systems other than ZupT (CorA1, CorA2, CorA3, PitA, HoxN, ZntB, MgtA, MgtB), and at least one further unknown system (19, 24, 25). These uptake systems, which are regulated only to a small extent in response to zinc ions, plus the efflux systems (mainly the PIB2-type ATPases ZntA and CadA for zinc), mediate a basic kinetic-flow equilibrium in the cytoplasm by uptake and efflux processes, and this flow equilibrium adjusts the cytoplasmic transition metal cation composition and concentration (5). The Czc system consists of the periplasmic metal efflux pump CzcCBA, the PIB4-type ATPase CzcP, and the periplasmic metal-binding proteins CzcE, CzcJ, and CzcI. CzcP removes loosely bound surplus zinc ions from the cytoplasm, and the periplasmic metal-binding proteins may sequester them, with CzcCBA then removing them from the periplasm to the outside (5). This process prevents reimport of metals from the periplasm to the cytoplasm by the nine uptake systems and allows a high level of zinc, cadmium, and cobalt resistance.
While Czc mediates periplasmic zinc homeostasis, the components of the Zur regulon, ZupT and the three CobWs, are responsible for cytoplasmic zinc homeostasis. ZupT production is induced by zinc starvation and is essential for the synthesis of Czc, so that ZupT serves as a zinc-regulated interface between Czc and the three CobWs, which interact with zinc-dependent proteins in a different manner. In this scenario, CobW2 would form the center of the zinc repository as a zinc buffer, and with the aid of GTPase activity, which responds to an unknown signal, switches between a nonbinding “closed” and a zinc-binding “open” dimer. CobW3 could measure the cytoplasmic zinc content by titrating the metal with its C-terminal histidine-rich region and use this information to control the basic metal transportome, specifically the low-specificity, high-rate metal importers of the MIT protein family. This prevents an overflow of CobW2 and the remaining zinc repository. Finally, CobW1 is proposed to deliver available metals to metal-promiscuous proteins to maintain sufficient zinc levels for strictly zinc-dependent proteins. This demonstrates that proteins of the COG0523 subfamily of G3E-loop GTPases are at the core of zinc homeostasis in C. metallidurans and perform different functions within the bacterium.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The C. metallidurans strains used in this study were wild-type CH34(pMOL28, pMOL30), its plasmid-free derivative strain AE104, and mutants (see Table S8 in the supplemental material) (16). Tris-buffered mineral salts medium containing 2 g sodium gluconate/liter (TMM) was used to cultivate these strains aerobically with shaking at 30°C (16). Analytical-grade salts of heavy metal chlorides were used to prepare 1 M stock solutions, which were sterilized by filtration. Solid Tris-buffered medium contained 20 g agar/liter.
Genetic techniques.
Standard molecular genetic techniques were used (58, 59). For conjugative gene transfer, overnight cultures of donor strain E. coli S17/1 (60) and of the C. metallidurans recipient strains grown at 30°C in Tris-buffered medium were mixed (1:1) and plated onto nutrient broth agar. After 2 days, the bacteria were suspended in TMM, diluted, and plated onto selective media as previously described (58).
All primer pairs used are listed in Table S8. Plasmid pECD1003 was used to construct deletion mutants. It is a derivative of plasmid pCM184 (61). These plasmids harbor a kanamycin resistance cassette flanked by loxP recognition sites. Plasmid pECD1003 additionally carries alterations of 5 bp at each loxP site. Using these mutant lox sequences, multiple gene deletions within the same genome are possible without interferences by secondary recombination events (62, 63). Fragments of 300 bp upstream and downstream of the target gene were amplified by PCR, cloned into vector pGEM T-Easy (Promega), sequenced, and further cloned into plasmid pECD1003. The resulting plasmids were used in a double-crossover recombination in C. metallidurans strains to replace the respective target gene by the kanamycin resistance cassette, which was subsequently also deleted by transient introduction of cre expression plasmid pCM157 (61). Cre recombinase is a site-specific recombinase from the phage P1 that catalyzes the in vivo excision of the kanamycin resistance cassette at the loxP recognition sites. The correct deletions of the respective transporter genes were verified by PCR. For construction of multiple-deletion strains, these steps were repeated. The resulting mutants carried a small open reading frame instead of the wild-type gene to prevent polarity effects.
Gene disruption.
Although the cobW2 gene could be interrupted by a single-crossover experiment, recombinant strains containing a deletion as a consequence of a double-crossover event could never be isolated. Therefore, cobW2 was inactivated by insertional mutagenesis. Its central portion was amplified by PCR from total DNA of strain AE104, cloned into plasmid pECD794 (pLO2lacZ), and conjugated (19). This procedure was also required for other ΔcobW interruptions as indicated.
β-Galactosidase assay and lacZ reporter constructions.
C. metallidurans cells with a lacZ reporter gene fusion were cultivated as a preculture in TMM containing 1.5 mg liter−1 kanamycin at 30°C and 250 rpm to early stationary phase and then diluted into fresh medium and incubated at 30°C. At a cell density of 60 Klett units, metal salts were added in different concentrations, and cells were incubated at 1,300 rpm in a neoLab DTS-2 shaker (neoLab, Heidelberg, Germany) for a further 3 h. The specific β-galactosidase activity was determined in permeabilized cells as previously published, with 1 U defined as the activity forming 1 nmol of o-nitrophenol min−1 at 30°C (64). The lacZ reporter gene was inserted into several target genes to construct reporter operon fusions or disruptions. This was done by a single-crossover recombination in C. metallidurans strains. A 300- to 400-bp PCR product of the 3′-end region or of the middle of the genes was amplified from total DNA of strain AE104 and the resulting fragments cloned into plasmid pECD794 (pLO2-lacZ) (19). The respective operon fusion cassettes were inserted directly downstream after the open reading frame of the target gene by conjugation and single-crossover recombination.
Dose-response growth curves in 96-well plates.
A preculture was incubated at 30°C and 250 rpm to the early stationary phase and then diluted 1:20 in fresh medium and incubated for 24 h at 30°C and 250 rpm to the late stationary phase. Twenty-four-hour cultures were used to inoculate parallel cultures with increasing metal concentrations in 96-well plates (Greiner, Germany). Cells were cultivated for 20 h at 30°C and 1,300 rpm in a neoLab DTS-2 shaker (neoLab, Heidelberg, Germany), and the optical density was determined at 600 nm as indicated in a Tecan Infinite 200 Pro reader (Tecan, Männersdorf, Switzerland). To calculate the IC50 value (the metal concentration that led to reduction of turbidity by half) and the corresponding b value (a measure of the slope of the sigmoidal dose-response curve), the data were adapted to the formula OD(c) = OD(0)/{1 + exp[(c − IC50)/b]}, which is a simplified version of a Hill-type equation as published previously (19, 65, 66). OD(c) is the turbidity at a given metal concentration, OD(0) is that with no added metal, and c is the metal concentration.
MIC.
The MIC was determined in triplicate as the lowest concentration inhibiting bacterial growth on solid TMM. A preculture was incubated at 30°C with shaking at 250 rpm to early stationary phase, diluted 1:20 in fresh medium, and incubated for 24 h at 30°C and 250 rpm. This 24-h culture was diluted 1:100 in fresh medium and used for streaking onto plates containing different concentrations of the respective metal salts. The plates were incubated at 30°C for 4 days, and cell growth was monitored.
RNA isolation.
C. metallidurans cells were cultivated as described above. At a cell turbidity of 100 Klett units, EDTA was added to a final concentration of 50 μM or not added. After a 10-min incubation at 30°C, the cells were rapidly harvested at room temperature and stored at –80°C. Total RNA was isolated with the RNeasy minikit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. DNase treatment was performed. To exclude experimental artifacts resulting from DNA contamination, only RNA that did not generate products in several PCRs with chromosomal and plasmid primers was used. The RNA concentration was determined photometrically, and RNA quality was checked on formamide gels (59) and measured as RNA integrity number (RIN) on an Agilent 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany). The published RNA-Seq data (43) (accession number SRP158096 in the SRA/NCBI database) were reanalyzed using the transcriptome analysis viewer TraV v2 (67). Transcriptional start sites were generated by Vertis Biotechnology (Freising, Germany).
RT-PCR.
For the RT reaction, 1 μg of total RNA and 0.1 μg hexamer primers were incubated at 65°C for 5 min and snap-cooled on ice. After addition of 0.5 mM (each) dATP, dGTP, dTTP, and dCTP, 20 mM dithiothreitol (DTT), and 100 U of reverse transcriptase (Superscript II) in reaction buffer (Thermo Fisher Scientific, Germany), reverse transcription proceeded for 10 min at room temperature, followed by 1 h at 50°C. After finishing the RT reaction, the enzyme was inactivated by incubation at 70°C for 10 min. One microliter of the resulting cDNA was amplified by PCR with 1 μM each primer and 1 U of Taq polymerase (Roche Diagnostics GmbH, Mannheim, Germany). As a control, rpoZ was used. A no-template control reaction was performed under conditions identical to those for the target genes. RT-PCRs from three independent biological samples were performed.
The resulting amplified DNA was separated on an agarose gel and stained using ethidium bromide. The gel was analyzed using ImageJ (68), with the background above each band subtracted from the signal to yield the band intensity. The intensity of the respective negative water control was subtracted from each value, which was subsequently divided by the value of the DNA positive control of the respective gel.
ICP-MS analysis.
To determine the metal content, cells were cultivated to 100 Klett units in TMM, and the different metal salts or EDTA was added. When the cells had reached the middle of their exponential phase of growth at 150 Klett units, 10 ml cell suspension was centrifuged for 30 min at 4,500 × g. Cells washed twice with 50 mM Tris-HCl buffer containing 10 mM EDTA (pH 7). The supernatant was discarded and the residual liquid carefully removed at each step. The pellet was suspended in concentrated 67% (wt/vol) HNO3 (trace metal grade; Normatom/Prolabo) and mineralized at 70°C for 2 h. Samples were diluted to a final concentration of 2% (wt/vol) nitric acid. The measurement was done as previously described (8, 28, 32).
For protein cofactor determination, the mineralization of a defined protein concentration with concentrated 67% (wt/vol) HNO3 occurred at 70°C for 2 h. Samples were diluted to a final concentration of 2% (wt/vol) nitric acid.
To all samples, indium was added as internal standard at a final concentration of 10 ppb. Elemental analysis was performed via inductively coupled plasma mass spectrometry (ICP-MS) using an ESI-sampler SC-2 (Elemental Scientific, Inc., Omaha, NE) and an X-Series II ICP-MS instrument (Thermo Fisher Scientific, Bremen, Germany) operating with a collision cell and flow rates of 5 ml · min−1 of He-H2 (93%:7%) (69), with an Ar carrier flow rate of 0.76 liter · min−1 and an Ar makeup flow rate of 15 liters · min−1. An external calibration curve was recorded with ICP-multielement standard solution XVI (Merck) in 2% nitric acid. The sample was introduced via a peristaltic pump and analyzed for its metal content. For blank measurement and quality/quantity thresholds, calculations based on German standard DIN32645 TMM were used.
Purification of His-tagged CobWs.
E. coli strain BL21-pLysS with pECD1239-41 (a pET28A derivative with a Tev cleavage site) was cultivated in a fermenter with fed batch of 1 M glucose in TB medium to an optical density of 3 at 600 nm. Expression of cobW genes was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and incubation was continued for 3 h at 30°C. Cells were harvested by centrifugation and disrupted by three passages through a French press (Gaulin Amicon, Silver Spring, MD). After centrifugation to remove cell debris, CobWs were purified using an Ni-NTA affinity chromatography column with an Äkta purification system (GE Healthcare, Freiburg, Germany) and washed with buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl) and buffer with increasing imidazole concentrations. Elution occurred with buffer plus 500 mM imidazole. To desalt the fractions, a PD10 column (GE Healthcare, Freiburg, Germany) with storage buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 5% glycerin) was used, and afterwards the proteins were concentrated by using VivaSpin6 or -20 columns (30,000 molecular weight cutoff [MWCO], polyethersulfone [PES] membrane; Sartorius, Göttingen, Germany). The protein concentration was determined by using a NanoDrop ND-1000 spectrophotometer with an extinction coefficient and the specific molecular weight calculated by using the ProtParam tool of the ExPASy-server (http://web.expasy.org/protparam/). Protein quality was analyzed with a 12.5% SDS gel stained with Coomassie brilliant blue and Western blotting (70). To evaluate the purity of the proteins, size exclusion chromatography with a buffer of 50 mM Tris-HCl (pH 8.0) and 150 mM NaCl was performed. For the size calculation, a kit for low-molecular-weight proteins was used (GE Healthcare, Freiburg, Germany). The different protein peaks were analyzed by native and SDS-PAGE.
Tev cleavage.
CobWs were purified with an N-terminal His tag, which was removed at the cleavage site (sequence motif, ENLYFQGH) by Tev protease. Therefore, both proteins in a 1:1 ratio were incubated for 24 h at 4°C under reducing conditions (50 mM Tris-HCl [pH 8.0], 0.5 mM EDTA, 1 mM DTT, 0.5% mercaptoethanol), and tag and the protease were removed by an additional Ni-NTA affinity chromatography step.
CD spectroscopy.
The secondary and tertiary structures of the proteins under different conditions were controlled by CD spectroscopy with a Jasco J-810 instrument (Jasco Europe S.R.L., Cremella, Italy). Samples were measured in 60 replicates at 20°C and 0.8 mg/ml for near-UV CD (320 to 250 nm) and far-UV CD (250 to 195 nm) with light paths of 1 cm and 0.1 mm, respectively. From the raw data, the molar ellipticity was calculated: εM = ε · M/(10 · d · c), where ε is ellipticity (millidegrees), M is molecular weight (g · mol−1, c is protein concentration (g · liter−1], and d is length of the light path (cm).
Metal incubation assay.
Metalation of CobWs was performed after the production of the apoproteins by adding a 10-fold excess of EDTA and incubating for 2 h on ice, and metal-EDTA complexes were removed over a PD10 column (the procedure was modified from that in reference 42). Success was verified by ICP-MS. The metalation was also performed with a 10-fold excess of ZnCl2 with or without GTP plus MgCl2 and subsequent removal of surplus zinc.
Zincon-zinc competition experiment.
The Zincon-zinc competition assay is based on the binding of zinc to the dye Zincon (2-carboxy-2′-hydroxy-5′-sulfoformazylbenzene), which is associated with the shift of the absorption maximum from 480 nm to 620 nm (71, 72). For this purpose, 10 μM concentrations of the produced apoproteins were used. Increasing ZnCl2 concentrations from 0 to 140 μM and 10 μM Zincon were added to the proteins. As a control, 10 μM EDTA was added to replace the apoproteins. Batches (20 μl) were filled with buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl). The spectra (230 to 1,000 nm) in 384-well microplates (Greiner Bio-One, Frickenhausen, Germany) were measured in a Tecan Infinite 200 instrument (Tecan Group, Switzerland).
Interaction assay.
For detection of specific interactions, 10 μg of recombinant Strep-tagged Zur or FolEIB2 was bound as bait protein in 96-well plates to 2 μg MagStrep type 3 XT magnetic beads (IBA Lifescience, Göttingen, Germany) for 30 min at 4°C, after washing them three times with buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl). The incubation with the prey protein CobWs was carried out for 3 h at 4°C with the optional addition of 500 μM MgCl2 and 10 μM GTP, 500 μM EDTA, or ZnCl2. Five washing steps with buffer and subsequent elution through 2× SDS sample buffers followed. The supernatants were separated in 12.5% SDS-polyacrylamide gels for analysis by Coomassie blue staining and Western blotting.
GTPase assay.
To measure GTPase activity, the GTPase-Glo assay (Promega, Mannheim, Germany) with a constant GTP concentration of 5 μM and different concentrations of the CobWs (0 to 50 μM) in the presence or absence of 50 μM ZnCl2 was used (73). The GTPase reaction occurred over 60 min at room temperature. Afterwards, the unhydrolyzed GTP was converted to ATP for 30 min. Subsequently, the ATP content was determined in a luminescence assay by light emission using a Tecan Spark reader (Tecan, Männersdorf, Switzerland). From the luminescence, the residual amount of GTP converted to ATP, and therefore the concentration of GTP hydrolyzed within 60 min, was calculated. Cleavage of 50% of the 5 μM GTP within 60 min by 10 μM CobW would represent a turnover number of 0.25 min−1 or 0.0042 s−1.
Statistics.
Student’s t test was used, but in most cases, the distance (D) value was used. D has been used several times previously (33, 36, 74). The D value is the absolute difference between the mean values divided by the sum of the deviations. It is a simple, more useful value than Student’s t test because nontouching deviation bars of two values (D > 1) at three repeats always means a statistically sound (≥ 95%) difference, provided the deviations are within a similar range. At n = 4 significance is ≥97.5%, at n = 5 it is ≥99% (significant), and at n = 8 it is ≥99.9% (highly significant).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Deutsche Forschungsgemeinschaft, grant Ni262/19-1.
We thank Grit Schleuder for skillful technical assistance. We thank Gary Sawers for critical comments on the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00192-19.
REFERENCES
- 1.Krężel A, Maret W. 2016. The biological inorganic chemistry of zinc ions. Arch Biochem Biophys 611:3–19. doi: 10.1016/j.abb.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Williams R. 2012. Zinc in evolution. J Inorg Biochem 111:104–109. doi: 10.1016/j.jinorgbio.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Andreini C, Banci L, Bertini I, Rosato A. 2006. Counting the zinc-proteins encoded in the human genome. J Proteome Res 5:196–201. doi: 10.1021/pr050361j. [DOI] [PubMed] [Google Scholar]
- 4.Capdevila DA, Wang JF, Giedroc DP. 2016. Bacterial strategies to maintain zinc metallostasis at the host-pathogen interface. J Biol Chem 291:20858–20868. doi: 10.1074/jbc.R116.742023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nies DH. 2016. The biological chemistry of the transition metal “transportome” of Cupriavidus metallidurans. Metallomics 8:481–507. doi: 10.1039/c5mt00320b. [DOI] [PubMed] [Google Scholar]
- 6.Busch W, Saier M. 2002. The transporter classification (TC) system. Crit Rev Biochem Mol Biol 37:287–337. doi: 10.1080/10409230290771528. [DOI] [PubMed] [Google Scholar]
- 7.Saier MHJ, Tran CV, Barabote RD. 2006. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucl Acids Res 34:D181–D186. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herzberg M, Bauer L, Nies DH. 2014. Deletion of the zupT gene for a zinc importer influences zinc pools in Cupriavidus metallidurans CH34. Metallomics 6:421–436. doi: 10.1039/c3mt00267e. [DOI] [PubMed] [Google Scholar]
- 9.Wu FYH, Huang WJ, Sinclair RB, Powers L. 1992. The structure of the zinc sites of Escherichia coli DNA-dependent RNA polymerase. J Biol Chem 267:25560–25567. [PubMed] [Google Scholar]
- 10.Markov D, Naryshkina T, Mustaev A, Severinov K. 1999. A zinc-binding site in the largest subunit of DNA-dependent RNA polymerase is involved in enzyme assembly. Genes Dev 13:2439–2448. doi: 10.1101/gad.13.18.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dressler C, Kües U, Nies DH, Friedrich B. 1991. Determinants encoding multiple metal resistance in newly isolated copper-resistant bacteria. Appl Environ Microbiol 57:3079–3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reith F, Rogers SL, McPhail DC, Webb D. 2006. Biomineralization of gold: biofilms on bacterioform gold. Science 313:233–236. doi: 10.1126/science.1125878. [DOI] [PubMed] [Google Scholar]
- 13.Wiesemann N, Bütof L, Herzberg M, Hause G, Berthold L, Etschmann B, Brugger J, Martinéz-Criado G, Dobritzsch D, Baginski S, Reith F, Nies DH. 2017. Synergistic toxicity of copper and gold compounds in Cupriavidus metallidurans. Appl Environ Microbiol 83:e01679-17. doi: 10.1128/AEM.01679-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janssen PJ, Van Houdt R, Moors H, Monsieurs P, Morin N, Michaux A, Benotmane MA, Leys N, Vallaeys T, Lapidus A, Monchy S, Medigue C, Taghavi S, McCorkle S, Dunn J, van der Lelie D, Mergeay M. 2010. The complete genome sequence of Cupriavidus metallidurans strain CH34, a master survivalist in harsh and anthropogenic environments. PLoS One 5:e10433. doi: 10.1371/journal.pone.0010433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dicenzo GC, Finan TM. 2017. The divided bacterial genome: structure, function, and evolution. Microbiol Mol Biol Rev 81:e00019-17. doi: 10.1128/MMBR.00019-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mergeay M, Nies D, Schlegel HG, Gerits J, Charles P, van Gijsegem F. 1985. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J Bacteriol 162:328–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nies DH. 2000. Heavy metal resistant bacteria as extremophiles: molecular physiology and biotechnological use of Ralstonia spec. CH34. Extremophiles 4:77–82. doi: 10.1007/s007920050140. [DOI] [PubMed] [Google Scholar]
- 18.Nies DH. 2013. RND-efflux pumps for metal cations, p 79–122. In Yu EW, Zhang Q, Brown MH (ed), Microbial efflux pumps: current research. Caister Academic Press, Norfolk, UK. [Google Scholar]
- 19.Scherer J, Nies DH. 2009. CzcP is a novel efflux system contributing to transition metal resistance in Cupriavidus metallidurans CH34. Mol Microbiol 73:601–621. doi: 10.1111/j.1365-2958.2009.06792.x. [DOI] [PubMed] [Google Scholar]
- 20.Legatzki A, Anton A, Grass G, Rensing C, Nies DH. 2003. Interplay of the Czc-system and two P-type ATPases in conferring metal resistance to Ralstonia metallidurans. J Bacteriol 185:4354–4361. doi: 10.1128/jb.185.15.4354-4361.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Legatzki A, Franke S, Lucke S, Hoffmann T, Anton A, Neumann D, Nies DH. 2003. First step towards a quantitative model describing Czc-mediated heavy metal resistance in Ralstonia metallidurans. Biodegradation 14:153–168. doi: 10.1023/A:1024043306888. [DOI] [PubMed] [Google Scholar]
- 22.Große C, Grass G, Anton A, Franke S, Navarrete Santos A, Lawley B, Brown NL, Nies DH. 1999. Transcriptional organization of the czc heavy metal homoeostasis determinant from Alcaligenes eutrophus. J Bacteriol 181:2385–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Lelie D, Schwuchow T, Schwidetzky U, Wuertz S, Baeyens W, Mergeay M, Nies DH. 1997. Two component regulatory system involved in transcriptional control of heavy metal homoeostasis in Alcaligenes eutrophus. Mol Microbiol 23:493–503. doi: 10.1046/j.1365-2958.1997.d01-1866.x. [DOI] [PubMed] [Google Scholar]
- 24.Herzberg M, Bauer L, Kirsten A, Nies DH. 2016. Interplay between seven secondary metal transport systems is required for full metal resistance of Cupriavidus metallidurans. Metallomics 8:313–326. doi: 10.1039/C5MT00295H. [DOI] [PubMed] [Google Scholar]
- 25.Grosse C, Herzberg M, Schüttau M, Nies DH. 2016. Characterization of the Δ7 mutant of Cupriavidus metallidurans with deletions of seven secondary metal uptake systems. mSystems 1:e00004-16. doi: 10.1128/mSystems.00004-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patzer SI, Hantke K. 1998. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol Microbiol 28:1199–1210. doi: 10.1046/j.1365-2958.1998.00883.x. [DOI] [PubMed] [Google Scholar]
- 27.Patzer SI, Hantke K. 2000. The zinc-responsive regulator Zur and its control of the znu gene cluster encoding the ZnuABC zinc uptake system in Escherichia coli. J Biol Chem 275:24321–24332. doi: 10.1074/jbc.M001775200. [DOI] [PubMed] [Google Scholar]
- 28.Kirsten A, Herzberg M, Voigt A, Seravalli J, Grass G, Scherer J, Nies DH. 2011. Contributions of five secondary metal uptake systems to metal homeostasis of Cupriavidus metallidurans CH34. J Bacteriol 193:4652–4663. doi: 10.1128/JB.05293-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt C, Schwarzenberger C, Große C, Nies DH. 2014. FurC regulates expression of zupT for the central zinc importer ZupT of Cupriavidus metallidurans. J Bacteriol 196:3461–3471. doi: 10.1128/JB.01713-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grass G, Wong MD, Rosen BP, Smith RL, Rensing C. 2002. ZupT Is a Zn(II) uptake system in Escherichia coli. J Bacteriol 184:864–866. doi: 10.1128/jb.184.3.864-866.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taudte N, Grass G. 2010. Point mutations change specificity and kinetics of metal uptake by ZupT from Escherichia coli. Biometals 23:643–656. doi: 10.1007/s10534-010-9319-z. [DOI] [PubMed] [Google Scholar]
- 32.Herzberg M, Dobritzsch D, Helm S, Baginsky S, Nies DH. 2014. The zinc repository of Cupriavidus metallidurans. Metallomics 6:2157–2165. doi: 10.1039/C4MT00171K. [DOI] [PubMed] [Google Scholar]
- 33.Bütof L, Schmidt-Vogler C, Herzberg M, Große C, Nies DH. 2017. The components of the unique Zur regulon of Cupriavidus metallidurans mediate cytoplasmic zinc handling. J Bacteriol 199:e00372-17. doi: 10.1128/JB.00372-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang CM, Christian T, Newberry KJ, Perona JJ, Hou YM. 2003. Zinc-mediated amino acid discrimination in cysteinyl-tRNA synthetase. J Mol Biol 327:911–917. doi: 10.1016/S0022-2836(03)00241-9. [DOI] [PubMed] [Google Scholar]
- 35.Zhang CM, Perona JJ, Hou YM. 2003. Amino acid discrimination by a highly differentiated metal center of an aminoacyl-tRNA synthetase. Biochemistry 42:10931–10937. doi: 10.1021/bi034812u. [DOI] [PubMed] [Google Scholar]
- 36.Herzberg M, Schüttau M, Reimers M, Große C, Hans-Günther-Schlegel H-G-S, Nies DH. 2015. Synthesis of nickel-iron hydrogenase in Cupriavidus metallidurans is controlled by metal-dependent silencing and un-silencing of genomic islands. Metallomics 7:632–649. doi: 10.1039/C4MT00297K. [DOI] [PubMed] [Google Scholar]
- 37.Leipe DD, Wolf YI, Koonin EV, Aravind L. 2002. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol 317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 38.Verstraeten N, Fauvart M, Versees W, Michiels J. 2011. The universally conserved prokaryotic GTPases. Microbiol Mol Biol Rev 75:507–542. doi: 10.1128/MMBR.00009-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haas CE, Rodionov DA, Kropat J, Malasarn D, Merchant SS, de Crecy-Lagard V. 2009. A subset of the diverse COG0523 family of putative metal chaperones is linked to zinc homeostasis in all kingdoms of life. BMC Genomics 10:470. doi: 10.1186/1471-2164-10-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lacasse MJ, Zamble DB. 2016. [NiFe]-hydrogenase maturation. Biochemistry 55:1689–1701. doi: 10.1021/acs.biochem.5b01328. [DOI] [PubMed] [Google Scholar]
- 41.Blaby-Haas CE, Flood JA, Crécy-Lagard V. d, Zamble DB. 2012. YeiR: a metal-binding GTPase from Escherichia coli involved in metal homeostasis. Metallomics 4:488–497. doi: 10.1039/c2mt20012k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sydor AM, Jost M, Ryan KS, Turo KE, Douglas CD, Drennan CL, Zamble DB. 2013. Metal binding properties of Escherichia coli YjiA, a member of the metal homeostasis-associated COG0523 family of GTPases. Biochemistry 52:1788–1801. doi: 10.1021/bi301600z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Große C, Poehlein A, Blank K, Schwarzenberger C, Schleuder G, Herzberg M, Nies DH. 2019. The third pillar of metal homeostasis in Cupriavidus metallidurans CH34: Preferences are controlled by extracytoplasmic functions sigma factors. Metallomics 11:291–316. doi: 10.1039/C8MT00299A. [DOI] [PubMed] [Google Scholar]
- 44.Böhm G, Muhr R, Jaenicke R. 1992. Quantitative analysis of protein far UV circular dichroism spectra by neural networks. Protein Eng 5:5651–5657. [DOI] [PubMed] [Google Scholar]
- 45.Kocyła A, Pomorski A, Krężel A. 2017. Molar absorption coefficients and stability constants of Zincon metal complexes for determination of metal ions and bioinorganic applications. J Inorg Biochem 176:53–65. doi: 10.1016/j.jinorgbio.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 46.Dawson RMC, Elliott DC, Elliott WH, Jones KM. 1969. Data for biochemical research, 2nd ed Clarendon Press, Oxford, UK. [Google Scholar]
- 47.Rebelo J, Auerbach G, Bader G, Bracher A, Nar H, Hosl C, Schramek N, Kaiser J, Bacher A, Huber R, Fischer M. 2003. Biosynthesis of pteridines. Reaction mechanism of GTP cyclohydrolase I. J Mol Biol 326:503–516. doi: 10.1016/S0022-2836(02)01303-7. [DOI] [PubMed] [Google Scholar]
- 48.Khil PP, Obmolova G, Teplyakov A, Howard AJ, Gilliland GL, Camerini-Otero RD. 2004. Crystal structure of the Escherichia coli YjiA protein suggests a GTP-dependent regulatory function. Proteins 54:371–374. doi: 10.1002/prot.10430. [DOI] [PubMed] [Google Scholar]
- 49.Kerby RL, Ludden PW, Roberts GP. 1997. In vivo nickel insertion into the carbon monoxide dehydrogenase of Rhodospirillum rubrum: molecular and physiological characterization of cooCTJ. J Bacteriol 179:2259–2266. doi: 10.1128/jb.179.7.2259-2266.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jeon WB, Cheng JJ, Ludden PW. 2001. Purification and characterization of membrane-associated CooC protein and its functional role in the insertion of nickel into carbon monoxide dehydrogenase from Rhodospirillum rubrum. J Biol Chem 276:38602–38609. doi: 10.1074/jbc.M104945200. [DOI] [PubMed] [Google Scholar]
- 51.Gregg CM, Goetzl S, Jeoung JH, Dobbek H. 2016. AcsF catalyzes the ATP-dependent insertion of nickel into the Ni, Ni-[4Fe4S] cluster of acetyl-CoA synthase. J Biol Chem 291:18129–18138. doi: 10.1074/jbc.M116.731638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jeoung JH, Giese T, Grunwald M, Dobbek H. 2009. CooC1 from Carboxydothermus hydrogenoformans is a nickel-binding ATPase. Biochemistry 48:11505–11513. doi: 10.1021/bi901443z. [DOI] [PubMed] [Google Scholar]
- 53.Hubbard PA, Padovani D, Labunska T, Mahlstedt SA, Banerjee R, Drennan CL. 2007. Crystal structure and mutagenesis of the metallochaperone MeaB—insight into the causes of methylmalonic aciduria. J Biol Chem 282:31308–31316. doi: 10.1074/jbc.M704850200. [DOI] [PubMed] [Google Scholar]
- 54.Gabriel SE, Miyagi F, Gaballa A, Helmann JD. 2008. Regulation of the Bacillus subtilis yciC gene and insights into the DNA-binding specificity of the zinc-sensing metalloregulator Zur. J Bacteriol 190:3482–3488. doi: 10.1128/JB.01978-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chaoprasid P, Dokpikul T, Johnrod J, Sirirakphaisarn S, Nookabkaew S, Sukchawalit R, Mongkolsuk S. 2016. Agrobacterium tumefaciens Zur regulates the high-affinity zinc uptake system TroCBA and the putative metal chaperone YciC, along with ZinT and ZnuABC, for survival under zinc-limiting conditions. Appl Environ Microbiol 82:3503–3514. doi: 10.1128/AEM.00299-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nairn BL, Lonergan ZR, Wang JF, Braymer JJ, Zhang YF, Calcutt MW, Lisher JP, Gilston BA, Chazin WJ, de Crecy-Lagard V, Giedroc DP, Skaar EP. 2016. The response of Acinetobacter baumannii to zinc starvation. Cell Host Microbe 19:826–836. doi: 10.1016/j.chom.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sankaran B, Bonnett SA, Shah K, Gabriel S, Reddy R, Schimmel P, Rodionov DA, de Crécy-Lagard V, Helmann JD, Iwata-Reuyl D, Swairjo MA. 2009. Zinc-independent folate biosynthesis: genetic, biochemical, and structural investigations reveal new metal dependence for GTP cyclohydrolase IB. J Bacteriol 191:6936–6949. doi: 10.1128/JB.00287-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nies D, Mergeay M, Friedrich B, Schlegel HG. 1987. Cloning of plasmid genes encoding resistance to cadmium, zinc, and cobalt in Alcaligenes eutrophus CH34. J Bacteriol 169:4865–4868. doi: 10.1128/jb.169.10.4865-4868.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 60.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Nat Biotechnol 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 61.Marx CJ, Lidstrom ME. 2002. Broad-host-range cre-lox system for antibiotic marker recycling in gram-negative bacteria. Biotechniques 33:1062–1067. doi: 10.2144/02335rr01. [DOI] [PubMed] [Google Scholar]
- 62.Suzuki N, Nonaka H, Tsuge Y, Inui M, Yukawa H. 2005. New multiple-deletion method for the Corynebacterium glutamicum genome, using a mutant lox sequence. Appl Environ Microbiol 71:8472–8480. doi: 10.1128/AEM.71.12.8472-8480.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Albert H, Dale EC, Lee E, Ow DW. 1995. Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J 7:649–659. doi: 10.1046/j.1365-313X.1995.7040649.x. [DOI] [PubMed] [Google Scholar]
- 64.Nies DH. 1992. CzcR and CzcD, gene products affecting regulation of resistance to cobalt, zinc and cadmium (czc system) in Alcaligenes eutrophus. J Bacteriol 174:8102–8110. doi: 10.1128/jb.174.24.8102-8110.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anton A, Weltrowski A, Haney JH, Franke S, Grass G, Rensing C, Nies DH. 2004. Characteristics of zinc transport by two bacterial cation diffusion facilitators from Ralstonia metallidurans and Escherichia coli. J Bacteriol 186:7499–7507. doi: 10.1128/JB.186.22.7499-7507.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pace CN, Scholtz MJ. 1997. Measuring the conformational stability of a protein, p 299–322. In Creighton T. (ed), Protein structure: a practical approach. IRL Press, Oxford, UK. [Google Scholar]
- 67.Dietrich S, Wiegand S, Liesegang H. 2014. TraV: a genome context sensitive transcriptome browser. PLoS One 9:e93677. doi: 10.1371/journal.pone.0093677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wagegg W, Braun V. 1981. Ferric citrate transport in Escherichia coli requires outer membrane receptor protein FecA. J Bacteriol 145:156–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weber K, Osborn M. 1969. Reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem 244:4406–4412. [PubMed] [Google Scholar]
- 71.Hilario E, Romero I, Celis H. 1990. Determination of the physicochemical constants and spectrophotometric characteristics of the metallochromic Zincon and its potential use in biological systems. J Biochem Biophys Methods 21:197–207. doi: 10.1016/0165-022X(90)90013-3. [DOI] [PubMed] [Google Scholar]
- 72.Sabel CE, Neureuther JM, Siemann S. 2010. A spectrophotometric method for the determination of zinc, copper, and cobalt ions in metalloproteins using Zincon. Anal Biochem 397:218–226. doi: 10.1016/j.ab.2009.10.037. [DOI] [PubMed] [Google Scholar]
- 73.Mondal S, Hsiao K, Goueli SA. 2015. A homogenous bioluminescent system for measuring GTPase, GTPase activating protein, and guanine nucleotide exchange factor activities. Assay Drug Dev Technol 13:444–455. doi: 10.1089/adt.2015.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wiesemann N, Mohr J, Grosse C, Herzberg M, Hause G, Reith F, Nies DH. 2013. Influence of copper resistance determinants on gold transformation by Cupriavidus metallidurans strain CH34. J Bacteriol 195:2298–2308. doi: 10.1128/JB.01951-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.